Abstract
Introduction
APOL1 G1 and G2 alleles have been associated with kidney-related outcomes in people living with HIV (PLHIV) of Black African origin. No APOL1-related kidney risk data have yet been reported in PLHIV in West Africa, where high APOL1 allele frequencies have been observed.
Methods
We collected clinical data from PLHIV followed in Burkina Faso (N = 413) and in the ANRS-12169/2LADY trial (Cameroon, Senegal, Burkina Faso, N = 369). APOL1 G1 and G2 risk variants were genotyped using TaqMan assays, and APOL1 high-risk (HR) genotype was defined by the carriage of 2 risk alleles.
Results
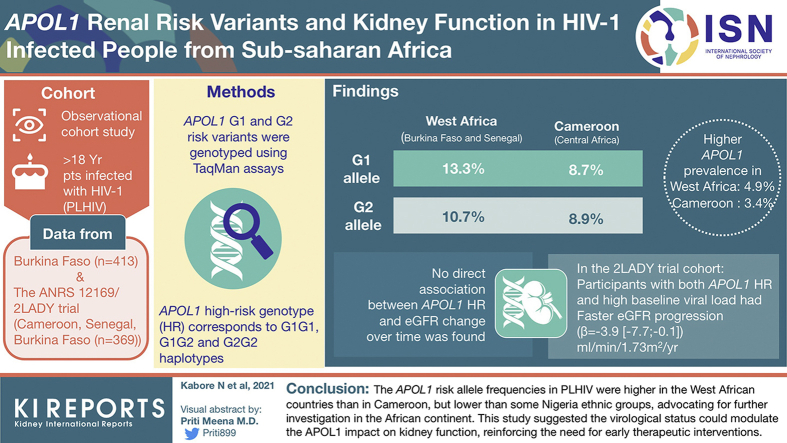
In West Africa (Burkina Faso and Senegal), the G1 and G2 allele frequencies were 13.3% and 10.7%, respectively. In Cameroon (Central Africa), G1 and G2 frequencies were 8.7% and 8.9%, respectively. APOL1 HR prevalence was 4.9% in West Africa and 3.4% in Cameroon. We found no direct association between APOL1 HR and estimated glomerular filtration rate (eGFR) change over time. Nevertheless, among the 2LADY cohort participants, those with both APOL1 HR and high baseline viral load had a faster eGFR progression (β = −3.9[−7.7 to −0.1] ml/min per 1.73 m2 per year, P < 0.05) than those with low-risk (LR) genotype and low viral load.
Conclusion
Overall, the APOL1 risk allele frequencies in PLHIV were higher in the West African countries than in Cameroon, but much lower than previously reported in some Nigeria ethnic groups, which strongly advocates for further investigation in the African continent. This study suggested that the virological status could modulate the APOL1 impact on kidney function, hence reinforcing the need for early therapeutic interventions.
Keywords: Africa, APOL1, Burkina Faso, eGFR, HIV, Kidney risk
Graphical abstract
The United Nations Program on HIV/AIDS estimated the number of PLHIV in 2019 at 38 (32.7–44.0) million worldwide, with Africa carrying two-thirds (25.7 million) of this burden, mainly in sub-Saharan Africa.1 Antiretroviral treatments (ARTs) have significantly reduced HIV-related morbidity and mortality; however, with increased life expectancy among PLHIV, noninfectious co-morbidities, such as cardiovascular, metabolic, and kidney diseases, have also increased. Chronic kidney disease (CKD) prevalence in PLHIV was estimated at 6.4% worldwide and 14.6% in West Africa.2 CKD occurrence increases by 2-fold the risk of death among PLHIV, and people of Black African ancestry are particularly affected.3,4 In the United States, African American PLHIV were 10 to 18 times more likely to develop HIV-associated nephropathy than their European counterparts.5,6 Two APOL1 coding alleles, termed G1 and G2 and found only on African-ancestry haplotypes, have been associated with a spectrum of CKD from HIV-associated nephropathy to focal segmental glomerular sclerosis, nondiabetic end-stage kidney disease, hypertension-attributed nephropathy, and increased proteinuria.7, 8, 9, 10, 11, 12 Notably, APOL1 HR genotypes are strongly associated with HIV-associated nephropathy in untreated PLHIV of African descent with odds ratios ranging from 29 to 89 in the United States and in South Africa, respectively.10, 11, 12 Moreover, a study in African American PLHIV revealed that patients with the HR genotypes experienced faster kidney function declines, with greater decline among those with a lack of sustained virological suppression.13
This study aimed to provide, for the first time, data on the distribution of APOL1 risk variants and their impact on kidney function among treated PLHIV in different settings from West and Central Africa, as high frequencies of these variants were previously reported in West Africa.14,15
Methods
We performed an observational cohort study. Data were collected from the following 2 cohorts: (i) a hospital cohort of PLHIV in Burkina Faso (day care unit [DCU] of Bobo-Dioulasso) and (ii) a cohort of HIV+ patients enrolled in a clinical trial (ANRS-12169/2LADY trial).
DCU Cohort
The first cohort is the DCU of the Centre Hospitalier Universitaire Sourô Sanou in Bobo-Dioulasso, Burkina Faso. The DCU, created in 2005, is part of the Infectious Disease Department and specialized in PLHIV care. The Ensemble pour une Solidarité Thérapeutique Hospitalière en Réseau hospital partnership initiative has supported the implementation of an electronic medical database used to monitor PLHIV care in DCU from 2007. Routine clinical follow-up visits were done every 6 months, and all clinical (general condition, symptoms, diagnosis, height, weight, blood pressure, body temperature), biological (blood count, CD4 count, glycemia, creatinine, cholesterol, triglycerides), and therapeutic (ART, cotrimoxazole prophylaxis, other treatments) data were recorded in real time by the physicians.
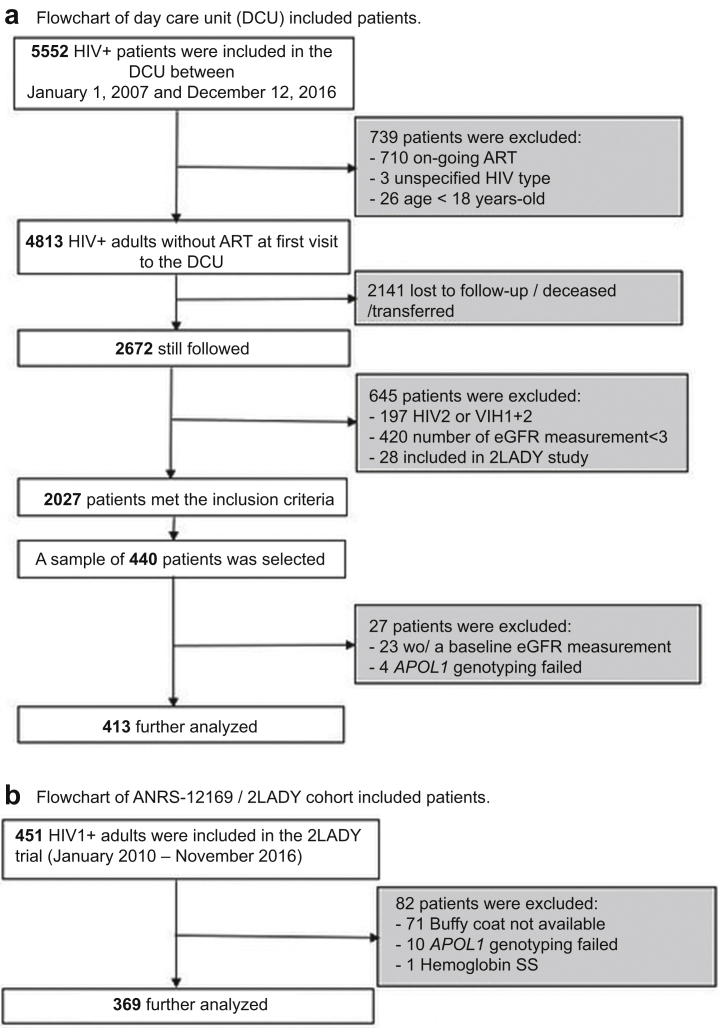
In this study, we included adult patients (≥18 years old) infected with HIV-1, followed in the cohort for at least 2 years, with at least 3 plasma measurements of creatinine (Figure 1a). Participants without creatinine measurement at baseline were excluded from the analyses. Patients were naive to any ART treatment at their first visit at the DCU between January 1, 2007, and December 31, 2016. For ART initiation, the 3 most prescribed treatment regimens were as follows: zidovudine (AZT) + lamivudine (3TC) + efavirenz (EFV) or nevirapine (NVP) (40.0%); tenofovir disoproxil fumarate (TDF) + emtricitabine (FTC) or lamivudine (TDF + FTC or TDF + 3TC) + EFV/NVP (31.9%); and stavudine (d4T) + 3TC + EFV/NVP (15.4%).16 Eligible patients who came for a routine consultation between January 1, 2018, and December 31, 2018, were invited to participate in the study. After informed written consent, a blood specimen was collected for the genetic analyses (N = 413).
Figure 1.
Study design for the (a) DCU and (b) 2LADY cohort study groups. The DCU patients were HIV+ adults initiating ART for the first time at baseline, who were subsequently followed for a median of 6.1 years—the subgroup of interest (n = 440) was notably selected for availability of kidney function-related data. The 2LADY patients were HIV+ adults whose first-line ART failed and who were enrolled in a second-line ART trial—they were monitored for a median of 4.8 years. All patients with available DNA were enrolled in this study. ART, Antiretroviral treatment; DCU, day care unit; eGFR, estimated glomerular filtration rate.
ANRS-12169/2LADY Cohort
The second cohort is from the ANRS-12169/2LADY trial,17 which aimed to evaluate the efficacy and safety of 3 ART combinations in PLHIV who failed a first-line treatment in Africa (Cameroon, Senegal, and Burkina Faso). The participants were adults (≥18 years old) failing first-line ART that did not contain TDF or a protease inhibitor. Their creatinine clearance (Cockcroft-Gault equation) was ≥50 ml/min at baseline. The 2LADY trial follow-up visits were scheduled at weeks 4, 12, 24, 36, and 48, and every 6 months thereafter, until the end of the study. Visits included clinical (general condition, height, weight, blood pressure, body temperature) and biological (blood count, creatinine, CD4 count, glycemia, cholesterol, triglycerides, proteinuria, phosphoremia, HIV viral load) evaluation. A total of 451 patients were included and randomized to receive either TDF + FTC + darunavir-ritonavir (DRVr), TDF + FTC + lopinavir-ritonavir (LPVr) or abacavir (ABC) + didanosine (ddI) + LPVr between January 2010 and September 2012 and were followed in a period of almost 5 years.18 Blood samples from patients were stored at −80 °C in the laboratory of the UMI 233 TransVIHMI laboratory in Montpellier, France. All consenting participants with a stored buffy coat blood sample and at least 3 creatinine measurements during the follow-up were included in this study (N = 369; Figure 1b).
Genetic Analyses
All genetic analyses were performed at the Molecular Genetics Epidemiology laboratory in Frederick, Maryland. DNA was extracted from blood samples and purified using the Qiagen Plasmid Midi kit. The genotyping of APOL1 G1 (rs73885319, p.S342G, and rs60910145, p.I384M) and G2 (rs71785313, p.N388Y389/–) was performed with TaqMan. The HR genotype is defined by the carriage of 2 risk alleles (G1/G1, G2/G2, or G1/G2) and the LR genotype by the carriage of 0 or 1 risk allele (G0/G0, G0/G1, or G0/G2).14
Estimation of GFR
The most widely used methods to measure serum creatinine are colorimetric and enzymatic methods on automated analyzer and isotope dilution-mass spectrometry. Creatinine methods based on automated colorimetric or enzymatic methods are generally used in clinical laboratories. The compensated Jaffe (colorometric) method was developed to minimize nonspecific interferences,19,20 but the enzymatic method reveals better analytical performance, better specificity with less interference, and better reproducibility.19 In this study, creatinine was assayed locally using isotope dilution-mass spectrometry for the 2LADY clinical trial participants and using the compensated Jaffe-based method implemented in routine medical follow-up for the DCU cohort participants.
According to the currently accepted standards, the eGFR was estimated using the CKD-EPI equation21 without the correction for race22 as recently revealed as the best estimate of kidney function in Black Africans.23, 24, 25, 26 CKD was defined as the persistence of eGFR <60 ml/min per 1.73 m2 in a 3-month period, corresponding to the G3a to G5 stage definition of the Kidney Disease Quality Outcome Initiative classification.27,28
Statistical Analyses
Baseline patient characteristics were compared by APOL1 risk status (HR vs. LR) using the Mann-Whitney test for continuous variables and the χ2 or the exact Fisher tests for categorical variables. These tests were also used to compare the West Africa and Cameroon cohorts.
We evaluated the association of the APOL1 genotype with eGFR at baseline using linear regression models and with annual eGFR change during follow-up using linear mixed regression models. Mixed models included random intercept and slope to account for correlation between repeated measurements of kidney function (eGFR). We constructed multivariable regressions (i.e., adjusted analyses) to account for potential confounding risk factors for development of kidney disease. These variables included age, hypertension, glycemia, CD4 count, and HIV viral load. eGFR at baseline was also included in the models evaluating eGFR changes.
We evaluated effect modification between APOL1 risk status and the immuno-virological status of the participants by evaluating interactions between APOL1 genotype and CD4 count at baseline, categorized as < or ≥200 cells/μl and between APOL1 and viral load at baseline categorized as < or ≥5 log/ml.
All statistical analyses were performed using the Stata software (version 15, Stata Corp., College Station, TX).
For power analysis, we used the R package “longpower.”29 Considering the APOL1 HR prevalences >20% reported in Nigeria, we assumed a prevalence of 20% for HR genotype in our study population.30 Therefore, a minimum of 350 patients (70 for HR group and 280 for LR group) was required to have a slope difference of 1 ml/min per 1.73 m2 per year in changes in eGFR between HR and LR groups, with an α risk = 0.05 and power ≥80%.
Results
Participant Characteristics
A total of 413 participants from the DCU cohort (Burkina Faso) and 369 patients from the 2LADY cohort (n = 293 in Yaoundé, Cameroon; n = 29 in Dakar, Senegal; and n = 47 in Bobo-Dioulasso, Burkina Faso) were included (Figure 1). The characteristics of the participants at baseline are presented in Supplementary Tables S1, S2, and S3. The DCU cohort consisted of 73.1% female participants, with median age (interquartile range) of 37 (31.0–43.4) years old, median eGFR of 99.3 (86.1–111.7) ml/min per 1.73 m2, and CD4 count of 202 (103–342) cells/μl. The proportion of eGFR <60 ml/min per 1.73 m2 was 1.9%, and that of hypertension was 6%. The median time to starting ART was 1.1 (0.6–5.0) months after the first visit in DCU. The 2LADY cohort consisted of 71.3% female participants, with median age of 38 (33–47) years old, median eGFR of 95.7 (80.9–111.2) ml/min per 1.73 m2, and CD4 count of 176 (79–288) cells/μl. The proportion of eGFR <60 ml/min per 1.73 m2 and hypertension were 3.0% and 7.3%, respectively. The median duration of first-line ART exposure was 4.3 (3.0–5.9) years. Overall, the 2LADY cohort participants were older (P < 0.001), had lower baseline CD4 count (P = 0.005), and lower baseline eGFR (P = 0.019) compared with the DCU participants.
Regarding the APOL1 genetic risk, there was no difference in the characteristic distribution between APOL1 HR and LR subgroups in both cohorts (Supplementary Tables S2 and S3).
Prevalence of APOL1 Risk Alleles
The Hardy-Weinberg equilibrium was verified for APOL1 genotype distribution in both cohorts (P > 0.05). In DCU, the G1 allele prevalence (95% CI) was 13.7% (11.5–16.2) and 10.8% (8.8–13.1) for the G2 allele (Supplementary Table S4A). In this cohort, 158 (38.3% [33.7–43.1]%) participants carried 1 risk allele and 22 of 413 (5.3% [3.5–8.0]%) carried 2 risk alleles (HR; Table 1). In 2LADY, the G1 and G2 allelic frequencies were 9.2% (7.3–11.5)% and 9.2% (7.3–11.5)%, respectively. The prevalence of APOL1 HR in the 2LADY cohort was 3.3% (1.9–5.6)% (12 of 369).
Table 1.
Distribution of participants according to the number of carried APOL1 renal risk alleles
| Study site | Number of APOL1 risk alleles carried |
||||
|---|---|---|---|---|---|
| 0 (G0/G0) |
1 (G0/G1 + G0/G2) |
2 (=HR) (G1/G1 + G2/G2 + G1/G2) |
|||
| n | n | P [CI95] (%) | n | P [CI95] (%) | |
| Day care unit cohort | 233 | 158 | 38.3 [33.7–43.1] | 22 | 5.3 [3.5–8.0] |
| 2LADY cohort | |||||
| All sites | 245 | 112 | 30.4 [25.9–35.3] | 12 | 3.3 [1.9–5.6] |
| Burkina Faso | 29 | 17 | 36.2 [23.5–51.1] | 1 | 2.1 [0.3–14.3] |
| Senegal | 16 | 12 | 41.4 [24.6–60.4] | 1 | 3.4 [0.2–22.3] |
| Cameroon | 200 | 83 | 28.3 [23.4–33.8] | 10 | 3.4 [1.8–6.2] |
| Burkina Fasoa | 262 | 175 | 38.0 [33.7–42.6] | 23 | 5.0 [3.3–7.4] |
| West Africab | 278 | 187 | 38.2 [34.0–42.6] | 24 | 4.9 [3.3–7.2] |
HR, high risk; P, prevalence.
The carriage of 0 or 1 risk allele defines the low-risk genotypes, when the carriage of 2 risk alleles defines the HR genotype. The detailed frequency for each genotype is provided in Supplementary Table 4B.
Burkina Faso = Burkina Faso from the 2LADY and day care unit cohorts combined;
West Africa = Burkina Faso + Senegal; Cameroon is representing Central Africa in our study.
When pooling data from both cohorts, the G1 prevalence was significantly higher in the West African sites (Burkina Faso and Senegal) compared with the Central African site in Cameroon (13.3% [11.3–15.6]% vs. 8.7% [6.7–11.3]%, P = 0.006) (Figure 2 and Supplementary Table S4A). The G2 allele was also more frequent in West Africa compared with Cameroon, but the difference was not statistically significant (10.7% [8.9–12.8]% vs. 8.9% [6.8–11.5]%, P = 0.235). Finally, there were 4.9% [3.3–7.2]% and 3.4% [1.8–6.2]% of the participants who carried the HR genotype in the West African sites and Cameroon, respectively (Table 1 and Supplementary Table S4B).
Figure 2.
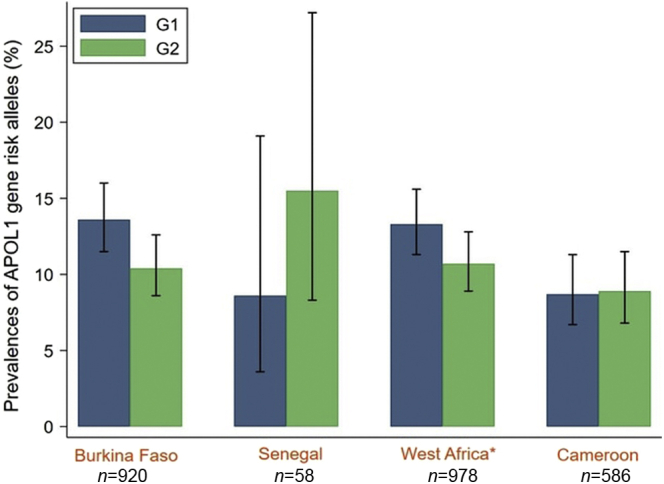
Allelic frequency of APOL1 risk variants in PLHIV in West Africa and Cameroon. ∗West Africa = Burkina Faso + Senegal; N, number of alleles (G0, G1, and G2). The bar graph displays the APOL1 risk alleles’ frequency (G1 in blue and G2 in green) with the corresponding 95% CIs. PLHIV, people living with HIV.
APOL1 Risk Alleles and Baseline eGFR
In the DCU cohort, where all individuals were ART naive at their first visit, there was no association between APOL1 HR and eGFR at baseline in the unadjusted analysis or after adjustment on age, hypertension, glycemia, and CD4 count (Table 2). Nevertheless, we found a significant interaction between APOL1 risk status and the immunosuppression status of the patients as reflected by their CD4 T-cell count level. In the subgroup with low CD4 counts (<200 cells/μl), APOL1 HR was associated with lower eGFR (β = −21.7 [−35.1 to −8.3] ml/min per 1.73 m2, P = 0.002) whereas eGFR was not associated with APOL1 risk genotype among participants with CD4 count ≥200 cells/μl (Figure 3).
Table 2.
Difference in baseline eGFR and in annual eGFR change according to APOL1 risk status
| Baseline and annual eGFR models | DCU cohort |
2LADY cohort |
||
|---|---|---|---|---|
| 0/1 allele (n = 391) | 2 alleles (n = 22) | 0/1 allele (n = 357) | 2 alleles (n = 12) | |
| Baseline eGFR (ml/min per 1.73 m2) | ||||
| Mean (95% CI) | 97.7 [95.9–99.6] | 94.0 [85.2–102.8] | 94.6 [92.6–96.6] | 101.0 [89.5–112.6] |
| Estimated difference in baseline eGFR, 2 vs. 0/1 alleles (95% CI)a | ||||
| Unadjusted | Reference | −3.7 [−11.7 to 4.2] | Reference | 6.5 [−4.6 to 17.6] |
| Adjusted on age, HTN, glycemia, and CD4 count | — | −4.2 [−11.9 to 3.4] | — | 8.5 [−1.7 to 18.7] |
| Adjusted on age, HTN, glycemia, and HIV viral load | — | — | — | 10.8 [0.8–20.8] |
| Adjusted on age, HTN, glycemia, CD4 count, and HIV viral load | — | — | — | 10.7 [0.8–20.7] |
| Annual change in eGFR (ml/min per 1.73 m2)b | ||||
| Mean (95% CI) | −0.8 [−1.0 to −0.6] | −0.7 [−2.2 to 0.7] | 2.0 [1.7–2.4] | 1.1 [−0.5 to 2.6] |
| Estimated difference in eGFR annual change, 2 vs. 0/1 alleles (95% CI)b | ||||
| Unadjusted | Reference | 0.2 [−0.8 to 1.2] | Reference | −1.0 [−3.1 to 1.1] |
| Adjustedc on age, HTN, glycemia, eGFR, and CD4 count | — | 0.0 [−1.1 to 1.2] | — | −1.2 [−3.4 to 1.0] |
| Adjustedc on age, HTN, glycemia, eGFR, CD4 count, and HIV viral load | — | — | — | −1.2 [−3.4 to 1.0] |
DCU, day care unit; eGFR, estimated glomerular filtration rate; HTN, hypertension.
At baseline, there was no difference in mean eGFR between low-risk (0/1 allele) and high-risk (2 alleles) genotypes in the DCU cohort. After adjusting for age, hypertension, glycemia, CD4 count, and HIV viral load, 2LADY patients with high-risk genotype had on average a 10.7 ml/min per 1.73 m2 higher GFR than those with a low-risk genotype. During follow-up, the annual eGFR change was not significantly different between high-risk and low-risk genotypes in both cohorts.
Linear regressions were used to obtain the coefficients.
Linear mixed model regressions were used to obtain the coefficients.
Adjustment variables were included using values recorded at baseline.
Figure 3.
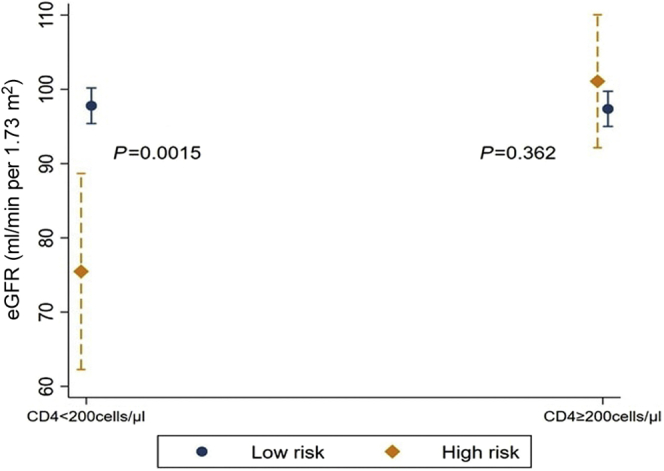
Baseline eGFR by CD4 levels and APOL1 risk status (DCU cohort). Mean baseline eGFR with the corresponding 95% CIs is presented for the DCU cohort’s participants according to baseline CD4 count (<200 cells/μl and ≥200 cells/μl) and to APOL1 risk status (high-risk [dark orange diamond] and low-risk [navy circle] genotypes). Predicted kidney function values were adjusted for age, glycemia, and hypertension. Mean baseline eGFR was significantly lower in the APOL1 high-risk group only for patients with low CD4 counts. DCU, day care unit; eGFR, estimated glomerular filtration rate.
In the 2LADY patients with a long history of ART, baseline eGFR was associated with APOL1 genotype in an unexpected direction, where patients with HR genotype had a higher mean eGFR than those with LR genotype (β = 10.7 [0.8–20.7] ml/min per 1.73 m2, P = 0.036 coh) (Table 2). In this ort, at baseline, a higher eGFR was also significantly associated with lower CD4 count and a higher viral load (Supplementary Table S5).
APOL1 Risk Alleles and Changes in eGFR During Follow-Up
The DCU participants were followed for a median duration of 6.1 (interquartile range: 4.2–8.2) years with an average of 1.9 measurements of creatinine per year. In the entire follow-up, the average eGFR decreased by 0.8 [0.6–1.0] ml/min per 1.73 m2 per year and 14 incident cases of CKD (5.7 [3.4–9.7] per 1000 person-years at risk), including 1 APOL1 HR genotype carrier, were reported (Supplementary Table S6). Neither the HR genotype, baseline CD4 count, nor the interaction between APOL1 and baseline CD4 count was associated with changes in eGFR over time.
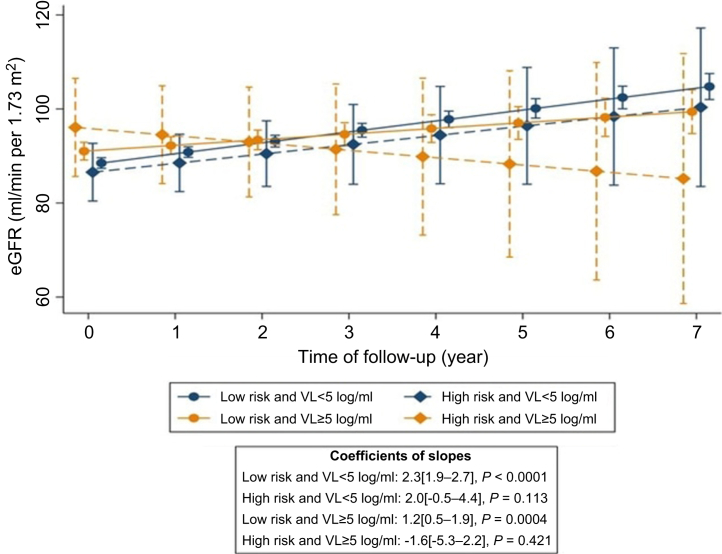
For the 2LADY participants, the median follow-up on second-line ART was 4.8 (interquartile range: 3.9–5.4) years with 3.1 measurements of creatinine per year on average. Over time, the eGFR increased by 2.0 (1.7–2.3) ml/min per 1.73 m2 per year and 16 incident cases of CKD (10.3 [6.3–16.8] per 1000 person-years at risk) were reported, all in the APOL1 LR subgroup (Supplementary Table S6). Neither the HR genotype, baseline CD4 count, nor the interaction between APOL1 and baseline CD4 count was associated with changes in eGFR. Nevertheless, high baseline HIV viral load (≥5 log/ml) was associated with faster eGFR progression (β = −1.2 [−2.0 to −0.4] ml/min per 1.73 m2 per year, P = 0.0018). In the subgroup with both high baseline HIV viral load and HR genotype, progression in eGFR was slower (β = −3.9 [−7.7 to −0.1] ml/min per 1.73 m2 per year, P = 0.046). These results held even after adjusting for baseline eGFR (Figure 4 and Supplementary Tables S7 and S8).
Figure 4.
Changes in eGFR over time by baseline HIV VL and APOL1 risk status (2LADY cohort). Predicted longitudinal eGFR with the corresponding 95% CIs is presented for the 2LADY cohort’s participants according to baseline HIV VL (<5 log/ml [navy color] and ≥5 log/ml [dark orange color]) and to APOL1 risk status (high-risk [diamond and dashed line] and low-risk [circle and solid line]). eGFR annual change was significantly faster in the APOL1 high-risk group exhibiting high baseline HIV VL. Predicted values were adjusted for the following baseline variables: age, eGFR, glycemia, hypertension, and CD4 count level (see Supplementary Table S8 for further details). eGFR, estimated glomerular filtration rate; VL, viral load.
Discussion
Prevalence of APOL1 Risk Alleles
In this report, we described for the first time the APOL1 renal risk allele frequencies in PLHIV from 3 countries of West (Burkina Faso and Senegal) and Central (Cameroon) Africa. The G1 and G2 allele frequencies were previously reported in various African populations.31,32 The reported frequencies were highly variable across studies, even within the same country, but the overall geographic distribution indicated higher frequencies of G1, and to a lesser extent of G2 alleles in West Africa.14,31 This study, reporting higher frequencies for G1 among PLHIV from Burkina Faso and Senegal (13.3%) than among PLHIV from Cameroon (8.7%), confirmed this geographic pattern. Nevertheless, the frequencies reported here in the West African sites were far below most observations within this region, which mainly came from 2 countries, Ghana and Nigeria, where the G1 allele frequencies exceeded 40% in several reports or subpopulations.31 Outside these 2 countries, there were few data available in West Africa and none in Burkina Faso, to our best knowledge. Further investigation is therefore necessary to better describe and understand the geographic distribution of the APOL1 risk variants in West African populations. Our data stood in the line of previous reports from Central Africa. We observed APOL1 risk allele frequencies (G1, 8.7% and G2, 8.9%) close to the previous studies among the general population in Cameroon (G1, 0.8%–16.4% and G2, 3.3%–12.3%)31 and among HIV-infected children from the Democratic Republic of Congo (G1, 13.5% and G2, 9.6%).32
Although the frequency of the APOL1 HR genotype was estimated at 14% in African Americans, very few data were available in Africa.14 In a pediatric population from the Democratic Republic of Congo, the HR prevalence was 5.7% in PLHIV and 7% in the general population.32 In a recent study on a cohort of HIV-positive Nigerian adults in northern of the country, the HR prevalence was 6.2%, with large variation by ethnic group (Hausa/Fulani = 2.1%, Igbo = 49.1%, and Yoruba = 14.5%).33 The HR prevalences in Cameroon (3.4%) or West Africa (4.9%) are close to these, but they are much lower than the 23% HR frequency reported in the Igbo group from Nigeria,30 hence limiting our study power for detecting genetic association. To date, Nigeria and to some extent Ghana look like the exceptions regarding APOL1 frequencies. Despite studies carried out in several Nigerian ethnic groups (i.e., the Yoruba, Esan, and Igbo), the APOL1 risk alleles and HR genotype prevalences remain constantly higher than any other tested country.31,34 The selective pressures responsible for the HR frequencies of G1 and to a lesser extent G2 among certain ethnic groups, primarily residing the eastern coastal regions of West Africa, remain to be elucidated.
APOL1, Immunosuppression Status, and Kidney Function
A few studies in African Americans have reported APOL1 HR genotype association with progressive loss of kidney function. In the African American Study of Kidney Disease and Hypertension, Parsa et al.35 found a more rapid decline in kidney function in individuals with the HR genotype compared with those with the LR genotype. Similarly, in another cohort study of young to middle-aged adults with preserved kidney function in the USA, HR genotype was associated with faster decline in eGFR by 0.38%.36 The decline in kidney function in the general population is similar by APOL1 genotype, but once renal injury occurs (onset of proteinuria), eGFR declines more rapidly in HR Black individuals. Similarly, HR Black individuals had the earliest onset of albuminuria compared with LR Black and White individuals.36 APOL1 HR genotypes seem to be specifically associated with renal injury that occurs more frequently and at a younger age.
In contrast with these American studies, we did not find a direct association between APOL1 HR genotype and decrease in kidney function over time, which could partly stem from unexpected limited study power (n = 22 and 12 HR patients in DCU and 2LADY, respectively). Even more surprising, we observed that the 2LADY cohort participants carrying the HR genotype had, at baseline, a higher eGFR than those with the LR genotype. In addition, we have found that severe immunosuppression (CD4 counts <200 cells/μl) and high HIV viral load (≥5 log/ml) were associated with higher baseline kidney function in the 2LADY cohort. These are unexpected results as most previous studies revealed that advanced HIV disease is accompanied by a deterioration in kidney function.5,37 Nevertheless, a similar counterintuitive association was reported in the DART cohort (where participants were symptomatic [World Health Organization disease stage ≥2] with CD4 cell counts <200 cells/μl at ART starting).38 These authors suggested that this result most likely reflected survivor and enrollment bias, that is, participants with both lower CD4 cell counts and poorer renal function were less likely to survive to meet enrollment criteria. Indeed, kidney impairment is associated with high mortality, both before and during ART.3, 4, 5 The 2LADY trial aimed to compare different second-line regimens among PLHIV failing first-line ART; participants had therefore to survive the pre-ART period and an unsuccessful first-line ART period. In addition, one of the 2LADY inclusion criteria in the trial was creatinine clearance >50 ml/min, which accentuated the selection bias.
Nevertheless, when investigating the interactions between APOL1 and immunologic or virological status in the 2 HIV cohorts with different study design, the results supported that the deleterious impact of the HR genotype on kidney function could be modulated by the immunologic or the virological status of the patients. In DCU, we found that HR participants initiating ART had lower baseline eGFR than participants with LR genotype, but only among those with CD4 count <200 cells/μl. It should be noted also that ART-naive PLHIV with CD4 count <200 cells/μl most often have very high HIV viral loads.39,40 Therefore, the observed association may be indirectly related to the virological status of patients. In 2LADY evaluating second-line ART strategies, APOL1 HR participants with a baseline viral load >5 log/ml had the poorest kidney function progression over time. This association between APOL1 and kidney function modulated by viral load levels has also previously been reported among HIV-infected men in USA. In that study, APOL1 HR was associated with a faster eGFR decline over time in men who did not have sustained viral load suppression.13
APOL1 and CKD
Many studies investigating the impact of APOL1 genotype on kidney function have focused on incident clinical outcomes (CKD or ESKD) rather than on the loss of kidney function.35,41 In addition, a Nigerian study found that HR carriers were associated with a higher occurrence (odds ratio = 4.8) of CKD in the general population.30 Owing to the unexpected low APOL1 allele frequency, this study was not powered to evaluate the risk of CKD progression according to the APOL1 genotype as both the numbers of HR carriers and CKD events were too low. Indeed, no case of CKD was recorded in HR genotype carriers in the 2LADY cohort and only 1 case was recorded in this group in DCU cohort. Yet, the incidence of CKD in the DCU and 2LADY cohorts (5.7 and 10.3 per 1000 person-years, respectively) was consistent with previous studies investigating kidney function among PLHIV in different countries of West Africa.42
Limitations of the Study
This study is the first to estimate the prevalence of the APOL1 HR variants among PLHIV in West Africa, where high allelic frequencies were previously reported in Nigeria and Ghana from the southwest coastal regions. With data from naive and long-term ART-exposed patients from 3 West and Central African cohorts, we were able to explore the association between kidney function and APOL1 genotype. Robust interpretation of the results was, however, compromised by a lack of statistical power, as the observed APOL1 HR frequencies in our settings (3.4%–5%) were far below the figures reported in previous studies from West Africa and USA in African Americans (15%–20%) and on which we based our sample size calculation. The 2 cohorts investigated here had different study designs, which strengthens similar conclusions but also makes some comparisons more challenging, as for example the lack of regular baseline viral load measurements in DCU. In addition, we cannot exclude a survivor bias in the study cohorts, when people with both lower immunity condition and poorer renal function would be less likely to survive and meet the study enrollment criteria. Furthermore, the sample size for Cameroon, a country from Central Africa, was smaller than that for the 2 West African countries (n = 293 vs. n = 489), calling for additional studies in the area. Finally, the study lacked baseline proteinuria data to better characterize kidney function outcomes beyond eGFR.
Conclusion
This first study on APOL1 in West African PLHIV provided a better picture of APOL1 prevalence in sub-Sahara African PLHIV. It underlined the particularity of Nigeria and Ghana in the region and strongly advocates for the urgent need in larger genetic studies in diverse populations and regions of Africa. Despite limited power, this study suggested a modulation of APOL1 kidney damage by HIV virological status, but larger cohort studies will be needed to validate this observation.
Disclosure
All the authors declared no competing interests.
Acknowledgments
This work is the result of a collaborative research between teams from Burkina Faso, France, and the United States. We thank Professor Eric Delaporte who accepted NFK in his laboratory in Montpellier, France, for his PhD thesis and who provided the necessary support for the achievement of this work. We also thank France Recherche Nord & Sud Sida-hiv Hépatites (ANRS) for the PhD scholarship (ANRS 12169 B98) and the HIV Research Trust for the scholarship that allowed NFK to do an internship at the Basic Research Laboratory, Frederick National Laboratory, Frederick, MD, USA. The project has been supported in part the National Institutes of Health and the National Cancer Institute Intramural Research Program (CAW and SL) and under contract HHSN26120080001E. The content of this publication does not necessarily reflect the view or policy of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the government. Finally, we thank all the patients whose data made this work possible. Some of the findings reported in this manuscript have been presented at Independent Communications Authority of South Africa 2019 in Kigali, Rwanda (poster: TUPEC212, French).
Author Contributions
NFK contributed to the data collection and study design, performed statistical analysis, and wrote the manuscript. AP, JZ, EBR, and VD contributed to the data collection, discussion, and reviewed/edited the manuscript. LC, AS, ABS, SED, SKS, CK, NFG, and NM contributed to the discussion and reviewed/edited the manuscript. SL and CW contributed to the study design, genetic analyses, discussion, and reviewed/edited the manuscript. AC contributed to the study design, statistical analysis, discussion, and reviewed/edited the manuscript. All authors have read and approved the final manuscript.
DATA AVAILABILITY STATEMENT
The data supporting the findings of this study cannot be made publicly available because patients did not consent to public sharing of their data. Access to data requires administrative authorization from the head of the Infectious Diseases Department of Souro Sanou University Hospite and the 2LADY/ANRS-12169 Scientific comity, after evaluation of the request. Researchers who wish to access some of the data from this study must address a request detailing the types of analyses they wish to perform. This request can be sent directly to the corresponding author at: sophie.limou@univ-nantes.fr; telephone: +33 244 768 271.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study, which involves participants from Cameroon, Burkina Faso, and Senegal, has been approved by the ethics committees of the 3 respective countries’ (“Comité d’éthique pour la recherche en santé” for Burkina Faso, “Comité national d’éthique pour la recherche en santé” for Senegal, and “Comité national d’éthique de la recherche pour la santé humaine” for Cameroon) upstream project initiation. Written consent was obtained from participants, and a waiver of consent was obtained from the ethics committee for participants in the 2LADY trial who were no longer alive at the time of this study.
Footnotes
Table S1. Baseline characteristics of all participants.
Table S2. Baseline characteristics of the day care unit (DCU) cohort participants stratified by APOL1 risk status.
Table S3. Baseline characteristics of the 2LADY cohort participants stratified by APOL1 risk status.
Table S4A. Prevalences of APOL1 risk alleles.
Table S4B. Distribution of APOL1 genotypes.
Table S5. Factors associated with eGFR in the 2LADY cohort at baseline.
Table S6. Follow-up data by study cohort.
Table S7. Baseline predictors of eGFR annual change in the 2LADY cohort.
Table S8. Baseline predictors of eGFR annual change in the 2LADY cohort by stratifying with the interaction between APOL1 and baseline HIV viral load.
Supplementary Material
Table S1. Baseline characteristics of all participants.
Table S2. Baseline characteristics of the day care unit (DCU) cohort participants stratified by APOL1 risk status.
Table S3. Baseline characteristics of the 2LADY cohort participants stratified by APOL1 risk status.
Table S4a. Prevalences of APOL1 risk alleles.
Table S4b. Distribution of APOL1 genotypes.
Table S5. Factors associated with eGFR in the 2LADY cohort at baseline.
Table S6. Follow-up data by study cohort.
Table S7. Baseline predictors of eGFR annual change in the 2LADY cohort.
Table S8. Baseline predictors of eGFR annual change in the 2LADY cohort by stratifying with the interaction between APOL1 and baseline HIV viral load.
References
- 1.UNAIDS-data 2019 UNAIDS. https://www.unaids.org/sites/default/files/media_asset/2019-UNAIDS-data_en.pdf Accessed September 24, 2019.
- 2.Ekrikpo U.E., Kengne A.P., Bello A.K., et al. Chronic kidney disease in the global adult HIV-infected population: a systematic review and meta-analysis. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardner L.I., Holmberg S.D., Williamson J.M., et al. Development of proteinuria or elevated serum creatinine and mortality in HIV-infected women. J Acquir Immune Defic Syndr. 2003;32:203–209. doi: 10.1097/00126334-200302010-00013. [DOI] [PubMed] [Google Scholar]
- 4.Choi A., Scherzer R., Bacchetti P., et al. Cystatin C, albuminuria, and 5-year all-cause mortality in HIV-infected persons. Am J Kidney Dis. 2010;56:872–882. doi: 10.1053/j.ajkd.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopp J.B., Winkler C. HIV-associated nephropathy in African Americans. Kidney Int Suppl. 2003;(83):S43–S49. doi: 10.1046/j.1523-1755.63.s83.10.x. [DOI] [PubMed] [Google Scholar]
- 6.Foy M.C., Estrella M.M., Lucas G.M., et al. Comparison of risk factors and outcomes in HIV immune complex kidney disease and HIV-associated nephropathy. Clin J Am Soc Nephrol. 2013;8:1524–1532. doi: 10.2215/CJN.10991012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fine D.M., Wasser W.G., Estrella M.M., et al. APOL1 risk variants predict histopathology and progression to ESRD in HIV-related kidney disease. J Am Soc Nephrol. 2012;23:343–350. doi: 10.1681/ASN.2011060562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estrella M.M., Wyatt C.M., Pearce C.L., et al. Host APOL1 genotype is independently associated with proteinuria in HIV infection. Kidney Int. 2013;84:834–840. doi: 10.1038/ki.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipkowitz M.S., Freedman B.I., Langefeld C.D., et al. Apolipoprotein L1 gene variants associate with hypertension-attributed nephropathy and the rate of kidney function decline in African Americans. Kidney Int. 2013;83:114–120. doi: 10.1038/ki.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopp J.B., Nelson G.W., Sampath K., et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22:2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasembeli A.N., Duarte R., Ramsay M., et al. APOL1 risk variants are strongly associated with HIV-associated nephropathy in Black South Africans. J Am Soc Nephrol. 2015;26:2882–2890. doi: 10.1681/ASN.2014050469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaboré N.F., Limou S. Balancing the genetic risk of APOL1 kidney disease variants. Nephrol Ther. 2019;15(suppl 1):S79–S84. doi: 10.1016/j.nephro.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Estrella M.M., Li M., Tin A., et al. The association between APOL1 risk alleles and longitudinal kidney function differs by HIV viral suppression status. Clin Infect Dis. 2015;60:646–652. doi: 10.1093/cid/ciu765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Limou S., Nelson G.W., Kopp J.B., Winkler C.A. APOL1 kidney risk alleles: population genetics and disease associations. Adv Chronic Kidney Dis. 2014;21:426–433. doi: 10.1053/j.ackd.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadkarni G.N., Gignoux C.R., Sorokin E.P., et al. Worldwide frequencies of APOL1 renal risk variants. N Engl J Med. 2018;379:2571–2572. doi: 10.1056/NEJMc1800748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaboré N.F., Poda A., Zoungrana J., et al. Chronic kidney disease and HIV in the era of antiretroviral treatment: findings from a 10-year cohort study in a west African setting. BMC Nephrol. 2019;20:155. doi: 10.1186/s12882-019-1335-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciaffi L., Koulla-Shiro S., Sawadogo A., et al. Efficacy and safety of three second-line antiretroviral regimens in HIV-infected patients in Africa. AIDS. 2015;29:1473–1481. doi: 10.1097/QAD.0000000000000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciaffi L., Koulla-Shiro S., Sawadogo A.B., et al. Boosted protease inhibitor monotherapy versus boosted protease inhibitor plus lamivudine dual therapy as second-line maintenance treatment for HIV-1-infected patients in sub-Saharan Africa (ANRS12 286/MOBIDIP): a multicentre, randomised, parallel, open-label, superiority trial. Lancet HIV. 2017;4:e384–e392. doi: 10.1016/S2352-3018(17)30069-3. [DOI] [PubMed] [Google Scholar]
- 19.Peake M., Whiting M. Measurement of serum creatinine – current status and future goals. Clin Biochem Rev. 2006;27:173–184. [PMC free article] [PubMed] [Google Scholar]
- 20.Delanghe J.R., Speeckaert M.M. Creatinine determination according to Jaffe-what does it stand for? NDT Plus. 2011;4:83–86. doi: 10.1093/ndtplus/sfq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levey A.S., Stevens L.A., Schmid C.H., et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glassock R.J., Warnock D.G., Delanaye P. The global burden of chronic kidney disease: estimates, variability and pitfalls. Nat Rev Nephrol. 2017;13:104–114. doi: 10.1038/nrneph.2016.163. [DOI] [PubMed] [Google Scholar]
- 23.Wyatt C.M., Schwartz G.J., Owino Ong’or W., et al. Estimating kidney function in HIV-infected adults in Kenya: comparison to a direct measure of glomerular filtration rate by iohexol clearance. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eastwood J.B., Kerry S.M., Plange-Rhule J., et al. Assessment of GFR by four methods in adults in Ashanti, Ghana: the need for an eGFR equation for lean African populations. Nephrol Dial. 2010;25:2178–2187. doi: 10.1093/ndt/gfp765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bukabau J.B., Sumaili E.K., Cavalier E., et al. Performance of glomerular filtration rate estimation equations in Congolese healthy adults: the inopportunity of the ethnic correction. PLoS One. 2018;13 doi: 10.1371/journal.pone.0193384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Deventer H.E., George J.A., Paiker J.E., Becker P.J., Katz I.J. Estimating glomerular filtration rate in Black South Africans by use of the modification of diet in renal disease and Cockcroft-Gault equations. Clin Chem. 2008;54:1197–1202. doi: 10.1373/clinchem.2007.099085. [DOI] [PubMed] [Google Scholar]
- 27.National Kidney Foundation K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002;39(suppl 1):S1–S266. [PubMed] [Google Scholar]
- 28.Chapter 1. Definition and classification of CKD. Kidney Int Suppl (2011) 2013;3:19–62. doi: 10.1038/kisup.2012.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diggle P., editor. Analysis of longitudinal data. Second Paperback Edition. Oxford University Press; 2013. [Google Scholar]
- 30.Ulasi I.I., Tzur S., Wasser W.G., et al. High population frequencies of APOL1 risk variants are associated with increased prevalence of non-diabetic chronic kidney disease in the Igbo people from south-eastern Nigeria. Nephron Clin Pract. 2013;123:123–128. doi: 10.1159/000353223. [DOI] [PubMed] [Google Scholar]
- 31.Cooper A., Ilboudo H., Alibu V.P., et al. APOL1 renal risk variants have contrasting resistance and susceptibility associations with African trypanosomiasis. ELife. 2017;6 doi: 10.7554/eLife.25461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ekulu P.M., Nkoy A.B., Betukumesu D.K., et al. APOL1 risk genotypes are associated with early kidney damage in children in sub-Saharan Africa. Kidney Int Rep. 2019;4:930–938. doi: 10.1016/j.ekir.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wudil U.J., Aliyu M.H., Prigmore H.L., et al. Apolipoprotein-1 risk variants and associated kidney phenotypes in an adult HIV cohort in Nigeria. Kidney Int. 2021;100:146–154. doi: 10.1016/j.kint.2021.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson R., Genovese G., Canon C., et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci U S A. 2014;111:E2130–E2139. doi: 10.1073/pnas.1400699111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsa A., Kao W.H.L., Xie D., et al. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med. 2013;369:2183–2196. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peralta C.A., Bibbins-Domingo K., Vittinghoff E., et al. APOL1 genotype and race differences in incident albuminuria and renal function decline. J Am Soc Nephrol. 2016;27:887–893. doi: 10.1681/ASN.2015020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyatt C.M., Klotman P.E. HIV-1 and HIV-associated nephropathy 25 years later. Clin J Am Soc Nephrol. 2007;2(suppl 1):S20–S24. doi: 10.2215/CJN.03561006. [DOI] [PubMed] [Google Scholar]
- 38.Reid A., Stöhr W., Walker A.S., et al. Severe renal dysfunction and risk factors associated with renal impairment in HIV-infected adults in Africa initiating antiretroviral therapy. Clin Infect Dis. 2008;46:1271–1281. doi: 10.1086/533468. [DOI] [PubMed] [Google Scholar]
- 39.Phillips A.N., Lampe F.C., Smith C.J., et al. Ongoing changes in HIV RNA levels during untreated HIV infection: implications for CD4 cell count depletion. AIDS. 2010;24:1561–1567. doi: 10.1097/QAD.0b013e32833a6056. [DOI] [PubMed] [Google Scholar]
- 40.Natural History Project Working Group for the Collaboration of Observational HIV Epidemiological Research Europe (COHERE) in EuroCoord Factors associated with short-term changes in HIV viral load and CD4(+) cell count in antiretroviral-naive individuals. AIDS. 2014;28:1351–1356. doi: 10.1097/QAD.0000000000000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foster M.C., Coresh J., Fornage M., et al. APOL1 variants associate with increased risk of CKD among African Americans. J Am Soc Nephrol. 2013;24:1484–1491. doi: 10.1681/ASN.2013010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poda A., Kabore N.F., Malateste K., et al. Validation of the D:A:D chronic kidney disease risk score in people living with HIV: the IeDEA West Africa Cohort Collaboration. HIV Med. 2021;22:113–121. doi: 10.1111/hiv.12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study cannot be made publicly available because patients did not consent to public sharing of their data. Access to data requires administrative authorization from the head of the Infectious Diseases Department of Souro Sanou University Hospite and the 2LADY/ANRS-12169 Scientific comity, after evaluation of the request. Researchers who wish to access some of the data from this study must address a request detailing the types of analyses they wish to perform. This request can be sent directly to the corresponding author at: sophie.limou@univ-nantes.fr; telephone: +33 244 768 271.