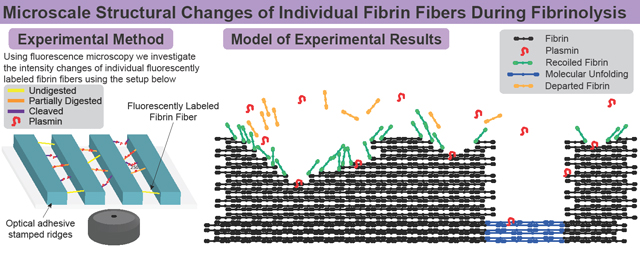
Abstract
Fibrinolysis is the enzymatic digestion of fibrin, the primary structural component in blood clots. Mechanisms of fibrin fiber digestion during lysis have long been debated and obtaining detailed structural knowledge of these processes is important for developing effective clinical approaches to treat ischemic stroke and pulmonary embolism. Using dynamic fluorescence microscopy, we studied the time-resolved digestion of individual fibrin fibers by the fibrinolytic enzyme plasmin. We found that plasmin molecules digest fibers along their entire lengths, but that the rates of digestion are non-uniform, resulting in cleavage at a single location along the fiber. Using mathematical modeling we estimated the rate of plasmin arrival at the fiber surface and the number of digestion sites on a fiber. We also investigated correlations between local fiber digestion rates, cleavage sites, and fiber properties such as initial thickness. Finally, we uncovered a previously unknown tension-dependent mechanism that pulls fibers apart during digestion. Taken together these results promote a paradigm shift in understanding mechanisms of fibrinolysis and underscore the need to consider fibrin tension when assessing fibrinolytic approaches.
Keywords: Fibrinogen, Hemostasis, Microscopy, Modeling, Plasmin
Graphical Abstract
1. Introduction
Wound healing requires a proper balance of hemostasis and fibrinolysis. Coagulation, the final step of the hemostatic process, results in the creation of a blood clot containing a fibrin mesh network that stems the flow of blood, while the fibrinolytic process digests the fibrin fibers composing the network, restoring vascular flow. Imbalance in either of these processes can lead to occluded blood vessels resulting in pathological conditions including myocardial infarction and ischemic stroke, which are leading causes of death and disability [1].
Fibrinolysis is a natural process involving a complicated system of promoters and inhibitors [2]. Plasmin, the primary enzyme responsible for the digestion of fibrin fibers, is activated from its inactive precursor plasminogen by either urokinase plasminogen activator (uPA) or tissue plasminogen activator (tPA).
One clinical approach to resolving an occluded blood vessel is to administer enzymes from outside the thrombus in a process called thrombolysis. While recombinant tPA is the standard thrombolytic agent used to treat strokes [3], a phase 1/2a safety study has shown potential for using plasmin as a thrombolytic [4]. Clinical studies have shown a limited time window for the safe administration of tPA treatments for stroke (within 4.5 hours after symptom onset) [5], and recanalization of the occluded blood vessel is often not achieved in thrombolytic treatments [6,7]. Thus, there is an important need for continued clinical assessments of safe thrombolytic approaches as well as basic biology studies to understand the mechanisms regulating fibrinolysis/thrombolysis.
In particular, there has been a dearth of studies aimed at elucidating the mechanisms by which fibrin fibers are digested. Fibrin fiber polymerization begins when thrombin cleaves specific residues in fibrinogen molecules, converting them to fibrin. Fibrin molecules then polymerize into half-staggered, double-stranded protofibrils, which laterally aggregate to form thicker fibers and a branched fibrin mesh. The conversion from fibrinogen to fibrin also exposes cryptic plasminogen, plasmin, and tPA binding sites [8,9]. However, once the binding sites emerge, the routes plasmin takes to digest the fibers and the structural and biophysical properties regulating fiber digestion are unresolved.
This knowledge gap remains, in part, because most fibrinolysis studies focus on the digestion of fibrin networks/clots and lack the spatial or temporal resolution to investigate fiber digestion. Previous studies have debated whether plasmin digests fibers along their entire length, leading to a uniform decrease in fiber diameter [10–12], or whether once the first plasmin molecule begins digesting at one site, feedback mechanisms promote direct digestion across the fiber leading to transverse cleavage [13–17]. A recent study suggested that fibers may swell during an initial phase of lysis as internal fiber associations are cut, after which the fiber fragments and is eventually cleaved through [18].
Formerly, we introduced the first assay to study the cleavage rates of individual fibers, but the assay lacked the spatial resolution to investigate mechanisms of fiber digestion [13,15]. In this study, we present an original approach to studying the mechanisms of individual fiber digestion using dynamic fluorescence microscopy. In our initial experiments we test whether fibers are uniformly digested or cleaved at one location and show that each fiber has multiple digestion sites. Next, we investigate the rate of digestion at each site and find that certain locations are digested faster than others, ultimately resulting in fiber cleavage at a single location. To understand whether our measured number of digestion sites is reasonable, we use mathematical models to estimate the number of plasmin molecules bound to the fiber at any time and find conditions that provide good agreement with experimental data. To determine the reason certain locations on the fiber are digested faster than others, we investigate correlations between local fiber digestion rates, cleavage sites, and fiber properties such as initial thickness. Importantly, we find that fibrin slides longitudinally along the fiber during digestion, suggesting that fibers are pulled apart. We show that inherent fiber tension [13,15,19,20] likely accounts for this observation and, therefore, plays an important role in regulating fibrinolysis. Taken together, these results resolve longstanding disagreements about fibrinolytic processes, reveal previously unknown mechanisms, and expose a need for a dramatic change in considering the role of fiber tension in regulating fibrinolytic outcomes.
2. Methods
2.1. Preparing and Imaging Samples
Fibrin fibers were polymerized between micropatterned ridges measuring 20 μm wide, 10 μm deep, and 20 μm apart as described previously [15]. Aliquots of unlabeled and ALEXA 488-labeled human fibrinogen were thawed from –80.0 °C and diluted to 2.0 mg/mL and 0.0308 mg/mL, respectively, using a HEPES Buffered Saline (HBS; 20mM HEPES, 150 mM NaCl, pH 7.4) and placed onto the ridged surface. Human alpha thrombin was added to the fibrinogen solution with final concentrations of 0.1 U/mL and 1 mg/mL, respectively, in HBS with 5mM Ca2+, and the solution polymerized for an hour at 37°C. Plasmin (final concentration 0.066 U/mL) was added to the samples right before imaging to initiate fibrinolysis.
2.2. Microscopy
Samples were placed face up on a Leica DMi8 epifluorescent microscope (Leica Microsystems Inc., Buffalo Grove, IL) and imaged with a 63x oil immersion objective. After the addition of plasmin, images were taken with a Leica DFC9000GT SCMOS 4 Megapixel monochrome camera with each image measuring 2048 pixels. After the addition of plasmin, images of 211 μm2-sized areas were taken every 2.5 seconds for 10 minutes, after which all fibers were lysed, leading to a maximum time error of ± 1.25 s.
2.3. Image Processing
Fibers were subject to strict exclusion/inclusion criteria for analysis (Supplement Sections A.1–A.5 and Supplemental Figures 1–5). Images were manually segmented into separate time series, each containing a single fiber. Background noise was subtracted using FIJI/ImageJ’s rolling ball method which was not found to significantly alter fiber intensity values [21]. Images were adjusted using FIJI/ImageJ’s “rotate” and “crop” so that fibers were horizontal and included all fiber signal with an additional 5 pixels of either ridge [22]. Processed images were imported into MATLAB (The Mathworks Inc., Natick, MA) as 2D matrices containing fiber fluorescence intensity values horizontally and vertically discretized into one-pixel increments. Since fiber intensity correlates with fiber width, the average intensity value of a pixel column was treated as apparent fiber width [23]. The apparent width at each location along a fiber at one time point will be referred to as a cross-sectional intensity profile (Supplement Section A.3 and Supplemental Figure 2). To look for correlations between fiber properties and for trends between fibers, we normalized each fiber’s intensity and length values (Supplement Section A.6 and Supplemental Figures 6&7).
2.4. Determining the Rate of Fiber Digestion
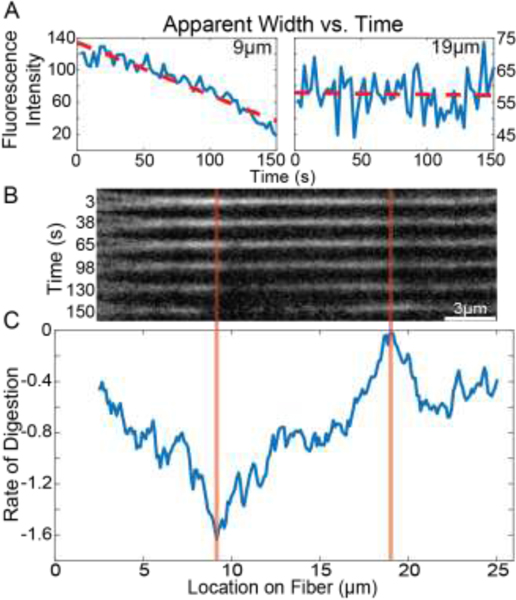
Linear fits were applied to the cross-sectional intensity profile values over time at each location along the fiber using MATLAB’s “polyfit” function and the resulting slope values correspond to the rate of change in cross-sectional intensity at that location. These digestion rates were plotted for each location on the fiber, representing the rate of change in the apparent fiber width over time versus location (Figure 1). In MATLAB a spline was fit to digestion rates and prominent minima of the fit function, interpreted as digestion sites, were found using the MATLAB “islocalmin” function (Supplement Section A.7 and Supplemental Figure 8A).
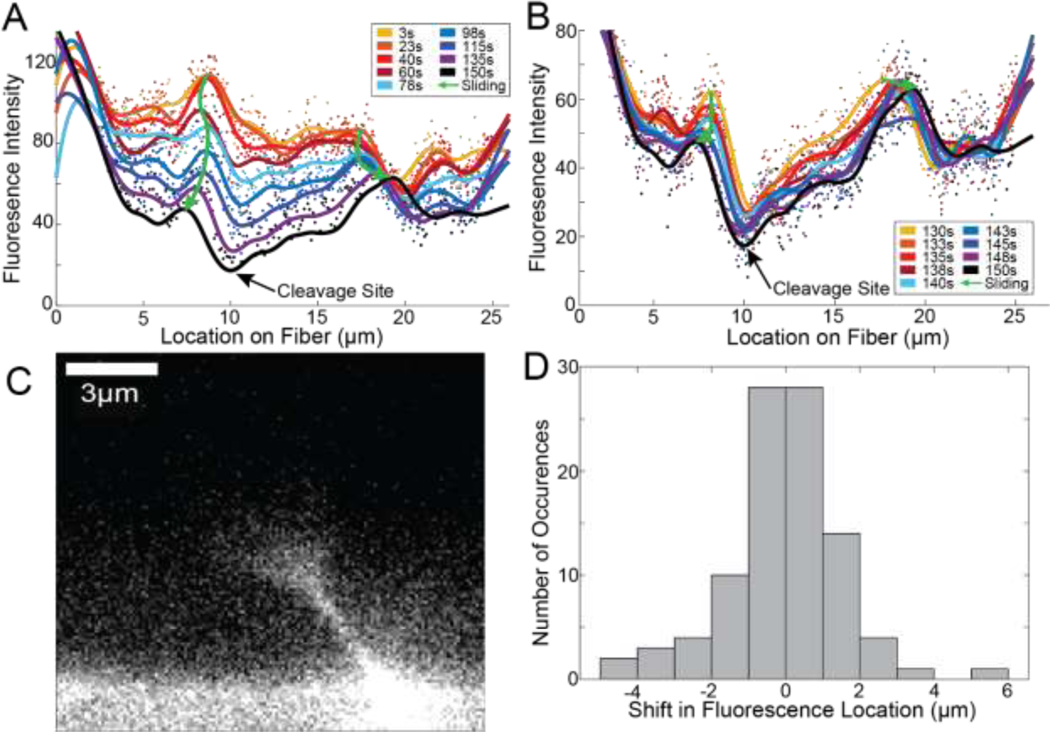
Figure 1: Fiber Fluorescence Intensity and Digestion Rates.
A) Plots showing the average cross-sectional fluorescence intensity as a function of time for representative locations. Locations of 9 μm and 19 μm are shown, which correspond to the locations marked by the orange lines in panel B. Red lines show linear fits to the data which allow a quantification of the average rate of change of the fluorescence intensity, and hence the average rate of change of fiber diameter. B) Snapshots of the fibrin fiber showing the fluorescence intensity along the fiber at different points in time. C) The average rate of change of fluorescence intensity at every location along the fiber. Rate of digestion was calculated at each location as the slope of the fluorescence vs time plot at that location (red lines in panel A).
2.5. Mathematical Model
We created a stochastic mathematical model, based on the experimental set-up described above, to estimate the time course of plasmin diffusion and binding (Supplement Section C). The model domain consisted of a fibrin free region through which plasmin could diffuse and four fibrin fibers to which plasmin could bind. At each time step, each plasmin molecule had a probability of diffusing, binding (if it was near a fiber), and unbinding (if it was bound). We recorded the location and bound/unbound status of each plasmin molecule at each time step, which allowed us to plot time courses of the number of bound plasmin molecules and to calculate the average time it took for plasmin to first bind to any fiber. We numerically solved a reaction diffusion master equation on a lattice using a kinetic Monte Carlo-like algorithm [24–26] with fixed time steps (Supplement Section C.5). The state of all plasmin molecules was updated at each fixed time step.
The model domain was a 3-dimensional square lattice (Supplemental Figure 12A), 4 nodes wide and 60 nodes tall, with lattice edges connecting adjacent horizontal and vertical nodes, and with one lattice edge coming out of the plane of the page at each node. To best mimic the experimental conditions, the length of each edge was 20 μm (corresponding to the length of a fibrin fiber), and nodes were 0.0727 μm wide (corresponding to the diameter of a fibrin fiber). The bottom four 3-dimensional edges (highlighted in pink in Supplemental Figure 12A), were assumed to be fibrin fibers, to which plasmin could bind. The remaining 652 edges, which we call “ghost edges”, were simply used to demarcate space and were used for plasmin diffusion as described below. For simplicity, the model considered a small 3-D volume that was only 1 edge thick in the direction coming out of the page with imposed periodic boundary conditions in that dimension (no-flux boundary conditions were used in the horizontal and vertical dimensions). This periodic slab of clot was assumed to run between edge midpoints, so the 1-edge depth was actually composed of 2 halves of 2 separate edges.
A fixed number of plasmin molecules (Supplement Section C.4) were randomly uniformly distributed on the ghost edges. For simplicity, we assumed every plasmin molecule was at the midpoint of the edge it occupied. At each time step, each plasmin molecule could diffuse to one of its 8 nearest neighbors (Supplemental Figure 12B, fewer neighbors if plasmin was on the boundary) with equal probability or remain on the current edge, and if the plasmin molecule was on an edge containing fibrin, it could bind with some probability. Bound molecules had an unbinding probability at each time step, and unbound molecules on a fibrin fiber had a binding probability. The model algorithm is outlined in detail in Supplement Section C.5.
3. Results
3.1. Digestion Rates
We interrogated the digestion rates at every location along the length of the fiber and refer to this as the change in apparent fiber width. Figure 1A shows representative plots of the cross-sectional intensity vs time at two locations fit with the linear trend line whose slope is the digestion rate. Figure 1C shows the rate of digestion at every location along the fiber. Digestion rates are not uniform along the fiber length, with some locations changing rapidly (more negative), while other locations have very little change in width (value closer to zero). This was true for all 60 fibers investigated with this approach.
3.2. Digestion and Cleavage Sites
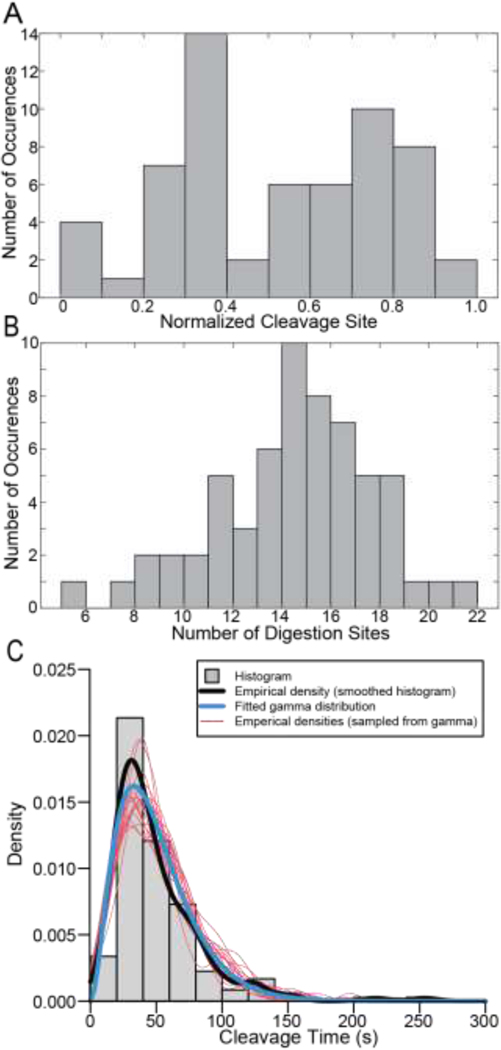
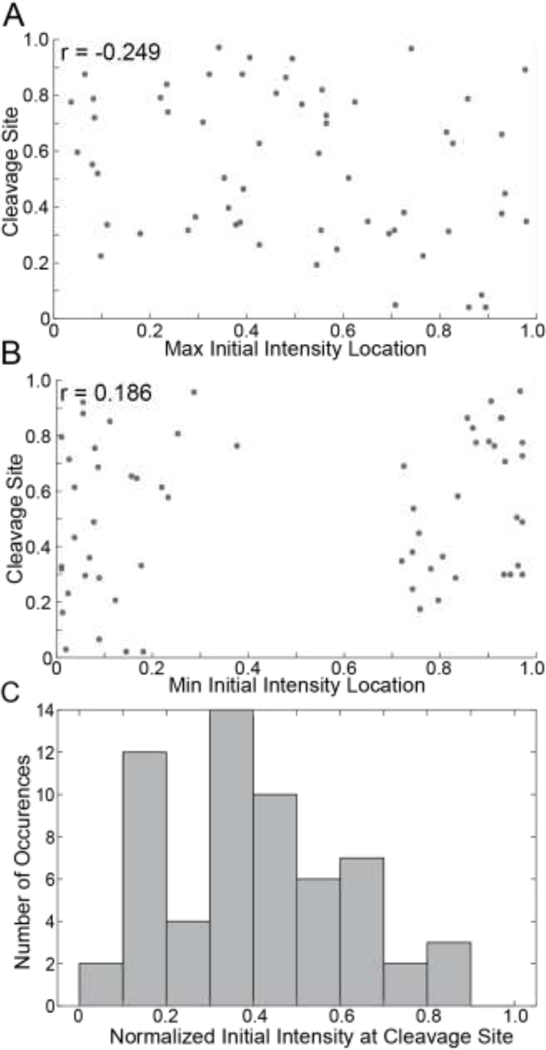
To interrogate trends in fiber cleavage locations we generated a histogram of the normalized cleavage site locations (Figure 2A) which shows that the cleavage site is not random, and that fibers were most likely to break 20–40% away from the fiber edges. We also investigated the number of digestion sites along the fiber (Figure 2B and Supplemental Figure 8) finding a mean number of 14 digestion sites with a range of 6 to 22 sites (Figure 2B).
Figure 2: Location of the Cleavage Site, Number of Distinct Digestion Sites and Cleavage Times.
A) Histogram showing the normalized location of cleavage sites of fibrin fibers. B) Histogram showing the number of digestion sites on a single fibrin fiber. 60 fibers were used for these analyses. Locations were normalized as described in Supplemental Section A.6 and Supplemental Figures 6&7. C) Histogram showing the time taken for fibers to be cleaved (gray), as well as an empirical density function (black) corresponding to the continuously measured cleavage times, fitted gamma distribution (blue), and empirical density functions calculated for twenty independent samples (each of size n = 178) from the fitted gamma distribution (red).
3.3. Cleavage Times
Every included fiber was digested at multiple locations along its length before being transversely cleaved into two distinct parts. To quantify the time it took plasmin to cleave the fiber after arriving, we analyzed each frame for any movement in the fiber indicating that lysis had begun. Starting with the first frame where any movement/digestion was observed, we then measured the number of frames until the fiber was cleaved. Using a frame rate of 2.5 s/frame, we then converted this into the cleavage time for the fiber. Cleavage times (sample mean = 49.782 s, sample SD = 34.311, n = 178) are fitted by a gamma distribution with shape parameter α = 2.956 and rate parameter β = 0.059, estimated simultaneously using the ‘glm()’ function in R [27]. The gamma distribution, a two-parameter distribution from the exponential family of distributions, has applications to event waiting times. The exponential distribution is a special case of the gamma distribution with a shape parameter α = 1, corresponding to the waiting time until the first occurrence of an event at rate β. The gamma distribution gives the waiting time until the αth event, when each event occurs independently at an exponential rate β. A possible interpretation to this fit by the gamma distribution is that cleavage is the result of an underlying process, events of which take place independently at a rate of β = 0.059 per second, of which approximately three events (α = 2.95) must occur before cleavage itself occurs. The fitted gamma distribution (blue) is superimposed on the raw empirical density (black) and histogram (gray) (Figure 2C). To assess the fit visually, we show empirical density functions for each of twenty simulated datasets sampled from the fitted distribution (red curves) (Figure 2C). Cleavage times were used for all 178 fibers with the only selection criterion being transection of the fiber.
3.4. Mathematical Model
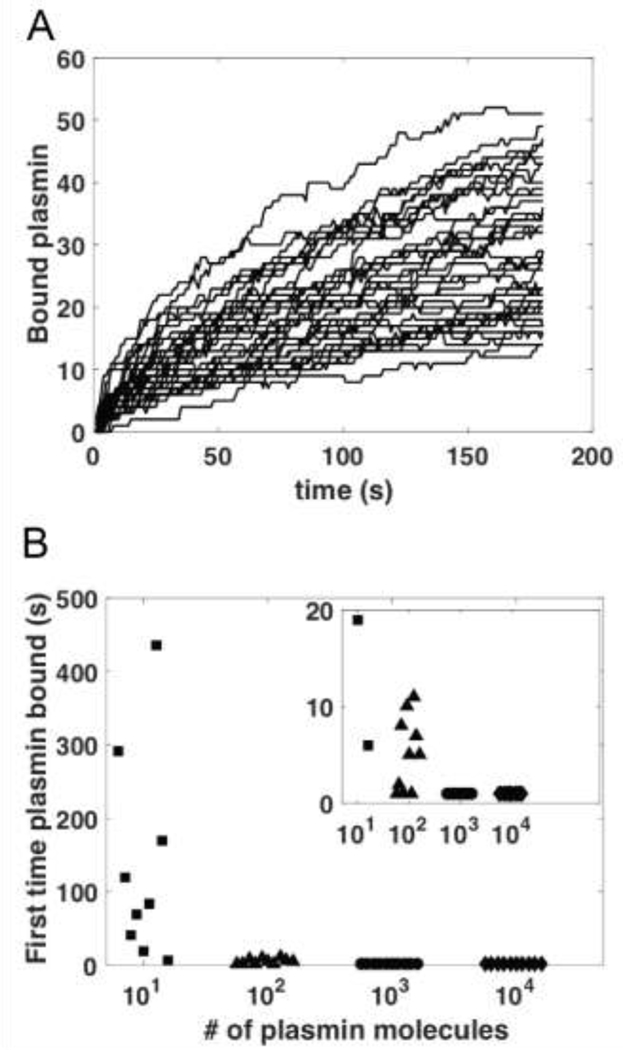
To better understand the number of experimentally identified digestion sites, we used our mathematical model to plot the time course of the number of bound plasmin molecules on 40 distinct fibers (Figure 3A). After 50 seconds (the experimental mean cleavage time), the model predicts between 5 and 27 bound plasmin molecules, with a mean of 14.25.
Figure 3: Mathematical Model Results.
A) Time course of plasmin binding to fibrin fibers. Individual curves show the data for the number of plasmin molecules bound to a distinct fiber as a function of time. Simulations (which included 4 fibrin fibers) were initialized with 1426 plasmin molecules randomly distributed throughout the domain and were repeated 10 times. B) The time at which plasmin first bound to any fiber, as a function of the order-of-magnitude number of plasmin molecules used in the simulation (details of plasmin number in Supplement Section C.4). 10 independent simulations were run for each plasmin amount. Inset is zoomed in version of larger figure. For both 103 and 104 plasmin molecules, plasmin first bound to a fiber within the first second in all 10 simulations.
The model was also used to study the mean first passage time [28,29], or how long it takes plasmin to first bind to a fiber (Figure 3B). We find that for model simulations initialized with 103 or more plasmin molecules uniformly distributed throughout the domain, the first plasmin binding always occurs within 1 second (Supplemental Figures 14&15). Reducing the number of plasmin molecules results in longer first binding times and more variability.
3.5. Correlations
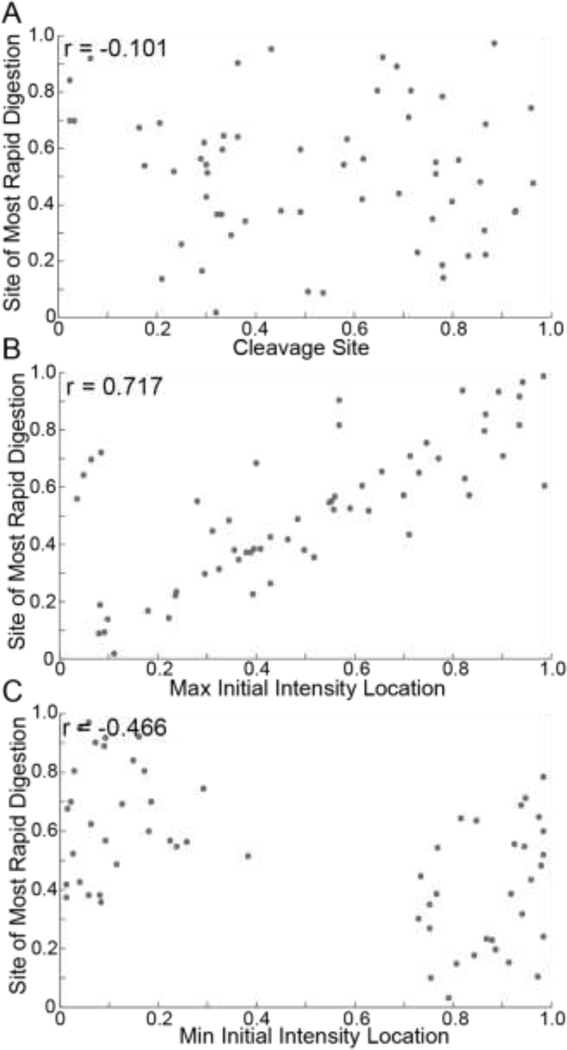
We investigated correlations between fiber characteristics and digestion rates. First, we find no significant relationship between the location of the maximum rate of digestion and the location of the cleavage site (Figure 4A). However, the site of the maximum rate of digestion and the location of the initially thickest part of the fiber (Figure 4B) shows a stronger correlation. A weak negative correlation exists between the maximum digestion rate and the initially thinnest part of the fiber (Figure 4C). Intriguingly, the initially thinnest part of the fiber never occurs in the middle, but rather in the 20% of the fiber closest to either end.
Figure 4: Relationships Between Fiber Characteristics and Digestion Rates.
A) Scatter plot of the site of most rapid digestion vs the cleavage site. B) Scatter plot of the site of most rapid digestion vs the location of maximum initial intensity. C) Scatter plot of the site of most rapid digestion vs the location of minimum initial intensity. 60 fibers were used for these analyses. All Pearson coefficients (r) were found with a 95% confidence interval. Locations were normalized as described in Supplemental Section A.6 and Supplemental Figures 6&7.
Additionally, we investigated correlations between fiber characteristics and the location of the cleavage site. We find very little correlation between the initially thickest and thinnest location of the fiber and the cleavage site (Figure 5A&B). To investigate the importance of initial fiber width, we created a distribution of the normalized initial fluorescence intensity values at the cleavage site of every fiber (Figure 5C). Cleavage sites most often correspond to thinner parts of the fiber, which have an initial relative width of roughly 20–40% of the maximum. Bins corresponding to the thickest (> 70%) parts of the fiber are relatively unpopulated. We also investigated the distance between the cleavage site and the location of the maximum initial fiber width (Supplemental Figure 10). While Figure 5A confirms there is no correlation between cleavage site location and maximum initial intensity, Supplemental Figure 10C additionally indicates that the relative distance between the cleavage site location and the location of maximum initial fiber width can be quite large. Thus, fibers are likely to be cleaved at thinner sites, but rarely the thinnest or thickest location on the fiber, and the cleavage sites can be close or far from the thickest part of the fiber.
Figure 5: Relationships Between Fiber Characteristics and Cleavage Sites.
A) Scatter plot of the site of initial maximum intensity vs cleavage site. B) Scatter plot of the site of initial minimum intensity vs cleavage site. All Pearson coefficients (r) were found with a 95% confidence interval. C) Histogram showing the normalized initial intensity at cleavage sites. 60 fibers were used for these analyses. Locations were normalized as described in Supplemental Section A.6 and Supplemental Figures 6&7.
3.6. Longitudinal Fluorescence Dynamics
Next, we analyzed trends in the cross-sectional fiber intensity at each location along the fiber during digestion. We observe that in the frame before digestion, the minimum in the intensity vs location plot corresponds with the observed cleavage site in the images (Figure 6A&B, black curve). In the frame after the cleavage event, the ends of 52% of fibers appear to form a V-shaped, cone-like pattern (Figure 6C). This conelike structure persists for an average of 13.5 and maximum of 45 seconds after cleavage.
Figure 6: Structural Evolution of a Representative Fiber.
A) Plot showing traces of the average cross-sectional fluorescence intensity for a representative fiber at multiple instances in time. Peaks in the traces change locations longitudinally along the fiber, termed fluorescence sliding. Two prominent peaks are marked with green arrows but are not the only fluorescence sliding occurring. B) Plot showing a zoom-in of the data shown in panel A but looking at the average cross-sectional fluorescence intensity for the last 9 timepoints before cleavage. C) The first frame after a fiber’s cleavage shows a cone-like structure, such as the one seen in this representative image, in 52% of fibers. D) Histogram showing the displacement of 95 peaks from 45 fibers in the last 20 seconds of their digestion. A negative shift represented a leftward movement, and a positive shift represented a rightward movement.
In analyzing time-series data (Figure 1B and Supplemental movies) it appears that fluorescence intensity slides longitudinally along the fiber during digestion with a majority of the sliding occurring towards the end of digestion. We tracked the location of peaks in the cross-sectional fluorescence intensity profiles over time during digestion (representative intensity curves in Figure 6A&B) and found that peaks shift over time, to the left and right. We tracked 95 peaks in 45 fibers over the last ~20s of digestion and found that most peaks shifted 1–2 μm but shifts up to 6 μm were observed (Figure 6D). Peaks typically shift away from digestion sites (Figure 6A&B). The initial direction of shifting does not determine the overall path of the intensity peak as shown by either green arrow in Figure 6.
4. Discussion
The results from this study help to resolve longstanding disagreements on mechanisms of fiber digestion, while also revealing new mechanisms that may play dramatic roles in regulating fibrinolysis. Previous studies, primarily utilizing fluorescence and scanning electron microscopy (SEM), indicated that fibers were transversely cleaved at a single point [13–16,30]. Hence, it was suggested that the formation of free C-terminal lysines during plasmin digestion creates a feedback mechanism, recruiting additional plasmin molecules to digestion sites and causing transverse cleavage at those sites [17,31]. Additionally, geometrical considerations suggest that plasmin molecules can crawl between binding sites on protofibrils, which would also promote transverse cleavage [17,32,33]. On the other hand, studies primarily using turbidity and atomic force microscopy (AFM) have suggested that the diameter of fibrin fibers decreases uniformly during lysis due to plasmin molecules binding and digesting the fiber radially along its entire length [10–12].
Our results indicate that digestion does occur along the entirety of a fibrin fiber, but that digestion rates are nonuniform. The apparent digestion rates are actually a result of two phenomena: digestion by plasmin and fibrin longitudinal sliding. The loss of cross-sectional fluorescence intensity in our experiments (Figure 1A & 6A&B) suggests that digestion by plasmin results in the loss of molecules or sections of protofibrils as plasmin severs their connections with the other parts of the fiber. The longitudinal shifting of the fluorescence intensity along the length of the fiber during digestion (Figure 6A&B) indicates that fibrin molecules also slide along the fiber. Both processes result in local thinning of the fiber and contribute to its eventual cleavage. These results validate both transverse cleavage and radial digestion as mechanisms contributing to the lysis of fibrin fibers, but also expose their individual incompleteness as comprehensive descriptions of the process. A wholistic account of the mechanisms of fiber digestion must include non-uniform digestion along the length of the fiber by multiple molecules, digestion across the fiber by individual molecules, and the longitudinal sliding of fibrin.
Our experimental setup had two advantages over previous studies, which allowed these insights. First, with respect to previous fluorescence-based studies of lysis, our time resolution (2.5s) was faster [30,32], allowing us to observe the digestion of single fibers. In addition, labeling our fibers with Alexa488 provided a more uniform fiber labeling than previous studies that used fluorescent microspheres [13,15,34], and likely resulted in less fiber elongation (data not shown). Second, unlike AFM-based experiments, where fibers are adsorbed to surfaces and sometimes dried and rehydrated [10,18], our fibers were suspended in solution, allowing plasmin to digest from all dimensions and fibrin to both leave and slide along the fiber during digestion. Adsorption would prevent some of the molecules from leaving the fiber and prevent the sliding of molecules along the fiber during digestion, which could explain why fibers appear to briefly “swell” during digestion in some AFM experiments [18].
The observation of fluorescence longitudinally sliding along the fiber was surprising and constitutes a previously unknown mechanism during digestion. Sliding often started slowly but became more prominent towards the end of digestion, and typically occurred as fluorescence signal moved away from digestion sites, in either direction. This sliding of signal clearly indicated that the fluorescently labeled fibrin was changing locations along the fiber. This could not be a result of plasmin digesting longitudinally down the fiber, or that would result in a loss of fluorescence, as opposed to a change in location of the fluorescence signal. Interestingly, sliding could start one direction, but then move back in another direction (left green arrow in Figure 6A). Because there are multiple digestion sites on the fiber, this is likely the result of competing digestion sites within the fiber shifting the distribution of tension along the fiber, causing the direction of sliding to change.
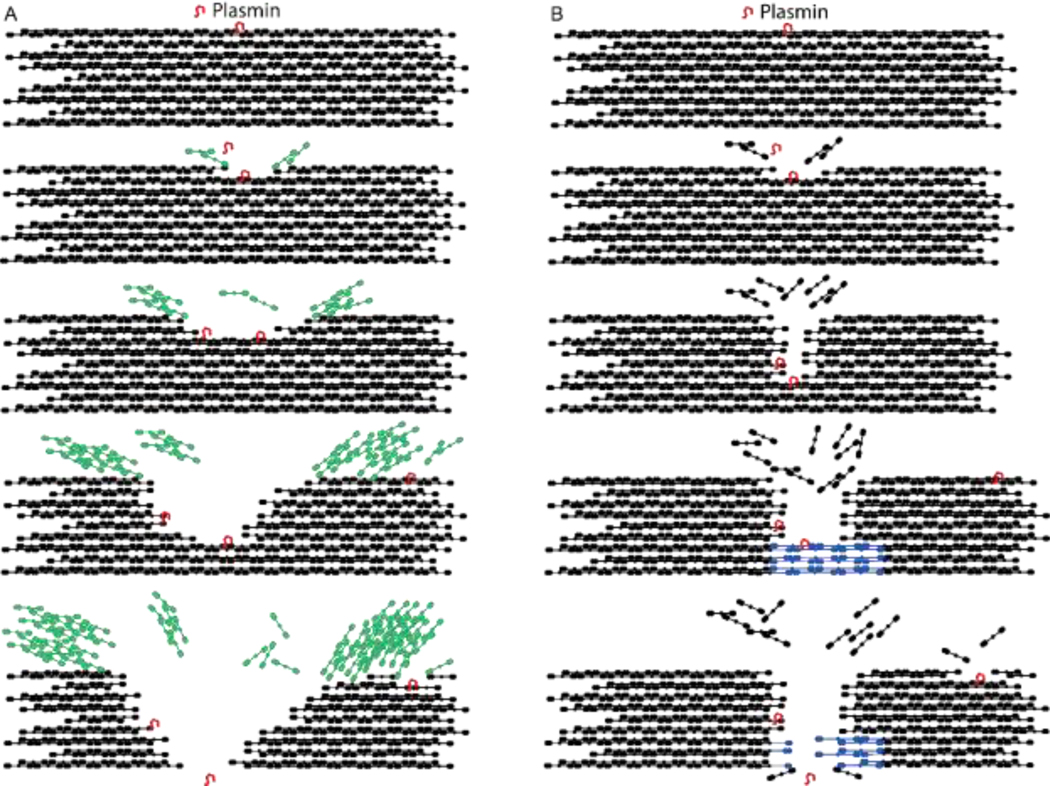
The mechanisms responsible for fibrin sliding likely depend on the inherent tension present in fibrin fibers [13,15]. Fibrin fibers polymerize into a state of tension, although the exact mechanisms responsible for the fiber tension have been debated [13,19,20]. The observation of cone-shaped ends of cleaved fibers (Figure 6C) is consistent with previously described “splayed ends” observed in EM studies of fibrinolysis [16] and indicates that protofibrils explode backward due to tension after being cut. Recent work indicates that inherent fiber tension plays a predominant role in regulating the clearance of fibrin from a region during fibrinolysis [15], and fluorescence sliding implies that tension regulates the digestion of individual fibers as well. We propose two possible mechanisms (models) that could account for tension-dependent fibrin sliding (Figure 7).
Figure 7: Models for Single Fiber Fibrinolysis.
A) Protofibril recoil model: Protofibrils, which are under tension in fibers, spring backwards as they are cleaved (in green), similar to how the strands of a tensed rope spring backward as they are cut. The springing backwards causes the fiber to appear as if it’s being pulled apart. B) Molecular stretching model: Protofibrils are under tension in fibers, and as protofibrils are cut, that tension is spread out across fewer and fewer protofibrils. Eventually the force on each protofibril is enough to unfold the remaining fibrin molecules (in blue) making up the protofibril, causing the fiber to slide apart. Here we show cut fibrin molecules leaving the fiber, rather than the protofibrils recoiling, to distinguish the mechanisms, but protofibril recoil could also occur concurrently with the molecular stretching model shown in panel B.
The first model (protofibril recoil, Figure 7A) is an analogy to a tensed rope, where, as the strands of the rope are cut, they spring backwards, leading to fiber “fraying”. Within a fiber, tensed protofibrils will also spring backwards as they are digested by plasmin, which is consistent with the observed cone-shaped fiber ends (Figure 6C) and would lead to the appearance of fibrin sliding along the fiber. SEM images showing a registry of fibrin molecules along the fiber [19,35], along with recent polymerization and structural models [36,37], suggest that interaction between molecules along the length of the fiber would limit the extent to which a protofibril could move backwards in any single recoil event. We investigated the distance of individual sliding events between consecutive frames (Supplemental Figure 11), but a clear trend was not present, perhaps because a frame rate of 2.5 s could not distinguish molecular scale events. If the protofibril recoil model is correct, this would constrain models of fiber tension, because sliding (recoil) occurs predominantly towards the end of digestion, and thus tension must be present in the fiber until it is fully cleaved. Models where tension originates from twisting of protofibrils around the fiber would suggest that tension is highest at thicker parts of the fiber and decreases during digestion.
The second model (molecular stretching, Figure 7B) relies on the observation that individual fibrin molecules can unfold under force [38–41] and assumes that the tension remains relatively constant as the fiber is digested. Since inherent fiber tension is distributed throughout the cross section of a fibrin fiber, as protofibrils are cut, the tension must be redistributed among the remaining protofibrils, increasing their individual tensions. Eventually the molecular tension will be high enough (~100 pN) to unfold fibrin molecules, causing the intact molecules to begin stretching out and resulting in the longitudinal sliding of fibrin.
Additional experiments are required to decipher the mechanisms of fibrin sliding and the role of tension in fiber digestion. Two ways that fibrin sliding could facilitate more rapid digestion are: 1) exposure of previously buried binding/cleavage sites to promote digestion of the remaining molecules and 2) clearance of fibrin away from transverse paths across the fiber, thereby removing adjacent binding sites which would otherwise “distract” plasmin from cutting across the fiber. The protofibril recoil mechanism would result in both exposure and clearance while the molecular stretching mechanism would only result in the latter. It is possible that both sliding mechanisms play a role, for example in the early stages of fiber degradation, the inherent tension in fibers may cause protofibrils to recoil away from the digestion sites, while towards the end of digestion, molecular unfolding could occur.
Both our experimental and computational results suggest that a 20-μm-long fiber will have on average 14 distinct digestion sites when exposed to 10 μL of ~1.43 μM plasmin solution. Additionally, model results show that at this plasmin concentration, the first plasmin molecule will bind to a fiber within the first second of the experiment, which is something that cannot be detected experimentally. The contrast between the experimental observation that sites between the fiber middle and edges have a higher probability of cleavage (Figure 2A) and the modeling results indicating that the first plasmin molecules will randomly bind anywhere along the fiber suggests that there may be something about these locations that make them predisposed to being cut. Future experiments should investigate possible mechanisms for this observation. Taken together, the modeling data underscore how discretely and quickly plasmin can act and provide the most fine-scale information to-date about the timing and action of plasmin on single fiber lysis.
Furthermore, our model is the first microscale model of plasmin-fibrin interaction, and it can be used to elucidate other factors affecting fibrinolysis, e.g., the impact of plasmin binding rate (Supplement Section C.6). Studies like these can be used to suggest ideal features of future therapeutics, and when combined with experimental data, to estimate unknown physiological rates. Coordinated microscale experimental and modeling work can be a powerful tool for uncovering mechanisms of lysis that are impossible to do with experimentation or computation alone. For example, coupling our model and experimental data allowed us to estimate the amount of plasmin (0.0001% of initial amount) that immediately infiltrates the entire experimental volume when a 10 μL droplet containing plasmin is added to an existing 10 μL droplet (Supplement Section C.4). The work presented here can also be extended to study – for the first time at the scale of individual fibers – the positive feedback mechanism by which plasmin degradation of fibrin exposes additional binding sites for lytic enzymes [17,32,33], thereby accelerating lysis. Our single fiber approach has the potential to uncover significant new information about this positive feedback and its implications for therapeutics.
Our data also allow a careful investigation into fiber properties that may govern digestion rates. The strongest correlation we found was between the most rapid digestion site and the initially thickest part of the fiber (Figure 4B). This is likely a consequence of the observed longitudinal sliding. When fibrin slides away from a digestion site, this will give the appearance in fluorescence data that a large amount of fibrin has digested at that location. Thus, the larger the initial intensity that slides, the larger the measured digestion rate at that location will be.
At first glance the lack of correlation between the site of most rapid digestion and the eventual cleavage site is surprising (Figure 4A). This indicates that while the tension dependent fibrin sliding is important in fiber thinning, additional factors contribute to fiber cleavage. Digestion sites occur at all points along the fibers (Supplemental Figure 8B). However, the cleavage sites on fibers typically occur in thinner parts of the fiber (46/60 fibers are cleaved in locations that were in the lower 50% of initial fiber thickness, Figure 5C), but only 2/60 fiber cleavage locations were in the lowest 10% of the initial thickness. In addition, there was little correlation between the initially thickest or thinnest parts of the fiber and the cleavage site (Figure 5A&B).
In these experiments we investigated the digestion of fibrin by plasmin, which is important given plasmin‟s potential use as a fibrinolytic [4]. The concentration of plasmin used (~1.43 μM) was similar to physiological plasminogen concentrations (2 μM) [42]. These concentrations were chosen, in part, to optimize imaging conditions, allowing fibers to be digested over the course of ~1 minute, enabling an observation of the lysis mechanisms while limiting the effects of photobleaching (Supplemental Figure 10). The local concentration of plasmin next to a fiber is unknown and depends on the balance of activators and inhibitors, so 1.43 μM is reasonable.
While these experiments are an exciting first step toward a deeper understanding of mechanisms of fiber digestion, future efforts should corroborate these results under more physiological conditions. For example, initiating lysis with tPA or uPA and plasminogen would enhance our understanding of the role of plasminogen activation during digestion. Moving from a 2-D experimental lysis model of individual fibers to a 3-D fibrin mesh will also be an important future step as interactions with other fibers will influence the lysis process in vivo [15,30]. There are also many other plasma proteins and cells that influence clot formation and lysis whose effects on fibrin digestion could be studied using this model. For instance, α2-antiplasmin and thrombin activatable fibrinolysis inhibitor (TAFIa) inhibit lysis by either inhibiting plasmin or by cleaving plasmin binding sites, respectively [43]. Understanding how these inhibitors, in addition to the influence of FXIIIa crosslinking, affect microscale lysis mechanisms is important.
The mathematical model, which in its present form only allows for (un)binding and diffusion of plasmin and does not account for cleavage of fibers, can be modified to include lysis and to reflect more physiological conditions. In particular, a version of the model already exists for lysis initiated by tPA [32], and both the tPA- and plasmin-initiated lysis models can be modified to include inhibitors. Experiments and the mathematical model can be iteratively adjusted and improved to explore further mechanisms of lysis under different physiological and pathological conditions.
Pathological conditions alter fibrinolysis and are important to understand mechanistically. One particularly relevant application in light of the global COVID-19 pandemic is the observation that the SARS-CoV-2 spike protein alters clot structure and hinders fibrinolysis [44]. Clots from COVID patients also contain “microclots” with increased α2-antiplasmin and amyloid deposits [45]. Fibrinogen and/or fibrin have been shown to participate in amyloid formation with other proteins including β-amyloid in Alzheimer‟s disease [46,47], serum amyloid A [48], and amylin fibrins in type 2 diabetes [49], often leading to hindered fibrinolysis, perhaps due to decreased binding of plasmin(ogen) [46] or altered clot structures. Fibrin has also been shown to form “plate” or “sheet” [20] structures, the presence of which are increased under amyloidogenic conditions [50]. The experimental approaches described herein could help to uncover the mechanisms responsible for aberrant lysis in many of these disorders by observing fiber digestion in conditions mimicking these pathological situations. We hypothesize that in amyloidic fibrin fibers, the fibrin sliding mechanism may be altered due to amyloid β-sheet structures locking fibrin molecules into place.
We conclude that studying the lysis of individual fibers reveals previously undiscovered mechanisms of fiber digestion. Fiber lysis is a balance of digestion by plasmin and tension-dependent fibrin sliding. The presence of fibrin sliding was an unexpected mechanism and adds to the growing body of evidence that inherent fiber tension plays important roles in regulating fibrinolysis [15]. The digestion mechanisms revealed in these studies provide a new framework for future biochemical and clinical studies investigating factors that alter blood clot digestion. This, in turn, will help to unravel the mystery of why up to 20% of clots are resistant to thrombolytic treatments [6] and inform the development of next-generation therapeutics.
Supplementary Material
Statement of Significance.
We developed a method for interrogating lysis of individual fibrin fibers, enabling the time-resolved observation of individual fiber digestion for the first time. Our results resolve longstanding disagreements about fibrinolytic processes and reveal previously unknown mechanisms that also play a role. Also, we developed the first microscale mathematical model of plasmin-fibrin interaction, which predicts the number of plasmin molecules on each fiber and can serve as a framework for investigating novel therapeutics.
Acknowledgements
NEH gratefully acknowledges support by Grant Number HL148842. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the NHLBI or NIH. BEB and SML gratefully acknowledge support from the University of Central Oklahoma College of Mathematics and Science CURE-STEM award. SRL gratefully acknowledges support from the ECU URCA and UR Mini awards.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- [1].Virani SS, Alonso A, Aparicio HJ, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Cheng S, Delling FN, Elkind MSV, Evenson KR, Ferguson JF, Gupta DK, Khan SS, Kissela BM, Knutson KL, Lee CD, Lewis TT, Liu J, Loop MS, Lutsey PL, Ma J, Mackey J, Martin SS, Matchar DB, Mussolino ME, Navaneethan SD, Perak AM, Roth GA, Samad Z, Satou GM, Schroeder EB, Shah SH, Shay CM, Stokes A, VanWagner LB, Wang N, Tsao CW, Heart Disease and Stroke Statistics-2021 Update: A Report From the American Heart Association, Circulation. 143 (2021) e254–e743. [DOI] [PubMed] [Google Scholar]
- [2].Hudson NE, Biophysical Mechanisms Mediating Fibrin Fiber Lysis, BioMed Research International. 2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tissue Plasminogen Activator for Acute Ischemic Stroke (Alteplase, Activase®),. [Google Scholar]
- [4].Mitchell Peter J., FRACP B. Yan, Brozman Miroslav, Marc Ribo, Marder V, Courtney KL, Saver Jeffrey L., FAHA, FAAN, FANA, Plasmin (Human) Administration in Acute Middle Cerebral Artery Ischemic Stroke: Phase 1/2a, Open-Label, Dose-Escalation, Safety Study, Journal of stroke and cerebrovascular diseases. 26 (2016) 308–320. [DOI] [PubMed] [Google Scholar]
- [5].Liu X, Beyond the Time Window of Intravenous Thrombolysis: Standing by or by Stenting? Interventional neurology. 1 (2012) 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kluft C, Sidelmann JJ, Gram JB, Assessing Safety of Thrombolytic Therapy, Semin Thromb Hemost. (2016). [DOI] [PubMed] [Google Scholar]
- [7].Wolpert SM, Bruckmann H, Greenlee R, Wechsler L, Pessin MS, Del Zoppo GJ, Neuroradiologic evaluation of patients with acute stroke treated with recombinant tissue plasminogen activator, American journal of neuroradiology : AJNR. 14 (1993) 3–13. [PMC free article] [PubMed] [Google Scholar]
- [8].Schielen WJG, Voskuilen M, Tesser GI, Nieuwenhuizen W, The sequence Aα (148–160) in fibrin, but not in fibrinogen, is accessible to monoclonal antibodies, Proceedings of the National Academy of Sciences of the United States of America. 86 (989) 8951–8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schielen WJ, Adams HP, van Leuven K, Voskuilen M, Tesser GI, Nieuwenhuizen W, The sequence gamma-(312–324) is a fibrin-specific epitope, Blood. 77 (1991) 2169–2173. [PubMed] [Google Scholar]
- [10].Blinc A, Magdic J, Fric J, Musevic I, Atomic force microscopy of fibrin networks and plasma clots during fibrinolysis, Fibrinolysis & Proteolysis. 14 (2000) 288–299. [Google Scholar]
- [11].Diamond SL, Anand S, Inner clot diffusion and permeation during fibrinolysis, Biophysical Journal. 65 (1993) 2622–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gabriel DA, Muga K, Boothroyd EM, The effect of fibrin structure on fibrinolysis, Journal of Biological Chemistry. 267 (1992) 24259–24263. [PubMed] [Google Scholar]
- [13].Bucay I, O’Brien E Tim 3, Wulfe SD, Superfine R, Wolberg AS, Falvo MR, Hudson NE, Physical determinants of fibrinolysis in single fibrin fibers, PloS one. 10 (2015) e0116350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Collet J, Lesty C, Montalescot G, Weisel JW, Dynamic changes of fibrin architecture during fibrin formation and intrinsic fibrinolysis of fibrin-rich clots, Journal of Biological Chemistry. 278 (2003) 21331–21335. [DOI] [PubMed] [Google Scholar]
- [15].Cone SJ, Fuquay AT, Litofsky JM, Dement TC, Carolan CA, Hudson NE, Inherent fibrin fiber tension propels mechanisms of network clearance during fibrinolysis, Acta Biomaterialia. 107 (2020) 164–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Veklich Y, Francis CW, White J, Weisel JW, Structural studies of fibrinolysis by electron microscopy, Blood. 92 (1998) 4721. [PubMed] [Google Scholar]
- [17].Weisel J, Litvinov R, The biochemical and physical process of fibrinolysis and effects of clot structure and stability on the lysis rate, Cardiovascular & Hematological Agents in Medicinal Chemistry. 6 (2008) 161–180. [DOI] [PubMed] [Google Scholar]
- [18].Feller T, Hársfalvi J, Csányi C, Kiss B, Kellermayer M, Plasmin-driven fibrinolysis in a quasi-two-dimensional nanoscale fibrin matrix, Journal of Structural Biology. 203 (2018) 273–280. [DOI] [PubMed] [Google Scholar]
- [19].Weisel JW, Nagaswami C, Makowski L, Twisting of fibrin fibers limits their radial growth, Proceedings of the National Academy of Sciences of the United States of America. 84 (1987) 8991–8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].O’Brien ET, Falvo MR, Millard D, Eastwood B, Taylor RM, Superfine R, Ultrathin self-assembled fibrin sheets, Proceedings of the National Academy of Sciences of the United States of America. 105 (2008) 19438–19443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sternberg SR, Biomedical Image Processing, IEEE Computer. 16 (1983) 22–34. [Google Scholar]
- [22].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis, Nature methods. 9 (2012) 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li W, Sigley J, Baker SR, Helms CC, Kinney MT, Pieters M, Brubaker PH, Cubcciotti R, Guthold M, Nonuniform Internal Structure of Fibrin Fibers: Protein Density and Bond Density Strongly Decrease with Increasing Diameter, BioMed Research International. 2017 (2017) 6385628–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gillespie DT, Hellander A, Petzold LR, Perspective: Stochastic algorithms for chemical kinetics, The Journal of chemical physics. 138 (2013) 170901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Isaacson SA, Peskin CS, Incorporating Diffusion in Complex Geometries into Stochastic Chemical Kinetics Simulations, SIAM journal on scientific computing. 28 (2006) 47–74. [Google Scholar]
- [26].Erban R, Chapman SJ, Stochastic Modelling of Reaction–Diffusion Processes, Cambridge University Press, Cambridge, 2019. [Google Scholar]
- [27].The R Core Team, R: A language and environment for statistical computing, R Foundation for Statistical Computing. (2021). [Google Scholar]
- [28].Polizzi NF, Therien MJ, Beratan DN, Mean First-Passage Times in Biology, Israel journal of chemistry. 56 (2016) 816–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Biswas S. Iyer, Zilman A, First-Passage Processes in Cellular Biology, in: Anonymous Advances in Chemical Physics, John Wiley & Sons, Inc, Hoboken, NJ, 2016, pp. 261–306. [Google Scholar]
- [30].Collet J, Park D, Lesty C, Soria J, Soria C, Montalescot G, Weisel J, Influence of fibrin network conformation and fibrin fiber diameter on fibrinolysis speed: Dynamic and structural approaches by confocal microscopy, Arteriosclerosis, Thrombosis, and Vascular Biology: Journal of the American Heart Association. 20 (2000) 1354–1361. [DOI] [PubMed] [Google Scholar]
- [31].Sakharov DV, Rijken DC, Superficial accumulation of plasminogen during plasma clot lysis, Circulation. 92 (1995) 1883–1890. [DOI] [PubMed] [Google Scholar]
- [32].Bannish BE, Chernysh IN, Keener JP, Fogelson AL, Weisel JW, Molecular and Physical Mechanisms of Fibrinolysis and Thrombolysis from Mathematical Modeling and Experiments, Scientific reports. 7 (2017) 6914–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Weisel JW, Veklich Y, Collet JP, Francis CW, Structural Studies of Fibrinolysis by Electron and Light Microscopy, Thrombosis and Haemostasis. 82 (1999) 277–282. [PubMed] [Google Scholar]
- [34].Li W, Li R, Lucioni T, Bonin K, Cho SS, Guthold M, Stretching single fibrin fibers hampers their lysis, Acta Biomaterialia. 60 (2017) 264–274. [DOI] [PubMed] [Google Scholar]
- [35].Voter WA, Lucaveche C, Blaurock AE, Erickson HP, Lateral packing of protofibrils in fibrin fibers and fibrinogen polymers, Biopolymers. 25 (1986) 2359–2373. [DOI] [PubMed] [Google Scholar]
- [36].Rocco M, Molteni M, Ponassi M, Giachi G, Frediani M, Koutsioubas A, Profumo A, Trevarin D, Cardinali B, Vachette P, Ferri F, Pérez J, A comprehensive mechanism of fibrin network formation involving early branching and delayed single- to double-strand transition from coupled time-resolved X-ray/light-scattering detection, Journal of the American Chemical Society. 136 (2014) 5376. [DOI] [PubMed] [Google Scholar]
- [37].Klykov O, van der Zwaan C, Heck AJR, Meijer AB, Scheltema RA, Missing regions within the molecular architecture of human fibrin clots structurally resolved by XL-MS and integrative structural modeling, Proceedings of the National Academy of Sciences - PNAS. 117 (2020) 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhmurov A, Brown AX, Litvinov R, Dima R, Weisel J, Barsegov V, Mechanism of Fibrin(ogen) Forced Unfolding, Structure. 19 (2011) 1615–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Averett LE, Schoenfisch MH, Akhremitchev BB, Gorkun OV, Kinetics of the Multistep Rupture of Fibrin „A-a‟ Polymerization Interactions Measured Using Atomic Force Microscopy, Biophysical Journal. 97 (2009) 2820–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Brown AEX, Litvinov RI, Discher DE, Weisel JW, Forced Unfolding of Coiled-Coils in Fibrinogen by Single-Molecule AFM, Biophysical Journal. 92 (2007) L39–L41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hudson NE, Ding F, Bucay I, O’Brien ET, Gorkun OV, Superfine R, Lord ST, Dokholyan NV, Falvo MR, Submillisecond elastic recoil reveals molecular origins of fibrin fiber mechanics, Biophysical Journal. 104 (2013) 2671–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Collen D, Tytgat G, Claeys H, Verstraete M, Wallén P, Metabolism of plasminogen in healthy subjects: effect of tranexamic acid, The Journal of Clinical Investigation. 51 (1972) 1310–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bannish BE, Hudson NE, The utility and potential of mathematical models in predicting fibrinolytic outcomes, Current opinion in biomedical engineering. 20 (2021) 100337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Grobbelaar L, Venter C, Vlok M, Ngoepe M, Laubscher G, Lourens P, Steenkamp J, Kell D, Pretorius E, SARS-CoV-2 spike protein S1 induces fibrin(ogen) resistant to fibrinolysis: implications for microclot formation in COVID-19, Bioscience reports. 41 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pretorius E, Vlok M, Venter C, Bezuidenhout JA, Laubscher GJ, Steenkamp J, Kell DB, Persistent clotting protein pathology in Long COVID/Post-Acute Sequelae of COVID-19 (PASC) is accompanied by increased levels of antiplasmin, Cardiovascular diabetology. 20 (2021) 1–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zamolodchikov D, Strickland S, Aβ delays fibrin clot lysis by altering fibrin structure and attenuating plasminogen binding to fibrin, Blood. 119 (2012) 3342–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cortes-Canteli M, Paul J, Norris EH, Bronstein R, Ahn HJ, Zamolodchikov D, Bhuvanendran S, Fenz KM, Strickland S, Fibrinogen and β-Amyloid Association Alters Thrombosis and Fibrinolysis: A Possible Contributing Factor to Alzheimer’s Disease, Neuron (Cambridge, Mass.). 66 (2010) 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Page MJ, Thomson GJA, Nunes JM, Engelbrecht A, Nell TA, Villiers de, Willem JS, de Beer MC, Engelbrecht L, Kell DB, Pretorius E, Serum amyloid A binds to fibrin(ogen), promoting fibrin amyloid formation, Scientific reports. 9 (2019) 3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pretorius E, Mbotwe S, Kell DB, Lipopolysaccharide-binding protein (LBP) reverses the amyloid state of fibrin seen in plasma of type 2 diabetics with cardiovascular co-morbidities, Scientific reports. 7 (2017) 9680–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Pretorius E, Mbotwe S, Bester J, Robinson CJ, Kell DB, Acute induction of anomalous and amyloidogenic blood clotting by molecular amplification of highly substoichiometric levels of bacterial lipopolysaccharide, Journal of the Royal Society interface. 13 (2016) 20160539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.