Abstract
SIRT6 belongs to the conserved NAD+-dependent deacetylase superfamily and mediates multiple biological and pathological processes. Targeting SIRT6 by allosteric modulators represents a novel direction for therapeutics, which can overcome the selectivity problem caused by the structural similarity of orthosteric sites among deacetylases. Here, developing a reversed allosteric strategy AlloReverse, we identified a cryptic allosteric site, Pocket Z, which was only induced by the bi-directional allosteric signal triggered upon orthosteric binding of NAD+. Based on Pocket Z, we discovered an SIRT6 allosteric inhibitor named JYQ-42. JYQ-42 selectively targets SIRT6 among other histone deacetylases and effectively inhibits SIRT6 deacetylation, with an IC50 of 2.33 μmol/L. JYQ-42 significantly suppresses SIRT6-mediated cancer cell migration and pro-inflammatory cytokine production. JYQ-42, to our knowledge, is the most potent and selective allosteric SIRT6 inhibitor. This study provides a novel strategy for allosteric drug design and will help in the challenging development of therapeutic agents that can selectively bind SIRT6.
KEY WORDS: SIRT6, Molecular dynamics simulations, Reversed allostery, Allosteric inhibitor, Pancreatic cancer, Cell migration, Cytokine production
Abbreviations: ADPr, ADP-ribose; BSA, bull serum albumin; CCK-8, Cell Counting Kit-8; DMSO, dimethyl sulfoxide; FBS, fetal bovine serum; FDL, Fluor de Lys; HDAC, histone deacetylase; H3K9, histone 3 lysine 9; H3K18, histone 3 lysine 18; H3K56, histone 3 lysine 56; HPLC, high-performance liquid chromatography; IPTG, isopropyl-β-d-thiogalactoside; IC50, half-maximum inhibitory concentration; MD, molecular dynamics; NAD+, nicotinamide adenine dinucleotide; NAM, nicotinamide; PBS, phosphate buffer saline; PMA, phorbol 12-myristate 13-acetate; PMSF, phenylmethanesulfonyl fluoride; RMSD, root-mean-square deviation; RT-qPCR, real-time quantitative PCR; SDS-PAGE, SDS-polyacrylamide gel electrophoresis; SIRT6, sirtuin 6
Graphical abstract
A novel allosteric Pocket Z within SIRT6 was induced and the first selective SIRT6 allosteric inhibitor JYQ-42 was rationally designed. JYQ-42 exhibits promising efficacy in pancreatic cancer cell lines.
1. Introduction
Sirtuin 6 (SIRT6) is a Sir2 homologue and a member of the sirtuin family of nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylases (HDACs)1. In mammals, the sirtuin family consists of seven members (SIRT1–7)2. These “magnificent seven” proteins have a plethora of cellular activities and play critical roles under both physiological and pathological conditions2, 3, 4. SIRT6 is involved in fatal events, such as aging, inflammation and carcinogenesis5, 6, 7. SIRT6 can catalyse deacetylation, mono-ADP-ribosylation and defatty–acylation reactions to execute its multiple functions, which largely depend on various conditions1,8, 9, 10, 11, 12. Remarkably, in vivo studies have revealed that under physiological conditions, SIRT6 mostly participates in deacetylation events13. SIRT6-mediated deacetylation events induce robust epigenetic changes in nucleosomes and chromatin and are thus of considerable significance.
SIRT6 consists of a large Rossmann fold for the binding of the cofactor NAD+ and a small domain containing a zinc ion binding motif, termed the zinc-binding domain (ZBD)14. Between these two domains, SIRT6 forms a long hydrophobic cleft for substrate peptide loading, facilitating deacetylation reactions14. SIRT6 shares a conserved catalytic domain and NAD+ binding site similar to those of other sirtuin proteins14. Their structural similarities lead to great difficulty in achieving selective modulation, which has therapeutic implications. Despite considerable efforts, only a few SIRT6 inhibitors have been reported to date, and these inhibitors do not have ideal efficacy or specificity15, 16, 17, 18, 19, 20, 21. Nicotinamide (NAM) is an endogenous inhibitor of all sirtuin members but has mild potency, and NAM-related pyrazinamide also exhibits a similar weak inhibitory effect22. On the other hand, although synthetic compounds such as the quercetin derivatives catechin gallate and gallocatechin gallate were established as the first series of potent SIRT6 small-molecule inhibitors, they frequently suffer from compromised selectivity23. Cyclic peptides represent another trend of SIRT6 inhibitors, but they are limited by poor specificity and low intracellular potency17. Therefore, developing SIRT6 inhibitors with high potency and optimal selectivity is of enormous interest in both academia and the pharmaceutical industry.
Allostery, or allosteric regulation, in which allosteric ligands fine-tune protein activity from topologically distinct allosteric sites, endows allosteric ligands with enhanced pharmacological performance, such as improved specificity. Allostery thus represents an innovative strategy for drug discovery, especially for notoriously intractable targets. Recently, through combined in silico, crystallographic, in vitro, and in vivo mouse studies, we discovered a first-in-class cellularly active allosteric SIRT6 activator, MDL-800, which showed promising potency and was implicated in the treatment of liver cancer, non-small-cell lung cancer and neural injury24.
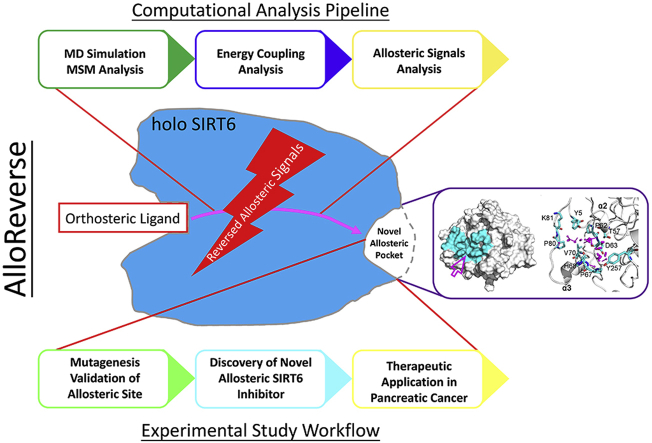
Here, based on a state-of-the-art reversed allosteric communication strategy AlloReverse, we predicted a novel, cryptic allosteric site, Pocket Z, in SIRT6 (Y5, P62, D63, P67, V70, P80, K81, Y257), which was only identified owing to the bidirectional perturbation induced by the orthosteric binding of NAD+, and confirmed its regulatory influence through mutagenesis. Based on this pocket, we carried out high-throughput screening and rational design and identified an SIRT6 allosteric inhibitor, JYQ-42, that was able to inhibit the deacetylation activity of SIRT6 in pancreatic cancer cell lines. Significantly, JYQ-42 uniquely inhibited SIRT6 among other sirtuins and HDAC enzymes. Most importantly, JYQ-42 markedly blocked the migration of pancreatic cancer cells and prevented cancer cell-induced local inflammation by hindering the production of IL6, TNF-α and IL8. In summary, we discovered a specific and cellularly active SIRT6 allosteric inhibitor, JYQ-42, with promising potential for targeting pancreatic cancer cells. This inhibitor represents a starting point for further SIRT6 allosteric inhibitor design. Moreover, our results provide a generic and robust strategy for the identification of allosteric modulators of other therapeutic proteins.
2. Materials and methods
2.1. Reagents and buffers
Antibiotics, imidazole, IPTG, phorbol 12-myristate 13-acetate (PMA), NAD+, IL8, ADPr and DMSO were purchased from Sigma. Cell culture medium, trypsin, PBS and FBS were purchased from Gbico. Buffers were used in this paper as follows: SIRT6 lysis buffer (1 × PBS, 0.5 mol/L NaCl, 5% glycerol, 1 mmol/L PMSF, pH 7.5); SIRT6 purification buffer (20 mmol/L Hepes, 250 mmol/L NaCl, 5% Glycerol, pH 7.0); ion exchange buffer (20 mmol/L Pipes, 30 mmol/L NaCl, 1 mmol/L DTT, pH 6.5; 20 mmol/L Pipes, 2 mol/L NaCl, 1 mmol/L DTT, pH 6.5); SIRT6 assay buffer (50 mmol/L Tris-HCl, 137 mmol/L NaCl, 2.7 mmol/L KCl, 1 mmol/L MgCl2, pH 8.0).
2.2. Construction of simulation systems
Human SIRT6 crystal structure previously solved by our lab in complex with substrate H3K9 myristoyl peptide, cofactor ADPr and allosteric modulator MDL-801 (PDB ID: 5Y2F) was downloaded from Protein Data Bank24. Apo-SIRT6 structure was obtained after deleting the substrate peptide, cofactor and allosteric compound within. To construct holo-SIRT6 system, the H3K9 myristoyl peptide was truncated and its 12-carbon aliphatic moiety was also removed to simulated the acetylated substrate. Cofactor ADPr was substituted with NAD+ based on the coordinates extracted from SIRT1‒NAD+ complex structure (PDB ID: 4I5I)25.
2.3. Docking simulations
AutoDockFR-AutoDock was exploited for molecular docking26. Apo SIRT6 and NAD+ molecular structures were first converted into PDBQT format and then loaded into AutoGridFR. Centroid of ADPr within structure was set for docking grid center generation with a 60 × 60 × 60 Å3 searching space based on a spacing value of 0.375 Å between every two grids. The atomic affinity maps retrieved from AutoGridFR were then input into AutoDockFR for ligand docking. 50 independent docking were run to maximize docking conformation sampling and the resulting poses were clustered based on a RMSD cutoff of 2 Å. Lowest energy docking poses from each cluster were then visually examined for the distance between the anomeric carbon within the nicotinamide ribose group in NAD+ and the acetyl oxygen in K9Ac. Furthermore, conformational rearrangement was allowed to facilitate the SN2 nucleophilic substitution reaction.
2.4. Classical MD (cMD) simulations
SIRT6 structure was modelled with the Amber ff14SB force field and zinc was treated using the cationic dummy atom approach27,28. In this approach, the four coordinated cysteine residues (Cys139, Cys142, Cys164, and Cys175) were modified to CYM. The distance and angle parameters for the zinc-mediated tetrahedral complex were obtained from the database27,28. A TIP3P truncated octahedral water (10 Å) was used for solvation with counterions added for neutralization. Another 0.15 mol/L NaCl was randomly added into the complex system to mimic physiological conditions.
Two rounds of energy minimization were run after preliminary preparation. Energy of water and ions within were first minimized for 5000 steps (2000 steepest descent followed by 3000 conjugate gradient minimization steps) with 500 kcal/(mol⋅Å2) position constraint on the protein backbone and ligand. Then, 4000 steepest descent and 6000 conjugate gradient cycles of minimization were performed for the whole complex system without any restriction. Afterwards, the simulation systems were heated to 300 K in 300 ps and then equilibrated for 700 ps under NVT conditions with a 10 kcal/(mol⋅Å2) constraint applied onto the protein and ligand structures. Finally, 50 ns cMD simulation was run with 2.0 fs timestep under 1 atm and 300 K conditions to calculate potential and dihedral energy for further accelerated MD (aMD) simulations. To modulate the system temperature, Langevin dynamics with 1 ps−1 collision frequency was applied, and the system pressure was regulated utilizing the Berendsen barostat with isotropic position scaling. During the cMD process, long-range electrostatic interactions were modelled with the particle mesh Ewald (PME) method29, with the cutoff for short-range electrostatic interactions and van der Waals force set to be 10 Å. Moreover, the SHAKE model was exploited to restrain covalent bonds involving hydrogen atoms29.
2.5. aMD simulations
After 50 ns cMD simulation, aMD was then carried out to enhance sampling and to cover a broader conformational space. During aMD process, “boost energy” was added to the simulation system to reduce energy barriers and facilitate easier transitions between different conformations within the free energy landscape30.
Systemic potential energy was calculated as Vr and the corresponding threshold was determined by the system size and its normal energy value as Ethresh in Eqs. (1), (2). During aMD, when the instantaneous potential energy was lower than Ethresh, it would be increased for ΔVr to reach . Otherwise, the original value Vr would be used.
| (1) |
| (2) |
where ΔVr is defined in Eq. (3) depending on Ethresh and the acceleration parameter α.
| (3) |
“Dual boost” protocol, which boosts both the potential and dihedral energy31, was applied during aMD based on Eqs. (4), (5), (6), (7). In Eqs. (4), (6), EthreshP represents the threshold for potential energy, and EthreshD stands for the threshold for dihedral energy; while in Eqs. (5), (7), αP and αD denote the acceleration parameters for potential and dihedral energy correspondingly. During cMD simulations, average total energy (Etot avg) and average dihedral energy (Edih avg) were calculated and then used for aMD. Natoms and Nresidues reflect the number of atoms and residues in the systems, respectively.
| (4) |
| (5) |
| (6) |
| (7) |
The final structures from cMD simulation trajectories were extracted as the initiating structures for aMD simulation and aMD was then run with random initial velocity for 500 ns For the two complex systems under study (apo versus holo SIRT6), to enhance sampling, 5 independent simulation replica were carried out respectively, retrieving a cumulative 2.5 μs timescale for each. Energetics calculation and systemic environmental parameter settings for aMD were consistent with the ones applied in cMD, and simulation trajectory analyses were carried with CPPTRAJ package from Amber.
2.6. Construction of MSM
Dihedral formed by α6 and α7 within Rossmann domain and the overall RMSD of ZBD were used as input for PyEMMA to carry out MSM analyses32. In brief, MSM models were constructed with aforementioned two structural feature descriptors and then validated for Markovian properties (please see next section). Next, “coordinate_cluster_kmeans” approach was exploited for clustering. The overall simulation trajectories were clustered into 200 microstates with maximum 200 rounds k-means iterations. “msm_estimate_markov_mode” function was then used for MSMs construction with a lag-time of 2 ns based on implied time-scale test results. Perron cluster analysis (PCCA+) model was employed to cluster 200 microstates into multiple metastable states, and they were further verified for Markovian properties by Chapman–Kolmogorov tests (please see next section).
For each metastable state, the corresponding representative structure was extracted from simulation trajectories utilizing the “coordinates_save_traj” plugin and the MDTraj package based on the similarity score Sij calculated by Eq. (8).
| (8) |
where dij reflects the RMSD comparing snapshot structures i and j, and dscale is the standard deviation of d. Through pair-wise iteration of each frame throughout the trajectories, the structure estimated with highest Sij was considered to be the representative conformation and clustered and extracted.
2.7. Validation of the MSM
Implied timescale test was carried out to test the Markovian properties of our simulation systems as well as the MSMs constructed thereof. The number of k-means centers and maximum iteration were first defined and then the corresponding multiple transition probability matrixes (TPMs) with various lag times were constructed. TPMs represent the transition probability among all microstates and they also determine the relaxation timescale or the implied timescale based on Eq. (9):
| (9) |
where τ denotes the lag time for TPMs, and λi stands for the ith eigenvalue corresponding to the TPM with lag time τ. When the microstate transition is relaxed for ith time, the corresponding implied timescale τi is thus calculated from Eq. (9).
Implied timescales originate from TPMs eigen decomposition, and the orders of eigenvalues reflect the sequence of transitions. Therefore, the first eigenvalue (λ1) corresponds to the slowest transition, and so on. Different τi with increasing τ were calculated for Markovian properties verification. When τi begins to stabilize as τ increases, that is, transitions among microstates become independent of lag times, it indicates that the corresponding MSM is Markovian33.
For implied timescale tests, changes of implied timescale were calculated against different lag times for both apo (Supporting Information Fig. S3A) and holo (Fig. S3B) systems, and the k-means cluster centers and the maximum iteration were both set at 200. As shown in Fig. S3A, the implied timescale curves for apo SIRT6 reach plateau at lag time of around 1 ns, while the ones for holo SIRT6 are converged at lag time of approximately 1.5 ns (Fig. S3B). Thus, lag time longer than 1.5 ns will retrieve MSM with Markovian property, and we used 2 ns to construct our models and for further analyses.
Exploiting PCCA + approach, we clustered the microstates from apo and holo systems into 2 and 4 MSM metastable states respectively. They further underwent Chapman–Kolmogorov tests shown in Supporting Information Fig. S4 to confirm that the MSM transition probability is within similar range to the practical transition process. In summary, the MSMs constructed based on SIRT6 simulation trajectories were again demonstrated to be Markovian and suitable for further analyses.
2.8. Energy coupling score calculation
Energy coupling scores for the predicted pockets were calculated using molecular mechanics generalized Born surface area (MM-GBSA) method based on the simulation trajectories for both apo and holo systems. Within the given pocket, the interaction energy was calculated for residues separated by at least three amino acids along the sequence based on Eq. (10):
| (10) |
where Eint implies the internal energy, Eeel represents the electrostatic energy, Evdw reflects the van der Waals energy, and Gpol and Gsas are the polar solvation energy and solvent-accessible surface energy, respectively.
Energy differences for residues within pockets between NAD+-bound (holo) and NAD+-unbound (apo) systems were utilized for energy coupling score calculations.
2.9. Prediction of potential allosteric sites of SIRT6 and virtual screening
Novel potential allosteric site was predicted based on our reversed allosteric model and the related hybrid computational and experimental pipeline29. Schrödinger suite 2009, v5.5 was then employed to virtually screen compound libraries. By targeting the predicted site, we screened more than 5.5 million compounds from 7 compound libraries including our labrotary's compound library. The 16 compounds with the top-ranked scores were chosen for further experiments.
2.10. Plasmid construction, protein expression and purification
All strains and plasmids used in this study are described in Supporting Information Table S3. 2 YT was used as rich medium, nutrient agar plates contained 1.5% (w/v) agar and supplemented with antibiotics as required. The antibiotic was used 50 μg/mL of kanamycin.
Site-directed mutagenesis of SIRT6 was performed with the corresponding primers Supporting Information Table S4, according to the protocols supplied with the KOD-Plus-Mutagenesis kit (Toyobo, SMK-101). For purification of SIRT6 and its mutants, all the constructed plasmids were introduced individually into E. coli strain BL21, and the resultant strains were grown in LB medium containing 100 μg/mL ampicillin at 37 °C. IPTG was added to a final concentration of 0.5 mmol/L at the time point when the absorbance of the culture at 600 nm reached 0.8. The culture was continuously incubated for 18 h at 16 °C. The cells were harvested by centrifugation and the protein was purified as described previously24.
2.11. Cell culture
HeLa cell line was purchased from ATCC. BXPC-3 and other pancreatic cancer cell lines were purchased from the Cell Resource Center of Shanghai Institute for Biological Sciences, Chinese Academy Science. BXPC-3 were maintained in RPMI1640 medium supplemented with 10% FBS, penicillin (50 units/mL), and streptomycin (50 μg/mL). Capan2 was maintained in McCoy's 5a medium supplemented with 10% FBS, penicillin (50 units/mL), and streptomycin (50 μg/mL). HeLa, MiaPaCa-2 were maintained in DMEM medium supplemented with 10% FBS, penicillin (50 units/mL), and streptomycin (50 μg/mL). The cells were cultured at 37 °C under 5% CO2. The cell lines were identified according to short tandem repeat (STR) analysis of up to 20 loci and compared with available published STR profiles (Shanghai Biowing Applied Biotechnology).
2.12. HDACs selectivity assays for JYQ-42
Selectivity assays for most HDACs were performed by the CEREP company according to CEREP standard assay protocols. Selectivity assays for SIRT5 and SIRT7 were performed as previously described method24. Briefly, SIRT5 (residues 34–302) and full-length SIRT7 were used for FDL deacetylation assays and the protocol for these assays were the same as those for SIRT6. IC50 values were calculated by fitting the data points with the dose–response function in GraphPad Prism v7 (GraphPad Software). Each experiment was performed independently at least three times.
2.13. Immunoblotting and antibodies
Pancreatic cancer cells were cultured with four concentrations of JYQ-42 (0, 10, 20, and 40 μmol/L) for 24 or 48 h. After treatment, the cells were collected and lysed with 1 × SDS loading buffer. Total proteins were resolved on 12% SDS-PAGE, transferred to PVDF membranes. 50 mmol/L Tris-HCl (pH 7.5) with 100 mmol/L NaCl, 0.5% (v/v) Tween-20 and 5% nonfat milk was used for blocking. 50 mmol/L Tris-HCl (pH 7.5) with 100 mmol/L NaCl and 1% BSA was used to prepare primary and secondary antibodies. An Immobilon Western Chemiluminescent HRP Substrate Kit (Merck Millipore) was used for detection of the protein bands and blots were scanned with G: BOX Chemi system (Syngene, Cambridge, UK).
Antibodies to the following proteins were used in the Western blots: HRP-linked anti-rabbit IgG (Cell Signaling, 7074P2; 1:20,000); HRP-linked anti-mouse IgG (Cell Signaling, 7076P2; 1:20,000); SIRT6 (Cell Signaling, 12486; 1:2000); Histone H3 (Abcam, ab10799; 1:2000); Histone H3 (acetyl K9) (Abcam, ab32129; 1:1000); Histone H3 (acetyl K18) (Abcam, ab1191; 1:1000); Histone H3 (acetyl K56) (Active Motif, 39281; 1:1000); β-actin (Proteintech Group, HRP-60008; 1:5000).
2.14. Migration assay with RTCA-DP
The migration assay was performed with the xCELLigence RTCA-DP system (Roche) as previously described methods with some modifications34. Briefly, 6 × 104 serum-starved pancreatic cancer cells were resuspended in 100 μL of serum-free medium which was added with PMA (30 ng/mL, final concentration) and a series of concentrations of JYQ-42 and then were added to the pre-equilibrated upper chamber of the xCELLigence CIM plate along with the bottom well plate containing complete medium which was added with PMA (30 ng/mL, final concentration), IL8 (10 ng/mL, final concentration) and a series of concentrations of JYQ-42 for migration. Cell index values were recorded every 15 min during the process. RTCA software v1.2 (Roche Applied Science) was used to calculate the slopes of the curves at various time points.
2.15. Statistical analysis
Statistical analyses were performed with GraphPad Prism software version 7.00 (GraphPad Software) and all data are represented as mean ± SD of at least three independent experiments. Differences between two groups were examined statistically as indicated (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001).
3. Results
3.1. Identification of a cryptic SIRT6 allosteric site
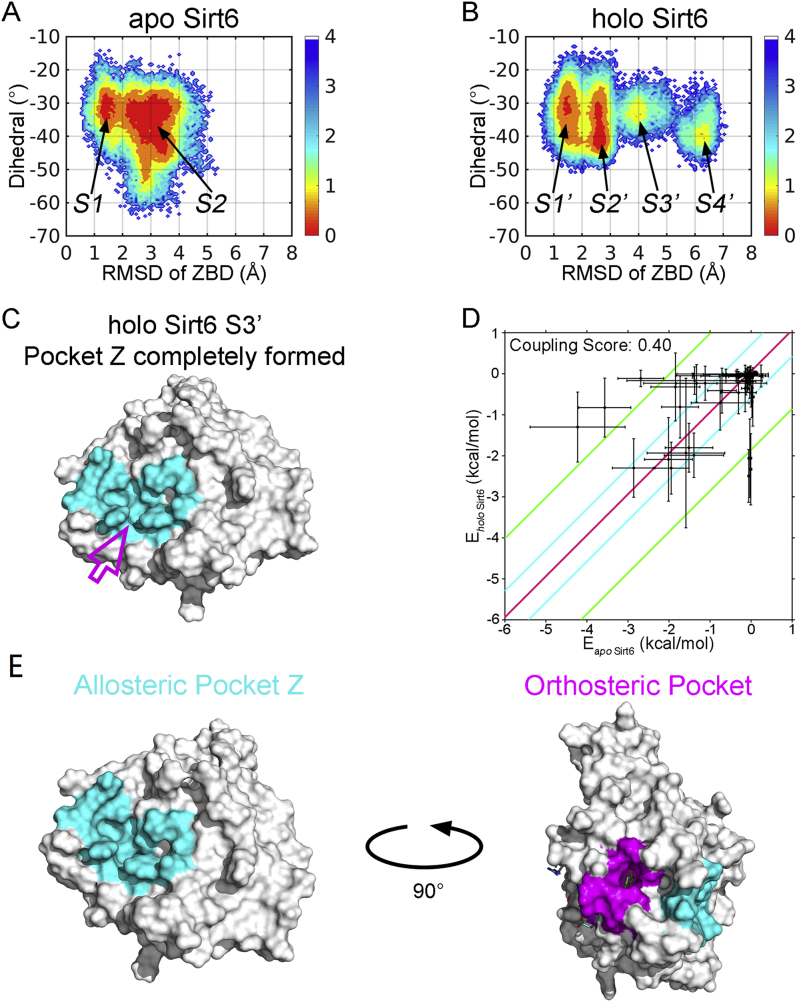
The discovery of allosteric modulators targeting SIRT6 has long been challenging29,35. Herein, the reversed allosteric strategy AlloReverse and the related hybrid pipeline developed by our lab were exploited to identify potential allosteric pockets in SIRT6 for further structure-based rational drug design (Supporting Information Fig. S1). Our bi-directional allosteric communication model by AlloReverse highlights the crosstalk between orthosteric perturbations and allosteric pockets. Thus, we first carried out large-scale accelerated MD (aMD) simulations of SIRT6 before and after ligand NAD+ loading to determine the conformational changes induced by orthosteric binding. To enhance sampling, 5 simulation replicates were performed for both apo (NAD+-unbound) and holo (NAD+-bound) systems. SIRT6 consists of eight α-helices and nine β-sheets, which form two major domains, the Rossmann domain and the zinc-binding domain (ZBD). The structural features of these domains were selected for conformational landscape analysis via simulations to investigate the dynamics of SIRT6. Dihedrals defined by α6 and α7 within the Rossmann domain and the overall RMSD of the ZBD were calculated as feature descriptors, and the trajectories of the MD simulation were projected onto their two-dimensional (2D) landscapes (Fig. 1A and B). Upon orthosteric binding, the structural ensemble of SIRT6 was significantly altered, as shown by the prominently shifted conformational landscapes. In the apo system, SIRT6 predominantly displayed two conformations, with free energy basins located at dihedrals of approximately −30° to −35° and RMSDs of approximately 1.4 and 3 Å, respectively (Fig. 1A). After NAD+ loading, the original two dominant conformations and their energy basins within the landscapes were both slightly reduced. Notably, two novel intermediate states emerged, with their dihedrals and RMSDs situated at approximately −28° and 4 Å and −40° and 6.2 Å, respectively (Fig. 1B). The significantly perturbed conformational landscapes of SIRT6 pinpointed the structural rearrangements induced by the reverse allosteric signals triggered by orthosteric binding of NAD+. These rearrangements may be responsible for the emergence of novel allosteric pockets.
Figure 1.
The novel allosteric Pocket Z of SIRT6 was identified by computational methods (A) Conformational landscape of apo SIRT6. (B) Conformational landscape of holo SIRT6. (C) The novel allosteric site Pocket Z in S3′ within the conformational ensemble of holo SIRT6. (D) Energy coupling analysis of Pocket Z. (E) Pocket Z is located topologically distal to the orthosteric NAD+ binding site.
Markov state models (MSMs), powerful tools to elucidate protein structural dynamics33, were next employed for a further in-depth investigation of the perturbed SIRT6 free energy landscape33,36. MSMs were first constructed for generating simulation trajectories of apo and holo systems using PyEMMA (please see the Supporting Information for details)32. Based on the conformational landscapes, in the apo system, two MSM metastable states (S1–S2) were identified, while in the holo system, four (S1′‒S4′) intermediate states were identified. Based on these structures, the Fpocket algorithm was used to predict the potential binding pocket, and indeed37,38, a novel druggable pocket, referred to as Pocket Z hereafter, was successfully identified in the S3′ structure within the holo system (Fig. 1C). Structural comparisons revealed that this cavity did not readily form in the apo system or in other metastable states in the holo complex (Supporting Information Fig. S2). These observations indicate that this Pocket Z is highly dynamic and emerges only in the presence of bidirectional allosteric signals that are fine-tuned by orthosteric perturbations. Pocket Z is also a cryptic pocket, which only transiently emerges within the holo SIRT6 conformational ensemble and remains hidden in other metastable states as well as in crystal structures (Fig. S2). Moreover, several state-of-the-art algorithms were utilized to evaluate and validate the allosteric potential of Pocket Z and its bi-directional crosstalk with orthosteric NAD+ sites. Based on the “energy coupling score” approach29, the coupling score of Pocket Z was calculated to be 0.40, significantly higher than the threshold of 0.25, implying its strong allosteric coupling (Fig. 1D). Additionally, residues within Pocket Z were uncovered as allosteric hotspots according to Ohm, a novel network-based allosteric methodology39, and had a relatively high allosteric coupling intensity (ACI) of 0.6. Furthermore, the allosteric free energy of Pocket Z within SIRT6 was found to be −1.15 by the AlloSigMa algorithm40, also indicating its considerable allosteric potential. These computational results all indicated the allosteric nature of Pocket Z and supported its topologically distal location relative to the orthosteric NAD+ binding pocket (Fig. 1E and Fig. S2).
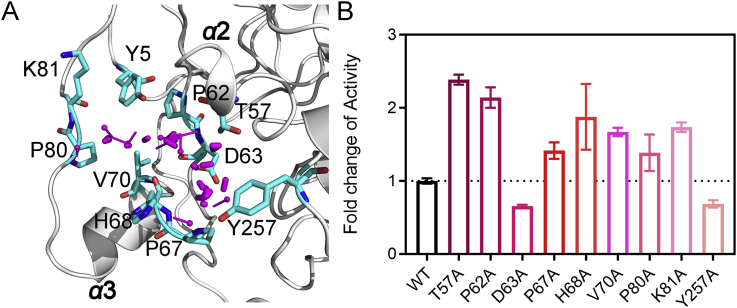
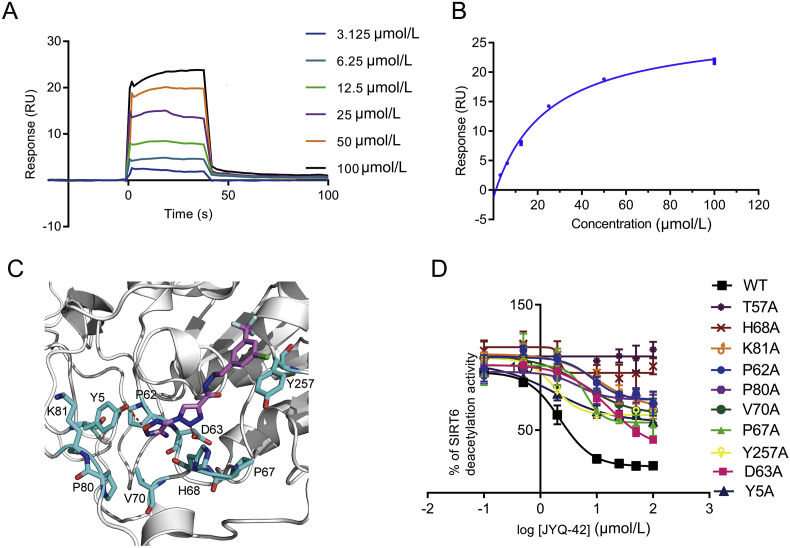
Next, structural insights into Pocket Z revealed that it is mainly composed of α2 and α3 (Fig. 2A), and to further verify predicted allosteric pocket's allosteric regulation of SIRT6 deacetylation, we individually mutated the key residues of the predicted pocket. A Fluor de Lys (FDL) assay was performed to quantify the effects of single point mutations (T57A, P62A, D63A, P67A, H68A, V70A, P80A, K81A, Y257A) on SIRT6 deacetylation activity towards an acetylated substrate peptide (Ac-RHKK-Ac-AMC, Fig. 2B). We found that mutations in the predicted Pocket Z greatly changed the deacetylation activity of SIRT6 (Fig. 2B). Most of the mutations increased the enzymatic activity of SIRT6 by 1.4- to 2.4-fold (Fig. 2B). Two mutations (D63A and Y257A) reduced the enzymatic activity of SIRT6 by approximately 1.5-fold (Fig. 2B). We also tested mutations outside of the predicted pocket and found that the mutations (E87A, V151A, K170A, P274A, D277A, P315A) outside of the predicted pocket did not change the enzymatic activity of SIRT6 (Supporting Information Fig. S5). Collectively, these results suggested that the predicted Pocket Z was an allosteric pocket.
Figure 2.
Validation of the predicted pocket in SIRT6. (A) Structural insights of Pocket Z. (B) Mutagenesis experiments of critical residues within Pocket Z. The deacetylation activity of different SIRT6 mutants was measured, and their relative activities were calculated as fold changes relative to that of the wild-type (WT) protein, which was set as 1. All assays were performed with at least three replicates, and the results are shown as the mean ± SD, n = 3 wells, from three independent experiments.
In summary, utilizing the latest reported reversed allosteric model, we successfully identified a novel potential allosteric pocket in SIRT6 that emerged only upon orthosteric binding of NAD+. This pocket is characterized by strong allosteric communication and its potential regulatory effects on SIRT6 and thus constitutes the structural basis for the discovery of other modulators.
3.2. Screening and optimization of SIRT6 allosteric inhibitors
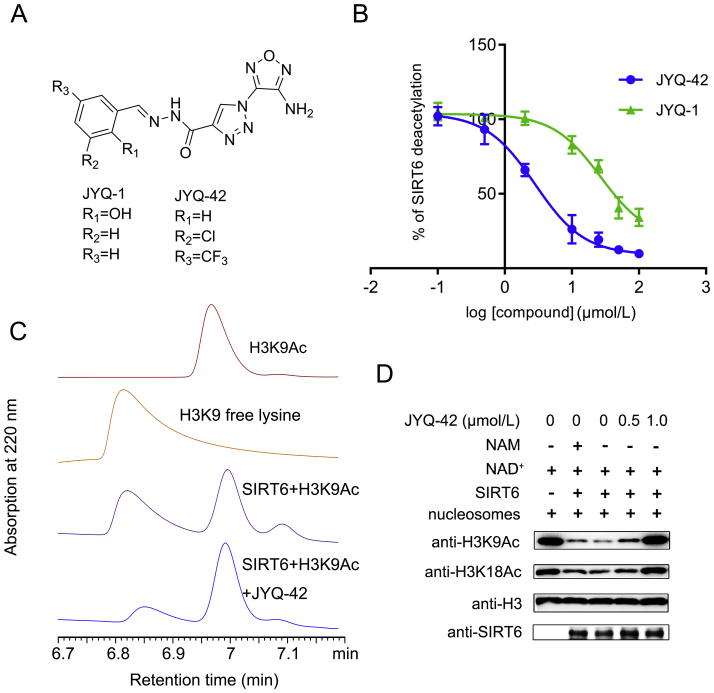
Based on the potential, novel allosteric Pocket Z, high-throughput virtual screening was carried out using the Schrӧdinger Suite. More than 2,000,000 compounds were docked in Pocket Z, and they were ranked based on their AlloFinder scores and then underwent visual inspection of their docking poses (Fig. S1). Finally, 16 compounds were selected for further experimental analysis. The effects of the compounds on SIRT6 deacetylation were evaluated using an FDL assay previously developed by our group24. These 16 compounds could all inhibit the deacetylation activity of SIRT6 in a dose-dependent manner (data not shown). The best inhibitory compound was JYQ-1, which had a half-maximal inhibitory concentration (IC50) value of 26.84 μmol/L and a 64.18% inhibition ratio at 50 μmol/L (Supporting Information Table S1, Fig. 3A and B). To guide further structure-based rational ligand design, JYQ-1 was docked in Pocket Z in SIRT6 to obtain further in-depth structural insights into its binding mode (Supporting Information Fig. S6). Within Pocket Z, JYQ-1 formed a π−alkyl interaction with P62 and a hydrogen bond with D63. The benzene ring of JYQ-1 displayed a π−π interaction with Y257 to facilitate its tight binding. Based on the docking results and the scaffold of JYQ-1, we performed chemical modifications of JYQ-1 to improve its inhibition of SIRT6 deacetylation (Supporting Information Scheme S1). We obtained 59 compounds, and their inhibition activity was tested by the FDL assay (Table S1). Among these compounds, the best inhibitor was JYQ-42, the structure of which was unambiguously established by single-crystal X-ray experiments (Fig. 3A, Supporting Information Fig. S7 and Table S2). The FDL assay showed that JYQ-42 strongly inhibited SIRT6 deacetylation activity in a dose-dependent manner, with a half-maximal inhibitory concentration (IC50) value of 2.33 ± 0.17 μmol/L and an 85.3% inhibition ratio at 50 μmol/L (Fig. 3B, Table S1).
Figure 3.
Discovery and biochemical characterization of JYQ-42 as an inhibitor of SIRT6 deacetylation. (A) Chemical structures of JYQ-1 and JYQ-42. (B) JYQ-42 can inhibit SIRT6 deacetylation on Ac-RHKK-Ac-AMC effectively. The inhibition activity of JYQ-42 on SIRT6 deacetylation was evaluated by the Fluor de Lys (FDL) assay. The IC50 of JYQ-42 was 2.33 ± 0.17 μmol/L, and the data are presented as the mean ± SD, n = 3 wells, from three independent experiments. (C) The effects of SIRT6 deacetylation on Ac-KQTARK-Ac-STGGWW-NH2 in the absence or presence of 50 μmol/L JYQ-42, as determined by HPLC. Data are representative of three independent experiments. (D) JYQ-42 inhibited SIRT6 deacetylation significantly on nucleosome substrates. Nucleosomes were extracted from HeLa cells, and the samples were analysed by Western blot analysis. Data are representative of three independent experiments.
To further confirm whether JYQ-42 also exhibited inhibitory impacts on other SIRT6 substrates, we measured the deacetylase activity of SIRT6 on another physiologically acetylated H3K9 peptide (Ac-KQTARK-Ac-STGGWW-NH2, H3K9Ac) in the presence of JYQ-42. The FDL assay showed that JYQ-42 effectively inhibited the SIRT6 deacetylation activity on H3K9Ac, with an IC50 value of 9.1 ± 0.21 μmol/L and a 90.7% inhibition ratio at 50 μmol/L (Supporting Information Fig. S8). High-performance liquid chromatography (HPLC) was performed to confirm the inhibition of deacetylation of the H3K9Ac peptide by JYQ-42. Compared with the FDL assay, HPLC can eliminate the interference caused by fluorescence and provide more definitive evidence. As Fig. 3C shows, in the absence of JYQ-42, SIRT6 effectively removed the acetyl group from H3K9Ac, and a considerable amount of H3K9-free lysine peptides were detected by HPLC. However, in the presence of JYQ-42, the amount of H3K9-free lysine peptides decreased prominently, which suggested that JYQ-42 could indeed effectively inhibit the deacetylation activity of SIRT6. SIRT6 displays poor deacetylase activity in vitro, as its deacetylase activity is nucleosome-dependent13. We then examined whether JYQ-42 more effectively inhibited SIRT6 deacetylase activity on nucleosomes than on substrate peptides. We extracted nucleosomes from HeLa cells, performed an in vitro nucleosome deacetylation assay and analysed the samples by Western blot analysis (Fig. 3D). SIRT6 efficiently removed the acetyl groups from H3K9Ac and H3K18Ac in nucleosomes in the presence of NAD+. The non-specific sirtuin inhibitor nicotinamide (NAM) was also tested as a reference for SIRT6 inhibition, and it only displayed weak inhibition of SIRT6 deacetylation at a concentration of 10 mmol/L. However, when treated with JYQ-42, concentrations as low as 0.5 μmol/L could inhibit the deacetylase activity of SIRT6 significantly, and at a concentration of 1 μmol/L JYQ-42, the deacetylase activity of SIRT6 was completely/totally inhibited (Fig. 3D).
These data suggested that JYQ-42 could indeed efficiently inhibit the deacetylation activity of SIRT6 and that its inhibition was more robust on more physiological substrates, such as nucleosomes. All these data demonstrated that JYQ-42 effectively inhibited SIRT6 deacetylation activity in vitro.
3.3. JYQ-42 inhibits SIRT6 through the newly identified allosteric exosite Pocket Z
To elucidate the structural mechanisms underlying the inhibition of SIRT6 by JYQ-42, we attempted to determine the structure of SIRT6 in complex with JYQ-42 using both X-ray and NMR methods, but neither method was successful. Hence, alternatively, we combined biophysical and docking methods to explore the structural mechanism of JYQ-42 inhibition of SIRT6 deacetylation. To confirm that JYQ-42 indeed binds to SIRT6, a surface plasmon resonance (SPR) assay was performed to evaluate the binding affinity of JYQ-42. HIS-SIRT6 was immobilized on a CM5 sensor chip, and the binding of JYQ-42 at concentrations ranging from 3.125 to 100 μmol/L was tested. SPR data showed that JYQ-42 bound to SIRT6 in a dose-dependent manner (Fig. 4A). Curve fitting of the sensorgrams showed that the Kd of JYQ-42 was 22.07 μmol/L, reflecting a high affinity of JYQ-42 for SIRT6 (Fig. 4B). In addition, we also used biolayer interferometry (Octet) assays to measure the ability of JYQ-42 to bind to immobilized SIRT6 (Supporting Information Fig. S9). The Octet assay results were consistent with the SPR results, revealing concentration-dependent binding of JYQ-42, with a Kd of 13.2 μmol/L. All these observations demonstrated that JYQ-42 directly binds to SIRT6 to inhibit its deacetylation activity.
Figure 4.
JYQ-42 is an allosteric inhibitor of SIRT6. (A) The binding effect of JYQ-42 on SIRT6 was analysed by surface plasmon resonance (SPR). Three independent experiments were performed. (B) SPR binding curves fitted with steady-state model showing the binding affinity of JYQ-42. The Kd value of JYQ-42 was 22.03 ± 3.11 μmol/L, and three independent experiments were performed. (C) Molecular docking of SIRT6 with JYQ-42. The SIRT6-ADPr crystal structure (PDB ID: 3K35) was used for docking, and the best JYQ-42 binding pose with the smallest Glide/IFD score or lowest energy was chosen for further analysis. (D) Effects of allosteric-site mutations located in the JYQ-42 binding pocket on the inhibition of SIRT6 deacetylation, as determined by FDL assays. Data are presented as the mean ± SD, n = 3 wells, from three independent experiments.
Next, we docked JYQ-42 into the Pocket Z of SIRT6. Based on our docked model, JYQ-42 bound to Pocket Z and made several critical interactions with adjacent residues to facilitate its binding and inhibition (Fig. 4C). Considering the plastic nature of allosteric pockets41,42, we further carried out MD simulation for docking optimizations, which retrieved a similar binding pose for JYQ-42 (Supporting Information Fig. S10). To verify this finding, we individually mutated the key residues in Pocket Z and performed FDL assays. In agreement with the results of the docking analysis, mutations of these key residues (Y5A, T57A, P62A, D63A, P67A, H68A, V70A, P80A, K81A, Y257A) led to dramatically decreased inhibition ratios at 50 μmol/L and increased IC50 values for JYQ-42. In particular, mutations of T57A and H68A completely disrupted JYQ-42 inhibition of SIRT6 deacetylation, suggesting that interactions between the SIRT6 mutants and JYQ-42 were disrupted (Fig. 4D). Collectively, our data demonstrated that JYQ-42 inhibited SIRT6 deacetylation through the newly identified allosteric exosite Pocket Z.
3.4. JYQ-42 is a non-competitive inhibitor with high selectivity for targeting SIRT6
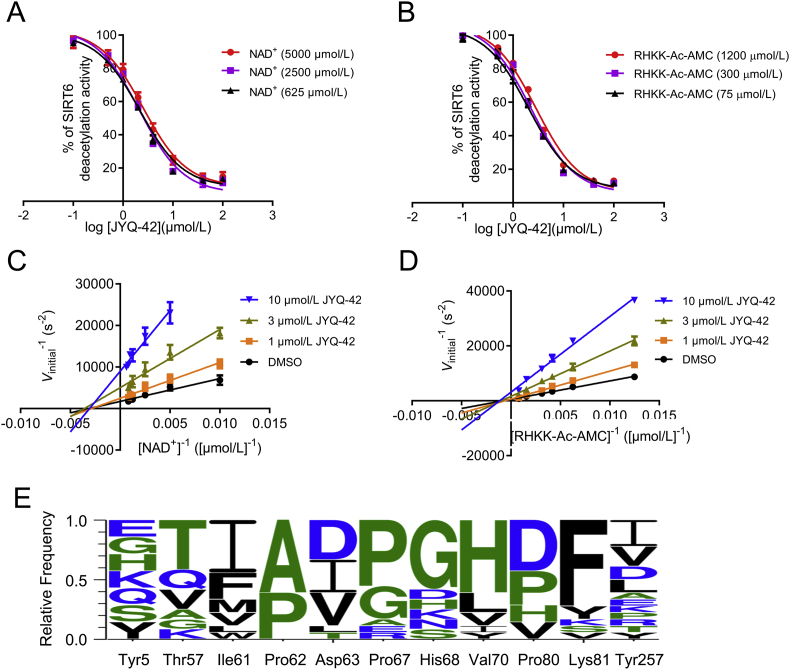
Docking and mutation analyses revealed that JYQ-42 binds to the novel allosteric cavity Pocket Z in SIRT6, and to biochemically confirm that JYQ-42 inhibits SIRT6 deacetylation through allostery, we performed competition assays between SIRT6 substrates and JYQ-42. As shown in Fig. 5A and B, increasing concentrations of NAD+ or the substrate peptide Ac-RHKK-Ac-AMC did not interfere with the inhibition by JYQ-42. Next, we performed an enzymatic kinetic assay to further elucidate the inhibition mode of JYQ-42. Lineweaver–Burk plot analysis showed that the inhibition curves crossed the horizontal axis (Fig. 5C and D). Kinetic assays showed that JYQ-42 did not change the Km but greatly decreased kcat and Vmax with increasing concentrations of JYQ-42 for both the acetylated substrate peptide and NAD+ (Table 1, Supporting Information Figs. S11 and S12). Consequently, the values of kcat/Km [L/(mol·s)] decreased significantly with increasing concentrations of JYQ-42 for both the acetylated substrate and NAD+. This finding suggests that JYQ-42 is a non-competitive inhibitor.
Figure 5.
JYQ-42 is a non-competitive inhibitor of SIRT6. (A) Competition assay between NAD+ and JYQ-42, as evaluated by the Fluor de Lys (FDL) assay. (B) Competition assay between Ac-RHKK-Ac-AMC and JYQ-42, as evaluated by the Fluor de Lys (FDL) assay. (C and D) Enzyme kinetics analysis of SIRT6 inhibition by JYQ-42 were performed with varying concentrations of NAD+(C) and Ac-RHKK-Ac-AMC (D) by HPLC. Double-reciprocal plots of the initial velocities (Lineweaver–Burk plots) showing non-competitive inhibition towards NAD+ and Ac-RHKK-Ac-AMC. Data are presented as the mean ± SD, n = 3 wells, from three independent experiments. (E) The sequence conservation of the allosteric pocket was analysed by using WebLogo62. The amino acids are coloured according to their chemical properties as follows: hydrophobic: black; hydrophilic: blue; neutral: green.
Table 1.
Kinetic parameters of SIRT6 deacetylation of Ac-RHKK-Ac-AMC in the absence or presence of 1, 3, and 10 μmol/L JYQ-42, as determined by HPLC.
| Compd. | Ac-RHKK-Ac-AMC |
NAD+ |
||||
|---|---|---|---|---|---|---|
| kcat (S−1) | Km(μmol/L) | kcat/Km [L/(mol.s)] | kcat (S−1) | Km(μmol/L) | kcat/Km [L/(mol.s)] | |
| DMSO | 55.59 × 10−4 | 12,146 | 0.46 | 2.36 × 10−4 | 240.3 | 0.98 |
| JYQ-42 (1 μmol/L) | 33.65 × 10−4 | 10,269 | 0.36 | 1.62 × 10−4 | 271.8 | 0.60 |
| JYQ-42 (3 μmol/L) | 22.12 × 10−4 | 12,437 | 0.18 | 8.57 × 10−5 | 257.2 | 0.33 |
| JYQ-42 (10 μmol/L) | 13.86 × 10−4 | 14,387 | 0.10 | 4.09 × 10−5 | 172.7 | 0.24 |
Data are presented as the mean ± SD from three independent experiments. The kcat and Km of NAD+ are apparent values because the concentration of Ac-RHKK-Ac-AMC was subsaturating (640 μmol/L).
SIRT6 is a member of the evolutionarily conserved sirtuin family of NAD+-dependent protein deacetylases. Historically, the structural similarity within the HDAC superfamily has led to considerable difficulty in developing specific modulators and has resulted in notorious off-target adverse/side effects. The selectivity of JYQ-42 for 17 HDACs, including other sirtuins, was thus tested.
As shown in Table 2, JYQ-42 exhibited no activity for most HDACs, including HDAC1 to HDAC11, SIRT3, SIRT5 and SIRT7. Although JYQ-42 showed mild inhibition of SIRT2, its efficacy was actually low, with a 38.6% inhibition ratio at 50 μmol/L and a half-maximal inhibitory concentration (IC50) value at 87.2 μmol/L, which was approximately 37-fold higher than that of SIRT6. Next, we analysed the conservation of the sequence features of the allosteric pocket among SIRT6 and other sirtuin family members. We extracted the residues from all the sirtuin family members that aligned with the residues that formed the allosteric pocket in SIRT6. We found that only P62 and P67 were relatively conserved, and amino acids at the other positions were less conserved, indicating the high selectivity of the allosteric pocket (Fig. 5E).
Table 2.
Target selectivity of JYQ-42 for HDACs and sirtuins.
| Enzyme | Effect | Activity IC50 (μmol/L)a |
|---|---|---|
| HDAC1 | ND | ND |
| HDAC2 | ND | ND |
| HDAC3 | ND | ND |
| HDAC4 | ND | ND |
| HDAC5 | ND | ND |
| HDAC6 | ND | ND |
| HDAC7 | ND | ND |
| HDAC8 | ND | ND |
| HDAC9 | ND | ND |
| HDAC10 | ND | ND |
| HDAC11 | ND | ND |
| SIRT1 | ND | ND |
| SIRT2 | Inhibition | 87.2 ± 1.1 |
| SIRT3 | ND | ND |
| SIRT5 | ND | ND |
| SIRT6 | Inhibition | 2.33 ± 0.17 |
| SIRT7 | ND | ND |
The activity values are calculated based on IC50 for the inhibition effect. ND had no detectable inhibitory effect on the targets in the presence of JYQ-42 at 100 μmol/L. Data are presented as the mean ± SD from three independent experiments.
Therefore, these data underscored that JYQ-42 was a highly selective deacetylase inhibitor targeting SIRT6, highlighting its promising pharmacological prospects.
3.5. JYQ-42 inhibits SIRT6 deacetylation activity in pancreatic cancer cells
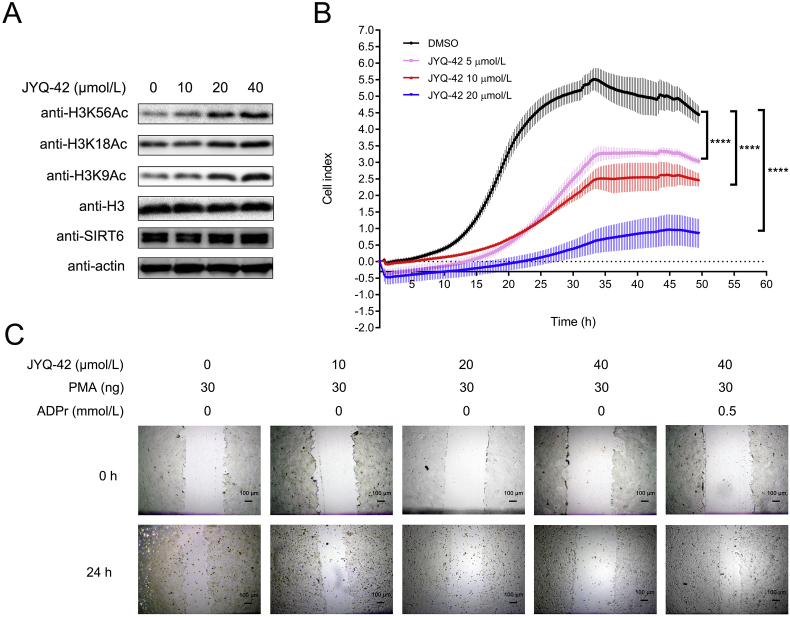
We have demonstrated that JYQ-42 inhibits SIRT6 deacetylation in vitro through allosteric effects. We next evaluated the potential of JYQ-42 to be used as a probe for the biological characterization and mechanistic investigation of SIRT6 in a cellular context. SIRT6 can deacetylate histone H3 at K9, K18, and K56 in cells9,11,12. Therefore, we first tested whether JYQ-42 could inhibit SIRT6 deacetylation in a plethora of pancreatic cancer cell lines. The BXPC-3 and MiaPaCa-2 cell lines were treated with different concentrations of JYQ-42, and their acetylation levels of H3K9, H3K18 and H3K56 were examined. We found that JYQ-42 increased the levels of H3K9Ac, H3K18Ac and H3K56Ac in a dose-dependent manner (Fig. 6A and Supporting Information Fig. S13). Therefore, our data demonstrated that JYQ-42 inhibited endogenous SIRT6 deacetylation activity in pancreatic cancer cells.
Figure 6.
JYQ-42 inhibits SIRT6 deacetylation and cell migration of pancreatic cancer cells. (A) Western blot analysis of SIRT6 and its substrates H3K9Ac, H3K18Ac and H3K56Ac in BXPC-3 cells treated with the indicated doses of JYQ-42 for 24 h. (B) Kinetic curves of the migration of BXPC-3 cells in the absence or presence of 5, 10, 20 μmol/L JYQ-42, as determined by xCELLigence RTCA-DP. Data are presented as the mean ± SD, n = 3 wells, from three independent experiments. ∗∗∗∗P < 0.0001, t-test (two-tailed and unpaired). (C) Representative images of wound-healing assays of BXPC-3 cells in the absence or presence of 10, 20, 40 μmol/L JYQ-42 obtained under phase contrast microscopy. Scale bar, 100 μm. The images shown are representative of triplicate experiments.
3.6. JYQ-42 inhibits cell migration of and cytokine production in pancreatic cancer cells
One of the oncogenic functions of SIRT6 in pancreatic cancer is its promotion of cell migration and related cancer metastasis. Through deacetylation of histone H3 at K9, SIRT6 can promote cell migration of and cytokine production in pancreatic cancer cells. Based on the aforementioned results, we thus investigated whether inhibition of SIRT6 deacetylation by JYQ-42 could block the migration of pancreatic cancer cells and probed their migratory properties. We used the xCELLigence Real-Time Cell Analysis (RTCA) system to obtain quantitative cell migration information. We followed the migration of BXPC-3 and MiaPaCa-2 cells exposed to DMSO and different concentrations of JYQ-42 in culture media and collected data at 15-min intervals. The cell index curve gradually reached a maximum under DMSO conditions, indicating that the untreated cells continued to migrate over time. However, upon addition of JYQ-42, the migration of pancreatic cancer cells was dramatically attenuated, even at concentrations as low as 5 μmol/L (Fig. 6B, Supporting Information Fig. S14). These RTCA results showed that JYQ-42 inhibited the migration of pancreatic cancer cells in a time- and dose-dependent manner (Fig. 6B, Fig. S14). Since it was plausible that the restricted cell migration might stem from compromised/decreased cell growth induced by JYQ-42, cell vitality and proliferation were thus assessed. We found that JYQ-42 had no significant impact on cell growth, even at an extremely high concentration of 20 μmol/L, which therefore ruled out the possibility that JYQ-42 inhibited cell migration by interfering with cell growth (Supporting Information Fig. S15).
We then performed a wound-healing assay to further determine the effect of JYQ-42 on cancer cell migration. Upon treatment with JYQ-42, the migration of BxPC-3 cells was significantly suppressed (the relative migration rate was suppressed by ~0.3 at 10 μmol/L JYQ-42, ~0.24 at 20 μmol/L JYQ-42, and ~0.10 at 40 μmol/L JYQ-42 compared to that of the control, ~0.60, Fig. 6C, Supporting Information Fig. S16). It has been previously reported that SIRT6 promotes cell migration by increasing the intracellular ADPr concentration43. Therefore, the inhibitory effect of JYQ-42 should be able to be rescued by ADPr co-treatment. As expected, in the presence of 0.5 mmol/L ADPr, even a high dose of JYQ-42 (40 μmol/L) had no influence on the migration of BXPC-3 cells (Fig. 6C, Fig. S16). These results demonstrated that JYQ-42 inhibited pancreatic cancer cell migration by blocking SIRT6-related intracellular levels of ADPr and downstream signalling, indicating the pharmacological effect on migration induced by JYQ-42 via SIRT6 inhibition.
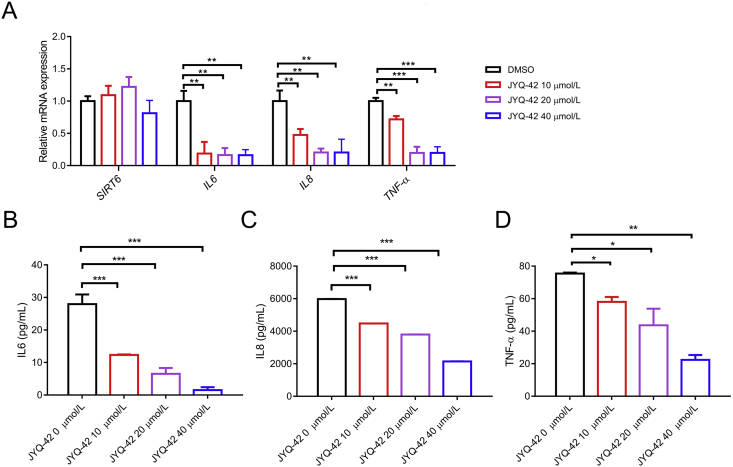
SIRT6 also confers advantages for pancreatic cancer progression by promoting pro-inflammatory cytokine production/secretion, which induce an inflammatory microenvironment supportive/beneficial to cancer development/carcinogenesis by enhancing angiogenesis. We thus investigated the influences of JYQ-42 on cytokine production and secretion. We first analysed the mRNA levels of cytokines in pancreatic cancer cells treated with JYQ-42 for 24 h. As shown in Fig. 7A, inhibitor treatment significantly reduced the expression of IL6, IL8 and TNF-α in BXPC-3 cells in a dose-dependent manner (Fig. 7A). Notably, the mRNA level of SIRT6 was not significantly changed upon JYQ-42 treatment, suggesting that it did not affect the expression of SIRT6. We next utilized cytokine ELISA kits to investigate cytokine secretion. Consistently, treatment with JYQ-42 for 24 h blocked the secretion levels of IL6, IL8 and TNF-α in BXPC-3 cells (Fig. 7B‒D) in a dose-dependent manner. These results showed that JYQ-42 reduced the expression and secretion of pro-inflammatory cytokines by pancreatic cancer cells by inhibiting the deacetylation activity of SIRT6, which represents a novel strategy for pancreatic cancer therapeutics.
Figure 7.
JYQ-42 inhibits the expression and secretion of inflammatory factors. (A) The SIRT6, IL6, IL8 and TNF-α mRNA levels in BXPC-3 cells in the absence or presence of 10, 20, 40 μmol/L JYQ-42 were measured by qPCR and quantified versus the GAPDH housekeeping gene. Data are presented as the mean ± SD, n = 3 wells, from three independent experiments. ∗∗P < 0.01, ∗∗∗P < 0.001, t-test (two-tailed and unpaired). (B–D) BXPC-3 cells were incubated for 24 h with 30 ng/mL PMA in the absence or presence of 10, 20, 40 μmol/L JYQ-42, and the concentrations of inflammatory factors (IL6, IL8, TNF-α) in supernatants were detected by ELISA. Data are presented as the mean ± SD, n = 3 wells, from three independent experiments. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001, t-test (two-tailed and unpaired).
4. Discussion
As a member of the mammalian sirtuin family, SIRT6 has been implicated in a myriad of biological processes, and drugging SIRT6 is drawing increasing attention as a strategy for treating SIRT6-mediated diseases, such as cancers and metabolic disorders4,44,45. Nevertheless, SIRT6 inevitably suffers from compromised selective targeting due to the highly conserved structural architecture of its catalytic site4,14,46. As an alternative, allosteric modulators, by targeting less conserved allosteric sites, provide an opportunity for specific binding to SIRT6 among other sirtuins24.
Using a state-of-the-art reversed allosteric communication model through a hybrid computational and experimental method, we identified a novel allosteric exosite Pocket Z in SIRT6 induced by the bidirectional signal stemming from orthosteric binding of NAD+. To the best of our knowledge, we identified the most potent SIRT6-selective small-molecule inhibitor, JYQ-42, and elucidated its pharmacological performance as well as its anti-pancreatic cancer activity.
With the significant progresses in biophysics and bioinformatics, computational exploration of allosteric sites has prominently boosted rational allosteric drug design47, 48, 49, 50, 51, 52, 53, 54, 55, 56. The reversed allosteric communication, namely orthosteric perturbations induce the emergence of allosteric sites, has been exemplified in the exchange protein activated by cAMP isoform 1 (EPAC1), 15-lipoxygenase and phosphoinositide-dependent protein kinase 1 (PDK1). Our developed computational method is the first attempt in exploiting reversed allostery theory to predict allosteric sites. This method can still undergo optimization. Calculation of energy coupling scores, which quantifies the strength of bi-directional allosteric communications, is the core of our computational pipeline. Such process is currently based on the MM-GBSA model provided within the Amber MD simulation package, providing an easy-to-use toolkit and retrieving favourable predictive outcomes. In the future, different algorithms such as energy matrix diagonalization methods could be incorporated into our pipeline to improve the predictive ability57, 58, 59, 60. Moreover, another direction is the characterization of detailed residual or even atomistic level signal pathways that the bi-directional allosteric signals propagate. With a plethora of emerging allosteric signal algorithms such as WISP and Ohm, this direction is ongoing in our lab, which will provide more thorough mechanistic insights towards the reversed allosterism theory and aid further enhancement of our reversed allostery-based methodology39,61.
Currently, one of the central unmet challenges of sirtuin modulator development is the inability of modulators to distinguish among structurally conserved sirtuins, SIRT1 to SIRT7, which unavoidably leads to off-target and adverse effects4,46. To date, few SIRT6 inhibitors with the desired efficacy and specificity have been reported15, 16, 17, 18, 19, 20. For example, the endogenous inhibitor nicotinamide (NAM) can inhibit all sirtuin members, and NAM-related pyrazinamide also exhibits similar inhibitory properties. Synthetic potent SIRT6 small-molecule inhibitors, such as the quercetin derivatives catechin gallate and gallocatechin gallate, exhibit compromised selectivity23. Cyclic peptide SRIT6 inhibitors suffer from poor specificity and low intracellular potency17. Our small-molecule JYQ-42 arguably showed both optimal specificity and ideal efficacy for SIRT6 relative to other sirtuins as well as members of the broader HDAC family, highlighting the pharmacological advantage of allosteric ligands. Most importantly, JYQ-42 not only inhibited the migration of pancreatic cancer cells but also inhibited pro-inflammatory cytokine secretion, both of which contributed to its potent anti-cancer effects.
5. Conclusions
In summary, based on reversed allosteric communication, we uncovered a novel, potential allosteric site in SIRT6 and discovered an allosteric small-molecule inhibitor with promising efficacy and selectivity as well as anti-cancer activity. The identified compound represents a starting point for the design of other SIRT6 allosteric inhibitors. Moreover, our study might provide a universal and applicable framework for the discovery of allosteric candidates for other therapeutic targets.
Acknowledgments
We thank Hao Hu and Minjia Tan at Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China for their help in examining the levels of acetylated peptides. This work was supported by the National Natural Science Foundation of China (81925034, 81903458, 22077082, 82003605, 81901423), the Innovation Program of Shanghai Municipal Education Commission (2019-01-07-00-01-E00036, China), Shanghai Science and Technology Innovation Fundation (19431901600, China), the Shanghai Health and Family Planning System Excellent Subject Leader and Excellent Young Medical Talents Training Program (2018BR12, China), Special Financial Grant of Postdoctoral Research Foundation of China (2019M660090).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2021.06.015.
Contributor Information
Shaoyong Lu, Email: lushaoyong@sjtu.edu.cn.
Jian Zhang, Email: jian.zhang@sjtu.edu.cn.
Author contributions
Qiufen Zhang, Yingyi Chen and Zhimin Huang designed the research. Qiufen Zhang, Zhimin Huang, Jiacheng Wei, Shaobo Ning, Li Feng, Jianrong Xu and Jun-Cheng Su performed the biological experiments of this work; Yingyi Chen, Yingqing Wei, Xiuyan Yang, Houwen Lin and Mingzhu Zhao are responsible for the design and synthesis of compounds. Shaoyong Lu, Duan Ni, Yuran Qiu, Kun Song, Zhengtian Yu and Xinyi Li performed the prediction of allosteric pocket. Qiufen Zhang, Yingyi Chen, Duan Ni and Shaoyong Lu wrote the manuscript. Jian Zhang and Shaoyong Lu supervised the project.
Conflicts of interest
The authors declare no conflicts of interest regarding this manuscript.
Appendix ASupplementary data
The following is the Supplementary data to this article:
References
- 1.Chang A.R., Ferrer C.M., Mostoslavsky R. SIRT6, a mammalian deacylase with multitasking abilities. Physiol Rev. 2020;100:145–169. doi: 10.1152/physrev.00030.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houtkooper R.H., Pirinen E., Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao E., Hou J., Ke X., Abbas M.N., Kausar S., Zhang L., et al. The roles of sirtuin family proteins in cancer progression. Cancers (Basel) 2019;11:1949. doi: 10.3390/cancers11121949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carafa V., Rotili D., Forgione M., Cuomo F., Serretiello E., Hailu G.S., et al. Sirtuin functions and modulation: from chemistry to the clinic. Clin Epigenet. 2016;8:61. doi: 10.1186/s13148-016-0224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tasselli L., Zheng W., Chua K.F. SIRT6: novel mechanisms and links to aging and disease. Trends Endocrinol Metabol. 2017;28:168–185. doi: 10.1016/j.tem.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saiyang X., Deng W., Qizhu T. Sirtuin 6: a potential therapeutic target for cardiovascular diseases. Pharmacol Res. 2021;163:105214. doi: 10.1016/j.phrs.2020.105214. [DOI] [PubMed] [Google Scholar]
- 7.He Y., Yang G., Yao F., Xian Y., Wang G., Chen L., et al. Sitagliptin inhibits vascular inflammation via the SIRT6-dependent signaling pathway. Int Immunopharm. 2019;75:105805. doi: 10.1016/j.intimp.2019.105805. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H., Khan S., Wang Y., Charron G., He B., Sebastian C., et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kokkonen P., Rahnasto-Rilla M., Mellini P., Jarho E., Lahtela-Kakkonen M., Kokkola T. Studying SIRT6 regulation using H3K56 based substrate and small molecules. Eur J Pharmaceut Sci. 2014;63:71–76. doi: 10.1016/j.ejps.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 10.Liszt G., Ford E., Kurtev M., Guarente L. Mouse Sir 2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 11.Michishita E., McCord R.A., Berber E., Kioi M., Padilla-Nash H., Damian M., et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492. doi: 10.1038/nature06736. U16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tasselli L., Xi Y.X., Zheng W., Tennen R.I., Odrowaz Z., Simeoni F., et al. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol. 2016;23:434–440. doi: 10.1038/nsmb.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gil R., Barth S., Kanfi Y., Cohen H.Y. SIRT6 exhibits nucleosome-dependent deacetylase activity. Nucleic Acids Res. 2013;41:8537–8545. doi: 10.1093/nar/gkt642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan P.W., Feldman J.L., Devries M.K., Dong A., Edwards A.M., Denu J.M. Structure and biochemical functions of SIRT6. J Biol Chem. 2011;286:14575–14587. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He B., Hu J., Zhang X.Y., Lin H.N. Thiomyristoyl peptides as cell-permeable Sirt6 inhibitors. Org Biomol Chem. 2014;12:7498–7502. doi: 10.1039/c4ob00860j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kokkonen P., Rahnasto-Rilla M., Kiviranta P.H., Huhtiniemi T., Laitinen T., Poso A., et al. Peptides and pseudopeptides as SIRT6 deacetylation inhibitors. ACS Med Chem Lett. 2012;3:969–974. doi: 10.1021/ml300139n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J., Zheng W. Cyclic peptide-based potent human SIRT6 inhibitors. Org Biomol Chem. 2016;14:5928–5935. doi: 10.1039/c5ob02339d. [DOI] [PubMed] [Google Scholar]
- 18.Parenti M.D., Grozio A., Bauer I., Galeno L., Damonte P., Millo E., et al. Discovery of novel and selective SIRT6 inhibitors. J Med Chem. 2014;57:4796–4804. doi: 10.1021/jm500487d. [DOI] [PubMed] [Google Scholar]
- 19.Rahnasto-Rilla M., Tyni J., Huovinen M., Jarho E., Kulikowicz T., Ravichandran S., et al. Natural polyphenols as sirtuin 6 modulators. Sci Rep. 2018;8:4163. doi: 10.1038/s41598-018-22388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sociali G., Galeno L., Parenti M.D., Grozio A., Bauer I., Passalacqua M., et al. Quinazolinedione SIRT6 inhibitors sensitize cancer cells to chemotherapeutics. Eur J Med Chem. 2015;102:530–539. doi: 10.1016/j.ejmech.2015.08.024. [DOI] [PubMed] [Google Scholar]
- 21.Wu J., Li D., Liu X., Li Q., He X., Wei J., et al. IDDB: a comprehensive resource featuring genes, variants and characteristics associated with infertility. Nucleic Acids Res. 2021;49:D1218–D1224. doi: 10.1093/nar/gkaa753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolivar B.E., Welch J.T. Studies of the binding of modest modulators of the human enzyme, sirtuin 6, by STD NMR. Chembiochem. 2017;18:931–940. doi: 10.1002/cbic.201600655. [DOI] [PubMed] [Google Scholar]
- 23.You W., Zheng W., Weiss S., Chua K.F., Steegborn C. Structural basis for the activation and inhibition of sirtuin 6 by quercetin and its derivatives. Sci Rep. 2019;9:19176. doi: 10.1038/s41598-019-55654-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Z., Zhao J., Deng W., Chen Y., Shang J., Song K., et al. Identification of a cellularly active SIRT6 allosteric activator. Nat Chem Biol. 2018;14:1118–1126. doi: 10.1038/s41589-018-0150-0. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X., Allison D., Condon B., Zhang F.Y., Gheyi T., Zhang A.P., et al. The 2.5 angstrom crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition. J Med Chem. 2013;56:963–969. doi: 10.1021/jm301431y. [DOI] [PubMed] [Google Scholar]
- 26.Ravindranath P.A., Forli S., Goodsell D.S., Olson A.J., Sanner M.F. AutoDockFR: advances in protein-ligand docking with explicitly specified binding site flexibility. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. Ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theor Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang Y.P. Novel zinc protein molecular dynamics simulations: steps toward antiangiogenesis for cancer treatment. J Mol Model. 1999;5:196–202. [Google Scholar]
- 29.Ni D., Wei J.C., He X.H., Rehman A.U., Li X.Y., Qiu Y.R., et al. Discovery of cryptic allosteric sites using reversed allosteric communication by a combined computational and experimental strategy. Chem Sci. 2021;12:464–476. doi: 10.1039/d0sc05131d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bucher D., Pierce L.C.T., McCammon J.A., Markwick P.R.L. On the use of accelerated molecular dynamics to enhance configurational sampling in ab initio simulations. J Chem Theor Comput. 2011;7:890–897. doi: 10.1021/ct100605v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao Y., Nichols S.E., Gasper P.M., Metzger V.T., McCammon J.A. Activation and dynamic network of the M2 muscarinic receptor. Proc Natl Acad Sci U S A. 2013;110:10982–10987. doi: 10.1073/pnas.1309755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scherer M.K., Trendelkamp-Schroer B., Paul F., Perez-Hernandez G., Hoffmann M., Plattner N., et al. PyEMMA 2: a software package for estimation, validation, and analysis of markov models. J Chem Theor Comput. 2015;11:5525–5542. doi: 10.1021/acs.jctc.5b00743. [DOI] [PubMed] [Google Scholar]
- 33.Husic B.E., Pande V.S. Markov state models: from an art to a science. J Am Chem Soc. 2018;140:2386–2396. doi: 10.1021/jacs.7b12191. [DOI] [PubMed] [Google Scholar]
- 34.Jiang H., Deng R., Yang X., Shang J., Lu S., Zhao Y., et al. Peptidomimetic inhibitors of APC−Asef interaction block colorectal cancer migration. Nat Chem Biol. 2017;13:994–1001. doi: 10.1038/nchembio.2442. [DOI] [PubMed] [Google Scholar]
- 35.Liu G., Chen H., Liu H., Zhang W., Zhou J. Emerging roles of SIRT6 in human diseases and its modulators. Med Res Rev. 2020;41:1089–1137. doi: 10.1002/med.21753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shukla D., Hernandez C.X., Weber J.K., Pande V.S. Markov state models provide insights into dynamic modulation of protein function. Accounts Chem Res. 2015;48:414–422. doi: 10.1021/ar5002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidtke P., Le Guilloux V., Maupetit J., Tuffery P. Fpocket: online tools for protein ensemble pocket detection and tracking. Nucleic Acids Res. 2010;38:W582–W589. doi: 10.1093/nar/gkq383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Guilloux V., Schmidtke P., Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinf. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., Jain A., McDonald L.R., Gambogi C., Lee A.L., Dokholyan N.V. Mapping allosteric communications within individual proteins. Nat Commun. 2020;11:3862. doi: 10.1038/s41467-020-17618-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan Z.W., Guarnera E., Tee W.V., Berezovsky I.N. AlloSigMA 2: paving the way to designing allosteric effectors and to exploring allosteric effects of mutations. Nucleic Acids Res. 2020;48:W116–W124. doi: 10.1093/nar/gkaa338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He X., Ni D., Lu S., Zhang J. Characteristics of allosteric proteins, sites, and modulators. Adv Exp Med Biol. 2019;1163:107–139. doi: 10.1007/978-981-13-8719-7_6. [DOI] [PubMed] [Google Scholar]
- 42.Spinello A., Martini S., Berti F., Pennati M., Pavlin M., Sgrignani J., et al. Rational design of allosteric modulators of the aromatase enzyme: an unprecedented therapeutic strategy to fight breast cancer. Eur J Med Chem. 2019;168:253–262. doi: 10.1016/j.ejmech.2019.02.045. [DOI] [PubMed] [Google Scholar]
- 43.Bauer I., Grozio A., Lasiglie D., Basile G., Sturla L., Magnone M., et al. The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. J Biol Chem. 2012;287:40924–40937. doi: 10.1074/jbc.M112.405837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong Z., Lei Q., Liu L., Cui H. Function of SIRT6 in tumor initiation and progression. Sheng Wu Gong Cheng Xue Bao. 2016;32:870–879. doi: 10.13345/j.cjb.150403. [DOI] [PubMed] [Google Scholar]
- 45.Etchegaray J.P., Zhong L., Mostoslavsky R. The histone deacetylase SIRT6: at the crossroads between epigenetics, metabolism and disease. Curr Top Med Chem. 2013;13:2991–3000. doi: 10.2174/15680266113136660213. [DOI] [PubMed] [Google Scholar]
- 46.Arrowsmith C.H., Bountra C., Fish P.V., Lee K., Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 47.Feher V.A., Durrant J.D., Van Wart A.T., Amaro R.E. Computational approaches to mapping allosteric pathways. Curr Opin Struct Biol. 2014;25:98–103. doi: 10.1016/j.sbi.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu S., He X., Ni D., Zhang J. Allosteric modulator discovery: from serendipity to structure-based design. J Med Chem. 2019;62:6405–6421. doi: 10.1021/acs.jmedchem.8b01749. [DOI] [PubMed] [Google Scholar]
- 49.Lu S., Shen Q., Zhang J. Allosteric methods and their applications: facilitating the discovery of allosteric drugs and the investigation of allosteric mechanisms. Acc Chem Res. 2019;52:492–500. doi: 10.1021/acs.accounts.8b00570. [DOI] [PubMed] [Google Scholar]
- 50.Lu S.Y., Ni D., Wang C.X., He X.H., Lin H.W., Wang Z., et al. Deactivation pathway of ras gtpase underlies conformational substates as targets for drug design. ACS Catal. 2019;9:7188–7196. [Google Scholar]
- 51.Qiu Y., Yin X., Li X., Wang Y., Fu Q., Huang R., et al. Untangling dual-targeting therapeutic mechanism of epidermal growth factor receptor (EGFR) based on reversed allosteric communication. Pharmaceutics. 2021;13:747. doi: 10.3390/pharmaceutics13050747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu S., Chen Y., Wei J., Zhao M., Ni D., He X., et al. Mechanism of allosteric activation of SIRT6 revealed by the action of rationally designed activators. Acta Pharm Sin B. 2021;11:1355–1361. doi: 10.1016/j.apsb.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boulton S., Selvaratnam R., Blondeau J.P., Lezoualc'h F., Melacini G. Mechanism of selective enzyme inhibition through uncompetitive regulation of an allosteric agonist. J Am Chem Soc. 2018;140:9624–9637. doi: 10.1021/jacs.8b05044. [DOI] [PubMed] [Google Scholar]
- 54.Meng H., Dai Z., Zhang W., Liu Y., Lai L. Molecular mechanism of 15-lipoxygenase allosteric activation and inhibition. Phys Chem Chem Phys. 2018;20:14785–14795. doi: 10.1039/c7cp08586a. [DOI] [PubMed] [Google Scholar]
- 55.Schulze J.O., Saladino G., Busschots K., Neimanis S., Suss E., Odadzic D., et al. Bidirectional allosteric communication between the ATP-binding site and the regulatory PIF pocket in PDK1 protein kinase. Cell Chem Biol. 2016;23:1193–1205. doi: 10.1016/j.chembiol.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 56.Meng H., McClendon C.L., Dai Z., Li K., Zhang X., He S., et al. Discovery of novel 15-lipoxygenase activators to shift the human arachidonic acid metabolic network toward inflammation resolution. J Med Chem. 2016;59:4202–4209. doi: 10.1021/acs.jmedchem.5b01011. [DOI] [PubMed] [Google Scholar]
- 57.Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun H., Li Y., Tian S., Xu L., Hou T. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys Chem Chem Phys. 2014;16:16719–16729. doi: 10.1039/c4cp01388c. [DOI] [PubMed] [Google Scholar]
- 59.Montefiori M., Pilotto S., Marabelli C., Moroni E., Ferraro M., Serapian S.A., et al. Impact of mutations on NPAC structural dynamics: mechanistic insights from MD simulations. J Chem Inf Model. 2019;59:3927–3937. doi: 10.1021/acs.jcim.9b00588. [DOI] [PubMed] [Google Scholar]
- 60.Serapian S.A., Marchetti F., Triveri A., Morra G., Meli M., Moroni E., et al. The answer lies in the energy: how simple atomistic molecular dynamics simulations may hold the key to epitope prediction on the fully glycosylated SARS-CoV-2 spike protein. J Phys Chem Lett. 2020;11:8084–8093. doi: 10.1021/acs.jpclett.0c02341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Wart A.T., Durrant J., Votapka L., Amaro R.E. Weighted implementation of suboptimal paths (WISP): an optimized algorithm and tool for dynamical network analysis. J Chem Theor Comput. 2014;10:511–517. doi: 10.1021/ct4008603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kannan S., Venkatachalam G., Lim H.H., Surana U., Verma C. Conformational landscape of the epidermal growth factor receptor kinase reveals a mutant specific allosteric pocket. Chem Sci. 2018;9:5212–5222. doi: 10.1039/c8sc01262h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.