Abstract
Eliciting antibodies to surface-exposed viral glycoproteins can generate protective responses that control and prevent future infections. Targeting conserved sites may reduce the likelihood of viral escape and limit the spread of related viruses with pandemic potential. Here we leverage rational immunogen design to focus humoral responses on conserved epitopes. Using glycan engineering and epitope scaffolding in boosting immunogens, we focus murine serum antibody responses to conserved receptor binding motif (RBM) and receptor binding domain (RBD) epitopes following severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike imprinting. Although all engineered immunogens elicit a robust SARS-CoV-2-neutralizing serum response, RBM-focusing immunogens exhibit increased potency against related sarbecoviruses, SARS-CoV, WIV1-CoV, RaTG13-CoV, and SHC014-CoV; structural characterization of representative antibodies defines a conserved epitope. RBM-focused sera confer protection against SARS-CoV-2 challenge. Thus, RBM focusing is a promising strategy to elicit breadth across emerging sarbecoviruses without compromising SARS-CoV-2 protection. These engineering strategies are adaptable to other viral glycoproteins for targeting conserved epitopes.
Keywords: immunogen design, glycan, immune focusing, SARS-CoV-2, coronavirus
Graphical abstract
Hauser et al. use structure-guided design to engineer SARS-CoV-2 immunogens that direct immune responses to conserved viral sites in the context of preexisting immunity. In mice, these immunogens elicit antibodies that potently neutralize related coronaviruses, including those of potential pandemic concern. Structural characterization of selected antibodies explains this observation.
Introduction
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; SARS-2) and the subsequent global pandemic have highlighted the disruptive threat posed by viruses for which humans have no prior immunity. Rapid vaccine development has led to an unprecedented number of candidates. Although differing in platform (e.g., mRNA, adenovirus, nanoparticle), the primary immunogen is the SARS-2 spike ectodomain (Amanat and Krammer, 2020). With the continued global spread of SARS-2 in conjunction with potential vaccinations, it is likely that a large proportion of the global population will eventually develop an immune response to SARS-2. However, even after potentially achieving herd immunity sufficient to slow its spread, SARS-2 evolution leading to variants that escape immunity as well as emerging novel coronaviruses with pandemic potential remain a concern. Elicited immunity to SARS-2 infection may not protect against emergent novel coronaviruses that are closely related to SARS-2 as well as SARS-2 variants (Chen et al., 2021; Garcia-Beltran et al., 2021a, 2021b; Martinez et al., 2021; Planas et al., 2021; Supasa et al., 2021; Wibmer et al., 2021; Zhou et al., 2021). It is therefore critical to develop vaccines that confer potential pan-coronavirus immunity.
Although we cannot readily predict which novel coronaviruses or SARS-2 variants will emerge, the coronavirus spike glycoprotein contains conserved sites that can be targeted proactively, leading to potentially broad immunity. A potential site is the angiotensin-converting enzyme 2 (ACE2) receptor binding motif (RBM) of the receptor binding domain (RBD) (Barnes et al., 2020; Ju et al., 2020; Piccoli et al., 2020). Several potently neutralizing RBM-directed antibodies that interfere with ACE2 binding are protective, and some can also neutralize related coronaviruses from the same sarbecovirus subgenus (Barnes et al., 2020; Hansen et al., 2020; Rappazzo et al., 2021; Wec et al., 2020; Wu et al., 2020b). Although the RBM does contain some broadly conserved epitopes, large portions vary between related sarbecoviruses (Rappazzo et al., 2021; Wec et al., 2020). Other conserved sites include RBD epitopes outside of the RBM that show remarkable conservation across the sarbecovirus subgenus and currently circulating SARS-2 variants; some antibodies that bind to these epitopes can also confer broad neutralization (Garcia-Beltran et al., 2021b; Pinto et al., 2020; Yuan et al., 2020). Thus, implementing rational design strategies aimed at directing the immune response to these conserved sites may reduce the likelihood of viral escape and lead to more broadly protective responses (He et al., 2021; Martinez et al., 2022; Pinto et al., 2020; Rappazzo et al., 2021; Wec et al., 2020).
Immunogen design approaches that can be leveraged to direct humoral immune responses include “masking” epitopes via engineering putative N-linked glycosylation (PNG) sites and epitope “scaffolding” to selectively present broadly protective epitopes (Bajic et al., 2019, 2020); these strategies have been used previously for the viral glycoproteins respiratory syncytial virus (RSV) F, influenza hemagglutinin, and HIV envelope proteins (Correia et al., 2014; Crispin et al., 2018; Ofek et al., 2010). Applying these approaches to the SARS-2 spike provides an opportunity to potentially improve serum neutralization potency, efficacy against variants, and cross-reactivity of antibody responses. Multimerized versions of the RBDs of several coronaviruses are potent immunogens (Cohen et al., 2021; Dai et al., 2020; Kang et al., 2021; Li et al., 2021; Ma et al., 2020; Saunders et al., 2021; Shinnakasu et al., 2021; Walls et al., 2020; Wang et al., 2021a). However, these engineered immunogens have only been tested as single immunization regimens or homologous prime/boost regimens in SARS-2-naïve animals.
SARS-2 spike-based boosters are now recommended for some mRNA vaccine recipients (Hause et al., 2021), and clinical trials of heterologous boosters with different SARS-2 variants are underway (e.g., ClinicalTrial.gov: NCT04889209 and NCT04713553). SARS-2 imprinting via infection or prior immunization therefore merits consideration as boosting immunizations are evaluated. The immunological effect of imprinting is especially notable in influenza re-infection or immunization, in which the antibody response often remains biased toward the influenza strain to which a human or model organism first developed humoral immunity; this “antigenic sin” can be detrimental to protection against influenza strains that are antigenically distinct from the imprinting strain (de St Groth and Webster, 1966; Jensen et al., 1956; Webster, 1966).
Here, we designed protein-based immunogens that used hyperglycosylation of the RBD and a “resurfacing” approach that grafts the RBM from SARS-2 onto heterologous coronavirus-based RBD scaffolds. We boosted with these immunogens to refocus the immune response in the context of SARS-2 spike imprinting. We immunized mice that were primed with SARS-2 spike as a surrogate for pre-existing immunity imprinted by vaccination or natural infection. We found that boosting with different regimens containing our engineered immunogens could selectively focus serum responses to the RBM or non-RBM epitopes. Importantly, even the RBM-focused response targets broadly conserved epitopes on related sarbecovirus RBDs. We isolated and structurally characterized antibodies targeting conserved and SARS-2-specific RBM epitopes, including a class of antibodies with broad sarbecovirus neutralization activity. The RBM-directed, immune-focused response is potently neutralizing, with breadth across SARS-2 variants and other coronaviruses without compromising SARS-2 neutralization or protection. Our data show how rationally designed immunogens can redirect immune responses to conserved coronavirus epitopes in the context of pre-existing immunity. These results could inform next-generation coronavirus vaccines.
Results
Epitope grafting of the SARS-2 RBM onto heterologous coronavirus scaffolds
The RBM of SARS-2 and the related sarbecoviruses SARS-CoV (SARS-1) and WIV1-CoV (WIV1) is a contiguous sequence spanning residues 437–508 (SARS-2 numbering) of the spike protein. To elicit RBM-specific responses only, we first evaluated whether the RBM itself could be expressed recombinantly in the absence of the rest of the RBD (Figure S1A). Although the SARS-2 RBM could indeed be expressed, it failed to engage the conformation-specific RBM-directed antibody B38 or bind to cell-surface-expressed ACE2 (Figure S1B). These results suggest that the RBM is conformationally flexible and that the RBD serves as a structural “scaffold” to stabilize the RBM in its binding-compatible conformation.
To circumvent the hurdle of de novo scaffold design to present the RBM, we tested whether heterologous sarbecovirus RBDs from SARS-1 and WIV1 and the more distantly related merbecovirus MERS-CoV (Middle East respiratory syndrome; MERS) could serve as scaffolds (Figure 1A); variations of this approach have been used previously to modulate ACE2 binding properties (Letko et al., 2020; Shang et al., 2020). The SARS-1, WIV1, and MERS RBDs share a pairwise amino acid identity with SARS-2 of 73.0%, 75.4%, and 19.5%, respectively. Despite both using ACE2 as a receptor, the RBM is less conserved for SARS-1 and WIV1, with only 49.3% and 52.1% identity, respectively; because MERS uses DPP4 as a receptor, its RBM shares no notable identity (Raj et al., 2013). Although we were unable to “resurface” the MERS RBD with the SARS-2 RBM, the related SARS-1 and WIV1 RBDs successfully accepted the RBM transfer. These resurfaced (rs) constructs, rsSARS-1 and rsWIV1, retained binding to the SARS-2 RBM-specific B38 antibody and efficiently engaged ACE2 (Figure S1C; Wu et al., 2020b). These data suggest that there are sequence and structural constraints within the RBD required for successful RBM grafting; such an approach may be facilitated by using CoV RBDs that use the same receptor for viral entry.
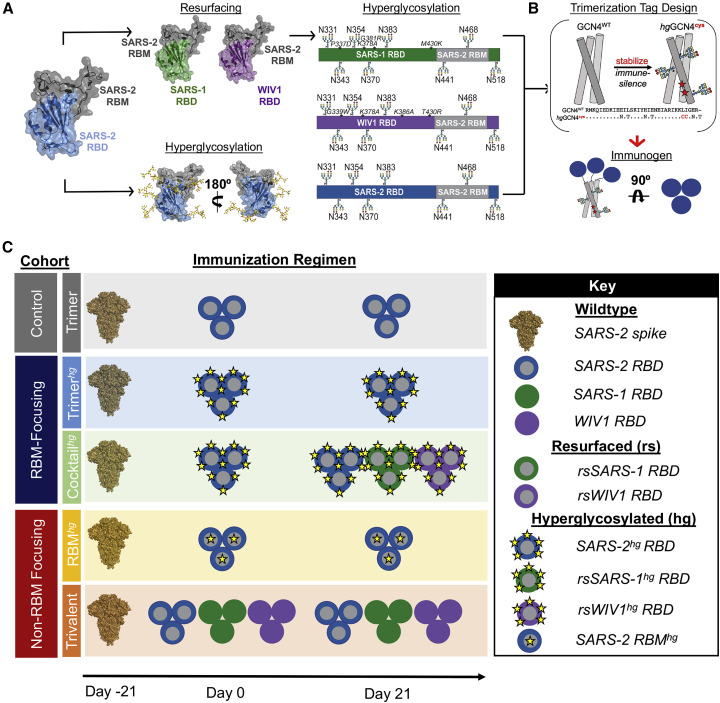
Figure 1.
Resurfacing and hyperglycosylation approaches for immune focusing
(A) Design schematics for resurfacing SARS-1 (rsSARS-1) and WIV1 (rsWIV1) scaffolds with the SARS-2 receptor binding motif (RBM) and for hyperglycosylating SARS-2 (blue), rsSARS-1 (green), and rsWIV1 (purple) receptor binding domains (RBDs). Non-native engineered glycans and native glycans are modeled; native SARS-2 RBD glycan at position 331 is omitted in the schematic. Although glycans at positions 441 and 468 technically fall within the RBM, they are on the sides of the RBD, as shown in the model. Mutations in the WIV1 and SARS-1 RBDs are shown in red and italicized in the linear diagram. All images were created using PDB: 6M0J.
(B) Design schematic for generating RBD trimers appended onto a cystine-stabilized (red stars) hyperglycosylated GCN4 tag (PDB: 6VSB).
(C) Schematic of immunization cohorts. The Trimer, Trimerhg, and Cocktailhg cohorts each contained 10 mice, and the Trivalent and RBMhg cohorts each contained 5 mice.
See also Figures S1 and S2.
Engineered glycans for epitope focusing
We next used these rsRBDs as templates for further modification using glycan engineering. This approach aimed to mask conserved, cross-reactive epitopes shared between the SARS-1, SARS-2, and WIV1 RBDs to further enhance potential immune focusing to the RBM. There are two evolutionarily conserved putative N-linked glycosylation sites (PNGs) at positions 331 and 343; SARS-1 and WIV1 have an additional conserved PNG at position 370 (SARS-2 numbering). To increase overall surface glycan density, we introduced novel PNGs onto wild-type SARS-2 as well as rsSARS-1 and rsWIV1 RBDs. Based on structural modeling and further biochemical validation, we identified 5 additional sites on rsSARS-1 and rsWIV1 and 6 on SARS-2. Including the native PNGs, all constructs had a total of 8 glycans (Figures 1A and S1D–S1G); we denote these hyperglycosylated (hg) constructs as SARS-2hg, rsSARS-1hg, and rsWIV1hg. We expressed these constructs in mammalian cells to ensure complex, heterogeneous glycosylation to maximize the glycan “shielding” effect. We subsequently characterized these constructs using the RBM-directed antibody B38, as well as ACE2 binding, to ensure that the engineered PNGs did not adversely affect the RBM conformation. The hyperglycosylated constructs were largely comparable in affinity for B38, with only an ∼2-fold decrease, and still effectively engaged ACE2 (Figure S1G). These results confirm a conformationally and functionally intact RBM.
Next we assessed whether the engineered PNGs abrogated binding to the sarbecovirus cross-reactive antibodies S309 and CR3022, which engage epitopes outside of the RBM (Pinto et al., 2020; Yuan et al., 2020). The CR3022 epitope between SARS-1 and WIV1 differs at only a single residue, whereas SARS-2 differs at 5 residues across the CR3022 and S309 epitopes (Wu et al., 2020a). Importantly, these epitope regions comprise a considerable portion of the non-RBM SARS-2 RBD, and, thus, any RBM focusing would require masking of these regions (Barnes et al., 2020; Pinto et al., 2020; Yuan et al., 2020). Although SARS-2hg effectively abrogated S309 and CR3022 binding, the engineered PNGs at the antibody:antigen interface on rsSARS-1hg and rsWIV1hg did not completely abrogate S309 and CR3022 binding. We therefore incorporated unique mutations on rsSARS-1hg and rsWIV1hg so that any potentially elicited antibodies would be less likely to cross-react between these two constructs. We found that K378A and the engineered glycan at residue 383 (SARS-2 numbering) abrogated CR3022 binding in rsSARS-1hg and rsWIV1hg (Figure S1F). For S309, mutation P337D in rsSARS-1hg and G339W in rsWIV1hg, in addition to glycans at residues 441 and 354 (SARS-2 numbering), were sufficient to disrupt binding (Figure S1F). We made two additional mutations, G381R, M430K on rsSARS-1hg and K386A, T430R on rsWIV1hg, to increase the antigenic distance between these scaffolds (Figure 1A).
Last, we wanted to determine whether hyperglycosylation could similarly focus the humoral immune response to non-RBM epitopes. We therefore engineered four novel glycans at positions 448, 475, 494, and 501 on the RBM of the wild-type SARS-2 RBD (RBMhg) (Figures S2A and S2B). The engineered PNGs effectively abrogate RBM-directed B38 antibody binding and engagement of ACE2 (Figure S2C). Because the engineered PNGs restrict binding of RBM-directed antibodies, this construct can also be used to assess RBM focusing compared with the wild-type SARS-2 RBD.
Design of a non-immunogenic trimerization tag for enhanced avidity
To increase avidity of our engineered immunogens while minimizing any off-target tag-specific responses, we designed a cysteine-stabilized and hyperglycsoylated variant of a GCN4 trimerization tag (hgGCN4cys) (Figure 1B; Sliepen et al., 2015). Although the two cysteines are within one subunit, they form an intermolecular disulfide with an adjacent subunit, allowing the RBDs to remain trimerized, whereas the tag is “immune silent.” We recombinantly expressed the engineered immunogens and wild-type RBD trimers in mammalian cells; the oligomeric state was confirmed using SDS-PAGE analysis under non-reducing conditions (Figures S2D–S2F). Antigenicity was assayed using Fab fragments of the conformation-specific antibodies CR3022 and/or B38 using biolayer interferometry; the RBD trimers had monovalent affinities comparable with the RBD monomers (Figure S2G). We also used the hgGCN4cys tag for the engineered hyperglycosylated and resurfaced immunogens (Figures S2H–S2J).
Cohorts and immunization regimens
We tested the immunogenicity and antigenicity of our designs and assessed their RBM and non-RBM immune-focusing properties in wild-type C57BL/6 mice (Figure 1C). Cohorts are color-coded in Figure 1C, and the same color coding is used throughout all subsequent figures. All cohorts were primed with SARS-2 spike to reflect pre-existing SARS-2 immunity. All protein immunizations were adjuvanted with Sigma adjuvant. The control cohort was boosted with wild-type (i.e., unmodified) SARS-2 RBD trimer (Figure 1C, Trimer cohort, gray). We divided our immune-focusing cohorts into RBM and non-RBM centered. For RBM immune-focusing, one cohort was boosted with SARS-2hg trimers (Figure 1C, Trimerhg cohort, light blue), and a second cohort was boosted first with SARS-2hg trimers followed by a second boost with a cocktail of rsSARS-1hg and rsWIV1hg (Figure 1C, Cocktailhg cohort, light green). For non-RBM immune focusing, one cohort was boosted with RBMhg trimers (Figure 1C, RBMhg, yellow), and a second was boosted with a cocktail of wild-type SARS-1, SARS-2, and WIV1 RBD trimers (Figure 1C, Trivalent cohort, orange).
The Trimerhg and RBMhg cohorts described above use hyperglycosylation as an immune focusing strategy, whereas the Cocktailhg cohort combines hyperglycosylation and resurfacing to enhance immune focusing to the RBM by reducing the prevalence of any non-RBM epitopes. The Trivalent cohort preferentially displays the conserved RBM across the wild-type SARS-2, SARS-1, and WIV1 RBDs; the majority of this conserved surface area falls outside of the RBM (Figure S2K).
Immune focusing of serum responses in the context of SARS-2 spike imprinting
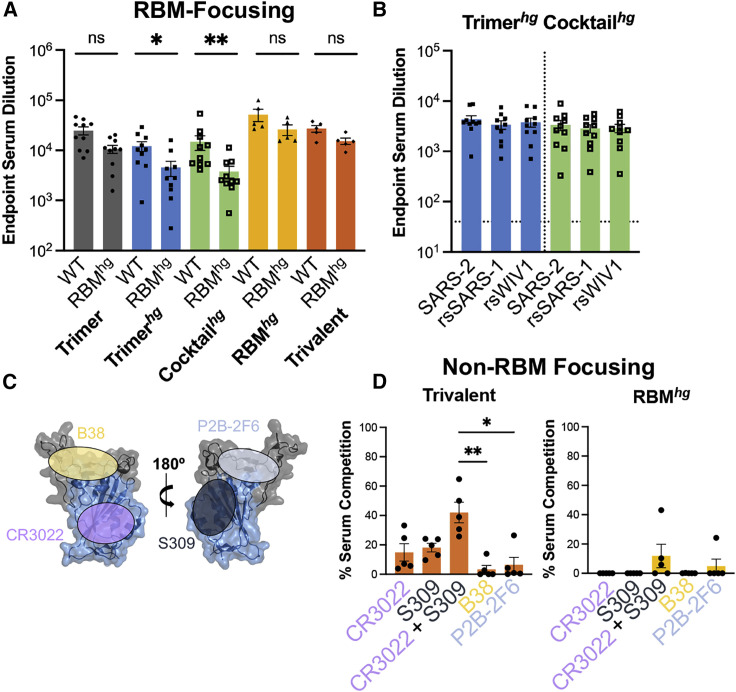
Across all cohorts, we observed a robust serum response to the wild-type SARS-2 RBD (Figures 2A and S3A–S3E) with minimal tag-directed responses (Figure S3F). To specifically assess RBM-directed responses, we compared serum ELISA titers with the wild-type SARS-2 RBD and our SARS-2 RBMhg RBD construct; the latter has glycans that occlude the RBM. The Trimerhg and Cocktailhg cohorts had a significant increase in serum antibody titers to the wild-type SARS-2 RBD relative to the SARS-2 RBMhg RBD; this contrasted with the Trimer, RBMhg, and Trivalent cohorts, for which this difference was not statistically significant (Figure 2A). For all mice, the titers to the SARS-2 RBMhg construct were lower than those to the wild-type SARS-2 RBD. Across the Trimerhg and Cocktailhg cohorts, the mean endpoint titer reduction to the SARS-2 RBMhg RBD relative to the wild-type SARS-2 RBD was ∼64%, which reflects a total or partial loss of affinity from antibodies because of steric interference by the RBMhg engineered glycans. The Cocktailhg cohort had a modest increase in RBM focusing relative to the Trimerhg cohort. This may be due to increasing the overall antigenic distance (i.e., sequence difference) between the WIV1 and SARS-1 RBDs relative to SARS-2 while maintaining the identical SARS-2 RBM epitope. We find that the Trimerhg and Cocktailhg cohorts had significantly lower titers to the SARS-1 and WIV1 RBDs than to the SARS-2 RBD (Figures S3B and S3C). This difference was most pronounced in the Cocktailhg cohort, suggesting that the hyperglycosylation and engineered mutations within the RBD dampened responses to conserved, cross-reactive epitopes present outside the RBM. Serum titers against the rsSARS-1 and rsWIV1 RBDs were comparable with the SARS-2 RBD, indicating that there is a minimal antibody response directed toward the wild-type SARS-1 and WIV1 RBD epitopes in comparison with the SARS-2 RBM (Figure 2B). We observed no significant glycan-dependent serum response in the Trimerhg or Cocktailhg cohort (Figure S3G). These data confirm an enhanced SARS-2 RBM-focused serum response elicited by our engineered immunogens.
Figure 2.
Assessing SARS-2 RBD immune focusing via serum analysis from cohorts
(A) Serum following immunizations was assayed by ELISA on day 35 with the wild-type SARS-2 RBD and RBMhg. Statistical significance was determined using a Mann-Whitney U test (∗p < 0.05, ∗∗p < 0.01). Bars represent mean ± SE.
(B) Day 35 serum samples from the Trimerhg and Cocktailhg cohorts showed significantly less binding to the SARS-1 and WIV1 RBDs compared with the SARS-2 RBD (Figures S3B and S3C). However, when assayed against rsSARS-1 and rsWIV1 RBDs, these sera no longer show statistically significant differences in binding compared with the SARS-2 RBD, as determined using a Kruskal-Wallis test with post hoc analysis using Dunn’s test corrected for multiple comparisons. Bars represent mean ± SE.
(C) Approximate locations of representative Ab epitopes from each of the four SARS-2 RBD-directed Ab classes (Barnes et al., 2020; PDB: 6M0J).
(D) Percent competition in ELISAs using day 35 mouse sera in the presence of competing IgGs versus a no-IgG control. SARS-2 RBD was the coating antigen. Statistical significance was determined using a Kruskal-Wallis test with post hoc analysis using Dunn’s test corrected for multiple comparisons, and pairwise comparisons pictured without bars were not significant (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001); ns, not significant.
See also Figure S3.
To determine whether the RBMhg and Trivalent cohorts successfully directed the immune response to non-RBM epitopes on the RBD, we performed serum competition by incubating RBD-coated ELISA plates with B38, P2B-2F6, CR3022, and S309 immunoglobulin G (IgG), representing each of the four previously defined “classes” of SARS-2 RBD epitopes (Figure 2C; Barnes et al., 2020). Indeed, the previously characterized CR3022 and S309 antibodies have footprints that together cover much of the conserved non-RBM region of the RBD, with buried surface area (BSA) of 917 Å2 and 795 Å2, respectively, in comparison with a BSA of 869 Å2 for ACE2 (Lan et al., 2020; Pinto et al., 2020; Yuan et al., 2020). We then assessed binding of mouse serum IgG (Figure 2D). Only the Trivalent cohort showed a significant increase in serum competition when the CR3022 and S309 IgGs were combined, suggesting that only this regimen could effectively focus on conserved non-RBM epitopes on the SARS-2 RBD.
Immunogen-elicited RBM-focused antibody responses potently neutralize sarbecoviruses
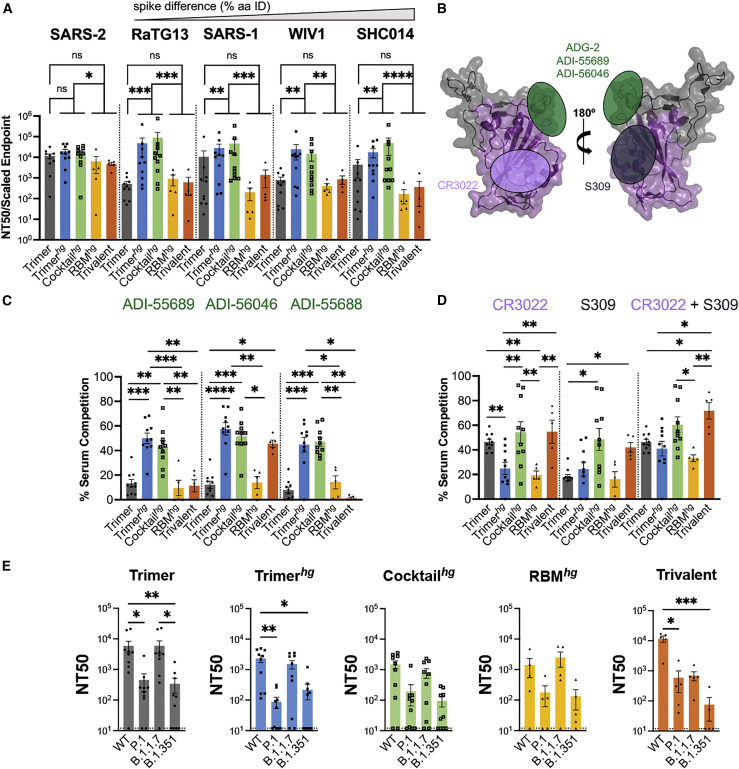
We next compared the neutralization potency (i.e., neutralization per unit of antigen-specific IgG) of all cohorts using SARS-1, SARS-2, WIV1, RaTG13-CoV (RaTG13), and SHC014-CoV (SHC014) pseudoviruses (Crawford et al., 2020; Garcia-Beltran et al., 2021a; Menachery et al., 2015; Shang et al., 2020). WIV1, RaTG13, and SHC014, in this instance, are broadly representative of possible future emerging sarbecoviruses with pandemic potential (Menachery et al., 2015, 2016; Shang et al., 2020). All cohorts elicited a potent SARS-2 neutralizing response. The RBM-focusing Trimerhg and Cocktailhg cohorts elicited a significantly more potent neutralizing response than the non-RBM focusing RBMhg and Trivalent cohorts. The Trimerhg and Cocktailhg cohorts also neutralized SARS-1-, WIV1-, RaTG13-, and SHC014-expressing pseudoviruses relative to the Trimer, RBMhg, and Trivalent cohorts (Figures 3A, S3A–S3E, and S3H). This is noteworthy for the Trimerhg cohort because it did not include any of these RBDs in the immunization regimen. Similarly, the Cocktailhg cohort neutralized RaTG13 and SHC014, neither of which was present in the immunogen. In contrast, the Trimer cohort was less potently neutralizing against RaTG13, SARS-1, WIV1, and SHC014, and the RBMhg cohort trended toward a loss in neutralization as well (Figures S4A and S4B). The Trivalent cohort also failed to neutralize SARS-1 and trended toward a loss against RaTG13, WIV1, and SHC014. Immune imprinting by priming with SARS-2 spike in the Trivalent cohort appears to have biased the subsequent serum antibody response, which shows significantly greater neutralization of SARS-2 compared with SARS-1 and WIV1 despite inclusion of all three components in the boosting immunizations. The neutralization patterns observed for the Trimer, RBMhg, and Trivalent cohorts are similar to those in humans and mice after SARS-2 infection or vaccination, in which sera show markedly reduced neutralization against related sarbecoviruses compared with SARS-2 (Garcia-Beltran et al., 2021a, 2021b; He et al., 2021).
Figure 3.
Potency and characterization of the SARS-like coronavirus neutralization response
(A) Day 35 serum from all mice was assayed for neutralization against SARS-2, RaTG13, SARS-1, WIV1, and SHC014 pseudoviruses (arranged in order of genetic similarity of the full-length spike to SARS-2). Neutralization potency was computed using scaled endpoint serum ELISA titers. Statistical significance was determined using a Kruskal-Wallis test with post hoc analysis using Dunn’s test corrected for multiple comparisons (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Bars represent mean ± SE.
(B) Approximate locations of representative Ab epitopes from the two non-RBM-directed SARS-2 RBD-directed Ab classes (Barnes et al., 2020) and ADG-2-like Abs on the WIV1 RBD (PDB: 6M0J).
(C and D) Ab competition ELISAs with WIV1 RBD as the coating antigen. Bars show the mean percent binding lost, with error bars representing the standard error of the mean. Comparisons were performed using a Kruskal-Wallis test with post hoc analysis using Dunn’s test corrected for multiple comparisons, and pairwise comparisons pictured without bars were not significant (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Bars represent mean ± SE.
(E) Day 35 serum was assayed against SARS-2 variant pseudoviruses for neutralization. Statistical significance was determined using a Kruskal-Wallis test with post hoc analysis using Dunn’s test corrected for multiple comparisons, and pairwise comparisons pictured without bars were not significant (∗p < 0.05, ∗∗p < 0.01). Bars represent mean ± SE.
See also Figure S4).
Receptor-binding-motif-focused antibody responses target a broadly conserved epitope
To epitope map cross-reactive, RBM-focused serum responses, we performed ELISA-based antibody competition using the cross-reactive antibodies CR3022, S309, ADI-55688, ADI-55689, and ADI-56046 with the WIV1 RBD (Figures 3B–3D). The latter three antibodies bind a conserved sarbecovirus RBM epitope also targeted by the antibody ADG-2, which is currently in clinical development and for which ADI-55688 is a precursor, as well as other antibodies with broad sarbecovirus neutralization (Martinez et al., 2022; Rappazzo et al., 2021; Wec et al., 2020). Competition ELISAs, which we used to broadly bin serum antibody epitopes, suggest that the cross-reactive WIV1-directed responses in the Trimerhg and Cocktailhg cohorts focus to the ADG-2-like epitope as well as to the CR3022 and S309 epitopes in the Cocktailhg cohort (Figures 3C and 3D). Thus, SARS-2hg, rsSARS-1hg, and rsWIV1hg RBDs can not only induce potent SARS-2 neutralizing antibodies but also cross-reactive antibodies that bind to a conserved RBM epitope.
Immune-focused responses neutralize variants of concern
Many SARS-2 variants of concern include mutations within the RBM, including B.1.1.7 (alpha), B.1.351 (beta), and P.1 (gamma), first detected in the United Kingdom, South Africa, and Brazil, respectively (Figure S4C). We evaluated how enhanced focusing to the RBM affected binding to these variants. Serum from the Cocktailhg cohort showed no significant loss of binding to the B.1.351 RBD compared with the wild-type SARS-2 RBD (Figure S4D). In contrast, the Trimer and Trimerhg cohorts had significant loss of binding; this parallels the observation of reduced serum binding from human subjects immunized with current SARS-2 vaccines (Chen et al., 2021; Garcia-Beltran et al., 2021b; Wang et al., 2021b; Zhou et al., 2021). The RBMhg and Trivalent cohorts showed no significant loss of binding, consistent with a non-RBM focused serum antibody response.
Additionally, we tested all cohort sera for neutralization against B.1.1.7-, B.1.351-, and P.1-expressing pseudoviruses. Although the Trimer and Trimerhg cohorts still neutralized all pseudoviruses to some degree, there was reduced neutralization of P.1 and B.1.351, consistent with our ELISA data. The Trivalent cohort also showed reduced neutralization of P.1 and B.1.351 despite maintaining binding to the B.1.351 RBD in ELISA. Although the Cocktailhg and RBMhg cohorts showed weaker neutralization of P.1 and B.1.351, this difference was not statistically significant (Figure 3E). Indeed, obscuring the RBM with glycans in our RBMhg cohort may have elicited neutralizing antibodies that are less sensitive to RBM mutations present in the variants, similar to other RBD-directed antibodies (e.g., CR3022/COVA1-16 and S309; Liu et al., 2020a; Pinto et al., 2020; Yuan et al., 2020). However, the RBM-directed elicited response from the Cocktailhg cohort still neutralized all variants. This may indicate that immune focusing to the RBM may allow greater recognition (i.e., accommodation) of mutations compared with the RBM-directed antibody response elicited by natural infection or current vaccines (Yuan et al., 2021; Zhou et al., 2021).
Additional multimerization does not improve SARS-2 neutralization or neutralization breadth
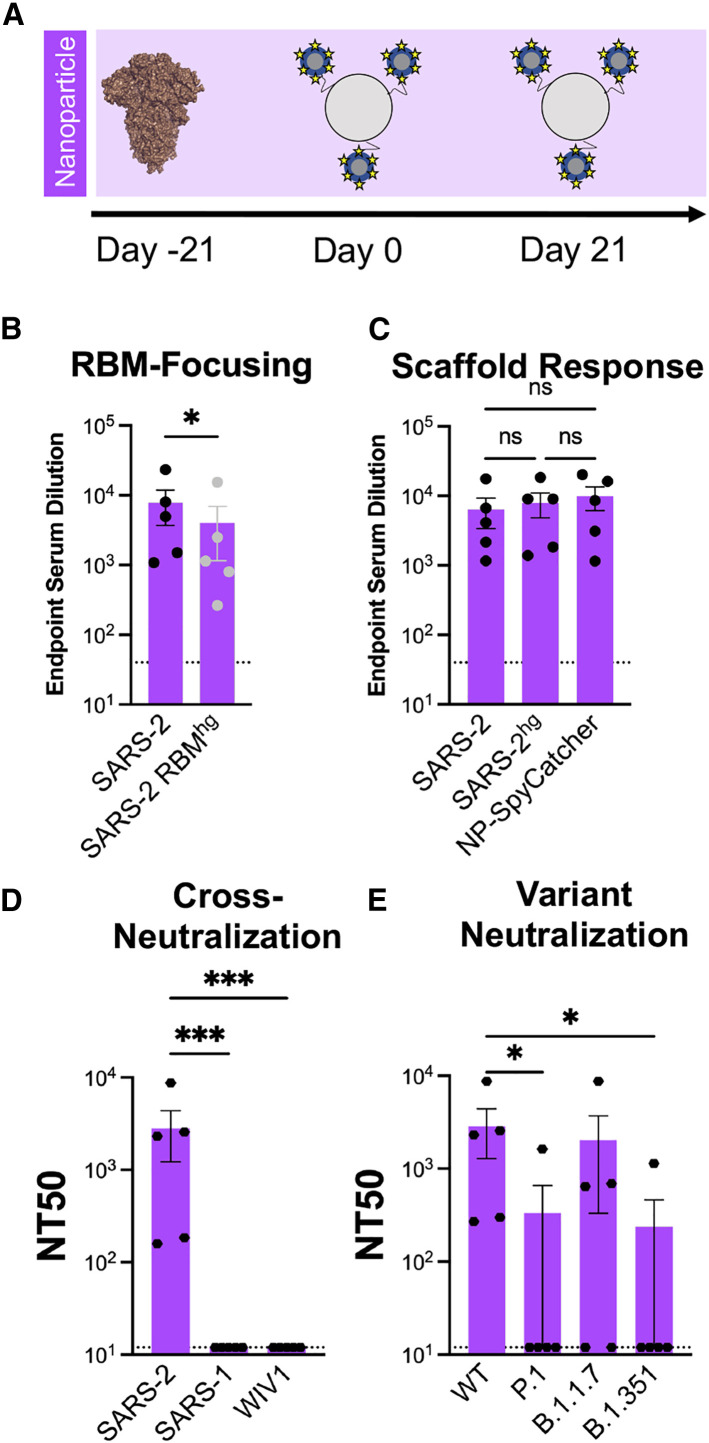
Nanoparticles are commonly used to enhance immunogenicity by increasing overall avidity (Dai et al., 2020); we therefore wanted to determine whether increasing the copy numbers of our engineered immunogens from 3 to 24 using ferritin-nanoparticles would improve overall immunogenicity and immune focusing. We covalently attached the SARS-2hg RBD to ferritin nanoparticles using SpyTag-SpyCatcher; this engineered RBD is the same as used in our Trimerhg cohort (Figure 4A; Zakeri et al., 2012). Using the same immunization regimen as in the Trimerhg cohort allowed direct comparison with antigenicity and immunogenicity because of valency. The nanoparticle immunogen did not elicit higher serum ELISA titers against the SARS-2 RBD (Figure S5A and S5B), maintained RBM-focusing (Figures 4B, 4C, and S5C) and had comparable SARS-2 pseudovirus neutralization titers (Figure S5D). However, nanoparticle-boosted mice had markedly lower neutralization titers against SARS-1 and WIV1 pseudoviruses (Figure 4D) and reduced neutralization of SARS-2 variants (Figures 4E and S5E). These data suggest that, at least for the SARS-2hg RBD, further multimerization using a ferritin nanoparticle confers minimal, if any, functional advantage.
Figure 4.
Immune response following boosting with ferritin nanoparticle multimerization of SARS-2hg
(A) Design schematic of a multimerized version of SARS-2hg using SpyTag-SpyCatcher conjugation to a ferritin nanoparticle.
(B) Serum following immunization was assayed in ELISA on day 35 with the wild-type SARS-2 RBD and RBMhg. Statistical significance was determined using Mann-Whitney U test (∗p < 0.05). Bars represent mean ± SE.
(C) Day 35 serum titers to the wild-type SARS-2 RBD were also compared with titers against SARS-2hg and the unconjugated ferritin nanoparticle-SpyCatcher fusion. A Kruskal-Wallis test was performed and detected no significant differences between the serum Ab responses to these three proteins. Bars represent mean ± SE.
(D) Day 35 serum was assayed for neutralization against SARS-2, SARS-1, and WIV1 pseudoviruses. Statistical significance was determined using a Kruskal-Wallis test with post hoc analysis using Dunn’s test corrected for multiple comparisons, and pairwise comparisons pictured without bars were not significant (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). Bars represent mean ± SE.
(E) Day 35 serum was assayed against SARS-2 variant pseudoviruses for neutralization. Statistical significance was determined using a Kruskal-Wallis test with post hoc analysis using Dunn’s test corrected for multiple comparisons, and pairwise comparisons pictured without bars were not significant (∗p < 0.05). Bars represent mean ± SE.
Isolated antibodies from expanded IgG+ B cell lineages include antibodies with broad neutralization of sarbecoviruses and variants of concern
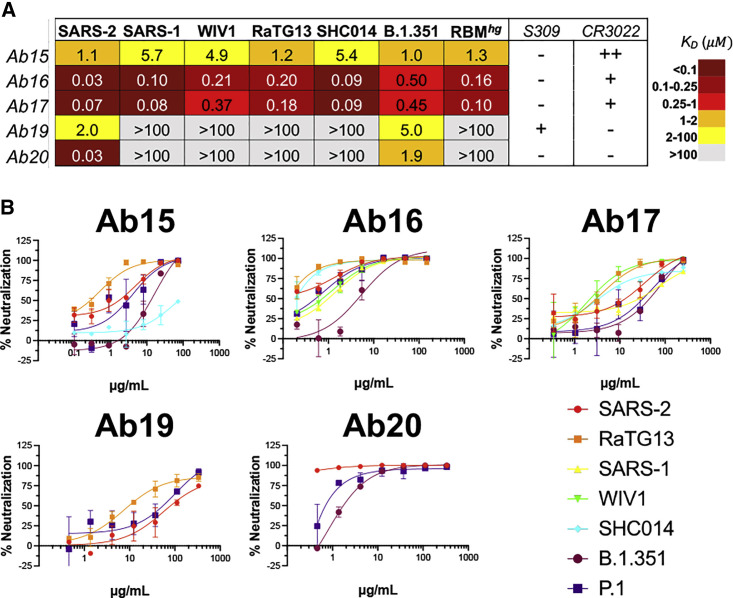
We next isolated a total of 85, 61, and 30 paired heavy- and light-chain sequences from SARS-2 RBD-specific IgG+ B cells from the Trimer, Trimerhg, and Cocktailhg cohorts, respectively (Figure S6A). Overall, there was a predominance of IGHV1-42 gene usage across all cohorts, but light-chain usage varied between the control Trimer cohort and the Trimerhg and Cocktailhg cohorts (Figures S6B and S6C). CDRH3 length was significantly longer in the Trimerhg cohort, with a median of 12 amino acids versus a median of 7 in the Trimer and Cocktailhg cohorts (Figure S6D). Median somatic hypermutation was relatively similar between the cohorts (Figure S6E). We chose 5 monoclonal antibodies from 4 different clonally related populations isolated from the Trimerhg cohort to express recombinantly for further characterization (Figures 5A and S6F–S6I; Table S1). The CDRH3s of these clonally related populations were not shared with any antibodies isolated from the control Trimer cohort. Antibody 19 (Ab19) and Ab20 were SARS-2 specific, did not bind the RBMhg construct, and did not compete with CR3022, suggesting a largely RBM-directed epitope. Ab15 and the clonally related Ab16 and Ab17 (Figure S6H) were exceptionally broad in their reactivity, engaging all coronavirus RBDs tested as well as the SARS-2 variant B.1.351 (Figure 5A). These Abs still bound the RBMhg construct and completely (Ab15) or partially (Ab16 and Ab17) competed with CR3022; affinities to the B.1.351 and RBMhg construct were between ∼2- and 20-fold lower than the affinity to the SARS-2 RBD. These data suggest a conserved epitope that partially overlaps the CR3022 epitope and the RBM, like that targeted by ADI-56046 (Wec et al., 2020; Figure S6J). In addition to their broad cross-reactivity, Ab16 and Ab17 neutralized SARS-2, RaTG13, SARS-1, WIV1, and SHC014 (Figure 5B). All Abs neutralized P.1, and all except Ab19 neutralized B.1.351. Although Ab19 has no detectable affinity for RaTG13 as an Fab (Figure 5A), it still neutralizes the pseudovirus as an IgG, suggesting that avidity is required. This indicates that RBM focusing can elicit Abs capable of broadly neutralizing both related SARS-2 variants of concern and diverse sarbecoviruses.
Figure 5.
SARS-2 RBD-directed B cell characteristics
Spleens were harvested on day 42, and SARS-2 RBD-directed IgG+ B cells were isolated via flow cytometry and sequenced.
(A) Abs representative of lineages that were expanded in RBM-focusing cohorts were expressed recombinantly as Fabs, and their binding was characterized using BLI.
(B) Pseudovirus neutralization for these Abs was also characterized.
See also Figure S6).
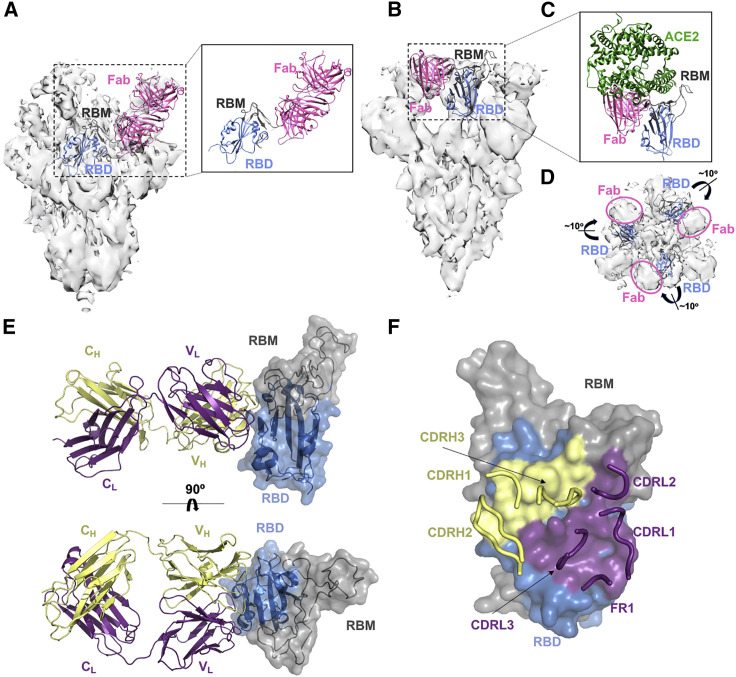
Structural characterization of broadly neutralizing Abs
To define the epitope targeted by these Abs, we first obtained a low-resolution (∼9.2-Å) cryoelectron microscopy (cryo-EM) structure of Ab20 in complex with the SARS-2 spike (Figure 6A). Ab20 appears to target an epitope on the upper loop of the SARS-2 RBM; its footprint will likely interfere with ACE2 binding and is consistent with the observation that Ab20 does not bind the RBMhg with appreciable affinity. Moreover, Ab20 has a significant reduction in affinity for the B.1.351 RBD relative to the wild-type SARS-2 RBD; mutation E484K within the RBM of B.1.351 appears to overlap with the Ab20 footprint (Figure S4C).
Figure 6.
Structural characterization of Abs from the RBM-focused immune response
(A) Low-resolution cryo-EM map with a model of Ab20 as Fab (pink) bound to the RBD (blue) with the RBM (gray) shown (the RBD is from PDB: 6VXX). For clarity, only a single RBD and Fab are shown.
(B) Low-resolution cryo-EM map with a model of Ab16 as Fab (pink) bound to the RBD (blue) with the RBM (gray) shown (the RBD is from PDB: 7DX9). For clarity, only a single RBD and Fab are shown.
(C) Model from (B) with docked ACE2 (from PDB: 6M0J).
(D) Cryo-EM map with 3 docked RBDs (blue, with RBMs in gray) from the “3 RBD up” spike in PDB: 7DX9. RBDs are shown as ribbons, and the Fab Ab16 was removed to show its density and the slight outward rotation of the RBD required to better fit the density compared with the docked model.
(E) Interface of the co-crystal structure of the SARS-2 RBD and the Ab17 heavy-chain (yellow) and light-chain (purple) complex.
(F) Surface of the SARS-2 RBD in contact with Ab17 heavy-chain (yellow) and light-chain (purple) residues.
See also Figure S7.
We next obtained a low-resolution (5.5-Å) cryo-EM structure of Ab16 in complex with SARS-2 spike (Figures 6B–6D) and determined a high-resolution crystal structure of the clonally related Ab17 in complex with the SARS-2 RBD (Figures 6E and 6F). In the cryo-EM structure with Ab16, the SARS-2 spike is in the “three RBD up” conformation, with density for each RBD to be occupied by a Fab. Consistent with the reactivity from biolayer interferometry (BLI), Ab16 appears to engage a conserved “class 4”-like epitope (explaining the observed competition with CR3022) that includes part of the RBM, and Ab16 will likely sterically interfere with ACE2 binding (Barnes et al., 2020; Figure 6C). The complex appears to show an outward rotation of the bound RBD relative to the previously characterized “three RBD up” (PDB: 7DX9) conformation (Figure 6D). This has been hypothesized previously to contribute to SARS-1 neutralization by CR3022, and it may contribute to broad neutralization by Ab16 and Ab17 as well (Yuan et al., 2020). The higher-resolution co-crystal structure of Ab17 in complex with the SARS-2 RBD provides a more complete view of the antigen-combining site of these clonally related Abs (Figure 6E; Table S2). The crystal structure confirms a “class 4” epitope with additional interactions extending into the RBM (Barnes et al., 2020).It overlaps with previously characterized conserved epitopes targeted by Abs with broad sarbecovirus neutralization activity: Ab ADI-56046 from a human donor and Abs K288.2 and K398.22 isolated from rhesus macaques (He et al., 2021; Wec et al., 2020). The overall footprint is large, with a BSA of 1,006 Å2, and includes interactions from CDR1–CDR3 from the heavy and light chains (Figure 6F). The Ab16 and Ab17 epitopes are also left unmasked in the Trimerhg and Cocktailhg cohort boosting immunogens (Figure 1A), allowing immune focusing to conserved broadly neutralizing epitopes and the SARS-2 RBM.
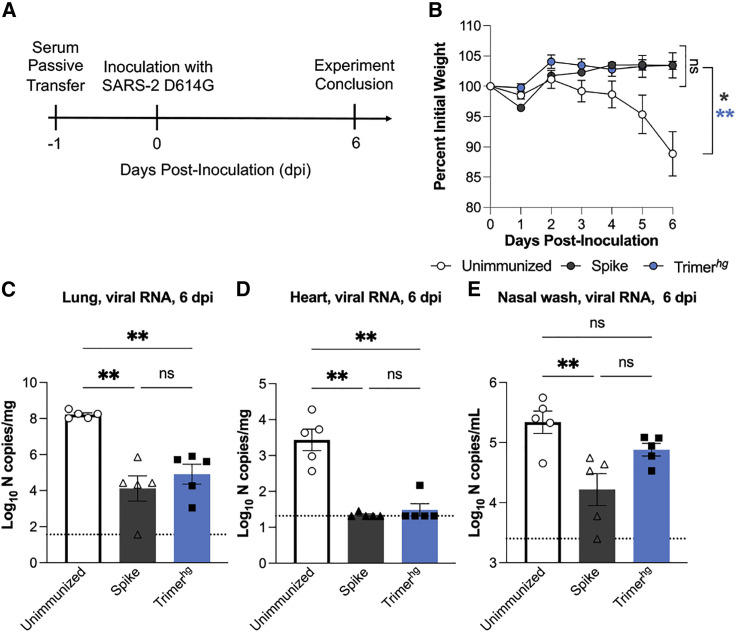
A SARS-2 RBM-focused serum response protects against SARS-2 infection
Given that an RBM-focused immune response appears to be potently cross-neutralizing, we evaluated whether refocusing the immune response toward the SARS-2 RBM following imprinting with the SARS-2 spike was not inferior to maintaining an immune response directed toward the full SARS-2 spike. We compared protection against infection in K18-hACE2 transgenic mice with a SARS-2 virus containing the D614G mutation following passive transfer of sera collected on day 35 from the Trimerhg cohort (Figure 7A; Winkler et al., 2020). For comparison, we used sera from mice that received SARS-2 spike protein (“Spike” cohort) three times, as well as unimmunized control mice (“Unimmunized” cohort). In comparison with the Unimmunized cohort, the Trimerhg and Spike immunization regimens conferred significant protection against weight loss (Figure 7B). We also compared the viral burden by analyzing viral RNA levels in the lungs, heart, and nasal washes 6 days after infection. In the lungs and heart, the Spike and Trimerhg sera conferred significant protection relative to the Unimmunized cohort (Figures 7C and 7D). In nasal washes, there was a significant reduction in viral burden in mice that received Spike sera relative to the Unimmunized cohort, but the difference among the mice that received the Trimerhg sera only trended toward significance (Figure 7E). Across all metrics of protection, there was not a significant difference between Spike sera and Trimerhg sera (Figures 7B–7E). These data suggest that the Trimerhg sera are relatively equivalent to the Spike sera in terms of protection conferred against severe SARS-2 infection and disease in mice and that refocusing the serum immune response toward the RBM may confer potent cross-neutralization of sarbecoviruses without compromising protection against SARS-2.
Figure 7.
SARS-2 protection studies in K18-hACE2 transgenic mice
(A) Schematic showing the passive transfer and SARS-2 D614G live virus challenge timeline.
(B) Following inoculation with SARS-2 D614G, each mouse was weighed daily. There were 5 mice in each of the 3 cohorts. The mean + SE for the three cohorts is shown at each time point. Cohorts were compared using an ordinary one-way ANOVA with Dunnett’s test of the area under the curve (∗p < 0.05, ∗∗p < 0.01).
(C–E) 6 days after inoculation, tissues were harvested, and viral RNA levels in the (C) lungs, (D) heart, and (E) nasal washes were assessed by qRT-PCR RNA. Cohorts were compared using a Kruskal-Wallis test with post hoc analysis using Dunn’s test corrected for multiple comparisons (∗p < 0.05, ∗∗p < 0.01).
Discussion
Our understanding of the durability of vaccine- or infection-elicited Ab responses to SARS-2 continues to evolve, but studies suggest that protection may wane over time (Brown et al., 2021; Nanduri et al., 2021; Pouwels et al., 2021; Rosenberg et al., 2021; Thomas et al., 2021). In comparison, data from seasonal coronaviruses, as well as SARS-1 and MERS, suggest that immunity wanes after several years and can vary in potency between individuals (Callow et al., 1990; Drosten et al., 2014; Hendley et al., 1972; Monto and Lim, 1974; Sariol and Perlman, 2020; Schmidt et al., 1986; Wu et al., 2007). Thus, if herd immunity is not achieved and/or antigenic drift of SARS-2 necessitates reformulation of current vaccines, then this may present an opportunity to “manage” immunity so that SARS-2 protection and broad sarbecovirus cross-neutralization are achieved.
Currently approved vaccines, particularly mRNA-based vaccines, provide some protection against SARS-2 variants of concern; however, it is unclear whether they will provide protection against emerging sarbecoviruses because some data show a substantial reduction in neutralization by vaccine-induced sera (Abu-Raddad et al., 2021; Collier et al., 2021; Edara et al., 2021; Shapiro et al., 2021; Wu et al., 2021). Although protein- and mRNA-based immunization strategies elicit SARS-2 and broad sarbecovirus immunity (Cohen et al., 2021; Martinez et al., 2021; Walls et al., 2021), it remains unknown whether these vaccine candidates can successfully focus immunity toward conserved cross-neutralizing epitopes in the context of SARS-2 spike imprinting. In contrast, our results suggest that, following initial SARS-2 exposure (e.g., vaccination or infection), subsequent boosting with immunogens engineered to focus to conserved epitopes could induce broad sarbecovirus immunity. Based on the data presented here, it would not occur at the expense of neutralizing activity against SARS-2. Although the RBM-focused sera and the non-RBM focused sera showed reduced neutralization potency against SARS-2 variants P.1 and B.1.351, this effect was ameliorated somewhat relative to sera from the control Trimer cohort.
In addition to its implications for shaping SARS-2 immunity following spike-based imprinting, our work also has implications for immunogen design more broadly. Monomeric as well as numerous SARS-2 multimerized RBD-based vaccine constructs have been published recently and are in various stages of preclinical and clinical testing (Cohen et al., 2021; Dai et al., 2020; Huang et al., 2020; Kang et al., 2021; Li et al., 2021; Liu et al., 2020b; Ma et al., 2020; Shinnakasu et al., 2021; Walls et al., 2020; Wang et al., 2021a; Yang et al., 2020). However, these multimerization platforms can give rise to a scaffold-specific Ab response, which has the potential to alter patterns of immune response. We saw no enhanced immunogenicity or immune focusing to the SARS-2 RBM when boosting with SARS-2hg-ferritin nanoparticles (Nanoparticle cohort) compared with the Trimerhg cohort. Sera from the Nanoparticle cohort demonstrated a reduced ability to neutralize related sarbecoviruses and SARS-2 variants of concern, suggesting that the ferritin nanoparticle-directed serum response may alter the SARS-2-directed Ab response. Our engineered hyperglycosylated, cysteine-stabilized GCN4 tag improves a previous hyperglycosylated version of the tag that already showed markedly reduced immunogenicity (Sliepen et al., 2015). In immunization regimens that aim to immune focus, multimerizing immunogens with non-immunogenic scaffolds may confer the benefits of increased avidity while contributing minimal additional epitopes that detrimentally alter patterns of immunodominance.
Our study also shows how to use structure-guided hyperglycosylation and resurfacing to modulate the immune response. We showed how the former strategy used in the Trimerhg and Cocktailhg cohorts could direct responses to the SARS-2 RBM. However, our findings suggest that shielding provided by engineered glycans may be imperfect. Comparing serum titers against RBMhg and SARS-2 RBDs, the Cocktailhg cohort appears to have increased RBM focusing relative to the Trimerhg cohort. This possibly suggests that boosting with rsSARS-1hg and rsWIV1hg, rather than SARS-2hg, results in increased focusing to the RBM. In other words, altering the epitopes presented in the hyperglycosylated, non-RBM portion of the RBD likely contributes to this effect because hyperglycosylation may not completely sterically occlude epitopes. Similarly, the RBMhg did not fully abrogate binding to some RBM-directed Abs, those that likely approach from angles different from B38 or ACE2—both used to characterize our RBMhg RBD construct. The observed differential binding to the RBMhg and SARS-2 RBD is only an approximation of the total RBM-directed Ab response. Nevertheless, our findings provide a framework on which future immunogen engineering efforts can build. These tools are broadly applicable to SARS-2 as well as other viruses, including other coronaviruses. Our results also suggest that leveraging multiple immunogen engineering approaches in combination can contribute to improved immune focusing.
Conclusions
Our results demonstrate immunogen design approaches that can be leveraged to refocus Ab responses following SARS-2 spike imprinting. These design strategies are not limited to coronaviruses and are adaptable to other viruses as a general approach to elicit protective responses to conserved epitopes. Refocusing to the SARS-2 RBM maintains protective SARS-2 neutralization while eliciting Ab responses that recognize emerging variants and potently neutralize coronaviruses with pandemic potential.
Limitations of the study
Achieving pan-variant and pan-sarbecovirus immunity in the context of pre-existing immunity to SARS-2 spike will be necessary; rationally designed immunogens that enable immune focusing to conserved sites is one possibility. Although our approach focused on RBD-based immunogens, other conserved regions within the N-terminal domain or elsewhere on spike may contribute to broad sarbecovirus immunity. SARS-2 pseudovirus neutralization assays may underestimate the contributions of Abs that target epitopes outside of the RBD, although it remains unclear to what extent this occurs when measuring a polyclonal serum response against other coronaviruses (Chen et al., 2021; Chi et al., 2020; Suryadevara et al., 2021). Still, the most potently neutralizing monoclonal Abs in humans elicited by natural infections or mRNA vaccines appear to target the RBD, emphasizing the importance of shaping the RBD-directed immune response with any potential future boosting immunizations (Greaney et al., 2021; Hansen et al., 2020; Jones et al., 2021; Rappazzo et al., 2021).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CR3022 | Produced in house (Yuan et al., 2020) | N/A |
| S309 | Produced in house (Pinto et al., 2020 | N/A |
| P2B-2F6 | Produced in house (Ju et al., 2020 | N/A |
| B38 | Produced in house (Wu et al., 2020a, 2020b) | N/A |
| ADI-56046 | Produced in house (Wec et al., 2020 | N/A |
| ADI-55689 | Produced in house (Wec et al., 2020) | N/A |
| ADI-55688 | Produced in house (Wec et al., 2020 | N/A |
| Ab15 | This paper | N/A |
| Ab16 | This paper | N/A |
| Ab17 | This paper | N/A |
| Ab19 | This paper | N/A |
| Ab20 | This paper | N/A |
| HRP-conjugated rabbit anti-mouse IgG antibody | Abcam | CAT#ab97046 |
| HRP-conjugated goat anti-mouse IgG, human/bovine/horse SP ads antibody | Southern Biotech | CAT#1013-05 |
| CD3-BV786 | BioLegend | CAT#100232 |
| CD19-BV421 | BioLegend | CAT#115549 |
| IgM-BV605 | BioLegend | CAT#406523 |
| IgG-PerCP/Cy5.5 | BioLegend | CAT#405314 |
| Bacterial and virus strains | ||
| Sarbecovirus pseudotyped viruses | Produced in house (Garcia-Beltran et al., 2021a, 2021b) | N/A |
| WA1/2020 SARS-CoV-2 strain with a D614G mutation | (Chen et al., 2021) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Monomeric and trimeric SARS-2, SARS-1, and WIV1 receptor binding domains | This paper | N/A |
| Monomeric SHC014 and RaTG13 receptor binding domains | This paper | N/A |
| SARS-2 two-proline stabilized spike ectodomain | Produced in house (Wrapp et al., 2020) | N/A |
| SARS-2hg monomer and trimer | This paper | N/A |
| rsSARS-1hg monomer and trimer | This paper | N/A |
| rsWIV1hg monomer and trimer | This paper | N/A |
| RBMhg monomer and trimer | This paper | N/A |
| Trimeric hemagglutinin head | This paper | N/A |
| Pierce HRV 3C protease | ThermoScientific | CAT#88946 |
| Sigma Adjuvant System | SigmaAldrich | CAT#S6322 |
| Streptavidin-PE conjugate | Invitrogen | CAT#12-4317-87 |
| Aqua Live/Dead amine-reactive dye | ThermoFisher | CAT#L34957 |
| Calcein, AM, cell-permeant dye | ThermoFisher | CAT#C3100MP |
| Streptavidin-APC/Cy7 conjugate | BioLegend | CAT#405208 |
| Streptavidin-BV650 conjugate | BioLegend | CAT#405232 |
| StrepTactin-PE conjugate | IBA Lifesciences | Item#6-5000-001 |
| StrepTactin-APC conjugate | IBA Lifesciences | Item#6-5010-001 |
| RNaseOUT | ThermoFisher | CAT#10777019 |
| TALON Metal Affinity Resin | Takara | CAT#635652 |
| Pierce Protein G Agarose | ThermoFisher | CAT#20399 |
| 1-Step ABTS substrate | ThermoFisher | Prod#37615 |
| Critical commercial assays | ||
| FAB2G Biosensors | ForteBio | Item#18-5125 |
| Ni-NTA Biosensors | ForteBio | Item#18-5101 |
| SuperScript IV VILO MasterMix | ThermoFisher | CAT#11756050 |
| Index crystal screen | Hampton Research | Item#HR2-144 |
| MagMax mirVana Total RNA isolation kit | ThermoFisher | CAT#A27828 |
| Deposited data | ||
| Atomic coordinates, Ab17 and SARS-2 RBD complex | Protein Data Bank | PDB 7TE1 |
| Electron density map, Ab16 and SARS-2 spike complex | Electron Microscopy Data Bank | EMD-24894 |
| Electron density map, Ab20 and SARS-2 spike complex | Electron Microscopy Data Bank | EMD-24895 |
| Ab15 variable heavy chain | Genbank | OM407408 |
| Ab16 variable heavy chain | Genbank | OM407409 |
| Ab17 variable heavy chain | Genbank | OM407410 |
| Ab19 variable heavy chain | Genbank | OM407411 |
| Ab20 variable heavy chain | Genbank | OM407412 |
| Ab15 variable light chain | Genbank | OM407413 |
| Ab16 variable light chain | Genbank | OM407414 |
| Ab17 variable light chain | Genbank | OM407415 |
| Ab19 variable light chain | Genbank | OM407416 |
| Ab20 variable light chain | Genbank | OM407417 |
| B cell receptor sequences | Genbank | See Table S4 for accession numbers |
| Experimental models: Cell lines | ||
| Human: FreeStyle 293F | Thermo Fisher | Cat#R79007; RRID: CVCL_D603 |
| Human: Expi293F | Thermo Fisher | Cat#A14527; RRID: CVCL_D615 |
| Human: HEK 293T-humanACE2 | (Moore et al., 2004) | |
| Human: HEK293S GnTI-/- | ATCC | ATCC CRL-3022 |
| Experimental models: Organisms/strains | ||
| C57BL/6 mice (strain: C57BL/6NCrl) | Charles River Laboratories | Strain code: 027 |
| K18-hACE2 C57BL/6J mice (strain: 2B6.Cg-Tg(K18-ACE2)2Prlmn/J) | Jackson Laboratory | Strain #034860 |
| Oligonucleotides | ||
| Murine B cell receptor sequencing primers | (Rohatgi et al., 2008; Tiller et al., 2009) | See Table S3 for oligo sequences |
| Software and algorithms | ||
| FlowJo v10 | TreeStar | https://www.flowjo.com; RRID: SCR_008520 |
| Prism v9 | GraphPad | https://www.graphpad.com; RRID: SCR_002798 |
| IMGT | International ImMunoGeneTics Information System | http://www.imgt.org; RRID: SCR_012780 |
| Cloanalyst | (Kepler et al., 2014) | https://www.bu.edu/computationalimmunology/research/software/ |
| RELION | (Scheres, 2012) | https://relion.readthedocs.io/en/release-3.1/ |
| Chimera | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera/ |
| PHASER | (McCoy et al., 2007) | http://www.phaser.cimr.cam.ac.uk/index.php/Phaser_Crystallographic_Software |
| PHENIX | (Adams et al., 2010) | https://www.phenix-online.org/ |
| COOT | (Emsley and Cowtan, 2004) | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PyMol | Schrödinger | http://www.pymol.org |
| MolProbity | (Chen et al., 2010) | http://molprobity.biochem.duke.edu |
| XDS | (Kabsch, 2010a, 2010b) | https://xds.mr.mpg.de |
| Other | ||
| Superdex 200 Increase 10/300 GL | GE Healthcare | Cytiva 28-9909-44 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Aaron G. Schmidt (aschmidt@crystal.harvard.edu).
Materials availability
All unique/stable reagents generated in this study will be made available on request, but we may require a payment and/or a completed materials transfer agreement if there is potential for commercial application. For non-commercial use, all unique/stable reagents generated in this study are available from the lead contact with a completed materials transfer agreement.
Experimental model and subject details
Mice
All immunizations were performed using female C57BL/6 mice (Charles River Laboratories, strain 027) aged 6-10 weeks. Immunization experiments were conducted with institutional IACUC approval (MGH protocol 2014N000252). Protection experiments were performed in K18-hACE2 C57BL/6J mice (strain: 2B6.Cg-Tg(K18-ACE2)2Prlmn/J) were obtained from Jackson Laboratory (034860).
Animal protection studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381–01).
Cell lines
FreeStyle 293F cells (Thermo Fisher Cat#R79007; RRID: CVCL_D603), Expi293F cells (Thermo Fisher Cat#A14527; RRID: CVCL_D615), and HEK293S GnTI-/- cells (ATCC CRL-3022) were cultured in accordance with the manufacturer's instructions. Human ACE2 expressing HEK 293T cells (Moore et al., 2004) were a gift from Nir Hacohen and Michael Farzan and were cultured in Dulbecco's Modified Eagle Medium (ThermoFisher) with 2% fetal bovine serum (Peak Serum FBS) and 1% penicillin-streptomycin at 10,000 U/mL (Gibco).
Method details
Immunogen and coating protein expression and purification
The SARS-CoV-2 (Genbank MN975262.1), SARS-CoV (Genbank ABD72970.1), WIV1-CoV (Genbank AGZ48828.1) RBDs were used as the basis for constructing these immunogens. To graft the SARS-2 RBM onto SARS-1 and WIV1 scaffolds to create the rsSARS-1 and rsWIV1 monomers, boundaries of SARS-2 residues 437– – 507 were used. All constructs were codon optimized by Integrated DNA Technologies and purchased as gblocks. Gblocks were then cloned into pVRC and sequence confirmed via Genewiz. Monomeric constructs for serum ELISA coating contained C-terminal HRV 3C-cleavable 8xHis and SBP tags. Trimeric constructs also included C-terminal HRV 3C-cleavable 8xHis tags, in addition to a hyperglycosylated GCN4 tag with two engineered C-terminal cystines modified from a previously published hyperglycosylated GCN4 tag (Sliepen et al., 2015). Dr. Jason McLellan at the University of Texas, Austin provided the spike plasmid, which contained a non-cleavable foldon trimerization domain in addition to C-terminal HRV 3C cleavable 6xHis and 2xStrep II tags (Wrapp et al., 2020).
Expi 293F cells (ThermoFisher) were used to express proteins. Transfections were performed with Expifectamine reagents per the manufacturer’s protocol. After 5-7 days, transfections were harvested and centrifuged for clarification. Cobalt-TALON resin (Takara) was used to perform immobilized metal affinity chromatography via the 8xHis tag. Proteins were eluted using imidazole, concentrated, and passed over a Superdex 200 Increase 10/300 GL (GE Healthcare) size exclusion column. Size exclusion chromatography was performed in PBS (Corning). For immunogens, HRV 3C protease (ThermoScientific) cleavage of affinity tags was performed prior to immunization. Cobalt-TALON resin was used for a repurification to remove the His-tagged HRV 3C protease, cleaved tag, and remaining uncleaved protein.
Fab and IgG expression and purification
The variable heavy and light chain genes for each antibody were codon optimized by Integrated DNA Technologies, purchased as gblocks, and cloned into pVRC constructs which already contained the appropriate constant domains as previously described (Schmidt et al., 2015a, 2015b). The Fab heavy chain vector contained a HRV 3C-cleavable 8xHis tag, and the IgG heavy chain vector contained HRV 3C-cleavable 8xHis and SBP tags. The same transfection and purification protocol as used for the immunogens and coating proteins was used for the Fabs and IgGs.
Biolayer interferometry
Biolayer interferometry (BLI) experiments were performed using a BLItz instrument (Fortebio) with FAB2G biosensors or Ni-NTA biosensors (Fortebio). All proteins were diluted in PBS. Fabs were immobilized to the biosensors, and coronavirus proteins were used as the analytes. To determine binding affinities, single-hit measurements were performed starting at 10 μM to calculate an approximate K D in order to evaluate which concentrations should be used for subsequent titrations. Measurements at a minimum of three additional concentrations were performed. Vendor-supplied software was used to generate a final K D estimate via a global fit model with a 1:1 binding isotherm.
Immunizations
All immunizations were performed using female C57BL/6 mice (Charles River Laboratories) aged 6-10 weeks. Mice received 20 μg of protein adjuvanted with 50% w/v Sigma adjuvant in 100 μL of inoculum via the intraperitoneal route. Following an initial prime (day −21), boosts occurred at days 0 and 21. Serum samples were collected for characterization on day 35 from all cohorts. All experiments were conducted with institutional IACUC approval (MGH protocol 2014N000252).
Serum ELISAs
Serum ELISAs were executed using 96-well, clear, flat-bottom, high bind microplates (Corning). These plates were coated with 100 μL of protein, which were adjusted to a concentration of 5 μg/mL (in PBS). Plates were incubated overnight at 4°C. After incubation, plates had their coating solution removed and were blocked using 1% BSA in PBS with 1% Tween. This was done for 60 minutes at room temperature. This blocking solution was removed, and sera was diluted 40-fold in PBS. A 5-fold serial dilution was then performed. CR3022 IgG, similarly serially diluted (5-fold) from a 5 μg/mL starting concentration, was used as a positive control. 40 μL of primary antibody solution was used per well. Following this, samples were incubated for 90 minutes at room temperature. Plates were washed three times using PBS-Tween. 150 μL of HRP-conjugated rabbit anti-mouse IgG antibody, sourced commercially from Abcam (at a 1:20,000 dilution in PBS), was used for the secondary incubation. Secondary incubation was performed for one hour, similarly at room temperature. Plates were subsequently washed three times using PBS-Tween. 1xABTS development solution (ThermoFisher) was used according to the manufacturer’s protocol. Development was abrogated after 30 minutes using a 1% SDS solution, and plates were read using a SectraMaxiD3 plate reader (Molecular Devices) for absorbance at 405 nm.
Competition ELISAs
A similar protocol to the serum ELISAs was used for the competition ELISAs. For the primary incubation, 40 μL of the relevant IgG at 1 μM was used at room temperature for 60 minutes. Mouse sera were then spiked in such that the final concentration of sera fell within the linear range for the serum ELISA titration curve for the respective coating antigen, and an additional 60 minutes of room temperature incubation occurred. After removing the primary solution, plates were washed three times with PBS-Tween. Secondary incubation consisted of HRP-conjugated goat anti-mouse IgG, human/bovine/horse SP ads antibody (Southern Biotech) at a concentration of 1:4000. The remaining ELISA procedure (secondary incubation, washing, developing) occurred as described for the serum ELISAs. Percent binding loss was calculated relative to a no IgG control. Negative percent binding loss values were set to zero for the purpose of visualizations.
ACE2 cell binding assay
ACE2 expressing 293T cells (Moore et al., 2004) (a kind gift from Nir Hacohen and Michael Farzan) were harvested. A wash was performed using PBS supplemented with 2% FBS. 200,000 cells were allocated to each labelling condition. Primary incubation occurred using 100 μL of 1 μM antigen in PBS on ice for 60 minutes. Two washes were performed with PBS supplemented with 2% FBS. Secondary incubation was performed using 50 μL of 1:200 streptavidin-PE (Invitrogen) on ice for 30 mins. Two washes were performed with PBS supplemented with 2% FBS, and then cells were resuspended in 100 μL of PBS supplemented with 2% FBS. A Stratedigm S1000Exi Flow Cytometer was used to perform flow cytometry. FlowJo (version 10) was used to analyze FCS files.
Pseudovirus neutralization assay
Serum neutralization against SARS-CoV-2, SARS-CoV, WIV1-CoV, RaTG13, and SHC014 was assayed using pseudotyped lentiviral particles expressing spike proteins described previously (Garcia-Beltran et al., 2021a). Transient transfection of 293T cells was used to generate lentiviral particles. Viral supernatant titers were measured using flow cytometry of 293T-ACE2 cells (Moore et al., 2004) and utilizing the HIV-1 p24CA antigen capture assay (Leidos Biomedical Research, Inc.). 384-well plates (Grenier) were used to perform assays on a Tecan Fluent Automated Workstation. For mouse sera, samples underwent primary dilutions of 1:3 or 1:9 followed by serial 3-fold dilutions. 20 μL each of sera and pseudovirus (125 infectious units) were loaded into each well. Plates were then incubated for 1 hour at room temperature. Following incubation, 10,000 293T-ACE2 cells (Moore et al., 2004) in 20 μL of media containing 15 μg/mL polybrene was introduced to each well. The plates were then further incubated at 37°C for 60–72 hours.
Cells were lysed using assay buffers described previously (Siebring-van Olst et al., 2013). Luciferase expression was quantified using a Spectramax L luminometer (Molecular Devices). Neutralization percentage for each concentration of serum was calculated by deducting background luminescence from cells-only sample wells and subsequently dividing by the luminescence of wells containing both virus and cells. Nonlinear regressions were fitted to the data using GraphPad Prism (version 9), allowing IC50 values to be calculated via the interpolated 50% inhibitory concentration. IC50 values were calculated with a neutralization values greater than or equal to 80% at maximum serum concentration for each sample. NT50 values were then calculated using the reciprocal of IC50 values. Serum neutralization potency values were calculated by dividing the NT50 against a particular pseudovirus by the endpoint titer against the respective RBD. For samples with NT50 values below the limit of detection, the lowest limit of detection across all neutralization assays was used as the NT50 value to calculate neutralization potency. This prevents a higher limit of detection from skewing neutralization potency results. Endpoint titers were normalized relative to a CR3022 IgG control, which was run in every serum ELISA. ELISA titers that were too low to calculate an endpoint titer were set to 40, which was the starting point for the serum dilutions.
In comparing NT50 values for the various cohorts across the wild-type and variant pseudoviruses, the lowest limit of detection across all neutralization assays performed for a given cohort was used for any NT50 values that fell below the limit of detection. This prevents a higher limit of detection in some assays from skewing the comparison results.
Flow cytometry
Single cell suspensions were generated from mouse spleens following isolation via straining through a 70 μm cell strainer. Treatment with ACK lysis buffer was performed to remove red blood cells, and cells were washed with PBS. Aqua Live/Dead amine-reactive dye (0.025 mg/mL) was first used to stain single cell suspensions. The following B and T cell staining panel of mouse-specific antibodies was then applied: CD3-BV786 (BioLegend), CD19-BV421 (BioLegend), IgM-BV605 (BioLegend), IgG-PerCP/Cy5.5 (BioLegend). Staining was performed using a previously described staining approach (Sangesland et al., 2019; Weaver et al., 2016).
SBP-tagged coronavirus proteins were labelled using streptavidin-conjugated flurophores as previously described (Kaneko et al., 2020). Briefly, a final conjugated probe concentration of 0.1 μg/mL was achieved following the addition of streptavidin conjugates to achieve a final molar ratio of probe to streptavidin valency of 1:1. This addition was performed in 5 increments with 20 minutes of incubation at 4°C with rotation in between. The coronavirus protein panel consisted of the following flurorescent probes: SARS-CoV-2 RBD-APC/Cy7 (streptavidin-APC/Cy7 from BioLegend), WIV1 RBD-BV650 (streptavidin-BV650 from BioLegend), SARS-CoV-2 spike-StreptTactin PE (StrepTactin PE from IBA Lifesciences), and SARS-CoV-2 spike-StreptTactin APC (StrepTactin APC from IBA Lifesciences).
A BD FACSAria Fusion cytometer (BD Biosciences) was used to perform flow cytometry. FlowJo (version 10) was used to analyze the resultant FCS files. Sorted cells were IgG+ B cells that were double-positive for SARS-CoV-2 spike and positive for the SARS-CoV-2 RBD.
B cell receptor sequencing
Cells were sorted into 96-well plates containing 4 μL of lysis buffer, consisting of 0.5X PBS, 10 mM DTT, and 4 units of RNaseOUT (ThermoFisher). Following sorting, plates were spun down at 3000 g for 1 minute and stored at −80°C. Plates were later thawed and a reverse transcriptase reaction was performed using the SuperScript IV VILO MasterMix (ThermoFisher) in a total volume of 20 μL according to the manufacturer's recommendations. Two rounds of PCR were then performed using previously published primers (Rohatgi et al., 2008; Tiller et al., 2009). Variable heavy and light chains were then sequenced via Sanger sequencing (Genewiz).
IMGT High V-Quest was used to analyze variable heavy and light chain sequences, and Cloanalyst was used to identify clonal lineages and to infer common ancestors in order to generate phylogenetic trees (Kepler et al., 2014). Data were plotted using Python and FigTree.
Cryo-EM grid preparation and image recording
Complexes of SARS-CoV-2 spike (6P) with Ab16 Fab or Ab20 Fab were formed by combining spike at 0.7 mg/mL with Fab at 0.6 mg/mL (three-fold excess of binding sites) in a buffer composed of 10 mM Tris pH 7.5 with 150 mM NaCl (Hsieh et al., 2020). Spike·Fab complexes were incubated for 30 minutes on ice before application to thick C-flat 1.2-1.3 400 Cu mesh grids (Protochips). Grids were glow discharged (PELCO easiGlow) for 30 seconds at 15 mA and prepared with a Gatan Cryoplunge 3 by applying 3.8 uL of sample and blotting for 4.0 seconds in the chamber maintained at a humidity between 88% and 92%. Images for Spike complexes with Ab16 or Ab20 were recorded on a Talos Arctica microscope operated at 200 keV with a Gatan K3 direct electron detector. Automated image acquisition was performed with Serial EM (Mastronarde, 2005).
Cryo-EM image analysis and 3D reconstruction and model fitting
Image analysis for was carried out in RELION as previously (Tong et al., 2021). Briefly, particles were extracted from motion-corrected micrographs and subjected to 2D classification, initial 3D model generation, 3D classification, and 3D refinement. Ab16 was C3 symmetric. CTF refinement was performed to correct beam tilt, trefoil, anisotropic magnification, and per particle defocus in RELION (Scheres, 2012). Bayesian polishing was also performed in RELION leading to a 6.6 Å reconstruction following 3D refinement. The final 3D refined map was sharpened with a B-factor of −297.5 Å2 resulting in a 5.5 Å resolution map as determined by the Fourier shell correlation (0.143 cutoff) (Figure S7A). Heavy and light chains of PDB entries 6LHQ and 4HC1 were aligned and extracted to make an initial model for the Fab. Spike with 3 RBD in the “up” conformation (PDB 7DX9) and model of Ab16 Fab were docked into the cryoEM map using Chimera (Figures 6B–6D).
Micrographs from Ab20 in complex with spike were processed as above for Ab16. Most particles exhibited C1 symmetry due to conformational heterotgeneity of the RBD relative to the S2 core of spike. Accordingly, particles with C3 symmetry were isolated by 3D classification for further refinement. CTF refinement was performed to correct beam tilt, trefoil, anisotropic magnification, and per particle defocus in RELION (Scheres, 2012). Bayesian polishing was also performed in RELION leading to a 10 Å reconstruction following 3D refinement. The final 3D refined map was sharpened with a B-factor of −768 Å2 resulting in a 9.2 Å resolution map as determined by the Fourier shell correlation (0.143 cutoff) (Figure S7B). Heavy and light chains of PDB entries 4L5F and 4HC1 were aligned and extracted to make an initial model for the Fab. Spike with 3 RBD in the “down” conformation (PDB 6VXX) and model of Ab20 Fab were docked into the cryoEM map using Chimera (Pettersen et al., 2004) (Figure 6A).
The final reconstructions for Ab16 (EMD-24894) and Ab20 (EMD-24895) in complex with SARS-CoV-2 spike were deposited in the Electron Microscopy Data Bank.
Crystallization
Ab17 and the SARS-2 RBD were incubated in a 1:1.2 molar ratio for 2 hours at 4°C. The resulting 1:1 complex was purified from excess SARS-2 RBD by gel filtration chromatography using a Superdex 200 Increase 10/300 GL (GE Healthcare) size exclusion column in 10 mM Tris-HCl, 150 mM NaCl, pH 7.5. The Ab17:SARS-2 RBD complex was concentrated to ∼13 mg/mL. Crystals grew in a hanging drop over a reservoir of 0.1 M HEPES pH 7.0% and 30% v/v Jeffamine® ED-2001 pH 7.0 (Index screen condition D3, Hampton Research). Crystals were harvested, cryoprotected with additional crystallization buffer supplemented with MPD, and flash cooled using liquid nitrogen.
Structure determination and refinement
Diffraction data were recorded at beamline 24-ID-E at the Advanced Photon source. Data were processed using XDS via the RAPD pipeline (Collaborative Computational Project, 1994; Evans, 2006; Evans, 2011; Evans and Murshudov, 2013; Kabsch, 2010a, b). Molecular replacement was performed using PHASER (McCoy et al., 2007). Refinement was performed using PHENIX, and model modifications were performed using COOT (Adams et al., 2010; Emsley and Cowtan, 2004; Terwilliger et al., 2008). For the search model, a VHVL comprised of PDB entries 4L5F and 4HCl, respectively, with the CDRH3, CDRH2, and CDRL3 removed along with the SARS-2 RBD from PDB 6M0J were used. The CDRH3, CDRH2, and CDRL3 were rebuilt de novo along with mutations in the VHVL using COOT, and refinement was performed using PHENIX. Refinement statistics are shown in Table S2. X-ray crystallography data were deposited in the Protein DataBank (7TE1).
Animal protection experiments
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381–01). Virus inoculations were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering.
K18-hACE2 C57BL/6J mice (strain: 2B6.Cg-Tg(K18-ACE2)2Prlmn/J) were obtained from Jackson Laboratory (034860). Mice were administered 200 μL of pooled immune sera via intraperitoneal injection. One day after transfer, mice were inoculated with 103 FFU of WA1/2020 SARS-CoV-2 strain with a D614G mutation via the intranasal route (Chen et al., 2021). Mice were monitored for weight loss, and daily weights were recorded. On day 6 post-inoculation, final weights were obtained, animals were sacrificed, and tissues were harvested.
Measurement of viral burden
Tissues from each mouse were weighed. Homogenization was performed with sterile zirconia beads using a MagNa Lyser instrument (Roche Life Sciences) in 1 mL of DMEM supplemented with 2% heat-inactivated fetal bovine serum. Clarification was performed via centrifugation at 10,000 rpm for 5 min. Samples were stored at −80°C. RNA extraction was performed using the MagMax mirVana Total RNA isolation kit (Thermo Scientific) in combination with a Kingfisher Flex extraction machine (Thermo Scientific). RT-qPCR was then used to determine viral RNA levels as previously described (Hassan et al., 2020). Viral RNA levels were normalized to tissue weight.
Quantification and statistical analysis
Curve fitting and statistical analyses were performed with GraphPad Prism (version 9). Non-parametric statistics were used throughout where feasible. Tests, numbers of animals, and statistical comparison groups are indicated in each of the Figure Legends. To compare multiple populations, the Kruskal-Wallis non-parametric ANOVA was used with post hoc analysis using Dunn's test for multiple comparisons. The Mann-Whitney U test was used to compare two populations without consideration for paired samples. The ratio-paired t-test was used to compare two populations with consideration for paired samples and evidence of normality. Analysis of weight change was determined by one-way ANOVA with Dunnett’s test of area under the curve. p values in ANOVA analyses were corrected for multiple comparisons. A p value <0.05 was considered significant.
Acknowledgments
We thank members of the Schmidt Laboratory for helpful discussions. We thank Catherine Jacob-Dolan for critical reading of the manuscript. We thank Dr. Jason McLellan from the University of Texas, Austin for the spike plasmid. We thank Nir Hacohen and Michael Farzan for the kind gift of the ACE2-expressing 293T cells. We thank the beamline staff at the Advanced Photon Source (APS) NE-CAT 24-ID for support. We acknowledge funding from NIH R01 grants AI146779 (to A.G.S.); AI124378, AI137057, and AI153098 (to D.L.); and AI157155 (to M.S.D.), and a Massachusetts Consortium on Pathogenesis Readiness (MassCPR) grant (to A.G.S.). This work was also supported by training grants NIGMS T32 GM007753 (to B.M.H. and T.M.C.), T32 AI007245 (to J.F.), F31 Al138368 (to M.S.), and F30 AI160908 (to B.M.H.). A.B.B. is supported by National Institutes for Drug Abuse (NIDA) Avenir New Innovator Award DP2DA040254, the MGH Transformative Scholars Program, as well as funding from the Charles H. Hood Foundation. This independent research was supported by the Gilead Sciences Research Scholars Program in HIV (to A.B.B.). J.B.C. is supported by a Helen Hay Whitney Foundation postdoctoral fellowship. This work is based on research conducted at the NE-CAT, which is funded by NIGMS (P30 GM124165). The Eiger 16M detector on 24-ID-E is funded by a NIH-ORIP HEI grant (S10OD021527). This research used resources of the APS, a DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract DE-AC02-06CH11357.
Author contributions
Conceptualization, B.M.H. and A.G.S.; methodology, B.M.H., E.C.L., I.W.W., J.B.C., T.M.C., A.B.B., D.L., and A.G.S.; investigation, B.M.H., M.S., K.J.S.D., E.C.L., J.F., T.K., I.W.W., J.B.C., and T.M.C.; writing – original draft, B.M.H. and A.G.S.; writing – review and editing, all authors; funding acquisition, M.S.D., A.B.B., D.L., and A.G.S.; supervision, M.S.D., A.B.B., D.L., and A.G.S.
Declaration of interests
B.M.H., T.M.C., and A.G.S. have filed a provisional patent for the described immunogens. M.S.D. is a consultant for Inbios, Vir Biotechnology, and Carnival Corporation and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Vir Biotechnology, Moderna, and Emergent BioSolutions.
Published: March 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110561.
Supplemental information
Data and code availability
-
•
CryoEM data have been deposited in the Electron Microscopy Data Base and are available as of the date of publication. X-ray crystallography data have been deposited in the Protein Data Bank (7TE1) and are available as of the date of publication. B cell receptor sequences have been deposited in Genbank. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abu-Raddad L.J., Chemaitelly H., Butt A.A., National Study Group for C.-V. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N. Engl. J. Med. 2021;385:187–189. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic G., Maron M.J., Adachi Y., Onodera T., McCarthy K.R., McGee C.E., Sempowski G.D., Takahashi Y., Kelsoe G., Kuraoka M., et al. Influenza antigen engineering focuses immune responses to a subdominant but broadly protective viral epitope. Cell Host Microbe. 2019;25:827–835.e26. doi: 10.1016/j.chom.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic G., Maron M.J., Caradonna T.M., Tian M., Mermelstein A., Fera D., Kelsoe G., Kuraoka M., Schmidt A.G. Structure-guided molecular grafting of a complex broadly neutralizing viral epitope. ACS Infect. Dis. 2020;6:1182–1191. doi: 10.1021/acsinfecdis.0c00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.M., Vostok J., Johnson H., Burns M., Gharpure R., Sami S., Sabo R.T., Hall N., Foreman A., Schubert P.L., et al. Outbreak of SARS-CoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings - Barnstable County, Massachusetts, July 2021. MMWR Morb. Mortal Wkly. Rep. 2021;70:1059–1062. doi: 10.15585/mmwr.mm7031e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow K.A., Parry H.F., Sergeant M., Tyrrell D.A. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990;105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr66. 2010:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.A., Gnanapragasam P.N.P., Lee Y.E., Hoffman P.R., Ou S., Kakutani L.M., Keeffe J.R., Wu H.J., Howarth M., West A.P., et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371:735–741. doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Collier D.A., De Marco A., Ferreira I., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- Correia B.E., Bates J.T., Loomis R.J., Baneyx G., Carrico C., Jardine J.G., Rupert P., Correnti C., Kalyuzhniy O., Vittal V., et al. Proof of principle for epitope-focused vaccine design. Nature. 2014;507:201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispin M., Ward A.B., Wilson I.A. Structure and immune recognition of the HIV glycan shield. Annu. Rev. Biophys. 2018;47:499–523. doi: 10.1146/annurev-biophys-060414-034156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Zheng T., Xu K., Han Y., Xu L., Huang E., An Y., Cheng Y., Li S., Liu M., et al. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell. 2020;182:722–733.e11. doi: 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de St Groth F., Webster R.G. Disquisitions of original antigenic sin. I. Evidence in man. J. Exp. Med. 1966;124:331–345. doi: 10.1084/jem.124.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Meyer B., Muller M.A., Corman V.M., Al-Masri M., Hossain R., Madani H., Sieberg A., Bosch B.J., Lattwein E., et al. Transmission of MERS-coronavirus in household contacts. N. Engl. J. Med. 2014;371:828–835. doi: 10.1056/NEJMoa1405858. [DOI] [PubMed] [Google Scholar]
- Edara V.V., Pinsky B.A., Suthar M.S., Lai L., Davis-Gardner M.E., Floyd K., Flowers M.W., Wrammert J., Hussaini L., Ciric C.R., et al. Infection and vaccine-induced neutralizing-antibody responses to the SARS-CoV-2 B.1.617 variants. N. Engl. J. Med. 2021;385:664–666. doi: 10.1056/NEJMc2107799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Evans P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., Astudillo M.G., Yang D., Miller T.E., Feldman J., Hauser B.M., Caradonna T.M., Clayton K.L., Nitido A.D., et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184:476–488.e11. doi: 10.1016/j.cell.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Gentles L.E., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Antibodies elicited by mRNA-1273 vaccination bind more broadly to the receptor binding domain than do those from SARS-CoV-2 infection. Sci. Transl. Med. 2021;13:eabi9915. doi: 10.1126/scitranslmed.abi9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753.e44. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause A.M., Baggs J., Gee J., Marquez P., Myers T.R., Shimabukuro T.T., Shay D.K. Safety monitoring of an additional dose of COVID-19 vaccine - United States, august 12-september 19, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1379–1384. doi: 10.15585/mmwr.mm7039e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W.T., Yuan M., Callaghan S., Musharrafieh R., Song G., Silva M., Beutler N., Lee W.H., Yong P., Torres J.L., et al. Broadly Neutralizing Antibodies to SARS-Related Viruses Can Be Readily Induced in Rhesus Macaques. bioRxiv. 2021 doi: 10.1101/2021.07.05.451222. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendley J.O., Fishburne H.B., Gwaltney J.M., Jr. Coronavirus infections in working adults. Eight-year study with 229 E and OC 43. Am. Rev. Respir. Dis. 1972;105:805–811. doi: 10.1164/arrd.1972.105.5.805. [DOI] [PubMed] [Google Scholar]
- Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.C., Zhou S., He X., Chiem K., Mabrouk M.T., Nissly R.H., Bird I.M., Strauss M., Sambhara S., Ortega J., et al. SARS-CoV-2 RBD neutralizing antibody induction is enhanced by particulate vaccination. Adv. Mater. 2020;32:e2005637. doi: 10.1002/adma.202005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K.E., Davenport F.M., Hennessy A.V., Francis T., Jr. Characterization of influenza antibodies by serum absorption. J. Exp. Med. 1956;104:199–209. doi: 10.1084/jem.104.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N., Kuo H.H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J., et al. Loss of bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.F., Sun C., Zhuang Z., Yuan R.Y., Zheng Q., Li J.P., Zhou P.P., Chen X.C., Liu Z., Zhang X., et al. Rapid development of SARS-CoV-2 spike protein receptor-binding domain self-assembled nanoparticle vaccine candidates. ACS Nano. 2021;15:2738–2752. doi: 10.1021/acsnano.0c08379. [DOI] [PubMed] [Google Scholar]
- Kepler T.B., Munshaw S., Wiehe K., Zhang R., Yu J.S., Woods C.W., Denny T.N., Tomaras G.D., Alam S.M., Moody M.A., et al. Reconstructing a B-cell clonal lineage. II. Mutation, selection, and affinity maturation. Front Immunol. 2014;5:170. doi: 10.3389/fimmu.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Guo L., Zheng H., Li J., Zhao X., Li J., Liang Y., Yang F., Zhao Y., Yang J., et al. Self-assembling nanoparticle vaccines displaying the receptor binding domain of SARS-CoV-2 elicit robust protective immune responses in rhesus monkeys. Bioconjug. Chem. 2021;32:1034–1046. doi: 10.1021/acs.bioconjchem.1c00208. [DOI] [PubMed] [Google Scholar]
- Liu H., Wu N.C., Yuan M., Bangaru S., Torres J.L., Caniels T.G., van Schooten J., Zhu X., Lee C.D., Brouwer P.J.M., et al. Cross-neutralization of a SARS-CoV-2 antibody to a functionally conserved site is mediated by avidity. Immunity. 2020;53:1272–1280.e75. doi: 10.1016/j.immuni.2020.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Xu W., Xia S., Gu C., Wang X., Wang Q., Zhou J., Wu Y., Cai X., Qu D., et al. RBD-Fc-based COVID-19 vaccine candidate induces highly potent SARS-CoV-2 neutralizing antibody response. Signal. Transduct. Target. Ther. 2020;5:282. doi: 10.1038/s41392-020-00402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zou F., Yu F., Li R., Yuan Y., Zhang Y., Zhang X., Deng J., Chen T., Song Z., et al. Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity. 2020;53:1315–1330.e19. doi: 10.1016/j.immuni.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D.R., Schaefer A., Leist S.R., De la Cruz G., West A., Atochina-Vasserman E.N., Lindesmith L.C., Pardi N., Parks R., Barr M., et al. Chimeric spike mRNA vaccines protect against sarbecovirus challenge in mice. Science. 2021;373:991–998. doi: 10.1126/science.abi4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D.R., Schafer A., Gobeil S., Li D., De la Cruz G., Parks R., Lu X., Barr M., Stalls V., Janowska K., et al. A broadly cross-reactive antibody neutralizes and protects against sarbecovirus challenge in mice. Sci. Transl. Med. 2022;14:eabj7125. doi: 10.1126/scitranslmed.abj7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F., et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R., et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. U S A. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monto A.S., Lim S.K. The Tecumseh study of respiratory illness. VI. Frequency of and relationship between outbreaks of coronavirus infection. J. Infect. Dis. 1974;129:271–276. doi: 10.1093/infdis/129.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Dorfman T., Li W., Wong S.K., Li Y., Kuhn J.H., Coderre J., Vasilieva N., Han Z., Greenough T.C., et al. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanduri S., Pilishvili T., Derado G., Soe M.M., Dollard P., Wu H., Li Q., Bagchi S., Dubendris H., Link-Gelles R., et al. Effectiveness of pfizer-BioNTech and Moderna vaccines in preventing SARS-CoV-2 infection among nursing home residents before and during widespread circulation of the SARS-CoV-2 B.1.617.2 (delta) variant - National Healthcare Safety Network, March 1-August 1, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1163–1166. doi: 10.15585/mmwr.mm7034e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek G., Guenaga F.J., Schief W.R., Skinner J., Baker D., Wyatt R., Kwong P.D. Elicitation of structure-specific antibodies by epitope scaffolds. Proc. Natl. Acad. Sci. U S A. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C., Buchrieser J., Rajah M.M., Bishop E., et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021;27:917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- Pouwels K.B., Pritchard E., Matthews P.L., Stoesser N., Eyre D.W., Vihta K., House T., Hay J., Bell J.I., Newton J.N., et al. Impact of delta on viral burden and vaccine effectiveness against new SARS-CoV-2 Infections in the UK. medRxiv. 2021 doi: 10.1101/2021.08.18.21262237. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi S., Ganju P., Sehgal D. Systematic design and testing of nested (RT-)PCR primers for specific amplification of mouse rearranged/expressed immunoglobulin variable region genes from small number of B cells. J. Immunol. Methods. 2008;339:205–219. doi: 10.1016/j.jim.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Rosenberg E.S., Holtgrave D.R., Dorabawila V., Conroy M., Greene D., Lutterloh E., Backenson B., Hoefer D., Morne J., Bauer U., et al. New COVID-19 cases and hospitalizations among adults, by vaccination status - New York, May 3-July 25, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1150–1155. doi: 10.15585/mmwr.mm7034e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangesland M., Ronsard L., Kazer S.W., Bals J., Boyoglu-Barnum S., Yousif A.S., Barnes R., Feldman J., Quirindongo-Crespo M., McTamney P.M., et al. Germline-encoded affinity for cognate antigen enables vaccine amplification of a human broadly neutralizing response against influenza virus. Immunity. 2019;51:735–749.e38. doi: 10.1016/j.immuni.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariol A., Perlman S. Lessons for COVID-19 immunity from other coronavirus infections. Immunity. 2020;53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K.O., Lee E., Parks R., Martinez D.R., Li D., Chen H., Edwards R.J., Gobeil S., Barr M., Mansouri K., et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021;594:553–559. doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A.G., Do K.T., McCarthy K.R., Kepler T.B., Liao H.X., Moody M.A., Haynes B.F., Harrison S.C. Immunogenic stimulus for germline precursors of antibodies that engage the influenza hemagglutinin receptor-binding site. Cell Rep. 2015;13:2842–2850. doi: 10.1016/j.celrep.2015.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A.G., Therkelsen M.D., Stewart S., Kepler T.B., Liao H.X., Moody M.A., Haynes B.F., Harrison S.C. Viral receptor-binding site antibodies with diverse germline origins. Cell. 2015;161:1026–1034. doi: 10.1016/j.cell.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O.W., Allan I.D., Cooney M.K., Foy H.M., Fox J.P. Rises in titers of antibody to human coronaviruses OC43 and 229E in Seattle families during 1975-1979. Am. J. Epidemiol. 1986;123:862–868. doi: 10.1093/oxfordjournals.aje.a114315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J., Dean N.E., Madewell Z.J., Yang Y., Halloran M.E., Longini I. Efficacy estimates for various COVID-19 vaccines: What we know from the literature and reports. medRxiv. 2021 doi: 10.1101/2021.05.20.21257461. Preprint at. [DOI] [Google Scholar]
- Shinnakasu R., Sakakibara S., Yamamoto H., Wang P.H., Moriyama S., Sax N., Ono C., Yamanaka A., Adachi Y., Onodera T., et al. Glycan engineering of the SARS-CoV-2 receptor-binding domain elicits cross-neutralizing antibodies for SARS-related viruses. J. Exp. Med. 2021;218:e20211003. doi: 10.1084/jem.20211003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebring-van Olst E., Vermeulen C., de Menezes R.X., Howell M., Smit E.F., van Beusechem V.W. Affordable luciferase reporter assay for cell-based high-throughput screening. J. Biomol. Screen. 2013;18:453–461. doi: 10.1177/1087057112465184. [DOI] [PubMed] [Google Scholar]
- Sliepen K., van Montfort T., Melchers M., Isik G., Sanders R.W. Immunosilencing a highly immunogenic protein trimerization domain. J. Biol. Chem. 2015;290:7436–7442. doi: 10.1074/jbc.M114.620534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Nutalai R., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211.e7. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadevara N., Shrihari S., Gilchuk P., VanBlargan L.A., Binshtein E., Zost S.J., Nargi R.S., Sutton R.E., Winkler E.S., Chen E.C., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331.e15. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Grosse-Kunstleve R.W., Afonine P.V., Moriarty N.W., Zwart P.H., Hung L.W., Read R.J., Adams P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S.J., Moreira E.D., Jr., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Marc G.P., Polack F.P., Zerbini C., et al. Six month safety and efficacy of the BNT162b2 mRNA COVID-19 Vaccine. medRxiv. 2021 doi: 10.1101/2021.07.28.21261159. Preprint at. [DOI] [Google Scholar]
- Tiller T., Busse C.E., Wardemann H. Cloning and expression of murine Ig genes from single B cells. J. Immunol. Methods. 2009;350:183–193. doi: 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Tong P., Gautam A., Windsor I.W., Travers M., Chen Y., Garcia N., Whiteman N.B., McKay L.G.A., Storm N., Malsick L.E., et al. Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike. Cell. 2021;184:4969–4980.e15. doi: 10.1016/j.cell.2021.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Fiala B., Schafer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O'Connor M.A., Chen C., et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382.e17. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Miranda M.C., Schafer A., Pham M.N., Greaney A., Arunachalam P.S., Navarro M.J., Tortorici M.A., Rogers K., O'Connor M.A., et al. Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell. 2021;184:5432–5447.e16. doi: 10.1016/j.cell.2021.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Huang B., Zhu Y., Tan W., Zhu M. Ferritin nanoparticle-based SARS-CoV-2 RBD vaccine induces a persistent antibody response and long-term memory in mice. Cell Mol. Immunol. 2021;18:749–751. doi: 10.1038/s41423-021-00643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver G.C., Villar R.F., Kanekiyo M., Nabel G.J., Mascola J.R., Lingwood D. In vitro reconstitution of B cell receptor-antigen interactions to evaluate potential vaccine candidates. Nat. Protoc. 2016;11:193–213. doi: 10.1038/nprot.2016.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G. Original antigenic sin in ferrets: the response to sequential infections with influenza viruses. J. Immunol. 1966;97:177–183. [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., et al. Serum neutralizing activity elicited by mRNA-1273 vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.P., Wang N.C., Chang Y.H., Tian X.Y., Na D.Y., Zhang L.Y., Zheng L., Lan T., Wang L.F., Liang G.D. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 2007;13:1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.C., Yuan M., Bangaru S., Huang D., Zhu X., Lee C.D., Turner H.L., Peng L., Yang L., Burton D.R., et al. A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody. PLoS Pathog. 2020;16:e1009089. doi: 10.1371/journal.ppat.1009089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y., et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- Yuan M., Huang D., Lee C.D., Wu N.C., Jackson A.M., Zhu X., Liu H., Peng L., van Gils M.J., Sanders R.W., et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373:818–823. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakeri B., Fierer J.O., Celik E., Chittock E.C., Schwarz-Linek U., Moy V.T., Howarth M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U S A. 2012;109:E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of Escape of SARS-CoV-2 variant B.1.351 from natural and vaccine induced sera. Cell. 2021;184:2348–2361. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
CryoEM data have been deposited in the Electron Microscopy Data Base and are available as of the date of publication. X-ray crystallography data have been deposited in the Protein Data Bank (7TE1) and are available as of the date of publication. B cell receptor sequences have been deposited in Genbank. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.