Summary
Hippocampal place cells underlie spatial navigation and memory. Remarkably, CA1 pyramidal neurons can form new place fields within a single trial by undergoing rapid plasticity. However, local feedback circuits likely restrict the rapid recruitment of individual neurons into ensemble representations. This interaction between circuit dynamics and rapid feature coding remains unexplored. Here we developed “all-optical” approaches combining novel optogenetic induction of rapidly forming place fields with 2-photon activity imaging during spatial navigation in mice. We find that induction efficacy depends strongly on the density of co-activated neurons. Place fields can be reliably induced in single cells, but induction fails during co-activation of larger subpopulations due to local circuit constraints imposed by recurrent inhibition. Temporary relief of local inhibition permits the simultaneous induction of place fields in larger ensembles. We demonstrate the behavioral implications of these dynamics, showing that our ensemble place field induction protocol can enhance subsequent spatial association learning.
eTOC
Rolotti, Ahmed, Szoboszlay, et al. develop “all-optical” strategies to induce rapidly forming place fields in mouse CA1 and find that local feedback inhibition restricts recruitment of neurons into ensemble representations. Temporary relief of local inhibition permits the simultaneous induction of place fields in larger ensembles to enhance subsequent association learning.
Introduction
Pyramidal cells in area CA1 (CA1PCs) of the hippocampus develop spatially restricted firing fields (place fields, PFs) as animals explore their environments (O’Keefe & Dostrovsky, 1971; Ekstrom et al., 2003). These “place cells” are thought to form the basis for a cognitive map that supports navigation and associative learning, and may be disrupted with aging and neuropsychiatric disease states (Eichenbaum, 2000; Mallory & Giocomo, 2018; Lester et al., 2017; Kunz et al., 2015; Jun et al., 2020; Zaremba et al., 2017). These critical roles in memory formation have led to increasing attention to the precise synaptic and cellular mechanisms by which single CA1PCs form PFs (see Sheffield & Dombeck (2019); Magee & Grienberger (2020) for review). Intracellular recordings have shown that complex (‘burst’) spiking at the soma, a putative readout of a dendritic plateau potential, is sufficient to induce formation of a stable PF (Bittner et al., 2015). This process is mediated through a novel “behavioral timescale” synaptic plasticity (BTSP) mechanism (Bittner et al., 2017; Zhao et al., 2020; Milstein et al. 2021) which has unique characteristics that allow associations to be formed over longer timescales within a single trial, permitting rapid PF formation via the potentiation of spatially tuned intrahippocamapl inputs active within the BTSP kernel. But empirically, rapid formation of spontaneous PFs is relatively rare, so there must be additional circuit constraints that regulate this process, likely through the dense inhibitory feedback circuits present in CA1 (Klausberger & Somogyi, 2008; Bezaire & Soltesz, 2013). However, this potential connection between local inhibition and the permissibility of rapid PF formation has not been investigated.
Prior studies of rapid PF formation in the context of BTSP have studied either spontaneous plateauburst events, or those evoked through somatic electrical stimulation of single pyramidal neurons (Bittner et al., 2015, 2017; Diamantaki et al., 2018). Electrical stimulation has allowed experimenters to induce PFs at arbitrary locations as an animal navigates an environment, which has permitted careful analysis of BTSP and single cell dynamics following PF formation (Bittner et al., 2015; Zhao et al., 2020; Milstein et al. 2021). However, the technical limitations of in vivo whole cell recordings restrict this work to the study of single cells over relatively short (~ 1h) timescales. The ability to simultaneously stimulate many cells and longitudinally track their dynamics would broaden the scope for studying the emergence of rapid feature tuning and allow for finer-scale manipulations of the population code to modify cognitive representations and potentially bias behavior.
Along this line, recent work has simultaneously stimulated larger numbers of CA1PCs, but these experiments failed to induce PF formation in a location-specific manner (McKenzie et al., 2021; Robinson et al., 2020). It is possible that by stimulating many CA1PCs concurrently, these protocols recruit a high level of local inhibitory feedback that constrains the triggering of rapid plasticity processes such as BTSP that depend on very strong activation of CA1PCs. Indeed, it is known that the rate of endogenous PF formation can be scaled by the circuit in response to factors such as context, salient cues, and behavioral goals (Frank et al., 2004; Zaremba et al., 2017; Gauthier & Tank, 2018). We hypothesize that tuning the level of local inhibition could open windows of heightened excitability in which rapid PF formation can occur with greater prevalence to quickly build new representations.
To evaluate this hypothesis, we developed a variety of sparse cellular labeling and optogenetic targeting strategies to interrogate the local circuit regulation of rapid PF formation. Using an “all-optical” approach, we provide the first demonstration of bona fide optogenetic induction of PFs, likely via BTSP, in both single cells and CA1PC ensembles (opto-PF induction, opto-PFi). We use this method to track the long-term dynamics of induced PFs over days. We find that our opto-PFi approaches scale to small numbers of cells, but fail when too many CA1PCs are co-activated, due to recruitment of local feedback inhibition. This constraint can be partially ameliorated by direct suppression of local inhibition during opto-PFi. Finally, we use this strategy to drive artificial over-representation of a rewarded location in CA1PC ensembles and show that this can enhance subsequent behavioral association during a spatial reward-learning task.
Results
We first developed an “all-optical” approach by combining optogenetics and calcium imaging, allowing us to stimulate and record CA1PCs in a manner analogous to electrophysiological strategies for inducing BTSP in single neurons (Bittner et al., 2015). We used 2-photon (2p) microscopy-guided single cell electroporation to express an excitatory opsin and a fluorescent calcium reporter in individual cells (Kitamura et al., 2008) (Fig 1A). Small numbers of CA1PCs (n=3-5) were targeted and co-electroporated (see Methods) with plasmids coding for the high photocurrent, red-shifted excitatory opsin ChRmine (pCAGGS-ChRmine-mScarlet) and GCaMP7b (pCAGGS-GCaMP7b) (Marshel et al., 2019) (Fig 1A,B). We identified mice in which electroporations resulted in persistent expression of ChRmine and GCaMP7b in only 1-2 CA1PCs after 24-48h. These animals subsequently underwent opto-PFi while running on a cued linear treadmill (Fig 1C). 2p imaging of GCaMP7b fluorescence was used to track cellular dynamics across days. This head-fixed behavioral paradigm has been used extensively to investigate spatial coding across CA1PC populations (Danielson et al., 2016; Zaremba et al., 2017), as well as to induce PFs in single cells through BTSP (Bittner et al., 2015, 2017).
Figure 1: Optogenetic stimulation of isolated CA1PCs at a fixed location rapidly induces new PF formation.
(A) Top, Schematic of 2p-guided single cell electroporation. Bottom, Example electroporation of a single CA1PC in vivo. (B) Expression of GCaMP7b and ChRmine-mScarlet in a CA1PC ~ 48H after electroporation. Maximum intensity projection (Z-MIP; z-depth 58 um) shows expression in neurites as well as soma. (C) Left, Schema of circuit with CA1PCs (triangles) and local INs (blue circle) during widefield optogenetic stimulation. Right, Schematic of behavior apparatus and opto-PFi protocol; 1 sec widefield LED stimulation is triggered at a fixed spatial location (SZ) for 5 consecutive laps as mice run on a cued linear treadmill for randomly-delivered water rewards. (D) Left, peri-SZ fluorescence (z-scored) of an example CA1PC before (PRE), during (STIM), and after (POST) induction of a de novo PF, with next day (24H) follow-up. Within-session lap numbers are indicated next to each fluorescence trace. Highlighting indicates 1 second after SZ entry, red indicates LED on. Right, raster of the running-related activity (deconvolved events) of the same cell during first 20 laps of induction session and 24H follow-up session. Vertical line indicates SZ (position 0). Horizontal lines demarcate induction session epochs. (E) Mean tuning curve of all cells by session block, centered on SZ (vertical line). (F) Mean change in spatial firing activity from PRE to POST session blocks (centered on SZ, vertical line). (G) Left, activity centroid distance of cells to SZ (mean ± sem; PRE: 59.42 ± 8.84, POST 27.58 ± 8.44, 24H: 42.12 ± 8.95, 48H: 44.58 ± 12.46, 72H 46.37 ± 8.51; POST: p = 0.0383, One-sample Student’s t-test against null hypothesis of 48.5 cm). Colored points indicate cells with peri-SZ PF. Right, activity centroid distance shift toward SZ after induction protocol (POST - PRE mean ± sem; 31.84 ± 11.37, p = 0.0231, One-sample Student’s t-test against null hypothesis of 0 cm). (H) Left, within-PF tuning curve peak location relative to SZ for successfully induced (7/9) cells (−10.88 ± 2.16; p = 0.0023, One-sample Student’s t-test against null hypothesis of 0 cm). Right, correlation of induced PF width to peri-SZ velocity during first stimulation (pearson’s r=0.943, p = 0.0014). For (E-F) data is shown for all stimulated cells (n = 9 cells, 7 mice). For (H) data is shown only for cells with successfully induced peri-SZ PF (n = 7 cells, 6 mice). All boxes indicate median and interquartile range.
Head-fixed mice initially ran for a 30 min session on the self-propelled treadmill belt (~ 2m length) while foraging for water rewards. A single water reward was delivered at a random belt location on each lap. We attempted to optogenetically induce BTSP in labeled CA1PCs during this “induction session”. Cellular activity was continuously imaged across three contiguous epochs (Fig 1D): a baseline period of 10 laps (“PRE”); an induction period of 5 consecutive laps (“STIM”) where we delivered 1 sec of widefield LED photostimulation (620 nm, 30 Hz) through the microscope objective when the mouse traversed a fixed stimulation zone (SZ); and any additional laps the mouse ran during the remainder of the session (“POST”). To follow long-term dynamics of photostimulated CA1PCs, the same cells were serially imaged during 20 min sessions of random foraging on the same belt 24h, 48h, and 72h after the induction session.
During photostimulation, CA1PCs displayed reliable, short-latency responses to illumination (STIM, Fig 1D). While stimulated cells were typically weakly active during PRE, after STIM they showed strong, consistent firing around the SZ during POST for the remainder of the session (Fig 1D-F), except for 2/9 cells in which photostimulation failed to induce spatially tuned activity after STIM laps (Fig S1A). In remaining cells, spatially tuned activity anticipating the SZ rapidly emerged following the very first stimulation lap (STIM, Fig 1D,E, Fig S1B). This de novo spatial tuning (Fig 1F) often persisted across subsequent days, though the strength of this new representation declined over time (Fig 1E,G), consistent with the dynamics of spontaneously formed spatial representations (Ziv et al., 2013). We quantified the bias of activity toward the SZ by calculating the absolute distance of the activity centroid (the center of mass of deconvolved events along the belt) to the SZ (Fig 1G). While activity during PRE was no closer than chance (1/4 belt length, 48.5 cm), activity during POST was significantly shifted toward the SZ. In addition to this continuous measure of activity bias, we also asked whether stimulated cells formed a reliable PF near the SZ. We considered newly formed PFs during POST to be induced by the opto-PFi protocol if they overlapped the SZ. By this metric, 7/9 photostimulated cells displayed an induced PF during POST (Fig 1G). The number of these “induced cells” with PFs near the SZ decreased over the subsequent days to 5/9 at 24h and 48h, with 2/9 remaining at 72h (Fig 1G). Importantly, though we were not able to readout dendritic plateau potentials using our somatic calcium imaging approach, PFs of induced cells exhibited all other features characteristic of BTSP’s seconds-long, asymmetric plasticity kernel. First, the peak PF firing was shifted to a location before the SZ in all induced place cells (Fig 1H, left). This ~ 11 cm shift closely matched the shift previously demonstrated with whole-cell patch-clamp recordings (Bittner et al., 2017). Second, the PF widths showed a strong correlation to the mouse’s velocity around the first stimulation (Fig 1H, right), again matching prior electrophysiological data (Bittner et al., 2017). Together, these data show that strong optogenetic activation of isolated CA1PCs is sufficient to drive rapid PF formation within just a few trials, through a process highly consistent with BTSP. Importantly, these results also demonstrate that artificially induced PFs are stable on timescales of several days, similar to natural, spontaneously formed PFs (Ziv et al., 2013).
We next asked whether optogenetic induction of PFs scales to larger CA1PC populations. We virally expressed Cre-dependent ChRmine and GCaMP6f in CaMKIIα-Cre mice to achieve dense expression specifically in the CA1PC population (Ahmed et al., 2020; Dragatsis & Zeitlin, 2000) and subjected mice to our widefield optogenetic induction protocol (Figs 1C, S1C,D). However, photostimulation of this large population of CA1PCs resulted in epileptiform-like activity, consisting of calcium waves followed by suppression of cellular activity with slow recovery over minutes (Fig S1C), similar to previous descriptions (Farrell et al., 2020). We did not detect any bias in population activity toward the SZ upon recovery (Fig S1D).
In order to scale single-cell opto-PFi to multi-cell ensembles, we reasoned that sparse opsin expression across the population may be critical to avoid triggering epileptiform-like activity upon photostimulation. To achieve controlled, sparse opsin expression via a viral approach, we co-injected a tamoxifen-dependent Cre virus (CaMKII-Cre-ERT2) (Brandalise et al., 2020) with a Cre-dependent, high photocurrent red-shifted excitatory opsin (ChRmine or bReaChES; data pooled) (Marshel et al., 2019; Rajasethupathy et al., 2015). By titrating the cumulative dose of tamoxifen administered to each mouse, we could vary the fraction of CA1PCs expressing excitatory opsin (Fig 2D,H). Co-injection of GCaMP6f virus allowed for the monitoring of the entire population of CA1PCs in the FOV (Fig 2A,B).
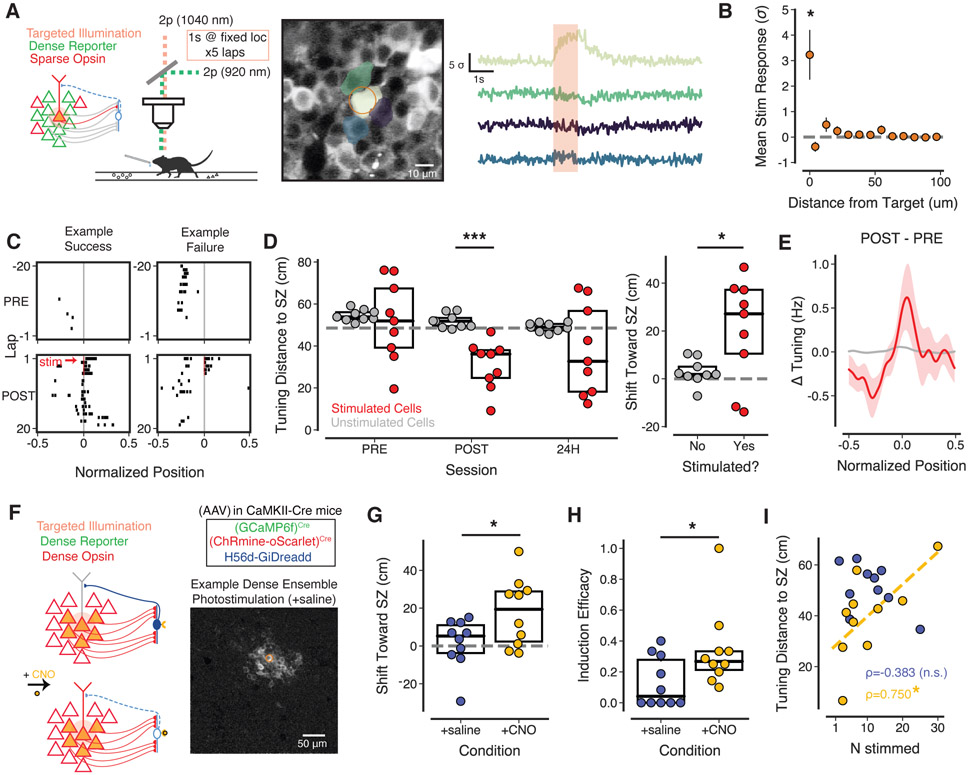
Figure 2: Efficacy of ensemble opto-PFi depends on the density of co-activated CA1PCs.
(A) Left, Viral strategy for sparse expression of excitatory opsins across CA1PCs. Right, Example FOV. (B) Left, schema of circuit activation during widefield photostimulation of sparse CA1PC subpopulation. Right, Ensemble opto-PFi and monitoring protocol. (C) Tuning profiles for stimulated cells with a PF in POST, sorted by session tuning peak. Heatmap intensity shows normalized event rate in each spatial bin, centered on SZ. (D) Mean activity centroid distance to SZ for photostimulated cells versus percent CA1PCs activated in FOV. Individual data points represent mean across cells for a single mouse FOV and coloring indicates median split used in E,F. Line shows linear fit (Spearman’s ρ = 0.556, p = 0.0165). (E) Left, mean activity centroid distance to SZ for stimulated cells by experiment group. Asterisks indicate significant difference from chance level (one sample Student’s t-test) or between groups (independent Student’s t-test). *p < 0.05, **p < 0.01 Right, mean shift toward SZ. mean ± sem, low: 27.46 ± 6.54; medium: 0.09 ± 4.42. p = 0.0032, independent Student’s t-test. (F) Induction efficacy, i.e., fraction of stimulated cells with induced peri-SZ PF. p = 0.0303, Mann-Whitney U test. (G) Left, Example FOV in stratum oriens with GCaMP6f expression in putative INs. Right, schema of imaging of IN and PC populations during randomly-administered photostimulation of CA1PC subpopulations of increasing size. (H) Fraction photostimulation-responsive PCs at each tamoxifen dose. Bars indicate mean ± sem. Spearman’s ρ = 0.5773, p = 0.0008, n = 6 mice. (I) Left, Mean response to photostimulation of all INs in example FOV across sessions with differing CA1PC co-activation densities (baselined to pre-photostimulation activity). n = 28 – 41 cells, 6 mice. Right, mean IN response (2s post-stim - 2s pre-stim) versus percent co-activated CA1PCs. Colored by mouse. Dashed line shows linear fit; Spearman’s ρ = 0.6549, p = 8.594×10−5. For all panels, shading indicates mean ± sem.
We then used our opto-PFi protocol (Figs 1C, 2B, S2B) to attempt to simultaneously induce PFs in CA1PC subpopulations of varying sizes. We found that stimulation of CA1PC subpopulations resulted in PF over-representation of the region around the SZ (Fig 2C). Overall, CA1PCs with optogenetically induced PFs were largely indistinguishable from CA1PCs with spontaneously formed PFs in terms of firing rates (Fig S3A,B) and spatial tuning metrics (Fig S3C,D). CA1PCs with artificially induced PFs were also equally as stable as those with spontaneous PFs, based on both 24 hour tuning correlation (Fig S3E) and 24 hour recurrence probability: of the 55 artificially induced place cells, 37 (or ~ 67%) retained a PF 24 hours later, compared to 2793 of the 3592 natural place cells (or ~ 77%), statistically indistinguishable turnover rates (Fisher’s exact test; p=0.0727). Relatedly, pair-wise reactivation during immobility was similar for spontaneous-induced place cell pairs relative to spontaneous-spontaneous pairs (Fig S3F), This indicated that artificially induced place cells were largely integrated into spontaneous neural assemblies representing the broader spatial map (Grosmark et al., 2021).
Interestingly, we found a significant correlation between the size of the stimulated subpopulation (expressed as percentage of the population of all identified CA1PCs) and the subpopulation’s mean activity centroid distance to the SZ during POST (Fig 2D), suggesting that stimulation of larger ensembles was less effective at biasing spatial activity towards the SZ. To examine this more closely, the 18 experiments were separated by a median split (2.86%) into low (0.75%-2.78%; range: 2-13 cells, mean: 4.78) and medium (2.93%-8.90%; range: 4-49 cells, mean: 22.67) density co-activation groups. Stimulated cells in low density mice became strongly biased toward the SZ after the induction protocol (POST epoch) and remained so up to at least 24h later (Fig 2E, S2C-F). They were significantly more biased than the medium density group at both of these time points, which showed no change in activity bias to the SZ compared to chance (Fig 2E) or compared to their neighboring unstimulated (primarily opsin-negative) cells (Fig S2C-F). We observed a similar difference between low and medium density subpopulations in the fraction of stimulated cells developing a novel peri-SZ PF (induction efficacy, Fig 2F, S2E). These data suggest that, at least in the case of a fixed linear environment without behaviorally-salient spatial cues, there is a strict limit on the size of the subpopulation capable of simultaneously forming a novel spatial representation around an arbitrary location.
This local circuit constraint is consistent with recent work showing that focal optogenetic stimulation of a relatively large population of CA1PCs fails to induce PFs near the stimulation location. Instead, stimulation led to an increased rate of nonspecific PF remapping (McKenzie et al., 2021; Robinson et al., 2020). We wondered if this process might also occur in parallel to our location-specific PF inductions. However, in our paradigm, with PRE and POST separated into different contextual exposures/behavioral episodes after a home cage rest period (Fig 2B), we observed a large degree of remapping even in opsin-negative control mice (Fig S3G), precluding the detection of any finer photostimulation-related remapping due to our limited sensitivity (Fig S3H).
Next, to further understand why rapid PF formation was successfully triggered in certain stimulated cells compared to others (Fig 2D-F), we examined the responses of photoactivated cells based on whether they developed induced PFs or not. We observed that the mean activity responses to photostimulation for CA1PCs with induced PFs was significantly higher than that of non-induced CA1PCs (Fig S3I), consistent with the dependence of rapid PF formation on strong cellular activation and bursting (Bittner et al., 2017). Differences in photostimulation responses across CA1PCs in our experiments could be due, at least in part, to factors in the local circuit architecture. When we examined the anatomical location of photoactivated cells, we found that induced CA1PCs had significantly fewer neighbors that were also co-activated during photostimulation (Fig S3J). These results indicate that CA1PCs with nearby neighbors that were co-activated at the SZ were less likely to develop induced PFs.
Such differences in PF induction outcomes could be due to strongly activated CA1PCs recruiting recurrent/lateral inhibition (Nakajima et al., 2021; Trouche et al., 2016; McKenzie et al., 2021) to inhibit other nearby CA1PCs via a competitive mechanism for neuronal assembly selection (Rao-Ruiz et al., 2019; Roux & Buzsaki, 2015). Though due to our recording configuration we could not differentiate between radial sublayer (superficial/deep) CA1PC populations, which are known to differ in burst propensity and inhibitory innervation (Soltesz & Losonczy, 2018; Lee et al., 2014), we examined the photostimulation responses of cells from each induction outcome group which had at least one co-activated CA1PC neighbor in a given radius. We observed a clear difference, with induced cells showing stronger activation, particularly when comparing cells with neighbors in close proximity (Fig S3K). The larger photoactivation responses in induced cells, despite co-activated neighbors, is consistent with a competitive process in which strongly stimulated CA1PCs recruit lateral inhibition to hyperpolarize surrounding cells. This would in turn reduce the level of reciprocal lateral inhibition the stimulated cell itself receives, effectively disinhibiting itself (Roux & Buzsaki, 2015; McKenzie, 2018; Rao-Ruiz et al., 2019). Alternatively, it may simply be the case that, in the context of strong recurrent/lateral inhibition from other nearby active CA1PCs, only those cells with the strongest burst spiking can trigger PF formation. In either case, these results raise the possibility that local inhibition might limit neuronal activation during synchronous firing and constrain the rapid formation of new PFs. However, we note that the scale on which we are able to detect influences between co-activated neighbors (~ 10μm, Fig S3J,K) is surprisingly small relative to the scale of interneuron axonal arborization (~ 100μm; Klausberger (2009); Müller & Remy (2014)), and warrants further investigation in future work designed to more directly examine any underlying neuroanatomical basis for these local functional interactions (Bezaire & Soltesz, 2013).
Nevertheless, given our overall findings of reduced opto-PFi efficacy with increased CA1PC co-activation density, we sought to directly measure how local inhibition might scale with increased co-activation of CA1PCs. We used our titratable sparse opsin expression approach to activate CA1PC ensembles of varying sizes while imaging the activity of putative interneurons (INs) in stratum oriens of CA1 (Figs 2I-K). Stimulating CA1 subpopulations of increasing size caused a linear scaling of local IN activity (Figs 2K, S2G). However, due to the nonlinear integration of calcium signals at INs’ higher baseline firing rates, it is difficult to determine whether the duration of this activation reflects a shift into a tonic firing mode or simply higher phasic activation (Arriaga & Han, 2017; Sheffield et al., 2017; Geiller et al., 2020). These results suggest that scaling recurrent/lateral inhibition (Rao-Ruiz et al., 2019; Bolding & Franks, 2018) could constrain induction of PFs across CA1PC subpopulations when local co-activation density exceeds particular thresholds.
To further investigate the role of recurrent/lateral inhibition in PF induction, we used 2p targeted photostimulation of CA1PCs to trigger either low (Fig 3A) or high (Fig 3F) density co-activation based on viral expression strategy (sparse vs. dense approaches). Targeted activation was carried out by repeatedly spiraling a photostimulation laser over a 15-20 μm region centered on an identified cell body (Rickgauer & Tank, 2009; Yang et al., 2018; Chettih & Harvey, 2019) for ~ 1s. Photostimulation of a single CA1PC, in a configuration with sparse excitatory opsin labeling, led to a rapid increase in activity in the targeted cell. Cells outside the photostimulation target were only weakly activated, if at all (Fig 3B).
Figure 3: CA1PC co-activation density limits on ensemble opto-PFi can be raised by suppressing local inhibition.
(A) Left, schema of circuit with sparse excitatory opsin expression and 2p-targeted photostimulation (see Methods). Right, Example 2p-target (orange ring) with spatial masks of targeted and neighboring cells overlaid. Mean responses to photostimulation (red shading) is shown for each cell. (B) Mean stimulation response amplitude for cells directly under target (leftmost point) or at evenly spaced distances from target. Bars indicate mean ± sem. (* p < 0.05, one-sample Student’s t-test against null hypothesis of 0). n = 1378 cells, 9 mice. (C) Event rasters for two example cells subjected to opto-PFi. PRE and POST sessions are shown for a cell successfully forming a new PF formation and another which only retained its previous PF. Red line indicates photostimulation laps. (D) Left, mean activity centroid distance to SZ for stimulated (red) and unstimulated (gray) cells. Dashed line indicates chance. Right, mean activity centroid shift toward SZ for stimulated and unstimulated cells in each FOV. Asterisks indicate significant difference between groups (paired Student’s t-test). *p < 0.05, **p < 0.01, ***p < 0.001. n = 9 mice. (E) Mean change in tuning curves from PRE to POST across stimulated and unstimulated cells. Shading indicates mean ± sem. n = 20 stimulated cells, n = 1358 unstimulated cells from 9 mice. (F) Left, schema of circuit with dense excitatory opsin labeling of CA1PCs and 2p-targeted stimulation (see Methods) before and after GiDREADD-mediated suppression of local IN activity with CNO. Right, Example baseline-subtracted FOV during 2p-targeted (orange ring) stimulation. Note the wide radius of co-activated cells outside the optical target. (G) Activity centroid shift toward SZ for each condition. +saline: 1.611 ± 4.15; +CNO: 17.91 ± 5.69; p = 0.0326, independent Student’s t-test. (H) Fraction stimulated cells with induced PF in each condition. +saline: 0.1312 ± 0.05; +CNO: 0.34 ± 0.08; p = 0.0216, Mann-Whitney U test. (I) Mean activity centroid distance to SZ for photostimulated cells without (blue) or with (yellow) GiDREADD activation versus number of cells photostimulated. Dashed line indicates linear fit. +saline: spearman’s ρ = −0.383, p = 0.2747; +CNO: spearman’s ρ = 0.750, p = 0.0125. n = 10 +saline experiments, 5 mice; n = 10 +CNO experiments, 4 mice. For D,G,I, individual data points represent mean across cells in a single FOV.
In this configuration, our opto-PFi protocol (Fig 2A) biased activity toward the SZ via the rapid formation of PFs lasting at least 24h (Fig 3C-E), similar to the low-density coactivation experiments using widefield photostimulation (Fig 2D-F). The increase in activity at the location immediately surrounding the SZ was specific to stimulated cells, with no change in unstimulated neighbors (Fig 3D,E). Interestingly, photostimulated cells also decreased their activity at distances farther away from the SZ (Fig 3E), in line with recent results demonstrating an inhibitory component of the BTSP kernel (Milstein et al. 2021). In the few cases where more than one cell was photoactivated (range 1-3, median 2), multiple cells formed PFs near the SZ (up to 3/3 in one case). Overall though, PF induction was less successful in this configuration than with widefield photostimulation/single cell electroporation, with only 42.6% of all 2p-photostimulated cells forming PFs near the SZ (versus 77.8%; see Fig 1), and only 5/9 induction experiments (versus 8/9) resulting in any of the stimulated cells forming a PF near the SZ. This is likely due to a combination of factors: weaker activation of individual cells (because of differences in photostimulation methods) and competition due to the recruitment of (weak) recurrent/lateral inhibition when multiple cells were co-activated.
In the wide-field stimulation experiments, co-activation of denser CA1 ensembles was associated with reduced PF induction efficacy (Fig 2D-F) and stronger recruitment of inhibition (Fig 2I-K). We next developed an approach to achieve dense co-activation via 2p-targeted photostimulation of CA1PCs under conditions of viral expression in a wider population of CA1PCs (as in Fig S1C,D). We conducted the same 2p-targeted photostimulation experiment as above (Fig 3A-E), but in a configuration where far more cells had the potential to be excited and in a regime that should not trigger epileptiform-like activity (Fig S1C,D). In addition, we injected a virus containing the inhibitory DREADD hM4Di targeted to INs via the h56D IN-specific promoter (h56D-GiDREADD) (Mehta et al., 2019), for chemogenetic suppression of CA1 INs upon administration of the DREADD ligand clozapine-N-oxide (CNO) (Fig 3F). We previously demonstrated that the h56D promoter provides selective access to nearly all mouse GABAergic neurons, with ~ 91% sensitivity and ~ 95% specificity (Mehta et al., 2019). Given the dense overlap of opsin-expressing cell bodies and neurites in the pyramidal layer and the limited spatial resolution of 2p excitation, 2p photostimulation targeted to individual CA1PC somata led to significant activation of a much larger neighborhood of CA1PCs (Fig 3F-G; Fig S4G), with cells as far as ~ 50μm outside the target radius of photostimulation being excited (range 2-25 stimulated cells; median 11.5). After our opto-PFi protocol in control conditions (intra-peritoneal administration of saline prior to PRE), 2p-photostimulated CA1PCs in this dense opsin labeling configuration no longer reliably shifted activity toward the SZ, with no increase in the rate of de novo PF formation near the SZ (Fig 3H).
On following days, mice were administered 10 mg/kg CNO intra-peritoneally ~ 20 minutes before the PRE session to suppress hM4di-expresssing INs. This led to a disinhibition of the activity of CA1PCs that lasted hours but recovered to baseline levels by the next day (Fig S4C,H) (Meira et al., 2018). The opto-PFi protocol was then carried out again at a new target region in the FOV, activated at a new SZ. hM4di-mediated suppression of INs largely restored the efficacy of opto-PFi (Fig 3G-I, S4J). Importantly though, even with partial suppression of inhibition, the bias of stimulated cells toward the SZ was still correlated with the number of CA1PCs activated in the FOV (Fig 3I). This argues against a general effect of CNO administration on excitability and is more consistent with competition between co-activated CA1PCs via mutual inhibition (Rao-Ruiz et al., 2019). In this case, while a partial relief of inhibition may be permissive for the development of feature selectivity in a small number of co-activated cells, stronger inhibition recruited by larger numbers of co-firing CA1PCs in the local neighborhood can likely still overcome this partial relief to prevent location-specific, rapid PF formation across the ensemble. Further arguing against a simple effect on CA1PC excitability, CNO did not enhance 2p photostimulation responses nor PFi efficacy in the sparse opsin expression configuration (Fig S4A-E). Overall, our data suggest that tuning local inhibition can have a significant impact on the efficacy of triggering rapid PF formation.
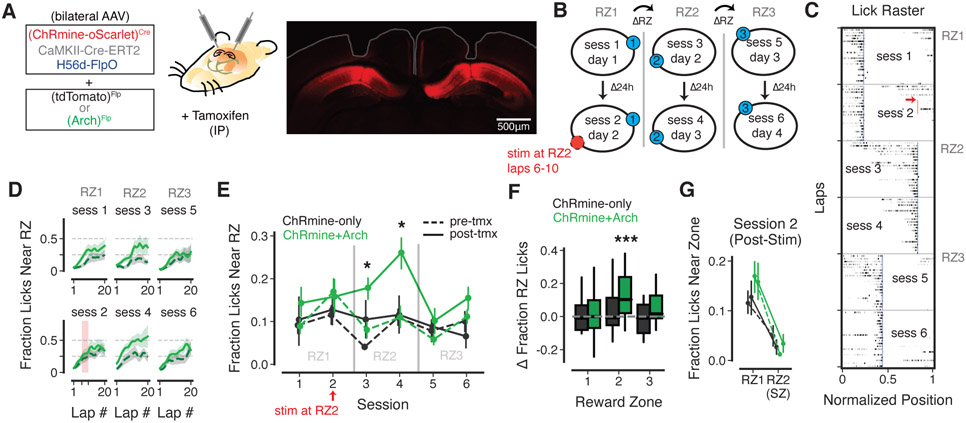
Finally, we asked whether applying our opto-PFi protocol in CA1PC subpopulations could be used to bias spatial learning and memory behavior. To examine this, we used our tamoxifen-induced ChRmine expression approach to label a subpopulation of CA1PCs across both hemispheres with excitatory opsin, while introducing the inhibitory opsin Arch (or a control protein) into CA1 INs using an intersectional approach (Fenno et al., 2020). We then implanted chronic optical fibers bilaterally over CA1 (Fig 4A) for subsequent optogenetic co-activation of CA1PCs expressing ChRmine with simultaneous optogenetic inhibition of local INs using a single 532 nm laser. We subjected mice to a head-fixed spatial reward learning task (Danielson et al., 2016; Zaremba et al., 2017), where mice had to learn the spatial location of hidden, lick-dependent rewards that were randomly shifted to a new reward zone (RZ) location every 24h (Fig 4B). During session 2 (24h recall of RZ1), we applied our opto-PFi protocol for 5 consecutive laps at a separate, randomly assigned SZ via photostimulation through the bilateral optical fiber cannulae (Fig 4A,B). The RZ was then moved on the very next session to the SZ location (RZ2, Fig 4B) to examine whether our optogenetic stimulation would affect learning of this new RZ where we had just applied our PFi protocol.
Figure 4: Ensemble opto-PFi while suppressing inhibition enhances subsequent goal oriented learning.
(A) Left, schematic of viral strategy for sparse, tamoxifen-dependent ChRmine expression in CA1PCs and for IN expression of Arch, with bilateral optical fiber implants. Right, post-hoc confirmation of ChRmine expression in dorsal CA1 (after tamoxifen administration) and optical fiber placement (white outline). (B) Schematic of goal-oriented learning task with photostimulation for opto-PFi. (C) Raster showing distribution of unrewarded licks during the first 20 laps of each session for an example mouse. Each row is an individual lap with lick rates normalized to the max lick rate at any spatial position on that lap. Blue vertical lines, RZ for each session. Red arrow and vertical line, SZ with photostimulation on laps 6-10. (D) Lap by lap fraction of unrewarded licks near the RZ for the first 20 laps of each session for ChRmine+Arch mice, pre- (dashed) and post- (solid) tamoxifen administration to induce ChRmine expression. Red shading shows laps when photostimulation is delivered at the SZ (RZ2) in session 2. Shading indicates mean ± sem. (E) Fraction of unrewarded licks near the RZ during each session for ChRmine-only (black) or ChRmine+Arch (green) mice, pre- (dashed) and post- (solid) tamoxifen administration to induce ChRmine expression. Gray lines demarcate RZs. ChRmine-only: n = 12 – 24 experiments per session, 8 mice; ChRmine+Arch: n = 23 – 24 experiments per session, 8 mice. Linear-mixed effects model for each session, fixed effects of opsin (ChRmine-only vs. ChRmine+Arch) and tamoxifen (pre vs. post), mouse as random effect. Asterisks indicate sessions with significant effects: Session 3: main effect of tamoxifen: z = 3.110, p = 0.002, 95% CI = (0.036, 0.160); Session 4: main effect of tamoxifen: z = 3.948, p = 7.9 × 10−5, 95% CI = (0.076, 0.226), effect of opsin x tamoxifen interaction: z = −2.312, p = 0.021, 95% CI = (−0.254, −0.02l). p > 0.05 for main effects and interaction for all other sessions. (F) Change in peri-RZ lick fraction from pre- to post-ChRmine expression for each belt/session in each RZ. Boxes indicate median and interquartile range, whiskers indicate 5th and 95th percentile. Asterisks indicate significant difference from null hypothesis of 0 (one-sample Student’s t-test with bonferonni correction for multiple comparisons); RZ2 ChRmine+Arch: p = 1.095 × 10−5. ChRmine-only: n = 24 – 30 mouse-session-belt pairs per RZ. ChRmine+Arch: n = 46 – 48 mouse-session-belt pairs per RZ. (G) Peri-zone lick fraction during session 2 post-stimulation laps for RZ1 and RZ2/SZ. Groups indicated by colors and dashes as in E.
Throughout each session, mice showed progressive learning of the reward location as evidenced by increasing fraction of licks near the RZ (Fig 4C,D). Prior to the induction of ChRmine expression in CA1PCs with tamoxifen (pre-tmx), neither Arch-expressing nor ChRmine-only control mice showed any changes in learning RZ2 (Fig 4D,E). This indicated that transient optogenetic inhibition of INs alone during our task had little effect on spatial reward learning. However, after administering 50 mg/kg tamoxifen (post-tmx) to induce expression of Cre-dependent ChRmine in ~ 3% of CA1PCs (Fig 2J), ChRmine+Arch-expressing mice now showed a significant enhancement in learning RZ2 (session 3) and 24h recall of this location (session 4) (Fig 4D-F). Consistent with an increase in spatially tuned representations specifically near the SZ with opto-PFi, these mice did not show enhanced learning of other locations on the belt when the reward was switched to another random location (RZ3) (Fig 4B-F). Moreover, control mice expressing ChRmine alone did not show significant learning enhancements, consistent with limited effectiveness of opto-PFi in CA1PC subpopulations where recurrent/lateral inhibition is intact (Figs 2,3). Finally, we observed that applying our opto-PFi stimulation alone was not sufficient to increase licking behavior near the SZ (Fig 4G), as ChRmine+Arch mice only developed increased licking near the opto-PFi location after the reward was moved to this location in session 3 (Fig 4C-F). This is consistent with opto-PFi resulting in enhanced learning of subsequent reward association at the opto-PFi triggering location. Taken together, our overall results show that successful triggering of opto-PFi across CA1PCs is greatly facilitated by transient suppression of inhibition and that this plasticity-triggering approach can be effectively used to rapidly augment spatial representations that enhance goal-directed associative learning.
Discussion
Prior experiments studying rapid PF formation through BTSP have relied on large somatic current injection to trigger dendritic plateaus (Bittner et al., 2015, 2017), which imposes many limits on experimental design. In our optical approach, we used excitatory opsins engineered for high photocurrent (Marshel et al., 2019; Rajasethupathy et al., 2015) to maximize depolarization. This likely accounts for our success in optogenetically inducing new PFs, compared to otherwise similar experimental configurations that did not trigger rapid, location-specific PF formation using lower photocurrent opsins (Rickgauer et al., 2014; Robinson et al., 2020). Though we are unable to unambiguously verify the triggering of plateau potentials in all induced cells using our functional calcium imaging approach, PFs induced via opto-PFi replicate the other principle characteristics of BTSP delineated in earlier electrophysiological studies (Bittner et al., 2015, 2017). Beyond this, we leveraged the capability to monitor neurons across days and found that experimentally induced PFs are functionally indistinguishable from spontaneously formed PFs, even at longer timescales (Fig S3A-F). It is important to note that, at least during novel environment exposures, plateau-associated complex spiking was not found to be strictly necessary for PF formation in single cell recordings (Cohen et al., 2017). However, recent work recording thousands of PFs in parallel finds enrichment of BTSP-like dynamics during the initial exposures to a new environment (Priestley et al., 2021). Thus, even if not strictly necessary for all field formation events, there may be a greater prevalence in recruitment of BTSP-like mechanisms than previously proposed.
Despite the success of our optical approach in small populations of neurons, we found that opto-PFi fails when extended to higher density co-activation of CA1PCs. Similarly, recent optogenetic experiments employing a comparatively weak stimulation of a dense subpopulation of CA1PCs also failed to induce new PFs at the stimulation location (McKenzie et al., 2021). However, our experiments identified local feedback inhibition as a major constraint on simultaneous PF formation across co-active CA1PC subpopulations. Our findings here are consistent with a tight excitation-inhibition balance to promote sparse coding (Rao-Ruiz et al., 2019; Deneve & Machens, 2016; Dalgleish et al., 2020), and support a model in which population activity itself regulates the rate of plasticity in response to changes in context, environmental salience, or behavioral demands (Milstein et al. 2021).
Given that hippocampal PFs are known to exhibit non-uniform tiling of environments in the presence of reward and salient stimuli during natural conditions (Dupret et al., 2010; Zaremba et al., 2017; Gauthier & Tank, 2018; Bourboulou et al., 2019; Kaufman et al., 2020), it is plausible that one way rapid PF formation is achieved is through the dynamic tuning of local inhibition to facilitate triggering of plasticity. Indeed, previous work has shown that switching between place cell ensembles is accompanied by a fine redistribution of IN firing activity (Trouche et al., 2016). Together with our work, this is consistent with the general notion that engagement of a selected subset of neurons in a given environment is associated with tight inhibitory control over other pyramidal cells (Hirase et al., 2001), which may occur through a competitive process between neighboring cells in the circuit (Rao-Ruiz et al., 2019). In the scope of our study here, we did not attempt to dissect the relative importance of different IN classes in constraining opto-PFi. Feedback INs, which are preferentially recruited during bursting (Pouille & Scanziani, 2004) and target the dendritic regions responsible for generating plateaus (Klausberger, 2009; Lovett-Barron et al., 2012), might be particularly well positioned for regulating PF formation. However, somatic inhibition (Sheffield et al., 2017; Pedrosa & Clopath, 2020) could also potentially constrain rapid place field formation, particularly in novel environments (Cohen et al., 2017). Furthermore, the activation of disinhibitory INs (Letzkus et al., 2015; Krabbe et al., 2019; Turi et al., 2019; Malik et al., 2021) could release inhibition of pyramidal cells across ensembles to exert a further layer of control over this process. Given the heterogeneity of inhibitory circuit elements and their compartment specific innervation patterns on CA1PCs (Pelkey et al., 2017; Klausberger & Somogyi, 2008), further investigation will be important to delineate the specific roles and recruitment patterns of various IN subtypes during PF induction and spontaneous formation in different environments and task demands.
Remarkably, we were able to enhance goal oriented learning using our combined photostimulation/disinhibition strategy to induce peri-SZ PFs across a sparse ensemble of CA1PCs. Our findings extend recent work on acute manipulations of spontaneously formed CA1 spatial representations, where activating specific ensembles can actively drive behavior (Robinson et al., 2020; Turi et al., 2019). Our opto-PFi approach for creating new, sparse representations could be used more generally to form novel, lasting associations that could influence behavior over longer timescales than those possible with acute circuit manipulations. Though we were only able to effectively induce peri-SZ PFs in relatively small fractions of CA1PCs (Figs 2,3), recent work has shown that manipulation of just a small fraction of CA1 place cells is sufficient for behavioral effects (Robinson et al., 2020), similar to the small numbers of cortical neurons required for sensory perception (Dalgleish et al., 2020). Consistent with this notion, we estimate based on measurements of pyramidal cell density in dorsal CA1 (Jinno & Kosaka, 2010), realistic modeling of optogenetic illumination volumes with the commercially available optical fibers we used (Yona et al., 2016), and high photosensitivity of ChRmine (Marshel et al., 2019; Chen et al., 2021), that we photostimulated ~ 120-160 CA1PCs across both hemispheres in these experiments. With at least 34 ± 8% opto-PFi efficacy with inhibition suppressed (Fig 3H), this would suggest that ~ 30-65 rapidly formed peri-SZ PFs are sufficient to enhance subsequent spatial reward learning at the SZ location. These estimates are in line with recent findings that activating as few as ~ 15 CA1 place cells can acutely bias behavior (Robinson et al., 2020).
Learning and recalling a change in reward location in a familiar environment may depend on the plasticity of PF maps, as evidenced by a correlation between reward zone over-representation (e.g., Fig 2C) and reward learning (Zaremba et al., 2017). We found that amplifying ensemble representations using our opto-PFi/disinhibition protocol could enhance learning of a rewarded location in the very next session, with the performance increase persisting at least 24h later. These results support a causal role for spatial over-representations in guiding behavior. As the introduction of an associated reward at the induction site was necessary to change behavior at that location (Fig 4F), rapid formation of PFs appears to allow for faster spatial association learning. This may occur by preempting or accelerating what would otherwise be a more gradual process through which PFs shift towards the reward location over multiple rounds of learning (Zaremba et al., 2017; Lee et al., 2006). It will be important in future studies to examine the role that other critical circuit elements, such as reward-related neuromodulation (Kaufman et al., 2020; Magee & Grienberger, 2020), may play in rapid PF formation and their impact on spatial learning, potentially through modulation of inhibitory feedback (Urban-Ciecko et al., 2018).
The strategies we develop here hold broad potential for illuminating fundamental questions regarding mechanisms of PF formation and feature tuning in individual neurons, as well as the role of long-term synaptic plasticity in modifying circuit computations and systems-level processing. As several emerging precision neuromodulation strategies aim to trigger long-term plasticity for therapeutic effects in neuropsychiatric disorders (Deng et al., 2020; Wang et al., 2020), our work highlights the importance of incorporating knowledge of constraints imposed by local circuit architecture and dynamics of target regions. Translation of strategies for transient calibration of local excitation-inhibition balance (Sohal & Rubenstein, 2019) may thus allow for more effective triggering of therapeutic neuroplasticity.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to the lead contact Attila Losonczy (al2856@columbia.edu).
Materials Availability
All unique resources generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and Code Availability
All data reported in this paper will be shared upon reasonable request to the lead contact.
All original code has been deposited at Zenodo and is publicly available on GitHub as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available upon reasonable request to the lead contact.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| rAAV1.Syn.Flex.GCaMP6f.WPRE.SV40 | This paper | Addgene 100833-AAV1 |
| rAAV8.Ef1α.DIO.ChRmine.oScarlet.WPRE | Karl Deisseroth (Marshel et al., 2019) | N/A |
| rAAV2/1.Ef1α.DIO.ChRmine.oScarlet.WPRE | This paper | N/A |
| rAAV8.nEF.Con/Foff.2.0.ChRmine-oScarlet | Karl Deisseroth Fenno et al., 2020 | N/A |
| rAAV1.Syn.GCaMP6f.WPRE.SV40 | This paper | Addgene 100837-AAV1 |
| rAAV2/9.Ef1α.FLEX.Breaches.tdTomato | Kaufman et al., 2020 | N/A |
| rAAV2/1.CaMKII.Cre.ERT2 | This paper | N/A |
| rAAV2/1.h56d.HA-hMD4(Gi).mCherry | This paper | N/A |
| rAAV2/1.h56d.FlpO | This paper | N/A |
| rAAV8.nEF.Coff/Fon.Arch3.3-eYFP | Karl Deisseroth (Fenno et al., 2020) | N/A |
| rAAV2/1.Syn.FRT.tdTomato | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Slc32a1-2A-FlpO-D knock-in (VGAT-FlpO): B6.Cg-Slc32a1tm1.1(flpo)Hze/J | Daigle et al., 2018 | Jackson Laboratory Strain 029591 |
| Mouse: R4Ag11: B6.Cg-Tg(CamKIIα-Cre)3Szi/J | Bina Santoro (Dragatsis and Zeitlin, 2000) | Jackson Laboratory Strain 027400 |
| Mouse: R1Ag5: B6.Cg-Tg(CamKIIα-Cre)2Szi/J | Bina Santoro (Dragatsis and Zeitlin, 2000) | Jackson Laboratory Strain 027310 |
| Recombinant DNA | ||
| pCAGGS-GCaMP7b | This paper | N/A |
| pCAGGS-ChRmine-mScarlet | This paper | N/A |
| Software and Algorithms | ||
| Suite2p | Pachitariu et al., 2017 | https://github.com/cortex-lab/Suite2P |
| OASIS | Friedrich et al., 2017 | https://github.com/j-friedrich/OASIS |
| Custom data processing and analysis code | This paper | https://dx.doi.org/10.5281/zenodo.5765716 |
| Python 2.7 | https://www.python.org | N/A |
| SIMA | Kaifosh et al., 2014 | https://github.com/losonczylab/sima |
Experimental Model and Subject Details
All experiments were conducted in accordance with the NIH guidelines and with the approval of the Columbia University Institutional Animal Care and Use Committee. Experiments were performed with adult (8-16 weeks) male and female C57Bl/6 mice (Jackson Laboratory), transgenic CaMKIIα-Cre mice on a C57Bl/6 background, where Cre is predominantly expressed in pyramidal neurons (R4Ag11 and R1Ag5 lines, Dragatsis and Zeitlin (2000); Jackson Laboratory, Stock No: 027400), and transgenic VGAT-Flp mice on a C575Bl/6 background, where Flp is predominantly expressed in interneurons (Slc32a1-2A-FlpO-D knock-in line, Daigle et al. (2018); Jackson Laboratory, Stock No: 029591).
Method Details
Behavior and Imaging
Surgical procedure
Viral delivery to hippocampal area CA1 and implantation of headposts and imaging cannulae were as described previously (Turi et al., 2019; Ahmed et al., 2020). Briefly, mice were anesthetized under isofluorane and viruses were delivered to dorsal CA1 by stereotactically injecting 50 nL (10 nL pulses) of rAAVs at three dorsoventral locations using a Nanoject syringe (−2.1 mm AP; −1.5 mm ML; −0.9, −1.05 and −1.2 mm DV relative to bregma). Mice were allowed to recover in their home cage for 3 days following virus delivery procedures. They were then surgically implanted with a custom metal headpost (stainless steel or titanium) along with an imaging window (diameter, 3.0 mm; height, 1.5 mm or 2.3 mm) over the left dorsal hippocampus. Imaging cannulae were constructed by adhering (Narland optical adhesive) a 3 mm glass coverslip (64-0720, Warner) to a cylindrical steel cannula. The imaging window surgical procedure was performed as detailed previously (Kaifosh et al., 2013; Lovett-Barron et al., 2014). Briefly, mice were anesthetized and the skull was exposed. A 3 mm hole was made in the skull over the virus injection site. Dura and cortical layers were gently removed under visual guidance while flushing with ice-cold cortex buffer. The imaging cannula was inserted through the surgical opening in the skull and secured so that external capsule fibers were visible through the cannula glass. Finally, the metal headpost was affixed to the skull with dental cement. For all surgeries, monitoring and analgesia (meloxicam as needed) was continued for 3 days postoperatively.
Single Cell Electroporation
Cannula implantation procedures were similar to as above, but custom-designed 316L stainless steel cannulae were used (InterPRO Additive Manufacturing group, CT, US). Height and diameter of the cannulae was 1.9 mm and 3.0 mm, respectively, with one side angled to allow for pipette insertion. The glass coverslip at the bottom of the cannulae had a 0.15 mm X 0.35 mm rectangular opening, and a 0.02 mm silicone layer was applied (J-B Weld clear silicone glue) to avoid direct exposure of the brain to the exterior.
To deliver the DNA plasmid constructs (pCAGGS-GCaMP7b and pCAGGS-ChRmine-mScarlet were both used at 50 ng/l concentration in the solution) into individual CA1 PCs, the following solution was used (ingredients in mM concentration): 155 K-gluconate, 10 KCl, 10 HEPES, 4 KOH (pH = 7.3 at 25 Celsius degrees, 316 mOsm), as well as Alexa Fluor 488 (0.33 mM, Invitrogen) for pipette visibility and electroporation success confirmation in the brain (see Figure 1A). Long-taper pipettes were pulled from borosilicate glass (Warner Instruments, MA; model #: G200-3) with a Zeitz DMZ puller (Zeits Instrumente Vertriebs GmbH, Germany). The pipettes had an open tip resistance of 2.5-6 MΩ when filled with the DNA plasmid mix solution.
Before the electroporation procedure, the external surface of the glass coverslip was washed multiple times with 0.1 M PBS, and the pipette filled with DNA plasmid solution was fixed at a 32-degree angle to lower into the brain. Slight positive pressure (~ 20 mBar) was applied to make sure the pipette is not clogged, diffusion of the Alexa Fluor 488 under 2p excitation at 920 nm and a stable open tip resistance were indicative. When the pipette touched the 0.02 mm silicone layer above the brain, the open tip resistance increased to 1-2 GΩ, which decreased again once the silicone layer was punctured through. Usually, the open tip resistance in the brain was slightly higher than in the PBS solution (on average 1-2 MΩ). Diagonally approaching, the PC layer of the dorsal CA1 hippocampal region was reached at an average of 150 μm below the surface of the brain. When a CA1 PC was in close contact with the pipette tip, a further 1-2 MΩ increase is experienced in the open tip resistance, and a 1 second-long 100 Hz 0.5 ms −5V pulse train was applied through an A.M.P.I ISO-Flex stimulus isolator (Israel). Stimulation protocol was generated in Clampex (version 10.7; Molecular Devices, CA), and applied through a Digidata 1550B A/D converter (Molecular Devices, CA) via a Dagan BVC-700A amplifier (Dagan Corporation, MN). Electroporation success was visually confirmed by the Alexa Fluor 488 hydrazide filling of the CA1 PCs, after that, the pipette was retracted. During one electroporation session, 3-5 neurons were electroporated per mouse. After a session, a 5-minute incubation period was allowed to let the cells recover, followed by visual inspection to reconfirm electroporation success (dying cells lose their fluorescence, most commonly by accidentally pulling out their nucleus when the pipette is retracted). 1-2 days after electroporation plasmid expression was checked, usually finding 1-2 expressing cells per brain, resulting in on average 30-40% electroporation success rate.
Viruses
We injected recombinant adeno-associated viruses (rAAVs) expressing constructs of interest (obtained from B. Zemelman or K. Deisseroth, unless indicated otherwise) unilaterally into dorsal CA1. For all viral labeling strategies, we used Cre-dependent ChRmine (Marshel et al., 2019) (AAV8.Ef1α.DIO.ChRmine.oScarlet.WPRE or AAV2/1.Ef1α.DIO.ChRmine.oScarlet.WPRE) for expression in CA1 pyramidal cells (see below). For interneuron imaging experiments, a Cre-ON/Flp-OFF ChRmine (AAV8.nEF.Con/Foff.2.0.ChRmine-oScarlet) (Fenno et al., 2020) was used in VGAT-Flp mice to specifically prevent direct stimulation of interneurons. For sparse excitatory opsin labeling experiments, we additionally used GCaMP6f (AAV1.Syn.GCaMP6f.WPRE.SV40), source Addgene), Cre-dependent bReaChES (Rajasethupathy et al., 2015) (AAV2/9.Ef1α.FLEX.Breaches.tdTomato) or ChRmine, and tamoxifen-inducible Cre under the excitatory CaMKIIα promoter (AAV2/1.CaMKII.Cre.ERT2) to limit and control opsin labeling and expression, in C57Bl/6 mice. For dense excitatory opsin labeling experiments, we used Cre-dependent GCaMP6f (AAV1.Syn.FLEX.GCaMP6f, source Addgene) along with Cre-dependent ChRmine and h56D-GiDREADD in CaMKIIα-Cre mice. For experiments involving suppression of inhibition, we used GiDREADD (AAV2/1.h56D.HAhMD4(Gi).mCherry) or FlpO (AAV2/1.h56D.FlpO) with Flp-dependent tdTomato (AAV2/1.Syn.FRT.tdTomato) or Flp-ON/Cre-OFF-Arch (AAV8.nEF.Coff/Fon.Arch3.3-eYFP) (Fenno et al., 2020), utilizing the h56D GABAergic cell promoter for targeting expression to interneurons (Mehta et al., 2019). For all experiments, we co-injected virus mixtures containing equal amounts of 3-5 of the individual rAAV constructs above (according to experiment type). For tamoxifen-dependent expression, 125μL of 5mg/mL tamoxifen (either ≥99% tamoxifen or ≥98% 4-hydroxytamoxifen; Sigma-Aldrich) was administered intra-peritoneally at least 48 hours before experiments.
Behavior
Head-fixed mice ran on 1.94 meter belts on a treadmill equipped with a water delivery port, lick sensor, and custom made electronics for recording licking behavior and spatial position on the belt, as previously described (Turi et al., 2019). Mice were first water restricted (~ 85-90% pre-deprivation weight) and trained to run on a cue-deficient burlap (single fabric) treadmill belt for a non-operantly delivered water reward over the course of 1–2 weeks. We applied a progressively restrictive water reward schedule, with mice initially receiving 12-15 randomly placed reward zones per lap and ultimately receiving 1 randomly placed reward zone per lap. Mice were habituated and trained for 20-80 min daily until they regularly ran at least one lap per minute.
For experimental imaging sessions, mice ran on cue-enriched multi-fabric belts (Turi et al., 2019) to forage for 1-2 randomly administered water rewards per lap. Water was non-operantly delivered once mice entered the reward zone on each lap and further delivered operantly for every subsequent lick inside the reward zone for a maximum of 2.5 sec. The reward zone position was changed randomly each lap to ensure there would be no contingency of reward probability on spatial position. Mice ran for 15 min “baseline” sessions, followed by a 20 min “induction” session after a > 15 min break. Mice then ran for 15 min follow up sessions at 24 hour intervals for up to 72 hours post-induction. In “induction” sessions, a “stimulation zone” was randomly assigned to a location on the cued belt. After mice ran 1 lap in the session, the stimulation zone was activated for 5 consecutive laps (location of induction zone held constant across laps). Immediately upon entering the stimulation zone during these laps, mice received optogenetic photostimulation for ~ 1 sec (see below). In SCE experiments the baseline and induction sessions were merged into a single 30 min session with 10 baseline laps and photostimulation occurring on laps 11-15. Upon completing recordings on a given belt, different target fields of view or cells for photostimulation were selected and mice repeated the cycle on a new cued belt.
For our goal oriented learning task, head-fixed mice learned the location of a 10 cm hidden reward zone (RZ) at a fixed spatial location on a cued belt (Turi et al., 2019). The RZ was set to be at least 10 cm away from the closest tactile cue, so that animals did not associate the cue with the reward. All spatial information was presented to the mice via the treadmill belt. For each experiment, mice ran up to 2 sessions per day for 4 consecutive days on a cued belt, with the RZ changing every 24 hours (after the first session of each day). Mice thus had to learn the new RZ during the second session of each day. On the first session of Day 2, an optogenetic stimulation protocol was administered through chronically-implanted optical fibers to trigger opto-PFi as described below. For all experiments across different behavioral paradigms, treadmill belts were uniformly cleaned with 70% ethanol between sessions to eliminate mouse excretions and to control for any localized odors.
2-photon microscopy
Mice were habituated to the imaging apparatus (e.g. microscope/objective, laser, sounds of resonant scanner and shutters) during the training period. All imaging was conducted using a 2-photon 8 kHz resonant scanning microscope (Bruker). For SCE and 2p-stimulation experiments, a 40x NIR water immersion objective (Nikon, 0.8 NA, 3.5mm working distance) was used. For all other LED-stimulation and control experiments, a 16x NIR water immersion objective (Nikon, 0.8 NA, 3.0mm WD) was used. To align the CA1 pyramidal layer with the horizontal two-photon imaging plane, we adjusted the angle of the mouse’s head using two goniometers (±10° range, Edmund Optics). For excitation, we used a 920 nm laser (50-100 mW at objective back aperture, Coherent) with power regulated by an electro-optic modulator (Pockels cell, Conoptics). Green and red fluorescence were separated with an emission filter cube set (green, HQ525/70m-2p; red, HQ607/45m-2p; 575dcxr, Chroma Technology) and detected with GaAsP (Hamamatsu, 7422P-40) and Multi-Alkali (Hamamatsu R3896; used for red signals in some experiments) photomultiplier tubes. A custom dual stage preamp (1.4 x 105 dB, Bruker) was used to amplify signals prior to digitization. All experiments were performed at 1-2x digital zoom, covering ~ 166-330 μm x 166-330 μm per imaging FOV (40x objective) and ~ 550-880 μm x 550-880 μm (16x objective). All data was collected at a resolution of 512 x 512 pixels at 30 Hz.
Optogenetic LED Stimulation
Wide-field optical stimulation (Figs 1,2,S1) to activate excitatory opsin (ChRmine or bReaChES) was performed as previously described (Turi et al., 2019) using an ultrafast and high-power collimated red LED (UHP-T-620-SR, Prizmatix) with an excitation cleanup filter (629/56 nm, Semrock) attached to the microscope camera port. Filtered LED output was combined with the imaging laser beam using a 775 nm shortpass multiphoton laser dichroic mirror (ZT775sp-2p, Chroma Technology) after the microscope scan lens. LED and laser light was then passed through a 561 nm long-pass primary dichroic mirror (Di02-R561-25x36, Semrock) to the objective back aperture while green fluorescence signals collected through the objective were reflected to the non-descanned detector pathway (see above). LED stimulation light pulses were generated with custom-made electronics gated by the blanking signal of the imaging beam Pockels cell, which activated the LED during Y-galvonometer flyback between every imaging frame. Photostimulation between imaging frames was important to avoid artifacts from LED light while collecting imaging data. This strategy effectively allowed for pulsed LED stimulation at a rate close to the imaging frame rate (30 Hz), at which ChRmine-expressing neurons maintain spike fidelity (Marshel et al., 2019). The average power of the pulsed LED was ~ 30 mW measured after the objective (irradiance ~ 54 mW/mm2 before cover glass). During induction experiments, pulsed LED stimulation for 1 sec was triggered upon entry of the mouse into the “induction zone” location on each lap of 5 consecutive laps as above. For random stimulation experiments, pulsed LED stimulation for 1 sec was triggered at a random time every 20-24s. As wide-field stimulation was applied to all cells in each FOV (experiments of Figs. 1,2), individual cells were not pre-selected for photostimulation but were excluded from analyses if they expressed PFs prior to stimulation that overlapped the SZ (X cm)
2-photon Targeted Optogenetic Stimulation
All 2p-targeted stimulation (Fig 3) and imaging experiments were conducted using a dual scanhead microscope (Ultima, Bruker) with resonant scanner-galvonometer pair in the imaging pathway (see above) and galvonometer-galvonometer pair in the photostimulation pathway. A 1040 nm femtosecond (140 fs pulses, 80 MHz repetition rate) fiber laser (Fidelity HP, Coherent) beam was routed to the photostimulation pathway of the dual scanhead system, independently of the imaging beam, and imaged onto close-coupled galvanometer mirrors (3 mm diameter) located at a conjugate plane to the microscope’s objective back aperture. The photostimulation beam power was modulated independently by a separate Pockels cell (Conoptics). Photostimulation and imaging beams were then combined after their respective scan mirrors with a 1030 nm short pass dichroic mirror (T1030SP, Chroma Technology) just prior to the microscope scan lens. The beams were then expanded through the microscope tube lens to overfill the 40x objective back aperture. Beam positions were co-aligned by imaging fluorescent targets and photostimulation galvonometer positions calibrated relative to the imaging scan mirrors. The angle of the mouse’s head was adjusted using goniometers (see above) to ensure that the glass coverslip was perpendicular to the laser beams to minimize distortion of the point-spread function.
Individual cells for photostimulation targeting in Fig 3 were pre-selected based on visual confirmation of both opsin-reporter expression and functional response to brief photostimuli on a separate uncued training belt composed of different fabric material than that used in experimental belts. Cells were not targeted based on absence/presence of PFs; experimenters had no prior knowledge of whether or not pre-selected cells would exhibit PFs on any given belt context (including the uncued training belt). However, cells that were found post-hoc to exhibit PFs overlapping the SZ during PRE were excluded from analyses just as in widefield LED experiments. Photostimulation of visually targeted CA1 cells or dense ensembles was triggered upon mice entering the induction zone on each lap for 5 consecutive induction laps (see above). Upon triggering, the photostimulation laser beam was unshuttered by the Pockels cell and spirally scanned (Rickgauer & Tank, 2009; Yang et al., 2018; Chettih & Harvey, 2019) for ~ 1 sec over a circular area slightly larger than a pyramidal cell body to account for brain motion in vivo (15-20 μm outer spiral diameter, with each stimulation consisting of 15 rotations of progressively increasing radius and the entire spiral repeatedly scanned 31 times). For inductions, photostimulation beam power was typically 80 mW (after objective) while imaging at depths >250 μm below the glass cover slip, but was adjusted according to imaging depth and stimulation efficacy (range ~ 60-120 mW). This stimulation protocol was the same for both single-cell targeted (Fig 3A-E) and ensemble co-activation (Fig 3F-I) experiments, with the only difference being the density of excitatory opsin labeling achieved using the viral approaches above. For 2-photon photostimulation experiments where we attempted to co-activate an ensemble of CA1PCs with dense labeling of excitatory opsin, we confirmed offline that multiple cells were co-activated in the imaging plane (see below). In 1/21 of these experiments, we could not confirm activation of > 1 cell in the imaging plane (only a single cell activated), and so this experiment was excluded from further analysis.
Chemogenetic Suppression of Inhibition
GiDREADD (hM4D(Gi)) was virally delivered to CA1 interneurons as described above. DREADD-expressing mice received an intraperitoneal injection of saline alone (control conditions) or 10 mg/kg CNO (in saline; Cayman Chemical) ~ 20-30 min prior to recording of the “baseline” PRE session. This allowed us to suppress inhibition for this session and during the subsequent “induction” (POST) session, with reversal of direct GiDREADD-mediated effects by the 24 hr follow-up timepoint as CNO is cleared (Meira et al., 2018).
Bilateral Optogenetic Manipulations During Goal Oriented Learning
Viruses containing a mixture of Tamoxifen-inducible Cre, Cre-dependent ChRmine, h56D-FlpO, and Flp-ON/Cre-OFF-Arch or Flp-dependent tdTomato (in ChRmine-only control experiments) were delivered bilaterally throughout the extent of dorsal CA1 (64 nL virus mixture at each location: ±2.1/±1.4, ±2.3/±1.5, and ±2.6/±1.5 at AP/ML coordinates relative to bregma with −0.9, −1.05 and −1.2 mm DV relative to brain surface) in wild-type mice, similar to that described above. Mice were then chronically implanted with bilateral optical fiber cannulae (200 μm core, 0.37 NA multimode fibers) above the CA1 injection sites immediately after virus delivery and stainless-steel headposts affixed to their skulls, as we previously described (Ahmed et al., 2020). A splitter patch cable (Thorlabs) was used to couple bilaterally implanted optical fibers to a 532 nm laser (50 mW, OptoEngine) for Arch and/or ChRmine activation while mice were head-fixed. All cables/connections were shielded to prevent light leak from laser stimuli. During the goal oriented learning paradigm, on session 2 of each condition, mice received 1 sec of 30 Hz bilateral 532 nm laser stimulation (20 mW, to otherwise match LED photostimulation irradiance parameters used above) at a randomly chosen stimulation zone (SZ) on laps 6-10. This SZ was at least 400 cm away from the RZ during that session. The optogenetic SZ was then used as the RZ for the subsequent session.
Quantification and Statistical Analysis
Image preprocessing
Imaging data were processed using the Suite2p (Pachitariu et al., 2016) and SIMA (Kaifosh et al., 2014) software packages. Motion correction was performed by concatenating all imaging sessions for a given field of view and using the Suite2p rigid motion correction strategy. Regions of interest (ROIs) and corresponding signals, corrected for neuropil contamination, were subsequently generated by Suite2p and visually inspected and manually curated to exclude non-somatic components using the graphical interface.
Neural data analysis
Signal processing and event detection
Deconvolved spikes were computed for each ROI for each imaging session according to the following procedure: the raw fluorescence trace was linearly detrended and negative outliers due to Suite2p neuropil subtraction artifacts were removed by replacing values 3 median absolute deviations (MADs) below the median with the median value. A baseline was calculated by smoothing the fluorescence trace with a Gaussian filter (std = 5 frames, window = 30s), calculating the rolling minimum of this smoothed trace, (window = 30s), and then calculating the rolling maximum of this last trace (window = 30). The resulting baseline-subtracted trace was used for stimulated cell detection, stimulation response calculation, and for display of example traces. A preliminary spike train was estimated by deconvolving the baseline-subtracted trace with OASIS (Friedrich et al., 2017) according to an AR1 model with ℓ1 penalty and pre-computed decay parameter (using 400 ms for the GCaMP6f decay time, 2s for GCaMP6s). Using this spike train, a noise threshold was identified by computing the median and MAD of the baseline-subtracted trace where no spikes were detected (i.e., putative noise-only time points). The noise threshold was defined as the median plus 3 MADs. A new spike train was then computed as before, now with the minimum spike size set explicitly to the noise threshold, and sparsity parameter, λ, set to 7.
Spatial Tuning Analysis
When evaluating the spatial tuning of PCs, we restricted our analysis to running-related epochs, defined as times with a minimum speed of 1 cm/sec. Position was discretized into 100 1.94cm bins. Deconvolved spikes were smoothed with a Gaussian filter (std = 1 frame) and binarized. The spatial tuning vector was calculated as , where si is the number of running frames with spikes occurring at position i and oi is the number of running frames acquired at position i (i.e. the occupancy). In order to assess the significance of the spatial selectivity, for each cell we generated a null tuning distribution by cyclically permuting the position vector (restricted to running frames) by a random offset and repeatedly recomputing the tuning vector. This process was repeated 1,000 times. The true and null tuning vectors were then smoothed with a Gaussian (std = 3 position bins).
Place fields were identified as at least 5 consecutive position bins above the 95th percentile of the null distribution, in which the cell fired on at least 15% of laps. The edges of each place field were then extended until they reached a value of at least half the normalized place field peak, and overlapping place fields merged. Reported widths of de novo place fields were calculated prior to this final extension/merging step to avoid overestimation due to merges with pre-existing place fields.
Stimulated Cell Detection
To avoid any potential for contamination from stimulation artifacts, cells were defined as stimulated based on their fluorescence during a 1s window prior to stimulation onsets (”pre-stim”) and a 0.5s window immediately after stimulation offsets (”post-stim”). Three criteria were used to define cells as stimulated: significance, reliability, and magnitude. (1) Significance: the mean peri-stimulation fluorescence was taken across all five stimulations to generate a peri-stimulation time histogram (PSTH), and the mean of the pre-stim period was subtracted from the mean of the post-stim period to generate a true value for the mean peri-stimulation change. A null distribution for this change was then generated by randomly drawing five frames (with the requirement that the frames have PSTHs that not overlap eachother or those of the true stimulations, and that they not extend beyond the bounds of the imaging session), and the same procedure of generating a null PSTH and calculating the post-”stim” minus pre-”stim” change was carried out. 1000 such null values were generated, and only cells whose true peri-stimulation change was at least in the 95th percentile of this distribution were considered. (2) Reliability: the baselined fluorescence trace was z-scored to frames with no detected events (see signal processing and event detection above), and only cells whose mean post-stim fluorescence was at least 1 standard deviation greater than the mean pre-stim fluorescence for at least three of the five stimulations were considered. This criterion was used to ensure that the cell was consistently responsive to stimulation. (3) Magnitude: Finally, we required that responses be meaningfully large by requiring that the mean of the post-stim period of the PSTH taken over all five stimulations was greater than 1.5 standard deviations above zero (i.e. baseline). In the targeted stimulation experiments, a fourth spatial distance criterion was applied by which only cells within 75 um of the stimulation radius were included, a conservative threshold based on the spatial extent of stimulation responses in the dense, inhibition-relieved condition (Fig S4). Cells that passed all three/four of these criteria were initially considered “stimulated”. Finally, to further avoid the possibility of cells being erroneously detected as stimulated because of pre-existing place field activity around the stimulation zone, any stimulated cell with a place field overlapping the SZ during baseline was excluded from analysis. For fair comparisons to activity changes in unstimulated cells, this baseline place field exclusion was also applied to unstimulated cells.
Remapping Analysis
The template-based remapping analysis was conducted as described in (McKenzie et al., 2021). Briefly, to get a remapping score for each cell, two templates were calculated: the mean spatial tuning vector during the last ten laps prior to stimulation (i.e. last ten laps of PRE), and the mean spatial tuning vector during the first ten laps after stimulation (i.e. laps 6-15 of POST). For each lap in those periods, we took the Pearson’s correlation between that lap’s smoothed (Gaussian, σ = 3) spatial tuning vector and each of the two smoothed templates, making sure to exclude that lap from the template calculation (e.g., lap 6 of POST is correlated to the full pre-stimulation template, but a post-stimulation template calculated only from laps 7-15). The remapping score was taken to be the difference between the mean pre-stimulation and post-stimulation template correlations for post-stimulation laps. As in (McKenzie et al., 2021), only cells with trial-to-trial spatial reliability (mean r > 0.25 with the pre-stimulation template for pre-stimulation laps and with the post-stimulation template for post-stimulation laps) were included.
Offline Correlation Analysis
To measure pairwise reactivation in Figure S3F, the distance between place field peaks during POST for pairs of place cells was compared to the Pearson’s correlation of those cells’ baselined fluorescence, restricted to periods of immobility during POST. The reported means across pairs within a mouse included all place cell pairs regardless of place field peak distance.
Statistics
Statistical details of experiments can be found in the figure legends. Statistical details of analysis methods are described in the corresponding sections above. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications.
Supplementary Material
Highlights.
Rapidly forming place fields can be optogenetically induced in mouse CA1 neurons
Feedback inhibition limits rapid place field induction to fewer principal neurons
Suppressing local inhibition allows induction of place fields in larger ensembles
Ensemble place field induction can enhance subsequent spatial association learning
Acknowledgments
We thank Coherent, Inc. (Santa Clara, CA) for providing a 18W Fidelity HP laser for testing and piloting of in vivo 2-photon targeted photostimulation experiments. We thank C. Ramakrishnan and K. Deisseroth for viral reagents used in this study, B. Vancura for assistance with histology, T. Mazidzoglou for technical assistance, and J.B. Priestley for critical comments on the manuscript. Some parts of behavior schemata created with BioRender.com. This work was supported by NIMH and NINDS grants (F31MH117892 to S.V.R.; K08MH113036 to M.S.A.; K99NS115984 to H.B.; U19NS104649 to D.S.P.; NS094330 to B.V.Z.; R21NS109753 and RO1NS067557 to F.P.; R01MH124047, R01MH124867, R01NS1121106, U01NS115530, and U19NS104590 to A.L.), the Leon Levy Foundation (to M.S.A.), the American Epilepsy Society (to F.T.S.), the McKnight Endowment Fund for Neuroscience (B.V.Z.), the Fondation Roger De Spoelberch (F.P.), the Nomis Foundation (F.P.), and the Kavli Foundation (to A.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
Inclusion and Diversity
We worked to ensure sex balance in the selection of non-human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
References
- Ahmed MS, Priestley JB, Castro A, Stefanini F, Solis Canales AS, Balough EM, Lavoie E, Mazzucato L, Fusi S & Losonczy A (2020), ‘Hippocampal network reorganization underlies the formation of a temporal association memory’, Neuron 107(2), 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriaga M & Han EB (2017), ‘Dedicated hippocampal inhibitory networks for locomotion and immobility’, J Neurosci 37(38), 9222–9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezaire MJ & Soltesz I (2013), ‘Quantitative assessment of ca1 local circuits: knowledge base for interneuron-pyramidal cell connectivity’, Hippocampus 23(9), 751–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner KC, Grienberger C, Vaidya SP, Milstein AD, Macklin JJ, Suh J, Tonegawa S & Magee JC (2015), ‘Conjunctive input processing drives feature selectivity in hippocampal CA1 neurons’, Nature Neuroscience 18(8), 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner KC, Milstein AD, Grienberger C, Romani S & Magee JC (2017), ‘Behavioral time scale synaptic plasticity underlies CA1 place fields’, Science 1036(September), 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolding KA & Franks KM (2018), ‘Recurrent cortical circuits implement concentration-invariant odor coding’, Science 361(6407). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourboulou R, Marti G, Michon F-X, El Feghaly E, Nouguier M, Robbe D, Koenig J & Epsztein J (2019), ‘Dynamic control of hippocampal spatial coding resolution by local visual cues’, Elife 8, e44487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandalise F, Kalmbach BE, Mehta P, Thornton O, Johnston D, Zemelman BV & Brager DH (2020), ‘Fragile x mental retardation protein bidirectionally controls dendritic ih in a cell type-specific manner between mouse hippocampus and prefrontal cortex’, J Neurosci 40(27), 5327–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Gore F, Nguyen QA, Ramakrishnan C, Patel S, Kim SH, Raffiee M, Kim YS, Hsueh B, Krook-Magnusson E, Soltesz I & Deisseroth K (2021), ‘Deep brain optogenetics without intracranial surgery’, Nat Biotechnol 39(2), 161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chettih SN & Harvey CD (2019), ‘Single-neuron perturbations reveal feature-specific competition in v1’, Nature 567(7748), 334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD, Bolstad M & Lee AK (2017), ‘Experience-dependent shaping of hippocampal CA1 intracellular activity in novel and familiar environments’, eLife pp. 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle TL, Madisen L, Hage TA, Valley MT, Knoblich U, Larsen RS, Takeno MM, Huang L, Gu H, Larsen R et al. (2018), ‘A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality’, Cell 174(2), 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgleish HW, Russell LE, Packer AM, Roth A, Gauld OM, Greenstreet F, Thompson EJ & Hausser M (2020), ‘How many neurons are sufficient for perception of cortical activity?’, Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson NB, Zaremba JD, Kaifosh P, Bowler J, Ladow M & Losonczy A (2016), ‘Sublayer-specific coding dynamics during spatial navigation and learning in hippocampal area ca1’, Neuron 91(3), 652–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneve S & Machens CK (2016), ‘Efficient codes and balanced networks’, Nat Neurosci 19(3), 375–82. [DOI] [PubMed] [Google Scholar]
- Deng ZD, Luber B, Balderston NL, Velez Afanador M, Noh MM, Thomas J, Altekruse WC, Exley SL, Awasthi S & Lisanby SH (2020), ‘Device-based modulation of neurocircuits as a therapeutic for psychiatric disorders’, Annu Rev Pharmacol Toxicol 60, 591–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamantaki M, Coletta S, Nasr K, Zeraati R, Laturnus S, Berens P, Preston-Ferrer P & Burgalossi A (2018), ‘Manipulating hippocampal place cell activity by single-cell stimulation in freely moving mice’, Cell Rep 23(1), 32–38. [DOI] [PubMed] [Google Scholar]
- Dragatsis I & Zeitlin S (2000), ‘CaMKII-cre transgene expression and recombination patterns in the mouse brain’, Genesis 26(2), 133–135. [DOI] [PubMed] [Google Scholar]
- Dupret D, O’Neill J, Pleydell-Bouverie B & Csicsvari J (2010), ‘The reorganization and reactivation of hippocampal maps predict spatial memory performance’, Nat Neurosci 13(8), 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenbaum H (2000), ‘A cortical–hippocampal system for declarative memory’, Nature Reviews Neuroscience 1(1), 41–50. [DOI] [PubMed] [Google Scholar]
- Ekstrom AD, Kahana MJ, Caplan JB, Fields TA, Isham EA, Newman EL & Fried I (2003), ‘Cellular networks underlying human spatial navigation’, Nature 425(6954), 184–8. [DOI] [PubMed] [Google Scholar]
- Farrell JS, Colangeli R, Dudok B, Wolff MD, Nguyen SL, Jackson J, Dickson CT, Soltesz I & Teskey GC (2020), ‘In vivo assessment of mechanisms underlying the neurovascular basis of postictal amnesia’, Sci Rep 10(1), 14992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno LE, Ramakrishnan C, Kim YS, Evans KE, Lo M, Vesuna S, Inoue M, Cheung KYM, Yuen E, Pichamoorthy N, Hong ASO & Deisseroth K (2020), ‘Comprehensive dual- and triple-feature intersectional single-vector delivery of diverse functional payloads to cells of behaving mammals’, Neuron 107(5), 836–853 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank LM, Stanley GB & Brown EN (2004), ‘Hippocampal plasticity across multiple days of exposure to novel environments’, Journal of Neuroscience 24(35), 7681–7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich J, Zhou P & Paninski L (2017), ‘Fast online deconvolution of calcium imaging data’, PLOS Computational Biology 13(3), e1005423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier JL & Tank DW (2018), ‘A Dedicated Population for Reward Coding in the Hippocampus’, Neuron 99(1), 179–193.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiller T, Vancura B, Terada S, Troullinou E, Chavlis S, Tsagkatakis G, Tsakalides P, Ocsai K, Poirazi P, Rozsa BJ & Losonczy A (2020), ‘Large-scale 3d two-photon imaging of molecularly identified ca1 interneuron dynamics in behaving mice’, Neuron 108(5), 968–983 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosmark AD, Sparks FT, Davis MJ & Losonczy A (2021), ‘Reactivation predicts the consolidation of unbiased long-term cognitive maps’, Nature Neuroscience . [DOI] [PubMed] [Google Scholar]
- Hirase H, Leinekugel X, Czurko A, Csicsvari J & Buzsaki G (2001), ‘Firing rates of hippocampal neurons are preserved during subsequent sleep episodes and modified by novel awake experience’, Proc Natl Acad Sci U S A 98(16), 9386–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinno S & Kosaka T (2010), ‘Stereological estimation of numerical densities of glutamatergic principal neurons in the mouse hippocampus’, Hippocampus 20(7), 829–40. [DOI] [PubMed] [Google Scholar]
- Jun H, Bramian A, Soma S, Saito T, Saido TC & Igarashi KM (2020), ‘Disrupted place cell remapping and impaired grid cells in a knockin model of alzheimer’s disease’, Neuron 107(6), 1095–1112 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaifosh P, Lovett-Barron M, Turi GF, Reardon TR & Losonczy A (2013), ‘Septohippocampal gabaergic signaling across multiple modalities in awake mice’, Nature Neurosci 16(9), 1182–1184. [DOI] [PubMed] [Google Scholar]
- Kaifosh P, Zaremba JD, Danielson NB & Losonczy A (2014), ‘SIMA: Python software for analysis of dynamic fluorescence imaging data’, Frontiers in Neuroinformatics 8(September), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman AM, Geiller T & Losonczy A (2020), ‘A role for the locus coeruleus in hippocampal ca1 place cell reorganization during spatial reward learning’, Neuron 105(6), 1018–1026 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Judkewitz B, Kano M, Denk W & Häusser M (2008), ‘Targeted patch-clamp recordings and single-cell electroporation of unlabeled neurons in vivo’, Nature methods 5(1), 61–68. [DOI] [PubMed] [Google Scholar]
- Klausberger T (2009), ‘Gabaergic interneurons targeting dendrites of pyramidal cells in the ca1 area of the hippocampus’, European Journal of Neuroscience 30(6), 947–957. [DOI] [PubMed] [Google Scholar]
- Klausberger T & Somogyi P (2008), ‘Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations’, Science 321(5885), 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krabbe S, Paradiso E, d’Aquin S, Bitterman Y, Courtin J, Xu C, Yonehara K, Markovic M, Muller C, Eichlisberger T, Grundemann J, Ferraguti F & Luthi A (2019), ‘Adaptive disinhibitory gating by vip interneurons permits associative learning’, Nat Neurosci 22(11), 1834–1843. [DOI] [PubMed] [Google Scholar]
- Kunz L, Schroder TN, Lee H, Montag C, Lachmann B, Sariyska R, Reuter M, Stirnberg R, Stocker T, Messing-Floeter PC, Fell J, Doeller CF & Axmacher N (2015), ‘Reduced grid-cell-like representations in adults at genetic risk for alzheimer’s disease’, Science 350(6259), 430–3. [DOI] [PubMed] [Google Scholar]
- Lee I, Griffin AL, Zilli EA, Eichenbaum H & Hasselmo ME (2006), ‘Gradual translocation of spatial correlates of neuronal firing in the hippocampus toward prospective reward locations’, Neuron 51(5), 639–50. [DOI] [PubMed] [Google Scholar]
- Lee SH, Marchionni I, Bezaire M, Varga C, Danielson N, Lovett-Barron M, Losonczy A & Soltesz I (2014), ‘Parvalbumin-positive basket cells differentiate among hippocampal pyramidal cells’, Neuron 82(5), 1129–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester AW, Moffat SD, Wiener JM, Barnes CA & Wolbers T (2017), ‘The aging navigational system’, Neuron 95(5), 1019–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzkus JJ, Wolff SB & Luthi A (2015), ‘Disinhibition, a circuit mechanism for associative learning and memory’, Neuron 88(2), 264–76. [DOI] [PubMed] [Google Scholar]
- Lovett-Barron M, Kaifosh P, Kheirbek MA, Danielson N, Zaremba JD, Reardon TR, Turi GF, Hen R, Zemelman BV & Losonczy A (2014), ‘Dendritic inhibition in the hippocampus supports fear learning’, Science 343(6173), 857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett-Barron M, Turi GF, Kaifosh P, Lee PH, Bolze F, Sun XH, Nicoud JF, Zemelman BV, Sternson SM & Losonczy A (2012), ‘Regulation of neuronal input transformations by tunable dendritic inhibition’, Nat Neurosci 15(3), 423–30, S1–3. [DOI] [PubMed] [Google Scholar]
- Magee JC & Grienberger C (2020), ‘Synaptic Plasticity Forms and Functions’, Annual Review of Neuroscience 10(7), 95–117. [DOI] [PubMed] [Google Scholar]
- Malik R, Li Y, Schamiloglu S & Sohal VS (2021), ‘Top-down control of hippocampal signal-to-noise by prefrontal long-range inhibition’, bioRxiv . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory CS & Giocomo LM (2018), ‘Heterogeneity in hippocampal place coding’, Current opinion in neurobiology 49, 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel JH, Kim YS, Machado TA, Quirin S, Benson B, Kadmon J, Raja C, Chibukhchyan A, Ramakrishnan C, Inoue M, Shane JC, McKnight DJ, Yoshizawa S, Kato HE, Ganguli S & Deisseroth K (2019), ‘Cortical layer-specific critical dynamics triggering perception’, Science 365(6453). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie S (2018), ‘Inhibition shapes the organization of hippocampal representations’, Hippocampus 28(9), 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie S, Huszar R, English DF, Kim K, Christensen F, Yoon E & Buzsaki G (2021), ‘Preexisting hippocampal network dynamics constrain optogenetically induced place fields’, Neuron 109(6), 1040–1054 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, Kreeger L, Wylie DC, Pattadkal JJ, Lusignan T, Davis MJ, Turi GF, Li WK, Whitmire MP, Chen Y, Kajs BL, Seidemann E, Priebe NJ, Losonczy A & Zemelman BV (2019), ‘Functional access to neuron subclasses in rodent and primate forebrain’, Cell Rep 26(10), 2818–2832 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meira T, Leroy F, Buss EW, Oliva A, Park J & Siegelbaum SA (2018), ‘A hippocampal circuit linking dorsal ca2 to ventral ca1 critical for social memory dynamics’, Nat Commun 9(1), 4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein Aaron D., et al. "Bidirectional synaptic plasticity rapidly modifies hippocampal representations". Elife 10 (2021): e73046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C & Remy S (2014), ‘Dendritic inhibition mediated by o-lm and bistratified interneurons in the hippocampus’, Frontiers in synaptic neuroscience 6, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima R, Laskaris N, Rhee JK, Baker BJ & Kosmidis EK (2021), ‘Gevi cell-type specific labelling and a manifold learning approach provide evidence for lateral inhibition at the population level in the mouse hippocampal ca1 area’, Eur J Neurosci 53(9), 3019–3038. [DOI] [PubMed] [Google Scholar]
- O’Keefe J & Dostrovsky J (1971), ‘The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat’, Brain Research 34, 171–175. [DOI] [PubMed] [Google Scholar]
- Pachitariu M, Stringer C, Schröder S, Dipoppa M, Rossi LF, Carandini M & Harris KD (2016), ‘Suite2p : beyond 10,000 neurons with standard two-photon microscopy’, Biorxiv pp. 1–14. [Google Scholar]
- Pedrosa V & Clopath C (2020), ‘The interplay between somatic and dendritic inhibition promotes the emergence and stabilization of place fields’, PLoS computational biology 16(7), e1007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkey KA, Chittajallu R, Craig MT, Tricoire L, Wester JC & McBain CJ (2017), ‘Hippocampal gabaergic inhibitory interneurons’, Physiological reviews 97(4), 1619–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouille F & Scanziani M (2004), ‘Routing of spike series by dynamic circuits in the hippocampus’, Nature 429(6993), 717–723. [DOI] [PubMed] [Google Scholar]
- Priestley JB, Bowler JC, Rolotti SV, Fusi S & Losonczy A (2021), ‘Signatures of rapid synaptic learning in the hippocampus during novel experiences’, bioRxiv p. 2021.07.02.450956. [Google Scholar]
- Rajasethupathy P, Sankaran S, Marshel JH, Kim CK, Ferenczi E, Lee SY, Berndt A, Ramakrishnan C, Jaffe A, Lo M, Liston C & Deisseroth K (2015), ‘Projections from neocortex mediate top-down control of memory retrieval’, Nature 526(7575), 653–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao-Ruiz P, Yu J, Kushner SA & Josselyn SA (2019), ‘Neuronal competition: microcircuit mechanisms define the sparsity of the engram’, Curr Opin Neurobiol 54, 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer JP, Deisseroth K & Tank DW (2014), ‘Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields’, Nat Neurosci 17(12), 1816–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer JP & Tank DW (2009), ‘Two-photon excitation of channelrhodopsin-2 at saturation’, Proc Natl Acad Sci U S A 106(35), 15025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NTM, Descamps LAL, Russell LE, Buchholz MO, Bicknell BA, Antonov GK, Lau JYN, Nutbrown R, Schmidt-Hieber C & Hausser M (2020), ‘Targeted activation of hippocampal place cells drives memory-guided spatial behavior’, Cell 183(7), 2041–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux L & Buzsaki G (2015), ‘Tasks for inhibitory interneurons in intact brain circuits’, Neuropharmacology 88, 10–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield ME, Adoff MD & Dombeck DA (2017), ‘Increased prevalence of calcium transients across the dendritic arbor during place field formation’, Neuron 96(2), 490–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield ME & Dombeck DA (2019), ‘Dendritic mechanisms of hippocampal place field formation’, Current opinion in neurobiology 54, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS & Rubenstein JLR (2019), ‘Excitation-inhibition balance as a framework for investigating mechanisms in neuropsychiatric disorders’, Mol Psychiatry 24(9), 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltesz I & Losonczy A (2018), ‘Ca1 pyramidal cell diversity enabling parallel information processing in the hippocampus’, Nat Neurosci 21(4), 484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche S, Perestenko PV, van de Ven GM, Bratley CT, McNamara CG, Campo-Urriza N, Black SL, Reijmers LG & Dupret D (2016), ‘Recoding a cocaine-place memory engram to a neutral engram in the hippocampus’, Nat Neurosci 19(4), 564–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turi GF, Li WK, Chavlis S, Pandi I, O’Hare J, Priestley JB, Grosmark AD, Liao Z, Ladow M, Zhang JF, Zemelman BV, Poirazi P & Losonczy A (2019), ‘Vasoactive intestinal polypeptide-expressing interneurons in the hippocampus support goal-oriented spatial learning’, Neuron 101(6), 1150–1165 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban-Ciecko J, Jouhanneau JS, Myal SE, Poulet JFA & Barth AL (2018), ‘Precisely timed nicotinic activation drives sst inhibition in neocortical circuits’, Neuron 97(3), 611–625 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JB, Di Ianni T, Vyas DB, Huang Z, Park S, Hosseini-Nassab N, Aryal M & Airan RD (2020), ‘Focused ultrasound for noninvasive, focal pharmacologic neurointervention’, Front Neurosci 14, 675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Carrillo-Reid L, Bando Y, Peterka DS & Yuste R (2018), ‘Simultaneous two-photon imaging and two-photon optogenetics of cortical circuits in three dimensions’, Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona G, Meitav N, Kahn I & Shoham S (2016), ‘Realistic numerical and analytical modeling of light scattering in brain tissue for optogenetic applications(1,2,3)’, eNeuro 3(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba JD, Diamantopoulou A, Danielson NB, Grosmark AD, Kaifosh PW, Bowler JC, Liao Z, Sparks FT, Gogos JA & Losonczy A (2017), ‘Impaired hippocampal place cell dynamics in a mouse model of the 22q11.2 deletion’, Nat Neurosci 20(11), 1612–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wang Y, Spruston N & Magee JC (2020), ‘Membrane potential dynamics underlying context-dependent sensory responses in the hippocampus’, Nature Neuroscience 23(7), 881–891. [DOI] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A & Schnitzer MJ (2013), ‘Long-term dynamics of ca1 hippocampal place codes’, Nat Neurosci 16(3), 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared upon reasonable request to the lead contact.
All original code has been deposited at Zenodo and is publicly available on GitHub as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available upon reasonable request to the lead contact.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| rAAV1.Syn.Flex.GCaMP6f.WPRE.SV40 | This paper | Addgene 100833-AAV1 |
| rAAV8.Ef1α.DIO.ChRmine.oScarlet.WPRE | Karl Deisseroth (Marshel et al., 2019) | N/A |
| rAAV2/1.Ef1α.DIO.ChRmine.oScarlet.WPRE | This paper | N/A |
| rAAV8.nEF.Con/Foff.2.0.ChRmine-oScarlet | Karl Deisseroth Fenno et al., 2020 | N/A |
| rAAV1.Syn.GCaMP6f.WPRE.SV40 | This paper | Addgene 100837-AAV1 |
| rAAV2/9.Ef1α.FLEX.Breaches.tdTomato | Kaufman et al., 2020 | N/A |
| rAAV2/1.CaMKII.Cre.ERT2 | This paper | N/A |
| rAAV2/1.h56d.HA-hMD4(Gi).mCherry | This paper | N/A |
| rAAV2/1.h56d.FlpO | This paper | N/A |
| rAAV8.nEF.Coff/Fon.Arch3.3-eYFP | Karl Deisseroth (Fenno et al., 2020) | N/A |
| rAAV2/1.Syn.FRT.tdTomato | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Slc32a1-2A-FlpO-D knock-in (VGAT-FlpO): B6.Cg-Slc32a1tm1.1(flpo)Hze/J | Daigle et al., 2018 | Jackson Laboratory Strain 029591 |
| Mouse: R4Ag11: B6.Cg-Tg(CamKIIα-Cre)3Szi/J | Bina Santoro (Dragatsis and Zeitlin, 2000) | Jackson Laboratory Strain 027400 |
| Mouse: R1Ag5: B6.Cg-Tg(CamKIIα-Cre)2Szi/J | Bina Santoro (Dragatsis and Zeitlin, 2000) | Jackson Laboratory Strain 027310 |
| Recombinant DNA | ||
| pCAGGS-GCaMP7b | This paper | N/A |
| pCAGGS-ChRmine-mScarlet | This paper | N/A |
| Software and Algorithms | ||
| Suite2p | Pachitariu et al., 2017 | https://github.com/cortex-lab/Suite2P |
| OASIS | Friedrich et al., 2017 | https://github.com/j-friedrich/OASIS |
| Custom data processing and analysis code | This paper | https://dx.doi.org/10.5281/zenodo.5765716 |
| Python 2.7 | https://www.python.org | N/A |
| SIMA | Kaifosh et al., 2014 | https://github.com/losonczylab/sima |