Summary
Very few drugs in clinical practice feature the chemical diversity, narrow therapeutic window, unique route of administration, and reversible cognitive effects of volatile anesthetics. The correlation between their hydrophobicity and their potency and the increasing amount of evidence suggesting that anesthetics exert their action on transmembrane proteins, justifies the investigation of their effects on phospholipid bilayers at the molecular level, given the strong functional and structural link between transmembrane proteins and the surrounding lipid matrix. Molecular dynamics simulations of a model lipid bilayer in the presence of ethylene, desflurane, methoxyflurane, and the nonimmobilizer 1,2-dichlorohexafluorocyclobutane (also called F6 or 2N) at different concentrations highlight the structural consequences of VA partitioning in the lipid phase, with a decrease of lipid order and bilayer thickness, an increase in overall lipid lateral mobility and area-per-lipid, and a marked reduction in the mechanical stiffness of the membrane, that strongly correlates with the compounds' hydrophobicity.
Subject areas: molecular physiology, membrane architecture, cell biology, structural biology
Graphical abstract
Highlights
-
•
Molecular simulations of lipid bilayer interaction with volatile anesthetics
-
•
Comparison of volatile anesthetics' and nonimmobilizers' effects on lipid bilayers
-
•
Ligand-dependent partitioning of the compounds in the lipid phase
-
•
Effects on bilayer thickness, stiffness, and lipid order upon ligand partitioning
Molecular physiology; Membrane architecture; Cell biology; Structural biology
Introduction
Volatile anesthetics (VAs) are a diverse set of compounds routinely used in medical practice to induce and/or sustain a reversible state of suspended consciousness, analgesia, and amnesia. Despite the fact that modern surgery would hardly be imaginable without such compounds, little is known about their mechanism of action, especially at the molecular level. This is partly due to the high chemical and physical diversity of available VAs, which range from single-atom gases such as Xenon, to more complex molecules such as halogen-substituted ethers and even steroids. In the past decades, several different theories of anesthetic action have been proposed with the aim of explaining anesthetic behavior despite this lack of structural similarity, starting from the Meyer-Overton correlation between the lipid solubility of VAs and their clinical potency (in terms of Minimum Alveolar Concentration, MAC). This theory paved the way toward what is known as the lipid theory, which postulates that the main mechanism of action of anesthetics lies in the alteration of the structure of lipid bilayers—in particular cell membranes—in a non-specific fashion (Meyer, 1937). Some shortcomings of this hypothesis, including the lack of any anesthetic effect of other lipid-altering factors, e.g. temperature, steered the interest of research around anesthesia toward finding specific molecular targets—i.e. proteins—which could explain the clinical effects of VAs. Indeed, an increasing amount of evidence points toward ion channels located in the CNS as relevant targets for anesthetics, starting from the works of Franks and Lieb (Franks and Lieb, 1984, 1994). A detailed review of molecular targets of anesthetics can be found in Campagna et al. (2003). Interestingly, despite the increasing evidence of interactions with ion channels, the exact mechanism of action remains unclear, and researchers failed to agree on the most relevant effectors of anesthesia at clinically relevant VA concentrations. In most works, the two aforementioned approaches to explaining anesthesia—the lipid theory and the receptor theory—are largely regarded as irreconcilable. What seems often overlooked, however, is the intimate connection between transmembrane receptors such as ion channels and their surrounding lipid environment, which highlights the duality, rather than the contrast, of the two theories. Indeed, the membrane-spanning portions of integral membrane proteins are known to be affected by the surrounding lipids, so that the conformational characteristics of specific sections of the transmembrane regions may change in response to alterations of the lipid bilayer. It has been shown, for example, that bilayer thickness can directly influence protein activity (De Planque and Killian, 2003; Mouritsen and Bloom, 1993). Conversely, there is increasing experimental evidence that the presence of proteins embedded in the membrane has profound effects on the latter’|s stabilization, mediated mainly by hydrophobic interactions (Dumas et al., 1999). At higher scales, the reciprocal interaction of the membrane’|s lipid environment and embedded proteins has also been shown to be mediated by so-called lipid rafts (Lingwood and Simons, 2010), which are sub-μm domains of spatially organized lipids, typically sphingomyelin and cholesterol (Allen et al., 2006; Levental et al., 2011; Moon et al., 2017).
It appears thus entirely reasonable that an interaction of increasingly hydrophobic compounds, such as VAs, within biological membranes might have significant effects on membrane organization and structure, but at the same time this cannot happen without altering the energetic landscape of the interactions between membranes and transmembrane receptors. The idea that small solutes such as VAs bear the potential of altering the mechanics and thermodynamics of the lipid bilayer, with possible consequences on the dynamics of embedded proteins, was already introduced in the work of Cantor (1999), who elegantly discussed the possible relevance of lateral pressure profiles within the lipid bilayer and suggested the mechanistic link between anesthetics, the lipid bilayer, and embedded ion channels (Cantor, 1997). Indeed, earlier molecular dynamics simulations by Huang et al. had predicted a possible structural effect of anesthetics within the phospholipid bilayer, in the form of an increased lateral diffusion of lipids and an increase in the overall fluidity of the bilayer (Huang and Bertaccini, 1995). More recently, following earlier speculations suggesting a role of lipid rafts in anesthesia (Gray et al., 2013; Lee, 1976; Lerner, 1997), Pavel et al. demonstrated a membrane-mediated effect of anesthetics, whereby the anesthetic-induced alteration of lipid raft organization is able to modulate the sensitivity of channel proteins to anesthetics (Pavel et al., 2020). In addition to these considerations, the direct effect of anesthetics on transmembrane receptors might be exerted within the transmembrane portion of the receptors rather than on the intracellular or extracellular domains alone and might thus be connected to the ability of compounds to partition inside the membrane and laterally diffuse within the lipid phase prior to interacting directly with cryptic, hydrophobic sites on the target. As a matter of fact, compounds that are more soluble in oil-like media, as is the case for VAs as shown by the Meyer-Overton correlation, tend to partition inside the membrane rather than in aqueous solutions, and vice versa.
In the context of investigating the properties of lipid bilayers, a vast literature exists exploring the behavior of model phospholipid membranes in different physical contexts and the structural and functional link between membranes and embedded proteins and peptides. Indeed, it is well known that the structure of phospholipid bilayers has strong functional consequences (Zhuang et al., 2014). The structural parameters usually reported in both experimental and computational studies include (a) the area per lipid (APL), which can be calculated from molecular densities or geometrically from the membrane patch surface; (b) the bilayer thickness δ, which is directly related to the APL; (c) deuterium order parameters (SCD), which provide quantitative evidence of lipid chain order and the membrane rigidity resulting from this; (d) direct measures of the mechanical characteristics of the membrane, such as the bilayer bending modulus (Kc). Owing to the limitations, both methodological and economical, of experimental settings aimed at investigating such properties for a vast array of model membranes in different physical and biochemical contexts, computational approaches such as molecular dynamics (MD) have proven a valuable tool for exploring and rationalizing the structural characteristics and interaction phenomena within model bilayers at the molecular level. While a great number of computational investigations employed single-component lipid patches (Grasso et al., 2018; Huang and Bertaccini, 1995; Tang and Xu, 2002), mostly of phosphatidylcholines (PCs) or phosphatidylethanolamines (PEs), recent advances in lipid force fields (Dickson et al., 2012, 2014; Jämbeck and Lyubartsev, 2012; Klauda et al., 2010; Pluhackova et al., 2016) and the increasing power of computational resources have paved the way for the simulation of complex, composite bilayers formed by multiple lipid species, and varying cholesterol concentrations, both at all-atom (AA) and coarse-grained (CG) resolutions (Ingólfsson et al., 2017 and references therein).
With this in mind, the present work focuses on investigating the interaction between volatile anesthetics and a composite model mammalian cell membrane through the use of computational molecular modeling, to explore the effects of VAs on lipid bilayers. With the goal of exploring the effect of a chemically and physically diverse set of hydrophobic compounds spanning a wide range of clinical potencies, we carried out simulations with desflurane (2-(difluoromethoxy)-1,1,1,2-tetrafluoroethane), a fluorinated ether with a MAC of 6% (Hudson et al., 2019), methoxyflurane (2,2-dichloro-1,1-difluoro-1-methoxyethane), a potent halogenated methyl ethyl ether with a MAC value of 0.16%, now largely abandoned in the light of its nephrotoxicity (Mazze, 1971), and ethylene, which is only mildly anesthetic with a MAC value of 67% (Miller et al., 1969). Simulations were also carried out with F6 (1,2-dichlorohexafluorocyclobutane), a widely investigated nonimmobilizer which does not follow the Meyer-Overton correlation in that it does not induce complete anesthesia as would be expected from its lipophilicity, but it has been demonstrated to induce amnesia (Eger et al., 2001; Perouansky et al., 2007; Taylor et al., 1999).
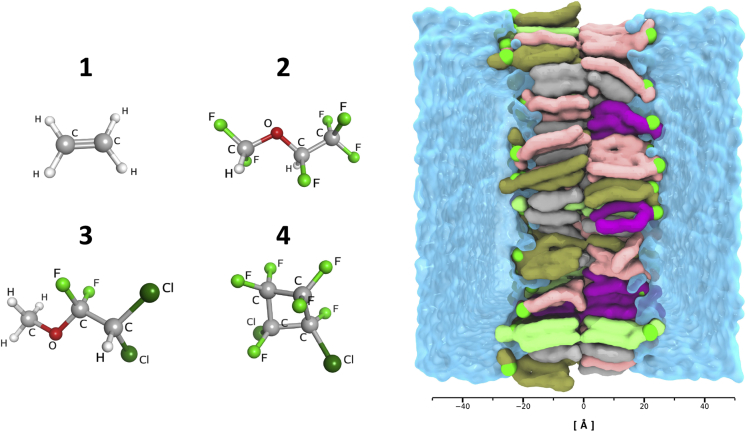
A graphical summary of the model membrane and of the simulated VAs is reported in Figure 1.
Figure 1.
Visual overview of the simulated systems
Left: visualization of the three simulated VAs (1) ethylene, (2) desflurane, (3) methoxyflurane, and the nonimmobilizer (4) F6. Right: visualization of the membrane system in its explicit TIP3P water box with ions and ligands omitted for clarity. P atoms highlighted in green, POPC lipids in pink, cholesterol in light gray, POPE in purple, POPS in dark green, PSM in bright green. Length scale in Ångstrom reported below for reference, centered at the membrane core region.
Results
Potent VAs alter the membrane structure upon partitioning
To quantitatively assess both the quality of the membrane model itself and the effect of volatile anesthetics on overall membrane structure, the geometric area per lipid (gAPL) and bilayer thickness (δ) were evaluated and are reported for all systems in detail in Table 1. The former is a crucial parameter influencing lipid diffusion profiles, lipid chain order, and overall membrane elastic properties. It also represents a metric to assess the reached equilibrium of membrane simulations, along with the closely related bilayer thickness. The control simulation without any ligands yielded an average gAPL of 42.89 Å2 (95% CI: 42.83–42.95 Å2) and an average bilayer thickness of 46.85 Å (95% CI: 46.81–46.89 Å), and proved to be consistent both with previous computational studies of membranes with similar lipid composition and comparable cholesterol content (Klähn and Zacharias, 2013; Saeedimasine et al., 2019; Shahane et al., 2019b) and with experimental data on cholesterol-enriched membranes (Maulik and Shipley, 1996), although it is to be noted that bilayer thickness heavily depends on the specific bilayer composition (Li et al., 2012) and experimental data of membranes with the exact lipid composition of the present model is, to the best of our knowledge, not available. Nevertheless, the reduced gAPL and δ values are consistent with the high cholesterol content (∼34%) inducing membrane condensation, as demonstrated in earlier literature (Hofsäß et al., 2003; Leftin et al., 2014; Meyer and Smit, 2009; McIntosh, 1978).
Table 1.
Average geometrical area per lipid, bilayer thickness, and frequency of water permeation for all simulated systems.
| System | gAPL [Å2] | Bilayer Thickness (δ) [Å] | Water permeation frequency [H2O/μs] |
|---|---|---|---|
| C | 42.89 [42.83–42.95] | 46.85 [46.81–46.89] | 16 |
| E12.5 | 43.40 [43.36–43.44] | 46.77 [46.71–46.83] | 40 |
| E25 | 43.90 [43.80–44.00] | 46.74 [46.68–46.80] | 37 |
| E50 | 45.16 [45.12–45.20] | 46.52 [46.46–46.58] | 112 |
| D12.5 | 44.93 [44.81–45.05] | 46.17 [46.03–46.31] | 75 |
| D25 | 46.47 [46.41–46.53] | 45.91 [45.89–45.93] | 163 |
| D50 | 48.18 [46.49–49.87] | 45.67 [45.45–45.89] | 285 |
| M12.5 | 45.12 [45.08–45.16] | 45.88 [45.87–45.89] | 64 |
| M25 | 46.86 [46.83–46.89] | 45.40 [45.38–45.42] | 160 |
| M50 | 48.53 [46.92–50.14] | 45.16 [44.89–45.43] | 391 |
| F6 12.5 | 44.95 [44.81–45.09] | 45.97 [45.89–46.05] | 93 |
| F6 25 | 46.85 [46.65–47.05] | 45.39 [45.28–45.50] | 206 |
| F6 50 | 46.44 [45.30–47.58] | 45.52 [45.09–45.95] | 155 |
95% CIs are reported in square brackets for block-averaged quantities.
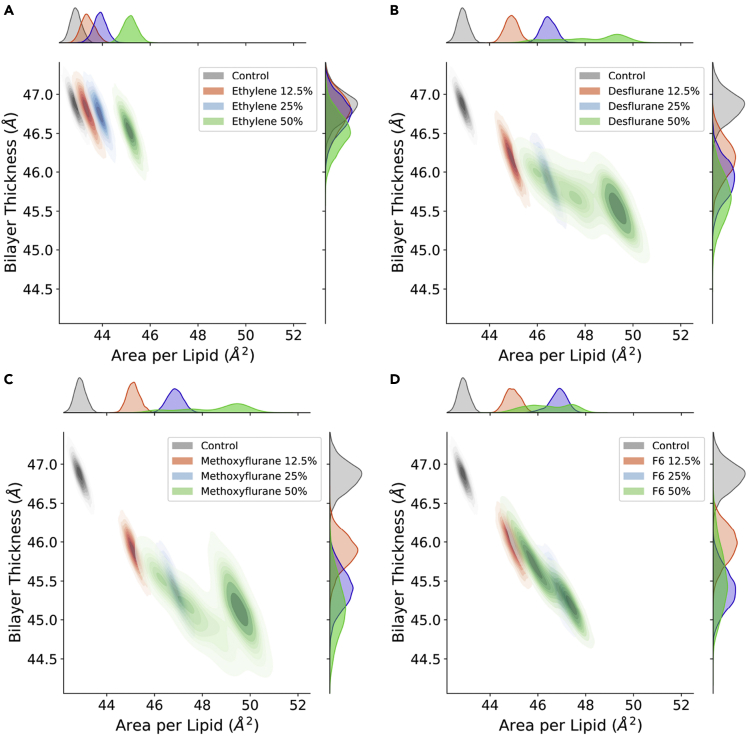
Figure 2 shows the effect of increasing ligand concentrations on both gAPL and thickness. In the case of ethylene (Figure 2A), no significant effect of ligand concentration on bilayer thickness is observed (from 46.85 Å to 46.52 Å), with only a mild increase in area per lipid, which reaches 45.16 Å2 with 50% ethylene. Desflurane (Figure 2B) and methoxyflurane (Figure 2C) on the other hand induce a marked reduction in bilayer thickness down to 45.67 Å with 50% desflurane and 45.16 Å with 50% methoxyflurane, despite the steric hindrance of the high number of ligand molecules partitioned within the membrane. At the same time, these two anesthetics induce a marked increase in gAPL, up to 48.18 Å2 and 48.53 Å2 for systems with 50% desflurane and methoxyflurane, respectively. Overall, the latter two ligands induce a progressive reduction of membrane thickness, along with a lateral spreading of the lipids on the xy plane, both in a fashion proportional to ligand concentration. This effect is totally absent for ethylene concentrations up to 25%, with only a mild increase in gAPL induced at 50% and no measurable thickness reduction effect. These results are in agreement with earlier computational studies reporting a significant lateral expansion and simultaneous thickness contraction induced in lipid membranes by halothane, another VA, over a wide range of molar fractions (Koubi et al., 2000; Pickholz et al., 2005; Tu et al., 1998). Lastly, simulations with the nonimmobilizer F6 (Figure 2D) highlight a reduction in bilayer thickness (from 46.85 Å to 45.52 Å with 50% F6) comparable to the simulations with desflurane and methoxyflurane, whereas the increase in gAPL is more subdued at higher concentrations, reaching at most 46.44 Å2 with 50% F6.
Figure 2.
Distribution of the bilayer thickness (δ) and geometric area per lipid (gAPL)
(A–D)Control simulation vs. ethylene (A), desflurane (B), methoxyflurane (C), and F6 (D) at increasing concentrations. Marginal axes show the individual data distributions collected in the last 750 ns of the simulations. Control simulation without anesthetics shown in gray, 12.5% concentration in red, 25% in blue, and 50% in green.
The increase in gAPL induced by ligand partitioning came alongside an increase in spontaneous water permeation through the membrane, reported as the number of water molecules crossing the bilayer per microsecond in Table 1: throughout the control simulation, a water permeation frequency of 16 water molecules/μs was observed, whereas this frequency increased to up to 285 molecules/μs and 391 molecules/μs in the case of 50% desflurane and 50% methoxyflurane, respectively. Conversely, just as for gAPL and bilayer thickness, more subdued differences were observed with ethylene, with at most 112 molecules/μs at the highest concentration of 50%. Throughout the simulations with F6, a permeation frequency of up to 206 molecules/μs was observed at 25% simulated fraction, with a slightly lower frequency of 155 molecules/μs at 50% concentration, consistent with the trends of gAPL and bilayer thickness. Despite the increase in spontaneous permeation frequency with increasing ligand concentrations, no pore formation was observed throughout the whole set of simulations, with no disruption of the overall structural integrity of the bilayer.
Anesthetics and nonimmobilizers are predicted to have specific localization areas within the bilayer
The partitioning of ligands inside the lipid bilayer not only plays a crucial role in ligand-receptor interaction with transmembrane proteins (Vauquelin and Packeu, 2009) but can also significantly alter the bilayer’|'s structural and mechanical properties (Koubi et al., 2000; Tsuchiya and Mizogami, 2013; Tu et al., 1998; Yamamoto et al., 2012). The analysis of the density distributions of the different membrane components and of the ligands along the z coordinate highlights a marked tendency of the four ligands to partition inside the bilayer in specific hydrophobic regions.
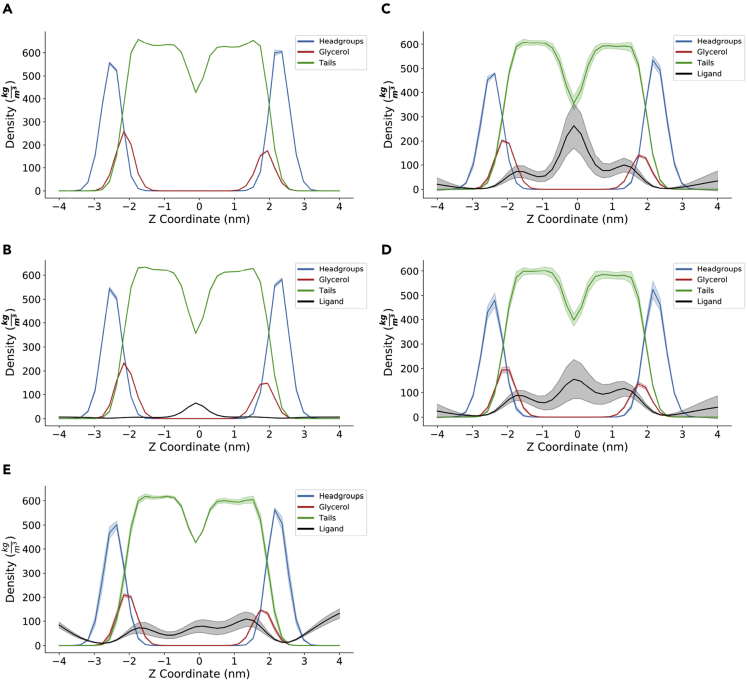
Figure 3 reports the density distributions for the control simulation (Figure 3A) and the simulations at the highest concentration of ethylene (Figure 3B), desflurane (Figure 3C), methoxyflurane (Figure 3D), and F6 (Figure 3E). The corresponding plots for 12.5% and 25% ligand concentrations, which highlight the same qualitative distribution pattern, are reported in Figures S1 and S2, respectively. For desflurane and methoxyflurane, three main areas of localization clearly emerge: the main peak is located at the bilayer center, corresponding to the minimum of lipid tail density. This is consistent with the hydrophobic nature of these compounds, and explains why the massive ligand partitioning inside the membrane does not result in a simultaneous increase in bilayer thickness, as would be expected by the effect of steric hindrance and molecular volume alone. Indeed, owing to the low lipid tail density in the membrane core, resulting in less occupied molecular volume, many freely diffusing hydrophobic species are known to temporarily localize in this region, including cholesterol during flip-flop transitions (Bennett et al., 2009). The secondary peaks on the other hand are located near the membrane-water interface, immediately below the glycerol groups. This is in agreement with earlier computational findings by Pohorille et al., who predicted this very area of localization to be involved in the molecular mechanism of anesthesia (Christophe Chipot et al., 1997; Pohorille et al., 1996, 1998). Interactions of volatile anesthetics near the water-lipid interface region have also been reported in the past by Tang and Xu, who employed MD simulations to evaluate the effect of halothane on a gramicidin A channel protein embedded in a DMPC bilayer (Tang and Xu, 2002). While these earlier simulations employed more simplistic membrane models composed of a single lipid type, and investigated remarkably lower timescales, the localization near the water/lipid interface is herein predicted to partially occur also in our composite, cholesterol-enriched membrane model, albeit not as predominantly as the localization at the membrane core. On the contrary, in the case of F6, the localization at the interface appeared comparable to that at the membrane core, resulting in a different density pattern with respect to the other compounds, with no predominant peak at the membrane core. These findings are consistent with the different effects observed for F6 on gAPL with respect to the VAs.
Figure 3.
Density distributions of lipid headgroups (blue), glycerol backbone (red), lipid tails (green), and anesthetics (black)
(A–C) control simulation, (B) with 50% ethylene, (C) with 50% desflurane, (D) with 50% methoxyflurane, and (E) with 50% F6. Shaded colors represent 95% CIs.
Quantitative measures of the tendency of ligands to reside inside the lipid bilayer with respect to the aqueous solvent are reported in literature in the form of either ligand equilibrium partition coefficients (Vauquelin and Packeu, 2009)—usually calculated as the ratio between the ligand concentration in the solvent and the concentration within the membrane—or directly as molar (Herold et al., 2017) or molal (Seeman, 1972) ligand concentration inside the membrane. Whatever the metric, these quantities depend, among others, on the chemical and physical nature of the ligand itself, in particular, its hydrophobicity and the presence of hydrophilic moieties, on the temperature of the membrane, i.e. its phase state, and on the membrane cholesterol concentration (Vauquelin and Packeu, 2009).
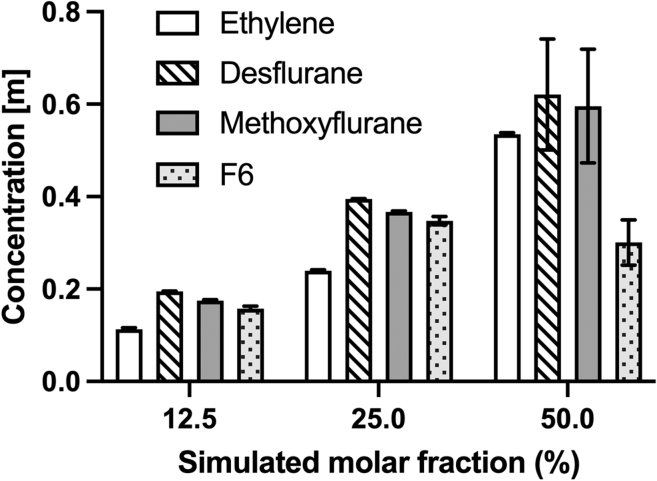
To provide a direct quantitative measurement of the amount of ligand able to dissolve into the membrane, Figure 4 reports the molal concentration reached by the four simulated ligands within the lipid bilayer. Results confirm that the concentration of ligands inside the membrane increases with increasing amounts of simulated ligand molecules, as expected by the physical characteristics of these compounds. One notable exception is represented by F6 (dotted bars in Figure 4), whose concentration inside the bilayer is comparable to that of the other compounds at 12.5% and 25% simulations, reaching up to 0.35 mol/kg (at 25% simulated molar fraction, 95% CI 0.33–0.37), but showing no further increase in the case of 50% simulations, plateauing at 0.30 mol/kg (95% CI 0.20–0.40) and with a considerable amount of ligand aggregating in the water phase without entering the membrane. Also, the analysis of ligand concentration inside the membrane highlights that ethylene (white bars in Figure 4) also partitioned inside the bilayer, albeit at lower rates in the 12.5% and 25% simulations. Conversely, when simulated at 50% molar fraction, the reached concentration (0.535 mol/kg, 95% CI: 0.529–0.541) is comparable to the one of desflurane (patterned bars in Figure 4, 0.621 mol/kg, 95% CI: 0.385–0.856) and methoxyflurane (shaded bars in Figure 4, 0.596 mol/kg, 95% CI: 0.355–0.837). Also, it is worth noting how the considerable number of ligands present at 50% molar fraction leads to greater fluctuations in ligand partitioning in the case of the latter two ligands, but not in the case of ethylene. This is a consequence of the key differences in behavior between ethylene and the other simulated ligands: firstly, ethylene does not form aggregates in the water phase even at 50% concentration as opposed to the other three ligands. Indeed, desflurane and methoxyflurane are observed to enter the membrane in the form of aggregates of up to tens of molecules, while F6 forms aggregates at 50% concentration that are partially unable to enter the bilayer and remain in the water phase throughout the simulations, resulting in lower overall membrane partitioning (see dotted bars in Figure 4). Secondly, ethylene did not show the secondary localization areas below the glycerol groups inside the membrane (see Figure 3), which are instead present for the other three ligands, but rather preferably positions itself at the membrane core, making ligand exchange between the membrane and the water phase less frequent.
Figure 4.
Molal concentration of the four simulated ligands inside the bilayer
Concentrations calculated as number of moles of anesthetic per kilogram of membrane. Error bars on the histograms represent the error estimate after block averaging.
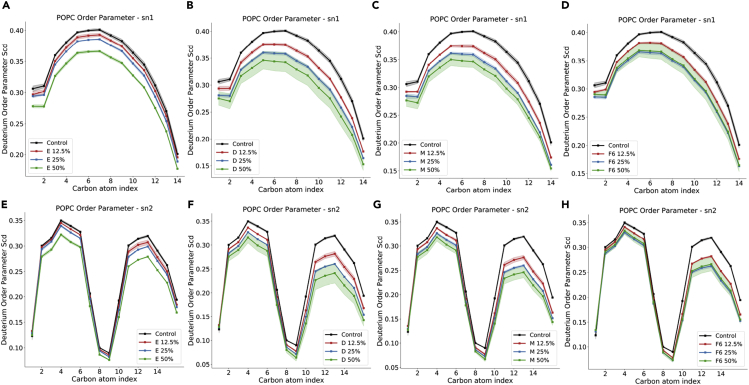
VAs and F6 decrease lipid chain order already at 12.5% molar fraction
Deuterium order parameters SCD represent a quantitative measurement of lipid packing and provide insights into the mobility of the hydrophobic chains. Data for POPC from the control simulation without ligands (Figure 5, blue lines) are in good agreement with recently published results of compositionally similar, cholesterol- and sphingomyelin-enriched POPC/POPE membranes (Saeedimasine et al., 2019), and confirms the membrane-ordering effect induced by cholesterol. Conversely, in the presence of desflurane (Figures 5B and 5F) and methoxyflurane (Figures 5C and 5G), the mechanical consequence of ligand partitioning within the hydrophobic core as well as below the glycerol groups is a reduction in acyl chain order parameters, with a trend proportional to the ligand concentration (Figure 5). This behavior is also present in the simulations with F6 (Figures 5D and 5H), with the exception of simulations at 50% molar fraction, where the effect of the ligand on lipid chain order is comparable within error to that at 25% concentration. This is coherent with the finding that there are no remarkable differences in the concentration reached by F6 within the bilayer at 25% and 50% simulated molar fraction (see results above), hence a comparable effect on lipid packing is not unexpected.
Figure 5.
Lipid tail order parameters for POPC sn1 (top row) and sn2 (bottom row) chains, with different ligands
(A–H) and (E) ethylene; (B) and (F) desflurane; (C) and (G) methoxyflurane; (D) and (H) F6. For the corresponding data for POPE, POPS, and PSM, see Supplementary Information. Shaded intervals correspond to 95% CIs.
The effect on lipid order is more subdued in the case of ethylene (Figures 5A and 5D), where the decrease in SCD is particularly evident only at 50% concentration, with only marginal reductions (<0.01) at lower ligand concentrations. These trends, reported in Figure 5 for POPC, are analogous for the other lipid species included in the employed membrane model (see Supplementary Information), and hint at a membrane-destabilizing effect of ligand partitioning, with consequences on overall bilayer mechanics.
Desflurane, methoxyflurane, and F6 decrease membrane bending rigidity in a concentration-dependent manner
In the light of the ligands' tendency to partition inside the lipid bilayer, and of the structural consequences thereof observed by the analysis of area per lipid, bilayer thickness and acyl chain order parameters, a more specific quantification of the bilayer’s mechanical characteristics was carried out by directly determining the bilayer bending modulus using a previously proposed methodology relying on the analysis of lipid splay.
The bilayer bending modulus for the control simulation is 88.80 kT (95% CI: 87.16–90.44), and while a direct comparison with other computational and experimental studies is often not trivial due to the differences in membrane composition, temperature, and methodology, this result is remarkably consistent with earlier studies of membranes with similar cholesterol content (around 0.3 mole fraction) which induces structural condensation of the lipid phase yielding a considerable increase in membrane stiffness and a shift toward the liquid-ordered phase (Khelashvili et al., 2013; Subczynski et al., 2017). Furthermore, the obtained value for the control simulation agrees with earlier literature reporting experimentally determined stiffness values for plasma membrane vesicles (PMVs, Kc = 99.75 kT), which are representative systems of the pure plasma membrane in vitro (see (Pontes et al., 2013) and references therein).
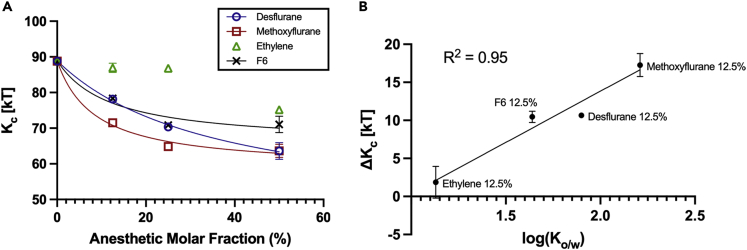
The trend of reduction of bilayer bending stiffness at increasing anesthetic concentrations is visible in Figure 6A and Table 2. At 12.5% anesthetic concentration, the presence of desflurane, methoxyflurane, and F6 leads to a reduction in monolayer bending stiffness by 12.01%, 19.44%, and 11.78%, respectively, compared to a mere 2.10% reduction with ethylene. At 25% anesthetic concentration, the bending stiffness is reduced by 20.80% and 26.95% by desflurane and methoxyflurane, respectively, and by 20.20% with F6, compared to a limited 2.48% reduction caused by ethylene. Lastly, in the simulations with 50% anesthetic molar fraction, the bending stiffness is reduced by 28.38% with desflurane and 28.30% by methoxyflurane, while the effect of F6 remains again comparable to the 25% simulation, yielding a reduction of the bilayer bending modulus of 19.98%. Only at this higher concentration does ethylene lead to a noticeable reduction in bending stiffness by 15.32%. This is consistent with order parameter results, which showed a decrease in lipid tail packing in the presence of ethylene only at 50% concentration (see Figure 5).
Figure 6.
Effect of ligands on membrane stiffness
(A) Bilayer bending modulus in kT units for the different systems. Control system represented as 0% ligand concentration. Error bars represent the error estimate after block averaging, omitted when smaller than the datapoint for clarity.
(B) Correlation between anesthetic lipophilicity (in terms of the logarithm of the octanol/water partition coefficient, log(Ko/w)) and the decrease in membrane bending modulus in kT units, ΔKc. Error bars represent the error estimate after block averaging, omitted when smaller than the datapoint for clarity.
Table 2.
Bilayer bending modulus Kc in kT units and reduction of Kc with respect to control simulation, ΔKc, for each simulated system.
| System | Kc [kT] | ΔKc |
|---|---|---|
| C | 88.80 [87.16–90.44] | – |
| E12.5 | 86.94 [84.42–89.44] | 1.86 |
| E25 | 86.60 [85.14–88.06] | 2.20 |
| E50 | 75.20 [73.98–76.42] | 13.60 |
| D12.5 | 78.14 [76.94–79.32] | 10.66 |
| D25 | 70.34 [68.82–71.84] | 18.46 |
| D50 | 63.60 [59.06–68.14] | 25.20 |
| M12.5 | 71.54 [69.92–73.14] | 17.26 |
| M25 | 64.86 [64.58–65.14] | 23.94 |
| M50 | 63.66 [60.04–67.30] | 25.14 |
| F6 12.5 | 78.33 [76.85–79.82] | 10.47 |
| F6 25 | 70.87 [69.94–71.80] | 17.93 |
| F6 50 | 71.07 [66.61–75.52] | 17.73 |
95% CIs reported in square brackets.
Overall, the trends in reduction in membrane bending stiffness are consistent with the hydrophobicity of these compounds. From the analysis of the data from the 12.5% concentration simulations, which is the closest to clinical concentrations, a linear relationship emerges between the lipophilicity of the ligands—quantified by the octanol/water partition coefficient log(Ko/w)—and the reduction in bilayer stiffness (ΔKc) observed in simulations (R2 = 0.95, Figure 6B). Interestingly, this relationship seems to hold true also for F6, which is not an anesthetic but a convulsant with amnesic properties, supporting the hypothesis that the alteration of bilayer mechanics might not be per se the mechanistic cause of anesthesia, but might be implicated in some of the effects caused by these compounds, especially at supra-clinical concentrations.
Discussion
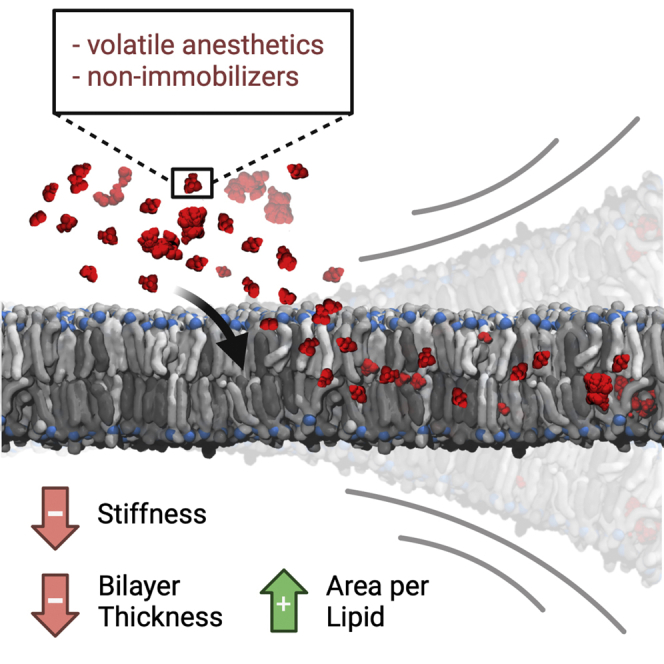
In the present work, we employed long all-atom molecular dynamics simulations to assess the structural effects of the volatile anesthetics desflurane, methoxyflurane, ethylene, a low-potency control, and the nonimmobilizer F6 on a model composite lipid bilayer composed of POPC, POPE, POPS, PSM, and cholesterol. Anesthetics rapidly partition inside the bilayer, reaching intra-membrane concentrations of approximately 0.6 molal, while F6 is unable to reach concentrations higher than 0.3 molal even when simulated at 50% ligand/lipid molar fraction. Desflurane and methoxyflurane preferentially localize at the membrane core region and immediately below the glycerol groups of the bilayer, with structural consequences on both area per lipid and bilayer thickness. Indeed, the partitioning of ligands causes a contraction in bilayer thickness while at the same time reducing lateral condensation and causing an increase in area per lipid and in spontaneous water permeation, albeit, with no pore formation or disruption of overall membrane integrity. The convulsant F6 shows a different localization pattern within the membrane, with preferential interaction below the lipid/water interface and a less prominent residency at the membrane core region, but with similar structural effects with respect to the aforementioned VAs. The structural rearrangement of the membrane has direct consequences on its mechanical properties, as testified by a progressive reduction in lipid hydrocarbon chain packing. The reduced energetic cost of splaying adjacent lipid tails caused by ligand partitioning leads to a reduction in bilayer bending rigidity in a fashion proportional to ligand concentration. These structural effects are not observed for ethylene at a molar ratio of up to 0.25 with respect to the lipids, with only marginal effects at 0.5 M ratio. Consistently with these considerations, ethylene also constitutes the least hydrophobic among the three studied VAs. It is to be underlined how the nonimmobilizer F6 caused a comparable reduction in bilayer bending rigidity despite its lack of potency as a general anesthetic. Hence, also bearing in mind that the simulated concentrations are above the typical concentrations reached in clinical settings, these findings shed light on important aspects of anesthetic-membrane interactions. Firstly, the two potent VAs and the nonimmobilizer F6 studied herein have, even at the smallest studied concentration, the capacity to alter the energetic landscape of a model mammalian lipid bilayer, which results in profound changes of its mechanical characteristics in terms of a marked reduction in bending stiffness and an overall shift toward a liquid-disordered phase, as shown by the reduction in thickness, the increase in APL, the increase in spontaneous water permeation, and the reduction of lipid chain order. This effect appears as antagonistic to the role of cholesterol, which induces instead a shift toward the liquid-ordered phase and an overall increase in membrane rigidity (Subczynski et al., 2017). Interestingly, VAs and cholesterol seem to have instead a similar effect in the context of lipid raft microdomains, whose number and size have been recently shown to increase with both anesthetics and cholesterol (Pavel et al., 2020). Given the fundamental role of the cell membrane not only in overall cell mechanics and structural stability but also in the function of several transmembrane proteins, including important ion channels thought to be directly involved in anesthesia or its side effects (Bertaccini, 2010; Bertaccini et al., 2013; Franks and Lieb, 1994; Herold and Hemmings, 2012; Yamakura et al., 2003), it appears entirely reasonable that bilayer alterations might be, directly or indirectly, involved in some of the effects exerted by VAs and F6, in the same way in which cholesterol is a crucial modulator of membrane mechanics and essential for many membrane functions. As a matter of fact, a hybrid protein/lipid mechanism based on the alteration of the physics of the lipid membrane has been recently proposed by Pavel et al., who described and demonstrated in vivo the indirect effect of volatile anesthetics on membrane-embedded channel proteins by means of an alteration of sphingomyelin lipid rafts (Pavel et al., 2020). Despite failing to highlight any effect of VAs on pure DOPC liposomes, employed as a model system of the pure membrane, the research provided further evidence for the key role of membrane biophysics in the molecular mechanisms of anesthetics, and supports the speculation that anesthetics directly interact with the phospholipid membrane, with diversified effects not only at different time and length scales, e.g. on local lipid arrangement vs. on larger-scale lipid microdomains, but also at different concentrations, e.g. clinical vs. supra-clinical. Indeed, not only does the alteration of the surrounding lipid environment bear the potential of altering the function of channel proteins, e.g. by modifying the energetic cost of key functional motions, but the rapid partitioning of VAs into the hydrophobic core might also be an essential prerequisite for anesthetics to reach cryptic hydrophobic binding sites of such proteins within regions embedded in the membrane, which are inaccessible from the external water phase. In this sense, the findings reported herein do not clash with earlier evidence of a direct action of anesthetics on ion channels (John Mihic et al., 1997; Mascia et al., 2000), which is still debated to be the final mechanism of action causative of anesthesia. Instead, the computational predictions provide a quantification of the interaction between VAs and the lipid phase and the mechanical alterations of the latter at increasing VA concentrations. This mechanism might thus be necessary, but arguably not sufficient, for a compound to exhibit anesthetic potency, thereby explaining both the Meyer-Overton correlation and outliers thereof such as nonimmobilizers, featuring considerable hydrophobicity but low to no anesthetic potency. This is confirmed by analyzing the effect on bilayer mechanics of the nonimmobilizer F6, which is herein predicted to alter membrane behavior in a similar manner to potent anesthetics. This further suggests that the alteration of the lipid membrane per se is unlikely to be the sole mechanistic cause of anesthesia as a whole. Rather, it might be a biophysical mechanism involved in some of the effects that are exerted both by anesthetic agents and nonimmobilizers such as F6, which has been shown, e.g. to induce convulsions and amnesia in vivo. Also, the direct action on membrane mechanics might rather provide a mechanistic basis to explain the side effects of anesthetics, which arise at higher concentrations and are in common with convulsants (Koblin et al., 1981; Modica et al., 1990). Indeed, given the exacerbation of the alteration of bilayer structure and mechanics predicted herein at such higher concentrations—0.25 and 0.5 mole fractions—it is reasonable that such a mechanism might be involved in the molecular basis of the side effects of VAs at supra-clinical concentrations.
Conclusions
The molecular mechanisms of general anesthesia are to this day an unsolved medical puzzle. While recent literature generally considers transmembrane proteins as the main functional target of volatile anesthetics, the Meyer-Overton correlation clearly hints at the ability of these compounds to interact with the lipid bilayer of cell membranes, even if the final functional action is not exerted directly on the membrane itself. Long molecular dynamics simulations of the three VAs ethylene, desflurane, methoxyflurane, and of the nonimmobilizer F6 confirm the strong tendency of these ligands to partition within the hydrophobic environment of a model membrane, and allowed to quantify the structural effects this determines: a reduction in bilayer thickness, a decrease in lipid chain order, and a reduction of membrane stiffness, with a trend proportional to the amount of partitioned ligands. Given the strong correlation observed between the compounds' lipophilicity and the reduction in the membrane bending modulus caused by their inclusion within the membrane, it appears that the phospholipid membrane might be a key component in determining some of the effects of anesthetics on channel proteins, by altering their structural and mechanical characteristics in the presence of VAs with possible consequences on embedded protein function and on the intracellular link between the membrane and the cytoskeleton. Moreover, the remarkable tendency to dissolve in the lipid phase followed by lateral diffusion within the membrane might be an essential step to reach key functional hydrophobic binding pockets in transmembrane proteins, which would be inaccessible from the aqueous solvent, such as some transmembrane domains which have been shown to bind anesthetics (Mascia et al., 2000). These considerations are well in line not only with the strong relationship between potency and hydrophobicity but also with the most recent theories indicating ion channels as ultimate targets for general anesthetics, and pointing at the lipid environment of the membrane as a first transducer of anesthetic action (Pavel et al., 2020). At the same time, the functional distinction between general anesthetics and compounds without any anesthetic effect but high lipophilicity, such as F6, might involve processes and molecular players downstream of the interaction with the membrane. This concept highlights how the lipid-centered and the protein-centered theories of anesthetic action are not, in fact, irreconcilable, but might rather be two aspects of a composite mechanism, which sees the interaction with the lipid membrane as a necessary but perhaps not sufficient condition. A more thorough analysis of how this occurs and to which extent, especially as to where the discrimination between general anesthetics and non-anesthetic Meyer-Overton outliers takes place, as well as of the effect of the membrane alteration on the cytoskeleton linked at the intracellular interface, certainly warrants further computational and experimental investigations, and seems well worth pursuing further.
Limitations of the study
-
•
While the membrane model employed in this work is a multi-component membrane which accounts for the major lipid constituents of mammalian cell membranes, it still represents a simplified representation, especially in the context of neural membranes which include several types of different phosphatidylcholines, phosphatidylethanolamines, sphingomyelins, phosphatidylserines, glycolipids, cerebrosides, and phosphatidylinositols, just to name a few. Building increasingly realistic models of cellular membranes is an active topic of research and requires major computational efforts, often demanding the use of coarse-grained modeling and extended parameter validations to accurately capture the physical and chemical characteristics of the simulated species.
-
•
The present work focuses on the effects of three different VAs of different chemical structure and spanning a wide range of clinical potencies. However, several other VAs exist that were not included in the present work, and are very well worth investigating in further studies. Also, we herein included a compound that would be expected to have high potency as an anesthetic based on its hydrophobicity and structural similarity to actual VAs, but actually lacks any anesthetic effect, namely F6. Given the comparable effect of this compound on pure membrane mechanics, further investigations are needed to explore downstream events (e.g. the interaction with transmembrane proteins) that would ultimately set apart potent anesthetics from hydrophobic nonimmobilizers and other similar negative controls.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| GROMACS version 2020.4 | (Abraham et al., 2015) | https://manual.gromacs.org/ |
| CHARMM-GUI Membrane Builder | (Jo et al., 2009) | https://charmm-gui.org/?doc=input |
| MDAnalysis version 2.0.0 | Michaud-Agrawal et al. (2011) | https://www.mdanalysis.org |
| Python version 3.7 | (Van Rossum and Drake, 2009) | https://www.python.org/downloads/release/python-370/ |
| OpenStructure version 2.3 | (Biasini et al., 2013) | https://openstructure.org |
| VMD Version 1.9.3 | (Humphrey et al., 1996) | https://www.ks.uiuc.edu/Research/vmd/ |
| Methodology for determining elastic properties of lipid assemblies from MD simulations | (Johner et al., 2016) | https://doi.org/10.1186/s12859-016-1003-z |
Resource availability
Lead contact
Further information and requests for resources should be directed to Lead Contact, prof. Marco A. Deriu (marco.deriu@polito.it).
Materials availability
This study did not generate any novel reagents and all materials used in this study are reported either the main text or in the Supplemental Information.
Method details
System setup
To overcome the intrinsic simplifications of single-component bilayers, and to account for the presence of cholesterol, which has a well-documented ordering effect on membranes (Róg et al., 2009) with profound consequences on their mechanical properties (Leftin et al., 2014; Needham and Nunn, 1990), we chose to simulate a composite asymmetrical lipid patch representative of the mammalian cell membrane, as first described by Zachowski (1993) (Zachowski, 1993) and employed in computational studies by Klähn and Zacharias (2013) and, more recently, Shahane et at (Shahane et al., 2019b), composed of POPC (1,2-palmitoyl-oleoyl-sn-glycero-3-phosphocholine), POPE (1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine), POPS (1,2-palmitoyl-oleoyl-sn-glycero-3-phosphoserine), PSM (palmitoylsphingomyelin) and Cholesterol (CHOL). The detailed amounts of the lipids in the two leaflets are reported in the following table:
Table. Number of different lipid molecules in the two leaflets of the model mammalian membrane
| Lipid | Inner Leaflet | Outer Leaflet | Total |
|---|---|---|---|
| POPC | 40 | 106 | 146 |
| POPE | 132 | 34 | 166 |
| POPS | 82 | 8 | 90 |
| PSM | 10 | 116 | 126 |
| CHOL | 136 | 136 | 272 |
| Total | 400 | 400 | 800 |
Bilayer systems were assembled using the Membrane Builder (Jo et al., 2007, 2009; Wu et al., 2014) tool of CHARMM-GUI (Jo et al., 2008), with a fixed number of 50 TIP3P waters per lipid to ensure adequate lipid hydration even at higher ligand concentrations, and a physiological NaCl concentration of 0.15M. In addition to the control simulation without any anesthetic, different systems were set up by randomly inserting desflurane, methoxyflurane, ethylene and F6 (1,2-Dichlorohexafluorocyclobutane) respectively in the surrounding aqueous solvent at 12.5%, 25 and 50% anesthetic/lipid molar ratios, for a total of 10 simulated systems, using the insert-molecules tool of GROMACS 2020.4 (Abraham et al., 2015). The higher concentrations (25%, 50%), while not intended to be representative of clinical concentrations, were included to enhance the sampling of the lipid-anesthetic interaction and to accelerate ligand partitioning, as seen in previously published studies (Arvayo-Zatarain et al., 2019; Koubi et al., 2000; Mojumdar and Lyubartsev, 2010). The 12.5% concentration on the other hand is more representative of clinical scenarios, with the molar ratio of e.g. Halothane at MAC being in the range of 5% (McCarthy et al., 2017) to 14% (Franks and Lieb, 1979). The detailed composition of each simulated system is reported in the table below.
Table. Components of each simulation system
| System | Short Name | Lipids | Water molecules | Cl- ions | Na+ ions | VA molecules | Total Molecules |
|---|---|---|---|---|---|---|---|
| Control | C | 800 | 40000 | 96 | 186 | 0 | 41082 |
| Ethylene 12.5% | E12.5 | 800 | 39754 | 96 | 186 | 100 | 40936 |
| Ethylene 25% | E25 | 800 | 39527 | 96 | 186 | 200 | 40809 |
| Ethylene 50% | E50 | 800 | 39062 | 96 | 186 | 400 | 40544 |
| Desflurane 12.5% | D12.5 | 800 | 39414 | 96 | 186 | 100 | 40596 |
| Desflurane 25% | D25 | 800 | 38892 | 96 | 186 | 200 | 40174 |
| Desflurane 50% | D50 | 800 | 37749 | 96 | 186 | 400 | 39231 |
| Methoxyflurane 12.5% | M12.5 | 800 | 39210 | 96 | 186 | 100 | 40392 |
| Methoxyflurane 25% | M25 | 800 | 38484 | 96 | 186 | 200 | 39766 |
| Methoxyflurane 50% | M50 | 800 | 36755 | 96 | 186 | 400 | 38237 |
| F6 12.5% | F6 12.5 | 800 | 39149 | 96 | 186 | 100 | 40331 |
| F6 25% | F6 25 | 800 | 38381 | 96 | 186 | 200 | 39663 |
| F6 50% | F6 50 | 800 | 36486 | 96 | 186 | 400 | 37968 |
Simulation protocol
Simulations were carried out in GROMACS 2020.4 (Abraham et al., 2015) using the CHARMM36 force field (Klauda et al., 2010), which is well-validated for membrane simulations over a wide range of lipid compositions (Zhuang et al., 2014), according to the following protocol: after an initial 5000-step energy minimization, systems were equilibrated stepwise with gradually decreasing harmonic restraints (from 1,000 to 0 kJ × mol-1 × nm-1), first in the NVT ensemble for 250 ps with a conservative timestep of 1 fs, using the Berendsen thermostat with a coupling time constant of 1 ps and a reference temperature of 303.15K, which is above the phase-transition temperature for the studied lipid mixture, and subsequently in the NPT ensemble for 125 ps with the same 1 fs timestep, followed by a further simulation of 375 ps with a 2 fs timestep, using the Berendsen thermostat with the same parameters as before and the Berendsen barostat (Berendsen et al., 1984) with semi-isotropic pressure coupling at 1 atm with a coupling time constant of 5 ps. Overall, systems underwent 750 ps of equilibration, and were subsequently simulated for production runs for a total of 1 μs each in the NPT ensemble, using the Nosé-Hoover thermostat (Nosé, 1998) with a time constant of 1 ps and a reference temperature of 303.15K, and the Parrinello-Rahman barostat (Parrinello and Rahman, 1981), with semi-isotropic pressure coupling at 1 atm with a time constant of 5 ps. Bonds involving hydrogens were constrained using the LINCS algorithm (Hess et al., 1997), while the Particle Mesh Ewald (PME) algorithm (Ewald, 1921) was used for electrostatics, with a cutoff radius of 1.2 nm, and a cutoff of 1.2 nm was used for van der Waals interactions, with a force-switch modifier from 1.0 to 1.2 nm. The first 250 ns of the production MD runs were regarded as additional structural equilibration, while the remaining 750 ns were used for the subsequent analyses described below, in line with previous literature regarding the computational simulation of biological lipid bilayers (Shahane et al., 2019a; 2019b). Properties were sampled every 200 ps, unless otherwise specified. Molecular visualizations were generated using the VMD software package (Humphrey et al., 1996).
Structural analyses
Geometric Area-per-Lipid (gAPL), Bilayer Thickness (δ) and water permeation were calculated using the MDAnalysis (Michaud-Agrawal et al., 2011) library for Python (Van Rossum and Drake, 2009). Briefly, the gAPL was calculated as the xy area of the simulation box divided by the number of lipids in each membrane leaflet (N = 400) and is reported in Å2. To calculate the bilayer thickness, the position of all P atoms of each leaflet was extracted and their average z coordinate calculated for each leaflet. Bilayer thickness was calculated as the distance between the avg. z coordinates the P atom cloud. Water permeation events were calculated by tracking individual water molecules throughout the simulation. Density distribution profiles along the z coordinate were calculated using the gmx density tool. Acyl chain deuterium order parameters, SCD, for the sn1 and sn2 chains of each lipid were calculated to directly quantify structural effects on the packing of the membranes’ hydrophobic core. Order parameters were calculated following Equation (1), using the gmx order tool:
| (Equation 1) |
where θ is defined as the angle between the bilayer normal and the vector C-D between the given carbon atom and the bound hydrogen atom, as sampled from the equilibrium MD simulations (Piggot et al., 2017). Unsaturated lipid chains were accounted for following the methodology described in Pluhackova et al. (2016).
To quantify the tendency of ligands to partition inside the bilayer, which can bear profound consequences on protein-ligand interaction affinity and kinetics on transmembrane protein targets, the ligand molal concentration inside the lipid bilayer was calculated as follows. MDAnalysis was used to extract the number of ligand molecules whose center-of-mass z coordinate lied between the two P-atom point clouds, i.e. between the two layers delimiting each leaflet’s boundary. These ligands were regarded as being embedded inside the membrane. The remainder of the ligands was considered outside of the membrane. The molality of the anesthetics inside the membrane was calculated as number of moles of embedded ligands divided by the total weight of the membrane in kg.
To calculate the bilayer bending modulus, Kc, for each simulated system, the methodology proposed by Khelashvili and colleagues (Khelashvili et al., 2013) was employed, leveraging on the relationship between the splay modulus, χ12, and the macroscopic bending modulus: in this approach, an improved ability of adjacent lipids to change the reciprocal orientation of their hydrophobic tails with respect to the local membrane normal, which is quantified by their splay angle (α), is associated to a decreased membrane bending rigidity. Briefly, this approach first calculates the Potential of Mean Force (PMF) of the distribution of splay angles sampled during equilibrium MD simulations, normalized with respect to the probability distribution of a non-interacting particle system (Khelashvili et al., 2010), denoted here P0(α), as shown in Equation (2):
| (Equation 2) |
where T represents the system temperature and kB the Boltzmann constant. The overall splay modulus, which is linked to the bilayer bending modulus as:
| (Equation 3) |
can be calculated by means of a quadratic fit of the PMF obtained from Equation (2) (Fošnarič et al., 2006; Watson et al., 2011).
In the present work, we employed the python implementation previously demonstrated by Johner et al. (2016) to first extend the trajectories to neighboring periodic images, followed by wrapping the trajectory around the central unit cell and re-aligning. Finally, the provided python modules were used to calculate the tilt and splay angle distributions for all lipids and subsequently extract the membrane elastic properties of interest following the above-mentioned methodology. We refer to (Johner et al., 2016) and references therein for a more complete theoretical background of the methodology and details on the python implementation relying on the OpenStructure (Biasini et al., 2013) toolkit.
Quantification and statistical analysis
For a more accurate estimation of the error of the sampled properties (Flyvbjerg and Petersen, 1989; Grossfield et al., 2019; Grossfield and Zuckerman, 2009; Nicholls, 2014), the equilibrium part of the MD simulations (i.e. the last 750 ns) was further divided into 250-ns long trajectory blocks, in line with previous literature reporting findings in μs-long MD simulations of complex lipid membranes (Shahane et al., 2019a; 2019b). First, the block average of each structural property was calculated for each block as the arithmetic mean of the data points of the given property p within the block:
| (Equation 4) |
where denotes the mean within the j-th block of property p and Nb is the number of samples composing the j-th block. The final estimate of the ensemble average of the given property p is given by the arithmetic mean of the block averages:
| (Equation 5) |
where n is the total number of blocks. Then, the experimental standard deviation of the mean, , of each property was calculated as:
| (Equation 6) |
where is the arithmetic mean of a given property over the j-th block and n is the number of blocks. Finally, the estimate of the standard deviation is given by:
| (Equation 7) |
This quantity is the reported standard deviation, represented as error bars on the plots, and was also used to calculate 95% confidence intervals which are reported throughout the text and in shaded colors on the plots, unless where explicitly specified.
Acknowledgments
We acknowledge the CINECA award under the ISCRA initiative, for the availability of high-performance computing resources and support.
Author contributions
Conceptualization, MD, MC, and JAT; Methodology, EAZ; Formal analysis, EAZ and MAD; Investigation, EAZ; Writing – Original Draft, EAZ; Writing – Review & Editing, all authors; Visualization, EAZ; Supervision, MD, MC, and JAT.
Declaration of interests
The authors declare no competing interests.
Published: March 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103946.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindah E. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Allen J.A., Halverson-Tamboli R.A., Rasenick M.M. Lipid raft microdomains and neurotransmitter signalling. Nat. Rev. Neurosci. 2006;828:128–140. doi: 10.1038/nrn2059. [DOI] [PubMed] [Google Scholar]
- Arvayo-Zatarain J.A., Favela-Rosales F., Contreras-Aburto C., Urrutia-Bañuelos E., Maldonado A. Molecular dynamics simulation study of the effect of halothane on mixed DPPC/DPPE phospholipid membranes. J. Mol. Model. 2019;25:1–10. doi: 10.1007/s00894-018-3890-6. [DOI] [PubMed] [Google Scholar]
- Bennett W.F.D., MacCallum J.L., Hinner M.J., Marrink S.J., Tieleman D.P. Molecular view of cholesterol flip-flop and chemical potential in different membrane environments. J. Am. Chem. Soc. 2009;131:12714–12720. doi: 10.1021/ja903529f/suppl_file/ja903529f_si_001.pdf. [DOI] [PubMed] [Google Scholar]
- Berendsen H.J.C., Postma J.P.M., Van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- Bertaccini E.J. The molecular mechanisms of anesthetic action: updates and cutting edge developments from the field of molecular modeling. Pharmaceuticals. 2010;3:2178–2196. doi: 10.3390/ph3072178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertaccini E.J., Yoluk O., Lindahl E.R., Trudell J.R. Assessment of homology templates and an anesthetic binding site within the γ-aminobutyric acid receptor. Anesthesiology. 2013;119:1087–1095. doi: 10.1097/ALN.0B013E31829E47E3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M., Schmidt T., Bienert S., Mariani V., Studer G., Haas J., Johner N., Schenk A.D., Philippsen A., Schwede T. OpenStructure: an integrated software framework for computational structural biology. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013;69:701–709. doi: 10.1107/s0907444913007051/ic5090sup2.txt. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna J.A., Miller K.W., Forman S.A. Mechanisms of actions of inhaled anesthetics. N. Engl. J. Med. 2003;348:2110–2124. doi: 10.1056/nejmra021261. [DOI] [PubMed] [Google Scholar]
- Cantor R.S. Lipid composition and the lateral pressure profile in bilayers. Biophys. J. 1999;76:2625–2639. doi: 10.1016/S0006-3495(99)77415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor R.S. The lateral pressure profile in membranes: a physical mechanism of general anesthesia. Biochemistry. 1997;36:2339–2344. doi: 10.1021/BI9627323. [DOI] [PubMed] [Google Scholar]
- Chipot Christophe, Wilson Michael A., Pohorille∗ Andrew. Interactions of anesthetics with the water−hexane interface. a molecular dynamics study. J. Phys. Chem. B. 1997;101:782–791. doi: 10.1021/JP961513O. [DOI] [PubMed] [Google Scholar]
- De Planque M.R.R., Killian J.A. Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring (Review) Mol. Membr. Biol. 2003;20:271–284. doi: 10.1080/09687680310001605352. [DOI] [PubMed] [Google Scholar]
- Dickson C.J., Madej B.D., Skjevik Å.A., Betz R.M., Teigen K., Gould I.R., Walker R.C. Lipid14: the amber lipid force field. J. Chem. Theor. Comput. 2014;10:865–879. doi: 10.1021/CT4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson C.J., Rosso L., Betz R.M., Walker R.C., Gould I.R. GAFFlipid: a general amber force field for the accurate molecular dynamics simulation of phospholipid. Soft Matter. 2012;8:9617–9627. doi: 10.1039/C2SM26007G. [DOI] [Google Scholar]
- Dumas F., Lebrun M.C., Tocanne J.F. Is the protein/lipid hydrophobic matching principle relevant to membrane organization and functions? FEBS Lett. 1999;458:271–277. doi: 10.1016/S0014-5793(99)01148-5. [DOI] [PubMed] [Google Scholar]
- Eger E.I., Halsey M.J., Koblin D.D., Laster M.J., Ionescu P., Königsberger K., Fan R., Nguyen B.V., Hudlicky T. The convulsant and anesthetic properties of cis-trans isomers of 1,2-dichlorohexafluorocyclobutane and 1,2-dichloroethylene. Anesth. Analg. 2001;93:922–927. doi: 10.1097/00000539-200110000-00025. [DOI] [PubMed] [Google Scholar]
- Ewald P. Die Berechnung optischer und elektrostatischer Gitterpotentiale. Ann. Phys. 1921;369:253–287. doi: 10.1002/ANDP.19213690304. [DOI] [Google Scholar]
- Flyvbjerg H., Petersen H.G. Error estimates on averages of correlated data. J. Chem. Phys. 1989;91:461–466. doi: 10.1063/1.457480. [DOI] [Google Scholar]
- Fošnarič M., Iglič A., May S. Influence of rigid inclusions on the bending elasticity of a lipid membrane. Phys. Rev. E. 2006;74:051503. doi: 10.1103/PhysRevE.74.051503. [DOI] [PubMed] [Google Scholar]
- Franks N.P., Lieb W.R. Molecular and cellular mechanisms of general anaesthesia. Nat. 1994;367:607–614. doi: 10.1038/367607a0. [DOI] [PubMed] [Google Scholar]
- Franks N.P., Lieb W.R. Do general anaesthetics act by competitive binding to specific receptors? Nat. 1984;310:599–601. doi: 10.1038/310599a0. [DOI] [PubMed] [Google Scholar]
- Franks N.P., Lieb W.R. The structure of lipid bilayers and the effects of general anaesthetics: an X-ray and neutron diffraction study. J. Mol. Biol. 1979;133:469–500. doi: 10.1016/0022-2836(79)90403-0. [DOI] [PubMed] [Google Scholar]
- Grasso G., Muscat S., Rebella M., Morbiducci U., Audenino A., Danani A., Deriu M.A. Cell penetrating peptide modulation of membrane biomechanics by molecular dynamics. J. Biomech. 2018;73:137–144. doi: 10.1016/j.jbiomech.2018.03.036. [DOI] [PubMed] [Google Scholar]
- Gray E., Karslake J., Machta B.B., Veatch S.L. Liquid general anesthetics lower critical temperatures in plasma membrane vesicles. Biophys. J. 2013;105:2751–2759. doi: 10.1016/j.bpj.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossfield A., Patrone P.N., Roe D.R., Schultz A.J., Siderius D., Zuckerman D.M. Best practices for quantification of uncertainty and sampling quality in molecular simulations [Article v1.0] Living J. Comput. Mol. Sci. 2019;1:1–24. doi: 10.33011/livecoms.1.1.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossfield A., Zuckerman D.M. Chapter 2 quantifying uncertainty and sampling quality in biomolecular simulations. Annu. Rep. Comput. Chem. 2009;5:23–48. doi: 10.1016/S1574-1400(09)00502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold K.F., Hemmings H.C. Sodium channels as targets for volatile anesthetics. Front. Pharmacol. 2012;3:50. doi: 10.3389/fphar.2012.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold K.F., Sanford R.L., Lee W., Andersen O.S., Hemmings H.C. Clinical concentrations of chemically diverse general anesthetics minimally affect lipid bilayer properties. Proc. Natl. Acad. Sci. U S A. 2017;114:3109–3114. doi: 10.1073/pnas.1611717114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:14631472. doi: 10.1002/(SICI)1096-987X. [DOI] [Google Scholar]
- Hofsäß C., Lindahl E., Edholm O. Molecular dynamics simulations of phospholipid bilayers with cholesterol. Biophys. J. 2003;84:2192. doi: 10.1016/S0006-3495(03)75025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Bertaccini E. Molecular dynamics simulation of anesthetic-phospholipid bilayer interactions. J. Biomol. Struct. Dyn. 1995;12:725–754. doi: 10.1080/07391102.1995.10508773. [DOI] [PubMed] [Google Scholar]
- Hudson A.E., Herold K.F., Hemmings H.C., Jr. Pharmacology and Physiology for Anesthesia. Foundations and Clinical Application. 2nd. Elsevier; 2019. Pharmacology of Inhaled Anesthetics; pp. 217–240. [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Ingólfsson H.I., Carpenter T.S., Bhatia H., Bremer P.T., Marrink S.J., Lightstone F.C. Computational lipidomics of the neuronal plasma membrane. Biophys. J. 2017;113:2271–2280. doi: 10.1016/j.bpj.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jämbeck J.P.M., Lyubartsev A.P. Derivation and systematic validation of a refined all-atom force field for phosphatidylcholine lipids. J. Phys. Chem. B. 2012;116:3164–3179. doi: 10.1021/jp212503e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim T., Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Jo S., Lim J.B., Klauda J.B., Im W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John Mihic S., Ye Q., Wick M.J., Koltchine V.V., Krasowski M.D., Finn S.E., Mascia M.P., Fernando Valenzuela C., Hanson K.K., Greenblatt E.P., et al. Sites of alcohol and volatile anaesthetic action on GABA(A) and glycine receptors. Nature. 1997;389:385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
- Johner N., Harries D., Khelashvili G. Implementation of a methodology for determining elastic properties of lipid assemblies from molecular dynamics simulations. BMC Bioinformatics. 2016;17:1–11. doi: 10.1186/s12859-016-1003-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelashvili G., Kollmitzer B., Heftberger P., Pabst G., Harries D. Calculating the bending modulus for multicomponent lipid membranes in different thermodynamic phases. J. Chem. Theor. Comput. 2013;9:3866–3871. doi: 10.1021/ct400492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelashvili G., Pabst G., Harries D. Cholesterol orientation and tilt modulus in DMPC bilayers. J. Phys. Chem. B. 2010;114:7524–7534. doi: 10.1021/jp101889k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klähn M., Zacharias M. Transformations in plasma membranes of cancerous cells and resulting consequences for cation insertion studied with molecular dynamics. Phys. Chem. Chem. Phys. 2013;15:14427–14441. doi: 10.1039/c3cp52085d. [DOI] [PubMed] [Google Scholar]
- Klauda J.B., Venable R.M., Freites J.A., O’Connor J.W., Tobias D.J., Mondragon-Ramirez C., Vorobyov I., MacKerell A.D., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblin D.D., Eger E.I., Johnson B.H., Collins P., Terrell R.C., Speers L. Are convulsant gases also anesthetics? Anesth. Analg. 1981;60:464–470. doi: 10.1213/00000539-198107000-00002. [DOI] [PubMed] [Google Scholar]
- Koubi L., Tarek M., Klein M.L., Scharf D. Distribution of halothane in a dipalmitoylphosphatidylcholine bilayer from molecular dynamics calculations. Biophys. J. 2000;78:800–811. doi: 10.1016/S0006-3495(00)76637-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.G. Model for action of local anaesthetics. Nat. 1976;262:545–548. doi: 10.1038/262545a0. [DOI] [PubMed] [Google Scholar]
- Leftin A., Molugu T.R., Job C., Beyer K., Brown M.F. Area per lipid and cholesterol interactions in membranes from separated local-field 13C NMR spectroscopy. Biophys. J. 2014;107:2274–2286. doi: 10.1016/J.BPJ.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner R.A. A hypothesis about the endogenous analogue of general anesthesia. Proc. Natl. Acad. Sci. U S A. 1997;94:13375–13377. doi: 10.1073/PNAS.94.25.13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I., Grzybek M., Simons K. Raft domains of variable properties and compositions in plasma membrane vesicles. Proc. Natl. Acad. Sci. U S A. 2011;108:11411–11416. doi: 10.1073/PNAS.1105996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.B., Vorobyov I., Allen T.W. The role of membrane thickness in charged protein–lipid interactions. Biochim. Biophys. Acta - Biomembr. 2012;1818:135–145. doi: 10.1016/J.BBAMEM.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/SCIENCE.1174621/ASSET/2C0E3756-BFCD-450C-8749-7EE77CABA75A/ASSETS/GRAPHIC/327_46_F3. [DOI] [PubMed] [Google Scholar]
- Mascia M.P., Trudell J.R., Harris R.A. Specific binding sites for alcohols and anesthetics on ligand-gated ion channels. Proc. Natl. Acad. Sci. U S A. 2000;97:9305–9310. doi: 10.1073/PNAS.160128797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulik P.R., Shipley G.G. Interactions of N-stearoyl sphingomyelin with cholesterol and dipalmitoylphosphatidylcholine in bilayer membranes. Biophys. J. 1996;70:2256–2265. doi: 10.1016/S0006-3495(96)79791-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazze R.I. Renal dysfunction associated with methoxyflurane anesthesia. JAMA. 1971;216:278. doi: 10.1001/jama.1971.03180280032006. [DOI] [PubMed] [Google Scholar]
- McCarthy N.L.C., Brooks N.J., Tyler A.I.I., ElGamacy M., Welche P.R.L., Payne M.C., Chau P.L. A combined X-ray scattering and simulation study of halothane in membranes at raised pressures. Chem. Phys. Lett. 2017;671:21–27. doi: 10.1016/J.CPLETT.2016.12.041. [DOI] [Google Scholar]
- Meyer F., Smit B. Effect of cholesterol on the structure of a phospholipid bilayer. Proc. Natl. Acad. Sci. U S A. 2009;106:3654–3658. doi: 10.1073/PNAS.0809959106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.H. Contributions to the theory of narcosis. Trans. Faraday Soc. 1937;33:1062–1064. doi: 10.1039/TF9373301062. [DOI] [Google Scholar]
- Michaud-Agrawal N., Denning E.J., Woolf T.B., Beckstein O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.D., Wahrenbrock E.A., Schroeder C.F., Knipstein T.W., Eger E.I., Buechel D.R. Ethylene--halothane anesthesia: addition or synergism? Anesthesiology. 1969;31:301–304. doi: 10.1097/00000542-196910000-00002. [DOI] [PubMed] [Google Scholar]
- Modica P.A., Templehoff R., White P.F. Pro- and anticonvulsant effects of anesthetics (Part II) Anesth. Analg. 1990;70:433–444. doi: 10.1213/00000539-199004000-00016. [DOI] [PubMed] [Google Scholar]
- Mojumdar E.H., Lyubartsev A.P. Molecular dynamics simulations of local anesthetic articaine in a lipid bilayer. Biophys. Chem. 2010;153:27–35. doi: 10.1016/j.bpc.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Moon S., Yan R., Kenny S.J., Shyu Y., Xiang L., Li W., Xu K. Spectrally resolved, functional super-resolution microscopy reveals nanoscale compositional heterogeneity in live-cell membranes. J. Am. Chem. Soc. 2017;139:10944–10947. doi: 10.1021/JACS.7B03846/SUPPL_FILE/JA7B03846_SI_004. [DOI] [PubMed] [Google Scholar]
- Mouritsen O.G., Bloom M. Models of lipid-protein interactions in membranes. Annu. Rev. Biophys. Biomol. Struct. 1993;22:145–171. doi: 10.1146/annurev.bb.22.060193.001045. [DOI] [PubMed] [Google Scholar]
- Needham D., Nunn R.S. Elastic deformation and failure of lipid bilayer membranes containing cholesterol. Biophys. J. 1990;58:997–1009. doi: 10.1016/S0006-3495(90)82444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A. Confidence limits, error bars and method comparison in molecular modeling. part 1: the calculation of confidence intervals. J. Comput. Aided. Mol. Des. 2014;28:887–918. doi: 10.1007/s10822-014-9753-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1998;81:511. doi: 10.1063/1.447334. [DOI] [Google Scholar]
- Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- Pavel M.A., Petersen E.N., Wang H., Lerner R.A., Hansen S.B. Studies on the mechanism of general anesthesia. Proc. Natl. Acad. Sci. U S A. 2020;117:13757–13766. doi: 10.1073/pnas.2004259117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perouansky M., Hentschke H., Perkins M., Pearce R.A. Amnesic concentrations of the nonimmobilizer 1,2-dichlorohexafluorocyclobutane (F6, 2N) and isoflurane alter hippocampal θ oscillations in Vivo. Anesthesiology. 2007;106:1168–1176. doi: 10.1097/01.ANES.0000267600.09764.AF. [DOI] [PubMed] [Google Scholar]
- Pickholz M., Saiz L., Klein M.L. Concentration effects of volatile anesthetics on the properties of model membranes: a coarse-grain approach. Biophys. J. 2005;88:1524–1534. doi: 10.1529/biophysj.104.044354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot T.J., Allison J.R., Sessions R.B., Essex J.W. On the calculation of acyl chain order parameters from lipid simulations. J. Chem. Theor. Comput. 2017;13:5683–5696. doi: 10.1021/acs.jctc.7b00643. [DOI] [PubMed] [Google Scholar]
- Pluhackova K., Kirsch S.A., Han J., Sun L., Jiang Z., Unruh T., Böckmann R.A. A critical comparison of biomembrane force fields: structure and dynamics of model DMPC, POPC, and POPE bilayers. J. Phys. Chem. B. 2016;120:3888–3903. doi: 10.1021/ACS.JPCB.6B01870/SUPPL_FILE/JP6B01870_SI_001.PDF. [DOI] [PubMed] [Google Scholar]
- Pohorille A., Cieplak P., Wilson M.A. Interactions of anesthetics with the membrane-water interface. Chem. Phys. 1996;204:337–345. doi: 10.1016/0301-0104(95)00292-8. [DOI] [PubMed] [Google Scholar]
- Pohorille A., Wilson M.A., New M.H., Chipot C. Concentrations of anesthetics across the water–membrane interface; the meyer–overton hypothesis revisited. Toxicol. Lett. 1998;100:421–430. doi: 10.1016/S0378-4274(98)00216-1. [DOI] [PubMed] [Google Scholar]
- Pontes B., Ayala Y., Fonseca A.C.C., Romão L.F., Amaral R.F., Salgado L.T., Lima F.R., Farina M., Viana N.B., Moura-Neto V., Nussenzveig H.M. Membrane elastic properties and cell function. PLoS One. 2013;8:e67708. doi: 10.1371/journal.pone.0067708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Róg T., Pasenkiewicz-Gierula M., Vattulainen I., Karttunen M. Ordering effects of cholesterol and its analogues. Biochim. Biophys. Acta - Biomembr. 2009;1788:97–121. doi: 10.1016/j.bbamem.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Saeedimasine M., Montanino A., Kleiven S., Villa A. Role of lipid composition on the structural and mechanical features of axonal membranes: a molecular simulation study. Sci. Rep. 2019;91 9:1–12. doi: 10.1038/s41598-019-44318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman P. The membrane actions of anesthetics and tranquilizers. Pharmacol. Rev. 1972;24:583–655. [PubMed] [Google Scholar]
- Shahane G., Ding W., Palaiokostas M., Azevedo H.S., Orsi M. Interaction of antimicrobial lipopeptides with bacterial lipid bilayers. J. Membr. Biol. 2019;252:317–329. doi: 10.1007/s00232-019-00068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahane G., Ding W., Palaiokostas M., Orsi M. Physical properties of model biological lipid bilayers: insights from all-atom molecular dynamics simulations. J. Mol. Model. 2019;25:1–13. doi: 10.1007/s00894-019-3964-0. [DOI] [PubMed] [Google Scholar]
- Subczynski W.K., Pasenkiewicz-Gierula M., Widomska J., Mainali L., Raguz M. High cholesterol/low cholesterol: effects in biological membranes Review. Cell Biochem. Biophys. 2017;75:369. doi: 10.1007/S12013-017-0792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang P., Xu Y. Large-scale molecular dynamics simulations of general anesthetic effects on the ion channel in the fully hydrated membrane: the implication of molecular mechanisms of general anesthesia. Proc. Natl. Acad. Sci. U S A. 2002;99:16035–16040. doi: 10.1073/PNAS.252522299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D.M., Eger E.I., Bickler P.E. Halothane, but not the nonimmobilizers perfluoropentane and 1,2-dichlorohexafluorocyclobutane, depresses synaptic transmission in hippocampal CA1 neurons in rats. Anesth. Analg. 1999;89:1040. doi: 10.1213/00000539-199910000-00041. [DOI] [PubMed] [Google Scholar]
- McIntosh T.J. The effect of cholesterol on the structure of phosphatidylcholine bilayers. Biochim. Biophys. Acta. 1978;513:43–58. doi: 10.1016/0005-2736(78)90110-4. [DOI] [PubMed] [Google Scholar]
- Tsuchiya H., Mizogami M. Interaction of local anesthetics with biomembranes consisting of phospholipids and cholesterol: mechanistic and clinical implications for anesthetic and cardiotoxic effects. Anesthesiol. Res. Pract. 2013;2013:297141. doi: 10.1155/2013/297141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu K., Tarek M., Klein M.L., Scharf D. Effects of anesthetics on the structure of a phospholipid bilayer: molecular dynamics investigation of halothane in the hydrated liquid crystal phase of dipalmitoylphosphatidylcholine. Biophys. J. 1998;75:2123–2134. doi: 10.1016/S0006-3495(98)77655-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rossum G., Drake F.L. Python 3 Reference Manual. CreateSpace; Scotts Valley, CA: 2009. [Google Scholar]
- Vauquelin G., Packeu A. Ligands, their receptors and plasma membranes. Mol. Cell. Endocrinol. 2009;311:1–10. doi: 10.1016/j.mce.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M.C., Penev E.S., Welch P.M., Brown F.L.H. Thermal fluctuations in shape, thickness, and molecular orientation in lipid bilayers. J. Chem. Phys. 2011;135:244701. doi: 10.1063/1.3660673. [DOI] [PubMed] [Google Scholar]
- Wu E.L., Cheng X., Jo S., Rui H., Song K.C., Dávila-Contreras E.M., Qi Y., Lee J., Monje-Galvan V., Venable R.M., et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakura T., Bertaccini E., Trudell J.R., Harris R.A. Anesthetics and ion channels: molecular models and sites of Action1. 2003. [DOI] [PubMed]
- Yamamoto E., Akimoto T., Shimizu H., Hirano Y., Yasui M., Yasuoka K. Diffusive nature of xenon anesthetic changes properties of a lipid bilayer: molecular dynamics simulations. J. Phys. Chem. B. 2012;116:8989–8995. doi: 10.1021/jp303330c. [DOI] [PubMed] [Google Scholar]
- Zachowski A. Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem. J. 1993;294:1–14. doi: 10.1042/bj2940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Makover J.R., Im W., Klauda J.B. A systematic molecular dynamics simulation study of temperature dependent bilayer structural properties. Biochim. Biophys. Acta - Biomembr. 2014;1838:2520–2529. doi: 10.1016/j.bbamem.2014.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.