Abstract
Elevated lipoprotein(a) [Lp(a)] is a risk factor for coronary heart disease (CHD), but there are few studies on the prediction of future cardiovascular events by Lp(a) and its LPA single nucleotide polymorphisms (SNPs). The aim of this study was to investigate whether elevated Lp(a) and its SNPs can predict cardiovascular events. We evaluated whether Lp(a) and LPA SNPs rs6415084 and rs12194138 were associated with the incidence rate and severity of CHD. All participants were followed up for 5 years. Elevated Lp(a) is an independent risk factor for the risk and severity of CHD (CHD group vs. control group: OR = 1.793, 95% CI: 1.053–2.882, p = 0.043; multiple-vessel disease group vs. single-vessel disease group: OR = 1.941, 95% CI: 1.113–3.242, p = 0.027; high GS group vs. low GS group: OR = 2.641, 95% CI: 1.102–7.436, p = 0.040). Both LPA SNPs were risk factors for CHD, and were positively associated with the severity of CHD (LPA SNPs rs6415084: CHD group vs. control group: OR = 1.577, 95% CI: 1.105–1.989, p = 0.004; multiple-vessel disease group vs. single-vessel disease group: OR = 1.613, 95% CI: 1.076–2.641, p = 0.030; high GS group vs. low GS group: OR = 1.580, 95% CI: 1.088–2.429, p = 0.024; LPA SNPs rs12194138: CHD group vs. control group: OR = 1.475, 95% CI: 1.040–3.002, p = 0.035; multiple-vessel disease group vs. single-vessel disease group: OR = 2.274, 95% CI: 1.060–5.148, p = 0.038; high GS group vs. low GS group: OR = 2.067, 95% CI: 1.101–4.647, p = 0.021). After 5 years of follow-up, elevated Lp(a) and LPA SNPs rs6415084 and rs12194138 can independently predict cardiovascular events. The increase of serum Lp(a) and LPA SNPs rs6415084 and rs12194138 are associated with increased prevalence and severity of CHD, and can independently predict cardiovascular events.
Subject terms: Predictive markers, Coronary artery disease and stable angina
Introduction
Cardiovascular disease has the highest incidence rate and mortality rate in the world1–4. Over the past decades, a large number of studies have reported its possible risk factors, such as diabetes, hypertension, dyslipidemia, and smoking, in order to early assess the risk of cardiovascular disease5–8. At present, large epidemiological and genetic studies have provided strong evidences that lipoprotein(a) [Lp(a)] is a causal risk factor for coronary heart disease (CHD)9–11.
Lp(a) is a lipoprotein synthesized from the liver. It is an LDL-like particle that consists of an apolipoprotein(a) moiety linked to one molecule of apolipoprotein B100 via a disulfide bond12. Like low density lipoprotein cholesterol (LDL-C), Lp(a) can accumulate in the subendothelial space, leading to progressive atherosclerosis13. It has also been shown to produce more signaling, enhance its atherosclerotic ability. Lp(a) can induce systemic inflammatory response, thrombosis and promote oxidation14.
Plasma Lp (a) level is mainly determined by the LPA gene variation encoding apolipoprotein (a)15. Individual Lp(a) levels range from < 0.1 mg/dl to > 200 mg/dl, and it was highly heritable16. LPA is the major gene controlling this quantitative and co-dominantly expressed trait in all populations3. In recent years, many single nucleotide polymorphisms (SNPs) have been found in LPA17,18, such as rs6415084 and rs12194138, were closely associated with Lp(a) levels.
The relationship between Lp(a) levels and SNPs and the risk and severity of CHD as well as future recurrent cardiac events have been less studied, but so far they seem to be weak. We aimed to systematically assess whether Lp(a) and two LPA SNPs are associated with the occurrence and severity of CHD and long-term cardiovascular events.
Methods
Study population
We enrolled 2766 Han Chinese subjects (1614 males, 1152 females) from Peking University People’s Hospital and Peking University International Hospital from May 2013 to September 2015. There were 1665 patients (993 males, 672 females) with CHD and 1101 subjects (621 males, 480 females) in the control group. The follow-up procedures were performed by experienced nurses or doctors every 6 months via telephone or face-to-face interviews. Major cardiovascular events (MACEs) are divided into cardiovascular mortality, non-fatal myocardial infarction (MI), non-fatal stroke, heart failure, hospitalized unstable angina and non-coronary heart disease patients diagnosed as coronary heart disease. The longest follow-up time is 5 years.
The diagnostic criteria for patients with CHD were based on the coronary angiography (CAG) performed in our institution, and defined as at least one major coronary artery occlusion or stenosis of more than 50% and the severity of CHD was evaluated by the Gensini score (GS). CHD patients were divided into three groups according to their GS: low GS (GS ≤ 25), intermediate GS (GS: 26–41) and high GS (GS ≥ 42). The control group who all received coronary computed tomographic angiography (CTA) was selected from the physical examination center during the same time period, and included individuals without CHD.
All patients diagnosed as type 2 diabetes were selected based on the criteria set by the American Diabetes Association: (1) self-reporting to the clinician that he/she has a history of type 2 diabetes, (2) under current treatment of oral hypoglycemic medicine or insulin, (3) repeated fasting plasma glucose (FPG) greater than 7.0 mmol/L, or (4) glycated hemoglobin A1c (HbA1c) ≥ 6.5%.
Exclusion criteria included: (1) percutaneous coronary intervention within the previous three months, (2) acute coronary syndrome within the previous six months, (3), history of coronary artery bypass operation, (4) chronic heart failure, cardiomyopathy, valvular heart disease, (5) pulmonary heart disease, (6) severe liver and kidney dysfunction, or (7) any known inflammatory or infectious disease, or confirmed or suspected cancer.
The present study complied with the Declaration of Helsinki and was approved by the Peking University People’s Hospital Research Ethics Committee. Informed written consents were obtained from all patients enrolled in this study.
Measurements of lipoprotein(a) and other biomarkers
Blood samples were collected in the morning after at least 12 h of fasting. All measurements were performed within 6 h. Lp(a) in the serum samples was measured using latex enhanced immunoturbidimetry. The Lp(a) detection kit (Roche Inc., Germany) was used to determine the precipitation at 800/660 nm using latex particles coated with anti Lp(a) antibody to allow agglutination with human lipoprotein. FPG, homocysteine (HCY), hypersensitive C-reactive protein (hs-CRP), serum lipid profiles, including triglycerides (TG), total cholesterol (TC), LDL-C, and high density lipoprotein cholesterol (HDL-C), were analyzed with a Beckman AU5832 analyzer (Beckman Coulter Inc., USA). Apolipoproteins A-1 (apoA1) and B (apoB) were measured by immunoturbidimetry (Daiichi Pure Chemicals Co., Ltd., Tokyo). Direct quantitative analysis of small dense low-density lipoprotein cholesterol (sdLDL-C) assay was done using sdLDL-C reagent kits (Denka Seiken Co., Ltd. Japan). The HbA1c was determined with high-performance liquid chromatography (Trinity Biotech Inc., USA).
DNA extraction and genotyping
According to the manufacturer's instructions, genomic DNA was extracted from 3.5 ml EDTA anticoagulant blood collection vessel (Becton, Dickinson and company, USA) using puregene DNA separation kit (TianGen Biotech, Beijing, China). The incidence of LPA variant rs6415084 or rs12194138 was determined by gene sequencing (TsingKe Biological Technology, Beijing, China). All Sanger sequencing data shall be visually inspected by three experienced operators in our laboratory.
Statistical analyses
All subjects were matched using propensity score matching, including age, sex, body mass index (BMI), diabetes, hypertension, smoking, consumers of alcohol. All data were tested by one sample Kolmogorov–Smirnov test to determine whether the distribution of quantitative variables was normal. The normally distributed data were reported as means ± SD, and the differences between various groups were compared by the Student’s t test. The abnormally distributed continuous variables were reported as medians (interquartile range), and the differences between various groups were compared by the Mann–Whitney U test. The χ2 test was used to examine the Hardy–Weinberg equilibrium for each variant and to compare the distribution of allele and genotype frequencies between CHD patients and control subjects. The association of Lp(a) and SNPs with the presence and severity of CHD was analyzed using multivariate logistic regression adjusted for age, sex, BMI, diabetes, hypertension, smoking, consumers of alcohol, FPG, HbA1c, ApoB, ApoA1, TC, TG, HDL-C, hs-CRP, and HCY. The Kaplan–Meier method was used to estimate the event-free survival rates among groups. The significant level of all statistical tests was p < 0.05. SPSS 22.0 for Windows (SPSS Inc., USA) and GraphPad Prism 7 (GraphPad Software Inc., USA) were employed for the statistical analyses.
Ethics approval and consent to participate
The present study complied with the Declaration of Helsinki and was approved by the Hospital Research Ethics Committee, and written informed consent was obtained from all patients.
Results
Baseline characteristics of the study group
The recruitment scheme of all subjects is shown in Fig. 1.
Figure 1.
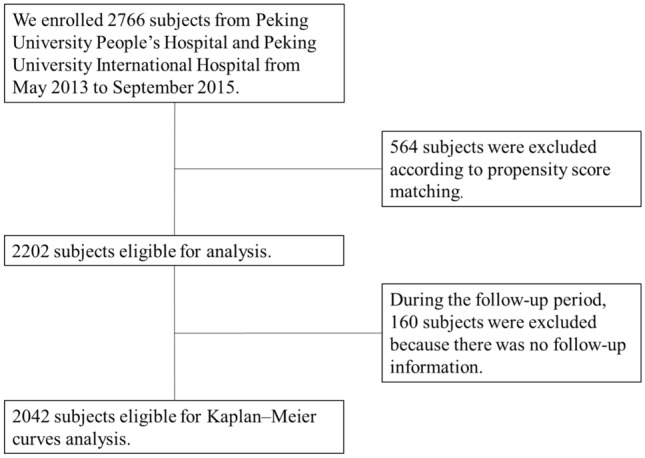
The flow chart of the patients’ selection process.
Baseline clinical data of all subjects (Table 1). There was no significant difference in age, sex, BMI, diabetes, hypertension, smoking, consumers of alcohol, FPG, HbA1c, TC and TG between CHD group and control group. The levels of ApoB, LDL-C, sdLDL-C, hs-CRP, Hcy and Lp(a) [39.71 (48.25) nmol/L vs. 38.17 (33.34) nmol/L, p = 0.005] in CHD group were significantly higher than those in the control group, whereas the apoA1 and HDL-C levels were lower. The distribution of LPA the two SNP genotypes were both significantly different between the CHD group and control group (Table 1). Rs6415084 (CT/TT) and rs12194138 (AT/TT) genotypes were more prone to CHD than CC and AA genotypes (17.25% vs. 11.44%, p. < 0.001; 6.81% vs. 4.54%, p = 0.027).
Table 1.
Baseline characteristics of all subjects.
| Variables | Total | CHD group | Control group | p value |
|---|---|---|---|---|
| N (%) | 2202 | 1101 (50.00%) | 1101 (50.00%) | – |
| Age (years) | 54.28 ± 11.01 | 54.98 ± 10.19 | 53.53 ± 11.32 | 0.187 |
| Male (%) | 1281 (58.17%) | 660 (59.95%) | 621 (56.40%) | 0.101 |
| BMI (kg/m2) | 26.05 ± 3.10 | 26.14 ± 2.92 | 25.83 ± 3.22 | 0.181 |
| Diabetes | 569 (25.84%) | 301 (27.34%) | 268 (24.34%) | 0.119 |
| Hypertension | 1531 (69.53%) | 781 (70.94%) | 750 (68.12%) | 0.165 |
| Smoking | 612 (27.79%) | 289 (26.25%) | 323 (29.34%) | 0.116 |
| Consumers of alcohol | 669 (30.38%) | 345 (31.34%) | 324 (29.43%) | 0.354 |
| Laboratory variables | ||||
| FPG (mmol/L) | 6.15 ± 2.02 | 6.22 ± 2.07 | 6.06 ± 1.81 | 0.224 |
| HbA1c (%) | 5.93 ± 1.41 | 6.03 ± 1.49 | 5.75 ± 1.40 | 0.142 |
| ApoB (mg/dL) | 82.05 ± 28.54 | 88.55 ± 29.71 | 76.65 ± 28.12 | < 0.001 |
| ApoA1 (mg/dL) | 148.78 ± 32.98 | 130.22 ± 30.13 | 175.85 ± 34.85 | < 0.001 |
| Total cholesterol (mmol/L) | 4.43 (1.36) | 4.51 (1.50) | 4.39 (0.96) | 0.155 |
| Triglycerides (mmol/L) | 1.33 (0.92) | 1.35 (0.88) | 1.32 (0.95) | 0.469 |
| HDL-C (mmol/L) | 1.06 (0.47) | 0.86 (0.31) | 1.32 (0.40) | < 0.001 |
| LDL-C (mmol/L) | 2.68 (0.87) | 2.79 (1.36) | 2.65 (0.63) | < 0.001 |
| sdLDL-C (mmol/L) | 0.73 (0.35) | 0.77 (0.48) | 0.71 (0.31) | 0.002 |
| hs-CRP (mg/L) | 2.43 (3.27) | 4.01 (3.82) | 2.10(1.56) | 0.021 |
| HCY (μmol/L) | 10.68 (6.97) | 12.16 (10.02) | 9.58 (5.48) | < 0.001 |
| Lp(a) (nmol/L) | 39.33 (38.49) | 39.71 (48.25) | 38.17 (33.34) | 0.005 |
| Minor allele frequency, N (%) | ||||
| SNP rs6415084 | 316 (14.35%) | 190 (17.25%) | 126 (11.44%) | < 0.001 |
| SNP rs12194138 | 125 (5.68%) | 75 (6.81%) | 50 (4.54%) | 0.027 |
Data are reported as means ± SD or n(%), median (interquartile ranges). SD: Standard deviation.
Statistical analysis was performed with the student t test or Mann–Whitney U test and with Chi-square test for categorical variables.
BMI Body mass index, HbA1c Hemoglobin A1c, apoB Apolipoprotein B, apoA1 Apolipoprotein A1, HDL-C High density lipoprotein cholesterol, LDL-C Low density lipoprotein cholesterol, sdLDL-C Small dense low-density lipoprotein cholesterol, Hs-CRP Hypersensitive C-reactive protein, HCY Homocysteine, Lp(a) Lipoprotein(a).
The serum Lp (a) level and the prevalence of LPA SNPs were associated with the severity of CHD
The patients with CHD were then classified into single-vessel (n = 287), two-vessel (n = 310), and multiple-vessel disease (n = 504) groups (Table 2). We found a significant higher in the Lp(a) levels in multiple-vessel diseases group [31.40 (41.32) nmol/L vs. 40.09 (49.55) nmol/L vs. 45.23 (49.01) nmol/L, p < 0.001]. The distribution of LPA SNPs is shown (Table 2) and both of them were significantly different among multiple-vessel diseases groups. In patients with CHD, rs6415084 (CT/TT) genotype carriers are more prone to multiple-vessel obstruction (13.59% vs. 18.84% vs. 20.83%, p = 0.014). Rs12194138 (AT/TT) genotype carriers are more likely to have more than two-vessel of obstruction (3.14% vs. 10.00% vs. 6.94%, p = 0.004).
Table 2.
Baseline characteristics of patients with multi vessel coronary artery disease.
| Variables | 1 vessel | 2 vessels | ≥ 3 vessels | p value |
|---|---|---|---|---|
| N (%) | 287 (26.07%) | 310 (28.16%) | 504 (45.78%) | – |
| Age (years) | 54.52 ± 11.01 | 55.74 ± 11.32 | 54.41 ± 10.27 | 0.592 |
| Male (%) | 169 (58.89%) | 181 (58.39%) | 310 (61.51%) | 0.619 |
| BMI (kg/m2) | 25.92 ± 3.15 | 26.04 ± 3.38 | 26.28 ± 3.61 | 0.197 |
| Diabetes | 76 (26.48%) | 89 (28.71%) | 136 (26.98%) | 0.806 |
| Hypertension | 189 (65.85%) | 226 (72.90%) | 366 (72.62%) | 0.088 |
| Smoking | 88 (30.66%) | 84 (27.10%) | 117 (23.21%) | 0.067 |
| Consumers of alcohol | 81 (28.22%) | 96 (30.97%) | 168 (33.33%) | 0.325 |
| Laboratory variables | ||||
| FPG (mmol/L) | 6.52 ± 2.31 | 5.98 ± 2.06 | 5.91 ± 2.04 | 0.080 |
| HbA1c (%) | 6.14 ± 1.43 | 6.01 ± 1.35 | 5.97 ± 1.42 | 0.202 |
| ApoB (mg/dL) | 69.45 ± 27.23 | 85.53 ± 21.70 | 102.19 ± 28.79*# | < 0.001 |
| ApoA1 (mg/dL) | 135.42 ± 25.40 | 122.68 ± 29.34* | 119.14 ± 32.53* | < 0.001 |
| Total cholesterol (mmol/L) | 3.66 (1.31) | 4.24 (1.31)* | 5.21 (1.82)*# | < 0.001 |
| Triglycerides (mmol/L) | 1.31 (0.76) | 1.32 (0.58) | 1.41 (0.94) | 0.325 |
| HDL-C (mmol/L) | 0.88 (0.35) | 0.84 (0.30) | 0.86 (0.27) | 0.378 |
| LDL-C (mmol/L) | 2.26 (1.12) | 2.70 (0.99)* | 3.38 (1.41)*# | < 0.001 |
| sdLDL-C (mmol/L) | 0.59 (0.23) | 0.74 (0.26)* | 1.04 ( 0.51)*# | < 0.001 |
| hs-CRP (mg/L) | 4.59 (5.13) | 4.10 (6.33) | 3.10 (5.97)*# | 0.001 |
| HCY (μmol/L) | 11.85 (8.08) | 12.75 (12.74) | 12.17 (12.25) | 0.271 |
| Lp(a) (nmol/L) | 31.40 (41.32) | 40.09 (49.55)* | 45.23 (49.01)*# | < 0.001 |
| Minor allele frequency, N (%) | ||||
| SNP rs6415084 | 39 (13.59%) | 46 (18.84%) | 105 (20.83%)*# | 0.014 |
| SNP rs12194138 | 9 (3.14%) | 31 (10.00%)* | 35(6.94%)* | 0.004 |
Data are reported as means ± SD or n (%), median (interquartile ranges). SD: Standard deviation.
Statistical analysis was performed with the ANOVA or Kruskal–Wall test and and with Chi-square test for categorical variables.
BMI Body mass index, HbA1c Hemoglobin A1c, apoB Apolipoprotein B, apoA1 Apolipoprotein A1, HDL-C High density lipoprotein cholesterol, LDL-C Low density lipoprotein cholesterol, sdLDL-C Small dense low-density lipoprotein cholesterol, Hs-CRP Hypersensitive C-reactive protein, HCY Homocysteine, Lp(a) Lipoprotein(a).
*p < 0.05 compared with the 1 vessel group.
#p < 0.05 compared with the 2 vessels group.
The patients were also divided into three groups based on the GS tercile: low GS (≤ 25, n = 378), intermediate GS (26–41, n = 354), and high GS (≥ 42, n = 369) group (Table 3). The results showed that the level of serum Lp(a) in high GS group and Intermediate GS were significantly higher than Low GS groups [32.72 (41.33) nmol/L vs. 42.41 (56.80) nmol/L vs. 47.63 (58.48) nmol/L, p < 0.001]. Similarly, the number of LPA SNPs rs6415084 (CT/TT) and rs12194138 (AT/TT) genotype carriers in the high GS group was significantly higher than Low GS groups (14.29% vs. 16.38% vs. 21.14%, p = 0.040; 4.50% vs. 6.50% vs. 9.49%, p = 0.025).
Table 3.
Baseline characteristics of Gensini score in patients with coronary heart disease.
| Variables | Low GS | Intermediate GS | High GS | p value |
|---|---|---|---|---|
| N (%) | 378 (34.33%) | 354 (32.15%) | 369 (33.51%) | – |
| Age (years) | 54.84 ± 10.34 | 56.05 ± 10.14 | 54.71 ± 10.06 | 0.491 |
| Male (%) | 213 (56.35%) | 222(62.71%) | 225 (60.98%) | 0.189 |
| BMI (kg/m2) | 26.11 ± 3.25 | 25.98 ± 3.13 | 26.42 ± 3.33 | 0.452 |
| Diabetes | 117 (30.95%) | 87 (24.58%) | 97 (26.29%) | 0.132 |
| Hypertension | 257 (67.99%) | 262 (74.01%) | 262 (71.00%) | 0.200 |
| Smoking | 106 (28.04%) | 100 (28.25%) | 83 (22.49%) | 0.132 |
| Consumers of alcohol | 107 (28.31%) | 124 (35.03%) | 114 (30.89%) | 0.135 |
| Laboratory variables | ||||
| FPG (mmol/L) | 6.32 ± 2.51 | 6.06 ± 2.37 | 5.98 ± 1.91 | 0.486 |
| HbA1c (%) | 6.18 ± 1.43 | 5.95 ± 1.42 | 6.04 ± 1.35 | 0.174 |
| ApoB (mg/dL) | 73.66 ± 27.06 | 88.54 ± 22.02* | 105.31 ± 30.52*# | < 0.001 |
| ApoA1 (mg/dL) | 126.83 ± 29.37 | 123.47 ± 31.10 | 132.42 ± 27.07 | 0.057 |
| Total cholesterol (mmol/L) | 3.97 (1.19) | 4.45 (1.34)* | 5.22 (1.94)*# | < 0.001 |
| Triglycerides (mmol/L) | 1.15 (0.76) | 1.23 (0.57) | 1.59 (1.01)*# | < 0.001 |
| HDL-C (mmol/L) | 0.90 (0.33) | 0.87 (0.29) | 0.83 (0.25) | 0.332 |
| LDL-C (mmol/L) | 2.46 (1.29) | 2.77 (0.95)* | 3.54 (1.41)*# | < 0.001 |
| sdLDL-C (mmol/L) | 0.58 (0.62) | 0.80 (0.50)* | 1.09 (1.05)*# | < 0.001 |
| hs-CRP (mg/L) | 2.65 (6.01) | 4.06 (7.16) | 3.90 (7.12) | 0.643 |
| HCY (μmol/L) | 11.86 (11.14) | 12.61 (11.02) | 12.99 (12.78) | 0.138 |
| Lp(a) (nmol/L) | 32.72 (41.33) | 42.41 (56.80)* | 47.63 (58.48)* | < 0.001 |
| Minor allele frequency, N (%) | ||||
| SNP rs6415084 | 54 (14.29%) | 58 (16.38%) | 78 (21.14%)* | 0.040 |
| SNP rs12194138 | 17 (4.50%) | 23 (6.50%) | 35 (9.49%)* | 0.025 |
Data are reported as means ± SD or n(%), median (interquartile ranges). SD: Standard deviation.
Statistical analysis was performed with the ANOVA or Kruskal–Wall test and and with Chi-square test for categorical variables.
BMI Body mass index, HbA1c Hemoglobin A1c, apoB Apolipoprotein B, apoA1 Apolipoprotein A1, HDL-C High density lipoprotein cholesterol, LDL-C Low density lipoprotein cholesterol, sdLDL-C Small dense low-density lipoprotein cholesterol, Hs-CRP Hypersensitive C-reactive protein, HCY Homocysteine, Lp(a) Lipoprotein(a).
*p < 0.05 compared with the Low GS group.
#p < 0.05 compared with the Intermediate GS group.
The effect of two SNPs on serum Lp(a) levels in Chinese Han people
The power values of SNPs rs6415084 and rs12194138 were 99% and 90%. The allele frequencies of SNPs rs6415084 and rs12194138 were 7.59% and 2.93%. The frequencies of the two tested variants did not deviate significantly in all subjects from the Hardy–Weinberg equilibrium: rs6415084, F = 2.630, p = 0.105; rs12194138, F = 2.502, p = 0.113.
Table 4 shows the relationship between LPA SNPs and serum Lp(a) levels. In LPA SNP rs6415084, the Lp(a) levels of rs6415084 (CC) genotype, rs6415084 (CT) genotype and rs6415084 (TT) genotype were different in all participants and CHD group (p < 0.001). In the control group, the Lp(a) levels of rs6415084 (CT/TT) genotype were higher than those of rs6415084 (CC) genotype (p < 0.001). In LPA SNP rs12194138, the Lp(a) levels of rs12194138 (AT/TT) genotype were higher than that of rs12194138 (AA) genotype in different groups (p < 0.001; p = 0.009; p < 0.001).
Table 4.
Serum Lp(a) levels in all subjects carrying different LPA SNP genotypes.
| rs6415084 | All subjects | p | ||
|---|---|---|---|---|
| CC (N = 1886) | CT (N = 298) | TT (N = 18) | ||
| Lp(a) nmol/L | 35.85 (31.33) | 72.75 (78.20)* | 157.91 (133.20)*# | < 0.001 |
| CHD group | ||||
| CC (N = 911) | CT (N = 175) | TT (N = 15) | ||
| Lp(a) nmol/L | 37.74 (38.06) | 81.83 (99.47)* | 195.38 (61.87)*# | < 0.001 |
| Control group | ||||
| CC (N = 975) | CT (N = 123) | TT (N = 3) | ||
| Lp(a) nmol/L | 34.16 (27.16) | 70.16 (38.62)* | 83.53 ( 36.28)* | < 0.001 |
| rs12194138 | All subjects | p | ||
|---|---|---|---|---|
| AA (N = 2077) | AT (N = 121) | TT (N = 4) | ||
| Lp(a) nmol/L | 38.10 (38.44) | 70.08 (32.47)* | 168.12 (119.13)*# | < 0.001 |
| CHD group | ||||
| AA (N = 1026) | AT (N = 71) | TT (N = 4) | ||
| Lp(a) nmol/L | 39.35 (50.53) | 70.07 (33.57)* | 168.12 (119.13)*# | 0.009 |
| Control group | ||||
| AA (N = 1051) | AT (N = 50) | TT (N = 0) | ||
| Lp(a) nmol/L | 36.84 (30.46) | 70.92 (36.84)* | – | < 0.001 |
*p < 0.05 compared with the rs6415084 (CC) or rs12194138 (AA) group.
#p < 0.05 compared with the rs6415084 (CT) or rs12194138 (AT) group.
Serum Lp (a) levels and LPA SNPs were associated with the risk of CHD.
In order to explore whether Lp(a) and LPA SNPs increase the risk of CHD, we conducted univariate and multivariate logistic regression analysis. All participants were divided into four groups according to the Lp(a) quartile level, and the presence and severity of CHD in individuals with different Lp(a) levels were assessed. In univariate logistic regression analysis, Lp(a) level was positively associated with the presence and severity of CHD (CHD group vs. control group: OR = 1.921, 95% CI: 1.102–3.121, p = 0.011; multiple-vessel disease group vs. single-vessel disease group: OR = 3.309, 95% CI: 2.293–5.030, p < 0.001; high GS group vs. low GS group: OR = 3.201, 95% CI: 1.234–8.09, p = 0.017) (Table 5). The multiple logistic regression analysis adjusted for age, sex, BMI, diabetes, hypertension, smoking, consumers of alcohol, FPG, HbA1c, ApoB, ApoA1, TC, TG, HDL-C, hs-CRP, and HCY, the level of Lp(a) remained to be independently associated with the presence and severity of CHD (CHD group vs. control group: OR = 1.793, 95% CI: 1.053–2.882, p = 0.043; multiple-vessel disease group vs. single-vessel disease group: OR = 1.941, 95% CI: 1.113–3.242, p = 0.027; high GS group vs. low GS group: OR = 2.641, 95% CI: 1.102–7.436, p = 0.040).
Table 5.
Odd ratios of CHD, multiple-vessel disease and high GS in relation to quartiles of lipoprotein(a).
| Variables | Lp(a), nmol/L | |||
|---|---|---|---|---|
| < 24.99 | 24.99–38.65 | 38.65–62.47 | > 62.47 | |
| CHD | ||||
| Model 1a | ||||
| Odds ratio (95% CI) | 1.00 (Ref.) | 1.735 (0.931–2.877) | 1.827 (1.063–2.832) | 1.921 (1.102–3.121) |
| p value | – | 0.106 | 0.024 | 0.011 |
| Model 2b | ||||
| Odds ratio (95% CI) | 1.00 (Ref.) | 1.628 (0.621–3.214) | 1.744 (0.798–2.012) | 1.843 (1.058–2.732) |
| p value | – | 0.658 | 0.195 | 0.041 |
| Model 3c | ||||
| Odds ratio (95% CI) | 1.00 (Ref.) | 1.552 (0.658–2.661) | 1.667 (0.723–2.985) | 1.793 (1.053–2.882) |
| p value | – | 0.505 | 0.449 | 0.043 |
| Multiple-vessel disease | ||||
| Model 1a | ||||
| Odds ratio (95% CI) | 1.00 (Ref.) | 2.364 (1.345–4.155) | 3.039 (1.506–6.132) | 3.309 (2.293–5.030) |
| p value | – | 0.003 | 0.002 | < 0.001 |
| Model 2b | ||||
| Odds ratio (95% CI) | 1.00 (Ref.) | 1.766 (0.660–4.725) | 2.050 (0.720–5.836) | 2.275 (1.318–3.927) |
| p value | – | 0.257 | 0.179 | 0.003 |
| Model 3c | ||||
| Odds ratio (95% CI) | 1.00 (Ref.) | 1.714 (0.754–4.821) | 1.890 (0.705–5.091) | 1.941 (1.113–3.242) |
| p value | – | 0.267 | 0.206 | 0.027 |
| High GS | ||||
| Model 1a | ||||
| Odds ratio (95% CI) | 1.00 (Ref.) | 2.321 (0.865–7.932) | 2.943 (1.133–6.543) | 3.201 (1.234–8.309) |
| p value | – | 0.101 | 0.043 | 0.017 |
| Model 2b | ||||
| Odds ratio (95% CI) | 1.00 (Ref.) | 2.358 (0.877–7.726) | 2.727 (0.861–7.446) | 2.766 (1.159–7.102) |
| p value | – | 0.125 | 0.105 | 0.021 |
| Model 3c | ||||
| Odds ratio (95% CI) | 1.00 (Ref.) | 1.784 (0.728–4.401) | 2.025 (0.968–4.421) | 2.641 (1.102–7.436) |
| p value | – | 0.205 | 0.095 | 0.040 |
Lp(a) Lipoprotein(a), GS Gensini score, CHD Coronary heart disease, CI Confidence interval.
aUnivariate model.
bAdjusted for age, sex, body mass index, diabetes, hypertension, smoking, consumers of alcohol.
cAdditionally adjusted for hemoglobin A1c, fasting plasma glucose, apolipoprotein B, apolipoprotein A1, total cholesterol, triglycerides, high density lipoprotein cholesterol, hypersensitive C-reactive protein, and homocysteine.
Both of the LPA SNPs genotypes and allele frequency distributions are shown in Table 6. There were significant differences in genotype (rs6415084: CC vs. CT/TT; rs12194138: AA vs. AT/TT) and allele frequency (rs6415084: C vs. T; rs12194138: A vs. T) between CHD group and control group, multi vessel disease group and single vessel disease group, high GS group and low GS group (p < 0.05 for both). After adjusting for age, sex, BMI, diabetes, hypertension, smoking, consumers of alcohol, FPG, HbA1c, ApoB, ApoA1, TC, TG, HDL-C, hs-CRP and HCY were adjusted by multiple logistic regression analysis, it was found that both LPA SNPs rs6415084 (CT/TT ) and rs12194138 (AT/TT) were risk factors for CHD, and were positively associated with the severity of CHD (LPA SNPs rs6415084: CHD group vs. control group: OR = 1.577, 95% CI: 1.105–1.989, p = 0.004; multiple-vessel disease group vs. single-vessel disease group: OR = 1.613, 95% CI: 1.076–2.641, p = 0.030; high GS group vs. low GS group: OR = 1.580, 95% CI: 1.088–2.429, p = 0.024; LPA SNPs rs12194138: CHD group vs. control group: OR = 1.475, 95% CI: 1.040–3.002, p = 0.035; multiple-vessel disease group vs. single-vessel disease group: OR = 2.274, 95% CI: 1.060–5.148, p = 0.038; high GS group vs. low GS group: OR = 2.067, 95% CI: 1.101–4.647, p = 0.021). These data together indicated that the level of Lp(a) and the prevalence of LPA SNPs rs6415084 (CT/TT) and rs12194138 (AT/TT) is positively correlate with the severity of CHD.
Table 6.
Relationship between two SNPs and coronary heart disease and its severity.
| SNP | Groups | Genotype, n (%) | M ↔ m | Mm + mm ↔ MM | |||
|---|---|---|---|---|---|---|---|
| MM | Mm | mm | Crude OR (95% Cl) | Crude OR (95% Cl) | Adjusted OR (95% Cl) | ||
| CHD | |||||||
| rs6415084 | Control group | 975 (88.6) | 123 (11.2) | 3 (0.2) | 1.650 (1.312–2.073) | 1.614 (1.266–2.057) | 1.577 (1.105–1.989) |
| CHD group | 911 (82.7) | 175 (15.9) | 15 (1.4) | p < 0.001 | p < 0.001 | p = 0.004 | |
| rs12194138 | Control group | 1051 (95.4) | 50 (4.6) | 0 (0.0) | 1.602 (1.118–2.294) | 1.537 (1.063–2.220) | 1.475 (1.040–3.002) |
| CHD group | 1026 (93.2) | 71 (6.4) | 4 (0.4) | p = 0.012 | p = 0.027 | p = 0.035 | |
| Multiple-vessel disease | |||||||
| rs6415084 | 1 vessel | 248 (86.4) | 39 (13.6) | 0 (0.0) | 1.698 (1.161–2.483) | 1.673 (1.107–2.466) | 1.613 (1.076–2.641) |
| ≥ 3 vessels | 399 (79.2) | 99 (19.6) | 6 (1.2) | p = 0.006 | p = 0.013 | p = 0.030 | |
| rs12194138 | 1 vessel | 278 (96.9) | 9 (3.1) | 0 (0.0) | 2.392 (1.146–4.993) | 2.305 (1.092–4.867) | 2.274 (1.060–5.148) |
| ≥ 3 vessels | 469 (93.1) | 33 (6.5) | 2 (0.4) | p = 0.019 | p = 0.024 | p = 0.038 | |
| High GS | |||||||
| rs6415084 | Low GS | 324 (85.7) | 54 (14.3) | 0 (0.0) | 1.670 (1.167–2.389) | 1.608 (1.098–2.355) | 1.580 (1.088–2.429) |
| High GS | 291 (78.9) | 72 (19.5) | 6 (1.6) | p = 0.005 | p = 0.016 | p = 0.024 | |
| rs12194138 | Low GS | 361 (95.5) | 17 (4.5) | 0 (0.0) | 2.360 (1.320–4.220) | 2.225 (1.223–4.047) | 2.067 (1.101–4.647) |
| High GS | 334 (90.5) | 32 (8.7) | 3 (0.8) | p = 0.004 | p = 0.009 | p = 0.021 | |
Crude OR was determined by χ2 test, cases versus control subjects.
Adjusted OR was obtained on multivariate logistic regression after controlling for age, sex, body mass index, diabetes, hypertension, smoking, consumers of alcohol, hemoglobin A1c, fasting plasma glucose, apolipoprotein B, apolipoprotein A1, total cholesterol, triglycerides, high density lipoprotein cholesterol, hypersensitive C-reactive protein, and homocysteine.
M = C and m = T for single nucleotide polymorphism (SNP) rs6415084; M = A and m = T for single nucleotide polymorphism (SNP) rs12194138.
CHD Coronary heart disease, CI Confidence interval.
Lp(a) levels, the prevalence of LPA SNPs and cardiovascular outcomes
A total of 2042 individuals participated in the follow-up. During the follow-up period, 293 (14.35%) individuals had MACEs. All 2042 individuals were divided into high Lp(a) group and low Lp(a) group according to the median Lp(a), and Kaplan–Meier analysis was performed. The results showed that the incidence of MACEs in high Lp(a) group was higher than that in low Lp(a) group in all subjects (Fig. 2A) (HR = 1.459, 95% CI: 1.160–1.835, p = 0.001). In control group and CHD group, the results were the same (Figs. 3A, 4A) (HR = 1.443, 95% CI: 1.009–2.062, p = 0.044; HR = 1.473, 95% CI: 1.091–1.987, p = 0.011). The CHD and the control group were divided into two groups according to the median Lp(a), and the relationship between Lp(a) level and MACEs risk was studied. As shown in Tables 7 and 8, the total incidence of MACEs in the high Lp(a) group was significantly higher than that in the low Lp(a) group, both in the control group and the CHD group. These data indicate that Lp(a) level is associated with MACEs of all subjects.
Figure 2.
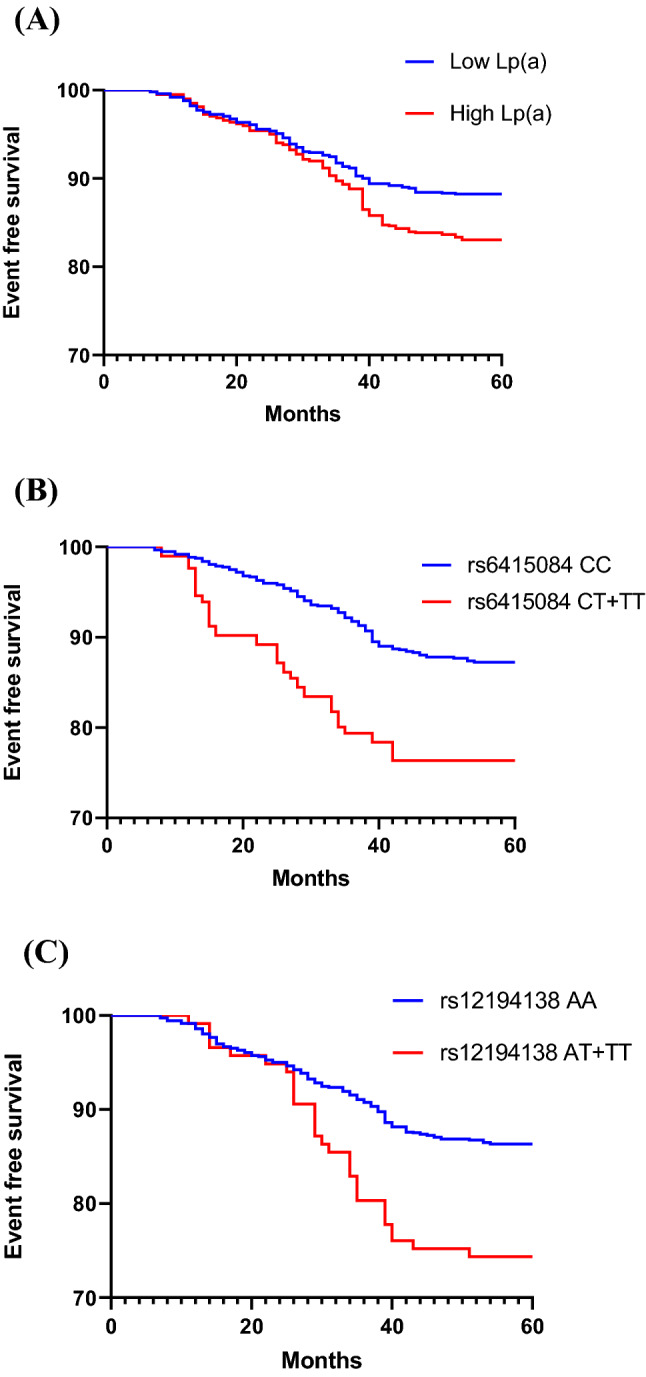
(A) Kaplan–Meier curves according to median value of Lp(a) (p = 0.001) in all subjects. (B, C) The cardiovascular event curves of SNPs rs6415084 (CT + TT) and rs12194138 (AT + TT) carriers were compared with SNPs rs6415084 (CC) and rs12194138 (AA) carriers (p < 0.001; p = 0.001) in all subjects.
Figure 3.
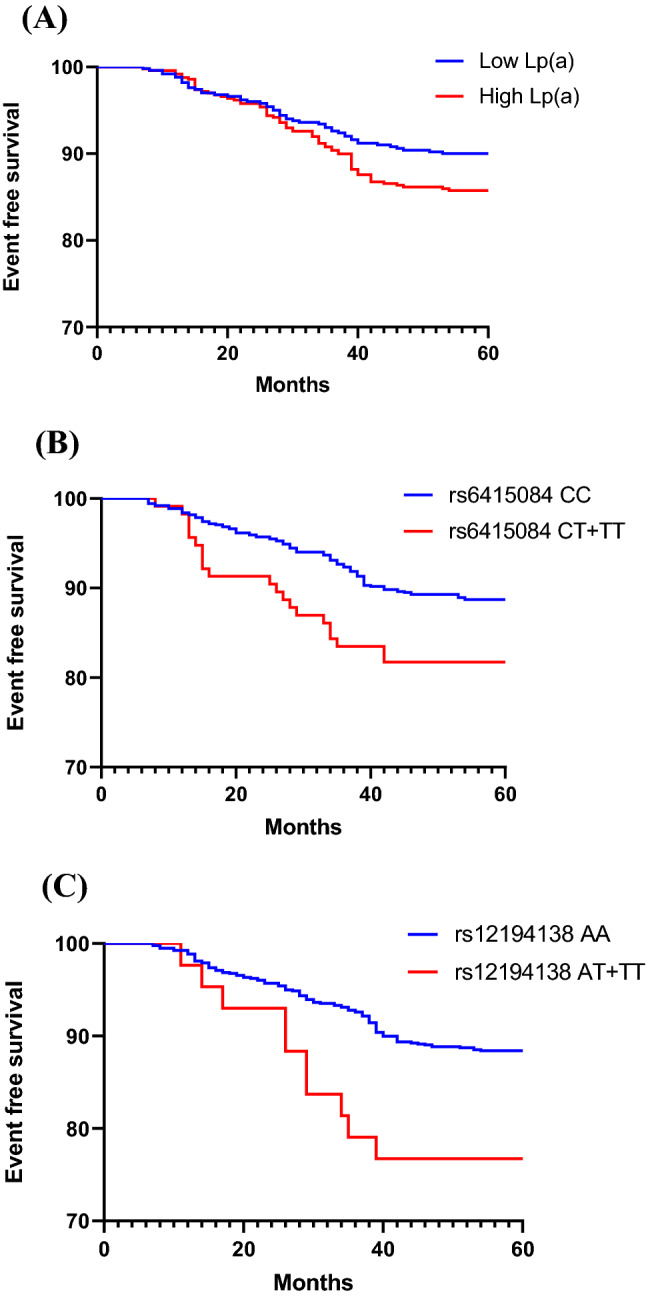
(A) Kaplan–Meier curves according to median value of Lp(a) (p = 0.044) in control group. (B, C) The cardiovascular event curves of SNPs rs6415084 (CT + TT) and rs12194138 (AT + TT) carriers were compared with SNPs rs6415084 (CC) and rs12194138 (AA) carriers (p = 0.022; p = 0.016) in control group.
Figure 4.
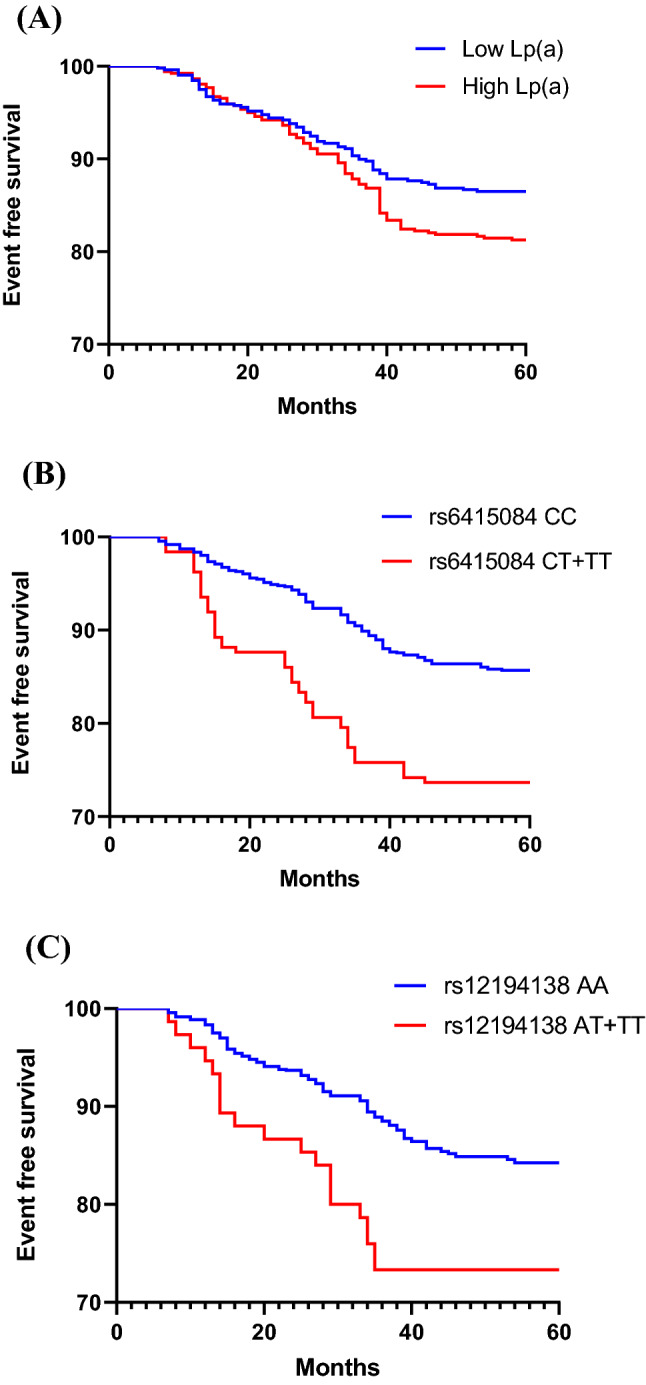
(A) Kaplan–Meier curves according to median value of Lp(a) (p = 0.011) in CHD patients. (B, C) The cardiovascular event curves of SNPs rs6415084 (CT + TT) and rs12194138 (AT + TT) carriers were compared with SNPs rs6415084 (CC) and rs12194138 (AA) carriers in CHD patients (p < 0.001; p = 0.007).
Table 7.
The relationship between Lp (a) level and the occurrence of MACEs in control group.
| MACEs | Low Lp(a) (n = 502) | High Lp(a) (n = 499) | p |
|---|---|---|---|
| CHD | 24 (4.78%) | 33 (6.61%) | 0.222 |
| Cardiovascular deaths | 0 (0.00%) | 3 (0.60%) | 0.124 |
| Non-fatal MI | 7 (1.39%) | 11 (2.20%) | 0.354 |
| Non-fatal strokes | 6 (1.20%) | 5 (1.00%) | 1.000 |
| Heart failure | 3 (0.60%) | 3 (0.60%) | 1.000 |
| Hospitalized unstable angina | 10 (1.99%) | 16 (3.21%) | 0.240 |
| Total | 50 (9.96%) | 71 (14.23%) | 0.042 |
Statistical analysis was performed with Chi-square test for categorical variables.
Lp(a) Lipoprotein(a), CHD Coronary heart disease, MACEs Major cardiovascular events.
Table 8.
The relationship between Lp(a) level and the occurrence of MACEs in CHD patients.
| MACEs | Low Lp(a) (n = 518) | High Lp(a) (n = 523) | p |
|---|---|---|---|
| Cardiovascular deaths | 9 (1.74%) | 12 (2.29%) | 0.660 |
| Non-fatal MI | 30 (5.79%) | 45 (8.60%) | 0.093 |
| Non-fatal strokes | 13 (2.51%) | 17 (3.25%) | 0.579 |
| Heart failure | 8 (1.54%) | 13 (2.49%) | 0.379 |
| Hospitalized unstable angina | 10 (1.93%) | 15 (2.87%) | 0.419 |
| Total | 70 (13.51%) | 102 (19.50%) | 0.010 |
Statistical analysis was performed with Chi-square test for categorical variables.
Lp(a) Lipoprotein(a), CHD Coronary heart disease, MACEs Major cardiovascular events.
Further study of LPA SNPs and cardiovascular outcomes showed that MACEs were significantly higher in individuals with LPA SNPs rs6415084 (CT/TT) and rs12194138 (AT/TT) genotype carriers than CC and AA genotype carriers in all subjects (Fig. 2B,C) (HR = 2.499, 95% CI: 1.783–3.502, p < 0.001; HR = 2.565, 95% CI: 1.545–4.257, p = 0.001). In the control group, the results were consistent with the test results of all subjects (Fig. 3B,C) (HR = 1.949, 95% CI: 1.099–3.456, p = 0.022; HR = 3.087, 95% CI: 1.238–7.698, p = 0.016). The results were the same in the CHD group (Fig. 4B,C) (HR = 2.441, 95% CI: 1.628–3.661, p < 0.001; HR = 2.298, 95% CI: 1.254–4.211, p = 0.007). As shown in Tables 9 and 10, the total incidence of MACEs in LPA SNPs rs6415084 (CT/TT) and rs12194138 (AT/TT) genotype carriers was significantly higher than that in CC and AA genotype carriers, and there were differences in the non-fatal MI and hospitalized unstable angina of SNPs rs6415084 (CT/TT vs. CC) in the CHD group in addition to the total incidence. These results indicate that the LPA SNPs rs6415084 (CT/TT) and rs12194138 (AT/TT) genotype are related to the occurrence of cardiovascular events in the future.
Table 9.
SNPs rs6415084 and rs12194138 were associated with MACEs in control group.
| MACEs | SNP rs6415084 | p | SNP rs12194138 | p | ||
|---|---|---|---|---|---|---|
| MM (886) | Mm + mm (115) | MM (958) | Mm + mm (43) | |||
| CHD | 47 (5.30%) | 10 (8.70%) | 0.137 | 52 (5.43%) | 5 (11.63%) | 0.092 |
| Cardiovascular deaths | 2 (0.23%) | 1 (0.87%) | 0.307 | 3 (0.31%) | 0 (0.00%) | 1.000 |
| Non-fatal MI | 17 (1.92%) | 1 (0.87%) | 0.711 | 16 (1.67%) | 2 (4.65%) | 0.179 |
| Non-fatal strokes | 10 (1.13%) | 1 (0.87%) | 1.000 | 11 (1.15%) | 0 (0.00%) | 1.000 |
| Heart failure | 4 (0.45%) | 2 (1.74%) | 0.144 | 5 (0.52%) | 1 (2.33%) | 0.232 |
| Hospitalized unstable angina | 20 (2.26%) | 6 (5.22%) | 0.108 | 24 (2.51%) | 2 (4.65%) | 0.308 |
| Total | 100 (11.29%) | 21 (18.26%) | 0.046 | 111 (11.59%) | 10 (23.26%) | 0.030 |
M = C and m = T for single nucleotide polymorphism (SNP) rs6415084; M = A and m = T for single nucleotide polymorphism (SNP) rs12194138.
Table 10.
SNPs rs6415084 and rs12194138 were associated with MACEs in CHD patients.
| MACEs | SNP rs6415084 | p | SNP rs12194138 | p | ||
|---|---|---|---|---|---|---|
| MM (860) | Mm + mm (181) | MM (967) | Mm + mm (74) | |||
| Cardiovascular deaths | 15 (1.74%) | 6 (3.31%) | 0.237 | 19 (1.96%) | 1 (1.35%) | 1.000 |
| Non-fatal MI | 56 (6.51%) | 21 (11.60%) | 0.027 | 67 (6.93%) | 8 (10.81%) | 0.238 |
| Non-fatal strokes | 21 (2.44%) | 8 (4.42%) | 0.141 | 26 (2.69%) | 4 (5.41%) | 0.159 |
| Heart failure | 13 (1.51%) | 5 (2.76%) | 0.221 | 16 (1.65%) | 3 (4.05%) | 0.147 |
| Hospitalized unstable angina | 18 (2.09%) | 9 (4.97%) | 0.037 | 24 (2.48%) | 4 (5.41%) | 0.132 |
| Total | 123 (14.30%) | 49 (27.07%) | < 0.001 | 152 (15.72%) | 20 (27.03%) | 0.022 |
M = C and m = T for single nucleotide polymorphism (SNP) rs6415084; M = A and m = T for single nucleotide polymorphism (SNP) rs12194138.
Discussion
Recently, the role of Lp(a) in cardiovascular diseases has attracted more and more attention15,19,20. The association between Lp(a) and CHD, which is independent of traditional cardiovascular risk factors, has been known for many years4,21. It is based on findings mainly from studies of healthy participants in the general population and investigations of patients with CHD22. This study demonstrates that Lp(a) level and the prevalence of LPA SNPs are associated with the risk and severity of CHD, and LPA SNPs rs6415084 and rs12194138 are significantly associated with serum Lp(a) levels. That the increase of Lp(a) level was the key variable to predict CHD risk, and the role of LPA SNPs rs6415084, rs12194138 and Lp(a) level in predicting CHD risk was clarified. Most of the previous data on LPA and Lp(a) were studied in Caucasian populations in Europe23–25. However, there are few studies on the relationship between LPA and Lp(a) and the risk of coronary heart disease in Chinese Han population. At the same time, our study also predicted the occurrence of MACEs between rs6415084 and rs12194138 in Chinese Han population, so as to better study the role of Lp(a) level and LPA SNPs in predicting future MACEs of different nationalities.
We demonstrated that the rs6415084 and rs12194138 polymorphisms were significantly more frequent in patients with CHD than in healthy subjects. A higher risk of CHD was observed for the rs6415084 CT/TT and the rs12194138 AT/TT heterozygotes and homozygotes polymorphism carriers. The study of Lee et al.15, showed that the LPA SNPs was associated with the size of apolipoprotein (a) isoforms and the serum level of Lp(a) in different ethnicity. Clarke et al.3 showed that LPA SNPs rs10455872 and rs3798220 were strongly associated with increased level of Lp(a), a reduced copy number in LPA, and a small Lp(a) size. Lanktree et al.14 reported that SNPs rs6415084, located in the 5′ haplotype block and associated with KIV-2 copy number. Tolbus et al.18 reported that LPA SNPs rs12194138 were not associated with KIV-2 number of repeats. The other study including that identified SNPs in the LPA gene that had an association with the risk of CAD in diabetic patients26 (OR = 1.25, 95CI: 1.19–1.31, p = 3.92E-21). The results of Sang-Rok Lee et al. support our findings on the LPA SNPs and CHD.
Consistent with the role of Lp(a) in predicting future MACEs, it can be concluded that higher Lp(a) levels are primarily important to CHD risk prediction27,28. The increase of plasma Lp(a) level can promote thrombosis, and there is a correlation between Lp(a) and atherosclerotic stenosis. The same results are presented in some recent research reports29.
At present, most of the studies on LPA SNPs focus on the risk of CHD and atherosclerosis30. The prediction of future MACEs by SNPs is only seen in a few articles, and most of them focus on the European population31. Therefore, our study of the prediction of MACEs by two SNPs rs6415084 and rs12194138 in Chinese Han population, in order to better study the role of Lp(a) level and LPA SNPs in predicting future MACEs in different ethnicity. Our results are similar to those predicted by Gudbjartsson et al.1, for patients with diabetes mellitus with CHD in Iceland. CHD is a multifactorial disease, the combination of genetic variation and environmental factors may lead to phenotypic variation32. In our study, elevated Lp(a) level is an independent MACEs predictor; and some specific LPA SNPs variations may cause the increase of serum Lp(a), which also explains that SNPs rs6415084 and rs12194138 are independent MACEs predictors. Some studies have shown that LPA SNPs variation can increase Lp(a) level, but there is no direct correlation between LPA SNPs and MACEs3,33. The reason for this difference was not fully understood, but the discrepancy between the results of the studies may be caused by a variety of confounding factors, such as different population characteristics, study design, disease status, or confounding variables. Therefore, we used a large number of Han Chinese in this study. We not only found that serum Lp(a) levels and LPA SNPs variants were associated with the risk and severity of coronary heart disease, but they were independent predictors of MACEs.
Serum Lp(a) level is largely determined by the variation of LPA in many populations. Genetic variation of LPA is directly related to the risk of cardiovascular disease. The reason and mechanism of LPA variants rs6415084 and rs12194138 and Lp(a) level increase remains unclear2,14. The mechanism of increased Lp (a) lipoprotein level increasing the risk of coronary heart disease is unclear, which may involve LDL lipoprotein cholesterol34, inhibition of plasminogen to plasmin35, inhibition of tissue factor expression36, or carrying pro-inflammatory oxidized phospholipids37. From our results, we found that the level of Lp(a) is positively associated with the risk and severity of CHD.
Besides, the present study has several limitations. First, only two centers were involved in the research, which might have led to selective biases in the data results, and some of the conclusions should be verified in larger multicenter studies. Second, we have only studied two SNPs and the power value of SNPs rs12194138 is low. In our future work, we will increase more SNPs and sample size. In addition, CAG results lack of centralized core laboratory for angiography analysis. However, our data still provide the incidence rate and severity of CHD increase with the increase of serum Lp(a) level. In addition, the increased serum Lp(a) level and SNPs rs6415084 and rs12194138 variants will also increase the risk of cardiovascular events in the future.
Conclusion
In conclusion, our data support the association of elevated serum Lp(a) levels and LPA SNPs rs6415084, rs3798220 variants with the risk and severity of coronary heart disease, and the prediction of future cardiovascular events.
Acknowledgements
The authors thank all the staff and participants of this study for their important contributions.
Author contributions
J.-X.G., J.H., S.-S.L., M.J.: the conception and design of the study. L.-H.Z., M.Y., Y.L., A.-M.Z., Y.Y., N.Z.: acquisition of data, analysis and interpretation of data. J.-X.G., J.H., S.-S.L. and M.J.: drafting the article and revising it critically for important intellectual content. J.-X.G., S.-S.L., M.S., and M.J.: final approval of the version to be submitted.
Funding
This study was supported by the National Natural Science Foundation of China (81870196).
Data availability
The datasets used and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jun-Xu Gu, Juan Huang and Shan-Shan Li.
Contributor Information
Mei Jia, Email: jiamei00718@sina.com.
Ming Su, Email: suming28@163.com.
References
- 1.Gudbjartsson DF, et al. Lipoprotein(a) concentration and risks of cardiovascular disease and diabetes. J. Am. Coll. Cardiol. 2019;74:2982–2994. doi: 10.1016/j.jacc.2019.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Gencer B, Kronenberg F, Stroes ES, Mach F. Lipoprotein(a): The revenant. Eur. Heart J. 2017;38:1553–1560. doi: 10.1093/eurheartj/ehx033. [DOI] [PubMed] [Google Scholar]
- 3.Clarke R, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 4.Stefanutti C, et al. Lipoprotein(a) concentration, genetic variants, apo(a) isoform size, and cellular cholesterol efflux in patients with elevated Lp(a) and coronary heart disease submitted or not to lipoprotein apheresis: An Italian case-control multicenter study on Lp(a) J. Clin. Lipidol. 2020;14:487–497. doi: 10.1016/j.jacl.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Thanassoulis G, et al. Genetic associations with valvular calcification and aortic stenosis. N. Engl. J. Med. 2013;368:503–512. doi: 10.1056/NEJMoa1109034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsimikas S, Stroes ESG. The dedicated “Lp(a) clinic”: A concept whose time has arrived? Atherosclerosis. 2020;300:1–9. doi: 10.1016/j.atherosclerosis.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Xu N, et al. Lipoprotein(a) levels are associated with coronary severity but not with outcomes in Chinese patients underwent percutaneous coronary intervention. Nutr. Metab. Cardiovasc. Dis. 2020;30:265–273. doi: 10.1016/j.numecd.2019.09.020. [DOI] [PubMed] [Google Scholar]
- 8.Virani SS, et al. Heart disease and stroke statistics-2020 update: A report from the American Heart Association. Circulation. 2020;141:e139–e596. doi: 10.1161/CIR.0000000000000757. [DOI] [PubMed] [Google Scholar]
- 9.Arnold M, et al. Lipoprotein(a) is associated with large artery atherosclerosis stroke aetiology and stroke recurrence among patients below the age of 60 years: results from the BIOSIGNAL study. Eur. Heart J. 2021 doi: 10.1093/eurheartj/ehab081. [DOI] [PubMed] [Google Scholar]
- 10.Di Maio S, et al. Investigation of a nonsense mutation located in the complex KIV-2 copy number variation region of apolipoprotein(a) in 10,910 individuals. Genome Med. 2020;12:74. doi: 10.1186/s13073-020-00771-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coassin S, et al. A comprehensive map of single-base polymorphisms in the hypervariable LPA kringle IV type 2 copy number variation region. J. Lipid Res. 2019;60:186–199. doi: 10.1194/jlr.M090381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waldeyer C, et al. Lipoprotein(a) and the risk of cardiovascular disease in the European population: Results from the BiomarCaRE consortium. Eur. Heart J. 2017;38:2490–2498. doi: 10.1093/eurheartj/ehx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiffman D, et al. Single variants can explain the association between coronary heart disease and haplotypes in the apolipoprotein(a) locus. Atherosclerosis. 2010;212:193–196. doi: 10.1016/j.atherosclerosis.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Lanktree MB, Anand SS, Yusuf S, Hegele RA, Investigators S. Comprehensive analysis of genomic variation in the LPA locus and its relationship to plasma lipoprotein(a) in South Asians, Chinese, and European Caucasians. Circ. Cardiovasc. Genet. 2010;3:39–46. doi: 10.1161/CIRCGENETICS.109.907642. [DOI] [PubMed] [Google Scholar]
- 15.Lee SR, et al. LPA gene, ethnicity, and cardiovascular events. Circulation. 2017;135:251–263. doi: 10.1161/CIRCULATIONAHA.116.024611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalifa M, et al. Lack of association of rs3798220 with small apolipoprotein(a) isoforms and high lipoprotein(a) levels in East and Southeast Asians. Atherosclerosis. 2015;242:521–528. doi: 10.1016/j.atherosclerosis.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 17.Egana-Gorrono L, et al. Association study of lipoprotein(a) genetic markers, traditional risk factors, and coronary heart disease in HIV-1-infected patients. Front. Immunol. 2012;3:367. doi: 10.3389/fimmu.2012.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolbus A, et al. Kringle IV type 2, not low lipoprotein(a), as a cause of diabetes: A novel genetic approach using SNPs associated selectively with lipoprotein(a) concentrations or with kringle IV type 2 repeats. Clin Chem. 2017;63:1866–1876. doi: 10.1373/clinchem.2017.277103. [DOI] [PubMed] [Google Scholar]
- 19.Lasek-Bal A, et al. The association of SNPs located in the CDKN2B-AS1 and LPA genes with carotid artery stenosis and atherogenic stroke. Front. Neurol. 2019;10:1170. doi: 10.3389/fneur.2019.01170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh SS, et al. Lipoprotein(a) plasma levels are not associated with incident microvascular complications in type 2 diabetes mellitus. Diabetologia. 2020;63:1248–1257. doi: 10.1007/s00125-020-05120-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waissi F, et al. Elevated Lp(a) (lipoprotein[a]) levels increase risk of 30-day major adverse cardiovascular events in patients following carotid endarterectomy. Stroke. 2020;51:2972–2982. doi: 10.1161/STROKEAHA.120.030616. [DOI] [PubMed] [Google Scholar]
- 22.Zewinger S, et al. Relations between lipoprotein(a) concentrations, LPA genetic variants, and the risk of mortality in patients with established coronary heart disease: A molecular and genetic association study. Lancet Diabetes Endocrinol. 2017;5:534–543. doi: 10.1016/S2213-8587(17)30096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capoulade R, et al. ApoCIII-Lp(a) complexes in conjunction with Lp(a)-OxPL predict rapid progression of aortic stenosis. Heart. 2020;106:738–745. doi: 10.1136/heartjnl-2019-315840. [DOI] [PubMed] [Google Scholar]
- 24.Akao H, et al. KIF6, LPA, TAS2R50, and VAMP8 genetic variation, low density lipoprotein cholesterol lowering response to pravastatin, and heart disease risk reduction in the elderly. Atherosclerosis. 2012;220:456–462. doi: 10.1016/j.atherosclerosis.2011.11.037. [DOI] [PubMed] [Google Scholar]
- 25.Welsh P, et al. Lipoprotein(a) and cardiovascular disease: Prediction, attributable risk fraction, and estimating benefits from novel interventions. Eur. J. Prev. Cardiol. 2020 doi: 10.1093/eurjpc/zwaa063. [DOI] [PubMed] [Google Scholar]
- 26.van Iperen EP, et al. Genetic analysis of emerging risk factors in coronary artery disease. Atherosclerosis. 2016;254:35–41. doi: 10.1016/j.atherosclerosis.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Paquette M, Bernard S, Baass A. SLC22A3 is associated with lipoprotein (a) concentration and cardiovascular disease in familial hypercholesterolemia. Clin. Biochem. 2019;66:44–48. doi: 10.1016/j.clinbiochem.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Xu N, et al. Impact of lipoprotein(a) on long-term (mean 6.2 years) outcomes in patients with three-vessel coronary artery disease. Am. J. Cardiol. 2020;125:528–533. doi: 10.1016/j.amjcard.2019.10.037. [DOI] [PubMed] [Google Scholar]
- 29.Farukhi ZM, Mora S. Lifelong low Lp(a) levels: Genetics give a green light? Eur. Heart J. 2021;42:1157–1159. doi: 10.1093/eurheartj/ehaa1112. [DOI] [PubMed] [Google Scholar]
- 30.Mu-Han-Ha-Li DL, Zhai TY, Ling Y, Gao X. LPA kringle IV type 2 is associated with type 2 diabetes in a Chinese population with very high cardiovascular risk. J. Lipid Res. 2018;59:884–891. doi: 10.1194/jlr.P082792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noureen A, Fresser F, Utermann G, Schmidt K. Sequence variation within the KIV-2 copy number polymorphism of the human LPA gene in African, Asian, and European populations. PLoS ONE. 2015;10:e0121582. doi: 10.1371/journal.pone.0121582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deo RC, et al. Single-nucleotide polymorphisms in LPA explain most of the ancestry-specific variation in Lp(a) levels in African Americans. PLoS ONE. 2011;6:e14581. doi: 10.1371/journal.pone.0014581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li ZG, et al. Lack of association between lipoprotein(a) genetic variants and subsequent cardiovascular events in Chinese Han patients with coronary artery disease after percutaneous coronary intervention. Lipids Health Dis. 2013;12:127. doi: 10.1186/1476-511X-12-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grainger DJ, Kemp PR, Liu AC, Lawn RM, Metcalfe JC. Activation of transforming growth factor-beta is inhibited in transgenic apolipoprotein(a) mice. Nature. 1994;370:460–462. doi: 10.1038/370460a0. [DOI] [PubMed] [Google Scholar]
- 36.Caplice NM, et al. Lipoprotein (a) binds and inactivates tissue factor pathway inhibitor: A novel link between lipoproteins and thrombosis. Blood. 2001;98:2980–2987. doi: 10.1182/blood.v98.10.2980. [DOI] [PubMed] [Google Scholar]
- 37.Tsimikas S, et al. Oxidized phospholipids, Lp(a) lipoprotein, and coronary artery disease. N. Engl. J. Med. 2005;353:46–57. doi: 10.1056/NEJMoa043175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.


