Abstract
Evolution of clear cell renal cell carcinoma is guided by dysregulation of hypoxia-inducible transcription factor (HIF) pathways following loss of the von Hippel-Lindau tumor suppressor protein. Renal cell carcinoma (RCC)-associated polymorphisms influence HIF–DNA interactions at enhancers of important oncogenes thereby modulating the risk of developing renal cancer. A strong signal of genome-wide association with RCC was determined for the single nucleotide polymorphism (SNP) rs4903064, located on chr14q.24.2 within an intron of DPF3, encoding for Double PHD Fingers 3, a member of chromatin remodeling complexes; however, it is unclear how the risk allele operates in renal cells. In this study, we used tissue specimens and primary renal cells from a large cohort of RCC patients to examine the function of this polymorphism. In clear cell renal cell carcinoma tissue, isolated tumor cells as well as in primary renal tubular cells, in which HIF was stabilized, we determined genotype-specific increases of DPF3 mRNA levels and identified that the risk SNP resides in an active enhancer region, creating a novel HIF-binding motif. We then confirmed allele-specific HIF binding to this locus using chromatin immunoprecipitation of HIF subunits. Consequentially, HIF-mediated DPF3 regulation was dependent on the presence of the risk allele. Finally, we show that DPF3 deletion in proximal tubular cells retarded cell growth, indicating potential roles for DPF3 in cell proliferation. Our analyses suggest that the HIF pathway differentially operates on a SNP-induced hypoxia-response element at 14q24.2, thereby affecting DPF3 expression, which increases the risk of developing renal cancer.
Keywords: renal cancer, polymorphism, HIF, enhancer, ATAC-seq
Abbreviations: ATAC, Assay for Transposase-Accessible Chromatin; BHLHE41, Basic Helix-Loop-Helix Family Member E41; CCC, Comprehensive Cancer Center; CCND1, Cyclin D1; ccRCC, Clear cell renal cell carcinoma; ChIP, Chromatin immunoprecipitation; DMOG, Dimethyloxalylglycine; DPF3, Double PHD Fingers 3; EGLN3, Egl-9 Family Hypoxia Inducible Factor 3; FAIRE, Formaldehyde-assisted isolation of regulatory elements; GLUT3, Glucose transporter 3; HIF, Hypoxia-inducible transcription factor; HRE, Hypoxia-response element; huPTC, Human urinary primary tubular cells; KIRC, Kidney Renal Clear Cell Carcinoma; PBRM1, Polybromo 1; SNP, single nucleotide polymorphism; TCGA, The Cancer Genome Atlas; TMA, Tumor microarray; VHL, von Hippel-Lindau
Development of clear cell renal cell carcinoma (ccRCC) is characterized by loss of function of the von Hippel-Lindau (VHL) tumor suppressor protein (1). Advanced tumors harbor several additional somatic mutations including frequent deletions of genes coding for chromatin modifying enzymes, but timing the order of events has pinpointed loss of VHL as the truncal mutation in ccRCC (2, 3, 4). Dysfunction of VHL leads to stabilization of the alpha subunits of hypoxia-inducible transcription factors (HIF). This can be observed already in early cancerous lesions in the kidney and triggers a broad pseudo hypoxic transcriptional program (5). The function of HIF in ccRCC has been well studied, but several aspects are still under debate. For example, data from genetic analyses and xenograft studies suggest that HIF-1α slows tumor progression, whereas HIF-2α may promote tumor growth (6, 7). This hypothesis is challenged by recent data generated in an animal model of ccRCC in which both isoforms appear to promote renal tumor development, assigning an even stronger oncogenic effect to HIF-1α than to HIF-2α (8). The presence of stage- or environmental-related effects of HIF action, which may result in deleterious or protective signals, may explain these observations. In addition, deletion of VHL in renal tubular cells leads to cell senescence indicating that additional mechanisms must operate to promote tumor formation beside HIF activation (9). In this respect, modulation of the HIF pathway by either intrinsic or extrinsic factors can cause substantial changes in the HIF transcriptional output and potentially mediate tumor progression. Genetic predisposition represents one such important intrinsic factor, and hence several germline polymorphisms have been linked to renal cancer development (10, 11, 12, 13). Functional analyses of several of these loci suggest that a great part of disease heritability in renal cancer may be explained by modulation of the quality and quantity of the HIF signaling. In this context, these polymorphisms interfere with HIF–DNA interactions at enhancers of important oncogenes such as MYC or CCND1 or may directly influence expression of HIF isoforms, e.g., genetic variation at the EPAS1 locus, which encodes for HIF-2α (11, 12, 14, 15, 16). Therefore, genetic predisposition provides an important prerequisite for analyzing HIF action in ccRCC development and may shed light on stage-specific events that guide renal tumor evolution from early lesions to full-blown tumors. Moreover, some of the effects of polymorphisms on HIF signaling have been reproduced in nondiseased renal tubular cells suggesting that additional cell type–specific mechanisms, such as epigenetic predisposition, or lineage-specific factors may operate on the HIF pathway (10, 11).
In a recent meta-analysis, one of the strongest genome-wide association study signals (p = 2.2 × 10−24) in RCC patients has been detected for rs4903064, which is located in an intronic region of the DPF3 gene at chr14q24.2 (14). Double PHD fingers 3 (DPF3) is predominantly expressed in muscle and brain tissue, but bulk and single cell RNAseq experiments have detected DPF3 expression in the kidney and tubular cells, respectively (proteinatals.org). This protein is a member of the BAF subfamily of SWI/SNF chromatin-remodeling complexes and has been shown to interact with acetylated residues of histone H3 by its double PHD finger domain during initiation of transcription (17). Increased expression of DPF3 is observed in patients with chronic lymphocytic leukemia, but little is known about the regulation of DPF3 expression in other tissues or malignancies such as the kidney or renal cancer (18). So far, genotype-expression analyses from RCC cohorts suggest that the risk allele of rs4906034 is associated with increased DPF3 expression in renal cancer, but the underlying mechanism is unknown (14).
Here we present a thorough functional analysis of how DPF3 expression is subjected to regulation by the HIF pathway in renal cells. We provide evidence that the risk allele of rs4906034 generates a hypoxia-response element (HRE) that allows HIF–DNA interactions at a ccRCC-associated intronic enhancer of DPF3 expression.
Results
Genotype- and allele-specific regulation of DPF3 expression in ccRCC
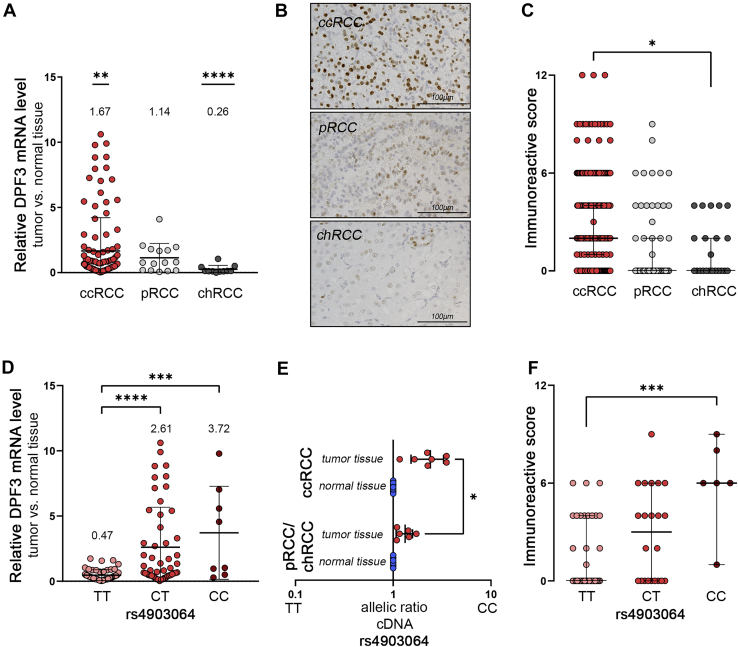
The RCC-associated single nucleotide polymorphism (SNP) rs4903064 is located in the third intron of the DPF3 gene indicating that genetic variation at this position could affect expression of DPF3 in renal cancer. To test for differential expression of DPF3 in cancer versus normal kidney tissue and in different renal cancer subtypes, we first measured expression of DPF3 mRNA in a series of 137 (111 clear cell, 15 papillary, and 11 chromophobe) renal cancer and corresponding normal tissue specimens collected at the Comprehensive Cancer Center (CCC) Erlangen-EMN. We detected increased expression of DPF3 specifically in ccRCC when compared to normal tissue (Fig. 1A). Expression of DPF3 was unchanged in papillary RCC and significantly reduced in chromophobe RCC. Overexpression of DPF3 in ccRCC is in line with data from the The Cancer Genome Atlas (TCGA) kidney renal clear cell carcinoma (KIRC) cohort when compared to other cancers including papillary and chromophobe RCC (Fig. S1). In order to examine whether augmented RNA levels also result in increased DPF3 protein expression, we established immunohistochemistry for DPF3 and evaluated DPF3 immunoreactivity in a tumor microarray (TMA) comprising 453 renal tumors. Confirming the results of our mRNA analysis, we observed overexpression of DPF3 protein in the nucleus of tumor cells in ccRCC samples when compared to the other two main renal cancer phenotypes (Fig. 1, B and C). As DPF3 is upregulated specifically in ccRCC, we hypothesized that this relates to the underlying genotype at rs4903064 (14). To explore this, we genotyped DNA samples from our CCC Erlangen-EMN cohort for rs4903064. As expected, we observed an enrichment of ccRCC patients carrying the risk allele C when compared to genotyping data from the 1000 Genomes EUR cohort as a control (Fig. S2A). Stratifying individuals in our ccRCC cohort according to the rs4903064 genotype revealed significant higher levels of DPF3 mRNA in tumors from individuals carrying at least 1 C allele (Fig. 1D). Levels of expression of an independent ccRCC-associated gene GLUT3 were not affected by the rs4903064 genotype (Fig. S2B). In individuals with non-ccRCC tumors, we could neither detect enrichment of the C allele nor a genotype-expression correlation for rs4903064 and DPF3 in the tumors (Fig. S2, A and C). To corroborate the finding that it is the risk allele that is expressed preferentially in ccRCC, we directly measured the ratio of the two alleles in pre-mRNA of DPF3 in samples from normal and tumor tissue from patients that were heterozygous for rs4903064 using a commercially available genotyping assay. We detected a significant imbalance of DPF3 pre-mRNA toward the C-allele in tumor samples from ccRCC patients when compared to normal tissue or tumor tissue from non-ccRCC patients (Fig. 1E). In accordance with these results, we measured genotype-dependent DPF3 protein expression when analyzing the immunoreactive scores (IRSs) in a subcohort of the TMA, for which genotyping results were available (Fig. 1F). From these results, we conclude that expression of DPF3 is increased in tumors from ccRCC patients carrying the risk allele C of rs4903064.
Figure 1.
Expression of DPF3 in renal cancer.A, relative expression levels of DPF3 compared to corresponding normal tissue in 111 clear cell (ccRCC), 15 papillary (pRCC), and 11 chromophobe (chRCC) tumors. Mean fold change is indicated above the values for each tumor entity. One sample t test versus values from normal tissue, ∗∗p < 0.01, ∗∗∗p < 0.001. Bars indicate mean fold change ± SD. B, immunostaining for DPF3 in the different renal tumor subtypes indicates increased nuclear staining for DPF3 in ccRCC. C, immunoreactivity scores for 306 clear cell, 44 papillary, and 22 chromophobe tumors. Bars indicate median, error bars are the 95% confidence interval. Mann–Whitney test, ∗p < 0.05. D, relative mRNA expression levels of DPF3 in ccRCC samples from (A) stratified for the rs4903064 genotype (T = protective, C = risk allele). Bars indicate mean fold expression ± SD. One-way ANOVA, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. E, allelic ratio for rs4903064 in pre-mRNA isolated from the indicated tissues (n = 7 for ccRCC, n = 6 for non-ccRCC). Each dot represents the mean of one individual determined in triplicates. Bars indicate mean values ± SD. ∗, t test, p < 0.05. F, immunoreactivity score for DPF3 in a selection of ccRCC sections from (C) stratified according to the rs4903064 genotype (TT n = 34, CT n = 22, CC n = 6). Bars indicate median, error bars are the 95% confidence interval. Mann–Whitney test, ∗∗∗p < 0.001. ccRCC, clear cell renal cell carcinoma; DPF3, double PHD fingers 3.
rs4903064 resides in an open chromatin region and creates a HIF-responsive element
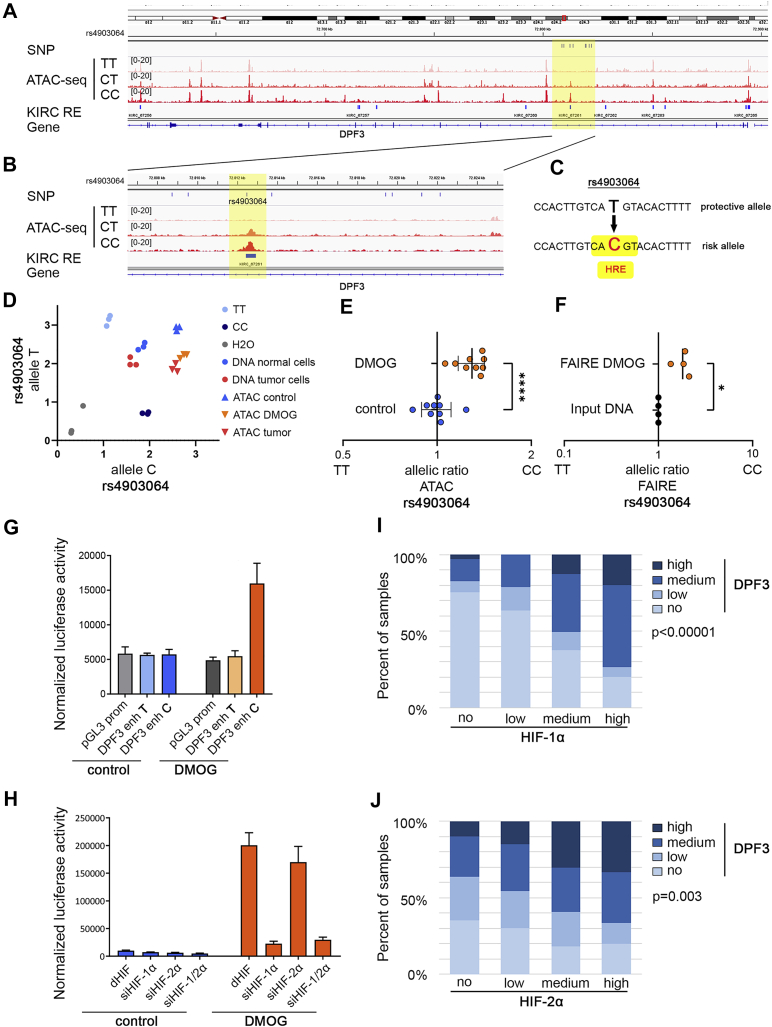
To gain further insights into how the SNP rs4903064 or related SNPs might interfere with DPF3 expression, we correlated genetic information and epigenetic data from assay for transposase-accessible chromatin (ATAC)-seq experiments, which we conducted in primary renal tumor cells. We first defined SNPs that are in high linkage disequilibrium (r2 ≥ 0.8) with rs4903064 in the EUR 1000 Genomes cohort. This resulted in seven additional SNPs (Table S1). Interestingly, out of the eight SNPs in total, only rs4903064 overlapped a region of accessible chromatin in primary renal tumor cells defined by our ATAC-seq data (Fig. 2A). We also observed that accessibility of this region is accentuated in individuals carrying the risk allele C (Fig. 2B). Furthermore, a recent comprehensive analysis of ATAC-seq data from the TCGA consortium defined this site as a regulatory region in ccRCC (KIRC_67261, Fig. 2B) (19). Thus, our findings suggest that the SNP rs4903064 resides in a regulatory DNA element, which might act as an enhancer of DPF3 expression. Upon closer inspection of the sequence around rs4903064, we observed that the risk allele C introduces a novel hypoxia response element HRE (CATGT > CACGT) into the regulatory element, thus creating a putative HIF-binding site (Fig. 2C). We were interested whether HIF was involved in generating the effect of differentially open chromatin at the regulatory element. Therefore, we resorted to ATAC and formaldehyde-assisted isolation of regulatory elements (FAIRE) samples from primary renal tumor and tubular cells isolated from individuals heterozygous for rs4903064 and performed allele-specific qPCR analyses. This revealed an allelic imbalance of chromatin accessibility toward the risk allele C at rs4903064 in tumor cells and tubular cells treated with the HIF-stabilizer dimethyloxalylglycine (DMOG) when compared to nontreated control tubular cells or input DNA (Fig. 2, D and E). Using FAIRE samples, we detected a similar shift to the C allele in open chromatin in tubular cells with stabilized HIF (Fig. 2F). These data indicate that accessibility to rs4903064 is modified upon HIF stabilization and that this effect is present in both, ccRCC and noncancerous tubular cells.
Figure 2.
rs4903064 resides in a renal cancer–associated regulatory element.A, ATAC-seq tracks from primary renal cancer cells at the DPF3 locus. The top track indicates SNPs in high LD with rs4903064 (r2 > 0.8). ATAC-seq was performed in cells from three individuals with different rs4903064 genotypes (TT, CT, CC). KIRC RE indicates regulatory elements defined by ATAC-seq in ccRCC TCGA samples. B, zoomed in view of (A). SNP rs4903064 resides in open chromatin and in KIRC_67261. Please note that open chromatin is only detected in cells carrying the risk allele C. C, sequences of the two alleles at rs4903064. The risk allele C creates a putative hypoxia-responsive element (HRE). D, allele-specific qPCR assay for rs4903064 indicates a shift toward the risk allele C in DNA fragments captured by ATAC experiments in tumor cells (red triangle) or tubular cells exposed to 1 mM DMOG (orange triangle) when compared to input DNA or DNA from ATAC experiments in untreated tubular cells. Cells were isolated from an individual heterozygous for rs4903064. E, allelic ratio of rs4903064 in ATAC samples from tubular cells isolated from three individuals heterozygous for the SNP performed in triplicates. Cells were exposed to 1 mM DMOG for 16 h or left untreated. Bars indicate mean values ± SD. t test, ∗∗∗∗p < 0.0001. F, allelic ratio of rs4903064 in FAIRE samples from cells of four heterozygous individuals. Cells were exposed to 1 mM DMOG for 16 h, and values were normalized to corresponding input DNA. The mean value for each individual is shown. Bars indicate mean values ± SD. ∗one sample t test, p < 0.05. G, reporter assay in HeLa cells using the pGL3 promoter vector with or without a 415 bp sequence covering the protective or the risk allele of rs4903064. Cells were treated with 1 mM DMOG for 16 h as indicated. Bars indicate mean values ± SD of one experiment performed in triplicates. H, HeLa cells were depleted for the indicated HIF-α isoforms using siRNA and transfected with the reporter plasmid including the risk allele. Cells were exposed to 1 mM DMOG for 16 h. Reporter activity was normalized to the activity of co-transfected β-galactosidase. Bars indicate mean values ± SD of one experiment performed in triplicates. I and J, DPF3 protein levels are high in HIF-positive ccRCC tumors. A total of 330 ccRCC tumors were stratified according to the immunoreactive score for HIF-1α, HIF-2α, and DPF3 staining in no, low, medium, or strong staining. Fractions of DPF3 signals are shown for each subgroup of HIF staining. Pearson χ2-test; p = 1.8 × 10−8 for HIF-1α (I) and p = 0.003 for HIF-2α (J). ATAC, Assay for Transposase-Accessible Chromatin; ccRCC, clear cell renal cell carcinoma; DMOG, dimethyloxalylglycine; DPF3, double PHD fingers 3; FAIRE, formaldehyde-assisted isolation of regulatory elements; HIF, hypoxia-inducible transcription factor; KIRC, kidney renal clear cell carcinoma; SNP, single nucleotide polymorphism.
To test for a differential regulatory role of the enhancer, we ligated 415 base pair sequences covering the enhancer region with the different alleles of rs4903064 into the pGL3 promoter vector and performed reporter assays in two cell lines (HeLa and MCF-7). Reporter activity was not modified by the variants under control conditions (Figs. 2G and S3A). Given the strong effect of HIF stabilization on allele-specific chromatin configuration, we also exposed the transfected cells to the HIF-stabilizer DMOG and analyzed reporter activity. Strikingly, reporter activity increased in cells transfected with plasmids carrying the risk allele C and the putative HIF-binding site (Figs. 2G and S3A). To confirm that the effect was mediated via HIF, we repeated transfection of the reporter plasmids in HeLa and MCF-7 cells, which were depleted for the different HIF isoforms using siRNA. This experiment revealed that the increase in reporter activity upon DMOG stimulation was dependent on the presence of HIF-1α (Figs. 2H and S3, B–D). Our data indicate that HIF controls expression of DPF3 in ccRCC via a regulatory element harboring an HRE created by the risk allele of rs4903064. To test for an association of DPF3 protein levels with HIF immunoreactivity in ccRCC tumors, we compared staining results for DPF3, HIF-1α, and HIF-2α in the TMA. We detected a significant correlation of HIF-1α and DPF3 immunoreactivity in ccRCC samples from the TMA supporting a possible regulatory function of HIF-1α on DPF3 expression (Pearson χ2-test; p < 0.00001, Fig. 2I). Correlation with HIF-2α staining was also significant but somewhat weaker (Pearson χ2-test; p = 0.003, Fig. 2J).
Allele-specific interaction of HIF with the DPF3 enhancer
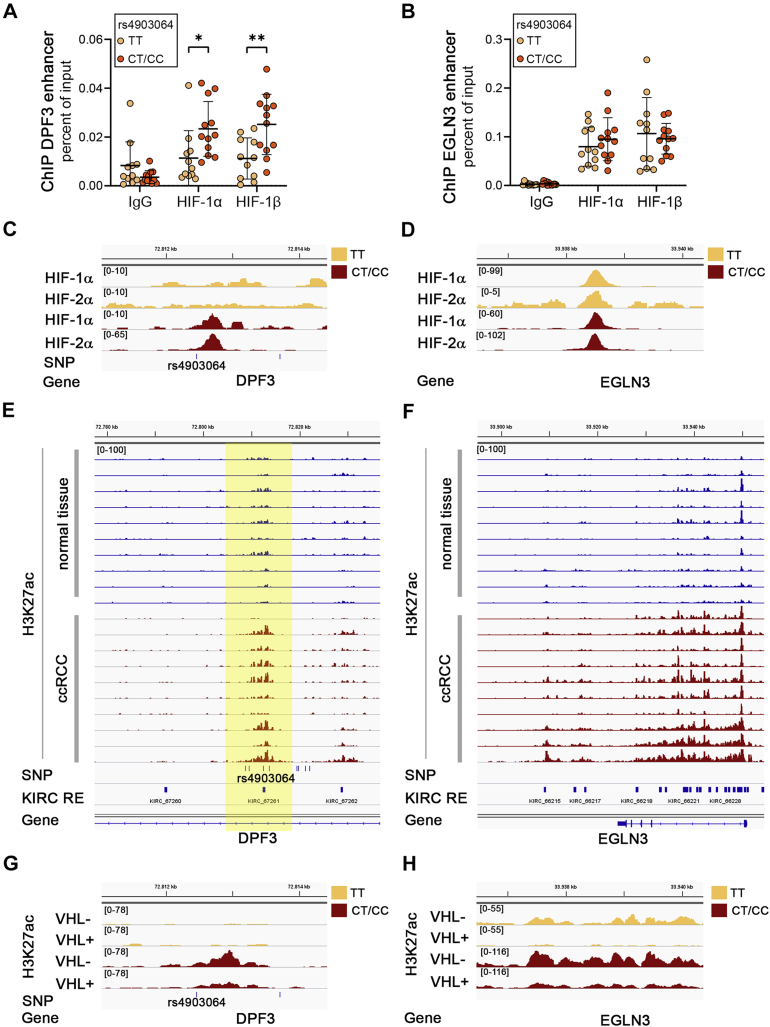
Next, we asked whether HIF directly interacts with the chromatin at the risk allele of rs4903064. We performed chromatin immunoprecipitations in primary renal tubular cells from 23 individuals with different rs4903064 allelic background (TT:11, CT:11, CC:1). The results indeed indicated interactions of HIF with this site and increased binding of HIF to the risk allele (Fig. 3A). As a control, we measured binding of HIF to the well-documented Egl-9 family hypoxia inducible factor 3 (EGLN3) enhancer and did not detect genotype-specific effects (Fig. 3B). A similar result was observed when analyzing DNA fragments captured with HIF antibodies in isolated primary tumor cells by qPCR (Fig. S4). We also reanalyzed publicly available HIF and H3K27ac chromatin immunoprecipitation (ChIP)-seq datasets published by Yao et al. and Smythies et al. from renal tumor cells and tissues for differential activity at this site (20, 21). In line with above observations, we did not observe HIF interactions at this site in the RCC4 cell line, which is homozygous for the T allele (Fig. 3C). In contrast, in HIF–ChIP sequencing data from an isolated tumor cell, we observed binding of both HIF-α subunits to the enhancer (Fig. 3C). Though we do not know the exact genotype of the donor of these cells, we exclusively detected the risk allele C in sequencing reads of the HIF ChIP-seq covering rs4903064 (data not shown). DNA interactions of HIF at the EGLN3 enhancer were comparable between the cell lines (Fig. 3D) (22, 23). Corroborating the finding that the HIF-binding site at KIRC_67261 coincides with a ccRCC-associated enhancer, we observed increased levels of the active chromatin marker H3K27ac at this site in tumors from VHL-defective ccRCC patients compared to normal tissue (Fig. 3E). This effect was similar to that seen at the EGLN3 locus (Fig. 3F).
Figure 3.
HIF binding to the RCC risk allele.A, HIF ChIP qPCR results for the DPF3 locus. Experiments were performed from cells isolated from 11 TT, 11 CT, and 1 CC individuals. Bars indicate mean values ± SD. Student’s t test, ∗p < 0.05, ∗∗p < 0.01. B, binding of HIF to the control locus at EGLN3 is not affected by the rs4903064 genotype. Bars indicate mean values ± SD. C, HIF ChIP-seq tracks in two different ccRCC cell lines at the rs4903064-associated enhancer (TT: RCC4 and CC: individual 40,911,432 from Yao et al.). HIF binding is only detectable in cells from a patient carrying the C allele (red, HIF-1α: 12 × C from 12 reads in total, HIF-2α 99 × C from 99 reads in total). D, same tracks as in (C) covering the EGLN3 control locus. E and F, H3K27ac ChIP-seq tracks from corresponding normal (blue) and ccRCC (red) tissue indicates increased activity of the disease-associated regulatory element at DPF3 (E, highlighted in yellow) and of the EGLN3 locus (F) in tumors. G, H3K27ac ChIP-seq tracks for two different ccRCC cell lines at the rs4903064-associated enhancer (TT: 786-O and CC: individual 40,911,432 from Yao et al.). Levels of the active enhancer mark H3K27ac are high in the individual carrying the C allele and reduced upon VHL re-expression. H, same tracks as in (G) covering the EGLN3 control locus. Note that H3K27ac levels are reduced upon VHL re-expression in both cell lines. ccRCC, clear cell renal cell carcinoma; ChIP, chromatin immunoprecipitation; DPF3, double PHD fingers 3; EGLN3, Egl-9 family hypoxia inducible factor 3; HIF, hypoxia-inducible transcription factor; KIRC, kidney renal clear cell carcinoma; SNP, single nucleotide polymorphism; VHL, von Hippel-Lindau.
We also reanalyzed sequencing data from Yao et al. (20). who carried out H3K27ac ChIP in renal cancer cells re-expressing empty vector or VHL to reduce HIF levels (Fig. 3, G and H). This indicated that in the individual with a CT/CC genotype, VHL expression reduced the level of chromatin activity at the DPF3 HIF-binding site suggesting that reduced HIF interactions led to decreased enhancer activity. Of note, signals for H3K27ac, a marker for active chromatin, were substantially higher in tumor cells from the CT or CC individual (Fig. 3G). At the control enhancer site within the EGLN3 gene, we observed comparable signals for H3K27ac between the different genotypes, which decreased upon VHL re-expression irrespective of the underlying rs4903064 genotype (Fig. 3H).
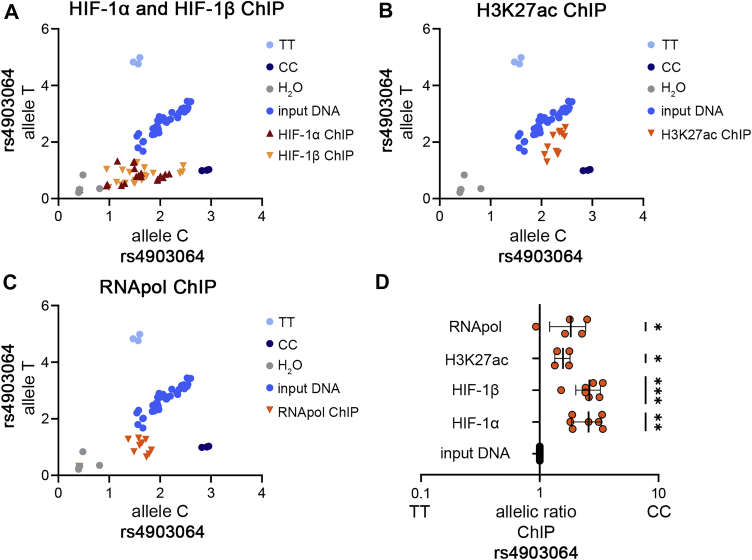
To confirm that rs4903064 modifies HIF-binding and enhancer activity in ccRCC progenitor cells, we also examined allele-specific interactions of HIF, H3K27ac, and RNApol2 in primary tubular cells heterozygous for the SNP. This revealed preferential interaction of the risk allele with HIF and RNApol2 as well as increased levels of H3K27ac at the risk allele upon HIF stimulation (Fig. 4, A–D). These findings further underline that HIF interacts with the risk allele of rs4903064 and activates the corresponding enhancer and its interaction with the transcriptional machinery.
Figure 4.
Allele specific interactions at rs4903064.A–C, allele-specific qPCR on ChIP samples generated from primary tubular cells exposed to 1 mM DMOG for 16 h. A, HIF ChIP indicates enrichment for the C allele in DNA fragments captured with HIF-1α or HIF-1β antibodies. n = 7 individuals. B, H3k27ac as a marker for active chromatin is enriched at the C allele. n = 4 individuals. C, RNApol2 is enriched at the C allele of rs4903064 indicating increased interaction with the transcriptional machinery from this allele. n = 4 individuals. D, quantification of the results in (A–C). The allelic ratio is shifted to the risk allele C, which indicated increased interaction with HIF and the transcriptional machinery as well as increased activity of this allele. Values are mean ± SD. One sample t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. ChIP, chromatin immunoprecipitation; DMOG, dimethyloxalylglycine; HIF, hypoxia-inducible transcription factor.
Allele-specific regulation of DPF3 in renal cells
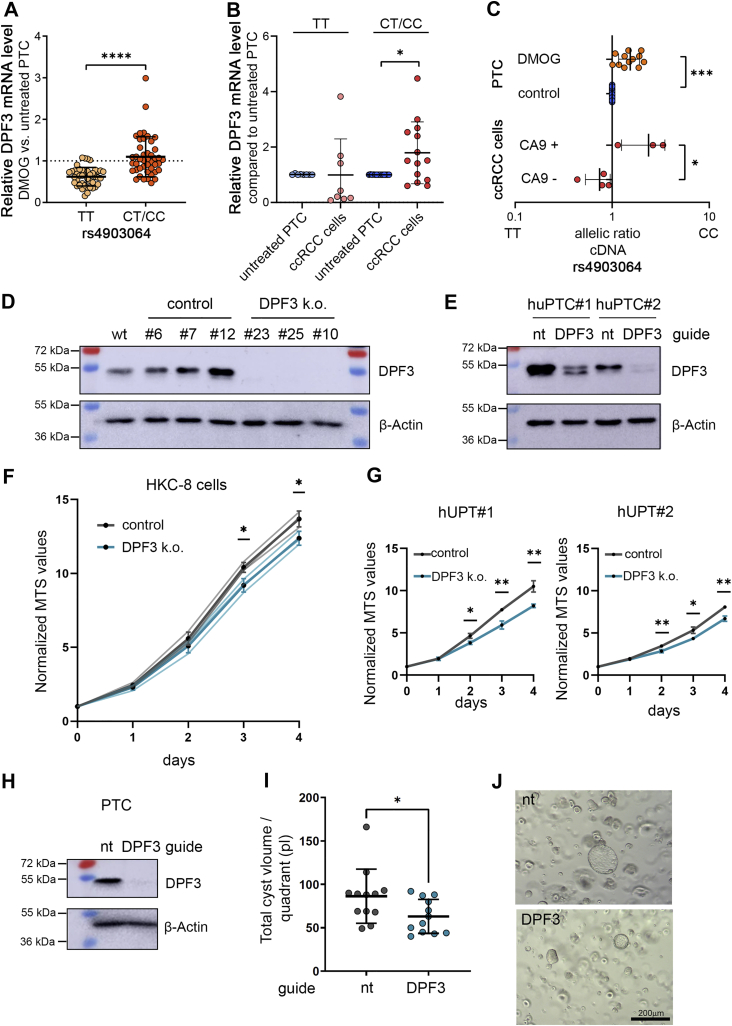
To examine functional consequences of these interactions on DPF3 expression, we measured DPF3 mRNA in primary tubular cells from our large cohort of patients (n = 86). We determined that exposure of these cells to the HIF stabilizer DMOG increased DPF3 expression in individuals carrying the risk allele C when compared to cells from individuals homozygous for the protective allele T (Fig. 5A). We did not observe HIF- or SNP-dependent regulation of genes located the broader genomic neighborhood (+/-0.5 Mb) of DPF3 related to this enhancer indicating that DPF3 is the sole target (Fig. S5). Of note, stabilization of HIF led to reduced expression of DPF3 in TT individuals, suggesting that additional regulatory mechanisms might operate on DPF3 expression when HIF becomes stabilized. Whether the rs4903064-associated enhancer also mediates this effect is unclear. Therefore, we examined chromatin configuration at the enhancer and expression of DPF3 upon DMOG stimulation in a variety of nonrenal cells. In ENCODE FAIRE-seq data, chromatin at the DPF3 enhancer has only very limited accessibility in other tissues and cell lines when compared to tubular cells (Fig. S6A). However, downregulation of DPF3 upon HIF stabilization was measured in a variety of the cell lines tested (Fig. S6B). Together with the findings from our reporter assays, in which HIF stabilization did not regulate reporter activity in cells transfected with plasmids with the protective allele, this suggests that reduction of DPF3 expression after HIF stabilization in TT individuals appears not to be mediated via the RCC-associated enhancer. Similar effects of the rs4903064 genotype on DPF3 mRNA levels were measured in explanted tumor cells when compared to untreated tubular cells (Fig. 5B). We expanded our analyses on DPF3 expression to allele-specific qPCR assays in primary tubular and ccRCC cells. We confirmed preferential expression of the risk allele in pre-mRNA from primary tubular cells exposed to DMOG compared to untreated cells. Since we consider HIF-1α as the main driver of DPF3 expression and some of the isolated tumor cells might have lost HIF-1α due to somatic mutation, we used carbonic anhydrase 9 mRNA expression as a bona fide marker for the activity of HIF-1α in these cells. Accordingly, only in cells with high levels of carbonic anhydrase 9 expression (compared to normal primary tubular cells of the same individual), we detected an allelic imbalance in DPF3 pre-mRNA expression toward the C allele. Thus, we confirmed that allele-specific regulation of DPF3 by HIF-1α is intrinsic to renal tubular derived cells and conserved in ccRCC cells.
Figure 5.
Regulation of DPF3 expression and effect of DPF3 on tubular cell growth.A, relative expression levels of DPF3 in primary renal tubular cells exposed to 1 mM of DMOG for 16 h. Samples were stratified according to the rs4903064 genotype (TT = 42, CT/CC = 44). Values were normalized to expression levels of the housekeeping gene HPRT and to values from corresponding untreated cells. Each dot represents the mean value from cells of one individual. qPCR was performed in duplicates per individual. Bars indicate mean values ± SD. Student’s t test, ∗∗∗∗p < 0.0001. B, DPF3 expression levels in isolated tumor cells compared to corresponding untreated tubular cells from 8 (TT) and 14 (CT/CC) individuals, respectively. Bars indicate mean values ± SD. One sample t test, ∗p < 0.05. C, top, allelic ratios of rs4903064 in cells from heterozygous individuals exposed to DMOG 1 mM or left untreated. One sample t test, ∗∗∗ p < 0.001. Bottom, allelic ratios of rs4903064 in ccRCC cells with high (CA9+) or low (CA9−) CA9 expression as a marker for HIF-1α activity. Values are mean from technical triplicates, and each dot represents values derived from one individual. Bars indicate mean values ± SD. t test, ∗ p < 0.05. D, Western blot for DPF3 and beta actin from lysates of different clones of HKC-8 cells with or without defective DPF3. E, Western blot for DPF3 and beta actin from lysates of two different huPTC lines transfected either with nontargeting (nt) guides or guides targeting DPF3. F, proliferation assays of the clones of cells indicated in (D). Dark lines and dots indicate the mean value from the three different clones of cells, which were tested in biological triplicates. Light gray and light blue lines indicate individual values for the three different clones of cells. G, proliferation assay from cells indicated in (E). H, Western blot for DPF3 and beta actin from lysates of primary renal tubular cells (PTC) transfected either with nontargeting (nt) guides or guides targeting DPF3. Cells were harvested at day 3 after transfection. I, cyst volume of PTC 7 days after plating in a Matrigel matrix (n = 3 wells per condition, 4 quadrants per well). J, representative images of the cysts in the Matrigel matrix. Values are mean ± SD. t test, ∗p < 0.05, ∗∗p < 0.01 for F, G, and I. CA9, carbonic anhydrase 9; ccRCC, clear cell renal cell carcinoma; DMOG, dimethyloxalylglycine; DPF3, double PHD fingers 3; huPTC, human urinary primary tubular cells.
Growth of renal tubular cells depends on DPF3
To gain more insights into how DPF3 might impinge on ccRCC development, we used CRISPR/Cas9–mediated knock-out of DPF3 in proximal renal tubular cells, from which ccRCC arise (Fig. 5, D and E). We generated clones of cells defective for DPF3 from the HKC-8 cell line (Fig. 5D) and used three different clones of cells to perform cell proliferation analyses. Cells with defective DPF3 expression grew slower than control clones of the respective cells suggesting that alterations in DPF3 expression in proximal tubular cells influence cell proliferation (Fig. 5F). Similar results were detected in pools of two proximal tubular cell lines (human urinary primary tubular cells [huPTCs]) derived from the urine. Pools of these cells transfected with guides targeting DPF3 displayed a reduced proliferation rate when compared to cells transfected with nontargeting control guides (Fig. 5G). In order to examine the effect of DPF3 manipulation on 3D-growth, we established Matrigel experiments for primary tubular cells (Fig. 5, H–J). We depleted cells for DPF3 by using CRISPR/Cas9-technology and seeded them into a Matrigel matrix. These cells form cyst-like structures in the 3D format. We used automated measurement of the perimeter of these cysts and extrapolated for volume at day 7 after plating. In concordance with the results from the 2D growth experiments, we detected a significant reduction in cystic growth in cells with a DPF3 knock-out indicating that DPF3 is relevant for spatial growth of these cells.
Taken together, we here link an RCC-associated SNP localized in the DPF3 gene with VHL-HIF regulation. Binding of HIF to the ccRCC-associated enhancer depends on the presence of the risk allele of rs4903064, which creates a de novo HRE, and leads to increased expression of DPF3, which has proproliferative effects in tubular cells.
Discussion
Our findings reveal that the risk allele of rs4903064 creates an HRE within an intronic enhancer region of the DPF3 gene. Consequently, HIF can bind to this region and activate DPF3 mRNA expression in a genotype-specific manner. Though increases in DPF3 mRNA are rather small, our data provide additional support for a model, in which genetic variation impacts on the transcriptional output of the HIF pathway, which in sum contributes substantially to development of ccRCC (24). Of note, these polymorphisms often reside in nonprotein-coding regions with accessibility restricted to renal tubular–derived cells (10, 11). In line with this, rs4903064 associates with an enhancer, which has also been identified as KIRC_67261 in a comprehensive TCGA ATAC-seq analysis (19). We observed striking differences in chromatin accessibility between genotypes and alleles in ccRCC cells and tubular cells. This effect appeared to be dependent on the presence of the C allele, i.e., a functional HRE, and stabilization of HIF. Therefore, this observation supports the hypothesis that HIF–DNA interaction is directly responsible for opening up chromatin at this site. This mechanism is unusual, since previous reports including ours have shown that HIF preferentially binds to chromatin at sites which are already accessible (25, 26). Whether additional mechanisms such as lineage-specific transcription factors guide this effect at the DPF3 locus and whether other loci behave similarly needs to be elucidated. The hypothesis of involvement of lineage-specific factors would be supported by the fact that the enhancer has only limited accessibility in nonrenal cells (Fig. S6A).
The finding that the effect of DPF3 regulation by HIF is present in undiseased tubular cells suggests that DPF3 dysregulation may contribute to malign transformation in the earliest cancerous VHL-lesions. Our data acquired in proliferation assays using proximal renal tubular cell lines and freshly isolated tubular cells imply that DPF3 has the potential to affect growth of these cells. This is in concordance with a recent study, in which overexpression of DPF3 promoted cell growth in the immortalized proximal tubular cell line HK2 (27). However, it remains to be shown whether DPF3 dysregulation is fundamentally involved throughout cancer progression or restricted to certain stages of tumor development. The fact that HIF-1α is able to activate DPF3 expression from the RCC risk allele is in line with an earlier observation from the BHLHE41 locus, at which BHLHE41 expression is also induced by the HIF-1α isoform, which preferentially binds to the risk allele (10). That HIF-1α operates at RCC risk loci is in contrast to the common hypothesis that HIF-1α acts as a tumor suppressor in ccRCC (6). In this respect, recent data from an animal model support a driver role for HIF-1α in tumor development (8). In addition, it is important to mention that deletions within chromosome 14 harboring both HIF1A and DPF3 are frequent in ccRCC (Fig. S7). The significance of this observation for the role of DPF3 and the interplay of HIF-1α with DPF3 in progressed stages of ccRCC is unclear but again may point to a relevance of DPF3 in early stages of the disease. We also propose that HIF-1α action is stage-specific and that it could indeed have protumorigenic effects in early stages of ccRCC. This hypothesis needs to be further explored in human models of renal cancer evolution.
We provide evidence for HIF-dependent upregulation of DPF3. However, in general, we observed reduced expression in nonrenal cells or in renal cells from individuals with a TT genotype upon HIF stabilization. This effect was rescued in cells with CT or CC genotypes. The observation that downregulation of DPF3 also occurs in a variety of nonrenal cells in which the regulatory element is not accessible suggests that the inhibitory effect of DMOG on DPF3 mRNA expression is not mediated via this element (Fig. S6). In line with this, we did not observe significant regulation of the reporter gene by the protective variant and HIF in the luciferase reporter assays. This indicates that at least two regulatory mechanisms operate on DPF3 expression upon HIF stabilization and that only the rs4903064-associated enhancer causes the stimulatory effect.
A recent analysis on gender-specific effects of RCC-associated polymorphisms has uncovered that rs4903064 contributes to RCC development especially in females (28). To explore this effect in our cohort, we stratified the data from expression analyses in primary tubular cells to gender of the donors but did not observe differences in DPF3 expression levels between males and females (Fig. S8). Our cohort might be too small to identify such gender-specific effects in DPF3 expression. However, this could also suggest that other mechanisms, probably independent of HIF and possibly outside the kidney, might be involved in generating the gender-specific effect.
The consequences of DPF3 dysregulation especially on the composition and function of chromatin remodeling complexes may influence renal cancer biology. It is possible that similar to mutations in other SWI/SNF complex members, upregulation of DPF3 interferes with proper function of the chromatin remodeling machinery. Interestingly, a comprehensive mass spectrometry analysis of interactions between SWI/SNF members in HAP1 cells revealed that DPF3 competes with BAF180 (encoded by PBRM1) in remodeling complexes (29). It is tempting to speculate that displacement of BAF180 from the complex might have similar effects as mutations in PBRM1 to promote renal cancer progression. In this regard, inactivation of BAF180 has been shown to increase expression of HIF target genes in ccRCC directly linking the chromatin remodeling machinery to the HIF pathway (30). However, we did not observe a difference in regulation of DPF3 mRNA expression when we stratified samples of isolated tumor cells according to the BAF180 protein status indicating that HIF-dependent DPF3 expression is not dependent on BAF180 in these cells (Fig. S9).
Our data on DPF3 gene regulation have defined the SNP-associated element as an enhancer, which interacts with the regulatory potential of the HIF system in an allele-specific manner in renal tubular cells. This provides a plausible explanation for the RCC risk caused by the rs4903064 and a solid basis for further projects to interrogate the interplay between HIF signaling and chromatin remodelers in the development of ccRCC.
Experimental procedures
Healthy kidney and tumor tissue samples
Healthy human kidney cortical tissue and tumor tissue from patients undergoing tumor nephrectomy was provided by the CCC Erlangen-EMN at the Universitätsklinikum Erlangen. The local ethics committee at the University of Erlangen-Nürnberg approved use of the tissue, and each patient gave informed consent. Tumor and normal kidney samples were examined and diagnosed by an expert pathologist. From our cohort, we used tumor tissue and corresponding normal kidney tissue from 111 clear cell, 15 papillary, and 11 chromophobe renal cancer patients for RNA analysis. Fresh frozen tissue was used to isolate total RNA using the peqGOLD Total RNA Kit (VWR Peqlab) and DNA using phenol/chloroform extraction.
Tumor microarray
Four hundred fifty-three renal tumors and corresponding normal renal tissue were collected from the archives of the Department of Pathology, University of Erlangen-Nürnberg, for tissue microarray construction. Details on TMA composition and tumor characteristics have been published previously (31). In short, archived formalin-fixed paraffin-embedded tissues were reclassified according to the 2004 World Health Organization classification of renal tumors and the 2002 TNM classification by two uropathologists. One representative punch of each tumor was transferred to a new block. Specimens were collected in accordance with the World Medical Association Declaration of Helsinki.
Immunohistochemistry
Immunostainings for HIF-1α and HIF-2α were conducted on paraffin-embedded tissue arrays as described earlier (11). For DPF3, we used a rabbit monoclonal antibody against DPF3 (E7F7N, #82788, Cell Signaling) at a dilution of 1:5000 and an antigen retrieval procedure (Dako, Agilent Technologies). Stainings were analyzed independently by two researchers and scored based on intensity of nuclear staining (0–4) and percent of positive cells (0–100%) using an IRS according to Remmele and Stegner (32). For comparison of the IRS, HIF and DPF3 stainings were divided into four categories: no (IRS = 0), low (IRS 1–3), intermediate (IRS 4–8), or high (9, 10, 11, 12) levels of immunoreactivity.
Cell culture
Most cell lines were purchased from ATCC. Caki and Kelly cells were a gift from C. Warnecke. U-87 cells were a gift from F. Müller. huPTCs were generated in the laboratory of M. Wiesener and immortalized using transfection with the SV40 large T antigen. HuPTCs were cultured in REBM medium (CC-3191, Lonza). huPTCs were maintained in REBMTM Basal Medium (CC-3191, Lonza) supplemented with REGMTM SingleQuotsTM supplements (CC-4127, Lonza), 100 U/ml penicillin, and 100 μg/ml streptomycin. huPTC#1 and huPTC#2 express high levels of NCAD, a marker for proximal renal epithelial cells. Cell lines were regularly tested for mycoplasma infection and were cultured as reported earlier (11). For U-87 cells, we used Dulbecco’s modified Eagle’s Medium high glucose, 40 U/ml penicillin, 40 μg/ml streptomycin, and 10% fetal calf serum. Healthy human kidney cortical tissue or tumor tissue from patients undergoing tumor nephrectomy was used for primary cell isolation as described previously (33). Each patient gave informed consent, and the use of human tissue was approved by the local ethics committee at the University of Erlangen-Nürnberg. Samples were minced, digested with collagenase II (Gibco, Thermo Fisher Scientific), and sieved through 100 μm and 70 μm filters. Primary human tubular cell cultures were maintained in Dulbecco’s modified Eagle’s medium/Ham’s F-12 supplemented with 2.5% (day 1) or 0% (day 2 onwards) fetal calf serum, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin, 5 μg/ml insulin, 5 μg/ml transferrin, and 5 ng/ml selenium (Sigma Aldrich), triiodothyronine (T3, Sigma Aldrich) 10 ng/ml, hydrocortisone 1 mg/ml, epidermal growth factor 100 μg/ml (Peprotech). Epithelial origin was confirmed by immunocytochemistry for N- and E-cadherin. Subconfluent cell cultures were exposed to 1 mM DMOG (Cayman Chemicals) as indicated to induce HIF.
Chromatin immunoprecipitation
ChIP experiments were performed as previously described using antibodies directed against HIF-1α (Cay10006421, rabbit polyclonal, Cayman Chemicals), HIF-1β (NB100–110, rabbit polyclonal, Novus Biologicals), RNApol 2 (SC-899, rabbit polyclonal, Santa Cruz), or H3K27ac (ab4729, rabbit polyclonal, Abcam) (11, 25). Nonimmunized rabbit serum or purified normal rabbit IgG (12–370, Merck Millipore) were used as negative controls as appropriate. ChIP-qPCR was conducted as described earlier using primers provided in Table S2 (34).
siRNA transfection
siRNAs targeting HIF-1α and HIF-2α subunits and control siRNA against drosophila HIF have been previously described (35). siRNAs were transfected at a final concentration of 40 nM using Saint red (Synvolux Therapeutics) transfection reagent.
RNA isolation
RNA from cells or tissue was isolated using peqGold total RNA kit (VWR Peqlab) according to the manufacturer's protocol and transcribed into cDNA using the high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific). qPCRs were performed on a StepOnePlus real-time PCR system (Thermo Fisher Scientific). For expression primers, please see (Table S2).
Formaldehyde-assisted isolation of regulatory elements
FAIRE experiments were performed according to Giresi et al., with the following modifications: crosslinking was performed for 5 min at room temperature using 1% formaldehyde, and DNA isolation was performed using three rounds of phenol chloroform extraction (10, 36).
Genotyping and allele-specific assays
We used a TaqMan assay (C_27898699_10, Thermo Fisher Scientific) to genotype for rs4903064. For allele-specific assays, DNA from ATAC, FAIRE, and ChIP experiments as well as cDNA was used. Genomic DNA from untreated samples from the same experiments was used in serial dilutions as an input control. DNA from cells homozygous for each allele was used as positive control. Data were analyzed using the TaqMan Genotyper Software V1.3 (Thermo Fisher Scientific). The mean ratio of minor allele/major allele (FAM/VIC) for the heterozygous input DNA derived from different dilutions was arbitrarily set to 1, and the ratios of DNA from assays (ATAC, FAIRE, ChIP, or cDNA) were normalized to the input DNA ratios. Allelic ratios of cDNA from DMOG-treated cells or tumor tissue were compared to the allelic ratio of the corresponding untreated or normal tissue control.
Genome editing
For genome editing of HKC-8 cells, gRNAs directed against the second exon of DPF3 were designed according to algorithms provided by the Zhang lab (Table S2). Guides were cloned into CRISPR/Cas9 plasmids (p × 458, #48138 Addgene). A total of 5 × 105 cells were transfected with 2 μg plasmid (4 different guides, 500 ng per guide) using Lipofectamine 3000 (Thermo Fisher Scientific). Single clones of cells were generated by dilution. For mutation screens, genomic DNA of each clone was isolated by phenol–chloroform extraction, and the region targeted by CRISPR/Cas9 was amplified by PCR. PCR products were resolved in a 15% nondenaturating polyacrylamide gel to detect indel mutations (34). Genomic DNA of clones with putative indel mutations was amplified by PCR, cloned into a pGL3-Basic vector (Promega), and analyzed by Sanger sequencing (for primers please see Table S2). Knock-out of DPF3 was verified by Western blotting. Knock-out of DPF3 in hUPT1 and hUPT2 cells was performed by transfecting the TrueCut Cas9 Protein (Thermo Fisher Scientific) together with a DPF3 targeting guide (Sigma Aldrich). A knock-out score was determined using the ICE method (ICE v2 CRISPR Analysis Tool, Synthego) and was 77% for hUPT1 and 92% for hUPT2.
Proliferation assay
Cells were plated in triplicates at 2 × 103 in 96-well tissue culture plates in 100 μl medium. An MTS CellTiter 96AQueous One Solution Cell Proliferation Assay (Promega) was conducted after 4, 24, 48, 72, or 96 h according to the manufacturer’s instructions (Promega). The number of viable cells was quantified by measuring absorbance at 490 nm using an ELISA Reader (TECAN infinite F50, TECAN) and analyzing software Magellan V7.0 (TECAN). Relative proliferation was calculated by normalizing to blank values and values measured 4 h after seeding (=day 0).
Matrigel assays
Three days after transfection of the CRISPR/Cas9 protein, cell pools treated with sgRNA targeting DPF3 or the nontarget control guides were used for the 3D-assay (3 wells per condition). In each well of a 48-well tissue culture plate, a mixture consisting of 93 μl Matrigel matrix (Standard Formulation, Corning) and 31 μl cell culture medium was dispensed and incubated for 30 min at 37 °C. Meanwhile, knockout and nontarget cell pools were trypsinized and counted. For each well, 10,000 cells were centrifuged and resuspended in 25 μl cell culture medium and 75 μl Matrigel matrix. Next, the suspension was added on top on the gel layer described above to avoid migration of cells to the bottom of the well. After incubation for 30 min at 37 °C, 1000 μl of cell culture medium was added. Medium was exchanged every 2 to 3 days. After 7 days, images were taken from each of the four quadrants per well using the Zeiss Primo Vert microscope and Zeiss Axiocam 105 camera (Zeiss Microscopy GmbH). Diameters of all captured spherical cysts were analyzed using ImageJ, and spherical cyst volumes were calculated according to 4/3πr3 (REF Nature communications).
Reporter assays
Four hundred fifteen base pair sequences covering the variant rs4903064 (hg38; chr14:72,812,429–72,812,843) were PCR amplified from genomic DNA of an individual heterozygous for the SNP and ligated into the pGL3 promoter vector (Promega) using KPNI and NHEI restriction sites. Sequences were verified using Sanger sequencing. Transfections of plasmids and a control β-galactosidase reporter were performed in HeLa or MCF-7 cells using Lipofectamine3000 (Thermo Fisher Scientific) or X-tremeGENEHP (Roche Diagnostics) DNA transfection reagent. Cells were stimulated with DMOG, applicable 16 h before harvest. Luciferase activity in extracts was measured using a Luciferase reporter assay system (Promega). Values were normalized to β-galactosidase activity in the same cell lysates. Sequences of primers are provided in Fig. S2.
Western blotting
Cells were lysed in UREA/SDS buffer, and proteins were resolved by SDS-PAGE. After transferring the proteins onto polyvinylidene difluoride membranes, proteins were detected using a rabbit polyclonal anti-HIF-1α antibody (Cay10006421, rabbit polyclonal, Cayman Chemicals) or a goat polyclonal HIF-2α antibody (AF2997, R&D Systems). We further used a rabbit monoclonal anti-DPF3 antibody (E7F7N, #82788, Cell Signaling), a mouse monoclonal anti-beta actin antibody (A3854, Sigma Aldrich), and horseradish peroxidase-conjugated secondary antibodies as applicable (Dako, Agilent Technologies).
Data analysis
Statistical analyses for RNA expression were performed using the Student’s t test, the one-sample t test comparing the mean with a hypothetical value of 1, an ordinary one-way ANOVA, or the Mann–Whitney test if applicable using GraphPadPrism, version 9.0.2 (GraphPad Software Inc). Staining intensities were compared using a Pearson χ2-test.
Assay for transposase accessible chromatin sequencing
Primary renal cancer cells from three individuals were grown under standard culture conditions and harvested at 80% confluency. Cells were trypsinized, manually counted, and 60,000 cells were directly subjected to the Omni-ATAC protocol as described by Corces et al. (37). Libraries were prepared using the Illumina Tagment DNA TDE1 Enzyme and Buffer Kit (Illumina) and purified using the DNA Clean and Concentrator-5 Kit (Zymo Research) according to the manufacturer’s recommendations. Following purification, library fragments were amplified by using the NEB Next High-Fidelity 2X PCR Master Mix (New England Biolabs) and custom made indexed/barcoded Nextera PCR primers (Sigma Aldrich) [as published by Corces et al.] (37). Libraries were amplified for a total of 8 to 12 cycles. Library clean-up and double-sided size selection was performed by using AMPure XP beads (Beckman Coulter GmbH) according to the manufacturer’s protocol. Fragment size and DNA concentration of the libraries were determined by electrophoresis using the Agilent 2100 Bioanalyzer (Agilent). Equimolar concentrations of barcoded libraries were pooled prior to sequencing.
Sequencing data analysis
Barcorded amplicons from ATAC-Library preparation were sequenced on a HiSeq 2500 (Illumina) platform to a 2 × 100 pair-ended format. Reads were quality filtered according to the standard Illumina pipeline and de-multiplexed. Fastq files were generated. Illumina adapter sequences were trimmed using Trim Galore (0.6.6). Reads were aligned against human reference genome (hg38) using Bowtie2 (2.3.4.1). SAMtools (1.8) removed aligned reads with a Mapping Quality (MAPQ) below 30 and those mapped to ChrM. PCR-duplicates reads were excluded via Picard (http://broadinstitute.github.io/picard/) (2.25.2). The Tn5 offset (“+” stranded +4 bp, “-” stranded -5 bp) were corrected in R (4.0.1) by converting the BAM files into a Genomic Ranges object via Rsamtools (2.4.0) “scanbam” and corrected by ATACseqQC (1.12.5) “shiftGAlignmentsList”. Technical replicates from the samples were merged using SAMtools. We excluded ENCODE blacklisted regions from merged files and generated normalized BigWigs for visualization using BamCoverage (3.3.2). For ChIP-seq, raw sequencing files in fastq format were generated via fastq-dump (2.8.0). Adapter sequences were trimmed using Trim Galore. Reads were mapped to the hg38 version of the human genome using Burrows-Wheeler Aligner (BWA-mem; 0.7.17-r1188). Subsequently, we filtered out reads with MAPQ of <30 and nonmitochondrial. Files were visualized using the Integrative Genomics Viewer.
Data availability
We used the following publicly available datasets: GSE86095 (ChIP-seq from renal cancer cells and tissue), GSE120887 (RCC4 HIF-ChIP-seq), GSE101064 (FAIRE-seq in primary renal tubular cells). Sequencing data were re-analyzed and mapped to hg38 as outlined above. KIRC regulatory elements from TCGA were downloaded from https://gdc.cancer.gov/about-data/publications/ATACseq-AWG. ATAC-sequencing data and RNA-sequencing data are available upon reasonable request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare no competing financial interests.
Acknowledgments
We thank Stephanie Palffy, Astrid Ebenau-Eggers, Barbara Teschemacher, Johanna Stöckert, and Margot Rehm for excellent technical assistance. We thank the Core Unit for Next Generation Sequencing and the Comprehensive Cancer Center Erlangen-EMN at the Universitätsklinikum Erlangen for their support. J. P. performed parts of this work in fulfillment of the requirements for obtaining the degree “Dr. med.” This study was funded by the Interdisciplinary Centre for Clinical Research (D35) at the Universitätsklinikum Erlangen and the German Research Foundation (Projektnummer 387509280; SFB 1350).
Author contributions
J. S. conceptualization; J. S. writing-original draft; J. S. supervision; J. S. funding acquisition, J. S., J. P., S. N., S. G., R. K., C. S., and A. K. data curation; J. P., S. N., and S. G. investigation; R. K. software; C. S. formal analysis; M. W., M. S., A. H., and B. W. resources; J. S., J. P., S. N., S. G., R. K., C. S, A. K., M. W., M. S., A. H., and B. W. writing-review and editing.
Edited by Eric Fearon
Supporting information
References
- 1.Gossage L., Eisen T., Maher E.R. VHL, the story of a tumour suppressor gene. Nat. Rev. Cancer. 2015;15:55–64. doi: 10.1038/nrc3844. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell T.J., Turajlic S., Rowan A., Nicol D., Farmery J.H.R., O'Brien T., Martincorena I., Tarpey P., Angelopoulos N., Yates L.R., Butler A.P., Raine K., Stewart G.D., Challacombe B., Fernando A., et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx renal. Cell. 2018;173:611–623.e17. doi: 10.1016/j.cell.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varela I., Tarpey P., Raine K., Huang D., Ong C.K., Stephens P., Davies H., Jones D., Lin M.L., Teague J., Bignell G., Butler A., Cho J., Dalgliesh G.L., Galappaththige D., et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandriota S.J., Turner K.J., Davies D.R., Murray P.G., Morgan N.V., Sowter H.M., Wykoff C.C., Maher E.R., Harris A.L., Ratcliffe P.J., Maxwell P.H. HIF activation identifies early lesions in VHL kidneys: Evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- 6.Shen C., Beroukhim R., Schumacher S.E., Zhou J., Chang M., Signoretti S., Kaelin W.G., Jr. Genetic and functional studies implicate HIF1alpha as a 14q kidney cancer suppressor gene. Cancer Discov. 2011;1:222–235. doi: 10.1158/2159-8290.CD-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raval R.R., Lau K.W., Tran M.G., Sowter H.M., Mandriota S.J., Li J.L., Pugh C.W., Maxwell P.H., Harris A.L., Ratcliffe P.J. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol. Cell. Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoefflin R., Harlander S., Schafer S., Metzger P., Kuo F., Schonenberger D., Adlesic M., Peighambari A., Seidel P., Chen C.Y., Consenza-Contreras M., Jud A., Lahrmann B., Grabe N., Heide D., et al. HIF-1alpha and HIF-2alpha differently regulate tumour development and inflammation of clear cell renal cell carcinoma in mice. Nat. Commun. 2020;11:4111. doi: 10.1038/s41467-020-17873-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young A.P., Kaelin W.G., Jr. Senescence triggered by the loss of the VHL tumor suppressor. Cell Cycle. 2008;7:1709–1712. doi: 10.4161/cc.7.12.6124. [DOI] [PubMed] [Google Scholar]
- 10.Grampp S., Schmid V., Salama R., Lauer V., Kranz F., Platt J.L., Smythies J., Choudhry H., Goppelt-Struebe M., Ratcliffe P.J., Mole D.R., Schodel J. Multiple renal cancer susceptibility polymorphisms modulate the HIF pathway. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grampp S., Platt J.L., Lauer V., Salama R., Kranz F., Neumann V.K., Wach S., Stohr C., Hartmann A., Eckardt K.U., Ratcliffe P.J., Mole D.R., Schodel J. Genetic variation at the 8q24.21 renal cancer susceptibility locus affects HIF binding to a MYC enhancer. Nat. Commun. 2016;7:13183. doi: 10.1038/ncomms13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schodel J., Bardella C., Sciesielski L.K., Brown J.M., Pugh C.W., Buckle V., Tomlinson I.P., Ratcliffe P.J., Mole D.R. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat. Genet. 2012;44:420–425. doi: 10.1038/ng.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmid V., Lafleur V.N., Lombardi O., Li R., Salama R., Colli L., Choudhry H., Chanock S., Ratcliffe P.J., Mole D.R. Co-incidence of RCC-susceptibility polymorphisms with HIF cis-acting sequences supports a pathway tuning model of cancer. Sci. Rep. 2019;9:18768. doi: 10.1038/s41598-019-55098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scelo G., Purdue M.P., Brown K.M., Johansson M., Wang Z., Eckel-Passow J.E., Ye Y., Hofmann J.N., Choi J., Foll M., Gaborieau V., Machiela M.J., Colli L.M., Li P., Sampson J.N., et al. Genome-wide association study identifies multiple risk loci for renal cell carcinoma. Nat. Commun. 2017;8:15724. doi: 10.1038/ncomms15724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han S.S., Yeager M., Moore L.E., Wei M.H., Pfeiffer R., Toure O., Purdue M.P., Johansson M., Scelo G., Chung C.C., Gaborieau V., Zaridze D., Schwartz K., Szeszenia-Dabrowska N., Davis F., et al. The chromosome 2p21 region harbors a complex genetic architecture for association with risk for renal cell carcinoma. Hum. Mol. Genet. 2012;21:1190–1200. doi: 10.1093/hmg/ddr551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purdue M.P., Johansson M., Zelenika D., Toro J.R., Scelo G., Moore L.E., Prokhortchouk E., Wu X., Kiemeney L.A., Gaborieau V., Jacobs K.B., Chow W.H., Zaridze D., Matveev V., Lubinski J., et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat. Genet. 2011;43:60–65. doi: 10.1038/ng.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng L., Zhang Q., Li S., Plotnikov A.N., Walsh M.J., Zhou M.M. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466:258–262. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theodorou M., Speletas M., Mamara A., Papachristopoulou G., Lazou V., Scorilas A., Katsantoni E. Identification of a STAT5 target gene, Dpf3, provides novel insights in chronic lymphocytic leukemia. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corces M.R., Granja J.M., Shams S., Louie B.H., Seoane J.A., Zhou W., Silva T.C., Groeneveld C., Wong C.K., Cho S.W., Satpathy A.T., Mumbach M.R., Hoadley K.A., Robertson A.G., Sheffield N.C., et al. The chromatin accessibility landscape of primary human cancers. Science. 2018;362 doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao X., Tan J., Lim K.J., Koh J., Ooi W.F., Li Z., Huang D., Xing M., Chan Y.S., Qu J.Z., Tay S.T., Wijaya G., Lam Y.N., Hong J.H., Lee-Lim A.P., et al. VHL deficiency drives enhancer activation of oncogenes in clear cell renal cell carcinoma. Cancer Discov. 2017;7:1284–1305. doi: 10.1158/2159-8290.CD-17-0375. [DOI] [PubMed] [Google Scholar]
- 21.Smythies J.A., Sun M., Masson N., Salama R., Simpson P.D., Murray E., Neumann V., Cockman M.E., Choudhry H., Ratcliffe P.J., Mole D.R. Inherent DNA-binding specificities of the HIF-1alpha and HIF-2alpha transcription factors in chromatin. EMBO Rep. 2019;20 doi: 10.15252/embr.201846401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau K.W., Tian Y.M., Raval R.R., Ratcliffe P.J., Pugh C.W. Target gene selectivity of hypoxia-inducible factor-alpha in renal cancer cells is conveyed by post-DNA-binding mechanisms. Br. J. Cancer. 2007;96:1284–1292. doi: 10.1038/sj.bjc.6603675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pescador N., Cuevas Y., Naranjo S., Alcaide M., Villar D., Landazuri M.O., Del Peso L. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochem. J. 2005;390:189–197. doi: 10.1042/BJ20042121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castro-Giner F., Ratcliffe P., Tomlinson I. The mini-driver model of polygenic cancer evolution. Nat. Rev. Cancer. 2015;15:680–685. doi: 10.1038/nrc3999. [DOI] [PubMed] [Google Scholar]
- 25.Schodel J., Oikonomopoulos S., Ragoussis J., Pugh C.W., Ratcliffe P.J., Mole D.R. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117:e207–e217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia X., Kung A.L. Preferential binding of HIF-1 to transcriptionally active loci determines cell-type specific response to hypoxia. Genome Biol. 2009;10 doi: 10.1186/gb-2009-10-10-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colli L.M., Jessop L., Myers T.A., Camp S.Y., Machiela M.J., Choi J., Cunha R., Onabajo O., Mills G.C., Schmid V., Brodie S.A., Delattre O., Mole D.R., Purdue M.P., Yu K., et al. Altered regulation of DPF3, a member of the SWI/SNF complexes, underlies the 14q24 renal cancer susceptibility locus. Am. J. Hum. Genet. 2021;108:1590–1610. doi: 10.1016/j.ajhg.2021.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laskar R.S., Muller D.C., Li P., Machiela M.J., Ye Y., Gaborieau V., Foll M., Hofmann J.N., Colli L., Sampson J.N., Wang Z., Bacq-Daian D., Boland A., Abedi-Ardekani B., Durand G., et al. Sex specific associations in genome wide association analysis of renal cell carcinoma. Eur. J. Hum. Genet. 2019;27:1589–1598. doi: 10.1038/s41431-019-0455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schick S., Rendeiro A.F., Runggatscher K., Ringler A., Boidol B., Hinkel M., Majek P., Vulliard L., Penz T., Parapatics K., Schmidl C., Menche J., Boehmelt G., Petronczki M., Muller A.C., et al. Systematic characterization of BAF mutations provides insights into intracomplex synthetic lethalities in human cancers. Nat Genet. 2019;51:1399–1410. doi: 10.1038/s41588-019-0477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao W., Li W., Xiao T., Liu X.S., Kaelin W.G., Jr. Inactivation of the PBRM1 tumor suppressor gene amplifies the HIF-response in VHL-/- clear cell renal carcinoma. Proc. Natl. Acad. Sci. U. S. A. 2017;114:1027–1032. doi: 10.1073/pnas.1619726114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jasinski-Bergner S., Stoehr C., Bukur J., Massa C., Braun J., Huttelmaier S., Spath V., Wartenberg R., Legal W., Taubert H., Wach S., Wullich B., Hartmann A., Seliger B. Clinical relevance of miR-mediated HLA-G regulation and the associated immune cell infiltration in renal cell carcinoma. Oncoimmunology. 2015;4 doi: 10.1080/2162402X.2015.1008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Remmele W., Stegner H.E. [Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue] Pathologe. 1987;8:138–140. [PubMed] [Google Scholar]
- 33.Keller C., Kroening S., Zuehlke J., Kunath F., Krueger B., Goppelt-Struebe M. Distinct mesenchymal alterations in N-cadherin and E-cadherin positive primary renal epithelial cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauer V., Grampp S., Platt J., Lafleur V., Lombardi O., Choudhry H., Kranz F., Hartmann A., Wullich B., Yamamoto A., Coleman M.L., Ratcliffe P.J., Mole D.R., Schodel J. Hypoxia drives glucose transporter 3 expression through hypoxia-inducible transcription factor (HIF)-mediated induction of the long noncoding RNA NICI. J. Biol. Chem. 2020;295:4065–4078. doi: 10.1074/jbc.RA119.009827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warnecke C., Zaborowska Z., Kurreck J., Erdmann V.A., Frei U., Wiesener M., Eckardt K.U. Differentiating the functional role of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha (EPAS-1) by the use of RNA interference: Erythropoietin is a HIF-2alpha target gene in Hep3B and kelly cells. FASEB J. 2004;18:1462–1464. doi: 10.1096/fj.04-1640fje. [DOI] [PubMed] [Google Scholar]
- 36.Giresi P.G., Lieb J.D. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corces M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B., Kathiria A., Cho S.W., Mumbach M.R., Carter A.C., Kasowski M., et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 2017;14:959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We used the following publicly available datasets: GSE86095 (ChIP-seq from renal cancer cells and tissue), GSE120887 (RCC4 HIF-ChIP-seq), GSE101064 (FAIRE-seq in primary renal tubular cells). Sequencing data were re-analyzed and mapped to hg38 as outlined above. KIRC regulatory elements from TCGA were downloaded from https://gdc.cancer.gov/about-data/publications/ATACseq-AWG. ATAC-sequencing data and RNA-sequencing data are available upon reasonable request.