Abstract
Mycobacterium tuberculosis is the etiologic agent of tuberculosis and can be accurately detected by laboratories using commercial genetic tests. Nontuberculosis mycobacteria (NTM) causing other mycobacterioses can be difficult to identify. The identification processes are confounded by an increasing diversity of newly characterized NTM species. The ubiquitous nature of NTM, combined with their potential to be opportunistic pathogens in immunocompromised as well as nonimmunodeficient patients, further complicates the problem of their identification. Since clinical case management varies depending on the etiologic agent, laboratories must identify the species in a timely manner. However, only a few identification methods can detect the species diversity within the Mycobacterium genus. Over the last decade, high-performance liquid chromatography analysis of the mycolic acids has become an accepted method for identification of mycobacteria. In this review, we assess its development and usefulness as an identification technique for Mycobacterium species.
INTRODUCTION
Early strategies employed to identify species of the Mycobacterium genus have included observations of staining properties of bacilli, cultural morphology, biochemical tests, and, rarely, the inoculation of susceptible animals with live bacilli for observation of animal pathogenicity. These tests were designed to discriminate among mycobacteria involved in disease and were directed toward detecting Mycobacterium tuberculosis, Mycobacterium bovis, Mycobacterium avium, or saprophytes (57–59). When other Mycobacterium species were recognized as infectious agents, it was obvious that additional differentiation criteria were needed. A classification system based on pigmentation and growth rate was introduced to define the occurrence of “atypical” (a term presently in disfavor in mycobacterial nomenclature) strains and their relationship to the Mycobacterium species perceived as pathogenic (80, 95). The “slow growers” were defined as having visible growth in >7 days and were categorized in the following groups: group I, photochromogens; group II, scotochromogens; and group III, nonphotochromogens. “Rapid growers” were defined as having visible growth in <7 days, and they were designated group IV. Although this simple system is not used as extensively now, its longevity is demonstrated by references in publications and frequent communications between mycobacteriologists. However, these simple designations are not practical or sufficient for defining species within the Mycobacterium genus.
In related efforts, members of the International Working Group on Mycobacterial Taxonomy (IWGMT) made significant contributions to mycobacterial identification and taxonomy. Their collaborative studies evaluated various groups of mycobacteria and related genera, defined variation in members of a given species, and proposed selected tests for routine species identification. These extensive studies, involving numeric taxonomy, clarified the phenetic integrity of the Mycobacterium genus (42, 106) and provided a practical biochemical identification scheme for clinically important species of Mycobacterium (107, 108). During this time, just over 20 of the known 54 species were regarded as potentially pathogenic, and the recommended tests appeared applicable for identification of these species (55). Eventually, as the number of species increased, the resulting taxonomical complexity caused ambiguities in the interpretation of biochemical test results due to their reduced discrimination ability.
Over the years, the unequivocal identification of M. tuberculosis and species of clinical interest has dominated the taxonomy of the mycobacteria (106, 111). This emphasis on identification of the most commonly recovered species by clinical laboratories prompted a development of genetic probes (Gen-Probe, San Diego, Calif.) for their detection. Laboratories using these genetic tests rarely misidentified species for which the tests were designed. However, tests were not developed for the majority of the mycobacteria, because they were not considered a serious threat to the public health, inasmuch as the majority of the species were infrequently isolated and were never transmitted from person to person. On occasion, these species produced serious (even fatal) infections, especially in patients with a reduced immune response. In these cases, rapid and accurate identification of the clinical species can be beneficial for effective medical intervention.
In this report, we will examine the development, introduction, and effectiveness of high-performance liquid chromatography (HPLC) analysis of the mycolic acids for chemotaxonomic classification and rapid identification of Mycobacterium species. (For a historical overview of liquid chromatography leading to the development of HPLC, the reader is referred to reference 49.) It is not within the scope of this review to examine every method proposed or currently used for the identification of the mycobacteria.
(Some of the chromatograms shown in this review were presented at the 96th General Meeting of the American Society for Microbiology by L. S. Guthertz, P. S. Duffey, and W. R. Butler.)
LABORATORY
HPLC principles and instrumentation originated in the field of chemistry during the mid-1960s and found widespread application in inorganic, organic, and biochemical areas (49). Clinical chemistry sections exploited the versatility of the method for rapid separation and identification of compounds for drug analysis (26, 38, 49). In 1985, scientists at the Centers for Disease Control and Prevention (CDC) proposed the use of HPLC as an aid in mycobacterial classification (10). In 1989, the procedure was incorporated into the regiment of tests at the CDC Mycobacteriology Reference Laboratory, and in 1990, it was offered as a standard test for identification of Mycobacterium species (40).
HPLC analysis of mycolic acids for species identification offered a reprieve from the traditional time-consuming identification process (55, 84). An isolate submitted on culture could be analyzed in hours by HPLC, compared to weeks or longer for routine methods. However, the suitability of the method should be evaluated prior to incorporation into the laboratory. HPLC is considered a sophisticated procedure compared to other laboratory methods, and a dedicated, highly trained operator is required (W. Gross, personal communication). Instrumentation is costly compared to conventional methods. Laboratories that detect acid-fast bacilli only by smears may not have the capacity to develop a proficiency with the HPLC method. However, many high-throughput laboratories that are proficient in all aspects of mycobacteriology have successfully incorporated HPLC into their methodologies (14, 21, 29, 45, 52, 93, 99). A significant challenge for laboratory personnel lies in developing expertise in the visual interpretation of chromatographic patterns for species determination. In this report, 63 chromatographic patterns representative of 73 known mycobacteria species are presented and may help with this difficulty.
MYCOLIC ACIDS
The analysis of lipid fractions has contributed significantly to the knowledge of Mycobacterium species. The abundance of lipid constituents in mycobacterial cell walls made them classic candidates for early chemical investigations. Of intense interest to researchers was a difficult-to-purify wax fraction, initially termed “unsaponifiable wax” (90). Isolated after prolonged saponification, it was clarified as an alcohol-insoluble, hydroxy acid of high molecular weight. The saponified wax fraction was chemically stable and was named “mycolic acid” following its isolation from the original H37 strain of M. tuberculosis. Complete saponification required 80 h of reflux in methanolic potassium hydroxide and benzene. Chemically, the mycolic acid fraction stained acid fast, contained hydroxy and methoxy groups with approximately 88-carbon chain lengths, and displayed a pyrolysis product with 26 carbons when heated at 300 to 350°C (90). Further structural studies confirmed that the hydroxy group of mycolic acid was in a β-position to the carboxyl group and thus defined the mycolic acids as high-molecular-weight β-hydroxy fatty acids with a long α-side chain (2). Additional studies demonstrated that the structural character of the mycolic acids was a mycolic acid-arabinogalactan-mucopeptide complex, an essential part of the cell wall whose abundance was not adversely affected by culture conditions (53, 61). This consistent production of mycolic acids was noted to be a stable phenotypic property for a species (31, 42, 54, 69).
The diagnostic value of the mycolic acids was quickly recognized by early chemical investigators who characterize the different mycolic acid-containing genera (1, 32, 61). The chemical complexity of members of the different taxa was demonstrated by gas-liquid chromatography, and single-dimension, thin-layer chromatography (1D-TLC) analysis of mycolic acid as methyl esters (32, 60). Methanolysates of mycobacterial mycolic acids were differentiated in 1D-TLC by production of multispot patterns, in contrast to single-spot patterns produced by species in related taxa (65, 69). Mycobacterium species were also chemically distinguished from those of other genera by the presence of C22 to C26 products from gas-liquid chromatography pyrolysis of mycolic acid methyl esters (32, 61, 69). Mycobacterium species were rapidly distinguished from Nocardia or Rhodococcus species by precipitation of mycobacterial mycolic acids from a solution of ether by the addition of alcohol, whereas the mycolic acids from the other genera remained soluble (54).
In addition, it was shown that mycobacteria and other mycolic acid-containing taxa could be distinguished using 2D-TLC of mycolic esters as whole-organism methanolysates (61, 65). This method was combined with mass spectrometry (MS) and used to identify and chemically characterize the functional groups of the mycolic acids, thus demonstrating the species-specific nature of the mycolates. Extensive studies described mycobacterial mycolic acids as phenotypically stable chemical characters, having a discontinuous distribution with chemotaxonomic potential (68, 69). Because mycolic acids were confined in related taxa and not widely distributed in nature, they were ideal for discrimination studies (70). The impressive work done with 2D-TLC and MS advanced the knowledge of the structure and complexity of mycobacterial mycolic acids. Analysis of 50 species of mycobacteria by 2D-TLC revealed that the mycobacteria were separated into 11 structural groups based on the composition of their mycolic acids, and the analysis was suggested as applicable in clinical laboratories for rapid identification of mycobacteria (69). These comprehensive studies defined the value of mycolic acid data in mycobacterial systematics and led to the recommendation that minimal descriptions for a new species of Mycobacterium include information on the chemical composition of mycolic acids (62). Presently, all the mycolic acid-containing genera can be distinguished by the length of their mycolic acid carbon chains. They include Corynebacterium, C20 to C38; Dietzia, C34 to C38; Rhodococcus, C34 to C52; Nocardia, C40 to C60; Gordonia, C48 to C66; Williamsia, C50 to C56; Skermania, C50 to C64; Tsukamurella, C64 to C78; and Mycobacterium, C60 to C90.
HIGH-PERFORMANCE LIQUID CHROMATOGRAPHY
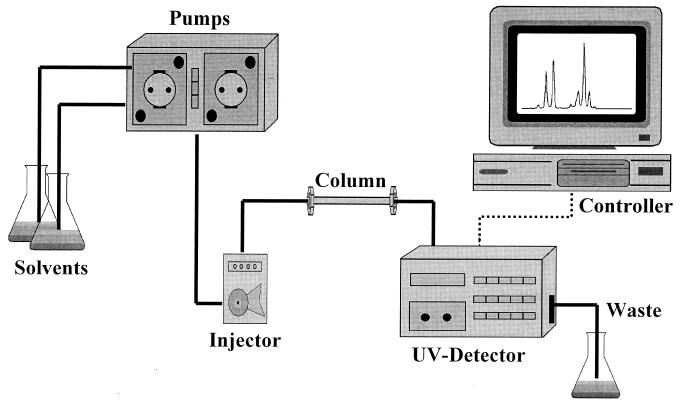
In the mid to late 1970s, the newly developed method was referred to as high-pressure liquid chromatography. The pressure was produced by pumps which forced a sample suspended in a liquid through a solid stationary phase, enclosed in a metal column, resulting in the rapid fractionation of sample components. The separated components were distinguished by a detector and depicted as peaks by a recorder (Fig. 1). The peaks were defined by specific elution, emergence, or retention times. Component fractionation was shown to be a complex intermixture of chemical reactions and physical factors occurring between the sample, the mobile phase, and the solid stationary phase. (For a review of the basic technique of liquid chromatography, the reader is referred to reference 113.)
FIG. 1.
Representative HPLC system with dual pumps for gradient elution of solvents.
HPLC separation methods permitted researchers interested in the biosynthesis and biological properties of the mycolic acids to fractionate and collect “pure” compounds and confirmed earlier TLC and nuclear magnetic resonance data of chemical structure (73). The sensitivity of mycolic acid detection was increased by derivatization to UV-adsorbing p-bromophenacyl esters (PBPA), (73, 89). Subsequent studies used multiple step methods with TLC, MS, and reverse-phase chromatography to achieve fractionation of discrete components of the mycolic acids (89, 92). These reverse-phase chromatography procedures utilized high-efficiency C18, microparticle-bonded stationary-phase columns with mixtures of organic solvents. Chromatographic methods combined reverse-phase HPLC and MS analysis of cell wall extracts to demonstrated the chemical nature of mycolic acids from M. tuberculosis strain H37Ra (73, 92, 112). Fractions were separated into chemically homologous mycolic acid series, consisting of 24 fractions from species in the M. tuberculosis complex and Mycobacterium smegmatis (92). It was noted that the mycolic acids separated in the reverse-phase column by a combination of factors, including chemical functional groups, polarity, and hydrocarbon chain length (89, 91, 92). In prior studies, similar results had been demonstrated with shorter-carbon-chain fatty acids, derivatized with chemically analogous phenacyl derivatives which had demonstrated an increase in retention times with longer carbon chains but a decrease in retention times with greater unsaturation (5, 30, 51, 79, 112). The separation properties of the mycolic acids were used to resolve an ongoing debate regarding the similarity of Mycobacterium gordonae and Mycobacterium leprae (67). In an elaborate two-step HPLC procedure combined with TLC, the presence of specific methoxymycolates was demonstrated in M. gordonae and was absent in M. leprae; thus, the close relationship was not supported. It was reported that reverse-phase HPLC profiles for α-mycolate, methoxymycolate, and ketomycolate chemical functional groups demonstrated increasing retention times with increased carbon chain length. Moreover, it was noted that the mycolic acids were not completely chemically resolved by HPLC when subjected only to reverse-phase partition chromatography (73, 89). Consequently, chromatographic patterns were published of mycolic acids for mycobacteria that demonstrated extensive overlapping of the chemical fractions (73, 89, 92). The recognition of pattern differences between the published chromatograms for members of the M. tuberculosis complex (slow-grower) and M. smegmatis (rapid-grower) by one of us (W.R.B.) provided the stimulus for further research with the HPLC instrument as a tool for species detection. These and other chemical studies demonstrated the separation capabilities of the various methods and the discriminatory power of HPLC.
APPLICATION OF HIGH-PERFORMANCE LIQUID CHROMATOGRAPHY FOR IDENTIFICATION OF MYCOBACTERIA
Phenacyl esters of fatty acids were separated with a single-step, C18 reverse-phase HPLC method utilizing UV detection (UV-HPLC) (30). The procedure employed a dicyclohexyl-18-crown-6 ether (crown ether) catalyst for a rapid (30-min) derivatization of long-chain fatty acids detectable in the nanogram range. The use of this derivative provided a sensitive detection method for complete separation of mycolic acids from mycobacteria into structural classes with silica and reverse-phase modes (73, 92). Moreover, complete chemical separation of the mycolic acids was not necessary for species detection, since the reverse-phase pattern with gradient elution appeared distinctive for M. smegmatis, BCG, M. bovis, and M. tuberculosis (89).
The reverse-phase UV-HPLC method was used with a modified gradient elution system of chloroform-methanol to differentiate 13 Mycobacterium species into chromatographically related groups (10). Within each chromatographic group, distinctive mycolic acid patterns were used for species classification of mycobacteria. However, the mycolic acid isolation procedure was cumbersome and involved multiple chloroform extractions. Moreover, the reflux-condensation apparatus used for sample derivatization was time-consuming; consequently, few samples could be analyzed in several days.
These early UV-HPLC investigations supported prior studies done with mycolic acids which included the differentiation of Corynebacterium, Rhodococcus, Nocardia, and Mycobacterium according to the carbon chain lengths of the mycolic acids (25, 69) and separation of Rhodococcus species into two groups composed of mycolic acids with either short or long carbon chains (1, 17, 65). Investigators eventually demonstrated that the species with the long carbon chains were a unique group and transferred them to the revived genus Gordona (88, 104). Also, in agreement with prior methods, the separation of mycobacteria from other mycolic acid-containing bacteria by UV-HPLC analysis of PBPA esters of mycolic acids was shown to result from an attraction of the different carbon chain lengths to the column's nonpolar stationary phase, which resulted in patterns with different retention times (11, 17, 91).
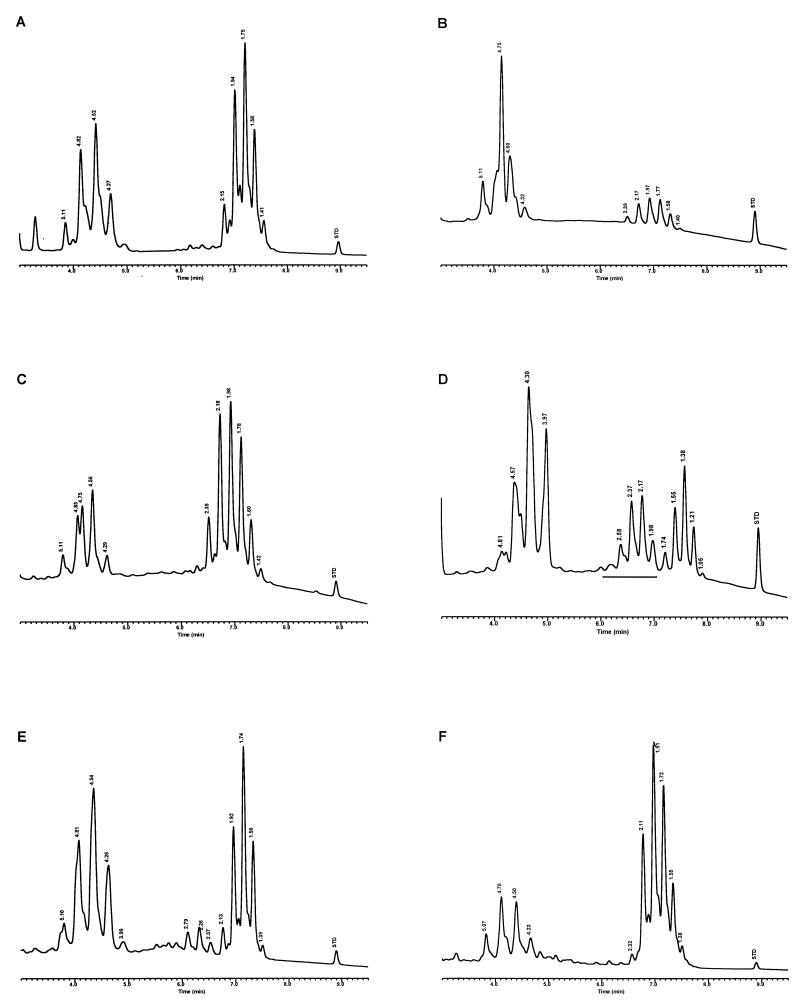
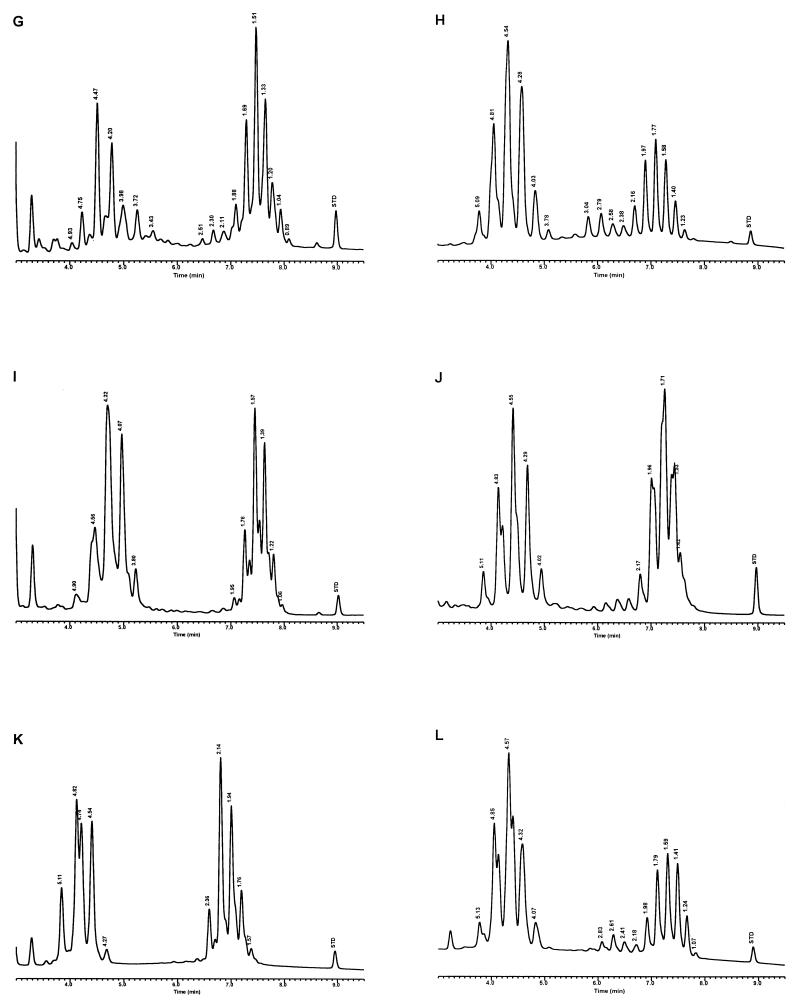
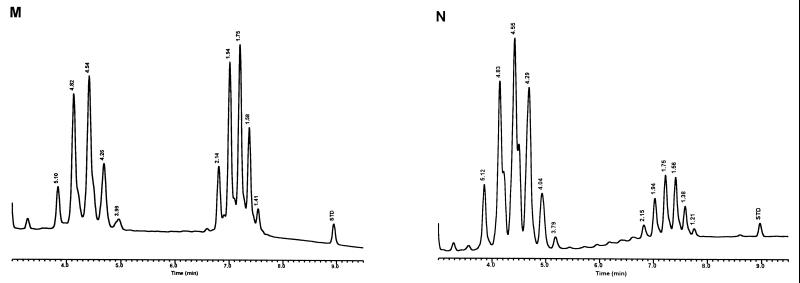
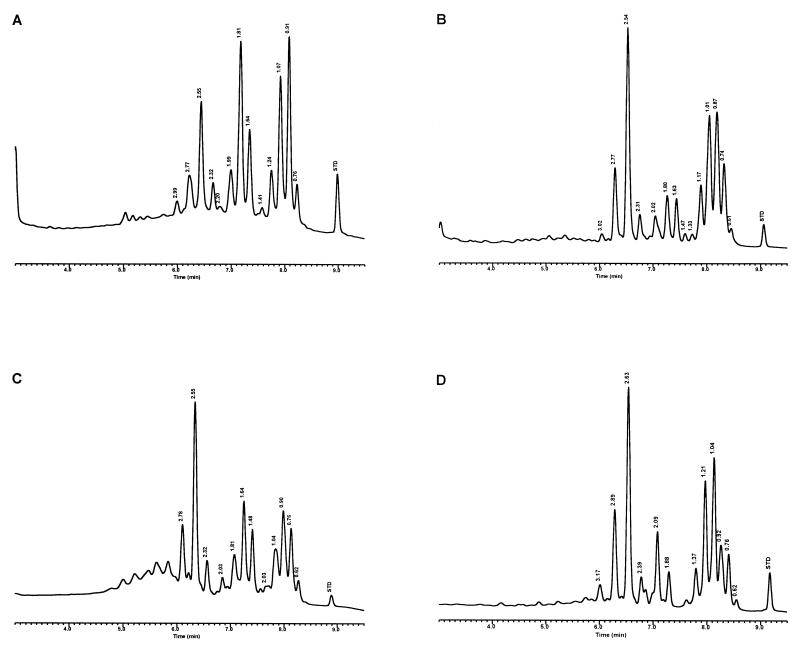
Modifications were made to the UV-HPLC method in studies with species of slow-growing pathogenic mycobacteria and the nonpathogenic M. gordonae (15). Whole cells of mycobacteria, grown on Löwenstein-Jensen (L-J) slants, were removed either by using a transfer loop or by washing the cells with 25% solution of potassium hydroxide in 50% ethanol and analyzed solely by reverse-phase UV-HPLC. The cells were saponified in glass tubes for 18 h at 85°C, and the extracted mycolic acids were derivatized for 45 min at 85°C to produce the UV-absorbing PBPA esters. The mycolic acids were fractionated with a 5-μm-particle-size, reverse-phase C18 column using a gradient elution system that was equilibrated for 13 min in chloroform-methanol (10:90, vol/vol), followed by a 1-min adjustment to chloroform-methanol (25:75, vol/vol), which rapidly eluted the more polar sample components, and a final 20-min linear gradient of chloroform-methanol (70:30, vol/vol) to elute the mycolic acids. Distinctive mycolic acid patterns consisted of a single cluster of contiguous peaks for M. tuberculosis, Mycobacterium kansasii, and the nonpathogenic M. gordonae (Fig. 3). A detection sensitivity for the method was defined with M. kansasii at 2.5 × 106 CFU. The reproducibility of the method for the single contiguous peak pattern was demonstrated by examination of the mycolic acid pattern for a continuously growing culture of M. tuberculosis sampled periodically for 3 weeks. Although slight variations in peak heights and times were noted, the basic pattern did not change. In contrast to these single peak clusters, Mycobacterium avium and Mycobacterium intracellulare produced disunited double-clustered peak patterns (see Fig. 8). These early studies required 20 h to identify a species (15).
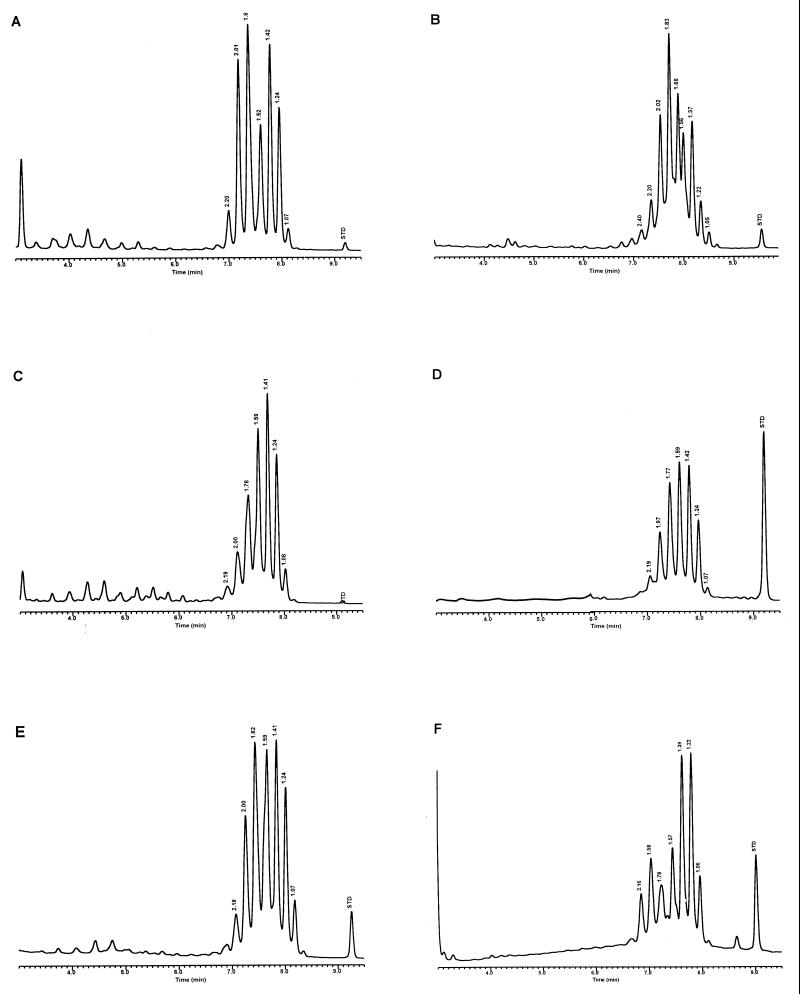
FIG. 3.
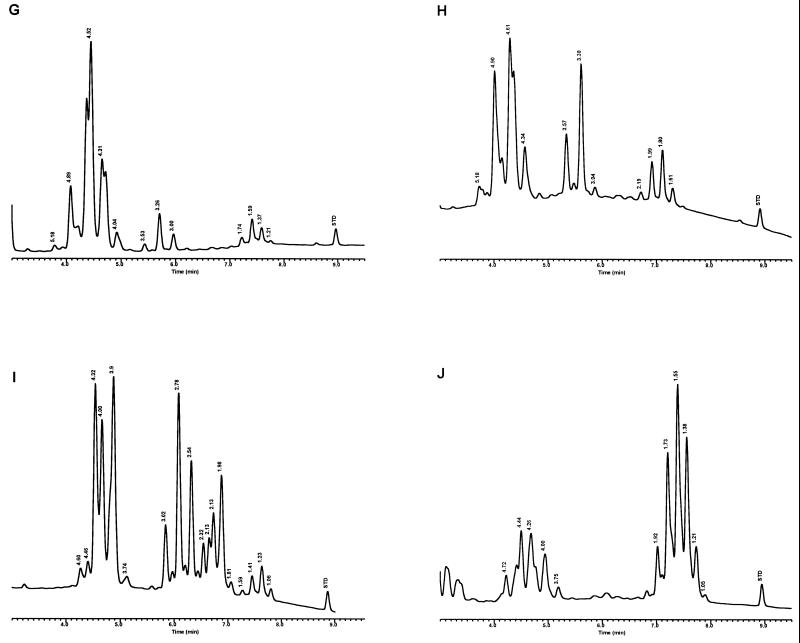
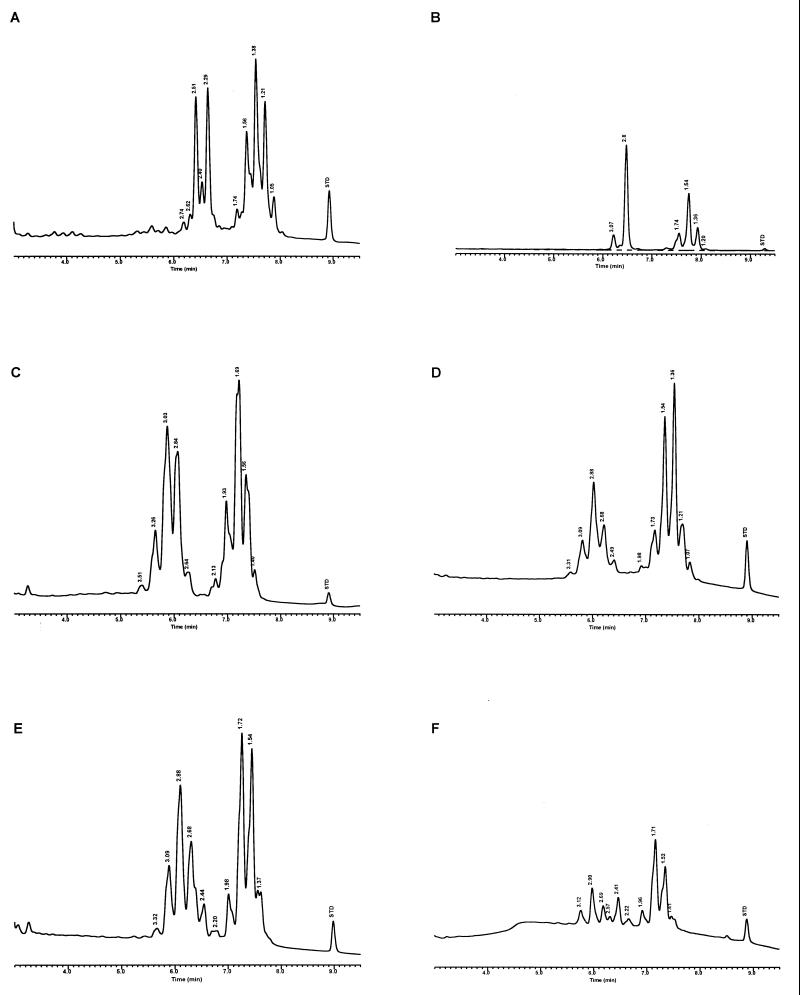
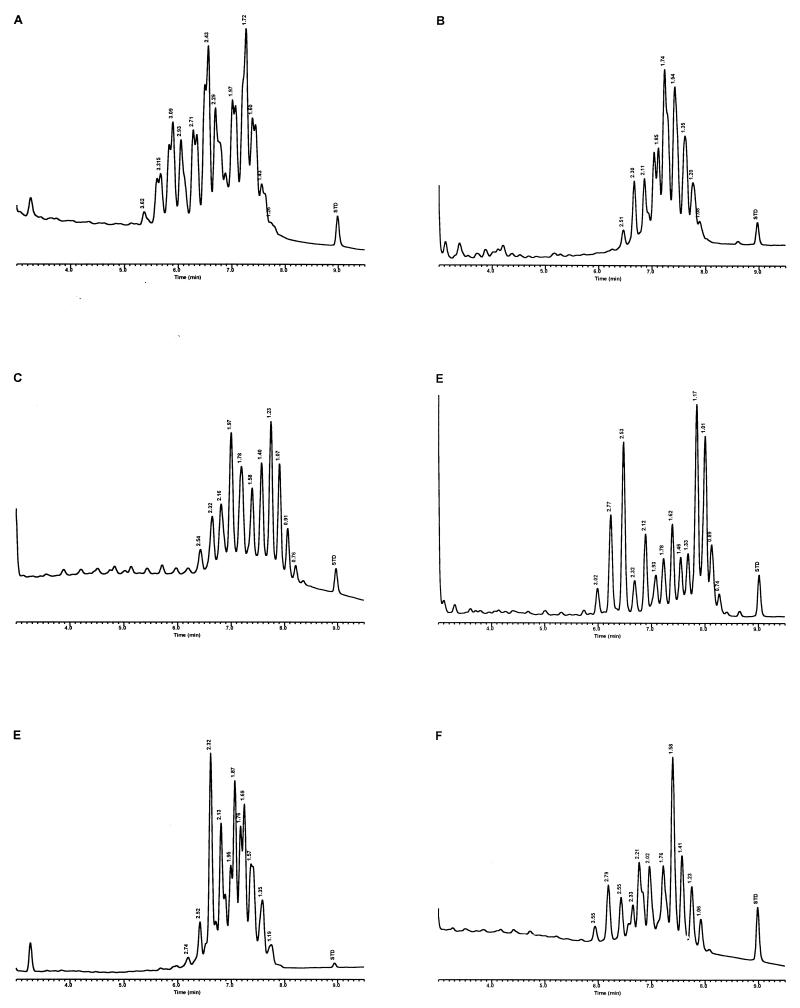
Characteristic UV-HPLC chromatograms of Mycobacterium species with late-emerging, simple, single-cluster peak patterns. (A) M. asiaticum ATCC 25276T; (B) M. bovis BCG Pasteur; (C) M. gastri ATCC 15754T; (D) M. gordonae ATCC 14470T, UV-HPLC chromotype I; (E) M. kansasii, ATCC 12478T; (F) Mycobacterium leprae, ‘armadillo’; (G) M. tuberculosis complex (includes M. africanum ATCC 25420T, M. bovis ATCC 19210T, “M. canettii” NZM 217/94, M. caprae CIP 105776T, M. microti ATCC 19422T, and M. tuberculosis ATCC 27294T); (H) M. szulgai ATCC 35799T.
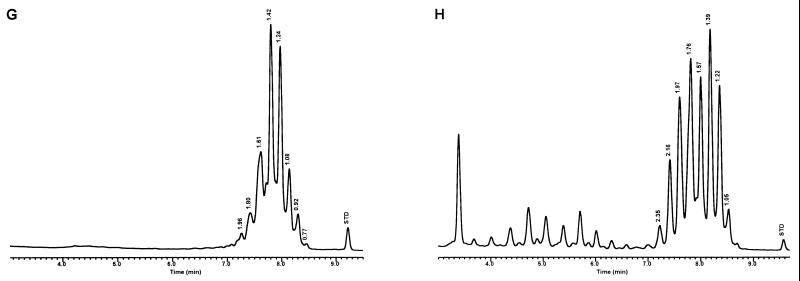
FIG. 8.
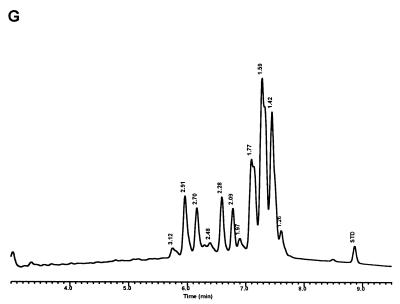
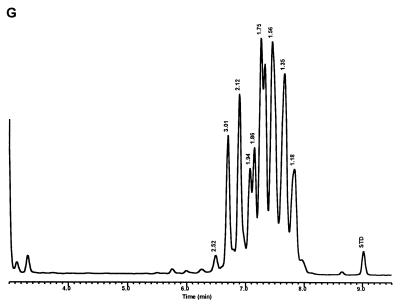
Characteristic UV-HPLC chromatograms of Mycobacterium species that have widely separated, double-peak clusters with prominent peaks in the early cluster that emerge after 5.0 min. (A) M. avium ATCC 19698T; (B) Mycobacterium branderi ATCC 51789T; (C) M. celatum ATCC 51131T; (D) M. gordonae ATCC 35759 (UV-HPLC chromotype II); (E) M. interjectum ATCC 51457T (underlined peaks are not present in all strains); (F) M. intracellulare ATCC 13950T; (G) M. scrofulaceum ATCC 19981T; (H) M. terrae ATCC 15755T; (I) M. xenopi ATCC 19250.
Additional improvements to the method were made in a study with rapidly growing mycobacteria of clinical significance. It was shown that organisms could be killed and fatty acids could be saponified in the autoclave for 1 h at 121°C. Mycolic acids were extracted, derivatized, and processed in 3 h (16). Similar but distinctive chromatographic patterns were developed for Mycobacterium chelonae and Mycobacterium abscessus under controlled growth conditions. On the other hand, 122 strains of the Mycobacterium fortuitum complex and M. smegmatis could not be separated under these conditions. The stability of the mycolic acid patterns demonstrated by UV-HPLC for slow-growing mycobacteria was in contrast to that of rapidly growing mycobacteria, which were affected by culture conditions, especially temperature (16). Related studies of rapidly growing mycobacteria demonstrated either an elongation or a shortening of mycolic acids as an adaptive response to changes in temperature, although the effects appeared to be species specific (3, 96).
During the study with rapid-growing mycobacteria, a proprietary high-molecular-weight standard synthesized by Ribi ImmunoChem (now Corixa Corp.), Hamilton, Mont., was used for the first time as an internal standard to determine reproducible relative retention times (RRT) with a standard deviation of ±0.01 to 0.09 min for individual peaks (16). This standard was used to devise an identification scheme to compare peak height ratios, calculated for peaks from different chromatograms with identical RRTs. The identification scheme required manual calculation of values and was time-consuming but correctly identified 36 isolates of M. chelonae and 24 of 25 isolates of M. abscessus (16). A subsequent 10-step dichotomous peak height differentiations scheme was reported for slow-growing species and tested at two different laboratories. At one laboratory, 129 isolates of the following species were correctly identified: Mycobacterium asiaticum, M. bovis, BCG attenuated variants of M. bovis, M. tuberculosis, Mycobacterium gastri, M. gordonae, M. kansasii, Mycobacterium marinum, and Mycobacterium szulgai. The other laboratory identified 661 of 670 different strains of these same species. The misidentifications occurred with two species, M. kansasii and M. gordonae. Overall, 790 strains (98.6%) were correctly identified by both laboratories (14).
Final modifications of the chromatographic conditions were evaluated by two laboratories with closely related, slow-growing Mycobacterium species (19). The robustness of the dichotomous identification scheme and UV-HPLC were challenged with mycolic acid extracts from M. avium, M. intracellulare, Mycobacterium scrofulaceum, Mycobacterium xenopi, and some phenotypically confusing M. gordonae. A gradient system of methanol-methylene chloride with a constant solvent flow of 1.5 ml/min was programmed to change from polar to nonpolar as follows. Initially, the column was equilibrated at 98%-2% (vol/vol), and then it was changed linearly after injection for 1 min to 80%-20% (vol/vol); thereafter, the solvents were changed linearly for 9 min to a final composition of 35%-65% (vol/vol). Final column reequilibration was carried out for 2 min at the beginning solvent concentration, for an analysis time of 12 min. The by-products of the derivatization procedure and the cellular short-carbon-chain fatty acids eluted rapidly at the beginning of the separation and did not interfere with the elution of the mycolic acids. Identifications with the flow chart scheme compared with results from control biochemical tests for M. avium, M. intracellulare, and M. scrofulaceum were 97.9, 97.5, and 89.2%, respectively. All strains of M. xenopi and M. gordonae were correctly identified. These refinements produced a standard method and permitted a single Mycobacterium isolate received by the laboratory as a fully grown culture to be identified in approximately 2 h (19).
STANDARD METHOD FOR PREPARATION OF MYCOLIC ACIDS
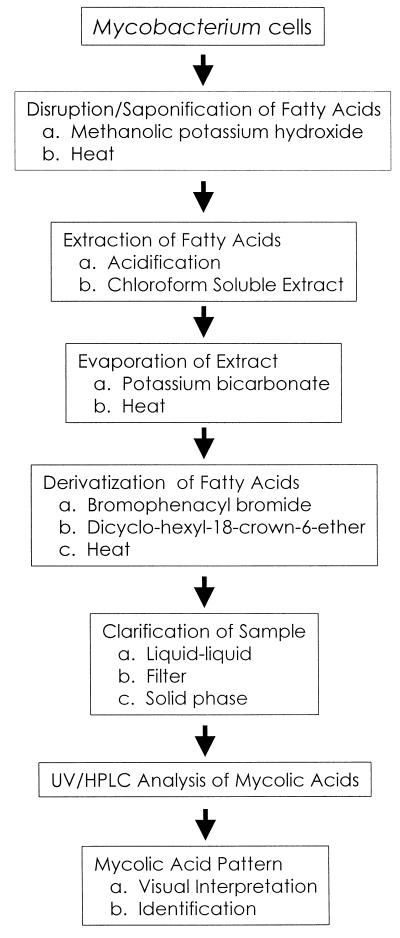
The sample preparation protocol consisted of cell harvesting, saponification, extraction, derivatization, and a cleanup or clarification step to generate mycolic acids amenable to detection by UV-HPLC (Fig. 2). The procedure involved a whole-cell saponification, acidification, and methanolic extraction of all cellular fatty acids, including the mycolic acids. Mycobacteria were removed from a Loẅenstein-Jensen slant with a sterile polyester swab and placed in 2 ml of saponification reagent composed of 25% potassium hydroxide in a water-methanol (1:1, vol/vol) mixture. The suspension was vigorously mixed and autoclaved for 1 h at 121°C. After the mixture had cooled to room temperature, 2 ml of chloroform was added, and then 1.5 ml of acidification reagent, a mixture of water and concentrated hydrochloric acid (1:1, vol/vol) was added slowly. This solution was mixed, and the layers were separated. The lower, organic layer was transferred with a Pasteur pipette to a new tube and evaporated in a heat block at 85 to 105°C with a stream of air. The commercial derivatization reagent was suspended in acetonitrile and consisted of PBPA and a crown ether catalyst (30). A mycolic acid sample was derivatized to PBPA esters by adding 20 mg of potassium bicarbonate, 50 μl of derivatization reagent, and 1.0 ml of chloroform. The tubes were secured with Teflon-lined screw caps and heated at 85 to 105°C for 20 min. Samples were cooled and then clarified by a choice of methods, including either filtration through a 0.45-μm-pore-size nylon 66 membrane filter (10, 15), solid-phase extraction (27), or liquid partitioning with an acidified aqueous methanolic solution (19). Derivatization of the mycolates to UV-absorbing PBPA esters resulted in a stable compound which could be stored for several months in the dark at 4°C. Specific details of the method are presented in standardization manuals (12, 13).
FIG. 2.
Flowchart for isolation and detection of mycolic acids.
DIFFERENTIATION OF MYCOBACTERIUM SPECIES BY UV–HIGH-PERFORMANCE LIQUID CHROMATOGRAPHY
The reproducibility of mycolic acid patterns from mycobacteria has been established by laboratory studies conducted under the auspices of the IWGMT. The first study employed molecular, biochemical, serologic, and chemotaxonomic methods in an attempt to identify closely related species suspected of being M. avium, M. intracellulare, or M. scrofulaceum (110). In fact, the taxonomical complexity of the study group was demonstrated when all the methods were combined and a consensus could not be reached for identification of four of the isolates. Overall, UV-HPLC results agreed with results from T-catalase serology for 86% of the isolates but were not as good as the 94% agreement between nucleic acid tests and T-catalase serology. The UV-HPLC result was comparable to the 84% result for laboratories using panels of sera that corresponded to the serovars represented in the study (110). Another IWGMT study with problematic phenotypic clusters of slowly growing mycobacteria demonstrated species-specific chromatographic patterns for Mycobacterium malmoense, Mycobacterium interjectum, and Mycobacterium simiae. However, some M. avium complex (MAC) species appeared to be misidentified when chromatographic results were compared with the molecular data. It was suggested that when chromatographic techniques were used for identifying MAC species, the results be supplemented with biochemical tests (109).
This relationship between UV-HPLC and standard biochemical tests was compared in a year-long study with 502 cultures of Mycobacterium and demonstrated a 97.2% agreement (45). Additionally, chromatographic patterns were compared with DNA probe results for 111 cultures that were designated MAC strains, and they demonstrated an agreement of 98.2%. Also, in this study, a positive reaction with a MAC genetic probe for a single isolate was clarified as a mixed culture by UV-HPLC. In another study UV-HPLC, biochemicals, and genetic probe results were compared in routine use for 18 months (93). UV-HPLC identified 96.1% of the 1,103 strains tested. Biochemical and/or DNA probes identified 98.9% of the strains. The chromatographic method accurately identified M. chelonae, M. xenopi, Mycobacterium terrae complex, M. simiae, and the rarely encountered species Mycobacterium haemophilum, M. malmoense, Mycobacterium shimoidei, and Mycobacterium fallax. UV-HPLC results were compared to genetic probe results and were 100% specific and 98.9% sensitive for M. tuberculosis complex. UV-HPLC identified 250 isolates as MAC, which was in 98.7% agreement with results obtained with genetic probes. Thirty-one strains were identified by UV-HPLC as M. scrofulaceum, and 90.3% of these strains were negative for MAC by genetic probes. Moreover, the accuracy of the genetic probe tests for M. tuberculosis complex and MAC species was confirmed for discrepant strains that produced negative results by detection of other species of nontuberculous mycobacteria (NTM) with UV-HPLC (4, 83).
Detection of unknown mycobacteria by UV-HPLC supports the belief that many species of mycobacteria remain uncharacterized. Unusual mycobacteria have been recovered from AIDS patients in liquid media from a BACTEC (Becton Dickinson Diagnostic Instrument Systems, Sparks, Md.) instrument (22, 85). The identity of the isolates was problematic because they failed to grow when subcultured on conventional solid medium used for mycobacteria. Dysgonic growth obtained in Middlebrook 7H11 medium supplemented with mycobactin J produced a mycolic acid pattern with three separate peak clusters (22). Similar mycolic acids were also detected in Bactec 12B vials by adding oleate-albumin-dextrose-catalase and by increasing the incubation 3 to 5 days beyond the final 999 growth index determination (21, 48, 85). In both studies, a triple-clustered mycolic acid pattern similar to that for M. simiae was reported but the chromatographic data were not definitive for a known species. Investigators performing genetic sequencing analysis clarified the mycobacteria as a novel species, Mycobacterium genavense (22, 78). Another unique chromatographic pattern was reported from an outbreak of peritonitis involving the use of dialysis machines (105). The biochemical reactions resembled those of M. chelonae; consequently, the isolates were referred to as M. chelonae-like organism. However, the UV-HPLC pattern and drug susceptibility characterizations were distinctive, which implied a novel species. Ultimately, the M. chelonae-like organism group was described as a new species, Mycobacterium mucogenicum (87, 105). Other characterization studies of new species have included mycolic acid patterns from UV-HPLC analysis in their initial description and include Mycobacterium celatum (18), Mycobacterium heidelbergense (46), Mycobacterium goodii (8), Mycobacterium septicum (82, 105), and Mycobacterium wolinsky (8). Also, Mycobacterium triplex and Mycobacterium tusciae were initially recognized by their UV-HPLC mycolic acid patterns (34, 101). Novel mycolic acid patterns are easily detected by UV-HPLC. However, when mycolic acid patterns from undefined isolates resemble those of known species, differentiation may be difficult (74). This was demonstrated in a characterization study with a suspected unknown environmental saprophyte with a mycolic acid pattern that was similar to those of several known rapid growers; this organism was later shown to be a variant of Mycobacterium austroafricanum by molecular methods (7).
Infrequent isolation of fastidious or uncommon mycobacteria does not offer laboratory personnel the opportunity to develop the expertise needed for their identification. Development of mycolic acid patterns can provide the results necessary to make a final decision. Such was the case for unusual isolates from cutaneous lesions, a lymph node and eye of a patient with AIDS, and a cervical lymph node of a 3-year-old child, which were suspected to be M. haemophilum (94). This initial identification was based on low growth temperature and a requirement for iron. However, the identification was suspect because the organism was rarely isolated from patients and the incidence of infection was unknown for this laboratory. The species was confirmed by comparison of the isolate's mycolic acid pattern to reference patterns by UV-HPLC similarity.
In a restricted study, the etiologic agents for six cases of chronic tenosynovitis of the hand were all identified as Mycobacterium nonchromogenicum, a member of the M. terrae complex, by UV-HPLC (77). Standard biochemical tests were reported unreliable for routine discrimination of members of this complex (77, 97). The final identification was made by a concurrence of the mycolic acid pattern with a susceptibility to the drug ofloxacin (77, 87). Interestingly, mycolic acid patterns were recently correlated with drug susceptibility results for strains of M. tuberculosis (36).
Initially, representative chromatographic patterns of known Mycobacterium species were determined by examination of type strains from American Type Culture Collection or Collection de I'Institut Pasteur grown with standardized growth conditions. However, chromatographic pattern variation was demonstrated when multiple strains of a species were examined. Normally, this pattern variation was in the form of minor peak height differences and did not affect the overall appearance of the chromatogram (15). Infrequently, known species that normally produced multiple-cluster mycolic acid patterns would develop a pattern with a cluster of peaks dramatically reduced in height or altogether absent (13). For example, M. interjectum strains produced patterns with a single cluster of peaks that differed from the biphasic pattern for which the species was known (64, 100). It was speculated that the discrepant patterns resulted from analyzing variants of the same species but with different mycolic acid patterns. A similar phenomenon had been reported for 14 of 63 isolates of M. gordonae, except that the variant pattern was biphasic compared to the normal single-cluster pattern (20). The frequency of mycolic acid patterns with dramatic differences from the standard species pattern are uncommon. However, when they occur, they appear stable for the isolate and may reflect a biological diversity within the species. In most cases, the UV-HPLC result can help avoid an incorrect identification on the basis of a gross pattern incompatibility with the reference pattern (78, 98, 99). This diversity of mycolic acid patterns demonstrated a need to develop a comprehensive Mycobacterium library of mycolic acid patterns for use with this method. A study of 23 species frequently encountered in clinical samples by five laboratories, using 350 well-characterized strains, established representative patterns for each species (12, 13).
COMPUTER USE WITH UV–HIGH-PERFORMANCE LIQUID CHROMATOGRAPHY
To improve on what has been perceived as an intrinsic subjectiveness of the manual identification schemes, automated chemometric methods of analysis using pattern recognition software were evaluated (75, 76). A multivariate method of analysis that simultaneously compiled the test data was used to compare the routinely analyzed results to a control set of mycolic acid samples with a preassigned species category. The basis of the pattern recognition method was a measure of similarity based on the K Nearest Neighbors (KNN) algorithm, part of Pirouette, a commercially available software package (Informetrix, Seattle, Wash.). The algorithm found to have the greatest accuracy for species prediction was a four-level, 15 KNN decision model. When tested with a validation test set of 549 cultures representing 27 species, the multilevel KNN approach accurately identified 92% of the species. The results were improved to 97% by combining the MAC-M. scrofulaceum prediction results into a complex. The performance of the model for the 27 species was as follows: 14 species were identified at 100%, 4 species were identified at 90% to 95%, 5 species were identified at 70 to 80%, and 4 species were identified at less than 70%. In general, the individual accuracy of the test was promising for some species but the initial objective of obtaining greater accuracy than that obtained with the manual method was not achieved. Mistakes by the software appeared to result from subsets of heterogenous species or intraspecies variation in a species, which had not been represented in the control set due to an insufficient number of test specimens. Support for this assumption was provided when incorrectly identified specimens from a validation set were reincorporated into a preassigned category, so that the computer model would then recognize the variation. Upon repeating the analysis, the computer model recognized the samples correctly (75). Further verification was demonstrated in an analysis which combined the commercial software with an independently generated “library” of mycolic acid patterns from 45 species of mycobacteria that were used to define the preassigned species categories (39). In an effort to increase the accuracy of the test, individual peak identification tables designed to compensate for variable ranges in the elution of the internal standard were included. For 1,155 strains representing 18 species of slowly growing mycobacteria, 97% were correctly identified. For 168 rapidly growing mycobacteria, 93% were correctly identified. It was verified that incorrect identifications occurred when an unknown sample's profile represented a chromatographic pattern variation for the species which was not incorporated in the software's preassigned category.
A related approach for computer pattern recognition was designed with commercially available software spread sheets and macro commands (52). The method used a calibration algorithm with a peak-naming table and pattern recognition for detection of different Mycobacterium complexes, including M. tuberculosis complex, MAC, and a user-defined MAC-M. scrofulaceum complex. Raw chromatographic data were imported into the spreadsheet and subjected to automated routine calculations. Calibrated peak retention times and peak height ratios were calculated for an unknown, and the results were compared to files in the Mycobacterium control set. This creative method standardized the elution time of the peaks from unknown patterns to both an internal high-molecular-weight standard and an external standard of pooled, derivatized mycolic acids extracted from the type strain of M. avium, ATCC 19705. The program demonstrated a sensitivity and specificity of 94.3 and 100%, respectively, for the MAC complex and 99 and 100%, respectively, for the M. tuberculosis complex (52).
The use of software classification models with the UV-HPLC method for automated prediction of mycobacteria species has limitations. Reliance on software alone will result in misidentification due to the inadequacy of the library database, which exists as a static entity and thus forces identification of an unknown sample. This is complicated by an instability of peak retention times caused by the coemergence of multiple peaks, which is a problem for the software's rigid identification times for peak location. For software to be used for identification, the system must be constantly updated as variants are recognized and new species are characterized. Presently, it is recommended that an identification be made by manual examination of the chromatogram and visual comparison of the pattern to those of reference chromatograms (12, 13).
MYCOLIC ACID PATTERN RECOGNITION AND MYCOBACTERIUM SPECIES IDENTIFICATION
Chromatographic pattern reproducibility and visual pattern interpretation are related factors, and a laboratory's expertise in species identification will be consistent with the experience of the chromatographer. A prerequisite for routine UV-HPLC analysis is adherence to defined chromatographic principles, including verifying the stability of the system by analysis of mycolic acid control samples for confirmation of analysis times (12, 13).
The visual pattern recognition method employs only chromatographic criteria, although when available, other identification test results should be included in the decision-making processes. The initial step for identifying a species is determining the overall complexity and number of mycolic acid peak clusters. These clusters may consist of a few peaks or many peaks and are further defined as single-, double-, and triple- or multiple-peak clusters. The range of time of elution between multiple-peak groups and the positions of the peaks are determined as RRTs to an internal standard. The RRT for a peak may also be adjusted by comparison to an external mycobacterial mycolic acid peak (13, 52). Usually, species with gross pattern differences are quickly eliminated, and the pattern of the unknown is compared to the remaining reference patterns (Fig. 3 to 10). When patterns are similar, the relationship of peak heights of major diagnostic peaks must be determined (16, 19, 52). If a reference pattern matches the unknown pattern, the species is identified; if not, the strain is reported as an “unidentified NTM.” Although this designation is not a specific identification, it reflects the detection of mycobacterial mycolic acids and could be of clinical value.
FIG. 10.
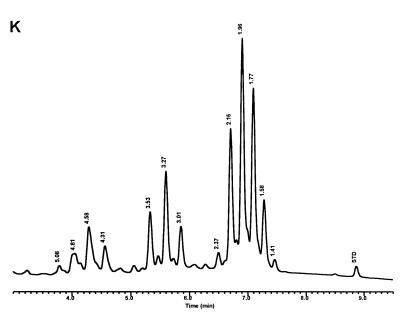
Characteristic UV-HPLC chromatograms of Mycobacterium species that have triple-peak clusters with peaks in the early cluster that emerge before 5.0 min. (A) Mycobacterium alvei ATCC 51304T; (B) Mycobacterium aurum ATCC 25793; (C) M. austroafricanum ATCC 33464T; (D) Mycobacterium chubuense ATCC 27278T; (E) Mycobacterium cookii ATCC 49103T; (F) Mycobacterium duvalii ATCC 43910T; (G) Mycobacterium obuense ATCC 27023T; (H) Mycobacterium parafortuitum ATCC 19686T; (I) M. shimoidei ATCC 27964; (J) M. tokaiense ATCC 27282T; (K) M. vaccae ATCC 15483T.
Recently, the occurrence of NTM species reported by state laboratories to the CDC Public Health Laboratory Information System (PHLIS) was tabulated and summarized for 1993 through 1996. The report can be viewed at the World Wide Web site http://www.cdc.gov/ncidod/dastlr/TB/TBarchive.htM. It should be noted that CDC personnel did not verify the identification methods or the identity of the species. The PHLIS report reflected the data as they were received by CDC, but the incidence of species was consistent with prior reports (59, 106). However, sampling errors associated with these data were recently demonstrated in an examination of the reports for M. malmoense (9).
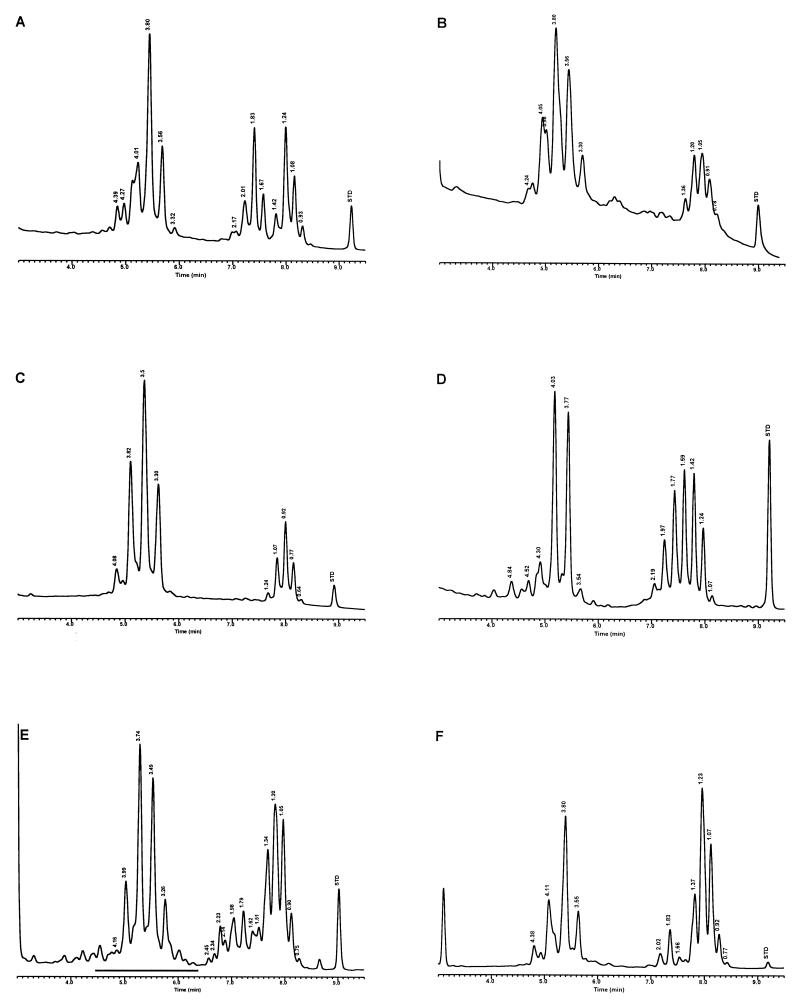
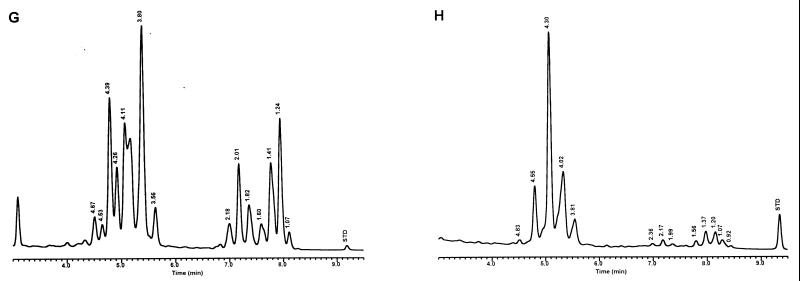
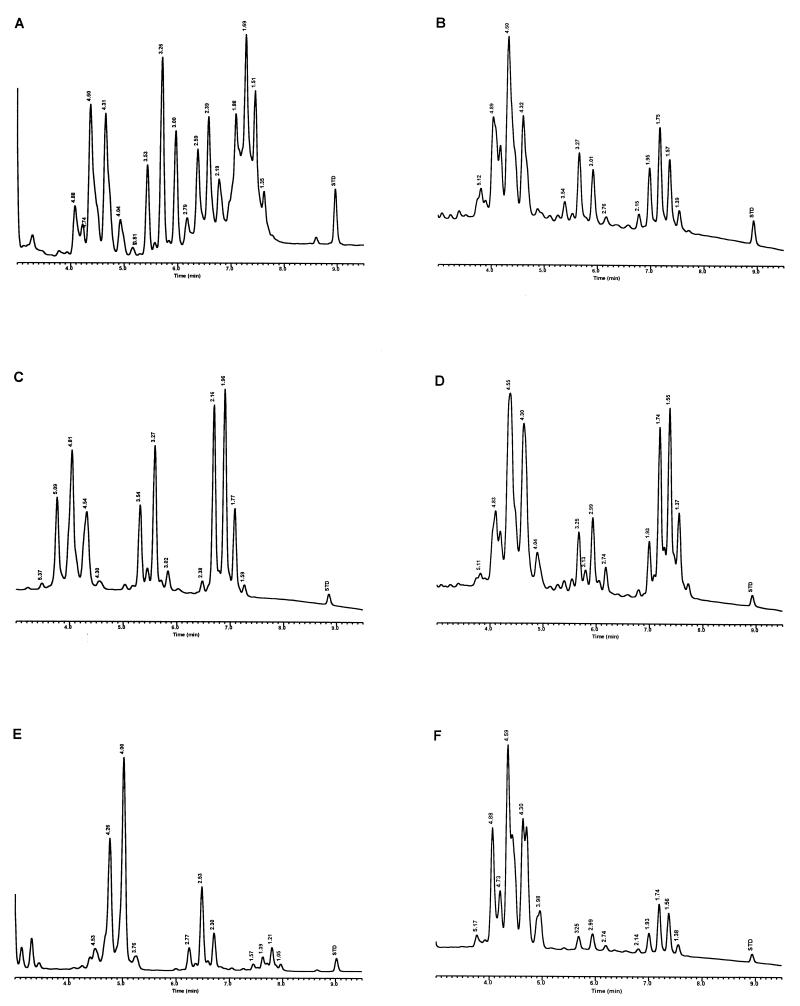
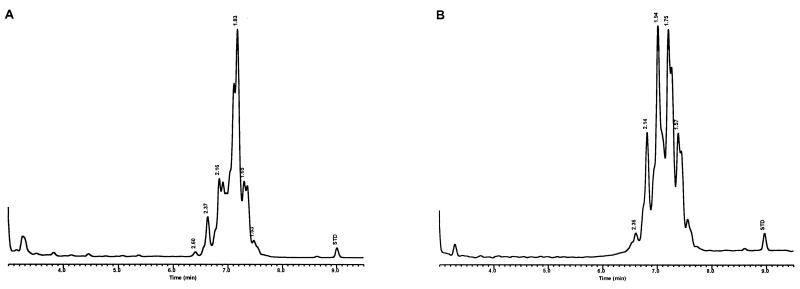
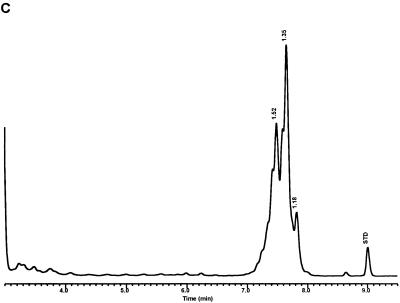
For the PHLIS analysis, a total of 82,475 NTM species reports were examined, of which 11,161 were reported as unknown or were not identified to the species level. For those specifically identified, the species reported and the number of reports were compared with a representative mycolic acid chromatogram with RRT's and were as follows: M. avium, 34,633 (Fig. 8A); M. gordonae, 18,440 (Fig. 3D); M. fortuitum, 5,601 (Fig. 6E); M. kansasii, 2,784 (Fig. 3E); M. terrae, 2,053 (Fig. 8H); M. chelonae, 1,789 (Fig. 6B); M. xenopi, 1,218 (Fig. 8I); M. intracellulare, 1,081 (Fig. 8F); Mycobacterium flavescens, 843 (Fig. 7D); M. simiae, 760 (Fig. 9C); M. marinum, 635 (Fig. 4E); M. scrofulaceum, 616 (Fig. 8G); M. szulgai, 226 (Fig. 3H); M. asiaticum, 107 (Fig. 3A); M. malmoense, 101 (Fig. 4D); M. abscessus, 90 (Fig. 6B); M. smegmatis, 49 (Fig. 6E); BCG variant of M. bovis, 36 (Fig. 3B); Mycobacterium vaccae, 32 (Fig. 10K); M. gastri, 31 (Fig. 3C); M. mucogenicum, 26 (Fig. 7J); M. haemophilum, 25 (Fig. 4B); M. nonchromogenicum, 20 (Fig. 8H); Mycobacterium peregrinum, 14 (Fig. 6E); M. interjectum, 9 (Fig. 8E); M. fallax, 7 (Fig. 5B); M. genavense, 4 (Fig. 9A); Mycobacterium neoaurum, 4 (Fig. 7K); M. celatum 3 (Fig. 8C); Mycobacterium ulcerans 3 (Fig. 4G); Mycobacterium triviale, 2 (Fig. 5C); and M. shimoidei, 2 (Fig. 10I). Species reported once during the reporting period were Mycobacterium aurum (Fig. 10B), Mycobacterium farcinogenes (Fig. 6D), Mycobacterium hiberniae (Fig. 7G), Mycobacterium intermedium (Fig. 4C), Mycobacterium senegalense (Fig. 6G), Mycobacterium thermoresistibile (Fig. 4F), and Mycobacterium tokaiense (Fig. 10J). The remaining chromatograms shown in Fig. 3 to 10 represent known species which were not reported by state health departments to CDC. However, the epidemiologic picture of NTM infection is reported to be changing, and it is possible that the etiologic significance of these species will also change (71).
FIG. 6.
Characteristic UV-HPLC chromatograms of Mycobacterium species with two-peak clusters that emerge late and close together. (A) Mycobacterium agri ATCC 27406T; (B) M. chelonae ATCC 35751 (includes M. abscessus ATCC 19977T); (C) Mycobacterium chitae ATCC 19629; (D) Mycobacterium farcinogenes ATCC 35753T; (E) M. fortuitum complex (includes M. goodii ATCC 700504T, M. peregrinum ATCC 14467T, M. smegmatis ATCC 14468, and M. wolinskyi ATCC 700010T); (F) M. porcinum, ATCC 33775T; (G) M. senegalense ATCC 35796.
FIG. 7.
Characteristic UV-HPLC chromatograms of Mycobacterium species that have widely separated, double-peak clusters with prominent peaks in the early cluster that emerge prior to 5.0 min. (A) Mycobacterium aichiense ATCC 27280T; (B) Mycobacterium chlorophenolicum ATCC 49826T; (C) Mycobacterium diernhoferi ATCC 19340T; (D) M. flavescens ATCC 14474T (underlined peaks are not present in all strains); (E) Mycobacterium gadium ATCC 27726T; (F) Mycobacterium gilvum ATCC 43909; (G) M. hiberniae ATCC 49874T; (H) Mycobacterium komossense ATCC 33013T; (I) Mycobacterium madagascariense ATCC 49865T; (J) M. mucogenicum ATCC 49651; (K) M. neoaurum ATCC 25795T; (L) Mycobacterium phlei ATCC 11758T; (M) Mycobacterium rhodesiae ATCC 27024T; (N) Mycobacterium sphagni ATCC 33027.
FIG. 9.
Characteristic UV-HPLC images of Mycobacterium species that have triple-peak clusters, close together that emerge later than 6.0 min. (A) M. genavense CDC 892120; (B) Mycobacterium lentiflavum 2186/92; (C) M. simiae ATCC 25275T; (D) M. triplex ATCC 700071T.
FIG. 4.
Characteristic UV-HPLC chromatograms of Mycobacterium species with late-emerging, complex, single-cluster peak patterns. (A) Mycobacterium confluentis ATCC 49920T; (B) M. haemophilum ATCC 29548T; (C) M. intermedium ATCC 51848T; (D) M. malmoense ATCC 29571T; (E) M. marinum CDC 875; (F) M. thermoresistibile CDC 7305; (G) M. ulcerans ATCC 19423T.
FIG. 5.
Characteristic UV-HPLC chromatograms of Mycobacterium species with late-emerging, simple, single-cluster peak patterns with unresolved shoulder peaks. (A) Mycobacterium brumae ATCC 51384T; (B) M. fallax ATCC 35219T; (C) M. triviale ATCC 23292T.
The PHLIS report excluded members of the M. tuberculosis complex because the Division of Tuberculosis Elimination, CDC, annually reports the verified cases of tuberculosis as part of the tuberculosis surveillance system. However, the M. tuberculosis complex chromatographic pattern is illustrated by the production of a stable, late-emerging, simple, mycolic acid peak cluster (Fig. 3G). It has been reported that BCG attenuated strains of M. bovis can be differentiated from the complex by this method (35). However, recently a confirmed isolate of a drug-resistant M. tuberculosis isolate from a foreign patient produced a mycolic acid pattern that was confused with the BCG pattern. A study is being conducted to examine this discrepancy.
FLUORESCENT DERIVATIVES FOR MYCOLIC ACID ANALYSIS
The most commonly used derivatization reagents for long-carbon-chain fatty acids are UV-adsorbing derivatives; however, the detection sensitivity can be improved by the use of fluorescence-labeling compounds. Theoretically, fluorescence detection could increase the sensitivity 10 to 1,000 times. A possible application for liquid chromatography separations was proposed when short-carbon-chain fatty acids were labeled with 4-bromomethyl-7-methoxycoumarin (Br-Mmc) and used for TLC analysis for detection of picomolar amounts (28). However, the production of 7-methoxy derivatives was tedious, and in gradient liquid chromatography the fluorescence yield varied and was solvent dependent (63). Moreover, carboxylic acid Br-Mmc derivatives had a lower detection sensitivity, and the fluorescence intensity of the label was affected by the type of carboxylic acid. Based on the assumption that the smaller the molecular size of the labeling reagent, the better the separation of the labeled carboxylic acids, an alternate fluorescence-labeling reagent, 4-bromomethyl-7-acetoxycoumarin (Br-Mac), was suggested for femtomolar detection (103). However, Br-Mmc derivatives were demonstrated to be comparable in yield and sensitivity to Br-Mmc derivatives. Subsequent derivatization of C1 to C5 short-carbon-chain fatty acids with 4-bromomethyl-6,7-dimethoxycoumarin (coumarin) was shown to be suitable for gradient elution with a reverse-phase column (33). In actual application, in two different studies the mycolic acid coumarin derivative produced 20 and 80-fold increases in sensitivity compared to the UV-absorbing PBPA derivative (47, 86). An advantage of using the fluorescence detection was that only a portion of a single colony was required, compared to a transfer loop full of cells for the UV method. Other fluorimetric reagents used to produce mycolic acid derivatives for identification of clinical mycobacteria were 3-bromomethyl-7-methoxy-1,4-benzoxazin-2-1 and 4-bromomethyl-7-acetoxycoumarin (47). They were shown to have 16 to 26.6 times greater sensitivity, respectively, for detection of mycobacterial mycolic acids than did the UV-adsorbing PBPA reagent. All three fluorophores produced mycolic acid patterns similar to the patterns for PBPA derivatives and were suggested for optimization of the method for clinical mycobacterial species identification.
An early demonstration of the use of a coumarin derivative with mycobacterial mycolic acids and reverse-phase HPLC was the detection of M. tuberculosis and M. paratuberculosis, the causative agent of Johne's disease (23). C48 to C76 mycolic acids were separated with a gradient mobile phase of chloroform and acetyl nitrile. The fluorescent derivative displayed an excitation at 285 nm and emission at 340 nm, with characteristic, coemerging homologous mycolic acid peaks. The author speculated that the mycolic acid pattern for M. paratuberculosis would be useful in detecting this bacterium in the feces of infected cattle. In another multiple-step chromatographic study, normal and reverse-phase HPLC produced highly characteristic profiles for M. tuberculosis with mycolic acids as anthrylmethy esters from sputum samples (66).
The sensitivity of automated identification of M. tuberculosis complex and MAC from BACTEC 12B cultures and from fluorochrome-stained smear-positive sputum specimens has been described (52). The attractiveness of this method to recognize mycobacteria sooner than conventional methods was demonstrated by a definitive identification of M. tuberculosis complex within 24 h of receiving specimens from individuals who were shedding a large number of bacilli. The fluorescence patterns were noted to be comparable with those from the standard UV-HPLC method.
The advantage of an increased sensitivity of detection for fluorescence detection was recently demonstrated by chromatographic separation criteria for some rapidly growing mycobacteria associated with wound infections, which had not been reported with UV-HPLC (8). However, the increased sensitivity of fluorescence detection requires careful manipulation of samples to prevent cross-contamination, and carryover of mycolic acids from previously analyzed samples must be cautiously monitored. It may also be necessary to use solid-phase extraction to remove undesirable by-products and contaminants to improve the signal-to-noise ratio (27). However, the potential for concomitantly reducing the cell mass and increasing the sensitivity, thus reducing the time required for identification, could substantially improve the chromatographic identification method.
UV–HIGH-PERFORMANCE LIQUID CHROMATOGRAPHY AND MOLECULAR METHODS
The conventional tests used in the clinical laboratory cannot identify many of the newly described NTM species, and newer, rapid methods must be employed (6, 50). A problem with some new identification methods is they may not be affordable or amenable to routine laboratory schedules. Even so, chemotaxonomic and molecular identification methods reduce the turnaround times and are more accurate for discriminating species. The intrinsic nature of genetic methods for the analysis of nucleic material has resulted in a variety of methods that may involve combinations of PCR amplification, oligonucleotide hybridization probes (specific or array), restriction enzyme digestion, and sequence analysis. The molecular method of choice for detecting M. tuberculosis is a single, specific genetic probe. Several of the methods mentioned above have been proposed for molecular identification of NTM, but a consensus method is not in use. Here, a comparison is made to the molecular method frequently reported for classifying NTM, i.e., PCR amplification and direct DNA sequencing of portions of the 16S rRNA gene (6, 56).
DNA sequence analysis of the 16S rRNA gene can be performed manually or with automated sequencers. Manual sequencing is inexpensive to perform, but the data are subject to base-calling errors, and depending on the method, radioactive by-products may be produced. For routine use, the automated method is preferred, but it is expensive. However, the data from automated sequencers are virtually free of base-calling errors and the method does not produce hazardous by-products. UV-HPLC produces hazardous by-products consisting of mixed organic solvents, including chloroform, methanol, and methylene chloride. UV-HPLC sample processing is rapid and easy, but for reproducibility of chromatographic patterns, standardized conditions of growth should be used. Molecular methods of analysis do not require standardized growth conditions but generally do require more hands-on time for sample processing. Although none of these methods requires viable cells, UV-HPLC requires more cell biomass than the molecular methods do. Molecular methods have a greater detection sensitivity than the UV-HPLC method. Both UV-HPLC and automated sequencing methods require expensive, sophisticated instrumentation. Although automated sequencing instrumentation is about twice as costly to install and maintain as chromatographic instruments. A single sample can be processed in 2 h by UV-HPLC, and automated sequencing requires >8 h. UV-HPLC and manual sequencing are inexpensive to run on a daily basis, while automated sequencing is expensive. The reported cost of UV-HPLC per sample is approximately $3.00 (45, 52, 93). Identifications of species using sequence analysis are made by comparison to international sequence databases or in-house references. Identifications of species with mycolic acids are made by comparison to in-house databases of reference patterns.
The support system for analyzing sequencing data is extensive and impressive, with easy-to-use commercial software and exhaustive World Wide Web sites. The only interactive Web site for support of the chromatographic method specifically for identification of mycobacteria is maintained by the HPLC Users Group at http://hplc.cjb.net. Both methods represent potentially rapid and reliable techniques for recognition of known and unknown mycobacteria and complement each other for identification of Mycobacterium species (8, 18, 34, 46, 81, 82, 101, 102).
INSTRUMENT CERTIFICATION AND INSPECTIONS
Chromatograph manufacturers are intensely concerned with the quality and performance of their products. Vendors constantly incorporate recent technological advances into the manufacture and operation of their instruments. In the 11 years since the UV-HPLC method was first described, instrument and column reliability has improved substantially. A major improvement was the replacement of manual controllers by computers which provided a visual, easy-to-use platform for direct control of all of the chromatographic features for solvent composition, delivery, and program development. Any computerized system capable of producing a controlled solvent gradient can be programmed with the UV-HPLC method. The components of an instrument have a finite life, and in high-throughput laboratories it would be prudent to maintain a commercial service contract. However, instrument dependability is excellent, and routine maintenance is normally all that is required. Since the quality of the chromatographic pattern is affected by the performance of the instrument, routine quality control is essential. When components are replaced, a reference standard, usually a designated species of Mycobacterium, must be routinely used to verify the reference conditions for analysis (12, 13).
In the United States, laboratories considering using HPLC should have the proficiency of a Level II or higher mycobacteriology laboratory before instituting the method in order to satisfy the Clinical Laboratory Improvement Act (CLIA) of 1988 (55). It should be noted that CLIA inspectors consider UV-HPLC to be a high complex diagnostic procedure, and the operator is accountable for records of method validation, quality control, and routine use of the system.
HPLC USERS GROUP
The HPLC Users Group was formed during the 1992 General Meeting of the American Society for Microbiology in New Orleans, La., by a group of mycobacteriologists who were using UV-HPLC and were concerned with the different laboratory techniques employed to analyze mycolic acids for identification of mycobacteria (L. S. Guthertz, Letter, ASM News 58:467, 1992). Afterwards, a survey was conducted for laboratories using UV-HPLC, and it demonstrated a variety of instruments and methods in use and a definite need for standardization. However, even with a lack of standardization, it was found that all laboratories produced comparable UV-HPLC patterns for similar species, attesting to the robustness of the method. After an extensive standardization study with five laboratories, the HPLC Users Group, in cooperation with CDC, electronically published two manuals, Standardized Method for HPLC Identification of Mycobacteria and the Mycolic Acid Pattern Standards for HPLC Identification of Mycobacteria. These manuals are available at the CDC Web page http://www.cdc.gov/ncidod/dastlr/TB/TB_HPLC.htm.
CONCLUSION
The identification of Mycobacterium species by UV-HPLC has been documented as a rapid, reliable method by numerous laboratories. The versatility of this method for detection of M. tuberculosis, a wide range of known or unknown NTM species, and other mycolic acid-containing genera by a single test offers a significant advantage for the laboratory.
REFERENCES
- 1.Alshamaony L, Goodfellow M, Minnikin D E. Free mycolic acids as criteria in the classification of Nocardia and the “rhodochrous” complex. J Gen Microbiol. 1976;92:188–199. doi: 10.1099/00221287-92-1-188. [DOI] [PubMed] [Google Scholar]
- 2.Asselineau J, Lederer E. Structure of the mycolic acids of mycobacteria. Nature (London) 1950;166:782–783. doi: 10.1038/166782a0. [DOI] [PubMed] [Google Scholar]
- 3.Baba T, Kaneda K, Kusunose E, Kusunose M, Yano I. Thermally adaptive changes of mycolic acids in Mycobacterium smegmatis. J Biochem. 1989;106:81–86. doi: 10.1093/oxfordjournals.jbchem.a122825. [DOI] [PubMed] [Google Scholar]
- 4.Body B A, Warren N G, Spicer A, Henderson D, Chery M. Use of Gen-Probe and Bactec for rapid isolation and identification of mycobacteria: correlation of probe results with growth index. Am J Clin Pathol. 1990;93:415–420. doi: 10.1093/ajcp/93.3.415. [DOI] [PubMed] [Google Scholar]
- 5.Borch R F. Separation of long chain fatty acids as phenacyl esters by high pressure liquid chromatography. Anal Chem. 1975;47:2437–2439. doi: 10.1021/ac60364a037. [DOI] [PubMed] [Google Scholar]
- 6.Böttger E C. Approaches for identification of microorganisms: despite longer experience with fatty acid profiles, DNA-based analysis offers several advantages. ASM News. 1996;62:247–250. [Google Scholar]
- 7.Böttger E C, Kirschner P, Springer B, Zumft W. Mycobacteria degrading polycyclic aromatic hydrocarbons. Int J Sys Bacteriol. 1997;47:247. [Google Scholar]
- 8.Brown B A, Springer B, Steingrube V A, Wilson R W, Pfyffer G E, Garcia M J, Menendez M C, Rodriguez-Salgado B, Jost K C, Jr, Chiu S H, Onyi G O, Böttger E C, Wallace R J., Jr Mycobacterium wolinskyi sp. nov. and Mycobacterium goodii sp. nov., two new rapidly growing species related to Mycobacterium smegmatis and associated with human wound infections: a cooperative study from the International Working Group on Mycobacterial Taxonomy. Int J Syst Bacteriol. 1999;49:1493–1511. doi: 10.1099/00207713-49-4-1493. [DOI] [PubMed] [Google Scholar]
- 9.Buchholz U T, McNeil M M, Keyes L E, Good R C. Mycobacterium malmoense infections in the United States, January 1993 through June 1995. Clin Infect Dis. 1998;27:551–558. doi: 10.1086/514702. [DOI] [PubMed] [Google Scholar]
- 10.Butler W R. Reverse phase high performance liquid chromatography separation of mycolic acids as para-bromophenacyl esters as an aid in mycobacterial classification. Masters Thesis. Atlanta: Georgia State University; 1985. [Google Scholar]
- 11.Butler W R, Ahearn D G, Kilburn J O. High-performance liquid chromatography of mycolic acids as a tool in the identification of Corynebacterium, Nocardia, Rhodococcus, and Mycobacterium species. J Clin Microbiol. 1986;23:182–185. doi: 10.1128/jcm.23.1.182-185.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler W R, Floyd M M, Silcox V, Cage G, Desmond E, Duffey P S, Guthertz L S, Gross W M, Jost K C, Jr, Ramos L S, Thibert L, Warren N. Standardized method for HPLC identification of mycobacteria. Atlanta, Ga: Centers for Disease Control and Prevention; 1996. [Google Scholar]
- 13.Butler W R, Floyd M M, Silcox V, Cage G, Desmond E, Duffey P S, Guthertz L S, Gross W M, Jost K C, Jr, Ramos L S, Thibert L, Warren N. Mycolic acid pattern standards for HPLC identification of mycobacteria. Atlanta, Ga: Centers for Disease Control and Prevention; 1999. [Google Scholar]
- 14.Butler W R, Jost K C, Jr, Kilburn J O. Identification of mycobacteria by high-performance liquid chromatography. J Clin Microbiol. 1991;29:2468–2472. doi: 10.1128/jcm.29.11.2468-2472.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butler W R, Kilburn J O. Identification of major slowly growing pathogenic mycobacteria and Mycobacterium gordonae by high-performance liquid chromatography of their mycolic acids. J Clin Microbiol. 1988;26:50–53. doi: 10.1128/jcm.26.1.50-53.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler W R, Kilburn J O. High-performance liquid chromatography patterns of mycolic acids as criteria for identification of Mycobacterium chelonae, Mycobacterium fortuitum, and Mycobacterium smegmatis. J Clin Microbiol. 1990;29:2094–2098. doi: 10.1128/jcm.28.9.2094-2098.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler W R, Kilburn J O, Kubica G P. High-performance liquid chromatography analysis of mycolic acids as an aid in laboratory identification of Rhodococcus and Nocardia species. J Clin Microbiol. 1987;25:2126–2131. doi: 10.1128/jcm.25.11.2126-2131.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler W R, O'connor S P, Yakrus M A, Smithwick R W, Plikaytis B B, Moss C W, Floyd M M, Woodley C L, Kilburn J O, Vadney F S, Gross W M. Mycobacterium celatum sp. nov. Int J Syst Bacteriol. 1993;43:539–548. doi: 10.1099/00207713-43-3-539. [DOI] [PubMed] [Google Scholar]
- 19.Butler W R, Thibert L, Kilburn J O. Identification of Mycobacterium avium complex strains and some similar species by high-performance liquid chromatography. J Clin Microbiol. 1992;30:2698–2704. doi: 10.1128/jcm.30.10.2698-2704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cage G D. High-performance liquid chromatography patterns of Mycobacterium gordonae mycolic acids. J Clin Microbiol. 1992;30:2402–2407. doi: 10.1128/jcm.30.9.2402-2407.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cage G D. Direct identification of Mycobacterium species in BACTEC 7H12B medium by use of high-performance liquid chromatography. J Clin Microbiol. 1994;32:521–524. doi: 10.1128/jcm.32.2.521-524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coyle M B, Carlson L C, Wallis C K, Leonard R B, Raisys V A, Kilburn J O, Samadpour M, Böttger E C. Laboratory aspects of “Mycobacterium genavense,” a proposed species isolated from AIDS patients. J Clin Microbiol. 1992;30:3206–3212. doi: 10.1128/jcm.30.12.3206-3212.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craig M A, Burdett R G. Separation by high-performance liquid chromatography of fatty acids from Mycobacterium paratuberculosis. In: Merkal R S, editor. Proceedings of the International Colloquium on research in paratuberculosis. Ames, Iowa: The National Animal Disease Center; 1983. pp. 2–8. [Google Scholar]
- 24.Crawford J T. Development of rapid techniques for identification of M. avium infections. Res Microbiol. 1994;145:13–17. doi: 10.1016/0923-2508(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 25.De Briel D, Couderc F, Riegel P, Jehl F, Minck R. High-performance liquid chromatography of corynomycolic acids as a tool in identification of Corynebacterium species and related organisms. J Clin Microbiol. 1992;30:1407–1417. doi: 10.1128/jcm.30.6.1407-1417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dixon P F, Stoll M S, Lim C K. High pressure liquid chromatography in clinical chemistry. Ann Clin Biochem. 1976;13:409–432. doi: 10.1177/000456327601300131. [DOI] [PubMed] [Google Scholar]
- 27.Duffey P S, Guthertz L S, Evans G C. Improved rapid identification of mycobacteria by combining solid-phase extraction with high-performance liquid chromatography analysis of BACTEC cultures. J Clin Microbiol. 1996;34:1939–1943. doi: 10.1128/jcm.34.8.1939-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dünges W. 4-Bromomethyl-7-methoxycoumarin as a new fluorescence label for fatty acids. Anal Chem. 1977;49:442–445. doi: 10.1021/ac50011a028. [DOI] [PubMed] [Google Scholar]
- 29.Dupree W G, Bradford H B., Jr New developments in mycobacteria identification: public health laboratory modernization. J La State Med Soc. 1992;144:379–382. [PubMed] [Google Scholar]
- 30.Durst H D, Milano M, Kikta E J, Jr, Connelly S A, Grushka E. Phenacyl esters of fatty acids via crown ether catalysts for enhanced ultraviolet detection in liquid chromatography. Anal Chem. 1975;47:1797–1801. doi: 10.1021/ac60361a025. [DOI] [PubMed] [Google Scholar]
- 31.Embley T M, Stackebrandt E. The molecular phylogeny and systematics of the actinomycetes. Annu Rev Microbiol. 1994;48:257–289. doi: 10.1146/annurev.mi.48.100194.001353. [DOI] [PubMed] [Google Scholar]
- 32.Etémadi A-H. The use of pyrolysis gas chromatography and mass spectroscopy in the study of the structure of mycolic acids. J Gas Chromatogr. 1967;5:447–456. [Google Scholar]
- 33.Farinotti R, Siard P, Bourson J, Kirkiacharian S, Valeur B, Mahuzier G. 4-Bromomethyl-6,7-dimethoxycoumarin as a fluorescent label for carboxylic acids in chromatographic detection. J Chromatogr. 1983;269:81–90. [Google Scholar]
- 34.Floyd M M, Guthertz L S, Silcox V A, Duffey P S, Jang Y, Desmond E P, Crawford J T, Butler W R. Characterization of a SAV organism and proposal of Mycobacterium triplex sp. nov. J Clin Microbiol. 1996;34:2963–2967. doi: 10.1128/jcm.34.12.2963-2967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Floyd M M, Silcox V A, Jones W D, Jr, Butler W R, Kilburn J O. Separation of Mycobacterium bovis BCG from Mycobacterium tuberculosis and Mycobacterium bovis by using high-performance liquid chromatography of mycolic acids. J Clin Microbiol. 1992;30:1327–1330. doi: 10.1128/jcm.30.5.1327-1330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garza-González E, Guerrero-Olazarán M, Tijerina-Menchaca R, Viader-Salvadó J M. Determination of drug susceptibility of Mycobacterium tuberculosis through mycolic acid analysis. J Clin Microbiol. 1997;35:1287–1289. doi: 10.1128/jcm.35.5.1287-1289.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garza-González E, Guerrero-Olazarán M, Tijerina-Menchaca R, Viader-Salvadó J M. Identification of mycobacteria by mycolic acid pattern. Arch Med Res. 1998;29:303–306. [PubMed] [Google Scholar]
- 38.Gerson B. HPLC monitoring improves drug therapy. Lab World. 1981;1:43–45. [Google Scholar]
- 39.Glickman S E, Kilburn J O, Butler W R, Ramos L S. Rapid identification of mycolic acid patterns of mycobacteria by high-performance liquid chromatography using pattern recognition software and a Mycobacterium library. J Clin Microbiol. 1994;32:740–745. doi: 10.1128/jcm.32.3.740-745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Good R. New culture identification procedure initiated in CDC Mycobacteriology Lab. TB notes, Summer. Atlanta, Ga: Division of TB Control, Centers for Disease Control; 1990. p. 13. [Google Scholar]
- 41.Good R C. The genus Mycobacterium—medical. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes—a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. II. New York, N.Y: Springer-Verlag; 1992. pp. 1238–1270. [Google Scholar]
- 42.Goodfellow M, Minnikin D E. Circumscription of the genus. In: Kubica G P, Wayne L G, editors. The mycobacteria: a sourcebook. New York, N.Y: Marcel Dekker, Inc; 1984. pp. 1–24. [Google Scholar]
- 43.Reference deleted.
- 44.Reference deleted.
- 45.Guthertz L S, Lim S D, Jang Y, Duffey P S. Curvilinear-gradient high-performance liquid chromatography for identification of mycobacteria. J Clin Microbiol. 1993;31:1876–1881. doi: 10.1128/jcm.31.7.1876-1881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haas W H, Butler W R, Kirschner P, Plikaytis B B, Coyle M B, Amthor B, Steigerwalt A G, Brenner D J, Salfinger M, Crawford J T, Böttger E C, Bremer H J. A new agent of mycobacterial lymphadenitis in children: Mycobacterium heidelbergense sp. nov. J Clin Microbiol. 1997;35:3203–3209. doi: 10.1128/jcm.35.12.3203-3209.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagen S R, Thompson J D. Analysis of mycolic acids by high-performance liquid chromatography and fluorimetric detection implication for the identification of mycobacteria in clinical samples. J Chromatogr. 1995;692:167–172. doi: 10.1016/0021-9673(94)00743-s. [DOI] [PubMed] [Google Scholar]
- 48.Heifets L B, Good R C. Current laboratory methods for the diagnosis of tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C.: American Society for Microbiology; 1994. p. 100. [Google Scholar]
- 49.Horváth C. High-performance liquid chromatography: advances and perspectives. New York, N.Y: Academic Press, Inc.; 1980. [Google Scholar]
- 50.Huebner R E, Good R C, Tokars J I. Current practices in mycobacteriology: results of a survey of state public health laboratories. J Clin Microbiol. 1993;31:771–775. doi: 10.1128/jcm.31.4.771-775.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jordi H C. Separation of long and short chain fatty acids as naphthacyl and substituted phenacyl esters by high performance liquid chromatography. J Liquid Chromatogr. 1978;1:215–230. [Google Scholar]
- 52.Jost K C, Dunbar D F, Barth S S, Headley V L, Elliot L B. Identification of Mycobacterium tuberculosis and M. avium complex directly from smear-positive sputum specimens and Bactec 12B cultures by high-performance liquid chromatography with fluorescence detection and computer-driven pattern recognition models. J Clin Microbiol. 1995;33:1270–1277. doi: 10.1128/jcm.33.5.1270-1277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanetsuna F. Chemical analyses of mycobacterial cell walls. Biochim Biophys Acta. 1968;158:130–143. doi: 10.1016/0304-4165(68)90080-9. [DOI] [PubMed] [Google Scholar]
- 54.Kanetsuna F, Bartoli A. A simple chemical method to differentiate Mycobacterium from Nocardia. J Gen Microbiol. 1972;70:209–212. doi: 10.1099/00221287-70-2-209. [DOI] [PubMed] [Google Scholar]
- 55.Kent P T, Kubica G P. Public health mycobacteriology: a guide for the level III laboratory. Atlanta, Ga: Centers for Disease Control, U.S. Department of Health and Human Services; 1985. [Google Scholar]
- 56.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange F-C, Böttger E C. Geotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical microbiology. J Clin Microbiol. 1993;31:2882–2889. doi: 10.1128/jcm.31.11.2882-2889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kubica G P. Differential identification of mycobacteria. VII. Key features for identification of clinically significant mycobacteria. Am Rev Respir Dis. 1973;107:9–21. doi: 10.1164/arrd.1973.107.1.9. [DOI] [PubMed] [Google Scholar]
- 58.Kubica G P. Classification and nomenclature of the mycobacteria. Ann Microbiol (Inst Pasteur) 1978;129:7–12. [PubMed] [Google Scholar]
- 59.Kubica G P. Clinical microbiology. In: Kubica G P, Wayne L G, editors. The mycobacteria: a sourcebook. New York, N.Y: Marcel Dekker, Inc; 1984. pp. 133–175. [Google Scholar]
- 60.Lanéelle G. Nature des acides mycoliques de Mycobacterium paratuberculosis: application de la chromatographie sur couche mince a leur fractionnement. C R Hebd Seances Acad Sci Paris. 1963;257:781–783. [PubMed] [Google Scholar]
- 61.Lechevalier M P, Horan A C, Lechevalier H. Lipid composition in the classification of nocardiae and mycobacteria. J Bacteriol. 1971;105:313–318. doi: 10.1128/jb.105.1.313-318.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lévy-Frébault V V, Portaels F. Proposed minimal standards for the genus Mycobacterium and for description of new slowly growing Mycobacterium species. Int J Syst Bacteriol. 1992;42:315–323. doi: 10.1099/00207713-42-2-315. [DOI] [PubMed] [Google Scholar]
- 63.Lloyd J B F. Phenanthrimidazoles as fluorescent derivatives in the analysis of fatty acids by high-performance liquid chromatography. J Chromatogr. 1980;189:359–373. [Google Scholar]
- 64.Lumb R, Goodwin A, Ratcliff R, Stapledon R, Holland A, Bastian I. Phenotypic and molecular characterization of three clinical isolates of Mycobacterium interjectum. J Clin Microbiol. 1997;35:2782–2785. doi: 10.1128/jcm.35.11.2782-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Minnikin D E, Alshamaony L, Goodfellow M. Differentiation of Mycobacterium, Nocardia, and related taxa by thin-layer chromatographic analysis of whole-organism methanolysates. J Gen Microbiol. 1975;88:200–204. doi: 10.1099/00221287-88-1-200. [DOI] [PubMed] [Google Scholar]
- 66.Minnikin D E, Bolton R C, Hartmann S, Besra G S, Jenkins P A, Mallet A I, Wilkins E, Lawson A M, Ridell M. An integrated procedure for the direct detection of characteristic lipids in tuberculosis patients. Ann Soc Belg Med Trop. 1993;73:13–24. [PubMed] [Google Scholar]
- 67.Minnikin D E, Dobson G, Goodfellow M, Draper P, Magnusson M. Quantitative comparison of the mycolic and fatty acid compositions of Mycobacterium leprae and Mycobacterium gordonae. J Gen Microbiol. 1985;131:2013–2021. doi: 10.1099/00221287-131-8-2013. [DOI] [PubMed] [Google Scholar]
- 68.Minnikin D E, Dobson G, Parlet J H, Datta A K, Minnikin S M, Goodfellow M. Analysis of mycobacteria mycolic acids. In: Klein R, Schmitz B, editors. Topics in lipid research: from structural elucidation to biological function. London, United Kingdom: Royal Society of Chemistry; 1986. pp. 139–143. [Google Scholar]
- 69.Minnikin D E, Goodfellow M. Lipid composition in the classification and identification of acid-fast bacteria. In: Goodfellow M, Board R G, editors. Microbiological classification and identification. London, United Kingdom: Academic Press, Ltd.; 1980. pp. 189–256. [PubMed] [Google Scholar]
- 70.Minnikin D E, Minnikin S M, Parlett J H, Goodfellow M, Magnusson M. Mycolic acid patterns of some species of Mycobacterium. Arch Microbiol. 1984;139:225–231. doi: 10.1007/BF00402005. [DOI] [PubMed] [Google Scholar]
- 71.O'Brien J R, Geiter J L, Snider D E., Jr The epidemiology of nontuberculous mycobacterial diseases in the United States: results from a national survey. Am Rev Respir Dis. 1987;135:1007–1014. doi: 10.1164/arrd.1987.135.5.1007. [DOI] [PubMed] [Google Scholar]
- 72.Pei P T, Kossa W C, Ramachandran S, Henly R S. High pressure reverse phase liquid chromatography of fatty acid p-bromophenacyl esters. Lipids. 1976;11:814–816. doi: 10.1007/BF02533409. [DOI] [PubMed] [Google Scholar]
- 73.Qureshi N, Takayama K, Jordi H C, Schnoes H K. Characterization of the purified components of a new homologous series of α-mycolic acids from Mycobacterium tuberculosis H37Ra. J Biol Chem. 1978;253:5411–5417. [PubMed] [Google Scholar]
- 74.Rafii F, Butler W R, Cerniglia C E. Differentiation of a rapidly growing, scotochromogenic, polycyclic-aromatic-hydrocarbon-metabolizing strain of Mycobacterium sp. from other known Mycobacterium species. Arch Microbiol. 1992;157:512–520. [Google Scholar]
- 75.Ramos L S. Classification of mycobacteria by HPLC and pattern recognition. AmBiotechnol Lab. 1992;10:27–32. [PubMed] [Google Scholar]
- 76.Ramos L S. Characterization of mycobacteria species by HPLC and pattern recognition. J Chromatogr Sci. 1994;32:219–227. doi: 10.1093/chromsci/32.6.219. [DOI] [PubMed] [Google Scholar]
- 77.Ridderhof J C, Wallace R J, Jr, Kilburn J O, Butler W R, Warren N G, Tsukamura M, Steele L C, Wong E S. Chronic tenosynovitis of the hand due to Mycobacterium nonchromogenicum: use of high-perofrmance liquid chromatography for identification of isolates. Rev Infect Dis. 1991;13:857–864. doi: 10.1093/clinids/13.5.857. [DOI] [PubMed] [Google Scholar]
- 78.Ritter, D., L. C. Carlson, B. K. Logan, L. S. Ramos, J. O. Kilburn, and M. B. Coyle. Differentiation of Mycobacterium genavense and Mycobacterium simiae by automated mycolic acid analysis with high-performance liquid chromatography. J. Clin. Microbiol. 34:2004–2006. [DOI] [PMC free article] [PubMed]
- 79.Roggero J P, Coen S V. Isocratic separation of fatty acid derivatives by reversed phase liquid chromatography. Influence of the solvent on selectivity and rules for elution order. J Liquid Chromatogr. 1981;4:1817–1829. [Google Scholar]
- 80.Runyon E H. Anonymous mycobacteria in pulmonary disease. Med Clin North Am. 1959;43:273–290. doi: 10.1016/s0025-7125(16)34193-1. [DOI] [PubMed] [Google Scholar]
- 81.Rynkiewicz D L, Cage G D, Butler W R, Ampel N M. Clinical and microbiological assessment of Mycobacterium simiae isolates from a single laboratory in southern Arizona. Clin Infect Dis. 1997;26:625–630. doi: 10.1086/514573. [DOI] [PubMed] [Google Scholar]
- 82.Schinsky M F, McNeil M M, Whitney A M, Steigerwalt A G, Lasker B A, Floyd M M, Hogg G G, Brenner D J, Brown J M. Mycobacterium septicum sp. nov., a new rapidly growing species associated with catheter-related bacteraemia. Int J Syst Evol Microbiol. 2000;50:575–581. doi: 10.1099/00207713-50-2-575. [DOI] [PubMed] [Google Scholar]
- 83.Sherman I, Harrrington N, Rothrock A, George H. Use of a cutoff range in identifying mycobacteria by the gen-probe rapid diagnostic system. J Clin Microbiol. 1989;27:241–244. doi: 10.1128/jcm.27.2.241-244.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shinnick T M, Good R C. Diagnostic mycobacteriology laboratory practices. Clin Infect Dis. 1995;21:291–299. doi: 10.1093/clinids/21.2.291. [DOI] [PubMed] [Google Scholar]
- 85.Siddiqi S H, Laszlo A, Butler W R, Kilburn J O. Bacteriologic investigations of unusual mycobacteria isolated from immunocompromised patients. Diagn Microbiol Infect Dis. 1993;16:321–323. doi: 10.1016/0732-8893(93)90083-j. [DOI] [PubMed] [Google Scholar]
- 86.Simon V A, Hale Y, Taylor A. Identification of Mycobacterium tuberculosis and related organisms by HPLC with fluorescence detection. LC-GC. 1993;11:444–448. [Google Scholar]
- 87.Springer B, Böttger E C, Kirschner P, Wallace R J., Jr Phylogeny of the Mycobacterium chelonae-like organism based on partial sequencing of the 16S rRNA gene and proposal of Mycobacterium mucogenicum sp. nov. Int J Syst Bacteriol. 1995;45:262–267. doi: 10.1099/00207713-45-2-262. [DOI] [PubMed] [Google Scholar]
- 88.Stackebrandt E, Smida J, Collins M D. Evidence of phylogenetic heterogeneity within the genus Rhodococcus; revival of the genus Gordona (Tsukamura) J Gen Appl Microbiol. 1988;34:341–348. [Google Scholar]
- 89.Steck P A, Schwartz B A, Rosendahl M S, Gray G R. Mycolic acids: a reinvestigation. J Biol Chem. 1978;253:5625–5629. [PubMed] [Google Scholar]
- 90.Stodola F H, Lesuk A, Anderson R J. The chemistry of the lipids of tubercle bacilli. LIV. The isolation and properties of mycolic acid. J Biol Chem. 1938;126:505–513. [Google Scholar]
- 91.Takayama K, Jordi H C, Benson F. Separation of fatty acids as their p-bromophenacyl esters on a C30-bonded silica column by high performance liquid chromatography. J Liq Chromatogr. 1980;3:61–69. [Google Scholar]
- 92.Takayama K, Qureshi N, Jordi H C, Schnoes H K. Separation of homologous series of mycolic acids from Mycobacterium tuberculosis H37Ra by high performance liquid chromatography. Chromatogr Sci. 1979;10:91–101. [Google Scholar]
- 93.Thibert L, LaPierre S. Routine application of high-performance liquid chromatography for identification of mycobacteria. J Clin Microbiol. 1993;31:1759–1763. doi: 10.1128/jcm.31.7.1759-1763.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thibert L, Lebel F, Martineau B. Two cases of Mycobacterium haemophilum infection in Canada. J Clin Microbiol. 1990;28:621–623. doi: 10.1128/jcm.28.3.621-623.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Timpe A, Runyon E H. The relationship of “atypical” acid-fast bacteria to human disease. J Lab Clin Med. 1954;44:202–209. [PubMed] [Google Scholar]
- 96.Toriyama S, Yano I, Masui M, Kusunose E, Kusunose M, Akimori N. Regulation of cell wall mycolic acid biosynthesis in acid-fast bacteria. I. Temperature-induced changes in mycolic acid molecular species and related compounds in Mycobacterium phlei. J Biochem. 1980;88:211–221. [PubMed] [Google Scholar]
- 97.Torkko P, Suutari M, Suomalainen S, Paulin L, Larsson L, Katila M L. Separation among species of Mycobacterium terrae complex by lipid analyses: comparison with biochemical tests and 16S rRNA sequencing. J Clin Microbiol. 1998;36:499–505. doi: 10.1128/jcm.36.2.499-505.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tortoli E, Bartoloni A. High-performance liquid chromatography and identification of mycobacteria. Rev Med Microbiol. 1996;7:207–219. [Google Scholar]
- 99.Tortoli E, Bartoloni A, Burrini C, Mantella A, Simonetti M T. Utility of high-performance liquid chromatography for identification of mycobacterial species rarely encountered in clinical laboratories. Eur J Clin Microbiol Infect Dis. 1995;14:240–243. doi: 10.1007/BF02310365. [DOI] [PubMed] [Google Scholar]
- 100.Tortoli E, Kirschner P, Bartoloni A, Burrini C, Manfrin V, Mantella A, Scagnelli M, Scarparo C, Simonetti M T, Böttger E C. Isolation of an unusual Mycobacterium from an AIDS patient. J Clin Microbiol. 1996;34:2316–2319. doi: 10.1128/jcm.34.9.2316-2319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tortoli E, Kroppenstedt R M, Bartoloni A, Caroli G, Jan I, Pawlowski J, Emler S. Mycobacterium tusciae sp. nov. Int J Syst Bacteriol. 1999;49:1839–1844. doi: 10.1099/00207713-49-4-1839. [DOI] [PubMed] [Google Scholar]
- 102.Tortoli E, Piersimoni C, Kirschner P, Bartoloni A, Burrini C, Lacchini C, Mantella A, Muzzi G, Tosi C P, Penati V, Scarparo C, Simonetti M T, Bottger E C. Characterization of mycobacterial isolates phylogenetically related to, but different from, Mycobacterium simiae. J Clin Microbiol. 1997;35:697–702. doi: 10.1128/jcm.35.3.697-702.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tsuchiya H, Hayashi T, Naruse H, Takagi N. High-performance liquid chromatography of carboxylic acids using 4-bromomethyl-7-acetoxycoumarin as fluorescence reagent. J Chromatogr. 1982;234:121–130. doi: 10.1016/s0378-4347(00)81849-6. [DOI] [PubMed] [Google Scholar]
- 104.Tsukamura M. Proposal of a new genus, Gordona, for slightly acid-fast organisms occurring in sputa of patients with pulmonary disease and in soil. J Gen Microbiol. 1971;68:15–26. doi: 10.1099/00221287-68-1-15. [DOI] [PubMed] [Google Scholar]
- 105.Wallace R J, Jr, Silcox V A, Tsukamura M, Brown B A, Kilburn J O, Butler W R, Onyi G. Clinical significance, biochemical features, and susceptibility patterns of sporadic isolates of the Mycobacterium chelonae-like organism. J Clin Microbiol. 1993;31:3231–3239. doi: 10.1128/jcm.31.12.3231-3239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wayne L G. Mycobacterial speciation. In: Kubica G P, Wayne L G, editors. The mycobacteria: a sourcebook. New York, N.Y: Marcel Dekker, Inc; 1984. pp. 25–65. [Google Scholar]
- 107.Wayne L G, Engbaek H C, Engel H W B, Froman S, Gross W, Hawkins J, Käppler W, Karlson A G, Kleeberg H H, Krasnow I, Kubica G P, McDurmont C, Nel E E, Pattyn S R, Schröder K H, Showalter S, Tárnok I, Tsukamura M, Vergmann B, Wolinsky E. Highly reproducible techniques for use in systematic bacteriology in the genus Mycobacterium: tests for pigment, urease, resistance to sodium chloride, hydrolysis of Tween 80 and β-galactosidase. Int J Syst Bacteriol. 1974;24:412–419. [Google Scholar]
- 108.Wayne L G, Engel H W B, Grassi C, Gross W, Hawkins J, Jenkins P A, Käppler W, Kleeberg W H, Krasnow I, Nel E E, Pattyn S R, Richards P A, Showalter S, Slosarek M, Szabo I, Tárnok I, Tsukamura M, Vergmann B, Wolinsky E. Highly reproducible techniques for use in systematic bacteriology in the genus Mycobacterium: tests for niacin and catalase and for resistance to isoziazid, thiophene 2-carboxylic hydrazide, hydroxylamine and p-nitrobenzoate. Int J Syst Bacteriol. 1976;26:311–318. [Google Scholar]
- 109.Wayne L G, Good R C, Böttger E C, Butler R, Dorsch M, Ezaki T, Gross W, Jonas V, Kilburn J, Kirschner P, Krichevsky M I, Ridell M, Shinnick T M, Springer B, Stackebrandt E, Tarnok I, Tarnok Z, Tasaka H, Vincent V, Warren N G, Knott C A, Johnson R. Sematide- and chemotaxonomy-based analyses of some problematic phenotypic clusters of slowly growing mycobacteria: a cooperative study of the International Working Group on Mycobacterial Taxonomy. Int J Syst Bacteriol. 1996;46:280–297. doi: 10.1099/00207713-46-1-280. [DOI] [PubMed] [Google Scholar]
- 110.Wayne L G, Good R C, Tsang A, Butler R, Dawson D, Groothuis D, Gross W, Hawkins J, Kilburn J, Kubin M, Schröder K H, Silcox V A, Smith C, Thorel M-F, Woodly C, Yakrus M A. Serovar determination and molecular taxonomic correlation in Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum: a cooperative study of the International Working Group on Mycobacterial Taxonomy. Int J Syst Bacteriol. 1993;43:482–489. doi: 10.1099/00207713-43-3-482. [DOI] [PubMed] [Google Scholar]
- 111.Wolinsky E. Nontuberculous mycobacteria and associated diseases. In: Kuciba G P, Wayne L G, editors. The mycobacteria: a sourcebook. New York, N.Y: Marcel Dekker, Inc; 1984. pp. 1141–1207. [Google Scholar]
- 112.Wong M Y H, Steck P A, Gray G R. The major mycolic acids of Mycobacterium smegmatis: characterization of their homologous series. J Biol Chem. 1979;254:5734–5740. [PubMed] [Google Scholar]
- 113.Yost R W, Ettre L S, Conlon R D. Practical liquid chromatography: an introduction. Norwalk, Conn: Perkin-Elmer Corp.; 1980. [Google Scholar]