Abstract
Methicillin-resistant Staphylococcus aureus (MRSA), biofilms, and persisters are three major factors leading to recurrent and recalcitrant implant infections. Although antibiotics are still the primary treatment for chronic implant infections in clinical, only few drugs are effective in clearing persisters and formed biofilms. Here, felodipine, a dihydropyridine calcium channel blocker, was reported for the first time to have antibacterial effects against MRSA, biofilm, and persisters. Even after continuous exposure to sub-lethal concentrations of felodipine, bacteria are less likely to develop resistance. Besides, low doses of felodipine enhances the antibacterial activity of gentamicin by inhibiting the expression of protein associated with aminoglycoside resistance (aacA-aphD). Next, biofilm eradication test and persisters killing assay suggested felodipine has an excellent bactericidal effect against formed biofilms and persisters. Furthermore, the result of protein profiling, and quantitative metabonomics analysis indicated felodipine reduce MRSA virulence (agrABC), biofilm formation and TCA cycle. Then, molecular docking showed felodipine inhibit the growth of persisters by binding to the H pocket of ClpP protease, which could lead to substantial protein degradation. Furthermore, murine infection models suggested felodipine in combination with gentamicin alleviate bacterial burden and inflammatory response. In conclusion, low dose of felodipine might be a promising agent for biomaterial delivery to enhance aminoglycosides efficacy against implant infections caused by MRSA, biofilm, and persisters.
Keywords: Aminoglycosides, Felodipine, Persisters, Methicillin-resistant Staphylococcus aureus, Implant infection
Graphical abstract
Highlights
-
•
Felodipine inhibits MRSA gene expression associated with aminoglycoside resistance and biofilm formation.
-
•
Felodipine eradicates formed biofilm and persisters on the surface of implants.
-
•
Felodipine induces proteolysis of MRSA and decreases energy metabolism.
-
•
Felodipine in combination with gentamicin alleviates murine periprosthetic joint infection.
1. Introduction
With the increased use of indwelling medical devices, implant infections have become one of the most severe post-surgery complications. Device-associated infections have been found to account for 25.6% of all healthcare-associated infections in the USA [1]. According to research, the average rate of infection in joint prosthesis is about 2% [2]. Besides, a report by the National Institutes of Health also pointed out that biofilms account for more than 80% of microbial infections in humans [3]. Once an implant infection occurs, patients must undergo multiple surgical debridement's and prolonged antibiotic treatments, and they also face an increased risk of disability and mortality [2]. The most common causal pathogens of implant infections are methicillin-resistant Staphylococcus aureus (MRSA), methicillin-sensitive Staphylococcus aureus (MSSA), methicillin-resistant Staphylococcus epidermidis (MRSE), and methicillin-sensitive Staphylococcus epidermidis (MSSE) [4]. Since MRSA contains multiple antibiotic resistance genes, it is tolerant to almost all conventional antibiotics, including aminoglycosides [5].
Currently, although many new treatments for drug-resistant infections have been developed, only few drugs are clinically available to remove persisters and biofilms from the surface of implants [6]. It has been reported that even the use of 100-fold minimum inhibition concentration of vancomycin, a first-line antibiotic for drug-resistant Staphylococcus aureus, was not effective in removing persisters and biofilms [7]. Because biofilms are heterogeneous structures consisting of bacterial cells enclosed within a self-produced matrix of hydrated extracellular polymeric substances [8], they can effectively reduce the penetration of antibiotics and evade being killed by immune cells. Compared to their planktonic counterparts, the removal of biofilms often requires 1000 times the concentration of antibiotics [9]. In addition, due to insufficient oxygen levels and nutrient supplies in the deep layers of biofilms, a small fraction of transiently antibiotic-tolerant phenotypical variants called persisters have emerged [10]. Persisters are a subpopulation of dormant bacteria that can survive under high doses of antibiotics and resume growth after the antibiotic pressure drops [11].
Nowadays, antibiotics are still the main approach to clinical treatment of chronic implant infections, although many antimicrobial approaches have been designed [[12], [13], [14]]. However, owing to the arduous process of developing entirely new antibiotics, few compound have been approved for the treatment of biofilm infections during the last 20 years [15,16]. Therefore, we urgently need a strategy that can accelerate the development of antibiotics to catch up with the pace at which resistance mutations are being developed by bacteria. Drug repurposing screens and synergistic drug combinations are useful alternatives for the treatment of severe infections caused by drug-resistant bacteria, biofilm and persisters [17,18]. This strategy helps to avoid attrition in clinical trials due to drug toxicity and unfavorable issues in pharmacokinetics, but also enhances the antibacterial activity of conventional antibiotics [19], such as aminoglycosides.
Therefore, we performed phenotype-based screening of a preclinical/clinical compound library containing more than 3000 compounds. After excluding drugs with significant cytotoxicity towards mammalian cells in vitro, we found a compound with an excellent antibacterial effect against MRSA, namely felodipine. Felodipine is an FDA-approved, dihydropyridine class of calcium channel blocker (CCB), which has been well studied in patients with essential hypertension [20]. Compared with other available CCBs, felodipine is one of the most vascular selective drugs, and has no negative inotropic effect at clinically administered doses [21]. Additionally, felodipine has been found to induce autophagy in mouse brains when used as a treatment for neurodegenerative diseases [22].
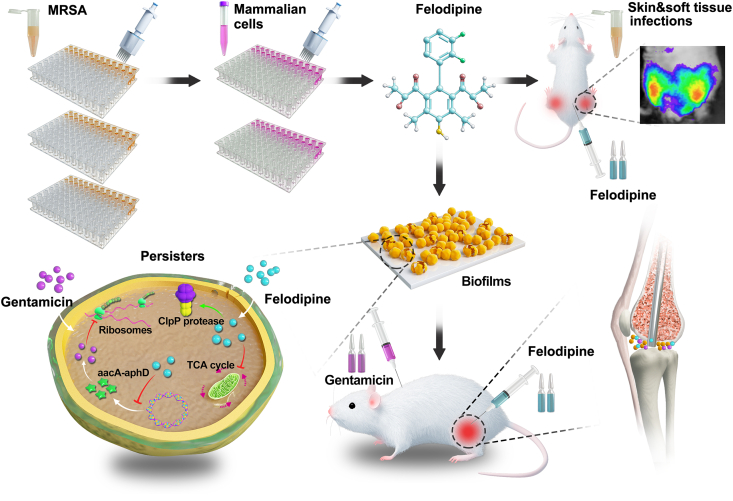
In this study, felodipine was reported for the first time to have antibacterial effects against MRSA, MRSE and persisters. Even low doses of felodipine could re-sensitize MRSA to aminoglycoside antibiotics. As shown in Fig. 1, the result of RNA sequencing, proteomics profiling, and quantitative metabonomic analysis demonstrated that felodipine could inhibit the TCA cycle and translation of aminoglycoside modifying enzyme (aacA-aphD). Therefore, felodipine has a synergistic effect with gentamicin against MRSA. Next, felodipine could bind to the caseinolytic protease proteolytic subunit (ClpP) and activate its catalytic activity to regulate biofilm formation and persisters. Furthermore, the result of murine skin and soft tissue infection models and periprosthetic joint infection models also proved that felodipine combined with gentamicin ameliorate implant infections and inflammatory response.
Fig. 1.
Scheme illustration of drug repurposing screens and synergistic drug-combination. Felodipine in combination with gentamicin alleviate chronic implant infections caused by biofilms and persisters. The proposed mechanism showed that felodipine activate ClpP protease and reduce the protein level of aminoglycosides modifying enzyme (aacA-aphD).
2. Methods
2.1. Bacterial strains and growth conditions
The MSSA (ATCC 25923), MRSA (ATCC 43300), and MRSE (ATCC 35984) strains were obtained from the American Type Culture Collection. Bacteria were added to trypticase soy broth (TSB; Hopebio; Qingdao, China) and incubated at 37 °C overnight with shaking. Subsequently, the bacterial suspensions were diluted 1:10,000 times with fresh TSB, and then incubation was continued for 4–6 h to allow them to reach exponential growth phases.
2.2. Antimicrobial agents and chemicals
Vancomycin and gentamicin were purchased from Sigma-Aldrich (St. Louis, MO, USA). Preclinical/Clinical Compound Library and felodipine were purchased from Selleck Chemicals (Houston, TX, USA). The LIVE/DEAD™ BacLight™ Bacterial Viability Kit and Membrane Potential Kit were purchased from Thermo Fisher Scientific (USA), while the Microbial Viability Assay Kit, Biofilm Formation Assay Kit, and Biofilm TestPiece Assay Kit were purchased from Dojindo (Japan).
2.3. Antimicrobial susceptibility testing
The minimal inhibitory concentrations (MICs) and minimum bactericidal concentrations (MBCs) for the agents used in this study were examined using the standard microdilution method following the guidelines outlined by the Clinical and Laboratory Standards Institute [23]. Briefly, log-phased bacteria were resuspended to approximately 5 × 105 CFU/mL using Mueller-Hinton broth (MHB). Next, 2-fold serial dilutions of the agent (64 μg/mL- 0.06 μg/mL) were prepared with a final volume of 100 μL. Then 96-well plates were incubated at 37 °C for 24 h, and then the bacterial viability was examined using the Microbial Viability Assay Kit (DOJINDO, Japan). The MIC value was determined as the lowest concentration of the agent that completely inhibited the visible growth of bacteria. Then, 10 μL of bacterial suspension (≥MIC value) was passaged onto an antibiotic-free tryptic soy agar (TSA) plate and incubated at 37 °C for 24 h before counting the number of viable colonies. MBC indicates the lowest concentration of an agent that is bactericidal to ≥99.9% of the initial inoculum.
2.4. Time kill assay
Time-kill experiments against MRSA (ATCC 43300) and MRSE (ATCC 35984) were performed as previously described [24]. Briefly, 50 μL of felodipine was added to the wells of a 96-well plate. Then, 50 μL of log-phased bacterial suspension (1 × 106 CFU/mL) was added to the above well and incubated at 37 °C. At specific time points, the bacterial suspensions were serially diluted with PBS and inoculated onto TSA plates. After incubation overnight at 37 °C, the number of viable bacteria remaining in the original culture was calculated by counting the bacterial colonies.
2.5. Resistance induction assay
The frequency of resistance mutations was examined by sequential passaging over a 20-day period, as previously described [25]. Briefly, 2-fold serial dilutions of felodipine and vancomycin (control) were added to 96-well plates containing 100 μL of log-phased MRSA (ATCC 43300) suspension (105 CFU/mL). After incubation at 37 °C for 24 h, the OD value at 600 nm was measured using a spectrophotometer (BioTek, VT, USA). Bacterial growth was defined as an OD600 of ≥0.1. For the following day's MIC assay, 10 μL of 1/2 MIC for the MRSA suspension was diluted with 10 mL of fresh TSB medium and incubated overnight at 37 °C. The remaining bacterial cultures were stored in 20% glycerol at −80 °C.
2.6. Flow cytometry
The viability and quantity of the bacterial populations were examined using flow cytometry, as previously described [26]. Briefly, log-phased bacteria (106 CFU/mL) were treated with different concentrations of felodipine at 37 °C and 220 rpm for 6 h. Next, the bacterial suspension was stained with the LIVE/DEAD™ BacLight™ Bacterial Viability Kit for 15 min according to the manufacturer's instructions. After washing twice with sterile saline, the samples were detected using a flow cytometer with FITC and PI channels (Beckman Coulter, USA). Data were analyzed using FlowJo software (TreeStar, USA).
2.7. RNA extraction and real time-PCR
Total RNA was extracted from MRSA (ATCC 43300) using the RNeasy Mini Kit (Qiagen, Frankfurt, Germany) according to the manufacturer's instructions. Next, a total of 500 ng of purified RNA was reverse transcribed into cDNA using an RT Master Kit (Takara, Shiga, Japan). Subsequently, the PCR reaction mixture was prepared using the TB Green Premix Ex Taq™ Kit (Takara, Shiga, Japan) following the manufacturer's instructions, and real-time PCR was performed in triplicate using the QuantStudio™ 7 Flex system (Applied Biosystems, Norwalk, USA). The amplification parameters were set as follows: initial denaturation at 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 45 s. The expression of 16s rRNA was used as a standard to analyze the changes in other genes using the comparative CT method. The primers used for the PCR are listed in Table S4 and were synthesized by Sangon Biotech (Shanghai, China).
2.8. RNA sequencing
The transcriptome expression levels of MRSA (ATCC 43300) were analyzed after treatment with felodipine (4 μg/mL) or 0.1% DMSO (control). Briefly, three independent biological replicates of the MRSA cultures were treated for 8 h. Next, bacterial suspensions were precipitated by centrifugation (5000×g, 5 min) and RNA sequencing was performed using the Illumina TruSeq RNA sample prep kit and HiSeq 4000 SBS kit (Illumina, Inc.). After sequencing, differentially expressed genes between samples were analyzed using edgeR software. A false discovery rate (FDR) < 0.05 and |log2Fold change | ≥ 1 were the criteria used to define significant differentially expressed genes.
2.9. Antibiotic synergy assay
The synergistic activity between felodipine and conventional antibiotics was determined using a checkerboard assay, as previously described [6]. Briefly, 2-fold serial dilutions of each compound in the appropriate range of concentrations were combined with 2-fold serial dilutions of felodipine to form an 8 × 8 matrix in a 96-well plate. Conventional antibacterial drugs were diluted along the longitudinal direction, whereas felodipine was diluted along the lateral direction. After incubation at 37 °C for 18 h, the OD value of each well was measured using a spectrophotometer at 600 nm. An OD600 nm ≥ 0.1 was defined as bacterial growth. The fractional inhibitory concentration index (FICI) of two compounds (A and B) was calculated as follows: FICI = (MIC of compound A in combination/MIC of compound A alone) + (MIC of compound B in combination/MIC of compound B alone). The interaction between the two compounds was interpreted as follows: FICI ≤0.5 indicates “synergy”; 0.5 < FICI ≤4 indicates “no interaction; ” FICI >4 indicates “antagonism”.
2.10. Gentamicin-Texas red uptake
Gentamicin-Texas Red was prepared as described previously [27]. Briefly, Texas Red-succinimidyl ester (Invitrogen) was dissolved in high-quality anhydrous N, N-dimethylformamide to a final concentration of 20 mg/mL. Gentamicin was dissolved in a K2CO3 solution (100 mM, pH = 8.5) to give a final concentration of 10 mg/mL. Next, 10 μL of Texas Red was slowly added to 350 μL of the gentamicin solution and incubated at 4 °C for the conjugation reaction. Then, the Gentamicin-Texas red conjugate from the reaction was added to the bacterial suspension to a final concentration of 50 μg/mL and incubated at 37 °C for 4 h. Gentamicin uptake was measured by washing bacteria twice with sterile PBS and analyzed using flow cytometry with mCherry voltage.
2.10.1. Scanning electron microscopy
The morphological changes of the bacteria after the felodipine treatment were observed using SEM according to a previously described method [28]. Briefly, log-phased bacterial cultures (106 CFU/mL) were incubated with felodipine (8 μg/mL), the combination of 1/8 MIC felodipine (1 μg/mL) and 1/8 MIC gentamicin (4 μg/mL) or 0.1% DMSO (control) at 37 °C for 8 h. Then, the bacterial suspension was centrifuged (5000×g, 5 min) and gently washed twice with PBS. Next, the bacterial precipitates were fixed with 2.5% glutaraldehyde (Solarbio, Beijing, China) for 24 h at 4 °C. Subsequently, the bacteria were dehydrated with an alcohol gradient series (40%, 50%, 60%, 70%, 80%, 90%, and 100%; 10 min each). After the sample was sprayed with gold, the bacterial morphology was observed using a scanning electron microscope (Hitachi S-4800, Japan).
2.11. Transmission electron microscopy
The ultrastructural changes in the bacteria after felodipine treatment were examined using Transmission electron microscope (TEM), as previously described [29]. Briefly, log-phased bacterial cultures were treated with felodipine (8 μg/mL), a combination of 1/8 MIC felodipine (1 μg/mL) and 1/8 MIC gentamicin (4 μg/mL), or 0.1% DMSO (control) for 8 h at 37 °C. Next, the bacterial suspension was centrifuged and resuspended in 1 mL of a fixative solution consisting of 2.5% glutaraldehyde and 5% formaldehyde. Then, fixed bacteria were washed three times with 0.1 M cacodylate buffer and post-fixed with 1% osmium tetroxide for 1 h. The bacteria were washed three times with water and further dehydrated with an alcohol gradient series (10 min each: 50%, 60%, 70%, 80%, 90%, and 100%). Subsequently, the samples were infiltrated with Epon™ resin and polymerized at 75 °C for 48 h. Ultrathin sections were cut with a diamond knife, picked up on a copper grid, and stained with lead citrate. Micrographs of the cells were observed using a JEM 1011 TEM (JEOL, Tokyo, Japan).
2.12. Membrane potential assay
A membrane potential kit (B34950, Thermo Fisher) was used to assess changes in the proton-motive force following the manufacturer's instructions. Briefly, 106 CFU/mL of log phase bacteria were incubated with CCCP (positive control), felodipine (8 μg/mL), or a combination of 1/8 MIC felodipine (1 μg/mL) and 1/8 MIC gentamicin (4 μg/mL) for 1 h. The bacterial suspensions were then collected, resuspended in sterile PBS containing DiOC2 (30 μM), and stained at 37 °C for 30 min. The stained bacteria were detected by flow cytometry and CLSM. When bacteria have a large membrane potential, DiOC2 dye molecules self-associate at higher cytosolic concentrations and fluoresce at 630 nm (red). When the membrane potential is disrupted, the dye fluoresces at 530 nm (green).
2.13. Membrane permeability assay
The bacterial membrane permeability was examined as previously described [30]. Briefly, log-phased MRSA (ATCC 43300) were washed three times with PBS and adjusted to 107 CFU/mL. Next, SYTOX green dye (Molecular Probes, MA, USA) was added to the diluted bacterial suspension to a final concentration of 5 μM and incubated for 30 min at 37 °C in the dark. Then, 50 μL of the bacterial/SYTOX green mixture was added to 50 μL of the indicated concentrations of felodipine solution in black, clear-bottom, 96-well plates (Corning, NY, USA). The fluorescence of the suspension was monitored continuously using a spectrophotometer (BioTek, ex = 485 nm, em = 525 nm). All biological experiments were performed in triplicate.
2.14. Membrane fluidity assay
Bacterial membrane fluidity was examined according to a previously described method [7]. Briefly, Laurdan was added to the log-phased bacterial cultures at a final concentration of 10 μM. After incubation at 37 °C in the dark for 10 min, the stained MRSA (ATCC 43300) were harvested by centrifugation and washed 4 times with pre-warmed buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 0.2% glucose, 1% DMF). Next, 100 μL of bacterial culture was added to each well of a black, clear-bottom, 96-well plate, which containing twice the indicated concentration of felodipine. Then, the Laurdan fluorescence intensities were measured after 1 h incubation at 37 °C in the dark using a spectrophotometer (excitation: 350 nm, emission: 435 nm and 490 nm). Laurdan GP = (I435–I490)/(I435+I490).
2.15. Minimum biofilm inhibitory concentration (MBIC) testing
The MBIC of felodipine was examined using the Biofilm Formation Assay Kit following the manufacturer's instructions. Briefly, log-phased bacterial cultures were collected and adjusted to approximately 107 CFU/mL in MHB. Next, felodipine was 2-fold serially diluted with MHB containing bacteria to a concentration of 128–0.125 μg/mL. A 180-μL mixture was added to each well of a 96-well plate and covered with a 96-peg lid (DOJINDO, Japan). After incubation at 37 °C for 24 h, the 96-peg lid was gently washed twice with sterile saline and stained with crystal violet solution for 30 min. To further quantify the biofilm mass, crystal violet was dissolved in ethanol and the OD at 595 nm was measured using a spectrophotometer. MBIC is the minimum concentration of the antibiotic required to inhibit bacterial biofilm formation, and it can be determined by observing the color changes on the peg lid.
2.16. Eradication of biofilm on titanium discs
To further explore the ability of felodipine or gentamicin to remove the biofilms from the surfaces of the metal implants, the Biofilm TestPiece Assay Kit was used following the manufacturer's instructions. Briefly, 1.8 mL/well of log-phased bacterial culture was added to a 24-well plate. The test piece holder with loaded titanium discs (commercially Ti6Al4V) was placed on a 24-well plate and incubated at 37 °C for 48 h to allow for biofilm formation. Subsequently, the titanium discs were washed twice with sterile saline and incubated with indicated concentrations of felodipine or gentamicin for 24 h at 37 °C. After gently washing once with sterile saline, the titanium discs were incubated with crystal violet solution for 30 min. To further quantify the biofilm mass, the crystal violet on the surface of the titanium discs was dissolved in ethanol and the OD values at 595 nm for the suspensions were measured using a spectrophotometer. Besides, the number of bacteria within the biofilm was calculated using spread plate method.
2.17. Persisters killing assay
According to a previously described method, stationary-phased bacteria were used to model persisters [25]. Briefly, MRSA and MRSE persisters were generated by incubating bacterial cultures at 37 °C, 220 rpm for 24h. Then, the stationary phased bacteria were centrifuged and washed three times with PBS buffer. Next, 107 CFU/mL of persisters suspension was added to the wells of a 96-well plate, which containing the indicated concentration of felodipine or gentamicin. After incubation at 37 °C, 220 rpm for specified times, 50 μL suspensions were harvested and serially diluted with sterile saline. Then, 10 μL of each dilution was spot plated in duplicate onto TSA plate and incubated at 37 °C for 24h. The colonies were counted to calculate the number of bacteria remaining in the original culture.
2.18. Data-independent acquisition-based SWATH mass spectrometry test
After treatment with 1/2 MIC felodipine (4 μg/mL) or DMSO (control) for 8 h, proteins were extracted from MRSA (ATCC 43300) strains for quantitative proteomic analysis. For each group three independent biological replicates of each strain were used. Bacterial precipitates were collected by centrifugation (10,000×g, 10 min, 4 °C) when the OD600 reached 0.6. Then, an Easy-nLC 1200 system coupled to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, Germany) was applied. Mass spectrometry experiments were performed according to a label-free strategy in the SWATH data-independent acquisition mode. For MS1, 3e6 ions were accumulated in the Orbitrap cell over a maximum injection time of 100 ms and scanned at a resolution of 120,000 at half maximum from 350 to 1650 m/z. SWATH spectra were identified by comparison with a reference spectral library obtained from traditional data-dependent acquisition experiments. Differentially expressed proteins were considered to meet the following conditions: foldchange >1.2-fold and p-value < 0.05. The UniProt database (http://www.uniprot.org/) was used to analyze the functionality of the differentially expressed proteins.
2.19. Parallel reaction monitoring (PRM) assay
To further validate the protein levels inhibited by felodipine, 30 proteins that were identified as candidate targets were examined using the PRM assay. The PRM method includes a complete MS1 scan and a targeted MS2 scan with the following setup parameters: The MS1 scan was collected from 350 to 1650 m/z at a resolution of 120,000 (AGC target: 3e6; maximum injection time: 100 ms), and the MS2 scans were collected from 200 to 2000 m/z at a resolution of 30,000 (AGC target: 1e5, maximum injection time: 80 ms). The targeted peptides were isolated within a 1.2 m/z window and fragmented by high-energy collisional dissociation using a normalized collision energy of 27. The raw data were imported to the Skyline software, and the quantification results were manually inspected for each peptide of the targeted proteins.
2.20. Metabolomic analysis
The intracellular metabolites of MRSA (ATCC 43300) were extracted as previously described [31]. Briefly, after treatment with 1/2 MIC felodipine (4 μg/mL) or DMSO for 8 h, the bacterial culture was centrifuged at 140,00×g for 5 min. Next, the precipitate was washed three times with PBS and rapidly frozen with liquid nitrogen to quench the metabolism. Six biological replicates from each group were prepared for metabolon-based energy metabolism detection using a UHPLC (1290 Infinity LC, Agilent Technologies) coupled to a QTRAP (AB SCIEX 5500). For LC-MS analysis, 100 μL of acetonitrile/water (1:1, v/v) was added to the dried metabolites and vortexed vigorously for 1 min. After centrifugation at 140,00×g for 15 min, the supernatants were collected for LC-MS analysis.
2.21. Molecular docking
The chemical structure of felodipine (PubChem CID:3333) was prepared using LigPrep 3.4 software. (Schrödinger, USA). The crystal structure of Staphylococcus aureus caseinolytic protease P (ClpP) (PDB ID: 5W18) was prepared using the Protein Preparation Wizard software (Schrödinger, USA). The H pocket region of ClpP was selected to generate the receptor grid. In silico molecular docking was performed using the standard precision scoring function of Glide 5.5 software (Schrödinger, USA), and the best pose with a docking score of −3.73 kcal/mol was chosen for the interaction analysis and structural figures were generated using UCSF Chimera software.
2.22. Cytotoxicity assay
For determine the cytotoxicity of felodipine at its MIC level, the viability of HepG2 (ATCC HB-8065), human keratinocytes (HaCaT) and rat bone marrow mesenchymal stem cells (rBMSC) were examined as previously described [[32], [33], [34]]. Briefly, 100 μL of HepG2, HaCaT and rBMSC cells were inoculated into a 96-well plate at a density of 1.0 × 104/well and incubated at 37 °C 5% CO2 for 24 h to allow cells to adhere to the plate. Next, a 2-fold serial dilution of felodipine (ranging from 64 μg/ml to 0 μg/ml) were added to the 96-well plates and incubated for another 24 h. Then, the viability of mammalian cells was examined using Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) and the Viability/Cytotoxicity Kit (Invitrogen, CA, USA) following the manufacturer's instructions. Fluorescence microscopy (Nikon, Japan) was used to visualize the labeled cells. Live cells were stained with Calcein-AM (green), while dead cells were stained with Ethidium homodimer-1 (EthD-1; red).
2.23. Hemolysis assay
The hemolytic activity of felodipine was determined using a previously described procedure with slight modifications [29]. Fresh rat venal blood containing heparin was centrifuged (2000×g, 20 min) and the precipitated erythrocytes were washed five times with PBS (1:1). The erythrocytes were diluted to 5% with PBS, and 100 μL was added to 100 μL of two-fold serial dilutions of felodipine in PBS, 0.2% DMSO (negative control), or 1% Triton-X 100 (positive control) in a 96-well plate. The plate was incubated at 37 °C for 1 h and then centrifuged at 1000×g for 10 min. The supernatant (100 μL) was transferred to a new 96-well plate, and the absorbance was measured at 540 nm using a spectrophotometer. Percentage hemolysis was calculated using the following equation: (OD540nm of felodipine − OD540nm of DMSO)/(OD540nm of TritonX-100 − OD540nm of DMSO) × 100.
2.24. Murine skin and soft tissue infection model
All in vivo animal experimental procedures were approved and performed in accordance with the guidelines of the Animal Ethics Committee of Renji Hospital, Shanghai Jiao Tong University School of Medicine, China. To examine the antibacterial effects of felodipine alone, skin and soft tissue infections caused by S. aureus were conducted following a previously described protocol [[35], [36], [37]] with some modifications. Briefly, eight-week-old female CD1 ICR outbred mice (25–30 g) were purchased from Shanghai JieSiJie Laboratory Animal Co., Ltd. and randomly divided into four groups: 1) vehicle, 2) gentamicin, 3) low-dose felodipine, and 4) high-dose felodipine. To observe the dermonecrotic infection in real-time, we used the bioluminescent Staphylococcus aureus Xen29 (PerkinElmer), which possesses a stable copy of the Photorhabdus luminescens luxABCDE operon in its bacterial chromosome. Next, the log-phased bacterial culture was mixed with an equal volume of autoclaved Cytodex 3 microcarrier beads (Sigma, St. Louis, USA) in PBS buffer. Microbeads were used as a foreign material to ensure localized and uniform lesions in dermonecrotic infections [38]. After general anesthesia with ketamine and xylazine, the mouse flanks were shaved, sterilized, and 200 μL of the suspension (1 × 106 CFU/mL) was injected subcutaneously into each flank. Next, the mice were administered a single dose of gentamicin (80 mg/kg, s.c.), low dose felodipine (20 mg/kg, s.c.), high dose felodipine (40 mg/kg, s.c.), or saline after 2 h of infection. Subsequently, treatment was administered every 24 h for 7 days.
2.24.1. Abscess evaluation
Magnitude: After infection, the abscess area and volume were measured every other day in each mouse flank. The lesion site length (l) and width (w) were measured to quantify the abscess area (cm2). Then, the abscess volume (cm3) was calculated using the formula for a spherical ellipsoid: [v = (π/6) × l × w2]. Next, to noninvasively monitor bacterial burden, bioluminescence images were detected using an in vivo imaging system (IVIS; Caliper Life Sciences). Luminescence signals from regions of interest of each infection site were quantified using the IVIS Living Image software, where the total flux (number of photons/s) was calculated.
2.24.2. CFU determination
Seven days after the infection, the mice were euthanized, and each flank was aseptically dissected. The abscesses were then removed and prepared for quantitative culture. After individually homogenizing and serially diluting the suspensions in sterile PBS, the dilutions were spread onto TSA plates. Then, the plates were incubated at 37 °C for 24 h and the resulting colonies were enumerated as Log10 CFU/abscess.
2.24.3. Histological evaluation
Seven days after the infection, histological staining of the skin tissue was performed to assess the inflammatory response and bacterial load around the abscess. Briefly, aseptically dissected skin specimens were rapidly fixed in 4% paraformaldehyde, dehydrated in gradient ethanol and embedded in paraffin. After staining with hematoxylin and eosin (H&E), Gram and immunohistochemistry (IL-6, TNF-α), tissue sections were observed using a microscope (Nikon, Japan).
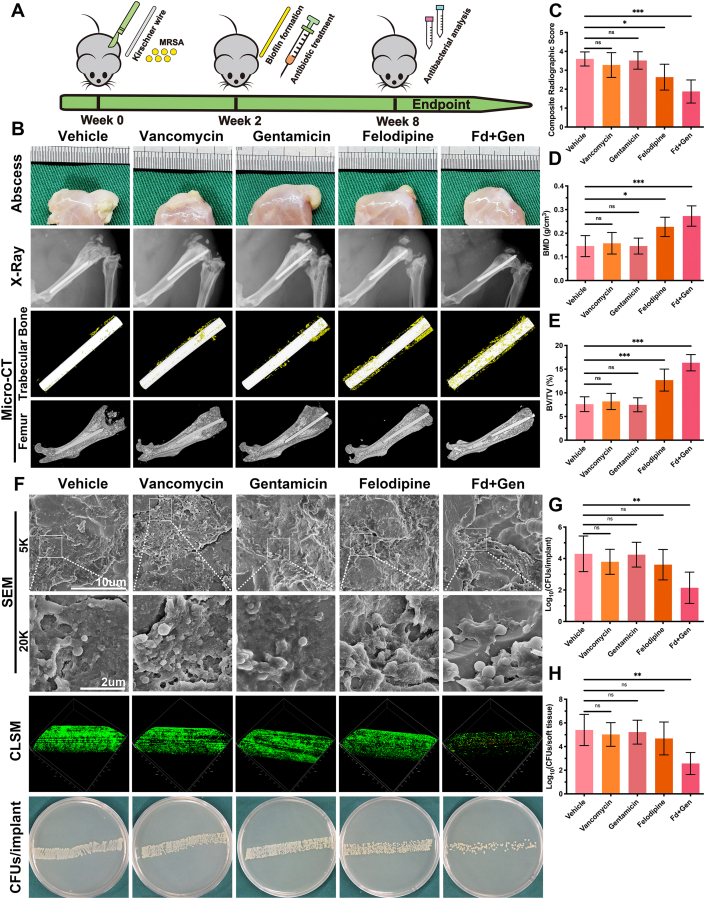
2.25. Murine periprosthetic joint infection model
To further examine the antibacterial effects of the felodipine and gentamicin against MRSA, persisters, and biofilm, a murine periprosthetic joint infection model was established as previously described [39], with some modifications. Briefly, 8-week-old CD1 ICR male mice were obtained from Shanghai JieSiJie Laboratory Animal Co., Ltd. and randomly assigned to 5 groups: 1) vehicle; 2) vancomycin; 3) gentamicin; 4) felodipine; and 5) felodipine + gentamicin. After anesthetization with ketamine and xylazine, the right knee of the mice was shaved and sterilized with iodophor. A vertical incision was made along the medial parapatellar, and the distal part of the femur was revealed after the patella was dislocated laterally. Next, the femoral medullary canal was reamed with a needle, and sterilized titanium Kirschner wire (0.5 × 9 mm) was inserted in a retrograde fashion with 1 mm protruding into the knee. Subsequently, 2 μL of MRSA suspension (103 CFU) was inoculated onto the exposed Kirschner wire in the knee, and the incision was sutured layer by layer. After a 2-week infection period to allow for biofilm formation on the Kirschner wire, a single dose of vancomycin (80 mg/kg, i.p.), gentamicin (80 mg/kg, s.c.), felodipine (40 mg/kg, s.c.), felodipine (40 mg/kg, s.c.) + gentamicin (80 mg/kg, s.c.), or saline was administered. Antibiotic therapy was administered every 24 h until 8-weeks after the surgery.
2.25.1. Radiographic evaluation
To assess the changes in bone mass, the right legs of the mice were examined using an X-ray machine and high-resolution micro-CT scanner (SkyScan 1072; Kontich, Belgium) as previously described [[40], [41], [42]]. Radiographic analysis was based on a previously described criterion [43] and scored by an observer blinded to the treatment groups. The evidence for evaluation included the following five aspects: (i) periosteal elevation, (ii) widening of the bone shaft, (iii) architectural deformation, (iv) soft tissue deformation, and (v) new bone formation. Each aspect was scored on a five-point scale (0–4), where 4 represented the most severe.
2.25.2. CFU determination
To examine the bacterial burden, the Kirschner wire and soft tissue surrounding the right knee were separately removed and immersed in sterile saline. Next, SEM and CLSM were conducted to observe the bacterial viability and biofilm distribution on the implant surface, respectively. Then, the soft tissue was individually homogenized and the Kirschner wire was sonicated for 10 min to detach the adherent bacteria. After serial dilution of the bacterial suspension with saline, 10 μL of bacterial suspension was spread onto TSA plates. Next, the plates were incubated at 37 °C for 24 h, and the number of bacteria was counted.
2.25.3. Histological evaluation
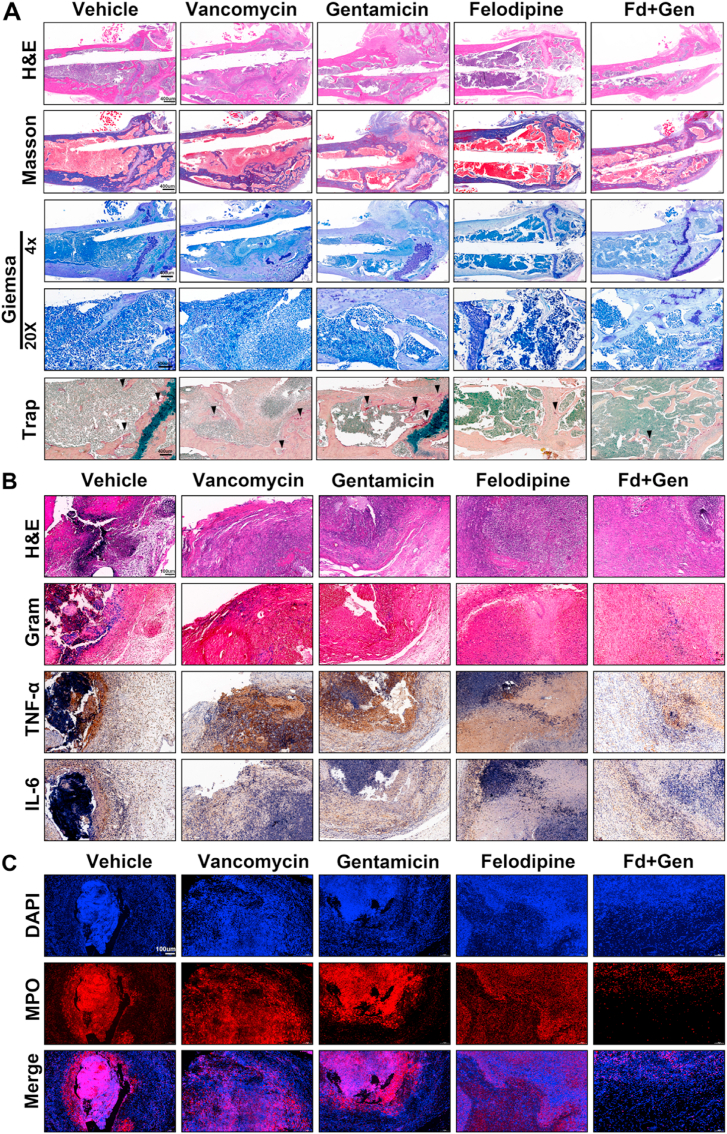
To further examine the inflammation level surrounding the infected joint, tissue sections and immunohistochemical staining were performed. Briefly, the harvested soft tissues and femurs were separately fixed in 4% paraformaldehyde for 48 h. Then the femur was decalcified with 10% EDTA solution (Servicebio, Wuhan, China) for 2 weeks and dehydrated with an alcohol gradient. After embedding in paraffin, the samples were cut at 5 μm intervals and stained with H&E, Masson, Gram, TRAP, Giemsa and immunohistochemistry (IL-6, TNF-α).
2.26. Statistical analysis
The results were analyzed using Prism 8 (GraphPad Software Inc., CA, USA), and the data are presented as the mean ± standard deviation (SD). Statistical significance was determined using an unpaired two-tailed Student's t-test when there were only two groups or by one-way ANOVA with Dunnett's or Tukey's post-hoc test if there were more than two groups. Differences between groups were considered significant at p < 0.05. (*p < 0.05, **p < 0.01, ***p < 0.001).
3. Result
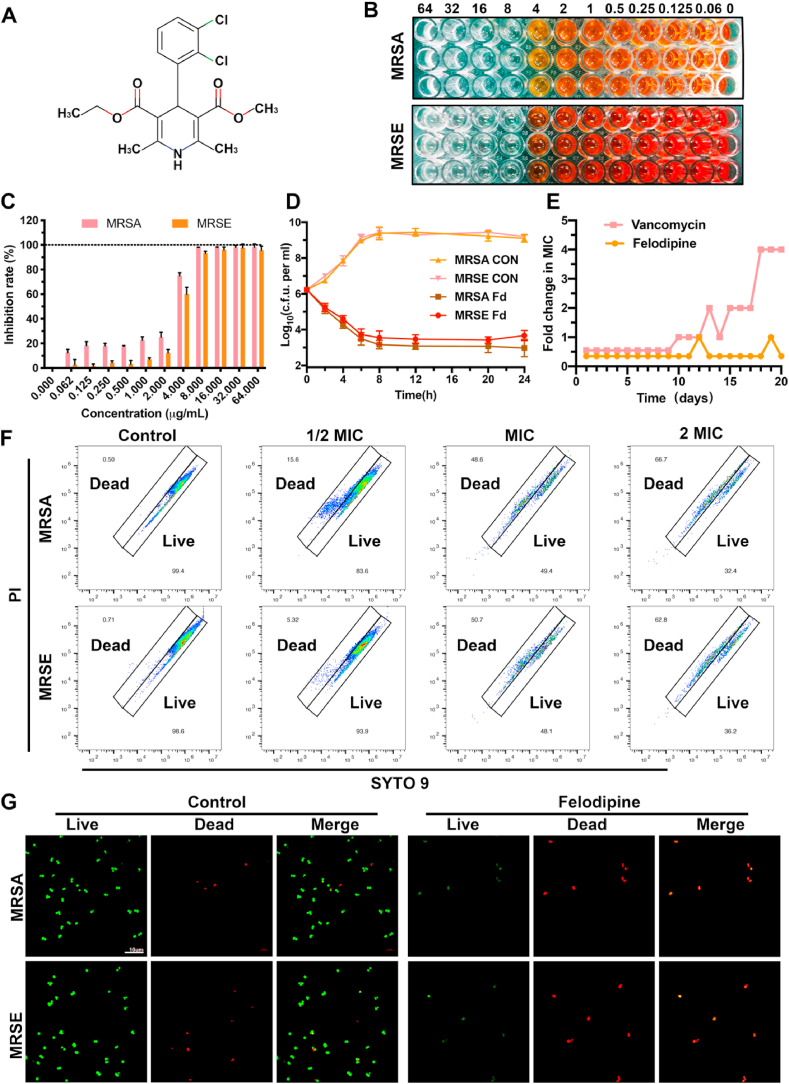
3.1. Felodipine inhibits the growth of MRSA and MRSE
The chemical structure of felodipine is shown in Fig. 2A. To further examine the antibacterial efficacy of felodipine against MSSA, MRSA, and MRSE, antimicrobial susceptibility testing was conducted. As shown in Table S1, the MIC and MBC of felodipine against MSSA, MRSA, and MRSE is 8 μg/mL and 16 μg/mL, respectively. This suggests that the antibacterial efficacy of felodipine may not be affected by the mode of bacterial resistance. However, felodipine failed to inhibit the growth of E. coli, which might be attributed to the specific extracellular membrane of Gram-negative bacteria. Next, the result of bacterial viability assay suggested that felodipine at 8 μg/ml could effectively reduce the viability of MRSE and MRSA (Fig. 2B–C). Besides, other drugs belonging to the CCB category were also examined for their antimicrobial potential, and the results are shown in Table S2. We found that felodipine had better antibacterial properties against MSSA, MRSA and MRSE compared to other drugs. Next, to examine the bactericidal kinetics of felodipine over 24 h, a time-kill kinetics assay was conducted, and the results revealed that the bacterial growth of MRSA and MRSE were both inhibited after 8 h of felodipine treatment (Fig. 2D). Additionally, resistance mutations are also an important factor that affect the antimicrobial efficacy of felodipine. By incubating MRSA at sublethal concentrations for 20 days, a lower frequency of resistance mutations was found in the felodipine solution when compared with the vancomycin group (Fig. 2E). Furthermore, the live/dead ratios of bacteria were also analyzed by flow cytometry and confocal laser scanning microscopy (CLSM), respectively. As shown in Fig. 2F–G, 2-fold MIC of felodipine induced more bacterial death when compared to other groups.
Fig. 2.
Felodipine inhibits the growth of MRSA and MRSE. (A) Chemical structure of felodipine. (B) After treated with felodipine for 24h, the bacterial viability was determined by monitoring the color intensity of WST formazan dye. (C) The bacterial inhibition rate of felodipine was determined by a colorimetric method. (D) Time-kill kinetics assay of felodipine at MIC level against MRSA and MRSE. The detection limit of the experiment was ≥ 102 CFU/mL. CON, Control; Fd, Felodipine. (E) Resistance development of MRSA after serial passaging in the presence of sub-lethal concentrations of felodipine. Vancomycin served as the control. Each point represents the average value of three independent trials. (F) After treated with different concentration of felodipine for 6h, the live/dead ratios of bacteria were examined by flow cytometry. (G) Representative confocal laser scanning microscopy (CLSM) images of bacteria after treatment with felodipine at MIC level for 6h. Live bacteria are stained green and dead bacteria are stained red.
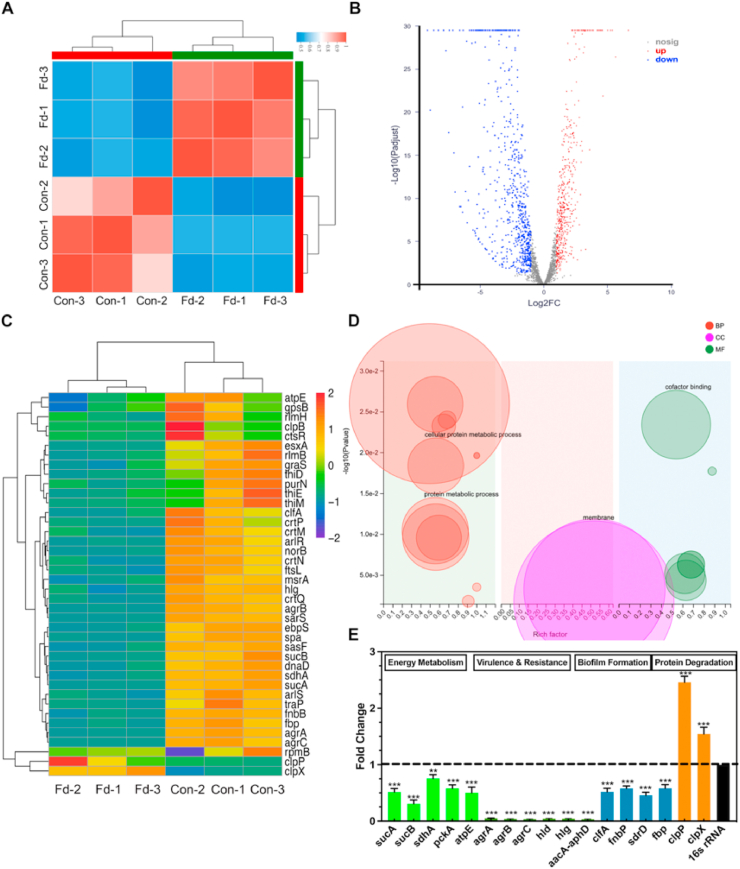
3.2. Felodipine inhibits gene expression associated with aminoglycoside resistance and biofilm formation
After treatment with felodipine, the transcriptome changes in the MRSA were examined using RNA sequencing to explore its antibacterial mechanisms. The results of the correlation analysis between the samples indicates that felodipine-treated MRSA had good biological reproducibility (Fig. 3A). As shown in Fig. 3B, the distribution of the differentially expressed genes (DEGs) was presented in the volcano map. A total of 1162 DEGs were identified after the felodipine treatment, of which 501 were upregulated and 661 were downregulated. Then, the DEGs were cluster analyzed; a heatmap include some of the important DEGs is presented in Fig. 3C. Besides, the gene ontology enrichment analysis of the DEGs showed that felodipine could affect the Go terms associated with the biological process (protein metabolic process), cellular component (membrane), and molecular function (Fig. 3D). Next, RT-PCR was performed to validate the result of RNA-seq. As shown in Fig. 3E, felodipine could reduce the gene expression associated with energy metabolism (sucA, sucB, sdhA, pckA, atpE), biofilm formation (fbp, clfA, fnbP, sdrD), aminoglycoside resistance (aacA-aphD), and bacterial virulence (agrA, agrB, agrC, hld, hlg) and increase the gene expression associated with protein degradation (clpX, clpP).
Fig. 3.
Changes in the transcriptome of MRSA treated with felodipine or DMSO (control). (A) Heatmap of correlation between samples. The different colors of the squares represent the different correlation coefficients of the two samples. (B) Volcano map of differentially expressed genes (DEGs). Each dot represents a specific gene. Red indicates significant upregulation, blue indicates significant downregulation, and gray indicates non-significant differential expression. (C) Clustering heatmap of some important DEGs. The shade of color indicates the amount of gene expression. Each group contains data for three independent samples. (D) Go enrichment analysis of DEGs. Each bubble represents a GO Term. The size of the bubble is proportional to the number of genes enriched to the GO Term. The different colors represent the three major classifications of GO. BP = biological process; CC = cellular component; MF = molecular function. (E) Validation of gene expression by RT-PCR. Felodipine inhibited the gene expression associated with energy metabolism, biofilm formation, aminoglycosides resistance, and bacterial virulence. And felodipine increased the gene expression associated protein degradation. Data analysis was performed using the comparative CT method, with 16S rRNA serving as the comparator. The results are presented as fold-changes relative to the control, which was set to a value of 1. Data are expressed as the mean ± SD. n = 3; **p < 0.01; ***p < 0.001.
3.3. Felodipine enhance gentamicin efficacy against MRSA and MRSE
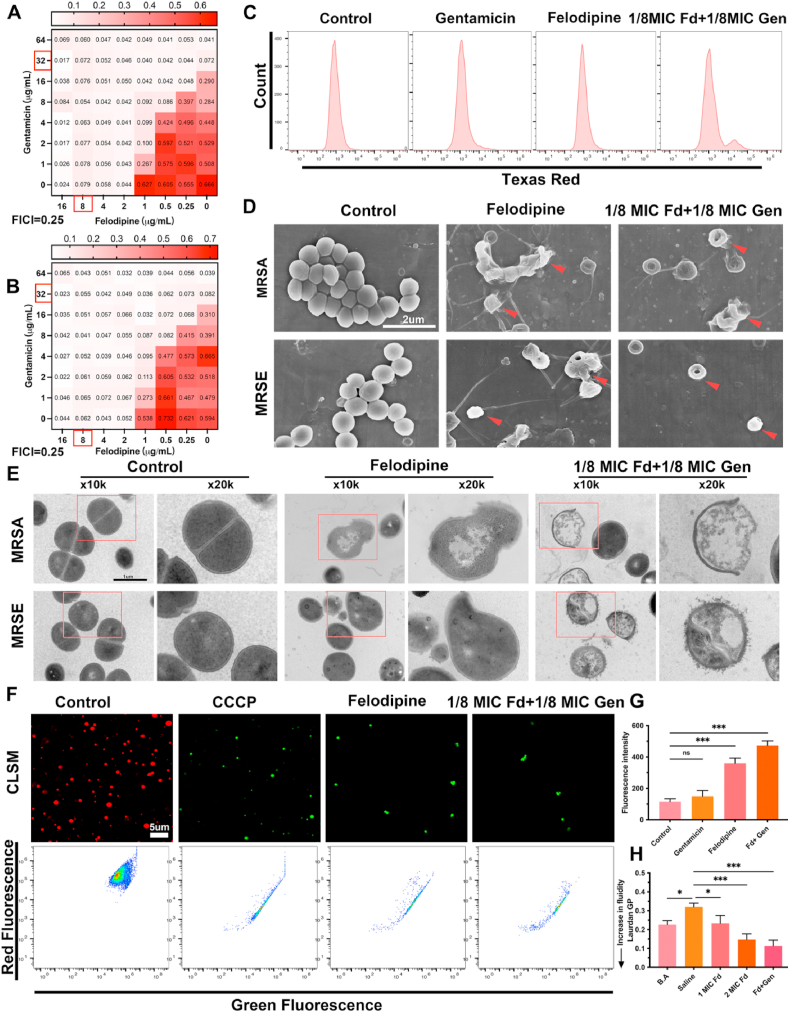
To further examine the synergistic effect of felodipine with aminoglycosides, we performed a broth microdilution checkerboard assay. As shown in Fig. 4A–B and Table S3, the value of the fractional inhibitory concentration index (FICI) was 0.25, which means that felodipine has a synergistic antibacterial effect with gentamicin. Only 1/8 MIC of felodipine (1 μg/mL) combined with 1/8 MIC of gentamicin (4 μg/mL) could inhibit the proliferation of MRSA and MRSE. Similar results were also found in other drugs belonging to aminoglycosides. This synergistic effect not only reduces the dose of both drugs, but also minimizes the side effects to some extent. Next, to investigate whether felodipine affects the uptake of gentamicin by MRSA, the Gentamicin-Texas Red conjugate was synthesized. As shown in Fig. 4C, low doses of felodipine could increase the uptake of gentamicin by the MRSA.
Fig. 4.
Felodipine enhance antibacterial efficacy of gentamicin and disrupt the ultrastructure of bacteria. (A) Synergy testing of felodipine and gentamicin against MRSA. OD600nm was measured after 18h incubation at 37 °C and fractional inhibitory concentration index (FICI) value is 0.25. Synergy, FICI ≤0.5; no interaction, 0.5 < FICI ≤4; antagonism, FICI >4. Experiments were independently repeated twice. (B) Synergy testing of felodipine and gentamicin against MRSE and the FICI value is 0.25. (C) After incubated with 1MIC felodipine, 1/8 MIC felodipine (Fd) combined with 1/8 MIC gentamicin (Gen), the Gentamicin-Texas red uptake was detected using flow cytometry. (D) After incubated with 1MIC felodipine (8 μg/mL), 1/8 MIC felodipine (Fd) and 1/8 MIC gentamicin (Gen), the morphological changes of bacteria were observed using SEM. Red arrowheads indicate bacteria with damaged structure. (E) The ultrastructure changes of bacteria were observed using TEM. The experiment was repeated three times. (F) MRSA membrane potential was examined by detecting the change of fluorescent dye, DiOC2(3), using CLSM and flow cytometer. DiOC2(3) exhibits green fluorescence in all bacterial cells, but in healthy cells its fluorescence shift to red. The proton ionophore CCCP served as positive control. (G) Bacterial membrane permeability was assessed by monitoring the uptake of SYTOX Green dye using a spectrophotometer. Data are expressed as the mean ± SD; n = 3; ***p < 0.001. (H) The membrane fluidity of MRSA was evaluated based on Laurdan generalized polarization (Laurdan GP). The benzyl alcohol (50 mM) was used as a positive control. Data are expressed as the mean ± SD; n = 3; ***p < 0.001.
3.4. Felodipine combined with gentamicin disrupt ultrastructure of MRSA and MRSE
To observe the ultrastructural changes of MRSA and MRSE treated with felodipine, SEM was performed (Fig. 4D). The bacteria in the control group showed a regular, round, and intact surface, while the bacteria in the drug treatment group had a collapsed and irregular shape. Next, the internal structures of the bacteria were observed using TEM (Fig. 4E). In the control group, the bacteria showed a circular and smooth shape with a clear septal midline within the nascent septum. However, the bacteria treated with felodipine showed “balloon-like” changes, accompanied by disruptions of the bacterial wall and membrane integrity. Next, the bacterial membrane potential was detected using the carbocyanine dye DiOC2(3), which exhibits green fluorescence in all bacteria, but shifts to a red fluorescence as the dye becomes more concentrated due to larger membrane potentials. As shown in Fig. 4F and Fig. S1, low dose of felodipine in combination with gentamicin destroyed the membrane potential of MRSA, like the positive control CCCP. Then, to further verify the membrane permeability of MRSA, SYTOX Green nucleic acid stain was used (Fig. 4G). In contrast to gentamicin alone group, 1/8 MIC felodipine combined with 1/8 MIC gentamicin exhibited a higher fluorescence intensity. Furthermore, to examine the effect of felodipine on bacterial membrane fluidity, Laurdan GP was used. As shown in Fig. 4H, 1/8 MIC felodipine combined with 1/8 MIC gentamicin could increase the fluidity of bacterial membrane, which facilitate the removal of persisters.
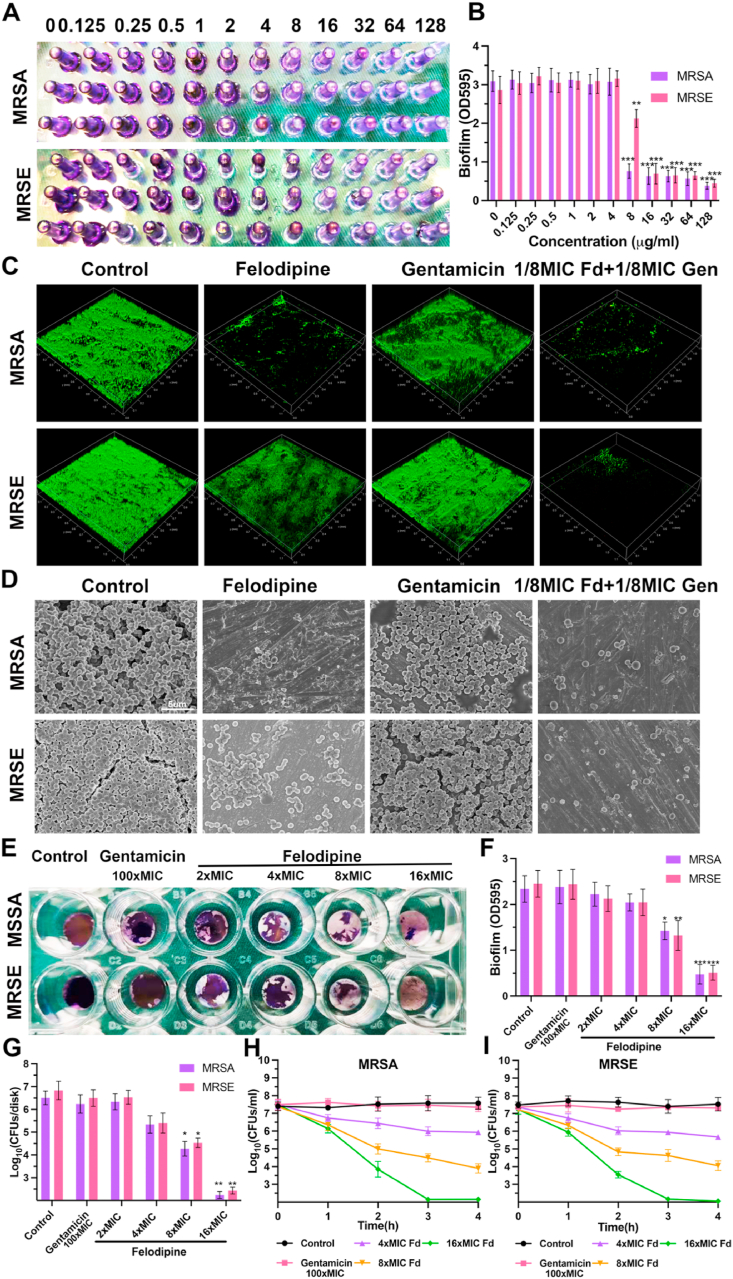
3.5. Felodipine combined with gentamicin against biofilm
Biofilm and persisters are another important factor associated with chronic implant infections. Thus, minimum biofilm inhibitory concentration (MBIC) testing was performed to evaluate the anti-biofilm efficacy of felodipine. As shown in Fig. 5A–B, felodipine at 8 μg/mL could reduce MRSA adhesion and prevent biofilm formation. While the MBIC of the felodipine for MRSE was 16 μg/mL, probably due to the strong adherence ability of Staphylococcus epidermidis. Next, to further investigate whether felodipine cold inhibit biofilm formation on the surface of medical implants (Ti6Al4V disks), live/dead bacterial staining was performed. As shown in Fig. 5C, only 1/8 MIC felodipine combined with gentamicin could effectively reduce the viability of biofilm. Besides, the result of SEM also suggests that 1/8 MIC of felodipine in combination with 1/8 MIC gentamicin could reduce the adherence of biofilm (Fig. 5D). Then, to determine whether felodipine could remove the established biofilm from Ti6Al4V disks, crystal violet staining was performed. As shown in Fig. 5E–F, 16-fold MIC of felodipine has better efficacy against established biofilms. In contrast, 100-fold MIC of gentamicin did not show the same effect. Besides, the bacterial counts within biofilms also showed the same trend (Fig. 5G). Next, persisters killing assay was performed to evaluate the bactericidal effect of felodipine against dormant bacteria. As shown in Fig. 5H–I, felodipine at its 16-fold MIC level significantly reduce the number of persisters. However, 100-fold MIC of gentamicin did not show the same antibacterial effect against persisters.
Fig. 5.
Felodipine combined with gentamicin against biofilm. (A) Minimum biofilm inhibitory concentration (MBIC) testing of felodipine against MRSA and MRSE. Biofilms was stained with crystal violet. (B) Biofilm mass was quantified by measuring sample absorbance at 595 nm using a spectrophotometer. Data are expressed as the mean ± SD; n = 3; ***p < 0.001. (C) After treated with felodipine or gentamicin, the biofilm formation on the surface of medical implant (Ti6Al4V disks) was stained with a bacterial viability kit and detected by CLSM. (D) SEM was conducted to observe the effect of felodipine or gentamicin in preventing biofilm formation. (E) Crystal violet staining was applied to examine the antibacterial efficacy of felodipine against established biofilms on the surface of implants. (F) Biofilms were quantified by measuring the absorbance of the samples at 595 nm using spectrophotometer. Data are expressed as the mean ± SD; n = 3; ***p < 0.001. (G) After treatment with different concentration of felodipine or gentamicin, the number of bacteria within the established biofilm was enumerated by the spreading plate method. Data are expressed as the mean ± SD; n = 3; ***p < 0.001. (H) After treatment with different concentration of felodipine or gentamicin, the number of MRSA persisters was counted at indicated time. (I) After treatment with felodipine or gentamicin, the number of MRSE persisters was counted at indicated time.
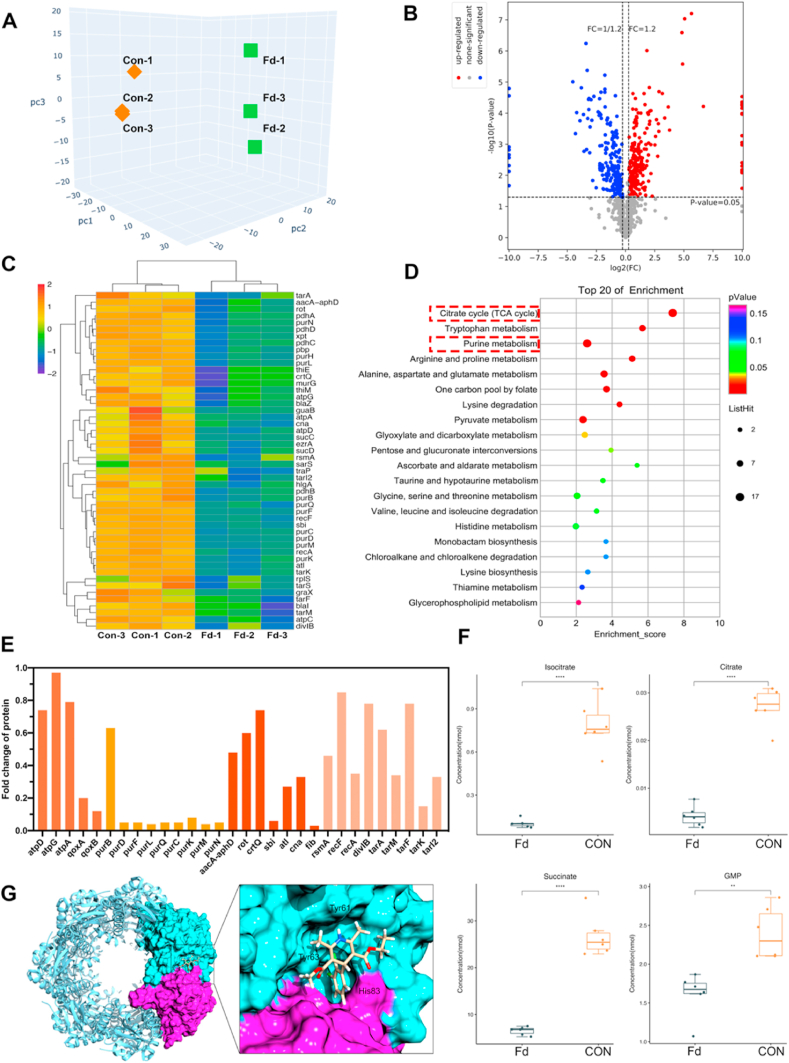
3.6. Felodipine induces proteolysis of MRSA and decreases energy metabolism
To further validate the antibacterial mechanism of felodipine from protein level, data-independent acquisition-based SWATH-MS was performed. As shown in Fig. 6A, the result of PCA analysis suggested that samples within the felodipine treatment group had good reproducibility. Next, the distribution of differentially expressed proteins (DEPs) was presented in the volcano plot (Fig. 6B). After treatment with felodipine, a total of 441 DEPs were identified, of which 192 were down-regulated and 249 were up-regulated. Then, after cluster analysis of some DEPs, we found felodipine could reduce the levels of aminoglycoside resistance proteins (aacA-aphD) and inhibit the production of virulence proteins (sbi; rot), which have a similar trend with the results of RNA-seq (Fig. 6C). Besides, KEGG enrichment analysis indicated that felodipine could inhibit the TCA cycle pathway that is a crucial component of energy metabolism (Fig. 6D). Meanwhile, the purine metabolism pathway associated with persisters infection was also inhibited. Next, to further validate the proteomics results, parallel reaction monitoring (PRM) was applied to quantify the levels of targeted DEPs. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD029484. As shown in Fig. 6E, the result of PRM indicated felodipine could reduce the level of protein associated with energy metabolism, aminoglycoside resistance, and biofilm formation, which showed consistent trends with results of data-independent acquisition mass spectrometry. Then, the TCA cycle of MRSA was further examined using targeted metabolomics. As shown in Fig. 6F and Fig. S2, compared with the control group, MRSA in the felodpine treated group had lower energy metabolites (isocitrate, citrate, succinate, GMP), which associated with TCA cycle. Besides, to determine whether felodipine could regulate the growth of persisters by activating ClpP protease, molecular docking was conducted. The structural mode in Fig. 6G and Fig. S3 suggested that felodipine could bind to the H pocket of ClpP protease and activate its hydrolysis which induce persisters death.
Fig. 6.
Felodipine induces proteolysis of MRSA and decreases energy metabolism. (A) PCA analysis of samples from each group. The relationship between samples is presented in different dimensions. Each point represents one replicate of a treatment group, with different colors for different groups. The samples within the felodipine treatment group had good similarity. (B) The distribution of differentially expressed proteins (DEPs) are presented in volcano plot. Blue dots indicate down-regulated proteins, red dots indicate up-regulated proteins. (C) Cluster heat map of some important DEPs. The protein level of aacA-aphD was reduced after treatment with felodipine. (D) KEGG pathway enrichment analysis of DEPs. The ordinate is the top 20 pathways that are significantly enriched. Pathways associated with TCA cycle, purine metabolism, arginine and proline metabolism were significantly influenced. (E) Parallel reaction monitoring was conducted to quantify the expression levels of some DEPs associated with energy metabolism, biofilm formation, aminoglycosides resistance, bacterial virulence. (F) Metabolon-based energy metabolism detection of MRSA after treatment with felodipine for 8h. Box plot shows the distribution of metabolites for each sample. Each group contains 6 biological replicates of the sample. **p < 0.01. ***p < 0.001. (G) Structural model of the ClpP complexed with Felodipine. In the close-up view, felodipine binds to the H pockets [Protein Data Bank (PDB) ID: 5 W18]. The amino acid residues involved includes Tyr61, Tyr63, and His83.
3.7. Felodipine at its MIC value has acceptable safety to mammalian cells
To evaluate the toxicity of felodipine on mammalian cells in vitro, the viability of cultured cells was examined using LIVE/DEAD staining and CCK-8 assay. As shown in Fig. S4A, felodipine at its MIC level did not induce significant cell death in contrast to control group. Besides, the evaluation of half maximal inhibitory concentration (IC50) also indicated that mammalian cells have good tolerance to MIC level of felodipine (Fig. S4B). Furthermore, EdU cell proliferation kit was used to determine the proliferation rate of mammalian cells after treated with felodipine at MIC level (Fig. S4C). As we can see, the number of green cells within the felodipine treated group did not show a significant decrease compared to the control group. Next, the result of hemolysis assay also indicated that felodipine at its MIC value did not induce erythrocyte lysis (Fig. S5).
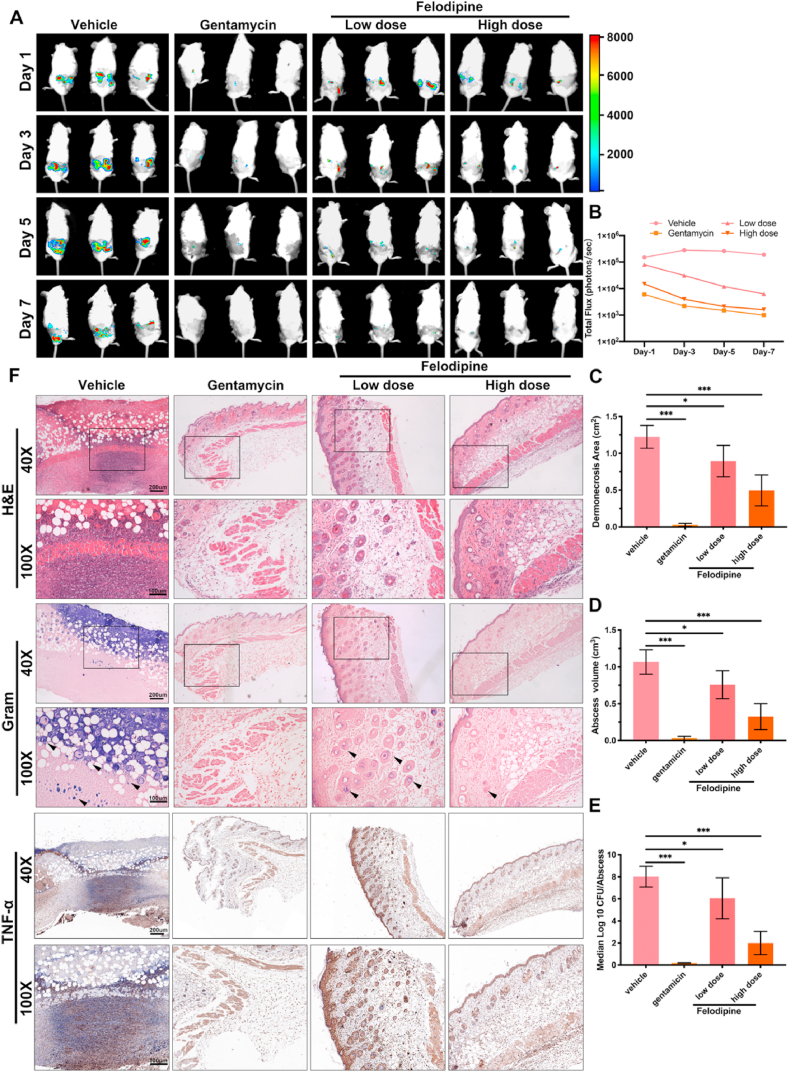
3.8. Felodipine reduces murine skin and soft tissue infections
To explore the antibacterial effects of felodipine in vivo, a mouse skin and soft tissue infection model caused by bioluminescent Staphylococcus aureus Xen29 was constructed. As shown in Fig. 7A–B, A high dose of felodipine was found to significantly reduce the fluorescence intensity at the site of infection when compared to the low doses of felodipine. Furthermore, when compared to vehicle group, both groups of felodipine alleviated the dermonecrotic area and abscess volume after 7 days of infection (Fig. 7C–D). After homogenization of infected tissues, bacterial counts also showed that felodipine reduced the number of viable bacteria in contrast to the vehicle group (Fig. 7E). Additionally, hematoxylin and eosin (H&E), Gram staining, and immunohistochemistry (IL-6, TNF-α) were conducted to detect the inflammatory responses and bacterial loads. A high dose of felodipine was found to significantly reduce the number of inflammatory cells and bacteria around the infected tissues (Fig. 7F).
Fig. 7.
Felodipine reduces murine skin and soft tissue infections caused by bioluminescent Staphylococcus aureus Xen29. (A) Bioluminescence images were observed using an in vivo imaging system after treated with low dose of felodipine (20 mg/kg, s.c.), high dose of felodipine (40 mg/kg, s.c.) or gentamicin (80 mg/kg, s.c.). (B) Luminescence signals from regions of interest of each infection sites were quantified with the IVIS Living Image software. (C) After 7 days of treatment, the dermonecrosis areas were measured. Data are expressed as mean ± SD; n = 10; *p < 0.05; ***p < 0.001. (D) After 7 days of treatment, the abscess volume was measured. Data are expressed as the mean ± SD; n = 10; *p < 0.05; ***p < 0.001. (E) After 7 days of treatment, the number of bacteria within the abscesses was quantified. Data are expressed as the means ± SD; n = 10; ns, not significant; *p < 0.05; **p < 0.01. ***p < 0.001. (F) After 7 days of treatment, histological staining of infected tissues was conducted to evaluate the inflammatory response and bacterial loads. S. aureus was stained purple in Gram-stained tissues.
3.9. Felodipine in combination with gentamicin alleviates murine periprosthetic joint infection
To evaluate the antibacterial effects of felodipine combined with gentamicin against chronic biofilm infection, murine periprosthetic joint infection caused by MRSA was constructed. The schematic diagram is shown in Fig. 8A. X-ray analysis revealed that chronic biofilm infection in the vehicle group caused a significant periosteal reaction and osteolysis, which led to the sinking of the Ti6Al4V rods (Fig. 8B–C). Next, the trabecular bone around the implant and cortical bone parameters were analyzed using micro-CT. When compared with the groups treated with vancomycin or gentamicin alone, felodipine combined with gentamicin reduced the osteolysis caused by the biofilm infection and maintained a higher bone mineral density and bone volume fraction (BV/TV) (Fig. 8D–E). Next, the bacteria on the surface of the Ti6Al4V rods were examined using SEM and CLSM. As shown in Fig. 8F, the combination of felodipine and gentamicin could reduce the adhesion and viability of MRSA on the surface of implants. However, felodipine alone did not show a superior antibiofilm effect than that of vancomycin alone or gentamicin alone. Furthermore, the bacterial counts showed that felodipine combined with gentamicin significantly reduced the number of bacteria per implant and per tissue (Fig. 8G–H). Additionally, histopathological analysis was conducted to evaluate bacterial load and inflammatory response from tissue level. As shown in Fig. 9A–C, femur and soft tissues in the combination group showed less bacterial load, inflammatory response, and neutrophil infiltration than those in the single drug group. Meanwhile, the histological staining of the heart, liver, spleen, lungs, and kidneys further suggested that the combination of felodipine and gentamicin did not cause severe organ failure (Fig. S6).
Fig. 8.
Felodipine in combination with gentamicin alleviates murine periprosthetic joint infection. (A) Schematic diagram of the treatment of implant infections in mice. (B) After treatment with felodipine (40 mg/kg, s.c.), gentamicin (80 mg/kg, s.c.), or the combination of felodipine (40 mg/kg, s.c.) and gentamicin (80 mg/kg, s.c.), the infected joint was examined using X-ray and Micro-CT to evaluate periosteal reaction, osteolysis and the position of Ti6Al4V rod. (C) Radiographic scores of infected joints. The evidence for scoring includes five aspects. Each aspect was scored on a five-point scale (0–4), where 4 represented the most severe. (D) Bone mineral density and (E) bone volume fraction (BV/TV) were measured using Micro-CT. Data are expressed as the means ± SD; n = 10; ns, not significant; **p < 0.01, ***p < 0.001. (F) Bacterial morphology and biofilm viability on the surface of Ti6Al4V rod were detected by SEM and CLSM. (G) Distribution of CFU density per implant was determined by spread plate method. Data are expressed as the means ± SD; n = 10; *p < 0.05; ***p < 0.001. (H) Distribution of CFU density per soft tissue was determined by spread plate method. Data are expressed as the means ± SD; n = 10; ns, not significant; **p < 0.01, ***p < 0.001.
Fig. 9.
Histological evaluation of infected femur and soft tissues. (A) After decalcifying the femur, H&E and Masson staining were used to evaluate the inflammatory response of the bone. Giemsa and Trap staining were used to observe the number of bacteria and osteoclasts, respectively. (B) Soft tissues around the infected joint were stained with H&E and Gram for evaluating bacteria load. MRSA was stained purple. Immunohistochemical staining of TNF-alpha, and IL-6 was performed to examine the inflammatory response within soft tissues. (C) Immunofluorescence staining of myeloperoxidase (MPO) was conducted to examine the neutrophil activity within soft tissues.
4. Discussion
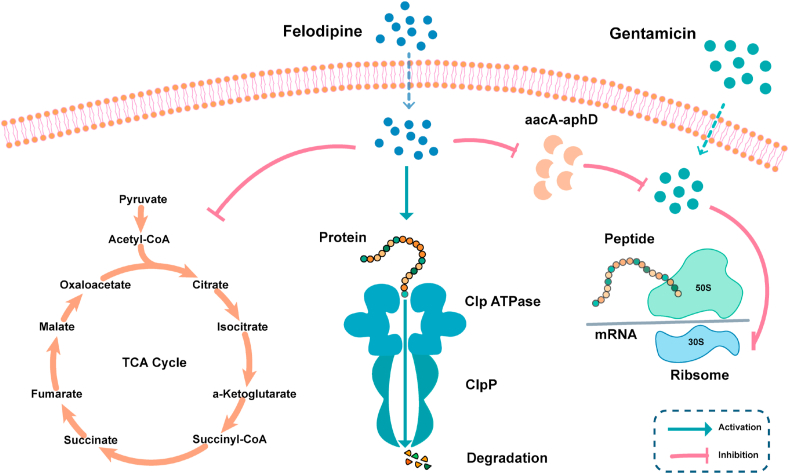
Currently, antibiotic-resistant bacteria, biofilms, and persisters are serious concerns when considering implant infections that must be addressed. This has been made difficult, however, by the arduous and costly processes involved in developing novel antibiotics which has consequently contributed to the reduction in clinically available antibiotics [16,44]. Drug repurposing screens and synergistic drug combinations offer a creative strategy by which to develop treatments for drug-resistant bacterial infections. In this study, we report for the first time that felodipine, a dihydropyridine calcium antagonist, has antibacterial effects against MRSA, biofilm and persisters. Even after continuous exposure to sub-lethal concentrations of felodipine, bacteria are less likely to develop resistance mutations to felodipine. As shown in Fig. 10, a low dose of felodipine could reduce the protein level associated with aminoglycoside resistance (aacA-aphD), thereby enhancing the antibacterial activity of gentamicin against MRSA, and reducing its MIC value to 1/8. Furthermore, felodipine could bind to the H pocket of the ClpP protease and activate its ability to regulate the growth of persisters. Additionally, a murine infection model indicated that felodipine combined with gentamicin ameliorated implant infections caused by MRSA.
Fig. 10.
Schematic diagram showing the antibacterial mechanism of felodipine in combination with gentamicin against MRSA and persisters. Low dose of felodipine reduce the TCA cycle and protein level of aacA-aphD. Besides, it binds to the active site of ClpP subunit, leading to protein degradation.
In recent years, clinically isolated bacteria have become frequently resistant to conventional antibiotics [45]. According to research, up to 46.7% of Staphylococcus aureus isolates are methicillin resistant in the United States [46]. Although high doses of vancomycin are a last resort that can eradicate clinically resistant bacteria, the emergence of vancomycin-resistant Staphylococcus aureus (VRSA) has further undermined the effectiveness of conventional antibiotics [47]. In this study, low doses of felodipine inhibited the gene expression of aacA-aphD and had a synergistic antibacterial effect with gentamicin. Compared to antibiotics or small molecule inhibitors, this effect not only enhanced the antibacterial effect of felodipine and gentamicin, but also reduced the doses of both drugs and side effects at the same time. Since the pharmacokinetics, pharmacodynamics, and safety of felodipine in humans are already known, the time and financial investment required for drug development could be reduced. Furthermore, felodipine could also inhibit the growth of MRSA through multiple targets, which can effectively prevent drug-resistant mutations.
Biofilm formation and persisters emergence are important factors associated with recalcitrant or recurrent implant infections. According to research, the removal of the biofilm of S. aureus requires a 600-fold increase in the concentration of sodium hypochlorite, one of the most effective antibacterial agents, when compared to the planktonic bacteria of S. aureus [48]. Furthermore, the treatment of chronic biofilm-associated infections in clinical settings often requires multiple surgical debridement's and a high dose of antibiotics. This not only causes many serious side effects for patients, such as disability, but also leads to high medical costs and extended hospital stays. In this study, we found that felodipine has a similar role to that of acyldepsipeptide antibiotic ADEP4, which is a potent activator of the ClpP protease. The felodipine-activated ClpP protease exhibits nonspecific protease activity, cleaves essential proteins, and regulates virulence, resistance, and persistence of MRSA infection. In contrast to other antibiotics that act on metabolically active bacteria, felodipine could effectively eliminate dormant persisters by regulating the metabolic processes of MRSA through ClpP protease.
Periprosthetic joint infections are a catastrophic complication after arthroplasty and lead to an elevated risk of disability in patients. According to research, more than 25% of revisions are attributed to periprosthetic joint infections, and this is expected to increase [49]. Although antibiotic-loaded bone cement has been used in primary total knee arthroplasty, its ability to reduce periprosthetic joint infections remains controversial [50]. It is estimated that up to 40% of S. epidermidis and 32% of S. aureus strains isolated from implant infections are resistant to gentamicin, which is a common antibiotic loaded in bone cement [51,52]. Therefore, using gentamicin-loaded bone cement alone is no longer effective in preventing implant infections caused by drug-resistant staphylococci. In future clinical practice, felodipine combined with gentamicin might be an effective regime reducing the incidence of periprosthetic joint infections. This synergistic effect not only eradicates drug-resistant bacteria and biofilm infections, but also reduces the dosage and side effects of an individual drug. When performing primary total knee arthroplasty in patients with high risk of infection, we can attempt to locally use gentamicin-loaded bone cement combined with felodipine to achieve intra-articular concentrations exceeding the MIC thresholds. In addition, felodipine and gentamicin could be loaded into bioactive materials for targeted bacterial killing to further improve the antibacterial effect against MRSA and MRSE.
As regards the clinical problem of recalcitrant biofilm-associated infections, the combination of gentamicin and felodipine has been shown to be effective in removing the biofilm from the implant surface. It has been reported that vancomycin could not effectively eradicate biofilms even after using 100-fold MIC [7]. However, only 16-fold MIC level of felodipine could effectively remove biofilm, which provides a novel approach for the treatment of biofilm-associated infections. In the future clinical practice, gentamicin and felodipine coatings can be prepared to prevent biofilm formation on the implant surface. For implants that have already formed biofilms, we can locally inject nanoparticles loaded with both felodipine and gentamicin to remove the biofilm more effectively in a targeted manner [53]. In addition, drug-carrying nanorobots with penetrating function can also be used to increase the concentration of drugs within biofilms, thus further enhancing the efficacy of removing biofilms [54]. Of course, more drug-loaded bioactive materials might be developed to enhance the anti-biofilm efficacy of felodipine, such as 3D printed biomaterials [37].
Although the phenomenon and mechanisms of felodipine against MRSA have been explored from different perspectives, there are still some shortcomings that need to be addressed in future studies. First, the antibacterial effects of felodipine have not been examined in human tissues, and thus relevant clinical trials are required to investigate its antibacterial effects in combination with gentamicin. Second, the antibacterial efficacy of felodipine alone were not superior to vancomycin. Thus, the chemical structure of felodipine should be modified by medicinal chemistry to enhance its activity against drug-resistant bacteria. Furthermore, the specific antibacterial mechanism of felodipine is still needed to explore using CRISPR genetic editing. Therefore, we will evaluate the antibacterial effect of felodipine against MRSA which has a ClpP protease mutation in the future.
5. Conclusion
In conclusion, felodipine, a dihydropyridine calcium antagonist, was reported for the first time to have antibacterial effects against MRSA, MRSE and persisters. Bacteria were less likely to develop resistance mutations to felodipine, even after continued exposure to sub-lethal concentrations. Next, low dose of felodipine inhibited the gene expression associated with aminoglycoside resistance (aacA-aphD), thereby enhancing the antibacterial activity of gentamicin, and reducing its MIC value to 1/8. Besides, felodipine could also inhibited the TCA cycle and activate ClpP protease by binding to its H pocket, which regulated the growth of biofilm and persisters. Then, murine infection models were conducted to suggest that felodipine combined with gentamicin could alleviate implant infections caused by MRSA and persisters. Taken together, felodipine is a promising compound against MRSA, MRSE, and persisters. The multi-target antibacterial effects of felodipine might have more advantages than traditional antibiotics in the treatment of periprosthetic joint infections.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
CRediT authorship contribution statement
Shutao Zhang: Conceptualization, Methodology, Investigation, Writing – original draft, Data curation. Xinhua Qu: Conceptualization, Methodology, Validation, Investigation, Writing – review & editing, Data curation. Juyang Jiao: Methodology, Investigation, Data curation. Haozheng Tang: Investigation, Methodology. Minqi Wang: Investigation, Methodology, Data curation. You Wang: Resources, Supervision. Hongtao Yang: Supervision, Formal analysis, Writing – review & editing. Weien Yuan: Formal analysis, Methodology, Writing – review & editing. Bing Yue: Conceptualization, Methodology, Resources, Supervision, Project administration, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
S.T. Z. and X.H.Q. contributed equally to this work. This work was supported by the National Natural Science Foundation of China (Grant No. 82172464, 82172453, 81972086); National Key Research and Development Project of China (Grant No. 2020YFC1107500, 2020YFC1107503); The Shanghai Rising-Star Program (21QA1405500); Shanghai “Rising Stars of Medical Talent” Youth Development Program (Youth Medical Talents - Specialist Program) (Grant No. 2019–72); “Technology Innovation Action Plan” Key Project of Shanghai Science and Technology Commission (Grant No. 19411962800); Shanghai municipal education commission—Gaofeng clinical medicine grant support (Grant No. 20161423); NSFC Advancing Targeted Projects (RJTJ-JX-005, RJTJ22-RC-011).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.11.019.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Magill S.S., Edwards J.R., Bamberg W., Beldavs Z.G., Dumyati G., Kainer M.A., Lynfield R., Maloney M., McAllister-Hollod L., Nadle J., Ray S.M., Thompson D.L., Wilson L.E., Fridkin S.K. I. Emerging infections Program healthcare-associated, T. Antimicrobial use prevalence survey, multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 2014;370(13):1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darouiche R.O. Treatment of infections associated with surgical implants. N. Engl. J. Med. 2004;350(14):1422–1429. doi: 10.1056/NEJMra035415. [DOI] [PubMed] [Google Scholar]
- 3.Davies D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003;2(2):114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 4.Pulido L., Ghanem E., Joshi A., Purtill J.J., Parvizi J. Periprosthetic joint infection: the incidence, timing, and predisposing factors. Clin. Orthop. Relat. Res. 2008;466(7):1710–1715. doi: 10.1007/s11999-008-0209-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner N.A., Sharma-Kuinkel B.K., Maskarinec S.A., Eichenberger E.M., Shah P.P., Carugati M., Holland T.L., Fowler V.G., Jr. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat. Rev. Microbiol. 2019;17(4):203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y., Jia Y., Yang K., Li R., Xiao X., Zhu K., Wang Z. Metformin restores tetracyclines susceptibility against multidrug resistant bacteria. Adv. Sci. 2020;7(12):1902227. doi: 10.1002/advs.201902227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim W., Zou G., Hari T.P.A., Wilt I.K., Zhu W., Galle N., Faizi H.A., Hendricks G.L., Tori K., Pan W., Huang X., Steele A.D., Csatary E.E., Dekarske M.M., Rosen J.L., Ribeiro N.Q., Lee K., Port J., Fuchs B.B., Vlahovska P.M., Wuest W.M., Gao H., Ausubel F.M., Mylonakis E. A selective membrane-targeting repurposed antibiotic with activity against persistent methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 2019;116(33):16529–16534. doi: 10.1073/pnas.1904700116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart P.S., Franklin M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008;6(3):199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 9.Olsen I. Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34(5):877–886. doi: 10.1007/s10096-015-2323-z. [DOI] [PubMed] [Google Scholar]
- 10.Fisher R.A., Gollan B., Helaine S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017;15(8):453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 11.Defraine V., Fauvart M., Michiels J. Fighting bacterial persistence: current and emerging anti-persister strategies and therapeutics. Drug Resist. Updates. 2018;38:12–26. doi: 10.1016/j.drup.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Ventola C.L. The antibiotic resistance crisis: part 2: management strategies and new agents. P T. 2015;40(5):344–352. [PMC free article] [PubMed] [Google Scholar]
- 13.Fu J., Li Y., Zhang Y., Liang Y., Zheng Y., Li Z., Zhu S., Li C., Cui Z., Wu S. An engineered pseudo-macrophage for rapid treatment of bacteria-infected osteomyelitis via microwave-excited anti-infection and immunoregulation. Adv. Mater. 2021;33(41) doi: 10.1002/adma.202102926. [DOI] [PubMed] [Google Scholar]
- 14.Zeng J.K., Wang Y.T., Sun Z.Y., Chang H.S., Cao M., Zhao J., Lin K.L., Xie Y.Z. A novel biocompatible PDA/IR820/DAP coating for antibiotic/photodynamic/photothermal triple therapy to inhibit and eliminate Staphylococcus aureus bio film. Chem. Eng. J. 2020;394 [Google Scholar]
- 15.Brown D. Antibiotic resistance breakers: can repurposed drugs fill the antibiotic discovery void? Nat. Rev. Drug Discov. 2015;14(12):821–832. doi: 10.1038/nrd4675. [DOI] [PubMed] [Google Scholar]
- 16.Payne D.J., Gwynn M.N., Holmes D.J., Pompliano D.L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007;6(1):29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 17.Zheng W., Sun W., Simeonov A. Drug repurposing screens and synergistic drug-combinations for infectious diseases. Br. J. Pharmacol. 2018;175(2):181–191. doi: 10.1111/bph.13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Law G.L., Tisoncik-Go J., Korth M.J., Katze M.G. Drug repurposing: a better approach for infectious disease drug discovery? Curr. Opin. Immunol. 2013;25(5):588–592. doi: 10.1016/j.coi.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyers M., Wright G.D. Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 2019;17(3):141–155. doi: 10.1038/s41579-018-0141-x. [DOI] [PubMed] [Google Scholar]
- 20.Todd P.A., Faulds D. Felodipine. A review of the pharmacology and therapeutic use of the extended release formulation in cardiovascular disorders. Drugs. 1992;44(2):251–277. doi: 10.2165/00003495-199244020-00008. [DOI] [PubMed] [Google Scholar]
- 21.Bansal A.B., Khandelwal G. 2021. Felodipine, StatPearls. Treasure Island (FL) [PubMed] [Google Scholar]
- 22.Siddiqi F.H., Menzies F.M., Lopez A., Stamatakou E., Karabiyik C., Ureshino R., Ricketts T., Jimenez-Sanchez M., Esteban M.A., Lai L., Tortorella M.D., Luo Z., Liu H., Metzakopian E., Fernandes H.J.R., Bassett A., Karran E., Miller B.L., Fleming A., Rubinsztein D.C. Felodipine induces autophagy in mouse brains with pharmacokinetics amenable to repurposing. Nat. Commun. 2019;10(1):1817. doi: 10.1038/s41467-019-09494-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.CLSI . Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. eleventh ed. 2018. [Google Scholar]
- 24.Blaskovich M.A.T., Hansford K.A., Gong Y., Butler M.S., Muldoon C., Huang J.X., Ramu S., Silva A.B., Cheng M., Kavanagh A.M., Ziora Z., Premraj R., Lindahl F., Bradford T.A., Lee J.C., Karoli T., Pelingon R., Edwards D.J., Amado M., Elliott A.G., Phetsang W., Daud N.H., Deecke J.E., Sidjabat H.E., Ramaologa S., Zuegg J., Betley J.R., Beevers A.P.G., Smith R.A.G., Roberts J.A., Paterson D.L., Cooper M.A. Protein-inspired antibiotics active against vancomycin- and daptomycin-resistant bacteria. Nat. Commun. 2018;9(1):22. doi: 10.1038/s41467-017-02123-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim W., Zhu W., Hendricks G.L., Van Tyne D., Steele A.D., Keohane C.E., Fricke N., Conery A.L., Shen S., Pan W., Lee K., Rajamuthiah R., Fuchs B.B., Vlahovska P.M., Wuest W.M., Gilmore M.S., Gao H., Ausubel F.M., Mylonakis E. A new class of synthetic retinoid antibiotics effective against bacterial persisters. Nature. 2018;556(7699):103–107. doi: 10.1038/nature26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng T.W., Chan W.L., Lai K.M. Influence of membrane fatty acid composition and fluidity on airborne survival of Escherichia coli. Appl. Microbiol. Biotechnol. 2018;102(7):3327–3336. doi: 10.1007/s00253-018-8826-7. [DOI] [PubMed] [Google Scholar]
- 27.Sandoval R., Leiser J., Molitoris B.A. Aminoglycoside antibiotics traffic to the Golgi complex in LLC-PK1 cells. J. Am. Soc. Nephrol. 1998;9(2):167–174. doi: 10.1681/ASN.V92167. [DOI] [PubMed] [Google Scholar]
- 28.Zhang S., Tang H., Wang Y., Nie B., Yang H., Yuan W., Qu X., Yue B. Antibacterial and antibiofilm effects of flufenamic acid against methicillin-resistant Staphylococcus aureus. Pharmacol. Res. 2020;160:105067. doi: 10.1016/j.phrs.2020.105067. [DOI] [PubMed] [Google Scholar]
- 29.Le P., Kunold E., Macsics R., Rox K., Jennings M.C., Ugur I., Reinecke M., Chaves-Moreno D., Hackl M.W., Fetzer C., Mandl F.A.M., Lehmann J., Korotkov V.S., Hacker S.M., Kuster B., Antes I., Pieper D.H., Rohde M., Wuest W.M., Medina E., Sieber S.A. Repurposing human kinase inhibitors to create an antibiotic active against drug-resistant Staphylococcus aureus, persisters and biofilms. Nat. Chem. 2020;12(2):145–158. doi: 10.1038/s41557-019-0378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim W., Zou G., Pan W., Fricke N., Faizi H.A., Kim S.M., Khader R., Li S., Lee K., Escorba I., Vlahovska P.M., Gao H., Ausubel F.M., Mylonakis E. The neutrally charged diarylurea compound PQ401 kills antibiotic-resistant and antibiotic-tolerant Staphylococcus aureus. mBio. 2020;11(3) doi: 10.1128/mBio.01140-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng W., Fu T., Zhang Z., Jiang X., Xie J., Sun H., Hu P., Ren H., Zhou P., Liu Q., Long Q. L-lysine potentiates aminoglycosides against Acinetobacter baumannii via regulation of proton motive force and antibiotics uptake. Emerg. Microb. Infect. 2020;9(1):639–650. doi: 10.1080/22221751.2020.1740611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song K., Huang M., Shi Q., Du T., Cao Y. Cultivation and identification of rat bone marrow-derived mesenchymal stem cells. Mol. Med. Rep. 2014;10(2):755–760. doi: 10.3892/mmr.2014.2264. [DOI] [PubMed] [Google Scholar]
- 33.Zhang S., Qu X., Tang H., Wang Y., Yang H., Yuan W., Yue B. Diclofenac resensitizes methicillin-resistant Staphylococcus aureus to beta-lactams and prevents implant infections. Adv. Sci. 2021;8(13):2100681. doi: 10.1002/advs.202100681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qu X., Yang H., Jia B., Wang M., Yue B., Zheng Y., Dai K. Zinc alloy-based bone internal fixation screw with antibacterial and anti-osteolytic properties. Bioact. Mater. 2021;6(12):4607–4624. doi: 10.1016/j.bioactmat.2021.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan L.C., Chaili S., Filler S.G., Barr K., Wang H., Kupferwasser D., Edwards J.E., Jr., Xiong Y.Q., Ibrahim A.S., Miller L.S., Schmidt C.S., Hennessey J.P., Jr., Yeaman M.R. Nonredundant roles of interleukin-17a (IL-17A) and IL-22 in murine host defense against cutaneous and hematogenous infection due to methicillin-resistant Staphylococcus aureus. Infect. Immun. 2015;83(11):4427–4437. doi: 10.1128/IAI.01061-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeaman M.R., Filler S.G., Chaili S., Barr K., Wang H., Kupferwasser D., Hennessey J.P., Jr., Fu Y., Schmidt C.S., Edwards J.E., Jr., Xiong Y.Q., Ibrahim A.S. Mechanisms of NDV-3 vaccine efficacy in MRSA skin versus invasive infection. Proc. Natl. Acad. Sci. U. S. A. 2014;111(51):E5555–E5563. doi: 10.1073/pnas.1415610111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M., Li H., Yang Y., Yuan K., Zhou F., Liu H., Zhou Q., Yang S., Tang T. A 3D-bioprinted scaffold with doxycycline-controlled BMP2-expressing cells for inducing bone regeneration and inhibiting bacterial infection. Bioact. Mater. 2021;6(5):1318–1329. doi: 10.1016/j.bioactmat.2020.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bunce C., Wheeler L., Reed G., Musser J., Barg N. Murine model of cutaneous infection with gram-positive cocci. Infect. Immun. 1992;60(7):2636–2640. doi: 10.1128/iai.60.7.2636-2640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson J.M., Saini V., Ashbaugh A.G., Miller R.J., Ordonez A.A., Ortines R.V., Wang Y., Sterling R.S., Jain S.K., Miller L.S. Oral-only linezolid-rifampin is highly effective compared with other antibiotics for periprosthetic joint infection study of a mouse model. J. Bone Jt. Surg. Am. 2017;99(8):656–665. doi: 10.2106/JBJS.16.01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang S., Li H., Tang H., Huo S., Nie B., Qu X., Yue B. Felodipine blocks osteoclast differentiation and ameliorates estrogen-dependent bone loss in mice by modulating p38 signaling pathway. Exp. Cell Res. 2020;387(2):111800. doi: 10.1016/j.yexcr.2019.111800. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y., Teng W., Zhang Z., Zhou X., Ye Y., Lin P., Liu A., Wu Y., Li B., Zhang C., Yang X., Li W., Yu X., Gou Z., Ye Z. A trilogy antimicrobial strategy for multiple infections of orthopedic implants throughout their life cycle. Bioact. Mater. 2021;6(7):1853–1866. doi: 10.1016/j.bioactmat.2020.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao C., Zhu W., Xiang Y., Zhu Y., Shen J., Liu X., Wu S., Cheung K.M.C., Yeung K.W.K. Enhanced near-infrared photocatalytic eradication of MRSA biofilms and osseointegration using oxide perovskite-based P-N heterojunction. Adv. Sci. 2021;8(15) doi: 10.1002/advs.202002211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gillaspy A.F., Hickmon S.G., Skinner R.A., Thomas J.R., Nelson C.L., Smeltzer M.S. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 1995;63(9):3373–3380. doi: 10.1128/iai.63.9.3373-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes D., Karlen A. Discovery and preclinical development of new antibiotics. Ups. J. Med. Sci. 2014;119(2):162–169. doi: 10.3109/03009734.2014.896437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossolini G.M., Arena F., Pecile P., Pollini S. Update on the antibiotic resistance crisis. Curr. Opin. Pharmacol. 2014;18:56–60. doi: 10.1016/j.coph.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Garvin K.L., Hinrichs S.H., Urban J.A. Emerging antibiotic-resistant bacteria. Their treatment in total joint arthroplasty. Clin. Orthop. Relat. Res. 1999;369:110–123. [PubMed] [Google Scholar]
- 47.Howe R.A., Bowker K.E., Walsh T.R., Feest T.G., MacGowan A.P. Vancomycin-resistant Staphylococcus aureus. Lancet. 1998;351(9102):602. doi: 10.1016/S0140-6736(05)78597-4. [DOI] [PubMed] [Google Scholar]
- 48.Luppens S.B., Reij M.W., van der Heijden R.W., Rombouts F.M., Abee T. Development of a standard test to assess the resistance of Staphylococcus aureus biofilm cells to disinfectants. Appl. Environ. Microbiol. 2002;68(9):4194–4200. doi: 10.1128/AEM.68.9.4194-4200.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kapadia B.H., Berg R.A., Daley J.A., Fritz J., Bhave A., Mont M.A. Periprosthetic joint infection. Lancet. 2016;387(10016):386–394. doi: 10.1016/S0140-6736(14)61798-0. [DOI] [PubMed] [Google Scholar]
- 50.Namba R.S., Prentice H.A., Paxton E.W., Hinman A.D., Kelly M.P. Commercially prepared antibiotic-loaded bone cement and infection risk following cemented primary total knee arthroplasty. J. Bone Jt. Surg. Am. 2020;102(22):1930–1938. doi: 10.2106/JBJS.19.01440. [DOI] [PubMed] [Google Scholar]
- 51.Campoccia D., Montanaro L., von Eiff C., Pirini V., Ravaioli S., Becker K., Arciola C.R. Cluster analysis of ribotyping profiles of Staphylococcus epidermidis isolates recovered from foreign body-associated orthopedic infections. J. Biomed. Mater. Res. 2009;88(3):664–672. doi: 10.1002/jbm.a.32090. [DOI] [PubMed] [Google Scholar]
- 52.Campoccia D., Baldassarri L., Pirini V., Ravaioli S., Montanaro L., Arciola C.R. Molecular epidemiology of Staphylococcus aureus from implant orthopaedic infections: ribotypes, agr polymorphism, leukocidal toxins and antibiotic resistance. Biomaterials. 2008;29(30):4108–4116. doi: 10.1016/j.biomaterials.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 53.Hajipour M.J., Saei A.A., Walker E.D., Conley B., Omidi Y., Lee K.B., Mahmoudi M. Nanotechnology for targeted detection and removal of bacteria: opportunities and challenges. Adv. Sci. 2021;8(21) doi: 10.1002/advs.202100556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayorga-Martinez C.C., Zelenka J., Grmela J., Michalkova H., Ruml T., Mares J., Pumera M. Swarming aqua sperm micromotors for active bacterial biofilms removal in confined spaces. Adv. Sci. 2021;8(19) doi: 10.1002/advs.202101301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).