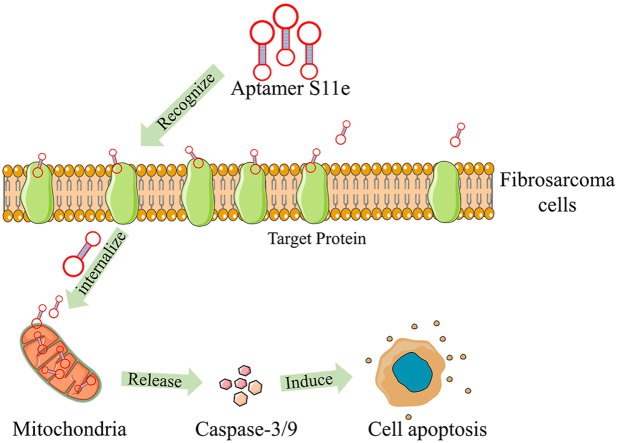
Abstract
Fibrosarcoma is a serious malignant mesenchymal tumor with strong invasiveness, high recurrence, and poor prognosis. Currently, surgical resection is the main treatment for fibrosarcoma. However, due to the lack of specific biomarkers, the inability to accurately diagnose fibrosarcoma can lead to sub-optimal surgical outcomes and decreased survival. Here, we seek to address this translational barrier and we show that DNA aptamer S11e was able to recognize fibrosarcoma cells (HT1080) but not human embryonic lung fibroblast cells with Kd values in the nanomolar range. In addition, we found that S11e discerned tumors in HT1080 xenograft mouse models and tumor tissues from fibrosarcoma patients. Furthermore, we demonstrated that S11e internalized into HT1080 cells independent of the lysosome pathway and located in mitochondria. Moreover, we revealed that S11e promoted the apoptosis of HT1080 cells and inhibited HT1080 cell migration. Finally, we investigated the biologically functional cellular target of S11e using a mass spectrometry approach, and identified that Diablo/SMAC protein is a cellular binding protein of S11e, by interacting to which S11e inhibited HT1080 cell migration and invasion. Taken together, these results provide the evidence that S11e may be useful for early diagnosis, targeted therapy, and prognostication of fibrosarcoma.
Keywords: Fibrosarcoma, Aptamer, Mitochondria, Apoptosis, Migration
Graphical abstract
Highlights
-
•
S11e specifically targets fibroscarcoma cells and could be a novel tool for the early diagnosis and therapy of fibrosarcoma.
-
•
S11e can be internalized into HT1080 fibrosarcoma cells and located in mitochondria, which induces the apoptosis of the cells.
-
•
S11e inhibits fibrosarcoma cell proliferation and migration via binding to Diablo/SMAC protein in mitochondria.
1. Introduction
In 2002, the World Health Organization (WHO) defined fibrosarcoma as a cancer consisting of fibroblasts and variable collagen [1]. Fibrosarcoma is a type of malignant neoplasm, originating from mesenchymal cells; it can occur in every part of human body with fibrous tissue and can occur at any age. Fibrosarcoma is mostly highly invasive and has a high recurrence rate. Lung is the most frequent site of fibrosarcoma metastasis, followed by the skeletal structure, lymph nodes, brain, and subcutaneous tissue [2]. Infant fibrosarcoma occurs in children under 5 years of age. Adult fibrosarcoma mostly occurs in adults aged 30–60 years, with a high degree of malignancy and prone to metastasis [3]. The incidence rate of fibrosarcoma in males and females is roughly equal. While surgical resection is the primary treatment, residual satellite lesions are the main cause of fibrosarcoma recurrence. However, multiple operations may lead to the progression of tumors. At present, fibrosarcoma lacks clinical pathological probes or biomarkers for its early stage [4]. Therefore, to potentially address this clinical need, it is important to explore the molecular probes or biomarkers for the early diagnosis, targeted therapy, and accurate prognosis of fibrosarcoma patients.
Aptamers, which are usually screened by systematic evolution of ligands by exponential enrichment (SELEX), are single-stranded (ss) DNA or RNA sequences [5]. Aptamers are able to bind to specific targets with high affinity by forming a specific secondary or tertiary structure. Compared with antibodies, aptamers have good thermal stability, low immunogenicity, and high adaptability to a variety of targets, including peptides, proteins, cells, bacteria, etc. [[6], [7], [8]]. Aptamers have been studied in biomarker discovery, and anti-tumor drug delivery [9,10]. At present, 11 aptamers have entered the clinical trial stage, including pegaptanib, E10030, ARC1905, REG1, ARC1779, NU172, BAX499, AS1411, NOX-A12, and NOX-H94 [[11], [12], [13], [14], [15], [16], [17], [18], [19]]. Pegaptanib, which is the first aptamer drug and has been approved by The United States Food and Drug Administration (FDA), is used to treat age-related macular degeneration (AMD) [20]. REG1, ARC1779, NU172, and BAX499 are four aptamers for coagulation. AS1411 has been demonstrated to exhibit anti-tumor effects for a variety of cancer types, such as breast cancer, kidney cancer, and lung cancer [21,22]. NOX-A12 is another anti-tumor aptamer to treat chronic lymphoblastic leukemia, and multiple myeloma [18]. NOX-H94 and NOX-E36 are used to manage disease-induced inflammation [14,23].
Apoptosis is a conserved process of cell metabolism that plays a critical role in diseases. Apoptosis stimuli can lead to the release of several cell death inducers from the mitochondria, including cytochrome c and SMAC (also known as DIABLO). Cytochrome c promotes the formation of a cytosolic protein complex that activates an initiator caspase, caspase-9. Diablo/SMAC is a mitochondrial intermembrane protein, which is also an activator of caspase [24]. They have a short region of similarity at their N-termini through which they can interact with inhibitors of apoptosis (IAP) proteins, and antagonizing IAP inhibition of caspases to promote cell death, as well as affect tumor cell cycle and other pathways in cancer, which show that Diablo/SMAC may be an important player and potential therapeutic target in tumor therapy.
In order to solve the current problems of cell-SELEX, we have applied a non-selective strategy to obtain an aptamer S11e to specifically recognize fibrosarcoma cells. Our work provides compelling evidence that S11e is a promising tool for fibrosarcoma diagnosis and therapy and opens the door as a paradigm for developing new strategies for fibrosarcoma therapy.
2. Materials and methods
2.1. Aptamer synthesis and labeling
All of the ssDNA aptamers appeared in the experiments were synthesized and labeled by Sangon Biological Technology Co., Ltd (Shanghai, China) and Hippo Biological Technology Co. Ltd (Huzhou, China).
2.2. Cell culture
Cell lines were cultured in sterile environment containing 5% CO2 at 37 °C. HT1080 (Fibrosarcoma cells) (CAS, cell bank Shanghai, China) and MRC-5 (Human embryonic lung fibroblast cell) (CAS) were cultured in MEM medium supplemented with 10% fetal bovine serum (FBS). U-2 OS (Osteosarcoma cells) (CAS) cells were cultured in McCoy's 5a Medium Modified supplemented with 10% FBS.
2.3. Aptamer binding detection
Washing buffer (WB) was Dulbecco's phosphate buffered saline (D-PBS) mixed with 5 mM MgCl2 and 4.5 g/L glucose. Then, 0.1 mg/mL yeast tRNA and 1 mg/L BSA were added into the WB to form the binding buffer (BB).
HT1080 or MRC-5 cells were cultured in 75-mm cell dishes. When the cell density reached 90%, cells were washed with D-PBS and digested with 0.2% EDTA. 3 × 105 cells were collected in a 1.5 mL Eppendorf (EP) tube and incubated with 250 nM FITC-labeled ssDNAs in BB for 1 h. Samples were washed with WB and the fluorescence signals were detected by BD FACSVerseTM Flow Cytometer (BD Biosciences, USA). All experiments in this study were repeated three times.
2.4. Confocal microscopy imaging
Cells were cultured in optical dishes (NEST Biological Technology Co., Ltd, China) for 24 h. Then, the cells were incubated with Cy5-labeled aptamers for 1 h. Before further assays, cells were washed several times with WB. The binding of aptamers to cells was monitored by confocal microscopy (Zeiss LSM510) and images were analyzed using FV10-ASW version 3.1 software. All experiments were repeated three times.
2.5. Target type determination
HT1080 cells were cultured in 35-mm cell dishes. After culturing and reaching a density of 90%, the cells were washed with WB. Then, 200 μL of 0.1 mg/mL of proteinase K or 0.25% trypsin was added for 1 min, and the complete medium was added to terminate the digestion. These cells were pipetted with WB and centrifuged at 1200 rpm for 4 min, and incubated for 1 h at 4 °C after 250 nM FITC-labeled aptamers were added. After the incubation, these cells were washed with WB and the fluorescence signals were detected by flow cytometry. The experiments were repeated three times.
2.6. Affinity analysis
The affinity analysis was conducted following the previously reported methods [25]. 3 × 105 HT1080 cells were incubated with a concentration gradient of FITC-labeled aptamers or Library at 4 °C for 1 h. The fluorescence intensity was tested by flow cytometry. The average fluorescence intensity was subtracted from that of the cells incubated with Library, and the curve of Kd was obtained from the equation of Y = Bmax X/(Kd + X) (Y: relative fluorescence intensity; X: aptamer concentration). Finally, results were plotted with Graph Pad Prism 7.0. All experiments were repeated three times.
2.7. Colocalization experiment
HT1080 cells were seeded into a 35-mm optical vessel. After incubation at 37 °C for 24 h, 500 μL of Cy5-labeled aptamer was appended at a total concentration of 250 nM. After incubation for 1 h, the supernatant was removed and 500 μL of Mito-tracker (C1048, Beyotime) solution or lysosome-tracker (C1046, Beyotime) was added at a final concentration of 200 nM at 37 °C for 30 min. Fluorescence signals were detected by Zeiss LSM510. All experiments were repeated three times.
2.8. In vivo fluorescence imaging
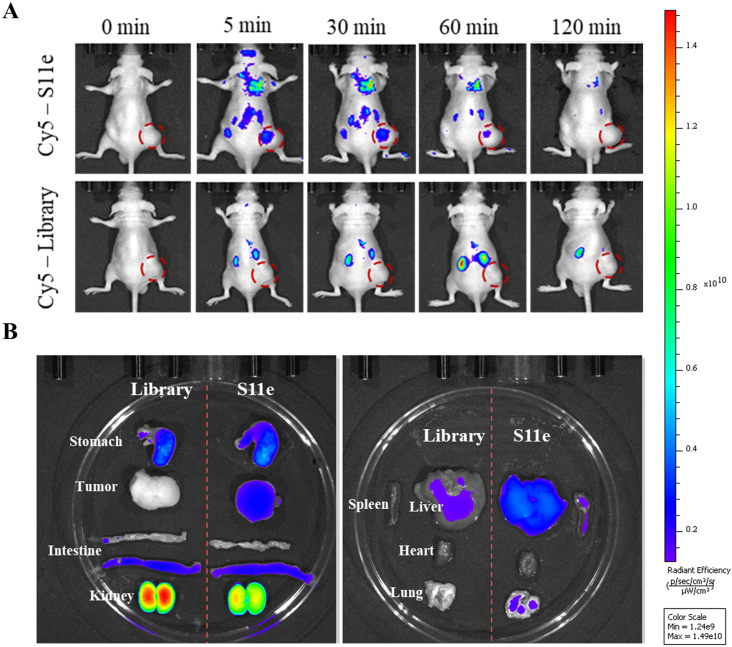
5–6 weeks old male athymic BALB/c (BALB/c-nude) mice were purchased from Hunan SLAC Experimental Animal Co., Ltd. 5 × 106 HT1080 cells were injected subcutaneously into the back of nude mice. As soon as the tumor volume reached a diameter of 1.0 cm, 100 μL phosphorothioate (PS) modified Cy5-S11e (Cy5-S11e-PS) or PS modified Cy5-Library (Cy5-Library-PS) was injected into mice via tail vein, and the final concentration was 5.5 μM. At 5 min, 30 min, 1 h, 2 h, 3 h, fluorescent images were collected by the In vivo Imaging System (IVIS) Lumina II imaging system (Caliper Life Science, USA). After fluorescence imaging analysis, tumor-bearing mice injected with Cy5-S11e-PS or Cy5-Library-PS were sacrificed and dissected. Tumor tissue and visceral organs including spleen, kidneys, liver, heart and lung were then imaged. All animal experiments were performed in compliance with the regulations of Institutional Animal Care and Use and were approved by Hunan Experimental Animal Center. The mice experiments were repeated three times.
2.9. Human tissue microarray staining
The slides of fibrosarcoma tissue array (SO2084) were purchased from Xi'an Elina Biotechnology Co., Ltd. These slides were preheated at 60 °C for 30 min and then immersed in xylene for 30 min. They were sequentially immersed in a gradually decreasing ethanol solution for rehydration (100%, 95%, 90%, 80%, and 70%) at 5 min intervals. They were washed with WB, and placed in 0.01 M citrate (pH 6.0), heat for 20 min. After naturally cooled down at room temperature, they were treated with pre-cooled blocking buffer (BB, 20% FBS and 0.1 mg/mL salmon sperm DNA) for 1 h. They were then incubated with 250 nM Cy5-labeled aptamer S11e and Library for 1 h at 4 °C, separately. Finally, the fluorescence images were captured by a Panoramic SCAN (3DHISTECH, Spain). Fluorescence intensity was calculated and marked as negative (–, <30,000), weak (+, 30,000–60000), moderate (++, 60,000–100000) or strong (+++, >100,000), respectively.
2.10. Cytotoxicity assay
Cells were seeded in 96-well plates at 5000 cells/well. After the cells were attached, a concentration gradient of Library or S11e was added and cultured at 37 °C for 24 h, 48 h, and 72 h, respectively. After supernatant removal, these wells were washed with D-PBS and then CCK-8 solution was added. After incubating at 37 °C for 1 h, the plates were then detected by Microplate Reader Synergy 2 (BioTek, USA). All experiments were repeated three times.
2.11. Scarification test
HT1080 cells were seeded into 6-well plates for 24 h. Then, the cells in monolayer were vertically scratched. The floating cells were removed by D-PBS and cultured at 37 °C with 8 μM Library or S11e. Images were taken daily through an inverted microscope (EVOS f1, AMG Corporation, USA).
2.12. Transwell assay
HT1080 were seeded into 24-well plates, then 5 μM ssDNA was added and incubated with the cells for 0 h, 12 h, and 24 h. Transwell chamber (Merck Millipore Ltd, MCEP24H48) was placed in a new 24-well plate, and then solidified in an incubator for 4 h after Matrigel addition. 2 × 104 aptamer-treated cells were added into the chamber (200 μL cell suspension + 800 μL complete medium), and cultured for another 24 h. Cells were then fixed with 95% ethanol for 20 min and stained with Giemsa for 10 min. Resulting cells were photographed under an inverted microscope (EVOS f1, AMG Corporation, USA).
2.13. Western blotting
HT1080 cells were seeded in 75-mm dishes and incubated for 24 h. When the cells reached 70–80% confluency, they were washed with D-PBS and harvested by centrifugation. Then, the RIPA buffer (P0013B, Beyotime) with Protease Inhibitor Cocktail (4,693,159,001, Roche) was added to lyse cells. Proteins were separated using 10% SDS polyacrylamide gels (P2012, NCM Biological Technology Co. Ltd) and transferred to PVDF membranes (Merck Millipore Ltd, ISEQ00010). Membranes were then incubated with primary antibodies at 4 °C overnight. The followings are the antibodies used in this study: anti-poly (ADP-ribose) polymerase (PARP) antibody (A19596, Company ABclonal Inc.), anti-caspase-3 antibody (sc-56053, Santa Cruz Biotechnology), anti-caspase-9 (sc-56076, Santa Cruz Biotechnology) antibody, anti-Diablo/SMAC antibody (AF789-SP, Santa Cruz Biotechnology). After washing three times with Tris-buffered saline with 0.1% Tween 20 (TBST), goat anti-rabbit secondary antibody (A9169, Sigma) was added to the membrane. The signal was detected with WesternBright™ ECL (K-12045-D50, Advansta) and visualized by ChemiDoc™ XRS Imager (Bio-Rad, USA).
2.14. Apoptosis assay
1 × 105 HT1080 cells were cultivated in a 35-mm dish, and unlabeled S11e was added to the cells. After 72 h incubation at 37 °C, the supernatant was transferred to a 15 mL centrifuge tube. Cells were treated with EDTA-free trypsin and transferred to the same centrifuge tube. After centrifuged at 1300 rpm for 5 min. Cell apoptosis was analyzed using Annexin V-FITC (C1062 M, Beyotime) according to the manufacturer's protocol. Fluorescent signals were detected by flow cytometry. Apoptosis assay was repeated three times.
2.15. Mass spectrometry (MS) analysis
Because the modification of biotin at the 5-terminal of S11e may decrease its binding efficiency, we used flow cytometry to test the binding ability of biotin-modified S11e to HT1080 cells. The result showed that biotin-modified aptamer could still bind HT1080 cells with high affinity. Next, HT1080 cells were washed three times with pre-cooled PBS buffer and incubated with 400 nM biotin-modified S11e or biotin-modified Library in 4 mL of BB containing protease inhibitors (0.1 mM PMSF) at 37 °C for 1 h. After incubation, the plasma membrane proteins of the HT 1080 cells were isolated by SM-005 Minute Plasma Membrane Protein Isolation and Cell Fractionation Kit (Invent biotech corporation, USA). Subsequently, the protein-Library or protein-S11e complex was captured by incubating with 2 mg (200 μL) of streptavidin beads at 4 °C for 45 min. After a collection by centrifugation, the proteins were eluted by heating in 30 μL of loading buffer and analyzed by SDS- PAGE gel and silver staining. The aptamer-purified protein bands were excised for digestion in situ and analyzed by liquid chromatography (LC)-MS/MS (APTBIO Co., Ltd, China).
2.16. Aptamer-based pull-down assay
The protein samples were prepared in the same manner as described for MS analysis. The captured proteins mixed with 40 μL of protein loading buffer (2 × ). The solution was heated at 95 °C for 5 min to denature the proteins. After centrifugation at 5500 rpm, 4 °C for 5 min, the solution was subjected to 12% SDS-page gel electrophoresis. Then, the gels were stained with Silver Stain Kit (Beyotime, Biotech P0017S) and scanned using a gel imaging system (Microtek, Bio-6000).
2.17. Lentivirus-mediated RNA interference
Lentivirus particles harboring a short hairpin RNA (shRNA) targeting Diablo/SMAC (shRNA1, 5ʹ-GATCCGCGTTGATTGAAGCTATTACTTTCAAGAGAAGTAATAGCTTCAATCAACGCTTTTTTA-3ʹ; shRNA2, 5ʹ-GATCCGCGCAGATCAGGCCTCTATAATTCAAGAGATTATAGAGGCCTGATCTGCGCTTTTTTA-3ʹ; shRNA3 5ʹ-GATCCGCTGGCAGAAGCACAGATAGATTCAAGAGATCTATCTGTGCTTCTGCCAGCTTTTTTA-3ʹ) or a scramble negative control shRNA (5ʹ-GATCCGCGGCTTTTTTA-3ʹ), were purchased from Vigene Biosciences (Jinan, China). HT1080 cells were incubated with lentivirus particles for 24 h and subsequent selection was performed with puromycin treatment for 4 days.
2.18. Statistical analysis
All statistical analyses were conducted with Graph Pad Prism 7.0. Statistical significance analyses were performed with the t tests and p < 0.05 was deemed significant.
3. Results
3.1. Identification of ssDNA aptamers against fibrosarcoma cells HT1080
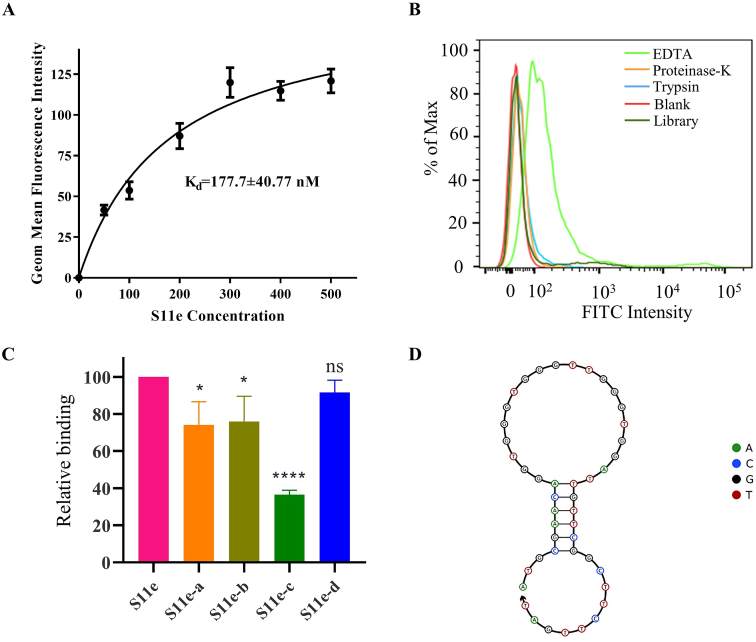
Aptamers are generally obtained by time-consuming SELEX, with multiple rounds of screening and analysis. We here used our previously found aptamers with known sequences as a pool for our non-SELEX strategies (The sequences are listed in Table S1). The binding abilities of all aptamers in this pool to HT1080 cells were tested by flow cytometry. As a result, we successfully identified an aptamer S11e [26] that can bind to HT1080 cells (Fig. S1). We found that the equilibrium dissociation constant (Kd) was 177.7 ± 40.77 nM, indicating its high affinity for HT1080 cells (Fig. 1A). To determine which type of target molecule S11e binds to, HT1080 cells were treated with 0.2% EDTA, proteinase K or trypsin, respectively, and then incubated with S11e at 4 °C for 1 h. The fluorescence intensity was measured by flow cytometry. The result showed that the affinity of S11e binding to HT1080 cells decreased after treated with proteinase K or trypsin, 0.2% EDTA was used as control (Fig. 1B), suggesting that the S11e targeting molecule may be a protein.
Fig. 1.
Study on the characteristics of S11e. (A) The Kd of S11e for HT1080 cells was measured by flow cytometry. (B) After incubating with proteinase K, trypsin or EDTA, the binding of S11e to HT1080 cells was measured by flow cytometry. A Library with randomized aptamer sequences was used as a negative control (each sequence in this Library includes a randomized sequence region and two prime regions). Blank: unstained negative control. (C) Bar graph showing the binding of four truncated S11e sequences to HT1080 cells analyzed by flow cytometry. The quantified fluorescence intensities were shown by mean ± standard deviation calculated from multiple results. (Compared to full length S11e: *p < 0.05; ***p < 0.001; ns, not significant). (D) S11e′s secondary structure was predicted by The Nucleic Acid Package (NUPACK).
3.2. Sequence optimization
The aptamers with fewer nucleotides, i.e., truncated aptamers, are more cost-efficient for chemical synthesis. They may have higher tissue permeability and lower immunogenicity than their original forms [27]. As shown in Table S2, four truncated sequences of S11e-a, S11e-b, S11e-c, and S11e-d were synthesized by deleting 5′ and 3′ bases of the aptamer S11e (45 nt, 1–45). The individual binding of each shortened aptamer to HT1080 cells was detected by flow cytometry. S11e-a (4–35 nt), S11e-b (4–30 nt) and S11e-d (6–30 nt) partially retained their ability to bind HT1080 cells, while S11e-c (4–22 nt) lost the ability (Fig. 1C).
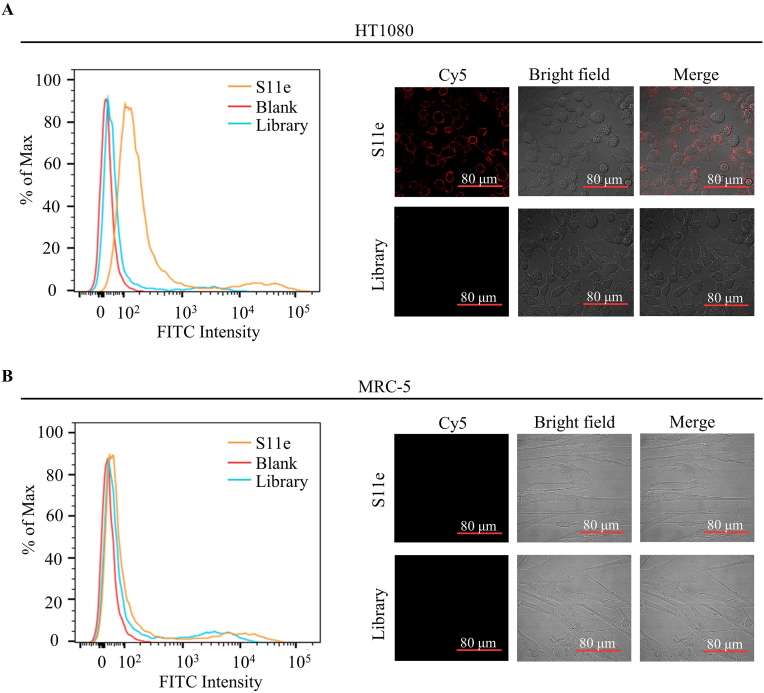
The specificity of aptamers binding to tumor cells is a prerequisite for the potential clinical application. We tested the binding selectivity of S11e to HT1080 cells and MRC-5 cells (a negative control), respectively. These cells were treated with FITC-labeled S11e at 4 °C for 1 h and fluorescence intensity was detected by flow cytometry (Fig. 2A and B). The result showed that S11e binds to HT1080 cells but not MRC-5 cells (Fig. S3). Confocal images of HT1080 cells and MRC-5 cells incubated with Cy5-labeled S11e further demonstrated that S11e only binds to HT1080 cells, suggesting its binding selectivity to tumor cells (Fig. 2B).
Fig. 2.
Identification of a DNA aptamer that can specifically bind to fibrosarcoma cells. (A) HT1080 cells were incubated with FITC-labeled S11e at 4 °C, the binding ability was analyzed by flow cytometry. The Cy5-labeled S11e (250 nM) binding to HT1080 cells was displayed by confocal microscopy, and Library: randomized sequences used as a control. Blank: unstained negative control. (B) Similarly, the ability of S11e binding to MRC-5 was monitored by flow cytometry at 4 °C. Images were captured by confocal microscopy.
3.3. Internalization and intracellular localization
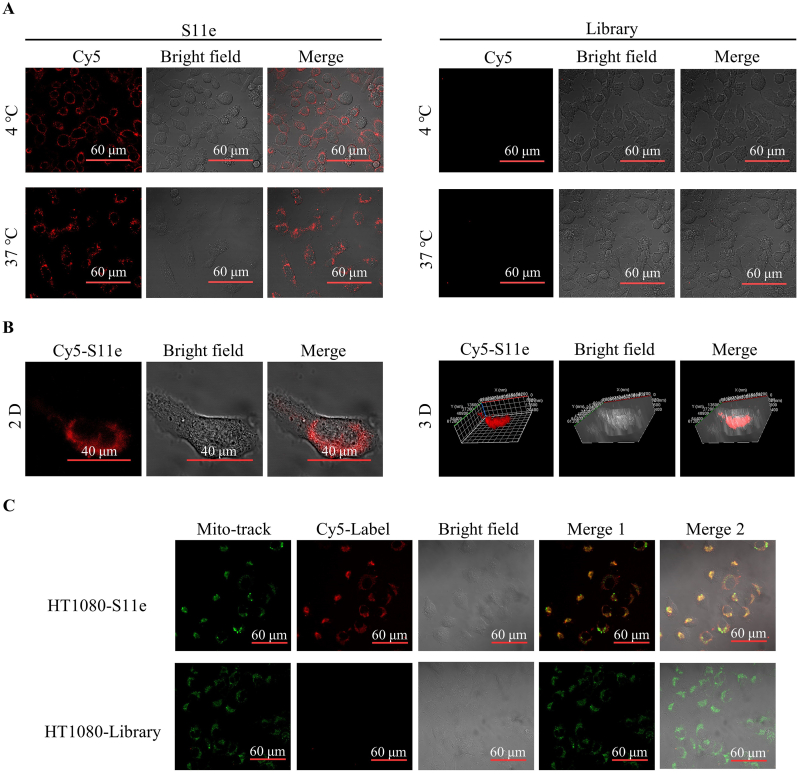
To address the cellular localization of S11e after binding to fibrosarcoma cells, we conducted confocal experiment. HT1080 cells were treated with Cy5-labeled S11e for 1 h at 4 °C or 37 °C, respectively. The results showed that the fluorescence signals of S11e mainly appeared on the cell membrane at 4 °C, while most of the fluorescent signals of S11e were in the cytoplasm at 37 °C (Fig. 3A). It was further confirmed with the three-dimensional (3D) image captured by confocal Z-axis scanning (Fig. 3B). In contrast, no fluorescent signals were detected in MRC-5 cells at either 4 °C or 37 °C (Fig. S2). Together, these results indicated that S11e can be internalized into HT1080 cells specifically.
Fig. 3.
Internalization of S11e in HT1080 cells. (A) HT1080 cells were incubated with Cy5-labeled S11e at 4 °C or 37 °C, then images were captured by confocal microscopy. (B) After incubating with Cy5-labeled S11e at 37 °C, the two-dimensional (2D) and three-dimensional (3D) images were captured by confocal microscopy. (C) Co-localization of the Cy5-S11e (red) and Mito-tracker (green).
In addition, we explored the internalization of S11e by colocalization analysis. Lysosome-tracker and Cy5-labeled S11e were co-incubated with HT1080 cells for 30 min, respectively, the fluorescent signals of lysosome-tracker and Cy5-labeled S11e were not overlapped (Fig. S5). After mitochondrial fluorescent probes Mito-tracker and Cy5-labeled S11e were used to co-incubate with HT1080 cells for 30 min, the fluorescent signals of Mito-tracker and Cy5-labeled S11e were partially overlapped. However, they are both completely overlapped for 1 h-incubation with HT1080 cells at 4 °C (Fig. 3C). We further test whether or not mitochondrial fluorescent probes are co-localized with Cy5-labeled S11e in MRC-5 cells. The results showed that only signals of mitochondrial fluorescent probes (green) were detected (Fig. S4), indicating no colocalization. Taken together, these results suggest that S11e can locate at the mitochondria in HT1080 cells. Our results also reveal that S11e can escape to mitochondria successfully without lysosomal internalization, making S11e an excellent potential carrier that can protect itself from degradation by various digestive enzymes in lysosomes.
3.4. In vivo fluorescence imaging
The ability of aptamers to recognize tumors in vivo is also important for clinical application. The stability of an aptamer affects its physiological condition, which is an important character to assess whether the aptamer can be clinically applied. However, when unmodified aptamers enter cells in vivo, they can be easily degraded by nucleases. Therefore, we modified oligonucleotides of aptamers to strengthen their resistance to nuclease degradation. Five bases at the 5′ end and five bases at the 3′ end of S11e and Library sequences were phosphorothioate modified. As shown in Fig. S6, the modified S11e sustained a relatively high concentration after 24 h in the blood serum. Then S11e was injected into mice via tail vein.
After the injection of S11e for 5 min, the fluorescence signal of S11e increased in the tumor sites, indicating that S11e aptamers accumulate rapidly in tumors. The fluorescence signal disappeared 2 h after injection, indicating that S11e can be excreted through kidney, liver and other metabolic organs (Fig. 4A). While after the injection of Library for 2 h, no obvious fluorescent signal was observed at the tumor sites, indicating that Library does not accumulate in tumors in vivo. The biological distribution of Cy5-labeled S11e and Cy5-labeled Library in mice was photographed 30 min after injection (Fig. 4B). Compared with the control group, mice injected with Cy5-labeled S11e showed accumulation of fluorescent signals in tumors. At the same time, fluorescent signals were observed in the kidney, liver, stomach and large intestine of the control group and the experimental group. Because of metabolism and rich of vessels, aptamers would induce some extent of wide distribution. These results suggest that S11e has the ability to target fibrosarcoma in vivo.
Fig. 4.
Fluorescence Imaging of S11e in xenografted mouse models. (A) Fibrosarcoma tumor-bearing mice were intravenously injected with Cy5-labeled S11e (upper panel) or Cy5-labeled randomized Library (lower panel). Fluorescence images were taken by in vivo imaging system. (B) Optical and fluorescent images of organs isolated from fibrosarcoma tumor-bearing mice at 30 min after injection.
3.5. Imaging fibrosarcoma tissue array
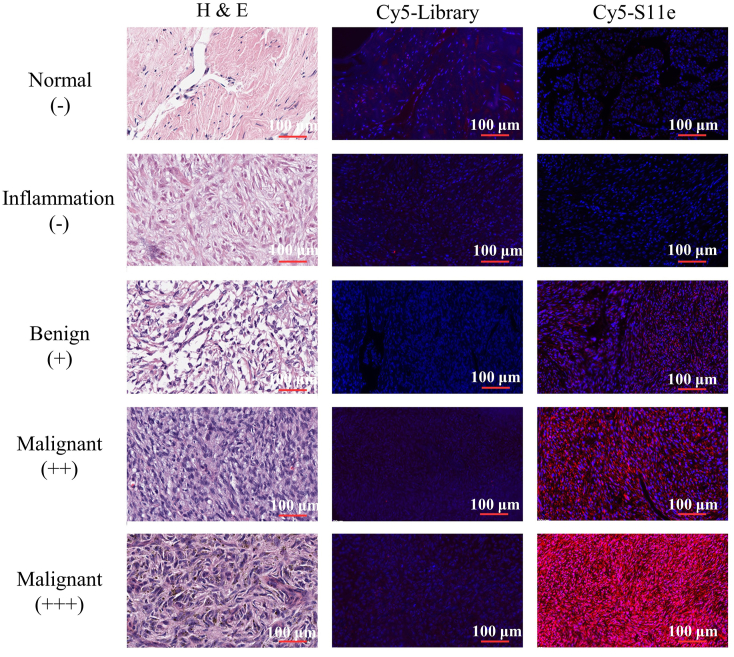
Above experiments have shown that S11e is able to recognize HT1080 fibrosarcoma cells in vivo and in vitro. Next, we speculated that S11e could also recognize tumor tissues of fibrosarcoma. Cy5-labeled S11e and Cy5-labeled Library were incubated with fibrous histopathological slides, respectively. These slides contain 20 malignant fibrous histiocytoma tissues, 20 malignant fibrosarcoma tissues, 25 Dermatofibrosarcoma protuberances, 1 fibroxanthoma tissues, 4 myofibroma tissues, 4 fibrous histiocytoma tissues, 24 fibrous tissues, 4 fasciitis tissues and 2 normal tissues. The fluorescence results obtained by scanning with a Panoramic SCAN (3DHISTECH) are shown in Fig. 5. There was no red Cy5 fluorescence signal in normal tissues, while slight red fluorescence signal in benign cancer tissues and strong red fluorescence signal in malignant cancer tissues were observed. These results suggest that S11e can recognize fibrosarcoma tissue in vitro. Statistical analysis of fluorescence signals obtained from the combination of S11e and Library on tissue microarray were summarized in Table S3. This analysis reveals that S11e has little recognition ability to normal tissues, mild recognition ability to inflammatory tissues (25%), moderate recognition ability to benign cancer tissues (45%) and the strongest recognition ability to malignant cancer tissues (74%).
Fig. 5.
S11e recognizes tumor cells in patient tissue samples. Fibrosarcoma tissue array were incubated with Cy5-labeled S11e or randomized Library. Fluorescence signals were detected with Panoramic scan.
3.6. Biological function test
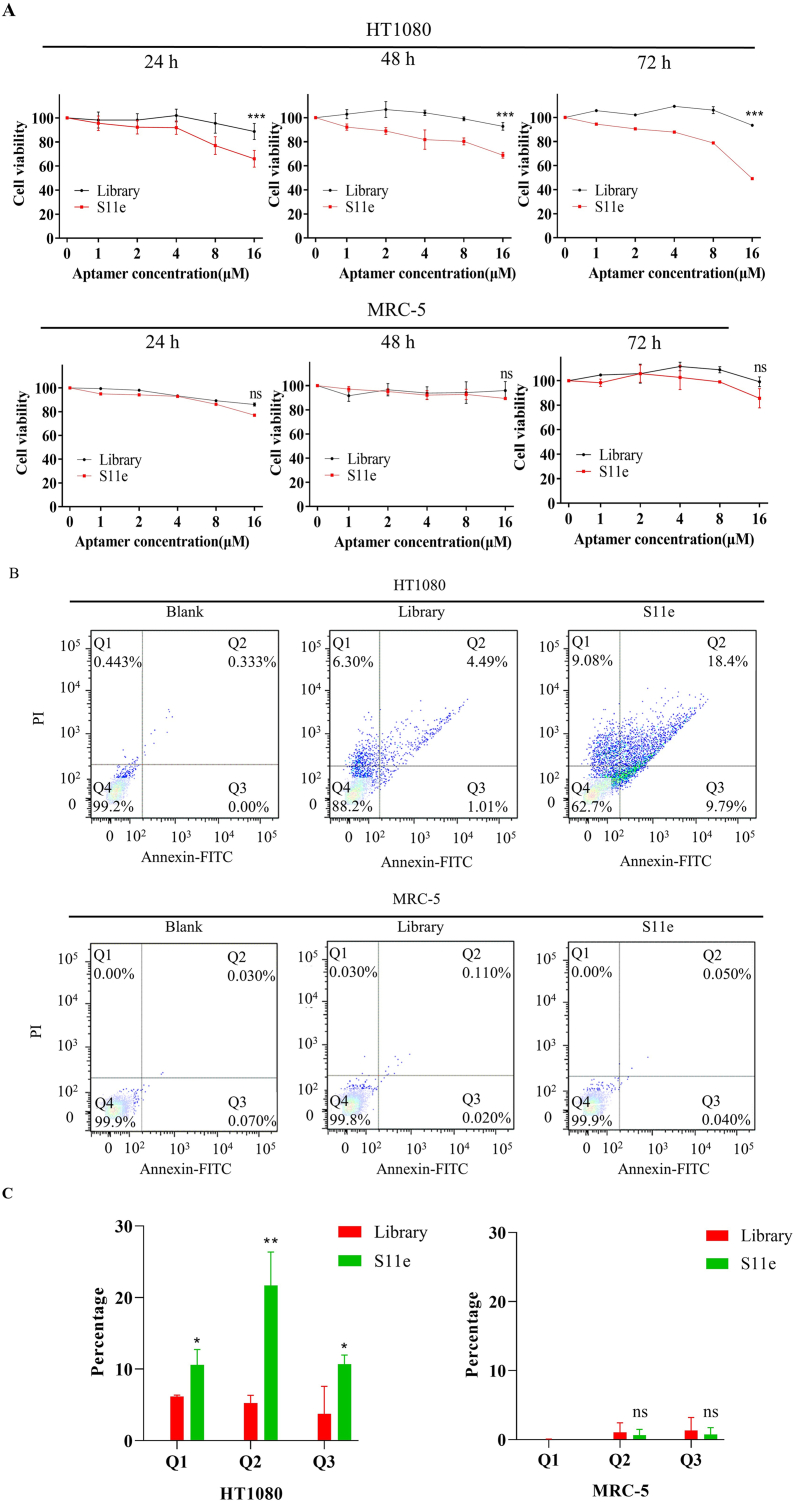
To investigate the biological function of S11e, cytotoxicity experiments were carried out. The 16 μM S11e and Library were added to HT1080 cells and treated for 24 h, 48 h and 72 h at 37 °C, respectively. After 24 h, the viability of HT1080 cells treated with S11e was lower than that of HT1080 cells treated with Library.
After 48 h, the viability of HT1080 cells in S11e group decreased by about 25% compared with that in Library group. The viability of HT1080 cells was 49.08% or 93.42% after treated with S11e or Library, respectively, suggesting that S11e but not Library has a cytotoxic effect on HT1080 cells (Fig. 6A).
Fig. 6.
S11e promotes apoptosis of fibrosarcoma cells. (A) HT1080 and MRC-5 cells were incubated with the indicated 16 μM concentrations of S11e or randomized Library. After adding CCK-8, the cytotoxicity of 24 h, 48 h, and 72 h treatment was detected by microplate reader (Comparison, S11e vs Library: ***p < 0.001; ns, not significant). (B) Detection of apoptotic effects of aptamers on HT1080 cells and MRC-5 cells by flow cytometry. (C) Statistical evaluation of apoptotic flow cytometry results (Comparison, S11e vs Library: *p < 0.05; **p < 0.01; ns, not significant).
As shown in Fig. 6B, the apoptosis was detected by flow cytometry using an Annexin V-FITC Apoptosis Detection Kit. The early apoptosis and late apoptosis ratios of HT1080 cells line treated with S11e increased to 9.08% and 9.79%, respectively, which were much higher than that of blank (0.443%, 0.00%) or Library (6.30, 1.01%). However, the early apoptosis and late apoptosis ratios of MRC-5 cells line treated with S11e were only 0.00% and 0.04%, respectively, which showed no significant difference compared with blank (0.00%, 0.07%) or Library (0.03%, 0.02%). Collectively, the results suggest that S11e effectively induces apoptosis in HT1080 cells indicating S11e′s potential therapeutic effect against tumor cells.
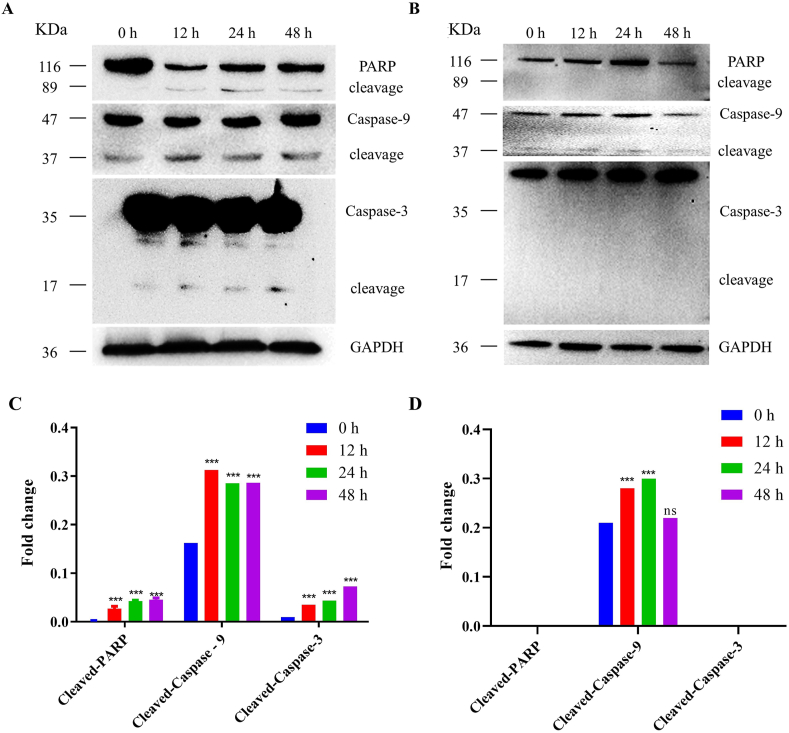
Next, we studied the pathway of S11e inducing apoptosis in HT1080 cells via Western blotting experiment. As shown in Fig. 7, the expression of cleaved-Caspase-9 (37 kDa), cleaved-Caspase-3 (17 kDa) and cleaved-PARP (89 kDa) increased in HT1080 cells after incubation with S11e. When Caspase-9 is activated, it activates Caspase-3, which further cleaves PARP, a key enzyme in apoptosis, and directly induces apoptosis. These results suggest that S11e specifically induces apoptosis through mitochondrial-mediated endogenous apoptotic pathway.
Fig. 7.
S11e regulates mitochondrial-mediated endogenous apoptotic pathway in HT1080 cells. Western blotting was used to analyze the expression of cleaved-PARP, cleaved-Caspase-9 and cleaved– Caspase-3 in HT1080 cells treated with S11e (A) or Library (B) for 0 h, 12 h, 24 h, 48 h. Quantified data of Western blotting results in An under treatment by S11e (C) and Library (D) were analyzed statistically (Compared to 0 h: ***p < 0.001; ns, not significant).
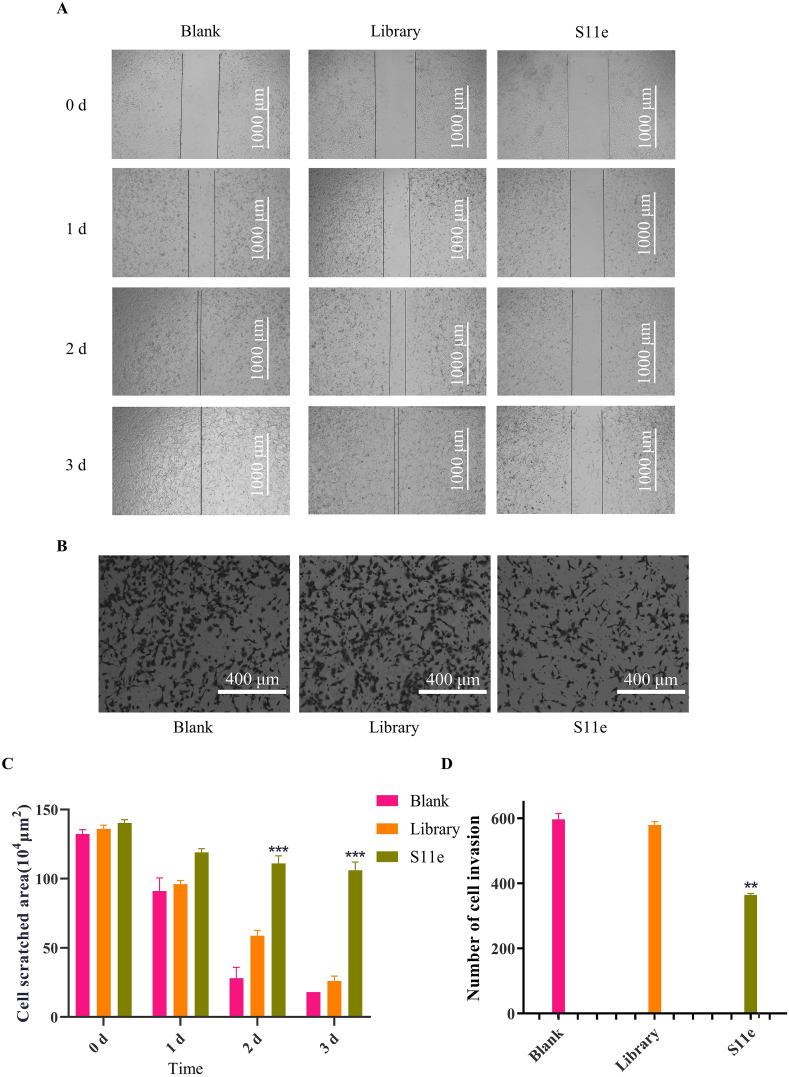
HT1080 is a highly metastatic cell line, which is often used in cell migration and invasion studies. We used scratch test to measure the effect of S11e on tumor cell migration. Scratches of HT1080 cells treated with medium healed completely at the second day. Scratches of HT1080 cells treated with Library healed 43% at the second day and almost healed completely at the third day. Scratches treated with S11e did not heal significantly even after 3 days (Fig. 8A). These results suggest that S11e significantly inhibits the migration of HT1080 cells (Fig. 8C).
Fig. 8.
S11e inhibits the migration and invasion of HT1080 cells. (A) The cell migration was monitored for 3 days, the representative images captured by an inverted microscope were shown here. (B) The number of invasive cells in a transwell chamber after treatment with S11e and Library for 24 h was observed by an inverted microscope. The representative images were shown here. (C) Statistically analyzed data of A (Comparison, S11e vs Library: ***p < 0.001). (D) Statistically analyzed data of B (Comparison, S11e vs Library: **p < 0.01; ***p < 0.001).
We also conducted cell invasion test experiments (Fig. 8B). In HT1080 cells treated with S11e, the number of cells crossing the transwell chamber (363.5) was significantly lower than that in cells treated with culture medium (596.5) or Library (579.5), which proved that S11e could inhibit the invasion of HT1080 tumor cells (Fig. 8D).
3.7. Identification of S11e-binding protein through Mass Spectrometry Analysis
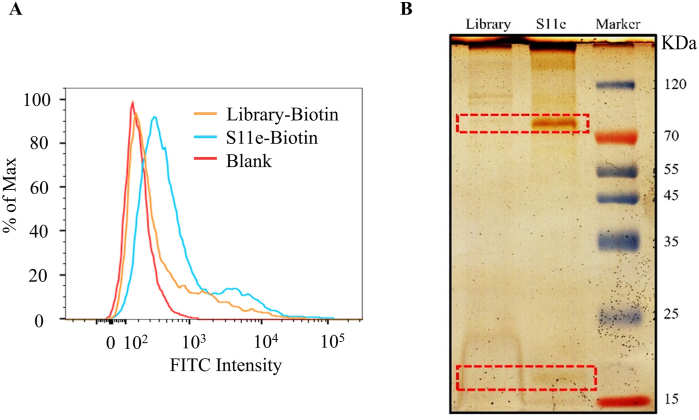
We next focused on the identification of functional cellular proteins that S11e binds to. HT1080 cells were incubated with S11e for 1 h. Total membrane proteins of HT1080 cells were extracted and incubated with biotin-labeled S11e or biotin-labeled Library, respectively. After analysis via flow cytometry assay, we first confirmed that biotin modification of S11e did not affect its binding to HT1080 cells (Fig. 9A). The binding complex was then separated by using streptavidin-coated agarose gel beads and analyzed through SDS-PAGE gel. Intriguingly, two protein bands with molecular weights of ∼85 kDa and ∼20 kDa were specifically detected in S11e treated cells, but not in Library-treated cells (Fig. 9B). The protein bands were then cut, trypsin digested and subjected to LC-MS/MS QSTAR analysis. The MS results showed a list of protein hits, which were shown in Table S4. Among these candidates, we focused on proteins with molecular weight between 15-25 kDa and 70–90 kDa, and then ranked them with the score ratio of S11e to Library. We found that Diablo/SMAC (21 kDa), which can be released from the mitochondrial intermembrane space to cytosol after an apoptotic trigger, ranked high compared to other hits. Therefore, we speculated Diablo/SMAC as a potential binding target of S11e.
Fig. 9.
S11e-binding proteins were analyzed by mass spectrometry. (A) The binding of biotin-modified S11e and Library to HT1080 cells was measured by flow cytometry. (B) Silver-stained SDS-PAGE was used to analyze aptamer target purification. Lane Library, protein captured with Library-treated sample; Lane S11e, protein captured with S11e-treated sample.
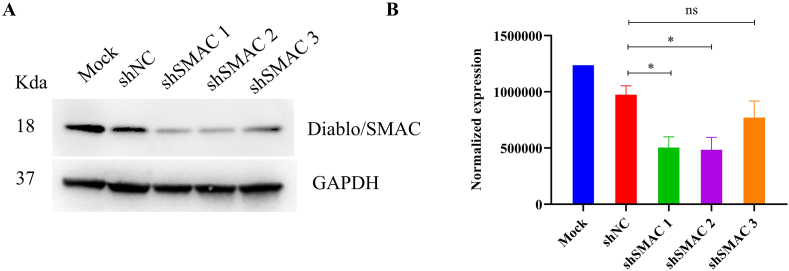
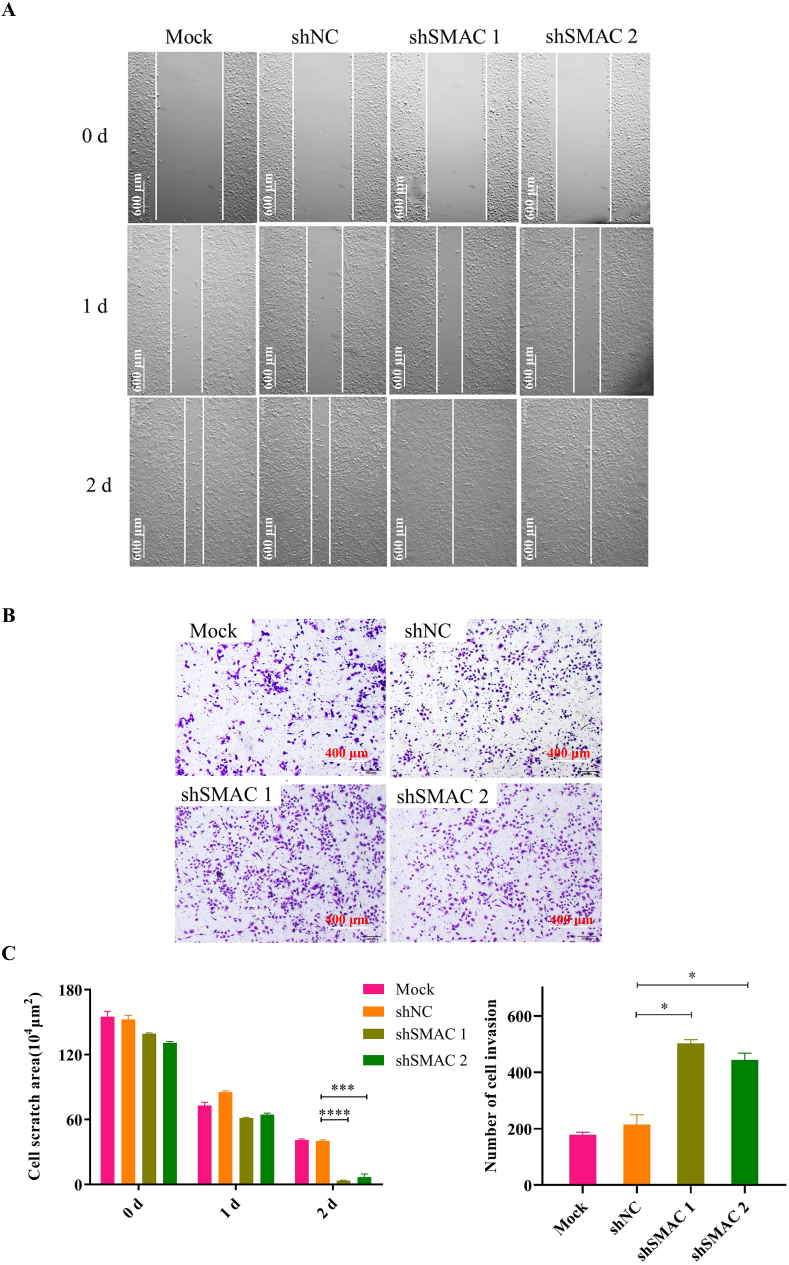
To further confirm Diablo/SMAC is a functional binding target of S11e, we performed a loss-of-function analysis using three independent Diablo/SMAC-specific shRNAs. As shown in Fig. 10, shSMAC 1 and shSMAC 2 showed more significant knockdown effects on Diablo/SMAC compared to the control samples (Mock or negative control shRNA). We also conducted cell scratch test and invasion assay of SMAC silenced HT1080 cells treated with S11e (Fig. 11A and B). The results showed that the inhibition of cell migration and invasion in HT1080 cells by S11e was profoundly abolished in samples treated with shSMAC 1 and shSMAC 2. This result supports that SMAC is a functional binding target of S11e in fibrosarcoma cells.
Fig. 10.
Knockdown effects of Diablo/SMAC shRNAs. (A) HT1080 cells were infected with the indicated shRNAs. The infected cell extracts were analyzed using Western blotting with anti-Diablo/SMAC antibody. (B) Bar graph showing quantification of (A). (Comparison, shSMAC vs shNC: *p < 0.05; ns, not significant). NC: negative control.
Fig. 11.
Diablo/SMAC is identified as a functional target for S11e. After HT1080 cells were treated with the indicated shRNAs (shSMAC 1 and shSMAC 2), cell scratch test and invasion assay were conducted. The representative images of scratch test (A) and invasion assay (B) were shown. (C) Bar graphs showing quantification of triplicate results of A and B, respectively (Comparison, shSMAC vs shNC: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). NC: negative control.
4. Discussion
In this study, with a non-selective strategy using a known sequence pool of ssDNA aptamers, we identified that S11e, which was previously obtained by SELEX for NSCLC [26], can bind to fibrosarcoma HT1080 cells but not to human embryonic lung fibroblast MRC-5 cells. Also, S11e can recognize fibrosarcoma patient tissues in vitro and accumulate in the xenograft tumors in mice. Therefore, we report here an aptamer, i.e., S11e, that specifically recognizes fibrosarcoma.
Some aptamers are reported to be degraded by lysosome after entering the cytoplasm [28]. Interestingly, we found in this study that S11e located in mitochondria but not lysosome via fluorescence confocal microscopy imaging technique. We speculated that S11e may locate in mitochondria through ATP/ADP or other non-lysosomal transport pathways. It makes S11e a promising tool for targeting mitochondrial in cellular instead of conventional mitochondrial targeting strategies, which can protect the loading-drugs from being degraded by lysosomes [29]. However, the internalization mechanism of S11e still remains to be further studied.
In recent years, studies have shown that mitochondria play an important role in cell apoptosis [30]. When stimulated by apoptotic signals, mitochondria release cytochrome c into the cytoplasm and bind to Apaf-L to initiate the caspase cascade reaction. Cytochrome c/Apaf-1 complex activates caspase-9, which in turn activates caspase-3 to induce apoptosis [31]. We found that Caspase-9, Caspase-3 and PARP were cleaved in S11e-treated HT1080 cells via Western blotting analysis. Diablo/SMAC is closely related with apoptotic pathways in mitochondria [32], we hypothesize that Diablo/SMAC is involved in the apoptotic function of S11e. However, Diablo/SMAC is immature in inner mitochondria and the mechanism that S11e induces apoptosis via Diablo/SMAC will be an intriguing question to be further studied.
Migration and invasion of malignant tumor cells is the obstacle for cancer treatment. The results of toxicity test and scratch test showed that S11e also could promote cell apoptosis and inhibit migration. Moreover, after Diablo/SMAC knockdown, the S11e inhibited migration and invasion was abolished. However, further studies may explore the correlation between Diablo/SMAC and migration-related pathways such as Akt pathway [33], which may provide new insights regarding the use of S11e as an effective prognostic tool in the clinical treatment of fibrosarcoma.
To sum up, the bio-functional aptamer with both in vivo and in vitro is rarely obtained by conventional cell-SELEX or protein-SELEX. Herein, we show that S11e exhibits high binding affinity and specificity to fibrosarcoma HT1080 cells. In addition, S11e is internalized into HT1080 cells independent of lysosome pathway and located in mitochondria. Furthermore, S11e promotes the apoptosis and inhibits the migration of HT1080 cells. Finally, we demonstrate that Diablo/SMAC protein is a biologically functional cellular target of S11e, suggesting that Diablo/SMAC is a potential therapeutic target for fibrosarcoma.
CRediT authorship contribution statement
Yunyi Liu: Investigation, Methodology, Formal analysis, Validation, Writing – original draft, Data curation. Cheng Peng: Investigation, Methodology, Formal analysis, Validation, Writing – original draft, Data curation. Hui Zhang: Investigation, Methodology, Software, Writing – review & editing. Juan Li: Investigation, Methodology, Software, Writing – review & editing. Hailong Ou: Investigation, Methodology, Software, Writing – review & editing. Yang Sun: Investigation, Methodology, Software, Writing – review & editing. Chaoqi Wen: Investigation, Methodology, Software, Writing – review & editing. Dan Qi: Formal analysis, Writing – original draft, Supervision, Writing – review & editing. Xiaoxiao Hu: Conceptualization, Resources, Data curation, Writing – original draft, Supervision, Writing – review & editing, Funding acquisition. Erxi Wu: Conceptualization, Resources, Data curation, Writing – original draft, Supervision, Writing – review & editing, Funding acquisition. Weihong Tan: Conceptualization, Resources, Data curation, Writing – original draft, Supervision, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors have no competing interest to declare.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31701249, 31970692, 81803068, 91953000, 21827811), Changsha Science and Technology Plan (kq2004009), Corbett Estate Fund for Cancer Research, the Hunan Provincial Key Area R&D Program (2019SK2201), Hunan Science and Technology Creative Program (2017XK2103), and the National Key R&D Program of China (2020YFA0909000). The authors would like to thank Euni Wu for the language editing of this manuscript.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.10.011.
Contributor Information
Xiaoxiao Hu, Email: xxhu@hnu.edu.cn.
Erxi Wu, Email: erxi.wu@bswhealth.org.
Appendix ASupplementary data
The following are the Supplementary data to this article:
References
- 1.Murphey M.D. World Health Organization classification of bone and soft tissue tumors: modifications and implications for radiologists. Semin. Muscoskel. Radiol. 2007;11:201–214. doi: 10.1055/s-2008-1038310. [DOI] [PubMed] [Google Scholar]
- 2.Meral O., Uysal H. Comparative proteomic analysis of fibrosarcoma and skin fibroblast cell lines. Tumour Biol. 2015;36:561–567. doi: 10.1007/s13277-014-2672-8. [DOI] [PubMed] [Google Scholar]
- 3.Toro J.R., Travis L.B., Wu H.J., Zhu K., Fletcher C.D., Devesa S.S. Incidence patterns of soft tissue sarcomas, regardless of primary site, in the surveillance, epidemiology and end results program, 1978-2001: an analysis of 26,758 cases. Int. J. Cancer. 2006;119:2922–2930. doi: 10.1002/ijc.22239. [DOI] [PubMed] [Google Scholar]
- 4.Augsburger D., Nelson P.J., Kalinski T., Udelnow A., Knösel T., Hofstetter M., Qin J.W., Wang Y., Gupta A.S., Bonifatius S., Li M., Bruns C.J., Zhao Y. Current diagnostics and treatment of fibrosarcoma -perspectives for future therapeutic targets and strategies. Oncotarget. 2017;8:104638–104653. doi: 10.18632/oncotarget.20136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X., Chen J., Wu M., Zhao J.X. Aptamers: active targeting ligands for cancer diagnosis and therapy. Theranostics. 2015;5:322–344. doi: 10.7150/thno.10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Civit L., Taghdisi S.M., Jonczyk A., Haßel S.K., Gröber C., Blank M., Stunden H.J., Beyer M., Schultze J., Latz E., Mayer G. Systematic evaluation of cell-SELEX enriched aptamers binding to breast cancer cells. Biochimie. 2018;145:53–62. doi: 10.1016/j.biochi.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Qu H., Csordas A.T., Wang J., Oh S.S., Eisenstein M.S., Soh H.T. Rapid and label-free strategy to isolate aptamers for metal ions. ACS Nano. 2016;10:7558–7565. doi: 10.1021/acsnano.6b02558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gopinath S.C., Sakamaki Y., Kawasaki K., Kumar P.K. An efficient RNA aptamer against human influenza B virus hemagglutinin. J. Biochem. 2006;139:837–846. doi: 10.1093/jb/mvj095. [DOI] [PubMed] [Google Scholar]
- 9.Xuan W., Xia Y., Li T., Wang L., Liu Y., Tan W. Molecular self-assembly of bioorthogonal aptamer-prodrug conjugate micelles for hydrogen peroxide and pH-independent cancer chemodynamic therapy. J. Am. Chem. Soc. 2020;142:937–944. doi: 10.1021/jacs.9b10755. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y., Ou H., Pei X., Jiang B., Ma Y., Liu N., Wen C., Peng C., Hu X. Chemo-drug controlled-release strategies of nanocarrier in the development of cancer therapeutics. Curr. Med. Chem. 2020 doi: 10.2174/0929867327666200605153919. [DOI] [PubMed] [Google Scholar]
- 11.Jaffe G.J., Eliott D., Wells J.A., Prenner J.L., Papp A., Patel S. A phase 1 study of intravitreous E10030 in combination with ranibizumab in neovascular age-related macular degeneration. Ophthalmology. 2016;123:78–85. doi: 10.1016/j.ophtha.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Lao Y.H., Phua K.K., Leong K.W. Aptamer nanomedicine for cancer therapeutics: barriers and potential for translation. ACS Nano. 2015;9:2235–2254. doi: 10.1021/nn507494p. [DOI] [PubMed] [Google Scholar]
- 13.Rosenberg J.E., Bambury R.M., Van Allen E.M., Drabkin H.A., Lara P.N., Jr., Harzstark A.L., Wagle N., Figlin R.A., Smith G.W., Garraway L.A., Choueiri T., Erlandsson F., Laber D.A. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest. N. Drugs. 2014;32:178–187. doi: 10.1007/s10637-013-0045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwoebel F., van Eijk L.T., Zboralski D., Sell S., Buchner K., Maasch C., Purschke W.G., Humphrey M., Zöllner S., Eulberg D., Morich F., Pickkers P., Klussmann S. The effects of the anti-hepcidin Spiegelmer NOX-H94 on inflammation-induced anemia in cynomolgus monkeys. Blood. 2013;121:2311–2315. doi: 10.1182/blood-2012-09-456756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung E., Landa G. Update on current and future novel therapies for dry age-related macular degeneration. Expet Rev. Clin. Pharmacol. 2013;6:565–579. doi: 10.1586/17512433.2013.829645. [DOI] [PubMed] [Google Scholar]
- 16.Jilma B., Paulinska P., Jilma-Stohlawetz P., Gilbert J.C., Hutabarat R., Knöbl P. A randomised pilot trial of the anti-von Willebrand factor aptamer ARC1779 in patients with type 2b von Willebrand disease. Thromb. Haemostasis. 2010;104:563–570. doi: 10.1160/th10-01-0027. [DOI] [PubMed] [Google Scholar]
- 17.Morita Y., Leslie M., Kameyama H., Volk D.E., Tanaka T. Aptamer therapeutics in cancer: current and future. Cancers. 2018;10 doi: 10.3390/cancers10030080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoellenriegel J., Zboralski D., Maasch C., Rosin N.Y., Wierda W.G., Keating M.J., Kruschinski A., Burger J.A. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood. 2014;123:1032–1039. doi: 10.1182/blood-2013-03-493924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Troisi R., Napolitano V., Spiridonova V., Russo Krauss I., Sica F. Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer. Nucleic Acids Res. 2018;46:12177–12185. doi: 10.1093/nar/gky990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng E.W., Shima D.T., Calias P., Cunningham E.T., Jr., Guyer D.R., Adamis A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe T., Hirano K., Takahashi A., Yamaguchi K., Beppu M., Fujiki H., Suganuma M. Nucleolin on the cell surface as a new molecular target for gastric cancer treatment. Biol. Pharm. Bull. 2010;33:796–803. doi: 10.1248/bpb.33.796. [DOI] [PubMed] [Google Scholar]
- 22.Soundararajan S., Chen W., Spicer E.K., Courtenay-Luck N., Fernandes D.J. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68:2358–2365. doi: 10.1158/0008-5472.Can-07-5723. [DOI] [PubMed] [Google Scholar]
- 23.Maasch C., Buchner K., Eulberg D., Vonhoff S., Klussmann S. Physicochemical stability of NOX-E36, a 40mer L-RNA (Spiegelmer) for therapeutic applications. Nucleic Acids Symp. Ser. 2008:61–62. doi: 10.1093/nass/nrn031. [DOI] [PubMed] [Google Scholar]
- 24.Verhagen A.M., Vaux D.L. Cell death regulation by the mammalian IAP antagonist Diablo/Smac. Apoptosis. 2002;7:163–166. doi: 10.1023/a:1014318615955. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H., Wang Z., Xie L., Zhang Y., Deng T., Li J., Liu J., Xiong W., Zhang L., Zhang L., Peng B., He L., Ye M., Hu X., Tan W. Molecular recognition and in-vitro-targeted inhibition of renal cell carcinoma using a DNA aptamer. Mol. Ther. Nucleic Acids. 2018;12:758–768. doi: 10.1016/j.omtn.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Z., Xu L., Shi X., Tan W., Fang X., Shangguan D. Recognition of subtype non-small cell lung cancer by DNA aptamers selected from living cells. Analyst. 2009;134:1808–1814. doi: 10.1039/b904476k. [DOI] [PubMed] [Google Scholar]
- 27.Karlsen K.K., Wengel J. Locked nucleic acid and aptamers. Nucleic Acid Therapeut. 2012;22:366–370. doi: 10.1089/nat.2012.0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong W., Pu Y., Tan W., Liu J., Liao J., Liu B., Chen K., Yu B., Hu Y., Deng Y., Zhang J., Liu H. Identification and application of an aptamer targeting papillary thyroid carcinoma using tissue-SELEX. Anal. Chem. 2019;91:8289–8297. doi: 10.1021/acs.analchem.9b01000. [DOI] [PubMed] [Google Scholar]
- 29.Fulda S., Galluzzi L., Kroemer G. Targeting mitochondria for cancer therapy. Nat. Rev. Drug Discov. 2010;9:447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 30.Chan D.C. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Su Y.C., Wu J.L., Hong J.R. Betanodavirus up-regulates chaperone GRP78 via ER stress: roles of GRP78 in viral replication and host mitochondria-mediated cell death. Apoptosis. 2011;16:272–287. doi: 10.1007/s10495-010-0565-x. [DOI] [PubMed] [Google Scholar]
- 32.Shoshan-Barmatz V., Keinan N., Abu-Hamad S., Tyomkin D., Aram L. Apoptosis is regulated by the VDAC1 N-terminal region and by VDAC oligomerization: release of cytochrome c, AIF and Smac/Diablo. Biochim. Biophys. Acta. 2010;1797:1281–1291. doi: 10.1016/j.bbabio.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Goetze S., Bungenstock A., Czupalla C., Eilers F., Stawowy P., Kintscher U., Spencer-Hänsch C., Graf K., Nürnberg B., Law R.E., Fleck E., Gräfe M. Leptin induces endothelial cell migration through Akt, which is inhibited by PPARgamma-ligands. Hypertension. 2002;40:748–754. doi: 10.1161/01.hyp.0000035522.63647.d3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.