Abstract
Aging has been demonstrated to play vital roles in the prognosis and treatment efficacy of cancers, including lung adenocarcinoma (LUAD). This novel study aimed to construct an aging‐related risk signature to evaluate the prognosis and immunogenicity of LUAD. Transcriptomic profiles and clinical information were collected from a total of 2518 LUAD patients from 12 independent cohorts. The risk signature was developed by combining specific gene expression with the corresponding regression coefficients. One cohort treated with the immune checkpoint inhibitor (ICI) was also used. Subsequently, a risk signature was developed based on 21 aging‐related genes. LUAD patients with low‐risk scores exhibited improved survival outcomes in both the discovery and validation cohorts. Further immunology analysis revealed elevated lymphocyte infiltration, decreased infiltration of immune‐suppressive cells, immune response‐related pathways, and favorable ICI predictor enrichment in the low‐risk subgroup. Genomic mutation exploration indicated the enhanced mutation burden and higher mutation rates in significantly driver genes of TP53, KEAP1, SMARCA4, and RBM10 were enriched in patients with a low‐risk signature. In the immunotherapeutic cohort, it was observed that low‐risk aging scores were markedly associated with prolonged ICI prognosis. Overall, the estimated aging signature proved capable of evaluating the prognosis, tumor microenvironment, and immunogenicity, which further provided clues for tailoring prognosis prediction and immunotherapy strategies, apart from promoting individualized treatment plans for LUAD patients.
Keywords: aging risk signature, immunotherapy efficacy, lung adenocarcinoma, predictive indicator, prognosis, tumor immunogenicity
Aging‐related signature predicts favorable outcome and immunogenicity in lung adenocarcinoma.

Abbreviations
- COSMIC
Catalogue of Somatic Mutations in Cancer
- GEO
Gene Expression Omnibus
- GSEA
gene set enrichment analysis
- ICI
immune checkpoint inhibitor
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- NB
neoantigen burden
- NSCLC
non–small‐cell lung cancer
- SMGs
significantly mutated genes
- ssGSEA
single sample GSEA
- TCGA
The Cancer Genome Atlas
- TCIA
The Cancer Immunome Atlas
- TMB
Tumor mutation burden
1. INTRODUCTION
Based on data obtained from the latest cancer statistics report, NSCLC is the major cause of cancer‐related morbidity and mortality. 1 It primarily comprises 2 histology subtypes, namely, LUAD and LUSC. 2 Amongst these, LUAD accounts for approximately 55% of all NSCLC patients. 3 Although improved clinical treatment strategies, such as combined treatment and targeted therapy (eg, tyrosine kinase inhibitors), have been developed for LUAD, the 5‐y survival rate remains quite poor. 4 , 5 Therefore, more effective and robust indicators with respect to prognosis evaluation are urgently needed to identify patients who are suited to distinct medication programs.
The emergence of ICI therapy has dramatically prolonged the survival outcomes of several advanced cancers, including melanoma, 6 renal cancer, 7 and NSCLC. 8 Moreover, ICI treatment has become a pillar of LUAD clinical practice, alongside conventional surgery, chemotherapy, and targeted treatment. At present, blockading the immune checkpoints of programmed cell death protein 1 (PD‐1) or its ligand PD‐L1 is the best described immunotherapy method 9 and is fast becoming the routine first‐line treatment strategy for NSCLC patients. 10 Despite the marked therapeutic advantage of ICI agents observed in both clinical trials and real‐world populations, the biggest limitation of this treatment modality is that only a subset of patients could obtain clinical benefits from such immune treatment. 11 Therefore, novel immunotherapeutic efficacy determinants that could be used for selecting patients who are suitable for ICI treatment need to be identified.
Age is a well known risk biomarker for most diseases, including human cancers. In addition to age itself, its related genomic molecular features have also been revealed to be associated with disease progression and survival. Several recent studies have reported that specific genes (eg, APOE 12 and FOXO3 12 , 13 ), genomic regions (eg, 5q33.3 14 ), and multiple single‐nucleotide polymorphisms 15 , 16 are linked to longevity. It is difficult to dissect and investigate aging because of its complex interactions with numerous factors, including genome, environment, and age‐relevant diseases. 17 To comprehensively depict the transcriptomic landscape of aging, Peters et al 17 conducted a population‐based large‐scale transcriptomic analysis and determined aging‐related genes.
In this study, in total, 2518 LUAD cases from 12 independent cohorts were curated from publicly available data sources to construct and validate an aging‐related risk signature. To investigate the potential mechanisms underlying the identified risk signature, multi‐dimensional immunology analyses were conducted, and the results demonstrated the significant capabilities of this signature in evaluating the tumor microenvironment. Findings gleaned from our work may provide helpful clues for prognosis assessment and immunotherapy prediction in LUAD patients.
2. MATERIALS AND METHODS
2.1. Publicly available LUAD cohorts, immunotherapy cohort, and aging‐relevant genes
The GEO and TCGA databases were searched to obtain eligible LUAD samples with both gene expression profiles and clinical characteristics. Samples without transcriptome data/follow‐up information or those diagnosed as other lung cancer types were excluded from the candidate list. Finally, in total, 2518 samples were acquired for subsequent analysis, including TCGA (N = 499), GSE72094 (N = 398), GSE68465 (N = 442), GSE50081 (N = 127), GSE42127 (N = 132), GSE41271 (N = 181), GSE31210 (N = 226), GSE30219 (N = 85), GSE13213 (N = 117), GSE26939 (N = 115), GSE11969 (N = 90), and GSE81089 (N = 106). GSE50081, GSE31210, and GSE30219 were integrated as the GEO‐meta cohort 1 due to the fact that they shared the same microarray platform (ie, Affymetrix Human Genome U133 Plus 2.0 Array). Similarly, GSE42127 and GSE41271, which were both detected using the Illumina HumanWG‐6 v3.0 expression BeadChip, were combined into the GEO‐meta cohort 2. The ComBat function embedded in the R sva package 18 was applied to eliminate batch effects from distinct cohorts. Among the included LUAD datasets, as TCGA had the largest sample size with 499 patients, it was regarded as the discovery cohort and used for establishing the risk signature. The detailed dataset information and microarray platforms of all the LUAD samples are shown in Table S1. In total, 348 urothelial cancer samples treated with the anti‐PD‐L1 agent (ie, atezolizumab) in the IMvigor210 cohort 19 were curated to further investigate the relationship between the identified risk signature and immunotherapy prognosis. The complete clinical features, survival information, and gene expression profiles of the immunotherapy cohort were acquired from http://research‐pub.gene.com/IMvigor210CoreBiologies. A comprehensive list of 1433 aging‐related genes was obtained from a recently published study, 17 that provided a transcriptional landscape of age.
2.2. Construction of the aging‐related risk signature
All 1433 aging‐related genes were subjected to univariate Cox regression analysis based on the gene expression data of the LUAD discovery cohort to identify the genes involved in prognosis risk. Thereafter, prognosis‐related aging genes were subjected to the Lasso‐Cox regression model (implemented by R glmnet package 20 ) to determine the genes that contributed the most to survival. According to the Lasso coefficient profile, the optimal gene combination can be identified to construct the risk signature. By combining specific gene expression levels with their corresponding regression coefficients weighted by the Lasso analysis, risk cores for each LUAD patient could be calculated. The detailed formula was as follows: risk score = . All patients were stratified into low‐risk and high‐risk subgroups with the median risk score as the cutoff value. Based on this risk stratification, the association of the determined aging risk signature with LUAD survival outcomes was evaluated in both the discovery and validation cohorts.
2.3. Estimation of tumor‐infiltrating immune cells and immune checkpoints
To elucidate the distinct immune infiltration levels in low‐risk vs high‐risk subgroups, evaluation of the abundance of 28 tumor‐infiltrating immune cell phenotypes, which were recently reported by Charoentong et al 21 study was performed. The 28 immunocytes were divided into the following 3 categories to analyze their distinct immune functions: anti‐tumor, pro‐tumor, and intermediate tumor immunocytes. The feature genes for each immune cell subtype are listed in Table S2. Moreover, the CIBERSORT method is capable of calculating the abundance of 22 human lymphocyte subtypes with 547 feature genes from the leukocyte gene signature matrix, named LM22. 22 The present study used both approaches to obtain mutual‐validation results.
In addition, a complete list of immune checkpoint genes was collected from a previously published immunogenomic study. 23 In the discovery LUAD cohort, the checkpoint gene VISTA was not observed due to the diverse sequencing platform. Therefore, we compared the distinct expression of 33 genes in the low‐risk and high‐risk subgroups.
2.4. Quantification of the enrichment of immunotherapy and immunogenicity predictors
Several recently reported immune‐related signatures have been demonstrated to be associated with immunogenicity and immunotherapy responses. Therefore, this study curated 6 representative signatures as follows: (a) T cell‐inflammation signature, which is composed of 18 genes involved in a T cell‐activated tumor microenvironment and linked with efficacy to pembrolizumab 24 ; (b) cytolytic activity 25 ; (c) immune cell signature, which indicates the overall immunocyte infiltration level in the microenvironment 26 ; (d) cytokines and chemokines 27 ; (e) immune signaling molecules 27 ; and (f) immune cell subsets, which reflect the infiltrating abundance of T cells, B cells and natural killer (NK) cells. 27 The gene panels used for evaluating the enrichment scores of each immune signature are presented in Table S3.
2.5. GSEA and ssGSEA
GSEA was applied to infer the dysregulated pathways of distinct risk subgroups. The t values extracted from the differential analysis results of genome‐wide expression (performed by the limma package 28 ) were regarded as the input variable for the ‘fgsea’ function implemented by the R fgsea package (https://github.com/ctlab/fgsea). Subsequently, signaling pathways derived from the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases were used as annotation circuits. The enriched pathway plot was obtained using the clusterProfiler package. 29 To evaluate the enrichment of specific gene panels involved in distinct immune cell subtypes and signatures, the single sample GSEA (ssGSEA) method within the GSVA package 30 was used to calculate the enrichment scores of each signature for each LUAD sample.
2.6. Extraction of the mutational signatures operative in the genome
The SignatureAnalyzer software 31 was used for the detection of mutational signatures present in the genome, on the basis of mutation profiles from TCGA LUAD cohort. Bayesian non‐negative matrix factorization is the core algorithm of SignatureAnalyzer and was used to optimally determine the number of mutational signatures. Specifically, mutation portrait matrix A was decomposed into 2 non‐negative matrices W and H (ie, A ≈ W × H ), in which W reflects the identified mutational signatures and H indicates the corresponding mutational activities. The determined mutational signatures were subsequently compared with the 30 well annotated signatures derived from the COSMIC database, 32 according to the coefficient of cosine similarity.
2.7. Identification of the SMGs
The MutSigCV algorithm 33 was used to identify SMGs in LUAD samples. One SMG could be included in the final list if it met the following 3 criteria: statistically significant, expressed in TCGA LUAD, and an encyclopedia of cell lines. 34 According to the principle proposed by the Kandoth et al 35 study, a gene was considered to be expressed if it had 3 or more reads in 75% or more of the samples. Mutational patterns of the identified SMGs in distinct subgroups were illustrated using the maftools package. 36
2.8. Acquisition and definition of the mutational burden
TMB was regarded as the total nonsynonymous mutation count per megabase based on somatic mutation data obtained from TCGA cohort. In addition, the NB was defined as the total neoantigen count of each sample. The neoantigen data of 486 LUAD samples were acquired from TCIA project (https://www.tcia.at/patients). The detailed neoantigen calculation is shown in Supplementary Methods. Taking into account the crucial roles of DNA maintenance genes (eg, TP53, POLE, and BRCA1/2) in genomic stability, the mutations of the above genes were incorporated into the multivariate logistic regression model to adjust the confounding factors and obtain a real connection of the constructed aging signature with the mutational burden.
2.9. Statistical analysis
R software (version 4.0.2) was used to perform the related analyses. Heatmap exhibition of determined aging genes in distinct risk subpopulations was completed using the pheatmap package. Moreover, the Kaplan‐Meier approach was applied to draw survival curves, and the significant difference was assessed via the log‐rank test. Multivariate regression models within the forestmodel package were used to rule out biased factors and to acquire a controlled link. The relationship of continuous and categorical variables with 2 risk subgroups was evaluated using the Wilcoxon rank‐sum test and Fisher exact test, separately.
3. RESULTS
3.1. Identification of the aging risk signature
As mentioned above, in total, 2518 LUAD patients were acquired after excluding samples without the requisite transcriptome expression data or prognosis information. As the LUAD cohort from TCGA harbors the largest sample size (N = 499) and detailed clinicopathologic characteristics, it was regarded as the discovery cohort to determine the aging risk signature that is linked with the survival outcome and immune infiltration of LUAD patients. Univariate Cox regression analysis of the curated 1433 aging‐related genes was performed using gene expression profiles from TCGA cohort. The results suggested that 256 genes were prognostic (all P < .05; Table S4). Subsequently, the Lasso‐Cox regression model was used with 10‐fold cross‐validation to determine the aging genes with the greatest contribution to LUAD prognosis. The Lasso coefficient profile between the log (λ) and the gene combination number is illustrated in Figure 1A. It was observed that the minimized partial likelihood deviance could be obtained when the combination number was 21 (Figure 1B). Therefore, we selected 21 aging‐related genes to construct a risk signature for LUAD prognosis evaluation.
FIGURE 1.
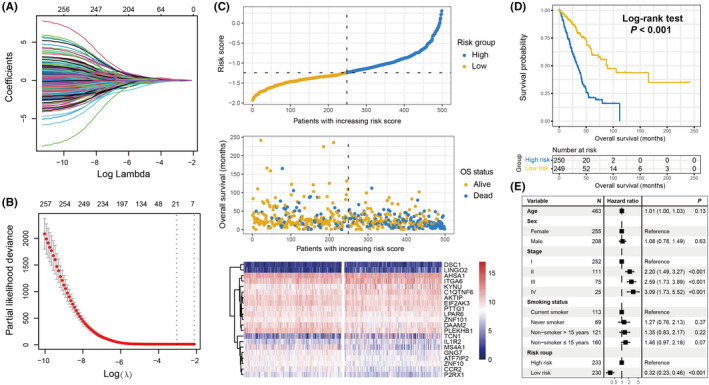
Development of the aging risk signature and its prognostic ability. A, Lasso coefficient profiles of the 256 prognostic aging genes in TCGA discovery cohort. B, Partial likelihood deviance of distinct variable combinations demonstrated by the Lasso‐Cox model. The red dots indicated the detailed partial likelihood of deviance values, the gray lines indicated the standard error (SE), the 2 vertical dotted lines on the left and right indicated the optimal gene combination with minimum criteria and 1 − SE criteria, separately. C, LUAD patients were partitioned into low‐risk and high‐risk subgroups with median risk score as the cutoff value. Distinct survival status and time were compared in low‐risk vs high‐risk subgroups. Heatmap representation of the different expression of the determined 21 aging‐related genes in 2 risk subpopulations. D, Kaplan‐Meier survival curves stratified by distinct risk patients in the discovery cohort. E, Association of the identified risk signature with LUAD prognosis in the multivariate Cox regression model with clinical confounding factors (ie, age, sex, stage, and smoking status) taken into account
The aforementioned 21 genes comprised ZNF101, PLEKHB1, P2RX1, EIF2AK3, LPAR6, ATF7IP2, MS4A1, CCR2, ZNF10, AKTIP, GNG7, DAAM2, PTTG1, IL1R2, KYNU, TCN1, ITGA6, AHSA1, DSC1, LINGO2, and C1QTNF6. The survival contribution coefficients for LUAD patients in TCGA cohort are depicted in Table S5. The present study established a risk signature to evaluate the risk score for each LUAD patient (Figure 1C) based on the linear integration between the identified 21 gene expression values and their corresponding regression coefficients obtained via the Lasso‐Cox model. The risk association plot of the evaluated risk scores, along with survival times and outcomes, is shown in Figure 1C. The expression levels of the identified 21 genes in distinct risk subgroups have also been illustrated with a heatmap (Figure 1C).
To explore the prognostic ability of the constructed risk signature, LUAD patients from the discovery cohort were classified into low‐risk (N = 249) and high‐risk (N = 250) subgroups with the median risk value as the cutoff value. It was observed that low‐risk patients harbored a significantly improved survival outcome compared with the high‐risk patients (log‐rank test P < .001; Figure 1D). This association remained significant even after adjusting for age, sex, stage, and smoking status in the multivariate Cox regression model (HR: 0.32, 95% CI: 0.23‐0.46, P < .001; Figure 1E).
3.2. Validation of the aging risk signature
To corroborate the prognostic roles of the determined aging signature, this study used 8 other LUAD cohorts. In 4 GEO cohorts of GSE72094, GSE68465, GEO‐meta cohort 1, and GEO‐meta cohort 2, it was observed that patients with low‐risk signature scores had significantly prolonged survival outcomes compared with high‐risk patients (log‐rank test P < .001 for GSE72094, P = .008 for GSE68465, and P < .001 for GEO‐meta cohorts 1 and 2; Figure 2A,C,E,G). Multivariate Cox regression models with clinical confounders (eg, age, sex, stage, grade, and smoking status) taken into consideration further confirmed the independent prognostic efficacy of the aging risk signature (GSE72094, HR, 0.42, 95% CI, 0.28‐0.63, P < .001; GSE68465, HR, 0.71, 95% CI, 0.54‐0.93, P = .012; GEO‐meta cohort 1, HR, 0.54, 95% CI, 0.37‐0.79, P = .002; GEO‐meta cohort 2, HR, 0.49, 95% CI, 0.33‐0.73, P < .001; Figure 2B,D,F,H). Moreover, clinical correlation analysis revealed that the elevated signature risk scores were significantly enriched in the LUAD patients with advanced clinical stage (TCGA, GSE72094, GEO‐meta cohorts 1 and 2, all P < .05; Figure S1A–D) or worse histological grade (GSE68465 cohort, Kruskal‐Wallis H test, P < .001; Figure S1E).
FIGURE 2.
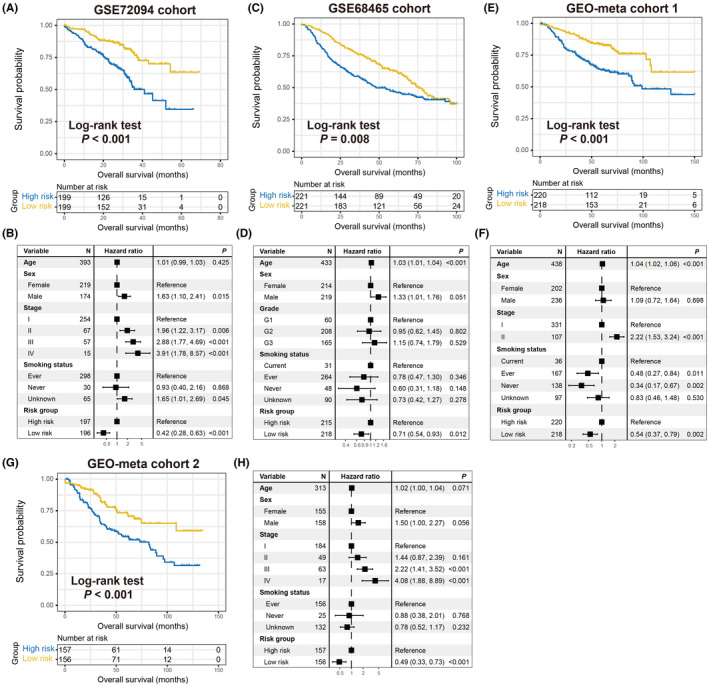
Validation of the prognostic capacity of the constructed risk signature. Kaplan‐Meier survival curves divided into low‐risk and high‐risk LUAD subgroups in (A) GSE72094, (C) GSE68468, (E) GEO‐meta cohort 1, and (G) GEO‐meta cohort 2. Multivariate Cox models of the connections between aging risk signature and LUAD survival outcome were conducted in (B) GSE72094, (D) GSE68468, (F) GEO‐meta cohort 1, and (H) GEO‐meta cohort 2
In the remaining GEO LUAD cohorts of GSE13213 (N = 117), GSE26939 (N = 115), GSE11969 (N = 90), and GSE81089 (N = 106), the results demonstrated that the trends of preferable survival outcomes were observed in patients with lower risk scores, although they did not attain statistical significance (log‐rank test, all P > .05; Figure S2). This may be attributed to the relatively small sample size.
3.3. Correlation of the identified aging signature with favorable immune infiltration and tumor immunogenicity
Recently, several studies have revealed that aging and its related molecular features are associated with cancer immune response and regulation. 37 , 38 Therefore, it was hypothesized that the identified risk signature may modulate immunocyte infiltration and signaling pathways related to immunogenicity. A heatmap based on the ssGSEA method was constructed to visualize the distinct infiltrating abundance of 28 lymphocyte subtypes between the low‐risk and high‐risk subgroups in the discovery cohort (Figure 3A). The results indicated that anti‐tumor immunocyte subtypes, such as activated CD8 T cells, effector memory CD8 T cells, NK cells, and type 1/17 T helper cells were markedly infiltrated in the low‐risk signature LUAD patients (all P < .05). Consistently, decreased infiltration of immune‐suppressive lymphocytes, such as CD56 dim NK cells and type 2 T helper cells was observed in the low‐risk subgroup (both P < .05). In addition, the infiltration of activated/immature B cells, eosinophils, mast cells, monocytes, and T follicular helper cells, which belong to the intermediate immunocyte group, was also significantly enriched in the low‐risk patients. Moreover, an immune infiltration analysis was also performed using the CIBERSORT algorithm, and similarly, the preferable lymphocyte infiltration represented by CD8 T cells was noticed in the low‐risk group (Figure S3).
FIGURE 3.
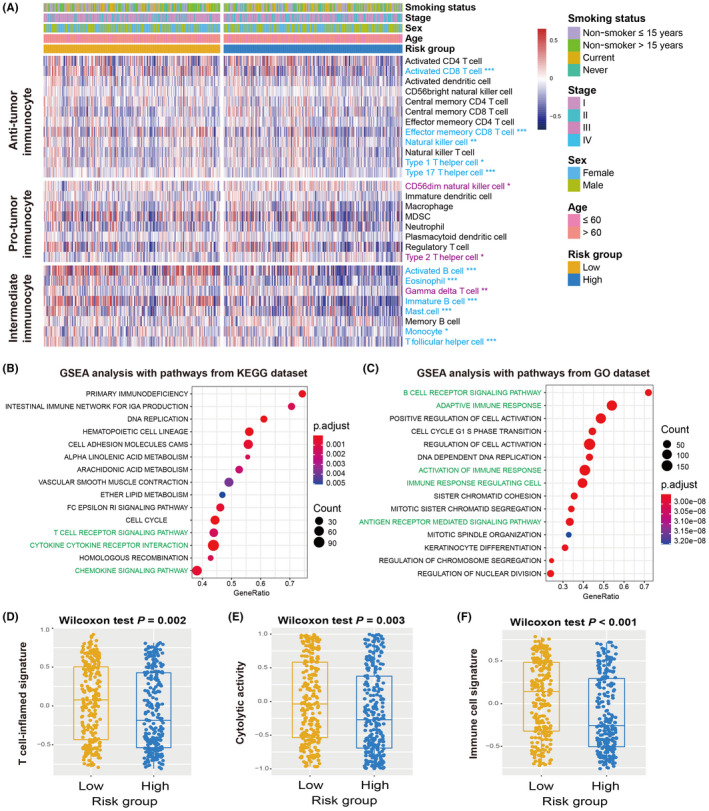
Association of the aging risk signature with immune infiltration and immunogenicity. A, Infiltrating abundance of distinct immunocyte in low‐risk vs high‐risk LUAD subgroups. Immunocyte highlighted with blue indicated its infiltration was significantly elevated in low‐risk patients, whereas the purple indicated the infiltration was significantly decreased in low‐risk patients. GSEA analysis of low‐risk patients with annotation pathways from (B) KEGG and (C) GO databases. Pathways highlighted with green were immune response‐related pathways. Distinct enrichment distribution of (D) T cell‐inflammation signature, (E) cytolytic activity signature, and (F) immune cell signature in low‐risk and high‐risk LUAD patients. *P < .05, **P < .01, ***P < .001
GSEA against KEGG/GO databases and LUAD transcriptomic expression profiles was applied to explore the signaling circuits associated with the aging risk signature. Consequently, it was observed that the immune response‐related pathways in the KEGG database (eg, T cell receptor signaling pathway, cytokine receptor interaction, and chemokine signaling pathway) and GO database (eg, activation of immune response, adaptive immune response, antigen receptor mediated signaling pathway, and B cell receptor signaling pathway) were significantly present in LUAD patients with low‐risk scores (Figure 3B,C).
Several molecular signatures concerning immunotherapy efficacy, immunogenicity, as well as immune cell status, were curated and their distinct enrichment was evaluated according to risk subgroups. Patients with low‐risk signature scores harbored significantly elevated enrichment of the T cell‐inflammation signature, cytolytic activity, and immune cell signature compared to high‐risk patients (Wilcoxon rank‐sum test, all P < .01; Figure 3D–F). In addition, a markedly higher enrichment of cytokine/chemokine signature, immune signaling molecules, and immune cell subset signature was observed in this low‐risk subgroup (Wilcoxon rank‐sum test, all P < .01; Figure S4).
The distinct expression of comprehensive immune checkpoint genes in the low‐risk and high‐risk subgroups was assessed. The results demonstrated that most of the genes (eg, PD‐L1, PD‐1, and CTLA‐4) exhibited enhanced expression in patients with the low‐risk aging signature (all P < .05; Figure S5).
3.4. Genomic features associated with the identified aging signature
The genomic mutational burden has been demonstrated to play a vital role in cancer progression and clinical immunotherapy response. 39 , 40 Therefore, the present study evaluated the association of the aging risk signature with TMB and NB. Based on the genomic mutation data obtained from TCGA cohort, we calculated the TMB for each LUAD sample and observed that patients with a low‐risk signature had a significantly elevated TMB compared with the high‐risk group (Wilcoxon rank‐sum test, P = .008; Figure 4A). Subsequently, using the neoantigen data from the TCIA project, a consistent result between the low‐risk signature and higher NB was observed (Wilcoxon rank‐sum test, P = .021; Figure 4B).
FIGURE 4.
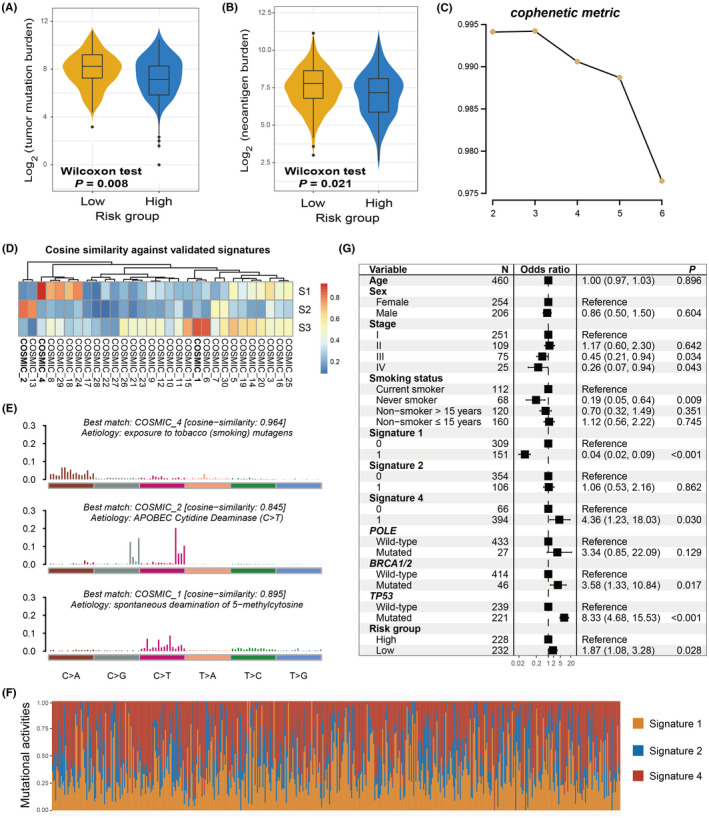
The constructed risk signature predictive of mutational burden in the discovery cohort. Associations of the identified aging signature with (A) TMB and (B) NB. C, The cophenetic metric plot was generated during the process of extracting LUAD mutational signatures. D, The extracted 3 mutational signatures vs well annotated COSMIC signatures with the cosine similarity. E, The detailed mutational patterns of the extracted 3 mutational signatures. F, Distinct mutational activities of 3 signatures across all LUAD patients. G, Multivariate logistic regression model was performed with age, sex, stage, smoking status, extracted mutational signatures, and DNA repair gene mutations taken into consideration to elucidate the link between aging risk signature and tumor mutation burden
Mutational signatures, which are characterized by distinct combinatorial patterns of nucleotide substitutions, have been shown to contribute to mutation burden and immune treatment outcomes. Therefore, we extracted potential mutational signatures present in LUAD samples by decomposing the nucleotide substitution matrix using the non‐negative matrix factorization (NMF) method. According to the cophenetic metric profile, 3 mutational signatures were found to potentially exist (Figure 4C). After comparing them with the well validated signatures in the COSMIC database and using the cosine similarity (Figure 4D), 3 mutational signatures were finally determined as signature 1 (initiated by spontaneous deamination of 5‐methylcytosine), signature 2 (attributed to the activity of the AID/APOBEC family), and signature 4 (associated with smoking). Their distinct mutation patterns are depicted in Figure 4E. Three mutational signature activities across all LUAD patients were evaluated and are presented in Figure 4F and Table S6. Further analysis revealed that low‐risk signature scores were markedly correlated with increased mutational activities of signature 1 (P = .029; Figure S6A) and decreased activities of signature 2 (P = .008; Figure S6B). No significant difference was observed between the 2 risk subgroups in relation to signature 4 (P = .412; Figure S6C).
To investigate whether the higher TMB of low‐risk patients was influenced by other confounding factors, clinical characteristics (ie, age, sex, stage, and smoking status), extracted mutational signatures (ie, signatures 1, 2, and 4), and mutations in genome maintenance regulators (eg, POLE, BRCA1/2, and TP53) were incorporated into the multivariate logistic regression model. To this end, the association of the low‐risk aging signature with the increased TMB remained significant (OR: 1.87, 95% CI: 1.08‐3.28, P = .028; Figure 4G).
In total, 23 SMGs were identified based on the genomic mutational profile of the discovery cohort. The SMG waterfall plot of low‐risk vs high‐risk subgroups (Figure 5) revealed a significantly distinct mutation rate in TP53 (104 of 247 [42.1%] vs 131 of 248 [52.8%]; P = .019), KEAP1 (31 of 247 [12.6%] vs 58 of 248 [23.4%]; P = .002), SMARCA4 (13 of 247 [5.2%] vs 33 of 248 [13.3%]; P = .003), and RBM10 (12 of 247 [4.9%] vs 24 of 248 [9.7%]; P = .048). The association between low‐risk patients and lower TP53 mutation frequency was also observed in 2 validation cohorts (GSE72094: 17.3% vs 30.9%, P = .002; GSE13213: 20.7% vs 44.8%, P = .009; Figure S7A,B). Nevertheless, in the GSE11969 cohort with the smallest sample size (N = 90), no significant difference in TP53 mutation rate was observed between the 2 risk subpopulations (26.7% vs 37.8%, P = .367; Figure S7C). Other SMG mutation data were unavailable for the above 3 LUAD validation datasets.
FIGURE 5.
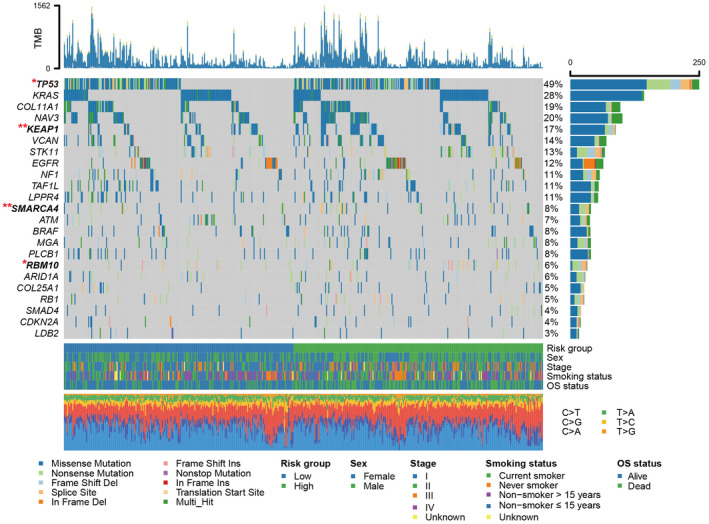
Waterfall plot illustration of the SMGs extracted from LUAD mutation profiles in low‐risk vs high‐risk subgroups. The left panel represents the gene symbols, the upper panel suggests the nonsynonymous mutation burden for each sample, the middle plot indicates detailed mutational patterns of the included SMGs with distinct mutation types colored distinctly, the right panel represents the mutation frequency of each SMG, and the bottom panel shows risk group information, clinical features, and base substitution types. SMGs highlighted with bold were significantly differentially mutated between 2 risk subgroups. *P < .05, **P < .01
3.5. The identified aging risk signature for evaluating immunotherapy prognosis
Considering that the constructed aging signature was demonstrated to be associated with tumor microenvironment and immunogenicity, it was hypothesized that this risk signature may contribute to the ICI prognosis. Therefore, an advanced urothelial cancer anti‐PD‐L1 immunotherapeutic cohort (Imvigor210) with transcriptome data and clinical information was used to explore the association of the identified risk signature with ICI response and outcome. The results suggested that patients with low‐risk aging signature scores exhibited a significantly improved ICI survival outcome compared with patients with high‐risk scores (log‐rank test P = .017; Figure 6A). Moreover, this association remained significant after adjusting for sex, ECOG score, smoking history, and immune phenotype in the multivariate Cox regression model (HR: 0.75, 95% CI: 0.57‐0.97, P = .028; Figure 6B). Further analysis demonstrated that more ICI responders were enriched in the low‐risk subgroup (30.0% vs 16.5%, P = .006; Figure 6C). In addition, an elevated proportion of patients with disease control was also found in the low‐risk group (52.1% vs 36.7%, P = .009; Figure 6D). Finally, an immunology analysis was performed with the gene expression profile of this ICI cohort. Consistent with the aforementioned results, a significantly increase in the enrichment of immune‐related signatures (all P < .01; Figure 6E), immune response‐relevant signaling pathways from KEGG and GO databases (Figure 6F,G), as well as favorable immune cell infiltration (Figure S8) were observed in patients with the low‐risk aging signature.
FIGURE 6.
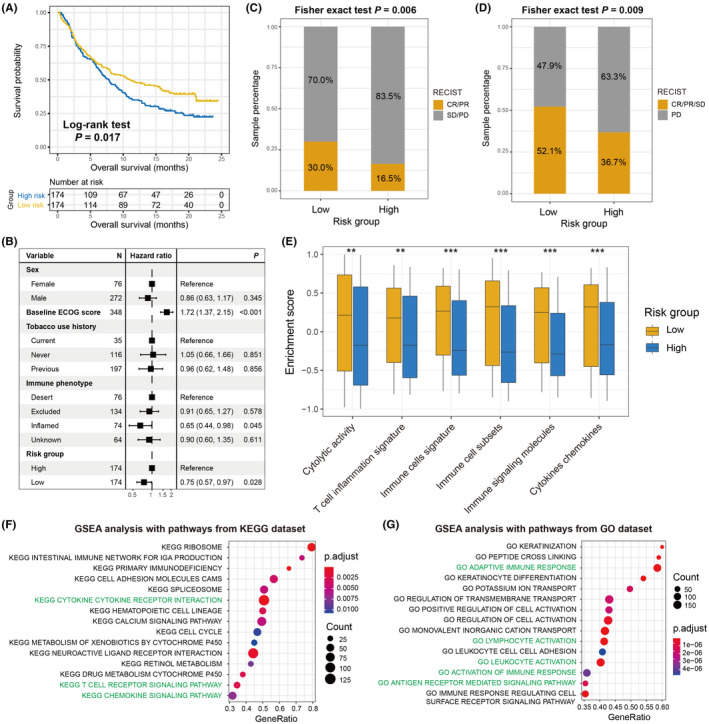
The constructed aging risk signature for evaluating ICI treatment efficacy. A, ICI prognosis difference was noticed between low‐risk and high‐risk subgroups under the urothelial cancer (UC) immunotherapy cohort. B, Multivariate‐adjusted Cox analysis was conducted to obtain the real association of aging risk signature with ICI survival outcome. C, Distinct objective response rate and (D) disease control rate were observed in 2 risk subpopulations. E, Quantification of the enrichment of immune response or immunotherapy efficacy‐relevant signatures in low‐risk and high‐risk subgroups with the data from the UC cohort. GSEA analysis of low‐risk patients with annotation pathways from (F) KEGG and (G) GO databases. *P < .05, **P < .01, ***P < .001
4. DISCUSSION
By integrating transcriptomic data and clinical information from multiple independent LUAD cohorts, this study developed and verified an aging‐related risk signature for survival prediction and immunogenicity evaluation. A forte of our study is that the risk signature was constructed based on a larger LUAD population and validated with distinct dimensions. Further prospective research is necessary, but the results from this work indicate the potential roles of the aging risk signature in LUAD survival outcome, immune microenvironment, and ICI treatment efficacy.
Mutational signatures are the fingerprints of endogenous and exogenous factors that represent distinct mutational patterns. 41 The aging‐related mutational signature 1 was recently reported to be associated with weaker immune infiltration and worse survival outcomes in triple‐negative breast cancer and prostate cancer, 42 suggesting its potential implication in immune treatment response. Furthermore, Chong et al 43 leveraged genomic mutation data from melanoma and NSCLC patients treated with ICI agents and observed that patients with this aging mutational signature exhibited an inferior prognosis in both tumors. In the present study, distinct from mutation‐specific indicators, an aging‐related risk signature was developed based on the transcriptomic level, and it was elucidated that this signature was predictive of survival status and immune infiltration.
Higher TMB, NB, and immunogenicity were demonstrated to be associated with better survival outcomes in both early and late NSCLC, although the use of TMB for early NSCLC is considered controversial. To rule out the possibility that the improved LUAD outcome was induced by the above factors, multivariate Cox regression analyses were performed with these factors taken into account (Figure S9A), along with interaction analyses of the aging signature with TMB, NB, and 6 immune‐related signatures using Cox regression (Figure S9B), as well as mutational burden stratification analyses (Figure S9C–F) using TCGA cohort. The associations between the aging signature and improved LUAD outcomes derived from the above analyses were consistent. These results suggested that the determined aging risk signature is an independent prognostic biomarker for LUAD patients.
Of the 21 identified aging‐related genes, 4 (ie, P2RX1, CCR2, PTTG1, and MS4A1) were also found to be linked to cancer immune response or evasion. Wang et al 44 discovered that a subset of P2RX1‐deficient neutrophils was an immunosuppressive factor that induces the evasion of anti‐tumor immunity in metastatic pancreatic tumors. The CCR2 deletion in cancer cells was established to associate with decreased tumor proliferation and improved survival in a mouse model, which was due to the enhanced infiltration of cytotoxic T cells and upregulation of MHC‐I members. 45 Furthermore, a recent study reported that PTTG1 knockdown could increase the ionizing radiation‐triggered T cell immune response via the TGF‐β1/SMAD3 circuit and further ameliorate the immunosuppressive tumor microenvironment. 46 The elevated expression of MS4A1 was demonstrated to be associated with a favorable prognosis as well as efficacious anti‐PD‐1 treatment, due to its positive association with T cell immunity. 47 These findings further support the predictive role of our constructed risk signature in immunocyte infiltration and ICI responses.
Based on recent findings, worse clinical outcomes were observed in lung cancer patients with mutations in TP53, 48 KEAP1, 49 SMARCA4, 50 or RBM10. 51 In this study, the low‐risk LUAD patients harbored lower mutational frequencies of the above 4 genes, which was consistent with the association between low‐risk scores and improved prognosis. Nonetheless, several studies have also reported the positive roles of TP53 and SMARCA4 mutations in evaluating favorable ICI treatment responses. 50 , 52 , 53 High‐risk patients with elevated mutation rates of TP53 and SMARCA4 nevertheless exhibited inferior ICI outcomes, indicating that more immunosuppressive factors (eg, CD56dim NK cells and type 2 T helper cells) were present in this subgroup, which inhibited the ICI promotion roles derived from the gene mutations. Therefore, further in‐depth studies are required to achieve a conclusive understanding of the same.
To elucidate the clinical implications of identified aging signature in LUAD, the present study examined its association with well known prognostic or immunologic indicators. Taking into consideration the significant correlations between this signature and multiple indicators, it is proposed that the aging risk signature may be a potential biomarker for evaluating LUAD prognosis and immunogenicity. Nevertheless, there are certain disadvantages posed by previously reported indicators. For instance, the crucial roles of TMB have been demonstrated in cancer prognosis and immune response. 54 , 55 However, different sequencing platforms, lack of accurate threshold definition, and whole‐exome sequencing costs hinder the broad utilization of TMB in clinical practice. 56 For another biomarker, PD‐L1, its high protein expression has been reported to be associated with a better immunotherapy response in NSCLC. 57 , 58 However, some patients with PD‐L1‐negative cancer also exhibited a favorable treatment response. 59 These findings suggest that novel and robust biomarkers are necessary, and our established aging signature may be a potential candidate for LUAD patients.
There are several limitations to this study. First, the transcriptomic gene expression data of LUAD patients were obtained from publicly accessible cohorts, which may introduce biases in data analysis between distinct datasets. Second, the somatic mutational data were used only from TCGA cohort and lacked relevant mutation‐level validation. Moreover, the potential mechanisms underlying the connections between the aging risk signature and multiple immunologic indicators remain unclear and require further experimental validation. Finally, the identified aging signature was associated with survival outcomes in both the non‐ICI as well as the ICI cohorts. This suggests that this signature might solely be a prognostic biomarker, rather than a predictive biomarker. Therefore, LUAD cohorts with both transcriptomic data and immunotherapy information as well as clinical trials are necessary to further validate the ICI predictive roles of the aging signature.
In summary, using the aging‐related transcriptomic profiles based on a larger LUAD population, the present study developed a risk signature to assess survival outcome, immunogenicity, and ICI treatment efficacy. The identified aging risk signature may be considered a potential biomarker for LUAD prognosis evaluation and immunotherapy response prediction.
DISCLOSURE
The authors have no conflict of interest.
Supporting information
Fig S1‐S9
Table S1‐S6
Supplementary Material
ACKNOWLEDGMENTS
QHW thanks WJZ for her company over the past 9 y and for giving a cute baby.
Zhang W, Li Y, Lyu J, et al. An aging‐related signature predicts favorable outcome and immunogenicity in lung adenocarcinoma. Cancer Sci. 2022;113:891–903. doi: 10.1111/cas.15254
Wenjing Zhang and Yuting Li contributed equally to this work.
Funding information
This study was supported by the Shandong Provincial Youth Innovation Team Development Plan of Colleges and Universities (No. 2019‐6‐156, Lu‐Jiao), National Natural Science Foundation of China (Nos. 81872719 and 81803337), Provincial Natural Science Foundation of Shandong Province (No. ZR201807090257), and National Bureau of Statistics Foundation Project (No. 2018LY79)
DATA AVAILABILITY STATEMENT
All data used in this study are acquired from publicly available cohorts.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209‐249. [DOI] [PubMed] [Google Scholar]
- 2. Gridelli C, Rossi A, Carbone DP, et al. Non‐small‐cell lung cancer. Nat Rev Dis Primers. 2015;1:15009. [DOI] [PubMed] [Google Scholar]
- 3. Shukla S, Evans JR, Malik R, et al. Development of a RNA‐seq based prognostic signature in lung adenocarcinoma. J Natl Cancer Inst. 2017;109:djw200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pao W, Girard N. New driver mutations in non‐small‐cell lung cancer. Lancet Oncol. 2011;12:175‐180. [DOI] [PubMed] [Google Scholar]
- 5. Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363‐385. [DOI] [PubMed] [Google Scholar]
- 6. Carlino MS, Larkin J, Long GV. Immune checkpoint inhibitors in melanoma. Lancet. 2021;398:1002‐1014. [DOI] [PubMed] [Google Scholar]
- 7. Zambrana F, Carril‐Ajuria L, Gomez de Liano A, et al. Complete response and renal cell carcinoma in the immunotherapy era: the paradox of good news. Cancer Treat Rev. 2021;99:102239. [DOI] [PubMed] [Google Scholar]
- 8. Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398:535‐554. [DOI] [PubMed] [Google Scholar]
- 9. Zhang C, Zhang Z, Li F, et al. Large‐scale analysis reveals the specific clinical and immune features of B7–H3 in glioma. Oncoimmunology. 2018;7:e1461304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reck M, Rodriguez‐Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD‐L1‐positive non‐small‐cell lung cancer. N Engl J Med. 2016;375:1823‐1833. [DOI] [PubMed] [Google Scholar]
- 11. Sharma P, Hu‐Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707‐723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Broer L, Buchman AS, Deelen J, et al. GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J Gerontol A Biol Sci Med Sci. 2015;70:110‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anselmi CV, Malovini A, Roncarati R, et al. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95‐104. [DOI] [PubMed] [Google Scholar]
- 14. Giuliani C, Garagnani P, Franceschi C. Genetics of human longevity within an eco‐evolutionary nature‐nurture framework. Circ Res. 2018;123:745‐772. [DOI] [PubMed] [Google Scholar]
- 15. Eicher JD, Landowski C, Stackhouse B, et al. GRASP v2.0: an update on the Genome‐Wide Repository of Associations between SNPs and phenotypes. Nucleic Acids Res. 2015;43:D799‐D804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP‐trait associations. Nucleic Acids Res. 2014;42:D1001‐D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peters MJ, Joehanes R, Pilling LC, et al. The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high‐throughput experiments. Bioinformatics. 2012;28:882‐883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mariathasan S, Turley SJ, Nickles D, et al. TGFbeta attenuates tumour response to PD‐L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1‐22. [PMC free article] [PubMed] [Google Scholar]
- 21. Charoentong P, Finotello F, Angelova M, et al. Pan‐cancer immunogenomic analyses reveal genotype‐immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248‐262. [DOI] [PubMed] [Google Scholar]
- 22. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ye Y, Jing Y, Li L, et al. Sex‐associated molecular differences for cancer immunotherapy. Nat Commun. 2020;11:1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ayers M, Lunceford J, Nebozhyn M, et al. IFN‐gamma‐related mRNA profile predicts clinical response to PD‐1 blockade. J Clin Investig. 2017;127:2930‐2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cancer Genome Atlas N . Genomic classification of cutaneous melanoma. Cell. 2015;161:1681‐1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (N Y). 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA‐seq data. BMC Bioinformatics. 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim J, Mouw KW, Polak P, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet. 2016;48:600‐606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alexandrov LB, Nik‐Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415‐421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer‐associated genes. Nature. 2013;499:214‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klijn C, Durinck S, Stawiski EW, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33:306‐312. [DOI] [PubMed] [Google Scholar]
- 35. Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333‐339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747‐1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang Z, Chen B, Liu X, et al. Effects of sex and aging on the immune cell landscape as assessed by single‐cell transcriptomic analysis. Proc Natl Acad Sci USA. 2021;118:e2023216118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fane M, Weeraratna AT. How the ageing microenvironment influences tumour progression. Nat Rev Cancer. 2020;20:89‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klempner SJ, Fabrizio D, Bane S, et al. Tumor mutational burden as a predictive biomarker for response to immune checkpoint inhibitors: a review of current evidence. Oncologist. 2020;25:e147‐e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim YA, Wojtowicz D, Sarto Basso R, et al. Network‐based approaches elucidate differences within APOBEC and clock‐like signatures in breast cancer. Genome Med. 2020;12:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen H, Chong W, Yang X, et al. Age‐related mutational signature negatively associated with immune activity and survival outcome in triple‐negative breast cancer. Oncoimmunology. 2020;9:1788252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chong W, Wang Z, Shang L, et al. Association of clock‐like mutational signature with immune checkpoint inhibitor outcome in patients with melanoma and NSCLC. Mol Ther Nucleic Acids. 2021;23:89‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang X, Hu LP, Qin WT, et al. Identification of a subset of immunosuppressive P2RX1‐negative neutrophils in pancreatic cancer liver metastasis. Nat Commun. 2021;12:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fein MR, He XY, Almeida AS, et al. Cancer cell CCR2 orchestrates suppression of the adaptive immune response. J Exp Med. 2020;217:e20181551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Z, Cao K, Hou Y, et al. PTTG1 knockdown enhances radiation‐induced antitumour immunity in lung adenocarcinoma. Life Sci. 2021;277:119594. [DOI] [PubMed] [Google Scholar]
- 47. Mudd TW Jr, Lu C, Klement JD, Liu K. MS4A1 expression and function in T cells in the colorectal cancer tumor microenvironment. Cell Immunol. 2021;360:104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jiao XD, Qin BD, You P, Cai J, Zang YS. The prognostic value of TP53 and its correlation with EGFR mutation in advanced non‐small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer. 2018;123:70‐75. [DOI] [PubMed] [Google Scholar]
- 49. Scalera S, Mazzotta M, Corleone G, et al. KEAP1 and TP53 frame genomic, evolutionary and immunological subtypes of lung adenocarcinoma with different sensitivity to immunotherapy. J Thorac Oncol. 2021;16(12):2065‐2077. [DOI] [PubMed] [Google Scholar]
- 50. Schoenfeld AJ, Bandlamudi C, Lavery JA, et al. The genomic landscape of SMARCA4 alterations and associations with outcomes in patients with lung cancer. Clin Cancer Res. 2020;26:5701‐5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li Z, Xue Q, Xu J, Zhang P, Ding B. The role of RBM10 mutations in the development, treatment, and prognosis of lung adenocarcinoma. Cell Cycle. 2020;19:2918‐2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dong ZY, Zhong WZ, Zhang XC, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD‐1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23:3012‐3024. [DOI] [PubMed] [Google Scholar]
- 53. Assoun S, Theou‐Anton N, Nguenang M, et al. Association of TP53 mutations with response and longer survival under immune checkpoint inhibitors in advanced non‐small‐cell lung cancer. Lung Cancer. 2019;132:65‐71. [DOI] [PubMed] [Google Scholar]
- 54. Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yi M, Jiao D, Xu H, et al. Biomarkers for predicting efficacy of PD‐1/PD‐L1 inhibitors. Mol Cancer. 2018;17:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jia Q, Wang J, He N, He J, Zhu B. Titin mutation associated with responsiveness to checkpoint blockades in solid tumors. JCI Insight. 2019;4:e127901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shen X, Zhao B. Efficacy of PD‐1 or PD‐L1 inhibitors and PD‐L1 expression status in cancer: meta‐analysis. BMJ. 2018;362:k3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bodor JN, Boumber Y, Borghaei H. Biomarkers for immune checkpoint inhibition in non‐small cell lung cancer (NSCLC). Cancer. 2020;126:260‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor‐based immunotherapy. Lancet Oncol. 2016;17:e542‐e551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S9
Table S1‐S6
Supplementary Material
Data Availability Statement
All data used in this study are acquired from publicly available cohorts.


