Summary
Identifying new pathways that regulate mammalian regeneration is challenging due to the paucity of in vivo screening approaches. We employed pooled CRISPR knockout and activation screening in the regenerating liver to evaluate 165 chromatin regulatory proteins. Both screens identified imitation-SWI chromatin remodeling components Baz2a and Baz2b, not previously implicated in regeneration. In vivo sgRNA, siRNA, and knockout strategies against either paralog confirmed increased regeneration. Distinct BAZ2-specific bromodomain inhibitors GSK2801 and BAZ2-ICR resulted in accelerated liver healing after diverse injuries. Inhibitor treated mice also exhibited improved healing in an inflammatory bowel disease model, suggesting multi-tissue applicability. Transcriptomics on regenerating livers showed increases in ribosomal and cell cycle mRNAs. Surprisingly, CRISPRa screening to define mechanisms showed that overproducing Rpl10a or Rpl24 was sufficient to drive regeneration, while Rpl24 haploinsufficiency was rate limiting for BAZ2 inhibition mediated regeneration. The discovery of regenerative roles for imitation-SWI components provides immediate strategies to enhance tissue repair.
eTOC statement
New regulators of tissue regeneration are challenging to identify in mammalian models. Here, pooled in vivo CRISPR screening in the liver identified imitation-SWI chromatin remodeling components Baz2a and Baz2b as regeneration suppressors. BAZ2-specific bromodomain inhibitors resulted in accelerated liver and intestinal healing, effects mediated through increased protein synthesis during regeneration.
Graphical Abstract

Introduction
Regeneration occurs when lost or diseased tissues are replaced through cell growth, division, and differentiation (Goldman and Poss, 2020). In humans, impaired regeneration contributes to poor outcomes after acute trauma, tissue damage, or organ transplant, as well as in chronic diseases such as liver cirrhosis (Cordero-Espinoza and Huch, 2018; Forbes and Newsome, 2016; Liang et al., 2021), inflammatory bowel disease (IBD) (Nagahama et al., 2018), and diabetes (Wei et al., 2018). Limited regeneration can also restrict the ability to perform aggressive surgical, radiotherapeutic, or chemotherapeutic interventions for cancer, which often depend on robust regeneration for organ recovery. Regenerative therapies could expand the number of patients that could receive anti-cancer treatments and improve the outcomes for those that do.
Studies of species that exhibit scarless wound healing abilities have hinted at the central role of epigenetic regulation. Zebrafish and newts regenerate in part through tissue de-differentiation, by which regenerating cells revert to the more plastic, earlier stages of development to replenish lost body parts (Goldman and Poss, 2020). Once regeneration nears completion, cells must be able to cease proliferation and faithfully reacquire differentiated states. We reasoned that the epigenome must be reconstructed by chromatin associated proteins to permit the transcriptional reprogramming required to toggle between regenerating and differentiated states (Maki and Kimura, 2013). Because epigenetic factors may play permissive, rather than constitutive roles in driving proliferation (Kang et al., 2016; Sun et al., 2016; Wang et al., 2020, 2019), targeting them may be less likely to induce diseases of persistent proliferation.
A major barrier to discovering new mechanisms that facilitate mammalian regeneration is the difficulty in identifying relevant genes in tissue culture systems, which provide poor surrogates for regenerative medicine. Currently, in vitro studies of regeneration are increasingly dependent on promising organoid platforms (Aloia et al., 2019; Planas-Paz et al., 2019). However, studies of tissue regeneration ultimately depend on in vivo systems that most accurately model growth after tissue injury. Here, we employed in vivo CRISPR screening approaches to identify pathways that promote regeneration across multiple tissues.
Results
In vivo CRISPR screening identified Baz2 paralogs as regulators of tissue regeneration
To identify the chromatin-associated genes that could be perturbed to increase tissue regeneration, we employed in vivo gain- and loss-of-function CRISPR screening in the liver. This was performed in fumarylacetoacetate hydrolase knockout (Fah KO) mice that experience chronic liver damage due to a deficiency in FAH, an enzyme that catabolizes tyrosine intermediates. Liver related mortality is prevented with a drug called nitisinone (NTBC) (Grompe et al., 1995; Overturf et al., 1996). In untreated Fah KO livers, there is a strong selection for FAH expressing hepatocytes that repopulate after correction of the Fah gene defect (Wuestefeld et al., 2013; Zahm et al., 2020). We used a transposable element containing Cas9, an sgRNA against any gene of interest, and Fah (as a selection marker) for targeted gene deletion (CRISPR KO: Figure 1A). Separately, we also engineered a transposable element to turn on genes of interest using dCas9 fused with the transcriptional activator VP64 (dCas9-VP64; CRISPRa: Figure 1B) (Gilbert et al., 2014; Liao et al., 2017; Wangensteen et al., 2018). These transposon plasmids, along with Sleeping Beauty Transposase (SB100), were delivered via hydrodynamic transfection (HDT) and integrated into the genomes of hepatocytes that received them (Chen and Calvisi, 2014) (Figure 1C, D).
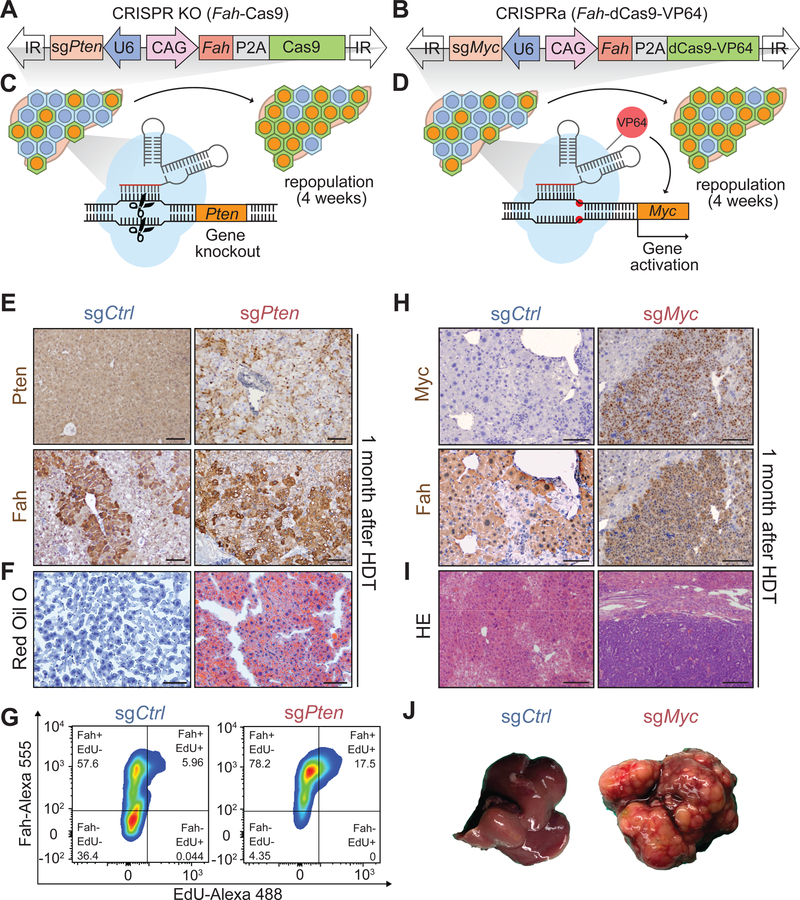
Figure 1. Sleeping Beauty transposon mediated in vivo CRISPR KO and CRISPRa systems can effectively delete and overexpress individual genes in the liver.
A. Transposon plasmid used for gene deletion and repopulation of Fah KO mouse livers.
B. Transposon plasmid used for gene activation.
C. Schema for generating Pten KO clones in regenerating Fah KO livers.
D. Schema for the Myc activation experiment in regenerating Fah KO livers.
E. Representative immunohistochemical (IHC) staining showing PTEN-, FAH+ hepatocytes 1 month after HDT (scale bar = 100 μm). This was performed in n = 3, 3 replicate mice.
F. Oil Red O staining showing the extent of liver steatosis 1 month after HDT (scale bar = 100 μm; representative of n = 3, 3 mice).
G. Flow cytometry analysis on perfused hepatocytes from mice that had received transposon with control or Pten targeting sgRNAs 1 month prior. The plot is representative of n = 3, 3 replicate mice.
H. Representative IHC staining showing Myc overexpressing, FAH+ hepatocytes 1 month after HDT (scale bar = 100 μm). The experiment was performed in n = 3, 3 replicate mice.
I. H&E images showing tumors in livers receiving sgRNA targeting the Myc promoter (scale bar = 200 μm).
J. Gross liver appearance 1 month after HDT. Tumors are in all 3 livers receiving sgRNA targeting the Myc promoter.
We first tested CRISPR KO and CRISPRa approaches for individual genes. We previously used the CRISPR KO system to delete Pten in the mouse liver (Zhu et al., 2019), and here we further characterized this approach (Figure 1A, C). Livers receiving Pten targeting sgRNAs accumulated PTEN protein deficient hepatocytes that also expressed FAH (Figure 1E) (Xue et al., 2014). After 4 weeks of Pten deficient clone expansion under Fah selection, livers became fatty, mimicking Cre-LoxP Pten models (Figure 1F) (Stiles et al., 2004). Pten deleted hepatocytes also exhibited increased proliferation when FAH+ and EdU+ hepatocytes were quantified using flow cytometry (Figure 1G). To examine gain-of-function phenotypes, we tested the CRISPRa system (Figure 1B, D). Livers receiving Myc promoter targeting sgRNAs exhibited Myc overexpression within one week. Myc overexpressing livers showed increased proliferation and transformed into cancer within 4 weeks (Figure 1H–J). Thus, these in vivo CRISPR systems could detect genes that drive increased proliferation, a property of increased regeneration, but were also able to read-out hepatic steatosis and cancer formation, undesirable consequences of dysregulated regeneration.
We then generated complimentary KO and activation libraries targeting the functional domains of 165 druggable epigenetic factors, previously screened by the Vakoc group (Shi et al., 2015). These libraries included 905 KO sgRNAs and 861 CRISPRa sgRNAs (Supplemental Table S1), and were delivered into Fah KO mice via HDT such that each mouse received the entire libraries for either gene KO or activation (Figure 2A). After NTBC was withdrawn, clones of transfected and FAH-rescued hepatocytes bearing distinct sgRNAs would compete during liver regeneration and sgRNAs that conferred a selective advantage or disadvantage in regenerating hepatocytes would become over or under-represented. Four weeks later, sgRNA repopulation was quantified by deep sequencing.
Figure 2. Pooled in vivo CRISPR screening identified Baz2a and Baz2b as suppressors of liver regeneration.
A. Schema of in vivo CRISPR KO and activation screens.
B. Genes identified in the CRISPR KO and CRISPR activation screens. The genes with larger fonts were significant in both screens and had log2 fold change in opposing directions. The red dots indicate genes that inhibit whereas the blue dots indicate genes that promote clonal expansion (n = 5 for both screens).
C. Representative composite IF images showing FAH+ and FAH; BrdU double+ hepatocytes 18 days after HDT of sleeping beauty transposons containing five distinct sgRNAs targeting either Baz2a or Baz2b (See Figure S1B). Top panels are images of whole liver scans, and the bottom panels are close up images (scale bar top = 2000 μm and bottom = 50 μm).
D. Percentage of FAH+ and FAH+; EdU+ hepatocytes from flow cytometry analysis of perfused hepatocytes after 18 days of liver repopulation (n = 5, 5, 5 mice). Individual flow cytometry plots are shown in Figure S1C.
E. Representative composite IF images showing FAH+ and FAH; BrdU double+ hepatocytes 1 month after HDT of sleeping beauty transposon expressing mouse Fah cDNA and human BAZ2A or BAZ2B cDNA (See Figure S1D; scale bar = 100 μm).
F. Percentage of FAH+ area over total tissue area taken from liver images of mice in E (n = 5, 5, 5 mice). Small black dots represent FAH+ areas from single 10X images. Larger dots represent FAH+ areas of each mouse averaged from five images.
CRISPR KO screening identified multiple enriched sgRNAs against Baz2a, Baz2b, Mll1, Kmt2a, Kdm4c, Kdm6b, Kdm5b, and Kdm3a (Figure 2B, Supplemental Table S1), while CRISPRa screening identified multiple depleted sgRNAs against Baz2a, Baz2b, Kdm6b and Kdm3a across replicate mice (Figure 2B, Supplemental Table S1). Imitation-SWI/SNF genes Baz2a and Baz2b, and histone demethylase genes Kdm6b and Kdm3a, were the four genes that scored in both gain and loss-of-function screens. To further assess these candidates, we examined mRNA expression in regenerating livers after 70% partial hepatectomy (hereafter called PHx). While the expression of Kdm6b and Kdm3a remained the same at all time points after PHx, Baz2a and Baz2b expression decreased starting 2 hours and nadired between 8–40 hours after PHx (Figure S1A). This suggested that Baz2 suppression is physiologically important for regeneration.
Validation of Baz2a and Baz2b as suppressors of liver regeneration
The interaction between BAZ2 proteins with the SMARCA5 enzyme defines the nucleolar remodeling complex (NoRC), one of five subtypes of imitation-SWI complexes that uses ATP hydrolysis to remodel nucleosomes (Anosova et al., 2015; Santoro et al., 2009; Savić et al., 2014; Zhou et al., 2009; Zillner et al., 2013). These simple two component complexes are distinct from the 10–15 component SWI/SNF complexes previously identified as regeneration regulators (Sun et al., 2016). We focused on the imitation-SWI subunits and sought to confirm that Baz2a and/or Baz2b suppress regeneration. To delete Baz2 genes individually, we delivered a transposon co-expressing Cas9 and FAH along with non-targeting (Fah-sgCtrl), Baz2a targeting (Fah-sgBaz2a), or Baz2b targeting sgRNAs (Fah-sgBaz2b) into Fah KO mice (Figure S1B). Each transposon was packaged with five distinct sgRNAs per gene to ensure effective deletion. NTBC was withdrawn and liver repopulation ensued for 18 days. sgBaz2a and sgBaz2b treated mice had larger FAH+ repopulating clones and higher fractions of EdU/FAH double+ hepatocytes (Figure 2C, D and Figure S1C). We did not observe steatosis or tumor formation, as was seen with Pten loss or Myc overexpression. Consistent with the loss-of-function studies, overexpression of human BAZ2A and BAZ2B inhibited liver repopulation in Fah KO mice (Figure S1D), resulting in smaller FAH+ clones (Figure 2E, F).
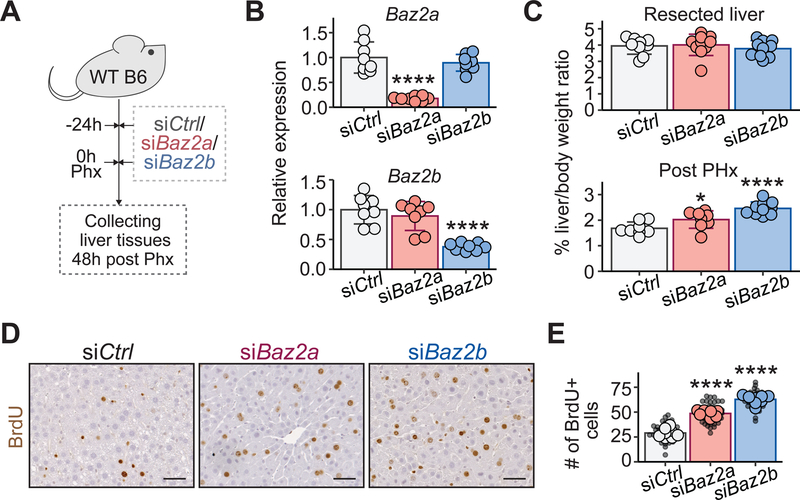
To assess the screening results using liver-wide knockdown rather than mosaic clonal deletion, we treated WT mice with in vivo siRNAs against either Baz2a or Baz2b prior to 70% PHx (Figure 3A). In vivo grade siRNAs, packaged in lipid nanoparticles (LNPs) and delivered intravenously into mice, effectively suppressed Baz2a and Baz2b mRNA (Figure 3B). Knockdown of either gene caused higher liver to body mass ratios (Figure 3C) and more proliferating cells after PHx (Figure 3D, E). Notably, neither Baz2a or Baz2b knockdown changed the expression of tyrosine metabolism enzymes (Figure S1E), reducing the possibility that the regenerative advantage in Fah KO mice was due to influences on tyrosine metabolism.
Figure 3. In vivo siRNA knockdown of Baz2a or Baz2b increased liver regeneration after 70% PHx.
A. Schema for the in vivo siRNA experiments. Two treatments of siRNAs packaged in 5A2-SC8 LNPs (4 mg/kg siRNA) were given 24 hours before surgery and at the time of surgery.
B. Relative mRNA levels of Baz2a or Baz2b measured 72 hours after the first dose of siRNA (n = 8, 8, 8 mice).
C. Resected liver mass/body mass ratios 24 hours after siRNA treatment and regenerated liver mass/body mass ratios 48 hours after 70% PHx (Basal: n = 9,10,10 mice; post-PHx: n = 8,8,10 mice).
D. IHC staining for BrdU incorporation 48 hours after 70% PHx. 100 mg/kg BrdU was given 3 hours before tissue collection (scale bar = 50 μm).
E. Quantification of BrdU incorporation from D (n = 8, 8, 8 mice). Small black dots represent the number of BrdU+ cells from a single 10X image. Larger dots represent the number of BrdU+ cells in each mouse averaged from five images.
Baz2b knockout mice are healthy and exhibit enhanced tissue regeneration
Deleting either Baz2 paralog caused increased regeneration phenotypes in the liver, but deleting Baz2b resulted in larger effects (Figure 2D). Although both paralogs were suppressed during PHx induced regeneration, Baz2b transcript levels decreased more (Figure S1A). Moreover, BAZ2B but not BAZ2A expression was positively correlated with human aging, which has been associated with diminished regenerative capacity (Lu et al., 2004; Yuan et al., 2020; Zhang et al., 2013). Altogether, these observations suggested that Baz2b could have a larger impact on regeneration. To test the effects of Baz2b loss, we generated whole body Baz2b knockout (KO) mice using CRISPR mediated deletion of exon 13, which is within the essential Methyl-CpG binding (MBD) domain (Anosova et al., 2015) (Figure S2A). Exon 13 deletion was predicted to result in a frame-shifted transcript with multiple premature stop codons. A qPCR assay targeting the deleted exon showed a reduction of this mRNA product in KO livers (Figure S2B). Baz2b KO mice were born in the expected mendelian ratios (194 total pups: 43 WT, 106 Het, 45 KO), and showed no significant differences in body weight (Figure S2C), liver/body weight ratio (Figure S2D), liver proliferation (Figure S2E), and overall appearance. Baz2b KO mice also had normal liver function tests (AST/ALT) (Figure S2F) as well as normal differentiation, cell cycle, and CYP450 gene expression (Figure S2G). Notably, KO mice had normal organ appearance and mass (Figure S2H–I), and did not develop abnormal liver histology (Figure S2J), even at 1.5 years of age. Importantly, no spontaneous tumorigenesis was observed. This suggested that prolonged BAZ2B inhibition would be therapeutically tolerated, and showed that there was no constitutive proliferation without tissue injury.
After 70% PHx, Baz2b KO livers exhibited increased regenerated mass (Figure 4A), proliferating cells (Figure 4B), and cell cycle gene expression (Figure S2K). Notably, increases in regeneration were not associated with persistent proliferation or dedifferentiation, as cell cycle and differentiation related expression in KO mice returned to baseline levels 1 month after PHx (Figure 4C, D). To test if Baz2b deletion could inhibit hepatotoxin mediated liver damage in addition to increasing regeneration, we exposed mice to carbon tetrachloride (CCl4), which causes centrilobular necrosis and a proliferative response in surviving hepatocytes. After a single dose of CCl4, KO vs. WT mice exhibited reduced serum markers of liver damage, less centrilobular necrosis, and increased post-injury proliferation (Figure 4E–H). Overall, Baz2b deficient mice exhibited improved regeneration but did not show overt signs of long-term health problems (cancer, steatosis, liver dysfunction).
Figure 4. Baz2b KO mice exhibited enhanced tissue regeneration.
A. The liver to body weight ratios of resected livers at the time of 70% PHx and regenerated livers from Baz2b WT and KO mice (n = 8, 8 mice) 48 hours after surgery.
B. IHC images and quantification of BrdU+ hepatocytes from WT and KO mice 48 hours after PHx (scale bar = 50 μm; n = 8, 8 mice). Smaller black dots represent the number of BrdU+ cells for a single 10X image. Larger dots represent the number of BrdU+ cells for each mouse averaged from five images.
C. Cell cycle gene expression in livers of WT and Baz2b−/− mice 1 month after 70% PHx as measured by qPCR (n = 5, 5 mice). PHx was performed at 8 weeks of age.
D. Differentiation related gene expression in livers of 8-week-old WT and Baz2b−/− 1 month after 70% PHx as measured by qPCR (n = 5, 5 mice).
E. Schema of acute CCl4 injury experiment. Baz2b−/− and littermate WT mice were injected with one dose of CCl4. AST and ALT levels were measured 24 hours after CCl4 injection, and liver tissues were collected 48 hours after injection.
F. AST and ALT levels observed 24 hours after CCl4 injection.
G. Representative H&E images taken 48 hours after one dose of CCl4 and quantification of necrotic areas (scale bar = 200 μm; n = 8, 7 mice). Smaller black dots represent the necrotic areas from single 5X images. Larger dots represent the necrotic areas for each mouse averaged from five images.
H. IHC staining and quantification of Ki67+ hepatocytes from WT and KO mice 48 hours after CCl4 (Left scale bar = 100 μm; right scale bar = 50 μm; n = 8, 7 mice). Smaller black dots represent the number of Ki67+ cells from single 10X images. Larger dots represent the number of Ki67+ cells for each mouse averaged from ten images.
Small molecule BAZ2 inhibition accelerates tissue regeneration
We asked if pharmacologic BAZ2 inhibition might also confer a tissue repair advantage. Fortuitously, two independent selective acetyl-lysine competitive inhibitors of BAZ2A and BAZ2B were previously generated (Chen et al., 2016; Drouin et al., 2015). These inhibitor scaffolds are chemically distinct, both disrupt the interaction between BAZ2 bromodomains and acetylated histones in biochemical assays, and both dissociate imitation-SWI complexes from chromatin in cell based assays (Chen et al., 2016; Drouin et al., 2015). GSK2801 binds to BAZ2 bromodomains with dissociation constants (KD) of 257 nM (BAZ2A) and 136 nM (BAZ2B), while BAZ2-ICR has KD’s of 109 nM (BAZ2A) and 170 nM (BAZ2B). Both inhibitors show high specificity for BAZ2A and BAZ2B, but GSK2801 is known to have off-target interactions. GSK2801 is partnered with a control compound called GSK8573 that shares the same off-target, BRD9, but does not inhibit BAZ2 proteins. AlphaScreen assays confirmed that GSK2801 and BAZ2-ICR bind to BAZ2B bromodomains with an IC50 of 0.65 μM and 0.26 μM, respectively, while GSK8575 did not bind to the BAZ2B bromodomain (Figure S3A). Moreover, we found that GSK2801 significantly disrupts chromatin binding of Flag-tagged BAZ2A or BAZ2B by using chromatin immunoprecipitation sequencing (ChIP-seq) (Figure S3B).
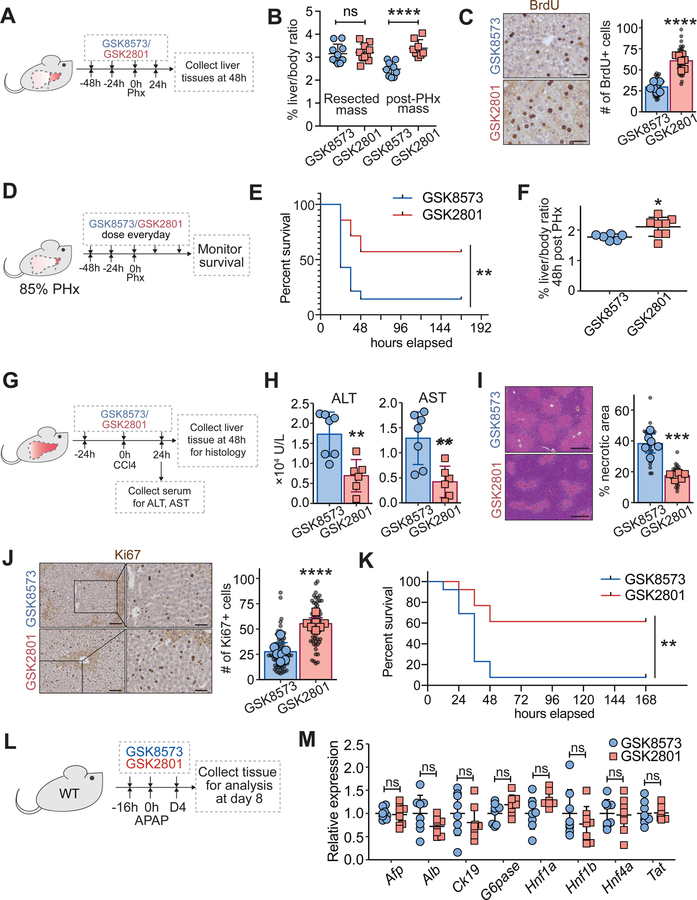
Consistent with the genetic data, mice given GSK2801 vs. GSK8573 for a total of 5 days did not exhibit liver overgrowth, damage, altered histology, or CYP450 expression (Figure S3C–F). To test if chemical BAZ2A/B inhibition could increase regeneration, we treated WT mice with two doses of GSK2801 prior to PHx (Figure 5A). This led to an increase in liver mass recovery and BrdU incorporation 48 hours after surgery (Figure 5B, C). Next, we performed a time course experiment in which GSK8573 or GSK2801 was dosed immediately after surgery (Figure S4A). GSK2801 caused an increase in liver mass recovery at 48 and 72 hours (Figure S4B), showing that BAZ2 inhibition causes rapid effects on regeneration even when drug is given after injury. Associated with this was increased BrdU incorporation and cell cycle gene expression (Figure S4C, D). We then asked if BAZ2 inhibition could improve outcomes for larger liver resections. We performed 85% PHx, where all lobes except for the right inferior and caudate lobes were removed surgically (Figure 5D). While only 14% (2/14) of control mice survived beyond 48 hours after surgery, 57% (8/14) of mice given GSK2801 survived (Figure 5E). GSK2801 treated mice also exhibited increased liver mass 48 hours after surgery (Figure 5F).
Figure 5. Chemical inhibition of BAZ2 resulted in increased liver regeneration.
A. Schema of an experiment to test BAZ2 inhibition prior to surgical liver resection. WT B6 mice were given GSK2801 or GSK8573 control (30 mg/kg IP daily). 70% PHx was performed 48 hours after the first dose and livers were harvested 48 hours after surgery.
B. Liver to body weight ratios of pre-treated mice at 0 and 48 hours after 70% PHx (pre-PHx: n = 10, 10 mice; post-PHx: n = 9, 8 mice).
C. IHC images and quantification of BrdU+ hepatocytes 48 hours after surgery (scale bar = 50 μm; n = 7, 7 mice). Smaller black dots represent the number of BrdU+ cells from a single 10X image. Larger dots represent the number of BrdU+ cells for each mouse averaged from five images.
D. Schema of an experiment to test BAZ2 inhibition in a more severe hepatectomy model. WT B6 mice were given GSK2801 or GSK8573. 85% PHx was performed 48 hours after the first dose and moribund mice were euthanized.
E. Kaplan-Meier survival curve of WT mice receiving GSK8573 or GSK2801 after 85% PHx (n = 14, 14 mice).
F. Liver-to-body weight ratios 48 hours after 85% PHx (n = 6, 8 mice).
G. Schema for CCl4 injury after BAZ2 inhibition. WT B6 mice were injected with GSK2801 or GSK8573. One dose of CCl4 was given one day after drug treatment. Serum ALT and AST were measured. Livers were collected 48 hours after CCl4.
H. Serum AST and ALT measured 24 hours after CCl4 (n = 7, 6 mice).
I. H&E staining and hepatic necrosis quantification 48 hours after CCl4 injection (scale bar = 200 μm; n = 6, 6 mice). Smaller black dots represent the necrotic areas from single 5X images. Larger dots represent the necrotic areas for each mouse averaged from five images.
J. IHC staining and Ki67 quantification 48 hours after CCl4 injection (scale bar (left) = 100 μm, (right) = 50 μm; n = 7, 7 mice). Smaller black dots represent the number of Ki67+ cells in a single 10X image. Larger dots represent the number of Ki67+ cells for each mouse averaged from ten images.
K. Kaplan-Meier survival curve of WT mice receiving GSK8573 or GSK2801 (30 mg/kg per day × 2 days) 4 hours after a single dose of APAP (500 mg/kg x1 IP; n = 13, 13 mice).
L. Schema for the APAP experiment in L, M. 16 hours prior to APAP (300 mg/kg), GSK8573 or GSK2801 at 30 mg/kg per day was initiated and lasted for 4 days. 8 days after APAP, liver tissues were collected.
M. Differentiation related gene expression 8 days after APAP as measured by qPCR (n = 8, 8 mice).
To test if chemical BAZ2A/B inhibition could prevent hepatocyte damage in addition to increasing regeneration, WT mice were treated with inhibitors prior to CCl4 exposure (Figure 5G). 24 hours after CCl4, BAZ2 inhibited mice were protected from tissue necrosis and showed increased proliferation (Figure 5H–J). To further test the idea that these phenotypes were due to the on-target effects of GSK2801, we also evaluated BAZ2-ICR, a structurally dissimilar BAZ2 inhibitor. This inhibitor also protected against CCl4 induced damage and increased proliferation (Figure S4E, F). To examine the importance of BAZ2 inhibitors in another clinically relevant scenario, we exposed mice to toxic doses of acetaminophen (APAP), which is a common cause of lethal acute liver failure in humans. 8% (1/13) of mice exposed to APAP and then dosed with control GSK8573 survived, while 62% (8/13) of mice dosed with GSK2801 survived (Figure 5K). Notably, livers treated with BAZ2 inhibitors functioned normally without detectable differences in histology, proliferation, or differentiation 8 days after APAP (Figure 5L, M and Figure S4G), when injury has been resolved via regeneration. These beneficial effects were not likely due to altered metabolism since APAP was given prior to BAZ2 inhibitors, and CYP450 expression did not change substantially after BAZ2 inhibition (Figure S3F). In the CCl4 injury experiments, tissue protective and pro-regenerative effects were likely operating in tandem to improve outcomes in both genetic and drug contexts. Overall, pharmacological inhibition of BAZ2 could improve regenerative responses and survival outcomes in the setting of clinically-relevant injuries and life threatening drug poisonings.
Small molecule BAZ2 inhibition enhances colonic regeneration in the setting of colitis
It was unknown if regeneration pathways identified in the liver could have important effects in other tissues, so we asked if BAZ2 inhibition could also promote intestinal regeneration. Indeed, GSK2801 promoted intestinal recovery in a model of inflammatory bowel disease. Ulcerative colitis is effectively modeled with Dextran Sodium Sulfate (DSS) salt, a branched glucan that induces intestinal inflammation and colitis in rodents when given in the drinking water (Tian et al., 2019; Zaki et al., 2010). 3% DSS treated mice were also given GSK2801 or GSK8573 (Figure S5A). Control mice began to lose weight 5 days post DSS initiation, while inhibitor treated mice remained within 10% of their original weight until day 8 (Figure S5B). In addition, control vs. BAZ2 inhibitor treated mice had more gastrointestinal bleeding and diarrhea, hallmarks of colitis (Figure S5C). Furthermore, control treated mice had shortened colons, a phenotype associated with increased inflammation and tissue damage (Figure S5D). Blinded histologic analysis of tissue damage, ulceration, inflammation, and involved surface area showed that GSK8573 treated mice had more severe disease (Figure S5E, S5F). In addition, BrdU incorporation in colonic crypts on day 8 after oral DSS treatment was increased by ~50% in BAZ2 inhibitor-treated animals (Figure S5G, S5H), suggesting that BAZ2 inhibition increased epithelial healing in inflammatory bowel disease.
CRISPRa screening identified effectors downstream of BAZ2 inhibition
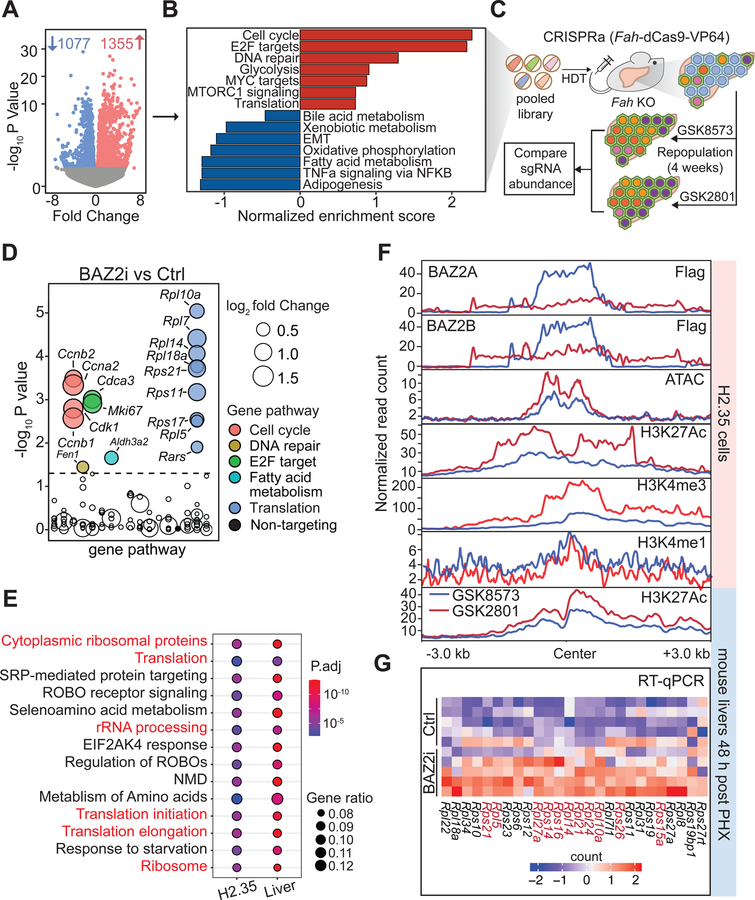
To examine the mechanisms underpinning regeneration phenotypes, we assessed transcriptional changes on regenerating livers 48 hours after PHx, with and without BAZ2 inhibition. RNA-seq revealed 1355 up-regulated and 1077 down-regulated transcripts (p-values < 0.05; Figure 6A; Supplemental Table S2). To identify pathways associated with BAZ2 inhibition, we performed Gene Set Enrichment Analysis (GSEA) on differentially expressed genes. Pathways related to cell cycle, translation, E2F targets, MYC targets, mTORC signaling, and glycolysis were upregulated, while those involved in fatty acid, bile acid, and xenobiotic metabolism were downregulated (Figure 6B and Supplemental Table S3). This is consistent with previous findings that the regenerating liver activates cell cycle genes and suppresses metabolic and biosynthetic pathways (Sun et al., 2016; Wang et al., 2018, 2020; White et al., 2005).
Figure 6. Ribosomal components are epigenetic targets of BAZ2 proteins.
A. Volcano plot showing differentially expressed genes from RNA-seq of 48h post-PHx mouse livers after BAZ2 inhibition. The differentially expressed genes were identified with a p-value < 0.05. Samples were obtained from the mice in Figure 5A.
B. Gene Set Enrichment Analysis (GSEA) on differentially expressed genes identified from A.
C. Schema for CRISPRa screening to identify pathways responsible for the pro-regeneration phenotypes associated with BAZ2 inhibition.
D. CRISPRa screening results of mice treated with GSK2801 or GSK8573 that underwent 4 weeks of repopulation. The x-axis shows the alphabetically-ordered gene pathways that each gene target belongs to.
E. Shared enriched pathways that were identified in both H2.35 cells and regenerating mouse livers. Pathways related to ribosomal biogenesis and translation are highlighted in red font. Pathway enrichment analysis is shown in Figure S6E, G and all pathways are listed in Supplemental Table S3.
F. Metaplots of normalized read densities around a 3 kb window flanking all peaks associated with 26 ribosome related genes bound by either Flag-BAZ2A or Flag-BAZ2B in H2.35 cells or regenerating livers treated with GSK8573 or GSK2801. For ChIP-seq of histone marks and ATAC-seq, the plots show the normalized results combined from three biological replicates. For ChIP-seq of Flag-BAZ2A and Flag-BAZ2B, the normalized plots were drawn from single biological replicates.
G. Heatmap showing qPCR analysis on ribosomal proteins in regenerating livers collected from Figure 5A. Red font highlights the genes with an p-value from student’s t-test < 0.05.
To isolate the functionally relevant changes downstream of BAZ2 inhibition, we performed in vivo CRISPRa screening on 111 differentially expressed genes selected from RNA-seq that are associated with GSEA identified pathways (Supplemental Table S1). After a CRISPRa library containing sgRNAs (≥5 unique sgRNAs/gene) was delivered into Fah KO livers via HDT, NTBC was withdrawn to induce injury. Mice were also randomized to GSK8573 or GSK2801 and allowed to regenerate for 4 weeks (Figure 6C). We reasoned that comparing the sgRNA frequencies +/− BAZ2 inhibition would allow us to hone in on mechanisms specific to BAZ2. In the presence of BAZ2 inhibition, sgRNAs that cause overexpression of regeneration promoting genes would experience less positive selection because the genes they target would already be activated by GSK2801. Thus, BAZ2 selective effects could be separated from BAZ2 independent ones. The sgRNAs with the largest differences in abundance between BAZ2-inhibited and control livers targeted genes involved in cell cycle progression (Ccnb1, Ccna2, Ccnb2, Cdk1, Cdca3, Mki67) or protein synthesis (Rpl10a, Rpl7, Rpl14, Rpl18a, Rps21, Rps11, Rps17, Rpl5, Rars) (Figure 6D, Supplemental Table S1). In fact, 9 out of 17 translation-related genes screened showed up as positive hits, which accounts for 52.9% of all hits (P < 0.01, Fisher’s exact test).
BAZ2 proteins regulate protein synthesis via chromatin remodeling
We sought to determine if the genes identified in CRISPRa screening were direct vs. indirect effectors of BAZ2. While BAZ2A-containing NoRCs are known to suppress ribosomal RNA (rRNA) transcription by recruiting histone deacetylases, histone methyltransferases, and DNA methyltransferases to rRNA promoters (Dalcher et al., 2020; Gu et al., 2015; Santoro et al., 2002; Zhou et al., 2002), it is less clear what other targets are remodeled by BAZ proteins. To define direct mechanisms, we examined the genome-wide BAZ2A and BAZ2B binding by ChIP-seq in mouse liver H2.35 cells transfected with Flag-tagged human BAZ2A or BAZ2B (Supplemental Table S2), and identified 1403 and 536 binding sites for BAZ2A and BAZ2B, respectively. Since GSK2801 can block BAZ2A/B’s interactions with chromatin (Figure S3B), we assessed changes in chromatin accessibility (ATAC-seq) and markers of histone activation (H3K27ac, H3K4me1, H3K4me3 ChIP-seq) resulting from BAZ2 inhibition. While global patterns of chromatin accessibility and H3K27ac were unperturbed (Figure S6A–C), a subset of genes directly bound by either BAZ2A and BAZ2B showed substantial changes (149 genes shown in Figure S6D and Supplemental Table S2). Enrichment analysis of these genes highlighted translational control and ribosome biogenesis pathways, among others (Figure 6E, Figure S6E). To determine if these targets were also relevant in vivo, we intersected H3K27ac ChIP-seq and RNA-seq data from regenerating livers, with and without BAZ2 inhibition (286 genes shown in Figure S6F and Supplemental Table S2). Consistent with data from H2.35 cells, ribosome and translation related pathways were the most enriched pathways in vivo (Figure 6E, Figure S6G). Given that both CRISPRa screening and epigenomic analysis identified translational machinery and ribosomal biogenesis, we focused on the 26 ribosomal protein encoding genes bound by either BAZ2A or BAZ2B. After BAZ2 inhibition, these loci showed increased chromatin accessibility, H3K27ac, and H3K4me3, in vitro and in vivo (Figure 6F). Because H3K4me1 levels did not change, BAZ2A and BAZ2B likely regulate ribosomal proteins through promoter interactions as opposed to distal enhancer ones. qPCR also verified the upregulation of these 26 ribosomal genes in regenerating livers (Figure 6G). These data show that BAZ2 proteins repress ribosome-related gene expression through chromatin remodeling.
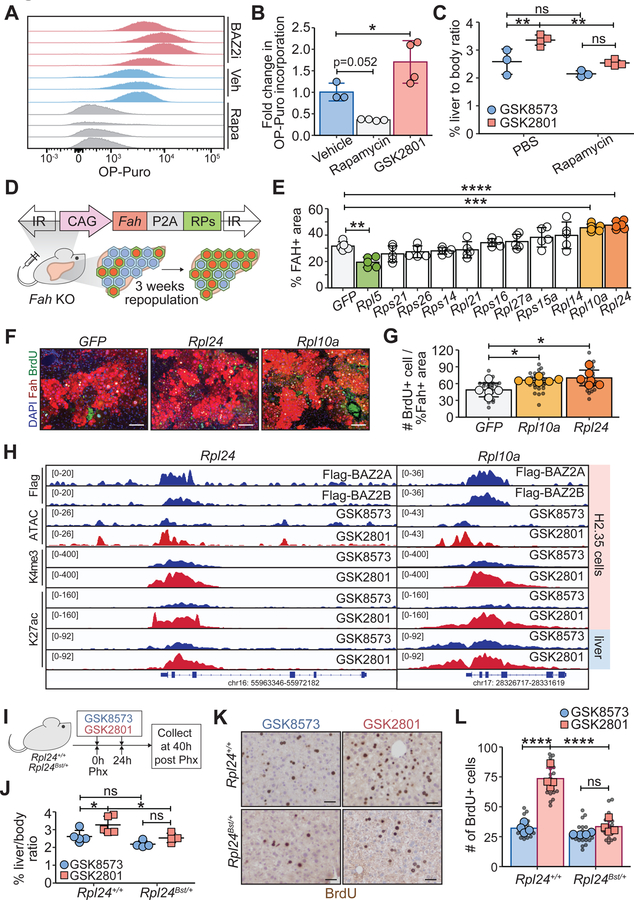
We asked if BAZ2 proteins actually mediate changes in protein synthesis. To directly measure rates of de novo protein production, mice were injected with O-propargyl-puromycin (OP-Puro), an alkyne analog of puromycin that is incorporated into nascent peptides and thus estimates global protein synthesis rates (Signer et al., 2014). Using flow cytometry to quantify OP-puro in 48h post-PHx livers, we found that inhibiting BAZ2 led to increased protein synthesis (Figure 7A, B). We next asked if protein synthesis was rate limiting for BAZ2 inhibition mediated regeneration. We dosed wild-type mice with 2.0 mg/kg rapamycin, a known inhibitor of protein synthesis (Fingar and Blenis, 2004; Terada et al., 1994), +/− BAZ2 inhibitor, while subjecting the mice to 70% PHx. Mice treated with rapamycin were able to undergo regeneration after PHx (Figure 7C and Figure S7A), but were limited in their ability to rapidly increase protein synthesis in response to injury (Figure 7A, B). By measuring liver mass (Figure 7C) and the number of cycling hepatocytes (Figure S7A), we found that BAZ2 inhibition did not enhance regeneration in rapamycin treated mice, suggesting the ability to upregulate protein synthesis was necessary to drive the pro-regeneration phenotypes (Figure 7C, Figure S7A).
Figure 7. Enhanced regeneration with BAZ2 inhibition was caused by increased protein synthesis rates.
A. Flow cytometry analysis of OP-puro incorporation in 48h-post-PHx mouse livers treated with vehicle, GSK2801, or rapamycin.
B. OP-puro quantification in hepatocytes (n = 3, 4, 4 mice).
C. Liver to body weight ratios 48 hours after 70% PHx (n = 3, 4, 3, 4 mice) in mice treated with PBS or 2.0 mg/kg of rapamycin. Small molecules were given to mice at the time of surgery and dosed every day x2 before tissue collection.
D. Schema showing the effect of overexpressing individual ribosomal proteins on hepatocyte repopulation in Fah KO mice.
E. Percent FAH+ area in ribosomal protein overexpression experiments from Figure S7B. FAH+ areas in each mouse were averaged from five 10X images.
F. Representative composite IF images of FAH and BrdU in mice with Rpl10a or Rpl24 overexpression (scale bar = 100 μm).
G. Number of BrdU+ cells per FAH+ percent area (n = 5, 6, 6 mice). Smaller black dots represent values from single 10X images. Larger dots represent average values from five images.
H. Normalized ChIP-seq tracks showing BAZ2 protein binding, chromatin accessibility, H3K4me3, and H3K27ac for Rpl24 and Rpl10a loci in H2.35 cells and in regenerating livers.
I. Schema of the experiment to determine the effect of reduced protein synthesis on BAZ2 inhibition. Two doses of GSK8573 or GSK2801 were given to Rpl24+/+ and Rpl24Bst/+ mice before and after surgery.
J. Liver to body weight ratios of Rpl24+/+ and Rpl24Bst/+ mice receiving GSK8573 and GSK2801, 40 hours after PHx (Rpl24+/+: n = 5, 5 mice; Rpl24Bst/+: n = 4, 4 mice).
K. BrdU staining 40 hours after PHx (scale bar = 50 μm).
L. Quantification of BrdU incorporation from K (Rpl24+/+: n = 4, 4 mice; Rpl24Bst/+: n = 4, 4 mice). Smaller black dots represent the number of BrdU+ cells from each 10X image. Larger dots represent the number of BrdU+ hepatocytes for each mouse averaged from five images.
Ribosomal proteins are sufficient and necessary for BAZ2 regenerative enhancement
Mechanistic CRISPRa screening showed that upregulation of individual ribosomal genes might be in part responsible for improved regeneration after BAZ2 inhibition. To probe the sufficiency of individual ribosomal genes in regenerative enhancement, we generated 11 transposons designed to overexpress FAH and ribosomal proteins observed to be bound by BAZ2A and BAZ2B and most significantly upregulated in BAZ2 inhibited livers (Figure 6G). We delivered these 11 transposons individually into Fah KO mice using HDT and allowed for 3 weeks of repopulation (Figure 7D). While most overexpressed ribosomal proteins did not affect liver repopulation (Figure S7B and Figure 7E), Rpl10a and Rpl24 accelerated repopulation, resulting in larger, proliferating FAH+ clones (Figure 7F, G). Both genes were bound by BAZ2A and BAZ2B, and their respective loci showed increased chromatin accessibility, H3K4me3, H3K27ac, and mRNA expression upon BAZ2 inhibition (Figure 7H).
To test the necessity of ribosomal genes for regeneration, we used a mouse with a hypomorphic allele of Rpl24 (Oliver et al., 2004). Rpl24Bst/+ mice develop normally, have normal liver function, and are able to undergo regeneration after PHx. We injected WT Rpl24+/+ and heterozygous Rpl24Bst/+ mice with GSK8573 or GSK2801 daily, then PHx was performed 2 days later (Figure 7I). Rpl24+/+ and Rpl24Bst/+ mice showed similar degrees of liver regeneration in the absence of BAZ2 inhibition, but GSK2801 was only able to drive enhanced regeneration in Rpl24+/+ mice (Figure 7J–L). Similarly, Rpl24Bst/+ mice partially rescued the transcriptional upregulation of cell cycle regulated genes induced by BAZ2 inhibition (Figure S7C), suggesting the ability to quickly upregulate translation to enhance regeneration is partially mediated by Rpl24.
Discussion
BAZ2 inhibition could be effective against liver failure induced by drugs, for accelerating recovery after surgery, or for improving donor or recipient outcomes after split liver transplants. BAZ2 inhibition may also be effective for IBD, which has multiple therapeutic strategies for reducing inflammation, but fewer strategies for promoting wound healing of refractory ulcers (Lindemans et al., 2015; Taniguchi et al., 2015). A key question is whether or not increasing regeneration equates to enforced proliferation, which could be achieved with the manipulation of any constitutive cell cycle regulator. This is not the case, since enforcing regeneration in the absence of tissue loss would create problems such as tissue overgrowth or malignancy. Neither BAZ2 inhibitor treated nor Baz2b KO mice exhibited overgrowth or spontaneous cancer development at steady state, thus there was no inappropriate regenerative drive without injury. In multiple cases, the suppression of chromatin remodelers does not constitutively drive proliferation (Gaub et al., 2010; Sun et al., 2016; Wang et al., 2019). However, targeting chromatin remodelers can be permissive of accelerated growth when appropriate regenerative signals are induced.
Increased ribosomal biogenesis is a known response to tissue injury (Chaudhuri and Lieberman, 1968; Loeb and Yeung, 1975; Nikolov et al., 1987; Scornik, 1974). Ribosomal abundance also displays circadian oscillation (Sinturel et al., 2017), and the kinetics of hepatocyte cell cycle entry after PHx correlates with these fluctuations (Matsuo et al., 2003). Despite the expectation that high protein synthesis rates might be essential for most types of regeneration, our findings support the tissue-specific importance of translational control. Low rates of protein synthesis are required to maintain long term metabolic homeostasis in blood and skin stem cells, even in the setting of rapid cell division (Signer et al., 2014; Blanco et al., 2016). This context specificity could explain why BAZ2 inhibition was unable to improve dorsal skin or ear hole wound healing (data not shown), but was effective in gastrointestinal wound healing. These observations are reminiscent of the fact that Myc overexpressing, but not Tp53 deficient, cancers are dependent on rapid increases in protein synthesis (Barna et al., 2008). Interestingly, overexpressing individual ribosomal subunits was alone sufficient to promote regeneration, supporting the possibility that subunit upregulation (Ebright et al., 2020), in addition to haploinsufficiency (Gregory et al., 2019; Khajuria et al., 2018), has meaningful physiological influences. It is also possible that BAZ2 inhibition provided just enough of a protein synthesis increase, or in just the right target genes, to drive increased regeneration without causing proteostasis-induced apoptosis or transformation.
Yuan et. al. found that Baz-2 knockdown caused lifespan extension and reduced features of neurological aging in C. elegans (Yuan et al., 2020). They also showed that older Baz2b deficient mice had reduced body weight and improved cognitive function. It is unknown if these phenotypes are linked to the regenerative phenotypes through shared molecular mechanisms such as translational control, or if differences in regenerative capacity have long term influences on neurologic function in aging. Collectively, our findings suggest that BAZ2 proteins represent promising therapeutic targets for several important disease processes.
Limitations of the Study
Experiments were not performed in human tissues, so it is unclear if regeneration will be enhanced to the same degree in clinical settings. We have not determined if BAZ2 inhibition would improve regeneration in cirrhotic livers, a clinical setting in need of therapeutic strategies. Most of the ChIP-seq analysis was performed in cell lines, and the finding that Rpl24 and Rpl10a were direct BAZ2 targets was obtained in immortalized mouse hepatocytes. This raises the possibility that in vivo and in vitro targets could be different. While increased liver regeneration is likely due to increased protein synthesis, we have not shown that this is true for the colitis model. It is possible that other mechanisms are responsible for improved intestinal healing.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources should be directed to and will be provided by the Lead Contact, Hao Zhu (hao.zhu@utsouthwestern.edu).
Materials availability statement
Plasmids, mouse lines, and other unique reagents generated in this study will be distributed upon request after completion of relevant material transfer agreements.
Data and code availability
RNA-seq and ChIP-seq data have been deposited at GEO and are publicly available. Accession numbers are listed in the key resources table. The analysed results are in Supplemental Table S2. Raw data for all the graphs in the figures are available in Supplemental Table S5. Microscopy data reported in this paper will be shared by the lead contact upon request.
There is no original code in this paper.
Any additional information required to reanalyze the data in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-PTEN | Cell Signaling Technology | Cat#9559; RRID: AB_390810 |
| Rabbit monoclonal anti c-Myc | Abcam | Cat#32072; RRID: AB_731658 |
| Rabbit polyclonal anti-FAH | Yecuris | Cat#20-0034 |
| Rat monoclonal anti-BrdU | Abcam | Cat#6326; RRID: AB_305426 |
| Mouse monoclonal anti-BrdU | BD Biosciences | Cat#555627; RRID: AB_10015222 |
| Rabbit polyclonal anti-Ki67 | Abcam | Cat#15580; RRID: AB_443209 |
| Rabbit monoclonal anti-E-cadherin | Cell Signaling Technology | Cat#3195; RRID: AB_2291471 |
| Rabbit polyclonal anti-H3K27ac | Abcam | Cat#4729; RRID: AB_2118291 |
| Rabbit polyclonal anti-H3K4me3 | Abcam | Cat#8580; RRID: AB_306649 |
| Rabbit polyclonal anti-H3K4me1 | Abcam | Cat#8895; RRID: AB_306847 |
| Mouse monoclonal anti-Flag | Sigma Aldrich | Cat#F1804; RRID: AB_262044 |
| Rabbit polyclonal anti-HNF4a | Santa Cruz Biotechnology | Cat#8987; RRID: AB_2116913 |
| Goat anti Rabbit IgG, Alexa Fluor 594 conjugated | Invitrogen | Cat#A-11012; RRID: AB_2534079 |
| Goat anti Rat IgG, Alexa Fluor 488 conjugated | Invitrogen | Cat#A-11006; RRID: AB_141373 |
| Goat anti Mouse IgG2a, Alexa Fluor 594 conjugated | Invitrogen | Cat#A-21135; RRID: AB_2535774 |
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| CuRx Nitisinone (NTBC) | Yecuris | Cat #20-0027 |
| 5-ethynyl-2’-deoxyuridine (EdU) | Abcam | Cat #146186 |
| 5-Bromo-2'-deoxyuridine (BrdU) | Sigma Aldrich | Cat #B5002 |
| Carbon tetrachloride (CCl4) | Sigma Aldrich | Cat #289116 |
| Saponin | Sigma Aldrich | Cat #47036 |
| GSK8583 | WuXi AppTec | N/A |
| GSK2801 | WuXi AppTec | N/A |
| BAZ2-ICR | Sigma Aldrich | Cat #SML1276 |
| Dextran sulfate sodium (DSS) | TdB Consultancy | Cat #DB001 |
| O-Propargyl-Puromycin (OP-Puro) | Cayman Chemical | Cat #15134 |
| Rapamycin | Selleck Chemicals | Cat #S1039 |
| Acetaminophen | Sigma Aldrich | Cat #A7085 |
| DAPI Solution | Thermo Scientific | Cat #62248 |
| UltraPure Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v) | Invitrogen | Cat #15593031 |
| TRIzol Reagent | Invitrogen | Cat #15596018 |
| 4% Paraformaldehyde Solution in PBS | Alfa Aesar | Cat #J19943K2 |
| Formaldehyde solution | Sigma Aldrich | Cat #F8775 |
| Hematoxylin QS | Vector Laboratories | Cat #NC9788954 |
| Fetal Bovine Serum | Sigma Aldrich | Cat #F4135 |
| Dulbecco’s High Glucose Modified Eagles Medium | Cytiva Hyclone | Cat #SH30022.FS |
| Dexamethasone | Sigma Aldrich | Cat #4902 |
| Penicillin Streptomycin 100X solution | Cytiva Hyclone | Cat #SV30010 |
| Critical commercial assays | ||
| VECTASTAIN Elite ABC-HRP Kit (Rabbit IgG) | Vector Laboratories | Cat #PK-6101 |
| VECTASTAIN Elite ABC-HRP kit (Rat IgG) | Vector Laboratories | Cat #PK-6104 |
| M.O.M Elite Immunodetection Kit | Vector Laboratories | Cat #PK-2200 |
| Q5 High-Fidelity DNA Polymerase | New England BioLabs | Cat #M0515 |
| Click-iT Edu Alexa Fluor 488 Flow Cytometry Kit | Invitrogen | Cat #C10425 |
| Alexa Fluor 555 Antibody Labeling Kit | Invitrogen | Cat #A20187 |
| Click-iT Cell Reaction Buffer Kit | Invitrogen | Cat #C10269 |
| Alexa Fluor 555 Azide | Invitrogen | Cat #A20012 |
| Liver Perfusion Medium | Gibco | Cat #17701038 |
| Liver Digest Medium | Gibco | Cat #17703034 |
| Hepatocyte Wash Medium | Gibco | Cat #17704024 |
| AST Activity Assay Kit | Sigma Aldrich | Cat #MAK055 |
| ALT Activity Assay Kit | Sigma Aldrich | Cat #MAK052 |
| iScript Reverse Transcription Supermix | Bio-Rad Laboratories | Cat #1708840 |
| iTaq Universal SYBR Green Supermix | Bio-Rad Laboratories | Cat #1725124 |
| ColoScreen ES Lab Pack | Helena Laboratories | Cat #5086 |
| Ovation Mouse RNA-Seq System | Nugen | Cat #0348-32 |
| RNeasy Mini Kit | Qiagen | Cat #74104 |
| QIAquick PCR Purification Kit | Qiagen | Cat #28104 |
| NEBNext Ultra II DNA Library Prep Kit | New England BioLabs | Cat #7645 |
| NEBNext High-Fidelity 2X PCR Master Mix | New England BioLabs | Cat #M0544 |
| Illumina Tagment DNA Enzyme and Buffer | Illumina | Cat #20034197 |
| Qubit dsDNA BR Assay Kit | Invitrogen | Cat #Q32850 |
| BAZ2B inhibitor AlphaScreen Kit | BPS Bioscience | Cat #32600 |
| NEBNext Multiplex Oligos for Illumina (Set 1) | New England BioLabs | Cat #7335 |
| NEBNext Multiplex Oligos for Illumina (Set 2) | New England BioLabs | Cat #7500 |
| NEBNext Multiplex Oligos for Illumina (Set 3) | New England BioLabs | Cat #7710 |
| NEBNext Multiplex Oligos for Illumina (Set 4) | New England BioLabs | Cat #7730 |
| Deposited data | ||
| Raw and analyzed RNA-seq and ChIP-seq data | This paper | GEO: GSE183502 |
| Experimental models: Cell lines | ||
| Mouse: H2.35 cells | ATCC | CRL-1995 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratories | JAX: 000664 |
| Mouse: CD1 IGS | Charles River Laboratories | N/A |
| Mouse: Rpl24Bst/+ on C57BL/6J | Oliver et al., 2004 | RRID:MGI:5295283 |
| Mouse: Fah KO on C57BL/6J | This paper | N/A |
| Mouse: Rag1 KO; Fah KO; Il2rg KO on C57BI/6 | Yecuris | Cat #10-0001 |
| Mouse: Baz2b KO on 129/C57BL/6J mixed background | This paper | N/A |
| Oligonucleotides | ||
| Primers for qRT-PCR: listed in Table S4 | This paper; Feng et al., 2012 | Table S4 |
| Primer for ATAC-seq library prep: listed in Table S4 | This paper | Table S4 |
| sgRNA sequences for screening validation: listed in Table S4 | Shi et al., 2015 | Table S4 |
| CRISPR sgRNA sequence against mouse Pten | Xue et al., 2014 | Table S4 |
| CRISPRa sgRNA sequence against mouse c-Myc | Wangensteen et al., 2018 | Table S4 |
| In vivo siRNAs for mouse Baz2a | Life Technologies | Cat #s100272 |
| In vivo siRNAs for mouse Baz2b | Life Technologies | Cat #s129008 |
| Primers for screening library: listed in Table S4 | This paper | Table S4 |
| Recombinant DNA | ||
| Fah-Cas9 | This paper | N/A |
| Fah-dCas9-VP64 | This paper | N/A |
| Fah-Cas9-sgBaz2a | This paper | N/A |
| Fah-Cas9-sgBaz2b | This paper | N/A |
| Fah-BAZ2A | This paper | N/A |
| Fah-BAZ2B | This paper | N/A |
| Fah-Cas9-sgPten | This paper | N/A |
| Fah-GFP | This paper | N/A |
| Fah-dCas9-VP64-sgMyc | This paper | N/A |
| pCMV(CAT)T7-SB100 | Mátés et al., 2009 | pCMV(CAT)T7-SB100 Addgene Cat #34879 |
| FAHIG | Wuestefeld et al., 2013 | N/A |
| Software and algorithms | ||
| HISAT | Kim et al., 2015 | http://www.ccb.jhu.edu/software/hisat/index.shtml |
| HTSeq | Anders et al., 2015 | https://htseq.readthedocs.io/en/master/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| BWA aligner | Li and Durbin. 2009 | http://bio-bwa.sourceforge.net/ |
| MACS | Zhang et al. 2008 | https://orionedocumentation.readthedocs.io/en/latest/chip_seq/ngs_peak_calling/macs14.html |
| DiffBind | Stark and Brown, 2011 Ross-Innes et al., 2012 | http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| deepTools | Ramirez et al., 2016 | https://deeptools.readthedocs.io/en/develop/ |
| UCSC | Kuhn et al., 2013 | https://genome.ucsc.edu/util.html |
| Gene Set Enrichment Analysis | Subramanian et al., 2005 Mootha et al., 2003 | https://www.gseamsigdb.org/gsea/index.jsp |
| clusterProfiler | Yu et al., 2012 Wu et al., 2021 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| Other | ||
| Agencourt Ampure XP Beads | Beckman Coulter | Cat #A63881 |
| Pierce Protein A/G Magnetic Beads | Thermo Scientific | Cat #26162 |
| BD FACSAria Fusion Flow Cytometer | BD Biosciences | N/A |
| Nickel Chelate AlphaLISA Acceptor Beads | Perkin Elmer | Cat #AL108C |
| Streptavidin-conjugated donor beads | Perkin Elmer | Cat #6760002 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
WT B6 (C57BL/6/J) mice were purchased from Jackson Laboratory (JAX: 000664) and WT CD1 mice were purchased from Charles River (CD-1 IGS). Fah KO mice were outbred from FRG mice purchased from Yecuris (#10–0001), and were backcrossed for at least six generations onto the C57BL/6 background. Fah KO mice were maintained on 7.5 μg/mL nitisinone (Yecuris #20–0027) until HDT was performed at 8–10 weeks of age. Baz2b KO mice were generated using Easi-CRISPR in the Children’s Research Institute Mouse Genome Engineering Core. Genotyping was performed with FullLength-F (5’-GGGTAGGAGCAAGATCACAAGTGACA-3’) and FullLength-R (5’-ATACATATATGAATTTACTGTAGTTGCTTTCAG-3’) to distinguish Het from WT, and Inside-F (5’-TATGCTTATAAACATAGATACAAATCAATCT-3’) and Inside-R (5’-AACCACTCCCACGTCAGAACTACTGGA-3’) to distinguish KO from Het. Rpl24Bst/+ mice (JAX: 000516) were backcrossed for at least eight generations onto the C57BL/6 background and were provided by Dr. Sean Morrison. Rpl24Bst/+ mice were crossed to CD-1 female mice one time and the offspring were used for Figure 7. Most experiments were performed on male mice; Figure 7E–G and Supplemental Figure S2 employed both male and female mice and H2.35 cells were derived from female mice. All mice were maintained in a specific pathogen free (SPF) facility. All experiments were done in an age and sex matched fashion. All studies were approved by the Institutional Animal Care and Use Committee (IACUC) of UT Southwestern.
Cell Lines
H2.35 (ATCC CRL-1995, original source: BALB/c female mouse) immortalized hepatocytes were cultured in DMEM (Cytiva Hyclone #SH30022.FS) supplemented with 4% (vol/vol) FBS (Sigma Aldrich #F4135), 1x Pen/Strep solution (Cytiva Hyclone #SV30010), and 200nM Dexamethasone (Sigma Aldrich #4902). Cells were cultured at 37°C and 5% CO2 in a humidified incubator. H2.35 cells were not authenticated after purchase from ATCC.
METHOD DETAILS
Plasmid and sgRNA library construction.
The transposon plasmids used for CRISPR KO screening (FAH-Cas9) and CRISPRa screening (FAH-dCas9-VP64) were modified from p/T FAHIG, which was originally provided by Dr. Lars Zender (Wuestefeld et al., 2013). The IRES-GFP sequence in p/T FAHIG was replaced by P2A-Cas9 or P2A-dCas9-VP64 after removing the Fah stop codon. Scaffold-linker-U6 was added using the MluI enzyme. Individual sgRNAs for knockout screening were extracted from the Vakoc chromatin domain library (Shi et al., 2015) (Supplemental Table S1). sgRNAs for activation screening were extracted from mCRISPRa-v2 (Addgene #83996) (Supplemental Table S1). The preparation of sgRNA libraries was described in (Canver et al., 2018) and subcloned to maintain a 1000-fold representation of colonies/number of sgRNAs. sgRNA oligo arrays were synthesized by GenScript.
In vivo CRISPR screening.
5 μg of library plasmid and 2 μg of SB100 (Addgene #34879) were resuspended in 0.9% NaCl solution to a final volume of 10% of mouse body weight, and hydrodynamically injected into 8–10 week old Fah KO mice through the tail vein. Nitisinone water was withdrawn immediately after HDT. One month after HDT, livers were collected. In liquid nitrogen, whole livers were ground using a mortar and pestle into a powder as finely as possible. Half of the ground liver tissue was stored in −80°C as backup material. The other half was suspended in 10 mL of digestion buffer (100 mM NaCl, 10 mM Tris-Cl (pH 8.0), 25 mM EDTA (pH 8.0), 0.5% SDS, and 0.1 mg/mL proteinase K) and digested overnight at 65°C. After digestion, 1 μg/mL DNase-free Rnase was added and incubated for 1 hour at 37°C to remove residual RNA. Then, 10 mL of digested solution was split into two: one 5 mL aliquot was frozen in −80°C and another 5 mL was processed as follows. 5 mL of the digested solution was aliquoted into multiple 1.5 mL tubes. Genomic DNA extraction was performed with the addition of an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) (Invitrogen #15593031), centrifugation at room temperature at 13000 rpm, followed by pooling of the upper aqueous phase from the aliquots, and then ethanol precipitation. The DNA concentration was determined using the Qubit dsDNA BR assay kit (ThermoFisher #Q32850). Two-step PCR was performed for the DNA sequencing preparation. For the first-round of PCR (PCR1), the amount of input genomic DNA was calculated for each sample in order to achieve at least 1000x coverage of the library, and the number of PCR reactions needed was calculated accordingly. In one 50 μL PCR1 reaction, in addition to adding Q5 DNA polymerase and 5x buffer (NEB #M0491), 1 μg of genomic DNA, 200 μM of dNTP mix, 0.5 μM of Lib-PCR-F and Lib-PCR-R primer were also added. 10, 15, 20, 25 PCR cycles were performed for PCR1, respectively. 30 μL of PCR1 product was run on a 2% agarose gel. The number of cycles that resulted in the faintest but still visible band was chosen. Then, the PCR1 products were combined. 1 μL of combined PCR1 was used as the template for the second round of PCR (PCR2). In a 50 μL PCR2 reaction, 0.5 μM barcoded primers, 200 μM of dNTP mix, Q5 DNA polymerase, and 5x buffer were added. Again, the minimum number of cycles (~15 cycles in our case) for PCR2 reaction was performed, followed by gel extraction. The DNA concentration of gel purified PCR2 product was determined with the Qubit dsDNA BR assay kit. PCR amplicon libraries for deep sequencing were adapted from (Canver et al., 2018). The original transposon library used for the HDT injection was sequenced as a control. All primer sequences are provided in Supplemental Table S4. The library was prepared to ensure 1000-fold coverage. All samples were sequenced on an Illumina HiSeq500 with 75 bp single-end reads. ~1–2 million reads were sequenced per library. For all in vivo screens, the log2 fold change for a gene was averaged from the log2 fold change of individual sgRNAs. The p-values were calculated using MAGeCK. Hits were selected with a p-value < 0.05 and an absolute fold change > 0.25. For CRISPRa screening to identify pathways responsible for the pro-regeneration phenotypes associated with BAZ2 inhibition, differentially regulated pathways were identified from differentially expressed genes found from RNA-seq in regenerating mouse livers treated with either GSK8573 or GSK2801. 6–18 differentially expressed genes within each pathway were selected as candidates. 5–8 sgRNAs targeting the promoters of each candidate gene were cloned into the Fah-dCas9-VP64 plasmid. A pooled sgRNA library of these plasmids was delivered via HDT into Fah KO mice. The mice were then randomized into two groups: one receiving GSK2801 and the other receiving GSK8573. Four weeks after HDT, livers were collected and sgRNA abundance was determined by deep sequencing.
In vivo siRNA experiments.
siRNAs (Baz2a: Life Technologies s100272; Baz2b: Life Technologies s119008) were first validated in vitro and the best of 3 individual siRNAs for Baz2a and Baz2b were selected for in vivo experiments. In vivo siRNAs were packaged in 5A2-SC8 LNPs (4 mg/kg siRNA) as previously described (Cheng et al., 2020; Zhang et al., 2018a, 2018b; Zhou et al., 2016). An ethanol solution of 5A2-SC8, DSPC, cholesterol, and DMG-PEG2000 (molar ratio of 50:38:10:2) was rapidly combined with acidic solutions of siRNA to yield a final weight ratio of 25:1 (5A2-SC8:siRNA). LNP packaged siRNAs were administered intravenously through the tail vein. One dose was delivered 24 hours prior to, and the second dose at the time of 70% PHx.
Partial Hepatectomy experiments.
The 70% PHx surgery was performed as described previously (Mitchell and Willenbring, 2008). The left lateral lobe and the median lobe were surgically removed while mice were under isoflurane anesthesia. The 85% PHx was performed by removing the median, left lateral and right inferior lobes. The PHx experiment in Figure 5E was blinded whereby one person injected the drug and the second person performed the surgery without knowing the identity of the treatment groups. Moribund mice (defined by > 20% weight loss, body condition scores of < 2, immobility, or behaviors associated with being severely withdrawn) were euthanized to minimize pain and distress. BrdU (Sigma Aldrich #B5002) was injected IP at a dose of 100 mg/kg 3 hours before tissue collection.
Liver injury experiments using chemical toxins.
GSK2801 and GSK8573 were synthesized by WuXi AppTec. GSK2801 or GSK8573 were first dissolved in DMSO and further diluted 1:8 in 0.5% sodium carboxymethyl cellulose. BAZ2-ICR (Sigma Aldrich #SML1276) was first dissolved in DMSO and further diluted 1:10 in PBS. CCl4 (Sigma Aldrich #289116) was diluted 1:10 in corn oil, and 100 μL was injected IP in 8–10 week old mice. Blood was drawn for LFTs 24 hours after CCl4 injection and Liver tissue was harvested 48 hours after the CCl4 injection. For APAP, a single dose was given IP in 10% DMSO in PBS at a dose of either 300 or 500 mg/kg, depending on the experiment. DMSO was used to increase the solubility of APAP.
Liver function tests.
Blood samples were obtained retro-orbitally in heparinized tubes, transferred into 1.5 mL tubes and centrifuged at 2000 g for 15 min at 4°C. The supernatants after centrifugation (plasma) were analyzed for liver damage by AST and ALT activity kits (Sigma Aldrich #MAK055 and #MAK052). Albumin was measured by the UTSW metabolism core.
DSS colitis modeling.
Mice were treated with 3% (w/v) DSS (Molecular mass 36–40 kDa; TdB Consultancy #DB001) dissolved in sterile, distilled water. Mice were given DSS water for 5 days, followed by normal water for 3 days. Body weights were measured daily up to day 8. Stool consistency and the presence of occult blood were scored on day 5 and day 8. Stool consistency scores were determined as follows: 0 = well-formed pellets, 1 = semi-formed stools that did not adhere to the anus, 2 = semi-formed stools that adhered to the anus, 3 = liquid stools that adhered to the anus. Bleeding scores were determined using ColoScreen ES Lab Pack (Helena Laboratories #5086) as follows: 0 = no blood by using hemoccult, 1 = positive hemoccult, 2 = blood traces in stool visible, 3 = gross rectal bleeding. Colitis pathology was analyzed and scored by P.G., a board certified clinical pathologist with expertise in liver and intestinal diseases. Tissue damage in DSS colitis was graded in the following categories: Epithelial damage (0 = normal; 1 = loss of goblet cells; 2 = loss of goblet cells in large areas; 3 = loss of crypts; 4 = loss of crypts in large areas); inflammatory cell infiltration (0 = no infiltration; 1 = infiltration around crypt bases; 2 = infiltration spreading to muscularis mucosa; 3 = extensive infiltration in the muscularis mucosa with abundant edema; 4 = infiltration spreading to submucosa; 5 = transmural infiltration); erosions vs. ulcers (0 = none; 1 = erosions only; 2 = ulcers +/−erosions); number of ulcers and the size of ulcers (1 = small ulcers; 2 = large ulcers). The histology scores were calculated as the sum of scores from each category.
Histology and immunohistochemistry (IHC).
Tissues were fixed overnight at 4°C in 4% paraformaldehyde (PFA; Alfa Aesar #J19943K2) and then paraffin-sectioned (UTSW Histo Pathology Core). Standard HE staining was also performed by this core. Paraffin-sectioned slides were deparaffinized in xylene and rehydrated in 100%, 90%, 80%, 70%, 50%, 30% ethanol and deionized water. Antigen retrieval was performed in 10 mM Sodium Citrate buffer (pH 6.0) with 0.05% Tween 20 at a sub-boiling temperature for 20 minutes in a microwave. After cooling down, slides were immersed in 3% hydrogen peroxide in methanol to block endogenous peroxidase activity. IHC was then performed using VECTASTAIN Elite ABC-HRP kits (Vector Laboratories #PL-6101, #PK-2200; #PK-6104) as described in the manufacturer’s instructions. The following primary antibodies and dilutions were used for IHC staining: FAH (Yecuris #20–0042; 1:500); PTEN (Cell Signaling #9559; 1:250); Ki67 (Abcam #15580; 1:250); c-Myc (Abcam #32072; 1:50); BrdU (Abcam #6326; 1:100 or BD Biosciences #555627; 1:250). The slides were counterstained with hematoxylin (Vector Laboratories #NC9788954).
For frozen-sectioned slides, tissues were fixed overnight in 4% paraformaldehyde (PFA; Alfa Aesar #J19943K2) at 4°C followed by 30% sucrose at 4°C for at least 6 hours until the tissues sank to the bottom. The tissues were frozen sectioned at a thickness of 10 μm. The slides were blocked with 5% BSA with 0.25% Triton X100 and incubated with primary antibody in 5% BSA with 0.25% Triton X100 at 4°C overnight. After washing with PBST for 3 times, slides were incubated with the appropriate secondary antibody diluted 1:100 in 5% BSA with 0.25% and DAPI (Thermo Scientific #62248) diluted 1:2000 for 1 hour at room temperature. The following primary antibodies were used for immunofluorescence staining: FAH (Yecuris #20–0042; 1:1000); BrdU (Abcam #6326; 1:50); E-cadherin (CST #3195; 1:200). The following secondary antibodies were used for immunofluorescence staining: Goat anti Rabbit IgG, Alexa Fluor 594 conjugated (Invitrogen #A-11012; 1:100); Goat anti Rat IgG, Alexa Fluor 488 conjugated (Invitrogen #A-11006; 1:100).
Determining the half-maximal inhibitory concentration for BAZ2 Inhibitors.
The IC50 for GSK8573 and GSK2801 toward acetylated histone peptide and BAZ2B bromodomain was measured with the BAZ2B inhibitor AlphaScreen Kit (BPS Bioscience #32600) as described in the manufacturer’s instructions. Specifically, every reaction contains 2.5 μL 3x BRD Homogeneous Assay Buffer 3, 1 μL Bromodomain Ligand 2, 3 μL water, 1 μL inhibitor at the appropriate concentrations. BAZ2B Bromodomain protein was diluted in 1x BRD Homogeneous Assay Buffer 3 to a final concentration of 200 ng/μL, and 2.5 μL of diluted BAZ2B protein in 1x BRD Homogeneous Assay Buffer 3 was added to initiate the reaction. After incubating at room temperature for 1 hour, 10 μL of Nickel Chelate AlphaLISA Acceptor Bead (Perkin Elmer #AL108C) diluted 1:200 in 1x BRD Homogeneous Detection Buffer 3 was added. After incubating at room temperature for 30 minutes, 10 μL of Streptavidin-conjugated donor beads (Perkin Elmer #6760002) diluted 1:100 in 1x BRD Homogeneous Detection Buffer 3 was added. After incubating at room temperature for 30 minutes, the plate was read. IC50 values were determined by fitting the normalized alpha reads with the Origin 6.0 program (OriginLab Corporation).
Hepatocyte isolation.
Mice were anesthetized and livers were perfused through the intrahepatic vena cava first with Liver Perfusion Medium (Gibco #17701038) followed by Liver Digest Medium (Gibco #17703034). Livers were excised in Liver Digest Medium, and the liver cell suspension was filtered through a 70 μM strainer. An equal amount of Hepatocyte Wash Medium (Gibco #17704024) was added to the liver cell suspension, which was centrifuged at 50 g for 5 minutes to collect hepatocytes.
Quantifying repopulating and proliferating hepatocytes by flow cytometry.
EdU (50 mg/kg; Abcam #146186) was injected IP daily starting three days before tissue collection. Hepatocytes were collected by liver perfusion. 2 × 106 cells were fixed in 0.5 mL of 1% PFA in PBS for 15 minutes on ice. Cells were washed in ice-cold PBS, then permeabilized in 200 μL PBS supplemented with 3% fetal bovine serum (Sigma Aldrich #F4135) and 0.1% saponin (Sigma Aldrich #47036) for 5 minutes at room temperature. FAH antibody (Yecuris #20–0042) was conjugated with Alexa Fluor 555 using the Alexa Fluor 555 Antibody Labeling Kit (Invitrogen #A20187) as described in the manufacturer’s instructions. This antibody was incubated with permeabilized cells in 200 μL PBS supplemented with 3% FBS and 0.1% saponin at 4°C overnight. EdU was stained using the Click-iT EdU Alexa Fluor 488 Flow Cytometry kit (Invitrogen #C10425). After the reaction, cells were washed and suspended in PBS supplemented with 3% FBS and 0.1% saponin, then analyzed by flow cytometry on a BD FACSAria Fusion Flow Cytometer.
Protein synthesis measurements.
Vehicle, Rapamycin (2.0 mg/kg IP), or GSK2801 (30 mg/kg IP) were given at the time of 70% PHx and 24 hours after. 47 hours after PHx, OP-puro (9.5 mg/kg, pH 6.4–6.6 in PBS; Cayman Chemical #15134) was injected IP. One hour after injection, hepatocytes were collected by liver perfusion-digestion as described above. 2 × 106 cells were fixed in 0.5 ml of 1% PFA in PBS for 15 minutes on ice. Cells were washed in ice-cold PBS, then permeabilized in 200 μL PBS supplemented with 3% FBS and 0.1% saponin (Sigma Aldrich #47036) for 5 minutes at room temperature. The azide-alkyne cycloaddition was performed using the Click-iT Cell Reaction Buffer Kit (Invitrogen #C10269) and the azide was conjugated to Alexa Fluor 555 (Invitrogen #20012) at a final concentration of 5 μM. After a 30 minute reaction, the cells were washed twice in PBS supplemented with 3% FBS and 0.1% saponin, and analyzed by flow cytometry.
RNA extraction and qRT-PCR.
Liver total RNA was isolated using TRIzol (Invitrogen #15596018) followed by purification using the RNeasy Mini kit (Qiagen #74104). For qRT-PCR, cDNA synthesis was performed with 1 μg of total RNA using the iScript RT Supermix (BioRad #1708840) in a total of 20 μL. Each cDNA sample (20 μL) was diluted to a total volume of 800 μL, and 5 μL of this was combined with primers and iTaq Universal SYBR green Supermix (BioRad #1725121) in 12 μL reactions. mRNA levels were normalized to β-actin mRNA expression. For data analysis, we assessed relative mRNA levels from control and experimental mice using the delta-delta approach. We first normalized the expression of each gene to the expression of β-actin mRNA in the same sample. Three technical replicates were used for each individual mouse. Each normalized expression in the experimental group was divided by the average normalized expression from the control group to get the fold change in expression. qRT-PCR primer sequences are listed in Supplemental Table S4 (Feng et al., 2012).
RNA-seq.
Liver total RNA was isolated using TRIzol (Invitrogen #15596018) followed by purification using the RNeasy Mini kit (Qiagen #74104). 500 ng of RNA for each sample was used to prepare RNA-seq library using Nugen Ovation Mouse RNA-seq System (Nugen #0348–32). The concentration of the final DNA product was quantified with the Qubit dsDNA BR Assay kit, then diluted to a concentration of 10 nM. 75 bp single-end sequencing was performed on an Illumina NextSeq500 at the UTSW Children’s Research Institute Sequencing Facility. Reads were mapped to the mouse reference genome GRCm38 (mm10) using HISAT (version 2.1.0) aligner with –rna-strandness F (Kim et al., 2015). Mapped reads were counted using HTSeq (version 0.6.1) to obtain raw read counts for each gene (Anders et al., 2015). Differential gene expression analysis was performed using the R package DESeq2 (version 1.36.0) (Love et al., 2014). Genes with a p-value < 0.05 were designated as differentially expressed genes between groups.
ChIP-seq.
For ChIP-seq, H2.35 cells were plated into 15 cm culture dishes, and were treated with either GSK2801 or GSK8573 (500 nM) for three days, or transiently transfected with BAZ2A or BAZ2B expressing plasmids. After three days of treatment, 37% formaldehyde (Sigma Aldrich #F8775) was added to culture plates to a final concentration of 1% and incubated for 5 minutes at room temperature with shaking. After incubation, 2.5 M glycine was added to a final concentration of 125 mM for 5 minutes at room temperature with shaking. Cells were washed twice with ice-cold PBS and were scraped into 5 mL PBS (with 2X protease inhibitors). Cells were collected by centrifugation at 1500 rpm for 5 minutes at 4°C. Per 40 million cells, 1 mL of sonication buffer (0.1% SDS, 1% Triton X-100, 10 mM Tris-HCL, 1 mM EDTA, 0,1% NaDOC, 0.25% Sarkosyl, 1X protease inhibitor) was added directly to the cell pellets. Cell lysates were sonicated to shear DNA to 250–500 bp for histones and 500–1000 bp for BAZ2 proteins. 5 M NaCl was added to chromatin to a final concentration of 0.3 M. Primary antibody was incubated with chromatin overnight at 4°C with rotation. The next day, 30–60 μL of pre-washed Pierce Protein A/G Magnetic Beads (Thermo Scientific #26162) were added to each IP sample, and incubated at 4°C for > 3 hours with rotation to collect chromatin/antibody. The magnetic beads were collected with a magnet and washed twice with 1 mL RIPA 0 buffer (0.1% SDS, 1% Triton X-100, 10 mM Tris-HCL, 1 mM EDTA, 0.1% NaDOC), twice with 1 mL of RIPA 0.3 buffer (0.1% SDS, 1% Triton X-100, 10 mM Tris-HCL, 1 mM EDTA, 0.1% NaDOC, 0.3 M NaCl), twice with 1 mL LiCl wash buffer (250 mM LiCl, 0.5% NP-40, 0.5% NaDOC, 1 mM EDTA, 10 mM Tris-HCl) and twice with 1 mL TE buffer (10 mM Tris-HCl, 1 mM EDTA). DNA was eluted from washed magnetic beads with incubation of SDS elution buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl) overnight at 65°C. The remaining RNA and protein residues were removed by incubating with 3 μL of 0.5 μg/μL RNaseA at 37°C for 30 minutes and 3 μL of 20 mg/mL Proteinase K at 37°C for 2 hours. ChIP DNA was recovered using the QIAquick PCR Purification Kit (Qiagen #74104). The ChIP-seq library was prepared using the NEBNext Ultra II DNA Library Prep Kit (New England BioLabs #7645) and NEBNext Multiplex Oligos for Illumina (New England BioLabs #7335, #7500, #7710, #7730). The final DNA product was quantified with the Qubit dsDNA BR Assay kit (Invitrogen #Q32850) and brought to a final concentration of 10 nM.
For ChIP-seq in mouse liver, post-PHx livers were collected (see Figure 5A). 0.5 g of frozen liver tissues were minced into small pieces and cross-linked in 1% formaldehyde for 20 minutes at room temperature with shaking. After incubation, 2.5 M glycine was added to cells to a final concentration of 125 mM for 5 minutes at room temperature with shaking. Liver tissues were homogenized with dounce homogenizer, and then filtered with a 10 μm cell strainer. Nuclei were collected by centrifuging at 2000 rpm for 5 minutes at 4°C. ChIP and subsequent library preparation was performed as described in the above in vitro ChIP-seq protocol.
75 bp single-end sequencing was performed on an Illumina NextSeq500 (UTSW Children’s Research Institute Sequencing Facility). Single-end sequencing reads were mapped to the mouse reference genome GRCm38 (mm10) using BWA aligner (version 0.7.5) with default parameters (Li and Durbin, 2009). Peak calling was performed using MACS (version 1.4.2) with --nomodel --p 1e-5 (Zhang et al., 2008). Differentially bound peaks were identified with DiffBind (version 3.13) in R (Ross-Innes et al., 2012; Stark and Brown, 2011). The peaks were annotated with the annotatedPeaks.pl command from HOMER (version 4.9) to associate peaks to their nearest genes (Heinz et al., 2010). BigWig files were generated using the bamCoverage command from deeptools (version 3.5.0) with --smoothLength 50 --binSize 5 and --scaleFactor (Ramírez et al., 2016). The scaleFactors were normalization factors calculated from DiffBind based on the number of all mapped reads that fall into the called peak regions. ChIP-seq heatmaps and profile plots were generated with computeMatrix followed by the plotHeatmap command from deeptools (version 3.5.0). Merged BigWig files were generated from three biological replicates with the bigWigMerge command followed by the bedGraphToBigWig command from UCSC (Kuhn et al., 2013).
ATAC-seq.
H2.35 cells were treated with either GSK2801 or GSK8573 (500 nM) for three days before collection. H2.35 cells were lysed with lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40) for 10 minutes at 4°C, and centrifuged at 2500 rpm for 10 minutes at 4°C to collect nuclei. The tagmentation reaction was performed on 50K nuclei with Illumina Tagment DNA Enzyme and Buffer (Illumina #20034197) as described in the manufacturer’ instructions for 1 hour at 37°C. Tagmented DNA was purified with the QIAquick PCR Purification Kit (Qiagen #74104) and was amplified with the NEBNext High-Fidelity 2X PCR Master Mix (New England BioLabs #M0544) with primers listed in Supplemental Table S4. Amplified DNA was recovered with Agencourt Ampure XP beads (Beckman Coulter #A63881) so that the final size was ~500 bp. The concentration of the final DNA product was quantified with the Qubit dsDNA BR Assay kit to a final concentration of 10 nM. 75 bp single-end sequencing was performed on an Illumina NextSeq500. Single-end sequencing reads were mapped to the mouse reference genome GRCm38 (mm10) using BWA aligner (version 0.7.5) with default parameters (Li and Durbin, 2009). Peak calling was performed using MACS (version 1.4.2) with --nomodel --p 1e-5 (Zhang et al., 2008). The differentially bound peaks were identified with DiffBind (version 3.13) in R (Ross-Innes et al., 2012; Stark and Brown, 2011). The peaks were annotated with the annotatedPeaks.pl command from HOMER (version 4.9) to associate peaks to their nearest genes (Heinz et al., 2010). BigWig files were generated using the bamCoverage command from deeptools (version 3.5.0) with --smoothLength 50 --binSize 5 and --scaleFactor (Ramírez et al., 2016). The scaleFactors were normalization factors calculated from DiffBind based on the number of all mapped reads that fall into the called peak regions. The ChIP-seq heatmap and profile plot were generated with computeMatrix followed by the plotHeatmap command from deeptools (version 3.5.0). Merged BigWig files were generated from three biological replicates with the bigWigMerge command followed by the bedGraphToBigWig command from UCSC (Kuhn et al., 2013).
Gene Set Enrichment and pathway enrichment analysis.
To perform GSEA analysis, fold change was used for each gene and GSEA was then performed against desired gene sets using default parameters (Mootha et al., 2003; Subramanian et al., 2005). Pathway enrichment analysis was performed using the enricher function in clusterProfiler in R (Wu et al., 2021). All GSEA and pathway enrichment genes are included in Supplemental Table S3.
QUANTIFICATION AND STATISTICAL ANALYSIS
For all figure panels, each larger dot is an individual biological replicate from one mouse, and data is presented as mean ± standard deviation (SD) unless otherwise noted. For Figure 2D and 2F, significance was assessed by One-Way ANOVA followed by Tukey’s multiple comparisons test. For Figure 3B, 3C, 3E, 7B, 7E and 7G, significance was assessed by One-Way ANOVA followed by Dunnet’s multiple comparisons tests. For Figure 4A, 4B, 4F, 4G, 4H, 5B, 5C, 5F, 5H, 5I, and 5J, significance was assessed with Student’s t-tests. For Figure 4C, 4D, and 5M, significance was assessed by multiple t-tests corrected with the Holm-Sidak method. For Figure 5E and 5K, significance was assessed by Mantel-Cox tests. For Figure 7C, significance was assessed by Two-Way ANOVA followed by Tukey’s multiple comparisons tests. For Figure 7J and 7L, significance was assessed by Two-Way ANOVA followed by Sidak’s multiple comparisons tests. Statistical testing used in the supplemental figures are described in the legends. Statistical significance is displayed as p<0.05 (*), p<0.01 (**), p<0.001 (***), p<0.0001 (****). When indicated in the legends or methods, experimental blinding was performed. For all IHC and IF staining quantifications, the staining was performed on all the mice in each experiment. The pictures were taken at the appropriate resolution (for necrosis analysis, the pictures were taken at 5X; for other data, pictures were taken at 10X), and the average quantification from 5 or 10 images was used as an individual data point, unless stated otherwise. If the tissue size was not big enough for 5 or 10 pictures, then the maximum number of pictures that covered the entire tissue section was used. All image quantification was performed in imageJ. For quantification of Ki67+ and BrdU+ cells, the images were converted into grayscale. A threshold was added to the image with the MaxEntropy option, and the number of cells was counted using analyze particles with size larger than 50 pixel2, and circularity greater than 0.70. For FAH+ area, the images were converted into grayscale. A threshold was added to the image with the default setting, and FAH+ area was measured. Raw data and statistical analysis are included in Supplemental Table S5.
Supplementary Material
Supplemental Table S1: In vivo CRISPR screening data. Related to Figure 2 and Figure 6.
Sheet 1. sgRNA sequences included in the epigenetic CRISPRa screen. sgRNAs against 165 genes and 11 controls were extracted from the Mouse Genome-wide CRISPRa-v2 Library (Addgene #83996).
Sheet 2. Results of the epigenetic CRISPRa screen.
Sheet 3. sgRNA sequences included in the epigenetic CRISPR KO screen. sgRNAs against 165 genes and 1 control were extracted from (Shi et al., 2015).
Sheet 4. Results of the epigenetic CRISPR KO screen.
Sheet 5. sgRNA sequences included in the mechanism related CRISPRa screen. sgRNAs against 110 genes and 46 controls were designed with CRISPick.
Sheet 6. Results of the mechanism related CRISPRa screen.
Supplemental Table S2: Transcriptomic and epigenomic sequencing results. Related to Figure 6 and Figure 7.
Sheet 1. DESeq2 results for RNA-seq in 48h post-PHx mouse livers treated with GSK8573 or GSK2801.
Sheet 2. DiffBind analysis for H3K27ac ChIP-seq in H2.35 cells treated with GSK8573 or GSK2801.
Sheet 3. DiffBind analysis for H3K27ac ChIP-seq in 48h post-PHx mouse livers treated with GSK8573 or GSK2801.
Sheet 4. DiffBind analysis for ATAC-seq in H2.35 cells treated with GSK8573 or GSK2801.
Sheet 5. DiffBind analysis for H3K4me3 ChIP-seq in H2.35 cells treated with GSK8573 or GSK2801.
Sheet 6. DiffBind analysis for H3K4me1 ChIP-seq in H2.35 cells treated with GSK8573 or GSK2801.
Sheet 7. MACS results of Flag-BAZ2A in H2.35 cells overexpressing Flag-tagged BAZ2A.
Sheet 8. MACS results of Flag-BAZ2B in H2.35 cells overexpressing Flag-tagged BAZ2B.
Supplemental Table S3: Genes included in GSEA analysis and pathway enrichment analysis and results. Related to Figure 6.
Sheet 1–125. Genes included in the identified GSEA and pathway enrichment analysis.
Sheet 126. Pathway enrichment analysis from overlapped genes in H2.35 cells.
Sheet 127. Pathway enrichment analysis from overlapped genes in 48h post-PHx livers. Sheet 128. GSEA results on differentially expressed genes identified from RNA-seq.
Supplemental Table S4: Primer sequences for qPCR analysis, CRISPR editing, and sequencing. Related to Figures 1–7.
Highlights.
In vivo CRISPR screening identified BAZ2 proteins, suppressors of liver regeneration.
Two distinct small molecule inhibitors of BAZ2 accelerated tissue regeneration.
BAZ2 inhibition increased ribosomal components and protein synthesis in regeneration.
Ribosomal components are sufficient and necessary for regenerative enhancement.
Acknowledgements
We thank J. Xu, M. Buszczak, M. Kharas, and A. Hsieh for constructive comments; C. Lewis (UTSW Tissue Procurement Service) and J. Shelton (UTSW Histo Pathology Core) for histopathology; and J. Xu (CRI Sequencing Core) for sequencing. H.Z. is supported by the Pollack Foundation, NIH R01 grants (DK111588, DK125396, CA251928), an Emerging Leader Award from The Mark Foundation for Cancer Research (#21-003-ELA), and a Stand Up To Cancer Innovative Research Grant (SU2C-AACR-IRG 10-16). The Zhu lab and Y.L are supported by an Innovation Award from the Moody Medical Research Institute. D.J.S. acknowledges financial support from NIH R01 EB025192-01A1 and Cancer Prevention and Research Institute of Texas (CPRIT) RP190251. J.M.R. acknowledges financial support from the Welch Foundation (I-1612).
Footnotes
Declaration of Interests
HZ has a sponsored research agreement with Alnylam Pharmaceuticals, consults for Flagship Pioneering, and serves on the SAB of Ubiquitix. These interests are not directly related to the contents of this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aloia L, McKie MA, Vernaz G, Cordero-Espinoza L, Aleksieva N, van den Ameele J, Antonica F, Font-Cunill B, Raven A, Aiese Cigliano R, et al. (2019). Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration. Nat. Cell Biol 21, 1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anosova I, Melnik S, Tripsianes K, Kateb F, Grummt I, and Sattler M (2015). A novel RNA binding surface of the TAM domain of TIP5/BAZ2A mediates epigenetic regulation of rRNA genes. Nucleic Acids Res. 43, 5208–5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, and Ruggero D (2008). Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456, 971–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortés-Garrido R, Gkatza N, et al. (2016). Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Haeussler M, Bauer DE, Orkin SH, Sanjana NE, Shalem O, Yuan G-C, Zhang F, Concordet J-P, and Pinello L (2018). Integrated design, execution, and analysis of arrayed and pooled CRISPR genome-editing experiments. Nat. Protoc 13, 946–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S, and Lieberman I (1968). Control of Ribosome Synthesis in Normal and Regenerating Liver. J Biol Chem 243. [PubMed] [Google Scholar]
- Cheng Q, Wei T, Farbiak L, Johnson LT, Dilliard SA, and Siegwart DJ (2020). Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol 15, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, and Calvisi DF (2014). Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am. J. Pathol 184, 912–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Chaikuad A, Bamborough P, Bantscheff M, Bountra C, Chung C-W, Fedorov O, Grandi P, Jung D, Lesniak R, et al. (2016). Discovery and characterization of GSK2801, a selective chemical probe for the bromodomains BAZ2A and BAZ2B. J. Med. Chem 59, 1410–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero-Espinoza L, and Huch M (2018). The balancing act of the liver: tissue regeneration versus fibrosis. J. Clin. Invest 128, 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalcher D, Tan JY, Bersaglieri C, Peña-Hernández R, Vollenweider E, Zeyen S, Schmid MW, Bianchi V, Butz S, Roganowicz M, et al. (2020). BAZ2A safeguards genome architecture of ground-state pluripotent stem cells. EMBO J. 39, e105606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin L, McGrath S, Vidler LR, Chaikuad A, Monteiro O, Tallant C, Philpott M, Rogers C, Fedorov O, Liu M, et al. (2015). Structure enabled design of BAZ2-ICR, a chemical probe targeting the bromodomains of BAZ2A and BAZ2B. J. Med. Chem. 58, 2553–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright RY, Lee S, Wittner BS, Niederhoffer KL, Nicholson BT, Bardia A, Truesdell S, Wiley DF, Wesley B, Li S, et al. (2020). Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science 367, 1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng GJ, Cotta W, Wei XQ, Poetz O, Evans R, Jardé T, Reed K, Meniel V, Williams GT, Clarke AR, et al. (2012). Conditional disruption of Axin1 leads to development of liver tumors in mice. Gastroenterology 143, 1650–1659. [DOI] [PubMed] [Google Scholar]
- Fingar DC, and Blenis J (2004). Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23, 3151–3171. [DOI] [PubMed] [Google Scholar]
- Forbes SJ, and Newsome PN (2016). Liver regeneration - mechanisms and models to clinical application. Nat. Rev. Gastroenterol. Hepatol 13, 473–485. [DOI] [PubMed] [Google Scholar]
- Gaub P, Tedeschi A, Puttagunta R, Nguyen T, Schmandke A, and Di Giovanni S (2010). HDAC inhibition promotes neuronal outgrowth and counteracts growth cone collapse through CBP/p300 and P/CAF-dependent p53 acetylation. Cell Death Differ. 17, 1392–1408. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. (2014). Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman JA, and Poss KD (2020). Gene regulatory programmes of tissue regeneration. Nat. Rev. Genet 21, 511–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory B, Rahman N, Bommakanti A, Shamsuzzaman M, Thapa M, Lescure A, Zengel JM, and Lindahl L (2019). The small and large ribosomal subunits depend on each other for stability and accumulation. Life Sci. Alliance 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompe M, Lindstedt S, al-Dhalimy M, Kennaway NG, Papaconstantinou J, Torres-Ramos CA, Ou CN, and Finegold M (1995). Pharmacological correction of neonatal lethal hepatic dysfunction in a murine model of hereditary tyrosinaemia type I. Nat. Genet 10, 453–460. [DOI] [PubMed] [Google Scholar]
- Gu L, Frommel SC, Oakes CC, Simon R, Grupp K, Gerig CY, Bär D, Robinson MD, Baer C, Weiss M, et al. (2015). BAZ2A (TIP5) is involved in epigenetic alterations in prostate cancer and its overexpression predicts disease recurrence. Nat. Genet 47, 22–30. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, Gemberling M, Goldman JA, Black BL, and Poss KD (2016). Modulation of tissue repair by regeneration enhancer elements. Nature 532, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, Abdulhay NJ, Specht H, Keshishian H, Mani DR, et al. (2018). Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell 173, 90–103.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Haussler D, and Kent WJ (2013). The UCSC genome browser and associated tools. Brief. Bioinformatics 14, 144–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang R, Lin Y-H, and Zhu H (2021). Genetic and cellular contributions to liver regeneration. Cold Spring Harb. Perspect. Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-K, Hatanaka F, Araoka T, Reddy P, Wu M-Z, Sui Y, Yamauchi T, Sakurai M, O’Keefe DD, Núñez-Delicado E, et al. (2017). In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell 171, 1495–1507.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemans CA, Calafiore M, Mertelsmann AM, O’Connor MH, Dudakov JA, Jenq RR, Velardi E, Young LF, Smith OM, Lawrence G, et al. (2015). Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 528, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb JN, and Yeung LL (1975). Synthesis and degradation of ribosomal RNA in regenerating liver. J. Exp. Med 142, 575–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao S-Y, Li C, Kohane I, Chan J, and Yankner BA (2004). Gene regulation and DNA damage in the ageing human brain. Nature 429, 883–891. [DOI] [PubMed] [Google Scholar]
- Maki N, and Kimura H (2013). Epigenetics and regeneration. Curr. Top. Microbiol. Immunol 367, 237–252. [DOI] [PubMed] [Google Scholar]
- Mátés L, Chuah MKL, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, et al. (2009). Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet 41, 753–761. [DOI] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, and Okamura H (2003). Control mechanism of the circadian clock for timing of cell division in vivo. Science 302, 255–259. [DOI] [PubMed] [Google Scholar]
- Mitchell C, and Willenbring H (2008). A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat. Protoc 3, 1167–1170. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet 34, 267–273. [DOI] [PubMed] [Google Scholar]
- Nagahama Y, Shimoda M, Mao G, Singh SK, Kozakai Y, Sun X, Motooka D, Nakamura S, Tanaka H, Satoh T, et al. (2018). Regnase-1 controls colon epithelial regeneration via regulation of mTOR and purine metabolism. Proc Natl Acad Sci USA 115, 11036–11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov EN, Dineva BB, Dabeva MD, and Nikolov TK (1987). Turnover of ribosomal proteins in regenerating rat liver after partial hepatectomy. Int. J. Biochem 19, 159–163. [DOI] [PubMed] [Google Scholar]
- Oliver ER, Saunders TL, Tarlé SA, and Glaser T (2004). Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development 131, 3907–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou CN, Finegold M, and Grompe M (1996). Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat. Genet 12, 266–273. [DOI] [PubMed] [Google Scholar]
- Planas-Paz L, Sun T, Pikiolek M, Cochran NR, Bergling S, Orsini V, Yang Z, Sigoillot F, Jetzer J, Syed M, et al. (2019). YAP, but Not RSPO-LGR4/5, Signaling in Biliary Epithelial Cells Promotes a Ductular Reaction in Response to Liver Injury. Cell Stem Cell 25, 39–53.e10. [DOI] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Li J, and Grummt I (2002). The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet 32, 393–396. [DOI] [PubMed] [Google Scholar]
- Santoro R, Lienemann P, and Fussenegger M (2009). Epigenetic engineering of ribosomal RNA genes enhances protein production. PLoS ONE 4, e6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savić N, Bär D, Leone S, Frommel SC, Weber FA, Vollenweider E, Ferrari E, Ziegler U, Kaech A, Shakhova O, et al. (2014). lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 15, 720–734. [DOI] [PubMed] [Google Scholar]
- Scornik OA (1974). In vivo rate of translation by ribosomes of normal and regenerating liver. J. Biol. Chem 249, 3876–3883. [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, and Vakoc CR (2015). Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol 33, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RAJ, Magee JA, Salic A, and Morrison SJ (2014). Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinturel F, Gerber A, Mauvoisin D, Wang J, Gatfield D, Stubblefield JJ, Green CB, Gachon F, and Schibler U (2017). Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell 169, 651–663.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark R, and Brown GD DiffBind: differential binding analysis of ChIP-Seq peak data. Bioconductor. [Google Scholar]
- Stiles B, Wang Y, Stahl A, Bassilian S, Lee WP, Kim Y-J, Sherwin R, Devaskar S, Lesche R, Magnuson MA, et al. (2004). Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity [corrected]. Proc Natl Acad Sci USA 101, 2082–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Chuang J-C, Kanchwala M, Wu L, Celen C, Li L, Liang H, Zhang S, Maples T, Nguyen LH, et al. (2016). Suppression of the SWI/SNF component arid1a promotes mammalian regeneration. Cell Stem Cell 18, 456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Wu L-W, Grivennikov SI, de Jong PR, Lian I, Yu F-X, Wang K, Ho SB, Boland BS, Chang JT, et al. (2015). A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature 519, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada N, Patel HR, Takase K, Kohno K, Nairn AC, and Gelfand EW (1994). Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc Natl Acad Sci USA 91, 11477–11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Xu J, Li Y, Zhao R, Du S, Lv C, Wu W, Liu R, Sheng X, Song Y, et al. (2019). MicroRNA-31 Reduces Inflammatory Signaling and Promotes Regeneration in Colon Epithelium, and Delivery of Mimics in Microspheres Reduces Colitis in Mice. Gastroenterology 156, 2281–2296.e6. [DOI] [PubMed] [Google Scholar]
- Wangensteen KJ, Wang YJ, Dou Z, Wang AW, Mosleh-Shirazi E, Horlbeck MA, Gilbert LA, Weissman JS, Berger SL, and Kaestner KH (2018). Combinatorial genetics in liver repopulation and carcinogenesis with a in vivo CRISPR activation platform. Hepatology 68, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AW, Wangensteen KJ, Wang YJ, Zahm AM, Moss NG, Erez N, and Kaestner KH (2018). TRAP-seq identifies cystine/glutamate antiporter as a driver of recovery from liver injury. J. Clin. Invest 128, 2297–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AW, Wang YJ, Zahm AM, Morgan AR, Wangensteen KJ, and Kaestner KH (2020). The dynamic chromatin architecture of the regenerating liver. Cell. Mol. Gastroenterol. Hepatol 9, 121–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang C, Hasson D, Desai A, SenBanerjee S, Magnani E, Ukomadu C, Lujambio A, Bernstein E, and Sadler KC (2019). Epigenetic compensation promotes liver regeneration. Dev. Cell 50, 43–56.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Yoshihara E, He N, Hah N, Fan W, Pinto AFM, Huddy T, Wang Y, Ross B, Estepa G, et al. (2018). Vitamin D switches BAF complexes to protect β cells. Cell 173, 1135–1149.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P, Brestelli JE, Kaestner KH, and Greenbaum LE (2005). Identification of transcriptional networks during liver regeneration. J. Biol. Chem 280, 3715–3722. [DOI] [PubMed] [Google Scholar]
- Wuestefeld T, Pesic M, Rudalska R, Dauch D, Longerich T, Kang T-W, Yevsa T, Heinzmann F, Hoenicke L, Hohmeyer A, et al. (2013). A Direct in vivo RNAi screen identifies MKK4 as a key regulator of liver regeneration. Cell 153, 389–401. [DOI] [PubMed] [Google Scholar]
- Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (N Y) 2, 100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, et al. (2014). CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Chang S-Y, Yin S-G, Liu Z-Y, Cheng X, Liu X-J, Jiang Q, Gao G, Lin D-Y, Kang X-L, et al. (2020). Two conserved epigenetic regulators prevent healthy ageing. Nature 579, 118–122. [DOI] [PubMed] [Google Scholar]
- Zahm AM, Wang AW, Wang YJ, Schug J, Wangensteen KJ, and Kaestner KH (2020). A High-Content Screen Identifies MicroRNAs That Regulate Liver Repopulation After Injury in Mice. Gastroenterology 158, 1044–1057.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, and Kanneganti T-D (2010). The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 32, 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea L-G, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, et al. (2013). Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Nguyen LH, Zhou K, Tu H-C, Sehgal A, Nassour I, Li L, Gopal P, Goodman J, Singal AG, et al. (2018a). Knockdown of anillin actin binding protein blocks cytokinesis in hepatocytes and reduces liver tumor development in mice without affecting regeneration. Gastroenterology 154, 1421–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhou K, Luo X, Li L, Tu H-C, Sehgal A, Nguyen LH, Zhang Y, Gopal P, Tarlow BD, et al. (2018b). The Polyploid State Plays a Tumor-Suppressive Role in the Liver. Dev. Cell 44, 447–459.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Nguyen LH, Miller JB, Yan Y, Kos P, Xiong H, Li L, Hao J, Minnig JT, Zhu H, et al. (2016). Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model. Proc Natl Acad Sci USA 113, 520–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Santoro R, and Grummt I (2002). The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 21, 4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Schmitz K-M, Mayer C, Yuan X, Akhtar A, and Grummt I (2009). Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat. Cell Biol 11, 1010–1016. [DOI] [PubMed] [Google Scholar]
- Zhu M, Lu T, Jia Y, Luo X, Gopal P, Li L, Odewole M, Renteria V, Singal AG, Jang Y, et al. (2019). Somatic mutations increase hepatic clonal fitness and regeneration in chronic liver disease. Cell 177, 608–621.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillner K, Filarsky M, Rachow K, Weinberger M, Längst G, and Németh A (2013). Large-scale organization of ribosomal DNA chromatin is regulated by Tip5. Nucleic Acids Res. 41, 5251–5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1: In vivo CRISPR screening data. Related to Figure 2 and Figure 6.
Sheet 1. sgRNA sequences included in the epigenetic CRISPRa screen. sgRNAs against 165 genes and 11 controls were extracted from the Mouse Genome-wide CRISPRa-v2 Library (Addgene #83996).
Sheet 2. Results of the epigenetic CRISPRa screen.
Sheet 3. sgRNA sequences included in the epigenetic CRISPR KO screen. sgRNAs against 165 genes and 1 control were extracted from (Shi et al., 2015).
Sheet 4. Results of the epigenetic CRISPR KO screen.
Sheet 5. sgRNA sequences included in the mechanism related CRISPRa screen. sgRNAs against 110 genes and 46 controls were designed with CRISPick.
Sheet 6. Results of the mechanism related CRISPRa screen.
Supplemental Table S2: Transcriptomic and epigenomic sequencing results. Related to Figure 6 and Figure 7.
Sheet 1. DESeq2 results for RNA-seq in 48h post-PHx mouse livers treated with GSK8573 or GSK2801.
Sheet 2. DiffBind analysis for H3K27ac ChIP-seq in H2.35 cells treated with GSK8573 or GSK2801.
Sheet 3. DiffBind analysis for H3K27ac ChIP-seq in 48h post-PHx mouse livers treated with GSK8573 or GSK2801.
Sheet 4. DiffBind analysis for ATAC-seq in H2.35 cells treated with GSK8573 or GSK2801.
Sheet 5. DiffBind analysis for H3K4me3 ChIP-seq in H2.35 cells treated with GSK8573 or GSK2801.
Sheet 6. DiffBind analysis for H3K4me1 ChIP-seq in H2.35 cells treated with GSK8573 or GSK2801.
Sheet 7. MACS results of Flag-BAZ2A in H2.35 cells overexpressing Flag-tagged BAZ2A.
Sheet 8. MACS results of Flag-BAZ2B in H2.35 cells overexpressing Flag-tagged BAZ2B.
Supplemental Table S3: Genes included in GSEA analysis and pathway enrichment analysis and results. Related to Figure 6.
Sheet 1–125. Genes included in the identified GSEA and pathway enrichment analysis.
Sheet 126. Pathway enrichment analysis from overlapped genes in H2.35 cells.
Sheet 127. Pathway enrichment analysis from overlapped genes in 48h post-PHx livers. Sheet 128. GSEA results on differentially expressed genes identified from RNA-seq.
Supplemental Table S4: Primer sequences for qPCR analysis, CRISPR editing, and sequencing. Related to Figures 1–7.
Data Availability Statement
RNA-seq and ChIP-seq data have been deposited at GEO and are publicly available. Accession numbers are listed in the key resources table. The analysed results are in Supplemental Table S2. Raw data for all the graphs in the figures are available in Supplemental Table S5. Microscopy data reported in this paper will be shared by the lead contact upon request.
There is no original code in this paper.
Any additional information required to reanalyze the data in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-PTEN | Cell Signaling Technology | Cat#9559; RRID: AB_390810 |
| Rabbit monoclonal anti c-Myc | Abcam | Cat#32072; RRID: AB_731658 |
| Rabbit polyclonal anti-FAH | Yecuris | Cat#20-0034 |
| Rat monoclonal anti-BrdU | Abcam | Cat#6326; RRID: AB_305426 |
| Mouse monoclonal anti-BrdU | BD Biosciences | Cat#555627; RRID: AB_10015222 |
| Rabbit polyclonal anti-Ki67 | Abcam | Cat#15580; RRID: AB_443209 |
| Rabbit monoclonal anti-E-cadherin | Cell Signaling Technology | Cat#3195; RRID: AB_2291471 |
| Rabbit polyclonal anti-H3K27ac | Abcam | Cat#4729; RRID: AB_2118291 |
| Rabbit polyclonal anti-H3K4me3 | Abcam | Cat#8580; RRID: AB_306649 |
| Rabbit polyclonal anti-H3K4me1 | Abcam | Cat#8895; RRID: AB_306847 |
| Mouse monoclonal anti-Flag | Sigma Aldrich | Cat#F1804; RRID: AB_262044 |
| Rabbit polyclonal anti-HNF4a | Santa Cruz Biotechnology | Cat#8987; RRID: AB_2116913 |
| Goat anti Rabbit IgG, Alexa Fluor 594 conjugated | Invitrogen | Cat#A-11012; RRID: AB_2534079 |
| Goat anti Rat IgG, Alexa Fluor 488 conjugated | Invitrogen | Cat#A-11006; RRID: AB_141373 |
| Goat anti Mouse IgG2a, Alexa Fluor 594 conjugated | Invitrogen | Cat#A-21135; RRID: AB_2535774 |
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| CuRx Nitisinone (NTBC) | Yecuris | Cat #20-0027 |
| 5-ethynyl-2’-deoxyuridine (EdU) | Abcam | Cat #146186 |
| 5-Bromo-2'-deoxyuridine (BrdU) | Sigma Aldrich | Cat #B5002 |
| Carbon tetrachloride (CCl4) | Sigma Aldrich | Cat #289116 |
| Saponin | Sigma Aldrich | Cat #47036 |
| GSK8583 | WuXi AppTec | N/A |
| GSK2801 | WuXi AppTec | N/A |
| BAZ2-ICR | Sigma Aldrich | Cat #SML1276 |
| Dextran sulfate sodium (DSS) | TdB Consultancy | Cat #DB001 |
| O-Propargyl-Puromycin (OP-Puro) | Cayman Chemical | Cat #15134 |
| Rapamycin | Selleck Chemicals | Cat #S1039 |
| Acetaminophen | Sigma Aldrich | Cat #A7085 |
| DAPI Solution | Thermo Scientific | Cat #62248 |
| UltraPure Phenol:Chloroform:Isoamyl Alcohol (25:24:1, v/v) | Invitrogen | Cat #15593031 |
| TRIzol Reagent | Invitrogen | Cat #15596018 |
| 4% Paraformaldehyde Solution in PBS | Alfa Aesar | Cat #J19943K2 |
| Formaldehyde solution | Sigma Aldrich | Cat #F8775 |
| Hematoxylin QS | Vector Laboratories | Cat #NC9788954 |
| Fetal Bovine Serum | Sigma Aldrich | Cat #F4135 |
| Dulbecco’s High Glucose Modified Eagles Medium | Cytiva Hyclone | Cat #SH30022.FS |
| Dexamethasone | Sigma Aldrich | Cat #4902 |
| Penicillin Streptomycin 100X solution | Cytiva Hyclone | Cat #SV30010 |
| Critical commercial assays | ||
| VECTASTAIN Elite ABC-HRP Kit (Rabbit IgG) | Vector Laboratories | Cat #PK-6101 |
| VECTASTAIN Elite ABC-HRP kit (Rat IgG) | Vector Laboratories | Cat #PK-6104 |
| M.O.M Elite Immunodetection Kit | Vector Laboratories | Cat #PK-2200 |
| Q5 High-Fidelity DNA Polymerase | New England BioLabs | Cat #M0515 |
| Click-iT Edu Alexa Fluor 488 Flow Cytometry Kit | Invitrogen | Cat #C10425 |
| Alexa Fluor 555 Antibody Labeling Kit | Invitrogen | Cat #A20187 |
| Click-iT Cell Reaction Buffer Kit | Invitrogen | Cat #C10269 |
| Alexa Fluor 555 Azide | Invitrogen | Cat #A20012 |
| Liver Perfusion Medium | Gibco | Cat #17701038 |
| Liver Digest Medium | Gibco | Cat #17703034 |
| Hepatocyte Wash Medium | Gibco | Cat #17704024 |
| AST Activity Assay Kit | Sigma Aldrich | Cat #MAK055 |
| ALT Activity Assay Kit | Sigma Aldrich | Cat #MAK052 |
| iScript Reverse Transcription Supermix | Bio-Rad Laboratories | Cat #1708840 |
| iTaq Universal SYBR Green Supermix | Bio-Rad Laboratories | Cat #1725124 |
| ColoScreen ES Lab Pack | Helena Laboratories | Cat #5086 |
| Ovation Mouse RNA-Seq System | Nugen | Cat #0348-32 |
| RNeasy Mini Kit | Qiagen | Cat #74104 |
| QIAquick PCR Purification Kit | Qiagen | Cat #28104 |
| NEBNext Ultra II DNA Library Prep Kit | New England BioLabs | Cat #7645 |
| NEBNext High-Fidelity 2X PCR Master Mix | New England BioLabs | Cat #M0544 |
| Illumina Tagment DNA Enzyme and Buffer | Illumina | Cat #20034197 |
| Qubit dsDNA BR Assay Kit | Invitrogen | Cat #Q32850 |
| BAZ2B inhibitor AlphaScreen Kit | BPS Bioscience | Cat #32600 |
| NEBNext Multiplex Oligos for Illumina (Set 1) | New England BioLabs | Cat #7335 |
| NEBNext Multiplex Oligos for Illumina (Set 2) | New England BioLabs | Cat #7500 |
| NEBNext Multiplex Oligos for Illumina (Set 3) | New England BioLabs | Cat #7710 |
| NEBNext Multiplex Oligos for Illumina (Set 4) | New England BioLabs | Cat #7730 |
| Deposited data | ||
| Raw and analyzed RNA-seq and ChIP-seq data | This paper | GEO: GSE183502 |
| Experimental models: Cell lines | ||
| Mouse: H2.35 cells | ATCC | CRL-1995 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratories | JAX: 000664 |
| Mouse: CD1 IGS | Charles River Laboratories | N/A |
| Mouse: Rpl24Bst/+ on C57BL/6J | Oliver et al., 2004 | RRID:MGI:5295283 |
| Mouse: Fah KO on C57BL/6J | This paper | N/A |
| Mouse: Rag1 KO; Fah KO; Il2rg KO on C57BI/6 | Yecuris | Cat #10-0001 |
| Mouse: Baz2b KO on 129/C57BL/6J mixed background | This paper | N/A |
| Oligonucleotides | ||
| Primers for qRT-PCR: listed in Table S4 | This paper; Feng et al., 2012 | Table S4 |
| Primer for ATAC-seq library prep: listed in Table S4 | This paper | Table S4 |
| sgRNA sequences for screening validation: listed in Table S4 | Shi et al., 2015 | Table S4 |
| CRISPR sgRNA sequence against mouse Pten | Xue et al., 2014 | Table S4 |
| CRISPRa sgRNA sequence against mouse c-Myc | Wangensteen et al., 2018 | Table S4 |
| In vivo siRNAs for mouse Baz2a | Life Technologies | Cat #s100272 |
| In vivo siRNAs for mouse Baz2b | Life Technologies | Cat #s129008 |
| Primers for screening library: listed in Table S4 | This paper | Table S4 |
| Recombinant DNA | ||
| Fah-Cas9 | This paper | N/A |
| Fah-dCas9-VP64 | This paper | N/A |
| Fah-Cas9-sgBaz2a | This paper | N/A |
| Fah-Cas9-sgBaz2b | This paper | N/A |
| Fah-BAZ2A | This paper | N/A |
| Fah-BAZ2B | This paper | N/A |
| Fah-Cas9-sgPten | This paper | N/A |
| Fah-GFP | This paper | N/A |
| Fah-dCas9-VP64-sgMyc | This paper | N/A |
| pCMV(CAT)T7-SB100 | Mátés et al., 2009 | pCMV(CAT)T7-SB100 Addgene Cat #34879 |
| FAHIG | Wuestefeld et al., 2013 | N/A |
| Software and algorithms | ||
| HISAT | Kim et al., 2015 | http://www.ccb.jhu.edu/software/hisat/index.shtml |
| HTSeq | Anders et al., 2015 | https://htseq.readthedocs.io/en/master/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| BWA aligner | Li and Durbin. 2009 | http://bio-bwa.sourceforge.net/ |
| MACS | Zhang et al. 2008 | https://orionedocumentation.readthedocs.io/en/latest/chip_seq/ngs_peak_calling/macs14.html |
| DiffBind | Stark and Brown, 2011 Ross-Innes et al., 2012 | http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| deepTools | Ramirez et al., 2016 | https://deeptools.readthedocs.io/en/develop/ |
| UCSC | Kuhn et al., 2013 | https://genome.ucsc.edu/util.html |
| Gene Set Enrichment Analysis | Subramanian et al., 2005 Mootha et al., 2003 | https://www.gseamsigdb.org/gsea/index.jsp |
| clusterProfiler | Yu et al., 2012 Wu et al., 2021 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| Other | ||
| Agencourt Ampure XP Beads | Beckman Coulter | Cat #A63881 |
| Pierce Protein A/G Magnetic Beads | Thermo Scientific | Cat #26162 |
| BD FACSAria Fusion Flow Cytometer | BD Biosciences | N/A |
| Nickel Chelate AlphaLISA Acceptor Beads | Perkin Elmer | Cat #AL108C |
| Streptavidin-conjugated donor beads | Perkin Elmer | Cat #6760002 |