Abstract
Thirty-two clofazimine derivatives, of which twenty-two were new, were synthesized and evaluated for their antiviral effects against both rabies virus and pseudo-typed SARS-CoV-2, taking clofazimine (1) as the lead. Among them, compound 15f bearing 4-methoxy-2-pyridyl at the N5-position showed superior or comparable antiviral activities to lead 1, with the EC50 values of 1.45 μM and 14.6 μM and the SI values of 223 and 6.1, respectively. Compound 15f inhibited rabies and SARS-CoV-2 by targeting G or S protein to block membrane fusion, as well as binding to L protein or nsp13 to inhibit intracellular biosynthesis respectively, and thus synergistically exerted a broad-spectrum antiviral effect. The results provided useful scientific data for the development of clofazimine derivatives into a new class of broad-spectrum antiviral candidates.
Keywords: Clofazimine analogues, Rabies virus, Structure–activity relationship, SARS-CoV-2, Broad-spectrum
Graphical abstract
1. Introduction
Rabies virus, a negative-stranded RNA virus, causes fatal brain damage and other systematic symptoms, which leads to a mortality rate of nearly 100% [[1], [2], [3]]. To date, pre- or post-exposure prophylaxis, along with rabies immunoglobulin (RIG) is the only available way to protect the infected crowd [3]. However, multiple administrations of rabies vaccine and the public's lack of awareness hampere their in-time protection against the rabies virus [[4], [5], [6]], and an average of 60,000 people die from rabies each year globally t [2]. The most favored Milwaukee protocol (M therapy) and its modified regimens obtained disappointing clinical outcomes [7,8]. Up to now, no drug has been approved for rabies treatment albeit with multiple efforts on drug discovery. Therefore, it is of great significance to find effective drugs for the treatment of rabies virus infection.
Recently, we found that riminophenazine alkaloid clofazimine (1, Fig. 1 ), an anti-leprosy and anti-tuberculous drug for clinical use, exhibited a good anti-rabies efficacy by affecting the g-glycoprotein (G protein)-mediated membrane fusion process [9]. At the meantime, Yuen's group discovered that it also effectively inhibits the replication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) both in vitro and in vivo, by interfering with spike glycoprotein (S protein)-mediated cell fusion and viral helicase activity [10]. These results indicated that 1 had a broad-spectrum antiviral activity, and deserved further investigation. However, highly lipophilic compound 1 upon therapeutic administration accumulates in lipid-rich tissues, and produces the most documented the red skin-coloring side effect [11], which might be alleviated by the reduction on lipophilicity [12,13].
Fig. 1.
The chemical structure, antiviral-activity and modification strategy of 1.
Therefore, in the present study, taking 1 as the lead compound, several series of clofazimine analogues with reduced ClogP values were synthesized and evaluated for anti-rabies as well as anti-pseudo-typed SARS-CoV-2 (pSARS-CoV-2) activities. The anti-rabies structure–activity relationship (SAR) analysis, safety profile and the dual-target antiviral mechanism exploration of the key compound were presented.
2. Result and discussion
2.1. Chemistry
A total of 32 clofazimine derivatives, of which 22 were new, were prepared as depicted in Scheme 1, Scheme 2 respectively. The synthesis of compounds 6a, 6b, 7a–c, 7l–o and 15b were reported previously [[13], [14], [15], [16], [17], [18], [19]].
Scheme 1.
Reagents and conditions: (a) DFDNB, Et3N, ethanol, rt; (b) Substituted aniline or benzylamine or 5-amino-2-methoxypyridine, Et3N, THF, 64 °C; (c) Zn/acetic acid, ethanol, rt; (d) Air, ethanol, rt; (e) R2NH2, acetic acid, dioxane, 101 °C.
Scheme 2.
Reagents and conditions: (a) 2-Fluoronitrobenzene, Et3N or NaHCO3, DMF, 90–100 °C; (b) 10% Pd/C, H2, ethanol, rt; (c) DFDNB, Et3N, ethanol, rt; (d) R1NH2, Et3N, THF, 64 °C; (e) 10% Pd/C, H2, methanol/THF, rt; (f) Air, ethanol/CH2Cl2, rt; (g) R2NH2, acetic acid, dioxane, 101 °C.
As shown in Scheme 1, nucleophilic substitution of starting material 2a or 2b with equimolar 1,5-difluoro-2,4-dinitrobenzene (DFDNB) achieved 3a or 3b, which underwent a subsequent nucleophilic substitution reaction with various of amines, such as anilines, benzylamines or 5-amino-2-methoxypyridine, to give key intermediates 4a–m in 78–95% yields. Reduction of nitro groups in 4a–m by zinc powder in acetic acid and a subsequent oxidative cyclization under ambient condition achieved riminophenazines 6a–m in yields of 24–55%. Finally, in the presence of acetic acid, the condensation of 6a–m with varied amines led to target compounds 7a–q in yields of 35–64%.
As described in Scheme 2, the nucleophilic substitution between 8a or 8b at 90–100 °C in DMF achieved 9a or 9b in yields of 55–61% instead of the conventional fusing reaction operation [13], followed by a reduction via Pd–C hydrogenation to yield key intermediate 10a or 10b in good yields. Next, following a similar five-step procedure as shown in Scheme 1, including nucleophilic substitution with DFDNB, nitro reduction, nucleophilic substitution with varied anilines, nitro reduction via Pd–C hydrogenation and ambient oxidation, compounds 14a–g were gained in yields of 46–80%. Finally, products 15a–j were acquired via condensation of compounds 14a–g with amines in the presence of acetic acid in yields of 48–80%. The products were isolated for the flash column chromatography with petroleum ether/CH2Cl2 or CH2Cl2/ethyl acetate as eluent.
2.2. SAR for anti-rabies activity of the target compounds in BSR cells
All target compounds were measured for their in vitro anti-rabies activities in BSR cells using rapid fluorescent focus inhibition test (RFFIT) assay constructed in our lab, taking 1 as the control [9]. The combination of the EC50 and selectivity index (SI, CC50/EC50 ratio) values of each compound was taken as the evaluation reference for antiviral potency. The structures and anti-rabies activities of all the target compounds were listed in Table 1 .
Table 1.
Anti-rabies activity and cytotoxicity of all the target compounds.
| R | R1 | R2 | CC50(μM)a | EC50 (μM)b | SIc | ClogPd | |
|---|---|---|---|---|---|---|---|
| 1 | Cl | 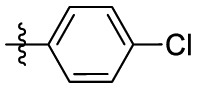 |
 |
323 | 2.28 | 141 | 5.39 |
| 6a | Cl | 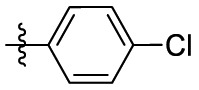 |
H | 35.4 | 20.6 | 1.7 | 4.50 |
| 6b | Cl | 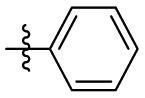 |
H | 35.4 | 20.6 | 1.7 | 3.94 |
| 6d | Cl | 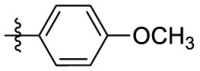 |
H | 106 | 106 | 1.0 | 3.82 |
| 6e | Cl | 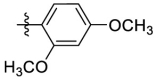 |
H | 35.4 | 35.4 | 1.0 | 3.69 |
| 6l | H |  |
H | 35.4 | 35.4 | 1.0 | 4.71 |
| 7a | Cl | 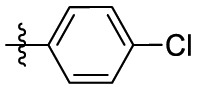 |
 |
323 | 22.7 | 14.2 | 5.03 |
| 7b | Cl |  |
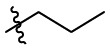 |
323 | 24.9 | 13.0 | 5.56 |
| 7c | Cl | 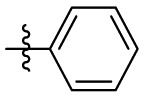 |
 |
185 | 8.44 | 21.9 | 4.83 |
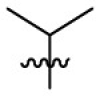
| 7d | Cl | 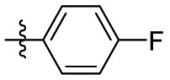 |
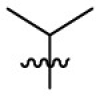 |
281 | 4.56 | 61.7 | 4.99 |
| 7e | Cl | 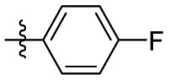 |
 |
323 | 6.86 | 47.1 | 4.63 |
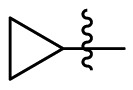
| 7f | Cl | 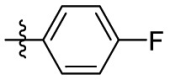 |
 |
54.5 | 1.67 | 32.6 | 5.16 |
| 7g | Cl | 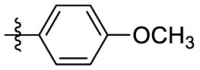 |
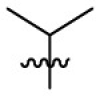 |
130 | 6.86 | 19.0 | 4.71 |
| 7h | Cl |  |
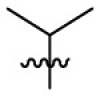 |
120 | 6.86 | 17.6 | 4.58 |
| 7i | Cl | 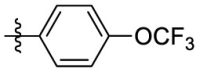 |
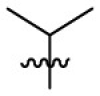 |
323 | 1.77 | 182 | 6.36 |
| 7j | Cl | 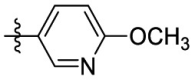 |
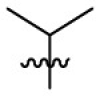 |
35.4 | 2.29 | 15.5 | 4.09 |
| 7k | Cl | 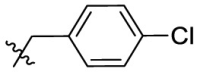 |
 |
93.1 | 0.76 | 123 | 5.60 |
| 7l | H | 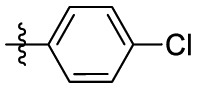 |
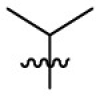 |
42.6 | 1.90 | 22.4 | 4.83 |
| 7m | H | 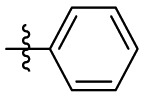 |
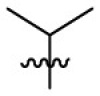 |
323 | 10.7 | 30.1 | 4.28 |
| 7n | H | 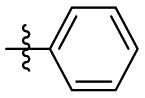 |
 |
14.4 | 0.76 | 19.0 | 4.44 |
| 7o | H | 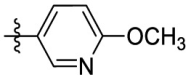 |
 |
107 | 5.03 | 21.2 | 3.53 |
| 7p | H | 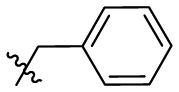 |
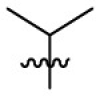 |
107 | 1.40 | 76.4 | 4.49 |
| 7q | H | 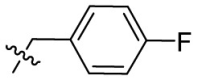 |
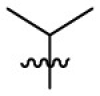 |
17.3 | 1.90 | 9.1 | 4.64 |
| 15a | F | 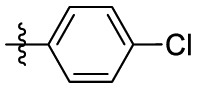 |
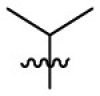 |
223 | 4.02 | 55.6 | 4.99 |
| 15b | F | 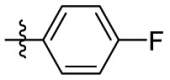 |
 |
314 | 2.29 | 134 | 4.59 |
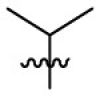
| 15c | OCH3 | 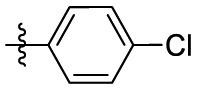 |
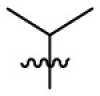 |
107 | 5.03 | 21.3 | 4.09 |
| 15d | OCH3 | 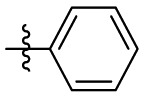 |
 |
323 | 12.3 | 26.3 | 3.53 |
| 15e | OCH3 | 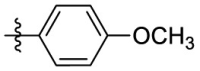 |
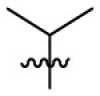 |
323 | 13.2 | 24.5 | 3.40 |
| 15f | OCH3 | 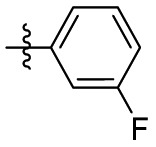 |
 |
323 | 1.45 | 223 | 3.69 |
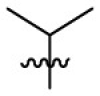
| 15g | OCH3 | 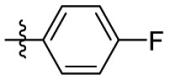 |
 |
323 | 1.41 | 229 | 3.69 |
| 15h | OCH3 | 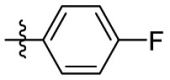 |
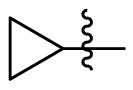 |
107 | 13.2 | 8.1 | 3.33 |
| 15i | OCH3 | 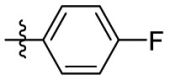 |
 |
107 | 1.40 | 76.4 | 3.86 |
| 15j | OCH3 | 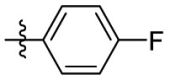 |
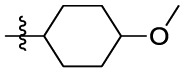 |
84.4 | 2.55 | 33.1 | 3.71 |
Cytotoxic concentration required to inhibit BSR cell growth by 50%.
Concentration required to inhibit rabies virus growth by 50%.
Selectivity Index values equaled to CC50/EC50.
Calculated by Chemdraw 14.0 (CambridgeSoft).
Initially, different substituents were introduced on the imine N atom at C-3 position to generate compounds 6a (H), 7a (cyclopropyl) and 7b (n-propyl), and their anti-rabies activity displayed declined activity. It was speculated that isopropyl group on the imine N atom might be beneficial for activity.
Next, the 3-isopropyl imine was retained, and the p-chloroaniline moiety attached on C-2 position was replaced by different anilines, 6-methoxy-3-pyridylamine or phenylethylamine, to generate compounds 7c, 7d and 7g–k. Most of them gave decreased activity to varying degrees, except that compounds 7i with p-trifluoromethoxyaniline and 7k with p-chlorobenzylamine displayed comparable activities to the lead 1, with the IC50 values of 1.77 μM and 0.76 μM and SI values of 182 and 123, respectively. Then, the 3-isopropyl imine was kept, modification on the N-5 substituents was carried out, and p-chlorophenyl moiety was altered into phenyl, p-fluorophenyl or 6-methoxy-3-pyridylamine, and the corresponding analogues 7l, 7m, 7o–q and 15a–g were generated and evaluated. Among them, compounds 7l, 7p and 7q bearing a p-chloroaniline, benzylamine and p-fluorobenzylamine on their C-2 position gave elevated antiviral activity with the IC50 values of 1.40, 1.40 and 1.90 μM respectively, as well as obviously increased cytotoxicity. Compounds 15f and 15g displayed the highest antiviral activity, with IC50 value of 1.45 μM and 1.41 μM, and SI value of 223 and 229, respectively. Meanwhile, the replacement of 3-isopropyl imine with imine, cyclopropyl imine, propyl imine or 4-methoxycyclohexyl imine led to decreased activity as in all cases of compounds 6b, 6d, 6e, 6l, 7e, 7f, 7n and 15h–j, and 3-isopropyl imine was thus considered as a beneficial moiety for the anti-rabies activity.
2.3. The anti-pSARS-CoV-2 activity of the target compounds in Huh7 cells
Meanwhile, the anti-SARS-CoV-2 activities of target clofazimine derivatives were evaluated on an S protein-based pSARS-CoV-2 model in Huh 7 cells constructed in our lab [20,21], taking 1 as the positive control. As indicated in Table 2 , the anti-pSARS-CoV-2 activity tendency of most compounds was similar to the anti-rabies tendency as disclosed above, and compounds 7m, 15f and 15g gave elevated activities compared to 1. Especially, compounds 15f and 15g gave the EC50 values of 14.6 μM and <10 μM, and SI values of 6.1 and >8.9, respectively, suggesting that 15f and 15g might own a broad-spectrum antiviral activity against rabies viruses and pSARS-Cov-2, worthy of the further investigation.
Table 2.
Anti-pSARS-CoV-2 activity and cytotoxicity of all the target compounds.
| Code |
pSARS-CoV-2 |
Code |
pSARS-CoV-2 |
||||
|---|---|---|---|---|---|---|---|
| CC50(μM)a | EC50 (μM)b | SIc | CC50(μM)a | EC50 (μM)b | SIc | ||
| 1 | 44.7 | 10 | 4.5 | 7l | 16.1 | <10 | >1.6 |
| 6a | NTd | NT | NT | 7m | 177 | 17.2 | 10.2 |
| 6b | <10 | <10 | / | 7n | 35.6 | <10 | >3.5 |
| 6d | >267 | 154 | >1.7 | 7o | 22.7 | 13.5 | 1.7 |
| 6e | 16.6 | 16.1 | 1.0 | 7p | <10 | <10 | / |
| 6l | NT | NT | NT | 7q | 29.6 | <10 | >2.9 |
| 7a | >267 | 141 | >1.9 | 15a | 17.0 | 29.6 | 0.6 |
| 7b | 107 | 39.3 | 2.7 | 15b | 267 | 81.1 | 3.3 |
| 7c | 154 | 40.9 | 3.8 | 15c | 51.3 | <10 | >5.1 |
| 7d | 40.6 | 14.5 | 2.8 | 15d | <10 | <10 | / |
| 7e | 16.7 | 13.5 | 1.2 | 15e | 17.1 | <10 | >1.7 |
| 7f | 14.9 | 10.3 | 1.5 | 15f | 88.9 | 14.6 | 6.1 |
| 7g | 88.9 | 16.6 | 5.4 | 15g | 88.9 | <10 | >8.9 |
| 7h | 13.52 | <10 | >1.3 | 15h | 88.9 | 33.1 | 2.7 |
| 7i | 38.37 | 9.98 | 3.8 | 15i | <10 | <10 | / |
| 7j | >267 | 153 | >1.7 | 15j | 51.3 | <10 | >5.1 |
| 7k | 16.1 | 12.4 | 1.3 | ||||
Cytotoxic concentration required to inhibit Huh 7 cell growth by 50%.
Concentration required to reduce 50% of the virus level in Huh 7 cell.
Selectivity Index values equaled to CC50/EC50.
NT: not tested.
2.4. The safety profiles of compounds 15g and 15f
To evaluate the safety profiles of 15g and 15f, an acute toxicity test was performed in Kunming mice. Both compounds were given orally in a single-dosing experiment at 0, 200, 400, 600 and 800 mg•kg−1, respectively. The mice were closely monitored for 7 days, and all the surviving mice had glossy hair, fleshy body and good appetite. Therefore, the median lethal dose (LD50) value for both 15g and 15f was over 800 mg•kg−1, indicating their good safety profiles in vivo.
2.5. Mechanism of action of the key compounds
2.5.1. Time of addition assay
In order to figure out the precise step of rabies proliferation that is blocked by the key compounds, we first evaluated their anti-rabies activity using a time-of-addition assay in a single infectious cycle in BSR cells. As illustrated in Fig. 2 A, compounds 15f or 15g was incubated into BSR cells at 1 h before rabies challenge virus standard (CVS) strain infection (−1 h), during infection (0 h), or 1 h post infection (+1 h), taking compound 1 as the positive control and DMSO as a negative control. As shown in Fig. 2B, both compounds 15f and 15g exerted strong inhibitory effects when added 1 h post infection at both 4 °C and 37 °C, indicating that they interfered with the viral membrane fusion and intracellular biosynthesis, similar to the lead 1. The results suggested that they might have dual-target or multi-target mechanism against rabies virus, and the inhibitory behavior of compound 15f was highly similar to that of the lead 1.
Fig. 2.
A single-cycle time-of-addition experiment was conducted with rabies CVS strain. A) Compound 15f, 15g or 1 (25 μM) was added and left for 1 h before infection (−1 h), during infection (0 h), and for 1 h after infection (+1 h). After incubated for 23 h, inhibition of rabies infection was detected as a decrease in luciferase signal. B) Inhibition of rabies CVS infection by a test compound was detected as a decrease in fluorescence activity, and error bars indicate standard deviations. The experiment was repeated for 3 times.
2.5.2. Dual-target mechanism of compound 15f against rabies virus
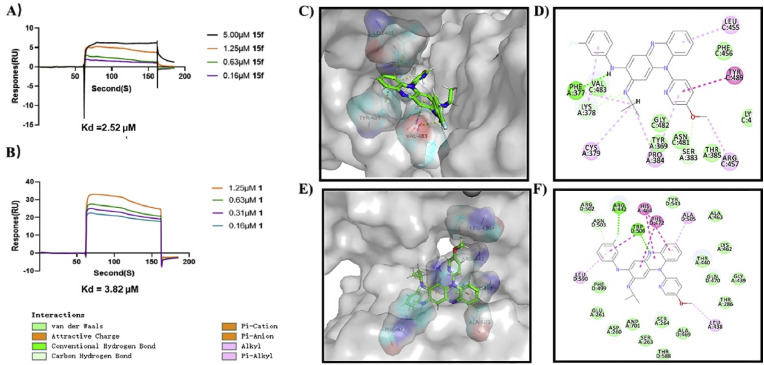
Our earlier molecular docking analysis indicated that compound 1 might target rabies G protein to interfere with viral membrane fusion [9], therefore, the recombinant rabies G protein was prepared, and surface plasmon resonance (SPR) assay was conducted to further confirm the direct interaction between rabies G protein and compound 15f. As depicted in Fig. 3 A and B, it displayed the concentration-dependent binding with immobilized G protein with Kd value of 0.28 μM, while 1 gave the Kd value of 0.73 μM. Then, molecular docking analysis between rabies G protein (PDB ID: 6LGW) and 15f was performed by Discovery Studio 4.5. As disclosed in Fig. 3C and D, van der waals force and hydrophobic interactions contributed to the strong interaction between 15f and G protein, and gave the Libdock scores of 106.1. These results demonstrated that compound 15f blocked rabies viral membrane fusion by binding to G protein.
Fig. 3.
A, B) SPR sensorgrams obtained on a rabies virus glycoprotein-coated chip at different concentrations of 15f and 1. C–F) Molecular docking models of 15f bound to rabies virus glycoprotein (PDB ID: 6LGW) or rabies L protein (PDB ID: 6EUB) by Discovery Studio 4.5. C, E) Solid surface map of the interaction pockets with compound 15f. D, F) 2D modes of interaction pockets with compound 15f. Ligand is colored by element type (N, blue; O, red; F, light blue), key bonds are indicated by dashed lines between the atoms involved, and the colors of key bonds and residues are shown according to the interaction mode.
Among all 5 proteins expressed by rabies RNA genome, L protein plays the vital role in conducting the RdRp activity in rabies virus [22]. Therefore, molecular docking analysis between compound 15f with lyssavirus L protein (PDB ID: 6EUB) was launched by Discovery Studio 4.5, taking 1 as the control. It gave a high Libdock score of 106.7, while 1 gave that of 104.7. As disclosed in Fig. 3E and F and Supplementary Figs. S1A and S1B, the multiple hydrogen bonds with Leu1835, Ile1843 and Gln1844 between 15f and L protein might contribute to the higher affinity of 15f over 1, suggesting that compound 15f might directly bind to rabies L protein, and thus inhibit the viral biosynthesis. Our results indicated that compound 15f might exert the anti-rabies efficiency by simultaneously targeting G protein and L protein in the membrane fusion and intracellular biosynthesis stages respectively, consistent with the above time-of-addition results.
2.5.3. Dual-target mechanism of compound 15f against SARS-CoV-2
Similarly, the direct interaction between recombinant SARS-CoV-2 S protein and 15f was conducted by SPR assay, taking 1 as the control. As demonstrated in Fig. 4 A and B, compound 15f could directly bind to immobilized SARS-CoV-2 S protein in a concentration-dependent manner, and gave the Kd value of 2.52 μM, while 1 gave the that of 3.82 μM. The molecular docking analysis between compound 15f and SARS-CoV-2 S protein (PDB ID: 6VSB) was performed (Fig. 4C and D), taking 1 as the control (Supplementary Figs. S1C and S1D). Compound 15f gave the Libdock scores of 124.1, while lead 1 gave the score of 118.6. As disclosed in Fig. 4C and D, besides the hydrogen bond between their N2 atoms with Phe377 and Val483 respectively, they fit in a T-shaped cavity of SARS-CoV-2 S protein, which was accomplished by the planar tricyclic scaffold and the pyridine or benzene moiety respectively. And the residues Thr385, Lys458, Arg547 and Ser383 might mainly contribute to strong val der waals force between 15f and the target S protein.
Fig. 4.
A, B) SPR sensorgrams obtained on a SARS-CoV-2 S protein-coated chip at different concentrations of 15f and 1. C–F) Molecular docking models of 15f bound to SARS-CoV-2 S protein (PDB ID: 6VSB) or nsp13(PDB ID: 7NN0) by Discovery Studio 4.5. C, E) Solid surface map of the interaction pockets with compound 15f. D, F) 2D modes of interaction pockets with compound 15f. Ligand is colored by element type (N, blue; O, red; F, light blue), key bonds are indicated by dashed lines between the atoms involved, and the colors of key bonds and residues are shown according to the interaction mode.
Since lead 1 inhibited the unwinding activity of nsp13 in the SARS-CoV-2 viral biosynthesis [10], the direct interaction between nsp13 (PDB ID: 7NN0) and compound 15f was predicted by docking analysis (Fig. 4E and F), taking 1 as the control (Supplementary Figs. S1E and S1F). Compounds 15f and 1 gave reasonable Libdock scores of 110.0 and 103.0, respectively. Apparently, they fit well in the same cavity, and formed a common hydrogen bond between the N10 atom and Trp506, but compound 15f also formed a second hydrogen bond between fluorine atom and Arg272, which might contribute to its higher affinity. Besides, both of them formed π-π stacking effect with His464 and Phe472, indicating that 15f might directly target nsp13 to inhibit SARS-CoV-2 biosynthesis.
As shown in Fig. 5 , our results suggested that clofazimine derivative 15f might simultaneously bind to the rabies G protein in viral membrane fusion, as well as L protein in viral biosynthesis stage, and synergistically exert the anti-rabies activity. Similarly, it could also simultaneously target SARS-CoV-2 S protein in viral membrane fusion, and nsp13 in viral biosynthesis to inhibit the proliferation of SARS-CoV-2, and thus shed light on the discovery of a new class of broad-spectrum antiviral agent through a dual-target mechanism.
Fig. 5.
The dual-target mechanism of compound 15f against rabies and SARS-CoV-2.
3. Conclusions
To sum up, 32 clofazimine derivatives, of which 22 were new, were designed, synthesized and evaluated for their antiviral activity against rabies and pSARS-CoV-2 viruses, taking 1 as the lead. Compound 15f showed potent antiviral activities with the IC50 values of 1.45 μM and 14.6 μM and the SI values of 223 and 6.1, respectively, superior or comparable to those of lead 1. Additionally, it gave a good safety profile with the oral LD50 value of over 800 mg•kg1. Compound 15f affected the rabies and SARS-CoV-2 proliferation by targeting G or S protein to block the viral membrane fusion, as well as binding to L protein or nsp13 to inhibit viral biosynthesis, respectively, and thus synergistically exerted antiviral effect. And our study provided useful scientific data for the development of clofazimine derivatives as a new class of broad-spectrum antiviral candidates with a dual-target antiviral mechanism, worthy of further investigation.
4. Experimental section
4.1. Chemistry
4.1.1. General
All commercial reagents and solvents were obtained from the commercial provider and used without further purification. Melting points (mp) were obtained with a MP90 melting point apparatus (Mettler-Toledo, Greifensee, Switzerland). 1H NMR and 13C NMR spectra were recorded on a Bruker Avance III 500 or II600 spectrometers (Varian, San Francisco, USA) in DMSO‑d 6 with Me4Si as the internal standard. ESI high-resolution mass spectra (HRMS) were recorded on an Autospec Ultima-TOF spectrometer (Micromass UK Ltd., Manchester, U.K.). Flash chromatography over silica gel was performed on Combiflash Rf 200 (Teledyne, Nebraska, USA). Compounds 6a, 6b, 7a–c, 7l–o and 15b were synthesized and identified by mp or 1H NMR [[13], [14], [15], [16], [17], [18], [19]].
4.1.2. General procedures for synthesis of compounds 6a–h
To a solution of refined N-(4-clorophenyl)-1,2-benzenediamone (2a, 45.9 mmol) in ethanol (120 mL), DFDNB (1.1 equiv) and TEA (1.1 equiv) were added successively, and the mixture was stirred at rt for 3 h. The formed precipitate was filtered, and the solid was washed with water (50 mL x 2) and ethanol (60 mL x 2) successively to give the intermediate 3a as an orange solid in a 97% yield. To a solution of 3a (2.5 mmol) in THF (16 mL), TEA (1 equiv.) and substituted aniline (1.1 equiv) were added. The reaction mixture was warmed to 60–70 °C and stirred overnight. The solvent was removed in vacuo, and the residue was slurried with ethanol (20 mL). The formed suspension was filtered, and the solid was washed with water and ethanol to give 4a–h as orange solid in yields of 73–93%.
To a suspension of 4a–h (2.0 mmol) in ethanol (20 mL) and acetic acid (6 mL), zinc powder (30 equiv) was added portionwise. The mixture was stirred at rt until the color turned to light green, filtered, and washed with ethanol and DCM. The filtrate was concentrated, water was added to the residue and made alkaline by ammonia. The formed precipitate was filtered and washed with water to gain the intermediates 5a–h, which was suspended or dissolved in ethanol (20 mL) and then was stirred under air overnight. The precipitation was filtered to give 6a–h, which was purified by flash chromatography using a gradient of CH2Cl2/CH3OH as the eluent.
4.1.2.1. 2-p-Methoxyanilino-3-imino-5-p-chlorophenyl-3,5-dihydrophenazine (6d)
Yield: 48%; red solid; mp: 217–219 °C; 1H NMR (500 MHz, CDCl3) δ 7.72–7.69 (3H, m, Ar–H), 7.31–7.29 (2H, m, Ar–H), 7.29–7.27 (2H, m, Ar–H), 7.22–7.18 (1H, m, Ar–H), 7.18–7.13 (1H, m, Ar–H), 6.95–6.92 (2H, m, Ar–H), 6.75 (1H, s, 1-H), 6.50 (1H, d, J = 8.2 Hz, Ar–H), 5.19 (1H, s, 4-H) 3.83 (3H, s, OMe); 13C NMR (126 MHz, CDCl3) δ 161.4, 156.6, 150.8, 145.0, 135.9, 135.8, 135.5, 134.0, 132.3, 131.7 (2), 130.9, 130.4 (2), 128.3, 127.7, 124.6 (2), 123.1, 114.7 (2), 114.0, 98.8, 96.8, 55.6; HRMS: calcd for C25H20ClON4 [M+H]+ 427.1320, found: 427.1318.
4.1.2.2. 2-(2,4-Dimethoxyanilino)-3-imino-5-p-chlorophenyl-3,5-dihydrophenazine(6e)
Yield: 35%; red solid; mp: 186–188 °C; 1H NMR (500 MHz, CDCl3) δ 8.37–8.18 (2H, m, NH), 7.72–7.66 (3H, m, Ar–H), 7.50 (1H, d, J = 8.6 Hz, Ar–H), 7.30–7.26 (2H, m, Ar–H), 7.20–7.16 (1H, m, Ar–H), 7.15–7.11 (1H, m, Ar–H), 6.81 (1H, s, 1-H), 6.55–6.53 (1H, m, Ar–H), 6.53–6.50 (1H, m, Ar–H), 6.49–6.46 (1H, m, Ar–H), 5.16 (1H, s, 4-H), 3.84 (3H, s, OMe), 3.83 (3H, s, OMe); 13C NMR (126 MHz, CDCl3) δ 161.7, 156.9, 152.7, 151.0, 144.1, 135.9, 135.7, 135.5, 133.9, 131.6 (2), 131.0, 130.4 (2), 128.2, 127.5, 123.0, 122.4, 122.1, 113.9, 103.7, 99.4, 98.9, 97.1, 55.7, 55.6; HRMS: calcd for C26H22O2ClN4 [M+H]+ 457.1426, found: 457.1426.
4.1.3. General procedures for synthesis of compounds 6i–m
The title compound was prepared following a similar procedure in 4.1.2, taking 2-aminodiphenylamine (2b) as the starting material.
4.1.3.1. 2-Benzyl-3-imino-5-phenyl-3,5-dihydrophenazine (6l)
Yield: 36%; red solid; mp: 186–188 °C; 1H NMR (500 MHz, DMSO‑d 6) δ 9.28 (1H, br s, NH), 7.78–7.73 (2H, m, Ar–H), 7.69–7.65 (1H, m, Ar–H), 7.56–7.53 (1H, m, Ar–H), 7.45–7.42 (2H, m, Ar–H), 7.39–7.33 (5H, m, Ar–H), 7.28–7.23 (1H, m, Ar–H), 7.14–7.10 (2H, m, Ar–H), 6.34–6.30 (1H, m, NH), 5.95 (1H, s, 1-H), 5.17 (1H, s, 4-H), 4.51 (2H, d, J = 5.4 Hz, CH 2Ar); 13C NMR (126 MHz, DMSO‑d 6) δ 159.7, 150.2, 147.3, 138.7, 137.1, 134.8, 132.9, 131.2 (2), 130.9, 129.6, 128.7(2), 128.4 (2), 127.1, 126.9 (3), 126.8, 122.1, 113.7, 96.4, 96.3, 45.4; HRMS: calcd for C25H21N4 [M+H]+ 377.1761, found: 377.1760.
4.1.4. General procedures for synthesis of compounds 7d–k, 7o and 7p
To a suspension of 6a–m (0.24 mmol) in dioxane (3 mL), acetic acid (2 drops) and different amine (10 or 20 equiv) were added, and then the reaction mixture stirred at 101 °C for 5 h before evaporation in vacuo. The residue was partitioned between CH2Cl2 (20 mL) and water (20 mL), then the water phase was extracted with CH2Cl2 (20 mL). The combined organic phases were washed with water (30 mL x 3) and brine (40 mL) successively, dried over anhydrous Na2SO4, filtered and evaporated. The residue was purified over by flash chromatography using a gradient of petroleum ether/CH2Cl2 as the eluent, and gave the title compounds.
4.1.4.1. 2-p-Fluoroanilino-3-isopropylimino-5-p-chlorophenyl-3,5-dihydrophenazine (7d)
Yield: 57%; red solid; mp: 210–212 °C; 1H NMR (400 MHz, CDCl3) δ 7.73–7.65 (3H, m, Ar–H), 7.34–7.28 (4H, m, Ar–H), 7.18–7.09 (2H, m, Ar–H), 7.09–7.04 (2H, m, Ar–H), 6.71 (1H, s, 1-CH), 6.44 (1H, d, J = 7.9 Hz, Ar–H), 5.27 (1H, s, 4-CH), 3.52–3.42 (1H, m, CHMe2), 1.09 (6H, d, J = 6.2 Hz, CHMe 2); 13C NMR (101 MHz, CDCl3) δ 160.4, 158.0, 151.2, 150.5, 144.8, 136.2, 135.9, 135.6, 134.8, 131.7 (2), 131.4, 130.5 (2), 128.1, 127.3, 124.2, 124.1, 122.9, 116.1, 115.9, 113.8, 98.2, 89.1, 49.4, 23.6 (2); HRMS: calcd for C27H23N4ClF [M+H]+ 457.1590, found: 457.1589.
4.1.4.2. 2-p-Fluoroanilino-3-cyclopropylimino-5-p-chlorophenyl-3,5-dihydrophena zine (7e)
Yield: 55%; red solid; mp: 215–217 °C; 1H NMR (400 MHz, CDCl3) δ 7.72–7.67 (2H, m, Ar–H), 7.67–7.63 (1H, m, Ar–H), 7.35–7.30 (2H, m, Ar–H), 7.30–7.26 (2H, m, Ar–H), 7.18–7.03 (4H, m, Ar–H), 6.65 (1H, s, 1-H), 6.44–6.40 (1H, m, Ar–H), 5.54 (1H, s, 4-H), 2.79–2.71 (1H, m, CHCH2CH2), 0.92–0.76 (4H, m, CHCH 2 CH 2); 13C NMR (101 MHz, CDCl3) δ 159.3 (d, J = 242.0 Hz, 1C–F), 152.5, 151.4, 144.6, 136.2, 135.8, 135.6, 134.6, 131.7 (3), 131.5, 130.6 (2), 128.1, 127.3, 124.4, 124.3, 122.9, 116.1 (2) (d, J = 23.0 Hz, 2C–F), 113.8, 98.2, 89.4, 32.9, 10.0 (2); HRMS: calcd for C27H21N4ClF [M+H]+ 455.1433, found: 455.1433.
4.1.4.3. 2-p-Fluoroanilino-3-propylimino-5-p-chlorophenyl-3,5-dihydrophenazine (7f)
Yield: 56%; red solid; mp: 176–178 °C; 1H NMR (400 MHz, CDCl3) δ 7.73–7.67 (3H, m, Ar–H), 7.33–7.28 (4H, m, Ar–H), 7.20–7.11 (2H, m, Ar–H), 7.10–7.04 (2H, m, Ar–H), 6.72 (1H, s, 1-H), 6.45 (1H, d, J = 7.7 Hz, Ar–H), 5.27 (1H, s, 4-H), 3.11 (2H, t, J = 6.8 Hz, CH 2CH2CH3), 1.72–1.63 (2H, m, CH2 CH 2CH3), 0.94 (3H, t, J = 7.4 Hz, CH2CH2 CH 3); 13C NMR (101 MHz, CDCl3) δ 159.3 (d, J = 242.0 Hz, 1C–F), 152.4, 151.2, 144.7, 136.1, 135.8, 135.7, 134.7, 131.7 (3), 131.3, 130.4 (2), 128.2, 127.4, 124.3, 124.2, 123.0, 116.1 (2) (d, J = 23.0 Hz, 2C–F), 113.9, 98.1, 89.1, 52.0, 24.3, 12.3; HRMS: calcd for C27H23N4ClF [M+H]+ 457.1590, found: 457.1590.
4.1.4.4. 2-p-Methoxyanilino-3-isopropylimino-5-p-chlorophenyl-3,5-dihydrophenazine (7g)
Yield: 54%; red solid; mp: 178–180 °C; 1H NMR (500 MHz, CDCl3) δ 7.73–7.68 (2H, m, Ar–H), 7.65 (1H, d, J = 7.7 Hz, Ar–H), 7.31–7.27 (4H, m, Ar–H), 7.15 (1H, m, Ar–H), 7.09 (1H, m, Ar–H), 6.94–6.89 (2H, m, Ar–H), 6.66 (1H, s, 1-H), 6.43 (1H, d, J = 8.1 Hz, Ar–H), 5.27 (1H, s, 4-H), 3.83 (3H, s, OMe), 3.50–3.43 (1H, m, CHMe2), 1.09 (6H, d, J = 6.2 Hz, CHMe 2); 13C NMR (126 MHz, CDCl3) δ 156.3, 151.2, 150.6, 145.5, 136.2, 135.7, 135.5, 134.7, 132.7, 131.6 (2), 131.3, 130.5 (2), 127.9, 126.9, 124.5 (2), 122.8, 114.5 (2), 113.8, 97.5, 89.0, 55.6, 49.3, 23.6 (2); HRMS: calcd for C28H26ON4Cl [M+H]+ 469.1790, found: 469.1791.
4.1.4.5. 2-(2,4-Dimethoxyanilino)-3-isopropylimino-5-p-chlorophenyl-3,5-dihydro phenazine (7h)
Yield: 46%; red solid; mp: 190–192 °C; 1H NMR (500 MHz, CDCl3) δ 7.72–7.67 (2H, m, Ar–H), 7.65 (1H, d, J = 8.0 Hz, Ar–H), 7.46 (1H, m, Ar–H), 7.31–7.27 (2H, m, Ar–H), 7.17–7.11 (1H, m, Ar–H), 7.11–7.05 (1H, m, Ar–H), 6.69 (1H, s, 1-H), 6.53 (1H, s, Ar–H), 6.50 (1H, d, J = 8.5 Hz, Ar–H), 6.42 (1H, d, J = 7.7 Hz, Ar–H), 5.27 (1H, s, 4-H), 3.83 (6H, s, OMe, OMe), 3.52–3.43 (1H, m, CHMe2), 1.10 (6H, d, J = 6.2 Hz, CHMe 2); 13C NMR (101 MHz, CDCl3) δ 156.9, 153.1, 151.6, 151.0, 145.1, 136.5, 135.9, 135.6, 134.8, 131.7 (2), 130.7 (2), 128.0, 126.9, 123.0, 122.8, 122.7, 113.9, 110.1, 104.0, 99.6, 98.0, 89.4, 56.0, 55.8, 49.5, 23.7 (2); HRMS: calcd for C29H28O2N4Cl [M+H]+ 499.1895, found: 499.1892.
4.1.4.6. 2-p-Trifluoromethoxyanilino-3-isopropylimino-5-p-chlorophenyl-3,5-dihydro phenazine (7i)
Yield: 63%; red solid; mp: 200–202 °C; 1H NMR (400 MHz, CDCl3) δ 7.73–7.67 (3H, m, Ar–H), 7.39–7.34 (2H, m, Ar–H), 7.32–7.27 (2H, m, Ar–H), 7.24–7.19 (2H, m, Ar–H), 7.19–7.09 (2H, m, Ar–H), 6.82 (1H, s, 1-H), 6.46–6.42 (1H, m, Ar–H), 5.28 (1H, s, 4-H), 3.51–3.41 (1H, m, CHMe2), 1.09 (6H, d, J = 6.2 Hz, CHMe 2); 13C NMR (101 MHz, CDCl3) δ 151.1, 150.4, 144.7 (2), 144.0, 138.8, 136.1, 135.7, 135.6, 134.8, 131.7 (2), 131.5, 130.5 (2), 128.2, 127.6, 123.0, 122.9 (2), 122.2 (2), 113.9, 99.0, 89.1, 49.4, 23.6 (2); HRMS: calcd for C28H23ON4ClF3 [M+H]+ 523.1507, found: 523.1509.
4.1.4.7. 2-(6-Methoxy-3-pyridylamino)-3-isopropylimino-5-p-chlorophenyl-3,5-di hydrophenazine (7j)
Yield: 45%; red solid; mp: 176–178 °C; 1H NMR (500 MHz, CDCl3) δ 8.15 (1H, d, J = 2.2 Hz, Ar–H), 7.73–7.68 (2H, m, Ar–H), 7.67–7.62 (2H, m, Ar–H), 7.32–7.27 (2H, m, Ar–H), 7.18–7.14 (1H, m, Ar–H), 7.13–7.08 (1H, m, Ar–H), 6.79 (1H, d, J = 8.7 Hz, Ar–H), 6.49 (1H, s, 1-H), 6.44 (1H, d, J = 8.1 Hz, Ar–H), 5.28 (1H, s, 4-H), 3.96 (3H, s, Me), 3.53–3.41 (1H, m, CHMe2), 1.10 (6H, d, J = 6.2 Hz, CHMe 2); 13C NMR (126 MHz, CDCl3) δ 161.2, 151.0, 150.5, 146.0, 142.5, 136.1, 135.6 (2), 135.2, 134.8, 131.7 (2), 131.3, 130.4 (2), 130.1, 128.1, 127.1, 122.9, 113.8, 111.0, 98.0, 89.0, 53.7, 49.4, 23.6 (2); HRMS: calcd for C27H25ON5Cl [M+H]+ 470.1742, found: 470.1741.
4.1.4.8. 2-p-Chlorobenzylamino-3-isopropylimino-5-p-chlorophenyl-3,5-dihydrophena zine (7k)
Yield: 64%; red solid; mp: 176–178 °C; 1H NMR (400 MHz, CDCl3) δ 7.70–7.66 (2H, m, Ar–H), 7.63 (1H, dd, J = 7.9, 1.4 Hz, Ar–H), 7.32–7.26 (6H, m, Ar–H), 7.16–7.11 (1H, m, Ar–H), 7.10–7.04 (1H, m, Ar–H), 6.41 (1H, d, J = 8.2 Hz, Ar–H), 6.06 (1H, s, 1-H), 5.23 (1H, s, 4-H), 4.47 (2H, s, CH 2Ar), 3.48–3.39 (1H, m, CHMe2), 1.05 (6H, d, J = 6.2 Hz, CHMe 2); 13C NMR (101 MHz, CDCl3) δ 151.1, 150.5, 147.9, 136.6, 136.3, 135.6, 135.5, 134.6, 132.9, 131.6 (2), 131.2, 130.5 (2), 128.8 (2), 128.5 (2), 127.8, 126.8, 122.7, 113.7, 96.5, 89.0, 49.3, 46.1, 23.6 (2); HRMS: calcd for C28H25N4Cl2 [M+H]+ 487.1451, found: 487.1449.
4.1.4.9. 2-Benzylamino-3-isopropylimino-5-phenyl-3,5-dihydrophenazine (7p)
Yield: 64%; red solid; mp: 199–201 °C; 1H NMR (400 MHz, CDCl3) δ 7.72–7.67 (2H, m, Ar–H), 7.66–7.59 (2H, m, Ar–H), 7.38–7.29 (7H, m, Ar–H), 7.16–7.10 (1H, m, Ar–H), 7.09–7.03 (1H, m, Ar–H), 6.44 (1H, d, J = 7.7 Hz, Ar–H), 6.14 (1H, s, 1-CH), 5.23 (1H, s, 4-CH), 4.50 (2H, s, CH 2Ar), 3.42–3.33 (1H, m, CHMe2), 1.02 (6H, d, J = 6.2 Hz, CHMe 2); 13C NMR (126 MHz, DMSO‑d 6) δ 149.7, 145.3, 142.2, 138.5, 137.4, 136.0, 134.6, 131.1 (3), 130.8, 129.3, 128.9, 128.5 (2), 127.5 (2), 127.3 (2), 127.2, 127.1, 117.0, 100.3, 89.6, 46.2, 45.9, 20.6 (2); HRMS: calcd for C28H27N4 [M+H]+ 419.2230, found: 419.2231.
4.1.4.10. 2-p-Fluorobenzylamino-3-isopropylimino-5-phenyl-3,5-dihydrophenazine (7q)
Yield: 61%; red solid; mp: 166–168 °C; 1H NMR (500 MHz, CDCl3) δ 7.73–7.68 (2H, m, Ar–H), 7.67–7.60 (2H, m, Ar–H), 7.34–7.29 (4H, m, Ar–H), 7.13 (1H, t, J = 7.3 Hz, Ar–H), 7.09–7.00 (3H, m, Ar–H), 6.45 (1H, d, J = 8.1 Hz, Ar–H), 6.10 (1H, s, 1-CH), 5.22 (1H, s, 4-CH), 4.47 (2H, s, CH 2Ar), 3.41–3.33 (1H, m, CHMe2), 1.02 (6H, d, J = 6.3 Hz, CHMe 2); 13C NMR (126 MHz, CDCl3) δ 162.0 (d, J = 244.0 Hz, 1C–F), 151.1, 150.7, 147.9, 137.8, 135.6, 134.8, 133.8, 131.5, 131.2 (2), 129.6, 128.9 (2), 128.8, 128.7, 127.6, 126.7, 122.5, 115.5 (2) (d, J = 22.0 Hz, 2C–F), 114.0, 96.3, 89.1, 49.2, 46.0, 23.5 (2); HRMS: calcd for C28H26N4F [M+H]+ 437.2136, found: 437.2136.
4.1.5. Synthesis of 2-p-fluoroanilino-3-isopropylimino-5-p-chlorophenyl-3,5-dihydrophenazine 15a
To a solution of 2-fluoronitrobenzene (21.0 mmol) in DMF (35 mL), 8a (1.1 equiv) and TEA (1.5 equiv) were added. The reaction mixture was warmed to 90 °C and stirred for 14 h the mixture was cooled to rt and partitioned between ethyl acetate (50 mL) and water (50 mL), and the water phase was extracted with ethyl acetate. The combined organic phases were washed with 2 N HCl (60 mL x 2) and brine (80 mL x 2), dried over anhydrous Na2SO4, filtered and evaporated. The residue recrystallized in ethanol to give 9a. Yield: 61%; red solid; 1H NMR (500 MHz, DMSO‑d 6) δ 9.36 (1H, br s,NH), 8.12 (1H, dd, J = 8.5, 1.5 Hz, Ar–H), 7.53–7.46 (1H, m, Ar–H), 7.39–7.35 (2H, m, Ar–H), 7.30–7.24 (2H, m, Ar–H), 7.07–7.04 (1H, m, Ar–H), 6.89–6.84 (1H, m, Ar–H).
Intermediate 9a (12.9 mmol) and 10% Pd/C (0.2 equiv) were suspended in ethanol (90 mL), and the mixture was shaken under hydrogen atmosphere for 6 h. The mixture was then filtered to achieve a solution of crude 10a in ethanol. Following similar procedures in 4.1.2 and 4.1.4 subsequently, 2a was replaced by 10a, the compound 15a was gained. Yield: 48%; red solid; mp: 180–182 °C; 1H NMR (400 MHz, CDCl3) δ 7.69 (1H, dd, J = 7.7, 1.4 Hz, Ar–H), 7.45–7.38 (2H, m, Ar–H), 7.35–7.27 (6H, m, Ar–H), 7.20–7.09 (2H, m, Ar–H), 6.83 (1H, s, 1-CH), 6.45 (1H, d, J = 8.1 Hz, Ar–H), 5.26 (1H, s, 4-CH), 3.49–3.39 (1H, m, CHMe2), 1.08 (6H, d, J = 6.2 Hz, CHMe 2); 13C NMR (101 MHz, CDCl3) δ 162.8, 151.1, 150.5, 143.7, 138.7, 135.7, 135.1, 133.5, 131.7, 130.9, 130.8, 129.3 (2), 128.2 (2), 127.5, 122.9, 122.8 (2), 118.6, 118.4, 113.9, 99.1, 89.1, 49.4, 23.6 (2); HRMS: calcd for C27H23N4ClF [M+H]+ 457.1590, found: 457.1590.
4.1.6. Synthesis of 2-p-chloroanilino-3-isopropylimino-5-(6-methoxy-3-pyridinyl)-3,5- dihydrophenazine 15c
To a solution of 2-fluoronitrobenzene (70.9 mmol) in DMF (70 mL), 8b (1.0 equiv) and NaHCO3 (1.2 equiv) were added. The reaction was stirred for 48 h at 100 °C before it was diluted with ethyl acetate (50 mL) and water (50 mL), and the water phase was extracted two times with ethyl acetate. The combined organic phases were washed with brine (100 mL x 2), dried over anhydrous Na2SO4, filtered and concentrated. The residue recrystallized in petroleum ether/ethanol to give 9b. Yield: 55%; red solid; 1H NMR (500 MHz, DMSO‑d 6) δ 9.35 (1H, br s, NH), 8.16 (1H, d, J = 2.7 Hz, Ar–H), 8.12 (1H, dd, J = 8.5, 1.5 Hz, Ar–H), 7.70 (1H, dd, J = 8.7, 2.7 Hz, Ar–H), 7.49–7.45 (1H, m, Ar–H), 6.93–6.87 (2H, m, Ar–H), 6.85–6.81 (1H, m, Ar–H), 3.88 (3H, s, OMe).
Intermediate 9b (12.9 mmol) and 10% Pd/C (0.2 equiv) were suspended in ethanol (90 mL), and the mixture was shaken under hydrogen atmosphere for 6 h. The mixture was then filtered to achieve a solution of 10b, which was taken as starting material instead of 2a, following similar procedures in 4.1.2 and 4.1.4 subsequently, to achieve 15c. Yield: 55%; red solid; mp: 199–201 °C; 1H NMR (400 MHz, CDCl3) δ 8.17 (1H, d, J = 2.5 Hz, Ar–H), 7.68 (1H, dd, J = 7.7, 1.3 Hz, Ar–H), 7.57 (1H, dd, J = 8.7, 2.6 Hz, Ar–H), 7.34–7.28 (4H, m, Ar–H), 7.20–7.10 (2H, m, Ar–H), 7.08 (1H, d, J = 8.7 Hz, Ar–H), 6.83 (1H, s, 1-CH), 6.49 (1H, d, J = 7.8 Hz, Ar–H), 5.32 (1H, s, 4-CH), 4.09 (3H, s, OMe), 3.55–3.44 (1H, m, CHMe2), 1.13–1.07 (6H, m, CHMe 2); 13C NMR (101 MHz, CDCl3) δ 164.3, 151.2, 150.4, 147.6, 143.7, 139.4, 138.6, 135.7, 135.4, 132.0, 129.3 (2), 128.3, 128.2, 127.8, 127.6, 123.0, 122.9 (2), 113.8, 113.5, 99.1, 89.2, 54.2, 49.4, 23.7, 23.5; HRMS: calcd for C27H25ClN5O [M+H]+ 470.1742, found: 470.1743.
4.1.7. General synthesis of compounds 15d–j
Following a similar procedure in preparing 4a–h in 4.1.2, the reaction of 10b with corresponding intermediates 12c–g. Then, the mixture of 12c–g (1 g) and 10% Pd/C (0.2 equiv.) in methanol (30 mL) and THF (30 mL) was shaken overnight under hydrogen atmosphere. The reaction mixture was filtered, and the filtrate was concentrated in vacuo. The residue suspended or dissolved in ethanol/CH2Cl2, and then stood still under air overnight. The precipitate was filtered, and then washed with ethanol to give compounds 14c–g respectively, in total yields of 64–65%.
Following similar procedures in 4.1.4, the reaction of 14c–g with amines achieved the title compounds 15d–j.
4.1.7.1. 2-Anilino-3-isopropylimino-5-(6-methoxy-3-pyridinyl)-3,5-dihydrophenazine (15d)
Yield: 50%; red solid; mp: 214–216 °C; 1H NMR (500 MHz, CDCl3) δ 11.03 (1H, s, NH), 8.29 (1H, s, Ar–H), 8.17 (1H, d, J = 8.2 Hz, Ar–H), 7.72 (1H, d, J = 7.0 Hz, Ar–H), 7.63 (1H, t, J = 7.6 Hz, Ar–H), 7.59–7.53 (4H, m, Ar–H), 7.45–7.39 (2H, m, Ar–H), 7.23–7.19 (2H, m, Ar–H), 7.02 (1H, d, J = 8.5 Hz, Ar–H), 5.87 (1H, s, 1-CH), 4.15 (3H, s, OMe), 3.75–3.66 (1H, m, CHMe2), 1.50 (6H, d, J = 11.6 Hz, CHMe 2); 13C NMR (126 MHz, CDCl3) δ 165.5, 152.2, 146.1, 145.3, 142.8, 138.6, 137.7, 137.2, 136.5, 130.8, 129.8, 129.6 (2), 129.0, 127.4, 126.3, 125.7, 123.9 (2), 116.2, 113.9, 102.5, 89.7, 54.7, 48.2, 21.2, 21.1; HRMS: calcd for C27H26ON5 [M+H]+ 436.2132, found:436.2131.
4.1.7.2. 2-p-Methoxyanilino-3-isopropylimino-5-(6-methoxy-3-pyridinyl)-3,5-dihydro phenazine (15e)
Yield: 51%; red solid; mp: 164–166 °C; 1H NMR (500 MHz, CDCl3) δ 8.17 (1H, d, J = 2.3 Hz, Ar–H), 7.66 (1H, d, J = 7.7 Hz, Ar–H), 7.57 (1H, dd, J = 8.7, 2.5 Hz, Ar–H), 7.30–7.27 (2H, m, Ar–H), 7.16 (1H, t, J = 7.3 Hz, Ar–H), 7.11 (1H, d, J = 7.5 Hz, Ar–H), 7.08 (1H, d, J = 8.6 Hz, Ar–H), 6.94–6.89 (2H, m, Ar–H), 6.66 (1H, s, 1-CH), 6.48 (1H, d, J = 8.1 Hz, Ar–H), 5.32 (1H, s, 4-CH), 4.09 (3H, s, OMe), 3.83 (3H, s, OMe), 3.54–3.48 (1H, m, CHMe2), 1.10 (6H, dd, J = 10.2, 6.3 Hz, CHMe 2); 13C NMR (126 MHz, CDCl3) δ 164.2, 156.4, 151.3, 150.5, 147.6, 145.5, 139.5, 135.8, 135.3, 132.7, 131.8, 128.0, 127.9, 127.0, 124.5 (2), 122.8, 114.5 (2), 113.7, 113.4, 97.6, 89.1, 55.6, 54.1, 49.4, 23.7, 23.6; HRMS: calcd for C28H28N5O2 [M + H]+ 466.2238, found: 466.2238.
4.1.7.3. 2-m-Fluoroanilino-3-isopropylimino-5-(6-methoxy-3-pyridinyl)-3,5-dihydro
phenazine (15f) Yield: 57%; red solid; mp: 209–211 °C; 1H NMR (400 MHz, CDCl3) δ 8.17 (1H, d, J = 2.6 Hz, Ar–H), 7.70 (1H, dd, J = 7.7, 1.6 Hz, Ar–H), 7.57 (1H, dd, J = 8.7, 2.7 Hz, Ar–H), 7.34–7.27 (1H, m, Ar–H), 7.20–7.07 (5H, m, Ar–H), 6.92 (1H, s, 1-CH), 6.80–6.74 (1H, m, Ar–H), 6.49 (1H, d, J = 8.0 Hz, Ar–H), 5.33 (1H, s, 4-CH), 4.09 (3H, s, OMe), 3.56–3.45 (1H, m, CHMe2), 1.12–1.08 (6H, m, CHMe 2); 13C NMR (101 MHz, CDCl3) δ 164.3 (d, J = 244.0 Hz, 1C–F), 163.4, 151.2, 150.3, 147.6, 143.3, 141.8, 139.5, 135.7, 135.4, 132.0, 130.0, 128.3, 127.8, 127.7, 123.0, 116.9, 113.8, 113.5, 109.9 (d, J = 21.0 Hz, 2C–F), 108.3 (d, J = 24.0 Hz, 2CF), 99.8, 89.2, 54.2, 49.4, 23.7, 23.5; HRMS: calcd for C27H25FN5O [M+H]+ 454.2038, found: 454.2040.
4.1.7.4. 2-p-Fluoroanilino-3-isopropylimino-5-(6-methoxy-3-pyridinyl)-3,5-dihydro phenazine (15g)
Yield: 55%; red solid; mp: 210–212 °C; 1H NMR (500 MHz, CDCl3) δ 8.17 (1H, d, J = 2.6 Hz, Ar–H), 7.67 (1H, dd, J = 7.8, 1.4 Hz, Ar–H), 7.57 (1H, dd, J = 8.7, 2.6 Hz, Ar–H), 7.34–7.29 (2H, m, Ar–H), 7.18–7.05 (5H, m, Ar–H), 6.71 (1H, s, 1-CH), 6.49 (1H, d, J = 8.1 Hz, Ar–H), 5.33 (1H, s, 4-CH), 4.09 (3H, s, OMe), 3.56–3.47 (1H, m, CHMe2), 1.10 (6H, dd, J = 10.3, 6.3 Hz, CHMe 2); 13C NMR (126 MHz, CDCl3) δ 164.3, 159.2 (d, J = 244.4 Hz, 1C–F), 151.2, 150.4, 147.6, 144.8, 139.5, 135.9, 135.7, 135.3, 131.9, 128.2, 127.8, 127.4, 124.2 (2), 122.9, 116.0 (2) (d, J = 22.5 Hz, 2C–F), 113.8, 113.4, 98.2, 89.1, 54.2, 49.4, 23.7, 23.5; HRMS: calcd for C27H25FN5O [M+H]+ 454.2038, found: 454.2036.
4.1.7.5. 2-p-Fluoroanilino-3-cyclopropylimino-5-(6-methoxy-3-pyridinyl)-3,5-dihydro phenazine (15h)
Yield: 57%; red solid; mp: 211–213 °C; 1H NMR (400 MHz, CDCl3) δ 8.18 (1H, d, J = 2.5 Hz, Ar–H), 7.65 (1H, dd, J = 7.7, 1.5 Hz, Ar–H), 7.59 (1H, dd, J = 8.7, 2.7 Hz, Ar–H), 7.30–7.26 (2H, m, Ar–H), 7.18–7.10 (2H, m, Ar–H), 7.09–7.03 (3H, m, Ar–H), 6.65 (1H, s, 1-CH), 6.47 (1H, d, J = 8.0 Hz, Ar–H), 5.59 (1H, s, 4-CH), 4.08 (3H, s, OMe), 2.82–2.75 (1H, m, CHCH2CH2), 0.93–0.87 (2H, m, CHCH 2CH2), 0.82–0.78 (2H, m, CHCH2 CH 2); 13C NMR (101 MHz, CDCl3) δ 164.3, 159.3 (d, J = 244.4 Hz, 1C–F), 152.5, 151.4, 147.7, 144.7, 139.6, 135.8 (2), 135.1, 132.0, 128.1, 127.8, 127.4, 124.5, 124.4, 122.9, 116.2 (2) (d, J = 27.5 Hz, 2C–F), 113.7, 113.5, 98.2, 89.6, 54.1, 33.0, 10.0 (2); HRMS: calcd for C27H23FN5O [M+H]+ 452.1881, found: 452.1881.
4.1.7.6. 2-p-Fluoroanilino-3-propylimino-5-(6-methoxy-3-pyridinyl)-3,5-dihydro phenazine (15i)
Yield: 49%; red solid; mp: 186–188 °C; 1H NMR (400 MHz, CDCl3) δ 8.17 (1H, d, J = 2.3 Hz, Ar–H), 7.69 (1H, d, J = 6.7 Hz, Ar–H), 7.57 (1H, dd, J = 8.7, 2.6 Hz, Ar–H), 7.35–7.28 (2H, m, Ar–H), 7.22–7.12 (2H, m, Ar–H), 7.11–7.04 (3H, m, Ar–H), 6.72 (1H, s, 1-CH), 6.51 (1H, d, J = 7.0 Hz, Ar–H), 5.31 (1H, s, 4-CH), 4.09 (3H, s, OMe), 3.14 (2H, t, J = 5.8 Hz, CH 2CH2CH3) 1.73–1.65 (2H, m, CH2 CH 2CH3), 0.95 (3H, t, J = 7.4 Hz, CH2CH2 CH 3); 13C NMR (101 MHz, CDCl3) δ 164.3, 159.3 (d, J = 244.4 Hz, 1C–F), 152.4, 151.2, 147.5, 144.7, 139.4, 135.8 (2), 135.3, 131.8, 128.2, 127.8, 127.5, 124.3 (2) (d, J = 8.0 Hz, 3C–F), 123.1, 116.1 (2) (d, J = 25.0 Hz, 2C–F), 113.9, 113.5, 98.2, 89.2, 54.2, 52.0, 24.3, 12.3; HRMS: calcd for C27H25FN5O [M+H]+ 454.2038, found: 454.2035.
4.1.7.7. 2-p-Fluoroanilino-3-(4-methoxycyclohexylimino)-5-(6-methoxy-3-pyridinyl)-3,5 -dihydrophenazine (15j)
Yield: 54%; orange solid; mp: 176–178 °C; 1H NMR (400 MHz, CDCl3) δ 8.16 (1H, d, J = 2.4 Hz, Ar–H), 7.70–7.65 (1H, m, Ar–H), 7.56 (1H, dd, J = 8.7, 2.6 Hz, Ar–H), 7.33–7.28 (2H, m, Ar–H), 7.20–7.11 (2H, m, Ar–H), 7.10–7.04 (3H, m, Ar–H), 6.72 (1H, s, 1-H), 6.53 (1H, d, J = 7.5 Hz, Ar–H), 5.29 (1H, s, 4-H), 4.08 (3H, s, OMe), 3.36 (3H, s, OMe), 3.24–3.10 (2H, m, N–CH, O–CH), 2.12–2.03 (2H, m, CH2), 1.76–1.66 (2H, m, CH2), 1.47–1.37 (2H, m, CH2), 1.24–1.14 (2H, m, CH2); 13C NMR (101 MHz, CDCl3) δ 164.4, 159.3 (d, J = 242.0 Hz, 1C–F),151.2, 151.1, 147.5, 144.7, 139.3, 135.8, 135.7, 135.3, 131.8, 128.2, 127.7, 127.5, 124.2 (2), 123.0, 116.2 (2) (d, J = 22.0 Hz, 2C–F), 113.8, 113.3, 98.3, 89.1, 57.3, 55.9, 54.2, 31.2, 31.2, 30.0 (2); HRMS: calcd for C31H31FN5O2 [M+H]+ 524.2456, found: 524.2456.
4.2. Biology
4.2.1. Cell lines, plasmids, and viruses
BSR and 293T cells were cultured in 5% CO2 at 37 °C in high glucose Dulbecco's Modified Eagle's Medium (HyClone, South Logan, UT) supplemented with 10% FBS (Gibco, Carlsbad, CA), and 1% penicillin–streptomycin solution (Gibco, Carlsbad, CA). Cells were passaged every 2 days. Virulent rabies virus CVS strain was adapted by BSR (variant strain of BHK) cells, and stored at −70 °C. The pSARS-Cov-2 virus was constructed as described previously [20].
4.2.2. Anti-rabies activity evaluation
Following a similar RFFIT method as described previously [9], serial dilutions of test compounds, starting from 200 μM were incubated with 50 μL of CVS (20, 000 FFU/well) in duplicate at 37 °C for 1 h, then mixed with BSR cells (1 × 106/ml) and subsequently incubated for 24 h. Then, after being fixed with pre-chilled 80% acetone, the cells were stained with fluorescein isothiocyanate (FITC)-conjugated anti-rabies N monoclonal antibody (Fujirebio Diagnostics, Malvern, PA), and the fluorescent intensity was recorded by a fluorescence microscope (Olympus, Tokyo, Japan). The EC50 value was determined from the dose-response curve.
4.2.3. Anti-pSARS-CoV-2 activity evaluation
Following a similar method as described previously [20,21], serial dilutions of test compounds, starting from 200 μM were incubated with 1.3 × 104 TCID50 of pSARS-CoV-2 at 37 °C for 1 h, then mixed with Huh 7 cells (2000 cells/well) and subsequently incubated for 24 h. The bioluminescence intensities were recorded to calculate EC50 value of each compound as described previously [20].
4.2.4. Cytotoxicity testing
Serial dilutions of test compounds, starting from 200 μM were incubated with BSR cells or 293T cells in 96-well plates, then 50 μL of medium was added instead of virus. After being incubated at 37 °C for 24 h, the cell viability was recorded by an Ensight microplate luminometer (PerkinElmer, Singapore), and the CC50 values of test compounds were calculate by GraphPad Prism 6 (San Diego, CA).
4.2.5. Acute toxicity
Female Kunming mice with weight of 20.0 ± 1.0 g were obtained from the Institute of Laboratory Animal Science (Beijing, China). The experiment was performed in accordance with the guidelines established by the Institute of Medicinal Biotechnology, Peking Union Medical College and Chinese Academy of Medical Science for the care and use of laboratory animals. The mice were fed with regular rodent chow and housed in an air-conditioned room. The mice were randomly divided into different groups with 6 mice each. Each compound was given orally in a single-dosing experiment at 0, 200, 400, 600 or 800 mg•kg−1 (ddH2O as control), respectively. The mice were closely monitored for 7 days. Body weight, behavior change and survival were monitored.
4.2.6. Time of addition assay
The “time-of-addition” experiment was performed as described previously [9]. Specifically, compounds 15f and 15g were applied as the test compound, and 1 was used as positive control. Data were analyzed based on three replicates.
4.2.7. Molecular modeling analysis
The protein crystal structures were obtained from the RCSB Protein Data Bank (https://www.rcsb.org). Discovery Studio 4.5 software (BIOVIA, San Diego, California, USA) was applied for both ligand and protein preparation using the default settings. Libdock program was employed and Libdock score function was applied to evaluate the affinity of the receptor-ligand interaction. The docking conformation with the highest score was chosen to inspect the binding modes.
4.2.8. SPR
Rabies virus G protein (20–459 AA residues) with a GST-His tag was cloned into pET28a vector and grown in E. coli BL21 (DE3) cells. The bacteria were grown at 37 °C until the OD600 value of the liquid reaches 0.6–0.8 following induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The protein was then purified using the Ni2+-loaded HiTrap Chelating System (GE Healthcare, US) according to the manufacturer's instructions. SARS-CoV-2 S recombinant protein was purchased from Sino biological Inc (Beijing, China). The S series CM5 sensor chip and amine coupling Kit were purchased from GE Healthcare (Uppsala, Sweden). The PBS-P buffer was filtered through a 0.45 μm membrane filter and degassed before use. SPR analysis was performed on a Biacore™ T200 system (GE Healthcare, US) and PBS-P buffer was used as sample and running buffer. The target G or S protein was covalently immobilized into the CM5 sensor chip using an amine coupling kit in a sodium acetate solution of pH 4.0. Compounds 15f and 1 were diluted in a series of concentrations in PBS-P buffer, and flowed through the fixed protein at a rate of 30 μL/min, with a contact time of 60 s and a dissociation time of 120 s. Data were processed by subtracting responses from the reference flow cell and from the blank cycles. All data were analyzed by the kinetic model in the Biacore™ T200 Evaluation Software 2.0 (GE Healthcare, US), and Kd was applied to evaluate the binding affinity.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the CAMS Innovation Fund for Medical Sciences (2021–12M-1-048) and National Natural Science Foundation of China (81872912).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2022.114209.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Schnell M.J., McGettigan J.P., Wirblich C., Papaneri A. The cell biology of rabies virus: using stealth to reach the brain. Nat. Rev. Microbiol. 2010;8:51–61. doi: 10.1038/nrmicro2260. [DOI] [PubMed] [Google Scholar]
- 2.Mi Z., Zhao L., Sun M., Gao T., Wang Y., Sui B., Li Y. Overexpression of interleukin-33 in recombinant rabies virus enhances innate and humoral immune responses through activation of dendritic cell-germinal center reactions. Vaccines (Basel) 2021;10:34. doi: 10.3390/vaccines10010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO/Department of control of neglected tropical diseases, WHO expert consultation on rabies. Third report, World Health Organ. Tech. Rep. 2018:1–141. back cover. [Google Scholar]
- 4.Wentworth D., Hampson K., Thumbi S.M., Mwatondo A., Wambura G., Chng N.R. A social justice perspective on access to human rabies vaccines. Vaccine. 2019;37(Suppl 1):A3–A5. doi: 10.1016/j.vaccine.2019.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gholami A., Shirzadi M.R., Asouri M., Farahtaj F., Mostafavi E., Gharibzadeh S., Pourmozafari J., Nabavi M., Nazari F., Bahrami F. Seroconversion after three doses of intramuscular rabies vaccine as a post-exposure treatment. Virus Res. 2020;278:197883. doi: 10.1016/j.virusres.2020.197883. [DOI] [PubMed] [Google Scholar]
- 6.Tran C.H., Afriyie D.O., Pham T.N., Otsu S., Urabe M., Dang A.D., Tran H.G.T., Nguyen H.V., Le H.T., Nguyen H.T.T. Rabies post-exposure prophylaxis initiation and adherence among patients in Vietnam, 2014-2016. Vaccine. 2019;37(Suppl 1):A54–A63. doi: 10.1016/j.vaccine.2019.01.030. [DOI] [PubMed] [Google Scholar]
- 7.Jackson A.C. Update on rabies diagnosis and treatment. Curr. Infect. Dis. Rep. 2019;11:296–301. doi: 10.1007/s11908-009-0044-0. [DOI] [PubMed] [Google Scholar]
- 8.Daniel G., Julia A. Overview, prevention, and treatment of rabies. Mayo Clin. Proc. 2004;79:671–676. doi: 10.4065/79.5.671. [DOI] [PubMed] [Google Scholar]
- 9.Wu J., Cao S., Lei S., Liu Q., Li Y., Yu Y., Xie H., Li Q., Zhao X., Chen R., Huang W., Xiao X., Yu Y., Song D., Li Y., Wang Y. Clofazimine: a promising inhibitor of rabies virus. Front. Pharmacol. 2021;12:598241. doi: 10.3389/fphar.2021.598241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan S., Yin X., Meng X., Chan J.F., Ye Z.W., Riva L., Pache L., Chan C.C., Lai P.M., Chan C.C., Poon V.K., Lee A.C., Matsunaga N., Pu Y., Yuen C.K., Cao J., Liang R., Tang K., Sheng L., Du Y., Xu W., Lau C.Y., Sit K.Y., Au W.K., Wang R., Zhang Y.Y., Tang Y.D., Clausen T.M., Pihl J., Oh J., Sze K.H., Zhang A.J., Chu H H., Kok K.H., Wang D D., Cai X.H., Esko J.D., Hung I.F., Li R.A., Chen H., Sun H., Jin D.Y., Sun R., Chanda S.K., Yuen K.Y. Clofazimine broadly inhibits coronaviruses including SARS-CoV-2. Nature. 2021;593:418–423. doi: 10.1038/s41586-021-03431-4. [DOI] [PubMed] [Google Scholar]
- 11.Murashov M.D., LaLone V., Rzeczycki P.M., Keswani R.K., Yoon G.S., Sud S., Rajeswaran W., Larsen S., Stringer K.A., Rosania G.R. The physicochemical basis of clofazimine-induced skin pigmentation. J. Invest. Dermatol. 2018;138:697–703. doi: 10.1016/j.jid.2017.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koval A., Bassanini I., Xu J., Tonelli M., Boido V., Sparatore F., Amant F., Annibali D., Leucci E., Sparatore A., Katanaev V.L. Optimization of the clofazimine structure leads to a highly water-soluble C3-aminopyridinyl riminophenazine endowed with improved anti-Wnt and anti-cancer activity in vitro and in vivo. Eur. J. Med. Chem. 2021;222:113562. doi: 10.1016/j.ejmech.2021.113562. [DOI] [PubMed] [Google Scholar]
- 13.Zhang D., Lu Y., Liu K., Liu B., Wang J., Zhang G., Zhang H., Liu Y., Wang B., Zheng M., Fu L., Hou Y., Gong N., Lv Y., Li C., Cooper C.B., Upton A.M., Yin D., Ma Z., Huang H. Identification of less lipophilic riminophenazine derivatives for the treatment of drug-resistant tuberculosis. J. Med. Chem. 2012;55:8409–8417. doi: 10.1021/jm300828h. [DOI] [PubMed] [Google Scholar]
- 14.Bassanini I., Parapini S., Basilico N., Sparatore A. Novel hydrophilic riminophenazines as potent antiprotozoal agents. ChemMedChem. 2019;14:1940–1949. doi: 10.1002/cmdc.201900522. [DOI] [PubMed] [Google Scholar]
- 15.Rubin H., Yano T., Kassovska-Bratinova S., Schechter N., Teh J.S., Winkler J. U.S. Pat. Appl. Publ.; 2014. Novel Therapeutic Agents. US 20140371228 A1. [Google Scholar]
- 16.Medlen C.E., Anderson R., O'Sullivan J.F.G. Use of a riminophenazine for treating MDR resistance. Eur. Pat. Appl. 1995 EP 676201 A2. [Google Scholar]
- 17.Liu K., Cooper C.B., Huang H., Li C., Liu B., Liu Y., Ma Z., Wang J., Yin D., Zhang D., Zhang G., Zhang H. Riminophenazines with 2-(heteroaryl)amino substituents and their anti-microbial activity. PCT Int. Appl. 2012 WO 2012003190 A1. [Google Scholar]
- 18.Savage J.E., O'Sullivan J.F., Zeis B.M., Anderson R. Investigation of the structural properties of dihydrophenazines which contribute to their pro-oxidative interactions with human phagocytes. J. Antimicrob. Chemother. 1989;23:691–700. doi: 10.1093/jac/23.5.691. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., Xie H., Liu Q., Cao S., Huang W., Nie J., Zhao C. Faming Zhuanli Shenqing; 2018. Application of Imino Phenazines as Rabies Virus Inhibitors. CN 108853106A. [Google Scholar]
- 20.Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Fan C., Huang W., Xu M., Wang Y. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microb. Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K., Wu J.J., Zhang X., Zeng Q.X., Zhang N., Huang W.J., Tang S., Wang Y.X., Kong W.J., Wang Y.C., Li Y.H., Song D.Q. Discovery and evolution of 12N-substituted aloperine derivatives as anti-SARS-CoV-2 agents through targeting late entry stage. Bioorg. Chem. 2021;115:105196. doi: 10.1016/j.bioorg.2021.105196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zandi F., Goshadrou F., Meyfour A., Vaziri B. Rabies infection: an overview of lyssavirus-host protein interactions. Iran. Biomed. J. 2021;25:226–242. doi: 10.52547/ibj.25.4.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.