Abstract
The ongoing outbreak of the COVID-19 has highlighted the importance of the pandemic prevention and control. A rapid and sensitive antigen assay is crucial in diagnosing and curbing pandemic. Here, we report a novel surface plasmon resonance biosensor based on laser heterodyne feedback interferometry for the detection of SARS-CoV-2 spike antigen, which is achieved by detecting the tiny difference in refractive index between different antigen concentrations. The biosensor converts the refractive index changes at the sensing unit into the intensity changes of light through surface plasmon resonance, achieving label-free and real-time detection of biological samples. Moreover, the gain amplification effect of the laser heterodyne feedback interferometry further improved the sensitivity of this biosensor. The biosensor can rapidly respond to continuous and periodic changes in the refractive index with a high resolution of 3.75 × 10−8 RIU, demonstrating the repeatability of the biosensor. Afterwards, the biosensor is immobilized by the anti-SARS-CoV-2 spike monoclonal antibodies, thus realizing the specific recognition of the antigen. The biosensor exhibited a high sensitivity towards the concentration of the antigen with a linear dynamic range of five orders of magnitude and a resolution of 0.08 pg/mL. These results indicate that this principle can be used as a rapid diagnostic method for COVID-19 antigens without sample labelling.
Keywords: Optical biosensor, Surface plasmon resonance, Laser feedback, Refractive index sensing, Label free approach, SARS-CoV-2
1. Introduction
Coronavirus disease 2019 (COVID-19), which began in late 2019, is caused by the newly discovered severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Gorbalenya et al., 2020; Lu et al., 2020; Wu et al., 2020), and the growing trend of infected cases is not yet under control. COVID-19 was officially announced as a pandemic by the World Health Organization (WHO) on March 12, 2020 (WHO, 2020). As of October 24, 2021, more than 243 million cases of COVID-19 have been confirmed globally, with 4.9 million deaths (WHO, 2021). Since there are no specific drugs for COVID-19, early diagnosis and management are critical to containing the outbreak. Real-time reverse transcription-polymerase chain reaction (RT-PCR) has been widely used to identify the COVID-19 and has become the gold standard for the diagnosis of SARS-CoV-2 infection (Corman et al., 2020; Noh et al., 2017). However, molecular diagnosis using real-time RT-PCR requires high manpower, long processing time, many reagents, and bulky instruments, which cannot rapidly test all suspected cases during a full-scale outbreak. Hence, highly-sensitive biosensors that can rapidly detect viral antigens in clinical samples without processing steps are urgently required for the accurate diagnosis of COVID-19.
Antigen detection has a short turnaround time and is amenable to point-of-care diagnostic methodologies. Relevant studies have been reported to date. For example, an ultrasensitive graphene field-effect transistor (Gr-FET) biosensor was recently reported for the simple and rapid detection of SARS-CoV-2 spike protein S1 (Zhang et al., 2020). Detection relies on the highly specific interaction between the SARS-CoV-2 spike protein S1 and the SARS-CoV-2 spike S1 subunit protein antibody (CSAb) or human angiotensin-converting enzyme 2 (ACE2)-functionalized graphene surface. This Gr-FET biosensor can identify the SARS-CoV-2 spike protein S1 within 2 min at a limit of detection of 0.2 pM. Seo et al. (2020) developed a graphene-based FET immunosensor functionalized with the anti-SARS-CoV-2 spike antibody. This reported COVID-19 immunosensor detects spike proteins without preprocessing of the samples and can detect SARS-CoV-2 when RNA is present at 2.42 × 102 copies/mL. Although FET biosensors have shown promise for applications in COVID-19 diagnosis, they still suffer from shortcomings, such as insufficient immunity to electromagnetic interference and no mechanism to eliminate noise. Optical methods can solve these problems.
Among the many potentially useful optical diagnostic methods currently available for viral antigen detection, surface plasmon resonance (SPR)-based biosensing devices are advantageous because of their capability for real-time, label-free detection and high reproducibility (Altintas, 2018; Homola, 2008; Liu et al., 2021; Xue et al., 2019). SPR is a strong photon-driven coherent oscillation of the surface conduction electrons that may exist at the interface of two media with dielectric constants of opposite signs, such as a metal film and dielectric medium. Owing to the enhanced light-matter interaction, a change in the refractive index (RI) of the tiny layer of the dielectric medium close to the interface can alter the coupling condition, which can be observed as a change in one of the characteristics of the light measured, such as the angle, wavelength, intensity, and phase (Homola et al., 1999; Patil et al., 2019; Shalabney and Abdulhalim, 2011). Compared to other conventional immunoassays, such as enzyme-linked immunosorbent assay (ELISA) (Sato et al., 2014), magnetic nanoparticle detection (Chang et al., 2006), and fluorescence in situ hybridization (FISH) (Levsky and Singer, 2003), SPR biosensors eliminate the complicated target-labeling processes and sophisticated instrumentation, which are expensive and time-consuming. In 2014, Choi et al. (2014) developed an SPR biosensor for human hepatitis B virus detection without any label on the sensor surface, and the detection limit was estimated to be less than 10 pg/mL. Loo et al. (2015) achieved the rapid detection of the microRNA miR-29a-3p from human subjects infected with influenza virus H1N1 with a detection limit of 1 nM using an SPR platform in 2015. These studies show that SPR biosensors can be applied for real-time and label-free virus detection. However, the sensitivity of such biosensors needs to be improved to meet the requirements of clinical testing, such as COVID-19 diagnosis.
Herein, we propose a new principle for highly-sensitive SPR biosensor based on advanced laser heterodyne feedback interferometry (LHFI) to further improve the sensitivity of this kind of optical biosensor, which has not been reported to the best of our knowledge. Laser feedback interferometry (LFI) has attracted considerable attention because of its self-alignment, simple structure, and high sensitivity (Otsuka, 2015; Taimre et al., 2015; Zhu et al., 2017). LFI occurs when the output light of the laser partially returns to the laser resonator, which induces intensity and phase modulation of the laser. Specifically, when the frequency of the output light is shifted before the light returns to the resonator, the sensing signal can resonate with the relaxation oscillation of the laser and be enhanced spontaneously—up to 106 in some cases (Lacot et al., 1999; Otsuka, 2011; Xu et al., 2021a). This remarkable enhancement makes it effective for increasing the intensity changes in the light caused by the RI changes when the biosensors excite SPR, thus greatly improving the sensitivity of this type of biosensors. Therefore, this pioneering biosensor can discriminate small differences in the RI of different samples in real time. In the case of SARS-CoV-2, the spike glycoprotein is the key antigen, and its conserved S2 subunit contributes to viral entry by mediating host-viral membrane fusion (Fan et al., 2020). Hence, we immobilized the anti-SARS-CoV-2 spike monoclonal antibodies (MAbs) onto the biosensor surface to enable the specific recognition of the SARS-CoV-2 spike antigen. The proposed biosensor showed an extremely low detection limit for the target antigen, demonstrating the successful fabrication of a COVID-19 SPR biosensor based on LHFI and its potential utility in rapid COVID-19 diagnosis.
2. Setup and methods
2.1. Materials
0.1% poly-L-lysine (PLL) aqueous solution and de-ionized water were purchased from Zhejiang Lianshuo Biological Technology Co., Ltd. 7.5% bovine serum albumin (BSA) solution and Dulbecco's phosphate-buffered saline (DPBS) solution were obtained from Sigma-Aldrich, Inc. Anti-SARS-CoV-2 spike monoclonal antibodies (MAbs) and SARS-CoV-2 spike antigens were purchased from Sino Biological, Inc.
2.2. Experimental setup
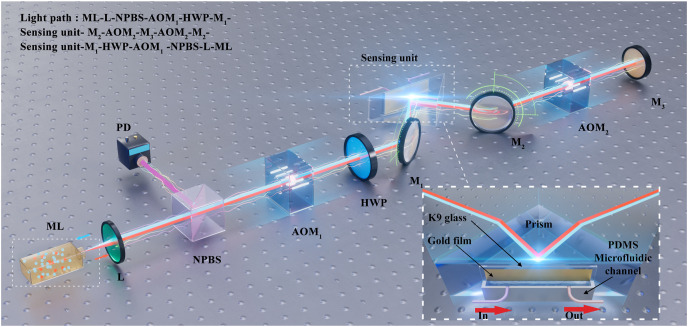
The schematic representation of the biosensor is shown in Fig. 1 . The solid microchip laser works in a single-longitudinal mode with a wavelength of 1064 nm. The output light of the laser is collimated via the lens (L) and divided into two beams by a non-polarization beam splitter (NPBS). The reflected light is detected by a photodetector (PD) and transferred to the electrical signals. The transmitted linearly polarized light passes through an acousto-optic modulator (AOM1) and is diffracted at +1 order to obtain a frequency shift of ω 1 = 70 MHz, which is the driving frequency of AOM1. The linearly polarized light is then converted into p-polarized light by a half-wave plate (HWP). The p-polarized light is reflected by a mirror (M1) and focused on the sensing unit at the center of its microfluidic channel, where SPR occurs. The biosensor converts the refractive index (RI) changes into light intensity changes. The inset of Fig. 1 shows the schematic of the sensing unit, which consists of a polydimethylsiloxane (PDMS) microfluidic channel/gold film/K9 glass sandwich structure on the prism. After being reflected by a mirror (M2), the light passes through another acousto-optic modulator (AOM2) and is diffracted at −1 order to get a frequency shift of ω 2 = 70.5 MHz, which is the driving frequency of AOM2. Finally, the light is reflected by a mirror (M3) and returns to the laser resonator via the original light path. Thus, the light returning to the laser resonator undergoes a frequency shift of (see Section 2.3 for the details of the differential frequency shift), generating a laser heterodyne feedback signal. The feedback signal modulates the laser output power, which can be expressed as (Zhu et al., 2019):
where denotes the intensity modulation of the laser, is the free-running laser intensity, is the effective coupling coefficient of feedback, and represent the fixed phase and the phase related to the external cavity length, respectively. is the gain coefficient and can reach 106 in the microchip laser used in this study (Lacot et al., 2001; Zhu et al., 2019), which means that it could improve the intensity change of the biosensor caused by a minor RI change.
Fig. 1.
Schematic diagram of the biosensor. ML: microchip laser; L: lens; NPBS: non-polarization beam splitter; PD: photodetector; AOM1, AOM2: acousto-optic modulators; HWP: half-wave plate; M1, M2, M3: mirrors. Inset: the schematic diagram of the sensing unit, in which the red arrows represent the in and out of the samples. PDMS: polydimethylsiloxane. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.3. Differential frequency shift
In the laser heterodyne feedback interferometry of the microchip laser used in this study, the gain coefficient can be deduced according to the laser rate equation as (Lacot et al., 2001; Wan et al., 2007; Zhu et al., 2019):
where is the laser cavity decay rate, is the normalized pumping rate, is the decay rate of the population inversion, represents the relaxation oscillation frequency of the laser.
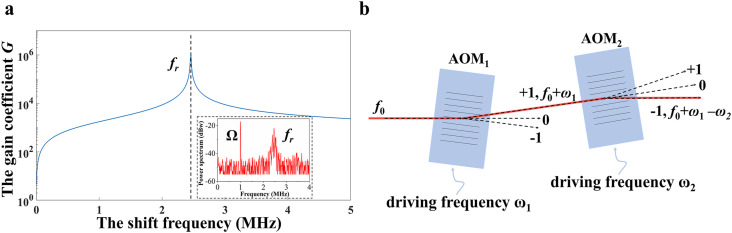
The simulation results based on the parameters of the microchip laser indicate that the closer the shift frequency () is to the relaxation oscillation frequency of the laser (), the larger the gain coefficient is, as shown in Fig. 2 a. The relaxation oscillation frequency of the laser is 2.46 MHz, which is determined by the parameters of the microchip laser. However, when the shift frequency is too close to the relaxation oscillation frequency, the spectrum will be in a state of chaos because of the extremely strong nonlinear effect (Ko et al., 2001; Lacot et al., 2016), which is not suitable for measurement. Therefore, the shift frequency is set to 1 MHz. However, the working frequency of the acousto-optic modulator cannot be as low as a few megahertz; thus, two AOMs are employed to obtain a low differential frequency shift (Xu et al., 2021b) to achieve a large gain coefficient . Fig. 2b shows the schematic diagram of the differential frequency shift module. The light with frequency passes through AOM1, where it is diffracted at +1 order, and the frequency changes to (ω 1 = 70 MHz is the driving frequency of AOM1). The light then passes through AOM2 and is diffracted at −1 order such that the frequency is (ω 2 = 70.5 MHz is the driving frequency of AOM2). In this biosensor, the light is reflected by a mirror and diffracted again at −1 order in AOM2 and at +1 order in AOM1. Consequently, the light returning to the laser resonator has a frequency of . By choosing an appropriate differential shift frequency, high sensitivity can be obtained in this biosensor. (Fig. 2a, inset: is adjusted to be 1 MHz).
Fig. 2.
Characteristics of laser heterodyne feedback interferometry. a The relationship between the gain coefficient and the shift frequency through the simulation. Inset: the power spectrum of the laser measured; b Schematic diagram of the differential frequency shift module using two AOMs. AOM1, AOM2: acousto-optic modulators.
2.4. Fabrication of sensing unit
The schematic of the sensing unit is shown in the inset of Fig. 1. The sensing unit is a polydimethylsiloxane (PDMS) microfluidic channel/gold film/K9 glass sandwich structure on a prism, which is fabricated through several steps. First, we design a mould to create the PDMS microfluidic channel and punch a hole on the left and right sides above the channel for the inside and outside of the samples. The gold film (GF) is then deposited on K9 glass by electron beam evaporation (EBE), and the thickness of the GF is close to 50 nm. Next, the PDMS microfluidic channel and K9 glass are exposed to oxygen plasma, which can change the surface chemistry of the PDMS microfluidic channel and K9 glass by oxidizing them. The oxidized layer of the PDMS microfluidic channel can now adhere to the K9 glass. Finally, the sandwich structure is combined with the prism through the matching liquid.
3. Results and discussion
3.1. Refractive index (RI) sensing
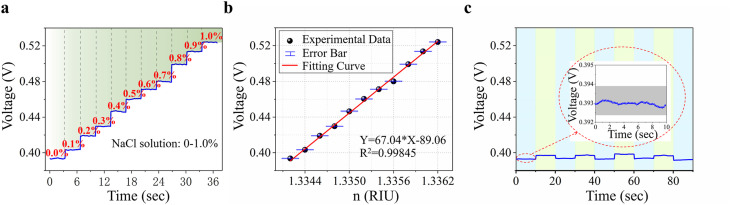
Because the antigen detection of the biosensor essentially responds to the RI changes caused by the molecular binding of the antibody-antigen at the sensing unit, we first conduct a series of RI sensing experiments with different concentrations of sodium chloride (NaCl) solution to evaluate the resolution of the biosensor to the RI changes. Different low concentrations of NaCl solution are detected using the LHFI-based SPR biosensor. Fig. 3 a shows the real-time responses of the biosensor to NaCl solutions with concentrations ranging from 0% to 1.0%. Calculating the average voltage and RI corresponding to the solution of each concentration, the fitted curve indicates that the biosensor has a good linear response, as shown in Fig. 3b. As evidence of the stability and repeatability of the biosensor, a continuous and periodic change in pure water and 0.003% NaCl solution has been clearly detected (Fig. 3c).
Fig. 3.
Results of RI sensing. a Signal of different NaCl solution concentrations; b Fitting curve of the response to different concentrations of NaCl solution corresponding to a. The error bar is the standard deviation of the voltage corresponding to each concentration; c Signal of pure water and 0.003% NaCl solution. The data in the blue background corresponds to pure water, and the data in the green background corresponds to 0.003% NaCl. Inset: the voltage fluctuation of the biosensor corresponding to pure water. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Sensitivity and resolution, which are two of the most important parameters in RI sensing, are calculated using the following formulas:
where S is the sensitivity of RI sensing, is the resolution of RI sensing, and is the response voltage difference in RI sensing between water and 0.003% NaCl solution (n = n water+0.000006), which is approximately 6.40 mV (Fig. 3c). is the standard deviation of the voltage corresponding to pure water, and its value is 0.04 mV (inset of Fig. 3c). The sensitivity is calculated to be 1.07 × 106 mV/RIU and the noise-limited resolution is calculated to be 3.75 × 10−8 RIU, which is better than most reported results. (Giorgini et al., 2013; Li et al., 2020; Mataji-Kojouri et al., 2020; Park and Park, 2021; Wu et al., 2021; Zhu et al., 2021), showing that this biosensor is capable of detecting the viral antigen.
3.2. Modification processes of the biosensor
For better sensing performance and specific recognition of the SARS-CoV-2 spike antigens, as in many previous studies (Li et al., 2015; Ramadan et al., 2021; Yesilkoy et al., 2018), the biosensor is modified by the anti-SARS-CoV-2 spike monoclonal antibodies (MAbs) to promote the effectiveness of specific binding events on the surface of the biosensor. In the sensing unit of the biosensor, the free-standing GF is immersed in 0.1% PLL aqueous solution positive charged for 2 h at room temperature, enhancing the immobilization of the anti-SARS-CoV-2 spike MAbs. The de-ionized water is then used to wash the GF thrice to remove the PLL aqueous solution. Subsequently, the biosensor is incubated at 4 °C overnight in the highly purified anti-SARS-CoV-2 spike MAbs solution at a concentration of 50 μg/mL and thoroughly rinsed with DPBS to wash out excess antibodies. Afterwards, the 7.5% BSA solution is diluted to 3% with DPBS before use. Finally, the biosensor is incubated in 3% BSA solution for 4 h at 4 °C to block nonspecific binding sites and washed again with DPBS (Li et al., 2015). After these surface modifications, the label-free SARS-CoV-2 spike antigen biosensor is ready for detection.
3.3. Characterization of the biosensor after modification
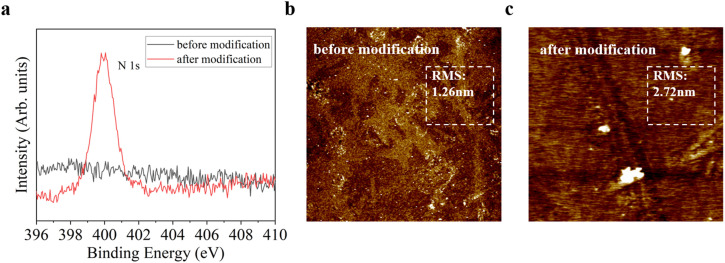
To confirm that the biosensor is successfully immobilized by the MAbs, we obtain the surface analysis data of the biosensor by the X-ray photoelectron spectrometer (XPS) and the atomic force microscopy (AFM). The elemental composition on the surface of the biosensor is analyzed by XPS (Thermo Fisher, ESCALAB 250Xi). The survey data shows that the atomic percentage of N increased from 3.33% to 12.30% after modification. Fig. 4 a indicates the comparison of the N 1s peak intensity before and after modification. Initially, the biosensor exhibited no meaningful N 1s peak (black line in Fig. 4a), whereas the N 1s peak was observed in the modified sample (red line in Fig. 4a). The increase in the intensity of the N 1s peak proves the success of the surface modification. Next, we characterize the surface morphology of the biosensor using AFM (Bruker, Dimension FastScan). The surface roughness (RMS) increases slightly from ∼1.26 nm to ∼2.72 nm after modification (Fig. 4b and c). These results suggest the successful modification of the antibodies, which can be used as a support for the SARS-CoV-2 spike antigen detection.
Fig. 4.
Surface analysis of the biosensor using the X-ray photoelectron spectrometer (XPS) and the atomic force microscopy (AFM). a XPS survey data (N 1s peak); b AFM image of the biosensor surface (before modification); c AFM image of the biosensor surface (after modification).
3.4. SARS-CoV-2 spike antigen detection
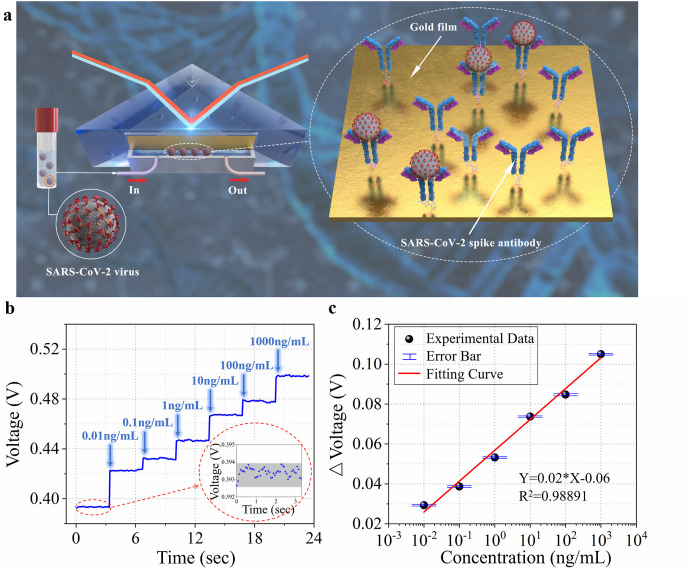
To investigate the performance of the biosensor in the SARS-CoV-2 spike antigen detection, we evaluate the real-time dynamic response of the biosensor to different concentrations of spike antigen, and the schematic is shown in Fig. 5 a. The specific binding of the SARS-CoV-2 spike antigens and anti-SARS-CoV-2 spike monoclonal antibodies is a single event and cannot be repeated; therefore, we cannot introduce repeatability in a single experiment. Notably, the biosensor can also be reused if the antibodies bound to the antigens are removed and new antibodies are immobilized onto the biosensor surface. First, a DPBS solution is introduced into the biosensor, and the corresponding voltage value is recorded as the baseline. Subsequently, the prepared SARS-CoV-2 spike antigen samples of different concentrations (0.01, 0.1, 1, 10, 100, and 1000 ng/mL) are used for immunoassays. The different voltage values at each concentration are displayed and recorded in real time (Fig. 5b). Taking the voltage value corresponding to DPBS solution as the reference, we calculate the voltage changes related to the concentrations of the SARS-CoV-2 spike antigen samples. The logistic function-fitted curve shows the linear dynamic range of five orders of magnitude between the voltage changes and concentrations, from 0.01 ng/mL to 1000 ng/mL, as shown in Fig. 5c. The noise-limited resolution of this biosensor toward the SARS-CoV-2 spike antigen, which is defined as the concentration at which the voltage change is greater than the standard deviation of the noise level (0.25 mV, as shown in the inset of Fig. 5b), is calculated to be 0.08 pg/mL. This indicates that the proposed biosensor can detect antigens at a relatively low concentration without sample labelling and has the potential to work as a rapid clinical SARS-CoV-2 spike antigen detection diagnostic and a detailed comparison of the proposed biosensor with other SARS-CoV-2 biosensors is presented in Table 1 .
Fig. 5.
Detection of the SARS-CoV-2 spike antigen. a Schematic illustration of the sensing unit; b Signal of different concentrations of SARS-CoV-2 spike antigen. Inset: the voltage fluctuation of the biosensor corresponding to the blank sample; c Response voltage changes as a function of solution concentrations and the fitting curve. The error bar is the standard deviation of the voltage change corresponding to each concentration.
Table 1.
Comparison of the SARS-CoV-2 biosensors.
| Analyte | Detection method | Limit of detection | Detection time | Ref. |
|---|---|---|---|---|
| SARS-CoV-2 spike protein | Graphene field-effect transistor (Gr-FET) | 0.2 pM | ∼2 min | Zhang et al. (2020) |
| SARS-CoV-2 nucleocapsid antibody | SPR | 1 μg/mL | <15 min | Djaileb et al. (2020) |
| SARS-CoV-2 RNA | DNA nanoscaffold hybrid chain reaction | 0.96 pM | ∼10 min | Jiao et al. (2020) |
| SARS-CoV-2 sequences | Localized SPR with plasmonic photothermal effect | 0.22 pM | / | Qiu et al. (2020) |
| SARS-CoV-2 nucleocapsid antigen | colorimetric assay based on gold nanoparticles | 0.18 ng/μL | <10 min | Moitra et al. (2020) |
| SARS-CoV-2 spike antigen | SPR based on LHFI | 0.08 pg/mL | <1 min | This paper |
The biosensor intrinsically responds to RI changes, and samples that can induce RI changes can, in principle, be detected. Testing the SARS-CoV-2 spike antigens is currently the only initial validation of the practical application of the biosensor. By processing the sensing unit in different ways, the biosensor can detect more diverse targets such as other antigens of the SARS-CoV-2.
4. Conclusions
A novel surface plasmon resonance biosensor based on laser heterodyne feedback interferometry has been successfully developed. A minor difference between the RI of water and that of 0.003% sodium chloride solution can be distinguished in real time by the biosensor, and the resolution is calculated to be 3.75 × 10−8 RIU. Specifically, we apply the biosensor to detect the SARS-CoV-2 spike antigen. After modification, the biosensor realizes the detection of SARS-CoV-2 spike antigen with a remarkable linear dynamic range of five orders of magnitude, from 0.01 ng/mL to 1000 ng/mL with a resolution as low as 0.08 pg/mL. Therefore, the proposed highly-sensitive, real-time, and label-free biosensor can potentially provide a reliable diagnostic platform and relieve the pressure on RT-PCR testing during the outbreak period of COVID-19.
Because the biosensor only has one optical path, it is susceptible to environmental disturbances and fluctuations in laser power. Moreover, it needs to work efficiently in a relatively stable environment, which affects its sensing performance. In future work, we would like to reuse the sensing unit by chemical treatment, thus greatly reducing the cost and fabrication time, and we will design a compensation optical path to further improve the resolution of this biosensor.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Zongren Dai and Yidong Tan conceived the biosensor and the experiments. Zongren Dai modified the biosensor and prepared the materials. Zongren Dai and Xin Xu built up the experimental system. Zongren Dai, Xin Xu, Yifan Wang, and Mingfang Li conceived the data analysis. Kaiming Zhou, Lin Zhang, and Yidong Tan gave assistance in discussing and analyzing the experimental results. Zongren Dai and Yidong Tan wrote the manuscript, which was then revised by all authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (51961130387), the Royal Society Newton Advanced Fellowship (NAF/R1/191072), the National Science Fund for Excellent Young Scholars (51722506) and Initiative Scientific Research Program of State Key Lab of Precision Measurement Technology and Instrument, Tsinghua University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114163.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Altintas Z. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-29585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.L., et al. Microelectron. Eng. 2006;83:1630–1633. [Google Scholar]
- Choi Y.H., et al. Biosens. Bioelectron. 2014;56:286–294. doi: 10.1016/j.bios.2014.01.035. [DOI] [PubMed] [Google Scholar]
- Corman V.M., et al. Euro Surveill. 2020;25:23–30. doi: 10.2807/1560-7917.ES.2020.25.21.2001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djaileb A., et al. 2020. Chemrxiv.org. 0-13. [Google Scholar]
- Fan X.Y., et al. Nat. Commun. 2020;11:1–10. [Google Scholar]
- Giorgini A., et al. Opt Lett. 2013;38:1951–1953. doi: 10.1364/OL.38.001951. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., et al. Nature Microbiology. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homola J. Chem. Rev. 2008;108:462–493. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- Homola J., et al. Sensor. Actuator. B Chem. 1999;54:3–15. [Google Scholar]
- Jiao J., et al. Biosens. Bioelectron. 2020;167 doi: 10.1016/j.bios.2020.112479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.Y., et al. Phys. Rev. Lett. 2001;86:4025. doi: 10.1103/PhysRevLett.86.4025. [DOI] [PubMed] [Google Scholar]
- Lacot E., et al. Opt Lett. 1999;24:744–746. doi: 10.1364/ol.24.000744. [DOI] [PubMed] [Google Scholar]
- Lacot E., et al. Phys. Rev. 2001;64 [Google Scholar]
- Lacot E., et al. Phys. Rev. 2016;94 [Google Scholar]
- Levsky J.M., Singer R.H. J. Cell Sci. 2003;116:2833–2838. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- Li P., et al. Biosens. Bioelectron. 2015;72:168–174. doi: 10.1016/j.bios.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Li T.S., et al. Sensors. 2020;20:741. [Google Scholar]
- Liu L.H., et al. Light Sci. Appl. 2021;10:1–14. doi: 10.1038/s41377-021-00618-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo J.F.C., et al. Analyst. 2015;140:4566–4575. doi: 10.1039/c5an00679a. [DOI] [PubMed] [Google Scholar]
- Lu R.J., et al. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mataji-Kojouri A., et al. ACS Nano. 2020;14:8518–8527. doi: 10.1021/acsnano.0c02797. [DOI] [PubMed] [Google Scholar]
- Moitra P., et al. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh J.Y., et al. Arch. Virol. 2017;162:1617–1623. doi: 10.1007/s00705-017-3281-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka K. Sensors. 2011;11:2195–2245. doi: 10.3390/s110202195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka K. Opt Lett. 2015;40:4603–4606. doi: 10.1364/OL.40.004603. [DOI] [PubMed] [Google Scholar]
- Park G.C., Park K. Sci. Rep. 2021;11:1–10. doi: 10.1038/s41598-021-98654-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil P.O., et al. Biosens. Bioelectron. 2019;139 doi: 10.1016/j.bios.2019.111324. [DOI] [PubMed] [Google Scholar]
- Qiu G.Y., et al. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Ramadan S., et al. ACS Appl. Mater. Interfaces. 2021;13:7854–7864. doi: 10.1021/acsami.0c18293. [DOI] [PubMed] [Google Scholar]
- Sato T., et al. Cancer Medicine. 2014;3:1377–1384. doi: 10.1002/cam4.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., et al. ACS Nano. 2020;14:12257–12258. doi: 10.1021/acsnano.0c06726. [DOI] [PubMed] [Google Scholar]
- Shalabney A., Abdulhalim I. Laser Photon. Rev. 2011;5:571–606. [Google Scholar]
- Taimre T., et al. Adv. Opt Photon. 2015;7:570–631. [Google Scholar]
- Wan X.J., et al. Opt Lett. 2007;32:367–369. doi: 10.1364/ol.32.000367. [DOI] [PubMed] [Google Scholar]
- WHO . 2020. Coronavirus Disease 2019: Situation Report-52. [Google Scholar]
- WHO . 2021. Coronavirus Disease 2019: Weekly Epidemiological Update on COVID-19. [Google Scholar]
- Wu F., et al. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., et al. Opt Express. 2021;29:10939–10948. doi: 10.1364/OE.422701. [DOI] [PubMed] [Google Scholar]
- Xu X., et al. 2021. arXiv Preprint arXiv:2105.13561. [Google Scholar]
- Xu Z., et al. Opt Laser. Eng. 2021;141 [Google Scholar]
- Xue T.Y., et al. Nat. Commun. 2019;10:1–9. [Google Scholar]
- Yesilkoy F., et al. Light Sci. Appl. 2018;7 doi: 10.1038/lsa.2017.152. 17152-17152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.Y., et al. 2020. arXiv Preprint arXiv:2003.12529. [Google Scholar]
- Zhu K.Y., et al. Appl. Sci. 2019;9:109. [Google Scholar]
- Zhu K.Y., et al. Optica. 2017;4:729–735. [Google Scholar]
- Zhu Z.D., et al. Opt Express. 2021;29:19034–19048. doi: 10.1364/OE.424995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.