Abstract
Maintenance of systemic homeostasis by kidney requires the coordinated response of diverse cell types. Use of single-cell RNA sequencing (scRNAseq) for patient samples remains fraught with difficulties. The ability to characterize immune and parenchymal cells during transplant rejection (some of which may be present at very small numbers due to injury) will be invaluable in defining transplant pathologies. Herein, we present feasibility data for multiplexing approach for droplet scRNAseq (Mux-Seq). Mux-Seq has the potential to minimize experimental batch biases and variations. Explant tissues from 6 normal and 2 transplant recipients after multiple rejection episodes leading to nephrectomy were pooled for Mux-Seq. Subsequently, a computational tool, Demuxlet was applied for demultiplexing the individual cells. Each sample was also applied individually in single microfluidic run (singleplex). We show that data from Mux-Seq correlated highly with singleplex (Pearson coefficient 0.982). Both are able to identify many known kidney cell types including immune cells. Trajectory analysis of proximal tubule and endothelial cells demonstrates separation between healthy and injured kidney from transplant explant suggesting various stages of differentiation. This study provides the technical groundwork for understanding the pathogenesis of alloimmune injury and host tissue response in transplant rejections leading to graft failure in clinical setting.
Introduction
The kidney is a highly complex organ with each functional unit-nephron consisting of more than 14 distinct cell types [1]. A highly coordinated response from the diverse cell types of different segments of the nephron are critical for renal function and maintenance of whole-body homeostasis. Therefore, detailed characterization of each cell type within the kidney is crucial towards understanding the molecular alterations that are associated with disease conditions. Knowledge regarding the transcriptional landscape in kidney has come largely from microarray and bulk RNA sequencing (RNAseq), technologies that reflect the average gene expression across thousands of kidney cells, without considering the fluctuations in cell specific gene expression [2]. Single cell RNA sequencing (scRNAseq) is now being harnessed for understanding the cellular diversity of different organ systems as part of the Human Cell Atlas initiative [3], with the deliverable of accurately interpreting cell-specific gene expression data, identify known and new cell types or subtypes involved in disease progression, and follow expression mapped cellular transition states. Successful application of scRNAseq to human and mice kidney enabled critical understanding of their composition and identification of novel cellular types [4–6]. We chose to focus on applying scRNAseq to human kidney, because bulk transcriptomic profiling has been extensively studied to predict outcomes in transplantation with several limitations. At present, there are two scRNAseq studies on solid organ transplants from the Humphreys group. First, Wu et al characterized single-cell transcriptomes from one healthy adult kidney and one transplant biopsy to define the inflammatory response during mixed rejection but as stated it is based on a single biopsy [7]. Second, Malone et al have utilized single nucleotide variations coupled with single cell RNA sequencing to define the donor versus recipient immune population in rejecting kidney transplant [8]. These shows that comprehensive scRNAseq of human solid organ is feasible and endorse the need for technical optimization and improved molecular characterization of such tissues.
In this study, we have optimized kidney cell dissociation methods and applied a multiplexed droplet single-cell sequencing approach (Mux-Seq) for scRNAseq and demonstrated its advantages over singleplex scRNASeq with regards to reducing experimental cost and variation. We present a data analysis pipeline that evaluates thresholds for quality assessment of kidney tissue data, inclusive of mitochondrial content and doublet rates. Mux-Seq uses Demuxlet, a software tool which utilizes the natural genetic variation among individuals to assign the identity of each cell to the individual source [9]. We show that Mux-Seq successfully identifies known renal cell types including immune cells and has high correlation with the gene expression output obtained by conventional scRNAseq methods, supporting its use as a viable method for large batches of samples. Our study also allowed improved understanding of the cellular states and molecular dynamics of kidney health and elucidate the divergence of existing cells and evolution of new cell-subsets in transplant explants removed after rejection.
Materials and Methods
Human kidney samples –
A total of 8 human kidney samples were obtained for this study - four from the University of California San Francisco (UCSF) and four from the University of Michigan under institutional review board-approved protocols. All samples were dissected from tumor-free regions of nephrectomy (native or transplant explant post multiple rejections). Samples consisted of four males, and four females. The ages of the patients ranged from 57–71 (mean 64.5, SD 5.1). All the tissue samples were processed and stored following the same method. Sample metadata is stored in Table S1.
Transplant explants:
Two explant tissues from nephrectomies performed due to transplant failure after multiple rejection episodes were included in the study. Both tissues had interstitial and tubular inflammation with preserved tubules and glomeruli. There are characteristic features of chronic antibody mediated rejection (ABMR) (Figure S1). Explant 2582 was a living unrelated kidney transplant in a Caucasian female induced with Simulect. The nephrectomy was performed 62 months post-transplant after 2 biopsy confirmed ABMR episodes at year 4 and year 5 post-transplant. The explant biopsy 2582 was confirmed by pathology to have C4d negative ABMR and TCMR 1A, 20% interstitial fibrosis and tubular atrophy (IFTA) and moderate arteriosclerosis (Figure S1, explant A). The estimated glomerular filtration rate (eGFR) at transplant nephrectomy was 13.
The explant 2584 was from a deceased donor renal transplant in an African American male (induced with Thymoglobulin), obtained at 22 months post-transplant, with repeated non-adherence. There were 3 biopsy confirmed rejection episodes beginning in the first post-transplant year. In year 2, two rejection episodes with C4d negative acute ABMR, TCMR, severe IFTA (80%), focal global glomerulosclerosis (40%) and moderate arteriosclerosis (Figure S1, explant B) led to transplant nephrectomy. The eGFR at transplant nephrectomy was 4. Both patients received maintenance immunosuppression with steroids, mycophenolate mofetil and tacrolimus.
Single-Cell Preparation
Kidney biopsy tissue of ~7.5 mg (needle biopsy) or nephrectomy dissected tissue (chopped tissue) (was frozen in CryoStor cell cryopreservation media (Cat#C2874, Sigma, St. Louis, MO) and stored in liquid nitrogen until further processing. On the day of experiment, the cells were thawed, and single cell suspension was prepared by an optimized method described in detail in Supplementary Methods. Cell viability was assessed and cell populations with ≥50% viability score were used for the experiment. The description of tissues, number of cells applied to 10X and included in the final analysis are shown in Figure S2.
Single Nucleotide Polymorphism (SNP) array
Genomic DNA was isolated from kidney samples using QIAamp DNA Mini Kit (Cat# 51104, Qiagen, Hilden, Germany), obtaining ~30ng of DNA from 5000 cells. DNA was used for genotyping using SNP arrays (OmniExpress Exome kit, Illumina).
Library Preparation and Sequencing
Libraries for single-cell RNAseq were prepared using the 10X Single Cell Immune Profiling Solution Kit according to the standard manufacturer protocol. For multiplexed RNAseq (Mux-Seq), 5,000 cells from 4 individual tissues of the cohort (2 transplant and 6 native nephrectomies) were mixed to create a pool of 20,000 cells and loaded onto each lane. Three such pools were run by mixing various combinations of individual samples. Samples and the cell numbers included in each pool are listed in Figure S2B. For Singleplex RNAseq, 5,000–20,000 cells per sample were loaded onto each lane individually. Cell numbers loaded onto 10X lane for each individual sample is listed in Figure S2B. Full length cDNA libraries were prepared by incubating GEMs in a thermocycler. Single-cell RNA libraries were sequenced on the Illumina NovaSeq S2 to a minimum sequencing depth of 50,000 reads/cell using the read lengths 26bp Read1, 8bp i7 Index, 91bp Read2.
Data was processed via the 10X Chromium 3’ v2 platform. Data matrices and barcode information were generated using the 10X Cell ranger version 2.1.1 software, aligned to the GRCh38 transcriptome. After 10X generated unfiltered barcodes, the EmptyDrops tool was used to identify true cells with an FDR threshold of 0.01. For both singleplex and Mux-Seq analyses, a threshold for 6,000 maximum gene count per cell (to eliminate doublets) and percentage of transcriptome containing mitochondrial reads at 50% was set. In order to demultiplex each pool used for Mux-Seq and identify the original sample source for each cell, the computational tool-Demuxlet was applied. The tool uses the SNP information generated from the Illumina OmniExpress SNP array unique to each kidney tissue. Based on the natural genetic variation of individual tissues, the tool determines the identity of each droplet containing a single cell. Furthermore, Demuxlet detects and removes droplets containing two or more cells as determined by the presence of SNPs originating from more than one genotype [9].
Data Analysis
The first phase of analysis was performed in R using the Seurat version 3 package [10]. Gene expression data was first normalized using the Seurat SCTransform normalization method. The harmony tool was then used to correct for batch differences between the kidneys from the two clinical sites. Linear dimension reduction was done by computing the top 50 principal components. These 50 principal components were then used for non-linear dimension reduction, shared nearest neighbor graph construction and Louvain clustering, and visualization via UMAP. The resolution parameter of the Seurat UMAP function, which controls the number of clusters produced, was set to 0.6.
Average expression of each gene was calculated for each cell population (cluster), then logged fold change was determined by taking the average expression of a single gene in one cluster against all other clusters. FDR-corrected p-values were calculated using the Wilcoxon rank-sum test. Genes with a minimum log fold change value of 0.5 and expressed in at least 25% of the cluster were included. Once a cluster’s identity was determined using a list of well-known marker genes (Table S2). For functional profiling of cell population clusters, gene set enrichment analysis (GSEA) was performed using the R bioconductor package clusterProfiler version 3.10.1. The Monocle 2 tool version 2.10.1 package in R was used for pseudotime computation and cell trajectory analysis [11]. The data associated with this project is available at the Data Lake of the Kidney Precision Medicine Project (www.kpmp.org).
Results
Normal and transplant kidney demonstrate variations in immune and major renal cell types.
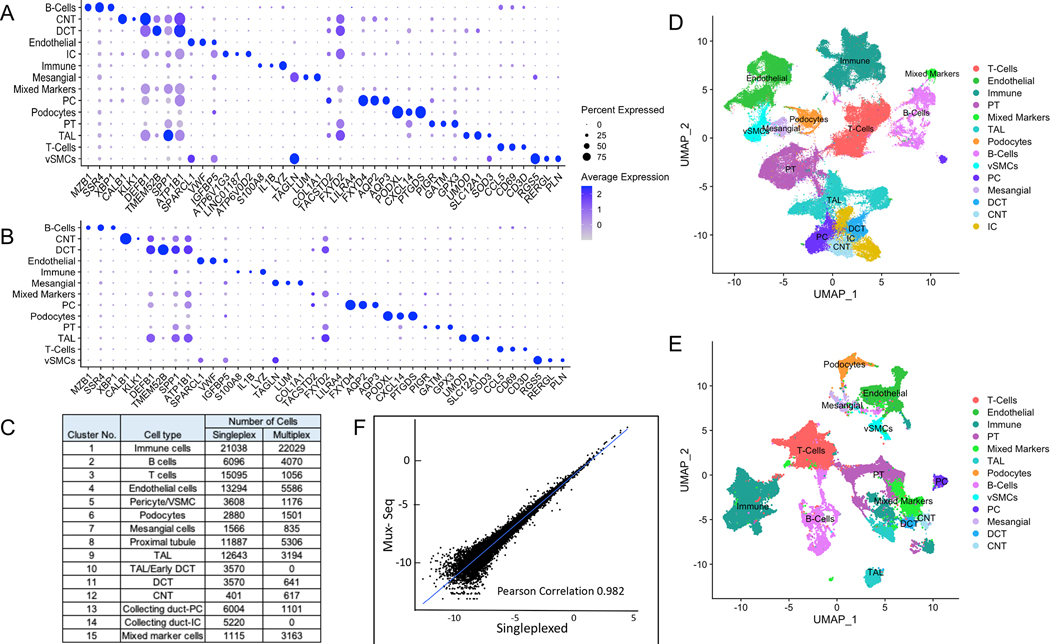
Kidney tissue from tumor-free regions of native nephrectomies to characterize normal kidney and transplant nephrectomies due to multiple rejections were subjected to single cell transcriptional profiling (10X Genomics platform) as described in materials and methods. A pipeline was established for one sample per microfluidic run (singleplex scRNAseq) as well as for pooled samples in one microfluidic run (Mux-Seq) using droplet single-cell RNA sequencing (Figure 1). In the singleplex RNAseq experiment, we obtained 106,760 cells after rigorous quality control that resolved into 38 distinct clusters. Based on the expression of previously established marker genes in the unique gene expression profile of each cluster (Table S2), we annotated the clusters and identified cells of all major types present in the kidney- renal epithelial cells including glomerular podocytes, proximal tubule cells (PT), thick ascending limb (TAL), distal convoluted tubule (DCT), connecting tubule (CNT), collecting duct (both principal and intercalated cells); endothelial cells; stromal cells; and immune cells including mononuclear phagocytes, neutrophils, T cells, and B cells (Figure 2C). The top three genes for each cell type based on their expression levels and abundance are represented by a dot plot in Figure 2A and the corresponding UMAP is displayed in Figure 2D.
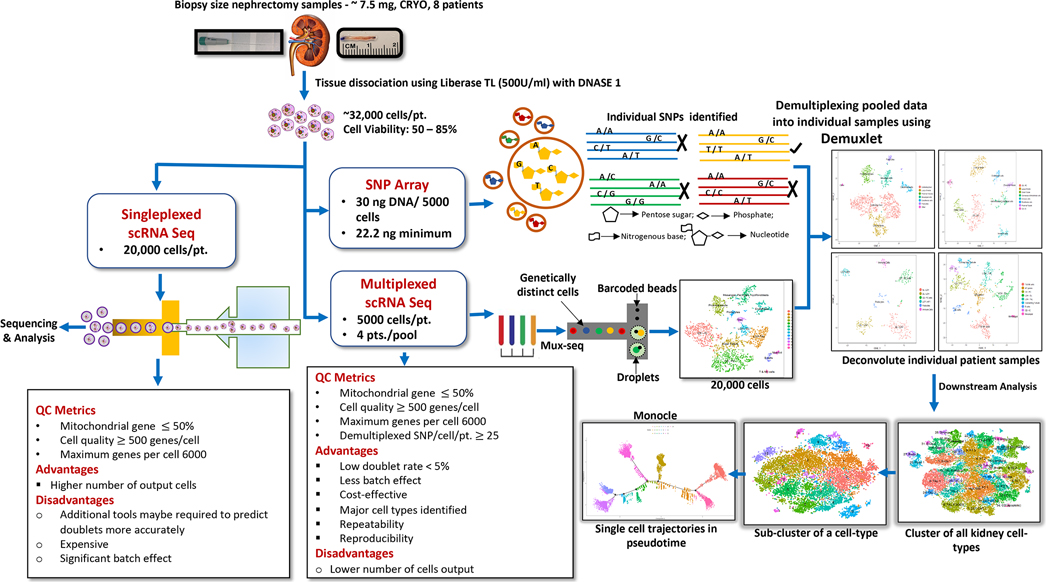
Figure 1. Overview of kidney tissue processing for droplet based scRNA-seq.
A pipeline indicating optimized protocol for cell isolation and data processing are shown for singleplex- scRNAseq as well as Mux-Seq.
Figure 2. Single cell transcriptomic analysis of human kidney by scRNAseq reveals distinct cell populations.
(A) Single cell transcriptomes from 8 nephrectomy tissues (2 transplant and 6 native) were analyzed by conventional singleplex scRNAseq. Unsupervised clustering of 106,760 cells resulting in 38 distinct cell populations. Dot plot of average gene expression values (log scale) of top 3 unique differentially expressed markers and percentage of cells expressing these markers in each cell type are shown. (B) 50,275 single cell transcriptomes from 2 transplant and 6 native nephrectomy tissues were analyzed by pooling cells and conducting Mux-Seq. Unsupervised clustering resulted in 39 distinct cell populations. Dot plot of average gene expression values (log scale) of top 3 unique differentially expressed markers and percentage of cells expressing these markers in each cell type are shown. (C) Table listing cell type annotations and number of cells. (D) UMAP plot showing distinct cell populations identified in (C) from singleplex data analysis and clustering. (E) UMAP plot showing distinct cell populations identified in (C) for multiplex analysis. (F) Scatterplot of log transformed average gene expression obtained by Mux-Seq (on y-axis) and singleplex-scRNAseq (on x-axis). Only genes expressed in at least five cells in each assay were considered. Pearson coefficient = 0.982.
Next, we successfully utilized multiplexed single cell RNA sequencing (Mux-Seq) to pool cells from normal and transplant rejection tissue in one microfluidic run. Following the run, we leveraged the genotypes of individual samples to assign each cell to a corresponding individual using a bioinformatic tool “Demuxlet” [9]. In this manner we were able to identify the sample origin of 50, 275 pooled cells, as well as identify doublets using a minimum of 50 unique SNPs per sample. The overall doublet rate was estimated at 6.2% which is in line with other published studies [12]. As a result, Mux-Seq analysis of 50,275 high quality cells clustered into 39 distinct populations. Based on the unique gene expression profile of each cluster (Table S2), clusters were annotated for cell type (Figure 2C and 2E). The top three genes for each cluster based on their expression levels and abundance are represented by a dot plot in Figure 2B. As with singleplex scRNAseq, major renal parenchymal cell types as well as numerous immune cells including B cells, T cells, granulocytes, macrophages/monocytes and dendritic cells were identified by Mux-Seq. However, we were unable to identify intercalated cells in this population. Interestingly, there were clusters that contained markers of multiple cell types including epithelial cell markers as well as markers of immune cells/antigen presenting cells. We labeled them as mixed marker cells. All clusters resulting from singleplex-scRNAseq and Mux-Seq contained cells from multiple patients (Figure S3) indicating minimal batch effect. Direct comparison of expression profiles of 20,150 genes expressed in at least five cells for both methods showed high concordance between singleplex-scRNAseq and Mux-Seq (Pearson coefficient 0.982, Figure 2F). Furthermore, the expression profiles of kidney specific marker genes showed very high correlation between singleplex-scRNAseq and Mux-seq (Figure S4). Together these results show a highly reproducible cluster mapping with Mux-Seq protocol despite the smaller number of input cells per sample. As a further measure of quality control, we explored the association of the transcriptomic mitochondrial read content with kidney function by gene set enrichment analysis. As stated before, the cutoff for the mitochondrial gene percentage was set at 50% resulting in a mean of 26% mitochondrial content across the clusters (Figure S5A). Gene ontology pathway analysis of the 125 genes positively correlated with the mitochondrial gene percentage resulted in the enrichment of biological processes associated with metabolism and energy expenditure (Figure S5B).
Single-cell changes in transplant explants after rejection.
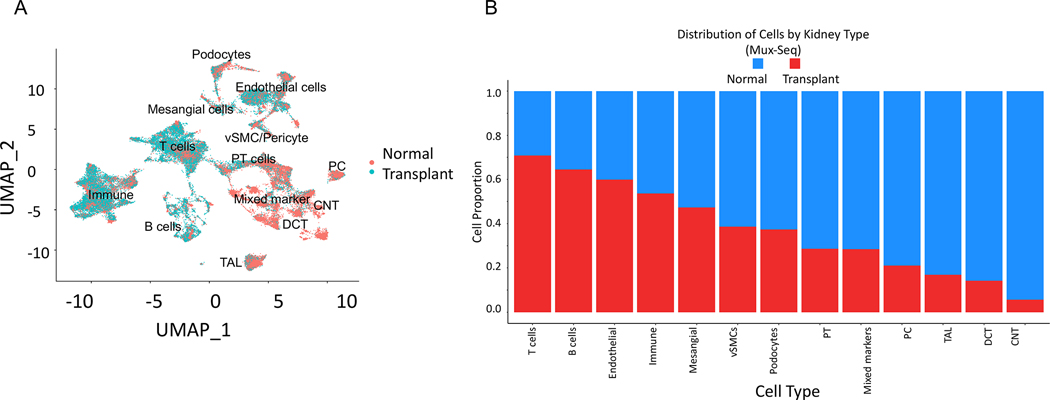
When we clustered all 50,275 cells based on their tissue source (normal versus transplant) we found that they clustered almost entirely separately suggesting an altered transcriptional profile (Figure 3A). Analysis of proportion of cells from normal and transplant explant tissues in each cluster revealed an enrichment of immune cells (both T and B cells) in cells from transplant explants with reduced abundance of renal cell types (connecting tubule and collecting duct cells, distal convoluted tubule cells, thick ascending limb cells, PT cells and podocytes) (Figure 3B). Pathway enrichment analysis with the top 100 upregulated genes in transplant explant samples compared to the normal identified several biological pathways including immune and cytokine responses as the most affected pathways. Recruitment of inflammatory cells from the circulation into the renal allograft is a key event in acute as well as chronic transplant rejection. Chemokines and chemokine receptors are crucial mediators for the infiltration of leukocytes into the allograft. Accordingly, we saw an upregulation of CCL5/RANTES in activated T /NK cell clusters (cluster 0, 11). We found upregulation of proinflammatory cytokines such IL1B, which may upregulate chemokines (CXCL3, CXCL10, CXCL8, CXCL2, CCL5, CCL3, CCL4) via transcription factors such as FOS [13, 14] cumulatively inducing immune activation. Moreover, genes including CD74 and PTPRC/CD45 (antigen presentation) [15], LGALS [16], immune activation (CD163) [17] and activation of granulocytes (SRGN) and monocytes (FCGR3A/CD16+ clusters 25, 26) [18, 19] are upregulated in cells from transplant explants compared with healthy cells. Downregulated genes in cells from transplant nephrectomies were involved in pathways associated with changes in oxidative phosphorylation (NDUFB9, ATP5I, NDUFB1, NDUFA4, NDUFA1, UQCRQ, NDUFA3, NDUFB2, NDUFC1) and reduced mitochondrial ATP synthesis (COX5B, COX7C) (Figure S6). Figure S7 shows comparable results from singleplex-scRNAseq and Mux-Seq analyses for cells form normal and transplant explants, with clearer representation of renal parenchymal and epithelial cell types in the Mux-Seq data.
Figure 3. Mux-Seq analysis shows distinct gene expression profiles for cells obtained from transplant nephrectomies versus normal tissue.
(A) UMAP plot of cells from 6 normal (25,470 cells) and 2 transplant nephrectomies (24,805 cells) shows separate clustering of transplant versus normal cells. Cells from the two groups are indicated by different colors (red, normal; blue, transplant). (B) Bar graph displays proportion of cells in each cluster from transplant versus normal tissue.
Pseudotime trajectory analysis of PT cells
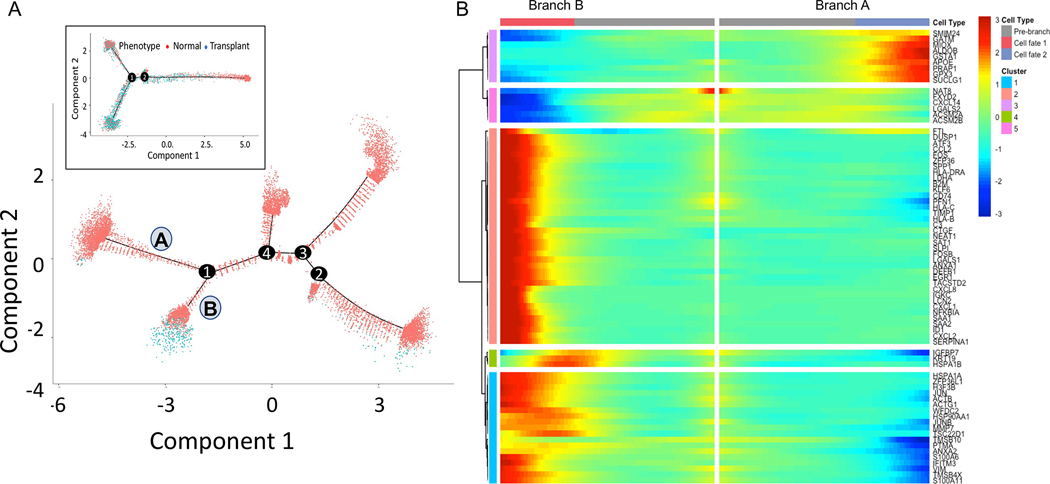
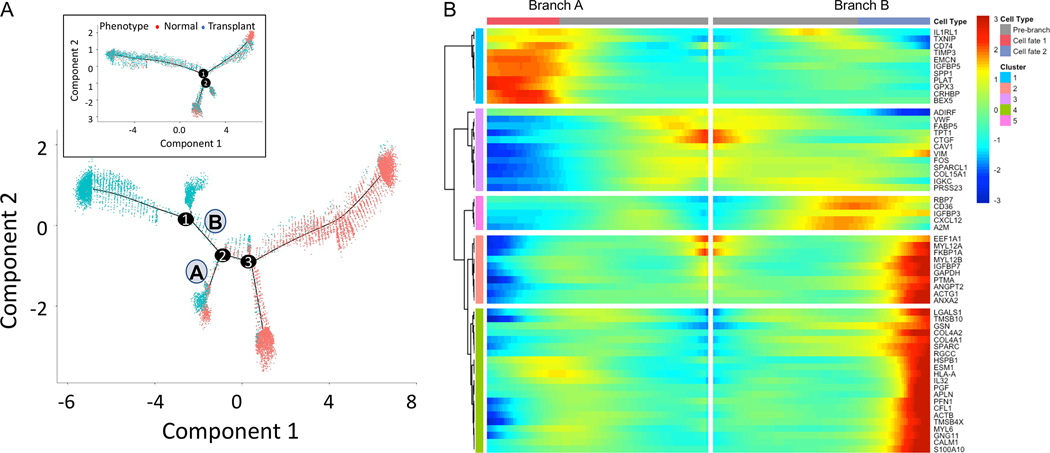
Renal PTs cells were distributed across five clusters of which two (cluster 20 and 32) could be identified as S1 segment cells with expression of SLC5A12, with significant reduction of PT cells in transplant rejection injury. To further understand this rejection injury response in PT cells, we performed cell trajectory analysis by reconstituting the PT cells from healthy and transplant explant tissues and performing pseudotime analysis. The analysis of both singleplex (11,925 normal and 397 transplant PT cells) and multiplex datasets (3,788 normal and 1,518 transplant PT cells) reveals a major branch point at 1 where normal cells separate from transplant explant cells (Figure 4A). The trajectories are very similar from the two datasets in that the transplant cells were separated from majority of normal cells in the pseudotime trajectory. There is an additional branch of normal cells for the singleplex dataset which we believe is the result of having significantly higher number of cells for analysis. Using monocle ordering we recognized that PT cells undergo changes along a continuous path in pseudotime starting from a population primarily composed of cells from normal tissue to intermediate to terminal cell states. Branchpoint 1 splits into two branches – A) PT cells predominantly from normal tissue (2,780 normal, 17 transplant), B) PT cells from a mixed population of healthy and transplant explants (1,219 normal and 289 transplant). We found that the branch B has low expression of canonical PT marker genes ALDOB, GATM, GPX3 [5] and higher expression of JUN, VIM, HSP genes [20], which are associated with injury and cell death (Figure 4B). In summary, PT cells from transplant explants progressively lose their characteristic PT markers and start expressing stress or injury markers. These findings strongly support the observation that healthy human PT epithelia undergo life cycles of spontaneous injury followed by homeostatic repair after a rejection episode.
Figure 4. Trajectory analysis of Proximal Tubule cells using Monocle pseudotime.
Cell trajectory plot shows all cells from singleplex ordered by A) Cellular phenotype. B) Heatmap of the top genes most significantly associated with pseudotime at branch point 1. Branch A has 2,780 cells from normal and only 17 cells from transplant explants. Branch B has a higher proportion of cells from transplant explants (1,219 normal and 289 transplant). Inset in A shows the cell trajectory with Mux-Seq dataset.
Endothelial cells separate into three different clusters.
Targeting of donor endothelial cells by the recipient immune response is a primary component of rejection. Both Mux-seq and conventional singleplex-scRNAseq identified endothelial clusters and had optimal representation of cells from normal as well as transplant tissue (Figure 3B, S7B). We identified three independent stable endothelial cell clusters (cluster 4, 6 and 24). Table 1 lists the top 20 genes in each cluster and their log fold change compared to all other clusters. Cluster 4 is unique for high expression of CTGF which is a mitogen expressed by vascular endothelial cells and is implicated in inflammation and fibrosis [21]. Cluster 24 displays high expression of CXCL12 which promotes lymphocyte and monocyte chemotaxis [22]. We also observe high expression of the CXCL12 ligand CXCR4 in T cells, monocytes and dendritic cells suggesting a ligand-receptor interaction.
Table 1:
Top 20 differentially expressed genes per cluster (indicated cluster versus remaining clusters) sorted by highest mean expression and log fold change (logFC).
| Cluster 4 | Cluster 6 | Cluster 24 | |||
|---|---|---|---|---|---|
| Gene | logFC | Gene | logFC | Gene | logFC |
| SPARCL1 | 1.27393354 | IGFBP5 | 1.84175832 | SPARC | 1.176654 |
| VWF | 1.24308996 | TIMP3 | 0.92492518 | SPARCL1 | 1.061858 |
| CTGF | 1.14718265 | IFI27 | 0.8979945 | RBP7 | 1.04657 |
| GNG11 | 0.83777438 | EMCN | 0.89418125 | CXCL12 | 1.010339 |
| SPARC | 0.83285705 | IFITM3 | 0.7606348 | PLPP1 | 1.00337 |
| HSPG2 | 0.81237852 | SLC9A3R2 | 0.60578072 | A2M | 0.98721 |
| MGP | 0.80623116 | TGFBR2 | 0.59745605 | FABP4 | 0.899181 |
| CAV1 | 0.79564635 | EPAS1 | 0.55182325 | IFI27 | 0.883899 |
| ACKR1 | 0.78097172 | CD74 | 0.54146685 | RGCC | 0.880663 |
| FKBP1A | 0.77075809 | GNG11 | 0.52146445 | GNG11 | 0.873019 |
| VIM | 0.76919635 | SPRY1 | 0.5164134 | VWF | 0.782918 |
| TM4SF1 | 0.70092886 | PLPP1 | 0.50376683 | COL4A1 | 0.744089 |
| IFITM3 | 0.65840752 | PLAT | 0.494256 | GSN | 0.717875 |
| IFI27 | 0.61486863 | HLA-E | 0.45238452 | TCF4 | 0.704507 |
| EDN1 | 0.60680967 | IL1RL1 | 0.44678486 | HSPG2 | 0.693016 |
| LGALS1 | 0.59000513 | HES1 | 0.44378708 | VIM | 0.667797 |
| CYR61 | 0.58527943 | TM4SF1 | 0.44064332 | COL15A1 | 0.650325 |
| RPS18 | 0.571311 | RNASE1 | 0.42499816 | IGFBP7 | 0.624117 |
| TPT1 | 0.57087518 | PLPP3 | 0.41930041 | CAV1 | 0.62274 |
| IGFBP7 | 0.56408824 | HLA-B | 0.41712389 | IFITM3 | 0.580499 |
To further understand the injury response in endothelial cells, we performed cell trajectory analysis by reconstituting the endothelial cells from healthy and transplant explants for both singleplex (Fig. 5A) and multiplex datasets (Fig. 5A, inset). We see from singleplex analysis that at branch point 2, endothelial cells from normal and transplant explant samples separate from a group of cells that are only derived from transplant rejection. Before branch point 2, combining cells from all states, we have 6,819 cells from normal and 1,186 cells from transplant explants. Following Branch B, there are 228 cells from normal and 4,002 cells from transplant. Branch A consists of a mix with 487 cells from normal and 527 cells from transplant explants. Similar to PT cells, we see more branches for endothelial cells using singleplex dataset. Heatmap of differentially expressed genes with significant change between before and after branch point 2 is shown in Figure 5B. According to their expression pattern, five clusters of genes can be visualized. Based on higher expression of genes such as CTGF and VWF we infer that the branch B leading to cell fate 2 refers to the branch with predominantly cells from transplants explanted after rejection. These results are consistent with the endothelial cell activation in transplant [16, 23].
Figure 5. Trajectory analysis of endothelial cells using Monocle pseudotime.
Cell trajectory plot shows all cells from singleplex ordered by A) Cellular phenotype. B) Heatmap of the genes most significantly associated with pseudotime at branch point 2. Before branch point 2, combining cells from all states, we have 6,819 cells from normal and 1,186 cells from transplant explants. Along Branch B, there are 228 cells from normal and 4,002 cells from transplant. Inset in A shows the cell trajectory with Mux-Seq dataset.
Discussion
In this focused study we profiled single cell transcriptomics of kidney cells using multiplexing approach and compared the results with that from singleplex scRNAseq. While doing this, we report multiple important findings. We further validate the multiplexing approach in analyzing kidney cells by scRNAseq. In this report, we provide protocols that optimize tissue processing for scRNAseq from limited amounts of human kidney tissue to mimic sample volumes that would normally be available during interrogation of clinical tissue samples from patients. We report high yield and viability after renal tissue dissociation to robustly classify single-cell heterogeneity of 8 kidney samples, provide a comparative analysis of robust kidney single-cell biology from Mux-Seq and singleplex scRNAseq and highlight the Mux-Seq approach for data deconvoluton by Demuxlet.
Though the scRNAseq technology for high throughput profiling of a large number of cells by droplet scRNAseq has substantially improved, the applicability of scRNAseq technology towards creating a kidney atlas has met several roadblocks. These hurdles include availability of clinical patient tissue, high costs of running multiple samples by scRNASeq, and run/batch variations—all of which can be readily addressed by applying the Mux-Seq methodology. Through the present study, we have demonstrated the efficacy of the first Mux-Seq data on human kidney samples and propose it as an ideal solution for handling limited human samples without loss of single-cell tissue biology. It is important to note that despite the lower number of input cells in Mux-seq we have been able to identify all major renal parenchymal cells and several immune populations as singleplex-scRNAseq. Indeed, we identified more immune cell types with Mux-seq than singleplex-RNAseq including plasmacytoid dendritic cells and Tregs.
Our study sheds light on the gene expression changes due to transplant rejections leading to graft failure and demonstrates the ability of Mux-Seq to efficiently distinguish between cells originating from normal kidney versus transplant tissue. We show that in renal parenchymal cell clusters, the proportion of cells from transplant nephrectomies are low while in immune cluster their proportion increases significantly (Figure 3B). Interestingly, we observed a better representation of parenchymal cells in transplant nephrectomy samples by Mux-Seq despite the lower number of input cells suggesting that Mux-Seq allows us to preserve the cell type distribution of the tissue.
In addition, we have further characterized the gene expression changes in PT and endothelial cells associated with transplant rejection injury. As previously known, there is a loss of the overall PT cell number in from transplant explant tissue in our dataset. Interestingly, we see a smooth transition of the normal phenotype of PT cells to a stressed phenotype that lacks the canonical markers like ALDOB, GPX3 and GATM during rejection (see Figure 4, Monocle analysis). We were able to confirm similar trajectories for PT cells using singleplex and Mux-Seq datasets. Another important cell type in alloimmune response during rejection is endothelial cells. We identified 3 clusters of endothelial cells consisting of cells from both normal and rejection tissues and after reconstituting them we performed cell trajectory analysis. Pseudotime analysis with Monocle showed that there was a transition from cells arising from normal kidney to cells from transplant rejection tissue (see Figure 5). We analyzed the branch point 2 which is where we see the maximal separation of cells from normal and transplant explant tissues. Cell Fate 2 is associated with very high expression of CAV1, COL4A1/A2, and higher levels of VIM compared to Cell Fate 1 (Figure 5B). Studies have suggested that upregulation of endothelial cell associated transcripts such as COL4A2 and VWF can predict the risk of ABMR [24]. The cells from rejection tissue also expressed markers of endothelial activation such as CTGF and CXCL12.
In summary, we have successfully established a pipeline to interrogate limited volume clinical specimens using the Mux-Seq and discovered potentially novel kidney cell subtype markers to explore the sub-cellular heterogeneity in the normal human kidney. This pilot, feasibility study, limited by small sample size, demonstrates the utility of Mux-Seq and Demuxlet for human kidney single cell profiling. In addition to Demuxlet, Cell Hashing has been developed for scRNAseq multiplexing, where cells are labeled with sample specific oligo-tagged antibodies against ubiquitous surface proteins [25]. One of the greatest advantages of Cell Hashing is the ability to multiplex samples of same genotype allowing in further cost reduction. In contrast, Cell Hashing does require an additional step for incubation of the antibodies in preparation of single cells. As cell viability remains a challenge for solid tissue scRNAseq, an additional step might not be desirable in certain experimental conditions. However, both approaches allow researchers to multiplex in a variety of experimental settings. To enable transition of scRNAseq technology to the patient bedside we need to ensure compatibility with clinical samples where tissue quantities by core needle biopsies have to be shared with pathology and are thus necessarily limited. We have ensured feasibility of getting reliable and biologically relevant single-cell data from complex human samples by using tissue sizes in the needle biopsy range. Mux-Seq allows us to establish an optimized pipeline to process and analyze multiple human kidney biopsy samples simultaneously in a cost effective and time efficient manner. Lowering cost and batch-effects by pooling samples while retaining valuable biological information is an important advancement toward the goal of one day making scRNAseq standard practice in the diagnostic workup of kidney biopsies.
Supplementary Material
Figure S1. Histology (H&E staining) of two transplant explants. These transplant kidneys have interstitial and tubular inflammation with preserved tubules and glomeruli. There are characteristic features of chronic antibody mediated rejection (ABMR). Explant 2582 was a living unrelated kidney transplant in a Caucasian female induced with Simulect. The nephrectomy was performed 62 months post-transplant, with 2 biopsy confirmed ABMR episodes at year 4 and year 5 post-transplant. The explant biopsy 2582 (A) was confirmed by pathology to have C4d negative ABMR and TCMR 1A, 20% interstitial fibrosis and tubular atrophy (IFTA) and moderate arteriosclerosis. The estimated glomerular filtration rate (eGFR) at transplant nephrectomy was 13. The explant biopsy 2584 (B) was from a deceased donor renal transplant in an African American male induced with Thymoglobulin, obtained at 22 months post-transplant, with repeated non-adherence. There were 3 biopsy confirmed rejection episodes beginning in the first post-transplant year. In year 2, the explant biopsy confirmed rejection with C4d negative acute ABMR, TCMR, severe IFTA (80%), focal global glomerulosclerosis (40%) and moderate arteriosclerosis. The eGFR at transplant nephrectomy was 4. Both patients received maintenance immunosuppression with steroids, mycophenolate mofetil and tacrolimus.
Figure S2. Images for the tissues used in the study. A) Each tissue was weighed and measured before processing for single cell isolation. B) Number to cells applied to 10X and the output cell number after QC included in the analysis for each sample are listed.
Figure S3. Distribution of cells in each cluster from individual patients shows minimal batch effect. (A) Bar graph shows the distribution of cells from each individual in 38 clusters identified by traditional singleplex-scRNAseq (B) Bar graph showing the distribution of cells from each individual in 39 clusters identified by Mux-Seq.
Figure S4. Gene expression profiles of kidney specific genes obtained by Mux-Seq correlate highly with that from singleplex-scRNAseq. Scatterplot of log transformed average gene expressions of top marker genes for renal parenchymal cells are depicted. Pearson coefficient = 0.973.
Figure S5: Expression of mitochondrial genes is correlated with renal function. (A) Violin plots of mitochondrial transcriptome content by cluster. The red line represents the 50% maximum threshold. (B) Results of gene ontology testing for significantly enriched biological processes. These were derived from the 125 genes that were positively correlated with mitochondrial gene percentage based on Pearson’s correlation (FDR adjusted p-value < 0.05).
Figure S6. Pathway analysis of upregulated and downregulated genes between normal and transplant rejection. A) Network graphic of top 100 upregulated genes in transplant compared to normal (top) and top ten biological pathways found to be enriched (bottom), B) Network graphic of top 100 downregulated genes in transplant compared to normal (top) and top ten biological pathways found to be enriched (bottom). TAL: Thick Ascending Limb, vSMCs: Vascular Smooth Muscle cells, PT: Proximal Tubule, PC: Principle Cells, CNT: Connecting tubule, DCT: Distal Connecting Tubule, IC: Intercalated Cells.
Figure S7. Singleplex-scRNAseq analysis shows distinct clustering of transplant versus normal cells but renal parenchymal cells are better represented by Mux-Seq analysis. (A) UMAP plot of cells from 5 normal (70,028 cells) and 2 transplant nephrectomies (36,732 cells) shows separate clustering of transplant versus normal cells after singleplex-scRNAseq. Cells from the two groups are indicated by different colors (red, normal; green, transplant). (B) Bar graph displays differences proportion of cells in each cluster from transplant versus normal tissue in singleplex-scRNAseq dataset (top) and Mux-Seq dataset (bottom).
Figure S8. RNA transcripts (A), gene count (B) and SNP (C) distribution across clusters for Mux-Seq.
Figure S9. RNA transcripts (A), and gene count (B) distribution across clusters for singleplex-scRNAseq dataset.
Acknowledgements/Funding
We acknowledge the assistance of Zoltan Laszik for providing the H&E staining images for the transplant explant tissue for this study and to Parhom Towfighi for assistance with sample collections. We acknowledge direct support for this study from the NIH/NIDDK KPMP UG3 -DK114937 (MS, TS, AS), with additional support for personnel on this project from NIH- KPMP UWSC11754 (MS), NIH NIAID-HIPC 3U19AI128913 (MS, PB, TS, SS) and NIDDK R01DK109720 (MS, PR, TS).
Abbreviations:
- Mux-Seq
Multiplex single cell RNA sequencing
- PT
Proximal tubule
- scRNAseq
Single cell RNA sequencing
- snRNAseq
Single nuclear RNA sequencing
- SNP
Single nucleotide polymorphism
Footnotes
Disclosure
The authors declare no conflicts of interest.
Supporting Information Statement
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Data Availability Statement
All data has been uploaded on KPMP website and will be available to the public upon publication.
References
- 1.Lee JW, Chou CL, and Knepper MA, Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes. J Am Soc Nephrol, 2015. 26(11): p. 2669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stewart BJ and Clatworthy MR, Applying single-cell technologies to clinical pathology: progress in nephropathology. J Pathol, 2020. 250(5): p. 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Regev A, et al. , The Human Cell Atlas. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potter SS, Single-cell RNA sequencing for the study of development, physiology and disease. Nat Rev Nephrol, 2018. 14(8): p. 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lake BB, et al. , A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat Commun, 2019. 10(1): p. 2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miao Z, et al. , Single cell regulatory landscape of the mouse kidney highlights cellular differentiation programs and disease targets. Nat Commun, 2021. 12(1): p. 2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu H, et al. , Single-Cell Transcriptomics of a Human Kidney Allograft Biopsy Specimen Defines a Diverse Inflammatory Response. J Am Soc Nephrol, 2018. 29(8): p. 2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malone AF, et al. , Harnessing Expressed Single Nucleotide Variation and Single Cell RNA Sequencing To Define Immune Cell Chimerism in the Rejecting Kidney Transplant. J Am Soc Nephrol, 2020. 31(9): p. 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang HM, et al. , Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol, 2018. 36(1): p. 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler A, et al. , Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol, 2018. 36(5): p. 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trapnell C, et al. , The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol, 2014. 32(4): p. 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePasquale EAK, et al. , DoubletDecon: Deconvoluting Doublets from Single-Cell RNA-Sequencing Data. Cell Rep, 2019. 29(6): p. 1718–1727 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung AC and Lan HY, Chemokines in renal injury. J Am Soc Nephrol, 2011. 22(5): p. 802–9. [DOI] [PubMed] [Google Scholar]
- 14.Zhou H, et al. , Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney Int, 2008. 74(5): p. 613–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng M, et al. , Bioinformatics analyses on the immune status of renal transplant patients, a systemic research of renal transplantation. BMC Med Genomics, 2020. 13(1): p. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimm PC, et al. , Clinical rejection is distinguished from subclinical rejection by increased infiltration by a population of activated macrophages. J Am Soc Nephrol, 1999. 10(7): p. 1582–9. [DOI] [PubMed] [Google Scholar]
- 17.Curnova L, et al. , Up-regulation of CD163 expression in subpopulations of blood monocytes after kidney allograft transplantation. Physiol Res, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vereyken EJ, et al. , A shift towards pro-inflammatory CD16+ monocyte subsets with preserved cytokine production potential after kidney transplantation. PLoS One, 2013. 8(7): p. e70152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Bosch TPP, et al. , Pretransplant Numbers of CD16(+) Monocytes as a Novel Biomarker to Predict Acute Rejection After Kidney Transplantation: A Pilot Study. Am J Transplant, 2017. 17(10): p. 2659–2667. [DOI] [PubMed] [Google Scholar]
- 20.Camara NO, Williams WW Jr., and Pacheco-Silva A, Proximal tubular dysfunction as an indicator of chronic graft dysfunction. Braz J Med Biol Res, 2009. 42(3): p. 229–36. [DOI] [PubMed] [Google Scholar]
- 21.Toda N, et al. , CTGF in kidney fibrosis and glomerulonephritis. Inflamm Regen, 2018. 38: p. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Z, et al. , Stromal cell-derived factor-1/CXC chemokine receptor 4 axis in injury repair and renal transplantation. J Int Med Res, 2019. 47(11): p. 5426–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cross AR, Glotz D, and Mooney N, The Role of the Endothelium during Antibody-Mediated Rejection: From Victim to Accomplice. Front Immunol, 2018. 9: p. 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sis B, et al. , Endothelial gene expression in kidney transplants with alloantibody indicates antibody-mediated damage despite lack of C4d staining. Am J Transplant, 2009. 9(10): p. 2312–23. [DOI] [PubMed] [Google Scholar]
- 25.Stoeckius M, et al. , Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol, 2018. 19(1): p. 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Histology (H&E staining) of two transplant explants. These transplant kidneys have interstitial and tubular inflammation with preserved tubules and glomeruli. There are characteristic features of chronic antibody mediated rejection (ABMR). Explant 2582 was a living unrelated kidney transplant in a Caucasian female induced with Simulect. The nephrectomy was performed 62 months post-transplant, with 2 biopsy confirmed ABMR episodes at year 4 and year 5 post-transplant. The explant biopsy 2582 (A) was confirmed by pathology to have C4d negative ABMR and TCMR 1A, 20% interstitial fibrosis and tubular atrophy (IFTA) and moderate arteriosclerosis. The estimated glomerular filtration rate (eGFR) at transplant nephrectomy was 13. The explant biopsy 2584 (B) was from a deceased donor renal transplant in an African American male induced with Thymoglobulin, obtained at 22 months post-transplant, with repeated non-adherence. There were 3 biopsy confirmed rejection episodes beginning in the first post-transplant year. In year 2, the explant biopsy confirmed rejection with C4d negative acute ABMR, TCMR, severe IFTA (80%), focal global glomerulosclerosis (40%) and moderate arteriosclerosis. The eGFR at transplant nephrectomy was 4. Both patients received maintenance immunosuppression with steroids, mycophenolate mofetil and tacrolimus.
Figure S2. Images for the tissues used in the study. A) Each tissue was weighed and measured before processing for single cell isolation. B) Number to cells applied to 10X and the output cell number after QC included in the analysis for each sample are listed.
Figure S3. Distribution of cells in each cluster from individual patients shows minimal batch effect. (A) Bar graph shows the distribution of cells from each individual in 38 clusters identified by traditional singleplex-scRNAseq (B) Bar graph showing the distribution of cells from each individual in 39 clusters identified by Mux-Seq.
Figure S4. Gene expression profiles of kidney specific genes obtained by Mux-Seq correlate highly with that from singleplex-scRNAseq. Scatterplot of log transformed average gene expressions of top marker genes for renal parenchymal cells are depicted. Pearson coefficient = 0.973.
Figure S5: Expression of mitochondrial genes is correlated with renal function. (A) Violin plots of mitochondrial transcriptome content by cluster. The red line represents the 50% maximum threshold. (B) Results of gene ontology testing for significantly enriched biological processes. These were derived from the 125 genes that were positively correlated with mitochondrial gene percentage based on Pearson’s correlation (FDR adjusted p-value < 0.05).
Figure S6. Pathway analysis of upregulated and downregulated genes between normal and transplant rejection. A) Network graphic of top 100 upregulated genes in transplant compared to normal (top) and top ten biological pathways found to be enriched (bottom), B) Network graphic of top 100 downregulated genes in transplant compared to normal (top) and top ten biological pathways found to be enriched (bottom). TAL: Thick Ascending Limb, vSMCs: Vascular Smooth Muscle cells, PT: Proximal Tubule, PC: Principle Cells, CNT: Connecting tubule, DCT: Distal Connecting Tubule, IC: Intercalated Cells.
Figure S7. Singleplex-scRNAseq analysis shows distinct clustering of transplant versus normal cells but renal parenchymal cells are better represented by Mux-Seq analysis. (A) UMAP plot of cells from 5 normal (70,028 cells) and 2 transplant nephrectomies (36,732 cells) shows separate clustering of transplant versus normal cells after singleplex-scRNAseq. Cells from the two groups are indicated by different colors (red, normal; green, transplant). (B) Bar graph displays differences proportion of cells in each cluster from transplant versus normal tissue in singleplex-scRNAseq dataset (top) and Mux-Seq dataset (bottom).
Figure S8. RNA transcripts (A), gene count (B) and SNP (C) distribution across clusters for Mux-Seq.
Figure S9. RNA transcripts (A), and gene count (B) distribution across clusters for singleplex-scRNAseq dataset.
Data Availability Statement
All data has been uploaded on KPMP website and will be available to the public upon publication.