Abstract
Purpose: Mutations leading to homologous recombination deficiency (HRD) increase the tumor sensitivity to platinum-based chemotherapy and PARP inhibitors. However, reversion mutations often develop conferring acquired drug resistance. There is still a lack of comprehensive investigation on HRR reversion mutations in large pan-cancer cohorts, especially in the Eastern Asian population. This study aims to characterize reversion mutations in homologous recombination repair (HRR)-related genes in a large cohort of Chinese pan-cancer patients.
Methods: Sequencing data from 23,375 patients across over 17 cancer types were retrospectively analyzed for pathogenic/likely pathogenic (P/LP) germline mutations in 15 HRR genes. Somatic mutations detected in tumor or circulating cell-free DNA predicted to restore the open reading frame of the deleterious allele were subsequently identified as reversion mutations.
Results: 654 cases out of 23,375 (2.8%) unselected pan-cancer patients were identified with HRR germline mutations. Secondary somatic mutations were further analyzed in their matched tumor/plasma samples. The overall frequency of reversion mutation was 1.7% (11/654). The reversion mutations occurred only in 3 out of the 15 HRR genes: BRCA1 (3.8%), BRCA2 (3.5%) and PALB2 (2.0%) from 11 patients (6 breast cancers, 1 ovarian cancer, 1 pancreatic cancer, 1 lung cancer and 2 breast and ovarian dual cancers). We identified total 25 reversion events (BRCA1, n=9; BRCA2, n=8; PALB2, n=8), including 12 pure deletions, 10 missense single nucleotide variants, 2 insertions and 1 splice site mutation. Besides, we detected microhomology length >1bp in seven out of the reversion deletions (58.3%), suggestive of microhomology-mediated end-joining (MMEJ) repair signature. Intriguingly, a positive correlation (r=0.85, p=0.001) between the length of deletion and the microhomology length was also observed. We obtained disease courses from 6/11 patients with reversion events. Four acquired reversions after the failure of the PARP inhibitor treatment. Two patients had somatic reversion mutations identified after progressing on platinum-based treatment.
Conclusion: This study comprehensively depicts the prevalence and characteristics of HRR reversion mutation of germline mutations in an unselected Chinese pan-cancer cohort. The reversion mutations predominantly occurred in BRCA1, BRCA2 and PALB2. The results revealed that reversion mutations frequently occurred after resistance to platinum-based chemotherapy and/or PARP inhibitor. Our study provides insight into the underlying mechanism of drug resistance in HRD tumors and suggests that monitoring HRR mutation status along the disease course could be beneficial especially for informing resistance mechanisms and guiding subsequent therapies.
Keywords: Homologous recombination repair, Reversion mutation, Next-generation sequencing, Platinum-based chemotherapy, PARP inhibitor
Introduction
Detecting and repairing DNA damage is pivotal for maintaining normal cell function and genomic stability. Homologous recombination repair (HRR) system, using the sister chromatid or homologous chromosome as the template to restore DNA, is the most fidelity mechanism in repairing double-stranded DNA breaks (DSBs) 1. The HRR-related mutations are predominantly present in the Fanconi anemia (FA)-BRCA pathway 2, 3. Those mutations, mainly frameshift, nonsense, splice site, and nonstart mutations, could lead to truncated protein translation. Dysfunction of the HRR mechanism confers the genomic instability that ultimately leads to carcinogenesis.
Germline mutations in HRR genes BRCA1/2 and PALB2 are known to associate with hereditary breast cancer and ovarian cancer, which have also been described in pancreatic cancer and prostate cancer. Due to synthetic lethality, patients harboring such mutations are more sensitive to DNA-damaging agents, such as platinum-based chemotherapy and poly ADP-ribose polymerase (PARP) inhibitors 4, 5. With the broader application of platinum-based chemotherapy and the newly approved PARP inhibitors, acquired resistance cases have being ever-increasingly reported 6-8. The HRR function recovery via acquiring secondary mutation is one of the major underlying mechanisms that mediate the resistance 5. Usually the observed secondary somatic mutations are close to the primary mutations, restore the open-reading frame (ORF) of the functional protein, and assist the cell in recovering HRR function 4, 9, 10.
Reversion mutations within multiple genes in the HRR pathway, including BRCA1, BRCA2, RAD51C, RAD51D, and PALB2, have been reported in ovarian, prostate, and breast carcinomas as a mechanism of acquired or primary resistance to platinum-based chemotherapy and PARP inhibitors 7-18. However, most studies either reported sporadic cases or recruited small cohorts mainly from Western populations. There is still a lack of comprehensive investigation on HRR reversion mutations in large pan-cancer cohorts, especially in the Eastern Asian population.
Here, we characterized the prevalence and spectrum of the reversion mutations in HRR genes across more than 17 different cancer types in a Chinese cohort of 23,375 patients. We also obtained treatment information and clinical outcomes from six cases with reversions to explore the heterogeneity in clinical response.
Methods
Patients and study design
We retrospectively recruited patients with solid tumors according to the following criteria: 1) underwent somatic genomic profiling from January 2019 to May 2020 in the Burning Rock LAVA Database; 2) also provided matched white blood cell (WBC) samples for sequencing in parallel for germline mutation filtration; 3) sequenced with targeted panel including 15 HRR-associated genes (BRCA1, BRCA2, ATM, PALB2, CHEK2, BRIP1, RAD51B, RAD51D, RAD54L, RAD51C, BARD1, FANCI, FANCL, CHEK1, CDK12). Pathogenic/likely pathogenic (P/LP) germline mutations in 15 HRR-associated genes were screened based on their WBC sequencing results. Frequencies of each mutation were determined for the total cohort, as well as for each cancer type (lung cancer, colorectal cancer, breast cancer, stomach/esophagus cancer, ovarian cancer, pancreatic cancer, sarcoma, liver cancer, cervical cancer, cancer of the biliary tract, endometrial adenocarcinoma, prostate cancer, kidney cancer, head-neck cancer, bladder cancer, malignant melanoma, glioma, and other cancer types). The study was approved by the Ethics Committee of Zhejiang Cancer Hospital. Patient's informed consent was waived due to the retrospective nature of the study. Procedures involving human participants in this study were complied with the Declaration of Helsinki.
Reversion detection in tumor and circulating cell-free DNA (cfDNA)
For patients identified with P/LP germline HRR mutations, secondary somatic mutations were further analyzed in their WBC matched tumor/plasma samples and their subsequently sequenced samples if applicable. The detected secondary mutations were filtered for potential reversion mutations and further divided into two sub-groups: confirmed and putative. A reversion mutation was classified as “confirmed” if i) it was present on the same sequencing read with the primary germline mutation and predicted to restore the ORF; or ii) both germline and somatic mutations occurred in or shared the same nucleobase(s). Other mutations, which were present in the same gene yet located far away from the germline mutations, were classified as “putative” ones.
Next-generation sequencing (NGS)
NGS were performed in Burning Rock Biotech, a College of American Pathologists (CAP)-accredited and Clinical Laboratory Improvement Amendments (CLIA)-certified clinical laboratory, as described previously 19, 20. In general, the overall procedure included DNA extraction, library construction, sequencing and data analysis. The cfDNA from plasma or genomic DNA from tumor samples were extracted using QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) following the manufacturing protocol. The DNA libraries were prepared with targeted enrichment by using one of the commercially available panels including 168 genes (Lung Plasma, Burning Rock Biotech, Guangzhou, China), 295 genes (OncoScreen, Burning Rock Biotech, Guangzhou, China), or 520 genes (OncoScreen Plus, Burning Rock Biotech, Guangzhou, China). All panels included the 15 HHR genes. Indexed libraries were subsequently sequenced on Illumina NextSeq 500 system (Illumina, Inc., Hayward, CA, USA) with paired-end reads.
Sequencing data analysis
The paired-end reads were mapped to the reference genome with Burrows-Wheeler aligner v0.7.10 21. The Genome Analysis Toolkit (GATK) v.3.2 22 and VarScan v.2.4.3 23 were employed for local realignment, variant calling and annotation. Variants with population frequencies of over 0.1% in the Exome Aggregation Consortium (ExAC), 1000 Genomes, dbSNP and ESP6500SI-V2 databases were grouped as single nucleotide polymorphisms (SNPs). ANNOVAR 24 (2016-02-01 release) and SnpEff v3.6 25 were used for the annotation of the remaining variants. All germline variants were manually annotated and categorized into five classes following the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) recommendations as follows: pathogenic (Class 5), likely pathogenic (Class 4), variants of uncertain significance (Class 3,) likely benign (Class 2) and benign (Class 1) 26. All variants classified as P/LP after manual curation were considered for further analysis, while variants classified as of unknown significance, likely benign or benign were disregarded.
Analysis of microhomologies
The presence of microhomologies in deletions was analyzed as previously described 27, 28. Briefly, if the nucleotides before a pure deletion were identical with the last nucleotides of the deleted sequence, the position of the identical nucleotides was adjusted. The number of contiguous nucleotides at the beginning of the deleted sequence that matched the sequence 3'-flanking the deletion was determined as the length of microhomology. Microhomologies of > 1 bp were defined to possess the signature of microhomology-mediated end-joining (MMEJ) repair.
Statistical analysis
Differences in proportion between groups were calculated using Fisher's exact test. The correlation between length of deletion and the microhomology length was analyzed by Pearson correlation. P-values <0.05 were considered statistically significant.
Results
Prevalence of germline mutations in HRR genes
We retrospectively evaluated the WBC samples of 23,375 solid tumors patients who underwent NGS. The overall cohort consisted of more than 17 cancer types, including lung cancer (n=17,029), colorectal cancer (n=1,282), breast cancer (n=1,024), stomach/esophagus cancer (n=732), ovarian cancer (n=656), pancreatic cancer (n=269; Table 1). Of the total population, the majority (82.5%) were late stage cancer patients (Table S1).
Table 1.
Next-generation sequencing by cancer type and germline mutations frequency.
| Cancer type | Patients analyzed | Patients with HRR germline mutations | Mutation frequency |
|---|---|---|---|
| Ovarian cancer | 656 | 93 | 14.2% |
| Breast cancer | 1024 | 100 | 9.8% |
| Endometrial adenocarcinoma | 169 | 11 | 6.5% |
| Pancreatic cancer | 269 | 16 | 5.9% |
| Prostate cancer | 158 | 7 | 4.4% |
| Cervical cancer | 189 | 8 | 4.2% |
| Malignant melanoma | 74 | 3 | 4.1% |
| Cancer of biliary tract | 201 | 8 | 4.0% |
| Liver cancer | 197 | 7 | 3.6% |
| Colorectal cancer | 1282 | 38 | 3.0% |
| Stomach/esophagus cancer | 732 | 18 | 2.5% |
| Head-neck cancer | 137 | 3 | 2.2% |
| Kidney cancer | 153 | 3 | 2.0% |
| Lung cancer | 17029 | 313 | 1.8% |
| Glioma | 76 | 1 | 1.3% |
| Sarcoma | 226 | 1 | 0.4% |
| Bladder cancer | 84 | 0 | 0.0% |
| Others | 809 | 31 | 3.8% |
| Total | 23375* | 654** | 2.8% |
* 90 patients with multiple primary cancers. ** 6 patients with two primary cancers: 3 cases of breast and ovarian dual cancers, 1 case of ovarian and cervical dual cancers, 1 case of ovarian and endometrial dual cancers, and 1 case of lung and cervical dual cancers.
A total of 663 P/LP germline mutations spanning 15 HRR genes were identified in 654 out of 23,375 (2.8%) pan-cancer patients, among which nine patients (2 with breast cancer, 2 with lung cancer, 2 with colorectal cancer and 3 with other cancer types) harbored dual germline mutations and the remaining all carried a single germline mutation. Across all investigated cancer types, ovarian cancer (14.2%, n=93) showed the highest prevalence of HRR germline mutations, followed by cancers of breast (9.8%, n=100), endometrium (6.5%, n=11), and pancreas (5.9%, n=16) (Table 1). No bladder cancer was detected with HRR P/LP germline mutation in this cohort.
Among the 15 HRR genes analyzed, germline mutations occurred predominantly in BRCA genes: 175 BRCA2 and 130 BRCA1 mutations were observed, comprising 26.4% (175/663) and 19.6% (130/663) of all HRR germline events, respectively. Other commonly mutated genes included ATM (n=67, 10.1%), RAD51D (n=58, 8.7%), PALB2 (n=49, 7.4%), CHEK2 (n=46, 6.9%), BRIP1 (n=35, 5.3%), and RAD54L (n=30, 4.5%) (Figure S1).
Comprehensive characterization of HRR reversion mutations
Among the 654 patients with germline mutations, 11 cases (1.7%) were identified with reversion mutations including six cases with breast cancer, one with ovarian cancer, one with pancreatic cancer, one with lung cancer and two with breast and ovarian dual primary cancers (Table 2, 3). Breast cancer exhibited the highest prevalence of reversion mutations (8.1%), followed by pancreatic (6.2%) and ovarian cancer (3.2%), while reversion events only occurred in 0.3% of lung cancers (Table 2). The reversion mutations occurred in 3 genes: BRCA1 (n=5), BRCA2 (n=5) and PALB2 (n=1), but not in any other HRR genes, ATM, CHEK2, BRIP1, RAD51B, RAD51D, RAD54L, RAD51C, BARD1, FANCI, FANCL, CHEK1, and CDK12, in our cohort. Among the three HRR genes with reversions, BRCA1 revealed the highest frequency of 3.8%, while 3.5% and 2.0% of BRCA2 and PALB2 germline mutations reverted, respectively (Table 2). PALB2 reversion mutation was only detected from one case with breast cancer.
Table 2.
Different HRR reversion mutations across different cancer types.
| Total HRR genes | BRCA1 | BRCA2 | PALB2 | Other HRR genes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cancer type | Patients with HRR germline mutations | Patients with reversions | Reversion frequency | Patients with HRR germline mutations | Patients with reversions | Reversion frequency | Patients with HRR germline mutations | Patients with reversions | Reversion frequency | Patients with HRR germline mutations | Patients with reversions | Reversion frequency | Patients with HRR germline mutations | Patients with reversions |
| Lung cancer | 313 | 1 | 0.3% | 28 | 0 | 0.0% | 75 | 1 | 1.3% | 22 | 0 | 0.0% | 188 | 0 |
| Breast cancer | 100 | 8 | 8.0% | 26 | 4 | 15.4% | 39 | 3 | 7.7% | 14 | 1 | 7.1% | 23 | 0 |
| Ovarian cancer | 94 | 3 | 3.2% | 59 | 3 | 5.1% | 22 | 0 | 0.0% | 0 | 0 | NaN | 13 | 0 |
| Pancreatic cancer | 16 | 1 | 6.2% | 2 | 0 | 0.0% | 8 | 1 | 12.5% | 1 | 0 | 0.0% | 5 | 0 |
| Total patient | 654* | 11** | 1.7% | 130*** | 5** | 3.8% | 144 | 5 | 3.5% | 49 | 1 | 2.0% | 305**** | 0 |
* 6 patients with multiple primary cancers. ** 2 patients with breast and ovarian dual primary cancers, both harboured BRCA1 reversion mutation. *** 4 patients with two primary cancers: 3 cases of breast and ovarian dual cancers, 1 case of ovarian and cervical dual cancers. **** 2 patients with two primary cancers: 1 case of ovarian and endometrial dual cancers, and 1 case of lung and cervical dual cancers.
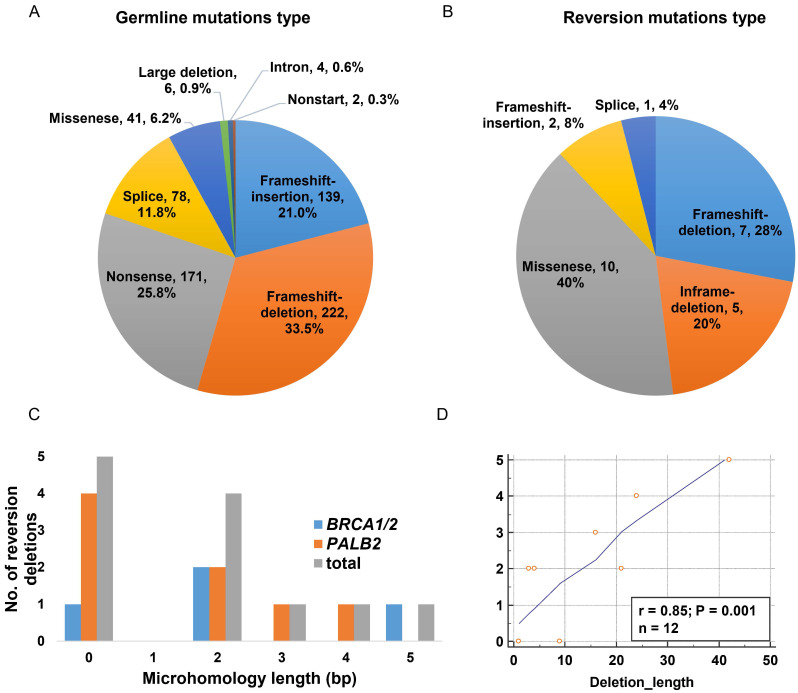
We further explored the propensity of HRR germline mutations to acquire reversion based on the mutation type. We observed that frameshift (insertion and deletion) and nonsense variants comprised of the vast majority of the overall P/LP germline mutations (54.5% and 25.8%, respectively), while missense and splice-site variants as well as large deletion, intron and nonstart mutations were observed with low frequencies (Figure 1A).
Figure 1.
Characterizing of reversion mutations. A. Variant type distribution of the overall germline mutation (n=663); B. Distribution of variant type in somatic reversion mutations (n=25); C. Distribution of microhomology length in somatic reversion deletions; D. The correlation of reversion deletion length and microhomology length.
The majority of the reversion mutations were pure deletions causing frameshift and in-frame mutations (48%, n=12, Figure 1B), ranging from 1bp to 42bp and mainly arising in PALB2 gene (n=8, Table 3). The missense single nucleotide variant (SNV) accounted for 40% (n=10) of reversion events. We further analyzed microhomologies surrounding the 12 reversion deletions and detected microhomology length >1bp in seven of them (58.3%), suggestive of MMEJ repair signature (Figure 1C). The remaining five deletion evens revealed no microhomology features. Intriguingly, we also observed a positive correlation (r=0.85, p=0.001) between the length of deletion and the microhomology length (Figure 1D).
Table 3.
List of 11 cases identified with somatic reversion mutation(s).
| Patient | Sex | Age | Cancer type | Clinical stage | Gene | Germline mutation | Somatic reversion mutations/status |
|---|---|---|---|---|---|---|---|
| P1 | F | 47 | Breast | IV | BRCA1 | c.2341G>T p.E781* | c.2307_2348del p.S770_I783del/confirmed c.2339_2347del p.Q780_I783delinsL/confirmed c.2343A>T p.E781Y/confirmedc.2342A>C p.E781S/confirmedc.2341G>C p.E781Q/confirmed |
| P2 | F | 42 | Breast/ovarian | IV(Br.)/II(ov.) | BRCA1 | c.3754_3755del p.L1252fs | c.3710_3711dup p.P1238fs/confirmed |
| P3 | F | 58 | Breast | IV | PALB2 | c.2480_2481del p.T827fs | c.2469_2492del p.C824_L831del/confirmedc.2466del p.Q822fs/confirmedc.2450_2465del p.T817fs/confirmedc.2457del p.E820fs/confirmedc.2339del p.G780fs/putativec.2322_2325del p.K774fs/putativec.2313del p.S771fs/putativec.2298_2301del p.C768fs/putative |
| P4 | F | 36 | Breast | IV | BRCA2 | c.8451T>A p.C2817* | c.8450G>C p.C2817S/confirmed |
| P5 | F | 39 | Breast | III | BRCA2 | c.2870del p.N957fs | c.2837_2838insG p.D946fs/confirmed |
| P6 | F | 46 | Breast | IV | BRCA1 | c.3436_3439del p.C1146fs | c.3430_3450del p.Q1144_P1150del/confirmed |
| P7 | F | 36 | Breast | IV | BRCA2 | c.8878C>T p.Q2960* | c.8878C>A p.Q2960K/confirmed |
| P8 | F | 53 | Breast/ovarian | IV | BRCA1 | c.66dup p.E23fs | c.67G>C p.E23Q/confirmed |
| P9 | F | 54 | Ovarian | IV | BRCA1 | c.4065_4068del p.N1355fs | c.4096G>A p.G1366S/confirmed |
| P10 | M | 73 | Pancreatic | IV | BRCA2 | c.3109C>T p.Q1037* | c.3109_3111del p.Q1037del/confirmedc.3109_3111delinsTAC p.Q1037Y/confirmedc.3109_3110delinsTC p.Q1037S/confirmedc.3109C>G p.Q1037E/confirmed |
| P11 | F | 63 | Lung | IV | BRCA2 | c.2979G>A p.W993* | c.2978G>C p.W993S/confirmed |
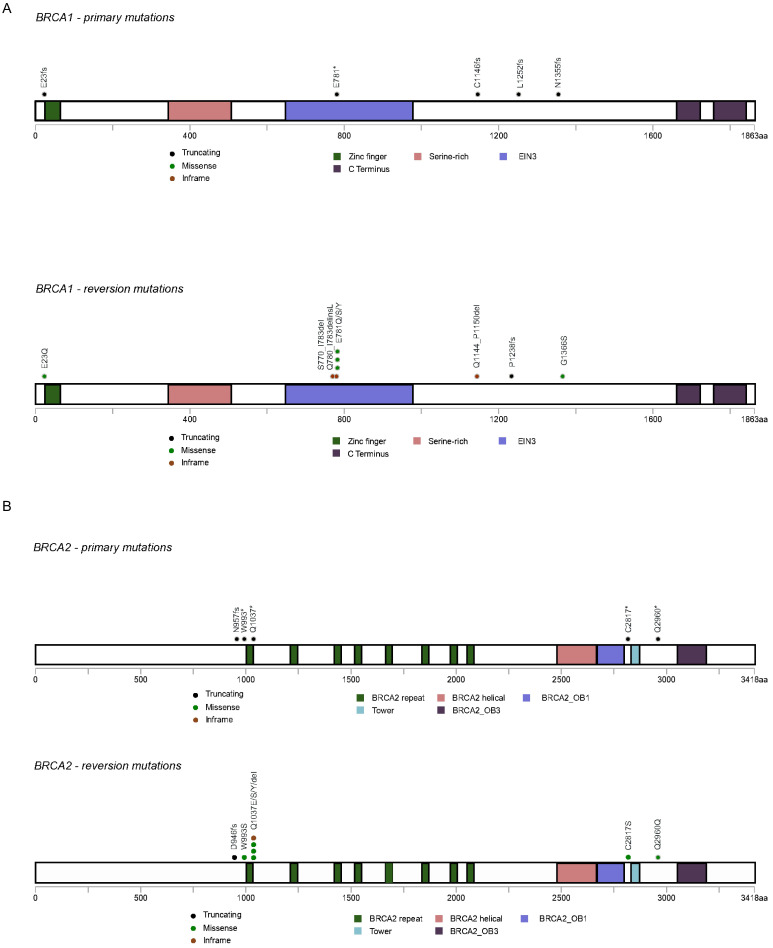
A total of 25 reversion mutations (21 confirmed and 4 putative) were found: nine within BRCA1, eight within BRCA2, and eight within PALB2. Eight out of the eleven patients acquired a single reversion mutation towards the primary germline mutation (Table 3); while Patient 1, 3 and 10 acquired multiple reversion mutations in the context of primary mutation BRCA1 p.E781*, PALB2 p.T827fs and BRCA2 p.Q1037*, respectively. For BRCA1 gene, reverted germline mutations were mainly located in the hotspot mutated regions in the sequence comprising exon 10 (n=4) and one was observed in exon 2 encoding the Zinc finger ring region (Figure 2A). For BRCA2 gene, reversion events were found within the region encoding BRCA2 repeats in exon 11 (n=3), exon 19 (n=1) and exon 22 (n=2) (Figure 2B). PALB2 reversion mutations were only observed in a single case located in exon 5.
Figure 2.
Distribution of reverted primary germline mutations and reversion mutations on BRCA1 and BRCA2. A. BRCA1; B. BRCA2.
HRR reversion mutations associated with resistance to platinum-based chemotherapy or PARP Inhibitor
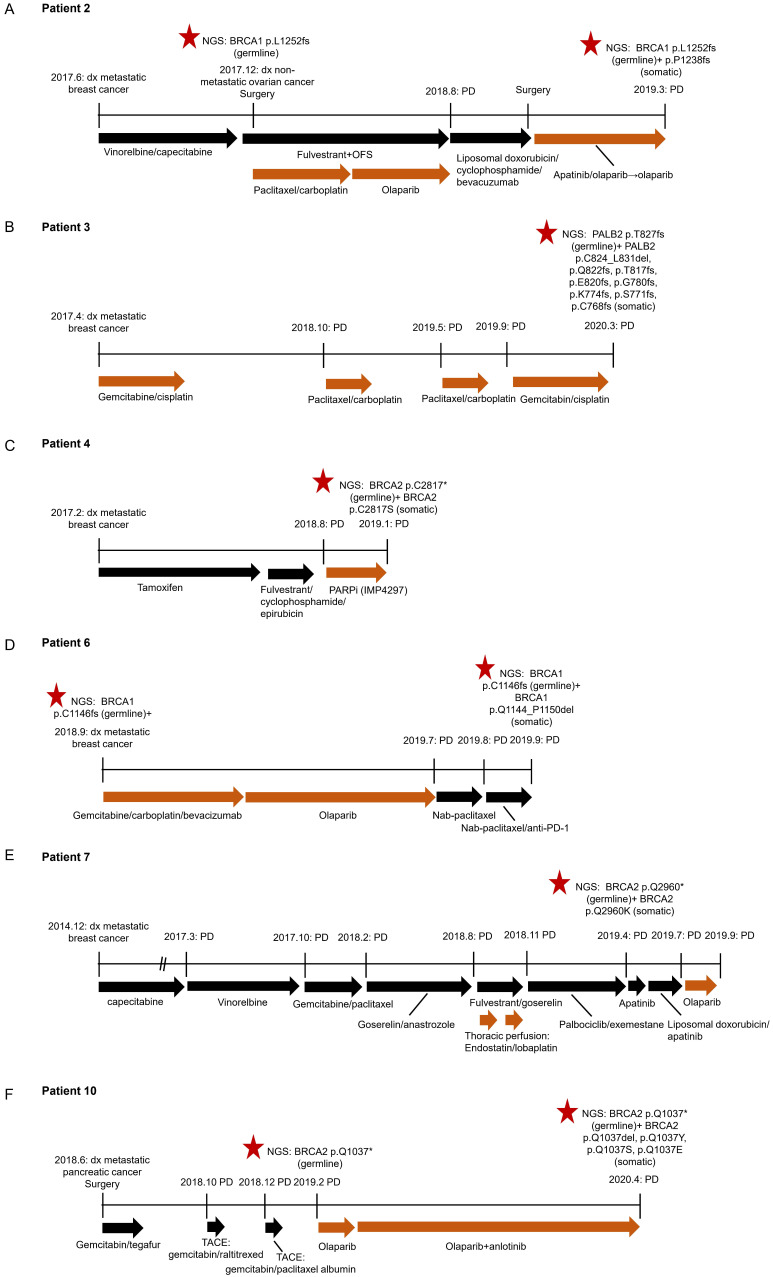
We obtained clinical characteristics and treatment history from six out of the eleven patients with reversion events with patients' consent (Figure 3). All of them received PARP inhibitor or platinum-therapy prior to reversion detection. Four patients (P2, P4, P6 and P10) acquired reversion mutations after the failure of the treatment with a PARP inhibitor. Two patients (P3 and P7) had somatic reversion mutations identified after progressing on platinum-based treatment, of whom P7 also showed primary resistance to the subsequent olaparib treatment. Our findings show that acquired reversion of HRR germline mutations indicated poor response to platinum-based therapy or PARP inhibition.
Figure 3.
Clinical courses of six patients with reversion mutations. A. Patient 2; B. Patient 3; C. Patient 4; D. Patient 6; E. Patient 7; F. Patient 10. dx, diagnosis. OFS, ovarian function suppression. SD, stable disease. PD, progressive disease.
Patient 2 was diagnosed with metastatic breast cancer (luminal B) in June 2017 at the age of 42 years. She subsequently received 6 cycles of vinorelbine and capecitabine combination, followed by fulvestrant and ovarian function suppression (OFS), and achieved stable disease (SD). After progressive disease (PD) in August 2018, the second-line treatment shifted to the combination of liposomal doxorubicin, cyclophosphamide and bevacizumab. Subsequently, the patient received palliative surgery and was treated with olaparib-based regimens based on the germline mutation BRCA1 p. L1252fs detected via NGS. After an SD lasting for 7 months, the disease progressed when a somatic reversion mutation BRCA1 p. P1238fs was identified. Of note, in the course of treatment for metastatic breast cancer, the patient was also detected with non-metastatic ovarian cancer in Dec 2017. She underwent resection and received olaparib treatment following paclitaxel+carboplatin for ovarian lesion (Figure 3A).
Patient 3 with metastatic triple-negative breast cancer (TNBC) received four lines of platinum-based chemotherapy. NGS performed at PD revealed a germline mutation PALB2 p. T827fs, accompanied by four confirmed somatic reversion mutations and 4 putative reversion mutations (Figure 3B).
Patient 4 was 36-year-old and diagnosed with stage IV breast cancer (luminal B) in February 2017. She received the treatment with tamoxifen, followed by the combination of fulvestrant, cyclophosphamide and epirubicin. Upon the disease progression in August 2018, she started receiving a PARP inhibitor (IMP4297) until January 2019. The germline mutation BRCA2 p. C2817* and somatic reversion mutation p. C2817S were identified upon PD (Figure 3C).
Patient 6, 46-year-old, was detected with TNBC in September 2018. NGS result suggested the presence of a germline mutation BRCA1 p. C1146fs. She received six cycles of carboplatin-based chemotherapy and achieved SD. Subsequently, olaparib was administrated as maintenance therapy and the disease remained stable until July 2019. Upon PD, the patient shifted to nab-paclitaxel and anti-PD-1 inhibitor. NGS was performed when the disease progressed again and indicated the emerging of somatic reversion mutation BRCA 1 p. Q1144_P1150del (Figure 3D).
Patient 7, who was diagnosed with metastatic breast cancer (luminal B), received three lines of chemotherapy followed by endocrine therapy with goserelin and anastrozole. Subsequently the treatment was switched to fulvestrant and goserelin. In the meantime, the patient also underwent thoracic perfusion with endostatin and lobaplatin. After the disease progressed, she received palbociclib and exemestane until April 2019 when the treatment was switched to apatinib. Meanwhile, NGS was performed and showed concomitant germline mutation BRCA2 p. Q2960* and somatic reversion mutation p. Q2960K. The patient then received olaparib treatment upon the progression of apatinib regimen but developed PD rapidly (Figure 3E).
Patient 10 was a 73-year-old male with metastatic pancreatic cancer. After surgery, the patient received treatment with gemcitabine and tegafur. After PD, he underwent two sequential transarterial chemoembolizations (TACEs) with gemcitabine+raltitrexed and gemcitabine +paclitaxel albumin, respectively. NGS screening indicated the presence of germline mutation of BRCA2 p. Q1037* in February 2019 after the disease progressed. Subsequently, the patient received the olaparib and anlotinib combination following the olaparib single agent and achieved a partial response with a PFS of 12 months. Four somatic reversion mutations in BRCA2 were identified upon the failure of olaparib regimen (Figure 3F).
Discussion
We carried out a retrospective study investigating reversion mutations in 15 HRR genes in a large cohort of Chinese pan-cancer patients across multiple cancer types. 654 cases out of 23,375 (2.8%) pan-cancer patients were identified with HRR germline mutations. We observed reversion mutations emerging in 1.7% (11/654) of the patients with HRR germline mutations. Specifically, the prevalence of reversion was 8.0%, 6.2%, 3.2% and 0.3% in breast cancer, pancreatic cancer, ovarian cancer and lung cancer, respectively. On the other hand, 3.8%, 3.5% and 2.0% of BRCA1, BRCA2 and PALB2 germline mutation patients were identified with reversion events, respectively. Our study is a large assessment of reversion mutation prevalence of 15 HRR germline mutations in Chinese patients. It is also one of the few studies to assess reversion mutations across multiple tumor types.
Of note, the lower incidence of reversion mutation observed in our cohort is largely attributable to that we did not screen for patients with resistance to platinum-based or PARPi treatment, which will tremendously enrich the patients with reversion events. A recent study identified reversion mutations in 12 out of 1,308 unselected germline or somatic BRCA1/2 mutant tumors from the MSKCC database 29, which is comparable with our study (1.7% vs 0.9%, P=0.118). Tobalina et al. reported a meta-analysis including 327 BRCA1/BRCA2 mutant patients with ovarian cancer, breast cancer, pancreatic cancer or prostate cancer, who progressed on platinum-based or PARP inhibitor treatment. They observed an overall reversion incidence of 26% and the prevalence varied from 20%-40% across different cancers except for prostate cancer that showed a high prevalence of 81.8%. The prevalence of reversion events was 22.0% and 30.7% in BRCA1 and BRCA2 in their study, respectively 27. We failed to identify any reversion event in our prostate cancer cohort, probably due to the small number of patients detected with germline primary mutations (n=8).
Interestingly, we discovered one case (P11) out of the 313 HRR germline-mutant lung cancers harboring a reversion mutation BRCA2 c.2978G>C p.W993S. Similarly, BRCA reversion mutations have recently been reported in two non-canonical BRCA-associated cancers: lung adenocarcinoma (n=1) and gastroesophageal junction adenocarcinoma (n=1), after the failure of platinum-based therapy 29. Particularly interesting is the two cases (P2 and P8) with multiple primary cancers, breast and ovary, out of the 11 patents with reversion mutations. Reversion mutations BRCA1 c.3710_3711dup p.P1238fs and BRCA1 c.67G>C p.E23Q were identified separately. It has been reported two primary cancers with breast-ovary is one of the most common (11.7%) cancer pairs in Chinese patients with multiple primary malignancies 30. For the breast cancer with BRCA1 and BRCA2 mutations, there is a strong association with an increased risk for a second breast or ovarian cancer 31.
Among the 25 reversion mutations we identified, 48% of the reversion mutations were pure deletions causing frameshift and in-frame mutations, followed by missense SNV (40%). This observation is in line with the finding that compared with missense and splice-site mutations, truncating pathogenic mutations in BRCA1/2 are more prone to revert 11, 28. Similarly, Tobalina et al. showed that most revision events were deletions in both BRCA1 (47/80) and BRCA2 (170/219). SNV was the second common type of reversions in BRCA1 (n=19), whereas insertion (n=18) and Indel (n=19) reversions occurred more frequently than SNV (n=8) in BRCA2 27. Moreover, the signature of MMEJ repair (microhomology length>1bp) was observed in 58.3% of reversion deletions in our study, compared with that of 30%-70% previously reported in BRCA1/2 deletions 27, 28. Collectively, these observations underscore the essential role of the MMEJ-driven repair mechanism and suggest the involvement of other DNA repairing processes in generating reversions. Interestingly, we observed a positive correlation between the deletion length and the microhomology length (Figure 1C), which was not seen by Tobalina et al. 27. The discordance may be partially due to the fact that we only evaluated reversion deletions, most of which occurred in PALB2, while Tobalina et al. analyzed both primary and reversion deletions in BRCA1/2. Nevertheless, the limited number of reversions identified in our study attenuates the strength of our finding, making it necessary to further investigate this correlation with an expanded sample size.
All the six patients with reversion mutations, whose disease courses were available in our study, underwent the treatment with a platinum-based regimen and/or PARP inhibitor prior to the identification of somatic reversion mutation. It is usually presumed that the reversion mutation emerging after platinum-based or PARP inhibitor treatment is induced by the treatment exerted pressure 4, 5. Lin et al. also reported BRCA reversion mutations from pretreatment samples in 18% platinum-refractory and 13% platinum-resistant high-grade ovarian carcinomas, which also predicted an inferior PARP inhibitor rucaparib survival 11, reserving the plausibility that reversion mutations could emerge primarily. In our study, patient 7, whose reversion mutation was identified after thoracic perfusion with lobaplatin, showed resistance to the following olaparib treatment. Conceivably, monitoring HRR mutation status along the disease course could be beneficial especially for informing resistance mechanisms and guiding subsequent therapies 32.
Our study has several limitations. Despite a large cohort encompassing over 17 cancer types recruited in the study, patients with lung cancer constituted the majority, resulting in a limited number of patients for most cancer types. Therefore, the capability of detecting reversions has been largely attenuated in these cancers (prostate cancer etc.). Moreover, due to the retrospective nature of the study, most patients lacked paired baseline/pretreatment and post-progression samples for genomic profiling. Thus we were only able to identify reversion events emerging on germline primary mutations while reversion on somatic primary mutation was not investigated in this study. Our study lacks of functional assay to elucidate the mechanisms and the direct impact of reversion mutations on cancer cells.
In the future, well-designed prospective studies that longitudinally monitor patients' mutation profile along the disease course will better track the emergence of reversion events and improve our understanding of the underlying mechanism, thus providing insight into the potential management of drug resistance in HRR-deficient tumors.
Conclusion
This retrospective study demonstrated that the reversion mutations were observed in three HRR-associated genes (BRCA1, BRCA2 and PALB2) with four cancer types (breast cancer, pancreatic cancer, ovarian cancer, and lung cancer) from this Chinese pan-cancer patient cohort. The reversion mutations frequently occurred after resistance to platinum-based chemotherapy and/or PARP inhibitor, and may predict poor outcome from ensuing PARP inhibition therapy. Therefore, monitoring HRR mutation status along the course of the disease could be beneficial especially to informing resistance mechanism and guiding subsequent therapies.
Supplementary Material
Supplementary figure and table.
Acknowledgments
The authors would like to thank Dr. Ting Hou, Luting Qiu, Dr. Lin Shao, Dr. Chunxiao Pan and Dr. Jianxing Xiang from Burning Rock Biotech for their helps in data analysis and manuscript writing.
Funding
This study was supported by the Key Research-Development Program of Zhejiang Province (2019C04001, 2020C04012), the Natural Science Foundation of Zhejiang Province, China (LY21H160005), the Education Department of Henan Province (15A320090) and the Social Development Research Project of Yinzhou District Ningbo (2017-59).
Abbreviations
- HRR
homologous recombination repair
- HRD
homologous recombination deficiency
- WBC
white blood cell
- P/LP
pathogenic/likely pathogenic
- cfDNA
cell-free DNA
- NGS
next-generation sequencing
- MMEJ
microhomology-mediated end-joining
- SNV
single nucleotide variant
References
- 1.Wright WD, Shah SS, Heyer WD. Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem. 2018;293:10524–35. doi: 10.1074/jbc.TM118.000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Andrea AD. Susceptibility pathways in Fanconi's anemia and breast cancer. N Engl J Med. 2010;362:1909–19. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kennedy RD, D'Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol. 2006;24:3799–808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- 4.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D'Andrea AD. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015;5:1137–54. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lord CJ, Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat Med. 2013;19:1381–8. doi: 10.1038/nm.3369. [DOI] [PubMed] [Google Scholar]
- 6.Waks AG, Cohen O, Kochupurakkal B, Kim D, Dunn CE, Buendia Buendia J. et al. Reversion and non-reversion mechanisms of resistance to PARP inhibitor or platinum chemotherapy in BRCA1/2-mutant metastatic breast cancer. Ann Oncol. 2020;31:590–8. doi: 10.1016/j.annonc.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao H, Liu S, Huang D, Han X, Wu X, Shao YW. et al. Acquired multiple secondary BRCA2 mutations upon PARPi resistance in a metastatic pancreatic cancer patient harboring a BRCA2 germline mutation. Am J Transl Res. 2020;12:612–7. [PMC free article] [PubMed] [Google Scholar]
- 8.Pan JN, Lei L, Ye WW, Wang XJ, Cao WM. BRCA1 Reversion Mutation Confers Resistance to Olaparib and Camrelizumab in a Patient with Breast Cancer Liver Metastasis. J Breast Cancer. 2021;24:474–80. doi: 10.4048/jbc.2021.24.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C. et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–20. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA. et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–5. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 11.Lin KK, Harrell MI, Oza AM, Oaknin A, Ray-Coquard I, Tinker AV. et al. BRCA Reversion Mutations in Circulating Tumor DNA Predict Primary and Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma. Cancer Discov. 2019;9:210–9. doi: 10.1158/2159-8290.CD-18-0715. [DOI] [PubMed] [Google Scholar]
- 12.Kondrashova O, Nguyen M, Shield-Artin K, Tinker AV, Teng NNH, Harrell MI. et al. Secondary Somatic Mutations Restoring RAD51C and RAD51D Associated with Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma. Cancer Discov. 2017;7:984–98. doi: 10.1158/2159-8290.CD-17-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quigley D, Alumkal JJ, Wyatt AW, Kothari V, Foye A, Lloyd P. et al. Analysis of Circulating Cell-Free DNA Identifies Multiclonal Heterogeneity of BRCA2 Reversion Mutations Associated with Resistance to PARP Inhibitors. Cancer Discov. 2017;7:999–1005. doi: 10.1158/2159-8290.CD-17-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmons AD, Nguyen M, Pintus E. Polyclonal BRCA2 mutations following carboplatin treatment confer resistance to the PARP inhibitor rucaparib in a patient with mCRPC: a case report. BMC Cancer. 2020;20:215. doi: 10.1186/s12885-020-6657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norquist B, Wurz KA, Pennil CC, Garcia R, Gross J, Sakai W. et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011;29:3008–15. doi: 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S. et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–94. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 17.Barber LJ, Sandhu S, Chen L, Campbell J, Kozarewa I, Fenwick K. et al. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J Pathol. 2013;229:422–9. doi: 10.1002/path.4140. [DOI] [PubMed] [Google Scholar]
- 18.Goodall J, Mateo J, Yuan W, Mossop H, Porta N, Miranda S. et al. Circulating Cell-Free DNA to Guide Prostate Cancer Treatment with PARP Inhibition. Cancer Discov. 2017;7:1006–17. doi: 10.1158/2159-8290.CD-17-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen B, Zhang G, Li X, Ren C, Wang Y, Li K. et al. Comparison of BRCA versus non-BRCA germline mutations and associated somatic mutation profiles in patients with unselected breast cancer. Aging (Albany NY) 2020;12:3140–55. doi: 10.18632/aging.102783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B, Zhang G, Wei G, Wang Y, Guo L, Lin J. et al. Heterogeneity of genomic profile in patients with HER2-positive breast cancer. Endocr Relat Cancer. 2020;27:153–62. doi: 10.1530/ERC-19-0414. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tobalina L, Armenia J, Irving E, O'Connor MJ, Forment JV. A meta-analysis of reversion mutations in BRCA genes identifies signatures of DNA end-joining repair mechanisms driving therapy resistance. Ann Oncol. 2021;32:103–12. doi: 10.1016/j.annonc.2020.10.470. [DOI] [PubMed] [Google Scholar]
- 28.Pettitt SJ, Frankum JR, Punta M, Lise S, Alexander J, Chen Y. et al. Clinical BRCA1/2 Reversion Analysis Identifies Hotspot Mutations and Predicted Neoantigens Associated with Therapy Resistance. Cancer Discov. 2020;10:1475–88. doi: 10.1158/2159-8290.CD-19-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murciano-Goroff YR, Schram AM, Rosen E, Janjigian YY, Berger MF, Donoghue M. et al. BRCA reversion mutations in a pan-cancer cohort to reveal BRCA-dependence in select noncanonical BRCA-mutant histologies. Journal of Clinical Oncology. 2021;39:3012. - [Google Scholar]
- 30.Chan GHJ, Ong PY, Low JJH, Kong HL, Ow SGW, Tan DSP. et al. Clinical genetic testing outcome with multi-gene panel in Asian patients with multiple primary cancers. Oncotarget. 2018;9:30649–60. doi: 10.18632/oncotarget.25769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogt A, Schmid S, Heinimann K, Frick H, Herrmann C, Cerny T. et al. Multiple primary tumours: challenges and approaches, a review. ESMO Open. 2017;2:e000172. doi: 10.1136/esmoopen-2017-000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abida W, Armenia J, Gopalan A, Brennan R, Walsh M, Barron D, Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis Oncol. 2017. 2017. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure and table.