Abstract
DNA methylation status correlates with clinical outcomes of anti‐epidermal growth factor receptor (EGFR) treatment. There is a strong need to develop a simple assay for measuring DNA methylation status for the clinical application of drug selection based on it. In this study, we collected data from 186 patients with metastatic colorectal cancer (mCRC) who had previously received anti‐EGFR treatment. We modified MethyLite to develop a novel assay to classify patients as having highly methylated colorectal cancer (HMCC) or low‐methylated colorectal cancer (LMCC) based on the methylation status of 16 CpG sites of tumor‐derived genomic DNA in the development cohort (n = 30). Clinical outcomes were then compared between the HMCC and LMCC groups in the validation cohort (n = 156). The results showed that HMCC had a significantly worse response rate (4.2% vs 33.3%; P = .004), progression‐free survival (median: 2.5 vs 6.6 mo, P < .001, hazard ratio [HR] = 0.22), and overall survival (median: 5.6 vs 15.5 mo, P < .001, HR = 0.23) than did LMCC in patients with RAS wild‐type mCRC who were refractory or intolerable to oxaliplatin‐ and irinotecan‐based chemotherapy (n = 101). The DNA methylation status was an independent predictive factor and a more accurate biomarker than was the primary site of anti‐EGFR treatment. In conclusion, our novel DNA methylation measurement assay based on MethyLight was simple and useful, suggesting its implementation as a complementary diagnostic tool in a clinical setting.
Keywords: anti‐EGFR antibodies, DNA methylation, metastatic CRC, MethyLight, predictive biomarker
In this study, we developed a novel diagnostic method that could classify mCRC cases as highly methylated colorectal cancer (HMCC) or low‐methylated colorectal cancer (LMCC) based on the methylation status of 16 CpG sites. We then compared the clinical outcomes of patients in the validation cohort between HMCC and LMCC groups. The primary results of our study are (1) HMCC was associated with a significantly worse response rate, progression‐free survival, and overall survival than LMCC for RAS wild‐type mCRC; (2) even in the right‐sided or left‐sided colon, LMCC had a better progression‐free survival (PFS) than HMCC; (3) DNA methylation status was an independent predictive factor of anti‐EGFR treatment and a more accurate biomarker than the primary tumor site. The novel DNA methylation assay we developed in this study was useful for predicting the clinical outcomes of anti‐EGFR treatment regardless of the primary tumor site, suggesting it can be used as a complementary diagnostic in a clinical setting.

Abbreviations
- CIMP
CpG island methylator phenotype
- CR
complete response
- DC
development cohort
- EGFR
epidermal growth factor receptor
- FFPE
formalin‐fixed paraffin‐embedded
- HMCC
highly methylated colorectal cancer
- HR
hazard ratio
- LMCC
low‐methylated colorectal cancer
- mCRC
metastatic colorectal cancer
- MeC‐mML
DNA methylation status assay of mCRC by mML
- ML
MethyLight
- mML
modified ML
- OS
overall survival
- PD
progressive disease
- PFS
progression‐free survival
- PR
partial response
- ROC
receiver operating characteristic
- RR
response rate
- SD
stable disease
- VC
validation cohort
1. INTRODUCTION
Colorectal cancer (CRC) has shown a significant increase in mortality in recent years, with 880 000 deaths reported worldwide as of 2018, making it the second most common neoplasm after lung cancer. 1 Molecular targeted therapies, such as anti‐EGFR antibodies, 2 , 3 have been introduced to treat metastatic CRC (mCRC). 4 As a result, the life expectancy of patients with mCRC is >30 mo. 5 , 6
Molecularly targeted agents are more effective than conventional cytotoxic chemotherapeutic agents and their efficacy is further increased when biomarkers are used to select appropriate treatment targets. 7
RAS (KRAS and NRAS) mutations are negative biomarkers for anti‐EGFR treatment resistance in mCRC. 8 , 9 However, several clinical trials have demonstrated that the RR to anti‐EGFR antibodies is ~30% 8 , 9 , 10 in patients with wild‐type RAS, suggesting that RAS mutations are not sufficient as predictive biomarkers.
Recently, it has become clear that the primary site of mCRC, in addition to the RAS mutation status, is associated with the efficacy of anti‐EGFR treatment, 11 , 12 , 13 and the strategy for selecting therapeutic agents based on the primary site is being incorporated into clinical settings. 14 , 15 , 16 However, both the primary site and the RAS mutation status are not associated with the efficacy of anti‐VEGF treatment. 17
We previously performed a genome‐wide DNA methylation analysis to identify novel biomarkers predicting the clinical outcomes of anti‐EGFR treatment. 18 We found a strong association between the comprehensive DNA methylation status and the therapeutic effect of anti‐EGFR antibodies, indicating that DNA methylation is a predictor of anti‐EGFR therapy efficacy. Specifically, highly methylated CRC (HMCC) was resistant to anti‐EGFR treatment, and its efficacy was comparable with that of RAS mutant CRC. In contrast, patients with low‐methylated CRC (LMCC) had significantly better outcomes compared with those of patients with RAS mutant CRC and HMCC. Multivariate analysis suggested that the DNA methylation status was a determinant of the efficacy of anti‐EGFR treatment independent of the RAS mutation status. An association between the DNA methylation status and anti‐EGFR antibody sensitivity has also been reported by Lee et al. 19 However, there are various challenges to be addressed in the clinical application of the clustering‐based classification method using genome‐wide DNA methylation data. Especially, clustering analysis is not the most accurate method to classify individual patients, because it is susceptible to being influenced by the analysis population. Therefore, the development of assays to conveniently measure the DNA methylation status in individual cases is of great significance.
This study aimed to develop an assay to easily determine the DNA methylation status and verify that its measurements correlate with the clinical outcomes of anti‐EGFR treatment in mCRC.
2. MATERIAL AND METHODS
2.1. Patients
This study included patients with mCRC who were refractory or intolerable to oxaliplatin‐ and irinotecan‐based chemotherapy and were treated with anti‐EGFR antibodies. Surgically resected primary tumors without preoperative chemotherapy were included in the analysis. Patients that overlapped with our previous study 18 were excluded from the development cohort and the validation cohort.
Informed consent was obtained from the case groups used for the extraction of the CpG site used to classify HMCC and LMCC and for the development of the assay (development cohort, DC), based on the “Development of a molecular biomarker for colorectal cancer” (UMIN000005490), which was approved by the Ethics Committee of the Tohoku University School of Medicine. Informed consent was obtained from the case group used to validate the accuracy of the developed assay in predicting the treatment effect of anti‐EGFR antibody drugs (validation cohort, VC), based on the “Prediction of the treatment effect of anti‐EGFR antibody drugs in colorectal cancer based on DNA methylation status and gene expression status” (UMIN000027296), which was approved by the Ethics Committee of the School of Medicine, Tohoku University School of Medicine.
Details are provided in Appendix S1.
2.2. Mutation analyses of KRAS, NRAS, and BRAF genes
Gene mutation analysis for codons 12 and 13 of the KRAS gene (KRAS status) was performed for all patients by the Sanger or Scorpion‐ARMS method. 20 The MEBGEN™ RASKET kit (MBL) 21 was used to identify mutations in exon 2 (codons 12 and 13), exon 3 (codons 59 and 61), and exon 4 (codons 117 and 146) of the KRAS and NRAS genes (RAS status) in patients who were available for analysis. Gene mutation analysis for codon 600 of the BRAF gene was performed in cases that were available for analysis by Sanger sequencing.
2.3. Genome‐wide DNA methylation analysis
Genome‐wide DNA methylation data obtained from our previous study 18 were used to identify the target CpG sites in the assay to be developed in this study. An exhaustive evaluation of the DNA methylation status was performed using the Illumina Infinium HumanMethylation450 BeadChip (Illumina). The BeadChips were scanned on a BeadStation and methylation measurements with a detection P‐value > .05 were excluded from the analysis. All samples achieved a CpG coverage >95%. The raw data were normalized using internal controls and GenomeStudio software. The methylation level [β value: intensity of the methylated signal/(intensity of the unmethylated signal + intensity of the methylated signal)] in each CpG site was calculated using the Methylation Module attached to the BeadStudio.
Details are provided in the Appendix S1.
2.4. Identification of target CpG sites in the DNA methylation status assay
Based on the genome‐wide DNA methylation status, mCRC was classified into HMCC and LMCC groups according to unsupervised clustering analyses. 18 CpG sites with a β value of <0.1 in the normal colon mucosa and LMCC and >0.3 in HMCC were selected. 22 Among these sites, a t test was performed between the LMCC and HMCC groups using a Welch t test, adjusted Bonferroni correction, and adjusted P‐value threshold of <1.0E‐05. The CpG sites showing statistically significant differences in β values between the 2 groups and those with an average β value in HMCC that was >0.35 higher than that in LMCC were selected as the regions for analysis using the DNA methylation status assay.
2.5. Development of the DNA methylation status assay of mCRC based on modified MethyLight
We had chosen MethyLight (ML) 23 as the primary experimental system for measuring the DNA methylation status. ML uses common primers for target amplification and specific probes for methylated and unmethylated target sequences. However, due to difficulties in maintaining the specificity of common primers that could amplify without being affected by methylation or unmethylation, we decided to use methylated sequence‐specific primers and probes for the target sequences at selected specific sites in our modified assay. The methylation status was qualitatively measured by comparing the amplification curve detected with the methylated probe of the target sequence with the amplification curve detected with the internal control, which does not contain CpG sites in the design regions of the primers and probes (modified MethyLight, mML).
For mML, forward and reverse primers and probes that were specific for methylation sequences at specific selected sites (as mentioned in Section 2.4) and for internal controls, which targeted the promoter region of the ACTB gene without CpG sites in the primer and probe regions, were designed. The genomic DNA of the tumor was subjected to sodium bisulfite treatment. After sodium bisulfite treatment, real‐time PCR reactions were performed using the aforementioned primer and probe sets. The difference between the Ct values obtained for the methylated sequence‐specific primer/probe and internal standard primer/probe sets was calculated, and the ΔCt value was determined. Twenty cases analyzed in a previously published paper 18 were measured by mML, and the calculated ΔCt values were compared with the β values calculated by the Infinium methylation assay. As a result, a ΔCt value of <4 showed strong concordance with a β value ≥0.3. Therefore, the site was judged to be methylation positive if the calculated ΔCt value was <4.
Details are described in the Supplementary Appendix.
2.6. Investigation of the effect of tumor purity on the measurement outcomes
The methylation status of samples with various tumor contents was measured by the DNA methylation status assay of mCRC by mML (MeC‐mML) with or without macro‐dissection. The classification results (HMCC or LMCC) were compared between matched samples with or without macro‐dissection.
2.7. Clinical outcomes and statistical analyses
The primary endpoint was PFS. The secondary endpoints were OS and RR.
Tumor reduction was analyzed using Response Evaluation Criteria in Solid Tumors (RECIST) v.1.1. The RR was calculated by dividing the total patients with CR and PR by the total number of patients.
The PFS was defined as the time from the date of anti‐EGFR treatment initiation to the date of tumor progression confirmed by imaging or clinically determined progression. OS was defined as the time from the date when anti‐EGFR treatment was initiated to the date of death.
Statistical analyses of patient demographic factors were performed using the chi‐square test, Wilcoxon test, and Fisher exact test. Analyses of patient background factors and anti‐EGFR treatment results were performed using JMP Pro14 software (SAS Institute Japan Co., Ltd.). Survival curves were constructed using the Kaplan‐Meier method, and differences between curves of the 2 subgroups were compared using the log‐rank test. Associations between patient background factors, including between the methylation status and PFS and OS, were used for univariate and multivariate analyses with a Cox proportion hazard model.
ROC curve analysis was performed to determine the cutoff value of methylation‐positive sites separating HMCC from LMCC using JMP Pro14 software (SAS Institute Japan Co., Ltd.). In this analysis, the variable was the number of methylation‐positive sites, and the number of positive sites that maximized the [sensitivity − (1 − specificity)] in determining a response case as LMCC was calculated as the cutoff value.
In statistical analysis, a P‐value of < .05 was considered statistically significant.
Details are provided in the Appendix S1.
3. RESULTS
3.1. Patients
This study included 186 mCRC patients treated with an anti‐cancer regimen, including anti‐EGFR antibodies. Among them, the first 30 patients were included in the DC to select CpG sites and develop an assay to classify the HMCC and LMCC groups. Another 156 patients were included in the VC to verify the accuracy of the developed assay in predicting the clinical outcomes of anti‐EGFR treatment. In the VC, all 156 patients were tested for KRAS status, and 13 patients had mutations in KRAS codons 12 or 13. In total, 122 patients in the VC were tested for RAS status in addition to KRAS status. As a result, 101 patients were determined to be RAS wild‐type and 21 patients were determined to be RAS mutant (Figure 1). None of the 21 patients had multiple RAS mutations.
FIGURE 1.
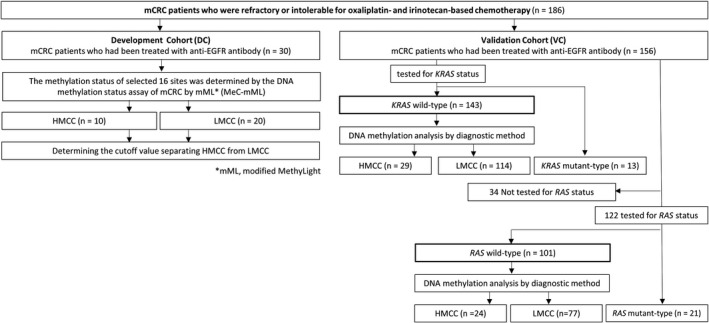
Study profile. EGFR, epidermal growth factor receptor; HMCC, highly methylated colorectal cancer; LMCC, low‐methylated colorectal cancer; mCRC, metastatic colorectal cancer
3.2. Identification of target sites for DNA methylation analysis and development of an assay to predict the clinical outcome of anti‐EGFR treatment based on DNA methylation status
In total, 2458 CpG sites matched the filtering criteria described in Section 2.3 and Supplementary Appendix. These sites involved 275 CpG sites with a mean β value <0.1 in LMCC and normal colonic mucosa and a mean β value >0.3 in HMCC. Among them, CpG sites with statistically significant differences in β values between LMCC and HMCC were identified. As a result, 16 sites were selected as target sites used for developing the assay (Table S1). The sequences of the primers and probes that were specific for methylation sequences at selected 16 sites and for internal controls are shown in Table S2.
The methylation status of these 16 sites was determined by MeC‐mML. To confirm whether the developed assay could accurately determine the methylation status, primer specificity was evaluated using fully methylated and fully unmethylated DNA for the 16 selected sites (n = 3, respectively). The presence or absence of methylation was accurately reflected in all 16 sites (Table S3). Therefore, the cutoff value was determined by ROC analysis in our preliminary test with the DC (Tables S4, S5; Figure S1). Therefore, we defined HMCC as ≥8 methylation‐positive sites and LMCC as ≤7 methylation‐positive sites out of the 16 sites. Finally, 10 cases of DC were classified as HMCC and 20 cases as LMCC (Table S6). Although 1 RAS mutant case was included in HMCC and 5 in LMCC, there were no significant differences between the 2 groups (Table S7) and there were no statistically significant differences in other patient background characteristics between the 2 groups (Table S7). The PFS in the LMCC group was significantly prolonged compared with that in the HMCC group (P = .013; Figure S2).
3.3. Measurement of methylation status by MeC‐mML and its comparison with patient backgrounds
The DNA methylation status of 156 patients in the VC was measured by MeC‐mML. As a result, 32 patients were classified into the HMCC group and 124 into the LMCC group.
Using specimens with different tumor content ratios of 20%‐80% in the VC (n = 40), we investigated whether the results obtained from the MeC‐mML differed based on tumor purity. The results showed that in 97.5% (39/40 pairs) of the matched samples, the results of the classification (HMCC or LMCC) were consistent when measured by MeC‐mML with or without macro‐dissection (Table S8). This result suggests that this assay was less sensitive to the tumor content of the samples.
Then, we compared patient backgrounds between the 2 groups (Table 1). The results revealed a significant difference in the primary site and BRAF status (P < .001). Especially, right‐sided CRC and BRAF mutant cases were more common in the HMCC group, whereas left‐sided CRC and BRAF wild‐type cases were more common in the LMCC group. RAS mutations were present in 6 HMCC cases and 15 LMCC cases, but there were no significant differences between the 2 groups. Comparisons of other factors demonstrated no significant differences between the 2 groups.
TABLE 1.
Comparison of patient backgrounds of the 2 groups in validation cohort
| Variable | All samples | HMCC* | LMCC* | P‐value | |||
|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | ||
| Total | 156 | 100 | 32 | 100 | 124 | 100 | |
| Gender | .54 * | ||||||
| Male | 114 | 73.1 | 22 | 68.8 | 92 | 74.2 | |
| Female | 42 | 26.9 | 10 | 31.2 | 32 | 25.8 | |
| Median age (range) | .12 + | ||||||
| 65.0 (33‐85) | 62.0 (39‐76) | 66.0 (33‐85) | |||||
| Primary site | <.001 * | ||||||
| Right‐sided | 46 | 29.5 | 22 | 68.8 | 24 | 19.4 | |
| Left‐sided | 110 | 70.5 | 10 | 31.2 | 100 | 80.6 | |
| Number of organs with metastasis | .12 + | ||||||
| 1 | 70 | 44.9 | 24 | 55.8 | 52 | 41.9 | |
| 2 | 68 | 43.6 | 16 | 37.2 | 56 | 45.2 | |
| ≥3 | 18 | 11.5 | 3 | 7 | 16 | 12.9 | |
| Number of previously administered treatment regimens | .60 + | ||||||
| 1 | 2 | 1.3 | 0 | 0 | 2 | 1.6 | |
| 2 | 112 | 71.8 | 28 | 87.5 | 84 | 67.7 | |
| ≥3 | 42 | 26.9 | 4 | 12.5 | 38 | 30.7 | |
| Anti‐EGFR antibody treatment regimen | .26 * | ||||||
| Cetuximab | 105 | 67.3 | 31 | 72.1 | 82 | 66.1 | |
| Monotherapy | 19 | 12.2 | 3 | 7 | 16 | 12.9 | |
| Combination | 86 | 55.1 | 28 | 65.1 | 66 | 53.2 | |
| Panitumumab | 51 | 32.7 | 12 | 27.9 | 42 | 33.8 | |
| Monotherapy | 23 | 14.7 | 3 | 7 | 21 | 16.9 | |
| Combination | 28 | 18 | 9 | 20.9 | 21 | 16.9 | |
| RAS status | .13 * | ||||||
| KRAS WT | 143 | 91.7 | 29 | 90.6 | 114 | 91.9 | |
| RAS WT | 101 | 64.7 | 24 | 75 | 77 | 62.1 | |
| RAS MT | 21 | 13.5 | 6 | 18.8 | 15 | 12.1 | |
| RAS unknown | 34 | 21.8 | 2 | 6.3 | 32 | 25.8 | |
| BRAF status | .007 ++ | ||||||
| BRAF WT | 60 | 38.5 | 21 | 65.6 | 39 | 31.5 | |
| BRAF MT | 8 | 5.1 | 7 | 21.9 | 1 | 0.8 | |
| BRAF unknown | 88 | 56.4 | 4 | 12.5 | 84 | 67.7 | |
HMCC and LMCC denotes highly methylated and low‐methylated colorectal cancer, respectively. WT and MT denotes wild‐type and mutant type, respectively.
Chi‐square (χ2) test.
Wilcoxon test.
Fisher exact test.
3.4. Comparison of the clinical outcomes of anti‐EGFR treatment based on the DNA methylation status in KRAS/RAS wild‐type mCRC
To determine differences in the clinical outcomes of anti‐EGFR treatment for mCRC patients who were refractory or intolerable to oxaliplatin‐ and irinotecan‐based chemotherapy according to methylation status as measured by MeC‐mML, we compared the RR, PFS, and OS among 3 groups: the KRAS wild‐type HMCC (n = 29), KRAS wild‐type LMCC (n = 114), and KRAS mutant (n = 13) groups. The KRAS wild‐type HMCC group had a significantly worse RR (3.4% vs 28.6%, P = .003), PFS (median: 2.5 vs 5.8 mo, P < .001; HR = 0.28), and OS (median: 5.6 vs 13.1 mo, P < .001; HR = 0.27) compared with those of the KRAS wild‐type LMCC group (Table S9, Figure 2A,B). There were no significant differences in clinical outcomes between the KRAS mutant group and the KRAS wild‐type HMCC group (Table S9, Figure 2A,B).
FIGURE 2.
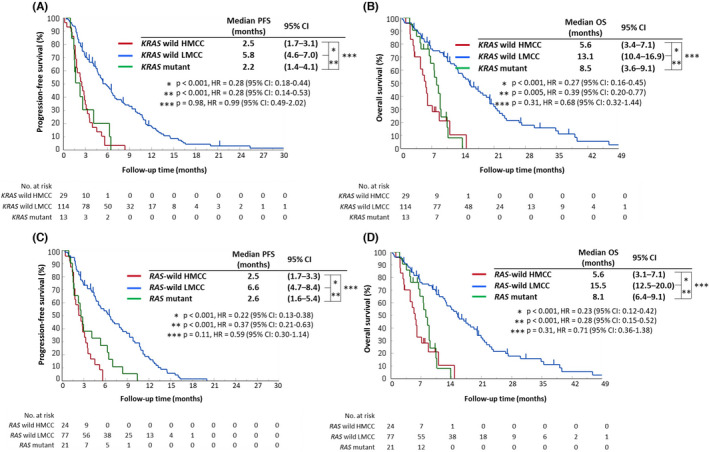
Kaplan‐Meier curves for progression‐free survival and overall survival after anti‐EGFR treatment according to DNA methylation and KRAS/RAS status. A, and B, Progression‐free survival (PFS) and overall survival (OS) after anti‐EGFR treatment according to the KRAS wild‐type HMCC group (red line, n = 29), KRAS wild‐type LMCC group (blue line, n = 114), and KRAS mutant group (green line, n = 13). C, and D, The PFS and OS after anti‐EGFR treatment in the RAS wild‐type HMCC group (red line, n = 24), RAS wild‐type LMCC group (blue line, n = 77), and RAS mutant group (green line, n = 21). The survival curves were generated by using the Kaplan‐Meier method, and the differences were assessed by the log‐rank test. CI, confidence interval; HMCC, highly methylated colorectal cancer; LMCC, low‐methylated colorectal cancer; OS, overall survival; PFS, progression‐free survival
The analysis included patients with a confirmed RAS genotype and compared anti‐EGFR treatment outcomes among the 3 groups: the RAS wild‐type HMCC (n = 24), RAS wild‐type LMCC (n = 77), and RAS mutant (n = 21) groups. The RR in the RAS wild‐type HMCC group was significantly lower compared with that in the RAS wild‐type LMCC group (4.2% vs 33.3%, P = .004; Table 2). In addition, PFS and OS in the RAS wild‐type HMCC group were significantly shorter compared with those in the RAS wild‐type LMCC group (median PFS: 2.5 vs 6.6 mo, P < .001; HR = 0.22; median OS: 5.6 vs 15.5 mo, P < .001, HR = 0.23; Figure 2C,D). Furthermore, the RR, PFS, and OS of the RAS wild‐type HMCC group were comparable with those of the RAS mutant group, which were significantly worse than those of the RAS wild‐type LMCC group (Table 2, Figure 2C,D).
TABLE 2.
Tumor response to anti‐EGFR treatment in RAS wild‐type mCRC
| Tumor response | All samples (n = 122) | RAS wild‐type HMCC (n = 24) | RAS wild‐type LMCC (n = 77) | RAS mutant (n = 21) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | P‐value | n | % | P‐value (vs LMCC) | |
| RR (%) | 22.9 | 4.2 | 33.3 | .004 | 5.3 | .01 | ||||
| CR | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| PR | 27 | 22.9 | 1 | 4.2 | 25 | 33.3 | 1 | 5.3 | ||
| SD | 43 | 36.4 | 7 | 29.2 | 30 | 40 | 6 | 31.6 | ||
| PD | 48 | 40.7 | 16 | 66.7 | 20 | 26.7 | 12 | 63.2 | ||
| NE | 4 | 0 | 2 | 2 | ||||||
Abbreviations: CR, complete response; NE, not evaluated; PD, progressive disease; PR, partial response; RR, response rate; SD, stable disease
Although the number of patients was limited (n = 67), PFS was compared in a group of patients with a confirmed RAS/BRAF status. The analysis included RAS/BRAF wild‐type HMCC (n = 15), RAS/BRAF wild‐type LMCC (n = 23), RAS mutant (n = 21), and BRAF mutant (n = 8) groups. Even when the analysis was limited to the RAS/BRAF wild‐type patients, the PFS in the RAS/BRAF wild‐type HMCC group was significantly shorter compared with that in the RAS/BRAF wild‐type LMCC group (median PFS: 3.1 vs 5.8 mo, P = .002; HR = 0.31; Figure 3).
FIGURE 3.
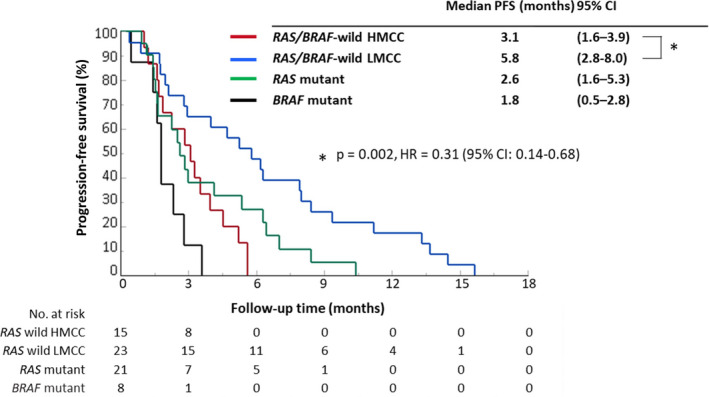
Kaplan‐Meier curves for progression‐free survival after anti‐EGFR treatment according to DNA methylation status and RAS/BRAF status. Progression‐free survival (PFS) after anti‐EGFR treatment in the RAS/BRAF wild‐type HMCC group (red line, n = 15), RAS/BRAF wild‐type LMCC group (blue line, n = 23), RAS mutant group (green line, n = 21), and BRAF mutant group (black line, n = 8) are shown. The survival curves were generated by using the Kaplan‐Meier method, and the differences were assessed by the log‐rank test. CI, confidence interval; HMCC, highly methylated colorectal cancer; LMCC, low‐methylated colorectal cancer; PFS, progression‐free survival
3.5. Investigation of factors contributing to the PFS and OS of patients receiving anti‐EGFR treatment
Univariate and multivariate analyses using Cox proportional hazard models were performed using the VC (n = 156) to identify the factors that contributed to PFS and OS in patients receiving anti‐EGFR treatment. Univariate analysis demonstrated that the methylation status measured by MeC‐mML, and primary site were statistically significant predictors of PFS (HR = 3.26, P < .001; HR = 1.82, P = .001; Table 3). Additionally, univariate analysis of OS after anti‐EGFR treatment demonstrated that the methylation status measured by MeC‐mML, gender, and primary site were predictive factors of OS (HR = 3.56, P < .001; HR = 0.56, P = .004: HR = 1.98, P < .001; Table 3). Multivariate analysis demonstrated that the DNA methylation status was the only independent predictor of PFS (HR = 2.83, P < .001; Table 3) and the DNA methylation status and gender were predictive factors of OS (HR = 2.78, P < .001; HR = 0.64, P = .02; Table 3).
TABLE 3.
Cox regression analysis of PFS and OS in anti‐EGFR treatment for mCRC (n = 156)
| PFS | OS | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||
| HR | HR | HR | HR | |||||
| Variable | (95% CI) | P‐value | (95% CI) | P‐value | (95% CI) | P‐value | (95% CI) | P‐value |
| Age (≧65 vs <65) | 0.83 (0.59‐1.16) | .27 | 0.94 (0.66‐1.34) | .74 | ||||
| Gender (Male vs Female) | 0.7 (0.48‐1.03) | .07 | 0.56 (0.38‐0.83) | .004 | 0.64 (0.43‐0.94) | .02 | ||
| Primary site (Right‐sided vs Left‐sided) | 1.82 (1.26‐2.61) | .001 | 1.3 (0.86‐1.97) | .22 | 1.98 (1.35‐2.91) | < .001 | 1.41 (0.91‐2.17) | .12 |
| Anti‐EGFR antibody treatment regimen (Combination vs Monotherapy) | 0.88 (0.60‐1.29) | .52 | 0.79 (0.53‐1.16) | .23 | ||||
| Methylation status (HMCC vs LMCC) | 3.26 (2.11‐5.05) | < .001 | 2.83 (1.73‐4.62) | < .001 | 3.56 (2.22‐5.70) | < .001 | 2.78 (1.65‐4.67) | < .001 |
Abbreviations: CI, confidence interval; EGFR, epidermal growth factor receptor; HR, hazard ratio; OS, overall survival; PFS, progression‐free survival.
To evaluate the confounding effect of RAS/BRAF and methylation status in predicting the clinical outcomes of anti‐EGFR treatments, we conducted univariate and multivariate analyses using Cox proportional hazard models in a group of patients with a confirmed RAS/BRAF status (n = 67). Univariate analysis demonstrated that methylation and BRAF status were statistically significant predictors of PFS (HR = 0.44, P < .004; HR = 0.32, P = .005; Table S10). Furthermore, multivariate analysis demonstrated that the DNA methylation and BRAF status were statistically significant predictors of PFS (HR = 0.51, P < .020; HR = 0.39, P = .029; Table S10).
To examine in detail the significance of the DNA methylation status and the primary site in predicting the therapeutic effects of anti‐EGFR antibodies, we classified 101 patients of RAS wild‐type CRC into 4 groups that combined their DNA methylation status as measured by MeC‐mML and the primary site, to compare the clinical outcomes of anti‐EGFR treatment. The HMCC group demonstrated a lower RR regardless of the location of the primary site (right‐sided: 0%; left‐sided: 14.3%; Table S11), whereas the LMCC group demonstrated a higher RR (right‐sided: 23.1%; left‐sided: 35.5%; Table S11). A comparison of PFS is provided in Figure 4(A). First, we compared the PFS of the LMCC and HMCC groups based on tumor site. For both right‐sided and left‐sided tumors, the LMCC group exhibited a significantly longer PFS than that in the HMCC group (right‐sided: median: 2.8 vs 5.0 mo, P = .004, HR = 0.30; left‐sided: median: 1.9 vs 6.8 mo, P < .001, HR = 0.20). However, when comparing the PFS between right‐sided and left‐sided tumors in the HMCC group and the LMCC group, respectively, there was no significant difference in survival duration based on primary site. Similar to the PFS results, the OS was significantly prolonged in the LMCC group compared with its rate in the HMCC group, regardless of the primary site (right‐sided: median: 5.3 vs 10.8 mo, P = .028, HR = 0.37; left‐sided: median: 6.0 vs 15.8 mo, P < .001, HR = 0.21; Figure 4B).
FIGURE 4.
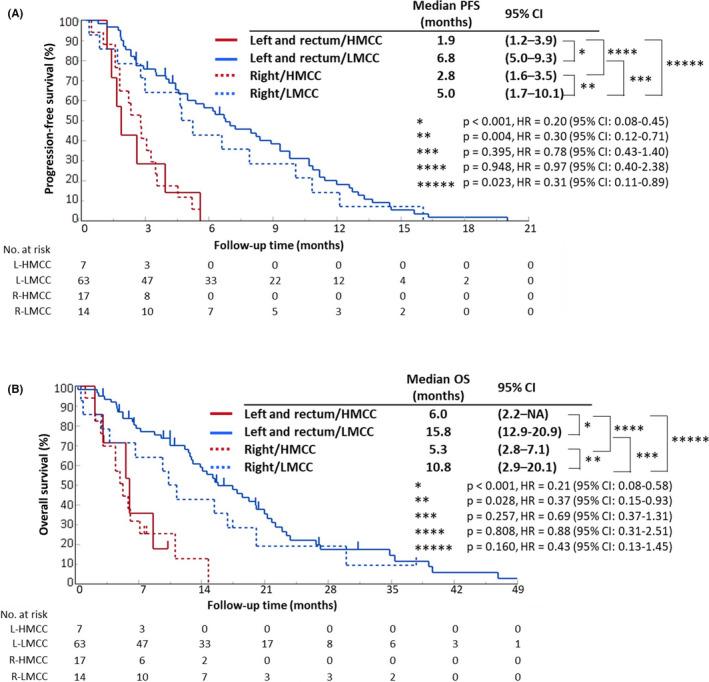
Kaplan‐Meier curves for progression‐free survival and overall survival after anti‐EGFR treatment according to DNA methylation status and the primary site. A, and B, Progression‐free survival (PFS) and overall survival (OS) after anti‐EGFR treatment in the left‐sided HMCC group (red solid‐line, n = 7), left‐sided LMCC group (blue solid‐line, n = 63), right‐sided HMCC group (red dotted‐line, n = 17), and right‐sided LMCC group (blue dotted‐line, n = 14). The survival curves were generated by the Kaplan‐Meier method, and the differences were assessed by the log‐rank test. CI, confidence interval; HMCC, highly methylated colorectal cancer; LMCC, low‐methylated colorectal cancer; OS, overall survival; PFS, progression‐free survival
4. DISCUSSION
In this study, we developed MeC‐mML, and confirmed the findings in our previous report 18 and the performance of this assay. The methylation classification of mCRC reported in our previous study was performed by unsupervised clustering analysis using genome‐wide DNA methylation data. 18 The most significant advantage of methylation classification is its ability to extract an anti‐EGFR treatment‐resistant group (ie, HMCC) from RAS wild‐type patients, and is a conventional marker for predicting treatment effects. However, there are various problems in the clinical application of the clustering‐based classification method using genome‐wide DNA methylation data. Limitations in obtaining methylation data include complicated procedures, higher costs, and time‐consuming processes. In addition, it is necessary to develop an analysis system to process a large amount of data, as the number of samples increases. Moreover, as mentioned in the Introduction, classification by clustering is susceptible to population influences.
Therefore, we aimed to reduce costs and obtain results efficiently by narrowing the CpG sites and using a simpler technique. The amount of data to be handled could also be reduced by necessity. Furthermore, the determination of methylation status of the site of interest should be based on the presence or absence of methylation (qualitative) rather than on the degree of methylation (quantitative), such that measurement results are less influenced by the tumor content of tissue samples. In addition, a cutoff value to classify HMCC and LMCC based on the number of methylation‐positive sites allows for a more objective classification of individual patients. Therefore, the MeC‐mML used in this study should have the advantages of simplicity, objectivity, and allowing for qualitative analysis.
The purpose of this DNA methylation status assay is to select a group of RAS wild‐type patients who are resistant to anti‐EGFR antibodies (ie, HMCC). The results of our previous genome‐wide DNA methylation analysis revealed that some of the frequently methylated sites in HMCC were not methylated in LMCC and normal colonic mucosa. 18 By examining the methylation status of these sites, it was deemed possible to identify HMCC regardless of normal colonic mucosal contamination. Finally, 16 CpG sites of interest that reflected the genome‐wide highly methylated status were identified for analysis (Table S1). The target region contains a mixture of genes located in the promoter region and the gene body. Some of these sites were in genes with a known function (Table S1), but no enrichment was detected for specific signaling pathways. From a review of the literature on methylation analysis, we modified MethyLight (mML) for our methylation assay.
The development of a novel assay to diagnose the DNA methylation status based on the mML assay was achieved, and its performance was tested using data from 156 patients with mCRC. The results revealed that the RR, PFS, and OS of KRAS and RAS wild‐type HMCCs were significantly worse compared with those of KRAS and RAS wild‐type LMCCs. It is important to note that there was no significant difference in clinical outcomes between the KRAS and RAS wild‐type HMCC and KRAS and RAS mutant groups, suggesting that the KRAS and RAS wild‐type HMCC group is as resistant to anti‐EGFR antibodies as is the KRAS and RAS mutant group. Because the current analysis was limited to patients who were refractory or intolerable to oxaliplatin‐ and irinotecan‐based chemotherapy, it directly reflects the treatment effect of anti‐EGFR antibodies with or without concomitant cytotoxic agents. These results indicated that MeC‐mML is useful for predicting the clinical outcomes of anti‐EGFR treatment in patients with KRAS or RAS wild‐type mCRC.
In addition to RAS mutations, BRAF mutations are also known to be associated with resistance to anti‐EGFR treatments. In this study, the rate of BRAF mutations was significantly higher in the HMCC group compared with that in the LMCC group, a trend consistent with previous reports on CIMP. 24 However, even in the analysis focusing on RAS/BRAF wild‐type cases, the PFS was significantly shorter in the HMCC group than it was in the LMCC group (Figure 3). Furthermore, the univariate and multivariate results suggested that methylation status is a predictor of treatment response independent from BRAF and RAS status (Table S10).
In our previous report, we explored the relationship between DNA methylation status and the primary site, and suggested that differences in susceptibility to anti‐EGFR antibodies based on the primary site could be explained by differences in DNA methylation status. 25 In this study, we examined the relationship between the DNA methylation status and primary site in 101 RAS wild‐type patients. When comparing the clinical outcomes of anti‐EGFR treatment in the 4 groups by considering the DNA methylation status and primary site, we found no significant difference in outcomes between right‐sided and left‐sided tumors within the HMCC or LMCC groups. In contrast, in patients with right‐sided colon cancer who are traditionally considered to be refractory to anti‐EGFR treatment, a comparison of clinical outcomes between the HMCC and LMCC groups demonstrated a significantly higher RR and prolonged PFS and OS in the LMCC group. In the treatment‐sensitive group of patients with left‐sided tumors, the HMCC group had significantly worse outcomes compared with those of the LMCC group. Notably, the PFS was significantly longer in the right‐sided LMCC group than it was in the left‐sided HMCC group (Figure 4A). Therefore, the HMCC group was similarly resistant to anti‐EGFR antibodies regardless of the primary site location, whereas the LMCC group was susceptible to anti‐EGFR antibodies regardless of the primary site location. Moreover, multivariate analyses demonstrated that the DNA methylation status measured by MeC‐mML is an independent predictor of PFS and OS for anti‐EGFR treatments. These results indicated that the difference in susceptibility to anti‐EGFR antibodies at the primary site may be explained by a difference in the distribution of patients with HMCC and LMCC in the right‐sided or left‐sided tumor groups.
As mentioned above, this study showed that DNA methylation status, as measured by MeC‐mML is a predictor of anti‐EGFR treatment response. Conversely, previous reports have shown that CIMP is a poor prognostic factor in mCRC. 26 , 27 In this cohort, as well as PFS, the OS was significantly shorter in HMCC than it was in LMCC, suggesting that DNA methylation status may act as a prognostic factor. As all patients analyzed in this study were treated with anti‐EGFR antibodies, it was difficult to determine whether the role of DNA methylation status in anti‐EGFR treatments was predominantly a predictor or a prognostic factor. Therefore, a clinical study is underway to examine the prognostic role of DNA methylation status in detail, using a cohort that includes patients who did or did not receive anti‐EGFR antibodies.
The MeC‐mML will lead to clinical applications and is currently being developed as an in vitro diagnostic agent, with plans to apply for approval as a complementary diagnostic tool.
There are a few limitations to this study. The analyses in this study mainly included patients who received anti‐EGFR antibodies as third‐line or later treatment. However, the association between the sidedness and the therapeutic effects of anti‐EGFR antibodies has been shown mainly when analyzing first‐line therapy. 11 , 12 Therefore, future validation in patients administered anti‐EGFR antibodies as first‐line therapy is needed.
Several sets of CIMP markers have been previously reported. 19 , 24 , 28 Therefore, the significance of MeC‐mML may become clearer by comparing its classification results with those based on existing CIMP markers. However, CIMP is known to be associated with poor prognosis, elderly age, right‐sidedness, and BRAF mutation. To compare the association of clinical outcomes of anti‐EGFR treatment with this assay and other existing classification methods, it is necessary to eliminate confounding factors. However, it was difficult to secure a sufficient number of cases for analysis in this study.
As mentioned above, previous studies have shown that HMCC has epidemiological and pathological features different from those of LMCC. 28 , 29 Additionally, because of their molecular characteristics, different factors may be involved in HMCC and LMCC tumor development. 30 The methylation of the CpG site of the genes encoding the EGFR ligands epiregulin/amphiregulin and decreased gene expression are thought to contribute to anti‐EGFR resistance in highly methylated mCRC. 19 Our analysis confirmed a similar trend (data not shown). However, even in patients with highly methylated mCRC without the downregulation of these genes, many patients are refractory to anti‐EGFR treatment. Therefore, it is desirable to elucidate the molecular mechanism of resistance to anti‐EGFR antibodies in HMCC. Our laboratory is currently working on an integrated analysis of ’omics data, such as gene mutations and gene expression, to achieve this goal.
In conclusion, our novel DNA methylation assay based on MethyLight will be a useful diagnostic test for predicting the clinical outcomes of anti‐EGFR treatment for mCRC regardless of the primary site.
DISCLOSURE
Dr. Takahashi has received research fund from Merck Biopharma. Professor Ishioka has received scholarship (incentive) endowments from Takeda, Daiichi‐Sankyo, Ono, Asahi‐Kasei Pharma, Taiho, Chugai, and Eisai. All remaining authors have declared no conflict of interest.
Supporting information
Fig S1‐S2
Table S1‐S11
Appendix S1
ACKNOWLEDGMENTS
The authors thank the patients and medical staff, especially Ms Yuan and Ms Junko Nakamura. This work was supported by a grant from the Project for Development of Innovative Research on Cancer Therapeutics (P‐DIRECT) (grant number 11110018) and the Project for Cancer Research and Therapeutic Evolution (P‐CREATE) by the Japan Agency for Medical Research and Development (grant number 16770660).
Ouchi K, Takahashi S, Okita A, et al. A modified MethyLight assay predicts the clinical outcomes of anti‐epidermal growth factor receptor treatment in metastatic colorectal cancer. Cancer Sci. 2022;113:1057–1068. doi: 10.1111/cas.15252
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Jonker DJ, O'Callaghan CJ, Karapetis CS, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040‐2048. [DOI] [PubMed] [Google Scholar]
- 3. Van Cutsem E, Peeters M, Siena S, et al. Open‐label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy‐refractory metastatic colorectal cancer. J Clin Oncol. 2007;25:1658‐1664. [DOI] [PubMed] [Google Scholar]
- 4. Martini G, Troiani T, Cardone C, et al. Present and future of metastatic colorectal cancer treatment: A review of new candidate targets. World J Gastroenterol. 2017;23:4675‐4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yamada Y, Denda T, Gamoh M, et al. S‐1 and irinotecan plus bevacizumab versus mFOLFOX6 or CapeOX plus bevacizumab as first‐line treatment in patients with metastatic colorectal cancer (TRICOLORE): a randomized, open‐label, phase III, noninferiority trial. Ann Oncol. 2018;29:624‐631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamazaki K, Nagase M, Tamagawa H, et al. Randomized phase III study of bevacizumab plus FOLFIRI and bevacizumab plus mFOLFOX6 as first‐line treatment for patients with metastatic colorectal cancer (WJOG4407G). Ann Oncol. 2016;27:1539‐1546. [DOI] [PubMed] [Google Scholar]
- 7. Amir E, Seruga B, Martinez‐Lopez J, et al. Oncogenic targets, magnitude of benefit, and market pricing of antineoplastic drugs. J Clin Oncol. 2011;29:2543‐2549. [DOI] [PubMed] [Google Scholar]
- 8. Ciardiello F, Lenz H‐J, Kohne C‐H, et al. Treatment outcome according to tumor RAS mutation status in CRYSTAL study patients with metastatic colorectal cancer (mCRC) randomized to FOLFIRI with/without cetuximab. J Clin Oncol. 2014;32. [Google Scholar]
- 9. Bokemeyer C, Kohne C‐H, Ciardiello F, et al. Treatment outcome according to tumor RAS mutation status in OPUS study patients with metastatic colorectal cancer (mCRC) randomized to FOLFOX4 with/without cetuximab. J Clin Oncol. 2014;32. [Google Scholar]
- 10. Schwartzberg LS, Rivera F, Karthaus M, et al. PEAK: a randomized, multicenter phase II study of panitumumab plus modified fluorouracil, leucovorin, and oxaliplatin (mFOLFOX6) or Bevacizumab plus mFOLFOX6 in patients with previously untreated, unresectable, wild‐type KRAS Exon 2 metastatic colorectal cancer. J Clin Oncol. 2014;32:2240‐2247. [DOI] [PubMed] [Google Scholar]
- 11. Holch JW, Ricard I, Stintzing S, Modest DP, Heinemann V. The relevance of primary tumour location in patients with metastatic colorectal cancer: a meta‐analysis of first‐line clinical trials. Eur J Cancer. 2017;70:87‐98. [DOI] [PubMed] [Google Scholar]
- 12. Tejpar S, Stintzing S, Ciardiello F, et al. Prognostic and predictive relevance of primary tumor location in patients with RAS wild‐type metastatic colorectal cancer: retrospective analyses of the CRYSTAL and FIRE‐3 trials. JAMA Oncol. 2017;3:194‐201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brulé SY, Jonker DJ, Karapetis CS, et al. Location of colon cancer (right‐sided versus left‐sided) as a prognostic factor and a predictor of benefit from cetuximab in NCIC CO.17. Eur J Cancer. 2015;51:1405‐1414. [DOI] [PubMed] [Google Scholar]
- 14. Yoshino T, Arnold D, Taniguchi H, et al. Pan‐Asian adapted ESMO consensus guidelines for the management of patients with metastatic colorectal cancer: a JSMO‐ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Ann Oncol. 2018;29:44‐70. [DOI] [PubMed] [Google Scholar]
- 15. Mahipal A, Grothey A. Role of biologics in first‐line treatment of colorectal cancer. J Oncol Pract. 2016;12:1219‐1228. [DOI] [PubMed] [Google Scholar]
- 16. Hashiguchi Y, Muro K, Saito Y, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int J Clin Oncol. 2020;25:1‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Denda T, Takashima A, Gamoh M, et al. Combination therapy of bevacizumab with either S‐1 and irinotecan or mFOLFOX6/CapeOX as first‐line treatment of metastatic colorectal cancer (TRICOLORE): exploratory analysis of RAS status and primary tumour location in a randomised, open‐label, phase III, non‐inferiority trial. Eur J Cancer. 2021;154:296‐306. [DOI] [PubMed] [Google Scholar]
- 18. Ouchi K, Takahashi S, Yamada Y, et al. DNA methylation status as a biomarker of anti‐epidermal growth factor receptor treatment for metastatic colorectal cancer. Cancer Sci. 2015;106:1722‐1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee MS, McGuffey EJ, Morris JS, et al. Association of CpG island methylator phenotype and EREG/AREG methylation and expression in colorectal cancer. Br J Cancer. 2016;114:1352‐1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thelwell N, Millington S, Solinas A, Booth J, Brown T. Mode of action and application of Scorpion primers to mutation detection. Nucleic Acids Res. 2000;28:3752‐3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yoshino T, Muro K, Yamaguchi K, et al. Clinical validation of a multiplex kit for RAS mutations in colorectal cancer: results of the RASKET (RAS KEy Testing) prospective, multicenter study. EBioMedicine. 2015;2:317‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saghafinia S, Mina M, Riggi N, Hanahan D, Ciriello G. Pan‐cancer landscape of aberrant DNA methylation across human tumors. Cell Rep. 2018;25:1066‐1080 e1068. [DOI] [PubMed] [Google Scholar]
- 23. Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high‐throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787‐793. [DOI] [PubMed] [Google Scholar]
- 25. Okita A, Takahashi S, Ouchi K, et al. Consensus molecular subtypes classification of colorectal cancer as a predictive factor for chemotherapeutic efficacy against metastatic colorectal cancer. Oncotarget. 2018;9:18698‐18711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Juo YY, Johnston FM, Zhang DY, et al. Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta‐analysis. Ann Oncol. 2014;25:2314‐2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cha Y, Kim K‐J, Han S‐W, et al. Adverse prognostic impact of the CpG island methylator phenotype in metastatic colorectal cancer. Br J Cancer. 2016;115:164‐171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000‐1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hinoue T, Weisenberger DJ, Lange CPE, et al. Genome‐scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. De Sousa E Melo F, Wang X, Jansen M, et al. Poor‐prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19:614‐618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S2
Table S1‐S11
Appendix S1


