Abstract
Triple-negative breast cancer (TNBC) is an aggressive type of breast cancer. High fibrosis, marked by increased collagen fibers, is widespread in TNBC and correlated with tumor progression. However, the molecular features of fibrosis and why it results in a poor prognosis remain poorly understood. Based on multiomics datasets of TNBC, we evaluated the pathological fibrosis grade of 344 samples for further analysis. Genomic, transcriptomic, and immune changes were analyzed among different subgroups of fibrosis. High fibrosis was an independent adverse prognosis predictor and had interactions with low stromal tumor-infiltrating lymphocytes. Genomic analysis identified copy number gains of 6p22.2–6p22.1 (TRIM27) and 20q13.33 (CDH4) as genomic hallmarks of tumors with high fibrosis. Transcriptome analysis revealed the transforming growth factor-beta pathway and hypoxia pathway were key pro-oncogenic pathways in tumors with high fibrosis. Moreover, we systematically evaluate the relationship between fibrosis and different kinds of immune and stromal cells. Tumors with high fibrosis were characterized by an immunosuppressive tumor microenvironment with limited immune cell infiltration and increased fibroblasts. This study proposes new insight into the genomic and transcriptomic alterations potentially driving fibrosis. Moreover, fibrosis is related to an immunosuppressive tumor microenvironment that contributes to the poor prognosis.
Keywords: triple-negative breast cancer, fibrosis, molecular features, fibrotic focus, tumor microenvironment, cancer associated fibroblast
Graphical abstract
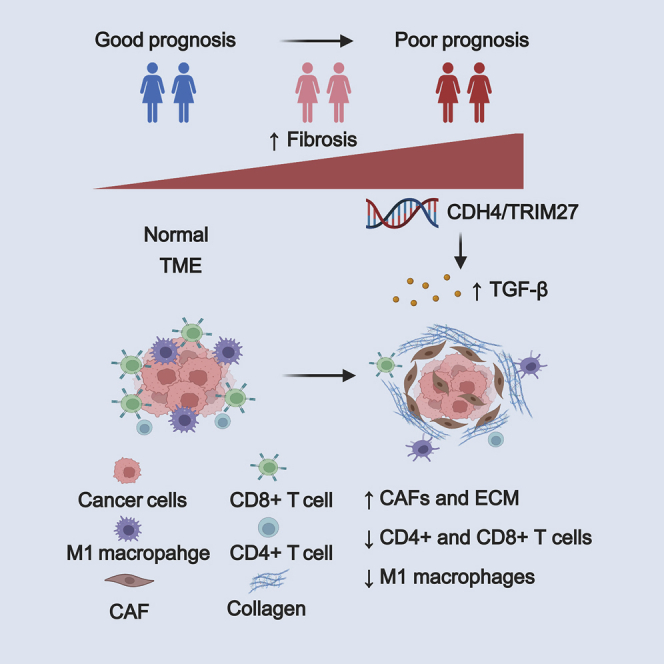
Fibrosis indicates a poor prognosis in triple-negative breast cancer (TNBC). With a large multi-omic TNBC cohort, Di et al. systematically revealed the genomic alterations and immunosuppressive microenvironment of fibrotic TNBCs. transforming growth factor (TGF)-β pathway might decrease immune cell infiltration and induce the immunosuppressive environment, which resulted in a poor prognosis for patients with TNBC.
Introduction
Triple-negative breast cancer (TNBC), defined by the absence of immunostaining for estrogen receptor and progesterone receptors and a lack of overexpression or amplification of human epidermal growth factor receptor 2, accounts for 15%–20% of breast cancer cases.1 TNBCs, which lack widely recognized targeted therapies, have high rates of distant recurrence and poor overall survival.2 TNBC is a heterogeneous disease, and previous studies by our group have classified TNBC into several molecular subtypes with distinct genomic alterations, transcriptomic changes, and tumor microenvironments.3 In addition to its molecular heterogeneity, there is marked heterogeneity in the pathological phenotype of TNBC. Previous research suggests that patients with TNBC with different histopathological characteristics exhibit different molecular features, prognoses, and treatment responses.4,5 Therefore, it is necessary to explore the molecular nature of TNBC combined with pathological features and genomic characteristics.
The fibrotic focus (FF) is a scar-like area in the center of a carcinoma and is regarded as a focus of exaggerated reactive tumor stroma formation.6 Previous studies indicated that the FF is significantly correlated with metastasis and a poor prognosis of invasive ductal carcinoma of the breast.7,8 Furthermore, some studies have tried to explore the mechanism by which FF leads to a poor prognosis. A high tumor angiogenesis ratio and hypoxia within tumors with FF have been proven to be aggravating factors in facilitating metastasis.9,10 Recently, some studies of tumor pathology have suggested an association between FF and the tumor microenvironment. A high level of CD68+, CD163+, or CD204+ tumor-associated macrophage infiltration was associated with the presence of FF in invasive ductal carcinoma.11 Yanai et al. found that the presence of myxoid changes and FF were significantly associated with low or intermediate tumor-infiltrating lymphocytes (TILs) in TNBC.12 However, previous studies have focused on only some specific immune cell subtypes, which might result in a biased understanding of the effects of FF on the TNBC microenvironment. More important, the molecular features of FF in TBNC remain unknown due to the lack of multiomics data from a large cohort.
In this study, we defined the fibrosis grade of TNBC based on the evaluation of FF. Collectively, we questioned whether the formation of fibrosis was driven by specific genomic or transcription events and the effects of fibrosis on the tumor microenvironment (TME). Multiomics data from the Fudan University Shanghai Cancer Center TNBC cohort with fibrosis evaluation were used to systematically analyze the prognostic significance, genomic features, and TME of fibrosis. This study suggested new perspectives in understanding the mechanism of how fibrosis resulted in a poor prognosis in TNBC.
Results
The landscape of fibrosis in TNBC
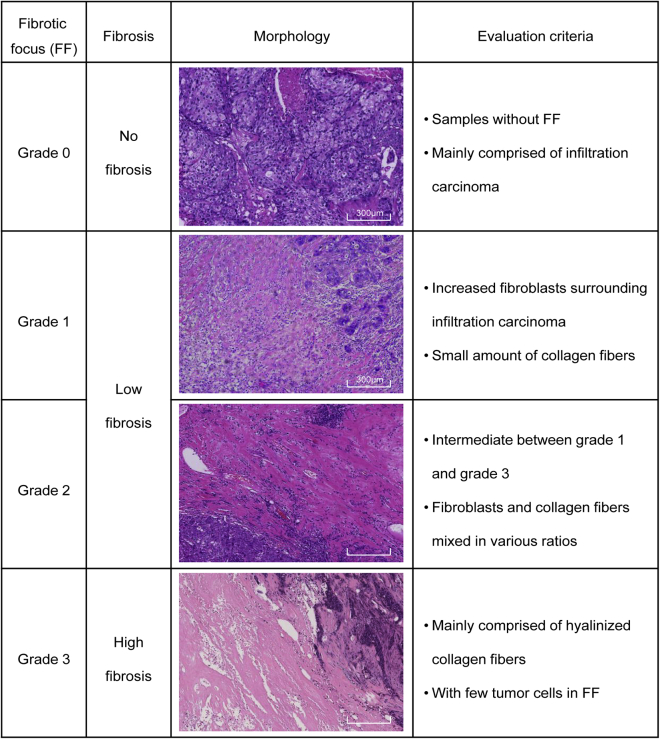
We first evaluated the FF of TNBC based on H&E sections. FF status was divided into four grades (grade 0–3) based on previously reported criteria.6,13 Representative images of different grades of fibrosis are presented in Figure 1. Samples rated as grade 0 (G0) were mostly composed of infiltrating carcinoma without FF. Samples rated as grade 1 (G1) had abundant fibroblasts arranged in a storiform pattern with FF. Samples rated as grade 2 (G2) consisted of intermediate fibroblasts mixed with collagen fibers. Samples rated as grade 3 (G3) consisted mainly of hyalinized collagen fibers and tumor cells were seldom seen in FF. According to the grade of FF, the fibrosis in TNBC was then classified into three groups as follows: no fibrosis (G0 FF), low fibrosis (G1–2 FF), and high fibrosis (G3 FF). Patients with grade 1 and grade 2 FF were grouped together as the low fibrosis group because there was no stringent distinction between the two groups pathologically.
Figure 1.
Morphology and definition of different grades of fibrosis in triple-negative breast cancer (TNBC)
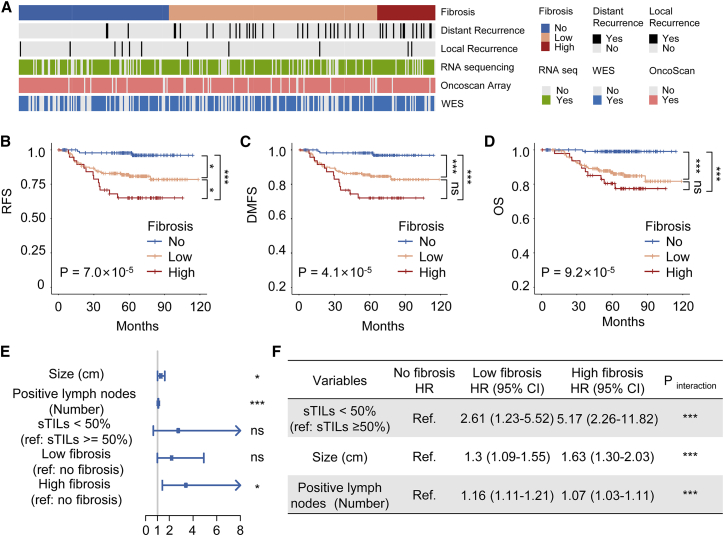
Detailed multiomics data of the cohort are summarized in Figure 2A. The whole cohort included 344 samples, 77% of which had RNA-seq data, 92% had OncoScan microarray copy number data, 65% had whole exome sequencing (WES) data. Forty-two patients of the whole cohort had a distant recurrence and 11 patients had a local recurrence. We first compared the clinicopathological characteristics among the different degrees of fibrosis (Table 1). Patients of different grades had similar clinicopathologic characteristics except for necrosis, which was found to be significantly associated with fibrosis (p < 0.001). Importantly, there was no significant association between fibrosis grades and the TNBC molecular subtypes (p = 0.353), indicating that fibrosis was a pathological feature evenly distributed among TNBC samples. Moreover, a survival analysis was conducted to explore the prognostic value of fibrosis grades. Patients with a high grade of fibrosis had significantly worse relapse-free survival (RFS) (log rank p = 2.2 × 10−5), distant metastasis-free survival (DMFS) (log rank p = 3.0 × 10−6) and overall survival (OS) (log rank p = 9.4 × 10−6) than the no fibrosis group (Figures 2B–2D).
Figure 2.
Prognosis analysis and interaction tests of fibrosis in TNBC
(A) Study cohort and omics data contained. (B–D) Kaplan-Meier curves for (B) RFS, (C) DMFS, and (D) OS. (E) Hazard ratios (HRs) and p values of the covariates in the multivariate Cox proportional hazards model for RFS. (F) Interaction tests and stratified analysis in patient subgroups of fibrosis. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns P ≥ 0.05. CI, confidence interval
Table 1.
Clinicopathological characteristics of patients with evaluation of fibrosis
| Clinical characters | No fibrosis (n = 124) | Low fibrosis (n = 172) | High fibrosis (n = 48) | p Value |
|---|---|---|---|---|
| Age (years) | 0.347 | |||
| ≥50 | 77 (62.1%) | 102 (59.3%) | 34 (70.8%) | |
| <50 | 47 (37.9%) | 70 (40.7%) | 14 (29.2%) | |
| Menopause | 0.826 | |||
| Premenopause | 47 (37.9%) | 62 (36.0%) | 14 (29.2%) | |
| Postmenopause | 76 (61.3%) | 108 (62.8%) | 34 (70.8%) | |
| Unknown | 1 (0.8%) | 2 (1.2%) | 0 (0.0%) | |
| PAM50 | 0.691 | |||
| Basal | 76 (61.3%) | 102 (59.3%) | 30 (62.5%) | |
| Other | 16 (12.9%) | 31 (18.0%) | 9 (18.8%) | |
| Unknown | 32 (25.8%) | 39 (22.7%) | 9 (18.8%) | |
| Histology | 0.220 | |||
| IDC | 115 (92.7%) | 164 (95.3%) | 47 (97.9%) | |
| ILC | 2 (1.6%) | 0 (0.0%) | 1 (2.1%) | |
| DCIS | 0 (0.0%) | 1 (0.6%) | 0 (0.0%) | |
| MC | 5 (4.0%) | 2 (1.2%) | 0 (0.0%) | |
| Others | 2 (1.6%) | 5 (2.9%) | 0 (0.0%) | |
| Grade | 0.226 | |||
| 2 | 25 (20.2%) | 31 (18.0%) | 6 (12.5%) | |
| 3 | 89 (71.8%) | 136 (79.1%) | 40 (83.3%) | |
| Unknown | 10 (8.1%) | 5 (2.9%) | 2 (4.2%) | |
| T stage | 0.101 | |||
| 1 | 58 (46.8%) | 55 (32.0%) | 18 (37.5%) | |
| 2 | 64 (51.6%) | 113 (65.7%) | 30 (62.5%) | |
| 3 | 2 (1.6%) | 4 (2.3%) | 0 (0.0%) | |
| N stage | 0.367 | |||
| 0 | 82 (66.1%) | 95 (55.2%) | 32 (66.7%) | |
| 1 | 32 (25.8%) | 49 (28.5%) | 11 (22.9%) | |
| 2 | 6 (4.8%) | 19 (11.0%) | 2 (4.2%) | |
| 3 | 3 (2.4%) | 7 (4.1%) | 3 (6.3%) | |
| Unknown | 1 (0.8%) | 2 (1.2%) | 0 (0.0%) | |
| Necrosis | <0.001 | |||
| 0 | 89 (71.8%) | 92 (53.5%) | 12 (25.0%) | |
| 1 | 35 (28.2%) | 80 (46.5%) | 36 (75.0%) | |
| Radiotherapy | 0.547 | |||
| Received | 94 (75.8%) | 121 (70.3%) | 36 (75.0%) | |
| Not received | 30 (24.2%) | 51 (29.7%) | 12 (25.0%) | |
| Chemotherapy | 0.267 | |||
| Received | 118 (95.2%) | 160 (93.0%) | 44 (91.7%) | |
| Not received | 1 (0.8%) | 6 (3.5%) | 2 (4.2%) | |
| Unknown | 5 (4.0%) | 3 (3.5%) | 2 (4.2%) | |
| Molecular subtype | 0.353 | |||
| BLIS | 33 (26.6%) | 54 (31.4%) | 19 (39.6%) | |
| IM | 29 (23.4%) | 29 (16.9%) | 6 (12.5%) | |
| LAR | 19 (15.3%) | 33 (19.2%) | 6 (12.5%) | |
| MES | 11 (8.9%) | 17 (9.9%) | 8 (16.7%) | |
| Unknown | 32 (25.8%) | 39 (22.7%) | 9 (18.8%) | |
Data are presented as number (percentage) of patients unless otherwise stated. Statistical analysis was performed with the Pearson's χ2 test. All statistical tests exclude the unknowns.
DCIS, ductal carcinoma in situ; IDC, Invasive ductal carcinoma; ILC, Invasive Lobular Carcinoma; MC, medullary carcinoma; BLIS, Basal-like and immune-suppressed subtype; IM, Immunomodulatory subtype; LAR, Luminal androgen receptor (LAR) subtype; MES, Mesenchymal-like (MES) subtype.
In addition, univariate and multivariate analyses were performed with the Cox regression model. The detailed results are given in Table 2. Multivariate analysis revealed that high fibrosis (hazard ratio [HR], 3.40; 95% confidence interval [CI], 1.40–8.28; p = 0.016), together with tumor size (HR, 1.28; 95% CI, 1.00–1.64; p = 0.048) and positive lymph nodes (HR, 1.07; 95% CI, 1.04–1.10; p < 0.001) predicted a worse RFS in TNBC (Figure 2E). Interestingly, we found that low stromal TILs (sTILs; <50%), which notably predicted a worse RFS in the univariate analysis (HR, 4.79; 95% CI, 1.17–19.68; p = 0.03), were not significant in the multivariate analysis (HR, 2.75; 95% CI, 0.64–11.77; p = 0.17). Next, we performed an interaction test to probe the effects of fibrosis on sTILs and other factors. With respect to the analyses stratified by fibrosis, there were statistically significant interactions between sTILs, positive lymph nodes, size and fibrosis (all p values for interaction < 0.001) (Figure 2F). These results showed that fibrosis was an independent prognostic factor for TNBC. Importantly, fibrosis might have complex effects on the proliferation and metastasis of cancer cells and the TME.
Table 2.
Univariate and multivariate Cox proportional hazard model for RFS
| Variables | Univariate |
Multivariate |
|||
|---|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | ||
| Ages (years) | 0.99 (0.97-1.02) | 0.656 | Not included | ||
| Tumor size (cm) | 1.37 (1.09–1.71) | 0.007 | 1.28 (1.00–1.64) | 0.048 | |
| Positive lymph nodes (number) | 1.08 (1.05–1.12) | <0.001 | 1.07 (1.04–1.10) | <0.001 | |
| PAM50 subtypes | Basal | Ref | Not included | ||
| Other | 1.20 (0.61–2.39) | 0.595 | |||
| Necrosis | 0 | Ref | Not included | ||
| 1 | 1.43 (0.83–2.45) | 0.194 | |||
| sTILs | ≥50% | Ref | |||
| <50% | 4.79 (1.17–19.68) | 0.03 | 2.75 (0.64–11.77) | 0.17 | |
| iTILs | 0.98 (0.9–1.01) | 0.197 | Not included | ||
| Fibrosis | No fibrosis | Ref | |||
| Low fibrosis | 2.37 (1.12–5.01) | 0.024 | 2.19 (0.98–4.91) | 0.057 | |
| High fibrosis | 4.52 (1.98–10.33) | 0.007 | 3.40 (1.40–8.28) | 0.016 | |
Genomic alterations in TNBCs with high fibrosis
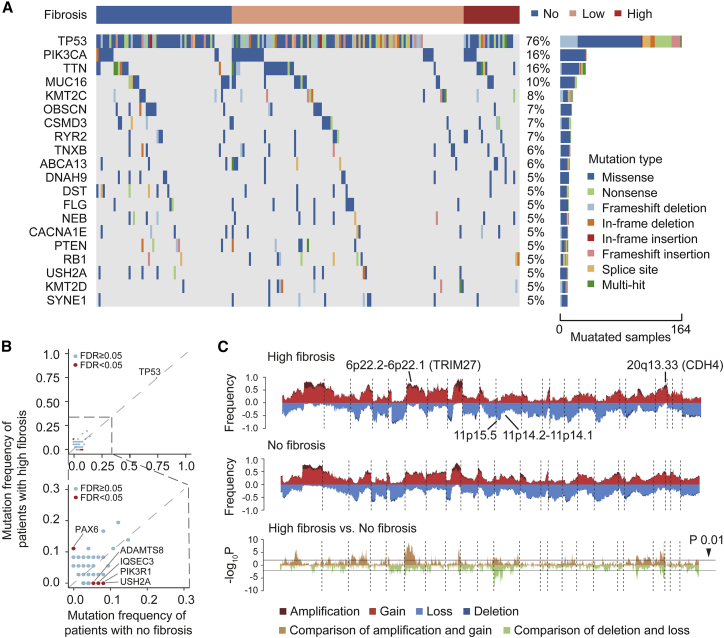
To describe the genomic alterations correlated with high fibrosis in TNBC, we analyzed copy number alterations and somatic mutations using data from the FUSCC cohort datasets. Generally, TNBCs with high fibrosis exhibited a mutation landscape similar to that of the no fibrosis group. Most frequently mutated genes in all TNBC samples included TP53 (76%), PIK3CA (16%), TTN (16%), MUC16 (10%), and KMT2C (8%) (Figure 3A). Compared with the no fibrosis group, the high fibrosis group had a lower frequency of PIK3R1 and USH2A mutations (both 0% vs 8%), and a higher frequency of PAX6 mutation (11% vs 0%) than the no fibrosis group (Figures 3B and Table S2).
Figure 3.
Genomic alterations of fibrotic TNBCs
(A) Oncoplot depicting the top 20 most frequently mutated genes and their mutation types per subgroups of fibrosis in TNBC. Different mutation types are shown in different colors and the percentage (%) of patients affected is shown in the paragraph. (B) Comparison of mutation frequency between the high fibrosis group and the no fibrosis group (calculated using Fisher's exact test). Significantly different mutated genes (mean mutation frequency >3% and a false discovery rate [FDR] of <0.05) are highlighted in red. (C) Comparison of the SCNVs between the high fibrosis group and the no fibrosis group. The top plot illustrates the frequency of the amplification (dark red), gain (light red), loss (light blue), and deletion (dark blue) of each gene in each group, and the bottom plot illustrates the –log10 p value of each gene when comparing the frequency of loss or deletion or gain or amplification between the high fibrosis group and the no fibrosis group.
We further explored the correlations between somatic copy number variations (SCNVs) and fibrosis. SCNVs at the level of genes identified by GISTIC analysis were summarized for each subgroup of fibrosis. SCNV analysis demonstrated that the amplifications or gains of 6p22.2–6p22.1 (TRIM27, 74% vs. 42%) and 20q13.33 (CDH4, 78% vs. 44%) were more frequent in the high fibrosis group than the no fibrosis group (all p values < .001) (Figures 3C and Table S3). Specific losses or deletions existing in the high fibrosis group included 11p15.5 (61% vs. 28%) and 11p14.2-11p14.1 (43% vs. 16%) (all p values < 0.001) (Figures 3C and Table S4). Copy number alterations might be the driver factors for high fibrosis. TRIM27 has been reported to activate the TGF-β signaling pathway and promote epithelial-mesenchymal transition (EMT).14, 15, 16, 17, 18 The CDH4 gene, encoding retinal cadherin (R-cadherin), is upregulated in a variety of sarcomas and has the function of activate Rac1 and c-Jun N-terminal kinase pathways, which are associated with many fibrotic diseases.19, 20, 21, 22 Overall, our analysis revealed significant differential genomic alterations in TNBCs with high fibrosis, which might drive the formation of fibrosis.
Transcriptome pathway enrichment analysis revealed the activation of TGF-β and hypoxia pathways in TNBCs with high fibrosis
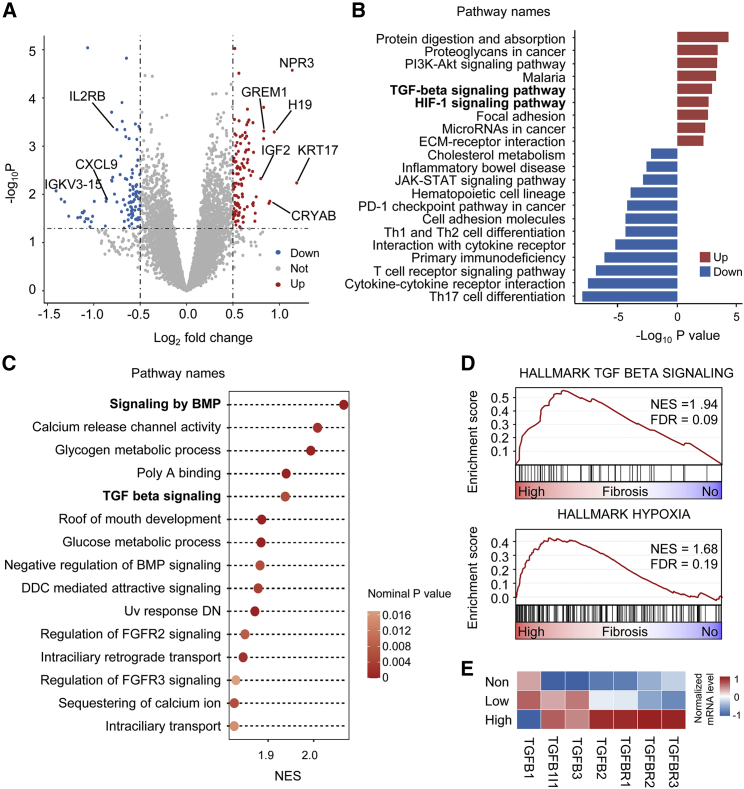
We used RNA-seq to explore aberrant transcription correlated with fibrosis grades. First, we identified 219 differentially expressed genes (DEGs), of which 112 were significantly upregulated and 107 were significantly downregulated in the high fibrosis group (Figure 4A, Table S5).
Figure 4.
TGF-β and hypoxia pathways were enriched in TNBCs with high fibrosis
(A) Volcano plot of differentially expressed genes between the high fibrosis group and the no fibrosis group. A total of 219 genes exhibited significant differential expression (p value < 0 05, |log2 (fold change) | > 0.5). (B) Upregulated and downregulated pathways in the high fibrosis group based on KEGG pathway enrichment analysis of differentially expressed genes. (C–D) Summary of GSEA results (C) and representative GSEA plots (D) enriched in the high fibrosis group versus the no fibrosis group. Significance is represented by the bubble color. NES, normalized enrichment score. (E) Gene expression levels of the TGF-β family and receptors of the three subgroups of fibrosis. ∗p < 0.05. FGFR2, fibroblast growth factor receptor 2.
The most significantly upregulated genes included KRT17, CRYAB, and GREM1, which have been reported to be related to the extracellular matrix (ECM) of tumors.23, 24, 25 To analyze the biological significance of abnormal gene transcription, we performed pathway enrichment analysis based on all DEGs. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis showed that the upregulated pathways in the high fibrosis group included the TGF-β signaling pathway, hypoxia-inducible factor 1 signaling pathway and the ECM-receptor interaction pathway (Figure 4B). Pathways downregulated in high fibrosis were the immune-related Th17 cell differentiation pathway, cytokine-cytokine receptor interaction and the T-cell receptor signaling pathway. Next, gene set enrichment analysis (GSEA) showed that the TGF-β, bone morphogenetic protein (BMP), and fibroblast growth factor receptor 2 signaling pathways were enriched in the high fibrosis group (Figure 4C). The GSEA results of two hallmark gene sets, TGF_BETA_SIGNALING and HALLMARK_HYPOXIA, are shown in the Figure 4D. Next, the gene expression levels of TGF-β and receptor family members were compared among subgroups of fibrosis. The expression of TGFB1I1 and TFGBR3 was significantly higher in the high fibrosis group (p = 0.03 and 0.04, respectively), and other molecules showed a trend toward upregulation, although it was not significant (Figure 4E). Overall, the TGF-β signaling pathway and hypoxia pathway were upregulated, while the immune-related pathways were downregulated in tumors with high fibrosis.
Immunosuppressive TME in TNBCs with high fibrosis
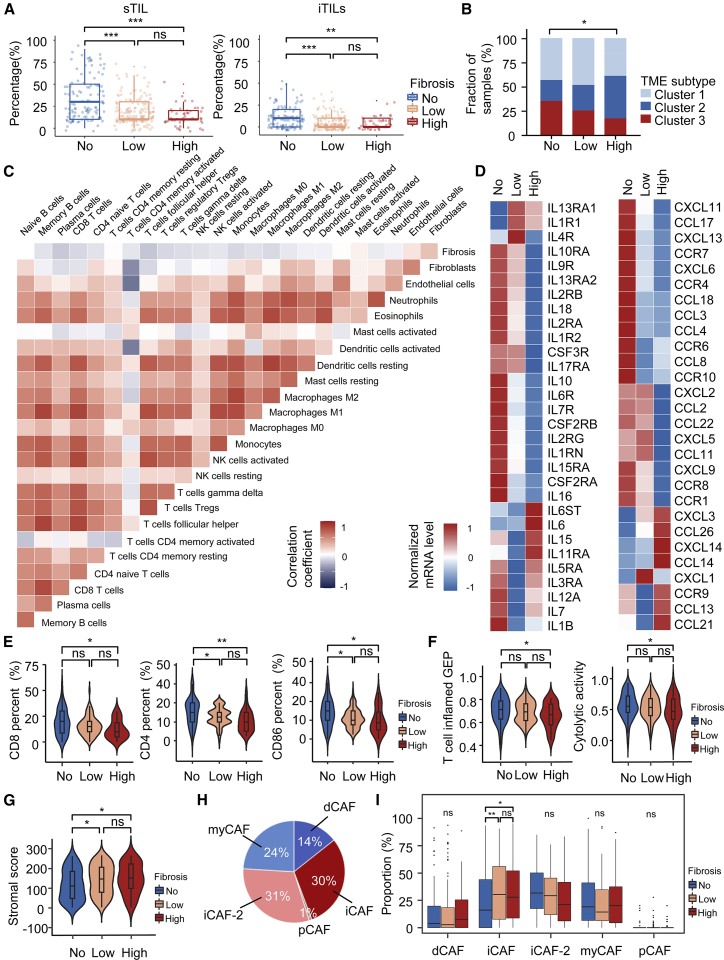
We further characterized the tumor immune microenvironment of different grades of fibrosis. First, sTILs and intratumoral TILs (iTILs) in different groups of fibrosis were compared. Both sTILs and iTILs were significantly higher in the no fibrosis group (all p values < 0.01) than the low and high fibrosis groups (Figure 5A, Table S1). We next analyzed the distribution of TME subtypes of TNBC, which was proposed by Xiao et al.26 before, among the subgroups of fibrosis. The no fibrosis group contained a higher proportion of patients in cluster 3 (the immune-inflamed cluster) than the high and low fibrosis groups (p = 0.018) (Figure 5B). Moreover, we used transcriptomic data and the ssGSEA algorithm to analyze the immune infiltration of 24 types of TME cells and their correlations with fibrosis. We found that the grade of fibrosis was negatively correlated with the infiltration of most immune cells, including follicular helper T cells (R = −0.21), CD8+ T cells (R = −0.15), and M1 macrophages (R = −0.15), but was positively correlated with fibroblasts (R = 0.25; all p values < 0.05) (Figure 5C). The expression of chemokines, interleukins, and their receptors also showed a downregulation trend in the high fibrosis group (Figure 5D). We evaluated the abundance of helper T cells, cytotoxic T cells, and M1 macrophages by immunohistochemical (IHC) staining of CD4, CD8, and CD86, respectively.27, 28, 29, 30, 31 The no fibrosis group had a significantly greater proportion of CD4 positive cells (15.92% vs. 10.29%; p = 0.0011), CD8-positive cells (19.3% vs. 12.12%; p = 0.0126), and CD86 positive cells (14.03% vs. 9.82%; p = 0.0120) than the high fibrosis group (Figure 5E). Representative IHC results were shown in Figure S1 and evaluation results of CD4, CD8, and CD86 were shown in Table S6. Next, we compared the immune signature scores among the three subgroups of fibrosis, including cytolytic activity and T-cell inflamed gene expression profile. The high fibrosis group showed a decrease in cytolytic activity (p = 0.049) and T-cell inflammation (p = 0.021) signatures compared with the no fibrosis group, which suggested that fibrosis was might be associated with T-cell hypofunction (Figure 5F). In addition to immune cells, stromal heterogeneity was analyzed among different subgroups of fibrosis. We used the ESTIMATE algorithm to calculate stromal scores, and our results showed that the stromal scores in tumors with no fibrosis were significantly lower than those in the low or high fibrosis groups (p = 0.019 and 0.047, respectively) (Figure 5G). To investigate cancer-associated fibroblast (CAF) subtypes in this cohort, we referred to the CIBERSORTx algorithm to predict the proportion of different kinds of CAF subtypes in each sample based on RNA-seq data. This method was proposed by Galbo et al., who used the CIBERSORTx algorithm to classify CAFs into five main subtypes: myofibroblast-like CAFs (pan-myCAFs), desmoplastic CAFs (pan-dCAFs), inflammatory-like CAFs (pan-iCAFs), pan-iCAFs-2, and proliferating CAFs (pan-pCAFs).32 Thirty-one percent of CAFs in this cohort were pan-iCAFs-2, 30% were pan-iCAFs, 24% were pan-myCAFs, 14% were pan-dCAFs, and 1% were pan-pCAFs (Figure 5H). Pan-iCAFs were significantly upregulated in the high fibrosis group compared with the no fibrosis group (32.2% vs. 24.0%; p = 0.041), while other pan-CAF subtypes did not show a significant difference among the three fibrosis groups (Figure 5I). Previous studies have revealed that iCAFs could inhibit the function of cytotoxic T cells and resulted in an immunosuppressive environment in TNBC, which is consistent with our study.33,34
Figure 5.
Immunosuppressive microenvironment induced by fibrosis
(A) TIL percentages of different subgroups of fibrosis. (B) Bar plots showing the distribution of the TNBC TME subtypes among the three subgroups of fibrosis. (C) Correlations between fibrosis and the abundance of 24 types of TME cells calculated through single-sample GSEA (ssGSEA). (D) Expression of immune-related secretory molecules, including chemokines (left) and interleukins (right), among the three subgroups of fibrosis. The p values were calculated by the Kruskal-Wallis test. (E) Comparison of IHC staining results for CD8, CD4, and CD86 among different subgroups of fibrosis. (F) Expression of two immune-related signatures, cytolytic activity and T cell inflamed gene expression profiles (GEPs) of three fibrotic subgroups. (G) Comparison of the stromal scores calculated by ESTIMATE algorithm of three subgroups of fibrosis. (H) Overall distribution of pan-CAF subtypes in the Fudan University Shanghai Cancer Center TNBC cohort. (I) Fraction of pan-CAF subtypes in different subgroups of fibrosis. (ns P ≥ 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Given the importance of immunomodulators in the TME, we questioned whether there existed a correlation between fibrosis and immunomodulators. Therefore, we compared the expression levels of immunomodulators, including costimulatory and coinhibitory molecules, among different grades of fibrosis. The expression of costimulatory (all p values > 0.05) and coinhibitory molecules (most p values > 0.05) in patients with no fibrosis was close to or higher than that in the high fibrosis group (Figure S2). These results suggested that fibrosis might affect antitumor immunity through mechanisms other than those affecting the expression of costimulatory or coinhibitory molecules. We inferred that TGF-β might inhibit the infiltration of antitumor immune cells, such as cytotoxic T cells and M1 macrophages and improve the activation of fibroblasts to produce more ECM, ultimately resulting in a fibrotic immunosuppressive TME.
Discussion
FF has been observed in breast cancer for more than 20 years. However, the mechanism of the formation of the FF and the reason why it predicts a poor outcome remain poorly understood. In this study, we defined fibrosis grades of TNBC based on the evaluation of FF. High fibrosis was an independent prognostic factor and it interacted with TILs in TNBC. Moreover, we explored the molecular features of fibrosis in TNBC at the genomic and transcriptomic levels. We propose that high fibrosis might promote the formation of an immunosuppressive TME through the activation of the TGF-β pathway and result in a poor prognosis in TNBCs. To the best of our knowledge, this study is the first systematic analysis of fibrosis in TNBC based on a dataset comprising a comprehensive pathological evaluation and multiomics data.
Taking advantage of the genomic and transcriptomic data, our study explored the molecular mechanism that might contribute to the formation of fibrosis. Gains of (TRIM27) and 20q13.33 (CDH4), together with losses of 11p.15.5 and 11p14.2-11p14.1 were significant characteristics of tumors with high fibrosis, which might be driver factors of fibrosis. The copy number gains of oncogenes such as TRIM27 and CDH4 have been reported to promotes the EMT or fibrosis.15,16,20 In addition, the losses of 11p15.5 have been confirmed to be associated with several malignancies, such as Wilms' tumor and rhabdomyosarcoma.35 In addition, genes upregulated in TNBCs with high fibrosis are potential driver alterations of fibrosis at the transcriptional level. KRT17, the most significant DEG, has been proven to promote the proliferation of liver cancer cells and EMT in hepatic fibrosis via TGF-β1 signaling.24 Insulin-like growth factor 2 (IGF2), upregulated in the high fibrosis group, are regulated by two different imprinting control regions located on 11p15.5.36 The loss of heterozygosity of 11p15.5 can lead to the overexpression of IGF2.37 IGF2 has been previously reported as a tumor-promoting gene and activates the function of fibroblasts in the tumor stroma.38, 39, 40 GREM1, also upregulated in the high fibrosis group, regulates stromal BMP signaling to promote colorectal carcinogenesis.23 These results support the hypothesis that copy number gains of (TRIM27) and 20q13.33 (CDH4), losses of 11p.15.5 and alterations of some key genes might drive the formation of fibrosis.
Our study also stressed the relationship among fibrosis, immunosuppressive TME, and a poor prognosis. Prognosis analyses revealed that fibrosis was an independent prognostic indicator and had interactions with low sTILs and positive lymph nodes. Interactions between fibrosis and low sTILs reflect that fibrosis has important effects on the components or status of TILs. In addition, immune-related pathways were downregulated in tumors with high fibrosis; thus, we analyzed the effects of fibrosis on the TME. Previous research has reported that there are fewer TILs in tumors in the presence of FF, which is in agreement with our results.12 However, TILs include various types of lymphocytes and, in addition to lymphocytes, many other TME cells, such as macrophages, natural killer cells, and fibroblasts, also play important roles in tumor immunity. Consequently, it is necessary to understand the characteristics of the TME in different grades of fibrosis from a holistic perspective. Our study systematically analyzed the correlations of immune and stromal cell infiltration with fibrosis. Additionally, we systematically compared the expression of immune-related genes and signatures between the high and low fibrosis groups.
Recently, three major immune phenotypes have been proposed in cancer, namely, the immune-inflamed, immune-excluded, and immune-desert phenotypes.41 Given the low expression of coinhibitory molecules in tumors with high fibrosis, highly fibrotic TNBC is more likely to be an immune-excluded cancer type. Greater quantities of ECM could be predicted as a consequence of activated fibroblasts, resulting in pathological collagen deposition. Excessive ECM might become a barrier preventing immune cell infiltration, and activated CAFs might collectively decrease the antitumor immune response.
Given that fibrosis is a simple and objective indicator to evaluate, we suppose that fibrosis has the clinical potential to stratify patients with TNBC. Due to the poor prognosis of high fibrosis in TNBC, optimized treatment strategies are necessary for this group. Our study revealed that the activation of the TGF-β pathway is a characteristic of tumors with high fibrosis. Previous studies have revealed that the TGF-β pathway regulates fibrotic process and promotes immune escape.42,43 TGF-β is able to activate or differentiate resident fibroblasts and endothelial cells into CAFs in the TME and suppress the function of immune cells such as cytotoxic CD8+ T cells or antitumor macrophages.44 The above theories present prospects for targeting TGF-β against TNBCs with high fibrosis. The efficacy of inhibiting of TGF-β using inhibitors of TGF-β receptors or ligands, as well as antisense oligonucleotides or small molecule receptor kinase inhibitors, needed to be verified in clinical trials.45,46 In addition, there remains a lack of predictive biomarkers to screen appropriate populations who can benefit from the blockade of TGF-β.47 Consequently, we need to explore whether fibrosis could become a candidate biomarker to predict the efficacy of TGF-β inhibitors.
Undeniably, this article has certain limitations. First, our analysis could only provide associations, and the speculations should be further confirmed. It is reassuring that previous studies are consistent with the key conclusions. The important role of TGF-β in the fibrosis and TME has been widely studied.48, 49, 50 Second, there are no external cohorts to validate our results. After extensive investigation, we found that multiomics data from large sample cohorts of TNBC generally lack the evaluation of FF or fibrosis. Therefore, it is necessary to promote the application of FF evaluation in the clinic and accumulate more omics data with the evaluation of FF. Additional experimental studies and the use of additional techniques like single-cell RNA-seq or mass cytometry are required to further investigate how fibrotic processes shape the TME.
In conclusion, our study identified fibrosis as an independent prognostic indicator and revealed the molecular features of fibrosis in TNBC. High fibrosis is characterized by TGF-β pathway activation and an immunosuppressive TME, which leads to a poor prognosis.
Materials and methods
Patient samples and datasets
Samples were retrospectively selected from 344 patients with TNBC from the Fudan University Shanghai Cancer Center TNBC multiomics cohort (The National Omics Data Encyclopedia:OEP000155).3 The FF of all patients were evaluated by experienced pathologists. A detailed evaluation process of FF was described in the following section. RNA-seq, WES, and somatic copy number alteration data were available for 264, 223, and 315 samples, respectively. The clinicopathological information for all patients were shown in Table S1. All samples were previously untreated primary breast cancers. All procedures were conducted in accordance with the Helsinki Agreement and with approval from the independent ethics committee/Institutional Review Board at Fudan University Shanghai Cancer Center Ethical Committee. Ethical approval and informed consent were obtained.
Histopathological analysis
Surgically resected specimens were fixed with formalin, sectioned, and stained with hematoxylin and eosin. Two certified pathologists independently evaluated all histopathological diagnoses. In case of a discordant outcome, the specimens were reviewed by a third independent expert pathologist. The presence of FF in breast cancer was evaluated according to the previously reported criteria: (1) grade 0, samples without FF; (2) grade 1, a small amount of collagen fibers with abundant fibroblasts in FF; (3) grade 3, mainly composed of hyalinized collagen fibers; and (4) grade 2, intermediate between grade 1 and 3, with fibroblasts and collagen fibers admixed in different proportions.6 The fibrosis grade of TNBC was classified into three grades depending on the result of the FF evaluation: (1) no fibrosis, samples without FF, corresponding with grade 0 FF; (2) low fibrosis, samples with grade 1 and 2 FF; and (3) high fibrosis, samples with grade 3 FF. Samples of grades 1 and 2 FF were grouped together as low fibrosis because there were no strict bounds between the two groups.
The evaluation of TILs
We evaluated TILs on hematoxylin and eosin-stained slides according to the International TIL Working Group guidelines.51 According to the guideline, TILs are defined as mononuclear immune cells that infiltrate in the tumor region (including lymphocytes and plasma cells, excluding polymorphonuclear leukocytes). The iTILs are defined as lymphocytes in tumor nests having cell-to-cell contact with no intervening stroma and directly interacting with carcinoma cells, while sTILs are located dispersed in the stroma between the carcinoma cells and do not directly contact carcinoma cells. The sTILs and iTILs were reported as the ratio of the area occupied by lymphocytes to the total area of the tumor region and recorded in 5% increments. Two experienced pathologists independently evaluated the histopathological results. Evaluation results of TILs were shown in the Table S1. We set a cutoff value of 50% in sTILs to define the high and low sTILs infiltrating groups.52
Immunohistochemistry
We performed immunohistochemical staining on paraffin-embedded sections (4 μm thick) of tumor specimens from the FUSCC cohort (n = 165) to evaluate the expression of CD4, CD8, and CD86. Sections were deparaffinized and rehydrated through a graded series of xylene-ethanol baths. Antigen retrieval was performed using a citrate buffer, pH 6.0, for 20 min at 95°C. Tissue sections were incubated with primary antibody for 1 h at room temperature. The antibodies were used in a 1:200 dilution for anti-CD4 (Clone EPR6855, Abcam), without dilution for anti-CD8 (clone SP57, Ventana), and 1:200 dilution for anti-CD86 (13395-1-AP, Proteintech). Visualization was performed using the Novolink Polymer Detection Systems (Leica Biosystems). Tissue sections were counterstained with hematoxylin, dehydrated, and mounted. The analysis was performed in a blinded fashion. Images were organized in folders identified by patient ID by one person and quantified by another. Results were recorded as percentage of IHC-stained cells and adjudicated by two pathologists.
Prognostic analysis of different grades of fibrosis
The median follow-up length was 60.5 months (interquartile range, 40.9–69.8 months). OS, RFS, and DMFS were taken as prognostic outcomes. RFS and DMFS were defined as the duration from the date of treatment to relapse or distant metastasis. The Kaplan-Meier method was used to estimate the survival curves for RFS, DMFS, and OS. Univariate and multivariate survival analyses were performed using a logistic regression model to analyze the prognostic significance of fibrosis grades. Age, tumor size, the number of positive lymph nodes, PAM50 subtypes, necrosis, sTILs, iTILs, and fibrosis grades were first analyzed in a univariate Cox proportional hazards model. All significant variables were then included as covariates in the multivariate Cox proportional hazards model. Interactions were tested with a threshold of p < 0.05, and, in cases of interactions, a stratified analysis was conducted. The prognostic analysis was performed using SPSS (IBM SPSS 22.0, SPSS Inc.).
Comparison of somatic mutations and SCNVs among the different grades of fibrosis
A mutation analysis was based on the WES data of the FUSCC cohort. The frequencies of mutations were compared between patients with high and no fibrosis by Fisher's exact test. Genes with mean mutation frequencies greater than 3% were included in the comparison. A p value of less than 0.05 after adjusting for mutation load was considered significant.
We adjusted the segmented copy number calls through the ASCAT algorithm by estimating ploidy and purity, and the gene level of the copy number variations (CNVs) was obtained by the GISTIC tool.53,54 The copy number status by sample was reported as −2 (deletion), −1 (loss), 0 (diploid status), 1 (gain), and 2 (amplification) levels. To identify specific copy number gains and losses associated with fibrosis, the comparison of CNV frequencies in patients with high fibrosis versus those with no fibrosis was performed using a two-tailed Fisher's exact test twice. The first comparison was gain centric, and thus, the frequencies of patients whose copy numbers were 1 or more in whole populations were compared between the two groups. The group-specific gains were defined as a continuous stretch of genes (arranged in genomic order) with an unadjusted p value of less than 0.01. Similarly, the second comparison was loss centric, and thus, the frequencies of patients whose copy numbers were −1 or less in whole populations were compared between the two groups. The group-specific losses were defined as a continuous stretch of genes (arranged in genomic order) with an unadjusted p value of less than 0.01.
Differential gene expression and pathway enrichment analysis
The R package limma was used to unveil differentially expressed genes (DEGs) between samples with high and no fibrosis.55 Any p values of less than 0.05 and |log2FC| of greater than 0.5 were considered to be statistically significant for DEGs. The DEGs were subjected to the KEGG pathway enrichment analysis. GSEA was performed using GSEA software (v4.0.3).56 Gene sets from the Molecular Signature Database were tested, and two thousand total permutations were used.57 Gene expression levels of the transforming growth factor-beta (TGF-β) signaling pathway were compared in the subgroups of fibrosis and the gene list was obtained from the ImmPort database (http://www.immport.org/).
Calculation of the microenvironment cell abundance
The abundance of immune infiltrating cells in the TNBC samples was quantified using the single sample GSEA algorithm with expression data.58 The gene sets comprised 354 genes that could predict the abundance of TME cells, including 22 immune cells, together with fibroblasts and endothelial cells.26 Spearman correlation analyses were used to compute correlation coefficients between the grades of fibrosis and the TME-infiltrating cells. Stromal scores were calculated by the ESTIMATE algorithm to analyze the infiltration levels of stromal cells in different grades of fibrosis.59 To investigate CAF subtypes in this cohort, we referred to the CIBERSORTx algorithm to predict the proportion of different kinds of CAF subtypes in each sample based on RNA-seq data.32,60 We downloaded single-cell RNA-seq datasets from the public database (GSE:GSE72056 and GSE103322) referred to the previous article of Galbo et al.32 to build a gene signature matrix file with pan-CAF clustering labels. And then we applied the gene signature matrix to impute the abundances of CAF subtypes in bulk RNA-seq data of the FUSCC cohort on the CIBERSORTx web server (https://cibersortx.stanford.edu/).
Immune signature scores and immune-related molecule comparisons between the different groups of fibrosis
The T-cell status was inferred by using two gene signatures characterizing T-cell inflammation status and T-cell cytolytic activity.61,62 Gene expression levels of chemokines, interleukins, costimulatory molecules, and coinhibitory molecules were compared between the different groups of fibrosis. A list of chemokines and interleukins was downloaded from the ImmPort database (http://www.immport.org/), and the gene list of the immunomodulators was obtained from a database of costimulatory and coinhibitory molecules (https://www.rndsystems.com/cn/research-area/co--stimulatory-and-co--inhibitory-molecules).
Statistical analysis
Differences between continuous variables were assessed by the Student t-test, Wilcoxon rank-sum test, and Kruskal-Wallis test. The Shapiro-Wilk test was first used to check whether the data followed a normal distribution. Categorical variables were compared using χ2 tests or the Fisher exact tests when needed. A permutation test was conducted when comparing gene mutation frequencies among different groups. Pearson or Spearman correlation was used for correlation analysis. All of the tests were two-sided, and a p value of less than 0.05 indicated significance unless otherwise stated. The false discovery rate correction was used in multiple tests to decrease the false-positive rates. Statistical analysis was performed using R software (version 3.6.3, http://www.R-project.org/).
Acknowledgments
This study was supported by grants from the National Key Research and Development Project of China (2020YFA0112304), the National Natural Science Foundation of China (82072922 and 82002792), the Shanghai Rising-Star Program (20YF1408600), the Clinical Research Plan of SHDC (SHDC2020CR4002, SHDC2020CR5005), and the Shanghai Key Laboratory of Breast Cancer (12DZ2260100). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors thank BIORENDER for the figures.
Author contributions
G.-H.D., Z.-M.S., Y.-Z.J., and Y.X. designed the study and contributed to writing the manuscript. J.-H.D. and S.Z. designed and performed the research, analyzed data, and wrote the manuscript. Y.X. and Y.-L.X. provided essential assistance and analyzed data. All authors read and approved the final manuscript.
Declaration of interests
The authors of this manuscript declare no conflicts of interest.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2022.02.003.
Contributor Information
Yi Xiao, Email: yixiao11@fudan.edu.cn.
Yi-Zhou Jiang, Email: yizhoujiang@fudan.edu.cn.
Gen-Hong Di, Email: genhongdi@163.com.
Supplemental information
References
- 1.Boyle P. Triple-negative breast cancer: epidemiological considerations and recommendations. Ann. Oncol. 2012;23 doi: 10.1093/annonc/mds187. vi7–12. [DOI] [PubMed] [Google Scholar]
- 2.Gruosso T., Gigoux M., Manem V.S.K., Bertos N., Zuo D., Perlitch I., Saleh S.M.I., Zhao H., Souleimanova M., Johnson R.M., et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J. Clin. Invest. 2019;129:1785–1800. doi: 10.1172/JCI96313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang Y.Z., Ma D., Suo C., Shi J., Xue M., Hu X., Xiao Y., Yu K.D., Liu Y.R., Yu Y., et al. Genomic and transcriptomic landscape of triple-negative breast cancers: subtypes and treatment strategies. Cancer Cell. 2019;35:428–440 e425. doi: 10.1016/j.ccell.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Pareja F., Geyer F.C., Marchio C., Burke K.A., Weigelt B., Reis-Filho J.S. Triple-negative breast cancer: the importance of molecular and histologic subtyping, and recognition of low-grade variants. NPJ Breast Cancer. 2016;2:16036. doi: 10.1038/npjbcancer.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdul Aziz A.A., Md Salleh M.S., Ankathil R. Clinicopathological and prognostic characteristics of Malaysian triple negative breast cancer patients undergoing TAC chemotherapy regimen. Int. J. Breast Cancer. 2020;2020:8424365. doi: 10.1155/2020/8424365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van den Eynden G.G., Colpaert C.G., Couvelard A., Pezzella F., Dirix L.Y., Vermeulen P.B., Van Marck E.A., Hasebe T. A fibrotic focus is a prognostic factor and a surrogate marker for hypoxia and (lymph)angiogenesis in breast cancer: review of the literature and proposal on the criteria of evaluation. Histopathology. 2007;51:440–451. doi: 10.1111/j.1365-2559.2007.02761.x. [DOI] [PubMed] [Google Scholar]
- 7.Mujtaba S.S., Ni Y.B., Tsang J.Y., Chan S.K., Yamaguchi R., Tanaka M., Tan P.H., Tse G.M. Fibrotic focus in breast carcinomas: relationship with prognostic parameters and biomarkers. Ann. Surg. Oncol. 2013;20:2842–2849. doi: 10.1245/s10434-013-2955-0. [DOI] [PubMed] [Google Scholar]
- 8.Ni Y.B., Tsang J.Y., Chan S.K., Tse G.M. A novel morphologic-molecular recurrence predictive model refines traditional prognostic tools for invasive breast carcinoma. Ann. Surg. Oncol. 2014;21:2928–2933. doi: 10.1245/s10434-014-3691-9. [DOI] [PubMed] [Google Scholar]
- 9.Colpaert C.G. The presence of a fibrotic focus in invasive breast carcinoma correlates with the expression of carbonic anhydrase IX and is a marker of hypoxia and poor prognosis. Breast Cancer Res. Treat. 2003;81:137–147. doi: 10.1023/A:1025702330207. [DOI] [PubMed] [Google Scholar]
- 10.Van den Eynden G.G., Smid M., Van Laere S.J., Colpaert C.G., Van der Auwera I., Bich T.X., van Dam P., den Bakker M.A., Dirix L.Y., Van Marck E.A., et al. Gene expression profiles associated with the presence of a fibrotic focus and the growth pattern in lymph node-negative breast cancer. Clin. Cancer Res. 2008;14:2944–2952. doi: 10.1158/1078-0432.CCR-07-4397. [DOI] [PubMed] [Google Scholar]
- 11.Shimada H., Hasebe T., Sugiyama M., Shibasaki S., Sugitani I., Ueda S., Gotoh Y., Yasuda M., Arai E., Osaki A., et al. Fibrotic focus: an important parameter for accurate prediction of a high level of tumor-associated macrophage infiltration in invasive ductal carcinoma of the breast. Pathol. Int. 2017;67:331–341. doi: 10.1111/pin.12550. [DOI] [PubMed] [Google Scholar]
- 12.Yanai H., Yoshikawa K., Ishida M., Tsuta K., Sekimoto M., Sugie T. Presence of myxoid stromal change and fibrotic focus in pathological examination are prognostic factors of triple-negative breast cancer: results from a retrospective single-center study. PLoS One. 2021;16:e0245725. doi: 10.1371/journal.pone.0245725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uematsu T. Triple-negative breast cancer : correlation between MR imaging and pathologic findings. Radiology. 2009;250:638–647. doi: 10.1148/radiol.2503081054. [DOI] [PubMed] [Google Scholar]
- 14.Zoumpoulidou G., Broceno C., Li H., Bird D., Thomas G., Mittnacht S. Role of the tripartite motif protein 27 in cancer development. J. Natl. Cancer Inst. 2012;104:941–952. doi: 10.1093/jnci/djs224. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H.X., Xu Z.S., Lin H., Li M., Xia T., Cui K., Wang S.Y., Li Y., Shu H.B., Wang Y.Y. TRIM27 mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis. Nat. Commun. 2018;9:3441. doi: 10.1038/s41467-018-05796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Feng Y., Ji D., Wang Q., Qian W., Wang S., Zhang Z., Ji B., Zhang C., Sun Y., et al. TRIM27 functions as an oncogene by activating epithelial-mesenchymal transition and p-AKT in colorectal cancer. Int. J. Oncol. 2018;53:620–632. doi: 10.3892/ijo.2018.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S. TRIM27 acts as an oncogene and regulates cell proliferation and metastasis in non-small cell lung cancer through SIX3-β-catenin signaling. Aging. 2020;12:25564–25580. doi: 10.18632/aging.104163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibbs Z.A., Reza L.C., Cheng C.C., Westcott J.M., McGlynn K., Whitehurst A.W. The testis protein ZNF165 is a SMAD3 cofactor that coordinates oncogenic TGFbeta signaling in triple-negative breast cancer. Elife. 2020;9:e57679. doi: 10.7554/eLife.57679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kucharczak J., Charrasse S., Comunale F., Zappulla J., Robert B., Teulon-Navarro I., Pelegrin A., Gauthier-Rouviere C. R-Cadherin expression inhibits myogenesis and induces myoblast transformation via Rac1 GTPase. Cancer Res. 2008;68:6559–6568. doi: 10.1158/0008-5472.CAN-08-0196. [DOI] [PubMed] [Google Scholar]
- 20.Tang Q., Lu J., Zou C., Shao Y., Chen Y., Narala S., Fang H., Xu H., Wang J., Shen J., et al. CDH4 is a novel determinant of osteosarcoma tumorigenesis and metastasis. Oncogene. 2018;37:3617–3630. doi: 10.1038/s41388-018-0231-2. [DOI] [PubMed] [Google Scholar]
- 21.Choi S.S., Sicklick J.K., Ma Q., Yang L., Huang J., Qi Y., Chen W., Li Y.X., Goldschmidt-Clermont P.J., Diehl A.M. Sustained activation of Rac1 in hepatic stellate cells promotes liver injury and fibrosis in mice. Hepatology. 2006;44:1267–1277. doi: 10.1002/hep.21375. [DOI] [PubMed] [Google Scholar]
- 22.Grynberg K., Ma F.Y., Nikolic-Paterson D.J. The JNK signaling pathway in renal fibrosis. Front Physiol. 2017;8:829. doi: 10.3389/fphys.2017.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi H., Gieniec K.A., Wright J.A., Wang T., Asai N., Mizutani Y., Lida T., Ando R., Suzuki N., Lannagan T.R.M., et al. The balance of stromal BMP signaling mediated by GREM1 and ISLR drives colorectal carcinogenesis. Gastroenterology. 2021;160:1224–1239 e1230. doi: 10.1053/j.gastro.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J., Ge S.-J., Feng H.-J., Wu S.-Z., Ji R., Huang W.-R., Huang W., Lu C.-H. KRT17 promotes the activation of HSCs via EMT in liver fibrosis. J. Clin. Transl. Hepatol. 2021;000:000. doi: 10.14218/JCTH.2021.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caporossi D., Parisi A., Fantini C., Grazioli E., Cerulli C., Dimauro I. AlphaB-crystallin and breast cancer: role and possible therapeutic strategies. Cell Stress Chaperones. 2021;26:19–28. doi: 10.1007/s12192-020-01175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao Y., Ma D., Zhao S., Suo C., Shi J., Xue M.Z., Ruan M., Wang H., Zhao J., Li Q., et al. Multi-omics profiling reveals distinct microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer. Clin. Cancer Res. 2019;25:5002–5014. doi: 10.1158/1078-0432.CCR-18-3524. [DOI] [PubMed] [Google Scholar]
- 27.Reuben A., Zhang J., Chiou S.-H., Gittelman R.M., Li J., Lee W.-C., Fujimoto J., Behrens C., Liu X., Wang F., et al. Comprehensive T cell repertoire characterization of non-small cell lung cancer. Nat. Commun. 2020;11:603. doi: 10.1038/s41467-019-14273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espinoza J.A., Jabeen S., Batra R., Papaleo E., Haakensen V., Timmermans Wielenga V., Møller Talman M.-L., Brunner N., Børresen-Dale A.-L., Gromov P., et al. Cytokine profiling of tumor interstitial fluid of the breast and its relationship with lymphocyte infiltration and clinicopathological characteristics. OncoImmunology. 2016;5:e1248015. doi: 10.1080/2162402X.2016.1248015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Souza A.W.S., van Timmeren M., Sanders J.S., Stegeman C., Heeringa P., Kallenberg C.G.M., Westra J. M2 macrophage is the predominant phenotype in airways inflammatory lesions in patients with granulomatosis with polyangiitis. Arthritis Res. Ther. 2017;19:100. doi: 10.1186/s13075-017-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y., Guan Z., Dai X., Shen Y., Wei Q., Ren L., Jiang J., Xiao Z., Jiang Y., Liu D., et al. Engineered macrophages as near-infrared light activated drug vectors for chemo-photodynamic therapy of primary and bone metastatic breast cancer. Nat. Commun. 2021;12:4310. doi: 10.1038/s41467-021-24564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee B.C., Lee J.Y., Kim J. Graphene quantum dots as anti-inflammatory therapy for colitis. Sci. Adv. 2021;6:eaaz2630. doi: 10.1126/sciadv.aaz2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galbo P.M., Jr., Zang X., Zheng D. Molecular features of cancer-associated fibroblast subtypes and their implication on cancer pathogenesis, prognosis, and immunotherapy resistance. Clin. Cancer Res. 2021;27:2636–2647. doi: 10.1158/1078-0432.CCR-20-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu S.Z., Roden D.L., Wang C., Holliday H., Harvey K., Cazet A.S., Murphy K.J., Pereira B., Al-Eryani G., Bartonicek N., et al. Stromal cell diversity associated with immune evasion in human triple-negative breast cancer. EMBO J. 2020;39:e104063. doi: 10.15252/embj.2019104063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kieffer Y., Hocine H.R., Gentric G., Pelon F., Bernard C., Bourachot B., Lameiras S., Albergante L., Bonneau C., Guyard A., et al. Single-cell analysis reveals fibroblast clusters linked to immunotherapy resistance in cancer. Cancer Discov. 2020;10:1330–1351. doi: 10.1158/2159-8290.CD-19-1384. [DOI] [PubMed] [Google Scholar]
- 35.Wesseler K., Kraft F., Eggermann T. Molecular and clinical opposite findings in 11p15.5 associated imprinting disorders: characterization of basic mechanisms to improve clinical management. Int. J. Mol. Sci. 2019;20:4219. doi: 10.3390/ijms20174219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Begemann M., Spengler S., Gogiel M. Clinical significance of copy number variations in the 11p15.5 imprinting control regions: new cases and review of the literature. J. Med. Genet. 2012;49:547–553. doi: 10.1136/jmedgenet-2012-100967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rainier S J.L., Dobry C.J., Ping A.J., Grundy P.E., Feinberg A.P. Relaxation of imprinted genes in human cancer. Nature. 1993;362:747–749. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 38.Mutgan A.C., Besikcioglu H.E., Wang S., Friess H., Ceyhan G.O., Demir I.E. Insulin/IGF-driven cancer cell-stroma crosstalk as a novel therapeutic target in pancreatic cancer. Mol. Cancer. 2018;17:66. doi: 10.1186/s12943-018-0806-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu W.W., Li B., Guan X.Y., Chung S.K., Wang Y., Yip Y.L., Law S.Y., Chan K.T., Lee N.P., Chan K.W., et al. Cancer cell-secreted IGF2 instigates fibroblasts and bone marrow-derived vascular progenitor cells to promote cancer progression. Nat. Commun. 2017;8:14399. doi: 10.1038/ncomms14399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Quetglas I., Pinyol R., Dauch D., Torrecilla S., Tovar V., Moeini A., Alsinet C., Portela A., Rodriguez-Carunchio L., Sole M., et al. IGF2 is up-regulated by epigenetic mechanisms in hepatocellular carcinomas and is an actionable oncogene product in experimental models. Gastroenterology. 2016;151:1192–1205. doi: 10.1053/j.gastro.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 41.Smyth M.J., Ngiow S.F., Ribas A., Teng M.W. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat. Rev. Clin. Oncol. 2016;13:143–158. doi: 10.1038/nrclinonc.2015.209. [DOI] [PubMed] [Google Scholar]
- 42.Meng X.M., Nikolic-Paterson D.J., Lan H.Y. TGF-beta: the master regulator of fibrosis. Nat. Rev. Nephrol. 2016;12:325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 43.Batlle E., Massague J. Transforming growth factor-beta signaling in immunity and cancer. Immunity. 2019;50:924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu S., Ren J., Ten Dijke P. Targeting TGFbeta signal transduction for cancer therapy. Signal Transduct Target Ther. 2021;6:8. doi: 10.1038/s41392-020-00436-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao H., Wei J., Sun J. Roles of TGF-beta signaling pathway in tumor microenvirionment and cancer therapy. Int. Immunopharmacol. 2020;89:107101. doi: 10.1016/j.intimp.2020.107101. [DOI] [PubMed] [Google Scholar]
- 46.Xue V.W., Chung J.Y., Cordoba C.A.G., Cheung A.H., Kang W., Lam E.W., Leung K.T., To K.F., Lan H.Y., Tang P.M. Transforming growth factor-beta: a multifunctional regulator of cancer immunity. Cancers (Basel) 2020;12:3099. doi: 10.3390/cancers12113099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colak S., Ten Dijke P. Targeting TGF-beta signaling in cancer. Trends Cancer. 2017;3:56–71. doi: 10.1016/j.trecan.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 48.Derynck R., Turley S.J., Akhurst R.J. TGFbeta biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 2021;18:9–34. doi: 10.1038/s41571-020-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caja L., Dituri F., Mancarella S., Caballero-Diaz D., Moustakas A., Giannelli G., Fabregat I. TGF-beta and the tissue microenvironment: relevance in fibrosis and cancer. Int. J. Mol. Sci. 2018;19:1294. doi: 10.3390/ijms19051294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morikawa M., Derynck R., Miyazono K. TGF-beta and the TGF-beta family: context-dependent roles in cell and tissue physiology. Cold Spring Harb. Perspect. Biol. 2016;8:a021873. doi: 10.1101/cshperspect.a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salgado R., Denkert C., Demaria S., Sirtaine N., Klauschen F., Pruneri G., Wienert S., Van den Eynden G., Baehner F.L., Penault-Llorca F., et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pruneri G., Vingiani A., Bagnardi V., Rotmensz N., De Rose A., Palazzo A., Colleoni A.M., Goldhirsch A., Viale G. Clinical validity of tumor-infiltrating lymphocytes analysis in patients with triple-negative breast cancer. Ann. Oncol. 2016;27:249–256. doi: 10.1093/annonc/mdv571. [DOI] [PubMed] [Google Scholar]
- 53.Van Loo P., Nordgard S.H., Lingjaerde O.C., Russnes H.G., Rye I.H., Sun W., Weigman V.J., Marynen P., Zetterberg A., Naume B., et al. Allele-specific copy number analysis of tumors. Proc. Natl. Acad. Sci. U S A. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian A T.P., Mootha V.K. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonja Hanzelmann R.C., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics. 2015;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshihara K., Shahmoradgoli M., Martinez E., Vegesna R., Kim H., Torres-Garcia W., Trevino V., Shen H., Laird P.W., Levine D.A., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R., Albright A., Cheng J.D., Kang S.P., Shankaran V., et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.