Abstract
The complex physiologic process of parturition includes the onset of labor, which requires the orchestrated stimulation of a common pathway involving uterine contractility, cervical ripening, and chorioamniotic membrane activation. However, the labor-specific processes taking place in these tissues have limited use as predictive biomarkers unless they can be probed in non-invasive samples, such as the peripheral blood. Herein, we utilized a transcriptomic dataset to assess labor-specific changes in the peripheral blood of women who delivered at term. We identified a set of genes that were differentially expressed with labor and enriched for immunological processes, and these gene expression changes were strongly correlated with results from prior studies, providing in silico validation of our findings. We then identified significant correlations between labor-specific transcriptomic changes in the maternal circulation and those detected in the chorioamniotic membranes, myometrium, and cervix of women at term, demonstrating that tissue-specific labor signatures are partly mirrored in the peripheral blood. Finally, we demonstrated a significant overlap between the peripheral blood transcriptomic changes in term parturition and those observed in asymptomatic women, prior to the diagnosis of preterm prelabor rupture of the membranes, who ultimately delivered preterm. Collectively, we provide evidence that the normal process of labor at term is characterized by a unique immunological expression signature, which may serve as a useful tool for assessing labor status and for potentially identifying women at risk for preterm birth.
Keywords: cervix, chorioamniotic membranes, fetal membranes, immune response, inflammation, myometrium, parturition, placenta, preterm labor, uterus
The maternal peripheral blood transcriptome undergoes labor-specific changes that correlate with those in the chorioamniotic membranes, myometrium, and cervix and partially overlap with those observed in asymptomatic women prior to preterm birth.
Introduction
Parturition is a complex physiologic process that allows the fetus to be delivered once it has matured enough to face extra-uterine life [1–3]. This process includes the onset of labor whose active phase leads to the vaginal delivery of the fetus [2–8]. Therefore, labor is a multi-stage process that requires the orchestrated stimulation of a common pathway involving uterine contractility, cervical ripening, and decidual/chorioamniotic membrane activation [2–7, 9]. Labor is also considered as a physiologic, sterile inflammatory process since most term-laboring women do not experience microbial invasion of the amniotic cavity [10, 11]. In line with such a concept, multiple investigations have demonstrated an increase in cellular and soluble immune mediators in the myometrium [12–24], cervix [12, 14–16, 25–34], decidua [14, 15, 35–44], and chorioamniotic membranes [14, 15, 36, 37, 39, 45–49] during spontaneous labor at term. However, the labor-specific mechanisms taking place in the reproductive and gestational tissues have limited use as predictive biomarkers, given the difficulty of obtaining biopsies from women with an ongoing pregnancy. Thus, there is a need to further explore labor-specific changes detectable in minimally invasive samples such as the peripheral blood.
In the last decade, high-dimensional techniques, such as transcriptomics, proteomics, and metabolomics, have been used to explore the complex and dynamic processes that are modulated in the maternal circulation prior to and during labor [50–52]. Single- and multi-omics studies have revealed distinct signatures that are closely associated with gestational age, including those related to immune cell types of interest, such as T cells, [53, 54] as well as those specific to the placenta and fetal liver [55]. Indeed, our recent study suggested that women who are in labor at term display a different RNA profile than that of non-laboring gestational age-matched controls, featuring an increased expression of T-cell-specific RNAs, among others [56]. Furthermore, integration of the maternal metabolome, proteome, and immunome has been utilized to predict the onset of active labor [57], and we recently assessed maternal transcriptomic and proteomic data for predicting gestational age as well as spontaneous preterm birth, using the Dialogue for Reverse Engineering Assessments and Methods (DREAM) crowdsourcing framework [58]. In this DREAM challenge, the prediction of gestational age was undertaken by using samples collected throughout gestation; thus, samples from women in labor at term were also included. However, the latter study did not assess the changes that occur in women who undergo spontaneous labor at term compared to those of gestational age-matched controls. Moreover, the question of how closely these systemic signatures mirror changes taking place in the reproductive and gestational tissues where the common pathway of labor occurs, such as the chorioamniotic membranes (including the decidua parietalis), myometrium, and cervix, remains unanswered.
Herein, we utilized the transcriptomic dataset featured in the DREAM Preterm Birth Prediction Challenge [58] to assess the changes in the maternal systemic transcriptome occurring in spontaneous labor at term. We have also investigated the extent to which maternal blood RNA changes that occur in term labor reflect those of human parturition in the chorioamniotic membranes [59], myometrium [60], and cervix [61]. Finally, we evaluated the overlap between the expression changes in spontaneous term labor and those observed in asymptomatic women, prior to the diagnosis of preterm prelabor rupture of the membranes (PPROM), who ultimately delivered preterm. The results herein may have implications for the development of non-invasive biomarkers to assess the progress of physiologic labor at term. Furthermore, the understanding of these changes may shed further light on pathological processes such as spontaneous preterm birth.
Materials and methods
Ethics
The collection of biological specimens and the use of clinical data were approved by the Institutional Review Boards of Wayne State University and NICHD. All participants provided written informed consent prior to the collection of samples.
Study population
Women who provided peripheral blood samples included in the transcriptomic analysis were enrolled in a prospective longitudinal study at the Center for Advanced Obstetrical Care and Research of the Perinatology Research Branch, NICHD/NIH/DHHS, the Detroit Medical Center, and the Wayne State University School of Medicine. Blood samples were collected longitudinally from the first or early second trimester until delivery, yet only data for the last available sample before delivery in women with spontaneous term labor as well as gestational age-matched samples from women not in labor were retained for the analysis reported in this study. Labor was defined by the presence of regular uterine contractions at a frequency of at least two contractions every 10 min with cervical changes resulting in delivery. Women with a multiple gestation or those who had a fetus affected with chromosomal and/or sonographic abnormalities were also excluded from this study. The first ultrasound scan during pregnancy was used to establish gestational age if this estimate was >7 days from the last menstrual period-based gestational age.
Maternal whole blood transcriptomics
Briefly, RNA was isolated from PAXgene Blood RNA collection tubes and was hybridized to Human Transcriptome Arrays 2.0, as previously described [62]. Exon-level gene expression data were generated from array images and were summarized for each ENTREZ gene ID with Robust Multi-array Average [63] implemented in the oligo package [64] by using a chip definition file from http://brainarray.mbni.med.umich.edu. Correction for potential batch effects was performed with the removeBatchEffect function of the limma [65] package in Bioconductor [66]. The transcriptomic dataset was previously deposited by the authors in the Gene Expression Omnibus super-series GSE149440. For each of the two groups included in the GEO series (TL: term in labor and TNL: term not in labor), only the last available samples from each patient were included in the analysis.
Differential expression analyses
Differences in gene expression between the TL and TNL groups were assessed based on linear models implemented in the limma package [67] in Bioconductor. Significant changes were based on a false discovery rate-adjusted p-value (q-value) < 0.25, provided that the fold change (FC) was at least 1.25-fold. Note that given the compression of the dynamic range of microarray-based intensity, even FCs <1.25 observed with this platform were previously validated [56]. Downstream analyses of the differentially expressed genes (DEGs) involved enrichment analysis via a hypergeometric test implemented in the GOstats package [68] to determine the over-representation of gene ontology [69] biological processes among the significant genes. Additional enrichment analyses were based on a hypergeometric test with pathway definitions extracted from the C2 collection of the MSigDB database. The C2 collection in MsigDB includes pathways from the Pathway Interaction Database [70], Kyoto Encyclopedia of Genes and Genomes [71], Reactome database [72], and WikiPathways [73], among other sources. The background list in the enrichment analyses featured all genes profiled on the microarray platform. A network representation of DEGs was performed by using the stringApp version (1.5.0) [74] in Cytoscape (version 3.7.2) [75]. The most interconnected sub-networks were displayed and nodes were annotated to significantly enriched biological processes. A false discovery rate-adjusted p-value (q-value) < 0.1 was applied in all enrichment analyses to infer significance and to allow direct comparisons to previous findings in preterm birth [58].
In silico validation of blood RNA changes in spontaneous labor at term
To assess the overlap of expression changes with term labor obtained in this study and the report of Tarca et al. [56], in which the blood transcriptomes of eight TL and eight TNL cases were contrasted, we calculated the Pearson correlation of log2FC (TL vs. TNL) between studies for all genes deemed significant in this study.
Correlation of whole blood RNA changes in spontaneous labor at term with previous reports on changes in gestational and reproductive tissues
The correlation between expression changes in fetal membranes with term labor (TL vs. TNL) reported in Haddad et al. [59] and those found in the current study was based on genes detected on both platforms regardless of their significance in the current study (n = 189, Table 2 of original publication). Similarly, we determined the extent to which term parturition, blood transcriptomic changes found herein reflected those reported in the human myometrium (462 genes) [60] and cervix (833 genes) [61]. For the two latter studies, the full lists of DEGs and FCs were obtained from the authors, and we retained only genes represented on microarray platform used in this study. Finally, to determine the relationship between the effects of term parturition on the maternal blood transcriptome and changes preceding a diagnosis of PPROM leading to preterm birth, we have correlated our data with the FCs of 402 RNAs dysregulated in asymptomatic patients at 17–33 weeks of gestation in two diverse populations [58]. Finally, the blood RNA changes preceding a diagnosis of PPROM (regardless of statistical significance) were correlated with those of DEGs with labor in term tissues, as described above for the blood term-labor signature. For each comparison, the proportion of RNAs with matching direction of change between the two studies was also reported. A Pearson correlation of p < 0.05 was deemed to be significant.
Identification of genes for which expression changes with term labor are different from those preceding preterm birth
Gene expression differences between the 21 TL and 28 TNL patients were contrasted with those observed between 30 patients sampled at 17–33 weeks of gestation, who subsequently were diagnosed with PPROM and delivered preterm, and gestational age-matched controls (n = 45, derived from the same 49 normal pregnancies used for the TL vs. TNL comparison). Moderated t-tests were used to assess the significance of the interaction. Significant interactions were based on a false discovery rate-adjusted p-value (q-value) < 0.25, provided that the FC was at least 1.25-fold.
Results
Labor involves immune inflammatory processes in the maternal circulation
The onset of labor requires the coordinated activation of different local and systemic pathways that will result in the delivery of the neonate [2–7, 9]. Thus, we first compared the transcriptomes in the peripheral blood between pregnant women at term with and without spontaneous labor by using microarrays. The clinical and demographic characteristics of the patients are shown in Table 1. Among the 49 women included in the study, the first ultrasound exam available to establish gestational age was performed before 14 weeks in 38 women and between 14 and 20 weeks in the remainder of 11 patients. There were no differences in maternal characteristics, gestational age at sampling, and birthweight percentiles between the two groups; yet, women in the TNL group delivered on average 5 days later (0.7 weeks) than those who were in labor (p < 0.001).
Table 1.
Clinical and demographic characteristics of women at term with or without labor
| TNL (n = 28) | TL (n = 21) | p | |
|---|---|---|---|
| Maternal age [years; median (IQR)]a | 25 (21–28.2) | 24 (22–28) | 0.97 |
| Body mass index [kg/m2; median (IQR)]a | 26.9 (24.1–34) | 24.8 (21.1–27.5) | 0.06 |
| Nulliparousb | 8/28 (28.6%) | 8/21 (38.1%) | 0.55 |
| Race/ethnicity | |||
| African-Americanb | 27/28 (96.4%) | 18/21 (85.7%) | 0.3 |
| Fetal sex (female)b | 16/28 (57.1%) | 11/21 (52.4%) | 0.78 |
| Gestational age at delivery [weeks; median (IQR)]a | 39.6 (39–40.6) | 38.9 (38.1–39.3) | <0.001 |
| Birthweight [g; median (IQR)]a | 3452.5 (3153.8–3596.2) | 3175 (2930–3380) | 0.004 |
| Birthweight percentile [%; median (IQR)]a | 62.4 (39.1–77.4) | 52.7 (29.2–71.3) | 0.29 |
| Gestational age at sampling [weeks; median (IQR)]a | 38.5 (38.0–39.0) | 38.9 (38.1–39.3) | 0.4 |
Data are given as medians (interquartile range, IQR) and percentages (n/N).
aWelch’s t-test.
bFisher’s exact test.
After comparing the expression profiles of 32 830 genes, we identified 103 DEGs, with 51 upregulated and 52 downregulated in women at TL compared to those without labor (q < 0.25, FC >1.25 for all DEGs) (Figure 1A and Table 2). The RNAs with differential expression in labor at term included not only mRNAs but also miRNAs and other non-coding genes (Figure 1A and Table 2). STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) analysis of DEGs revealed interconnected protein–protein interaction sub-networks, as displayed in Figure 1B. These genes corresponded to immunologic processes that were found to be enriched with labor at term (q < 0.1), namely, “immune effector process,” “immune response-regulating signaling pathway,” and “immune system process,” among others (Figure 1B and Table 3).
Figure 1.
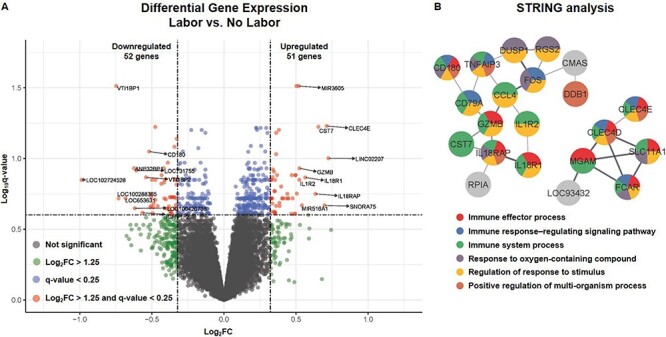
Differential gene expression and pathway enrichment in term labor. (A) Volcano plot showing differential gene expression in peripheral blood from women who underwent term labor compared to that of women who delivered at term without labor, where the Y-axis shows the –log10(q-value) and the X-axis shows the log2(FC). Differentially expressed genes (DEGs) were considered those with q < 0.25 and FC >1.25 (red dots). Blue dots indicate genes with a low q-value but low FC, green dots indicate genes with a high FC but high q-value, and gray dots indicate all other genes tested. (B) STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) analysis of the most interconnected protein–protein interaction sub-networks corresponding to DEGs, with nodes annotated to significantly enriched biological processes.
Table 2.
Genes differentially expressed in the peripheral blood in term labor compared to term without labor
| SYMBOL | ENTREZ | log2FC | p | Adj. p |
|---|---|---|---|---|
| DUSP1 | 1843 | 0.50 | 1.6E-06 | 0.031 |
| MIR3605 | 100500853 | 0.52 | 2.2E-06 | 0.031 |
| VTI1BP1 | 391559 | −0.75 | 2.8E-06 | 0.031 |
| CLEC4E | 26253 | 0.71 | 7.2E-06 | 0.059 |
| CST7 | 8530 | 0.66 | 1.1E-05 | 0.060 |
| RPL36P12 | 100271012 | −0.48 | 1.1E-05 | 0.060 |
| APMAP | 57136 | 0.38 | 2.6E-05 | 0.063 |
| TMCC3 | 57458 | 0.36 | 3.4E-05 | 0.066 |
| HSP90AB3P | 3327 | −0.33 | 4.0E-05 | 0.073 |
| SNORD101 | 594,837 | −0.34 | 0.0001 | 0.083 |
| CD180 | 4064 | −0.52 | 0.0001 | 0.089 |
| SLC37A3 | 84255 | 0.36 | 0.0001 | 0.093 |
| LINC02207 | 101927153 | 0.73 | 0.0001 | 0.100 |
| LOC100420707 | 100420707 | −0.39 | 0.0001 | 0.107 |
| ANP32BP1 | 646791 | −0.62 | 0.0002 | 0.117 |
| YBX3P1 | 440359 | −0.42 | 0.0002 | 0.117 |
| GZMB | 3002 | 0.52 | 0.0002 | 0.117 |
| SLC2A3P4 | 399495 | 0.34 | 0.0002 | 0.120 |
| LOC731755 | 731755 | −0.61 | 0.0002 | 0.122 |
| H2BC2P | 100288742 | 0.35 | 0.0002 | 0.124 |
| YBX3 | 8531 | −0.43 | 0.0002 | 0.131 |
| CCL4 | 6351 | 0.47 | 0.0002 | 0.131 |
| FOS | 2353 | 0.50 | 0.0002 | 0.131 |
| TNFAIP3 | 7128 | 0.37 | 0.0003 | 0.131 |
| RGS2 | 5997 | 0.50 | 0.0003 | 0.131 |
| TPST1 | 8460 | 0.34 | 0.0003 | 0.135 |
| VTI1BP2 | 389246 | −0.54 | 0.0003 | 0.136 |
| IL18R1 | 8809 | 0.56 | 0.0003 | 0.136 |
| RLIMP1 | 100533695 | −0.48 | 0.0003 | 0.142 |
| IGKJ5 | 28946 | −0.50 | 0.0004 | 0.142 |
| CAPRIN2 | 65981 | −0.41 | 0.0004 | 0.142 |
| ABCB10 | 23,456 | −0.34 | 0.0004 | 0.142 |
| REPS2 | 9185 | 0.33 | 0.0004 | 0.142 |
| IL1R2 | 7850 | 0.52 | 0.0004 | 0.142 |
| LOC102724528 | 102724528 | −0.99 | 0.0005 | 0.142 |
| C3orf86 | 102724231 | 0.33 | 0.0005 | 0.142 |
| LOC102723340 | 102723340 | 0.35 | 0.0005 | 0.142 |
| CDC16 | 8881 | −0.33 | 0.0005 | 0.142 |
| KAT2B | 8850 | −0.42 | 0.0005 | 0.148 |
| NAP1L4P1 | 728589 | −0.43 | 0.0006 | 0.151 |
| METTL7A | 25840 | −0.33 | 0.0006 | 0.153 |
| CRISPLD2 | 83716 | 0.40 | 0.0006 | 0.158 |
| SIGLEC5 | 8778 | 0.35 | 0.0007 | 0.170 |
| PTTG2 | 10744 | 0.35 | 0.0007 | 0.170 |
| IRS2 | 8660 | 0.51 | 0.0008 | 0.177 |
| HMGB2P1 | 729119 | 0.44 | 0.0008 | 0.178 |
| IL18RAP | 8807 | 0.64 | 0.0008 | 0.178 |
| IGKV1–17 | 28937 | −0.51 | 0.0008 | 0.178 |
| PIP5K1B | 8395 | −0.34 | 0.0010 | 0.187 |
| DNAJA4 | 55466 | −0.35 | 0.0010 | 0.188 |
| DDB1 | 1642 | −0.36 | 0.0010 | 0.188 |
| QRSL1P3 | 100422374 | −0.50 | 0.0010 | 0.188 |
| DCK | 1633 | −0.37 | 0.0010 | 0.188 |
| LSM12P1 | 653122 | −0.37 | 0.0011 | 0.188 |
| KCNJ15 | 3772 | 0.40 | 0.0011 | 0.191 |
| SAMSN1 | 64092 | 0.45 | 0.0012 | 0.191 |
| SNORA2C | 677815 | 0.43 | 0.0012 | 0.191 |
| MGAM2 | 93432 | 0.48 | 0.0012 | 0.191 |
| LOC653631 | 653631 | −0.68 | 0.0012 | 0.191 |
| GPR141 | 353345 | 0.33 | 0.0014 | 0.191 |
| NUTM2A-AS1 | 728190 | −0.33 | 0.0014 | 0.191 |
| MPZL3 | 196264 | 0.33 | 0.0014 | 0.191 |
| CYP4F3 | 4051 | 0.32 | 0.0014 | 0.191 |
| TPGS2 | 25941 | −0.41 | 0.0015 | 0.191 |
| F5 | 2153 | 0.34 | 0.0015 | 0.191 |
| MGAM | 8972 | 0.44 | 0.0015 | 0.191 |
| SLC26A8 | 116369 | 0.41 | 0.0015 | 0.191 |
| LOC100288365 | 100288365 | −0.73 | 0.0015 | 0.191 |
| RPIA | 22934 | −0.40 | 0.0015 | 0.194 |
| NTAN1P2 | 100420307 | −0.48 | 0.0016 | 0.196 |
| LOC101927851 | 101927851 | 0.38 | 0.0017 | 0.206 |
| SLC6A8 | 6535 | −0.49 | 0.0018 | 0.208 |
| LSMEM1 | 286006 | 0.33 | 0.0018 | 0.208 |
| ALDH5A1 | 7915 | −0.33 | 0.0019 | 0.213 |
| SNORA75 | 654321 | 0.70 | 0.0020 | 0.214 |
| MIR516A1 | 574498 | 0.54 | 0.0020 | 0.214 |
| VNN3 | 55350 | 0.36 | 0.0021 | 0.216 |
| NSRP1P1 | 100420557 | −0.50 | 0.0021 | 0.217 |
| LOC101927759 | 101927759 | 0.39 | 0.0021 | 0.217 |
| C9orf78 | 51759 | −0.46 | 0.0022 | 0.217 |
| YBX1P2 | 646531 | −0.38 | 0.0022 | 0.218 |
| LOC107986698 | 107986698 | −0.49 | 0.0023 | 0.222 |
| STRADBP1 | 389599 | −0.51 | 0.0023 | 0.222 |
| LOC100420738 | 100420738 | −0.62 | 0.0025 | 0.225 |
| CD79A | 973 | −0.41 | 0.0025 | 0.225 |
| TRAJ23 | 28732 | −0.34 | 0.0026 | 0.225 |
| IGHV5–51 | 28388 | −0.34 | 0.0026 | 0.226 |
| RPLP2P3 | 643949 | −0.40 | 0.0026 | 0.226 |
| MARCHF8 | 220972 | −0.38 | 0.0026 | 0.226 |
| FCAR | 2204 | 0.33 | 0.0029 | 0.237 |
| SESN3 | 143686 | −0.37 | 0.0029 | 0.238 |
| SLC11A1 | 6556 | 0.35 | 0.0030 | 0.238 |
| CMAS | 55907 | −0.37 | 0.0030 | 0.239 |
| GTF2H2C | 728340 | 0.35 | 0.0030 | 0.239 |
| IGHV1-2 | 28474 | −0.56 | 0.0031 | 0.242 |
| LOC101928044 | 101928044 | −0.49 | 0.0032 | 0.243 |
| PFKFB2 | 5208 | 0.35 | 0.0033 | 0.244 |
| MIR223 | 407008 | 0.36 | 0.0033 | 0.245 |
| RNF10 | 9921 | −0.36 | 0.0034 | 0.245 |
| CLEC4D | 338339 | 0.49 | 0.0035 | 0.245 |
| SCARNA10 | 692148 | 0.47 | 0.0035 | 0.246 |
| SNORA1 | 677792 | 0.49 | 0.0035 | 0.247 |
| GUCD1 | 83606 | −0.35 | 0.0036 | 0.249 |
Table 3.
Biological processes enriched in the peripheral blood in term labor compared to term without labor
| Biological process | Count | Size | Odds ratio | q | p | Significant in PPROMa |
|---|---|---|---|---|---|---|
| Leukocyte activation | 17 | 1288 | 4.8 | 0.001 | 1.44E-06 | 1 |
| Immune response | 22 | 2187 | 3.9 | 0.001 | 2.68E-06 | 1 |
| Cell activation | 17 | 1447 | 4.2 | 0.001 | 7.00E-06 | 1 |
| Immune system process | 26 | 3156 | 3.3 | 0.001 | 9.69E-06 | 1 |
| Immune effector process | 15 | 1253 | 4.2 | 0.002 | 2.21E-05 | 1 |
| Regulation of immune system process | 17 | 1580 | 3.8 | 0.002 | 2.22E-05 | 1 |
| Leukocyte mediated immunity | 12 | 850 | 4.8 | 0.003 | 3.49E-05 | 1 |
| Adaptive immune response | 10 | 621 | 5.3 | 0.004 | 5.74E-05 | 1 |
| Positive regulation of cellular carbohydrate metabolic process | 4 | 62 | 20.8 | 0.004 | 6.67E-05 | 0 |
| Inflammatory response | 11 | 787 | 4.6 | 0.005 | 8.46E-05 | 0 |
| Leukocyte activation involved in immune response | 10 | 709 | 4.6 | 0.009 | 0.000171 | 1 |
| Cell activation involved in immune response | 10 | 713 | 4.6 | 0.009 | 0.000179 | 1 |
| Positive regulation of carbohydrate metabolic process | 4 | 84 | 15 | 0.01 | 0.000218 | 0 |
| Lymphocyte activation | 10 | 737 | 4.4 | 0.01 | 0.000234 | 0 |
| Neutrophil activation | 8 | 497 | 5.2 | 0.013 | 0.000339 | 1 |
| Granulocyte activation | 8 | 504 | 5.1 | 0.014 | 0.000372 | 1 |
| Positive regulation of glucose metabolic process | 3 | 42 | 22.8 | 0.014 | 0.000432 | 0 |
| Myeloid leukocyte activation | 9 | 656 | 4.4 | 0.014 | 0.000457 | 1 |
| T cell cytokine production | 3 | 43 | 22.2 | 0.014 | 0.000464 | 0 |
| Lymphocyte activation involved in immune response | 5 | 185 | 8.4 | 0.014 | 0.000484 | 0 |
| T-helper 1 type immune response | 3 | 44 | 21.7 | 0.014 | 0.000496 | 1 |
| Regulation of immune response | 11 | 971 | 3.7 | 0.014 | 0.000522 | 1 |
| T cell activation involved in immune response | 4 | 108 | 11.6 | 0.014 | 0.000568 | 0 |
| B cell activation | 6 | 305 | 6.2 | 0.017 | 0.000701 | 0 |
| Regulation of glucose metabolic process | 4 | 120 | 10.4 | 0.02 | 0.000843 | 0 |
| Defense response | 15 | 1752 | 2.9 | 0.02 | 0.000907 | 0 |
| Lymphocyte mediated immunity | 6 | 328 | 5.7 | 0.022 | 0.001022 | 1 |
| Positive regulation of immune system process | 11 | 1058 | 3.4 | 0.022 | 0.001061 | 1 |
| Adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 6 | 331 | 5.7 | 0.022 | 0.001071 | 1 |
| Response to reactive oxygen species | 5 | 225 | 6.9 | 0.023 | 0.001167 | 0 |
| Regulation of mitotic sister chromatid separation | 3 | 61 | 15.3 | 0.024 | 0.001293 | 0 |
| Mitotic sister chromatid separation | 3 | 64 | 14.6 | 0.027 | 0.001485 | 0 |
| Neutrophil degranulation | 7 | 483 | 4.6 | 0.027 | 0.001499 | 1 |
| Neutrophil activation involved in immune response | 7 | 486 | 4.5 | 0.027 | 0.001553 | 1 |
| Regulation of chromosome separation | 3 | 66 | 14.1 | 0.027 | 0.001623 | 0 |
| Negative regulation of protein serine/threonine kinase activity | 4 | 144 | 8.6 | 0.027 | 0.001656 | 0 |
| Neutrophil mediated immunity | 7 | 497 | 4.4 | 0.028 | 0.001764 | 1 |
| Regulation of cellular carbohydrate metabolic process | 4 | 147 | 8.4 | 0.028 | 0.001786 | 0 |
| T cell differentiation involved in immune response | 3 | 70 | 13.3 | 0.028 | 0.001922 | 0 |
| Positive regulation of small molecule metabolic process | 4 | 150 | 8.2 | 0.028 | 0.001923 | 0 |
| Regulation of mitotic sister chromatid segregation | 3 | 72 | 12.9 | 0.03 | 0.002084 | 0 |
| Negative regulation of kinase activity | 5 | 264 | 5.8 | 0.033 | 0.002353 | 0 |
| Leukocyte degranulation | 7 | 533 | 4.1 | 0.036 | 0.002614 | 1 |
| Regulation of adaptive immune response | 4 | 166 | 7.4 | 0.037 | 0.002777 | 0 |
| Myeloid cell activation involved in immune response | 7 | 544 | 4 | 0.038 | 0.002929 | 1 |
| Myeloid leukocyte mediated immunity | 7 | 551 | 4 | 0.039 | 0.003144 | 1 |
| Response to bacterium | 8 | 704 | 3.6 | 0.039 | 0.00316 | 0 |
| Regulation of B cell activation | 4 | 173 | 7.1 | 0.039 | 0.003221 | 0 |
| Regulation of sister chromatid segregation | 3 | 84 | 11 | 0.039 | 0.003231 | 0 |
| Negative regulation of transferase activity | 5 | 291 | 5.3 | 0.041 | 0.003574 | 0 |
| Cellular carbohydrate metabolic process | 5 | 292 | 5.3 | 0.041 | 0.003627 | 0 |
| Positive regulation of immune response | 8 | 721 | 3.5 | 0.041 | 0.003653 | 1 |
| Monosaccharide metabolic process | 5 | 295 | 5.2 | 0.042 | 0.003788 | 0 |
| Regulation of response to external stimulus | 10 | 1081 | 2.9 | 0.046 | 0.004264 | 0 |
| Regulation of response to stimulus | 25 | 4308 | 2.1 | 0.046 | 0.004322 | 0 |
| Chromosome separation | 3 | 94 | 9.7 | 0.046 | 0.004434 | 0 |
| Negative regulation of inflammatory response | 4 | 190 | 6.4 | 0.046 | 0.004497 | 0 |
| Defense response to bacterium | 5 | 310 | 5 | 0.046 | 0.004671 | 0 |
| Leukocyte proliferation | 5 | 310 | 5 | 0.046 | 0.004671 | 0 |
| B cell proliferation | 3 | 97 | 9.4 | 0.047 | 0.004841 | 0 |
| Regulation of carbohydrate biosynthetic process | 3 | 98 | 9.3 | 0.048 | 0.004981 | 0 |
| Innate immune response | 9 | 943 | 3 | 0.052 | 0.005515 | 0 |
| T cell mediated immunity | 3 | 103 | 8.9 | 0.053 | 0.00572 | 1 |
| Cytokine production involved in immune response | 3 | 104 | 8.8 | 0.053 | 0.005875 | 0 |
| Immune response-regulating cell surface receptor signaling pathway | 6 | 468 | 4 | 0.053 | 0.005976 | 0 |
| Regulation of chromosome segregation | 3 | 105 | 8.7 | 0.053 | 0.006033 | 0 |
| Immune response-regulating signaling pathway | 6 | 471 | 3.9 | 0.053 | 0.006161 | 0 |
| Regulation of carbohydrate metabolic process | 4 | 208 | 5.9 | 0.053 | 0.00618 | 0 |
| Negative regulation of phosphorylation | 6 | 473 | 3.9 | 0.053 | 0.006286 | 0 |
| Regulated exocytosis | 8 | 793 | 3.2 | 0.054 | 0.006439 | 0 |
| Glucose metabolic process | 4 | 213 | 5.7 | 0.055 | 0.006713 | 0 |
| Defense response to other organism | 10 | 1154 | 2.7 | 0.055 | 0.006716 | 0 |
| Interferon-gamma production | 3 | 113 | 8.1 | 0.059 | 0.007387 | 0 |
| Fc receptor signaling pathway | 4 | 220 | 5.5 | 0.06 | 0.007509 | 0 |
| Carboxylic acid transport | 5 | 349 | 4.4 | 0.06 | 0.007639 | 0 |
| Organic acid transport | 5 | 352 | 4.3 | 0.061 | 0.007911 | 0 |
| Response to stress | 23 | 4047 | 2 | 0.066 | 0.008709 | 0 |
| Response to cytokine | 10 | 1203 | 2.6 | 0.066 | 0.008903 | 0 |
| Fc-gamma receptor signaling pathway | 3 | 121 | 7.5 | 0.066 | 0.008907 | 0 |
| Regulation of leukocyte chemotaxis | 3 | 124 | 7.3 | 0.07 | 0.009521 | 0 |
| Secretion by cell | 11 | 1420 | 2.5 | 0.073 | 0.010082 | 0 |
| Negative regulation of protein kinase activity | 4 | 242 | 5 | 0.075 | 0.010409 | 0 |
| Positive regulation of response to biotic stimulus | 4 | 248 | 4.9 | 0.079 | 0.01131 | 0 |
| Leukocyte differentiation | 6 | 537 | 3.4 | 0.079 | 0.01136 | 0 |
| Negative regulation of intracellular signal transduction | 6 | 543 | 3.4 | 0.083 | 0.011952 | 0 |
| Hexose metabolic process | 4 | 253 | 4.8 | 0.083 | 0.012098 | 0 |
| Response to hydrogen peroxide | 3 | 138 | 6.6 | 0.085 | 0.012708 | 0 |
| Monocarboxylic acid catabolic process | 3 | 138 | 6.6 | 0.085 | 0.012708 | 0 |
| Export from cell | 11 | 1471 | 2.4 | 0.085 | 0.012923 | 0 |
| Negative regulation of defense response | 4 | 261 | 4.6 | 0.088 | 0.013431 | 0 |
| Regulation of response to biotic stimulus | 5 | 404 | 3.8 | 0.089 | 0.013767 | 0 |
| Exocytosis | 8 | 910 | 2.7 | 0.09 | 0.014096 | 0 |
| Regulation of defense response | 7 | 735 | 2.9 | 0.091 | 0.014416 | 0 |
| Cellular response to cytokine stimulus | 9 | 1113 | 2.5 | 0.095 | 0.015419 | 0 |
| Carbohydrate transport | 3 | 149 | 6.1 | 0.095 | 0.015592 | 0 |
| Carboxylic acid transmembrane transport | 3 | 149 | 6.1 | 0.095 | 0.015592 | 0 |
| Regulation of leukocyte activation | 6 | 578 | 3.2 | 0.095 | 0.015842 | 0 |
| Organic acid transmembrane transport | 3 | 150 | 6 | 0.095 | 0.015871 | 0 |
| Negative regulation of cellular protein metabolic process | 9 | 1121 | 2.5 | 0.095 | 0.01609 | 0 |
| Regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 3 | 151 | 6 | 0.095 | 0.016153 | 0 |
| Response to other organism | 11 | 1524 | 2.3 | 0.095 | 0.016491 | 0 |
| Response to external biotic stimulus | 11 | 1526 | 2.3 | 0.095 | 0.016639 | 0 |
| Negative regulation of response to external stimulus | 5 | 424 | 3.6 | 0.095 | 0.016641 | 0 |
| Production of molecular mediator of immune response | 4 | 279 | 4.3 | 0.095 | 0.016764 | 0 |
| Lymphocyte proliferation | 4 | 280 | 4.3 | 0.095 | 0.016963 | 0 |
| Positive regulation of response to stimulus | 15 | 2392 | 2 | 0.095 | 0.01711 | 0 |
| Negative regulation of phosphate metabolic process | 6 | 591 | 3.1 | 0.095 | 0.017489 | 0 |
| Mononuclear cell proliferation | 4 | 283 | 4.3 | 0.095 | 0.017569 | 0 |
| Negative regulation of phosphorus metabolic process | 6 | 592 | 3.1 | 0.095 | 0.01762 | 0 |
| Import across plasma membrane | 3 | 157 | 5.7 | 0.095 | 0.017903 | 0 |
| Negative regulation of protein phosphorylation | 5 | 433 | 3.5 | 0.095 | 0.018056 | 0 |
| Immune response-activating cell surface receptor signaling pathway | 5 | 436 | 3.5 | 0.096 | 0.018545 | 0 |
| Immune response-activating signal transduction | 5 | 436 | 3.5 | 0.096 | 0.018545 | 0 |
| Secretion | 11 | 1557 | 2.2 | 0.097 | 0.019064 | 0 |
| Mitotic sister chromatid segregation | 3 | 161 | 5.6 | 0.097 | 0.019126 | 0 |
| Response to biotic stimulus | 11 | 1558 | 2.2 | 0.097 | 0.019146 | 0 |
| Cellular response to reactive oxygen species | 3 | 162 | 5.6 | 0.097 | 0.019439 | 0 |
| Cell surface receptor signaling pathway | 18 | 3122 | 1.9 | 0.097 | 0.019617 | 0 |
| Response to oxidative stress | 5 | 443 | 3.4 | 0.097 | 0.019719 | 0 |
| Regulation of mitotic nuclear division | 3 | 163 | 5.5 | 0.097 | 0.019755 | 0 |
| Cytokine-mediated signaling pathway | 7 | 787 | 2.7 | 0.098 | 0.020221 | 0 |
a0, not significant; 1, significant.
To assess the reliability of labor-related transcriptomic changes identified herein, we performed a comparative analysis between our current microarray results and a report of an independent, smaller, microarray-based study from the same population [56]. The changes in expression of the 103 DEGs determined herein were highly correlated with those derived from the available dataset (r = 0.83, p < 0.0001, 95% agreement in direction of change) (Figure 2), providing an in silico validation of the current findings. Together, these results support the inflammatory nature of the physiologic process of labor at term.
Figure 2.
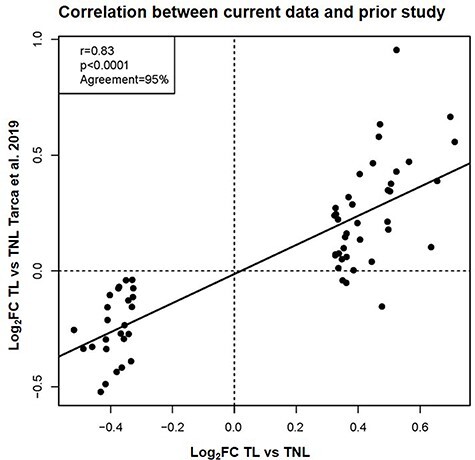
Correlation between labor-specific gene expression in the peripheral blood from the current study and a previous report. Scatterplot showing the correlation between the log2FC reported in this study and that from a previous report [56]. The Pearson correlation coefficient, p-value, and percentage of agreement on direction of gene expression FC are shown.
Labor-specific signatures in gestational and reproductive tissues implicated in the common pathway of labor correlate with those in peripheral blood
Labor is a complex process that requires the orchestrated stimulation of a common pathway involving uterine contractility, cervical ripening, and decidual/chorioamniotic membrane activation [2–7, 9]. Therefore, we next utilized previously published datasets to evaluate whether labor-specific signatures in the chorioamniotic membranes, myometrium, and cervix correlated with those in the peripheral blood of women with term labor. The differential expression of 189 genes reported as modulated during labor in the chorioamniotic membranes were correlated to those based on maternal peripheral transcriptomic data (r = 0.34, p < 0.0001, direction of change agreement 63%) (Figure 3A). Moreover, the differential expression of 462 genes reported as modulated during labor in the myometrium also correlated to those derived from the maternal circulation (r = 0.30, p < 0.0001, direction of change agreement 58%) (Figure 3B). Finally, a modest but significant correlation was observed between transcriptomic labor-related changes in the peripheral blood and those reported in the cervix (r = 0.2, p < 0.0001, direction of change agreement 58%) (Figure 3C). Collectively, these results suggest that some of the labor-related transcriptomic changes in the gestational and reproductive tissues are mirrored in the maternal circulation. These findings may have implications for the development of non-invasive biomarkers to monitor the physiologic process of labor at term.
Figure 3.
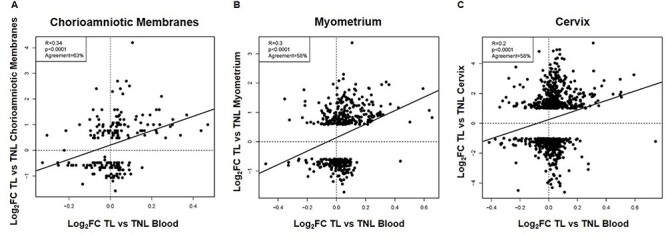
Correlation between labor-specific gene expression in the peripheral blood and that in the chorioamniotic membranes, myometrium, or cervix. Scatterplots showing the correlation between gene expression (log2FC) reported in this study and that in (A) the chorioamniotic membranes [59], (B) the myometrium [60], or (C) the cervix [61]. The Pearson correlation coefficients, p-values, and percentages of agreement on direction of gene expression FC are shown for each comparison.
Correlation between gene expression changes in term labor and those occurring prior to preterm birth
The understanding of the mechanisms implicated in the physiologic process of labor at term may provide insights into the pathologic processes that lead to preterm birth. During normal labor, the activation of the chorioamniotic membranes culminates with rupture of these tissues to allow delivery of the fetus [7]. The premature occurrence of this process is termed PPROM, which accounts for one-third of preterm birth cases [76]. Thus, we evaluated the overlap between the transcriptomic changes observed in the maternal circulation during labor at term and those observed in the peripheral blood of asymptomatic women, prior to the diagnosis of PPROM, who ultimately delivered preterm. This analysis was performed with samples collected at 17–22 weeks and at 27–33 weeks from cases and gestational age-matched controls. The transcriptomic changes of 402 RNAs preceding the diagnosis of PPROM (hereafter referred to as “PPROM signature”) were significantly correlated with those derived from the comparison between TL and TNL groups herein (r = 0.71, p < 0.0001, 86% agreement in direction of change) (Figure 4). Of note, among the 51 biological processes enriched in the PPROM signature [58], 26 were also significant in the spontaneous term parturition-derived signature (Table 3). Yet, 25 of these processes were not significantly enriched in labor at term, highlighting the fact that the sub-clinical processes leading to preterm birth are not merely the premature initiation of normal labor, but instead represent the occurrence of distinct pathological events culminating in the activation of one or more of the components of the common pathway of labor [7]. In addition, four of the five Reactome pathways we found herein to be associated with term parturition (innate immune system, neutrophil degranulation, signaling by interleukins, cytokine signaling in immune system, q < 0.05, and enrichment odds ratios >2.5 for all) were previously reported as enriched in the PPROM signature [58]. Taken together, these results suggest that there is a substantial overlap between the transcriptomic changes taking place in the circulation of women who ultimately undergo spontaneous preterm birth and of those observed during spontaneous labor at term.
Figure 4.
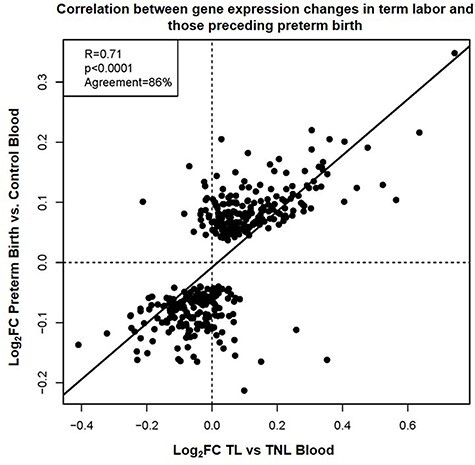
Correlation between gene expression changes in term labor and those preceding preterm birth. Scatterplot showing the correlation between gene expression (log2FC) derived from the comparison between women at term with and without labor and those from a previous report of asymptomatic women who subsequently underwent preterm birth associated with PPROM [58]. The Pearson correlation coefficient, p-value, and percentage of agreement on direction of gene expression FC are shown.
To fully appreciate the differential gene expression changes between the normal process of labor at term and those occurring prior to preterm birth, we performed a direct assessment of the transcriptomic changes that are specific to term labor but were not observed in asymptomatic women, prior to the diagnosis of PPROM, who ultimately delivered preterm using interaction tests (Supplementary Table S1). The majority of genes with a significant interaction test were dysregulated in term labor but not prior to preterm birth, including DDIT4 (DNA damage inducible transcript 4), DUSP1 (dual specificity phosphatase 1), TPST1 (tyrosylprotein sulfotransferase 1), FOS (Fos proto-oncogene, AP-1 transcription factor subunit), TNFAIP3 (TNF alpha induced protein 3), CCL4 (C-C motif chemokine ligand 4), and GZMB (granzyme B) (Supplementary Table S1). Pathway analysis revealed that these genes are involved in “cytokine signaling in immune system” and “innate immune system” as well as in multiple immune processes (enrichment q < 0.1, Supplementary Table S2). By contrast, transcripts that were decreased in term labor but not prior to preterm birth included RPL36P12 (ribosomal protein L36 pseudogene 12), HSP90AB3P (heat shock protein 90 alpha family class B member 3, pseudogene), CSNK2A3 (casein kinase 2 alpha 3), VTI1BP1 (vesicle transport through interaction with t-SNAREs 1B pseudogene 1), and CD180 (CD180 molecule), among others (Supplementary Table S1). A notable exception was KLRD1 (killer cell lectin like receptor D1), which displayed a slight increase (log2FC = 0.35) in term labor but a slight decrease (log2FC = −0.26) prior to preterm birth (Supplementary Table S1). These data highlight the differences between the transcriptional activity that accompanies the physiologic process of parturition at term and that preceding preterm birth.
Finally, we determined the correlation between gene expression changes preceding preterm birth in the maternal blood and those previously reported in tissues collected at term. Significant but modest correlations were also noted between the whole blood transcriptomic changes preceding the diagnosis of PPROM in women who ultimately delivered preterm [58] and those reported in the chorioamniotic membranes (r = 0.23, p = 0.002), myometrium (r = 0.19, p < 0.001), and cervix (r = 0.12, p < 0.001) from women with labor at term (Supplementary Figure S1). Of note, the changes derived from samples collected from women who ultimately delivered preterm are not necessarily specific to preterm birth since they are also observed in women with labor at term [as shown in Figure 4 and in the interaction analyses presented herein (Supplementary Tables S1 and S2)]. Yet, these results indicate that the whole blood, labor-derived transcriptomic signature reported herein has potential use as a “liquid biopsy” to identify women who are at risk for preterm birth.
Discussion
Herein, we utilized a large, previously generated, longitudinal dataset to assess labor-specific changes in the peripheral blood transcriptome of women at term. We identified a set of genes that were differentially expressed with labor and enriched for immunological processes. Further, we have shown a strong correlation between the reported expression changes and those reported in a prior study, providing an in silico validation of our findings. Notably, we also identified significant correlations between labor-specific transcriptomic changes in the maternal circulation and DEGs determined in the chorioamniotic membranes, myometrium, and cervix of women at term, demonstrating that the tissue-specific signatures are mirrored with those in the peripheral blood. Finally, we demonstrated a significant degree of overlap between the peripheral blood transcriptomic changes derived from women with labor at term and those obtained from asymptomatic women who subsequently were diagnosed with PPROM and delivered preterm. Therefore, the normal process of labor at term is characterized by an immunological expression signature, which is partially shared with that observed in women who ultimately underwent spontaneous preterm birth, and may potentially serve as a useful tool for assessing labor status and for potentially identifying women at risk for preterm birth.
Labor is a well-characterized state of system-wide physiologic inflammation, including not only the gestational and reproductive tissues but also extending to the maternal circulation. This systemic response includes the dynamic and intricate modulation of peripheral immune cells and their activation status [16, 77–80] as well as fluctuations in the soluble fraction [81–83], which includes cell-free fetal DNA [84–86]. Consistently, we have previously utilized maternal whole blood transcriptomics to identify immune signatures that are modified both throughout gestation and with labor [56, 62]. Notably, gene signatures corresponding to T cells, B cells, and CD71+ erythroid cells, among others, were found to follow unique expression patterns during normal pregnancy, with the T-cell and erythroid signatures increasing in late gestation [62]. In addition, a pilot study of women with or without labor at term revealed that a T-cell signature was overexpressed with labor [56], further emphasizing that this inflammatory process is reflected in the maternal peripheral blood transcriptome. Herein, we utilized exon-level resolution gene expression data to demonstrate labor-specific differences in the maternal blood transcriptome that are characterized by enrichment of immune-related processes, which is in line with previous findings. Importantly, our current data were strongly correlated with our prior pilot study [56], demonstrating agreement in terms of labor-specific gene expression changes. Together, these data may be useful for the development of tools to detect and monitor labor.
In the current study, we observed significant correlations between the labor-specific transcriptomic changes taking place in the maternal circulation and those previously reported in the chorioamniotic membranes [59], myometrium [60], and cervix [61]. Activation of the chorioamniotic membranes/decidua is a component of the common pathway of parturition [2–7, 9, 87], which is characterized by the influx of maternal immune cells that promote the physiologic inflammation required for labor [15, 39, 42, 47, 88]. A prior whole tissue microarray analysis of the chorioamniotic membranes revealed upregulated expression of genes that were enriched for processes such as “taxis” and “chemokine activity, cytokine activity or cytokine binding,” demonstrating an acute inflammatory signature specific to the process of labor [59]. These observations are consistent with a more recent microarray-based investigation of choriodecidual leukocytes from women with labor at term, which revealed that infiltrating immune cells displayed upregulated expression of gene networks associated with cell migration and chemotaxis, among others [89]. Given that the process of labor has been proposed to involve the recruitment of circulating immune cells to the gestational and reproductive tissues [15, 39, 42, 47, 88, 90], the above evidence provides a plausible explanation for the observed correlation between the chorioamniotic membrane transcriptome and that of the maternal peripheral blood reported herein.
The uterus, which includes the myometrium, is a contractile organ and key participant in the process of labor [2, 5–7]. Given its important functional role in delivery of the fetus, multiple investigations have explored the changes in gene expression that occur in this tissue using microarrays [60, 91–94], RNA-seq [95, 96], and epigenetic surveys [97]. Similar to the transcriptomic signatures observed in gestational tissues (e.g., the chorioamniotic membranes), enrichment of processes related to immune and inflammatory responses have been reported in the myometrium, such as cytokine and chemokine signaling pathways [60, 94, 95]. Multiple meta-analyses have further established the inflammatory nature of labor-specific changes in the myometrium [94, 98, 99]. Indeed, the latter study demonstrated high-confidence overlap of myometrial transcriptomic changes identified in multiple study populations, providing a labor-specific signature that included known components, such as cytokines as well as novel transcripts related to apoptosis and cell proliferation/differentiation [99]. Consistently, in this study, we identified a significant correlation between previously reported labor-specific gene expression changes in the myometrium and those found in the maternal blood herein, which included immune- and inflammation-related processes. Thus, given the impracticality of sampling the myometrium during pregnancy, the maternal peripheral blood transcriptome may represent a potential “liquid biopsy” to evaluate the changes occurring in the myometrium and other critical gestational and reproductive tissues.
The uterine cervix is the gatekeeper of pregnancy [100] and plays a key role in the maintenance of the conceptus in utero. The cervix evolves from a rigid and closed to a soft and dilated structure through the process of cervical remodeling [100–109]. Such a process occurs throughout pregnancy and culminates in cervical dilation during labor, thereby permitting the delivery of the neonate [100–107, 109]. During labor, multiple cellular and molecular mechanisms take place in the cervix [12, 14, 26, 30, 33, 34, 100, 104, 110], which include inflammatory processes as indicated by the upregulation of immune- and inflammation-related transcripts in this tissue [61, 111]. Notably, given that the transcripts upregulated in the ripened cervices of women at term without labor do not involve inflammatory processes [112], it is tempting to suggest that the process of labor itself shifts the cervical transcriptome from a non-inflammatory to an inflammatory status. This concept is consistent with the observed correlation between the labor-specific transcriptomic changes taking place in the cervix and those detected in the maternal circulation, as reported herein. Therefore, these findings highlight the potential use of transcriptomic signatures monitored in the peripheral blood as biomarkers to evaluate changes occurring in reproductive tissues such as the cervix.
When studying the mechanisms of parturition, it is tempting to hypothesize that preterm labor is merely the premature initiation of normal labor and that these two processes are otherwise similar. However, a growing body of evidence has demonstrated that preterm labor is a syndrome of multiple etiologies involving pathological processes that culminate in the activation of one or more components of the common pathway of labor [4, 6, 7]. In the current study, we found that 86% of the 402 RNAs dysregulated prior to diagnosis with PPROM in asymptomatic women also change in the same direction as it occurs in physiologic term labor. This result is even more striking when considering the fact that the PPROM signature was derived from two diverse cohorts (Detroit, MI, USA, and Calgary, Alberta, Canada), while the changes with term labor reported herein were derived from the same Detroit cohort. Moreover, one-half of the biological processes associated with gene expression changes in the peripheral blood of women with spontaneous labor at term were also significant in the PPROM signature. Differences in the enrichment analysis results for the PPROM signature (402 genes derived from two cohorts including the one in the current study) and the term-labor signature (103 genes derived from this study cohort) are driven by differences between phenotypes, unavoidable cohort differences, and the higher power for enrichment of the larger PPROM signature. Indeed, when interaction tests were used to assess if differences with term labor were different from those detected prior to diagnosis with PPROM using only data from the cohort in this study, we found term labor-specific changes but almost no PPROM-specific changes.
Our findings are consistent with a previous microarray study showing different tissue-specific transcriptomic profiles in women who delivered term or preterm in the presence or absence of labor [113]. The authors of the study suggested that pregnancy is maintained by the downregulation of chemokines at the maternal-fetal interface and that withdrawal of this downregulation leads to term birth, whereas the pathological activation of multiple inflammation-related pathways culminates in some cases of preterm birth [113], of which the latter concept has also been demonstrated in mice [114, 115]. In line with this concept, we recently utilized scRNA-seq to decipher the cellular landscape of human parturition in the chorioamniotic membranes and placental tissues and demonstrated that parturition-associated single-cell signatures can be monitored in the peripheral blood [116]. Notably, while specific single-cell signatures enriched in the peripheral blood of women with term labor were also observed in women with preterm labor (e.g., T-cell activation), other signatures were only enriched in either preterm labor (e.g., monocytes and macrophages) or term labor (NK cells), further emphasizing the differences between these two processes.
Collectively, these results indicate that labor is characterized by a transcriptomic signature in the maternal peripheral blood, which correlates with term labor-specific changes taking place in the chorioamniotic membranes, myometrium, and cervix. Importantly, the transcriptomic activity of several term labor-specific processes and pathways was also enriched in women who ultimately were diagnosed with PPROM and delivered preterm, demonstrating further the potential utility of these signatures as non-invasive disease biomarkers. Yet, further investigation is required to clinically validate such an approach.
Supplementary Material
Footnotes
† Grant Support: This research was supported, in part, by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), and, in part, with Federal funds from NICHD/NIH/DHHS under Contract No. HHSN275201300006C. RR has contributed to this work as part of his official duties as an employee of the United States Federal Government. NG-L and ALT were also supported by the Wayne State University Perinatal Initiative in Maternal, Perinatal and Child Health. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Nardhy Gomez-Lopez, Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Bethesda, MD, and Detroit, MI, USA; Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, USA; Department of Biochemistry, Microbiology, and Immunology, Wayne State University School of Medicine, Detroit, MI, USA.
Roberto Romero, Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Bethesda, MD, and Detroit, MI, USA; Department of Obstetrics and Gynecology, University of Michigan, Ann Arbor, MI, USA; Department of Epidemiology and Biostatistics, Michigan State University, East Lansing, MI, USA; Center for Molecular Medicine and Genetics, Wayne State University, Detroit, MI, USA; Detroit Medical Center, Detroit, MI, USA.
Jose Galaz, Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Bethesda, MD, and Detroit, MI, USA; Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, USA.
Gaurav Bhatti, Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Bethesda, MD, and Detroit, MI, USA; Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, USA.
Bogdan Done, Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Bethesda, MD, and Detroit, MI, USA; Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, USA.
Derek Miller, Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Bethesda, MD, and Detroit, MI, USA; Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, USA.
Corina Ghita, Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Bethesda, MD, and Detroit, MI, USA.
Kenichiro Motomura, Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Bethesda, MD, and Detroit, MI, USA; Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, USA.
Marcelo Farias-Jofre, Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Bethesda, MD, and Detroit, MI, USA; Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, USA.
Eunjung Jung, Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Bethesda, MD, and Detroit, MI, USA; Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, USA.
Roger Pique-Regi, Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Bethesda, MD, and Detroit, MI, USA; Center for Molecular Medicine and Genetics, Wayne State University, Detroit, MI, USA.
Sonia S Hassan, Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, USA; Office of Women’s Health, Integrative Biosciences Center, Wayne State University, Detroit, MI, USA; Department of Physiology, Wayne State University School of Medicine, Detroit, MI, USA.
Tinnakorn Chaiworapongsa, Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Bethesda, MD, and Detroit, MI, USA; Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, USA.
Adi L Tarca, Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS), Bethesda, MD, and Detroit, MI, USA; Department of Obstetrics and Gynecology, Wayne State University School of Medicine, Detroit, MI, USA; Department of Computer Science, Wayne State University College of Engineering, Detroit, MI, USA.
Data availability
The transcriptomic dataset utilized in this study was previously deposited by the authors in the Gene Expression Omnibus super-series GSE149440.
Authors’ contributions
A.L.T., R.R., and N.G-L. conceived and designed the study. N.G-L., A.L.T., R.R., J.G., D.M., and M.F-J. wrote the manuscript. A.L.T., G.B., B.D., C.G., and R.P-R. analyzed the data. K.M., E.J., S.S.H., and T.C. provided intellectual input. All authors approved the final version of the manuscript.
Conflict of interest
The authors have declared that no conflict of interest exists.
References
- 1. McLean M, Bisits A, Davies J, Woods R, Lowry P, Smith R. A placental clock controlling the length of human pregnancy. Nat Med 1995; 1:460–463. [DOI] [PubMed] [Google Scholar]
- 2. Smith R. Parturition. N Engl J Med 2007; 356:271–283. [DOI] [PubMed] [Google Scholar]
- 3. Cunningham FG, Leveno KJ, Bloom SL, Dashe JS, Hoffman BL, Casey BM, Spong CY. Physiology of labor. In: Williams Obstetrics, 25th ed. New York: McGraw-Hill Education; 2018. [Google Scholar]
- 4. Romero R, Mazor M, Munoz H, Gomez R, Galasso M, Sherer DM. The preterm labor syndrome. Ann N Y Acad Sci 1994; 734:414–429. [DOI] [PubMed] [Google Scholar]
- 5. Norwitz ER, Robinson JN, Challis JR. The control of labor. N Engl J Med 1999; 341:660–666. [DOI] [PubMed] [Google Scholar]
- 6. Romero R, Espinoza J, Kusanovic JP, Gotsch F, Hassan S, Erez O, Chaiworapongsa T, Mazor M. The preterm parturition syndrome. BJOG 2006; 113:17–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science 2014; 345:760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rosenbloom JI, Woolfolk CL, Wan L, Stout MJ, Tuuli MG, Macones GA, Cahill AG. The transition from latent to active labor and adverse obstetrical outcomes. Am J Obstet Gynecol 2019; 221:487.e481–487.e488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lopez BA. Mechanisms of labour—biochemical aspects. BJOG 2003; 110:39–45. [DOI] [PubMed] [Google Scholar]
- 10. Romero R, Nores J, Mazor M, Sepulveda W, Oyarzun E, Parra M, Insunza A, Montiel F, Behnke E, Cassell GH. Microbial invasion of the amniotic cavity during term labor. Prevalence and clinical significance. J Reprod Med 1993; 38:543–548. [PubMed] [Google Scholar]
- 11. Seong HS, Lee SE, Kang JH, Romero R, Yoon BH. The frequency of microbial invasion of the amniotic cavity and histologic chorioamnionitis in women at term with intact membranes in the presence or absence of labor. Am J Obstet Gynecol 2008; 199:e371–e375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mackler AM, Iezza G, Akin MR, McMillan P, Yellon SM. Macrophage trafficking in the uterus and cervix precedes parturition in the mouse. Biol Reprod 1999; 61:879–883. [DOI] [PubMed] [Google Scholar]
- 13. Thomson AJ, Telfer JF, Young A, Campbell S, Stewart CJ, Cameron IT, Greer IA, Norman JE. Leukocytes infiltrate the myometrium during human parturition: further evidence that labour is an inflammatory process. Hum Reprod 1999; 14:229–236. [PubMed] [Google Scholar]
- 14. Young A, Thomson AJ, Ledingham M, Jordan F, Greer IA, Norman JE. Immunolocalization of proinflammatory cytokines in myometrium, cervix, and fetal membranes during human parturition at term. Biol Reprod 2002; 66:445–449. [DOI] [PubMed] [Google Scholar]
- 15. Osman I, Young A, Ledingham MA, Thomson AJ, Jordan F, Greer IA, Norman JE. Leukocyte density and pro-inflammatory cytokine expression in human fetal membranes, decidua, cervix and myometrium before and during labour at term. Mol Hum Reprod 2003; 9:41–45. [DOI] [PubMed] [Google Scholar]
- 16. Yellon SM, Mackler AM, Kirby MA. The role of leukocyte traffic and activation in parturition. J Soc Gynecol Investig 2003; 10:323–338. [DOI] [PubMed] [Google Scholar]
- 17. Shynlova O, Tsui P, Dorogin A, Lye SJ. Monocyte chemoattractant protein-1 (CCL-2) integrates mechanical and endocrine signals that mediate term and preterm labor. J Immunol 2008; 181:1470–1479. [DOI] [PubMed] [Google Scholar]
- 18. Shynlova O, Tsui P, Jaffer S, Lye SJ. Integration of endocrine and mechanical signals in the regulation of myometrial functions during pregnancy and labour. Eur J Obstet Gynecol Reprod Biol 2009; 144:S2–S10. [DOI] [PubMed] [Google Scholar]
- 19. Shynlova O, Lee YH, Srikhajon K, Lye SJ. Physiologic uterine inflammation and labor onset: integration of endocrine and mechanical signals. Reprod Sci 2013; 20:154–167. [DOI] [PubMed] [Google Scholar]
- 20. Lombardi A, Makieva S, Rinaldi SF, Arcuri F, Petraglia F, Norman JE. Expression of matrix metalloproteinases in the mouse uterus and human myometrium during pregnancy, labor, and preterm labor. Reprod Sci 2018; 25:938–949. [DOI] [PubMed] [Google Scholar]
- 21. Ulrich CC, Arinze V, Wandscheer CB, Copley Salem C, Nabati C, Etezadi-Amoli N, Burkin HR. Matrix metalloproteinases 2 and 9 are elevated in human preterm laboring uterine myometrium and exacerbate uterine contractility. Biol Reprod 2019; 100:1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leimert KB, Messer A, Gray T, Fang X, Chemtob S, Olson DM. Maternal and fetal intrauterine tissue crosstalk promotes proinflammatory amplification and uterine transition. Biol Reprod 2019; 100:783–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leimert KB, Verstraeten BSE, Messer A, Nemati R, Blackadar K, Fang X, Robertson SA, Chemtob S, Olson DM. Cooperative effects of sequential PGF2alpha and IL-1beta on IL-6 and COX-2 expression in human myometrial cells. Biol Reprod 2019; 100:1370–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wendremaire M, Hadi T, Pezze M, Barrichon M, Lopez T, Neiers F, Sagot P, Garrido C, Lirussi F. Macrophage-induced reactive oxygen species promote myometrial contraction and labor-associated mechanisms. Biol Reprod 2020; 102:1326–1339. [DOI] [PubMed] [Google Scholar]
- 25. Liggins CG. Cervical ripening as an inflammatory reaction. In: Elwood DA, Andersson ABM (eds.), (eds.)Cervix in Pregnancy and Labour. Edinburgh: Churchill Livingstone; 1981: 1–9. [Google Scholar]
- 26. Bokström H, Brännström M, Alexandersson M, Norström A. Leukocyte subpopulations in the human uterine cervical stroma at early and term pregnancy. Hum Reprod 1997; 12:586–590. [DOI] [PubMed] [Google Scholar]
- 27. Kelly RW. Inflammatory mediators and cervical ripening. J Reprod Immunol 2002; 57:217–224. [DOI] [PubMed] [Google Scholar]
- 28. Sakamoto Y, Moran P, Bulmer JN, Searle RF, Robson SC. Macrophages and not granulocytes are involved in cervical ripening. J Reprod Immunol 2005; 66:161–173. [DOI] [PubMed] [Google Scholar]
- 29. Yellon SM, Ebner CA, Sugimoto Y. Parturition and recruitment of macrophages in cervix of mice lacking the prostaglandin F receptor. Biol Reprod 2008; 78:438–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Timmons BC, Fairhurst AM, Mahendroo MS. Temporal changes in myeloid cells in the cervix during pregnancy and parturition. J Immunol 2009; 182:2700–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yellon SM, Oshiro BT, Chhaya TY, Lechuga TJ, Dias RM, Burns AE, Force L, Apostolakis EM. Remodeling of the cervix and parturition in mice lacking the progesterone receptor B isoform. Biol Reprod 2011; 85:498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clyde LA, Lechuga TJ, Ebner CA, Burns AE, Kirby MA, Yellon SM. Transection of the pelvic or vagus nerve forestalls ripening of the cervix and delays birth in rats. Biol Reprod 2011; 84:587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Payne KJ, Clyde LA, Weldon AJ, Milford TA, Yellon SM. Residency and activation of myeloid cells during remodeling of the prepartum murine cervix. Biol Reprod 2012; 87:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Myers DA. The recruitment and activation of leukocytes into the immune cervix: further support that cervical remodeling involves an immune and inflammatory mechanism. Biol Reprod 2012; 87:107. [DOI] [PubMed] [Google Scholar]
- 35. Vince GS, Starkey PM, Jackson MC, Sargent IL, Redman CW. Flow cytometric characterisation of cell populations in human pregnancy decidua and isolation of decidual macrophages. J Immunol Methods 1990; 132:181–189. [DOI] [PubMed] [Google Scholar]
- 36. Fidel PL Jr, Romero R, Ramirez M, Cutright J, Edwin SS, LaMarche S, Cotton DB, Mitchell MD. Interleukin-1 receptor antagonist (IL-1ra) production by human amnion, chorion, and decidua. Am J Reprod Immunol 1994; 32:1–7. [DOI] [PubMed] [Google Scholar]
- 37. Keelan JA, Marvin KW, Sato TA, Coleman M, McCowan LM, Mitchell MD. Cytokine abundance in placental tissues: evidence of inflammatory activation in gestational membranes with term and preterm parturition. Am J Obstet Gynecol 1999; 181:1530–1536. [DOI] [PubMed] [Google Scholar]
- 38. Keski-Nisula L, Aalto ML, Katila ML, Kirkinen P. Intrauterine inflammation at term: a histopathologic study. Hum Pathol 2000; 31:841–846. [DOI] [PubMed] [Google Scholar]
- 39. Gomez-Lopez N, Estrada-Gutierrez G, Jimenez-Zamudio L, Vega-Sanchez R, Vadillo-Ortega F. Fetal membranes exhibit selective leukocyte chemotaxic activity during human labor. J Reprod Immunol 2009; 80:122–131. [DOI] [PubMed] [Google Scholar]
- 40. Gomez-Lopez N, Guilbert LJ, Olson DM. Invasion of the leukocytes into the fetal-maternal interface during pregnancy. J Leukoc Biol 2010; 88:625–633. [DOI] [PubMed] [Google Scholar]
- 41. Hamilton S, Oomomian Y, Stephen G, Shynlova O, Tower CL, Garrod A, Lye SJ, Jones RL. Macrophages infiltrate the human and rat decidua during term and preterm labor: evidence that decidual inflammation precedes labor. Biol Reprod 2012; 86:39. [DOI] [PubMed] [Google Scholar]
- 42. Gomez-Lopez N, Vega-Sanchez R, Castillo-Castrejon M, Romero R, Cubeiro-Arreola K, Vadillo-Ortega F. Evidence for a role for the adaptive immune response in human term parturition. Am J Reprod Immunol 2013; 69:212–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hamilton SA, Tower CL, Jones RL. Identification of chemokines associated with the recruitment of decidual leukocytes in human labour: potential novel targets for preterm labour. PLoS One 2013; 8:e56946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Castillo-Castrejon M, Meraz-Cruz N, Gomez-Lopez N, Flores-Pliego A, Beltran-Montoya J, Viveros-Alcaraz M, Vadillo-Ortega F. Choriodecidual cells from term human pregnancies show distinctive functional properties related to the induction of labor. Am J Reprod Immunol 2014; 71:86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lonergan M, Aponso D, Marvin KW, Helliwell RJ, Sato TA, Mitchell MD, Chaiwaropongsa T, Romero R, Keelan JA. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), TRAIL receptors, and the soluble receptor osteoprotegerin in human gestational membranes and amniotic fluid during pregnancy and labor at term and preterm. J Clin Endocrinol Metab 2003; 88:3835–3844. [DOI] [PubMed] [Google Scholar]
- 46. Esplin MS, Peltier MR, Hamblin S, Smith S, Fausett MB, Dildy GA, Branch DW, Silver RM, Adashi EY. Monocyte chemotactic protein-1 expression is increased in human gestational tissues during term and preterm labor. Placenta 2005; 26:661–671. [DOI] [PubMed] [Google Scholar]
- 47. Gomez-Lopez N, Vadillo-Perez L, Hernandez-Carbajal A, Godines-Enriquez M, Olson DM, Vadillo-Ortega F. Specific inflammatory microenvironments in the zones of the fetal membranes at term delivery. Am J Obstet Gynecol 2011; 205:e215–e224. [DOI] [PubMed] [Google Scholar]
- 48. Gomez-Lopez N, Vadillo-Perez L, Nessim S, Olson DM, Vadillo-Ortega F. Choriodecidua and amnion exhibit selective leukocyte chemotaxis during term human labor. Am J Obstet Gynecol 2011; 204:e369–e316. [DOI] [PubMed] [Google Scholar]
- 49. Lozovyy V, Richardson L, Saade G, Menon R. Progesterone receptor membrane components: key regulators of fetal membrane integrity. Biol Reprod 2021; 104:445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koh W, Pan W, Gawad C, Fan HC, Kerchner GA, Wyss-Coray T, Blumenfeld YJ, El-Sayed YY, Quake SR. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci U S A 2014; 111:7361–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tsui NB, Jiang P, Wong YF, Leung TY, Chan KC, Chiu RW, Sun H, Lo YM. Maternal plasma RNA sequencing for genome-wide transcriptomic profiling and identification of pregnancy-associated transcripts. Clin Chem 2014; 60:954–962. [DOI] [PubMed] [Google Scholar]
- 52. Liang L, Rasmussen MH, Piening B, Shen X, Chen S, Rost H, Snyder JK, Tibshirani R, Skotte L, Lee NC, Contrepois K, Feenstra Bet al. Metabolic dynamics and prediction of gestational age and time to delivery in pregnant women. Cell 2020; 181:1680–1692.e1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aghaeepour N, Ganio EA, McIlwain D, Tsai AS, Tingle M, Van Gassen S, Gaudilliere DK, Baca Q, McNeil L, Okada R, Ghaemi MS, Furman Det al. An immune clock of human pregnancy. Sci Immunol 2017; 2:eaan2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ghaemi MS, DiGiulio DB, Contrepois K, Callahan B, Ngo TTM, Lee-McMullen B, Lehallier B, Robaczewska A, McIlwain D, Rosenberg-Hasson Y, Wong RJ, Quaintance Cet al. Multiomics modeling of the immunome, transcriptome, microbiome, proteome and metabolome adaptations during human pregnancy. Bioinformatics 2019; 35:95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ngo TTM, Moufarrej MN, Rasmussen MH, Camunas-Soler J, Pan W, Okamoto J, Neff NF, Liu K, Wong RJ, Downes K, Tibshirani R, Shaw GMet al. Noninvasive blood tests for fetal development predict gestational age and preterm delivery. Science 2018; 360:1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tarca AL, Romero R, Xu Z, Gomez-Lopez N, Erez O, Hsu CD, Hassan SS, Carey VJ. Targeted expression profiling by RNA-Seq improves detection of cellular dynamics during pregnancy and identifies a role for T cells in term parturition. Sci Rep 2019; 9:848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stelzer IA, Ghaemi MS, Han X, Ando K, Hedou JJ, Feyaerts D, Peterson LS, Rumer KK, Tsai ES, Ganio EA, Gaudilliere DK, Tsai ASet al. Integrated trajectories of the maternal metabolome, proteome, and immunome predict labor onset. Sci Transl Med 2021; 13:eabd9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tarca AL, Pataki BÁ, Romero R, Sirota M, Guan Y, Kutum R, Gomez-Lopez N, Done B, Bhatti G, Yu T, Andreoletti G, Chaiworapongsa Tet al. Crowdsourcing assessment of maternal blood multi-omics for predicting gestational age and preterm birth. Cell Reports Medicine 2021; 2:100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Haddad R, Tromp G, Kuivaniemi H, Chaiworapongsa T, Kim YM, Mazor M, Romero R. Human spontaneous labor without histologic chorioamnionitis is characterized by an acute inflammation gene expression signature. Am J Obstet Gynecol 2006; 195:394:e391–e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mittal P, Romero R, Tarca AL, Gonzalez J, Draghici S, Xu Y, Dong Z, Nhan-Chang CL, Chaiworapongsa T, Lye S, Kusanovic JP, Lipovich Let al. Characterization of the myometrial transcriptome and biological pathways of spontaneous human labor at term. J Perinat Med 2010; 38:617–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hassan SS, Romero R, Haddad R, Hendler I, Khalek N, Tromp G, Diamond MP, Sorokin Y, Malone J Jr. The transcriptome of the uterine cervix before and after spontaneous term parturition. Am J Obstet Gynecol 2006; 195:778–786. [DOI] [PubMed] [Google Scholar]
- 62. Gomez-Lopez N, Romero R, Hassan SS, Bhatti G, Berry SM, Kusanovic JP, Pacora P, Tarca AL. The cellular transcriptome in the maternal circulation during normal pregnancy: a longitudinal study. Front Immunol 2019; 10:2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4:249–264. [DOI] [PubMed] [Google Scholar]
- 64. Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010; 26:2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S (eds.), Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; 2012: 397–420. [Google Scholar]
- 66. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn Tet al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics 2007; 23:257–258. [DOI] [PubMed] [Google Scholar]
- 69. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DPet al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the pathway interaction database. Nucleic Acids Res 2009; 37:D674–D679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 1999; 27:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, Sidiropoulos K, Cook J, Gillespie M, Haw R, Loney F, May Bet al. The reactome pathway knowledgebase. Nucleic Acids Res 2020; 48:D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Martens M, Ammar A, Riutta A, Waagmeester A, Slenter DN, Hanspers K, Miller RA, Digles D, Lopes EN, Ehrhart F, Dupuis LJ, Winckers LAet al. WikiPathways: connecting communities. Nucleic Acids Res 2021; 49:D613–D621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res 2019; 18:623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Otasek D, Morris JH, Boucas J, Pico AR, Demchak B. Cytoscape automation: empowering workflow-based network analysis. Genome Biol 2019; 20:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet 2008; 371:75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sacks GP, Studena K, Sargent K, Redman CW. Normal pregnancy and preeclampsia both produce inflammatory changes in peripheral blood leukocytes akin to those of sepsis. Am J Obstet Gynecol 1998; 179:80–86. [DOI] [PubMed] [Google Scholar]
- 78. Naccasha N, Gervasi MT, Chaiworapongsa T, Berman S, Yoon BH, Maymon E, Romero R. Phenotypic and metabolic characteristics of monocytes and granulocytes in normal pregnancy and maternal infection. Am J Obstet Gynecol 2001; 185:1118–1123. [DOI] [PubMed] [Google Scholar]
- 79. Bardou M, Hadi T, Mace G, Pesant M, Debermont J, Barrichon M, Wendremaire M, Laurent N, Sagot P, Lirussi F. Systemic increase in human maternal circulating CD14+CD16- MCP-1+ monocytes as a marker of labor. Am J Obstet Gynecol 2014; 210:e71–e79. [DOI] [PubMed] [Google Scholar]
- 80. Zhang J, Shynlova O, Sabra S, Bang A, Briollais L, Lye SJ. Immunophenotyping and activation status of maternal peripheral blood leukocytes during pregnancy and labour, both term and preterm. J Cell Mol Med 2017; 21:2386–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Unal ER, Cierny JT, Roedner C, Newman R, Goetzl L. Maternal inflammation in spontaneous term labor. Am J Obstet Gynecol 2011; 204:e221–e225. [DOI] [PubMed] [Google Scholar]
- 82. Cierny JT, Unal ER, Flood P, Rhee KY, Praktish A, Olson TH, Goetzl L. Maternal inflammatory markers and term labor performance. Am J Obstet Gynecol 2014; 210:e441–e446. [DOI] [PubMed] [Google Scholar]
- 83. Neal JL, Lamp JM, Lowe NK, Gillespie SL, Sinnott LT, McCarthy DO. Differences in inflammatory markers between nulliparous women admitted to hospitals in preactive vs active labor. Am J Obstet Gynecol 2015; 212:e61–e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ariga H, Ohto H, Busch MP, Imamura S, Watson R, Reed W, Lee TH. Kinetics of fetal cellular and cell-free DNA in the maternal circulation during and after pregnancy: implications for noninvasive prenatal diagnosis. Transfusion 2001; 41:1524–1530. [DOI] [PubMed] [Google Scholar]
- 85. Herrera CA, Stoerker J, Carlquist J, Stoddard GJ, Jackson M, Esplin S, Rose NC. Cell-free DNA, inflammation, and the initiation of spontaneous term labor. Am J Obstet Gynecol 2017; 217:583.e581–583.e588. [DOI] [PubMed] [Google Scholar]
- 86. Gomez-Lopez N, Romero R, Schwenkel G, Garcia-Flores V, Panaitescu B, Varrey A, Ayoub F, Hassan SS, Phillippe M. Cell-free fetal DNA increases prior to labor at term and in a subset of preterm births. Reprod Sci 2020; 27:218–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Harris LK, Benagiano M, D'Elios MM, Brosens I, Benagiano G. Placental bed research: II. Functional and immunological investigations of the placental bed. Am J Obstet Gynecol 2019; 221:457–469. [DOI] [PubMed] [Google Scholar]
- 88. Osman I, Young A, Jordan F, Greer IA, Norman JE. Leukocyte density and proinflammatory mediator expression in regional human fetal membranes and decidua before and during labor at term. J Soc Gynecol Investig 2006; 13:97–103. [DOI] [PubMed] [Google Scholar]
- 89. Arenas-Hernandez M, Gomez-Lopez N, Garcia-Flores V, Rangel-Escareno C, Alvarez-Salas LM, Martinez-Acuna N, Vazquez-Perez JA, Vega-Sanchez R. Choriodecidual leukocytes display a unique gene expression signature in spontaneous labor at term. Genes Immun 2019; 20:56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gomez-Lopez N, Olson D. (inventors). The Governors of the University of Alberta, assignee. Leukocyte activation and methods of use thereof. United States patent US9880152B2.
- 91. Esplin MS, Fausett MB, Peltier MR, Hamblin S, Silver RM, Branch DW, Adashi EY, Whiting D. The use of cDNA microarray to identify differentially expressed labor-associated genes within the human myometrium during labor. Am J Obstet Gynecol 2005; 193:404–413. [DOI] [PubMed] [Google Scholar]
- 92. Mittal P, Romero R, Tarca AL, Draghici S, Nhan-Chang CL, Chaiworapongsa T, Hotra J, Gomez R, Kusanovic JP, Lee DC, Kim CJ, Hassan SS. A molecular signature of an arrest of descent in human parturition. Am J Obstet Gynecol 2011; 204:e115–e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chaemsaithong P, Madan I, Romero R, Than NG, Tarca AL, Draghici S, Bhatti G, Yeo L, Mazor M, Kim CJ, Hassan SS, Chaiworapongsa T. Characterization of the myometrial transcriptome in women with an arrest of dilatation during labor. J Perinat Med 2013; 41:665–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sharp GC, Hutchinson JL, Hibbert N, Freeman TC, Saunders PT, Norman JE. Transcription analysis of the myometrium of labouring and non-labouring women. PLoS One 2016; 11:e0155413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chan YW, Berg HA, Moore JD, Quenby S, Blanks AM. Assessment of myometrial transcriptome changes associated with spontaneous human labour by high-throughput RNA-seq. Exp Physiol 2014; 99:510–524. [DOI] [PubMed] [Google Scholar]
- 96. Stanfield Z, Amini P, Wang J, Yi L, Tan H, Chance MR, Koyuturk M, Mesiano S. Interplay of transcriptional signaling by progesterone, cyclic AMP, and inflammation in myometrial cells: implications for the control of human parturition. Mol Hum Reprod 2019; 25:408–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Mitsuya K, Singh N, Sooranna SR, Johnson MR, Myatt L. Epigenetics of human myometrium: DNA methylation of genes encoding contraction-associated proteins in term and preterm labor. Biol Reprod 2014; 90:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lui S, Duval C, Farrokhnia F, Girard S, Harris LK, Tower CL, Stevens A, Jones RL. Delineating differential regulatory signatures of the human transcriptome in the choriodecidua and myometrium at term labor. Biol Reprod 2018; 98:422–436. [DOI] [PubMed] [Google Scholar]
- 99. Stanfield Z, Lai PF, Lei K, Johnson MR, Blanks AM, Romero R, Chance MR, Mesiano S, Koyuturk M. Myometrial transcriptional signatures of human parturition. Front Genet 2019; 10:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yellon SM. Immunobiology of cervix ripening. Front Immunol 2019; 10:3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Leppert PC. Anatomy and physiology of cervical ripening. Clin Obstet Gynecol 1995; 38:267–279. [DOI] [PubMed] [Google Scholar]
- 102. Word RA, Li XH, Hnat M, Carrick K. Dynamics of cervical remodeling during pregnancy and parturition: mechanisms and current concepts. Semin Reprod Med 2007; 25:69–79. [DOI] [PubMed] [Google Scholar]
- 103. Norman JE. Preterm labour. Cervical function and prematurity. Best Pract Res Clin Obstet Gynaecol 2007; 21:791–806. [DOI] [PubMed] [Google Scholar]
- 104. Timmons B, Akins M, Mahendroo M. Cervical remodeling during pregnancy and parturition. Trends Endocrinol Metab 2010; 21:353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Yellon SM. Contributions to the dynamics of cervix remodeling prior to term and preterm birth. Biol Reprod 2017; 96:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Yoshida K, Jayyosi C, Lee N, Mahendroo M, Myers KM. Mechanics of cervical remodelling: insights from rodent models of pregnancy. Interface Focus 2019; 9:20190026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Yellon SM, Greaves E, Heuerman AC, Dobyns AE, Norman JE. Effects of macrophage depletion on characteristics of cervix remodeling and pregnancy in CD11b-dtr mice. Biol Reprod 2019; 100:1386–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Qi W, Zhao P, Sun Z, Ma X, Wang H, Wu W, Wen Z, Kisrieva-Ware Z, Woodard PK, Wang Q, McKinstry RC, Cahill AGet al. Magnetic resonance diffusion tensor imaging of cervical microstructure in normal early and late pregnancy in vivo. Am J Obstet Gynecol 2021; 224:101.e101–101.e111. [DOI] [PubMed] [Google Scholar]
- 109. Chatterjee A, Saghian R, Dorogin A, Cahill LS, Sled JG, Lye S, Shynlova O. Combination of histochemical analyses and micro-MRI reveals regional changes of the murine cervix in preparation for labor. Sci Rep 2021; 11:4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yang J, Lai Y, Chen J, Lin B, Zhou B, Han X. Changes in alpha-7 nicotinic acetylcholine receptor and macrophage polarization state participate in the regulation of cervical remodeling in pregnant rats. Biol Reprod 2019; 101:950–960. [DOI] [PubMed] [Google Scholar]
- 111. Hassan SS, Romero R, Tarca AL, Draghici S, Pineles B, Bugrim A, Khalek N, Camacho N, Mittal P, Yoon BH, Espinoza J, Kim CJet al. Signature pathways identified from gene expression profiles in the human uterine cervix before and after spontaneous term parturition. Am J Obstet Gynecol 2007; 197:e251–e257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hassan SS, Romero R, Tarca AL, Nhan-Chang CL, Vaisbuch E, Erez O, Mittal P, Kusanovic JP, Mazaki-Tovi S, Yeo L, Draghici S, Kim JSet al. The transcriptome of cervical ripening in human pregnancy before the onset of labor at term: identification of novel molecular functions involved in this process. J Matern Fetal Neonatal Med 2009; 22:1183–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bukowski R, Sadovsky Y, Goodarzi H, Zhang H, Biggio JR, Varner M, Parry S, Xiao F, Esplin SM, Andrews W, Saade GR, Ilekis JVet al. Onset of human preterm and term birth is related to unique inflammatory transcriptome profiles at the maternal fetal interface. Peer J 2017; 5:e3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Migale R, Mac Intyre DA, Cacciatore S, Lee YS, Hagberg H, Herbert BR, Johnson MR, Peebles D, Waddington SN, Bennett PR. Modeling hormonal and inflammatory contributions to preterm and term labor using uterine temporal transcriptomics. BMC Med 2016; 14:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Willcockson AR, Nandu T, Liu CL, Nallasamy S, Kraus WL, Mahendroo M. Transcriptome signature identifies distinct cervical pathways induced in lipopolysaccharide-mediated preterm birth. Biol Reprod 2018; 98:408–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Pique-Regi R, Romero R, Tarca AL, Sendler ED, Xu Y, Garcia-Flores V, Leng Y, Luca F, Hassan SS, Gomez-Lopez N. Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife 2019; 8:e52004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptomic dataset utilized in this study was previously deposited by the authors in the Gene Expression Omnibus super-series GSE149440.


