Abstract
The α2A adrenergic receptor (α2AAR) is a G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptor that mediates important physiological functions in response to the endogenous neurotransmitters norepinephrine and epinephrine, as well as numerous chemically distinct drugs. However, the molecular mechanisms of drug actions remain poorly understood. Here, we report the cryo–electron microscopy structures of the human α2AAR-GoA complex bound to norepinephrine and three imidazoline derivatives (brimonidine, dexmedetomidine, and oxymetazoline). Together with mutagenesis and functional data, these structures provide important insights into the molecular basis of ligand recognition, activation, and signaling at the α2AAR. Further structural analyses uncover different molecular determinants between α2AAR and βARs for recognition of norepinephrine and key regions that determine the G protein coupling selectivity. Overall, our studies provide a framework for understanding the signal transduction of the adrenergic system at the atomic level, which will facilitate rational structure-based discovery of safer and more effective medications for α2AAR.
Active α2A adrenergic receptor structures revealed distinct agonist activation and facilitate design of therapeutics for α2AAR.
INTRODUCTION
The adrenergic receptors (adrenoceptors) are a class of G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptors (GPCRs) that mediate the physiological actions of the endogenous catecholamines norepinephrine and epinephrine (1, 2). There are nine distinct adrenoceptors in mammalian species, which are grouped into three main classes on the basis of their amino acid sequences and biological properties: α1 (α1A, α1B, and α1D), α2 (α2A, α2B, and α2C), and β (β1, β2, β3) adrenoceptors (2, 3). The α2 adrenoceptors modulate a wide range of physiological functions, including the heart rate, blood pressure, regulation of blood glucose, insulin homeostasis, and analgesia, and they are implicated in mediating the presynaptic feedback inhibition of neurotransmitter release from noradrenergic nerve terminals (4–6).
All three α2 subtypes are traditionally known to couple primarily to the Gi/o protein family and inhibit the activity of adenylyl cyclase, resulting in a decrease in intracellular cyclic adenosine monophosphate (cAMP) levels (4, 7). However, there is evidence suggesting that α adrenoceptors can also functionally couple to Gs (7–10). In addition to G proteins, α2 adrenoceptors are also subject to agonist-dependent phosphorylation by G protein-coupled receptor kinases (GRKs), followed by the coupling of β-arrestins, which, in turn, leads to receptor internalization and G protein–independent signaling (11–13). Recent pharmacological studies have characterized a number of biased agonists that preferentially activate one of the signaling pathways in favor of the others for several GPCRs, including the μ-opioid receptor, the angiotensin II receptor type 1, and the dopamine D2 receptor (14). Since activation of multiple signaling pathways may lead to functional profiles conferring both beneficial and adverse effects, biased agonists may markedly reduce side effects (15). In the past decades, numerous α2 agonists have been developed and characterized, and some of them have, for example, been used in anesthesia, in pain management, and for the treatment of hypertension (16, 17). Functional and pharmacological studies show that structurally different α2 agonists (e.g., catecholamines, imidazolines, and azepines) display distinct efficacies in the activation of G proteins (18, 19), while the signaling behavior of these α2 agonists toward the β-arrestin pathway remains poorly understood.
In addition to downstream signaling diversity, both α2 and β adrenoceptors respond to norepinephrine and epinephrine. While the molecular mechanisms of drug action (including norepinephrine and epinephrine) on β adrenoceptors (β1AR and β2AR) have been extensively investigated (20–25), relatively little is known about the structural basis of drug action and activation mechanism of the α2 adrenoceptors, with the only reported structures being the inactive α2A adrenergic receptor (α2AAR) and α2CAR and the active α2BAR–G protein complex structures (9, 26, 27). Here, we report the high-resolution cryo–electron microscopy (cryo-EM) structures of the α2AAR-GoA complex bound to four chemically different agonists. In combination with structure-guided mutagenesis and functional analysis, our studies provide a basis for understanding the molecular mechanisms of ligand recognition, activation, and signaling of the α2AAR using pathway-specific assays.
RESULTS
Cryo-EM structures of the α2AAR-GoA complex bound to norepinephrine and imidazoline agonists
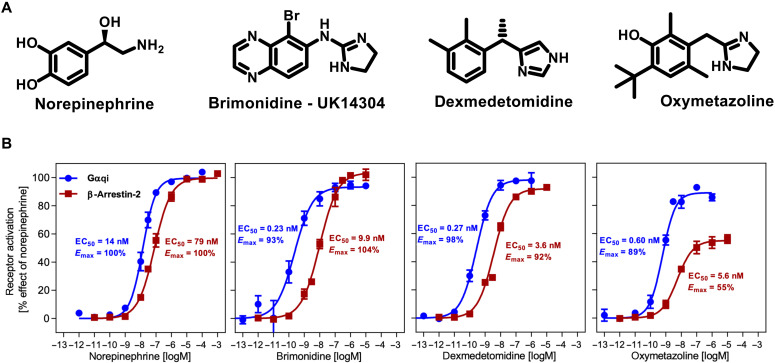
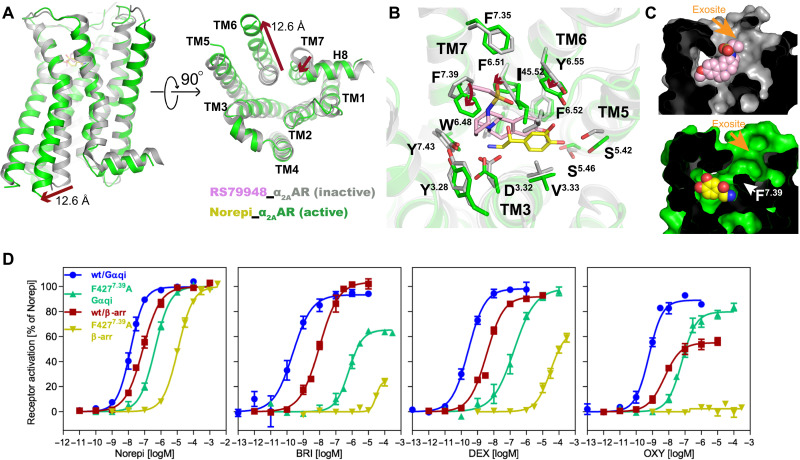
In this study, we determined the structure of the α2AAR-GoA complex bound to four chemically different agonists including the endogenous agonist norepinephrine and three imidazoline derivatives brimonidine, dexmedetomidine, and oxymetazoline (Fig. 1A; denoted as Norepi, BRI, DEX, and OXY, respectively). Both BRI and DEX are highly selective synthetic α2AAR agonists over the α1AAR subtype, but they have different therapeutic applications (16). BRI is commonly used to treat ocular hypertension and open-angle glaucoma, while DEX is an anxiety-reducing and pain-relieving drug, and most notable for its clinical use for providing sedation without compromising the airway and depressing respiration (16). OXY is generally available as a nasal decongestant that acts on both α2AAR and α1AAR. Previous functional and pharmacological studies of these α2AAR agonists have been largely restricted to the G protein–dependent signaling pathway, while very little is known regarding their signaling profiles in the β-arrestin pathway. Thus, we performed β-arrestin recruitment assays and compared the results to G protein activation by α2AAR in a well-established downstream signaling assay (IP-one) using a chimeric Gqi protein and Norepi as a reference agent (Fig. 1B and table S2). Our results show that all drugs display full agonist activity in the Gqi-mediated signaling, except for the slightly lower efficacy for OXY (89% of Norepi) (Fig. 1B). Of interest, OXY also shows only partial agonist properties of arrestin recruitment (55% Emax of Norepi), which is in contrast to the full agonist properties for the other three drugs, suggesting that OXY is a potential Gi/o-biased agonist at the α2AAR.
Fig. 1. Structure and function of α2AAR agonists.
(A) Chemical structures of norepinephrine (Norepi), brimonidine (BRI) or UK14304, dexmedetomidine (DEX), and oxymetazoline (OXY). (B) Concentration-response curves of different agonists toward G protein activation (blue curve) and β-arrestin-2 recruitment (red curve) measured by a cell-based Gqi-inositol phosphate accumulation assay and the PathHunter assay respectively. Data are presented as means ± SEM of 4 to 10 independent experiments with repeats in duplicate.
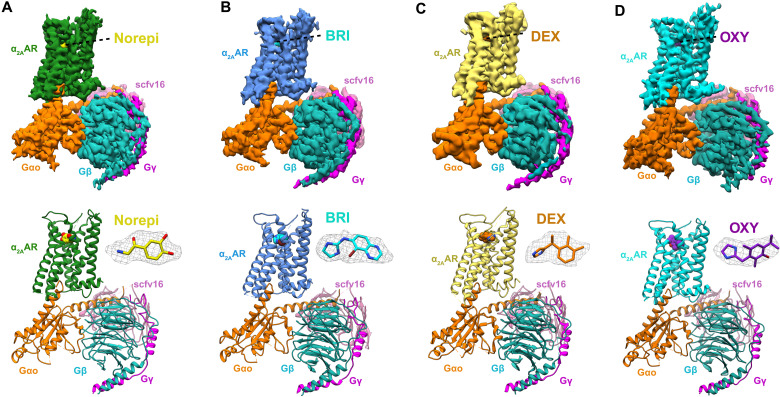
To gain insights into receptor recognition by different ligands, we assembled the complex of human α2AAR and the heterotrimeric GoA in the presence of the four different agonists. We kept the long intracellular loop 3 (ICL3) and used the wild-type receptor for our structural studies (fig. S1A). The complexes were prepared as described in Materials and Methods, and the agonist-bound α2AAR formed a stable nucleotide-free complex with GoA in the presence of an antibody fragment scFv16 (fig. S1B) (28). We obtained the cryo-EM maps of α2AAR-GoA-scFv16 in complex with the four different agonists with overall resolutions of 3.2 Å (Norepi), 3.0 Å (BRI), 3.6 Å (DEX), and 3.4 Å (OXY) (Fig. 2, figs. S2 and S3, and table S1). These maps allowed us to build the model of the transmembrane domain of the α2AAR, the GoA heterotrimer, and the scFv16. The maps also showed well-defined densities for each of the agonists in the orthosteric pocket (Fig. 2 and fig. S3). We did not observe clear densities for the first ~30 amino acids and the ICL3 of α2AAR, suggesting disordered conformations of these regions. The α-helical domain of Gαo was also poorly resolved because of its flexibility as observed in most cryo-EM structures of GPCR–G protein complexes (29, 30).
Fig. 2. Cryo-EM structures of Norepi, BRI-, DEX-, and OXY-bound α2AAR-GoA complexes.
Cryo-EM density maps and models of the α2AAR-GoA complex bound to Norepi (A), BRI (B), DEX (C), and OXY (D). The densities of the agonists (shown as sticks) are depicted as gray meshes. The maps are colored according to different subunits.
Orthosteric binding pocket of α2AAR
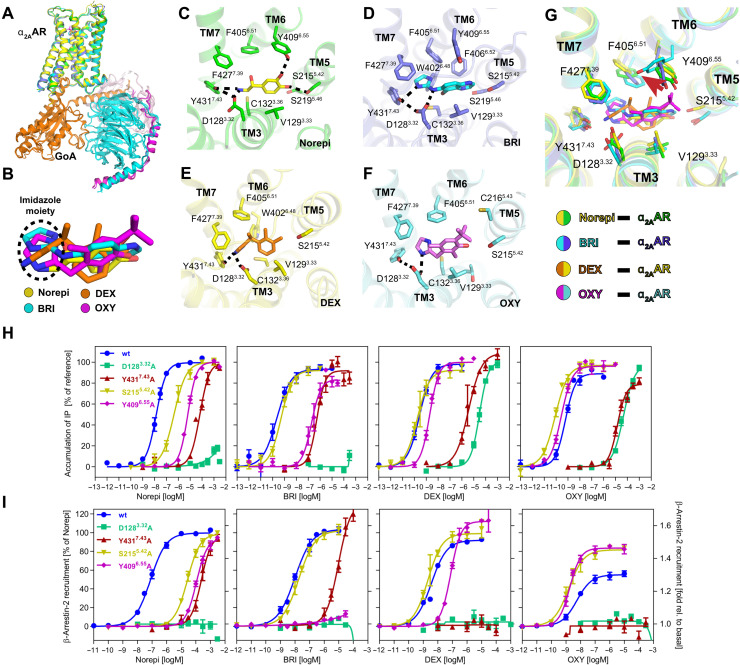
Despite the chemical diversity of the agonists, the α2AAR-GoA complexes bound to Norepi or the imidazoline agonists have very similar overall structures (Fig. 3A), and all four drugs occupy a similar pose in the orthosteric pocket (Fig. 3B). However, there are substantial differences for their interactions with surrounding residues. Figure 3 (C to G) shows detailed interactions of each agonist with the residues located in the α2AAR orthosteric pocket, which consists of the extracellular parts of TM3, TM5, TM6, and TM7.
Fig. 3. Orthosteric binding pocket of active α2AAR bound to different agonists.
(A) Alignment of four structures of α2AAR signaling complexes. (B) Superposition of Norepi, BRI, DEX, and OXY from cryo-EM structures. The imidazole moiety is highlighted by the dashed circle. (C to F) Detailed interactions of Norepi (C), BRI (D), DEX (E), and OXY (F) with α2AAR. Residues within 4 Å of agonist are shown in sticks. The polar interactions are indicated by black dashed lines. (G) Superposition of the α2AAR orthosteric binding pocket residues bound to four different agonists. (H and I) Concentration-response curves of different agonists for G protein activation (H) and β-arrestin-2 recruitment (I) for wild-type (wt) α2AAR and receptor mutants D1283.32A, Y4317.43A, S2155.42A, and Y4096.55A, respectively. Except for the activation of D1283.32A, which is normalized to DEX for G protein activation and relative to basal for arrestin recruitment, receptor activation is shown relative to the maximum effect of Norepi. Data are presented as means ± SEM of 3 to 11 independent experiments with repeats in duplicate.
Norepi binds to α2AAR primarily through two polar interaction networks. First, the amino-ethanol group of Norepi interacts with D1283.32 and Y4317.43 (superscript indicates Ballesteros-Weinstein numbering) via a salt bridge and a hydrogen bond (Fig. 3C). This polar interaction network with TM3 and TM7 is also observed for all three imidazoline agonists (Fig. 3, D to F). Consistently, mutations D1283.32A and Y4317.43A lead to a notable loss of activity for all drugs in both G protein signaling and arrestin recruitment (Fig. 3, H and I, and table S2). D3.32 is highly conserved in aminergic GPCRs, and structural and biochemical studies showed that it also plays pivotal roles in ligand recognition and activation of dopamine receptors and β adrenergic receptors (25, 31, 32). Notably, OXY can only form a direct interaction with D1283.32 but not Y4317.43 (Fig. 3F), resulting in a relatively weaker interaction with TM7. This may account for its partial agonism in arrestin recruitment (Figs. 1B and 3, H and I). Alanine replacement of Y4317.43 completely abolishes the arrestin recruitment induced by OXY and DEX, while it preserves G protein signaling at high ligand concentration, suggesting a potential role of Y4317.43 in mediating pathway-specific signaling bias of α2AAR (Fig. 3, H and I, and table S2).
Second, the para- and meta-phenolic hydroxyls of the Norepi phenyl ring form hydrogen bonds with S2155.42 and Y4096.55, respectively, which is in accordance with the recent docking pose (9). Consistently, mutation of S2155.42A and Y4096.55A markedly reduced the activity of Norepi (Fig. 3, H and I, and table S2). In contrast, BRI is not able to form stable hydrogen bonds with either S2155.42 or Y4096.55, in spite of the polar nitrogen atoms of the bicyclic aromatic moiety (Fig. 3D). Indeed, mutation S2155.42A has little effect on the activity of BRI. However, Y4096.55A reduces the EC50 by ~800-fold on Gqi signaling and completely abolished the arrestin recruitment induced by BRI (Fig. 3, H and I, and table S2). This is likely due to breakage of the π-π interactions between Y4096.55 and the bicyclic aromatic moiety of BRI. It is also possible that there is a water-mediated hydrogen bond between BRI and Y4096.55. Besides, the greater reduction of activity in arrestin recruitment than G protein activation for Norepi and BRI suggests that Y4096.55 also plays a potential role in regulating pathway-specific signaling of α2AAR.
The phenyl ring of DEX lacks the two hydroxyls of catecholamines; instead, only two methyl groups and no polar substituents are present. As a result, DEX interacts with TM5 and TM6 mainly through hydrophobic interactions (Fig. 3E), which is similar to what was observed in the α2BAR (27). Mutagenesis data show that S2155.42A has no influence for the potency of DEX, and Y4096.55A reduces its potency less compared with Norepi and BRI (Fig. 3, H and I, and table S2). Although OXY comprises a phenolic hydroxyl, it cannot form a stable hydrogen bond with S2155.42 (Fig. 3F). The Y4096.55 side chain displays a unique conformation in the OXY-bound structure by rotating around 70° toward TM7, likely due to the presence of the tert-butyl substituent of OXY (Fig. 3G). This unique bulky hydrophobic group may contribute to the partial agonist properties for arrestin recruitment of OXY because of the incompatibility with the hydrophilic properties of Y4096.55 and S2155.42. Indeed, both S2155.42A and Y4096.55A mutations lead to a slightly increased activity of OXY, in particular, the efficacy in arrestin recruitment is markedly enhanced, which is different from what is observed for other agonists (Fig. 3, H and I, and table S2). The agonist-dependent functional impacts of these mutations, particularly for Y4096.55, suggest important but complicated roles of these residues in modulating biased signaling of α2AAR. Notably, recent structural, functional, and computational studies also suggested that the residue in position 6.55 plays an important role in regulating bias for the β2AR (24, 33). It should be noted that the functional effects of these mutations on the α2AAR are not due to changes in receptor expression, since receptor mutants S2155.42A, Y4096.55A, and D1283.32A show surface expression comparable to wild type and expression is only slightly reduced for Y4317.43 (fig. S4).
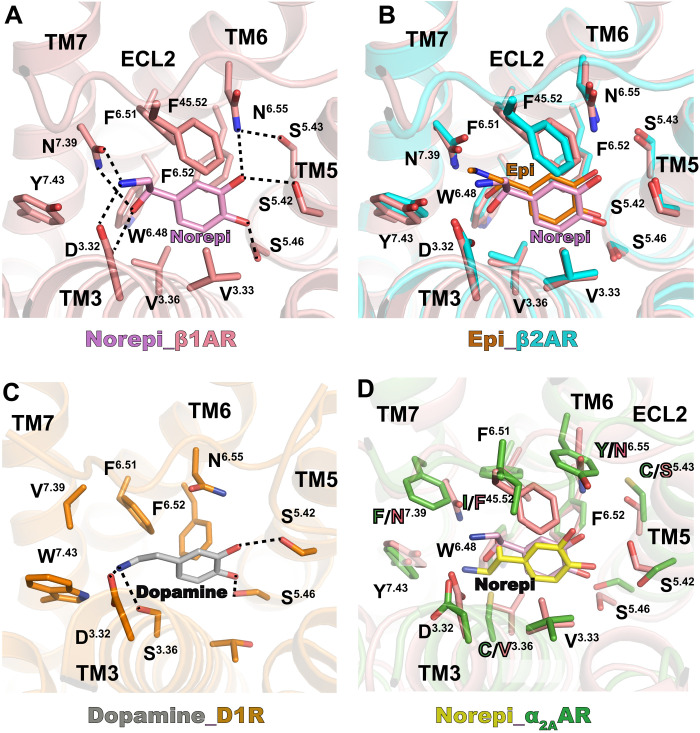
Recognition of Norepi by α2AAR and β adrenoceptors
Different levels of Norepi in human body could result in distinct physiological behaviors because of its highest affinity for α2 adrenoceptors and lowest affinity for β adrenoceptors (5, 6). It is therefore of interest to understand the molecular basis of Norepi recognition by the two adrenergic subfamilies that have opposite functional effects. Comparison of Norepi-bound β1AR and epinephrine-bound β2AR shows identical binding pockets (Fig. 4, A and B). Norepi interacts with β adrenoceptors mainly through two polar networks, similarly as observed in α2AAR (Figs. 3C and 4A). The hydrogen bonds with the conserved D3.32 and S5.42 are maintained in β adrenoceptors, while the para-hydroxyl of Norepi forms additional hydrogen bond with S5.46. This D3.32-S5.42-S5.46 motif is also found to be involved in polar interactions between dopamine and the D1 dopamine receptor (Fig. 4C) (32). Although S5.46 is not involved in the hydrogen-bonding interaction with Norepi in α2AAR, previous mutagenesis data showed that S5.46A markedly reduced the potency of epinephrine to inhibit forskolin-stimulated cAMP accumulation (4), suggesting a conserved role of the D3.32-S5.42-S5.46 motif also in the α2AAR.
Fig. 4. Comparison of Norepi binding for α2A and β adrenergic receptors.
(A) Detailed interactions of Norepi with β1AR [Protein Data Bank (PDB) code: 7BU6]. Residues within 4 Å of agonist are shown in sticks. The polar interactions are indicated by black dashed lines. (B) Superposition of orthosteric pockets of β1AR-Norepi and β2AR-Epi (epinephrine) (PDB code: 4LDO). (C) Detailed interactions of dopamine with D1R (PDB code: 7CKZ). The polar interactions are indicated by black dashed lines. (D) Superposition of orthosteric pockets of β1AR-Norepi and α2AAR-Norepi. Residues within 4 Å of agonist are shown in sticks.
On the other hand, the S5.43 in β adrenoceptors, which is a cysteine in α2AAR, participates in the polar network through hydrogen bonding with N6.55 (Fig, 4, A and B). The two aromatic residues in TM6 (Y6.55) and TM7 (F7.39) of α2AAR are also replaced with two asparagine residues (N6.55 and N7.39) in β adrenoceptors, both of which are involved in the polar interaction networks with Norepi (Fig. 4, A and B). Simulation studies showed that the direct interaction of the amino-ethanol of Norepi and other β adrenergic compounds with N7.39 was very prominent (33), while this polar interaction with residue 7.39 almost gets lost in α2AAR because of the sequence difference (Fig. 4D). By contrast, the amino-ethanol of Norepi forms direct interaction with Y7.43 in α2AAR (Fig. 3C), which is probably an analog to the polar interaction with N7.39 in β adrenoceptors. Notably, the two aromatic residues (Y6.55 and F7.39) in α2AAR, together with F6.51, F6.52, Y7.43, and W6.48, form an aromatic cage, which may play key roles in high-affinity Norepi binding (Fig. 4D) (9). Recent functional studies showed that the F7.39N mutation completely abolishes the activity of Norepi, and the Y6.55N mutation can substantially reduce the EC50 of Norepi for α2AAR (9). Moreover, the bulkier F45.52 in ECL2 of the β adrenoceptors is a smaller isoleucine in the α2AAR. Although I 45.52 makes weaker hydrophobic contacts with the phenyl ring of norepinephrine than F45.52, it may result in different conformational dynamics along the entrance pathway in α2AAR, contributing to the higher affinity of Norepi (22).
Activation of the α2AAR
Although the first inactive structure of the β adrenoceptor has been determined in 2007 (34), the inactive structures of α2 adrenoceptors were solved only recently (9, 26), allowing us to analyze the conformational changes of α2AAR from inactive to active states. The outward movement of TM6 (12.6 Å) and inward movement of TM7, which are hallmarks of GPCR activation (35, 36), also occur upon α2AAR activation (Fig. 5A). In the orthosteric pocket, the antagonist RS79948 seems to occupy a distinct pose compared with agonists (Fig. 5B). The antagonist RS79948 binds at a further outside position than Norepi and extends to an exosite to form interactions with the extracellular loops (Fig. 5, B and C). The side chains of several residues undergo large rearrangements upon receptor activation, especially for F4277.39 (Fig. 5B). The recent α2AAR structure bound to the partial agonist RES also shows similar changes in the orthosteric pocket (9). F4277.39 was proposed to serve as a switching lid of an aromatic cage (together with F6.51, F6.52, Y7.43, and W6.48) upon agonist binding (9). Indeed, F4277.39 displays similar active conformations when bound to different agonists (Fig. 3G), suggesting a pivotal role of the F4277.39 conformational change in receptor activation. Likewise, alanine replacement of F4277.39 markedly impaired the functions of all agonists, and the imidazoline drugs displayed greater reduction of activities than Norepi (Fig. 5D and table S2). This is consistent with the observation that F4277.39 has stronger contacts to the imidazoline ring than the amino-ethanol group of Norepi (Fig. 3G).
Fig. 5. Comparison of inactive and active α2AAR.
(A) Structural comparison of inactive α2AAR bound to RS79948 and active α2AAR bound to Norepi, with changes highlighted as red arrows. The distance is calculated between positions of Cα of residue 6.29 in TM6. (B) Conformational changes within the orthosteric pocket are shown from the extracellular side, with changes highlighted as red arrows. (C) Cross sections of α2AAR bound to antagonist and agonist are shown, with the interior in black and the exosite highlighted. (D) Concentration-response curves of F4277.39 mutant for different agonists toward G protein activation and β-arrestin-2 recruitment. Data are presented as means ± SEM of 4 to 10 independent experiments with repeats in duplicate.
Notably, the binding poses for agonists and antagonists are more overlapped for β adrenoceptors (fig. S5A), and most of the interactions with the receptor are identical for agonists and antagonists (20). Moreover, the residue at position 7.39 is a smaller asparagine in β adrenoceptors and shows similar side-chain conformations when bound to different ligands (fig. S5A). This asparagine disrupts the aromatic cage for β adrenoceptors (9). This disruption seems to allow a few β adrenoceptor agonists with a longer tail, such as salmeterol, to extend to an exosite in the extracellular vestibule, resulting in great subtype selectivity (fig. S5B) (24). Whether the extracellular site can be used for novel α2AAR agonist design needs further investigation.
In spite of these differences in conformational changes of the orthosteric pocket and in ligand recognition, the α2AAR shows similar activation-associated structural changes to the β adrenoceptors in residues that connect the orthosteric pocket to the cytoplasmic surface. The conformational rearrangements of several conserved microswitches, representing the PIF, NPxxY, and DRY motifs, are also similar to that of β adrenoceptors (fig. S5, C to E). These structural features suggest a conserved mechanism for the allosteric coupling between the orthosteric pocket and G protein coupling domain for adrenoceptors.
G protein coupling interface
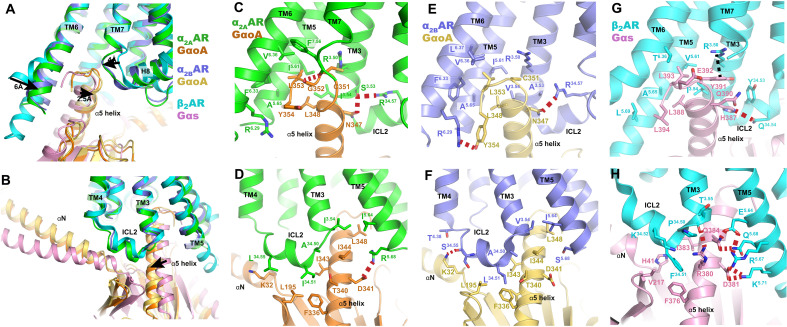
Given that α2 and β2 adrenoceptors have adverse physiological functions, with α2 inhibiting the activity of adenylyl cyclase via Gi/o and β2AR stimulating it via Gs, they have long served as model systems for studying G protein coupling selectivity. Figure 6 shows the comparison of the coupling interface for GoA with α2AR (α2AAR and α2BAR) and Gs with β2AR. Here, we used the Norepi-bound α2AR-GoA for comparison with other adrenoceptors, as the interfaces with GoA for different agonist-bound α2AR are almost identical (fig. S6A).
Fig. 6. G protein binding interface.
(A and B) Superposition of the G protein coupling interfaces of α2AAR-GoA, α2BAR-GoA, and β2AR-Gs complexes, using receptor for alignment. (C to H) Detailed interactions of α2AAR with Goα (C and D), α2BAR with Goα (E and F), and β2AR with Gsα (G and H). The polar interactions are indicated by red dashed lines.
The overall interaction interfaces for α2AAR-GoA and α2BAR-GoA are almost identical (Fig. 6, A and B). The major interaction interface is composed of hydrophobic residues I343G.H5.15, I344G.H5.16, L348G.H5.20, and L353G.H5.25 (superscript indicates CGN numbering system) of the Gαo cluster with surrounding hydrophobic residues from TM3, TM5, TM6, and ICL2. However, several differences in polar interactions with the GoA heterotrimer were observed between α2AAR and α2BAR. For example, the last amino acid Y354G.H5.26 of Gαo adopts a distinct side-chain conformation and forms a hydrogen bond with R6.29 in α2BAR. In contrast, this interaction is not observed in α2AAR (Fig. 6, C to F). A hydrogen bond was observed between the backbone carbonyl of F7.56 and G352G.H5.24 of Gαo. A similar polar interaction was observed between rhodopsin and Gi (37), while this interaction pattern is not present in α2BAR. In addition, N347G.H5.19 of Gαo participates in a hydrogen bond network with S3.53 and R34.57 in α2AAR. However, position 3.53 is an alanine in α2BAR and N347G.H5.19 of Gαo directly forms a hydrogen bond with R34.57. R5.68 in α2AAR, which is a serine in α2BAR, also forms a salt bridge with D341G.H5.13 of Gαo. On the other hand, both ICL1 and ICL2 of α2BAR form hydrogen bonds with the Gβ subunit and the αN helix via R12.48 and S34.55, respectively (Fig. 6F and fig. S6B), while these polar interactions were not observed in the α2AAR-GoA complex (fig. S6C). These distinct interactions between α2AAR-GoA and α2BAR-GoA suggest the versatility of Go coupling with GPCRs.
The comparison of α2ARs-GoA and β2AR-Gs complex structures shows that the relative positions for Gαo and Gαs are different, with the extreme C terminus of Gαo helix α5 shifting 2.5 Å toward TM7 and a large displacement of the αN helix. This is associated with the smaller outward displacement of TM6 and inward movement of TM7 in α2ARs, resulting in a smaller cavity formed by TM3/5/6/7 for GoA than for Gs (Fig. 6, A and B). This structural feature is also observed for several other Gi/o-coupled receptors and is believed to be one of the key determinants for the coupling specificity of Gi/o and Gs (29, 30, 37). However, the orientations of Gi/o proteins coupled to different receptors are also divergent. For example, the extreme C terminus of Gαi helix α5 of M2R and μOR shifts toward TM7, while the displacement of TM6 for M2R and μOR is even smaller than that of the α2AR (fig. S6D).
In contrast to the relatively dispersive and sparse polar interactions between α2ARs and GoA, hydrophilic residues R380G.H5.12, D381G.H5.13, and Q384 G.H5.16 on one side of the α5 helix of the Gαs protein form a cluster of hydrogen-bonding interactions with hydrophilic residues from TM3 and TM5 of β2AR (Fig. 6, G and H). Notably, because of the shift of the α5 helix of Gαo, its interaction with the TM5 of α2ARs is much weaker than that of the β2AR-Gs complex (Fig. 6, C to F). Therefore, the TM5 intracellular domain may be essential for the coupling of Gs but not for Gi/o. This observation is consistent with previous functional studies, showing that deletion of residues of the TM5 intracellular end of α2AAR entirely ablated the activation of Gs while the activity of Gi was retained (38). To further investigate the role of TM5 in the coupling specificity to Gi/o and Gs, we made chimeric constructs by swapping the intracellular domain of TM5 (positions 5.60 to 5.71) between α2AAR and β2AR. However, we did not observe a switch of coupling selectivity for the two chimeric receptors α2AAR- β2-TM5 and β2AR-α2A-TM5 (fig. S7). Although α2AAR- β2-TM5 showed decreased Gi coupling activity (~3-fold), the Gs activity also decreased substantially (~10-fold) compared with that of the wild-type α2AAR (fig. S7, B and C). By contrast, β2AR-α2A-TM5 showed markedly increased activity for both Gi and Gs coupling (over 100-fold) compared with that of the wild-type β2AR (fig. S7, B and C). These data suggest that the TM5 intracellular domain plays important roles in G protein coupling and that the amino acid sequence of α2AAR has stronger ability to couple with both G proteins than that of β2AR, despite the weaker interactions with G protein (Fig. 6, C to F). It is possible that there are intermediate conformational states of the α2AAR-GoA signaling complex where the TM5 forms strong interactions with the Gαo. Such an intermediate state has been observed in other GPCR–G protein signaling complex (39). Moreover, these data also suggest that the TM5 intracellular domain is not sufficient to determine the coupling specificity of α2AAR and β2AR. It is likely that TM5, TM6, and other regions may act simultaneously to determine the G protein coupling specificity and that a possible intermediate state may also play important roles in coupling specificity. It is worthy of note that these functional studies were performed with chimeric Gqi/s proteins and thus may not be able to fully recapitulate the natural behavior of Gi or Gs.
In addition to TM5 and TM6, another major difference between the α2ARs-GoA and β2AR-Gs structures lies in the interaction between ICL2 and the hydrophobic patch formed by the α5 and αN helices of the G protein. The small residue L/I34.51 in α2ARs is involved in weak hydrophobic interactions with L195G.S3.01, F336G.H5.08, and I343G.H5.15 from GoA, while in the case of β2AR, the bulkier F34.51 forms stronger interactions with Gs (Fig. 6, B, F, and H). These observations are similar to several other GPCR-Gi/o complex structures, suggesting that ICL2 may be less important for the coupling with Gi/o but is essential for Gs activation (29, 30, 40). Indeed, previous mutagenesis studies showed that F34.51A mutation can abolish the activity of β2AR to activate Gs, while this mutation has little effect on its Gi activity (41). Recent functional studies also showed that the I34.51A mutation of α2AAR can maintain the activation of Gi while abolishing Gs signaling (9).
DISCUSSION
Adrenoceptors are model systems for studying GPCR signaling and important drug targets for a wide variety of diseases. In more recent years, agonists of α2ARs have been found to have broad applications in the field of anesthesia and pain management because of their reduced propensity to induce respiratory depression. In the present study, we reported four cryo-EM structures of α2AAR signaling complexes bound to the endogenous agonist Norepi and three widely used imidazoline drugs. These structures, in combination with mutagenesis and functional studies, reveal different molecular determinants that recognize Norepi and imidazoline agonists, and key residues including Y4317.43, S2155.42, and particularly Y4096.55, that may lead to a signaling bias of OXY. By comparison with the inactive α2AAR, these structures also reveal a key aromatic switch involving F4277.39 that plays essential roles in receptor activation, which is consistent with recent structural and biochemical studies (9). Comparison with β adrenoceptors shows that the α2AAR shares a conserved mechanism for the allosteric coupling between the extracellular ligand binding pocket and the intracellular G protein binding site. However, the activation-associated structural changes in the orthosteric pocket differ between α2AAR and β adrenoceptors. There are also substantial differences in the molecular mechanism of Norepi recognition by α2 and β adrenoceptors because of several nonconserved residues such as F7.39 and Y6.55. These signaling complex structures also reveal key regions, including TM5, TM6, and ICL2, that may act together to determine the functional selectivity for Gi/o and Gs for α2ARs and β2AR, respectively. Nevertheless, the molecular mechanisms of signaling promiscuity at Gi/o and Gs for α2AR and β2AR are not well elucidated by the current structures. Further structural and biophysical studies on α2ARs-Gs and β2AR-Gi/o complexes are needed to better understand this dual G protein coupling effect of adrenoceptors. Together, our studies provide a structural framework for understanding the signal transduction of the adrenergic system and are expected to facilitate structure-based drug discovery targeting α2ARs.
MATERIALS AND METHODS
Expression and purification of α2AAR
The human wild-type α2AAR was cloned to pFastBac vector with an N-terminal FLAG tag and a C-terminal histidine tag to express in Sf9 insect cells. Recombinant baculovirus for insect cell expression was made using the Bac-to-Bac system. Sf9 cells were grown in SIM SF Medium (Sino Biological Inc.) and were infected with recombinant baculovirus containing the α2AAR gene at a density of 4 × 106 cells ml−1 in the presence of 5 μM rauwolscine. After 48 hours of infection at 27°C, the cells were spun down and cell pellets were stored at −80°C until use.
Thawed cell pellets were resuspended in hypotonic lysis buffer [10 mM tris, 1 mM EDTA, 5 μM rauwolscine, leupeptin (2.5 μg ml−1), and benzamidine (160 μg ml−1)]. Cell membranes were then spun down and solubilized with a buffer consisting of 20 mM Hepes (pH 7.5), 500 mM NaCl, 1% n-Dodecyl-b-D-Maltopyranoside (DDM), 0.2% sodium cholate, 0.03% Cholesteryl Hemisuccinate (CHS), 5 μM rauwolscine, leupeptin (2.5 μg ml−1), and benzamidine (160 μg ml−1). Nickel-NTA sepharose was added to the solubilized receptor and rotated for 2 hours at 4°C. The resin was spun down and washed in batch for three times with a buffer composed of 20 mM Hepes (pH 7.5), 500 mM NaCl, 0.1% DDM, 0.02% sodium cholate, 0.03% CHS, 5 μM rauwolscine, leupeptin (2.5 μg ml−1), and benzamidine (160 μg ml−1). The washed resin was poured into a glass column, and the receptor was eluted in the wash buffer supplemented with 250 mM imidazole.
The Ni-NTA chromatography–purified receptor was immobilized using anti-flag M1 affinity resin and was extensively washed with a buffer containing 20 mM Hepes (pH 7.5), 500 mM NaCl, 0.1% DDM, 0.02% sodium cholate, 0.003% CHS, 2 mM CaCl2, and supplemented with 100 μM agonist. The receptor was subsequently eluted with the same buffer supplemented with flag peptide (0.2 mg ml−1) and 5 mM EDTA. The eluted receptor (2 to 3 ml) was concentrated to 500 μl using a 50-kDa molecular weight cutoff Millipore concentrator and Sorvall Legend Micro 17 Microcentrifuge (Thermo Fisher Scientific). The concentrated protein was then loaded on a Superdex 200 size exclusion column (GE Healthcare) with a buffer containing 20 mM Hepes (pH 7.5), 500 mM NaCl, 0.1% DDM, 0.02% sodium cholate, 0.002% CHS, and 100 μM agonist. The monomeric peak fractions of the receptor were collected and concentrated to ~20 mg ml−1 and stored at −80°C until use.
Expression and purification of GoA and scFv16
The GoA protein, which represents the heterotrimeric complex of the guanine nucleotide–binding protein Go subunit α (Gαo), Gβ, and Gγ subunits, was expressed and purified as below. Human Gαo and Gβ1γ2 were synthesized in General Biology company (Hefei). Gαo was cloned into pFastBac vector, Gβ1 with 3C protease-cleavable 6xHis-tag, and Gγ2 was cloned into pFastBac_Dual vector. The GoA was expressed in HighFive insect cells grown in SIM HF Medium (Sino Biological Inc.). Cells were grown to a density of 3 million per milliliter and infected with Gαo and Gβ1γ2 baculovirus at a ratio of 10 to 20 ml liter−1 and 1 to 2 ml liter−1, respectively. After 48 hours of incubation, the infected cells were harvested by centrifugation and stored at −80°C until use.
Thawed cell pellets were resuspended in lysis buffer [10 mM tris (pH 7.5), 0.1 mM MgCl2, 5 mM β-mercaptoethanol (β-ME), 10 μM guanosine diphosphate (GDP), leupeptin (2.5 mg ml−1), and benzamidine (160 mg ml−1)] and stirred at room temperature for 15 min. Cell membranes were spun down and resuspended with solubilization buffer [20 mM Hepes (pH 7.5), 100 mM NaCl, 1% sodium cholate, 0.05% DDM, 5 mM MgCl2, 2 ml CIP, 5 mM β-ME, 15 mM imidazole, 10 μM GDP, leupeptin (2.5 mg ml−1), and benzamidine (160 mg ml−1)] using a Dounce homogenizer. The samples were stirred at 4°C for 40 min and then centrifuged for 30 min to remove insoluble debris.
Ni-NTA resin preequilibrated in solubilization buffer was added to the supernatant and shaken for 2 hours at 4°C. After incubation, the Ni-NTA resin was spun down and poured into a glass column and washed with 50 ml of solubilization buffer. The heterotrimeric GoA was gradually exchanged into E2 buffer [20 mM Hepes (pH 7.5), 50 mM NaCl, 0.1% DDM, 1 mM MgCl2, 5 mM β-ME, 10 μM GDP, leupeptin (2.5 mg ml−1), and benzamidine (160 mg ml−1)]. The protein was then eluted with E2 buffer supplemented with 250 mM imidazole.
The sample was then dephosphorylated by treating with 5 μl lambda phosphatase (supplemented with 1 mM MnCl2 for activity; New England Biolabs) and incubated at 4°C for 30 min. The Ni-NTA chromatography–purified GoA was further purified with a MonoQ column (GE Healthcare). The peak fractions of the MonoQ chromatography were collected and exchanged to E3 buffer [20 mM Hepes (pH 7.5), 100 mM NaCl, 0.1% DDM, 1 mM MgCl2, 10 μM GDP, and 50 μM Tirs(2-carboxyethy)phosphine (TCEP)] by repeated concentration and dilution using a 50-kDa molecular weight cutoff Millipore concentrator. The concentrated heterotrimeric GoA was aliquoted, flash frozen in liquid nitrogen, and stored at −80°C before use.
The scFv16 gene was cloned into pFastBac vector with a C-terminal histidine tag and expressed in secreted form in HighFive insect cells using the Bac-to-Bac system (Thermo Fisher Scientific). The protein was purified with Ni-NTA column followed by size exclusion chromatography using a Superdex 200 Increase 10/300GL column (GE Healthcare). The monomeric peak fractions were collected, concentrated, and stored at −80°C until use.
α2AAR–GoA complex formation
The complex of α2AAR with heterotrimeric GoA was formed in a buffer composed of 20 mM Hepes (pH 7.5), 100 mM NaCl, 0.1% DDM, 1 mM MgCl2, 10 μM GDP, and 100 μM agonist. The α2AAR-GoA complex was then treated with 50 U of apyrase (NEB) on ice overnight and exchanged on an anti-Flag M1 column into a buffer containing 20 mM Hepes (pH 7.5), 100 mM NaCl, 0.0075% lauryl maltose neopentyl glycol (MNG, NG310 Anatrace), 0.0025% GDN (GDN101, Anatrace), 0.001% CHS, 100 μM agonist, and 2 mM CaCl2 in a stepwise manner. After elution by adding 5 mM EDTA and Flag peptide (0.2 mg ml−1), the complex was concentrated and incubated with excess scFv16 for 1 hour on ice and then loaded onto Superdex 200 Increase 10/300GL column (GE Healthcare) with a running buffer of 20 mM Hepes (pH 7.5), 100 mM NaCl, 0.00075% MNG, 0.00025% GDN, 0.0001% CHS, and 100 μM agonist. The monomeric peak fraction of the α2AAR-GoA complex was collected and concentrated to ~10 mg/ml for cryo-EM.
Cryo-EM sample preparation and data collection
The gold film (42) (UltraAuFoil, 300 mesh, R1.2/1.3) or amorphous alloy film (43) (300 mesh, R1.2/1.3, Zhenjiang Lehua Electronic Technology Co. Ltd.) was glow discharged with air for 40 s at 15 mA at easiGlow Glow Discharge Cleaning System (PELCO, USA). Three microliters of purified complex sample was dropped onto the grid and then blotted for 3.5 s with blotting force 0 and plunged into liquid ethane cooled by liquid nitrogen using Vitrobot Mark IV (Thermo Fisher Scientific, USA). Cryo-EM datasets were collected with the 300-kV Titan Krios Gi3 microscope. The raw movies were collected by Gatan K3 BioQuantum Camera at magnification of 105,000, with a pixel size of 0.85 Å. Inelastically scattered electrons were excluded by a GIF Quantum energy filter (Gatan, USA) using a slit width of 20 eV. The movies were acquired with the defocus range of −1.0 to −2.0 μm with total exposure time of 2.5 s fragmented into 50 frames and with the dose rate from 17.36 to 17.65 e per pixel per second. SerialEM (44) was used for semiautomatic data acquisition.
Cryo-EM data processing, model building, and refinement
The image stacks were collected and subjected for motion correction using MotionCor2 (45). Contrast transfer function parameters were estimated by CTFFIND4 (46), implemented in RELION3.1 (47). Particles were auto-picked from micrographs by RELION and then subjected to two-dimensional (2D) classification using cryoSPARC (48). Selected particles with an appropriate 2D average from 2D classification were further subjected to 3D classification using RELION with an initial model of the α2BAR-GoA-scFv16 complex (27). Eventually, particles with high-resolution 3D average were selected from 3D classification, which result in a map with initial resolution of near atomic level by 3D auto-refinement. The refined particles were subjected to CTF refinement to update per-particle defocus and per-micrograph astigmatism. Bayesian polishing, which means local motion correction, was then performed on the refined particles. Another round of CTF refinement was performed on the shiny particles generated from polishing followed by another round of 3D auto-refinement and, lastly, yielded a series of final map with resolution from 3.0 to 3.6 Å after postprocessing determined by gold standard Fourier shell correlation using the 0.143 criterion. The local resolution map was calculated from the Bsoft package (49) using two unfiltered half maps.
Functional assays
The human wild-type α2AAR, β2AR, and respective receptor mutants or chimeras all carrying an N-terminal HA-signal sequence and a FLAG-tag (50) were cloned to pCDNA3.1 for G protein activation assays or fused to the ARMS2-PK2 sequence and cloned to pCMV (DiscoverX, Eurofins) for β-arrestin-2 recruitment assays, respectively, using polymerase chain reaction and Gibson Assembly (New England Biolabs) (51). Sequence integrity was verified by DNA sequencing (Eurofins Genomics).
The determination of receptor-mediated G protein signaling by wild-type and mutant receptors was performed applying an IP accumulation assay (IP-One HTRF, Cisbio, Codolet, France) according to the manufacturer’s protocol and in analogy to previously described protocols (52, 53). In brief, human embryonic kidney (HEK) 293T cells were cotransfected with the cDNA coding for α2AAR, α2AAR-D1283.32A, α2AAR-S2155.42A, α2AAR-Y4096.55A, α2AAR-F4277.39A, or α2AAR-Y4317.43A, α2AAR-β2-TM5, or β2AR-α2A-TM5, respectively, and the hybrid G protein Gαqi or Gαqs (Gαq protein with the last five amino acids at the C terminus replaced by the corresponding sequence of Gαi or Gαs, respectively; gift from The J. David Gladstone Institutes, San Francisco, CA) in a ratio of 1:2 and transferred into 384-well microplates. On the day of the experiment, incubation started by adding the agonists for 90 min (α2AAR, α2AAR-S2155.42A, and α2AAR-F4277.39A) or 150 min (α2AAR-D1283.32A, α2AAR-Y4096.55A, and α2AAR-Y4317.43A), or 120 min (for the direct comparison of α2AAR and β2AR with the α2AAR-β2-TM5 and β2AR-α2A-TM5 chimeras), respectively. Accumulation of second messenger was stopped by addition of the detection reagents (IP1-d2 conjugate and anti-IP1cryptate TB conjugate). After a further 60 min, time-resolved fluorescence resonance energy transfer (FRET) was measured using the Clariostar plate reader (BMG, Ortenberg, Germany). FRET signals were calculated as the ratio of emissions at 665 and 620 nm and ligand-induced changes in FRET (deltaFRET) normalized to the maximum effect of norepinephrine (100%) and vehicle (0%). For the mutant α2AAR-D1283.32A, the maximum effect of dexmedetomidine was used as 100%. Normalized concentration-response curves from 4 to 11 experiments each done in duplicates were analyzed using the algorithms for four-parameter nonlinear regression implemented in PRISM 8.0 (GraphPad Software, USA) to derive EC50 and Emax values.
Determination of receptor stimulated β-arrestin-2 recruitment was performed applying the PathHunter assay (DiscoverX, Birmingham, UK) measuring fragment complementation of β-galactosidase as described (53, 54). In detail, HEK293T cells were cotransfected with α2AAR wild-type or mutant receptor each fused to the ARMS2-PK2 fragment for enzyme complementation and GRK2 in a ratio of 1:1 and transferred into 384-well microplates. Measurement was started by incubating the cells with the agonists for 60 min (α2AAR, α2AAR-S2155.42A, α2AAR-Y4096.55A, α2AAR-F4277.39A, or α2AAR-Y4317.43A) or 150 min (α2AAR-D1283.32A), respectively. Chemoluminescence was monitored with a Clariostar plate reader and analyzed by normalizing the raw data relative to basal activity (0%) and the maximum effect of norepinephrine (100%). For the mutant α2AAR-D1283.32A, normalization was done by analyzing the raw data as fold change relative to basal (=100%). Three to nine individual experiments each done in duplicate were analyzed by nonlinear regression, applying the algorithms in Prism 8.0 (GraphPad, San Diego, CA) to get dose-response curves representing average EC50 and Emax values.
Receptor surface expression was analyzed using an enzyme-linked immunosorbent assay directed against the N-terminal FLAG-tag as previously described (55). In brief, HEK293T cells were diluted to a density of 2 × 105 cells ml−1 in Dulbecco’s modified Eagle’s medium/F12 supplemented with 10% fetal bovine serum and transfected with wild-type α2AAR, β2AR, or respective receptor mutants in pcDNA3.1 or pCMV using polyethyleneimine (PEI) at a 3:1 PEI/DNA ratio. Per 1.2 ml of cell suspension, 200 ng of α2AAR and 800 ng of single-stranded salmon sperm DNA (Sigma-Aldrich) were used. Fifty thousand cells per well were transferred to a 48-well plate coated with poly-d-lysine and incubated at 37°C and 5% CO2 for 48 hours. Cells were fixated using 4% paraformaldehyde (200 μl, 10 min, room temperature), washed once (wash buffer, 300 μl, 150 mM NaCl, 25 mM tris, pH 7.5), and blocked for 60 min [skim milk powder (30 g liter−1) in wash buffer], before anti-FLAG mouse immunoglobulin G (IgG; Sigma-Aldrich; 1:4000 in blocking solution, 200 μl) was added. After 60 min, wells were washed twice (300 μl) and blocked for a further 60 min, before 200 μl of anti-mouse rabbit IgG-HPR (Sigma-Aldrich; 1:20,000 in blocking solution) was added. After 60 min, cells were washed thrice, before the addition of substrate buffer (300 μl, 2.8 mM o-phenylenediamine in 35 mM citric acid, 66 mM Na2HPO4, pH 5.0). Reactions were kept in the dark for 5 to 10 min and stopped by addition of 1 M H2SO4 (200 μl). The resulting solution (150 μl) was transferred to clear, flat bottom 96-well plates, and absorption at 492 nm was measured with the Clariostar microplate reader. Data were normalized to the expression level of wild-type α2AAR (100%) and mock transfected HEK293T cells (0%), respectively. N = 5 independent experiments were performed, with each condition in triplicate.
Acknowledgments
We would like to thank the Kobilka Cryo-Electron Microscopy Center, The Chinese University of Hong Kong, Shenzhen, for our cryo–electron microscopy.
Funding: Y.D. is supported by grants from Science, Technology and Innovation Commission of Shenzhen Municipality (Project codes C10120210014 and C10120210142), and in part by the Kobilka Institute of Innovative Drug Discovery and Presidential Fellowship at the Chinese University of Hong Kong, Shenzhen. Z.L. is supported by the Kobilka Institute of Innovative Drug Discovery at the Chinese University of Hong Kong, Shenzhen. The work was supported by the DFG grant GRK 1910 (P.G.).
Author contributions: J.X. expressed and purified proteins, assembled the complex, made mutations for functional assays, performed the model building and refinement, analyzed the structures, and prepared the figures. S.C. prepared cryo-EM grids, collected cryo-EM images, processed cryo-EM data, and participated in the preparation of the supplementary figures and methods writing. H.H. and D.W. performed the G protein IP-one assay and β-arrestin recruitment assay, measured the receptor surface expression, and analyzed the functional data under the supervision of P.G. G.C. expressed and purified scFv16 proteins for complex assembling. Q.L. participated in the collection of cryo-EM images. Z.L. supervised all collection and processing of cryo-EM images. D.Y. initiated the project with Y.D. and designed the initial expression construct and established the receptor purification protocol. Y.D. provided overall project supervision. J.X. and Y.D. wrote the manuscript with contributions from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 and S2
REFERENCES AND NOTES
- 1.Link R., Daunt D., Barsh G., Chruscinski A., Kobilka B., Cloning of two mouse genes encoding alpha 2-adrenergic receptor subtypes and identification of a single amino acid in the mouse alpha 2-C10 homolog responsible for an interspecies variation in antagonist binding. Mol. Pharmacol. 42, 16–27 (1992). [PubMed] [Google Scholar]
- 2.Ruffolo R. R. Jr., Nichols A. J., Stadel J. M., Hieble J. P., Pharmacologic and therapeutic applications of alpha 2-adrenoceptor subtypes. Annu. Rev. Pharmacol. Toxicol. 32, 243–279 (1993). [DOI] [PubMed] [Google Scholar]
- 3.MacDonald E., Kobilka B. K., Scheinin M., Gene targeting - Homing in on α2-adrenoceptor-subtype function. Trends Pharmacol. Sci. 18, 211–219 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Wang C. D., Buck M. A., Fraser C. M., Site-directed mutagenesis of alpha 2A-adrenergic receptors: Identification of amino acids involved in ligand binding and receptor activation by agonists. Mol. Pharmacol. 40, 168–179 (1991). [PubMed] [Google Scholar]
- 5.Ramos B. P., Arnsten A. F. T., Adrenergic pharmacology and cognition: Focus on the prefrontal cortex. Pharmacol. Ther. 113, 523–536 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maletic V., Eramo A., Gwin K., Offord S. J., Duffy R. A., The role of norepinephrine and its α-adrenergic receptors in the pathophysiology and treatment of major depressive disorder and schizophrenia: A systematic review. Front. Psych. 8, 42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wade S. M., Lim W. K., Lan K.-L., Chung D. A., Nanamori M., Neubig R. R., Gi activator region of α2A-adrenergic receptors: Distinct basic residues mediate Gi versus Gs activation. Mol. Pharmacol. 56, 1005–1013 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Eason M. G., Kurose H., Holt B. D., Raymond J. R., Liggett S. B., Simultaneous coupling of alpha 2-adrenergic receptors to two G-proteins with opposing effects. Subtype-selective coupling of alpha 2C10, alpha 2C4, and alpha 2C2 adrenergic receptors to Gi and Gs. J. Biol. Chem. 267, 15795–15801 (1992). [PubMed] [Google Scholar]
- 9.Qu L., Zhou Q., Xu Y., Guo Y., Chen X., Yao D., Han G. W., Liu Z.-J., Stevens R. C., Zhong G., Wu D., Zhao S., Structural basis of the diversity of adrenergic receptors. Cell Rep. 29, 2929–2935.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Jasper J. R., Lesnick J. D., Chang L. K., Yamanishi S. S., Chang T. K., Hsu S. A., Daunt D. A., Bonhaus D. W., Eglen R. M., Ligand efficacy and potency at recombinant α2 adrenergic receptors: Agonist-mediated [35S]GTPgammaS binding. Biochem. Pharmacol. 55, 1035–1043 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Pao C. S., Benovic J. L., Structure/function analysis of α2A-adrenergic receptor interaction with G protein-coupled receptor kinase 2. J. Biol. Chem. 280, 11052–11058 (2005). [DOI] [PubMed] [Google Scholar]
- 12.DeGraff J. L., Gurevich V. V., Benovic J. L., The third intracellular loop of α2-adrenergic receptors determines subtype specificity of arrestin interaction. J. Biol. Chem. 277, 43247–43252 (2002). [DOI] [PubMed] [Google Scholar]
- 13.De Graff J. L., Gagnon A. W., Benovic J. L., Orsini M. J., Role of arrestins in endocytosis and signaling of α2-adrenergic receptor subtypes. J. Biol. Chem. 274, 11253–11259 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Tan L., Yan W., McCorvy J. D., Cheng J., Biased ligands of G protein-coupled receptors (GPCRs): Structure–functional selectivity relationships (SFSRs) and therapeutic potential. J. Med. Chem. 61, 9841–9878 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Wootten D., Christopoulos A., Marti-Solano M., Babu M. M., Sexton P. M., Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 19, 638–653 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Giovannitti J. A. Jr., Thoms S. M., Crawford J. J., Alpha-2 adrenergic receptor agonists: A review of current clinical applications. Anesth. Prog. 62, 31–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith H., Elliott J., Alpha2 receptors and agonists in pain management. Curr. Opin. Anaesthesiol. 14, 513–518 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Nikolaev V. O., Hoffmann C., Bunemann M., Lohse M. J., Vilardaga J. P., Molecular basis of partial agonism at the neurotransmitter alpha2A-adrenergic receptor and Gi-protein heterotrimer. J. Biol. Chem. 281, 24506–24511 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Eason M. G., Jacinto M. T., Liggett S. B., Contribution of ligand structure to activation of α2-adrenergic receptor subtype coupling to Gs. Mol. Pharmacol. 45, 696–702 (1994). [PubMed] [Google Scholar]
- 20.Rasmussen S. G. F., Choi H.-J., Fung J. J., Pardon E., Casarosa P., Chae P. S., De Vree B. T., Rosenbaum D. M., Thian F. S., Kobilka T. S., Schnapp A., Konetzki I., Sunahara R. K., Gellman S. H., Pautsch A., Steyaert J., Weis W. I., Kobilka B. K., Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen S. G. F., De Vree B. T., Zou Y., Kruse A. C., Chung K. Y., Kobilka T. S., Thian F. S., Chae P. S., Pardon E., Calinski D., Mathiesen J. M., Shah S. T. A., Lyons J. A., Caffrey M., Gellman S. H., Steyaert J., Skiniotis G., Weis W. I., Sunahara R. K., Kobilka B. K., Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu X., Kaindl J., Clark M. J., Hübner H., Hirata K., Sunahara R. K., Gmeiner P., Kobilka B. K., Liu X., Binding pathway determines norepinephrine selectivity for the human β1AR over β2AR. Cell Res. 31, 569–579 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warne T., Moukhametzianov R., Baker J. G., Nehmé R., Edwards P. C., Leslie A. G. W., Schertler G. F. X., Tate C. G., The structural basis for agonist and partial agonist action on a β1-adrenergic receptor. Nature 469, 241–244 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masureel M., Zou Y., Picard L. P., van der Westhuizen E., Mahoney J. P., Rodrigues J. P. G. L. M., Mildorf T. J., Dror R. O., Shaw D. E., Bouvier M., Pardon E., Steyaert J., Sunahara R. K., Weis W. I., Zhang C., Kobilka B. K., Structural insights into binding specificity, efficacy and bias of a β2AR partial agonist. Nat. Chem. Biol. 14, 1059–1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ring A. M., Manglik A., Kruse A. C., Enos M. D., Weis W. I., Christopher Garcia K., Kobilka B. K., Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nature 502, 575–579 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X., Xu Y., Qu L., Wu L., Han G. W., Guo Y., Wu Y., Zhou Q., Sun Q., Chu C., Yang J., Yang L., Wang Q., Yuan S., Wang L., Hu T., Tao H., Sun Y., Song Y., Hu L., Liu Z.-J., Stevens R. C., Zhao S., Wu D., Zhong G., Molecular mechanism for ligand recognition and subtype selectivity of α2C adrenergic receptor. Cell Rep. 29, 2936–2943.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Yuan D., Liu Z., Kaindl J., Maeda S., Zhao J., Sun X., Xu J., Gmeiner P., Wang H. W., Kobilka B. K., Activation of the α2B adrenoceptor by the sedative sympatholytic dexmedetomidine. Nat. Chem. Biol. 16, 507–512 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Maeda S., Koehl A., Matile H., Hu H., Hilger D., Schertler G. F. X., Manglik A., Skiniotis G., Dawson R. J. P., Kobilka B. K., Development of an antibody fragment that stabilizes GPCR/G-protein complexes. Nat. Commun. 9, 3712 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koehl A., Hu H., Maeda S., Zhang Y., Qu Q., Paggi J. M., Latorraca N. R., Hilger D., Dawson R., Matile H., Schertler G. F. X., Granier S., Weis W. I., Dror R. O., Manglik A., Skiniotis G., Kobilka B. K., Structure of the μ-opioid receptor–Gi protein complex. Nature 558, 547–552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishna Kumar K., Shalev-Benami M., Robertson M. J., Hu H., Banister S. D., Hollingsworth S. A., Latorraca N. R., Kato H. E., Hilger D., Maeda S., Weis W. I., Farrens D. L., Dror R. O., Malhotra S. V., Kobilka B. K., Skiniotis G., Structure of a signaling cannabinoid receptor 1-G protein complex. Cell 176, 448–458.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhuang Y., Xu P., Mao C., Wang L., Krumm B., Zhou X. E., Huang S., Liu H., Cheng X., Huang X. P., Shen D. D., Xu T., Liu Y. F., Wang Y., Guo J., Jiang Y., Jiang H., Melcher K., Roth B. L., Zhang Y., Zhang C., Xu H. E., Structural insights into the human D1 and D2 dopamine receptor signaling complexes. Cell 184, 931–942.e18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao P., Yan W., Gou L., Zhong Y. N., Kong L., Wu C., Wen X., Yuan Y., Cao S., Qu C., Yang X., Yang C. C., Xia A., Hu Z., Zhang Q., He Y. H., Zhang D. L., Zhang C., Hou G. H., Liu H., Zhu L., Fu P., Yang S., Rosenbaum D. M., Sun J. P., du Y., Zhang L., Yu X., Shao Z., Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes. Cell 184, 943–956.e18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanek M., Picard L.-P., Schmidt M. F., Kaindl J. M., Hübner H., Bouvier M., Weikert D., Gmeiner P., Hybridization of β-adrenergic agonists and antagonists confers G protein bias. J. Med. Chem. 62, 5111–5131 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Rosenbaum D. M., Cherezov V., Hanson M. A., Rasmussen S. G. F., Thian F. S., Kobilka T. S., Choi H.-J., Yao X.-J., Weis W. I., Stevens R. C., Kobilka B. K., GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science 318, 1266–1273 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Erlandson S. C., McMahon C., Kruse A. C., Structural basis for G protein-coupled receptor signaling. Annu. Rev. Biophys. 47, 1–18 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Kobilka B., Structural insights into the dynamic process of G-protein- coupled receptor activation. FASEB J. 27, (2013). [Google Scholar]
- 37.Kang Y., Kuybeda O., de Waal P. W., Mukherjee S., van Eps N., Dutka P., Zhou X. E., Bartesaghi A., Erramilli S., Morizumi T., Gu X., Yin Y., Liu P., Jiang Y., Meng X., Zhao G., Melcher K., Ernst O. P., Kossiakoff A. A., Subramaniam S., Xu H. E., Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature 558, 553–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eason M. G., Liggett S. B., Identification of a Gs coupling domain in the amino terminus of the third intracellular loop of the α2A-adrenergic receptor. J. Biol. Chem. 270, 24753–24760 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Kato H. E., Zhang Y., Hu H., Suomivuori C.-M., Kadji F. M. N., Aoki J., Krishna Kumar K., Fonseca R., Hilger D., Huang W., Latorraca N. R., Inoue A., Dror R. O., Kobilka B. K., Skiniotis G., Conformational transitions of a neurotensin receptor 1–Gi1 complex. Nature 572, 80–85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maeda S., Qu Q., Robertson M. J., Skiniotis G., Kobilka B. K., Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 364, 552–557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim H. R., Xu J., Maeda S., Duc N. M., Ahn D., du Y., Chung K. Y., Structural mechanism underlying primary and secondary coupling between GPCRs and the Gi/o family. Nat. Commun. 11, 3160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russo C. J., Passmore L. A., Ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang X., Zhang L., Wen Z., Chen H., Li S., Ji G., Yin C. C., Sun F., Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation. Prog. Biophys. Mol. Biol. 156, 3–13 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Heymann J. B., Guidelines for using Bsoft for high resolution reconstruction and validation of biomolecular structures from electron micrographs. Protein Sci. 27, 159–171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guan X. M., Kobilka T. S., Kobilka B. K., Enhancement of membrane insertion and function in a type IIIb membrane protein following introduction of a cleavable signal peptide. J. Biol. Chem. 267, 21995–21998 (1992). [PubMed] [Google Scholar]
- 51.Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A. III, Smith H. O., Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Xu J., Hu Y., Kaindl J., Risel P., Hübner H., Maeda S., Niu X., Li H., Gmeiner P., Jin C., Kobilka B. K., Conformational complexity and dynamics in a muscarinic receptor revealed by NMR spectroscopy. Mol. Cell 75, 53–65.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Liu H., Hofmann J., Fish I., Schaake B., Eitel K., Bartuschat A., Kaindl J., Rampp H., Banerjee A., Hübner H., Clark M. J., Vincent S. G., Fisher J. T., Heinrich M. R., Hirata K., Liu X., Sunahara R. K., Shoichet B. K., Kobilka B. K., Gmeiner P., Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists. Proc. Natl. Acad. Sci. U.S.A. 115, 12046–12050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X., Kaindl J., Korczynska M., Stößel A., Dengler D., Stanek M., Hübner H., Clark M. J., Mahoney J., Matt R. A., Xu X., Hirata K., Shoichet B. K., Sunahara R. K., Kobilka B. K., Gmeiner P., An allosteric modulator binds to a conformational hub in the β2 adrenergic receptor. Nat. Chem. Biol. 16, 749–755 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tabor A., Moller D., Hubner H., Kornhuber J., Gmeiner P., Visualization of ligand-induced dopamine D2S and D2L receptor internalization by TIRF microscopy. Sci. Rep. 7, 10894 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 and S2