Abstract
BACKGROUND:
Fine needle aspiration (FNA) is used to diagnose malignancies, recurrences, and metastases. The procedure is quick, well-tolerated, and can be facilitated by ultrasound guidance.
METHODS:
Here we describe our experience using serial FNAs to harvest cellular material during four clinical trials of immunotherapy by in situ vaccination in patients with low grade lymphoma.
RESULTS:
Two hundred and ninety-six FNA samples were collected from 44 patients spanning approximately six weeks for each patient. Samples were sufficient in quantity and quality to be analyzed by flow cytometry and/or single-cell mRNA sequencing. FNA samples yielded an average of 12 x 106 cells with a mean cellular viability of 86%. Material collected from the tumor lymph nodes differed significantly in proportions and phenotype of cellular populations compared to matched peripheral blood samples. Comparison of flow cytometry results obtained by FNA directly from the patient to that of an FNA performed ex vivo and a dissociation of the same lymph node after surgical excision confirmed that FNA sampling of the patient accurately represents the tumor and the microenvironment. Analysis of the FNA samples from immunotherapy-treated target lymph nodes vs nodes from non-treated tumor sites provided insight into the impact of specific immunotherapy regimens.
CONCLUSIONS:
This is the largest study describing the use of serial FNA sampling to harvest cellular material during immunotherapy clinical trials. The success of this technique opens the door for FNA sampling to expand significantly future investigation of the dynamic effects of investigational agents, be they immune therapies or targeted therapies.
Keywords: Fine-needle aspiration, FNA, clinical trial, immunotherapy, lymphoma
Precis:
Fine needle aspiration (FNA) is a dependable method of collecting samples directly from multiple tumor nodules sequentially over the course of immunotherapy regimes. These cellular samples can then be studied and profiled to track patient responses to therapy, and to study both the tumor and its microenvironment.
Background
In clinical practice, fine needle aspiration (FNA) is routinely used to easily and reliably sample and diagnose tumors using a combination of morphology, flow cytometry, ultrasound guidance, and additional ancillary studies.1-4 Now FNA is beginning to expand into the clinical trial setting.5-9 FNA offers the ability to directly sample tumors and their immune microenvironments with a procedure that is minimally disruptive to the tissue, cost-effective, and well-tolerated by patients.7,8 Serial FNA samples of target lesions can be collected over time to harvest cellular material that allows cellular and molecular profiling of changes occurring over the course of therapy.7,8,10,11 In comparison, excisional and/or core biopsy procedures remove or significantly disrupt the targeted tissue allowing for a one-time only harvest and are also costlier, more painful and inconvenient for patients. This study describes our collective experience using serial FNAs in the clinical trial setting to harvest cellular material from tumor sites to evaluate cellular and molecular changes during the course of low-grade lymphoma immunotherapy trials.
Methods
Sample Collection
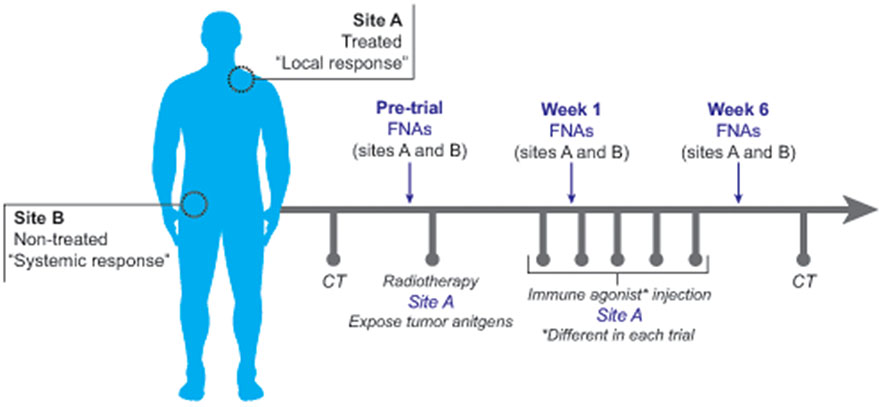
Over the course of four different Institutional Review Board-approved immunotherapy clinical trials of patients with low grade lymphomas, 296 serial FNAs were performed on 44 consented patients spanning a period of at least 6 weeks for each patient. Two nodal sites were designated at the beginning of therapy: (A) the treated site and (B) a site distal to the treatment site that could be used as an indicator of global immune effects of the local therapy (Figure 1). The two nodal sites were each sampled at three points in time: before, during and after completion of local therapy. FNAs were performed by experienced cytopathologists using 23-gauge needles with palpation, or an ultrasound directed approach.
Figure 1.
Overview of FNA methodology in clinical trials. Two FNA sites were identified in each patient including “Site A”, the treated site, and “Site B”, the non-treated site. Following pre-treatment CT scans, an initial pre-treatment FNA was performed at sites A and B. Low-dose radiation therapy followed by the first immunotherapy dose was then administered to Site A only. Eight days later, a second FNA was performed at sites A and B (Week 2). A third FNA was performed at sites A and B at week 6, subsequent to the final immunotherapy dose. CT scans were performed at intervals defined by each trial protocol. Each trial utilized different immune agonists or combinations of immune agonists.
Ultrasound guidance was used to sample small, or difficult to palpate lymph node targets.4,12-14 It proved important when collecting samples during these studies as: 1) the clinical teams wanted to biopsy smaller nodes that were detected on scans and were barely palpable or non-palpable, and 2) some of the target lymph nodes decreased in size and became non-palpable during the clinical trials. ROSE (rapid onsite evaluation) was not used in order to preserve as much cellular material as possible for analysis and cellular profiling. Instead of ROSE, ultrasound directed aspiration was optimized to identify needle placement within the lymph node cortex, and to avoid sampling the lymph node hilum, or piercing hilar vessels (which could result in blood dilution and reduced cellular sampling). With this ultrasound directed technique we replaced microscopic examination of the sample at the time of biopsy (ROSE) with strategic placement of the biopsy needle into the cortical aspects of the target nodes in an effort to maximize cellular sample yields.
Three FNA passes, targeting different regions of the tumor/ lymph node were performed during each biopsy sampling. Targeted nodes were predominantly located in inguinal, axillary, and neck regions. In some cases, peripheral blood samples were collected concurrently. In one case, an excisional biopsy was performed and an FNA was done ex vivo on the excised specimen using the same technique as in vivo to show that sampling by FNA is representative of the whole node. Most of the patients were diagnosed with follicular lymphoma, but the study also included patients with marginal zone lymphoma and chronic lymphocytic leukemia/small lymphocytic lymphoma.
Definition of sample adequacy
An “adequate” FNA sample was defined as one yielding at least 0.2 x 10^6 live cells which was enough to quantify and characterize the malignant tumor cells as well as the microenvironment by multi-color flow cytometry.
Sample processing and analysis
Red blood cells were lysed using High-Yield Lyse, Fixative-Free Lysing Solution (Life Technologies). Live cells were counted using trypan blue exclusion and single cell suspensions were stained and analyzed by flow cytometry. After gating live cells using an Aqua live/dead fixable stain (ThermoFisher Scientific), CD3 Alexa700, CD19 BV711, CD56 BV605, CD14 BV786 and CD33 FITC with CD11b PE-Cy7 antibodies were used to designate T cells, B cells, NK cells, macrophage/monocytes and granulocyte populations, respectively. The presence or absence of Lambda light chain was used to designate tumor vs. normal B cells, a strategy previously used by Frank, et al.7 Flow cytometry data acquisition and analysis were performed using an LSRII (Becton Dickinson Immunocytometry Systems) and web-based analysis software provided by Cytobank.org.
A split of the single-cell suspension used for flow was taken for 32 samples and subjected to single cell RNA sequencing, as previously described.8 Briefly, single cell suspensions were adjusted to target 10,000 cells per sample and processed using Chromium Single Cell 5' Library & Gel Bead Kit (10x Genomics) according to the manufacturer’s protocol. Single cell libraries were sequenced on Illumina Novaseq or Nextseq platforms, and processed using Cell Ranger (10X Genomics) with alignment to GRCh38. Basic pre-processing and clustering of the scRNA-seq data were performed using Seurat running on R framework.9 Categorical variables were compared using Fisher’s exact tests (significance cut-off of p<0.05). Sequencing metrics are provided in Table 3.
Table 3.
Metrics for next-generation single cell sequencing of 32 fine needle aspirate samples from lymphoma from pre- and post-treatment timepoints
| Pre-Treatment | Post-Treatment Timepoint 1 |
Post-Treatment Timepoint 2 |
|
|---|---|---|---|
| No. of Samples | 11 | 12 | 9 |
|
Cells per sample Avg. [range] |
7121 [3465-9548] | 6957 [2855-10,973] | 6866 [4646-10,403] |
|
Reads per cell Avg. (+/− std. dev.) |
63,433 (+/− 12,051) | 56,447 (+/− 16,684) | 61,305 (+/− 6505) |
|
Genes per sample Avg. (+/− std. dev.) |
21,164 (+/− 1137) | 20,501 (+/− 1341) | 20,486 (+/− 851) |
|
Genes per cell Avg. (+/− std. dev.) |
1,621 (+/− 355) | 1366 (+/− 415) | 1457 (+/− 196) |
|
Reads mapped to genome Avg. (+/− std. dev.) |
93.0% (+/− 2.3%) | 92.3% (+/− 3.6%) | 92.0% (+/− 1.5%) |
|
Fraction of reads in cells Avg. (+/− std. dev.) |
82.4% (+/− 9.0%) | 86.1% (+/− 4.8%) | 85.0% (+/− 7.0%) |
Samples were sequenced on Illumina Nextseq or Novaseq and processed using Cell Ranger (10x Genomics). Cells per sample indicate cell numbers detected by Cell Ranger.
Results
FNA is an excellent method of specimen procurement for clinical trials
Our collective experience over the course of these four clinical trials demonstrated that FNA allows for minimally invasive but quantitatively accurate and representative sampling of tumors in a way that is convenient for physicians as well as patients. A summary of the experienced advantages of FNA are listed in Table 1.
Table 1.
Advantages of fine-needle aspiration (FNA) using ultrasound-assisted adequacy assessment as a clinical trial methodology
| Advantage | Details |
|---|---|
| Preservation of targeted lesion and cell number | -Compared to biopsy, does not compromise tumor integrity, allowing continued monitoring of tumor response via imaging |
| Convenience for doctor and patient | -Uses transportable FNA kits and ultrasound equipment |
| -Does not waste time on smear preparation, staining, and adequacy evaluation on site | |
| Efficacy | -Can be used for multiple ongoing trials |
| -Facilitates serial sampling of the same target lesions | |
| -Allows sampling of directly treated and non-treated tumor nodules to evaluate systemic effects of therapy | |
| -Ultrasound allows visualization of lymph node hilum | |
| Cost-effectiveness | -Less expensive compared to interventional radiology or surgical biopsy techniques |
Safety of repeated sampling with FNA
Across the four clinical trials, no significant adverse events were observed during FNA sampling, and no adverse events attributed to the procedure were reported. This demonstrates the relative ease and success of collecting serial FNA samples during clinical trials.
Serial FNA samples are highly cellular and sufficient for multiple analyses
Across four clinical trials, cell yield from these FNA samples was more than adequate for the intended analyses (Table 2). Mean cell yield was 12x10^6 cells [range 0-139x10^6], and mean sample viability was 86% [range 0-100%]. FNAs from pre-treatment sites were successful in collecting at least 0.2x10^6 cells (2 flow cytometry tubes) in 94.2% of samples (82 of 87), and at least 0.1x10^6 cells (1 flow cytometry tube) in 96.6% (84 of 87). The success rate of collecting at least 0.1x10^6 cells (1 tube) across all trial timepoints was 92.6% (274 of 296 samples), and the success rate of collecting at least 0.2x10^6 cells (2 tubes) across timepoints was 88.9% (263 of 296 samples). Three samples in two patients showed zero yield and viability, which could be attributed to either cellular necrosis, treatment effect, and/or shrinking or anatomically difficult to target FNA site(s).
Table 2.
Cellularity yields from fine-needle aspiration sampling
| FNA sampling† |
Yield Cellularity | Viability | ||||
|---|---|---|---|---|---|---|
| Mean | Median | Range | Mean | Median | Range | |
| All nodules (n = 296) | 12 x 10^6 cells | 5 x 10^6 cells | 0 to 139 x 10^6 cells | 85% | 90% | 0% to >95% |
| Pre-treatment nodules only (n = 87) | 18 x 10^6 cells | 9 x 10^6 cells | 0 to 139 x 10^6 cells | 88% | 91% | 057% to >95% |
One FNA sampling procedure includes 3 passes with a 23-gauge needle
Comparison with biopsy: FNA samples accurately reflect the tumor and microenvironment
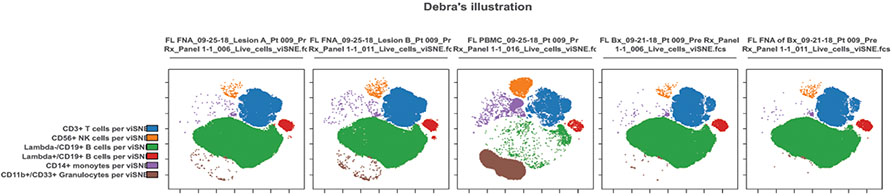
Characterization of the malignant tumor B cells, as well as infiltrating CD3 T cells, CD56 NK cells, monocytes and granulocytes by flow cytometry are shown for two FNAs from one patient prior to treatment (Figure 2A). The visualization of the multi-dimensional flow data by tSNE plots highlights the similarity between the two FNAs as well as the FNA performed on an excisional biopsy of a third site from the same patient and the single-cell suspension made from that same excisional biopsy (Figure 2B and C). Furthermore, there is a strong correlation between the T cells, NK cells and tumor B cells measured in the excisional biopsies and subsequent FNA performed on 4 different patients (Figure 3). All biopsies and corresponding FNA were done prior to treatment and were within 8 days of each other. In this single case, we show that FNA is a robust sampling technique that can obtain cellular material comparable to an excisional biopsy.
Figure 2.
Visualization of high-dimensional flow cytometry data analyzed by the dimensional-reduction algorithm, viSNE. Cells are clustered by surface phenotypes: CD3+ T cells (blue), CD56+ NK cells (orange), CD14+ macrophages or monocytes (purple) and CD33+ granulocytes (brown). CD19+ B cells are split between Lambda- tumor cells (green) and Lambda+ normal B cells (red). (A) Example of 2 separate FNA from 2 different sites of a follicular lymphoma patient. (B) Single-cell suspension of a surgical biopsy from the same patient. (C) FNA performed on the biopsy ex vivo and prior to dissociation. (D) Example of peripheral blood drawn at the same time as the FNA.
Figure 3.
Correlation of 3 major cell populations found in the excisional biopsies and FNA performed on 4 different patients. All biopsies and corresponding FNA were performed prior to treatment and within 8 days of each other. Data is expressed as percent of all live cells. Each color represents a different patient and each symbol represents a different cell population as noted in the legend.
Comparison with peripheral blood: FNA samples are significantly different
While FNA results matched the excisional biopsy results, significant differences were documented in the blood mononuclear samples, providing reassurance that the FNA samples were not significantly contaminated by blood, and highlighting the need to study intratumoral leukocyte populations, rather than only peripheral blood cells, for understanding anti-tumor immunity (Figure 2D).
Serial FNA samples yield excellent material for single-cell RNA sequencing
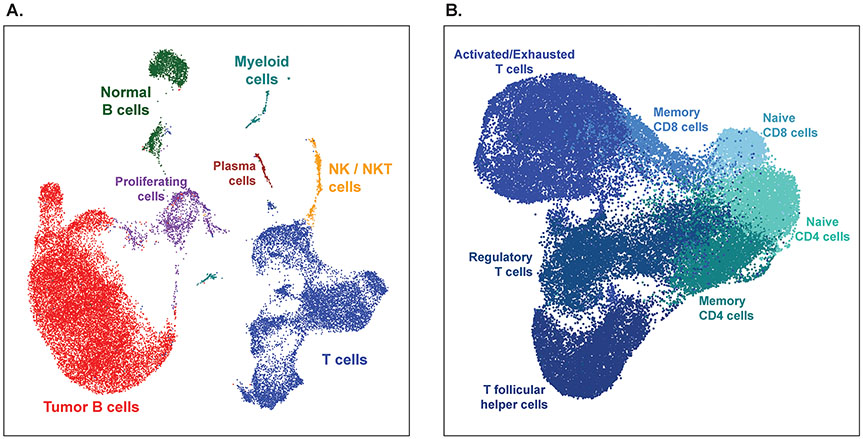
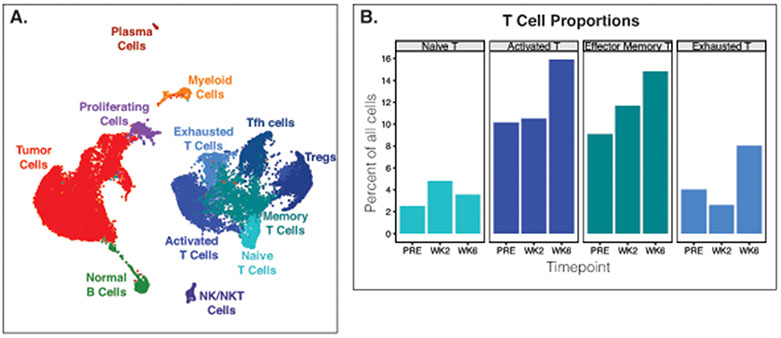
In addition to flow cytometry, many of these samples were analyzed by single cell RNA sequencing for high dimensional phenotyping. Table 3 shows the excellent sequencing metrics obtained from fresh FNA samples, particularly the high fraction of reads mapping to the genome. Single cell gene expression profiles of follicular lymphoma FNAs showed that even with only ~7000 cells analyzed per sample, malignant and normal B cells, large diverse populations of T cells, NK and NKT cells, myeloid cells, and plasma cells, along with proliferating cells could all be clearly resolved (Figure 4A). Additionally, changes in T-cell population proportions could be detected before and after therapy (Figure 4B).
Figure 4.
Fine needle aspirates yield high quality single cell sequencing data. (A) Dimensionality reduction plot showing cells clustered by single-cell gene expression profiles. Cells from 3 fine needle aspirates from the same tumor from 3 different timepoints from one patient are combined and clustered as one dataset. (B) Frequencies of tumor T cell populations before and after treatment. PRE, pre-treatment. WK2, week 2 post-treatment. WK6, week 6 post-treatment. Tfh cells, T follicular helper. Treg, Regulatory T cells. NK/NKT cells, natural killer/natural killer T cells.
Techniques for FNA yield optimization
We found that three needle passes with 23-gauge needles worked well for collecting adequate samples at most sites. The nodules at both sites A and B (Figure 1) had the potential to reduce in size as a result of the therapy during the treatments and become difficult to palpate and biopsy. We employed ultrasound and an ultrasound-directed biopsy technique (see methods) to assist in sampling these smaller and shrinking targets.
Discussion
This is the largest study yet describing FNA methodology and its efficacy in collecting samples directly from tumor nodules during clinical trials. The technique yields fresh, live cellular material that can be used in a wide array of immunologic, diagnostic, and molecular assays and studies. Our studies show excellent adequacy (97% of FNA samples pre-treatment were adequate, and 93% overall), good yield (12x106 cells), and well-maintained viability (86%).
We also describe in our methods the use of ultrasound guided FNA to help target and sample small and shrinking nodules while maintaining optimal cell sample collection.4,13-15 While a team in 2001 did report serial fine-needle sampling of metastatic melanoma samples, this study provides data for a concert of clinical trial studies, expands the discussion of the techniques used to acquire and analyze the samples, including single cell sequencing techniques, and references and demonstrates the sum of the results obtained from these techniques.12
Limitations of performing serial FNAs to support clinical trials of this type include: 1) the necessity to have well-trained cytopathologists readily available to see patients and perform FNA biopsies within the time schedule windows of the clinical trials, and 2) nodules selected for FNA biopsy can decrease in size after therapies begin, making sample collection more difficult and challenging. In addition, the treated lymph node targets often underwent degenerative change following therapy which limited the cellular viability of some of the samples from this site. Tumor site shrinkage and post treatment effects within the nodules most likely contributed to slightly lower cell yields in the post-therapy sites vs pre-treatment FNAs.
Another limitation of this study is that the FNA yields were compared with only one concurrent tissue core biopsy; although figure 3 does show good correlation of FNA vs tissue core collection over multiple samples, supporting the utility of this technique.
To date, the analysis of our samples has demonstrated significant differences in cell populations between tumor nodules and matched peripheral blood samples, changes in tumor microenvironment which correlate with clinical response, and unexpected heterogeneity between tumor sites in the same patient.7,8,12 FNA samples from these trials provided the cellular material for recently published findings, as well as ongoing studies. Studies by Frank et al7 demonstrated changes in T lymphocyte populations over time and a finding that low percent of CD4+ T-regulatory cells at baseline correlated with a better distal clinical response after trial therapy. Work by Haebe and Shree et al8 using single-cell RNA sequencing and flow cytometry demonstrated significant heterogeneity between sites A and B prior to any treatment in many patients with lymphoma; these differences may be intrinsic to the biology of the tumors and may influence prognosis and response to therapy. These two studies demonstrate the feasibility and immense potential of serial FNA sampling during clinical trials to collect cellular tissue samples for analysis and profiling.
Ongoing work with these samples to detect and define mechanisms of anti-tumor immune responses, study molecular heterogeneity, and link T-cell and B-cell receptor sequencing to observed phenotypes and clinical responses continues.
Conclusion
We describe in this manuscript a new, exciting window of opportunity to use FNA in research tissue sample collection, especially in ongoing trials requiring sampling over multiple time points. FNA can be an ideal harvesting technique as it allows direct collection from tumor nodules, preservation of sampled tissue targets, good patient tolerance, relatively low cost, is performed with easily portable equipment, and provides cellular material to support a broad range of research endevors.1,14,15
Funding Statement:
Research supported by the National Institute of Health, Rising Tide Foundation for Clinical Cancer Research, and the Leukemia and Lymphoma Society.
Footnotes
Conflict of Interest Disclosures: The authors made no disclosures.
Clinical Trial Registration Numbers: NCT02927964, NCT03410901, NCT02266147, NCT02254772, NCT02927964, NCT03410901
References
- 1.Paksoy N, Ozbek B. Cytopathologist-performed and ultrasound-guided fine needle aspiration cytology enhances diagnostic accuracy and avoids pitfalls: An overview of 20 years of personal experience with a selection of didactic cases. Cytojournal. 2018;15:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandhaus LM. Fine-needle aspiration cytology in the diagnosis of lymphoma. The next step. Am J Clin Pathol. 2000;113(5):623–627. [DOI] [PubMed] [Google Scholar]
- 3.Carmeci C, Jeffrey RB, McDougall IR, Nowels KW, Weigel RJ. Ultrasound-guided fine-needle aspiration biopsy of thyroid masses. Thyroid. 1998;8(4):283–289. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Liu C, Cao Y. The value of sonographically guided fine-needle aspiration in the diagnosis of small lymph. Chin Clin Oncol. 2020;9(2):12. [DOI] [PubMed] [Google Scholar]
- 5.Lizotte PH, Jones RE, Keogh L, et al. Fine needle aspirate flow cytometric phenotyping characterizes immunosuppressive nature of the mesothelioma microenvironment. Sci Rep. 2016;6:31745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manzo JL, Cuda J, Pantanowitz L, et al. Clinical trial cytology: Use of on-site evaluation of small biopsy and FNA samples for clinical trials and biomarker research studies. Cancer Cytopathol. 2018;126(7). [DOI] [PubMed] [Google Scholar]
- 7.Frank MJ, Reagan PM, Bartlett NL, et al. In Situ Vaccination with a TLR9 Agonist and Local Low-Dose Radiation Induces Systemic Responses in Untreated Indolent Lymphoma. Cancer Discov. 2018;8(10):1258–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haebe S, Shree T, Sathe A, et al. Single-cell analysis can define distinct evolution of tumor sites in follicular lymphoma. Blood. 2021;137(21):2869–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stuart T, Butler A, Hoffman P, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177(7):1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baloch ZW, Tam D, Langer J, Mandel S, LiVolsi VA, Gupta PK. Ultrasound-guided fine-needle aspiration biopsy of the thyroid: role of on-site assessment and multiple cytologic preparations. Diagn Cytopathol. 2000;23(6):425–429. [DOI] [PubMed] [Google Scholar]
- 11.Fleischman GM, Thorp BD, Difurio M, Hackman TG. Accuracy of Ultrasonography-Guided Fine-Needle Aspiration in Detecting Persistent Nodal Disease After Chemoradiotherapy. JAMA Otolaryngol Head Neck Surg. 2016;142(4):377–382. [DOI] [PubMed] [Google Scholar]
- 12.Fetsch PA, Steinberg SM, Riker AI, Marincola FM, Abati A. Melanoma antigen expression in serial fine-needle aspiration samples in patients with metastatic malignant melanoma participating in immunotherapy clinical trials: a preliminary look. Cancer. 2001;93(6):409–414. [DOI] [PubMed] [Google Scholar]
- 13.Rollins SD. Teaching FNA techniques and ultrasound guided FNA. Cancer Cytopathol. 2019;127(1):7–8. [DOI] [PubMed] [Google Scholar]
- 14.Lieu D Cytopathologist-performed ultrasound-guided fine-needle aspiration and core-needle biopsy: a prospective study of 500 consecutive cases. Diagn Cytopathol. 2008;36(5):317–324. [DOI] [PubMed] [Google Scholar]
- 15.Abele JS. The case for pathologist ultrasound-guided fine-needle aspiration biopsy. Cancer. 2008;114(6):463–468. [DOI] [PubMed] [Google Scholar]