Abstract
Tumor cells have unique metabolic programming that is biologically distinct from that of corresponding normal cells. Resetting tumor metabolic programming is a promising strategy to ameliorate drug resistance and improve the tumor microenvironment. Here, we show that carboxyamidotriazole (CAI), an anticancer drug, can function as a metabolic modulator that decreases glucose and lipid metabolism and increases the dependency of colon cancer cells on glutamine metabolism. CAI suppressed glucose and lipid metabolism utilization, causing inhibition of mitochondrial respiratory chain complex I, thus producing reactive oxygen species (ROS). In parallel, activation of the aryl hydrocarbon receptor (AhR) increased glutamine uptake via the transporter SLC1A5, which could activate the ROS-scavenging enzyme glutathione peroxidase. As a result, combined use of inhibitors of GLS/GDH1, CAI could effectively restrict colorectal cancer (CRC) energy metabolism. These data illuminate a new antitumor mechanism of CAI, suggesting a new strategy for CRC metabolic reprogramming treatment.
KEY WORDS: CAI, Glutaminolysis, Glutamine metabolism, AhR, Colorectal cancer metabolism, Mitochondrial oxidative stress, Redox homeostasis, Metabolic reprogramming
Abbreviations: 2-NBDG, glucalogue 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose; AhR, aryl hydrocarbon receptor; ATP, adenosine triphosphate; CAI, carboxyamidotriazole; CHIP, chromatin immunoprecipitation; CRC, colorectal cancer; DMF, 3′,4′-dimethoxyflavone; DNA, deoxyribonucleic acid; ECAR, extracellular acidification rate; FACS, flow cytometry; GDH1, glutamate dehydrogenase 1; GLS, glutaminase; GPx, glutathione peroxidase; GSH, glutathione; GSSG, oxidized glutathione; Kyn, kynurenine; Mito-Q, mitoquinone mesylate; MT, mito-TEMPO; OCR, oxygen consumption rate; TCA, tricarboxylic acid; α-KG, α-ketoglutarate
Graphical abstract
The cytostatic agent CAI suppresses glucose and lipid metabolism utilization and further increases tumour cell dependency on glutamine metabolism, suggesting a synergistic anticancer strategy with glutamine metabolic pathway inhibitors.
1. Introduction
Tumor cells proliferate quickly, evade immune surveillance, in some cases moving to other parts of the body, and resist drug attacks while also finding ways to overcome anticancer process. The biological characteristics of tumors are closely related to material and energy metabolism1,2. If we had a way to selectively control certain metabolic pathways in tumor cells, it would be possible to block the corresponding malignant phenotype. Tumor cells have biological characteristics that differ from those of normal cells3,4. They certainly do not faithfully adopt the metabolic patterns of corresponding normal cells and must change their ways. We call these metabolic changes “metabolic reprogramming”5,6. Moreover, metabolic pathways are reprogrammed and further shaped by the tumor microenvironment to meet the material and energy requirements of tumor growth7.
Tumor cells have a unique metabolic need for amino acids to support their rapid growth8,9. Amino acids are not only essential for protein synthesis but also a source of carbon and nitrogen in the synthesis of purine and pyrimidine nucleotides, amino sugars, and glutathione (GSH)8. To meet these needs, transporters that differ in substrate selectivity, kinetics, and ion dependence are employed10,11. Tumor cells must be intelligently selected to regulate specific transporters to meet their amino acid nutritional requirements. Glutamine is an essential amino acid that plays an important role in the metabolism of tumor cells12, 13, 14. Specifically, blocking tumor cell glutamine metabolism could disrupt tumor metabolism. If the “Warburg effect” of a tumor is paralysed, the nutrient deficiency of the tumor microenvironment can be reversed.
Carboxyamidotriazole (CAI) is a small-molecule compound with antitumor activity that has been demonstrated to inhibit the growth of a variety of cancer cell lines15,16. Intriguingly, recent evidence indicates that metabolic reprogramming appears in tumor cells after CAI treatment. However, whether CAI exerts a metabolism-modifying effect remains unexplored. In the present study, we provide evidence that CAI functions as a metabolic modulator, increasing tumor cell dependency on glutamine metabolism and tumor cell sensitivity to glutamine metabolic pathway inhibitors. On this basis, the mechanism of action was further confirmed.
2. Materials and methods
2.1. Cell lines and animals
The colon carcinoma tumor cell lines C26 and HCT116 were purchased from the China Center for Type Culture Collection (Beijing, China) and cultured in RPMI 1640 medium supplemented with 10% foetal bovine serum (FBS).
Female nude mice, BALB/c mice and NOD-SCID mice (6–8 weeks old, 18–22 g) were purchased from the Center of Medical Experimental Animals of the Chinese Academy of Medical Sciences (Beijing, China). All animal studies and procedures were approved by the Institutional Animal Care and Use Committee of Peking Union Medical College (registration number: ACUC-A02-2017-013). Safety evaluation of drugs was determined by haematoxylin and eosin (HE) staining.
2.2. Reagents
CAI was synthesized by the Institute of Materia Medica, Chinese Academy of Medical Sciences (Beijing, China). l-Glutamine-14C5, rotenone (RTN) and 3-nitropropionic acid (3-NPA) were purchased from Sigma–Aldrich (MO, USA). V-9302 and CB-839 were purchased from Selleck Chemicals (CA, USA). Mitoquinone mesylate, Mito-TEMPO and R162 was purchased from MedChemExpress (NJ, USA). MitoTracker Red, BODIPY FL C16 (4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-hexadecanoic acid), 2-NBDG and MitoSOX Red were purchased from Thermo Fisher (MA, USA).
2.3. Patient samples
Human colon cancer tissue specimens were obtained from patients at Peking Union Medical College Hospital. Ethical permission was granted by the Clinical Trial Ethics Committee of Peking Union Medical College Hospital, and written informed consent was obtained from the enrolled patients. The relevant clinical and histopathological data provided to the researchers were anonymized.
2.4. Seahorse XF24 bioenergetic assays
A Seahorse XF24 Extracellular Flux Analyser (Agilent Technologies, CA, USA) was used to measure the OCR and ECAR and perform a fuel flex test. Briefly, cells with or without CAI treatment were seeded in XF24 plates, and then the plates were detected according to the instructions of the XFp Cell Mito Stress Test Kit (103015-100), XFp Glycolysis Stress Test Kit (103020-100) or XFp Mito Fuel Flex Test Kit (103270-100). Data were assessed with XFe Wave Software. The cell number was quantified as described, and all data were normalized to the cell number.
2.5. RT-PCR
Total RNA was isolated from cells using the Pure RNA Extraction Kit (Bio Teke Corporation) and reverse transcribed into cDNA by using TransScript First-Strand cDNA Synthesis Supermix (TransGen Biotech Co., Beijing, China). The primer sequences are listed in Supporting Information Table S1. mRNA levels were normalized to the β-actin mRNA level. Real-time PCR was performed using an ABI StepOne Plus (Applied Biosystems, MA, USA). Values are reported as the mean ± standard error of mean (SEM) of three independent experiments performed in duplicate. Statistical comparisons among groups were performed using ANOVA followed by Fisher’s PLSD test. The differences in all parameters were considered statistically significant at a value of P < 0.05.
2.6. siRNA experiments
siRNAs and control siRNA were purchased from RiboBio and transfected into HCT116 cells using Lipofectamine 2000 Transfection Reagent (Invitrogen) at a final concentration of 100 nmol/L. Twenty-four hours after transfection, the cells were treated with the indicated drugs for an additional 24 h, and mRNA was detected by real-time PCR. Knockdown of the expression of the indicated genes was verified by real-time PCR.
2.7. Western blotting
Cells were collected and lysed with lysis buffer supplemented with a protease and phosphatase inhibitor cocktail (Thermo Fisher, MA, USA). Then, the proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted. The antibodies used are as follows: anti-SLC1A5 (SAB2108565, Sigma, MO, USA), anti-SLC7A5 (PA5-50485, Thermo Fisher, MA, USA), anti-glutamine (ab93434, Abcam, UK), anti-glutamate dehydrogenase (PA5-28301, Thermo Fisher, MA, USA), anti-caspase 3, anti-cleaved caspase 3, anti-caspase 6, anti-cleaved caspase 6, anti-caspase 7, anti-cleaved caspase 7, and anti-β-actin (Cell Signaling Technology, MA, USA). Secondary antibodies conjugated to horseradish peroxidase were subsequently incubated with the membranes, followed by enhanced chemiluminescence development (Thermo Fisher, MA, USA). Three independent experiments were performed for each analysis. Co-immunoprecipitation was performed using the Pierce Classic Magnetic IP/Co-IP Kit (Thermo Fisher, MA, USA). Anti-AhR (Abcam, Ab2769) and IgG isotype control (Cell Signaling Technology, 3900) antibodies were used.
2.8. Immunofluorescence
For immunofluorescence staining of cultured cells, HCT116 and C26 cells were plated on fibronectin (1 μg/mL)-coated coverslips, fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. The fixed cells were blocked in 5% BSA and stained using the mouse monoclonal anti-COX IV mitochondrial marker (ab33985, Abcam, UK) followed by donkey anti-mouse DyLight 594 (1:500) secondary antibodies (Invitrogen, CA, USA). Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI). Fluorescence was analyzed using a Leica laser scanning confocal microscope.
2.9. Intracellular metabolite level measurements
The intracellular levels of glutamine, glutamate, α-KG, citrate, isocitrate, succinate, fumarate, and malate were determined by using commercial kits (BioVision, CA, USA). After washing 2 times with cold PBS, 2 × 106 cells were homogenized in PBS. For the glutamine test, cells were cultured in glutamine-free medium for 6 h in advance to remove all endogenous glutamine. The supernatant was collected, and proteins were removed by using 3-KD Amicon Ultra Centrifugal Filters (Millipore, MA, USA). The flow-through containing the metabolites was used for the measurement of metabolite levels according to the manufacturer's instructions.
2.10. Enzymatic activity analysis
GLS, GDH and GPx enzyme activities were determined by using commercially available kits from BioVision according to the manufacturer’s instructions. The values were normalized to the protein concentration and then normalized to the corresponding control group value.
2.11. Glutamine uptake assay
Cells were washed with glutamine-free RPMI-1640 medium (Thermo Fisher, MA, USA) and incubated with 4 mmol/L 13C-glutamine (Sigma–Aldrich, MO, USA) in RPMI-1640 medium for 12 h. Then, the lysates were evaluated to determine the [13C] level using a scintillation counter to analyse relative glutamine uptake. Radioactivity was measured as counts per minute (CPM) and normalized to the protein concentration.
2.12. ATP content and the GSH/GSSG ratio
Following the indicated treatments, HCT116 and C26 cells were lyzed, and the ATP content in the supernatant was measured with an ATP Assay Kit (Beyotime Biotechnology, Shanghai, China). The values were normalized to the protein concentration. The GSH/GSSG ratio was determined using the Glutathione (GSH/GSSG/Total) Fluorometric Assay Kit (BioVision, CA, USA).
2.13. ROS detection
Cells were detached from plates after 48 h of treatment, washed twice with PBS buffer, and incubated with Mitosox for 20 min at 37 °C in the dark. The cells were washed and analyzed by using flow cytometry (FCM). FCM was performed using excitation/emission wavelengths of 510/580 nm for Mitosox.
2.14. Apoptosis detection
Cells were seeded and treated with the indicated drugs for 48 h, detached from the plates with Trypsin Solution without EDTA (Beyotime Biotechnology, Shanghai, China) and washed with cold PBS. The induction of apoptosis was measured by FCM using the Annexin V-PE/7-AAD Apoptosis Detection Kit (Yeasen Biotech, Shanghai, China). And for detection of DNA breakage, a TUNEL assay was performed following the protocol provided by the manufacturer (Yeasen Biotech).
2.15. Generation of knockout cell lines with CRISPR-Cas9 technology
For construction of stable-knockout HCT116 and C26 cell lines, the following sgRNAs targeting GLS1 were used: SGGFP, CACCGGGGCGAGGAGCTGTTC ACCG (sense) and AAACCGGTGAACAGCTCCTCGCCCC (antisense); GLS1-SGRNA1, GGACGCGTTTGGCAACAGCG (sense) and CGCTGTTGCCAAACGC GTCC (antisense); GLS1-SGRNA2, CGGGGAGACGGACGCGTTTG (sense) and CAAACGCGTCCGTCTCCCCG (antisense); Gls1-SGRNA1, GGGCCCCAAGGAC AGCCCGG (sense) and CCGGGCTGTCCTTGGGGCCC (antisense); Gls1-SGRNA2, CGACGCGTTCGGCAACAGCG (sense) and CGCTGTTGCCGAACGCGTCG (antisense). The following sgRNAs targeting GDH1 were used: GDH1-SGRNA1, CACCGCTACCTGGGCGAAGCGCTGT (sense) and AAACACAGCGCTTCGCC CAGGTAGC (antisense); GDH1-SGRNA2, CACCGGGGCGAAGCGCTGTTGC TGT (sense) and AAACACAGCAACAGCGCTTCGCCCC (antisense); Gdh1-SGRNA1, CACCGCCGTCTGGGCGAAGCGCTGC (sense) and AAACGCAGC GCTTCGCCCAGACGGC (antisense); Gdh1-SGRNA2, CACCGCGCTGCTGCTGT CCCGCGCC (sense) and AAACGGCGCGGGACAGCAGCAGCGC (antisense). These sgRNAs were cloned into the pL-CRISPR.EFS.GFP vector plasmid and transfected into HEK 293T cells together with the packaging plasmids psPAX2 and pMD2.G. Forty-eight hours later, the lentivirus was harvested and concentrated to infect C26 or HCT116 cells simultaneously treated with polybrene at a final concentration of 8 μg/mL. Two days later, GFP-positive cells were sorted by FCM using a BD Biosciences FACSAria III. The candidate knockout cells were verified by Western blotting.
2.16. Animal studies
Female BALB/c, nude mice or NOD-SCID mice (6 weeks old) were subcutaneously injected with appropriate amounts of the indicated tumor cells (C26 cells, HCT116 cells or patient tumor sample suspension) in the right flank. After the tumor size reached 5 mm × 5 mm, the mice were randomized into different groups (n = 6–10). Then, the mice in each group were treated with one of the following drug treatments for the indicated time: CAI (intragastric injection, 20 mg/kg/day); CB-839 (intragastric injection, 200 mg/kg once every 2 days); CAI + CB-839 (same injection regimens as those used for monotherapy); R162 (intragastric injection, 20 mg/kg/day); and CAI + R162 (same injection regimens as those used for monotherapy). The mice in the control group received an equal volume of saline as a mock treatment. Female BALB/c or nude mice (6 weeks old) were subcutaneously injected with a scramble control (left) or stable GLS1-silenced/stable GDH1-silenced (right) C26 or HCT116 cells in both flanks. When the xenograft volume reached approximately 5 mm × 5 mm, CAI (intragastric injection, 20 mg/kg/day) or vehicle treatment was begun. Tumor growth and mouse survival were recorded daily. Tumor volume was calculated according to Eq. (1):
| (1) |
2.17. ChIP-qPCR assay
HCT116 cells treated with 10 μmol/L CAI for 48 h were evaluated in a ChIP assay. The ChIP assay was performed using the ChIP-IT Express Chromatin Immunoprecipitation Kit (Active Motif, CA, USA) according to the manufacturer’s protocols. The antibodies used included anti-AhR (Abcam, Ab2769) and an IgG isotype control (Cell Signaling Technology, 3900). The SLC1A5 primer sequences are 5′-GGCATGCACGTGTCATCCTC-3′ (sense) and 5′-GGCATGCACGTGTCATCCTC-3′ (antisense). The SLC7A5 primer sequences are 5′-CAGGTCATCCTCTCATCCCTC-3′ (sense) and 5′-GCAGCCTCTCGTCAAGCCT-3′ (antisense). All sequences were designed to give amplicons <200 bp. The purified DNA (immunoprecipitated sample) was amplified by qPCR. The fold-enrichment method was used to normalize the ChIP-qPCR data.
2.18. Statistical analysis
The data shown are from one representative experiment of multiple independent experiments and reported as the mean ± SEM. All experiments were performed at least three times. The statistical significance of intergroup differences was determined by Student’s t-tests or one-way ANOVA followed by Dunnett’s t-test for multiple comparisons. P values less than 0.05 (P < 0.05) were considered significant. Analyses were conducted using GraphPad 6.0 software. Sample exclusion was never carried out.
3. Results
3.1. CAI predominantly regulates the metabolic phenotype in colon tumor cells
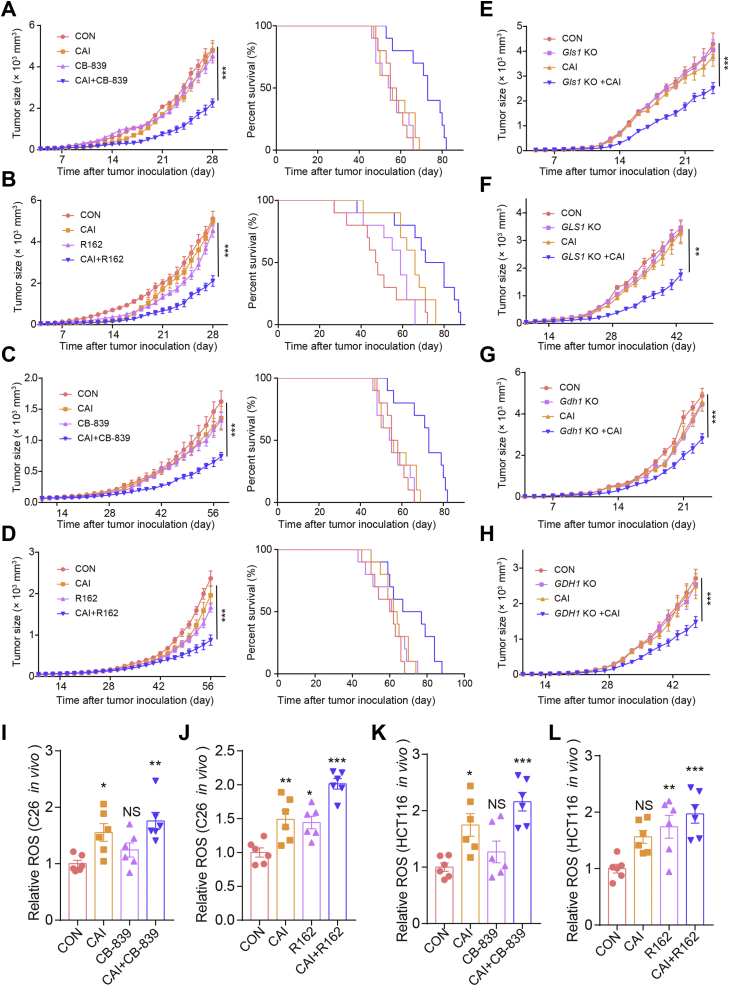
To better understand the regulatory effect of CAI on colorectal cancer (CRC) metabolism, we tested the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of HCT116 and C26 cells (Fig. 1A and B). We found that the OCR and ECAR decreased over time after CAI treatment. These impacts were apparent even over 2 h (Supporting Information Fig. S1A and S1B). Next, we tested whether CAI-induced alterations in glucose metabolism are mediated by absorption processes. We found that CAI did not affect the GLUT1 mRNA expression, but decrease the uptake of the glucalogue 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino)-2-deoxyglucose (2-NBDG) (Fig. 1C and Fig. S1C), suggesting that the inhibitory effect of CAI on glucose metabolism. We also identified that uptake of the green-fluorescing fatty acid C16-BODIPY induced by CAI was reduced, as was the upregulation of acetyl-CoA carboxylase phosphorylation (Fig. 1D and E and Fig. S1D). Intriguingly, under the same conditions, CAI caused a remarkable increase in MitoTracker staining and weak apoptosis in tumor cells (Fig. 1F–H and Fig. S1E–S1I), which suggested the majority of CRC tumor cells might be metabolically compensated under the stress of CAI. Additionally, the adenosine triphosphate (ATP) level in the CAI group was very low (Fig. 1I and Fig. S1J). This further confirmed that tumor cells adjusted metabolic patterns under CAI treatment. To further clarify the role of CAI in metabolism, we determined the dependency and flexibility of HCT116 and C26 cells to oxidize three fuel sources, glucose/glutamine/fatty acids, by measuring the real time mitochondrial respiration (OCR) of living cells in the presence or absence of fuel pathway inhibitors (Fig. 1J). The data suggest that CAI efficiently decreased the metabolism of glucose and long-chain fatty acids (LCFAs) in the mitochondria and increased the glutamine capacity. Importantly, glutamine starvation increased CAI-induced cell apoptosis (Fig. 1K and Fig. S1K), suggesting that CRC cells might initiate a new metabolic pattern that is more dependent on glutamine.
Figure 1.
CAI influenced tumor cells by changing the metabolic phenotype. (A) and (B) The OCR and ECAR in HCT116 and C26 cells were measured with or without 10 μmol/L CAI treatment for 48 h (n = 3). (C) HCT116 cells were grown in the presence or absence of CAI for 48 h. Glucose uptake was evaluated by measuring 2-NBDG uptake using flow cytometry (FACS) and the GLUT1 mRNA expression was detected by qPCR. (D) Flow cytometry plots show changes in the green-fluorescing fatty acid BODIPY FL C16. Representative histogram plots (left) and statistical histogram plots (right) are shown. (E) ACC and P-ACC levels were determined by Western blotting. (F) and (G) HCT116 cells were labelled with MitoTracker red and analyzed via flow cytometry and fluorescence microscopy. A representative image shows staining with an anti-mitochondria probe and DAPI. (H) Cell apoptosis was analysed by flow cytometry at 48 h after CAI treatment. (I) ATP levels in HCT116 cells were assessed after 48 h of incubation with or without CAI. (J) The Mito Fuel Flex test of HCT116 cells shows the dependence on and capacity for the oxidation of glucose, glutamine, or fatty acids before and after CAI treatment. (K) HCT116 cells were grown in culture medium, glucose-free medium, glutamine-free medium or serum-free medium for 6 h. Cell apoptosis was analyzed by flow cytometry. The data shown are representative of three independent experiments, and the error bars represent the mean ± SEM; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; NS, not significant (Student’s t-test).
3.2. CAI inhibits complex I and regulates cell metabolism or redox homeostasis in mitochondria of tumor cells
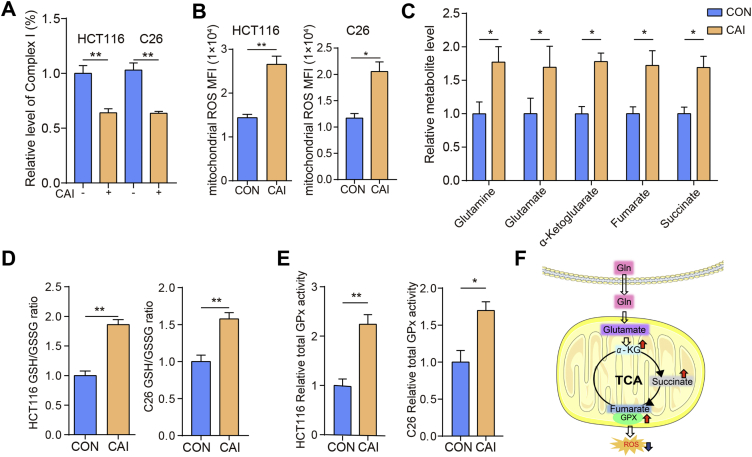
CAI restricts the production of ATP and causes metabolic changes in tumor cells, suggesting that mitochondria may be the key place where CAI works to interfere with cellular homeostasis. Therefore, we detected the activity of mitochondrial respiratory chain complexes. The activity of complex I was inhibited by CAI, inducing the production of the primary ROS superoxide (O2·–) from the mitochondrial electron transport chain (ETC, Fig. 2A and B)17. Unlike complex I, other complexes, except complex V, were not affected by CAI (Supporting Information Fig. S2A and S2B). CAI downregulated the activity of complex I, also known as ATP synthase18, consistent with the data shown in Fig. 1I and Fig. S1I. Obviously, CAI induced a large accumulation of ROS, but this did not lead to the expected large-scale cell apoptosis. Studies have suggested that ROS-induced apoptosis is also associated with decreased GSH levels and the loss of redox homeostasis19, 20, 21. Is the contradiction between ROS production and cell apoptosis related to changes in cell metabolism or redox homeostasis under CAI treatment? We further measured the levels of five key tricarboxylic acid (TCA) cycle metabolites. An increased cellular level of glutamine after CAI exposure was observed, as well as the abundance of glutamate, α-ketoglutarate (α-KG), fumarate and succinate (Fig. 2C). We also observed that CAI significantly increased the GSH levels in HCT116 and C26 cells (Fig. 2D). The enzymatic activity of total glutathione peroxidase (GPx) was significantly upregulated in tumor cells stimulated with CAI (Fig. 2E). These results indicate that CAI treatment might trigger glutamine uptake and catabolism in CRC tumor cells and achieve TCA cycle anaplerosis, which contributes to the production of α-KG and its downstream metabolites succinate and fumarate (Fig. 2F). TCA cycle anaplerosis from glutamine maintains the levels of ATP and GSH, plus the activated GPx, can prevent high ROS levels from causing chromosomal instability and improve cell viability to offset the suppressed effect of CAI.
Figure 2.
CAI inhibits complex I and regulates cell metabolism or redox homeostasis in mitochondria of tumor cells. HCT116 and C26 cells were treated with control or CAI (10 μmol/L) for 48 h. (A) Relative values of activity for complex I expressed relative to the control treatment activity level (100%). (B) The levels of ROS detected by measuring the Mitosox MFI with FCM. (C) The levels of intracellular glutamine, glutamate, α-KG, fumarate, and succinate were determined in the presence or absence of CAI. (D) and (E) The ratio of GSH to GSSG and GPx activity in HCT116 and C26 cells. (F) Proposed model for the role of CAI in ROS reduction way. The data shown are representative of three independent experiments, and the error bars represent the mean ± SEM; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
3.3. CAI induces metabolic changes in tumor cells by enhancing glutaminolysis
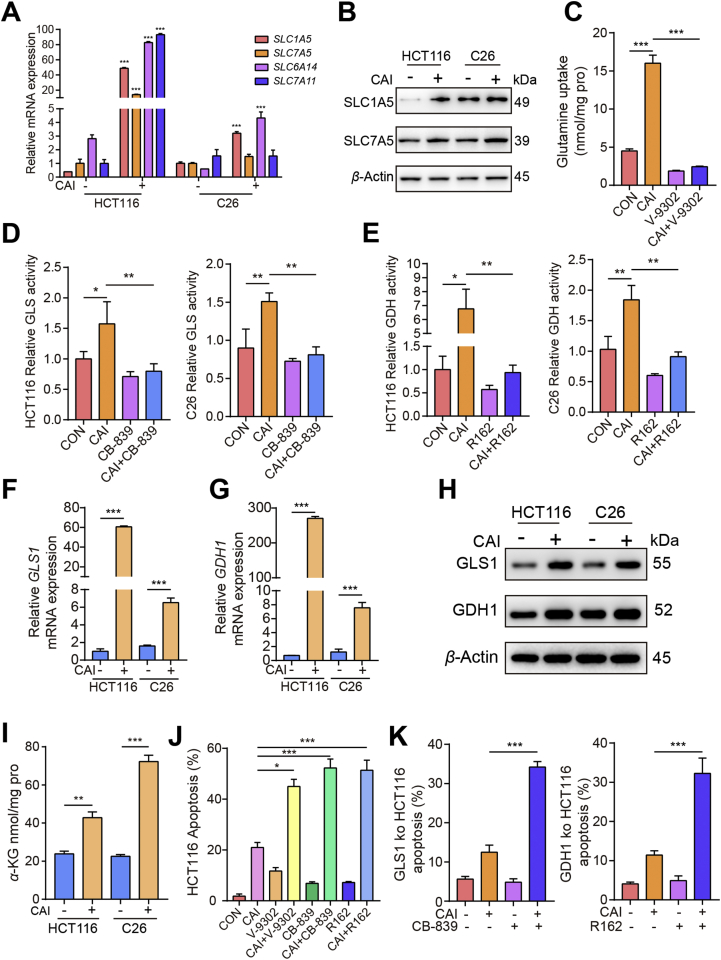
Next, we investigated how CAI regulates metabolism, leading to glutamine dependence. Genome-wide transcriptional profiling and metabolic analysis uncovered a potential glutaminolysis response in tumor cells exposed to CAI (Supporting Information Fig. S3A and S3B). Tumor cells rely on solute carrier superfamily transporters on the cell membrane to extract glutamine from the extracellular environment. Notably, CAI exposure significantly increased the expression of SLC7A5, SLC1A5, SLC6A14 and SLC7A11 (Fig. 3A), which each increase attributable to various aspects of glutamine transport. Although these four transporters were found to be expressed in tumor cells, the addition of CAI strikingly upregulated the mRNA and protein expression of the main transporters of glutamine, SLC1A5 and SLC7A5 (Fig. 3B and Fig. S3C). The results revealed that CAI also affects glutamine uptake. The SLC1A5 inhibitor V-930222 reduced glutamine basal levels and the CAI-induced accumulation of glutamine (Fig. 3C and Fig. S3D). After entering cells, glutamine is converted into glutamate by GLS, and glutamate is then converted into α-KG via the action of GDH123. Although there are different subtypes of GLS, most cancers, including CRCs, overexpress GLS1, which contributes to GLS activity in tumor cells24,25. Therefore, we assessed the impact of targeting GLS1. As shown in Fig. 3D, E, Fig. S3E and S3F, compared to control cells, CAI-treated cells had an increase in enzymatic activity in vitro and in vivo. In addition, the mRNA and protein levels of GLS1 and GDH1 were observed upon CAI treatment (Fig. 3F–H), which suggested that the elevated level of α-KG was caused by both increased uptake and enhanced utilization of glutamine (Fig. 3I). We speculate that the enhanced glutaminolysis may be a metabolic self-rescue response for the CRC tumor cells to survival from CAI treatment. Indeed, accompanied with CAI treatment, blocking SLC1A5 with the inhibitor V-9302, or inhibiting GLS with the inhibitor CB-839, or inhibiting GDH1 with the inhibitor R162, or knocking out GLS1/GDH1 expression in HCT116 or C26 cells resulted in greatly increased cell apoptosis (Fig. 3J, K, Fig. S3G and S3H).
Figure 3.
CAI induces metabolic changes in tumor cells by enhancing glutaminolysis. HCT116 and C26 cells were cultured in the presence or absence of 10 μmol/L CAI. (A) The relative mRNA abundance of the indicated amino acid transporters. (B) Protein abundance of two transporters SLC1A5 and SLC7A5. (C) The glutamine uptake in HCT116 cells was assayed by 13C-glutamine labelling in cells treated with vehicle, CAI (10 μmol/L), V-9302 (5 μmol/L) or CAI + V-9302 for 12 h. (D) and (E) Histograms show the relative activities of GLS1 and GDH1. (F) and (G) The mRNA expression of GLS1 and GDH1 in HCT116 and C26 cells. (H) Protein abundance of GSL1 and GDH1. (I) Cellular α-KG levels were quantified using the α-KG Assay Kit. (J) and (K) Tumor cell apoptosis was quantified by flow cytometry. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; Student’s t-test (A, F, G and I) or one-way ANOVA (C–E, J and K). The data represent the mean ± SEM of three independent experiments.
3.4. CAI-activated AhR facilitates glutamine import into tumor cells
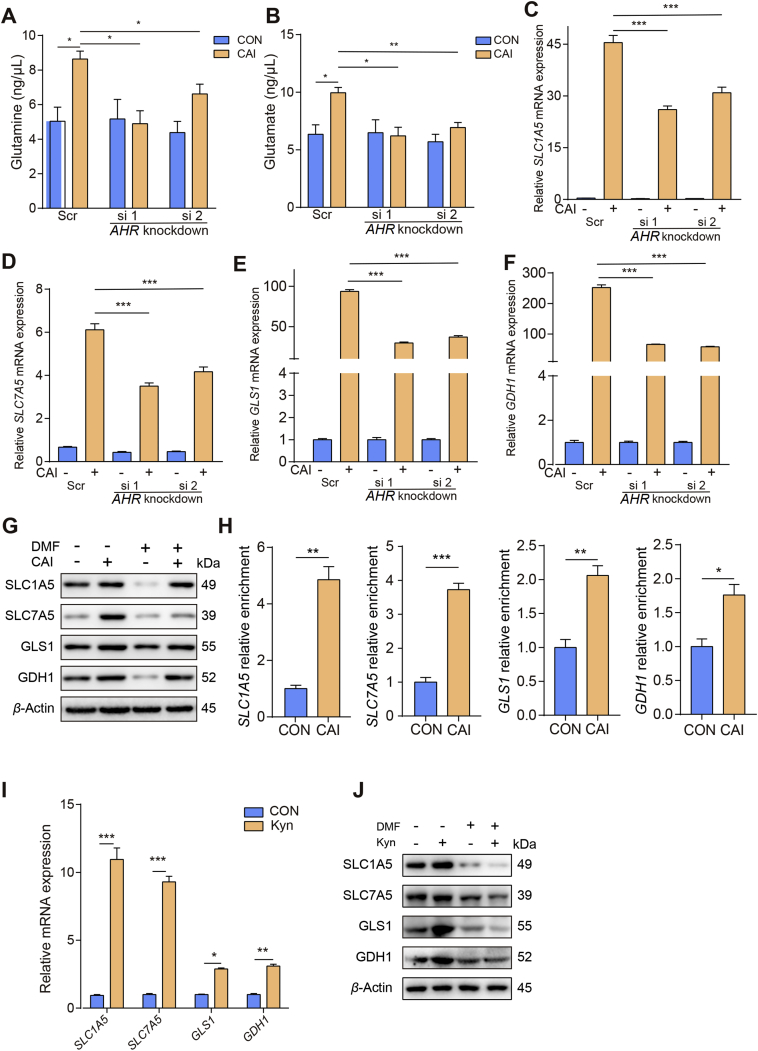
Next, we investigated the molecular mechanism by which CAI regulates glutaminolysis activity. We studied the proto-oncogene MYC, which is known to promote metabolic reprogramming by altering glutamine uptake and metabolism in cancer cells26. Surprisingly, no significant changes in MYC protein levels were observed upon CAI treatment (Supporting Information Fig. S4A and S4B). Considering our recent research findings that a key tryptophan-metabolizing enzyme regulated by CAI, IDO1, increased aryl hydrocarbon receptor (AhR) activity, we determined the role of AhR27. In line with the previous results CAI markedly upregulates the mRNA expression of CYP1A1 and CYP1B1 in HCT116 cells (Fig. S4C), suggesting that AhR was activated by CAI. We found that basal glutamine and glutamate levels were much higher after CAI treatment and that they could be downregulated by knocking down AHR expression (Fig. 4A and B). Despite the high consumption of glutamine induced by CAI treatment, the intracellular glutamine level was not reduced, suggesting that tumor cells might replenish their glutamine store via active transportation to maintain intracellular glutamine levels. Additionally, knocking down AHR expression or blocking AhR activity using 3′,4′-dimethoxyflavone (DMF) resulted in the downregulation of SLC1A5, SLC7A5, GLS1 and GDH1 expression (Fig. 4C–G and Fig. S4D). The chromatin immunoprecipitation (ChIP)-qPCR data show that CAI facilitated the binding of AhR to the promoter of SLC1A5, SLC7A5, GLS1 and GDH1 genes, and caused significant increase in their gene expressions (Fig. 4H). To further confirm that AhR-driven cancer cells rely on glutamine metabolism, we tested another endogenous AhR agonist, kynurenine (Kyn). Consistently, Kyn also upregulated the mRNA and protein expression of SLC1A5, SLC7A5, GLS1 and GDH1 (Fig. 4I and J). Together, these data suggest that CAI can upregulate glutaminolysis in tumor cells through induction and activation of AhR.
Figure 4.
CAI-activated AhR facilitates glutamine import into tumor cells. A scramble siRNA (Scr) or siRNAs against AHR were transfected into HCT116 cells for 24 h, and then the cells were treated with CAI (10 μmol/L) for 48 h. (A) and (B) Glutamine and glutamate concentrations were measured using appropriate assay kits. (C)–(F) SLC1A5, SLC7A5, GLS1 and GDH1 mRNA expression was determined by qPCR. (G) HCT116 cells were treated with PBS, CAI (10 μmol/L), DMF (20 μmol/L), or CAI/DMF for 48 h, protein expression was determined by Western blotting. (H) ChIP-qPCR analysis was performed with an anti-AhR antibody and SLC1A5, SLC7A5, GLS1 and GDH1 promotor-specific primers. Data are presented as values relative to the control group value. HCT116 cells were incubated with vehicle or Kyn (200 mmol/L), and mRNA expressions (I) of SLC1A5, SLC7A5, GLS1 and GDH1 was determined by qPCR. (J) Protein expression was determined by Western blotting. The data shown are representative of three independent experiments, and the error bars represent the mean ± SEM; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
3.5. Combining CAI with glutaminolysis inhibitor greatly promotes ROS production and cell apoptosis
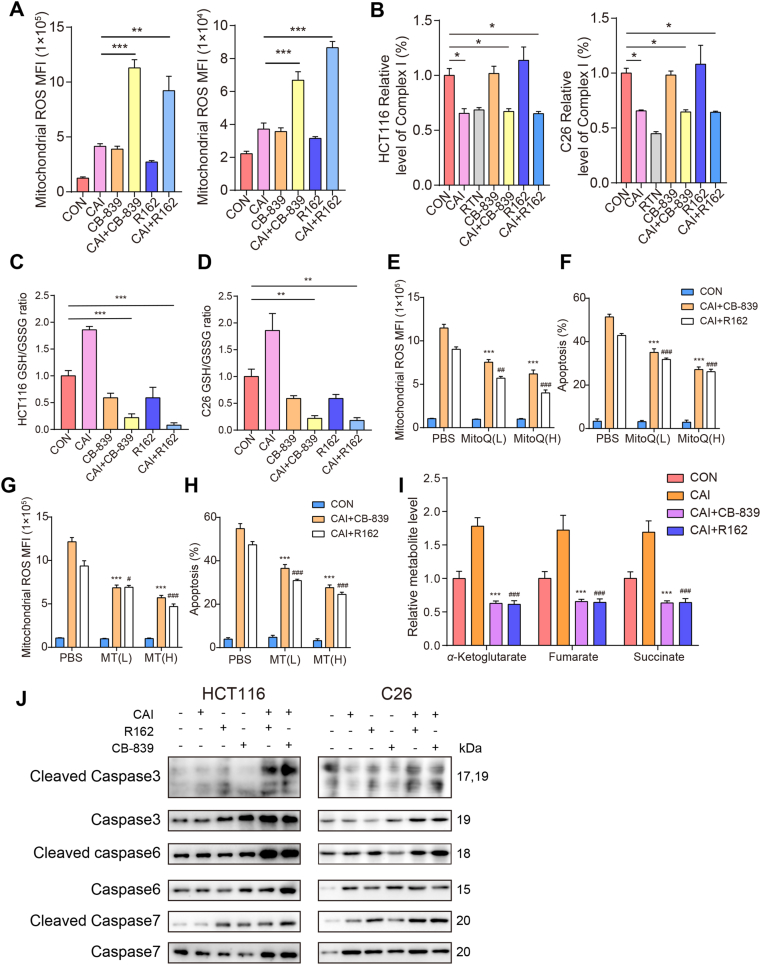
Increased ROS production has been detected in various cancers and shown to initiate oxidative stress-induced tumor cell death28,29. To clarify the reason for apoptosis, flow cytometry was conducted to detect the effect of CAI with glutaminolysis inhibitor on ROS and apoptosis. The results show that CAI, CB-839, or R162 alone slightly increased the generation of intracellular ROS, whereas ROS generation was significantly increased in the combination group (Fig. 5A). The activity of complex I was inhibited by CAI, RTN, CAI + CB-839 or CAI + R162, suggesting that CAI regulated ROS by increasing the production of O2·– (Fig. 5B). Consistent with the data shown in Fig. 4D, CAI increased the GSH levels in HCT116 and C26 cells, which was reversed by the GLS inhibitor CB-839 or GDH1 inhibitor R162 (Fig. 5C and D). To further verify the role of CAI in regulation of ROS levels, two specific scavengers of mitochondrial superoxide mitoquinone mesylate (MitoQ) and mito-TEMPO (MT) were used concomitantly with the indicated drugs. As shown in Fig. 5E–H and Fig. S5C–S5F, either MitoQ or MT could dose-dependently rescue cells from the enhanced ROS production and the synergistic apoptotic effect of the combination drugs, CAI + CB839 or CAI + R162. Compared with CAI-treated CRC cells, several TCA cycle intermediates generated from glutamine-based anaplerosis were decreased more than 70% in the combination drug-treated cells (Fig. 5I). Activation of caspases is one of the hallmarks of apoptosis and ultimately results in cell death30. It has been reported that the anti-tumor activity of glutamine inhibition is dependent on the caspase-dependent apoptotic pathway31. The levels of cleaved caspase 3, caspase 6, and caspase 7 were elevated in the combination group, indicating that cells underwent programmed cell death (Fig. 5J).
Figure 5.
Combined therapy with CAI and CB-839 or R162 targets tumor cell apoptosis dependent on ROS. HCT116 and C26 cells were treated with control, CAI (10 μmol/L), CB-839 (100 nmol/L), CAI + CB-839, R162 (20 μmol/L) or CAI + R162 for 48 h. (A) The levels of ROS detected by measuring the Mitosox MFI with FCM. (B) Relative values of activity for complex I expressed relative to the control treatment activity level (100%). RTN (0.5 μmol/L) was used as a positive control. (C) and (D) The ratio of GSH to GSSG in HCT116 cells and C26 cells with the indicated treatments. For A–D, ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. (E)–(H) HCT116 cells were treated with the selective mitochondrial ROS scavengers mitoquinone mesylate (MitoQ) and mito-TEMPO (MT) at both low and high doses for rescue. ROS levels (E and G) or cell apoptosis (F and H) after the combinations treatment were analysed by flow cytometry. ∗∗∗P<0.001 (compared with CAI + CB-839 in PBS); #P<0.05, ##P<0.01, ###P<0.001 (compared with CAI + R162 in PBS). (I) Intracellular levels of α-KG, fumarate, and succinate were determined. ∗∗∗P<0.001; ###P<0.001 (compared with CAI). The data are from 3 independent experiments. Graphs represent the mean ± SEM. (J) Western blots showing the expressions of caspase 3, cleaved caspase 3, caspase 6, cleaved caspase 6, caspase 7 and cleaved caspase 7 after 48 h of the indicated treatment.
3.6. Efficacy and safety of combining CAI and a GLS or GDH1 inhibitor to treat xenograft tumors
The above experiments show that increased ROS levels led to apoptosis. Here, we further verified this process in vivo. To address the antitumor effects of the combination treatments in vivo, we established xenograft tumor models in BALB/c mice injected with C26 cells and nude mice injected with HCT116 cells. As shown in Fig. 6A–D, CAI, CB-839, or R162 alone slightly inhibited tumor growth to a certain extent. The combinations of CAI and CB-839 or R162 were found to be superior to the corresponding single treatments, significantly repressing tumor growth and prolonging the survival of mice. In addition, when we knocked out GLS1 and injected normal or GLS1-knockout HCT116 or C26 cells into the two sides of mice, we found that GLS1 knockdown combined with CAI treatment significantly suppressed tumor growth (Fig. 6E and F). A similar advantage in terms of slowing tumor growth was also observed in GDH1-knockdown C26 or HCT116 cell tumor-bearing models (Fig. 6G and H). Again, this improved tumoral control was accompanied by increased ROS levels in vivo (Fig. 6I–L). For initial in vivo toxicity studies, tumor-bearing mice received the indicated drugs for 28 days. These chronic drug treatments did not result in significant histopathological differences between the vehicle-treated and drug-treated groups (Supporting Information Fig. S6A) or altered levels of blood aminotransferase, urea nitrogen and serum creatinine (Fig. S6B–S6E), suggesting the drug treatments had minimal toxicity in vivo. Moreover, the combined treatments dramatically attenuated cell viability in a group of CRC cell lines but not in the human intestinal mucosal tissue-sourced cell line CCC-HIE-2 for up to 48 h (Fig. S6F).
Figure 6.
Combining CAI and an inhibitor of GLS or GDH1 disrupts tumor growth. BALB/c and nude mice were subcutaneously injected with C26 (1 × 105 cells/mouse) or HCT116 (2 × 105 cells/mouse) tumor cells. When the tumor size was 5 mm × 5 mm, the tumor-bearing mice (n = 10 per group) were treated with PBS, CAI (20 mg/kg), CB-839 (200 mg/kg), or CAI/CB-839 or PBS, CAI, R162 (20 mg/kg), or CAI/R162 for 28 days. (A, B, E and G) C26 cell colorectal cancer model. (C, D, F and H) HCT116 cell melanoma model. (E and F) Stable GLS1-silenced or scrambled control (E) C26 cells (1 × 105 cells/mouse) or (F) HCT116 cells (2 × 105 cells/mouse) were injected (s.c.) into each side of the mice. (G and H) Stable GDH1-silenced or scrambled control (G) C26 cells (1 × 105 cells/mouse) or (H) HCT116 cells (2 × 105 cells/mouse) were injected (s.c.) into each side of the mice. Tumor growth was monitored. Relative ROS levels in C26 (I and J) and HCT116 (K and L) cells in vitro. Representative biological triplicates are shown as the mean ± SEM; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; NS, not significant, two-way ANOVA.
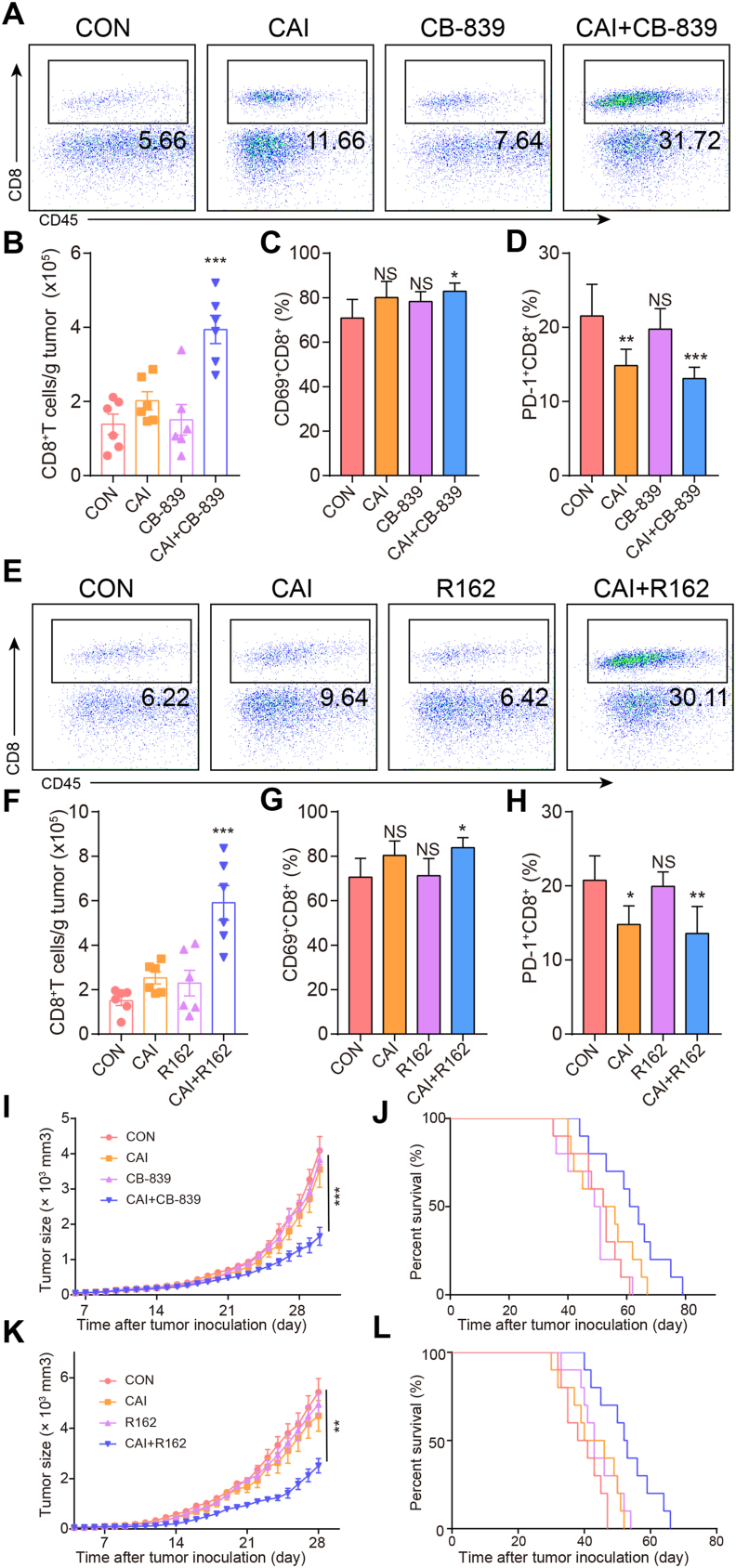
3.7. The proportion of cytotoxic CD8+ T cells in the tumor microenvironment can be increased by the combined therapy with CAI and CB-839 or R162 but is not dependent for the antitumor activity of combinations
In addition to tumor cells, the interactions between different cells in the tumor microenvironment shape the unique metabolic characteristics of the microenvironment and maintain tumor growth32,33. For comprehensively understanding the effects of the indicated drugs, we further detected the effects of the drugs or drug combinations on the proportion of other cell types in the tumor microenvironment that may affect the growth of tumor cells. It is worth mentioning that the upregulation of the CD8+ T cell levels in the combination groups was very obvious, along with the distinct differences in PD-1 expression and the CD69 level (Fig. 7A–H). To avoid the antitumor impact of CD8+ T cells, we subsequently assessed the antitumor activity of the indicated therapeutics in nude mice bearing C26 tumors. We found that combined treatment still enhanced tumor growth inhibition and produced therapeutic activity with prolonged survival (Fig. 7I–L). There were no significant differences in the numbers and typical functions of CD4+ T cells (Supporting Information Fig. S7A–S7C) and tumor-associated macrophages (TAMs, Fig. S7D and S7E) among the different groups of treated C26 tumor-bearing mice. The same was true for other cell types, including regulatory T cells (Tregs, Fig. S7F and S7G) and myeloid-derived suppressor cells (MDSCs, Fig. S7H and S7I). The results illustrate that the antitumor activities of the two combinations are principally aimed at tumor cells. Consistent with previous reports27, we confirmed that tumor cells and T cells are engaged in a fierce competition for nutrients in the tumor microenvironment, which affects the lethality of T cells and promotes the further development of cancer.
Figure 7.
The proportion of cytotoxic CD8+ T cells in the tumor microenvironment can be increased by the combined therapy with CAI and CB-839 or R162 but is not dependent for the combination’s antitumor activity. Tumors were harvested 14 days after the injection of 1 × 105 C26 cells into BALB/c mice (n = 6/per group) and analyzed by flow cytometry. (A) and (B) Representative flow cytometric analysis and quantification of CD8+ T cell numbers per gram of tumor tissue in different groups. (C) and (D) The percentages of CD69+CD8+ and PD-1+CD8+ T cells in the TIL population in the tumor microenvironment. (E)–(H) The same measurements for the evaluation of the effects of another drug combination (CAI and R162). (I)–(L) Antitumor activity of CAI, CAI + CB-839, or CAI + R162 in immunodeficient mice. Nude mice (n = 10) bearing 5 mm × 5 mm C26 cell tumors were treated with PBS, CAI (20 mg/kg), CB-839 (200 mg/kg), or CAI/CB-839 or PBS, CAI, R162 (20 mg/kg), or CAI/R162 for 21 days. The tumor growth curves (I and K) and survival curves (J and L) of tumor-bearing mice receiving various treatments. Data represent the mean ± SEM, n = 6.
3.8. Metabolic alterations triggered by CAI in primary human CRC cells
Next, we investigated whether the mechanism underlying CAI-induced metabolic reprogramming could be translated to human CRC. To this end, single CRC cells were isolated from CRC patients, and then these primary cells were treated with CAI. Consistently, these human primary CRC cells were also induced into glutamine dependency, as evidenced by their low OCR and ECAR and their Mito Fuel Flex Test changes (Fig. 8A–C). Furthermore, upon CAI treatment, these primary CRC cells also upregulated the expression of SLC1A5, SLC7A5, GLS and GDH1 and increased the total GPx activity and the GSH level (Fig. 8D–G). In line with increased ROS levels, apoptosis and the expression of cleaved caspase 3, caspase 6, and caspase 7 were markedly upregulated (Fig. 8H–J). Moreover, the ROS scavengers MitoQ and MT dose-dependently reversed the effects of CAI combined with CB839 or R162 (Supporting Information Fig. S8A–S8D). To further validate these results, NOD-SCID mice were inoculated with 100 μL of patient tumor tissue suspension to generate a patient-derived xenograft (PDX) model, followed by combined CAI and CB-839 or R162 treatment. We found that these combined treatments strikingly inhibited tumor growth and prolonged survival compared to control treatment (Fig. 8K and L). Moreover, increased in vivo ROS levels were observed in the combination therapy group (Fig. 8M and N). Together, these data suggest that CAI reset CRC metabolic programming through an AHR–SLC1A5/SLC7A5–GLS1–GDH1 pathway and that combining GLS or GDH1 inhibitor therapeutics with CAI can further potentiate the beneficial effect of CAI on the energy-deficient tumor microenvironment.
Figure 8.
Combined therapy with CAI and CB-839 or R162 exhibits enhanced antitumor activity against primary tumor cells. (A) and (B) The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of primary tumor cells were measured with or without 10 μmol/L CAI treatment for 48 h (n = 3). (C) Seahorse XFp Fuel Flex analysis shows the dependency on and flexibility of all 3 fuels after acute 10 μmol/L CAI treatment of primary tumor cells. The expression of the amino acid transporters GLS1 and GDH1 was determined by real-time PCR (D) and Western blotting (E) in 3 different primary tumor cell lines. (F) GPx activity levels in primary tumor cells were measured. (G) The ratio of GSH to GSSG was calculated. (H) The MFI of ROS in primary tumor cells was measured. (I) Tumor cell apoptosis was quantified by flow cytometry. (J) Western blots show the expression of caspase 3, cleaved caspase 3, caspase 6, cleaved caspase 6, caspase 7 and cleaved caspase 7 after 48 h of the indicated treatments. (K) and (L) NOD-SCID mice subcutaneously injected with 100 μL fragments of colorectal cancer tissue from patients were treated with PBS, CAI, R162, or CAI/R162 or PBS, CAI, CB-839, or CAI/CB-839 for 42 days. Tumor volumes were measured and calculated (left). Long-term survival was calculated by Kaplan–Meier analysis and analysed with the log-rank (Mantel–Cox) test (right). (M) and (N) The effects of in vivo treatment on ROS production were evaluated. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; NS, not significant, Student’s t-test (C, D, and F) or one-way ANOVA (G, H, I, M and N). The data represent the mean ± SEM.
4. Discussion
Here, we have identified a novel role for CAI, a tumor metabolism regulator, in regulating glutamine dependency. The present study reveals that besides showing a certain apoptosis-inducing effect in vitro CAI treatment induces metabolic changes by enhancing glutaminolysis for tumor cells to survival. Intra-tumor heterogeneity might play a role in poor responses of tumor cells to CAI therapy. The plasticity of metabolic pathways allows tumor cells to quickly switch adaptive mechanisms in the face of CAI stress. The metabolic reprogramming of tumor cells enables them to make the best use of nutrients in the microenvironment and synthesize nutrients that do not exist in the microenvironment for growth, thus gaining living space34. Furthermore, we demonstrated a crucial role for CAI in CRC metabolic reprogramming. We found that CAI promotes glutamine metabolism by activating the amino acid transporters SLC1A5 and SLC7A5 in a transcriptional manner and is associated with CRC cell survival and sensitivity to glutamine metabolism inhibitors.
We systematically dissected the metabolic alterations triggered by CAI in CRC cell lines through complex I inhibition and found that this inhibition resulted in an adaptive increase in glutamine metabolism. The significant finding of this study lies in the conclusion that AhR is primarily responsible for inducing glutamine import. Intrinsically, tumor cells express higher levels of AHR35,36, GLS124,37,38 and GDH139 as well as SLC1A540,41 than their differentiated counterparts. This means that tumor cells and normal cells have very different dependence on glutamine metabolism. In this case, CAI further activates AhR and increases the expression of SLC1A5, GLS and GDH1, highlighting the reprogrammed glutamine pathway as the more depended source of energy for tumor cells to survive. Thus, the combined therapy with CAI and CB-839 or R162 can bring a dramatic anti-tumor effect.
CAI has exhibited the ability to inhibit or kill tumor cells, which is used to explain the antitumor effect of CAI15,42. However, CAI is unable to kill large bulks of tumor cells, implying that tumor cells mobilize other processes to respond to the antitumor effects of CAI. Our studies show that CAI treatment could relatively specifically target glutaminolysis and that GLS or GDH1 expression were commonly upregulated in human CRC cells but not in proliferating nonmalignant human cells. On the one hand, combination treatments suppressed the energy metabolism of tumor cells with minimal toxicity. Additionally, the combined therapies potentiated immune reactions against tumors, which was not contradictory to the killing effect on tumor cells. Different changes in cell metabolism form the basis for treating cancer by producing a powerful antitumor response43,44. It could be expected that a new treatment approach combined with immune checkpoint inhibitors could alleviate the inhibition of T cells by cancer cells.
It has long been thought that the increased demand for glutamine in tumor cells is due solely to the role of glutamine in protein synthesis and its use as a source of nitrogen for the synthesis of purines and pyrimidines45. However, recent studies have shown that the roles of glutamine in tumor cells go well beyond these synthetic processes46. Increased metabolism of glutamine points to the accumulation of alpha-ketoglutaric acid47,48. α-Ketoglutaric acid is involved not only in the metabolic processes of anabolism and catabolism but also in the regulation of signalling pathways and epigenetic modification49. Importantly, inhibition of glutamine metabolism suggests that this therapy silences certain oncogenes50. Lactic acid derived from glutamine is exported by tumor cells to generate a transmembrane proton gradient, which provides driving force for tumor cells to enhance uptake of selective nutrients with proton-coupled transporters. Therefore, lactic acid might be an accomplice involved in glutamine transport.
Direct targeting of proto-oncogenes can achieve good therapeutic results. Some targeting strategies still meet with difficulties, such as a lack of directly targeted drugs or extreme difficulties in developing corresponding targeted drugs. As a result, the metabolic dependence mechanism of tumors should be approached from another perspective to restrain the malignant biological phenotype of tumors51,7. At the same time, rationally designed small-molecule combinations may also hold promise as adjunctive therapies for patients to develop directed therapeutic interventions or tumor prevention treatments aimed at metabolic pathways.
5. Conclusions
We identified that tumor cells adapt to CAI treatment by increasing reductive glutamine metabolism, which directly affects source selection for biosynthesis. Thus, a GLS or GDH1 inhibitor administered in combination with CAI specifically enhanced the beneficial effects of CAI-based cancer treatment. These findings support an antitumor role for CAI, making combined treatments promising therapies for CRC.
Acknowledgments
This study as supported by the National Natural Science Foundation of China (grants 81872897 and 81672966) and the CAMS Major Collaborative Innovation Project 2016-I2 M-1-011 (China).
Author contributions
Lei Guo conceived the project. Jing Shi, Lei Guo, and Dechang Zhang participated in the research design. Jing Shi, Hongting Gao and Yuqing Huang conducted the experiments. Rui Ju provided technical or material support. Jing Shi and Lei Guo performed the data analysis. Lei Guo and Jing Shi wrote the manuscript.
Conflicts of interest
The authors declare no competing financial interests.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.07.008.
Appendix ASupplementary data
The following is the Supplementary data to this article:
References
- 1.Kroemer G., Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Hsu P.P., Sabatini D.M. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBerardinis R.J., Lum J.J., Hatzivassiliou G., Thompson C.B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metabol. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 5.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci Adv. 2016;2 doi: 10.1126/sciadv.1600200. e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 7.Jin N., Bi A., Lan X., Xu J., Wang X., Liu Y., et al. Identification of metabolic vulnerabilities of receptor tyrosine kinases-driven cancer. Nat Commun. 2019;10:2701. doi: 10.1038/s41467-019-10427-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lukey M.J., Katt W.P., Cerione R.A. Targeting amino acid metabolism for cancer therapy. Drug Discov Today. 2017;22:796–804. doi: 10.1016/j.drudis.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vander Heiden M.G. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 10.Fotiadis D., Kanai Y., Palacin M. The SLC3 and SLC7 families of amino acid transporters. Mol Aspects Med. 2013;34:139–158. doi: 10.1016/j.mam.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 11.McCracken A.N., Edinger A.L. Nutrient transporters: the Achilles' heel of anabolism. Trends Endocrinol Metab. 2013;24:200–208. doi: 10.1016/j.tem.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altman B.J., Stine Z.E., Dang C.V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;10:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wise D.R., Thompson C.B. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hensley C.T., Wasti A.T., DeBerardinis R.J. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo L., Li Z.S., Wang H.L., Ye C.Y., Zhang D.C. Carboxyamido-triazole inhibits proliferation of human breast cancer cells via G2/M cell cycle arrest and apoptosis. Eur J Pharmacol. 2006;538:15–22. doi: 10.1016/j.ejphar.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 16.Chen C., Ju R., Shi J., Chen W., Sun F., Zhu L., et al. Carboxyamidotriazole synergizes with sorafenib to combat non-small cell lung cancer through inhibition of NANOG and aggravation of apoptosis. J Pharmacol Exp Therapeut. 2017;362:219–229. doi: 10.1124/jpet.117.240986. [DOI] [PubMed] [Google Scholar]
- 17.Zhao R.Z., Jiang S., Zhang L., Yu Z.B. Mitochondrial electron transport chain, ROS generation and uncoupling. Int J Mol Med. 2019;44:3–15. doi: 10.3892/ijmm.2019.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frechin M., Enkler L., Tetaud E., Laporte D., Senger B., Blancard C., et al. Expression of nuclear and mitochondrial genes encoding ATP synthase is synchronized by disassembly of a multisynthetase complex. Mol Cell. 2014;56:763–776. doi: 10.1016/j.molcel.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 19.Poprac P., Jomova K., Simunkova M., Kollar V., Rhodes C.J., Valko M. Targeting free radicals in oxidative stress-related human diseases. Trends Pharmacol Sci. 2017;38:592–607. doi: 10.1016/j.tips.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 20.Kou L., Sun R., Xiao S., Zheng Y., Chen Z., Cai A., et al. Ambidextrous approach to disrupt redox balance in tumor cells with increased ROS production and decreased GSH synthesis for cancer therapy. ACS Appl Mater Interfaces. 2019;11:26722–26730. doi: 10.1021/acsami.9b09784. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan L.B., Martinez-Garcia E., Nguyen H., Mullen A.R., Dufour E., Sudarshan S., et al. The proto-oncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Mol Cell. 2013;51:236–248. doi: 10.1016/j.molcel.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulte M.L., Fu A., Zhao P., Li J., Geng L., Smith S.T., et al. Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat Med. 2018;24:194–202. doi: 10.1038/nm.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovacevic Z. The pathway of glutamine and glutamate oxidation in isolated mitochondria from mammalian cells. Biochem J. 1971;125:757–763. doi: 10.1042/bj1250757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang Y., Stine Z.E., Xia J., Lu Y., O'Connor R.S., Altman B.J., et al. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J Clin Invest. 2015;125:2293–2306. doi: 10.1172/JCI75836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv C., Qu H., Zhu W., Xu K., Xu A., Jia B., et al. Low-dose paclitaxel inhibits tumor cell growth by regulating glutaminolysis in colorectal carcinoma cells. Front Pharmacol. 2017;8 doi: 10.3389/fphar.2017.00244. 244-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao P., Tchernyshyov I., Chang T.C., Lee Y.S., Kita K., Ochi T., et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi J., Chen C., Ju R., Wang Q., Li J., Guo L., et al. Carboxyamidotriazole combined with IDO1–Kyn–AhR pathway inhibitors profoundly enhances cancer immunotherapy. J Immunother Cancer. 2019;7:246. doi: 10.1186/s40425-019-0725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach?. Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 29.Perry G., Raina A.K., Nunomura A., Wataya T., Sayre L.M., Smith M.A. How important is oxidative damage? Lessons from Alzheimer's disease. Free Radic Biol Med. 2000;28:831–834. doi: 10.1016/s0891-5849(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 30.Taylor R.C., Cullen S.P., Martin S.J. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 31.Jacque N., Ronchetti A.M., Larrue C., Meunier G., Birsen R., Willems L., et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood. 2015;126:1346–1356. doi: 10.1182/blood-2015-01-621870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun L., Suo C., Li S.T., Zhang H., Gao P. Metabolic reprogramming for cancer cells and their microenvironment: beyond the Warburg effect. Biochim Biophys Acta Rev Cancer. 2018;1870:51–66. doi: 10.1016/j.bbcan.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Biswas S.K. Metabolic reprogramming of immune cells in cancer progression. Immunity. 2015;43:435–449. doi: 10.1016/j.immuni.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Boroughs L.K., DeBerardinis R.J. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheong J.E., Sun L. Targeting the IDO1/TDO2–KYN–AhR pathway for cancer immunotherapy—challenges and opportunities. Trends Pharmacol Sci. 2018;39:307–325. doi: 10.1016/j.tips.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y., Lv J., Liu J., Liang X., Jin X., Xie J., et al. STAT3/p53 pathway activation disrupts IFN-beta-induced dormancy in tumor-repopulating cells. J Clin Invest. 2018;128:1057–1073. doi: 10.1172/JCI96329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallipoli P., Giotopoulos G., Tzelepis K., Costa A.S.H., Vohra S., Medina-Perez P., et al. Glutaminolysis is a metabolic dependency in FLT3(ITD) acute myeloid leukemia unmasked by FLT3 tyrosine kinase inhibition. Blood. 2018;131:1639–1653. doi: 10.1182/blood-2017-12-820035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng S.J., Chen H.Y., Zeng Z., Deng S., Zhu S., Ye Z., et al. Nutrient stress-dysregulated antisense lncRNA GLS-AS impairs GLS-mediated metabolism and represses pancreatic cancer progression. Cancer Res. 2019;79:1398–1412. doi: 10.1158/0008-5472.CAN-18-0419. [DOI] [PubMed] [Google Scholar]
- 39.Jin L., Li D., Alesi G.N., Fan J., Kang H.B., Lu Z., et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer cell. 2015;27:257–270. doi: 10.1016/j.ccell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q., Beaumont K.A., Otte N.J., Font J., Bailey C.G., van Geldermalsen M., et al. Targeting glutamine transport to suppress melanoma cell growth. Int J Cancer. 2014;135:1060–1071. doi: 10.1002/ijc.28749. [DOI] [PubMed] [Google Scholar]
- 41.Hassanein M., Hoeksema M.D., Shiota M., Qian J., Harris B.K., Chen H., et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin Cancer Res. 2013;19:560–570. doi: 10.1158/1078-0432.CCR-12-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das M. Carboxyamidotriazole orotate in glioblastoma. Lancet Oncol. 2018;3 doi: 10.1016/S1470-2045(18)30347-4. S1470-2045(18)30347-4. [DOI] [PubMed] [Google Scholar]
- 43.Stienstra R., Netea-Maier R.T., Riksen N.P., Joosten L.A.B., Netea M.G. Specific and complex reprogramming of cellular metabolism in myeloid cells during innate immune responses. Cell Metabol. 2017;26:142–156. doi: 10.1016/j.cmet.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Vernieri C., Casola S., Foiani M., Pietrantonio F., de Braud F., Longo V. Targeting cancer metabolism: dietary and pharmacologic interventions. Cancer Discov. 2016;6:1315–1333. doi: 10.1158/2159-8290.CD-16-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu W., Pelicano H., Huang P. Cancer metabolism: is glutamine sweeter than glucose?. Cancer cell. 2010;18:199–200. doi: 10.1016/j.ccr.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeBerardinis R.J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu P.S., Wang H., Li X., Chao T., Teav T., Christen S., et al. α-Ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nature Immunol. 2017;18:985–994. doi: 10.1038/ni.3796. [DOI] [PubMed] [Google Scholar]
- 48.Wang X., Liu R., Qu X., Yu H., Chu H., Zhang Y., et al. α-Ketoglutarate-activated NF-κB signaling promotes compensatory glucose uptake and brain tumor development. Mol Cell. 2019;76:148–162.e7. doi: 10.1016/j.molcel.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Campbell S.L., Wellen K.E. Metabolic signaling to the nucleus in cancer. Mol Cell. 2018;71:398–408. doi: 10.1016/j.molcel.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 50.Stuart S.D., Schauble A., Gupta S., Kennedy A.D., Keppler B.R., Bingham P.M., et al. A strategically designed small molecule attacks alpha-ketoglutarate dehydrogenase in tumor cells through a redox process. Cancer Metab. 2014;2:4. doi: 10.1186/2049-3002-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mayers J.R., Vander Heiden M.G. Nature and nurture: what determines tumor metabolic phenotypes?. Cancer Res. 2017;77:3131–3134. doi: 10.1158/0008-5472.CAN-17-0165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.