Abstract
Background
Salmonella is an important zoonotic pathogen and is one of the main causes of foodborne outbreaks and infections in the European Union. Pigs are a significant reservoir and are frequently subclinical carriers of this organism. Salmonella can be shed in the faeces allowing infection to spread to other pigs, the environment, transport vehicles, lairages and other areas. Inadvertent spillage of gut contents during the slaughter process also leads to contamination. A pig Salmonella control programme has operated in Ireland since 2002 but many local surveys and an EUMS baseline survey in 2008 continued to indicate high levels of the organism in the pig sector. The objectives of this study were to generate updated information on the prevalence of Salmonella spp, in slaughter pigs and carcasses in Irish abattoirs. Five pigs from each of 164 herds were randomly sampled over a 14-week period during 2016. One sample from each of the five pigs of; caecal content, ileo-caecal lymph nodes and carcass swabs (pre-chill) were collected. The five caeca and lymph node samples from each herd were processed as one pool of caecal samples and one pool of lymph node samples, respectively, while the five carcass swabs were tested as individual samples. All isolates were characterised by serotyping and antimicrobial susceptibility.
Results
In total, 235 Salmonella spp. were isolated from 820 individual carcass swabs, 164 pooled lymph nodes and 164 caecal contents. Salmonella spp. were isolated from 54.3% of the caecal contents and from 31.7% of the ileo-caecal lymph node sample pools. A total of 11.5% of carcass-swab samples yielded Salmonella spp. S. Typhimurium 4,[5],12:i:1,2 or its monophasic variant 4,[5],12:i:-: predominated among isolates from all positive samples; accounting for 73% of lymph nodes, 68% of caecal contents and 56% of carcass swab isolates. S. London and S. Derby were the next most common isolated serotypes.
Conclusions
These results confirm continuing high levels of Salmonella in fattening pigs in Ireland although reductions in carcass contamination compared to previous surveys were noted. A high prevalence of Salmonella in lymph nodes suggests that it remains a significant problem pre slaughter and a challenge to abattoirs in adhering to process hygiene requirements. The high prevalence of monophasic S. Typhimurim 4,[5],12:i:-: is of serious concern.
Therefore, it is important to identify contributing factors in the dissemination of this pathogen in the pork industry in order to minimise the risk of human salmonellosis cases.
Keywords: Salmonella, Pigs, Ireland, Abattoirs, Carcass swabs, Serology, Caecal, Lymph nodes
Background
Salmonella is recognised as a major cause of food-borne illness in humans [1] with the poultry sector a major source. However, following the introduction of control programmes in EU Member States (EUMS), Salmonella levels in the poultry sector in EUMS have been greatly reduced as noted by EFSA and the ECDC and the focus of control in recent years has turned to the pig sector [2]. Pigs have been identified in a number of countries including Ireland as important carriers of Salmonella spp. and attribution studies show them to be an important source of human infection [2–5]. Pig meat is an important source of human infection [6] and it has been estimated that between 15 and 23% of all cases of human salmonellosis are related to the consumption of pork [2, 7, 8]. It should be noted that some Salmonella serotypes found in pigs are uncommonly associated with human infection whereas S. Typhimurium, and monophasic S. Typhimurium, important causes of human infection, account for about half of the isolates recovered from slaughter pigs across EUMS. Over 40% of pig Salmonella isolates that were serotyped and reported in 2019 were S. Typhimurium or monophasic S. Typhimurium [2, 9].
An EU-wide baseline study of Salmonella in slaughter pigs published in 2008 showed that 10.3% of slaughter pigs were Salmonella-positive, (based on testing of ileo-caecal lymph node samples), giving rise to concerns over risks to human health and the need to control and manage the disease and reduce that risk [10]. Salmonella prevalence in the pig sector varies across Europe; countries with long standing control programmes show lower infection levels in slaughter pigs [11]. A number of cost benefit studies have been undertaken to determine effective ways of introducing EU-wide controls [11] but to date there is no agreed strategy on control.
A legally defined Salmonella control programme has been operating in Ireland since 2002. Although the programme has been modified and refined on a number of occasions the basic principles have been retained. Herds are risk ranked on the basis of antibody tests on muscle juice samples [6] with individual abattoirs required to place additional controls on pigs processed from high-risk herds to minimise cross contamination of pigs and carcasses. Irish data from the EU baseline survey on the prevalence of Salmonella spp. in slaughter pigs in EUMS showed a Salmonella prevalence of 16.1% in ileo-caecal lymph nodes and 20% in carcass swabs, of which approximately 57% and 59% were S. Typhimurium, respectively [10]. A study of Salmonella in pigs at slaughter in Northern Ireland in 2002 showed the organism present in 31.4% of caecal contents and 40.0% of carcass swabs taken post-evisceration [12].
Serological test data show only limited changes to Salmonella antibody levels in Irish pigs since 2002 (DAFM unpublished). As the 2006–2007 baseline study was the last comprehensive study of Salmonella levels in Irish pigs, the current study was undertaken with the objective of updating baseline data of Salmonella in slaughter pig herds and carcasses, in order to obtain updated data to inform the future reviews of the National Salmonella Control Programme. Since completion of this survey, the results found have provided valuable updated information for a new “Pig Health Check” work programme initiated, since 2018, by Animal Health Ireland, which is a joint industry and government agency that works to improve animal health in farmed livestock in Ireland.
Methods
Bacteriology survey
The six main Irish pig abattoirs were recruited for the study and given targets for the number of herds to sample based on their annual throughput and number of suppliers. The sample collected was a convenience sample insofar as there were limitations both at the abattoir and in the laboratory governing the practicalities of sample collection and numbers that could be processed. Samples were collected from 164 of the total 300 commercial pig herds.
Five pigs from each of 164 herds (820 pigs in total) were randomly sampled by official veterinary inspectors at each abattoir over a 14-week period during 2016. From each of the five pigs, samples of caecal content, ileo-caecal lymph nodes and carcass swabs (pre-chilling) were collected. The five caeca and lymph node samples from each herd were processed as one pool of caecal samples and one pool of lymph node samples, respectively, while the five carcass swabs were tested as individual samples.
A standard protocol for the collection of samples was supplied to samplers with the logistics of collecting the samples in a suitable area away from the line being left to the discretion of the abattoir veterinary inspectors. Carcass swabs were collected pre-chilling using a commercial foam swab (Whirl–Pak®Speci-Sponge®Environmental Surface Sample Bags), approximately 100cm2 along the belly and extending down to the neck area on each side of the carcass (Chapter III of Regulation 2073/2005 (Microbiological Criteria). Samples were sent by overnight courier to the laboratory. Details of abattoir, the source of the pigs, the sampling date and time were entered onto a submission form, which accompanied the samples to the laboratory. Sampling commenced in early March 2016 and continued for a 14-week period.
Laboratory methods
Salmonella isolation was carried out using a modification of the ISO 6579:2002 protocol [13]. In brief, all samples, 25 g caecal content, 25 g lymph nodes and each carcass swab were processed using an initial enrichment step at a 1:10 dilution in buffered peptone water (BPW) (Oxoid CM509). Lymph node samples were first washed in alcohol and then air dried to eliminate surface contamination. Selective enrichment in MRSV agar (Lab M) was followed by plating on both xylose lysine deoxycholate agar (XLD) (E&O Laboratories, Bonnybridge, Scotland) and brilliant green agar (BGA) (E&O Labs). Salmonella suspected isolates were plated onto chromogenic agar (E&O). Serotyping was carried out as per the White-Kauffmann-Le Minor classification scheme [14].
DNA extraction and PCR confirmation of monophasic variant 4,[5],12:i:-:
PCR was performed on the 97 isolates for which conventional serotyping showed an incomplete antigenic formula that shared antigens with the S. Typhimurium formula 4,[5],12:i:1,2 i.e., for which formulas 4,[5],12:i:-: were obtained. Isolates were cultured overnight on Nutrient Agar (Lab M Ltd.,Lancashire, UK) at 37 °C. One isolated colony was suspended in 100 µl InstaGene™ Matrix (BioRad laboratories, USA) and DNA was extracted as per manufacturer’s instructions. A multiplex real-time PCR method for the identification and differentiation of S. Typhimurium and monophasic 4,[5],12:i:- was carried out using the method and primers as described by Prendergast et al. 2013 [15].
S. Typhimurium LT2 ATCC 29,946 was used as a positive control and Escherichia coli NCTC 9001 as a negative control.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was conducted on two hundred and thirty-five isolates using the Sensititre broth microdilution method for Salmonella, EUVSEC (Sensititre, TREK Diagnostic Systems Inc., Sussex, England) and in accordance with the Clinical and Laboratory Standards Institute (CLSI) methods [16]. Custom-made panels of 14 antimicrobial drugs at specified concentrations were configured in 96-well microtiter plates. The panel of antimicrobials and the cut-off values (mg/l) were in agreement with the EU commission implementing Decision of 12 November 2013 on the monitoring and reporting of antimicrobial resistance in zoonotic and commensal bacteria [17], as follows: Ampicillin (AMP) (1–64 mg/l), Cefotaxime (CTX) (0.25–4 mg/l), Ceftazidime (CAZ) (0.5–8 mg/l), Meropenem (MER) (0.03–16 mg/l), Nalidixic acid (NAL) (4–128 mg/l), Ciprofloxacin (CIP) (0.015–8 mg/l), Tetracycline (TET) (2–64 mg/l), Colistin (COL) (1–16 mg/l), Gentamicin (GEN) (0.5–32 mg/l), Trimethoprim (TMP) (0,25–32 mg/l), Sulfamethoxazole (SMX) (8–1024 mg/l), Chloramphenicol (CHL) (8–128 mg/l), Azithromycin (AZI) (2–64 mg/l), Tigecycline (TIG) (0.25–8 mg/l). The wells of the microtiter plates were manually read for bacterial growth using the Sensititre Vizion System (TREK disgnostic Systems). E. coli ATCC 25,922 was used as the quality control strain for this assay. The AMR result was designated as “Fully Susceptible” or by indicating the abbreviation of the antimicrobial to which the strain was resistant [18].
Data analysis
All data were entered into a Microsoft Excel (Microsoft Corporation) spread sheet with visual checks for accuracy made at the point of entry, and again at the end of the survey. After validation, data were transferred to, and all statistical analyses carried out using SAS version 9.4. Serology data for each herd sampled was taken from the national monitoring programme database and merged with the bacteriology data.
Monthly serological prevalence
The average serological prevalence per month was calculated by summing the total number of tests conducted and total number of positive samples within a month over the years 2010 to 2016. Estimates for January to June were based on 7 years of data and those for July to December were based on 6 years of data.
Herds with the highest and lowest serological prevalence
Serology tests carried out from July 2015 to June 2016 were used to rank herds according to the serology prevalence within the herd over this time period. Herds in the top and bottom 10% of herds, based on serology prevalence, were compared for the bacteriology results on lymph nodes, caeca and carcass swabs. A chi-squared test was used to compare herds in the top and bottom groups, except when the expected number in a cell was < 5 in which case a Fisher’s exact test was used.
Probability of a positive carcass swab
A logistic regression model was developed to model the probability of a herd having a positive carcass swab. The results of the following independent variables were tested within the model: caecal contents, lymph nodes, caecal contents /lymph nodes, the processing plant and the herd serological result. For the serological result the following time periods were considered for inclusion within the model: Jan to July 2016; June 2015 to July 2016 and Jan 2010 to July 2016. Initially each independent variable was tested within a univariable model. The best fitting time period to use for serological results within the multivariable model was determined by comparing the AIC (lowest value gave the better fit) of each time variable in the respective univariable model. Similarly, the choice of whether to include caecal and lymph results separately or combined was determined by the AIC of the univariable models. A backward selection procedure was used to eliminate terms from the model based on a likelihood ratio test.
Results
Salmonella spp. were isolated from the pooled caecal contents of 91 (55.5%, 95% confidence interval (CI) 46.3–62.0) of the 164 herds and from the ileo-caecal lymph nodes in 52 herds (31.7%, 95% confidence interval (CI) 24.8–39.5). A total of 94 (11.5%, 95% confidence interval (CI) 9.4–13.9) of the 820 carcass-swab samples yielded Salmonella spp. (Table 1).
Table 1.
Total number of animals sampled, and type and number of samples positive for Salmonella in 6 abattoirs
| Number of Positive | Number of Positive | ||||||
|---|---|---|---|---|---|---|---|
| Pooled Samples (%)a | Carcass Swabs (%)b | ||||||
| Abattoir | Herds | Animals | Caeca | Lymph nodes | Caeca/Lymph nodes | Positive herds | Positive pigs |
| A | 22 | 110 | 11 (50) | 5 (22.7) | 11 (50) | 6 (27.3) | 6 (5.5) |
| B | 30 | 150 | 17 (56.7) | 10 (33.3) | 17 (56.7) | 13 (43.3) | 25 (16.7) |
| C | 40 | 200 | 19 (47.5) | 8 (20) | 20 (50) | 7 (17.5) | 10 (5) |
| D | 26 | 130 | 14 (53.8) | 5 (19.2) | 15 (57.7) | 4 (15.4) | 5 (3.8) |
| E | 25 | 125 | 19 (76) | 15 (60) | 19 (76) | 8 (32) | 10 (8) |
| F | 21 | 105 | 11 (52.4) | 9 (42.9) | 12 (57.1) | 12 (57.1) | 38 (36.2) |
| Total | 164 | 820 | 91 (55.5) | 52 (31.7) | 97 (59.1) | 50 (30.5) | 94 (11.5) |
aPools of 5 samples were tested in the case of caecal and lymph node samples and thus each pooled sample represents the result for one herd
bCarcass swabs were tested individually; 5 samples were collected per herd
The distribution of Salmonella serotypes found and the abattoir, from which the positive samples were collected, is shown in Table 2. The most dominant serotype from each sample type was mST. Among the 52 Salmonella isolated from lymph nodes, 38 (73%) of these were either serotyped as S. Typhimurium i.e., 4,[5],12:i:1,2 or as its monophasic variant, mST 4,[5],12:i:-: with the remaining serotypes identified as S. London (4), S. Kimuenza (2), S. Derby (2), S. Infantis (1), S. Goldcoast (1), S. Enteritidis (1), S. Mbandaka (1) and S. Unnamed (2). The two S. Unnamed isolates from lymph nodes had antigenic formulas 4:-:2 and 6,7:-:5. Among the 91 Salmonella isolated from caecal contents, 61 (67%) were either serotyped as S. Typhimurium or as mST with the remaining serotypes identified as S. London (9), S. Derby (6), S. Kimuenza (4), S. Infantis (2), S. Anatum (2), S. Goldcoast (1), S. Enteritidis (1), S. Manhattan (1), S. Putten (1), S. Cubana (1) and S. Unnamed (2). The two unnamed isolates had antigenic formulas 4:-:2 and 6,8:-:5 (Table 2).
Table 2.
Main Salmonella serotypes isolated from *pooled caecal content and lymph node samples and individual carcass swabs collected in each of 6 abattoirs
| Number Positive (% of isolates from same sample type and same abattoir) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample Type | Abattoir | Total isolates per abattoir | Mono ST | ST | S. Kimuenza | S. London | S. Derby | S. Infantis | S. Anatum | Other | |
| *Lymph Node | A | n = 5 | 2 (4) | 1(2) | - | - | - | - | - | 2(4) | |
| B | n = 10 | 5(10) | 3(6) | - | - | - | 1(2) | - | 1(2) | ||
| C | n = 8 | 4(8) | 3(6) | - | - | - | - | - | 1(2) | ||
| D | n = 5 | 1(2) | 4(8) | - | - | - | - | - | - | ||
| E | n = 15 | 6(12) | 5(10) | - | 4(8) | - | - | - | - | ||
| F | n = 9 | 3(6) | 1(2) | 2(4) | - | 2(4) | - | - | 1(2) | ||
| *Caecal Contents | A | n = 11 | 4(4) | 4(4) | - | 1(1) | 1(1) | - | - | 1(1) | |
| B | n = 17 | 7(8) | 6(7) | 1(1) | - | - | 1(1) | - | 2(2) | ||
| C | n = 19 | 11(12) | 3(3) | - | - | 1(1) | - | 2(2) | 2(2) | ||
| D | n = 14 | 9(10) | 2(2) | - | 1(1) | 1(1) | 1(1) | - | - | ||
| E | n = 19 | 9(10) | 1(1) | - | 7(8) | 1(1) | - | - | 1(1) | ||
| F | n = 11 | 3(3) | 2(2) | 3(3) | - | 2(2) | - | - | 1(1) | ||
| Carcass Swabs | A | n = 6 | 6(6) | - | - | - | - | - | - | - | |
| B | n = 25 | 17(18) | 4(4) | 1(1) | - | 1(1) | - | - | 2(2) | ||
| C | n = 8 | 5(5) | - | - | - | 1(1) | - | - | 2(2) | ||
| D | n = 5 | - | 5(5) | - | - | - | - | - | - | ||
| E | n = 10 | 3(3) | 3(3) | - | 1(1) | 1(1) | - | - | 2(2) | ||
| F | n = 38 | 7(7) | 3(3) | 25(27) | - | 3(3) | - | - | - | ||
Ninety-two Salmonella were isolated from carcass swabs and among these, 53 (57%) were serotyped as S. Typhimurium or as mST with the remaining serotypes identified as S. Kimuenza (26), S. London (1), S. Derby (6), S. Gloucester (1), S. Rissen (1) and S. Unnamed (4). The four S. Unnamed isolates had antigenic formulas 4:-:2, 4:l,v:-:, 13:-:-: and 13:f,g:-: (Table 2).
Herds with the highest and lowest serological prevalence
Herds in the bottom 10% had an average serological prevalence of < 1.3% while those in the top 10% had an average prevalence of > 37.9%. There were no significant differences in any of the bacteriological results between herds in the top and bottom 10% serological prevalence groups (data not shown).
Probability of a positive carcass swab
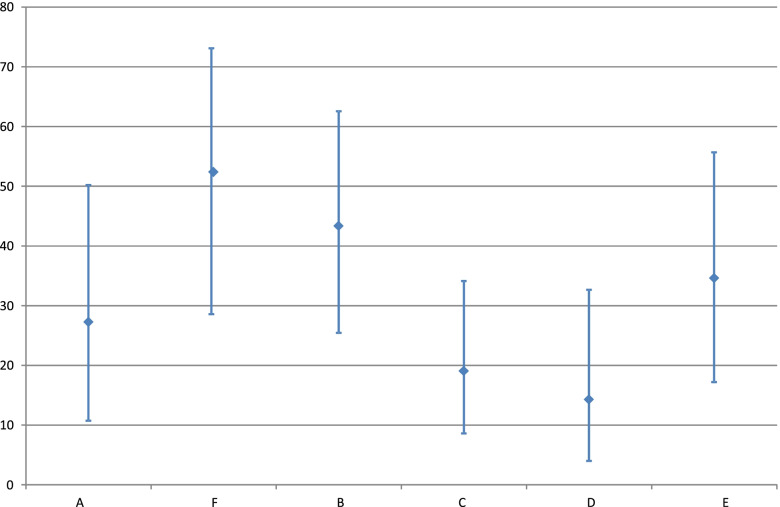
Figure 1 shows the proportion of herds from each of the 6 abattoirs with a positive carcass swab along with 95% confidence intervals. All of the confidence intervals overlap; Abattoir F had the highest proportion of positive carcass swabs and Abattoir D had the lowest.
Fig. 1.
Percentage of herds with a positive carcass swab by processing plant
The final logistic regression model developed to model the probability of a herd having a positive carcass swab contained just the lymph node variable; all other variables were not significant and were excluded from the final model. The odds of a herd having a positive carcass swab were 2.69 (95% CI 1.35–5.39) times higher when the herd had a positive lymph node result compared to herds with a negative lymph node result (P = 0.005).
Antimicrobial resistance
The antimicrobial resistance profiles of the two hundred and thirty-five isolates tested are shown in Table 3. Among the 102 mST 4 [5], 12:i:-, one isolate was resistant to 6 antimicrobials (AMP CHL GEN TET TIG TMP), 34 were resistant to 5 antimicrobials (AMP CHL GEN TET TMP), 22 were resistant to different combinations of 4 antimicrobials and 32 isolates displayed AMR profiles of AMP TET. In general, the AMR profiles of the 50 S. Typhimurium isolates differed from those of mST. The predominant pattern in mST isolates was resistance to AMP CHL GEN TET TMP whereas the most common pattern in the ST isolates was resistance to AMP CHL TET.
Table 3.
Antimicrobial resistance profiles of Salmonella (n = 235) from lymph nodes, caecal samples and carcass swabs
| AMR Profile | Monophasic variant | S. Typhimurium | S. Derby | Other serovars | Total |
|---|---|---|---|---|---|
| AMP CHL CIP GEN TET TIG TMP | 1 | 1 | |||
| AMP CHL GEN TET TIG TMP | 1 | 1 | |||
| AMP CHL GEN TET TMP | 34 | 3 | 2 | 39 | |
| CHL CIP TET TIG TMP | 1 | 1 | |||
| AMP CHL TET TIG TMP | 2 | 1 | 3 | ||
| CHL TET TIG TMP | 1 | 1 | |||
| AMP GEN TET TMP | 2 | 1 | 3 | ||
| AMP CHL TET TMP | 4 | 4 | 8 | ||
| AMP CHL CIP TET | 1 | 1 | |||
| AMP GEN TET | 15 | 1 | 1 | 17 | |
| AMP CHL TET | 14 | 1 | 15 | ||
| AMP TET TMP | 2 | 1 | 1 | 4 | |
| CIP NAL | 2 | 2 | |||
| TET TMP | 8 | 8 | |||
| CHL TET | 1 | 1 | |||
| AMP TET | 32 | 3 | 35 | ||
| TMP | 2 | 2 | |||
| TET | 2 | 5 | 3 | 2 | 12 |
| AMP | 5 | 8 | 13 | ||
| Fully Susceptible | 4 | 7 | 3 | 54 | 68 |
| Total number of isolates | 102 | 50 | 14 | 69 | 235 |
The 69 isolates of serotypes other than S. Typhimurium, mST, and S. Derby were grouped together as the majority of these were fully susceptible, i.e., 54 isolates were fully susceptible. Within this group of serotypes, eight were multi drug resistant displaying profiles of AMP CHL CIP GEN TET TIG TMP (1), AMP CHL GEN TET TMP (2), CHL CIP TET TIG TMP (1), AMP CHL TET TIG TMP (1), AMP GEN TET (1), AMP CHL TET (1) and AMP TET TMP (1); the two S. Goldcoast isolates showed AMR profiles of AMP CHL CIP GEN TET TIG TMP and CHL CIP TET TIG TMP. The three untypable 4:-:2: isolates were multi drug resistant with AMR profiles of AMP CHL GEN TET TMP, AMP CHL TET and AMP CHL TET TIG TMP. One untypable 6,8:-:5: isolate displayed an AMR profile of AMP, CHL, GEN, TET, TMP and one S. Gloucester isolate had an AMR profile of AMP, GEN, TET. The remaining 7 isolates consisting of two S. Anatum, one S. Rissen, one S. Kimuenza and three untypable isolates of antigenic formula 13:f,g:-, 13:-:-: and 4:l,v:-: displayed AMR profiles of CIP NAL, TET, AMP TET, AMP TET, AMP TET and TET, respectively.
Discussion
Salmonella spp. were isolated from 91 (55.5%) of pooled caecal contents and 52 (31.7%) of pooled ileo-caecal lymph nodes from 164 herds sampled, which represent about half of the commercial pig fattening herds in Ireland. The much lower rate of detection, 11.5% (94 of 820), in carcass-swab samples, is indicative of improvements in the effectiveness of process hygiene controls, despite the high levels of caecal and lymph node positive samples.
Salmonella contamination of carcasses is a key parameter used to measure the effectiveness of process hygiene criteria of the abattoirs. While the rate of Salmonella detected on carcasses in the present study was higher than the EU average of 10.3% which was reported in the baseline study [10], nevertheless, the reduction in Salmonella contamination of carcasses (to 11.5%) in this study compared to that of the baseline study (20%) is a positive development. This reduction was achieved despite a possibly increased Salmonella prevalence in the fattening pigs where carriage rates, as reflected by ileo-caecal lymph node Salmonella prevalence increased from 16.1% during the baseline survey to 31.7% in this study. A further reduction in the proportion of carcasses from which Salmonella spp. were detected to; 4.7%, 4.2% and 5.9% in the years 2018, 2019 and 2020, respectively, has been reported from results of official testing in accordance with Commission Regulation (EC) No. 218/2014 [2, 9, 19]. However, the results between the baseline and this present study are not directly comparable since the baseline survey refers to the prevalence from individual pigs whereas this study refers to prevalence in pooled samples from 5 pigs.
Isolation of Salmonella serotypes from the ileo-caecal lymph nodes is frequently considered to reflect Salmonella prevalence on farm; however short-term exposure in the lairage can also lead to lymph node positivity [20, 21]. The high prevalence in lymph nodes suggests that Salmonella remains a significant (and possibly an increasing) problem at farm level and in the lairage area and a challenge to abattoirs in adhering to process hygiene requirements.
Although many studies on Salmonella in pigs in EUMS have been published, the baseline study [10] provided a uniform approach in which methods of sampling and testing were standardised across all participating countries. Factors such as differences in the nature of the swabs used, the areas swabbed and in many cases the point at which the swabs were taken, must be considered, in any comparison of the carcass swab results in the present study to those found in other studies. All the sampling done in our study was undertaken under official veterinary supervision to ensure consistency of approach between different abattoirs. The results of the present study are not unlike those found in Northern Ireland and England although culture methods varied. During 2002, Salmonella spp. was isolated from 31.4% of caecal contents and 40.0% of pre chill carcass swabs in Northern Ireland [12]. The UK baseline study identified Salmonella in 21.2% of ileo-caecal lymph nodes and 13.5% of carcasses [1]. An earlier UK survey during 2000 [22] reported carriage of Salmonella in 23.0% of caecal contents. Further studies found Salmonella spp. in 23.4% of pigs sampled during 2003 [23]. A more recent study conducted in the UK during 2013 [24] reported a Salmonella prevalence of 30.5% in caecal contents and 9.6% of carcass swabs, which are very similar to the results of this present study (caecal contents = 31.7%; carcass swabs = 11.5%). In addition, process hygiene criterion monitoring data on Salmonella from pig carcasses for 2019, reported a prevalence of 3.15%, compared to 6.7% of Salmonella-positive samples from carcasses during 2006 / 2007 [10]. Other studies provide further information on the prevalence of Salmonella in pigs in the Republic of Ireland. A study during 2001 identified Salmonella in the caecum samples of 37% low risk herds and 66.6% higher risk herds [25]. Another study conducted during 2007, reported a Salmonella prevalence of 14.8% and 11.7% of lymph nodes and caecal contents respectively, and 10.2%, 3.9%, 1.8%, and 7.2% of pre-wash, post wash, post chill and belly-strip carcass swabs, respectively [26]. Bolton et al., 2013 [27] investigated the prevalence of Salmonella in pigs from birth to carcass and reported a prevalence of 27.5% and 5% of throat/rectal and carcass samples positive for Salmonella, respectively.
The results of this study shows that S. Typhimurium and mST predominated in all samples, accounting for 65% of all isolates, 67% of caecal content isolates, 73% of lymph node isolates and 57% of carcass swab isolates. The predominance of S. Typhimurium and mST reported among porcine Salmonella isolates here is similar to that reported in the EU baseline survey and other surveys in Ireland and elsewhere. However, while the predominance of S. Typhimurium and mST, collectively, has continued it is noteworthy that the relative proportion of isolates that are mST (n = 102) relative to S. Typhimurium (n = 50) has increased in recent years and mST is now widely seen across Europe as the more prevalent serotype in pigs and pigmeat [28–31]. Overall, the prevalence of mST relative to samples positive was 43.4% in this present study compared to 21.3% for S. Typhimurium which is very similar to that (39%) reported by Mueller-Doblies et al. [28] during 2014. It is also noteworthy that mST was more predominant than ST in the present study from each porcine sample type with 40.4%, 47.3% and 41.3% mST identified in lymph nodes, caecal contents and carcass swabs respectively compared to 32.7%, 19.8% and 16.3% positive for S. Typhimurium. During 2017, mST were among the most commonly reported serotypes isolated from pigs in the EU. Pigs and pig meat accounted for, 167 (37.4%) and 129 (22%) of mST isolates respectively. These results confirm that pigs are the main reservoir for mST and the results of the study reported here support this conclusion.
The levels of antimicrobial resistance detected, with high levels of resistance to ampicillin, chloramphenicol, streptomycin, sulphonamides, tetracycline and trimethoprim, in S. Typhimurium and mST are comparable to other studies [22, 32]. For all serotypes, resistance to nalidixic acid and ciprofloxacin (1.7%), was low compared to other studies where resistance in all Salmonella serotypes to nalidixic acid alone was reported to be 4.1% in GB and 6.5% Spain [22, 32]. In both of these previous studies nalidixic acid resistance was common in S. Typhimurium. A study conducted in the Republic of Ireland by Bolton et al. 2013 [27] reported a high level of resistance of S. Typhimurium to streptomycin, sulfonamides, tetracyclines and trimethoprim that are commonly used individually or in combination in veterinary medicine [33]. In this study, 94 (40%) of isolates were resistant to at least three or more antimicrobials, 41 (17.4%) of isolates were fully susceptible and 27 (11.5%) of isolates were resistant to only one antimicrobial. Among the 94 isolates resistant to three or more antimicrobials, 59 (62.7%) were mST, 27 (28.7%) were S. Typhimurium and 8 (8.5%) were a combination of other serotypes. These results provide more evidence that mST and S. Typhimurium have developed a high level of resistance in particular to older antibiotics which have been used over a long period of time in the pig industry. Antimicrobials, to which the highest frequencies of resistance detected in E. coli isolates from pigs have been reported previously to be tetracycline, trimethoprim/sulphamethoxazole and streptomycin, with the highest levels of resistance in weaned pigs [34]. The high level of multidrug resistance observed, particularly in mST is consistent with other research reporting this variant to be a host of multiple drug resistance [35].
Reducing Salmonella contamination of pig meat products is dependent on effective control on the farm and in the abattoir. S. Typhimurium is endemic on a significant proportion of Irish pig farms and further research is required to determine factors which have created a favourable environment for the establishment and persistence of this organism. However, progress in reducing Salmonella contamination of pork carcasses could likely be made in the short term by focussing on further improvements during the slaughter process [26].
Conclusions
The results of this study indicate a decrease in Salmonella contamination of carcasses in Ireland since the EU Baseline study in 2008, most likely due to improvements in hygiene control measures during processing. However, as Salmonella spp. isolates were recovered from a higher percentage of ileo-caecal lymph nodes, there remain a significant and possibly growing prevalence of Salmonella spp in fattening pigs. The high level of multidrug resistance in mST serotypes is a cause for concern. These results coupled with the results of the on-going serological monitoring of fattening pigs indicate that the control programme may be having little impact at farm level. The prevalence of Salmonella spp. in pigs and the difficulty in its control are well recognised and attempts for coordinated EUMS controls have not been feasible. The continuing high prevalence of Salmonella in fattening pigs should focus attention on the need for further controls at farm level and a re-examination of the current control programme.
Acknowledgements
The authors wish to acknowledge the management of the Department of Agriculture, Food and the Marine and its Backweston Laboratories, DAFM Veterinary and Agricultural Inspectorate in headquarters and at the pig processing abattoirs and the school of veterinary medicine, UCD. The authors also wish to acknowledge the laboratory help and support provided by Tony O’Brien.
Authors’ contributions
The study’s conception and design was done by J.E and F.L who also contributed to writing and editing the manuscript. A.D carried out all testing and contributed to writing the manuscript. D.P supervised the testing, interpreted data and contributed to writing and editing the manuscript. W.B reviewed and edited the final draft. D.M, G.M and M.G co-ordinated various testing activities. All authors read and approved the final manuscript.
Funding
N/A.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
N/A
Consent for publication
N/A
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Herikstad H, Motarjemi Y, Tauxe RV. Salmonella surveillance: a global survey of public health serotyping. Epidemiol Infect. 2002;129(1):1–8. doi: 10.1017/S0950268802006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.EFSA and ECDC European Food Safety Authority and European Centre for Disease Prevention and Control https://www.efsa.europa.eu/en/efsajournal/pub/6406. Accessed 27 Jan 2022 3. Hald T, Wingstrand A, Swanenburg M, von Altrock A, Thornberg BM. The occurrence and epidemiology of Salmonella in European pig slaughterhouses. Epidemiol Infect. 2003;131(3):1187–203. [DOI] [PMC free article] [PubMed]
- 3.Hald T, Wingstrand A, Swanenburg M, von Altrock A, Thornberg BM. The occurrence and epidemiology of Salmonella in European pig slaughterhouses. Epidemiol Infect. 2003;131(3):1187–1203. doi: 10.1017/S0950268803001171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prendergast DM, Duggan SJ, Gonzales-Barron U, Fanning S, Butler F, Cormican M, Duffy G. Prevalence, numbers and characteristics of Salmonella spp. on Irish retail pork. Int J Food Microbiol. 2009;131(2–3):233–9. doi: 10.1016/j.ijfoodmicro.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Prendergast DM, Duggan SJ, Fanning S, Cormican M, Gonzales-Barron U, Butler F, Duffy G. Prevalence and numbers of Salmonella spp. and Enterobacteriaceae on pork cuts in abattoirs in the Republic of Ireland. J Appl Microbiol. 2008;105(4):1209–19. doi: 10.1111/j.1365-2672.2008.03854.x. [DOI] [PubMed] [Google Scholar]
- 6.Alban L, Stege H, Dahl J. The new classification system for slaughter-pig herds in the Danish Salmonella surveillance-and-control program. Prev Vet Med. 2002;53(1–2):133–146. doi: 10.1016/S0167-5877(01)00270-7. [DOI] [PubMed] [Google Scholar]
- 7.Van Pelt W, Valkenburg S. Zoonoses and zoonotic agents in human, food, animals and feed in the Netherlands. Netherlands: Insp Heal Prot Vet Publ Heal Hague; 2001. [Google Scholar]
- 8.Duggan SJ, Mannion C, Prendergast DM, Leonard N, Fanning S, Gonzales-Barron U, Egan J, Butler F, Duffy G. Tracking the Salmonella status of pigs and pork from lairage through the slaughter process in the Republic of Ireland. J Food Prot. 2010;73(12):2148–2160. doi: 10.4315/0362-028X-73.12.2148. [DOI] [PubMed] [Google Scholar]
- 9.EFSA and ECDC European Food Safety Authority and European Centre for Disease Prevention and Control https://www.efsa.europa.eu/en/efsajournal/pub/5926. Accessed 27 Jan 2022.
- 10.EFSA. European Food Safety Authority. 2008 https://efsa.onlinelibrary.wiley.com/doi/10.2903/j.efsa.2008.206r. Accessed 27 Jan 2022.
- 11.EFSA. European Food Safety Authority. 2010 https://www.efsa.europa.eu/en/efsajournal/pub/1547. Accessed 27 Jan 2022.
- 12.McDowell SWJ, Porter R, Madden R, Cooper B, Neill SD. Salmonella in slaughter pigs in Northern Ireland: prevalence and use of statistical modelling to investigate sample and abattoir effects. Int J Food Microbiol. 2007;118(2):116–125. doi: 10.1016/j.ijfoodmicro.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Microbiology of the food chain — horizontal method for the detection, enumeration and serotyping of Salmonella — part 1: detection of Salmonella spp., ISO 6579–1:2017, International Organization for Standardization, Geneva. [DOI] [PubMed]
- 14.Popoff MY. Antigenic formulas of the Salmonella serovars. 8. Paris, France: WHO Collaborating Centre for Reference and Research on Salmonella; 2001. [Google Scholar]
- 15.Prendergast DM, Hand D, Nί Ghallchóir E, McCabe E, Fanning S, Griffin M, Egan J, Gutierrez M. A multiplex real-time PCR assay for the identification and differentiation of Salmonella enterica serovar Typhimurium and monophasic serovar 4,[5],12:i:–. Int J Food Microbiol. 2013;166(1):48–53. doi: 10.1016/j.ijfoodmicro.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. M07-A10. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approv Stand Ed CLSI Doc M07-A10. 2015;(10):1–87.
- 17.Official Journal of the European Union. Commission Implementing Decision2013/652/EU of 12 November 2013 on the monitoring and reporting of antimicrobial resistance in zonotic and commensal bacteria. L 303, 14.11.2013; 2013. p. 26–39. https://eur-europa.eu/LexUriServ/LexUriServ.do. Accessed 13 June 2019.
- 18.Ramovic E, Madigan G, McDonnell S, Griffin D, Bracken E, Ni Ghallchoir E, Quinless E, Galligan A, Egan J, Prendergast D.M. A pilot study using environmental screening to determine the prevalence of Mycobacterium avium subspecies paratuberculosis (MAP) and antimicrobial resistance (AMR) in Irish cattle herds. Ir Vet J. 2020;73(3). 10.1186/s13620-020-0156-2. [DOI] [PMC free article] [PubMed]
- 19.EFSA. European Food Safety Authority. 2021 https://efsa.onlinelibrary.wiley.com/doi/10.2903/j.efsa.2018.5500. Accessed 16 Feb 2021.
- 20.Hurd HS, McKean JD, Griffith RW, Wesley IV, Rostagno MH. Salmonella enterica infections in market swine with and without transport and holding. Appl Environ Microbiol. 2002;68(5):2376–2381. doi: 10.1128/AEM.68.5.2376-2381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boughton C, Egan J, Kelly G, Markey B, Leonard N. Rapid Infection of pigs following exposure to environments contaminated with different levels of salmonella typhimurium. Foodborne Pathog Dis. 2007;4(1):33–40. doi: 10.1089/fpd.2006.58. [DOI] [PubMed] [Google Scholar]
- 22.Davies RH, Dalziel R, Gibbens JC, Wilesmith JW, Ryan JMB, Evans SJ, Byrne C, Paiba GA, Pascoe SJ, Teale CJ. National survey for Salmonella in pigs, cattle and sheep at slaughter in Great Britain (1999–2000) J Appl Microbiol. 2004;96:750–760. doi: 10.1111/j.1365-2672.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- 23.Milnes AS, Stewart I, Clifton-Hadley FA, Davies RH, Newell DG, Sayers AR, Cheasty T, Cassar C, Ridley A, Cook AJ, Evans SJ, Teale CJ, Smith RP, McNally A, Toszeghy M, Futter R, Kay A, Paiba GA. Intestinal carriage of verocytotoxigenic Escherichia coli O157, Salmonella, thermophilic Campylobacter and Yersinia enterocolitica, in cattle, sheep and pigs at slaughter in Great Britain during 2003. Epidemiol Infect. 2008;136(6):739–751. doi: 10.1017/S0950268807009223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powell LF, Cheney TEA, Williamson S, Guy E, Smith RP, Davies RH. A prevalence study of Salmonella spp., Yersinia spp., Toxoplasma gondii and porcine reproductive and respiratory syndrome virus in UK pigs at slaughter. Epidemiol Infect. 2016;144(7):1538–49. doi: 10.1017/S0950268815002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quirke AM, Leonard N, Kelly G, Egan J, Lynch PB, Rowe T, Quinn PJ. Prevalence of Salmonella serotypes on pig carcasses from high- and low-risk herds slaughtered in three abattoirs. Berl Munch Tierarztl Wochenschr. 2001;114(9–10):360–362. [PubMed] [Google Scholar]
- 26.Mannion C, Fanning J, McLernon J, Lendrum L, Gutierrez M, Duggan S, Egan J. The role of transport, lairage and slaughter processes in the dissemination of Salmonella spp. in pigs in Ireland. Food Res Int. 2012;45(2):871–9. doi: 10.1016/j.foodres.2011.02.001. [DOI] [Google Scholar]
- 27.Bolton DJ, Ivory C, McDowell D. A study of salmonella in pigs from birth to carcass: serotypes, genotypes, antibiotic resistance and virulence profiles. Int J Food Microbiol. 2013;160(3):298–303. doi: 10.1016/j.ijfoodmicro.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Mueller-Doblies D, Speed K, Davies RH. A retrospective analysis of Salmonella serovars isolated from pigs in Great Britain between 1994 and 2010. Prev Vet Med. 2013;110(3–4):447–455. doi: 10.1016/j.prevetmed.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 29.Sun H, Wan Y, Du P, Bai L. The Epidemiology of monophasic salmonella typhimurium. Foodborne Pathog Dis. 2020;17(2):87–97. doi: 10.1089/fpd.2019.2676. [DOI] [PubMed] [Google Scholar]
- 30.Mandilara G, Sideroglou T, Chrysostomou A, Rentifis I, Papadopoulos T, Polemis M, Tzani M, Tryfinopoulou K, Mellou K. The rising burden of salmonellosis caused by monophasic salmonella typhimurium (1,4,[5],12:i:-) in Greece and new food vehicles. Antibiot. 2021;10(2):185. doi: 10.3390/antibiotics10020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Incau M, Salogni C, Giovannini S, Ruggeri J, Scali F, Tonni M, Formenti N, Guarneri F, Pasquali P, Alborali GL. Occurrence of salmonella typhimurium and its monophasic variant (4, [5],12:i:-) in healthy and clinically ill pigs in northern Italy. Porc Heal Manag. 2021;26(7):34. doi: 10.1186/s40813-021-00214-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agustín AI, Carramiñana JJ, Rota C, Herrera A. Antimicrobial resistance of Salmonella spp. from pigs at slaughter in Spain in 1993 and 2001. Lett Appl Microbiol. 2005;41(1):39–44. doi: 10.1111/j.1472-765X.2005.01710.x. [DOI] [PubMed] [Google Scholar]
- 33.Aragaw K, Molla B, Muckle A, Cole L, Wilkie E, Poppe C, Kleer J, Hildebrandt G. The characterization of Salmonella serovars isolated from apparently healthy slaughtered pigs at Addis Ababa abattoir. Ethiopia Prev Vet Med. 2007;82(3–4):252–261. doi: 10.1016/j.prevetmed.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Gibbons JF, Boland F, Egan J, Fanning S, Markey BK, Leonard FC. antimicrobial resistance of faecal escherichia coli isolates from pig farms with different durations of in-feed antimicrobial use. Zoonoses Public Health. 2016;63(3):241–250. doi: 10.1111/zph.12225. [DOI] [PubMed] [Google Scholar]
- 35.Ingle DJ, Ambrose RL, Baines S, Duchene S, Gonçalves da Silva A, Lee D, Jones M, Valcanis M, Taiaroa G, Ballard SA, Kirk MD, Howden BP, Pearson JS, Williamson DA. Evolutionary dynamics of multidrug resistant Salmonella enterica serovar 4,[5],12:i:- in Australia. Nat Commun. 2021;12(1):4786. doi: 10.1038/s41467-021-25073-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.