Abstract
Annatto (Bixa orellana) is a perennial shrub native to the Americas, and bixin, derived from its seeds, is a methoxylated apocarotenoid used as a food and cosmetic colorant. Two previous reports claimed to have isolated the carotenoid cleavage dioxygenase (CCD) responsible for the production of the putative precursor of bixin, the C24 apocarotenal bixin dialdehyde. We re-assessed the activity of six Bixa CCDs and found that none of them produced substantial amounts of bixin dialdehyde in Escherichia coli. Unexpectedly, BoCCD4-3 cleaved different carotenoids (lycopene, β-carotene, and zeaxanthin) to yield the C20 apocarotenal crocetin dialdehyde, the known precursor of crocins, which are glycosylated apocarotenoids accumulated in saffron stigmas. BoCCD4-3 lacks a recognizable transit peptide but localized to plastids, the main site of carotenoid accumulation in plant cells. Expression of BoCCD4-3 in Nicotiana benthamiana leaves (transient expression), tobacco (Nicotiana tabacum) leaves (chloroplast transformation, under the control of a synthetic riboswitch), and in conjunction with a saffron crocetin glycosyl transferase, in tomato (Solanum lycopersicum) fruits (nuclear transformation) led to high levels of crocin accumulation, reaching the highest levels (>100 µg/g dry weight) in tomato fruits, which also showed a crocin profile similar to that found in saffron, with highly glycosylated crocins as major compounds. Thus, while the bixin biosynthesis pathway remains unresolved, BoCCD4-3 can be used for the metabolic engineering of crocins in a wide range of different plant tissues.
A carotenoid cleavage dioxygenase from Bixa orellana elicits crocin production in Solanaceae.
Introduction
Carotenoids are C40 isoprenoid compounds acting as photoreceptors and photoprotectants in leaves, as precursors of signaling molecules such as ABA and strigolactones, and as pigments in fruits and flowers. Their cleavage products, apocarotenoids, are synthesized through the action of carotenoid cleavage dioxygenases (CCDs) endowed with different cleavage specificities and acting on different substrates (Giuliano et al., 2003a; Ahrazem et al., 2016). In plants, apocarotenoids have functions in both intra- and inter-plant signaling and as aromas and pigments (Giuliano et al., 2003a; Walter and Strack, 2011; Hou et al., 2016; D’Alessandro and Havaux, 2019; Moreno et al., 2020).
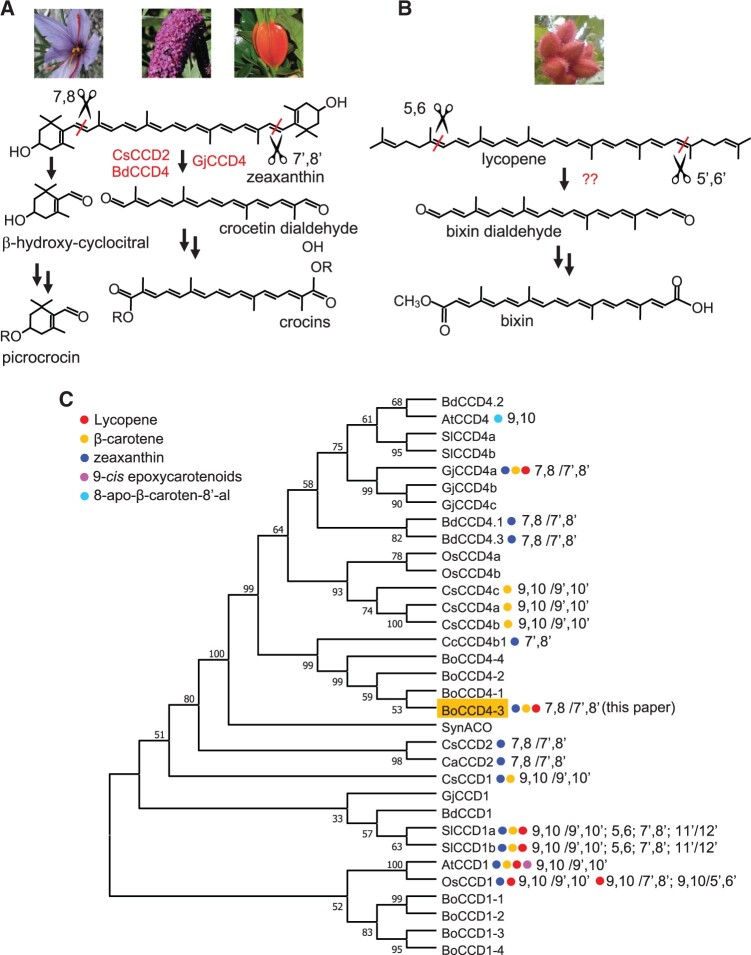
Apocarotenoid pigments are often accumulated at very high levels in non-photosynthetic tissues of different groups of flowering plants. Examples include citraurin, a C30 apocarotenal from tangerine (Citrus clementina) peel (Rodrigo et al., 2013); crocins, a complex mixture of glucosyl esters of the C20 apocarotenoid crocetin, accumulated in saffron (Crocus sativus stigmas) (Frusciante et al., 2014), Buddleja (Buddleja davidii) flowers (Ahrazem et al., 2017), and gardenia (Gardenia jasminoides) fruits (Xu et al., 2020; Figure 1, A) and bixin, a methoxy ester of the C24 apocarotenoid norbixin produced by annatto (Bixa orellana) seeds (Giuliano et al., 2003b; Rivera-Madrid et al., 2016; Figure 1, B).
Figure 1.
Apocarotenoid biosynthesis pathways in different plant species. A, Crocin biosynthesis pathways in C. sativus, B. davidii, and G. jasminoides. B, Proposed bixin biosynthesis pathway in B. orellana. C, Maximum-likelihood phylogenetic tree of CCDs from A. thaliana (At), B. orellana (Bo), B. davidii (Bd), C. clementina (Cc), C. sativus (Cs), C. ancyrensis (Ca), G. jasminoides (Gj), S. lycopersicum (Sl), O. sativa (Os), and Synechocystis sp. (Syn). The bootstrap consensus tree inferred from 500 replicates is shown. The percentage of replicate trees in which the associated CCDs clustered together in the bootstrap test is shown next to the branches and branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The carotenoid substrates, position of cleavage, and bootstrap values are indicated. Protein sequences are shown in Supplemental Figure S1.
Bixin and crocins are highly prized as food colorants, and, in the case of crocins, also for their potential health applications (Alavizadeh and Hosseinzadeh, 2014; Rivera-Madrid et al., 2016). Crocins confer the characteristic red color to the saffron spice, while its flavor and aroma are mainly due to the apocarotenoids picrocrocin and safranal. Crocins combine high antioxidant activity, water solubility, and very low toxicity and have been claimed to possess antioxidant, antitumoral, antidepressant, and retina-protecting properties (Falsini et al., 2010; Alavizadeh and Hosseinzadeh, 2014). Due to the high number of chiral centers, chemical synthesis of crocins is very difficult to achieve and the natural sources of these compounds are the saffron spice (composed of the hand-picked stigmas of saffron and dried fruits of Gardenia). Both sources contain 8%–10% (w/w) crocins. Although Gardenia fruits are used in traditional Chinese medicine, they are not permitted for use as food additives in the USA or the EU, due to their high content of geniposide and their consequent hepatotoxicity (Yamano et al., 1988).
The first step in crocin biosynthesis consists of the symmetric 7,8/7′,8′ cleavage of a carotenoid substrate by a CCD. The substrate and CCD are, respectively, zeaxanthin and CsCCD2 in C. sativus stigmas (Frusciante et al., 2014); zeaxanthin and BdCCD4.1/BdCCD4.3 in B. davidii (Ahrazem et al., 2017); lycopene, beta-carotene and zeaxanthin, and GjCCD4a in G. jasminoides (Xu et al., 2020; Figure 1). The cleavage produces crocetin dialdehyde, which is dehydrogenated and then glycosylated by dedicated aldehyde dehydrogenases and UDP-glucosyl transferases (Demurtas et al., 2018; Xu et al., 2020). In the case of saffron, the cleavage dehydrogenation, and glycosylation steps occur in the plastid, endoplasmic reticulum, and cytoplasm, respectively, while crocins are transported in the vacuole by ABC transporters (Demurtas et al., 2018).
Bixin has been reported to be generated by the symmetric 5,6/5′,6′ cleavage of lycopene by Lycopene Cleavage Dioxygenase (BoLCD), a member of the CCD4 subfamily (Bouvier et al., 2003; Figure 1). However, BoLCD is truncated at the N-terminus, losing one of the seven beta-propeller blades shown to be essential for CCD activity, and is not found in a recent transcriptome survey of B. orellana seeds and leaves (Cardenas-Conejo et al., 2015). In the same survey, a series of transcripts encoding CCD1 and CCD4 enzymes were identified. Three of them (BoCCD4-1, BoCCD4-3, and BoCCD1-1) had expression patterns consistent with bixin accumulation during seed development. In a later study by the same group, BoCCD1-1 and BoCCD4-3 were reported to cleave symmetrically lycopene at the 5,6/5′,6′ positions, yielding bixin dialdehyde (Carballo-Uicab et al., 2019).
To re-assess the role of different B. orellana CCDs in apocarotenoid biosynthesis, we expressed six BoCCD genes in Escherichia coli strains producing various carotenoids. To our surprise, none of the CCD enzymes was able to produce bixin dialdehyde in these conditions, while one of the proteins, BoCCD4-3, was able to cleave lycopene, β-carotene, and zeaxanthin at the 7,8/7′,8′ positions, producing crocetin dialdehyde. Heterologous expression of BoCCD4-3 in Nicotiana leaves and tomato (Solanum lycopersicum) fruits resulted in the production of high levels of crocins, confirming its suitability for the biotechnological production of these highly prized compounds.
Results
Identification and functional characterization of BoCCDs
The B. orellana transcriptome (Cardenas-Conejo et al., 2015) was re-analyzed for the presence of members of the CCD1 and CCD4 sub-families, using sequence identity with known CCDs. Four CCD1s (BoCCD1-1, BoCCD1-2, BoCCD1-3, and BoCCD1-4) and four CCD4s (BoCCD4-1, BoCCD4-2, BoCCD4-3, and BoCCD4-4) were identified, following the nomenclature of (Cardenas-Conejo et al. 2015; Supplemental Figure S1). No homolog of Crocus CsCCD2 or Bixa BoLCD was found in this search.
A phylogenetic tree of CCD protein sequences from several plant species and from Synechocystis spp. was inferred using the maximum-likelihood method (Figure 1, C). The BoCCD1 enzymes were closely related to each other. The closest enzymes to BoCCD1 enzymes were Arabidopsis and rice CCD1 which cleave carotenoids at the 9,10/9′,10′ positions, and at the 9,10/9′,10′, 9,10/7′,8′, and 9,10/5′,6′positions, respectively (Schwartz et al., 2001; Ilg et al., 2009). The BoCCD4 enzymes formed a cluster with Citrus CCD4b1, which cleaves zeaxanthin asymmetrically at the 7,8 position to yield citraurin, the main apocarotenoid in tangerine peel (Rodrigo et al., 2013).
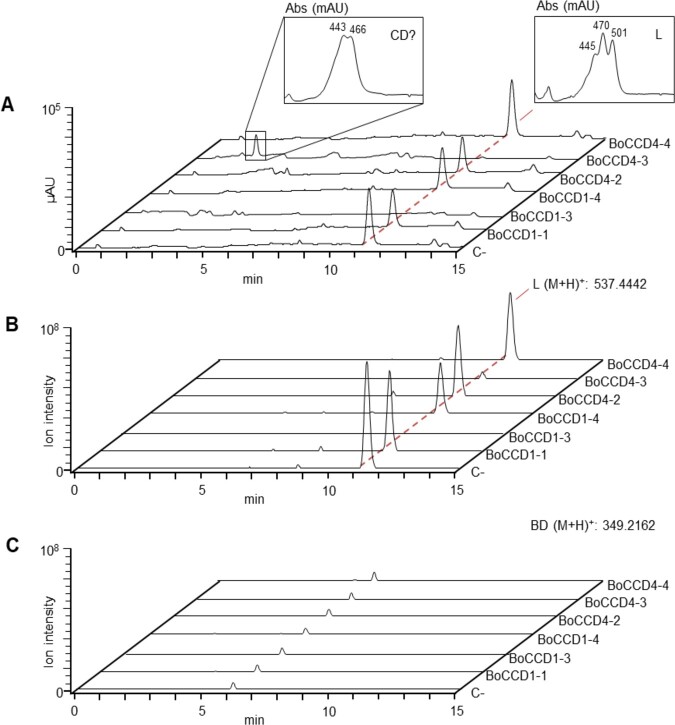
Six BoCCDs were cloned in the pTHIO-DAN1 expression vector, affording arabinose-inducible expression (Frusciante et al., 2014), and transformed into E. coli cells able to synthesize lycopene, β-carotene, or zeaxanthin, as described previously (Frusciante et al., 2014; Supplemental Figure S2). After induction of BoCCD expression with arabinose 0.2% (w/v) for 16 h at 20°C, carotenoids and apocarotenoid aldehydes were extracted and analyzed by Liquid Chromatography–Photodiode Array Spectroscopy–High Resolution Mass Spectrometry (LC–PDA–HRMS) (Frusciante et al., 2014). Four of the six BoCCDs tested did not show a major cleavage activity on any of the carotenoid substrates (Figure 2 and Supplemental Figures S3, S4). BoCCD4-3 was able to cleave all three substrates, producing a compound having a chromatographic mobility and an online spectrum reminiscent of crocetin dialdehyde, the C20 precursor of crocin biosynthesis in saffron stigmas (Frusciante et al., 2014). The second CCD, BoCCD1-3, was able to almost completely degrade all three carotenoid substrates without forming a major cleavage product (Figure 2 and Supplemental Figures S3, S4).
Figure 2.
Lack of bixin dialdehyde production in bacterio by different BoCCDs. HPLC–PDA–HRMS profiles of E. coli cells producing lycopene (L) and expressing various Bixa carotenoid dioxygenases, induced for 16 h at 20°C with arabinose (0.2%, w/v). Cells expressing the pTHIO-DAN empty vector are indicated with C-. A, Maximum absorbance UV–Vis chromatogram in the 250–700 nm range. Online spectra of the main peaks (L, lycopene and CD?, putative crocetin dialdehyde) are shown in the insets. µAU, Micro-absorbance units. B, Ion chromatograms of lycopene (L, M+H+: 537.4442 ± 2 ppm) extracted with the Xcalibur Software (Thermo Fisher Scientific). C, Ion chromatograms of Bixin Dialdehyde (BD, M+H+: 349.2162 ± 2 ppm) were extracted with the Xcalibur Software (Thermo Fisher Scientific).
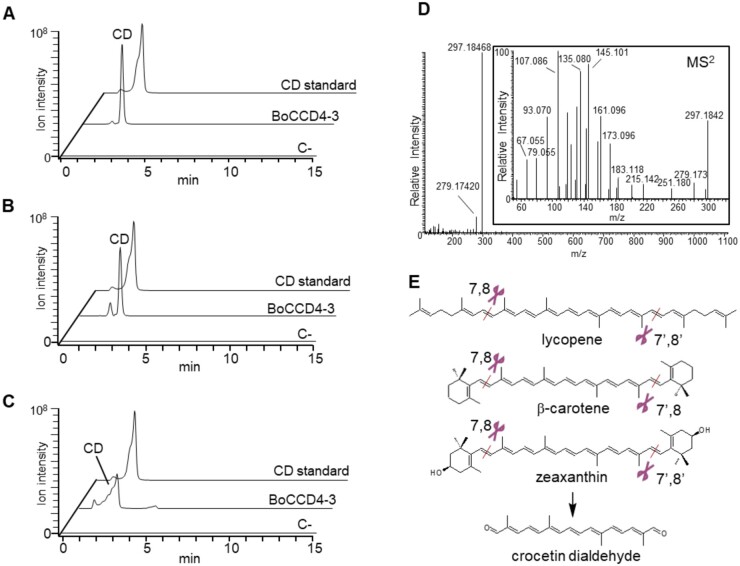
We confirmed the identity of the putative crocetin dialdehyde peak produced by BoCCD4-3 by full MS, MS2 analysis, and comigration with an authentic CD standard (Figure 3). This result is unexpected, since crocetin dialdehyde is generated by cleavage at the 7,8/7′,8′ positions. To verify the presence of trace amounts of additional compounds, derived by single or double cleavage at the 5,6, 7,8, or 9,10 positions, the corresponding masses were extracted from chromatograms of E. coli cells and expressing either the empty pTHIO-DAN vector or different BoCCDs. The results (Supplemental Table S1) confirm that, in our experimental conditions, the only cleavage produced in high amounts (ion intensity 107) is crocetin dialdehyde in cells expressing BoCCD4-3. Its intermediate products (apo-8′-lycopenal, β-apo-8′-carotenal, and β-citraurin) also appear at much lower ion intensities (103–104) in the same cells. In cells expressing BoCCD1-1 or BoCCD1-3, we found ions corresponding to 5′/6′ and/or 9′/10′ cleavage activities, but at very low ion intensities (103–104). A compound with an experimental mass compatible with that of bixin dialdehyde (349.2161) appears at very low ion intensity (104) in all cells, including those expressing the empty pTHIO-DAN vector. We conclude that this compound is not a specific product of the BoCCD activity, but rather an endogenous molecule present in trace amounts in E. coli extracts. Given the unavailability of an authentic standard, we are not able to determine its chemical structure.
Figure 3.
BoCCD4-3 cleaves completely lycopene, b-carotene, and zeaxanthin to produce crocetin dialdehyde in E. coli. A–C, LC–HRMS chromatograms of E. coli extracts accumulating lycopene (A), β-carotene (B), and zeaxanthin (C) after induction with 0.2% of arabinose (w/v) for 16 h at 20°C. The ion of crocetin dialdehyde CD (M+H)+: 297.1846 was extracted. Cells expressing BoCCD4-3 accumulate a compound that has both the accurate mass and the chromatographic mobility of the crocetin dialdehyde standard. D, MS spectrum and (inset) MS/MS spectrum of the crocetin dialdehyde peak produced by E. coli cells expressing BoCCD4-3. E, Cleavage reactions catalyzed by BoCCD4-3.
It was reported previously (Carballo-Uicab et al., 2019) that BoCCD1-1 and BoCCD4-3 are able to cleave lycopene in E. coli, with the formation of very low levels of bixin dialdehyde. We find several inconsistencies in those data: (1) the mass spectrum of the compound shown in Figure 8 of that paper does not resemble bixin dialdehyde: the experimental mass of both the main peak (m/z 348.2507) and the +1 peak (m/z 349.2404) is too distant from that of the bixin dialdehyde M + H adduct (m/z 349.2162), given the fact that they were produced with a high-resolution instrument; in the absence of an authentic bixin dialdehyde standard or of MS/MS data, its identification as bixin dialdehyde is dubious; (2) the full MS scans shown in Figure 8 span from m/z 340 to 550, so the M + H adduct of crocetin dialdehyde (m/z 297.1846) produced by BoCCD4-3 would have been missed.
Subcellular localization of BoCCD4-3 in N. benthamiana leaves
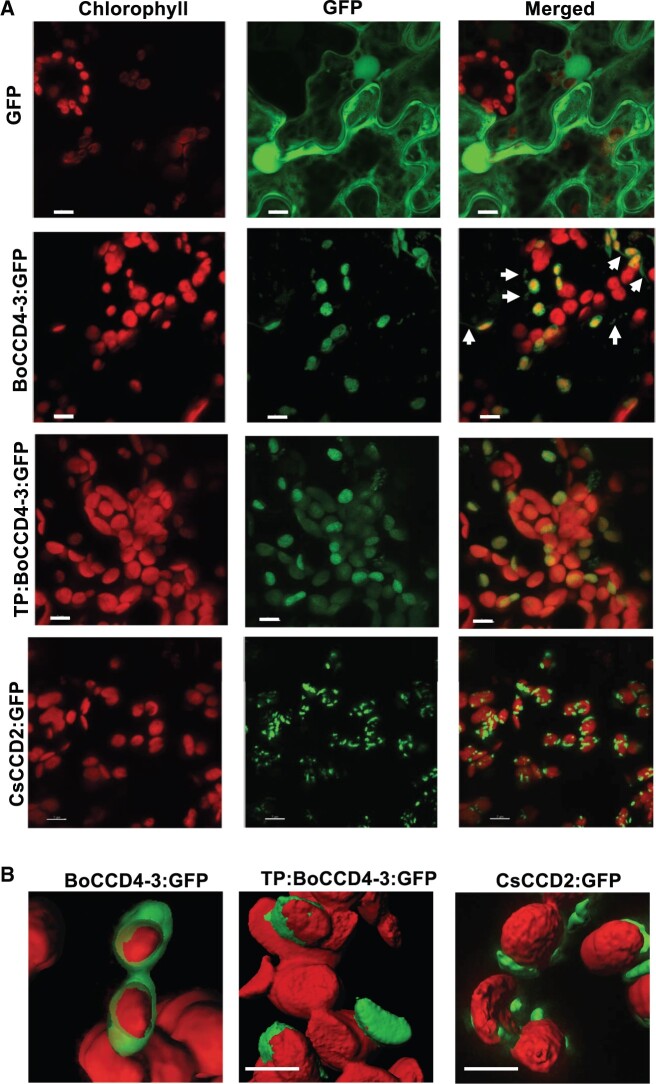
Despite the fact that its carotenoid substrates are presumably localized in plastids, BoCCD4-3 does not carry a recognizable transit peptide at its N-terminus (Supplemental Figure S5). Since up to 12% of chloroplast-localized proteins lack a recognizable transit peptide (Armbruster et al., 2009), we decided to verify experimentally the subcellular localization of BoCCD4-3 through C-terminal fusion to an enhanced Green Fluorescent Protein (GFP) and transient expression in N. benthamiana leaves, as described previously (Demurtas et al., 2018). The results (Figure 4) clearly show that the BoCCD4-3:GFP localizes to plastids like CsCCD2:GFP, which carries a recognizable transit peptide. However, while the latter shows a speckle-like localization to specific areas close to the plastid envelope, the BoCCD4-3:GFP fluorescence localizes to the stroma. This is suggested both by the diffuse labeling that surrounds the thylakoids, and by the presence of green fluorescent tubular-like protrusions extending into the cytoplasm, which probably represent stromules (Delfosse et al., 2015). This difference between the localization of the two proteins is not due to the presence versus absence of a plastid transit peptide, since the N-terminal fusion of the CsCCD2 transit peptide to BoCCD4-3 does not substantially change its sub-plastidial localization. We therefore hypothesize that this difference is due to a structural difference in the mature CsCCD2 and BoCCD4-3 proteins. Indeed, while both proteins carry a central hydrophobic domain, only the CsCCD2 domain has a considerable transmembrane probability (Supplemental Figure S5).
Figure 4.
Subcellular localization of BoCCD4-3 in N. benthamiana leaves. A, Confocal images of N. benthamiana leaves expressing (top to bottom): GFP, BoCCD4c:GFP, TP:BoCCD4c:GFP (BoCCD4c fused to the CsCCD2 transit peptide), and CsCCD2:GFP fusion proteins. Red (chlorophyll fluorescence), green (GFP fluorescence), and merged (overlap of chlorophyll and GFP fluorescence) are shown. The unfused GFP protein shows the typical cytoplasmic and nuclear localization. Both BoCCD4-3:GFP and CsCCD2:GFP localize to plastids. Scale bars: 7 μm. Arrows in the BoCCD4c:GFP composite image point at green fluorescent protrusions of the plastid stroma (stromules). B, 3D reconstruction of red and green fluorescence in plastids expressing BoCCD4-3:GFP, TP:BoCCD4-3:GFP, and CsCCD2:GFP. The latter localizes to plastid-associated speckles (Demurtas et al., 2018). Scale bars: 7 μm.
Heterologous expression of CsCCD2 and BoCCD4-3 in Nicotiana leaves and tomato fruits results in the production of apocarotenoids found in saffron stigmas
BoCCD4-3 shows a much broader substrate specificity than CsCCD2, cleaving lycopene, beta-carotene, and zeaxanthin. To compare the capacity of the two dioxygenases to cleave carotenoids in planta, we expressed them transiently in N. benthamiana leaves under the control of the CaMV 35S promoter. Four constructs were used, expressing different proteins: BoCCD4-3; TP:BoCCD4-3 (BoCCD4-3 fused to the CsCCD2 transit peptide); CsCCD2; and CsCCD2short (CsCCD2 lacking its transit peptide). The four constructs were agroinfiltrated in N. benthamiana leaves and leaf extracts were analyzed for the presence of compounds found in Bixa seeds or Crocus stigmas. No bixin-related compounds were detected in agroinfiltrated leaves, while several cis- and trans-isomers of crocetin and crocins were clearly detectable (Figure 5;Supplemental Figure S6, A and Supplemental Table S2). For both CsCCD2 and BoCCD2, the presence of the CCD2 transit peptide boosted apocarotenoid production, with the efficiency of the four constructs being TP:BoCCD4-3 > CsCCD2 > BoCCD4-3 > CsCCD2short. The higher efficiency of BoCCD4-3 is not surprising, given the fact that it is able to cleave β-carotene, which is abundant in leaves, while the CCD2 substrate, zeaxanthin, is present only in trace amounts. Interestingly, the CCD2 lacking the transit peptide gave some residual activity, in keeping with the crocetin accumulation detected in maize endosperm agroinfiltrated with the transit peptide-less CCD2 (Frusciante et al., 2014). The apocarotenoid profiles generated by the two CCDs differed also qualitatively: in leaves agroinfiltrated with CsCCD2 variants, crocetin and crocins with ≤2 glucose groups (Supplemental Figure S7) were, respectively, ≈14% and 70%–73% of total apocarotenoids, while crocins with ≥3 glucose groups were 12%–15%. The percentage of the latter increased up to 21% in leaves agroinfiltrated with BoCCD4-3, indicating that the latter CCD was able to induce synthesis of more highly glycosylated compounds (Supplemental Table S2). No crocins with ≥4 glucose groups were detected.
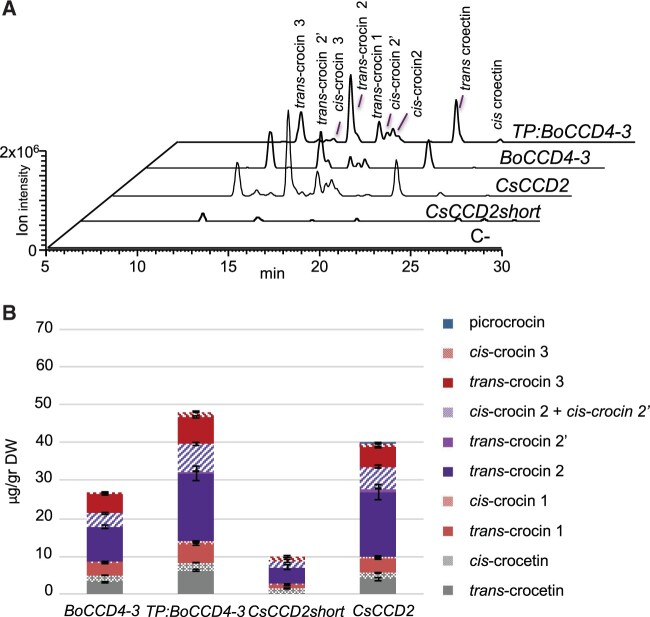
Figure 5.
Crocetin/crocin production in N. benthamiana leaves transiently expressing CsCCD2 or BoCCD4-3. A, Representative LC–HRMS chromatograms of the extracted accurate mass of crocetin (M+H+ 329.1747), generated from crocin fragmentation, in leaves expressing BoCCD4-3, TP:BoCCD4-3, CsCCD2, and CsCCD2short (CsCCD2 lacking the TP) and C- (empty vector). B, Quantification of crocetin and crocins in agroinfiltrated leaves. Data are the avg ± sd of three independent pools of agroinfiltrated plants. See Supplemental Table S3 for quantitative data. HPLC–PDA chromatograms are shown in Supplemental Figure S6, A.
Additionally, in leaves agroinfiltrated with CsCCD2, we observed the production of trace amounts of picrocrocin (Supplemental Table S2), an abundant apocarotenoid in saffron stigmas, which was not detected in BoCCD4-3-agroinfiltrated leaves. In saffron, picrocrocin is produced by the action of a specific glycosyltransferase named UGT709G1 (Diretto et al., 2019). However, its appearance in CsCCD2-transformed leaves is not surprising, since (1) picrocrocin is thought to originate from beta-hydroxy-cyclocitral (Figure 1), which is generated by the cleavage of zeaxanthin, the likely in planta substrate of CsCCD2, while the major substrate of BoCCD4-3 in leaves is beta-carotene, whose cleavage yields beta-cyclocitral, (2) the presence of endogenous glycosyltransferases in N. benthamiana leaves, able to synthesize picrocrocin, has been described previously (Demurtas et al., 2018; Marti et al., 2020).
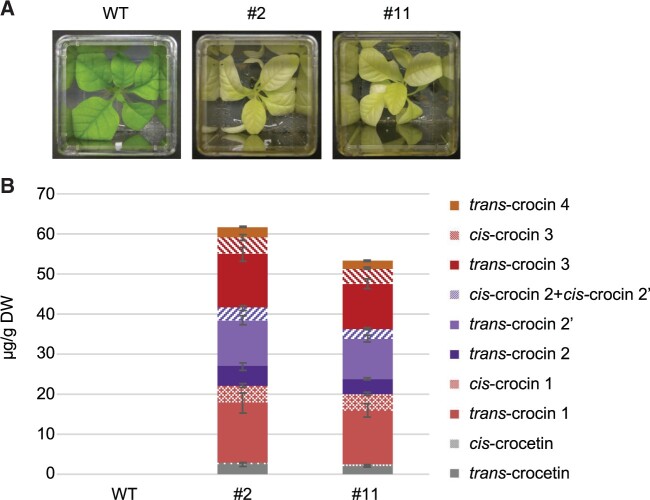
Next, we expressed the BoCCD4-3 protein in plastids by generating stable chloroplast-transformed (transplastomic) tobacco (Nicotiana tabacum) plants under the control of a theophylline-inducible, synthetic riboswitch. To this end, the coding sequence of BoCCD4 was codon optimized according to the preferred codon usage in the tobacco plastid genome, and the coding sequence was fused to the T7 RNA polymerase promoter and the 5′UTR from gene10 of bacteriophage T7. The expression cassette was integrated into a plastid transformation vector (Supplemental Figure S8, A) and used to transform, by biolistic transformation, a previously generated transplastomic tobacco line harboring a theophylline-inducible T7 RNA polymerase (T7RNAP; Emadpour et al., 2015; Supplemental Figure S8, A). Primary transplastomic clones were obtained by selection for spectinomycin resistance conferred by the chimeric aadA gene in the transformation vector (Svab and Maliga, 1993). Homoplasmic lines were selected by several additional rounds of regeneration on spectinomycin-containing regeneration medium (Bock, 2001; 2015) and presence of a uniform population of transformed plastid genomes was confirmed by Southern blot analysis (Supplemental Figure S8, B and C). Transplastomic BoCCD4-3 expressing plants showed a pale, slightly yellow/orange leaf phenotype when grown on synthetic medium, even in the absence of theophylline induction (Figure 6). When transferred to soil, the transplastomic plants were not able to survive, suggesting that they are unable to grow autotrophically. LC–PDA–HRMS analysis of the transplastomic plants confirmed the presence in leaves of crocetin and several crocins, including trans-crocin 4, which was the most highly glycosylated crocin and was not detected in N. benthamiana transient expression (Figure 6; Supplemental Figure S6, B and Supplemental Table S3). The apocarotenoid profile of tobacco transplastomic lines was more similar to that of saffron, with crocins with ≥3 glucose groups being ≈32% of the total apocarotenoids (Figure 6 and Supplemental Table S3).
Figure 6.
Crocetin/crocin production in transplastomic tobacco plants expressing BoCCD4-3. A, Visual phenotype of transplastomic lines #2 and #11 cultured in vitro. B, Quantitation of crocetin and crocins in young leaves from lines #2 and #11. Data are the avg ± sd of three biological replicates. See Supplemental Table S4 for quantitative data. HPLC–PDA chromatograms are shown in Supplemental Figure S6, B.
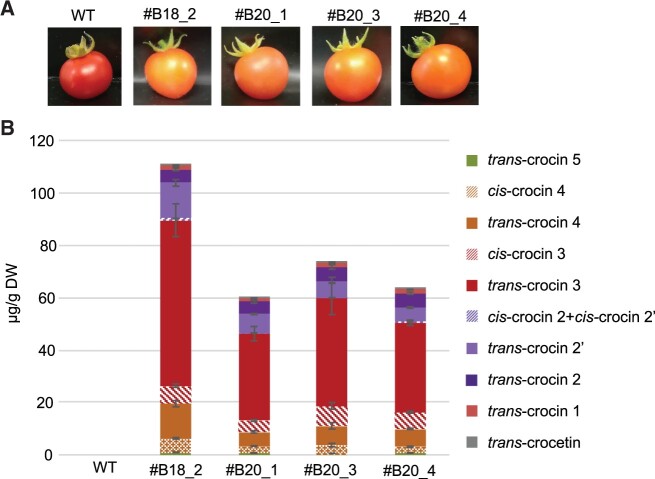
Finally, we performed combinatorial transformation of tomato (cv Microtom) plants with 35S:TP:BoCCD4-3 and 35S:CsUGT74AD1. The rationale behind this experiment is that tomato fruits accumulate very high levels of lycopene, a BoCCD4-3 substrate, while CsUGT74AD1 specifically catalyzes the glycosylation of crocetin (Demurtas et al., 2018). Two out of eight regenerated lines displayed a distinctive orange-colored fruit phenotype. These lines were characterized by PCR for the presence of the transgenes, and both were found to contain both the 35S:TP:BoCCD4-3 and 35S:CsUGT74AD1 transgenes (Supplemental Figure S9). Fruits from the T1 progeny displaying the orange phenotype were harvested and the crocin/crocetin content was determined. The crocin content of transgenic fruits was the highest of all the heterologous expression experiments (Figure 7; Supplemental Figure S6, C and Supplemental Table S4). Also, the qualitative profile of transgenic fruits was very different from that of agroinfected or transplastomic Nicotiana leaves, with crocetin representing ≤0.5% and crocins with ≥3 glucose groups representing ≈80% of total apocarotenoids, a percentage very similar to that of saffron extract (≈84%, Supplemental Table S4).
Figure 7.
Crocetin/crocin production in transgenic Microtom fruits expressing TP:BoCCD4-3 and CsUGT74AD1. A, Phenotypes of Microtom T1 transgenic fruits expressing TP:BoCCD4-3 + CsUGT74AD1. B, Quantification of crocetin and crocins in ripe fruits. Data are the avg ± sd of three biological replicates (fruits) from each line. HPLC–PDA chromatograms are shown in Supplemental Figure S6, C.
To verify if CCD4-3 was able to catalyze the synthesis of additional apocarotenoids in planta, we searched the chromatograms from transformed plant tissues for ions of known apocarotenoids. The only additional apocarotenoid was a compound with the brute formula C30H40O2, corresponding to citraurin, generated by a single 7,8 cleavage of lutein or zeaxanthin, respectively, and found in transformed tomato fruits (Supplemental Table S5).
Discussion
Convergent evolution of crocin biosynthesis in plants
Crocins are synthesized by plants as distant as Crocus (Iridaceae), Buddleja (Scrophulariaceae), and Gardenia (Rubiaceae). The evolutionary distance of these genera and the fact that different subfamilies of CCDs are involved in crocin biosynthesis (CCD2 in Crocus and CCD4 in Buddleja) strongly suggest that the crocin pathway has appeared multiple times during plant evolution, through parallel or convergent evolution (Stern, 2013). Convergent evolution is not a rare phenomenon in plant specialized metabolism: gene duplication and neofunctionalization of different N-methyltransferases has given rise to caffeine biosynthesis independently in coffee (Rubiaceae) and tea (Theaceae) (Denoeud et al., 2014).
The CCD1/CCD2 phylogeny shown in Figure 1, C is consistent with the accepted taxonomy: CCD1s from B. orellana, Arabidopsis thaliana, and B. davidii (dicots) are more similar to each other than to CCD1 of C. sativus (monocots). All enzymatically characterized CCDs within this clade possess 9,10/9′,10′ cleavage activity, with the exception of CCD2s, which instead perform 7,8/7′,8′ cleavage. The latter are well separated from CCD1s, confirming that they belong to a different subfamily of probable monophyletic origin (Frusciante et al., 2014). In contrast, the CCD4 clade shows multiple subclades, comprising enzymes from both monocots and dicots and with different cleavage specificities. The closest relative to BoCCD4s is CCD4b1 from C. clementina, which possesses 7,8 cleavage activity (Rodrigo et al., 2013). The CCD4s from B. davidii and from G. jasminoides, which possess 7,8/7′8′ cleavage activity, belong to subclades that are distant from that of BoCCD4.3. This suggests that the capacity to synthesize crocetin dialdehyde has evolved multiple times in the CCD4 subfamily, that is, is of polyphyletic origin. The recent sequencing of the G. jasminoides genome indicated that the crocin-synthesizing CCD in this species, GjCCD4a, evolved through recent tandem duplication of a CCD4 gene that is still in single copy in the closely related genus Coffea (Xu et al., 2020). None of the other CCD4s in Coffea or Gardenia exhibit 7,8/7′,8′ cleavage.
Metabolic engineering of crocin biosynthesis
In bacterio, BoCCD4-3 cleaves lycopene and, with lower efficiency β-carotene and zeaxanthin to yield crocetin dialdehyde. This broad substrate specificity is similar to the one exhibited by the Gardenia enzyme (GjCCD4a) (Xu et al., 2020) and contrasts with those of the Crocus and Buddleja enzymes (CsCCD2, BdCCD4.1, and BdCCD4.3) which cleave only zeaxanthin (Frusciante et al., 2014; Ahrazem et al., 2017; Marti et al., 2020). We took advantage of this broad substrate specificity to express BoCCD4-3 in Nicotiana leaves (containing mainly lutein and beta-carotene) and in tomato fruits that instead accumulate lycopene and beta-carotene. As observed before for CsCCD2, overexpression in Nicotiana leaves results in the accumulation of crocins (Demurtas et al., 2018; Marti et al., 2020). This result suggests that Nicotiana express endogenous ALDH and UGT enzymes catalyzing the dehydrogenation of crocetin dialdehyde and the glycosylation of crocetin. Since leaves are known to contain glycosylated apocarotenoids (Latari et al., 2015; Mi et al., 2019), it is possible that the enzymes mediating their synthesis are promiscuous and that they are recruited for crocetin/crocin biosynthesis in Nicotiana leaves expressing the appropriate CCD enzyme. A corollary of this hypothesis is that the synthesis of a given glycosylated apocarotenoid in leaves is largely controlled at the cleavage step. In agreement with this corollary, we observe the formation of picrocrocin in leaves overexpressing CsCCD2, which cleaves zeaxanthin to form the picrocrocin precursor 3-OH-β-cyclocitral. In contrast, BoCCD4-3 expression does not produce picrocrocin, despite the higher activity demonstrated by this enzyme in crocin formation. The likely BoCCD4-3 substrate in leaves is β-carotene, both because BoCCD4-3 cleaves β-carotene more efficiently than zeaxanthin in E. coli, and because β-carotene is much more abundant than zeaxanthin in N. benthamiana leaves. Cleavage of β-carotene would generate β-cyclocitral, which is not a picrocrocin precursor. Thus, the ratio of crocins (the main saffron color) to picrocrocin (the main flavor) can be manipulated through the expression of CCDs with different substrate specificities in tissues with different carotenoid composition.
BoCCD4-3 is plastid-localized in N. benthamiana leaves, despite the lack of a recognizable transit peptide. While CcCCD2 localizes to speckle-like structures, probably associated with the plastid envelope, BoCCD4-3 seems to have a stromal localization. This type of localization is not changed by the N-terminal fusion of the CsCCD2 transit peptide to BoCCD4-3, suggesting that the different sub-organellar localization of the two enzymes is due to determinants found in the mature proteins. Indeed, while both proteins have a central hydrophobic domain, CsCCD2 has a much higher transmembrane probability than BoCCD4-3 (Supplemental Figure S5). A “short” CsCCD2 lacking the transit peptide, and therefore showing a cytoplasmic localization (Frusciante, 2014, p. 31), was able to produce low levels of crocins when expressed in N. benthamiana leaves. This result suggests that CsCCD2 is able to cleave carotenoids localized in the outer plastid envelope (Markwell et al., 1992), and may explain the mechanism through which CCD1 enzymes, that are closely related to CsCCD2 and lack an identifiable transit peptide, access their carotenoid substrates.
Transplastomic expression of BoCCD4-3 results in the production of crocins, indicating that the enzyme expressed from the chloroplast genome is biologically active and able to access its carotenoid substrate. This is in agreement with previous work reporting the transplastomic expression of carotenoid biosynthesis genes in tomato and tobacco (Wurbs et al., 2007; Hasunuma et al., 2008). Given the broad substrate specificity of BoCCD4-3 and the fact that carotenoids are essential for photosynthetic activity, we expressed the gene under the control of the phage T7 promoter and transformed the expression cassette into a transplastomic recipient line harboring a T7 RNA polymerase gene controlled by a theophylline-inducible synthetic riboswitch (Verhounig et al., 2010; Emadpour et al., 2015). However, even in the absence of theophylline induction, transplastomic plants exhibited bleaching and were unable to grow on soil, indicating that some expression is occurring due to leakiness of the riboswitch. Leaky expression was confirmed by the accumulation of crocins in young leaves of these plants.
Transgenic expression in tomato of BoCCD4-3 in combination with CsUGT74AD1, a C. sativus UGT mediating the glycosylation of crocetin (Demurtas et al., 2018), results in fruits containing high levels (>100 µg/g dry weight [DW]) of crocins. Although the quantity of these crocins is still 1,000-fold lower than in saffron, their qualitative profile resembles very much that of the spice, in which crocins with ≥3 glucose groups represent >80% of total apocarotenoids. Glycosylation has a profound influence on the water solubility and hence on the bioavailability of crocins, and therefore the ability to modulate its levels is an important prerequisite for their production through metabolic engineering.
Tomatoes present many ideal characteristics for the production of carotenoid-derived nutritional supplements. They contain up to 1 mg/g DW of the BoCCD4-3 substrate, lycopene, and, contrary to tobacco leaves and Gardenia fruits, they are generally regarded as safe (GRAS). Tomato-derived products are an inexpensive source of nutraceuticals (mainly lycopene), are available both as spray-dried powder and as oleoresins, present very low toxicity even at very high doses, and a good stability and bioavailability profile, as exemplified by their use for the supplementation of aquaculture feed with transgenically produced ketocarotenoids (Nogueira et al., 2017). Worldwide tomato production stands at 180 M tons annually and thus, at the crocin levels reported here, converting 10% of this production to crocin-producing tomatoes would yield 180 tons of crocins, that is, 60-fold more than the total worldwide saffron production. In the transgenic lines reported here, only about 10% of fruit lycopene is converted into crocins, and some adverse leaf phenotypes are observed in the highest expressing lines. Thus, we believe that fruit crocin levels can be further improved through a combination of mutations increasing lycopene content, the use of different transgenes derived from Gardenia and saffron for boosting crocin biosynthesis and storage, and the use of fruit-specific promoters for avoiding adverse leaf phenotypes.
Conclusions and outlook
Our results open two major questions related to apocarotenoid biosynthesis in B. orellana: (1) what are the “in vivo” products, if any, of BoCCD4a and (2) what is the pathway that mediates bixin biosynthesis?
Regarding the first question, apocarotenoids derived from crocetin dialdehyde have not been reported in B. orellana seeds (Mercadante et al., 1997, 1999; Chiste et al., 2011). However, LC–HRMS analysis of non-polar extracts from B. orellana leaves and seeds revealed several peaks with an accurate mass compatible with that of apocarotenoids generated from a single or double 7/8 cleavage, including α- or β-citraurin and dimethyl-8,8′-diapocarotene-8,8′-dioate (dimethylcrocetin) (Supplemental Table S6 and Supplemental Figure S10). The full characterization of the structure of these compounds is outside the scope of this paper.
The second question relates to the bixin biosynthesis pathway in B. orellana, which remains unresolved. Two previous reports (Bouvier et al., 2003; Carballo-Uicab et al., 2019) claimed to have identified the Bixa CCD responsible for the formation of bixin dialdehyde. However, the BoLCD identified by Bouvier et al. (2003) is not present in Bixa transcriptome data (Cardenas-Conejo et al., 2015), and the data presented in this paper do not confirm the findings of Carballo-Uicab et al. (2019) that the main product of BoCCD1-3 and BoCCD4-3 is bixin dialdehyde. It is attractive to speculate that bixin biosynthesis is mediated by the action of a hitherto non-characterized CCD, or that, similar to what was discovered in strigolactone biosynthesis (Alder et al., 2012), an additional isomerase-like enzyme is needed for the synthesis of this apocarotenoid pigment.
Materials and methods
Bioinformatic analyses
An annatto (B. orellana) transcriptome (Cardenas-Conejo et al., 2015) was searched for contigs bearing homology to CCD1 and CCD4 enzymes using BLAST (Altschul et al., 1990). The obtained sequences were very similar, or identical, to those deposited in GeneBank by Rivera-Madrid and collaborators (Supplemental Table S1). For expression analysis, we used NGS data from Cardenas-Conejo et al. (2015) (accession number SRX1117606) and from the 1,000 Plants Project (Matasci et al., 2014) (accession number ERX2099513). Raw reads were cleaned from adapter sequences, low-quality residues, and very short length reads using Cutadapt with 20 as quality cut-off (Martin, 2011). Reads were aligned to the B. orellana transcriptome (Cardenas-Conejo et al., 2015) with BWA (Li and Durbin, 2009) with default options and expression values were estimated with Cufflinks (Trapnell et al., 2012). Phylogenetic trees were generated using MEGA X version 10.2.2 with the maximum-likelihood method (Kumar et al., 2016). Predictions of subcellular localization and hydrophobic profiles were obtained using the TargetP1.1 server (Emanuelsson et al., 2000) and the TMHMM 2.0 software (www.cbs.dtu.dk/services/TMHMM), respectively.
Gene synthesis and cloning
Gene sequences were synthesized by the Invitrogen GeneArt Gene Synthesis service (www.thermofisher.com/us/en/home/life-science/cloning/gene-synthesis/geneart-gene-synthesis.html) and cloned into pTHIO-Dan1 vector using Gibson assembly (Gibson et al., 2009). The vector was digested with the XbaI and KpnI restriction enzymes and the CCD CDS were amplified with Q5 High Fidelity DNA polymerase (NEB, Cat No. M0491S). Purified PCR fragments were then assembled obtaining pTHIO-Dan1-CCD1-1, pTHIO-Dan1-CCD1-3, pTHIO-Dan1-CCD1-4, pTHIO-Dan1-CCD4-2, pTHIO-Dan1-CCD4-3, and pTHIO-Dan1-CCD4-4 (Supplemental Figure S2) and re-sequenced before transformation into E. coli cells able to synthesize lycopene, β-carotene, or zeaxanthin (Frusciante et al., 2014). Primers used are listed in Supplemental Table S7. For in planta expression, BoCCD4-3 was C-terminally fused to the enhanced GFP (eGFP) in the pBI-eGFP vector using Gibson assembly, generating the pBI-CCD4.3:eGFP construct. To obtain TP:BoCCD4-3, the CsCCD2 transit peptide was isolated by PCR and cloned by Gibson assembly into pBI:BoCCD4-3:eGFP, generating pBI:TP:CCD4.3:eGFP. pBI:BoCCD4-3 and pBI:TP:CCD4.3 were also generated through Gibson assembly. CsUGT4AD1 was amplified by PCR from pTHIO-UGT4AD1 (Demurtas et al., 2018) and cloned in pBI digested by XbaI through Gibson assembly. All primers used are listed in Supplemental Table S7. The maps of the plasmids are shown in Supplemental Figure S2. All plasmids were checked by resequencing before transformation in Agrobacterium tumefaciens strain C58C1 (for agroinfiltration of Nicotiana benthamiana leaves) or EHA105 (for tomato [S. lycopersicum] transformation).
Expression in E. coli
Escherichia coli strains engineering to accumulate lycopene, β-carotene, and zeaxanthin were transformed with pTHIO-Dan1 vectors carrying the various CCDs (Frusciante et al., 2014). Overnight cultures were inoculated in 25 mL of LB medium containing 50 µg/mL of ampicillin and 25 µg/mL of kanamycin, grown at 37°C to an OD600 of 0.7 and induced with arabinose 0.2% (w/v) for 16 h at 20°C. Cells were pelleted and extracted as described previously (Frusciante et al., 2014) with slight modifications: pellets were resuspended with 4 mL of acetone, lysed on ice by 5 sonication at 10 Hz output (10 s each), and centrifuged at 6,000g for 10 min. The supernatant was dried and dissolved in 200 μL of ethyl acetate 100%, then centrifuged at 18,000g for 20 min and subjected to HPLC–PDA–HRMS analysis.
Expression in N. benthamiana
Nicotiana benthamiana were agroinfiltrated as described previously (Demurtas et al., 2018), using the RK19 silencing suppressor (Francisco et al., 2013). After 4 d, leaves were analyzed by a confocal laser-scanning microscope (Olympus FV1000 and Zeiss LSM880) as reported before (Demurtas et al., 2018). Images were acquired using a 40× oil immersion objective (N.A. 1,30) with optical zooming 1× or 3× and analyzed with the ImageJ software. For each construct, at least three N. benthamiana plants were infiltrated and observed. Representative images, including 3D reconstructions are shown in Figure 4. For functional characterization studies, leaves were infiltrated with C58C1 cells containing the eGFP-less constructs and pBI121 as negative control (Frusciante, 2014, p. 31), or with pBI121:CsCCD2 (Demurtas et al., 2018) and the RK19 silencing suppressor. To minimize variability, plants of the same size and developmental stage (4 weeks old) were selected, infiltrating only apical leaves. For each construct, leaves of four different plants were infiltrated, collected as a pool 4 d post infiltration, and stored at −80°C. Each experiment was repeated with three different batches of plants.
Construction of transgenic and transplastomic plants
Transgenic tomato plants were obtained as described previously (Qiu et al., 2007), using a mixture of three Agrobacterium (EHA105) strains, transformed with plasmids pBI:TP:BoCCD4-3 and pBI:CsUGT74AD1, respectively (Supplemental Figure S2). Double transformants were selected through PCR with the following primers: CCD4-3_for_1515/UGT74AD1_for_1080 and NosT_rev161 (Supplemental Table S5).
An expression cassette consisting of the T7 RNA polymerase promoter, the T7 gene10 leader sequence, the coding sequence of BoCCD4, and the terminator of the chloroplast atpA gene from Chlamydomonas reinhardtii was synthesized (GeneCust, Boynes, France). The coding sequence of CCD4.3 was codon optimized according to the codon usage of the tobacco (N. tabacum) chloroplast genome. The CCD4.3 expression cassette was integrated into a chloroplast transformation vector that contains a cloned fragment of the tobacco chloroplast genome from position 37,657 to 40,209 (Yukawa et al., 2005), to facilitate integration into the plastid genome by homologous recombination. The transformation vector contained a chimeric aadA gene, driven by the psaA promoter and the atpB 3′-UTR from Chlamydomonas, and was introduced into plastids of a previously produced recipient line that harbors a T7 RNA polymerase gene in the intergenic spacer between the ycf3 and psaA genes. Plastid transformation was performed by the biolistic protocol followed by selection for spectinomycin resistance (conferred by the aadA marker). Briefly, young leaves from aseptically grown tobacco (N. tabacum cv. Petit Havana) plants were bombarded with vector DNA-coated 0.6 μm gold particles with a helium-driven particle gun (PDS-1000/He, equipped with the Hepta adaptor; Bio-Rad). Primary spectinomycin-resistant lines were selected on regeneration medium containing 500 mg/L spectinomycin (Svab and Maliga, 1993). Several independent transplastomic lines were selected, and homoplasmic transplastomic lines were obtained by two to three additional cycles of plant regeneration in the presence of spectinomycin (500 mg/L). Plant DNA was isolated from frozen leaf tissue by a cetyltrimethylammoniumbromide (CTAB)-based method (Doyle and Doyle, 1990). Five micrograms of total DNA were digested with the restriction enzyme BamHI, separated by gel electrophoresis in 0.8% (w/v) agarose gels, and transferred onto Hybond nylon membranes (GE Healthcare) by capillary blotting. A 550-bp PCR product derived from the psaB coding region in the plastid DNA was amplified with the primer pair P7247 (5′-CCCAGAAAGAGGCTGGCCC-3′) and P7244 (5′-CCCAAGGGGCGGGAACTGC-3′), subsequently gel purified, labeled with [α-32P]dCTP by random priming, and used as an RFLP probe in Southern blot hybridizations to verify plastid transformation and to assess homoplasmy.
LC–HRMS analysis
HPLC separation, ionization, and MS parameters for the E. coli extracts were as described previously (Frusciante et al., 2014). Metabolites were identified on the basis of their absorbance spectra, accurate masses, and by comparison with authentic standards. For analysis of crocetin and crocins, Nicotiana leaves or tomato fruits were freeze-dried, and 10 mg of powder was ground with tungsten beads at 20 Hz for 2 min in a mixer mill (MM 400, Retsch). Samples were suspended in 750 µL of 75% (v/v) cold methanol spiked with 0.5 µg/mL formononetin (Sigma–Aldrich, Cat. No. 47752-25MG-F). Metabolites were extracted at room temperature by continuous agitation for 30 min in MM 400 at 20 Hz. Samples were centrifuged at 20,000g for 20 min, the supernatant was collected, filtered with HPLC filter tubes (0.45 µm pore size), and subjected to LC–PDA–HRMS analysis as described (Demurtas et al., 2018). Crocins were quantified by LC–MS, integrating the areas of M + H adduct plus those of fragmentation ions corresponding to deglucosylation products (due to in-source loss of glucose moieties). A calibration curve was established using a purified trans-crocin 3 standard obtained by preparative HPLC as described previously (Demurtas et al., 2019). The data were interpolated with crocin signal intensities in leaves and fruits, and metabolite quantification was performed using peak areas normalized to leaf DW.
Non-polar analyses of B. orellana leaves were carried out as reported previously (Sulli et al., 2017). Non-polar metabolites were extracted from 2 mg of lyophilized, homogeneous leaf tissue using 100% (v/v) methanol, chloroform, 50 mM Tris–HCl (1:2:1), spiked with 10 μg/mL dl-α-tocopherol acetate as internal standard. The organic extracts were dried with a Speed Vac concentrator and the residue was resuspended in ethyl acetate (100 μL). LC parameters were as reported previously (Sulli et al., 2017). Atmospheric pressure chemical ionization (APCI) parameters were as follows: nitrogen was used as sheath and auxiliary gas, set to 20 and 10 units, respectively. The vaporizer temperature was 300°C, the capillary temperature was 250°C, the discharge current was set to 5.5 μA, and S-lens RF level was set at 50. The acquisition was carried out in the 110/1600 m/z scan range, with the following parameters: resolution 70,000, microscan 1, AGC target 1e6, and maximum injection time 50. Full scan MS with data-dependent MS/MS fragmentation in both positive and negative ionization mode was used for metabolite identification.
Accession numbers
Sequence data from this article can be found in the GenBank under accession numbers of the BoCCDs are BK011288 (BoCCD1-1), KT359019 (BoCCD1-2), BK011289 (BoCCD1-3), BK011290 (BoCCD1-4), BK011291 (BoCCD4-1), BK011292 (BoCCD4-2), BK011293 (BoCCD4-3), and BK011294 (BoCCD4-4).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Accession numbers of plant CCDs and protein sequences of BoCCDs used to construct the phylogenetic tree in Figure 1.
Supplemental Figure S2. Constructs used for expression in bacteria or plants.
Supplemental Figure S3. In bacterio assays of different BoCCDs expressed in a β-carotene-accumulating E. coli strain.
Supplemental Figure S4. In bacterio assays of different BoCCDs expressed in a zeaxanthin-accumulating E. coli strain.
Supplemental Figure S5. Prediction of subcellular localization and of hydrophobic profiles of BoCCD4-3 and CsCCD2 proteins.
Supplemental Figure S6. HPLC–PDA analysis shows chromatographic profiles recorded at maximum absorbance of main apocarotenoids (440 nm) of plant tissues expressing BoCCD4-3.
Supplemental Figure S7. Structures of different saffron crocins.
Supplemental Figure S8. Generation of transplastomic plants expressing BoCCD4 from the plastid genome.
Supplemental Figure S9. PCR screening of regenerated T1 plants.
Supplemental Figure S10. Structures of non-polar apocarotenoids identified in B. orellana leaves and seeds.
Supplemental Table S1. Extraction of diagnostic ion masses produced in E. coli expressing lycopene, b-carotene and zeaxanthin + various BoCCDs.
Supplemental Table S2. Identification and quantification (μg/g DW) of apocarotenoids in N .benthamiana leaves transiently expressing various CCD constructs.
Supplemental Table S3. Identification and quantification (μg/g DW) of apocarotenoids in N. tabacum transplastomic lines.
Supplemental Table S4. Identification and quantification (μg/g DW) of crocins in T1 Microtom fruits expressing TP:BoCCD4.3 in combination with CsUGT74AD1 (#B18_2 and #B20_1,3,4).
Supplemental Table S5. Tentative identification of additional apocarotenoids in BoCCD4-3 expressing plants.
Supplemental Table S6. Apocarotenoids tentatively identified in B. orellana leaves (L) and seeds (S).
Supplemental Table S7. List of primers.
Supplementary Material
Acknowledgments
We thank E. Romano at the Centre of Advanced Microscopy “Patrizia Albertano,” U of Rome Tor Vergata, for the confocal images, L. Frigerio (U of Warwick) and J. Mathur (U of Guelph) for helpful comments on the sub-organellar localization of CsCCD2 and BoCCD4-3, Paola Ferrante (ENEA) for preparation of plant expression constructs, and P. Fraser and J. Enfissi (Royal Holloway U of London) for supplying Bixa leaves.
Funding
This work was supported by the European Commission (Project NEWCOTIANA, grant agreement 760331) and by COST action CA15136 (EUROCAROTEN).
Conflict of interest statement. The authors declare no conflict of interest.
S.F., O.C.D., M.S., P.M., and D.K. produced the data. S.F., O.C.D., M.S., G.D., G.A., and D.K. analyzed the data. G.G. conceived the project, coordinated the research, and wrote the manuscript with contributions from S.F. G.G. and R.B. designed the research. All authors reviewed the results and approved the final version of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is: Sarah Frusciante (sarah.frusciante@enea.it)
References
- Ahrazem O, Diretto G, Argandona J, Rubio-Moraga A, Julve JM, Orzaez D, Granell A, Gomez-Gomez L (2017) Evolutionarily distinct carotenoid cleavage dioxygenases are responsible for crocetin production in Buddleja davidii. J Exp Bot 68:4663–4677 [DOI] [PubMed] [Google Scholar]
- Ahrazem O, Gomez-Gomez L, Rodrigo MJ, Avalos J, Limon MC (2016) Carotenoid cleavage oxygenases from microbes and photosynthetic organisms: features and functions. Int J Mol Sci 17:1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavizadeh SH, Hosseinzadeh H (2014) Bioactivity assessment and toxicity of crocin: A comprehensive review. Food Chem Toxicol 64:65–80 [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S (2012) The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 335:1348–1351 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Hertle A, Makarenko E, Zuhlke J, Pribil M, Dietzmann A, Schliebner I, Aseeva E, Fenino E, Scharfenberg M. et al. (2009) Chloroplast proteins without cleavable transit peptides: rare exceptions or a major constituent of the chloroplast proteome? Mol Plant 2:1325–1335 [DOI] [PubMed] [Google Scholar]
- Bock R (2001) Transgenic plastids in basic research and plant biotechnology. J Mol Biol 312:425–438 [DOI] [PubMed] [Google Scholar]
- Bock R (2015) Engineering plastid genomes: Methods, tools, and applications in basic research and biotechnology. Annu Rev Plant Biol 66:211–241 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Dogbo O, Camara B (2003) Biosynthesis of the food and cosmetic plant pigment bixin (annatto). Science 300:2089–2091 [DOI] [PubMed] [Google Scholar]
- Carballo-Uicab VM, Cardenas-Conejo Y, Vallejo-Cardona AA, Aguilar-Espinosa M, Rodriguez-Campos J, Serrano-Posada H, Narvaez-Zapata JA, Vazquez-Flota F, Rivera-Madrid R (2019) Isolation and functional characterization of two dioxygenases putatively involved in bixin biosynthesis in annatto (Bixa orellana L.). PeerJ 7:e7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas-Conejo Y, Carballo-Uicab V, Lieberman M, Aguilar-Espinosa M, Comai L, Rivera-Madrid R (2015) De novo transcriptome sequencing in Bixa orellana to identify genes involved in methylerythritol phosphate, carotenoid and bixin biosynthesis. BMC Genomics 16:877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiste RC, Yamashita F, Gozzo FC, Mercadante AZ (2011) Simultaneous extraction and analysis by high performance liquid chromatography coupled to diode array and mass spectrometric detectors of bixin and phenolic compounds from annatto seeds. J Chromatogr A 1218:57–63 [DOI] [PubMed] [Google Scholar]
- D’Alessandro S, Havaux M (2019) Sensing beta-carotene oxidation in photosystem II to master plant stress tolerance. New Phytol 223:1776–1783 [DOI] [PubMed] [Google Scholar]
- Delfosse K, Wozny MR, Jaipargas EA, Barton KA, Anderson C, Mathur J (2015) Fluorescent protein aided insights on plastids and their extensions: A critical appraisal. Front Plant Sci 6:1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demurtas OC, de Brito Francisco R, Diretto G, Ferrante P, Frusciante S, Pietrella M, Aprea G, Borghi L, Feeney M, Frigerio L. et al. (2019) ABCC transporters mediate the vacuolar accumulation of crocins in saffron stigmas. Plant Cell 31:2789–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demurtas OC, Frusciante S, Ferrante P, Diretto G, Azad NH, Pietrella M, Aprea G, Taddei AR, Romano E, Mi J. et al. (2018) Candidate enzymes for saffron crocin biosynthesis are localized in multiple cellular compartments. Plant Physiol 177:990–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, Zheng C, Alberti A, Anthony F, Aprea G. et al. (2014) The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345:1181–1184 [DOI] [PubMed] [Google Scholar]
- Diretto G, Ahrazem O, Rubio-Moraga A, Fiore A, Sevi F, Argandona J, Gomez-Gomez L (2019) UGT709G1: A novel uridine diphosphate glycosyltransferase involved in the biosynthesis of picrocrocin, the precursor of safranal in saffron (Crocus sativus). New Phytol 224:725–740 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:39–40 [Google Scholar]
- Emadpour M, Karcher D, Bock R (2015) Boosting riboswitch efficiency by RNA amplification. Nucleic Acids Res 43:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016 [DOI] [PubMed] [Google Scholar]
- Falsini B, Piccardi M, Minnella A, Savastano C, Capoluongo E, Fadda A, Balestrazzi E, Maccarone R, Bisti S (2010) Influence of saffron supplementation on retinal flicker sensitivity in early age-related macular degeneration. Invest Ophthalmol Vis Sci 51:6118–6124 [DOI] [PubMed] [Google Scholar]
- Francisco RM, Regalado A, Ageorges A, Burla BJ, Bassin B, Eisenach C, Zarrouk O, Vialet S, Marlin T, Chaves MM. et al. (2013) ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell 25:1840–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frusciante S, Diretto G, Bruno M, Ferrante P, Pietrella M, Prado-Cabrero A, Rubio-Moraga A, Beyer P, Gomez-Gomez L, Al-Babili S. et al. (2014) Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc Natl Acad Sci USA 111:12246–12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345 [DOI] [PubMed] [Google Scholar]
- Giuliano G, Al-Babili S, von Lintig J (2003a) Carotenoid oxygenases: cleave it or leave it. Trends Plant Sci 8:145–149 [DOI] [PubMed] [Google Scholar]
- Giuliano G, Rosati C, Bramley PM (2003b) To dye or not to dye: biochemistry of annatto unveiled. Trends Biotechnol 21:513–516 [DOI] [PubMed] [Google Scholar]
- Hasunuma T, Miyazawa S, Yoshimura S, Shinzaki Y, Tomizawa K, Shindo K, Choi SK, Misawa N, Miyake C (2008) Biosynthesis of astaxanthin in tobacco leaves by transplastomic engineering. Plant J 55:857–868 [DOI] [PubMed] [Google Scholar]
- Hou X, Rivers J, Leon P, McQuinn RP, Pogson BJ (2016) Synthesis and function of apocarotenoid signals in plants. Trends Plant Sci 21:792–803 [DOI] [PubMed] [Google Scholar]
- Ilg A, Beyer P, Al-Babili S (2009) Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS J 276:736–747 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latari K, Wust F, Hubner M, Schaub P, Beisel KG, Matsubara S, Beyer P, Welsch R (2015) Tissue-specific apocarotenoid glycosylation contributes to carotenoid homeostasis in Arabidopsis leaves. Plant Physiol 168:1550–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwell J, Bruce BD, Keegstra K (1992) Isolation of a carotenoid-containing sub-membrane particle from the chloroplastic envelope outer membrane of pea (Pisum sativum). J Biol Chem 267:13933–13937 [PubMed] [Google Scholar]
- Marti M, Diretto G, Aragones V, Frusciante S, Ahrazem O, Gomez-Gomez L, Daros JA (2020) Efficient production of saffron crocins and picrocrocin in Nicotiana benthamiana using a virus-driven system. Metab Eng 61:238–250 [DOI] [PubMed] [Google Scholar]
- Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12 [Google Scholar]
- Matasci N, Hung LH, Yan Z, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M. et al. (2014) Data access for the 1,000 plants (1KP) project. Gigascience 3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercadante AZ, Steck A, Pfander H (1997) Isolation and identification of new apocarotenoids from annatto (Bixa orellana) seeds. J Agric Food Chem 45:1050–1054 [Google Scholar]
- Mercadante AZ, Steck A, Pfander H (1999) Three minor carotenoids from annatto (Bixa orellana) seeds. Phytochemistry 52:135–139 [Google Scholar]
- Mi J, Jia K-P, Balakrishna A, Wang JY, Al-Babili S (2019) An LC–MS profiling method reveals a route for apocarotene glycosylation and shows its induction by high light stress in Arabidopsis. Analyst 144:1197–1204 [DOI] [PubMed] [Google Scholar]
- Moreno JC, Mi J, Alagoz Y, Al-Babili S (2020) Plant apocarotenoids: from retrograde signaling to interspecific communication. Plant J 105:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira M, Enfissi EMA, Martinez Valenzuela ME, Menard GN, Driller RL, Eastmond PJ, Schuch W, Sandmann G, Fraser PD (2017) Engineering of tomato for the sustainable production of ketocarotenoids and its evaluation in aquaculture feed. Proc Natl Acad Sci USA 114:10876–10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Diretto G, Tavazza R, Giuliano G (2007) Improved protocol for agrobacterium mediated transformation of tomato and production of transgenic plants containing carotenoid biosynthetic gene CsZCD. Sci Hortic 112:172–175 [Google Scholar]
- Rivera-Madrid R, Aguilar-Espinosa M, Cardenas-Conejo Y, Garza-Caligaris LE (2016) Carotenoid derivates in achiote (Bixa orellana) seeds: Synthesis and health promoting properties. Front Plant Sci 7:1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo MJ, Alquezar B, Alos E, Medina V, Carmona L, Bruno M, Al-Babili S, Zacarias L (2013) A novel carotenoid cleavage activity involved in the biosynthesis of citrus fruit-specific apocarotenoid pigments. J Exp Bot 64:4461–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JA (2001) Characterization of a novel carotenoid cleavage dioxygenase from plants. J Biol Chem 276:25208–25211 [DOI] [PubMed] [Google Scholar]
- Stern DL (2013) The genetic causes of convergent evolution. Nat Rev Genet 14:751–764 [DOI] [PubMed] [Google Scholar]
- Sulli M, Mandolino G, Sturaro M, Onofri C, Diretto G, Parisi B, Giuliano G (2017) Molecular and biochemical characterization of a potato collection with contrasting tuber carotenoid content. PLoS ONE 12:e0184143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA 90:913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhounig A, Karcher D, Bock R (2010) Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc Natl Acad Sci USA 107:6204–6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MH, Strack D (2011) Carotenoids and their cleavage products: Biosynthesis and functions. Nat Prod Rep 28:663–692 [DOI] [PubMed] [Google Scholar]
- Wurbs D, Ruf S, Bock R (2007) Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. Plant J 49:276–288 [DOI] [PubMed] [Google Scholar]
- Xu Z, Pu X, Gao R, Demurtas OC, Fleck SJ, Richter M, He C, Ji A, Sun W, Kong J. et al. (2020) Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. BMC Biol 18:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Tsujimoto Y, Noda T, Shimizu M, Ohmori M, Morita S, Yamada A (1988) Hepatotoxicity of gardenia yellow color in rats. Toxicol Lett 44:177–182 [DOI] [PubMed] [Google Scholar]
- Yukawa M, Tsudzuki T, Sugiura M (2005) The 2005 version of the chloroplast DNA sequence from tobacco (Nicotiana tabacum). Plant Mol Biol Rep 23:359–365 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.