Abstract
Ex vivo expansion conditions used to generate T cells for immunotherapy are thought to adopt metabolic phenotypes that impede therapeutic efficacy in vivo. The comparison of five different culture media used for clinical T cell expansion revealed unique optima based on different output variables, including proliferation, differentiation, function, activation, and mitochondrial phenotypes. The extent of proliferation and function depended on the culture media rather than stimulation conditions. Moreover, the expanded T cell end products adapted their metabolism when switched to a different media formulation, as shown by glucose and glutamine uptake and patterns of glucose isotope labeling. However, adoption of these metabolic phenotypes was uncoupled to T cell function. Expanded T cell products cultured in ascites from ovarian cancer patients displayed suppressed mitochondrial activity and function irrespective of the ex vivo expansion media. Thus, ex vivo T cell expansion media have profound impacts on metabolism and function.
KEYWORDS: T cell expansion, cell-based immunotherapy, culture media, phenotype, metabolism, 13C tracer analysis
Graphical abstract

Cell therapies require expansion of T cells ex vivo, yet no universal medium formulation has been established for this purpose. We demonstrate that culture media impact T cell proliferation, metabolism, and function. The metabolic influences of media can condition T cell end products but are largely uncoupled to function.
Introduction
Cell-based immunotherapies are garnering considerable attention for their promise in treating human cancers. These therapies involve isolating and expanding autologous human tumor-infiltrating lymphocytes (TILs) or genetically modified chimeric antigen receptor (CAR) T cells. In particular, CAR T cell therapy has achieved exceptional response rates in hematological cancers.1,2 However, there are several barriers that impede the efficacy of cell-based therapies targeting solid tumors.3, 4, 5 These include tumor antigen escape, restricted intraepithelial trafficking, limited persistence, and an inhospitable tumor microenvironment (TME).6, 7, 8 Another contributing factor is a complex cell-manufacturing process that can generate products with undesirable metabolic phenotypes and ultimately hamper in vivo functionality.9, 10, 11 Culture methods that maintain metabolic profiles favoring central memory phenotypic subsets have been shown to highly correlate with improved clinical outcomes.12 A recent study identified medium-dependent transcriptional responses in several metabolic pathways during early activation, which may be crucial in programming T cells to a committed phenotype.13, 14, 15, 16, 17 At the present time, there has yet to be a universally accepted formulation to manufacture T cells, making it a major challenge not only to cross-compare different clinical trials but also to understand the metabolic parameters that may be responsible for the behavior of T cells post-transfer.
The first synthetic complex medium used for in vitro expansion of lymphocytes was RPMI 164018 and is widely used in the rapid expansion protocol (REP) of TILs.19 Improvements in formulations that incorporate defined serum components have led to media formulations, such as X-VIVO 15 and AIM-V. However, recent evidence has indicated that cells maintained under these non-physiological conditions activate different signaling and metabolic pathways compared with analogous cells generated in vivo.20 This has led to the generation of culture media that better recapitulate the metabolic composition of human plasma, such as Human Plasma-like Medium (HPLM)21 and others.22 However, these formulations may not provide all environmental cues required to induce and sustain robust expansion specific to T cells and thus necessitate further supplementation with human AB serum. In contrast, ImmunoCult-XF T cell Expansion Medium (ICM) is a serum-free, xeno-free medium that supports robust T cell expansion, while Physiologix XF hGFC xeno-free serum replacement improves CAR T cell replicative capacity and survival in vivo.23
Here, we investigated the influence of five cell culture media conditions on T cell metabolism, proliferation, differentiation, activation, and function (Table 1). Although each condition supported cell proliferation, the extent of expansion varied, as did differentiation, function, mitochondrial phenotypes, and metabolism. Many of these phenotypes were dictated by the media at the time of activation rather than the precise method used to activate the T cells. The medium that supported the greatest proliferation and function (CTL:AIM-V) showed a preference for glucose uptake over glutamine. Importantly, T cells expanded for 11 days were able to adopt this preference for glucose when switched to CTL:AIM-V for 24 h, irrespective of the media that was used for the initial expansion. However, the change in metabolism did not appear to be linked to increased function, as the switch to CTL:AIM-V differentially impacted tumor necrosis factor alpha (TNF-α) and interferon γ (IFNγ) production. Carbon tracing of labeled glucose revealed REP-expanded T cells having increased labeled lactate, whereas ICM expanded T cells displayed increased labeling of one-carbon metabolism and entry into the tricarboxylic acid (TCA) cycle. These patterns of labeling were media dependent, as switching ICM-expanded T cells into CTL:AIM-V medium reverted T cells back to higher lactate labeling, which was associated with an increase in CD25+ and PD-1+ populations. Notably, all five T cell products, when exposed to the ascites derived from ovarian cancer patients, manifested suppressed T cell function and mitochondrial activity. Thus, our studies highlight the multifaceted effects of cell culture media on the metabolic programs, phenotypes, and function of human T cells. Collectively, these data further bolster the notion that the choice of appropriate culture media requires careful consideration depending on the preferred type of T cells required for therapeutic intervention.
Table 1.
List of culture formulations and expansion conditions
| Condition | Abbreviation | Stimulation | Medium | Glucose (mM) | Glutamine (mM) |
|---|---|---|---|---|---|
| Rapid Expansion Protocol | REP | Soluble αCD3 and 50 Gy-irradiated PBMCs | AIM-V Medium and RPMI1640 (CTL:AIM-V) 5% human serum |
12.40 (SD ± 0.40) | 3.75 (SD ± 0.12) |
| StemCell | ICM | ImmunoCult Human CD3/CD28 T cell Activators | ImmunoCult-XF T cell Expansion Medium no human serum |
22 (SD ± 2.21) | 6.68 (SD ± 0.29) |
| Miltenyi | TAC | T cell CD3/CD28 TransAct | TextMACS Medium 3% human serum |
17 (SD ± 0.89) | 2.19 (SD ± 0.04) |
| Corning | CMM | plate-bound αCD3 and soluble αCD28 | Corning 88-581-CM Medium 3% human serum |
27.25 (SD ± 0.63) | 3.68 (SD ± 0.11) |
| Human Plasma-like Medium | HPLM | plate-bound αCD3 and soluble αCD28 | Human Plasma-like Medium 3% human serum |
5.92 (SD ± 0.33) | 0.76 (SD ± 0.04) |
Glucose and glutamine concentrations were measured using colorimetric assay.
Results
Expansion conditions skew proliferation, differentiation, and activation
To examine the influence of cell culture media on T cell products, five basal media were examined. A 1:1 mixture of RPMI 1640 and AIM-V (CTL:AIM-V), which is typically used to expand TILs in a REP19 was used as a control. As a comparison, we included ICM (STEMCELL Technologies), TexMACS (TAC) (Miltenyi Biotec), 88-581-CM Medium (CMM) (Corning Life Sciences), and HPLM (kindly provided by Dr. Jason Cantor), which has been developed specifically to recapitulate the human plasma environment in vitro.13 With the exception of ICM, all basal media were supplemented with human AB serum (Table 1). Enriched CD3+ T cells were isolated from apheresis products of six different healthy donors and expanded over the course of 12 days following the manufacturers' recommended protocols using their respective cell culture media. Unlike the report by Leney-Greene et al.,13 we were primarily interested in the longitudinal changes after expansion and the T cell phenotypes produced after 12–14 days, a time course that is similar to most clinically approved schemes used in manufacturing of T cells for cancer immunotherapy.
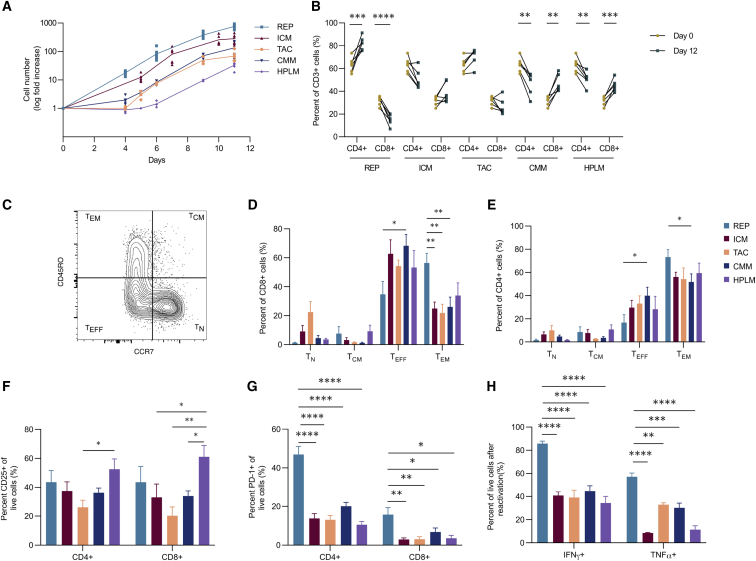
Given the clinical importance of maximizing the number of T cells in the expansion product, the proliferative capacities of all five culture medias were compared. Expansion differed significantly across all of the conditions ranging from 40- to 860-fold (Figure 1A). This difference in expansion appeared as early as day 4 and was maintained throughout the entire expansion, with the REP condition yielding the largest number of T cells. While the CMM and HPLM expansion conditions used the same stimulation method and differed only in defined composition, CMM mediated a 2-fold-greater expansion than HPLM. It has been reported that an equivalent ratio of CD4+ and CD8+ T cells in the final infusion product is associated with better clinical responses to CAR T cell therapy.24,25 Although each condition resulted in a variable percentage of CD4+ and CD8+ T cells, the CD4:CD8 T cell ratio within each condition was consistent across different healthy donors (Figure 1B). For example, the REP promoted a strong enrichment of CD4+ T cells and a concomitant reduction in CD8+ T cells that was significantly different than the other conditions (Figures 1B and S1A). Of note, the TAC and ICM expansion media did not appear to significantly alter the percentage of CD4+ and CD8+ T cells from day 0, whereas CMM and HPLM media generated more CD8+ T cells over the same 12-day period (Figure 1B).
Figure 1.
Expansion conditions skew proliferation, differentiation, and activation
(A–G) T cells from six healthy donors were expanded in five different conditions for 12 days: CTL:AIM-V (REP), ImmunoCult-XF (ICM), TexMACS (TAC), Complete Corning media (CMM), and Human Plasma-like Medium (HPLM). (A) Log fold increase in cell number throughout the 11-day expansion is shown. (B) Proportion of CD4+ and CD8+ T cells of live CD3+ cells, pre- and post-expansion from six healthy donors, is shown. (C–E) Representative plot (C) and tabulated data for CD8+ (D) and CD4+ (E) T cells following expansion are shown: naïve (TN), central memory (TCM), effector (TEFF), and effector memory (TEM). (F and G) Percentage of (F) CD25+ and (G) PD-1+ CD4+ and CD8+ T cells is shown. (H) Percentage of IFNγ+ and TNF-α+ cells after CD3/CD28 reactivation following expansion is shown (n = 3). Bar graphs represent mean of n = 6 (A–G) and (H) n = 3 + SEM from healthy donors. Statistical significance was calculated using a Student’s t test (B) or a one-way ANOVA (D–H); ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
The differentiation state of the post-expanded population correlates with long-term persistence of both TIL and CAR T cells and, to a large extent, tumor control.26,27 For instance, the skewing toward CD8+ T effector (TEFF) cells limits anti-tumor responses after adoptive cell therapy.28 On the other hand, adoptive transfer of CD8+ T central memory (TCM) cells has been shown to have a superior anti-tumor response than CD8+ T effector memory (TEM) cells.29 Across all of the conditions tested, expanded CD4+ and CD8+ T cells displayed a predominant TEFF and/or TEM phenotype, with the REP condition producing the highest percentage of TEM cells (Figures 1C–1E). Overall, the five media conditions generated few (<10%) CD4+ or CD8+ T cells displaying a TCM differentiated phenotype.
Clonal variation, marked by a diverse T cell receptor (TCR) Vβ repertoire, enables T cells to recognize a wide array of epitopes, including those from tumors. Selective expansion of tumor-reactive clones has been reported to correlate with clinical responses.30 It is unclear whether certain media conditions favor the outgrowth of specific clonotypes. Therefore, we profiled TCR Vβ repertoires using flow cytometry to assess population clonality for each condition. Overall, the Vβ spectratyping revealed that TCR diversity is largely maintained across all conditions (Figures S1B and S1C).
To determine whether culture conditions influenced T cell activation, we measured the expression of CD25 and CD137 by flow cytometry. Expression of CD137 was generally higher on CD8+ T cells than CD4+ T cells across all conditions (Figure S1D). Although the REP produced the lowest frequency of CD8+ T cells, it achieved the highest percentage of CD8+ CD137+ cells compared with all other conditions (Figure S1D). In contrast, CD25 expression was similar between CD4+ and CD8+ T cells, with the HPLM condition producing the highest proportion of CD25+ cells compared with other conditions (Figure 1F). Since CD25 is also a marker of CD4+ regulatory T (Treg) cells,31 we also measured the proportion of CD4+ CD25+ FoxP3+ Treg cells in each condition. Treg cells comprised <5% of the expanded T cells in all conditions (Figure S1E), indicating that the conditions used in this study do not support the proliferation of FoxP3+ Treg cells. We also evaluated the expression of PD-1 as an indicator of potential T cell exhaustion, since its expression on expanded CAR T cells is correlated with poorer patient outcomes.32 PD-1 expression was generally low on cells in all conditions except for T cells maintained in REP medium, where 50% of the CD4+ T cell population expressed PD-1 (Figure 1G). Moreover, upon reactivation, REP-expanded T cells displayed higher TNF-α and IFNγ production (Figure 1H). Collectively, these results establish that T cell products vary in proliferative capacity, differentiation (TEFF and TEM), function (TNF-α and IFNγ), and expression of exhaustion markers (PD-1).
Mitochondria mass, activity, and ROS vary across all conditions
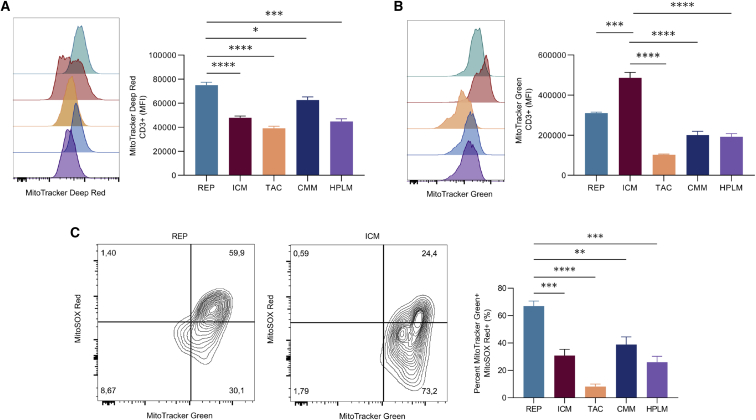
Mitochondrial activity and biomass have been recognized in both pre-clinical and clinical models to be indicators of improved CAR T cell therapeutic efficacy. Specifically, low mitochondrial activity and increased mitochondrial biogenesis supports T cell persistence and anti-tumor function in chronic lymphocyte leukemia patients.27 Therefore, we assessed the impact of expansion conditions on mitochondrial activity and mitochondrial mass as determined by flow cytometry analysis of cells stained with MitoTracker Deep Red and MitoTracker Green, respectively. Overall, there was no consistent pattern that emerged across conditions, although statistical differences were observed depending on the pairwise comparisons (Figures 2A, 2B, and S2A). We found that the REP-expanded T cells produced the highest cell number and also had the highest mitochondrial activity (Figure 2A). Notably, high mitochondrial activity is associated with a terminal differentiation state and poor tumor killing capacity.33 In contrast, T cells maintained in ICM had the second largest expansion with lower mitochondrial activity compared with the REP but had significantly higher mitochondrial mass compared with all other conditions (Figure 2B). The substantial increase in mitochondrial biogenesis observed in the ICM condition was not due to a difference in cell size (Figure S2B).
Figure 2.
T cell products exhibit different mitochondrial phenotypes
T cells from three healthy donors were expanded in five different conditions for 12 days. (A and B) Representative plot (left) and tabulated data (right) for median fluorescence intensity (MFI) of mitochondrial activity (MitoTracker Deep Red) and (B) mitochondrial mass (MitoTracker Green) are shown. (C) Representative plot (left) and tabulated data (right) for percentage of mitochondrial mass high and mitochondrial ROS high (MitoSOX Red) live populations are shown. Bar graphs represent mean of n = 3 + SEM from healthy donors. Statistical significance was calculated by one-way ANOVA (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001).
Cells with elevated levels of mitochondrial activity often result in elevated mitochondrial reactive oxygen species (ROS) production that can limit the cell’s functional capability in conditions of oxidative stress, such as those found in the TME. Therefore, we investigated how mitochondrial activity is associated with mitochondrial ROS for each expansion product. We determined that mitochondrial activity and mitochondrial ROS were positively correlated in all conditions, although at varying degrees (Figure S2C). In accordance with the elevated mitochondrial activity, REP-expanded T cells had the highest ROS production, where approximately 70% of mitochondria were positive for ROS (Figure 2C). In contrast to the REP, the ICM condition displayed higher mitochondrial mass and low levels of ROS (Figure 2C), while the conditions with the lowest proliferation and mitochondrial activity, TAC and HPLM, appeared to produce the lowest mitochondrial-specific ROS.
Cell culture media dictate T cell phenotypes
Although unique phenotypes emerged across the different expansion conditions, it was unclear whether these were due to the specific way by which T cells were activated or whether the culture media was responsible for regulating these cellular programs. It should be noted that T cells in the REP use soluble CD3 and allogenic irradiated feeder cells for activation. In contrast, while the ICM uses CD3/CD28 soluble tetrameric antibody complexes, the TAC uses a CD3/CD28 colloidal polymeric nanomatrix. Therefore, we compared how these three different T cell activators affect T cell phenotypes when cultured using only CTL:AIM-V. Consistent with our previous findings, expansion varied significantly across conditions, with the REP producing the greatest fold expansion (Figure S3A). The increased proportion of CD4+ to CD8+ T cells in the REP was found to be unique to the REP stimulation and not CTL:AIM-V medium (Figure S3B). However, in terms of T cell phenotype, we found that all of the activation methods produced phenotypes with similar mitochondrial mass and TNF-α+ populations when cultured in CTL:AIM-V (Figures S3C and S3D).
Given these findings, we performed a reciprocal experiment where we activated cells using the ICM or TAC stimulators and cultured the cells in different media (CTL:AIM-V, ICM, and TAC). As early as day 3 of expansion, we noticed considerable differences in the gross size of lymphoblasts across media, but not between the activation method used (Figure S3E). CTL:AIM-V displayed large lymphoblasts, ICM displayed an intermediate size, and TAC medium had smaller dispersed lymphoblasts. Proliferation was also found to be influenced by the culture media, with the CTL:AIM-V and ICM media promoting the greatest expansion, regardless of stimulation method used (Figures S3F and S3G). Consistent with our previous findings, ICM cells had increased mitochondrial mass, regardless of activation strategy used (Figure S3H). Furthermore, upon reactivation, the proportions of TNF-α+ cells differed, depending on the media used. The CTL:AIM-V medium produced the greatest percentage of functional T cells, regardless of the activation strategy used (Figure S3I). Collectively, these data imply that the cell culture media rather than the type of T cell stimulator contributes to the distinct T cell phenotypes.
The culture medium dictates nutrient uptake but is uncoupled from T cell function
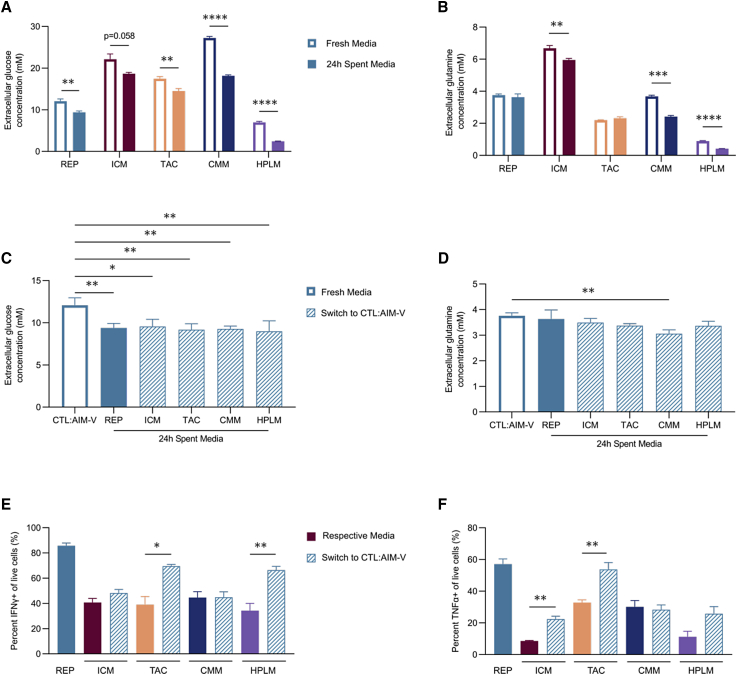
Media conditions are often proprietary, so the precise composition of basic nutrient levels is not available; however, certain components are amenable to investigation. Two key nutrients, glucose and glutamine, were assessed, as both are well studied in terms of their regulation of T cells.34, 35, 36 The concentrations of glucose and glutamine varied significantly across media, ranging from 5.92 to 27.25 mM and 0.76 to 6.68 mM, respectively (Table 1). Next, we examined the effect of different culture conditions on the uptake of these nutrients. All five conditions supported varying levels of both glucose and glutamine uptake (Figures 3A and 3B). Furthermore, glucose uptake appeared to be independent of proliferation, as both the highest (REP, ICM, and CMM) and lowest (HPLM) proliferative conditions had significant changes in glucose concentration compared with their respective fresh media condition (Figure 3A). The elevated nutrient uptake observed in HPLM-expanded cells may be due to the differing proliferative state at day 11 (Figure S4A). Despite containing supraphysiological level of glucose and glutamine, the REP-expanded T cells resulted in no significant change in glutamine concentration in the media over 24 h of culture, indicative of low glutamine uptake and a preference for glucose as a carbon source. In contrast, the CMM expansion condition produced significant differences in both glucose and glutamine concentrations in the media after 24 h (Figures 3A and 3B). To gain insight into whether the dependence on glucose in the REP was mediated by the cell culture media formulation, we expanded T cells using the respective four conditions and on day 11 replaced the media with the REP medium (CTL:AIM-V) for 24 h. This switch resulted in a significant change in glucose concentration in CTL:AIM-V with no concomitant change in glutamine across almost all expansion products (Figures 3C and 3D). Thus, this media replacement phenocopied the glucose metabolism of the REP-expanded condition in the CTL:AIM-V. Although there was a modest reduction in glutamine consumption by CMM-expanded T cells that had been switched to CTL:AIM-V, the shift to reduced glutamine uptake was the most dramatic of all conditions. These findings imply that T cells exhibit metabolic flexibility and adapt to their surrounding nutrient environments, regardless of their original metabolic state at the time of activation and/or expansion.
Figure 3.
Cell culture media dictate nutrient uptake but are uncoupled to T cell function
(A–D) T cells from three healthy donors were expanded in five different conditions for 11 days. Glucose and glutamine concentrations in the media were measured between days 11 and 12. (A and B) Extracellular (A) glucose and (B) glutamine concentrations in the fresh media (white bars) and spent media after culture for 24 h (solid bars) are shown. (C and D) On day 11, expanded T cells from all five conditions were switched to the REP media (CTL:AIM-V) for 24 h. Extracellular (C) glucose and (D) glutamine concentrations in fresh CTL:AIM-V (blue open), spent media after culture for 24 h with REP-expanded cells (blue closed) and the four other expansion products in CTL:AIM-V (dashed open) are shown. (E and F) On day 12, T cells from all five conditions underwent CD3/CD28 reactivation for 2 days in their respective conditions (solid bars) or were switched to CTL:AIM-V (blue dashed bars). Percentage of (E) IFNγ+ and (F) TNF-α+ cells is shown. Bar graphs represent mean n = 3 + SEM from healthy donors. Statistical significance was calculated by one-way ANOVA (A, B, E, and F) or Student’s t test (C and D); ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
To uncover whether the shift in metabolism in CTL:AIM-V was sufficient to increase function as observed in cells originally expanded in CTL:AIM-V, we reactivated all products in their respective media or in CTL:AIM-V for 2 days. Unlike our expansion findings (Figure S3I), T cell function was not universally influenced by the switch to CTL:AIM-V medium (Figures 3E and 3F). Although all T cell products adopted similar glucose and glutamine uptake in CTL:AIM-V, differences in IFNγ and TNF-α production were observed. While TAC significantly increased both IFNγ and TNF-α production in CTL:AIM-V, CMM cells displayed no change in either cytokine (Figures 3E and 3F). This demonstrates that, while metabolism adapts to the change in extracellular conditions, the inherited impacts of the initial expansion conditions on T cell function cannot be universally overcome by switching T cells into another medium.
Glucose utilization and phenotype are influenced by the culture medium
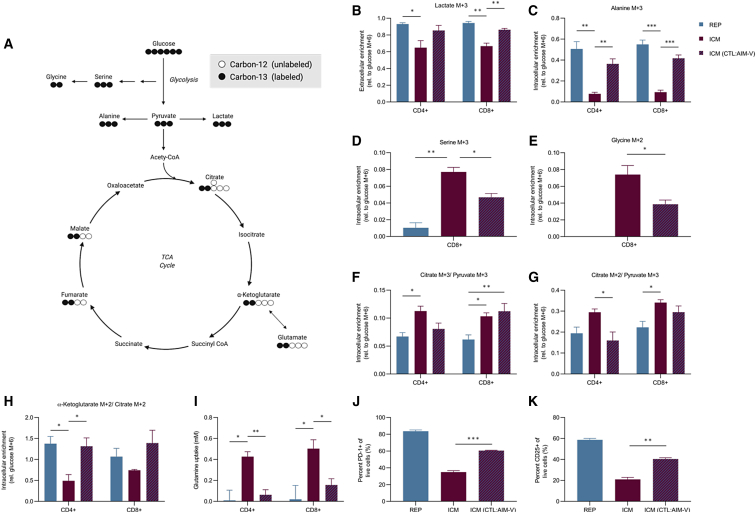
To gain further insight into the influence of media on T cell metabolism, we investigated the difference in glucose utilization pathways between the REP and ICM condition. These media produced the largest cell expansions across different modes of activation and supported similar glucose uptake levels, regardless of glucose concentrations in the media (Table 1; Figure S5A). Therefore, stable isotope labeling was performed using [U-13C]glucose to delineate how carbon utilization differed in REP and ICM conditions (Figure 4A). Current in vitro models suggest that the rate of glycolysis is matched to the rate of proliferation. Indeed, we found that aerobic glycolysis was active in both conditions, producing high extracellular and intracellular lactate M+3 fractions in both CD4+ and CD8+ T cells (Figures 4B and S5B). However, the REP condition produced significantly more extracellular lactate compared with ICM (Figure S5C), consistent with a more glycolytic and activated phenotype (Figures 1F and 1G). Furthermore, the ICM condition produced approximately 30% less extracellular glucose-derived lactate M+3, indicating that other carbon sources support lactate production (Figure 4B). To assess whether glucose utilization was dependent on the culture medium, the day 11 ICM-expanded T cell products were placed in CTL:AIM-V. After 24 h in CTL:AIM-V, the ICM-expanded T cell products did not increase in lactate production (Figure S5C); however, their carbon utilization pathways shifted, resulting in a significantly higher fraction of lactate M+3 that was similar to what was observed in the REP condition (Figure 4B). A similar enrichment in the 13C labeling pattern was observed for alanine M+3. There was a higher level of alanine M+3 enrichment in the REP and ICM switched to CTL:AIM-V compared with ICM cells in their respective condition (Figure 4C). Interestingly, ICM contains approximately seven times more alanine than CTL:AIM-V (Figure S5D), which was associated with higher intracellular alanine levels (Figure S5E). Therefore, the reduced fraction of alanine M+3 in the ICM condition could be due to a combination of increased de novo production and/or uptake.
Figure 4.
Culture media influences glucose utilization and the metabolic profile of T cells
(A) Schematic of [U-13C]glucose metabolism; circles represent the carbons for each metabolite. CoA, coenzyme A. (B–K) Three healthy donors were expanded in REP and ICM for 11 days. On day 11, CD4+ and CD8+ cells were isolated and incubated in the following [U-13C]glucose conditions for 24 h: REP cells in CTL:AIM-V (blue bar), ICM cells in ImmunoCult-XF (red bar), and ICM cells cultured in CTL:AIM-V (red and blue dashes). (B) Extracellular lactate M+3 relative to extracellular glucose M+6 enrichment is shown. (C) Intracellular alanine M+3 relative to intracellular glucose M+6 enrichment is shown. (D and E) CD8+ intracellular enrichment of (D) serine M+3 and (E) glycine M+2 relative to intracellular glucose M+6 enrichment is shown. (F and G) Pyruvate carboxylase activity (F; citrate M+3/pyruvate M+3) and pyruvate dehydrogenase activity (G; citrate M+2/pyruvate M+3) are shown. (H) Proportion of intracellular α-ketoglutarate M+2 to citrate M+2 enrichment is shown. (I) Glutamine uptake (mM) from days 11 to 12 in CD4+ and CD8+ T cells is shown. (J and K) Percentage of PD-1+ (J) and CD25+ (K) cells is shown. Data are shown as mean of n = 3 + SEM from healthy donors. Statistical significance was calculated by Student’s t tests (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001).
Due to the decrease in lactate and alanine labeling in ICM conditions, we interrogated other upstream glucose-derived metabolites. Serine is a non-essential amino acid that contributes to nucleotide biosynthesis. Recent evidence indicates that serine is necessary to support CD8+ T cell expansion and effector function.37,38 The [U-13C]glucose metabolite isotope analysis revealed a greater fractional enrichment of labeled serine and glycine in ICM-expanded T cells compared with REP-expanded T cells. More specifically, CD8+ T cells from the ICM expansion had significant enrichment of serine M+1 and M+3 and glycine M+2 compared with CD8+ T cells from the REP expansion (Figures 4D, 4E, and S5F). Although the relative serine labeling was reduced in ICM-expanded T cells that were switched to the CTL:AIM-V, there were detectable levels of serine M+3 enrichment. This suggests that, relative to the REP expansion conditions, ICM expansion conditions promote one-carbon metabolism pathways in CD8+ T cells and that changes in one-carbon metabolism may be less susceptible to fluctuations in the levels of extracellular nutrients.
At the peak of an effector response, T cells undergo a metabolic switch from glycolysis to oxidative phosphorylation (OXPHOS), supporting mitochondrial biogenesis and T cell memory development. Therefore, we assessed the fate of the [U-13C]glucose carbons into the mitochondria (Figure 4A). Citrate M+2/pyruvate M+3 and citrate M+3/pyruvate M+3 ratios serve as surrogates for pyruvate dehydrogenase (PDH) and pyruvate carboxylase (PC) activity, respectively.39,40 Between conditions, both CD4+ and CD8+ T cells from the ICM expansion had higher PC and PDH activity than CD4+ and CD8+ T cells from the REP expansion (Figures 4F and 4G). The ICM-mediated contribution of glucose-derived carbons into the TCA cycle is consistent with the observed increase in mitochondrial biogenesis (Figure 2B). However, CD4+ T cells seem to be more influenced by the switch to CTL:AIM-V condition than the CD8+ T cells that maintained the PDH and PC labeling patterns regardless of condition. Furthermore, the ICM-expanded T cells produced roughly 50% of α-ketoglutarate M+2 from citrate M+2 (Figure 4H). This reduction in α-ketoglutarate M+2 was corroborated by the reduction in glutamate M+2 and malate M+2 fractional enrichment in the ICM expansion condition (Figures S5G and S5H). The loss in α-ketoglutarate M+2 enrichment was likely due to entry of unlabeled carbons from glutamine catabolism, both of which were reversed in CTL:AIM-V conditions (Figures 4H and 4I). These results imply that REP and ICM expansion conditions differentially control the source of carbons entering the TCA cycle. ICM-expanded T cells direct glucose-derived carbons preferentially into citrate via PC and PDH and likely use glutaminolysis as an alternative carbon source to support synthesis of TCA cycle metabolites.
CTL:AIM-V medium supports increased glycolysis, glucose dependence, elevated mitochondrial activity, and ROS generation, characteristic hallmarks associated with T cell exhaustion. Indeed, the REP cells resulted in greater than 50% of the expanded T cells expressing PD-1 (Figure 1G). Therefore, we investigated whether conditioning ICM-expanded T cells in CTL:AIM-V would also influence exhaustion. On day 12, REP- and ICM-expanded T cells were reactivated in their respective conditions or in CTL:AIM-V for an additional 2 days. The REP-expanded T cells showed a significantly higher proportion of cells that were PD-1+ and CD25+ compared with ICM-expanded T cells (Figures 4J and 4K). However, the ICM-expanded cells that were reactivated in CTL:AIM-V conditions also significantly increased in activation and exhaustion markers, resembling the exhaustion state of the REP-expanded T cells (Figures 4J and 4K). This suggests that, while ICM-expanded T cells do not increase glycolysis when switched to CTL:AIM-V, the nutrient conditions in the CTL:AIM-V medium resulted in changes to both glucose and glutamine metabolism that were sufficient to impart alterations in activation and exhaustion phenotypes.
The tumor microenvironment imposes different metabolic constraints on T cell products
Given the observed changes in T cell metabolism and phenotypes when expanded T cells are placed into different media, it is possible that T cells manufactured under these parameters may also be affected by the TME. There is a growing appreciation that ex vivo clinical manufacturing conditions used to expand T cells for adoptive cell therapies may impact the effectiveness of the final T cell product when the infused T cells encounter the nutrient-depleted TME.41,42 To test this possibility, patient-derived ovarian cancer ascites fluid was used as a proxy due to its known immunosuppressive features. We expanded T cells in all five conditions and, on day 12 of the expansion, T cell products were cultured and reactivated in 100% ascites fluid for 2 days (Figure 5A). Four of the five expanded T cell conditions maintained their viability in the presence of ascites (Figure 5B). However, the REP-expanded T cells showed a consistent and noticeable decrease in viability compared with the remaining four conditions. This decrease in viability was not due to the environment, as reactivation of REP-expanded T cells in CTL:AIM-V also resulted in decreased viability (Figure S6A).
Figure 5.
T cell products exhibit different metabolic and functional phenotypes in the ascites tumor microenvironment
T cells from three healthy donors were expanded in five different conditions over 12 days. On day 12, T cell products were reactivated (CD3/CD28) in ascites, and T cell metabolism and function was assessed after 2 days. (A) Schematic of experimental timeline is shown. Created with BioRender.com. (B) Percentage of live cells in ascites supernatant is shown. (C) MFI of mitochondrial activity (MitoTracker Deep Red) in the ascites is shown. (D and E) Percentage of (D) PD-1 and (E) IFNγ+ and TNF-α+ cells in the ascites is shown. Data are shown as mean of n = 3 + SEM from healthy donors. Statistical significance was calculated by one-way ANOVA (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001).
The ascites environment has been reported to be immune suppressive, owing to reduced levels of critical nutrients, such as glucose and glutamine.43,44 Consistent with this, analysis of the ascites supernatant revealed significantly lower levels of both glucose and glutamine compared with levels in all of the media formulations, except for HPLM (Table 1; Figures S6B and S6C). We found that, despite containing physiological levels of glucose and glutamine, mitochondrial metabolism was lower in all of the expanded T cell products in ascites fluid (Figure S6D). However, similar to what was seen in the media conditions, the REP had the highest mitochondrial activity in ascites fluid compared with the other conditions (Figure 5C). Although the REP condition had high levels of PD-1 expression in the ascites across all conditions (Figure 5D), the expression of PD-1 was reduced in the ascites (Figure S6E). Since PD-1 is also a marker of activation, reduced PD-1 expression could be a reflection of the suppressed activation state of T cells in ascites. Indeed, the T cell activation marker CD25 was also reduced across conditions in the ascites, further supporting the contention that ascites is an immune-suppressive environment (Figure S6F). Consistent with the reduction in mitochondrial activity, expanded T cells cultured in ascites resulted in a decrease in IFNγ and TNF-α production, regardless of the expansion condition (Figures S6G and S6H). While the REP-expanded T cells produced the largest proportion of IFNγ+ cells, there was no significant difference in TNF-α expression across conditions in ascites fluid (Figure 5E).
Discussion
Clinical grade T cells are manufactured under stringent and defined conditions, yet to date, there is no universal media formulation or standardized protocols in the field. Moreover, the release criteria do not take into account how different expansion conditions alter T cell metabolism or how these conditions may impact T cell function post-infusion. Recent work has highlighted important differences in how T cells metabolize glucose in vitro versus in vivo.20 During CD8+ T cell responses to physiological infection, T cells adopt an oxidative metabolic phenotype with greater flux of glucose-derived carbons into serine and other biosynthetic pathways. This is consistent with another study showing that glucose pre-conditioning can metabolically enhance adoptive T cell anti-tumor immunity potentially by shifting cells into a temporary oxidative state.42 These studies suggest that the nutrient conditions during the ex vivo manufacturing could be tailored to enhance the fitness of T cells following infusion into patients.
We evaluated how five commonly used ex vivo expansion culturing conditions alter the metabolic state and phenotypes of CD4+ and CD8+ T cells. Across these conditions, there were pervasive differences in metabolism that resulted in shifts in differentiation and function. Although all conditions resulted in T cell proliferation, the extent of expansion was markedly different and ranged from 40- to 860-fold. However, above a certain threshold, the size of the CAR T cell product has not been shown to correlate with in vivo expansion or therapeutic success;25 thus, T cell phenotypes may be a better predictor of therapeutic efficacy.10 Nonetheless, we found that certain media formulations supported superior proliferation following the same activation and expansion protocol. However, the baseline concentration of glucose present in each media did not correlate with the degree of expansion. For example, under the same activation protocol, CTL:AIM-V and ICM media yielded the highest fold expansion despite the TAC containing a similar concentration of glucose (Table 1). Although REP and ICM T cells support a high degree of expansion, [U-13C]glucose isotope-tracing studies revealed that T cells expanded in the ICM culture medium had lower levels of extracellular lactate and lactate M+3 enrichment, suggesting that glycolysis was less engaged in these cells. The reduction in lactate was associated with a greater proportion of CD8+ TEFF cells and decreased the proportion of TEM. This observation was unexpected given that TEFF cells have been shown to be highly glycolytic and the transition to TEM cells is associated with a switch to oxidative metabolism. These results suggest that, during ex vivo expansion, metabolites other than glucose that are found in the ICM medium play a role supporting TEFF cell differentiation. It is also possible that the specific TCR stimulation method activates unique signaling programs that could contribute to the different metabolic responses. However, this was not observed in the CMM and HPLM conditions. Furthermore, this is independent from the TCR clonality, as no major biases were seen in the outgrowth of TCR diversity.
Due to the implications of mitochondrial metabolism in the context of CAR and TIL therapy, we assessed the mitochondrial phenotype produced from all five conditions. ICM-expanded T cells had significantly higher mitochondrial mass compared with all other conditions. This was surprising, as the majority of ICM cells had a TEFF phenotype and increases in mitochondrial biogenesis are commonly associated with T memory subsets. This may be due to the comparisons of TEFF and TEM within the same media, rather than comparing across media as performed here. In fact, we found that the ICM medium was responsible for the higher mitochondrial mass. The REP-expanded T cells also showed high mitochondrial activity, which correlated with increased production of mitochondrial-specific ROS. Although ROS production is required for T cell activation, elevated production of free radicals can induce mitochondrial stress and trigger cell apoptosis. This mitochondrial phenotype may explain the poor persistence in vivo and poor survival and cytotoxic efficacy under oxidative stress, an observation that has been previously reported for T cells expanded using the REP formulation.45 In contrast, the ICM medium produced cells with increased mitochondrial mass and reduced activity and ROS production compared with cells cultured under REP conditions. Based on previous clinical findings, a product with this mitochondrial profile may be most advantageous for cell-based therapies.27 However, in our immune-suppressive TME model used here, no expansion condition supported sustained mitochondrial activity in the presence of ascites fluid.
At present, REP expansions are largely restricted to the generation of expanded TIL products, while the use of CD3/CD28 activators are widely used for the manufacturing of CAR T cells. The majority of the expansions tested here incorporated CD3/CD28-based stimulation using different delivery methods: soluble tetrameric antibody complexes (ICM), colloidal polymeric nanomatrix (TAC), and plate-bound CD3 soluble CD28 (CMM and HPLM). In contrast, the REP involves stimulation with soluble CD3 and allogenic irradiated feeder cells as a source of co-stimulatory molecules. While CD3/CD28-based activation strategies have been suggested to produce comparable products,46,47 cell-to-cell interactions in the REP may contribute to the skewing of T cell differentiation.48 Indeed, our study found that the REP activation method uniquely produces a higher proportion of CD4+ T cells versus CD8+ T cells in the same culture. We also found variations in proliferation across activation methods. It should be mentioned that each expansion protocol requires users to subculture cells according to recommended manufacturers’ protocols. The timing of subculturing varies depending on the protocol and represents an important factor when considering the type of expansion for user applications.
To further understand how culture media influences T cell metabolism, we traced [U-13C]glucose utilization in T cells that were expanded using the REP and ICM condition, as well as ICM-expanded T cells, and subsequently switched into CTL:AIM-V. These results revealed that the elevated glycolysis labeling and reduction of TCA cycle intermediate labeling was media dependent. Of note, 13C enrichment was reduced in metabolites downstream of citrate M+2 in ICM-expanded T cells compared with REP T cells and ICM T cells switched in CTL:AIM-V for 24 h. This is likely due to the increased uptake of glutamine in ICM conditions, while REP and T cells switched to CTL:AIM-V medium preferentially utilize glucose over glutamine as a carbon source. These patterns were also associated with a corresponding increase in both intracellular and extracellular alanine M+3. Alanine has been shown to be essential for T cell activation and can be produced from a transamination reaction with pyruvate and glutamate via alanine aminotransferase. However, this increased proportion of fractional enrichment of alanine M+3 is also a reflection of the lower proportions of unlabeled extracellular and intracellular alanine in T cells expanded under CTL:AIM-V compared with T cells expanded under ICM medium. When ICM-expanded T cells were switched into CTL:AIM-V, the total intracellular alanine levels decreased (Figure S5E), supporting the idea that REP-expanded T cells may have reduced alanine uptake. This implies that exogenous alanine in the ICM medium is sufficient to meet cellular demands, while T cells expanded with the REP medium appear to require synthesis of alanine. However, further studies are needed to formally demonstrate this possible scenario.
Isotope-tracing studies also revealed metabolic pathways that are less susceptible to reprogramming due to fluctuations in extracellular nutrient conditions. For example, some metabolic phenotypes of the ICM-expanded T cells persisted even when the T cells were switched to CTL:AIM-V medium, including the increased PDH and PC activity in CD8+ T cells and enrichment of 13C to one-carbon metabolism pathways. These data provide an example of how T cells adapt their metabolism based on extracellular levels of nutrients and the plasticity of certain metabolic pathways. This also further supports the idea that some metabolic pathways may have a more permanent inheritability throughout the expansion, while the utilization of other metabolic pathways, such as glycolysis, may change rapidly depending on precise conditions at the time. It will be important to consider which metabolic pathways and cell phenotypes will be maintained after the expanded T cells are administered and subsequently traffic into the TME.
Lastly, we tested how all five expansion conditions supported T cell metabolism and function when transferred to patient-derived ovarian cancer ascites fluid, a known immunosuppressive TME. It is important to note that the mechanism of immunosuppression observed here in the ascites was not due to hypoxia or acidity, common immunosuppressive features of the solid TME (data not shown). Due to the elevated level of ROS and PD-1 expression in the REP-expanded T cells, we speculated that this state may contribute to activation-induced T cell death upon reactivation in ascites.49 As expected, the REP condition had significantly reduced T cell viability after reactivation compared with the other media conditions. Interestingly, glucose uptake was not suppressed across all conditions and actually increased in T cells expanded under the HPLM conditions. This result further highlights the notion that the ex vivo metabolism of T cells is not necessarily coupled to their functional behavior when subjected to the TME. The increase in glucose uptake could be associated with higher CD28 expression, as it has been shown that CD28 supports Akt activation.50 It is tempting to speculate that HPLM may be more suited for expansion of second or third generation CAR T cells that encode for CD28 co-stimulatory domains versus 4-1BB co-stimulatory domains. HPLM contains similar glucose levels to that found in ascites, suggesting that initial expansion of T cells in low-glucose conditions programmed T cells to subsist under environments where glucose levels are limited.42 On the other hand, mitochondrial activity was significantly suppressed across all conditions, which is commonly observed in the TME and could contribute to the observed suppression of T cell function. Similar to other reports, we observed that T cell products with elevated mitochondrial activity showed a tendency to have higher PD-1 expression.49 This demonstrates that the impact of the media formulations extends beyond nutrient uptake and glucose utilization in that they directly affect T cell activation.
In conclusion, we uncovered distinct metabolic programs activated by ex vivo clinical expansion protocols. The observed differences under various media formulations contributed to the skewed patterns of T cell differentiation and effector function, outputs that could be uncoupled with their metabolic profiles. Most T-cell-based immunotherapies focus on applications to improve cell expansion and to produce phenotypes that enable in vivo persistence and maximal cytolytic function. Here, we demonstrate that media formulations may influence metabolic fidelity of the final immunotherapeutic product. Further studies will be required to determine whether these metabolic states imparted by different ex vivo expansion conditions ultimately impact the in vivo anti-tumor ability of T cells and their long-term persistence.
Materials and methods
Cell culture reagents
Five culture media were used in this study. (1) Complete CTL:AIM-V medium, used in REP, consisted of equal parts of supplemented RPMI 1640 and AIM-V medium. To a 1× RPMI 1640 Medium basal, the following components were added: 2 mM L-Glutamine (Fisher Scientific), 10% heat-inactivated (56°C, 60 min) human AB serum (Sigma), 12.5 mM HEPES (Fisher), 1× penicillin streptomycin solution (Fisher), and 50 μM β-mercaptoethanol (Sigma). AIM-V Medium (Invitrogen) was supplemented with 20 mM HEPES and 2 mM L-glutamine (CTL:AIM-V). (2) ImmunoCult-XF T cell Expansion Medium (STEMCELL Technologies) was supplemented with 1× penicillin streptomycin solution (ICM). (3) TexMACS medium (Miltenyi) was supplemented with 3% heat-inactivated human AB serum (Sigma) and 1× penicillin streptomycin solution (TAC). (4) Corning medium consisted of Lymphocyte Serum-Free Medium KBM 581 (Corning) supplemented with 3% heat-inactivated human AB serum (CMM). (5) Basal HPLM (kindly provided by Dr. Jason Cantor)21 was prepared with four additional components (5 μM acetylcarnitine, 5 μM α-ketoglutarate, 5 μM malate, and 3 μM uridine), as recently reported,51 and then further supplemented with 3% heat-inactivated human AB serum (HPLM). All complete media were filtered through a 0.22-μM filter prior to use.
Patient ascites collection
Patient specimens and clinical data were obtained through the BC Cancer Tumor Tissue Repository, certified by the Canadian Tissue Repository Network. All specimens and clinical data were obtained with either informed written consent or a formal waiver of consent under protocols approved by the Research Ethics Board of BC Cancer and the University of British Columbia (H07-00463). Patient ascites were centrifuged at 1,500 rpm for 10 min at 4°C to pellet cells, and supernatant was frozen at −80°C. The preserved supernatants were thawed for the functional assays as described below.
T cell expansions
Peripheral blood mononuclear cells (PBMCs) were isolated from six human peripheral blood leukapheresis packs (STEMCELL Technologies) using Ficoll gradient density centrifugation. CD3+ T cells were isolated from cryopreserved PBMCs using human CD3 MicroBeads (Miltenyi) according to the manufacturer's instructions. Specific details about stimulation protocols used can be found in Table 1. All media were supplemented with 300 IU/mL interleukin-2 (IL-2) (Novartis) before cell culture. REP CD3+ T cells (1.0 × 105) were stimulated with 50-Gy-irradiated allogenic feeder PBMCs from two healthy donors (2.0 × 107) and soluble CD3 (30 ng/mL; OKT3) in complete CTL:AIM-V medium. Other expansion methods were seeded using 1.0 × 106 CD3+ T cells in 1 mL of media in a 48-well plate. ICM T cells were stimulated with ImmunoCult Human CD3/CD28 T cell activators (25 μL/mL; STEMCELL Technologies). TAC T cells were stimulated with T cell CD3/CD28 TransAct, human (10 μL/mL; Miltenyi Biotec). CMM and HPLM T cells were stimulated with plate-bound CD3 (5 μg/mL; OKT3) and soluble CD28 (2 μg/mL; 15EB). On days 2 to 3, the medium from TAC cells was replaced with fresh medium as per manufacturer’s instructions. T cells were first split either on day 3 (ICM) or day 4 (other conditions) of the expansion and subsequently split as needed to maintain a concentration of 100,000–600,000 cells/mL (ICM) or 500,000–1,000,000 cells/mL (other conditions). During expansion from day 0 to day 11, cells were isolated, diluted in trypan blue, and counted using a hemocytometer. Cell counts were measured throughout the expansion to ensure protocol-recommended densities were maintained. T cell reactivation in their respective condition, or ascites, was performed using plate-bound CD3 (5 μg/mL; OKT3) and soluble CD28 (2 μg/mL; 15EB) for 2 days.
Lymphoblast phenotype imaging
On day 3 of T cell expansion, activated T cells were imaged using the ZEISS Axiocam 208 microscope camera and the Labscope imaging application. Photos were saved at 500 pixels as a jpeg file.
Phenotypic and metabolic profiling by flow cytometry
Prior to expansion (day 0), cells were collected from CD3+ magnetic bead isolation. Cells were stained with viability dye (eFlour506, Thermo) diluted in PBS (Invitrogen) at 4°C for 15 min. Cells were stained with a panel of antibodies (Table S1) in flow cytometry staining buffer for 30 min at 4°C. After staining, cells were washed twice and resuspended in flow cytometry staining buffer prior to flow cytometry analysis. Following expansion, separate flow cytometry panels were used on day 12 for cell phenotyping and mitochondrial analysis (Table S1). For mitochondrial analysis, cells were stained with MitoTracker Deep Red (10 nM), MitoTracker Green (100 nM), or MitoSOX Red (2.5 μM) for 30 min at 37°C. Cells were then washed twice with PBS and then stained with viability dye at 4°C for 15 min. For cell phenotype analysis, cells were then washed with flow cytometry staining buffer before being fixed and permeabilized using Fixation/Permeabilization Solution Kit (BD Biosciences) as per the manufacturer's instructions. Cells were stained with a panel of antibodies (Table S1) in 1× BD Perm/Wash buffer (BD Biosciences) for 30 min at 4°C. After staining, cells were washed twice in 1× BD Perm/Wash buffer (BD Biosciences) and resuspended in flow cytometry staining buffer prior to flow cytometry analysis. Flow cytometry analysis was carried out using a Cytek Aurora spectral flow cytometer (3L-16V-14B-8R configuration). Data were unmixed using SpectroFlo Software (Cytek) and manually gated and analyzed using FlowJo v.10.6.1. Figures were created using GraphPad Prism 8.1.2. To assess phenotype, cells were classified as naïve (TN) (CCR7+CD45RO−), TEFF (CCR7−CD45RO−), TEM (CCR7−CD45RO+), or TCM (CCR7+CD45RO+).52 To assess effector function following expansion, on day 12 in their respective condition or ascites, T cells were reactivated for 2 days using plate-bound CD3 (5 μg/mL; OKT3) and soluble CD28 (2 μg/mL; 15EB). Six hours prior to staining, cells were treated with BD GolgiStop (1 μL/mL; BD Biosciences) to assess TNF-α and IFNγ production. Cells that underwent metabolic analysis (Table S1) with MitoTracker Deep Red (10 nM) were not treated with BD GolgiStop.
Vβ spectratyping by flow cytometry
To assess TCR diversity following expansion (day 12), cell surface TCR Vβ repertoires were profiled using the IOTest Beta Mark Kit (Beckman Coulter Genomics) as per the manufacturer’s guidelines. Cells were washed in PBS (Invitrogen) and blocked with Anti-Hu Fc Receptor Binding Inhibitor (eBioscience) for 10 min at room temperature. Cells were stained with a panel of antibodies (Table S1) in flow cytometry staining buffer with BD Horizon Brilliant Stain Buffer Plus (BD Biosciences) for 30 min at 4°C. Cell viability was assessed using the Zombie NIR Fixable Viability Kit (BioLegend) as per the manufacturer’s guidelines. After staining, cells were washed once in flow cytometry staining buffer and resuspended in flow cytometry staining buffer prior to flow cytometry analysis using a Cytek Aurora spectral flow cytometer (3L-16V-14B-8R configuration). Data were unmixed using SpectroFlo Software (Cytek) and manually gated and analyzed using FlowJo v.10.6.1, and figures were created using GraphPad Prism 8.1.2. Manual gating was carried out following the manufacturer’s guidelines (Beckman Coulter).
Isotope-tracing analysis by gas chromatography-mass spectrometry (GC-MS) using [U-13C]glucose
For metabolic labeling experiments, complete CTL:AIM-V labeling medium consisted of glucose-free CTL:AIM-V with 5% heat-inactivated human AB serum supplemented with 11 mM uniformly labeled 13C-glucose ([U-13C]glucose; Cambridge Isotope Laboratories). Glucose-free ImmunoCult-XF T cell Expansion Medium (STEMCELL Technologies) was supplemented with 24 mM [U-13C]glucose. On day 12 of expansion, CD4+ T cells maintained under CTL:AIM-V or ICM were isolated using human CD4 MicroBeads (Miltenyi) and a MACS LS magnetic column according to the manufacturer's instructions; CD8+ T cells were collected from the flowthrough. Separately, the cells were incubated in complete [U-13C]glucose CTL:AIM-V or complete [U-13C]glucose ICM medium supplemented with 300 IU/mL IL-2 (Novartis) for 24 h at 5% CO2 and 37°C. Metabolic steady state was confirmed on day 12 with 2-NBDG, and isotope steady state was confirmed at 24 h.
To prepare cells for GC-MS analysis, four million cells per condition were washed once with ice-cold saline solution (0.9% NaCl solution filtered through a 0.22-μm membrane filter) and resuspended in 1 mL of 50% (vol/vol) methanol (Sigma; 0.22 μm filtered) cooled at −80°C. Cells were snap frozen at −80°C for 20 min, and the resulting lysate/methanol mixtures were frozen in liquid nitrogen. Samples were thawed on ice, vortexed, and subject to three freeze-thaw cycles using liquid nitrogen and 37°C water bath. Samples were centrifuged at 10,000 rpm for 10 min at 4°C and then the metabolite-containing supernatant collected. The supernatant was evaporated until dry using a SpeedVac and no heat and stored at −80°C. Before running GC-MS, norvaline (1 μL) internal standard was added to each sample. Samples were resuspended in 40 μL pyridine containing methoxyamine (10 mg/mL), transferred to GC-MS vials, and heated at 70°C for 15 min, and 70 μL N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide (MTBSTFA) was added, samples vortexed, and heated at 70°C for 1 h before analyzing with GC-MS.
To prepare media for GC-MS analysis, cells were centrifuged and 1 mL of supernatant collected. The samples were frozen at −80°C, thawed on ice, and vortexed and then 25 μL was transferred to a borosilicate tube. Before running GC-MS, 400 μL of each methanol, chloroform, and Milli-Q purified water were added to samples, along with 1 μL of norvaline internal standard (total 1,226 μL). Samples were vortexed and centrifuged at 2,000 rpm for 5 min to separate phases. The aqueous phase was collected and evaporated until dry at 42°C using a SpeedVac. The samples were transferred to GC-MS vials and heated at 70°C for 15 min before 70 μL MTBSTFA was added. Samples were vortexed and heated at 70°C for 1 h before analyzing with GC-MS.
Metabolites were analyzed using an Agilent 6970 gas chromatograph and an Agilent 5973 mass selective detector as previously described.53 GC-MS data were analyzed using Chemstation (Agilent Technologies) and Skyline,54 and graphs were created using GraphPad Prism 8.1.2. The measured distribution of mass isotopomers was corrected for natural abundance of 13C.55 The fractional enrichment of isotopologues was then compared against the fractional enrichment of glucose M+6 for the respective condition. Metabolites with a fractional enrichment of below 0.05 were not considered a significant finding.
Separately, 600 μL media was collected from cells and frozen at −80°C. Glucose and glutamine concentrations in the media were measured using a NOVA BioProfile4 or by colorimetric assay (BioVision Technologies).
Data availability
Processed metabolomics data files are available at https://github.com/vicDRC/metaboData.git. Flow cytometry data will be deposited at Flow Repository (Figures 1, 2, and 3: FR-FCM-Z4TC, Figure 5: FR-FCM-Z4V4, Figure S1: FR-FCM-Z4V3, and Figure S3: FR-FCM-Z4UN).
Statistical analysis
Statistical analysis was carried out using GraphPad Prism 8.1.2. An unpaired Student’s t test was used when comparing means of two groups, and one-way ANOVA was used when comparing more than two groups. Differences were considered significant at ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Acknowledgments
This study was supported by the research grants to J.J.L. from the Canadian Institutes of Health Research (PJT 162279). S.K. is supported by a BioCanRx Studentship Award and BC Cancer Studentship Award. M.K. is supported by a University of Victoria Graduate Award. J.S. is supported by a Canadian Institutes of Health Research Banting and Best Doctoral Award. R.J.D. is supported by grants from the National Cancer Institute (R35CA22044901) and the Cancer Prevention and Research Institute of Texas (RP180778). J.R.C. is supported by the NIH/NCI (K22 CA225864), and K.S.H. is supported by a fellowship from the NIH (T32HG002760). The summary figure was created with Biorender.com.
Author contributions
S.M., S.K., and M.K.K. designed and performed experiments, analyzed the data, and wrote the manuscript. J.Smazynski and V.C. helped design and performed experiments. J.Sudderth and R.J.D. ran the mass spectrometry samples and analyzed the raw data. T.T. and A.D. irradiated the PBMCs. K.S.H., J.R.C., J.Y., and C.S. provided reagents and contributed to experimental design. J.J.L. conceived the project and wrote the manuscript.
Declaration of interests
R.J.D. is a member of the Scientific Advisory Boards of Vida Ventures and Agios Pharmaceuticals and is a founder of Atavistik Biosciences. J.R.C. is an inventor on a patent application for HPLM (PCT/US2017/061,377) assigned to the Whitehead Institute. C.S. is a Principal Scientist at STEMCELL Technologies. J.Y. is a Scientist at STEMCELL Technologies. STEMCELL Technologies provided reagents in kind for the study but were not involved in funding the study, performing experiments, or analyzing the data.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2022.02.004.
Supplemental information
References
- 1.Maude S.L., Laetsch T.W., Buechner J., Rives S., Boyer M., Bittencourt H., Bader P., Verneris M.R., Stefanski H.E., Myers G.D., et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018;378:439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neelapu S.S., Locke F.L., Bartlett N.L., Lekakis L.J., Miklos D.B., Jacobson C.A., Braunschweig I., Oluwole O.O., Siddiqi T., Lin Y., et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 2017;377:2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schubert M.-L., Hoffmann J.-M., Dreger P., Müller-Tidow C., Schmitt M. Chimeric antigen receptor transduced T cells: tuning up for the next generation. Int. J. Cancer. 2018;142:1738–1747. doi: 10.1002/ijc.31147. [DOI] [PubMed] [Google Scholar]
- 4.Martinez M., Moon E.K. CAR T cells for solid tumors: new strategies for finding, infiltrating, and surviving in the tumor microenvironment. Front. Immunol. 2019;10:128. doi: 10.3389/fimmu.2019.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radvanyi L.G., Bernatchez C., Zhang M., Fox P.S., Miller P., Chacon J., Wu R., Lizee G., Mahoney S., Alvarado G., et al. Specific lymphocyte subsets predict response to adoptive cell therapy using expanded autologous tumor-infiltrating lymphocytes in metastatic melanoma patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012;18:6758–6770. doi: 10.1158/1078-0432.CCR-12-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kilgour M.K., MacPherson S., Zacharias L.G., Ellis A.E., Sheldon R.D., Liu E.Y., Keyes S., Pauly B., Carleton G., Allard B., et al. 1-Methylnicotinamide is an immune regulatory metabolite in human ovarian cancer. Sci. Adv. 2021;7:eabe1174. doi: 10.1126/sciadv.abe1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marofi F., Motavalli R., Safonov V.A., Thangavelu L., Yumashev A.V., Alexander M., Shomali N., Chartrand M.S., Pathak Y., Jarahian M., et al. CAR T cells in solid tumors: challenges and opportunities. Stem Cell Res. Ther. 2021;12:81. doi: 10.1186/s13287-020-02128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Sullivan G.M., Velickovic Z.M., Keir M.W., Macpherson J.L., Rasko J.E.J. Cell and gene therapy manufacturing capabilities in Australia and New Zealand. Cytotherapy. 2019;21:1258–1273. doi: 10.1016/j.jcyt.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Stock S., Schmitt M., Sellner L. Optimizing manufacturing protocols of chimeric antigen receptor T cells for improved anticancer immunotherapy. Int. J. Mol. Sci. 2019;20:6223. doi: 10.3390/ijms20246223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geltink R.I.K., Kyle R.L., Pearce E.L. Unraveling the complex interplay between T cell metabolism and function. Annu. Rev. Immunol. 2018;36:461–488. doi: 10.1146/annurev-immunol-042617-053019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraietta J.A., Lacey S.F., Orlando E.J., Pruteanu-Malinici I., Gohil M., Lundh S., Boesteanu A.C., Wang Y., O’Connor R.S., Hwang W.-T., et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat. Med. 2018;24:563–571. doi: 10.1038/s41591-018-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leney-Greene M.A., Boddapati A.K., Su H.C., Cantor J.R., Lenardo M.J. Human plasma-like medium improves T lymphocyte activation. iScience. 2020;23:100759. doi: 10.1016/j.isci.2019.100759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cantor J.R. The rise of physiologic media. Trends Cell Biol. 2019;29:854–861. doi: 10.1016/j.tcb.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacPherson S., Kilgour M., Lum J.J. Understanding lymphocyte metabolism for use in cancer immunotherapy. FEBS J. 2018;285:2567–2578. doi: 10.1111/febs.14454. [DOI] [PubMed] [Google Scholar]
- 16.Fucà G., Reppel L., Landoni E., Savoldo B., Dotti G. Enhancing chimeric antigen receptor T cell efficacy in solid tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020;26:2444–2451. doi: 10.1158/1078-0432.CCR-19-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato K., Kondo M., Sakuta K., Hosoi A., Noji S., Sugiura M., Yoshida Y., Kakimi K. Impact of culture medium on the expansion of T cells for immunotherapy. Cytotherapy. 2009;11:936–946. doi: 10.3109/14653240903219114. [DOI] [PubMed] [Google Scholar]
- 18.Moore G.E., Gerner R.E., Franklin H.A. Culture of normal human leukocytes. JAMA. 1967;199:519–524. [PubMed] [Google Scholar]
- 19.Dudley M.E., Wunderlich J.R., Shelton T.E., Even J., Rosenberg S.A. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J. Immunother. 2003;26:332–342. doi: 10.1097/00002371-200307000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma E.H., Verway M.J., Johnson R.M., Roy D.G., Steadman M., Hayes S., Williams K.S., Sheldon R.D., Samborska B., Kosinski P.A., et al. Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8+ T cells. Immunity. 2019;51:856–870.e5. doi: 10.1016/j.immuni.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Cantor J.R., Abu-Remaileh M., Kanarek N., Freinkman E., Gao X., Louissaint A., Lewis C.A., Sabatini D.M. Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase. Cell. 2017;169:258–272.e17. doi: 10.1016/j.cell.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medvec A.R., Ecker C., Kong H., Winters E.A., Glover J., Varela-Rohena A., Riley J.L. Improved expansion and in vivo function of patient T cells by a serum-free medium. Mol. Ther. Methods Clin. Dev. 2017;8:65–74. doi: 10.1016/j.omtm.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghassemi S., Martinez-Becerra F.J., Master A.M., Richman S.A., Heo D., Leferovich J., Tu Y., García-Cañaveras J.C., Ayari A., Lu Y., et al. Enhancing chimeric antigen receptor T cell anti-tumor function through advanced media design. Mol. Ther. Methods Clin. Dev. 2020;18:595–606. doi: 10.1016/j.omtm.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., Chen S., Xiao W., Li W., Wang L., Yang S., Wang W., Xu L., Liao S., Liu W., et al. CAR-T cells targeting CLL-1 as an approach to treat acute myeloid leukemia. J. Hematol. Oncol.J Hematol. Oncol. 2018;11:7. doi: 10.1186/s13045-017-0553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sommermeyer D., Hudecek M., Kosasih P.L., Gogishvili T., Maloney D.G., Turtle C.J., Riddell S.R. Chimeric antigen receptor-modified T cells derived from defined CD8+ and CD4+ subsets confer superior antitumor reactivity in vivo. Leukemia. 2016;30:492–500. doi: 10.1038/leu.2015.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawalekar O.U., O’Connor R.S., Fraietta J.A., Guo L., McGettigan S.E., Posey A.D., Patel P.R., Guedan S., Scholler J., Keith B., et al. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity. 2016;44:380–390. doi: 10.1016/j.immuni.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 27.van Bruggen J.A.C., Martens A.W.J., Fraietta J.A., Hofland T., Tonino S.H., Eldering E., Levin M.-D., Siska P.J., Endstra S., Rathmell J.C., et al. Chronic lymphocytic leukemia cells impair mitochondrial fitness in CD8+ T cells and impede CAR T-cell efficacy. Blood. 2019;134:44–58. doi: 10.1182/blood.2018885863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gattinoni L., Klebanoff C.A., Palmer D.C., Wrzesinski C., Kerstann K., Yu Z., Finkelstein S.E., Theoret M.R., Rosenberg S.A., Restifo N.P. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. J. Clin. Invest. 2005;115:1616–1626. doi: 10.1172/JCI24480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klebanoff C.A., Gattinoni L., Torabi-Parizi P., Kerstann K., Cardones A.R., Finkelstein S.E., Palmer D.C., Antony P.A., Hwang S.T., Rosenberg S.A., et al. Central memory self/tumor-reactive CD8+ T cells confer superior antitumor immunity compared with effector memory T cells. Proc. Natl. Acad. Sci. U S A. 2005;102:9571. doi: 10.1073/pnas.0503726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandran S.S., Klebanoff C.A. T cell receptor-based cancer immunotherapy: emerging efficacy and pathways of resistance. Immunol. Rev. 2019;290:127–147. doi: 10.1111/imr.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitoiset F., Barbié M., Monneret G., Braudeau C., Pochard P., Pellegrin I., Trauet J., Labalette M., Klatzmann D., Rosenzwajg M. A standardized flow cytometry procedure for the monitoring of regulatory T cells in clinical trials. Cytometry B Clin. Cytom. 2018;94:777–782. doi: 10.1002/cyto.b.21622. [DOI] [PubMed] [Google Scholar]
- 32.Finney O.C., Brakke H., Rawlings-Rhea S., Hicks R., Doolittle D., Lopez M., Futrell B., Orentas R.J., Li D., Gardner R., et al. CD19 CAR T cell product and disease attributes predict leukemia remission durability. J. Clin. Invest. 2019;129:2123–2132. doi: 10.1172/JCI125423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sukumar M., Liu J., Mehta G.U., Patel S.J., Roychoudhuri R., Crompton J.G., Klebanoff C.A., Ji Y., Li P., Yu Z., et al. Mitochondrial membrane potential identifies cells with enhanced stemness for cellular therapy. Cell Metabol. 2016;23:63–76. doi: 10.1016/j.cmet.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang C.-H., Curtis J.D., Maggi L.B., Faubert B., Villarino A.V., O’Sullivan D., Huang S.C.-C., van der Windt G.J.W., Blagih J., Qiu J., et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blagih J., Coulombe F., Vincent E.E., Dupuy F., Galicia-Vázquez G., Yurchenko E., Raissi T.C., van der Windt G.J.W., Viollet B., Pearce E.L., et al. The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity. 2015;42:41–54. doi: 10.1016/j.immuni.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 36.Carr E.L., Kelman A., Wu G.S., Gopaul R., Senkevitch E., Aghvanyan A., Turay A.M., Frauwirth K.A. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma E.H., Bantug G., Griss T., Condotta S., Johnson R.M., Samborska B., Mainolfi N., Suri V., Guak H., Balmer M.L., et al. Serine is an essential metabolite for effector T cell expansion. Cell Metabol. 2017;25:345–357. doi: 10.1016/j.cmet.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 38.Ron-Harel N., Santos D., Ghergurovich J.M., Sage P.T., Reddy A., Lovitch S.B., Dephoure N., Satterstrom F.K., Sheffer M., Spinelli J.B., et al. Mitochondrial biogenesis and proteome remodeling promotes one carbon metabolism for T cell activation. Cell Metabol. 2016;24:104–117. doi: 10.1016/j.cmet.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Comte B., Vincent G., Bouchard B., Rosiers C.D. Probing the origin of acetyl-CoA and oxaloacetate entering the citric acid cycle from the 13 C labeling of citrate released by perfused rat hearts. J. Biol. Chem. 1997;272:26117–26124. doi: 10.1074/jbc.272.42.26117. [DOI] [PubMed] [Google Scholar]
- 40.Courtney K.D., Bezwada D., Mashimo T., Pichumani K., Vemireddy V., Funk A.M., Wimberly J., McNeil S.S., Kapur P., Lotan Y., et al. Isotope tracing of human clear cell renal cell carcinomas demonstrates suppressed glucose oxidation in vivo. Cell Metabol. 2018;28:793–800.e2. doi: 10.1016/j.cmet.2018.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sukumar M., Liu J., Ji Y., Subramanian M., Crompton J.G., Yu Z., Roychoudhuri R., Palmer D.C., Muranski P., Karoly E.D., et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. Invest. 2013;123:4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein Geltink R.I., Edwards-Hicks J., Apostolova P., O’Sullivan D., Sanin D.E., Patterson A.E., Puleston D.J., Ligthart N.A.M., Buescher J.M., Grzes K.M., et al. Metabolic conditioning of CD8 + effector T cells for adoptive cell therapy. Nat. Metabol. 2020;2:703–716. doi: 10.1038/s42255-020-0256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong Y., Yang J., Wang Y., Xue L., Wang J. Metabolic factors contribute to T-cell inhibition in the ovarian cancer ascites. Int. J. Cancer. 2020;147:1768–1777. doi: 10.1002/ijc.32990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S., Kim B., Song Y.S. Ascites modulates cancer cell behavior, contributing to tumor heterogeneity in ovarian cancer. Cancer Sci. 2016;107:1173–1178. doi: 10.1111/cas.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin C., Yu D., Hillerdal V., Wallgren A., Karlsson-Parra A., Essand M. Allogeneic lymphocyte-licensed DCs expand T cells with improved antitumor activity and resistance to oxidative stress and immunosuppressive factors. Mol. Ther. Methods Clin. Dev. 2014;1:14001. doi: 10.1038/mtm.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X., Rivière I. Clinical manufacturing of CAR T cells: foundation of a promising therapy. Mol. Ther. Oncolytics. 2016;3:16015. doi: 10.1038/mto.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casati A., Varghaei-Nahvi A., Feldman S.A., Assenmacher M., Rosenberg S.A., Dudley M.E., Scheffold A. Clinical-scale selection and viral transduction of human naïve and central memory CD8+ T cells for adoptive cell therapy of cancer patients. Cancer Immunol. Immunother. CII. 2013;62:1563–1573. doi: 10.1007/s00262-013-1459-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y., Kurlander R.J. Comparison of anti-CD3 and anti-CD28-coated beads with soluble anti-CD3 for expanding human T cells: differing impact on CD8 T cell phenotype and responsiveness to restimulation. J. Transl. Med. 2010;8:104. doi: 10.1186/1479-5876-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tkachev V., Goodell S., Opipari A.W., Hao L.-Y., Franchi L., Glick G.D., Ferrara J.L.M., Byersdorfer C.A. Programmed death-1 controls T cell survival by regulating oxidative metabolism. J. Immunol. 2015;194:5789–5800. doi: 10.4049/jimmunol.1402180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frauwirth K.A., Riley J.L., Harris M.H., Parry R.V., Rathmell J.C., Plas D.R., Elstrom R.L., June C.H., Thompson C.B. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 51.Rossiter N.J., Huggler K.S., Adelmann C.H., Keys H.R., Soens R.W., Sabatini D.M., Cantor J.R. CRISPR screens in physiologic medium reveal conditionally essential genes in human cells. Cell Metabol. 2021;33:1248–1263.e9. doi: 10.1016/j.cmet.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Golubovskaya V., Wu L. Different subsets of T cells, memory, effector functions, and CAR-T immunotherapy. Cancers. 2016;8:36. doi: 10.3390/cancers8030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mullen A.R., Wheaton W.W., Jin E.S., Chen P.-H., Sullivan L.B., Cheng T., Yang Y., Linehan W.M., Chandel N.S., DeBerardinis R.J. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams K.J., Pratt B., Bose N., Dubois L.G., St John-Williams L., Perrott K.M., Ky K., Kapahi P., Sharma V., MacCoss M.J., et al. Skyline for small molecules: a unifying software package for quantitative metabolomics. J. Proteome Res. 2020;19:1447–1458. doi: 10.1021/acs.jproteome.9b00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernandez C.A., Rosiers C.D., Previs S.F., David F., Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J. Mass Spectrom. 1996;31:255–262. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed metabolomics data files are available at https://github.com/vicDRC/metaboData.git. Flow cytometry data will be deposited at Flow Repository (Figures 1, 2, and 3: FR-FCM-Z4TC, Figure 5: FR-FCM-Z4V4, Figure S1: FR-FCM-Z4V3, and Figure S3: FR-FCM-Z4UN).