Abstract
Introduction
Contact lens (CL) wear has been reported to cause changes to the microbiome of the ocular surface. More insight into the alteration of this microenvironment can help to understand the pathogenesis of CL-related eye infections. Knowledge of the relationship between the CL wearer's behaviours and pathogens would help health care providers focus on each step of proper CL care. This study aims to determine the behaviours that might be associated with the community of bacteria on CL.
Methods
A cross-sectional design was performed using anonymous questionnaires to obtain demographic data and assess hygiene practices among volunteering wearers. The CLs used were collected to evaluate the prevalence of pathogenic bacteria associated with ocular infections by PCR and microbiota analysis.
Results
The bacterial microbiota study revealed a total of 19 genera and 26 isolated strains from 20 eligible CLs. Enterobacter, Staphylococcus, and Achromobacter were the main genus in this subject population. Staphylococcus pasteuri and Achromobacter agilis were the most common pathogens at 65% and 35%, respectively. Enterobacter mori, a nonpathogenic organism, was found to be the most predominant strain, accounting for 27.51% of the total bacterial constituents. The risk behaviour of CL wear that was significantly associated with A. agilis contamination was cleaning the CL case with tap water (P value = 0.04).
Conclusions
This is the first study focusing on the association between the culture selected microbial community on the CL surface and compehensive behavioural characteristics. Environmental contamination was the main source of microbes found on CL surfaces. An emphasis in patient education should be placed on careful handling during the CL care routine and managing the hygiene of the surroundings.
Keywords: Contact lens, Contact lens care behaviour, Bacterial microbiota, Ocular infections, Cuture-selected microbial community
Contact lens, Contact lens care behaviour, Bacterial microbiota, Ocular infections, Cuture-selected microbial community.
1. Introduction
The contact lens (CL) is a preferred choice for a large number of people for correcting refractive errors due to its ability to correct a wide range of refractive errors, ease of adaptation, and its practicality for an active lifestyle. Even in the absence of refractive errors, CLs are still a popular choice for cosmetics. More than 140 million people worldwide use CLs on a regular basis [1]; however, this may also come with complications, some of which are sight-threatening. Wearing a CL compromises the ocular surface in many ways, both from the lens itself and from unfavourable behaviours accompanied by CL wear. CL is a foreign body in the eye that can potentially foster pathogenic microorganisms.
Recently, over one million visits for keratitis and CL-related complications occurred each year in the USA [2], and one of the major important risk factors for microbial keratitis is the use of CLs [3]. Bacteria are the most common pathogen of CL-related eye infections [4, 5]. Both gram-negative and gram-positive bacteria, including Staphylococcus aureus, S. epidermidis, Klebsiella sp. Acinetobacter sp. and Pseudomonas aeruginosa, are recognized as the main bacterial pathogens of keratitis [6]. These organisms possessed the ability to attach and adhere to the CL surface [7, 8]. In particular, the biofilm formation of the bacteria promoted an interplay between specific properties of the CL surface and the organism [9].
Each type of CL material's unique chemical and physical properties, such as hydrophobicity, ionicity, and surface roughness, all contribute to the risk of infection. Interaction between the CL and the eye can lead to an altered state of the ocular surface [10]. The front and back surfaces demand more tears for covering, leading to dryness. The movement of the CL on the lubricant-deprived ocular surface puts the cornea at an even greater vulnerability to microabrasions [3]. Corneal oxygenation is reduced as the lens acts as a physical barrier, blocking normal tear-gas exchange. Prolonged wearing only worsens dryness and cellular hypoxia, making the cornea more susceptible to pathogens [11]. Environmental exposure to dust and water brings pathogenic bacteria into the already compromised corneal surface. Apart from the inevitable risk from the lens acting on the corneal surface, another modifiable but hazardous complication associated with CL wear is mainly due to human behaviour [12]. Mishandling of CLs can both compromise the ocular surface and bring microbial contamination into the CL care system and the eye. The majority of CL users fail to adhere to good CL care behaviours, an important risk factor for CL-related eye infection, putting 40.9 million CL wearers in the United States at risk for serious eye infections [7]. Poor CL hygiene accounts for 12–66% of CL-related eye infection cases [6]. Poor hygiene is an important problem that is perhaps underestimated. A surprising 50% of CL wearers are not compliant with simple hygiene, such as hand washing [13].
Regarding the many steps involved in cleaning, disinfecting, and storing reusable CLs, mistakes in different steps may affect bacterial contamination differently. The ability to identify the causative risk behaviour can greatly help eye care professionals and CL wearers to focus on the steps and important points to avoid CL-related eye infection. Normal conjunctival bacterial normal flora has traditionally been considered predominantly gram-positive, reflecting those found on the skin [14]. The scientific advancements of the second decade of this century has allowed researchers to overcome the limitations of traditional culture. Microbiome analysis allows a deeper understanding of the unique microbial community in each niche. The conjunctival core microbiota is composed of the genera Corynebacterium, Pseudomonas, Staphylococcus, Acinetobacter, Streptococcus, Millisia, Anaerococcus, Finegoldia, Simonsiella, and Veillonella [15]. Human microbiomes help to regulate the homeostasis of human health and disease. The imbalance of the normal gut microbiota causes many noninfectious diseases, such as inflammatory bowel disease (IBD), irritable bowel syndrome (IBS), and obesity [16, 17, 18]. From one study that investigated the ocular surface microbiota, a relationship between Streptococcus infection of the lens storage container and allergy symptoms related to CL wear was found [19].
Considering the available information, the ocular surface microbiome most likely plays a key role in maintaining ocular surface homeostasis, and its alteration could be linked to the development and progression of eye diseases. The purpose of this study was to clarify how CL care behaviours affect bacterial contamination by using microbiota analysis, which may help to reveal the causal relationship between microorganisms and behaviour. It would also allow health personnel to emphasize the importance of certain steps in CL care and would aid future studies in developing strategies to avoid such pitfalls. Furthermore, microbiome data reflect the state of the microenvironment, revealing changes in ocular microorganisms in persons wearing CLs that are a plausible cause for allergic symptoms [19]. A cross-sectional study was conducted to examine the relationship between risk behaviours and the main pathogens causing eye infections. The findings of this study provide more insights into the behaviour of Thai CL wearers that have been rarely studied.
2. Materials and methods
2.1. Study design and sample collection
A cross-sectional study was performed from November 2020 to March 2021. The study protocol was reviewed and approved by the Human Research Ethics Committee of Walailak University (WUEC-20-321-01) before the first volunteer was enrolled, in accordance with the tenets of the Declaration of Helsinki and with international restrictions on this study. A CL wearer was defined as a person who wore the CLs at least 5 days per week during the past month. The study population consisted of participants aged between 17 and 58 years who attended the ophthalmology clinic at Walailak University Hospital, Walailak University, and academic colleges in Nakhon Si Thammarat, Thailand. The subjects were recruited by a research assistant. Participants who were administered any topical medications of anti-allergic agents and/or antimicrobial agents within 2 months prior to initiation of the present study were excluded. The eligible lens wearers were advised to bring their CL with them for collection on the consultation day. Written informed consent was obtained from all subjects after the explanation of the study. Participants completed a validated, anonymous, self-administered questionnaire regarding personal demographic information, the use of CL behaviours, and CL hygienic practice. Furthermore, the CLs used were collected to study the characteristics of bacterial accumulation and community, which might be related to the personal hygiene of those individuals. The optometrist placed the CLs in the sterile CL storage case containing normal saline solution, along with the questionnaire, put these in the sample envelope and returned them to the laboratory on the same day.
2.2. Questionnaire
Each participant completed a 47-item, anonymous, standardized paper questionnaire, which provided demographic data and behaviour of CL wear. In total, 20 soft CLs were obtained from 20 CL wearers. The questionnaire was divided into 3 parts: personal information (5 items), CL-related behaviours (20 items), and assessment of hygienic practices (22 items). Demographic information was collected, including sex, age, educational level, underlying disease, and history of antimicrobial agent administration as exclusion criteria.
2.3. Laboratory sampling, bacterial isolation and DNA extraction
The CLs from the participants were obtained within 4 h of CL wear on the same day, received at the laboratory and aseptically transferred to a culture tube. The CLs were grown on brain heart infusion broth (BHI, HiMedia, India) at 37 °C for 48 h for recovery of bacterial cells. The culture was centrifuged at 10,000×g for 5 min at 4 °C, washed once with Tris-ethylenediaminetetraacetic acid (TE) buffer [10 mM Tris-HCl (pH 8.0), 1 mM ethylenediaminetetraacetic acid], and resuspended in 0.5 ml of TE buffer. An aliquot of 1 mL of all samples was centrifuged for 1 min at 15,000×g, and then the supernatant was discarded from the tubes for DNA extraction. Genomic DNA (gDNA) was extracted from the bacterial pellet in accordance with the manufacturer's protocol of the Presto™ Mini gDNA Bacteria Kit (Geneaid Biotech, Ltd., New Taipei City, Taiwan). The pellet was resuspended in 200 μL of lysozyme and incubated at 37 °C for 30 min. The supernatant was removed, 20 μL of proteinase K was added to the tube, and the tube was incubated at 60 °C for 10 min. DNA was lysed and bound to the GD column. The gDNA was washed and eluted in a collection tube. The purified gDNA was collected into one microcentrifuge tube and centrifuged at 15,000×g for 30 s. The concentration of gDNA was determined spectrophotometrically in a Nanodrop instrument and kept at -20 °C until library construction.
2.4. Sequencing of 16S rRNA gene and microbiota analysis
For each sample, 10 ng of precipitate was used to amplify the V3 and V4 region of the 16S rRNA gene following the procedure developed by Illumina MiSeq System, [Primer:16S Amplicon PCR Forward Primer = 5'TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG CCTACGGGNGGCWGCAG, 16S Amplicon PCR Reverse Primer = 5'GTCTCGTGGGCTC GGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC and adaptor sequences: Forward overhang: 5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-(locus specific sequence) Reverse overhang: 5′ GTCTCGTGGGCTCGGAGATGTGTATAA GAGACAG-(locus specific sequence)]; this method also used molecular barcodes to enable multiplex sequencing as previously described [20]. Paired-end sequencing (2 × 150 base pairs [bp]) of these amplicons was performed on a desktop sequencer (MiSeq; Illumina, Inc., San Diego, CA, USA). 16S rRNA gene pipeline data acquisition incorporated phylogenetic and alignment-based approaches to maximize data resolution. Read pairs were demultiplexed based on the unique molecular barcodes, and reads were merged using FlASH v1.2.11 [21] with at least a 50-bp overlap and no more than 1-bp mismatch. Merged sequences were clustered into operational taxonomic units (OTUs) at a similarity cut-off value of 97% using the CD-HIT-OTU program. An expected error rate of 0.5 was applied for quality filtering. We mapped OTUs to the rDnaTools database to determine taxonomies [22]. QIIME was used to cluster the operational taxonomic units (OTUs), and constructed an OTU table from the output files generated in the previous two steps for downstream analyses of alpha diversity (observed OTUs, Chao 1 estimator, and Shannon diversity index; confirm species diversity), beta diversity (unweighted and weighted UniFrac; visualize the community diversity), and taxonomic trends (at the phylum and genus level).
2.5. Scanning electron microscopy (SEM)
Bacterial accumulation on the used CLs was examined by SEM on the representative CL surface as the contamination subjects. The lenses were cut into two pieces and fixed in 2.5% glutaraldehyde/0.1 M cacodylate buffer pH 7.4, at 4 °C overnight. The sample was resuspended in 0.1 M cacodylate buffer pH 7.4 at 4 °C and secondarily fixed with 1% osmium tetroxide (OsO4) (OsO4; Electron Microscopy Sciences, Hatfield, PA) in cacodylate buffer for 1 h at room temperature. All samples were dehydrated in a graded ethanol series of 20%, 40%, 60%, 80%, 90%, and finally two changes of absolute ethanol [23]. The lenses were subjected to electron microscopy at the Center for Scientific and Technological Equipment (CSE), Walailak University, according to the following protocol. Samples were dried immediately, mounted on aluminium stubs, and sputter-coated with gold in the vacuum chamber of a Cressington 108 Auto Sputter Coater (Cressington Scientific Instruments, UK). Visualization was performed under a scanning electron microscope (Merlin Compact, Zeiss, Germany).
2.6. Statistical analysis
All statistical analyses were performed using SPSS software (version 23.0; SPSS Inc., Chicago, IL). The data were analyzed for both descriptive and inferential statistics. Continuous variables were described using the mean and standard deviation. Independent categorical variables were described using frequencies and expressed as percentages. For the continuous variables, a t-test (two groups) was used to compare the groups. The chi-square test was used to examine bivariate associations between independent variables and pathogen-related eye infections. A P value <0.05 was considered statistically significant for the group comparison.
3. Results
3.1. Demographic data and behaviour of the CL wearers
A total of 20 CL wearers were enrolled and completed the questionnaires in this study. A summary of the participants’ demographic data is shown in Table 1. All participants were female with a mean age of 35.2 years, ranging from 17-58 years. Forty-five percent of the participants had a postgraduate degree education. All subjects wore soft reusable CLs, with the majority wearing monthly disposable lenses and 80% of the participants wearing CLs every day. Although it is generally recommended not to wear CLs more than 8 h per day, this survey found that 90% of subjects wore CLs longer than the recommendation. All CLs were purchased from non-health care professionals. Most of the participants (95%) also bought CL care solutions from non-health care professionals. Moreover, data revealed that 35% had a history of eye infections, including keratitis, conjunctivitis, and blepharitis. The undesirable activities found in this study were skipping annual eye check-ups, wearing CLs in water (such as swimming, diving, and shower), exceeding the recommended planned replacement of CLs and storage cases, and applying eye makeup.
Table 1.
Participant's demographic data and behaviors of contact lens wear.
| Demographic data | N = 20 (%) |
|---|---|
| Sex | |
| Female | 20 (100%) |
| Age | |
| ≤18 | 3 (15%) |
| 19-30 | 3 (15%) |
| 31-40 | 8 (40%) |
| 40-50 | 5 (25%) |
| >50 | 1 (5%) |
| Underlying disease | |
| Yes | 4 (20%) |
| No | 16 (80%) |
| Educational level | |
| High school | 1 (5%) |
| Vocational/High vocational certificate | 3 (15%) |
| Graduate | 7 (35%) |
| Postgraduate |
9 (45%) |
| Behavior of contact lens wear | |
| Lens wear experience | |
| 1–5 years | 8 (40%) |
| 6–10 years | 2 (10%) |
| more than 10 years | 10 (50%) |
| Type of lenses | |
| Clear soft CL | 11(55%) |
| Cosmetic CL | 9 (45%) |
| Frequency of wear in a week | |
| 1–3 day | 0 |
| 4–6 day | 4 (20%) |
| Everyday | 16 (80%) |
| Duration of wear | |
| Less than 8 h | 2 (10%) |
| More than 8 h | 18 (90%) |
| Source of CL purchase | |
| Health care professionals | 0 |
| Non-health care professionals | 20 (100%) |
| Source of disinfecting solution purchase | |
| Health care professionals | 1 (5%) |
| Non-health care professionals | 19 (95%) |
| Symptoms associated with CL wear | |
| Dryness | 14 (70%) |
| Irritation | 10 (50%) |
| Tearing | 7 (35%) |
| Redness | 6 (30%) |
| Itchiness | 4 (20%) |
| Blurry vision | 2 (10%) |
| Discharge | 2 (10%) |
| History of eye infection associated with CL wear | |
| Yes | 7 (35%) |
| Keratitis | 3 (15%) |
| Conjunctivitis | 2 (10%) |
| Blepharitis or hordeolum | 2 (10%) |
| None | 13 (65%) |
| Sleep with CL in | |
| Yes | 9 (45%) |
| No | 11 (55%) |
| Sharing CL with others | |
| Yes | 0 |
| No | 20 (100%) |
| Exceed the recommended planned replacement | |
| Yes | 15 (75%) |
| No | 5 (25%) |
| Using expired CL solutions (opened for more than 3 months) | |
| Yes | 7 (35%) |
| No | 13 (65%) |
| Exposure to water during CL wear | |
| Yes | 19 (95%) |
| No | 1 (5%) |
| The use of eye drops in conjunction with CL | |
| Yes | 13 (65%) |
| No | 7 (35%) |
| First or second-hand smoker | |
| Yes | 1 (5%) |
| No | 19 (95%) |
| Time spent work with terminal screen per day | |
| Less than 12 h | 9 (45%) |
| More than 12 h | 11 (55%) |
| Exposure to air-conditioned environment | |
| Less than 12 h | 12 (60%) |
| More than 12 h | 8 (40%) |
| Use of makeup close to the eye | |
| Yes | 15 (75%) |
| No | 5 (25%) |
The CL wearers’ hygiene behaviour is demonstrated in Table 2. Most of the subjects had good practices in CL care, such as checking the expiration of the CL product and solutions, checking the side of the lens, and washing hands with soap before putting in and taking off the CL. Impressively, most performed the correct routine by the drop-rub-rinse regimen after wearing the CLs and even before. Moreover, they mostly followed the correct routine regarding the use of the CL care solution and its storage case, such as always renewing the cleaning solution, keeping the bottle clean, and not using the same CL case for more than 3 months. However, half of the participants continued to use lenses that had been dropped, which might have been contaminated, and had an improper CL case care regimen by cleaning them with tap water only.
Table 2.
CL wearers hygiene behaviors.
| CL care behaviors | N = 20 (%) |
|---|---|
| Always check expiration date and integrity of packaging before use | |
| Yes | 17 (85%) |
| No | 3 (15%) |
| Check for the correct side (inside-outside) before use | |
| Yes | 18 (90%) |
| No | 2 (10%) |
| Start inserting and removing the lens from the same eye | |
| Yes | 17 (85%) |
| No | 3 (15%) |
| Continued using the lens that had been dropped | |
| Yes | 10 (50%) |
| No | 10 (50%) |
| Hand wash before putting in the CLs | |
| With water only | 5 (25%) |
| With soap | 14 (70%) |
| Not done | 1 (5%) |
| Routine before putting in the CLs | |
| Rub the lenses | 1 (5%) |
| Rinse the lenses | 7 (35%) |
| Rub and rinse the lenses | 8 (40%) |
| No management | 4 (20%) |
| Hand wash before CLs removal | |
| With water only | 4 (20%) |
| With soap | 10 (50%) |
| Not done | 6 (30%) |
| Routine after CLs removal | |
| Rub the lenses | 1 (5%) |
| Rinse the lenses | 3 (15%) |
| Rub and rinse the lenses | 9 (45%) |
| None | 7 (35%) |
| Products used to clean the CL | |
| CL cleaning solution | 13 (65%) |
| NSS | 5 (25%) |
| Tap water | 2 (10%) |
| Soaking CLs in the cleaning solution for ≥6 h before reuse | |
| Yes | 19 (95%) |
| No | 1 (5%) |
| Fill CL case with fresh CL solution every day | |
| Yes | 20 (100%) |
| No | 0 |
| Topping off the old cleaning solution | |
| Yes | 7 (35%) |
| No | 13 (65%) |
| Close the cap of the cleaning solution tightly after use | |
| Yes | 15 (75%) |
| No | 5 (25%) |
| Keep using the same bottle of cleaning solution for more than 3 months | |
| Yes | 1 (5%) |
| No | 19 (95%) |
| Close the CL case tightly after use | |
| Yes | 19 (95%) |
| No | 1 (5%) |
| Keep using the same case for more than 3 months | |
| Yes | 4 (20%) |
| No | 16 (80%) |
| Clean the CL case with | |
| Water only | 9 (45%) |
| Water and soap | 5 (25%) |
| With CL solution | 6 (30%) |
| Not done | 0 |
| Clean the CL case daily | |
| Yes | 9 (45%) |
| No | 11 (55%) |
3.2. Culture selected bacterial community on CLs
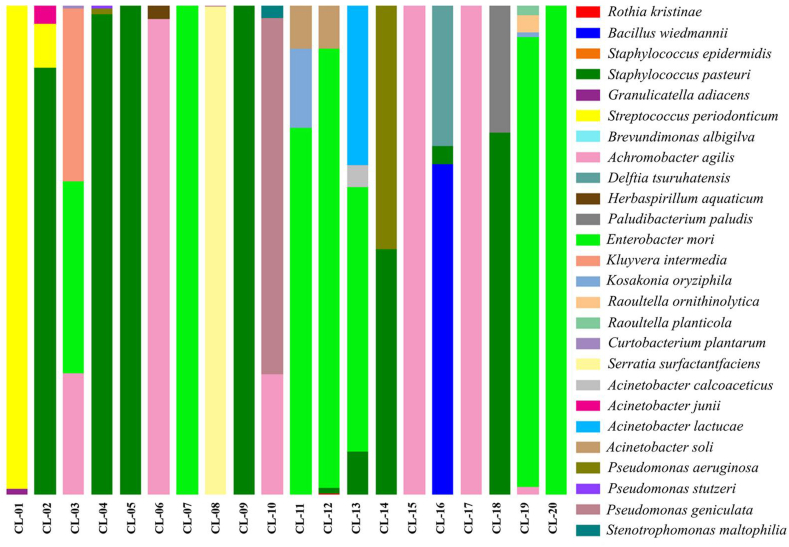
The 20 CL samples were coded as CL-1 to CL-20. The bacteria that were cultured from the CLs were detected and identified by microbiota analysis at the phylum to species level by 16S rRNA amplicon sequencing data. A bacterial microbiota study revealed that a total of 19 genus and 26 isolated trains were obtained from all CLs. Among the genus, Enterobacter, Staphylococcus, and Achromobacter were the most abundant representing 27.51%, 26.18% and 17.41% of total population, respectively. The isolated strains and CL care behaviours of each subject are shown in Table 3. The bacterial constituents of each sample are illustrated in Figure 1. The overall abundance of bacteria showed that Enterobacter mori, Staphylococcus pasteuri, and Achromobacter agilis were the 3 most predominant species, representing 27.51%, 26.17%, and 16.16% of the total population, respectively. Moreover, the bacterial isolates were justified as pathogens related to eye infections according to their contamination sources and background of causing the disease, as shown in Table 4. The main pathogens that were found in the present population were represented by 13 strains: Serratia surfactantfaciens (10%), Staphylococcus epidermidis (5%), S. pasteuri (65%), Stenotrophomonas maltophilia (5%), Pseudomonas aeruginosa (10%), Delftia tsuruhatensis (5%), Acinetobacter calcoaceticus (5%), Granulicatella adiacens (5%), Raoultella planticola (10%), R. ornithinolytica (5%), A. agilis (35%), Pseudomonas stutzeri (5%), and Acinetobacter junii (5%). Sources of contamination are normally residential water, rivers, soil, mud, and some plants [19, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41], which might be related to the CL care routines and nonhygenic environments. For contamination by nonpathogenic species, 13 strains were found: Brevundimonas albigilva, E. mori, Herbasprillum aquaticum, Kluyvera intermedia, Pseudomonas geniculata, Acinetobacter lactucae, Bacillus wiedmanni, Curtobacterium plantarum, Paludibacterium paludis, Kosakonia oryziphila, Rothia kristinae, Acinetobacter soli, and Streptococcus periodonticum.
Table 3.
Sample collection and culture-selected microbial community.
| Subject ID | Age, sex | Past-history of eye infection or symptoms | Behavior | Isolated species | % found on CL |
|---|---|---|---|---|---|
| CL-1 | 38, Female | Abscess |
|
S. periodonticum G. adiacens |
98.91 1.09 |
| CL-2 | 18, Female | Never |
|
S. pasteuri S. periodonticum A. junii S. epidermidis B. albigilva |
87.24 9.03 3.67 0.04 0.01 |
| CL-3 | 17, Female | Never |
|
E. mori K. intermedia A. agilis C. plantarum S. pasteuri |
39.10 35.49 24.83 0.53 0.04 |
| CL-4 | 17, Female | Never |
|
S. pasteuri P. aeruginosa P. stutzeri A. agilis |
98.21 1.19 0.53 0.07 |
| CL-5 | 46, Female | Keratitis |
|
S. pasteuri B. albigilva |
99.99 0.01 |
| CL-6 | 37, Female | Never |
|
A. agilis H. aquaticum B. albigilva S. periodonticum B. wiedmannii |
97.22 2.74 0.02 0.01 0.01 |
| CL-7 | 33, Female | Never |
|
E. mori B. albigilva |
99.99 0.01 |
| CL-8 | 27, Female | Conjunctivitis |
|
S. surfactantfaciens P. geniculata |
99.90 0.10 |
| CL-9 | 31, Female | Never |
|
S. pasteuri | 100.00 |
| CL-10 | 49, Female | Never |
|
P. geniculata A. agilis S. maltophilia E. mori S. pasteuri |
72.89 24.63 2.45 0.02 0.01 |
| CL-11 | 48, Female | Conjunctivitis |
|
E. mori K. oryziphila A. soli K. intermedia S. surfactantfaciens |
75.03 16.09 8.79 0.06 0.03 |
| CL-12 | 33, Female | Keratitis |
|
E. mori A. soli S. pasteuri R. kristinae |
89.93 8.71 1.24 0.13 |
| CL-13 | 33, Female | Abscess |
|
E. mori A. lactucae S. pasteuri A. calcoaceticus |
54.07 32.62 8.84 4.47 |
| CL-14 | 58, Female | Never |
|
S. pasteuri P. aeruginosa |
50.20 49.80 |
| CL-15 | 27, Female | Never |
|
A. agilis E. mori B. albigilva |
99.97 0.02 0.01 |
| CL-16 | 47, Female | Never |
|
B. wiedmannii D. tsuruhatensis S. pasteuri |
67.53 28.76 3.71 |
| CL-17 | 26, Female | Never |
|
A. agilis B. albigilva S. pasteuri E. mori |
99.95 0.03 0.01 0.01 |
| CL-18 | 50, Female | Never |
|
S. pasteuri P. paludis R. planticola E. mori |
73.95 26.03 0.01 0.01 |
| CL-19 | 39, Female | Never |
|
E. mori R. ornithinolytica R. planticola A. agilis K. oryziphila K. intermedia S. pasteuri |
92.00 3.47 2.05 1.46 0.89 0.10 0.02 |
| CL-20 | 31, Female | Keratitis |
|
E. mori B. albigilva |
99.97 0.03 |
Figure 1.
Stacked bar plots showing the percentage of bacterial populations as a taxonomic composition for each CL sample from the genus to species level. A total of 26 bacterial strains were identified in the subjects. Enterobacter mori, Staphylococcus pasteuri, and Achromobacter agilis were the 3 most predominant species, representing 27.51%, 26.17%, and 16.16% of the total population, respectively.
Table 4.
Pathogens related eye infections.
| Pathogens | % found in population (n = 20) | Source | References |
|---|---|---|---|
| S. surfactantfaciens | 10 | water and marine environments, contaminated soil, plants, animals, hospitalized patients | Grimont (2006) [24]; Su et al. (2016) [25] |
| S. epidermidis | 5 | human skin, upper respiratory tract | Du et al. (2021) [26] |
| S. pasteuri | 65 | drinking water, common skin flora, food products, air | Santoiemma et al. (2020) [27] |
| S. maltophilia | 5 | soil, sediment, wastewater, sputum | Ma et al. (2020) [28]; Al-Dhabi et al. (2021) [29] |
| P. aeruginosa | 10 | CLs, wet surfaces, chronic infection sites | Enzor et al. (2021) [30]; Riquelme et al. (2020) [31] |
| D. tsuruhatensis | 5 | soil, water, sludge, human microflora, CLs | Hotta et al. (2020) [19] |
| A. calcoaceticus | 5 | Soil, water | Roy et al. (2013) [32] |
| G. adiacens | 5 | human oral cavity, urogenital tract, gastrointestinal tract | Borroni (2002) [33] |
| R. planticola | 10 | vegetables, food, liquid soap | Vassallo et al. (2016) [34] |
| R. ornithinolytica | 5 | CLs, water, urine, wounds | Eguchi et al. (2017) [35]; |
| A. agilis | 35 | rivers, ponds, residential water sources, soil, mud, some plants | Price et al. (2020) [36] Agbaji et al. (2020) [37] Vandamme et al. (2016) [38] |
| P. stutzeri | 5 | soil, water | Gilardi (1972) [39]; Lalucat et al. (2006) [40] |
| A. junii | 5 | water, soil, animals | Broniek et al. (2014) [41] |
3.3. Risk behaviours of CL wear associated with eye infections
S. pasteuri and A. agilis were the most common organisms found from this studied population, at 65% and 35%, respectively. The two organisms were demonstrated to be representative of the main pathogens for evaluation of the association between CL wearers’ behaviour and eye infection. Chi-square factors that showed the significance of the data are presented in Table 5. The two risk behaviours of cleaning the CL case with tap water and a prior history of eye infection were shown to be statistically significant in this study, with P values of 0.04 and 0.01, respectively.
Table 5.
Risk behaviors related to main pathogens causing eye infections.
| Risk factor | N (%) |
Staphylococcus pasteuri |
P-value |
Achromobacter agilis |
P-value |
Enterobacter mori |
P-value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Yes N (%) | No N (%) | Yes N (%) | No N (%) | Yes N (%) | No N (%) | |||||
| Sleeping or napping in CLs | ||||||||||
| No | 11 (55) | 7 (53.85) | 4 (57.14) | 0.88 | 5 (71.43) | 6 (46.15) | 0.27 | 7 (63.64) | 4 (44.44) | 0.39 |
| Yes | 9 (45) | 6 (46.15) | 3 (42.86) | 2 (28.57) | 7 (53.85) | 4 (36.36) | 5 (55.56) | |||
| Exceed the recommended period of CL | ||||||||||
| No | 5 (25) | 2 (15.38) | 3 (42.86) | 0.17 | 0 (0) | 5 (38.46) | 0.058 | 3 (27.27) | 2 (22.22) | 0.75 |
| Yes | 15 (75) | 11 (84.62) | 4 (57.14) | 7 (100) | 8 (61.54) | 8 (72.73) | 7 (77.78) | |||
| Exceed the recommended period of CL solution | ||||||||||
| No | 13 (65) | 8 (61.54) | 5 (71.43) | 0.65 | 4 (57.14) | 9 (69.23) | 0.58 | 7 (63.64) | 6 (66.67) | 0.88 |
| Yes | 7 (35) | 5 (38.46) | 2 (28.57) | 3 (42.86) | 4 (30.77) | 4 (36.36) | 3 (33.33) | |||
| Wearing in water | ||||||||||
| No | 4 (20) | 1 (7.69) | 3 (42.86) | 0.06 | 2 (28.57) | 2 (15.38) | 0.48 | 2 (18.18) | 2 (22.22) | 0.82 |
| Yes | 16 (80) | 12 (92.31) | 4 (57.14) | 5 (71.43) | 11 (84.62) | 9 (81.82) | 7 (77.78) | |||
| Wearing in shower | ||||||||||
| No | 3 (15) | 1 (7.69) | 2 (28.57) | 0.21 | 1 (14.29) | 2 (15.38) | 0.94 | 3 (27.27) | 0 (0) | 0.08 |
| Yes | 17 (85) | 12 (92.31) | 5 (71.43) | 6 (85.71) | 11 (84.62) | 8 (72.73) | 9 (100) | |||
| Hand washing | ||||||||||
| No | 6 (30) | 4 (30.77) | 2 (28.57) | 0.91 | 3 (42.86) | 3 (23.08) | 0.35 | 2 (18.18) | 5 (55.56) | 0.49 |
| Yes | 14 (70) | 9 (69.23) | 5 (71.43) | 4 (57.14) | 10 (76.92) | 9 (81.82) | 4 (44.44) | |||
| With soap | ||||||||||
| No | 10 (50) | 7 (53.85) | 3 (42.86) | 0.63 | 5 (71.43) | 5 (38.46) | 0.16 | 5 (45.45) | 5 (55.56) | 0.65 |
| Yes | 10 (50) | 6 (46.15) | 4 (57.14) | 2 (28.57) | 8 (61.54) | 6 (54.55) | 4 (44.44) | |||
| Rub and rinse CL | ||||||||||
| No | 7 (35) | 6 (46.16) | 1 (14.29) | 0.15 | 3 (42.86) | 4 (30.77) | 0.58 | 2 (18.18) | 5 (55.56) | 0.08 |
| Yes | 13 (65) | 7 (53.84) | 6 (86.71) | 4 (57.14) | 9 (69.23) | 9 (81.82) | 4 (44.44) | |||
| Reuse old CL solution | ||||||||||
| No | 13 (65) | 7 (53.85) | 6 (85.71) | 0.15 | 4 (57.14) | 9 (69.23) | 0.58 | 7 (63.64) | 6 (66.67) | 0.88 |
| Yes | 7 (35) | 6 (46.15) | 1 (14.29) | 3 (42.86) | 4 (30.77) | 4 (36.36) | 3 (33.33) | |||
| Soaking CL with normal saline solution | ||||||||||
| No | 15 (75) | 10 (76.92) | 5 (71.43) | 0.78 | 4 (57.14) | 11 (84.62) | 0.17 | 8 (72.73) | 7 (77.78) | 0.79 |
| Yes | 5 (25) | 3 (23.08) | 2 (28.57) | 3 (42.86) | 2 (15.38) | 3 (27.27) | 2 (22.22) | |||
| Clean the CL case with tap water | ||||||||||
| No | 11 (55) | 6 (46.15) | 5 (71.43) | 0.27 | 6 (85.71) | 5 (38.46) | 0.04∗ | 7 (63.64) | 4 (44.44) | 0.39 |
| Yes | 9 (45) | 7 (53.85) | 2 (28.57) | 1 (14.29) | 8 (61.54) | 4 (36.36) | 5 (55.56) | |||
| History of eye infection | ||||||||||
| No | 13 (65) | 10 (76.92) | 3 (42.86) | 0.12 | 7 (100) | 6 (46.15) | 0.01∗ | 7 (63.64) | 6 (66.67) | 0.88 |
| Yes | 7 (35) | 3 (23.08) | 4 (57.14) | 0 (0) | 7 (53.85) | 4 (36.36) | 3 (33.33) | |||
Note: ∗A P value <0.05 was considered statistically significant from chi-square test.
3.4. Visualization of CLs
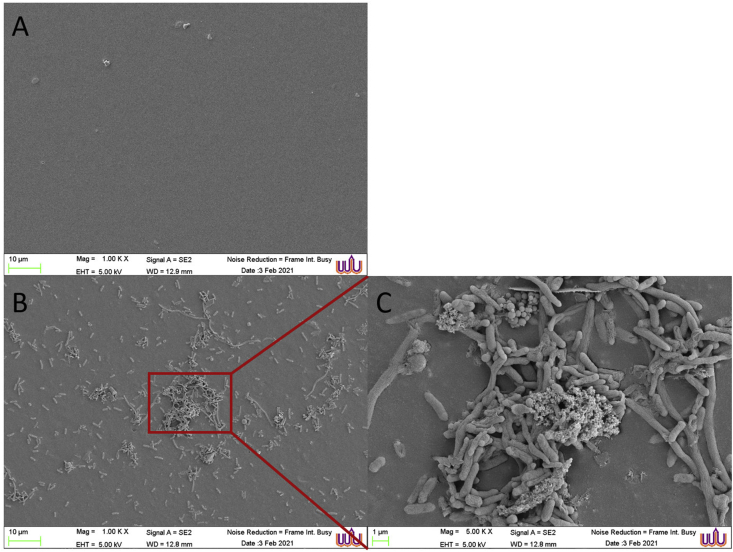
SEM was used to visualize the ultrastructure of bacterial accumulation on the CL surface. Figure 2A shows the surface of a CL that was taken from a control following the correct CL routine, such as hand washing with soap before applying and taking out the CL, not exceeding the recommended wearing period, and dropping-rubbing-rinsing the surface with CL care solution. This control was utilized as a reference for comparison with the CL from a subject with unfavourable behaviours (Figure 2B, C). The control CL shows a clear surface without bacterial attachment, whereas Figure 2B shows large amounts of bacterial adhesion. Additionally, the SEM photograph revealed typical biofilm morphologies and dense networks of cells arranged in multiple layers, forming microcolonies with the visible granular extracellular matrix of both gram-positive and gram-negative bacteria (Figure 2C).
Figure 2.
Scanning electron micrographs of bacterial accumulation on the CL surface. (A) A clear surface of a control CL with the correct care routine, such as hand washing with soap before applying and taking out the CL, not exceeding the recommended wearing period, and dropping-rubbing-rinsing the surface with CL care solution. (B,C) Markedly contaminated CL surface of a subject with unfavorable behaviours, at scale bars of 10 μm and 1 μm, respectively.
4. Discussion
The ocular surface is a newly described niche with the unique characteristics of microbiota. Its consortium comprises a ‘putative core’ of 12 bacterial genera consisting of Pseudomonas, Propionibacterium, Bradyrhizobium, Corynebacterium, Acinetobacter, Brevundimonas, Staphylococci, Aquabacterium, Sphingomonas, Streptococcus, Streptophyta, and Methylobacterium [42, 43, 44]. The members of this putative core are the permanent residents of the ocular surface, despite occasional changes and the introduction of other bacteria. Recently, alteration of the ocular microbiome associated with CLs has been reported [45]. The microbial community in the presence of CL wear was found to be more variable than the community from the normal ocular surface, reflecting more of the skin flora, with higher abundances of Methylobacterium, Lactobacillus, Acinetobacter, and Pseudomonas, while Haemophilus, Streptococcus, Staphylococcus, and Corynebacterium showed lower abundances. Despite the close contact of the CL and the ocular surface, the microbiota of the CL surface was markedly different from those of the conjunctiva. The major microbes isolated from CL were previously reported, including coagulase-negative staphylococci (CNS), Propionibacterium sp., and Corynebacterium sp [46]. Another study of alterations in soft CL wearers also detected the constituents of Streptococcus, Methylobacterium, and Acinetobacter in the bacterial microbiome data [45].
This present study provided more insight into the nature of the bacterial microbiota on CLs. The 20 collected CLs contained 19 genera and 26 strains of bacteria. Additionally, 13 of these strains have been recognized as the causative pathogens of ocular infections or CL-related infections. The species with the highest predominance was E. mori. This bacteria is not a major pathogen causing disease in humans but rather in plants. However, it was mentioned in a case report as causing otitis externa in a 59-year patient in Austria [47]. Since bacteria are usually found in the rhizosphere and sometimes cause plant diseases, the detection of this pathogen at high levels indicates the contamination of soil and water [47].
The most ubiquitous gram-positive bacterium found in this study was S. pasteuri, a pathogenic bacterium with a coagulase-negative reaction that normally colonizes human skin or acts as a contaminant in water, food products, and unsanitary environments [23]. Contamination can occur by inappropriate handling of the CL during the process of wearing and taking off. Another Staphylococcus sp. found in this study was S. epidermidis, which was found in 5% of the studied population. This was in contrast to results from a previous study, which suggested that Staphylococcus sp. established on ocular surfaces in healthy adults represents as much as 73% of the bacterial community, especially S. epidermidis [48]. Aside from the skin flora discussed above, the gram-negative bacteria A. agilis showed a high abundance in the bacterial microbiota constituents in this study, which indicated that the contamination might be from pollutants in the surroundings [36, 47]. The source of A. agilis was previously study and the strain was obtained from soil as a rhizobacterial flora [37, 38]. The microbiota on the CLs reflected external contamination much more than that of the conjunctiva. The sources of the constituents on the lens surfaces were mostly from water, soil, the oral cavity, and the urogenital tract. This difference gave a clearer picture of the microenvironment of a CL wearer, with a population reflecting the environmental contaminants on the CL surface. Furthermore, the association of bacterial contamination with CL wearer behaviour was determined. The results of this study emphasize the danger of using tap water to clean CL cases. CL wearers who used tap water to clean the CL case carried a significantly higher risk of A. agilis contamination (p = 0.04). The CL case was recently acknowledged as the bulk of microbial contamination. The accumulation of bacteria and biofilm formation on the case surface was a commonly susceptible part of the CL care system more than the CLs themselves [49, 50]. This study provided insight into factors that may be significant in maintaining lens case hygiene and explored some of the issues previously proven in the in vitro study that tap-water use was associated with the contamination rate of gram-negative bacteria, particularly the strains Pseudomonas sp., Stenotrophomonas maltophilia, and Achromobacter sp [50]. Moreover, the bacterial pathogens that were reported in this study are commonly found on human skin, the oral cavity, and the urogenital tract, reflecting the non-compliance of CL wearers with hand washing. Seventy percent of the subjects routinely washed their hands before putting in CLs, and only half of them did so before taking off their CLs. The fact that most of the CL care process and CL case drying occurred in restrooms may increase urogenital tract pathogen contamination into the CL care system. In agreement with previous research, this study again highlights the negative effect of improper CL behaviour. The five most common improper CL care practices in Thai CL wearers were wearing CL for longer than recommended, not changing the CL solution, swimming with CLs, rinsing CLs with tap water, and not washing hands before handing the CLs [51]. Although our result showed that cleaning the CL case with tap water is statistically significant associated with A. agilis contamination. The non-significance difference of other behavior might be due to the low number of participants. Further studies of greater sample size would be necessary to confirm these findings.
The summation of this information suggests poor hygiene or overlooked pitfalls in CL handling. Most CL wearers received less-than-adequate to no education regarding CL handling at their time of purchase in conjunction with the surprisingly low proportion of CL wearers who seek their CLs and CL care solutions from a health care provider. Thus, no professional advice or patient evaluation for potential risks was ever provided. Appropriate behaviour remains a crucial point for eye care professionals to emphasize with their patients for a safe CL-corrected vision.
Statement of Ethics
The study protocol was reviewed and approved by the Human Research Ethics Committee of Walailak University (WUEC-20-321-01) before the first volunteer was enrolled, in accordance with the tenets of the Declaration of Helsinki and with international restrictions on this study.
Declarations
Author contribution statement
Lunla Udomwech: Conceived and designed the experiments; Wrote the paper.
Kulwadee Karnjana and Juntamanee Jewboonchu: Performed the experiments; Analysed and interpreted the data.
Phisut Rattanathamma, Udomsak Narkkul and Jakkrit Juhong: Analysed and interpreted the data.
Auemphon Mordmuang: Performed the experiments; Analysed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by Walailak University (Grant Number WU-IRG-63-081, 2020).
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors are grateful to thank the Research Center of Excellence in Innovation on Essential Oil and Research Institute for Health Science, and the Research Center in Tropical Pathobiology, Walailak University, for their kind support and laboratory facilities.
References
- 1.Lim C.H.L., Stapleton F., Mehta J.S. Review of contact lens-related complications. Eye Contact Lens. 2018;44(Suppl 2):S1–s10. doi: 10.1097/ICL.0000000000000481. [DOI] [PubMed] [Google Scholar]
- 2.Collier S.A., Gronostaj M.P., MacGurn A.K., et al. Estimated burden of keratitis--United States, 2010. MMWR Morb. Mortal. Wkly. Rep. 2014;63(45):1027–1030. [PMC free article] [PubMed] [Google Scholar]
- 3.Eltis M. Contact-lens-related microbial keratitis: case report and review. J. Optometr. 2011;4(4):122–127. [Google Scholar]
- 4.Wu Y.T., Willcox M., Zhu H., Stapleton F. Contact lens hygiene compliance and lens case contamination: a review. Contact Lens Anterior Eye. 2015;38(5):307–316. doi: 10.1016/j.clae.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Zimmerman A.B., Nixon A.D., Rueff E.M. Contact lens associated microbial keratitis: practical considerations for the optometrist. Clin. Optom. 2016;8:1–12. doi: 10.2147/OPTO.S66424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stapleton F. Contact lens-related corneal infection in Australia. Clin. Exp. Optom. 2020;103(4):408–417. doi: 10.1111/cxo.13082. [DOI] [PubMed] [Google Scholar]
- 7.Giraldez M.J., Resua C.G., Lira M., et al. Contact lens hydrophobicity and roughness effects on bacterial adhesion. Optom. Vis. Sci. 2010;87(6):E426–E431. doi: 10.1097/OPX.0b013e3181da8656. [DOI] [PubMed] [Google Scholar]
- 8.Miller M.J., Ahearn D.G. Adherence of Pseudomonas aeruginosa to hydrophilic contact lenses and other substrata. J. Clin. Microbiol. 1987;25(8):1392–1397. doi: 10.1128/jcm.25.8.1392-1397.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dosler S., Hacioglu M., Yilmaz F.N., Oyardi O. Biofilm modelling on the contact lenses and comparison of the in vitro activities of multipurpose lens solutions and antibiotics. PeerJ. 2020;8:e9419. doi: 10.7717/peerj.9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mordmuang A., Udomwech L., Karnjana K. Influence of contact lens materials and cleaning procedures on bacterial adhesion and biofilm formation. Clin. Ophthalmol. 2021;15:2391–2402. doi: 10.2147/OPTH.S310862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleiszig S.M., Evans D.J. Pathogenesis of contact lens-associated microbial keratitis. Optom. Vis. Sci. 2010;87(4):225–232. doi: 10.1097/OPX.0b013e3181d408ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bui T.H., Cavanagh H.D., Robertson D.M. Patient compliance during contact lens wear: perceptions, awareness, and behavior. Eye Contact Lens. 2010;36(6):334–339. doi: 10.1097/ICL.0b013e3181f579f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fonn D., Jones L. Hand hygiene is linked to microbial keratitis and corneal inflammatory events. Contact Lens Anterior Eye. 2019;42(2):132–135. doi: 10.1016/j.clae.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 14.Ratnumnoi R., Keorochana N., Sontisombat C. Normal flora of conjunctiva and lid margin, as well as its antibiotic sensitivity, in patients undergoing cataract surgery at Phramongkutklao Hospital. Clin. Ophthalmol. 2017;11:237–241. doi: 10.2147/OPTH.S109247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrillo F., Pignataro D., Lavano M.A., et al. Current evidence on the ocular surface microbiota and related diseases. Microorganisms. 2020;8(7):1033. doi: 10.3390/microorganisms8071033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin J., Li Y., Cai Z., et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 17.Rinttilä T., Lyra A., Krogius-Kurikka L., Palva A. Real-time PCR analysis of enteric pathogens from fecal samples of irritable bowel syndrome subjects. Gut Pathog. 2011;3(1):6. doi: 10.1186/1757-4749-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swidsinski A., Weber J., Loening-Baucke V., et al. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 2005;43(7):3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hotta F., Eguchi H., Nakayama-Imaohji H., et al. Microbiome analysis of contact lens care solutions and tear fluids of contact lens wearers: possible involvement of streptococcal antigens in allergic symptoms related to contact lens wear. Int. J. Mol. Med. 2020;46(4):1367–1376. doi: 10.3892/ijmm.2020.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartram A.K., Lynch M.D., Stearns J.C., et al. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl. Environ. Microbiol. 2011;77(11):3846–3852. doi: 10.1128/AEM.02772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magoč T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schloss P.D., Westcott S.L., Ryabin T., et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiley L., Bridge D.R., Wiley L.A., et al. Bacterial biofilm diversity in contact lens-related disease: emerging role of Achromobacter, Stenotrophomonas, and Delftia. Invest. Ophthalmol. Vis. Sci. 2012;53(7):3896–3905. doi: 10.1167/iovs.11-8762. [DOI] [PubMed] [Google Scholar]
- 24.Grimont P.A., Grimont F. The genus serratia. Annu. Rev. Microbiol. 1978;32(1):221–248. doi: 10.1146/annurev.mi.32.100178.001253. [DOI] [PubMed] [Google Scholar]
- 25.Su C., Xiang Z., Liu Y., et al. Analysis of the genomic sequences and metabolites of Serratia surfactantfaciens sp. nov. YD25(T) that simultaneously produces prodigiosin and serrawettin W2. BMC Genom. 2016;17(1):865. doi: 10.1186/s12864-016-3171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du X., Larsen J., Li M., et al. Staphylococcus epidermidis clones express Staphylococcus aureus-type wall teichoic acid to shift from a commensal to pathogen lifestyle. Nat. Microbiol. 2021;6(6):757–768. doi: 10.1038/s41564-021-00913-z. [DOI] [PubMed] [Google Scholar]
- 27.Santoiemma P.P., Kalainov D.M., Mehta M.P., Bolon M.K. An unusual case of Staphylococcus pasteuri osteomyelitis. Infect. Dis. Rep. 2020;12(2):8523. doi: 10.4081/idr.2020.8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma J., Feng J., Shan Y., et al. Characteristic antimicrobial resistance of clinically isolated Stenotrophomonas maltophilia CYZ via complete genome sequence. J. Glob Antimicrob. Resist. 2020;23:186–193. doi: 10.1016/j.jgar.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 29.Al-Dhabi N.A., Esmail G.A., Alzeer A.F., Arasu M.V. Removal of nitrogen from wastewater of date processing industries using a Saudi Arabian mesophilic bacterium, Stenotrophomonas maltophilia Al-Dhabi-17 in sequencing batch reactor. Chemosphere. 2021;268:128636. doi: 10.1016/j.chemosphere.2020.128636. [DOI] [PubMed] [Google Scholar]
- 30.Enzor R., Bowers E.M.R., Perzia B., et al. Comparison of clinical features and treatment outcomes of Pseudomonas aeruginosa keratitis in contact lens and non-contact lens wearers. Am. J. Ophthalmol. 2021;227:1–11. doi: 10.1016/j.ajo.2021.02.024. [DOI] [PubMed] [Google Scholar]
- 31.Riquelme S.A., Liimatta K., Wong Fok Lung T., et al. Pseudomonas aeruginosa utilizes host-derived itaconate to redirect its metabolism to promote biofilm formation. Cell Metabol. 2020;31(6):1091–1096.e6. doi: 10.1016/j.cmet.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy R., Panigrahi P., Malathi J., et al. Endophthalmitis caused by Acinetobacter baumanni: a case series. Eye. 2013;27(3):450–452. doi: 10.1038/eye.2012.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borroni D. Granulicatella adiacens as an unusual cause of microbial keratitis: a metagenomic approach. Ocul. Immunol. Inflamm. 2021:1–2. doi: 10.1080/09273948.2021.1933066. [DOI] [PubMed] [Google Scholar]
- 34.Vassallo J., Vella M., Cassar R., Caruana P. Four cases of Raoultella planticola conjunctivitis. Eye. 2016;30(4):632–634. doi: 10.1038/eye.2015.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eguchi H., Hotta F., Kuwahara T., et al. Acute keratoconjunctivitis due to contamination of contact lens care solution with histamine-producing Raoultella species: a case report. Medicine (Baltim.) 2017;96(50) doi: 10.1097/MD.0000000000009310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price E.P., Soler Arango V., Kidd T.J., et al. Duplex real-time PCR assay for the simultaneous detection of Achromobacter xylosoxidans and Achromobacter spp. Microb. Genom. 2020;6(7) doi: 10.1099/mgen.0.000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agbaji J.E., Nwaichi E.O., Abu G.O. Optimization of bioremediation-cocktail for application in the eco-recovery of crude oil polluted soil. AAS Open Res. 2020;3:7. doi: 10.12688/aasopenres.13028.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandamme P.A., Peeters C., Inganäs E., et al. Taxonomic dissection of Achromobacter denitrificans Coenye et al. 2003 and proposal of Achromobacter agilis sp. nov., nom. rev., Achromobacter pestifer sp. nov., nom. rev., Achromobacter kerstersii sp. nov. and Achromobacter deleyi sp. nov. Int. J. Syst. Evol. Microbiol. 2016;66(9):3708–3717. doi: 10.1099/ijsem.0.001254. [DOI] [PubMed] [Google Scholar]
- 39.Gilardi G.L. Infrequently encountered Pseudomonas species causing infection in humans. Ann. Intern. Med. 1972;77(2):211–215. doi: 10.7326/0003-4819-77-2-211. [DOI] [PubMed] [Google Scholar]
- 40.Lalucat J., Bennasar A., Bosch R., et al. Biology of Pseudomonas stutzeri. Microbiol. Mol. Biol. Rev. 2006;70(2):510–547. doi: 10.1128/MMBR.00047-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broniek G., Langwińska-Wośko E., Szaflik J., Wróblewska M. Acinetobacter junii as an aetiological agent of corneal ulcer. Infection. 2014;42(6):1051–1053. doi: 10.1007/s15010-014-0647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong Q., Brulc J.M., Iovieno A., et al. Diversity of bacteria at healthy human conjunctiva. Invest. Ophthalmol. Vis. Sci. 2011;52(8):5408–5413. doi: 10.1167/iovs.10-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y., Yang B., Li W. Defining the normal core microbiome of conjunctival microbial communities. Clin. Microbiol. Infect. 2016;22(7):643. doi: 10.1016/j.cmi.2016.04.008. e7-.e12. [DOI] [PubMed] [Google Scholar]
- 44.Omar A., Wright J.B., Schultz G., et al. Microbial biofilms and chronic wounds. Microorganisms. 2017;5(1) doi: 10.3390/microorganisms5010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin H., Price K., Albert L., et al. Changes in the eye microbiota associated with contact lens wearing. mBio. 2016;7(2) doi: 10.1128/mBio.00198-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willcox M.D. Characterization of the normal microbiota of the ocular surface. Exp. Eye Res. 2013;117:99–105. doi: 10.1016/j.exer.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 47.Hartl R., Kerschner H., Gattringer R., et al. Whole-Genome analysis of a human Enterobacter mori isolate carrying a bla(IMI-2) carbapenemase in Austria. Microb. Drug Resist. 2019;25(1):94–96. doi: 10.1089/mdr.2018.0098. [DOI] [PubMed] [Google Scholar]
- 48.Wen X., Miao L., Deng Y., et al. The influence of age and sex on ocular surface microbiota in healthy adults. Invest. Ophthalmol. Vis. Sci. 2017;58(14):6030–6037. doi: 10.1167/iovs.17-22957. [DOI] [PubMed] [Google Scholar]
- 49.Wu Y.T., Zhu H., Willcox M., Stapleton F. The effectiveness of various cleaning regimens and current guidelines in contact lens case biofilm removal. Invest. Ophthalmol. Vis. Sci. 2011;52(8):5287–5292. doi: 10.1167/iovs.10-6785. [DOI] [PubMed] [Google Scholar]
- 50.Tilia D., Lazon de la Jara P., Zhu H., et al. The effect of compliance on contact lens case contamination. Optom. Vis. Sci. 2014;91(3):262–271. doi: 10.1097/OPX.0000000000000163. [DOI] [PubMed] [Google Scholar]
- 51.Supiyaphun C., Jongkhajornpong P. Contact lens use patterns, behavior and knowledge among University Students in Thailand. Clin. Ophthalmol. 2021;15:1249–1258. doi: 10.2147/OPTH.S304735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material/referenced in article.