Abstract
Parkin, an E3 ubiquitin ligase, plays a role in maintaining mitochondrial homeostasis through targeting damaged mitochondria for mitophagy. Accumulating evidence suggests that the acetylation modification of the key mitophagy machinery influences mitophagy level, but the underlying mechanism is poorly understood. Here, our study demonstrated that inhibition of histone deacetylase (HDAC) by treatment of HDACis activates mitophagy through mediating Parkin acetylation, leading to inhibition of cervical cancer cell proliferation. Bioinformatics analysis shows that Parkin expression is inversely correlated with HDAC2 expression in human cervical cancer, indicating the low acetylation level of Parkin. Using mass spectrometry, Parkin is identified to interact with two upstream molecules, acetylase acetyl-CoA acetyltransferase 1 (ACAT1) and deacetylase HDAC2. Under treatment of suberoylanilide hydroxamic acid (SAHA), Parkin is acetylated at lysine residues 129, 220 and 349, located in different domains of Parkin protein. In in vitro experiments, combined mutation of Parkin largely attenuate the interaction of Parkin with PTEN induced putative kinase 1 (PINK1) and the function of Parkin in mitophagy induction and tumor suppression. In tumor xenografts, the expression of mutant Parkin impairs the tumor suppressive effect of Parkin and decreases the anticancer activity of SAHA. Our results reveal an acetylation-dependent regulatory mechanism governing Parkin in mitophagy and cervical carcinogenesis, which offers a new mitophagy modulation strategy for cancer therapy.
KEY WORDS: Parkin, Mitophagy, ACAT1, HDAC2, Acetylation, Ubiquitination, Tumor suppression, Cervical cancer
Abbreviations: ACAT1, acetyl-CoA acetyltransferase 1; CCK-8, cell counting kit-8; COXⅣ, cytochrome c oxidase Ⅳ; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HDAC, histone deacetylase; HIF-1α, hypoxia inducible factor-1α; HSP60, heat shock protein 60 kDa; LC3, microtubule-associated proteins 1A/1B light chain 3; MS, mass spectrometry; MFN2, mitofusion 2; PINK1, PTEN induced putative kinase 1; PARK2, Parkin; ROS, reactive oxygen species; SAHA, suberoylanilide hydroxamic acid; TOMM20, translocase of outer mitochondrial membrane 20; TIM23, translocase of the inner membrane 23; TSA, trichostatin A; ULK1, unc-51 like autophagy activating kinase 1; VDAC1, voltage-dependent anion-selective channel protein 1
Graphical abstract
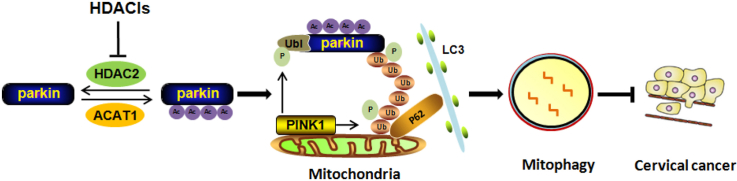
HDACis inhibit HDAC2 activity and cause Parkin acetylation in cervical cancer, which exerts an impact on Parkin-dependent mitophagy and tumor suppression. It reveals an acerylation-dependent mechanism regulating the tumor-suppressive function of Parkin.
1. Introduction
Mitophagy refers to the process of selective removal of damaged or dysfunctional mitochondria via the autophagy/lysosome pathway, which plays an important role in mitochondrial quality control. In the past decade, the molecular mechanisms underlying mitophagy have been extensively studied. The well-studied pathway is PINK1–Parkin mediated ubiquitin pathway1. Under normal condition, PINK1 is localized in mitochondrial inner membrane and cleaved by PARL protease2. Parkin is an E3 ubiquitin ligase and initially found to be closely related to Parkinson's disease3. Upon mitochondrial damage, PINK1 cleavage is inhibited and PINK1 is translocalized to mitochondrial outer membrane, where phosphorylates ubiquitin and Parkin at Ser654. After phosphorylation, Parkin is activated and ubiquitinates mitochondrial outer membrane proteins, which are further recognized by autophagy adaptor proteins5,6. Finally, they bind to microtubule-associated protein 1A/1B-light chain 3 (LC3) on the autophagosome membrane and are degraded by the lysosomal pathway7.
In addition to phosphorylation and ubiquitination modification, the key mitophagy machinery may undergo extensive acetylation modification8. Acetylation of mitochondrial proteins has been revealed to influence mitophagy process9. Under starvation, mitochondrial acetyltransferase GCN5-like protein 1 deletion diminishes mitochondrial protein acetylation and augments mitochondrial enrichment of autophagy mediators10. With the activation of transcription factor EB, mitochondrial degradation is enhanced11. Phosphorylation of ubiquitin leads to the activation of Parkin12 while acetylation of ubiquitin represses the formation and elongation of ubiquitin chains13. In addition, there are also many HDACs regulating mitophagy, which are aberrantly expressed in multiple cancers, including cervical cancer14. HDAC6 is recruited to mitochondria and positively regulates mitophagy upon mitochondrial damage15. Sirtuin 1 deletion increases production of reactive oxygen species (ROS) and recruits Parkin to mitochondria and induces mitophagy16. Other mitochondrial deacetylase Sirtuin 2 ablation leads to heat shock protein 70 (HSP70) acetylation and mitophagy induction17; Sirtuin 3 has also been found to associate with mitophagy process9,18. When Sirtuin inhibition, mitophagy is observed to be activated that is selective for depolarized mitochondria19. Given the regulatory effect of acetylation on mitophagy, it is necessary to investigate the acetylation of the key mitophagy machinery, which could offer a unique window of opportunity for the control of mitophagy-related diseases, such as cancer.
Increasing evidence demonstrate that dysfunction in mitophagy has a close connection with tumorigenesis and tumor development20. But the role of mitophagy in carcinogenesis remains largely unclear. On the one hand, mitophagy serves as tumor suppression by eliminating dysfunctional mitochondria21. Certain mitophagy receptors or adaptor proteins are tumor suppressor in cancer and their loss, mutation or functional changes result in tumorigenesis22,23. Parkin is downregulated in multiple tumors including cervical cancer24, which results in mitotic instability25 and also attenuates its regulatory role in glucose metabolism and antioxidant defense26. Loss of PINK1 expression activates hypoxia inducible factor-1α (HIF-1α)-dependent Warburg effect and inflammasome in cancer cells27. The key mitophagy receptor BCL-2 interacting protein 3 is epigenetically silenced28, leading to drug resistance while BCL-2 interacting protein 3L/NIX activates cell apoptosis29. FUN14 domain containing 1 is hypoxia-induced mitophagy receptor and suppresses tumor development through inhibiting inflammasome activation30. On the other hand, mitochondrial maintenance is crucial for cancer cell survival and mitophagy serves to eliminate the dysfunctional mitochondria to relieve stress21,31. In tumor microenvironment, HIF-1α-driven mitophagy leads to hypoxia adaption and cancer cell survival, promoting tumorigenesis32,33. Moreover, HIF-1α quenches mitochondrial biogenesis, which in turn stabilizes HIF-1α in a regulatory loop mechanism34. Hence, the study of mitophagy regulatory mechanisms in cancer-related events is of great significance, which is helpful for the discovery of novel mitophagy interventional strategies for cancer therapy.
In the present study, we investigated the acetylation modification of Parkin and observed that Parkin acetylation had an important impact on Parkin-dependent mitophagy and tumor suppression. Our results reveal an acerylation-dependent molecular mechanism regulating the function of Parkin in cancer. Using mass spectrometry, we successfully identify the upstream molecules controlling Parkin acetylation, which are ACAT1 and HDAC2, and the acetylation sites of Parkin are also identified and validated. Mutation assay reveals the importance of Parkin acetylation in mitophagy and tumor suppression, indicating the tumor suppressive role of mitophagy in cervical cancer. Thus, the acetylation of Parkin may be a novel target in mitophagy-modulated cancer therapy.
2. Materials and methods
2.1. Cell culture
HeLa, SiHA and HEK293 cells were obtained from ATCC (American Type Culture Collection). Cells were maintained in DMEM (Sigma, D1152) containing 10% fetal bovine serum (HyClone, SV30160.03) in a 5% CO2 atmosphere at 37 °C.
2.2. Reagents and antibodies
The antibodies used in our experiments included: ACAT1 (CST, 44276), acetylated-lysine (CST, 9441), BAX (Proteintech, 50599-2-Ig), BCL-2 (Proteintech, 12789-1-AP), COXⅣ (CST, 11967), cleaved-PARP-1 (CST, 5625), cleaved-caspase3 (CST, 9661), EP300 (Abcam, ab14984), FLAG (Sangon Biotech, D191041), anti-FLAG® M2 Affinity Gel (MERCK, F2426), GAPDH (Proteintech, 600004-1-lg), HDAC1 (Proteintech, 10197-1-AP), HDAC2 (Proteintech, 12922-3-AP), HDAC3 (Proteintech, 10255-1-AP), HSP60 (CST, 12165), HA (Sangon Biotech, D110004), Ki67 (Proteintech, 27309-1-AP), LC3 (MERCK, L7543), MFN2 (Proteintech, 12186-1-AP), PINK1 (CST, 6946), Parkin (Proteintech, 14060-1-AP; CST, 4211), Parkin (phospho-Ser65, Biorbyt, orb312554), P62 (MERCK, P0067), TOMM20 (CST, 42406), TIM23 (Santa Cruz Biotechnology, sc-514463), ubiquitin (Proteintech, 10201-2-AP), ULK1 (CST, 8054), VDAC1 (Proteintech, 10866-1-AP), α-tubulin (Sigma, T6199), β-actin (ABclonal, AC004).
The chemicals used in our experiments were: bafilomycin A1 (Sigma, B1793), chloroquine (MedChemExpress, HY-17589A), Hoechst (Beyotime, 33342), suberoylanilide hydroxamic acid (SAHA; MedChemExpress, HY-10221), and trichostatin A (TSA, MedChemExpress, HY-15144).
2.3. Small interfering RNA (siRNA) and plasmids transient transfection
The siRNAs targeting PINK1 (Santa Cruz Biotechnology, sc-44598), ULK1 (GenePharma, Shanghai), ACAT1 (Santa Cruz Biotechnology, sc-96390), HDAC1 (GenePharma, Shanghai), HDAC2 (Santa Cruz Biotechnology, sc-29345), HDAC3 (GenePharma, Shanghai) or the plasmids for Flag-Parkin, GFP-Parkin were transfected into HeLa or HEK293 cells using Lipofectmine™ 3000 Transfection Reagent (Invitrogen, L3000015) according to the manufacturer's protocols.
2.4. Western blotting
After indicated treatment, cells were collected by scraping and washed twice with PBS. Then, cells were lysed with RIPA Lysis Buffer (Beyotime, P0013). Protein concentrations in the supernatant were measured by a BCA Protein Assay Kit (Solarbio, PC0020). Equal amounts of proteins were separated by SDS-polyacrylamide gels and then electroblotted onto Immun-Blot® PVDF membrane (polyvinylidene fluoride, Bio-Rad, 1620184). After blocking with 5% skimmed milk, the membrane wad incubated with primary and secondary antibodies, respectively. Finally, the immunoblots were visualized with an ECL system.
2.5. Transmission electron microscopy assay
Cells were seeded into 5-cm Petridishes and treated with SAHA. After washing with PBS, cells were scraped and harvested with 4% glutaraldehyde and centrifuged at 4 °C. The precipitated cells were continued to fix with 4% glutaraldehyde for another 2 h at room temperature and stored at 4 °C. Electron photomicrographs of the HeLa cells were taken by Wuhan Goodbio Technology Co.
2.6. Immunofluorescence staining and confocal microscope
Cells were first seeded to Nunc™ Lab-Tek™ II Chamber Slide™ (Lab-Tek, NUNC, 155411). After the designated treatments, cells were fixed with 4% paraformaldehyde and permeabilized by 0.25% Triton X-100. After incubation with first antibody and fluorochrome-conjugated secondary antibody. Fluorescence intensity of cells was observed using Leika TCS SP5 Confocal (Leica MICROSYSTEMS, Germany) and representative cells were selected and photographed. ImageJ was used to analyze the colocalization.
2.7. Immunohistochemistry assay
Tumor tissue samples were embedded in paraffin and antigen retrieval was performed. Following the blockade of endogenous peroxidase activity, the samples were incubated with the primary antibodies of interest and the appropriate secondary antibodies and reacted with DAB Detection Kit (Servicebio, G1212). The immunoreactive staining of proteins in tumor tissue was detected using Immunofluorescence Microscopy (Nikon, Japan).
2.8. Mito-Keima assay
HeLa cells were transfected with the mKeima-Red-Mito-7 (Addgene, 56018) plasmid using Lipofectamine 3000 for 24 h and then treated with SAHA for another 12 h. The cells were imaged using Leika TCS SP5 Confocal (Ex = 550 nm, Em = 620 nm for acidic red fluorescence; Leica Microsystems, Germany).
2.9. Immunoprecipitation assay
Briefly, the cells were lysed on ice for 30 min with RIPA Lysis Buffer (Beyotime, P0013) with phosphatase and protease inhibitor. Cell lysates were incubated with indicated antibodies overnight with gentle rocking at 4 °C. The immunoprecipitates were then washed three times in IP buffer and the immunoprecipitated complexes were eluted by boiling in sample buffer. Immunoblotting was performed to analyze the precipitated proteins.
2.10. In vitro acetylation assay
The acetylation reaction was performed based on a reported protocol with modifications35. Briefly, in 30 μL of reaction mixture containing 20 mmol/L Tris-HCl, pH 8.0, 20% glycerol, 100 mmol/L KCl, 1 mmol/L dithiothreitol, 0.1 mmol/L EDTA, 2 μmol/L SAHA, 1 mmol/L PMSF, 100 μmol/L acetyl-CoA (Sigma, A2056), 200 ng ACAT1 recombinant protein and 100 ng of Parkin recombinant protein were added and incubated at 30 °C for 1 h. The reaction was stopped by adding SDS sample buffer. The samples were subjected to SDS-PAGE and analyzed using immunoblotting.
2.11. Mitochondria isolation and analysis
After indicated treatment, cells were harvested, resuspended in mitochondria isolation reagent, and transferred to Dounce Tissue Grinder. After homogenization, cellular mitochondria were isolated using Cell Mitochondria Isolation Kit (Beyotime, C3601) according to the manufacturer's instructions. The isolated mitochondria were lysed with RIPA Lysis Buffer (Beyotime, P0013). The sample was separated on 12% (w/v) SDS-PAGE gels through electrophoresis.
2.12. Mass spectrometry (MS) analysis
In this study, we used MS analysis for two objectives. First, to determine the interaction proteins with Parkin, HEK293 cells with Flag-Parkin overexpression were treated with SAHA for 12 h and anti-FLAG® M2 Affinity Gel was used for immunoprecipitation. Immunoprecipitation of FLAG-Parkin from cell lysates were performed as described36. Immunoprecipitates were washed with lysis buffer and resolved on an SDS-PAGE gel. And gel slices from SDS-PAGE separation of cell lysates were subjected to in-gel tryptic digestion. The resulting peptides were analyzed by LC–MS (Thermo Fisher Easy-nLC 1000, Thermo Fisher LTQ Obitrap ETD). The results are retrieved by MASCOT software according to the following conditions. After label-free relative quantitation, GO (gene ontology) analysis, KEGG (kyoto encyclopedia of genes and genomes) pathway analysis and IPA (ingenuity pathways) analysis were conducted to analyze the protein-interaction network among the identified proteins by LC–MS. The analysis was performed by Jiyun Biotech (Shanghai, China).
Second, to identify the acetylation sites of Parkin, HEK293 cells with Flag-Parkin overexpression were treated with SAHA and cell lysates were immunoprecipitated using anti-FLAG® M2 Affinity Gel. The proteomic sample preparation was performed as described previously. The peptides were analyzed with an MS system which comprised a Dionex Ultimate 3000 RSLC nano LC system (Thermo Fisher, Waltham, MA, USA) coupled to a Q-Exactive mass spectrometer (Thermo Fisher, Waltham, MA, USA). The UniProt human database was used for data searches using an in-house Mascot server (version 2.4.1, Matrix Science, Boston, MA, USA). Shortlisted acetylated peptides were further confirmed by manual inspection of MS/MS spectra to identify the exact acetylation sites.
2.13. Determination of mitochondrial membrane potential
JC-1 dye (Thermo Fisher SCIENTIFIC, T3168) is a membrane permeable dye to determine mitochondrial membrane potential. It can selectively enter the mitochondria where it reversibly changes color with membrane potential changing. Upon membrane polarization, the formation of JC-1 aggregates cause shifts in emitted light from 530 to 590 nm when excited at 488 nm. The treated cells were labeled with 10 μg/mL JC-1 dye and fluorescence was determined using BD FACSCanto™ II Cell Analyzer (BD Biosciences-US) or Immunofluorescence Microscopy (Nikon, Japan).
2.14. Mitochondrial ROS measurement
Cells were seeded in 12-well plate and treated with SAHA for the indicated times. Then, the cells were exposed to MitoSOX™ Red Mitochondrial Superoxide Indicator (Thermo Fisher Scientific, M36008, 5 μmol/L) for 30 min at 37 °C in the dark. Cells were harvested and prepared for measurement. BD FACSCanto™ II Cell Analyzer (BD Biosciences-US) was used to measure the fluorescence intensity (Ex/Em 510/580 nm).
2.15. Cell viability assay
Cell viability was measured using the cell counting kit-8 (CCK-8; Yeasen, 40203ES60) assay. Cells were first seeded in 96-well plates and allowed to attach overnight in a 5% CO2 incubator (Thermo Fisher Scientific). The cells were then treated with drugs as indicated. 10 μL CCK-8 was added to each well and incubated for 1–4 h at 37 °C. The absorbance at 450 nm was measured with a Thermo Scientific Multiskan Sky Microplate Spectrophotometer (Thermo Fisher Scientific).
2.16. In vivo xenograft tumor model
Four-week-old female BALB/c nude mice were purchased from the Institute of Zoology, Zhejiang Chinese Medical University. All experiments were performed in accordance with the official recommendations of the Chinese Zoological Society, and animals received humane care according to the criteria outlined in Guide for the Care and Use of Laboratory Animals. A suspension containing 2 × 106 HeLa cells with empty vector, wild-type or mutant Parkin expression, was subcutaneously injected into the right flanks of the nude mice. After 7 days, mice were randomly divided into five groups: Empty vector; Parkin-WT control and SAHA treatment; Parkin-KR control and SAHA treatment. Tumor-bearing mice were intraperitoneal injected with SAHA (50 mg/kg) or saline, respectively. The tumor dimensions were measured using a Vernier caliper twice per week. Mice were killed 30 days after inoculation, and xenograft tumors were removed and weighed.
2.17. Statistical analysis
All Western blotting data and image data presented were representative of 3 independent experiments. The numeric data were presented as mean ± standard deviation (SD) from 3 independent experiments. An analysis of variance and Student's t-test were used for comparison among groups. A P-value less than 0.05 was significant (∗P < 0.05, ∗∗∗P < 0.01, ∗∗∗P < 0.001) and a P-value higher than 0.05 was not statistically significant (#P > 0.05).
3. Results
3.1. HDACi SAHA activates mitochondrial-dependent apoptosis through inhibition of HDAC1/2/3 activity
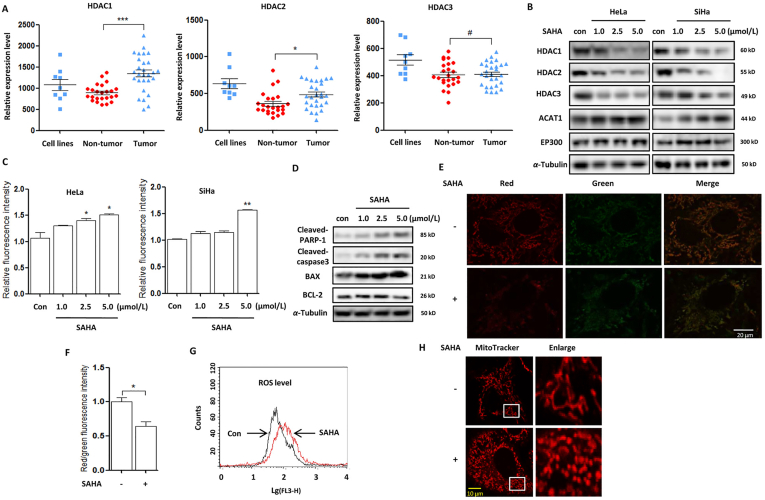
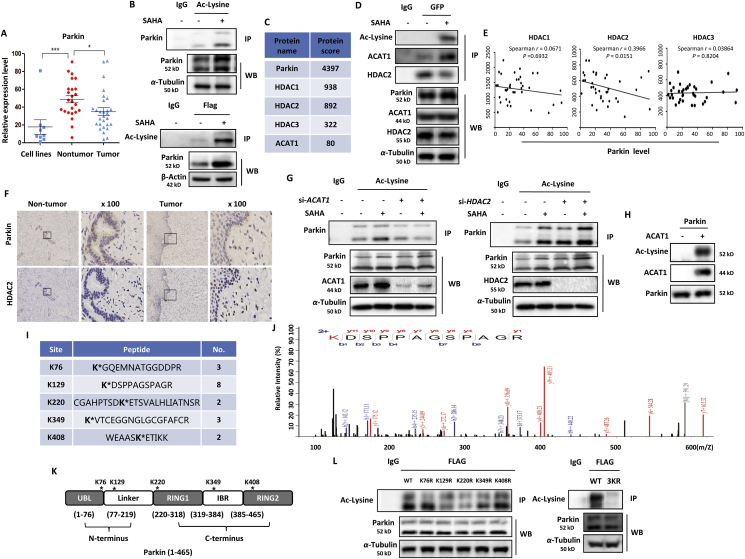
Cervical cancer cells are characterized with the expression of human papillomavirus oncoproteins E6 and E737. The oncogencity of E6 is mediated in part by targeting P53 and PDZ-family tumor suppressor proteins for rapid proteasomal degradation38, whereas E7 oncoprotein acts in part by co-operating with class I HDACs39,40. Oncomine database analysis shows that HDAC1/2 were significantly upregulated in human cervical cancer tissue while HDAC3 was only highly overexpressed in human cervical cancer cell lines (Fig. 1A), including C4-I, CaSki, C-33A, HT-3, SiHa, SW756, MS751, ME-180 and HeLa. In HeLa and SiHA cells, HDACis SAHA treatment decreased the expression of HDAC1/2/3 in a dose-dependent manner (Fig. 1B). Cell morphology changes showed the shrinked, round and low-density cells under SAHA treatment and more dead cells were observed with the dose increasing (Supporting Infromation Fig. S1A). We next analyzed the type of cell death and detected a significant increase of cell fluorescence using annexin V staining (Fig. 1C), indicating cell apoptosis. Meanwhile, Western blotting analysis also showed that SAHA treatment increased the expression of the pro-apoptotic proteins, including BAX, cleaved poly (ADP-ribose) polymerase 1 (PARP-1) and caspase3, and decreased the expression of anti-apoptotic protein BCL-2 (Fig. 1D).
Figure 1.
SAHA treatment decreases HDACs expression and induces mitochondria-dependent apoptosis. (A) Analysis of the expression levels of HDAC1/2/3 in human cervical cancer cell lines, tumor & nontumor tissues from Oncomine database. ∗P < 0.05, ∗∗∗P<0.001, #P>0.05. (B) HeLa and SiHA cells were treated with different doses of SAHA (1.0, 2.5, and 5.0 μmol/L) for 24 h as designated. Cells were harvested and lysed for Western blotting. α-Tubulin was used as loading control. (C) as in (B), cells were stained with the Annexin V, Pacific Blue™ conjugate and measured by BD FACS™. Cell fluorescence was calculated and statistically analysed. ∗P < 0.05, ∗∗P < 0.01. (D) Cell lysates were prepared from SAHA-treated SiHA cell and subjected to Western blotting analysis using antibodies against PARP-1 and caspase3. (E) and (F) SiHA cells were first treated with SAHA (5 μmol/L, 12 h) and then stained with 2 μmol/L JC-1 dye for 15 min at 37 °C. Fluorescence of J-aggregates (red) and J-monomers (green) was examined either by confocal microscope (scale bar = 10 μm) or BD FACS™. The ratio of J-aggregates to J-monomers was calculated as indicated and statistically analysed. ∗P < 0.05. (G) as in (F), mitochondrial ROS was detected by mitochondrial superoxide indicator (5 μmol/L, 30 min) and flow cytometry graph was shown. (H) as in (E), SAHA-treated cells were stained using MitoTracker™ Red CMXRos (100 nmol/L, 15 min) for mitochondrial length. Confocal Microscope was performed for evaluation. Scale bar = 10 μm.
As is well established, a variety of key events in apoptosis occur in the mitochondria41, including the release of caspase activators, the participation of pro- and anti-apoptotic BCL-2 family proteins. In SAHA-treated cells, JC-1 staining showed the reduction of red fluorescence (J-aggregate formation, Fig. 1E, F) and the ratio of J-aggregates to J-monomers was significantly decreased, indicating a lower mitochondrial polarization. In addition, we also evaluated mitochondrial oxidation using flow cytometry and found that there was more mitochondrial ROS production under SAHA treatment (Fig. 1G). Consequently, mitochondria could be damaged due to oxidative stress and we performed the mitochondria-selective probe MitoTracker staining and observed the decrease of mitochondria length and excessive mitochondria fragmentation (Fig. 1H).
3.2. SAHA treatment induces mitophagy in human cervical cancer cells
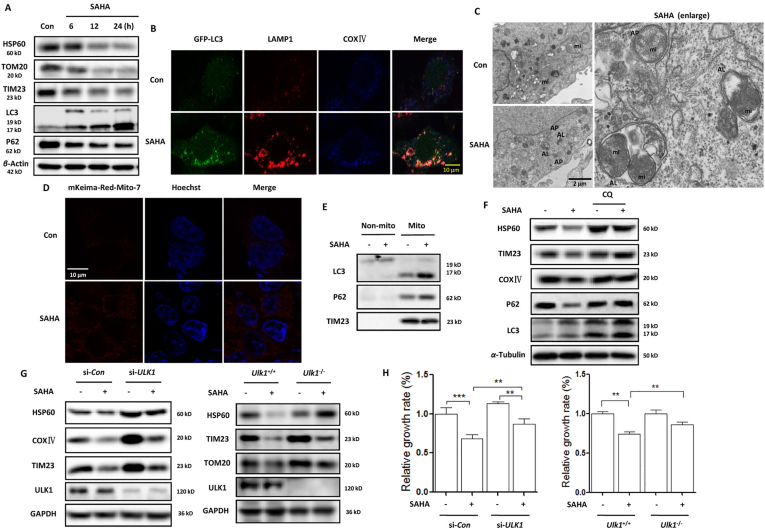
Our previous studies42 demonstrated that HDACis activate autophagy and autophagy inhibition enhanced the therapeutic efficacy of HDACis. Under SAHA treatment, there was too much mitochondria damage. To maintain mitochondrial quality control, mitophagy could be induced to remove malfunctioning or damaged mitochondria through the lysosome, which is also essential for normal cellular physiology43. We thus examined the level of mitophagy in SAHA-treated SiHA cells. As shown in Fig. 2A, Western blotting analysis shows that SAHA treatment decreased mitochondrial protein levels of heat shock protein 60 (HSP60), translocase of outer membrane 20 (TOMM20), and translocase of the inner membrane 23 (TIM23) through inducing autophagy, which was accompanied with the increase of LC3 protein level (autophagosome marker) and the decrease of P62 protein level (autophagy substrate). Under confocal microscope, we found that in HeLa cells stably expressing GFP-LC3, SAHA treatment increased the colocalization of GFP-LC3 with LAMP1 in mitochondria, indicating the enhanced formation of mitophagosomes and its fusion with lysosomes (Fig. 2B). Transmission electron microscopy revealed high electron-density substances and abnormal mitochondria surrounded by double membranes (autophagosomes) or inside the autolysosomes, indicating the presence of mitophagosomes (Fig. 2C). To further confirm the occurrence of mitophagy, mito-Keima, a pH-sensitive fluorescent protein35, was transfected into HeLa cells to determine mitochondrial movement from the cytoplasm to the lysosome. As shown in Fig. 2D and Fig. S1C, red spots appeared in the cytoplasm under SAHA or TSA treatment, indicating that mitochondria tend to form autolysosomes. In addition, we prepared the mitochondrial fractions of SAHA-treated SiHA cells and detected the increase of LC3 (autophagosome marker) and P62 (a selective autophagy adaptor) in mitochondria (Fig. 2E).
Figure 2.
Mitophagy induction occurs in SAHA-treated human cervical cancer cells. (A) SiHA cells were treated with 5 μmol/L SAHA for different time (6, 12, and 24 h). Cell lysates were then prepared and subjected to Western blotting analysis for mitochondrial proteins TOMM20, TIM23 and HSP60. α-Tubulin was used as loading control. (B) HeLa cells stably expressing GFP-LC3 were treated with SAHA (5 μmol/L, 12 h). After immunostaining with LAMP1 (red) and COXⅣ (blue), cells were examined under confocal microscope. Scale bar = 10 μm. (C) Transmission electron microscopy images of the ultrastructure of SiHA cells under SAHA treatment (scale bar = 2 μm). AP and AL refer to double-membrane autophagosomes or autolysosomes. (D) HeLa cells were transfected with the mKeima-Red-Mito-7 plasmid, followed by treatment with SAHA. The fluorescence was detected by confocal microscope. Scale bar = 10 μm. (E) Mitochondrial fractions were prepared from SAHA-treated SiHA cells and subjected to Western blotting analysis of LC3 and P62. TIM23 was used as a representative of mitochondrial fraction. (F) SiHA cells were treated with 5 μmol/LSAHA in the presence or absence of chloroquine (10 μmol/L) for 12 h. Then mitochondrial protein levels were detected by Western bolting with α-tubulin was used as loading control. (G) SiHA cells were first transfected with the ULK1-specific siRNA and then cells were treated with SAHA (5 μmol/L, 12 h, left panel). The same treatment was also performed in both Ulk1+/+ and Ulk1−/− MEFs. Western blotting was used to determine the mitochondrial protein levels. (H) as in (G), after SAHA treatment, 10 μL CCK-8 solution was added into each well of the plate and the absorbance was measured at 450 nm using microplate spectrophotometer. ∗∗P < 0.01,∗∗∗P < 0.001.
Pharmacologic or genetic inhibition of autophagy was also performed to further examine the mitophagy level change. As shown in Fig. 2F, Western blotting analysis shows that autophagy inhibitor chloroquine impaired the decrease of mitochondrial proteins, including HSP60, TIM23 and cytochrome c oxidase Ⅳ(COXⅣ). In the presence of lysosomal inhibitor bafilomycin A1, the increase of mitochondrial ROS generation by SAHA treatment was also impaired (Fig. S1B). Unc-51 like autophagy activating kinase 1 (ULK1) is critical for the induction of autophagy, which is upregulated and translocates to fragmented mitochondria and induces mitophagy in response to hypoxia or mitochondrial uncouplers44. Here, we knocked down ULK1 in SiHA cells and observed that SAHA treatment failed to diminish the level of mitochondrial proteins (Fig. 2G), including HSP60, TIM23 and mitofusion 2 (MFN2). The similar results were also shown in Ulk1 knockout cells, where SAHA treatment only led to a large reduction of mitochondrial proteins in Ulk1 wild-type cells. Moreover, knockdown or knockout of ULK1 attenuated the growth-inhibitory effect of SAHA on SiHA cells (Fig. 2H), suggesting the tumor-suppressive role of mitophagy.
3.3. The involvement of PINK1–Parkin signaling pathway in SAHA-induced mitophagy
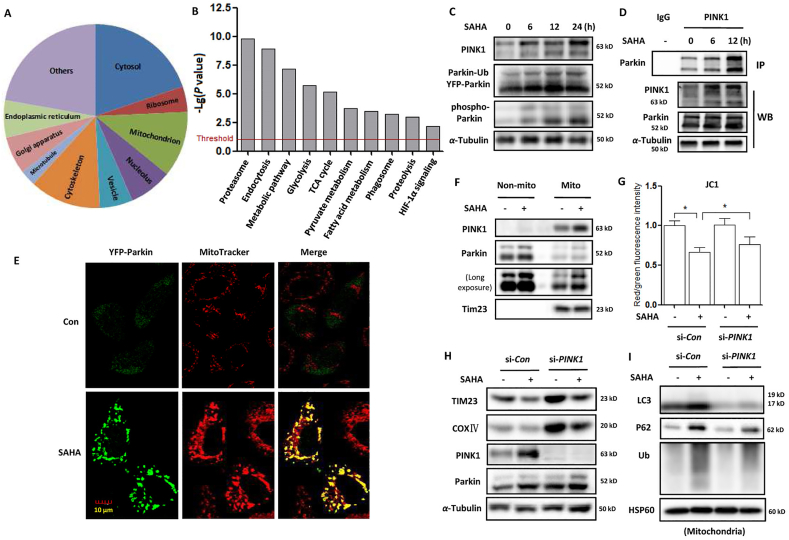
PINK1–Parkin mitophagy is a key mechanism to maintain mitochondrial quality control7. Upon loss of mitochondrial membrane potential, PINK1 and Parkin are activated to promote the proteasomal degradation of mitochondrial proteins and selective elimination of damaged mitochondria by autophagy31. To reveal the molecular mechanism of SAHA-induced mitophagy, we performed quantitative proteomics to identify the targets of Parkin. HEK293 cells with Flag-Parkin overexpression were treated with SAHA and cell lysates were then subjected to affinity enrichment using FLAG-fusion beads. MS (mass spectrometry) was used to pool and analyze the derived peptides and finally the target proteins were identified and quantified. In our study, 2619 proteins were identified as the specific targets of Parkin under SAHA treatment (Supporting Information Table S1). Subsequently, we performed GO (gene ontology) analysis of the Parkin targets under SAHA treatment. It was shown that these targets were broadly distributed in different parts of the cell, especially in the cytosol, nucleolus, mitochondria, ribosome, endoplasmic reticulum, Golgi apparatus etc. (Fig. 3A). KEGG (kyoto encyclopaedia of genes and genomes) analysis of pathways demonstrated that Parkin has multiple molecular functions (Fig. 3B), which are closely related with mitochondrial function, including proteasomal degradation, energy metabolism, phagosome formation, HIF-1α, etc. Among them, a large number of Parkin-targeted mitochondrial metabolic enzymes were listed (Supporting Information Table S2), which were involved in cellular energy metabolism, including glucose, amino acid, lipid etc. The above results indicate that Parkin may target and ubiquitinate mitochondrial proteins to initiate mitophagy.
Figure 3.
SAHA treatment activates the PINK1–Parkin signaling pathway and enhances Parkin-dependent mitophagy. SAHA-treated HEK293 cells with Flag-Parkin overexpression were lysed and subjected to immunoprecipitation using anti-FLAG® M2 affinity gel. Total 2619 proteins were profiled as target proteins of Parkin using LC–MS analysis. (A) GO analysis of cellular component distribution of the Parkin targets. (B) Top molecular and cellular functional classes to which the Parkin-targeted proteins are associated. (C) HeLa cells stably expressing YFP-Parkin were treated with 5 μmol/L SAHA for different time (6, 12, and 24 h). Cell lysates were prepared for Western blotting and the expression levels of PINK1 and Parkin were detected. (D) HeLa cells with Parkin expression were treated with 5 μmol/L SAHA at different time. Cell lysates were prepared and subjected to immunoprecipitation using anti-PINK1 antibody. The associated Parkin protein was detected using immunoblotting. (E) as in (D), after MitoTracker™ Red CMXRos staining, confocal microscope was performed to evaluate the localization of Parkin. Scale bar = 10 μm. (F) as in (D), mitochondrial fractions were prepared from SAHA-treated SiHA cells and subjected to Western blotting analysis. TIM23 was used as a representative of mitochondrial fraction. (G) SiHA cells were first transfected with the PINK1-specific siRNA and then cells were treated with SAHA (5 μmol/L, 12 h). After JC-1 dye staining, BD FACS™ was performed for evaluation and the ratio of J-aggregates to J-monomers was calculated and statistically analysed. ∗P < 0.05. (H) and (I) as in (G), SAHA-treated SiHA cells with or without PINK1 knockdown were either lysed or subjected to isolate mitochondria. Western blotting was used to determine the level of mitochondrial proteins (left panel) or autophagy and ubiquitin proteins (right panel).
In addition, we examined the effect of SAHA treatment on PINK1 expression and found that in HeLa cells expressing Parkin, SAHA treatment markedly increased the level of full-length PINK1 in a time-dependent manner (Fig. 3C). Meanwhile, the expression of phospho-Ser65 Parkin was also increased with time. An immunoprecipitation assay was also performed and revealed that SAHA treatment enhanced the interaction of PINK1 with Parkin (Fig. 3D). As a result, the recruitment of Parkin (green) to mitochondria (MitoTracker, red) was promoted in SAHA-treated cells (Fig. 3E), leading to the ubiquitination of mitochondrial proteins. Consistently, mitochondrial fractions from SAHA-treated cells showed the increased translocation of both PINK1 and Parkin proteins (Fig. 3F). These findings indicate that SAHA treatment stabilizes the expression of PINK1 at the mitochondrial surface and in turn recruits more Parkin to mitochondria.
To further confirm the role of the PINK1-dependent pathway in mitophagy induction by SAHA, we used siRNA to knock down PINK1 expression. As shown in Fig. 3G, SAHA treatment significantly increased mitochondrial depolarization, which was indicated by a decrease in the red/green fluorescence intensity ratio. When PINK1 was knocked down, depolarized regions indicated by the green fluorescence of the JC-1 monomers were decreased. The decrease of red to green fluorescence was also reversed with PINK1 knockdown (Fig. 3G). Confocal imaging also revealed that PINK1 knockdown attenuated the movement of mitochondria to the lysosome (Supporting Information Fig. S2).
Western blotting results showed that when PINK1 knockdown, the decrease of mitochondrial proteins COXIV and TIM23 levels following SAHA treatment was attenuated (Fig. 3H). In the mitochondrial fractions, knockdown of PINK1 also impaired the SAHA-induced increase in mitochondrial P62, LC3 and ubiquitin levels (Fig. 3I). These results indicate that SAHA treatment fails to induce mitophagy with PINK1 knockdown and SAHA-induced mitophagy is dependent on the PINK1 pathway.
3.4. SAHA treatment leads to Parkin acetylation through inhibition of HDAC2 activity
Parkin, a tumor suppressor, becomes inactivation due to loss of heterozygosity and plays an important role in human cervical cancer45,46. Oncomine database analysis also showed that Parkin was significantly downregulated in either human cervical cancer cells or cancer tissues (Fig. 4A). Parkin is mainly regulated by protein kinase PINK1 in a phosphorylation-dependent manner. Upon mitochondrial depolarization, PINK1 phosphorylates and activates both ubiquitin and Parkin at their respective Ser65 residues47,48. It is still not known whether there are other forms of post-translational modification in regulation of Parkin activity. Lysine acetylation is a reversible post-translational modification that plays a crucial role in regulating protein function, chromatin structure, and gene expression36,49. Thus, the acetylation level of Parkin was investigated in SAHA or TSA-treated HEK293 cells with Parkin overexpression using an immunoprecipitation assay. As shown in Fig. 4B and Supporting Information Fig. S3A and a significant increase in Parkin acetylation level was observed by SAHA or TSA treatment. The immunoprecipitated Parkin protein complex was further analyzed by MS and the associated acetylase and deacetylase were identified. As listed in Fig. 4C, ACAT1 and HDAC1/2/3 are revealed as the top candidates interacting with Parkin. It was further confirmed by immuoprecipitation with Parkin. As shown in Fig. 4D, SAHA treatment enhanced the interaction between ACAT1 and Parkin and attenuated the interaction between HDAC2 and Parkin.
Figure 4.
SAHA treatment inhibits HDAC2 activity and leads to Parkin acetylation. (A) Analysis of Parkin level in human cervical cancer cell lines, tumor and nontumor tissues from Oncomine database. ∗P < 0.05, ∗∗∗P < 0.001. (B) HEK293 cells were transiently transfected with Flag-Parkin and then treated with 2.5 μmol/L SAHA for 12 h. Cells were lysed and subjected to acetyl-lysine immunoprecipitation followed by immunoblotting for Parkin (above panel). In the below panel, cell lysates were immunoprecipitated with anti-FLAG® M2 affinity gel and the level of acetyl-lysine was detected by Western blotting. (C) as in (B), LC–MS was performed to analyze Parkin-interacting proteins and the associated acetylase and deacetylase were listed. (D) as in (B), cell lysates were subjected to FLAG immunoprecipitation and immunoblotting for acetyl-lysine, ACAT1 and HDAC2. (E) The correlation of Parkin level with HDAC1/2/3 levels was analyzed from gene expression profiling interactive analysis. (F) Immunohistochemistry analysis of Parkin and HDAC2 expression was performed in human cervical cancer tissue and noncancer tissue. (G) HEK293 cells were transiently transfected with Flag-Parkin, together with specific siRNA for ACAT1 or HDAC2. After SAHA treatment, cell lysates were prepared and immunoprecipitated with acetyl-lysine and immunoblotted for Parkin. (H) Human recombinant Parkin was incubated with recombinant ACAT1 in the presence of 100 μmol/L Ac-CoA for 1 h at 30 °C. Western blotting was performed for acetyl-lysine. (I) Identification of Parkin acetylation sites (K76, K129, K220, K349, and K408) using LC–MS analysis. Fragment ions containing acetylated lysine are denoted by ∗. (J) The LC–MS spectrum of the peptide contains the acetylated K129. Fragment ions containing acetylated lysine are denoted by red color. (K) Illustration of Parkin protein fragment domains with different acetylation sites of Parkin. (L) HEK293 cells were transfected with Flag-tagged WT Parkin or K76R, K129R, K220R, K349R, K408R mutant Parkin (left panel) and 3 KR mutant Parkin (K129/220/349R, right panel). After the FALG pulldown, the basal acetyl-lysine level was detected by immunoblotting.
In human cervical cancer, a negative correlation of Parkin with HDAC1/2/3 was revealed at the mRNA level from gene expression profiling interactive analysis, (Fig. 4E) and cBioPortal for Cancer Genomics (Fig. S3B). But only the correlation between Parkin and HDAC2 was significant, indicating that HDAC2 may be the upstream deacetylase of Parkin. Consistently, their inverse correlation was also observed at the protein level. In human cervical cancer tissue, HDAC2 protein was highly overexpressed while the expression level of Parkin was lower (Fig. 4F). But in non-cancer tissue, the opposite results were displayed. To further confirm this mechanism, we performed knockdown experiments with siRNA for ACAT1 or HDAC1/2/3 and then determined Parkin acetylation level in SAHA-treated cells. As expected, knockdown of ACAT1 resulted in decreased Parkin acetylation level under SAHA treatment (Fig. 4G). On the contrary, knockdown of HDAC2 resulted in a significant increase of Parkin acetylation level under SAHA treatment (Fig. 4G) while knockdown of HDAC1 only led to a slight increase of Parkin acetylation level and there was little change of Parkin acetylation level in HDAC3 knockdown cells (Fig. S3C). Moreover, the presence of ACAT1 can directly acetylate Parkin protein in an in vitro acetylation assay (Fig. 4H). These results demonstrate that ACAT1 and HDAC2 are closely involved in control of Parkin acetylation.
MS-based proteomic analysis was performed to identify the acetylation sites of Parkin protein36. To do this, ectopically expressed Parkin was immunoprecipitated from cells and then analyzed as described in Methods. Accordingly, we identified five high confident acetyl-lysine-containing peptides K(Ac)GQEMNATGGDDPR (pep_score = 70.56, pep_expect = 3.20E-07), K(Ac)DSPPAGSPAGR (pep_score = 83.53, pep_expect =8.00E-08), CGAHPTSDK(Ac)ETSVALHLIATNSR (pep_score = 46.27, pep_expect = 0.00083), K(Ac)VTCEGGNGLGCGFAFCR (pep_score = 35.07, pep_expect = 0.0017), WEAASK(Ac)ETIKK (pep_score = 38.84, pep_expect = 0.0041). They were mapped to regions containing K76, K129, K220, K349, and K408 on human Parkin (Fig. 4I, J, Supporting Information Fig. S4, the detailed MS spectra were shown in Supporting Information Table S3). All these acetylation sites within Parkin protein fragment domains were also illustrated in Fig. 4K. Next, we set to examine the biological importance of these acetylation sites by creating the mutants. When we mutated each lysine (K) to arginine (R), single mutation on K129, K220 and K349 resulted in a weak reduction in Parkin acetylation (Fig. 4L). More importantly, the combined mutations of all the 3 sites (Parkin-3KR) led to more evident decrease in the acetylation level of Parkin, indicating their significant impact on the regulation of Parkin acetylation.
3.5. Acetylation modification enhances Parkin-dependent mitophagy
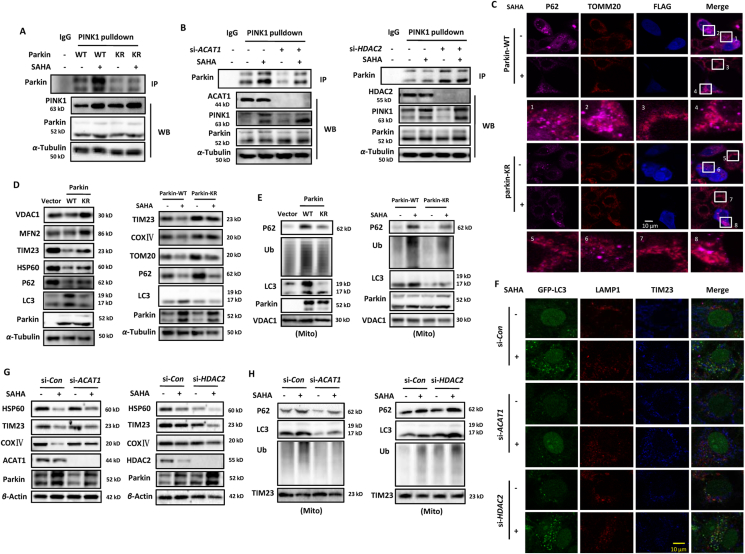
As an E3 ubiquitin ligase, Parkin mediates mitophagy as a downstream of PINK11. Following translocation to the mitochondrial surface, Parkin ubiquitinates numerous mitochondrial outer membrane proteins, which in turn recruit other proteins to mitochondria to initiate mitophagy50. Here, we first examined the effect of acetylation modification on the interaction of Parkin with its upstream protein PINK1. As shown in Fig. 5A, SAHA treatment increased the interacted wild-type Parkin level in PINK1 immunoprecipitated protein complexes. But in SAHA-treated mutant Parkin-expressing cells, the immunoprecipitated Parkin level was lower than that of wild-type Parkin. In addition, we also manipulated the acetylation level of Parkin indirectly through its upstream molecules. As presented in Fig. 4, ACAT1 and HDAC2 control the Parkin acetylation level together. Thus, HeLa cells with Parkin expression were interfered by siRNA for ACAT1 or HDAC2. As shown in Fig. 5B, ACAT1 knockdown attenuated the enhanced interaction of Parkin with PINK1 under SAHA treatment while an opposite effect was detected in HDAC2 knockdown cells, indicating that acetylation modification of Parkin has a positive role in the interacting with PINK1. It also represents a high phosphorylation level of Parkin with acetylation level increasing. Consequently, the colocalization of Parkin with mitochondria and P62 was increased by SAHA treatment, which was further enhanced in HDAC2 knockdown cells when compared to ACAT1 knockdown cells (Supporting Information Fig. S5A and S5B). It demonstrates that SAHA treatment results in the translocalization of Parkin into mitochondria through acetylating Parkin.
Figure 5.
Parkin acetylation is required for mitophagy induction by SAHA treatment. (A) HEK293 cells were transfected with WT or mutant Parkin, respectively. SAHA treatment (5 μmol/L, 12 h) was applied to cells and cell lysates were prepared and subjected to immunoprecipitation using anti-PINK1 antibody. The associated Parkin protein was detected using immunoblotting. (B) as in (A), HEK293 cells with Parkin expression were transfected with specific siRNA for ACAT1 or HDAC2, respectively. Immunoprecipitated Parkin was subjected to Western blotting analysis. (C) HeLa cells were transiently transfected with WT Parkin or mutant Parkin and then treated with SAHA (5 μmol/L, 12 h). Confocal imaging of P62 (pink) and TOMM20 (red) was performed in cells with or without Parkin overexpression (blue). Scale bar = 10 μm. (D) HeLa cells were transfected with empty vector, WT Parkin or mutant Parkin, respectively. Cell lysates were prepared for Western blotting analysis (left panel). On the right panel, SAHA treatment was given to cells with WT Parkin or mutant Parkin expression. (E) as in (D), mitochondrial fractions were isolated from the above cells and used for Western blotting analysis. VDAC1 was used as a representative of mitochondrial fraction. (F) SiHA cells were first transfected with specific siRNA for ACAT1 or HDAC2, and then treated with SAHA (5 μmol/L, 12 h). After fixation and permeabilization, cells were immunostained with LAMP1 (red) and TIM23 (blue), and examined under confocal microscope. Scale bar = 10 μm. (G) as in (F), after ACAT1 or HDAC2 knockdown, SiHA cells were given SAHA treatment. Western blotting was performed to determine mitochondrial proteins level. β-Actin was used as loading control. (H) as in (G), mitochondrial fractions were isolated and subjected to Western blotting analysis. TIM23 was used as a representative of mitochondrial fraction.
Next, we examined the regulatory effect of Parkin acetylation on mitophagy. As shown in Fig. 5C, Parkin overexpression led to more colocalization of autophagy adaptor P62 and mitochondrial protein TOMM20 and SAHA treatment further enhanced their colocalization, indicating the recruitment of ubiquitinated protein aggregates to autophagosomes. But in mutant Parkin-expressing cells, the colocalization of P62 and TOMM20 was less than that of wild-type Parkin. Even under SAHA treatment, only a slight increase of their colocalization was observed (Fig. 5C). Consistently, Western blotting analysis also showed Parkin mutation attenuated the clearance of mitochondrial proteins, including voltage dependent anion channel 1 (VDAC1), MFN2, TIM23, and HSP60 (Fig. 5D). Under SAHA treatment, damaged mitochondria were degraded much more in wild-type Parkin-expressing cells than that of mutant Parkin. Similarly, in mitochondrial fractions, wild-type Parkin caused more accumulation of autophagy proteins LC3, P62 and ubiquitin protein, but when Parkin mutation, their translocation into mitochondria was diminished (Fig. 5E). In response to SAHA treatment, wild-type Parkin overexpression resulted in more recruitment of autophagy and ubiquitin proteins into mitochondria than that of mutant Parkin. The above results demonstrate the participation of Parkin acetylation in mitophagy induction.
In addition, we also determined the positive regulation of Parkin acetylation in mitophagy through knocking down the Parkin upstream proteins. As shown in Fig. 5F and Fig. S5C, in HeLa cells expressing GFP-LC3, SAHA treatment significantly increased the colocalization of GFP-LC3, lysosomal associated membrane protein 1 and TIM23, indicating the enhanced fusion of mitophagosomes and lysosomes. But in ACAT1 knockdown cells, the increased formation of mitolysosomes was impaired under SAHA treatment. On the contrary, HDAC2 knockdown significantly increased the formation of mitolysosomes to degrade damaged mitochondria (Fig. 5F and Fig. S5C). Consistently, in ACAT1 knockdown cells, SAHA treatment resulted in decreased mitochondrial protein degradation, including HSP60, TIM23 and COXⅣ, while HDAC2 knockdown resulted in increased mitochondrial protein degradation under SAHA treatment (Fig. 5G). In mitochondrial fractions, ACAT1 knockdown attenuated the translocation of autophagy and ubiquitin proteins into mitochondria under SAHA treatment (Fig. 5H). But in HDAC2 knockdown cells, SAHA treatment resulted in increased translocation of autophagy and ubiquitin proteins into mitochondria. The above results demonstrate that ACAT1 and HDAC2 influence the mitophagy level through regulating Parkin acetylation.
3.6. Parkin acetylation is required for tumor suppression of SAHA treatment
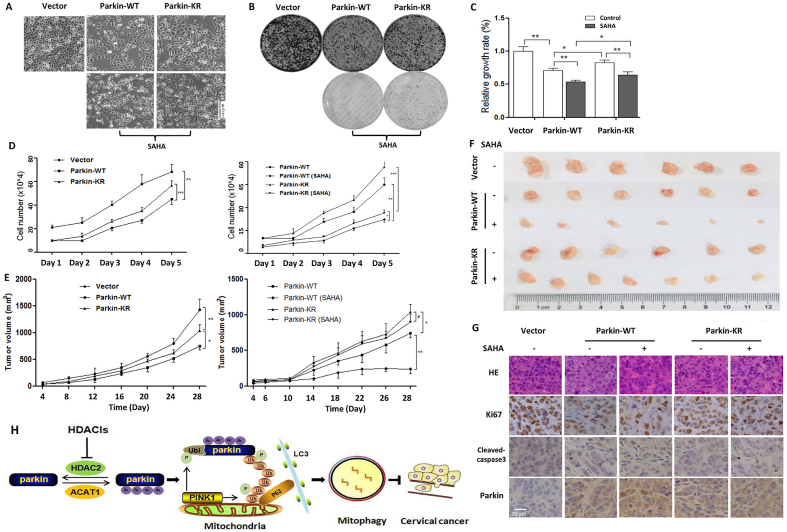
Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25–q2751. Indeed, Parkin overexpression was characterized with low-confluence cells when compared with control under the Phase Contrast Microscopy, indicating a slow cell proliferation (Fig. 6A, B). When the essential autophagy gene ATG7 knockdown, the inhibitory effect of Parkin on cell proliferation was abolished (Supporting Information Fig. S6), suggesting that Parkin-dependent mitophagy serves as tumor suppression. Under SAHA treatment, cell morphology changes showed the shrinked, round and low-density cells and more dead cells floating in the medium were also detected with Parkin expression. Quantitatively, the cell proliferation rate was also significantly decreased in CCK-8 assay (Fig. 6B). But when Parkin mutation, a slight increase of cell density was observed and SAHA treatment resulted in a weaker growth-inhibitory effect when compared with wild-type Parkin. In cell colony formation assay, wild-type Parkin overexpression resulted in a significant decrease in the formed colonies. The number and size of colonies were further decreased with SAHA-mediated Parkin acetylation (Fig. 6C). When Parkin mutation, the ability of colony formation was stronger than that of wild-type Parkin, either in the presence or absence of SAHA treatment. Moreover, cell growth curve was also drawn to detect the cell growth dynamics for consecutive 5 days. As shown in Fig. 6D, wild-type Parkin overexpression significantly decreased cell growth rate while acetylation-defective mutant attenuated the growth inhibitory effect of Parkin. Under SAHA treatment, either cells with wild-type or mutant Parkin expression grow slowly when compared with control, but cells with mutant Parkin expression still grew faster than that of wild-type Parkin (Fig. 6D). In addition, HDAC2 knockdown enhanced the cytotoxic activity of SAHA treatment in HeLa cells (Supporting Information Fig. S7), accompanied with shrinked and low-confluence cells. But in ACAT1 knockdown cells, the cytotoxic effect of SAHA treatment seemed to be weaker. The above results demonstrate that acetylation modification plays a positive role in tumor suppression of Parkin and SAHA treatment exerts the cytotoxic activity through Parkin acetylation.
Figure 6.
SAHA treatment exerts anticancer activity through mediating Parkin acetylation. (A) HeLa cells with WT or mutant Parkin expression were subjected to SAHA treatment (5 μmol/L, 24 h) and cell morphology was captured using a Phase contrast microscopy. Scale bar = 100 μm. (B) as in (A), cells were seeded into 96-well plate and treated with SAHA. CCK-8 solution was added into each well of the plate and the absorbance was measured at 450 nm using microplate spectrophotometer. (C) as in (A), under SAHA treatment, cell proliferation was determined by colony formation assay. After crystal violet staining, the formed colonies were photographed. (D) as in (A), cells were seeded into 6-well plate and cultured for consecutive 5 days. Each day, cells were trpsined and counted and cell growth curve was drawn accordingly. (E) 2 × 106 HeLa cells with empty vector, wild-type or mutant Parkin expression, was subcutaneously injected into the right flanks of the nude mice. Tumor-bearing mice were intraperitoneally injected with SAHA (50 mg/kg) or saline, respectively. Xenograft tumor volumes were measured twice per week and then quantified and statistically analyzed. (F) When mice execution, xenograft tumors from different groups were removed and typical images were captured. (G) Representative histopathological images of tumor sections were from hematoxylin and eosin staining. The expressions of Ki67, cleave-caspase3 and Parkin in tumor xenograft were detected using immunohistochemistry. The images were taken at 400 × magnification. (H) A schematic model of the acetylation of Parkin in mitophagy and tumor suppression by HDACis in human cervical cancer. #P > 0.05, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Based on the above results, xenograft model was used to further confirm the inhibitory-effect of Parkin acetylation on tumor growth in vivo. When HeLa cells with empty vector, wild-type or mutant Parkin expression were injected subcutaneously into the right flanks of nude mice, mice were divided into 3 groups. As shown in Fig. 6E, Parkin overexpression resulted in a significant tumor suppression when compared with empty vector. But with Parkin mutation, its tumor suppressive effect was markedly attenuated, indicating the importance of acetylation modification in anticancer activity of Parkin. One week after inoculation, tumor-bearing mice were given an intraperitoneal injection of SAHA. As shown in Fig. 6E, under wild-type Parkin expression, a significant difference in tumor suppression was observed between the SAHA-treated group and the PBS-treated control group. However, when Parkin mutation, there was no significant difference under SAHA treatment, suggesting that SAHA treatment exerts anticancer activity through acetylating Parkin. Three weeks after inoculation, all mice were sacrificed and tumor was removed from mice. As expected, a significant difference in the tumor size and weight was shown between Parkin overexpression group and empty vector group (Fig. 6F, Supporting Information Fig. S8). Under wild-type Parkin expression, SAHA treatment resulted in a further decrease of tumor size and weight. However, with Parkin mutation, the tumor-suppressive effect of Parkin was markedly attenuated, either in the presence or absence of SAHA treatment. It was also confirmed in the histopathological and immunohistochemical analysis. SAHA treatment decreased the expression level of Ki67 (cell proliferation marker) and increased the expression level of cleaved caspase3 (cell apoptosis marker) (Fig. 6G). HE staining showed less live cells, reduced cell sizes and more chromatin condensed under SAHA treatment when compared with control group. But this inhibitory effect was attenuated with Parkin mutation (Fig. 6G). These results indicate that Parkin acetylation is required for the tumor suppression of SAHA treatment.
4. Discussion
The association of mitochondrial protein acetylation with mitophagy initiation has been implied in many studies. More importantly, many acetylases and deacetylases are involved in mitophagy process and directly regulate the level of mitophagy. In the current study, we explored the acetylation modification of the key mitophagy machinery effector Parkin and revealed the importance of Parkin acetylation in mitophagy induction and tumor suppression. It may be a novel target for cervical cancer therapy and offers an opportunity to develop mitophagy-modulated anticancer drugs. The detailed molecular mechanism regulating Parkin acetylation is summarized in Fig. 6H.
Parkin, as a tumor suppressor, is downregulated in cervical carcinoma due to allelic loss of 6q25–q2744,52. Structurally, Parkin protein contains 5 domains, referring to UBL, RING1, RING2, IBR, Linker domain (Fig. 4K). Under normal condition, Parkin distributes in the cytosol and exists in a closed-inactive conformation through intraprotein domain–domain interactions53. Upon mitochondrial depolarization, Parkin is rapidly recruited to mitochondria and activated54. The activity of Parkin is mainly regulated by phosphorylation and the turnover of Parkin phosphorylation is very rapid. Many protein kinases have been revealed to interact with Parkin and phosphorylate it55, 56, 57, such as casein kinase-1, protein kinase A, protein kinase C, cyclin-dependent kinase 5, c-ABL, PINK1, etc. Phosphoamino acid analysis showed that Parkin phosphorylation occurred mainly on serine residues, including Ser65 (located in the UBL region), Ser101, Ser131, and Ser136 (located in the Linker region) as well as Ser296 and Ser378 (located in the RING–IBR–RING motif). The tyrosine residue 143 of Parkin is also phosphorylated by c-ABL55. After phosphorylation, the auto-ubiquitylation of Parkin is also inhibited, followed by the increase of Parkin protein stability. Among them, phosphorylation of Parkin by PINK1 is widely studied and plays an important role in mitophagy initiation58. On the one hand, PINK1 directly phosphorylates Parkin at Ser65 of the UBL domain4. On the other hand, it is mediated by pSer65–ubiquitin, which is phosphorylated ubiquitin at Ser65 by PINK159. pSer65–ubiquitin binds to RING1 domain of Parkin and changes the conformation of Parkin protein, leading to the translocation of Parkin from cytosol to mitochondria12. Here, we detected the increase of PINK1 interaction with Parkin and the phosphorylation level of Parkin under SAHA treatment (Fig. 3C and D). Meanwhile, the acetylation level of Parkin was also increased in SAHA-treated cells. These two forms of modification reciprocally resulted in the enhanced function of Parkin in mitophagy and tumor suppression. Undoubtedly, in response to SAHA treatment, the influence of Parkin acetylation is far more than the phosphorylation of Parkin, either on mitophagy or tumor suppression. When Parkin mutation, the interaction of PINK1 with Parkin was also attenuated under SAHA treatment (Fig. 5A and B), indicating the decrease of the phosphorylation level of Parkin. But we did not examine the regulatory effect of phosphorylation modification on the acetylation of Parkin.
Cervical cancer cells are characterized with the expression of the HPV oncoproteins E6 and E7. The oncogencity of E6 is mediated by targeting P53 and PDZ-family tumor suppressor proteins for proteasomal degradation while the E7 oncoprotein acts by co-operating with HDAC1/260. In addition, HDAC8 shows functional redundancy with HDAC6 and are highly overexpressed in cervical cancer, where deacetylaes α-tubulin and leads to cervical cancer proliferation and progression61. HDAC10 exerts an opposite effect and suppresses cervical cancer metastasis through expression inhibition of matrix metalloproteinases 2 and 962. Thus, class Ⅰ of HDAC family seems to be closely associated with cervical cancer development, including HDAC1/2/3 and -8. Indeed, Oncomine database analysis also demonstrated the upregulation of HDAC1/2/3 in human cervical cancer cells and tissues (Fig. 1A). As is known, HDACs are capable of suppressing the activity of tumor suppressors through deacetylation modification63, 64, 65. Here, it is speculated that aberrant expression of HDACs dacetylates tumor suppressor Parkin and leads to loss of function of Parkin and the development of cervical cancer. To reveal the underlying mechanism, we performed mass spectrometry to identify the acetylase and deacetylase interacting with Parkin protein. In Table S1, there are several acetylases associated with Parkin, including ACAT1, ACAT2, EP300, N-acetyltransferase 10, N-α-acetyltransferase 10, and N-α-acetyltransferase 30. The associated deacetylases included HDAC1/2/3 (class Ⅰ), HDAC4/6/7 (class Ⅱ) and sirtuin 1/2 (class Ⅲ). Due to low protein detection score, we only chose and analyzed the mitochondrial acetylase ACAT1 in controlling Parkin acetylation. For deacetylase, only HDAC1/2/3 had a high protein detection score and among them, only HDAC2 had a significant inverse correlation with Parkin level based on correlation analysis from GEPIA and cBioPortal for cancer genomics. Thus, HDAC2 was selected to study the regulation of Parkin acetylation. Although other molecules were not analyzed, we could not exclude their regulatory effect on Parkin acetylation and cervical cancer.
Metabolic reprogramming in tumors is now recognized as a hallmark of cancer, participating both in cancer development and cancer progression. Cancer cells develop global metabolic adaptations allowing them to survive in the low oxygen and nutrient tumor microenvironment23. Among these metabolic adaptations, one particular adaptation of cancer cell metabolism is the use of mitophagy to recycle the dysfunctional or damaged mitochondria in condition of metabolic stress66, demonstrating the protective effect of mitophagy. Similarly, in response to anticancer treatments, mitophagy also serves as a cell survival mechanism because inhibition of mitophagy pathways sensitizes cancer cells to death67,68, although the increased mitophagy level is accompanied with anticancer therapies. Knockdown of key mitophagy receptors, such as PINK1, FUN14 domain containing 1 or autophagy and beclin 1 regulator 1, increases the sensitivity of cancer cells to chemotherapy67,69. With the decrease of PINK1- or RAB9A-mediated mitophagy, cancer cells also become more sensitive to radiotherapy70. It seems that the induction of mitophagy exerts a function in modulating the cytotoxic effect of anti-cancer treatments23, leading to cancer cell survival. But in our study, mitophagy serves as a cell death mechanism, which is in contrast to the above findings. We guess that this discrepancy may be associated with the cellular context. In the current study, overexpression of Parkin increased the level of mitophagy and inhibited cervical cancer cell growth. When Parkin mutation or ATG7 knockdown, mitophagy induction was disrupted and the inhibitory effect of Parkin on cancer cell proliferation was also attenuated (Fig. S7). Similarly, under SAHA treatment, mitophagy level was increased and cervical cancer cell growth was inhibited. But ULK1 knockdown impaired the mitophagy induction by SAHA treatment and attenuated the growth-inhibitory effect on cervical cancer cells (Fig. 2G and H). To achieve the goal of anti-cervical cancer, the strategy of mitophagy enhancement should be taken and Parkin acetylation may be a novel target through HDAC inhibition. Collectively, our findings also provide a mechanistic rationale for the use of HDAC inhibitors in cancer therapeutics71, 72, 73. Thus, mitophagy modulation would be a promising approach in cervical cancer therapy with a great potential.
5. Conclusions
In human cervical cancer, in response to HDACis, tumor suppressor protein Parkin is acetylated with HDAC2 inhibition. Mass spectrometry analysis reveals the acetylation sites of Parkin protein and the important role of Parkin acetylation is also validated using mutation assay in Parkin-dependent mitophagy and tumor suppression. Our results reveal an acerylation-dependent molecular mechanism regulating the function of Parkin in cancer.
Acknowledgments
This study was supported by research grants from Zhejiang Provincial Natural Science Foundation (LR18H160002, China), National Natural Science Foundation of China (32070740), Zhejiang Provincial Program in Medicine and Health Sciences and Technology (2018KY010), Zhejiang Provincial Outstanding Talent Project of Ten Thousand Talents Program, Zhejiang Provincial Qianjiang Talents Program, Zhejiang Provincial High-Level Innovative Health Talents Program to Dr. Jianbin Zhang.
Author contributions
Xin Sun performed the experiments of Parkin acetylation identification and drafted the manuscript. Yuhan Shu examined the effect of Parkin acetylation on mitophagy. Guiqin Ye and Caixia Wu conducted the experiments on animals. Mengting Xu did statistical analysis in the experiments. Ruilan Gao gave constructive feedback and helped to revise the manuscript. Dongsheng Huang administered the project and provided the financial support. Jianbin Zhang proposed the project and took part in the project coordination.
Conflicts of interest
No potential conflicts of interest were disclosed.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.07.003.
Contributor Information
Dongsheng Huang, Email: dshuang@zju.edu.cn.
Jianbin Zhang, Email: zhangjianbin@hmc.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin S.M., Lazarou M., Wang C., Kane L.A., Narendra D.P., Youle R.J. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 4.Kondapalli C., Kazlauskaite A., Zhang N., Woodroof H.I., Campbell D.G., Gourlay R., et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan N.C., Salazar A.M., Pham A.H., Sweredoski M.J., Kolawa N.J., Graham R.L., et al. Broad activation of the ubiquitin–proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin S.M., Youle R.J. PINK1- and Parkin-mediated mitophagy at a glance. J Cell Sci. 2012;125:795–799. doi: 10.1242/jcs.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L., Qi H., Tang Y., Shen H.M. Post-translational modifications of key machinery in the control of mitophagy. Trends Biochem Sci. 2020;45:58–75. doi: 10.1016/j.tibs.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Auburger G., Gispert S., Jendrach M. Mitochondrial acetylation and genetic models of Parkinson's disease. Prog Mol Biol Transl Sci. 2014;127:155–182. doi: 10.1016/B978-0-12-394625-6.00006-4. [DOI] [PubMed] [Google Scholar]
- 10.Webster B.R., Scott I., Han K., Li J.H., Lu Z., Stevens M.V., et al. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J Cell Sci. 2013;126:4843–4849. doi: 10.1242/jcs.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott I., Webster B.R., Chan C.K., Okonkwo J.U., Han K., Sack M.N. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J Biol Chem. 2014;289:2864–2872. doi: 10.1074/jbc.M113.521641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiba-Fukushima K., Arano T., Matsumoto G., Inoshita T., Yoshida S., Ishihama Y., et al. Phosphorylation of mitochondrial polyubiquitin by PINK1 promotes Parkin mitochondrial tethering. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herhaus L., Dikic I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 2015;16:1071–1083. doi: 10.15252/embr.201540891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park J.S., Kim E.J., Kwon H.J., Hwang E.S., Namkoong S.E., Um S.J. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. Implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J Biol Chem. 2000;275:6764–6769. doi: 10.1074/jbc.275.10.6764. [DOI] [PubMed] [Google Scholar]
- 15.Lee J.Y., Nagano Y., Taylor J.P., Lim K.L., Yao T.P. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Sante G., Pestell T.G., Casimiro M.C., Bisetto S., Powell M.J., Lisanti M.P., et al. Loss of Sirt1 promotes prostatic intraepithelial neoplasia, reduces mitophagy, and delays PARK2 translocation to mitochondria. Am J Pathol. 2015;185:266–279. doi: 10.1016/j.ajpath.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun F., Jiang X., Wang X., Bao Y., Feng G., Liu H., et al. Vincristine ablation of Sirt2 induces cell apoptosis and mitophagy via Hsp70 acetylation in MDA-MB-231 cells. Biochem Pharmacol. 2019;162:142–153. doi: 10.1016/j.bcp.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 18.Qiao A., Wang K., Yuan Y., Guan Y., Ren X., Li L., et al. Sirt3-mediated mitophagy protects tumor cells against apoptosis under hypoxia. Oncotarget. 2016;7:43390–43400. doi: 10.18632/oncotarget.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song S.B., Jang S.Y., Kang H.T., Bie Wei, Jeoun U.W., Yoon G.S., et al. Modulation of mitochondrial membrane potential and ROS generation by nicotinamide in a nanner independent of SIRT1 and mitophagy. Mol Cells. 2017;40:503–514. doi: 10.14348/molcells.2017.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chourasia A.H., Boland M.L., Macleod K.F. Mitophagy and cancer. Cancer Metab. 2015;3:4. doi: 10.1186/s40170-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panigrahi D.P., Praharaj P.P., Bhol C.S., Mahapatra K.K., Patra S., Behera B.P., et al. The emerging, multifaceted role of mitophagy in cancer and cancer therapeutics. Semin Cancer Biol. 2020;66:45–58. doi: 10.1016/j.semcancer.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Drake L.E., Springer M.Z., Poole L.P., Kim C.J., Macleod K.F. Expanding perspectives on the significance of mitophagy in cancer. Semin Cancer Biol. 2017;47:110–124. doi: 10.1016/j.semcancer.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferro F., Servais S., Besson P., Roger S., Dumas J.F., Brisson L. Autophagy and mitophagy in cancer metabolic remodelling. Semin Cell Dev Biol. 2020;98:129–138. doi: 10.1016/j.semcdb.2019.05.029. [DOI] [PubMed] [Google Scholar]
- 24.Liu J., Zhang C., Hu W., Feng Z. Parkinson's disease-associated protein Parkin: an unusual player in cancer. Cancer Commun. 2018;38:40. doi: 10.1186/s40880-018-0314-z. (Lond) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikeuchi K., Marusawa H., Fujiwara M., Matsumoto Y., Endo Y., Watanabe T., et al. Attenuation of proteolysis-mediated cyclin E regulation by alternatively spliced Parkin in human colorectal cancers. Int J Cancer. 2009;125:2029–2035. doi: 10.1002/ijc.24565. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C., Lin M., Wu R., Wang X., Yang B., Levine A.J., et al. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci U S A. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agnihotri S., Golbourn B., Huang X., Remke M., Younger S., Cairns R.A., et al. PINK1 is a negative regulator of growth and the Warburg effect in glioblastoma. Cancer Res. 2016;76:4708–4719. doi: 10.1158/0008-5472.CAN-15-3079. [DOI] [PubMed] [Google Scholar]
- 28.He J., Pei L., Jiang H., Yang W., Chen J., Liang H. Chemoresistance of colorectal cancer to 5-fluorouracil is associated with silencing of the BNIP3 gene through aberrant methylation. J Cancer. 2017;8:1187–1196. doi: 10.7150/jca.18171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Passer B.J., Nancy-Portebois V., Amzallag N., Prieur S., Cans C., Climens A.R., et al. The p53-inducible TSAP6 gene product regulates apoptosis and the cell cycle and interacts with Nix and the Myt1 kinase. Proc Natl Acad Sci U S A. 2003;100:2284–2289. doi: 10.1073/pnas.0530298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W., Li Y., Siraj S., Jin H., Fan Y., Xinrong Yang X., et al. FUN14 domain-containing 1-mediated mitophagy suppresses hepatocarcinogenesis by inhibition of inflammasome activation in mice. Hepatology. 2019;69:604–621. doi: 10.1002/hep.30191. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J., Sun X., Wang L., Wong Y.K., Lee Y.M., Zhou C., et al. Artesunate-induced mitophagy alters cellular redox status. Redox Biol. 2018;19:263–273. doi: 10.1016/j.redox.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Band M., Joel A., Hernandez A., Avivi A. Hypoxia-induced BNIP3 expression and mitophagy: in vivo comparison of the rat and the hypoxia-tolerant mole rat, Spalax ehrenbergi. FASEB J. 2009;23:2327–2335. doi: 10.1096/fj.08-122978. [DOI] [PubMed] [Google Scholar]
- 33.Hervouet E., Cízková A., Cizkova A., Demont J., Vojtísková A., Pecina P., Franssen-van H., et al. HIF and reactive oxygen species regulate oxidative phosphorylation in cancer. Carcinogenesis. 2008;29:1528–1537. doi: 10.1093/carcin/bgn125. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H., Gao P., Fukuda R., Kumar G., Krishnamachary B., Zeller K.I., et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Lin S.Y., Li T.Y., Liu Q., Zhang C., Li X., Chen Y., et al. GSK3–TIP60–ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336:477–481. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- 36.Choudhary C., Kumar C., Gnad F., Nielsen M., Rehman M., Walther T., et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 37.Walboomers J.M., Jacobs M.V., Manos M.M., Bosch F.X., Kummer J.A., Shah K.V., et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 38.Werness B.A., Levine A.J., Howley P.M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 39.Brehm A., Nielsen S.J., Miska E.A., McCance D.J., Reid J.L., Bannister A.J., et al. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 1999;18:2449–2458. doi: 10.1093/emboj/18.9.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longworth M.S., Laimins L.A. The binding of histone deacetylases and the integrity of zinc finger-like motifs of the E7 protein are essential for the life cycle of human papillomavirus type 31. J Virol. 2004;78:3533–3541. doi: 10.1128/JVI.78.7.3533-3541.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green D.R., Reed J.C. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J., Ng S., Wang J., Zhou J., Tan S.H., Yang N., et al. Histone deacetylase inhibitors induce autophagy through FOXO1-dependent pathways. Autophagy. 2015;11:629–642. doi: 10.1080/15548627.2015.1023981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding W.X., Yin X.M. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012;393:547–564. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao N., Wang C., Hu N., Li Y., Liu M., Lei Y., et al. Inhibition of PINK1/Parkin-dependent mitophagy sensitizes multidrug-resistant cancer cells to B5G1, a new betulinic acid analog. Cell Death Dis. 2019;10:232. doi: 10.1038/s41419-019-1470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu W., Tian W., Hu Z., Chen G., Huang L., Li W., et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15:566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehdi S.J., Alam M.S., Batra S., Rizvi M.M. Allelic loss of 6q25-27, the PARKIN tumor suppressor gene locus, in cervical carcinoma. Med Oncol. 2011;28:1520–1526. doi: 10.1007/s12032-010-9633-x. [DOI] [PubMed] [Google Scholar]
- 47.Duplan E., Giaime E., Viotti J., Sévalle J., Corti O., Brice A., et al. ER-stress-associated functional link between Parkin and DJ-1 via a transcriptional cascade involving the tumor suppressor p53 and the spliced X-box binding protein XBP-1. J Cell Sci. 2013;126:2124–2133. doi: 10.1242/jcs.127340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 49.Kane L.A., Lazarou M., Fogel A.I., Li Y., Yamano K., Sarraf S.A., et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J., Wang J., Zhou Z., Park J.E., Wang L., Wu S., et al. Importance of TFEB acetylation in control of its transcriptional activity and lysosomal function in response to histone deacetylase inhibitors. Autophagy. 2018;14:1043–1059. doi: 10.1080/15548627.2018.1447290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geisler S., Holmström K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 52.Lee K., Lee M.H., Kang Y.W., Rhee K.J., Kim T.U., Kim Y.S. Parkin induces apoptotic cell death in TNF-alpha-treated cervical cancer cells. BMB Rep. 2012;45:526–531. doi: 10.5483/bmbrep.2012.45.9.104. [DOI] [PubMed] [Google Scholar]
- 53.Sauvé V., Sung G., Soya N., Kozlov G., Blaimschein N., Miotto L.S., et al. Mechanism of parkin activation by phosphorylation. Nat Struct Mol Biol. 2018;25:623–630. doi: 10.1038/s41594-018-0088-7. [DOI] [PubMed] [Google Scholar]
- 54.Gladkova C., Maslen S.L., Skehel J.M., Komander D. Mechanism of parkin activation by PINK1. Nature. 2018;559:410–414. doi: 10.1038/s41586-018-0224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ko H.S., Lee Y., Shin J.H., Karuppagounder S.S., Gadad B.S., Koleske A.J., et al. Phosphorylation by the c-Abl protein tyrosine kinase inhibits parkin's ubiquitination and protective function. Proc Natl Acad Sci U S A. 2010;107:16691–16696. doi: 10.1073/pnas.1006083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamamoto A., Friedlein A., Imai Y., Takahashi R., Kahle P.J., Haass C. Parkin phosphorylation and modulation of its E3 ubiquitin ligase activity. J Biol Chem. 2005;280:3390–3399. doi: 10.1074/jbc.M407724200. [DOI] [PubMed] [Google Scholar]
- 57.Avraham E., Rott R., Liani E., Szargel R., Engelender S. Phosphorylation of Parkin by the cyclin-dependent kinase 5 at the linker region modulates its ubiquitin-ligase activity and aggregation. J Biol Chem. 2007;282:12842–12850. doi: 10.1074/jbc.M608243200. [DOI] [PubMed] [Google Scholar]
- 58.Kim Y., Park J., Kim S., Song S., Kwon S.K., Lee S.H., et al. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun. 2008;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 59.Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D.G., Ritorto M.S., Hofmann K., et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin Z., Bazzaro M., Wang M.C., Chan K.C., Peng S., Roden R.B. Combination of proteasome and HDAC inhibitors for uterine cervical cancer treatment. Clin Cancer Res. 2009;15:570–577. doi: 10.1158/1078-0432.CCR-08-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanaja G.R., Ramulu H.G., Kalle A.M. Overexpressed HDAC8 in cervical cancer cells shows functional redundancy of tubulin deacetylation with HDAC6. Cell Commun Signal. 2018;16:20. doi: 10.1186/s12964-018-0231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song C., Zhu S., Wu C., Kang J. Histone deacetylase (HDAC) 10 suppresses cervical cancer metastasis through inhibition of matrix metalloproteinase (MMP) 2 and 9 expression. J Biol Chem. 2013;288:28021–28033. doi: 10.1074/jbc.M113.498758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee K.J., Lee K.Y., Lee Y.M. Downregulation of a tumor suppressor RECK by hypoxia through recruitment of HDAC1 and HIF-1alpha to reverse HRE site in the promoter. Biochim Biophys Acta. 2010;1803:608–616. doi: 10.1016/j.bbamcr.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 64.Juan L.J., Shia W.J., Chen M.H., Yang W.M., Seto E., Lin Y.S., et al. Histone deacetylases specifically down-regulate p53-dependent gene activation. J Biol Chem. 2000;275:20436–20443. doi: 10.1074/jbc.M000202200. [DOI] [PubMed] [Google Scholar]
- 65.Meng Z., Jia L.F., Gan Y.H. PTEN activation through K163 acetylation by inhibiting HDAC6 contributes to tumour inhibition. Oncogene. 2016;35:2333–2344. doi: 10.1038/onc.2015.293. [DOI] [PubMed] [Google Scholar]
- 66.Hjelmeland A., Zhang J. Metabolic, autophagic, and mitophagic activities in cancer initiation and progression. Biomed J. 2016;39:98–106. doi: 10.1016/j.bj.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abdrakhmanov A., Kulikov A.V., Luchkina E.A., Zhivotovsky B., Gogvadze V. Involvement of mitophagy in cisplatin-induced cell death regulation. Biol Chem. 2019;400:161–170. doi: 10.1515/hsz-2018-0210. [DOI] [PubMed] [Google Scholar]
- 68.Villa E., Proics E., Rubio-Patino C., Obba S., Zunino B., Bossowski J., et al. Parkin-independent mitophagy controls chemotherapeutic response in cancer cells. Cell Rep. 2017;20:2846–2859. doi: 10.1016/j.celrep.2017.08.087. [DOI] [PubMed] [Google Scholar]
- 69.Wei R., Cao J., Yao S. Matrine promotes liver cancer cell apoptosis by inhibiting mitophagy and PINK1/Parkin pathways. Cell Stress Chaperones. 2018;23:1295–1309. doi: 10.1007/s12192-018-0937-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Zheng R., Yao Q., Xie G., Du S., Ren C., Wang Y., et al. TAT-ODD-p53 enhances the radiosensitivity of hypoxic breast cancer cells by inhibiting Parkin-mediated mitophagy. Oncotarget. 2015;6:17417–17429. doi: 10.18632/oncotarget.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodriguez-Fonseca R.A., Sixto-Lopez Y., Fragoso-Vazquez M.J., Flores-Mejía R., Cabrera-Pérez L.C., Vázquez-Moctezuma I., et al. Design, synthesis and biological evaluation of a phenyl butyric acid derivative, N-(4-chlorophenyl)-4-phenylbutanamide: a HDAC6 inhibitor with anti-proliferative activity on cervix cancer and leukemia cells. Anticancer Agents Med Chem. 2017;17:1441–1454. doi: 10.2174/1871520617666170103092851. [DOI] [PubMed] [Google Scholar]
- 72.Huang Z., Peng S., Knoff J., Lee S., Yang B., Wu T., et al. Combination of proteasome and HDAC inhibitor enhances HPV16 E7-specific CD8+ T cell immune response and antitumor effects in a preclinical cervical cancer model. J Biomed Sci. 2015;22:7. doi: 10.1186/s12929-014-0111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li J., Bonifati S., Hristov G., Marttila T., Valmary-Degano S., Stanzel S., et al. Synergistic combination of valproic acid and oncolytic parvovirus H-1PV as a potential therapy against cervical and pancreatic carcinomas. EMBO Mol Med. 2013;5:1537–1555. doi: 10.1002/emmm.201302796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.