SUMMARY
Influenza A virus (IAV) is a human-infecting pathogen with a history of causing seasonal epidemics and on several occasions worldwide pandemics. Infection by IAV causes a dramatic decrease in host mRNA translation, whereas viral mRNAs are efficiently translated. The IAV mRNAs have a highly conserved 5ʹ-untranslated region (5ʹUTR) that is rich in adenosine residues. We show that the human polyadenylate binding protein 1 (PABP1) binds to the 5’UTR of the viral mRNAs. The interaction of PABP1 with the viral 5ʹUTR makes the translation of viral mRNAs more resistant to canonical cap-dependent translation inhibition than model mRNAs. Additionally, PABP1 bound to the viral 5ʹUTR can recruit eIF4G in an eIF4E-independent manner. These results indicate that PABP1 bound to the viral 5ʹUTR may promote eIF4E-independent translation initiation.
Keywords: Poly(A) binding protein, Eukaryotic Initiation Factor 4G, anisotropy, RT-qPCR, immunoprecipitation
INTRODUCTION
Influenza A virus (IAV) is a zoonotic pathogen capable of infecting the epithelial cells of the upper respiratory tract in humans. Infection by IAV can lead to acute respiratory distress such as coughing, sneezing and even pneumonia and be fatal to those that are most vulnerable in our society [1]. Seasonal influenza is the cause for an estimated 350,000 deaths worldwide each year while IAV pandemics have been known to cause millions of deaths on several occasions in the past century [2,3]. An estimated economic burden of $10.4 billion for direct medical costs and $87.1 billion for the total economic impact per year has been attributed to IAV in the United States alone [4]. These reasons among others make IAV an important subject for research into the mechanisms of infection and viral proliferation to better defend the global community from this disease.
A hallmark of efficient viral infection and proliferation is the ability to direct host resources and cellular machinery towards the production of new virions. Much of our understanding of how cells operate stem from how viruses modulate specific signaling pathways and host proteins. One of the most important mechanisms that a virus must influence is that of mRNA translation, since it is only through viral protein production that more viruses can be made. Host mRNAs have a 7-methyl guanosine cap structure (m7G cap) at the 5ʹ-end and a 3ʹ polyadenosine tail sequence. The m7G cap is recognized by the eukaryotic initiation factor 4E (eIF4E) while the poly(A) tail is bound by multiple polyadenylate binding proteins (PABP1) [5,6]. With both ends of the mRNA bound, subsequent initiation factors can assemble on the mRNA including the eukaryotic initiation factor 4G (eIF4G) that leads to the eventual recruitment of the 40S and 60S ribosomal subunits necessary for translating the mRNA sequence into a protein [5,6].
IAV has evolved its own mechanism to best take advantage of the translation mechanism in host cells. During the course of infection, IAV will release its eight segmented (−)-sense RNAs into the host cell which enter the nucleus. In the nucleus, the viral polymerase subunits bound to each RNA will begin to cleave off the first dozen or so nucleotides of host mRNAs, which include the 5ʹ-m7G cap [7–11]. This cap snatched sequence then serves as a primer to synthesize the (+)-sense mRNA and a final stuttering event on a poly(U) stretch allows for the synthesis of a 3ʹ poly(A) tail [12]. The viral mRNAs are then exported to the cytoplasm where they are believed to be translated in the canonical cap-dependent manner like host mRNAs.
To investigate the mechanism of translation of viral mRNAs, a previous study used RNA-seq and ribosome profiling techniques to monitor IAV mRNA levels and translation, respectively [13]. They observed a strong correlation between the viral mRNA levels and ribosome footprints indicating that viral transcripts are not preferentially translated compared to host mRNAs [13]. Nevertheless, these techniques cannot inform the precise mechanism by which translation initiation occurs on a given mRNA because ribosome profiling only measures the overall distribution of ribosomes on the mRNA as a proxy for translation. Furthermore, it has been suggested that IAV infection will stimulate the mTOR pathway and activate the 4E binding proteins (4EBP) which can sequester eIF4E, in a similar fashion, as found in the case of poliovirus and encephalomyocarditis virus (EMCV) infection [14]. Sequestration of eIF4E by 4EBP prevents association between eIF4E and eIF4G, thus inhibiting canonical cap-dependent translation initiation. This suggests the possibility that viral mRNAs are translated in a manner that can be independent of 5ʹ cap recognition by eIF4E.
In this study, we examine the IAV mRNA sequences found across different strains and show that PABP1 binds to the 5ʹUTR of all eight viral segments. We demonstrate using a cell-free protein synthesis system that the translation of reporter mRNAs containing the viral 5ʹUTR is more resistant to cap-dependent translation inhibition than host mRNAs. We further used immunoprecipitation of PABP1 in IAV-infected cells to demonstrate that PABP1 enriches the 5ʹUTRs of viral mRNAs. We propose that the recognition of the 5ʹUTR by PABP1 serves to recruit eIF4G and the subsequent initiation factors to stimulate the translation of viral mRNAs in IAV-infected cells.
RESULTS
5ʹUTR sequence is conserved across most IAV strains.
To better understand the mechanism by which IAV mRNA are so highly translated, we began by analyzing the sequence identity and conservation of the 5ʹUTRs of each of the eight segments across several strains. To do so, we analyzed all unique IAV sequences that are present in the Influenza Research Database (www.fludb.org) whose sequences begin with the universally conserved sequence: 5ʹ-AGCRAAAGC-3ʹ [15]. The database provided roughly 1000 – 4000 unique sequences for each viral segment. These sequences were separated by the 5ʹUTR length up to the AUG start codon and conservation was analyzed by LOGO (Figure 1A) [16,17]. The results exhibited a significant degree of conservation across the vast majority of IAV strains for each individual segment [18]. In the few cases where a nucleotide was not fully conserved, the alternative possible nucleotides were usually consistent as either a purine or a pyrimidine. Furthermore, the 5ʹUTRs are notably purine rich, primarily made up of adenosines with several segments containing stretches of adenosines (Figure 1B). Minor populations of specific segments (HA, NP, NA and M) with varying lengths were also highly conserved and purine rich (Figure S1). Mutation rates of IAV are known to be quite high and yet the high conservation found in the 5ʹUTRs of its mRNAs suggest an importance to this particular stretch of nucleotides [19–22].
Figure 1.

IAV 5ʹUTR Sequence Conservation. (A) LOGO analysis of the eight IAV 5ʹUTR sections found across strains. Analysis is based on DNA sequencing files and thus the RNA would have an uracil instead of a thymine. For each segment, the last three nucleotides (ATG) correspond to the mRNA start codon. Note that the M1 5ʹUTR is also used by M2, but for simplicity we refer to it as M1. A black line above ATG is used to indicate the start codon. (B) Estimated nucleotide percent representation per IAV 5ʹUTR. The sum of each individual nucleotide was calculated relative to the total number of nucleotides making up the 5ʹUTR. In cases where a position had low conservation, the most conserved nucleotide was chosen for that position and used as part of the calculation. The plot is separated based on individual nucleotide or purine versus pyrimidine prevalence.
PABP1 has a significant affinity for the M1 portion of the 5ʹUTR.
Given the richness in adenosines of these UTRs, we wondered whether PABP1, which is known to bind poly(A) and to a lesser extent poly(G) sequences will bind to the 5ʹUTR [23–25]. Previous studies have shown that during the cap snatching process the 5ʹUTR of the M1/M2 segment (referred to as M1 in the text) contains the first 12 nucleotides of the U2 snRNA [8–11]. We were curious therefore, to see how PABP1 would interact with an RNA segment of the M1 5ʹUTR with or without the cap-snatched sequence. Therefore, we purified recombinant human PABP1 and synthesized the RNA of the M1 5ʹUTR segment for A/WSN/1933 (H1N1) strain, with and without the U2 snRNA cap-snatched sequence (Figure 2A). An EMSA was performed to qualitatively analyze the binding of PABP1 to the M1 5ʹUTR and control RNAs. We observed a shift of the Poly(A)18 RNA and M1 5ʹUTR with and without the cap-snatch sequence in the presence of PABP1 (Figure 2B). PABP1 binds as a monomer and dimer to Poly(A)18 RNA, which was resolved by the EMSA [24,26]. A shift was also observed with the U2 snRNA cap-snatch sequence and PABP1, but to a lesser extent. No shift was observed with a single-stranded control RNA (ssCR1) in the presence of PABP1. We note that the electrophoretic mobility of the PABP1 bound RNAs is not entirely consistent with the size of the RNAs suggesting that the conformation of the PABP1•RNA complexes is affecting the extent of the shift in the EMSA. Nevertheless, our results indicate that PABP1 binds to the M1 5ʹUTR and to a lesser extent to the cap-snatched sequence. We next performed fluorescence anisotropy studies to quantitatively determine the binding affinity of PABP1 for M1 5ʹUTR. Fluorescence anisotropy studies showed that PABP1 binds to the M1 5ʹUTR with an equilibrium dissociation constant (KD) of 86 ± 7 nM, regardless of the presence of the cap snatch sequences (Figure 2C and Table S1). Despite the affinity being an order of magnitude weaker than PABP1’s affinity to poly(A) sequences, the binding affinity suggest biological relevance given the high concentration (~ 4 μM) of PABP1 found in the cell [27]. Absence of PABP1 binding to other control RNAs demonstrates the specificity it has for the M1 5ʹUTR sequence (Figures 2C and S2 and Table S2).
Figure 2.
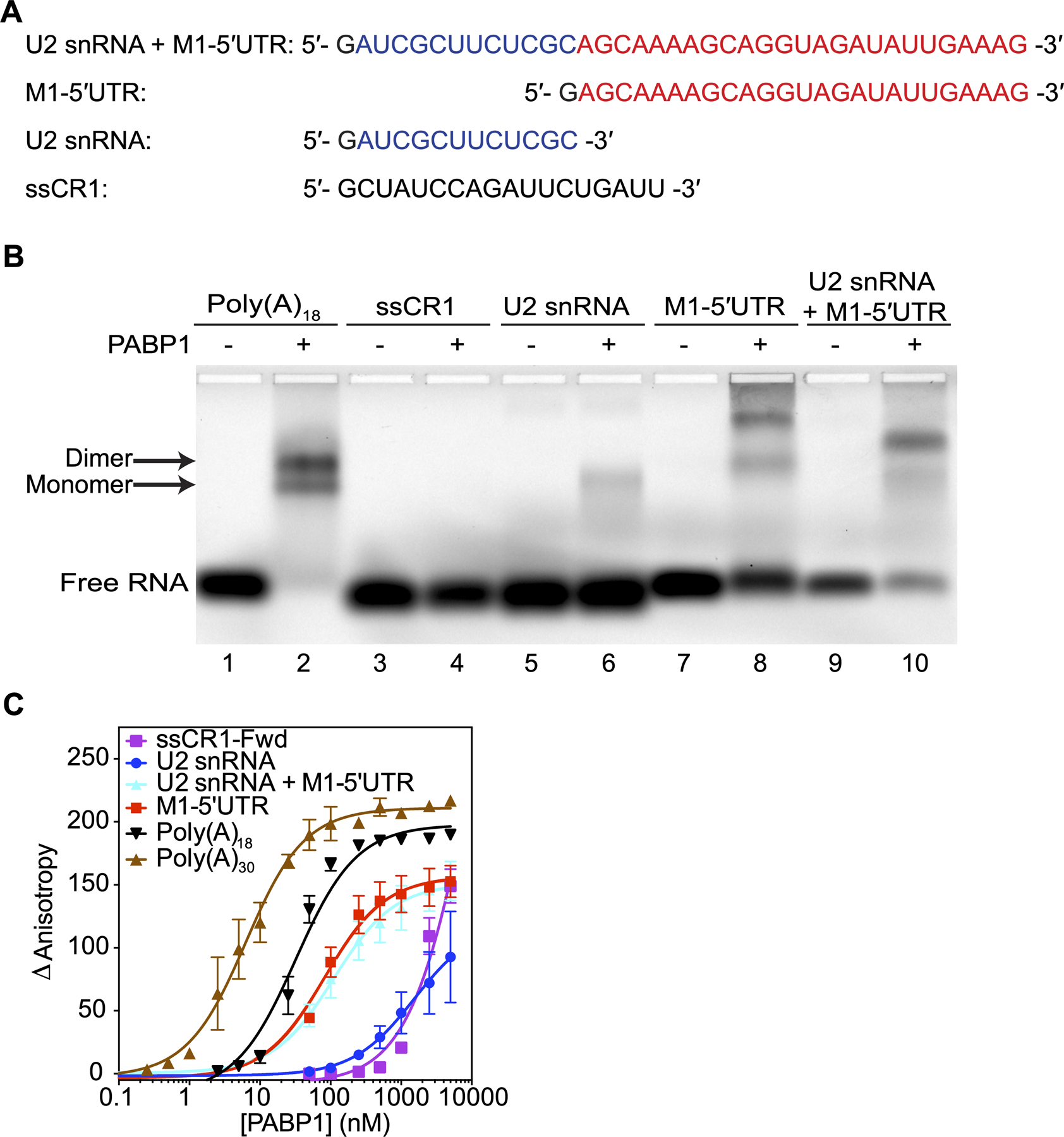
PABP1 binds to the 5ʹUTR of M1 mRNA. (A) The sequences used in the binding studies. The U2 snRNA (blue) and M1–5ʹUTR (red) sequences are color coded for visual clarity. The first guanosine residue (black) is due to T7 transcription and is not part of the native sequence. (B) EMSA assay comparing the binding of PABP1 to the sections that make up the M1 5ʹUTR RNA and control RNAs. Plus sign indicates 500 nM of PABP1 was added to the reaction. Minus sign indicates no PABP1 was added to the reaction. Arrow points to the shifted PABP1 monomer•Poly(A)18 complex and PABP1 dimer•Poly(A)18 complex. (C) Anisotropy assay of PABP1 binding to the sections that make up the M1 5ʹUTR RNA and control RNAs. The final concentration of the RNAs was 1 nM [Poly(A)18, Poly(A)30, and M1–5ʹUTR] and 10 nM [ssCR1-Fwd, U2 snRNA, and U2 snRNA+M1–5ʹUTR]. The final concentration of PABP1 was increased from 0 to 5 μM. The change in anisotropy is shown on the y-axis. The error bars represent the SEM from three independent experiments.
PABP1 has varying affinities for the eight IAV 5ʹUTRs.
We were curious then to see how PABP1 interacted with the 5’UTRs of the remaining IAV segments of A/WSN/1933 (H1N1). The seven 5ʹUTR RNAs were synthesized without the cap-snatch sequence and binding to PABP1 was analyzed using EMSA and fluorescence anisotropy. Our studies showed that PABP1 has varying affinities to each IAV 5ʹUTR ranging from 20 nM up to 1 μM (Figure 3 and Table S3). Furthermore, we measured the binding affinity PABP1 has to the eight 5ʹUTRs of two other human-infecting strains used most prevalently in the literature, namely A/Puerto Rico/8/34 (H1N1) and A/Udorn/307/1972 (H3N2) (Figures S3 and S4, Tables S4 and S5). Our studies showed that PABP1 binds to the 5ʹUTRs of all eight segments from the two additional IAV strains.
Figure 3.
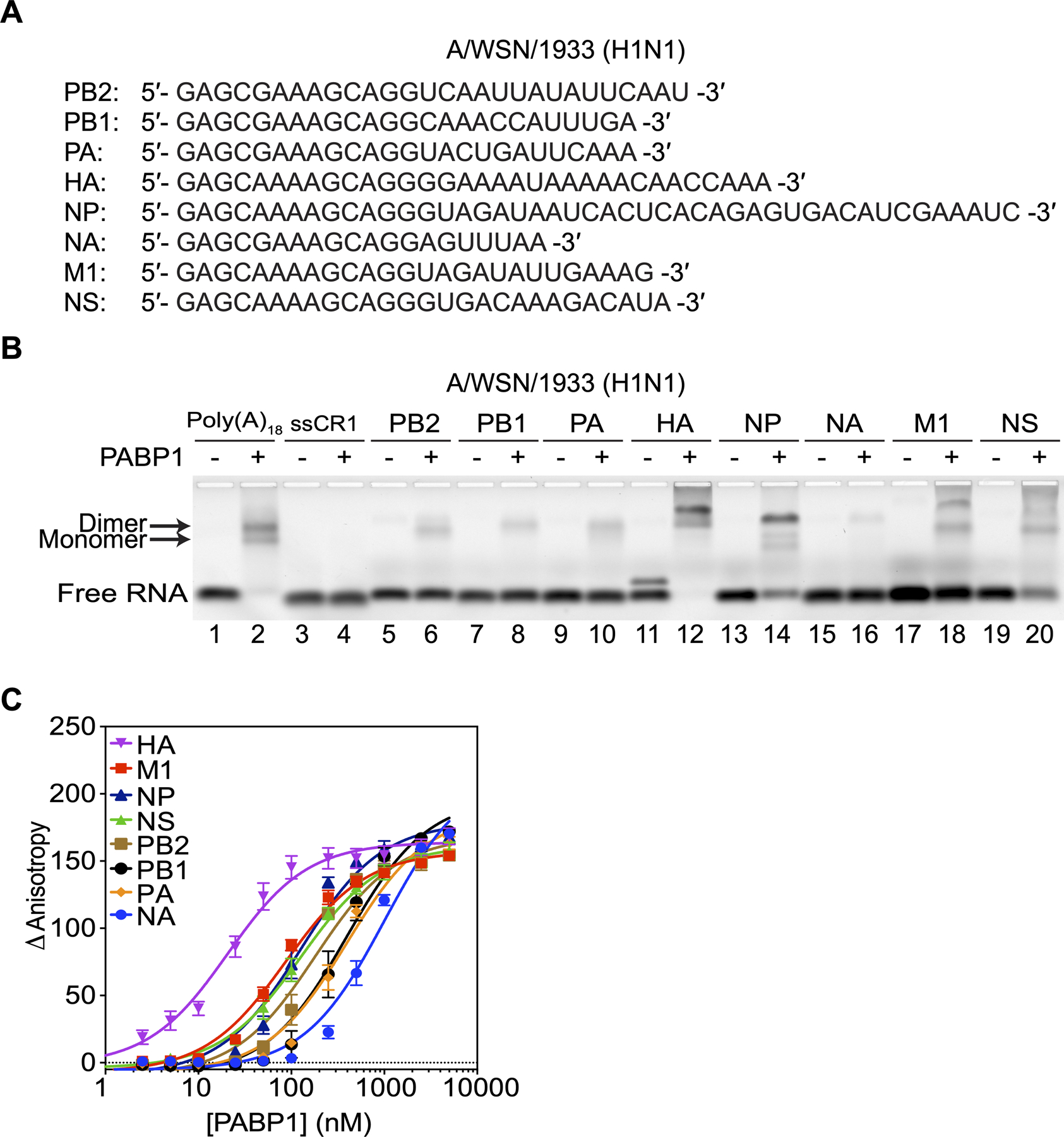
PABP1 binds to the 5ʹUTR of all eight segments of A/WSN/1933 (H1N1) IAV. (A) The sequences of the 5ʹUTRs of A/WSN/1933 (H1N1) used in the binding studies. The first guanosine residue (black) is due to T7 transcription and is not part of the native sequence. (B) EMSA assay comparing the binding of PABP1 to the different 5ʹUTR RNA of each IAV segment. Minus sign indicates no PABP1 was added to the reaction. Arrow points to the shifted PABP1•Poly(A)18 complexes. (C) Anisotropy assay to determine the binding affinity of PABP1 for the different 5ʹUTR RNA of each IAV segment. The final concentration of the RNAs was 1 nM, and the final concentration of PABP1 was increased from 0 to 5 μM. The change in anisotropy is shown on the y-axis. The error bars represent the SEM from three independent experiments.
Translation initiation with IAV 5ʹUTR are resistant to cap-dependent downregulation.
Given the evidence that PABP1 binds to the 5ʹUTRs with significant affinity, we set out to examine the biological relevance of this binding. We hypothesized that due to PABP1’s canonical role of initiating translation by binding to the 3ʹ end, perhaps it could do the same for viral mRNAs from the 5ʹ end. To test this, we took the viral 5ʹUTR sequences of M1 with and without the cap snatch sequence (shown in Figure 2A) and incorporated them upstream of a Renilla luciferase coding sequence with a poly(A)25 sequence at the 3ʹ end. The viral 5ʹUTR containing Renilla luciferase mRNAs were synthesized by in vitro transcription. Additionally, we made a control Renilla luciferase mRNA with the Kozak sequence (5ʹ-GCCACCAUG-3ʹ), which is known to be found in the 5ʹUTR of highly expressed mRNAs [28]. All mRNAs were capped at the 5ʹ end with a m7G cap using the vaccinia virus capping enzyme. We tested the baseline Renilla expression of these mRNAs in a HeLa cell-derived in vitro translation system (IVTS) and found that the IAV 5’UTR sequences do not confer any significant advantage compared to the Kozak driven mRNA (Figure 4A) [29]. This suggests that under normal cellular conditions viral mRNAs would not have any advantage over host mRNA with regard to translation.
Figure 4.
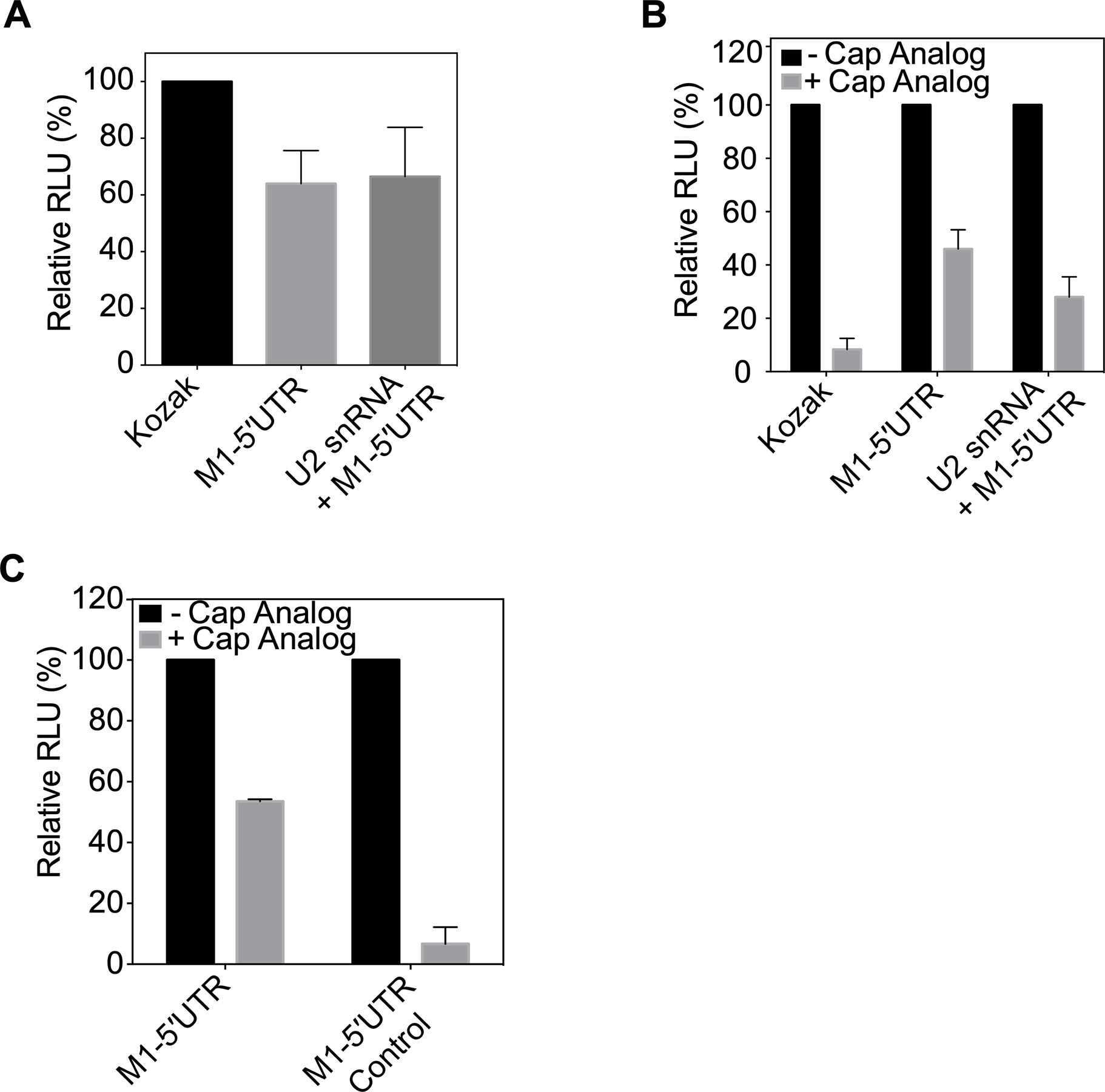
The IAV 5ʹUTR confer resistance to cap-dependent translation inhibition. (A) Translation of Kozak sequence driven Renilla luciferase mRNA, M1–5ʹUTR driven Renilla luciferase mRNA, and U2 snRNA + M1–5ʹUTR driven Renilla luciferase mRNA in HeLa lysate monitored by relative luminescence units (RLU). RNAs are capped at the 5’ end with m7G and have a 25-nucleotide long polyadenosine tail at the 3’ end. The RLU was normalized relative to the control Kozak driven Renilla mRNA translation. (B) Translation of Kozak sequence driven Renilla luciferase mRNA, M1–5ʹUTR driven Renilla luciferase mRNA, and U2 snRNA + M1–5ʹUTR driven Renilla luciferase mRNA in HeLa lysate in the presence or absence of 1 mM m7G cap analog. RNAs are capped at the 5ʹ-end with m7G and have a 25-nucleotide long polyadenosine tail at the 3ʹ-end. In each case, the RLU with the cap analog was normalized to the absence of cap analog arbitrarily set as 100%. (C) Translation of M1–5ʹUTR driven Renilla luciferase mRNA, and M1–5ʹUTR Control driven Renilla luciferase mRNA in HeLa lysate in the presence or absence of 1 mM m7G cap analog. RNAs are capped at the 5ʹ-end with m7G and have a 25-nucleotide long polyadenosine tail at the 3ʹ-end. The RLU in the presence of the cap analog was normalized relative to the signal in the absence of the cap analog arbitrarily set as 100%. The standard deviations from three experiments are shown.
Interestingly, previous studies have shown that the translation of influenza viral mRNA is independent of eIF4E activity [14,30–32]. We therefore determined how expression of the M1 5ʹUTR mRNAs handled shutoff of eIF4E driven translation initiation by including an m7G cap analog in the IVTS (Figures 4B and S5) [33]. Results show that 90% of the Kozak driven mRNA translation is inhibited, whereas only ~50% of the translation of the mRNA driven by the M1 5’UTR is affected by the presence of the analog. The same trend was found to be true when comparing the relative translation of the same mRNAs that were capped and poly(A) tailed to versions that contained no cap and a long randomized 3ʹUTR sequence (Figure S5B). This suggests, that while knockdown of cap-dependent translation creates an environment that decreases translation for all mRNAs, it does give mRNAs with the viral 5ʹUTR sequence a distinct advantage over the host mRNAs. We also used a bicistronic RNA where the varying 5ʹUTRs and the cricket-paralysis virus (CrPV) IRES drove the production of firefly and Renilla luciferase, respectively (Figure S5D). By measuring the production of firefly luciferase and normalizing it to the production of the CrPV IRES driven Renilla luciferase, we found that the downregulation of eIF4E inhibited translation of the Kozak driven mRNA more than the M1–5ʹUTR driven mRNA. In fact, the translation of the varying 5ʹUTR of the bicistronic mRNAs resemble those of the monocistronic mRNAs, indicating the importance of the 5ʹUTR sequence on its resistance to cap-dependent translation inhibition.
We wanted to see whether this translational advantage during eIF4E inhibition is also observed with the eight different 5ʹUTRs of the IAV segments. We thus incorporated the different 5ʹUTRs of each IAV viral segment upstream of the monocistronic Renilla mRNA reporter. The reporter mRNAs containing the 5ʹUTR of NS, HA, M1, and NP IAV segments were found to be more resistant to the presence of the cap analog compared to the Kozak sequence (Figure S5C). These UTRs also have the highest binding affinity for PABP1 (Table S3). Our results suggest that if eIF4E is sequestered or inhibited during the course of IAV infection, several of the viral mRNAs are capable of overcoming cap-dependent translation inhibition.
To further validate that the translational levels observed in the in vitro translation system is due to PABP1 binding to the 5ʹUTR, we compared the translational efficiency of an M1 5’UTR driven Renilla construct with a Renilla sequence whose 5ʹUTR is the complementary sequence of the M1 5ʹUTR (Control). Binding studies reveal that PABP1 does not bind with significant affinity to the M1 5ʹUTR Control (Figure S6). The translation assay showed that the M1–5ʹUTR Control driven mRNA is more susceptible to knocking down cap-dependent translation initiation than the M1–5ʹUTR driven mRNA sequence (Figure 4C).
eIF4G binds to PABP1•IAV 5ʹUTR complex.
During canonical cap-dependent translation initiation, PABP1 utilizes RRM1 and RRM2 to bind to the poly(A) tail and interacts with eIF4G bound to the 5ʹ-end of the mRNA. Studies have shown that eIF4G will bind to RRM2 and that this interaction is allosterically driven by PABP1 binding to its target RNA sequence [34]. For IAV to utilize PABP1 for translation initiation from the 5ʹ end of the mRNA, it may recruit eIF4G to the 5ʹ-end in an eIF4E independent manner. To test this hypothesis, we purified a fragment of eIF4G (88–653) which contains the PABP1 binding site and confirmed its ability to recognize PABP1 on a poly(A) sequence by EMSA (Figure 5A, indicated by blue circle). We then tested the binding of eIF4G to PABP1 bound to the M1 5ʹUTR by EMSA. At low concentration of PABP1, we observed the monomer of PABP1 bound to the M1 5ʹUTR (Figure 5B, indicated by the red circle in lane 3). While at higher concentration of PABP1, we observed the dimer of PABP1 bound to the M1 5ʹUTR (Figure 5B, indicated by the purple circle in lane 5). This is consistent with previous data showing that at high concentrations PABP1 binds as a dimer to RNA [24,26,35]. In both cases, the presence of eIF4G causes the PABP1 RNA complex to shift up even further (Figure 5B, indicated by the blue and green circles in lanes 4 and 6, respectively). In the absence of PABP1, eIF4G has weak affinity for the M1 5ʹUTR (Figure 5B, lane 2). However, in the presence of PABP1, eIF4G cannot bind to the M1 5ʹUTR because PABP1 binds with higher affinity to the RNA and it will cover about 30 nucleotides thereby blocking access to the 26 nucleotide M1 5ʹUTR [23]. Thus, PABP1 bound to the M1 5ʹUTR recruits eIF4G by protein-protein interaction. Our results suggest that PABP1 is capable of binding to the 5ʹ-end of the viral mRNA and recruiting eIF4G, which should be sufficient for the assembly of the subsequent initiation factors and the ribosome to commence translation.
Figure 5.
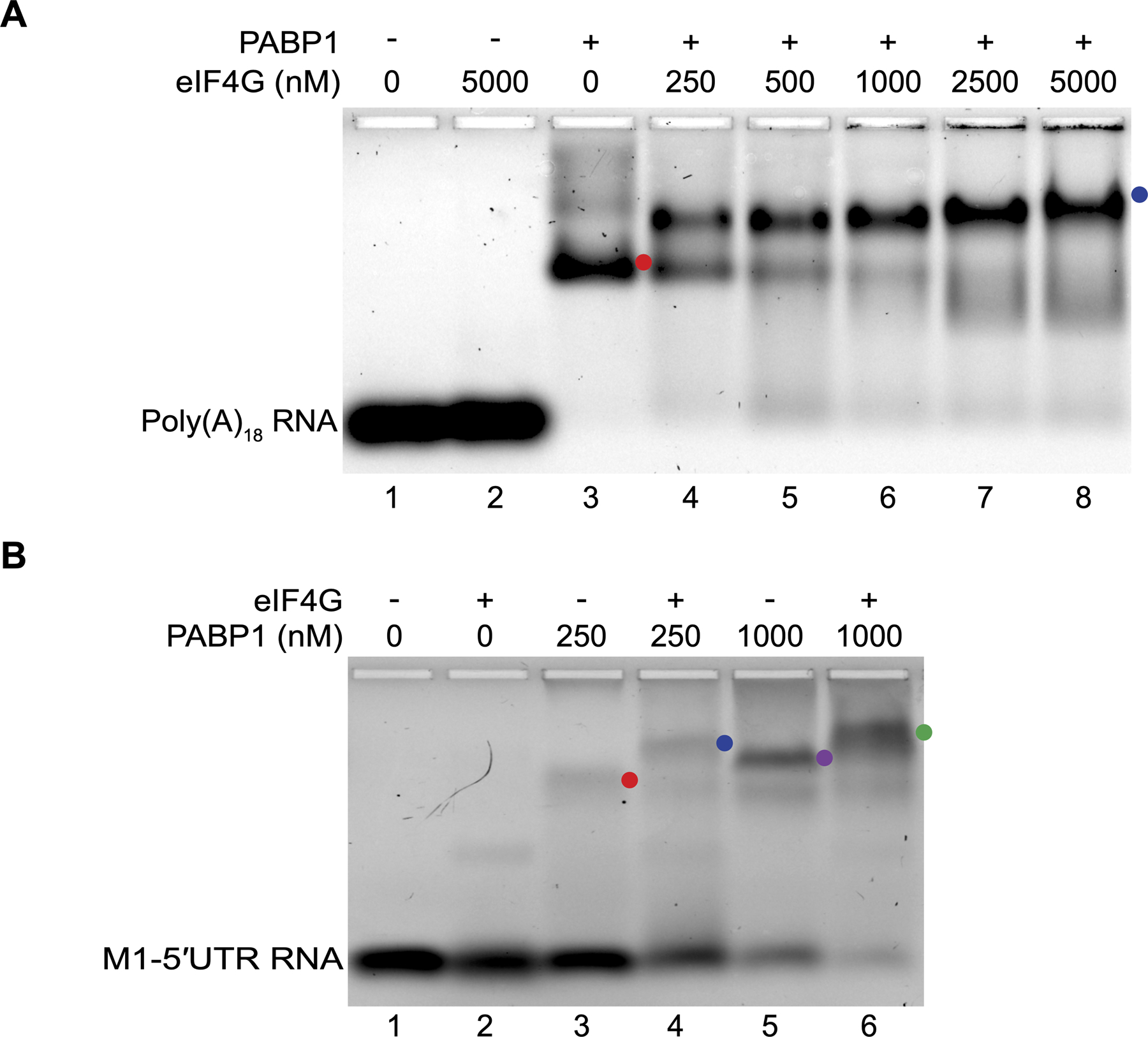
eIF4G is recruited by PABP1 to the 5ʹ-UTR of M1 mRNA. (A) EMSA assay monitoring the binding of PABP1 to poly(A)18 RNA in the presence of varying amounts of eIF4G. Minus sign indicates no PABP1 was added to the reaction. The plus sign indicates 250 nM PABP1 was added to the reaction. The red circle points to the shifted PABP1•poly(A)18 complex (lane 3) and the blue circle points to the PABP1•poly(A)18•eIF4G complex (lanes 4 to 8). (B) EMSA assay monitoring the binding of PABP1 to M1–5ʹUTR RNA in the presence or absence of eIF4G. Minus sign indicates no eIF4G was added to the reaction. The plus sign indicates 5 μM eIF4G was added to the reaction. The red circle points to the PABP1 monomer•M1–5ʹUTR complex (lane 3), the blue circle points to the PABP1 monomer•M1–5ʹUTR•eIF4G complex (lane 4), the purple circle points PABP1 dimer•M1–5ʹUTR complex (lane 5), and the green circle points to the PABP1 dimer•M1–5ʹUTR •eIF4G complex (lane 6).
PABP1 is enriched on IAV 5ʹUTR sequences in infected cells.
We used immunoprecipitation (IP) and quantitative reverse transcription PCR (RT-qPCR) to determine whether PABP1 would preferentially enrich RNA pools with the IAV 5ʹUTR. To this end, we took A549 cells that were infected for 24 hours with the A/Puerto Rico/8 (H1N1) virus (MOI = 0.5) and used formaldehyde to crosslink proteins bound to RNA sequences. We did not observe significant cell death and were able to recover more viral mRNAs for subsequent analysis under these conditions. We used sonication to lyse the cells and fragment the RNA to shortened sequences of 100 nt to 500 nt in length. Using an antibody against PABP1, we performed an IP reaction to pull down PABP1 and any RNAs it was bound to during infection. After isolating the RNAs by reversing the crosslinks, we used RT-qPCR to amplify different portions of the IAV segment to examine the relative preference PABP1 has for different portions of an mRNA (Figures 6A and 6B). Using the actin gene as a housekeeping control, we find that the PABP1 IP enriches the RNA pool with the 3ʹ-end of the mRNA by about 4-fold higher than the middle region. This trend is expected given that the 3ʹ end of the actin mRNA abuts the poly(A) tail and should be enriched during the IP. We also find that these results are consistent regardless of whether the cells were MOCK infected or IAV-infected. Furthermore, we examined the abundance of the 5ʹ-end of the actin mRNA in the IP to test whether the closed loop model of translation initiation would cause the IP to enrich the 5ʹ-ends of mRNAs (Figure 6C). We find that the relative enrichment of the 5ʹ mRNA ends are 5-fold less than the 3ʹ-ends indicating that the background we may encounter due to the closed loop model is very low [36].
Figure 6.
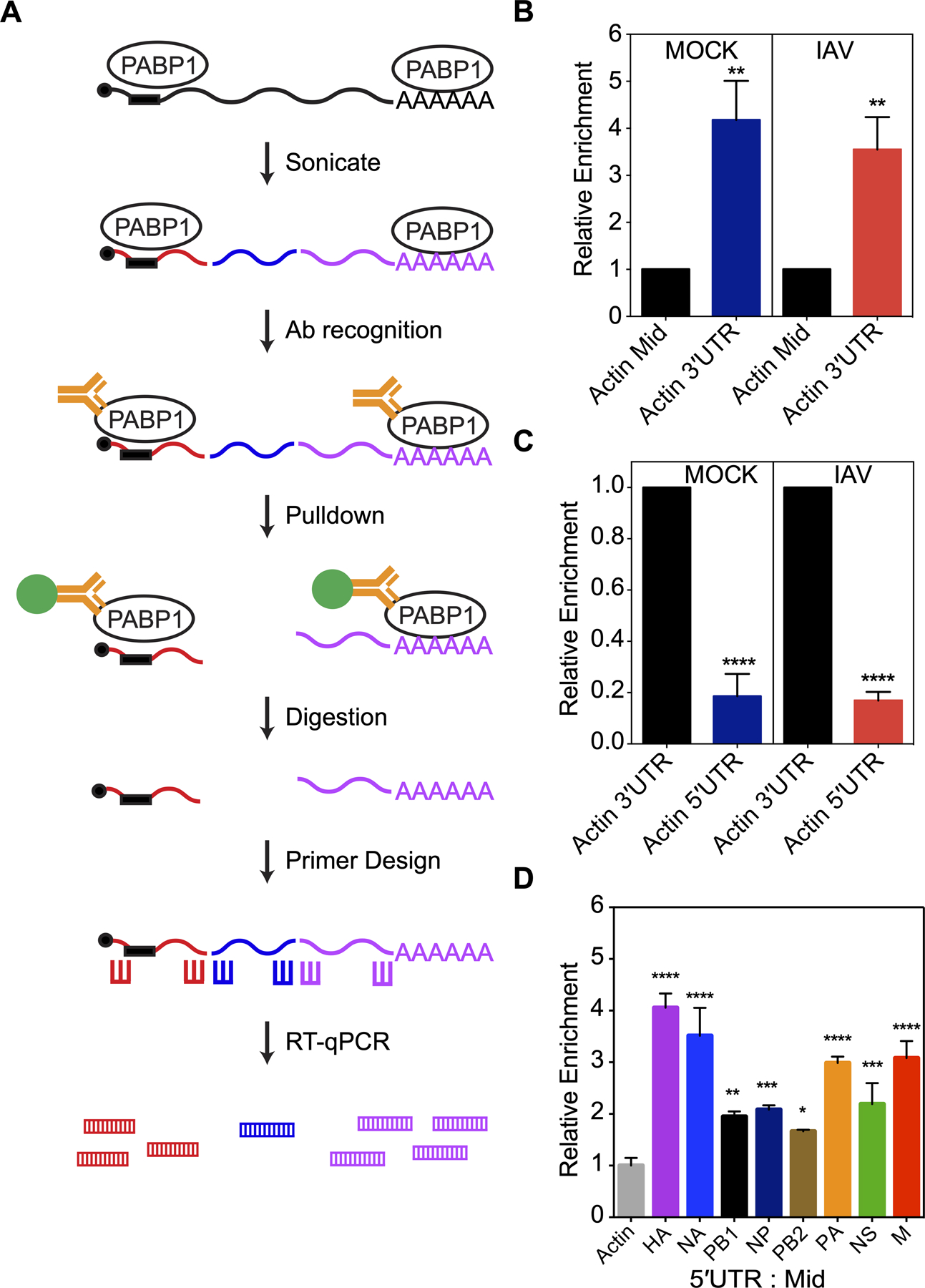
PABP1 pulldown from IAV-infected cells enriches for viral 5ʹUTR RNAs. (A) The workflow of the immunoprecipitation experiment from live IAV-infected cells. Briefly, lysate containing PABP1 crosslinked to RNA are sheared into fragments by the sonication step, followed by recognition of PABP1 by specific antibody and pulldown of the PABP1•RNA complex. Proteins are digested and total RNA fragments are purified. Primers for reverse transcription and RT-qPCR were designed to amplify the 5ʹ-, 3ʹ- and middle section of the mRNAs of interest. RT-qPCR was performed to determine the enrichment for the 5ʹ-, 3ʹ- and middle section of selected mRNAs. (B) Results of RT-qPCR comparing the enrichment of actin mRNA fragments from the 3ʹ-end (Actin 3ʹUTR) relative to the middle fragments (Actin Mid) set to 1. MOCK refers to cells that were mock infected while IAV refers to cells infected with A/Puerto Rico/8/34 (H1N1) IAV strain. (C) Results of RT-qPCR comparing the enrichment of actin mRNA fragments from the 5ʹ-end (Actin 5ʹUTR) relative to fragments from the 3ʹ-end (Actin 3ʹUTR) set to 1. MOCK refers to cells that were mock infected while IAV refers to cells infected with A/Puerto Rico/8/34 (H1N1) IAV strain. (D) Results of RT-qPCR comparing the enrichment of RNA fragments from the 5ʹ-end relative to fragments from the middle of IAV mRNAs. The data was normalized relative to the actin mRNA fragments from the 5ʹ-end over middle set to 1 and the error was propagated for all conditions. The cells were infected with A/Puerto Rico/8/34 (H1N1). The standard deviations from three biological replicates are shown. P < 0.05 is denoted with one asterisk, P < 0.01 is denoted with two asterisks, P < 0.001 is denoted with three asterisks, and P < 0.0001 is denoted with four asterisks.
Next, we analyzed the eight IAV mRNAs coding for HA, NA, PB1, NP, PB2, PA, NS and M by IP and RT-qPCR. Examination of the relative enrichment of the IAV mRNA 5ʹ ends shows a 2- to 4-fold increase over that of actin mRNA when normalized against the middle sections of the same mRNA (Figures 6D). We also compared the 5ʹ-end enrichment of tubulin mRNA, a different housekeeping gene, and found the relative differences to be similar (Figure S7). These results indicate that the IP of PABP1 is enriching the RNA pool with the 5ʹ-ends of each viral mRNA in line with what might be expected if PABP1 is bound to the 5ʹUTR.
DISCUSSION
Canonical eukaryotic translation initiation is an intricate process and involves more than ten initiation factors. One of the key steps during initiation is the recruitment of the 43S preinitiation complex to the 5ʹ-end of an mRNA by eIF4F (Figure 7). Mammalian eIF4F is composed of eIF4E, eIF4G, and eIF4A subunits. The eIF4E subunit binds to the m7G cap structure present at the 5ʹ-end of cellular mRNAs; therefore, it is responsible for the placement of eIF4F at the 5ʹ-end. The eIF4G subunit is a large scaffolding protein that interacts with eIF4E, eIF4A, mRNA, 43S preinitiation complex (via eIF3), and PABP1 bound to the 3ʹ-end of the mRNA. Finally, the eIF4A subunit is an RNA helicase that melts RNA structures in the 5ʹUTR, which facilitates the recruitment of the 43S preinitiation complex. Interestingly, the concentration of eIF4E in the cell is low and it is the limiting factor for translation initiation [6]. Additionally, the activity of eIF4E is regulated by a phosphorylation/dephosphorylation cycle and by 4EBP [6]. Thus, eIF4E serves as a regulatory hub for translation initiation.
Figure 7.
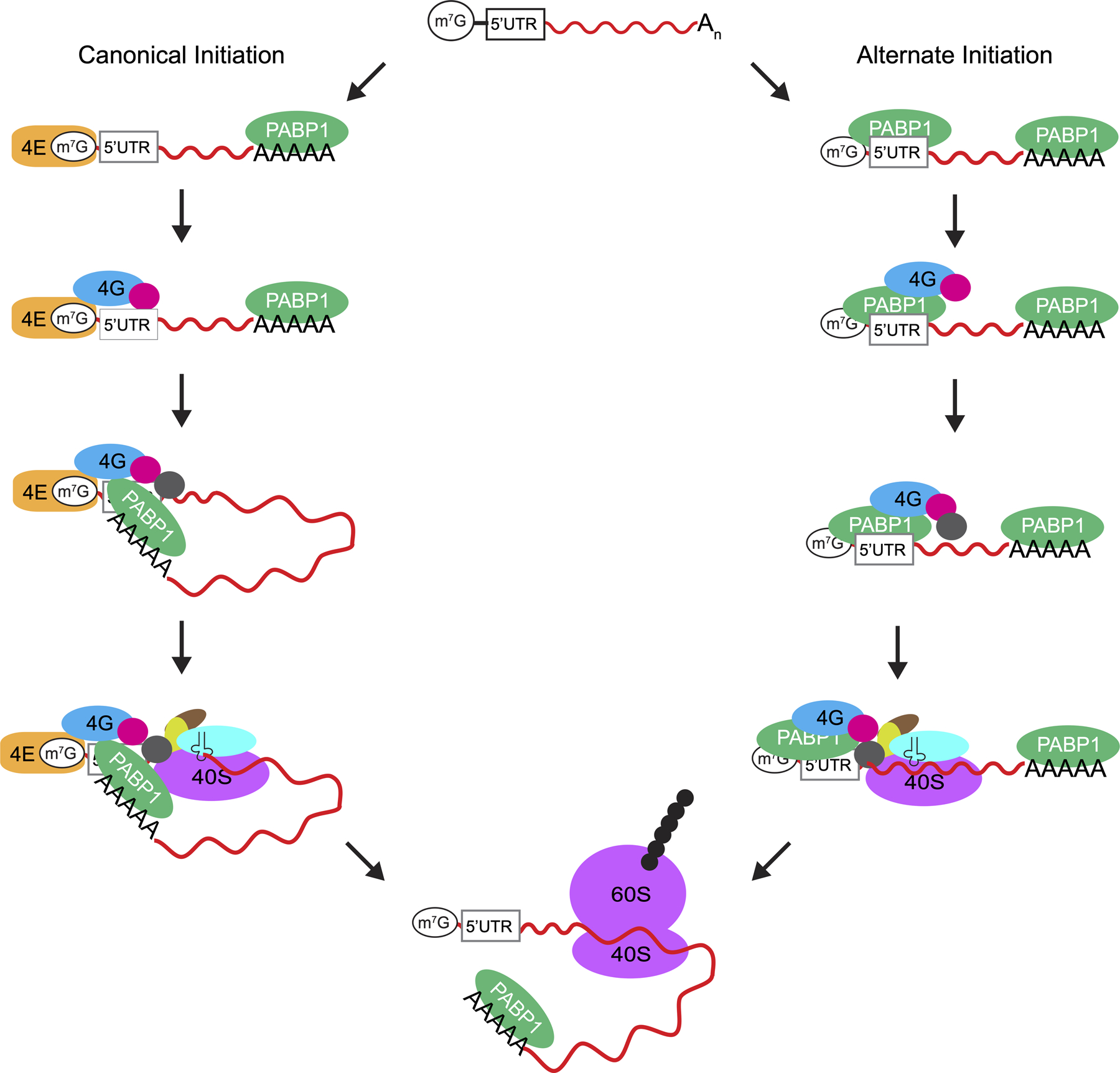
Models for translation initiation on IAV mRNAs. The IAV mRNAs have a m7G cap structure at the 5ʹ-end, a highly conserved 5ʹUTR and a poly(A) tail at the 3ʹ-end. During canonical translation initiation shown on the left, PABP1 binds to the 3ʹ poly(A) tail, and eIF4F consisting of eIF4E, eIF4G and eIF4A subunits binds to the m7G cap at the 5ʹ-end (for clarity the binding of eIF4E subunit to the m7G cap is shown as a separate step). This is followed by mRNA circularization by the interaction of eIF4G with PABP1 and the recruitment of the 43S preinitiation complex to the mRNA by eIF4F. In the alternate mechanism of initiation shown on the right, PABP1 binds both to the 3ʹ poly(A) tail and to the viral 5ʹUTR. PABP1 bound to the viral 5ʹUTR then recruits eIF4G and eIF4A. Finally, the 43S preinitiation complex is recruited by eIF4G to initiate translation in an eIF4E-independent manner.
Many viruses subvert canonical translation initiation in order to direct ribosome assembly onto viral mRNA sequences. For example, EMCV activates 4EBP and poliovirus encodes a protease that cleaves eIF4G [37]. Both strategies neutralize eIF4E-dependent translation initiation of capped host mRNAs. These viruses contain an IRES in the 5ʹUTR regions of their mRNA that directly recruit eIF4G in the case of EMCV or the cleaved eIF4G fragment in the case of poliovirus to proceed with translation initiation [38]. Many such IRES’s have been discovered in viral mRNAs whose ability to recruit initiation factors and the ribosome without the need for eIF4E give them a clear advantage over host mRNA translation [39]. To date no IRES-like activity for the IAV 5’UTR has been reported. However, numerous studies have shown that the IAV mRNAs are efficiently translated, whereas the translation of the host mRNAs is dramatically attenuated in infected cells [31,40–45]. This is explained by the inhibition of host RNA polymerase II, and the degradation of host mRNAs after viral infection [7,46–49]. Viral mRNAs escape the fate of the host mRNAs because they are transcribed by the viral polymerase, have a 5ʹ-cap obtained from host mRNAs, and a 3ʹ poly(A) tail added by a specialized process [50–59].
Even though the above-described processes reduce the amount of host mRNAs, there are still significant amounts of host mRNAs in the cytoplasm of infected cells, especially during the early phase of infection that will compete with viral mRNAs for translation [46]. More importantly, IAV infection activates the cellular stress response pathways, which will inhibit global translation [30,37,60–64]. One of the mechanisms for inhibiting global translation during the stress response is by the dephosphorylation of eIF4E and the activation of 4EBPs [30,37,60–64]. The dephosphorylation of eIF4E or the binding of 4EBPs to eIF4E reduces the activity of eIF4E and inhibit canonical cap-dependent mRNA translation [30,37,60–64]. However, many stress response mRNAs escape the inhibition of cap-dependent translation by recruiting PABP1 to their 5ʹ-UTR to initiate translation in an eIF4E-independent mechanism [33].
We propose that IAV mRNAs also use an eIF4E-independent mechanism for translation initiation to compete with cellular mRNAs under virus-induced stress conditions. Indeed, previous studies have shown that downregulating eIF4E during the course of IAV infection does not hinder the overall translation of the viral proteins [14,30–32]. These observations can be more fully explained by our findings since PABP1 recognition of the 5ʹUTR is capable of recruiting eIF4G and thus exclude the need for cap-recognition by eIF4E (Figure 7). Additionally, viral mRNA translation may be enhanced because eIF4E, which is the limiting factor for canonical translation initiation, is not needed for translation initiated by PABP1 binding to the 5ʹUTR.
The high sequence conservation of the 5ʹUTR comes as no major surprise considering other studies have also noted the conservation of both the 5ʹ- and 3ʹ-ends of IAV segments allow for the panhandle structure to form for the negative stranded RNA [65]. While this sequence is known to be important for viral replication, our studies suggest that the complementary sequence present in the IAV mRNAs may additionally be important for translation. The conservation of A-rich sequence identity in the 5ʹUTR may be to ensure favorable binding to PABP1 [66,67]. In summary, our unexpected discovery that PABP1 binds with high affinity to the conserved sequence present in the viral 5ʹUTR and recruits eIF4G to initiate translation is consistent with previous reports that IAV mRNA translation is resistant to eIF4E inhibition [14,30–32]. The alternative mechanism could be used to enhance viral mRNA translation in competition with host mRNAs especially when the activity of eIF4E is reduced [68]. More studies are needed to understand this alternative mechanism of translation initiation on viral mRNAs, which could be targeted to treat IAV infection.
METHODS
LOGO Analysis.
We performed a sequence search on www.fludb.org for all eight IAV segments individually [15]. Search results were filtered based on human-infecting IAV strains with duplicate sequences removed by the websites search parameter options. Minimum sequence lengths were also included to enrich results containing the respective 5ʹUTRs of each segment. Lengths are as follows; PB2 = 2341 nucleotides, PB1 = 2339 nucleotides, PA = 2233 nucleotides, HA = 1760 nucleotides, NP = 1565 nucleotides, NA = 1466 nucleotides, M1 = 1011 nucleotides, and NS = 889 nucleotides. Resulting sequences were trimmed after the first ATG of the sequence and the results were separated by length. Sequences were then analyzed using weblogo.berkeley.edu/logo.cgi.
Purification of Human PABP1.
Human PABP1 (GenBank accession no. BC015958) in the pANT7_cGST vector was purchased from DNASU. The PABP1 gene was subcloned into pMCSG26, which contains a C-terminal six-His tag [69,70]. E. coli Rosetta 2 (DE3) pLysS cells (Millipore) were transformed with pMCSG26-PABP1 construct. The cells were grown at 37 °C in LB/ampicillin/chloramphenicol to an OD600 of 0.6−0.8, cooled to 18 °C, and then induced with 0.25 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 12−18 h. Cells were pelleted, flash-frozen, and stored at −80 °C. Cells were resuspended in PABP1 lysis buffer [25 mM Tris (pH 7.5), 250 mM NaCl, 10% (v/v) glycerol, 8 mM DTT, 0.5 mM EDTA, 1 mM PMSF, 0.1% (v/v) Triton X-100, and 5 mM imidazole] and disrupted by sonication. The cell lysate was centrifuged at 20000 × g for 45 min at 4 °C. The supernatant was incubated with 4 mL of Ni-NTA beads for 15 min at 4 °C on a rotator. The slurry was poured over a column and washed with 50 mL of PABP1 wash buffer (lysis buffer with 20 mM imidazole and 1 mg/mL heparin sodium salt from porcine intestinal mucosa (Sigma)). Protein was eluted with PABP1 elution buffer [25 mM Tris (pH 7.5), 250 mM NaCl, 10% (v/v) glycerol, 8 mM DTT, 0.5 mM EDTA, and 250 mM imidazole]. Fractions were collected and concentrated using a 50K MWCO concentrator until the volume was 1 mL. The protein sample was filtered and further purified using a Superdex 16/60 200 pg column at a flow rate of 1 mL/min using PABP1 storage buffer [25 mM Tris (pH 7.5), 250 mM NaCl, 5% (v/v) glycerol, and 0.25 mM TCEP]. Sample peaks were collected and analyzed by 10% SDS−PAGE. Fractions free of nucleic acids, based on A280/A260 measurements, were pooled and concentrated using a 50K MWCO concentrator, aliquoted, and flash-frozen. Concentrations of purified proteins were determined by the Bradford assay (Bio-Rad).
Purification of Human eIF4G.
The human eIF4G1 (NP_937884.1) gene (coding from amino acid 88–653) was codon optimized for E. coli expression and purchased as a FragmentGENE (GENEWIZ). The gene coding for eIF4G1 88–653 was subcloned using NdeI and SapI sites into the pTXB1 vector (NEB) which contains a C-terminal Mxe GyrA Intein and a chitin binding domain. An additional threonine was inserted after D653 to enhance cleavage during purification.
The E. coli BL21 (DE3) Star cells (Novagen) were transformed with the pTXB1-eIF4G plasmid. Cells were grown overnight at 37 °C in a 5 ml LB starter culture supplemented with 100 μg/mL ampicillin. 1 L LB media containing 100 μg/mL ampicillin was inoculated with 3–5 ml of the overnight starter culture and grown at 37 °C to OD600 ~0.5. The culture was cooled to 30 °C then induced with 0.4 mM IPTG and grown at that temperature for 2.5 – 3 hrs. Cells were pelleted at 5000 RPM for 15 min at 4 °C and then stored at −80 °C.
Cells were resuspended in pTXB1 lysis buffer (20 mM HEPES, 100 mM KCl, 10% (v/v) glycerol, 1 mM PMSF, pH 8.5) then lysed by French Press. Lysate was clarified at 50,000 × g for 30 min at 4 °C then loaded into a pre-equilibrated column containing chitin resin (NEB) with a flow rate of ~0.5 ml/min at 4 °C. The resin was washed with 10 column volumes of pTXB1 lysis buffer, 10 column volumes of pTXB1 wash buffer (20 mM HEPES, 1 M KCl, 10% (v/v/) glycerol, 1 mg/ml heparin sodium salt, pH 8.5), and finally an additional 10 column volumes of pTXB1 lysis buffer. The resin was quickly washed with 3 column volumes of cleavage buffer (20 mM HEPES, 100 mM KCl, 10% (v/v) glycerol, 50 mM DTT, pH 8.5) before closing the column and incubating in cleavage buffer for 16–20 hrs at room temperature. Fractions were collected, analyzed by SDS-PAGE, then pooled and concentrated to ~5 ml in an Amicon Ultra Centrifugal Filter (Millipore). The protein was further purified on a Superdex 200 16/600 gel filtration column (GE Healthcare) in Storage Buffer (20 mM HEPES, 200 mM KCl, 10% (v/v) glycerol, 1 mM DTT, 0.1 mM EDTA, pH 7.5). Fractions were analyzed by SDS-PAGE and only the purest fractions were pooled, concentrated, aliquoted, flash-frozen, and stored at −80 °C.
RNAs for Fluorescence Anisotropy.
The poly(A)18, ssIAV (5ʹ-AGCAAAAGCAGG-3ʹ), and ssCR1 (5ʹ-GCUAUCCAGAUUCUGAUU-3ʹ) RNA with a fluorescein dye attached to the 3ʹ-end were purchased from GE Dharmacon. The dsRK1 with a fluorescein dye attached to the 5′-end was synthesized as two complementary RNAs: 5ʹ-FL-CCAUCCUCUACAGGCG-3ʹ and 5ʹ-FL-CGCCUGUAGAGGAUGG-3ʹ. All RNAs were deprotected and purified by denaturing urea−PAGE. All RNAs were resuspended in water, and their concentrations were determined by measuring the absorbance at 260 nm. All RNAs were stored at −80 °C in small aliquots. To make double-stranded RNA, equimolar amounts of sense and antisense RNAs were heated to 95 °C in 50 mM Tris (pH 8), 50 mM KCl, and 1 mM DTT for 2 min and then allowed to cool slowly to room temperature. The dsRNA was then aliquoted and stored at −80 °C.
RNA Transcription, Purification, and Capping.
RNAs not directly purchased were synthesized by in vitro transcription using either DNA oligonucleotides purchased from IDT or PCR products. Briefly, complementary sequences to the desired RNA sequence were designed with an additional 5ʹ-CCTATAGTGAGTCGTATTA-3ʹ sequence at the 3ʹ end that is complementary to the 18T7T sequence (5ʹ-TAATACGACTCACTATAG-3ʹ). All DNA oligonucleotides were purified by denaturing urea-PAGE and resuspended in RNase-free water. The quality and yield were assessed with a NanoDrop 2000c (ThermoFischer). The 18T7T was annealed to the oligonucleotide templates and used for in vitro transcription using T7 RNA polymerase to synthesize RNAs. All RNAs were purified by denaturing urea-PAGE. Each RNA was then purified via chloroform extraction followed by ethanol precipitation and resuspended in RNase-free water. The quality and yield were assessed by measuring A260 with a NanoDrop 2000c (ThermoFischer).
For long RNAs used for in vitro translation assays, the templates were synthesized by PCR. Briefly, the gene encoding the Renilla luciferase (RLuc) in the pRL-null vector (Promega) was modified with different sequences at the 5ʹ-end and a poly(A)25 was inserted at the 3ʹ-end of the gene by PCR. Similarly, the bicistronic construct encoding the Firefly luciferase and CrPV driven Renilla luciferase proteins in the pFR-CrPV vector was modified with different sequences at the 5ʹ-end and a poly(A)25 was inserted at the 3ʹ-end of the gene. The bicistronic constructs were amplified by PCR to add the T7 promoter sequence. The PCR products were used as templates for in vitro transcription by T7 RNA polymerase to synthesize the RNAs. RNAs were purified using the Monarch RNA Cleanup Kit (New England BioLabs) and resuspended in RNase-free water. RNA length and quality were checked using denaturing urea-PAGE and concentration was measured with a NanoDrop 2000c (ThermoFischer).
The mRNAs were 5ʹ-capped with 7-methylguanosine (m7G) using a homemade Vaccinia virus capping enzyme [71]. The mRNAs were first heated for 5 minutes at 65°C and then put on ice for 5 minutes. Capping was carried out for 2 hours at 37°C in 50 μL reactions with 1X capping buffer [50 mM Tris pH 8, 5 mM KCl, 1 mM MgCl2, and 1 mM DTT], 0.5 mM GTP, 0.1 mM SAM, and 10 μg homemade Vaccinia capping enzyme; the amount of mRNA substrate to be modified was limited to maintain a large stoichiometric excess of both GTP and SAM to ensure all mRNA molecules were capped. The 5ʹ m7G-capped mRNAs were then cleaned up using the Monarch RNA cleanup kit (New England Biolabs) and yield was assessed with a NanoDrop 2000c (ThermoFischer).
Fluorescence Anisotropy.
Fluorescence anisotropy studies were performed using a fixed concentration of fluorescein-labeled RNA and an increasing concentration of protein in anisotropy buffer [50 mM Tris (pH 8), 50 mM KCl, 50 ng/μL E. coli total tRNA, 1 mM DTT, and 0.01% (v/v) Tween 20] [72]. For the anisotropy experiments, the concentration of RNA was fixed at 1 nM, and the concentration of PABP1 was titrated from 0 to 5 μM. Samples were incubated for 1 h at room temperature and the anisotropy studies were performed using a Tecan Spark plate reader in a 96-well plate. The sample (final volume of 100 μL) was excited at 470 nm, and the polarized emission at 520 nm was measured with 10 nm band slits for both excitation and emission. The G-factor was determined using a control sample with fluorescein-labeled RNA. The anisotropy values were subtracted from their initial values, plotted, and fit to the following quadratic equation to determine KD as described previously [72,73]:
where [P+FL]/[FL] is the anisotropy value, [FL] is the fluorescently labeled species and [P] is the protein concentration. GraphPad Prism (GraphPad Software Inc.) was used to perform the curve fits. All experiments were performed a minimum of three times with different protein batches to ensure reproducibility.
Electrophoretic Mobility Shift Assay (EMSA).
An EMSA was performed by incubating fluorescein-labeled RNA [final concentrations of 100 nM] with protein in anisotropy buffer to a final volume of 11 μL at room temperature for 1 h. After incubation, 1.3 μL of ice-cold 50% (v/v) glycerol and xylene cyanol were added to the mix. The complexes were separated from unbound species by electrophoresis on a 0.7% nondenaturing gel using a SEAKEM GTG agarose solution (Lonza) made with 1× TBE buffer. Samples were separated at 4 °C in 1× TBE buffer for 1.5 h at a 66 V constant voltage. The gels were visualized by scanning with a FLA9500 Typhoon instrument using the Cy2 excitation laser at a 600 PMT voltage and 50 μm resolution. In cases where concentrations are not explicit, 500 nM PABP to 100 nM poly(A) RNA ratio was used. Gels were analyzed with ImageJ software [74].
In vitro translation assay.
HeLa extract and translation assays were performed as described previously [75]. Briefly, HeLa cells were cultured in several 10 cm or 15 cm tissue culture plates until > 95% confluency. Cells were trypsinized, collected, spun down, washed with PBS and spun down again. The cell pellet mass was measured in a falcon tube (~ 200 – 300 mg) and resuspended in an equal volume of lysis buffer (10 mM HEPES pH 7.6, 10 mM potassium acetate, 0.5 mM magnesium acetate, 5 mM dithiothreitol, 15 mM 2-aminopurine and 1 tablet of cOmplete Mini, EDTA free proteinase inhibitor tablet from Roche). Cells were then lysed with 25 strokes of a 2 mL dounce homogenizer and lysate was clarified via centrifugation. Supernatant was isolated and subjected to a 7-minute incubation at 25°C with 15 U/mL of micrococcal nuclease (New England Biolabs) and 0.75 mM calcium chloride. Reaction was stopped with the addition of 3 mM EGTA pH 7.0 and sample was aliquoted and stored in liquid nitrogen.
The in vitro translation reactions were carried out as recommended with some slight modifications [75]. Briefly, HeLa extract made up 40% of each reaction with the remaining volume made up of translation buffer [16 mM HEPES pH 7.6, 20 mM creatine phosphate, 0.1 μg/μL creatine kinase, 0.1 mM spermidine, 100 μM amino acid mix, 8 mM ATP and 500 μM GTP], 150 mM potassium acetate, 2.5 mM magnesium acetate and 15 nM RNA. Samples were incubated at 30°C for 1.5 hours and luminescence was measured with a Tecan Spark plate reader after adding 3 μM coelenterazine (Promega) for the monocistronic Renilla luciferase mRNAs. Samples of Firefly and Renilla luciferase bicistronic mRNAs were measured using the Dual Reporter Luciferase Assay System kit (Promega). All experiments were performed a minimum of three times with different lysate batches to ensure reproducibility.
Tissue culture, Virus propagation and IAV infection.
HeLa and A549 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% Fetal Bovine Serum (FBS) in an incubator set a 37°C with 5% CO2. A/Puerto Rico/8/34 (H1N1 ATCC-1469) virus was propagated in MDCK cells and transferred to DMEM supplemented with 25 mM HEPES buffer, 0.2% BSA Fraction V, 2 μg/mL TPCK-trypsin, and 1% penicillin/streptomycin (DMEM-IAV Media). Viral titers were determined via the hemagglutination test in Turkey red blood cells. MDCK cells were infected and used to determine the 50% tissue culture infective dose (TCID50) using the Spearman-Karber method.
Cells used for infection were carried out by passaging 106 A549 cells into a 10-cm dish for two days prior to infection with IAV. Infection was done at an MOI of 0.5 in DMEM-IAV Media and incubated for 1 hour prior to multiple washes with 1x PBS. A mock infected control plate was prepared in parallel. Cells were incubated in DMEM + 10% FBS for 24 hours before harvest. Briefly, media was aspirated and cells were washed twice with cold 1x PBS. A crosslinking solution made up of 0.1% (v/v) formaldehyde in 1x PBS solution was added to each plate and incubated at room temperature for ten minutes with gentle mixing. Afterwards glycine was added (final concentration of 125 mM) to each plate and incubated for five minutes at room temperature. Solution was removed from each plate and cells were washed twice with cold 1x PBS. Cells were harvested by scraping the plates in 1 mL of 1x PBS using a cell scraper and transferred to an Eppendorf tube. Cells were pelleted, supernatant was aspirated and pellet was stored at −80°C until ready for immunoprecipitation. Experiment was performed a minimum of three times.
PABP1 Immunoprecipitation.
Pulldown of PABP1 was performed by resuspending the A549 pellets in ice cold RIPA buffer [25 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.1% SDC and 1 mM EDTA]. Resuspended cells were sonicated on ice for 5 rounds 15 seconds on, 60 seconds off using a Branson Sonifier 450 equipped with a microtip and set at 50% Duty Cycle and an Output Control of 4. Samples were spun down, supernatant was transferred to washed Dynabeads Protein G Magnetic Beads (Invitrogen) with Rabbit IgG (Diagenode Cat. # c15410206) and placed on a rotator for 30 minutes at 4 °C. Aliquots were collected prior to the next stage to serve as an RNA Input control. Samples were separated from beads and added to anti-PABPC1 rabbit polyclonal antibody (Abcam Cat. # ab21060)) and placed on a rotator for 2 hours at 4 °C followed by washed Dynabeads Protein G Magnetic Beads for another hour. Supernatant was then discarded and beads were gently washed three times with RIPA buffer. Protein and RNA were eluted off with elution buffer [50 mM Tris-HCl pH 7.5, 5 mM EDTA, 1% SDS and 10 mM DTT] and samples were subjected to a proteinase K (New England Biolabs) treatment at 65 °C for 45 minutes. RNA was separated from beads and then subjected to the Monarch RNA cleanup kit (New England Biolabs) where yield and purity were checked via NanoDrop 2000c (ThermoFischer). Pulldown experiments were performed a minimum of three times with three different infection experiments to ensure reproducibility.
Quantitative RT-PCR.
For RT-qPCR, 10 ng of total RNA taken before and after the IP was reverse transcribed with gene specific primers for different sections of the target mRNAs. Briefly, three different reactions per target mRNA were performed using the following primers: HA-5ʹUTR-Rev: 5ʹ-CGTTGTGGCTGTCTTCGAGC-3ʹ, HA-Mid-Rev: 5ʹ-CTGGAAAGGGAGACTGCTGTTTATAGC-3ʹ, HA-3ʹUTR-Rev: 5ʹ- TCAGATGCATATTCTGCACTGCAAAGAT-3ʹ, Actin-5ʹUTR-Rev: 5ʹ-GCCCACATAGGAATCCTTCTGACC-3ʹ, Actin-Mid-Rev: 5ʹ-GGTACATGGTGGTGCCGCC-3ʹ, Actin-3ʹUTR-Rev: 5ʹ-TCATTTTTAAGGTGTGCACTTTTATTCAACTGG-3ʹ, Tubulin-5ʹUTR-Rev: 5ʹ-TGCATGTGTTAAAAGGCGCAGG-3ʹ, Tubulin-Mid-Rev: 5ʹ-AAGCAGTGATGGAGGACACAATCTG-3ʹ, Tubulin-3ʹUTR-Rev: 5ʹ-GACATTTAAAATGGAAACTTCAATTTTATTAACAATTTACGGC-3ʹ, NP-5ʹUTR-Rev: 5ʹ-AATCACTGAGTTTGAGTTCGGTGCA-3ʹ, NP-Mid-Rev: 5ʹ-CCGACCCTCTCAATATGAGTGCA-3ʹ, NP-3’UTR-Rev: 5ʹ-TTAATTGTCGTACTCCTCTGCATTGTCTC-3ʹ, NA-5ʹUTR-Rev: 5ʹ-TGATGTTTTGGTTGCATATTCCAGTATGGT-3ʹ, NA-Mid-Rev: 5ʹ-TGATTTAGTAACCTT CCCCTTTTCGATCTTG −3ʹ, NA-3ʹUTR-Rev: 5ʹ-CTACTTGTCAATGGTGAATGGCAACTC-3ʹ, PB1–5ʹUTR-Rev: 5ʹ-CTGAGTACTGATGTGTCCTGTTGACA-3ʹ, PB1-Mid-Rev: 5ʹ-TGAATCCCTTCATGATTGGGTGCA-3ʹ, PB1–3ʹUTR-Rev: 5ʹ-CTATTTTTGCCGTCTGAGCTCTTCAATG-3ʹ, NS-5ʹUTR-Rev: 5ʹ-TGC CTC ATC GGATTCTTCTTTCAGAATC-3ʹ, NS-Mid-Rev: 5ʹ-GGAAGAGAAGGCAATGGTGAAATTTCG-3ʹ, NS-3ʹUTR-Rev: 5ʹ-TAAATAAGCTGAAACGAGAAAGTTCTTATCTCTTGC-3ʹ, M-5ʹUTR-Rev: 5ʹ-CGTGAACACAAATCCTAAAATCCCCTTAGT-3ʹ, M-Mid-Rev: 5ʹ-CTCCATAGCCTTAGCTGTAGTGCTG-3ʹ, M-3ʹUTR-Rev: 5ʹ-TTACTCCAGCTCTATGCTGACAAAATGAC-3ʹ, PB2–5ʹUTR-Rev: 5ʹ-GCTGTAATTGGATATTTCATTGCCATCATCC-3ʹ, PB2-Mid-Rev: 5ʹ-GCTGATTCGCCCTATTGACGAAATTC-3ʹ, PB2–3ʹUTR-Rev: 5ʹ-AAACTATTCGACACTAATTGATGGCCATCC-3ʹ, PA-5ʹUTR-Rev: 5ʹ-TCTACGATTATTGACTCGCCTTGCTC-3ʹ, PA-Mid-Rev: 5ʹ-GCCCACTTTAGCTGACTTGTTTTCTTC-3ʹ, PA-3ʹUTR-Rev: 5ʹ-GGACAGTATGGATAGCAAATAGTAGCACTG-3. The reverse transcriptase used was Superscript III (Invitrogen) and reaction followed manufacturer’s protocol.
The RT-qPCR was performed on a Bio-Rad CFX Connect Real-Time System using specific primers for different sections of the target mRNAs. Briefly, RT-qPCR was performed with 2 μL of total cDNA from the above step using the Luna Universal RT-qPCR Master Mix (NEB) and specific primers according to the manufacturer’s protocol. The reverse primers used are the same as the aforementioned primers for reverse transcription. The forward primers are as follows:
HA-5ʹUTR-Fwd: 5ʹ-AGCAAAAGCAGGGGAAAATAAAAACAACC-3ʹ,
HA-Mid-Fwd: 5ʹ-GGAGGATGAACTATTACTGGACCTTGC-3ʹ,
HA-3ʹUTR-Fwd: 5ʹ-ATCAATGGGGATCTATCAGATTCTGGC-3ʹ,
Actin-5ʹUTR-Fwd: 5ʹ-ACAGAGCCTCGCCTTTGC-3ʹ,
Actin-Mid-Fwd: 5ʹ-AGCTGCCTGACGGCCAG-3ʹ,
Actin-3ʹUTR-Fwd: 5ʹ-TTTTAATCTTCGCCTTAATACTTTTTTATTTTGTTTTATTTTGAATGA-3ʹ,
Tubulin-5ʹUTR-Fwd: 5ʹ-CTAAAATGACAGCCTGGTTCAATGGG-3ʹ,
Tubulin-Mid-Fwd: 5ʹ-CCAGGTTTCCACAGCTGTAGTTGA-3ʹ,
Tubulin-3ʹUTR-Fwd: 5ʹ-GACATGGCTGCCCTTGAGAAG-3ʹ,
NP-5ʹUTR-Fwd: 5ʹ-AGCAAAAGCAGGGTAGATAATCACTCAC-3ʹ,
NP-Mid-Fwd: 5ʹ-GGTGAGAATGGACGAAAAACAAGAATTGC-3ʹ,
NP-3ʹUTR-Fwd: 5ʹ-ATCTGACATGAGGACCGAAATCATAAGG-3ʹ,
NA-5ʹUTR-Fwd: 5ʹ-GCAGGAGTTTAAAATGAATCCAAATCAGAAAATAATAACC-3ʹ, NA-Mid-Fwd: 5ʹ-GCAGTGGCTGTATTAAAATACAACGGC-3ʹ,
NA-3ʹUTR-Fwd: 5ʹ-GGAAGTTTCGTTCAACATCCTGAGC-3ʹ,
PB1–5ʹUTR-Fwd: 5ʹ-CAGGCAAACCATTTGAATGGATGTCAATC-3ʹ,
PB1-Mid-Fwd: 5ʹ-CTGCATCATTGAGCCCTGGAATG-3ʹ,
PB1–3ʹUTR-Fwd: 5ʹ-TGCTGCAATTTATTTGAAAAATTCTTCCCCAG-3ʹ, NS-5ʹUTR-Fwd: 5ʹ-AGCAAAAGCAGGGTGACAAAAACAT-3ʹ, NS-Mid-Fwd: 5ʹ-CATGCTCATACCCAAGCAGAAAGTG-3ʹ, NS-3ʹUTR-Fwd: 5ʹ-CTCCACTCACTCCAAAACAGAAACGA-3ʹ, M-5ʹUTR-Fwd: 5ʹ-AGCAAAAGCAGGTAGATATTGAAAGATGAGTC-3ʹ, M-Mid-Fwd: 5ʹ-CTCAGTTATTCTGCTGGTGCACTTG-3ʹ, M-3ʹUTR-Fwd: 5ʹ-AACGGTTCAAGTGATCCTCTCACTATTG-3ʹ, PB2–5ʹUTR-Fwd: 5ʹ-CGAAAGCAGGTC AATTATATTCAATATGGAAAGAATAAAAG-3ʹ, PB2-Mid-Fwd: 5ʹ-AGGGATATGAAGAGTTCACAATGGTTGG-3ʹ, PB2–3ʹUTR-Fwd: 5ʹ-GATTCCTCATTCTGGGCAAAGAAGACA-3ʹ, PA-5ʹUTR-Fwd: 5ʹ-TGATCCAAAATGGAAGATTTTGTGCGAC-3ʹ, PA-Mid-Fwd: 5ʹ-GGAACCCAATGTTGTTAAACCACACG-3ʹ, PA-3ʹUTR-Fwd: 5ʹ-GAAGGATTTTCAGCTGAATCAAGAAAACTGC-3ʹ. Reactions were run in duplicates and the 2-ΔΔCt was calculated by subtracting the Ct values of the samples post pulldown from pre-pulldown and relating different sections of a gene to each other before relating genes to each other. Note that primer efficiency for each pair of primers was validated according to recommended practices [76]. RT-qPCR was done on RNA from three biological replicates. GraphPad Prism was used to perform one-way ANOVA and t-test analysis to obtain P-values. P < 0.05 is denoted with one asterisk, P < 0.01 is denoted with two asterisks, and P < 0.0001 is denoted with four asterisks.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Ti Wu for writing the Python code to trim and filter the sequences downloaded from the Influenza Research Database. We would also like to thank Amy Pasquinelli and Angela Nicholson for sharing their PABP1 immunoprecipitation protocol and Brian Zid for comments on the manuscript. This work was supported by the National Institutes of Health (R01GM114261 and R35GM141864 to S.J.) and the Molecular Biophysics Training Grant funded by the National Institutes of Health (T32 GM008326).
REFERENCES
- [1].Ghebrehewet S, MacPherson P, Ho A, Influenza, BMJ. (2016) i6258. 10.1136/bmj.i6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, van Asten L, Pereira da Silva S, Aungkulanon S, Buchholz U, Widdowson M-A, Bresee JS, Azziz-Baumgartner E, Cheng P-Y, Dawood F, Foppa I, Olsen S, Haber M, Jeffers C, MacIntyre CR, Newall AT, Wood JG, Kundi M, Popow-Kraupp T, Ahmed M, Rahman M, Marinho F, Sotomayor Proschle CV, Vergara Mallegas N, Luzhao F, Sa L, Barbosa-Ramírez J, Sanchez DM, Gomez LA, Vargas XB, Herrera aBetsy Acosta, Llanés MJ, Fischer TK, Krause TG, Mølbak K, Nielsen J, Trebbien R, Bruno A, Ojeda J, Ramos H, an der Heiden M, del Carmen Castillo Signor L, Serrano CE, Bhardwaj R, Chadha M, Narayan V, Kosen S, Bromberg M, Glatman-Freedman A, Kaufman Z, Arima Y, Oishi K, Chaves S, Nyawanda B, Al-Jarallah RA, Kuri-Morales PA, Matus CR, Corona MEJ, Burmaa A, Darmaa O, Obtel M, Cherkaoui I, van den Wijngaard CC, van der Hoek W, Baker M, Bandaranayake D, Bissielo A, Huang S, Lopez L, Newbern C, Flem E, Grøneng GM, Hauge S, de Cosío FG, de Moltó Y, Castillo LM, Cabello MA, von Horoch M, Medina Osis J, Machado A, Nunes B, Rodrigues AP, Rodrigues E, Calomfirescu C, Lupulescu E, Popescu R, Popovici O, Bogdanovic D, Kostic M, Lazarevic K, Milosevic Z, Tiodorovic B, Chen M, Cutter J, Lee V, Lin R, Ma S, Cohen AL, Treurnicht F, Kim WJ, Delgado-Sanz C, de mateo Ontañón S, Larrauri A, León IL, Vallejo F, Born R, Junker C, Koch D, Chuang J-H, Huang W-T, Kuo H-W, Tsai Y-C, Bundhamcharoen K, Chittaganpitch M, Green HK, Pebody R, Goñi N, Chiparelli H, Brammer L, Mustaquim D, Estimates of global seasonal influenza-associated respiratory mortality: a modelling study, The Lancet. 391 (2018) 1285–1300. 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cozza V, Campbell H, Chang HH, Iuliano AD, Paget J, Patel NN, Reiner RC, Troeger C, Viboud C, Bresee JS, Fitzner J, Global Seasonal Influenza Mortality Estimates: A Comparison of 3 Different Approaches, Am. J. Epidemiol. 190 (2021) 718–727. 10.1093/aje/kwaa196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Molinari N-AM, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB, The annual impact of seasonal influenza in the US: Measuring disease burden and costs, Vaccine. 25 (2007) 5086–5096. 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- [5].Jackson RJ, Hellen CUT, Pestova TV, The mechanism of eukaryotic translation initiation and principles of its regulation, Nat. Rev. Mol. Cell Biol. 11 (2010) 113–127. 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pelletier J, Sonenberg N, The Organizing Principles of Eukaryotic Ribosome Recruitment, Annu. Rev. Biochem. 88 (2019) 307–335. 10.1146/annurev-biochem-013118-111042. [DOI] [PubMed] [Google Scholar]
- [7].Plotch SJ, Bouloy M, Ulmanen I, Krug RM, A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription, Cell. 23 (1981) 847–858. 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- [8].Gu W, Gallagher GR, Dai W, Liu P, Li R, Trombly MI, Gammon DB, Mello CC, Wang JP, Finberg RW, Influenza A virus preferentially snatches noncoding RNA caps, RNA. 21 (2015) 2067–2075. 10.1261/rna.054221.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Koppstein D, Ashour J, Bartel DP, Sequencing the cap-snatching repertoire of H1N1 influenza provides insight into the mechanism of viral transcription initiation, Nucleic Acids Res. 43 (2015) 5052–5064. 10.1093/nar/gkv333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sikora D, Rocheleau L, Brown EG, Pelchat M, Deep sequencing reveals the eight facets of the influenza A/HongKong/1/1968 (H3N2) virus cap-snatching process, Sci. Rep. 4 (2015) 6181. 10.1038/srep06181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sikora D, Rocheleau L, Brown EG, Pelchat M, Influenza A virus cap-snatches host RNAs based on their abundance early after infection, Virology. 509 (2017) 167–177. 10.1016/j.virol.2017.06.020. [DOI] [PubMed] [Google Scholar]
- [12].te Velthuis AJW, Fodor E, Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis, Nat. Rev. Microbiol. 14 (2016) 479–493. 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bercovich-Kinori A, Tai J, Gelbart IA, Shitrit A, Ben-Moshe S, Drori Y, Itzkovitz S, Mandelboim M, Stern-Ginossar N, A systematic view on influenza induced host shutoff, ELife. 5 (2016) e18311. 10.7554/eLife.18311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Burgui I, Yángüez E, Sonenberg N, Nieto A, Influenza Virus mRNA Translation Revisited: Is the eIF4E Cap-Binding Factor Required for Viral mRNA Translation?, J. Virol. 81 (2007) 12427–12438. 10.1128/JVI.01105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang Y, Aevermann BD, Anderson TK, Burke DF, Dauphin G, Gu Z, He S, Kumar S, Larsen CN, Lee AJ, Li X, Macken C, Mahaffey C, Pickett BE, Reardon B, Smith T, Stewart L, Suloway C, Sun G, Tong L, Vincent AL, Walters B, Zaremba S, Zhao H, Zhou L, Zmasek C, Klem EB, Scheuermann RH, Influenza Research Database: An integrated bioinformatics resource for influenza virus research, Nucleic Acids Res. 45 (2017) D466–D474. 10.1093/nar/gkw857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Crooks GE, WebLogo: A Sequence Logo Generator, Genome Res. 14 (2004) 1188–1190. 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schneider TD, Stephens RM, Sequence logos: a new way to display consensus sequences, Nucleic Acids Res. 18 (1990) 6097–6100. 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Furuse Y, Oshitani H, Evolution of the influenza A virus untranslated regions, Infect. Genet. Evol. 11 (2011) 1150–1154. 10.1016/j.meegid.2011.04.006. [DOI] [PubMed] [Google Scholar]
- [19].Parvin JD, Moscona A, Pan WT, Leider JM, Palese P, Measurement of the mutation rates of animal viruses: influenza A virus and poliovirus type 1., J. Virol. 59 (1986) 377–383. 10.1128/JVI.59.2.377-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nobusawa E, Sato K, Comparison of the Mutation Rates of Human Influenza A and B Viruses, J. Virol. 80 (2006) 3675–3678. 10.1128/JVI.80.7.3675-3678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC, The genomic and epidemiological dynamics of human influenza A virus, Nature. 453 (2008) 615–619. 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen R, Holmes EC, Avian Influenza Virus Exhibits Rapid Evolutionary Dynamics, Mol. Biol. Evol. 23 (2006) 2336–2341. 10.1093/molbev/msl102. [DOI] [PubMed] [Google Scholar]
- [23].Sachs AB, Davis RW, Kornberg RD, A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability, Mol. Cell. Biol. 7 (1987) 3268–3276. 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kühn U, Pieler T, Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction, J. Mol. Biol. 256 (1996) 20–30. 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- [25].Burd CG, Matunis EL, Dreyfuss G, The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities., Mol. Cell. Biol. 11 (1991) 3419–3424. 10.1128/MCB.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].de Rozières CM, Joseph S, Influenza A Virus NS1 Protein Binds as a Dimer to RNA-Free PABP1 but Not to the PABP1·Poly(A) RNA Complex, Biochemistry. 59 (2020) 4439–4448. 10.1021/acs.biochem.0c00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Görlach M, Burd CG, Dreyfuss G, The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity, Exp. Cell Res. 211 (1994) 400–407. 10.1006/excr.1994.1104. [DOI] [PubMed] [Google Scholar]
- [28].Kozak M, Structural features in eukaryotic mRNAs that modulate the initiation of translation, J. Biol. Chem. 266 (1991) 19867–19870. [PubMed] [Google Scholar]
- [29].Witherell G, In Vitro Translation Using HeLa Extract, Curr. Protoc. Cell Biol. 6 (2000). 10.1002/0471143030.cb1108s06. [DOI] [PubMed] [Google Scholar]
- [30].Feigenblum D, Schneider RJ, Modification of eukaryotic initiation factor 4F during infection by influenza virus, J. Virol. 67 (1993) 3027–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yángüez E, Nieto A, So similar, yet so different: Selective translation of capped and polyadenylated viral mRNAs in the influenza virus infected cell, Virus Res. 156 (2011) 1–12. 10.1016/j.virusres.2010.12.016. [DOI] [PubMed] [Google Scholar]
- [32].Yángüez E, Rodriguez P, Goodfellow I, Nieto A, Influenza virus polymerase confers independence of the cellular cap-binding factor eIF4E for viral mRNA translation, Virology. 422 (2012) 297–307. 10.1016/j.virol.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gilbert WV, Zhou K, Butler TK, Doudna JA, Cap-Independent Translation Is Required for Starvation-Induced Differentiation in Yeast, Science. 317 (2007) 1224–1227. 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- [34].Safaee N, Kozlov G, Noronha AM, Xie J, Wilds CJ, Gehring K, Interdomain Allostery Promotes Assembly of the Poly(A) mRNA Complex with PABP and eIF4G, Mol. Cell. 48 (2012) 375–386. 10.1016/j.molcel.2012.09.001. [DOI] [PubMed] [Google Scholar]
- [35].Deo RC, Bonanno JB, Sonenberg N, Burley SK, Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein, Cell. 98 (1999) 835–845. 10.1016/S0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- [36].Vicens Q, Kieft JS, Rissland OS, Revisiting the Closed-Loop Model and the Nature of mRNA 5′–3′ Communication, Mol. Cell. 72 (2018) 805–812. 10.1016/j.molcel.2018.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Walsh D, Mathews MB, Mohr I, Tinkering with translation: protein synthesis in virus-infected cells, Cold Spring Harb Perspect Biol. 5 (2013) a012351. 10.1101/cshperspect.a012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bushell M, Sarnow P, Hijacking the translation apparatus by RNA viruses, J. Cell Biol. 158 (2002) 395–399. 10.1083/jcb.200205044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Martinez-Salas E, Francisco-Velilla R, Fernandez-Chamorro J, Embarek AM, Insights into Structural and Mechanistic Features of Viral IRES Elements, Front. Microbiol. 8 (2018) 2629. 10.3389/fmicb.2017.02629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Katze MG, DeCorato D, Krug RM, Cellular mRNA translation is blocked at both initiation and elongation after infection by influenza virus or adenovirus, J Virol. 60 (1986) 1027–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Katze MG, Krug RM, Translational control in influenza virus-infected cells, Enzyme. 44 (1990) 265–77. [DOI] [PubMed] [Google Scholar]
- [42].Garfinkel MS, Katze MG, Translational control by influenza virus. Selective translation is mediated by sequences within the viral mRNA 5’-untranslated region, J Biol Chem. 268 (1993) 22223–6. [PubMed] [Google Scholar]
- [43].Enami K, Sato TA, Nakada S, Enami M, Influenza virus NS1 protein stimulates translation of the M1 protein, J Virol. 68 (1994) 1432–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].de la Luna S, Fortes P, Beloso A, Ortin J, Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs, J Virol. 69 (1995) 2427–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kash JC, Goodman AG, Korth MJ, Katze MG, Hijacking of the host-cell response and translational control during influenza virus infection, Virus Res. 119 (2006) 111–20. 10.1016/j.virusres.2005.10.013. [DOI] [PubMed] [Google Scholar]
- [46].Katze MG, Krug RM, Metabolism and expression of RNA polymerase II transcripts in influenza virus-infected cells, Mol Cell Biol. 4 (1984) 2198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Beloso A, Martinez C, Valcarcel J, Santaren JF, Ortin J, Degradation of cellular mRNA during influenza virus infection: its possible role in protein synthesis shutoff, J Gen Virol. 73 ( Pt 3) (1992) 575–81. [DOI] [PubMed] [Google Scholar]
- [48].Chen Z, Krug RM, Selective nuclear export of viral mRNAs in influenza-virus-infected cells, Trends Microbiol. 8 (2000) 376–83. [DOI] [PubMed] [Google Scholar]
- [49].Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P, An overlapping protein-coding region in influenza A virus segment 3 modulates the host response, Science. 337 (2012) 199–204. 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ortin J, Martin-Benito J, The RNA synthesis machinery of negative-stranded RNA viruses, Virology. 479–480C (2015) 532–544. 10.1016/j.virol.2015.03.018. [DOI] [PubMed] [Google Scholar]
- [51].Ulmanen I, Broni BA, Krug RM, Role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription, Proc Natl Acad Sci U A. 78 (1981) 7355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RW, Ortin J, Hart DJ, Cusack S, The structural basis for cap binding by influenza virus polymerase subunit PB2, Nat Struct Mol Biol. 15 (2008) 500–6. 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- [53].Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y, Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site, Nature. 458 (2009) 909–913. 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- [54].Blass D, Patzelt E, Kuechler E, Identification of the cap binding protein of influenza virus, Nucleic Acids Res. 10 (1982) 4803–4812. 10.1093/nar/10.15.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dias A, Bouvier D, Crépin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RWH, The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit, Nature. 458 (2009) 914–918. 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- [56].Pflug A, Lukarska M, Resa-Infante P, Reich S, Cusack S, Structural insights into RNA synthesis by the influenza virus transcription-replication machine, Virus Res. 234 (2017) 103–117. 10.1016/j.virusres.2017.01.013. [DOI] [PubMed] [Google Scholar]
- [57].Te Velthuis AJW, Fodor E, Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis, Nat. Rev. Microbiol. 14 (2016) 479–493. 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Poon LL, Pritlove DC, Fodor E, Brownlee GG, Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template, J Virol. 73 (1999) 3473–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pritlove DC, Poon LL, Fodor E, Sharps J, Brownlee GG, Polyadenylation of influenza virus mRNA transcribed in vitro from model virion RNA templates: requirement for 5’ conserved sequences, J. Virol. 72 (1998) 1280–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kleijn M, Vrins CL, Voorma HO, Thomas AA, Phosphorylation state of the cap-binding protein eIF4E during viral infection, Virology. 217 (1996) 486–494. 10.1006/viro.1996.0143. [DOI] [PubMed] [Google Scholar]
- [61].Joshi-Barve S, Rychlik W, Rhoads RE, Alteration of the major phosphorylation site of eukaryotic protein synthesis initiation factor 4E prevents its association with the 48 S initiation complex, J. Biol. Chem. 265 (1990) 2979–2983. [PubMed] [Google Scholar]
- [62].Huang JT, Schneider RJ, Adenovirus inhibition of cellular protein synthesis involves inactivation of cap-binding protein, Cell. 65 (1991) 271–280. [DOI] [PubMed] [Google Scholar]
- [63].Morley SJ, Rau M, Kay JE, Pain VM, Increased phosphorylation of eukaryotic initiation factor 4 alpha during early activation of T lymphocytes correlates with increased initiation factor 4F complex formation, Eur. J. Biochem. 218 (1993) 39–48. 10.1111/j.1432-1033.1993.tb18349.x. [DOI] [PubMed] [Google Scholar]
- [64].Kuss-Duerkop SK, Wang J, Mena I, White K, Metreveli G, Sakthivel R, Mata MA, Muñoz-Moreno R, Chen X, Krammer F, Diamond MS, Chen ZJ, García-Sastre A, Fontoura BMA, Influenza virus differentially activates mTORC1 and mTORC2 signaling to maximize late stage replication, PLOS Pathog. 13 (2017) e1006635. 10.1371/journal.ppat.1006635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Desselberger U, Racaniello VR, Zazra JJ, Palese P, The 3’ and 5’-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity, Gene. 8 (1980) 315–328. 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- [66].Xia X, MacKay V, Yao X, Wu J, Miura F, Ito T, Morris DR, Translation Initiation: A Regulatory Role for Poly(A) Tracts in Front of the AUG Codon in Saccharomyces cerevisiae, Genetics. 189 (2011) 469–478. 10.1534/genetics.111.132068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Shirokikh NE, Spirin AS, Poly(A) leader of eukaryotic mRNA bypasses the dependence of translation on initiation factors, Proc. Natl. Acad. Sci. 105 (2008) 10738–10743. 10.1073/pnas.0804940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gingras AC, Svitkin Y, Belsham GJ, Pause A, Sonenberg N, Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus., Proc. Natl. Acad. Sci. 93 (1996) 5578–5583. 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Eschenfeldt WH, Maltseva N, Stols L, Donnelly MI, Gu M, Nocek B, Tan K, Kim Y, Joachimiak A, Cleavable C-terminal His-tag vectors for structure determination, J. Struct. Funct. Genomics. 11 (2010) 31–39. 10.1007/s10969-010-9082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Eschenfeldt WH, Lucy S, Millard CS, Joachimiak A, Mark ID, A Family of LIC Vectors for High-Throughput Cloning and Purification of Proteins, in: Doyle SA (Ed.), High Throughput Protein Expr. Purif, Humana Press, Totowa, NJ, 2009: pp. 105–115. 10.1007/978-1-59745-196-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Fuchs A-L, Neu A, Sprangers R, A general method for rapid and cost-efficient large-scale production of 5′ capped RNA, RNA. 22 (2016) 1454–1466. 10.1261/rna.056614.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Cho EJ, Xia S, Ma L-C, Robertus J, Krug RM, Anslyn EV, Montelione GT, Ellington AD, Identification of Influenza Virus Inhibitors Targeting NS1A Utilizing Fluorescence Polarization–Based High-Throughput Assay, J. Biomol. Screen. 17 (2012) 448–459. 10.1177/1087057111431488. [DOI] [PubMed] [Google Scholar]
- [73].Pollard TD, A guide to simple and informative binding assays, Mol. Biol. Cell. 21 (2010) 4061–4067. 10.1091/mbc.E10-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis, Nat. Methods. 9 (2012) 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Rakotondrafara AM, Hentze MW, An efficient factor-depleted mammalian in vitro translation system, Nat. Protoc. 6 (2011) 563–571. 10.1038/nprot.2011.314. [DOI] [PubMed] [Google Scholar]
- [76].Nolan T, Hands RE, Bustin SA, Quantification of mRNA using real-time RT-PCR, Nat. Protoc. 1 (2006) 1559–1582. 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


