Abstract
Structural variants have a considerable impact on human genomic diversity. However, their evolutionary history remains mostly unexplored. Here, we developed a new method to identify potentially adaptive structural variants based on a similarity-based analysis that incorporates genotype frequency data from 26 populations simultaneously. Using this method, we analyzed 57,629 structural variants and identified 576 structural variants that show unusual population differentiation. Of these putatively adaptive structural variants, we further showed that 24 variants are multiallelic and overlap with coding sequences, and 20 variants are significantly associated with GWAS traits. Closer inspection of the haplotypic variation associated with these putatively adaptive and functional structural variants reveals deviations from neutral expectations due to: 1) population differentiation of rapidly evolving multiallelic variants, 2) incomplete sweeps, and 3) recent population-specific negative selection. Overall, our study provides new methodological insights, documents hundreds of putatively adaptive variants, and introduces evolutionary models that may better explain the complex evolution of structural variants.
Keywords: copy number variation, Denisovan, complex traits, neutrality test, population genetics
Introduction
Emerging technologies have recently revealed hundreds of thousands of genomic structural variants (SVs), including polymorphic duplications, deletions, inversions, and mobile transposable elements in the human genome (Hurles et al. 2008; Conrad et al. 2010; Pang et al. 2010; Mukamel et al. 2021). Unlike single-nucleotide variants, each SV affects a continuous block in the genome and thus is more likely to result in a phenotypic effect (Hurles et al. 2008; Weischenfeldt et al. 2013; Sudmant, Rausch, et al. 2015). Several SVs have been documented to have considerable effects on human disease and evolution (Dennis and Eichler 2016; Payer et al. 2017; Hsieh et al. 2019; Ho et al. 2020; Mukamel et al. 2021). Some of these functional variants reach >20% allele frequency in human populations, and some affect the copy number variation (CNV) of entire protein-coding genes (McCarroll et al. 2005; Handsaker et al. 2015).
The poster child for adaptive structural variation in humans is the CNV of the amylase gene. Several studies put forward evidence for positive selection of higher amylase gene copy numbers in the human lineage, and further in high starch-consuming human populations (Perry et al. 2007). Another striking example of potentially adaptive SVs is the deletion of LCE3B and LCE3C. This variant is one of the leading susceptibility markers to psoriasis (de Cid et al. 2009). This deletion was shown to be retained in the human lineage since Human–Neanderthal divergence under balancing selection (Pajic et al. 2016), arguably maintaining a balance between protection against pathogens and facilitating immune-mediated disorders. Recently, a genome-wide analysis identified several Neanderthal- and Denisovan-introgressed SVs that show strong signatures of adaptation (Hsieh et al. 2019; Yan et al. 2021). Collectively, these studies, along with others (see Saitou and Gokcumen [2020] for a detailed review), imply that several common SVs contribute to human phenotypic variation and may have evolved under diverse adaptive scenarios.
Despite the increasing appreciation of their role in human adaptive evolution, SVs have not been scrutinized as much as single-nucleotide variants due to technical difficulties. From a methodological perspective, SVs are more challenging to discover and genotype due to their localization in highly repetitive sections of the genome. In addition, they are generated through complex mutational mechanisms, often involving gene conversions and unequal recombination events (Kidd et al. 2010; Handsaker et al. 2011; Lupski 2015; Carvalho and Lupski 2016; Sekar et al. 2016). As a result, SVs are usually harbored by complex haplotypes, and they often are not tagged perfectly with flanking variants (Sudmant, Mallick, et al. 2015). The complexity of haplotypic architecture harboring SVs complicates analyses of neutrality and integration of SVs to genome-wide association studies. Most studies primarily interrogate single-nucleotide variants and SVs are often considered if only a “tag” single-nucleotide variant can be found. For example, our work has resolved the complex evolutionary history of the common deletion of the metabolizing GSTM1 gene (Saitou, Satta, and Gokcumen 2018). Locus-specific studies showed a strong association between this deletion and bladder cancer (The GSTM1 deletion is the risk allele, P = 4 × 10−11) (Rothman et al. 2010). A haplotype-based analysis of the locus suggested that this deletion has formed multiple times through independent mutation events and undergone gene conversion events (Saitou, Satta, Gokcumen, et al. 2018). Thus, due to the lack of single-nucleotide variants tagging this deletion, most genome-wide association studies and traditional selection scans did not include this deletion. Investigating individual haplotypes that harbor the deletion led us to identify one particular haplotype associated with the deletion that has been subject to a recent selective sweep in the East Asian populations (Saitou, Satta, and Gokcumen 2018).
A second factor that complicates the evolutionary study of SVs is that some are multiallelic (Quinlan and Hall 2012; Handsaker et al. 2015). For example, the haptoglobin locus harbors two large multiallelic and recurrent SVs that are not tagged by any single-nucleotide variant. Only after careful, locus-specific resolution of haplotypic variation were they shown to be associated with cholesterol levels (Boettger et al. 2016). Similarly, AMY1 (Perry et al. 2007), as we noted above, and DMBT1 (Polley et al. 2015) loci harbor multiallelic structural variations that were associated with dietary and metabolic traits. However, even for amylase gene CNV, arguably the best-studied SV in the human genome from an evolutionary perspective, the timing and existence of putative adaptive forces remain elusive (Mathieson and Mathieson 2018). In fact, SVs are often consciously left out from most selection scans along with segmental duplications and other repetitive regions due to the complications that we described above (Schrider and Kern 2017). In sum, we argue that the full impact of SVs on human evolution has not been understood and may explain some of the most exciting, yet to be described, adaptive variation in humans.
Given the complexity of haplotypes that harbor a considerable number of SVs, measures that depend on accurate genotyping of haplotypic variation, such as allele frequency spectra (e.g., Tajima’s D; Tajima 1993) or linkage-disequilibrium/homozygosity (e.g., iHS, Voight et al. 2006; XP-EHH, Sabeti et al. 2007) are often underpowered. Instead, direct population differentiation metrics may be the most appropriate and unbiased way to identify putatively adaptive SVs among human populations. Population differentiation-based methods are robust to haplotype disruption due to gene conversion, recurrence, or the presence of multiple alleles. Most studies that identify adaptive SVs have employed population differentiation-based methods (Redon et al. 2006; Xue et al. 2008; Sudmant, Mallick, et al. 2015; Almarri et al. 2020; Bergström et al. 2020). Deviations from expected allele frequency distribution can provide information on several types of selection (positive, negative, or stabilizing), and differential selection with complex histories (selection on standing variation, recent geography-specific negative selection, oscillating selective forces such as dynamic environmental change; Key et al. 2014). This is important because it has been shown that “classical” sweeps were rare (Hernandez et al. 2011) and selection on standing variants are likely to be the major force of human genomic adaptation (Schrider and Kern 2017), as recently shown for multiple alleles shaping skin color (Crawford et al. 2017; Martin et al. 2017).
To measure the population differentiation of genetic variants, FST statistics (Weir and Cockerham 1984) and VST statistics for CNVs (Redon et al. 2006) are commonly used. More recent research has developed methods to compare multiple populations, primarily for admixture analysis (e.g., F3 statistics; Reich et al. 2009). However, these methods can only compare two or three populations to each other. Recently, (Duforet-Frebourg et al. 2016) developed a PCA-based method to identify single-nucleotide variants with population differentiation by analyzing ten populations simultaneously, confirming well-known targets for positive selection, and discovered new candidate genes. Here, we developed a new, similarity-based method to identify adaptive SVs with unusual allele frequency distribution with which one can analyze: 1) multiallelic variants and 2) the distribution of genotype frequency in multiple populations collectively.
Results and Discussion
Structural Variants with Unusual Population Differentiation
Inspired by the emerging work that integrates all available population differentiation information to understand demographic and adaptive trends (Duforet-Frebourg et al. 2016), we developed a new method based on the Bhattacharyya similarity metric specifically to identify putatively adaptive outliers among SVs (Bhattacharyya 1943; Materials and Methods, fig. 1A–F, and supplementary fig. S1, Supplementary Material online).
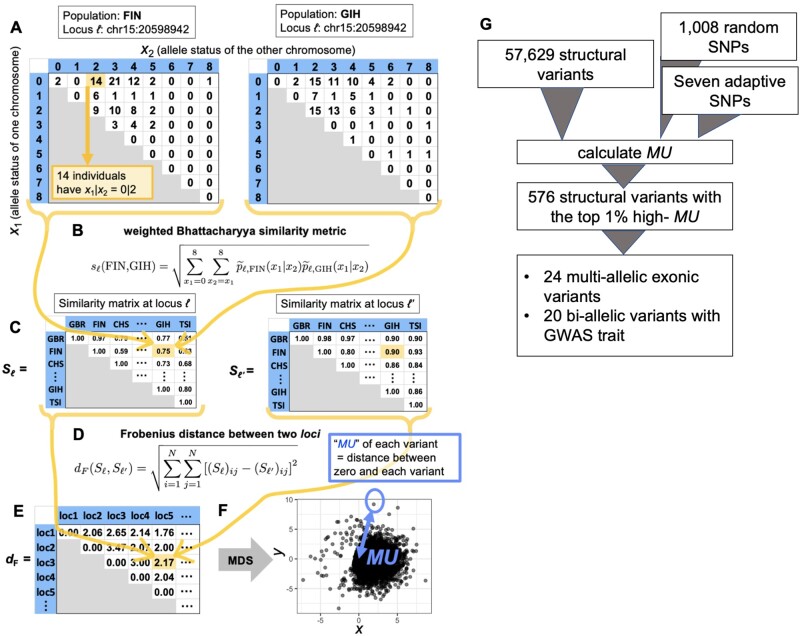
Fig. 1.
An overview of the calculation of “MU” (see Materials and Methods for details). (A) For each population and each locus, we have a 9×9 matrix representing the genotype. The row and column of the matrix represent one of the two chromosomes each. The cells contain the frequencies of specific genotype combinations. (B) We calculate the similarity in the 9×9 matrices exemplified in panel A between each pair of populations. The similarity value ranges between 0 and 1. (C) In this manner, for each locus, we obtain a 26×26 matrix representing the similarity between different pairs of populations. The diagonal entries of the similarity matrix are equal to 1 because any population is identical to itself, yielding the largest possible value of the similarity, which is 1. (D) We calculate the distance between the 26×26 matrices for each pair of loci. (E) In this manner, we obtain a distance matrix representing the distance between the different pairs of loci. (F) We carry out the MDS to project the obtained distance matrix into the 2D embedding space. Each circle represents a locus. The distance between the locus and the origin in the embedding space defines MU for each locus. (G) The general results from our pipeline.
Briefly, we characterize each locus (ℓ) as an N×N similarity matrix Sℓ based on the genotype frequency of the N = 26 populations in the 1000 Genomes Project phase 3 data set (Sudmant, Rausch, et al. 2015). We measure a modified Bhattacharyya similarity metric between each pair of populations based on the transformed probability distribution (for the original Bhattacharyya metric, see Bhattacharyya 1943; Cha and Srihari 2002). To increase the sensitivity to identify the population differentiation of variants with many alleles, we use a weighted variant of the Bhattacharyya similarity metric. Overall, we analyzed M = 58,644 variants, including 57,629 SVs, 1,008 uniformly randomly chosen single-nucleotide polymorphisms (SNPs), as well as seven SNPs that were reported to be under adaptive evolution (six SNPs under positive selection and one SNP under balancing selection) (Norton et al. 2006; Mou et al. 2008; Kimura et al. 2009; Smith et al. 2009; Basu Mallick et al. 2013; Ding et al. 2013; Ko et al. 2013; Wilde et al. 2014; Wu et al. 2016; Deng and Xu 2018). We constructed a distance matrix for each of the M loci and compared the similarity matrix Sℓ across all these loci (ℓ = 1, …, M). We define the distance between Sℓ (at one locus) and Sℓ′ (at another locus) by the Frobenius norm, denoted by dF. The M×M Frobenius distance matrix, denoted by F, tabulates the difference between each pair of loci, and its (ℓ, ℓ′) entry is given by dF(Sℓ, Sℓ′). These steps provided us with a matrix indicating how SVs relate to each other based on their global genotype frequency distribution.
We assumed, based on previous literature (Conrad et al. 2010), that the majority of SVs will be evolving under neutrality or near neutrality. Therefore, population differentiation should primarily be driven by genetic drift. We then reasoned that SVs that have unusual allele frequency distribution among the 26 populations compared with the genome-wide observations are likely to have evolved under nonneutral conditions. To visualize the relationships between SVs based on their global allele frequency distribution, we ran a multidimensional scaling (MDS) algorithm. To empirically measure these relationships, we calculated the distance between the origin and each variant in the MDS space and defined it as “Measure of Unusualness (MU),” or degree of the unusual allele frequency distribution (Materials and Methods, fig. 1F). This measure informs on the unusualness of global population differentiation of a given SV, as compared with the entirety of the data set.
Simulation and Empirical Confirmation
To validate the accuracy and sensitivity, we conducted forward simulations using SLiM 3.6 (Messer 2013; Haller and Messer 2019) (Materials and Methods). We modeled stepwise copy number gains or losses in each locus under different mutation rates/selection coefficients in three populations (YRI, CEU, and CHB) for which the demographic parameters were previously established (Gravel et al. 2011). In each simulation, we generated 2,970 potentially variable neutral loci, and 10×3 potentially variable loci under population-specific selection in each population (with a range of selection coefficients). We used these data to calculate MU and assess the accuracy and sensitivity of our approach (supplementary fig. S2, Supplementary Material online). Our results suggest that our approach is unable to distinguish between drift and selection if the mutation rate is higher than 10−7 mutations per locus per generation. Further, we found that the population for which the selection is acting is an important parameter in determining the power and accuracy of MU. Specifically, we found that if the selection is acting on the YRI population, our power to detect selection increases, possibly because the effect of drift is lower in African populations due to their higher effective population sizes (Tenesa et al. 2007). Although these simulations are useful in the general assessment of our approach, they have two major limitations. First, we were not able to simulate MU for all 26 populations because the demographic parameters for most of these populations were not established. Thus, it is likely that MU is more powerful when applied to a larger number of populations. Second, the mutation rates of SVs are highly variable (Lin and Gokcumen 2019). Thus, without having a better sense of the mutation rates of SVs, it is difficult to assess the power of MU for the whole range of SVs in the human genome. Thus, we argue that an empirical comparison to known variants with well-established population-selection signatures may be currently a better benchmark than simulation-based methods for understanding the power of MU.
To assess the accuracy of our approach empirically, we calculated MU for 1,008 random SNPs (supplementary table S1, Supplementary Material online; Materials and Methods). We reasoned that the control SNPs will provide an additional marker set whereby differentiation is determined by near-neutral forces. Using this data set, we confirmed that the distribution of MU measured for SVs is not significantly different from that measured for the uniformly randomly chosen SNPs from the 1000 Genome Phase 3 data set (Sudmant, Rausch, et al. 2015) (P = 0.88, Mann–Whitney test, supplementary fig. S3A, Supplementary Material online), suggesting that similar to single-nucleotide variants, the overall distribution of MU for SVs is neutral-like. We replicated this analysis using 5,000 neutral SNPs reported in (Pouyet et al. 2018) (supplementary fig. S3B, Supplementary Material online) and found similar results (P = 0.41, Mann–Whitney test, supplementary fig. S3C, Supplementary Material online).
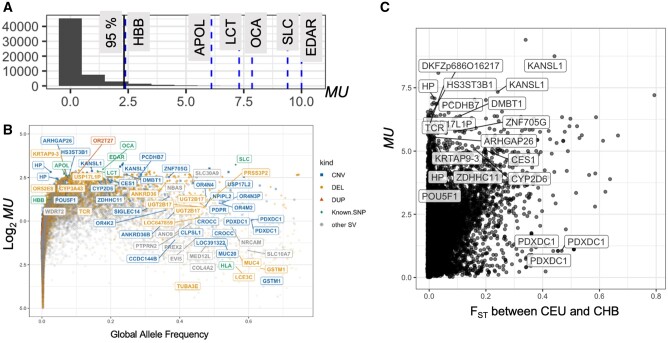
Next, we investigated the sensitivity of our method by measuring MU from six SNPs that have repeatedly been reported to be under population-specific positive selection and their functional relevance was well established (Materials and Methods). Thus, they provide an appropriate gold standard to test the sensitivity of our method. We found that all of these six positively selected SNPs were shown to be in the top 5% of the MU distribution (fig. 2A), improving our confidence in our method. In addition to those six SNPs, we also included in our analysis rs1129740, which resides in the HLA locus and has been one of the handfuls of variants in humans that are thought to have evolved under long-term balancing selection (Teixeira et al. 2015). This allowed us to observe how MU behaves for such variants even though MU was not designed to test balancing selection. We found that this nonsynonymous mutation shows low MU values when considering its allele frequency (fig. 2B). We found it noteworthy that a SV, LCE3BC gene deletion, that we speculated previously to have evolved under balancing selection (Pajic et al. 2016) shows similarly low MU values despite their high allele frequency (fig. 2B). Thus, it is plausible that high allele frequency SVs that may have been evolving under balancing selection may exhibit unusually low MU values (supplementary table S2, Supplementary Material online).
Fig. 2.
An overview of the study pipeline and results. (A) The histogram of MU of the 1,008 randomly selected SNPs and the six positively selected SNPs that were found to be under selection in previous studies (see Materials and Methods). The latter group was indicated by blue vertical dashed lines on the histogram, and the genes affected by these variants were labeled. The 95th percentile of the distribution was marked by a black vertical line. (B) The relationship between allele frequency and MU. The horizontal axis indicates the global alternative allele frequency. The vertical axis indicates the logarithm of the MU value. The exonic SVs with MU>5 or global allele frequency >0.25 are labeled, as well as the six positively selected SNPs and one HLA SNP which was shown to have evolved under balancing selection. Some gene names are shown multiple times (e.g., HP, KANSP1, and PDXDC1); this happens because multiple SVs overlapping these genes were reported in the 1000 Genomes Project Phase 3 data set. Colors represent different types of variants. The abbreviation is from the 1000 Genome Project phase 3 SVs data set. CNV, copy number variants (multiallelic variants); DEL, deletion; DUP, duplication, Known; SNP, SNPs from previous studies (see Materials and Methods); Other SVs, insertion, inversion, Alu, Long interspersed nuclear element, SINE-VNTR retrotransposons. (C) Comparison of FST (Weir and Cockerham 1984) between CEU and CHB populations and MU. Biallelic SVs with FST (between CEU and CHB) >0.4 and MU>5 were labeled. The shade in blue represents the density of the SVs (see supplementary fig. S4, Supplementary Material online).
To understand the differences between more traditional methods of measuring population differentiation and our method, we compared MU with direct allele frequency-based FST between representative continental populations (fig. 2C and supplementary fig. S4, Supplementary Material online). As expected, we found a significant correlation between these two measures (Spearman rank correlation coefficient >0.49 and P < 10−15 for all comparisons). However, we also found notable discrepancies. We noted 125 variants that are in the top 1st percentile for MU, but show FST < 0.2 in any pairwise comparison of European (CEU), East Asian (CHB), and African (YRI) populations (supplementary table S3, Supplementary Material online). Closer inspection of these variants suggests that MU captures multiallelic variations and within-continent variation, which fell below the detection threshold of standard pairwise population comparisons. In addition, we noted SVs that have large values of FST but do not stand out in terms of MU. When we investigated the allele frequency distribution of SVs with high FST (>0.25) but low MU (<1), we consistently observed a clinal distribution of allele frequencies across the continents (supplementary fig. S5, Supplementary Material online), likely due to serial founder effects that define human genetic variation as described previously (Ramachandran et al. 2005). In other words, we argue that variants that have high among-continental differences that the standard FST detects often reflect the effects of major bottleneck/drift events, such as out-of-Africa migrations, rather than adaptive sweeps. That suggests that a gradual population differentiation may not lead to a high MU value. Instead, our method is sensitive to deviations from such expected clinal allele frequency changes, including unusually low or high allele frequency in a single population as compared with its neighbors. This is an advantage of our method because local ecological and cultural variation often underlies adaptive evolution in humans (Rees et al. 2020). Thus, our method shows promise in capturing hundreds of novel putatively adaptive variants that have not been captured by traditional SNP-based pairwise population comparisons.
MU Identifies Dozens of SVs Invisible to Traditional Selection Scans
There are several outstanding questions concerning the enrichment of specific properties of adaptive SVs, including their functional relevance, the mutation mechanisms through which the variants are generated, and their size distribution. However, there are tremendous technical biases inherent in the short-read sequencing-based characterization of these variants, especially concerning extremely high false-negative rates in the discovery of certain types of SVs, such as tandem and dispersed duplications and inversions (Kronenberg et al. 2015). Thus, instead of searching for general trends in our data set (e.g., adaptively evolving SVs are larger or smaller than neutrally evolving ones), we focused on resolving the evolutionary forces shaping individual SVs with functional implications.
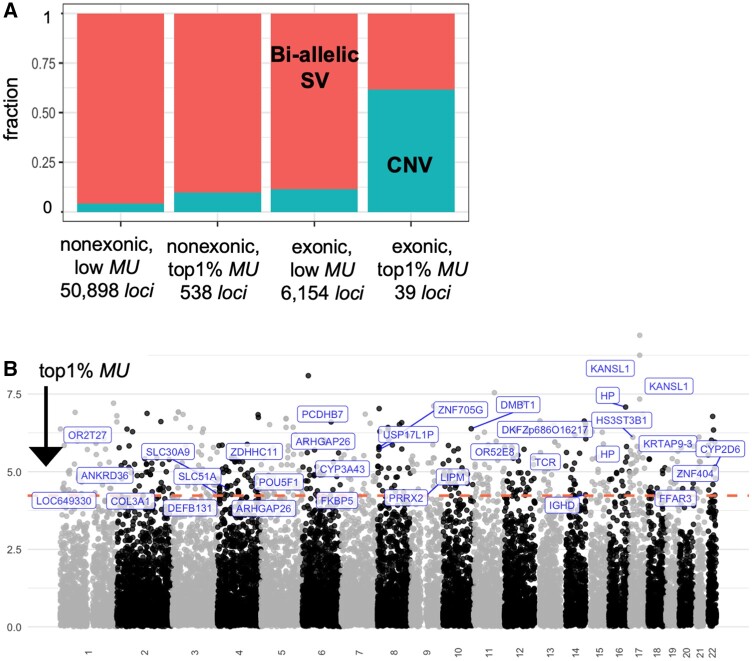
In this spirit, we first investigated SVs that overlap with coding sequences. We identified 39 SVs with the top 1% MU value that contain one or more entire exon (fig. 3A and supplementary table S4, Supplementary Material online). Regardless, many of these exonic SVs were associated with metabolic traits and diseases in previous locus-specific analyses and include members of cytochrome p450 (CYP3A43, CYP2D6), solute carrier (SLC30A9, SLC51A), olfactory receptor (OR2T27, OR52E8) gene families. For example, DMBT1 gene copy number was noted for its population differentiation and associated with dietary subsistence strategies (Polley et al. 2015). Similarly, the CNV affecting the CES1 (Zhu and Markowitz 2013), CYP2D6 (Candiotti et al. 2005), HS3ST3B1 (Kim et al. 2010), and SULT1 (Hebbring et al. 2008) are associated with differences in metabolizing of xenobiotic substances, primarily described within a pharmacogenomics context. Interestingly, we found that 24 (∼65%) of these exonic SVs are multiallelic (fig. 3B and table 1), more than five times higher than genome-wide expectations (P = 0.0005, χ2 test). We found that intervals that overlap with multiallelic SVs are enriched for “defense response to Gram-negative bacterium” function (FDR Q value = 1.09×10−3), concordant with previous literature linking adaptive SVs with immune-related functions.
Fig. 3.
Unusually distributed structural variants with exonic contents. (A) The relative ratio of bi-allelic structural variants and multiallelic copy number variants in each grouping based on exonic overlap (structural variants that contain at least one exon vs. those that do not) and MU values (top 1% vs. the rest). Bi-allelic SV: biallelic structural variants. CNV: multiallelic copy number variants with three or more alleles. (B) Manhattan plots of MU of structural variants. The horizontal axis shows the chromosomal location of structural variants, and the vertical axis shows the MU value. The exonic variants with the top 1% MU (>4.23) were labeled.
Table 1.
Twenty-Four Multiallelic Exonic SVs (CNV, Copy Number Variation in fig. 2C) with the Top 1% MU (>4.23).
| ID | MU | chr | Start | End | Size | Allele Freq. | Gene Name | OMIM |
|---|---|---|---|---|---|---|---|---|
| esv3640680; esv3640681 | 8.75 | chr17 | 44230893 | 44262697 | 31,804 | 0.22723629 | KANSL1 | Chromatin Modification |
| esv3640677; esv3640678 | 7.34 | chr17 | 44165338 | 44211686 | 46,348 | 0.1453675 | KANSL1 | Chromatin Modification |
| esv3638992; esv3638993; esv3638994; esv3638995; esv3638996; esv3638997 | 7.08 | chr16 | 72094527 | 72110961 | 16,434 | 0.02515976 | HP | Glycoprotein |
| esv3606964; esv3606965; esv3606966; esv3606967; esv3606968; esv3606969; esv3606970 | 6.43 | chr5 | 140554408 | 140558942 | 4,534 | 0.287539932 | PCDHB7 | Protocadherin |
| esv3624777; esv3624778; esv3624779; esv3624780 | 6.38 | chr10 | 124344431 | 124353237 | 8,806 | 0.24580733 | DMBT1 | Brain Tumor |
| esv3640025; esv3640026 | 6.11 | chr17 | 14224374 | 14483419 | 259,045 | 0.078474481 | HS3ST3B1 | Heparan Sulfate |
| esv3616116; esv3616117; esv3616118; esv3616119 | 5.78 | chr8 | 7212582 | 7227421 | 14,839 | 0.36841027 | ZNF705G | Zing Finger |
| esv3607012; esv3607013 | 5.56 | chr5 | 142263109 | 142447062 | 183,953 | 0.074480842 | ARHGAP26 | Leukemia |
| esv3638989; esv3638990; esv3638991 | 5.33 | chr16 | 72080868 | 72098986 | 18,118 | 0.04572683 | HP | Glycoprotein |
| esv3603782; esv3603783; esv3603784; esv3603785 | 5.22 | chr5 | 814446 | 825367 | 10,921 | 0.18250742 | ZDHHC11 | Zing Finger |
| esv3647809; esv3647810; esv3647811; esv3647812 | 5.09 | chr22 | 42523949 | 42533891 | 9,942 | 0.160942323 | CYP2D6 | Metabolism |
| esv3608531; esv3608532 | 5.07 | chr6 | 31131451 | 31272307 | 140,856 | 0.069289142 | POU5F1 | Transcription Factor |
| esv3638688; esv3638689; esv3638690 | 5.04 | chr16 | 55832207 | 55864521 | 32,314 | 0.202276441 | CES1 | Metabolism |
| esv3624140; esv3624141 | 4.98 | chr10 | 90551092 | 90632203 | 81,111 | 0.061900981 | LIPM | Signal Peptide |
| esv3638686; esv3638687 | 4.69 | chr16 | 55798890 | 55822423 | 23,533 | 0.20127776 | CES1P1 | Metabolism |
| esv3621839; esv3621840 | 4.68 | chr9 | 132463983 | 132648102 | 184,119 | 0.062899361 | PRRX2 | Homeobox |
| esv3644233; esv3644234 | 4.61 | chr19 | 35851718 | 35863310 | 11,592 | 0.12879353 | FFAR3 | Fatty Acid |
| esv3585247; esv3585248; esv3585249 | 4.50 | chr1 | 12901370 | 12921250 | 19,880 | 0.149361003 | LOC649330 | Unknown |
| esv3608684; esv3608685 | 4.46 | chr6 | 35521984 | 35568895 | 46,911 | 0.03674116 | FKBP5 | Binding |
| esv3638338; esv3638339; esv3638340; esv3638341; esv3638342 | 4.40 | chr16 | 28614507 | 28626916 | 12,409 | 0.26936938 | SULT1A1 | Metabolism |
| esv3641584; esv3641585 | 4.38 | chr18 | 3200017 | 3415245 | 215,228 | 0.064696481 | MYOM1 | Muscle |
| esv3599276; esv3599277 | 4.30 | chr3 | 195954431 | 196022808 | 68,377 | 0.060303542 | SLC51A | Solute Carrier |
| esv3599572; esv3599573; esv3599574 | 4.26 | chr4 | 9418201 | 9457405 | 39,204 | 0.05271566 | DEFB131 | Defensin |
| esv3607010; esv3607011 | 4.24 | chr5 | 142174919 | 142260351 | 85,432 | 0.061701281 | ARHGAP26 | Leukemia |
Note.—Variant information is retrieved from the 1000 Genomes Project phase 3 data set (Sudmant, Rausch, et al. 2015). Start and End refer to the starting and ending locations of variants on the chromosome, respectively. We described the gene name if the SV contains one or more entire exon(s) of UCSC Genes. Gene function was retrieved from OMIM (https://www.omim.org/).
The exonic SVs with the highest MU have been invisible to previous genome-wide association studies and selection scans. We argue that this is primarily because the current GWAS pipelines interrogate single-nucleotide variants. Single-nucleotide variants may not tag multiallelic SVs due to gene conversion, recurrence, and potential genotyping errors, as discussed in the introduction. This phenomenon was diligently dissected by Boettger et al. (Boettger et al. 2016) for the haptoglobin (HP) locus, which harbors two recurrent and multiallelic exonic CNVs that we found to show unusually high MU. They described a novel way to use a combination of single-nucleotide variants to impute these SVs in the locus. Their reanalysis of the genome-wide association studies revealed a previously hidden association between CNV affecting HP gene function and blood cholesterol levels. Based on our results, we argue that the effects of multiallelic SVs on human evolution and phenotypic variation remain underappreciated.
Multiallelic SVs are often genotyped inaccurately (Mills et al. 2011). If such inaccuracies are systemic to given populations, it may lead to errors in identifying spurious genome-wide signals pertaining to population differentiation (Anderson-Trocmé et al. 2020). Thus, some of our observations of unusual allele frequency distribution may be due to such batch effects and genotyping errors inherent in the 1000 Genomes Phase 3 data set. To address this issue, we used mrCaNaVaR (https://github.com/BilkentCompGen/mrcanavar, last accessed March 23, 2020) to estimate the copy number of individual genes using new high-coverage (∼30×) sequencing data from the same samples in the 1000 Genomes Phase 3 data set (Byrska-Bishop et al. 2021). These new gene copy number estimates are different from the SV calls from the 1000 Genomes in three ways. First, the gene copy number is estimated for individual samples and no population-level SV discovery or genotyping was performed, eliminating batch effects. Second, it provides continuous values of normalized read depth, rather than discrete categories of different types of SVs as was reported from the 1000 Genomes Phase 3 data set. This allows us to measure, for each gene, the distance between arbitrary two populations directly by the earth mover’s distance, which directly uses the difference between the normalized read-depth value for an individual in one population and that for an individual in the other population. In this manner, we can avoid technical biases in the 1000 Genomes Phase 3 data set introduced by the categorization of SVs into discrete types (Materials and Methods). Third, this new data set allowed us to estimate CNVs in genes that were not discovered by the conservative discovery pipeline of 1000 Genomes Phase 3.
This approach allowed us to conduct a parallel assessment of the MU approach in detecting putatively adaptive SVs (supplementary table S5 and fig. S6, Supplementary Material online). We were able to assess 21 copy number variable genes (see Materials and Methods) that we identified to show unusually high MU in our original pipeline and found that 8 (∼38%) and 13 (∼62%) of these also have unusually high MU values in the mrCaNaVaR database at the 99th and 95th percentile, respectively. These include CNVs of KANSL1, HP, and DMBT1 genes with well-described likely adaptive functions. We individually investigated the remaining genes for which the CNV showed disparate MU percentiles in our original analysis and mrCaNaVaR analysis. We found that all of them are large (18–259 kb) and relatively rare (<7.5% global allele frequency) variants that fall into regions rich in segmental duplications. These regions are prone to both genotyping errors and recurrence. In addition, using the mrCaNaVaR data set, we were able to identify several additional candidates exonic SVs, including AMY1, SIGLEC14, and multiple CCL genes, among others, that were not included in the 1000 Genomes Phase 3 SV data set but were noted because of their relevance to human evolution (Hollox et al. 2022). Thus, our results confirm that the MU provides a robust and reliable approach to identify putatively adaptive SVs. However, genotyping errors are a considerable factor in determining the false-positive and -negative rates in our approach and we argue that it is imperative to conduct follow-up analyses of the candidate adaptive SVs to validate deviations from neutrality at the haplotype level. We provide examples for such analysis below.
Resolving the Haplotypes of Putatively Adaptive SVs
The complex evolutionary dynamics of SVs often do not fit classical population genetics expectations, such as complete classical sweeps. Thus, we argue that careful investigation of the evolutionary histories of a few examples can provide valuable insights that can later be generalized at the genome-wide scale. Therefore, we wanted to resolve the haplotypes that harbor SVs in order to investigate functional associations, coalescence times, and signatures of selection concerning these variants in more detail. There are 344 biallelic SVs that are in the 1st percentile in terms of MU (>4.23) and have strong linkage disequilibrium with nearby single-nucleotide variants (R2 > 0.95) (supplementary table S6, Supplementary Material online).
Among these, we identified 20 haplotypes that are significantly associated with phenotypes (nominal P < 10−9; GWAS Atlas; https://atlas.ctglab.nl/; last accessed March 23, 2020) (table 2). The selected 20 loci provided us with a means to further investigate the evolutionary and functional effects of SVs that show unusual geographical distribution.
Table 2.
Structural Variants with the Top 1% MU Value and Observed Phenotypic Effects through Tag SNP (R2>0.95) in GWAS Atlas.
| ID | esv number | Trait | MU | Exon | Afr. Freq. | EAsia. Freq. | Eur. Freq. | Selection |
|---|---|---|---|---|---|---|---|---|
| 1 | esv3592798 | Age started wearing glasses or contact lenses | 4.91 | 0.08620 | 0.6478 | 0.4245 | Positive | |
| 2 | esv3587652 | White blood cells | 5.92 | 0.08090 | 0.5913 | 0.3956 | Positive | |
| 3 | esv3647661 | Height, waist–hip ratio (adjusted for BMI) | 6.78 | 0.07190 | 0.5238 | 0.6809 | Positive | |
| 4 | esv3590788 | Hot drink temperature | 5.14 | 0.04920 | 0.5317 | 0.3231 | Positive | |
| 5 | esv3634671 | Height, fat, vertical cup-disc ratio, weight, sexual maturity | 4.60 | 0.02800 | 0.4573 | 0.2227 | Positive | |
| 6 | esv3626213 | BMI, alcohol intake, neuroticism, walking pace | 5.23 | 0.08400 | 0.4375 | 0.4314 | Positive | |
| 7 | esv3597888 | Total bilirubin, schizophrenia, worry | 6.38 | 0.05750 | 0.7857 | 0.4493 | Positive | |
| 8 | esv3629184 | Heart pulse rate, morning person | 5.44 | 0.02800 | 0.0347 | 0.3827 | Positive | |
| 9 | esv3629182 | Heart pulse rate, morning person | 5.44 | 0.02800 | 0.0347 | 0.3827 | ||
| 10 | esv3629434 | Hematocrit, hemoglobin concentration, red blood cell count | 6.35 | 0.01890 | 0.0109 | 0.326 | ||
| 11 | esv3591931 | Hair type, platelet volume | 4.88 | 0.23220 | 0.0238 | 0.4016 | Positive | |
| 12 | esv3616249 | Heel bone mineral density, platelet, red cell | 6.30 | 0.74050 | 0.0208 | 0.3887 | Negative | |
| 13 | esv3639698 | Height | 4.85 | 0.03100 | 0.4851 | 0.2823 | ||
| 14 | esv3640666 | Red blood cell, bone mineral density, baldness, brain volume, | 4.62 | ARL17A | 0.01510 | 0.001 | 0.2416 | |
| 15 | esv3636701 | Heel bone mineral density | 4.24 | 0.08700 | 0.623 | 0.1421 | ||
| 16 | esv3625336 | Water intake | 4.33 | 0.07870 | 0.3194 | 0.4523 | ||
| 17 | esv3609042 | Gamma-glutamyl-transferase | 5.70 | 0.09080 | 0.5089 | 0.674 | ||
| 18 | esv3636957 | FEV1/FVC ratio, impedance, height | 5.90 | 0.06880 | 0.3056 | 0.6759 | ||
| 19 | esv3642017a | Height | 4.29 | 0.69820 | 0.2817 | 0.0984 | Positive | |
| 20 | esv3587631 | White blood cells | 8.50 | 0.52120 | 0.002 | 0 | Negative |
Note.—All the SVs in the table are deletions. ID is the same as figure 4. We described the GWAS traits in the “Trait” column if the SV shows phenotypic effects through tag SNPs on GWAS Atlas. We described the gene name in the “Exon” column if the SV contains one or more entire exons of UCSC Genes. “Afr/EAsia/Eur.freq” are the frequency of the alternative allele in each population, Africa, East Asia, and Europe. The “Selection” column describes estimated natural selection based on the neutrality tests (fig. 4). Specifically, “negative” indicates cases where we found recent selection favoring the ancestral allele, whereas “positive” indicates cases where we found recent selection favoring the derived alleles.
This “deletion (esv3642017)” found in the 1000 Genomes samples as compared with the reference genome is actually a derived insertion that happens to be represented in the reference genome (Anagnou et al. 1988; Conrad et al. 2010; Schrider et al. 2013). Thus, even though the putative action of selection is on the nondeleted haplotypes, given that this haplotype carries the derived allele, we categorized the selection as “positive” in this case. Two variants highlighted in yellow are discussed specifically as examples. Green color gradient indicates the allele frequency in each population.
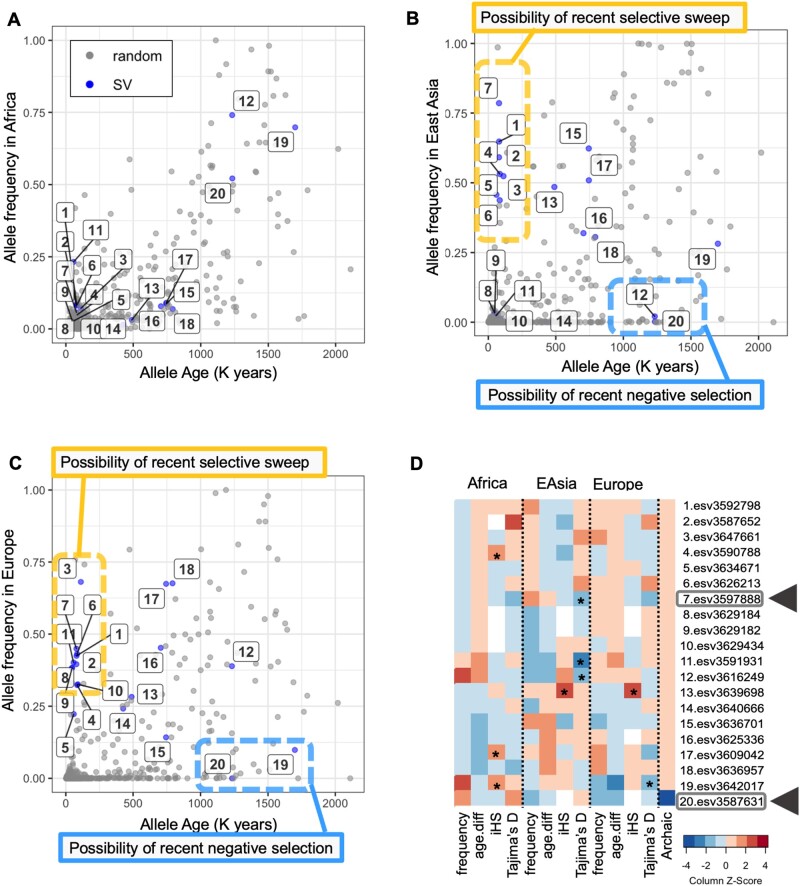
Using the linked haplotypic variation for the 20 SVs, we retrieved allele ages from the Human Genome Dating database (Albers and McVean 2020) (https://human.genome.dating; last accessed, March 3, 2020). Under neutrality, an allele’s age is expected to positively correlate with its allele frequency (Patterson 2005). Given that we are explicitly investigating variants that are putatively evolving under population-specific adaptive forces, we expect deviations from this expectation. Figure 4A–C shows the estimated age of the allele and its frequency in European, East Asian, and African populations, respectively. If a variant has emerged recently, but its frequency is common (left upper side) in a given population, it suggests a potential recent selective sweep (i.e., a new allele is rapidly favored and increases its frequency). In contrast, if a variant is old and its frequency is rare, these are candidates for recent negative selection against the allele in that particular population. To more formally interrogate this line of inquiry, we calculated how long it takes for a new allele to reach a given frequency in each population under neutrality using formula (15) in (Kimura and Ohta 1973) assuming previously published demographic parameters (Schaffner et al. 2005) (supplementary fig. S7, Supplementary Material online). In a manner similar to allele frequency expectations, the age estimate of a variant older than the neutral estimation may suggest a faster increase in allele frequency and a recent selective sweep (fig. 4A–C). In parallel, we calculated Tajima’s D scores of 5 kb upstream and downstream regions of the 20 SVs of interest and the iHS scores of the tag single-nucleotide variants of the target SVs (Materials and Methods). We summarized these values in figure 4D.
Fig. 4.
Neutrality tests on the 20 SVs with phenotypic effects. (A–C) Age and frequency of the SVs in African (A), East Asian (B), and European (C) populations. The variant’s ID is the same as table 2. The blue dots represent the tag SNPs associated with the 20 SVs. The gray dots represent 599 random SNPs. The horizontal axis shows the age of allele in the Human Genome Dating database; the vertical axis is the alternative allele frequency in each population. (D) A heatmap summarizing neutrality tests of the 20 SVs in each population. It shows allele frequency, the allele age difference between haplotype-based estimation in the Human Genome Dating database (Albers and McVean 2020) and neutral expectation based on the allele frequency (Kimura and Ohta 1973), iHS value, Tajima’s D, and if the allele is observed in the Denisovan genome. (We did not observe these variants in the Neanderthal genomes.) Warmer colors indicate higher values. Similarly, for the allele age difference, colder and warmer colors show that the allele is older and newer than neutral expectation, respectively. For the archaic genome, blue shows that the allele is shared with the Denisovan. The asterisk indicates that the value is less than five percentile or more than 95 percentile when compared with at least 500 uniformly randomly selected variants or windows across the genome (see Materials and Methods for detail). Two variants highlighted by the arrows (7 and 20) are discussed specifically as examples.
We found that the flanking haplotypes of putatively adaptive SVs predicted by MU do not show consistent trends of haplotypic variation, extended homozygosity, or population differentiation. Rather, our observations fit the emerging consensus in evolutionary genomics that the adaptive SVs are shaped by complex evolutionary trajectories that change over time and space (Mérot et al. 2020). As an example of the complicated nature of the evolutionary histories of adaptive SVs, we highlight esv3642017. This variant is recorded as a deletion compared with the reference genome in the 1000 Genomes Phase 3 data set. However, a closer inspection reveals that this variant is a human-specific retro-insertion of the DHFR gene (Anagnou et al. 1988; Conrad et al. 2010; Schrider et al. 2013). The haplotype that harbors this insertion is associated with decreased height (P < 10−16). Even though deletion seems to be predominantly found in Africa, the derived retrogene inserted is predominantly found in Eurasia. The locus that harbors the insertion shows unusually low Tajima’s D in the European population and unusually low genetic diversity in another European-ancestry cohort as reported in Schrider et al. (2013), which altogether suggest a Eurasian-specific sweep of a recent insertion. Based on such locus-specific analyses, we identified incomplete population-specific sweeps and recent population-specific negative selection as the two main drivers for shaping the allele frequency distribution of putatively adaptive SVs.
Incomplete, Population-Specific Sweeps: The Example of the Propionyl-CoA Carboxylase Gene
The classical scenario for population-specific adaptive evolution is characterized by: 1) high frequency of the variant in the specific population compared with other populations, 2) deviations in the site frequency spectrum suggesting rapid expansion of the selected allele, resulting in an excess of rare variants in the locus, 3) lower than expected allele age, and 4) long haplotype homozygosity suggesting rapid expansion of the selected allele (Rees et al. 2020). We look for signatures of this scenario among the 20 haplotypes that we highlight because they harbor SVs with unusual allele frequency distributions (MU in the 1st percentile, >4.23) and because they are associated with GWAS traits (table 2). We found that 12 (60%) of them fit the scenario of a recent population-specific adaptive sweep (fig. 4D).
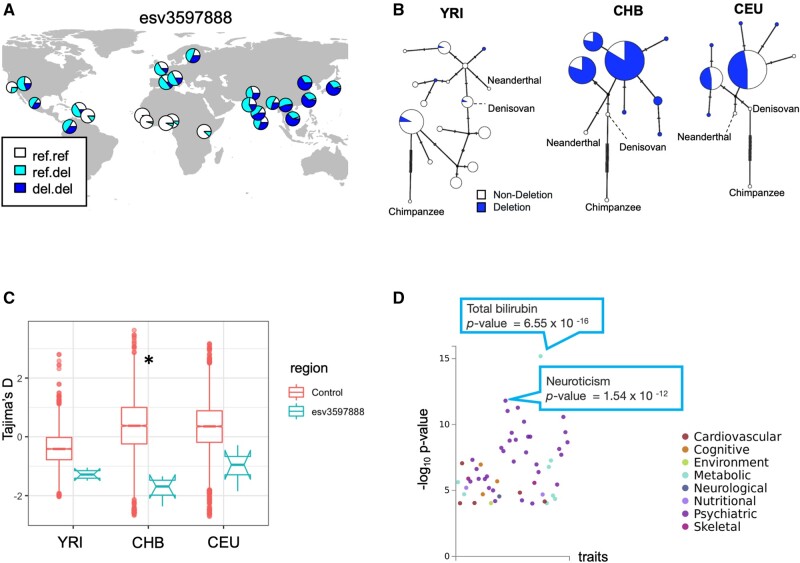
The haplotype harboring esv3597888 provides an informative example of the population-specific incomplete sweep scenario. The haplotype has a lower than 5% allele frequency in most African populations but reaches near 75% allele frequency in East Asian populations (fig. 5A). Further, the Median Joining network of the haplotypic variation in this locus shows a dramatic reduction of haplotypic diversity beyond the expected reduction due to drift in the East Asian population as compared with the African population, which is consistent with a recent selective sweep (fig. 5B). The Tajima’s D values retrieved from the flanking sequences of the deletion are lower than genome-wide expectations in all three continental populations (fig. 5C). Last but not least, the estimated age of the allele is much more recent than what is expected based on its frequency, especially in the East Asian population (fig. 4B and supplementary fig. S7, Supplementary Material online). Collectively, these results suggest a recent selective sweep in Eurasian populations. However, even for this locus, not all the neutrality tests capture this sweep. For example, in a traditionally defined recent sweep, we expect to find high iHS values. Instead, for this locus, the iHS is relatively low, mirroring the surprisingly high overall haplotypic variation in this locus. Regardless, esv3597888 remains one of the best candidates for a derived SV that has recently been swept to higher allele frequency in a population-specific manner.
Fig. 5.
The evolutionary analysis of esv3597888, the deletion overlapping the intronic region of the PCC gene. (A) The geographic distribution of the esv3597888. (B) Haplotype networks constructed from the 5 kb upstream and downstream sequences from the esv3597888 location of three modern human populations (Yoruban [YRI], Han Chinese [CHB], and European [CEU]), the Altai Neanderthal sequence (Prüfer et al. 2014), and the Denisovan sequence (Reich et al. 2010) that are mapped to hg19 reference genome, and the chimpanzee reference genome (panTro4). The haplotypes that harbor the deletion are indicated by white and those that do not are indicated by blue. (C) Tajima’s D value in the 5 kb upstream and downstream regions of esv3597888 (10 kb in total) (Tajima 1993). Asterisk shows that Tajima’s D of the esv3597888 flanking region is lower than the bottom five percentile of Tajima’s D of 5,000 random regions. The asterisk shows that the mean value of esv3597888 tag region is lower than the five percentile of the control region. (D) The PheWAS result of rs556788, which tags esv3597888 (supplementary table S4, Supplementary Material online). Each dot indicates a trait. The vertical axis shows the −log10 P value of the association between the genotype and phenotype. The color indicates the phenotype category in GWAS ATLAS.
The phenotypic effects of the haplotype harboring this variant further support the potential adaptive relevance of esv3597888. This 5.4-kb deletion overlaps with the intronic region of the propionyl-CoA carboxylase (PCC) gene, which encodes for an enzyme that metabolizes specific amino acids and lipid species (Wongkittichote et al. 2017). The haplotype harboring esv3597888 (tagged by rs556788) is associated with the expression of the PCCB gene in the adrenal gland (P = 7.8×10−10 in the GTEx Analysis Release V8; GTEx Consortium 2013). Moreover, the haplotype is associated with total bilirubin, a cardiometabolic signaling molecule (P = 6.6 × 10−16), and neuroticism (P = 1.5 × 10−12) (fig. 5D). We argue that it is likely that the 5.4-kb deletion esv3597888 is the causal variant in these associations, given its size and that it overlaps with a well-documented binding site for the abundant transcription factor CTCF (supplementary fig. S8, Supplementary Material online). The haplotype has pleiotropic functional effects, and thus the exact reasons why it confers an adaptive advantage in East Asia particularly remains to be seen.
Recent Population-Specific Negative Selection: The DAP3 Gene
Among the haplotypes that we highlighted, we noticed that some show unexpectedly low allele frequency in Eurasian populations compared with the expectation based on their estimated age. We hypothesize that these haplotypes, and by proxy the SVs that they harbor, may have been subjected to recent, population-specific negative selection, favoring the ancestral allele. Variations that have emerged early in human evolution and remain in extant populations are often found at high allele frequencies in all extant human populations under neutrality (Lin et al. 2015). Thus, if recent negative selection on the derived SV is acting in a population-specific manner, we expect to observe in that population: 1) an unusual reduction in allele frequency of the variant that cannot be explained by drift alone and 2) a shift in the allele frequency spectrum toward rare variants in the locus.
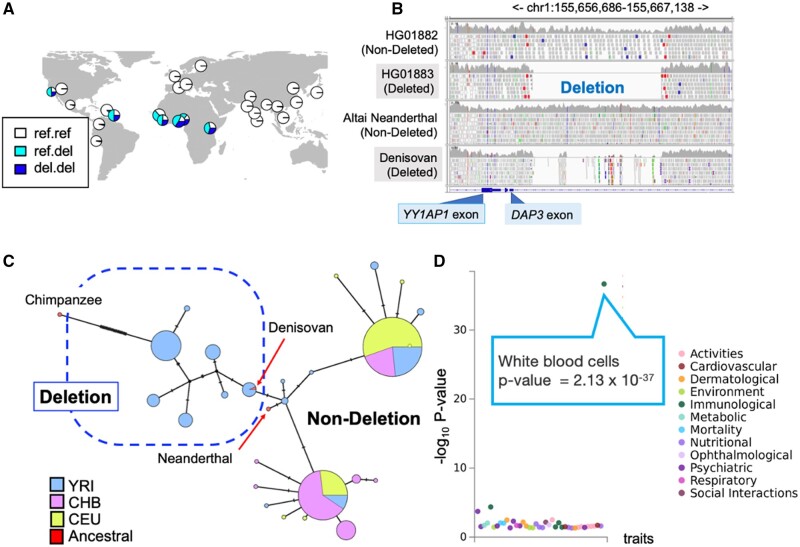
A striking example of population-specific negative selection is provided by the haplotypes harboring esv3587631, which shows one of the highest MU values in the genome (MU = 8.50). This deletion is the major allele (i.e., >50% allele frequency) in most sub-Saharan African populations but almost absent in non-African populations (fig. 6A). Human Genome Dating database estimates the age of a single-nucleotide variant tagging esv3587631 to be 1.1–1.3 My old (Albers and McVean 2020). Thus, the deletion has emerged prior to human–Neanderthal divergence. Consistent with this result, we found that Denisovan but not Altai Neanderthal carries this deletion (fig. 6B). The haplotype network showed that the haplotypes harboring the deletion are similar to those from archaic hominins, consistent with our observation that this deletion is present in archaic human genomes (fig. 6C). Collectively, it is clear that the deletion has evolved before Human Neanderthal divergence and increased in allele frequency in African populations to more than 75%, harbored by diverse haplotypes. However, none of the haplotypes that harbor the deletion is found in Eurasian populations. Furthermore, the locus shows significantly negative Tajima’s D values in the East Asian population, further supporting nonneutral forces acting on the deletion (fig. 4D).
Fig. 6.
The evolutionary analysis of esv3587631, the intronic deletion polymorphism of the death-associated protein gene3 (DAP3). (A) The geographic distribution of the esv3587631. (B) esv3587631 in modern and ancient hominin genomes. These Integrated Genome Browser snapshots show the genome assembly (Hg19) of a human with the ancestral, homozygous nondeleted genotype and another with a homozygous deleted genotype that shows no reads mapping to the deletion region (top two rows). Similarly, sequences from Neanderthal and Denisovan genomes were mapped to this region. The Denisovan genome shows a clear signature of the deletion with breakpoints indistinguishable from the deletion observed in modern humans. (C) A haplotype network of three modern human populations, Yoruba (YRI), Han Chinese (CHB), European (CEU), as well as Altai Neanderthal and the Denisovan, and Chimpanzee, constructed from the flanking sequences from the esv3587631 location. (D) The Phewas result of rs348195, the tag SNP of esv3587631 at GWAS Atlas. Each dot indicates a trait and the y axis shows the −log10 P value of the association between the genotype and phenotype.
Functionally, this ∼4.8-kb deletion overlaps with one of the introns of the well-studied and highly conserved DAP3 gene (supplementary fig. S8, Supplementary Material online). DAP3 is a mitoribosome protein that regulates apoptosis at the cellular level and is linked to multiple developmental, immune-related, and biomedically relevant phenotypes at the organismal level (Greber and Ban 2016; Kim et al. 2017). Specifically, the deletion overlaps with a conserved regulatory region comprising multiple transcription factor binding sites. Consistent with these observations, the haplotypes harboring the deletion (tag variant, rs348195) were strongly associated with the increased expression of the DAP3 gene in various tissues (P < 10−6), with the effect size exceeding 0.4 in some cases. Moreover, the deletion (through the analysis of tag SNP rs348195) is strongly associated with decreased levels of white blood cells (nominal P = 2.1×10−37, fig. 6D). Collectively, these results are consistent with a scenario where an ancient deletion variant that has been either neutral or beneficial in African populations has become detrimental to fitness in Eurasian populations, perhaps due to adaptive constraints concerning immune function.
Conclusion
Although several putatively adaptive SVs have been reported in previous studies, a genome-wide selection scan of SVs has remained challenging. In this study, we built a network-based analysis of population differentiation among 26 populations in the 1000 Genome Project data set to identify putatively adaptive SVs including multiallelic variants. Our method assumes that drift is the major force that shapes the distributions of genomic variants among human populations as articulated by others (Ramachandran et al. 2005; Coop et al. 2009). In identifying the most common allele frequency distribution combinations across the 26 populations, our method parallels the recent variant-centric integrative analysis method proposed by Biddanda et al. (2020). We argue that such direct, empirical scrutiny of the geographical distribution of variants will provide a valuable and relatively unbiased picture of demographic and nonneutral trends that shape human genetic variation.
Our method is designed to identify SVs with population differentiation that deviate from neutral expectations without any a priori adaptive model. It identified hundreds of putatively adaptive SVs with unusual genotype frequency distributions in humans. The majority of these SVs were hidden from traditional selection scans which mainly focus only on single-nucleotide variants. Our study identified 24 putatively adaptive exonic multiallelic SVs, the majority of which were not discussed within an adaptive context in humans. In addition to incomplete sweeps of derived SVs, we found that recent population-specific negative selection is a considerable force shaping the geographic distribution of functional SVs in humans. Overall, our study supports the emerging notion that SVs significantly contribute to nonneutral and biomedically relevant phenotypic variation in humans (Radke and Lee 2015; Mérot et al. 2020) and highlight specific trajectories underlying the evolution of such variants.
From an evolutionary genomics perspective, the prominence of exonic multiallelic CNVs among the putatively adaptive SVs is not surprising. Cross-species analyses have repeatedly revealed the outsized role of recurrent gain and losses in gene families in shaping phenotypic characteristics in a variety of species, with recurrent evolution of caffeine in plants (Denoeud et al. 2014), salivary amylase in mammals (Pajic et al. 2019), and venom in snakes (Casewell et al. 2020) providing notable examples. Moreover, studies in humans reported that multiallelic CNVs have seven times more effect on gene dosage than the combined effect of biallelic deletions and duplications (Handsaker et al. 2015). The same multiallelic SVs, however, are hidden in the majority of GWAS and selection analyses. Multiallelic variants are not necessarily tagged by nearby single-nucleotide variants, and they often reside in the genomic regions with enriched segmental duplications where identifying variants can be problematic. Thus, we expect that better genotyping of multiallelic SVs with long-read sequencing platforms will dramatically increase our ability to identify multiallelic SVs and their previously unknown adaptive roles.
A surprising result from our study is the identification of recent negative selection favoring ancestral alleles as a notable force determining the allele frequency distribution of putatively adaptive SVs. Selective sweeps are often thought to increase the allele frequency of the derived and not ancestral variant. In this work, we found that at least 10% of the putative adaptive SVs show recent sweeps favoring the ancestral allele. It is plausible that recent human adaptive evolution involves repeated adaptation to similar environmental conditions across time and geography as reported in (Bergey et al. 2018). Thus, an ancestral adaptive variant that confers a smaller fitness advantage than the derived variant may become adaptively beneficial again if environmental pressures revert back to an earlier state. This scenario is particularly applicable to immune system-related traits within the context of an evolutionary arms race as articulated previously (Key et al. 2014). Similarly, adaptive landscapes concerning metabolic traits have drastically changed multiple times for human populations due to technological advances (e.g., agricultural transition) (Hancock et al. 2010) and migrations to new ecologies (e.g., arctic populations) (Marciniak and Perry 2017). Thus, under the assumption that neither the ancestral nor derived alleles are fixed, it is not surprising that ancestral SVs are favored in certain geographies and instances. Such cases will appear as negative selection against the derived allele. We reported in detail one such case involving the exonic deletion of the growth hormone receptor in another study (Saitou et al. 2021). The current study identifies several other cases, suggesting that recent, geography-specific negative selection is a considerable force shaping allele frequency distribution and population differentiation of functional SVs.
There are caveats to our study and to the investigation of adaptive SVs in general. First, it is clear from existing literature that the current data sets suffer from significant false-positive rates, potentially missing up to 80% of the SVs (Mahmoud et al. 2019). Moreover, current technologies can discover certain types of SVs (e.g., large biallelic deletions) much more sensitively than other types of variants (e.g., duplications, inversions). It is telling that one of the SVs most relevant to human evolution, amylase CNV, are not cataloged by the 1000 Genomes Phase 3 data set because of alignment issues in the locus. Even when such multiallelic variants are discovered, it is not uncommon that their exact genotypes (e.g., exact copy number) may not be accurately documented. Second, the genotyping platforms commonly used in genetic association studies mostly focus on biallelic single-nucleotide variants only. In fact, even this study, which is aware of these limitations, highlighted biallelic variants, for which the haplotype can be readily resolved, and thus trait associations can be investigated. The true contribution of most SVs, including multiallelic variants, to phenotypic variation, remains mostly unknown. Third, most SV maps, including the data set we use in our study are not an ideal representation of human variation. For example, a more powerful and adequate sampling would involve hypothesis-driven efforts where specific adaptive pressures are in mind (Scheinfeldt and Tishkoff 2013; Rees et al. 2020). Further, ascertainment bias in GWAS studies (Sirugo et al. 2019), which still comprise primarily European cohorts, limits our power to link evolutionary trends shaping the SV allele frequency distributions to their functional effects. Overall, the current picture of the evolutionary effects of SVs, including those revealed in this study, remains incomplete and should be treated as a theoretical and methodological framework for future studies with more comprehensive data sets. We believe that as long-read sequencing-based discovery and later genotyping become affordable, the full impact of SVs on human evolution and diversity will be better revealed.
Materials and Methods
1000 Genomes Phase 3 Data Set
As the input data set, we used 1000 Genome Project phase 3 data sets (Sudmant, Rausch, et al. 2015) for the following three reasons. First, the genotyping is based on whole-genome sequencing and multiple detection methods such as Delly (Rausch et al. 2012), which combines short insert paired-ends, long-range mate pairs, and split-read alignments, and GenomeSTRiP (Handsaker et al. 2011), which uses read depth and read pairs for SV identification to improve accuracy. Thus, this data set provides a highly accurate SV genotype. Second, it contains approximately 100 individuals from each population. Therefore, one can increase the power to detect geographically differentiated SVs due to population-specific adaptation by assessing deviations from expected population differentiation. Third, it provides phased genotype information not only of the SVs but also of the SNPs from the same individuals. This allows us to apply our methods for identifying population differentiation to known SNPs to assess their performance and to carry out the subsequent haplotype-based analysis on a subset of SVs.
Preprocessing SVs and Selection of Known SNPs in the Analysis
We selected 57,629 autosomal SVs with annotations in the 1000 Genomes project phase 3 data set (Sudmant, Rausch, et al. 2015) since variants in sex chromosomes are differently described from autosomes due to the smaller number of the observed number of chromosomes and cannot be analyzed in the same pipeline as autosomal variants. As controls, we also used 1,008 uniformly randomly selected single-nucleotide variants from the same data set and six single-nucleotide variants that have undergone putative natural selection, including rs334 in HBB (Ding et al. 2013), rs73885319 in APOL (Ko et al. 2013), rs4988235 in LCT (Smith et al. 2009), rs12913832 in OCA (Wilde et al. 2014), rs3827760 in EDAR, which is common in East Asian populations and associated with hair and dental traits (Mou et al. 2008; Kimura et al. 2009; Wu et al. 2016), rs1426654 in SLC24A5, which is associated with skin color (Norton et al. 2006; Basu Mallick et al. 2013; Deng and Xu 2018). In addition to these SNPs, we included rs1129740 that falls into HLA-DQA1, which is one of the few variants in the human genome that showed classical signatures of balancing selection (Teixeira et al. 2015). This HLA allele showed unusually low MU (0.22) despite the global allele frequency of 0.52 (fig. 2).
To verify that 1,008 randomly chosen SNPs indeed represent a neutral data set, we redid our analysis with 5,000 SNPs shown to be evolving under near-neutrality by Pouyet et al. (2018). Briefly, we calculated MU in the same manner as our previous analysis, where we include all the SVs and the 5,000 neutral SNPs. We found that this reanalysis did not change our results. Specifically, we found that the MU values calculated with the additional SNP data set remain nearly identical to those that were calculated with our initial data set (Spearman’s correlation = 0.997) (supplementary fig. S3B, Supplementary Material online). Further, we replicated our finding that the MU values for all SVs are not significantly different from those calculated for the neutral SNPs (P = 0.41, Mann–Whitney test, supplementary fig. S3C, Supplementary Material online), indicating that the majority of SVs are evolving neutrally.
Calculation of the MU and MDS Plot
We characterize each locus ℓ as an N×N similarity matrix, denoted by Sℓ, where N = 26 is the number of populations in the 1000 Genome Project Phase 3 data set (fig. 1A). The entries of matrix Sℓ represent the similarity between pairs of populations in terms of the frequency of each allele at a locus. Specifically, for each locus ℓ and population i, the genotype count is given by the 1000 Genome Project Phase 3 data set. In general, variant call format (VCF), genotype (i.e., the allele status of a pair of chromosomes of one individual) is denoted by x1|x2. Genotypes 1|0 and 0|1 in general VCF are effectively the same and mean that one individual has one reference (i.e., 0) allele and one alternative (i.e., 1) allele at the locus. Therefore, we summarized both 1|0 and 0|1 into 0|1 in the following analysis. The maximum (alternative + reference) allele number at a locus was nine, in which case the allele number ranges from 0 to 8 (supplementary fig. S9, Supplementary Material online). Therefore, in general, we summarized x1|x2 and x2|x1 into x1|x2, where x1, x2 = 0,1, …, 8 and x1 ≤ x2. We denote the frequency of genotype x1|x2 at locus ℓ and population i by pℓ,i(x1|x2), where . Note that . To increase the sensitivity to identify the population differentiation of multiallelic variants (i.e., variants with more than two alleles), especially, with large CNV (such as a multiallelic variant with copy number one to eight, even if the frequency of copy number eight is rare), we use a weighted variant of the Bhattacharyya similarity metric, which modifies the Bhattacharyya similarity metric (Bhattacharyya 1943; Cha and Srihari 2002), as follows (fig. 1B).
First, we transform the original distribution, to , where:
| (1) |
and
| (2) |
This transformation magnifies the frequency of genotype and its differences between populations at large x1 and x2 values (i.e., large CNV). Second, we measure the Bhattacharyya metric between each pair of populations i and j based on the transformed probability distribution, that is:
| (3) |
If the two distributions and are identical, then sℓ(i,j) = 1, which is the largest value of sℓ(i, j). In this manner, for each locus ℓ, we obtain an N×N similarity matrix Sℓ whose (i, j) entry is given by equation (3).
To analyze the organization of M = 58,644 loci (which is composed of 57,629 SVs, 1,008 random SNPs, and seven SNPs under adaptive evolution), we constructed a distance matrix for the M loci by comparing similarity matrix Sℓ across the loci. We define the distance between Sℓ and Sℓ′ using the Frobenius norm, denoted by dF, which is given by
| (4) |
Note that . Also note that one can replace the summation by without loss of generality because the similarity matrix Sℓ is symmetric and all of its diagonal elements are equal to 1. The M×M Frobenius distance matrix, denoted by F, tabulates the difference between each pair of loci, and its (ℓ, ℓ′) entry is given by dF(Sℓ,Sℓ′). Finally, we ran a MDS algorithm on F to map out relationships between the M loci on a 2D space. We used the Python package manifold, which is part of scikit-learn (Pedregosa et al. 2011), to estimate the MDS. To empirically measure these relationships, we calculated the distance between the origin and each variant in the MDS space and defined it as “MU,” or the degree of unusual allele frequency distribution (Materials and Methods, fig. 1). This measure informs on the unusualness of global population differentiation of a given SV, as compared with the entirety of the data set. We only used one initial condition for the MDS due to the long time required for the computation. The analyses have been done on the server of the University at Buffalo Center for Computational Research (http://www.buffalo.edu/ccr.html).
Simulations and MU Calculations
To model the evolution of CNV in humans, we modified recipe 14.11 (modeling microsatellites) and recipe 5.4 (model of human evolution) in SLiM 3.6 (Haller and Messer 2019). We modeled stepwise copy number gains or losses in each locus under different mutation rates/selection coefficients in three populations (YRI, CEU, and CHB) for which the demographic parameters were previously established (Gravel et al. 2011). In each simulation, we generated 2,970 potentially variable neutral loci, and 10×3 potentially variable loci under population-specific selection in each population (with a range of selection coefficients), respectively. All the variable loci were initially neutral, but at generation 78084, just after the split of CEU and CHB, the ten loci became adaptive in each population. We used 0.5, 0.05, 0.005 as selection coefficient and 10−5, 10−6, 10−7 for mutation rate. We also ran simulations with the mutation rate 10−8, however, most of them did not produce polymorphic sites. So we did not eventually use the mutation rate of 10−8. We simulated each model 100 times per condition (three mutation rates×three selection coefficients). The allowed copy number range was 1–8. The script can be found on our GitHub webpage (https://github.com/mariesaitou/Network_humanpop_SV).
mrCaNaVaR Copy Number Estimates and MU Calculations
We used mrCaNaVaR (https://github.com/BilkentCompGen/mrcanavar, last accessed March 23, 2020) to estimate the normalized read depth for each gene in the human genome using high-coverage (∼30×) sequencing data (Byrska-Bishop et al. 2021) available for the same samples we used in our original analysis. mrCaNaVaR remaps short reads promiscuously and measures the normalized read depth of intervals across the genome. Using this approach, we calculated the read depth of each gene across the human genome and use this data to calculate the MU values for each human gene as follows. First, we discarded the genes on chrM (mitochondrial DNA), chrX, chrY, which left us 36,486 genes. We denote by yg,p,i the read depth associated with gene g (g = 1, …, 36,486) in the ith individual in population p (p = 1, …, 26), from the 1000 Genome phase 3 data set. Second, for each gene, we divided each yg,p,i by the average over all individuals and populations. We applied this normalization to enable a gene-to-gene comparison of the CNV in a single population and across multiple populations without being affected by the typical copy number, which substantially depends on the gene. We denote the normalized yg,p,i by ȳg,p,i. Third, for the given gene g, we calculated the distance between two populations by the earth mover’s distance (Levina and Bickel 2001; Pérez-Barbería et al. 2007) as follows. The distribution of the normalized copy number for population p is given by assigning probability 1/np to each value of ȳg,p,i that appears in the data, where np is the number of individuals in population p. If there are two individuals that have the same value of ȳg,p,i, for example, then the probability of this normalized copy number is 2/np. The earth mover’s distance between populations p and p′ is the minimal cost to move the distribution of the normalized copy number for population p to that for population p′. The cost of moving a unit probability mass from a location ȳg,p,i in one distribution (corresponding to population p) to another location in another distribution (corresponding to population p′) is given by . By calculating the earth mover’s distance between each pair of populations, for each gene g, we obtain a 26×26 distance matrix, denoted by Dg, which represents how similar/different each pair of populations is in terms of the copy number distribution. Fourth, we calculate the distance between each pair of genes using the Frobenius norm using equation (4), but with matrix Sl being replaced by matrix Dg. The remaining steps for calculating MU are the same as those for the 1000 Genome Project Phase 3 data set. The script is available on GitHub (https://github.com/mariesaitou/Network_humanpop_SV). For comparing the results from mrCaNaVaR calls and our original analysis, we identified 21 genes that are multiallelic SVs in the 1000 Genomes Phase 3 data set with high MU values (99th percentile) that are also presented in mrCaNaVaR analysis.
Allele Frequency, MU, and SVs
Since MU is related to alternative allele frequency by definition, we categorized the variants into four groups in terms of the allele frequency using ranges 1–0.25, 0.25–0.5, 0.5–0.75, and 0.75–1, and investigated the distribution of MU of the variants in each group. In intermediate allele frequency groups, multiallelic CNV had higher MU than SNPs (Wilcoxon test, P = 0.0021 and P = 0.00095 for allele frequency ranges 0.25–0.5 and 0.5–0.75, respectively). This result indicates that our methods may detect unusual population differentiation due to the excess of multicopy alleles than biallelic variants. In addition, we noticed that the small number of variants that have allele frequencies higher than 0.5 show smaller MU values than those variants that are less than 0.5. There is no mathematical reason for this observation. Thus, this observation may be due to various other reasons, including potential genotyping errors which may be increased among this group of variants. We want to further acknowledge here other studies that have employed tools to capture variation at the multiallelic SV locus (e.g., VST; Redon et al. 2006) and use thoughtful hypothesis-driven population sampling (Huerta-Sánchez et al. 2014). Regardless, the majority of genome-wide scans of selection employ biallelic locus and use available continental populations such as the 1000 Genomes data set and almost none captures variation across dozens of populations.
Functional Genomics Analysis
We retrieved exonic content from UCSC Genome Browser (http://genome.ucsc.edu/; UCSC Genes, table: knownCanonical) and examined if each SV contained one or more entire exon using bedtools (Quinlan and Hall 2010). To find functionally relevant loci, we first calculated linkage disequilibrium between the top 1% high MU biallelic SVs and neighboring regions (5 kb upstream and downstream) with vcftools (Danecek et al. 2011). We searched the resulting tag SNPs (r2 > 0.95) in GWAS Atlas Phewas database (https://atlas.ctglab.nl/PheWAS, last accessed March 23, 2020), defining that P < 10−9 as a statistically significant association. Of the 576 top 1% SVs in terms of MU, we found that 500 variants were biallelic, which were suitable for haplotype analysis. Among the 500 variants, 344 SVs showed R2 larger than 0.95 with neighboring variant(s). Of these 344 SVs, 20 of them have flanking tag SNPs that are significantly associated with phenotypic variation (fig. 2A and table 2). Further, we used the GTEx portal (GTEx Consortium 2013) for associating these SNPs to variation in gene expression levels (supplementary fig. S4, Supplementary Material online). We used ShinyGO (Ge et al. 2020) or GREAT (McLean et al. 2010) to conduct functional enrichment analysis for genes and intervals overlapping SVs with the whole genome as the background. These tools search for enrichment in multiple databases and provide multiple hypotheses-corrected false discovery rates.
Evolutionary Genomics Analysis of the Haplotypes Harboring Putatively Adaptive SVs
We used the 344 SVs that are among the top 1% in terms of MU and have flanking tag SNPs with R2 value larger than 0.95 for the following evolutionary analysis. We used the age estimates from Human Genome Dating database (Albers and McVean 2020) using tag SNPs as proxies to the adaptive SVs. This database documents the allele age estimates based on the analysis of pairwise haplotype identical tracts in 1000 Genomes (1000 Genomes Project Consortium et al. 2015) and Simons Genome Diversity Projects (Mallick et al. 2016) (https://human.genome.dating; last accessed, March 23, 2020). We also calculated how long it takes for a new allele to reach a given frequency in each population under neutrality using equation (15) of (Kimura and Ohta 1973). For this calculation, we used demographic models for each population detailed in (Schaffner et al. 2005) (fig. 3D).
For haplotype-level population genetics measures, we targeted 5 kb upstream and downstream regions of the SVs and calculated Tajima’s D scores of the tag SNP of the target SVs using VCFTools (Danecek et al. 2011). We retrieved the iHS score from the 1000 Genomes Selection Browser (Pybus et al. 2014). For comparative purposes, we calculated the same scores for ∼500 random regions generated with bedtools (Quinlan and Hall 2010) across the genome. Ancient human (Altai Neanderthal and Denisovan) genomic bam files are published on the Max-Planck Institute website (https://www.eva.mpg.de/index.html, last accessed March 23, 2020) (Reich et al. 2010; Prüfer et al. 2014). We used samtool-based (Li et al. 2009) read-depth analysis to genotype deletions in archaic genomes (fig. 5B). We generated haplotype networks using VCFtoTree (Xu et al. 2017) and POPArt (Clement et al. 2002; Leigh and Bryant 2015) by Minimum Spanning Network method (Bandelt et al. 1999).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dr John Novembre and Dr Simen Rød Sandve for the careful reading of this manuscript. N.M. acknowledges support from Japan Science and Technology Agency (JST) Moonshot R&D (Grant No. JPMJMS2021). O.G. acknowledges support from the National Science Foundation (Grant No. 2123284). We thank Can Alkan for generously running mrCaNaVaR to estimate copy numbers. We also thank Fanny Pouyet for generously providing us with a high-recombination region map.
Data Availability
For the data visualization, we used Rstudio (v1.2.1335), R(v3.5.3), and ggplot2 (Wickham 2009). The script underlying this article are available in https://github.com/mariesaitou/Network_humanpop_SV, and the datasets from Sudmant, Rausch, et al. (2015), can be accessed with ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/integrated_sv_map/.
Contributor Information
Marie Saitou, Department of Biological Sciences, University at Buffalo, State University of New York, Buffalo, NY, USA; Section of Genetic Medicine, Department of Medicine, The University of Chicago, Chicago, IL, USA.
Naoki Masuda, Department of Mathematics, University at Buffalo, State University of New York, Buffalo, NY, USA; Computational and Data-Enabled Science and Engineering Program, University at Buffalo, State University of New York, Buffalo, NY, USA.
Omer Gokcumen, Department of Biological Sciences, University at Buffalo, State University of New York, Buffalo, NY, USA.
References
- 1000 Genomes Project Consortium, et al. 2015. A global reference for human genetic variation. Nature 526:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers PK, McVean G. 2020. Dating genomic variants and shared ancestry in population-scale sequencing data. PLoS Biol. 18:e3000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almarri MA, Bergström A, Prado-Martinez J, Yang F, Fu B, Dunham AS, Chen Y, Hurles ME, Tyler-Smith C, Xue Y. 2020. Population structure, stratification, and introgression of human structural variation. Cell 182:189–199. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnou NP, Antonarakis SE, O’Brien SJ, Modi WS, Nienhuis AW. 1988. Chromosomal localization and racial distribution of the polymorphic human dihydrofolate reductase pseudogene (DHFRP1). Am J Hum Genet. 42:345–352. [PMC free article] [PubMed] [Google Scholar]
- Anderson-Trocmé L, Farouni R, Bourgey M, Kamatani Y, Higasa K, Seo J-S, Kim C, Matsuda F, Gravel S. 2020. Legacy data confound genomics studies. Mol Biol Evol. 37:2–10. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16(1):37–48. [DOI] [PubMed] [Google Scholar]
- Basu Mallick C, Iliescu FM, Möls M, Hill S, Tamang R, Chaubey G, Goto R, Ho SYW, Gallego Romero I, Crivellaro F, et al. 2013. The light skin allele of SLC24A5 in South Asians and Europeans shares identity by descent. PLoS Genet. 9:e1003912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergey CM, Lopez M, Harrison GF, Patin E, Cohen JA, Quintana-Murci L, Barreiro LB, Perry GH. 2018. Polygenic adaptation and convergent evolution on growth and cardiac genetic pathways in African and Asian rainforest hunter-gatherers. Proc Natl Acad Sci U S A. 115(48):E11256–E11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström A, McCarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, Chen Y, Felkel S, Hallast P, Kamm J, et al. 2020. Science 367(6484):eaay5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A. 1943. On a measure of divergence between two statistical populations defined by their probability distributions. Bull Calcutta Math Soc. 35:99–109. [Google Scholar]
- Biddanda A, Rice DP, Novembre J. 2020. A variant-centric perspective on geographic patterns of human allele frequency variation. eLife 9:e60107. [DOI] [PMC free article] [PubMed]
- Boettger LM, Salem RM, Handsaker RE, Peloso GM, Kathiresan S, Hirschhorn JN, McCarroll SA. 2016. Recurring exon deletions in the HP (haptoglobin) gene contribute to lower blood cholesterol levels. Nat Genet. 48(4):359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrska-Bishop M, Evani US, Zhao X, Basile AO, Abel HJ, Regier AA, Corvelo A, Clarke WE, Musunuri R, Nagulapalli K, et al. 2021. High coverage whole genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. bioRxiv. 2021.02.06.430068. doi: 10.1101/2021.02.06.430068. [DOI] [PMC free article] [PubMed]
- Candiotti KA, Birnbach DJ, Lubarsky DA, Nhuch F, Kamat A, Koch WH, Nikoloff M, Wu L, Andrews D. 2005. The impact of pharmacogenomics on postoperative nausea and vomiting: do CYP2D6 allele copy number and polymorphisms affect the success or failure of ondansetron prophylaxis? Anesthesiology 102(3):543–549. [DOI] [PubMed] [Google Scholar]
- Carvalho CMB, Lupski JR. 2016. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet. 17(4):224–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell NR, Jackson TNW, Laustsen AH, Sunagar K. 2020. Causes and consequences of snake venom variation. Trends Pharmacol Sci. 41(8):570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha S-H, Srihari SN. 2002. On measuring the distance between histograms. Pattern Recognit. 35(6):1355–1370. [Google Scholar]
- Clement M, Snell Q, Walker P, Posada D, Crandall K. 2002. TCS: estimating gene genealogies. Parallel and Distributed Processing Symposium, International. Vol. 2. New York City: IEEE. p. 184. Ft. Lauderdale, FL, USA. [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. 2010. Origins and functional impact of copy number variation in the human genome. Nature 464(7289):704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Pickrell JK, Novembre J, Kudaravalli S, Li J, Absher D, Myers RM, Cavalli-Sforza LL, Feldman MW, Pritchard JK, et al. 2009. The role of geography in human adaptation. PLoS Genet. 5(6):e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NG, Kelly DE, Hansen MEB, Beltrame MH, Fan S, Bowman SL, Jewett E, Ranciaro A, Thompson S, Lo Y, et al. 2017. Loci associated with skin pigmentation identified in African populations. Science 358(6365):eaan8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cid R, Riveira-Munoz E, Zeeuwen PLJM, Robarge J, Liao W, Dannhauser EN, Giardina E, Stuart PE, Nair R, Helms C, et al. 2009. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 41(2):211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Xu S. 2018. Adaptation of human skin color in various populations. Hereditas 155:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MY, Eichler EE. 2016. Human adaptation and evolution by segmental duplication. Curr Opin Genet Dev. 41:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, Zheng C, Alberti A, Anthony F, Aprea G, et al. 2014. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345:1181–1184. [DOI] [PubMed] [Google Scholar]
- Ding K, de Andrade M, Manolio TA, Crawford DC, Rasmussen-Torvik LJ, Ritchie MD, Denny JC, Masys DR, Jouni H, Pachecho JA, et al. 2013. Genetic variants that confer resistance to malaria are associated with red blood cell traits in African-Americans: an electronic medical record-based genome-wide association study. G3 (Bethesda) 3:1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duforet-Frebourg N, Luu K, Laval G, Bazin E, Blum MGB. 2016. Detecting genomic signatures of natural selection with principal component analysis: application to the 1000 Genomes Data. Mol Biol Evol. 33(4):1082–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge SX, Jung D, Yao R. 2020. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36(8):2628–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, Clark AG, Yu F, Gibbs RA, Bustamante CD, 1000 Genomes Project. 2011. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci U S A. 108(29):11983–11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Ban N. 2016. Structure and function of the mitochondrial ribosome. Annu Rev Biochem. 85:103–132. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium. 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller BC, Messer PW. 2019. SLiM 3: forward genetic simulations beyond the Wright–Fisher model. Mol Biol Evol. 36(3):632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Ehler E, Alkorta-Aranburu G, Beall C, Gebremedhin A, Sukernik R, Utermann G, Pritchard J, Coop G, et al. 2010. Colloquium paper: human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc Natl Acad Sci U S A. 107(Suppl 2):8924–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handsaker RE, Korn JM, Nemesh J, McCarroll SA. 2011. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet. 43(3):269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handsaker RE, Van Doren V, Berman JR, Genovese G, Kashin S, Boettger LM, McCarroll SA. 2015. Large multiallelic copy number variations in humans. Nat Genet. 47(3):296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbring SJ, Moyer AM, Weinshilboum RM. 2008. Sulfotransferase gene copy number variation: pharmacogenetics and function. Cytogenet Genome Res. 123(1–4):205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, Kelley JL, Elyashiv E, Melton SC, Auton A, McVean G, Sella G, Przeworski M, 1000 Genomes Project. 2011. Classic selective sweeps were rare in recent human evolution. Science 331(6019):920–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollox EJ, Zuccherato LW, Tucci S. 2022. Genome structural variation in human evolution. Trends Genet. 38(1):45–58. [DOI] [PubMed] [Google Scholar]
- Ho SS, Urban AE, Mills RE. 2020. Structural variation in the sequencing era. Nat Rev Genet. 21(3):171–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P, Vollger MR, Dang V, Porubsky D, Baker C, Cantsilieris S, Hoekzema K, Lewis AP, Munson KM, Sorensen M, et al. 2019. Adaptive archaic introgression of copy number variants and the discovery of previously unknown human genes. Science 366(6463):eaax2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Sánchez E, Jin X Asan, Bianba Z, Peter BM, Vinckenbosch N, Liang Y, Yi X, He M, Somel M, et al. 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512:194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Dermitzakis ET, Tyler-Smith C. 2008. The functional impact of structural variation in humans. Trends Genet. 24(5):238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key FM, Teixeira JC, de Filippo C, Andrés AM. 2014. Advantageous diversity maintained by balancing selection in humans. Curr Opin Genet Dev. 29:45–51. [DOI] [PubMed] [Google Scholar]
- Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, Malig M, Kallicki J, Kaul R, Wilson RK, Eichler EE, et al. 2010. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143(5):837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-J, Maiti P, Barrientos A. 2017. Mitochondrial ribosomes in cancer. Semin Cancer Biol. 47:67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Cho S, Yu J, Sung S, Kim H. 2010. Analysis of copy number variation in 8,842 Korean individuals reveals 39 genes associated with hepatic biomarkers AST and ALT. BMB Rep. 43(8):547–553. [DOI] [PubMed] [Google Scholar]
- Kimura M, Ohta T. 1973. The age of a neutral mutant persisting in a finite population. Genetics 75(1):199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura R, Yamaguchi T, Takeda M, Kondo O, Toma T, Haneji K, Hanihara T, Matsukusa H, Kawamura S, Maki K, et al. 2009. A common variation in EDAR is a genetic determinant of shovel-shaped incisors. Am J Hum Genet. 85(4):528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko W-Y, Rajan P, Gomez F, Scheinfeldt L, An P, Winkler CA, Froment A, Nyambo TB, Omar SA, Wambebe C, et al. 2013. Identifying Darwinian selection acting on different human APOL1 variants among diverse African populations. Am J Hum Genet. 93(1):54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg ZN, Osborne EJ, Cone KR, Kennedy BJ, Domyan ET, Shapiro MD, Elde NC, Yandell M. 2015. Wham: identifying structural variants of biological consequence. PLoS Comput Biol. 11(12):e1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JW, Bryant D. 2015. popart: full-feature software for haplotype network construction. Methods Ecol Evol. 6(9):1110–1116. [Google Scholar]
- Levina E, Bickel P. 2001. The Earth Mover’s distance is the Mallows distance: some insights from statistics. Proceedings Eighth IEEE International Conference on Computer Vision. ICCV 2001.Vol. 2. New York City: IEEE Institute of Electrical and Electronics Engineers. p. 251–256. Vancouver, BC, Canada. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-L, Gokcumen O. 2019. Fine-scale characterization of genomic structural variation in the human genome reveals adaptive and biomedically relevant hotspots. Genome Biol Evol. 11(4):1136–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-L, Pavlidis P, Karakoc E, Ajay J, Gokcumen O. 2015. The evolution and functional impact of human deletion variants shared with archaic hominin genomes. Mol Biol Evol. 32(4):1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. 2015. Structural variation mutagenesis of the human genome: impact on disease and evolution. Environ Mol Mutagen. 56(5):419–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud M, Gobet N, Cruz-Dávalos DI, Mounier N, Dessimoz C, Sedlazeck FJ. 2019. Structural variant calling: the long and the short of it. Genome Biol. 20(1):246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao M, Chennagiri N, Nordenfelt S, Tandon A, et al. 2016. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538(7624):201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak S, Perry GH. 2017. Harnessing ancient genomes to study the history of human adaptation. Nat Rev Genet. 18(11):659–674. [DOI] [PubMed] [Google Scholar]
- Martin AR, Lin M, Granka JM, Myrick JW, Liu X, Sockell A, Atkinson EG, Werely CJ, Möller M, Sandhu MS, et al. 2017. An unexpectedly complex architecture for skin pigmentation in Africans. Cell 171(6):1340–1353. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson S, Mathieson I. 2018. FADS1 and the timing of human adaptation to agriculture. Mol Biol Evol. 35(12):2957–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Hadnott TN, Perry GH. 2005. Common deletion polymorphisms in the human genome. Nature 38:86–92. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 28(5):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérot C, Oomen RA, Tigano A, Wellenreuther M. 2020. A roadmap for understanding the evolutionary significance of structural genomic variation. Trends Ecol Evol. 35(7):561–572. [DOI] [PubMed] [Google Scholar]
- Messer PW. 2013. SLiM: simulating evolution with selection and linkage. Genetics 194(4):1037–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. 2011. Mapping copy number variation by population-scale genome sequencing. Nature 470(7332):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou C, Thomason HA, Willan PM, Clowes C, Harris WE, Drew CF, Dixon J, Dixon MJ, Headon DJ. 2008. Enhanced ectodysplasin-A receptor (EDAR) signaling alters multiple fiber characteristics to produce the East Asian hair form. Hum Mutat. 29(12):1405–1411. [DOI] [PubMed] [Google Scholar]
- Mukamel RE, Handsaker RE, Sherman MA, Barton AR, Zheng Y, McCarroll SA, Loh P-R. 2021. Protein-coding repeat polymorphisms strongly shape diverse human phenotypes. Science 373(6562):1499–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton HL, Kittles RA, Parra E, McKeigue P, Mao X, Cheng K, Canfield VA, Bradley DG, McEvoy B, Shriver MD, et al. 2006. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol. 24(3):710–722. [DOI] [PubMed] [Google Scholar]
- Pajic P, Lin Y-L, Xu D, Gokcumen O. 2016. The psoriasis-associated deletion of late cornified envelope genes LCE3B and LCE3C has been maintained under balancing selection since Human Denisovan divergence. BMC Evol Biol. 16(1):265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic P, Pavlidis P, Dean K, Neznanova L, Romano R-A, Garneau D, Daugherity E, Globig A, Ruhl S, Gokcumen O, et al. 2019. Independent amylase gene copy number bursts correlate with dietary preferences in mammals. eLife 8:e44628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang AW, MacDonald JR, Pinto D, Wei J, Rafiq MA, Conrad DF, Park H, Hurles ME, Lee C, Venter JC, et al. 2010. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 11(5):R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson NJ. 2005. How old is the most recent ancestor of two copies of an allele? Genetics 169(2):1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer LM, Steranka JP, Yang WR, Kryatova M, Medabalimi S, Ardeljan D, Liu C, Boeke JD, Avramopoulos D, Burns KH, et al. 2017. Structural variants caused by Alu insertions are associated with risks for many human diseases. Proc Natl Acad Sci U S A. 114(20):E3984–E3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res. 12:2825–2830. [Google Scholar]
- Pérez-Barbería FJ, Shultz S, Dunbar RIM. 2007. Evidence for coevolution of sociality and relative brain size in three orders of mammals. Evolution 61(12):2811–2821. [DOI] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 39(10):1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley S, Louzada S, Forni D, Sironi M, Balaskas T, Hains DS, Yang F, Hollox EJ. 2015. Evolution of the rapidly mutating human salivary agglutinin gene (DMBT1) and population subsistence strategy. Proc Natl Acad Sci U S A. 112(16):5105–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyet F, Aeschbacher S, Thiéry A, Excoffier L. 2018. Background selection and biased gene conversion affect more than 95% of the human genome and bias demographic inferences. Elife 7:e36317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505(7481):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus M, Dall’Olio GM, Luisi P, Uzkudun M, Carreño-Torres A, Pavlidis P, Laayouni H, Bertranpetit J, Engelken J. 2014. 1000 Genomes Selection Browser 1.0: a genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Res. 42(Database issue):D903–D909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2012. Characterizing complex structural variation in germline and somatic genomes. Trends Genet. 28(1):43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke DW, Lee C. 2015. Adaptive potential of genomic structural variation in human and mammalian evolution. Brief Funct Genomics. 14(5):358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. 2005. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci U S A. 102(44):15942–15947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28(18):i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. 2006. Global variation in copy number in the human genome. Nature 444(7118):444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees JS, Castellano S, Andrés AM. 2020. The genomics of human local adaptation. Trends Genet. 36(6):415–428. [DOI] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PLF, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468(7327):1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. 2009. Reconstructing Indian population history. Nature 461(7263):489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, Figueroa JD, Real FX, Van Den Berg D, Matullo G, Baris D, et al. 2010. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 42(11):978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449(7164):913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Gokcumen O. 2020. An evolutionary perspective on the impact of genomic copy number variation on human health. J Mol Evol. 88(1):104–119. [DOI] [PubMed] [Google Scholar]
- Saitou M, Resendez S, Pradhan AJ, Wu F, Lie NC, Hall NJ, Zhu Q, Reinholdt L, Satta Y, Speidel L, et al. 2021. Sex-specific phenotypic effects and evolutionary history of an ancient polymorphic deletion of the human growth hormone receptor. Sci Adv. 7(39):eabi4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Satta Y, Gokcumen O. 2018. Complex haplotypes of GSTM1 gene deletions harbor signatures of a selective sweep in East Asian populations. G3 (Bethesda) 8(9):2953–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Satta Y, Gokcumen O, Ishida T. 2018. Complex evolution of the GSTM gene family involves sharing of GSTM1 deletion polymorphism in humans and chimpanzees. BMC Genomics 19(1):293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D. 2005. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 15(11):1576–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeldt LB, Tishkoff SA. 2013. Recent human adaptation: genomic approaches, interpretation and insights. Nat Rev Genet. 14(10):692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Kern AD. 2017. Soft sweeps are the dominant mode of adaptation in the human genome. Mol Biol Evol. 34(8):1863–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Navarro FCP, Galante PAF, Parmigiani RB, Camargo AA, Hahn MW, de Souza SJ. 2013. Gene copy-number polymorphism caused by retrotransposition in humans. PLoS Genet. 9(1):e1003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, et al. 2016. Schizophrenia risk from complex variation of complement component 4. Nature 530(7589):177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirugo G, Williams SM, Tishkoff SA. 2019. The missing diversity in human genetic studies. Cell 177(4):1080. [DOI] [PubMed] [Google Scholar]
- Smith GD, Lawlor DA, Timpson NJ, Baban J, Kiessling M, Day INM, Ebrahim S. 2009. Lactase persistence-related genetic variant: population substructure and health outcomes. Eur J Hum Genet. 17(3):357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Mallick S, Nelson BJ, Hormozdiari F, Krumm N, Huddleston J, Coe BP, Baker C, Nordenfelt S, Bamshad M, et al. 2015. Global diversity, population stratification, and selection of human copy-number variation. Science 349(6253):aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH-Y, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526(7571):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1993. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135(2):599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira JC, de Filippo C, Weihmann A, Meneu JR, Racimo F, Dannemann M, Nickel B, Fischer A, Halbwax M, Andre C, et al. 2015. Long-term balancing selection in LAD1 maintains a missense trans-species polymorphism in humans, chimpanzees, and bonobos. Mol Biol Evol. 32(5):1186–1196. [DOI] [PubMed] [Google Scholar]
- Tenesa A, Navarro P, Hayes BJ, Duffy DL, Clarke GM, Goddard ME, Visscher PM. 2007. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 17(4):520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4(4):e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370. [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J, Symmons O, Spitz F, Korbel JO. 2013. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet. 14(2):125–138. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2009. Ggplot2: elegant graphics for data analysis. 2nd ed. New York City: Springer Publishing Company, Incorporated. [Google Scholar]
- Wilde S, Timpson A, Kirsanow K, Kaiser E, Kayser M, Unterländer M, Hollfelder N, Potekhina ID, Schier W, Thomas MG, et al. 2014. Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proc Natl Acad Sci U S A. 111(13):4832–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongkittichote P, Ah Mew N, Chapman KA. 2017. Propionyl-CoA carboxylase – a review. Mol Genet Metab. 122(4):145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tan J, Yang Y, Peng Q, Zhang M, Li J, Lu D, Liu Y, Lou H, Feng Q, et al. 2016. Genome-wide scans reveal variants at EDAR predominantly affecting hair straightness in Han Chinese and Uyghur populations. Hum Genet. 135(11):1279–1286. [DOI] [PubMed] [Google Scholar]
- Xu D, Jaber Y, Pavlidis P, Gokcumen O. 2017. VCFtoTree: a user-friendly tool to construct locus-specific alignments and phylogenies from thousands of anthropologically relevant genome sequences. BMC Bioinformatics 18(1):426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Sun D, Daly A, Yang F, Zhou X, Zhao M, Huang N, Zerjal T, Lee C, Carter NP, et al. 2008. Adaptive evolution of UGT2B17 copy-number variation. Am J Hum Genet. 83(3):337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan SM, Sherman RM, Taylor DJ, Nair DR, Bortvin AN, Schatz MC, McCoy RC. 2021. Local adaptation and archaic introgression shape global diversity at human structural variant loci. eLife 10:e67615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H-J, Markowitz JS. 2013. Carboxylesterase 1 (CES1) genetic polymorphisms and oseltamivir activation. Eur J Clin Pharmacol. 69(3):733–734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For the data visualization, we used Rstudio (v1.2.1335), R(v3.5.3), and ggplot2 (Wickham 2009). The script underlying this article are available in https://github.com/mariesaitou/Network_humanpop_SV, and the datasets from Sudmant, Rausch, et al. (2015), can be accessed with ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/integrated_sv_map/.