Abstract
ATP7A and ATP7B, two homologous copper-transporting P1B-type ATPases, play crucial roles in cellular copper homeostasis, and mutations cause Menkes and Wilson diseases, respectively. ATP7A/B contains a P-type ATPase core consisting of a membrane transport domain and three cytoplasmic domains, the A, P, and N domains, and a unique amino terminus comprising six consecutive metal-binding domains. Here, we present a cryo–electron microscopy structure of frog ATP7B in a copper-free state. Interacting with both the A and P domains, the metal-binding domains are poised to exert copper-dependent regulation of ATP hydrolysis coupled to transmembrane copper transport. A ring of negatively charged residues lines the cytoplasmic copper entrance that is presumably gated by a conserved basic residue sitting at the center. Within the membrane, a network of copper-coordinating ligands delineates a stepwise copper transport pathway. This work provides the first glimpse into the structure and function of ATP7 proteins and facilitates understanding of disease mechanisms and development of rational therapies.
Cryo-EM reveals the structure of the Wilson disease copper transporter ATP7B.
INTRODUCTION
Ubiquitously found in all kingdoms of life, P-type adenosine triphosphatases (ATPases) use adenosine 5′-triphosphate (ATP) hydrolysis to actively pump substrates across cell membranes (1). In humans, two Cu+-transporting P-type ATPases, ATP7A and ATP7B, are key players in maintaining cellular copper homeostasis (2–4). Dysregulation of cellular copper levels is deleterious, as copper functions as an essential cofactor for numerous enzymes, while excess copper is cytotoxic (2–4). Human ATP7A and ATP7B, which share 56% sequence identity, differ in tissue expression, cellular trafficking, and physiological functions (2–4). ATP7A is important for copper uptake in the intestine, and dysfunction causes Menkes disease characterized by systemic copper deficit (5, 6). By contrast, ATP7B is most abundantly expressed in hepatocytes to excrete excess copper into bile, and consequently, missense mutations give rise to Wilson disease that is represented by hepatic copper overload and toxicity (7, 8).
The P-type ATPases, which constitute a large superfamily of membrane pumps that catalyze the transmembrane transport of diverse substrates, all contain a common structural core composed of a transmembrane domain (TMD), an actuator (A) domain, a phosphorylation (P) domain, and a nucleotide-binding (N) domain (1). Substrate transport is actively driven by conformational changes associated with phosphorylation and dephosphorylation of a conserved aspartate residue in the P domain, which result in alternating access of substrate binding sites in the TMD within the membrane (1). The stepwise transport process is described by the Post-Albers cycle with distinct functional states, including high-affinity E1 and low-affinity E2 states for substrate as well as phosphoenzyme intermediate states E1P and E2P (9, 10).
ATP7A and ATP7B belong to the prevalent P1B subfamily of heavy metal–transporting ATPases that are widespread in both prokaryotes and eukaryotes (11). Crystal structures of a prokaryotic homolog from Legionella pneumophila, LpCopA, have delineated the core structure of a P1B ATPase and suggested a potential Cu+ transport pathway across cell membranes (12, 13). The structure and ion permeation mechanism appear to differ substantially from those of the prototypical P2 ATPase subfamily exemplified by the best characterized sarcoplasmic reticulum Ca2+-ATPase (SERCA1a) (14–17). The LpCopA TMD has two additional P1B-specific transmembrane helices, a long and curved MA helix and a kinked MB helix with an amphipathic helical segment at the cytoplasmic membrane interface, that are not present in the canonical SERCA1a (12, 13). In addition to the common P-type ATPase core pieces comprising the TMD and A, P, and N domains, P1B-ATPases contain one or multiple metal-binding domains (MBDs) in the N terminus (11, 18, 19). The prokaryotic LpCopA acquires a single MBD, which was not resolved in the crystal structures (12, 13). The N terminus is elaborated in higher eukaryotic Cu+-ATPases, which contain six consecutive MBDs (MBD1 to MBD6) that receive Cu+ from a cytoplasmic Cu+ chaperone antioxidant protein 1 (ATOX1) and participate in subsequent Cu+ transport across the membrane (19–24). While MBD1 to MBD4 are not strictly required and appear to play a regulatory role, either MBD5 or MBD6 is necessary for Cu+ transport (25–27). ATP7A/B additionally contains a conserved C-terminal region absent in LpCopA that appears to communicate with the N-terminal domains to regulate copper-stimulated trafficking (28). These extra segments at both the N and C termini render ATP7A/B much larger in size than the prokaryotic counterparts and confer sophisticated Cu+-dependent regulation mechanisms of protein targeting, cellular trafficking, and post-translational modification (25–28), which remain poorly understood.
To better understand the unique features and physiological and pathological properties of ATP7 proteins, here, we describe a cryo–electron microscopy (cryo-EM) structure of ATP7B from Xenopus tropicalis determined at an overall resolution of ~3.2 Å. The structure reveals a conserved Cu+-ATPase core surrounded by the N-terminal MBDs and the C-terminal regulatory domain and delineates a plausible Cu+ transport pathway lined by a network of amino acids serving as Cu+ ligands in the membrane. Meanwhile, the first atomic model of a vertebrate Cu+-ATPase provides a molecular blueprint for understanding hundreds of missense mutations associated with devastating Menkes and Wilson diseases and facilitates rational design of therapeutic interventions.
RESULTS
Structure determination
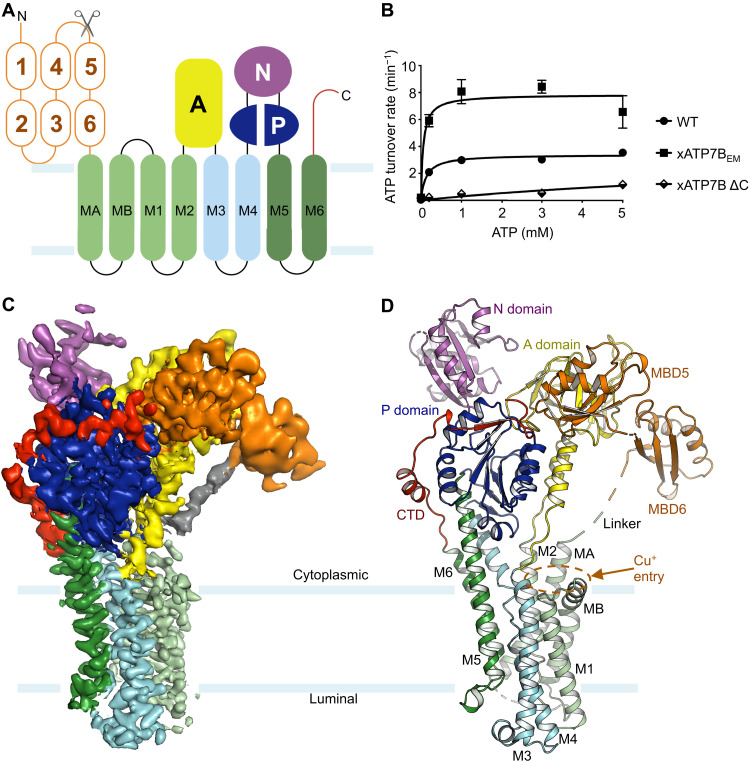
We expressed and purified the full-length X. tropicalis ATP7B protein consisting of 1467 amino acids, which shares 67% sequence identity with the human ortholog (fig. S1). We observed protein degradation likely owing to the flexible linkage of the six N-terminal MBDs. Nuclear magnetic resonance (NMR) spectroscopy studies have indicated that the individual MBDs are loosely connected like beads on a string except for MBD5 and MBD6 forming a compact unit preceding the first TM helix (21, 22, 29). Thus, for structural studies, we removed the first four of six MBD domains (MBD1 to MBD4) that are not essential for Cu+ transport (25–27, 30). The truncated construct, which we termed xATP7BEM, consists of amino acids 481 to 1467 with intact MBD5 and MBD6 and C terminus. The resulting construct has a predicted molecular weight of ~107 kDa (Fig. 1A). Consistent with previous studies (25–27), purified xATP7BEM lacking MBD1 to MBD4 retains ATPase activity (Fig. 1B). Furthermore, the ATP turnover rate of xATP7BEM (7.8 min−1) is higher than that of the full length (3.4 min−1) (Fig. 1B). This observation supports the notion that MBD1 to MBD4 negatively regulate ATPase activity before stimulation by cytoplasmic Cu+ chaperone ATOX1 (31).
Fig. 1. Cryo-EM structure of xATP7B.
(A) Membrane topology and domain organization of ATP7B. ATP7B contains six N-terminal MBDs (orange), a TMD with eight TM helices (cyan and green), and four cytoplasmic domains including A (yellow), P (blue), N (magenta), and C-terminal (red) domains. The MBD1 to MBD4 deletion construct used for cryo-EM analysis is indicated by a pair of scissors. The cyan lines mark the approximate membrane boundaries. (B) ATP turnover rates for the full-length wild type (WT; 3.4 ± 0.1 min−1), xATP7BEM (7.8 ± 0.5 min−1), and C-terminal deletion (xATP7B ΔC) using the inactive mutant (D1024A) as the baseline for subtraction. Data are means ± SD (three independent measurements). For comparison, the maximum turnover rate of LpCopA was ~11.7 min−1 (12). (C) Cryo-EM density map. The TMD, cytoplasmic domains (MBD5 and MBD6, CTD, and A, P, and N domains), and the MBD6-TMD linker are indicated. (D) Structure of xATP7BEM. The TM helices (MA, MB, and M1 to M6) and cytoplasmic domains are indicated.
We purified xATP7BEM to homogeneity and determined the cryo-EM structure at an overall resolution of ~3.2 Å (Fig. 1C, fig. S2, and table S1). The cryo-EM densities are of high quality for the TMD and A and P domains, allowing unambiguous model building (Fig. 1C and fig. S3). The densities were less well resolved for the cytoplasmic N domain, and thus the crystal structure of the prokaryotic LpCopA [Protein Data Bank identification (PDB ID): 3RFU] was used to guide model building in this region (12). Three-dimensional variability analysis (3DVA) revealed that the densities corresponding to MBD5 to MBD6, which were relatively weak in the consensus reconstruction, were present in only a subset of the particles. After 3D classification, we obtained two subsets, one in which MBD5 and MBD6 were ordered (state 1) and one in which the MBD5 and MBD6 densities were absent (state 2). To model MBD5 and MBD6, we docked the crystal structure of the isolated human MBD4 (PDB ID: 6A71) into the state 1 density map and then manually adjusted (32). Assignment of MBD5 and MBD6 was further corroborated by the visible densities for the MBD5-MBD6 and MBD6-MA linkers (Fig. 1C). The final atomic model, which was refined to good stereochemistry, fits well into the density maps (Fig. 1, C and D, fig. S3, and table S1).
Overall structure of ATP7B
The xATP7BEM structure was determined in the presence of Mg2+ and AlF4− and in the absence of the substrate ions (Cu+), thus presumably representing an occluded, Cu+-released E2-Pi transition state in the Post-Albers cycle. Mg2+ and AlF4− are bound at the conserved phosphorylation site constituted by the invariant “DKTG” motif from the P domain and the “TGE” loop from the A domain (fig. S4A), which is analogous to the observation in the crystal structure of LpCopA captured in the E2-Pi state (12). The assignment of the functional state is further supported by the global structural agreement between xATP7BEM and LpCopA (fig. S4B) (12).
The TMD comprises eight TM helices that closely resemble those of LpCopA, including the two P1B-specific helices (MA and MB) preceding the canonical six TM helices (M1 to M6) shared by P-type ATPases (Fig. 1, C and D). The first TM helix, MA, is connected to MBD6 via a partially ordered linker that is ~20 Å in length. The MB helix contains a sharp kink, enabled by two consecutive glycine residues that are conserved among Cu+-ATPases (fig. S1). Consequently, the second helical segment in MB runs approximately parallel to the membrane. A short linker follows MB and connects to the common core architecture of a P-type ATPase, which sequentially consists of M1, M2, the A domain, M3, M4, the P and N domains, M5, and M6.
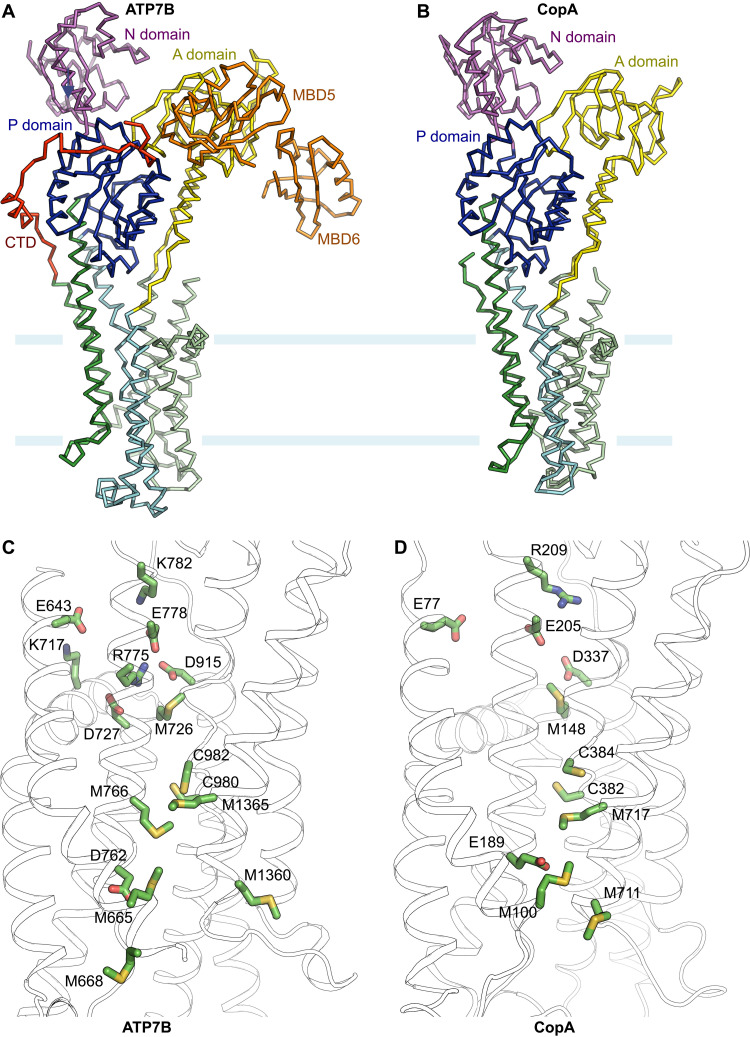
Unique to eukaryotic Cu+-ATPases, the cytoplasmic N terminus contains six MBDs, which confer exquisite regulation mechanisms (19). The cryo-EM structure reveals that the last two MBDs (MBD5 and MBD6) are distantly positioned from the lipid bilayer and pack against both the A and P domains in one state and uncoupled from the transporter in the other (Fig. 1, C and D, and fig. S2). The locations of MBD5 and MBD6, clearly resolved in the density map of state 1, differ substantially from the previously suggested positions of the single N-terminal MBD in LpCopA (12, 33). In addition, in contrast to LpCopA, eukaryotic ATP7 transporters contain a C-terminal tail following the TMD, which comprises ~90 amino acids. In the xATP7BEM structure, the C terminus emerging from M6 wraps around the P domain and reaches MBD5 (Fig. 1, C and D), thus forming an integral structural unit specific for eukaryotic ATP7 proteins. Together, it appears that both the extra N- and C-terminal regions constitute peripheral structural domains that surround the P and A domains, presumably providing further regulatory mechanisms through direct interdomain interactions (Fig. 1D).
Transmembrane Cu+ pathway
The overall arrangement of the cytoplasmic domains and, in particular, the positions of the obligatory MBDs (MBD5 or MBD6) indicate that the Cu+ entryway is most likely constituted by the cytoplasmic interface created by MA, MB, and M2 (Figs. 1, C and D, and 2A), which aligns with the proposed entry site of LpCopA (12, 13). The Cu+-free structure, together with the distal placement of MBD5 and MBD6 above the lipid bilayer, prevents visualization of how Cu+ is transferred from the MBDs to the membrane entrance. Notably, the poorly ordered linker connecting MBD6 and MA (Fig. 1C) makes no contacts with other cytoplasmic domains. Together with the MBDs being disordered in state 2, it appears that the flexible linker may allow the long-range movement of MBDs necessary for Cu+ delivery to this entry site.
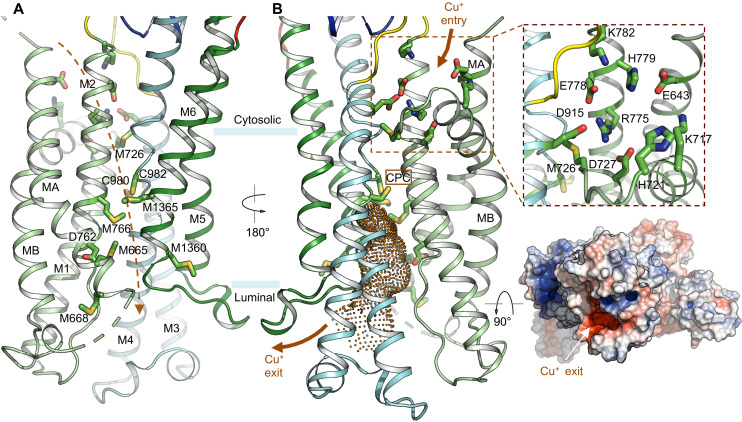
Fig. 2. Cu+ transport pathway.
(A and B) Two views of the presumed Cu+ transport pathway. Critical residues lining the Cu+ permeation path from the cytosol to the lumen are highlighted. The dashed line indicates the approximate Cu+ path in (A). The cytoplasmic Cu+ entry site is highlighted in (B). The invariant CPC motif is indicated. Pore calculation indicates that the Cu+ exit pathway is accessible from the luminal side (E2-Pi state). Surface electrostatic potential (red, −5 kT/e; white, neutral; and blue, +5 kT/e) is viewed from the luminal side.
To illuminate the potential Cu+ transport pathway across the membrane, we mapped the conserved Cu+-binding residues, including cysteine, methionine, and histidine as well as charged amino acids in the transmembrane region (Fig. 2A). These residues delineate a virtually complete route allowing Cu+ movement across the lipid bilayer from the cytoplasmic entry to the luminal exit. The presumed cytoplasmic Cu+ entry site comprises multiple conserved charged residues (Fig. 2B). A ring of negatively charged residues, including E643, D727, E778, and D915, appears to form a cation-binding site. Notably, two conserved histidine residues in the vicinity, H721 and H779, likely facilitate Cu+ entry by serving as additional Cu+ ligands. In the absence of Cu+ ions, a conserved arginine residue, R775, occupies the center of the acidic ring and seals the cytoplasmic side of the entryway. The negative electrostatic potential of the acidic ring is partially balanced by nearby lysine residues K717 and K782. Therefore, Cu+ entering this acidic binding site would likely displace the side chain of R775, which appears to be critical in stabilizing the occluded-to-the-cytoplasm conformation and thus functions as a cytoplasmic gate facilitating Cu+ occlusion. Consistent with this proposed role of R775, a single missense mutation of the corresponding residue in human ATP7B (R778L) represents one of the most prevalent of the more than 700 Wilson disease mutations that have been identified (8, 34).
M726, immediately beneath R775 in the proposed transport pathway, demarcates the cytoplasmic and intramembranous binding sites and may facilitate Cu+ movement further into the lipid bilayer (Fig. 2). In the cryo-EM structure, the Cα-Cα distances between M726 in M1 and cysteine residues (C980 and C982) in the invariant “CPC” motif in M4 are ~10 Å. Presumably, conformational changes in M1 and M4 associated with ATP hydrolysis could bring M726, C980, and C982 together to bind Cu+ through Cu-S coordination with trigonal geometry that is selective for Cu+ over Cu2+ ions (35–37). A network of Cu+-binding residues, including M1365, M766, D762, M665, M1360, and M668, lines the transmembrane ion path reaching the luminal side. These Cu+ ligands seem to be meticulously arranged to relay the bound Cu+ ions through a series of ligand exchange reactions to deliver Cu+ to the lumen. An analogous mechanism of Cu+ translocation has been suggested for the structurally unrelated CusA heavy metal efflux pump, in which a network of methionine residues lines the transmembrane path (38, 39). Estimation of the water-accessible pore dimension in the transmembrane region indicates that the permeation pathway below the CPC site is sufficiently wide for Cu+ conduction (Fig. 2B) (40). In addition, the negative electrostatic potential on the luminal side may contribute to the directionality of Cu+ movement to the lumen through electrostatic attraction (Fig. 2B). These analyses indicate that the xATP7BEM structure in the E2-Pi conformational state is occluded to the cytoplasm but open to the lumen, consistent with an open ion-release pathway suggested by crystal structures and molecular dynamics simulations of LpCopA in the E2P and E2-Pi states (12, 13).
N-terminal MBDs
Cu+-ATPases from different organisms share the core catalytic pieces but have a varying number of MBDs in the N terminus (11). The prokaryotic Cu+-ATPases contain a single MBD, and the lower eukaryotic ones have two or four, whereas the higher eukaryotic transporters comprise six consecutive MBDs. NMR studies of individual MBDs have revealed a conserved ferredoxin-like βαββαβ fold consisting of ~70 amino acids with an invariant “MxCxxC” Cu+-binding motif (11, 21, 41–43). The ATP7 transporters engage the MBDs to receive Cu+ ions from cytosolic Cu+ chaperones such as ATOX1, which shares the same βαββαβ structural fold, and to relay Cu+ ions to the intramembranous transport pathway (19–24). However, the overall structural arrangement of the MBDs in the intact transporters and the mechanisms of Cu+ delivery and Cu+-dependent regulation of transport activity via these MBDs remain unknown. Mutagenesis studies of ATP7B have demonstrated that either MBD5 or MBD6, but not MBD1 to MBD4, is required for Cu+ transport activity (25–27), suggesting that MBD1 to MBD4 play a regulatory role, whereas MBD5 or MBD6 is essential for Cu+ relay to the membrane entry site. Consistently, purified xATP7BEM lacking MBD1 to MBD4 showed increased ATPase activity compared with the full-length enzyme (Fig. 1B).
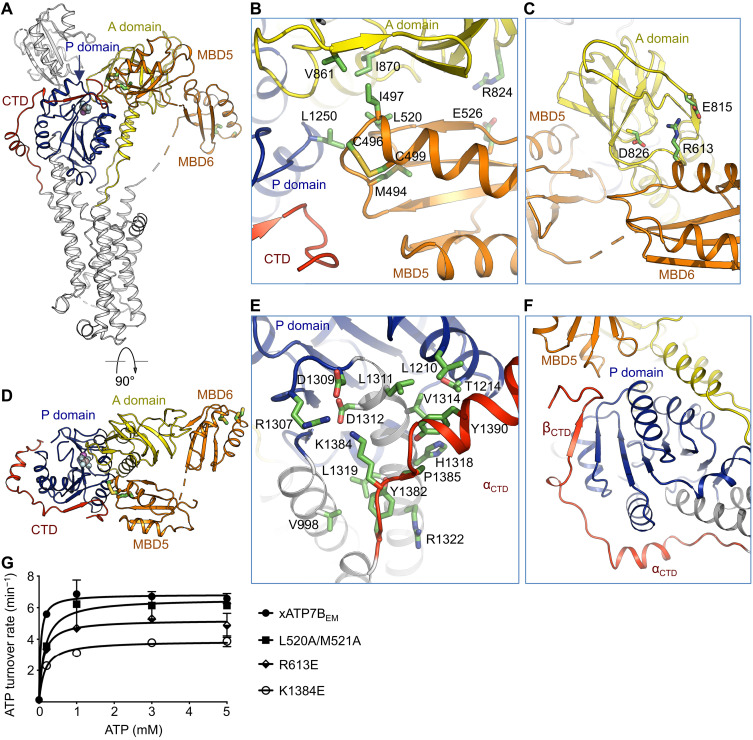
In the xATP7BEM structure, MBD5 simultaneously contacts the A, P, and C-terminal domains (Fig. 3A). Notably, the Cu+-binding motif, which consists of M494, C496, and C499, is precisely located at the multidomain junction (Fig. 3B). In the vicinity of the Cu+-binding site, MBD5 and the A and P domains are held together by a hydrophobic interface comprising I497 and L520 of MBD5, V861 and I870 of the A domain, and L1250 of the P domain. In addition, E526 of MBD5 and R824 of the A domain form salt bridges. In contrast to MBD5 making extensive interdomain interactions, MBD6 appears to be rather detached from the core pieces, only contacting the A domain via salt bridge interactions involving R613, E815, and D826 (Fig. 3, C and D).
Fig. 3. CTD and the N-terminal MBDs.
(A) Overall structure highlighting positions of the CTD and the N-terminal MBD5 and MBD6. AlF4− bound in the P domain and the presumed Cu+-binding motifs in MBD5 and MBD6 are highlighted. (B) MBD5 interacts with both the A and P domains and contacts the CTD. (C) Interface between MBD6 and the A domain. (D) Orthogonal view as in (A) to illustrate domain organization of MBD5 and MBD6, CTD, and the A and P domains. (E) The α helix in the C-terminal tail αCTD interacts with the P domain. (F) The C-terminal β strand βCTD extends the β sheet of the P domain. (G) ATP turnover rates for xATP7BEM and mutants weakening interdomain interactions. Data are means ± SD (three independent measurements).
In the P-type ATPases, conformational changes in the cytoplasmic domains (A, P, and N domains) driven by ATP hydrolysis are coupled with ion movement in the TMD (1). In the cryo-EM structure, MBD5 interacts with multiple cytoplasmic domains via its Cu+-binding motif and nearby residues. This observation indicates that MBD5 could engage Cu+-dependent regulation of structural rearrangements of the cytoplasmic domains and, consequently, Cu+ movement in the TMD. Both Cu+-binding motifs in MBD5 and MBD6 are distant from the cytoplasmic Cu+ entry point as estimated by the Cα-Cα distances between R775 and C496 (~50 Å) and between R775 and C572 (~56 Å). Therefore, if MBD5 or MBD6 participates in delivering Cu+ to the membrane entry site, long-range domain movements in MBD5 or MBD6 are expected to occur.
C-terminal tail
Following the TMD, the eukaryotic ATP7 transporters contain a C-terminal tail that is absent in prokaryotic homologs (fig. S1). The C-terminal tail forms intimate interactions with the P domain (Fig. 3, D and E). The short C-terminal helix αCTD makes multiple contacts with the P domain (Fig. 3E). Furthermore, the C-terminal tail donates an extra β strand (βCTD) that complements the β sheet of the P domain (Fig. 3F). In addition, the distal C terminus continues toward the N-terminal MBD5, joining the multidomain interface involving MBD5 and the A and P domains (Fig. 3F). Together, the C-terminal tail appears to be an integral structural component extending the cytoplasmic headpieces. Presumably, the C-terminal tail could impart additional regulatory roles specific for the eukaryotic Cu+-ATPases through direct interactions with MBD5 and the P domain.
Yeast complementation assays with ATP7B have demonstrated that the C terminus is critical for function and communicates with the N terminus as assessed by phosphoproteomics studies (28, 44), in which Cu+ binding at the N terminus resulted in hyperphosphorylation of the C-terminal sites. The xATP7BEM structure provides direct evidence that the N and C termini physically interact and thus offers a straightforward explanation for the observed long-range cross-talk between the N and C termini. To further probe the functional role of the C-terminal tail, we removed this portion beginning at K1384, and the resulting construct (xATP7BΔC) essentially eliminated the ATPase activity (Fig. 1B). In accordance with previous studies (44), our structural and functional data demonstrated the obligatory role of the C-terminal tail that is unique in eukaryotic Cu+-ATPases. In contrast, in P4-ATPases, an analogous C-terminal tail wraps around the cytosolic headpieces to confer an autoinhibitory role by blocking ATP binding and constricting domain movement (45–47).
In addition to a conserved Cu+-ATPase core, our vertebrate ATP7B structure reveals previously unknown structural elements at both the N and C termini (Fig. 3). To further validate these structural observations, we introduced mutations in the N-terminal MBD5 (L520A/M521A) and MBD6 (R613E) domains and the C-terminal tail (K1384E) on the xATP7BEM background, which presumably weaken domain-domain interactions with the catalytic core, and measured the ATPase activity (Fig. 3G). Consistently, all these mutants demonstrated a reduced ATP turnover rate, thus indicating the involvement of these structural elements in regulation of transport activity through interactions with the cytosolic headpieces. Notably, the charge reversal mutation (K1384E) in the C-terminal tail resulted in the most pronounced reduction of the ATP turnover rate.
Wilson disease mutations
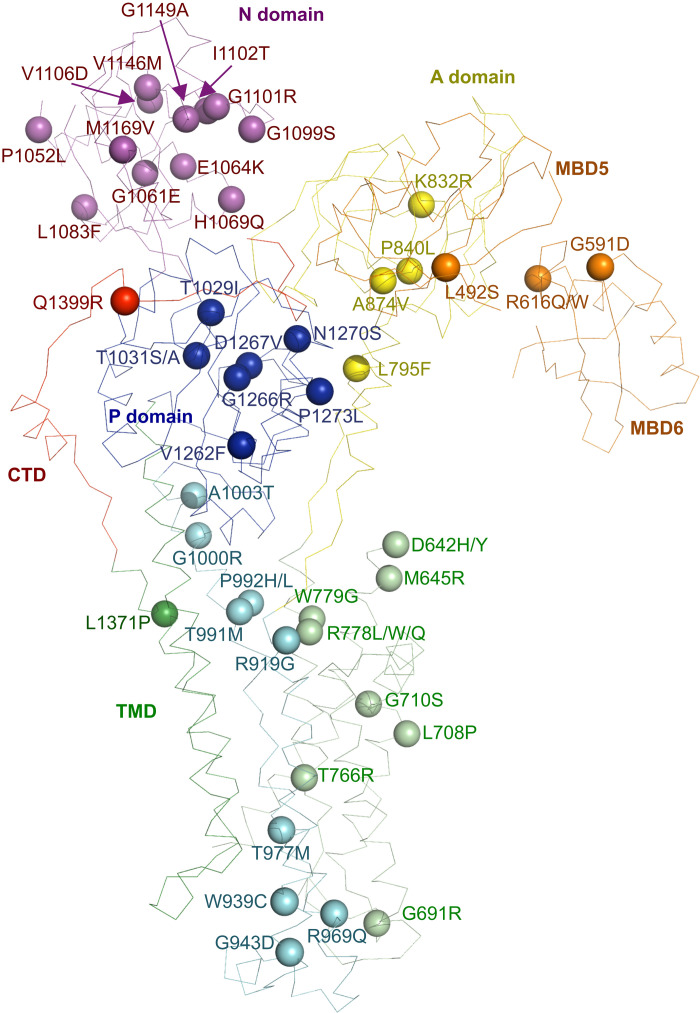
Our cryo-EM structure provides a molecular blueprint for understanding Wilson disease mutations. We mapped known missense human mutations of Wilson disease onto the cryo-EM structure of frog ATP7B and labeled these disease mutations according to the human ATP7B sequence for consistency with the Wilson disease database (Fig. 4) (48). Most of the known disease mutations are located within the structurally resolved region of the full-length protein, including the TMD and cytosolic A, P, and N domains. This reinforces the engagement of each of these domains during the transport cycle driven by ATP hydrolysis. Notably, several mutations occur in structural elements located at the N-terminal MBDs and the obligatory C-terminal tail that interact with the cytosolic headpieces. In particular, the human mutation R616Q/W (corresponding to R613 in frog ATP7B) would disrupt salt bridge interactions between MBD6 and the A domain (Fig. 3C). Another disease mutation Q1399R (equivalent to Q1405 in frog ATP7B) is located at the interface between the C-terminal tail and the P domain. These observations further strengthen the critical roles of these peripheral regulatory components.
Fig. 4. Wilson disease mutations.
The Wilson disease mutations are mapped onto the xATP7BEM structure. Annotation of disease mutations is based on the human protein sequence.
DISCUSSION
This work now reveals the first high-resolution structure of a vertebrate Cu+-ATPase, which delineates the shared and distinct features among Cu+-ATPases and provides a molecular blueprint for further understanding numerous Wilson disease mutations. The Cu+-ATPases from eukaryotes and prokaryotes share a core structure consisting of the A, P, and N domains and a P1B-specific TMD with eight TM helices (Fig. 5). In contrast to prokaryotic Cu+-ATPases, the eukaryotic counterparts contain a greater number of N-terminal MBDs and an extended C-terminal tail. These extra structural components surround the conserved cytosolic headpieces of the Cu+-ATPase apparatus and form extensive interdomain packing interactions. Both the N-terminal MBDs and C-terminal tail appear to be strategically positioned to exert additional regulatory roles by interfering with conformational changes in cytosolic headpieces, which are necessary for coupling ATP hydrolysis to TMD movements that facilitate active Cu+ transport across the membrane. Furthermore, structural organization of these domains indicates that the regulatory roles are poised to be modulated by Cu+ binding to MBDs.
Fig. 5. Structural comparison of the eukaryotic and prokaryotic Cu+-ATPases.
(A) Ribbon representation of xATP7BEM. (B) Ribbon representation of LpCopA (PDB: 3RFU). Individual domains are in the same colors as in (A). (C) Putative transmembrane Cu+ pathway in xATP7B. (D) Transmembrane Cu+ pathway in LpCopA. Conserved residues presumably involved in Cu+ coordination are highlighted.
The arrangement of TM helices is conserved between prokaryotic and eukaryotic Cu+-ATPases (root mean square deviation of the Cα atoms between eight TM helices is ~1.6 Å). Mapping of potential Cu+ ligands in the TMD, including cysteine, methionine, and charged amino acids, suggests a similar stepwise shuttle mechanism for Cu+ transport as well as an analogous “platform” entry site to accommodate the N-terminal MBDs or a cytosolic chaperone for Cu+ transfer (Fig. 5 and fig. S5) (12). Presumably, structural rearrangements, associated with Cu+ loading to the MBDs and ATPase activity in the cytosolic domains, drive unidirectional Cu+ movement from the cytosolic entry site through the intramembranous binding sites and to the luminal side. Notably, the wide exit tunnel to the lumen observed in the presumed E2-Pi state supports a unique ion-release mechanism for Cu+-ATPases as previously suggested (13). The Cu+-coordinating amino acids seem to be delicately arranged within the membrane to relay Cu+ via ligand exchange reactions. In comparison with the prokaryotic Cu+-ATPases, the eukaryotic counterparts constitute a more refined transmembrane Cu+ path that involves additional Cu+-binding amino acids such as M766 and M668 (Fig. 5). Notably, the eukaryotic ATP7 transporters have evolved to engage a conserved basic residue R775, which corresponds to glutamine at the equivalent position in LpCopA (fig. S1), as the cytoplasmic gatekeeper for Cu+ entry. Coincidently, an amino acid substitution at this position in human ATP7B (R778L) gives rise to a prevalent Wilson disease mutation (8, 34).
MATERIALS AND METHODS
Cloning, expression, and purification of ATP7B
DNA encoding X. tropicalis ATP7B was codon-optimized, synthesized (Bio Basic), and served as the template for subcloning. Deletion of the N-terminal 480 amino acids corresponding to MBD1 to MBD4 (xATP7BEM) improved biochemical stability and decreased protein degradation and thus was suitable for structural studies. DNA was polymerase chain reaction–amplified and ligated into the 5′ Xho I and 3′ Eco RI cloning sites of a modified pPICZ-B vector (Invitrogen) under a methanol-inducible promoter. The expression construct contains a C-terminal PreScission protease cleavage site linked to a green fluorescent protein (GFP)–His10 tag. The plasmid was linearized by Pme I and transformed into Pichia pastoris for expression.
Yeast cells were disrupted by milling (Retsch MM400) and resuspended in lysis buffer containing 50 mM Hepes (pH 7.0), 150 mM NaCl, 5 mM MgCl2, 0.5 mM adenosine 5′-diphosphate (ADP), 10 mM NaF, and 1 mM AlCl3 supplemented with protease inhibitors [leupeptin (2.5 μg/ml), pepstatin A (1 μg/ml), 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (100 μg/ml), aprotinin (3 μg/ml), 1 mM benzamidine, and 200 μM phenylmethylsulfonyl fluoride] and deoxyribonuclease I. Cell membranes were solubilized in 1% (w/v) lauryl maltose neopentyl glycol (Anatrace) and 0.1% cholesteryl hemisuccinate for 2 hours with stirring at 4°C. Solubilized ATP7B protein was separated from the insoluble fraction by centrifugation for 1 hour at 30,000g and incubated with 3 ml of TALON metal affinity resin (Clontech) for 3 hours at 4°C. Resin was then washed with 10 column volumes of 20 mM Hepes (pH 7.0), 150 mM NaCl, 5 mM MgCl2, 0.5 mM ADP, 10 mM NaF, 1 mM AlCl3, 10 mM imidazole, and 85 μM glyco-diosgenin (GDN; Anatrace). Bound protein was eluted with buffer containing 200 mM imidazole and further incubated with anti-GFP nanobody bound to Glutathione Sepharose 4B resin (GE Healthcare) for another 2 hours at 4°C. Unbound protein was washed away with 10 column volumes of wash buffer containing 20 mM Hepes (pH 7.0), 150 mM NaCl, 5 mM MgCl2, 0.5 mM ADP, 10 mM NaF, 1 mM AlCl3, and 85 μM GDN. The C-terminal GFP-His10 tag was removed by digestion with PreScission protease on the column at 4°C overnight. The flowthrough from the column was then concentrated using an Amicon Ultra-15 centrifugal filter unit (molecular weight cutoff 100 kDa) and injected into a Superose 6 Increase 10/300 gel filtration column (GE Healthcare) in 20 mM Hepes (pH 7.0), 150 mM NaCl, 5 mM MgCl2, 0.5 mM ADP, 10 mM NaF, 1 mM AlCl3, and 40 μM GDN. The peak fractions were collected and concentrated to ~7 mg ml−1 for cryo-EM experiments.
Cryo-EM sample preparation and imaging
Four microliters of purified xATP7BEM protein at ~7 mg ml−1 was added to Au 400 mesh Quantifoil R1.2/1.3 holey carbon grids (Quantifoil) that had been glow-discharged for 60 s. After a 20-s incubation, the grids were blotted for 2 s with a blot force of 0 and rapidly plunged into liquid nitrogen–cooled liquid ethane using a FEI Vitrobot Mark IV (FEI Thermo Fisher Scientific). A total of 3439 images were recorded in super-resolution mode at a nominal margination of 22,500× on a Titan Krios (Thermo Fisher Scientific) operating at 300 kV equipped with a K2 Summit (Gatan). The calibrated super-resolution pixel size was 0.544 Å/pixel. The dose rate was eight electrons per pixel per second for 8 s using 0.2-s frames, and images were acquired with a defocus range of −1.2 to −2.5 μm using SerialEM (49).
Image processing and map calculation
The super-resolution movies were gain-corrected, Fourier-cropped to a pixel size of 1.088 Å/pixel, and aligned using whole frame and local motion correction using MotionCor2 (50). Contrast transfer function (CTF) parameters were estimated using CTFFIND 4.1.10 (51). Particles were automatically selected using Laplacian of Gaussian autopicking in Relion 3.0 (52). False-positive selections were eliminated using iterative heterogeneous classification in cryoSPARC v3 (53) using models generated from the ab initio algorithm in cryoSPARC v3. After Bayesian polishing in Relion and global and local CTF refinement in cryoSPARC v3, the consensus refinement from 257,208 particles achieved a resolution of 3.19 Å using nonuniform refinement in cryoSPARC v3 (54). 3DVA in cryoSPARC revealed the presence of two distinct states, one in which MBD5 and MBD6 were resolved (state 1) and one in which the density for MBD5 and MBD6 was absent (state 2). Heterogeneous classification using the maps generated by 3DVA yielded a stack of 138,790 particles for state 1 and 118,418 particles for state 2, which achieved resolutions of 3.32 and 3.49 Å, respectively. All reconstructions were subsequently subjected to density modification in Phenix using a soft mask (55).
Model building and coordinate refinement
The structure of LpCopA (PDB: 3RFU) was manually docked into the density modified consensus map in UCSF Chimera (56) and then rebuilt in Coot (57). The structure of human MBD4 (PDB: 6A71) was docked in the densities corresponding to MBD5 and MDB6 in state 1 using UCSF Chimera and then rigid-body fit and manually rebuilt in Coot (32). The manually built models were refined in ISOLDE (58) and by using real-space refinement in Phenix with geometric and Ramachandran restraints (59). The refined consensus model was docked into the state 2 structure, and the missing MBDs were removed from the model before real space refinement in Phenix.
ATPase assay
All xATP7B constructs used in the ATPase assay were expressed and purified following the procedure described for xATP7BEM, except that NaF and AlCl3 were removed in the lysis and wash buffers, and the gel filtration buffer contained 20 mM Hepes (pH 7.0), 150 mM NaCl, and 40 μM GDN. Peak fractions were collected and concentrated to ~1 to 2 mg ml−1 for the ATPase assay. Concentrated protein was immediately flash-frozen and stored in −80°C until further usage. ATPase activity was measured using Bismuth detection method (12, 60). Measurements were conducted with equal amounts of protein (1 μM) for each construct. In a total reaction volume of 50 μl, protein was mixed with the ATPase assay buffer [40 mM Mops-KOH (pH 6.8), 150 mM NaCl, 5 mM KCl, 5 mM MgCl2, 20 mM (NH4)2SO4, 20 mM cysteine, 5 mM NaN3, 0.25 mM Na2MoO4, and 80 μM GDN]. Ammonium tetrathiomolybdate (TTM; 10 μM) was added to each reaction mix to eliminate free copper. An increasing amount of ATP was added to initiate the reaction, and the mixture was incubated at 37°C for 15 min. Reactions were stopped by addition of 75 μl of freshly made quench solution [2.86% ascorbic acid, 1 M HCl, 0.48% (NH4)2MoO4, and 2.86% SDS] and then incubated at room temperature for 8 min. Last, 125 μl of stabilizing solutions (3.5% bismuth citrate, 1 M HCl, and 3.5% sodium citrate) was added to the mixture and incubated for 30 min at room temperature. Absorbance was measured at 710 nm using Cytation 5 (BioTek) plate reader. The background from blank (assay buffer with TTM) was subtracted from each data point to retrieve the actual reads. The amount of inorganic phosphate released was determined by using the K2HPO4 standard as described previously (60). All measurements were performed in three replicates. The kinetic parameters were deduced from the Michaelis-Menten equation using Prism (www.graphpad.com). We have not been able to establish in vitro Cu+-stimulated ATPase activity, because the purified frog ATP7B protein was prone to aggregation with the addition of Cu+.
Acknowledgments
We thank M. de la Cruz at the MSKCC Richard Rifkind Center for Cryo-EM for assistance with data collection and the MSKCC HPC group for assistance with data processing.
Funding: This work was supported in part by NIH R01 grant NS109307 (to P.Y.), NIH-NCI Cancer Center Support Grant (P30 CA008748), the Josie Robertson Investigators Program, and the Searle Scholars Program (to R.K.H.).
Author contributions: R.M.B., Z.D., and S.R. performed biochemical preparations and ATPase assays. S.O. and R.K.H. collected cryo-EM data and determined the structure. P.Y. designed and supervised the project. All authors analyzed the results. R.K.H. and P.Y. prepared the manuscript with input from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The cryo-EM maps of xATP7BEM have been deposited to the Electron Microscopy Data Bank with accession codes EMD-25137 (consensus), EMD-25138 (state 1), and EMD-25139 (state 2). Atomic coordinates of the xATP7BEM structure have been deposited to the Protein Data Bank with accession codes 7SI3 (consensus), 7SI6 (state 1), and 7SI7 (state 2). The expression plasmids of ATP7B can be provided by Washington University pending scientific review and a completed material transfer agreement. Requests for the expression plasmids should be submitted to P.Y. (yuanp@wustl.edu).
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Table S1
REFERENCES AND NOTES
- 1.Dyla M., Kjærgaard M., Poulsen H., Nissen P., Structure and mechanism of P-type ATPase ion pumps. Annu. Rev. Biochem. 89, 583–603 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Gupta A., Lutsenko S., Human copper transporters: Mechanism, role in human diseases and therapeutic potential. Future Med. Chem. 1, 1125–1142 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim B.-E., Nevitt T., Thiele D. J., Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 4, 176–185 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Madsen E., Gitlin J. D., Copper and iron disorders of the brain. Annu. Rev. Neurosci. 30, 317–337 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Kaler S. G., Inborn errors of copper metabolism. Handb. Clin. Neurol. 113, 1745–1754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tümer Z., An overview and update of ATP7a mutations leading to menkes disease and occipital horn syndrome. Hum. Mutat. 34, 417–429 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Das S. K., Ray K., Wilson’s disease: An update. Nat. Clin. Pract. Neurol. 2, 482–493 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Lutsenko S., Modifying factors and phenotypic diversity in Wilson’s disease. Ann. N. Y. Acad. Sci. 1315, 56–63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albers R. W., Biochemical aspects of active transport. Annu. Rev. Biochem. 36, 727–756 (1967). [DOI] [PubMed] [Google Scholar]
- 10.Post R. L., Hegyvary C., Kume S., Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 247, 6530–6540 (1972). [PubMed] [Google Scholar]
- 11.Argüello J. M., Eren E., González-Guerrero M., The structure and function of heavy metal transport P1B-ATPases. BioMetals 20, 233–248 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Gourdon P., Liu X. Y., Skjørringe T., Morth J. P., Møller L. B., Pedersen B. P., Nissen P., Crystal structure of a copper-transporting PIB-type ATPase. Nature 475, 59–64 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Andersson M., Mattle D., Sitsel O., Klymchuk T., Nielsen A. M., Møller L. B., White S. H., Nissen P., Gourdon P., Copper-transporting P-type ATPases use a unique ion-release pathway. Nat. Struct. Mol. Biol. 21, 43–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toyoshima C., Nakasako M., Nomura H., Ogawa H., Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405, 647–655 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Toyoshima C., Structural aspects of ion pumping by Ca2+-ATPase of sarcoplasmic reticulum. Arch. Biochem. Biophys. 476, 3–11 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Møller J. V., Olesen C., Winther A. M. L., Nissen P., The sarcoplasmic Ca2+-ATPase: Design of a perfect chemi-osmotic pump. Q. Rev. Biophys. 43, 501–566 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Dyla M., Hansen S. B., Nissen P., Kjaergaard M., Structural dynamics of P-type ATPase ion pumps. Biochem. Soc. Trans. 47, 1247–1257 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Williams L. E., Mills R. F., P1B-ATPases–An ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 10, 491–502 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Yu C. H., Dolgova N. V., Dmitriev O. Y., Dynamics of the metal binding domains and regulation of the human copper transporters ATP7B and ATP7A. IUBMB Life 69, 226–235 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Rosenzweig A. C., Wernimont A. K., Huffman D. L., Lamb A. L., O’Halloran T. V., Structural basis for copper transfer by the metallochaperone for the Menkes/Wilson disease proteins. Nat. Struct. Biol. 7, 766–771 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Achila D., Banci L., Bertini I., Bunce J., Ciofi-Baffoni S., Huffman D. L., Structure of human Wilson protein domains 5 and 6 and their interplay with domain 4 and the copper chaperone HAH1 in copper uptake. Proc. Natl. Acad. Sci. U.S.A. 103, 5729–5734 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banci L., Bertini I., Cantini F., Massagni C., Migliardi M., Rosato A., An NMR study of the interaction of the N-terminal cytoplasmic tail of the Wilson disease protein with copper (I)-HAH1. J. Biol. Chem. 284, 9354–9360 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Granillo A., Crespo A., Estrin D. A., Wittung-Stafshede P., Copper-transfer mechanism from the human chaperone Atox1 to a metal-binding domain of Wilson disease protein. J. Phys. Chem. B 114, 3698–3706 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Pufahl R. A., Singer C. P., Peariso K. L., Lin S. J., Schmidt P. J., Fahrni C. J., Culotta V. C., Penner-Hahn J. E., O’Halloran T. V., Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science 278, 853–856 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Cater M. A., Forbes J., La Fontaine S., Cox D., Mercer J. F. B., Intracellular trafficking of the human Wilson protein: The role of the six N-terminal metal-binding sites. Biochem. J. 380, 805–813 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huster D., Lutsenko S., The distinct roles of the N-terminal copper-binding sites in regulation of catalytic activity of the Wilson’s disease protein. J. Biol. Chem. 278, 32212–32218 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Guo Y., Nyasae L., Braiterman L. T., Hubbard A. L., NH2-terminal signals in ATP7B Cu-ATPase mediate its Cu-dependent anterograde traffic in polarized hepatic cells. Am. J. Physiol. - Gastrointest. Liver Physiol. 289, G904–G916 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Braiterman L. T., Gupta A., Chaerkady R., Cole R. N., Hubbard A. L., Communication between the N and C termini is required for copper-stimulated Ser/Thr phosphorylation of Cu(I)-ATPase (ATP7B). J. Biol. Chem. 290, 8803–8819 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fatemi N., Korzhnev D. M., Velyvis A., Sarkar B., Forman-Kay J. D., NMR characterization of copper-binding domains 4-6 of ATP7B. Biochemistry 49, 8468–8477 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Jayakanthan S., Braiterman L. T., Hasan N. M., Unger V. M., Lutsenko S., Human copper transporter ATP7B (Wilson disease protein) forms stable dimers in vitro and in cells. J. Biol. Chem. 292, 18760–18774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu C. H., Yang N., Bothe J., Tonelli M., Nokhrin S., Dolgova N. V., Braiterman L., Lutsenko S., Dmitriev O. Y., The metal chaperone Atox1 regulates the activity of the human copper transporter ATP7B by modulating domain dynamics. J. Biol. Chem. 292, 18169–18177 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang T., Chen W., Sheng Y., Yuan S., Tang Q., Li G., Huang G., Su J., Zhang X., Zang J., Liu Y., Tetrathiomolybdate induces dimerization of the metal-binding domain of ATPase and inhibits platination of the protein. Nat. Commun. 10, 186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C. C., Rice W. J., Stokes D. L., Structure of a copper pump suggests a regulatory role for its metal-binding domain. Structure 16, 976–985 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Członkowska A., Litwin T., Dusek P., Ferenci P., Lutsenko S., Medici V., Rybakowski J. K., Weiss K. H., Schilsky M. L., Wilson disease. Nat. Rev. Dis. Prim. 4, 21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su C.-C., Long F., Zimmermann M. T., Rajashankar K. R., Jernigan R. L., Yu E. W., Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature 470, 558–562 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubino J. T., Franz K. J., Coordination chemistry of copper proteins: How nature handles a toxic cargo for essential function. J. Inorg. Biochem. 107, 129–143 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Ren F., Logeman B. L., Zhang X., Liu Y., Thiele D. J., Yuan P., X-ray structures of the high-affinity copper transporter Ctr1. Nat. Commun. 10, 1386 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long F., Su C. C., Zimmermann M. T., Boyken S. E., Rajashankar K. R., Jernigan R. L., Yu E. W., Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467, 484–488 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moseng M. A., Lyu M., Pipatpolkai T., Glaza P., Emerson C. C., Stewart P. L., Stansfeld P. J., Yu E. W., Cryo-EM structures of CusA reveal a mechanism of metal-ion export. MBio 12, e00452-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Gitschier J., Moffat B., Reilly D., Wood W. I., Fairbrother W. J., Solution structure of the fourth metal-binding domain from the Menkes copper-transporting ATPase. Nat. Struct. Biol. 5, 47–54 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Banci L., Bertini I., Del Conte R., D’Onofrio M., Rosato A., Solution structure and backbone dynamics of the Cu(I) and apo forms of the second metal-binding domain of the menkes protein ATP7A. Biochemistry 43, 3396–3403 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Banci L., Bertini I., Cantini F., Rosenzweig A. C., Yatsunyk L. A., Metal binding domains 3 and 4 of the wilson disease protein: Solution structure and interaction with the copper(I) chaperone HAH1. Biochemistry 47, 7423–7429 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsi G., Cullen L. M., Glerum D. M., Cox D. W., Functional assessment of the carboxy-terminus of the Wilson disease copper-transporting ATPase, ATP7B. Genomics 83, 473–481 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Hiraizumi M., Yamashita K., Nishizawa T., Nureki O., Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Science 365, 1149–1155 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Timcenko M., Lyons J. A., Januliene D., Ulstrup J. J., Dieudonné T., Montigny C., Ash M. R., Karlsen J. L., Boesen T., Kühlbrandt W., Lenoir G., Moeller A., Nissen P., Structure and autoregulation of a P4-ATPase lipid flippase. Nature 571, 366–370 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Bai L., Kovach A., You Q., Hsu H.-C., Zhao G., Li H., Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p. Nat. Commun. 10, 4142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar M., Gaharwar U., Paul S., Poojary M., Pandhare K., Scaria V., BK B., WilsonGen a comprehensive clinically annotated genomic variant resource for Wilson’s Disease. Sci. Rep. 10, 9037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheres S. H. W., Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 579, 125–157 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Punjani A., Zhang H., Fleet D. J., Non-uniform refinement: Adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Terwilliger T. C., Sobolev O. V., Afonine P. V., Adams P. D., Read R. J., Density modification of cryo-EM maps. Acta Crystallogr. Sect. D Struct. Biol. 76, 912–925 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Croll T. I., ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. Sect. D Struct. Biol. 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX : A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cariani L., Thomas L., Brito J., Del Castillo J. R., Bismuth citrate in the quantification of inorganic phosphate and its utility in the determination of membrane-bound phosphatases. Anal. Biochem. 324, 79–83 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Table S1