Abstract
Circulating Ly6Chi monocytes often undergo cellular death upon exhaustion of their antibacterial effector functions, which limits their capacity for subsequent macrophage differentiation. This shrouds the understanding on how the host replaces the tissue-resident macrophage niche effectively during bacterial invasion to avert infection morbidity. Here, we show that proliferating transitional premonocytes (TpMos), an immediate precursor of mature Ly6Chi monocytes (MatMos), were mobilized into the periphery in response to acute bacterial infection and sepsis. TpMos were less susceptible to apoptosis and served as the main source of macrophage replenishment when MatMos were vulnerable toward bacteria-induced cellular death. Furthermore, TpMo and its derived macrophages contributed to host defense by balancing the proinflammatory cytokine response of MatMos. Consequently, adoptive transfer of TpMos improved the survival outcome of lethal sepsis. Our findings hence highlight a protective role for TpMos during bacterial infections and their contribution toward monocyte-derived macrophage heterogeneity in distinct disease outcomes.
TpMos serve as a source for the rapid replenishment of functionally distinct macrophages after bacteria-induced macrophage loss.
INTRODUCTION
Tissue-resident macrophages (TRMs) are predominantly derived from embryonic progenitors before birth and, in certain tissues, are able to maintain themselves with minimal monocyte input under the steady state (1–3). Because of their tissue residence, TRMs are poised to act as early immune sentinels in detecting and controlling invading microbes (4, 5). However, it is also increasingly clear that TRMs are prone to cellular death through pyroptosis or necroptosis in response to pathogen challenge (6–10). While a temporary depletion of the TRM niche may not be detrimental in the steady state, the lack of TRMs in the presence of pathogenic bacteria results in a gap in host defense and predisposes the host toward increased morbidity (11–13). Therefore, it is critical that circulating monocytes are recruited rapidly to occupy the niche and reinstate the peripheral immune response.
Monocytes are short-lived cells that are derived from a committed monocyte progenitor known as the common monocyte progenitor (cMoP) in the bone marrow (BM) (14, 15). cMoPs then undergo a transitional developmental stage to become transitional premonocytes (TpMos) before differentiating into mature classical Ly6Chi monocytes (MatMos) (16). During bacterial invasion, MatMos are recruited into the periphery and are required to perform two mutually exclusive functions (17). To control an infection, MatMos perform effector functions through production of cytokines, reactive oxidative species, and phagocytosis of bacteria. At the same time, monocytes are also required to differentiate into monocyte-derived macrophages (MDMs) to repopulate the empty niche vacated by dying TRMs. However, active monocyte differentiation is incompatible with simultaneous effector function as monocytes often undergo phagocytosis-induced cell death, a process that involves apoptosis after phagocytosis of bacteria and exhaustion of their antibacterial functions (18, 19). Therefore, it remains unclear whether the host has evolved strategies to balance the need of monocytes for immunosurveillance versus the replenishment of the TRM niche to sustain immune defense against invading bacteria.
Here, we show that in response to acute bacterial infection and sepsis, a distinct subpopulation of Ly6Chi monocytes, which we identify as TpMos (16), appeared in the periphery and served as an important source for macrophage replenishment when MatMos were vulnerable toward bacteria-induced cellular death. TpMos and their derived macrophages provided protection against sepsis by balancing the highly proinflammatory cytokine response of MatMos that contribute toward the cytokine storm in sepsis. Together, our results highlight a specialization of monocytic roles against bacterial invasion to balance the need for monocyte effector function versus macrophage replenishment to protect the host from infection morbidity.
RESULTS
A distinct proliferative subset of Ly6Chi monocytes emerges in the blood in response to acute bacterial infection and sepsis
Monocytes in the circulation are short-lived and have been established to be nonproliferative cells (16, 20, 21). Nevertheless, to repopulate an empty niche left behind by dying TRMs, monocytes are able to enter the cell cycle through local cues in the periphery (22, 23). However, this process requires monocytes to first infiltrate peripheral tissues to receive instructive cues for their proliferation and differentiation into macrophages. Since this process must be rapidly deployed during pathogenic infections and monocytes are susceptible to apoptosis upon exposure to bacteria (18, 19), it remains unclear whether the host has evolved other immune mechanisms to hasten this repopulation process. Specifically, it is unknown whether circulating Ly6Chi monocytes can be “remotely programmed” to proliferate before entering the tissue to provide a source of cells that can rapidly repopulate the niche. To address this question, we established an acute bacterial infection model that mimics bacterial peritonitis caused by Escherichia coli (24, 25) to determine how bacteria might influence the macrophages in the peritoneal niche and affect the subsequent monocyte response.
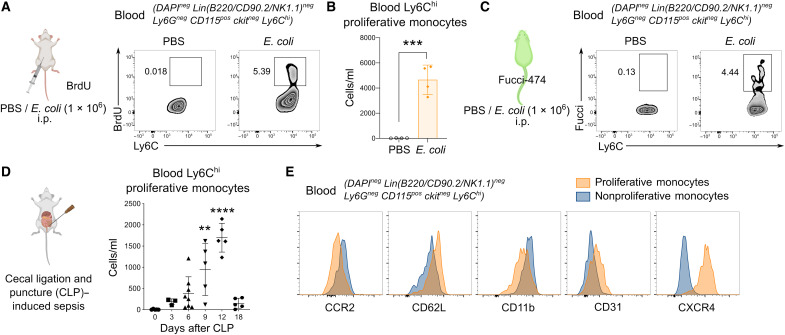
Upon E. coli infection, we noticed a population of proliferative Ly6Chi monocytes in the circulation that stained positive for bromodeoxyuridine (BrdU) (Fig. 1, A and B, and fig. S1), a thymidine analog that labels proliferating cells in the S phase. Administration of E. coli in Fucci-expressing mice, whereby cells in the S-G2-M phase of the cell cycle are labeled positive for green fluorescent protein (GFP) (26), further confirmed that these Ly6Chi monocytes were actively cycling in the circulation (Fig. 1C). Since bacterial peritonitis can often lead to sepsis and subsequent mortality, we also asked whether these proliferative Ly6Chi monocytes were present in the circulation during bacterial sepsis. To address this question, we used a model of sepsis using the cecal ligation and puncture (CLP) method (27), whereby feces from the cecum were expelled into the peritoneum, mimicking peritonitis and subsequent sepsis. Analysis of septic mice revealed a notable presence of proliferative Ly6Chi monocytes in the circulation that started to emerge at around day 6 of sepsis, peaked in numbers at day 12 after CLP, and disappeared from the blood upon resolution of the infection at day 18 (Fig. 1D). We further confirmed that these proliferative Ly6Chi monocytes are a distinct subset of cells that differed from nonproliferative Ly6Chi monocytes based on their phenotypic markers being CXCR4hi, CCR2lo, CD62Lhi, CD11blo, and CD31hi (Fig. 1E). Together, our study identifies the presence of a distinct subset of Ly6Chi monocytes that exhibits proliferative activity in the blood upon bacterial infection and sepsis.
Fig. 1. A distinct proliferative subset of Ly6Chi monocytes emerges in the peripheral blood during bacterial infection and sepsis.
(A to C) Proliferative Ly6Chi monocytes in the blood assessed by BrdU incorporation in vivo (A) and numbers quantified (B) or Fucci-474 mice (C) were administered E. coli or phosphate-buffered saline (PBS) intraperitoneally (i.p.) for 9 hours. Numbers in fluorescence-activated cell sorting (FACS) plots represent the percentage of positive cells. Results are expressed as means ± SD (n = 4) and representative of one of three experiments. ***P < 0.001 (Student’s t test). (D and E) Mice were subjected to CLP-induced sepsis, and proliferative Ly6Chi monocytes in the blood were quantified on the basis of BrdU incorporation at indicated time points (D). Results are expressed as means ± SD (n = 5 per group) and representative of one of three experiments. **P < 0.01, and ****P < 0.0001 [one-way analysis of variance (ANOVA)] compared to day 0. (E) Overlay of surface markers of blood proliferative versus nonproliferative Ly6Chi monocytes on day 12 of CLP-induced sepsis.
Emergence of proliferative Ly6Chi monocytes in the blood was CCR2 independent and occurred after bacteria-induced TRM loss
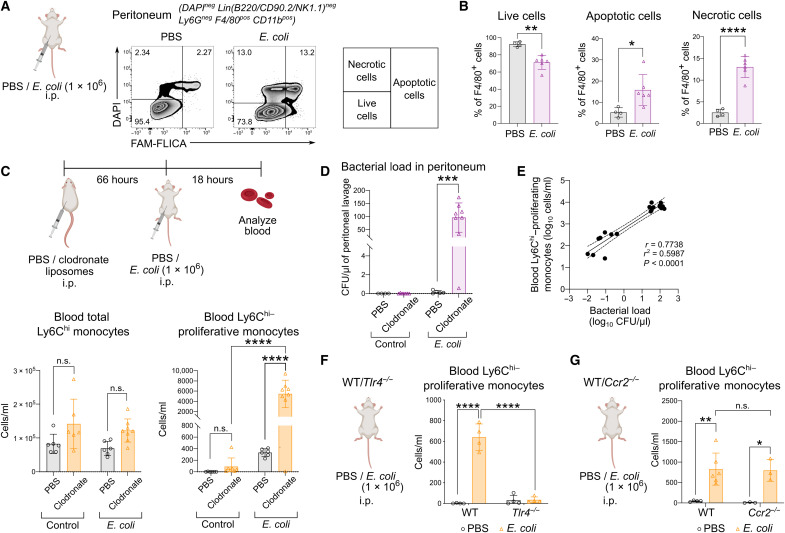
Since nonproliferative blood Ly6Chi monocytes were not detected in the steady state (Fig. 1, A and B), we next investigated the factors that might induce their presence in the circulation during infection. Analysis of the peritoneum compartment revealed increased apoptotic and necrotic peritoneal macrophages in E. coli–infected mice compared to the control (Fig. 2, A and B, and fig. S2). On the basis of these results, we hypothesized that the proliferative Ly6Chi monocytes in the circulation occurred in response to diminishing peripheral macrophages in the peritoneal niche. To examine this hypothesis, we depleted tissue macrophages in the peritoneal cavity using clodronate liposomes with a protocol that does not affect blood monocytes as described by others (Fig. 2C) (5, 28). Our results revealed that blood Ly6Chi–proliferative monocytes were not detected in the circulation after macrophage depletion alone (Fig. 2C). However, with both macrophage depletion followed by E. coli infection, Ly6Chi-proliferative monocytes in the circulation increased markedly in numbers, with E. coli–infected mice registering a 10-fold increase (Fig. 2C). In contrast, the number of total circulating Ly6Chi monocytes did not differ significantly regardless of macrophage depletion during E. coli infection (Fig. 2C). While these results demonstrated a reciprocal correlation between Ly6Chi-proliferative monocytes in the blood and a diminishing peripheral macrophage niche, these cells were detected only when the loss of macrophages occurred in E. coli infection (Fig. 2C), therefore suggesting that bacterial infection critically induces the emergence of blood Ly6Chi–proliferative monocytes. Since the absence of tissue macrophages would have lowered the clearance of bacteria in the periphery, we next determined whether there was a correlation between Ly6Chi-proliferative monocytes in the blood and an increase in bacterial burden. The increase in the number of blood Ly6Chi–proliferative monocytes after macrophage depletion was proportionate to the expansion of E. coli load in infected mice (Fig. 2, D and E). We further confirmed the bacterial signal as an important trigger for the emergence of blood Ly6Chi–proliferative monocytes as the presence of these cells was abolished in Tlr4−/− mice (Fig. 2F). However, in contrast to the current paradigm that activation of Toll-like receptor 4 (TLR4)–signaling triggers CCR2-dependent mobilization of Ly6Chi monocytes (29, 30), the presence of proliferative Ly6Chi monocytes was independent of CCR2 signaling as these cells were still present in similar numbers in both Ccr2−/− and wild-type (WT) mice (Fig. 2G). Together, we have identified a feedback loop that amplifies the emergence of Ly6Chi-proliferative monocytes in the blood in response to a surge in bacterial burden caused by a diminishing peripheral macrophage niche during infection.
Fig. 2. Emergence of Ly6Chi-proliferative monocytes was CCR2 independent and occurred after bacteria-induced TRM loss.
(A and B) Peritoneal macrophages of mice infected with E. coli or PBS were analyzed for live, apoptotic, and necrotic cells using FAM-FLICA and 4′,6-diamidino-2-phenylindole (DAPI) (A) and their percentage quantified (B) after 18 hours of infection. Numbers in FACS plots represent the percentage of positive cells. Results are expressed as means ± SD (n = 5) and representative of one of two experiments. *P < 0.05, **P < 0.01, and ****P < 0.0001 (Student’s t test). (C to E) Mice were administered clodronate or PBS liposomes intraperitoneally before infection with or without E. coli as indicated. (C) Total Ly6Chi monocytes (bottom left) and proliferative Ly6Chi monocytes (bottom right) in the blood were quantified. Results are expressed as means ± SD (n = 6) and representative of one of three experiments. n.s., not significant; ****P < 0.0001 (one-way ANOVA). (D) Quantification of the peritoneal bacterial load was quantified. Results are expressed as means ± SD (n = 6) and representative of one of three experiments. ***P < 0.001 (Student’s t test). (E) Correlation graph of blood proliferative Ly6Chi monocytes versus bacterial load. (F and G) WT and Tlr4−/− (F) or WT and Ccr2−/− mice (G) were administered E. coli or PBS intraperitoneally, and the blood was analyzed for proliferative Ly6Chi monocytes using BrdU incorporation in vivo after 18 hours. Results are expressed as means ± SD (n = 3 to 6) and representative of one of three experiments. *P < 0.05, **P < 0.01, and ****P < 0.0001 (one-way ANOVA).
Blood proliferative Ly6Chi monocytes were TpMos that have been mobilized from the BM
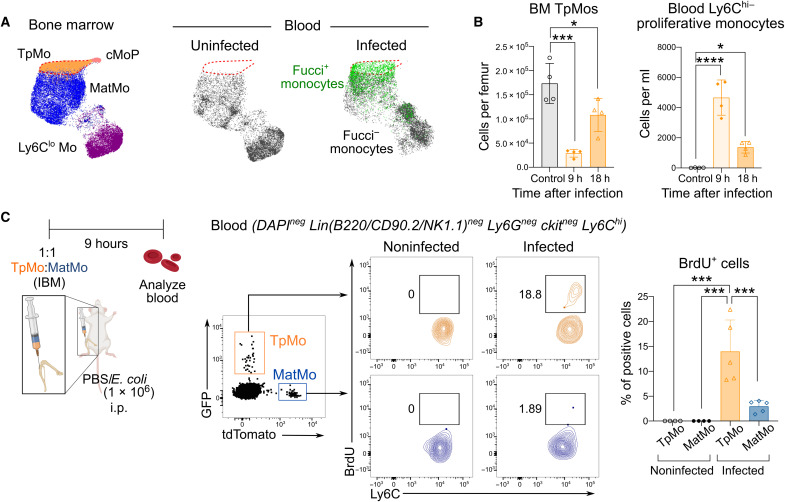
Since blood Ly6Chi–proliferative monocytes expressed a distinct repertoire of surface makers and were mobilized in a CCR2-independent manner, we next sought to investigate the origin of these cells. Phenotypic mapping of blood Ly6Chi–proliferative monocytes to BM monocytes and their progenitors revealed that these blood proliferative (Fucci+) Ly6Chi monocytes phenocopied TpMos that are found in the BM (Fig. 3A). Notably, TpMos are the immediate precursors of MatMos and have been previously shown to be in a constitutive proliferative state where they serve as a reservoir for the replenishment of MatMos in the BM (16). Analysis of the BM and blood after E. coli infection (Fig. 3B) revealed that the appearance of Ly6Chi-proliferative monocytes in the circulation corresponded reciprocally to a decrease in the number of TpMos in the BM, suggesting that BM TpMos could have been released into the circulation. To provide definitive proof that blood proliferative Ly6Chi monocytes were TpMos that have exited the BM into the circulation, we performed an adoptive cotransfer of fluorescent tagged TpMos (GFP-tagged) and MatMos (tdTomato-tagged) into the femoral BMs of recipient mice to track their egress from the BM into the circulation under the same experimental conditions. We demonstrated that in the absence of infection, both adoptively transferred TpMos and MatMos appeared in the circulation and were negative for BrdU staining, indicating that they egressed as nonproliferative cells (Fig. 3C). These findings are in line with our previous study whereby TpMos were shown to exit the cell cycle and differentiate into MatMos before entering the circulation in the steady state (16). During E. coli infection, both transferred TpMos and MatMos were also found to have egressed from the BM into the circulation (Fig. 3C). However, upon gating of these cells, only TpMos stained positively for BrdU (Fig. 3C). In contrast, MatMos egressed from the BM and appeared as nonproliferative cells, therefore suggesting that bacterial infection or TLR4 signaling does not cause the expansion/proliferation of circulating monocytes during infection (Fig. 3C). Hence, these results show that blood Ly6Chi–proliferative monocytes that emerged during E. coli infection were TpMos that have exited the BM into circulation. Our results hence highlight an atypical host response initiated by invading bacteria that triggered the mobilization of a monocyte precursor in the event of a compromised peripheral macrophage niche.
Fig. 3. Blood Ly6Chi–proliferating monocytes are TpMos that have been mobilized from the BM.
(A) Uniform Manifold Approximation and Projection (UMAP) analysis was performed on total monocytes from uninfected BM cells (left) total uninfected blood cells (middle), and infected blood cells (right). Parameters used for UMAP projection include Ly6C, CXCR4, CD49f, CD115, cKit, CD43, CX3CR1, and CD48. Monocyte subsets, including proliferative (Fucci+) Ly6Chi monocytes, were then manually gated and overlaid onto the UMAP space. (B) BM TpMos (left) and blood Ly6Chi–proliferative monocytes (right) were quantified after E. coli infection at indicated time points. Results are expressed as means ± SD (n = 5) and representative of one of two experiments. *P < 0.05, ***P < 0.001, and ****P < 0.0001 (one-way ANOVA). (C) GFP-tagged TpMos and tdTomato-tagged MatMos were resuspended in a 1:1 ratio and adoptively transferred as a single injection via the intrafemoral route [intra-BM (IBM)] into the femurs of donor mice. Donor mice were subsequently infected with or without E. coli and analyzed for GFP-tagged TpMos and tdTomato-tagged MatMos in the blood after 9 hours (left). These cells were subsequently gated and examined for BrdU incorporation in vivo (middle). Results (right) are expressed as means ± SD (n = 4 to 8 per group) and representative of one of three experiments. ***P < 0.001 (one-way ANOVA).
TpMos express effector genes distinct from MatMos in response to sepsis
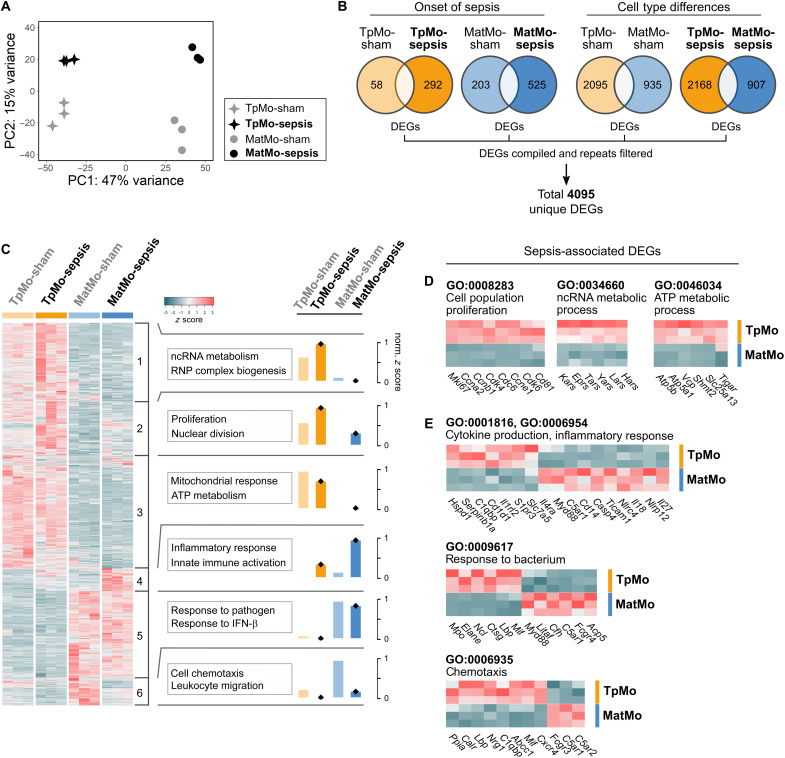
Since TpMos are absent in the circulation in the steady state (16), it is pertinent to ask how their recruitment into the periphery may result in differing functions from MatMos during sepsis. To address this question, we sorted these cells and compared their transcriptomic signatures via bulk RNA sequencing (RNA-seq). Principal components analysis (PCA) of TpMos and MatMos revealed that while sepsis altered both their transcriptomic signatures, they first clustered separately according to developmental stage (PC1, TpMo, or MatMo) before segregating according to whether sepsis was induced (PC2, sham, or sepsis) (Fig. 4A). As TpMos and MatMos were found to retain a substantial amount of their phenotype from each other in these settings, we next assessed how these cells may function differently during sepsis. To this end, we compiled the differentially expressed genes (DEGs) of TpMos and MatMos during sham and sepsis conditions and derived 4095 unique DEGs (Fig. 4B). We next performed unsupervised k-means clustering on these DEGs and identified six transcriptional clusters (Fig. 4C). Analyses of Gene Ontology (GO) revealed that TpMos were characterized by genes enriched in processes of noncoding RNA (ncRNA) metabolism, cell proliferation, and adenosine triphosphate (ATP) metabolism (clusters 1 to 3). On the contrary, MatMos were distinguished by enriched genes involved in immune inflammatory responses, pathogen response, and chemotaxis (clusters 4 to 6) (Fig. 4C). Analysis of genes revealed that TpMos retained their higher proliferative capacity compared to MatMos during sepsis based on up-regulation of Mki67 and several other cell proliferation markers (Fig. 4D). TpMos also displayed a distinct immunometabolic profile from up-regulated genes annotated to ncRNA- (GO:0034660) and ATP-metabolic (GO: 0046034) processes in comparison to MatMos (Fig. 4D). For example, TpMos were characterized by aminoacyl-tRNA synthetase genes such as Kars and Lars, which have been shown to be important for innate immune responses and regulation of cellular energetics via mammalian target of rapamycin 1 signaling, respectively (31, 32). In addition, TpMos also up-regulated Atp5a1 and Tigar genes (Fig. 4D) that have been described in the reprogramming of mitochondria activity and energy metabolism regulation to support efficient immune responses (33, 34). These findings hence suggest a metabolically robust phenotype for TpMos that may contribute toward enhanced responses against infections compared to MatMos.
Fig. 4. TpMos express distinct effector genes from MatMos in response to sepsis.
(A to E) TpMos and MatMos from sham and CLP-induced sepsis conditions on day 6 were sorted from the BM. (A) PCA of bulk RNA-seq data for BM TpMo and MatMo subsets across sham and sepsis conditions; TpMo-sepsis (black star, top left), MatMo-sepsis (black circle, top right), TpMo-sham (gray star, bottom left), and MatMo-sham (gray circle, bottom right). (B) Venn diagrams representing DEGs (up/down) between BM TpMo and MatMo subsets across sham and sepsis conditions; TpMo-sham (pale orange), TpMo-sepsis (orange), MatMo-sham (pale blue), and MatMo-sepsis (blue). All DEGs identified across four conditions were merged, and repetitive genes were excluded to obtain 4095 unique DEGs for gene expression analysis. (C) Heatmap of unique DEGs expression across subsets represented as z score. Gene clusters (1 to 6) were obtained by unsupervised k-means clustering and subjected to GO biological process enrichment analysis. Functional characterization of gene clusters was based on the top five GO terms obtained from analysis. Functional scores for each gene cluster across conditions were represented as normalized z score. RNP, ribonucleoprotein. (D) Cell population proliferation, ncRNA metabolic process, and ATP metabolic process (left to right) related gene expression between BM TpMo and MatMo subsets during sepsis. (E) Cytokine production/inflammatory response, response to bacterium, and chemotaxis (top to bottom) related gene expression between BM TpMo and MatMo subsets during sepsis.
We next investigated the inflammatory-related genes in TpMos compared to MatMos. MatMos were found to positively regulate inflammatory pathways via activation of the proinflammatory response and inflammasome-activated interleukin-1β (IL-1β) production, as annotated in the inflammatory response (GO:0006954) and cytokine production (GO:0001816) pathways (Fig. 4E). In contrast to MatMos, TpMos up-regulated genes such as Cd1d1, Il1rl2, and Serpinb1a that are either anti-inflammatory or are involved in dampening IL-1β production (35, 36). These results suggest that TpMos may have an attenuated inflammatory phenotype compared to MatMos. TpMos and MatMos also up-regulated distinct sets of genes in response to bacterium (GO:0009617) pathway (Fig. 4E). While MatMos up-regulated genes mainly associated with complement and bacterial sensing such as C5ar1, Cfh, and Myd88 (37, 38), TpMos up-regulated antimicrobial genes that involved cytotoxic immune serine proteases such as Ctsg, Elane, and Mpo (Fig. 4E), which are associated with direct killing of bacteria (39, 40). Last, TpMos and MatMos were also found to enrich differing genes in the chemotaxis (GO:0006935) pathway, with TpMos up-regulating genes such as Cxcr4, Mif, and Nrg1, and MatMos up-regulated genes such as C5ar1 and C5ar2 (Fig. 4E). Together, our results reveal distinct metabolic and effector transcriptional signatures between TpMos and MatMos during sepsis, suggesting that the direct mobilization of TpMos into the circulation may provide distinct effector responses in the periphery.
TpMos surpass MatMos in replenishing the macrophage niche
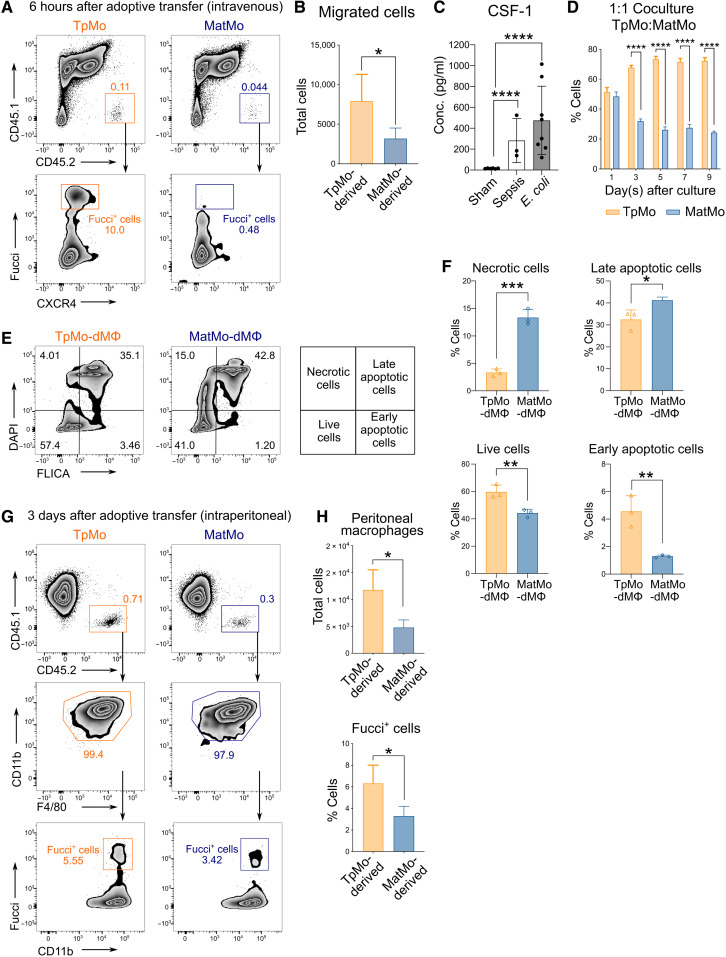
Our current results suggest that TpMos are transcriptomically distinct from MatMos during sepsis (Fig. 4). However, as immediate precursors of MatMos, it is unclear whether mobilized TpMos are merely replenishing their mature counterparts in the periphery or they have distinct effector functions that may modify the overall immune response. To seek clarity, we first evaluated the migratory properties of TpMos and MatMos by comparing their recruitment competency into the infection site. TpMos were found to down-regulate CXCR4 when they enter the peritoneum and were therefore challenging to trace upon their migration. To mitigate this issue, we adoptively transferred Fucci (CD45.2+) BM TpMos or MatMos intravenously into CD45.1 recipient septic mice. TpMos were present within the peritoneum 6 hours after intravenous transfer as detected by their Fucci+ signal (Fig. 5, A and B). Hence, these results suggest that TpMos are not inferior in their migratory properties compared to MatMos during inflammation.
Fig. 5. TpMos are more competent than MatMos in replenishing the macrophage niche.
(A and B) TpMos and MatMos from Fucci-474 CD45.2 mice were sorted and adoptively transferred intravenously into CLP-induced CD45.1 recipient mice. Analysis of peritoneal lavage (PL) of recipient mice for transferred cells was performed 6 hours later (A), and total migrated cells into peritoneum was quantified (B). Results are expressed as means ± SD (n = 4 to 6) and representative of one of three experiments. *P < 0.05 (Student’s t test). (C) Concentration of CSF-1 in the PL of mice that have undergone sham surgery versus CLP-induced sepsis and infection with E. coli. Results are expressed as means ± SD (n = 3 to 8) and representative of one of three experiments. ****P < 0.0001 (one-way ANOVA). (D) CD45.1 TpMos and CD45.2 MatMos were cocultured in 1:1 ratio in vitro with CSF-1 (20 ng/ml), and percentage of CD11b+ F4/80+ macrophages derived from each cell type were analyzed at indicated time points. Results are expressed as means ± SD and representative of one of three experiments. ****P < 0.0001 (Student’s t test). (E and F) TpMos and MatMos were cultured with CSF-1 (20 ng/ml) for 2 days, and percentage of live, early and late apoptotic and necrotic cells was quantified using FLICA Poly Caspase and DAPI via flow cytometry. Results are expressed as means ± SD (n = 3) and representative of one of three experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test). (G and H) TpMos and MatMos from Fucci-474 CD45.2 mice were sorted and adoptively transferred intraperitoneally into CD45.1 recipient mice. Analysis of adoptive transferred cells in the PL of recipient mice was performed 3 days later (G), and total number of CD11b+ F4/80+ macrophages (top) and percentage of Fucci signal in these transferred TpMo- or MatMo-dMΦs (bottom) were quantified (H). Results are expressed as means ± SD (n = 4 to 6) and representative of one of three experiments. *P < 0.05 (Student’s t test).
Monocytes are highly plastic cells known for their ability to differentiate into monocyte-derived cells with properties according to the environmental stimuli (41–43). Among the many peripheral stimuli that influence monocyte fate, colony stimulating factor 1 (CSF-1) is a secreted cytokine that is critical for monocyte differentiation into macrophages or other related cell types (44, 45). Notably, CSF-1 production is often increased during bacterial infections and has been shown to be critical for host defense by mediating survival and antimicrobial functions of mononuclear phagocytes (46, 47). We found that CSF-1 was increased during infection in septic and E. coli–infected mice compared to the controls as detected by enzyme-linked immunosorbent assay (ELISA) of peritoneal lavage (PL) (Fig. 5C). Therefore, we cultured sorted TpMos and MatMos in the presence of CSF-1 in vitro for 5 to 7 days to assess how these two cell types respond to CSF-1 as a local stimulus and determine their functional outcome. We noticed that TpMos generated more CD11b+ F4/80+ macrophages than MatMos at differing time points, plating densities and doses of CSF-1 (fig. S3, A to C). Coculture of TpMos and MatMos at a 1:1 ratio also resulted in a significantly higher percentage of macrophages derived from TpMos (Fig. 5D). Furthermore, TpMo-derived macrophages (TpMo-dMΦs) were more viable and less susceptible to cell death compared to MatMo-derived macrophages (MatMo-dMΦs) (Fig. 5, E and F). The ability of TpMos to surpass MatMos in macrophage generation was further validated in vivo when adoptively transferred TpMos or MatMos into the peritoneum of recipient mice resulted in increased TpMo-dMΦs that retained a higher percentage of Fucci signal (Fig. 5, G and H). These results hence reveal that in contrast to MatMos, TpMos are poised to replenish the macrophage niche rapidly in response to external stimuli such as CSF-1.
TpMo-dMΦs were distinct from MatMo-dMΦs
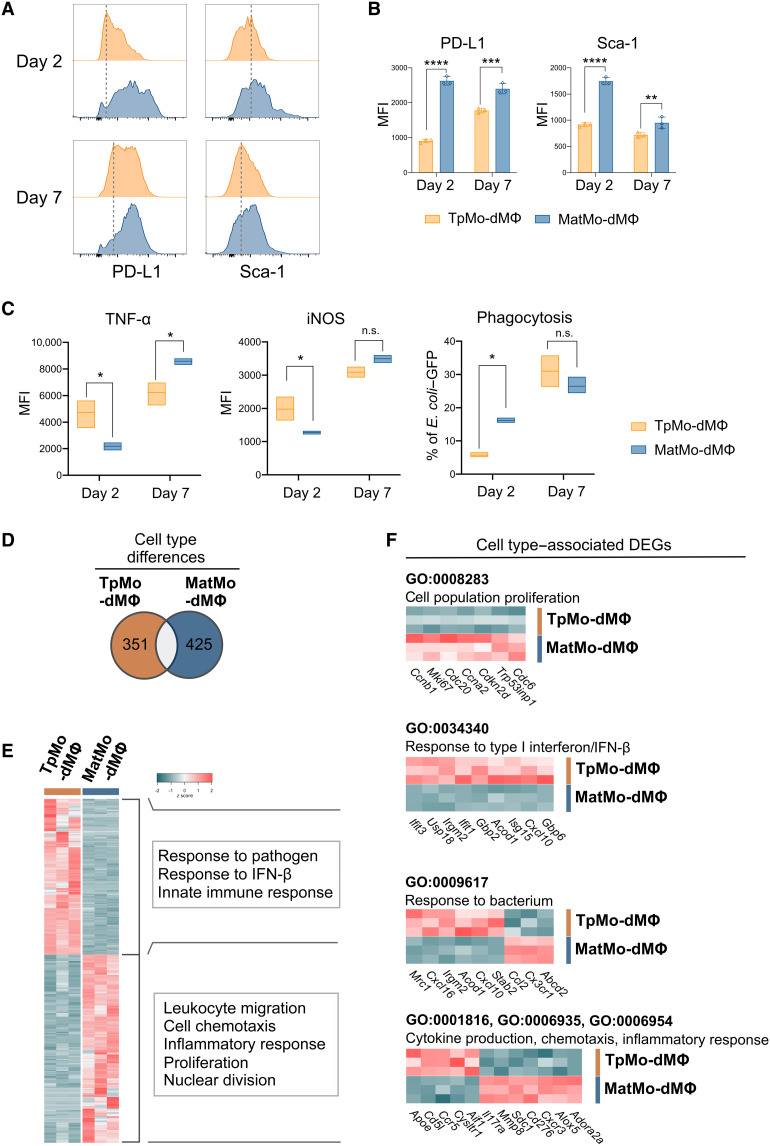
While our current results now indicate that TpMos are more competent at generating macrophages rapidly, it remains unclear whether TpMo-dMΦs would be functionally distinct from MatMo-dMΦs and whether they are able to alter the course of infection. To address this question, we first analyzed a panel of surface markers on TpMo-dMΦs and MatMo-dMΦs. TpMo-dMΦs were found to be phenotypically distinct from MatMo-dMΦs as they express distinct surface markers during the course of differentiation (fig. S4, A and B). In particular, TpMo-dMΦs displayed reduced expression of programmed cell death ligand 1 (PD-L1) and stem cell antigen-1 (Sca-1) compared to MatMo-dMΦs (Fig. 6, A and B). Upon exposure to bacteria, TpMo-dMΦs responded by preferentially expressing tumor necrosis factor–α (TNF-α) and inducible nitric oxide synthase (iNOS) instead of phagocytosis compared to MatMo-dMΦs at the early stages of differentiation (Fig. 6C). However, at day 7 after differentiation, both TpMo-dMΦs and MatMo-dMΦs appeared to converge in an equal ability to phagocytose bacteria and express iNOS, but MatMo-dMΦs expressed a higher level of TNF-α (Fig. 6C). These results hence suggest that TpMo-dMΦs and MatMo-dMΦs were not only phenotypically distinct but also functionally discrete in their response to bacteria at different stages of differentiation.
Fig. 6. TpMo-dMΦs are distinct from MatMo-dMΦs.
(A to C) TpMos and MatMos were cultured with CSF-1 (20 ng/ml) and analyzed at 2 or 7 days after culture for surface markers (A) with median fluorescence intensity (MFI) quantified (B) or incubated with E. coli–GFP for 3 hours before TNF-α and iNOS expression, as well as phagocytosis of E. coli–GFP, was quantified (C). Results are expressed as means ± SD and representative of one of three experiments. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (Student’s t test). (D to F) TpMo-dMΦs and MatMo-dMΦs 7 days after culture were analyzed by RNA-seq. (D) Venn diagram representing 776 DEGs between TpMo-dMΦ and MatMo-dMΦ subsets. (E) Heatmap of unique DEGs expression across subsets represented as z score. DEGs were subjected to GO biological process enrichment analysis. Functional characterization of DEGs was based on the top five GO terms obtained from analysis. (F) Cell population proliferation, response to type I interferon/IFN-β, response to bacterium, and cytokine production/chemotaxis/inflammatory response (top to bottom) related gene expression between TpMo-dMΦ and MatMo-dMΦ subsets.
To provide a deeper insight on how TpMo-dMΦs and MatMo-dMΦs may differ in their immune responses from each other, we performed bulk RNA-seq on these cells 7 days after their differentiation to examine their gene expression profiles. Comparison of their transcriptome profiles across 776 significant DEGs by GO enrichment analyses (Fig. 6D) revealed that TpMo-dMΦs were still transcriptomically distinct from MatMo-dMΦs and were characterized by enriched expression of genes involved in immune response to pathogens and interferon-β (IFN-β) (Fig. 6E). In contrast, MatMo-dMΦs were characterized by genes involved in inflammatory response, chemotaxis, and cell proliferation (Fig. 6E). At this time point, we found that MatMo-dMΦs up-regulated proliferative genes including Mki67 and Trp53inp1, while TpMo-dMΦs had already down-regulated these genes (Fig. 6F). These findings suggested that the increased macrophage numbers derived from TpMos compared to that of MatMos could possibly be due to a head start in the activation of proliferating genes in TpMos. We next analyzed the response of TpMo-dMΦs and MatMo-dMΦs toward bacterium. TpMo-dMΦs up-regulated genes such as Acod1, Cxcl10, and Irgm2 tagged under response to bacterium (GO:0009617) (Fig. 6F). Notably, Acod1 has been shown to promote the production of the metabolite, itaconate, involved in bacterial killing and suppression of proinflammatory cytokines (48). Furthermore, TpMo-dMΦs showed up-regulated Cxcl10 and Irgm2, which are genes associated with decreased bacterial burden and inhibition of inflammasome activation in septic mice (49, 50). Acod1, Cxcl10, and Irgm2 in TpMo-dMΦs were also found to have an enriched response toward type I IFN-β (GO:0034340) (Fig. 6F), which has been demonstrated in macrophages to promote efferocytosis and reprogramming toward a phenotype that favors inflammation resolution (51). Consistent with the attenuated inflammatory phenotype of TpMos described earlier (Fig. 4E), TpMo-dMΦs were also found to up-regulate genes including Apoe, Ccr5, and Cd5l (52–54) in comparison to MatMo-dMΦs (Fig. 6F). Together, these contrasting gene signatures propose a distinctive functional phenotype in addition to their differential kinetics, which distinguishes TpMo-dMΦs from MatMo-dMΦs.
TpMos protect against lethal sepsis by balancing the proinflammatory functions of MatMos
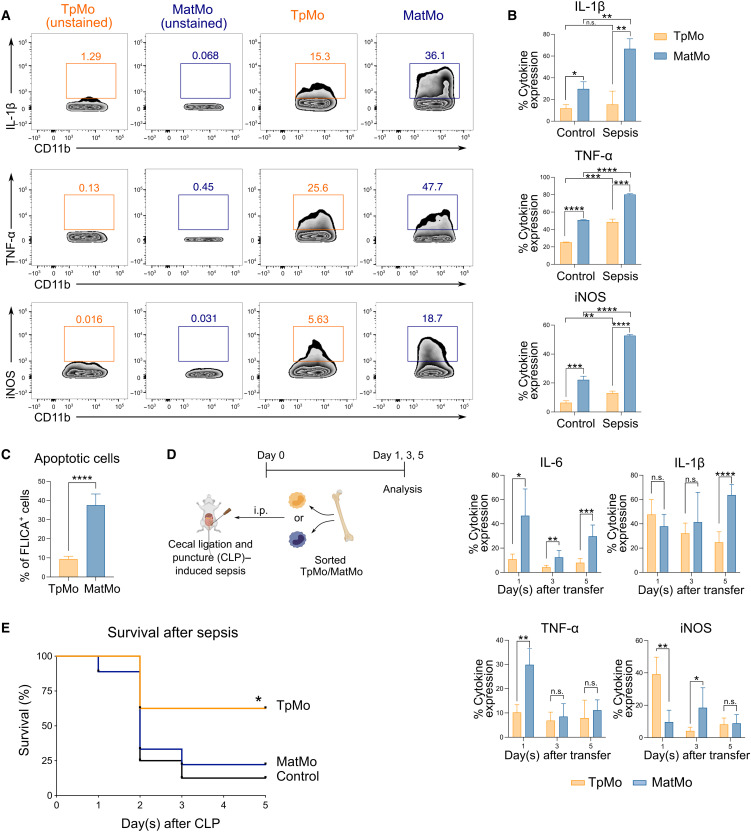
Given the divergent functions of TpMos versus MatMos in sepsis response and their distinct outcomes even after differentiation, we next determined how these two monocyte subsets would influence the outcome of sepsis. We found that at the steady state, MatMos reacted differently from TpMos by expressing significantly larger amounts of proinflammatory mediators such as IL-1β, TNF-α, and iNOS after stimulation with lipopolysaccharide (LPS) (Fig. 7, A and B). Furthermore, when stimulated with LPS, “sepsis-primed” MatMos isolated from CLP mice generated much more of these proinflammatory mediators (Fig. 7B) and were more sensitive to apoptotic cell death than their TpMo counterparts (Fig. 7C). These results hence suggest that TpMos and MatMos display functional differences that poise them toward distinct subsequent roles during infection.
Fig. 7. TpMos display protective functions during sepsis by balancing the proinflammatory functions of MatMos.
(A) TpMos and MatMos were sorted and analyzed for the expression of IL-1β, TNF-α, and iNOS after stimulation with LPS. (B) Control and CLP-induced septic BM TpMos and MatMos were stimulated with LPS and analyzed for the expression of IL-1β, TNF-α, and iNOS. (C) LPS-stimulated control BM TpMos and MatMos were analyzed for apoptotic cells using FLICA Poly Caspase. (D) TpMos and MatMos were sorted from CD45.1 mice and adoptively transferred into CLP-induced CD45.2 recipient mice via the intraperitoneal route. Analysis of PL of recipient mice for transferred cells was performed on days 1, 3, and 5 after adoptive transfer for the expression of IL-6, IL-1β, TNF-α, and iNOS. Results are expressed as means ± SD (n = 4 to 6) and representative of one of three experiments. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (Student’s t test). (E) TpMos or MatMos were sorted and adoptively transferred into recipient mice shortly after they were subjected to CLP. The mortality of these recipient mice was assessed using the Kaplan-Meier survival curve. Results are representative of one of three independent experiments (n = 10). *P < 0.05 (Mantel-Cox).
Since TpMos are direct precursors of MatMos and originate from the same cell lineage, we next performed an adoptive transfer of sorted TpMos and MatMos into septic mice to track their development/differentiation and determine key effector differences between these two cell types that would functionally influence the course of sepsis. We found that TpMo-dMΦs expressed lower levels of F4/80 compared to MatMo-dMΦs when analyzed 1 day after the adoptive transfer of these cells into the peritoneum of septic mice; however, this disparity diminished over time (fig. S5, A and B). In addition, while the expression of IL-1β, TNF-α, and iNOS differed between these two cell types at certain time points, MatMo-dMΦs consistently expressed higher levels of IL-6 throughout the course of infection (Fig. 7D). Hence, these results do not indicate any interswitching of phenotypes between TpMos and MatMos even under inflammatory conditions. Finally, we observed an increase in the survival rate when TpMos, but not MatMos, were transferred into septic mice (Fig. 7E). While these septic mice that received TpMos or MatMos did not display significant differences in bacteria load, aspartate aminotransferase/alanine aminotransferase (AST/ALT), and leukocyte numbers in the blood (fig. S6, A to C), we noticed distinct differences in the serum profile of these recipient mice (fig. S7, A and B). In particular, mice that received TpMos had an increase in chemokines such as CCL2, CCL3, and CXCL10 as well as IL-10 and IL-18 at day 1 after sepsis compared to mice that received MatMos (fig. S7A). Notably, the difference in IL-10 was detected significantly only at day 1 and not at later time points tested during sepsis (fig. S7B). Furthermore, TpMos may have modulated the production of IL-10 through other cell types as we did not detect significant levels of IL-10 in transferred TpMos themselves (fig. S7C). Therefore, these results reveal a protective role of TpMos for the host during bacterial sepsis by balancing the proinflammatory functions of MatMos.
DISCUSSION
In summary, our findings illustrate a previously unidentified protective monocyte response that occurs in response to bacterial invasion (Fig. 8). While monocyte precursors have been traditionally thought to function merely as cells for the replenishment of MatMos in the BM (14, 16, 55), our study highlights a paradigm shift from these findings by showing that proliferative TpMos can be mobilized into the periphery to partake in effector functions when the host’s immune response is at threat of being overwhelmed by a bacterial stimulus. In particular, the recruitment of TpMos into the periphery suggests an “on-demand” mobilizable source of proliferative monocytes that can replenish the macrophage niche rapidly. These findings also elucidate an outstanding conundrum of macrophage replenishment during bacterial infection when MatMos are susceptible to cell death upon exhaustion of their antibacterial functions (18, 19) and can only enter the cell cycle after a few days in the specific presence of local proliferative cues in the tissue site (22). Furthermore, TpMos responded differently from MatMos in response to local and bacterial stimuli by differentiating into phenotypically and functionally distinct macrophages. TpMos and their derived macrophages conferred protection against sepsis by serving as a reservoir of cells to equilibrate the antibacterial and inflammatory responses of MatMos. Together, we believe that our present work uncovers a host regulatory mechanism that balances the competing demands of monocytes during bacterial infections and contributes to the emerging paradigm of MDM heterogeneity in distinct disease outcomes.
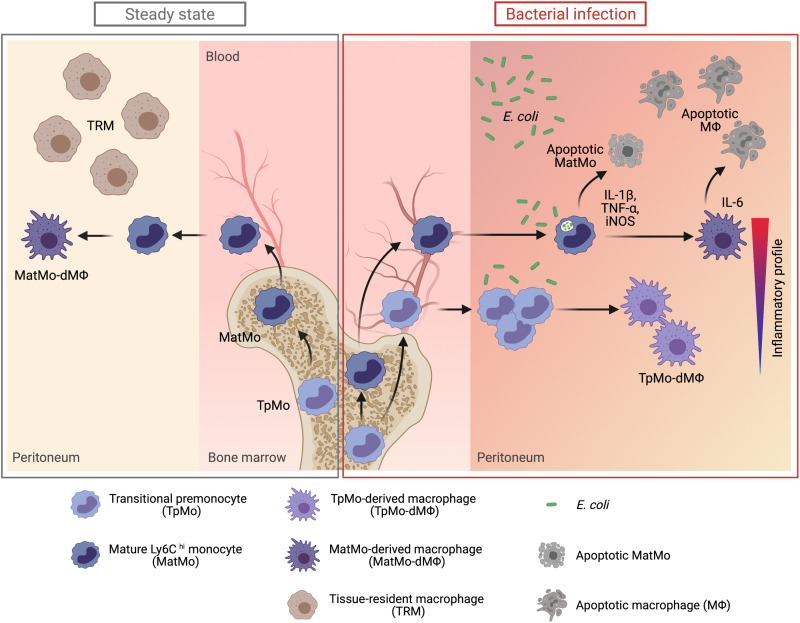
Fig. 8. The mobilization of TpMos into the periphery allows a diversification of monocytic roles to protect the host against bacterial infections.
During steady state, TpMos are immobilized in the BM where they serve as a reservoir of precursor cells to replenish the pool of MatMos in the circulation. Consequently, the replenishment of TRMs in the periphery are carried out solely by MatMos, giving rise to MatMo-dMΦs. However, upon bacterial infection, TRMs undergo rapid cellular death, which predisposes the host toward morbidity from an empty macrophage niche and the ensuing bacterial burden. This scenario results in competing demands placed on MatMos, which must now fulfil both antibacterial effector functions and macrophage replenishment roles simultaneously. To circumvent this challenge, TpMos are mobilized from the BM into the circulation to serve as the main source of macrophage replenishment when MatMos are vulnerable toward bacteria-induced cell death after the exhaustion of their effector functions. Furthermore, TpMos and their derived macrophages provided protection against sepsis by balancing the highly proinflammatory cytokine response of MatMos that contribute toward the cytokine storm in sepsis. Together, our findings highlight a specialization of monocytic roles against bacteria invasion through the mobilization of a monocyte precursor, which may be an evolutionary designed mechanism to balance the competing demands placed on the immune system in their arms race against pathogens. (Figure created with BioRender.com.)
During an infection, a surge in demand for monocytes is essential to replace bacteria-induced TRM loss and sustain the clearance of the infection (56, 57). This demand is first achieved through the mobilization of BM MatMos into the bloodstream through CCR2 signaling whereby CCL2 is expressed by BM mesenchymal Nestin+ stromal cells upon sensing of TLR ligands such as LPS in the circulation (30). At the same time, BM hematopoietic precursors are activated to proliferate and differentiate toward the myeloid lineage, in a process known as “emergency myelopoiesis” (58), to sustain the output of MatMos throughout the course of infection. These mechanisms are in line with a progressive layered defense that the immune system executes to control the pathogen response. Contrary to these findings, our data suggest a new working model whereby proliferative TpMos can be mobilized directly into the circulation in a CCR2-independent manner after a surge in peripheral bacterial burden. Notably, this response provides an important conceptual bridge in our understanding of the host response toward MDM replenishment during bacteria-induced TRM loss. Specifically, MatMos require a substantial duration to activate their proliferative genes by local stimuli in the tissue (22, 59, 60) and, during this period, are susceptible to cell death (19). In contrast to MatMos, our data revealed that proliferative TpMos are advantaged in the periphery as these cells are already in cell cycle before infiltrating into the tissue, which allows them to overcome the challenge of cell death by differentiating into macrophages rapidly. In support of this notion, our results have demonstrated that TpMos that have been exposed to local stimuli such as CSF-1 were more resistant to cell death and exhibited reduced phagocytosis of bacteria compared to MatMos during the early differentiation stages. Consequently, the mobilization of proliferative TpMos fulfills a critical gap in immune defense during bacteria invasion by providing a source of rapidly generated macrophages, while MatMos are occupied with antibacterial effector functions that predispose them to cell death (18). Our findings also indicate that the host has evolved strategies to bypass the hierarchal process of monocyte generation in the BM should the barrier of immune defense become overwhelmed.
It is increasingly perceived that monocytes can be preeducated in the BM to promote tissue-specific functions (61–66). Specifically, monocyte precursors can be primed to up-regulate differential phenotypic markers according to particular disease states, which leads to functionally distinct MatMos that leave the BM with enhanced tissue regulatory properties. Furthermore, monocyte progenitors have also been recently shown to give rise to multinucleated giant cells present in granuloma formation during mycobacteria infections (67). Our results demonstrating the mobilization of proliferative TpMos into the circulation during bacterial infections further highlights the BM as a critical player in modifying immune fate and outcome in the release of distinct subsets of immune cells. In particular, the egressed TpMos were found to be protective against sepsis, and these cells displayed properties distinct from MatMos upon exposure to local and bacterial stimuli. Notably, MatMos were found to be highly proinflammatory upon exposure to bacterial stimuli, resulting in a rapid production of cytokines that are important for the cytokine storm. This can be illustrated by their increased expression of IL-1β and TNF-α and their continued higher expression of IL-6 during the course of their differentiation compared to TpMos. In particular, septic mice that received an adoptive transfer of TpMos displayed a significant difference in IL-10 at day 1 of sepsis compared to mice that received MatMos. The presence of IL-10 in the early stages of sepsis have been shown to play a critical role in controlling the onset of irreversible septic shock (68, 69), suggesting that TpMos may exert their protective effects via IL-10 production. Since we were unable to identify detectable levels of IL-10 in transferred TpMos, it is feasible that TpMos may modulate IL-10 production by other cell types. We have also found that TpMo-dMΦs may also be beneficial to the host in avoiding excessive inflammation during bacterial infections as they displayed reduced expression of PD-L1, which has been shown to increase host survival during sepsis (70–72). While our functional and transcriptomic findings have demonstrated an attenuated inflammatory response for TpMos, it is unlikely that this will lead to an attenuated host response as our data have revealed that TpMos eventually up-regulate their proinflammatory cytokine response at later stages of macrophage differentiation, indicating that TpMos do not remain at an attenuated state throughout the course of infection. Rather, our data suggest that TpMos and MatMos likely give rise to macrophages on a continuum since both macrophage subsets are derived from monocyte subsets at different stages of maturity. Consequently, these findings further underscore the diversified yet complementary roles of TpMos and MatMos in the periphery to equilibrate the antibacterial and inflammation response during the course of infection. As there are currently no effective ways of targeting TpMos and MatMos exclusively without affecting the other subset due to their developmental relationship, understanding the exact impact of each individual subset by tracing their kinetics and development into macrophages in vivo in a single entity would require more sophisticated fate-mapping models in the future. Since monocytes have also been shown to differentiate into self-renewing functional macrophages that persist after infection (73–75), further research depicting the impact of TpMo-dMΦs in the periphery upon a secondary microbial challenge in vivo should be explored.
Together, our results resolve the longstanding and seemingly contradictory demands placed on circulatory monocytes during infections. Our model suggests that the host resolves this conundrum by mobilizing TpMos into the periphery, such that the bactericidal and macrophage replacement tasks can be specialized by MatMos and TpMos in the early stages of infection, respectively. The presence of TpMos in the periphery functions as a reservoir of cells in an early stage of differentiation that balances the antibacterial and inflammatory responses during infections. Our findings also demonstrate a rapid host response without the need to generate a new subset during the early stages of infection, supporting our hypothesis that the mobilization of TpMos represents an evolutionary designed mechanism to balance the competing demands placed on the immune system during their arms race against pathogens. In summary, we envision that our results will provide further insights into monocyte and macrophage heterogeneity during bacterial infections and may lead to the engineering of TpMo-mobilization strategies for future therapeutic intervention of sepsis.
MATERIALS AND METHODS
Study design
The objective of this study was to identify a monocyte mechanism that allows for an effective replenishment of macrophages during bacterial infections. To achieve this goal, we induced an acute bacterial infection with a pathogenic strain of E. coli and confirmed these findings in septic mice. We next profiled peripheral blood monocytes in these mice and identified a distinct subset of proliferative Ly6Chi monocytes in the circulation that was subsequently characterized to be TpMos (16). We next used a combination of in vivo adoptive transfer assays, transcriptomics, in vitro differentiation, and functional assays to investigate the role and function of TpMos upon their migration into the periphery. Experiments that included genetically modified mice were performed with littermate controls using both males and females. Animals were randomized, and survival outcome of sepsis was performed in a blinded fashion. Sample sizes varied according to the goal of each experiments and are correspondingly provided in each figure for every experimental setup in the figure legend. In general, the sample size of n = 3 to 10 mice per group was used and determined to be the optimal size for statistical analyses, and experiments were repeated independently. No animals were excluded from analyses.
Ethics statement
All experiments were performed under the approval of the Institutional Animal Care and Use Committee of the Biological Resource Center (BRC) under protocol #191437, in accordance with the guidelines of the Agri-Food and Veterinary Authority and the National Advisory Committee for Laboratory Animal Research of Singapore.
Experimental subjects
C57BL/6 (6 to 10 weeks old) mice were bred and maintained under specific pathogen-free conditions in the BRC of A*STAR (Agency for Science, Technology and Research), Singapore. Both males and females were used for experiments, and animals were gender- and age-matched in each experiment. CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ) mice and Ccr2−/− (B6.129S4-Ccr2tm1Ifc/J) mice were obtained from the Jackson Laboratory. Tlr4−/− (IMSR_OBS:4) mice were obtained from Oriental BioService. Fucci-474Bsi-Green (B6.B6D2-Tg(Fucci)474Bsi) mice were obtained from the RIKEN BioResource Center (Ibaraki, Japan) (76). LysM-GFP+ (Lyz2tm1.1Graf) mice were provided by T. Graf (Centre for Genomic Regulation, Barcelona, Spain). Ms4a3creRosa-tdTomatoflox mice were provided by F. Ginhoux (77). All transgenic mice were maintained on a C57BL/6 background.
E. coli infection model
E. coli strain (UTI89) was provided by S. Chen, A*STAR, Singapore. To prepare the bacteria for infection, UTI89 was cultured in lysogeny broth (LB) and incubated in a shaker incubator at 37°C. Bacteria were grown to mid-log phase [an optical density at 600 nm (OD600) of 0.4 to 0.6] and then washed twice with phosphate-buffered saline (PBS). Mice were infected intraperitoneally with 1 × 106 colony-forming units (CFU) E. coli in 200 μl of PBS. Control group was injected intraperitoneally with 200 μl of PBS. Mice were harvested 9 or 18 hours after infection. E. coli burden was measured by collecting the peritoneal fluid 18 hours after infection and diluted accordingly before culturing overnight on LB plates. Colonies were counted, and CFU were calculated the next day. Bar graphs were plotted as CFU per microliter of peritoneal fluid.
CLP-induced sepsis
Experimental procedures were performed as previously described (27). The peritoneal cavity was exposed under ketamine/xylazine anesthesia, and the cecum was exteriorized. Mid- and high-grade sepses were performed through 50 and 75% cecum ligation, respectively, using a nonabsorbable 7-0 suture. A 26 ½ –gauge needle was used to perforate the distal end of the cecum, and a small drop of feces was extruded through the puncture before being relocated back into the peritoneal cavity. The peritoneum was closed and subsequently treated with saline and buprenorphine via subcutaneous injection. Age- and gender-matched sham-operated controls were included for all procedures.
BrdU pulsing and proliferation assays
Mice were administered 1.5 mg of BrdU (BD Biosciences) via intraperitoneal injection for 30 min to assess their proliferative capacity. To detect BrdU incorporation, cells were surface-stained, fixed, permeabilized, and subjected to intracellular staining with fluorescein isothiocyanate– or anaphase-promoting complex–conjugated anti-BrdU antibody according to the manufacturer’s protocol (BrdU Flow kit, BD Biosciences) before analysis by flow cytometry. The Fucci-transgenic mouse model, which labels cells in the S-G2-M phase of the cell cycle with a green-emitting fluorescent protein, was also used to identify proliferating cells through flow cytometry.
Macrophage depletion
Clodronate liposomes suspension (LIPOSOMA) was administered intraperitoneally according to the recommended doses (100 μl of suspension/10 g of mouse weight), and E. coli infection was carried out 66 hours after. PBS liposomes suspension was injected as a control. Macrophages were depleted without affecting blood monocytes as previously described (5, 28).
Molecular biology
ELISA assay for CSF-1
To determine the levels of CSF-1 in the peritoneum, 3 ml of PBS was administered into the peritoneum using a 26 ½–gauge needle, and the PL was collected. The PL was spun down, and the supernatant was collected for ELISA. ELISA for CSF-1 was performed according to the manufacturer’s instructions (catalog no. MMC00, R&D systems).
AST and ALT
A total of 400 to 800 μl of blood was obtained from CLP-induced septic mice via an incision in the submandibular region using a 5-mm lancet. Collected blood was spun down to collect blood serum. Serum AST and ALT levels were measured according to the manufacturer’s instructions (AST, catalog no. K753, BioVision; AST, catalog no. MAK052, Sigma-Aldrich).
Tissue preparation for flow cytometry and sorting
A total of 200 μl of blood was obtained via an incision in the submandibular region using a 5-mm lancet and treated with commercial-grade red blood cell lysis buffer (eBioscience). Mice were euthanized by CO2 inhalation. To harvest peritoneal cavity cells, 3 ml of PBS with 2 mM EDTA was administered into the peritoneum using a 26 ½–gauge needle. The PL was collected, and the cell suspension was passed through a 70-μm nylon mesh. To collect BM cells, mouse femurs were flushed using a 23-gauge needle in PBS with 2 mM EDTA and 3% fetal bovine serum, and the effluent was passed through a 70-μm nylon mesh. Antibodies were purchased from eBioscience, BioLegend, BD Biosciences, or R&D Systems. Mouse cells were stained with the following antibodies: CCR2 (475301), CD3e (145-2C11), CD11b (M1-70), CD11c (N418), CD16/32 (2.4G2), CD31(390), CD43 (S7), CD45 (30-F11), CD45.1 (A20), CD45.2 (104), CD45R (B220) (RA3-6B2), CD62L (MEL-14), CD90.2 (53-2.1), CD106 (429/MVCAM.A), CD115 (AFS98), c-kit (2B8), CXCR4 (2B11), CX3CR1 (SA011F11), F4/80 (BM8), I-A/I-E (M5/114.15.2), Flt-3 (A2F10), Ly6C (HK1.4), Ly6G (1A8), NK1.1 (PK136), PD-L1 (10F.9G2), Sca-1 (D7), Siglec-F (E502440), and Tim-4 (54). Dead cells were identified and excluded using 4′,6-diamidino-2-phenylindole (DAPI) staining. Blood and BM monocytes were identified as Lin (B220/CD90.2/NK1.1)neg Ly6Gneg CD115pos. Monocytes were further divided into cMoPs (CD11bneg Ly6Chi ckitpos), TpMos (CD11blo Ly6Chi ckitneg CXCR4hi), and MatMos (CD11bpos Ly6Chi ckitneg CXCR4lo). Total PL macrophages were identified as CD45pos Lin (B220/CD90.2/NK1.1)neg CD11bpos F4/80pos. To check for cytokine expression, BM or peritoneal cells were stimulated with LPS for 3 hours (or 24 hours for iNOS expression) at 37°C and 5% CO2 in the presence of GolgiStop and GolgiPlug (BD Biosciences) in the medium at a dilution recommended by the manufacturer’s instructions; stained with relevant surface markers; fixed with fixation/permeabilization buffer (BD Biosciences) before staining with IL-1β (NJTEN3), IL-6 (MQ213A5), IL-10 (JES5-16E3), TNF-α (MP6-XT22), and iNOS/NOS2 (CXNFT); washed; and acquired via flow cytometry. Cells were acquired on BD LSR II flow cytometer using FACSDiva software, and data were subsequently analyzed with FlowJo software (Tree Star). The total number of cells collected was quantified using count beads (CountBright, Life Technologies) according to the manufacturer’s protocol. BM TpMos and MatMos were sorted using a BD FACSAria II sorter to achieve >98% purity.
In vitro differentiation and macrophage function assays
Sorted cells were plated according to indicated plating densities and cultured at 37°C and 5% CO2 in complete RPMI 1640 medium with 25 mM Hepes (HyClone), 10% fetal bovine serum (Serana), 2 mM l-glutamine (Invitrogen), and 100× penicillin-streptomycin (Invitrogen) with indicated CSF-1 (STEMCELL Technologies) concentrations for up to 9 days. Differentiated macrophages were harvested and stained for surface markers for analysis by flow cytometry. Apoptosis and necrosis of macrophages were measured using the FAM FLICA Poly Caspase kit (Bio-Rad) according to the manufacturer’s instructions. To investigate the functions of differentiated macrophages, DH5α E. coli–expressing GFP (78) was grown in LB medium overnight at 37°C to an optical density at 600 nm (OD600) of 1.5 to 1.8, at which point, the bacteria were diluted and grown for 1 to 2 hours to an OD600 of ~0.5 and were lastly washed twice with PBS. Seeded macrophages were incubated with bacteria at a ratio of 1:100 for 3 hours at 37°C and 5% CO2 in the presence of GolgiStop and GolgiPlug (BD Biosciences) in the medium at a dilution recommended by the manufacturer’s instructions. Upon harvesting the differentiated macrophages, cells were stained with relevant surface markers, fixed with fixation/permeabilization buffer (BD Biosciences) before staining with TNF-α (MP6-XT22) and iNOS/NOS2 (CXNFT), washed, and acquired via flow cytometry to determine the median fluorescence intensity (MFI) of TNF-α and iNOS and phagocytosis capacity of macrophages via percentage of positive E. coli–GFP.
Adoptive transfer of TpMos and MatMos
BM TpMos and MatMos sorted from WT or Fucci-474 mice were suspended in 200 μl of PBS and adoptively transferred through intravenous or intraperitoneal routes (as indicated) into CD45.1 recipient mice in steady state or after induction of sepsis. Recipient mice were euthanized at the time points indicated, and blood, BM, and PL were harvested, surface-stained, and analyzed by flow cytometry. For intra-BM (IBM) transfer of BM TpMos and MatMos, sorted 5 × 105 GFP+ TpMos and tdTomato+ MatMos were resuspended in a 1:1 ratio and transferred as a single injection into CD45.2+ mice as previously described (16). Briefly, recipient mice were anesthetized using ketamine/xylazine with legs shaven before transfer. Recipient tibia and blood were collected, surface-stained, and analyzed by flow cytometry 9 hours after bacterial infection.
Multiplex cytokine assay
Serum cytokine and chemokine levels were measured with Cytokine & Chemokine Convenience 36-Plex Mouse ProcartaPlex Panel 1A (Thermo Fisher Scientific) according to the manufacturer’s protocol. Plasma was incubated with fluorescent coded magnetic beads precoated with respective antibodies in a black 96-well clear-bottom plate overnight at 4°C. After incubation, plates were washed five times with wash buffer [PBS with 1% bovine serum albumin (Capricorn Scientific) and 0.05% Tween-20 (Promega)]. Sample antibody–bead complexes were incubated with biotinylated detection antibodies for 1 hour and washed five times with wash buffer. Subsequently, streptavidin–phycoerythrin (PE) was added and incubated for another 30 min. Plates were washed five times again, before sample antibody–bead complexes were resuspended in sheath fluid for acquisition on the FLEXMAP 3D (Luminex) using xPONENT 4.0 (Luminex) software. Data analysis was performed on Bio-Plex Manager 6.1.1 (Bio-Rad). Standard curves were generated with a 5-PL (5-parameter logistic) algorithm, reporting values for both MFI and concentration data.
Transcriptomics
BM TpMos, MatMos, TpMo-dMΦs, and MatMo-dMΦs were sorted or isolated as described above. Total RNA was extracted using Arcturus Picopure RNA Isolation kit according to the manufacturer’s protocol. All mouse RNA samples were analyzed on PerkinElmer Labchip GX system for quality assessment with an RNA integrity number (RIN) of >7.9. cDNA libraries preparation used 2 ng of total RNA and 1 μl of a 1:50,000 dilution of ERCC RNA Spike-In Controls (Ambion) using SMARTSeq v2 protocol (79), with modifications: (i) 20 μM template switching oligo (TSO) and (ii) 250 pg of cDNA with one-fifth reaction of an Illumina Nextera XT kit. Length distribution of the cDNA libraries was monitored using DNA High Sensitivity Reagent kit on the PerkinElmer Labchip. Samples were subjected to an indexed PE sequencing run of 2 × 51 cycles on an Illumina HiSeq 2500 Rapid mode (14 samples per lane).
RNA-seq data in FASTQ files were mapped to mouse genome build mm10 using the STAR alignment software. Mapped reads were counted using featureCounts (part of Subread package) based on the GENCODE M9 annotations. Raw counts were processed by log2 transformation using the Bioconductor DESeq2 v1.30.0 package, followed by performing PCA using prcomp function in R. Count per million (CPM) values were determined from raw counts using edgeR. Differential gene expression analysis was then performed on CPM values to identify DEGs. Only DEGs with an adjusted P value (false discovery rate) of less than 0.05 were represented. CPM values of DEGs were normalized and represented as z score for heatmap plotting. Unsupervised k-means clustering was performed on DEGs using a Euclidean distance measurement and ward.D2 algorithm method. Genes in each DEG cluster were characterized by GO enrichment biological processes using R package clusterProfiler v4.0.3 (80). Functional gene sets from the Mouse Genome Informatics database were used to annotate DEGs across all clusters. For the analysis of BM TpMos and MatMos in sham and sepsis, z scores of genes in each DEG cluster were averaged and normalized across conditions then represented in bar plots. For the GO enrichment analysis of TpMo-dMΦs and MatMo-dMΦs under response to IFN-β, additional gene annotation was referenced from Kumaran Satyanarayanan et al. (51).
Statistical analysis
Statistical analyses were performed using Prism software (GraphPad). Student’s t test (normal distribution) and one-way or two-way analysis of variance (ANOVA) were performed as appropriate. For correlation analysis, linear regression was used to generate the line of best fit, and Pearson’s correlation test was performed to generate P value. P < 0.05 was considered as statistically significant.
Acknowledgments
We thank the SIgN Flow cytometry team for assistance with cell sorting, SIgN Immunomonitoring Platform for expertise on the multiplex analysis of proteins, SIgN functional genomics team for expertise on transcriptomics, and the SIgN mouse core facility for technical help and support.
Funding: This research was funded by Singapore Immunology Network (SIgN) core funding and A*STAR, Singapore. F.G. is an EMBO YIP awardee and is supported by SIgN core funding and Singapore National Research Foundation Investigatorship (NRF2016NRF-NRF1001-02). L.G.N. is supported by SIgN core funding. S.Z.C. is supported by the Ministry of Health, Singapore’s National Medical Research Council under its Open Fund-Young Individual Research Grant (OFYIRG17may036), and A*STAR Career Development Award (192D8043). Y.C.T. is a graduate scholar supported by the Ministry of Education, Singapore. M.Y.C. is a graduate scholar supported by the SIgN-NTU-NUS scholarship, Singapore. SIgN Flow Cytometry facility is supported by National Research Foundation (NRF) Singapore under Shared Infrastructure Support (SIS) (NRF2017_SISFP09).
Author contributions: Conceptualization: L.G.N., J.L.D., and S.Z.C. Methodology: Y.C.T., M.Y.C., D.L., I.K., M.E., G.C.L., L.A.O.Y., S.L.C., L.G.N., J.L.D., and S.Z.C. Validation: Y.T., L.T., K.H.L., K.L., A.Y.J.C., N.B.S., Y.Y.H., and C.C.G. Investigation: Y.C.T., M.Y.C., D.L., M.E., C.K.M., and S.Z.C. Formal analysis: Y.C.T., M.Y.C., D.L., I.K., M.E., and S.Z.C. Writing: Y.C.T., M.Y.C., I.K., M.N., J.L.Y.L., L.G.N., J.L.D., and S.Z.C. Visualization: Y.C.T., M.Y.C., and I.K. Funding acquisition: L.G.N. Resources: H.C., T.C., W.Y., H.L.T., A.L., A.S.J., V.A., C.R., B.L., F.G., and S.L.C. Supervision: L.G.N., J.L.D., and S.Z.C.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data used for Figs. 4 and 6 are deposited in the NCBI Gene Expression Omnibus under the accession numbers GEO: GSE151682, GSE192448, and GSE192449.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
REFERENCES AND NOTES
- 1.Hashimoto D., Chow A., Noizat C., Teo P., Beasley M. B., Leboeuf M., Becker C. D., See P., Price J., Lucas D., Greter M., Mortha A., Boyer S. W., Forsberg E. C., Tanaka M., van Rooijen N., García-Sastre A., Stanley E. R., Ginhoux F., Frenette P. S., Merad M., Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perdiguero E. G., Geissmann F., The development and maintenance of resident macrophages. Nat. Immunol. 17, 2–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginhoux F., Guilliams M., Tissue-resident macrophage ontogeny and homeostasis. Immunity. 44, 439–449 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Epelman S., Lavine K. J., Randolph G. J., Origin and functions of tissue macrophages. Immunity 41, 21–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang N., Czepielewski R. S., Jarjour N. N., Erlich E. C., Esaulova E., Saunders B. T., Grover S. P., Cleuren A. C., Broze G. J., Edelson B. T., Mackman N., Zinselmeyer B. H., Randolph G. J., Expression of factor V by resident macrophages boosts host defense in the peritoneal cavity. J. Exp. Med. 216, 1291–1300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zychlinsky A., Prevost M. C., Sansonetti P. J., Shigella flexneri induces apoptosis in infected macrophages. Nature 358, 167–169 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Robinson N., McComb S., Mulligan R., Dudani R., Krishnan L., Sad S., Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat. Immunol. 13, 954–962 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Paolo N. C., Doronin K., Baldwin L. K., Papayannopoulou T., Shayakhmetov D. M., The transcription factor IRF3 triggers “defensive suicide” necrosis in response to viral and bacterial pathogens. Cell Rep. 3, 1840–1846 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blériot C., Dupuis T., Jouvion G., Eberl G., Disson O., Lecuit M., Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity 42, 145–158 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Lai S. M., Sheng J., Gupta P., Renia L., Duan K., Zolezzi F., Karjalainen K., Newell E. W., Ruedl C., Organ-specific fate, recruitment, and refilling dynamics of tissue-resident macrophages during blood-stage malaria. Cell Rep. 25, 3099–3109.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Ferrer M. F., Scharrig E., Charo N., Rípodas A. L., Drut R., Carrera Silva E. A., Nagel A., Nally J. E., Montes de Oca D. P., Schattner M., Gómez R. M., Macrophages and galectin 3 control bacterial burden in acute and subacute murine leptospirosis that determines chronic kidney fibrosis. Front. Cell. Infect. Microbiol. 8, 384 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hur D. G., Kurabi A., Lim H. W., Spriggs M., Pak K., Ryan A. F., Macrophage depletion in CCR2−/− mice delays bacterial clearance and enhances neutrophil infiltration in an acute otitis media model. J. Infect. Dis. 223, 333–341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H. H., Aslanyan L., Vidyasagar A., Brennan M. B., Tauber M. S., Carrillo-Sepulveda M. A., Dores M. R., Rigel N. W., Martinez L. R., Depletion of alveolar macrophages increases pulmonary neutrophil infiltration, tissue damage, and sepsis in a murine model of Acinetobacter baumannii pneumonia. Infect. Immun. 88, e00128-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hettinger J., Richards D. M., Hansson J., Barra M. M., Joschko A.-C., Krijgsveld J., Feuerer M., Origin of monocytes and macrophages in a committed progenitor. Nat. Immunol. 14, 821–830 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Kawamura S., Onai N., Miya F., Sato T., Tsunoda T., Kurabayashi K., Yotsumoto S., Kuroda S., Takenaka K., Akashi K., Ohteki T., Identification of a human clonogenic progenitor with strict monocyte differentiation potential: A counterpart of mouse cMoPs. Immunity 46, 835–848.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Chong S. Z., Evrard M., Devi S., Chen J., Lim J. Y., See P., Zhang Y., Adrover J. M., Lee B., Tan L., Li J. L. Y., Liong K. H., Phua C., Balachander A., Boey A., Liebl D., Tan S. M., Chan J. K. Y., Balabanian K., Harris J. E., Bianchini M., Weber C., Duchene J., Lum J., Poidinger M., Chen Q., Rénia L., Wang C.-I., Larbi A., Randolph G. J., Weninger W., Looney M. R., Krummel M. F., Biswas S. K., Ginhoux F., Hidalgo A., Bachelerie F., Ng L. G., CXCR4 identifies transitional bone marrow premonocytes that replenish the mature monocyte pool for peripheral responses. J. Exp. Med. 213, 2293–2314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A. Mildner, S. Yona, S. Jung, in Advances in Immunology, K. M. Murphy, M. Merad, Eds. (Academic Press, 2013), vol. 120 of Development and Function of Myeloid Subsets, pp. 69–103; https://sciencedirect.com/science/article/pii/B978012417028500003X. [DOI] [PubMed]
- 18.DeLeo F. R., Modulation of phagocyte apoptosis by bacterial pathogens. Apoptosis 9, 399–413 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Webster S. J., Daigneault M., Bewley M. A., Preston J. A., Marriott H. M., Walmsley S. R., Read R. C., Whyte M. K. B., Dockrell D. H., Distinct cell death programs in monocytes regulate innate responses following challenge with common causes of invasive bacterial disease. J Immunol. 185, 2968–2979 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Furth R., Cohn Z. A., The origin and kinetics of mononuclear phagocytes. J. Exp. Med. 128, 415–435 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robbins C. S., Hilgendorf I., Weber G. F., Theurl I., Iwamoto Y., Figueiredo J.-L., Gorbatov R., Sukhova G. K., Gerhardt L. M. S., Smyth D., Zavitz C. C. J., Shikatani E. A., Parsons M., van Rooijen N., Lin H. Y., Husain M., Libby P., Nahrendorf M., Weissleder R., Swirski F. K., Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat. Med. 19, 1166–1172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins S. J., Knipper J. A., Zaiss D. M. W., Local proliferation of monocytes. J. Leukoc. Biol. 107, 547–549 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Otero K., Turnbull I. R., Poliani P. L., Vermi W., Cerutti E., Aoshi T., Tassi I., Takai T., Stanley S. L., Miller M., Shaw A. S., Colonna M., Macrophage colony-stimulating factor induces the proliferation and survival of macrophages via a pathway involving DAP12 and β-catenin. Nat. Immunol. 10, 734–743 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson J. R., Russo T. A., Uropathogenic Escherichia coli as agents of diverse non–urinary tract extraintestinal infections. J. Infect. Dis. 186, 859–864 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Chen S. L., Hung C.-S., Xu J., Reigstad C. S., Magrini V., Sabo A., Blasiar D., Bieri T., Meyer R. R., Ozersky P., Armstrong J. R., Fulton R. S., Latreille J. P., Spieth J., Hooton T. M., Mardis E. R., Hultgren S. J., Gordon J. I., Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: A comparative genomics approach. Proc. Natl. Acad. Sci. U.S.A. 103, 5977–5982 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakaue-Sawano A., Hoshida T., Yo M., Takahashi R., Ohtawa K., Arai T., Takahashi E., Noda S., Miyoshi H., Miyawaki A., Visualizing developmentally programmed endoreplication in mammals using ubiquitin oscillators. Development 140, 4624–4632 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Rittirsch D., Huber-Lang M. S., Flierl M. A., Ward P. A., Immunodesign of experimental sepsis by cecal ligation and puncture. Nat. Protoc. 4, 31–36 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J., Kubes P., A reservoir of mature cavity macrophages that can rapidly invade visceral organs to affect tissue repair. Cell 165, 668–678 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Serbina N. V., Pamer E. G., Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat. Immunol. 7, 311–317 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Shi C., Jia T., Mendez-Ferrer S., Hohl T. M., Serbina N. V., Lipuma L., Leiner I., Li M. O., Frenette P. S., Pamer E. G., Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating Toll-like receptor ligands. Immunity 34, 590–601 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee M.-S., Kwon H., Nguyen L. T., Lee E.-Y., Lee C. Y., Choi S. H., Kim M. H., Shiga toxins trigger the secretion of lysyl-tRNA synthetase to enhance proinflammatory responses. J. Microbiol. Biotechnol. 26, 432–439 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Yoon M.-S., Son K., Arauz E., Han J. M., Kim S., Chen J., Leucyl-tRNA synthetase activates Vps34 in amino acid-sensing mTORC1 signaling. Cell Rep. 16, 1510–1517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y.-R., Kim J.-S., Yun J.-S., Kim S., Kim S. Y., Jang K., Yang C.-S., Toxoplasma gondii GRA8 induces ATP5A1–SIRT3-mediated mitochondrial metabolic resuscitation: A potential therapy for sepsis. Exp. Mol. Med. 50, e464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu J., Niu X., Chang J., Shao M., Peng L., Xi Y., Lin A., Wang C., Cui Q., Luo Y., Fan W., Chen Y., Sun Y., Guo W., Tan W., Lin D., Wu C., Metabolic remodeling by TIGAR overexpression is a therapeutic target in esophageal squamous-cell carcinoma. Theranostics 10, 3488–3502 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medina-Contreras O., Harusato A., Nishio H., Flannigan K. L., Ngo V., Leoni G., Neumann P.-A., Geem D., Lili L. N., Ramadas R. A., Chassaing B., Gewirtz A. T., Kohlmeier J. E., Parkos C. A., Towne J. E., Nusrat A., Denning T. L., Cutting edge: IL-36 receptor promotes resolution of intestinal damage. J. Immunol. 196, 34–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgener S. S., Leborgne N. G. F., Snipas S. J., Salvesen G. S., Bird P. I., Benarafa C., Cathepsin G inhibition by Serpinb1 and Serpinb6 prevents programmed necrosis in neutrophils and monocytes and reduces GSDMD-driven inflammation. Cell Reports. 27, 3646–3656.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rittirsch D., Flierl M. A., Nadeau B. A., Day D. E., Huber-Lang M., Mackay C. R., Zetoune F. S., Gerard N. P., Cianflone K., Köhl J., Gerard C., Sarma J. V., Ward P. A., Functional roles for C5a receptors in sepsis. Nat. Med. 14, 551–557 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia T., Leiner I., Dorothee G., Brandl K., Pamer E. G., MyD88 and type I interferon receptor-mediated chemokine induction and monocyte recruitment during listeria monocytogenes infection. J. Immunol.. 183, 1271–1278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinwede K., Maus R., Bohling J., Voedisch S., Braun A., Ochs M., Schmiedl A., Länger F., Gauthier F., Roes J., Welte T., Bange F. C., Niederweis M., Bühling F., Maus U. A., Cathepsin G and neutrophil elastase contribute to lung-protective immunity against mycobacterial infections in mice. J. Immunol. 188, 4476–4487 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Parker H., Albrett A. M., Kettle A. J., Winterbourn C. C., Myeloperoxidase associated with neutrophil extracellular traps is active and mediates bacterial killing in the presence of hydrogen peroxide. J. Leukoc. Biol. 91, 369–376 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Varol C., Mildner A., Jung S., Macrophages: Development and tissue specialization. Annu. Rev. Immunol. 33, 643–675 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Jakubzick C. V., Randolph G. J., Henson P. M., Monocyte differentiation and antigen-presenting functions. Nat. Rev. Immunol. 17, 349–362 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Guilliams M., Thierry G. R., Bonnardel J., Bajenoff M., Establishment and maintenance of the macrophage niche. Immunity 52, 434–451 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Stanley E. R., Cifone M., Heard P. M., Defendi V., Factors regulating macrophage production and growth: Identity of colony-stimulating factor and macrophage growth factor. J. Exp. Med. 143, 631–647 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tagliani E., Shi C., Nancy P., Tay C.-S., Pamer E. G., Erlebacher A., Coordinate regulation of tissue macrophage and dendritic cell population dynamics by CSF-1. J. Exp. Med. 208, 1901–1916 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bettina A., Zhang Z., Michels K., Cagnina R. E., Vincent I. S., Burdick M. D., Kadl A., Mehrad B., M-CSF mediates host defense during bacterial pneumonia by promoting the survival of lung and liver mononuclear phagocytes. J. Immunol. 196, 5047–5055 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kandalla P. K., Sarrazin S., Molawi K., Berruyer C., Redelberger D., Favel A., Bordi C., de Bentzmann S., Sieweke M. H., M-CSF improves protection against bacterial and fungal infections after hematopoietic stem/progenitor cell transplantation. J. Exp. Med. 213, 2269–2279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu R., Chen F., Wang N., Tang D., Kang R., ACOD1 in immunometabolism and disease. Cell. Mol. Immunol. 17, 822–833 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelly-Scumpia K. M., Scumpia P. O., Delano M. J., Weinstein J. S., Cuenca A. G., Wynn J. L., Moldawer L. L., Type I interferon signaling in hematopoietic cells is required for survival in mouse polymicrobial sepsis by regulating CXCL10. J. Exp. Med. 207, 319–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eren E., Planès R., Bagayoko S., Bordignon P.-J., Chaoui K., Hessel A., Santoni K., Pinilla M., Lagrange B., Burlet-Schiltz O., Howard J. C., Henry T., Yamamoto M., Meunier E., Irgm2 and Gate-16 cooperatively dampen Gram-negative bacteria-induced caspase-11 response. EMBO Rep. 21, e50829 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumaran Satyanarayanan S., El Kebir D., Soboh S., Butenko S., Sekheri M., Saadi J., Peled N., Assi S., Othman A., Schif-Zuck S., Feuermann Y., Barkan D., Sher N., Filep J. G., Ariel A., IFN-β is a macrophage-derived effector cytokine facilitating the resolution of bacterial inflammation. Nat. Commun. 10, 3471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobaczewski M., Xia Y., Bujak M., Gonzalez-Quesada C., Frangogiannis N. G., CCR5 signaling suppresses inflammation and reduces adverse remodeling of the infarcted heart, mediating recruitment of regulatory T cells. Am. J. Pathol. 176, 2177–2187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baitsch D., Bock H. H., Engel T., Telgmann R., Müller-Tidow C., Varga G., Bot M., Herz J., Robenek H., von Eckardstein A., Nofer J.-R., Apolipoprotein E induces antiinflammatory phenotype in macrophages. Arterioscler. Thromb. Vasc. Biol. 31, 1160–1168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanjurjo L., Aran G., Roher N., Valledor A. F., Sarrias M.-R., AIM/CD5L: A key protein in the control of immune homeostasis and inflammatory disease. J. Leukoc. Biol. 98, 173–184 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Ginhoux F., Jung S., Monocytes and macrophages: Developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14, 392–404 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Ginhoux F., Bleriot C., Lecuit M., Dying for a cause: Regulated necrosis of tissue-resident macrophages upon infection. Trends Immunol. 38, 693–695 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Serbina N. V., Jia T., Hohl T. M., Pamer E. G., Monocyte-mediated defense against microbial pathogens. Annu. Rev. Immunol. 26, 421–452 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schultze J. L., Mass E., Schlitzer A., Emerging principles in myelopoiesis at homeostasis and during infection and inflammation. Immunity 50, 288–301 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Dixit A., Bottek J., Beerlage A.-L., Schuettpelz J., Thiebes S., Brenzel A., Garbers C., Rose-John S., Mittrücker H.-W., Squire A., Engel D. R., Frontline science: Proliferation of Ly6C+ monocytes during urinary tract infections is regulated by IL-6 trans-signaling. J. Leukoc. Biol. 103, 13–22 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Pang J., Urao N., Koh T. J., Proliferation of Ly6C+ monocytes/macrophages contributes to their accumulation in mouse skin wounds. J. Leukoc. Biol. 107, 551–560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Askenase M. H., Han S.-J., Byrd A. L., Morais da Fonseca D., Bouladoux N., Wilhelm C., Konkel J. E., Hand T. W., Lacerda-Queiroz N., Su X., Trinchieri G., Grainger J. R., Belkaid Y., Bone-marrow-resident NK cells prime monocytes for regulatory function during infection. Immunity 42, 1130–1142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Satoh T., Nakagawa K., Sugihara F., Kuwahara R., Ashihara M., Yamane F., Minowa Y., Fukushima K., Ebina I., Yoshioka Y., Kumanogoh A., Akira S., Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature 541, 96–101 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Ikeda N., Asano K., Kikuchi K., Uchida Y., Ikegami H., Takagi R., Yotsumoto S., Shibuya T., Makino-Okamura C., Fukuyama H., Watanabe T., Ohmuraya M., Araki K., Nishitai G., Tanaka M., Emergence of immunoregulatory Ym1+Ly6Chi monocytes during recovery phase of tissue injury. Sci. Immunol. 3, eaat0207 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Cirovic B., de Bree L. C. J., Groh L., Blok B. A., Chan J., van der Velden W. J. F. M., Bremmers M. E. J., van Crevel R., Händler K., Picelli S., Schulte-Schrepping J., Klee K., Oosting M., Koeken V. A. C. M., van Ingen J., Li Y., Benn C. S., Schultze J. L., Joosten L. A. B., Curtis N., Netea M. G., Schlitzer A., BCG vaccination in humans elicits trained immunity via the hematopoietic progenitor compartment. Cell Host Microbe 28, 322–334.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giladi A., Wagner L. K., Li H., Dörr D., Medaglia C., Paul F., Shemer A., Jung S., Yona S., Mack M., Leutz A., Amit I., Mildner A., Cxcl10+ monocytes define a pathogenic subset in the central nervous system during autoimmune neuroinflammation. Nat. Immunol. 21, 525–534 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Netea M. G., Domínguez-Andrés J., Barreiro L. B., Chavakis T., Divangahi M., Fuchs E., Joosten L. A. B., van der Meer J. W. M., Mhlanga M. M., Mulder W. J. M., Riksen N. P., Schlitzer A., Schultze J. L., Stabell Benn C., Sun J. C., Xavier R. J., Latz E., Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 20, 375–388 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lösslein A. K., Lohrmann F., Scheuermann L., Gharun K., Neuber J., Kolter J., Forde A. J., Kleimeyer C., Poh Y. Y., Mack M., Triantafyllopoulou A., Dunlap M. D., Khader S. A., Seidl M., Hölscher A., Hölscher C., Guan X. L., Dorhoi A., Henneke P., Monocyte progenitors give rise to multinucleated giant cells. Nat. Commun. 12, 2027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Poll T., Marchant A., Buurman W. A., Berman L., Keogh C. V., Lazarus D. D., Nguyen L., Goldman M., Moldawer L. L., Lowry S. F., Endogenous IL-10 protects mice from death during septic peritonitis. J. Immunol. 155, 5397–5401 (1995). [PubMed] [Google Scholar]
- 69.Latifi S. Q., O’Riordan M. A., Levine A. D., Interleukin-10 controls the onset of irreversible septic shock. Infect. Immun. 70, 4441–4446 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakamori Y., Park E. J., Shimaoka M., Immune deregulation in sepsis and septic shock: Reversing immune paralysis by targeting PD-1/PD-L1 pathway. Front. Immunol. 11, 624279 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shindo Y., McDonough J. S., Chang K. C., Ramachandra M., Sasikumar P. G., Hotchkiss R. S., Anti-PD-L1 peptide improves survival in sepsis. J. Surg. Res. 208, 33–39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patil N. K., Guo Y., Luan L., Sherwood E. R., Targeting immune cell checkpoints during sepsis. Int. J. Mol. Sci. 18, 2413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van de Laar L., Saelens W., De Prijck S., Martens L., Scott C. L., Van Isterdael G., Hoffmann E., Beyaert R., Saeys Y., Lambrecht B. N., Guilliams M., Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity 44, 755–768 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Scott C. L., Zheng F., De Baetselier P., Martens L., Saeys Y., De Prijck S., Lippens S., Abels C., Schoonooghe S., Raes G., Devoogdt N., Lambrecht B. N., Beschin A., Guilliams M., Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 7, 10321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mariano L. L., Rousseau M., Varet H., Legendre R., Gentek R., Coronilla J. S., Bajenoff M., Perdiguero E. G., Ingersoll M. A., Functionally distinct resident macrophage subsets differentially shape responses to infection in the bladder. Sci. Adv. 6, eabc5739 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomura M., Sakaue-Sawano A., Mori Y., Takase-Utsugi M., Hata A., Ohtawa K., Kanagawa O., Miyawaki A., Contrasting quiescent G0 phase with mitotic cell cycling in the mouse immune system. PLOS ONE 8, e73801 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Z., Gu Y., Chakarov S., Bleriot C., Kwok I., Chen X., Shin A., Huang W., Dress R. J., Dutertre C.-A., Schlitzer A., Chen J., Ng L. G., Wang H., Liu Z., Su B., Ginhoux F., Fate mapping via Ms4a3-expression history traces monocyte-derived cells. Cell 178, 1509–1525.e19 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Chua R. Y. R., Wong S. H., SNX3 recruits to phagosomes and negatively regulates phagocytosis in dendritic cells. Immunology 139, 30–47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Picelli S., Faridani O. R., Björklund Å. K., Winberg G., Sagasser S., Sandberg R., Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 80.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7