Abstract
Mutations in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike receptor-binding domain (RBD) may alter viral host tropism and affect the activities of neutralizing antibodies. Here, we investigated 153 RBD mutants and 11 globally circulating variants of concern (VOCs) and variants of interest (VOIs) (including Omicron) for their antigenic changes and cross-species tropism in cells expressing 18 ACE2 orthologs. Several RBD mutations strengthened viral infectivity in cells expressing ACE2 orthologs of non-human animals, particularly those less susceptible to the ancestral strain. The mutations surrounding amino acids (aas) 439–448 and aa 484 are more likely to cause neutralization resistance. Strikingly, enhanced cross-species infection potential in the mouse and ferret, instead of the neutralization-escape scores of the mutations, account for the positive correlation with the cumulative prevalence of mutations in humans. These findings present insights for potential drivers of circulating SARS-CoV-2 variants and provide informative parameters for tracking and forecasting spreading mutations.
Keywords: SARS-CoV-2 variants, RBD mutations, cross-species tropism, neutralization resistance, variants of concern, variants of interest, ACE2 receptor
Graphical abstract

Zhang et al. show in vitro cross-species infectivity and neutralization-escape characteristics of 153 SARS-CoV-2 RBD mutants and 11 globally circulating VOCs and VOIs. They reveal an association between enhanced cross-species infection potential and the current cumulative prevalence of mutations, which can inform surveillance and forecasting of SARS-CoV-2 spike mutations.
Introduction
The emerging variants of the pandemic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) bring enormous challenges to public health (Lauring and Hodcroft, 2021). Mutations occurring in viral spike protein, particularly in the receptor-binding domain (RBD), may change the protein conformation and affect the interaction with the host ACE2 receptor. In addition, some RBD mutations may cause the cross-species spillover by introducing the ACE2-binding capacity to non-human species that SARS-CoV-2 was initially insusceptible. Moreover, because the RBD is the primary target of the neutralizing antibody (nAb) response elicited by natural infection or vaccine immunization, RBD mutations may confer nAb resistance and possibly increase the re-infection risk, reducing the effectiveness of vaccines based on the original virus.
Given the widespread and continuous evolution of SARS-CoV-2, multiple variants constantly emerged. So far, five variants of concern (VOCs) have been designated: Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529; World Health Organization, 2022; Centers for Disease Control and Prevention, 2022). Besides, numerous variants of interest (VOIs) were also identified, including the Epsilon (B.1.429), Eta (B.1.525), Iota (B.1.526), Kappa (B.1.617.1), et al. Most VOCs and VOIs harbor RBD mutations, such as the N501Y in the Alpha, Beta, Gamma, and Omicron; the E484K/Q/A in the Beta, Gamma, Kappa, and Omicron; and the T478K in the Delta and Omicron. Before Omicron's emergence, the K417N/E484K/N501Y-harbored Beta variant showed the most noticeable resistance to nAbs acquired from natural infection or vaccine immunization (Garcia-Beltran et al., 2021; Wang et al., 2021; Zhou et al., 2021). The Omicron (B.1.1.529), a recently documented and highly contagious VOC harboring ∼15 RBD mutations, is spreading rapidly worldwide and has become a severe threat due to its more aggressive immune evasion ability (Dejnirattisai et al., 2022; Hoffmann et al., 2021). Several studies have investigated the antigenic influence of representative RBD mutations. However, the fully antigenic landscapes of RBD mutants and their cross-species tropism remain to be explored. Exploring the underlying association between the biological impacts and the circulating prevalence of RBD mutations will be informative for forecasting the variants that may be prevalent in the future.
Based on the analyses of viral sequences deposited at the public SARS-CoV-2 genome database, we generated a panel of lentiviral-pseudotyped particles (LVpp) composed of 129 single-site RBD mutants, 24 double-site RBD (DM) mutants, and 11 VOCs and VOIs, including the newly emerged Omicron. The infection potentials of these variants in cells expressing various ACE2 from 18 animal species were evaluated. In addition, the neutralizing resistance of variants to human convalescent plasmas (HCPs) and vaccine- or vaccine-candidates-elicited antisera and monoclonal antibodies (mAbs) were also investigated. More importantly, the association among the cumulative frequencies of circulating RBD mutations and their cross-species infection potentials and neutralization escape characteristics was systematically analyzed.
Results
Impact of RBD mutations on the function of spike
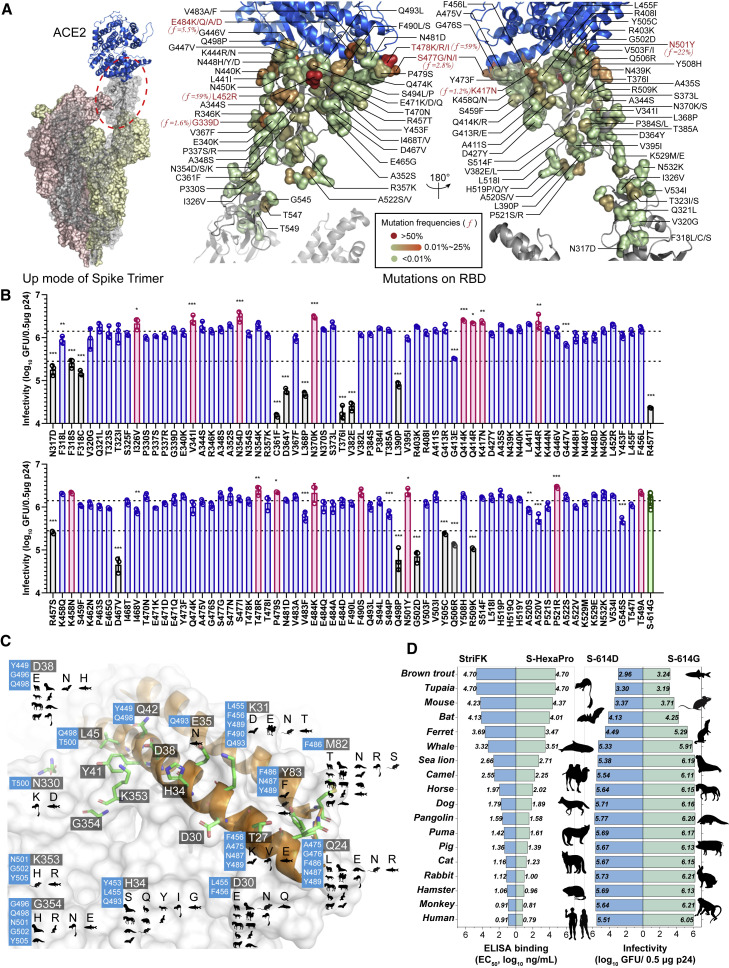
We initially tracked the RBD mutations since August 2020. Based on the analyses of viral spike gene variation annotations from the 2019nCoVR database of China National Center for Bioinformation (CNCB), a total of 129 RBD mutations were selected for further tests. The spatial locations and frequencies of these mutations in the spike/ACE2 complex structure are illustrated in Figure 1A. For LVpp productions, we generated the spike-expressing plasmids with single-site mutations in combination with D614G. The 18-amino-acid (aa) of spike C terminus was replaced with a HiBit bioluminescent tag for antibody-free detection (Figure S1A). Compared with the parental D614G (designated S-614G), we noted 17 mutants (13.2%) showed >50% spike-expressing reduction and 7 (5.4%) had notably decreased spike secretions (Figure S1B). In huACE2-H1299 cells, 17 RBD mutants showed >5-fold decreased infectivity to S-614G (Figure 1B, as indicated by black bars; p < 0.05 for each). In contrast, the I326V, V341I, N354D, N370K, Q414K/R, K417N, K444R, K458N, T478R, P479S, E484K, F490S, N501Y, P521R, and T549A showed a >1.5× increased infectivity (Figure 1B, as indicated by pink bars; p = 0.062 for K458N, p = 0.052 for F490S, p = 0.062 for E484K, p = 0.061 for T549A, and p < 0.05 for others).
Figure 1.
Profiles of RBD mutations and ACE2 orthologs involved in the study
(A) The spatial locations and frequencies of RBD mutations involved in the study in spike/ACE2 complex structure. The left panel shows the complex structure of ACE2 (in blue) and SARS-CoV-2 spike trimer in “up” mode. The spike trimer (PDB: 7KNB) is shown as surface mode, and the ACE2-binding spike monomer is rendered as a transparent surface and the interior cartoon model. The right panel presents zoom-in details of the RBD/ACE2 interface. The selected mutations are labeled and rendered as individual residue surfaces colored according to a diagram mapping the cumulative frequencies of mutations in all viral sequences (n = 6,619,244) in GISAID by January 1, 2021.
(B) Infectivities of LVpp bearing mutated spikes in huACE2-H1299 cells. Data are plotted as the mean ± SD of greater than or equal to three replicates. The p24-normalized (0.5 μg) infectious titer of LVpp is considered as a parameter indicating the infectivity. The upper broken line represents the average level of S-614G, whereas the lower broken line indicates the 5-fold decrease of infectivity of S-614G. Mutants with >1.5× increased infectivity are plotted in pink, and those with >5× decreased infectivity are plotted in black. Statistical significance of each mutant relative to S-614G is shown only when p < 0.05. ∗∗∗, p < 0.001; ∗∗, p < 0.01; ∗, p < 0.05
(C) Variation of ACE2 orthologs of 18 involved species based on human ACE2 structure (PDB: 6M17). RBD is shown as a white surface, and the critical residues of ACE2 for RBD/ACE2 interaction are shown as a sticks model with the two helix domains colored in orange. Critical residues are highlighted by a dark background followed by variations on other species listed by cartoon icons. The blue color boxes show the adjacent RBD residues that form contact. Silhouettes indicating the species were from PhyloPic.org and available under the Public Domain Dedication 1.0 license, with the exception of Physeter catodon (Chris Huh, Creative Commons Attribution-ShareAlike 3.0 Unported).
(D) The binding capabilities of spike proteins to rACE2 proteins of various species (left) and the infectivities of SARS-CoV-2 LVpp in cells expressing ACE2 orthologs (right).
See Figures S5A and S5B for further information.
Broad tropism of SARS-CoV-2 for both terrestrial and marine mammals
A total of 18 ACE2 orthologs from various species were selected to set a panel for assessment in mediating cellular entry of SARS-CoV-2 variants (Table S1). The key variation residues in these orthologs at the RBD/ACE2 binding interface are shown in Figure 1C. The H1299, a human-lung-tumor-derived cell with deficient endogenous ACE2 but that is TMPRSS2 expressing, was used to develop ACE2-reconstituted cells (Ou et al., 2021; Zhang et al., 2021). Exogenous reconstitution of human ACE2 could efficiently render the H1299 cells susceptible to SARS-CoV-2 LVpp (Zhang et al., 2021). Therefore, we generated 18 stable H1299 cell lines with exogenous expression of various ACE2 orthologs. Flow cytometry (fluorescence-activated cell sorting [FACS]) analyses demonstrated all cell lines had a >99% positive rate for integrated mRuby3 expression (Figure S2). The fluorescent spike probe (SARS-CoV-2-STG) (Zhang et al., 2021) showed efficient bindings with 13 of 18 cells, except those transfected with ACE2 of the ferret, horseshoe bat, mouse, brown trout, and tupaia, in either FACS (Figure S2) or imaging assays (Figure S3). Recombinant ACE2-Fc chimeric proteins (rACE2) of these orthologs were produced for biochemical analyses (Figure S4). The ELISA-binding capabilities of the rACE2 proteins to two recombinant SARS-CoV-2 S-ectodomain proteins (non-prefusion-stabilized StriFK and prefusion-stabilized S-HexaPro; Hsieh et al., 2020; Wu et al., 2021) are shown in Figure S5A. The rACE2 proteins of human, rhesus macaque, hamster, rabbit, cat, pig, puma, pangolin, and dog showed a half-maximal effective concentration (EC50) < 100 ng/mL for both S proteins, suggesting potent ACE2-spike bindings (Figure 1D, left panel). The LVpp infection assays demonstrated that most cell lines could efficiently support the ancestral virus entry (Figure 1D, right panel), except those with ACE2 of tupaia, horseshoe bat, ferret, mouse, and brown trout (Figure S5B). The ACE2 of two marine mammals, sea lion and whale, also efficiently rendered cells susceptible to SARS-CoV-2, despite only having moderate-to-low binding to S proteins. Overall, the binding capabilities of rACE2 proteins to S-ectodomain proteins positively correlate with the apparent viral infectivity in H1299 cells expressing the corresponding ACE2 (Figure 1D). An EC50 < 3.5 log10 ng/mL determined by rACE2-spike binding ELISA is likely to be a crucial requirement for efficient SARS-CoV-2 pseudovirus infection.
Impacts of RBD mutations on in vitro cross-species infection potentials
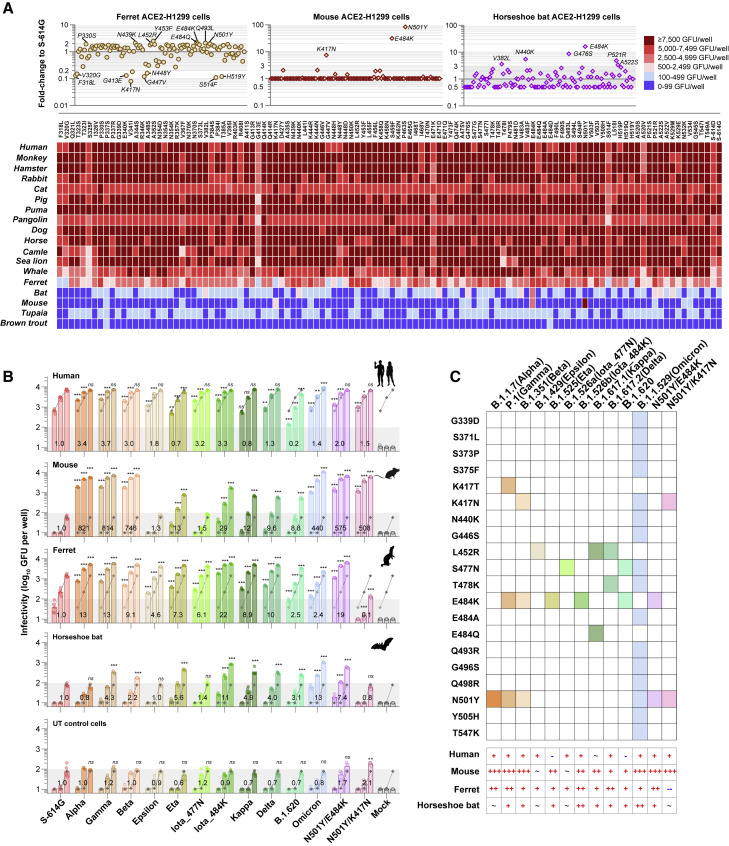
To determine host tropism influence of RBD mutations, we evaluated the susceptibilities of 18 ACE2-expressing H1299 cells to 112 RBD mutants (17 infectivity-decreased variants were excluded). In cells expressing ACE2 orthologs that can support efficient infection of the ancestral virus, most of the mutants preserved comparable infectivity to S-614G (<2-fold changes; Figures 2A and S5C). Of note, the susceptibility of H1299 cells with ferret ACE2 (feACE2) was quite sensitive to RBD mutations (Figure 2A). The P330S, N439K, L452R, Y453F, E484K/Q, Q493L, and N501Y exhibited an ∼2-fold increased infection performance to S-614G in feACE2 cells (Figure 2A). In cells expressing mouse ACE2 (muACE2), the K417N, E484K, and N501Y conferred dramatically increased infectivity (Figure 2A). Moreover, the V382L, N440K, G476S, E484K, P521R, and A522S presented a ≥3× increased infection performance to S-614G in horseshoe bat ACE2 (hbACE2) reconstituted cells, and E484K had a more emphatic effect than others (Figures 2A and S5C). The T470N, S477G, T478R, F490S, N501Y, H519P, N532K, and T549A showed ≥3× infection improvements in the whale ACE2-expressing cells (Figures 2A and S5C). By contrast, in cells with ACE2 of tupaia (tuACE2) or brown trout (btACE2), no mutation showed a markedly promoting effect for viral infection (Figure 2A).
Figure 2.
Cross-species infection potentials of SARS-CoV-2 spike variants
(A) Infection performance of 112 LVpps bearing single-site RBD mutated spike in H1299-expressing ACE2 orthologs. A heatmap (the lower panel) shows the cross-species infection performance (green fluorescence units [GFUs]/well at a virus inocula of 10 ng p24) of RBD mutants in cells expressing 18 ACE2 orthologs. The infection performance (relative to S-614G) of RBD mutants in the ferret, mouse, and horseshoe bat ACE2-expressed cells are shown in the upper panel, whereas those in other cells are displayed in Figure S5C.
(B) The infectivities of VOC and VOI LVpps in cells expressing ACE2 orthologs of human, mouse, ferret, and horseshoe bat. Viruses were tested at three doses (0.2, 1, and 5 ng p24, from left to right columns in each group). Data are mean ± SD of greater than or equal to three replicates (six for S-614G; four for Kappa, Delta, B.1.620, and Omicron; and three for others). The numbers labeled on the bars indicate the average fold change of the variant relative to the S-614G, which is calculated at 0.2 ng p24 for huACE2 cells, at 1 ng p24 for feACE2 cells, and at 5 ng p24 for hbACE2, muACE2, and untransfected cells, respectively. As the dramatically increased infectivities of Alpha, Beta, Gamma, Omicron, N501Y/E484K, and N501Y/K417N in muACE2 cells, their infection performance at 5 ng p24 is calculated as the value at 0.2 ng p24 multiplied by 25 for comparisons with other variants. The gray broken lines under the columns show the average levels of the S-614G. Dark shadows indicate the lower limit of quantification (100 GFUs/well). Silhouettes indicating the species were from PhyloPic.org and available under the Public Domain Dedication 1.0 license. Mock, uninfected control (no virus, buffer only); ∗∗∗, p < 0.001; ∗∗, p < 0.01; ∗, p < 0.05; ns, not significant (p > 0.05).
(C) Summary schematics of the RBD mutations presented in spikes of VOCs and VOIs and their influence on infectivity in various cells. +, 1–10× increase, p < 0.05; ++, 10–100× increase, p < 0.05; +++, ≥100× increase, p < 0.05; ∼, no significant change; -, 1–10× decrease, p < 0.05; --, ≥10× decrease, p < 0.05.
We next assessed 11 VOCs and VOIs (Figures 2B and 2C), including the newly documented Omicron, for their infectivities in cells expressing ACE2 orthologs of human, mouse, ferret, horseshoe bat, and tupaia. In huACE2 cells, the Alpha, Beta, Gamma, Epsilon, Iota, Delta, and Omicron variants, as well as the N501Y/E484K and N501Y/K417N DM mutants, showed 1.3–3.7× higher increase (p < 0.001) in infectivity than S-614G (Figure 2B). In contrast, the LVpp of Eta and B.1.620 displayed reduced infectivity (p < 0.001; Figure 2B). In agreement with the single-site RBD mutant tests, variants harboring the N501Y, particularly for the Alpha, Beta, Gamma, Omicron, and two DM mutants, exhibited a dramatically and over 100-fold increased infectivity in muACE2 cells (p < 0.001; Figure 2B). The Eta, Iota_E484K, and B.1.620, carrying E484K, but not N501Y, displayed approximately 10–30× increased infectivity in muACE2 cells. These findings suggested that the N501Y played the most conspicuous contribution in rendering muACE2 cells susceptible to SARS-CoV-2. Moreover, the E484K also had an enhanced effect on the viral infectivity in muACE2 cells, even though it was less efficient than the N501Y. Interestingly, the Kappa (with L452R/E484Q) and Delta (with L452R/T478K) variants also showed significantly increased infectivity in muACE2 cells (Figure 2B). However, neither L452R, T478K, nor E484Q exhibited such effect in the single-site mutant tests (Figure 2A). Therefore, it was possibly attributed to a potential synergistic effect of the dual mutations of L452R/E484Q or L452R/T478K. Moreover, the feACE2 cells showed significantly increased (2.4–22× increase to S-614G and p < 0.001 for each comparison) susceptibilities to 11 tested VOCs and VOIs and the N501Y/E484K mutant but were less susceptible to the N501Y/K417N (Figures 2B and 2C). In hbACE2 cells (Figure 2C), the infectivity of Beta, Gamma, Eta, Iota_E484K, Kappa, Delta, B.1.620, Omicron, and N501Y/E484K was slightly increased (2.2–13× increase to S-614G; p < 0.001). In contrast, no significant infectivity change was observed for the Alpha, Epsilon, Iota_S477N, and N501Y/K417N (p > 0.05). Notably, in the hbACE-H1299 cells, enhanced infectivity of the Omicron variant may be attributed to the N440K (Figure 2A). As a control, untransfected H1299 cells only showed weak infection signals, even at the highest virus dose tested (Figure 2B). Compared with the control cells, none of the tested variants presented improved infectivity in tuACE2 or btACE2 cells (Figure S5D).
Overall, these results suggest that several circulating RBD mutants have enhanced infectivity in cells that expressed ACE2 orthologs of non-human animals, particularly for the mouse and ferret. The increased infection potentials of RBD mutants in animals may expand the host range of SARS-CoV-2 and possibly cause cross-species spillover.
Antigenic characterization for variants by mAbs
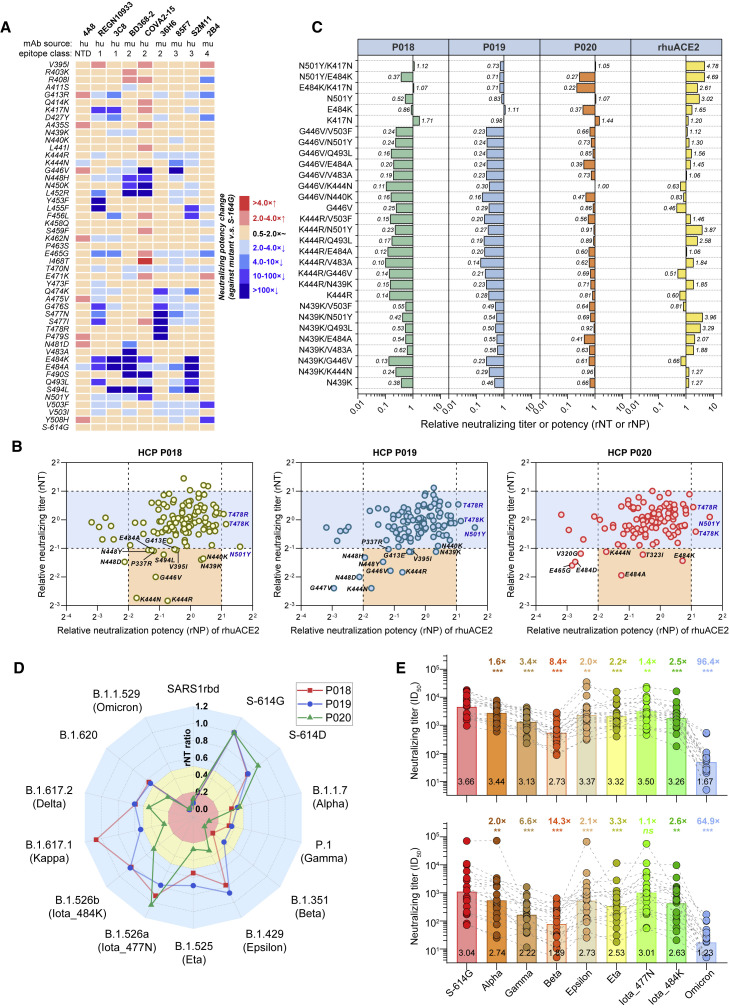
To investigate the mutations-related influence to antibodies induced by ancestral spike derivates, we first tested the neutralizing activities of 9 mAbs against 48 mutants with mutations in the receptor-binding motif (RBM) (Figure 3A). Among these mAbs, the 4A8 is an N-terminal domain (NTD)-targeting nAb, and others are RBD-targeting nAbs. The 4A8, COVA2-15, BD368-2, and S2M11 were reported human mAbs isolated from patients who have recovered from coronavirus disease 2019 (COVID-19) (Brouwer et al., 2020; Cao et al., 2020; Chi et al., 2020; Tortorici et al., 2020); the REGN10933 was isolated from humanized mice (Hansen et al., 2020); and the 3C8, 36H6, 2B4, and 85F7 were mouse mAbs developed in our lab (Wu et al., 2022; Zhang et al., 2021). According to the recognizing modes of these mAbs, the REGN10933 and 3C8 are class 1 nAbs; BD368-2, COVA2-15, and 36H6 are class 2 nAbs; 85F7 and S2M11 are class 3 nAbs; and 2B4 is a class 4 nAb (Figure 3A; Barnes et al., 2020; Wu et al., 2022). The potencies of these mAbs were quantified as half-maximal inhibitory concentration (IC50) values in neutralizing assay (Figure S6). A >4× relative neutralizing potency (rNP) decrease (>4× IC50 increase to that against S-614G) was considered a marked immune escape. As the 4A8 is an NTD-targeting mAb, little change was observed in its activity against tested mutants (Figure S6A). The E484K caused 26× to >100× rNP decrease on the REGEN1093, 3C8, BD368-2, COVA2-15, and S2M11 but had little influence on the 85F7, 36H6, and 2B4. The E484A mutation showed a similar impact as E484K on the activities of nAbs, with two exceptions on COVA2-15 (<4× decrease) and 85F7 (4.5× decrease). The N501Y caused a 14× rNP decrease for the COVA2-15 but had little impairment on other mAbs. The K417N reduced the activities of REGN10933 and 3C8 by 42× and 12×, respectively. Besides E484A/K, the mutations surrounding aa 484, including N481D (for BD368-2), V483A (for BD368-2), F490S (for COVA2-15, BD368-2, and S2M11), Q493L (for S2M11 and REGN10933), and S494L (for COVA2-15, BD368-2, S2M11, 3C8, and 85F7) also conferred obvious resistance to the mAbs. The mutations in aas 444–456 region, including the K444N (for 85F7), G446V (for COVA2-15 and 85F7), N448H (for COVA2-15 and BD368-2), N450K (for COVA2-15 and BD368-2), L452R (for COVA2-15, BD368-2, and REGN10933), Y453F (for REGN10933), L455F (for S2M11 and REGN10933), and F456L (for S2M11 and 3C8), could also cause an obvious neutralization decrease in tested mAbs (Figures 2 and S6). The 36H6, a potent nAb recognizing aas 470–490, was only sensitive to Q474K, G476S, S477N/I, T478R, and P479S. The 2B4 mAb, showing comparable neutralization potencies for SARS-CoV-2 and SARS-CoV (Zhang et al., 2021), slightly lost activity against R408I, G413R, D427Y, and E465G and were dramatically attenuated by V503F and Y508H. Overall, these tests reveal the mutations in aas 444–456 and aas 484–494 are more likely to lead to neutralization escape from RBD-targeting mAbs.
Figure 3.
Antigenic characterization for variants by mAbs and HCPs
(A) Heatmap shows changes of IC50 of mAbs against 48 RBM mutants. The heatmap generated on the IC50 fold changes to S-614G control of each mAb (see Figure S6) against each mutant.
(B and C) Influence of (B) 112 single-site RBD mutants (17 infectivity-decreased variants are excluded) and (C) 24 DM mutants on neutralization of three representative HCPs and rhuACE2.
(D) A spider plot shows rNT of P018, P019, and P020 against VOCs and VOIs.
(E) Comparisons of the NT of HCPs from patients (n = 20, the upper panel) and ASI cases (n = 20, the lower panel) against VOCs and VOIs. For 17 subjects with longitudinal samples, only the data of their first samples are used. The black numbers exhibited within the bars are the GMT (at log10) of HCPs against each variant. The colored numbers show the GMT fold decrease for each variant to S-614G. ∗∗∗, p < 0.001; ∗∗, p < 0.01; ∗, p < 0.05; ns, not significant (p > 0.05).
Antigenic characterization for RBD mutants by HCPs
We next tested 3 HCPs (P018, P019, and P020) against 112 infectivity-preserved RBD mutants for their neutralizing titer (NT) changes. The three HCPs are derived from an early COVID-19 cohort (Table S2) collected during the first wave of COVID-19 in China (Chang et al., 2021; Zhang et al., 2021). Their cross-neutralization activities against mutants were determined as the relative neutralizing titers (rNTs) to control (the NT ratio of against mutant to that against S-614G). Among all mutants, the neutralizing potency of human rACE2 (rhuACE2) showed a >2-fold increase against the N501Y and T478K/R, suggesting that the three mutants may have a higher-binding-capability to huACE2 (Figure 3B). We focused on the mutations leading to >50% neutralization decrease of the HCPs, as shown in Figure 3B. Overall, the P018 and P019 exhibited a similar cross-neutralization pattern (R2 = 0.52; p < 0.001) to most variants, but the P020 had a different antigenic profile (Figure S7A). Mutations surrounding aas 439–448, such as N439K, N440K, K444N/R, G446V, and N448D/H/Y, caused more marked neutralization reduction on P018 and P019 but less influence on P020. In contrast, the E484A/K caused more loss on the activity of P020 than that on P018/P019 (Figures 3B and 3C).
We further assessed the 3 HCPs against 24 DM mutants, which carried combined mutations with dominant effects in changing cross-species tropism or causing neutralization resistance. The N439K/G446V, K444R/V483A, G446V/N440K, and G446V/V483A caused a near-to-over 5× NT decrease on the P018 and P019, whereas only slightly decreased the activity of P020. On the contrary, the E484K/K417N weakened the NT of P020 by about 4× but caused minor impairment for P018 and P019. Against the N501Y/E484K, the P019 showed a more delicate NT decrease than the P018 and P020. The most consistent outcome fell on the G446V/E484A, which decreased the NT by 2.5–5× for all three HCPs (Figure 3C). These data suggest that aas 439–448 and aa 484 are two essential antigenic regions, where the neutralization activities of HCP antibodies are markedly affected by mutations surrounding these sites.
Cross-neutralization profiles of the 3 HCPs against 11 VOCs and VOIs were also characterized (Figures 3D and S7B). Remarkably, in neutralizing the Omicron, P018 and P019 completely lost their activities (half-maximal inhibitory dilution [ID50] < 20), whereas P020 showed a detectable but quite low titer (ID50 = 26). The Omicron variant caused dramatic NT attenuation for nearly or over 100× for the three HCPs, which were even more marked than that induced by the SARS1rbd (a chimeric variant with SARS-CoV RBD). For others, the P020 appeared to be more sensitive than the other two to E484K-harboring variants, including Beta, Gamma, Eta, Iota_E484K, and B.1.620. For example, the P020 showed 11–15× NT decrease against the Beta and Gamma, whereas the P018 and P019 presented approximately 3–5× NT reduction against the two variants. Moreover, the L452R-harboring Epsilon, Kappa, and Delta caused a more arresting NT decrease in P020 than P018 or P019. By contrast, the rNT of the three HCPs decreased to 0.3–0.4 against Alpha and altered little against Iota_477N (Figure 3D). To avoid potential bias resulting from the three HCP samples, we additionally measured other samples in our COVID-19 cohort, which was composed of patients recovered from COVID-19 (n = 20) and individuals with past asymptomatic infections (ASIs) (n = 20; Chang et al., 2021; Zhang et al., 2021). The characteristics of the involved subjects are summarized in Table S2. Due to the limited sample volume, we just tested 8 of 11 involved VOC and VOI strains (not including the Kappa, Delta, and B.1.620). Against various variants, the cross-neutralization profiles of longitudinal HCPs from the same subject were overall consistent (Figure S7C) for either patients or ASIs. However, some discrepancies were also noted in rare cases, particularly those (like A007; Figure S7C) during the immunoglobulin M (IgM)-seroclearance period. The Omicron showed the most striking resistance among all variants, which caused a 96.4× and 64.9× nAb geometric mean titer (GMT) decrease in patients and ASIs (Figure 3E). The E484K-harboring Beta, Gamma, Eta, and Iota_E484K variants attenuated the nAb GMT by 8.4–14.3×, 3.4–6.6×, 2.2–3.3×, and 2.5–2.6×, respectively. The remaining Alpha, Epsilon, and Iota_S477N variants had less impact (∼<2× decrease). Overall, HCPs either from patients or ASI presented similar cross-neutralization profiles, and there was no significant difference on rNT against most of these variants between the two groups (Figure S8A). According to the cross-neutralizing patterns, all tested HCPs could be divided into six groups (Figure S8B). Of note, 21 of 40 (52.5%) were classified into group 1, including the P020. The P018 and P019 were categorized into a distinct group 5. As shown in Figure S8C, against tested variants, the NT changes of P018 (R2 = 0.741; p = 0.003), P019 (R2 = 0.634; p = 0.01), and P020 (R2 = 0.906; p < 0.001) significantly correlated with the nAb GMT changes of the cohort. These data suggest the cross-variants neutralizing characteristics of the three HCPs, particularly for the P018 and P020, can partially represent the overall antigenic profile of HCPs from the early COVID-10 wave.
Influence of variants on nAbs elicited by vaccination in animals
We further investigated the impacts of VOCs, VOIs, and DM mutants with a noticeable HCP neutralization resistance on nAbs elicited by recombinant subunit vaccine in mice and monkeys. The immunized animals are derived from our previous studies, including four monkeys and six mice vaccinated with spike protein (StriFK) and two monkeys and six mice immunized with recombinant RBD protein (Wu et al., 2021). We first tested sera of these animals collected at 2 weeks after three-shot immunizations. The spike elicited higher nAb titers in both animals than RBD (Figure 4A), suggesting the former was more immunogenic than the latter. Against the Omicron and SARS1rbd variants, spike-elicited antisera displayed significantly (p < 0.05) higher rNT than that elicited by RBD (Figure S9A). However, nAbs elicited by the two antigens showed comparable rNTs to other variants (Figures 4B and S9A). For inter-animals comparison, mice showed significantly higher rNT to 8 of 19 variants than monkeys (p < 0.05; Figure S9A). In both species, the cross-neutralizing profiles of vaccine-elicited antisera were more similar to human HCP P020 (Figure 4C) instead of P018 and P019 (Figures S9B and S9C), and monkeys showed better correlation on rNT against variants with that of P020 than mice (Figure 4C). Consistent with findings using HCPs, the Omicron led to the most striking nAb resistance, which caused a 15× and 26× nAb GMT decrease in monkeys and mice (Figure 4A). Besides Omicron, vaccine-elicited nAbs in both animals showed more attenuated neutralization against the E484A/K-harboring variants (Beta, Gamma, Iota_484K, B.1.620, and G446V/E484A) than others. Notably, these sera displayed relatively higher NT to the VOCs and VOIs than HCPs; therefore, we speculated the three-shot immunization might be a possible explanation for these differences. To validate this, we further tested the sera of spike-immunized animals before the third-dose administration (2 weeks after second shot) against seven variants. As the results show in Figures 4D and 4E, the third boost generated more nAb increase against six of seven tested variants (except Delta) than against S-614G in both animals. Moreover, the median rNTs of animals’ antisera significantly (p < 0.05) raised from two to three doses in neutralizing the Beta, Gamma, Kappa, Omicron, and G446V/E484A variants. These data appeared to be consistent with the findings that the three-dose mRNA vaccinations help to elicit cross-variants neutralization antibodies in humans (Garcia-Beltran et al., 2022a; Nemet et al., 2021). However, even after the third booster, the nAbs of either humans or animals against the antigenic-distinct variants, such as Omicron, were still significantly lower than others by several folds.
Figure 4.
Cross-neutralization profiles against spike variants of sera from recombinant-vaccine-immunized animals
(A) Comparisons of the NTs of sera from recombinant spike or RBD proteins immunized monkeys (n = 6) and mice (n = 12). Sera were collected at 2 weeks after three doses of vaccination. The numbers labeled under the spots are the GMTs (at log10). Blue shadows represent the range of 50%–200% (within 2-fold changes) of the nAb GMT against D614G (as the white line indicated). The numbers labeled on the top show the GMT decrease fold for a variant to S-614G (only statistically significant changes are shown).∗∗∗, p < 0.001; ∗∗, p < 0.01; ∗, p < 0.05.
(B) Comparison of the cross-neutralizing activities of RBD- (x axis) and spike-elicited (y axis) polyclonal nAbs against variants in mice (left) and monkeys (right) receiving three doses of vaccination. The rNT was calculated as its ID50 ratio against a variant to the S-614G for each sample.
(C–E) Comparison of the cross-neutralizing activities of HCP P020 (x axis) and vaccine-elicited (spike and RBD-based vaccine pooled) animal’s sera (y axis) against spike variants. For both (B) and (C), the data are plotted as mean ± SEM. Comparisons of serum NT (D) and rNT (E) of animals against the Gamma, Beta, Kappa, Delta, (B)1.620, Omicron, and G446V/E484A variants at 2 weeks after two- or three-dose spike-based vaccinations are shown. For (D), the median fold increase in neutralization for each variant from two to three doses of immunization is shown. For (E), data from mice and monkeys are pooled for analyses; the numbers show the median rNT fold changes. ∗∗∗, p < 0.001; ∗∗, p < 0.01; ∗, p < 0.05; ns, not significant.
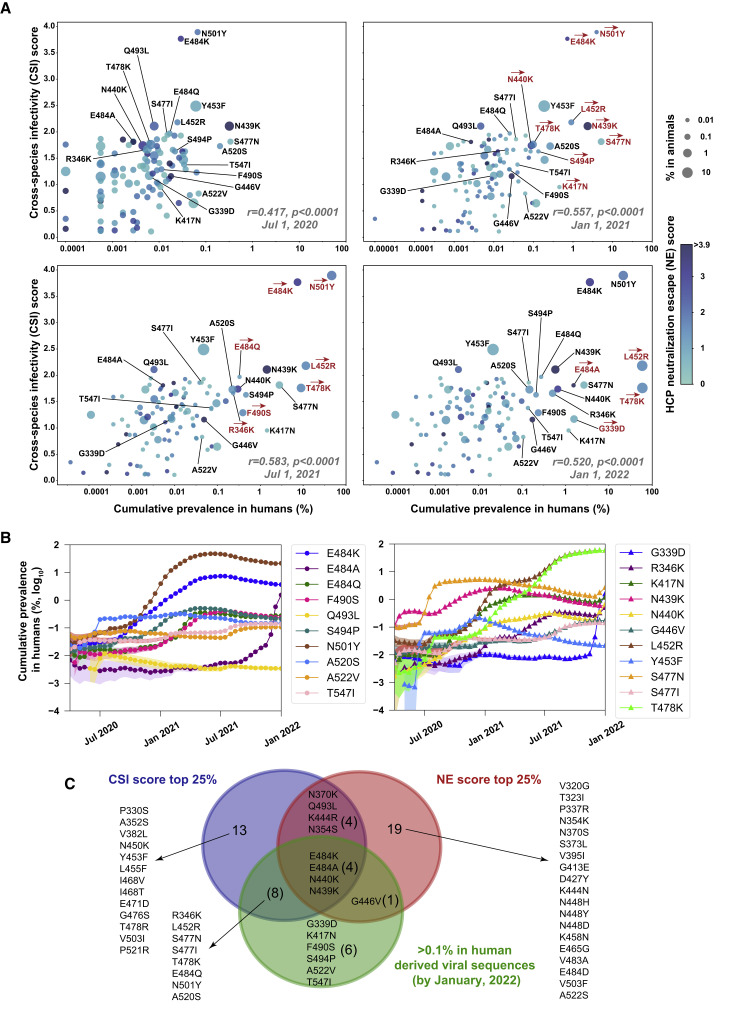
Associations between the circulating frequencies and the in vitro phenotypic characteristics of RBD mutations
To test whether the circulating frequencies of RBD mutations associate with their in vitro phenotypic characteristics, we calculated the frequencies of RBD mutations from all SARS-CoV-2 sequences in GISAID by January 1, 2022. We analyzed how the RBD mutations were associated with their cumulative frequencies from two perspectives, the one considering the cross-species breakout potentials and the other concerning the neutralization escape of HCPs. By four different time points of July 1, 2020; January 1, 2021; July 1, 2021; and January 1, 2022, the multivariate regression analyses demonstrated relative infectivity changes (fold change to S-614G) caused by RBD mutations in feACE2 cells were independently associated with their cumulative prevalences (Table S3). This association was statistically significant in either viral sequence derived from humans or non-human animals (Table S3). Moreover, the relative infectivities of RBD mutants in muACE2 cells were also independently associated with their cumulative prevalences when analyzing the summarized viral sequences in GISAID at time points after 2021 (Table S3). In contrast, no significant association was noted between the mutation prevalences and their influence on viral infectivity in huACE2-H1299 cells or the neutralizing activities of HCPs at all time points we analyzed (Table S3).
Following these findings, we developed a cross-species infectivity (CSI) score (based on the relative infectivities in feACE2 and muACE2 cells) to characterize the cross-species spillover possibility of mutation and a neutralization-escape (NE) score (based on rNTs of P018 and P020) representing the mutation-related immune-escape potential. Statistically, the CSI scores of mutations positively correlated (p < 0.0001) with their cumulative prevalences in human-derived viral sequences summarized by July 1, 2020 (r = 0.417); January 1, 2021 (r = 0.557); July 1, 2021 (r = 0.583); and January 1, 2022 (r = 0.520; Figure 5A), whereas the NE scores were not (p > 0.05). Time-series analyses revealed the correlation coefficient (r) between the CSI scores and the cumulative prevalences of mutations presented an increasing trend from 2020 to 2021 (Figure S10A). The dynamic changes of frequencies of mutations with a CSI score ≥2.0 or cumulative prevalence ≥0.1% (by January 1, 2022) are shown in Figure 5B. Notably, the T478K (cumulative prevalence = 59.4%), L452R (58.9%), N501Y (21.6%), E484K (3.67%), and S477N (2.68%) were the top five frequent mutations in human-derived sequences, simultaneously having relatively higher (>1.5) CSI scores (1.76–3.90). Among 19 RBD mutations with a cumulative prevalence ≥0.1%, 12 (63%) had a CSI score at the top 25% (≥1.66; Figure 5C). These results reveal the statistical association between increased infection potentials in ferret or mouse ACE2-expressing cells and the cumulative frequencies of RBD mutations, suggesting a possible role of cross-species spillover in the spread of a mutated virus in humans and animals.
Figure 5.
Associations between the cumulative frequencies and the in vitro phenotypic characteristics of circulating RBD mutations
(A) Multiple-variable bubble charts show associations among the CSI scores, NE scores, and the cumulative frequencies in humans and animals of RBD mutations at four different time points. A total of 112 RBD mutations were analyzed (17 mutations with markedly reduced infectivity were excluded). The r values showing at the lower right corner of each panel were calculated using a univariate linear regression model to correlate the CSI scores and the cumulative frequencies in humans of mutations. Red arrows illustrate mutations (also labeled in red) with >5-fold increased cumulative frequencies in humans compared with the former time point.
(B) Dynamic changes of cumulative frequencies in humans of RBD mutations with a CSI score ≥2.0 or cumulative prevalence (by January 1, 2022) ≥0.1% over time. The colored shadow indicates a 95% confidential interval.
(C) Venn diagram depicting the overlap among mutations with the top 25% CSI score (≥1.66; n = 29), with the top 25% NE score (≥1.83; n = 28), and with ≥0.1% cumulative prevalence (n = 19).
Discussion
We provide a comprehensive evaluation of the infection potentials of circulating SARS-CoV-2 variants on cells expressing various ACE2 orthologs. Consistent with previous studies, our data suggest that the SARS-CoV-2 may have a broad tropism for terrestrial and marine mammals (Conceicao et al., 2020; Liu et al., 2021). Although there was no direct evidence to support the infection and pathogenicity of SARS-CoV-2 in aquatic mammals, several studies indicated the presence of SARS-CoV-2 in natural water systems receiving untreated human sewage, which should be considered a threat to mammals living in the river and sea (Audino et al., 2021). Among species we tested, the ACE2 of ferret, horseshoe bat, mouse, tupaia, and brown trout showed weak or no binding capability to spike proteins; they were also less efficient or inefficient in mediating ancestral SARS-CoV-2 infection (Figure 1D). However, several RBD mutations can increase the viral infectivity in cells with these ACE2 orthologs, particularly for muACE2, feACE2, and hbACE2. The mouse and ferret are two species that should be concerned, as the former has vast populations and close contact with humans and the latter was a domestic animal farmed on a large scale as mink. The mouse is naturally resistant to the ancestral SARS-CoV-2 due to the absent binding between muACE2 and the viral spike. However, the K417N, E484K, and N501Y can markedly improve the pseudovirus infectivity of SARS-COV-2 in muACE2 cells (Figure 2). The effect of N501Y in rendering mice susceptible to SARS-CoV-2 was previously found in developing the mouse-adapted strain (Gu et al., 2020). A recent study also suggested the N501Y and E484K and their combination effect in promoting the pseudovirus entry in muACE2 cells (Li et al., 2021). The three mutations are commonly present in several VOCs and VOIs. Therefore, it is not surprising to observe the dramatically enhanced infectivity in muACE2 cells of N501Y- or E484K-harboring VOCs and VOIs. Notably, some recently published or preprinted studies also showed the increased susceptibility of the mouse to the Alpha, Beta, and Gamma variants (Montagutelli et al., 2021). For the Omicron, some preliminary in vivo studies suggested it just caused attenuated disease in mice (Halfmann et al., 2022), but both our and other studies demonstrated its extended mouse ACE2 usage in cell cultures (Hoffmann et al., 2021; Peacock et al., 2022). As Omicron also appears to cause milder pulmonary illness in humans, its attenuated pathogenicity in mice may be attributed to restrictions in post-entry steps.
Unlike mice, ferrets can support the ancestral SARS-CoV-2 infection when challenged with high-dose inoculum, but viral replications are limited in the upper respiratory tract (Ryan et al., 2021; Sawatzki et al., 2021; Shi et al., 2020). To our data, the ferret ACE2 has a low binding capability and is less efficient in mediating cellular entry of the ancestral virus, but several RBD mutants showed increased infectivity in feACE2 cells (Figure 2A). Both mink and ferret belong to the Mustelidae family and share highly homologous ACE2, with the only differences at two positions (aa 286 and aa 548) that are not responsible for interaction with the spike. As early as April 2020, SARS-CoV-2 outbreaks in farmed minks caused by the human-derived virus were reported in Europe (Oreshkova et al., 2020), warning that viral spillover from humans to minks and its back transmission to humans is possible and likely to happen (Koopmans, 2021; Oude Munnink et al., 2021). The mink-related “cluster 5” variant carries Y453F mutation, which showed infection enhancement in feACE2 cells and may improve viral fitness in the minks. Our pseudovirus data in feACE2 cells (Figure 2C) suggest the increased infectivities of VOCs and VOIs in ferrets and also possibly in minks. Strikingly, several RBD mutations with noticeable effects to enhance viral infectivity in muACE2 or feACE2 cells are found to have a high cumulative prevalence in the GISAID SARS-CoV-2 database. These findings suggest the cross-species transmission potentials of currently circulating variants. The extended infectivity potentials in originally insusceptible animals may enlarge zoonotic viral reservoirs and bring a new infection source, highlighting the possible contribution of variant-associated human-to-animal spillover and animal-to-human back transmission in the accelerating variants spread.
Previous studies have concerned the challenges of new variants escaping nAbs or cellular immunity elicited by natural infection or vaccine immunizations (Garcia-Beltran et al., 2021; Li et al., 2021; Motozono et al., 2021; Wang et al., 2021). Our data provide a systematical antigenic landscape for prospective estimations of the immune escape possibility of continually emerging variants harboring various RBD mutations. Mutations surrounding aas 439–448 and aa 484 are most likely to cause neutralization resistance to antibodies acquired from vaccinations or natural infections. The N439K, N440K, K444R/N, G446V, G447V, N448Y/D/H, and E484K/A should be considered the most worrisome mutations that are more refractory to polyclonal nAbs of HCPs than others, and some of them are also resistant to several mAbs (Figure 3). The VOCs and VOIs with these RBD mutations usually present increased immune evasion. Currently circulating Omicron variant, which carries critical mutations in both aas 439–448 (N440K and G446S) and aa 484 (E484A), caused dozens of folds neutralization decreases for human HCPs and vaccine-induced antibodies (Figures 3E and 4). Similar to that observed in humans (Choi et al., 2021; Garcia-Beltran et al., 2022a), our comparison in mice and monkeys immunized with two or three doses of ancestral spike-based vaccine suggests additional boost could partially improve nAb response against immune-escape variants, including Omicron. It should be noted that the presented animal experiments followed a short-interval immunization procedure that is different from that administered in humans. However, for either our animal data or results from humans who received long-interval vaccine boosters (Ai et al., 2022; Dejnirattisai et al., 2022; Garcia-Beltran et al., 2022b; Gruell et al., 2022), the nAbs titers elicited by three-dose vaccinations were still markedly lower to neutralize the Omicron than those against other variants. These results highlight developing next-generation COVID-19 vaccines may be required to provide complementary antigenic coverage for the immune-evasive variants. The antigenic landscapes of circulating RBD mutations offer prospective insights to designing new antigens of next-generation COVID-19 vaccines. Notably, our subsequent studies following this study developed lineage-mosaic and mutation-patched spike antigens, which could elicit high titers of broad-spectrum antibodies to neutralize various VOCs and VOIs, including Omicron (Wu et al., 2022).
The SARS-CoV-2 pandemic is continually evolving, with more and more emerged lineages harboring various mutations spreading worldwide. An interesting issue is how a mutant will be more widespread than the most common strain. Mutation-associated viral fitness improvements are essential to rapidly transmitting a variant in humans. The famous D614G, with enhanced infectivity and transmission potentials, has become globally dominant now (Hou et al., 2020; Plante et al., 2021). Remarkably, all five VOCs showed increased infection potentials in muACE2- or feACE2-expressing cells (Figure 2). A key finding of our study is that the increased infection potentials of single-site RBD mutants in non-human animals, particularly for the ferrets and mice, positively correlate with their cumulative prevalence (Figure 5). This association provides suggestive evidence supporting the zoonotic intermediate hosts may play a role in facilitating the selection and spread of SARS-CoV-2 variants, highlighting the importance of further surveillance of viral mutations in humans and animals. Timely assessment for the cross-species infection potentials of newly emerged mutants using the ACE2-cell panel developed in our study may enable predicting their spread possibilities. Because of the absence of statistical linkage between the NE scores and the prevalences of RBD mutations (Figure 5; Table S3), the immune-escape characteristics of currently prevalent variants are more likely to be incidental. However, mutations having both CSI and NE at high scores may confer a competitive advantage for viral variant transmission and possibly appear in emerging VOCs.
In summary, our study provides comprehensive landscapes regarding the cross-species tropism breakout possibilities and the antigenic changes of circulating SARS-CoV-2 variants. These findings highlight the antigenic drift and the possible cross-species spillover driven by viral genetic changes and will guide the prediction and surveillance of SARS-CoV-2 spike mutations.
Limitations of the study
Some limitations should be noted for our study. First, we performed infection and neutralization assays using pseudoviruses instead of authentic SARS-CoV-2 variants. Although the performance of our system has been well validated (Chang et al., 2021; Zhang et al., 2021), it can only measure the entry function of the mutated spikes in an ACE2-dependent manner. On the other hand, we just evaluated the cross-species infection potentials of variants in human cells with exogenous expressions of animals’ ACE2 orthologs, but the actual infections and transmissions of SARS-CoV-2 in animals may be dependent on multiple and complicated factors. Moreover, with the continuous evolution of the SARS-CoV-2, characterizations of some newly emerged mutations not included in this study certainly require further evaluation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human NTD mAb 4A8 | (Chi et al., 2020) | N/A |
| Human RBD mAb COVA2-15 | (Brouwer et al., 2020) | N/A |
| Human RBD mAb BD368-2 | (Cao et al., 2020) | N/A |
| Human RBD mAb S2M11 | (Tortorici et al., 2020) | N/A |
| Human RBD mAb REGN10933 | (Hansen et al., 2020) | N/A |
| Mouse RBD mAb 36H6 | (Zhang et al., 2021) | N/A |
| Mouse RBD mAb 3C8 | (Zhang et al., 2021) | N/A |
| Mouse RBD mAb 2B4 | (Zhang et al., 2021) | N/A |
| Mouse RBD mAb 85F7 | (Wu et al., 2022) | N/A |
| Biological samples | ||
| Human COVID-19 convalescent plasmas | (Chang et al., 2021; Zhang et al., 2021) | Table S2 |
| Recombinant spike-protein immunized animal sera | (Wu et al., 2021) | N/A |
| Recombinant RBD-protein immunized animal sera | (Wu et al., 2021) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| rACE2 protein of human | This study | N/A |
| rACE2 protein of monkey | This study | N/A |
| rACE2 protein of tupaia | This study | N/A |
| rACE2 protein of cat | This study | N/A |
| rACE2 protein of puma | This study | N/A |
| rACE2 protein of dog | This study | N/A |
| rACE2 protein of ferret | This study | N/A |
| rACE2 protein of rabbit | This study | N/A |
| rACE2 protein of hamster | This study | N/A |
| rACE2 protein of mouse | This study | N/A |
| rACE2 protein of horse | This study | N/A |
| rACE2 protein of camel | This study | N/A |
| rACE2 protein of pig | This study | N/A |
| rACE2 protein of pangolin | This study | N/A |
| rACE2 protein of horseshoe bat | This study | N/A |
| rACE2 protein of whale | This study | N/A |
| rACE2 protein of sea lion | This study | N/A |
| rACE2 protein of brown trout | This study | N/A |
| SARS-CoV2-STG | (Zhang et al., 2021) | N/A |
| StriFK protein | (Wu et al., 2021) | N/A |
| S-HexaPro protein | (Hsieh et al., 2020) | N/A |
| Critical commercial assays | ||
| Nano Glo HiBiT Extracellular Detection System | Promega | Cat# N2421 |
| Nano Glo HiBiT Lytic Detection System | Promega | Cat# N3040 |
| HIV-4 Ag/Ab ELISA kit | Beijing Wantai BioPharm | Cat# S20080008 |
| Lipofectamine® 3000 Transfection Reagent | Thermo Scientific | Cat# L3000-015 |
| ExpiFectamine™ CHO Transfection Kit | Thermo Scientific | Cat# A29129 |
| Ni Sepharose 6 Fast Flow | Cytiva | Cat# 17-5318-03 |
| MabSelect SuRe resin | Cytiva | Cat# 17-5474-02 |
| Experimental models: Cell lines | ||
| ExpiCHO-S cells | Thermo Scientific | Cat# A29127; RRID: CVCL_5J31 |
| H1299 cells | ATCC | Cat# CRL-5803; RRID: CVCL_0060 |
| HEK293T/17 cells | ATCC | Cat# CRL-11268; RRID: CVCL_1926 |
| huACE2-H1299 (human ACE2) | (Zhang et al., 2021) | N/A |
| moACE2-H1299 (monkey ACE2) | This study | N/A |
| tuACE2-H1299 (tupaia ACE2) | This study | N/A |
| catACE2-H1299 (cat ACE2) | This study | N/A |
| puACE2-H1299 (puma ACE2) | This study | N/A |
| dogACE2-H1299 (dog ACE2) | This study | N/A |
| feACE2-H1299 (ferret ACE2) | This study | N/A |
| rabACE2-H1299 (rabbit ACE2) | This study | N/A |
| haACE2-H1299 (hamster ACE2) | This study | N/A |
| muACE2-H1299 (mouse ACE2) | This study | N/A |
| hoACE2-H1299 (horse ACE2) | This study | N/A |
| camACE2-H1299 (camel ACE2) | This study | N/A |
| pigACE2-H1299 (pig ACE2) | This study | N/A |
| pagACE2-H1299 (pangolin ACE2) | This study | N/A |
| hbACE2-H1299 (horseshoe bat ACE2) | This study | N/A |
| whACE2-H1299 (whale ACE2) | This study | N/A |
| slionACE2-H1299 (sea lion ACE2) | This study | N/A |
| btACE2-H1299 (brown trout ACE2) | This study | N/A |
| Recombinant DNA | ||
| EIRBsMie-dSwtG(S614G) | This study | N/A |
| EIRBsMie-dSwtD(S614D) | This study | N/A |
| EIRBsMie-SARS1rbd | This study | N/A |
| Plasmid panel: EIRBsMie-dSwtG based single-site RBD mutants (n=129) | This study | N/A |
| Plasmid panel: EIRBsMie-dSwtG based double-site RBD mutants (n=24) | This study | N/A |
| ACE2 orthologs cDNA | GenBank | Table S1 |
| Plasmid panel: pLVEF1αIHRB-ACE2 orthologs panel (n=18) | This study | N/A |
| EIRBsMie-dSuk1201G(Alpha/B.1.1.7) | This study | N/A |
| EIRBsMie-dSBR1216(Gamma/P.1) | This study | N/A |
| EIRBsMie-dSSA118(Beta/B.1.351) | This study | N/A |
| EIRBsMie-dSCAL20C (Epsilon/B.1.429) | This study | N/A |
| EIRBsMie-dSuk1188 (Eta/B.1.525) | This study | N/A |
| EIRBsMie-dSus1123A (Iota_477N/B.1.526a) | This study | N/A |
| EIRBsMie-dSus1123B (Iota_484K/B.1.526b) | This study | N/A |
| EIRBsMie-dSIND288 (Kappa/B.1.617.1) | This study | N/A |
| EIRBsMie-dSIND682 (Delta/B.1.617.2) | This study | N/A |
| EIRBsMie-dSOLV639 (B.1.620) | This study | N/A |
| EIRBsMie-dSSA668 (Omicron/B.1.1.529) | This study | N/A |
| psPAX2 | Addgene | RRID: Addgene_12260 |
| pLVEF1αmNG | (Zhang et al., 2021) | N/A |
| Software and algorithms | ||
| Columbus Analysis system (version 2.5.0) | PerkinElmer | https://www.perkinelmer.com/ |
| GraphPad Prism (version 8.0.1/9.0.0) | Graphpad | https://www.graphpad.com/ |
| FlowJo (version 10.6.0) | FlowJo | https://www.flowjo.com/ |
| Origin 2021 | OriginLab | https://www.originlab.com/2021 |
| Leica LAS X (Version 3.5.5) | Leica | https://www.leicamicrosystems.com |
| SAS (version 9.4) | SAS | https://www.sas.com/ |
| Python (version 3.9.6) | Python | https://www.python.org/downloads/release/python-396/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ningshao Xia (nsxia@xmu.edu.cn).
Materials availability
All unique/stable materials and reagents generated in this study are available for non-commercial usage upon a completed material transfer agreement (MTA).
Experimental model and subject details
Human COVID-19 convalescent plasma
The HCP samples (n = 61) from patients with confirmed COVID-19 (n = 20) and people with past asymptomatic infections (n = 20) were used for antigenic analyses (Figure 3). All HCP samples were collected during the early wave of the pandemic and were positive for total RBD antibody, RBD-IgG, and neutralizing antibody (Chang et al., 2021; Zhang et al., 2021). The detailed information of these cases is shown in Table S2. Longitudinal samples (≥2-time points for each case) with a median follow-up of 44-day were available for 17 subjects (5 patients and 12 ASI), and single-time-point samples were available for others. The study was approved (number: SPHIRB-202101) by the institutional review board of the School of Public Health of Xiamen University following the Declaration of Helsinki, and written informed consent was obtained.
Vaccine-immunized animal sera
Sera from 6 monkeys and 12 mice with recombinant protein-based vaccine immunizations were enrolled (Figure 4). These animals were derived from our previous study (Wu et al., 2021). Among the monkeys, 4 were immunized with the StriFK-based (trimeric S-ectodomain) vaccine (20 μg per dose), and the other 2 were immunized with RBD-based vaccine (20 μg per dose) following an immunization schedule of one priming dose at week 0 plus two boosters at weeks 2 and 6. For mice, half (n = 6) were with StriFK-based vaccine (1 μg per dose), and the other half were immunized with RBD-based vaccine (1 μg per dose) following an immunization schedule of one priming dose at week 0 plus two boosters at weeks 2 and 4. Animal studies were conducted in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals under the approval of the Institutional Animal Care and Use Committee of Xiamen University (mouse) or JOINN Labora-tories, Inc (monkey).
Cell lines
The HEK293T/17 and H1299 cells were Dulbecco's modified Eagle medium (Sigma, D6429) supplemented with 10% fetal bovine serum (Thermo Scientific, 10099-141) and 0.1 mM non-essential amino acids (Thermo Scientific,1140-050). All cell lines (H1299-derived) expressing ACE2 orthologs were cultured with the same medium but supplemented with blasticidin (10μg/mL). The HEK293T/17, H1299, and ACE2-expressing H1299 cells were at 37°C and 5% CO2 in a humidified incubator. The ExpiCHO-S cells were cultured with ExpiCHO™ Expression Medium (Thermo Scientific) in a stackable CO2 incubator shaker.
Method details
Analysis and selection of RBD mutations among circulating SARS-CoV-2 isolates
We initially analyzed circulating RBD mutations based on the variation annotation of 2019 Novel Coronavirus Resources (2019nCoVR) at CNCB-NGDC (https://ngdc.cncb.ac.cn/ncov/) on Aug 2020 (Zhao et al., 2020). The complete genome sequences used for generating the variation annotation table were obtained from CNGBdb, GenBank, GISAID, GWH, and NMDC databases. First, the non-human derived or low-quality sequences were removed. In addition, each degenerate base in the genome was substituted with corresponding bases for keeping every possible amino acid mutation in every record which shared the isolate count number. Finally, a total of 129 mutations were selected for functional analysis (Figure 1A), according to the following refining criteria: (i) detected in more than two independent sequences; (ii) mutations in epitopes of reported neutralizing antibodies; (iii) amino acid substitution to that identical with SARS-CoV.
The prevalence of the mutations was calculated based on the protein sequences of the spike in the GISAID databases (accessed on January 4, 2022). The sequence set was downloaded from the GISAID website directly and aligned to the reference spike protein sequence (spike sequence from EPI_ISL_402124). The species origination and collection date were extracted from the FASTA title for further filtering. The records with collection dates specifying only the year were excluded, while the date specifying only the year and month were assumed to have occurred on the 15th of that month. The counts and frequencies of mutations at each selected RBD site were then computed using this sequence dataset. In calculating the frequencies of mutations, the cumulative prevalence value was set as a minimum value when the cumulative count from GISAID was zero. Thus, in detail, zero in human-derived or total viral sequence dataset, the prevalence calculation was set as 10−6 for time points in 2020 and 10−7 in 2021, whereas zero in the animal-derived viral sequence dataset calculation was as 10−3 for time point earlier than October 1, 2020, and 10−4 for the else.
Productions of pseudoviruses bearing SARS-CoV-2 spike variants
Plasmids containing spike variant-expressing cassettes were generated by site-directed site-specific mutagenesis on a mammalian vector (EIRBsMie-dSwtD, containing codon-optimized spike gene from MN908947.3). The 18aa from the C-terminus of the spike was replaced with a HiBit bioluminescent tag (14aa, GSGVSGWRLFKKIS) for detection using split-NanoLuc assay (Promega) (Dixon et al., 2016). To our data, the HiBit-fusion didn't influence the production and infection of pseudovirus. The plasmids for expressing D614G mutated spike (EIRBsMie-dSwtG) were constructed as the parental vector for other variants. A plasmid designated SARS1rbd, encoding a chimeric spike of SARS-CoV-2 with SARS-CoV RBD replacement, was also constructed as a control. The spike expression cassettes of VOC/VOI variants (Figure 2B), including Alpha/B.1.1.7 (referring EPI_ISL_601443), Beta/B.1.351 (referring EPI_ISL_700428), Gamma/P.1 (referring EPI_ISL_792680), Epsilon/B.1.429 (referring EPI_ISL_873881), Eta/B.1.525 (referring EPI_ISL_762449), Iota_477N/B.1.526a (referring EPI_ISL_995145), Iota_484K/B.1.526b (EPI_ISL_1009654), Kappa/B.1.617.1 (referring EPI_ISL_1595904), Delta/B.1.617.2 (referring EPI_ISL_1662451), B.1.620 (referring EPI_ISL_1620228), and Omicron/B.1.1.529 (referring EPI_ISL_6704867) were also generated via site-specific mutagenesis on the EIRBsMie-dSwtG. For productions of SARS-CoV-2 variants-bearing LVpp, the spike-expressing plasmid, the lentiviral packing plasmid of psPAX2, and the mNeonGreen reporter vector (pLVEF1αmNG) were co-transfected into 293T cells using Lipofectamine® 3000 (Thermo Scientific). Two days after transfection, the supernatants were collected and filtrated by a 0.45-μm pore size filter. Both supernatants and cell lysates were subjected to spike protein detections using Nano-Glo® HiBiT luminescent detection system (Promega). Moreover, the p24 concentrations of viral stocks (supernatants) were determined by using an HIV-4 Ag/Ab ELISA kit (Wantai, Beijing, China) in calibrating with the p24 quantitative standards (Beijing Controls & Standards Biotechnology Inc., China). The p24 concentrations of viral stocks were expressed as U/mL (1 U = 50 pg of p24 antigen). Aliquot viral stocks (supernatants) were stored at −80°C freezer until use.
Recombinant expressions of ACE2 orthologs, viral spike proteins, and nAbs
The reference cDNA of 18 ACE2 orthologs (Table S1) were synthesized (Generalbiol, Anhui, China). Recombinant proteins of ACE2-Fc (ACE2 extracellular domain fused with mouse IgG1 Fc) chimeric proteins were expressed and purified as previously described (Lei et al., 2020). Two recombinant SARS-CoV-2 S-ectodomain proteins (non-prefusion-stabilized StriFK and prefusion-stabilized S-HexaPro) were also prepared as described (Hsieh et al., 2020; Zhang et al., 2021). The encoding sequences of heaven chain and light chain of reported nAbs of 4A8, COVA2-15, BD368–2, S2M11, and REGN10933 were synthesized (Generalbiol, Anhui, China) in a human-IgG1 backbone for recombinant expression. The mouse RBD-targeting nAbs of 36H6, 3C8, 2B4, and 85F7 were generated via mouse hybridoma technology as previously described (Zhang et al., 2021). The variable regions of these mouse mAbs were obtained via RT-PCR using mRNA isolated from the corresponding hybridoma cells and were cloned into the expressing vector in chimeric with human IgG1 backbone. All expressions of recombinant proteins and antibodies were performed by using ExpiCHO-S cells (Thermo Scientific). In brief, the plasmids were transfected into ExpiCHO-S cells using ExpiFectamine CHO transfection kit (Thermo Scientific, A29129). Subsequently, transfected cells were cultured in a stackable CO2 incubator shaker (Kühner AG, SMX1503C) for 7-day. The rACE2 proteins and antibodies were purified from cell culture supernatants using MabSelect SuRe resin (Cytiva), whereas the spike proteins were purified using Ni Sepharose 6 Fast Flow (Cytiva).
Generation and characterization of cell lines expressing ACE2 orthologs
The H1299 cell line stably expressing human ACE2 was developed and described in our previous study. Other cell lines expressing non-human ACE2 orthologs were generated via transduction by lentiviral vectors. Briefly, the cDNA of ACE2 orthologs were synthesized (Generalbiol, Anhui, China) and cloned into a pLVEF1αIHRB vector containing an IRES-driven expression cassette of H2BmRuby3-P2A-BsR (Zhang et al., 2021). The lentiviruses carrying full-length ACE2 orthologs (driven by an EF1α promoter) were produced in 293T cells. Subsequently, stably-transduced cells were enriched via FACS cell sorting (by activated red fluorescence of H2BmRuby3) and blasticidin selection.
Infection and neutralization assays based on pseudoviruses
For LVpp infection assays, H1299 cells expressing ACE2 orthologs were seeded in a 96-well plate with an optically clear bottom (CellCarrier-96 Black, PerkinElmer) at 6×103 cells/well. After 16–18 h of culture, the cells were incubated with viral stock dilutions (60 μL+60 μL fresh culture medium) at 37°C in a CO2 incubator. Two-day later, the fluorescence images of the cells were acquired by using Opera Phenix high-content imaging system (PerkinElmer). The mNeonGreen (infection-reporter) activated cell number of each well was determined and expressed as the green-fluorescent unit per well (GFU/well). For determinations of infectious titers of pseudoviruses, 2-fold serial dilutions of viral stock were prepared and measured by infection assays. To our experience, the LVpp infection assay has a linear dynamic range of about 50-5,000 GFU/well. The infectious titers of viral stocks were calculated by the log-log regression model. For comparisons of the infectivities of pseudoviruses bearing various spikes, the infectious titers (GFU/mL) or infection performance (GFU/well) of variants were normalized with their p24 concentrations.
For neutralization tests, serially-diluted samples (rACE2 proteins, HCPs, mAbs, or immunized animal sera) were pre-incubated with an LVpp inoculum (2,500 GFU/well) for 1 h. Then, the mixtures were incubated with the huACE2-H1299 cells that pre-seeded in 96-well cell culture plates. Following the abovementioned infection assay and imaging analysis procedures, the infection performance of each well was obtained and the neutralizing activity was calculated. The ID50 or IC50 was determined by the 4PL regression on GraphPad Prism v8.0 as previously described (Chang et al., 2021).
Measurements of the ELISA-binding capability between rACE2 and spike proteins
For measurements of the binding capabilities between rACE2 and spike proteins (Figures 1D and S5A), ELISA plates were coated with StriFK or S-HexaPro at 200 ng/well. A series of 3-fold series dilutions that ranged from 10,000 to 0.056 ng/mL for each rACE2 protein were prepared. For the assay, 100 μL of rACE2 dilutions were added to the well and incubated for 1-h at 37°C, followed by washing and reaction with the goat anti-mouse IgG (HRP) (Wantai, Beijing, China). Following a further 1-h incubation, the plates were washed five times and then reacted with chromogen solution (100 μL/well). Finally, the reaction was stopped by adding 50 μL of 2 M H2SO4, and the OD value (OD450-630) was measured on an absorbance plate reader. Three replicates were done for each concentration.
Quantification and statistical analysis
Data visualization and graph generation were conducted using GraphPad Prism version 8.0.1 or version 9.0.0, Origin 2021, or Python. GraphPad Prism or SAS 9.4 was used for statistical analysis. The Student’s t-test or Mann-Whitney U test was used to compare continuous variables between two groups according to the data distribution (normal or non-normal distribution). The one-way ANOVA test was applied to analyze differences among groups. The calculations of 95% confidence interval of ratios were calculated based on the Poisson distribution. The cross-species infectivity score (CSI score) of the mutation is defined by the sum of relative infection performance (RI, fold-change to S-614G) in the feACE2 and muACE2–H1299 cells: CSI = (RI feACE2) + log10 (RI muACE2).
The HCP neutralizing escape score of the mutation (NE score) is defined by the rNT of two different HCP samples (P018 and P020): NE = (rNT P018 × rNT P019)-1.
The association between two variables was evaluated by the Pearson correlation method and univariate linear regression model. In addition, multiple linear regression was used to explore the independent association factors of the cumulative prevalence of mutations in human- or animal-derived viral sequences. p values less than 0.05 were considered to be statistically significant. If not otherwise stated, the mean or median value of 3 technical replicates was used for quantification assays. ∗∗∗, p < 0.001; ∗∗, p < 0.01; ∗, p < 0.05; ns, not significant (p > 0.05)
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81902057, U1905205, and 81871316), the Science and Technology Major Project of Xiamen (3502Z2021YJ013), and the CAMS Innovation Fund for Medical Sciences (no. 2019RU022).
Author contributions
Conceptualization, Y.Z., Q.Y., T.C., Y.G., and N.X.; methodology, Y.Z., M.W., Y.W., H.Y., and Q.Y.; investigation, Y.Z., M.W., Y.W., J.W., Y. Hong, Y. Huang, L.Y., J.M., K.W., S.W., Y. Shi, Z.W., H.G., J.X., C.Y., J.Y., J.C., Y.L., B.F., M.L., and P.G.; formal analysis, H.Y., Y. Huang, Z.H., Y. Su, and Q.Y.; writing – original draft, Y.Z., M.W., H.Y., T.C., and Q.Y.; writing – review & editing, T.C., Q.Y., Y.C., T.Z., J.Z., H.Z., Y.G., and N.X.; funding acquisition, Y.Z., Q.Y., and N.X.; resources, Y.Z., Q.Y., T.C., and H.Y.; supervision, H.Y., Q.Y., T.C., Y.G., and N.X.
Declaration of interests
The authors declare no competing interests.
Published: March 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110558.
Supplemental information
Data and code availability
This paper does not report original code. Original images data from Figure S3 and S4 have been deposited on Mendeley at https://doi.org/10.17632/67k5byngdx.1. Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
References
- Ai J., Zhang H., Zhang Y., Lin K., Zhang Y., Wu J., Wan Y., Huang Y., Song J., Fu Z., et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg. Microbes Infect. 2022;11:337–343. doi: 10.1080/22221751.2021.2022440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audino T., Grattarola C., Centelleghe C., Peletto S., Giorda F., Florio C.L., Caramelli M., Bozzetta E., Mazzariol S., Guardo G.D., et al. SARS-CoV-2, a threat to marine mammals? A study from Italian seawaters. Animals. 2021;11:1663. doi: 10.3390/ani11061663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention SARS-CoV-2 Variant Classifications and Definitions. 2022. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html
- Chang L., Hou W., Zhao L., Zhang Y., Wang Y., Wu L., Xu T., Wang L., Wang J., Ma J., et al. The prevalence of antibodies to SARS-CoV-2 among blood donors in China. Nat. Commun. 2021;12:1383. doi: 10.1038/s41467-021-21503-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi A., Koch M., Wu K., Chu L., Ma L., Hill A., Nunna N., Huang W., Oestreicher J., Colpitts T., et al. Safety and immunogenicity of SARS-CoV-2 variant mRNA vaccine boosters in healthy adults: an interim analysis. Nat. Med. 2021;27:2025–2031. doi: 10.1038/s41591-021-01527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceicao C., Thakur N., Human S., Kelly J.T., Logan L., Bialy D., Bhat S., Stevenson-Leggett P., Zagrajek A.K., Hollinghurst P., et al. The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins. Plos Biol. 2020;18:e3001016. doi: 10.1371/journal.pbio.3001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022 doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon A.S., Schwinn M.K., Hall M.P., Zimmerman K., Otto P., Lubben T.H., Butler B.L., Binkowski B.F., Machleidt T., Kirkland T.A., et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 2016;11:400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e2379. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022 doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St. Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022 doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., Kurth F., Sander L.E., Klein F. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat. Med. 2022 doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., Darling T.L., Joshi A., Loeber S., Singh G., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022 doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kruger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.S., Winkler M.S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2021 doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schafer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M. SARS-CoV-2 and the human-animal interface: outbreaks on mink farms. Lancet Infect Dis. 2021;21:18–19. doi: 10.1016/S1473-3099(20)30912-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2-what do they mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- Lei C., Qian K., Li T., Zhang S., Fu W., Ding M., Hu S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun. 2020;11:2070. doi: 10.1038/s41467-020-16048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., Zhang Y., Li T., Liu S., Zhang M., et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362–2371.e2369. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hu G., Wang Y., Ren W., Zhao X., Ji F., Zhu Y., Feng F., Gong M., Ju X., et al. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagutelli X., Prot M., Levillayer L., Salazar E.B., Jouvion G., Conquet L., Donati F., Albert M., Gambaro F., Behillil S., et al. The B1.351 and P.1 variants extend SARS-CoV-2 host range to mice. bioRxiv. 2021 doi: 10.1101/2021.03.18.436013. Preprint at. [DOI] [Google Scholar]
- Motozono C., Toyoda M., Zahradnik J., Saito A., Nasser H., Tan T.S., Ngare I., Kimura I., Uriu K., Kosugi Y., et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29:1124–1136.e1111. doi: 10.1016/j.chom.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemet I., Kliker L., Lustig Y., Zuckerman N., Erster O., Cohen C., Kreiss Y., Alroy-Preis S., Regev-Yochay G., Mendelson E., Mandelboim M. Third BNT162b2 vaccination neutralization of SARS-CoV-2 Omicron infection. N. Engl. J. Med. 2021 doi: 10.1056/NEJMc2119358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreshkova N., Molenaar R.J., Vreman S., Harders F., Oude Munnink B.B., Hakze-van der Honing R.W., Gerhards N., Tolsma P., Bouwstra R., Sikkema R.S., et al. SARS-CoV-2 infection in farmed minks, the Netherlands, Euro Surveill. 2020;25 doi: 10.2807/1560-7917\ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou T., Mou H., Zhang L., Ojha A., Choe H., Farzan M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 2021;17:e1009212. doi: 10.1371/journal.ppat.1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T.P., Brown J.C., Zhou J., Thakur N., Newman J., Kugathasan R., Sukhova K., Kaforou M., Bailey D., Barclay W.S. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. Preprint at bioRxiv. 2022 doi: 10.1101/2021.12.31.474653. [DOI] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.A., Bewley K.R., Fotheringham S.A., Slack G.S., Brown P., Hall Y., Wand N.I., Marriott A.C., Cavell B.E., Tree J.A., et al. Dose-dependent response to infection with SARS-CoV-2 in the ferret model and evidence of protective immunity. Nat. Commun. 2021;12:81. doi: 10.1038/s41467-020-20439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawatzki K., Hill N.J., Puryear W.B., Foss A.D., Stone J.J., Runstadler J.A. Host barriers to SARS-CoV-2 demonstrated by ferrets in a high-exposure domestic setting. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2025601118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- World Health Organization Tracking SARS-CoV-2 Variants. 2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ [PubMed]
- Wu Y., Huang X., Yuan L., Wang S., Zhang Y., Xiong H., Chen R., Ma J., Qi R., Nie M., et al. A recombinant spike protein subunit vaccine confers protective immunity against SARS-CoV-2 infection and transmission in hamsters. Sci. Transl. Med. 2021:eabg1143. doi: 10.1126/scitranslmed.abg1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang S., Zhang Y., Yuan L., Zheng Q., Wei M., Shi Y., Wang Z., Ma J., Wang K., et al. Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine. bioRxiv. 2022 doi: 10.1101/2022.01.25.477789. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang S., Wu Y., Hou W., Yuan L., Shen C., Wang J., Ye J., Zheng Q., Ma J., et al. Virus-free and live-cell visualizing SARS-CoV-2 cell entry for studies of neutralizing antibodies and compound inhibitors. Small Methods. 2021;5:2001031. doi: 10.1002/smtd.202001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.M., Song S.H., Chen M.L., Zou D., Ma L.N., Ma Y.K., Li R.J., Hao L.L., Li C.P., Tian D.M., et al. The 2019 novel coronavirus resource. Yi Chuan. 2020;42:212–221. doi: 10.16288/j.yczz.20-030. [DOI] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e2346. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper does not report original code. Original images data from Figure S3 and S4 have been deposited on Mendeley at https://doi.org/10.17632/67k5byngdx.1. Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.