Abstract
Objectives
Colorectal cancer (CRC) is one of the most common solid tumors worldwide, but its diagnosis and treatment are limited. The objectives of our study were to compare the metabolic differences between CRC patients and healthy controls (HC), and to identify potential biomarkers in the serum that can be used for early diagnosis and as effective therapeutic targets. The aim was to provide a new direction for CRC predictive, preventive, and personalized medicine (PPPM).
Methods
In this study, CRC patients (n = 30) and HC (n = 30) were recruited. Serum metabolites were assayed using an ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF/MS) technology. Subsequently, CRC cell lines (HCT116 and HCT8) were treated with metabolites to verify their function. Key targets were identified by molecular docking, thermal shift assay, and protein overexpression/inhibition experiments. The inhibitory effect of celastrol on tumor growth was also assessed, which included IC50 analysis, nude mice xenografting, molecular docking, protein overexpression/inhibition experiments, and network pharmacology technology.
Results
In the CRC group, 15 serum metabolites were significantly different in comparison with the HC group. The level of glycodeoxycholic acid (GDCA) was positively correlated with CRC and showed high sensitivity and specificity for the clinical diagnostic reference (AUC = 0.825). In vitro findings showed that GDCA promoted the proliferation and migration of CRC cell lines (HCT116 and HCT8), and Poly(ADP-ribose) polymerase-1 (PARP-1) was identified as one of the key targets of GDCA. The IC50 of celastrol in HCT116 cells was 121.1 nM, and the anticancer effect of celastrol was supported by in vivo experiments. Based on the potential of GDCA in PPPM, PARP-1 was found to be significantly correlated with the anticancer functions of celastrol.
Conclusion
These findings suggest that GDCA is an abnormally produced metabolite of CRC, which may provide an innovative molecular biomarker for the predictive identification and targeted prevention of CRC. In addition, PARP-1 was found to be an important target of GDCA that promotes CRC; therefore, celastrol may be a potential targeted therapy for CRC via its effects on PARP-1. Taken together, the pathophysiology and progress of tumor molecules mediated by changes in metabolite content provide a new perspective for predictive, preventive, and personalized medical of clinical cancer patients based on the target of metabolites in vivo.
Clinical trials registration number: ChiCTR2000039410.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13167-021-00269-8.
Keywords: Predictive preventive personalized medicine, Metabolomics, UPLC-Q-TOF/MS, Colorectal cancer, Serum, Glycodeoxycholic acid, Therapeutic targets, Celastrol, Poly(ADP-ribose) polymerase-1
Introduction
CRC is a global challenge in healthcare
Cancer can occur in almost any organ or tissue of the human body, and its mortality and incidence rate are increasing yearly. According to the latest World Cancer Report (2020) of the World Health Organization (WHO), the number of global cancer cases could increase by 60% in the next two decades [1]. Globally, it is estimated that in 2018, 880,792 people died of colorectal cancer (CRC) and 1.85 million CRC cases were diagnosed [2]. CRC is a progressive disease caused by hereditary factors and unhealthy lifestyles; for instance, obesity, poor diet, lack of exercise, alcohol, and smoking are risk factors for CRC [3–5].
The relevance of predictive, preventive, and personalized medicine (PPPM) for the current study
Cancer is a disease with high mortality rates, and the occurrence and development of cancer are complicated processes. CRC is the third most common cause of cancer mortality worldwide. There are great differences in patients’ clinical results and treatment responses, and the use of standard treatment options is still limited [6]. During the development of PPPM, more cancer-related screening and diagnostic technologies, predictive markers, and treatment outcome have been proposed, leading to implementation personalized medicine and better drug development solutions, and then improved the efficiency of patient management [7]. Therefore, these preventive and predictive treatment methods will become the focus of anticancer and promote the overall management of cancer, so as to facilitate the provision of accurate and personalized medicine for patients in the future [2, 8]. The rapid development of “omics” technology has pursued the research and development of cancer mechanisms; this technology can provide accurate screening markers through multi-parameter systematic strategy, rapidly find and accurately diagnose each cancer type, and accurately and effectively predict and prevent cancer [7, 9]. In addition, it clarifies the complex pathogenesis of cancer from different levels of molecules (DNA, RNA, protein, and metabolite), and provide personalized medicine at different omics levels, and truly apply PPPM to cancer research [10].
Metabolomics is a tool for prediction and targeted treatment of CRC
At present, surgical resection, radiotherapy, and chemotherapy are the most common treatment options for CRC. The accuracy of screen test for CRC varies depending on the tumor site. For instance, colonoscopy, sigmoidoscopy, fecal occult blood test, fecal DNA, and CT colonography have certain limitations regarding sensitivity, specificity, invasiveness, or specific preparations [11–13]. Predictive markers, such as CEA, BRAF, DPYD, KRAS, and other genes that serve as CRC tumor biomarkers, are insufficient screening markers for CRC [7].
Metabolomics, a new omics discipline in systems biology, can provide information on changes caused by environmental factors, genetic variation and regulation, intestinal microbiota, and enzyme activity [14–16]. Metabolic changes are some of the most important characteristics of tumor cells; therefore, metabolomics can be used to study changes in tumor markers [17–19]. At present, commonly used CRC biomarkers are derived from tumors, blood, urine, and feces, and can act as prognostic, predictive, and diagnostic markers for different disease states, point to the potential molecular mechanisms of the disease, and lead to the development of new therapeutic targets and methods [20]. Yachida et al. [21] detected CRC metabolites by fecal metabolomics analysis. This study included 406 subjects and monitored branched-chain amino acids and phenylalanine levels, which increased significantly in intramucosal carcinoma; their findings also showed that deoxycholate increased in multiple polypoid adenomas and intramucosal carcinoma. Ma et al. [22] detected 24 metabolites related to CRC stage via serum metabolomics and found that the serum level of benzoic acid was negatively correlated with CRC stage; benzoic acid also showed excellent diagnostic ability in the discovery cohort. A serum metabolomics study by Wu et al. [23] showed that succinic acid is a cancer progression factor that polarizes macrophages into tumor-related macrophages via succinic acid receptor signal transduction to promote tumor progression and metastasis. Together, these studies show that metabolomics is a rapid, accurate, and noninvasive method. Metabolites are closely related to biological phenotypes. Tumor metabolites reflect the chemicals existing in the tumor itself, which is helpful to understand the pathological process of cancer. It is of great significance for early diagnosis, prognosis diagnosis and treatment guidance, and monitoring of recurrence and metastasis. In addition, tumor metabolites can help to provide personalized medicine for patients and have a potential role in the discovery of new therapeutic targets/drugs.
Personalization of medical services in CRC management
Many bioactive compounds have been found during the clinical use of traditional medicinal plants [24]. These naturally occurring compounds have attracted great attention recently because of their high safety, targeting ability, and involvement in key signaling pathways [25]. Phytochemicals have become a hot topic in cancer research because of their anticancer activity. Phytochemicals play a significant role in antioxidant activity, carcinogen detoxification, and DNA repair, and can enhance the effects of chemotherapy and influence survival rate [24]. For example, flavonoids exist within traditional Chinese medicine and can be used as anticancer agents via their functions in inhibiting the Warburg effect, which is mediated by the regulation of specific enzymes involved in aerobic glycolysis and transporters required for glucose uptake [26]. Apigenin (AP) can inhibit the epithelial–mesenchymal transition of colon cancer cells via the NF-kB/Snail signaling pathway [27]. Additionally, AP is regarded as a potential allosteric inhibitor of PKM2, thereby affecting glycolysis [28]. Celastrol can inhibit CRC cell growth by promoting β-catenin degradation via the HSF1-LKB1-AMPKα-YAP pathway [29]. In addition, celastrol combined with tumor necrosis factor-related apoptosis-inducing ligand (TRAIL/Apo-2L) exhibits significant synergistic anticancer activities [30]. In conclusion, natural substances demonstrate great potential for CRC personalized medicine.
Methods
Chemicals and reagents
Chemicals used for UPLC-Q-TOF/MS
Analytical grade acetonitrile (Merck, Germany), formic acid (Thermo Fisher, America), methanol (Oceanpak, Sweden), purified water (Watsons, China) UPLC-Q-TOF/MS analysis.
Reagents used for cell experiments
Antibody against PARP-1(1:10,000 times dilution) (catalog number: 13389) was from Proteintech Group. RPMI 1640 medium (with glutamine) was purchased from Gibco (catalog number: No. 8120357). The CRC cell lines (HCT116 and HCT8) and human normal colorectal mucosal cells (FHC) were purchased from BeNa Culture Collection. Both cells were cultured in an RPMI-1640 (with L-glutamine) medium with 10% fetal bovine serum (FBS, catalog number: QT08773) and maintained with 5% CO2 atmosphere at 37 ℃.
Sample preparation and metabolomics assay
Participants
Colorectal cancer patients were enrolled from the second hospital of Tianjin Medical University, Tianjin, China, and healthy individuals (healthy controls, HC group) were recruited from the same hospital from 2019 to 2020. The inclusion criteria for the CRC group were age between 30 and 80 years and diagnosis by pathological tissue biopsy. The HC group were participants who were systemically healthy without the gastrointestinal disease. In total, 60 subjects were enrolled in this study, including 30 CRC patients and 30 age- and sex-matched healthy individuals in the disease (Table 1). Ethical approval was obtained from the Research Ethics Committee of the second hospital of Tianjin Medical University (approval No. KY2019K144).
Table 1.
Information of the study participants
| CRC | NC | ||||
|---|---|---|---|---|---|
| Gender | Female | Male | Female | Male | |
|
Age (years) |
Mean | 63.89 | 62.48 | 64.33 | 62.86 |
| Range | 42–80 | 47–75 | 48–80 | 44–77 | |
| Number | 30 | 9 | 21 | 9 | 21 |
| Stage | I | 2 | 11 | ||
| II | 4 | 1 | |||
| III | 1 | 3 | |||
| IV | 2 | 6 | |||
Sample preparation
Peripheral blood was collected in vacutainer tubes from all participants. The blood samples were congealed at room temperature for not more than 1 h and centrifuged for 15 min at 4000 rpm at 4 ℃. The serum was split charging in clean cryogenic vials and stored in − 80℃. For metabolite extraction from serum, 100 μL of sample was mixed with 400 μL of extraction solution. The mixture was vortexed for 1 min and sonicated for 10 min in an ice-water bath, following centrifuged at 14,000 rpm for 15 min at 4 ℃ and finally, dried with nitrogen, and re-dissolved in methanol: water (50:50, V/V). The QC sample contains all sample information for testing.
UPLC conditions
Serum metabolomics analysis was performed using a UPLC-Q-TOF/MS System (Waters, USA) coupled with a UPLC Waters ACQUITY UPLC BEH C18 (2.1 mm × 100 mm, 1.7 μM). Mobile phase A consisted of 0.1% formic acid in water and mobile phase B was 0.1% formic acid in methanol. The elution gradient was set as follows: 0–0.5 min, 3% B; 0.5–7.5 min, 3–80% B; 7.5–8 min, 80–98% B; 8–13 min, 98% B; 13–13.5 min, 98–100%B; 13.5–17 min, 100%B; 17–18 min, 100–3%B; 18–25 min, 3%B. The flow rate was 0.35 mL/min, and the injected volume was 5 μL. Mass spectrometer (MS) was performed on a Waters Micro mass Q/TOF micro Synapt High Definition Mass Spectrometer. Electrospray ionization source was used for mass spectrometric detection in negative mode, and the m/z range was set to 50–1000 with a mass window of 0.05 Da.
Data processing
For the metabolomics analysis, MS raw data files were converted to mzXML format using MassLynx V4.1 software and processed with R software for peak detection, extraction, alignment, and integration. The missing values in the raw data were filled by average value. Additionally, the overall normalization method was used for data analysis. To test the instrument precision, method repeatability, and sample stability, the relative standard derivation (RSD) of the peaks in the QC samples < 30% account for more than 80% was qualified. The multivariate analysis, principal component analysis (PCA), and orthogonal projections to latent structures analysis (OPLS-DA) were performed using SIMCA-P software (version 14.1). According to the variable importance in the projection (VIP) obtained in the OPLS-DA analysis, the candidate biomarkers of difference were preliminarily screened. The metabolites with VIP > 1 and p < 0.05 in the t-test were considered significantly different. Moreover, the OPLS-DA model quality was evaluated with standard parameters (R2Y and Q2) by 200 permutations. The m/z value is used to retrieve candidate metabolites in HMDB database (http:www.hmdb.ca/) and confirm metabolites through MS/MS analysis and literature.
In order to visualize the diagnostic performance of metabolites, we further drew heat map, cluster analytic hierarchy process, and receiver operating characteristic curve (ROC curve).
In vitro experiment
Cell proliferation assay
The HCT116, HCT8, and FHC cells were seeded in 6-well plates (2000 cell/well) and treated with GDCA (0 μM, 2 μM, 5 μM, and 10 μM). The HCT116 cell was seeded in 96-well plates (2000 cell/well) and treated with celastrol (0 nM, 10 nM, 20 nM, 50 nM, 100 nM, 200 nM, 500 nM, and 1000 nM). Relative cell proliferation was determined by cell counting at 4 days after being seeded and the percentage cell proliferation of the control.
Colony formation assay
HCT116 cells were plated into a 6-well plate incubated with GDCA (0 μM, 10 μM). We renew the GDCA containing medium every 3 days. Following 14 days of incubation, colonies were fixed with methanol solution and stained with 0.5% crystal violet aqueous. Cells were subsequently washed with PBS, dried, imaged, and counted the number of colonies.
Wound healing assay
Wound healing assay was used to detect the ability of CRC migration. The HCT116 cell was seeded in 6-well plates. When cells were grown to approximately 90% confluency, an artificial wound was created with a 200-μL pipette tip, and then treated with metabolite (0 μM, 10 μM) for 24 h. The cells were then cultured in fresh medium without FBS. To visualize wound healing, measured the gap distance of the wound on day 0 and after 24 h using Image-J software.
Identification of key targets
Network Pharmacology
First, the metabolite and celastrol structure were obtained in PubChem database (https://pubchem.ncbi.nlm.nih.gov). The PharmMapper database (http://www.lilab-ecust.cn/pharmmapper/) and STP database (http://www.swisstargetprediction.ch) were used to target prediction by the metabolite and celastrol structure. The targets of CRC were obtained in the CTD database (http://ctdbase.org/). The celastrol target and CRC-related gene set was used to construct PPI network by using STRING database (https://string-db.org/). The PPI network from STRING was then imported into Cytoscape 3.7.2 to construct a compound‐target-disease network. The David database (https://david.ncifcrf.gov) and Metascape database (http://metascape.org/) were used to performed enrichment analysis, including gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Potential personalized treatment mechanisms are revealed from biological processes (BP), cellular components (CC), molecular functions (MF), and key pathways.
Molecular docking
The receptor protein coded by the selected gene was searched in the UniProt database (https://www.uniprot.org/), and the 3D structure of the protein was downloaded at RCSB PDB database (https://www.rcsb.org/). BIOVIA Discovery Studio 2017 R2 (DS) software performed the dehydration of the receptor protein, and exported the 3D structure by minimizing energy. Finally, dock the receptor protein with the small molecule ligands by CDOCKER semi-flexible.
In vitro thermal shift assay
GDCA and cell protein extract were incubated on ice in PCR tube for 30 min (compared with dimethyl sulfoxide), then transferred to PCR machine, heated at 34.9 to 73 ℃ for 3 min, and cooled at room temperature for 20 min. This was followed by 14,000 rpm at 4℃ for 20 min in order to separate the soluble fraction from the pellet. Finally, the supernatant was transferred to a new tube and analyzed by SDS-PAGE and Western blot. In order to better match the combination of protein and metabolites in vivo, the same concentration of metabolites was applied to cells for 24 h. Based on the previous experimental results, the cell protein extract was treated with the same treatment method from the temperature of 60.9 to 76.2 ℃.
shRNA-mediated gene silencing and viral infection
The shRNA plasmids targeting PARP-1 were purchased from Transheep Biological Corporation. To establish stable knockdown cells, the HEK293T cells were transfected with lentiviral shRNA constructs plus with viral packaging plasmids (psPAX2 and pMD2.G). The viral supernatant was collected and filtered through 0.45 μM filter after 3 days transfection. Then, the HCT116 cells were transduced by lentivirus and selected with 2 μg/mL puromycin. The knockdown efficacy was determined by western blot analysis.
Overexpression of PARP-1 in HCT116 cells
Flag-PARP1 plasmid was purchased from Hunan Fenghui Biotechnology Corporation. For transient transfections, cells were grown to 80% confluency and transfected with plasmids using Polyethylenimine (PEI) Transfection Reagent (Polysciences, USA) according to the manufacturer’s protocol.
Tumor formation in nude mice
All animal care and experimental procedures were carried out in accordance with the recommendation of the Animal Care Ethics and Use Committee of Nankai University and approved by this Committee. Five-week-old BALB/c nude female mice were purchased from Vital River Laboratories (Beijing, China). Mice were housed at a constant room temperature with a 12:12 h light/dark cycle and fed a standard rodent diet and given water ad lib. Mice were randomly divided into two experimental groups (n = 7): control (NC) and celastrol treatment groups (celastrol group). HCT116 cells were injected subcutaneously into the right flank of all mice at 1 × 107 cells in PBS. After day 16 of cell injection, treatment of mice with celastrol at 3 mg/kg by intraperitoneal injection was started. Control mice received PBS vehicle control. Tumor volumes were determined by measuring length (L) and width (W) and calculating volume (V = 0.5 × L × W2) at the indicated time points. Treatments were maintained for 15 days. Mice were sacrificed, and tumor tissues were removed, weighed, and photographed.
Statistical analysis
The data were analyzed by SPSS software (ver. 25.0). Statistical analysis was performed with a t-test or one-way ANOVA. Data were presented as mean ± standard deviation. Error bars indicate ± SD. *p < 0.05, **p < 0.001, ***p < 0.0001.
Results
Changes of differential metabolites in CRC patients
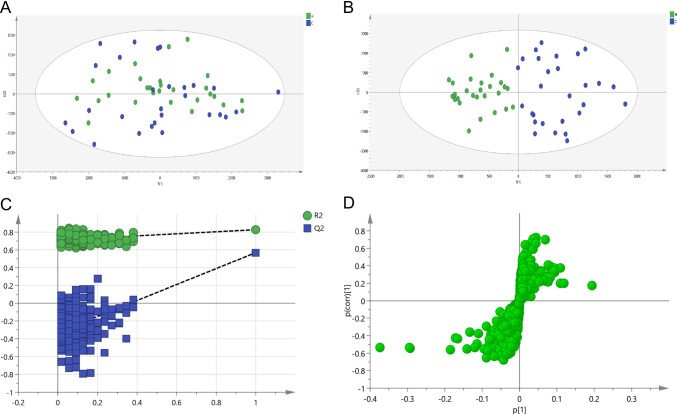
In methodology study, the established method has good reproducibility, the sample is stable within 48 h, and the instrument precision is good, which can be used for subsequent metabolomics experiments. In the negative ionization mode, the base peak intensity chromatogram of serum in the QC sample is shown in SFig. 1. Multivariate statistical analysis (PCA and OPLS-DA) was performed to comprehensively evaluate the metabolic spectrum of CRC. PCA (Fig. 1A) is used to evaluate the reproducibility of data and eliminate outlier samples, but the plot of PCA shows that the inter-group differences of samples cannot be well distinguished. Then, the OPLS-DA model (Fig. 1B) was constructed, R2Y = 0.736 and Q2 = 0.493, demonstrating that the model was no overfitting (Fig. 1C). The S-load diagram (Fig. 1D) shows that the metabolites contribute greatly to the differentiation between groups, and the farther away from the distant point, the greater the VIP value. Finally, 15 differential metabolites (Table 2) were screened, including 9 downregulated and 6 upregulated (Spectrum fragment information in STable 1).
Fig. 1.
Non-targeted metabolomics profiling analysis for human serum. A PCA score plot; B OPLS-DA score plot; C permutation test (n = 200) of the OPLS-DA model; D validation plots
Table 2.
15 differential metabolite information
| Compound | TR (min) | m/z | Molecular Formula | m/z Obsd | ppm | Trend | Fold change |
|---|---|---|---|---|---|---|---|
| N(2)-phenylacetyl-L-glutaminate | 4.6561 | 263.1029 | C13H16N2O4 | 263.1032 | 1.14 | ↓** | 0.77 |
| Phosphoribosyl formamidocarboxamide | 4.6956 | 411.0549 | C10H15N4O9P | 411.0553 | 0.97 | ↓** | 0.79 |
| Chorismate | 5.6357 | 225.0397 | C10H10O6 | 225.0399 | 0.89 | ↑** | 1.44 |
| Prenyl glucoside | 5.7165 | 165.0544 | C11H20O6 | 247.1182 | 1.62 | ↑** | 2.09 |
| Desaminotyrosine | 5.6951 | 247.1178 | C9H10O3 | 165.0552 | 4.85 | ↓** | 0.23 |
| Indolelactic acid | 5.794 | 204.0646 | C11H11NO3 | 204.0661 | 7.35 | ↓** | 0.76 |
| 2-hydroxy-3-(2,3,4-trimethoxyphenyl)propanoic acid | 6.3562 | 255.0863 | C12H16O6 | 255.0869 | 2.35 | ↑** | 1.06 |
| 3-Hydroxyanthranilic acid | 7.1356 | 152.034 | C7H7NO3 | 152.0348 | 5.26 | ↓** | 0.64 |
| Glycodeoxycholic acid | 9.4667 | 448.3063 | C26H43NO5 | 448.3063 | 0 | ↑** | 1.34 |
| LysoPE(18:1(9Z)/0:0) | 10.3918 | 478.2929 | C23H46NO7P | 478.2934 | 1.04 | ↓** | 0.80 |
| (R)-3-Hydroxy-hexadecanoic acid | 10.4043 | 271.2272 | C16H32O3 | 271.2273 | 0.37 | ↑** | 1.39 |
| LysoPC(20:2(11Z,14Z)/0:0) | 10.4742 | 592.3606 | C28H54NO7P | 592.3614 | 1.35 | ↓** | 0.82 |
| Dihomolinoleic acid | 10.6535 | 279.2319 | C18H32O2 | 279.2324 | 1.79 | ↓** | 0.84 |
| Calcidiol | 10.8594 | 445.3313 | C27H44O2 | 445.3318 | 1.12 | ↑** | 1.60 |
| Glyceraldehyde 3-phosphate | 10.9793 | 168.9895 | C3H7O6P | 168.9902 | 4.14 | ↓** | 0.69 |
(**: p < 0.01; ↓, down-regulation; ↑, upregulation)
Glycodeoxycholic acid (GDCA) has a significant effect on the development of colorectal cancer
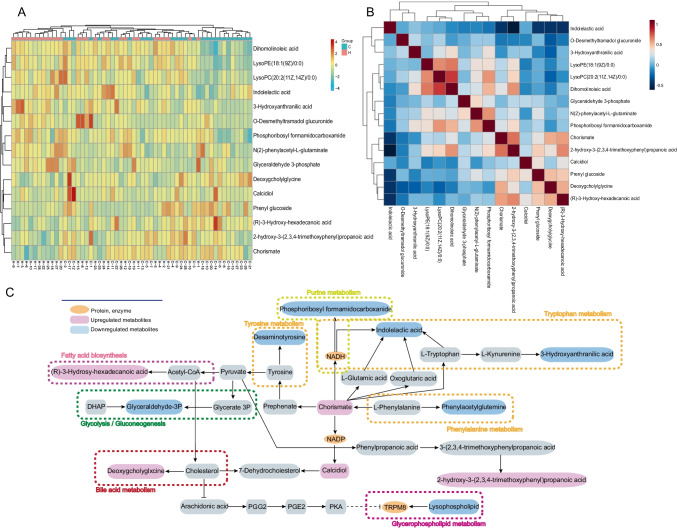
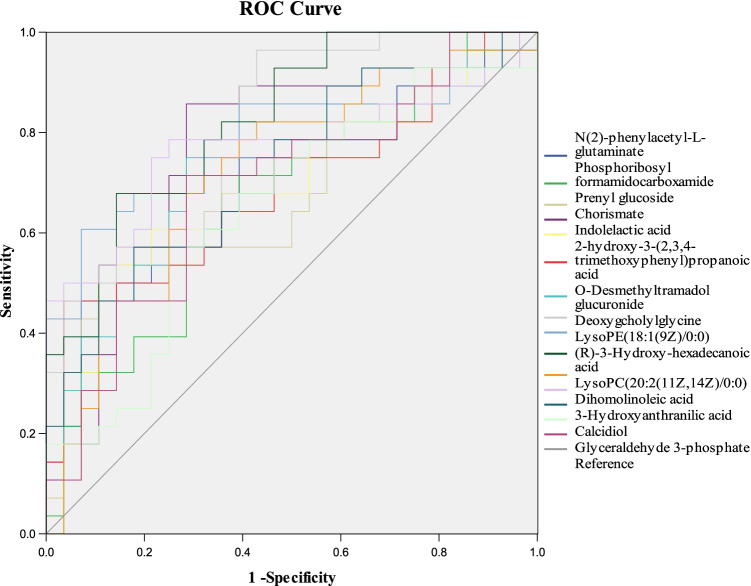
The heatmap was used to classify the upregulated and downregulated differential metabolites in patients with CRC (Fig. 2A). Correlation analysis (Fig. 2B) is helpful to explore the relationship between metabolites, and lay a foundation for subsequent metabolic pathway analysis. The results show that (R)-3-Hydroxy-hexadecanoic acid, prenyl glucoside, chorismate, and GDCA had a certain correlation; glycerophospholipids were associated with dihomolinoleic acid. Additionally, a KEGG pathway analysis (Fig. 2C) was performed to explore the enriched metabolic pathways associated with CRC initiation and progression. The results showed that the main pathways involved were amino acid metabolism, bile acid metabolism, fatty acid biosynthesis pathway, purine metabolism, glycolysis/gluconeogenesis, and glycerol phospholipid metabolism. To assess the clinical diagnostic ability of the metabolites, the area under curve (AUC) (Fig. 3) was employed to identify 15 metabolites of serum. Among them, the AUC of GDCA reached 0.825, despite its content change was not the highest in the 15 metabolites. Meanwhile, recent studies have reported that bile acid metabolism is a risk factor in the occurrence and progression of CRC, especially the secondary bile acid produced by microorganisms can promote the occurrence and development of intestinal tumors [31–33]. Kühn Tilman et al. [34] have shown that GDCA is highly expressed in CRC, and secondary bile acids contribute to colorectal cancer promotion, but the mechanism has not been elucidated.
Fig. 2.
Identification of the differential metabolomics profiles by clustering, correlation and pathways analysis. A Heatmap of differentially expressed metabolites; B correlation analysis of metabolites; C metabolic pathways of 15 different metabolites
Fig. 3.
ROC curve of 15 differential metabolites for distinguishing the CRC group from the healthy group
GDCA can promote the proliferation of colorectal cancer cells
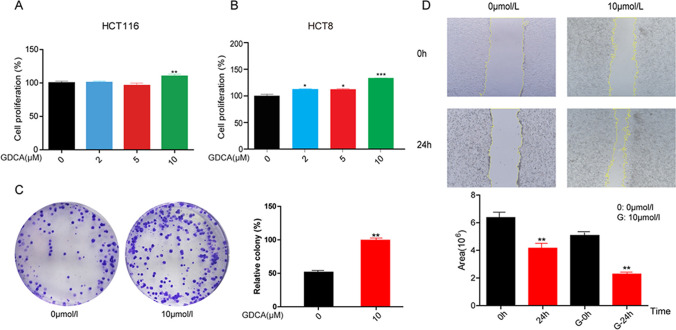
According to the results of metabolomics, GDCA may play a promoting role in the occurrence and development of colorectal cancer. In order to verify this hypothesis, a series of in vitro experiments were carried out. Firstly, the effect of GDCA on tumorigenesis was measured with cell proliferation assay after GDCA (0 μM, 2 μM, 5 μM, 10 μM) treatment in HCT116, HCT8, and FHC cells. Cell proliferation assay observed that GDCA promotes cell proliferation of HCT116 (Fig. 4A) and HCT8 (Fig. 4B) cells in concentration of 5 μM and 10 μM; however, GDCA had no effect on FHC cell viability (SFig 2). To identify the impact of GDCA on basic cell properties, we performed a colony formation assay and wound healing assay in the HCT116 cell line. The colony formation assay evaluates the effect of GDCA on colony formation of the HCT116 cells. Ten micrometers of GDCA strongly promotes clonogenicity of HCT116 cells (Fig. 4C). Furthermore, wound healing assay found that GDCA significantly promoted cell migration of cells HCT116 in a concentration of 10 μM (Fig. 4D). These results suggest that GDCA contributes to the proliferation of CRC cells and plays a significant role in the progression of CRC.
Fig. 4.
GDCA can promote the proliferation of CRC cells. A–B GDCA promotes CRC cell lines (HCT116 and HCT8) proliferation; C–D GDCA strongly promotes clonogenicity of HCT116 cells; E GDCA promoted cell migration of HCT116 cell line. Three independent experiments were conducted for each assay (*p < 0.05, **p < 0.01, ***p < 0.001)
PARP-1 may be a key target for GDCA to promote the proliferation of CRC cells
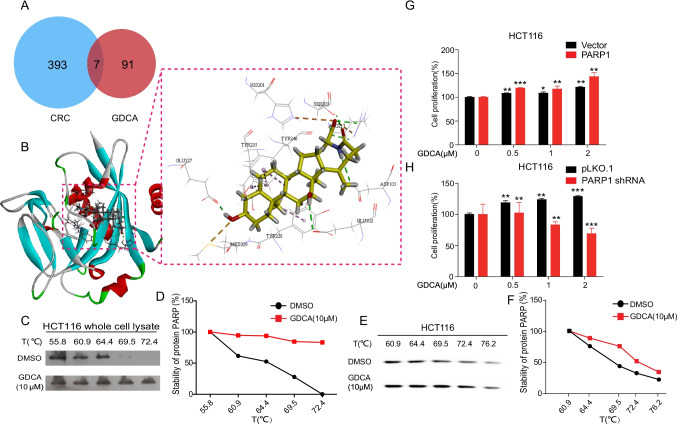
To develop the mechanism of GDCA on CRC cell proliferation promotion, the key targets of GDCA were screened by network pharmacology technology (GDCA and CRC targets information in STable 2). By taking an intersection of the GDCA targets and CRC-related genes, the 7 common targets were finally obtained (Fig. 5A), which are PARP-1, AKT1, PPARG, EGFR, ADH1B, BMP2, and SRC. The common targets conducted molecular docking with GDCA, to screen the targets that can produce the best binding mode with GDCA. Subsequently, molecular docking results indicated that GDCA could easily enter and bind to the active pocket of the PARP-1 protein (PDB:5DS3) as shown in Fig. 5B. At the same time, Nosho K et al. [35] found that mRNA overexpression of PARP-1 was detected in 64 (70.3%) of 91 CRC tumors. Other studies have shown that the differential expression of PARP-1 in CRC is 1.08 to 1.28 fold [36].
Fig. 5.
The expression of PARP-1 played a key role in the promotion of CRC cell proliferation by GDCA. A Intersection analysis between GDCA and CRC targets; B molecular docking results of GDCA with PARP-1; C–F thermal shift assay was measured the combination effect of GDCA and PARP-1; G–H sensitivity of PARP-1 overexpression/knockdown cells to GDCA treatment. Three independent experiments were conducted for each assay (*p < 0.05,**p < 0.01, ***p < 0.001)
The docking results are analyzed as follows: oxygen atoms interact with amino acid residues Ser904, Phe897, and His862 to form carbon-hydrogen bonds. The compound also forms a hydrophobic interaction with residues Ala898, Ser864, Asn868, Leu877, and ILe895, etc., and the hydrophobic interaction and π-π stacking interaction provide a strong van der Waals force for the compound. Oxygen atoms are easy to obtain electrons, and hydroxyl groups are electron-donating, so it is reasonable for Gly863, Arg878, and Gly876 to bond with oxygen atoms. The C = O group in the compound will also form a hydrogen bond with Tyr907 to establish a fourth hydrogen bond force. His862 is a weak alkaline and forms an attractive group with carboxyl groups. His862, Tyr896, and Arg878 form Pi-Alkyl with the rings in the compound. At the same time, it has been reported that there are three critical H-bond forces between Gly863 and Ser904, the two key amino acid residues in PARP-1 inhibitors, through the crystal structure and molecular simulation of PARP-1, so we supposed that GDCA binds to PARP-1 to promote the occurrence and development of CRC [37]. The analysis of this binding pattern helps to understand the possible mechanism of GDCA promoting CRC through PARP-1. Based on the above experimental results, it is a hypothesis that PARP-1 may be one of the key targets for GDCA to promote CRC.
In order to verify the results of molecular docking experiment, the thermal shift assay was used to check the binding effect of PARP-1 and GDCA. The results showed that with the increase of temperature, the unchain temperature of the GDCA group was higher than that of the control group, which indicated that GDCA was directly bound to PARP-1 protein (Fig. 5C–F). In addition, we found that the PARP1 overexpression cells were sensitive to GDCA treatment, whereas PARP1 knockdown cells were resistant to GDCA treatment (Fig. 5G, H). Overall, these results suggest that GDCA can promote CRC progression by targeting PARP-1.
Celastrol may become a new therapeutic agent for CRC
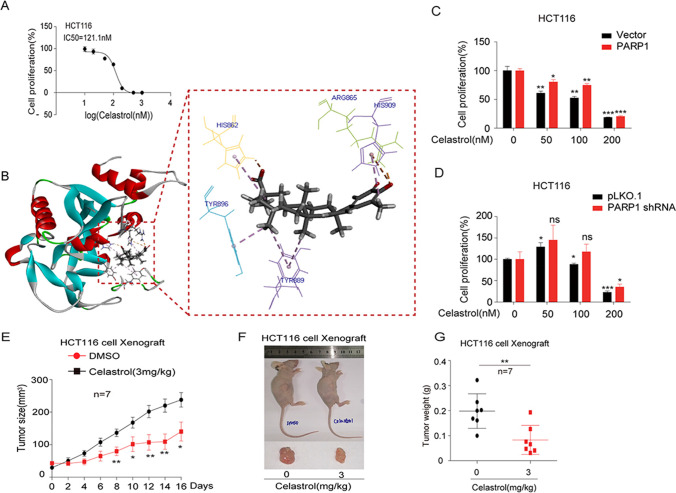
Celastrol is a main active ingredient derived from the traditional Chinese medicine Tripterygium wilfordii and has potential anticancer activity. The anticancer effect of celastrol was measured with cell proliferation assay after celastrol (0–1000 nM) treatment in HCT116 cells. The IC50 of celastrol was 121.1 nM in HCT116 cells (Fig. 6A). In order to explore whether celastrol inhibits CRC through PARP-1, we also do molecular docking verification (Fig. 6B) between celastrol and PARP-1. Compared with GDCA, most amino acid residues have reproducibility. The hydrogen on N atom in the imidazolium ring in histidine (His) is easy to leave in the form of hydrogen ions, so that celastrol and His909 form Pi-Anion interaction with the formation of electrostatic force. Form a salt bridge with Arg865 and His862. There are aromatic rings in amino acid residues His862, Tyr889, Tyr896, and His909 that interact with celastrol to form Pi-Alkyl. In addition, both Gly894, Ile895, Leu877, Agr878, Ile872, Ala880, and Gly888 formed different degrees of van der Waals forces with celastrol. Furthermore, we found that the PARP-1 overexpression cells were resistant to GDCA treatment, whereas PARP-1 knockdown cells were sensitive to GDCA treatment (Fig. 6C, D). Based on these results, this study attempted to evaluate the above results in a xenograft mouse model. Remarkably, mice treated with celastrol showed attenuated tumor growth compared with untreated mice (Fig. 6E). The overall size and weight of the tumors in the celastrol-treated groups were obviously lower than the model group (Fig. 6F, G). There was no significant difference in mean body weight between celastrol-treated and untreated mice throughout the treatment schedule (SFig 3). Collectively, these data indicated that celastrol inhibits CRC tumor growth in vitro and in vivo.
Fig. 6.
Celastrol may become a new therapeutic agent for CRC. A The IC50 of celastrol; B molecular docking results of Celastrol with PARP-1; C–D sensitivity of PARP-1 overexpression/knockdown cells to celastrol treatment; E attenuated tumor growth in celastrol treated mice; F–G the overall size and weight of the tumors in nude mice. Three independent experiments were conducted for each assay (*p < 0.05, **p < 0.01, ***p < 0.001)
Mechanisms of celastrol therapy for CRC
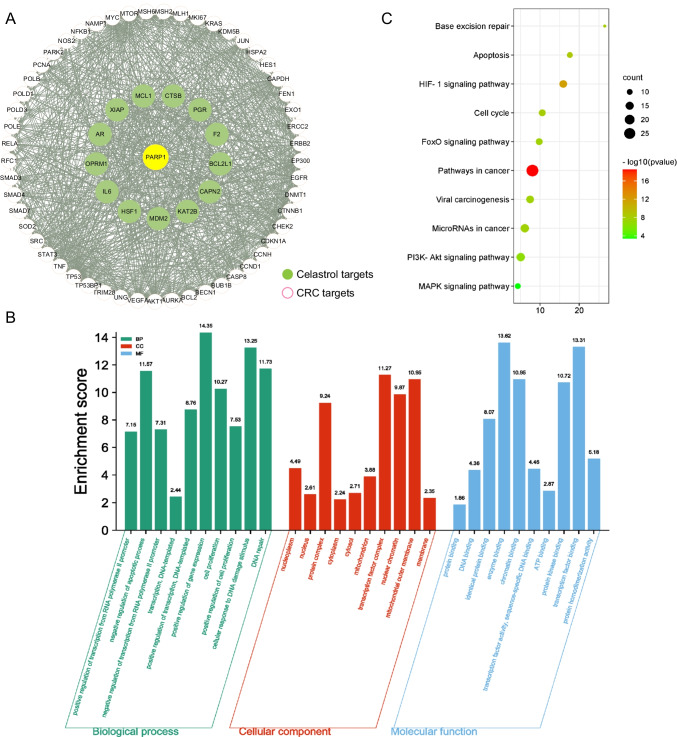
After discovering that celastrol can treat CRC through PARP-1, a target interaction network was constructed by network pharmacology technology (Targets information in STable 2). There are 66 CRC targets interacting with PARP-1, which include 13 celastrol targets. Then, visualized the target interaction network (Fig. 7A) with 67 nodes and 900 edges by using Cytoscape 3.7.2 (Nodes information in STable 3). GO enrichment analysis was used to discover the underlying BP, CC, and MF of the 67 target genes. By setting the filter as adjusted p-value cutoff: 0.01, we obtained 220 significant enriched GO terms. The top 10 terms of BP, CC, and MF are illustrated in Fig. 7B. The GO terms suggested that these target genes played an essential role in apoptotic, DNA damage, and transcription regulation (GO terms statistical information in S Table 4). KEGG enrichment analysis was performed to discover those pathways enriched by the 67 target genes. The filter was also set as an adjusted p-value cutoff: 0.01. A total of 96 KEGG pathways were significantly enriched, which showed that these target genes affected the pathways of cancer, HIF-1 signaling pathway and PI3K-Akt signaling pathway, as well as a series of important pathological processes such as apoptosis. The bubble plot of the most significant 10 KEGG pathways was shown in Fig. 7C. The pathways statistics can be acquired in the STable 4.
Fig. 7.
The mechanisms of celastrol therapy for CRC. A The target interaction network for 66 targets; B GO functional enrichment analysis; C KEGG enrichment analysis
Discussion
PPPM concept in the current study
Due to the lack of special clinical manifestations in early stage of CRC, leading to difficulty in diagnosis, the incidence rate is increasing year by year [38]. Early screening can reduce the incidence rate and mortality of CRC through early detection and treatment. However, no matter what screening methods are used, screening is both cost-effective and risky [39]. Therefore, it is urgent to find a non-invasive screening tool with high sensitivity and specificity. In addition, during cancer treatment, there are individual differences in treatment effect (gene, gender, race, lifestyle, environmental factors, etc.), involving each stage of prediction/prevention, early diagnosis/treatment, and late diagnosis/treatment [3]. Therefore, it is necessary to carry out personalized medical treatment according to the personal characteristics of patients in the process of treatment. The occurrence of cancer results from the combined action of the imbalance of DNA, RNA, protein, and metabolites; the multi-omics technology can connect these different levels of molecules, which is helpful for researchers to understand cancer biology and may contribute to cancer prevention, targeted therapy, and drug development [10, 40]. Different from other omics, metabolomics can be used as an indicators of physiological or pathological status, by exploring the overall changes in the process of tumor occurrence and development, clarifying the etiology, screening biomarkers that can provide early diagnosis, staging, evaluating curative effect and prediction of prognosis, and formulate effective personalized medical treatment strategies [10, 41]. Therefore, through the research of metabolites in cancer and the functional analysis of these metabolites, we can fully understand the differences of individual biological systems, potential therapeutic targets, and chemicals for targeted therapy, and promote personalized medicine [7]. This is consistent with the goal of cancer PPPM [10].
Achievements in the current study
The results of this study indicate that GDCA can potentially enhance DNA damage repair in cells via its association with PARP-1 targets, which can promote CRC progression. Therefore, PARP-1 may be used as a targeted therapy. In addition, this study found that celastrol has a role in the multi-dimensional prevention and control of CRC via PARP-1. These results demonstrate the ability to apply metabolomics in personalized medicine management based on PPPM. In this study, a total of 15 differential metabolites were found that were related to abnormal energy metabolism, such as amino acid metabolism, bile acid (BA) metabolism and synthesis, purine metabolism, glycolysis/gluconeogenesis, and glycerol phospholipid metabolism [42–44]. These metabolic pathways are necessary to maintain normal cell function and biological survival. Once cancer cells continue to proliferate and nutrient supply is insufficient, the content of metabolites changes significantly in the process of tumor occurrence and development. Of the measured metabolites, GDCA was positively correlated with CRC and had a high clinical diagnostic effect; therefore, GDCA could act as a biomarker for the prediction and diagnosis of CRC. Since the actual tumor-promoting effect of deoxycholic acid in 1939, the cancer incidence rate and mortality with BA have aroused people’s concern [45]. The intestine is the main place for the distribution, absorption, and excretion of BAs in the body. Some primary bile acids are biotransformed through flora in the colon, and secondary bile acids are also partially reabsorbed during colonic transport [46]. Studies have shown that some genotoxic secondary bile acid accumulation can directly cause DNA damage, protein damage, mucosal cell proliferation, and inflammation which promote colon cancer [31, 34, 47, 48]. Farnesol X receptor (FXR) is a key regulator of BA homeostasis, and its function impairment is related to higher BA concentration and tumor-promoting phenotype [49, 50]. Several prospective studies of CRC have found that the pre-diagnosis level of GDCA is positively correlated with the risk of CRC [34, 51, 52]. The results of this study also demonstrate the correlation between serum GDCA concentration and CRC, and further verify the pathogenic role of GDCA in CRC.
At present, many candidate biomarkers with potential clinical value have been found; however, their biological functions and their physiological and pathological significance in human biological systems, as well as related mechanisms, have not been studied. With the use of metabolomics, this study comprehensively employed a variety of technical means, including molecular and cell biology techniques, bioinformatics, and molecular simulation, to screen potential targets for individualized prevention and medical treatment. This study found that PARP-1 is related to the occurrence and development of CRC. PARP-1 is an enzyme located in the nucleus that can catalyze the synthesis of poly ADP-ribose on protein substrates by binding to DNA damage sites, leading to the recruitment of additional DNA repair proteins to the damaged site to complete DNA damage repair [10]. In recent years, PARP-1 has attracted extensive attention as a potential cancer therapeutic target [40, 41]. For instance, PARP-1 expression in breast cancer is correlated with tumor grade and can independently predict poor prognosis in invasive breast cancer patients [44]. Schiewer et al. showed that the activity of PARP-1 was enhanced in a model of advanced prostate cancer, which was closely related to disease progression, and they suggested that targeting PARP-1 could effectively inhibit tumor growth [48]. Previous studies have found that PARP-1 is overexpression in CRC patients and can promote CRC tumor growth by triggering inflammation and activating the IL6-STAT3-cyclin D1 axis [53, 54]. Oxidative stress, bacterial products (LPS), and inflammatory cytokines (IL-1, TNFα) can activate PARP-1 and nuclear factor-κB (NF-κB), the latter of which participates in inflammation as well as carcinogenesis [55–57]. Therefore, PARP-1 inhibition is of great significance for cancer treatment, and PARP-1 may be used as a protein biomarker for future treatment, such as breast cancer, pancreatic cancer, ovarian cancer, and prostate cancer [58, 59]. Studies have found that the increase of PARP-1 protein occurs in many tumor types, suggesting that it is involved in the carcinogenic process and may have prognostic value [60]. In addition, the liver is the most common metastatic site in the occurrence and development of CRC [61]. At present, it has been reported that fatty acid binding protein 6 (FABP6), which acts as an intracellular transporter of BA, is highly expressed in primary CRC and adenomas compared with the normal epithelium, but significantly decreased in lymph node metastasis [62]. Through organoid transcriptional analyses, the signature genes changed in liver metastasis of primary tumors and CRC and SOX2 inducible knockdown weakens the growth of invasion, proliferation, and liver metastasis. It is proved that SOX2 is associated with colorectal cancer progression and may serve as a potential prognostic biomarker and therapeutic target [63]. In this experiment, PARP-1 can participate in tumor invasion and metastasis through different molecular mechanisms (e.g., hypoxia, epithelial-mesenchymal transition, and angiogenesis) [64, 65]. Inhibition of PARP can counteract the ability of melanoma cells to metastasize to the lung, which is related to the downregulation of the intermediary filament vimentin in both endothelial cells and melanoma cells, which led to a reversion of mesenchymal phenotype of the two cell types and prevented malignant melanoma cells from developing vasculogenic mimicry [66]. Moreover, PARP-1 has been reported as a biomarker of poor prognosis for stage II–III colon cancer. PARP-1 should be included in the evaluation index when selecting treatment strategies [67]. In summary, these data suggest that the occurrence and development of CRC are related to GDCA and PARP-1. As a direct target of GDCA, PARP-1 expression is regulated by the content of GDCA, which affects the occurrence, development, metastasis, and prognosis of CRC. Therefore, we recommend that GDCA and PARP-1 can be saved as evaluation indexes in the prevention, diagnosis, and treatment of CRC.
Personalization of medical services in CRC management
Cancer is a major factor threatening human health and life safety. Extensive studies have identified numerous dietary metabolites and phytochemicals with chemopreventive potential [68]. Phytochemicals widely exist in nature; phytochemicals, plant extracts, and plant-based diets are the main sources for the preventive effects on cancer development, and cancer progression and metastasis [24, 69]. There are many natural plant metabolites with anti-CRC effects, including terpenoids, alkaloids, saponins, flavonoids, and polysaccharides [70]. These anticancer phytochemicals have multi-targeting potential because they can regulate multiple pathways to affect each stage of tumor growth. Phytochemicals may prevent or treat cancer by improving immunity, regulating specific signaling pathways, inhibiting angiogenesis, cell cycle arrest, and inducing apoptosis and mitochondrial/DNA damage [24, 26, 71–74].
Celastrol, a natural chemical derived from Tripterygium wilfordii, has shown important antioxidant and anti-inflammatory activities. It can inhibit the secretion of pro-inflammatory cytokines and inhibit tumor proliferation, growth, and metastasis in various cancer models [75]. In this study, we found that the therapeutic effect of celastrol in CRC was related to its action on the PARP-1 protein, resulting in DNA damage, affecting cancer cell cycle, inhibiting angiogenesis and cancer cell apoptosis. The main pathways involved in apoptosis are the PI3K-Akt signaling pathway, MAPK signaling pathway, HIF-1 signaling pathway, and the mitochondrial endogenous signaling pathway, among others. Celastrol promoted the association of an ATP–cytochrome C complex to activate Casp-9 and its downstream caspase, Casp-3, by regulating the key factor Bcl-2 [76]. In addition, the increase in Bax resulted in the activation of Casp-3. In the early stage of caspase-dependent apoptosis, activated caspases cleave PARP-1 into fragments, enabling cancer cell apoptosis [77]. Conversely, celastrol may also inhibit CRC cell proliferation via the NF-κB signaling pathway in a dose- and time-dependent manner, by upregulating p21 expression, NF-κB decreases expression, inducing apoptosis, and increasing casp-9/-3 activities [78]. Celastrol may induce anti-CRC effects via the mTOR signaling cascade, which involves several proteins, including Akt, mTOR, and S6K, and its effects may also be related to angiogenesis inhibition [79]. In addition, triptolide can reduce inflammatory mediators (TNF-α, IL-1β, IL-6, COX-2, and iNOS) and NF-κB inactivates and inhibits the inflammatory response; EMT may also be suppressed [80]. Celastrol upregulated HSF1 expression, enhanced LKB1 transcriptional activity, activated AMPKα and YAP, and promoted β-catenin degradation via the ubiquitin–proteasome system to inhibit CRC cell growth in vitro and in vivo [28]. Therefore, celastrol is a natural, active tumor drug that has shown great potential for targeted CRC therapy.
Conclusion and expert recommendations
There have been many studies on the metabolomics of CRC [21, 81, 82]; however, it has no study that could be traced about carry out personalized medicine by verifying the function of metabolites, discover key targets. Thus, the present study was conducted to compare the significantly altered metabolites between two groups, the HC and CRC groups, and to determine the potential of personalized medicine management via metabolites. In the present study, we found that GDCA may have potential as a biomarker for the identification of CRC and has prediction and prevention potential in the occurrence and development of CRC. Given the findings, PARP-1 serves as GDCA target protein which provides a scientific basis and theoretical support for the discovery of effective therapeutic targets for CRC. We also showed that celastrol can inhibit CRC via PARP-1, and the attributing mechanisms may be related to the PI3K-Akt signaling pathway, HIF-1 signaling pathway, cell cycle, apoptosis, or DNA damage. Therefore, functional metabolomics can provide a new perspective for the medical management of CRC based on the concept of PPPM.
We recommend the use of metabonomics techniques in PPPM-related studies of cancer. Metabolomics can quickly and accurately inform on physiological and pathological processes. The discovery of a large number of potential biomarkers and therapeutic targets has produced short-term benefits in the process of anti-cancer, and metabonomics will be a powerful tool to fight cancer in the future. However, there is a lack of research on the biological functions and the related physiological and pathological significance of differential metabolites. It remains a challenge to achieve highly sensitive early diagnosis, personalized drugs based on metabolite spectrum, and long-term benefits of improving the survival rate of cancer patients. Tumor metabolic status is a key factor affecting tumor metastasis and development. By exploring the biological function of different metabolites, we can determine key intervention targets for manipulating tumor metabolism, which is an important aspect of personalized medicine management and targeted preventive measures for cancer patients [83]. In future phytochemical research, we should pay attention to the clinical therapeutic dose. High doses may not be metabolized efficiently in the human body and may cause toxic organ damage [84]. In the clinical setting, in addition to considering anticancer effects, it can be used to reverse drug resistance, enhance curative effects, alleviate side effects following radiotherapy/chemotherapy, enhance immunity and anti-metastasis, and prevent recurrence in combination with surgery. In addition, the bioavailability of phytochemicals should be considered, and new dosage levels may be used to improve treatment. The abovementioned considerations will help to inform cancer prediction, prevention, and personalized medicine based on biomarkers in omics studies. Moreover, they will aid to systematically clarify the molecular mechanisms to find new therapeutic targets and drugs.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file 1 Figure 1: The base peak intensity (BPI) chromatogram of serum in the QC sample. (ai 481 KB)
Supplementary file 2 Figure 2: Effect of GDCA on the activity of FHC cell line. Cell viability was measured with MTT assay in FHC cells. (ai 175 KB)
Supplementary file 3 Figure 3: Anticancer effect of celastrol on CRC. A: The docking results of PARP-1 protein (PDB:5DS3) with celastro (2D). B: Weight changes of xenograft mice. (ai 200 KB)
Abbreviations
- PPPM
Predictive, preventive, and personalized medicine
- CRC
Colorectal cancer
- GDCA
Glycodeoxycholic acid
- PARP-1
Protein poly (ADP-ribose) polymerase-1
- PCA
Principal component analysis
- OPLS-DA
Orthogonal partial least squares discriminant analysis
- ROC
Receiver operating characteristic
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GO
Gene Ontology
Author contribution
SZ and YL conceived the study and designed the experiments. YY and CY performed most experiments and analyzed the data. YY, CY, YW, SS and LL assisted with cells culture. GS and JH collected clinical samples. YY, SS, and CB assisted with UPLC-Q-TOF/MS analysis. YM and MS performed animal experiments. CS, SZ, and YL performed writing. The author(s) read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81573825, 81902826, 81672781 and 81973356) and Tianjin Development Program for Innovation and Entrepreneurship (20190617), the Fundamental Research Funds for the Central Universities, Nankai University (3206054, 91923101, 63213082 and 92122017), and the National Key R&D Program of China (No. 2018YFC2002000).
National Natural Science Foundation of China,81573825,Yubo Li,81902826,Shuai Zhang,81672781,Shuai Zhang,81973356,Shuai Zhang,Tianjin Development Program for Innovation and Entrepreneurship,20190617,Yubo Li,Fundamental Research Funds for the Central Universities,3206054,Changliang Shan,91923101,Changliang Shan,63213082,Changliang Shan,92122017,Changliang Shan,National Key R&D Program of China,2018YFC2002000,Changliang Shan
Data availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
All software applications used are included in this article.
Declarations
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Institutional Ethics Committee of the Second Hospital of Tianjin Medical University (Ethical approval number: KY2019K144) and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Consent to participate
Written informed consent was obtained from individual or guardian participants.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yu Yuan and Chenxin Yang contributed equally to this work
Contributor Information
Shuai Zhang, Email: shuaizhang@tjutcm.edu.cn.
Yubo Li, Email: yuboli1@163.com.
References
- 1.Edited by Wild CP, Weiderpass E, Stewart BW. World Cancer Report: Cancer Research for Cancer Prevention
- 2.Sharma R. An examination of colorectal cancer burden by socioeconomic status: evidence from GLOBOCAN 2018. EPMA J. 2019;11(1):95–117. doi: 10.1007/s13167-019-00185-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akimoto N, Ugai T, Zhong R, Hamada T, Fujiyoshi K, Giannakis M, et al. Rising incidence of early-onset colorectal cancer-a call to action. Nat Rev Clin Oncol. 2021;18(4):230–243. doi: 10.1038/s41571-020-00445-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reynolds A, Mann J, Cummings J, Winter N, Mete E, Te Morenga L. Carbohydrate quality and human health: a series of systematic reviews and meta-analyses. Lancet. 2019;393(10170):434–445. doi: 10.1016/S0140-6736(18)31809-9. [DOI] [PubMed] [Google Scholar]
- 5.Kerr J, Anderson C, Lippman SM. Physical activity, sedentary behaviour, diet, and cancer: an update and emerging new evidence. Lancet Oncol. 2017;18(8):e457–e471. doi: 10.1016/S1470-2045(17)30411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sveen A, Kopetz S, Lothe RA. Biomarker-guided therapy for colorectal cancer: strength in complexity. Nat Rev Clin Oncol. 2020;17(1):11–32. doi: 10.1038/s41571-019-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grech G, Zhan X, Yoo BC, Bubnov R, Hagan S, Danesi R, et al. EPMA position paper in cancer: current overview and future perspectives. EPMA J. 2015;6(1):9. doi: 10.1186/s13167-015-0030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Mouradov D, Wang X, Jorissen RN, Chambers MC, Zimmerman LJ, et al. Colorectal cancer cell line proteomes are representative of primary tumors and predict drug sensitivity. Gastroenterology. 2017;153(4):1082–1095. doi: 10.1053/j.gastro.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aydin B, Caliskan A, Arga KY. Overview of omics biomarkers in pituitary neuroendocrine tumors to design future diagnosis and treatment strategies. EPMA J. 2021;12:383–401. doi: 10.1007/s13167-021-00246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu M, Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018;9(1):77–102. doi: 10.1007/s13167-018-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hull MA, Rees CJ, Sharp L, Koo S. A risk-stratified approach to colorectal cancer prevention and diagnosis. Nat Rev Gastroenterol Hepatol. 2020;17(12):773–780. doi: 10.1038/s41575-020-00368-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging. 2016;11:967–976. doi: 10.2147/CIA.S109285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin B, Lieberman DA, McFarland B, Andrews KS, Brooks D, Bond J, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134(5):1570–1595. doi: 10.1053/j.gastro.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Needham BD, Adame MD, Serena G, Rose DR, Preston GM, Conrad MC, et al. Plasma and fecal metabolite profiles in autism spectrum disorder. Biol Psychiatry. 2021;89(5):451–462. doi: 10.1016/j.biopsych.2020.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danczak RE, Chu RK, Fansler SJ, Goldman AE, Graham EB, Tfaily MM, et al. Using metacommunity ecology to understand environmental metabolomes. Nat Commun. 2020;11(1):6369. doi: 10.1038/s41467-020-19989-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ericksen RE, Lim SL, McDonnell E, Shuen WH, Vadiveloo M, White PJ, et al. Loss of BCAA catabolism during carcinogenesis enhances mTORC1 activity and promotes tumor development and progression. Cell Metab. 2019;29(5):1151–1165.e6. doi: 10.1016/j.cmet.2018.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaymak I, Williams KS, Cantor JR, Jones RG. Immunometabolic interplay in the tumor microenvironment. Cancer Cell. 2021;39(1):28–37. doi: 10.1016/j.ccell.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamaidi I, Zhang L, Kim N, Wang MH, Iclozan C, Fang B, et al. Sirt2 inhibition enhances metabolic fitness and effector functions of tumor-reactive T cells. Cell Metab. 2020;32(3):420–436.e12. doi: 10.1016/j.cmet.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peitzsch C, Gorodetska I, Klusa D, Shi Q, Alves TC, Pantel K, et al. Metabolic regulation of prostate cancer heterogeneity and plasticity. Semin Cancer Biol. 2020;S1044–579X(20)30261–3. [DOI] [PubMed]
- 20.Monedeiro F, Monedeiro-Milanowski M, Ligor T, Buszewski B. A review of GC-based analysis of non-invasive biomarkers of colorectal cancer and related pathways. J Clin Med. 2020;9(10):3191. doi: 10.3390/jcm9103191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25(6):968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 22.Uchiyama K, Yagi N, Mizushima K, Higashimura Y, Hirai Y, Okayama T, et al. Serum metabolomics analysis for early detection of colorectal cancer. J Gastroenterol. 2017;52(6):677–694. doi: 10.1007/s00535-016-1261-6. [DOI] [PubMed] [Google Scholar]
- 23.Wu JY, Huang TW, Hsieh YT, Wang YF, Yen CC, Lee GL, et al. Cancer-derived succinate promotes macrophage polarization and cancer metastasis via succinate receptor. Mol Cell. 2020;77(2):213–227.e5. doi: 10.1016/j.molcel.2019.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Koklesova L, Liskova A, Samec M, Qaradakhi T, Zulli A, Smejkal K, et al. Genoprotective activities of plant natural substances in cancer and chemopreventive strategies in the context of 3P medicine. EPMA J. 2020;11(2):261–287. doi: 10.1007/s13167-020-00210-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dandawate PR, Subramaniam D, Jensen RA, Anant S. Targeting cancer stem cells and signaling pathways by phytochemicals: novel approach for breast cancer therapy. Semin Cancer Biol. 2016;40–41:192–208. doi: 10.1016/j.semcancer.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samec M, Liskova A, Koklesova L, Samuel SM, Zhai K, Buhrmann C, et al. Flavonoids against the Warburg phenotype-concepts of predictive, preventive and personalised medicine to cut the Gordian knot of cancer cell metabolism. EPMA J. 2020;11(3):377–398. doi: 10.1007/s13167-020-00217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong J, Shen Y, Zhang Z, Hu Y, Zhang X, Han L. Apigenin inhibits epithelial-mesenchymal transition of human colon cancer cells through NF-κB/Snail signaling pathway. Biosci Rep. 2019;39(5):BSR20190452. [DOI] [PMC free article] [PubMed]
- 28.Wang S, Ma K, Zhou C, Wang Y, Hu G, Chen L, et al. LKB1 and YAP phosphorylation play important roles in Celastrol-induced β-catenin degradation in colorectal cancer. Ther Adv Med Oncol. 2019;11:1758835919843736. doi: 10.1177/1758835919843736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu H, Liu XW, Ding WJ, Xu DQ, Zhao YC, Lu W, et al. Up-regulation of death receptor 4 and 5 by celastrol enhances the anti-cancer activity of TRAIL/Apo-2L. Cancer Lett. 2010;297(2):155–164. doi: 10.1016/j.canlet.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 30.Sun C, Li T, Song X, Huang L, Zang Q, Xu J, et al. Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc Natl Acad Sci USA. 2019;116(1):52–57. doi: 10.1073/pnas.1808950116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen PB, Black AS, Sobel AL, Zhao Y, Mukherjee P, Molparia B, et al. Directed remodeling of the mouse gut microbiome inhibits the development of atherosclerosis. Nat Biotechnol. 2020;38(11):1288–1297. doi: 10.1038/s41587-020-0549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17(4):223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 33.Funabashi M, Grove TL, Wang M, Varma Y, McFadden ME, Brown LC, et al. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature. 2020;582(7813):566–570. doi: 10.1038/s41586-020-2396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kühn T, Stepien M, López-Nogueroles M, Damms-Machado A, Sookthai D, Johnson T, et al. Prediagnostic plasma bile acid levels and colon cancer risk: a prospective study. J Natl Cancer Inst. 2020;112(5):516–524. doi: 10.1093/jnci/djz166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nosho K, Yamamoto H, Mikami M, Taniguchi H, Takahashi T, Adachi Y, et al. Overexpression of poly(ADP-ribose) polymerase-1 (PARP-1) in the early stage of colorectal carcinogenesis. Eur J Cancer. 2006;42(14):2374–2381. doi: 10.1016/j.ejca.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 36.Slyskova J, Korenkova V, Collins AR, Prochazka P, Vodickova L, Svec J, et al. Functional, genetic, and epigenetic aspects of base and nucleotide excision repair in colorectal carcinomas. Clin Cancer Res. 2012;18(21):5878–5887. doi: 10.1158/1078-0432.CCR-12-1380. [DOI] [PubMed] [Google Scholar]
- 37.Elmasry GF, Aly EE, Awadallah FM, El-Moghazy SM. Design and synthesis of novel PARP-1 inhibitors based on pyridopyridazinone scaffold. Bioorg Chem. 2019;87:655–666. doi: 10.1016/j.bioorg.2019.03.068. [DOI] [PubMed] [Google Scholar]
- 38.Kanth P, Inadomi JM. Screening and prevention of colorectal cancer. BMJ. 2021;374:n1855. [DOI] [PubMed]
- 39.Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16(12):713–732. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 40.Menyhárt O, Győrffy B. Multi-omics approaches in cancer research with applications in tumor subtyping, prognosis, and diagnosis. Comput Struct Biotechnol J. 2021;19:949–960. doi: 10.1016/j.csbj.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang L, Sun F, Wang H, Hu Z. Metabolomics, metabolic flux analysis and cancer pharmacology. Pharmacol Ther. 2021;224:107827. doi: 10.1016/j.pharmthera.2021.107827. [DOI] [PubMed] [Google Scholar]
- 42.Hao Y, Li D, Xu Y, Ouyang J, Wang Y, Zhang Y, et al. Investigation of lipid metabolism dysregulation and the effects on immune microenvironments in pan-cancer using multiple omics data. BMC Bioinformatics. 2019;20(Suppl 7):195. doi: 10.1186/s12859-019-2734-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng C, Geng F, Cheng X, Guo D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun (Lond) 2018;38(1):27. doi: 10.1186/s40880-018-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snaebjornsson MT, Janaki-Raman S, Schulze A. Greasing the wheels of the cancer machine: the role of lipid metabolism in cancer. Cell Metab. 2020;31(1):62–76. doi: 10.1016/j.cmet.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 45.Costarelli V. Bile acids as possible human carcinogens: new tricks from an old dog. Int J Food Sci Nutr. 2009;60(Suppl 6):116–125. doi: 10.1080/09637480902970967. [DOI] [PubMed] [Google Scholar]
- 46.Ocvirk S, O'Keefe SJ. Influence of bile acids on colorectal cancer risk: potential mechanisms mediated by diet - gut microbiota interactions. Curr Nutr Rep. 2017;6(4):315–322. doi: 10.1007/s13668-017-0219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity. 2019;50(2):432–445.e7. doi: 10.1016/j.immuni.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan X, Jin Y, Chen G, Ma X, Zhang L. Gut microbiota dysbiosis drives the development of colorectal cancer. Digestion. 2021;102(4):508–515. doi: 10.1159/000508328. [DOI] [PubMed] [Google Scholar]
- 49.Yu J, Li S, Guo J, Xu Z, Zheng J, Sun X. Farnesoid X receptor antagonizes Wnt/β-catenin signaling in colorectal tumorigenesis. Cell Death Dis. 2020;11(8):640. doi: 10.1038/s41419-020-02819-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gadaleta RM, Garcia-Irigoyen O, Moschetta A. Bile acids and colon cancer: is FXR the solution of the conundrum? Mol Aspects Med. 2017;56:66–74. doi: 10.1016/j.mam.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 51.Mamianetti A, Garrido D, Carducci CN, Vescina MC. Fecal bile acid excretion profile in gallstone patients. Medicina (B Aires) 1999;59(3):269–273. [PubMed] [Google Scholar]
- 52.Cross AJ, Moore SC, Boca S, Huang WY, Xiong X, Stolzenberg-Solomon R, et al. A prospective study of serum metabolites and colorectal cancer risk. Cancer. 2014;120(19):3049–3057. doi: 10.1002/cncr.28799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kam TI, Mao X, Park H, Chou SC, Karuppagounder SS, Umanah GE, et al. Poly(ADP-ribose) drives pathologic α-synuclein neurodegeneration in Parkinson’s disease. Science. 2018;362(6414):eaat8407. [DOI] [PMC free article] [PubMed]
- 54.Martín-Oliva D, O'Valle F, Muñoz-Gámez JA, Valenzuela MT, Nuñez MI, Aguilar M, et al. Crosstalk between PARP-1 and NF-kappaB modulates the promotion of skin neoplasia. Oncogene. 2004;23(31):5275–5283. doi: 10.1038/sj.onc.1207696. [DOI] [PubMed] [Google Scholar]
- 55.Chan EM, Shibue T, McFarland JM, Gaeta B, Ghandi M, Dumont N, et al. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature. 2019;568(7753):551–556. doi: 10.1038/s41586-019-1102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ali T, Kim MO. Melatonin ameliorates amyloid beta-induced memory deficits, tau hyperphosphorylation and neurodegeneration via PI3/Akt/GSk3β pathway in the mouse hippocampus. J Pineal Res. 2015;59(1):47–59. doi: 10.1111/jpi.12238. [DOI] [PubMed] [Google Scholar]
- 57.Pazzaglia S, Pioli C. Multifaceted role of PARP-1 in DNA repair and inflammation: pathological and therapeutic implications in cancer and non-cancer diseases. Cells. 2019;9(1):41. doi: 10.3390/cells9010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rojo F, García-Parra J, Zazo S, Tusquets I, Ferrer-Lozano J, Menendez S, et al. Nuclear PARP-1 protein overexpression is associated with poor overall survival in early breast cancer. Ann Oncol. 2012;23(5):1156–1164. doi: 10.1093/annonc/mdr361. [DOI] [PubMed] [Google Scholar]
- 59.Schiewer MJ, Goodwin JF, Han S, Brenner JC, Augello MA, Dean JL, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2(12):1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dörsam B, Seiwert N, Foersch S, Stroh S, Nagel G, Begaliew D, et al. PARP-1 protects against colorectal tumor induction, but promotes inflammation-driven colorectal tumor progression. Proc Natl Acad Sci U S A. 2018;115(17):E4061–E4070. doi: 10.1073/pnas.1712345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan SR, Chen ZY, Zhao K, Liu YC, Wang PY. Clinical research progress on disappearing colorectal liver metastases. Zhonghua Wei Chang Wai Ke Za Zhi. 2021;24(11):1028–1034. doi: 10.3760/cma.j.cn441530-20201210-00657. [DOI] [PubMed] [Google Scholar]
- 62.Ohmachi T, Inoue H, Mimori K, Tanaka F, Sasaki A, Kanda T, et al. Fatty acid binding protein 6 is overexpressed in colorectal cancer. Clin Cancer Res. 2006;12(17):5090–5095. doi: 10.1158/1078-0432.CCR-05-2045. [DOI] [PubMed] [Google Scholar]
- 63.Li H, Dai W, Xia X, Wang R, Zhao J, Han L, et al. Modeling tumor development and metastasis using paired organoids derived from patients with colorectal cancer liver metastases. J Hematol Oncol. 2020;13(1):119. doi: 10.1186/s13045-020-00957-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodríguez MI, Majuelos-Melguizo J, Martí Martín-Consuegra JM, Ruiz de Almodóvar M, López-Rivas A, Javier Oliver F. Deciphering the insights of poly(ADP-ribosylation) in tumor progression. Med Res Rev. 2015;35(4):678–697. [DOI] [PubMed]
- 65.Rodríguez MI, González-Flores A, Dantzer F, Collard J, de Herreros AG, Oliver FJ. Poly(ADP-ribose)-dependent regulation of Snail1 protein stability. Oncogene. 2011;30(42):4365–4372. doi: 10.1038/onc.2011.153. [DOI] [PubMed] [Google Scholar]
- 66.Rodríguez MI, Peralta-Leal A, O'Valle F, Rodriguez-Vargas JM, Gonzalez-Flores A, Majuelos-Melguizo J, et al. 2013 PARP-1 regulates metastatic melanoma through modulation of vimentin-induced malignant transformation PLoS Genet 9 (6) e1003531 [DOI] [PMC free article] [PubMed]
- 67.Abdelrahman AE, Ibrahim DA, El-Azony A, Alnagar AA, Ibrahim A. ERCC1, PARP-1, and AQP1 as predictive biomarkers in colon cancer patients receiving adjuvant chemotherapy. Cancer Biomark. 2020;27(2):251–264. doi: 10.3233/CBM-190994. [DOI] [PubMed] [Google Scholar]
- 68.Umar A, Dunn BK, Greenwald P. Future directions in cancer prevention. Nat Rev Cancer. 2012;12(12):835–848. doi: 10.1038/nrc3397. [DOI] [PubMed] [Google Scholar]
- 69.Amin AR, Kucuk O, Khuri FR, Shin DM. Perspectives for cancer prevention with natural compounds. J Clin Oncol. 2009;27(16):2712–2725. doi: 10.1200/JCO.2008.20.6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang XM, Yang ZJ, Xie Q, Zhang ZK, Zhang H, Ma JY 2019 Natural products for treating colorectal cancer: a mechanistic review Biomed Pharmacother 117 109142 [DOI] [PubMed]
- 71.Pang G, Wang F, Zhang LW. Dose matters: direct killing or immunoregulatory effects of natural polysaccharides in cancer treatment. Carbohydr Polym. 2018;195:243–256. doi: 10.1016/j.carbpol.2018.04.100. [DOI] [PubMed] [Google Scholar]
- 72.Huang M, Lu JJ, Huang MQ, Bao JL, Chen XP, Wang YT. Terpenoids: natural products for cancer therapy. Expert Opin Investig Drugs. 2012;21(12):1801–1818. doi: 10.1517/13543784.2012.727395. [DOI] [PubMed] [Google Scholar]
- 73.Khan H, Ullah H, Castilho PCMF, Gomila AS, D'Onofrio G, Filosa R, et al. Targeting NF-κB signaling pathway in cancer by dietary polyphenols. Crit Rev Food Sci Nutr. 2020;60(16):2790–2800. doi: 10.1080/10408398.2019.1661827. [DOI] [PubMed] [Google Scholar]
- 74.Efferth T, Oesch F. Repurposing of plant alkaloids for cancer therapy: pharmacology and toxicology. Semin Cancer Biol. 2021;68:143–163. doi: 10.1016/j.semcancer.2019.12.010. [DOI] [PubMed] [Google Scholar]
- 75.Liu X, Zhao P, Wang X, Wang L, Zhu Y, Song Y, et al. Celastrol mediates autophagy and apoptosis via the ROS/JNK and Akt/mTOR signaling pathways in glioma cells. J Exp Clin Cancer Res. 2019;38(1):184. doi: 10.1186/s13046-019-1173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang YL, Zhou YX, Zhou D, Zuo YX, Jia YP. Celastrol in the inhibition of neovascularization. Chinese Journal of Oncology. 2003;25(5):429–432. [PubMed] [Google Scholar]
- 77.Xu ZY, Shi JF, Xian J, Zhang C, Zhang JM. Research progress on anti-tumor mechanism of celastrol alone and in combination. Chinese Traditional and Herbal Drugs. 2021;52(14):4372–4375. [Google Scholar]
- 78.Zhang H, Li J, Li G, Wang S. Effects of celastrol on enhancing apoptosis of ovarian cancer cells via the downregulation of microRNA-21 and the suppression of the PI3K/Akt-NF-κB signaling pathway in an in vitro model of ovarian carcinoma. Mol Med Rep. 2016;14(6):5363–5368. doi: 10.3892/mmr.2016.5894. [DOI] [PubMed] [Google Scholar]
- 79.Pang X, Yi Z, Zhang J, Lu B, Sung B, Qu W, et al. Celastrol suppresses angiogenesis-mediated tumor growth through inhibition of AKT/mammalian target of rapamycin pathway. Cancer Res. 2010;70(5):1951–1959. doi: 10.1158/0008-5472.CAN-09-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin L, Sun Y, Wang D, Zheng S, Zhang J, Zheng C. Celastrol ameliorates ulcerative colitis-related colorectal cancer in mice via suppressing inflammatory responses and epithelial-mesenchymal transition. Front Pharmacol. 2016;6:320. doi: 10.3389/fphar.2015.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, Felding BH, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21(6):891–897. doi: 10.1016/j.cmet.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen F, Dai X, Zhou CC, Li KX, Zhang YJ, Lou XY, et al. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut. 2021; gutjnl-2020–323476. [DOI] [PMC free article] [PubMed]
- 83.Zhang L, Wei TT, Li Y, Li J, Fan Y, Huang FQ, et al. Functional metabolomics characterizes a key role for n-acetylneuraminic acid in coronary artery diseases. Circulation. 2018;137(13):1374–1390. doi: 10.1161/CIRCULATIONAHA.117.031139. [DOI] [PubMed] [Google Scholar]
- 84.Liu Chang-xiao. Scientific understanding of toxicity and safety of Chinese medicines. Chinese Herbal Medicines. 2018;10(2):107. doi: 10.1016/j.chmed.2018.04.001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file 1 Figure 1: The base peak intensity (BPI) chromatogram of serum in the QC sample. (ai 481 KB)
Supplementary file 2 Figure 2: Effect of GDCA on the activity of FHC cell line. Cell viability was measured with MTT assay in FHC cells. (ai 175 KB)
Supplementary file 3 Figure 3: Anticancer effect of celastrol on CRC. A: The docking results of PARP-1 protein (PDB:5DS3) with celastro (2D). B: Weight changes of xenograft mice. (ai 200 KB)
Data Availability Statement
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
All software applications used are included in this article.