Abstract
Rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) are chronic autoimmune diseases that result from the combined influence of genetic and environmental factors that promotes the loss of tolerance to cellular components. The complexity of these diseases converts them into a major challenge at the diagnostic and treatment level. Therefore, it is convenient to implement the use of tools for a better understanding of the physiopathology of these diseases to propose reliable biomarkers. The “omics” disciplines like metabolomics and lipidomics allow to study RA and SLE in a higher degree of detail since they evaluate the metabolites and metabolic pathways involved in disease pathogenesis. This review has compiled the information of metabolomics and lipidomics studies where samples obtained from RA and SLE patients were evaluated to find the metabolites and pathways differences between patients and healthy controls. In both diseases, there is a decrease in several amino acids and oxidative stress-related metabolites like glutathione. These findings may be useful for functional metabolomics studies aiming to reprogram the metabolism in a disease setting to recover normal immune cell homeostasis and function.
Keywords: Rheumatoid arthritis, Systemic lupus erythematosus, Autoimmune tautology, Metabolomics, Lipidomics, Metabolites, Metabolic pathways
Highlights
-
•
Metabolomics and lipidomics allow to explore the metabolic pathways associated with RA and SLE.
-
•
Shared altered metabolites in RA and SLE support the autoimmune tautology hypothesis.
-
•
Metabolic alterations could be used as potential targets in translational medicine.
1. Introduction
The immune system functions to defend the cells and tissues of our organism from diverse threats, mainly those imposed by infectious agents [1]. However, under the influence of genetic and environmental factors, it may be directed against self-components in what is known as autoimmunity. The prevalence of autoimmune diseases in the population is approximately 7–9%, and they mostly affect women in the prime of their life causing significant morbidity and mortality [2]. These diseases exhibit clinical heterogeneity, but they share several clinical signs and symptoms. This is the result of the combination of genetic and environmental factors that generate similar physiopathologic mechanisms indicating they have a common origin in what is known as autoimmune tautology [3]. They can be classified into organ-specific diseases like type I diabetes and multiple sclerosis, and systemic such as rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) [2].
RA is a chronic and autoimmune inflammatory disease that is known for its joint symptoms, although multiple additional organ systems are known to be involved, including the pulmonary, cardiovascular, ocular, and cutaneous [4]. The pathogenesis of RA is influenced by the citrullination or other post-translational modifications of proteins in the lung and other mucosal sites [5]. This generates neoantigens that bind to major histocompatibility complex molecules and are presented to T cells, which in turn stimulate B cells to synthesize a range of autoantibodies, including rheumatoid factor (RF) and ACPA (anti-citrullinated protein antibodies) among others [6].
SLE is also a chronic and autoimmune inflammatory disease with a complex clinical picture that includes a broad spectrum of symptoms and diverse organ involvement [7,8]. In the pathogenesis of SLE, it is thought that apoptotic cells and neutrophil extracellular traps (NETs) are the sources of nuclear antigens that promote the production of type I interferon (IFN) by plasmacytoid dendritic cells [7]. The type I IFN released stimulates conventional dendritic cells for the presentation of the nuclear antigens to T cells, which support B cell production of autoantibodies. The SLE autoantibodies form immune complexes whose deposition in the tissues causes injury [7,9].
Despite the increased understanding of the basic mechanisms of autoimmune diseases like RA and SLE, they continue to be challenging because their diagnosis relies on the judgment of experienced clinicians recognizing the set of characteristic symptoms, signs, and laboratory findings, instead of a gold standard test [10]. Additionally, the conventional serologic tests used to support diagnosis and disease monitoring in RA (RF and ACPA) and SLE (anti-nuclear antibodies (ANA), anti-double-stranded DNA antibodies (anti-dsDNA), and complement levels) are of limited sensitivity and specificity [8]. It has been also recognized the unpredictable course of these diseases, with periods of relapse and remission that make their treatment and follow-up difficult [8]. Thus, reliable biomarkers for the diagnosis, classification, and treatment of both diseases are needed [8]. In the search of biomarkers, the “omics” disciplines provide a comprehensive approach for the understanding of the pathophysiological processes of complex diseases and can be conducted at any level of the gene expression sequence: genes, messenger RNA, proteins, and metabolites [8,11]. Metabolomics is the newest “omics”, and it aims to measure the complete set of metabolites in a biological sample [12].
Metabolomics allows the evaluation of the metabolic pathways that influence the pathogenesis of autoimmune diseases like RA and SLE [[13], [14], [15]]. There is knowledge about the configuration of immune cells, but the effect of systemic metabolism on immune cell function has not been sufficiently explored. Metabolic organs like the liver, pancreas, kidney, gut, and adipose tissue may redirect metabolites to alternative routes affecting systemic metabolic responses, but also altering immune cell function through the regulation of nutrient availability and delivering signaling metabolites [16]. For this reason, emerges the possibility that the metabolic pathways used by immune cells in RA and SLE can be targeted as a therapeutic strategy to shut down the inappropriate immune responses [17]. Here, we performed a comparative analysis of metabolomic data identifying the metabolic alterations associated with RA and SLE to give a general overview and an update on this topic.
2. Metabolomics
Metabolites are the substrates and products of metabolism that drive essential cellular functions, such as energy production and storage, signal transduction, and apoptosis [18]. Metabolites are present in the body from endogenous sources as the intermediates of the metabolic pathways, but also exogenous sources like the diet, the metabolism of the gut microbiota, and the drugs being taken by a patient [19,20]. For this reason, they have become a direct signature of biochemical activities and a tool for the analysis of the specific response of organisms to environmental stimuli, that might be easier to correlate with phenotypes [14,21]. Metabolites have a wide range of functions in the cell and the organism, so the implementation of metabolomic approaches identifying metabolites and metabolic pathways associated with particular phenotypes results useful in understanding their physiological roles, as this knowledge is integrated with functional and mechanistic biological studies [18].
To perform metabolomics analysis, it is important to establish whether the experimental approach is meant to be targeted or untargeted. The untargeted analysis measures in each sample as many metabolites as possible, thereby providing information to propose hypotheses regarding metabolite changes in a disease setting. On the other hand, the targeted approach evaluates a predefined set of metabolites and therefore is driven by a specific biochemical question that encompasses the intermediates of just one or more related pathways of interest [18,21].
Regardless of the approach, metabolomics analysis is performed through technologies such as mass spectrometry (MS) and nuclear magnetic resonance (NMR) [14]. MS is an analytical platform that provides high sensitivity and reproducibility. It measures the mass-to-charge ratio (m/z) of ions that are formed by inducing the ionization of neutral species, and it allows to determine their identity. The sample containing a mixture of metabolites is introduced into the mass spectrometer either directly or preceded by a separation step through liquid chromatography (LC), gas chromatography (GC), or capillary electrophoresis (CE) [18]. In addition, NMR is a spectroscopic analysis technique that provides information about the molecular structure and concentration of the metabolites in a complex mixture through the measurement of the energy emitted by the atom nuclei that builds up a molecule once a magnetic field has been applied [22,23].
Metabolomics experiments generate highly complex data sets with hundreds or thousands of metabolic features or possible compounds, so that proper data handling and processing may have a positive impact on the quality of the results [19]. The first step is data pre-processing, which aims to extract information about metabolite ions from the raw MS data to detect the chromatographic peak features corresponding to the metabolite ions along with the quantification data. Initially, the identity of these metabolite ions is unknown, but after the analysis of their LC or GC retention time, along with the isotope distribution, accurate mass and fragment ion patterns in MS/MS helps to identify them. Then, univariate and multivariate statistical algorithms are applied to find the metabolite ions that are significantly changed between control and experimental groups [24].
Metabolomics can provide important information relating to the pathogenesis of autoimmune diseases since it makes a further understanding of the exposome, which is the sum of external (i.e. toxic substances, air pollution, lifestyle, water quality, tobacco, alcohol) and internal exposures (i.e microbiome and genetics) that shapes the immune system and confers risk or protection for the development of these diseases [25]. Metabolomics can explain the molecular mechanisms behind the specific phenotype of a disease and provide specific metabolic profiles or signatures [14,25,26].
Several studies have focused on the identification of metabolites and metabolic pathways associated with RA and SLE, but they are still not well characterized. The most commonly used specimens are serum/plasma, urine, and fecal extracts because of their low invasiveness and the huge number of metabolites that they contain [22]. The characterization of metabolic alterations might be useful in the discovery of new biomarkers to improve the early diagnosis of these diseases, the classification of patients, the prediction of response to specific therapies, and treatment personalization [27]. Biomarkers proposed through metabolomics can be physiopathologically relevant molecules involved in the development of the disease, since their alterations may occur during immune cell activation [28,29]. The area of immunometabolism explains how the alterations of immune function relate to changes in intracellular metabolic pathways. The study of immunometabolism along with metabolomics helps to understand the association between metabolism and inflammation [13].
3. Immunometabolism
The energy and chemical components provided by metabolism are important for all the biological processes including immunity, so the findings in this area have allowed the emergence of the new field of immunometabolism [30]. During immune responses, the immune cells suffer functional alterations that impose metabolic stresses that are efficiently managed by their ability to reprogram their cellular metabolism [31]. This reprogramming uses different metabolic pathways that allow the production of energy to support survival and to generate the biosynthetic intermediates needed for cellular growth and proliferation [28].
Autoimmune diseases result from the activation of both innate and adaptive immune effector cells and often are combined with a systemic inflammatory syndrome [17,32]. Cells like neutrophils are involved in acute inflammation, while macrophages and dendritic cells promote the activation of T and B lymphocytes. The metabolism of these effector cells is tightly regulated and upon activation they undergo metabolic reprogramming [17]. Metabolic pathways like glycolysis, TCA cycle, electron transport chain, oxidative phosphorylation, lipid metabolism, uptake and biosynthesis of amino acids, and cell death pathways have been recognized as relevant for the development of autoimmune rheumatic diseases [33]. Each of these pathways has a unique role in the immune cells and is regulated to fulfill cellular needs. Glycolysis is dominant in the metabolism of rapidly proliferating cells because despite having a relatively inefficient ATP production it allows to redirect intermediate products to pathways for the synthesis of nucleotides, amino acids, and fatty acids. On the other hand, the TCA cycle and oxidative phosphorylation are highly efficient in ATP generation and are used by non-proliferative cells whose primary need is energy [28].
4. Metabolites and metabolic pathways associated with RA
The potential of metabolomics to distinguish RA patients from healthy controls has been evaluated. Most of the studies analyzed the metabolites through MS coupled with GC or LC in serum or plasma samples, but a couple of them used synovial fluid. Most of the patients had active RA because the disease activity score 28 (DAS28) was higher than 3.2, and also established RA because the disease duration was longer than one year. However, Sasaki et al. and Yang et al. included patients with inactive disease (DAS28 less than 3.2) [34,35], while Madsen et al. and Young et al. involved patients with early RA (less than one year of disease duration) [36,37]. The treatments received by the patients consisted typically of prednisolone and conventional DMARDs (disease-modifying anti-rheumatic drugs) such as hydroxychloroquine, methotrexate, sulfasalazine, and leflunomide. In some studies, the patients were additionally receiving biological therapies (TNF-α and IL-6 antagonists) [27,36,38] and NSAIDs (non-steroidal anti-inflammatory drugs) [35,37]. Table 1 summarizes the results about the metabolites and pathways that are different between both study groups.
Table 1.
Altered metabolites and metabolic pathways in RA. The metabolites and pathways altered in RA with respect to healthy controls were extracted from the references listed.
| Sample | Analytical platform | Metabolite differences RA vs healthy controls | Pathways differences RA vs healthy controls | References |
|---|---|---|---|---|
| Serum | GC-MS and UPLC-MS |
Decreased: Histidine, Threonic acid, methionine, cholesterol, asparagine, threonine. Increased: Glyceric acid, d-ribofuranose, Hypoxanthine. |
Aminoacid metabolism, Nucleotide synthesis | [36] |
| 1H-NMR |
Decreased: valine, isoleucine, lactate, alanine, creatinine, GPC (sn-glycero-3-phosphocholine) +APC (acetyl phosphocholine), and histidine. Increased: 3-hydroxyisobutyrate, acetate, NAC (N-acetylated glycoprotein), acetoacetate, and acetone. |
Propanoate metabolism, the synthesis and degradation of ketone bodies, valine, leucine and isoleucine degradation, glycolysis, gluconeogenesis, pyruvate metabolism, and glycerophospholipid metabolism | [27] | |
| GC-MS |
Decreased: Glucose, leucine, serine, isoleucine, valine, threonine, alanine, methionine, proline, tyrosine, phenylalanine, pyroglutamate, urea, urate, 2-ketoisocaproate, 3-methyl-2-oxovalerate, 1,5-anhydrosorbitol. Increased: Pyruvate, citrate, cholesterol, glycerol, palmitelaidate, oleate, trans-9-octadecenoate, cis-5,8,11-eicosatrienoate, docosahexaenoate, eicosanoate, hexadecanoate, arachidonate, mannose, ribose, scyllo-inositol. |
Glycolysis, TCA cycle, Urea cycle, Amino acid metabolism, Fatty acid metabolism | [39] | |
| UPLC-HRMS |
Decreased: capric acid, bilirubin. Increased: 4-methoxyphenylacetic acid, glutamic acid, argininosuccinic acid, l-leucine, l-phenylalanine, l-tryptophan, l-proline, glyceraldehyde, fumaric acid, cholesterol. |
Protein biosynthesis, Glutathione metabolism, Alpha-linolenic acid and linoleic acid metabolism, Biotin metabolism. | [40] | |
| Plasma | CE-Q-TOFMS |
Decreased: histidine, methionine, serine, 1-methylnicotinamide, gamma-butyrobetaine, Cysteine-glutathione disulfide–Divalent, N-acetylleucine, azelaic acid. Increased: 2-hydroxybutyric acid, lactic acid, mucic acid, N,N-dimethylglycine, asymmetric dimethylarginine, pelargonic acid, threonic acid, gluconic acid, 3-methylhistidine, glucuronic acid, glyceric acid, phenylalanine, tyrosine, glutamic acid, glycerol-3-phosphate, pyruvic acid |
Glycolysis, amino acid metabolism | [34] |
| LC-MS |
Decreased: dihydroceramides (dhCer 22:0, dhCer 24:0) alkylphosphatidylethanolamine (PE(O-36:4)), Alkenylphosphatidylethanolamines (PE(P-36:4)), PE(P-38:5), PE (P- 40:5), PE(P-40:4), phosphatidylserines (PS 36:1, PS 38:4, and PS 40:6). Increased: lysophosphatidylinositol (LPI 18:2) |
Lipid metabolism | [38] | |
| Synovial fluid | GC-TOF MS |
Decreased: LDL-lipids, alanine, methylguanidine. Increased: 3-hydroxybutyrate, lactate, acetylglycine, taurine, glucose. |
Lipolysis, lactic acid fermentation | [37] |
| GC-TOF MS |
Decreased: valine, citric acid, gluconic lactone, glucose, glucose-1P, mannose, 5-Methoxytryptamine, ribitol. Increased: Lactic acid, beta-mannosylglycerate, carnitine, diglycerol, pipecolinic acid. |
Tryptophan metabolism, lysine degradation, TCA cycle, pentose phosphate pathway, glycolysis, lysine degradation, valine, leucine and isoleucine degradation, fructose, and mannose metabolism. | [35] |
Abbreviations: 1H-NMR (Proton nuclear magnetic resonance), GC-MS (Gas chromatography-mass spectrometry), LC-MS (Liquid chromatography-mass spectrometry, UPLC-MS (Ultra performance liquid chromatography-mass spectrometry), UPLC-HRMS (Ultra performance Liquid Chromatography-High Resolution Mass Spectrometry), CE-Q-TOFMS (capillary electrophoresis-quadrupole-time-of-flight mass spectrometry), GC-TOF MS (gas chromatography-time-of-flight mass spectrometry).
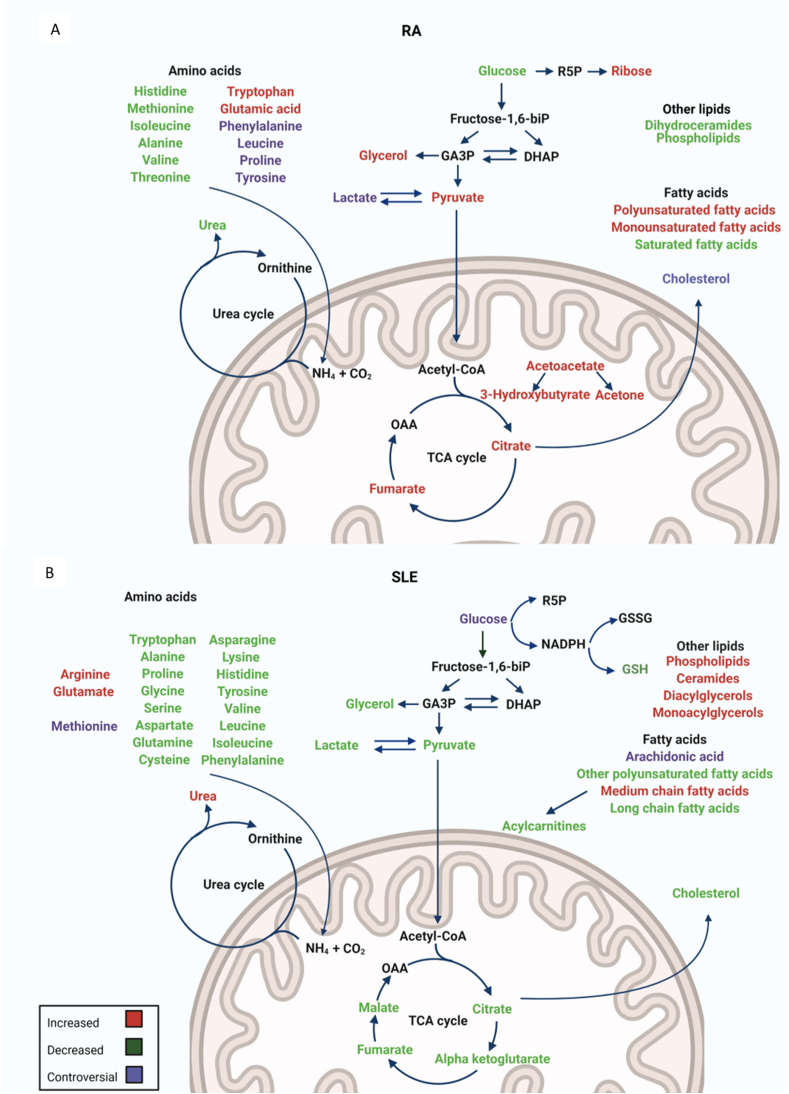
The first results that will be discussed are those obtained in serum or plasma. However, it is worth noting that serum metabolites can be affected by multiple cells and tissues [17]. The results showed a decrease in glucose and elevation of pyruvate in RA patients [34,39]. There was also an increase of ribose suggesting the up-regulation of the pentose phosphate pathway (PPP) [39]. In addition, lactate levels were decreased according to Zabek et al. but increased for Sasaki et al. [27,34]. Intermediates of the TCA cycle such as citrate and fumaric acid were increased in RA patients [39,40]. Many amino acids such as histidine, methionine, isoleucine, alanine, valine, serine, and threonine were consistently diminished across the studies [27,34,36,39,40], while tryptophan and glutamic acid were elevated [34,40]. Leucine and proline were decreased in the study from Zhou et al. but showed an increase in the results from Li et al. [39,40]. Likewise, tyrosine was elevated for Sasaki et al. and diminished for Zhou et al. [34,39]. Another controversy was found regarding phenylalanine levels since Li et al. and Sasaki et al. showed an elevation in RA patients but Zhou et al. demonstrated its diminution [34,39,40]. The study by Zhou et al. evidenced low levels of urea [39].
Regarding the lipid profile, the cholesterol levels were increased in RA patients in the studies from Zhou et al. and Li et al., however, they were decreased in the results from Madsen et al. [36,39,40]. Levels of polyunsaturated and monounsaturated fatty acids were higher in the study by Zhou et al., while a saturated fatty acid such as capric acid showed lower levels in the study by Li et al. [39,40]. The results from Zabek et al. and Young et al. displayed an up-regulation of metabolic intermediates from ketone bodies synthesis like 3-hydroxybutyrate, acetate, acetoacetate, and acetone in RA patients [27,37]. The study by Fang et al. evaluated 24 lipid classes and subclasses in patients with RA and healthy controls so that five were different among them: dihydroceramides, alkylphosphatidylethanolamine, alkenylphosphatidylethanolamine, lysophosphatidylinositol, and phosphatidylserine. Within those five classes, 36 lipid species were measured and 11 were different between RA patients and controls [38].
There were alterations in the metabolites associated with oxidative stress and inflammation. Threonic acid, which has a role in oxidative stress was increased in RA patients according to Sasaki et al. but decreased in these patients for Madsen et al. The levels of N-acetylated glycoproteins were increased in RA patients, and these are markers of inflammation [27,36].
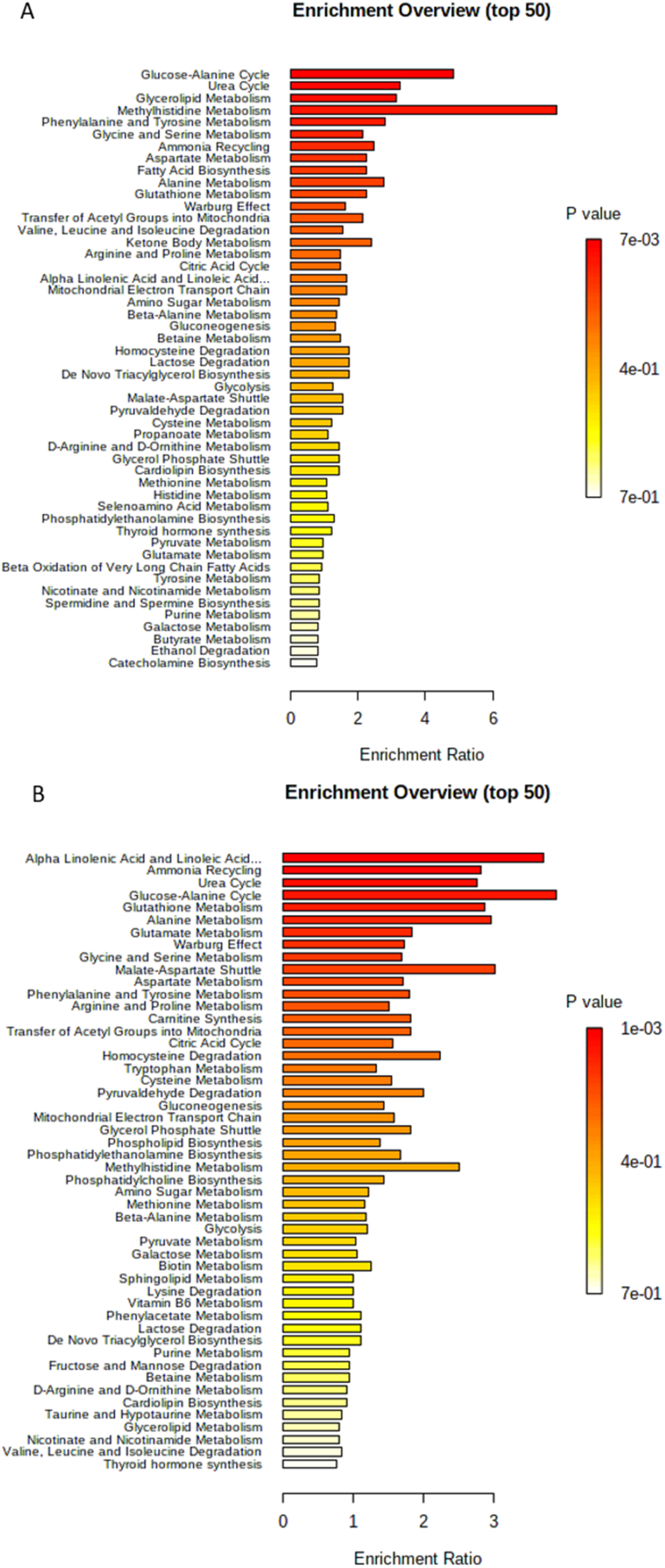
In Fig. 1A there is a summary of the metabolites that were found altered in RA patients compared to healthy controls. The over-representation analysis in Fig. 2A presents the enrichment ratio of metabolic pathways in RA. The top five enriched pathways are the glucose-alanine cycle, urea cycle, glycerolipid metabolism, methylhistidine metabolism, and phenylalanine and tyrosine metabolism.
Fig. 1.
Altered metabolites in RA and SLE. The information of the metabolites increased or decreased with respect to healthy controls was extracted from the articles listed in Table 1 for RA (A) and Table 2 for SLE (B). Created with BioRender.com.
Fig. 2.
Over representation analysis (ORA) of pathway enrichment in RA (A) and SLE (B). The ORA was performed with the software MetaboAnalyst 4.0. The software uses the list of metabolites that were decreased or increased in RA and evaluates whether a metabolite set from a pathway is represented more than expected by chance within the compound list with a hypergeometric test that provides p values adjusted for multiple testing.
The disease-specific relevant cells in RA are T lymphocytes and macrophages, and it has been shown that these cells present immunometabolic abnormalities [41]. These cells in RA display phenotypic traits such as aberrant proliferation and commitment to proinflammatory effector functions, which may impose substantial metabolic demands. RA T cells have low ATP, high NADPH levels and shunt glucose into the PPP, while macrophages present high glucose uptake and high levels of ATP and mitochondrial reactive oxygen species (ROS) [42]. In plasma, it was observed an increase in ribose levels that might be evidence of the PPP increase in RA cells.
Two metabolomic studies employed synovial fluid samples. The glucose levels were decreased in the results from Yang et al., while the study from Young et al. showed the opposite. However, the increase in lactate levels in both studies might indicate that lactic acid fermentation is up-regulated in RA patients, and therefore glycolysis is also occurring [35,37]. Citric acid was diminished and that relates to a down-regulation of the TCA cycle and also of ATP production since this pathway is central to the aerobic oxidation of fats, carbohydrates, and amino acids [35]. The reports about amino acid changes show that valine and alanine are decreased in RA patients [35,37]. The ketone body 3-hydroxybutyrate was elevated in RA patients [37]. Additionally, there was a decrease in LDL-lipids and acylcarnitines [35,37]. These last results about amino acids diminution and 3-hydroxybutyrate increase coincide with those shown for serum in RA patients.
A different research interest focused on the discovery of disease activity-related metabolic alterations, which might provide a potential tool to better classify RA patients. Two studies aimed to distinguish active RA from inactive RA at the metabolic profile level. The study by Yang et al. showed an elevation of glucose and diminution of lactate in synovial fluid of active RA patients compared to inactive RA and healthy controls [35]. Years later, Sasaki et al. made a deeper characterization of the metabolic profiles distinguishing active RA from inactive RA, and they identified 9 metabolites in plasma and 15 in urine significantly different between both groups of patients. A multiple logistic regression model effectively allowed the discrimination between active and inactive patients [34]. Another study aimed to differentiate RA patients with moderate and highly active disease. It was found that in synovial fluid seven metabolites showed a positive correlation with DAS28-ESR (DAS28– erythrocyte sedimentation rate) and were increased in highly active RA patients, while five were negatively correlated with DAS28-ESR and were elevated in moderately active RA. The metabolic alterations found, especially those related to amino acid metabolism may be useful candidates for monitoring disease activity in RA [43].
There have been efforts to elucidate the metabolic profile associated with the response of the patients to biologic agents like rituximab, TNF inhibitors (TNFi), and abatacept. The study by Sweeney et al. aimed to determine if the metabolic profile of serum samples from RA patients might be able to predict their response to rituximab. Before treatment, there were differences in glycine, serine, and threonine metabolism and glycerol and choline. These differences were not observed after treatment but instead, there were changes in pyruvate and lactate metabolism, TCA cycle metabolites, and the glycerophospholipid pathways. These data indicate that the global metabolite profiles of responders and non-responders are distinctive before and after treatment with rituximab [44]. The study of Zabek et al. evaluated the response to the TNFis etanercept and adalimumab, so that serum metabolites were evaluated at baseline and three months after treatment. Thirteen metabolites were able to evidence the treatment response in these patients, however, the persistence of the inflammatory process in those who have been treated was still a hallmark in their metabolome that separates them from the healthy controls [27]. On the other hand, the study by Takahashi et al. aimed to predict the response to a broader range of TNFis (etanercept, golimumab, infliximab, certolizumab pegol, and adalimumab) and abatacept. The results showed that betonicine, glycerol 3-phosphate, N-acetylalanine, hexanoic acid, and taurine are associated with TNFi response in RA patients, while citric acid, quinic acid, and 3-aminobutyric acid levels are related to abatacept response [45]. Notably, there were no shared metabolites between the TNFi responders from both previous studies. The study by Teitsma et al. aimed to identify relevant metabolites and pathways associated with sustained drug-free remission status (sDFR) after treatment with tocilizumab, methotrexate, or their combination in DMARD-naïve patients with early RA. There were different metabolic profiles for each treatment strategy. For the tocilizumab plus methotrexate strategy the most significantly altered pathway was histidine metabolism, in the tocilizumab-only treatment it was arachidonic acid metabolism, and in the methotrexate treatment, it was arginine and proline metabolism [46].
Another important approach in several studies looks for metabolic profiles discriminating RA from other diseases whose signs and symptoms are similar, and therefore constitute a source of confusion for a proper diagnosis. The study by Kim et al. was the first that identified potential biomarkers for distinguishing RA from other inflammatory arthritis such as ankylosing spondylitis, Behςet's disease, and gout. It was proposed a set of 20 metabolites where several amino acids (lysine, asparagine, tyrosine, tryptophan, and glutamine) and metabolites like succinate and citrulline were elevated in RA, while there was a decrease of major fatty acids such as isopalmitic acid, myristic acid and palmitoleic acid showing alterations in fatty acid metabolism [47]. Additionally, a couple of studies compared the metabolic profile of patients with RA and psoriatic arthritis (PsoA). The results obtained by Madsen et al. showed that histidine decrease was one of the most reliable metabolic markers for RA. It was remarkable that larger metabolic differences existed between RA and PsoA patients than among RA and healthy controls [36]. The study from Souto-Carneiro et al. aimed to identify metabolic differences between RA patients that were seronegative (negRA) and PsoA patients since the differential diagnosis between both conditions is difficult. 24 metabolites were identified and quantified and among them, there were nine that had significantly different concentrations between both groups: alanine, leucine, phenylalanine, threonine, valine, acetate, choline, creatine, and lactate. There were lipid groups that also resulted statistically different between both patients comparing their ratio relative to lipid methyl group: lipid β-methylenes, lipid α-methylenes, and lipid polyunsaturated allylic methylenes [48]. The study by Łuczaj et al. investigated the plasma phospholipid profile discriminating RA from Lyme arthritis and found that lipid species belonging to the classes of phosphatidylethanolamines and phosphatidylinositols could differentiate between both diseases [49].
Finally, there was a set of studies with varied interests which answered some different types of inquiries regarding metabolomics in RA. For example, the study of Young et al. evaluated the metabolic profile in RA patients that may be associated with inflammatory markers. It showed that thirteen metabolites (choline, LDL lipids, lactate, acetylglycine, glucose, methylguanidine, methylhistidine, cholesterol, taurine, threonine, fatty acids, methylxanthine, homocysteine) in two groups of early RA patients were strongly associated with C-reactive protein levels, so that they may predict the inflammatory phenotype in these patients [37]. The study by Surowiec et al. aimed to identify biochemical patterns in plasma of pre-symptomatic individuals who subsequently developed RA. The pre-symptomatic individuals were characterized by a profile with lower levels of acylcarnitines and fatty acids, but higher levels of lysophosphatidylcholines, metabolites from tryptophan metabolism, and hypoxanthine. There were other lipids elevated in the pre-symptomatic individuals like phosphatidylcholines and sphingomyelins. These results might be related to the appearance of inflammation and oxidative stress [50]. The research by Narasimhan evaluated the correlation between serum metabolomic signatures and synovial gene expression markers. The results showed two metabolite signatures, while one of them correlated with markers of B cells, the other was associated with plasma cell biology markers. The discriminant analysis revealed that the combination of two or three metabolites can distinguish between high or low gene expression levels of synovial TNF and CD3E. Serum metabolic profiling by NMR can be used for predicting pathogenic pathways in the inflamed synovium of RA patients [51].
5. Metabolites and metabolic pathways associated with SLE
There have been efforts to identify the metabolic profiles that discriminate SLE patients from healthy controls. Most of the studies analyzed the metabolites found in serum or plasma, but there were also results from other samples like peripheral blood lymphocytes (PBL), feces, and urine. The analytical platform most employed was MS coupled to GC or LC. The patients who participated in these studies were on an active phase of the disease since they had a SLEDAI (Systemic Lupus Erythematosus Disease Activity Index) score higher than 4. However, many of them also included patients with inactive disease (they presented SLEDAI score less than 4) [[52], [53], [54], [55]]. The patients were prescribed DMARDs like methotrexate, hydroxychloroquine, azathioprine, cyclophosphamide, mycophenolate mofetil, and leflunomide. Interestingly, the study by Åkesson et al. included a group of patients without treatment [56]. The results showing the metabolite and pathways differences between both study groups are shown in Table 2.
Table 2.
Altered metabolites and metabolic pathways in RA. The metabolites and pathways altered in RA with respect to healthy controls were extracted from the references listed.
| Sample | Analytical platform | Metabolite differences SLE vs healthy controls | Pathways differences SLE vs healthy controls | References |
|---|---|---|---|---|
| Serum | 1H-NMR |
Decreased: valine, tyrosine, phenylalanine, lysine, isoleucine, histidine, glutamine, alanine, citrate, creatinine, creatine, pyruvate, HDL, cholesterol, glycerol, formate. Increased: N-acetyl glycoprotein, VLDL, and LDL. |
Energy metabolism, amino acid metabolism, lipid metabolism, and purine nucleotide metabolism | [59] |
| GC-MS and LC MS |
Decreased: 1,2 propanediol, 3-hydroxybutyrate, Alpha ketoglutarate, citrate, G3P, lactate, malate, pyruvate, phosphocholine, essential polyunsaturated fatty acids (PUFA), long chain FA, acyl carnitines, GSH, methionine, cysteine, choline, pyridoxate, vitamin B6. Increased: medium chain FA, 9-HODE, 13-HODE, LTB4, 5-HETE, gamma-glutamyl peptides. |
Glycolysis, Krebs cycle, beta oxidation, lipid peroxidation, oxidative stress, essential fatty acids | [52] | |
| GC-MS |
Decreased: amino acids (tryptophan, alanine, proline, glycine, serine, threonine, aspartate, glutamine, asparagine, lysine, histidine, tyrosine, valine, leucine, isoleucine), fructose, mannose, glucose, gluconic acid-lactone, glycerol, oleic acid, arachidonic acid, fumarate, aminomalonate, threonate, alpha tocopherol. Increased: methionine, glutamate, cystine, 1-monopalmitin, 1-monolinolein, 1-monoolein, 2-hydroxyisobutyrate. |
Amino acid turnover or protein biosynthesis, galactose metabolism, glutathione metabolism, Krebs cycle | [65] | |
| NMR |
Decreased in SLE (with and without nephritis): amino acids (leucine, valine, alanine, glycine, proline), citrate, choline, lactate. Increased (with and without nephritis): Glucose and N-acetyl glycoprotein. |
Amino acid metabolism, TCA cycle, lactic acid fermentation, glycolysis | [58] | |
| GC-MS |
Decreased: Lysine, fumaric acid, malic acid, methionine, tyrosine, alanine, asparagine, threonic acid, histidine, lactic acid, cysteine, citric acid, tryptophan. Increased: Urea, cystine, threonine, glucose |
Oxidative stress, urea cycle, glycolysis, tryptophan metabolism, dopamine, and serotonin biosynthesis, kynurenine pathway | [57] | |
| MS |
Decreased: dihydroceramide, alkylphosphatidylethanolamine, alkenylphosphatidylethanolamine, phosphatidylserine. Increased: lysophosphatidylinositol |
Lipids metabolism | [55] | |
| HPLC-MS |
Decreased: acylcarnitines, caffeine, hydrocortisone, itaconic acid and serotonin. Increased: ceramides, phosphatidylethanolamine, ether phosphatidylcholine, diacylglycerol, sphingomyelin (SM), arachidonic acid, amino acids (arginine, l-glutamic acid, l-histidine), drug metabolites, 2-coumaric acid, acetylcholine, beta-guanidino propionic acid, xanthine, inosine, galacturonic acid, rac-glycerol 3 phosphate and trimethylamine N-oxide (TMAO) |
Aminoacyl-tRNA biosynthesis, sphingolipid metabolism, nitrogen metabolism, cyanoamino acid metabolism, caffeine metabolism, alanine/aspartate and glutamate metabolism, and methane metabolism. | [54] | |
| Plasma | GC-MS |
Decreased: caproic, caprylic, linoleic, stearic, behenic, lignoceric, arachidonic and hexacosanoic acid. Increased: myristic, palmitoleic, oleic and eicosanoic acid. |
Free fatty acids | [60] |
| Plasma and urine | GC-MS, LC-MS, NMR | Increased: Kynurenine, quinolinic acid, trigonelline (urine). | Kynurenine metabolism | [56] |
| Urine | GC-MS | Increased: Valine, leucine, 3-Hydroxyisobutyrate, fumarate, malate, cystine, pyroglutamarate, cysteine, threonate, uracil, pseudouridine, xanthine, urate, p-Cresol, 2-Hydroxyisobutyrate, tryptophan, glyceric acid, Myo-inositol, 2,3-dihydroxybutyrate, 2,4-dihydroxybutyrate, 3,4-dihydroxybutyrate, 3,4,5-trihidroxypentanoic acid glutarate. | Energy metabolism, oxidative stress, nucleotide metabolism, gut microbiome derived metabolism | [65] |
| Peripheral blood lymphocytes | GC-MS and LC-MS |
Decreased: Cysteine and inosine. Increased: Kynurenine, methionine sulfoxide, cystine, OAA, PEP, DHAP, 3 PG, R5P, Guanine, Guanosine, GDP, dGDP, AMP, ADP, cytosine, dCTP, PHE. |
Pentose phosphate pathway, glycolysis, starch, purine, and pyrimidine metabolism | [63] |
| Feces | UHPLC-MS |
Decreased: adenosine, adenosine 5′-diphosphate (ADP), D-Alaninyl-d-alanine (D-Ala-D-Ala), lauryl diethanolamide, sulfoquinovosyl diacylglyceride (SQDG) 26:5, thiamine pyrophosphate, trigonelline, and mucic acid. Increased: proline, l-tyrosine, l-methionine, l-asparagine, dl-pipecolinic acid, Glycyl-l-proline, xanthurenic acid, kynurenic acid, l-carnosine, monoacylglycerol (MG) 22:6, MG 16:5, lysophosphatidylethanolamine (lysoPE) 16:0, lysophosphatidylcholine (lysoPC) 22:5, phosphatidylglycerol (PG) 27:2 and 1,2-dioleoyl-rac-glycerol |
Aminoacyl-tRNA biosynthesis, thiamine metabolism, nitrogen metabolism, tryptophan metabolism, and cyanoamino acid metabolism | [64] |
Abbreviations: 1H-NMR (Proton nuclear magnetic resonance), GC-MS (Gas chromatography-mass spectrometry), NMR (Nuclear magnetic resonance), LC-MS (Liquid chromatography-mass spectrometry), UHPLC-MS (Ultra high-performance liquid chromatography-mass spectrometry), HPLC-MS (High-performance liquid chromatography-mass spectrometry).
The evidence regarding glucose levels is conflicting since Yan et al. showed a decrease in SLE while Bengtsson et al. and Guleria et al. showed an increase [53,57,58]. Pyruvate and lactate were diminished [52,[57], [58], [59]]. With regards to metabolites of the TCA cycle, there was a diminution in SLE of citrate, alpha-ketoglutarate, fumarate, and malate [52,53,58,59]. The levels of several amino acids (tryptophan, alanine, proline, glycine, serine, aspartate, glutamine, asparagine, lysine, histidine, tyrosine, valine, leucine, isoleucine, and phenylalanine) were lower in SLE according to many studies [53,[57], [58], [59]]. Nevertheless, the results from Li et al. and Yan et al. showed an elevation in amino acids like arginine and glutamate [53,54]. Methionine is an amino acid whose levels in SLE patients are controversial since Bengtsson and Wu et al. reported a decrease but Yan et al. showed an increase [52,53,57]. The diminished levels of tryptophan are involved in its degradation that results in metabolites such as kynerunine, which appeared increased in SLE patients [56]. There was an elevation of urea in SLE patients [57].
Concerning the lipid profile in SLE, there was a diminution of HDL and cholesterol along with an increase in LDL and VLDL [59]. The study by Wu et al. reported in SLE patients decreased levels of long-chain fatty acids and polyunsaturated fatty acids (PUFA), while there was an increase in medium-chain fatty acids [52]. There were lower levels of glycerol in SLE patients according to a couple of studies [53,59]. The results from Yan et al. showed higher levels of monoacylglycerols like 1-monopalmitin, 1-monolinolein, and 1-monoolein in SLE patients. However, there were lower levels of arachidonic acid and oleic acid [53]. The dropped levels of arachidonic acid might be related to the increased amount of its downstream product leukotriene B4, which is a lipid mediator with important immune functions [52]. Shin et al. also reported significantly decreased levels of arachidonic acid in SLE patients, but also of other free fatty acids: caproic, caprylic, linoleic, stearic, behenic, lignoceric, and hexacosanoic acids. However, there were free fatty acids that were significantly higher in SLE: myristic, palmitoleic, oleic, and eicosanoic acids [60]. Li et al. reported an increase in arachidonic acid, but also in ceramides and phospholipids like phosphatidylethanolamine, sphingomyelin, and diacylglycerol in SLE patients [54]. A different study using a shotgun lipidomics approach showed an alteration in diacyl phosphatidylethanolamines and ceramides in SLE patients, so that some lipid species were increased while others were decreased compared to healthy controls [55]. Li et al. and Wu et al. agreed on the diminution of acylcarnitines [52,54].
In SLE patients there was evidence of oxidative stress shown by the decrease of GSH, cysteine, and the increase of cystine [52,53,57]. Threonate has also a major role in oxidative stress and damage and it was elevated in SLE patients [34,53,57]. Another metabolite elevated in SLE and that is related to oxidative stress is xanthine, whose production catalyzed by xanthine oxidase is accompanied by the generation of reactive oxygen species [54]. Wu et al. reported elevated levels in SLE of 9-HODE and 13-HODE, which are lipid peroxidation products that indicate the loss of integrity of the cell membranes [52]. There was also an increase in N-acetyl glycoproteins, which are mainly acute phase proteins that are expressed during inflammation and immune responses [58,59].
Fig. 1B shows the summary of the serum and plasma metabolites that were found increased or decreased in SLE patients compared with healthy controls. The over-representation analysis in Fig. 2B presents the enrichment ratio of metabolic pathways in SLE. The top 5 enriched pathways are alpha-linolenic acid and linoleic acid metabolism, ammonia recycling, urea cycle, glucose-alanine cycle, and glutathione metabolism.
One of the best characterized populations in SLE are CD4+ T cells. These cells present dysfunction in cellular metabolism, which is characterized by increased oxidative phosphorylation and glycolysis [61]. Despite the hyperactivity of the electron transport chain complex I, ATP production is decreased in these T cells, leading to the release of ROS and an elevation of mitochondrial transmembrane potential that leads to cell death [61,62]. It was also found that kynurenine was one of the most elevated metabolites, while cysteine was the most depleted [63]. There is also depletion of the antioxidant glutathione in T cells [61]. Therefore, tryptophan metabolism and oxidative stress are enriched in T lymphocytes of SLE patients. Oxidative stress can promote autoimmunity by modulating signal transduction and cytokine production [63]. In plasma, there were also hallmarks of oxidative stress like the decrease of GSH and cysteine, and accumulation of kynurenine was also observed.
Along with the metabolomics research in serum comparing SLE patients and healthy controls, there were also some studies exploring other samples obtained from SLE patients, like urine and feces. The study by Zhang et al. investigated the metabolites in fecal samples of SLE patients and healthy controls. There were elevated levels of xanthurenic acid and kynurenic acid in SLE patients, showing once again the rise in the tryptophan metabolism pathway. Amino acids like proline and l-asparagine were increased in SLE patients. Furthermore, in SLE patients there was a diminution of trigonelline and thiamine pyrophosphate, which are intermediates of the vitamin B metabolism that may play a role in the persistence of microbes. There were also imbalances in the lipid metabolism of SLE patients, which was evidenced by the decrease of polyunsaturated lipids like monoacylglycerol 22:6, monoacylglycerol 16:5, lysophosphatidylcholine 22:5, and phosphatidylglycerol 27:2 [64]. On the other hand, the characterization of the urinary metabolic profile associated with SLE resulted in 23 up-regulated metabolites. Among them, valine, leucine, 3-hydroxyisobutyrate, fumarate, and malate are related to alterations in energy metabolism pathways like the TCA cycle, while cysteine, pyroglutamate, cysteine and, threonate have a role in oxidative stress. There were also elevated intermediates of purine metabolism like uracil and xanthine. Interestingly, there were high levels of p-cresol and 2-hydroxyisobutyrate, which are metabolites generated through gut microbial amino acid metabolism [65].
There was a set of studies that aimed to evaluate the metabolic factors associated with different disease manifestations. The study of Åkesson et al. aimed to determine in patients with SLE if there was an association of metabolites from the kynurenine pathway with fatigue and depression, and it resulted that some of these metabolites (tryptophan, kynurenine, and quinolinic acid) are significantly altered in patients with SLE-related fatigue compared to healthy controls [56]. On the other hand, the study by Guleria et al. was the first report on serum metabolic profiling to distinguish lupus nephritis from lupus without nephritis. It showed that elevated levels of lipoproteins and lipids, but decreased acetate are characteristic of lupus nephritis patients. This makes sense since acetate diminishes as it constitutes the building block of fatty acids and cholesterol [58]. On the other hand, the study by Yan et al. used metabolomics to find new potential disease activity-related biomarkers, so that it was found evidence of differences in six metabolites (glutamate and 2- hydroxyisobutyrate, citrate, glycerol, linoleic acid, and propylparaben) between patients with active and inactive disease. These metabolites along with C3 and C4 levels reached an AUC of 0.917 in ROC curve analysis, and this allows to propose them as additional diagnostic markers for inactive and active SLE [53].
Additionally, some studies evaluated the metabolic features associated with response to treatment. The study by Pego-Reigosa et al. investigated the role of rituximab treatment in the improvement of the lipid profile in patients with active SLE. The results showed that lower levels of HDL cholesterol and increased triglycerides went back to normal after the B cell depletion, and this correlated with a decrease in the activity of the disease. From this finding, we learned that rituximab acts positively on the lipid profile, which is a risk factor for atherosclerosis, and may reduce the development of this complication of SLE [66]. A different study by Pearl et al. examined the metabolic impact of the treatment of SLE patients with the amino acid precursor N-acetylcysteine. This treatment reduced kynurenine while enhancing NADPH levels in peripheral blood lymphocytes of SLE patients in vitro, so that the pathogenic scenario where kynurenine accumulation is a potential contributor to mTOR activation, was reversed. Therefore, kynurenine is a metabolic target of the action of NAC in patients with SLE [63].
A couple of studies explored the metabolic profiling of serum samples of SLE patients to discriminate them against individuals suffering from other autoimmune diseases. The study by Bengtsson et al. compared SLE with primary Sjögren's syndrome (pSS) and systemic sclerosis (SSc), and it was found that there are metabolites significantly different between SLE and the other autoimmune diseases. In SLE there was a more pronounced diminution of amino acids than in SSc, and a decrease in organic acids compared to pSS. Notably, even though these diseases share clinical pictures, their metabolic differences with healthy controls do not overlap [57]. This lack of overlap largely means that the metabolic profiles reported for SLE and pSS are specific for each disease. The research by Wu et al. aimed to determine the specificity of the metabolic markers proposed for SLE by examining them in RA patients. The results showed that in RA patients there were alterations in leukotriene B4, MDA, and glutathione, but to a lesser extent than those found in SLE patients. However, the ROC analysis evidenced that these markers were effective in discriminating SLE from RA [52]. Conversely, the results reported by Ouyang et al. demonstrated that the changes in metabolic profiles observed for SLE patients were not as marked as those observed with the RA group [59]. Despite this inconsistency found, both studies suggest that SLE and RA may share to some degree their metabolic profile.
6. Overlapping metabolic factors between RA, SLE, and other autoimmune diseases
There is more evidence identifying the shared metabolic characteristics across autoimmune diseases, which makes sense since autoimmune diseases share physiopathologic mechanisms, genetic and environmental factors due to autoimmune tautology. For example, the study by Bellocchi et al. evaluated the intestinal microbiome and plasma metabolome in patients with SLE, primary antiphospholipid syndrome (PAPS), Sjögren's syndrome (SS), and undifferentiated connective tissue disease (UCTD). In the autoimmune diseases compared to healthy controls, there was evidence of an increase in bacteria type correlated with an inflammatory phenotype, and so there was a decrease in tolerogenic bacteria producing short-chain fatty acids. The metabolomic analysis consistently showed that autoimmune diseases have a specific metabolic profile; however, it was not a unifying pattern able to discriminate autoimmune diseases from controls. Furthermore, an interaction between intestinal microbiota and metabolism in patients with autoimmune diseases is suggested by the correlation between microbial genera and metabolites [67]. In addition, the study by Blackmore et al. aimed to explore the metabolomic profile overlap found between myasthenia gravis (MG) and RA. There were 20 significantly changed metabolites shared by MG and RA, which were related to phenylalanine metabolism, tyrosine metabolism, ubiquinone, and another terpenoid-quinone biosynthesis, and pyruvate metabolism. Again, since there are shared metabolites between MG and RA, these are not specific to any of the diseases [68].
7. Conclusions and perspectives
The power of metabolomics extends from biomarker discovery and stratification of patients to the prediction of response to specific therapies and personalized medicine and is a helpful tool for the understanding of the mechanisms that underlie the phenotypes specific for RA and SLE [18]. Here we described the metabolites and metabolic pathways identified in patients with these autoimmune diseases. There are some common features found across the studies for each disease and some differences related to the population studied, the disease activity, the genetic background, and the context of each individual that can influence the metabolic profiles. In agreement with the autoimmune tautology respect the common origin and physiopathologic mechanisms of autoimmune diseases, we found in this review that RA and SLE shared some metabolic alterations; however, there are unique metabolites changes described only for RA or SLE. In RA and SLE there is a decrease of several amino acids, but in SLE there have been found more amino acids reduced than in RA. Another important feature shared among both diseases is the increase of metabolites related to oxidative stress. Nevertheless, in SLE there have been more reports regarding metabolites associated with oxidative stress, which leads us to think that this response is observed more often in SLE patients. In SLE patients it is remarkable the tryptophan metabolism that generates kynurenine, which is a metabolite important for immune cell activation. On the other hand, in RA patients there is an increase in ribose showing the up-regulation of PPP and biosynthetic pathways.
The functional effects of metabolites depend on their abundance in the tissue microenvironment, the transport mechanisms they use, and the diversity of cells that generates and use them. A metabolite can reflect changes and can be increased because of its excessive production and accumulation, while the decreased levels may be the result of its constant expenditure and insufficient generation. The potential clinical translation of the knowledge that has been gathered about metabolic pathways in autoimmune diseases will need functional metabolomics studies to evaluate the effect of metabolic products in cellular homeostasis [42]. While the discovery metabolomics studies measure the up and down-regulation of metabolites and address to the identification of biomarkers, the functional metabolomics studies are aimed at the potential to reprogram a metabolome through the identification of metabolites able to modulate the immune mechanisms associated with the disease pathogenesis, providing the framework for reprogramming metabolomics [69].
Funding
During the preparation of the manuscript, we received financial support from Minciencias Colombia ID PRY 120389666081. Grant number 830-2018.
Authors contributions
Nancy P. Duarte-Delgado: Writing - original draft preparation. Mónica P. Cala, Alfonso Barreto and Luz-Stella Rodríguez: Writing - review & editing.
Submission declaration
The manuscript has not been published or submitted elsewhere. It has been read and approved by all the authors.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Luz-Stella Rodriguez C. reports financial support was provided by Minciencias. ID PRY 120389666081.
Acknowledgments
We would like to thank Manuel A. Franco and Juana Angel from Instituto de Genética Humana of Pontificia Universidad Javeriana for their critical review of the manuscript and helpful comments.
References
- 1.Aribi M. Immunopathogenesis and Immune-Based Therapy for Selected Autoimmune Disorders. 2017. Introductory chapter: immune system dysfunction and autoimmune diseases; pp. 1–9. [Google Scholar]
- 2.Theofilopoulos A.N., Kono D.H., Baccala R. The multiple pathways to autoimmunity. Nat. Immunol. 2018;18(7):716–724. doi: 10.1038/ni.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anaya J.-M. The autoimmune tautology. Arthritis Res. Ther. 2010;12(147):6–8. doi: 10.1186/ar3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deane K.D., Holers V.M. The natural history of rheumatoid arthritis. Clin Ther [Internet] 2019;41(7):1256–1269. doi: 10.1016/j.clinthera.2019.04.028. Available from: [DOI] [PubMed] [Google Scholar]
- 5.Firestein G., Mcinnes I.B., Jolla L. Immunopathogenesis of rheumatoid arthritis. 2018;46(2):183–196. doi: 10.1016/j.immuni.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smolen J.S., Aletaha D., Barton A., Burmester G.R., Emery P., Firestein G.S., et al. Rheumatoid arthritis. Nat Rev Dis Prim [Internet] 2018;4(8):1–23. doi: 10.1038/nrdp.2018.1. Available from: [DOI] [PubMed] [Google Scholar]
- 7.Tsokos G.C., Lo M.S., Reis P.C., Sullivan K.E. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat Rev Rheumatol [Internet] 2016;12(12):716–730. doi: 10.1038/nrrheum.2016.186. Available from: [DOI] [PubMed] [Google Scholar]
- 8.Vasquez-Canizares N., Wahezi D., Putterman C., Einstein A. Diagnostic and prognostic tests in systemic lupus erythematosus. Best Pract. Res. Clin. Rheumatol. 2018;31(3):351–363. doi: 10.1016/j.berh.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaul A., Gordon C., Crow M.K., Touma Z., Urowitz M.B., Vollenhoven R Van, et al. Systemic lupus erythematosus. Nat. Publ. Gr. [Internet] 2016;2(June):1–22. doi: 10.1038/nrdp.2016.39. Available from: [DOI] [PubMed] [Google Scholar]
- 10.Yazdany J., Era M.D. Eighth Edi. Dubois' Lupus Erythematosus and Related Syndromes. Elsevier Inc.; 2013. Chapter 1 – definition and classification of lupus and lupus-related disorders [internet] pp. 1–7. Available from: [DOI] [Google Scholar]
- 11.Arriens C., Mohan C. Systemic lupus erythematosus diagnostics in the ‘ omics ’ era. Int. J. Clin. Rheumatol. 2013;8(6):671–687. doi: 10.2217/ijr.13.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riekeberg E., Powers R. New frontiers in metabolomics: from measurement to insight. F1000 Res. 2017;6(1148) doi: 10.12688/f1000research.11495.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta L., Ahmed S., Jain A., Misra R. 2018. Emerging Role of Metabolomics in Rheumatology; pp. 1468–1477. June. [DOI] [PubMed] [Google Scholar]
- 14.Priori R., Scrivo R., Brandt J., Valerio M., Casadei L., Valesini G., et al. Metabolomics in rheumatic diseases: the potential of an emerging methodology for improved patient diagnosis, prognosis, and treatment efficacy. Autoimmun. Rev. [Internet] 2013;12(10):1022–1030. doi: 10.1016/j.autrev.2013.04.002. Available from: [DOI] [PubMed] [Google Scholar]
- 15.Guma M., Tiziani S., Firestein G.S. Vol. 12. 2016. pp. 269–281. (Metabolomics In Rheumatic Diseases: Desperately Seeking Biomarkers). 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norata G.D., Caligiuri G., Chavakis T., Matarese G., Netea M.G., Nicoletti A., et al. The cellular and molecular basis of translational immunometabolism. Immun. Rev. [Internet] 2015;43(3):421–434. doi: 10.1016/j.immuni.2015.08.023. Available from: [DOI] [PubMed] [Google Scholar]
- 17.Rhoads J.P., Rathmell J.C. Fine tuning of immune metabolism for the treatment of rheumatic diseases. Nat. Rev. Rheumatol. 2018;13(5):313–320. doi: 10.1038/nrrheum.2017.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson C.H., Ivanisevic J., Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell. Biol. [Internet] 2016;17(7):1–9. doi: 10.1038/nrm.2016.25. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudzik D., Barbas-Bernardos C., García A., Barbas C. Quality assurance procedures for mass spectrometry untargeted metabolomics. a review. J. Pharm. Biomed. Anal. [Internet. 2018;147:149–173. doi: 10.1016/j.jpba.2017.07.044. Available from: [DOI] [PubMed] [Google Scholar]
- 20.Barnes S., Benton H.P., Casazza K., Cooper S.J., Cui X., Du X., et al. Training in metabolomics research. I. Designing the experiment, collecting and extracting samples and generating metabolomics data. J. Mass Spectrom. 2016;51(7):461–475. doi: 10.1002/jms.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patti G., Yanes O., Siuzdak G. Metabolomics: the apogee of the omic trilogy. Nat. Rev. Mol. Cell Biol. 2013;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang J., Zhu L., Lu J., Zhang X. Application of metabolomics in autoimmune diseases : insight into biomarkers and pathology. J. Neuroimmunol. [Internet] 2015;279(1):25–32. doi: 10.1016/j.jneuroim.2015.01.001. Available from: [DOI] [PubMed] [Google Scholar]
- 23.Julià A., Alonso A., Marsal S. Metabolomics in rheumatic diseases. Int. J. Clin. Rheumtol. 2014;9:353–369. [Google Scholar]
- 24.Du X., Engler J., Kabarowski J.H., Li S., Pathmasiri W. Training in metabolomics research. II. Processing and statistical analysis of metabolomics data, metabolite identification, pathway analysis, applications of metabolomics and its future. J. Mass Spectrom. 2017;51(8):535–548. doi: 10.1002/jms.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anaya J.M., Restrepo-Jiménez P., Ramírez-Santana C. The autoimmune ecology: an update. Curr. Opin. Rheumatol. 2018;30(4):350–360. doi: 10.1097/BOR.0000000000000498. [DOI] [PubMed] [Google Scholar]
- 26.Guma M., Tiziani S., Firestein G.S., Jolla L. Metabolomics in rheumatic diseases: desperately seeking biomarkers. Nat. Rev. Rheumatol. 2016;12(5):269–281. doi: 10.1038/nrrheum.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zabek A., Swierkot J., Malak A., Zawadzka I., Deja S., Bogunia-Kubik K., et al. Analysis Application of 1 H NMR-based serum metabolomic studies for monitoring female patients with rheumatoid arthritis. J. Pharm. Biomed. Anal. [Internet] 2016;117:544–550. doi: 10.1016/j.jpba.2015.10.007. Available from: [DOI] [PubMed] [Google Scholar]
- 28.O'Neill L.A.J., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim H., Suh C. Biomarkers for systemic lupus erythematosus: an update. Int. J. Clin. Rheumatol. 2015;10:195–204. [Google Scholar]
- 30.Wang A., Luan H.H., Medzhitov R. An evolutionary perspective on immunometabolism. Science. 2019;80:3932. doi: 10.1126/science.aar3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loftus R.M., Finlay D.K. Immunometabolism : cellular metabolism turns immune regulator. J. Biol. Chem. 2016;291(1):1–10. doi: 10.1074/jbc.R115.693903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z., Matteson E.L., Goronzy J.J., Weyand C.M. T-cell metabolism in autoimmune disease. Arthritis Res. Ther. 2015;17(29):1–10. doi: 10.1186/s13075-015-0542-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perl A. Vol. 69. 2017. pp. 2259–2270. (Metabolic Control Of Immune System Activation In Rheumatic Diseases). 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasaki C., Hiraishi T., Oku T., Okuma K., Suzumura K., Hashimoto M., et al. Metabolomic approach to the exploration of biomarkers associated with disease activity in rheumatoid arthritis. PLoS One. 2019;14(7) doi: 10.1371/journal.pone.0219400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X.Y., Zheng K Di, Lin K., Zheng G., Zou H., Wang J.M. Energy metabolism disorder as a contributing factor of rheumatoid arthritis : a comparative proteomic and metabolomic study. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0132695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madsen R.K., Lundstedt T., Gabrielsson J., Sennbro C., Alenius G., Moritz T., et al. Diagnostic properties of metabolic perturbations in rheumatoid arthritis. Arthritis Res. Ther. [Internet] 2011;13(1):R19. doi: 10.1186/ar3243. http://arthritis-research.com/content/13/1/R19 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young S.P., Kapoor S.R., Viant M.R., Byrne J.J., Filer A., Buckley C.D., et al. The impact of inflammation on metabolomic profiles in patients with arthritis. Arthritis Rheum. 2013;65(8):2015–2023. doi: 10.1002/art.38021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang L., Mundra P.A., Fan F., Galvin A., Weir J.M., Wong G., et al. Plasma lipidomic profiling in patients with rheumatoid arthritis. Metabolomics. 2016;12(8):1–10. [Google Scholar]
- 39.Zhou J., Chen J., Hu C., Xie Z., Li H., Wei S., et al. Exploration of the serum metabolite signature in patients with rheumatoid arthritis using gas chromatography–mass spectrometry. J. Pharm. Biomed. Anal. [Internet. 2016;127:60–67. doi: 10.1016/j.jpba.2016.02.004. Available from: [DOI] [PubMed] [Google Scholar]
- 40.Li J., Che N., Xu L., Zhang Q., Wang Q., Tan W., et al. LC-MS-based serum metabolomics reveals a distinctive signature in patients with rheumatoid arthritis. Clin. Rheumatol. 2018;37(6):1493–1502. doi: 10.1007/s10067-018-4021-6. [DOI] [PubMed] [Google Scholar]
- 41.Weyand C.M., Zeisbrich M. Metabolic signatures of T-cells and macrophages in rheumatoid arthritis. Curr. Opin. Rheumatol. 2018:112–120. doi: 10.1016/j.coi.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weyand C.M., Goronzy J.J. Immunometabolism in early and late stages of rheumatoid arthritis. Nat Rev Rheumatol [Internet] 2017;13(5):291–301. doi: 10.1038/nrrheum.2017.49. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahn J.K., Kim J., Cheong Y.E., Kim K.H., Cha H. vols. 500–7. 2020. (Variation in the Synovial Fluid Metabolome According to Disease Activity of Rheumatoid Arthritis). [PubMed] [Google Scholar]
- 44.Sweeney S.R., Kavanaugh A., Lodi A., Wang B., Boyle D., Tiziani S., et al. Metabolomic profiling predicts outcome of rituximab therapy in rheumatoid arthritis. RMD Open. 2016;2(e000289) doi: 10.1136/rmdopen-2016-000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi S., Saegusa J., Onishi A., Morinobu A. Biomarkers identified by serum metabolomic analysis to predict biologic treatment response in rheumatoid arthritis patients. Rheumatology. 2019;58(12):2153–2161. doi: 10.1093/rheumatology/kez199. [DOI] [PubMed] [Google Scholar]
- 46.Teitsma X.M., Yang W., Jacobs J.W.G., Pethö-Schramm A., Borm M.E.A., Harms A.C., et al. Baseline metabolic profiles of early rheumatoid arthritis patients achieving sustained drug-free remission after initiating treat-to-target tocilizumab, methotrexate, or the combination: insights from systems biology. Arthritis Res. Ther. 2018;20(1):1–11. doi: 10.1186/s13075-018-1729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim S., Hwang J., Xuan J., Jung Y.H., Cha H., Kim K.H. Global metabolite profiling of synovial fluid for the specific diagnosis of rheumatoid arthritis from other inflammatory arthritis. PLoS One. 2014;9(6):1–9. doi: 10.1371/journal.pone.0097501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Souto-Carneiro M., Tóth L., Behnisch R., Urbach K., Lorenz M., Klika K.D., et al. Differences in the serum metabolome and lipidome identify potential biomarkers for seronegative rheumatoid arthritis versus psoriatic arthritis. Ann. Rheum. Dis. 2020;79:499–506. doi: 10.1136/annrheumdis-2019-216374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Łuczaj W., Moniuszko-Malinowska A., Domingues P., Domingues M.R., Gindzienska-Sieskiewicz E., Skrzydlewska E. Plasma lipidomic profile signature of rheumatoid arthritis versus Lyme arthritis patients. Arch. Biochem. Biophys. 2018;654(July):105–114. doi: 10.1016/j.abb.2018.07.021. [DOI] [PubMed] [Google Scholar]
- 50.Surowiec I., Ärlestig L., Rantapää-Dahlqvist S., Trygg J. Metabolite and lipid profiling of biobank plasma samples collected prior to onset of rheumatoid arthritis. PLoS One. 2016;11(10):1–14. doi: 10.1371/journal.pone.0164196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Narasimhan R., Coras R., Rosenthal S.B., Sweeney S.R., Lodi A., Tiziani S., et al. Serum metabolomic profiling predicts synovial gene expression in rheumatoid arthritis. Arthritis Res. Ther. 2018;20 doi: 10.1186/s13075-018-1655-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu T., Xie C., Han J., Ye Y., Weiel J., Li Q., et al. Metabolic disturbances associated with systemic lupus erythematosus. PLoS One. 2012;7(6):1–9. doi: 10.1371/journal.pone.0037210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan B., Huang J., Zhang C., Hu X., Gao M., Shi A., et al. Serum metabolomic profiling in patients with systemic lupus erythematosus by GC/MS. Mod. Rheumatol. 2016;7595(26):6. doi: 10.3109/14397595.2016.1158895. [DOI] [PubMed] [Google Scholar]
- 54.Li Y., Liang L., Deng X., Zhong L. Lipidomic and metabolomic profiling reveals novel candidate biomarkers in active systemic lupus erythematosus. Int. J. Clin. Exp. Pathol. 2019;12(3):857–866. [PMC free article] [PubMed] [Google Scholar]
- 55.Lu L., Hu C., Zhao Y., He L., Zhou J., Li H., et al. Shotgun lipidomics revealed altered profiles of serum lipids in systemic lupus erythematosus closely associated with disease activity. Biomolecules. 2018;8(4) doi: 10.3390/biom8040105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Åkesson K., Pettersson S., Ståhl S., Surowiec I., Hedenström M., Eketjäll S., et al. 2018. Kynurenine Pathway Is Altered in Patients with SLE and Associated with Severe Fatigue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bengtsson A.A., Trygg J., Wuttge D.M., Sturfelt G., Theander E., Surowiec I., et al. Metabolic profiling of systemic lupus erythematosus and comparison with primary sjögren ’ s syndrome and systemic sclerosis. PLoS One. 2016;11(7) doi: 10.1371/journal.pone.0159384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guleria A., Pratap A., Dubey D., Rawat A., Chaurasia S., Sukesh E., et al. NMR based serum metabolomics reveals a distinctive signature in patients with Lupus Nephritis. Sci. Rep. 2016;6(35309) doi: 10.1038/srep35309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ouyang X., Dai Y., Wen J.L., Wang L.X.H. NMR-based metabolomic study of metabolic profiling for systemic lupus erythematosus. Lupus. 2011;20:1411–1420. doi: 10.1177/0961203311418707. [DOI] [PubMed] [Google Scholar]
- 60.Shin T.H., Kim H.A., Jung J.Y., Baek W.Y., Lee H.S., Park H.J., et al. Analysis of the free fatty acid metabolome in the plasma of patients with systemic lupus erythematosus and fever. Metabolomics [Internet] 2018;14(1) doi: 10.1007/s11306-017-1308-6. 0. Available from: [DOI] [PubMed] [Google Scholar]
- 61.Morel L. Immunometabolism in systemic lupus erythematosus. Nat Rev Rheumatol [Internet] 2017;13(5):280–290. doi: 10.1038/nrrheum.2017.43. Available from: [DOI] [PubMed] [Google Scholar]
- 62.Li W., Sivakumar R., Titov A.A., Choi S.-C., Morel L. Metabolic factors that contribute to lupus pathogenesis. Crit. Rev. Immunol. 2016;36(1):75–98. doi: 10.1615/CritRevImmunol.2016017164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perl A., Hanczko R., Oaks Z., Kelly R., Borsuk R., Asara J.M., et al. Comprehensive metabolome analyses reveal N -acetylcysteine- responsive accumulation of kynurenine in systemic lupus erythematosus : implications for activation of the mechanistic target of rapamycin. Metabolomics [Internet] 2015;11:1157–1174. doi: 10.1007/s11306-015-0772-0. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Q., Yin X., Wang H., Wu X., Li X., Li Y., et al. Fecal metabolomics and potential biomarkers for systemic lupus erythematosus. Front. Immunol. 2019;10(976) doi: 10.3389/fimmu.2019.00976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan B., Huang J., Dong F., Yang L., Huang C., Gao M., et al. Urinary metabolomic study of systemic lupus erythematosus based on gas chromatography/mass spectrometry. Biomed. Chromatogr. 2016;30(11):1877–1881. doi: 10.1002/bmc.3734. Nov 1. [DOI] [PubMed] [Google Scholar]
- 66.Pego-Reigosa J.M., Lu T.Y., Fontanillo M.F., del Campo-Pérez V., Rahman A., Isenberg D.A. Long-term improvement of lipid profile in patients with refractory systemic lupus erythematosus treated with B-cell depletion therapy : a retrospective observational study. Rheumatology. 2010;49:691–696. doi: 10.1093/rheumatology/kep446. [DOI] [PubMed] [Google Scholar]
- 67.Bellocchi C., Á Fern, Montanelli G., Vigone B., Santaniello A., Quirantes-pin R., et al. Identification of a shared microbiomic and metabolomic profile in systemic autoimmune diseases. J. Clin. Med. 2019;8(1291):1–15. doi: 10.3390/jcm8091291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blackmore D., Li L., Wang N., Maksymowych W., Yacyshyn E., Siddiqi Z.A. Metabolomic profile overlap in prototypical autoimmune humoral disease: a comparison of myasthenia gravis and rheumatoid arthritis. Metabolomics [Internet] 2020;16(1):10. doi: 10.1007/s11306-019-1625-z. Available from: [DOI] [PubMed] [Google Scholar]
- 69.Peng B., Li H., Peng X.X. Functional metabolomics: from biomarker discovery to metabolome reprogramming. Protein Cell. 2015;6(9):628–637. doi: 10.1007/s13238-015-0185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]