Abstract
Dopaminergic neurons are critical to movement, mood, addiction, and stress. Current techniques for generating dopaminergic neurons from human induced pluripotent stem cells (hiPSCs) yield heterogenous cell populations with variable purity and inconsistent reproducibility between donors, hiPSC clones, and experiments. Here, we report the rapid (5 weeks) and efficient (~90%) induction of induced dopaminergic neurons (iDANs) through transient overexpression of lineage-promoting transcription factors combined with stringent selection across five donors. We observe maturation-dependent increase in dopamine synthesis and electrophysiological properties consistent with midbrain dopaminergic neuron identity, such as slow-rising after hyperpolarization potentials, an action potential duration of ~3ms, tonic sub-threshold oscillatory activity, and spontaneous burst firing at a frequency of ~1.0-1.75 Hz. Transcriptome analysis reveals robust expression of genes involved in fetal midbrain dopaminergic neuron identity. Specifically expressed genes in iDANs, as well as those from isogenic induced GABAergic and glutamatergic neurons, were enriched in loci conferring heritability for cannabis use disorder, schizophrenia, and bipolar disorder; however, each neuronal subtype demonstrated subtype-specific heritability enrichments in biologically relevant pathways, and iDANs alone were uniquely enriched in autism spectrum disorder risk loci. Therefore, iDANs provide a critical tool for modeling midbrain dopaminergic neuron development and dysfunction in psychiatric disease.
INTRODUCTION
Dopaminergic neurotransmission regulates human behavior, motivation, affect, and cognition (Schultz, 2007). Dysfunction of dopaminergic neurons is importantly involved in the pathogenesis of neurological and psychiatric disorders such as Parkinson disease (Meder, Herz, Rowe, Lehéricy, & Siebner, 2019), substance use disorders (Volkow, Wise, & Baler, 2017), and psychosis (Grace & Gomes, 2019).
Human induced pluripotent stem cell (hiPSC) models provide an approach to generate large numbers of disease-relevant cell types and investigate disease processes at the cellular and molecular level, thus enabling the functional characterization of pathways and risk factors through genetic, pharmacologic, and physiological manipulations not possible in the relevant in vivo tissues (LaMarca, Powell, Akbarian, & Brennand, 2018; Powell, O’Shea, Shannon, Akbarian, & Brennand, 2020). Current methods to generate dopaminergic neurons in vitro either recapitulate key aspects of neurodevelopment through sequential application of small molecules and growth factors (Kriks, Shim, Piao, Ganat, Wakeman, Xie, et al., 2011) or overexpress exogenous transcription factors known to induce dopaminergic neuron identity (Addis, Hsu, Wright, Dichter, Coulter, & Gearhart, 2011; Caiazzo, Dell’Anno, Dvoretskova, Lazarevic, Taverna, Leo, et al., 2011; Mahajani, Raina, Fokken, Kügler, & Bähr, 2019; Pfisterer, Kirkeby, Torper, Wood, Nelander, Dufour, et al., 2011; Theka, Caiazzo, Dvoretskova, Leo, Ungaro, Curreli, et al., 2013). A remaining limitation of current techniques is the high degree of variable reproducibility between hiPSC donor lines and investigator groups. The most widely utilized technique8 employs sequential additional of small molecules and protein factor combinations to the media based upon known developmental pathways; however, studies have reported highly inconsistent yields ranging from ~8% to >90% (Awad, Panicker, Deranieh, Srikanth, Brown, Voit, et al., 2017; Beevers, Lai, Collins, Booth, Zambon, Parkkinen, et al., 2017; Ishikawa, Imamura, Kondo, Koshiba, Hara, Ichinose, et al., 2016; Sheng, Filichia, Shick, Preston, Phillips, Cooperman, et al., 2016; Sundberg, Bogetofte, Lawson, Jansson, Smith, Astradsson, et al., 2013). Consequently, many differentiations result in heterogenous cell populations (Fernandes, Patikas, Foskolou, Field, Park, Byrne, et al., 2020; la Manno, Gyllborg, Codeluppi, Nishimura, Salto, Zeisel, et al., 2016) and also include non-dopaminergic cells that are poorly characterized and of unknown relevance to the model system.
Here, we report the reliable induction of dopaminergic neurons from hiPSCs by transient overexpression of ASCL1, LMX1B, and NURR1 (Caiazzo, Dell’Anno, Dvoretskova, Lazarevic, Taverna, Leo, et al., 2011) (ALN) combined with antibiotic selection via a single doxycycline-inducible lentiviral vector, achieving a median percent purity across five independent donors of 92%. These induced dopaminergic neurons (iDANs) express genes consistent with midbrain regional patterning and dopaminergic neuron identity, demonstrate maturation-dependent dopamine synthesis, and electrophysiological hallmarks of in vivo dopaminergic neuron activity. Transcriptomic analyses of iDANs and post-mortem midbrain tissues provide further evidence of a fetal midbrain dopaminergic neuron identity of iDANs. Finally, specifically expressed genes in iDANs and isogenic induced GABAergic and glutamatergic neurons uncovered enrichment in risk loci for several psychiatric disorders with evidence of neuronal subtype- and disorder-specific enrichments among biologically relevant pathways.
MATERIALS AND METHODS
Human Induced Pluripotent Stem Cell Culture:
All hiPSCs were derived by sendai viral OKSM reprogramming of dermal fibroblasts obtained from control donors in a previous cohort (Hoffman, Hartley, Flaherty, Ladran, Gochman, Ruderfer, et al., 2017). hiPSCs were maintained in StemFlex media (Gibco, #A3349401) supplemented with Antibiotic-Antimycotic (Gibco, #15240062) on Matrigel-coated (Corning, #354230) plates and passaged at 80-90% confluency with 0.5mM EDTA (Life Technologies, #15575-020) every four to seven days for a maximum of 10 passages; no hiPSCs were cultured beyond passage 30. Routine cytogenetic analysis at WiCell confirmed normal karyotype of all donor lines. Donor meta-data are included in Supplementary Table 1A.
Post-Mortem Samples and Tissue-Processing:
Post-mortem sample processing, dissection, and fluorescence-activated nuclei-sorting (FANS) have been described previously (Espeso-Gil, Halene, Bendl, Kassim, ben Hutta, Iskhakova, et al., 2020). In brief, substantia nigra pars compacta (SNpc) along with surrounding regions of the ventral tegmental area (VTA) were dissected from adult brains with a post-mortem interval of less than 24 hours. All donors were controls in an ongoing cohort study and did not have any known psychiatric illnesses. Donor meta-data are included in Supplementary Table 1B, and representative quality-control data are shown in Supplementary Figure 1.
TetO-ALN-PuroR Vector:
We cloned a puromycin-resistance gene (PuroR) into a publicly available (Addgene: 43918) lentivirus vector encoding tetO-ASCL1-LMX1B-NURR1 (“ALN”). PuroR was inserted at the 3’ end of NURR1 and separated by a 2A peptide sequence.
Lentivirus Production:
Third-generation lentiviruses for pUBIQ-rtTA (Addgene 20342), tetO-ASCL1-LMX1B-NURR1-PuroR, tetO-ASCL1-PuroR (Addgene 97329), tetO-DLX2-HygroR (Addgene 97330), and tetO-Ngn2-PuroR-GFP (Addgene 79823) were generated via polyethylenimine (PEI, Polysciences, #23966-2)-mediated transfection of human embryonic kidney 293T (HEK293T) cells using existing protocols(Tiscornia, Singer, & Verma, 2006).
Production of Induced Dopaminergic Neurons (iDANs):
hiPSCs were harvested via incubation in Accutase Cell Detachment Solution (Innovative Cell Technologies, #AT104), quenched with DMEM (Gibco, #11965092), and centrifuged at RT at 800g for five minutes. Cell pellets were gently resuspended in StemFlex (Gibco, #A334901) supplemented with 10μM ROCK Inhibitor (StemCell Technologies, #72307) and counted with a Countess machine from Thermo Fisher Scientific (#AMQAX1000); the proportion of living cells was estimated by exclusion of Trypan Blue Solution, 0.4% (Gibco, #15250061). The cell suspension was then diluted in StemFlex with ROCK inhibitor to a concentration of 1 x 106 cells/mL and mixed with 50μL aliquots of both tetO-ALN-PuroR and pUBIQ-rtTA viruses tittered at an estimated 1 x 107 IU/mL using a qPCR Lentivirus Titration Kit (Applied Biological Materials, #LV900). hiPSCs were plated on Matrigel-coated plates and incubated at 37°C with virus overnight. The following day, DIV1, the media was aspirated and replaced with Induction Media (see Supplementary Note 1) with 1.0μg/mL doxycycline (Sigma, #D9891). 1.0μg/mL puromycin (Sigma, #7255) was added the following day on DIV2. Media was changed on DIV3 if substantial cell death was present. Beginning on DIV5, media consisted of Induction Media with 1.0μg/mL doxycycline, 1.0μg/mL puromycin, and 2.0μM arabinosylcytosine (Sigma, #C6645) (“Ara-C”) added to inhibit the proliferation of non-neuronal cells. On DIV7, cells were dissociated and replated in Induction Media supplemented with 1.0μg/mL doxycycline, 1.0μg/mL puromycin, 2.0μM Ara-C, and 10μM ROCK inhibitor on plates double-coated with 0.1% polyethylenimine (PEI) and 80μg/mL Matrigel. The following day (DIV8), media was replaced with Induction Media supplemented with 1.0μg/mL doxycycline and 2.0μM Ara-C. Ara-C was continued until DIV9, and doxycycline until DIV14, at which time the media was switched to Neuron Media. Half media changes were made every other day until the time of harvest (e.g., DIV35 for RNA-sequencing). See Supplementary Note 1 for media recipes, a more detailed protocol, and trouble-shooting information.
Production of Induced GABAergic Neurons (iGANs):
iGANs were generated from two hiPSC donors (C-1 and C-2) via transduction with two separate doxycycline-inducible lentivirus vectors encoding tetO-ASCL1-PuroR and tetO-DLX2-HygroR according to Yang et al., 2017 (Yang, Chanda, Marro, Ng, Janas, Haag, et al., 2017), with slight modification; also see detailed method available in Protocol Exchange (Yang, Chanda, Südhof, & Wernig, 2017). In brief, hiPSCs were harvested in Accutase, dissociated into a single-cell solution, quenched in DMEM, pelleted via centrifugation for five minutes at 800g, and resuspended in StemFlex with 10μM ROCK Inhibitor. Volumetric equivalents of tetO-ASCL1-PuroR, tetO-DLX2-HygroR, and pUBIQ-rtTA were added to the suspension, mixed gently by inversion, dispensed onto Matrigel-coated plates, and incubated overnight at 37°C. The next day, media was changed to Induction Media (identical recipe to that used for iDAN generation) with 1.0μg/mL doxycycline (DIV1). 1.0μg/mL puromycin and 250μg/mL hygromycin (Thermo, #10687010) were added the next day (DIV2) and continued for four days. We included 4.0μM Ara-C in the media from DIV4-8. Cells were harvested, dissociated, and replated on 0.1% PEI and 80μg/mL Matrigel-coated plates around DIV5-7. Media was switched to Neuron Media on DIV14, and doxycycline was withdrawn at that time. Half media changes were performed every other day from DIV14 until the time of harvest at DIV42 for the samples used for RNA-sequencing library-generation. Importantly, these inhibitory neurons are reported to be predominantly SST+, with some double staining for CB+ and/or CR+, and little PV+ staining, thus indicating somewhat of a GABAergic neuron subtype-specific (Yang, Chanda, Marro, Ng, Janas, Haag, et al., 2017) identity for iGANs.
Production of Induced Glutamatergic Neurons (iGLUTs):
iGLUTs, which most closely resemble fetal cortical pyramidal neurons (Miskinyte Id, Hansen, Monni, Lam, Bengzon, Lindvall, et al., 2018), were generated via transduction of hiPSCs (donors C-1 and C-2) with tetO-NGN2-eGFP-PuroR (Ho, Hartley, TCW, Beaumont, Stafford, Slesinger, et al., 2016; Zhang, Pak, Han, Ahlenius, Zhang, Chanda, et al., 2013) using the same protocol steps used to produce iGANs, with the exception that cells were dissociated and replated on DIV3 or DIV4 and matured until DIV21, at which time they were harvested for RNA extraction and RNA-sequencing library-generation.
RNA Extraction, Purification, and Quantification:
Media was aspirated, and the cells were washed twice with PBS. Samples were lysed with TRIzol Reagent (Thermo, #15596026). RNA was extracted and purified using the Direct-zol RNA miniprep kit with in-column DNAse treatment (Zymo Research, #R2051). Purified RNA was eluted in UltraPure water and stored at −80°C until needed for rt-qPCR or RNA-sequencing library-preparation. RNA concentration was determined by running the samples on a Qubit 3 Fluorometer (Invitrogen, #Q33216) with the Qubit RNA HS Assay Kit (Thermo, #Q32852).
Reverse-Transcription Quantitative qPCR (rt-qPCR):
50ng of RNA per each sample was loaded into a 384-well plate and quantified using the Power SYBR Green RNA-to-Ct 1-Step Kit (Thermo, #4389986). Reverse transcription and quantitative PCR took place on a QuantStudio 5 Real-Time PCR System (Thermo, #28570). Forward and reverse primer sequences for each mRNA are provided in Supplementary Table 2. Relative transcript abundance levels were quantified using the ΔΔ-Ct method (Schmittgen & Livak, 2008), with normalization of RNA input to ACTB as a loading control. For each gene and timepoint shown in Figure 2, data from samples generated from multiple donors and batches were pooled to better capture any donor- and batch-related variance in the true expression value of the gene. For each gene, a one-way ANOVA with Tukey post-hoc testing was utilized to test for differences in expression level at each of the three time points (DIV0 (hiPSCs), DIV14, and DIV35) using the aov and TukeyHSD functions in R.
Figure 2:
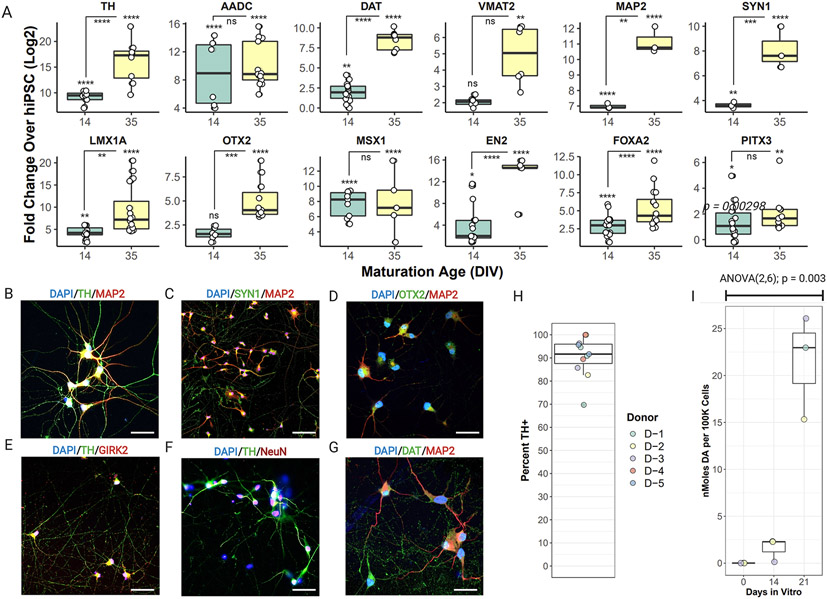
Marker gene expression, purity, and dopamine production in iDANs. (A) Fold-change above hiPSCs in the expression of TH, AADC, DAT, VMAT2, MAP2, SYN1, LMX1A, OTX2, MSX1, EN2, FOXA2, and PITX3. A maturation-dependent increase in expression was seen for the majority of genes. Confocal images of immunocytochemical staining of DIV35 iDANs with (B) TH and MAP2, (C) SYN1 and MAP2, (D) OTX2 and MAP2, (E) TH and GIRK2, (F) TH and NEUN, and (G) DAT and MAP2. (H) Across five donors with replicate experiments, a median of 92% of all cells are TH+. (I) Maturation-dependent increase in dopamine biosynthesis, with all three independent donors tested showing dopamine production by DIV21. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns = not significant. DIV = “days in vitro”.
Immunocytochemistry:
We adapted a protocol from one of our previous reports (Barretto, Zhang, Powell, Fernando, Zhang, Flaherty, et al., 2020). At DIV7, immature iDANs were split onto glass coverslips in a 24-well plate and matured until the desired timepoint. At the time of harvest, the media was aspirated from each well followed by two washes with PBS. 500μL of 4% para-formaldehyde solution (Electron Microscopy Sciences, #15170) in PBS was added to each well and incubated at room temperature for 10 minutes followed by three PBS washes. 500μL of a blocking solution consisting of PBS with 5% donkey serum (Jackson, #017-000-121) and 0.1% Triton X-100 (Sigma, #T8787) was added to the wells and incubated for one hour at RT. After two PBS washes, diluted primary antibodies were added in 5% donkey serum and 0.1% Tween-20 (Boston BioProducts, #IBB-181X) in PBS and incubated overnight at 4°C. The solution was then removed, and the wells were washed three times with PBS, followed by addition of secondary antibodies diluted in PBS and incubation in the dark for two hours. Finally, the secondary antibody solution was removed, the wells were washed three times with PBS, and 500μL of 0.5μg/mL DAPI (Sigma, #D9542) in PBS was added to the wells for a ten-minute incubation to stain cellular nuclei. Coverslips were carefully transferred to glass slides (Fisher Scientific, #12-544-7) and fixated using AquaPolymount (Polysciences Inc., #18606-20). See Supplementary Table 3 for primary and second antibody information. The percentage of cells positive for TH was determined by manual counting of all DAPI+ nuclei that also stained TH+ in random views of confocal images (containing at least 15 cells) taken across two or more separate experiments among five independent donors.
Post-Mortem Brain Sample Preparation and Fluorescence-Activated Nuclei-Sorting:
Frozen, but not fixed, tissue samples were homogenized in ice-cold lysis buffer and the resulting homogenate was mixed with a sucrose solution and ultra-centrifuged for one hour. The pellet was then resuspended and incubated with primary antibodies targeting NeuN (EMD Millipore, MAB377X; pre-conjugated with Alexa 488) and Nurr1 (Santa Cruz Biotechnology, sc-990), the latter of which had been incubated with Alexa Fluor 647 fluorochrome (Thermo Fisher, A27040) for one hour prior. After incubating in primary antibodies for two hours and DAPI (4′,6-diamidine-2′-phenylindole dihydrochloride, Sigma Aldrich) for the last ten minutes, the nuclei suspension was processed on a FACSAria flow cytometry sorter.
Nuclear RNA-Sequencing of Post-Mortem Midbrain Samples:
Nuclear RNA-sequencing libraries from NeuN+/Nurr1+ and NeuN−/Nurr1− nuclei were prepared as described previously (Espeso-Gil, Halene, Bendl, Kassim, ben Hutta, Iskhakova, et al., 2020). In summary, nuclei were lysed in TRIzol LS Reagent (Thermo Fisher, #10296028) and mixed with an equal volume of 100% ethanol. DNA-digestion and RNA-extraction were performed using the Zymo-Spin IC Column from the Direct-zol RNA MicroPrep kit (Zymo Research, R2060) per the manufacturer’s instructions. The yield and quality of the resulting RNA samples were assessed with the Agilent Bioanalyzer using an Agilent RNA 6000 Pico kit (Agilent, #5067-1513). Ribosomal rRNA-depleted RNA-sequencing libraries were prepared using the SMARTer Stranded RNA-Seq kit (Clontech, #634836) according to the manufacturer’s instructions with the following specifications: (a) RNA was fragmented at 94°C for three minutes; (b) after index-annealing with the Illumina indexing primer set (Illumina, #20020492), 12 PCR cycles were used for cDNA-amplification. Libraries were subsequently purified using a 1:1 volumetric ratio of SPRI beads (Beckman Coulter Life Sciences, #B23318) to remove primer dimers and enrich for a target library size of ~300bp, which was confirmed on the Agilent Bioanalyzer.
Whole-Cell RNA-Sequencing Library-Preparation and Sequencing of In Vitro Samples:
Strand-specific, rRNA-depleted RNA-sequencing libraries were prepared from 100-1,000ng RNA per sample using the KAPA RNA HyperPrep Kit with RiboErase (HMR) (Roche, #KK8560). RNA-fragmentation was performed at 94°C for six minutes and 10 PCR cycles were used during library amplification with TruSeq single-index adapters (Illumina, #20020492). Final library concentrations were quantified with both Qubit fluorometric quantification (DNA dsDNA HS kit, Thermo, #Q32851) and the KAPA Library Quantification kit (Kapa Biosystems, #KK4873). The samples were run on an Agilent 2100 Bioanalyzer (Agilent #G2939BA) with the High Sensitivity DNA Kit (Agilent, #5067-4626) to confirm the appropriate distribution of fragment sizes and the absence of significant artifactual contaminants. 150 base-pair paired-end sequencing was performed on a NovaSeq 6000 system to a desired depth of ~50,000,000 reads per library.
RNA-Sequencing Snalysis:
Library-Processing: Raw data were aligned to the genome (GRCh38) using STAR v2.7.0 (Dobin, Davis, Schlesinger, Drenkow, Zaleski, Jha, et al., 2013). Counts per gene were extracted using the featureCounts (Liao, Smyth, & Shi, 2014) function in the Rsubread package (Liao, Smyth, & Shi, 2019). Sample count matrices were processed in R version 4.0.2 using the limma (Ritchie, Phipson, Wu, Hu, Law, Shi, et al., 2015) and edgeR (McCarthy, Chen, & Smyth, 2012; Robinson, McCarthy, & Smyth, 2009) packages. For the dataset consisting of iDANs, hiPSCs, midbrain NeuN+/Nurr1+ nuclei (midbrain dopaminergic neurons, or “MDNs”), and midbrain NeuN−/Nurr1− nuclei (“non-MDNs”), lowly expressed genes were filtered out using the filterByExpr function in edgeR, leading to a reduction in the total number of ensemble gene IDs from 58,037 to 16,641. Library log2(CPM) distributions were normalized with the trimmed mean of M-values method (Robinson & Oshlack, 2010). Heteroscedasticity was removed from the data using the voom function of limma, and linear models were fit for cell type contrasts of interest with weights generated using empirical Bayes moderation (Smyth, 2004). The results of library-processing and quality controls are shown in Supplementary Figure 2.
Derivation of Differentially and Specifically Expressed Genes: “Differentially expressed genes (DEGs)” were defined as genes with a log2(fold change in CPM) of at least 1.0 and that surpassed a significance threshold of p < 0.05 after correcting for multiple testing with the Benjamini-Hochberg procedure in the decideTests function in limma. The in vitro RNA-sequencing dataset consisting of hiPSCs, iDANs, iGANs, and iGLUTs was processed identically to the dataset with post-mortem samples but with the following two changes: (1) we did not filter out lowly expressed genes in order to capture all genes with potential specificity for a given cell type, and (2) we derived “specifically expressed genes (SEGs)”, instead of DEGs, by first contrasting the expression of all genes in each of the four cell types to the expression levels in the other three; SEGs were defined as genes in the top 10th percentile of t-statistics for each cell type (Supplementary Table 4), as in previous reports (Agarwal, Sandor, Volpato, Caffrey, Monzón-Sandoval, Bowden, et al., 2020; Finucane, Reshef, Anttila, Slowikowski, Gusev, Byrnes, Conet al., 2018). Genes with the highest 10% of t-statistics were deemed “specifically expressed,” with the caveat that there is indeed some degree of overlap in SEGs; accordingly, specifically expressed genes are not exclusively expressed in one cell type alone.
Gene-Set Overrepresentation Analyses (GSOA): We used clusterProfiler (Yu, Wang, Han, & He, 2012) to perform GSOA on (i) DEGs in the dataset consisting of hiPSCs, iDANs, MDNs, and non-MDNs, and (ii) SEGs with enrichment in psychiatric disorder heritability from the in vitro dataset. Tested gene sets included KEGG pathways (Kanehisa, Furumichi, Tanabe, Sato, & Morishima, 2017) and Gene Ontology Biological Processes (Ashburner, Ball, Blake, Botstein, Butler, Cherry, et al., 2000; Carbon, Douglass, Good, Unni, Harris, Mungall, et al., 2021) (BP). We used an FDR threshold of q < 0.05, and significance values are indicated throughout the text by the corresponding FDR q values. Significant results were visualized with tile plots, with the magnitude of the −log(FDR q value) represented by the height of the tile. Enrichment maps showing relationships among pathways/processes based upon overlapping genes were created using the emapplot function in clusterProfiler after reducing the degree of redundancy in terms using semantic similarity analysis (Yu, 2020) with GOSemSim (Yu, Li, Qin, Bo, Wu, & Wang, 2010). Gene-concept network plots for the top enriched pathways were created with the cnetplot function of clusterProfiler to create gene networks.
Competitive Gene-Set Testing for Enrichment in Cell Type Identity SEGs: A competitive gene-set testing procedure was conducted using Correlation-Adjusted Mean Rank gene-set test (CAMERA) (Wu & Smyth, 2012) on hiPSCs, iDANs, and the post-mortem tissue type DEGs to test for enrichment among specifically expressed genes in previously published datasets for brain cell types. We defined these brain cell type SEGs as those genes with the top 1% of specificity in the K1 and K2 cell types reported by Skene et al. (2018) (Skene, Bryois, Bakken, Breen, Crowley, Gaspar, et al., 2018) and those reported by La Manno et al (2016) (la Manno, Gyllborg, Codeluppi, Nishimura, Salto, Zeisel, et al., 2016) on developing midbrain cell types (Supplementary Table 5, A-C). The full results are shown without a specific FDR threshold cutoff.
Correlation Analyses of SEG Enrichments in Psychiatric Risk Loci: We calculated the spearman correlation coefficients in gene-level z-scores for those neuronal subtype-specific SEG sets with enrichment in any psychiatric disorder. A correlation matrix was generated in R using the corr.test function of the psych package, with adjustment of p values with Bonferroni correction; results with an adjusted p value < 0.05 were considered significant.
Assessment of In Vitro Neuron Subtype Heritability Enrichment of Psychiatric Risk Loci:
We intersected cell-type-specific expression patterns with genetic risk of 11 specified psychiatric disorders (attention-deficit/hyperactivity disorder (Demontis, Walters, Martin, Mattheisen, Als, Agerbo, et al., 2019) (ADHD), anorexia nervosa (Duncan, Yilmaz, Gaspar, Walters, Goldstein, Anttila, et al., 2017) (AN), autism spectrum disorder (Grove, Ripke, Als, Mattheisen, Walters, Won, et al., 2019) (ASD), alcohol dependence (Walters, Polimanti, Johnson, McClintick, Adams, Adkins, et al., 2018) (AUD), bipolar disorder (Mullins, Forstner, O’Connell, Coombes, Coleman, Qiao, et al., 2020) (BIP), cannabis use disorder (Johnson, Demontis, Thorgeirsson, Walters, Polimanti, Hatoum, et al., 2020) (CUD), major depressive disorder (Howard, Adams, Clarke, Hafferty, Gibson, Shirali, et al., 2019) (MDD), obsessive-compulsive disorder (Arnold, Askland, Barlassina, Bellodi, Bienvenu, Black, et al., 2018) (OCD), post-traumatic stress disorder (Nievergelt, Maihofer, Klengel, Atkinson, Chen, Choi, et al., 2019) (PTSD), and schizophrenia (Ripke, Walters, & O’Donovan, 2020) (SCZ), as well as Cross Disorder (Cross-Disorder Group of the Psychiatric Genomics Consortium, Lee, Anttila, Won, Feng, Rosenthal, et al., 2019) ((CxD) GWAS summary statistics), along with Alzheimer disease (Marioni, Harris, Zhang, McRae, Hagenaars, Hill, et al., 2018) (AD) and Parkinson disease (Nalls, Blauwendraat, Vallerga, Heilbron, Bandres-Ciga, Chang, et al., 2019) (PD) to identify disorder-relevant cell types (Supplementary Table 6). We performed these cell-type association analyses using multi-marker analysis of genomic annotation (MAGMA) (de Leeuw, Mooij, Heskes, & Posthuma, 2015). Four gene sets were defined by the protein-coding genes present in curated lists of SEGs for hiPSCs, iDANs, iGANs, and iGLUTs. Using MAGMA, SNPs were mapped to genes based on the corresponding build files for each GWAS summary dataset using the default method, snp-wise=mean (a test of the mean SNP association). A competitive gene set analysis was then used to test enrichment in genetic risk for a disorder across the four cell-type-specific gene sets with an adjusted p-value threshold of < 0.05.
Multi-Electrode Array (MEA):
Commercially available human astrocytes (HA; Sciencell, #1800; isolated from fetal female brain) were thawed and seeded onto matrigel-coated 100mm culture dish in commercial astrocyte medium (Sciencell, #1801) and expanded three passages in Astrocyte medium. Upon confluency, cells were detached, pelleted, and resuspended with Astrocyte medium supplemented with Antibiotic-Antimycotic (Anti/Anti; Thermo Fisher Scientific, #15240062) and split as 1 x 105 cells per well on matrigel-coated 48-well CytoView MEA plates (Axion Biosystems). HAs were fed by full medium change with the Brainphys medium (2% FBS + Anti/Anti) + 2.0μM Ara-C. At DIV7, iDANs were split on the HAs with neuron media supplemented with 2% FBS by gently detaching them with Accutase for one hour, centrifuging (1000g x 5 mins), and resuspending in Neuron Media supplemented with 2% FBS and 5μM ROCK Inhibitor. After counting cells with a Countess machine, iDANs were seeded on the astrocyte culture (1 x 105 cells/well). The media was changed the next day to neuronal medium supplemented with 0.5% FBS and 2μM Ara-C. Half media changes were performed twice a week, one day before MEA measurement. 2.0μM Ara-C treatment was discontinued after one week. MEA plates were measured twice a week on a Maestro Multi-Electron Array system (Axion Biosystems) at 37°C starting on DIV21 of iDAN differentiation. For each measurement, plates were equilibrated in the machine for five minutes followed by 10-minute recordings, with spontaneous neural real-time configuration at a threshold of 5.5. The plates were measured until week six of neuronal maturation followed by batch processing of files and analysis of compiled statistics.
Electrophysiology:
Neurons from two donors (C-1 and C-2) were plated on acid-etched coverslips and co-cultured with human fetal astrocytes in Neuron Media to promote maturation (Bardy, van den Hurk, Eames, Marchand, Hernandez, Kellogg, et al., 2015). Recordings were performed at five to six weeks after induction. Coverslips were transferred to a bath filled with modified aCSF solution, adapted from a mouse slice electrophysiology protocol (Rifkin, Moss, & Slesinger, 2017) containing NaCl 119mM, D-glucose 11mM, NaHCO3 26.2mM, KCl 2.5mM, MgCl2 1.3mM, NaH2PO4 1mM, CaCl2 2.5mM (pH adjusted to 7.3 with HCl). Glass microelectrodes of 4.0 – 4.6MΩ resistance were filled with an internal solution of 140mM Potassium D-Gluconate, 4mM NaCl, 2mM MgCl2 x 6-H2O, 1.1mM EGTA, 5mM HEPES, 2mM Na2ATP, 5mM NaCreatinePO4, and 0.6mM Na3GTP (chemicals obtained from Sigma-Aldrich). All solutions were ~ 295mOsm. Whole-cell currents were recorded with an Axopatch 200B amplifier with application of manual series-resistance and capacitance compensation, filtered at 10kHz for current-clamp and 1kHz for voltage-clamp, and digitized at 20kHz and 10kHz, respectively, with the 1550 Digidata digitizer (Molecular Devices). For current-clamp recordings, a holding current was applied to set the resting potential to −65mV and 1s current steps were applied in 0.02nA increments. Spontaneous activity was measured in I=0 current-clamp mode. For voltage-clamp recordings, voltage steps (200ms) were applied in 10mV increments from a holding voltage of −80mV to 50mV. All voltage measurements were corrected for a calculated junction potential of −16.1mV. Data were collected and analyzed using Molecular Devices pClamp 11 software and with custom-made routines written in R.
Dopamine ELISA:
For whole-cell dopamine ELISA, cells were harvested with Accutase and spun down for five minutes at room temperate at 1000g. The media supernatants were completely aspirated, and the cell pellets were flash frozen in liquid nitrogen. The ELISA was carried out using the Dopamine Research ELISA Kit from ALPCO (#17-DOPHU-E01-RES) according to the manufacturer’s instructions, beginning with cell lysis in homogenization solution in a Dounce homogenizer. Each sample was split into three technical triplicates. Absorbance at 450nm was measured on a Varioskan LUX multielectrode microplate reader (Thermo, #VLBL00D0). A non-parametric local regression curve was fit to the values of the standards using the loess function in R, with the absorbance as the predictor variable and log2-transformed concentration (nM) as the response variable. Concentrations of the samples were extrapolated from the sample absorbances using the predict function in R using the regression model fitted to the standards.
RESULTS
Transient overexpression of ASCL1, LMX1B, and NURR1 induces dopaminergic neurons
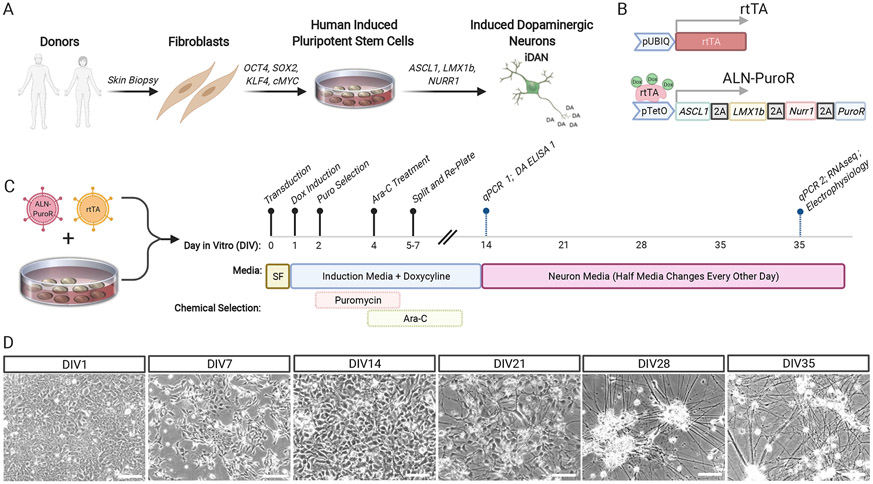
Previous reports demonstrated that overexpression of ASCL1, LMX1A, and NURR1 (also known as NR4A2) in human fibroblasts (Caiazzo, Dell’Anno, Dvoretskova, Lazarevic, Taverna, Leo, et al., 2011) and hiPSCs (Theka, Caiazzo, Dvoretskova, Leo, Ungaro, Curreli, et al., 2013) could result in dopaminergic neurons with low yields limited to ~5% and ~33% purity, respectively. We designed an improved vector for induction of dopaminergic neurons (iDANs) from hiPSCs (Fig. 1A) that incorporated antibiotic selection (tetO-ASCL1-LMX1B-NURR1-PuroR, “ALN-PuroR”) (Fig. 1B). Doxycycline was administered until 14 days in vitro (DIV) (Fig. 1C), while selection with puromycin occurred from DIV2 to DIV6, and Ara-C was added from ~DIV4-6 to eliminate residual mitotic cells (see Supplementary Note 1). Early neuronal processes appeared at DIV7, and iDANs were matured for 35 days, at which time they show extensive branching and lengthy processes (Fig. 1D).
Figure 1:
Production of induced dopaminergic neurons with ASCL1, LMX1B, and NURR1 transduction. (A) Schematic showing overall process of producing iDANs from hiPSCs. (B) Cartoon illustrating key features of the tetO-ALN-PuroR and rtTA vectors. (C) Timeline of iDAN generation, beginning with transduction of hiPSCs at DIV0 and ending with sample harvesting; “SF” stands for StemFlex media. (D) Weekly brightfield images showing progressive development of neuronal morphology in iDANs. White scale bars = 50 μm.
Across five independent donor lines, qPCR revealed that DIV14 iDANs showed increased expression of the neuronal genes MAP2 and SYN1, as well as TH, the rate-limiting enzyme in dopamine biosynthesis, and AADC, which converts L-DOPA to dopamine (Fig. 2A). They likewise showed robust expression of the midbrain marker genes LMX1A, MSX1, EN2, FOXA2, and PITX3, and a modest increase in expression of the dopamine transporter (DAT) and VMAT2 (Fig. 2A). By DIV35, TH, LMX1A, OTX2, EN2, DAT, and FOXA2 expression further increased (Fig. 2A). At the protein level, DIV35 iDANs co-expressed TH with MAP2 (Fig. 2B), SYN1 (Fig. 2C), NEUN (Fig. 2F), DAT (Fig. 2E), as well as the midbrain marker OTX2 (Fig. 2D) and GIRK2, an inwardly rectifying potassium channel that mediates D2R stimulation (Beckstead, Grandy, Wickman, & Williams, 2004) (Fig. 2E).
A median of ~92% of cells were positively stained for TH across five donors, with two or more replicate experiments per donor (Fig. 2H). Moreover, ELISA analysis of iDANs from three donors confirmed a maturation-dependent increase in total dopamine biosynthesis (ANOVA (2,6) = 20.78; p = 0.0030); by DIV21, dopamine production across the three donors was significantly elevated relative to DIV0 (p = 0.0017) and DIV14 (p = 0.018) (Fig. 2I). Altogether, transduction with ALN-PuroR leads to robust induction of >90% dopaminergic neurons (range: 70-98%), showing widespread dopaminergic marker gene expression and dopamine biosynthesis in a maturation-dependent manner.
iDANs show physiological hallmarks of in vivo dopaminergic neurons
Across two independent donors, multi-electrode array recordings revealed increasing burst frequency (Hz), weighted mean firing rate (Hz), network burst frequency (Hz), and the fraction of active electrodes with bursting activity across maturation (Supplementary Figure 3). In contrast, burst duration, network spike duration, and spikes per network burst remained steady across maturation, in agreement with previous MEA analyses of developing primary rodent cortical neurons in vitro (Cotterill, Hall, Wallace, Mundy, Eglen, & Shafer, 2016).
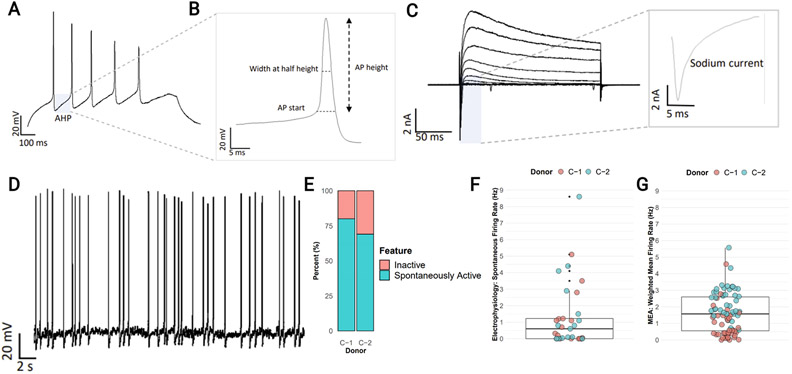
With the same two donors, we examined the intrinsic excitability of iDANs using patch-clamp electrophysiology. After approximately five weeks of induction, iDANs exhibited regenerative action potentials in response to current injections (Fig. 3A), with a notable slow after-hyperpolarization potential (AHP) (Fig. 3B) typical of dopaminergic neurons (Nedergaard, 2004). The action potential width of 3ms was similar to that reported for primate/rodent DA neurons (Nedergaard, 1999). We also observed prominent voltage-gated sodium and potassium currents but not Ih inward currents (Fig. 3C). The cell capacitance for iDANs (20 ± 8pF) was smaller than in rodents (Bean, 2007). iDANs exhibited spontaneous activity at resting membrane potentials (Fig. 3E), with some showing continuous tonic-like firing (Fig. 3D). We compared the distributions of spontaneous firing for both patch-clamp and MEA assays and observed a median frequency of about 1.0 – 1.75Hz (Fig. 3F, G). The basic neuronal properties (e.g., capacitance, resting potential) and firing behavior were indistinguishable between the two donor lines, highlighting the replicability of this induction method across individuals (Supplementary Table 7). Altogether, iDANs displayed many of the electrophysiological hallmarks of their in vivo midbrain dopaminergic neuron counterparts.
Figure 3:
Electrophysiological characterization of iDANs. (A) A representative voltage trace shows evoked action potentials with current injection step (.02 pA). Highlighted area (grey box) illustrates the slow after-hyperpolarization potential (AHP). (B) Enlarged view of a representative iDAN action potential, illustrating threshold, height, and duration (width at half-height) measurements. (C) Representative traces of voltage-gated potassium and sodium currents evoked by voltage steps from −80 mV to 50 mV. Inset shows inward sodium current at −50 mV. (D) Example of spontaneous tonic firing at resting potential (Vm = −53 mV). (E) Proportion of spontaneously active neurons by donor (N=18 C-1, N=15 C-2). (F,G) Comparison of spontaneous firing rates measured by whole-cell patch-clamp with a mean frequency of 1.28 Hz (F, cells (n)= 18,15 for C-1 and C-2, respectively) and by multielectrode array (MEA), with a median weighted mean firing rate (WMFR) of 1.64 (G, wells (n) = 39, 50).
iDANs exhibit a fetal midbrain dopaminergic neuron transcriptomic profile
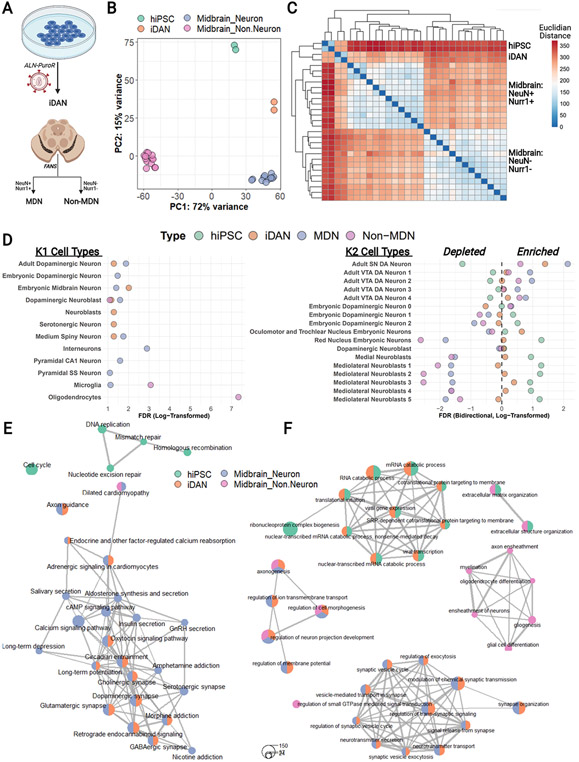
To benchmark iDANs to a reference in vivo dataset, we conducted an RNA-sequencing analysis of neurons and non-neuronal cells sorted from post-mortem midbrain (Fig. 4A), comparing them to midbrain dopaminergic neurons (NeuN+/Nurr1+ nuclei, “MDNs”) and midbrain non-dopaminergic neurons (NeuN−/Nurr1− nuclei, “Non-MDNs”). Principal component analysis of total gene expression across all samples revealed distinct clustering by cell type, with iDANs aligning with MDNs on PC1, which accounted for 72% of the total variance (Fig. 4B). Hierarchical clustering separated hiPSCs and iDANs from the post-mortem samples but also demonstrated greater relatedness of iDANs to MDNs than non-MDNs (Fig. 4C).
Figure 4:
Transcriptomic analysis of iDANs. (A) Schematic showing the generation iDANs and post-mortem samples with fluorescence-activated nuclear sorting from human midbrain. (B) Principal component analysis on PC1 and PC2 shows strong clustering of samples by cell type, with iDANs aligning with post-mortem midbrain dopamine neurons (MDNs) on PC1. (C) Hierarchical clustering of RNA-seq samples by Euclidean distances between transcriptomic profiles (all 16, 641 genes), revealing sample relatedness by cell type. (D) Results of competitive gene set testing for enrichment in specifically expressed genes from the K1 (left) and K2 (right) cell types from Skene et al., 2018 (Skene, Bryois, Bakken, Breen, Crowley, Gaspar, et al., 2018). Left: for the K1 results, hiPSC data points are omitted for graphical purposes, and only enrichments in the positive direction are shown; full results are contained in Supplementary Figure 3. Right: selected results for dopaminergic lineage cell types from the K2 datasets, with enrichments in the “down” direction represented by log-transformed FDR q values multiplied by −1. The full results including all 149 K2 cell types are shown in Supplementary Figure 4. (E) Enrichment maps depicting top KEGG pathways enriched among cell-type-specific gene expression. Individual pathways are shown as circular “nodes”, with the node color indicating cell type and node size representing the number of genes within the pathway node overlapping the specifically expressed genes for that cell type. Pathway nodes are connected by edges to form networks based upon overlapping genes. Spaces between pathway networks and free-standing nodes are not biologically meaningful, as individual networks were arranged for graphical purposes. (F) Same as in E, but with nodes representing Gene Ontology Biological Processes instead of KEGG pathways.
We generated DEGs from our in vitro and in vivo cell types and performed a competitive gene set testing procedure to explore potential enrichments in established brain cell-type-specific marker genes (Fig. 4D, Supplementary Figure 4). Among a group of 24 cell-type-specific marker gene lists (Skene, Bryois, Bakken, Breen, Crowley, Gaspar, et al., 2018) (Supplementary Table 5A), MDNs showed strong enrichment in the positive direction for several cell types including “Interneuron” (FDR q = 1.27 x 10−3) and “Adult Dopaminergic Neuron” (FDR q = 0.0133). While the magnitude of iDAN enrichment in “Adult Dopaminergic Neuron” (FDR q = 0.0516) was somewhat lower than that observed for MDNs, iDANs were most highly enriched in “Embryonic Midbrain Neurons” (FDR q = 9.69 x 10−3). In contrast, the non-MDNs showed high enrichment in the gene sets specific to “Oligodendrocytes” (FDR q = 4.68 x 10−8) and “Microglia” (FDR q = 8.42 x 10−4). We then expanded the analysis to a more refined set of 149 specific cell types (Skene, Bryois, Bakken, Breen, Crowley, Gaspar, et al., 2018) (Supplementary Table 5B), each of which belong to one of the 24 broader cell type classifications in the first dataset (Supplementary Figure 5). Among the cell types belonging to the dopaminergic neuron lineage, both MDNs (FDR q = 7.38 x 10−3) and iDANs (FDR q = 0.0398) were most enriched in “Adult Substantia Nigra Neurons”, with iDANs showing additional positive enrichment in early developmental cell types. With a third dataset (la Manno, Gyllborg, Codeluppi, Nishimura, Salto, Zeisel, et al., 2016), this time derived from early developmental midbrain cell types (Supplementary Table 5C), competitive gene set testing confirmed that iDANs were most strongly enriched in specifically expressed genes defining early dopaminergic neurons and midbrain progenitor cells (Supplementary Figure 6).
Finally, we conducted gene set overrepresentation analyses (GSOA) to evaluate the biological relevance of cell-type-specific gene expression. Broadly, enriched terms were consistent with the known identities and functions of the respective cell type (Supplementary Table 8). While hiPSC gene expression was enriched in KEGG pathways (Kanehisa, Furumichi, Tanabe, Sato, & Morishima, 2017) involved in the cell cycle (e.g., “Cell Cycle”, q = 4.08 x 10−10), both MDNs and iDANs were enriched in pathways with clear links to dopaminergic neuron biology, such as “Dopaminergic Synapse” (q = 0.000163 in iDANs; q = 7.08 x 10−11 in MDNs), “Long-Term Potentiation” (q = 0.030 in iDANs; q = 2.16 x 10−6 in MDNs) and “Morphine Addiction” (q = 0.010 in iDANs; q = 3.41 x 10−10 in MDNs) (Fig. 4E). These findings were corroborated by additional enrichment in a network of Gene Ontology Biological Processes (Ashburner, Ball, Blake, Botstein, Butler, Cherry, et al., 2000; Carbon, Douglass, Good, Unni, Harris, Mungall, et al., 2021) related to synaptic structure (e.g., “Synapse Organization” (q = 1.30 x 10−21 in iDANs; q = 3.54 x 10−21 in MDNs)) and neurotransmission (e.g., “Dopamine Secretion” (q = 3.16 x 10−6 in iDANs; q = 0.0060 in MDNs)), whereas non-MDNs were enriched in processes pertaining to glial cells (e.g., “Glial Cell Differentiation”, q = 1.49 x 10−8; “Myelination”, q = 1.71 x 10−6) (Fig 4F). In total, these results support a fetal-like midbrain dopaminergic neuron identity for iDANs.
Differential enrichment of induced dopaminergic, GABAergic, and glutamatergic neurons in psychiatric risk genes
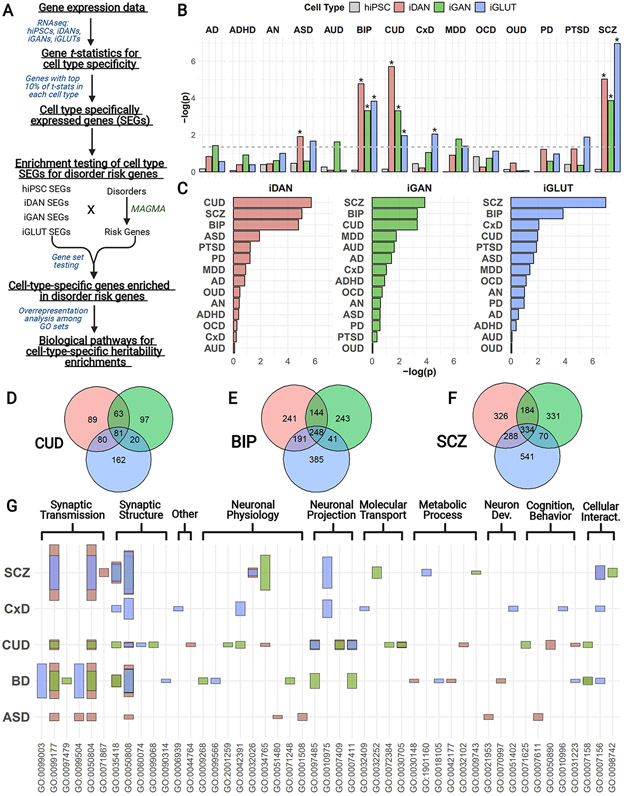
The observation that GWAS-based risk variants for genetically complex disease capture commonly occurring polymorphisms (mean allele frequency > 0.01) (Wray, Lee, Mehta, Vinkhuyzen, Dudbridge, & Middeldorp, 2014) implies that molecular studies of risk factor mechanisms conducted on tissues from non-diseased donors are relevant to the disease biology of the trait under study (Huckins, Dobbyn, Ruderfer, Hoffman, Wang, Pardiñas, et al., 2019; Schrode, Ho, Yamamuro, Dobbyn, Huckins, Matos, et al., 2019; Wray, Lee, Mehta, Vinkhuyzen, Dudbridge, & Middeldorp, 2014; Wray, Wijmenga, Sullivan, Yang, & Visscher, 2018). For this reason, we next sought to test whether hiPSC-derived neuronal subtype-specific gene expression was enriched in psychiatric disease risk loci. After generating isogenic iDANs, iGANs, and iGLUTs and calculating those SEGs most specifically expressed in each induced neuronal subtype, we applied (MAGMA) (de Leeuw, Mooij, Heskes, & Posthuma, 2015) to test for the enrichment of in vitro cell type SEGs among an array of psychiatric disorders (Arnold, Askland, Barlassina, Bellodi, Bienvenu, Black, et al., 2018; Demontis, Walters, Martin, Mattheisen, Als, Agerbo, et al., 2019; Duncan, Yilmaz, Gaspar, Walters, Goldstein, Anttila, et al., 2017; Grove, Ripke, Als, Mattheisen, Walters, Won, et al., 2019; Howard, Adams, Clarke, Hafferty, Gibson, Shirali, et al., 2019; Johnson, Demontis, Thorgeirsson, Walters, Polimanti, Hatoum, et al., 2020; Mullins, Forstner, O’Connell, Coombes, Coleman, Qiao, et al., 2020; Nievergelt, Maihofer, Klengel, Atkinson, Chen, Choi, et al., 2019; Polimanti, Walters, Johnson, McClintick, Adkins, Adkins, et al., 2020; Ripke, Walters, & O’Donovan, 2020; Walters, Polimanti, Johnson, McClintick, Adams, Adkins, et al., 2018) (Fig. 5A), as well as a set of pleiotropic loci implicated in a cross-disorder (CxD) analysis of eight psychiatric conditions (Cross-Disorder Group of the Psychiatric Genomics Consortium, Lee, Anttila, Won, Feng, Rosenthal, et al., 2019). Alzheimer disease (Marioni, Harris, Zhang, McRae, Hagenaars, Hill, et al., 2018) (AD) and Parkinson disease (Nalls, Blauwendraat, Vallerga, Heilbron, Bandres-Ciga, Chang, et al., 2019) (PD) were included as brain-related but non-psychiatric disorders. SEGs for iDANs, iGANs, and iGLUTs were significantly enriched in risk genes for cannabis use disorder (CUD; iDAN: p = 1.94 x 10−6; iGAN: p = 4.89 x 10−4; iGLUT: p = 0.0109), bipolar disorder (BIP; iDAN: p = 1.68 x 10−5, iGAN: p = 4.82 x 10−4; iGLUT: p = 1.48 x 10−4), and schizophrenia (SCZ; iDAN: p = 9.32 x 10−6, iGAN: p = 1.37 x 10−4; iGLUT: p = 1.10 x 10−7), with iDANs showing additional enrichment in autism spectrum disorder (ASD; p = 0.0122) and iGLUTs showing enrichment in the cross-disorder pleiotropic loci (CxD; p = 0.0090) (Fig 5B). As expected, hiPSCs were not enriched in any condition (Supplementary Table 10A). Overall, iDANs were most enriched in CUD, while both iGANs and iGLUTs were most enriched in SCZ (Fig. 5C), consistent with emerging data that glutamatergic and GABAergic neurons are particularly impacted by SCZ genetic risk loci (de Jonge, Vinkers, Hulshoff Pol, Marsman, et al., 2017; Hauberg, Creus-Muncunill, Bendl, Kozlenkov, Zeng, Corwin, et al., 2020; Ragland, Maddock, Hurtado, Tanase, Lesh, Niendam, et al., 2019; Skene, Bryois, Bakken, Breen, Crowley, Gaspar, et al., 2018). Further supporting our findings of heritability enrichment for iDAN SEGs, we also found that SEGs from our post-mortem MDNs were similarly enriched in risk genes for BIP (p = 5.82 x 10−6), SCZ (p = 0.00074), and CUD (0.016), along with an additional enrichment in ADHD (p = 0.012) (Supplementary Figure 7).
Figure 5:
Neuronal subtype heritability enrichment for psychiatric disorders among hiPSC-derived neurons. (A) Analysis workflow for identification of biological pathways implicated in neuronal subtype-specific heritability enrichments in psychiatric disorders (Finucane, Reshef, Anttila, Slowikowski, Gusev, Byrnes, et al., 2018). (B) Gene set testing results utilizing MAGMA-derived risk genes and in vitro cell type SEGs for hiPSCs, iDANs, iGANs, and iGLUTs. Dashed line indicates nominal significance (p < 0.05), while * indicates those SEG sets that were significantly enriched after correction for multiple testing. Alzheimer disease = AD; attention-deficit/hyperactivity disorder = ADHD; anorexia nervosa = AN; autism spectrum disorder = ASD; alcohol dependence = AUD; bipolar disorder = BIP; cannabis use disorder = CUD; cross-disorder pleiotropic loci = CxD; major depressive disorder = MDD; obsessive-compulsive disorder = OCD; opioid use disorder = OUD; Parkinson disease = PD; post-traumatic stress disorder = PTSD; schizophrenia = SCZ. (C) Plot showing the results of MAGMA by each cell type group. Overlap among significantly enriched neuronal subtype-specific genes among (D) CUD, (E) BIP, and (F) SCZ. (G) Results from gene set overrepresentation analyses of significantly enriched gene sets and their involvement in biological processes. GO sets are grouped into meta categories; the set names corresponding to each GO ID number are listed in the full results in Supplementary Table 11. Bar colors indicate neuronal subtype, and bar height represents the magnitude −log(FDR q values).
For each disorder with enrichment among one or more hiPSC-derived neuronal subtypes, we queried the biological relevance of the significantly enriched genes (Supplementary Table 10B-F). Across all disorders and neuronal subtypes, we found consistent enrichment among biological processes related to synaptic transmission and structure (Fig. 5G and Supplementary Table 11A); however, there was also significant neuronal subtype specificity among enriched pathways within the individual disorders (Supplementary Table 11B-F). In SCZ, for example, only iDAN SEGs were overrepresented in processes such as “Monoamine Response” (q = 3.83 x 10−3) and “Response to Auditory Stimulus” (q = 1.41 x 10−3), while only iGAN SEGs were overrepresented in the term “Auditory Behavior” (q = 4.27 x 10−3), and iGLUT SEGs were uniquely overrepresented in the term “Cognition” (q = 5.13 x 10−9) (Supplementary Figure 8). Further illustration of differential enrichment of induced neuronal subtypes in BIP and CUD are shown in Supplementary Figures 9 and 10, respectively.
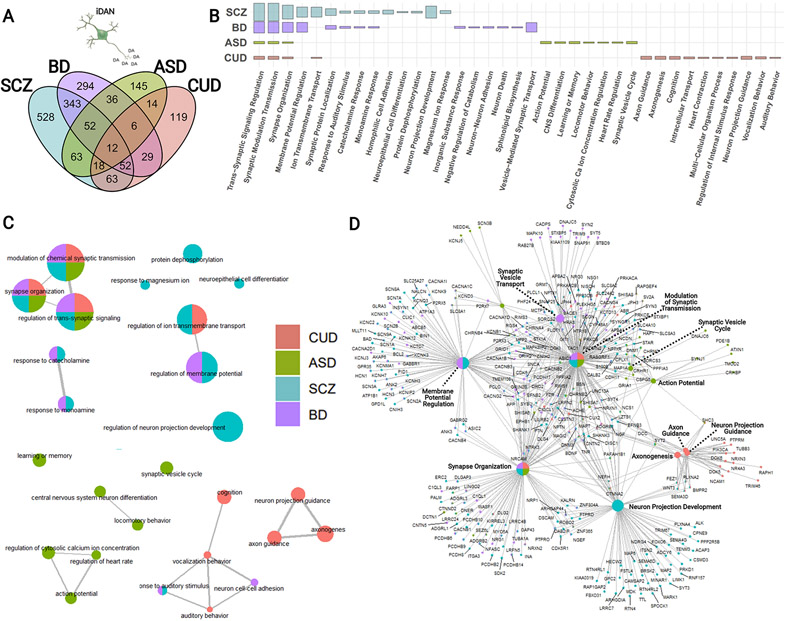
Having shown that unique subtypes of induced neurons are enriched in both shared and subtype-specific pathways within three major psychiatric disorders, we next assessed the extent to which gene expression specific to a given neuronal subtype is differentially enriched across disorders. Specifically expressed genes in iDANs demonstrated heritability enrichment in SCZ, BIP, CUD, and ASD (Fig. 6A). Intriguingly, iDAN SEGs enriched in ASD heritability, for example, were uniquely overrepresented in biological process such as “CNS Differentiation” (q = 7.31 x 10−3) and “Learning or Memory” (q = 1.31 x 10−3) (Fig. 6B), while those implicated in CUD risk alone formed a distinct pathway network related to neuron projection guidance (e.g., “Axon Guidance”, q = 8.08 x 10−5) (Fig. 6C, D). Significantly enriched iGAN genes, in contrast, were overrepresented in a neuron projection development network in both BIP and CUD (Supplementary Figure 11 and Supplementary Table 11H). The shared versus psychiatric disorder-specific pathway enrichments for iGLUTs are shown in Supplementary Figure 12, including a unique overrepresentation of iGLUT SEGs involved in “Synaptic Vesicle Cycle” in BIP (q = 7.25 x 10−15), for example. Furthermore, the biological processes implicated among iDAN SEGs enriched in ASD heritability and those implicated among iGLUT SEGs enriched in the cross-disorder loci are shown in Supplementary Figure 13 and Supplementary Figure 14, respectively, with the gene-level summary statistics listed in Supplementary Table 12. Overall, we observed neuronal cell-type-specific enrichment of risk variants for major psychiatric disorders among biological processes important for or unique to the respective neuronal subtype.
Figure 6:
Biological interpretation of iDAN-specific gene expression with differential enrichment in genetic loci associated with schizophrenia (SCZ), bipolar disorder (BIP), autism spectrum disorder (ASD), and cannabis use disorder (CUD). (A) Venn diagram illustrating the overlap in significantly enriched iDAN SEGs among the four disorders. (B) Summary results from GSOA of enriched iDAN SEGs by disorder; height of the bar represents the magnitude of −log(FDR q values). (C) Enrichment map of top biological processes overrepresented among disorder-specific iDAN SEGs. Circle (node) colors indicate the disorder, and multiple colors within a node correspond to pathways shared among the indicated conditions. Node size represents the number of iDAN SEGs represented in the particular pathway. Pathway nodes are connected by edges to other nodes with shared genes to form pathway networks. The spatial arrangement between networks and free-standing nodes is not biologically meaningful and was adjusted for graphical purposes. (D) Gene-concept network plot depicting relationships between significantly enriched genes as a function of node membership among the top enriched pathways in iDANs. As in C, node size indicates the number of iDAN SEGs found within the GO set; colors of nodes and individual gene dots represent disorder involvement.
Finally, we investigated the extent to which correlation in genetic risk between psychiatric disorders was reflected in neuronal subtype-specific enrichment in psychiatric heritability. Broadly, disorder, rather than cell type, was the strongest driver of gene-level correlations in psychiatric heritability enrichments (Supplementary Figure 15; full results in Supplementary Table 13A,B). Correlations between neuronal subtypes within each disorder were generally the lowest between iGANs and iGLUTs; for example, in CUD, the coefficient of correlation between iGANs and iGLUTs was r = 0.33 (p = 5.16 x 10−152), but r = 0.45 (p = 1.20 x 10−306) between iGANs and iDANs and r = 0.46 (p = 4.40 x 10−313) between iDANs and iGLUTs. A similar pattern was observed for SCZ and BIP (Supplementary Figure 15). For a given neuronal subtype, however, correlations across disorders were higher between SCZ and BIP than for any other disorder pair: for iDANs, the correlation between SCZ and BIP was r = 0.24 (p = 2.76 x 10−81) but only r = 0.070 (p = 5.97 x 10−8) between SCZ and ASD and r = 0.071 (p = 3.61 x 10−8) between SCZ and CUD, for example. These results suggest that the shared genetic risk architecture between SCZ and BIP (Anttila, Bulik-Sullivan, Finucane, Walters, Bras, Duncan, et al., 2018; Lee, Ripke, Neale, Faraone, Purcell, Perlis, et al., 2013; Ruderfer, Ripke, McQuillin, Boocock, Stahl, Pavlides, et al., 2018) is reflected in the heritability enrichments obtained from hiPSC-derived neuronal subtypes.
DISCUSSION
Transient overexpression of just three transcription factors, ASCL1, LMX1B, and NURR1, yields homogenous populations of induced dopaminergic neurons. iDANs demonstrate a maturation-dependent increase in the expression of several marker genes of midbrain dopaminergic neuron identity and develop dopamine biosynthesis capabilities by DIV21. Relative to previous reports (Caiazzo, Dell’Anno, Dvoretskova, Lazarevic, Taverna, Leo, et al., 2011; Theka, Caiazzo, Dvoretskova, Leo, Ungaro, Curreli, et al., 2013), we demonstrate a substantial improvement in the yield and purity of iDANs. Moreover, we employed five independent donors to calculate percent purity, and note that the inter-donor variability (70-98%) was narrower than in a previous study that reported values ranging from 13-65% across four donors (Mahajani, Raina, Fokken, Kügler, & Bähr, 2019). Our approach, which combined all three transcription factors and the selection cassette into a single doxycycline-inducible vector, ensured that the expression levels of all three transcription factors did not vary considerably relative to one another, as opposed to other methods where individual vectors deliver each transgene separately.
Although DIV35 iDANs are spontaneously active and exhibit hallmark physiological properties of in vivo MDNs, we did not detect Ih currents. These hyperpolarization-activated inwardly rectifying currents are mediated by HCN channel activity (Chu & Zhen, 2010). We found that HCN1, HCN2, HCN3, and HCN4 were all expressed at high levels in iDANs (Supplementary Figure 16), and so it remains unclear why iDANs lack Ih currents. This likely reflects the immature nature of iDANs compared to adult MDNs, as Ih currents do not typically develop until the early post-natal period (Picken Bahrey & Moody, 2003) in rodents. iDAN cell capacitance, another measure of maturity, was 20(±8) pF, comparable to values recorded in iGLUT neurons (22 ±1 pF)(Rosa, Dhingra, Uysal, Mendis, Loeffler, Elsen, et al., 2020) and human second trimester neurons (range of 18.5 ±2.5 pF to 24.8 ±3.5 pF) (Moore, Filipovic, Mo, Rasband, Zecevic, & Antic, 2009). Likewise, competitive gene set testing also indicated global gene expression patterns consistent with an early neurodevelopmental, midbrain dopaminergic neuron phenotype. This is unsurprising, given previous reports that other types of hiPSCs-derived neurons most closely resemble human fetal neurons (Brennand, Savas, Kim, Tran, Simone, Hashimoto-Torii, et al., 2015; Nehme, Zuccaro, Ghosh, Li, Sherwood, Pietilainen, et al., 2018), specifically those at 16-24 post-conception weeks (Rosa, Dhingra, Uysal, Mendis, Loeffler, Elsen, et al., 2020). This likely makes iDANs more suitable for studies of mechanisms related to psychiatric disease risk and onset, rather than phenotypes observed in mature, adult neurons that occur with late-stage disease.
Isogenic neuronal populations are uniquely suited for CRISPR-based functional genomic studies of subtype specific mechanisms across psychiatric disorders (Powell, Gregory, Akbarian, Brennand, 2017). While CUD, BIP, and SCZ risk loci were enriched for unique subsets of specifically expressed genes in iDANs, iGANs, and iGLUTs, ASD was only enriched in iDAN SEGs. Critically, our MAGMA-based enrichment analyses conducted with post-mortem DA neurons recapitulated our CUD, BIP, and SCZ results obtained for iDANs (Supplementary Figure 7). Prior studies of cell-type-specific heritability enrichments in schizophrenia have yielded similar, but not identical, results. An analysis employing single-cell RNA-sequencing of numerous brain cell types found that specifically expressed genes in cortical interneurons, pyramidal neurons, and medium spiny neurons, but not dopaminergic neurons or their progenitors, were enriched in schizophrenia heritability (Skene, Bryois, Bakken, Breen, Crowley, Gaspar, et al., 2018); importantly, our iGANs and iGLUTs most closely resemble fetal cortical inhibitory (Yang, Chanda, Marro, Ng, Janas, Haag, et al., 2017) and cortical pyramidal neurons (Miskinyte Id, Hansen, Monni, Lam, Bengzon, Lindvall, et al., 2018), respectively. The discrepancy in dopaminergic associations may reflect gene expression differences between the dopaminergic cell types profiled in Skene et al. (Skene, Bryois, Bakken, Breen, Crowley, Gaspar, et al., 2018) and the iDANs and MDNs in our study, different computational approaches for identifying disease heritability enrichments, and/or different RNA-sequencing methods (single-cell RNA-sequencing versus bulk RNA-sequencing). To our knowledge, similar cell-type-specific gene expression studies of heritability enrichment for bipolar disorder and cannabis use disorder have not been conducted to date; nevertheless, the current literature strongly supports a mechanistic role for glutamatergic, GABAergic, and dopaminergic neurons in both cannabis use disorder (Connor, Stjepanović, le Foll, Hoch, Budney, & Hall, 2021; Johnson, Demontis, Thorgeirsson, Walters, Polimanti, Hatoum, et al., 2020; Pasman, Verweij, Gerring, Stringer, Sanchez-Roige, Treur, et al., 2018) and bipolar disorder (Ashok, Marques, Jauhar, Nour, Goodwin, Young, et al., 2017; Cherlyn, Woon, Liu, Ong, Tsai, & Sim, 2010; Kim, Santos, Gage, & Marchetto, 2017; Stahl, Breen, Forstner, McQuillin, Ripke, Trubetskoy, et al., 2019). Altogether, the concordant results obtained from our hiPSC-based studies and existing data on cell types implicated in psychiatric risk add additional evidence that hiPSC-derived neurons enable modeling of true disease processes identified from studies of human in vivo tissues.
Genes enriched in psychiatric disease heritability in all three neuronal subtypes were overrepresented among biological pathways involved in synaptic structure and neurotransmission, implicating broad disruption of these processes in psychiatric disease. These results are highly consistent with prior studies (Ripke, Walters, & O’Donovan, 2020) identifying biologically relevant pathways in these conditions, which further supports the utility of hiPSC-derived neurons for modeling bona fide disease mechanisms. Here, different neuronal subtypes captured different aspects of disease biology; furthermore, within a given neuronal subtype, unique pathways were implicated in different disorders. Thus, we posit that each induced neuronal subtype captures both shared and distinct aspects of heritability enrichment that correspond to specific biological pathways that drive disease risk across psychiatric disorders. Consistent with the shared genetic architecture of schizophrenia and bipolar disorder (Anttila, Bulik-Sullivan, Finucane, Walters, Bras, Duncan, et al., 2018; Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ruderfer, Ripke, McQuillin, Boocock, Stahl, et al., 2018; Lee, Ripke, Neale, Faraone, Purcell, Perlis, et al., 2013; Ruderfer, Ripke, McQuillin, Boocock, Stahl, Pavlides, et al., 2018), cross-disorder correlations were far greater between SCZ and BIP then between any other pair of conditions, although this effect may be somewhat inflated by overlapping control groups in SCZ and BIP GWA studies (Mullins, Forstner, O’Connell, Coombes, Coleman, Qiao, et al., 2020; Ripke, Walters, & O’Donovan, 2020; Stahl, Breen, Forstner, McQuillin, Ripke, Trubetskoy, et al., 2019). We also note that GWA studies of SCZ, BIP, and CUD are among the better-powered of those included in this study (Supplementary Table 6), and future research expanding sample sizes for the other disorders (e.g., obsessive-compulsive disorder, opioid use disorder, anorexia nervosa) may facilitate discoveries about enrichment of loci in cell-type-specific gene expression. Moving forward, it will be critical to evaluate the functional effects of risk loci on gene expression and activity in specific subtypes of neurons in order to understand the mechanisms by which (epi)genetic variation adversely impacts brain phenotypes (Powell, O’Shea, Brennand, Akbariab, 2021).
Supplementary Material
Acknowledgements:
This research was supported by R01MH106056, U01DA047880, R01DA048279, 6R56MH101454. Figures in this manuscript were created with Biorender.com. The authors wish to thank Rachel Oren for helpful feedback on an earlier version of this manuscript, Dana Infante for assistance in sample collection, and Dr. Stefano Marenco, Dr. Barbara Lipska, and Dr. Pavan Auluck and their staff in the Human Brain Collection Core at the National Institutes of Health for providing postmortem brain tissues.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors report that they have no conflicts of interest to disclose.
Code availability: Available from the authors upon request.
DATA AVAILABILITY STATEMENT
The source data described in this manuscript are available via the PsychENCODE Knowledge Portal (https://psychencode.synapse.org/). The PsychENCODE Knowledge Portal is a platform for accessing data, analyses, and tools generated through grants funded by the National Institute of Mental Health (NIMH) PsychENCODE program. Data is available for general research use according to the following requirements for data access and data attribution: (https://psychencode.synapse.org/DataAccess). For access to content described in this manuscript see: http://doi.org/10.7303/syn25500352
REFERENCES
- Addis RC, Hsu FC, Wright RL, Dichter MA, Coulter DA, & Gearhart JD (2011). Efficient conversion of astrocytes to functional midbrain dopaminergic neurons using a single polycistronic vector. PLoS ONE. 10.1371/journal.pone.0028719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal D, Sandor C, Volpato V, Caffrey TM, Monzón-Sandoval J, Bowden R, et al. (2020). A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nature Communications. 10.1038/s41467-020-17876-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, Duncan L, et al. (2018). Analysis of shared heritability in common disorders of the brain. Science. 10.1126/science.aap8757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold PD, Askland KD, Barlassina C, Bellodi L, Bienvenu OJ, Black D, et al. (2018). Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Molecular Psychiatry. 10.1038/mp.2017.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry, et al. (2000). Gene ontology: Tool for the unification of biology. In Nature Genetics. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok AH, Marques TR, Jauhar S, Nour MM, Goodwin GM, Young AH, et al. (2017). The dopamine hypothesis of bipolar affective disorder: The state of the art and implications for treatment. In Molecular Psychiatry (Vol. 22, Issue 5, pp. 666–679). Nature Publishing Group. 10.1038/mp.2017.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad O, Panicker LM, Deranieh RM, Srikanth MP, Brown RA, Voit A, et al. (2017). Altered Differentiation Potential of Gaucher’s Disease iPSC Neuronal Progenitors due to Wnt/β-Catenin Downregulation. Stem Cell Reports. 10.1016/j.stemcr.2017.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy C, van den Hurk M, Eames T, Marchand C, Hernandez R. v., Kellogg M, et al. (2015). Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proceedings of the National Academy of Sciences of the United States of America. 10.1073/pnas.1504393112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N, Zhang H, Powell SK, Fernando MB, Zhang S, et al. (2020). ASCL1-and DLX2-induced GABAergic neurons from hiPSC-derived NPCs. 10.1016/j.jneumeth.2019.108548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean BP (2007). The action potential in mammalian central neurons. In Nature Reviews Neuroscience. 10.1038/nrn2148 [DOI] [PubMed] [Google Scholar]
- Beckstead MJ, Grandy DK, Wickman K, & Williams JT (2004). Vesicular dopamine release elicits an inhibitory postsynaptic current in midbrain dopamine neurons. Neuron. 10.1016/j.neuron.2004.05.019 [DOI] [PubMed] [Google Scholar]
- Beevers JE, Lai MC, Collins E, Booth HDE, Zambon F, Parkkinen L, et al. (2017). MAPT Genetic Variation and Neuronal Maturity Alter Isoform Expression Affecting Axonal Transport in iPSC-Derived Dopamine Neurons. Stem Cell Reports. 10.1016/j.stemcr.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ruderfer DM, Ripke S, McQuillin A, Boocock J, Stahl EA, et al. (2018). Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell, 173(7), 1705–1715.e16. 10.1016/j.cell.2018.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, et al. (2015). Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Molecular Psychiatry. 10.1038/mp.2014.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, et al. (2011). Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. In Nature. 10.1038/nature10284 [DOI] [PubMed] [Google Scholar]
- Carbon S, Douglass E, Good BM, Unni DR, Harris NL, Mungall CJ, et al. (2021). The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Research. 10.1093/nar/gkaa1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherlyn SYT, Woon PS, Liu JJ, Ong WY, Tsai GC, & Sim K (2010). Genetic association studies of glutamate, GABA and related genes in schizophrenia and bipolar disorder: A decade of advance. In Neuroscience and Biobehavioral Reviews (Vol. 34, Issue 6, pp. 958–977). Pergamon. 10.1016/j.neubiorev.2010.01.002 [DOI] [PubMed] [Google Scholar]
- Chu HY, & Zhen X (2010). Hyperpolarization-activated, cyclic nucleotide-gated (HCN) channels in the regulation of midbrain dopamine systems. In Acta Pharmacologica Sinica. 10.1038/aps.2010.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor JP, Stjepanović D, le Foll B, Hoch E, Budney AJ, & Hall WD (2021). Cannabis use and cannabis use disorder. In Nature Reviews Disease Primers (Vol. 7, Issue 1, pp. 1–24). Nature Research. 10.1038/s41572-021-00247-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium, Lee PH, Anttila V, Won H, Feng Y-CA, Rosenthal J, et al. (2019). Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell, 179(7), 1469–1482.e11. 10.1016/j.cell.2019.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotterill E, Hall D, Wallace K, Mundy WR, Eglen SJ, & Shafer TJ (2016). Characterization of early cortical neural network development in multiwell microelectrode array plates. Journal of Biomolecular Screening. 10.1177/1087057116640520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge JC, Vinkers CH, Hulshoff Pol HE, & Marsman A (2017). GABAergic mechanisms in schizophrenia: Linking postmortem and In vivo studies. In Frontiers in Psychiatry. 10.3389/fpsyt.2017.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, & Posthuma D (2015). MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Computational Biology. 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. (2019). Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nature Genetics. 10.1038/s41588-018-0269-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L, Yilmaz Z, Gaspar H, Walters R, Goldstein J, Anttila V, et al. (2017). Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. American Journal of Psychiatry. 10.1176/appi.ajp.2017.16121402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeso-Gil S, Halene T, Bendl J, Kassim B, ben Hutta G, Iskhakova M, et al. (2020). A chromosomal connectome for psychiatric and metabolic risk variants in adult dopaminergic neurons. Genome Medicine. 10.1186/s13073-020-0715-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes HJR, Patikas N, Foskolou S, Field SF, Park JE, Byrne ML, et al. (2020). Single-Cell Transcriptomics of Parkinson’s Disease Human In Vitro Models Reveals Dopamine Neuron-Specific Stress Responses. Cell Reports. 10.1016/j.celrep.2020.108263 [DOI] [PubMed] [Google Scholar]
- Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, et al. (2018). Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nature Genetics, 50(4), 621–629. 10.1038/s41588-018-0081-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace AA, & Gomes F. v. (2019). The Circuitry of Dopamine System Regulation and its Disruption in Schizophrenia: Insights Into Treatment and Prevention. Schizophrenia Bulletin, 45(1), 148–157. 10.1093/schbul/sbx199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. (2019). Identification of common genetic risk variants for autism spectrum disorder. Nature Genetics. 10.1038/s41588-019-0344-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauberg ME, Creus-Muncunill J, Bendl J, Kozlenkov A, Zeng B, Corwin C, et al. (2020). Common schizophrenia risk variants are enriched in open chromatin regions of human glutamatergic neurons. Nature Communications. 10.1038/s41467-020-19319-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S-M, Hartley BJ, TCW J, Beaumont M, Stafford K, Slesinger PA, et al. (2016). Rapid Ngn2-induction of excitatory neurons from hiPSC-derived neural progenitor cells HHS Public Access. Methods, 101, 113–124. 10.1016/j.ymeth.2015.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GE, Hartley BJ, Flaherty E, Ladran I, Gochman P, Ruderfer DM, et al. (2017). Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nature Communications, 8(1), 2225. 10.1038/s41467-017-02330-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, et al. (2019). Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nature Neuroscience. 10.1038/s41593-018-0326-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckins LM, Dobbyn A, Ruderfer DM, Hoffman G, Wang W, Pardiñas AF, et al. (2019). Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nature Genetics. 10.1038/s41588-019-0364-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Imamura K, Kondo T, Koshiba Y, Hara S, Ichinose H, et al. (2016). Genetic and pharmacological correction of aberrant dopamine synthesis using patient iPSCs with BH4 metabolism disorders. Human Molecular Genetics. 10.1093/hmg/ddw339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EC, Demontis D, Thorgeirsson TE, Walters RK, Polimanti R, Hatoum AS, et al. (2020). A large-scale genome-wide association study meta-analysis of cannabis use disorder. The Lancet Psychiatry. 10.1016/S2215-0366(20)30339-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, & Morishima K (2017). KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Santos R, Gage FH, & Marchetto MC (2017). Molecular mechanisms of bipolar disorder: Progress made and future challenges. In Frontiers in Cellular Neuroscience (Vol. 11). Frontiers Research Foundation. 10.3389/fncel.2017.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, et al. (2011). Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 10.1038/nature10648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, et al. (2016). Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell. 10.1016/j.cell.2016.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMarca EA, Powell SK, Akbarian S, & Brennand KJ (2018). Modeling neuropsychiatric and neurodegenerative diseases with induced pluripotent stem cells. Frontiers in Pediatrics, 6. 10.3389/fped.2018.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Alhamdoosh M, Su S, Dong X, Tian L, Smyth GK, et al. RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR [version 3; peer review: 3 approved]. F1000Research 2018, 5:1408 ( 10.12688/f1000research.9005.3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone S. v., Purcell SM, Perlis RH, et al. (2013). Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nature Genetics. 10.1038/ng.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, & Shi W (2014). FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, & Shi W (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Research. 10.1093/nar/gkz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajani S, Raina A, Fokken C, Kügler S, & Bähr M (2019). Homogenous generation of dopaminergic neurons from multiple hiPSC lines by transient expression of transcription factors. Cell Death and Disease. 10.1038/s41419-019-2133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Harris SE, Zhang Q, McRae AF, Hagenaars SP, Hill WD, et al. (2018). GWAS on family history of Alzheimer’s disease. Translational Psychiatry. 10.1038/s41398-018-0150-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, & Smyth GK (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Research. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder D, Herz DM, Rowe JB, Lehéricy S, & Siebner HR (2019). The role of dopamine in the brain - lessons learned from Parkinson’s disease. In NeuroImage. 10.1016/j.neuroimage.2018.11.021 [DOI] [PubMed] [Google Scholar]
- Miskinyte Id G, Hansen MG, Monni E, Lam M, Bengzon J, Lindvall O, et al. (2018). Transcription factor programming of human ES cells generates functional neurons expressing both upper and deep layer cortical markers. 10.1371/journal.pone.0204688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AR, Filipovic R, Mo Z, Rasband MN, Zecevic N, & Antic SD (2009). Electrical excitability of early neurons in the human cerebral cortex during the second trimester of gestation. Cerebral Cortex. 10.1093/cercor/bhn206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. (2020). Genome-wide association study of over 40,000 bipolar disorder cases provides novel biological insights. In medRxiv. 10.1101/2020.09.17.20187054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, et al. (2019). Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. The Lancet Neurology. 10.1016/S1474-4422(19)30320-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard S (1999). Regulation of action potential size and excitability in substantia nigra compacta neurons: Sensitivity to 4-aminopyridine. Journal of Neurophysiology. 10.1152/jn.1999.82.6.2903 [DOI] [PubMed] [Google Scholar]
- Nedergaard S (2004). A Ca2+-independent slow afterhyperpolarization in substantia nigra compacta neurons. Neuroscience. 10.1016/j.neuroscience.2004.02.030 [DOI] [PubMed] [Google Scholar]
- Nehme R, Zuccaro E, Ghosh SD, Li C, Sherwood JL, Pietilainen O, et al. (2018). Combining NGN2 Programming with Developmental Patterning Generates Human Excitatory Neurons with NMDAR-Mediated Synaptic Transmission. Cell Reports. 10.1016/j.celrep.2018.04.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen CY, Choi KW, et al. (2019). International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nature Communications. 10.1038/s41467-019-12576-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasman JA, Verweij KJH, Gerring Z, Stringer S, Sanchez-Roige S, Treur JL, et al. (2018). GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nature Neuroscience, 21(9), 1161–1170. 10.1038/s41593-018-0206-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, et al. (2011). Direct conversion of human fibroblasts to dopaminergic neurons. Proceedings of the National Academy of Sciences of the United States of America. 10.1073/pnas.1105135108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picken Bahrey HL, & Moody WJ (2003). Early development of voltage-gated ion currents and firing properties in neurons of the mouse cerebral cortex. Journal of Neurophysiology. 10.1152/jn.00972.2002 [DOI] [PubMed] [Google Scholar]
- Polimanti R, Walters RK, Johnson EC, McClintick JN, Adkins AE, Adkins DE, et al. (2020). Leveraging genome-wide data to investigate differences between opioid use vs. opioid dependence in 41,176 individuals from the Psychiatric Genomics Consortium. Molecular Psychiatry. 10.1038/s41380-020-0677-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell SK, Gregory J, Akbarian S, & Brennand KJ (2017). Application of CRISPR/Cas9 to the study of brain development and neuropsychiatric disease. Molecular and Cellular Neuroscience, 82. 10.1016/j.mcn.2017.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell SK, O’Shea C, Brennand KJ, & Akbarian S (2021). Parsing the Functional Impact of Noncoding Genetic Variants in the Brain Epigenome. In Biological Psychiatry (Vol. 89, Issue 1, pp. 65–75). Elsevier Inc. 10.1016/j.biopsych.2020.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell SK, O’Shea CP, Shannon SR, Akbarian S, & Brennand KJ (2020). Investigation of Schizophrenia with Human Induced Pluripotent Stem Cells. 155–206. 10.1007/978-3-030-45493-7_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland JD, Maddock RJ, Hurtado MY, Tanase C, Lesh TA, Niendam TA, et al. (2019). Disrupted GABAergic facilitation of working memory performance in people with schizophrenia. NeuroImage. Clinical, 25, 102127. 10.1016/j.nicl.2019.102127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin RA, Moss SJ, & Slesinger PA (2017). G Protein-Gated Potassium Channels: A Link to Drug Addiction. In Trends in Pharmacological Sciences. 10.1016/j.tips.2017.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Walters JTR, & O’Donovan MC (2020). Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. In medRxiv. 10.1101/2020.09.12.20192922 [DOI] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, & Smyth GK (2009). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, & Oshlack A (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology. 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa F, Dhingra A, Uysal B, Mendis GDC, Loeffler H, Elsen G, et al. (2020). In Vitro Differentiated Human Stem Cell-Derived Neurons Reproduce Synaptic Synchronicity Arising during Neurodevelopment. Stem Cell Reports. 10.1016/j.stemcr.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer DM, Ripke S, McQuillin A, Boocock J, Stahl EA, Pavlides JMW, et al. (2018). Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell. 10.1016/j.cell.2018.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, & Livak KJ (2008). Analyzing real-time PCR data by the comparative CT method. Nature Protocols. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Schrode N, Ho S-M, Yamamuro K, Dobbyn A, Huckins L, Matos MR, et al. (2019). Synergistic effects of common schizophrenia risk variants. Nature Genetics, 51(10), 1475–1485. 10.1038/s41588-019-0497-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz W (2007). Multiple dopamine functions at different time courses. In Annual Review of Neuroscience. 10.1146/annurev.neuro.28.061604.135722 [DOI] [PubMed] [Google Scholar]
- Sheng Y, Filichia E, Shick E, Preston KL, Phillips KA, Cooperman L, et al. (2016). Using iPSC-derived human DA neurons from opioid-dependent subjects to study dopamine dynamics. Brain and Behavior. 10.1002/brb3.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar HA, et al. (2018). Genetic identification of brain cell types underlying schizophrenia. Nature Genetics. 10.1038/s41588-018-0129-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. (2019). Genome-wide association study identifies 30 loci associated with bipolar disorder. Nature Genetics. 10.1038/s41588-019-0397-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg M, Bogetofte H, Lawson T, Jansson J, Smith G, Astradsson A, et al. (2013). Improved cell therapy protocols for Parkinson’s disease based on differentiation efficiency and safety of hESC-, hiPSC-, and non-human primate iPSC-derived dopaminergic neurons. Stem Cells. 10.1002/stem.1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theka I, Caiazzo M, Dvoretskova E, Leo D, Ungaro F, Curreli S, et al. (2013). Rapid Generation of Functional Dopaminergic Neurons From Human Induced Pluripotent Stem Cells Through a Single-Step Procedure Using Cell Lineage Transcription Factors. STEM CELLS Translational Medicine. 10.5966/sctm.2012-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G, Singer O, & Verma IM (2006). Production and purification of lentiviral vectors. Nature Protocols. 10.1038/nprot.2006.37 [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wise RA, & Baler R (2017). The dopamine motive system: Implications for drug and food addiction. In Nature Reviews Neuroscience. 10.1038/nrn.2017.130 [DOI] [PubMed] [Google Scholar]
- Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, et al. (2018). Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nature Neuroscience. 10.1038/s41593-018-0275-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Lee SH, Mehta D, Vinkhuyzen AAE, Dudbridge F, & Middeldorp CM (2014). Research Review: Polygenic methods and their application to psychiatric traits. In Journal of Child Psychology and Psychiatry and Allied Disciplines (Vol. 55, Issue 10, pp. 1068–1087). Blackwell Publishing Ltd. 10.1111/jcpp.12295 [DOI] [PubMed] [Google Scholar]
- Wray NR, Wijmenga C, Sullivan PF, Yang J, & Visscher PM (2018). Common Disease Is More Complex Than Implied by the Core Gene Omnigenic Model. In Cell. 10.1016/j.cell.2018.05.051 [DOI] [PubMed] [Google Scholar]
- Wu D, & Smyth GK (2012). Camera: A competitive gene set test accounting for inter-gene correlation. Nucleic Acids Research. 10.1093/nar/gks461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Chanda S, Marro S, Ng YH, Janas JA, Haag D, et al. (2017). Generation of pure GABAergic neurons by transcription factor programming. Nature Methods. 10.1038/nmeth.4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Chanda S, Südhof T, & Wernig M (2017). Generation of pure GABAergic neurons by transcription factor programming. Protocol Exchange. 10.1038/protex.2017.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G (2020). Gene ontology semantic similarity analysis using GOSemSim. In Methods in Molecular Biology. 10.1007/978-1-0716-0301-7_11 [DOI] [PubMed] [Google Scholar]
- Yu G, Li F, Qin Y, Bo X, Wu Y, & Wang S (2010). GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 10.1093/bioinformatics/btq064 [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, & He QY (2012). ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS A Journal of Integrative Biology, 16(5), 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pak CH, Han Y, Ahlenius H, Zhang Z, Chanda S, et al. (2013). Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 10.1016/j.neuron.2013.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source data described in this manuscript are available via the PsychENCODE Knowledge Portal (https://psychencode.synapse.org/). The PsychENCODE Knowledge Portal is a platform for accessing data, analyses, and tools generated through grants funded by the National Institute of Mental Health (NIMH) PsychENCODE program. Data is available for general research use according to the following requirements for data access and data attribution: (https://psychencode.synapse.org/DataAccess). For access to content described in this manuscript see: http://doi.org/10.7303/syn25500352