Abstract
During Escherichia coli cell division, an intracellular complex of cell division proteins known as the Z-ring assembles at midcell during early division and serves as the site of constriction. While the predominant protein in the Z-ring is the widely conserved tubulin homolog FtsZ, the actin homolog FtsA tethers the Z-ring scaffold to the cytoplasmic membrane by binding to FtsZ. While FtsZ is known to function as a dynamic, polymerized GTPase, the assembly state of its partner, FtsA, and the role of ATP are still unclear. We report that a substitution mutation in the FtsA ATP-binding site impairs ATP hydrolysis, phospholipid vesicle remodeling in vitro, and Z-ring assembly in vivo. We demonstrate by transmission electron microscopy and Förster Resonance Energy Transfer that a truncated FtsA variant, FtsA(ΔMTS) lacking a C-terminal membrane targeting sequence, self assembles into ATP-dependent filaments. These filaments coassemble with FtsZ polymers but are destabilized by unassembled FtsZ. These findings suggest a model wherein ATP binding drives FtsA polymerization and membrane remodeling at the lipid surface, and FtsA polymerization is coregulated with FtsZ polymerization. We conclude that the coordinated assembly of FtsZ and FtsA polymers may serve as a key checkpoint in division that triggers cell wall synthesis and division progression.
Keywords: FtsZ-ring, cytokinesis, ATPase, actin, copolymer
Abbreviations: GMPCPP, guanylyl-(alpha, beta)-methylene-diphosphonate; LS, light scatter; MTS, membrane targeting sequence; PL, phospholipid; SUV, small unilamellar vesicle; TEM, transmission electron microscopy
Cell division in bacteria occurs through the organized and concerted efforts of many proteins that temporally localize to the division site and execute a physical process to divide a single cell into two discrete cells. The architectural remodeling that occurs at the site of septation during division in Escherichia coli initiates with the assembly of the Z-ring at midcell. The most conserved and essential protein of the divisome, FtsZ, forms highly dynamic polymers that locate to the center of the cell and may serve as a scaffold to facilitate cell constriction and septum formation (1). FtsZ polymers are unable to bind to the inner membrane of the cell directly, and two proteins, FtsA and the less conserved ZipA, serve as membrane tethers for FtsZ (2, 3, 4, 5, 6, 7). FtsA is a highly conserved actin homolog that interacts directly with the FtsZ C-terminus (7, 8). FtsA binds to the inner membrane through a conserved C-terminal amphipathic helix that is required for in vivo function (4). Although FtsA hydrolyzes ATP rapidly in vitro in a reaction that requires phospholipids (PLs) (7), the cellular role or requirement for ATP by FtsA has not been clearly elucidated. In addition to its ATP-hydrolyzing activity, FtsA remodels and tubulates liposomes, destabilizes FtsZ polymers in vitro, and modifies FtsZ dynamics in a reconstituted system (7, 9). FtsA and FtsZ act as the first of at least 12 proteins that localize to midcell during the early steps of division in E. coli (10, 11, 12, 13). FtsA interacts with many other cell division proteins directly including ZipA, FtsI, FtsEX, and, most notably, FtsN (14, 15, 16, 17).The interaction between FtsA and FtsN has been suggested to serve as a trigger to engage FtsQBL, which activates FtsWI complex for peptidoglycan synthesis (18).
In eukaryotes, actin polymerizes intracellularly to provide a networked cytoskeletal structure, aids in protein locomotion, trafficking, and cell division, and is the basis for muscle contraction in higher eukaryotes (19, 20, 21, 22). Notable bacterial actin homologs include the plasmid segregation protein ParM, cell shape determining protein MreB, and cell division protein FtsA (23, 24, 25). FtsA contains similar domain organization to actin; however, the 1C domain of FtsA is in a different orientation as the 1B domain of actin and participates in direct interactions with later stage cell division proteins (8, 14, 15, 16, 17, 24) (Fig. 1A). This domain change has significant implications for the structure of putative FtsA filaments. Indeed, crystallization of an FtsA dimer from Thermotoga maritima shows that dimerization appears to occur through the 1A and 2B domains near the active site interacting with the 1C and 2A domains on a subsequent protomer, whereas MreB self-interacts similarly to actin through repeated binding of domains 2B and 1B with 2A (8, 24). Actin polymerization is dependent on ATP but is also modulated by ionic strength and divalent cations; in addition, actin assembly is regulated by a suite of other eukaryotic proteins (26, 27, 28, 29). Recently, high resolution cryo-EM conformational studies have revealed major structural changes for ATP-bound actin in a polymer, whereas minor, but notable, changes take place upon ATP hydrolysis and phosphate release (30, 31). Although actin filament formation has been well studied, direct confirmation of E. coli FtsA polymerization has been elusive. After early reports of E. coli FtsA self-interaction from yeast two-hybrid studies (32), Pichoff and Lutkenhaus reported that a Gfp-FtsA fusion protein lacking the C-terminal membrane targeting sequence (MTS), Gfp-FtsA(Δ15), formed fluorescent rod-like structures in the cytoplasm (4), and mutants defective for rod formation were also defective for self-interaction in a yeast two-hybrid system (33). Interestingly, cells expressing mutant FtsA proteins defective for self-interaction and overexpression of FtsN bypass the requirement for ZipA, as well as FtsEX, suggesting that these other cell division proteins may help promote monomerization of FtsA in vivo (17, 34, 35, 36, 37). In vitro FtsA protofilament-like structures have been observed by transmission electron microscopy (TEM) on lipid monolayers of FtsA from T. maritima (8) and E. coli (38, 39) and in solution of FtsA from Streptococcus pneumoniae (40) and Vibrio cholerae (41). Only S. pneumoniae FtsA has shown ATP-dependent polymerization (40). Moreover, mechanistic detail into FtsA polymerization, depolymerization, and the ATP cycle remains undetermined.
Figure 1.
FtsA(E14R), which contains a mutation close to the predicted active site, is defective for function in vivo.A, Escherichia coli FtsA (residues 8–385) modeled onto Thermotoga maritima FtsA (pdb: 4A2B) (8) using Swiss-Model and overlayed in blue. Amino acid Glu14 (cyan) and ATPγS are shown as stick models. Mg2+ divalent cation (gold) is shown as a Corey-Pauling-Koltun (CPK) model. Domains 1A, 1C, 2A, and 2B are indicated. B, confocal fluorescence and differential interference contrast (DIC) microscopy of E. coli MG1655 araECP cells expressing plasmid-encoded Gfp-FtsA, Gfp-FtsA(E14R), Gfp-FtsA(ΔMTS), or Gfp-FtsA(E14R/ΔMTS). The cells were grown as described in Experimental procedures. The scale bars represent 2 μm. C, log dilutions of cultured E. coli MCA12 (ftsA12) cells expressing FtsA or FtsA mutant proteins [FtsA(E14R), FtsA(ΔMTS), FtsA(E14R/ΔMTS) from a plasmid and grown overnight at permissive (30 °C) and restrictive (42 °C) temperatures, where indicated, on LB agar plates containing ampicillin (100 μg ml−1). The data shown is representative of three replicates. MTS, membrane targeting sequence.
Several models have been proposed to resolve the precise individual roles for FtsZ and FtsA during cell division, including but not limited to scaffolds, constriction machines, and spatiotemporal organizers. While there is a large body of evidence to suggest that FtsZ assembles into tubulin-like protofilaments in vivo, there is scant evidence to demonstrate that E. coli FtsA also polymerizes as part of its functional role. Furthermore, both FtsZ and FtsA localize to a nascent Z-ring in vivo at the same time, thus creating a paradox about the initial step of divisome assembly (42). Here, we demonstrate that an E. coli FtsA variant lacking the C-terminal MTS, FtsA(ΔMTS), forms linear polymers in the presence of ATP. Furthermore, substitution of a critical residue in the predicted active site of FtsA leads to impaired ATP hydrolysis, reduced FtsZ interaction in vitro, and major functional defects in vivo. Our results show that FtsA polymerization is coregulated with FtsZ polymerization. This model suggests that the FtsZ polymerization state is coordinated with FtsA, and the FtsA/FtsZ coassembled state would then be communicated to inner membrane protein FtsN. We predict that other cell division proteins such as ZipA or FtsEX may contribute to regulating the FtsA polymerization state and, therefore, this initial step in division (17, 43, 44, 45). After interacting with FtsA, FtsN would then activate FtsQBL and FtsWI for cell wall synthesis (18, 45).
Results
FtsA(E14R) is defective for PL remodeling and FtsZ interactions
In E. coli FtsA, which is modeled on the crystal structure of T. maritima FtsA (pdb: 4A2B) (8), Glu14 is near Mg2+ in the active site (Fig. 1A). Therefore, we predicted that E14 could be important for FtsA function, possibly through nucleotide sensing or hydrolysis activity. In an alignment of E. coli FtsA with hexokinase, Hsc70-like proteins, and several actins, this residue is highly conserved as a negatively charged amino acid (46). To determine if Glu14 is important for FtsA function in E. coli, we mutagenized Glu14 to Arg and evaluated this mutant protein, FtsA(E14R), in vivo and in vitro. We first tested if the FtsA substitution mutant, constructed as a Gfp fusion protein, localizes to Z-rings in vivo. We used a plasmid that encodes a Gfp-FtsA fusion protein (pSEB293) (4), then mutagenized ftsA and expressed Gfp-FtsA variants in E. coli MG1655, which also contains a constitutively expressed arabinose transporter, araECP (Figs. 1B and S1A), to reduce cell to cell variability in expression of arabinose-inducible genes (Table 1). As expected, Gfp-FtsA exhibited a fluorescence localization pattern consistent with a division ring at midcell (Fig. 1B). Cells with the plasmid encoding Gfp-FtsA(E14R) contained solely diffuse fluorescence, and we detected no ring or foci formation in these cells (Fig. 1B). FtsA contains a MTS at its C-terminus that localizes FtsA to the cytoplasmic membrane at the divisome in vivo (4, 47) and directly recruits FtsA to PLs in vitro (7). The MTS penetrates into the nonpolar core of the lipid bilayer, and this insertion is required for rapid FtsA ATP hydrolysis in vitro (7). Deletion of the MTS from FtsA or removal of PLs from the reactions containing WT FtsA abrogates rapid ATPase activity, suggesting that membrane association is critical for FtsA enzyme function and regulation (7). We constructed and expressed Gfp-FtsA(ΔMTS) and confirmed that it was unable to localize to a ring but instead formed long, narrow foci adjacent to the cell membrane, which have been previously reported (4, 44, 48). The E14R mutation was introduced into the plasmid encoding Gfp-FtsA(ΔMTS) to construct Gfp-FtsA(E14R/ΔMTS). In contrast to the long, narrow foci exhibited by Gfp-FtsA(ΔMTS), we instead observed diffuse cytoplasmic fluorescence (Fig. 1B). The fluorescent rod-like structures of Gfp-FtsA(ΔMTS) observed in vivo were previously hypothesized to report FtsA(ΔMTS) self-interaction, and this phenotype has been used to identify self-interaction deficient FtsA mutants in vivo (4, 33). Our results suggest that introduction of the E14R mutation reduces or prevents self-association of Gfp-FtsA(ΔMTS) (Fig. 1B).
Table 1.
E. coli strains and plasmids
| Strain or plasmid | Relevant genotype | Source, reference, or construction |
|---|---|---|
| Strain | ||
| MG1655 | LAM- -rph1 | (63) |
| MG1655 araECP | MG1655 (ΔaraEp kan PCP18-araE) | (51, 64) |
| BL21(ℷde3) | F- ompT gal dcm lon hsdSB(rB- mB-) | EMD Millipore |
| ℷ(de3[lacI lacUV5-T7 gene 1 Ind1 sam7 nin5]) | ||
| MCA12 | F-, [araD139]B/r, leu-260::Tn10, ftsA12(ts), | (49) |
| Δ(argF-lac)169, λ−, e14-, flhD5301, Δ(fruK-yeiR) | ||
| 725(fruA25), relA1, rpsL150(strR), rbsR22, | ||
| Δ(fimB-fimE)632(::IS1), deoC1 | ||
| MCA27 | F-, [araD139]B/r, leu-260::Tn10, ftsA27(ts), | (49) |
| Δ(argF-lac)169, λ−, e14-, flhD5301, Δ(fruK-yeiR) | ||
| 725(fruA25), relA1, rpsL150(strR), rbsR22, | ||
| Δ(fimB-fimE)632(::IS1), deoC1 | ||
| Plasmid | ||
| pET-24b | kan, PT7 promoter | EMD Millipore |
| pET-ftsA | kan | (7) |
| pET-ftsA(ΔMTS) | kan | (7) |
| pET-ftsA(E14R) | kan | This study |
| pET-ftsZ | kan | (58) |
| pET-H6-Gfp-ftsZ | kan | (51) |
| pSEB293 | amp Para::Gfp-ftsA | (4, 33) |
| pBAD-Gfp-ftsA(E14R) | amp Para::Gfp-ftsA(E14R) | This study |
| pBAD-Gfp-ftsA(ΔMTS) | amp Para::Gfp-ftsA(ΔMTS) | This study |
| pBAD-Gfp-ftsA(ΔMTS/E14R) | amp Para::Gfp-ftsA(ΔMTS/E14R) | This study |
| pQE9 | amp PT5 promoter | (65) |
| pQE9-ftsA | amp | This study |
| pQE9-ftsA(E14R) | amp | This study |
| pQE9-ftsA(ΔMTS) | amp | This study |
| pQE9-ftsA(ΔMTS/E14R) | amp | This study |
Next, to determine if FtsA(E14R) supports division in vivo, we used the two temperature-sensitive strains of E. coli MCA12 and MCA27 (49). MCA12 contains a chromosomal ftsA mutation that substitutes amino acid Ala188 with Val, and MCA27 contains a chromosomal ftsA mutation that substitutes amino acid Ser195 with Pro (49), and each mutation in FtsA prevents growth of these strains at high temperatures (i.e., 42 °C) while still allowing growth at permissive temperatures (i.e., 30 °C). Cultures of temperature-sensitive strains containing plasmids encoding WT FtsA and FtsA(E14R) were grown at 30 °C to log phase (A600 0.4 A.U.), diluted and spotted onto LB agar plates containing ampicillin ,and grown overnight at the permissive (30 °C) and restrictive (42 °C) temperatures. We observed that the strains containing the vector (pQE-9) were unable to grow at the restrictive temperature (Figs. 1C and S1B). However, growth at high temperature was restored in both strains by the introduction of a plasmid containing ftsA (Figs. 1C and S1B). A plasmid encoding FtsA(E14R) did not support the growth of either strain at the restrictive temperature, although all strains grew at the permissive temperature (Figs. 1C and S1B). A plasmid encoding FtsA(ΔMTS) also did not support growth at the restrictive temperature, which is in agreement with a previous report (4). Finally, a plasmid containing both the E14R and ΔMTS mutations in ftsA was unable to restore growth at the restrictive temperature (Figs. 1C and S1B). Together, our results indicate that not only does FtsA(E14R) show no localization to the Z-ring and disrupted rod formation as a fluorescent fusion protein, an expression plasmid encoding FtsA(E14R) is also unable to complement temperature-sensitive strains in vivo suggesting that this protein has major functional defects.
To investigate why FtsA(E14R) is defective for function and localization in vivo, we purified FtsA(E14R) to determine if it is also defective for known FtsA activities in vitro. The failure to assemble into foci-like structures adjacent to the cell membrane, or at a division ring, could suggest that the protein is impaired for self-interaction, PL binding, or ATP hydrolysis. To determine if FtsA(E14R) is capable of robust ATP hydrolysis similar to WT FtsA, we measured the ATPase activity and determined that the rate of ATP hydrolysis by FtsA(E14R) is 60% slower than WT FtsA under the condition tested (12 and 33 ATP per min, respectively) (Fig. 2A). Based on the proximity of the side chain to the predicted hydrolysis site (Fig. 1A), we suspected that replacement of the conserved Glu near the active site with Arg could impair substrate interaction or activity as a result of introduction of the longer, positively charged side chain. Therefore, we measured Vmax and Km values for FtsA WT and mutant proteins (Fig. S1C). The Vmax values for FtsA and FtsA(E14R) were calculated to be 41.53 ± 1.76 min−1 and 26.51 ± 1.35 min−1, respectively. However, Km values for FtsA and FtsA(E14R) were not significantly different (0.358 ± 0.053 mM and 0.423 ± 0.073 mM, respectively), suggesting that although FtsA(E14R) binds and hydrolyzes ATP with a similar Km as WT FtsA, it is not in a conformation associated with maximal enzymatic activity.
Figure 2.
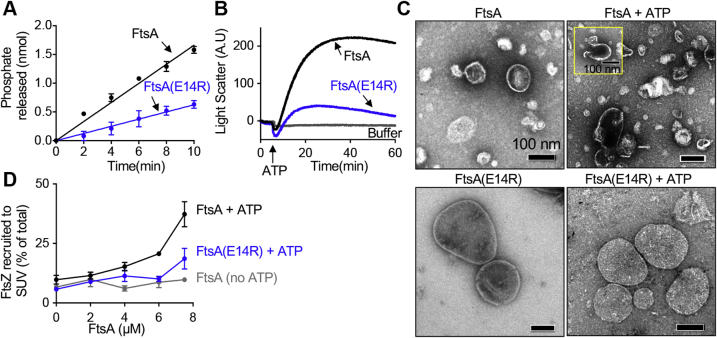
FtsA(E14R) is impaired for in vitro function.A, hydrolysis of ATP by FtsA (1 μM) and FtsA(E14R) (1 μM) with time as described in Experimental procedures. B, PL reorganization by FtsA (2 μM) and FtsA(E14R) (2 μM) were monitored by 90º angle light scatter. A baseline signal was measured for 5 min, and ATP was added and reactions were monitored for 60 min. C, FtsA (4 μM) and FtsA(E14R) (4 μM) incubated in the presence and absence of ATP and visualized by TEM. The scale bars represent 100 nm. Yellow box inset shows vesicle image from a replicate reaction. D, PL recruitment assay with reaction mixtures of FtsZ (6 μM) with GTP were pre-assembled and added to preincubated mixtures of FtsA or FtsA(E14R) (0–8 μM), SUV’s (250 μg ml−1), and ATP and fractionated by low-speed centrifugation. FtsA and FtsA(E14R) used in (A–D) contain copurifying PLs. The data from (A) and (D) are an average of three replicates and represented as mean ± SEM, and the data from (B) is representative of three replicates. Pellets and supernatants for (D) were visualized by SDS-PAGE and quantified by densitometry. PL, phospholipid; SUVs, small unilamellar vesicles; TEM, transmission electron microscopy.
Rapid ATP hydrolysis by FtsA requires PLs, and FtsA binds to PLs efficiently in vitro (7). To confirm that FtsA(E14R) is not defective for PL binding, we incubated FtsA(E14R) with small unilamellar vesicles (SUVs) prepared from E. coli membrane PLs and then collected the SUVs and bound protein by centrifugation. We determined that FtsA(E14R) was recruited to SUVs comparably to WT FtsA in the absence and presence of ATP (Fig. S1D). We previously reported that full-length FtsA copurifies with E. coli PLs, therefore we measured the amount of copurifying PLs with FtsA(E14R) and detected similar levels to those detected in WT FtsA, and they are not appreciably present in FtsA(ΔMTS) preparations (Fig. S1E).
In the presence of ATP, FtsA remodels PL architecture and induces tubulation of vesicles, which has been observed by TEM and 90º light scatter (LS) (7). Tubulation of vesicles is consistent with the polymerization of FtsA on the surface of a vesicle. To determine if FtsA(E14R) is defective for PL remodeling (i.e., vesicle tubulation), we used LS to detect large changes in complexes after addition of ATP. Under the conditions tested, FtsA produced a large change to the LS signal in response to the addition of ATP; however, FtsA(E14R) was unable to produce a similar LS increase as WT FtsA, which was 5-fold higher in magnitude (Fig. 2B). This suggests that FtsA(E14R) is defective for PL remodeling, yet, interestingly, it binds to PLs and hydrolyzes ATP, although at a slower rate than WT FtsA (Figs. 2A and S1, C and D). Next, we directly visualized FtsA-associated vesicles by TEM to determine if protrusions and tubulations associated with lipid remodeling are produced upon incubation of FtsA(E14R) with ATP (Fig. 2C). While we noted a large homogeneous population of PL vesicles in reactions with FtsA(E14R), addition of ATP produced no large observable effect on the architecture of the vesicles, and no rod-like tubulated structures were detected (Fig. 2C). Similar to a previous report, the addition of ATP to reactions containing WT FtsA and PLs resulted in large tubulated protrusions of the vesicles and short rod-like structures (Fig. 2C) (7). Across the high-resolution images collected for FtsA of 200 vesicles, we also observed 196 short tubules in the presence of ATP, but not when ATP was omitted. In contrast, in images collected for FtsA(E14R) of 200 vesicles, we observed seven short tubules with ATP, considerably fewer than WT FtsA, and tubules were not present when ATP was omitted. Together, these results indicate that FtsA(E14R) is unable to remodel vesicle architecture, which suggests a defect in assembly or conformational organization. Furthermore, the results are consistent with a model in which Glu14 relays information between the adjacent active site Mg2+ and regions of the protein that mediate self-interaction. This is further supported by the observation that Gfp-FtsA(E14R/ΔMTS) fails to produce long self-associated structures similar to Gfp-FtsA(ΔMTS) in vivo.
Finally, to determine if FtsA(E14R) is perturbed for binding to and recruitment of FtsZ, we tested direct binding in a PL recruitment assay. We performed low-speed sedimentation assays of FtsA and FtsA(E14R) with SUVs and ATP, where indicated. We observed that FtsA recruited FtsZ to PLs in a concentration- and ATP-dependent manner, demonstrating that ATP enhances FtsA recruitment of FtsZ to PLs, consistent with our previous report (7). We observed that FtsA(E14R) was defective for the recruitment of FtsZ to PLs in the presence of ATP compared to WT FtsA (Fig. 2D). These results show that FtsA(E14R) is impaired for the interaction with FtsZ, rapid ATP hydrolysis, and ATP-dependent PL remodeling.
Purified E. coli FtsA(ΔMTS) polymerizes in an ATP-dependent manner
E. coli FtsA(ΔMTS) is defective for ATP hydrolysis (7) (Fig. S1C), and Gfp-FtsA(ΔMTS) forms large, rod-like foci in vivo suggestive of polymer bundles (43). Therefore, we hypothesized that FtsA(ΔMTS) may be competent for assembly of stable actin-like polymers (50) that slowly turnover ATP and may be trapped in the polymer conformation. To test this, we purified FtsA(ΔMTS) and incubated it with and without ATP, then visualized the reactions by negative staining and TEM. In the presence of FtsA(ΔMTS) and ATP, we observed long, linear filaments consistent with single-stranded polymers (Fig. 3A). The polymers were highly varied in length and approximately 65 Å in width. In the absence of ATP, we did not observe filaments, polymers, or other observable structures (Fig. 3A). Therefore, FtsA(ΔMTS) polymerizes with ATP into long filaments.
Figure 3.
FtsA(ΔMTS) assembles into linear polymers in the presence of ATP.A, FtsA(ΔMTS) (12 μM) was incubated with (i and ii) and without (iii) ATP, and the reactions were visualized by TEM as described in Experimental procedures. The scale bars represent 100 nm. B, cartoon representation of FRET assembly reaction. AF594-labeled FtsA(ΔMTS) (donor) and AF647-labeled FtsA(ΔMTS) (acceptor) was mixed with FtsA(ΔMTS) and the reactions were excited at the donor excitation wavelength and scanned for the emission of each fluorophore. Close proximity of the donor fluorophore to the acceptor fluorophore causes a large increase in acceptor emission fluorescence, as occurs by the addition of ATP. C, FRET using increasing concentrations of Alexa Fluor 647-labeled FtsA(ΔMTS) (2.5–25 pmol) were mixed with a fixed concentration of Alexa Fluor 594-labeled FtsA(ΔMTS) (100 pmol) and FtsA(ΔMTS) for a total of 600 pmol. The reactions were monitored after addition of ATP as described in Experimental procedures. D, FRET reactions were assembled and monitored as in (C) but without the addition of ATP. E, FRET emission of Alexa Fluor 647-labeled FtsA(ΔMTS) (25 pmol) mixed with Alexa Fluor 594-labeled FtsA(ΔMTS) (100 pmol) and FtsA(ΔMTS) (475 pmol) was monitored after addition of ATP, ADP, ATP with EDTA, ATPγS, or no nucleotide addition. In (C–E), fluorescence values were corrected by subtraction of donor signal without acceptor. MTS, membrane targeting sequence; TEM, transmission electron microscopy.
Next, we developed a fluorescence-based self-assembly assay (FRET) to more quantitatively compare FtsA(ΔMTS)–FtsA(ΔMTS) interactions. We labeled two populations of FtsA(ΔMTS), one with Alexa fluor 594 (AF594; donor) and another with Alexa fluor 647 (AF647; acceptor), to measure energy transfer from donor to acceptor fluorophore during ATP-dependent assembly (Figs. 3B and S2A). We incubated donor-labeled FtsA(ΔMTS) with acceptor-labeled FtsA(ΔMTS) and observed a large emission signal in the presence of ATP (Figs. 3C and S2B), along with a corresponding decrease in donor-emission near 625 nm. The emission amplitudes that we measured titrated with the amount of acceptor-labeled FtsA(ΔMTS) in each reaction. We repeated this experiment in the absence of ATP and detected low level emission of the acceptor-labeled FtsA(ΔMTS), even at high acceptor concentration, compared to the emission observed in the presence of ATP (Figs. 3D and S2, B and C). Together, these results demonstrate that ATP enables the energy transfer between donor- and acceptor-labeled populations of FtsA(ΔMTS), consistent with FtsA(ΔMTS) self-interaction occurring under conditions that promote polymerization as detected by TEM (Fig. 3A).
Next, we compared the acceptor FRET signal associated with various nucleotide analogs and observed that ATP supported energy transfer most efficiently (Fig. 3E). The ATP-dependent emission signal was reduced in the presence of EDTA, which would prevent Mg2+ from binding in the active site, and the emission was also lower with ADP, compared to ATP. Neither ATPγS nor AMP-PNP supported energy transfer over the nucleotide-free control (Fig. 3E). Next, if ATP induces the assembly of stable FtsA(ΔMTS) polymers, then we hypothesized that we would be able to collect polymers by high-speed centrifugation. We incubated increasing amounts of FtsA(ΔMTS) with and without ATP, collected pellet and supernatant fractions after centrifugation at 160,000g for 30 min, and analyzed both fractions by SDS-PAGE. We observed that FtsA(ΔMTS) fractionated predominantly with the pellet (>50%) in the presence of ATP and predominantly with the supernatant (>95 %) when ATP was omitted (Fig. S2, D and E). Moreover, the relative amount of FtsA(ΔMTS) in the pellet increased with the total FtsA(ΔMTS) concentration in the reaction, suggesting that assembly efficiency increases with FtsA(ΔMTS) concentration. We compared the relative abilities of several nucleotides to support the polymerization of FtsA(ΔMTS) in the sedimentation assay and observed that ATP yielded the largest amount of pellet-associated FtsA(ΔMTS) (50.0%), and this amount was reduced by EDTA (16.5%) or with ADP (22.4%), ATPγS (19.5%), or AMP-PNP (<5%) (Fig. S2F). Finally, we used 90º LS to compare polymerization efficiency of FtsA(ΔMTS) at different temperatures, protein concentrations, and nucleotides. We observed that at 37 °C, addition of ATP produced a rapid increase in LS, which plateaued after approximately 10 min, and the amplitude change upon addition of ATP was reduced at lower temperatures (30 °C and 23 °C) (Fig. S2G). The amplitude of the LS change with ATP also increased with FtsA(ΔMTS) concentration up to 20 mM (Fig. S2H). Finally, the addition of ADP to FtsA(ΔMTS) also led to a modest increase in LS amplitude; however, it was ∼ 60% lower than the increase obtained with ATP (Fig. S2I), which is consistent with results from sedimentation assays (Fig. S2F). Addition of EDTA to the assembly reactions prevented the amplitude increase associated with both ATP and ADP, indicating that Mg2+ association is likely important for polymerization (Fig. S2I). Together, these results demonstrate that FtsA(ΔMTS) self-interacts in the presence of ATP and Mg2+ to form actin-like polymers.
FtsA(ΔMTS) polymers bind directly to FtsZ polymers but are destabilized by nonpolymerized FtsZ
To determine if polymers of FtsA(ΔMTS) stably interact with polymers of FtsZ, we developed a bead recruitment assay to overcome limitations associated with interpreting cosedimentation results of two polymer subassemblies. In this assay, we incubated a 6x-His-Gfp-tagged chimeric FtsZ with guanylyl-(alpha, beta)-methylene-diphosphonate (GMPCPP), a GTP analog that promotes the assembly of stable FtsZ polymers. We pre-assembled GMPCPP-stabilized polymers of FtsZ (H6-Gfp-FtsZ) and then recruited the polymers to the surface of a cobalt bead, which has an average diameter of 55 μm (Fig. 4A). H6-Gfp-FtsZ has previously been reported to hydrolyze GTP and polymerize in vitro, similar to WT FtsZ (51). Next, we pre-assembled FtsA(ΔMTS), labeled with AF594, with ATP to induce polymerization and then tested for recruitment to cobalt beads in the presence and absence of FtsZ and GMPCPP (Fig. 4B). We observed bundles of FtsZ and FtsA(ΔMTS) fluorescence in the presence of nucleotides GMPCPP and ATP, respectively (Fig. 4Bi). However, H6-Gfp-FtsZ with GMPCPP localized very differently when FtsA(ΔMTS) was omitted and instead localized along the bead surface with no visible clusters (Fig. 4Bii). Fluorescence microscopy does not resolve single-stranded polymers but does distinguish soluble protein, as diffuse fluorescence, from foci, which is suggestive of higher order assembly architecture. This was confirmed in a control assay in which fluorescent bundles were not observable by microscopy for either FtsZ or FtsA(ΔMTS) when GMPCPP or ATP were omitted, respectively (Fig. S3, A and B). Together, incubation of pre-assembled H6-Gfp-FtsZ polymers with FtsA(ΔMTS) polymers resulted in the colocalization of FtsA(ΔMTS) to the bead surface in the presence of His6-Gfp-FtsZ (i) but not when H6-Gfp-FtsZ was omitted (Fig. 4, Bi and Biii). Therefore, under conditions that promote polymerization of both proteins (i.e., with nucleotides GMPCPP and ATP), H6-Gfp-FtsZ recruits FtsA(ΔMTS) to the bead surface, suggesting that both polymer subassemblies interact directly.
Figure 4.
Coassembly of FtsA(ΔMTS) and FtsZ polymers.A, cartoon schematic of the recruitment of FtsA(ΔMTS)594 polymers by His6-Gfp-FtsZ complexes on cobalt beads. B, fluorescence and DIC microscopy of the reaction mixtures of (i) H6-Gfp-FtsZ(green), FtsZ, GMPCPP, and cobalt beads added to the reactions of AF594 FtsA(ΔMTS) (red), FtsA(ΔMTS), and ATP; (ii) H6-Gfp-FtsZ(green), FtsZ, GMPCPP, and cobalt beads; (iii) AF594 FtsA(ΔMTS) (red), FtsA(ΔMTS), ATP, and cobalt beads were assembled and visualized as described in Experimental procedures. The scale bars represent 1 μm for (i) and 5 μm for (ii) and (iii). C, FtsZ (8 μM) and GMPCPP added to pre-assembled reaction mixtures of FtsA(ΔMTS) (8 μM), where indicated, and ATP were visualized by TEM as described in Experimental procedures. Yellow box highlights areas enlarged for detail. DIC, differential interference contrast; GMPCPP, guanylyl-(alpha, beta)-methylene-diphosphonate; MTS, membrane targeting sequence; TEM, transmission electron microscopy.
To elucidate the in vitro protein architecture in mixtures of FtsA(ΔMTS) polymers, assembled with ATP, and FtsZ polymers, assembled with GMPCPP, we visualized polymers alone and together by TEM. We observed that GMPCPP-stabilized FtsZ polymers were long, single-stranded and approximately 40 to 45 Å in width, consistent with previous reports (7) (Fig. 4C). In contrast, FtsA(ΔMTS) polymers were visibly wider, approximately 60 to 75 Å in width, and also long and single-stranded (Figs. 3A and 4C). When FtsA(ΔMTS) and FtsZ polymers were incubated together and then visualized by TEM, we observed wide filaments (100–120 Å) and a population of polymers with a distinct paired-filament morphology. These doublet polymers also had a width of approximately 100 to 120 Å (Fig. 4C) and were composed of two aligned polymers of widths (∼ 60 Å and 40 Å) (Fig. 4C). The morphological features of the paired filament are consistent with an FtsZ polymer aligned with and bound to an FtsA(ΔMTS) polymer.
Next, we interrogated if FtsZ has a direct effect on the propensity for or degree of FtsA polymerization. Direct visualization of polymers is nonquantitative, therefore, to test if FtsZ modifies ATP-dependent assembly of FtsA(ΔMTS), we used the fluorescence-based FtsA(ΔMTS) subunit interaction FRET assay. We observed that addition of FtsZ (8 μM) reduced the fluorescence emission of acceptor-labeled FtsA(ΔMTS) by ∼ 40% under the conditions tested and to a lesser extent at lower FtsZ concentration (4 μM) (Fig. 5A) However, we did not observe reduced FRET emission at 675 nm by FtsA(ΔMTS) when FtsZ was pre-assembled with GMPCPP or GTP and then included in the reaction (Fig. S3C).
Figure 5.
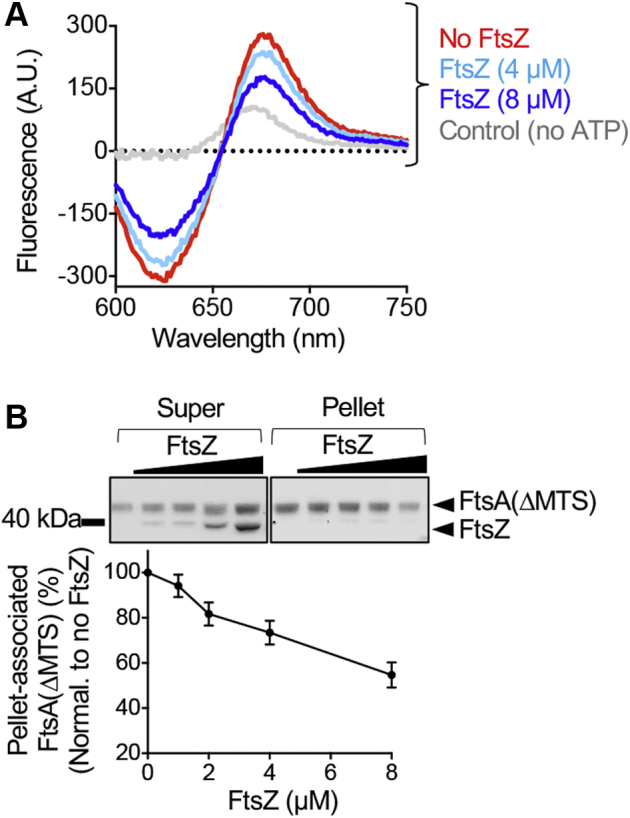
FtsA(ΔMTS) polymers are regulated by FtsZ.A, FRET emission of FtsA(ΔMTS) assembly reactions without FtsZ (red) or with FtsZ (8 μM, blue; 4 μM, light blue). The reactions contained Alexa Fluor 647-labeled FtsA(ΔMTS) (25 pmol), Alexa Fluor 594-labeled FtsA(ΔMTS) (100 pmol), and FtsA(ΔMTS) (475 pmol), and fluorescence emission was monitored after the addition of ATP as described in Experimental procedures. FtsA(ΔMTS) without ATP is shown as a control (gray). B, FtsZ (0–8 μM) was added to pre-assembled reaction mixtures of FtsA(ΔMTS) (8 μM) and ATP, incubated for 10 min, fractionated by ultracentrifugation, and analyzed by SDS-PAGE and densitometry. The graph shown displays an average of three replicates represented as mean ± SEM. MTS, membrane targeting sequence.
The reduced FtsA(ΔMTS) acceptor fluorescence with ATP and in the presence of FtsZ (nonpolymerized) suggests that FtsZ destabilizes the FtsA(ΔMTS) self-interaction, which would lead to disassembly of FtsA(ΔMTS) polymers. To test this directly, we performed high speed sedimentation assays to collect ATP-stimulated FtsA(ΔMTS) polymers and titrated FtsZ concentration. First, we pre-assembled FtsA(ΔMTS) (8 μM) with ATP (4 mM) and then added increasing concentrations of FtsZ (0–8 μM). After incubation for 10 min, FtsA(ΔMTS) polymers were collected by high-speed centrifugation, and then supernatants and pellets were analyzed by SDS-PAGE for the presence of FtsA(ΔMTS) and FtsZ. We observed that although we titrated the amount of FtsZ in the reaction, we failed to recover FtsZ in the sedimentation pellet with FtsA(ΔMTS) and ATP (Fig. 5B). In addition, the amount of FtsA(ΔMTS) fractionating with the pellets decreased as the FtsZ concentration in the reaction increased (Fig. 5B), with approximately 40% less FtsA(ΔMTS) in the pellet with FtsZ. This suggests that FtsZ, without GTP, destabilizes FtsA(ΔMTS) polymers assembled with ATP and is in agreement with the observed FRET result (Fig. 5A). Together, these results show that unassembled FtsZ destabilizes FtsA(ΔMTS) polymers and further suggest that stable FtsZ polymers and FtsA(ΔMTS) polymers align as a paired filament.
Discussion
Here, we demonstrate that in vitro FtsA forms linear ATP-dependent polymers using the truncated E. coli FtsA variant FtsA(ΔMTS) that are clearly observable by TEM (Fig. 3A) and in direct biophysical assays (Figs. 3C and S2, B–I). Most surprisingly, we also observed that FtsZ induces disassembly of FtsA(ΔMTS) polymers in the absence of GTP, but not when GTP or GMPCPP is present (Figs. 5, A and B, and S3C), and FtsZ coassembles with FtsA(ΔMTS) polymers in the presence of GMPCPP, suggesting that FtsZ polymers facilitate FtsA polymerization, and depolymerized FtsZ facilitates FtsA depolymerization. Moreover, this model further suggests that FtsA and FtsZ copolymers laterally associate and, in the presence of the MTS of FtsA, assemble at the membrane. This is consistent with the suggestion that in vivo, FtsA and FtsZ polymers assemble at midcell together and exhibit treadmilling (52). It has been proposed that FtsZ-interacting proteins, such as ZapA, modify the spatial order and dynamics of membrane-associated FtsZ bundles, but would not directly affect treadmilling (53), reflecting that the treadmilling behavior is likely inherent to FtsZ and FtsA nucleotide cycling.
While PL engagement is essential for rapid ATP hydrolysis by FtsA in vitro, the precise steps of nucleotide binding, hydrolysis and release, and how those steps regulate FtsA conformation remain to be determined. Interestingly, substitution of Glu14 with Arg, which is adjacent to the Mg2+ in the model of the active site, does not completely abolish ATPase activity (Figs. 2A and S1C), suggesting that, while Glu14 plays a role, it is not the major catalytic residue involved in hydrolysis or release of the ATP gamma phosphate. This role may well be played by D210, which is also ∼ 3 to 4 Å from the gamma phosphate in structural models. While catalysis by human and yeast actin has remained enigmatic, of particular importance are residues Q137, D154, and D11 with consideration of Q137 as supporting critical activation of a water molecule (54). Interestingly, actin residues D11, D154, and Q137 correspond to E14, D210, and A188, respectively, in E. coli FtsA (46). The minor substitution mutation A188V in the chromosome underlies the thermosensitive growth defect of the temperature-sensitive strain ftsA(12), suggesting that A188 is essential for growth under some conditions (49). Asp210 in E. coli FtsA has also been reported to be critical for function in vivo (33, 43).
FtsA(E14R) is defective for recruiting FtsZ polymers to SUVs in vitro and fails to form rings in vivo. Although FtsA(E14R) binds to SUV’s, in the presence of ATP it fails to induce lipid tubulation, a hallmark of polymerization and introduction of the E14R mutation in Gfp-FtsA(ΔMTS) prevents the accumulation of cytoplasmic foci in cells. These results have several implications. First, ATP hydrolysis does not appear to absolutely require polymerization, which is reminiscent of differing hydrolysis rates of G-actin (globular) and F-actin (filamentous) (55) and is in contrast to the mechanism for hydrolysis by FtsZ, where residues from adjacent protomers are required for enzymatic activity at a dimer interface (56). If FtsA(E14R) can engage PLs and hydrolyze ATP, then why is it defective for polymerization? It is likely that Glu14 regulates and/or stabilizes a key conformational transition that facilitates polymerization and thus stabilizes a direct FtsZ interaction, which must also be coordinated with PL engagement. This implies that FtsA therefore undergoes multiple conformational transitions, with full activity elicited by engagement of both PLs and ATP.
During cell division, we predict that the role of FtsA nucleotide binding is to regulate FtsA polymerization on the membrane and facilitate stable copolymerization with FtsZ polymers, in such a manner as to localize the periplasmic cell division proteins to a position adjacent to the cytoplasmic divisome. Coregulated assembly and disassembly of FtsZ and FtsA polymers, and therefore GTP and ATP cycling, would set the pace of the internal dynamic ring, enabling FtsZ and FtsA to treadmill in coordinated manner, and trigger initiation of peptidoglycan synthesis. We predict that other cell division proteins, such as ZipA or FtsEX, may also contribute to regulating the FtsA polymerization state and have been implicated in other studies (17, 43, 44, 45). Once recruited by FtsA, FtsN would then activate FtsQBL and FtsWI for cell wall synthesis (18, 45).
Experimental procedures
Bacterial strains and plasmid construction
The strains and plasmids used in this study are listed in Table 1. The ftsA gene was amplified from pET-ftsA and cloned into pQE9 using BamHI and an engineered NdeI restriction site. Site-specific mutants in ftsA were constructed by site-directed mutagenesis of ftsA expression plasmids using the QuikChange II XL mutagenesis system (Agilent) and confirmed by sequencing. pQE-9-ftsA was mutagenized to create FtsA(E14R), FtsA(ΔMTS), and FtsA(E14R/ΔMTS). FtsA(ΔMTS) was created by inserting a TAA stop codon at amino acid position 405 in ftsA. Plasmid pSEB293 (4), containing gfp-ftsA, was mutagenized to create Gfp-FtsA(E14R), Gfp-FtsA(ΔMTS), and Gfp-FtsA(E14R/ΔMTS). pET-ftsA was mutagenized to create FtsA(E14R) and FtsA(ΔMTS).
Protein purification and ATP hydrolysis assays
FtsA, FtsA(E14R), and FtsA(ΔMTS) were purified as previously reported (7). FtsZ and H6-Gfp-FtsZ were purified as previously reported (57, 58). ATP hydrolysis assays were performed by measuring the amount of inorganic phosphate released at 37 °C in reaction mixtures (25 μl) with a reaction buffer containing 50 mM Tris–HCl (pH 7.5), 150 mM KCl, and 10 mM MgCl2, FtsA, FtsA(E14R), or FtsA(ΔMTS) (1 μM) and ATP (4 mM) using Biomol Green (Enzo Life Sciences) and comparing to a phosphate standard curve. Protein concentrations refer to FtsA monomers and FtsZ monomers.
Transmission electron microscopy
Reactions of reaction buffer, FtsA(ΔMTS) (12 μM), FtsA (4 μM), FtsA(E14R) (4 μM), and ATP (4 mM), where indicated, were incubated for 5 min at 23 °C, applied to a 300-mesh carbon/formvar coated grid, fixed with glutaraldehyde (2.5%), and negatively stained with uranyl acetate (2%). The samples were imaged by TEM using a JEM-2100 80 Kev instrument.
Förster resonance energy transfer assays
Several Cys residues in FtsA(ΔMTS) are surface exposed when mapped on the structural model of E. coli FtsA (Fig. S2A). Two populations of FtsA(ΔMTS) were labeled with Alexa Fluor 594 (donor) and Alexa Fluor 647 (acceptor) according to the manufacturer’s protocol (ThermoFisher) to a degree of labeling of 0.5 to 2.0 dyes per protomer. This widely used donor-acceptor pair has a Förster radius of 79 Å, which corresponds closely to a distance equivalent to two adjacent FtsA protomers, with each protomer being approximately 40 Å in diameter. To monitor energy transfer, the reaction mixtures (80 μl) were incubated in an assembly buffer containing 50 mM Tris–HCl (pH 7.5), 150 mM KCl, 4 mM MgCl2, and, where indicated, AF594-labeled FtsA(ΔMTS) (100 pmol), AF647-labeled FtsA(ΔMTS) (0–25 pmol), FtsA(ΔMTS) (475–500 pmol), FtsZ (4–8 μM), and EDTA (15 mM) for 5 min at 37 °C. Fluorescence emission was scanned from 600 to 800 nm at an excitation wavelength of 590 nm. To compare nucleotides, ATP (4 mM), ADP (4 mM), or ATPγS (4 mM) were added, where indicated, and the reactions were incubated at 37 °C for 10 min, then emission spectra were collected. To correct for background and off-peak nonspecific donor signal, equivalent reactions omitting acceptor fluorophore were scanned and the curves were subtracted as indicated.
Assembly and recruitment assays
To measure FtsA(ΔMTS) polymer formation by ultracentrifugation, reaction mixtures (25 μl) containing assembly buffer, FtsA(ΔMTS) (0–12 μM), and ATP (4 mM), where indicated, were assembled. Reactions were incubated at 23 °C for 10 min and, where indicated, FtsZ (0–8 μM), or FtsZ (0–8 μM) preassembled with GTP (4 mM) and a nucleotide regenerating system containing acetate kinase (25 μg ml−1) and acetyl phosphate (15 mM), or GMPCPP (0.2 mM), were added. The reactions with FtsZ were carried out in assembly buffer with 10 mM MgCl2 instead of 4 mM MgCl2. The reactions were centrifuged at 160,000g for 30 min in a Beckman TLA 120.1 rotor. Pellets and supernatants were resuspended in equal volumes, analyzed by SDS-PAGE and Coomassie staining, and quantified by densitometry using NIH ImageJ.
To analyze PL recruitment of FtsZ polymers by FtsA, the reaction mixtures were assembled containing reaction buffer, FtsZ (6 μM), GTP (2 mM), a nucleotide regenerating system containing acetate kinase (25 μg ml−1) and acetyl phosphate (15 mM), were incubated for 3 min at 23 °C and added to preassembled reactions containing FtsA or FtsA(E14R) (0–8 μM), SUVs (250 μg ml−1), and ATP (4 mM). Small unilamellar vesicles from E. coli extracts (Avanti Polar lipids) were prepared as described previously (7). The reactions were incubated for an additional 10 min at 30 °C and centrifuged at 21,130g for 15 min to pellet vesicles. Pellets and supernatants were resuspended in equal volumes, analyzed by SDS-PAGE and Coomassie staining, and quantified by densitometry using NIH ImageJ.
Fluorescence microscopy
To observe fluorescence of FtsA in vivo, MG1655 araECP-containing plasmid pSEB293 (4) encoding Gfp-FtsA or variants Gfp-FtsA(E14R), Gfp-FtsA(ΔMTS), or Gfp-FtsA(E14R/ΔMTS) were grown overnight on solid LB media containing ampicillin (100 μg ml−1), restreaked, and grown for 6 h at 30 °C. Cells containing plasmid expressing Gfp-FtsA(ΔMTS) or Gfp-FtsA(E14R/ΔMTS) were restreaked onto LB agar plates supplemented with 0.001% L-arabinose. The cells were harvested from the plate, resuspended in PBS (phosphate buffered saline), applied to 5% agarose gel pads containing M9 minimal media supplemented with 0.2% glucose, and a coverslip was added. Samples were visualized with a Zeiss LSM 700 confocal fluorescence microscope with excitation and emission at 488/508 nm. Where indicated, a Nomarski prism was used to acquire differential interference contrast images. All the images were captured on an AxioCam digital camera with ZEN 2012 software.
To observe fluorescent FtsA(ΔMTS) and FtsZ polymers, H6-Gfp-FtsZ and FtsZ were mixed in a 1:1 ratio for a final concentration of 8 μM in assembly buffer with GMPCPP (0.2 mM) and resuspended Talon Cobalt beads (Takara) and added, where indicated, to pre-assembled reactions of Alexa Fluor 594 labeled FtsA(ΔMTS) mixed in a 1:1 ratio with FtsA(ΔMTS) for a final concentration of 8 μM and ATP (4 mM), where indicated. The reactions were imaged by epifluorescence microscopy using excitation and emission wavelengths of 488 and 508 nm respectively for Gfp (green channel) and 594 and 617 nm respectively for Alexa Fluor 594 (red channel) and where indicated, a Nomarski prism was used to acquire differential interference contrast images. Lipid concentrations of FtsA and mutant proteins were measured using the lipophilic dye probe FM 4 to 64 as described previously (7).
Temperature-sensitive growth assays for function in vivo
To assay for temperature-sensitive growth, MCA12 or MCA27 (49) containing plasmid pQE9, pQE9-ftsA, or pQE9-ftsA mutagenized to pQE9-ftsA(E14R), pQE9-ftsA(ΔMTS), or pQE9-ftsA(E14R/ΔMTS) were grown overnight at 30 °C in liquid LB media supplemented with ampicillin (100 μg ml−1), diluted to an A600 of 0.1 in LB media, grown to an A600 of 0.4 at 30 °C, spot plated log dilutions onto LB with ampicillin agar plates, and grown overnight at 30 or 42 °C, where indicated.
For immunoblotting of Gfp-FtsA, MG1655 araECP-containing plasmid pSEB293 (4) encoding Gfp-FtsA, or Gfp-FtsA mutagenized to Gfp-FtsA(E14R), Gfp-FtsA(ΔMTS), or Gfp-FtsA(E14R/ΔMTS) was grown in liquid LB medium supplemented with ampicillin (100 μg ml−1) and for cells expressing Gfp-FtsA(ΔMTS), or Gfp-FtsA(E14R/ΔMTS) supplemented with 0.001% L-arabinose, to an A600 of 0.8 at 30 °C. Proteins were precipitated with 15% trichloroacetic acid (Sigma-Aldrich) for 30 min at 4 °C. Suspensions were then centrifuged at 5000g for 10 min at 4 °C. Pellets were isolated and washed with acetone for 10 min at 4 °C followed by centrifugation at 10,000g for 10 min at 4 °C. 0.2 M NaOH was added to the pellets and then resuspended in buffer consisting of 20 mM Tris (pH 7.5) and 2% SDS, and a bicinchoninic acid protein determination was carried out. Equal concentrations of lysate (5 μg) were analyzed by reducing SDS-PAGE and transferred to a nitrocellulose membrane (Invitrogen). The membranes were washed with tris buffered saline (pH 7.6) and Tween-20 (0.05%), blocked for 2 h with 2% (w/v) bovine serum albumin, and probed with rabbit Gfp polyclonal antibody serum and goat anti-rabbit IgG coupled with horseradish peroxidase. Gfp-FtsA and mutant proteins were visualized using Pierce ECL Western blotting substrate.
Light scattering assays
To monitor PL remodeling-dependent LS, reaction mixtures (80 μl) containing reaction buffer and FtsA or FtsA(E14R) (2 μM) were monitored for 5 min to collect a baseline, then ATP (4 mM) or buffer was added, and the reactions were monitored for an additional 60 min. To monitor FtsA(ΔMTS) polymerization-dependent LS, reaction mixtures (80 μl) containing assembly buffer, where indicated, EDTA (15 mM) and FtsA(ΔMTS) (0–20 μM) were monitored at 37 °C unless otherwise indicated for 5 min to collect a baseline, then ATP (4 mM), ADP (4 mM), or buffer was added, and the reactions were monitored for an additional 30 min.
Structural modeling
The amino acid sequence of E. coli FtsA was modeled onto the coordinates of T. maritima FtsA crystallized with ATPγS (pdb: 4A2B) (8) by target-template alignment using ProMod3 by the Swiss-Model homology modeling server (59, 60, 61, 62).
Statistical information
Where indicated, values are reported as mean value plus or minus standard error generated from at least three independent replicates.
Data availability
All data pertinent to this work are contained within this article or available upon request. For requests, please contact Jodi Camberg at the University of Rhode Island, cambergj@uri.edu.
Supporting information
This article contains supporting information (8).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Janet Atoyan for sequencing and microscopy assistance, Katie Nelson for technical assistance, Cathy Trebino, Colby Ferreira, Eric DiBiasio, Negar Rahmani, and Ben Piraino for helpful suggestions and edits. Microscopy and sequencing were performed at the Rhode Island Genomics and Sequencing Center, supported in part by the National Science Foundation (MRI Grant No. DBI-0215393 and EPSCoR Grant No. 0554548 & EPS-1004057), the US Department of Agriculture (Grant Nos. 2002-34438-12688, 2003-34438-13111, and 2008-34438-19246), and the University of Rhode Island. The TEM data was acquired at the RI Consortium for Nanoscience and Nanotechnology, a URI College of Engineering core facility partially funded by the National Science Foundation EPSCoR, Cooperative Agreement #OIA-1655221.
Author contributions
J. J. M., J. C., and J. L. C. conceptualization; J. J. M., J. C., and J. L. C. methodology; J. J. M. and J. L. C. formal analysis; J. J. M. and J. L. C. writing–original draft; J. J. M., J. C., and J. L. C. writing–review and editing; J. C. resources; J. L. C. visualization; J. L. C. supervision; J. L. C. funding acquisition.
Funding and additional information
Research reported in this publication was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM118927 to J. L. C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the authors' respective institutions.
Edited by Enrique De La Cruz
Supporting information
References
- 1.Haeusser D.P., Margolin W. Splitsville: Structural and functional insights into the dynamic bacterial Z ring. Nat. Rev. Microbiol. 2016;14:305–319. doi: 10.1038/nrmicro.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z., Mukherjee A., Lutkenhaus J. Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol. 1999;31:1853–1861. doi: 10.1046/j.1365-2958.1999.01322.x. [DOI] [PubMed] [Google Scholar]
- 3.Pichoff S., Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pichoff S., Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 5.Mosyak L., Zhang Y., Glasfeld E., Haney S., Stahl M., Seehra J., Somers W.S. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000;19:3179–3191. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hale C.A., de Boer P.A. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 7.Conti J., Viola M.G., Camberg J.L. FtsA reshapes membrane architecture and remodels the Z-ring in Escherichia coli. Mol. Microbiol. 2018;107:558–576. doi: 10.1111/mmi.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szwedziak P., Wang Q., Freund S.M., Lowe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loose M., Mitchison T.J. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du S., Lutkenhaus J. Assembly and activation of the Escherichia coli divisome. Mol. Microbiol. 2017;105:177–187. doi: 10.1111/mmi.13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsang M.J., Bernhardt T.G. Guiding divisome assembly and controlling its activity. Curr. Opin. Microbiol. 2015;24:60–65. doi: 10.1016/j.mib.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du S., Lutkenhaus J. At the heart of bacterial cytokinesis: The Z ring. Trends Microbiol. 2019;27:781–791. doi: 10.1016/j.tim.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soderstrom B., Mirzadeh K., Toddo S., von Heijne G., Skoglund U., Daley D.O. Coordinated disassembly of the divisome complex in Escherichia coli. Mol. Microbiol. 2016;101:425–438. doi: 10.1111/mmi.13400. [DOI] [PubMed] [Google Scholar]
- 14.Busiek K.K., Eraso J.M., Wang Y., Margolin W. The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J. Bacteriol. 2012;194:1989–2000. doi: 10.1128/JB.06683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbin B.D., Geissler B., Sadasivam M., Margolin W. Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J. Bacteriol. 2004;186:7736–7744. doi: 10.1128/JB.186.22.7736-7744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vega D.E., Margolin W. Direct interaction between the two Z ring membrane anchors FtsA and ZipA. J. Bacteriol. 2019;201 doi: 10.1128/JB.00579-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du S., Pichoff S., Lutkenhaus J. FtsEX acts on FtsA to regulate divisome assembly and activity. Proc. Natl. Acad. Sci. U. S. A. 2016;113 doi: 10.1073/pnas.1606656113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park K.T., Du S., Lutkenhaus J. Essential role for FtsL in activation of septal peptidoglycan synthesis. mBio. 2020;11 doi: 10.1128/mBio.03012-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominguez R., Holmes K.C. Actin structure and function. Ann. Rev. Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanson J., Lowy J. Molecular basis of contractility in muscle. Br. Med. Bull. 1965;21:264–271. doi: 10.1093/oxfordjournals.bmb.a070407. [DOI] [PubMed] [Google Scholar]
- 21.Kilmartin J.V., Adams A.E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollard T.D., Cooper J.A. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaevitz J.W., Gitai Z. The structure and function of bacterial actin homologs. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Ent F., Lowe J. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 2000;19:5300–5307. doi: 10.1093/emboj/19.20.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoddard P.R., Williams T.A., Garner E., Baum B. Evolution of polymer formation within the actin superfamily. Mol. Biol. Cell. 2017;28:2461–2469. doi: 10.1091/mbc.E15-11-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper J.A., Buhle E.L., Jr., Walker S.B., Tsong T.Y., Pollard T.D. Kinetic evidence for a monomer activation step in actin polymerization. Biochemistry. 1983;22:2193–2202. doi: 10.1021/bi00278a021. [DOI] [PubMed] [Google Scholar]
- 27.Gershman L.C., Newman J., Selden L.A., Estes J.E. Bound-cation exchange affects the lag phase in actin polymerization. Biochemistry. 1984;23:2199–2203. doi: 10.1021/bi00305a015. [DOI] [PubMed] [Google Scholar]
- 28.Gurung R., Yadav R., Brungardt J.G., Orlova A., Egelman E.H., Beck M.R. Actin polymerization is stimulated by actin cross-linking protein palladin. Biochem. J. 2016;473:383–396. doi: 10.1042/BJ20151050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostrowska Z., Moraczewska J. Cofilin - a protein controlling dynamics of actin filaments. Postepy Hig. Med. Dosw. (Online) 2017;71:339–351. doi: 10.5604/01.3001.0010.3818. [DOI] [PubMed] [Google Scholar]
- 30.Merino F., Pospich S., Funk J., Wagner T., Kullmer F., Arndt H.D., Bieling P., Raunser S. Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM. Nat. Struct. Mol. Biol. 2018;25:528–537. doi: 10.1038/s41594-018-0074-0. [DOI] [PubMed] [Google Scholar]
- 31.Chou S.Z., Pollard T.D. Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc. Natl. Acad. Sci. U. S. A. 2019;116:4265–4274. doi: 10.1073/pnas.1807028115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan K., Pearce K.H., Payne D.J. A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA-FtsZ interaction in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 2000;270:387–392. doi: 10.1006/bbrc.2000.2439. [DOI] [PubMed] [Google Scholar]
- 33.Pichoff S., Lutkenhaus J. Identification of a region of FtsA required for interaction with FtsZ. Mol. Microbiol. 2007;64:1129–1138. doi: 10.1111/j.1365-2958.2007.05735.x. [DOI] [PubMed] [Google Scholar]
- 34.Geissler B., Shiomi D., Margolin W. The ftsA∗ gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology (Reading) 2007;153:814–825. doi: 10.1099/mic.0.2006/001834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geissler B., Elraheb D., Margolin W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernard C.S., Sadasivam M., Shiomi D., Margolin W. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol. Microbiol. 2007;64:1289–1305. doi: 10.1111/j.1365-2958.2007.05738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pichoff S., Du S., Lutkenhaus J. Disruption of divisome assembly rescued by FtsN-FtsA interaction in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E6855–E6862. doi: 10.1073/pnas.1806450115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krupka M., Rowlett V.W., Morado D., Vitrac H., Schoenemann K., Liu J., Margolin W. Escherichia coli FtsA forms lipid-bound minirings that antagonize lateral interactions between FtsZ protofilaments. Nat. Commun. 2017;8:15957. doi: 10.1038/ncomms15957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martos A., Monterroso B., Zorrilla S., Reija B., Alfonso C., Mingorance J., Rivas G., Jimenez M. Isolation, characterization and lipid-binding properties of the recalcitrant FtsA division protein from Escherichia coli. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krupka M., Cabre E.J., Jimenez M., Rivas G., Rico A.I., Vicente M. Role of the FtsA C terminus as a switch for polymerization and membrane association. mBio. 2014;5 doi: 10.1128/mBio.02221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nag D., Chatterjee A., Chakrabarti G. FtsA-FtsZ interaction in Vibrio cholerae causes conformational change of FtsA resulting in inhibition of ATP hydrolysis and polymerization. Int. J. Biol. Macromol. 2020;142:18–32. doi: 10.1016/j.ijbiomac.2019.11.217. [DOI] [PubMed] [Google Scholar]
- 42.Rueda S., Vicente M., Mingorance J. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J. Bacteriol. 2003;185:3344–3351. doi: 10.1128/JB.185.11.3344-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pichoff S., Shen B., Sullivan B., Lutkenhaus J. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA's self-interaction competes with its ability to recruit downstream division proteins. Mol. Microbiol. 2012;83:151–167. doi: 10.1111/j.1365-2958.2011.07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pichoff S., Du S., Lutkenhaus J. The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA-FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring. Mol. Microbiol. 2015;95:971–987. doi: 10.1111/mmi.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du S., Pichoff S., Lutkenhaus J. Roles of ATP hydrolysis by FtsEX and interaction with FtsA in regulation of septal peptidoglycan synthesis and hydrolysis. mBio. 2020;11 doi: 10.1128/mBio.01247-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bork P., Sander C., Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yim L., Vandenbussche G., Mingorance J., Rueda S., Casanova M., Ruysschaert J.M., Vicente M. Role of the carboxy terminus of Escherichia coli FtsA in self-interaction and cell division. J. Bacteriol. 2000;182:6366–6373. doi: 10.1128/jb.182.22.6366-6373.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gayda R.C., Henk M.C., Leong D. C-shaped cells caused by expression of an ftsA mutation in Escherichia coli. J. Bacteriol. 1992;174:5362–5370. doi: 10.1128/jb.174.16.5362-5370.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai K., Xu Y., Lutkenhaus J. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts) J. Bacteriol. 1993;175:3790–3797. doi: 10.1128/jb.175.12.3790-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fowler W.E., Aebi U. A consistent picture of the actin filament related to the orientation of the actin molecule. J. Cell Biol. 1983;97:264–269. doi: 10.1083/jcb.97.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viola M.G., LaBreck C.J., Conti J., Camberg J.L. Proteolysis-dependent remodeling of the tubulin homolog FtsZ at the division septum in Escherichia coli. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bisson-Filho A.W., Hsu Y.P., Squyres G.R., Kuru E., Wu F., Jukes C., Sun Y., Dekker C., Holden S., VanNieuwenhze M.S., Brun Y.V., Garner E.C. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science. 2017;355:739–743. doi: 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caldas P., Lopez-Pelegrin M., Pearce D.J.G., Budanur N.B., Brugues J., Loose M. Cooperative ordering of treadmilling filaments in cytoskeletal networks of FtsZ and its crosslinker ZapA. Nat. Commun. 2019;10:5744. doi: 10.1038/s41467-019-13702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vorobiev S., Strokopytov B., Drubin D.G., Frieden C., Ono S., Condeelis J., Rubenstein P.A., Almo S.C. The structure of nonvertebrate actin: Implications for the ATP hydrolytic mechanism. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5760–5765. doi: 10.1073/pnas.0832273100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCullagh M., Saunders M.G., Voth G.A. Unraveling the mystery of ATP hydrolysis in actin filaments. J. Am. Chem. Soc. 2014;136:13053–13058. doi: 10.1021/ja507169f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsui T., Han X., Yu J., Yao M., Tanaka I. Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop. J. Biol. Chem. 2014;289:3501–3509. doi: 10.1074/jbc.M113.514901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camberg J.L., Viola M.G., Rea L., Hoskins J.R., Wickner S. Location of dual sites in E. coli FtsZ important for degradation by ClpXP; one at the C-terminus and one in the disordered linker. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Camberg J.L., Hoskins J.R., Wickner S. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc. Natl. Acad. Sci. U. S. A. 2009;106:10614–10619. doi: 10.1073/pnas.0904886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guex N., Peitsch M.C., Schwede T. Automated comparative protein structure modeling with SWISS-model and Swiss-PdbViewer: A historical perspective. Electrophoresis. 2009;30:S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 61.Bienert S., Waterhouse A., de Beer T.A., Tauriello G., Studer G., Bordoli L., Schwede T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017;45:D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blattner F.R., Plunkett G., 3rd, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Gregor J., Davis N.W., Kirkpatrick H.A., Goeden M.A., Rose D.J., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 64.Khlebnikov A., Datsenko K.A., Skaug T., Wanner B.L., Keasling J.D. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology (Reading) 2001;147:3241–3247. doi: 10.1099/00221287-147-12-3241. [DOI] [PubMed] [Google Scholar]
- 65.Meyer H.H., Shorter J.G., Seemann J., Pappin D., Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data pertinent to this work are contained within this article or available upon request. For requests, please contact Jodi Camberg at the University of Rhode Island, cambergj@uri.edu.