Abstract
Predicting and understanding plant responses to perturbations require integrating the interactions between nutritional sources, genes, cell metabolism, and physiology in the same model. This can be achieved using metabolic modeling calibrated by experimental data. In this study, we developed a multi-organ metabolic model of a tomato (Solanum lycopersicum) plant during vegetative growth, named Virtual Young TOmato Plant (VYTOP) that combines genome-scale metabolic models of leaf, stem and root and integrates experimental data acquired from metabolomics and high-throughput phenotyping of tomato plants. It is composed of 6,689 reactions and 6,326 metabolites. We validated VYTOP predictions on five independent use cases. The model correctly predicted that glutamine is the main organic nutrient of xylem sap. The model estimated quantitatively how stem photosynthetic contribution impacts exchanges between the different organs. The model was also able to predict how nitrogen limitation affects vegetative growth and the metabolic behavior of transgenic tomato lines with altered expression of core metabolic enzymes. The integration of different components, such as a metabolic model, physiological constraints, and experimental data, generates a powerful predictive tool to study plant behavior, which will be useful for several other applications, such as plant metabolic engineering or plant nutrition.
A multi-organ metabolic model provides insight into tomato plant functions, such as organic xylem metabolites, the role of the stem, and the response to environmental or genetic perturbation.
Introduction
Systems biology can predict and explain how a whole organism responds to a stimulus or a perturbation and how different components of the organism are connected. In particular, one way to unravel physiological mechanisms is the use of constraint-based metabolic modeling. Constraint-based metabolic modeling relies on genome-scale metabolic networks, which gather all the identified metabolic reactions of an organism according to its genomic sequences and current metabolic knowledge summarized in databases (Gu et al., 2019). It predicts and quantifies the metabolic pathways used, thanks to mathematical constraints that encompass physiological states, such as the uptake rate of nutrients present in the environment. Initially developed for unicellular organisms, constraint-based metabolic modeling has been extended to diverse plant species such as Arabidopsis thaliana (Mintz-Oron et al., 2012; Gomes de Oliveira Dal’Molin et al., 2015; Shaw and Cheung, 2018), Medicago truncatula (Pfau et al., 2018), barley (Hordeum vulgare; Grafahrend-Belau et al., 2013), Steraria viridis (Shaw and Cheung, 2019), and soybean (Glycine max; Moreira et al., 2019). These studies analyzed mechanisms such as the effect of the nitrogen (N) source (Arnold and Nikoloski, 2014; Arnold et al., 2015; Gomes de Oliveira Dal’Molin et al., 2015; Seaver et al., 2018), the N fixation by symbiotic bacteria (Pfau et al., 2018), the effect of diurnal cycles (Gomes de Oliveira Dal’Molin et al., 2015; Shaw and Cheung, 2018), or the genetic modifications to perform in order to enhance a product of interest such as vitamin E (Mintz-Oron et al., 2012; Saha et al., 2011). Some of these studies were supported with labeling data, such as Robaina-Estévez et al. (2017), which studied the metabolic differences between guard cells and mesophyll cells. For a full review of all models of plants developed so far, refer to Gerlin et al. (2021) and Shaw and Cheung (2020).
The representation of the multi-organ structure of plants through modeling is challenging (Clark et al., 2020), but is required to better understand plant metabolism. To model exchanges between organs, a global exchange pool is usually used (Gomes de Oliveira Dal’Molin et al., 2015; Shaw and Cheung, 2018), but the veracity of the exchanged fluxes has not been studied. Another major limitation is the lack of experimental calibrations since most of the methodologies gather heterogeneous data from literature, issued from multiple species and different growth scenarios, which could imply bias and inaccuracies. In addition, tomato (Solanum lycopersicum), a major agricultural crop, has not benefited from the advances in multi-organ metabolic models, as only a genome-scale metabolic model of leaf cell has been published (Yuan et al., 2016), as well as a metabolic model of the fruit (Colombié et al., 2017, 2015; Li et al., 2020).
In this study, we developed a multi-organ metabolic model of tomato during vegetative growth, in order to provide to the plant research community a comprehensive tool adapted for many biological questions such as the impact of environmental factors on the early development of the tomato plant, the exchange fluxes of matter between organs, or the prediction of the reactions to be modified to obtain a desired phenotype. This model, named Virtual Young TOmato Plant (VYTOP), includes leaf, stem, root and, contrary to existing multi-organ models, dissociated xylem and phloem compartments. It is able to simulate all the major metabolic reactions used in the plant and the sink/source relationships between organs. The model was calibrated with homogeneous data from experiments we performed, using an automated phenotyping platform, gathering both physiological data (growth and transpiration) and metabolomics (xylem sap chemistry and organ biomass composition). We chose five independent use cases to validate the usefulness of our model for a broad range of biological questions: (1) prediction of core metabolic fluxes in each tomato organ; (2) contribution of stem to photosynthesis; (3) organic xylem composition; (4) impact of N limitation on growth; and (5) predictions of metabolic changes in transgenic lines. Beyond its use to model the fluxes between tomato organs, this model could serve as a template for modeling other plants of interest as well as predicting and understanding the impact of diverse perturbations.
Results and Discussion
VYTOP, a multi-organ metabolic model
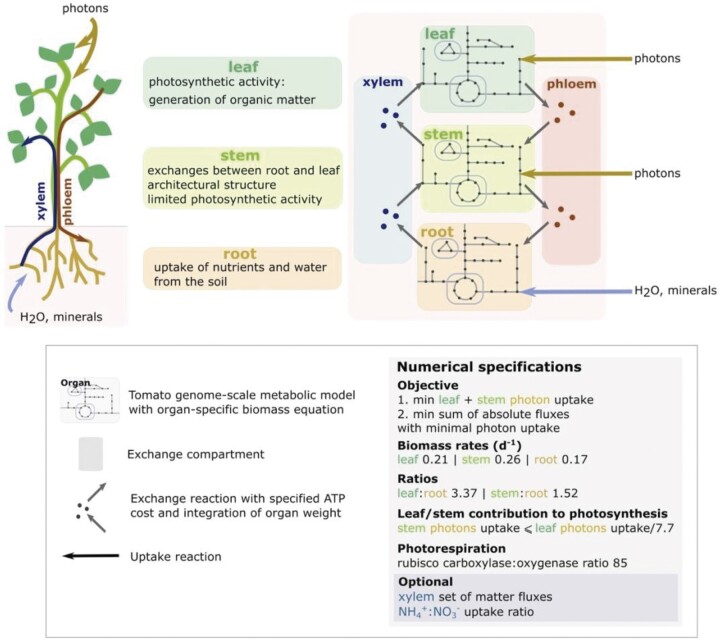
The multi-organ metabolic model, named VYTOP (Figure 1) was built by aggregating genome-scale metabolic models of each organ. The published tomato leaf genome-scale metabolic model iHY3410 (Yuan et al., 2016) was used as a starting point. We curated the model to integrate missing pathways such as the catabolism of different metabolites (lysine, ethanol [ETOH], isoleucine, beta-alanine, leucine, and cysteine). As the iHY3410 metabolic model identifiers (IDs) mostly followed the MetaCyc IDs nomenclature (Caspi et al., 2016), we converted them into the Biochemical Genetic and Genomic format (BiGG) (King et al., 2016) using a semi-automatic conversion framework. BiGG is widely used for genome-scale metabolic models from bacteria to microalgae and humans (Norsigian et al., 2020). The converted model was also manually curated to achieve mass balance for carbon (C), N, oxygen (O2), phosphate, and sulfur for each reaction. We name this tomato metabolic model Sl2183; it includes 2,183 reactions, 2,097 metabolites and 3,433 genes. It is available in different formats: table format (Supplemental File S1), Systems Biology Markup Language (SBML) format (Supplemental File S2), in the database BioModels (Glont et al., 2018) under the ID MODEL2111120001, and in the MetExplore database to enable pathway visualization and omics mapping (Cottret et al., 2018) https://metexplore.toulouse.inrae.fr/metexplore2/?idBioSource=6353.
Figure 1.
Generation and calibration of VYTOP based on a tomato genome-scale metabolic model. Top left panel: role of each organ as modeled in VYTOP. Top right: Schematic view of the different organs and exchange compartments as modeled in VYTOP. Bottom: legend, numerical constraints, and objective functions used in VYTOP.
To transpose this genome-scale metabolic model into a multi-organ model, we considered the whole plant as three main organs (Figure 1). Each organ was modeled with a replicate of the Sl2183 genome-scale metabolic model with an organ-specific biomass equation. In addition, each organ has specific physiological roles: photosynthesis/organic matter production in leaf, partial photosynthesis in stem, and minerals/water uptake in root represented by accurate physiological constraints. The transport tissues xylem and phloem were defined as exchange compartments: xylem represents exchanges from root to leaf and phloem represents exchanges from leaf to root. Stem can exchange with both compartments in the two directions: uptake or contribution to xylem and phloem. Leaf and stem assimilate photons (i.e. light) while root assimilates water and minerals. An ATP cost was set up for transport reactions between organs and xylem/phloem to include the transfer costs. The final VYTOP model includes 6,689 reactions and 6,326 metabolites.
VYTOP calibration with physiological and metabolic data
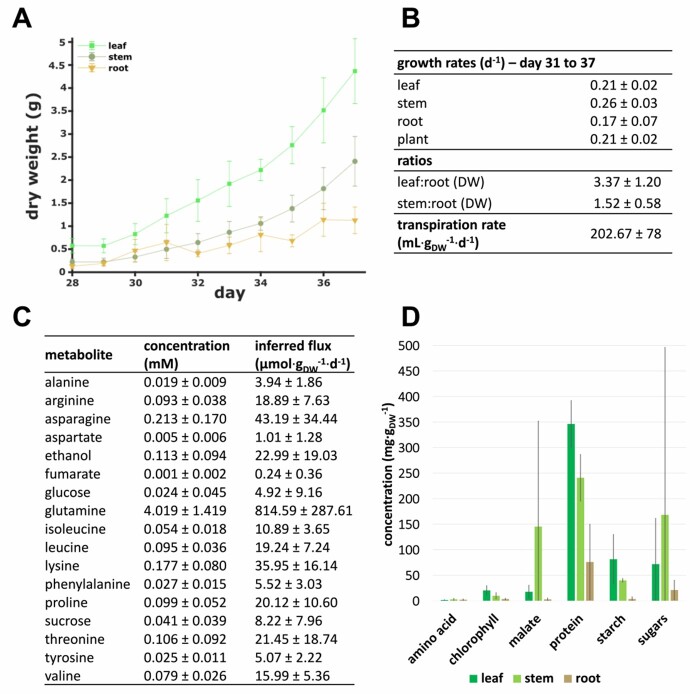
To calibrate VYTOP, we performed physiological and biochemical measurements (Figure 2) on 90 tomato plants. The growth of 90 plants was monitored on an automatic phenotyping facility, which allowed automatic monitoring of plant transpiration rates. Sampling of six plants per day was performed during 9 d, starting at 28 d after seeding. Xylem organic metabolite concentrations and organ weights were determined on sampled plants. The biomass composition of each organ was measured in another experiment performed in the same environmental conditions, with the same tomato variety and a similar plant age.
Figure 2.
Measurement of physiological and biochemical parameters on 4-week-old tomato plants for calibration of VYTOP. A, Organ dry weight evolution between Days 31 and 37. The weight (mean ± standard deviation) of six plants was measured by day. B, Growth rate of each organ (from Days 31 to 37), dry weight ratios between organs (from Days 29 to 37), and transpiration rate (from Days 31 to 37). Values (mean ± standard deviation) are based on three replicates for growth rate, 54 plants for dry weights ratios and on 42 plants for transpiration rate. C, Average organic xylem sap composition. Concentrations (mean ± standard deviation) are based on 33 samples analyzed using 1H NMR. d, Organ composition. Measurements are based on at least 19 samples per organ. Mean weights are plotted and bars indicate standard deviation.
A weight dataset of sampled plants was used to measure the growth rate for each organ, the organ dry weight ratios (leaf:root and stem:root) (Figure 2, A and B), and the fresh:dry weight ratio for each organ (Supplemental Figure S1). The growth was considered as exponential between Days 31 and 37 after sowing (Supplemental Figure S1) and regressions of fresh/dry, leaf/root, stem/root weights showed high correlation coefficients (Supplemental Figure S1). Growth rates were used in VYTOP to constrain biomass fluxes; organ ratios were implemented in exchange reactions to appropriately represent the difference of weight between the three organs (Figure 1). Transpiration was constant per mass unit, and was converted into a transpiration rate (Figure 2B). The transpiration rate was further used to compute organic xylem metabolite fluxes (see “Materials and methods”).
We measured organ biomass composition on fresh samples of growing tomato (Figure 2D). As expected, the leaf accumulates the highest proportion of starch and has the highest chlorophyll content. The data obtained was completed with data for lipids, minerals, (hemi)cellulose, and lignin from the literature to generate accurate organ-specific biomass equations (Supplemental File S1). For each organ, the biomass equation yields 1 g of dry weight biomass (Supplementary File S1).
In comparison with the Yuan et al. model iHY3410 (2016), our updated biomass equation for leaf had a C:N ratio closer to the one observed from experimental data (6.5) in a high CO2 level and high N provided as a nitrate (NO3−):ammonium () mixture (Royer et al., 2013): 5.48 in VYTOP versus 18.52 in iHY3410.
Quantification of organic metabolites concentration in xylem sap (Figure 2C) was performed using Nuclear Magnetic Resonance (NMR). Glutamine was the major metabolite in xylem sap (4.019 ± 1.419 mM). Fifteen other organic molecules were detected: 11 amino acids, 2 sugars, ETOH, and fumarate (FUM), whose concentrations were much lower than that of glutamine (<0.250 mM). Organic metabolic fluxes in xylem were computed using these concentration data and the transpiration rate of plants (see “Materials and methods”).
We also gathered data from literature to choose which compounds are present in the phloem compartment (Hijaz and Killiny, 2014) and to estimate photosynthetic activity in the stem. Former experimental results indicated that photosynthesis in the stem is limited due to less exposed surface to light (Hetherington et al., 1998). According to this study, the leaf:stem ratio of contribution to photosynthesis is around 7.7 (considering here the petioles as stems), and this value was added in the model as a constraint. The : uptake ratio can be optionally constrained in the model to take into account different fertilizer conditions. Since the fertilizer used for the experiments contained both and and we did not measure the uptake rate of each N source, the ratio was unconstrained for our simulations except for Use Case 4.
Model framework of VYTOP validated by experimental data
We used FBA (Orth et al., 2010), a constraint-based modeling approach to compute the metabolic fluxes in VYTOP. FBA requires a quasi-steady-state approximation (QSSA) in the whole system. This approximation is commonly accepted for bacterial growth modeling, but to extend the approach to plant vegetative growth, we checked if QSSA was still reasonable. We examined:
the daily weight ratios between organs and observed no substantial discrepancies between days (Supplemental Figure S1),
the organic chemical composition of xylem, and also observed no important daily variation (Supplemental Figure S2),
the three organ growths, which followed, as expected, an exponential growth (Supplemental Figure S1),
the biomass composition of each organ, and found that it had no substantial daily variation (Supplemental Figure S3).
These experimental observations performed over a 10-d period proved that we could reasonably extend QSSA to a tomato plant at the vegetative growth stage. We did not take into account the day/night variation as we measured plant growth on the scale of a day and not of the hour, considering that our simulation results represented an overall average of the metabolic fluxes over a 24-h period (Bénard et al., 2015).
Simulation of metabolic fluxes using FBA implies defining a linear optimization problem: the formulation of linear mathematical constraints (equalities or inequalities) and a linear objective function (maximization or minimization of one or several fluxes). The choice of this objective function is still debated for plants (Sweetlove and George Ratcliffe, 2011; Collakova et al., 2012). We assumed that plant metabolism is regulated to favor the most efficient use of light uptake and C fixation to grow, which was translated into minimization of the photon uptake flux, while growth was constrained via the biomass flux value. In addition, we assumed that plant metabolism was also efficient in the use of enzymes and solved a second optimization problem with objective function minimization of the sum of all the model’s fluxes (absolute value). This second simulation was also performed to avoid stoichiometric balanced cycles. The resulting fluxes of this double optimization problem were analyzed in the following sections.
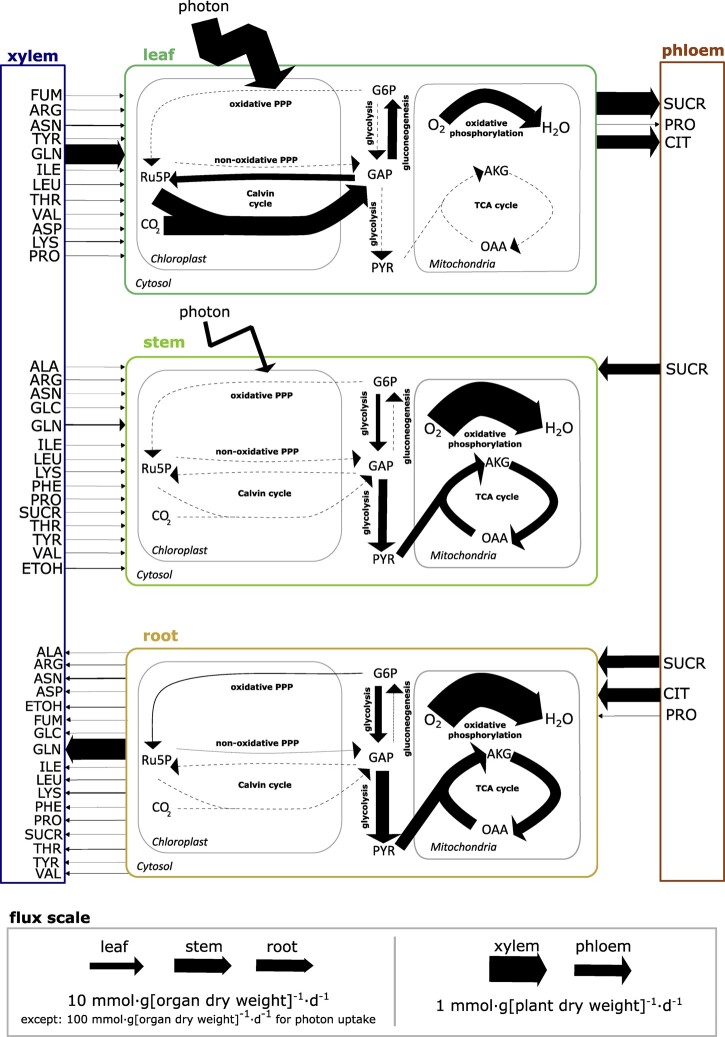
Use Case 1: VYTOP predicts metabolic fluxes in the whole plant
Figure 3 depicts the central metabolic fluxes obtained through modeling (Supplemental File S3). We used flux variability analysis (FVA) on the central metabolic fluxes to estimate how much they were constrained and if many alternative pathways existed. FVA provides for each reaction the minimum and maximum flux values that still sustain the optimal solution found in FBA (Mahadevan and Schilling, 2003). We found no variation on the fluxes obtained, except for some reactions of glycolysis (pyruvate [PYR] kinase and phosphofructokinase) that can either be performed in the chloroplast or in the cytosol (Supplemental File S3).
Figure 3.
Core C metabolic fluxes of a tomato plant at the vegetative growth stage predicted by VYTOP, with xylem fluxes constrained from experimental data. Each pathway flux was estimated based on a reaction flux value representative of the pathway (listed in Supplemental File S3). OAA, oxaloacetate; GAP, glyceraldehyde 3-phosphate; G6P, glucose-6-phosphate; Ru5P, ribulose-5-phosphate. Other abbreviations follow standard abbreviations for amino acids.
The analyses established that core metabolic fluxes are consistent with the current knowledge on plant physiology:
Metabolic activity is more intense in leaves than in stems and roots. This makes sense since leaves generate the major part of organic matter necessary for whole plant growth. In particular, there is intense photosynthetic activity and organic matter production in leaves with major fluxes in the Calvin cycle, while this pathway is not used in the root and is poorly used in the stem.
The generated organic matter is primarily converted into sugars (sucrose [SUCR], glucose [GLC], and fructose), either transformed into starch or transferred to the stem and root through the phloem flux. Most of the organic matter (>50% of leaf assimilated C) is used directly by the leaf, as it is the organ with the highest weight (Figure 2).
Stems need a lower SUCR uptake flux compared to roots (40% per gram of stem versus 60% per gram of root), because it has some photosynthetic activity allowing the production of part of the needed C and energy (as ATP and NADPH). Yet, the stem is still dependent on leaf C production.
Glycolysis and the TCA cycle have high fluxes in the stem and root, allowing the generation of energy and biomass precursors from phloem SUCR and citrate.
Previous multi-organ metabolic models of plants were developed (Shaw and Cheung, 2020). The term multi-organ model refers here to models that represent several plant organs with different roles for each organ and with metabolite exchanges between each organ and not to tissue-specific models which simulate one type of organ at a time. For the multi-organ metabolic models developed so far, the calibration of constraints (organ ratio, growth rate, and authorized fluxes) to accurately predict the behavior of a plant organ remains rudimentary and does not rely on extensive biochemical and physiological data. A question that also arises for these models is how to represent accurately metabolism differences between each organ. To this end, several frameworks were developed and rely on different strategies. Some of these frameworks such as the model of Arabidopsis developed by Mintz-Oron et al. (2012) used -omics data (RNAseq, proteomics, and metabolomics) to take into account organ/tissue specificities. The initial methodology consisted of pruning inappropriate reactions using an expression threshold on RNAseq data to decide whether or not the reaction takes place in the organ/tissue, and therefore should give a closer biological relevance. However, it is limited by the fact that the transcription does not include subsequent regulation layers (translation, posttranslational modifications, and enzyme activation), which can be particularly important for central C metabolism (de Groot et al., 2007). Also, choosing this expression threshold can be sometimes arbitrary, particularly for nonmodel plants. Other frameworks were developed since, which minimize these drawbacks, such as GIMME-like, iMAT-like, and MBA-like methods and the RegrEx method (Robaina Estévez and Nikoloski, 2014; Scheunemann et al., 2018). Even if thresholds can still be used in some of these methods, pruning is not as strict as the initial method since only a minimization of the discrepancy between the model and -omics data is performed. Finally, another multi-organ plant model relies on defined constraints with a global source–sink macroscopic model representing the allocation of C between organs (Grafahrend-Belau et al., 2013).
In our approach, we chose not to constrain each organ using -omics data, to allow all the possible chemical reactions a priori. To constrain VYTOP, we chose instead to use an important set of external physiological constraints (growth rates, ratios, well-calibrated biomass, and photosynthetic contributions) to impose organ-specific and sink/source metabolic behaviors in the different organs, which is closer to the methodology developed for barley. The C fluxes distribution in central metabolism obtained in VYTOP demonstrated that this approach, relying only on physiological constraints, allows an overall good prediction of metabolic fluxes between the different organs of a whole plant, despite the omission of -omics data. Its disadvantage is that it relies on the acquisition of physiological (and ideally metabolic) data that are not always available (whereas numerous sets of RNAseq data are available). However, it advantageously removes some of the methodological questions required to build -omics-based metabolic models (Richelle et al., 2019). In the future, it would be interesting to compare the predictions of VYTOP with models exploiting -omics data in order to reveal more precisely the advantages and drawbacks of each methodology.
Use Case 2: VYTOP assesses the impact of physiological changes on the plant
We used VYTOP to assess the influence of the stem on plant metabolism, namely its contribution to photosynthesis (Figure 4) and the effect of the stem/whole plant ratio (Figure 5).
Figure 4.
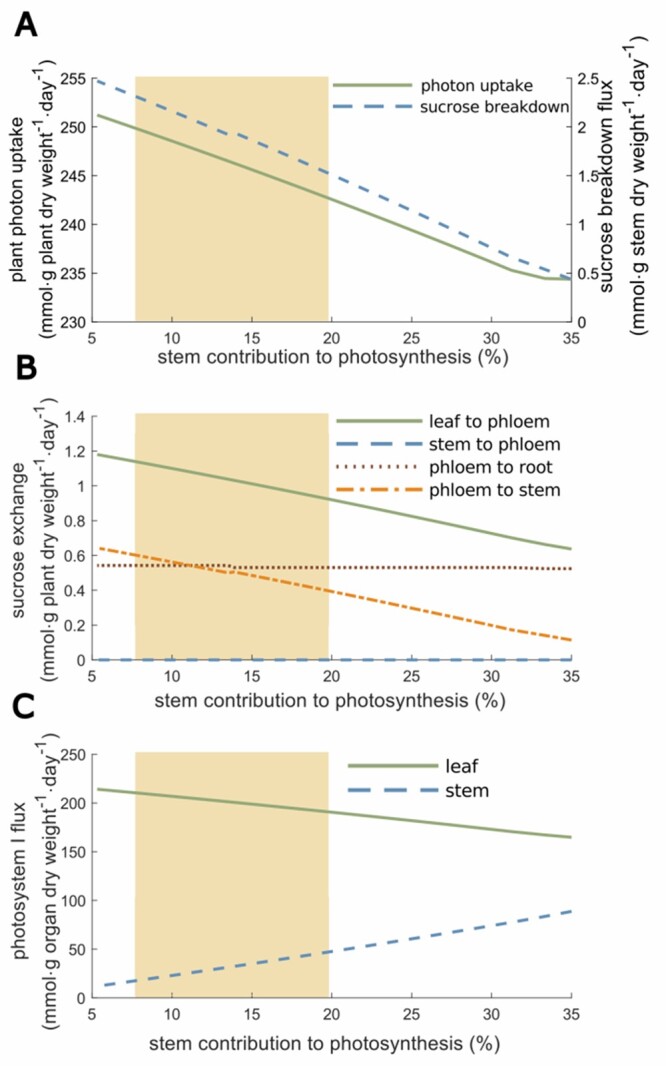
Effect of different leaf:stem photosynthetic capabilities on photon uptake, SUCR exchange, and photosystem I. Results are presented in percentage of stem contribution to photosynthesis. A, Effect of stem contribution to photosynthesis on plant photon uptake and SUCR breakdown fluxes. B, Effect of stem contribution to photosynthesis on SUCR exchange reactions between organs. C, Effect of stem contribution to photosynthesis on the photosystem I flux. The yellow area represents the range of experimentally observed values: mean ± standard deviation.
Figure 5.
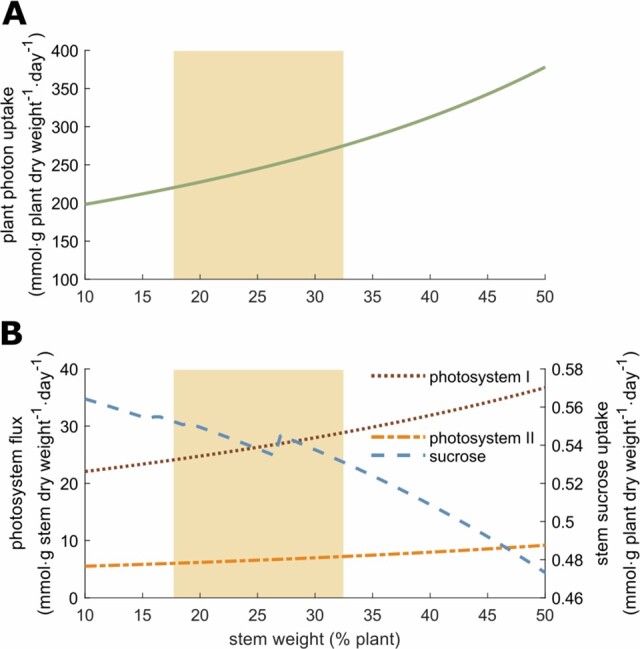
Effect of stem weight on photon uptake, the stem SUCR uptake flux, and the stem photosynthesis. A, Effect of stem weight on global plant photon uptake. B, Effect of the stem weight on the stem SUCR uptake flux and the stem photosystems I and II fluxes. The yellow area represents the range of experimentally observed values: mean ± standard deviation.
The contribution of the stem to photosynthesis represents the ability to perform photosynthesis comparatively to the leaf and depends on the surface of the stem (Hetherington et al., 1998). The model shows that this contribution strongly affects photon demand since the latter decreases rapidly while the stem photosynthesis capacity increases (Figure 4). In agreement with this observation, photosystem I metabolic flux strongly decreases in the leaf while it increases in the stem, as it becomes more advantageous to perform photosynthesis in the stem (also true for photosystem II, see Supplemental Figure S4). We also observed a proportional decrease of leaf export to phloem and stem uptake from phloem. However, there is no C contribution of the stem to phloem, because the range of values shown here did not allow sufficient photosynthetic activity of the stem to support both stem and root growth. Yet, a SUCR flux from stem to phloem appeared when stem contribution to photosynthesis exceeds 40% (Supplemental Text). Simulations also showed that increasing the stem weight proportion in the plant impacted progressively the photon demand (Figure 5). In this case, the plant stem plays a role of sink for C sources, which progressively becomes a burden affecting the photon demand (i.e. synthesis of C sources).
VYTOP does not integrate the architectural role of the stem in plant growth, enabling a better access to light that counterbalances its cost as sink of matter. A model integrating both plant geometry and metabolic fluxes would therefore be of great interest to study the tradeoff between the architectural role and the C sink, but remains a challenging issue.
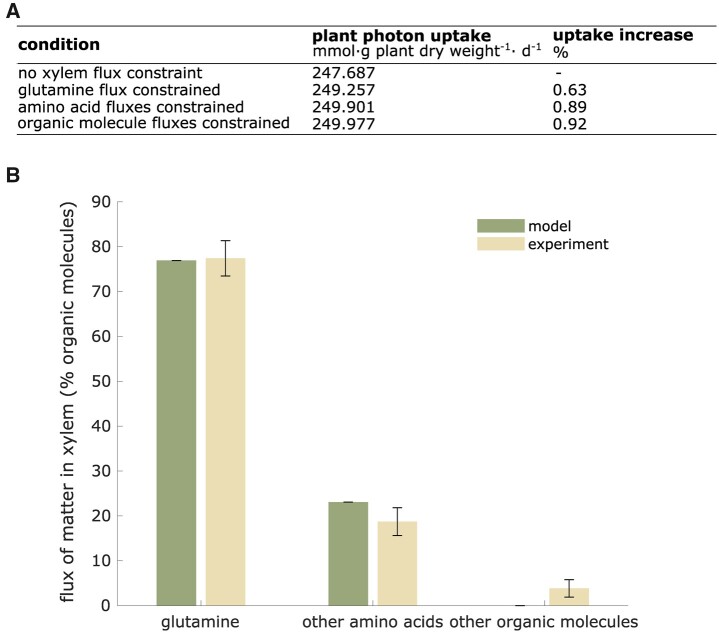
Use Case 3: VYTOP infers the relative composition of organic metabolites of tomato xylem sap by applying physiological constraints
To evaluate the relevance of VYTOP on exchange fluxes between organs, we simulated plant metabolic fluxes with the experimentally measured organic xylem fluxes on the root to xylem export reactions as additional constraints in the model. We analyzed how these additional constraints impacted the global photon demand (Figure 6A). We observed a very low effect on photon demand since photon demand increased by 0.63% if a constraint is set on glutamine, the major organic molecule in concentration. Photon demand increased by 0.92% if a constraint was set for each metabolite measured experimentally. Therefore, experimental values do not disturb much the photon demand. This emphasized that VYTOP is a consistent model of matter exchange between plant organs.
Figure 6.
Prediction of the relative composition of organic metabolites in tomato xylem fluxes by VYTOP. A, Impact of the addition of constraints on organic xylem fluxes (set from experimental data) on plant photon demand. B, Comparison of predicted fluxes by VYTOP and experimental fluxes of organic matter in xylem sap. Modeling results were obtained from a simulation without organic xylem flux constraint. Error bars indicate FVA range for the modeled distribution and mean ± standard deviation range for the experimental distribution (sample size: 33 plants).
In a second analysis, we compared the VYTOP composition of organic xylem fluxes to the experimental data (Figure 6B) with, this time, no constraints on root to xylem exports. The global features of xylem organic chemistry are well predicted by VYTOP: predominance (around 80%) of glutamine, a central metabolite found in xylem sap from diverse plant species is inferred by VYTOP. VYTOP also predicts the presence of other amino acids, in accordance with reported experimental data (Andersen and Brodbeck, 1989; Zuluaga et al., 2013; Anguita-Maeso et al., 2021). FVA results (Supplemental File S3) showed no variability for the different organic fluxes predicted, as represented in Figure 6B by the absence of error bars. FVA thus confirmed that VYTOP predicts the importance of glutamine in xylem sap, as observed by metabolomics experiments.
With only global physiological constraints, specific biomass equation, and resource use optimization (photon minimization and sum of flux minimization; Figures 1 and 2), VYTOP efficiently predicts transport of metabolites to aerial parts. This result shows that the organic composition of xylem sap is probably driven by plant physiology and resource optimization. The predominance of glutamine in both the model and experiments is remarkable. Glutamine is, with glutamate, the precursor of other amino acids and can be directly used as a C source since it is directly connected to the TCA cycle. Transporting a central metabolite, branched with both central metabolism and amino acid biosynthesis, thus appears more advantageous and robust. The predominance of glutamine over glutamate in xylem sap may be due to the acidic property of the latter, which could be deleterious at high concentrations. The presence of other amino acids is also a conserved property of xylem sap. We hypothesize that this redundancy of N sources could bring robustness to plant metabolism.
Several other organic molecules (organic acids, sugars, and ETOH) were observed experimentally but not predicted by the presented flux distribution (Figure 6B). This must be due to a certain level of exchange and porosity between compartments that exists in nature and is not taken into account in the model, such as possible exchange fluxes between xylem and phloem via stem cells. Furthermore, an ascending phloem is observed in tomato (Bonnemain, 1980), contributing to the observation of sugars in xylem sap. The presence of ETOH may reflect an O2 limitation in certain tomato cells, a local phenomenon not depicted in the model. Organic molecules can also act as shuttles for some ions, such as citrate for iron (Rellán-Álvarez et al., 2010). VYTOP does not represent these complex behaviors and provides a simplified and more schematized view of matter fluxes, nevertheless consistent for around 95% of the organic xylem sap content.
Use Case 4: VYTOP predicts the impact of nutritional limitation on the tomato growth rate
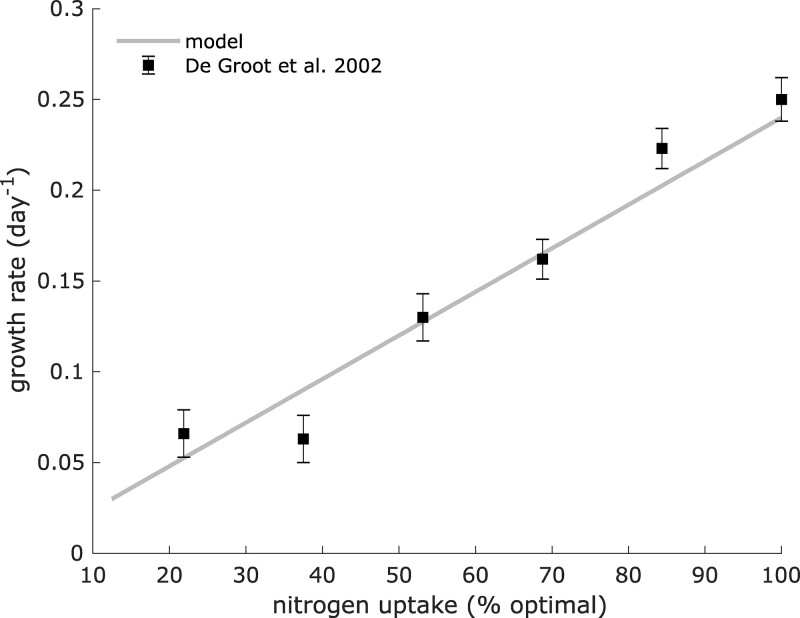
We used VYTOP to predict how a whole tomato plant responds to changes in a nutrient resource. De Groot et al. (2002) analyzed how the tomato growth rate was affected by N limitation. They monitored growth rates associated with different N supplies (Supplemental File S3), which revealed a progressive decrease of plant growth rate (Figure 7). To simulate the impact of N limitation in VYTOP, we used the N uptake flux and photon uptake computed in optimal conditions as constraints and progressively decreased the N uptake flux as in De Groot et al. (2002), upon the same photon availability. The simulation was performed with only as the N source, as in the experimental study. We assumed as a new objective function the maximization of leaf biomass growth, with a constant root/leaf and stem/leaf ratio, obtained from our experimental results in N replete conditions. The simulation predicted a linear decrease of growth rate, which followed the same pattern as for the experiment, with very close values. For example, both VYTOP and the experimental data showed a reduction of the plant growth rate from 0.24 to around 0.13/day when the N content is reduced by 50%. Thus, VYTOP agrees with the experimentally observed impact of N limitation on tomato plant growth.
Figure 7.
Impact of N limitation on plant growth. Experimental data were taken from De Groot et al. (2002) and are represented as mean ± standard deviation (sample size: 6). Growth rates were predicted using VYTOP model with a limited N uptake flux.
Use Case 5: VYTOP predicts and helps to understand the physiology and metabolism of transgenic tomato lines
We assessed how efficiently VYTOP can predict the effect of a genetic perturbation on plant physiology (i.e. mutant behavior). We selected four studies presenting transgenic tomato lines. Three lines express a fragment of a TCA cycle gene in the antisense orientation, resulting in a reduced enzymatic activity of, respectively, mitochondrial citrate synthase (mCS; Sienkiewicz-Porzucek et al., 2008), mitochondrial succinyl-CoA ligase (SCoAL; Studart-Guimarães et al., 2007) and mitochondrial alpha-ketoglutarate (AKG) dehydrogrenase (AKGDH; Araújo et al., 2012). The fourth line is a SUCR phosphate synthase (SPS) gene knockout (C.M Rick TGRC https://tgrc.ucdavis.edu/Data/Acc/GenRepeater.aspx?Gene=Sus). We modeled these transgenic lines with VYTOP by forcing the matter flux to zero for the knockout line or performing simulation at 50% of the optimal flux for attenuated lines, in agreement with the activity reduction presented in the three studies. We then analyzed the impact of these changes on metabolic fluxes (Table 1).
Table 1.
Effects of transgenic tomato lines on plant physiology and metabolism and comparison between experimentally observed effects and VYTOP predictions
| Enzyme | Activity | Effect | VYTOP prediction | Consistency |
|---|---|---|---|---|
| mCS | Attenuated | Minor effect on growth | Optimal growth achieved | + |
| mCS | Attenuated | Unaltered leaf photosynthesis | <1.5% increase | + |
| mCS | Attenuated | Enhanced CSp activity | CSp activation | + |
| mCS | Attenuated | Reduction of metabolism | No change | − |
| SCoAL | Attenuated | Minor effect on growth | Optimal growth achieved | + |
| SCoAL | Attenuated | Unaltered leaf photosynthesis | <1.5% increase | + |
| SCoAL | Attenuated | Evidences of GABA shunt use | GABA shunt activation | + |
| AKGDH | Attenuated | Unaltered leaf photosynthesis | <1.5% flux increase | + |
| AKGDH | Attenuated | Evidences of GABA shunt use | GABA shunt activation | + |
| AKGDH | Attenuated | Minor effect on growth | Optimal growth achieved | + |
| SPS | Suppressed | Lethal | Growth infeasible | + |
Experimental results for mCS, SCoAL, AKGDH, and SPS are, respectively, from Sienkiewicz-Porzucek et al. (2008), Studart-Guimarães et al. (2007), Araújo et al. (2012), and C.M Rick TGRC (https://tgrc.ucdavis.edu/Data/Acc/GenRepeater.aspx?Gene=Sus).
One major experimental result on the three lines with attenuated activity of the TCA cycle is that the impact on vegetative growth and young tomato physiology appears to be weak, with slightly reduced growth and photosynthetic activity similar to the wild-type line (Studart-Guimarães et al., 2007; Sienkiewicz-Porzucek et al., 2008; Araújo et al., 2012). Similarly, using VYTOP, we found that plant growth was the same. We also analyzed photosynthesis flux predicted in the model and found only a very limited impact (<1.5% of flux increase) compared to the wild-type model.
In the different experimental studies, authors suggested that there was a bypass to compensate the enzyme deficiencies in the transgenic plants. In line with this hypothesis, VYTOP predicted an activation of peroxisomal citrate synthase (pCS) flux for the line attenuated in mCS. The experimental study on the same line confirms this prediction since an enhanced activity of pCS is observed. Similarly, for the lines attenuated in mSCoAL and mAKGDH activities, VYTOP predicts the activation of three reactions: gamma-amino-butyric acid (GABA)-Transaminase, Succinic Semialdehyde Dehydrogenase, and Glutamate Decarboxylase, constituting the GABA shunt. This pathway can generate succinate from AKG without the use of AKGDH and SCoAL. Interestingly, metabolomics samplings and 14CO2 in the experimental studies also point toward the use of this pathway.
Conversely, no bypass exists after SPS gene knockout, as its silencing led to no growth in the resulting homozygous line. VYTOP also predicts this dependence as no growth can be obtained with silenced SPS flux. Thus, VYTOP seems to be an adequate tool to predict the alternative pathways that could be used to bypass a bottleneck in a transgenic line, or to predict the essentiality of some other reactions and the absence of alternative pathways.
VYTOP could thus be both a predictive tool to anticipate the effect of genetic engineering before generating a transgenic line, or a way to understand and analyze a phenotype observed in the transgenic line. However, it is worth noting that metabolism is often tightly linked with regulation, and that some effects are not predictable by VYTOP yet. As an example, the model could not predict the reduction of metabolism observed in plants with attenuated mCS activity, and the authors suggested that it was due to posttranslational regulation. The development of hybrid metabolic-regulation models is challenging, particularly because regulation is difficult to predict and reconstruct at a genome-scale level.
Conclusion
Modeling the complex interactions between nutritional sources, genes, energetic requirements, and exchange of matter is an ongoing challenge in plant systems biology. In this study, we developed a modeling framework that integrates all these components, based on a genome-scale metabolic model and physiological characterizations. The resulting tomato model VYTOP predicts consistent flux distribution and global behavior for each organ without constraining the internal fluxes, highlighting that the methodology used is relevant to build organ-specific models. We demonstrated that VYTOP gave consistent results in five different use cases: prediction of core metabolic fluxes and exchanges, role of the stem in the plant, analysis of the organic xylem sap composition, estimation of nutrient limitation impact on growth, and impact of genetic modifications on metabolism and physiology. This tool should be useful to guide genetic/metabolic engineering of plants as well as to understand at a system level how plants respond to nutritional variations. Other modeling approaches conducted on tomato linked plant architecture and physiology to environmental variations such as temperature (Cieslak et al., 2016; Pradal et al., 2008, 2015). It will be interesting to combine these macroscopic models with VYTOP. In addition, VYTOP could be extended to other tomato plant lifecycle stages, by adding metabolic models for flower and/or fruit. Adding other metabolic models for organisms interacting with the plant, such as fungi or bacteria (Peyraud et al., 2017), is also a promising perspective to explore plant interactions with other organisms.
Materials and methods
Experimental procedures
Plant cultures and automatic phenotyping
Tomato seeds (S. lycopersicum M82) were grown in soil (SB2, Proveen, The Netherlands) supplemented with Osmocote coated fertilizer at a rate of 4 g L−1. Osmocote coated fertilizer contained nitric N (5.3% w/v), ammoniacal N (6.7% w/v), 7% of phosphorus pentoxide (w/v), and 19% of potassium oxide (w/v).
Seeds were germinated in a growth chamber (26°C, 67% Relative Humidity, 12-h LED light per day). Around 100 plantlets were transplanted in individual plastic pots (8 × 8 cm) 8 d after sowing. Sixteen days after, 90 young plants were chosen and repotted in 3-L pots until the end of the experiment. Foam cover discs were placed on each pot to limit physical evaporation. The plants were loaded on the Phenoserre robot facility of the Toulouse Plant Microbe Phenotyping infrastructure. Twelve-hour-light per day at 28°C and 50% humidity were programmed.
All the plants were watered with 100 mL 3 times on the loading day and weighed, in order to define a well-watered target weight. The daily transpiration was determined as the weight differences between two consecutive days, at the time of watering. Transpiration rate was deduced as following: transpiration rate at Day i (mL·g[dry weight]−1·day−1) = transpiration at Day i (mL·day−1)/plant dry weight at Day i (g[dry weight]). Temperature, hygrometry, and light intensity were recorded during the whole experiment.
Collection and preparation of plant samples
Six plants were removed each day from the conveyor belt for samplings during nine consecutive days, starting 4 d after the loading on the robot. For these plants, stems were cut just above the cotyledons node, rinsed with ∼1 mL of water and the upcoming xylem sap was harvested by repeated pipetting and collection into Eppendorf tubes placed on ice. The tubes were placed at −80°C for further quantitative NMR analyses.
Quantitative NMR analyses
The xylem saps were analyzed by 1D 1H NMR on MetaToul analytics platform (UMR5504, UMR792, CNRS, INRAE, INSA 135 Avenue de Rangueil 31077 Toulouse Cedex 04, France), using the Bruker Avance 800 MHz equipped with an ATMA 5-mm cryoprobe. Each xylem sap sample was centrifuged to remove the residues (5 min, 13520 RCF, Hettich Mikro 200 centrifuge), then placed in 3-mm NMR tubes. TSP-d4 standard (Sodium 3-(trimethylsilyl)(1-13C,2H4)propanoate) was used as a reference. The pH 6.0 phosphate buffer was used to standardize the chemical shifts among samples. Acquisition conditions were as follows: 30° pulse angle, 20.0287 ppm spectral width, 64 scans per acquisition for a total scan time of ∼8 min per sample, and zgpr30 water presaturation sequence. The samples were kept at a temperature of 280 K (6.85°C) all along the analysis. Resonances of metabolites were manually integrated and the concentrations were calculated based on the number of equivalent protons for each integrated signal and on the TSP final concentration.
Xylem sampling and NMR analyses only give the concentration of a metabolite (in mmol·mL−1) at a given time. However, xylem sap is a continuous flow. Using the transpiration of the plant, and assuming a constant concentration of the metabolite in the xylem on a 24-h basis (validated throughout our experiment, see “Model framework of VYTOP is validated by experimental data”), the flow of metabolite per day can be estimated using the following formulae:
Metabolite flux (in mmol·g dry weight−1·day−1) = Metabolite concentration (in mmol·mL−1)· transpiration rate (mL·g dry weight−1.day−1).
Biochemical analyses of metabolites
The different organs of each plant were collected separately (stems, leaves, and roots) on another experiment performed on the same platform and plant variety and close plant age (4-week old). Approximately 300 mg fresh weight for each collected organ was frozen in liquid N and stored at −20°C for further biochemical analyses.
Quantifications of metabolites were performed at the High-Throughput Metabolic phenotyping (HiTMe) platform (INRAE-IBVM-71 avenue E. Bourlaux-CS 20032-33882 Villenave d'Ornon Cedex, France). The plants samples, previously frozen in liquid N, were ground to a powder using liquid N to avoid thawing. A quantity of 20 ± 10 mg of each was weighted in previously frozen Micronic tubes. Free amino acids, GLC, fructose, malate, proteins, starch, SUCR, and chlorophylls in leaves, stems, and roots were quantified as described in (Biais et al., 2014). Briefly, ethanolic extracts from every sample were obtained using three consecutive incubations of the frozen ground powder aliquots. ETOH 80% v/v with HEPES/KOH 10 mM pH 6 buffer was used for the first two incubations, and ETOH 50% v/v with HEPES/KOH 10 mM pH 6 buffer was used for the third. Supernatants were pooled and used for the quantification of chlorophylls, GLC, fructose, SUCR, malate, and free amino acids. Pellets were used for the determination of protein and starch contents. The extracts and pellets were stored at −20°C between each quantification. For each sample, chlorophylls were quantified by measuring optical densities at 645 and 665 nm on a mix of 50 µL of extract supplemented with 150 µL of analytics grade ETOH. Amino acids were quantified using the fluorescamine method. Excitation wavelength was 405 nm and emission was measured at 485 nm. The proteins were quantified using Bradford reagent. Starch was quantified in GLC equivalent after full pellet digestion in an oven at 37°C for 18 h. For the other analytes cited above, the NADH/NADPH appearance was measured, and the analytes were quantified using a 1:1 stoichiometric coefficient.
In silico procedures
Tomato metabolic network curation and conversion
The tomato metabolic model iHY3410 published by Yuan et al. (2016) was used as a starting point. The authors provided the tomato metabolic network in two formats: table (.xlsx) and SBML (Hucka et al., 2003). Some reaction directions were different in the SBML and table files, and some metabolites (proton and water) were missing in the table file. Thus, we performed a first curation step to merge the files and select the appropriate reaction directions.
Then, we converted the metabolic network IDs (metabolites and reactions) to BiGG nomenclature. The advantage of BiGG nomenclature is that it is usually more explicit for the analysis as the IDs are usually an abbreviation of the metabolite/reaction names, while in MetaCyc ID it is often a generic acronym (CPD for metabolites, RXN for reactions) and a number. The web tool Semi Automatic Metabolic Identifier Reconciliation (Peyraud et al., 2016) was used to find the appropriate BiGG IDs for metabolites and reactions. The correspondences between MetaCyc and BiGG IDs are provided in the final metabolic network in table format (Supplemental File S1). For some metabolites and reactions, no BiGG ID was found even after manual searches in the BiGG database, probably because they are specific to plants. New BiGG IDs following the usual nomenclature were created for 1,094 reactions and 758 metabolites (see Supplemental File S1). The SBML file (Hucka et al., 2003) was generated using Met4j (https://forgemia.inra.fr/metexplore/met4j) and ModelPolisher (Römer et al., 2016).
The resulting metabolic model was tested on autotrophic growth and heterotrophic growth (using FBA, see “Computational Simulations”) on different organic substrates before generating a multi-organ metabolic model. Growth was achievable with CO2 + light, starch, SUCR, GLC, fructose, glycerol, glutamate, glutamine, AKG, FUM, asparagine, alanine, and isocitrate as C sources. Other organic (glutamine, glutamate, aspartate, asparagine, alanine, proline, histidine, leucine, methionine, lysine, and cysteine) and inorganic ( and ) elements can be used as N sources. No catabolic reactions were found for the assimilation of lysine, ETOH, isoleucine, beta-alanine, leucine, and cysteine as C sources whereas they were experimentally observed in the xylem in amount superior to biomass assimilation, suggesting that they could be catabolized. Thus, we searched the catabolic pathways of these amino acids in plants and incorporated them in the network, using BLAST to find the orthologous proteins in tomato. Twenty-one reactions, linked to 34 genes were added to the model iHY3410. Fifteen reactions had a Gene-Protein-Reaction (GPR) complete with an e-value superior to 5e-131 (Supplemental File S1), 6 had none. After this second curation step, we computed the mass balance (on C, N, phosphate, sulfur, and O2) and manually curated the reactions with an incorrect C, N, phosphate, sulfur, and O2 balance. We generated a final tomato metabolic network Sl2183. The model is available in MetExplore (Cottret et al., 2018)https://metexplore.toulouse.inrae.fr/metexplore2/?idBioSource=6353, in the database BioModels under the ID MODEL2111120001 (Glont et al., 2018) and in a github repository https://github.com/lgerlin/slyc-metabolic-model/.
VYTOP construction
To generate VYTOP (see flow chart in Supplemental Figure S5), we built organ-specific metabolic models for leaf, stem, and root. To this end, organ growth rates and organ-specific biomass equations were determined from our experiments and implemented as constraints in the simulations. We generated an in-house script to parse the metabolic network Sl2183 (SBML file) and triplicated it to represent three organs. In the generated model, each organ is represented as a “metacompartment”: metabolite and reaction IDs have a final letter (l, s and r for leaf, stem and root, respectively) to indicate their organ location (e.g. R_NAD2 reaction becomes R_NAD2_l, R_NAD2_s, R_NAD2_r and M_gln_L_c becomes M_gln_L_c_l, M_gln_L_c_s, M_gln_L_c_r).
Our in-house script also generates exchange reactions to represent transfers between organs and exchange compartments (represented with the letters xyl for xylem and phl for phloem), such as 1 M_gln_L_c_r → n M_gln_L_xyl and 1 M_gln_L_xyl → m M_gln_L_c_l. The list of metabolites authorized to be exchanged in xylem and phloem respectively is available in Supplemental File S1. n and m represent the mass ratios between the organs and the whole plant, in order to take into account the different weights of organs and have quantitative predictions at the whole plant level. For example, for transfer from the root to the xylem, 1 µmol.g−1 root dry weight per day will generate n = 1/(1 + 1.52 + 3.37) = 0.1697 µmol.g−1 plant dry weight per day in xylem, and then 1 µmol.g−1 plant dry weight per day in xylem will generate m = (1 + 1.52 + 3.37)/3.37 = 1.74 µmol.g−1 leaf dry weight per day. Finally, a transport cost (ATP cost) of 0.5 was added in the model: transporting k µmol.g−1 organ dry weight per day would consume k·(transport cost) µmol.g−1 organ dry weigh per day of ATP. Performing several simulations of Use Case 3 with different values of ATP transport cost, we found that a transport cost between 0.02 and 1.0 mol of ATP per mol of exchanged molecule, which appears reasonable, provided the most consistent predictions in agreement with experimental data, while use Case 4 is not impacted by the different sets of values tested (Supplemental Text).
Physiological constraints were added to impose different physiological roles in organs.
Uptake of minerals and water is not allowed in leaf and stem, while it is allowed with no limitation in roots (assuming they are not limiting factors for well-watered plants and adequate nutritional supply in the soil).
Photon uptake was allowed in leaf and stem but not in root.
The : uptake ratio can be constrained. Few data are available to predict the balance between and in root uptake. We analyzed the impact of varying the parameter in Supplemental Text. The ratio was not constrained in our simulations, excepted use Case 4, to agree with the experimental data.
Photosynthesis is limited in stems by its reduced exchange surface compared to leaves (Hetherington et al., 1998). We then introduced a ratio (leaf/stem) of contribution to photosynthesis, to limit the ability of the stem to perform photosynthesis: photosynthesis in stems is constrained to be inferior or equal to photosynthesis in leaves/surface ratio.
Photorespiration was modeled by imposing a ratio between rubisco carboxylase and oxygenase fluxes. The ratio was set to 85, which represents the mean ratio of enzymatic specificity for CO2 versus O2 for C3 plants, according to experimental data gathered by Tcherkez et al. (2006).
Finally, ATP maintenance, which represents the global cost in a cell for nonmetabolic processes (such as housekeeping functions), was kept at the value used by Yuan et al. (2016) as no organ-specific ATP maintenance values are available.
Computational simulations
Modeling of whole tomato plant metabolism was performed using constrained-based modeling, with the FBA methodology (Orth et al., 2010). Briefly, FBA relies on (1) the QSSA in the biological system modeled, (2) the formulation of a biologically relevant objective: flux(es) to minimize/maximize, and (3) eventually additional physiological constraints. The solution of FBA will be the optimal matter fluxes (regarding the objective) in a metabolic network assuming QSSA and respecting the constraints. Use of QSSA in a whole tomato plant is justified by our experimental results (see part “Model framework of VYTOP is validated by experimental data” of the “Results and Discussion”). We decided to define the minimization of total photon uptake as our objective. The photon uptake flux obtained from this simulation was then integrated as a new constraint in the model, and FBA was run a second time with the objective of minimizing the sum of absolute fluxes. The use of these two objectives is discussed in part “Model framework of VYTOP is validated by experimental data” of “Results and Discussion”.
FBA was run with or without the integration of experimentally measured organic xylem fluxes, and with varying stem proportions, surface ratios, and transport costs.
FVA (Mahadevan and Schilling, 2003) was also performed. This extension of the FBA aims at finding alternative solutions of the one generated by FBA, as its flux distribution is usually not unique. It consists of imposing the optimal objective values found in FBA (here both photon uptake and sum of absolute fluxes) as additional constraints, and determining the minimum and maximum flux that can carry each reaction in these conditions.
Simulations were performed with Python version 3.5 scripts, the open access libraries lxml, pandas and the linear programming solver CPLEX Python API, developed by IBM and free for academic institutions. Scripts are available at https://github.com/lgerlin/slyc-metabolic-model.
Accession numbers
The tomato genome used in this article is ITAG version 4.0 and can be found on the Sol Genomic Networks website (ftp://ftp.solgenomics.net/tomato_genome/annotation/ITAG4.0_release/).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Additional physiological data.
Supplemental Figure S2. Additional xylem metabolomics data.
Supplemental Figure S3. Additional organ metabolomics data.
Supplemental Figure S4. Additional representation of model responses to plant physical variations.
Supplemental Figure S5. Flow chart of the multi-organ modeling pipeline.
Supplemental Text. Additional analyses (effects of ATP cost variation, Impact of physiological changes on the plant at high percentages, Impact of the uptake ratio between nitrates and ammonium, Stoichiometric Balance Cycles).
Supplemental File S1. Table of tomato plant genome-scale metabolic network Sl2183.
Supplemental File S2. SBML file of tomato plant genome-scale metabolic network Sl2183, SBML Level 3 version 1.
Supplemental File S3. Table with modeling results.
Supplementary Material
Acknowledgments
We thank Toulouse Plant Microbe Phenotyping (TPMP) platform (Castanet-Tolosan, France) and its staff Nemo Peeters, Felicià Maviane Macia, and Fabrice Devoilles for their technical support in plant cultures and imaging, as well as HiTMe platform (Villenave d’Ornon, France) for biochemical analysis of organ metabolites. We acknowledge MetaToul (Metabolomics & Fluxomics Facitilies, Toulouse, France, www.metatoul.fr) platform, which is part of the MetaboHUB-ANR-11-INBS-0010 national infrastructure (www.metabohub.fr) and its staff Cécilia Berges, Edern Cahoreau, and Lindsay Peyriga for access to NMR facilities. We also thank Sophie Colombié for helpful discussions and feedbacks on the manuscript.
Data availability
All data supporting the findings of this study are available within the paper and within its supplementary materials published online.
Funding
L.G. and A.E. were, respectively, funded by a grant from the French Ministry of Higher Education and Research and by the French Laboratory of Excellence TULIP (ANR-10-LABX-41 and ANR-11-IDEX-0002-02). The study was funded by the French Laboratory of Excellence (LABEX) project TULIP (ANR-10-LABX-41 and ANR-11-IDEX-0002-02). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest statement: None declared.
C.B. conceived the project and designed the experiments; C.B. and L.C. supervised the construction, calibration, and validation of the model; L.G. constructed, calibrated, and validated the model; C.B. and S.G. supervised the experiments; A.E. and L.G. performed the phenotypic experiments and A.E. performed the metabolomics analysis; L.G., A.E., and C.B. analyzed the data; L.G. wrote the draft article; C.B., L.C., and S.G. supervised and completed the writing. C.B. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Caroline Baroukh (caroline.baroukh@inrae.fr).
References
- Anguita-Maeso M, Haro C, Montes-Borrego M, De La Fuente L, Navas-Cortés JA, Landa BB (2021) Metabolomic, Ionomic and Microbial Characterization of Olive Xylem Sap Reveals Differences According to Plant Age and Genotype. Agronomy 11: 1179 [Google Scholar]
- Araújo WL, Tohge T, Osorio S, Lohse M, Balbo I, Krahnert I, Sienkiewicz-Porzucek A, Usadel B, Nunes-Nesi A, Fernie AR (2012) Antisense inhibition of the 2-oxoglutarate dehydrogenase complex in tomato demonstrates its importance for plant respiration and during leaf senescence and fruit maturation. Plant Cell 24: 2328–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold A, Nikoloski Z (2014) Bottom-up metabolic reconstruction of arabidopsis and its application to determining the metabolic costs of enzyme production. Plant Physiol 165: 1380–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold A, Sajitz-Hermstein M, Nikoloski Z (2015) Effects of varying nitrogen sources on amino acid synthesis costs in Arabidopsis thaliana under different light and carbon-source conditions. PLoS One 10: 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénard C, Bernillon S, Biais B, Osorio S, Maucourt M, Ballias P, Deborde C, Colombié S, Cabasson C, Moing A, et al. (2015) Metabolomic profiling in tomato reveals diel compositional changes in fruit affected by source-sink relationships. J Exp Bot 66: 3391–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biais B, Bénard C, Beauvoit B, Colombié S, Prodhomme D, Ménard G, Bernillon S, Gehl B, Gautier H, Ballias P, et al. (2014) Remarkable reproducibility of enzyme activity profiles in tomato fruits grown under contrasting environments provides a roadmap for studies of fruit metabolism. Plant Physiol 164: 1204–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnemain JL (1980) Microautoradiography as a tool for the recognition of phloem transport. Bericht Deutsch Bot Gesellsch 93: 99–107 [Google Scholar]
- Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Karp PD. et al. (2016) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 44: D471–D480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieslak M, Cheddadi I, Boudon F, Baldazzi V, Génard M, Godin C, Bertin N (2016) Integrating physiology and architecture in models of fruit expansion. Front Plant Sci 7: 1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TJ, Guo L, Morgan J, Schwender J (2020) Modeling plant metabolism: from network reconstruction to mechanistic models. Ann Rev Plant Biol 71:303–326 [DOI] [PubMed] [Google Scholar]
- Collakova E, Yen JY, Senger RS (2012) Are we ready for genome-scale modeling in plants? Plant Sci 191–192: 53–70 [DOI] [PubMed] [Google Scholar]
- Colombié S, Beauvoit B, Nazaret C, Bénard C, Vercambre G, Le Gall S, Biais B, Cabasson C, Maucourt M, Gibon Y, et al. (2017) Respiration climacteric in tomato fruits elucidated by constraint-based modelling. New Phytologist 213: 1726–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombié S, Nazaret C, Bénard C, Biais B, Mengin V, Solé M, Fouillen L, Dieuaide-Noubhani M, Mazat JP, Gibon Y, et al. (2015) Modelling central metabolic fluxes by constraint-based optimization reveals metabolic reprogramming of developing Solanum lycopersicum (tomato) fruit. Plant J 81: 24–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottret L, Frainay C, Chazalviel M, Cabanettes F, Gloaguen Y, Camenen E, Merlet B, Heux S, Portais JC, Jourdan F, et al. (2018) MetExplore: collaborative edition and exploration of metabolic networks. Nucleic Acids Res 46: W495–W502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot CC, Marcelis LFM, Van den Boogaard R, Lambers H (2002) Interactive effects of nitrogen and irradiance on growth and partitioning of dry mass and nitrogen in young tomato plants. Funct Plant Biol 29: 1319–1328 [DOI] [PubMed] [Google Scholar]
- de Groot MJL, Daran-Lapujade P, van Breukelen B, Knijnenburg TA, de Hulster EAF, Reinders MJT, Pronk JT, Hec AJR, Slijper M (2007) Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post-transcriptional regulation of key cellular processes. Microbiology 153: 3864–3878 [DOI] [PubMed] [Google Scholar]
- Gerlin L, Frainay C, Jourdan F, Baroukh C, Prigent S (2021) Plant genome-scale metabolic networks. Advances in Botanical Research, Vol. 98. Academic Press Inc., Cambridge, MA, pp 237–270 [Google Scholar]
- Glont M, Nguyen TVN, Graesslin M, Hälke R, Ali R, Schramm J, Wimalaratne SM, Kothamachu VB, Rodriguez N, Hermjakob H, et al. (2018) BioModels: expanding horizons to include more modelling approaches and formats. Nucleic Acids Res 46: D1248–D1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes de Oliveira Dal’Molin C, Quek LE, Saa PA, Nielsen LK (2015) A multi-tissue genome-scale metabolic modeling framework for the analysis of whole plant systems. Front Plant Sci 6: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafahrend-Belau E, Junker A, Eschenröder A, Müller J, Schreiber F, Junker BH (2013) Multiscale metabolic modeling: dynamic flux balance analysis on a whole-plant scale. Plant Physiol 163: 637–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Kim GB, Kim WJ, Kim HU, Lee SY (2019) Current status and applications of genome-scale metabolic models. Genome Biol 20: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington SE, Smillie RM, Davies WJ (1998) Photosynthetic activities of vegetative and fruiting tissues of tomato. J Exp Bot 49: 1173–1181 [Google Scholar]
- Hijaz F, Killiny N (2014) Collection and chemical composition of phloem sap from Citrus sinensis L. Osbeck (sweet orange). PLoS One 9: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Wang J, et al. (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19: 524–531 [DOI] [PubMed] [Google Scholar]
- King ZA, Lu J, Dräger A, Miller P, Federowicz S, Lerman JA, Ebrahim A, Palsson BO, Lewis NE (2016) BiGG models: a platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res 44: D515–D522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen Y, Zhou L, You S, Deng H, Chen Y, Alseekh S, Yuan Y, Fu R, Zhang Y, et al. (2020) MicroTom metabolic network: rewiring tomato metabolic regulatory network throughout the growth cycle. Mol Plant 13: 1203–1218 [DOI] [PubMed] [Google Scholar]
- Mahadevan R, Schilling CH (2003) The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng 5: 264–276 [DOI] [PubMed] [Google Scholar]
- Mintz-Oron S, Meir S, Malitsky S, Ruppin E, Aharoni A, Shlomi T (2012) Reconstruction of Arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc Natl Acad Sci 109: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira TB, Shaw R, Luo X, Ganguly O, Kim HS, Coelho LGF, Cheung CYM, Williams TC R. (2019) A genome-scale metabolic model of soybean (Glycine max) highlights metabolic fluxes in seedlings. Plant Physiol 180: 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norsigian CJ, Pusarla N, McConn JL, Yurkovich JT, Dräger A, Palsson BO, King Z (2020) BiGG Models 2020: multi-strain genome-scale models and expansion across the phylogenetic tree. Nucleic Acids Res 48: D402–D406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JJD, Thiele I, Palsson BBØ (2010) What is flux balance analysis? Nat Biotechnol 28: 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyraud R, Cottret L, Marmiesse L, Gouzy J, Genin S (2016) A resource allocation trade-off between virulence and proliferation drives metabolic versatility in the plant pathogen Ralstonia solanacearum. PLoS Pathog 12: e1005939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyraud R, Dubiella U, Barbacci A, Genin S, Raffaele S, Roby D (2017) Advances on plant-pathogen interactions from molecular toward systems biology perspectives. Plant J 90: 720–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfau T, Christian N, Masakapalli SK, Sweetlove LJ, Poolman MG, Ebenhöh O (2018) The intertwined metabolism during symbiotic nitrogen fixation elucidated by metabolic modelling. Sci Rep 8: 12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradal C, Dufour-Kowalski S, Boudon F, Fournier C, Godin C (2008) OpenAlea: a visual programming and component-based software platform for plant modelling. Funct Plant Biol 35: 751. [DOI] [PubMed] [Google Scholar]
- Pradal C, Fournier C, Valduriez P, Cohen-Boulakia S (2015) OpenAlea: scientific workflows combining data analysis and simulation. ACM International Conference Proceeding Series. Association for Computing Machinery, New York, NY, pp 1–6
- Rellán-Álvarez R, Giner-Martínez-Sierra J, Orduna J, Orera I, Rodríguez-Castrilln JÁ, García-Alonso JI, Abadía J, Álvarez-Fernández A (2010) Identification of a tri-Iron(III), tri-citrate complex in the xylem sap of iron-deficient tomato resupplied with iron: new insights into plant iron long-distance transport. Plant Cell Physiol 51: 91–102 [DOI] [PubMed] [Google Scholar]
- Richelle A, Joshi C, Lewis NE (2019) Assessing key decisions for transcriptomic data integration in biochemical networks. PLoS Computat Biol 15: e1007185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaina-Estévez S, Daloso DM, Zhang Y, Fernie AR, Nikoloski Z (2017) Resolving the central metabolism of Arabidopsis guard cells. Sci Rep 7: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaina Estévez S, Nikoloski Z (2014) Generalized framework for context-specific metabolic model extraction methods. Front Plant Sci 5: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer M, Eichner J, Dräger A, Wrzodek C, Wrzodek F, Zell A (2016) ZBIT bioinformatics toolbox: a web-platform for systems biology and expression data analysis. PLoS One 11: e0149263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer M, Larbat R, Le Bot J, Adamowicz S, Robin C (2013) Is the C:N ratio a reliable indicator of C allocation to primary and defence-related metabolisms in tomato? Phytochemistry 88: 25–33 [DOI] [PubMed] [Google Scholar]
- Saha R, Suthers PF, Maranas CD (2011) Zea mays irs1563: a comprehensive genome-scale metabolic reconstruction of maize metabolism. PLoS One 6: e21784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheunemann M, Brady SM, Nikoloski Z (2018) Integration of large-scale data for extraction of integrated Arabidopsis root cell-type specific models. Sci Rep 8: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver SMD, Lerma-Ortiz C, Conrad N, Mikaili A, Sreedasyam A, Hanson AD, Henry CS (2018) PlantSEED enables automated annotation and reconstruction of plant primary metabolism with improved compartmentalization and comparative consistency. Plant J 95: 1102–1113 [DOI] [PubMed] [Google Scholar]
- Shaw R, Cheung CYM (2018) A dynamic multi-tissue flux balance model captures carbon and nitrogen metabolism and optimal resource partitioning during Arabidopsis growth. Front Plant Sci 9: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R, Cheung CYM (2020) Multi - tissue to whole plant metabolic modelling. Cell Mol Life Sci 77: 489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R, Maurice Cheung CY (2019) A mass and charge balanced metabolic model of Setaria viridis revealed mechanisms of proton balancing in C4 plants. BMC Bioinform 20: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienkiewicz-Porzucek A, Nunes-Nesi A, Sulpice R, Lisec J, Centeno DC, Carillo P, Leisse A, Urbanczyk-Wochniak E, Fernie AR (2008) Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth. Plant Physiol 147: 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studart-Guimarães C, Fait A, Nunes-Nesi A, Carrari F, Usadel B, Fernie AR (2007) Reduced expression of succinyl-coenzyme a ligase can be compensated for by up-regulation of the γ-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol 145: 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, George Ratcliffe R (2011) Flux-balance modeling of plant metabolism. Front Plant Sci 2: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez GGB, Farquhar GD, Andrews TJ (2006) Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc Natl Acad Sci USA 103: 7246–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Cheung CYM, Poolman MG, Hilbers PAJJ, van Riel NAWW (2016) A genome-scale metabolic network reconstruction of tomato (Solanum lycopersicum L.) and its application to photorespiratory metabolism. Plant J 85: 239–304 [DOI] [PubMed] [Google Scholar]
- Zuluaga AP, Puigvert M, Valls M (2013) Novel plant inputs influencing Ralstonia solanacearum during infection. Front Microbiol 4: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary materials published online.