Abstract
Pharmacological activation of the xenobiotic-sensing nuclear receptors pregnane X receptor (PXR) and constitutive androstane receptor (CAR) is well-known to increase drug metabolism and reduce inflammation. Little is known regarding their physiological functions on the gut microbiome. In this study, we discovered bivalent hormetic functions of PXR/CAR modulating the richness of the gut microbiome using genetically engineered mice. The absence of PXR or CAR increased microbial richness, and absence of both receptors synergistically increased microbial richness. PXR and CAR deficiency increased the pro-inflammatory bacteria Helicobacteraceae and Helicobacter. Deficiency in both PXR and CAR increased the relative abundance of Lactobacillus, which has bile salt hydrolase activity, corresponding to decreased primary taurine-conjugated bile acids (BAs) in feces, which may lead to higher internal burden of taurine and unconjugated BAs, both of which are linked to inflammation, oxidative stress, and cytotoxicity. The basal effect of PXR/CAR on the gut microbiome was distinct from pharmacological and toxicological activation of these receptors. Common PXR/CAR-targeted bacteria were identified, the majority of which were suppressed by these receptors. hPXR-TG mice had a distinct microbial profile as compared to wild-type mice. This study is the first to unveil the basal functions of PXR and CAR on the gut microbiome.
Key words: PXR, CAR, Gut microbiome, Bile acids, Inflammation, Mice, Nuclear receptor, Feces
Abbreviations: BA, bile acid; BSH, bile salt hydrolase; CA, cholic acid; CAR, constitutive androstane receptor; CDCA, chenodeoxycholic acid; CITCO, 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime; CV, conventional; CYP, cytochrome P450; DCA, deoxycholic acid; EGF, epidermal growth factor; GF, germ free; GLP-1, glucagon-like peptide-1; GM-CSF, granulocyte-macrophage colony-stimulating factor; HDCA, hyodeoxycholic acid; hPXR-TG, humanized PXR transgenic; IBD, inflammatory bowel disease; IFNγ, interferon-gamma; IL, interleukin; PA, indole-3 propionic acid; IS, internal standards; LCA, lithocholic acid; LC–MS/MS, liquid chromatography–tandem mass spectrometry; MCA, muricholic acid; MCP-1, monocyte chemoattractant protein-1; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; NSAID, non-steroidal anti-inflammatory drug; OH, hydroxylated; OTUs, operational taxonomy units; PBDEs, polybrominated diphenyl ethers; PCBs, polychlorinated biphenyls; PCoA, Principle Coordinate Analysis; PiCRUSt, Phylogenetic Investigation of Communities by Reconstruction of Observed States; PXR, pregnane X receptor; QIIME, Quantitative Insights Into Microbial Ecology; SCFAs, short-chain fatty acids; SNP, single-nucleotide polymorphism; SPF, specific-pathogen-free; T-, taurine conjugated; TCPOBOP, 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene, 3,3′,5,5′-Tetrachloro-1,4-bis(pyridyloxy)benzene; TGR-5, Takeda G-protein-coupled receptor 5; TLR4, toll-like receptor 4; TNF, tumor necrosis factor; UDCA, ursodeoxycholic acid; T, wild type; YAP, yes-associated protein
Graphical abstract
Contrary to the reported anti-inflammatory functions of PXR and CAR following their pharmacological activations, basal PXR and CAR activities are necessary for immune surveillance through modulating the gut microbiota.
1. Introduction
The gut microbiome has a variety of effects on the intermediary metabolism of the host. One important function of the gut microbiome is bile acid (BA) metabolism. In humans, primary BAs are synthesized from cholesterol in the liver, and are then conjugated with taurine (T) or glycine. In mice, T-conjugated BAs are abundant. In the intestine, bacteria can perform deconjugation, dehydroxylation, and epimerization reactions, generating more hydrophobic and thus more toxic BA profiles1. The microbial enzymes bile salt hydrolase and bile acid 7α-dehydroxylase catalyze BA deconjugation and secondary BA synthesis reactions, respectively2. Specific secondary BAs are more potent activators of the host Takeda G-protein-coupled receptor 5 (TGR-5) than primary BAs, and can promote thermogenesis and energy expenditure of the host3. At exceedingly high concentrations, unconjugated secondary BAs are considered more toxic than primary BAs and are implicated in cholestatic liver injury, inflammation, and cancer4. Specifically, unconjugated BAs (both primary and secondary) produce a more prominent increase in pro-inflammatory cytokines from hepatocytes during cholestatic liver injury5. Microbially-derived BAs are also known to contribute to inflammatory bowel disease (IBD)6 and host immune surveillance by directly modulating the balance of T helper 17 (TH17) and regulator T (Treg) cells7. This highlights the importance of the gut microbiome and BAs on immune surveillance.
Gut microbiota also contribute to xenobiotic metabolism. The absence of gut microbiota in mice alters the expression of xenobiotic-processing genes, such as those for cytochrome P450 enzymes (CYPs) and other Phase I oxidases8. The absence of gut microbiota in mice also alters the host metabolism of polybrominated diphenyl ethers (PBDEs), and modulates the PBDE-mediated differential regulation of xenobiotic-processing genes9. Therefore, the gut microbiome is an important regulator of host xenobiotic biotransformation pathways.
The well-known host xenobiotic-sensing nuclear receptors PXR and CAR are highly expressed in the liver and intestine. Upon ligand activation, PXR and CAR up-regulate certain drug-metabolizing enzymes and efflux transporters as a compensatory mechanism against xenobiotic insult10,11. PXR and CAR share many target genes, such as the drug-metabolizing enzyme cytochrome P450 3A4 (CYP3A4) in humans12. PXR and CAR can be activated by environmental chemicals such as specific congeners of the PBDE flame retardants and non-coplanar polychlorinated biphenyls (PCBs)13,14, endogenous ligands such as steroids and BAs15, as well as their prototypical ligands (pregnenolone 16α-carbonitrile [PCN] for mouse PXR and 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene, 3,3′,5,5′-tetrachloro-1,4-bis(pyridyloxy)benzene [TCPOBOP]) for mouse CAR, or rifampicin for human PXR and 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime [CITCO] for human CAR)16,17. A wide variety of bacterial metabolites in the colonic lumen activate PXR and CAR, such as the tryptophan metabolite indole-3-propionic acid (IPA) for PXR and the secondary BAs deoxycholic acid (DCA) and lithocholic acid (LCA) for PXR and CAR target genes18. In livers of germ-free mice (GF), the prototypical PXR-target genes Cyp3a11 and Cyp3a44 mRNAs were decreased compared to conventional (CV) mice with healthy intestinal microbiota19. This relationship has been explored in another study where specific-pathogen-free (SPF) mice had higher cytochrome P450 isozyme expression with accompanying higher PXR and CAR expression than GF mice, due to increased LCA20. Therefore, the presence of the gut microbiome affects the expression of PXR- and CAR-target genes, the receptors themselves, and the activity of PXR and CAR through modulating their activators.
Recent studies unveiled that PXR and CAR have anti-inflammatory functions. For example, pharmacological activation of PXR can play an anti-inflammatory role in the prevention of inflammatory bowel disease (IBD) by inhibiting the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) transcription factor21, as well as NF-κB-targeted pro-inflammatory response genes22. The activation of PXR via the microbial metabolite IPA, which is produced from Clostridium sporogenes, decreases intestinal permeability and maintains gut barrier functions26. PXR activation also inhibits inflammation by inhibiting the toll-like receptor 4 (TLR4) pathway, thus preventing the overproduction of cytokines. Inhibition of the TLR4 pathway results in resistance to Listeria monocytogenes infection compared to PXR-null mice23. PXR inhibits the TLR4 pathway by decreasing the stability of TLR4 mRNA and may also repress TLR4 gene transcription24,25. This TLR4 up-regulation due to PXR deficiency can also cause leaky gut physiology26, which can be resolved by PXR activation via microbial metabolites as previously described27.
Pharmacological and toxicological exposures can affect the composition of the gut microbiome. IPA activates mouse PXR, leading to the down-regulation of the TLR pathway and pro-inflammatory tumor necrosis factor α (TNFα)26. Mice treated with statins gained weight, had increased members of the S24-7 family (now Muribaculaceae28), had up-regulation of PXR-target genes, reduced butyrate, and increased DCA. These effects of statins were found to be PXR-dependent29. In mice, pharmacological activation of PXR and CAR by their prototypical ligands PCN and TCPOBOP affects the composition of the gut microbiome in part by down-regulating certain BA-metabolizing bacteria in the intestine30. Furthermore, mice orally-gavaged with PBDEs, which are PXR and CAR activators, had increased Akkermansia muciniphila and Allobaculum spp., as well as unconjugated secondary BAs13. In another study, mice dosed with PCBs, which can also activate PXR and CAR, had increased A. muciniphila, Clostridium scindens, Enterococcus sp., and Prevotella sp. as well as serum BAs31. In summary, pharmacological and toxicological activation of PXR and CAR can alter the composition of the gut microbiome and the production of distinct microbial metabolites.
While a wealth of literature has demonstrated that the gut microbiome affects PXR and CAR through microbial metabolites, and that pharmacological and toxicological PXR activation affects the composition of the gut microbiome, no studies have been conducted to examine the basal physiological functions of PXR and CAR on the gut microbiome and to compare the potential differences between basal functions of these nuclear receptors and pharmacological/toxicological activation conditions. Considering the fact that genetic polymorphisms of PXR and CAR genes in humans associated with decreased PXR and CAR activity are correlated with human diseases such as liver injury32, and gut microbiome is an important molecular target for xenobiotic metabolism and nutrient homeostasis, it is important to characterize the basal regulation of gut microbiome following PXR and CAR inhibition. The present study focuses on defining the basal regulation of the microbial milieu within the intestines in mice expressing or lacking species-specific receptors, PXR/CAR.
2. Materials and methods
2.1. Chemicals and reagents
E.Z.N.A.® Genomic DNA Isolation Kits were purchased from Omega Bio-Tek (Norcross, GA, USA). The following deuterated internal standards (IS) were used: deuterated (d4)-DCA (CDN Isotopes; CAS No: 112076-61-6), d4-glycocholic acid (GCA) (CDN Isotopes; CAS No.: 1201918-15-1), d4-chenodeoxycholic acid (CDCA) (CDN Isotopes; CAS No.: 99102-69-9), d4-cholic acid (CA) (TRC, Canada; Cat No.: C432603), d4-GCDCA (Iso Sciences, CAS No.: 1201918-16-2), d4 LCA (PubChem CID of LCA: 9903, Steraloids). In total, 19 primary and secondary BAs were quantified with their T-conjugated forms, namely taurine-conjugated cholic acid (T-CA), T-α muricholic acid (T-αMCA), T-β muricholic acid (T-βMCA), T-ω muricholic acid (T-ωMCA), T-chenodeoxycholic acid (T-CDCA), T-ursodeoxycholic acid (T-UDCA), T-hyodeoxycholic acid (T-HDCA), T-deoxycholic acid (T-DCA), T-lithocholic acid (T-LCA), α-muricholic acid (αMCA), β-muricholic acid (βMCA), cholic acid (CA), chenodeoxycholic acid (CDCA), ursodeoxycholic acid (UDCA), ω-muricholic acid (ωMCA), murideoxycholic acid (MDCA), hyodeoxycholic acid (HDCA), deoxycholic acid (DCA), and lithocholic acid (LCA). CA, CDCA, DCA, and LCA were purchased from Sigma–Aldrich (St. Louis, MO, USA); αMCA, βMCA were purchased from Steraloids (Newport, Rhode Island, USA). ωMCA and T-ωMCA was a kind gift from Dr. Daniel Raftery's laboratory at the University of Washington Northwest Metabolomics Research Center. Other BAs were kindly obtained from the University of Kansas Medical Center. Agilent ZORBAX Eclipse Plus C18 columns were purchased from Agilent Technologies (Santa Clara, CA, USA). The samples were eluted using gradient mobile phases of A (10 mmol/L ammonium acetate in 20% acetonitrile) and B (10 mmol/L ammonium acetate in 80% acetonitrile). The column temperature was set at 45 °C and the sample tray temperature was maintained at 9 °C. MS/MS spectra were produced using the negative ionization mode. The UPLC–MS/MS operating parameters are shown in Supporting Information Table S1. All other chemicals and reagents, unless indicated otherwise, were purchased from Sigma–Aldrich.
2.2. Animals
C57BL/6J wild type (WT) breeders were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) and then bred in-house (n = 10, 5 per sex). The knockout mice were all in C57BL/6 background and have been backcrossed at least 10-generations to achieve the homogeneity of the genetic background. Specifically, PXR-null mice were generated and backcrossed into a C57BL/6 background as described previously33, and pups were obtained from in-house breeders (n = 10, 5 per sex). CAR-null mice were generated by Tularik Inc. (South San Francisco, CA, USA) as described previously34, obtained from University of Kansas Medical Center (Kansas City, KS, USA), and pups were obtained from in-house breeders (n = 10, 5 per sex). PXR-CAR-null male and female mice were generated by crossing PXR-null and CAR-null mice (n = 10, 5 per sex). FVB/NJ WT male and female mice were purchased from the Jackson Laboratory (aged 21 days upon arrival, (n = 10, 5 per sex). They were acclimated for at least 9 days within the animal facilities before experiments. Male and female humanized PXR (hPXR) breeders in the FVB/NJ background were a generous gift from Frank Gonzalez (National Cancer Institute, Bethesda, MD, USA) and were bred in-house (n = 10, 5 per sex) and the characterization of this mouse model has been published before35. Specifically, this model was produced by bacterial artificial chromosome (BAC) transgenesis, in which the transgene contains the complete human PXR gene and the 5′- and 3′-flanking sequences. The BAC clone was linearized by restriction enzyme digestion (P1-Sce) and purified before microinjection into fertilized FVB/N mouse eggs. Mice resulting from this breeding step that were positive for the human PXR transgene by PCR analysis were bred further with PXR-null mice. Mice positive for the human PXR transgene and the PXR null allele, as determined by PCR genotyping, were designated as PXR-humanized transgenic (hPXR) mice. Mice heterozygous for the hPXR transgene were interbred to generate a homozygous line. For the present study, the homozygous status of hPXR-TG mice was confirmed by breeding for multiple generations and all litters were tested positive for hPXR-TG by PCR genotyping (data not shown).
All mice were individually housed at weaning age in the animal facility at the University of Washington according to the Association for Assessment and Accreditation of Laboratory Animal Care International guidelines (https://aaalac.org/resources/theguide.cfm). The rationale of single housing was to rule out co-housing effects such as difference in calorie intake and social stress. This is especially important for the male mice because co-housing frequently leads to injurious fighting and the dominating male has a natural advantage to the nutrient resources as compared to the other cage mates. All mice were exposed to laboratory autoclaved rodent diet (LabDiet #5010, LabDiet, St. Louis, MO, USA), non-acidified autoclaved water, and autoclaved Enrich-N’Pure bedding (Andersons, Maumee, OH, USA). All studies were approved by the Institutional Animal Care and Use Committee at the University of Washington. Twenty-four-hour fecal samples were collected from mice at adolescent age (1-month of age) and adult age (2–2.5 months of age).
2.3. DNA isolation
Total DNA was isolated from frozen fecal samples using E.Z.N.A.® Genomic DNA Isolation Kits (Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer's protocol, and the concentration was quantified using a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). The 16S rRNA gene sequencing was performed by Novogene Corporation (Sacramento, CA, USA; 250 bp paired-end, n = 5 per group). Selected bacteria within the Lactobacillus genus and the DNA encoding bile salt hydrolase (Bsh) were confirmed by qPCR.
2.4. BA quantification
Approximately 50 mg of fecal samples were homogenized in 1 mL of high-performance liquid chromatography (HPLC) grade nano pure H2O. 10 μL of internal standard (IS) was added to 200 μL of fecal sample homogenate, mixed, and equilibrated on ice for 5–10 min. 1.5 mL of ice-cold alkaline acetonitrile (5% ammonia in acetonitrile) was added to the homogenate, which was then vortexed vigorously and shaken continuously for 1 h at room temperature. The mixture was then centrifuged at 12,000×g for 15 min at 4 °C, and the supernatant was collected into 5 mL glass tubes. The pellet was re-suspended in 750 μL of HPLC grade methanol, shaken for 5 min, and centrifuged again at 15,000×g for 20 min. The two supernatants obtained were combined, evaporated under vacuum (45 °C) for 4 h, and reconstituted in 100 μL of 50% methanol. The suspension was transferred into a 0.2 μm Costar Spin-X HPLC microcentrifuge filter (purchased from Corning Inc., Corning, NY, USA), and centrifuged at 12,000×g for 10 min. 19 BAs were quantified using liquid chromatography-tandem mass spectrometry (LC–MS/MS), at negative ionization mode. Standard and different quality control samples were extracted using the similar sample preparation procedure described above.
2.5. Cytokine quantification
Approximately 20 mg of fecal samples from C57BL/6 and WT FVB/NJ mice (adolescent and adult ages, males and females, n = 5 per group) were mixed with phosphate buffered saline (PBS; pH = 7.2 supplemented with 1 mmol/L phenylmethylsulfonyl fluoride (PMSF) [final concentration] and 1 × protease inhibitor cocktail [Sigma–Aldrich, catalog No.: P8340]) to a final concentration of 100 mg/mL, and were homogenized by vortex shaker and centrifuged at 10,000×g at 4 °C for 15 min. The supernatant was collected and diluted 1:1 in the PBS solution described above. The cytokines were quantified using the Mouse Cytokine Array Pro-inflammatory Focused 10-plex (MDF10; Eve Technologies Corp., Calgary, Alberta, Canada) per manufacturer's instructions. The 10 cytokines that were determined include: granulocyte-macrophage colony-stimulating factor (GM-CSF), interferon-gamma (IFNγ), interleukin 1 beta (IL-1β), interleukin 2 (IL-2), interleukin 4 (IL-4), interleukin 6 (IL-6), interleukin 10 (IL-10), interleukin 12 (IL-12p70), monocyte chemoattractant protein-1 (MCP-1), and tumor necrosis factor (TNF-α). The ten cytokines were simultaneously quantified in a multiplex panel using a MILLIPLEX Mouse Cytokine/Chemokine 10-Plex Kit (Millipore, St. Charles, MO, USA) according to the manufacturer's protocol, and was performed using the Luminex™ 100 system (Luminex, Austin, TX, USA). The assay sensitivities of these markers range from 0.4 to 10.9 pg/mL for the 10-Plex. Individual analyte values are available in the MILLIPLEX protocol.
2.6. Total RNA isolation
Total RNA was extracted from frozen livers using RNAzol Bee reagent (Tel-Test Inc., Friendswood, TX, USA) per the manufacturer's protocol. RNA concentration was quantified using a NanoDrop Spectrophotometer (Thermo Scientific, Wilmington, DE, USA) at 260 nm. The integrity of each RNA sample was evaluated by formaldehyde agarose gel electrophoresis to visualize the 18S and 28S rRNA bands.
2.7. Reverse transcription (RT)-quantitative polymerase chain reaction (qPCR) quantification of cytokine genes
The total RNAs of mouse livers were reverse-transcribed into cDNAs using the High Capacity cDNA Reverse Transcription Kit (Life Technologies, CA, USA). The resulting cDNA products were amplified by qPCR, using the Sso Advanced Universal SYBR Green Supermix in a Bio-Rad CFX384 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The primers for all qPCR reactions were synthesized by Integrated DNA Technologies (Coralville, IA, USA), and primer sequences are shown in Supporting Information Table S2. Data are expressed as % of the expression of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (Gapdh).
2.8. Comparison among physiological activation of PXR/CAR, pharmacological activation of PXR/CAR, and toxicological activation of PXR/CAR, through public database inquiry
To illustrate the potential differences between the physiological roles of PXR/CAR vs. ligand effect (pharmacological activation of PXR/CAR), or vs. toxicant effect (toxicological activation of PXR/CAR), we compared the data from the present study with the literature.
For comparison with ligand effects, we utilized a previous 16S ribosomal DNA (rDNA) sequencing dataset where adult C57BL/6J male mice were orally gavaged with the prototypical PXR ligand PCN (75 mg/kg) or the prototypical CAR ligand TCPOBOP (3 mg/kg) once daily for 4 consecutive days (vehicle: corn oil, 10 mL/kg)30. The 4-day dosing regimen for PCN and TCPOBOP was designed to compare our findings with the previous literature using the same dosing regimen, which resulted in up-regulation of the prototypical target genes of PXR (Cyp3a11) and TCPOBOP (Cyp2b10), respectively11,16,36. Differentially regulated taxa from the previous study (PCN vs. corn oil, TCPOBOP vs. corn oil)30 were compared with the differentially regulated taxa from the present study in age-matched adult C57BL/6 males (PXR-null vs. WT, and CAR-null vs. WT). The filtering criteria included: average % operational taxonomic units (OTUs) across all groups > 0.001% in each study, and P-value <0.1.
For comparison with toxicant effects, we utilized a previous 16S rDNA sequencing dataset where adult C57BL/6J male mice were orally gavaged with the environmental pollutants PBDEs (BDE-47 or BDE-99, 100 μmol/kg) once daily for 4 consecutive days (vehicle: corn oil, 10 mL/kg)9,13,37. The 4-day dosing regimen of BDE-47 and BDE-99 was chosen to compare our findings with the literature regarding the modulation of host P450s in liver and intestine as well as gut microbiome9,13,37,38. To note, these PBDE congers are known to be PXR and CAR activators9,38,39. Differentially regulated taxa from the previous study (BDE-47 vs. corn oil, and BDE-99 vs. corn oil)9,13,37 were compared with the differentially regulated taxa from the present study in age-matched adult C57BL/6 males (PXR-null vs. WT, and CAR-null vs. WT). The filtering criteria included: average % OTUs across all groups >0.001% in each study, and P-value <0.1.
For comparison with a separate study which measured toxicant effects, we utilized a previous 16S rDNA sequencing dataset where adult C57BL/6J female mice were orally administered the environmental pollutants Fox River PCB mixture (6 mg/kg) once daily for 3 consecutive days (vehicle: corn oil, 10 mL/kg)31,40. The 3-day dosing regimen of Fox River PCB mixture was chosen to compare our findings with the previous studies regarding the regulation of host P450s in liver as well as gut microbiome31,40. The Fox River PCB mixture includes 4 Aroclors, namely Aroclor 1242 (35% of total weight), Aroclor 1248 (35%), Aroclor 1254 (15%), and Aroclor 1260 (15%). As we have previously characterized, this PCB mixture can up-regulate the prototypical aryl hydrocarbon receptor (AhR) target genes (Cyp1a1 and 1a2), CAR-target genes Cy2b10 and 2c50, and PXR target genes Cyp3a16 and 3a41a31,40. Differentially regulated taxa from the previous study (PCBs vs. corn oil)31,40 were compared with the differentially regulated taxa from the present study in age-matched adult C57BL/6 females (PXR-null vs. WT, and CAR-null vs. WT). The filtering criteria included: average % OTUs across all groups >0.001% in each study, and P-value <0.1.
2.9. Data analysis
Analysis of FASTQ files was conducted using various python scripts in Quantitative Insights Into Microbial Ecology (QIIME)41, including de-multiplexing, quality filtering, OTU picking, as well as alpha- and beta-diversity determinations. Metagenome functional content was predicted using Phylogenetic Investigation of Communities by Reconstruction of Observed States (PICRUSt)42. Line plots representing the alpha diversity of each group were generated using ggplot2 (v 3.0.0) in R. Three-dimensional Principle Coordinate Analysis (PCoA) plots (beta diversity) were generated using the weighted UniFrac diversity metric in Emperor (Gigascience). OTUs were visualized using stacked bar plots generated in SigmaPlot (Systat Software, Inc.). Hierarchical clustering dendrograms (Ward's minimum variance method, distance scale) of the top significantly abundant taxa (abundance >0.005%, P < 0.05) were generated using gplots (v 3.0.1) and RColorBrewer (v 1.1-2) in R. Bar plots representing cytokine concentration, BA concentration, and SCFA concentration were generated using ggplot2 (v 3.0.0) in R. Correlation matrices representing SCFA and taxa associations were generated using reshape2 (v 1.4.3) and ggplot2 (v 3.0.0) in R.
Asterisks (∗) represent significant differences between WT and nuclear receptor-gene-null mice (P < 0.05), as well as WT and hPXR-TG mice. Statistically significant differences among WT, PXR-null, CAR-null, and PXR-CAR-null mice (all in C57BL/6 background) were determined using one-way ANOVA followed by Duncan's post hoc test in R using the DescTools package (v 0.99.26) or SPSS (IBM SPSS Statistics).
2.10. Experimental design
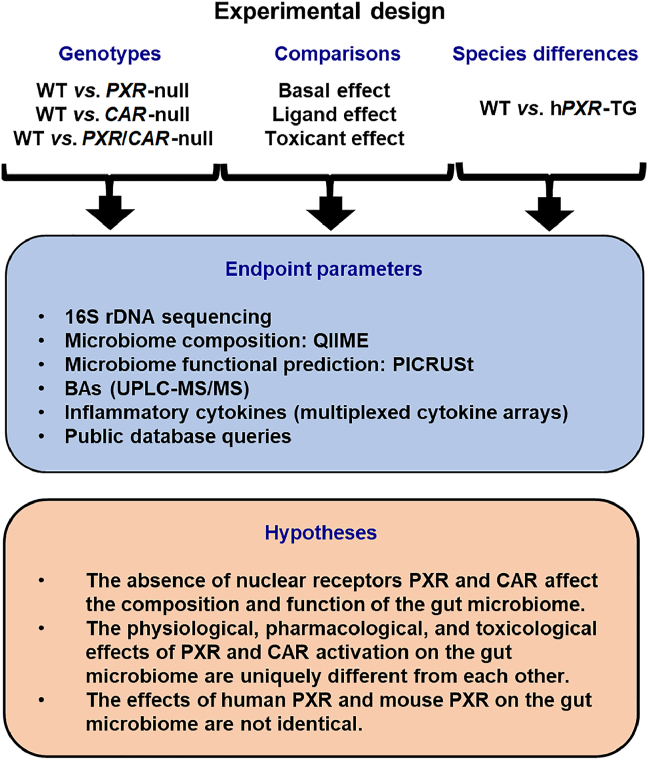
The overall experimental design is shown in Fig. 1. Three experimental settings were designed to model the effect of host PXR/CAR genetics on the gut microbiome. In Study 1, we focused on the necessity of the host PXR and CAR on the basal regulation of gut microbiome, using PXR-null, CAR-null, and PXR-CAR-double null mice (all in C57BL/6 background). In Study 2, we performed a systematic comparison between the effects of basal functions of PXR/CAR and the ligand-mediated pharmacological activation of PXR and CAR on microbiome, as well as between the effects of basal functions of PXR/CAR and the toxicological activation of PXR/CAR on microbiome. In Study 3, we compared the gut microbiomes between the mice that carry mPXR vs. mice that carry hPXR (both in FVB background), to determine how species-differences of this host drug–receptor modulate the gut microbiome. Due to the markedly reduced pregnancy rate and litter size of the hCAR-TG colony and the lack of availability of hCAR-TG in other genetic backgrounds, the comparison of hCAR vs. mCar was not part of the scope of the present study.
Figure 1.
Schematic of the experimental design of this study. [Study 1] To determine the necessity of the host PXR and CAR genes on modulating the compositions and functions of the gut microbiome under physiological conditions, fecal samples were collected from WT, PXR-null, CAR-null, and PXR-CAR-null (all in C57BL/6 background) male and female mice of adolescent and adult ages (n = 5 of each group). [Study 2] To compare the physiological roles of PXR and CAR with ligand-mediated activation and toxicant-mediated activation of these nuclear receptors, data from the present study were compared with publicly inquired databases of the same genetic background (C57BL/6), sexes, and age. For ligand effect, 16S rDNA sequencing data were retrieved from mice that were orally exposed to the prototypical PXR ligand PCN or the prototypical CAR ligand TCPOBOP30. For toxicant effect, 16S rDNA sequencing data were retrieved from mice that were orally exposed to PBDEs9,13,37 or PCBs31,40 [Study 3] To compare the role of mouse and human PXR genes on the composition and function of the gut microbiome, fecal samples were collected from WT and hPXR-TG (both in FVB background) male and female mice of adolescent and adult ages (n = 5 of each group). Due to poor breeding capacity of hCAR-TG mice in C57BL/6 background and the lack of access of hCAR-TG mice in the FVB background, the comparison of mouse and human CAR genes on gut microbiome was not determined in this study. Fecal samples were collected after 24 h, and 16S rDNA gene sequencing was conducted by amplifying the hypervariable V4 region. Analysis of FASTQ files was conducted using various python scripts in Quantitative Insights Into Microbial Ecology (QIIME)41, including de-multiplexing, quality filtering, operational taxonomy unit (OTU) picking, as well as alpha- and beta-diversity determinations. Metagenome functional content was predicted using Phylogenetic Investigation of Communities by Reconstruction of Observed States (PICRUSt)42. BAs were quantified using liquid chromatography–tandem mass spectrometry (LC–MS/MS). Cytokines were quantified using the Mouse Cytokine Array Pro-inflammatory Focused 10-Plex (MDF10; Eve Technologies Corp., Calgary, Alberta, Canada). For Study 1, we hypothesize that the absence of PXR and CAR affect the composition and function of the gut microbiome. For Study 2, we hypothesize that the physiological, pharmacological, and toxicological effects of PXR and CAR activation on gut microbiome are uniquely different from each other. For Study 3, we hypothesize that the effects of human PXR and mouse PXR on the gut microbiome are not identical.
3. Results
3.1. Understanding the necessity of host nuclear receptors PXR and CAR on the constitutive regulation of the gut microbiome
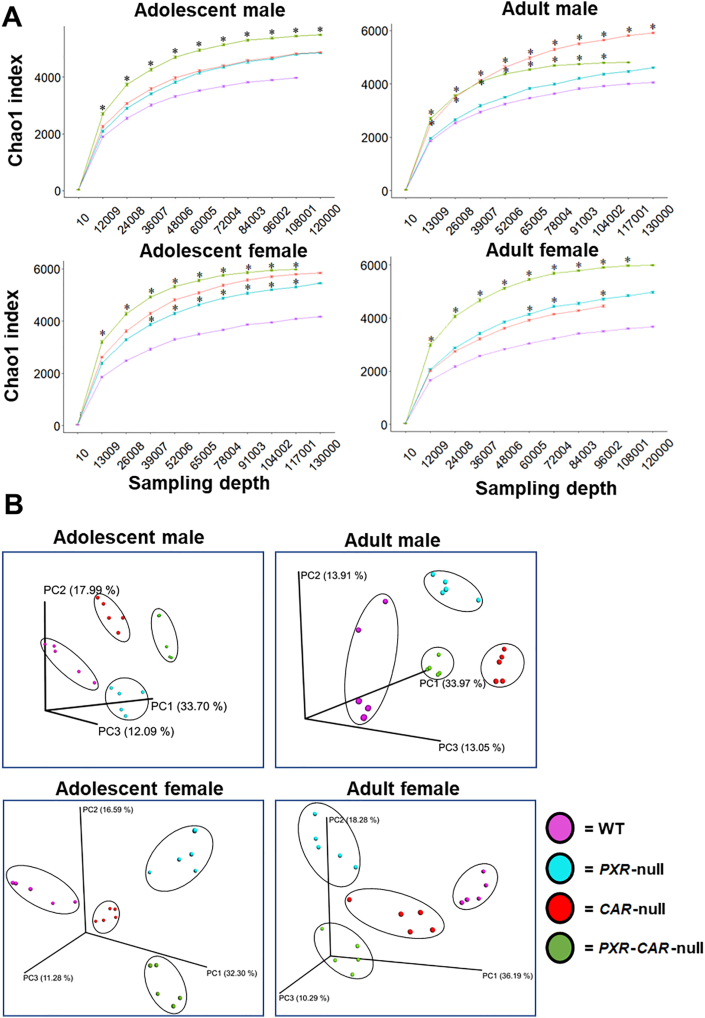
16S rRNA gene sequencing was conducted on feces collected over a 24-h period of adolescent and adult aged WT, PXR-null, CAR-null, and PXR-CAR-double null male and female mice (all in the C57BL/6 background, n = 5 per group), to determine the necessity of host nuclear receptors on the composition and function of the gut microbiome. As shown in Fig. 2A, the alpha diversity of PXR and CAR single or double knockout mice tended to have greater richness than WT controls in all groups, and this trend was especially prominent for PXR-CAR double null mice. The only exception was in adult males; CAR-null mice had the highest richness as compared to the other genotypes. Increased diversity of the gut microbiome due to absence of PXR or CAR has also been shown elsewhere43. As shown in Fig. 2B, regarding the beta diversity, all four genotypes of mice (WT, PXR-null, CAR-null, and PXR-CAR-double null) exhibited distinct separations among their microbial communities at both ages and sexes. The microbial separations indicate that PXR and CAR are essential and unique modulators of the gut microbiome.
Figure 2.
Alpha and beta diversities of WT, PXR-null, CAR-null, and PXR-CAR-null mice. (A) Mean (SE) alpha diversity of gut microbiota within WT, PXR-null, CAR-null, and PXR-CAR-null mice. Line plots are generated using the R package ggplot. Asterisks (∗) represent statistically significant differences compared to WT mice (one-way ANOVA followed by Duncan's post hoc test, P < 0.05). (B) Principal components analysis (PCA) plots showing the beta diversities of adolescent male, adult male, adolescent female, and adult female WT, PXR-null, CAR-null, and PXR-CAR-null mice.
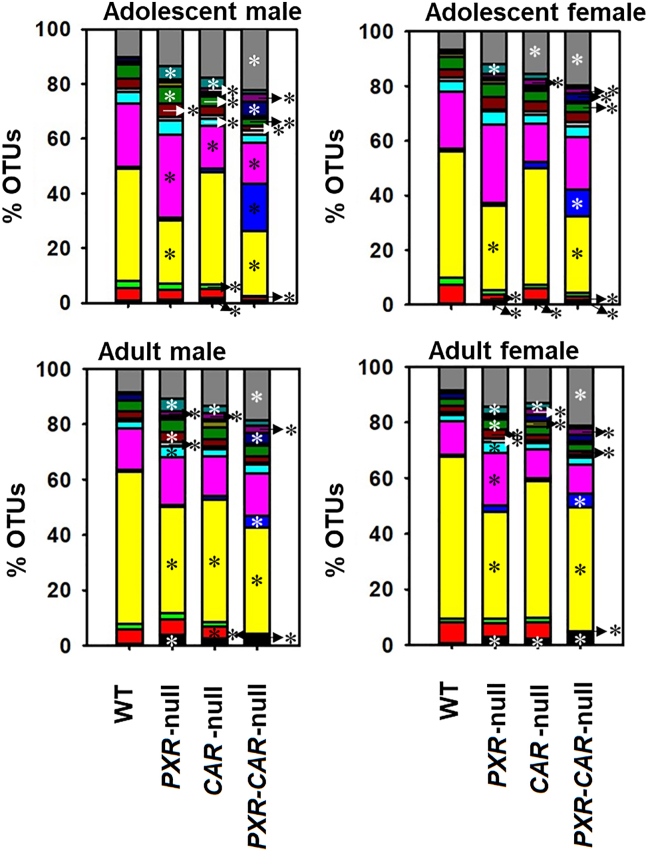
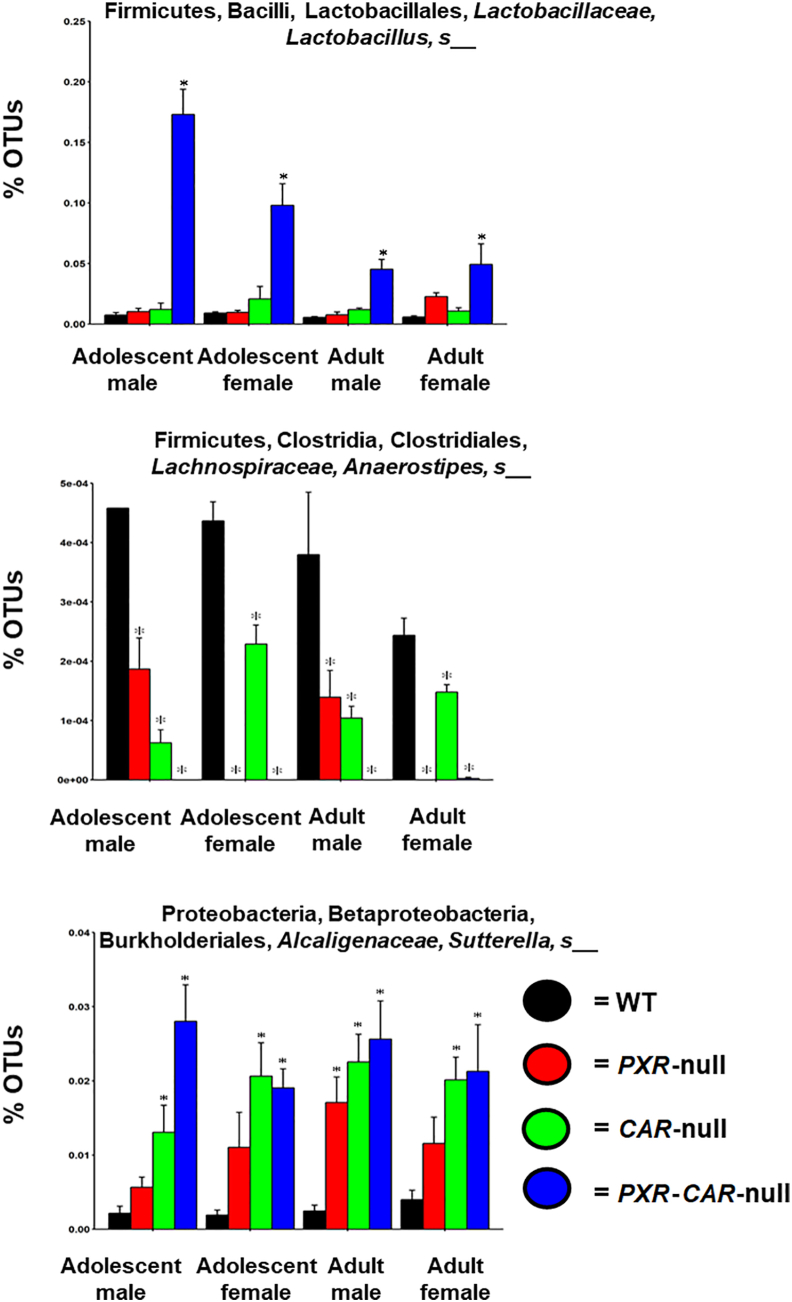
Individual variations contributing to these differences are displayed in the heatmap in Supporting Information Fig. S1. In total, 63 taxa were significantly different between WT and PXR-null, CAR-null, and PXR-CAR-null mice (Fig. S1). Fig. 3 shows the microbial compositional changes at L7 (species level) of the top 15 most abundant bacteria as quantified by the percentage of OTUs for the four genotypes of mice (note: the 15th category (Other) includes all other taxa summed together; figure legend located in Supporting Information Fig. S13). To note, for the taxa that are not differentiable at the species level, the most specific level is reported (e.g., the genus or family level). In all exposure groups, the predominant phyla were Bacteroidetes, Firmicutes, and Proteobacteria. Bacteria in the family Helicobacteraceae, which are positively correlated with IBD44, increased in relative abundance in PXR-null samples in all groups, as well as both male CAR-null age groups, and CAR-null adult female samples. Most notably, as shown in Fig. 4, Lactobacillus, which is known to carry bile salt hydrolase activity for BA-deconjugation to generate unconjugated BAs, which are more toxic than the conjugated BAs at high concentrations45,46, was markedly increased in PXR/CAR-double null mice, and this bacteria also tended to be higher in single receptor gene null mice. To note, Lactobacillus is known to be anti-inflammatory for probiotic use47,48; however, it is shown to be pro-inflammatory endogenously49,50. The pro-inflammatory Sutterella51, which is enriched in IBD, was increased in PXR- and CAR-null mice and further increased in the double knockout mice. To note, Sutterella is also known to be bile-resistant52. The pro-inflammatory helicobacter (a less abundant taxon) was also higher in the absence of PXR and CAR (Fig. S1). Corresponding to the increase in the relative abundance of these potentially harmful taxa, the relative abundance of the anti-inflammatory A. muciniphila was lower in the absence of PXR and CAR (Fig. S1); the relative abundance of a taxon in the S24-7 (Muribaculaceae) family was lower in all PXR-null and PXR-CAR-double null groups compared to WT feces, as well as in adult CAR-null males; Muribaculaceae have been shown to be lower during colitis53. Anaerostipes sp. decreased in relative abundance in all nuclear receptor-deficient mice of both ages and sexes. This microbe is anti-inflammatory through its production of butyrate54. In contrast, Allobaculum sp., which is a microbial biomarker that is inversely associated with obesity55, was higher in all PXR-CAR-null groups, except for adult females. This suggests that the lack of PXR and CAR together may also have certain beneficial effects against obesity.
Figure 3.
Percentage OTUs (L7) of the fecal microbiome among WT, PXR-null, CAR-null, and PXR-CAR-null mice. Stacked bar charts illustrate the mean percentages of differentially regulated taxa in fecal samples in WT, PXR-null, CAR-null, and PXR-CAR-null male and female, adolescent and adult mice. The top 14 differentially abundant taxa in each group were plotted and all other detected taxa were summed together to form the category labeled as “Other”. Asterisks (∗) represent statistically significant differences compared to WT mice (one-way ANOVA followed by Duncan's post hoc test, P < 0.05).
Figure 4.
Percentage OTUs of Lactobacillus sp., Anaerostipes sp., and Sutterella sp. Individual bar plots of mean (SE) percentage OTUs of Lactobacillus sp., Anaerostipes sp., and Sutterella sp., are generated by the R package ggplot2. Asterisks (∗) represent statistically significant differences compared to WT mice (one-way ANOVA followed by Duncan's post hoc test, P < 0.05).
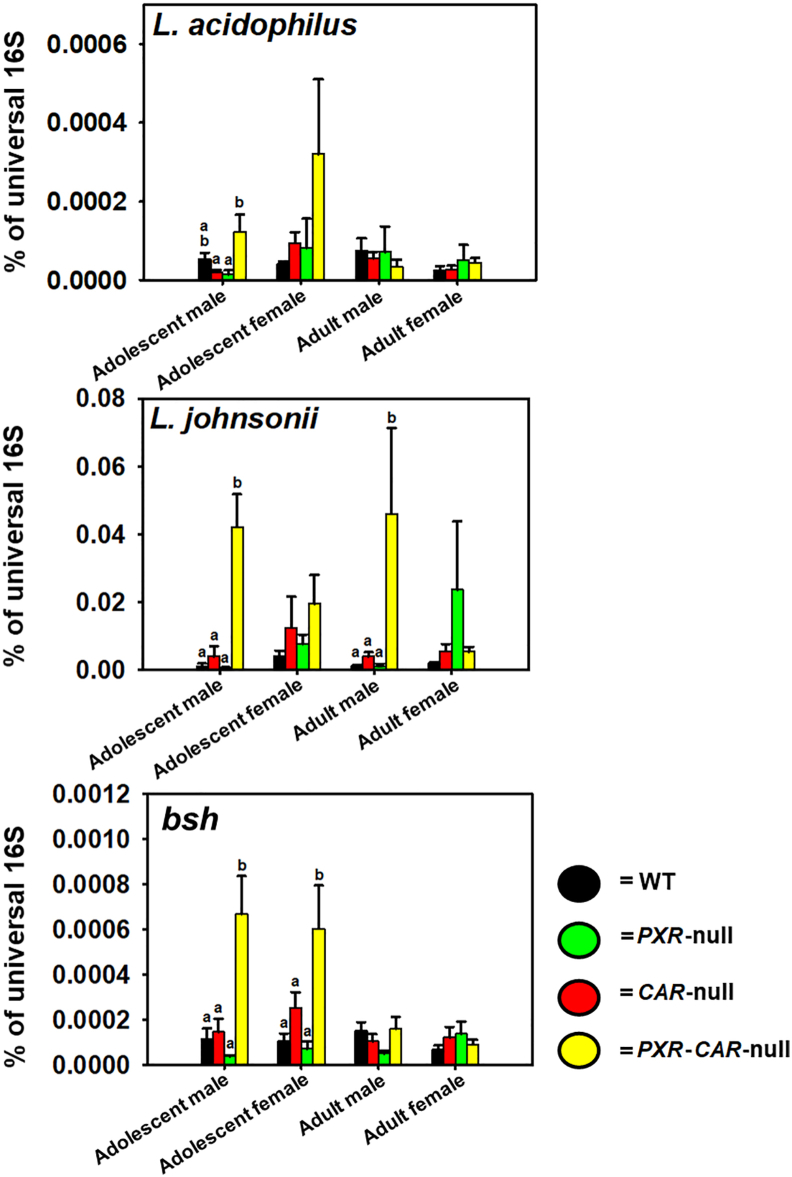
As shown in Fig. 530,56, we used qPCR to quantify levels of Lactobacillus; in addition, we also quantified the bile salt hydrolase DNA abundance in these samples. Among all the 7 Lactobacillus species that we examined, L. acidophilus, L. johnsonii, L. bulgaricus, and L. paracasei were present in the fecal microbiome, among which L. acidophilus and L. johnsonii are known to have bile salt hydrolase activities46,57. L. bulgaricus and L. paracasei are not known to have BSH activities and their abundance was very low (data not shown). Compared to the other genotypes, fecal L. acidophilus tended to be higher in PXR-CAR-null mice in adolescent age (both sexes), whereas L. johnsonii was higher in male PXR-CAR-null mice at adolescent and adult age. L. acidophilus tended to be higher in CAR-null than WT in adolescent females. L. johnsonii tended to be higher in CAR-null and PXR-CAR-null than WT in adolescent females, as well as higher in PXR-null than WT in adult females, although statistical significance was not achieved. The DNA encoding bsh was higher in feces of PXR-CAR-null mice than the other genotypes in adolescent age.
Figure 5.
qPCR analysis of the BSH-expressing L. acidophilus and L. johnsonii as well as the DNA encoding BSH. The primer sequences were described as we reported before30,56 and in Table S2. All primers were synthesized by Integrated DNA Technologies. The abundance of the genomic DNA encoding the bacterial 16S rRNAs in the intestinal content of mice was determined by quantitative polymerase chain reaction (qPCR) using a CFX384 Real-Time PCR Detection System. Results are expressed as the mean (SE) delta–delta cycle value (calculated as 2ˆ[−(Cq − average reference Cq)]) of the quantitative PCR as compared with the universal bacteria, per nanogram of DNA from the intestinal content. (a) and (b) represent statistically significant differences compared to post hoc groups. Treatment groups that are not statistically different are labeled with the same letter.
The functional predictions of the gut microbiome in the absence of PXR and CAR (figure not shown) were done using PICRUSt. Females exhibited markedly more functional content predictions (188 in adolescents, and 170 in adults) than their male counterparts at both developmental ages sampled (8 in adolescents, and two in adults). In both female ages, absence of PXR caused an increase in the greatest number of pathways, with the opposite being true for CAR-null samples. The PXR-CAR-null group appeared to respond to combinatorial effects of the absence of PXR or CAR, as no change in the abundance of these pathways was seen for these females. The opposite is true for the samples from male mice in that PXR-CAR-null in both ages, and CAR-null in adult males had a marked increase in pathways compared to PXR-null and WT samples. Therefore, sex and host nuclear receptor status affect the function of the gut microbiome in mice.
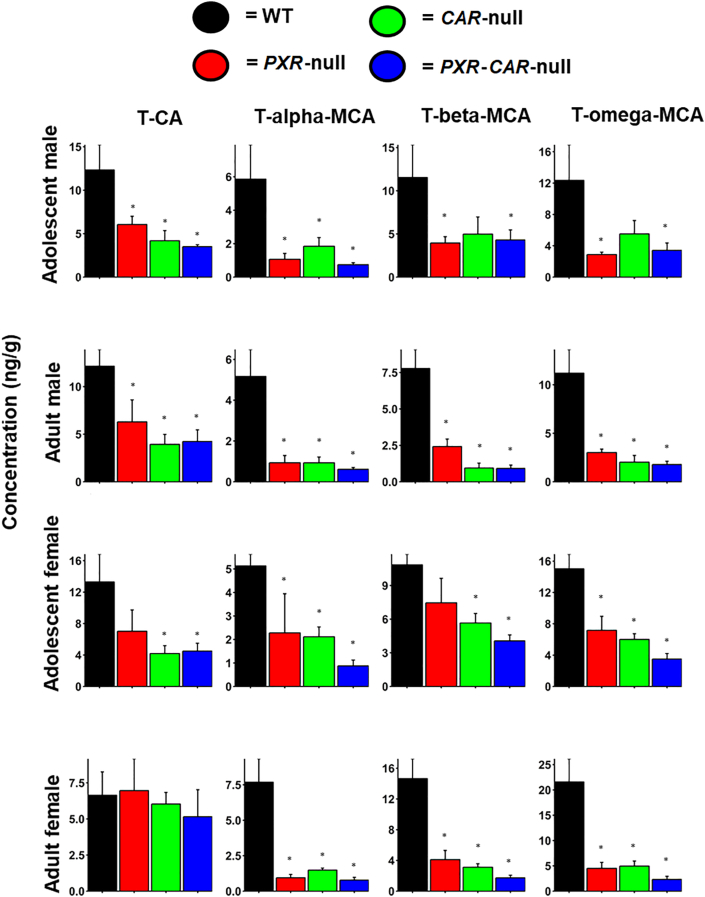
Because there was a marked increase in the BA-deconjugating Lactobacillus in the PXR/CAR-double null mice (and to a lesser extent tended to be higher in the single null mice), we hypothesized that this would result in a reduction in conjugated BAs in feces of the PXR/CAR-null mice. To test this hypothesis, we conducted LC–MS-based targeted metabolomics of all major BAs in mice (Fig. 6 and Supporting Information Figs. S2–S5). As expected, as shown in Fig. 6, the most abundant conjugated BAs in mice, namely T-CA, T-αMCA, T-βMCA, and T-ωMCA, tended to be lower in feces of the PXR/CAR single and double null mice, and the trend was most predominant in the PXR/CAR double null mice. Thus the qPCR data on the bsh-expressing bacteria and the bsh DNA (Fig. 5) at least partly explained the reduction in taurine-conjugated bile acids in observed in the feces of the receptor null mice. Other bacteria that express Bsh from different DNA sequences, which were not probed in the present study due to lack of a complete reference sequences of the microbes at this time in history, may also contribute to the increased deconjugation reactions. Other minor T-conjugated secondary BAs, such as T-HDCA T-UDCA, were also decreased in the knockout mice in an age and sex-dependent manner (Figs. S2–S5). We also observed a slight increase in the major unconjugated secondary BA LCA in adolescent PXR/CAR double null mice of both sexes (Figs. S2 and S4). However, several other unconjugated secondary BAs were lower in adult PXR/CAR null mice. It is speculated that enhanced BA de-conjugation may prime the host for inflammation related diseases5.
Figure 6.
BA concentrations in WT, PXR-null, CAR-null, and PXR-CAR-null mice. Mean (SE) fecal BA concentrations (ng/g) were quantified by LC–MS/MS as described in Materials and methods. Asterisks (∗) represent statistically significant differences compared to WT mice (one-way ANOVA followed by Duncan's post hoc test, P < 0.05).
3.2. Comparison between the effects of physiological and pharmacological activation of PXR and CAR on the gut microbiome
To compare the effects of physiological and pharmacological activation of PXR and CAR on the gut microbiome, we examined the following two datasets using age-, sex- and genetic background matched mice (adult C57BL/6J males) with the goal to seek novel bacterial targets of PXR and CAR: 1) 16S rDNA sequencing data of nuclear receptor gene null mice vs. WT mice (present study); and 2) 16S rDNA sequencing data of prototypical PXR ligand (PCN) and CAR ligand (TCPOBOP) treated WT mice30. A microbe that is consistently regulated under both basal conditions and under receptor-activated conditions is considered to be a PXR/CAR target with high confidence. As shown in Supporting Information Fig. S6A, there were 2 taxa that were differentially regulated in both conditions, among which Dorea sp. was suppressed by the basal presence of PXR, and was further decreased by PCN, suggesting that this bacterium is a PXR target and is suppressed by this nuclear receptor. Conversely, the constitutive abundance of Blautia sp. depends on the basal presence of PXR, whereas PCN suppressed its abundance, suggesting that PXR has context-specific duality between basal and pharmacological activation conditions.
As shown in Fig. S6B, there were 3 taxa that were differentially regulated in both conditions, among which Coprobacillus sp. and a member of the Peptococcaceae family were suppressed by the basal presence of CAR, and were further decreased by TCPOBOP, suggesting that these bacteria are CAR-targets and are suppressed by this nuclear receptor. Additionally, there was an unassigned taxon that was increased by both CAR deficiency and CAR ligand, suggesting that CAR also has context-specific duality between basal and pharmacological activation conditions.
3.3. Comparison between the effects of physiological and toxicological activation of PXR and CAR on the gut microbiome
To compare the roles of basal PXR/CAR vs. toxicological activation of PXR/CAR by known activators on the gut microbiome, we examined the following two datasets using age-, sex- and genetic background matched mice (adult C57BL/6J males): 1) 16S rDNA sequencing data of nuclear receptor gene null mice vs. WT mice (present study); and 2) 16S rDNA sequencing data of PBDEs-exposed WT mice9,13,37. To note, the two diet-enriched PBDE congers, namely BDE-47 and BDE-99, are known activators of PXR and CAR as evidenced by the up-regulation of prototypical PXR and CAR target genes9,38,39. As shown in Supporting Information Fig. S7A, there were 3 taxa that were commonly regulated by the basal presence of PXR and BDE-47, and 4 taxa that were commonly regulated by the basal presence of PXR and BDE-99. Interestingly, Dorea sp. which was a PXR-suppressed target under both basal and pharmacological activation conditions (Fig. S6A), was also consistently suppressed by both PBDE congeners, indicating its down-regulation is mediated by PXR and not by off-target effects of the toxicants. In addition, a member of the Mogibacteriaceae family was consistently suppressed by basal presence of PXR and both PBDE congeners, whereas a member of the Lachnospiraceae family was consistently suppressed by basal presence of PXR and BDE-47, whereas Clostridium sp. and Adlercreutzia sp. were consistently suppressed by basal presence of PXR and BDE-99. These observations suggest that these bacteria are also common targets of PXR under both basal and toxicological activation conditions.
As shown in Fig. S7B, there were 4 taxa that were commonly regulated by the basal presence of CAR and BDE-47, and 2 taxa that were commonly regulated by the basal presence of CAR and BDE-99. Interestingly, Lactobacillus sp. was suppressed by CAR under both basal conditions and following exposure to both PBDE congeners, indicating that this bacterium is a CAR target. In addition, Prevotella sp. and a member of the Clostridiaceae family were commonly suppressed by the basal presence of CAR and BDE-47, and Adlercreutzia sp. was commonly suppressed by the basal presence of CAR and BDE-99, suggesting that the down-regulation of these taxa may also act through CAR.
In a separate comparison to examine the basal effect vs. the effect of toxicological activation of PXR and CAR on the gut microbiome, we examined the following two datasets using age-, sex- and genetic background matched mice (adult C57BL/6J females): 1) 16S rDNA sequencing data of nuclear receptor gene null mice vs. WT mice (present study); and 2) 16S rDNA sequencing data of Fox River PCB mixture exposed WT mice. Notably, at the given dose (6 mg/kg), this PCB mixture activates PXR and CAR as evidenced by up-regulation of their prototypical target genes40. As shown in Supporting Information Fig. S8A, between PXR-null vs. WT and PCB vs. vehicle (corn oil) conditions, 9 taxa were commonly regulated, among which a member of the Ruminococcaceae family was consistently suppressed by PXR under basal and PCB-exposed conditions, whereas Streptococcus sp. and Anaeroplasma sp. were consistently induced by PXR under basal and PCB-exposed conditions, indicating that these taxa are PXR targets (Fig. S8A). Between CAR-null vs. WT and PCB vs. vehicle (corn oil conditions), 7 taxa were commonly regulated, among which a member of the Ruminococcaceae family and an unassigned taxon were consistently suppressed by CAR under both basal and PCB-exposed conditions, suggesting that this bacterium is a CAR target (Fig. S8B). There were several taxa that were regulated in opposite directions between the basal presence of PXR/CAR and PCB-exposed conditions, suggesting either context-specific duality of the nuclear receptors or off-target effect of PCBs.
In summary, we have performed a systematic comparison among PXR and CAR targeted bacteria under basal, pharmacological, and toxicological conditions in mice, and identified bacteria that are commonly regulated under basal and at least one of the two receptor-activated conditions, suggesting that these bacteria are novel targets of these host receptors. The bacteria commonly regulated across PXR-null vs. WT and CAR-null vs. WT comparisons are shown in Supporting Information Table S3. Although we do not know at this point whether the changes in these bacteria were due directly to the immediate effects of PXR/CAR absence/activation, a clear PXR/CAR dependency in their regulation was observed.
3.4. Comparison between mouse and human PXR in regulating the gut microbiome
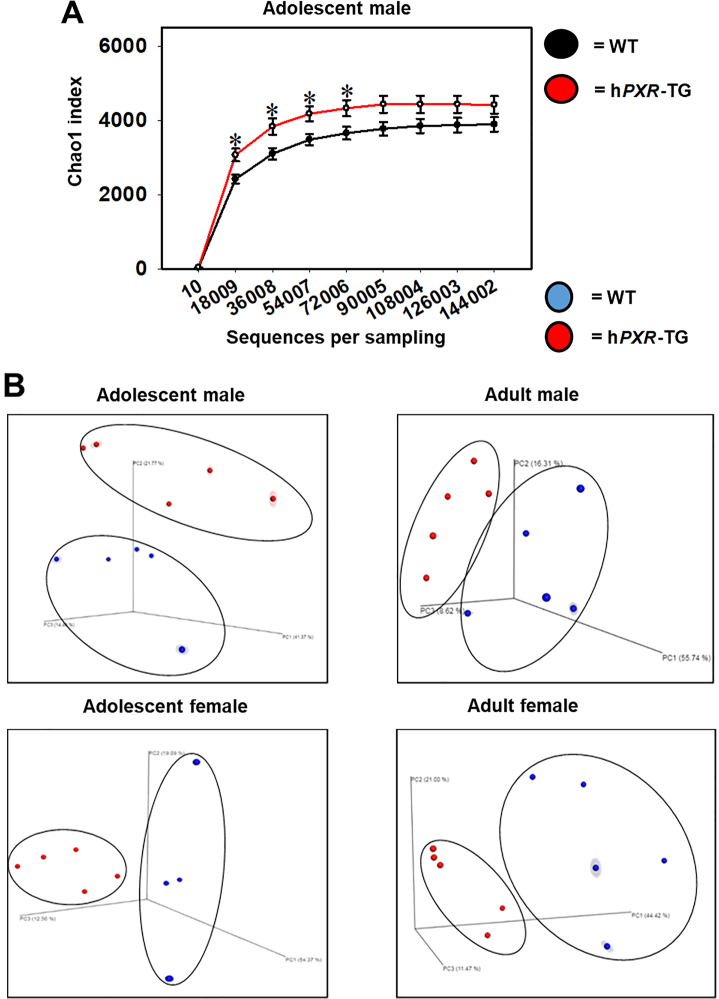
To compare the role of mouse and human PXR genes on the composition and function of the gut microbiome, 16S rRNA gene sequencing was conducted on feces collected over a 24-h period of adolescent and adult aged wild type (WT) and hPXR-TG male and female mice (n = 5 per group). Both of these two mouse strains were in the FVB genetic background. The overall microbial richness was similar between WT and hPXR-TG mice, except for adolescent males, as evidenced by a higher microbial richness in hPXR-TG mice (Fig. 7A). As shown in Fig. 7B, in all 4 comparisons, hPXR-TG and WT mice exhibited distinct separations between their microbial communities as measured by beta diversity (weighted uniFrac).
Figure 7.
Alpha and beta diversities of WT and hPXR-TG mice. (A) Mean (SE) alpha diversity of gut microbiota within WT and hPXR-TG FVB/NJ adolescent male mice. Line plots were generated using SigmaPlot. Asterisks (∗) represent statistically significant differences compared to WT mice (t-test, P < 0.05). (B) Principal components analysis (PCA) plots showing the beta diversities of adolescent male, adult male, adolescent female, and adult female WT and hPXR-TG mice.
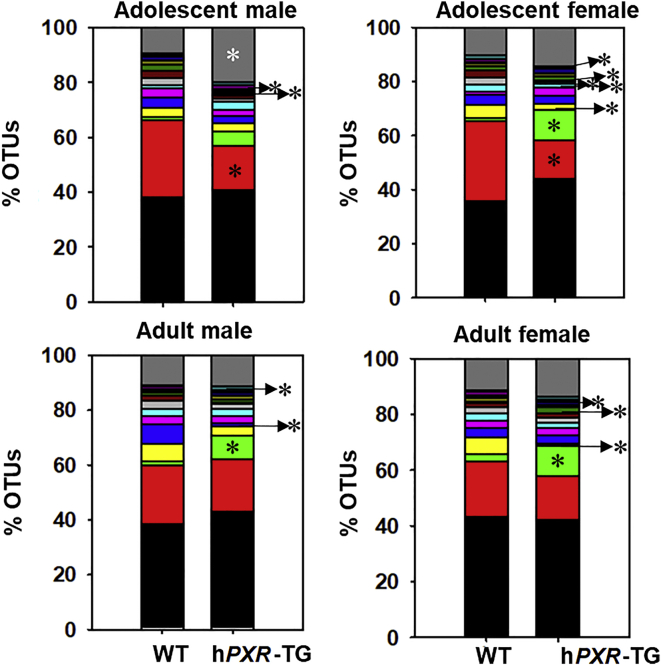
In total, 27 taxa with abundance above 0.00005 were significantly different between WT and hPXR-TG mice (Supporting Information Fig. S9). Fig. 8 shows the top 15 species level compositional changes of the gut microbiome as quantified by % OTUs (figure legend located in Supporting Information Fig. S14). In all groups, the predominant phyla were Bacteroidetes, Firmicutes, Tenericutes, and Verrucomicrobia, plus Proteobacteria in both adult groups and adolescent males. The most predominant difference was a relative increase in Prevotella sp. in hPXR-TG mice.
Figure 8.
Percentage OTUs of WT and hPXR-TG mice. Stacked bar charts illustrate the mean percentages of differentially regulated taxa in fecal samples in WT and hPXR-TG male and female, adolescent and adult mice. The top 14 differentially abundant taxa in each group were plotted and all other detected taxa were summed together to form the Other category. Asterisks (∗) represent statistically significant differences compared to WT mice (t-test, P < 0.05).
As shown in Supporting Information Fig. S10, PICRUSt was used to predict the effect of host nuclear receptor genotype on the metagenomic functional content of the gut microbiome in mice. Adult male hPXR-TG mice were the only group with altered functional pathways. These 21 pathways are: lysine degradation, styrene degradation, aminobenzoate degradation, sulfur metabolism, limonene and pinene degradation, atrazine degradation, biosynthesis of siderophore group nonribosomal peptides, biosynthesis of unsaturated fatty acids, arachidonic acid metabolism, retinol metabolism, chlorocyclohexane and chlorobenzene degradation, bacterial invasion of epithelial cells, proximal tubule bicarbonate reclamation, metabolism of cofactors and vitamins, ion channels, flavonoid biosynthesis, cytochrome P450-mediated xenobiotic metabolism, caprolactam degradation, tryptophan metabolism, and cell motility and secretion. Notably, as a group, all of these pathways decreased in the adult male hPXR-TG mice, with some individual variation. Therefore, species specificity of PXR (i.e., mPxr vs. hPXR) affects the predicted functional differences of the gut microbiome, in a sex- and age-specific manner.
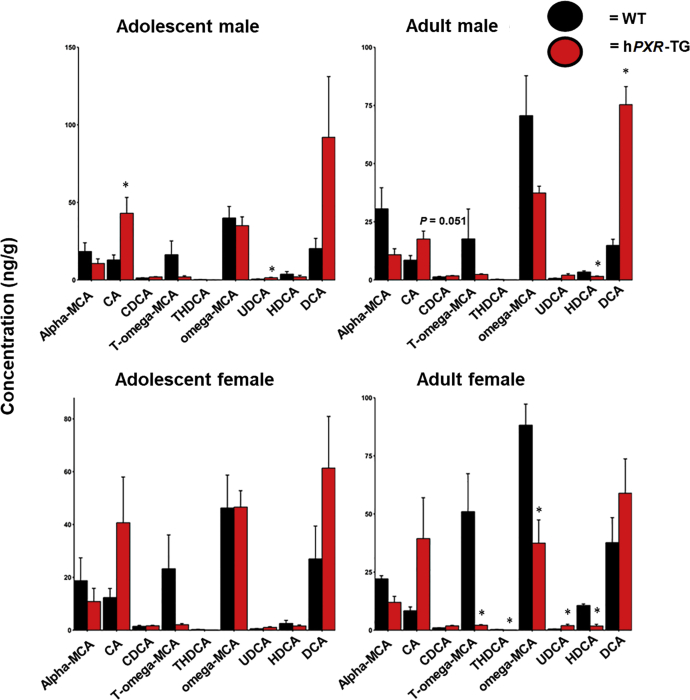
As shown in Fig. 9, BA profiles were different between WT and hPXR-TG mice. Specifically, the largest increase in relative concentration occurred with the secondary BA DCA in hPXR-TG adult male mice, and this trend was seen in the other groups as well. The primary BA CA, which is the precursor of DCA, also tended to be higher in hPXR-TG mice at all 4 comparisons. Conversely, the major secondary BA T-ωMCA tended to be lower in hPXR-TG mice in all 4 comparisons, and its unconjugated form ωMCA also tended to be lower in female hPXR-TG mice. Regarding other minor BAs in feces, HDCA was lower in both hPXR-TG adult mice groups, with this trend observed in the adolescent mice, and UDCA was higher in adolescent male and adult female hPXR-TG mice, with this trend continuing in the other groups as well.
Figure 9.
BA concentrations in WT and hPXR-TG FVB/NJ mice. Bar plots of mean (SE) BA concentrations (ng/g) in WT and hPXR-TG mice as generated by the R package ggplot2. BAs were quantified by LC–MS/MS as described in Materials and methods. Asterisks (∗) represent statistically significant differences compared to WT mice (t-test, P < 0.05).
4. Discussion
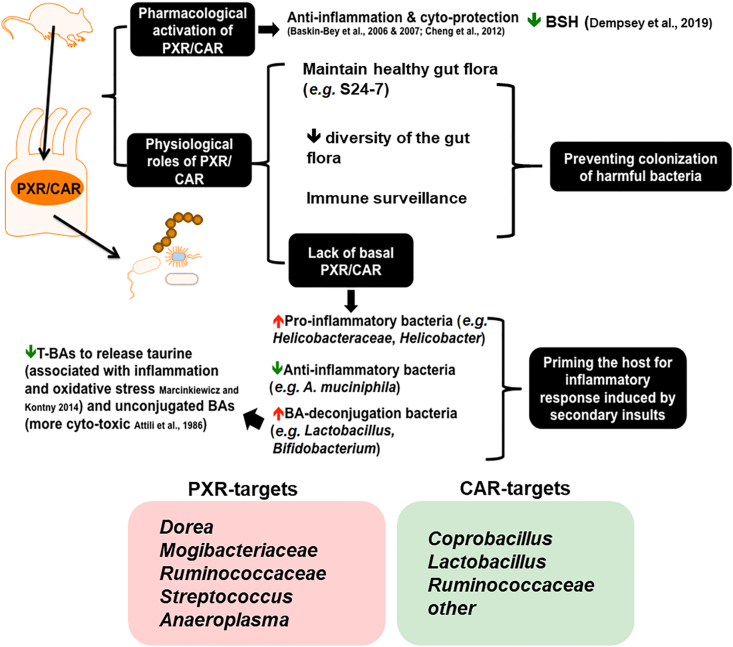
As summarized in Fig. 10, through three independent experimental settings, the present study has determined that host genetics and differences in the xenobiotic-sensing nuclear receptors PXR and CAR profoundly influence the composition and predicted functions of the gut microbiome, and there is an apparent association between increased bsh-expressing bacteria and reduced conjugated BAs. Specifically, although at the descriptive level at this point, our study suggests that the presence of the drug receptors PXR and CAR prevents the bloom of other types of bacteria that contribute to the richness of the gut microbiome, likely serving a protective mechanism against opportunistic bacteria that may be harmful to the host. The absence of PXR and CAR also led to an increase in the Lactobacillus genus, corresponding to reduced taurine-conjugated BAs. Interestingly, through a comprehensive comparison between the present study and published datasets, we showed that the physiological, pharmacological, and toxicological activation of PXR produces mostly distinct effects on gut microbiome. We identified several intestinal bacteria that are PXR/CAR targets commonly targeted under both basal and receptor-activated conditions, including Dorea sp., a member of the Mogibacteriaceae family, a member of the Ruminococcaceae family, Streptococcus sp., and Anaeroplasma sp. for PXR, as well as Coprobacillus sp., Lactobacillus sp., and a member of the Ruminococcaceae family for CAR. Context-specific duality of PXR and CAR in regulating gut microbiome has also been observed. Lastly, species differences in PXR (mPXR vs. hPXR) also profoundly altered the gut microbiome, including higher Prevotella sp. as well as lower A. muciniphila, both of which are hallmarks of inflammatory bowel disease58,59, thus caution is warranted when using WT mouse models to study PXR and inflammation, as mice (with mPXR) may be more resistant to GI inflammation than humans (with hPXR).
Figure 10.
A schematic illustrating the key findings of the study. There is a bivalent hormetic relationship between PXR/CAR levels and microbial richness, and PXR/CAR interacts with the gut microbiome to modulate immune surveillance and BA metabolism of the host. Physiological, pharmacological, and toxicological activation of PXR and CAR produce distinct effects on the gut microbiome; most taxa are not commonly regulated; however, common receptor-targets have been identified, and context-specific duality of PXR and CAR is noted.
Regarding the effect of PXR and CAR on the basal regulation of gut microbiome, observations on microbial compositional and metabolite changes in nuclear receptor gene null mice have provided new insights into the necessity of these host xenobiotic-sensing nuclear receptors on the regulation of gut microbiome under physiological conditions. Especially, the markedly higher proportion of Lactobacillus genus in the PXR-CAR-double null mice consistently corresponds to a marked decrease in major T-BAs in feces. Many species and strains in the Lactobacillus genus carry bsh that deconjugates the taurine and glycine molecules from BAs45,46,56,57,60, 61, 62, 63, 64; in mice, the predominant effect is expected to be T-BA deconjugation, because taurine conjugation is a predominant pathway in mice over glycine conjugation. We have previously demonstrated that pharmacological activation of PXR and CAR leads to decreased gene abundance of the BSH in intestinal content30. Findings from the present study further support the inhibitory roles of PXR and CAR in the microbial BSH activities.
Lactobacillus tended to be higher in PXR- and/or CAR-single null mice, although statistical significance was not achieved, whereas the major T-BAs were significantly lower in single receptor gene null mice. This indicates that other bacteria that remain to be characterized may also contribute to T-BA deconjugation in PXR- and CAR-null mice. Although most abundant T-BAs were lower in the PXR-null, CAR-null, and PXR-CAR-double null mice, the unconjugated BAs did not increase in feces, except for the adolescent females, where there was an apparent increase in unconjugated αMCA, βMCA in all three null mouse genotypes, as well as an increase in CA in PXR-null mice, and these unconjugated BAs are all major primary BAs synthesized from the liver. Conversely, certain unconjugated secondary BAs were lower in the receptor gene null mice, and this pattern was especially prominent in the adult age for all the three null genotypes. This indicates that the absence of the host PXR and/or CAR receptor may negatively impact microbial dehydroxylase activities, either via lowering the bacteria that carry the dehydroxylase genes, inhibiting the enzyme activities, or altering the intestinal environment through mechanisms that remain to be characterized.
In the present study, we observed a profound decrease in fecal T-βMCA, which is a known potent antagonist of the major BA-receptor farnesoid X receptor (FXR)65, in PXR-null, CAR-null, and PXR/CAR double null mice. The decrease in T-βMCA, likely mediated by deconjugation mediated by gut microbiota especially in the PXR/CAR double null mice, is expected to relieve FXR inhibition and favors increased FXR activity66. Enhanced FXR signaling due to lack of antagonist may inhibit hepatic BA synthesis to further reinforce the lower BA output during PXR/CAR deficiency, and may contribute to the increased alpha diversity of the gut microbiome due to lack of the bacteriostatic effect of BAs. Conversely, activation of BA synthesis has been noted in FXR-null mice, due to the induction of PXR and CAR67. Enhanced FXR signaling may also promote fatty acid and carbohydrate metabolism68,69 in the absence of PXR and CAR. T-UDCA was down-regulated in feces of PXR-null and CAR-null mice, and it is known to be a TGR-5 agonist with anti-inflammatory effects70. Therefore, the decrease in T-UDCA may further augment GI inflammation together with the pro-inflammatory microbial signatures observed from the 16S rDNA sequencing experiment.
As shown in Supporting Information Fig. S16, we have quantified the mRNA expression in liver using RT-qPCR on the major bile acid synthesis enzymes, namely Cyp7a1, Cyp8b1, Cyp7b1, and Cyp27a1. Cyp27a1 and Cyp8b1 mRNAs were not altered by PXR and CAR deficiency (data not shown). Cyp7b1 mRNA was also not altered by the absence of PXR or CAR, although sex difference was noted, which was a male pre-dominant gene expression as reported before71. Cyp7a1 mRNA was down-regulated by the absence of PXR and CAR in males. The down-regulation of host Cyp7a1 mRNA in males may also contribute to decreased fecal output of conjugated BAs, but should not be the major player, because the decrease in these fecal BAs was observed in all groups, and that the other BA-synthetic enzymes were only minimally influenced by the absence of PXR and CAR.
This study also examined the species differences of PXR on the gut microbiome, by comparing humanized-PXR-transgenic (hPXR-TG) mice and WT mice with murine PXR, both within the FVB/NJ background. Even though murine PXR and human PXR share roughly 82% DNA and 77% protein identity, differences in gut microbiome composition were observed between the two mouse genotypes (NCBI HomoloGene).
While higher microbial richness (alpha diversity, Chao1 index) was only observed in adolescent male hPXR-TG mice compared to WT mice, fecal microbial communities were strikingly separated (beta diversity) between the PXR species types in both ages and sexes (Fig. 7B). While Paraprevotellaceae Prevotella sp. was responsible for the largest increase in relative abundance in hPXR-TG mice compared to WT, multiple other taxa increased as well, including AF12 sp. Interestingly, a member of the Christensenellaceae family and Desulfovibrio sp. increased in relative abundance only in adolescent hPXR-TG mice of both sexes, and Parabacteroides sp., a member of the Bacteroidales order, and a member of the YS2 order increased only in adult hPXR-TG mice of both sexes, possibly showing an age-mediated species effect of PXR on gut microbiota. Taxa decreased in hPXR-TG mice as well, most notably including a member of the RF39 order (class Mollicutes) and A. muciniphila. An age effect presented as well in that a member of the Clostridiales order and Anaeroplasma sp. decreased solely in adolescent hPXR-TG mice.
A member of the AF12 genus also increased in relative abundance in all groups of hPXR-TG mice. AF12 sp. is an under-characterized microbe, with some literature finding a correlation between higher abundance and lower body weight, although some evidence is conflicting72, 73, 74, 75. Additionally, AF12 sp. has been observed to increase in relative abundance after dietary supplementation of the herbal remedies daikenchuto and the Lingzhi mushroom (Ganoderma lucidum)76,77. In particular, in Meneses et al.77, the increase in the relative abundance of AF12 after Lingzhi mushroom consumption by C57BL/6 mice was followed by lowered cholesterol and greater excretion of fecal bile acids. This study observed increased levels of cholic acid (CA) and deoxycholic acid (DCA) in hPXR-TG mice, which may be related to the increase in AF12 sp.
Lastly, A. muciniphila decreased in relative abundance in hPXR-TG mice. A. muciniphila is a notable bacterial species with mucin-degrading abilities, which has been linked to anti-inflammatory function in liver injury and in diseases such as inflammatory bowel disease (IBD), potentially through its ability to improve an injured gut barrier78, 79, 80, 81. Therefore, this decrease in A. muciniphila may lead to susceptibility for hPXR-TG mice to IBD. Interestingly, hPXR-TG mice are used to study IBD, such as in Dou et al.117, where hPXR-TG mice were treated with dextran sulfate sodium (DSS) to induce colitis, and then treated with isorhamnetin, a PXR agonist in order to study its effects on ameliorating IBD. The propensity for hPXR-TG mice to already be predisposed to colitis through decreased A. muciniphila before DSS treatment throws into question the mechanisms by which PXR agonist IBD treatments work—do they treat the underlying A. muciniphila under-abundance or the damage caused by DSS; or both? In another study, Cheng et al.118 treated hPXR-TG mice with rifaximin, a potent PXR agonist used to treat IBD, and found that chronic exposure to rifaximin causes hepatic steatosis, compared to treated WT and treated PXR-null mice. As noted above, hPXR-TG mice are low in A. muciniphila, potentially increasing their susceptibility to inflammation and liver injury, and so the results in that study may be due to the presence of hPXR rather than rifaximin.
DCA is a bacterially derived secondary bile acid, which has been correlated with colon cancer82. DCA increased in hPXR-TG mice, possibly indicating that human PXR increases susceptibility to colon cancer compared to its mouse equivalent. However, it was recently shown that the basal levels of DCA reduces intestinal inflammation in a mouse model of colitis83, therefore, the net outcome of the rise in DCA in hPXR-TG mice requires further investigation. Interestingly, CA was also increased in hPXR-TG mice, possibly indicating that DCA was formed via bacterial deconjugation mechanisms. This proposed mechanism is further confirmed by the observation that the conjugated form of CA, taurocholic acid (T-CA), was unchanged, indicating CA and DCA were not increased through de novo synthesis in the liver.
Just as humans have inter-individual and inter-population genetic differences, strains of mice also differ genetically. Multiple studies have explored the effects of a treatment on the gut microbiota of C57BL/6 and FVB/NJ mice, but to date relatively less is known regarding the basal regulation of the gut microbiome in these mice. The present study creates a model for how host genetic differences affect the composition and function of the gut microbiome using fecal samples as proxy from the two most widely used laboratory strains of mice.
Higher microbial richness (alpha diversity, Chao1 index) was observed in FVB/NJ mice compared to C57BL/6 mice in both ages and sexes (Supporting Information Fig. S11A), and these strains of mice exhibited distinctly separated communities of microbiota in all groups as well (Supporting Information Fig. S11B). Some of the largest difference in relative abundance of bacteria was due to the lower abundance of Lactococcus in FVB/NJ mice in all groups, accompanied by an increase in other taxa such as Acinetobacter sp., and Eubacterium dolichum (Fig. S12A). Certain taxa in the Lactococcus genus, such as Lactococcus lactis NCD0 2118 and L. lactis NK34, are known to be anti-inflammatory during colitis84,85. The colonization of one species in the Acinetobacter genus, namely Acinetobacter baumannii, is known to be facilitated by secretory IgA, the principal humoral immune defense of mucosal surfaces86. Coincidentally, E. dolichum is a high secretory immunoglobulin A (IgA)-coated pro-inflammatory bacterium, and is detectable in feces of patients with ulcerative colitis; in addition, germ-free mice colonized with this bacterium have increased susceptibility to chemical-initiated colitis87. Together these microbial signatures indicate that FVB/NJ mice have higher basal immune-reactivity in the intestine than C57BL/6 mice. This is supported by higher pro-inflammatory cytokines in the feces of FVB/NJ mice (Fig. S12B).
The majority of the species changed between the two strains have been associated with inflammation. For example, Prevotellaceae Prevotella sp., which decreased in relative abundance in FVB/NJ mice, promotes mucosal inflammation by stimulating the Th17 immune response via epithelial cell production of IL-8, IL-6, and CCL2088. Prevotellaceae Prevotella sp. has been further implicated in IBD through its ability to exacerbate DSS-induced colitis via activation of the inflammasome89. The genus Enterococcus, which also decreased in these mice, has been implicated in infection and increasing susceptibility to IBD90. Carnobacterium sp., which is found in both extreme temperature and animal environments, potentially contains mucin-degrading abilities91,92. The genus Lactococcus is well known for its anti-inflammatory capabilities, has been shown to ameliorate colitis, and also protects the liver from inflammation84. Several taxa were also higher in FVB/NJ mice. Anaeroplasma sp., which potentially has anti-inflammatory function by inducing the anti-inflammatory cytokine TGF-β, has also been associated with existing in lower relative abundance in hypercholesterolemia subjects leading to an unfavorable lipid profile93. The order RF32 has been correlated with damaged histopathology and colonic inflammation in mice with colitis94. Acinetobacter sp. has also been correlated with inflammation through the induced signaling of TLR2 and TLR4, which activate the immune response95. E. dolichum has been shown to produce propionate, which potentially contributes to the inflammatory disease relapsing polychondritis (RP) by continuously stimulating intestinal regulatory T (Treg) cells to produce IL-10, leading to hyporesponsiveness of the Treg cells to mitogen stimulation96. In addition to this, E. dolichum has been associated with Western diets, and frailty97, 98, 99, 100. Therefore, the strain differences including differing basal expression levels of PXR and CAR, affect the abundance of inflammation-related bacteria.
Due to the inflammation-related bacterial changes, we decided to measure cytokine levels in these mice. Overall, cytokines increased in relative concentration and abundance in FVB/NJ mice. Interestingly, the largest increase was exhibited by MCP-1, which serves to regulate the migration and infiltration of monocytes, memory T lymphocytes, and natural killer (NK) cells to sites of inflammation, both within mice and humans101. IL-10, IL-12, and IL-6 increased in relative concentration in FVB/NJ mice as well. The increase in the anti-inflammatory cytokine IL-10 is potentially due to the concomitant increase in IL-12, because IL-10 has been shown to be induced during inflammation to suppress IL-12102, indicating a compensatory response. Notably, the cytokines which increased in FVB/NJ mice are affiliated with a type-1 inflammation response. While IL-4 is typically implicated in type-2 inflammation, such as during the pathogenesis of asthma103, IL-4 also increases during type-1 inflammation104. The elevation of the type-1 inflammation response accompanied by the aforementioned bacteria potentially indicates a response to inflammation within the FVB/NJ mice. Whether this type-1 response is caused by LPS from the bacteria, or some other mechanism requires further testing. Since this is an animal study, all variation in experimental parameters was restrained to the strains themselves, potentially indicating a genomic difference-mediated effect. FVB/NJ mice contain the G protein-coupled receptor 84 (GPR84) deletion, which leads to accumulation of triglycerides and kidney fibrosis105. In fact, GPR84 mRNA expression is elevated during inflammation, and activation of this receptor led to lowered levels of IL-12, IL-6, and MCP-1 in a study by Gagnon et al.105, indicating a protective response by this receptor. This effect would explain the phenomenon observed in this study.
It is important to consider the confounding factors such as batches of mice, caging, and chow for studies on microbiome. Therefore, in the present study, all of the mice were in the same genetic background for the same comparative studies (C57BL/6 for the knockout studies and the inducer studies, and FVB for the humanized PXR study). In addition, the majority of the mice have been backcrossed for at least 10 generations before use. The WT FVB/NJ mice were purchased from the Jackson Laboratory and acclimated for 30 days before use in this study. While the transport of mice may have an effect on the gut microbiota, acclimation to the housing facility after shipment has been shown to complete with stabilization of the gut microbiota after 4 weeks106,107. Still, an effect from shipment of the WT FVB/NJ mice should be considered as a potential confounding factor. To prevent other confounding factors, the breeding colonies of the other genotypes have been maintained in the same animal facility for at least 3 years before their offspring were used for the present study. All of the mice used in this study were singly housed to rule out the co-housing effect such as difference in calorie intake and social stress. This is especially important for the male mice because co-housing frequently leads to injurious fighting and the dominating male has a natural advantage to the nutrient resources as compared to the other cage mates. This procedure has also been discussed in a previous review article108, which concluded that responses to single- and group-housing are highly context-dependent. Since nutrition is a critical confounding factor for the gut microbiome in the present study, singly housing the mice is highly important.
It is known in humans that host genetics may play an important role in modulating the constitutive levels of gut microbiome leading to altered metabolic phenotypes109. It is also known that genetic background and vendor source may lead to variations in the composition of the fecal microbiota of inbred mice110. In the present study, two genetic backgrounds of mice were used, namely C57BL/6, from which the PXR-null, CAR-null, and PXR-CAR-double-null mice were generated, as well as FVB/NJ, from which the hPXR-TG mice were generated. There are basal differences between C57BL/6 WT and FVB/NJ WT, including lower alpha diversity in C57BL/6 WT (Fig. S11A), and a distinct separation of the gut microbiome configuration between the two mouse strains at both developmental ages sampled (Fig. S12B). Specifically, Lactococcus was exclusively present in the fecal microbiome of C57BL/6 WT, whereas Acinetobacter and E. dolichum are exclusively present in the fecal microbiome of FVB/NJ WT mice (Fig. S12A). In addition, FVB/NJ mice generally had higher fecal cytokine levels than age- and sex-matched C57BL/6 WT mice (Fig. S11B). The reason that we are not able to generate hPXR-TG mice in the same genetic background as the knockout mice is that hPXR-TG mice do not breed well in the C57BL/6 background, and that our two main focuses of the study regarding 1) determining the necessity of PXR/CAR on gut microbiome and 2) comparing the effect of the human PXR vs. mouse PXR on gut microbiome are independent from each other. Therefore, two WT controls have been used in the present study, namely the C57BL/6 WT for the knockout mice and the FVB/NJ WT for the hPXR-TG mice.
To probe for host inflammation marks in the absence of PXR and/or CAR, we used liver as a proximal organ and examined the basal expression of several inflammatory cytokines in adult WT, PXR-null, and CAR-null mice. Interestingly, corresponding to a pro-inflammatory microbial signature in the intestine, there was an increase (or a tendency to increase) in IL-6, which is known to be produced during inflammation to play a key role in acute phase response towards chronic inflammation111, in IL-2, which is a potent inducer of T cell signaling112, as well as in IL-12p35, which contributes to T helper (Th) cell polarization and Th1 T-cell differentiation113 (Fig. S15). Therefore, the microbial signature not only impacts the GI tract locally, but may impact other important organs such as the liver.
A limitation in the present study is not being able to compare the effect of hPXR activation and mPXR activation on the gut microbiome under pharmacological or toxicological conditions. There are no published datasets on pharmacological activation of human PXR in hPXR-TG mice with a focus on the gut microbiome for us to compare with our dataset on pharmacological activation of mouse PXR (by PCN)13. While the primary focus of the present manuscript is the basal regulation of the gut microbiome by PXR and CAR under physiological conditions, we will address the issue in our future directions.
5. Conclusions
Well-known as xenobiotic-sensing receptors, pharmacologically activated PXR and CAR have been shown to have anti-inflammatory and cytoprotective effects114, 115, 116, as well as lead to a reduction in the BA-deconjugation enzyme bsh and lower hepatic secondary BAs30. The present study has been the first to establish the physiological functions of the host PXR and CAR on the compositions of the gut microbiome as well as the microbial functions related to basal immune surveillance and BA metabolism. Specifically, we showed that lack of PXR and CAR individually and synergistically increased the richness of the gut microbiome, also accompanied by a pro-inflammatory microbial signature. This suggests that the presence of PXR/CAR is necessary in preventing the disproportional blooming of certain commensal bacteria, which can become pathogenic if they escape their original niche. Lack of both PXR and CAR also reduced BA-deconjugating bacteria and levels of major taurine-conjugated BAs in feces, suggesting increased hydrophobicity of the internal load of unconjugated BAs and the co-substrate taurine, which may further prime the host for inflammatory response induced by secondary insults. The basal roles of PXR/CAR in gut microbiome regulation are distinct from the pharmacological and toxicological activations of PXR/CAR, highlighting the context-specific functions of these host nuclear receptors. PXR/CAR targeted intestinal bacteria have also been identified, and most of them were suppressed by these host receptors. Finally, mPXR and hPXR have different effects on the gut microbiome. Taken together, the bivalent hormetic functions of PXR and CAR under physiological conditions highlight the novel roles of these host drug receptors in modulating the host immune surveillance and BA metabolism through targeting the gut microbiome.
Acknowledgments
The authors would like to thank the GNAC technical support for the germ-free mouse experiment, Mr. Brian High and John Yokum from UW DEOHS IT team for providing server access, and the members of Dr. Cui laboratory for their help in tissue collection and revising the manuscript. This study is supported by National Institutes of Health (NIH, USA) grant ES025708, ES030197, GM111381, ES031098, the University of Washington Center for Exposures, Diseases, Genomics, and Environment, USA [P30 ES0007033], and the Murphy Endowment, USA; The Peer Reviewed Medical Research Program – Investigator Initiated Research Award under Award No. W81XWH-17-1-0479; NIH grants (CA 222469, USA).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.07.022.
Contributor Information
Haiwei Gu, Email: haiweigu@asu.edu.
Sridhar Mani, Email: sridhar.mani@einsteunmed.org.
Julia Yue Cui, Email: juliacui@uw.edu.
Author contributions
Participated in research design: Mallory Little and Julia Yue Cui; Conducted experiments: Mallory Little, Moumita Dutta, Hao Li, Adam Matson, Xiaojian Shi, Haiwei Gu, Sridhar Mani, Julia Yue Cui, Gabby Mascarinas, Bruk Molla, and Kris Weigel; Contributed new reagents or analytic tools: Haiwei Gu, Sridhar Mani, and Julia Yue Cui; Performed data analysis: Mallory Little, Moumita Dutta, Hao Li, Adam Matson, and Xiaojian Shi; Wrote or contribute to the writing of the manuscript: Mallory Little, Moumita Dutta, Hao Li, Adam Matson, Xiaojian Shi, Haiwei Gu, Sridhar Mani, and Julia Yue Cui.
Conflicts of interest
The authors declare that they have no competing interests.
Availability of data and material
The datasets generated during the current study are available in the Dryad database with the Accession Number https://doi.org/10.5061/dryad.5mkkwh72v.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Klaassen C.D., Cui J.Y. Review: mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids. Drug Metab Dispos. 2015;43:1505–1521. doi: 10.1124/dmd.115.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ridlon J.M., Kang D.J., Hylemon P.B., Bajaj J.S. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe M., Houten S.M., Mataki C., Christoffolete M.A., Kim B.W., Sato H., et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 4.Li T., Apte U. Bile acid metabolism and signaling in cholestasis, inflammation, and cancer. Adv Pharmacol. 2015;74:263–302. doi: 10.1016/bs.apha.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen K., Jaeschke H., Copple B.L. Bile acids induce inflammatory genes in hepatocytes: a novel mechanism of inflammation during obstructive cholestasis. Am J Pathol. 2011;178:175–186. doi: 10.1016/j.ajpath.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinken A., Ravcheev D.A., Baldini F., Heirendt L., Fleming R.M.T., Thiele I. Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome. 2019;7:75. doi: 10.1186/s40168-019-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hang S., Paik D., Yao L., Kim E., Trinath J., Lu J., et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature. 2019;576:143–148. doi: 10.1038/s41586-019-1785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Z.D., Selwyn F.P., Cui J.Y., Klaassen C.D. RNA-Seq profiling of intestinal expression of xenobiotic processing genes in germ-free mice. Drug Metab Dispos. 2017;45:1225–1238. doi: 10.1124/dmd.117.077313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C.Y., Lee S., Cade S., Kuo L.J., Schultz I.R., Bhatt D.K., et al. Novel interactions between gut microbiome and host drug-processing genes modify the hepatic metabolism of the environmental chemicals polybrominated diphenyl ethers. Drug Metab Dispos. 2017;45:1197–1214. doi: 10.1124/dmd.117.077024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oladimeji P.O., Chen T. PXR: more than just a master xenobiotic receptor. Mol Pharmacol. 2018;93:119–127. doi: 10.1124/mol.117.110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aleksunes L.M., Klaassen C.D. Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARalpha-, and Nrf2-null mice. Drug Metab Dispos. 2012;40:1366–1379. doi: 10.1124/dmd.112.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee M., Robbins D., Chen T. Targeting xenobiotic receptors PXR and CAR in human diseases. Drug Discov Today. 2015;20:618–628. doi: 10.1016/j.drudis.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C.Y., Dempsey J.L., Wang D., Lee S., Weigel K.M., Fei Q., et al. PBDEs altered gut microbiome and bile acid homeostasis in male C57BL/6 mice. Drug Metab Dispos. 2018;46:1226–1240. doi: 10.1124/dmd.118.081547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pencikova K., Svrzkova L., Strapacova S., Neca J., Bartonkova I., Dvorak Z., et al. In vitro profiling of toxic effects of prominent environmental lower-chlorinated PCB congeners linked with endocrine disruption and tumor promotion. Environ Pollut. 2018;237:473–486. doi: 10.1016/j.envpol.2018.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou H., Hylemon P.B. Bile acids are nutrient signaling hormones. Steroids. 2014;86:62–68. doi: 10.1016/j.steroids.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui J.Y., Klaassen C.D. RNA-Seq reveals common and unique PXR- and CAR-target gene signatures in the mouse liver transcriptome. Biochim Biophys Acta. 2016;1859:1198–1217. doi: 10.1016/j.bbagrm.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang A., Li C.Y., Kelly E.J., Sheppard L., Cui J.Y. Transcriptomic profiling of PBDE-exposed HepaRG cells unveils critical lncRNA- PCG pairs involved in intermediary metabolism. PLoS One. 2020;15 doi: 10.1371/journal.pone.0224644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhutia Y.D., Ogura J., Sivaprakasam S., Ganapathy V. Gut microbiome and colon cancer: role of bacterial metabolites and their molecular targets in the host. Curr Colorectal Cancer Rep. 2017;13:111–118. doi: 10.1007/s11888-017-0362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selwyn F.P., Cheng S.L., Bammler T.K., Prasad B., Vrana M., Klaassen C., et al. Developmental regulation of drug-processing genes in livers of germ-free mice. Toxicol Sci. 2015;147:84–103. doi: 10.1093/toxsci/kfv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toda T., Saito N., Ikarashi N., Ito K., Yamamoto M., Ishige A., et al. Intestinal flora induces the expression of Cyp3a in the mouse liver. Xenobiotica. 2009;39:323–334. doi: 10.1080/00498250802651984. [DOI] [PubMed] [Google Scholar]
- 21.Shah Y.M., Ma X., Morimura K., Kim I., Gonzalez F.J. Pregnane X receptor activation ameliorates DSS-induced inflammatory bowel disease via inhibition of NF-kappaB target gene expression. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1114–G1122. doi: 10.1152/ajpgi.00528.2006. [DOI] [PubMed] [Google Scholar]
- 22.Xu C., Huang M., Bi H. PXR- and CAR-mediated herbal effect on human diseases. Biochim Biophys Acta. 2016;1859:1121–1129. doi: 10.1016/j.bbagrm.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Qiu Z., Cervantes J.L., Cicek B.B., Mukherjee S., Venkatesh M., Maher L.A., et al. Pregnane X receptor regulates pathogen-induced inflammation and host defense against an intracellular bacterial infection through toll-like receptor 4. Sci Rep. 2016;6:31936. doi: 10.1038/srep31936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ranhotra H.S., Flannigan K.L., Brave M., Mukherjee S., Lukin D.J., Hirota S.A., et al. Xenobiotic receptor-mediated regulation of intestinal barrier function and innate immunity. Nucl Recept Res. 2016;3:101199. doi: 10.11131/2016/101199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang K., Mukherjee S., DesMarais V., Albanese J.M., Rafti E., Draghi Ii A., et al. Targeting the PXR-TLR4 signaling pathway to reduce intestinal inflammation in an experimental model of necrotizing enterocolitis. Pediatr Res. 2018;83:1031–1040. doi: 10.1038/pr.2018.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A.P., et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma N., Guo P., Zhang J., He T., Kim S.W., Zhang G., et al. Nutrients mediate intestinal bacteria-mucosal immune crosstalk. Front Immunol. 2018;9:5. doi: 10.3389/fimmu.2018.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagkouvardos I., Lesker T.R., Hitch T.C.A., Galvez E.J.C., Smit N., Neuhaus K., et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome. 2019;7:28. doi: 10.1186/s40168-019-0637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caparros-Martin J.A., Lareu R.R., Ramsay J.P., Peplies J., Reen F.J., Headlam H.A., et al. Statin therapy causes gut dysbiosis in mice through a PXR-dependent mechanism. Microbiome. 2017;5:95. doi: 10.1186/s40168-017-0312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dempsey J.L., Wang D., Siginir G., Fei Q., Raftery D., Gu H., et al. Pharmacological activation of PXR and CAR downregulates distinct bile acid-metabolizing intestinal bacteria and alters bile acid homeostasis. Toxicol Sci. 2019;168:40–60. doi: 10.1093/toxsci/kfy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng S.L., Li X., Lehmler H.J., Phillips B., Shen D., Cui J.Y. Gut microbiota modulates interactions between polychlorinated biphenyls and bile acid homeostasis. Toxicol Sci. 2018;166:269–287. doi: 10.1093/toxsci/kfy208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Xiang X., Huang W.W., Sandford A.J., Wu S.Q., Zhang M.M., et al. Association of PXR and CAR polymorphisms and antituberculosis drug-induced hepatotoxicity. Sci Rep. 2019;9:2217. doi: 10.1038/s41598-018-38452-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staudinger J.L., Goodwin B., Jones S.A., Hawkins-Brown D., MacKenzie K.I., LaTour A., et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ueda A., Hamadeh H.K., Webb H.K., Yamamoto Y., Sueyoshi T., Afshari C.A., et al. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Ma X., Shah Y., Cheung C., Guo G.L., Feigenbaum L., Krausz K.W., et al. The PREgnane X receptor gene-humanized mouse: a model for investigating drug-drug interactions mediated by cytochromes P450 3A. Drug Metab Dispos. 2007;35:194–200. doi: 10.1124/dmd.106.012831. [DOI] [PubMed] [Google Scholar]
- 36.Li C.Y., Renaud H.J., Klaassen C.D., Cui J.Y. Age-specific regulation of drug-processing genes in mouse liver by ligands of xenobiotic-sensing transcription factors. Drug Metab Dispos. 2016;44:1038–1049. doi: 10.1124/dmd.115.066639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scoville D.K., Li C.Y., Wang D., Dempsey J.L., Raftery D., Mani S., et al. Polybrominated diphenyl ethers and gut microbiome modulate metabolic syndrome-related aqueous metabolites in mice. Drug Metab Dispos. 2019;47:928–940. doi: 10.1124/dmd.119.086538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pacyniak E.K., Cheng X., Cunningham M.L., Crofton K., Klaassen C.D., Guo G.L. The flame retardants, polybrominated diphenyl ethers, are pregnane X receptor activators. Toxicol Sci. 2007;97:94–102. doi: 10.1093/toxsci/kfm025. [DOI] [PubMed] [Google Scholar]
- 39.Sueyoshi T., Li L., Wang H., Moore R., Kodavanti P.R., Lehmler H.J., et al. Flame retardant BDE-47 effectively activates nuclear receptor CAR in human primary hepatocytes. Toxicol Sci. 2014;137:292–302. doi: 10.1093/toxsci/kft243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim J.J., Li X., Lehmler H.J., Wang D., Gu H., Cui J.Y. Gut microbiome critically impacts PCB-induced changes in metabolic fingerprints and the hepatic transcriptome in mice. Toxicol Sci. 2020;177:168–187. doi: 10.1093/toxsci/kfaa090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wahlang B., Alexander N.C., 2nd, Li X., Rouchka E.C., Kirpich I.A., Cave M.C. Polychlorinated biphenyls altered gut microbiome in CAR and PXR knockout mice exhibiting toxicant-associated steatohepatitis. Toxicol Rep. 2021;8:536–547. doi: 10.1016/j.toxrep.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L., Day A., McKenzie G., Mitchell H. Nongastric Helicobacter species detected in the intestinal tract of children. J Clin Microbiol. 2006;44:2276–2279. doi: 10.1128/JCM.02017-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Flaherty S., Briner Crawley A., Theriot C.M., Barrangou R. The Lactobacillus bile salt hydrolase repertoire reveals niche-specific adaptation. mSphere. 2018;3 doi: 10.1128/mSphere.00140-18. e00140–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ridlon J.M., Kang D.J., Hylemon P.B. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 47.Oliveira M., Bosco N., Perruisseau G., Nicolas J., Segura-Roggero I., Duboux S., et al. Lactobacillus paracasei reduces intestinal inflammation in adoptive transfer mouse model of experimental colitis. Clin Dev Immunol. 2011;2011:807483. doi: 10.1155/2011/807483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D.H., Kim S., Lee J.H., Kim J.H., Che X., Ma H.W., et al. Lactobacillus acidophilus suppresses intestinal inflammation by inhibiting endoplasmic reticulum stress. J Gastroenterol Hepatol. 2019;34:178–185. doi: 10.1111/jgh.14362. [DOI] [PubMed] [Google Scholar]
- 49.Chetwin E., Manhanzva M.T., Abrahams A.G., Froissart R., Gamieldien H., Jaspan H., et al. Antimicrobial and inflammatory properties of South African clinical Lactobacillus isolates and vaginal probiotics. Sci Rep. 2019;9:1917. doi: 10.1038/s41598-018-38253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rocha-Ramirez L.M., Perez-Solano R.A., Castanon-Alonso S.L., Moreno Guerrero S.S., Ramirez Pacheco A., Garcia Garibay M., et al. Probiotic Lactobacillus strains stimulate the inflammatory response and activate human macrophages. J Immunol Res. 2017;2017:4607491. doi: 10.1155/2017/4607491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hiippala K., Kainulainen V., Kalliomaki M., Arkkila P., Satokari R. Mucosal prevalence and interactions with the epithelium indicate commensalism of. Sutterella spp. Front Microbiol. 2016;7:1706. doi: 10.3389/fmicb.2016.01706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams B.L., Hornig M., Parekh T., Lipkin W.I. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. mBio. 2012;3 doi: 10.1128/mBio.00261-11. e00261–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osaka T., Moriyama E., Arai S., Date Y., Yagi J., Kikuchi J., et al. Meta-analysis of fecal microbiota and metabolites in experimental colitic mice during the inflammatory and healing phases. Nutrients. 2017;9:1329. doi: 10.3390/nu9121329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kant R., Rasinkangas P., Satokari R., Pietila T.E., Palva A. Genome sequence of the butyrate-producing anaerobic bacterium Anaerostipes hadrus PEL 85. Genome Announc. 2015;3 doi: 10.1128/genomeA.00224-15. e00224–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ravussin Y., Koren O., Spor A., LeDuc C., Gutman R., Stombaugh J., et al. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity (Silver Spring) 2012;20:738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Selwyn F.P., Cheng S.L., Klaassen C.D., Cui J.Y. Regulation of hepatic drug-metabolizing enzymes in germ-free mice by conventionalization and probiotics. Drug Metab Dispos. 2016;44:262–274. doi: 10.1124/dmd.115.067504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allain T., Chaouch S., Thomas M., Vallee I., Buret A.G., Langella P., et al. Bile-salt-hydrolases from the probiotic strain Lactobacillus johnsonii La1 mediate anti-giardial activity in vitro and in vivo. Front Microbiol. 2017;8:2707. doi: 10.3389/fmicb.2017.02707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Png C.W., Linden S.K., Gilshenan K.S., Zoetendal E.G., McSweeney C.S., Sly L.I., et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 59.Olbjorn C., Cvancarova Smastuen M., Thiis-Evensen E., Nakstad B., Vatn M.H., Jahnsen J., et al. Fecal microbiota profiles in treatment-naive pediatric inflammatory bowel disease—associations with disease phenotype, treatment, and outcome. Clin Exp Gastroenterol. 2019;12:37–49. doi: 10.2147/CEG.S186235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chae J.P., Valeriano V.D., Kim G.B., Kang D.K. Molecular cloning, characterization and comparison of bile salt hydrolases from Lactobacillus johnsonii PF01. J Appl Microbiol. 2013;114:121–133. doi: 10.1111/jam.12027. [DOI] [PubMed] [Google Scholar]
- 61.De Smet I., Van Hoorde L., Vande Woestyne M., Christiaens H., Verstraete W. Significance of bile salt hydrolytic activities of Lactobacilli. J Appl Bacteriol. 1995;79:292–301. doi: 10.1111/j.1365-2672.1995.tb03140.x. [DOI] [PubMed] [Google Scholar]
- 62.Kumar R.S., Brannigan J.A., Prabhune A.A., Pundle A.V., Dodson G.G., Dodson E.J., et al. Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin V acylase. J Biol Chem. 2006;281:32516–32525. doi: 10.1074/jbc.M604172200. [DOI] [PubMed] [Google Scholar]
- 63.Foley M.H., O'Flaherty S., Barrangou R., Theriot C.M. Bile salt hydrolases: gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allain T., Chaouch S., Thomas M., Travers M.A., Valle I., Langella P., et al. Bile salt hydrolase activities: a novel target to screen anti-giardia Lactobacilli?. Front Microbiol. 2018;9:89. doi: 10.3389/fmicb.2018.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shyng S.L. Targeting the gut microbiota-FXR signaling axis for glycemic control: does a dietary supplement work magic?. Diabetes. 2017;66:571–573. doi: 10.2337/dbi16-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sayin S.I., Wahlstrom A., Felin J., Jantti S., Marschall H.U., Bamberg K., et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metabol. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 67.Paraiso I.L., Tran T.Q., Magana A.A., Kundu P., Choi J., Maier C.S., et al. Xanthohumol ameliorates diet-induced liver dysfunction via farnesoid X receptor-dependent and independent signaling. Front Pharmacol. 2021;12:643857. doi: 10.3389/fphar.2021.643857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teodoro J.S., Rolo A.P., Palmeira C.M. Hepatic FXR: key regulator of whole-body energy metabolism. Trends Endocrinol Metabol. 2011;22:458–466. doi: 10.1016/j.tem.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 69.Thomas A.M., Hart S.N., Kong B., Fang J., Zhong X.B., Guo G.L. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology. 2010;51:1410–1419. doi: 10.1002/hep.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yanguas-Casas N., Barreda-Manso M.A., Nieto-Sampedro M., Romero-Ramirez L. TUDCA: an agonist of the bile acid receptor GPBAR1/TGR5 with anti-inflammatory effects in microglial cells. J Cell Physiol. 2017;232:2231–2245. doi: 10.1002/jcp.25742. [DOI] [PubMed] [Google Scholar]
- 71.Lu Y.F., Jin T., Xu Y., Zhang D., Wu Q., Zhang Y.K., et al. Sex differences in the circadian variation of cytochrome p450 genes and corresponding nuclear receptors in mouse liver. Chronobiol Int. 2013;30:1135–1143. doi: 10.3109/07420528.2013.805762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lai Z.L., Tseng C.H., Ho H.J., Cheung C.K.Y., Lin J.Y., Chen Y.J., et al. Fecal microbiota transplantation confers beneficial metabolic effects of diet and exercise on diet-induced obese mice. Sci Rep. 2018;8:15625. doi: 10.1038/s41598-018-33893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garcia-Mazcorro J.F., Mills D., Noratto G. Molecular exploration of fecal microbiome in quinoa-supplemented obese mice. FEMS Microbiol Ecol. 2016;92 doi: 10.1093/femsec/fiw089. [DOI] [PubMed] [Google Scholar]
- 74.Chen M., Liao Z., Lu B., Wang M., Lin L., Zhang S., et al. Huang-Lian-Jie-Du-Decoction ameliorates hyperglycemia and insulin resistant in association with gut microbiota modulation. Front Microbiol. 2018;9:2380. doi: 10.3389/fmicb.2018.02380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alteber Z., Sharbi-Yunger A., Pevsner-Fischer M., Blat D., Roitman L., Tzehoval E., et al. The anti-inflammatory IFITM genes ameliorate colitis and partially protect from tumorigenesis by changing immunity and microbiota. Immunol Cell Biol. 2018;96:284–297. doi: 10.1111/imcb.12000. [DOI] [PubMed] [Google Scholar]
- 76.Miyoshi J., Nobutani K., Musch M.W., Ringus D.L., Hubert N.A., Yamamoto M., et al. Time-, sex-, and dose-dependent alterations of the gut microbiota by consumption of dietary daikenchuto (TU-100) Evid Based Complement Alternat Med. 2018;2018:7415975. doi: 10.1155/2018/7415975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meneses M.E., Martinez-Carrera D., Torres N., Sanchez-Tapia M., Aguilar-Lopez M., Morales P., et al. Hypocholesterolemic properties and prebiotic effects of Mexican Ganoderma lucidum in C57BL/6 mice. PLoS One. 2016;11 doi: 10.1371/journal.pone.0159631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ring C., Klopfleisch R., Dahlke K., Basic M., Bleich A., Blaut M. Akkermansia muciniphila strain ATCC BAA-835 does not promote short-term intestinal inflammation in gnotobiotic interleukin-10-deficient mice. Gut Microb. 2019;10:188–203. doi: 10.1080/19490976.2018.1511663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu W., Lv L., Shi D., Ye J., Fang D., Guo F., et al. Protective effect of Akkermansia muciniphila against immune-mediated liver injury in a mouse model. Front Microbiol. 2017;8:1804. doi: 10.3389/fmicb.2017.01804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao S., Liu W., Wang J., Shi J., Sun Y., Wang W., et al. Akkermansia muciniphila improves metabolic profiles by reducing inflammation in chow diet-fed mice. J Mol Endocrinol. 2017;58:1–14. doi: 10.1530/JME-16-0054. [DOI] [PubMed] [Google Scholar]
- 81.Reunanen J., Kainulainen V., Huuskonen L., Ottman N., Belzer C., Huhtinen H., et al. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl Environ Microbiol. 2015;81:3655–3662. doi: 10.1128/AEM.04050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ajouz H., Mukherji D., Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol. 2014;12:164. doi: 10.1186/1477-7819-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sinha S.R., Haileselassie Y., Nguyen L.P., Tropini C., Wang M., Becker L.S., et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe. 2020;27:659–670. doi: 10.1016/j.chom.2020.01.021. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luerce T.D., Gomes-Santos A.C., Rocha C.S., Moreira T.G., Cruz D.N., Lemos L., et al. Anti-inflammatory effects of Lactococcus lactis NCDO 2118 during the remission period of chemically induced colitis. Gut Pathog. 2014;6:33. doi: 10.1186/1757-4749-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Han K.J., Lee N.K., Park H., Paik H.D. Anticancer and anti-inflammatory activity of probiotic Lactococcus lactis NK34. J Microbiol Biotechnol. 2015;25:1697–1701. doi: 10.4014/jmb.1503.03033. [DOI] [PubMed] [Google Scholar]
- 86.Ketter P.M., Yu J.J., Guentzel M.N., May H.C., Gupta R., Eppinger M., et al. Acinetobacter baumannii gastrointestinal colonization is facilitated by secretory IgA which is reductively dissociated by bacterial thioredoxin A. mBio. 2018;9 doi: 10.1128/mBio.01298-18. e01298–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Palm N.W., de Zoete M.R., Cullen T.W., Barry N.A., Stefanowski J., Hao L., et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Larsen J.M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151:363–374. doi: 10.1111/imm.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Elinav E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Golinska E., Tomusiak A., Gosiewski T., Wiecek G., Machul A., Mikolajczyk D., et al. Virulence factors of Enterococcus strains isolated from patients with inflammatory bowel disease. World J Gastroenterol. 2013;19:3562–3572. doi: 10.3748/wjg.v19.i23.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Iskandar C.F., Borges F., Taminiau B., Daube G., Zagorec M., Remenant B., et al. Comparative genomic analysis reveals ecological differentiation in the genus Carnobacterium. Front Microbiol. 2017;8:357. doi: 10.3389/fmicb.2017.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jena P.K., Sheng L., Liu H.X., Kalanetra K.M., Mirsoian A., Murphy W.J., et al. Western diet-induced dysbiosis in farnesoid X receptor knockout mice causes persistent hepatic inflammation after antibiotic treatment. Am J Pathol. 2017;187:1800–1813. doi: 10.1016/j.ajpath.2017.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Granado-Serrano A.B., Martin-Gari M., Sanchez V., Riart Solans M., Berdun R., Ludwig I.A., et al. Faecal bacterial and short-chain fatty acids signature in hypercholesterolemia. Sci Rep. 2019;9:1772. doi: 10.1038/s41598-019-38874-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Castro-Mejia J., Jakesevic M., Krych L., Nielsen D.S., Hansen L.H., Sondergaard B.C., et al. Treatment with a monoclonal anti-IL-12p40 antibody induces substantial gut microbiota changes in an experimental colitis model. Gastroenterol Res Pract. 2016;2016:4953120. doi: 10.1155/2016/4953120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Erridge C., Moncayo-Nieto O.L., Morgan R., Young M., Poxton I.R. Acinetobacter baumannii lipopolysaccharides are potent stimulators of human monocyte activation via Toll-like receptor 4 signalling. J Med Microbiol. 2007;56:165–171. doi: 10.1099/jmm.0.46823-0. [DOI] [PubMed] [Google Scholar]
- 96.Shimizu J., Kubota T., Takada E., Takai K., Fujiwara N., Arimitsu N., et al. Propionate-producing bacteria in the intestine may associate with skewed responses of IL10-producing regulatory T cells in patients with relapsing polychondritis. PLoS One. 2018;13 doi: 10.1371/journal.pone.0203657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brown K., DeCoffe D., Molcan E., Gibson D.L. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients. 2012;4:1095–1119. doi: 10.3390/nu4081095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pallister T., Jackson M.A., Martin T.C., Glastonbury C.A., Jennings A., Beaumont M., et al. Untangling the relationship between diet and visceral fat mass through blood metabolomics and gut microbiome profiling. Int J Obes. 2017;41:1106–1113. doi: 10.1038/ijo.2017.70. (Lond) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Turnbaugh P.J., Backhed F., Fulton L., Gordon J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jackson M.A., Jeffery I.B., Beaumont M., Bell J.T., Clark A.G., Ley R.E., et al. Signatures of early frailty in the gut microbiota. Genome Med. 2016;8:8. doi: 10.1186/s13073-016-0262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Deshmane S.L., Kremlev S., Amini S., Sawaya B.E. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29:313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rahim S.S., Khan N., Boddupalli C.S., Hasnain S.E., Mukhopadhyay S. Interleukin-10 (IL-10) mediated suppression of IL-12 production in RAW 264.7 cells also involves c-rel transcription factor. Immunology. 2005;114:313–321. doi: 10.1111/j.1365-2567.2005.02107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dunican E.M., Fahy J.V. The role of type 2 inflammation in the pathogenesis of asthma exacerbations. Ann Am Thorac Soc. 2015;12 Suppl 2:S144–S149. doi: 10.1513/AnnalsATS.201506-377AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Matson A.P., Zhu L., Lingenheld E.G., Schramm C.M., Clark R.B., Selander D.M., et al. Maternal transmission of resistance to development of allergic airway disease. J Immunol. 2007;179:1282–1291. doi: 10.4049/jimmunol.179.2.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gagnon L., Leduc M., Thibodeau J.F., Zhang M.Z., Grouix B., Sarra-Bournet F., et al. A newly discovered antifibrotic pathway regulated by two fatty acid receptors: GPR40 and GPR84. Am J Pathol. 2018;188:1132–1148. doi: 10.1016/j.ajpath.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 106.Montonye D.R., Ericsson A.C., Busi S.B., Lutz C., Wardwell K., Franklin C.L. Acclimation and institutionalization of the mouse microbiota following transportation. Front Microbiol. 2018;9:1085. doi: 10.3389/fmicb.2018.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Caruso R., Ono M., Bunker M.E., Nunez G., Inohara N. Dynamic and asymmetric changes of the microbial communities after cohousing in laboratory mice. Cell Rep. 2019;27:3401–3412. doi: 10.1016/j.celrep.2019.05.042. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kappel S., Hawkins P., Mendl M.T. To group or not to group? Good practice for housing male laboratory mice. Animals (Basel) 2017;7:88. doi: 10.3390/ani7120088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R., et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ericsson A.C., Davis J.W., Spollen W., Bivens N., Givan S., Hagan C.E., et al. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One. 2015;10 doi: 10.1371/journal.pone.0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gabay C. Interleukin-6 and chronic inflammation. Arthritis Res Ther. 2006;2 Suppl 8:S3. doi: 10.1186/ar1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hoyer K.K., Dooms H., Barron L., Abbas A.K. Interleukin-2 in the development and control of inflammatory disease. Immunol Rev. 2008;226:19–28. doi: 10.1111/j.1600-065X.2008.00697.x. [DOI] [PubMed] [Google Scholar]
- 113.Neurath M.F. IL-12 family members in experimental colitis. Mucosal Immunol. 2008;1 Suppl 1:S28–S30. doi: 10.1038/mi.2008.45. [DOI] [PubMed] [Google Scholar]
- 114.Baskin-Bey E.S., Anan A., Isomoto H., Bronk S.F., Gores G.J. Constitutive androstane receptor agonist, TCPOBOP, attenuates steatohepatitis in the methionine choline-deficient diet-fed mouse. World J Gastroenterol. 2007;13:5635–5641. doi: 10.3748/wjg.v13.i42.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baskin-Bey E.S., Huang W., Ishimura N., Isomoto H., Bronk S.F., Braley K., et al. Constitutive androstane receptor (CAR) ligand, TCPOBOP, attenuates Fas-induced murine liver injury by altering Bcl-2 proteins. Hepatology. 2006;44:252–262. doi: 10.1002/hep.21236. [DOI] [PubMed] [Google Scholar]
- 116.Cheng J., Shah Y.M., Gonzalez F.J. Pregnane X receptor as a target for treatment of inflammatory bowel disorders. Trends Pharmacol Sci. 2012;33:323–330. doi: 10.1016/j.tips.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dou W., Zhang J., Li H., Kortagere S., Sun K., Ding L., et al. Plant flavonol isorhamnetin attenuates chemically induced inflammatory bowel disease via a PXR-dependent pathway. J Nutr Biochem. 2014;25:923–933. doi: 10.1016/j.jnutbio.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cheng J., Krausz K.W., Tanaka N., Gonzalez F.J. Chronic exposure to rifaximin causes hepatic steatosis in pregnane X receptor-humanized mice. Toxicol Sci. 2012;129:456–468. doi: 10.1093/toxsci/kfs211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.