Abstract
Tumor dormancy is a stage in which residual cancer cells remain inactive, but regrowth of dormant cancer cells contributes to recurrence. The complex ecosystem in cancer that promotes cell survival and the factors that eventually overcome growth constraints and result in proliferation remain to be fully elucidated. Doing so may provide new insights and help identify novel strategies to prolong cancer dormancy and prevent disease recurrence. To dissect the molecular pathways and the microenvironments involved in regulation of dormancy, we utilized a novel immunocompetent transgenic model to study minimal residual disease and relapse. This model revealed a significant reorganization of cancer cell structures, stroma, and immune cells with cancer cells showing dormant cell signatures. Single-cell RNA sequencing uncovered remodeling of myeloid and lymphoid compartments. Additionally, the Jagged-1/Notch signaling pathway was shown to regulate many aspects of tumorigenesis, including stem cell development, epithelial-mesenchymal transition, and immune cell homeostasis during minimal residual disease. Treatment with an anti-Jagged-1 antibody inhibited the Jagged-1/Notch signaling pathway in tumor cells and the microenvironment, delaying tumor recurrence. These findings uncover a cascade of regulatory changes in the microenvironment during dormancy and identify a therapeutic strategy to undercut these changes.
Keywords: Minimal Residual Disease, Dormancy, Tumor Immune Microenvironment, Notch Signaling Pathway, Single Cell Analysis
Introduction
Tumor relapse and metastasis are a major cause of death in women with breast cancer (BC). By the time of primary tumor diagnosis, cancer may have already metastasized, and patients have either developed metastatic tumors or dormant disseminated tumor cells (DTCs). DTCs have been detected even at the early pre-invasive stages of tumor evolution (1). Tumor dormancy is a specific stage in which residual cells remain inactive and therapy resistant- possibly due to quiescence, niche protection, or both (2). Even after five years of adjuvant endocrine therapy, BC recurrence continues to occur steadily between 5–20 years and is strongly correlated with DTCs in lymph nodes and tumor grades (1).
The role of the microenvironment in regulating the survival and proliferation of residual cells following therapy is understudied and a very limited number of microenvironmental factors have been shown to promote dormancy. Microenvironment niches have been implicated in either a conversion to cells with characteristics of cancer stem cells (CSCs) or better survival of CSCs as compared to non-CSCs at the metastatic sites (3–7). The sprouting microvascular endothelial niche has been implicated in dormancy and survival of breast cancer cells (BCCs) at the metastatic sites and that inhibition of Notch signaling was shown to reduce neovascular tips, and therefore to suppress tumor cell outgrowth (8, 9). The majority of these studies used established cell lines. Various studies suggest that neutrophils awaken tumor cells from dormancy and facilitate therapeutic resistance. Neutrophils have been found in conjunction with circulating tumor cells (CTCs) in patient samples and exerted higher survival capabilities and induced proliferation of CTCs while in circulation (10). Neutrophils are a major component of the lung premetastatic niche (11) and awaken dormant cells in part by releasing neutrophil extracellular traps (NETs) and mediating microenvironment extra-cellular-matrix (ECM) remodeling (12).
A conditional mouse model for Her2/neu-driven mammary tumors has been used to demonstrate that following Her2 removal Notch signaling becomes activated in a subset of residual cells resulting in accelerated tumor recurrence (13). Furthermore, following Her2 removal, TNFα/NFκB signaling led to elevated CCL5 expression. CCL5 promoted tumor recurrence by recruiting CCR5-expressing macrophages, which contributed to collagen deposition in residual tumors (14). Inhibition of CCL5, however, was not sufficient to prolong the time to relapse (14). One limitation of these studies is that they were performed in immunocompromised hosts.
Here, we report generation of a Wnt/iFGFRGFP (iFGFRGFP) tumor model to study cell signaling and the tumor microenvironment during minimal residual disease (MRD) and recurrence. One advantage of this model is that these mice have an intact immune system and that the GFP reporter facilitates the isolation and characterization of the small number of residual cancer cells in a tolerized host. We generated four stages of tumorigenesis: primary, MRD (dormant), long-term dormant (long-term DT), and recurrent and performed single-cell RNA sequencing (scRNA-seq) and immuno-phenotyping to unveil the changes in microenvironment.
Materials and Methods
Animals, tumor transplant, and treatments
Sterile surgical procedure and mouse handling were in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the AAALAC accredited Baylor College of Medicine Institutional Animal Care and Use Committee (IACUC # AN-504).
MMTV-Wnt1/iFGRR1 (Wnt-iR1) mice were generated and characterized previously (15). The GFP mice (FVB.Cg-Tg(CAG-EGFP)B5Nagy/J- Stock No:003516), kindly provided by Dr. Michael Lewis (Baylor College of Medicine, Houston, TX), were bred with Wnt-iR1 bigenic mice to generate GFP+ tumors and tumor chunks were cryopreserved in 90%FBS+10%DMSO for future experiments. The GFP+ tumors were then transplanted into tolerized Glow-Head (GH) recipients (FVB.rGH-Luc-GFP) kindly provided by Dr. Chi-Ping Day (NIH/NCI) (16).
Tumor pieces 1–2 mm3 in size were surgically transplanted into the cleared fat-pad of 5-week-old GH mice. The fat-clearing and tumor transplantation technique were performed by modifying a procedure described previously (17). Briefly, instead of a traditional Y-shaped incision, a 4mm medial semi-circle incision was made around the 4th MG teat; e.g., for the left (viewer’s) gland cut the right side of the nipple. The skin was detached by a sterile cotton swab and the fat-pad was pulled out by a narrow-head forceps (WPI-500451, World Precision Instruments) holding the tissue close to the lymph node. This part of tissue contained the epithelial cells that were to be removed. As we cauterized the major vessels and finally the tissue containing the lymph node, we retracted out the gland. While still holding on to the tissue, the forceps was placed on the body outside the cut. A very small tumor piece was placed into the fat-pad. The tissue holding on to the forceps was finally cut and the wound was closed using 7mm clips. Pre- and post-surgery procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals.
One day after tumor transplantation, mice were injected intraperitoneally (i.p) with the B/B homodimerizer (dimerizer- Clontech) every 3 days until tumors reached about 4–5mm in diameter. At that point tumors grew independently of the dimerizer. Once tumors reached about 7–9mm in diameter, mice were treated with the FGFR inhibitor NVP-BGJ398 (BGJ) (18) for 14 days by oral gavage (50–810-47, Fisher Scientific) and then drug treatment was stopped for another 14 days. At this timepoint, depending on the experiment, mice were: 1) sacrificed for residual tissue collection, or 2) left untreated for recurrence, or 3) treated with the α-Jagged-1(Jag1) blocking antibodies kindly provided by Dr. Christian Siebel (Genentech, San Francisco, CA), or α-IgG2b (BE0086, BioXcell).
Mice were treated with α-Jag1 or IgG control (15mg/kg) once a week for 5 weeks and then treatment was stopped. Tumors were measured by a digital caliper and once they reached 5mm in diameter, they were designated as “recurrent”.
Tissue harvest and generation of single cells
Before tissue harvest, each mouse was perfused by 20ml of Heparin/PBS solution (10 U/ml Heparin in PBS) using the infusion pump (14–831-200, Fisher Scientific) set to 4 ml/min. Small pieces of tumor, lung, liver, and spleen were stored in 10% formalin for histopathology and the remainder of the tumors was immediately prepped for single cell isolation using the gentleMACS Dissociators and mouse Tumor Dissociation kit (130–096-730, Miltenyi Biotec) for 30 min. Red blood cell lysis was performed for 5 min (555899, BD Pharm Lyse) and after washing cells in PBS+2%FBS, cells were frozen in 90%FBS+10% DMSO for future experiments.
Cytokine Array
Live single cells that were frozen in 90%FBS+10%DMSO described in the tissue harvest section were thawed and pooled to perform the Mouse Cytokine Antibody Array (ab133993, Abcam), according to the manufacture’s protocol. Single cells were lysed and 250μg protein were used on each membrane. This assay only measured the intra-cellular cytokines as the excreted cytokines were washed off during the single cell prep. Membranes were developed by SuperSignal West Femto (34095, Thermo Scientific) and scanned on Amersham Imager 600 imagers (29083463, GE). Image was quantified by ImageJ.
Antibodies for immunofluorescence (IF) and immunohistochemistry (IHC)
Mouse tissues were collected and fixed in 10% formalin-neutral buffer. Paraffin embedding and Hematoxylin and Eosin (H&E) staining were performed at BCM Breast Center Histopathology Core. Antibodies: GFP (1:500, ab13970, Abcam), Ki67 (1:200, ab15580, Abcam), p-P27 (1:100, 71–7700, Zymed), Jagged-1 (1:100, ab7771), 2 different clones of S100A8 (1:5000, MAB3059, R&D Systems) used for mouse tissue and MRP8 (S100A8) (1:400, EPR3554, Abcam) used for human samples, CD4 (1:200, EPR19514, Abcam), CD8 (1:200, EPR21769, Abcam), F4–80 (1:200, CI:A3–1, Invitrogen), FoxP3 (1:200, FJK-16S, eBioscience), CD3 (1:400, A045229–2, Agilent), CD45 (1:200, 103102, Biolegend), and DAPI (R37606, Invitrogen). H&E and IHC images were scanned and quantified by Aperio ImageScope 11.2.0.780 (Aperio Technologies). IF images were scanned on an Axioscan and analyzed by using Zen Blue 3.1 (Zeiss). Human patient samples were purchased from Oregon Health and Science University (SR0494).
Immunophenotyping by Flow Cytometry
Live single cells from primary tumors that were frozen 90%FBS+10%DMSO described in the tissue harvest section were thawed and washed with PBS+2%FBS (FACS buffer) for flow cytometry analysis. Cells (2×106) were blocked in Fc block (553142, BD) and True-Stain Monocyte Blocker (426102, Biolegend) and stained for live (Live/Dead Aqua, L34957, Invitrogen) on ice for 30 min. After washing, cells were stained with antibodies on ice for 30 min in the dark: myeloid panel (Cd45, Cd11c, Cd11b, F4–80, Ly6c, Ly6g, PDCA-1, MHC2), lymphoid panel (CD45, CD3e, γσTCR, CD4, CD8α, CXCR3, Granzyme B, PD-1, CD19, CD49b).
For the myeloid panel, cells were fixed in BD Cytofix (BDB554655) and saved in the FACS buffer. For the lymphoid panel, cells were stained for FoxP3 by using the Foxp3/Transcription Factor Staining Buffer Set (00–5523-00 and 00–8333, eBioscience). After staining, cells were kept in FACS buffer at 4oC to be analyzed on a BD LSRFortessa the next day. Data were analyzed by FlowJo (version 10.6.2).
10X Chromium library preparation and sequencing
Single-cell 3’ RNA sequencing was performed at the Single Cell Genomics Core at Baylor College of Medicine. After thawing frozen cells (recovery of about 40–50% of cells), live single cells (DAPI- R37606, Fisher Scientific) were sorted on BD FACSAria I to obtain cells with the highest purity. A single cell suspension was prepared at 1000 cells per μl in DMEM+10%FBS. The single cell 3’ RNAseq library was prepared according to the Chromium single cell 3’ reagent kit version 2 (120234 & 120235, 10x Genomics) for figure 2 and version 3 for figure 6. Briefly, single cell suspension was loaded on a Chromium controller (10x Genomics) to generate single cell GEMS (Gel Beads-In-Emulsions) where full length cDNA was synthesized and barcoded. Subsequently the GEMs were broken and cDNAs from each single cell were pooled. Following cleanup using Dynabeads MyOne Silane Beads (370020, Thermo Fisher), cDNA was amplified by PCR. The amplified product was fragmented to optimal size before end-repair, A-tailing, and adaptor ligation. Finally, PCR was carried out to produce the library.
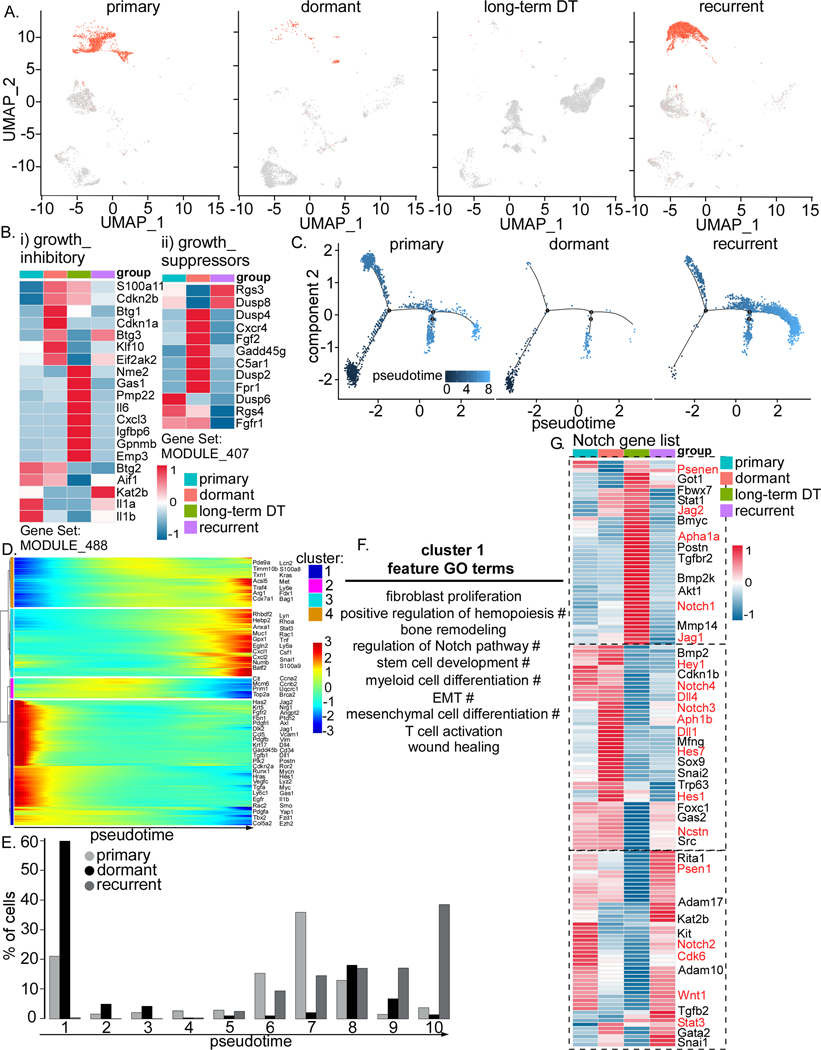
Figure 2. Tumor cells become quiescent (dormant) when regressed.
A) UMAP projection of GFP+ cells in Wnt/iFGFRGFP tumors at primary, dormant, long-term DT, and recurrence. B) Heatmap showing expression of growth inhibitory (i) and growth suppressor (ii) genes in different tumor samples. C) Trajectory analysis of cancer cells in 3 tumor samples, primary, dormant and recurrence. D) Heatmap showing 4 clusters of genes differentially expressed along the trajectory in C. E) Bar graph with 10 bins of pseudotime showing cells in the dormant stage mainly emerge in the early stage (bin #1). F) Feature Gene Ontology (GO) terms enriched in cluster1. The # symbol designates pathways regulated by Jagged-1 and Notch. G) Heatmap showing expression of the Notch pathway in different tumor samples. Known Notch pathway genes are highlighted in red.
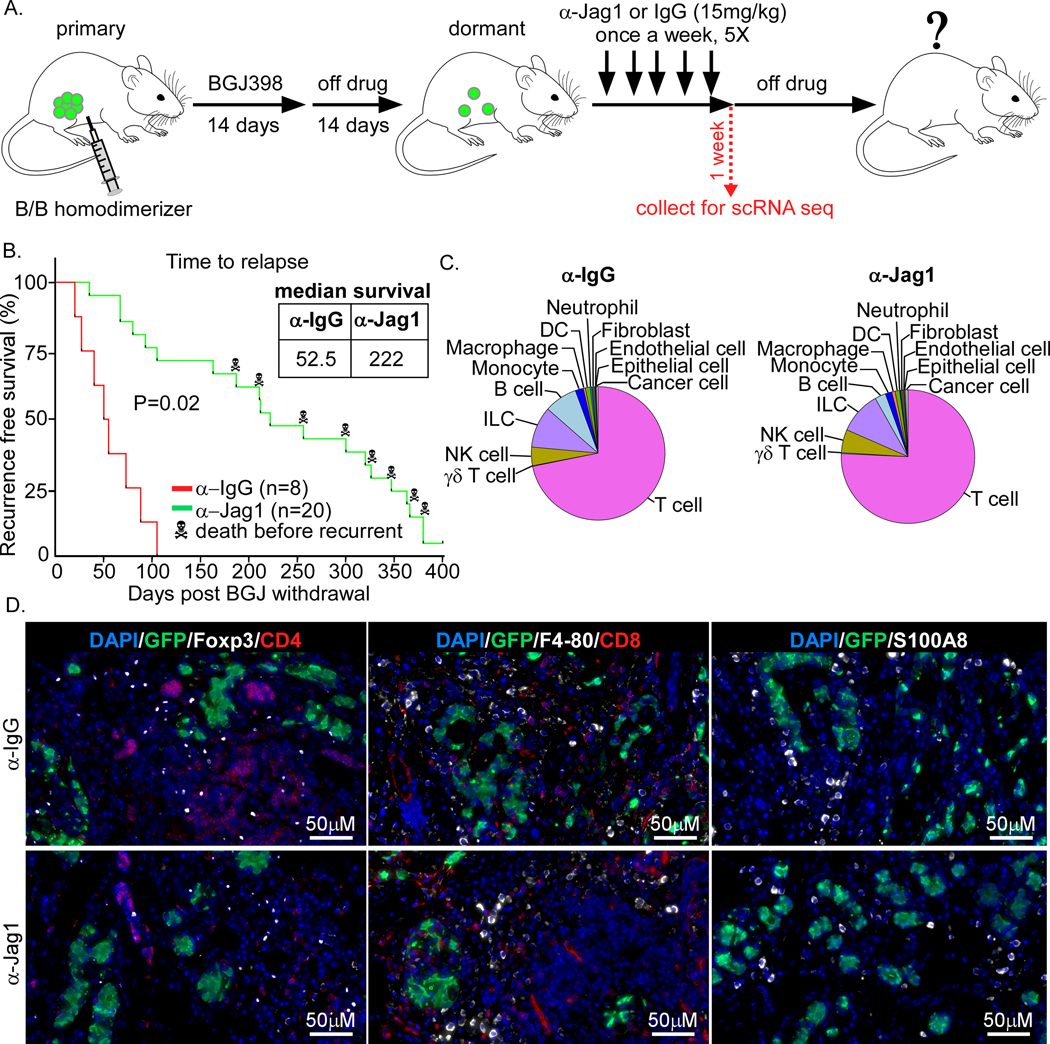
Figure 6. Anti-Jagged-1 blockade delays tumor recurrence.
A) Workflow of tumor generation, treatment with the anti-Jagged-1 (α-Jag1) blockade, and tumor harvest for scRNA seq. Tumors were treated with α-Jag1 or the control α-IgG (15mg/kg) once a week for 5 weeks and then treatment was stopped. The majority of tumors were called “recurrent” when they reached 5mm in diameter (12 out of 20 total mice). B) Survival graph showing α-Jag1 delays tumor recurrence, median survival of 222 days for α-Jag1 vs. 52.5 days for α-IgG. A few older mice died of unknown cases (8 out of 20 total mice). The p-value from a log rank test is shown. C) Pie chart of different cell types after α-IgG or α-Jag1 treatments. D) IF staining of cancer cells (GFP), CD4 and CD8 T cells, Tregs (Foxp3), macrophages, and MDSCs
Sequencing was performed at Novogene using HiSeq (HiSeq 4000) and Novaseq (NovaSeq 6000, S4 lane). Sequencing strategy for the Novaseq S4 and HiSeq platform on these runs was PE150 (paired-end 150bp) with an approximate data output of 800Gb (roughly 2.6B reads per lane) and 110Gb (roughly 360–400M reads per lane), respectively.
Single cell RNA sequencing data analysis
Raw sequencing reads were aligned to mouse mm10 reference genome following Cell Ranger v.3.1.0 pipeline. To ensure the quality, only the cells that express at least 200 genes and less than 10% mitochondrial genes and the genes expressed in less than 3 cells were retained for downstream analysis. In addition, suspected doublets with the cells expressing more than 4000 genes (3000 for α-IgG) were also filtered out. In each sample, cells were annotated by comparison to the reference datasets of pure mouse immune cells from the Immunologic Genome Project using SingleR package (19, 20). The GFP sequence was added to the reference gene (21). We noticed that some cancer cells that were annotated as “cancer cells”, lost their GFP expression. Therefore, for these analyses we used the population that was annotated as “cancer cells”, as opposed to GFP+ alone. The identity of the cells from closely related cell types, e.g., T cells and NKT cells, and NK cells and ILC cells, was refined based on their expression profiles of canonical markers (22–24). Due to the low number, the cancer cells in the long-term DT were only included in a limited analysis.
Because of the lack of an apparent batch effect, the first four samples (primary, dormant, long-term DT, and recurrent) and the second two samples (α-IgG and α-Jag1 treated samples) were directly merged for downstream analysis, respectively, using Seurat v.3 (25). To unveil the underlying cell transition within both cancer cells and immune cells, we performed single cell trajectory analysis for cancer cells, monocyte/macrophages and T cells from the first four samples, respectively, using Monocle v. 2.8.0 (26). The high dimensional data were reduced into a two-dimensional subspace using DDRTree algorithm, and the pseudotime and state were defined by orderCells function. Genes whose expression was significantly changed along the trajectory were identified using differentialGeneTest function with an q-value < 0.001. GO analysis was implemented on each cluster of differentially expressed genes (DEGs) using clusterProfiler (27). GO terms were considered as significantly enriched if adjusted p-value < 0.01.
Human breast cancer microarray datasets
Publicly available microarray data for patients (n=1911) from eleven human primary breast cancer data sets were downloaded from NCBI GEO (GSE9893; GSE7390; GSE22226; GSE4922; GSE1378; GSE1379; GSE5327; GSE2740; GSE21653; GSE11121; GSE2990; GSE2034) along with the corresponding clinical annotations. Data was first log2 transformed. Principle component analysis (PCA) was performed on the gene expression signature, and then k-means method was applied to cluster the samples into two category. The association between sample category and relapse-free survival was estimated using the hazard ratio (HR) from Cox proportional hazards regression model.
Statistics
Graphs were generated using GraphPad Prism 8 and R. To calculate recurrence, days were counted from the day when the treatment stopped to the day when recurrent tumors reached 5mm in diameter and median survival was measured. The logrank test was performed to measure the significance level for the difference in tumor recurrence between α-Jag1 and α-Jag1 treatment.
Data availability
ScRNA-seq gene expression data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE171464).
Results
Generation of Wnt/iFGFRGFP tumors to study cell signaling and tumor microenvironment during dormant MRD and recurrence
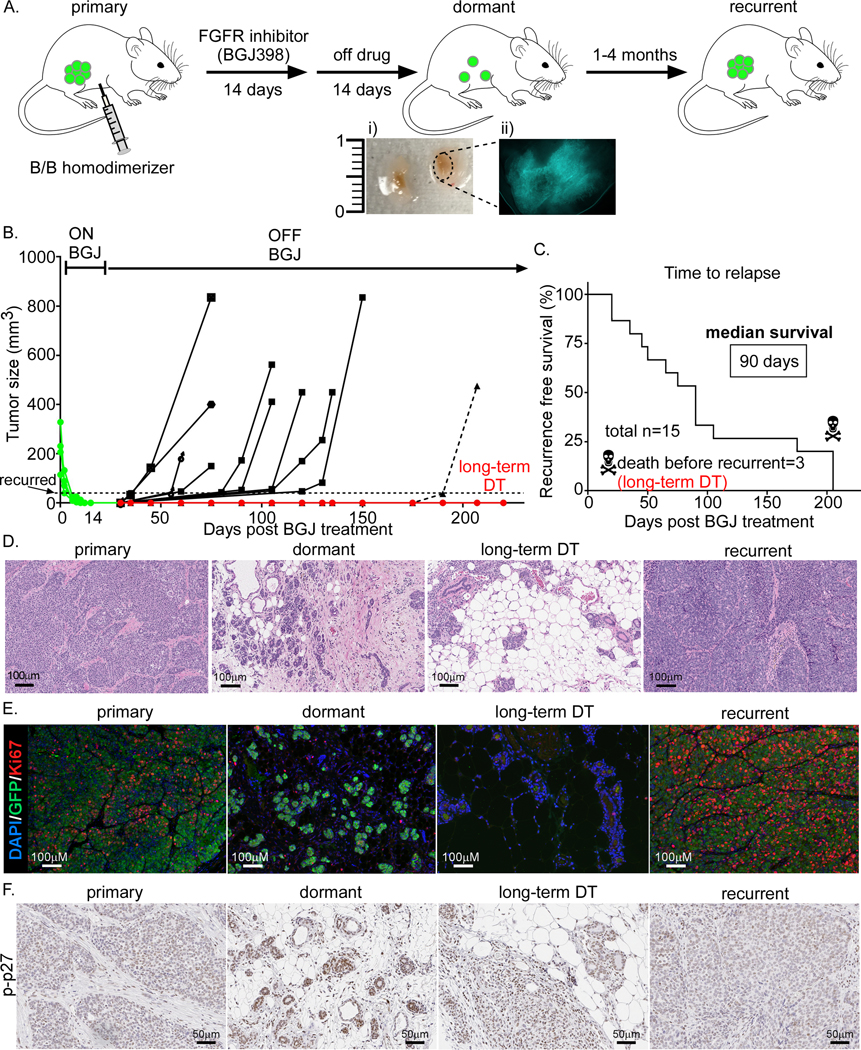
Previously, bigenic iFGFR transgenic mice developed in our laboratory and treated with a FGFR inhibitor provided a new model to study MRD and recurrence (18). In this syngeneic model, tumor regression induced tumor stroma remodeling and a decrease in myeloid derived suppressor cells (MDCSs). When tumors recurred epidermal growth factor receptor (EGFR) signaling was upregulated and MDSCs increased (18). However, using this model it was not feasible to isolate and characterize the small number of residual cancer cells because they lacked a reporter gene. To facilitate their isolation and to begin to elucidate the mechanisms involved in MRD, we generated spontaneous GFP+ tumors from iFGFRGFP mice. The GFP+ tumors were then transplanted into tolerized Glow-Head (GH) recipients (Figure 1A) (16). Briefly, as described previously (18), GFP+ tumor chunks were transplanted into the cleared fat-pad of the GH-mice and treated initially with a dimerizer to activate FGFR signaling until they were able to grow independently of the dimerizer. Mice bearing these large tumors then were treated with the FGFR inhibitor NVP-BGJ398 (BGJ) (18) for 14 days, and subsequently drug treatment was stopped for another two weeks. We hypothesized that under these conditions the residual cells would not be affected by the immediate cytotoxic effect of BGJ treatment. At this timepoint, depending on the experiment, mice were either: 1) sacrificed for MRD studies (Figure 1A; i and ii), or 2) left untreated to monitor for recurrence. Tumor recurrence was measured once tumors reached 5mm in diameter (Figure 1B and C). Median survival was 90 days (1–4 months) consistent with previously published results using non-GFP labeled cells (18). In three instances there were no detectable recurrences and these samples were designated as long-term DT (Figure 1B and C, in red). We characterized four stages of tumorigenesis: primary, MRD (dormant), long-term DT, and recurrent. Analysis of primary and recurrent tumors indicated that they had a similar histology, while the regressed lesions had increased stroma and immune cell infiltration (Figure 1D). Interestingly, in the majority of the cases when tumors regressed, the epithelial cells formed duct-shape structures which either lacked detectable or expressed low levels of Ki67 (Figure 1E), but expressed a high level of phosphorylated-p27 (p-p27) (Figure 1F). GFP+ cells were undetectable by immunohistochemistry in long-term DT due to very low number of remaining cells (Figure 1E). Thus, we utilized this improved iFGFRGFP tumor model to elucidate the properties of MRD that exhibited decreased proliferation and increased immune cell infiltration and stroma.
Figure 1. Generation of Wnt/iFGFRGFP tumors to study cell signaling and tumor microenvironment during dormant MRD and recurrence.
A) Workflow of tumor generation for various stages including primary, dormant, and recurrent. Activation of iFGFR1 and growth of transplanted Wnt/iFGFRGFP tumors were stimulated by dimerizer treatment. Once primary tumors reached 8–10mm in diameter, mice were treated with BGJ398 for 14 days. Regressed tumors (image Ai and Aii showing GFP expression) were off drug treatment for another 14 days and then collected. A separate cohort was used to study recurrence after 1–4 months from BGJ398 treatment. Tumors were collected when they reached 1.5cm in diameter both at the primary and recurrent stage. B) Growth curve for “recurrent” tumors. Tumors were designated as “recurrent” when they reached 5mm in diameter. Three lesions that remained dormant for more than 6 months were harvested and labeled long-term DT. C) Survival graph showing mice with recurrent tumors same as in B (12 out of 15 total mice). The 3 long-term DT lesions were collected without recurrence. D) H&E staining, D) Immuno-fluorescent (IF) staining of Ki67 and GFP, and E) Immuno-histochemistry (IHC) staining of phosphorylated-p27 (p-p27) at different stages.
Tumor cells become quiescent (dormant) during MRD
We next performed scRNA-seq analysis to investigate the transcriptional profiles of MRD (Figure S1A and S1B) and assess if the cells become dormant or are proliferating, as well as to identify any changes in their microenvironment. To ensure that we captured and characterized the dormant stage, we only selected lesions for further analysis that were negative for Ki67 in GFP+ tumor cells. Similar to immunohistochemistry analysis of GFP (Figure 1E), GFP+ cancer cells decreased when regressed and more dramatically in long-term DT (Figure 2A). As indicated by the analysis of gene expression related to growth inhibition (MODULE_488, Gene Set Enrichment Analysis (GSEA)) and growth suppression (MODULE_407, GSEA) (28), we observed that both MRD and long-term DT cells represented a quiescent state (Figure 2B, i and ii). Furthermore, we also detected the elevated RNA expression of dormancy markers including Bhlhe41 (Dec2), Cdkn1b (p27), Zfp281, Nr2f1, Gas1, and Gas6 (29–33) in both MRD and/or long-term DT cells (Figure S1C). However, we did not detect any RNA expression of β-galactosidase- a senescence marker- in either the MRD or long-term DT cells. Moreover, senescence is by definition irreversible leading to cell death and clearance by innate immune cells whereas dormancy is reversible and can lead to recurrence (34). Additionally, dormant cells activate variable degrees of pluripotency programs (34). GFP+ cells are present in all regressed lesions and eventually recur within 1–4 months, which argues against senescence, i.e a reversible process. Additionally, comparing dormant to long-term DT, we found that these two states express different markers of quiescence and dormancy, suggesting that these two stages were a result of dormancy, but not senescence, hence these were designated as dormant.
We next applied trajectory analysis to determine the different status between dormant cells and the primary and recurrent cells (Figure 2C). Pseudotemporal gene expression analysis identified four clusters of genes that were differentially expressed (Figure 2D). Cluster1, which consists of the genes more highly expressed at the early stage of trajectory, was enriched in the dormant cells (Figure 2E). Cluster3 and 4, which were highly expressed at the far end of the trajectory were enriched in the primary and recurrent cells (Figure 2E). Gene Ontology (GO) enrichment analysis showed that Cluster1 genes are involved in remodeling of stroma and immune cells and recruitment of both myeloid and lymphoid cells (Figure 2F). Genes from Cluster2 predominantly function in DNA replication and the cell cycle, while Clusters 3 and 4 are enriched for innate immunity and response to virus, and metabolic pathways, respectively (Table S1).
We also noticed that regulation of the Notch, Hippo, and Wnt signaling pathways are significantly enriched in Cluster1 (Figure 2D and F) but we focused on Notch signaling pathway. Previously, we showed that MDSCs increase when tumors recur (18) and that MDSCs directly regulate tumor initiating cells through regulating the Notch pathway (35). In addition, tumors can induce Jagged ligands in MDSCs through NFκB-p65 (36). Inhibiting MDSCs by using an anti-Jagged immunotherapy overcomes T cell tolerance and enhances T-cell immunotherapy (36). Interestingly, we found that during dormancy, Jagged-1 not only participates in the Notch signaling pathway, but also regulates a variety of other biological processes including hemopoiesis, myeloid cell differentiation, stem cell (SC) development, mesenchymal cell differentiation, and epithelial-mesenchymal transition (EMT) (Figure 2F, annotated by #). Both Jagged-1 and −2 were expressed in dormant and long-term dormant (Figure 2G). Moreover, different Notch signaling genes were found to be involved in the regulation of the four stages of tumorigenesis (Figure 2G). For example, Notch2 is most likely involved in the regulation of primary and recurrent cells, while Notch3 and 4 may regulate dormant cells, and Notch1 may regulate long-term DT cells. The different subunits of the γ-secretase complex, presenilin-1 (Psen1), nicastrin (Ncstn), anterior pharynx-defective 1 (APH-1), and presenilin enhancer 2 (Psenen), were also shown to be differentially expressed at these different stages (Figure 2G). Altogether, these results suggest that when tumor cells become quiescent, therefore dormant, they have enhanced Notch signaling.
Residual cells compromise different classes of mammary gland stem cells
Murine mammary gland (MG) stem/progenitor cells have been identified previously at different stages of mammary gland development. These include fetal mammary SCs that peak in late embryogenesis and their subsequent differentiation to adult basal and luminal SCs (37). Because the dormant cells were quiescent, we assessed any similarities to these different classes of MG stem/progenitor cells. Interestingly, primary and recurrent- compared to dormant and long-term dormant- cells were found to be enriched for the features of luminal progenitors (Figure S1D, i), whereas dormant cells were enriched for the signatures observed in fetal and basal progenitors as well as mature luminal cells (Figure S1D, ii, iii and iv). Moreover, to further investigate response differences among the subgroups of cancer cells to the BGJ treatment, we performed unsupervised clustering on cancer cells and identified eight clusters in the combined samples (Figure S1E, CC1–8). There was a substantial change in the composition of the different clusters among the primary, dormant and recurrent stages (Figure S1F). Primary tumors mainly consist of CC2, 3, 4 and 7, which were related to metabolic pathways (2), remodeling of stroma and T cell activation (3), DNA replication and cell cycle (4), and regulation of both myeloid and lymphoid cells (7) (Table S2). The major clusters observed in dormant lesions were CC3, 4, 5, 6 and 8, where CC6 and 8 seemed to appear de novo and did not exist in the primary tumor or were presumably present at an extremely low abundance. The GO analysis showed that CC5 and 8 participate in recruitment of immune cells both myeloid and lymphoid, and gland regeneration, respectively (Table S2). CC6 was of a great interest that it exhibited the most diverse functions including remodeling of stroma and immune cells and recruitment of both myeloid and lymphoid cells, metabolic pathways, DNA and RNA biosynthetic processes, regulation of SC differentiation, and gland regeneration (Table S2). In addition, Jagged-1 regulated many pathways in this cluster. Recurrent tumors primarily consisted of CC1, 2, 4 and 7, where the feature genes in CC1 are enriched for regulation of myeloid cells and gland development (Table S2).
These results suggest that there either may be rare cell subpopulations or perhaps a transition into a de novo population that becomes the major repopulating cluster after cells become dormant. This is contrary to the concept that all the clusters regress identically with treatment resulting in a similar probability of recurrence.
Residual cells have similar gene signatures with patient residual disease
To determine whether the gene signatures of the dormant stage were associated with human breast residual disease, we compared the expression profiles of dormant-overexpressed genes in residual BC following neoadjuvant targeted therapy from two public microarray datasets (GSE10281 (38) and GSE21974 (39)) to their corresponding pre-treated samples. We found that signature genes of residual cells were more highly expressed in patient tumors with residual disease (Figure S2A and B). For example, CCL5, which can promote BC recurrence through macrophage recruitment in residual tumors (14) was shown to be upregulated in the dormant stage as compared to primary and recurrent cells, and was also more highly expressed in the residual tumor samples. However, we did not find any association of the dormant gene signature with recurrence-free survival in the primary tumors of BC patients (Figure S2C).
Because MDSCs increase during recurrence and may regulate Notch signaling pathway through Jagged-1, we sought to identify Jagged-1 expressing MDSCs in patient samples. We performed Immunofluorescent (IF) staining of Jagged-1 and S100A8 (a MDSC marker) in infiltrating ductal carcinoma (IDC) and adjacent areas with fibrocystic changes (FCC) and found that in both sites, MDSCs express Jagged-1(Figure S2D). Additionally, some cancer cells in IDC also express Jagged-1 (Figure S2D). Interestingly, we found S100A8 expression in the cluster4 which consists of primarily recurrent cells, as well as some primary and dormant cells (Figure 2D).
Immune cells and microenvironment change during tumor dormancy
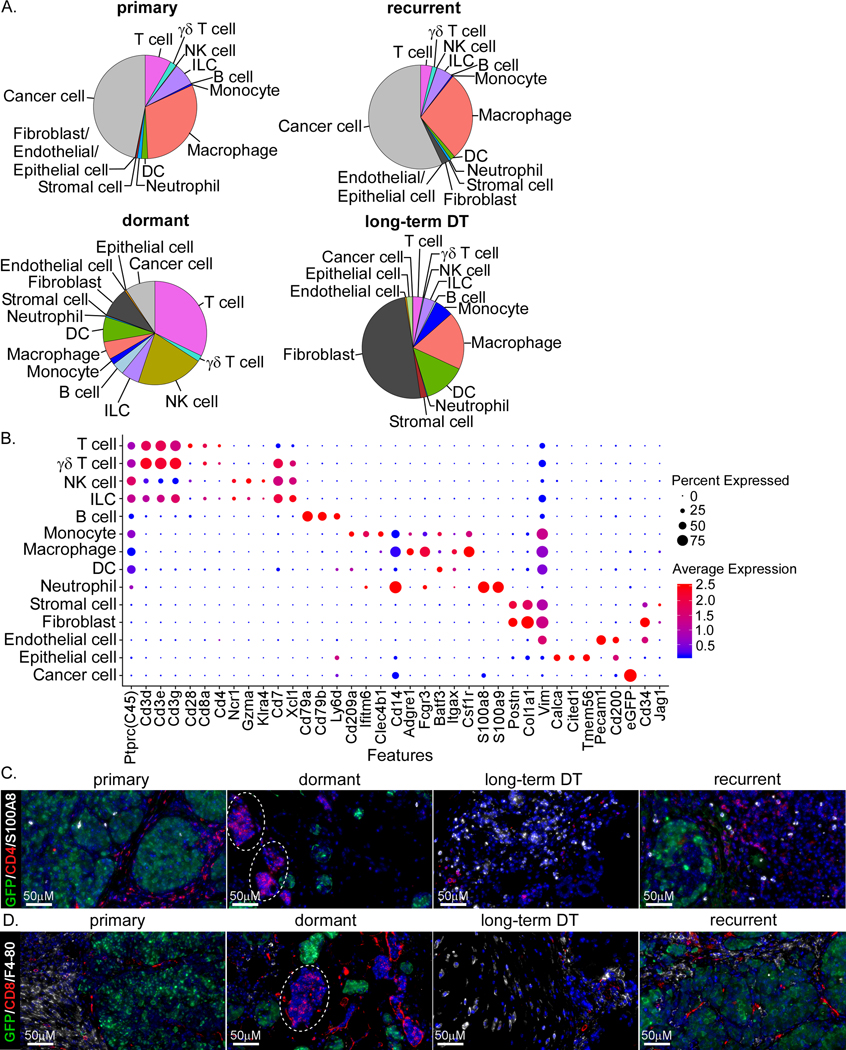
Because this mouse model has an intact immune system we also were able to investigate the transcriptional changes in the stroma and immune cells during dormancy (Figure 3A and B). Both primary and recurrent tumors have very similar microenvironments with a small decrease in T cells and a minor increase in fibroblasts. In contrast, the dormant cells showed substantial differences in the abundance of stroma and immune cell populations, including an increase in T cells, NK cells, B cells, monocytes, dendritic cells (DCs), fibroblasts, and endothelial cells and a decrease in macrophages and neutrophils (Figure 3A). In long-term DT cells, however, we found a decrease in T cells, NK cells and B cells, as compared to dormant cells, and an increase in DCs, monocytes, macrophages, stromal cells and fibroblasts (Figure 3A), suggesting that the microenvironment during progression from dormant to long-term DT might have become more immunosuppressive. These results obtained from scRNA-seq were confirmed by immunofluorescent (IF) detection of some of the known markers for these immune cells (Figure 3C and D). Interestingly, in the dormant stage, we found small aggregates of T cells (CD4+ and CD8+; Figure 3C and D, dotted line circles). Consistent with the scRNA-seq data, we found macrophages (F4/80) decreased in the dormant stage as compared to other stages (Figure 3D). To validate these results, we performed flow cytometry analysis of six frozen primary tumors using canonical markers for different immune cells grouped into myeloid or lymphoid panels (Figure S3A and B). Using functional markers, we identified a population of T regulatory cells (Tregs), CD4 T cells that are not Th1 activated CXCR3+, and CD8 T cells that are not cytotoxic (Granzyme B+) (Figure S3C) (40, 41). Moreover, we measured the cytokines expressed in cells (intracellular) from different samples and found that CCL5 and IL2 were expressed in the dormant stage (Figure S3D), again suggesting an increased recruitment and proliferation of immune cells (42, 43).
Figure 3. Immune cells and microenvironment change during tumor dormancy.
A) Pie chart showing contributions of major distinct cell types in four tumor samples. B) Dot plot of canonical marker genes for different cell types. C and D) Staining of some immune cell markers, CD4+ T cells (CD4), CD8+ T cells (CD8), macrophages (F4–80), and MDSCs (S100A8), and GFP in different tumor samples.
In addition, we identified a population of CD34+ cells (Figure 3B), which exhibits transcriptional signatures primarily of fibroblasts and endothelial cells. CD34+ fibroblast-like stromal cells can be recruited to the tumor reactive stroma and are involved in wound healing, antigen presenting, and the secretion of various types of collagen and cytokines (44–46). These cells also had increased expression of Jagged-1 as compared to other populations (Figure 3B). Together, these results show that numerous immune and stroma cells are being remodeled during dormancy and various cytokines are expressed at this stage that cause recruitment and proliferation of immune cells.
Dormancy is associated with remodeling of the myeloid compartment
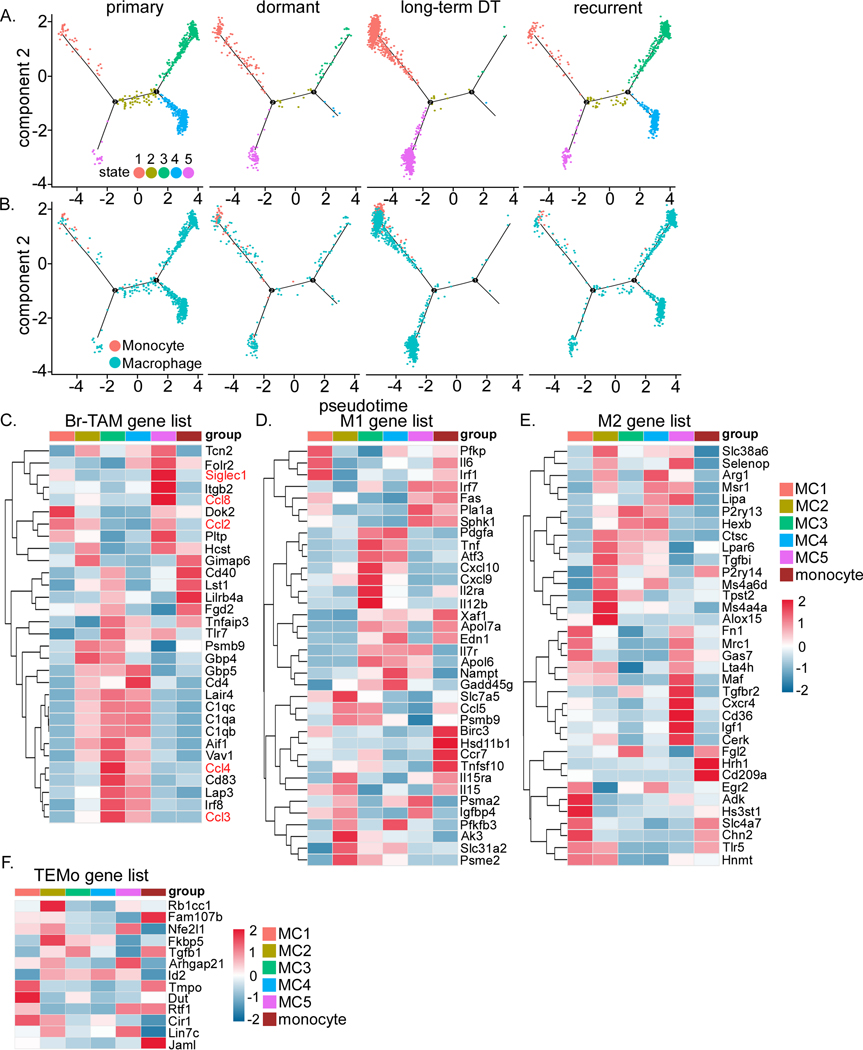
To obtain a better understanding of the functions of myeloid cells during dormancy, we first performed a trajectory analysis on monocytes and macrophages. This identified five distinct states of macrophages (MC1–5), where MC1 and MC5 are more prominent in the dormant and long-term DT samples (Figure 4A). Interestingly, we observed that monocytes are more prominent in the dormant and long-term DT stages (Figure 4B), suggesting an increase of monocyte recruitment. Macrophages display different polarization phenotypes often referring to M1 and M2, where M2 is immune-suppressive (47). To characterize the functions of the five distinct states of macrophages, we profiled the expression of the feature genes of M1, M2, BC associated macrophages (Br-TAM), and tumor-educated monocytes (TEMo) (48) (Figure 4C–F). In general, a mixed phenotype was observed in all macrophage states. Dormant and long-term DT-prominent MC1 grouped together with monocytes at the far left of the trajectory (the early stage of pseudotime), suggesting that MC1 may be less differentiated. In addition, MC1 showed increased expression of genes characteristic of M2 and TEMo (Figure 4E and F) but not Br-TAM (Figure 4C). This suggests that dormant and long-term DT might maintain a higher level of immuno-suppressive innate immunity. Comparatively, the primary- and recurrent-prominent macrophages, MC3 and MC4, showed a Br-TAM phenotype, and were more characteristic of M1 than M2 macrophages (Figure 4C–E). However, although they express the Br-TAM gene signature, they did not express the two suggested TAM markers SIGLEC1 and CCL8 (48), but instead expressed CCL3 and CCL4- known to regulate TAMs (Figure 4C) (49, 50). The MC5 state was the only dormant- and long-term DT-prominent population that showed Br-TAM features and expressed SIGLEC1 and CCL8 (Figure 4C). MC2 showed some expression of all phenotypes but more of M2, TEMo and Br-TAM, in that order (Figure 4C–F), suggesting a transient state. Furthermore, monocytes did not display TEMo features (Figure 4F). Thus, these results demonstrate that there is a spectrum of macrophage and monocyte phenotypes rather than canonical M1-M2 polarization during these four stages of tumorigenesis.
Figure 4. Dormancy is associated with remodeling of the myeloid compartment.
A) Trajectory analysis of macrophage-monocyte showing 5 distinct states. B) Trajectory analysis of macrophage-monocyte showing monocytes to be primarily recruited during dormancy and long-term DT. C to F) Heatmaps showing expression of genes involved in C) Breast tumor-associated-macrophage (Br-TAM), D) M1, E) M2, and F) Tumor-educated-monocytes (TEMo) in the 5 macrophage states (MC1 to 5) and monocytes.
MDSCs are a heterogenous population of neutrophil- and monocyte-like myeloid cells that play immunosuppressive roles (denoted as G- and M-MDSCs, respectively) (51). MDSCs promote tumor progression by various mechanisms including suppressing immune cells, supporting tumor initiating cells (TICs), and enhancing angiogenesis and metastasis (52). G-MDSCs characterized in BC models have a distinct gene signature as compared to normal neutrophils (51). In our studies, we found that neutrophils display high expression of the signature genes of BC-associated MDSCs (Br-MDSCs; combining the G- and M-MDSC signature genes) (Figure S4A). In contrast, monocytes did not express many Br-MDSC genes (Figure S4A). Furthermore, CD84, a suggested G-MDSCs marker (51), was not expressed in these neutrophils, but was detected on some macrophage states (Figure S4A).
Our previous studies showed that MDSCs decreased during dormancy and increased during recurrence (18) and that MDSCs regulated TICs in part through activating Notch (35). Here, we found a similar expression of tumor resident neutrophils as compared to Br-MDSCs (Figure S4A). In addition, we demonstrated that MDSCs expressed Jagged-1 that increased during long-term DT and recurrence (Figure S4B). Comparing the expression of Jagged-1 in circulating monocytes and neutrophils from primary and recurrent to non-tumor bearing mice showed that both monocytes and neutrophils in recurrent tumors expressed Jagged-1 (Figure S4C).
Dormancy is associated with remodeling of the lymphoid compartment
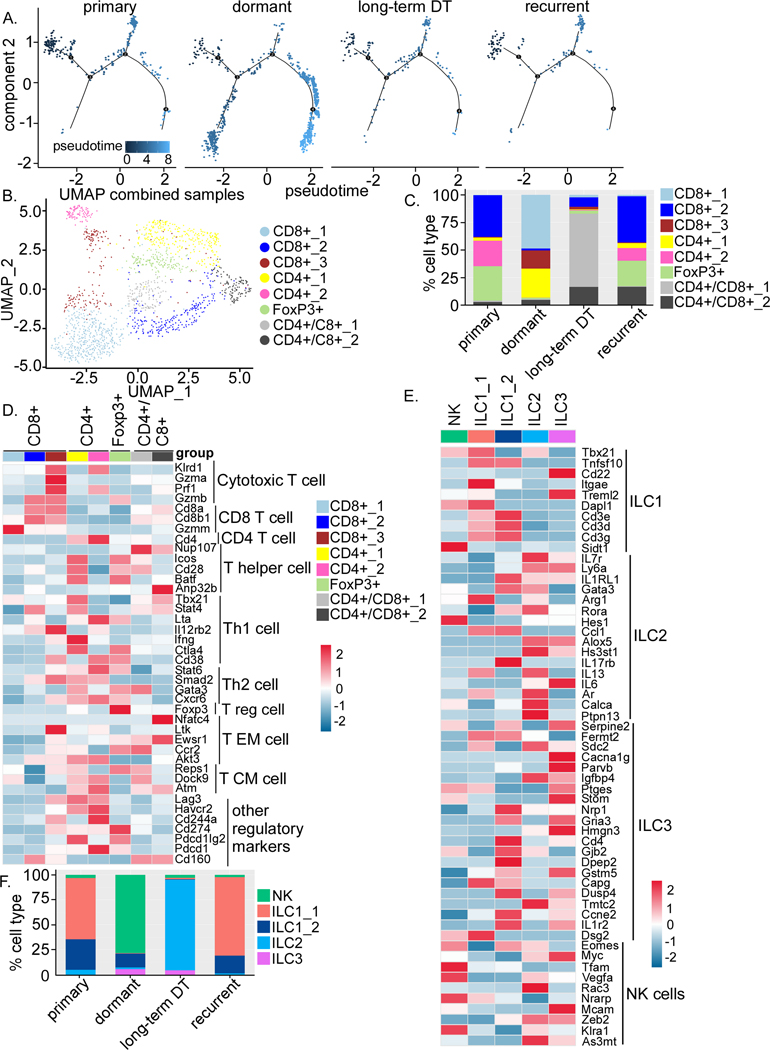
Utilizing IF staining and scRNA-seq, we also observed an increase of T cells during dormancy (Figure 3A, C and D). Trajectory analysis for T cells revealed that T cells are elevated in the dormant site as compared to primary, long-term DT and recurrent stages (Figure 5A). Moreover, the composition of the subclusters of CD4 and CD8 T cells changed during each stage of tumorigenesis (Figure 5B and C). Specifically, Tregs decreased during dormancy and long-term DT as compared to primary and recurrent tumors (Figure 5C), while total CD8 T cells increased (Figure 5C). According to the expression profiles of canonical markers (53), the majority of these CD8 T cells (CD8+_1) were not cytotoxic (Granzyme A-ve/B-ve and perforin-ve) (Figure 5D). The CD8+_2 population, which was decreased during dormancy, showed the most activated and non-exhausted phenotype (Figure 5C and D). The CD8+_3 population that was increased during dormancy resembled exhausted T cells because of the expression of Lag3, Cd244a, Cd274(PD-L1), Pdcd1 (PD-1), and Cd160 (Figure 5C and D) (53, 54). Similarly, we observed that CD4 T cells also exhibit potentially different functions among samples (Figure 5C and D). CD4 T cells during dormancy (CD4+_1) have Th1 activity, whereas CD4 T cells in primary tumors (CD4+_2) exhibit Th2 activity with increased expression of Lag3, Havcr2 (Tim3), Cd244a, Cd274, and Pdcd1 (Figure 5D). The majority of T cells in long-term DT are CD4+/C8+ T cells (Figure 5D). These results suggest that overall T cell functions may favor a more anti-tumorigenic state in dormant samples because of a decrease in Tregs and Th2 cells and an increase in CD8 T cells and Th1 cells. However, since most of CD8 T cells appear not to be not fully activated/cytotoxic (Granzyme A-ve/ B-ve and perforin-ve) or are already exhausted, cancer cells may have escaped from T cell immunity.
Figure 5. Dormancy is associated with remodeling of the lymphoid compartment.
A) Trajectory analysis of T cells at different stages. B) UMAP plot of T cells demonstrating distinct clusters. C) Bar graph quantifying the percentage of different clusters within T cells at different stages. D) Heatmap showing expression of known genes involved in different subtype of T cells and some regulatory genes. E) Heatmaps showing expression of NK cells and 4 distinct clusters of ILCs. F) Bar graph showing the changes in the percentage of NK cells and 4 distinct clusters of ILCs in different tumor samples.
Innate lymphoid cells (ILCs) are innate counterparts of T cells and are classified into ILC1, ILC2, ILC3, and NK cells. However, they have not been well characterized in cancers. Using a previously published gene lists for ILCs (22), we found various clusters of ILCs that resembled ILC1, 2, 3, and NK cells (Figure 5E). Although the total abundance of ILCs were similar, the proportion of these different types of ILCs changed among the four samples (Figure 5F). NK cells increased during dormancy compared to the primary, long-term DT and recurrent stages, while ILC1 decreased during both dormancy and long-term DT (Figure 5F). ILC2 decreased during dormancy but drastically increased during long-term DT and ILC3 increased during both dormancy and long-term DT as compared to primary and recurrent (Figure 5F). Increased NK cells and decreased ILC2 during dormancy suggest an anti-tumor function of ILCs. The function of ILC1 and ILC3 remain unclear because of their dual roles in tumors.
Anti-Jagged-1 blockade delays tumor recurrence and has a pleotropic effect on immune cells
The scRNA-seq data suggested a role of Jagged-Notch signaling pathway in dormancy. Notch signaling has been shown to accelerate tumor recurrence (13) and regulate endothelial-dormant cell interaction (8). Notch signaling is also implicated in MDSC-mediated regulation of TICs (35). Given that MDSCs increase during long-term DT and recurrence, we next investigated the effect of an anti-Jagged-1 (α-Jag1) blockade on tumor dormancy. We induced tumor dormancy using a protocol identical to the cohort described previously (Figure 6A). Two weeks after BGJ treatment was stopped, mice were divided into α-Jag1 or control (α-IgG) groups. Mice were treated with α-Jag1 once a week for a total of five weeks, and then treatment was stopped. Recurrence was reported once tumors reached 5mm in diameter (Figure 6A). Compared to the control group, the α-Jag1 blockade markedly delayed tumor recurrence (median survival=222 days in α-Jag1 vs. 52.5 days in α-IgG) (Figure 6B).
We repeated this experiment in a third cohort, collected samples one week after the cessation of the treatment (Figure 6A), and performed scRNA-seq on α-IgG and α-Jag1 treated samples (Figure S5A–C). Similar to the first cohort, we also observed substantial changes in immune and stroma cells in α-Jag1 and α-IgG treated samples (Figure 6C). Specifically, macrophages, NK cells, T cells, and γσT cells increased in the α-Jag1 treated group compared to α-IgG, while monocytes, neutrophils, and B cells decreased (Figure 6C). The expression of some of these immune cells was validated by IF staining of tissues for canonical markers of macrophages, MDSCs, CD4 or CD8 T cells, and Tregs in α-Jag1 and α-IgG treated samples (Figure 6D).
Additionally, we identified three subclusters of CD4 and CD8 T cells and a cluster of Treg cells in both sample types (Figure S5D). Treg cells were found to be slightly decreased while CD4 and CD8 T cells were increased overall in the α-Jag1 as compared to the α-IgG treatment group (Figure S5D). Two of the CD8 subclusters (CD8+_2 and CD8+_3) showed a cytotoxic phenotype (Granzyme B+), with CD8+_3 showing some exhaustion features (Figure S5E). The proportion of CD4+_2, which has a Th1 phenotype, slightly increased following α-Jag1 treatment (Figure S5D and S5E).
Altogether, these results show that the effect of α-Jag1 treatment on delaying recurrence is most likely due to a pleotropic effect on immune cells and possibly cancer and endothelial cells. α-Jag1 treatment is associated with the increase of NK cells, macrophages, and CD4 and CD8 T cells and the decrease of monocytes, neutrophils, B cells, and Tregs.
Discussion
This is the first time, to our knowledge, that a comprehensive characterization of stroma and immune cells has been analyzed during dormant MRD as summarized in Figure 7. To understand the complex ecosystem that promotes dormant cell survival and remodeling of microenvironment, we utilized a novel transgenic model, iFGFR1GFP with an intact immune system, to study MRD and relapse following treatment with an FGFR inhibitor. We demonstrated that Ki67-ve MRD have gene expression profiles similar to growth-arrested cells with dormancy markers including Cdkn1b, Zfp281, Nr2f1, and Gas6. Dormancy is accompanied by remodeling of stroma including endothelial cells and fibroblasts and immune cells including recruitment of both myeloid and lymphoid cells, the majority of which show immunosuppressive activity or are not fully functional and cytotoxic (Figure 7).
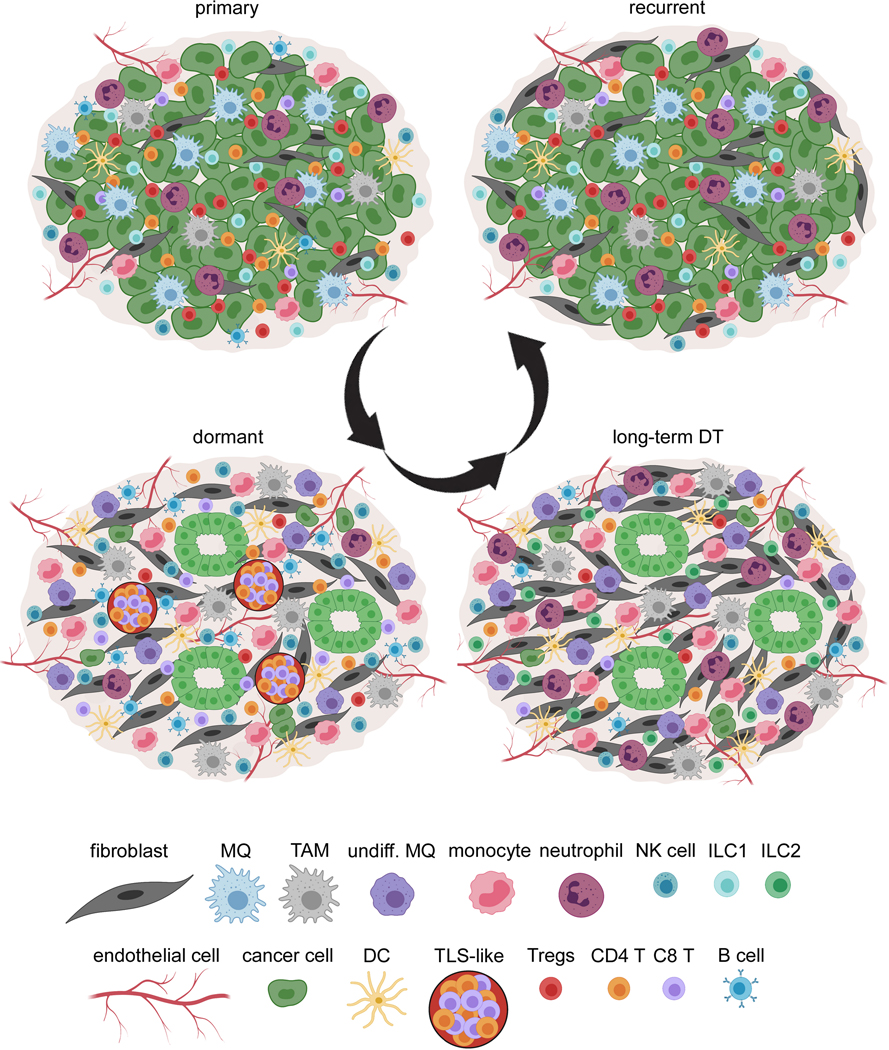
Figure 7. Remodeling of cancer cell populations, stroma, and immune cells during dormancy.
There is a dramatic remodeling of cancer cells, stroma, and immune cells during tumor dormancy and long-term DT, as well as ECM which have been described previously (18). Cancer cells organize into duct-shape structures and remain quiescent. During dormancy, there is an increase of T cells (CD8 T cells) and TLS-like bodies, B cells, NK cells, monocytes, undifferentiated macrophages (undiff. MQ) and TAMs, DCs, fibroblasts, and endothelial cells and there is a decrease of differentiated MQ (MQ), neutrophils (or MDSCs), and Tregs. During long-term dormancy, cancer cells evade immune surveillance by dramatically decreasing T cells, B cells, and NK cells, and increasing monocytes, neutrophils, undiff. MQ, TAMs, ILC2s, fibroblasts, and endothelial cells. When tumors relapse, they acquire an almost similar immune environment compared to the primary cancer with increased numbers of differentiated MQ, neutrophils, ILC1, and T regs and decreased numbers of active CD8 T cells, NK cells, and DCs.
Interestingly, different groups of MG progenitor cells were detected during dormancy and recurrence perhaps as a function of either de novo reprogramming or by transition from other cell types. Thus, it does not appear that during dormancy the FGFR inhibitor treatment results only in tumor de-bulking and that similar cell populations exist throughout these different stages of tumorigenesis. Given that different MG progenitors appear during dormancy, we hypothesize that the CC6 cells identified by scRNA analyses may be the major group of progenitor cells that give rise to the recurrent tumors, but this will have to be established by lineage tracing experiments. Interestingly, a recent study has also demonstrated that dormant residual tumor cells express a mammary SC signature which is only capable of forming recurrent tumors but not primary tumors, suggesting that distinct TICs may give rise to primary or recurrent tumors (55).
Jagged-1 has been shown to regulate many aspects of tumorigenesis including SC development, EMT, and immune cells homeostasis during dormancy and may be involved in the CC6 cells. The pleiotropic effects of Jagged-1 expression in cancer cells, fibroblasts, endothelial cells, and myeloid cells, may all account for the marked effect of α-Jag1 treatment to increase dormancy and delay recurrence.
This study focused on some myeloid cell types including monocytes, macrophages, neutrophils and NK cells. The increase of immune-suppressive myeloid cells may explain the lack of T cell activity. Although we saw an increase of neutrophils (MDSCs) during long-term dormant stage by immune-histochemistry analysis, there was a discrepancy when we analyzed scRNA-seq. This can be explained by the fact that MDSCs decrease viability during cryopreservation (56, 57).
Although we saw an increase of T cells and the appearance of T cell aggregates in during dormancy which appear to resemble a tertiary lymphoid structure (TLS) (58), most of CD8 T cells seem not fully activated/cytotoxic (Granzyme A-ve/B-ve and perforin-ve) or are already exhausted. Cytotoxic NK cells also increased but dormant cancer cells may eventually find an opportunity to evade immune surveillance including NK cells (7) and T cell immunity as suggested by the decrease of NK and CD8 T cells in long-term DT. Interestingly, a distinct population of CD8 T cells (CD39+PD-1+), were discovered to inhibit the outgrowth of lung mets and mediate the dormancy of DTCs in a mouse model but also was unable to fully eradicate all of the cancer cells due to expression of checkpoint molecules (59).
We treated mice once weekly for only five weeks with an α-Jag1 blockade antibody during dormancy, but notwithstanding we observed a significant sustained delay of tumor relapse. These results suggest that this brief window of treatment during dormancy was sufficient both for the survival of dormant cancer cells, and also to inhibit the changes observed in both the stroma and immune cells. Treatment with α-Jag1 had a pleotropic effect on immune cells. It induced an anti-tumor immunity by increasing macrophages, NK cells, and CD8 T cells (Granzyme B+ and perforin+) and decreased myeloid cells including monocytes and neutrophils, as well as B cells and Tregs. The GO analysis of residual tumor cells suggests that there are other potential targets including those of Notch pathway signaling (Jagged-2, DLL1, DLL4, and etc.) and Hippo, and Wnt signaling pathways that can be evaluated to target dormant cells in the future.
Overall, our study demonstrates that there is significant remodeling of both the stroma and immune cells during dormant MRD. The ongoing immune surveillance process recruits a mixture of immune-active (T cells and NK cells) and suppressive (T regs and myeloid cells) cells. In long-term dormancy however, cancer cells can evade the immune surveillance and the immune-suppressive cells become the prominent population. Cancer (stem) cells can also transition into a different state. The Notch signaling pathway plays an important role in regulating both the cancer and immune cells and targeting this pathway, provides a promising therapeutic strategy to inhibit tumor relapse. In future studies it will be important to determine if similar mechanisms are employed in the metastatic microenvironment. Additionally, we did not find any association between our dormant gene signature and survival of patients with BC. One caveat is that gene expression data obtained from all of these studies are from tumor chunks as opposed to single cell analysis so there is no resolution to specifically assess the dormant signature in cancer cells. The other caveat is that gene expression data are only from primary tumors with their survival metadata, and not regressed tumors. As our study suggests, the CC6 is a rare or de novo population that appears during dormancy with the potential to contribute to tumor relapse. Without the availability of single cell and spatial transcriptomic data from patient samples to specifically identify these populations during MRD, we are not able to interrogate patient survival by only investigating primary samples. Future studies using single cell analysis of both human primary and MRD will be required to address the relevance of these mouse tumor models.
Supplementary Material
Statement of Significance.
Single-cell RNA-seq analysis reveals dormancy-associated changes in immune and stromal cells and demonstrates a rationale to pursue Jagged-1/Notch pathway inhibition as a viable therapeutic strategy to reduce disease recurrence.
Acknowledgments
We would like to thank Dr. Chris Siebel for kindly providing anti-Jagged-1 blockade antibody and helpful suggestions, Dr. Chi-Ping Day (NIH/NCI) for providing the Glow-Head mice and Dr. Michael Lewis for providing the GFP mouse, Dr. Yumei Li for prepping single cell libraries, Dr. Brian Gibson for training in mouse surgery techniques and protocol preparation, Dr. Alejandro Contreras for providing access to Aperio slide scanner at the Department of Translational Molecular Pathology-MD Anderson, Dr. Susan Hilsenbeck for helping with statistical power analysis, Dr. Rainer Lanz for helping with some data analysis, a Dr. Brian Ruffell for helping with the flow cytometry, and Dr. Lisa Coussens for advice about some data analysis.
We acknowledge the joint participation of the Diana and Adrienne Helis Malvin Medical Research Foundation and Baylor College of Medicine. This project was supported by grant CA016303 (jmr), the Quantitative Science Shared Resource in the Duncan Comprehensive Cancer Center (CA125123), the Cytometry and Cell Sorting Core at Baylor College of Medicine (BCM) with funding from the CPRIT Core Facility Support Award (CPRIT-RP180672), the NIH (CA125123, RR024574, S10 OD025251) and the assistance of Joel M. Sederstrom, the RNA In Situ Hybridization Core at BCM, which is, in part, supported by a Shared Instrumentation grant from the NIH (1S10OD016167) and the assistance of Dr. Cecilia Ljungberg. Single Cell 3’ RNAseq was performed at the Single Cell Genomics Core at BCM, which is partially supported by the NIH shared instrument grants (S10OD023469, S10OD025240) and P30EY002520 to Dr. Rui Chen. This work benefitted from data assembled by the ImmGen consortium (20).
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7(11):834–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014;14(9):611–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks MD, Burness ML, Wicha MS. Therapeutic Implications of Cellular Heterogeneity and Plasticity in Breast Cancer. Cell Stem Cell. 2015;17(3):260–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarmiento-Castro A, Caamano-Gutierrez E, Sims AH, Hull NJ, James MI, Santiago-Gomez A, et al. Increased Expression of Interleukin-1 Receptor Characterizes Anti-estrogen-Resistant ALDH(+) Breast Cancer Stem Cells. Stem Cell Reports. 2020;15(2):307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481(7379):85–9. [DOI] [PubMed] [Google Scholar]
- 6.Cao Z, Scandura JM, Inghirami GG, Shido K, Ding BS, Rafii S. Molecular Checkpoint Decisions Made by Subverted Vascular Niche Transform Indolent Tumor Cells into Chemoresistant Cancer Stem Cells. Cancer Cell. 2017;31(1):110–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malladi S, Macalinao DG, Jin X, He L, Basnet H, Zou Y, et al. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell. 2016;165(1):45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H, et al. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol. 2013;15(7):807–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goddard ET, Bozic I, Riddell SR, Ghajar CM. Dormant tumour cells, their niches and the influence of immunity. Nat Cell Biol. 2018;20(11):1240–9. [DOI] [PubMed] [Google Scholar]
- 10.Szczerba BM, Castro-Giner F, Vetter M, Krol I, Gkountela S, Landin J, et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566(7745):553–7. [DOI] [PubMed] [Google Scholar]
- 11.Kowanetz M, Wu X, Lee J, Tan M, Hagenbeek T, Qu X, et al. Granulocyte-colony stimulating factor promotes lung metastasis through mobilization of Ly6G+Ly6C+ granulocytes. Proc Natl Acad Sci U S A. 2010;107(50):21248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science. 2018;361(6409). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abravanel DL, Belka GK, Pan TC, Pant DK, Collins MA, Sterner CJ, et al. Notch promotes recurrence of dormant tumor cells following HER2/neu-targeted therapy. J Clin Invest. 2015;125(6):2484–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walens A, DiMarco AV, Lupo R, Kroger BR, Damrauer JS, Alvarez JV. CCL5 promotes breast cancer recurrence through macrophage recruitment in residual tumors. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pond AC, Herschkowitz JI, Schwertfeger KL, Welm B, Zhang Y, York B, et al. Fibroblast growth factor receptor signaling dramatically accelerates tumorigenesis and enhances oncoprotein translation in the mouse mammary tumor virus-Wnt-1 mouse model of breast cancer. Cancer Res. 2010;70(12):4868–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Day CP, Carter J, Weaver Ohler Z, Bonomi C, El Meskini R, Martin P, et al. “Glowing head” mice: a genetic tool enabling reliable preclinical image-based evaluation of cancers in immunocompetent allografts. PLoS One. 2014;9(11):e109956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tavera-Mendoza LE, Brown M. A less invasive method for orthotopic injection of breast cancer cells into the mouse mammary gland. Lab Anim. 2017;51(1):85–8. [DOI] [PubMed] [Google Scholar]
- 18.Holdman XB, Welte T, Rajapakshe K, Pond A, Coarfa C, Mo Q, et al. Upregulation of EGFR signaling is correlated with tumor stroma remodeling and tumor recurrence in FGFR1-driven breast cancer. Breast Cancer Res. 2015;17:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heng TS, Painter MW, Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9(10):1091–4. [DOI] [PubMed] [Google Scholar]
- 21.Hadjantonakis AK, Gertsenstein M, Ikawa M, Okabe M, Nagy A. Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech Dev. 1998;76(1–2):79–90. [DOI] [PubMed] [Google Scholar]
- 22.Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16(3):306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, et al. Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity. 2019;50(5):1317–34 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He D, Wang D, Lu P, Yang N, Xue Z, Zhu X, et al. Single-cell RNA sequencing reveals heterogeneous tumor and immune cell populations in early-stage lung adenocarcinomas harboring EGFR mutations. Oncogene. 2021;40(2):355–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177(7):1888–902 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taichman RS, Patel LR, Bedenis R, Wang J, Weidner S, Schumann T, et al. GAS6 receptor status is associated with dormancy and bone metastatic tumor formation. PLoS One. 2013;8(4):e61873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sosa MS, Parikh F, Maia AG, Estrada Y, Bosch A, Bragado P, et al. NR2F1 controls tumour cell dormancy via SOX9- and RARbeta-driven quiescence programmes. Nat Commun. 2015;6:6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bragado P, Estrada Y, Parikh F, Krause S, Capobianco C, Farina HG, et al. TGF-beta2 dictates disseminated tumour cell fate in target organs through TGF-beta-RIII and p38alpha/beta signalling. Nat Cell Biol. 2013;15(11):1351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linde N, Fluegen G, Aguirre-Ghiso JA. The Relationship Between Dormant Cancer Cells and Their Microenvironment. Adv Cancer Res. 2016;132:45–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92(20):9363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Risson E, Nobre AR, Maguer-Satta V, Aguirre-Ghiso JA. The current paradigm and challenges ahead for the dormancy of disseminated tumor cells. Nat Cancer. 2020;1(7):672–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welte T, Kim IS, Tian L, Gao X, Wang H, Li J, et al. Oncogenic mTOR signalling recruits myeloid-derived suppressor cells to promote tumour initiation. Nat Cell Biol. 2016;18(6):632–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sierra RA, Trillo-Tinoco J, Mohamed E, Yu L, Achyut BR, Arbab A, et al. Anti-Jagged Immunotherapy Inhibits MDSCs and Overcomes Tumor-Induced Tolerance. Cancer Res. 2017;77(20):5628–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung CY, Ma Z, Dravis C, Preissl S, Poirion O, Luna G, et al. Single-Cell Chromatin Analysis of Mammary Gland Development Reveals Cell-State Transcriptional Regulators and Lineage Relationships. Cell Rep. 2019;29(2):495–510 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106(33):13820–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stickeler E, Pils D, Klar M, Orlowsk-Volk M, Zur Hausen A, Jager M, et al. Basal-like molecular subtype and HER4 up-regulation and response to neoadjuvant chemotherapy in breast cancer. Oncol Rep. 2011;26(4):1037–45. [DOI] [PubMed] [Google Scholar]
- 40.Groom JR, Luster AD. CXCR3 in T cell function. Exp Cell Res. 2011;317(5):620–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruffell B, Au A, Rugo HS, Esserman LJ, Hwang ES, Coussens LM. Leukocyte composition of human breast cancer. Proc Natl Acad Sci U S A. 2012;109(8):2796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aldinucci D, Colombatti A. The inflammatory chemokine CCL5 and cancer progression. Mediators Inflamm. 2014;2014:292376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol. 2012;12(3):180–90. [DOI] [PubMed] [Google Scholar]
- 44.Quan TE, Cowper S, Wu SP, Bockenstedt LK, Bucala R. Circulating fibrocytes: collagen-secreting cells of the peripheral blood. Int J Biochem Cell Biol. 2004;36(4):598–606. [DOI] [PubMed] [Google Scholar]
- 45.San Martin R, Barron DA, Tuxhorn JA, Ressler SJ, Hayward SW, Shen X, et al. Recruitment of CD34(+) fibroblasts in tumor-associated reactive stroma: the reactive microvasculature hypothesis. Am J Pathol. 2014;184(6):1860–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barth PJ, Westhoff CC. CD34+ fibrocytes: morphology, histogenesis and function. Curr Stem Cell Res Ther. 2007;2(3):221–7. [DOI] [PubMed] [Google Scholar]
- 47.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177(10):7303–11. [DOI] [PubMed] [Google Scholar]
- 48.Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, et al. Human Tumor-Associated Macrophage and Monocyte Transcriptional Landscapes Reveal Cancer-Specific Reprogramming, Biomarkers, and Therapeutic Targets. Cancer Cell. 2019;35(4):588–602 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitamura T, Qian BZ, Soong D, Cassetta L, Noy R, Sugano G, et al. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J Exp Med. 2015;212(7):1043–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De la Fuente Lopez M, Landskron G, Parada D, Dubois-Camacho K, Simian D, Martinez M, et al. The relationship between chemokines CCL2, CCL3, and CCL4 with the tumor microenvironment and tumor-associated macrophage markers in colorectal cancer. Tumour Biol. 2018;40(11):1010428318810059. [DOI] [PubMed] [Google Scholar]
- 51.Alshetaiwi H, Pervolarakis N, McIntyre LL, Ma D, Nguyen Q, Rath JA, et al. Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. Sci Immunol. 2020;5(44). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janghorban M, Xin L, Rosen JM, Zhang XH. Notch Signaling as a Regulator of the Tumor Immune Response: To Target or Not To Target? Front Immunol. 2018;9:1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khalsa JK, Cheng N, Keegan J, Chaudry A, Driver J, Bi WL, et al. Immune phenotyping of diverse syngeneic murine brain tumors identifies immunologically distinct types. Nat Commun. 2020;11(1):3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saleh R, Taha RZ, Toor SM, Sasidharan Nair V, Murshed K, Khawar M, et al. Expression of immune checkpoints and T cell exhaustion markers in early and advanced stages of colorectal cancer. Cancer Immunol Immunother. 2020;69(10):1989–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruth JR, Pant DK, Pan TC, Seidel HE, Baksh SC, Keister BA, et al. Cellular dormancy in minimal residual disease following targeted therapy. Breast Cancer Res. 2021;23(1):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kotsakis A, Harasymczuk M, Schilling B, Georgoulias V, Argiris A, Whiteside TL. Myeloid-derived suppressor cell measurements in fresh and cryopreserved blood samples. J Immunol Methods. 2012;381(1–2):14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grutzner E, Stirner R, Arenz L, Athanasoulia AP, Schrodl K, Berking C, et al. Kinetics of human myeloid-derived suppressor cells after blood draw. J Transl Med. 2016;14:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pages F, Galon J, Dieu-Nosjean MC, Tartour E, Sautes-Fridman C, Fridman WH. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29(8):1093–102. [DOI] [PubMed] [Google Scholar]
- 59.Tallon de Lara P, Castanon H, Vermeer M, Nunez N, Silina K, Sobottka B, et al. CD39(+)PD-1(+)CD8(+) T cells mediate metastatic dormancy in breast cancer. Nat Commun. 2021;12(1):769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ScRNA-seq gene expression data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE171464).