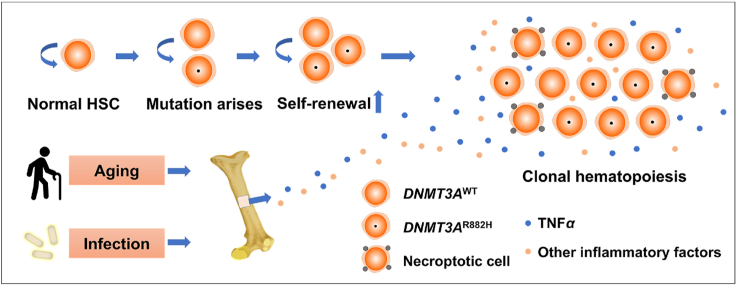
Abstract
Aging-elevated DNMT3A R882H-driven clonal hematopoiesis (CH) is a risk factor for myeloid malignancies remission and overall survival. Although some studies were conducted to investigate this phenomenon, the exact mechanism is still under debate. In this study, we observed that DNMT3A R878H bone marrow cells (human allele: DNMT3A R882H) displayed enhanced reconstitution capacity in aged bone marrow milieu and upon inflammatory insult. DNMT3A R878H protects hematopoietic stem and progenitor cells from the damage induced by chronic inflammation, especially TNFα insults. Mechanistically, we identified that RIPK1–RIPK3–MLKL-mediated necroptosis signaling was compromised in R878H cells in response to proliferation stress and TNFα insults. Briefly, we elucidated the molecular mechanism driving DNMT3A R878H-based clonal hematopoiesis, which raises clinical value for treating DNMT3A R882H-driven clonal hematopoiesis and myeloid malignancies with aging.
KEY WORDS: Aging, Inflammation, DNMT3A R882H, Clonal hematopoiesis, Hematopoietic stem cells, Necroptosis, TNFα
Graphical abstract
With aging, hematopoietic stem cells accumulate DNMT3A mutation with necroptosis resistance which results in clonal hematopoiesis, and this process can be aggravated by inflammation challenge.
1. Introduction
Hematopoietic stem cell (HSC) generates all blood cells throughout the lifespan and certain mutations within it leads to clonal hematopoiesis with aging, which is a risk factor for many diseases, especially blood malignancies1, 2, 3, 4, 5. DNMT3A (DNA methyltransferase 3A) is the most frequently mutated gene2 and several elegant studies have intensively studied the role of DNMT3A in hematopoietic stem cells and hematopoietic malignancies, which shows that deficiency of DNMT3A results in increased self-renewal and malignant transformation of hematopoietic stem cells6,7. Mechanistically, deficiency of DNMT3A immortalizes HSCs by losing focal DNA methylation of key regions wherein self-renewal genes located8, predominantly at hematopoietic enhancer regions9. A different perspective of research shows that an extended region of low methylation (canyons) borders, wherein genes related to human leukemia are enriched, is eroded in the absence of DNMT3A10.
The aforementioned studies investigated the function of DNMT3A by using clinical samples and knockout mouse models. While, DNMT3A mutations, especially the amino acid R882, are associated with poor overall survival of leukemia patients5,11,12, and exhibit resistance to anthracycline13, aclarubicin14 and other cytotoxic chemotherapy15. Mechanistically, although several groups have revealed that the dominant-negative effect of DNMT3A R882H16 and this mutation resulted in the increase of leukemogenesis-related genes17, 18, 19, the molecular mechanism is still not fully understood.
In this study, we observed that aging-elevated inflammatory milieu, mainly TNFα signaling, fostered R878H cells expansion. Meanwhile, hematopoietic stem and progenitor cells with DNMT3A R878H mutation exhibited compromised response to lipopolysaccharide (LPS)-induced inflammation stress and withstood the impairment triggered by chronic inflammatory and TNFα insults. Furthermore, it is notable that RIPK1–RIPK3–MLKL mediated necroptosis pathway was compromised in R878H cells in response to transplantation stress and TNFα insults. This finding provides a possible mechanism to explain why DNMT3A R882H-driven clonal expansion increases with age, and necroptosis may be a promising target for treating DNMT3A R882H-based clonal hematopoiesis and leukemia patients with DNMT3A R882 mutation.
2. Materials and methods
2.1. Generation of Dnmt3afl-R878H/+ mice
Dnmt3afl-R878H/+ mice were generated in Cyagen Biosciences Inc. (Guangzhou, China). Targeting vectors were kindly provided by Dr. Saijuan Chen from Shanghai Jiao Tong University (Shanghai, China). The Dnmt3a R878H knock-in construct was generated via the recombineering method. Briefly, approximately 17.5 kb of genomic DNA surrounding Dnmt3a exon 23 was subcloned into a targeting vector (Fig. 1A). The linearized vector was subsequently delivered to ES cells (C57BL/6) via electroporation, followed by drug selection, PCR screening, and Southern blot confirmation. After gaining 99 drug-resistant clones, 4 potentially targeted clones were confirmed, 4 of which were expanded for Southern blotting. After confirming correctly targeted ES clones, some clones were selected for blastocyst microinjection, followed by chimera production. F0 generation was confirmed as germline-transmitted via crossbreeding with wile-type. Dnmt3afl-R878H/+ and Dnmt3a+/+ littermate mice were genotyped by PCR with primers Dnmt3a-FP (5′-GCCAGTATAGATGCCTGTGAGGT-3′) and Dnmt3a-RP (5′-TGCCTCTTGGATGTGCTCTACAG-3′) using the following parameters: 94 °C for 3 min, followed by 35 cycles of 94 °C for 20 s, 60 °C for 20 s, 72 °C for 20 s and 72 °C for 5 min to final extension. PCR product for WT and Mutant band is 260 bp and 316 bp, respectively. Expression of the R878H mutation (that results in an arginine to histidine substitution at amino acid position 878, DNMT3A R878H) was confirmed by cDNA sequencing using the following primer: 5′-GGAGTGTGAATCTCAAAGCTGGGAT-3′.
Figure 1.
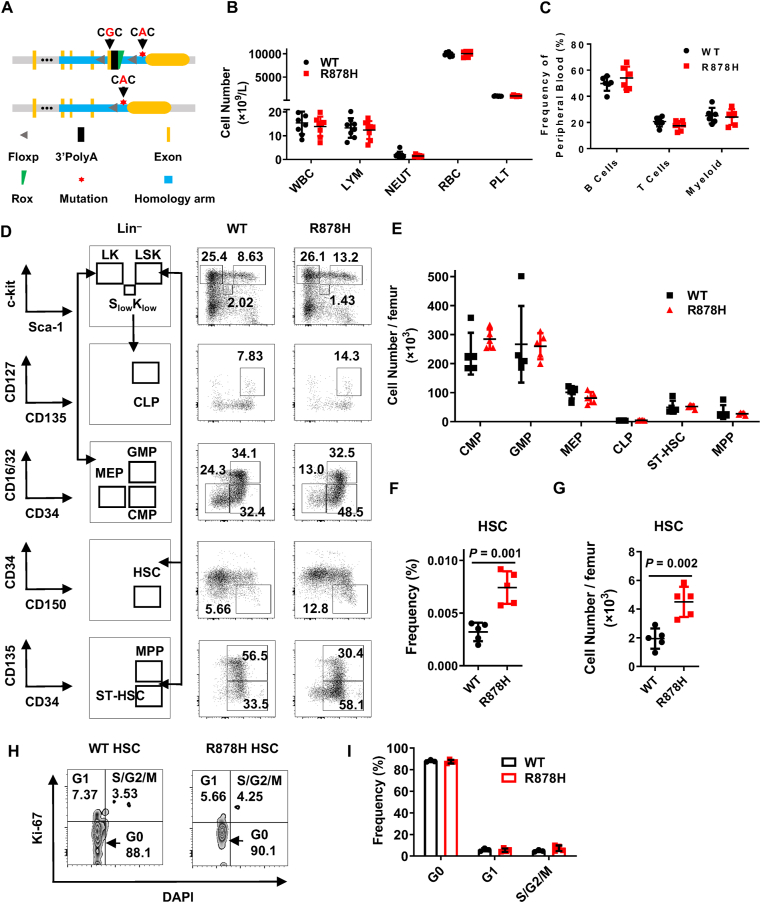
DNMT3A R878H mutation expands the HSC compartment. (A) The schematic diagram depicts the targeting strategy to generate the DNMT3A R878H conditional knockin mouse. Asterisk representing guanine at the 2633 site in the exon 23 of Dnmt3a was replaced by an adenine to encode R878H mutation. (B) The histogram shows the count of white blood cell (WBC), lymphocyte (LYM), neutrophil (NEUT), red blood cell (RBC) and platelet (PLT) between WT (Dnmt3afl-R878H/+, Vav-icre–) and R878H mice (Dnmt3afl-R878H/+, Vav-icre+; 4 months). n = 6 mice per group from two independent experiments. (C) Six 4-month old R878H mice and six age-matched WT mice were analyzed for myeloid, B and T cells. The histogram displays the frequency of B cells (B220+), T cells (CD3+) and myeloid (CD11b+) in the peripheral blood (PB). (D) Representative plots depict the gating strategies for HSC (Lineage– SCA-1+ c-KIT+ CD150+ CD34–), CMP (Lineage– SCA-1– c-KIT+ CD16/32– CD34+), GMP (Lineage– SCA-1– c-KIT+ CD16/32+ CD34+), MEP (Lineage– SCA-1– c-KIT+ CD16/32– CD34–), CLP (Lineage– SCA-1low c-KITlow CD135+ CD127+), ST-HSC (Lineage– SCA-1+ c-KIT+ CD135– CD34+) and MPP (Lineage– SCA-1+ c-KIT+ CD135+ CD34+) cells analysis in the bone marrow (BM) of WT and R878H mice. (E)–(G) 4 months old R878H mice and age-matched WT mice were analyzed for HSC and progenitors. (E) The histogram exhibits the absolute number of CMPs, GMPs, MEPs, CLPs, ST-HSCs and MPPs in the BM of WT and R878H mice. The histograms depict the frequency (F) and absolute number (G) of HSCs in the BM of WT and R878H mice. n = 5 mice per group from two independent experiments. (H)–(I) Cell cycle analysis of HSCs in six-month-old WT and R878H mice using Ki67 and DAPI. Representative flow cytometry plots (H) and the frequency of cells in each phase (I) are shown. WT, n = 3; R878H, n = 4. All data above are shown as mean ± SD.
When Dnmt3afl-R878H/+ mice exposed to Cre recombinase leads to an inversion of the mutant exon inserted and then with an excision reaction to remove the WT exon and one loxP site, and another loxP site, fixing the inversion in the right place (Fig. 1A). To achieve hematopoietic-specifically mutant mice, Dnmt3afl-R878H/+ mice were crossed to Vav-iCre mice. In all experiments, mice were heterozygous for both the Dnmt3aR878H allele and the Vav-iCre allele (Dnmt3aR878H/+Vav-iCre+ or Dnmt3aR878H/+).
Mlkl–/– mice20 were kindly provided by Dr. Xiaodong Wang (National Institute of Biological Sciences). Ripk1K45A mice21 and Ripk3Δ/Δ mice22 were kindly provided by Dr. Haibing Zhang (Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences).
All mice were housed in specific-pathogen-free, AAALAC-accredited animal care facilities at the Laboratory Animal Research Center, Tsinghua University and all procedures were approved by the Institutional Animal Care and Use Committee of Tsinghua University.
2.2. Flow cytometric analysis and cell sorting
Cells were suspended in HBSS+ buffer (D-Hank's buffer containing 2% fetal bovine serum, 1% penicillin/streptomycin and 1% HEPES) and then stained with fluorochrome-labeled antibodies. Flow cytometric analysis data were collected with a BD LSRFortessa SORP flow cytometer (BD Biosciences) and analyzed using FlowJo Software (Becton, Dickinson and Company). Cell sorting was performed by BD Influx (BD Biosciences) and the target fraction was sorted into HBSS+ buffer. Non-lysed bone marrow (BM) cells were applied for analysis of hematopoietic stem and progenitor cells (HSPC) (antibodies containing Lin-APC/Cy7 cocktail, c-KIT APC, SCA-1 PE/Cy7, CD150 PE, CD34 AF700, CD127 BV421, CD16/32 FITC and CD135 PE-CF594) and lineage (antibodies against MAC1, GR1, B220, CD3). Chimerism analysis in mature cells from peripheral blood (PB) was lysed by ACK buffer (NH4Cl, 150 mmol/L; KHCO3, 10 mmol/L; Na2EDTA, 0.1 mmol/L; adjust the pH to 7.2–7.4) and subsequently subjected to flow cytometer after staining with fluorochrome-conjugated antibodies (antibodies containing MAC1, B220, CD3, CD45.1 and CD45.2). A detailed list of antibodies is provided in the Supporting Information Table S1.
2.3. Competitive bone marrow transplantation
For the competitive bone marrow transplantation assay. 5 × 105 whole BM cells were freshly isolated from WT or R878H mice (CD45.2, C57BL/6J) and injected intravenously (i.v.) into lethally irradiated (10 Gy) WT recipient mice (CD45.1/2, F1 generated by mating CD45.1 with CD45.2 mice, C57BL/6J mice) together with 5 × 105 WT (CD45.1, C57BL/6J mice) whole BM competitor cells. Peripheral blood from the recipient was analyzed for donor-derived chimerism (myeloid, B, and T cells) monthly. The antibodies combination (CD3, B220, CD11b, CD45.1 CD45.2) were used to evaluate peripheral blood chimerism. For the young and aged recipient transplantation assay, 1 × 105 WT or R878H mice (CD45.2, C57BL/6J) total BM cells were transplanted into 2 or 15 months lethally irradiated (10 Gy) WT recipients (CD45.1, C57BL/6J mice) together with 5 × 105 competitor cells (CD45.1/2, C57BL/6J mice) and the chimera in peripheral blood was evaluated every month until the 4th month.
2.4. In vitro treatments
For the HSC in vitro culture and TNFα treatment assay, 100 HSCs (Lineage– SCA-1+ c-KIT+ CD34– CD150+) were sorted from either WT or R878H mice and cultured in 100 μL SFEM medium (supplemented with 30 ng/mL SCF, 30 ng/mL TPO and 100 U/mL penicillin–streptomycin) for 12 h. Then all cultures were treated with various combinations of TNFα (200 ng/mL), RIPA56 (20 μmol/L) and resibufogenin (20 μmol/L) for 7 days. Subsequently, phenotypic “HSCs” (CD150+ CD48– SCA-1+ c-KIT+) were counted by flow cytometry after staining with various combinations of indicated antibodies.
2.5. In vivo treatments
For the clonal hematopoiesis mouse model, 1 × 105 total BM cells either from WT or R878H mice (CD45.2, C57BL/6J) were transplanted into lethally irradiated recipients (CD45.1/2, C57BL/6J mice) mixed with 5 × 105 competitor cells (CD45.1, C57BL/6J mice). One month later, LPS (0.8 mg/kg) was administrated to the recipients intraperitoneally every two days for 15 doses, and the chimera of peripheral blood was evaluated every month until the 5th month. In the acute inflammatory insult experiment, WT or R878H mice were intraperitoneally injected with LPS (0.8 mg/kg) or an equal volume of PBS for a single dose and mature cells from peripheral blood together with HSPCs from BM of the indicated mice were analyzed 48 h later.
2.6. Cytokine antibody array
Femurs and tibias of young and aged recipient mice reconstituted by WT cells were isolated and flushed with a total of 150 μL PBS using an insulin syringe with a 29G needle. Then, centrifuge at 500×g for 8 min to remove cells. Supernatants were further purified by centrifuging at 14,000×g for 15 min to remove cell debris. Samples were dispensed into single-use aliquots and stored at −80 °C until use. Each sample was diluted 40-fold before cytokine proteins in the samples were determined by a mouse Antibody Arrays Kit with a glass slide-based antibody cytokine array containing 200 proteins (GSM-CAA-4000, RayBiotech) according to the manufacturer's instructions.
2.7. Hematological cell counts
Hematologic parameters in PB after tail bleeding were analyzed by Auto Hematology Analyzer BC-5000 (MINDRAY). BM cells were harvested from one femur and suspended in HBSS+ buffer on ice before evaluating by Vi-CELL Cell Counter (Beckman).
2.8. Western blotting
1 × 106 c-KIT+ BM cells from either WT or R878H mice were purified by magnetic beads enrichment (Miltenyi Biotec) after staining with c-KIT-APC antibody and cultured in 500 μL SFEM medium (supplemented with 30 ng/mL SCF, 30 ng/mL TPO and 100 U/mL penicillin–streptomycin) for 12 h. Then half of the cultures were treated with TSZ (T, 50 ng/mL TNFα; S, 100 nmol/L SMAC mimetic; Z, 20 μmol/L z-VAD) for 1 h. Cells were harvested and lysed with 150 μL NETN buffer (100 mmol/L NaCl, 20 mmol/L Tris-HCl, pH 8.0, 1 mmol/L EDTA, 0.5 mmol/L PMSF and 0.5% Nonidet P-40) on ice for 10 min. Lysis was completed by sonication and subsequently centrifuged at 14,000×g for 5 min. The supernatant was mixed with 2 × loading buffer and then boiled for 6 min. Samples were resolved on 10% SDS-PAGE followed by transferring onto a PVDF membrane (BioRad), and then the membrane was blocked by 5% skim milk in TBST buffer before incubating with indicated primary antibodies.
2.9. Quantitative real-time PCR
Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions from 2 × 104 donor-derived CD45.2 LSK (Lineage– SCA-1+ c-KIT+) cells sorted from BM of the fourth month aged recipients after whole BM transplantation. The concentration of RNA was normalized and cDNA was synthesized by PrimeScript RT reagent Kit (Takara, Cat. # RR047A). Acquired cDNA was analyzed by PowerUp SYBR Green mix (Applied Biosystems, Cat. # A25780) with indicated primers on a QuantStudio-3 Real-time PCR System (Applied Biosystems). The full primer information is listed in Supporting Information Table S2.
2.10. Statistical analysis
All data are shown as mean ± SD. Two-tailed unpaired Student's t-test was used for statistical significance analysis after testing for normal distribution and data were plotted using GraphPad Prism 6 software.
3. Results
3.1. DNMT3A R878H mutation expands the HSC compartment
To investigate the functional role of DNMT3A R878H in HSCs, Dnmt3afl-R878H/+ mice (Fig. 1A, Supporting Information Fig. S1A and S1B) was constructed and then crossed with Vav-iCre mice to generate DNMT3A R878H heterozygous mice (Dnmt3aR878H/+), wherein DNMT3A R878H occurs mainly in hematopoietic cells (hereafter named R878H mice). To evaluate the influence of DNMT3A R878H on the production of blood cells, we performed a complete blood count assay for R878H mice and littermate controls. The results revealed that the white blood cell (WBC), lymphocyte (LYM), neutrophil (NEUT), red blood cell (RBC) and platelet (PLT) of R878H mice kept static compared with controls (Fig. 1B). Moreover, the bone marrow cellularity of R878H mice was indistinguishable from their littermate controls (Fig. S1C). We then sought to investigate the hematopoietic composition in peripheral blood and bone marrow of R878H mice by flow cytometry. The results show that the frequency of lineage cells, including B cells, T cells and myeloid cells, in peripheral blood (PB) and bone marrow (BM) of R878H mice was indistinguishable from controls (Fig. 1C, Fig. S1D and S1E). Furthermore, the absolute number and frequency of CMP (common myeloid progenitor), GMP (granulocyte-macrophage progenitors), CLP (common lymphoid progenitors), MEP (megakaryocyte-erythroid progenitors), ST-HSC (short-term HSC) and MPP (multipotent progenitor cell) of R878H mice holds static (Fig. 1D, E and Fig. S1F). While, both the frequency and absolute number of HSCs in R878H mice are significantly larger than WT mice (Fig. 1D–G), which is consistent with a previous report that DNMT3A R878H mutation expands HSC compartment23. Cell cycle analysis showed that DNMT3A R878H mutation does not alter the quiescent state of HSCs (Fig. 1H and I).
3.2. Aged bone marrow microenvironment promotes DNMT3A R878H-based clonal hematopoiesis
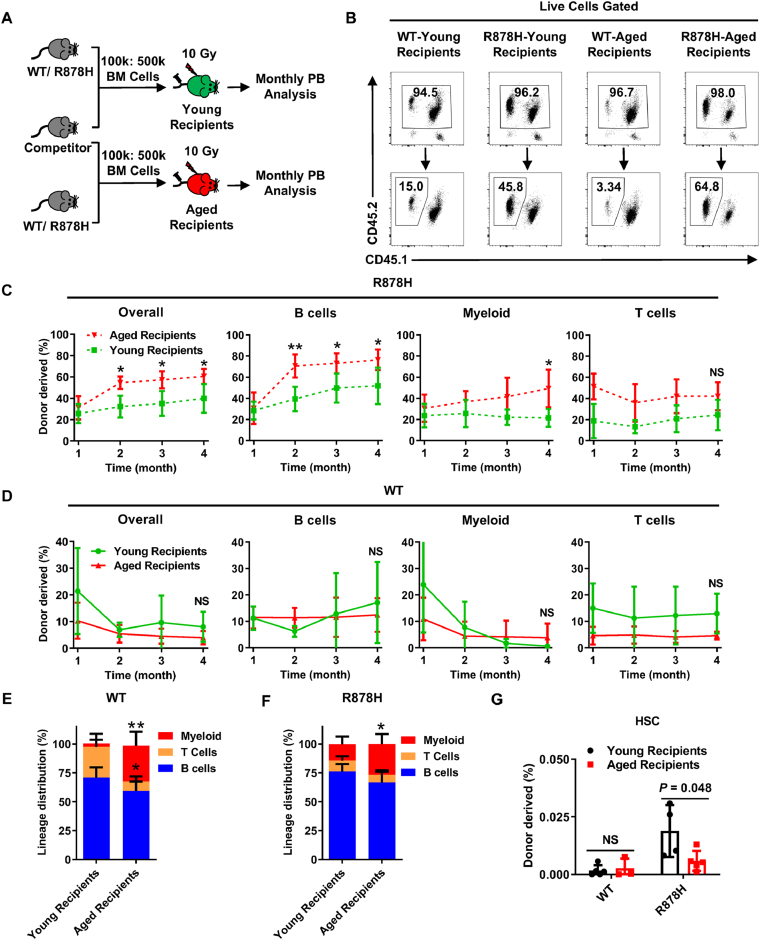
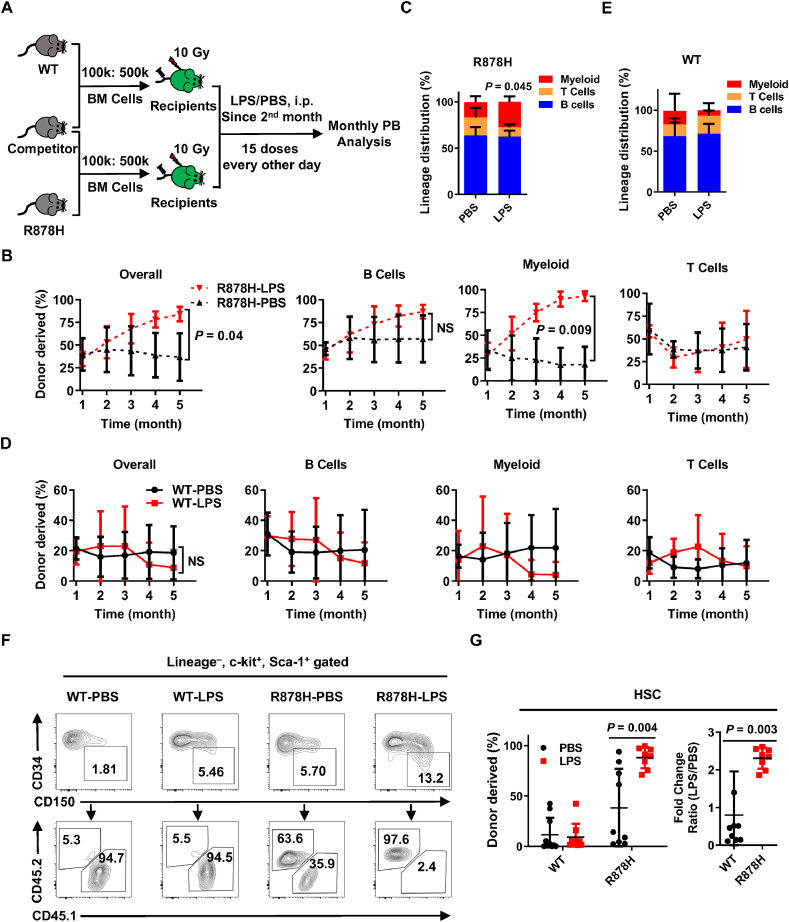
Several recent studies have revealed that human DNMT3A R882H (mouse allele: DNMT3A R878H) is a driver mutation and leads to clonal hematopoiesis with age2, 3, 4, therefore we attempted to speculate that aged microenvironment might promote R878H cells expansion. To test this hypothesis, we transplanted 1 × 105 either R878H or WT total bone marrow cells into lethally irradiated young (2-month) and aged (15-month) recipients together with 5 × 105 competitor cells and the chimera in peripheral blood was evaluated every month until the 4th month (Fig. 2A). The result show that the reconstitution capacity of R878H cells in aged recipients is significantly higher than that in young recipients (60.6 ± 7.1% vs. 39.9 ± 13.5%) (Fig. 2B and C and Supporting Information Fig. S2A), while WT cells exhibit no difference between young and aged recipients (8.0 ± 5.6% vs. 4.0 ± 2.5%) (Fig. 2D), indicating that aged bone marrow milieu fosters R878H cells. Moreover, we observed significantly increased differentiation bias towards myeloid lineage of both R878H and WT cells in aged recipients (Fig. 2E, F and Fig. S2B), which is consistent with previous conclusions that aged bone marrow environment promotes the differentiation bias to myeloid lineage24,25. We then analyzed the donor-derived HSCs in the bone marrow of recipients. The results reveal that the percentage of WT-derived HSCs between young and aged recipients shows no difference, but the percentage of R878H-derived HSCs in aged recipients is significantly lower than that in young recipients (Fig. 2G), which suggests that aged bone marrow promotes the differentiation of R878H HSCs and then facilitates DNMT3A R878H-based clonal hematopoiesis.
Figure 2.
Aged bone marrow microenvironment promotes DNMT3A R878H-based clonal hematopoiesis. (A)–(F) Freshly isolated 1 × 105 BM cells (CD45.2) from 2 months old R878H mice or age-matched WT mice were transplanted into lethally irradiated young (2 months) or aged (15 months) recipients (CD45.1) together with 5 × 105 competitor cells (CD45.1/2). Chimera in the peripheral blood was evaluated every month until the fourth month. (A) Experimental design. (B) Representative flow cytometry plots displaying the frequencies of donor-derived cells in the PB of young or aged recipient mice transplanted with the indicated bone marrow cells at the 4th month after transplantation (n = 4–5 mice per group). (C) and (D) These line plots depict the percentage of indicated donor-derived cells in the PB of young or aged recipients carrying R878H (C) or WT (D) BM cells. (E) and (F) The histograms show the lineage distribution of myeloid, T cells and B cells among donor-derived cells in the PB of young or aged recipients carrying WT (E) or R878H (F) BM cells at the 4th month after transplantation. n = 4–5 mice per group from two independent experiments. (G) The histogram displays the percentage of WT/R878H-derived HSCs in young recipients (black) and aged recipients (red) (n = 4–5 mice per group). All data above are shown as mean ± SD; ∗P < 0.05, ∗∗P < 0.01, NS represents no significance.
3.3. DNMT3A R878H blunts the response of hematopoietic stem and progenitor cells upon inflammation challenge
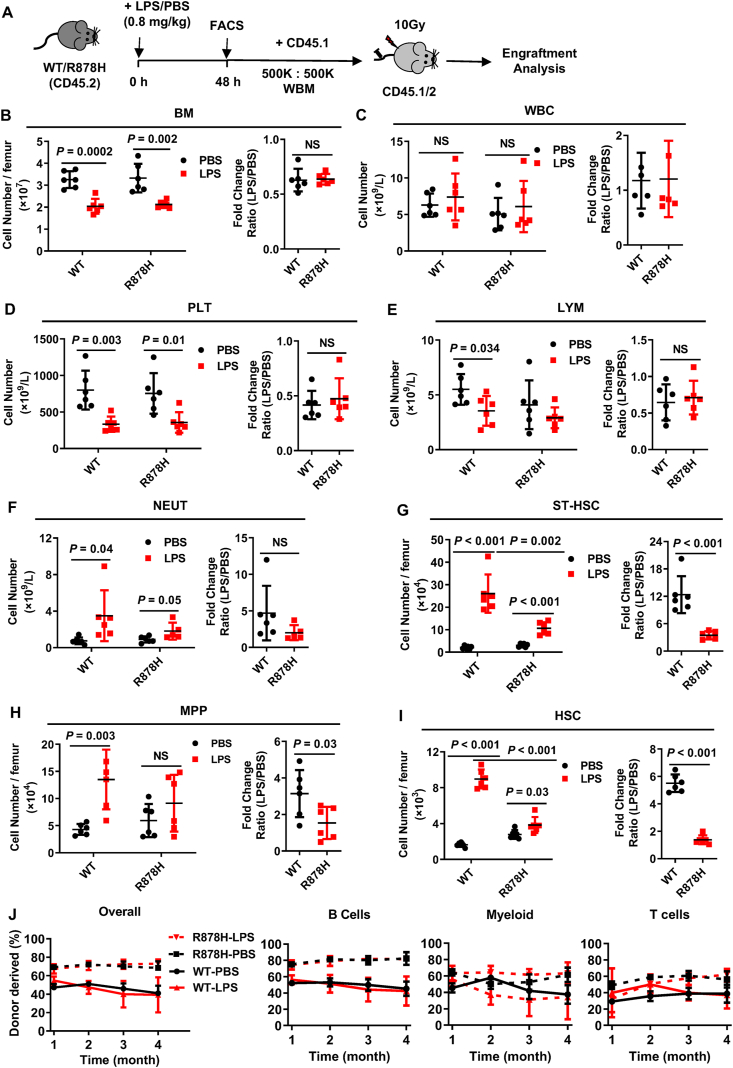
It has been known for several decades that aging is frequently accompanied by a chronic, low-grade inflammation, which is called “inflammaging” and it modulates both the aging process and age-related diseases26,27. Then, we wondered whether aging-elevated inflammation promotes DNMT3A R878H-based clonal hematopoiesis. To test this hypothesis, we challenged R878H and WT mice by a single dose of LPS intraperitoneally (Fig. 3A), which is frequently utilized as an acute inflammation model28. Forty-eight hours later, bone marrow cellularity was significantly decreased by 44% in R878H mice and 30% in WT mice separately (Fig. 3B). Blood count assay revealed that WBC and RBC of both R878H and WT mice are comparable in response to the LPS challenge (Fig. 3C and Supporting Information Fig. S3A). While, PLT were significantly decreased by ∼50% in both R878H and WT mice (Fig. 3D). It is notable that the number of LYM declines significantly in WT mice, but not in R878H mice (Fig. 3E). Furthermore, NEUT increases significantly after LPS insults in both WT and R878H mice (Fig. 3F). However, the fold change (the ratio of LPS-treated group to PBS-treated group) of the above cell populations between WT and R878H mice exhibits no difference, indicating that DNMT3A R878H does not affect the production of mature blood cells upon acute inflammatory challenge.
Figure 3.
DNMT3A R878H blunts the response of hematopoietic stem cells upon inflammation challenge. (A) A model was used to evaluate the response of R878H mice to acute inflammatory challenges. The schematic diagram showing the experimental design: WT and R878H mice were challenged by LPS (0.8 mg/kg) or an equal volume of PBS intraperitoneally and these mice were analyzed 48 h later. 5 × 105 LPS/PBS-treated WT or R878H BM cells (CD45.2) together with 5 × 105 competitor cells (CD45.1) were transplanted into lethally irradiated recipients (CD45.1/2). Chimerism in the PB was analyzed monthly. (B)–(F) The histograms show the cell number (left) of BM cells (B), WBC (C), PLT (D), LYM (E), NEUT (F), and the fold change (LPS/PBS, right) of WT and R878H mice upon LPS (red) or PBS (black) treatment (n = 6 mice per group from 4 independent experiments). (G)–(I) The left histograms display the absolute cell number of ST-HSC (G), MPP (H), HSC (I) in the BM of the indicated mice treated with a single dose (0.8 mg/kg) of LPS or PBS. The right histograms depict a ratio of the absolute cell number between LPS and PBS groups in the indicated mice from different populations of hematopoietic cells (n = 6 mice/group from 4 independent experiments). (J) These line plots show the percentage of donor-derived overall and B cells, T cells, myeloid within PB cells of the indicated recipients at each time point (n = 4–6 mice per group from 2 independent experiments). All data above are shown as mean ± SD; NS represents no significance.
We next sought to analyze the lineage composition in PB and BM of both R878H and WT mice upon LPS challenge. We observed that both B and T cells in PB and BM of both mice obviously dropped, while the myeloid cells in PB and BM of both mice were increased, wherein the increase in PB is larger than that in BM (Fig. S3B and S3C). We then calculated the fold change, and the results show that there is no difference between R878H and WT mice in response to the LPS challenge (Fig. S3B and S3C, the rightmost data of each graph).
Given that HSPCs generate all lineages, we then set out to analyze HSPCs of R878H and WT mice in response to the LPS challenge. We observed that CMP, GMP, MEP and CLP significantly dropped in both mice and the fold change exhibited no difference between WT and R878H mice (Fig. S3D–S3G). It is notable that ST-HSC and MPP of both mice expanded significantly upon the LPS challenge, while the expansion in R878H mice was less pronounced than that in WT mice (Fig. 3G and H). More interesting is that the LPS challenge results in an increase in the number of both WT HSCs (from 1630 ± 237 to 8967 ± 1051) and R878H HSCs (from 2778 ± 493 to 3834 ± 892), while the fold change in R878H mice was significantly lower than that in WT mice (Fig. 3I). These data imply that DNMT3A R878H mutation blunts the response of hematopoietic stem and multipotent progenitor cells, but not the downstream progenitor or terminally differentiated cells, upon inflammatory stress.
To further investigate the function of R878H and WT bone marrow cells in response to acute LPS challenge, 5 × 105 total bone marrow cells isolated from either LPS/PBS-treated R878H or LPS/PBS-treated WT mice were transplanted into lethally irradiated recipients together with 5 × 105 competitor cells and the chimera in peripheral blood was checked monthly until the fourth month (Fig. 3A). The results show that the reconstitution ability of LPS-treated R878H and WT bone marrow cells exhibited no difference from that of the control group, indicating that acute inflammatory stress did not impair the reconstitution capacity of bone marrow cells of both mice (Fig. 3J). This result is consistent with a recent study showing that Escherichia coli-induced inflammation accelerates hematopoietic stem and progenitor cells proliferation, but preserves the functional HSCs in the bone marrow29.
3.4. LPS-induced chronic inflammation promotes DNMT3A R878H-driven clonal hematopoiesis
Given that a previous study reported that repeated activation of HSCs out of their dormant state provoked the attrition of normal HSCs30,31, and our data show that DNMT3A R878H mutation prevented HSCs from LPS-induced activation (Fig. 3I), we then speculated that R878H HSCs might protect HSCs from chronic inflammatory damage. To test this hypothesis, we transplanted 1 × 105 freshly isolated total bone marrow cells from either WT or R878H mice into lethally irradiated recipients together with 5 × 105 competitor cells. One month later, we challenged the recipients with LPS every two days intraperitoneally for one month, which is frequently utilized as a chronic inflammation model28, and the chimera of peripheral blood and bone marrow was evaluated every month until the 5th month (Fig. 4A). The result shows that R878H cells significantly outcompeted the competitor cells upon chronic LPS challenge compared to PBS-treated ones (70.0 ± 29.0% vs. 36.7 ± 26.2%) and the difference mainly stemmed from the myeloid lineage (Fig. 4B and Supporting Information Fig. S4A). The mature hematopoietic lineage distribution of R878H cells exhibited significant differentiation skewing towards myeloid lineage in response to the LPS challenge (Fig. 4C). While both reconstitution capacity and lineage distribution of WT cells showed no difference between LPS and PBS group (Fig. 4D, E and Fig. S4A).
Figure 4.
LPS-induced chronic inflammation promotes DNMT3A R878H-driven clonal hematopoiesis. (A) Schematic diagram of the experimental procedure is designed for generating chimeric mice simulating clonal expansion of Dnmt3aR878H/+ cells upon inflammation challenges. WT or R878H BM cells (CD45.2) together with competitor cells (CD45.1) at a ratio of 1 to 5 (1 × 105:5 × 105) were transplanted into lethally irradiated recipients (CD45.1/2). One month post-transplantation, the recipients were subjected to sustaining treatment with LPS (0.8 mg/kg injected i.p. every other day for 15 doses) or an equal volume of PBS. Chimerism in the PB was analyzed monthly until the fifth month. (B) These line plots show the frequencies of R878H-derived overall and B cells, T cells, myeloid within PB cells at the indicated time point in response to LPS (red) and PBS (black) treatment. (C) The histogram displays the lineage distribution of R878H-derived T, B and myeloid in peripheral blood at the 5th month post-transplantation. (D) These line plots depict the percentage of WT-derived overall and B cells, T cells, myeloid within PB cells at indicated the time point in response to LPS (red) and PBS (black) treatment. (E) The histogram exhibits the lineage distribution of WT-derived T, B and myeloid in peripheral blood at the 5th month post-transplantation. (F) Representative flow cytometry plots depict the gating strategy and the percentage of donor-derived HSC (CD34– CD150+ LSK) in the indicated recipients at the fifth month after transplantation. (G) The histograms show the frequency of WT/R878H-derived HSCs in response to LPS (red) and PBS (black) treatment; the right histogram shows the fold change of HSCs (WT: black; R878H: red) in response to LPS treatment. n = 5–7 mice per group from two independent experiments, the data are shown as mean ± SD; NS represents no significance.
To find out the reason why R878H-carrying cells exhibit differentiation bias towards myeloid cells in response to LPS treatment, we analyzed donor-derived myeloid progenitors and HSC in the bone marrow of recipient mice five months after transplantation. We observed that all myeloid progenitors (CMP, GMP and MEP), multiple potential progenitors (CD34+ LSK) and HSCs were increased significantly upon LPS challenge in the R878H group rather than the WT group (Fig. 4F, G, Fig. S4B and S4C), which could explain the expansion of R878H-derived myeloid cells during aging. Briefly, the above data suggest that DNMT3A R878H-carrying HSPCs resist chronic inflammation-induced damage, and furthermore result in DNMT3A R878H-driven clonal hematopoiesis.
3.5. Elevated-TNFα in aged bone marrow promotes DNMT3A R878H-based clonal hematopoiesis
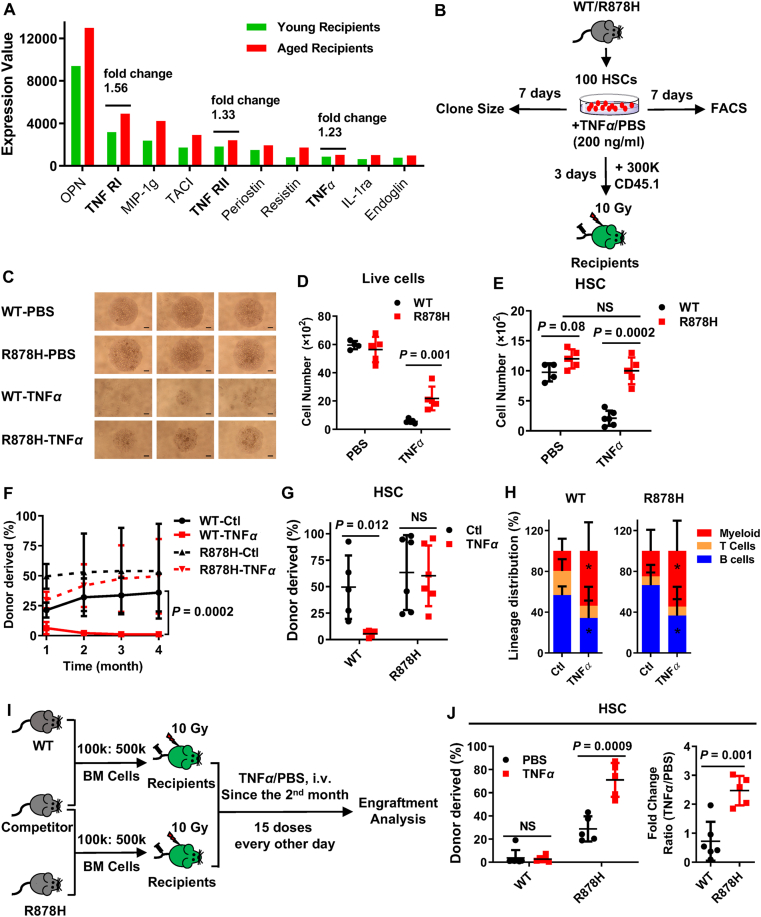
Given that R878H-carrying HSCs resist inflammation-induced damage (Fig. 4G), and that aged bone marrow milieu facilitate the proliferative advantage of R878H cells (Fig. 2C), and that aged bone marrow is accompanied by several elevated pro-inflammatory cytokines32,33, we then attempted to speculate that inflammatory factor(s) in aged bone marrow might promote the clonal expansion of R878H cells.
To identify such inflammatory factor(s) in the aged microenvironment, we conducted cytokine array assays with bone marrow aspirates of young and aged recipients reconstituted by WT cells (Fig. 2A, detailed in Supporting Information Table S3). The results show that the levels of several pro-inflammatory cytokines are upregulated in aged recipients (Fig. 5A, Supporting Information Fig. S5A and S5B), especially the TNFα signaling (Fig. S5C). Given that TNFα is one of the major inflammatory cytokines produced by hematopoietic cells upon LPS challenge34, we speculated that TNFα may be the main factor driving R878H-based clonal hematopoiesis. To test this hypothesis, 100 freshly isolated R878H and WT HSCs were treated with TNFα and the clone size was estimated 7 days later (Fig. 5B). The result show that TNFα strongly depleted WT cells, while DNMT3A R878H compromised the impairment in response to TNFα insults (Fig. 5C and D). Then, we counted phenotypic “HSCs” by this combination: CD150+ CD48– SCA-1+ c-KIT+, and found that the number of WT HSCs decreased by 75% upon TNFα insults, while the number of R878H HSCs held static in comparison to PBS-treated ones (Fig. 5E and Fig. S5D). These data suggest that DNMT3A R878H prevents the impairment of HSCs induced by TNFα.
Figure 5.
Elevated-TNFα in aged bone marrow promotes DNMT3A R878H-based clonal hematopoiesis. (A) The pooled bone marrow aspirates of young recipients and aged recipients reconstituted by WT cells (Fig. 2A) were subjected to a mouse cytokine antibody array. Cytokines were identified as upregulated candidates in aged recipients with the criteria that fold change between aged recipients and young recipients are greater than 1.2, while fold change lower than 0.83 is considered as downregulated cytokines. The histogram depicts the top 10 cytokines upregulated in the bone marrow aspirates of aged recipients which are ranked by the expression levels. Each sample was pooled from 3 mice for each group. (B)–(H) 100 HSCs (CD34– CD150+ LSK) were freshly isolated from either WT or R878H mice (CD45.2), and the HSCs were cultured in vitro in the presence or absence of TNFα (200 ng/mL) for 3 or 7 days (all HSCs were cultured in SFEM medium supplemented with 30 ng/mL SCF, 30 ng/mL TPO and 100 U/mL Penicillin–Streptomycin). Three days later, each culture was mixed with 3 × 105 competitor cells (CD45.1) and transplanted into lethally irradiated recipients (CD45.1/2). Chimera in the peripheral blood was evaluated for 4 months. (B) Experimental design. (C) On the 7th day, cell growth was monitored by a light microscopy, and images were acquired under an inverted microscope (Olympus CKX41). Scale bar, 0.5 mm. (D) and (E) The number of live cells (D, DAPI–) and HSCs (E, CD150+ CD48– c-KIT+ SCA-1+) was analyzed on days 7 (n = 5). (F) These line plots show the percentage of donor-derived cells within PB cells at the indicated time point after transplantation. (G) The histogram depicts the frequency of donor-derived HSC (CD34– CD150+ LSK) in the indicated recipients at the fourth month post-transplantation. (H) The histograms exhibit the lineage distribution of WT-derived or R878H-derived T, B and myeloid cells in peripheral blood at the 4th month post-transplantation. n = 5–7 mice per group from two independent experiments, the data are shown as mean ± SD, ∗P < 0.05, NS represents no significance. (I) and (J) Experiment design for TNFα treatment in vivo. 1 × 105 freshly isolated total bone marrow cells from either 2–4 months old WT or R878H mice (CD45.2) together with 5 × 105 competitor cells from age-matched WT mice (CD45.1) were transplanted into lethally irradiated recipients (CD45.1/2). One month later, the recipients were challenged with TNFα (100 μg/kg) every other day intravenously for one month, and donor-derived HSCs were evaluated at the 5th month. (I) Schematic diagram of the experimental procedure. (J) The left histogram shows the number of WT/R878H-derived HSCs in response to TNFα (red) and PBS (black) treatment; the right histogram shows the fold change of HSCs (WT: black; R878H: red) in response to TNFα treatment. n = 5–7 mice per group, the data are shown as mean ± SD, NS represents no significance.
To evaluate whether TNFα affects the self-renewal and differentiation capacity of R878H HSCs, 100 R878H and WT HSCs were treated with TNFα for 3 days. Then, each culture was mixed with 3 × 105 competitor cells and transplanted into lethally irradiated recipients. The results showed that TNFα-treated WT HSCs completely lose their reconstitution capacity, while the reconstitution ability of R878H HSCs is not affected by TNFα (Fig. 5F). Consistent with this finding, we observed that the WT-derived HSCs are almost depleted in recipients when their donor HSCs were pretreated with TNFα, but R878H-derived HSCs are not disturbed by TNFα (Fig. 5G), suggesting that TNFα impairs WT HSCs but does not affect the reconstitution capacityR878H HSCs. However, we observed significantly increased differentiation bias towards myeloid lineage but decreased differentiation into B cell lineage of both TNFα-treated R878H and WT HSCs (Fig. 5H), indicating that TNFα promotes the differentiation bias to the myeloid lineage of both WT and R878H HSCs which is in line with the phenotype we observed in aged recipients.
Now that R878H HSCs acquire survival advantage over WT HSCs upon TNFα insults, we wondered whether this effect can directly result in R878H-based clonal hematopoiesis. To test this hypothesis, 1 × 105 freshly isolated total bone marrow cells from either WT or R878H mice together with 5 × 105 competitor cells were transplanted into lethally irradiated recipients. One month later, we challenged the recipients with TNFα (100 μg/kg) every other day intravenously for one month, and donor-derived HSCs in the bone marrow were evaluated at the 5th month (Fig. 5I). We observed that R878H-derived HSCs significantly outcompeted the competitor cells upon chronic TNFα insults compared to PBS-treated ones (71.0 ± 14.6% vs. 28.7 ± 10.9%) but not the WT-derived HSCs (Fig. 5J), indicating TNFα facilitates R878H-driven clonal hematopoiesis.
These data suggest that DNMT3A R878H counteracts the damage induced by TNFα to HSCs, which furthermore indicated that aging-elevated TNFα promotes DNMT3A R878H-based clonal hematopoiesis.
3.6. DNMT3A R878H prevents the activation of necroptosis in HSPCs upon TNFα challenge
To explore the potential mechanism that DNMT3A R878H compromised the TNFα-induced damage to HSCs, we examined the genes regulated by TNFα, including NF-κB, apoptosis and necroptosis related genes35. Firstly, we transplanted 1 × 105 bone marrow cells freshly isolated from either R878H mice or wild-type mice into lethally irradiated aged recipients together with 5 × 105 young competitor cells. Four months later, 2 × 104 donor-derived LSK cells (Lineage– SCA-1+ cKIT+) were isolated to evaluate TNFα-regulated genes (Fig. 6A, primers see Supporting Information Table S2). The results reveal that NF-κB pathway genes remained undisturbed in R878H cells (Supporting Information Fig. S6A), while the apoptosis-related genes (Fadd and Casp8) and necroptosis genes (Ripk1 and Ripk3) were significantly decreased (Fig. 6B and C), suggesting that apoptosis and necroptosis pathways may be compromised in R878H cells.
Figure 6.
DNMT3A R878H prevents the activation of necroptosis in HSPCs upon TNFα challenge. (A) Experimental procedure to evaluate the expression of TNF signaling genes: 1 × 105 bone marrow cells freshly isolated from either R878H mice or wild-type mice (CD45.2) were transplanted into lethally irradiated aged recipients (CD45.1) together with 5 × 105 young competitor cells (CD45.1/2). Four months after transplantation, 2 × 104 donor-derived LSK cells (CD45+ LIN– c-KIT+ SCA-1+) were isolated for qRT-PCR analysis. (B) and (C) The histogram showing the relative expression of apoptosis-related genes (B) and necroptosis-associated genes (C) in the indicated cells. (D) and (E) Experiments were designed to evaluate the response of R878H HSPCs upon apoptotic and necroptotic stress. (D) The schematic diagram showing the procedure for in vitro TSZ or TS treatment in the indicated experiment (S, 100 nmol/L SMAC mimetic; T, 50 ng/mL TNFα; Z, 20 μmol/L z-VAD). To determine the percentage of cell death upon TS or TSZ treatment, LSK cells were isolated freshly from WT or R878H mice and cultured with the indicated chemicals for 6 h, and the resulting cells were stained with DAPI and the dead cells were determined by flow cytometry. (E) The histogram depicts the percentage of cell death in LSK cells upon TS, TSZ and TSZ plus RIPA56 (RIPA56: 20 μmol/L) treatment (n = 3–4 per group from 2 independent experiments). (F) and (G) c-KIT+ bone marrow cells were freshly isolated from WT or R878H mice and cultured in SFEM medium overnight. Then half of the cultures were treated with TSZ for 1 h, and the resulting cells were harvested for Western blotting analysis. (F) Experimental design. (G) Representative Western blot showing the expression of RIPK1, RIPK3, MLKL and phosphorylated MLKL in c-KIT+ cells upon indicated treatment (three biological replicates). (H) and (I) 100 HSCs (CD34– CD150+ LSK) were freshly isolated from either WT or R878H mice, and the HSCs were cultured in vitro with the indicated treatment for 7 days (all HSCs were cultured in SFEM medium supplemented with 30 ng/mL SCF, 30 ng/mL TPO and 100 U/mL penicillin–streptomycin; TNFα: 200 ng/mL, RIPA56: 20 μmol/L, RES: 20 μmol/L). (H) Experimental design. (I) The histogram shows the number of HSCs (CD150+ CD48– c-KIT+ SCA-1+) analyzed on days 7. (J) and (K) An in vivo assay was applied to evaluate the role of necroptosis in R878H mice. (J) The cartoon shows the procedure for repeated in vivo TNFα injections in WT, Dnmt3aR878H/+, Ripk1K45A, Ripk3Δ/Δ and Mlkl–/– mice (TNFα: 5 μg/mouse), intravenously. All of these mice were analyzed 48 h later. (K) The histogram depicts the absolute number of HSC (CD34– CD150+ LSK) in the bone marrow of indicated genotypic mice treated with TNFα (n = 3 mice per group from 2 independent experiments). All data above are shown as mean ± SD, NS represents no significance.
To further investigate whether R878H cells acquire growth advantage by resisting necroptosis or apoptosis, we forced WT and R878H LSK cells to undergo apoptosis (induced by the combination of TNFα and SMAC mimetic, abbreviated as “TS”)36 and necroptosis (induced by the combination of TNFα, SMAC mimetic, and a pan-caspase inhibitor Z-VAD-FMK, abbreviated as “TSZ”)37 for 6 h (Fig. 6D). We observed that there is no difference between WT and R878H LSKs in the TS-treated group, indicating that apoptosis is not the factor resulting in the growth advantage of R878H cells. While R878H LSKs exhibited significantly less cell death upon TSZ stimulation (Fig. 6E). Furthermore, the cell death of both R878H and WT LSKs decreased by about 60% upon TSZ and RIPA56 (RIPK1 inhibitor) treatment, which suggests that both R878H and WT LSKs undergo cell death through RIPK1-mediated necroptosis (Fig. 6E). These data indicate that LSK cells carrying DNMT3A R878H compromised TSZ-induced necroptosis rather than TS-induced apoptosis.
To further verify this observation, we evaluated the expression of RIPK1, RIPK3, MLKL and phosphorylated MLKL (p-MLKL) in c-KIT+ bone marrow cells of R878H and WT mice upon TSZ treatment (Fig. 6F). The result show that RIPK1 and RIPK3 decreased in R878H cells, which is consistent with the trend we observed in mRNA levels (Fig. 6G). Furthermore, we detected decreased p-MLKL, the final executioner of necroptosis, in R878H cells upon TSZ treatment (Fig. 6G), which confirmed that the necroptosis signaling is compromised in R878H cells.
The above result revealed that R878H cells, but not WT cells, acquire survival advantage by dampening TNFα-activated necroptosis. To further confirm this observation, we treated WT and R878H HSCs with various combinations of TNFα, RIPA56 (necroptosis inhibitor38) and resibufogenin (RES, necroptosis activator39) (Fig. 6H). The result show that necroptosis inhibition fully restores WT HSCs survival from TNFα insults, and then resulted in no difference between WT and R878H HSCs, indicating that TNFα impairs WT HSCs by activating necroptosis and necroptosis may be the target of R878H HSCs to resist TNFα cytotoxicity (Fig. 6I, Fig. S6B and S6C). When necroptosis signaling was activated by RES, we observed that the number of HSCs in both WT and R878H groups dropped significantly compared to the control group and TNFα-treated groups (Fig. 6I). Additionally, no difference was observed between WT and R878H HSCs in response to TNFα plus RES treatment, indicating that necroptosis activation abrogates the resistance of R878H HSCs to TNFα cytotoxicity (Fig. 6I, Fig. S6B and S6C).
To further verify this observation in vivo, we challenged two-month old WT, Dnmt3aR878H/+, Ripk1K45A, Ripk3Δ/Δ and Mlkl–/– mice with 3 doses of TNFα (5 μg/mouse, intravenous injection) every 12 h and analyzed them 48 h later (Fig. 6J). The results show that TNFα injection depletes HSCs in WT mice to about half of the PBS-treated counterpart, while the number of HSCs in Dnmt3aR878H, Ripk1K45A, Ripk3Δ/Δ and Mlkl–/– mice keeps static (Fig. 6K). The above data suggest that DNMT3A R878H protected HSCs from inflammatory insult by modulating necroptosis signaling.
Collectively, these results suggest that DNMT3A R878H regulates HSC survival upon TNFα stress by inhibiting necroptosis.
4. Discussions
In this study, we found that the necroptosis signaling was compromised in DNMT3A R878H cells, which resulted in enhanced reconstitution capacity of R878H HSCs upon aging-elevated inflammation. This process promotes the expansion of R878H cells in the aged bone marrow milieu, and furthermore leads to clonal hematopoiesis with aging. Similarly, a previous study revealed that IFNγ signaling promotes Dnmt3a–/– clonal expansion40. In addition, TNFα promotes the clonal expansion of JAK2 V617F myeloproliferative neoplasm cells41, and LPS-induced inflammation promotes the clonal hematopoiesis of Tet2−/− cells28. All these results reveal an important scientific finding that inflammation may be a key factor resulting in the occurrence of clonal hematopoiesis with aging.
A previous study showed that mice transplanted with OPN-treated aged HSCs exhibit an almost twofold reduction of HSCs frequency42, indicating that OPN may serve as a positive regulator promoting HSC differentiation. It is notable that the expression of OPN is increased in the bone marrow of aged recipients (Fig. 5A). Therefore, it is possible that aging-elevated inflammatory factors, such as TNFα promotes the clonal expansion of R878H HSCs, and at the same time, other aging-elevated factors in the aged bone marrow microenvironment, such as OPN, may promote the differentiation of R878H HSCs, which finally results in the clonal expansion of R878H cells in age recipients. Further studies to untangle the complexity between the expansion and differentiation of R878H HSCs are worth exploring, which may pave the way for the treatment of DNMT3A R882-based clonal hematopoiesis and hematopoietic malignancies.
We found that the RIPK1–RIPK3–MLKL-mediated necroptosis pathway is compromised in the HSPCs of DNMT3A R878H, and treatment with necroptosis agonists can restore this process. Therefore, the inhibition of necroptosis may be an internal mechanism that promotes the growth advantage of DNMT3A R878H HSCs. A recent study suggested that the functional decline of WT HSC induced by chronic inflammation can be reversed by blocking the necroptosis pathway43, indicating that necroptosis may be an important form of cell death in HSC upon inflammatory insults. However, it is not clear whether the expression level of necroptosis genes in R878H HSPCs is related to their DNA methylation status. A previous report showed that the promoter region of Ripk3 was hypomethylated in DNMT3A R878H LSK cells by conducting a genome-wide DNA methylation profile13. Other studies revealed that the expression of RIPK3 is negatively correlated with the methylation status in its promoter44 and methylation-mediated silencing of RIPK3 is observed in cancer cell lines45. Based on the above conclusion, the expression of RIPK3 should be increased in DNMT3A R878H LSK cells. While we observed the expression of RIPK3 was decreased in R878H HSPCs. Therefore, we attempted to speculate that the expression of Ripk3 and other necroptosis genes might not be directly regulated by DNA methylation. A previous study showed that DNMT3A R882 mutant protein preferentially interacts with polycomb repressive complex 1 (PRC1), and then results in gene silencing46. It is conceivable that the necroptotic proteins may be regulated by PRC1, which results in the reduction of necroptotic proteins.
Given that R878H cells compromised the necroptosis pathway to withstand the inflammatory stress, we tried to overexpress Ripk1 or Ripk3 in R878H HSPCs. Both WT and R878H cells transduced with lentiviruses expressing Ripk1 and Ripk3 failed to give any discernible reconstitution (data not shown), indicating that excessive activation of necroptosis results in cell death.
In patients, DNMT3A R882 mutations are more frequently observed in MDS or AML than clonal hematopoiesis47. In addition, a recent study showed that single-mutant clones carrying the DNMT3A R882 missense mutation were less frequently detected than non-R882 DNMT3A missense mutant initiating clones48. According to these clinical data, DNMT3A R882-inhibited necroptosis may contribute to leukemogenesis in addition to promoting clonal hematopoiesis.
Given that the DNMT3A R882 mutations occur in a variety of leukemia5, and patients with the mutations have an inferior outcome after chemotherapy13. Thus, developing small molecule inhibitors targeting DNMT3A R882 mutations will have important clinical significance, which is focused on by another project of ours.
5. Conclusions
We found that the up-regulated TNFα signaling in the aged bone marrow microenvironment is one of the major factors that promote DNMT3A R878H-driven clonal expansion of HSCs by preventing necroptosis upon TNFα and LPS-induced chronic inflammatory stimulation. Our findings provide the evidence that inflammatory factor TNFα may play an important role in the initiation and progression of DNMT3A R882 mutant clonal hematopoiesis, leukemia and the related disease.
Acknowledgments
We thank Dr. Saijuan Chen and Dr. Zhu Chen from Shanghai Jiao Tong University for the generous offer of DNMT3A R878H plasmid. We thank Dr. Xiaodong Wang (National Institute of Biological Sciences) for providing Mlkl–/– mice and Dr. Haibing Zhang (Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for providing Ripk1K45A and Ripk3Δ/Δ mice. We thank Zhiyuan Zhang (National Institute of Biological Sciences) for providing RIPA56. We thank the Beijing Advanced Innovation Center for Structural Biology and the Tsinghua-Peking Center for Life Sciences for facility and financial support. This work was supported by grant numbers 2018YFA0800200, 2017YFA0104000, 91849106, Z200022, Z181100001818005 and 81870118 to Jianwei Wang from the National Key R&D Program of China or the Beijing Municipal Science & Technology Commission and the National Natural Science Foundation of China.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.09.015.
Contributor Information
Hong Jiang, Email: jianghong961106@zju.edu.cn.
Jianwei Wang, Email: jianweiwang@mail.tsinghua.edu.cn.
Author contributions
Conceptualization, Jianwei Wang and Hong Jiang; Methodology, Jianwei Wang, Hong Jiang, Wei He, and Min Liao; Investigation, Min Liao, Ruiqing Chen, Yang Yang, Hanqing He, Liqian Xu, and Yuxuan Jiang; Formal analysis, Jianwei Wang and Hong Jiang; Resources, Zhenxing Guo; Writing, Jianwei Wang and Min Liao; Funding acquisition, Jianwei Wang; Supervision, Jianwei Wang.
Conflicts of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genovese G., Kahler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie M., Lu C., Wang J., McLellan M.D., Johnson K.J., Wendl M.C., et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L., Rau R., Goodell M.A. DNMT3A in haematological malignancies. Nat Rev Cancer. 2015;15:152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Challen G.A., Sun D., Jeong M., Luo M., Jelinek J., Berg J.S., et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayle A., Yang L., Rodriguez B., Zhou T., Chang E., Curry C.V., et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125:629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong M., Park H.J., Celik H., Ostrander E.L., Reyes J.M., Guzman A., et al. Loss of Dnmt3a immortalizes hematopoietic stem cells in vivo. Cell Rep. 2018;23:1–10. doi: 10.1016/j.celrep.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L., Rodriguez B., Mayle A., Park H.J., Lin X., Luo M., et al. DNMT3A loss drives enhancer hypomethylation in FLT3-ITD-associated leukemias. Cancer Cell. 2016;29:922–934. doi: 10.1016/j.ccell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong M., Sun D., Luo M., Huang Y., Challen G.A., Rodriguez B., et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat Genet. 2014;46:17–23. doi: 10.1038/ng.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley T.J., Ding L., Walter M.J., McLellan M.D., Lamprecht T., Larson D.E., et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spencer D.H., Russler-Germain D.A., Ketkar S., Helton N.M., Lamprecht T.L., Fulton R.S., et al. CpG island hypermethylation mediated by DNMT3A is a consequence of AML progression. Cell. 2017;168:801–816. doi: 10.1016/j.cell.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guryanova O.A., Shank K., Spitzer B., Luciani L., Koche R.P., Garrett-Bakelman F.E., et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med. 2016;22:1488–1495. doi: 10.1038/nm.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan X.Q., Chen P., Du Y.X., Zhu K.W., Zhang D.Y., Yan H., et al. Influence of DNMT3A R882 mutations on AML prognosis determined by the allele ratio in Chinese patients. J Transl Med. 2019;17 doi: 10.1186/s12967-019-1959-3. 220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shlush L.I., Zandi S., Mitchell A., Chen W.C., Brandwein J.M., Gupta V., et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russler-Germain D.A., Spencer D.H., Young M.A., Lamprecht T.L., Miller C.A., Fulton R., et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25:442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J., Wang Y.Y., Dai Y.J., Zhang W., Zhang W.N., Xiong S.M., et al. DNMT3A Arg882 mutation drives chronic myelomonocytic leukemia through disturbing gene expression/DNA methylation in hematopoietic cells. Proc Natl Acad Sci U S A. 2014;111:2620–2625. doi: 10.1073/pnas.1400150111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai Y.J., Wang Y.Y., Huang J.Y., Xia L., Shi X.D., Xu J., et al. Conditional knockin of Dnmt3a R878H initiates acute myeloid leukemia with mTOR pathway involvement. Proc Natl Acad Sci U S A. 2017;114:5237–5242. doi: 10.1073/pnas.1703476114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu R., Wang J., Ren Z., Yin J., Wang Y., Cai L., et al. A model system for studying the DNMT3A hotspot mutation (DNMT3AR882) demonstrates a causal relationship between its dominant-negative effect and leukemogenesis. Cancer Res. 2019;79:3583–3594. doi: 10.1158/0008-5472.CAN-18-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li D., Meng L., Xu T., Su Y., Liu X., Zhang Z., et al. RIPK1–RIPK3–MLKL-dependent necrosis promotes the aging of mouse male reproductive system. Elife. 2017;6 doi: 10.7554/eLife.27692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Fan C., Zhang Y., Yu X., Wu X., Zhang X., et al. RIP1 kinase activity-dependent roles in embryonic development of Fadd-deficient mice. Cell Death Differ. 2017;24:1459–1469. doi: 10.1038/cdd.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Q., Yu X., Zhang H., Liu Y., Zhang X., Wu X., et al. RIPK3 mediates necroptosis during embryonic development and postnatal inflammation in Fadd-deficient mice. Cell Rep. 2017;19:798–808. doi: 10.1016/j.celrep.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Loberg M.A., Bell R.K., Goodwin L.O., Eudy E., Miles L.A., SanMiguel J.M., et al. Sequentially inducible mouse models reveal that Npm1 mutation causes malignant transformation of Dnmt3a-mutant clonal hematopoiesis. Leukemia. 2019;33:1635–1649. doi: 10.1038/s41375-018-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ju Z., Jiang H., Jaworski M., Rathinam C., Gompf A., Klein C., et al. Telomere dysfunction induces environmental alterations limiting hematopoietic stem cell function and engraftment. Nat Med. 2007;13:742–747. doi: 10.1038/nm1578. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z., Amro E.M., Becker F., Holzer M., Rasa S.M.M., Njeru S.N., et al. Cohesin-mediated NF-kappaB signaling limits hematopoietic stem cell self-renewal in aging and inflammation. J Exp Med. 2019;216:152–175. doi: 10.1084/jem.20181505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franceschi C., Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69(S1):S4–S9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- 27.Kovtonyuk L.V., Fritsch K., Feng X., Manz M.G., Takizawa H. Inflamm-aging of hematopoiesis, hematopoietic stem cells, and the bone marrow microenvironment. Front Immunol. 2016;7:502. doi: 10.3389/fimmu.2016.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai Z., Kotzin J.J., Ramdas B., Chen S., Nelanuthala S., Palam L.R., et al. Inhibition of inflammatory signaling in Tet2 mutant preleukemic cells mitigates stress-induced abnormalities and clonal hematopoiesis. Cell Stem Cell. 2018;23:833–849. doi: 10.1016/j.stem.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., Karatepe K., Chiewchengchol D., Zhu H., Guo R., Liu P., et al. Bacteria-induced acute inflammation does not reduce the long-term reconstitution capacity of bone marrow hematopoietic stem cells. Front Immunol. 2020;11:626. doi: 10.3389/fimmu.2020.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esplin B.L., Shimazu T., Welner R.S., Garrett K.P., Nie L., Zhang Q., et al. Chronic exposure to a TLR ligand injures hematopoietic stem cells. J Immunol. 2011;186:5367–5375. doi: 10.4049/jimmunol.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matatall K.A., Jeong M., Chen S., Sun D., Chen F., Mo Q., et al. Chronic infection depletes hematopoietic stem cells through stress-induced terminal differentiation. Cell Rep. 2016;17:2584–2595. doi: 10.1016/j.celrep.2016.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao M., Wang J. Mechanisms of hematopoietic stem cell ageing and targets for hematopoietic tumour prevention. Adv Exp Med Biol. 2018;1086:117–140. doi: 10.1007/978-981-13-1117-8_8. [DOI] [PubMed] [Google Scholar]
- 33.He H., Xu P., Zhang X., Liao M., Dong Q., Cong T., et al. Aging-induced IL27Ra signaling impairs hematopoietic stem cells. Blood. 2020;136:183–198. doi: 10.1182/blood.2019003910. [DOI] [PubMed] [Google Scholar]
- 34.Chen C., Liu Y., Liu Y., Zheng P. Mammalian target of rapamycin activation underlies HSC defects in autoimmune disease and inflammation in mice. J Clin Invest. 2010;120:4091–4101. doi: 10.1172/JCI43873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenner D., Blaser H., Mak T.W. Regulation of tumour necrosis factor signalling: live or let die. Nat Rev Immunol. 2015;15:362–374. doi: 10.1038/nri3834. [DOI] [PubMed] [Google Scholar]
- 36.Li L., Thomas R.M., Suzuki H., De Brabander J.K., Wang X., Harran P.G. A small molecule Smac mimic potentiates TRAIL- and TNFalpha-mediated cell death. Science. 2004;305:1471–1474. doi: 10.1126/science.1098231. [DOI] [PubMed] [Google Scholar]
- 37.Li D., Xu T., Cao Y., Wang H., Li L., Chen S., et al. A cytosolic heat shock protein 90 and cochaperone CDC37 complex is required for RIP3 activation during necroptosis. Proc Natl Acad Sci U S A. 2015;112:5017–5022. doi: 10.1073/pnas.1505244112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren Y., Su Y., Sun L., He S., Meng L., Liao D., et al. Discovery of a highly potent, selective, and metabolically stable inhibitor of receptor-interacting protein 1 (RIP1) for the treatment of systemic inflammatory response syndrome. J Med Chem. 2017;60:972–986. doi: 10.1021/acs.jmedchem.6b01196. [DOI] [PubMed] [Google Scholar]
- 39.Han Q., Ma Y., Wang H., Dai Y., Chen C., Liu Y., et al. Resibufogenin suppresses colorectal cancer growth and metastasis through RIP3-mediated necroptosis. J Transl Med. 2018;16:201. doi: 10.1186/s12967-018-1580-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hormaechea-Agulla D., Matatall K.A., Le D.T., Kain B., Long X., Kus P., et al. Chronic infection drives Dnmt3a-loss-of-function clonal hematopoiesis via IFNγ signaling. Cell Stem Cell. 2021;28:1428–1442. doi: 10.1016/j.stem.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fleischman A.G., Aichberger K.J., Luty S.B., Bumm T.G., Petersen C.L., Doratotaj S., et al. TNFalpha facilitates clonal expansion of JAK2V617F positive cells in myeloproliferative neoplasms. Blood. 2011;118:6392–6398. doi: 10.1182/blood-2011-04-348144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guidi N., Sacma M., Ständker L., Soller K., Marka G., Eiwen K., et al. Osteopontin attenuates aging-associated phenotypes of hematopoietic stem cells. EMBO J. 2017;36:840–853. doi: 10.15252/embj.201694969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamashita M., Passegué E. TNF-α coordinates hematopoietic stem cell survival and myeloid regeneration. Cell Stem Cell. 2019;25:357–372. doi: 10.1016/j.stem.2019.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z., Jiang B., Wang Y., Ni H., Zhang J., Xia J., et al. 2-HG inhibits necroptosis by stimulating DNMT1-dependent hypermethylation of the RIP3 promoter. Cell Rep. 2017;19:1846–1857. doi: 10.1016/j.celrep.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 45.Koo G.B., Morgan M.J., Lee D.G., Kim W.J., Yoon J.H., Koo J.S., et al. Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res. 2015;25:707–725. doi: 10.1038/cr.2015.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koya J., Kataoka K., Sato T., Bando M., Kato Y., Tsuruta-Kishino T., et al. DNMT3A R882 mutants interact with polycomb proteins to block haematopoietic stem and leukaemic cell differentiation. Nat Commun. 2016;7:10924. doi: 10.1038/ncomms10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buscarlet M., Provost S., Zada Y.F., Barhdadi A., Bourgoin V., Lepine G., et al. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood. 2017;130:753–762. doi: 10.1182/blood-2017-04-777029. [DOI] [PubMed] [Google Scholar]
- 48.Miles L.A., Bowman R.L., Merlinsky T.R., Csete I.S., Ooi A.T., Durruthy-Durruthy R., et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature. 2020;587:477–482. doi: 10.1038/s41586-020-2864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.