Abstract
Cellular function is critically regulated through degradation of substrates by the proteasome. To enable direct analysis of naturally cleaved proteasomal peptides under physiological conditions, we developed mass spectrometry analysis of proteolytic peptides (MAPP), a method for proteasomal footprinting that allows capture, isolation and analysis of proteasome-cleaved peptides. Application of MAPP to cancer cell lines as well as primary immune cells reveals dynamic modulation of the cellular degradome in response to various stimuli, such as pro-inflammatory signals. Further, we demonstrate analysis of minute amounts of clinical samples by studying cells from peripheral blood of patients with systemic lupus erythematosus (SLE). We find increased degradation of histones in patient immune cells, which suggests a role for aberrant proteasomal degradation in the pathophysiology of SLE. Taken together, MAPP offers a broadly applicable method to facilitate the study of the cellular degradation landscape in various cellular conditions and diseases involving changes in proteasomal degradation, including protein aggregation diseases, autoimmunity and cancer.
The mammalian proteasome is estimated to cleave ~70% of all intracellular proteins1 and is increasingly recognized as a dynamic complex that modulates cellular function in health and disease2. Proteins that have been ubiquitinated as a signal for degradation3, as well as intrinsically disordered proteins4, are targeted for destruction by cellular proteasomes. The proteasome may also be involved in removing damaged proteins, generating peptides for antigen presentation and activating proteins5–7. However, the regulatory principles targeting specific substrates to proteasomal degradation and their cleavage products are still poorly understood. Mass spectrometry (MS) allows quantitative measurement of protein abundance8–11, including ubiquitinated species12, and many technologies have been developed for determining the half-life of proteins by examining changes in protein abundance11,13–15. However, current proteomic methods are still hindered by the large dynamic range of cellular protein abundance as well as the diversity of modified protein species16,17. Moreover, there is no available method for direct capture and analysis of proteasome-cleaved peptides from cells.
Here, we established MAPP, which identifies proteasome-cleaved peptides that are captured inside or near cellular proteasomes. It relies on reversible crosslinking together with immunoprecipitation of cellular proteasomes prior to reverse-phase isolation and elution of captured peptides. The method inhibits proteases throughout sample processing to ensure retention of native peptide characteristics. We use MAPP to show that inflammatory cytokines can sharply alter the proteolytic landscape in vitro, and we identify both known and new proteasome targets following stimulation. We demonstrate analysis of human clinical samples using as little as 75 micrograms of cellular extract from patients with SLE. An altered histone degradation pattern in these samples suggests that aberrant proteasomal activity or target selectivity may be involved in disease pathophysiology.
Results
Establishing MAPP
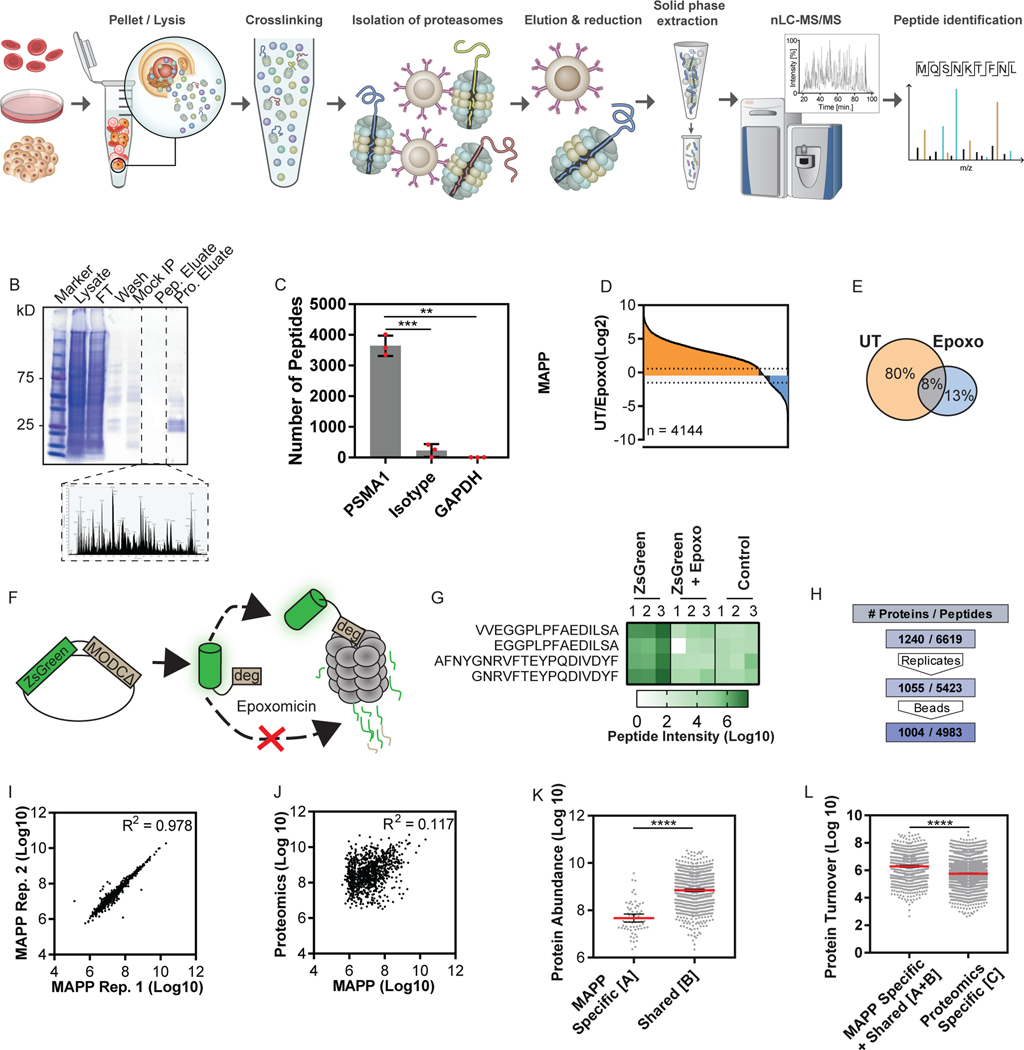
The method is illustrated in Figure 1A. First, we immunoprecipitated proteasomes from HEK293 cells using an antibody against PSMA1, a 20S proteasome subunit (Figure 1A and Supplementary Figure 1A). We further verified that the precipitated proteasomes contained both the 20S and 19S subunits by MS and Western blot (Supplementary Figure 1B, Supplementary Data 1). Endopeptidase inhibitors were used to inhibit further peptide processing (Supplementary Figure 1C). We used reversible crosslinking to immunoprecipitate associated peptides along with proteasomes, and peptides were then eluted, separated from proteins, and analyzed by MS (Figure 1B). Immunoprecipitation with an antibody against a different subunit of the 20S proteasome yielded peptides which strongly correlated with those from the PSMA1 pull-down (Supplementary Figure 1D; R2 = 0.738). In contrast, immunoprecipitation with an isotype-matched antibody or an antibody with a different cytosolic complex specificity resulted in a significant reduction in detected peptides (Figure 1C, Supplementary Figure 1E), suggesting that the isolated peptides were cleaved by proteasomes.
Figure 1. Establishing Proteasomal Profiling by Mass Spectrometry Analysis of Proteolytic Peptides (MAPP).
(A) MAPP involves immunoprecipitation of cellular proteasomes after cross-linking. Captured peptides are separated from the proteins and further subjected to mass spectrometry analysis. (B) Immunoprecipitated proteasomes were eluted (Pro. Eluate), separated from co-purified peptides (Pep. Eluate) and separated on SDS-PAGE or analyzed by mass spectrometry. Mock IP: Empty beads. FT: flow through. Wash: IP wash. (C) MAPP using antibodies against PSMA1, an isotype antibody control or GAPDH (Two tailed Welch’s corrected t-test, **P=0.0028, ***P=0.0003; Bars: mean ± standard deviation). (D + E) The fold changes in intensity of peptides identified by MAPP between untreated (UT) and Epoxomicin treated (1uM, 4 hours) cells were ranked. Orange: 2-fold or greater increase, Blue: 2-fold or greater decrease. Of the 4144 peptides identified by MAPP, 3302 peptides (D), or 80% the total (E), were reduced by at least 2 fold in intensity. (F) Schematic representation of the ZsGreen model. (G) Peptide intensity of ZsGreen peptides significantly changed under epoxomicin treatment. The graph shows area under the curve (intensity). Two-way ANOVA (p=0.0036, two degrees of freedom). (H) Thousands of identified peptides were filtered for identification in 2/3 replicates in one biological condition and at least 2-fold intensity above mock IP (beads). (I) High correlation of MAPP protein intensities between two biological replicates (n=1394 proteins, Log 10 transformed LFQ intensities; R2=0.978). (J) Poor correlation between the abundance of proteins identified by MAPP or standard proteomics (n = 979 proteins, Log 10 transformed LFQ intensities; R2=0.117). (K+L) Proteins identified by: solely MAPP ([A]), both standard proteomics and MAPP (shared [B]) and solely standard proteomics ([C]; Supplementary Figure 5A). Protein abundance was inferred from deep proteomics of HEK293 cells 31,32. The average abundance for [A] is significantly lower than ([B]; Log 10 transformed LFQ intensities, Mann Whitney s****p <0.0001). Line: Mean error bars: 95% confidence intervals. n=65 or 779 proteins ([A] or ([B], respectively) (K). The amount of protein degraded per hour (Protein Turnover) was inferred for proteins identified by MAPP and standard proteomics from turnover rates 18, 33 (Supplementary Figure 5B). The average protein turnover for [A + B] was significantly lower than ([C]; Log 10 transformed, Two tailed Mann Whitney test ****p <0.0001). Line: Mean error bars: 95% confidence intervals. n=785 or 2509 proteins (MAPP [A+B] and proteome specific [C], respectively (L).
To verify whether the isolated peptides were indeed products of proteasomal cleavage, we compared peptides that were identified upon proteasome inhibition (1 μM of epoxomicin or 50nM of velcade for 4 hours) to those identified in untreated cells. 80% of the peptides identified by MAPP (3302 peptides), were reduced by 2-fold or more in intensity after inhibition of the proteasome with epoxomicin (Figure 1D-E, Supplementary Figure 2A). Similarly, 70% were reduced by 2-fold or more after inhibition with velcade (Supplementary Figure 2B). We also confirmed that immunoprecipitation of the proteasome subunit was equally efficient after proteasome inhibition (Supplementary Figure 2C) and that no decrease in total peptide abundance occurred when translation was inhibited (Supplementary Figure 2D). While these data suggest that most peptides are products of proteasomal cleavage, we cannot exclude the possibility that MAPP-identified peptides may not entirely derive from the proteasome.
We evaluated whether we could detect the degradation of an expected proteasome substrate using MAPP. Using the pZsProSensor construct, in which ZsGreen is fused to a well-characterized degron that targets the protein for proteasomal degradation we found that the ZsGreen protein is continuously targeted for degradation in a proteasome-dependent manner (Figure 1F,G). ZsGreen peptides were absent in cells that were treated with a proteasome inhibitor or in non-transfected cells (Figure 1G, Supplementary Figure 3A), further demonstrating that MAPP identifies peptides derived from cellular proteasomes.
We then proceeded to determine the sequences and abundance of the extracted peptides and identified 4983 peptides that were at least 2-fold more abundant than in mock immunoprecipitation from similar extracts (Figure 1H, Supplementary Figure 4A, Supplementary Data 2), and which were assigned to 1004 unique proteins. The protein intensity measurements using MAPP were reproducible under steady state conditions (Figure 1I; R2= 0.978). 78% of these proteins were also identified in a parallel bottom-up proteomic analysis of the same samples (hereafter referred to as standard proteomics; Supplementary Figure 4B). For proteins identified by both standard proteomics and MAPP, we found no correlation between inferred degradation and cellular protein abundance (Figure 1J; R2= 0.117).
We compared the half-life and abundance of proteins identified by MAPP with existing datasets of protein abundance and turnover rates (Supplementary Figure 5A-B). Proteins identified solely by MAPP were significantly lower on average in abundance than proteins detected by both methods (Figure 1K and Supplementary Figure 5C). By inferring the amount of proteins degraded per hour using an external dataset18, we found that proteins identified by MAPP have on average more copies degraded per hour compared to proteins identified exclusively by standard proteomics (Figure 1L and Supplementary Figure 5D). These data suggest that MAPP measures actively degraded proteins independent of general protein abundance.
MAPP retains information on cleavage patterns of proteasomal peptides
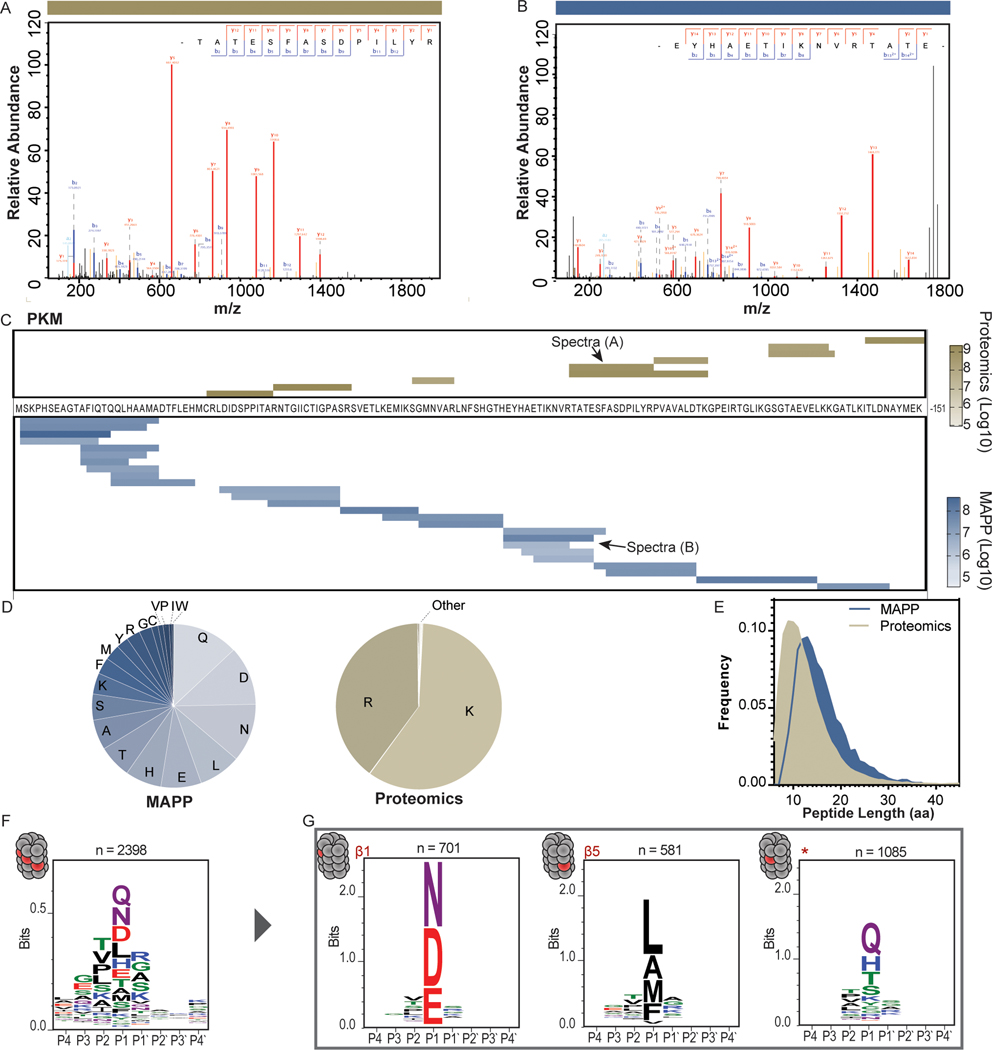
We examined cleavage properties of the peptides identified by MAPP and determined that MAPP-identified spectra and peptide species for a given protein differed from those identified by standard proteomics (Figure 2A-C). In general, MAPP-detected peptides were slightly enriched at C termini of the identified proteins (Supplementary Figure 5E). Further, MAPP included all types of C-terminal residues while tryptic peptides were highly enriched for K and R C-terminal residues as expected (Figure 2D). However, the amino acid composition of MAPP-identified peptides largely agreed with that of peptides cleaved by tryptic digest (Supplementary Figure 5F; R2= 0.763), suggesting that our protocol did not enrich for any specific peptide composition. Additionally, the average peptide length was greater for peptides identified by MAPP than by standard proteomics (Figure 2E).
Figure 2. MAPP-identified peptides reveal properties of endogenous proteasome cleavage.
(A+B) A sample MS/MS spectrum of a peptide from Pyruvate kinase (PKM) identified by standard proteomics following tryptic digest (A) or by MAPP (B). (C) The peptides identified by standard proteomics (top) or MAPP (bottom) are shown aligned to the PKM sequence (residues 1 – 151). Color scale indicates peptide abundance (Log10 transformed intensities). (D) Standard proteomics (right, n = 33,747 peptides) identifies peptides with arginine or lysine as the carboxy-terminal residue. By contrast, MAPP (left, n = 6,311 peptides) is not constrained by tryptic cleavage and captures peptides with almost every residue in the carboxy-terminal position. (E) Distribution of the lengths (number of residues) of peptides identified by MAPP (n = 6,311 peptides) and standard proteomics (n = 33,747 peptides). Peptides identified by MAPP are significantly longer than those identified by standard proteomics (Two tailed Mann-Whitney test, P<0.0001). (F) The amino acid motif surrounding the peptide cleavage site (P4 – P4’; n = 2396 unique cleavage sites). (G) Clustering of the peptide cleavage sites based on amino acid properties reveals three distinct motifs: a motif enriched for negatively charged residues (left, n = 701 unique cleavage sites), a motif enriched for hydrophobic residues (center, n = 581 unique cleavage sites), and a non-canonical motif (*) enriched for polar residues (right, n = 1085 unique cleavage sites).
We aligned peptide sequences to host proteins and identified the four amino acids adjacent to the peptide cleavage site at either end (P4-P’4 of the peptide N terminus and C terminus). We defined these 8 amino acid sequences as the cleavage motifs and extracted this information for all identified peptides (Figure 2F). We then clustered these cleavage motifs according to a distance matrix based on amino acid composition and properties and compared to the known cleavage patterns of the three catalytic subunits of the proteasome: PSMB1 (β1, caspase-like), PSMB2 (β2, trypsin-like) and PSMB5 (β5, chymotrypsin-like) 19,20. We found cleavage patterns that matched those expected for the β1 and β5 catalytic subunits (Figure 2G). Interestingly, the third cluster reflected a different motif from the tryptic-like cleavage of β2, which may be due to biological or technical reasons and is subject to further evaluation.
Identification of dynamic degradome changes in response to inflammatory stimuli
To study the proteasomal response to inflammation, we stimulated HEK293 cells with TNF-α and IFN-γ for 24 hours and then analyzed the cells with MAPP and standard proteomics in parallel. Using PANTHER we found that both approaches identified proteins with a wide range of protein classes (Supplementary Figure 6A) suggesting that targets that were degraded in response to the pro-inflammatory signals were not limited to a specific protein category. In 17 out of these 23 protein categories, over 70% of the proteins identified were the same using both methods (Supplementary Figure 6B). Proteins below this threshold included transcription factors, which exhibit rapid turnover, and receptors and transporters, which are membrane-bound and hard to extract, plausibly explaining their limited detection with standard proteomics techniques 21. Notably, MAPP identified a higher percentage of proteins that had significantly changed in intensity (Supplementary Figure 6C). While 24% of the proteins identified by MAPP differed in intensity between untreated (UT) and TNF-α and IFN-γ (T+I) treated conditions, only 6% changed in standard proteomics (Supplementary Figure 6D). Moreover, 131 proteins that did not change in abundance by more than 2-fold using standard proteomics did exhibit intensity changes by MAPP (Supplementary Data 3). These proteins included subunits of different cellular complexes as well as members of the ubiquitin pathway (Supplementary Figure 6E-G). These results suggest that proteasomal degradation is heterogeneous, with several subpopulations existing for different proteins. While this snapshot of the active degradome may not predict the half-life of bulk protein populations, it may identify a protein subpopulation that is highly transient or difficult to detect by current MS analyses.
We also evaluated more rapid proteasomal responses to TNF-α and IFN-γ stimulation for 1, 2 or 6 hours (Supplementary Figure 7A). Of the 504 proteins identified by MAPP in these conditions, 219 had a 2-fold or greater change of intensity at one or more of the time points (Supplementary Figure 7B). Of these, 127 were clustered into four distinct groups based on their intensity pattern upon treatment (Supplementary Figure 7C, Supplementary Data 4). 8 of these proteins were previously reported to be involved in the cellular response to inflammation, including HIF1A, ZFAND5 (ZNF216) and CNBP (Supplementary Figure 7D-E). While standard proteomics identified comparable abundance trends for both ZFAND5 and CNBP, neither passed the 2-fold intensity change cut-off using that approach. ER resident proteins, chaperone proteins and transcription factors were among the protein groups that changed in intensity following stimulation (Supplementary Figure 7F), and these groups may include targets relevant to the inflammatory response.
Primary T cells undergo significant proteolytic changes following PD-1 blockade
To determine whether MAPP had broader applicability, we profiled primary human T cells in response to in vitro α-CD3/CD28 stimulation followed by checkpoint blockade using the α-PD-1 monoclonal antibody Nivolumab. T cell exhaustion was determined by decreased IL-7R expression and increase in PD-1 expression (Supplementary Figure 8A). T cells were left untreated or treated for 30 minutes or 6 hours with Nivolumab before MAPP analysis. PD-1 blockade resulted in substantial degradation changes for 168 of the approximately 1300 proteins identified by MAPP (Supplementary Figure 8B-D and Supplementary Data 5). We identified proteins involved in T cell activation and proliferation, actin cytoskeleton organization components and members of the ubiquitin pathway (Supplementary Figure 8E, Supplementary Data 6). We also observed the degradation of key regulators of T-cell signaling, including NF-kB signaling pathway components such as NFKB1, RELA, and PAK2 (Supplementary Figure 8F). Although the clinical relevance of these putative targets requires further investigation, our data suggest that changes in the proteolytic landscape in response to specific treatment may be detected in primary immune cells early after treatment.
PBMCs from SLE patients have a distinct proteolytic signature
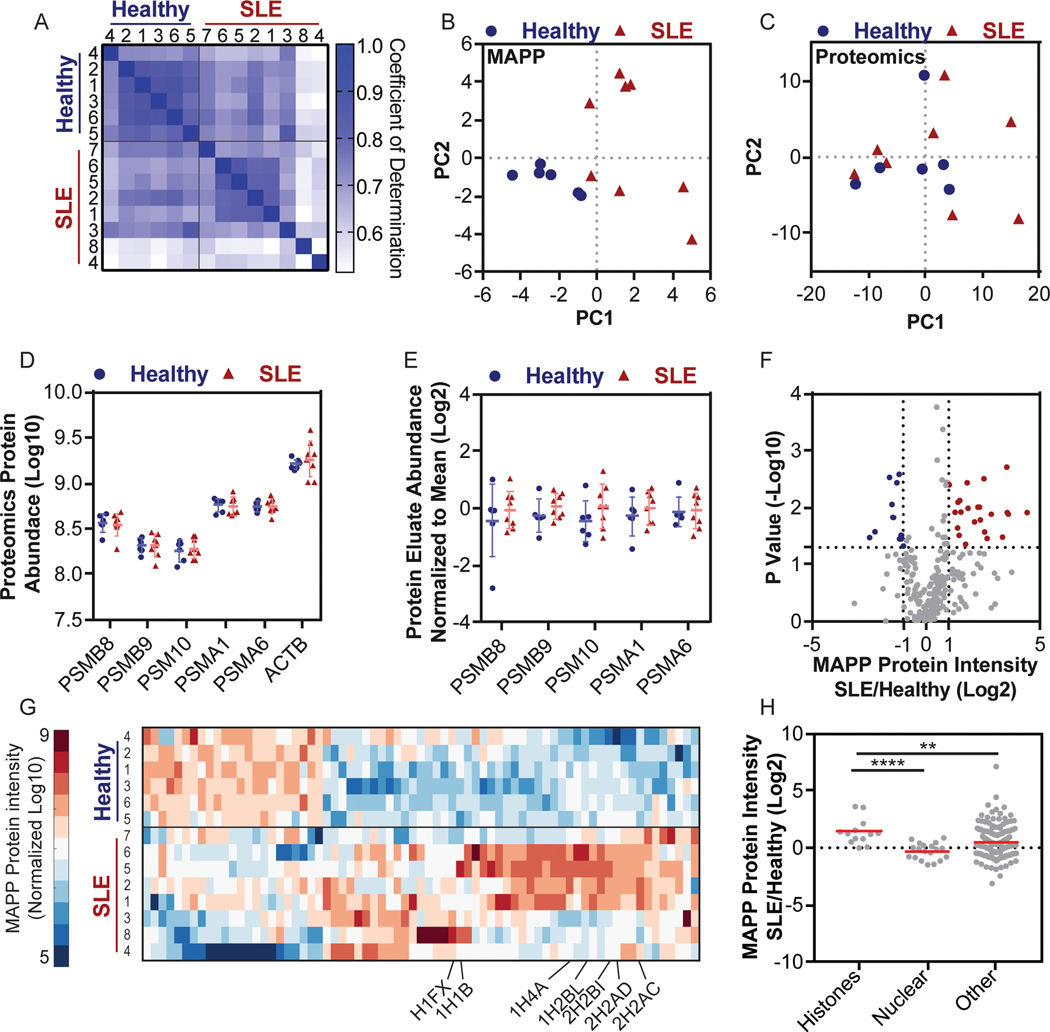
While our results demonstrated that defined in vitro stimuli could elicit degradation changes that were detectable by MAPP, it was unclear whether MAPP could deconvolute degradomes from more complex samples. Given the crucial role of proteasomal degradation antigen presentation and autoimmunity 22 we sought to examine changes in the cellular degradome in SLE. To this end, we purified peripheral blood mononuclear cells (PBMCs) from 6 healthy individuals and 8 SLE patients (Supplementary Data 7). In spite of the low protein extract yields (approximately 75 μg per sample), we identified 508 peptides, from 100 inferred proteins, per sample (Supplementary Data 8). 85% of proteins derived from healthy and SLE-patient samples were shared, with the latter exhibiting more variable degradation patterns (Figure 3A). Notably, principle component analysis using MAPP-identified proteins could effectively segregate healthy and SLE PBMCs while standard proteomics-identified proteins could not (Figure 3B-C). This difference could not be attributed to the number of proteins identified, changes in proteasome levels or variation in proteasome isolation efficiencies between the two approaches (Figure 3D-E, Supplementary Figure 9).
Figure 3. MAPP profiles distinguish between PBMCs from SLE patients and Healthy individuals.
PBMCs were purified from SLE patients and healthy individuals and analyzed by both MAPP and standard proteomics. (A) Matrix of the pairwise coefficient of determination in each sample of healthy individuals (n = 6) or SLE patients (n = 8). (B + C) Principal component analysis of protein intensities from PBMCs of healthy individuals or SLE patients, as identified by MAPP (B) or standard proteomics (C). (D) The abundance of proteasome subunits or actin (ACTB) in PBMCs was determined by standard proteomics (Log 10 transformed LFQ intensities; error bars: mean ±standard deviation). (E) The abundance of the indicated proteasome subunits in the protein fraction of the proteasome immunoprecipitate. Each subunit was normalized to the mean abundance (Log 2 normalized ratio of LFQ intensities; Bars: mean ± standard deviation). (F) Volcano plot of MAPP-identified proteins in PBMCs from healthy individuals or SLE patients (Log2 normalized ratio of LFQ intensities; -Log10 transformed P values). Two tailed t test, no correction for multiple comparisons. (G) Supervised clustering of proteins whose abundance changed by 2-fold or more between MAPP of healthy individuals and SLE patients (Log 10 transformed LFQ intensities standardized around 0, city block distance function). Identified histones are labeled. (H) Higher levels of histones are identified by MAPP of PBMCs from SLE patients, compared to healthy individuals (Histones lane, SLE/healthy). The ratio of histones in MAPP of SLE patients compared to healthy individuals is significantly higher than the ratio of other MAPP-identified proteins (Bootstrapped Mann-Whitney, P =0.0042; Log 2 transformed ratio of LFQ intensities), including other nuclear proteins (Two tailed Mann-Whitney, P < 0.0001, n = 13 (histones) n = 19 (nuclear) n = 202 (other) proteins. Bars: mean ± standard deviation.
Based on these results, we hypothesized that the differences in proteolytic profiles between healthy individuals and SLE patients may derive from altered substrate selectivity. Although differentially expressed proteins identified by MAPP were typically of higher intensity in SLE samples (Figure 3F), there was no correlation between intensity and protein abundance (Supplementary Figure 10A-B). This suggested that the proteolytic profile differences were not due to sampling bias. The 71 differentially degraded proteins included 7 histone variants whose degradation was increased 2-fold or more in SLE PBMCs (Figure 3G). In contrast, total histone proteins did not differ between healthy and SLE PBMCs based on standard proteomics (Supplementary Figure 10C). Examining this result in peptide resolution revealed higher intensity and more peptide species for histones in SLE PBMCs (Supplementary Figure 10D-E). Notably, the significant increase in histone degradation in SLE PBMCs as identified by MAPP was not observed in the general protein pool nor in other nuclear proteins (Figure 3H), which indicates that the increased histone degradation was not merely reflecting cellular content spillage or cell death.
Proteasomal degradation of histone proteins is known to increase following oxidative damage 23,24. We speculated that high levels of reactive oxygen species (ROS), which are typically found in SLE patient cells 25, may induce histone protein degradation. We used Gene Set Enrichment Analysis (GSEA) to examine whether the observed changes in histone degradation may be connected to altered ROS production, and identified that oxidative phosphorylation was significantly reduced in the SLE samples (Supplementary Figure 11). Indeed, impaired oxidative phosphorylation is known to induce abnormally high ROS26,27. Collectively, these data suggest that altered proteolytic profiles, including abnormal degradation of histone proteins, may be associated with SLE pathophysiology.
Discussion
MAPP combines reversible crosslinking and immunoprecipitation with MS to capture and analyze nascent peptides cleaved by cellular proteasomes. MAPP can be used to study protein cleavage patterns and proteasomal regulation induced by defined stimuli, which until now have been studied in purified reactions using model substrates. We show that MAPP identifies proteolytic changes not found by standard proteomics, including substantial histone degradation in PBMCs from SLE patients as compared to healthy patients. Moreover, we show that MAPP may be useful for clinical profiling by demonstrating its ability to separate healthy individuals from SLE patients. Our results suggest that proteasomal processing of histones, and potentially other auto-antigens, may generate auto-reactive epitopes. Presentation of such proteasome-cleaved peptides may thus provoke antigenic or immunogenic responses even prior to cell death. Future studies on the peptides identified in SLE patients may clarify the role of aberrant proteasome activity or substrate selectivity in breaking tolerance. Longitudinal analyses of the degradome during different SLE disease stages, including in response to treatment, could also provide more insights into the role of proteasomal processing in SLE.
Although standard proteomics achieves deeper coverage of the cellular proteome, MAPP enables the detection of less abundant proteins and proteins undergoing rapid turnover. While it does not appear that MAPP has a sampling bias, we cannot exclude the possibility that peptide proximity to the proteasome or their residency may impact their detection as previous studies have shown that non-catalytic modifier sites of proteasomes may be bound by peptides28–30. Areas for further development of MAPP include standardization of sample processing, labeling techniques, and kinetic analyses.
Given the broad importance of proteasomal degradation, we expect MAPP to provide unique insights into numerous diseases and physiological states. MAPP can be used in profiling multiple types of specimens, including primary cells and in a variety of contexts, from basic cell biology to clinical applications, and may identify new proteasomal regulators or therapeutic targets. Since proteasomes are key for antigen presentation, MAPP may be utilized to study the regulatory steps from peptide cleavage to HLA presentation. Thus, we envision that MAPP will expand our current understanding of proteasome function and also serve as a discovery tool to enable future research directions in cellular proteostasis control.
Methods
Cell culture and treatments
HEK293 cells were grown in DMEM supplemented with 10% fetal bovine serum, 1% Penicillin/streptomycin and L-glutamine (2mM) (Biological industries) at 37°C with 5% CO2. Cells were treated with 20 ng/mL TNF-α and IFN-γ (Peprotech) for the indicated period of time, or 100 μg/ml Cycloheximide (SIGMA) for 2 hours. Epoxomicin was used at a concentration of 1 μM for 4 hours and Velcade at a concentration of 50 nM for 4 hours. pZsProSensor 34,35 (Clontech, Mountain View, CA, USA) was transfected to HEK293 cells using lipofectamine 2000 (Thermo Scientific, USA).
Purification of proteasome complexes
Proteasomes were purified from approximately 5 × 107 cells. Cells were lysed with 25mM HEPES pH7.4, 10% glycerol, 5mm MgCl2, 1 mm ATP, 1:400 Protease Inhibitors Mixture (Calbiochem), homogenized by freeze-thaw cycles and passed through a 25 gauge needle. The lysates were cleared by 30 minutes centrifugation at 21,130 × g at 4 degrees. Lysate were treated with 1 mM PMSF (SIGMA), 2 mM 1,10-phenanthroline (SIGMA), and cross-linked with 0.5mM DSP (ThermoFisher) 30 min at room temperature, and quenched in 20 mM Tris-HCl pH 7.4, 5 mM L-Cysteine for 10min at room temperature. For immunoprecipitation, the lysates were then incubated with antibodies on protein-G sepharose beads (Santa Cruz biotechnology) against either PSMA1, PSMA4, GAPDH, mouse IgG1k isotype control, or empty beads as mock, and eluted with 50mM DTT for 30 minutes at 370C followed by addition of 0.5% trifluoroacetic acid (TFA). Aliquots of each elution fraction were analyzed by SDS-PAGE to evaluate yield and purity.
Purification and concentration of proteasome peptides
A critical step in our procedure is the separation of peptides from the proteins that are eluted in the proteasomal pull-down. MAPP analyzes endogenously cleaved peptides, while the proteasome complex and associated proteins are physically excluded. Immunoprecipitated proteasomes and their encompassed peptides were loaded on tC18 cartridges (Waters, Milford, MA) that were prewashed with 80% acetonitrile (ACN) in 0.1% TFA followed by wash with 0.1% TFA only. After loading, the cartridges were washed with 0.1% TFA. Peptides were eluted with 30% ACN in 0.1% TFA. Protein fractions were eluted with 80% ACN in 0.1% TFA.
Antibodies for western blot and immunoprecipitation
Primary antibodies: PSMA1 (hybridoma kindly provided by Tanaka KG, The University of Tokyo). PSMA4 (Santa Cruz, sc-271297), PSMB7 (Santa Cruz, sc-58410); PSMA1–7 (Enzo, BML-PW8195–0025), PSMB5 (Enzo, bml-pw8895–0025); PSMD6 (Abcam), PSMD11 (GAPDH, ab99414), GAPDH (Abcam, ab8245); IgG1k Isotype Antibody (BioLegend, BLG-400139), 20S core subunits (Enzo, bml-pw8895–0025). Secondary antibodies: Goat α-mouse HRP; Goat α-Rabbit HRP (Jackson labs).
Purification of CD8+ T cells and ex-vivo culture
Primary human CD8+ T cells were separated from healthy leucocyte samples, obtained from Blood Service Center (MDA, Israel), by centrifugation over Lymphoprep (Axis-Shield) and positive selection using CD8+ magnetic microbeads (Milteny Biotic). The purity of separated cell populations was determined by three-color flow cytometry (PerCP/Cy5.5 α-human CD3 Antibody, PE α-human CD8a Antibody and FITC α-human CD4 Antibody, Biolegend) using LSRII. PD-1 and IL-7Rα levels were examined by flow cytometry 36–38 (APC α-human CD279 (PD-1) and PerCP/Cy5.5 α-human CD127 (IL-7Rα), Biolegend). Purified CD8+ T cells were re-suspended in complete RPMI 1640 (Biological Industries) in the presence of 5% Human Serum, 1% L-Glutamine, 1% Penicillin/Streptomycin, 1% non-essential amino acids and 0.5% β-Mercaptoethanol. Purified CD8+ T cells were then stimulated in sterile 6-well culture plates (Corning) pre-coated with LEAF Purified α-human CD3 and LEAF Purified α-human CD28 (Biolegend) for three days. To induce exhaustion, the A375 melanoma cell-line was treated with IFN-γ (200U/ml) for 24 hours and co-cultured with the stimulated T-cells in a 1:2 ratio, respectively, for 4 days. Conditions for T-cell exhaustion were determined by measuring surface PD-1 and IL-7Rα levels by flow cytometry. Cells were divided into two groups, one group was treated with a monoclonal IgG4 α-human PD-1 antibody (Nivolumab, a gift from Dr. Jair Bar, Sheba Medical Center, Israel) for 0.5h or 6h, and a second group that was not treated with the α-human PD-1 antibody and served as control.
Human samples, inclusion criteria, study approval, and preparation.
Blood from individuals with diagnosis of SLE, or healthy controls, was obtained at the Clinical Center, NIH and the Rheumatology Clinic, University of Washington Medical Center. This human research study has been approved by the NIDDK/ NIAMS IRB board at the National Institutes of Health. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study. No identifiable images of human research participants are included in the manuscript. SLE individuals fulfilled ACR revised diagnostic criteria39. We chose for analysis patients that had a low SLEDAI (3–4) and for which no leukopenia was recorded to reduce the possibility of indirect effects due to cell death. PBMC fractions were purified from approximately 60ml of whole blood using Sepmate (StemCell Technologies) and immediately frozen in liquid nitrogen. Frozen PBMCs were resuspended in PBS, lysed and processed as described in the Purification of proteasome complexes section above. Approximately 75μg of protein was taken for MAPP.
Mass spectrometry sample processing and analysis
Each type of experiment, MAPP, tryptic digest of whole lysate or the protein eluate fractions were analyzed using the appropriate, optimal method. In all cases, samples were subjected to nanoflow liquid chromatography (nanoAcquity, Waters, USA) coupled to high resolution tandem mass spectrometry (Q Exactive, Thermo Scientific, USA). The detailed sample preparation and LC-MSMS acquisition parameters are listed in Supplementary Note 1.
Mass spectrometry data analysis
Raw data was analyzed by the MaxQuant software (version 1.5.3.3040) with the default parameters for the analysis of the proteasomal peptides, except for the following: unspecific enzyme, label free quantitation (LFQ) minimum ratio count of 1, minimum peptide length for unspecific search of 6, maximum peptide length for unspecific search of 40, and match between runs enabled. A stringent false discovery rate (FDR) of 1% was applied for peptide identification (in accordance with the FDR reported in a previous peptidomics study41). For the analysis of tryptic digests the default parameters were set, apart from minimum peptide length of 6. Masses were searched against the human proteome database from UniprotKB (last update on 02.2016).
Bioinformatics analysis and label-free quantitation
Peptides resulting from MaxQuant were initially filtered to remove reverse sequences and known mass spectrometry contaminants. To reduce ambiguity, we allowed peptides that had at least two valid LFQ intensities out of three independent biological replicates, and included razor peptides which belong to a unique MaxQuant “protein group”. To eliminate peptides which non-specifically attached to the beads in the mock control, we only considered peptides which were detected at least two folds higher in abundance than in the mock control (Fig. S4A).
MAPP protein intensities were inferred using MaxQuant. For graphical representation, intensities were log transformed, and using Python v3.6 zero intensity were imputed to a random value chosen from a normal distribution of 0.3 the standard deviation and downshifted 1.8 standard deviations 42. Statistical analyses were performed using Excel v2016, R v3.4.2, GraphPad Prism v 7.04 and MATLAB v2016a. For bootstrapping analysis, the p value was obtained against a background distribution drawn from a subset of the full dataset 10,000 times.
Protein annotation and gene ontology was done using the PANTHER classification system Version 10.043, Gene Set Enrichment Analysis (GSEA)44, GeneAnalytics, Corum45,46 and AmiGo 2 v2.4.26. Protein Network visualization was done using Cytoscape v3.4.047. Protein abundance data was mined from MaxQB 31,32. For hierarchical clustering we used city block distance unless noted otherwise.
Protein Turnover Rate Calculations
Protein half-lives were inferred from Larance et al.18 using calculations adapted from Schwanhäusser et al.33 as follows:
R(t)= The relative abundance of protein ‘P’ in time point ‘t’ is determined by protein level in the CHX treated sample divided by its level in the DMSO-treated sample, in the same time point, such that:
The relative abundance after 6 hours treatment of CHX may be defined as: where δ denotes the decay/degradation rate of the protein
when :
We can assume that at t=0, R(0)=1 as the protein abundance in CHX and DMSO at the ‘0’ time point should be equal.
Thus:
iBAQ intensities of PDMSO were used as a proxy for steady-state abundance and therefore the amount of protein degraded per hour (protein turnover) may be estimated by multiplying δ with protein abundance.
Motif clustering
Peptide sequences were aligned to host protein sequence to identify the 4 amino acids before and after the cleavage site (P4-P’4 of the peptide N terminus and C terminus). 1063 of the 9,996 potential cleavage sites were located in the N or C terminus of the proteins and were thus not considered. The remaining cleavage sites were filtered according to the following criteria: 1. Peptide search score above 80; 2. Cleavage sites that share a C terminus of one peptide with the N terminus of another peptide; 3. The sum of peptide intensities that share a specific cleavage site exceeded 10% of the protein maximal cleavage site intensity. This retrieved 2398 cleavage sites that were clustered according to a distance function based on a substitution matrix developed by Yoo et al. 48 (Matrix - BPAS0707) on positions P1 and P2. The number of clusters was set to three according to the number of catalytic units of the proteasome.
Statistics and Reproducibility
All experiments were performed in three individual replicates, unless otherwise mentioned. For each experiment, all compared conditions were analyzed by Mass Spectrometry at the same time. Comparative analysis of PSMA1 and PSMA4 was performed once. For the PBMC study, 6 samples from healthy individuals and 8 samples from SLE patients were analyzed. The samples were processed independently and analyzed by Mass Spectrometry at the same time to maintain comparability across samples and reduce batch effects.
Data Availability.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository49 with the dataset identifiers (PXD009092, PXD009360, PXD007263, PXD009181, PXD009438, PXD009112, PXD009508).
Supplementary Material
Acknowledgements
We thank Keiji Tanaka and Shigeo Murata for the α-PSMA1 hybridoma; Jair Bar and Iris Kamer for α-PD-1 monoclonal antibody Nivolumab. We thank Members of the Merbl lab and Michal Sharon for critically reviewing the manuscript and Shalev Itzkovitz, Manor Askenazi, Shai Shen-Orr, Ido Amit and Eran Elinav for scientific advice. Meital Kupervaser and Alon Savidor for analytic support. This project has received funding from Merck KGaA; the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 677748). The I-CORE Program of the Planning and Budgeting Committee and The Israel Science Foundation (grant No. 12/7751); Rothschild Caesarea; Gruber Peter&Patricia (Gruber award); Adiel Eleanor; Carla Hunter and Andre Schub (YM). The Intramural Research Program at NIAMS/NIH ZIA AR041199 (CCR and MJK). Y.M. is the incumbent Leonard and Carol Berall Career Development Chair.
Footnotes
Competing Financial Interests.
The authors declare no competing interests.
Ed summ:
Proteasomal footprinting reveals changes in the active cellular degradome
References
- 1.Rock KL et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–71 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Schmidt M. & Finley D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 1843, 13–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hershko A. & Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem 61, 761–807 (1992). [DOI] [PubMed] [Google Scholar]
- 4.Finley D, Chen X. & Walters KJ Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem. Sci. 41, 77–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rock KL, York IA, Saric T. & Goldberg AL Protein degradation and the generation of MHC class I-presented peptides. Adv. Immunol. 80, 1–70 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Rock KL, Reits E. & Neefjes J. Present Yourself! By MHC Class I and MHC Class II Molecules. Trends Immunol. 37, 724–737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rape M. & Jentsch S. Taking a bite: proteasomal protein processing. Nat. Cell Biol. 4, E113––E116 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Aebersold R. & Mann M. Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Doherty MK, Hammond DE, Clague MJ, Gaskell SJ & Beynon RJ Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J Proteome Res 8, 104–112 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Yen H-CS, Xu Q, Chou DM, Zhao Z. & Elledge SJ Global Protein Stability Profiling in Mammalian Cells. Science (80-. ). 322, 918–923 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Savitski MM et al. Multiplexed Proteome Dynamics Profiling Reveals Mechanisms Controlling Protein Homeostasis. Cell 173, 260–274.e25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathieson T. et al. Systematic analysis of protein turnover in primary cells. Nat. Commun. 9, 689 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McShane E. et al. Kinetic Analysis of Protein Stability Reveals Age-Dependent Degradation. Cell 167, 803–815.e21 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Eden E. et al. Proteome half-life dynamics in living human cells. Science (80-. ) 331, 764–768 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Mann M. & Kelleher NL Precision proteomics: the case for high resolution and high mass accuracy. Proc. Natl. Acad. Sci. U. S. A. 105, 18132–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aebersold R. et al. How many human proteoforms are there? Nat. Chem. Biol. 14, 206–214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larance M, Ahmad Y, Kirkwood KJ, Ly T. & Lamond AI Global Subcellular Characterization of Protein Degradation Using Quantitative Proteomics. Mol. Cell. Proteomics 12, 638–650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardozo C, Vinitsky A, Hidalgo MC, Michaud C. & Orlowski MA 3,4-dichloroisocoumarin-resistant component of the multicatalytic proteinase complex. Biochemistry 31, 7373–80 (1992). [DOI] [PubMed] [Google Scholar]
- 20.Nussbaum AK et al. Cleavage motifs of the yeast 20S proteasome beta subunits deduced from digests of enolase 1. Proc. Natl. Acad. Sci. U. S. A. 95, 12504–12509 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou F. et al. Genome-scale proteome quantification by DEEP SEQ mass spectrometry. Nat. Commun. 4, 2171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feist E, Burmester G-R & Krüger E. The proteasome — victim or culprit in autoimmunity. Clin. Immunol. 172, 83–89 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Ullrich O. et al. Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proc. Natl. Acad. Sci. U. S. A. 96, 6223–8 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cervantes-Laurean D, Roberts MJ, Jacobson EL & Jacobson MK Nuclear proteasome activation and degradation of carboxymethylated histones in human keratinocytes following glyoxal treatment. Free Radic. Biol. Med. 38, 786–795 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Perl A. Oxidative stress in the pathology and treatment of systemic lupus erythematosus. Nat. Rev. Rheumatol. 9, 674–686 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang CH, Wu SB, Wu Y, W. Y. Oxidative stress response elicited by mitochondrial dysfunction: Implication in the pathophysiology of aging. Exp Biol Med (Maywood). 238, 450–460 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Cadenas E. & Davies KJ Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 29, 222–30 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Kisselev AF, Kaganovich D. & Goldberg AL Binding of Hydrophobic Peptides to Several Non-catalytic Sites Promotes Peptide Hydrolysis by All Active Sites of 20 S Proteasomes. J. Biol. Chem. 277, 22260–22270 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Schmidtke G, Emch S, Groettrup M. & Holzhütter H-G Evidence for the Existence of a Non-catalytic Modifier Site of Peptide Hydrolysis by the 20 S Proteasome. J. Biol. Chem. 275, 22056–22063 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Liepe J. et al. Quantitative time-resolved analysis reveals intricate, differential regulation of standard- and immuno-proteasomes. Elife 4, e07545 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaab C, Geiger T, Stoehr G, Cox J. & Mann M. Analysis of High Accuracy, Quantitative Proteomics Data in the MaxQB Database. Mol. Cell. Proteomics 11, M111.014068 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geiger T, Wehner A, Schaab C, Cox J. & Mann M. Comparative Proteomic Analysis of Eleven Common Cell Lines Reveals Ubiquitous but Varying Expression of Most Proteins. Mol. Cell. Proteomics 11, M111.014050 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwanhäusser B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Murakami Y. et al. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360, 597–599 (1992). [DOI] [PubMed] [Google Scholar]
- 35.Vlashi E. et al. In Vivo Imaging, Tracking, and Targeting of Cancer Stem Cells. JNCI J. Natl. Cancer Inst. 101, 350–359 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philips GK & Atkins M. Therapeutic uses of anti-{PD}−1 and anti-{PD}-L1 antibodies. Int. Immunol. 27, 39–46 (2014). [DOI] [PubMed] [Google Scholar]
- 37.McKinney EF, Lee JC, Jayne DRW, Lyons PA & Smith KGC T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 523, 612–616 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang W, Chen PW, Li H, Alizadeh H. & Niederkorn JY PD-L1: PD-1 Interaction Contributes to the Functional Suppression of T-Cell Responses to Human Uveal Melanoma Cells In Vitro. Investig. Opthalmology Vis. Sci. 49, 2518 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan EM et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 25, 1271–7 (1982). [DOI] [PubMed] [Google Scholar]
- 40.Cox J. & Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ & Mann M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol Cell Proteomics 14, 658–673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyanova S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Mi H. The {PANTHER} database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 33, D284––D288 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–50 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruepp A. et al. CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 36, D646–50 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruepp A. et al. CORUM: the comprehensive resource of mammalian protein complexes—2009. Nucleic Acids Res. 38, D497–D501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shannon P. et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository49 with the dataset identifiers (PXD009092, PXD009360, PXD007263, PXD009181, PXD009438, PXD009112, PXD009508).