Abstract
The unexpected appearance and global spread of COVID-19 create significant difficulties for healthcare systems and present an unusual challenge for the fast discovery of medicines to combat this fatal disease. Screening metallodrugs libraries from the medicinal inorganic chemistry society may expand the studied ‘chemical space’ and improve the probability of discovering effective anti-COVID drugs, including polyoxometalates. POMs are an oxygen-rich family of inorganic cluster systems that have previously been tested for antiviral action against different types of viruses. Human angiotensin-converting enzyme 2 (ACE2), human transmembrane protease serine 2 (TMPRSS2), and the SARS-CoV-2 spike glycoprotein are required for host cell-mediated viral entrance. Targeting these proteins demonstrates potential possibilities for preventing infections and transmissions in the initial stage. As a result, POMs with known antiviral effects were investigated for this purpose using molecular docking and dynamic simulations. This research shows that POMs can prevent SARS CoV-2 from entering cells by blocking TMPRSS2, which SARS-CoV-2 uses for spike glycoprotein priming. They may also engage with ACE2 and the spike glycoprotein and disrupt their binding by blocking the active sites. We think that a thorough investigation of POMs as possible anti-COVID-19 drugs will provide significant opportunities.
Keywords: POMs, COVID-19, Protein, Docking, Dynamic, Infection
Graphical abstract
1. Introduction
A novel coronavirus, named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the latest pandemic in the series of other infectious diseases that poses an unprecedented challenge for the rapid explore of drugs against this deadly virus. Iran, which reported two fatalities due COVID-19 50 days following China on Feb 18, 2020, is among the nations dealing with the highest number of instances of COVID-19 infections and consequent deaths [1]. Even with the promising results of vaccines developed in the world, it is well recognized that the need for additional modalities is essential, due to the sheer enormity of the problem [2]. Therefore, both experimental and computational approaches are employed to investigate appropriate drugs from the library of FDA-approved drugs against this deadly virus. SARS-CoV-2 is an RNA virus that encapsulated inside a membrane envelope which has proteins like spikes that sticking out from its surface. These proteins are surface exposed and are involved in virus entry into the host cells [3]. The spike glycoprotein of SARS-CoV-2 contains two subunits: S1 subunit that contains the receptor binding domain (RBD) [4], and the S2 subunit (carryout the union of viral and host cell membrane). These subunits are responsible for binding to the host cell angiotensin-converting enzyme 2 peptidase domain (PD) and ensuring membrane fusion with the host cell, respectively [5]. ACE2 is an enzyme that is found on the outer membranes of the intestines, lung cells, kidneys, arteries and heart and is a primary target for CoVs. Interaction of surface spike glycoproteins of CoVs with this enzyme facilitates the virus entry into the host cell [6]. At first, the spike glycoprotein of SARS-CoV-2 is cleaved by host cell transmembrane protease serine 2 (TMPRSS2) [7,8], then ACE2 is hijacked by cleaved spike glycoprotein as an entry point to host cell [9]. This process provides the entry of the viral RNA genome into the host cell and increases the SARS-CoV-2 chance of human to human transmission [10]. Alternative therapeutic options must be investigated when a novel severe disease emerges for which there are no viable medical therapies, such as COVID-19. Metals and metallic compounds have long fascinated physicians for their intriguing, almost ‘magic’ characteristics and have therefore played a key role in the pioneering days of advanced pharmacology beginning in the late 19th to early 20th century. Notably, over the last 40 years, attention in metal-based drugs has grown significantly, due mainly to the enormous successful treatment of cisplatin, which the FDA first approved in 1978, and has greatly contributed to a fairly large and active scientific community working in the field of inorganic medicinal chemistry. The distinct chemical and biological characteristics of different metal centers, in certain instances, non-physiological metals, can be used for medicinal purposes [11,12]. POMs are inorganic metal-oxide assemblies presenting great interest properties in medicine including, a wide range of possible structures, metal-combination and size, as well as large redox chemistry and high solubility in water and the low toxicity toward the human body. Therefore, POMs were examined to produce inorganic-drugs against bacteria, tumor cells, and viruses. Computational chemistry is a helpful way to illuminate POM complex chemistry and its structure [13]. Molecular docking and molecular dynamics simulation are two major computational methods that provide great applicability in pharmaceutical research industries in search of novel drugs in a short duration time and assist experimental studies [14]. POMs were reported to have anti-tumor, -bacterial and - viral activities. In the antibacterial and antiviral actions, suppression of translation/transcription processes and inhibition of viral binding to the host cell and/or penetration have been observed. There is no organized study on the inhibition of the coronavirus by POMs to the best of our knowledge. The primary aim of this research is to investigate the ability of POMs to engage with and block the cleavage function of TMPRSS2, which may prevent COVs from binding and fusion to cells, thus preventing cell infection as well as viral multiplication. We examined POMs further using in silico calculations to see how effective they were in inhibiting spike glycoprotein binding to the ACE2 receptor and, as a result, inhibiting COVID-19 progression in an infected host. Our group thinks that extensive investigation of POMs as possible anti-COVID-19 drugs will provide significant possibilities; grounds for this belief are provided below.
2. Methods
2.1. Molecular docking studies
2.1.1. Receptors and ligand preparation
In this section, we selected an α-Keggin structure of POMs [SiW12O40]−4 to perform molecular docking simulations and evaluate the binding affinity and interaction of the POMs to TMPRSS2 and SARS-CoV-2 chimeric receptor-binding domains complexed with ACE2 (ACE2-RBD). Such computational study helps us to assess the potential of POMs in preventing SARS-CoV-2 cell entry. AutoDock Vina [15] with MGL tools 1.5.4 was used to perform blind docking calculations. AutoDock Vina runs faster than AutoDock software and makes more precise docking calculations [16]. The TMPRSS2 (PDB ID: 2OQ5) and ACE2-RBD (PDB ID: 6LZG) were obtained from the Protein Data Bank, and inhibitors were selected and removed. AutoDock Tools (v.1.5.4) was used to prepare the receptors. MGL Tools (v.1.5.4) combined non-polar hydrogen atoms and added Kollman charged atoms to protein crystal structures. TMPRSS2and ACE2-RBD coordinate files were then saved in PDBQT format. The 2OQ5 was surrounded in a 52 × 46 × 44 Å box direction with a grid spacing of 1.00 Å and grid set centers of −1.87, 18.03, 17.47 Å, while the 6LZG was surrounded in a 64 × 74 × 110 box direction with a grid spacing of 1.00 Å and grid set centers of −25.41, 18.43, and −6.37. The structures of POM (PDB ID: 6Y7O) were obtained from protein data bank. MGL Tools (v.1.5.4) was then used to save the POM in PDBQT format, and docked results were visualized through the BIOVIA Discovery Studio software. The following are the methods for achieving rigid docking of POM. Initially, during the simulation process, the rigid form of POM is accurately preserved [17]. Secondly, to improve electrostatic interaction, MULLIKEN charges are allocated to each atom within POM [17]. Lastly, for tungsten and silicon, efficient “dummy” atoms are employed. In this study, iodine is used in place of tungsten, and carbon is used in place of silicon. The atomic replacement method's concept is described in Hill and colleagues' paper [18]. Optimization of the POM was performed using Gaussian 09 software package at the semi-empirical PM6 method [19,20]. We hypothesized that POMs with antiviral properties may disrupt the cell entry of CoVs and consequently prevent viral replication, transmissibility, and pathogenicity. Hence, we examined the virtual interaction of the POMs to the TMPRSS2 and ACE2 -RBD.
2.2. Molecular dynamics (MD) simulations
The molecular dynamic simulations of POM were investigated by using GROMACS software in the presence of TMPRSS2 and spike receptor domain complexed with ACE2 proteins. Classical MD simulations [21,22] were conducted using the CHARMM27 force field [23,24]. TIP3P water [25] was used to solve the free protein and protein-ligand complexes in the cubic box with periodic boundary conditions in three directions. The solutes were positioned in the box's center, with a minimum distance of 1.0 nm between the solutes' surface and the box. Na+ ions were added to the system to neutralize the charge. The systems were balanced at 300 K and 1.0 bar after energy minimization through the steepest descent method. The Parrinello-Rahman barostat was used to maintain a pressure of 1.0 bar, and a temperature of 300 K was maintained using a modified Berendsen thermostat. The LINCS algorithm enabled the computation of bond lengths, while the particle-mesh Ewald scheme (PME) was used to compute long-range electrostatic forces (grid spacing 0.16 nm) [26]. The short-range nonbonded interactions were computed using cutoff ratios of 1.0 nm for van der Waals and Coulomb potentials. Finally, a 50 ns MD simulation with a time step of 2 fs was performed with random generation of velocities through a Maxwell distribution.
2.3. Interaction analysis by MM-PBSA binding energy
Using the Molecular Mechanics Poisson Boltzmann Surface Area (MM-PBSA) technique, the binding free energy of protein-ligand systems was calculated. This in silico method was a combined energy system described by the binding free energy, composed of electrostatic, SASA, van der Waals, and polar solvation energies. The MM-PBSA binding free energies were calculated using the GROMACS script g_mmpbsa [27]. Using the MM-PBSA method, the following equation was employed to determine the binding free energy of the interacting proteins:
| ΔGbinding = Gcomplex - (Greceptor + Gligand) |
ΔG binding denotes the total binding energy of the protein-ligand complexes; Greceptor and Gligand denote the binding energy of the free receptor and unbounded ligand, respectively.
3. Results
3.1. Molecular docking simulation
While significant efforts are being made to develop drugs and vaccines against COVID-19, only a few available therapeutic agents are currently available. In this regard, molecular knowledge of the virus increases quickly, and potential druggable targets are being discovered and profiled [28]. Several inorganic medicines are still used in medical care today for a few particular applications in which they serve important and irreplaceable functions, combining exceptional effectiveness with tolerable toxicity [29,30]. The most notable example is the widespread utilization of cisplatin as well as its analogs in chemotherapy; despite their significant systemic toxicity, platinum medications are believed to be included in approximately 50% of present chemotherapeutic regimens for treatment for cancer [30]. Because of the high severity of cancer, their pertinent toxicity could be tolerated on the foundation of a cost/benefit balance. In light of these reasons, we strongly urge the worldwide scientific community to investigate the potential of metallic compounds in pharmaceutical industries for COVID-19 treatments in a systematic and timely manner. It is critical to address toxicity concerns since metal-based medicines are usually recognized to cause significant adverse effects. The various metal centers have distinct chemical characteristics which could be successfully taken advantage of against specific pharmacological targets and diseases; thereby, some inorganic drugs are already in medical usage with significant functions due to the emergence of particular pharmacological effects that may not be achieved with conventional organic drugs. As a result, we may anticipate that POMs will make a major contribution to the combat against COVID-19. However, the safety statement should contain at least two types of toxicity evaluation, acute and systemic toxicity, in which a broad range of models and methods, in vivo, in vitro, or even in silico, must be used. Polyoxometalates (POMs) are oxygen rich class of inorganic cluster systems that the antiviral activity of them has previously been examined against various types of viruses. These negatively charged clusters with three-dimensional structures perform remarkable biological activities, with low toxicity and high efficacy. POMs were reported to have in vitro antiviral activities against different virus types. many polyoxotungstates, in particular, K7[PTi2W10O40]·6H2O (PM-19), [PriNH3]6H[PTi2W10O38(O2)2]·H2O (PM-523), K9H5[(GeTi3W9O37)2O3]·16H2O (PM-504), [Me3NH]8- [(SiNb3W9O37)2O3] (JM2820), K13[Ce(SiW11O39)2]·26H2O (JM1590), K6[BGa(H2O)W11O39]·15H2O (JM2766), K7[P2W17(NbO2)O61], K10[Fe4(H2O)2- (PW9O34)2].xH2O (HS-058), and K7[P2W17NbO62], demonstrated in vitro activity against HIV and other RNA viruses [31]. It has been found that, POM93 Cs2K4Na [SiW9Nb3O40] demonstrated antiviral activity against influenza virus, HSV-1, HSV-2, HIV-1, and HBV. This kind of POM localizes on the cell surface, and nonspecifically masked cellular receptors for viral entry [32]. The mechanism of action of POM-4960 is inhibition of virus attachment and subsequently penetration [33]. POM with Keggin-type structures was shown to have activity against human immuno-deficiency virus types 1 and 2 [31,[34], [35], [36], [37], [38]], herpes simplex viruses [39], thymidine kinase deficient herpes simplex virus [38], togaviruses [31], measles virus [40], HCMV [38], parainfluenza virus [40], rhabdovirus [31], RSV [38,40,41] influenza viruses [40,41] arenaviruses [31] and other retroviruses [31]. The surface of the cell membrane is surrounded by negatively charged polymers. The virus is electrostatically attracted to the cellular membranes and uses the HA molecule to bind to a particular receptor [42]. Negatively charged POMs are believed to be disrupting the mechanism and binding the virions to their receptors non-specifically [33,43]. POMs with vanadium or titanium were shown to inhibit several RNA viruses, including Paramyxoviridae (respiratory syncytial virus), Flaviviridae (Dengue virus), Lentiviridae (human immunodeficiency virus type 1) and Orthomyxoviridae (influenza virus type A) [44]. The suppression of virus-cell interaction and syncytium formation between HIV-infected cells has been ascribed to the process of anti-HIV suppression for most POM compounds [31,37,45]. Similar chemicals blocked the binding of an HIV-specific gp120 monoclonal antibody to the gp120 protein [31], and the anti-HIV action was ascribed to POM binding of gp120, which prevented virus-mediated fusion and cell-to-cell transmission. Envelope glycoprotein GP120 is a glycoprotein exposed on the surface of the HIV envelope and is required for viral attachment to particular cell surface receptors and virus entry into cells. PM-1001 was shown to significantly inhibit the binding of viral gp-120 antibodies [46]. The enveloped virus such as Ebola, HIV-1, Influenza and the SARS-CoV-2 enter to the cells through their spike-like glycoprotein [47]. One effective approach to fight SARS-CoV-2 infections is to prevent the virus from entering host cells by inhibiting the molecular machinery involved in this key phase [[48], [49], [50], [51]]. Pradhan et al. [52] investigated the 2019-nCoV spike glycoprotein and discovered many intriguing results. This group discovered four separate inserts in the 2019-nCoV spike glycoprotein, which have not been found in any other COVID-19 case reported to date, and all four inserts in the 2019-nCoV mapped to short segments of amino acids in the HIV-1 gp120 and Gag proteins, across all annotated virus proteins in the NCBI database. These proteins are required for viruses to detect and attach to host cells, as well as for viral assembly [53]. Because surface proteins are important for host tropism, alterations in these proteins indicate an alteration in the virus's host specificity. Based on these results, we can conclude that POMs may inhibit the attachment of SARS-CoV-2 to angiotensin converting enzyme-2 receptors by a conformational change in the structural spike protein of SARS-CoV-2. In other words, the anti-covid-19 activity may be attributed to POM binding of spike protein, and blocking virus-mediated fusion and cell to cell spread of the virus. It is very important to investigate the inhibition potency of POMs to SARS-CoV-2. 1- Whether POMs interact with SARS-CoV-2; 2- What the interaction mechanism should be; 3- It is of great important to evaluate molecular mechanisms when studying problems related to POMs. Hu et al. [54], evaluated the α-Keggin type POM isomers including [α −1,2- PTi2W10O40] 7-, [α −1,4-PTi2W10O40] 7-, [α −1,5-PTi2W10O40] 7-, [α −1,6-PTi2W10O40]7-, [α −1,11-PTi2W10O40] 7- against main protease of SARS-CoV by molecular modeling. The results of this study indicated that, POMs bind with the main protease of SARS-CoV in the active site region with high affinity. Employing POMs at high doses might be toxic to fight against SARS-CoV-2, an issue that there is a major concern about it. Therefore, low toxicity POMs are highly desirable to achieve an equivalent antiviral capability. Several additional factors should be taken into account throughout the POMs anti-Covid-19 clinical trials: (1) verifying the known safety profile of POMs and determining the minimal dosage and frequency of administration needed to achieve optimum anti-Covid-19 effectiveness (2) Identifying individuals who should not receive POMs and outlining contraindications for co-administered medications. (3) Identification of any short-term and long-term negative effects that may occur as a result of POM treatment. Considering this fact that POMs can be used as an antiviral agent and have shown to have good biocompatibility, they can be promising candidates against COVID-19. However, there are still very few anti-viral experiments using POMs against COVID-19. More studies are needed to confirm the role of POMs in COVID-19 patients. We hope that POMs can play a key role in combating against COVID-19 and for infection prevention and control in the near future. However a major challenge is their safety profile in vivo and large-scale manufacture.
Table 1, Table 2 represent the obtained results from docking analysis of the interaction of POM with TMPRSS2, and ACE2-RBD. The obtained binding free energy for [SiW12O40]−4 was −9.4, and −11.4 kcal/mol toward the TMPRSS2, and ACE2-RBD, respectively. These negative values of binding free energy authenticate the strong interaction of [SiW12O40]−4 with TMPRSS2, and ACE2-RBD. In other words, [SiW12O40]−4 interacts efficiently with TMPRSS2, and ACE2-RBD and it makes a stable complex with them. The nature of interactions, atoms involved in bonding with POM and bond lengths are shown in Table 1, Table 2. The amino acid residues of TMPRSS2 and ACE2-RBD that interacted with POM are demonstrated in Fig. 1, Fig. 2 . The POM + ACE2-RBD forms hydrogen bonds with twelve amino acids, which are Asp30, Ala387, Asn33, His34, Arg393, and Pro389, from ACE2 and Glu406, Arg403, Arg408, Gln409, Lys417 and Gly416 of the spike glycoprotein (RBD). Also, for the POM + ACE2-RBD system, there are electrostatic interactions between Asp30 from ACE2 and Asp30, Asp405, and Glu416 of the spike glycoprotein (RBD) and the POM. Six amino acids Arg41, His57, Thr62, His96, Ser195, and Thr61 are involved in the formation of hydrogen bonding between the POM and TMPRSS2. As can be seen, [SiW12O40]−4 has stronger interactions and makes a more stable complex with ACE2-RBD compared to TMPRSS2as revealed by its lower value of binding free energy. The crucial step for the fusion of SARS-CoV-2 and host cell membranes is the activation and cleavage of spike glycoprotein of SARS-CoV-2 by host protease TMPRSS2 [55,56]. Therefore, as reported already, we can inhibit cell entry of SARS-CoV by blocking the activity of TMPRSS2 [57]. The association of POMs with TMPRSS2 may restrict the functionality of the S1/S2 site in the spike glycoprotein [58], limiting the fusion of viral and human cellular membrane, and suggests that POMs may serve as SARS-CoV-2 cell entry inhibitors. Lung with the massive surface area and the vast distribution of ACE2 in human alveolar epithelial cells is the vulnerable target organ for SARS-CoV. Thus, the investigation of compounds that interact with ACE2, and hinder viral entry into the cells by blocking this receptor and halting transmissibility and pathogenicity is an important field for research. The docking method showed that the POM might prevent SARS-CoV-2/ACE-2 interaction and viral entry because it had a high binding affinity at the ACE-2–RBD complex interface. Overall, computational studies indicate that POMs may limit CoV cell entry by decreasing the TMPRSS2 used by SARS-CoV-2 for spike glycoprotein priming and interrupting the interaction of SARS-CoV-2 spike protein with the human ACE-2 receptor. However, their systemic effects should be further examined in suitable ex vivo human organ culture or organoids, animal models, or clinical trials.
Table 1.
Predicted bonds between interacting atoms of POM and TMPRSS2.
| S. No. | Amino acid | Amino acid atom | POM atom | Distance | Nature of interaction |
|---|---|---|---|---|---|
| 1 | ARG41 | H-Donor | O(H-Acceptor) | 2.23 | Hydrogen Bond |
| 2 | ARG41 | H-Donor | O(H-Acceptor) | 2.48 | Hydrogen Bond |
| 3 | ARG41 | H-Donor | O(H-Acceptor) | 2.21 | Hydrogen Bond |
| 4 | HIS57 | H-Donor | O(H-Acceptor) | 2.43 | Hydrogen Bond |
| 5 | THR62 | H-Donor | O(H-Acceptor) | 2.09 | Hydrogen Bond |
| 6 | HIS96 | H-Donor | O(H-Acceptor) | 3.07 | Hydrogen Bond |
| 7 | SER195 | H-Donor | O(H-Acceptor) | 2.23 | Hydrogen Bond |
| 8 | HIS57 | H-Donor | O(H-Acceptor) | 2.93 | Hydrogen Bond |
| 9 | THR61 | H-Donor | O(H-Acceptor) | 3.12 | Hydrogen Bond |
| 10 | HIS57 | Pi-Orbitals | O(H-Donor) | 3.94 | Hydrogen Bond |
Table 2.
Predicted bonds between interacting atoms of POM and ACE2-RBD.
| S. No. | Amino acid | Amino acid atom | POM atom | Distance | Nature of interaction |
|---|---|---|---|---|---|
| 1 | A:ASP30 | Negative | W(Positive) | 4.31 | Electrostatic |
| 2 | B:ASP405 | Negative | W(Positive) | 5.59 | Electrostatic |
| 3 | A:ASP30 | Negative | W(Positive) | 5.02 | Electrostatic |
| 4 | B:GLU406 | Negative | W(Positive) | 4.44 | Electrostatic |
| 5 | B:GLU406 | Negative | W(Positive) | 5.52 | Electrostatic |
| 6 | B:GLU406 | H-Acceptor | O(H-Donor) | 3.24 | Hydrogen Bond |
| 7 | A:ASP30 | H-Acceptor | O(H-Donor) | 3.22 | Hydrogen Bond |
| 8 | A:ALA387 | H-Acceptor | O(H-Donor) | 3.01 | Hydrogen Bond |
| 9 | A:ASN33 | H-Donor | O(H-Acceptor) | 3.07 | Hydrogen Bond |
| 10 | A:HIS34 | H-Donor | O(H-Acceptor) | 2.67 | Hydrogen Bond |
| 11 | A:HIS34 | H-Donor | O(H-Acceptor) | 2.49 | Hydrogen Bond |
| 12 | A:ARG393 | H-Donor | O(H-Acceptor) | 2.62 | Hydrogen Bond |
| 13 | B:ARG403 | H-Donor | O(H-Acceptor) | 2.53 | Hydrogen Bond |
| 14 | B:ARG403 | H-Donor | O(H-Acceptor) | 2.45 | Hydrogen Bond |
| 15 | B:ARG403 | H-Donor | O(H-Acceptor) | 2.36 | Hydrogen Bond |
| 16 | B:ARG408 | H-Donor | O(H-Acceptor) | 2.10 | Hydrogen Bond |
| 17 | B:ARG408 | H-Donor | O(H-Acceptor) | 2.56 | Hydrogen Bond |
| 18 | B:ARG408 | H-Donor | O(H-Acceptor) | 2.58 | Hydrogen Bond |
| 19 | B:ARG408 | H-Donor | O(H-Acceptor) | 1.89 | Hydrogen Bond |
| 20 | B:GLN409 | H-Donor | O(H-Acceptor) | 2.15 | Hydrogen Bond |
| 21 | B:GLN409 | H-Donor | O(H-Acceptor) | 3.09 | Hydrogen Bond |
| 22 | B:LYS417 | H-Donor | O(H-Acceptor) | 2.86 | Hydrogen Bond |
| 23 | A:PRO389 | H-Donor | O(H-Acceptor) | 3.45 | Hydrogen Bond |
| 24 | B:GLY416 | H-Donor | O(H-Acceptor) | 3.30 | Hydrogen Bond |
Fig. 1.
Molecular docking perspective of POM-TMPRSS2.
Fig. 2.
Molecular docking perspective of POM-(ACE2-RBD).
3.2. Molecular dynamics simulations
3.2.1. Radius of gyration (Rg)
The radius of gyration (Rg) of both TMPRSS2-POM and ACE2-RBD-POM complexes quantifies the molecule's overall extension during a 50ns MD run (Fig. 3, Fig. 4 ). A low Rg value demonstrates better structural entirety and folding treatment [59]. Throughout the 50ns MD simulation, two complexes maintain a stable mean Rg of 1.65 nm for 2OQ5-POM and 3.12 nm for 6LZG-POM. Likewise, the obtained Rg value for the unbound 2OQ5 and 6LZG was 1.66 and 3.13 nm. This further shows that the proteins gained more stability upon the binding of the ligand. During simulation, a slight enhancement in the Rg value of the 6LZG-POM complex was observed, indicating its structural integrity. The MD simulation results entirely support that POM forms stable complexes with 2OQ5 and 6LZG, indicating its inhibitory properties for the TMPRSS2 and ACE2-RBD receptor, respectively [60].
Fig. 3.
Radius of gyration (Rg) for TMPRSS2 and POM-TMPRSS2 during 50 ns MD simulation.
Fig. 4.
Radius of gyration (Rg) for ACE2-RBD and POM-(ACE2-RBD) during 50 ns MD simulation.
3.2.2. RMSD
RMSD analysis revealed insights and structural changes in the protein that confirm the protein's stability and equilibrium during simulation. The RMSD plot of the backbone atoms for the 2OQ5, 2OQ5-POM and 6LZG, 6LZG-POM is shown in Fig. 5, Fig. 6 A, respectively. RMSD was calculated for the unbound proteins and proteins-POM structures that converged during the 50ns MD simulation. The average RMSD of 2OQ5-POM was found to be 0.37 Å, and for the unbound 2OQ5 was 0.39 Å. Likewise, the obtained average RMSD for 6LZG-POM and 6LZG was 0.36 Å, and 0.37, respectively. If the RMSD value is less than 1.5 Å, it is considered to be good and acceptable. But, with the value of more than 3 Å for RMSD, it is clearly rejected. RMSD values are used to find the stability of the receptors with and without ligands and also to study the conformational changes of the receptors [61]. The low RMSD values indicated that POM was stable in MD simulations with proteins [62].
Fig. 5.
(A) RMSD plots for TMPRSS2 and POM-TMPRSS2, (B) RMSF plots for TMPRSS2 and POM-TMPRSS2 during 50 ns MD simulation.
Fig. 6.
(A) RMSD plots for ACE2-RBD and POM-(ACE2-RBD), (B) RMSF plots for ACE2-RBD and POM-(ACE2-RBD) during 50 ns MD simulation.
3.2.3. RMSF
RMSF analysis was used to determine the flexibility of the total protein concerning its average structure. Low RMSF values demonstrated narrowed movements, whereas high RMSF values demonstrated increased flexibility [63]. Ligand binding poses energy, and interaction has a direct correlation with residual fluctuation (RMSF) values. The RMSF plots for 2OQ5, 2OQ5-POM and 6LZG, 6LZG-POM are shown in Fig. 5, Fig. 6B, respectively. The RMSF values for the 2OQ5-POM and 6LZG-POM complexes were extremely low; as a result, they exhibited minimal movement, indicating that both complexes were stable. RMSF values were 0.30 and 0.29 Å on average for 2OQ5 and 2OQ5-POM during a 50 ns simulation. Likewise, the RMSF values for 6LZG and 6LZG-POM were 0.29 and 0.29 Å. During the simulation, it was observed that Lys187 in the loop region of 2OQ5 was more fluctuated than those in the alpha-helix and beta-sheet regions. This indicated that the protein remained stable throughout the 50ns simulation period [64]. Except for the loop region, the residues 102 and 562 in the alpha-helix region of 6LZG exhibited significant variation up to 0.40 Å and 0.45 Å, respectively. Overall, the RMSF values for both proteins indicated that the complexes 2OQ5-POM and 6LZG-POM were stable [65].
3.2.4. Hydrogen bond interactions
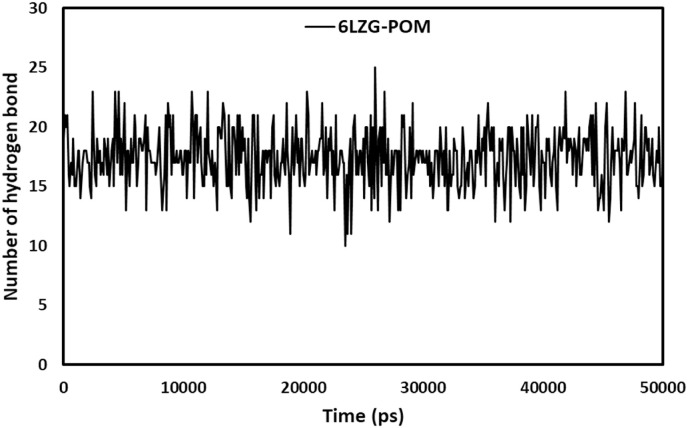
Hydrogen bond formation plays an important role in the stabilization of protein-ligand complex structure by minimizing the energy of the system. The hydrogen bond interactions of the complexes were calculated to validate the affinity of the POM to inhibit the proteins. Protein-ligand hydrogen bonding pattern were studied in bound POM with TMPRSS2 and ACE2-RBD. Fig. 7, Fig. 8 show the number of hydrogen bonds versus time during the simulation. The average H-bonding in 2OQ5-POM and 6LZG-POM was 9.05 and 17.41 during the 50 ns simulation. Overall, the H-bonding patterns in 2OQ5-POM and 6LZG-POM interactions showed an energetically favorable and stable complex formation [66].
Fig. 7.
Number of hydrogen bonds for POM-TMPRSS2 during 50 ns MD simulation.
Fig. 8.
Number of hydrogen bonds for POM-(ACE2-RBD) during 50 ns MD simulation.
3.3. MM-PBSA binding free energy
The average free binding energy of the 2OQ5-POM and 6LZG-POM was computed by a python script MmPbSaStat.py (Table 3 ). We computed the average free binding energy and its standard deviation/error of the files, which were obtained from g_mmpbsa. The binding free energy can acceptably illustrate the durability of the linking ligand receptor, which is an essential parameter of evaluation in drug discovery. The lesser the binding energy, the better is the binding of the ligand and protein [67]. Except for the polar solvation energy, the favorable contribution of van der Waals, SASA, and electrostatic energies were devoted to the binding of POM with TMPRSS2 and ACE2-RBD. Van der Waals energy's augmentation to the overall binding free energy was higher upon the electrostatic contribution energy. The binding free energy value of 2OQ5-POM and 6LZG-POM were found to be −103.65 ± 0.16 and −151.94 ± 2.19 kJ/mol, respectively, indicating the studied POM has unique interactions within the 2OQ5 and 6LZG proteins. Fig. 9 delineates the plot of the binding energy against time graphs for 2OQ5-POM and 6LZG-POM. The above findings suggest that POM is a potential candidate in inhibiting the TMPRSS2 and ACE2-RBD receptors.
Table 3.
Binding free energy (MM-PBSA) calculations for 2OQ5-POM and 6LZG-POM.
| system | ΔEvan der Waal (kJ/mol) | ΔEElectrostatic (kJ/mol) | ΔEPolar solvation (kJ/mol) | ΔESASA (kJ/mol) | ΔEBinding (kJ/mol) |
|---|---|---|---|---|---|
| 2OQ5-POM | −123.19 ± 1.15 | −22.17 ± 0.65 | 57.94 ± 1.36 | −16.23 ± 0.17 | −103.65 ± 0.16 |
| 6LZG-POM | −203.15 ± 4.56 | −42.85 ± 2.08 | 120.38 ± 8.65 | −26.32 ± 1.15 | −151.94 ± 2.19 |
Fig. 9.
Graphical representation of the Delta_E_Binding free energy (A) POM-TMPRSS2 and (B) POM-(ACE2-RBD) during 50 ns MD simulation.
4. Conclusions
This study found that POMs may prevent COVs from entering the cells by blocking the host cell serine protease TMPRSS2, which SARS-CoV-2 uses for spike glycoprotein priming. They may also engage with the S protein and ACE2 and disrupt their binding by blocking the active sites. The GROMACS software was used to simulate the interaction of POM with TMPRSS2 and ACE2-RBD. Furthermore, the radius of gyration of TMPRSS2-POM and ACE2-RBD-POM has values that indicate the systems' stability. Moreover, low RMSF and RMSD values indicate that POM is stable in the presence of proteins during MD simulation. The average H-bonding in 2OQ5-POM and 6LZG-POM was 9.05 and 17.41 during the 50 ns simulation. The binding free energy values of 2OQ5-POM and 6LZG-POM were found to be −103.65 ± 0.16 and −151.94 ± 2.19 kJ/mol, respectively, indicating the studied POM has unique interactions within the mentioned proteins. Finally, the POMs may be tested in vivo as a potent and promising anti-COVID-19 candidate. Because of the wide range of antiviral action, and based on our computational results, it is evident to consider POMs as a great source of new medicinally helpful drugs; consequently, we highly suggest that POMs be included as much as possible in new drug discovery screening programs. This may be especially important in the pursuit of new medications for treating COVID-19 illness, a severe and quickly spreading disease for which no medical treatments are currently available and are desperately required.
Data availability
Because the majority of work is performed using licensed software, these are not transferable. Additional scripts written by members of the group may be made available upon request.
Code availability
Most of the work is done with licensed software therefore those are not transferable. Other scripts written in the group could be made available on request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors thank the Razi University Research Council for support of this work.
References
- 1.Zhan C., Tse C.K., Fu Y., Lai Z., Zhang H. Modeling and prediction of the 2019 coronavirus disease spreading in China incorporating human migration data. PLoS One. 2020;15 doi: 10.1371/journal.pone.0241171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lokhande K.B., Banerjee T., Swamy K.V., Ghosh P., Deshpande M. An in silico scientific basis for LL‐37 as a therapeutic for Covid‐19. Proteins. 2021 doi: 10.1002/prot.26198. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lokhande K.B., Apte G.R., Shrivastava A., Singh A., Pal J.K., Swamy K.V., Gupta R.K. Sensing the interactions between carbohydrate-binding agents and N-linked glycans of SARS-CoV-2 spike glycoprotein using molecular docking and simulation studies. J. Biomol. Struct. 2020:1–19. doi: 10.1080/07391102.2020.1851303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 5.Tong T.R. Drug targets in severe acute respiratory syndrome (SARS) virus and other coronavirus infections. Infect Disord Drug Targets. 2009;9:223–245. doi: 10.2174/187152609787847659. [DOI] [PubMed] [Google Scholar]
- 6.Pulakuntla S., Lokhande K.B., Padmavathi P., Pal M., Swamy K.V., Sadasivam J., Singh S.A., Aramgam S.L., Reddy V.D. Mutational analysis in international isolates and drug repurposing against SARS-CoV-2 spike protein: molecular docking and simulation approach. Virus Dis. 2021;32:690–702. doi: 10.1007/s13337-021-00720-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pöhlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miodragović Đ., Merlino A., Swindell E.P., Bogachkov A., Ahn R.W., Abuhadba S., Ferraro G., Marzo T., Mazar A.P., Messori L. Arsenoplatin-1 is a dual pharmacophore anticancer agent. J Am Chem Soc. 2019;141:6453–6457. doi: 10.1021/jacs.8b13681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mjos K.D., Orvig C. Metallodrugs in medicinal inorganic chemistry. Chem Rev. 2014;114:4540–4563. doi: 10.1021/cr400460s. [DOI] [PubMed] [Google Scholar]
- 13.Poblet J.M., López X., Bo C. Ab initio and DFT modelling of complex materials: towards the understanding of electronic and magnetic properties of polyoxometalates. Soc. Rev. 2003;32:297–308. doi: 10.1039/b109928k. [DOI] [PubMed] [Google Scholar]
- 14.Lokhande K., Nawani N., Venkateswara S.K., Pawar S. Biflavonoids from Rhus succedanea as probable natural inhibitors against SARS-CoV-2: a molecular docking and molecular dynamics approach. J. Biomol. Struct. 2020:1–13. doi: 10.1080/07391102.2020.1858165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao Q., Lu S., Huang C., Su W., Zhou S., Sheng J., Huang S. An electrochemical chiral sensor based on amino-functionalized graphene quantum dots/β-cyclodextrin modified glassy carbon electrode for enantioselective detection of tryptophan isomers. J Iran Chem Soc. 2017;14:1957–1970. [Google Scholar]
- 17.Guan W., Yan L., Su Z., Liu S., Zhang M., Wang X. Electronic properties and stability of dititaniumIV substituted α-Keggin polyoxotungstate with heteroatom phosphorus by DFT. Inorg Chem. 2005;44:100–107. doi: 10.1021/ic049830u. [DOI] [PubMed] [Google Scholar]
- 18.Judd D.A., Nettles J.H., Nevins N., Snyder J.P., Liotta D.C., Tang J., Ermolieff J., Schinazi R.F., Hill C.L. Polyoxometalate HIV-1 protease inhibitors. A new mode of protease inhibition. J Am Chem Soc. 2001;123:886–897. doi: 10.1021/ja001809e. [DOI] [PubMed] [Google Scholar]
- 19.Frisch A. 2009. Gaussian 09W reference, wallingford, USA; p. 25. [Google Scholar]
- 20.Stewart J.J. Optimization of parameters for semiempirical methods V: modification of NDDO approximations and application to 70 elements. J Mol Model. 2007;13:1173–1213. doi: 10.1007/s00894-007-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berendsen H.J., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43–56. [Google Scholar]
- 22.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software. 2015;1:19–25. [Google Scholar]
- 23.Bjelkmar P., Larsson P., Cuendet M.A., Hess B., Lindahl E. Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J Chem Theor Comput. 2010;6:459–466. doi: 10.1021/ct900549r. [DOI] [PubMed] [Google Scholar]
- 24.Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I. CHARMM general force field: a force field for drug‐like molecules compatible with the CHARMM all‐atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 26.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅ log (N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 27.Kumari R., Kumar R., Consortium O.S.D.D., Lynn A. g_mmpbsa A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 28.Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., Kumar S., Bhattacharyya A., Kumar H., Bansal S. Drug targets for corona virus: a systematic review. Indian J Pharmacol. 2020;52:56. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barry N.P., Sadler P.J. Exploration of the medical periodic table: towards new targets. Chem. Commun. 2013;49:5106–5131. doi: 10.1039/c3cc41143e. [DOI] [PubMed] [Google Scholar]
- 30.Johnstone T.C., Suntharalingam K., Lippard S.J. The next generation of platinum drugs: targeted Pt (II) agents, nanoparticle delivery, and Pt (IV) prodrugs. Chem Rev. 2016;116:3436–3486. doi: 10.1021/acs.chemrev.5b00597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto N., Schols D., De Clercq E., Debyser Z., Pauwels R., Balzarini J., Nakashima H., Baba M., Hosoya M., Snoeck R. Mechanism of anti-human immunodeficiency virus action of polyoxometalates, a class of broad-spectrum antiviral agents. Mol Pharmacol. 1992;42:1109–1117. [PubMed] [Google Scholar]
- 32.Wang J., Liu Y., Xu K., Qi Y., Zhong J., Zhang K., Li J., Wang E., Wu Z., Kang Z. Broad-spectrum antiviral property of polyoxometalate localized on a cell surface. ACS Appl Mater Interfaces. 2014;6:9785–9789. doi: 10.1021/am502193f. [DOI] [PubMed] [Google Scholar]
- 33.Hosseini S.M., Amini E., Kheiri M.T., Mehrbod P., Shahidi M., Zabihi E. Anti-influenza activity of a novel polyoxometalate derivative (POM-4960) IJMCM. 2012;1:21. [PMC free article] [PubMed] [Google Scholar]
- 34.Hill C., Hartnup M., Faraj M., Weeks M., Prosser-McArtha C., Brown R., Jr., Kadkhodayan M. SAR; 1990. Polyoxometalates as inorganic anti-HIV-1 compounds. [Google Scholar]
- 35.Inouye Y., Tokutake Y., Yoshida T., Yamamoto A., Yamase T., Nakamura S. Antiviral activity of polyoxomolybdoeuropate PM-104 against human immunodeficiency virus type 1 1. Chem Pharm Bull. 1991;39:1638–1640. doi: 10.1248/cpb.39.1638. [DOI] [PubMed] [Google Scholar]
- 36.Inouye Y., Tokutake Y., Kunihara J., Yoshida T., Yamase T., Nakata A., Nakamura S. Suppressive effect of polyoxometalates on the cytopathogenicity of human immunodeficiency virus type 1 (HIV-1) in vitro and their inhibitory activity against HIV-1 reverse transcriptase. Nakamura. 1992;40:805–807. doi: 10.1248/cpb.40.805. [DOI] [PubMed] [Google Scholar]
- 37.Take Y., Tokutake Y., Inouye Y., Yoshida T., Yamamoto A., Yamase T., Nakamura S. Inhibition of proliferation of human immunodeficiency virus type 1 by novel heteropolyoxotungstates in vitro. Antivir Res. 1991;15:113–124. doi: 10.1016/0166-3542(91)90029-q. [DOI] [PubMed] [Google Scholar]
- 38.Ikeda S., Neyts J., Yamamoto N., Murrer B., Theobald B., Bossard G., Henson G., Abrams M., Picker D., De Clercq E. In vitro activity of a novel series of polyoxosilicotungstates against human myxo-, herpes-and retroviruses. Antivir Chem Chemother. 1993;4:253–262. [Google Scholar]
- 39.Fukuma M., Seto Y., Yamase T. In vitro antiviral activity of polyoxotungstate (PM-19) and other polyoxometalates against herpes simplex virus. Antivir Res. 1991;16:327–339. doi: 10.1016/0166-3542(91)90047-u. [DOI] [PubMed] [Google Scholar]
- 40.Shigeta S., Mori S., Watanabe J., Yamase T., Hill C., Schinazi R. Anti-influenzavirus-activities of polyoxometalates. Antivir Res. 1995;26 A298-A298. [Google Scholar]
- 41.Shigeta S., Mori S., Watanabe J., Baba M., Khenkin A., Hill C., Schinazi R. In vitro antimyxovirus and anti-human immunodeficiency virus activities of polyoxometalates. Antivir Chem Chemother. 1995;6:114–122. [Google Scholar]
- 42.Rhule J.T., Hill C.L., Judd D.A., Schinazi R.F. Polyoxometalates in medicine. Chem Rev. 1998;98:327–358. doi: 10.1021/cr960396q. [DOI] [PubMed] [Google Scholar]
- 43.Witvrouw M., Weigold H., Pannecouque C., Schols D., De Clercq E., Holan G. Potent anti-HIV (type 1 and type 2) activity of polyoxometalates: structure− activity relationship and mechanism of action. J Med Chem. 2000;43:778–783. doi: 10.1021/jm980263s. [DOI] [PubMed] [Google Scholar]
- 44.Shigeta S., Mori S., Kodama E., Kodama J., Takahashi K., Yamase T. Broad spectrum anti-RNA virus activities of titanium and vanadium substituted polyoxotungstates. Antivir Res. 2003;58:265–271. doi: 10.1016/s0166-3542(03)00009-3. [DOI] [PubMed] [Google Scholar]
- 45.Hill C.L., Weeks M.S., Schinazi R.F. Anti-HIV-1 activity, toxicity, and stability studies of representative structural families of polyoxometalates. J Med Chem. 1990;33:2767–2772. doi: 10.1021/jm00172a014. [DOI] [PubMed] [Google Scholar]
- 46.Yamase T. Anti-tumor,-viral, and-bacterial activities of polyoxometalates for realizing an inorganic drug. J Mater Chem. 2005;15:4773–4782. [Google Scholar]
- 47.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057. doi: 10.1016/j.cell.2020.09.033. e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joshi S., Joshi M., Degani M.S. Tackling SARS-CoV-2: proposed targets and repurposed drugs. Future Med Chem. 2020;12:1579–1601. doi: 10.4155/fmc-2020-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tiwari V., Beer J.C., Sankaranarayanan N.V., Swanson-Mungerson M., Desai U.R. Discovering small-molecule therapeutics against SARS-CoV-2. Drug Discov. 2020;25:1535–1544. doi: 10.1016/j.drudis.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whisenant J., Burgess K. Blocking coronavirus 19 infection via the SARS-CoV-2 spike protein: initial steps. ACS Med Chem Lett. 2020;11:1076–1078. doi: 10.1021/acsmedchemlett.0c00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiu S., Dick A., Ju H., Mirzaie S., Abdi F., Cocklin S., Zhan P., Liu X. Inhibitors of SARS-CoV-2 entry: current and future opportunities. J Med Chem. 2020;63:12256–12274. doi: 10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pradhan P., Pandey A.K., Mishra A., Gupta P., Tripathi P.K., Menon M.B., Gomes J., Vivekanandan P., Kundu B. BioRxiv; 2020. Uncanny similarity of unique inserts in the 2019-nCoV spike protein to HIV-1 gp120 and Gag. [Google Scholar]
- 53.Beniac D.R., Andonov A., Grudeski E., Booth T.F. Architecture of the SARS coronavirus prefusion spike. Nat Struct Biol. 2006;13:751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu D., Shao C., Guan W., Su Z., Sun J. Studies on the interactions of Ti-containing polyoxometalates (POMs) with SARS-CoV 3CLpro by molecular modeling. J Inorg Biochem. 2007;101:89–94. doi: 10.1016/j.jinorgbio.2006.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glowacka I., Bertram S., Mü ller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffmann M., Kleine-Weber H., Schroeder S., Krü ger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol. 2012;86:6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Erva R.R., Rajulapati S.B., Durthi C.P., Bhatia M., Pola M. Molecular dynamic simulations of Escherichia coli L-asparaginase to illuminate its role in deamination of asparagine and glutamine residues. 3 Biotech. 2016;6:2. doi: 10.1007/s13205-015-0339-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haider S., Barakat A., Ul-Haq Z. Discovery of potential chemical probe as inhibitors of CXCL12 using ligand-based virtual screening and molecular dynamic simulation. Molecules. 2020;25:4829. doi: 10.3390/molecules25204829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar D., Kumari K., Vishvakarma V.K., Jayaraj A., Kumar D., Ramappa V.K., Patel R., Kumar V., Dass S.K., Chandra R. Promising inhibitors of main protease of novel corona virus to prevent the spread of COVID-19 using docking and molecular dynamics simulation. J. Biomol. Struct. 2021;39:4671–4685. doi: 10.1080/07391102.2020.1779131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shahabadi N., Zendehcheshm S., Mahdavi M., Khademi F. Inhibitory activity of FDA-approved drugs cetilistat, abiraterone, diiodohydroxyquinoline, bexarotene, remdesivir, and hydroxychloroquine on COVID-19 main protease and human ACE2 receptor: a comparative in silico approach. IMU. 2021;26:100745. doi: 10.1016/j.imu.2021.100745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Priya R., Sumitha R., Doss C.G.P., Rajasekaran C., Babu S., Seenivasan R., Siva R.J.P.m. Molecular docking and molecular dynamics to identify a novel human immunodeficiency virus inhibitor from alkaloids of Toddalia asiatica. Phcog Mag. 2015;11:S414. doi: 10.4103/0973-1296.168947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prasanth D., Murahari M., Chandramohan V., Panda S.P., Atmakuri L.R., Guntupalli C. In silico identification of potential inhibitors from Cinnamon against main protease and spike glycoprotein of SARS CoV-2. J. Biomol. Struct. 2020:1–15. doi: 10.1080/07391102.2020.1779129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Basit A., Ali T., Rehman S.U. Truncated human angiotensin converting enzyme 2; a potential inhibitor of SARS-CoV-2 spike glycoprotein and potent COVID-19 therapeutic agent. J. Biomol. Struct. 2020:1–10. doi: 10.1080/07391102.2020.1768150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gupta S., Singh A.K., Kushwaha P.P., Prajapati K.S., Shuaib M., Senapati S., Kumar S. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J. Biomol. Struct. 2021;39:4334–4345. doi: 10.1080/07391102.2020.1776157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. 2021;39:3449–3458. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Because the majority of work is performed using licensed software, these are not transferable. Additional scripts written by members of the group may be made available upon request.