Abstract
SARS-CoV-2 Omicron strain emergence raised concerns that its enhanced infectivity is partly due to altered spread/contamination modalities. We therefore sampled high-contact surfaces and air in close proximity to patients who were verified as infected with the Omicron strain, using identical protocols applied to sample patients positive to the original or Alpha strains. Cumulatively, for all 3 strains, viral RNA was detected in 90 of 168 surfaces and 6 of 49 air samples (mean cycle threshold [Ct]=35.2±2.5). No infective virus was identified. No significant differences in prevalence were found between strains.
Keywords: SARS-CoV-2, Omicron, Alpha, original, surface contamination, air samples
SARS-CoV-2 was shown to be transmitted mainly by respiratory droplets and direct contact with contagious individuals. Previously, we characterized this transmissibility through contaminated inanimate surfaces, which is indirect contact (Ben-Shmuel et al., 2020). Recently, emergence of the Omicron (B.1.1.529) variant caused a global surge in new cases, rapidly spreading while seemingly causing an altered, less severe disease (Araf et al., 2022; Kannan et al., 2021). The infection rate raised concerns regarding the variant's mode of spread in the populace. Thus, debate has arisen whether Omicron's increased infectivity is due to altered contamination/persistence on surfaces and/or the gain of airborne transmissibility (Riediker et al., 2022; Wong et al., 2022). We therefore executed a contamination sampling campaign in proximity to patients positive for the original, Alpha (B.1.1.7), or Omicron SARS-CoV-2 strains.
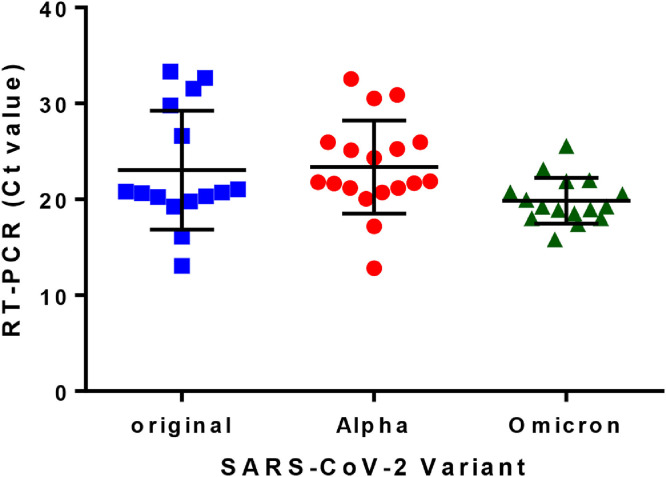
Omicron-infected patients (n=16) were sampled during January 2022 in a hospital isolation ward (n=12, mild-to-severe condition) and in a quarantine hotel (n=4, mild condition). Original strain– or Alpha variant–infected patients (n=15 and n=18, respectively) were sampled during February 2021 in a hospital ward (mild-to-severe condition). Inclusion criteria were newly hospitalized patients (within 24-48 hours of RT-PCR) and high upper respiratory viral loads (by reverse transcriptase–PCR [RT-PCR], 12-36 hours before sampling). Patients were not masked and were in bed during the entire sampling process. As shown (Figure 1 ), all 3 groups were comparable, with statistically similar mean Ct values of 23.1 (13.1-33.3), 23.3 (12.1-32.5), and 21.1 (17.4-26.8) for patients infected with the original, Alpha, or Omicron strains, respectively. Surface samples were taken from high-contact objects in close proximity to each patient (bedrails, bedside tables, and patients’ monitors). Each surface was swabbed with 3 sterile 6” applicators, sampling an area of 20 × 20 cm, pooled into a 15 ml tube containing 2 ml viral transfer medium (Minimal Essential Media [MEM] supplemented with 2% fetal calf serum [FCS] and 200 Units/ml Penicillin, 0.2% streptomycin, and 25 units/ml Nystatin). Air sampling was performed using an MD8 air sampler (Sartorius, Gottingen, Germany) equipped with 3.0 µm gelatin membranes, at 50 liter/min for 20 minutes (1000 liters per sample), positioned facing the patient, 1-1.5 m away. After sampling, each gelatin membrane was immediately dissolved in a 50 ml tube containing 10 ml viral transfer medium. Cold-chain transport of samples was maintained (4-8°C), with sample processing performed within 2-3 hours of sampling. RNA extraction, RT-PCR assays, and virus infectivity cytopathic effect (CPE) assays were performed as previously described (Ben-Shmuel et al., 2020).
Figure 1.
Patient viral load. Upper respiratory SARS-CoV-2 viral load in patients as determined by RT-PCR of nasal and mouth swabs (original strain n= 15, Alpha strain n=18, Omicron strain n=16). Data symbols present individual patient Ct values from 12-36 hours before sampling; black bars present patients’ group mean±SD. Statistical analysis carried by Prism 6 for Windows (GraphPad Software, San Diego, California, USA). Ct, cycle threshold; RT-PCR, reverse transcriptase–PCR.
Viral RNA was detectable on 60% of high-contact surfaces near Omicron-infected patients (table 1 ). Contamination, inferred by Ct values, was relatively low (Ct=35.2±2.5). For the original and Alpha strains, each group included ventilated patients (original 6/15, Alpha 6/18); similar levels of contamination were detected, at a slightly lower prevalence (46%), not a statistically significant difference (p=0.29). Patient monitors were sampled to assess medical staff contamination via gloves. Viral RNA was detected on 68%, 52%, and 41% of the patients’ monitors for patients infected with the original, Alpha, and Omicron strains, respectively (statistically insignificant, p=0.26). Contamination levels (by Ct) were similar for all strains. None of the surfaces sampled contained viable infectious virus (0/168, table 1). Taken together, high-contact surface and monitor contamination rates were virtually identical for all strains (55.6%, 49.2%, and 56.7% for the original, Alpha, and Omicron strains, respectively). Finally, we detected traces of viral RNA in 3 of 16 air-sample filters next to Omicron-infected patients, 2 of 18 filters next to Alpha-infected patients, and 1 of 15 filters next to original strain–infected patients. These differences were also statistically insignificant (p=0.59). All air samples were found to be free from infective virus (0/49, table 1).
Table 1.
SARS-CoV-2 contamination on surfaces and air samples in proximity to patients with COVID-19
| SARS-CoV-2 strain | Sample type | description | Positive/total samples(%) | Mean Ct value(range) | Infective virus/total samples(%) |
|---|---|---|---|---|---|
| Original strain | Surface samples(400 cm2) | Bed rails, bedside table | 12/26 (46%) | 33.3 (31.7-35.5) | 0/26 (0%) |
| Patient's monitor | 13/19 (68%) | 33.4 (30.1-36.2) | 0/19 (0%) | ||
| Air Sampling(1 m3) | 1-1.5 m in front of the patient | 1/15 (6%) | 35.1 (35.1) | 0/15 (0%) | |
| Alpha (B.1.1.7) | Surface samples(400 cm2) | Bed rails, bedside table | 20/42 (47%) | 31.6 (24.4-36.3) | 0/42 (0%) |
| Patient's monitor | 11/21 (52%) | 33.2 (26.4-37.1) | 0/21 (0%) | ||
| Air Sampling(1 m3) | 1-1.5 m in front of the patient | 2/18 (11%) | 34.2 (32.1-36.3) | 0/18 (0%) | |
| Omicron (B.1.1.529) | surface samples(400 cm2) | Bed rails, bedside table | 29/48 (60%) | 35.2 (29.5-39.5) | 0/48 (0%) |
| Patient's monitor | 5/12 (41%) | 36.7 (34.5-38.6) | 0/12 (0%) | ||
| Air Sampling(1 m3) | 1-1.5 m in front of the patient | 3/16 (19%) | 36.7 (36.3-37.1) | 0/16 (0%) | |
| Total | surface samples (400 cm2) | 90/168 (53%) | 0/168 (0%) | ||
| Air samples(1 m3) | 6/49 (12%) | 0/49 (0%) |
Abbreviation: Ct, cycle threshold.
This study aimed to test whether the Omicron variant is better at contaminating surfaces, surviving on real-world surfaces, or being an airborne infective pathogen. Although surface contamination was substantial for all strains (90/168 RT-PCR positive samples, 53%), viral viability in the clinical setting was negligible, with no samples containing cultivable virus. The rates and Ct values observed for contaminated samples were similar between variants. Compared with this relatively high rate, air sample contamination was both significantly less frequent and much lower: 12% contamination, mean Ct value of 35.6 (32.1-37.1). Differences between strains were statistically insignificant. Viable SARS-CoV-2 was not isolated from any air sample. Our results show that in terms of contamination, viability, and stability on surfaces and in air samples, the Omicron variant is remarkably similar to previous COVID-19 strains (original and Alpha strains). This correlates with previous studies conducted by us (Ben-Shmuel et al., 2020) and others (Lane et al., 2021; Ong et al., 2021; Thakar et al., 2021; Winslow et al., 2021) in healthcare settings, isolation units, quarantine hotels, and schools. Moreover, it was recently shown that SARS-CoV-2 significantly loses viability, with near complete viability loss occurring within 2-5 minutes, in controlled laboratory aerosolized dispersion at normal relative humidity (Oswin et al., 2022).
This preliminary work suggests that Omicron's increased transmissibility does not result from acquiring airborne infectivity, higher environmental contamination, or better resilience on surfaces. Alternatively, plausible explanations may include the mutated spike protein's higher affinity toward the angiotensin-converting enzyme 2 (ACE2) receptor and lower immune recognition. In addition, enhanced viral loads in clinically milder patients facilitate spread by having more infective, less symptomatic individuals in the community for longer durations before diagnosis and isolation.
Acknowledgments
Conflict of interest
None declared.
Funding statement
No external funding was received.
Ethical approval
The study was approved by the ethics committee of Rambam hospital and and the Institutional Biosafety Committee at Israel Institute for Biological Research.
References
- Araf Y, Akter F, Tang Y, Fatemi R, Parvers S, Zheng C, et al. Omicron variant of SARS-CoV-2: genomics, transmissibility, and respones to the current COVID-19 vaccines. J Med Vrol. 2022;94(5):1825–1832. doi: 10.1002/jmv.27588. Epub 2022 Jan 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shmuel A, Brosh-Nissimov T, Glinert I, Bar-David E, Sittner A, Poni R, et al. Detection and infectivity potential of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) environmental contamination in isolation units and quarantine facilities. Clinical Microbiology and Infection. 2020;26:1658–16662. doi: 10.1016/j.cmi.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S, Shaik P, Ali S, Sheeza A. Omicron (B.1.1.529)- variant of concern- molecular profile and epidemiology: a mini review. Eur Rev Med Pharmacol Sci. 2021;25:8019–8022. doi: 10.26355/eurrev_202112_27653. [DOI] [PubMed] [Google Scholar]
- Lane M, Brownsword E, Babiker A, Ingersoll J, Waggoner J, Ayers M, et al. Bioaerosol sampling for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in referral center withcritical ill coronavirus disease 2019 (COVID-19) patients march-may 2020. Clinical Infectious Diseases. 2021;73:1790–1794. doi: 10.1093/cid/ciaa1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S, Tan Y, Coleman K, Tan B, Leo Y, Wang D, et al. Lack of viable severe acute respiratory coronavirus 2 (SARS-CoV-2) among PCR-positive air samples from hospital rooms and community isolation facilities. Infection Control & Hospital Epidemiology. 2021;42:1327–1332. doi: 10.1017/ice.2021.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswin H, Haddrell A, Otero-Fernandez M, Mann J, Cogan T, Hilditch T, et al. The dynamics of SARS-CoV-2 infectivity with changes in aerosol microenviorment. MedRxiv. 2022 doi: 10.1101/2022.01.08.22268944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riediker M, Briceno-Ayala L, Ichihara G, Albani D, Poffet D, Tsai D, et al. Higher viral load and infectivity increase risk of aerosol transmission for Delta and Omicron variants of SARS-CoV-2. Swiss Med Wkly. 2022;152:w30133. doi: 10.4414/smw.2022.w30133. [DOI] [PubMed] [Google Scholar]
- Thakar A, Dutkiewicz S, Hoffman T, Joyce P, Shah V. Absence of SARS-CoV-2 in the air and on the surfaces within the school environment. J Med Microbiol. 2021;70 doi: 10.1099/jmm.0.001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow R, Zhou J, Windle E, Nur I, Lall R, Ji C, et al. SARS-CoV-2 environmental contamination from hospitalised patients with COVID-29 reciving aerosol-generating procedures. Thorax. 2021;0:1–9. doi: 10.1136/thoraxjnl-2021-218035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Au A, Chen H, Yuen L, Lung D, Chu A, et al. Transmition of Omicron (B.1.1.529) - SARS-Cov-2 variant of concern in a designed quarantine hotel for travelers: a challange of elimination strategy of COVID-19. THe LANCET Regional Health Western Pacific. 2022 doi: 10.1016/j.lanwpc.2021.100360. published online Dec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]