Abstract
Objective
Skeletal muscle is a heterogeneous and dynamic tissue that adapts to functional demands and substrate availability by modulating muscle fiber size and type. The concept of muscle fiber type relates to its contractile (slow or fast) and metabolic (glycolytic or oxidative) properties. Here, we tested whether disruptions in muscle oxidative catabolism are sufficient to prompt parallel adaptations in energetics and contractile protein composition.
Methods
Mice with defective mitochondrial long-chain fatty acid oxidation (mLCFAO) in the skeletal muscle due to loss of carnitine palmitoyltransferase 2 (Cpt2Sk−/−) were used to model a shift in muscle macronutrient catabolism. Glycolytic and oxidative muscles of Cpt2Sk−/− mice and control littermates were compared for the expression of energy metabolism-related proteins, mitochondrial respiratory capacity, and myosin heavy chain isoform composition.
Results
Differences in bioenergetics and macronutrient utilization in response to energy demands between control muscles were intrinsic to the mitochondria, allowing for a clear distinction of muscle types. Loss of CPT2 ablated mLCFAO and resulted in mitochondrial biogenesis occurring most predominantly in oxidative muscle fibers. The metabolism-related proteomic signature of Cpt2Sk−/− oxidative muscle more closely resembled that of glycolytic muscle than of control oxidative muscle. Respectively, intrinsic substrate-supported mitochondrial respiration of CPT2 deficient oxidative muscles shifted to closely match that of glycolytic muscles. Despite this shift in mitochondrial metabolism, CPT2 deletion did not result in contractile-based fiber type switching according to myosin heavy chain composition analysis.
Conclusion
The loss of mitochondrial long-chain fatty acid oxidation elicits an adaptive response involving conversion of oxidative muscle toward a metabolic profile that resembles a glycolytic muscle, but this is not accompanied by changes in myosin heavy chain isoforms. These data suggest that shifts in muscle catabolism are not sufficient to drive shifts in the contractile apparatus but are sufficient to drive adaptive changes in metabolic properties.
Keywords: Skeletal muscle, Fatty acid oxidation, Carnitine palmitoyltransferase 2, Mitochondrial biogenesis, Bioenergetics, Fiber-typing
Abbreviations: CPT, Carnitine palmitoyltranferase; mLCFAO, Mitochondrial long-chain fatty acid oxidation; MyHC, Myosin Heavy Chain; LCAC, Long-chain acylcarnitine; PC, Palmitoylcarnitine
Highlights
-
•
Fuel oxidation in glycolytic compared to oxidative muscles are different and intrinsic to the mitochondria.
-
•
Muscle CPT2 loss elicits fiber-type dependent mitochondrial biogenesis.
-
•
Muscle CPT2 loss elicits an oxidative-to-glycolytic shift in mitochondrial and metabolic properties.
-
•
Muscle CPT2 loss does not alter myosin heavy chain isoform composition.
-
•
CPT2 deficient muscles demonstrate a metabolic-contractile apparatus mismatch.
1. Introduction
Skeletal muscles are composed of numerous distinct muscle fibers that exist in a diverse array within a given muscle. A muscle's fiber composition dictates the characteristics of its contractile dynamics, and this will in turn determine the muscle's contribution to whole body locomotion. The concept of fiber type relates to a combination of both contractile (slow- or fast-twitch) and metabolic (glycolytic or oxidative) properties [1]. From the many contractile proteins within skeletal muscle, myosin is a major characterizing component due to its critical role as a molecular motor. Specifically, myosin heavy chain (MyHC) isoforms are widely used to classify muscle fiber types based on contractile velocity and fatigue resistance. In adult mammalian muscles, MyHC-based characterization identifies two major categories: slow-twitch fibers composed of MyHC-β/1 and fast-twitch fibers composed of either MyHC-2A, MyHC-2B or MyHC-2X. The metabolic properties of skeletal muscle encompass an important characteristic for the tissues' ability to cope with sudden and extreme changes in energy demands and substrate availability. Transitioning between rest and intense physical activity, for example, acutely increases muscle ATP requirements by 100-fold [1]. In order to meet such versatile demands, the skeletal muscle relies on a unique set of resources to quickly and efficiently accelerate ATP production: creatine kinase (CK) activity, substrate level phosphorylation during glycolysis, and aerobic metabolism via mitochondrial oxidative phosphorylation. In different muscles, the relative contribution of these three processes to ATP production is highly distinct and linked to its contractile properties. Thus, in slow-twitch fibers, energy is provided mainly by mitochondrial oxidative metabolism, while in fast-twitch fibers there is a greater contribution of glycolysis towards ATP generation. Within a muscle fiber, metabolism is thought to match contractile apparatus to allow for form-fit-function based physiology. Based on this, fibers can be further classified as slow-oxidative (type 1 fibers), fast-oxidative-glycolytic (type 2A), fast-glycolytic (type 2B) and fast-intermediate (type 2X) [2].
It is important to remember though that despite this a priori compartmentalized classification, muscle fibers are highly dynamic and can adapt to new functional demands and substrate availability (muscle plasticity). These adaptations are predicted to tightly couple contractile apparatus to energy metabolism. Indeed, damage to mitochondrial DNA results in changes in contractile apparatus, suggesting that mitochondrial function is a regulator of skeletal muscle MyHC fiber type [3]. Yet, how primary shifts in muscle macronutrient catabolic activity in the absence of overall mitochondrial disfunction impact adaptations of the contractile apparatus is not well understood. In the present study, we used a model of defective mitochondrial long-chain fatty acid oxidation (mLCFAO) to investigate the consequences of macronutrient catabolic imbalance on the contraction-metabolism coupling of skeletal muscle. Ablation of mLCFAO was induced by loss of carnitine palmitoyltransferase 2 in the skeletal muscle of mice (Cpt2Sk−/−) [4,5]. CPT2 is a critical and irreplaceable enzyme for mLCFAO, and its absence results in the accumulation of the enzymatic substrate long-chain acylcarnitines [5].
Here, we hypothesized that the halt of mLCFAO in muscle-specific CPT2 knockout mice would drive a shift in mitochondrial energy metabolism towards non-fatty acid substrates, similar to the metabolic profile of glycolytic muscles, and that this metabolic change would be accompanied by a coupled shift towards predominantly glycolytic myosin heavy chain isoforms. Thus, muscle plasticity during mLCFAO deficiency would drive an adaptive reduction of oxidative-prone fibers and an increase of glycolytically active fibers to allow for effective contraction-metabolism coupling. Therefore, we profiled the proteome, the MyHC isoform expression, and the bioenergetic metabolic capacity of skeletal muscle in both control and Cpt2Sk−/− mice, distinguishing between predominantly glycolytic and predominantly oxidative muscles. CPT2-deficient glycolytic muscles had proteomic profiles and substrate-supported mitochondrial respiration capabilities similar to controls; however, in oxidative soleus muscles, Cpt2Sk−/− proteome and mitochondrial bioenergetics shifted toward that of the glycolytic muscles. Surprisingly, the metabolic shift of CPT2-deficient oxidative muscle was not accompanied by myosin heavy chain isoform switch. Together, these data suggest that mLCFAO-deficient muscles adapt their energy metabolism in a fiber type-specific manner but that this adaptation is not coupled to changes in fiber type contractile components.
2. Materials and methods
2.1. Animal model
CPT2 skeletal muscle-specific conditionally deficient mice (Cpt2Sk−/−) were generated using the human alpha-skeletal actin promoter (The Jackson Laboratory; Stock No: 006149) as described [5,6]. Littermates lacking the Cre gene were used as controls. Mice were given free access to water and standard chow (PicoLab 5053, Lab Diets) in pathogen-free housing under 12-h light–dark cycles. All procedures were performed in 3 to 8-month-old mice and were approved by the Institutional Animal Care and Use Committee of East Carolina University (Assurance A3469-01).
2.2. Exercise
Four-month-old male mice acclimated to the treadmill for two consecutive days with 5-min intervals of no speed, walking speed (3 m/min), and at low running speed (10 m/min). The high-intensity exercise protocol consisted of 1-min running intervals starting at 10 m/min, increasing by 5 m/min, with 2 min rest in between each interval, until exhaustion. Gentle running encouragement was used [7]. All acclimation and running sessions were performed during the dark cycle.
2.3. Histology and immunohistochemistry
Skeletal muscles from 5-month-old male mice were collected immediately after euthanasia, covered with a thin layer of Optimal Cutting Temperature compound (O.C.T., Fisher Scientific), flash frozen in liquid nitrogen-chilled isopentane, and stored at −80 °C until further processing. Serial cross-sections of 10 μm thickness were obtained, and immunohistochemical detection of myosin heavy chain (MyHC) isoforms and of Cytochrome C Oxidase (COX IV) was performed as previously described [8]. Briefly, sections were fixed in 4% paraformaldehyde (PFA), ice-cold, permeabilized in 0.3% Triton X-100, blocked with 5% goat serum (G9023; Sigma-Millipore), and incubated overnight with a primary antibody mix against MyHC-β (type I fibers; Developmental Studies Hybridoma Bank BA-F8), MyHC-2A (type IIa fibers; DSHB SC-71), MyHC-2B (type IIb fibers; DSHB BF–F3), and dystrophin (PA5-32388). MyHC-2X (type IIx fibers) were identified by absence of staining when all previous antibodies were used on a given muscle section. The primary antibody against COX IV was from Cell Signaling (4844). The following secondary antibodies were used to visualize the different targets: Alexa Fluor 350 IgG2b (A-21140, Life Technologies) for MyHC I, Alexa Fluor 488 IgG1 (A-21121) for MyHC IIa, Alexa Fluor 546 IgM (A-21045) for MyHC IIb, and Alexa fluor 647 goat anti-mouse IgG1 (A-21236) for dystrophin and COX IV. Gomori Trichrome stain (Engel-Cunningham modification) was performed in serial cross-sections as described [9,10], with modifications. Briefly, sections were fixed in 4% PFA, counterstained with Mayer's hematoxylin, immersed in modified Gomori Trichrome stain (Chromotrope 2R + Fast Green + Phosphotungstic acid) for 20 min, differentiated in 0.2% acetic acid, then dehydrated and mounted. Images were acquired using EVOS FL auto microscope and software (Life Technologies, Inc.) and fiber-type distribution was quantified using ImageJ software (National Institutes of Health). A total of 3 biological replicates were analyzed per muscle type and genotype. For each of the biological replicates, a total of 3 (EDL) or 4 (Soleus) 20X images spanning the entirety of the muscle cross-section were analyzed and quantitated for MyHC fiber-typing. Images were coded and randomized for blinded analyses.
2.4. Mitochondrial oxidation assays
Oroboros® high-resolution respirometry was performed on freshly isolated mitochondria from skeletal muscle of 3-month-old female mice similar to previous reports [11,12]. Specifically, mitochondria were isolated from white quadriceps, tibialis anterior, and soleus muscles by differential centrifugation in KMEM buffer (KCl 100 mM, MOPS 50 mM, EGTA 1 mM, MgSO4 5 mM, BSA 0.2% pH 7.1). Yield was assessed by protein concentration using the BCA method [13]. Maximum respiration was measured in ATP containing buffer Z (K-MES 105 mM, KCl 30 mM, KH2PO4 10 mM, MgCl2 5 mM, EGTA 1 mM, BSA 2.5 g/L, pH 7.1), using 25–50 μg of isolated mitochondria in each chamber. After baseline respiration was established, malate (2 mM, Sigma M1296) was added to maintain the TCA cycle flux. Substrates were added in the following concentrations: octanoylcarnitine 0.2 mM (Sigma 50,892), palmitoylcarnitine 0.02 mM (Sigma P1645), pyruvate 5 mM (Combi-Blocks, QA1116), or succinate 10 mM (Fisher BP336) plus rotenone 0.005 mM (Sigma R8875). A modified version of the creatine kinase clamp technique was used to determine oxygen consumption in response to changes in ATP free energy, as described [11]. Briefly, the respiration assay buffer Z was supplemented with 20 U/mL of creatine kinase (CK), 5 mM ATP, and 1 mM Tris-Phosphocreatine (PCr) to stimulate maximal demand for ATP synthesis. Sequential additions of PCr to 6, 9, 15, 21, 24, and 30 mM allowed for gradual reduction of the ATP demand state. Calculation of the ΔGATP at each titration point was performed with the following online bioenergetics calculator tool, described in reference [12]. The relationship between ΔGATP and JO2 was plotted, and the slope of the linear portion was calculated to serve as the conductance measure of the system under the specified respiratory substrate conditions. Due to the small mass of soleus muscle (≈8 mg) and to secure sufficient isolated mitochondria for parallel assessment of all substrates and conditions, muscles from 3 to 4 littermate mice with the same genotype were pooled and considered as one biological replicate. A total of 4–6 biological replicates were analyzed per muscle type and genotype.
2.5. Acylcarnitine and metabolite analysis
Acylcarnitine profiling of TA skeletal muscle tissue from 4-month-old male mice was performed as described [5,6]. Briefly, lipids were extracted from tissues using Bligh and Dyer Method [14], dried, resuspended, and injected directly via a micro-autosampler (G1377A) into a QQQ6410 triple quadrupole mass spectrometer (Agilent Technologies, San Jose, CA) operated in the positive ion mode and equipped with ESI ion source. Data were collected in multiple reaction monitoring mode [6,15,16] and ion intensities acquired by an in-house script were further normalized to sample protein concentration and expressed as a percent of total ion counts. Metabolites from skeletal muscle of 6-month-old male mice were retrieved from the polar phase of the extraction method described above, dried, resuspended, and injected directly into a QQQ6410 triple quadrupole mass spectrometer (Agilent Technologies, San Jose, CA) using the instrument conditions described previously [41] and the MRMs listed at Table S1. This is a first pass exploratory method with direct injection. Therefore, the attributions are tentative, and the molecular structure of the MRMs presenting higher intensity compared to the blank sample was not confirmed.
2.6. Mass spectrometry lipid imaging
Lipid imaging was performed in skeletal muscle from 4-month-old male mice by nanospray desorption electrospray ionization (nano-DESI) as described [18]. Briefly, 10-μm-thick muscle sections were mounted onto a glass slide and stored at −80 °C prior to analysis. Nano-DESI MSI experiments were performed on a Q Exactive HF Orbitrap mass spectrometer (Thermo Electron, Bremen, Germany) equipped with a custom-designed nano-DESI source. A shear-force probe was used to maintain a constant distance between the sample and probe. A 9:1 (v/v) methanol:water mixture was used as solvent at a flow rate of 0.5 μL/min. Additionally, the solvent contained 200 nM LPC (19:0) and 5 μM C5-carnitine internal standards. Ionization was achieved by applying a 3.5 kV potential to the solvent syringe needle. The heated capillary inlet was held at 30 V and 200 °C. Mass spectra were acquired in positive mode in the range of m/z 150–1000 with a mass resolution of m/Δm = 60,000 at 412 m/z. Imaging data were acquired at a scan rate of 40 μm/s and a step between the lines of 30 μm. Each line scan was collected as an individual file. Data processing was performed using Peak-by-Peak software (Spectroswiss, Lausanne, Switzerland). Two biological replicates were analyzed per genotype.
2.7. Lipid peroxidation assay
Malondialdehyde (MDA), an indicator of lipid peroxidation, was measured using a commercially available colorimetric assay (10009055, Cayman Chemical). Briefly, Tibialis Anterior muscles from 6-month-old female mice were homogenized in ice-cold lysis buffer and centrifuged, then the supernatant was recovered. Samples and standards were mixed with TBA solution and boiled for 60 min. Reaction was stopped, samples were cleared by centrifugation, and absorbance at 535 nm was assessed using a plate reader. A total of 6 biological replicates were used per genotype.
2.8. Gene expression analysis
For real-time PCR, RNA was isolated from skeletal muscle of 6-month-old female mice using Trizol (Invitrogen), and RNA was converted to cDNA (Applied Biosystems High-Capacity cDNA RT Kit) and used for SYBR Green (Bio-Rad)-based real-time PCR using an Applied Biosystems QuantStudio3 Real-time PCR system. Results of detection with specific target primers were normalized to housekeeping gene (Rpl22) and expressed as arbitrary units of 2-ΔCT. A total of 6 biological replicates were utilized per muscle type and genotype. Average CT values were determined from 3 technical replicates per each biological replicate.
For mitochondrial DNA (mtDNA) analysis, total DNA was isolated from skeletal muscle of 5-month-old male mice using a QiAmp DNA Mini kit (Quiagen, #51304), and 10 μg per reaction were used as a template for SYBR Green (Bio-Rad)-based real-time PCR. Mitochondrial target genes were NADH Dehydrogenase 1 (Nd1) [19], and Mito1 [20] with H19 were used as nuclear DNA control. A total of 6 biological replicates were utilized per muscle type and genotype. Average CT values from 3 technical replicates per each biological replicate were obtained and used to determine the mitochondrial DNA content relative to nuclear DNA [21].
2.9. Protein analysis
For immunoblotting, skeletal muscle was harvested in the fed state immediately after euthanasia from 6-month-old male mice (anti-CPT2 and anti-OXPHOS) and 6-month-old female mice (anti-AMPK). Tissue homogenates were performed in sucrose media (10 mM Tris, 1 mM EDTA, 250 mM sucrose) with protease and phosphatase inhibitors using a hand-held mechanic homogenizer (Tissue Tearor, Thermo Fisher Scientific). Protein concentration was measured by BCA assay, and samples were equally loaded (20 μg protein/lane) onto 10 or 12% SDS-polyacrylamide gels. After electrophoresis, gel was transferred to nitrocellulose membrane and stained with Ponceau S for total protein visualization. Next it was blocked with 5% milk-TBST for 1 h, incubated with primary antibody (1:1,000–1:2,000) against CPT2 (Millipore, ABS85), total OXPHOS cocktail (Abcam, MS604-300), total AMPKα (Cell Signaling, 5831), or phosphorylated AMPKα T172 (Cell Signaling, 2535), and finally washed and incubated with secondary antibodies (1:10,000) conjugated to IR dye 800CW or 680LT (LiCor). Proteins were visualized with Odyssey and quantified using Image Studio (LiCor). A total of 3–6 biological replicates were utilized per muscle type and genotype.
For native gel electrophoresis, skeletal muscle was harvested after 1–2 h of fasting, and mitochondria was isolated as described for oxidation assays. Then, 200 μg of isolated mitochondria from white quadriceps and soleus muscle were lysed by mechanical trituration in 1X sample Buffer (Invitrogen, BN2003) supplemented with protease inhibitor (cOmplete mini, Roche) and 6% digitonin (Sigma, D141). Lysates were incubated on ice for 20 min and then centrifugated at 20,000×g. Clear supernatants were recovered, and protein concentration was measured again by BCA assay (Thermo, 23,225). Next, 30 μg of the recovered mitochondrial protein was mixed with G-250 sample additive (Invitrogene, BN2004) at 1.5% final concentration and loaded into a 4–16% BisTris gel (BN1004BOX10). Electrophoresis was performed at 150 V for 3.5 h at 4 °C. Upon completion, the gel was fixed in 40% methanol (Fisher, 194,449) + 10% acetic acid (Fisher A38-212) for 15 min, destained in 8% acetic acid until the desired background level was achieved, and imaged using the ChemiDoc™ Touch system (BioRad). A total of 3 biological replicates were analyzed per muscle type and genotype.
For TMT quantitative proteomics, soleus and EDL muscles from 5-month-old male mice were lysed in ice-cold 8 M Urea Lysis Buffer (8 M urea in 50 mM Tris, pH 8.0, 40 mM NaCl, 2 mM CaCl2, 1x cOmplete ULTRA mini EDTA-free protease inhibitor tablet), as described [11]. Three freeze–thaw cycles were performed, and samples were disrupted by sonication with a probe sonicator in three 5s bursts set at an amplitude of 30 (Q Sonica, Newtown, CT). Samples were centrifuged at 10,000×g for 10 min at 4 °C. Protein concentration was determined by BCA, and equal amounts of protein (200 μg, adjusted to 2.5 mg/mL with Urea Lysis Buffer) were reduced with 5 mM DTT at 32 °C for 30 min, cooled to room temperature, and then alkylated with 15 mM iodoacetamide for 30 min in the dark. Unreacted iodoacetamide was quenched by the addition of DTT up to 15 mM. Initial digestion was performed with Lys C (Thermo Fisher) 1:100w-w; 2 μg enzyme per 200 μg protein) for 4 h at 32 °C. Following dilution to 1.5M urea with 50 mM Tris (pH 8.0), 30 mM NaCl, 5 mM CaCl2, the samples were digested overnight with trypsin (Promega, Madison, WI) 50:1 w/w, protein:enzyme at 32 °C. Samples were acidified to 0.5% TFA and centrifuged at 10,000×g for 10 min at 4 °C to pellet insoluble material. Supernatant containing soluble peptides was desalted on a 50 mg tC18 SEP-PAK solid phase extraction column (Waters, Milford, MA) and eluted (500 μL 25% acetonitrile/0.1% TFA and 2 × 500 μL 50% acetonitrile/0.1% TFA). The 1.5 mL eluate was frozen and dried down via SpeedVac.
TMT labeling was performed as described [22]. Dried down peptides were re-suspended in 100 μL of 200 mM triethylammonium bicarbonate (TEAB), mixed with a unique 10-plex Tandem Mass Tag (TMT) reagent (0.8 mg re-suspended in 50 μL100% acetonitrile), and shaken for 4 h at room temperature (Thermo Fisher). A total of 3 × 10-plex kits were used. Following quenching with 0.8 μL 50% hydroxylamine, samples were frozen and placed in SpeedVac. Samples were re-suspended in ∼1 mL of 0.5% TFA and again subjected to solid phase extraction but with a 10 mg tC18 SEP-PAK SPE column (Waters). The multiplexed peptide sample was subjected to high pH reversed phase fractionation (Thermo Fisher; Catalog #84868). In this protocol, peptides (100 μg) are loaded onto a pH-resistant resin and then desalted with water washing combined with low-speed centrifugation. A step-gradient of increasing acetonitrile concentration in a high-pH elution solution is then applied to columns to elute bound peptides into 8 fractions. Following elution, fractions were frozen and placed in SpeedVac.
nLC-MS/MS was performed as described [22]. Peptide fractions were suspended in 0.1% formic acid at a concentration of 0.25 μg/μL, following peptide quantification (ThermoFisher). All samples were subjected to nanoLC-MS/MS analysis using an UltiMate 3000 RSLCnano system (Thermo Fisher) coupled to a Q Exactive PlusHybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher) via nanoelectrospray ionization source. For each injection of 4 μL (1 μg), the sample was first trapped on an Acclaim PepMap 100 20 mm × 0.075 mm trapping column (Thermo Fisher) 5 μl/min at 98/2 v/v water/acetonitrile with 0.1% formic acid, after which the analytical separation was performed over a 90-min gradient (flow rate of 300 nl/min) of 3%–30% acetonitrile using a 2 μm EASY-Spray PepMap RSLC C18 75 μm × 250 mm column (Thermo Fisher) with a column temperature of 55 °C. MS1 was performed at 70,000 resolution, with an AGC target of 1 × 106 ions and a maximum IT of 60 ms. MS2 spectra were collected by data-dependent acquisition (DDA) of the top 20 most abundant precursor ions with a charge greater than 1 per MS1 scan, with dynamic exclusion enabled for 30 s. Precursor ions were filtered with a 1.0 m/z isolation window and fragmented with a normalized collision energy of 30. MS2 scans were performed at 17,500 resolution, AGC target of 1 × 105 ions, and a maximum IT of 60 ms.
Proteome Discoverer 2.2 (PDv2.2) was used for raw data analysis, with default search parameters including oxidation (15.995 Da on M) as a variable modification and carbamidomethyl (57.021 Da on C) and TMT6plex (229.163 Da on peptide N-term and K) as fixed modifications, and 2 missed cleavages (full trypsin specificity). Data were searched against both the full mouse proteome, as well as the Mito Carta 3.0 database [23] and mitochondrial gene/protein lists from Stenton et al. [24] and Murayama et al. [25]. Peptide spectrum matches (PSMs) were filtered to a 1% FDR. PSMs were grouped to unique peptides while maintaining a 1% FDR at the peptide level. Peptides were grouped to proteins using the rules of strict parsimony, and proteins were filtered to 1% FDR using the Protein FDR Validator node of PD2.2. MS2 reporter ion intensities for all PSMs having co-isolation interference below 0.5 (50% of the ion current in the isolation window) and an average S/N > 10 for reporter ions were added up together at the peptide and protein levels. Imputation was performed via low abundance resampling. The protein group tab in the PDv2.2 results was exported as tab delimited.txt. files and statistically analyzed based on a previously described workflow [22]. First, M2 reporter (TMT) intensities were added up together for each TMT channel, and each channel's sum was divided by the average of all channels' sums, resulting in channel-specific loading control normalization factors to correct for any deviation from equal protein input in the 10-plex experiments. Reporter intensities for proteins were divided by the loading control normalization factors for each respective TMT channel. All loading control-normalized reporter intensities were converted to log2 space, and the average value from the ten samples per kit was subtracted from each sample-specific measurement to normalize the relative measurements to the mean of each kit. Data from each kit were then combined for statistical comparisons. For comparisons, condition average, standard deviation, p-value (p, two-tailed student's t-test, assuming equal variance), and adjusted p-value (Padjusted, Benjamini Hochberg FDR correction) were calculated [26,27]. For protein-level quantification, only Master Proteins—or the most statistically significant protein representing a group of parsimonious proteins containing common peptides identified at 1% FDR—were used for quantitative comparison. Venn diagrams were generated using the free online tool jvenn [28]. Pathway analysis was performed using Reactome [29]. A total of 6 biological replicates were analyzed per muscle type and genotype. The entire proteome data set has been made available online using accession number ‘PXD030896’ for Proteome Xchange [30] and accession number ‘JPST001445’ for jPOST Repository [31].
2.10. Statistics
Data are presented as mean ± SEM, unless otherwise specified. Statistical analysis and figures were generated using Excel or GraphPad Prism version 8.0.0 for Windows (GraphPad Software) except for proteomics data. Data were compared using unpaired Student's t-test and one-way or two-way ANOVA followed by multiple comparison analysis. Significance level was set at P < 0.05.
3. Results
3.1. Muscle fiber-type composition is a major determinant in fatty acid-supported mitochondrial respiration and responsiveness to increased energetic demands
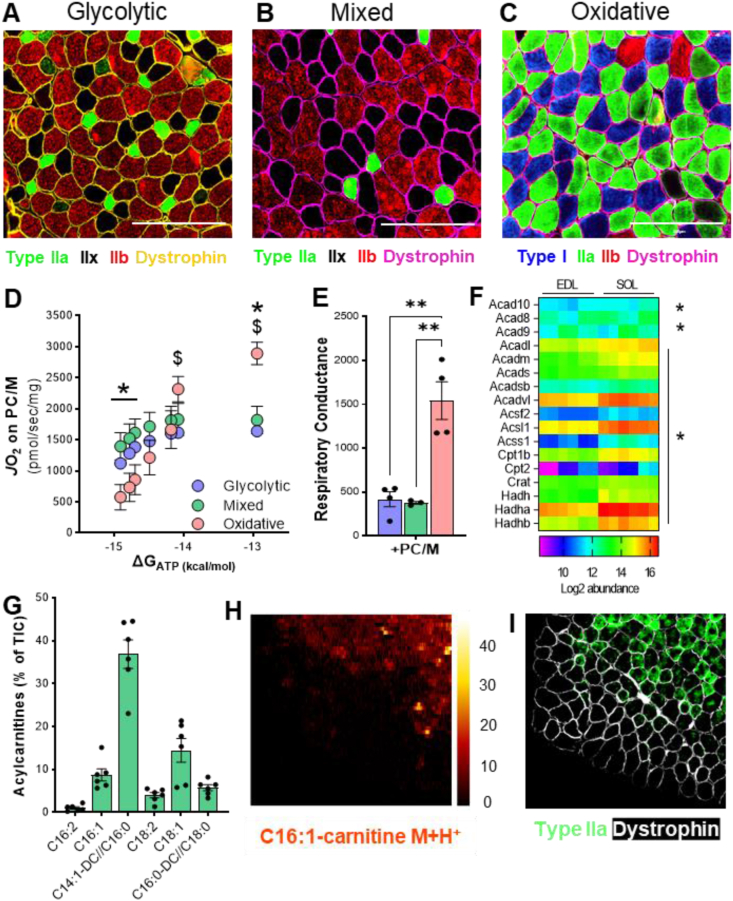
Understanding the consequences of mitochondrial long-chain fatty acid oxidation deficiency in skeletal muscle requires establishing first the physiological contribution of these lipid substrates to energy production across muscle types. Here, glycolytic (extensor digitorum longus, EDL), mixed (tibialis anterior, TA), and oxidative (Soleus) muscles were assessed. These muscles were investigated due to their known, distinct fiber-type composition relative to one another [32]. We confirmed co-existence of different fiber types within these muscles via fluorescence-based immunohistochemical detection of myosin heavy chain (MyHC) isoforms [8,9]. The EDL muscle was rich in the expression of MyHC-2B, hallmarking fast-twitch glycolytic type IIb fibers (Figure 1A). The TA muscle was composed of an approximately equal combination of the glycolytic Type IIb and the metabolically intermediate MyHC-2X-expressing Type IIx fibers (Figure 1B). The soleus muscle was highly enriched with the expression of MyHC-β and MyHC-2A proteins, representing slow, oxidative Type I and fast, oxidative-glycolytic Type IIa fibers, respectively (Figure 1C). Based upon MyHC expression patterns, these data confirm that EDL and soleus muscles are at opposite ends of the fiber-type spectrum, from predominately fast-glycolytic to predominately slow-oxidative, respectively, with TA as an intermediate muscle.
Figure 1.
Muscle fiber type composition is a major determinant in fatty acid-supported mitochondrial respiration. (A), (B), (C) Representative imaging of muscle fiber typing via Myosin Heavy Chain (MyHC) immunohistochemistry in predominately glycolytic (EDL), mixed (TA), and oxidative (Soleus) muscles. Magnification 20X. Scale bar 200 μm. (D) Mitochondrial respiration (JO2) across a continuum of ATP free energy (ΔGATP) and (E) respiratory conductance in predominantly glycolytic, mixed, and oxidative muscles energized with palmitoylcarnitine/malate (PC/M). (F) Heatmap of abundance of proteins involved in mitochondrial fatty acid oxidation in glycolytic (EDL) and oxidative (SOL) muscles. (G) Relative abundance of long-chain acylcarnitines (LCACs) in mixed TA muscle. (H) Digital reconstitution of mass spectroscopy-based lipid scanning (nano-DESI) of palmitoleyl-carnitine (C16:1) in mixed TA muscle and (I) immunostaining for myosin heavy chain type 2A (green) and dystrophin (white) on consecutive muscle section. For (D), (E), and (G), data is presented as Mean ± SEM; n = 3–6 and ∗P ≤ 0.05 by 2-way ANOVA. For (F), data is presented as log2 abundance; n = 6 and ∗P ≤ 0.05 of adjusted p-value calculated by Benjamini Hochberg FDR correction.
Next, we compared mitochondrial oxidative capacity between these muscles when long-chain fatty acids are provided as energetic substrate. To do so, oxygen consumption rates (JO2), as indicative of mitochondrial respiration, were measured in isolated mitochondria using high resolution respirometry in the presence of palmitoylcarnitine (PC), with a high concentration of malate (M; >2 mM) to facilitate exchange of metabolites and TCA cycle intermediates between mitochondria and assay buffer. The assay was conducted under a creatine kinase (CK) clamp to facilitate the exploration of mitochondrial oxidative metabolism and responsiveness across a range of physiological energy demands [12]. This clamp takes advantage of the physiological function of CK enzyme which converts extra-mitochondrial ADP to ATP when phosphocreatine (PCr) is available. Thus, by manipulating the levels of phosphocreatine in the respiration assay buffer, it is possible to alter the ATP/ADP ratio and ATP free energy levels; this in turn regulates mitochondrial respiration.
At resting energy demands, represented by DeltaG_ATP = −14.91 and generated by increasing the ATP/ADP ratio with high phosphocreatine, the rate of mitochondrial respiration on long-chain fatty acid palmitoylcarnitine was not different between glycolytic and mixed muscles as indicated by similar JO2 (Figure 1D). However, respiratory rate at resting conditions of the oxidative soleus muscle with palmitoylcarnitine was significantly lower than for mixed muscle (Figure1D). This was surprising given that oxidative muscles are known to rely on fatty acid oxidation for energy production more than on glycolytic or mixed counterparts [33].
To assess the intrinsic mitochondrial response to increased energy demands, respiration on palmitoylcarnitine was measured on a continuum of free energy, from low energy demand (DeltaG_ATP = −14.91) to high energy demand (DeltaG_ATP = −12.94). Here, although glycolytic and mixed muscles presented with higher palmitoylcarnitine-supported respiration when energy demands were low compared to oxidative muscle, this rate was not significantly increased at higher energy demands (Figure 1D). Then, the slope of the linear portion of the relationship between the two variables was calculated to generate a measure of respiratory conductance. This calculated respiratory conductance serves as a measure of OXPHOS pathway sensitivity to changes in the system's free energy, such that a steeper slope reflects higher responsiveness of the mitochondria to a given substrate over the range of given free energy conditions. Here, neither glycolytic nor mixed muscles demonstrated a significant increase in conductance for palmitoylcarnitine, suggesting a minimal ability of glycolytic and mixed muscles to accelerate fatty acid oxidation with increased energy demands (Figure 1E). Conversely, the oxidative soleus muscle energized with palmitoylcarnitine exhibited a ∼5-fold increase in mitochondrial respiration (Figure 1D) and a ∼3-fold increase in respiratory conductance (Figure 1E) in response to increased energy demands. These data suggest intrinsic differences in the composition and/or regulation of fatty acid oxidative metabolism between glycolytic and oxidative muscles. In agreement, significant enrichment of mLCFAO-related proteins was observed in oxidative compared to glycolytic muscle (Figure 1F). Specifically, medium-chain acyl-CoA dehydrogenase (ACADM; 2-fold), long-chain acyl-CoA dehydrogenase (ACADL; 1.6-fold), long-chain acyl-CoA synthetase family member 1 (ACSL1; 1.9-fold), short-chain acyl-CoA synthetase family member 1 (ACSS1; 2.6-fold), carnitine palmitoyltransferase 1B (CPT1B; 1.7-fold), carnitine palmitoyltransferase 2 (CPT2; 1.6-fold), and hydroxyacyl-CoA dehydrogenase subunit B (HADHA-B; 2-fold) were all significantly more abundant in oxidative muscle (Figure 1F).
Next, we sought to determine whether differential reliance on lipid catabolism for energy production occurred in a fiber-type dependent manner by assessing the spatial distribution of long-chain acylcarnitines (LCACs), which are indicators of mLCFAO flux. Mixed TA muscle demonstrated high relative abundance of LCACs particularly C16, C18:1. and C16:1 (Figure 1G) which were subsequently targeted for visualization by mass spectroscopy-based imaging (nano-DESI). Here, the presence, distribution, and abundance of LCACs in different fibers of TA muscle were determined based on the differential puncta-like signaling the lipid imaging (Figure 1H). Comparison of the distribution pattern of LCAC C16:1, obtained by nano-DESI tissue scanning, and of fiber types, determined by immunohistochemistry in serial sections. Revealed that oxidative fibers marked by type IIa MyHC protein had higher intensity for LCACs signal compared to glycolytic fibers marked by type IIb MyHC (Figure 1I). These data allowed visualization of the metabolic distinctions between the heterogeneous fibers within a given muscle. The differences in spatial distribution of long-chain acylcarnitines suggest that metabolic activity can be strikingly distinct between two adjacent muscle fibers. Furthermore, it can be used to distinguish muscle fiber types from a metabolic perspective.
Taken together, these data suggest that differences in fatty acid oxidative capabilities between glycolytic and oxidative muscles are intrinsic to the mitochondria. The significantly high conductance observed in oxidative fiber-enriched muscles when energized with palmitoylcarnitine and the spatial distribution of long-chain acylcarnitines suggests that long-chain fatty acids are a critical substrate for oxidative muscle fibers, perhaps more significantly under high energy demands.
3.2. Loss of carnitine palmitoyltransferase 2 in skeletal muscle prevents mLCFAO and induces mitochondrial biogenesis
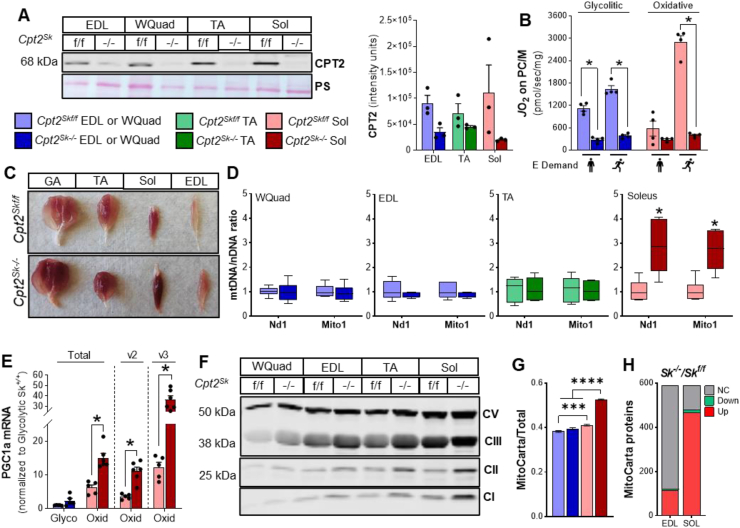
To determine how impairing muscle metabolic capacity might disproportionately impact muscles with different fiber-type composition, we targeted CPT2, an irreplaceable enzyme in the mLCFAO pathway. In agreement with our proteomic data (Figure 1F), we found a differential amount of CPT2 protein between muscles with higher expression in oxidative (soleus) compared to mixed (TA) and glycolytic (EDL and white Quadriceps (WQuad) muscles (Figure 2A). Muscle-specific genetic targeting of Cpt2 using floxed-conditional knockout mice (Cpt2Sk−/−) [4,5] effectively depleted CPT2 protein in glycolytic, mixed, and oxidative muscles (Figure 2A). To confirm that loss of CPT2 compromises mLCFAO in Cpt2Sk−/− muscles, fatty acid-supported oxygen consumption in isolated mitochondria was measured. Respiration rates for the long-chain fatty acid substrate palmitoylcarnitine were decreased in both glycolytic and oxidative muscles by 77% and 86%, respectively (Figure 2B), and in agreement with our previous reports [4,5].
Figure 2.
Loss of carnitine palmitoyltransferase 2 in skeletal muscle prevents mLCFAO and induces mitochondrial biogenesis. (A) Western blot for CPT2 on glycolytic (EDL and White Quadriceps), mixed (TA), and oxidative (Soleus) muscle homogenates (representative image and quantitation). (B) Mitochondrial respiration (JO2) at minimum (standing icon) and maximum (running icon) energy demands in predominantly glycolytic and oxidative muscles energized with palmitoylcarnitine/malate (PC/M). (C) Representative image of Gastrocnemius (GA), Tibialis Anterior (TA), Soleus (Sol), and Extensor Digitorium Longus muscles from control (top row) and Cpt2Sk−/− (bottom row) mice. (D) Quantitation of mitochondrial DNA (mtDNA) by levels of NADH Dehydrogenase 1 (Nd1), and Mito1 relative to nuclear DNA (nDNA) in different muscles of control and Cpt2Sk−/− mice. (E) Relative mRNA levels of PCG1α total, variant 2 (v2), and variant 3 (v3) in glycolytic and oxidative muscles from control and Cpt2Sk−/− mice. (F) Western blot for individual subunits of the mitochondrial OXPHOS complexes. (G) Enrichment of MitoCarta 3.0 positive proteins relative to the muscle total proteome as determined by quantitative proteomics in glycolytic (EDL) and oxidative (Soleus) muscle. (H) Differential expression of MitoCarta 3.0 positive proteins between control and Cpt2Sk−/− in glycolytic (EDL) and oxidative (SOL) muscles. Data is presented as Mean ± SEM except for (D) which also displays min and max values. For (B), (D), and (E), n = 4–6 and ∗P ≤ 0.05 by 2-way ANOVA. For (G), n = 6 and ∗P ≤ 0.05 by 1-way ANOVA.
Mitochondrial biogenesis is a common response to limitations in mitochondrial metabolic capacity [34]. Macroscopic inspection of muscles demonstrated increased red hues in Cpt2Sk−/− animals (Figure 2C). To determine whether the increased muscle redness was related to changes in mitochondrial content, mitochondrial DNA was quantified to reveal a ∼2.5-fold increase in Cpt2Sk−/− oxidative soleus compared to control, but no changes were observed in glycolytic or mixed muscles (Figure 2D). In agreement, Cpt2Sk−/− oxidative, but not glycolytic, muscles showed a significant increase in the expression of PGC1α, a master regulator of mitochondrial biogenesis (Figure 2E). Specifically, mRNA levels of PGC1α isoforms 2 and 3, which are known to regulate fatty acid oxidation, and mitochondrial biogenesis genes [35] were elevated in soleus muscle of Cpt2Sk−/− mice (Figure 2E). In further agreement, AMPK activation drives mitochondrial biogenesis, and western blotting demonstrated increases in the activated phosphorylation status of AMPK (Figure S1) and in the expression of several individual proteins of the oxidative phosphorylation (OXPHOS) complexes I through V in mixed and oxidative muscles of Cpt2Sk−/− mice but to a lesser extent in glycolytic muscles (Figure 2F).
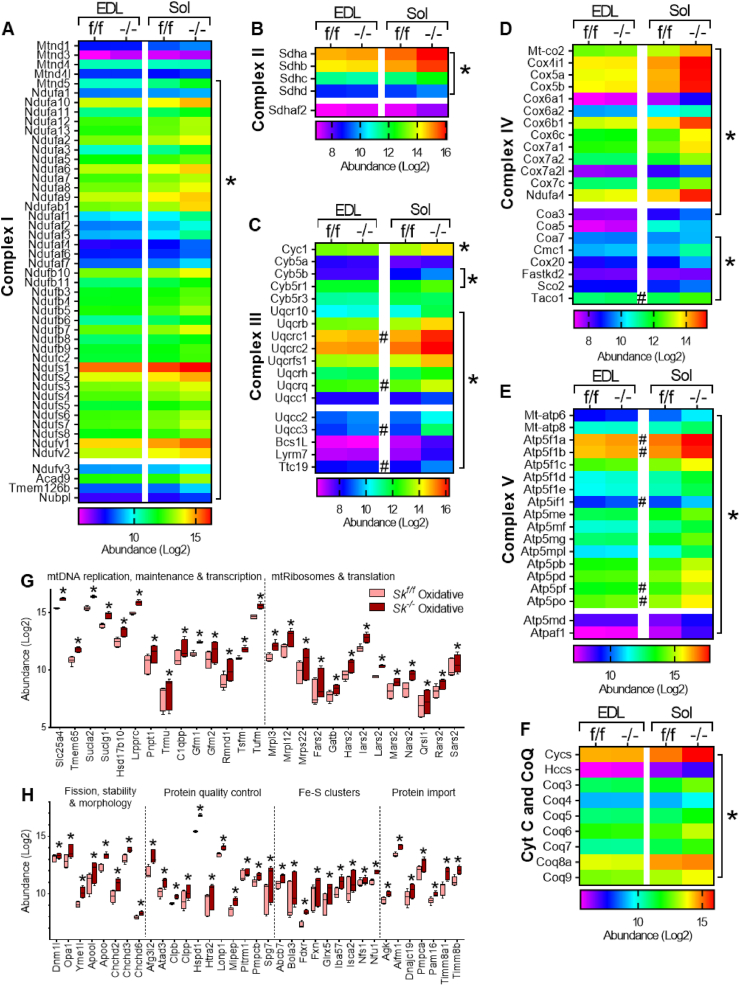
An untargeted comprehensive inquiry of the muscle proteome was next performed via whole tissue proteomics. A total of 2581 proteins were identified and quantitated in glycolytic and oxidative muscles of both control and Cpt2Sk−/− mice. Of those, 588 were identified as mitochondrial targets according to the MitoCarta 3.0 database [23]. Comparison of the summed abundance of mitochondrial proteins relative to total protein abundance between muscles confirmed intrinsically higher mitochondrial enrichment in control oxidative soleus compared to control glycolytic EDL (Figure 2G). Upon muscle-specific CPT2 deletion, mitochondrial protein abundance was not different between genotypes in glycolytic muscles (Figure 2G). However, in Cpt2-deficient oxidative muscles, the abundance of mitochondrial proteins was ∼30% higher compared to controls (Figure 2G). Furthermore, only 21% (122 of the 588) of mitochondrial proteins were differentially expressed (q value < 0.1) in Cpt2Sk−/− glycolytic muscles, whereas in Cpt2Sk−/− oxidative muscle, a vast majority—82% (481 of the 588)—were significantly different relative to control muscle (Figure 2H). Of the altered mitochondrial proteins in oxidative muscle, only 3% were downregulated, and 97% were upregulated (Figure 2H). Detailed assessment of the mitochondrial proteome revealed that 93% of the identified individual subunits and assembly factors of the OXPHOS complexes I through V (111 out of 119 protein targets) were significantly increased in Cpt2Sk−/− oxidative muscle compared to control (Figure 3A–F). Contrastingly, in Cpt2Sk−/− glycolytic muscle, only 8% of these OXPHOS-related proteins were upregulated compared to control (Figure 3A–F), suggesting that the mitochondrial biogenesis response to CPT2 deletion is predominately muscle-type specific. Like the OXPHOS expression response, only oxidative Cpt2-deficient muscle presented with a significant increase in expression of most proteins needed to support mitochondrial biogenesis such as mtDNA maintenance and transcription (Figure 3G), mitochondrial dynamics and morphology, protein quality control and import, and synthesis of iron-sulphur clusters (Figure 3H).
Figure 3.
CPT2 deficiency triggers major mitochondrial response in a muscle-type-specific manner. (A), (B), (C), (D), (E), (F) Heatmaps of expression abundance of proteins related to electron transport chain complexes subunits and assembly factors in glycolytic (EDL) and oxidative (Sol) muscles of control and Cpt2Sk−/− mice. (G), (H) Abundance of proteins involved in mitochondrial processes in control and Cpt2Sk−/− oxidative muscle. For (A) to (F), data is presented as an average of 6 biological replicates. For (G) and (H), data is presented as Median with Min and Max values; n = 6. #Adjusted p-value P ≤ 0.05 for EDL between genotypes and ∗ adjusted p-value P ≤ 0.05 for Soleus between genotypes as calculated by Benjamini Hochberg FDR correction.
Together these data suggest that upon CPT2 deletion, mitochondrial biogenesis and upregulation of proteins directly involved in oxidative phosphorylation and mitochondrial function are muscle-specific, with a predominantly robust response in oxidative muscles.
3.3. Mitochondrial biogenesis in response to CPT2 deficiency occurs in a fiber-type specific manner
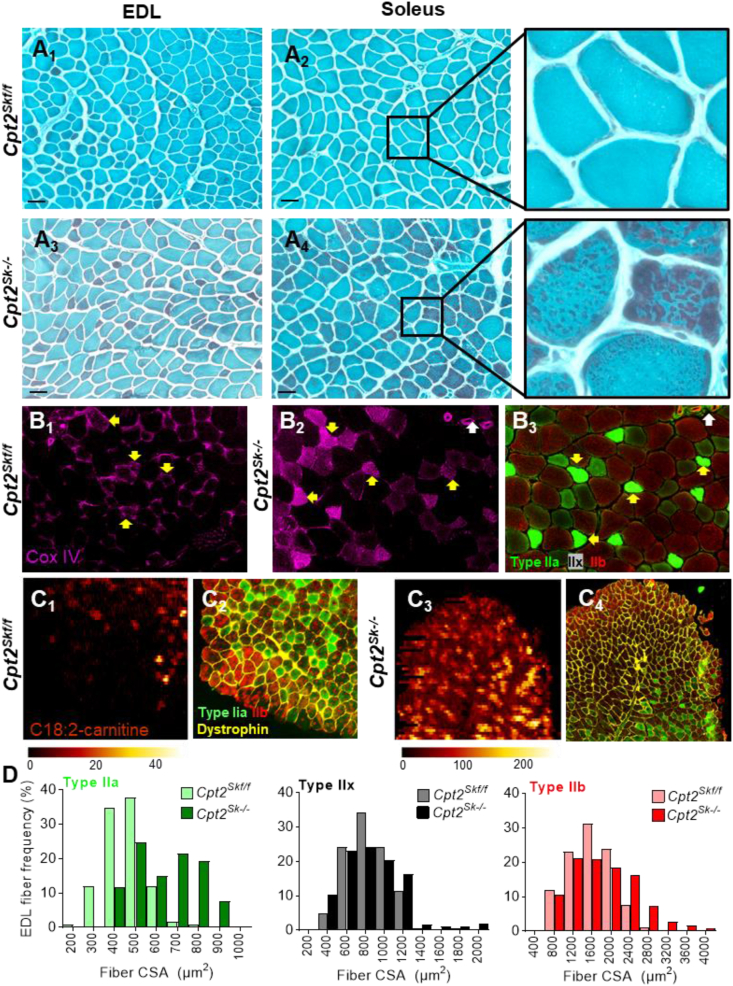
Next, we sought to assess the muscle fiber-type specific response to mitochondrial biogenesis by performing modified Gomori stain in conjunction with traditional myosin heavy chain immunostaining in glycolytic (EDL) and oxidative (Soleus) muscles. Typically, physiological abundance of muscle mitochondria is not visible by Gomori stain; however, when mitochondrial biogenesis is elevated, mitochondria can be detected as red aggregates throughout the myofiber cross-section. In control muscles, mitochondria were not visible (Figure 4A and Figure S2); by contrast, CPT2-deficient muscles presented with red-positive staining at regions pertaining to both the subsarcolemmal and intermyofibrillar compartments, suggesting a high degree of mitochondrial proliferation (Figure 4A and Figure S2). Although affected myofibers could be observed in both muscles, the number and extent of highly stained, Gomori-positive fibers were far greater in oxidative Soleus compared to glycolytic EDL muscle. When comparing individual fibers of the same muscle, the Cpt2Sk−/− soleus presented with a high degree of differential staining, suggesting that the intensity of mitochondrial biogenesis occurred in a fiber-type specific manner. The fiber-type-specific increase in mitochondrial abundance in Cpt2Sk−/− compared to control was confirmed in the glycolytic EDL muscle by a highly sensitive immunostaining against the mitochondrial OXPHOS subunit Cytochrome c oxidase (COX IV) (Figure 4B). The heavily COX IV-stained fibers in the EDL of CPT2-deficient mice overlapped with slow-oxidative type IIa fibers on serial sections, whereas fast-glycolytic type IIb myofibers appeared relatively unstained (Figure 4B), indicating a fiber-type-specific mitochondrial biogenic response in favor of oxidative fibers.
Figure 4.
Differential muscle mitochondrial biogenesis, acylcarnitine accumulation, and fiber enlargement in Cpt2Sk−/−mice is fiber-type-specific. (A1 to A4) Representative Gomori staining of mitochondria (red) in muscle fibers (light blue) of glycolytic (EDL) and oxidative (Soleus) muscles for both genotypes. Far-right column contains magnified images of A2 and A4. Magnification 20X. Scale bar = 100 μm. (B) cytochrome c oxidase protein (COX IV) immunodetection of (B1) control and (B2) Cpt2Sk−/− EDL muscle and (B3) matched myosin heavy chain immunodetection of Cpt2Sk−/− EDL. White arrows indicate the same anatomic structure in B2 and B3. Yellow arrows indicate type IIa myofibers. (C) Digital reconstitution of mass spectroscopy-based lipid scanning (nano-DESI) of linoleoyl-carnitine (C18:2) and matched myosin heavy chain immunodetection of Control (C1,2) and Cpt2Sk−/− (C3,4) TA muscle. (D) Distribution of fiber cross-sectional area (CSA) of control and Cpt2Sk−/− EDL analyzed for Type IIa, IIx, and IIb fibers independently. Data is presented here as the mean of 3 biological replicates per fiber type and genotype.
To assess the fiber-type-specific response to LCAC accumulation, a hallmark of CPT2 deficiency in skeletal muscle [5], nanoDESI lipid imaging was employed. Imaging of LCACs revealed that in the CPT2-deficient TA muscle, the long-chain acylcarnitine linoleoyl-carnitine (C18:2) preferentially accumulated in the more oxidative type IIa fibers (Figure 4C). Due to the increase in mitochondrial abundance and LCAC accumulation, we next questioned whether excessive oxidative stress could be occurring in the Cpt2Sk−/− muscle. Levels of malondialdehyde (MDA), a marker of lipid-derived oxidative stress, were not different between control and Cpt2Sk−/− mixed muscle. We next questioned whether fiber size was impacted. Quantitative analysis of a cross-sectional area (CSA) by fiber type in EDL muscle revealed an overall shift towards larger fibers in Cpt2Sk−/− compared to control, an effect that was most pronounced among oxidative type IIa fibers (Figure 4D). These data suggest that in CPT2-deficient muscles mitochondrial biogenesis, LCAC accumulation, and changes in fiber size occur predominately in oxidative fibers.
Together, these data suggest that mLCFAO-deficient muscles due to CPT2 deletion undergo a significant wave of mitochondrial proliferation and LCACs accumulation in a fiber-type-specific manner. Specifically, slow-oxidative type I and small fast-oxidative-glycolytic type IIa fibers become heavily loaded with mitochondria and LCACs in the absence of CPT2, which in turn increases fiber size. In the oxidative muscles of Cpt2Sk−/− mice, enhanced mitochondrial biogenesis was regulated in part by the master transcriptional regulator PGC1α and was accompanied by upregulation of most bioenergetic-related proteins, such as the mitochondrial OXPHOS complexes.
3.4. CPT2 deficiency shifts oxidative muscles towards a glycolytic-like metabolic proteome
Given the distinct mitochondrial biogenic response in oxidative compared to glycolytic muscles of Cpt2Sk−/− mice, we next assessed native conformation and assembly of OXPHOS complexes which are known to be unique between glycolytic and oxidative muscles [27]. As expected, a distinctive pattern of band thickness and molecular weight-based migration, visualized by native gel electrophoresis, was evident between control glycolytic and oxidative muscle mitochondria (Figure 5A). For glycolytic muscle, the band separation was not different between control and Cpt2Sk−/− (Figure 5A). However, for oxidative muscle, the Cpt2Sk−/− muscle presented an electrophoretic pattern that was distinct from control oxidative muscle but was nearly identical to glycolytic muscle (Figure 5A). These data suggest that the absence of CPT2 in oxidative muscle not only promotes mitochondrial biogenesis but also alters the mitochondrial proteome and OXPHOS conformation toward a more glycolytic-like phenotype.
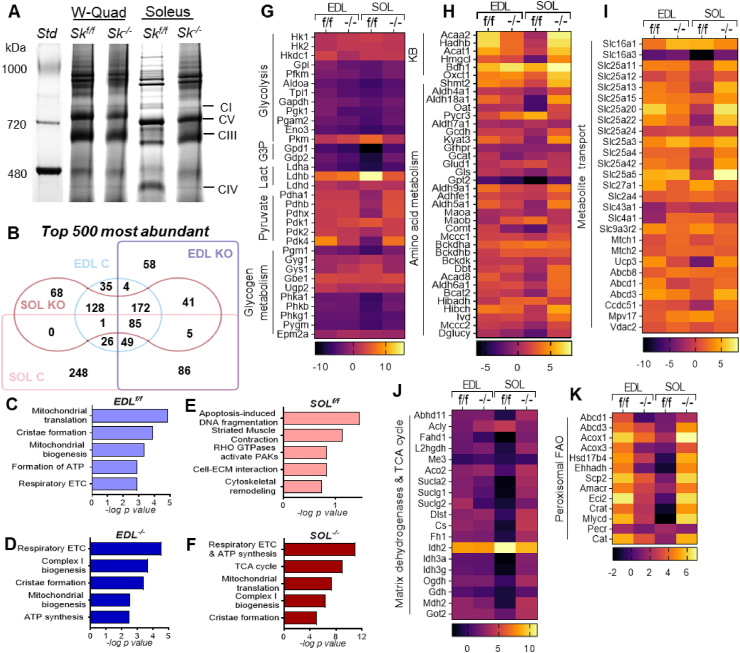
Figure 5.
Loss of CPT2 shifts oxidative muscle proteome towards a glycolytic-like protein signature. (A) Representative image of Native-PAGE of mitochondria lysates to demonstrate OXPHOS complexes in control and Cpt2Sk−/− muscles. (B) Venn diagram demonstrating overlap among the top-500 most abundant proteins in glycolytic (EDL) and oxidative (Soleus) muscle from control and Cpt2Sk−/− mice. (C–F) Pathway analysis for the top-500 most abundant proteins arranged by -log p value. (G–K) Heatmaps of abundance of proteins relative to the tissue mean related to macronutrient catabolism and metabolite transport in glycolytic (EDL) and oxidative (SOL) muscles of control and Cpt2Sk−/− mice; data is presented as average of 6 biological replicates per muscle type and genotype.
Out of the 30,000 genes present in the mouse genome, ≈1000 are differentially expressed between different muscle fiber types [36]. We calculated the abundance of individual proteins relative to the tissue mean to identify the most abundant targets and established a top-500 most abundant protein signature for each sample. Comparison of the top-500 most abundant proteins between muscle types showed that glycolytic and oxidative muscles shared a total of 161 of these 500 proteins (32%) (Figure 5B). However, the Cpt2Sk−/− glycolytic and oxidative muscles shared a total of 303 proteins (61%), double that of the controls (Figure 5B). Furthermore, oxidative Cpt2Sk−/− and glycolytic control muscles had 384 (77%) proteins in common, which was 2.4-fold more than those common between control glycolytic and oxidative muscles. These data suggest that the oxidative Cpt2Sk−/− muscle protein signature was more similar to glycolytic muscle than it was to counterpart control oxidative muscle. Next, comparison between genotypes revealed that glycolytic muscles from control and Cpt2Sk−/− mice shared a majority, 310 (62%), of common proteins; however, oxidative control and Cpt2Sk−/− muscle had far less in common at 91 (18%) (Figure 5B). These data suggest that CPT2-deficiency in mitochondria of oxidative muscles impacts the overall tissue proteome, causing a shift towards a more glycolytic-like protein signature.
Functional analysis of these top-500 most abundant proteins provided further insight on how glycolytic and oxidative muscles are differentially affected by lack of mLCFAO due to CPT2 loss. The most significant biological pathways in glycolytic EDL muscle of both the control and the Cpt2Sk−/− mice included mitochondrial biogenesis, cristae formation, respiratory electron transport chain, and ATP synthesis (Figure 5C,D and Figure S4). This suggests that in glycolytic muscles of both genotypes, the relative abundance of the mitochondrial machinery is significant and contributes largely to the whole-muscle proteome. On the contrary, highly abundant signature pathways in control Soleus muscle involved muscle contraction, cell-extracellular matrix interaction, and cytoskeletal remodeling (Figure 5E and Figure S4), highlighting the relevance of sarcomere-related structures in oxidative muscles that work to produce long-lasting contractions. Moreover, Cpt2Sk−/− oxidative muscle diverged from its control counterpart to show mitochondrial-centered pathways that were of high similarity to glycolytic muscles (Figure 5F). This data suggests that in CPT2-deficient oxidative muscles, relative abundance and pathway profile of the protein signature shifts towards the glycolytic muscle phenotype.
Next, we questioned whether this shift in the oxidative muscle proteome included specific elements of the metabolic machinery necessary for energy production. Analysis of quantitative proteomics data revealed that the expression pattern of targets related to macronutrient catabolism and energy provision in oxidative Cpt2Sk−/− muscle did not resemble that of the control oxidative muscle, but rather that the oxidative muscle proteome was remarkably similar to control glycolytic muscle (Figure 5G–J). Specifically, the relative abundance of several enzymes involved in the oxidative metabolism of glucose, lactate, pyruvate (Figure 5G), ketone bodies, and amino acids (Figure 5H) in Cpt2Sk−/− oxidative muscle resembled glycolytic muscle. Likewise, several mitochondrial matrix dehydrogenases, TCA cycle enzymes (Figure 5I), and metabolite transporters (Figure 5J) in Cpt2Sk−/− soleus muscle showed relative abundance levels comparable with glycolytic muscle. According to muscle metabolomics, minimal alterations in metabolites were observed between genotypes, except for increased glucose and galactose levels in Cpt2Sk−/− oxidative muscles, suggesting higher carbohydrate use (Figure S5). Among TCA cycle intermediates, succinate was significantly lower in Cpt2Sk−/− muscles, especially in oxidative muscle (Figure S5), possibly representing higher flux through complex II due to higher abundance of the SDH protein subunits (Figure 3B) and/or to the loss of competition that normally exists between mLCFAO and SDH for the transfer of electrons to Coenzyme Q. Together, these data demonstrate that upon CPT2 deletion, muscles experience an oxidative-to-glycolytic proteome shift, specifically increasing the abundance of targets involved in glucose, ketones, and amino acid catabolism.
3.5. Shifts in glycolytic metabolism of Cpt2Sk−/− oxidative muscle were not accompanied by shifts in myosin heavy chain
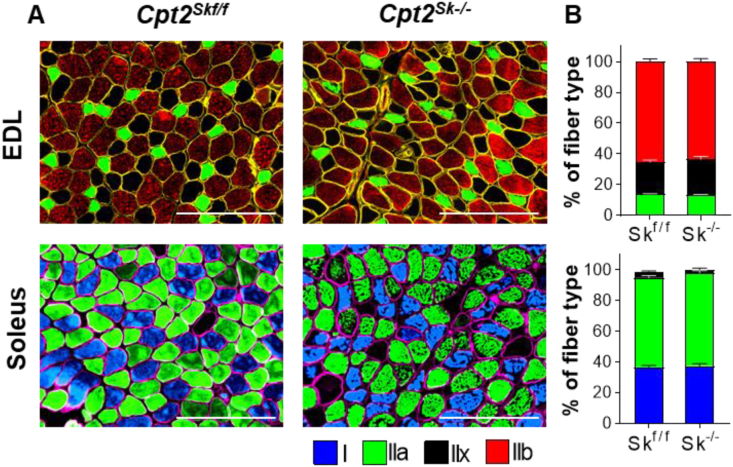
The structure and molecular composition of a muscle fiber is shaped by the type of mechanical work the muscle is required to perform [2,37]. As such, training and fitness regulate both mitochondrial content and specific sarcomere characteristics. One of the major determinants of fiber contractile capacity is myosin, wherein myosin heavy chain (MyHC) isoforms differentially distributed across fiber types allow for a straightforward distinction of the contractile properties of individual myofibers. Here, immunohistochemical detection of MyHC isoforms in the EDL, TA, and soleus muscles was performed to determine whether a deficit in fatty acid oxidation and a shift in proteome markers from oxidative toward glycolytic would also result in changes in the expression of the contractile proteins. Despite the loss of mLCFAO and the proteomic shifts observed in Cpt2Sk−/− oxidative muscles, the relative composition of the MyHC isoforms was not different compared to control EDL or Soleus muscles (Figure 6, Figs. S6, S7). These results show that loss of mLCFAO capacity in skeletal muscle via deletion of CPT2 does not trigger fiber type switching as defined by changes in myosin heavy chain isoform composition.
Figure 6.
CPT2 deficiency does not induce myosin heavy chain isoform switching. (A) Representative images and (B) quantification of control and Cpt2Sk−/− muscles by immunofluorescent detection of myosin heavy chain isoforms: MyHC-β (Type I, blue), MyHC-2B (Type IIb, red), MyHC-2A (Type IIa, green), or no stain (Type IIx, black). Myofiber perimeter is demonstrated with dystrophin (yellow or pink). EDL, extensor digitorum longus. Data are presented as mean ± SEM. N = 3. Magnification 20X. Scale bar = 200 μm.
3.6. mLCFAO deficiency causes metabolic reprogramming of mitochondria in oxidative muscles towards a glycolytic phenotype
Next, we wanted to determine how the intrinsic differences in the proteome of glycolytic and oxidative muscles would reflect the capacity to utilize different substrates for energy production. High-resolution respirometry was performed on isolated mitochondria from glycolytic and oxidative muscles under an array of energy substrates at rest and at high energy demand states. Respiration rates on all substrates tested, i.e., pyruvate, medium-chain octanoylcarnitine, long-chain palmitoylcarnitine, and succinate, measured along with the ETC Complex I inhibitor rotenone (Rot) to capture complex II metabolism independent of complex I, were significantly lower in oxidative muscle compared to glycolytic of control mice at rest conditions (Figure 7A). These data suggest that mitochondria from oxidative muscle are intrinsically built to cover low energy demands with less respiratory effort than mitochondria from glycolytic muscle, which could be related to the lower amount of ATP required to produce tension in slow-twitch, oxidative myofibers [38]. Comparison between respiratory rates across different substrates revealed that succinate oxidative capacity was highest in both glycolytic and oxidative muscle mitochondria at resting conditions (Figure 7A).
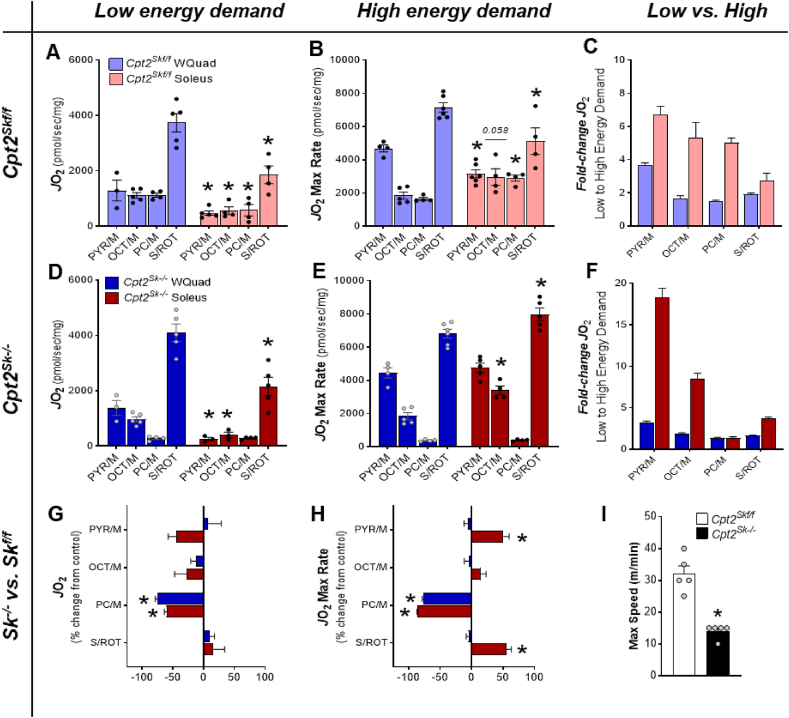
Figure 7.
Bioenergetic adaptations in Cpt2Sk−/−oxidative muscles result in a glycolytic metabolic phenotype. (A) Maximum rates of substrate-supported oxygen consumption (JO2) in isolated mitochondria from control glycolytic and oxidative muscles under low energy demands; PC = palmitoylcarnitine, OCT = octanoylcarnitine, PYR = pyruvate, S = succinate, M = malate, and ROT = rotenone. (B) Same as A but under high energy demands. (C) Oxygen consumption fold-change from low to high energy demands in control glycolytic and oxidative muscle mitochondria. (D) Same as A for mitochondria isolated from Cpt2Sk−/− glycolytic and oxidative muscles. (E) Same as D under high energy demands. (F) Same as C for Cpt2Sk−/− mitochondria. (G) Comparison of mitochondrial oxygen consumption rates at low energy demands between genotypes for glycolytic and oxidative muscles presented as Cpt2Sk−/− percentage change from control. (H) Same as G under high energy demands. (I) Max speed reached during a high-intensity treadmill-based exercise test. All data are presented as Mean ± SEM. For (A) to (F), n = 3–6 and ∗P ≤ 0.05 by T-test between glycolytic and oxidative muscle mitochondria for each substrate within the same genotype. For (G) and (H), n = 3–6 and ∗P ≤ 0.05 by T-test between control and Cpt2Sk−/− muscle mitochondria for each substrate within the same energy demand. For (I) n = 5 and ∗P ≤ 0.05 by T-test between genotypes.
It has been proposed that transitioning from rest state to contraction can increase ATP consumption in muscle by up to ≈100 fold [1]. Thus, mitochondria were challenged with increased energy demands by keeping phosphocreatine levels low to in turn decrease ATP/ADP ratio to drive mitochondrial ATP synthesis. Oxygen consumption (JO2) in this condition was considered the maximum mitochondrial respiration rate (JO2 Max Rate). In mitochondria from glycolytic control muscle, the maximum respiration rates were highest when energized with succinate, followed by pyruvate and the medium- and long-chain acylcarnitines, which elicited respiratory rates 74% and 77% lower than succinate, respectively (Figure 7B). In mitochondria from oxidative control muscle, succinate-supported respiration also provided the highest maximum rate (Figure 7B), yet respiration on pyruvate and on acylcarnitines were equal and only 30% lower than that for succinate (Figure 7B). Taken together, these data suggest that glycolytic muscles are primed to utilize succinate and pyruvate to support mitochondrial respiration, while medium- and long-chain fatty acids contribute to a lesser extent to energy production. Although succinate was also the major contributor to max respiration in oxidative muscles, acylcarnitines elicit remarkably higher respiration rates, ∼1.6-fold higher, compared to glycolytic mitochondria. On the contrary, pyruvate-supported respiration in oxidative muscle was 30% lower than in glycolytic control muscle (Figure 7B). Interestingly, in oxidative but not in control glycolytic muscles, oxygen consumption rates on pyruvate, octanoylcarnitine, and palmitoylcarnitine under high energy demand were similar (Figure 7B). Comparison of mitochondrial respiratory rates from low to high energy demands with any given substrate demonstrated that oxidative control muscle mitochondria have a greater response to high energy demands compared to glycolytic muscle mitochondria as shown by the 5 to 7-fold increase in oxygen consumption when ATP free energy decreased (Figure 7C). Taken together, these data suggest that while glycolytic muscles are better equipped to utilize substrates more directly linked to carbohydrates and proteins, oxidative muscle mitochondria rely more on fatty acids as energetic substrates. These data also suggest a more robust capacity of oxidative muscles to respond to increasing energy demands.
Next, we questioned whether the glycolytic-like proteome shift would better position the Cpt2Sk−/− oxidative muscle to utilize non-fatty acid fuel sources. At low and high energy demands, mitochondria from glycolytic control, and Cpt2Sk−/− muscle had a similar substrate-supported respiration pattern, suggesting that loss of CPT2 in glycolytic muscles did not impact glycolytic mitochondrial respiration (Figure 7D,G). However, significant differences between genotypes were demonstrated in oxidative muscle mitochondria, particularly in response to high energy demands. Specifically, Cpt2Sk−/− oxidative muscle mitochondria, compared to controls, had higher maximum respiration rates for pyruvate and succinate by 50% and 80%, respectively (Figure 7E,H). This change in pyruvate utilization represented an 18-fold increase in oxygen consumption going from low to high energy demands in the Cpt2Sk−/−, compared to a 3-fold increase in control oxidative soleus mitochondria (Figure 7F). In agreement with the proteomics data, Cpt2Sk−/− mitochondria from oxidative muscle had nearly identical max respiration rates to mitochondria from glycolytic muscle for pyruvate and succinate (Figure 7E). Taken together these data suggest that in Cpt2Sk−/− mice, unlike in control, the mitochondria from oxidative and glycolytic muscles have a high degree of metabolic similarity. Specifically, in the absence of mLCFAO, oxidative muscle mitochondria increased reliance on pyruvate and succinate as energetic substrates to levels comparable with glycolytic muscle. Despite these adaptations, CPT2-deficient mice present with exercise intolerance as demonstrated by poor treadmill performance (Figure 7I). Thus, while the metabolic adaptations made by Cpt2Sk−/− muscles may be sufficient to support low intensity physical activities such as cage exploration, feeding, and grooming, they are inadequate to support mid-high intensity exercise in the absence of mLCFAO.
Taken together, these data suggest that when substrate-supported max respiratory capacity (JO2) and sensitivity are analyzed as a whole, the overall metabolic profile of Cpt2Sk−/− mitochondria isolated from oxidative muscle reflects that of glycolytic mitochondria. Thus, impairments in fatty acid oxidation will shift energy metabolism without changing myosin heavy chain structure, resulting in a metabolic-contractile mismatch. Ultimately, these data suggest that shifts in substrate-supported metabolism are not sufficient to drive corresponding shifts in the contractile apparatus.
4. Discussion
The net energy production upon oxidation of palmitate, a common and abundant dietary fatty acid, is roughly 129 ATP equivalents, whereas complete glucose oxidation yields approximately 32 ATP equivalents. Thus, on a purely molecule-to-molecule comparison, fatty acids have far greater capacity than glucose to provide energy for cellular processes. The loss of mLCFAO capacity in tissues that are highly dependent on fatty acids as substrates, such as the heart and oxidative skeletal muscles, is bound to have significant metabolic consequences. As such, the loss of CPT2 in the heart drives the reversion of mature adult cardiomyocytes into their fetal genetic program [6], in which mLCFAO is low and glucose oxidation predominates. In a similar fashion, the loss of CPT2 in skeletal muscle might be expected to drive a shift in myofibers that would favor utilization of non-lipid substrates for energy production.
To understand the impact of mLCFAO deficiency in different muscles, we separately characterized glycolytic and oxidative muscles for whole-tissue proteome and mitochondrial metabolic profiles, and as a result, we identified significant distinctions between these muscle types. Notably, our dataset showed that among the whole-tissue proteome, oxidative muscle had relatively lower contribution of mitochondrial proteins compared to glycolytic. This was surprising because traditionally, oxidative muscles are described as red-colored tissue, heavily loaded with mitochondria, in contrast to white, glycolytic muscles which are perceived to be lower in mitochondrial content [39]. We report here that oxidative muscles had a more contraction-centered proteome, while in glycolytic muscles the most abundant proteins relative to whole-muscle were related to mitochondrial biogenesis and mitochondrial function. This agrees with glycolytic muscles producing high power contractions in a short period of time, which puts them at risk of achieving ATP consumption rates higher than regeneration rates [1]. Therefore, glycolytic muscles utilize a combination of mitochondrial metabolism and glycolysis to meet ATP demands [1]. Although our data does not discredit the mitochondrial abundance in oxidative muscles, it does question whether a mitocentric view of red muscles is appropriate.
Mitochondrial biogenesis is a natural response to energetic stress; thus, the robust mitochondrial biogenesis that took place in Cpt2Sk−/− muscles, particularly those of an oxidative nature, was likely compensatory to maintain functional capacity in an energetically compromised condition. Furthermore, we have shown here that upon CPT2 deletion there is a significant activation of the AMPK-PGC1a axis and its downstream effects in the skeletal muscle, suggesting some degree of parallelism between our Cpt2Sk−/− model and endurance training. However, the increased mitochondrial content in the face of preserved proportion of slow-twitch myofibers in Cpt2Sk−/− mice did not translate into improved muscle performance in the absence of mLCFAO. Muscle histology and lipid imaging revealed accumulation of mitochondria and spatial distribution of LCAC, and that was most pronounced in oxidative muscle fibers and exacerbated by loss of CPT2. It is possible that the sheer overload of LCACs and mitochondrial reticulum in Cpt2Sk−/− oxidative fibers could interfere with normal muscle function and structure. Mechanisms governing the response of the contractile apparatus in relation to the degree of mitochondrial content have not been extensively investigated. In addition to greater mitochondrial content in Cpt2Sk−/− muscles, further adaptive processes occurred including increased mitochondrial oxidative capacity for non-fatty acid substrates. Particularly, CPT2 deletion in oxidative muscles forced a shift in metabolism towards a glycolytic phenotype demonstrated by a higher contribution of mitochondria to whole-muscle proteome and a substrate-supported respiration that favored carbohydrates and amino acids for energy production to levels comparable with glycolytic muscles. This oxidative-to-glycolytic metabolic shift was also evident in the distinctive OXPHOS complex assembly pattern on native gels.
Despite the metabolic adaptation of oxidative muscle towards a glycolytic phenotype in Cpt2Sk−/− mice, we did not observe a matched conversion of MyHC isoforms, suggesting that mLCFAO deficiency due to CPT2 deletion is not sufficient to induce a concerted metabolic and contractile fiber type switching. This is contrary to what has been reported when mLCFAO is increased in skeletal muscle by overexpressing CPT1 which results in glycolytic muscles increasing reliance on fatty acids as fuel along with a matched switch towards oxidative MyHC isoforms [40]. Furthermore, numerous physiological and pathological conditions have been shown to result in adaptations of both metabolic and contractile facets in a seemingly synchronous manner. Specifically, exercise training, the overexpression or loss of PGC1alpha or PPARdelta, and defects in mitochondrial DNA are all reported to alter levels of oxidative metabolism in concert with changes in MyHC isoforms (41). Consequently, MyHCs are considered the best marker to characterize fiber types, with the assumption that metabolism is matched with MyHC isoforms to provide optimal function (42). There are conditions where metabolism and contractile units are not changed in concert; specifically, variables in exercise training modalities (43), atrophy (44), high-fat diet feeding (44), and genetic predispositions that impact mitochondrial metabolism such as those presented here result in mismatches between metabolism-contractile coupling.
A direct consequence of CPT2 deletion in skeletal muscle is the significant accumulation of long-chain acylcarnitines (LCACs) as we have reported [5]. There is speculation that LCAC accumulation interferes with the muscle bioenergetics by eliciting direct mitochondrial dysfunction, independent of mtFAO-derived energetic deficit [[31], [32], [33]]. However, in the case of Cpt2Sk−/− mice, long-chain acylcarnitine accumulation did not result in discernible defects in mitochondrial metabolism. Indeed, the mitochondrial respiration rates for non-fatty acid substrates were increased in the Cpt2Sk−/− oxidative muscle, suggesting that LCACs may not directly impair muscle mitochondrial metabolic capacity. Considering the germline deletion nature of the Cre model used here, it is possible that some of these effects are the result of developmentally chronic adaptations and that acute reductions in mLCFAO with acute fluctuations in LCACs could result in different outcomes. One additional potential caveat is in the fact that Cre-expressing non-floxed mice were not evaluated in the current study.
In summary, we report here clear distinctions in the mitochondrial metabolic profile across a range of substrates and energy demands between oxidative and glycolytic muscles that condition the response to mLCFAO deficiency. While mitochondrial function itself did not appear to be overtly impaired upon CPT2 deletion, the metabolic rates, proteome, and OXPHOS complex assembly of Cpt2Sk−/− oxidative soleus muscle matched tightly to the ones of glycolytic muscle and strikingly mismatched with those of control soleus muscle. These changes in energy metabolism were not accompanied with the corresponding MyHC-type switch, confirming that standard fiber-type assessment by MyHCs alone could mask unappreciated differential metabolic underpinnings. The consequences of a mismatch between energy metabolism and MyHC isoform expression for muscle physiology remain to be explored.
Author contribution statement
Andrea S. Pereyra: Conceptualization, Methodology, Investigation, Analysis, Writing-Original Draft, Visualization.
Chien-Te Lin: Investigation, Analysis.
Daniela Mesa Sanchez: Investigation, Visualization.
Julia Laskin: Investigation, Visualization, Resources.
Espen E. Spangenburg: Resources, Methodology.
P. Darrell Neufer: Resources, Writing-Review & Editing.
Kelsey Fisher-Wellman: Analysis, Methodology, Resources, Writing-Review & Editing.
Jessica M. Ellis: Conceptualization, Methodology, Investigation, Analysis, Writing-Original Draft, Visualization, Resources, Supervision, Funding acquisition.
Acknowledgements
We thank Dr. Michael J. Wolfgang for generously gifting the CPT2 floxed mice. We thank Mia Ferry and Arvind Rajan for technical assistance with histology and image analysis. We thank Dr. Christina Ferreira (Bindley Bioscience Center, Purdue University, IN, USA) for lipidomics and metabolomics technical assistance. This work was supported by the U.S. Public Health Services grants PDN-R01DK110656 and R01AG069679; EES-R61AR078100; and JME-R01DK125812.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101456.
Contributor Information
Andrea S. Pereyra, Email: pereyraa18@ecu.edu.
Jessica M. Ellis, Email: ellisje18@ecu.edu.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Multimedia component 1
References
- 1.Blaauw B., Schiaffino S., Reggiani C. Mechanisms modulating skeletal muscle phenotype. Comprehensive Physiology. 2013;3(4):1645–1687. doi: 10.1002/cphy.c130009. [DOI] [PubMed] [Google Scholar]
- 2.Schiaffino S., Reggiani C. Fiber types in mammalian skeletal muscles. Physiological Reviews. 2011;91(4):1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 3.Venhoff N., Lebrecht D., Pfeifer D., Venhoff A.C., Bissé E., Kirschner J., et al. Muscle-fiber transdifferentiation in an experimental model of respiratory chain myopathy. Arthritis Research and Therapy. 2012;14(5):R233. doi: 10.1186/ar4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereyra AS, Harris KL, Soepriatna AH, Waterbury QA, Bharathi SS, Zhang Y, et al. Octanoate is differentially metabolized in liver and muscle and fails to rescue cardiomyopathy in CPT2 deficiency. The Journal of Lipid Research. 2021:100069. [DOI] [PMC free article] [PubMed]
- 5.Pereyra A.S., Rajan A., Ferreira C.R., Ellis J.M. Loss of muscle carnitine palmitoyltransferase 2 prevents diet-induced obesity and insulin resistance despite long-chain acylcarnitine accumulation. Cell Reports. 2020;33(6):108374. doi: 10.1016/j.celrep.2020.108374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pereyra A.S., Hasek L.Y., Harris K.L., Berman A.G., Damen F.W., Goergen C.J., et al. Loss of cardiac carnitine palmitoyltransferase 2 results in rapamycin-resistant, acetylation-independent hypertrophy. Journal of Biological Chemistry. 2017;292(45):18443–18456. doi: 10.1074/jbc.M117.800839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloemberg D., Quadrilatero J. Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0035273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hulland T.J. Selected histochemical and histopathological methods. Canadian Veterinary Journal. 1967;8(2):51. [Google Scholar]
- 9.Sheehan D.C.H.B.B. Battelle Press; Columbus, Ohio: 1987. Theory and practice of histotechnology. [Google Scholar]
- 10.McLaughlin K.L., Hagen J.T., Coalson H.S., Nelson M.A.M., Kew K.A., Wooten A.R., et al. Novel approach to quantify mitochondrial content and intrinsic bioenergetic efficiency across organs. Scientific Reports. 2020;10(1):17599. doi: 10.1038/s41598-020-74718-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher-Wellman K.H., Davidson M.T., Narowski T.M., Lin C.-T., Koves T.R., Muoio D.M. Mitochondrial diagnostics: a multiplexed assay platform for comprehensive assessment of mitochondrial energy fluxes. Cell Reports. 2018;24(13):3593–3606. doi: 10.1016/j.celrep.2018.08.091. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg E.J., Buddo K.A., McLaughlin K.L., Fernandez R.F., Pereyra A.S., Psaltis C.E., et al. Tissue-specific characterization of mitochondrial branched-chain keto acid oxidation using a multiplexed assay platform. Biochemical Journal. 2019;476(10):1521–1537. doi: 10.1042/BCJ20190182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 14.Franco J., Ferreira C., Paschoal Sobreira T.J., Sundberg J.P., HogenEsch H. Profiling of epidermal lipids in a mouse model of dermatitis: identification of potential biomarkers. PLoS One. 2018;13(4) doi: 10.1371/journal.pone.0196595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Lima C.B., Ferreira C.R., Milazzotto M.P., Sobreira T.J.P., Vireque A.A., Cooks R.G. Comprehensive lipid profiling of early stage oocytes and embryos by MRM profiling. Journal of Mass Spectrometry. 2018;53(12):1247–1252. doi: 10.1002/jms.4301. [DOI] [PubMed] [Google Scholar]
- 16.Yin R., Burnum-Johnson K.E., Sun X., Dey S.K., Laskin J. High spatial resolution imaging of biological tissues using nanospray desorption electrospray ionization mass spectrometry. Nature Protocols. 2019;14(12):3445–3470. doi: 10.1038/s41596-019-0237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malik A.N., Czajka A., Cunningham P. Accurate quantification of mouse mitochondrial DNA without co-amplification of nuclear mitochondrial insertion sequences. Mitochondrion. 2016;29:59–64. doi: 10.1016/j.mito.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Leuthner T.C., Hartman J.H., Ryde I.T., Meyer J.N. In: Mitochondrial regulation: methods and protocols. Palmeira C.M., Rolo A.P., editors. Springer US; New York, NY: 2021. pp. 91–111. [Google Scholar]
- 20.Nelson M.A., McLaughlin K.L., Hagen J.T., Coalson H.S., Schmidt C., Kassai M., et al. Intrinsic OXPHOS limitations underlie cellular bioenergetics in leukemia. Elife. 2021;10 doi: 10.7554/eLife.63104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rath S., Sharma R., Gupta R., Ast T., Chan C., Durham T.J., et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Research. 2021;49(D1):D1541. doi: 10.1093/nar/gkaa1011. d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stenton S.L., Prokisch H. Genetics of mitochondrial diseases: identifying mutations to help diagnosis. EBioMedicine. 2020;56:102784. doi: 10.1016/j.ebiom.2020.102784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murayama K., Shimura M., Liu Z., Okazaki Y., Ohtake A. Recent topics: the diagnosis, molecular genesis, and treatment of mitochondrial diseases. Journal of Human Genetics. 2019;64(2):113–125. doi: 10.1038/s10038-018-0528-6. [DOI] [PubMed] [Google Scholar]
- 24.Lesack K., Naugler C. An open-source software program for performing Bonferroni and related corrections for multiple comparisons. Journal of Pathology Informatics. 2011;2:52. doi: 10.4103/2153-3539.91130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korthauer K., Kimes P.K., Duvallet C., Reyes A., Subramanian A., Teng M., et al. A practical guide to methods controlling false discoveries in computational biology. Genome Biology. 2019;20(1):118. doi: 10.1186/s13059-019-1716-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bardou P., Mariette J., Escudié F., Djemiel C., Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics. 2014;15(1):293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., et al. The reactome pathway knowledgebase. Nucleic Acids Research. 2019;48(D1):D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kammoun M., Cassar-Malek I., Meunier B., Picard B. A simplified immunohistochemical classification of skeletal muscle fibres in mouse. European Journal of Histochemistry. 2014;58(2):2254. doi: 10.4081/ejh.2014.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pande S., Blanchaer M. Carbohydrate and fat in energy metabolism of red and white muscle. American Journal of Physiology-Legacy Content. 1971;220(2):549–553. doi: 10.1152/ajplegacy.1971.220.2.549. [DOI] [PubMed] [Google Scholar]
- 30.Zong H., Ren J.M., Young L.H., Pypaert M., Mu J., Birnbaum M.J., et al. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(25):15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miura S., Kai Y., Kamei Y., Ezaki O. Isoform-specific increases in murine skeletal muscle peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) mRNA in response to beta2-adrenergic receptor activation and exercise. Endocrinology. 2008;149(9):4527–4533. doi: 10.1210/en.2008-0466. [DOI] [PubMed] [Google Scholar]
- 32.Chemello F., Bean C., Cancellara P., Laveder P., Reggiani C., Lanfranchi G. Microgenomic analysis in skeletal muscle: expression signatures of individual fast and slow myofibers. PLoS One. 2011;6(2) doi: 10.1371/journal.pone.0016807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiaffino S., Hanzlíková V., Pierobon S. Relations between structure and function in rat skeletal muscle fibers. The Journal of Cell Biology. 1970;47(1):107–119. doi: 10.1083/jcb.47.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bottinelli R., Canepari M., Reggiani C., Stienen G.J. Myofibrillar ATPase activity during isometric contraction and isomyosin composition in rat single skinned muscle fibres. The Journal of Physiology. 1994;481(3):663–675. doi: 10.1113/jphysiol.1994.sp020472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvarado Rigault M.Y., Blanchaer M.C. Respiration and oxidative phosphorylation by mitochondria of red and white skeletal muscle. Canadian Journal of Biochemistry. 1970;48(1):27–32. doi: 10.1139/o70-005. [DOI] [PubMed] [Google Scholar]
- 36.Hénique C., Mansouri A., Vavrova E., Lenoir V., Ferry A., Esnous C., et al. Increasing mitochondrial muscle fatty acid oxidation induces skeletal muscle remodeling toward an oxidative phenotype. The FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology. 2015;29(6):2473–2483. doi: 10.1096/fj.14-257717. [DOI] [PubMed] [Google Scholar]
- 37.Zhang L., Zhou Y., Wu W., Hou L., Chen H., Zuo B., et al. Skeletal muscle-specific overexpression of PGC-1α induces fiber-type conversion through enhanced mitochondrial respiration and fatty acid oxidation in mice and pigs. International Journal of Biological Sciences. 2017;13(9):1152–1162. doi: 10.7150/ijbs.20132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kallabis S., Abraham L., Müller S., Dzialas V., Türk C., Wiederstein J.L., et al. High-throughput proteomics fiber typing (ProFiT) for comprehensive characterization of single skeletal muscle fibers. Skeletal Muscle. 2020;10(1):7. doi: 10.1186/s13395-020-00226-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pengam M., Amérand A., Simon B., Guernec A., Inizan M., Moisan C. How do exercise training variables stimulate processes related to mitochondrial biogenesis in slow and fast trout muscle fibres? Experimental Physiology. 2021;106(4):938–957. doi: 10.1113/EP089231. [DOI] [PubMed] [Google Scholar]
- 40.Lang F., Khaghani S., Türk C., Wiederstein J.L., Hölper S., Piller T., et al. Single muscle fiber proteomics reveals distinct protein changes in slow and fast fibers during muscle atrophy. Journal of Proteome Research. 2018;17(10):3333–3347. doi: 10.1021/acs.jproteome.8b00093. [DOI] [PubMed] [Google Scholar]
- 41.Edwards M.E., Marasco C.A., Schock T.B., Sobreira T.J.P., Ferreira C.R., Cooks R.G. Exploratory analysis using MRM profiling mass spectrometry of a candidate metabolomics sample for testing system suitability. International Journal of Mass Spectrometry. 2021;468:17. 116663, In press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1