The authors wish to correct the transcript numbering of CCDC188. The outdated transcript NM_001243537 was incorrectly used in the published paper. This should have been NM_001365892. The authors therefore update the description of the CCDC188 mutation to c.937C>T (p.Arg313*).
The corrected Table 1, Figure 1, Supplementary Materials Table S7 and Fig. S2 are presented as below. Allele frequency in gnomAD was also updated according to NM_001365892 in Table 1 and Supplementary Materials Table S7. The revision does not change the conclusions of this paper.
Table 1.
Five homozygous rare variants in five novel candidate genes identified in four RP probands.
| Genes | Chromosome | Reference | Patient | Variant | Nucleotide | Amino acid | Expression & interaction§ |
Allele frequency in gnomAD |
Allele frequency of other LoF variants |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| position | transcript | ID | number | change | Change | Retina | IRD genes | All | EA | control | gnomAD | |
| ENSA | chr01: 150599935 | NM_207168 | ZOCRP0009 | M1 | c.191delG | p.Gly64Alafs*2 | Ninth | OFD1 | 1/251460 | 1/18394 | 2/9456 | 22/282912 |
| DACT2 | chr06: 168710973 | NM_214462 | ZOCRP0156 | M2 | c.533C>A | p.Ser178* | Second | NA | NA | NA | 4/9456 | 144/282912 |
| EXTL2 | chr01: 101343065 | NM_001439 | ZOCRP0156 | M3 | c.400C>T | p.Arg134* | Highest | NA | 3/247450 | 3/18342 | 2/9456 | 37/282912 |
| DDR1 | chr06: 30864446 | NM_001202523 | ZOCRP0830 | M4 | c.1616dupT | p.Pro540Alafs*15 | Fifth | CCT2, BBS10 | NA | NA | 0/9456 | 178/282912 |
| CCDC188 | chr22: 20136745 | NM_001365892 | ZOCRP0559 | M5 | c.937C>T | p.Arg313* | NA | NA | 16/118792 | 0/10622 | 0/9456 | 62/282912 |
Notes: EA, East Asian; gnomAD, genome aggregation database; IRD, inherited retinal degeneration; LoF, loss-of-function; M, mutation; NA, not available.
§Information based on GeneCards where expression in retina ranked by SAGE.
All these variants were not present in Exome Variant Server and 1000 Genomes.
No homozygous LoF variants in these five genes were found in 1000 Genomes.
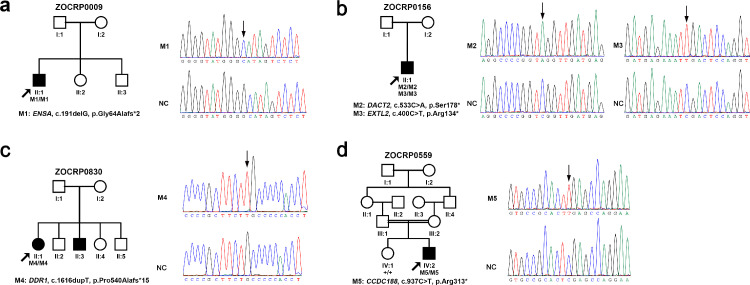
Figure 1.
Pedigrees and sequences of the five homozygous loss-of-function variants in ENSA, DACT2, EXTL2, DDR1, and CCDC188. The genotypes of all probands and available family members are shown below each individual. The blackened symbols represent affected individuals. Mx, mutant alleles; +, wild-type allele.
The authors would like to apologise for any inconvenience caused.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2022.103913.
Appendix. Supplementary materials
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.