Summary
Maintenance of energy balance is essential for overall organismal health. Mammals have evolved complex regulatory mechanisms that control energy intake and expenditure. Traditionally, studies have focused on understanding the role of macronutrient physiology in energy balance. In the present study, we examined the role of the essential micronutrient iron in regulating energy balance. We found that a short course of dietary iron caused a negative energy balance resulting in a severe whole body wasting phenotype. This disruption in energy balance was because of impaired intestinal nutrient absorption. In response to dietary iron-induced negative energy balance, adipose triglyceride lipase (ATGL) was necessary for wasting of subcutaneous white adipose tissue and lipid mobilization. Fat-specific ATGL deficiency protected mice from fat wasting, but caused a severe cachectic response in mice when fed iron. Our work reveals a mechanism for micronutrient control of lipolysis that is necessary for regulating mammalian energy balance.
Subject areas: Physiology, Cellular physiology
Graphical abstract
Highlights
-
•
Iron causes negative energy balance by disrupting intestinal nutrient absorption
-
•
ATGL promotes lipolysis in response to iron-induced negative energy balance
-
•
ATGL protects from cachexia during dietary iron excess
Physiology; Cellular physiology
Introduction
Mammals have evolved complex homeostatic control mechanisms to maintain their energy balance within a defined set point. These include mechanisms that regulate nutrient acquisition, nutrient utilization, and energy storage, to achieve a balance between energy intake and energy expenditure. Energy expenditure is a function of multiple factors including basal metabolism, adaptive thermogenesis, and physical activity (Argilés et al., 2014; Spiegelman and Flier, 2001). Basal metabolism of an organism involves processes within the organism that are necessary to sustain life. Adaptive thermogenesis describes energy that is dissipated in the form of heat when animals consume a meal or when they adapt to a cold environment. Physical activity involves voluntary movements by the organism. Homeostatic control of energy balance is maintained through the interactions between genetic and environmental factors. Disruptions in these interactions that ultimately lead to energy imbalances will manifest as changes in energy storage (Spiegelman and Flier, 2001). Animals that have an energy intake that exceeds energy expenditure will have a net gain, resulting in increased energy storage. If animals have an energy intake that is less than energy expenditure, there will be a net loss, resulting in a loss of fat and other energy stores. The ability to maintain energy balance within the homeostatic set point is critical as disruptions to the balance and resulting changes in energy storage may render the organism more susceptible to diseases. For example, the increased adiposity in obesity predisposes individuals to cardiovascular disease and type 2 diabetes (O’|'Neill and O’|'Driscoll, 2015; Tune et al., 2017). Extreme causes of net energy loss can result in dangerously low body fat content that predisposes animals to infections, cardiovascular damage, and musculoskeletal issues (Carrero et al., 2013; Dobner and Kaser, 2018; Rausch et al., 2021). Thus understanding the genetic and environmental factors that regulate energy balance is necessary for our understanding of the metabolic basis of disease.
The essential micronutrient iron has emerged as a critical regulator of body weight and energy balance. In children and adults, there is a greater prevalence of iron deficiency in both overweight and obese individuals (Ikeda et al., 2013). The causality of iron in the regulation of adipose tissue stores has been demonstrated in animal studies. Iron deficient diet causes increased adiposity in rats and excess dietary iron causes a reduction in body fat mass in rodents (Yook et al., 2019). The possible underlying mechanisms for iron mediated control of energy balance and the resulting changes to fat storage are likely complex and multifactorial. For example, iron status affects energy intake by regulating food consumption. Iron deficiency is associated with appetite loss that can be reversed with iron supplementation possibly through its effects on the satiety hormone leptin (Gao et al., 2015; Lawless et al., 1994; Stoltzfus et al., 2004). Iron has dramatic effects on multiple aspects of the gastrointestinal tract that will influence nutrient absorption and affect the overall energy balance of the individual. For example, changes in luminal iron concentration will lead to changes in the composition of the intestinal microbiome that can lead to functional changes that control nutrient harvesting and uptake (Yilmaz and Li, 2018). Iron can also change the inflammatory state of digestive organs that will affect the ability of an individual to absorb nutrients (Niepel et al., 2018). Iron has also been shown to regulate systemic responses that control the ability of the small intestine to regulate nutrient absorption (Sanchez et al., 2018). The critical role of iron for providing adequate levels of oxygen will control an individual’s capacity to endure the amount and rigor of physical activity. Individuals that are anemic experience fatigue, reducing the amount of physical activity they undertake. Furthermore, individuals who experience low iron without anemia have impaired adaptation to aerobic activity (Brownlie et al., 2004). Lastly, as iron regulates various aspects of energy metabolism including cellular respiration, the ability of an organism to carry out their basic basal metabolic functions will also be dependent on the iron status of the individual.
The changes in energy balance and storage controlled by the iron status of an individual will result in physiological responses involved in energy substrate mobilization, storage, and breakdown. White adipose tissue (WAT) serves as the major storage depot for lipids, where excess lipid species are stored as triacylglycerol in lipid droplets. Under conditions of increased energetic demand or in response to stress stimuli, triacylglycerol is mobilized from WAT to meet organismal energy demands that are not met by diet (Duncan et al., 2007). Triacylglycerides (TG) in lipid droplets are hydrolyzed into diacylglycerol (DG) by adipose triglyceride lipase (ATGL) and monoacylglycerol (MG)/free fatty acids and glycerol by hormone-sensitive lipase (HSL). FFA and glycerol are then released into circulation where they can be utilized by peripheral tissues. ATGL and HSL are considered essential lipolytic enzymes and are negatively regulated by insulin and IGF-1 signaling through the PKA/PKC signaling cascade (Degerman et al., 1998; Wijkander et al., 1998). In vitro, it was shown that iron and transferrin from serum can induce lipolysis in an acute time frame and is independent of PKA and PKC signaling (Rumberger et al., 2004). More recently, it was shown that the elevated levels of fatty acid mobilization, adipose tissue HSL and ATGL in obese women correlated with elevated levels of ferritin, suggesting that in vivo, elevated iron stores correlates with increased fatty acid mobilization (Ryan et al., 2018). Although iron has an established correlative relationship with adipose tissue physiology, a causative role for iron in regulating lipid mobilization in vivo and how this relates to iron mediated changes in energy balance, as well as the underlying mechanisms for these processes remain unknown.
In the present study, we examined the role of the essential micronutrient iron in regulating energy balance. Using a dietary model of iron overload, we found that an acute course of excess dietary iron leads to a negative energy balance and profound whole body wasting in mice. The negative energy balance was because of iron dependent intestinal nutrient malabsorption. To meet energetic demands, we found that iron caused increased lipid utilization associated with increased lipolysis, lipid mobilization, and fat wasting. Using a transgenic mouse model, we found that lipolysis was dependent on ATGL activity specifically in adipose tissue. Animals lacking ATGL in adipose tissue shifted to carbohydrate utilization when fed dietary iron and exhibited a severe cachectic response because of the intestinal nutrient malabsorption. Our work highlights the multifaceted role of iron regulation of organismal metabolism and provides an in vivo mechanism for micronutrient control of lipolysis that is necessary for regulating mammalian energy balance.
Results
Iron rich diet causes negative energy balance and nutrient malabsorption
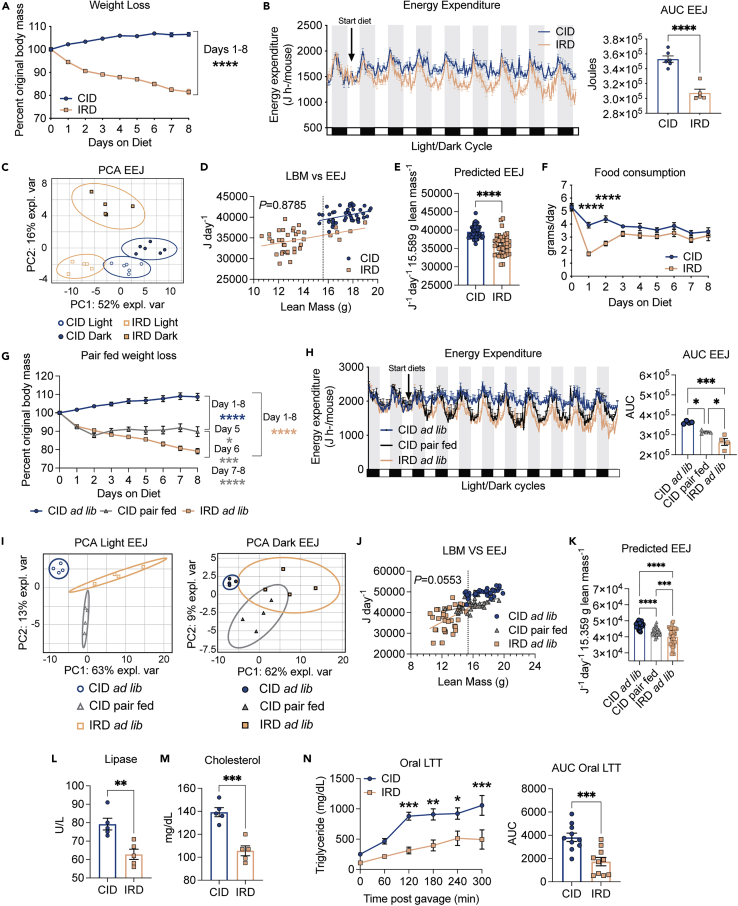
Although it is well established that iron regulates organismal metabolism and energy balance, the underlying mechanisms regulating these processes are not completely known. We established a dietary model to determine the mechanistic basis for the effects of an acute course of surplus dietary iron on organismal metabolism and energy balance. Carbonyl iron is a dietary iron supplement that has reduced inflammatory effects on the gastrointestinal tract relative to iron sulfate-based alternatives (Devasthali et al., 1991). We fed 6-week-old C57BL/6 mice an iron rich diet (IRD) containing 2% carbonyl iron or a nutrient matched control iron diet (CID) and found that while CID-fed mice exhibited an expected slight increase in whole body weight over the course of the experiment, IRD-fed mice exhibited a drastic whole-body wasting phenotype, with mice losing approximately 20% of total body weight by day 8 post diet initiation (Figures 1A and S1A). Net negative energy balance caused by the IRD can be because of an increase in energy expenditure and/or a decrease in energy intake. To determine the effects of dietary iron on energy expenditure, we fed 6-week-old C57BL/6 mice CID or IRD for nine days and housed them in the Comprehensive Lab Animal Monitoring System (CLAMS) to measure gas exchange volumes and rates continuously over the experimental course (mice were allowed to acclimate for a day before introduction of the diets). We observed a drastic continuous decrease in energy expenditure of mice following introduction of the IRD (Figures 1B and S1B). Principal component analysis (PCA) of light/dark cycle EEJ in CID and IRD-fed mice revealed a distinct clustering of samples according to light cycle and diet, suggesting that patterns of energy expenditure are impacted by diet and light cycle (Figure 1C). The clustering is supported by nonoverlapping 95% CIs (ellipses) (Figure 1C). As energy expenditure is positively correlated with body mass, we performed a regression-based analysis of daily energy expenditure as a function of daily total body mass (TMB) or daily lean body mass (LBM) over the eight day dietary regimen (Figures 1D and S1C). We found that the relationship between TBM and energy expenditure significantly depended on diet and that energy expenditure increased significantly with TBM on both CID and IRD (Figure S1C). Energy expenditure increased significantly with increased LBM and the influence of LBM on energy expenditure was equivalent for mice fed CID or IRD (Figure 1D). Interestingly, ANCOVA analysis revealed that IRD resulted in decreased energy expenditure as a function of mouse LBM relative to mice fed CID, suggesting that IRD does not cause negative energy balance by increasing energy expenditure (Figure 1E). Changes in activity level and thermogenesis can contribute to differences in energy expenditure. We observed no significant difference in total activity levels, but PCA of light/dark cycle activity levels revealed distinct clustering of dark cycle activity, suggesting that activity patterns are impacted by diet during the dark cycle (Figures S1D and S1E). In addition, mice fed IRD exhibited a decrease in core body temperature over the course of the dietary regimen, suggesting that reduced energy expenditure caused by excess dietary iron may in part be because of a decrease in thermogenesis (Figure S1F).
Figure 1.
Iron rich diet causes negative energy balance and nutrient malabsorption
Six-week old C57BL/6 males were given control diet (CID) or 2% carbonyl iron diet (IRD) and individually housed in static or metabolic cages in the Comprehensive Laboratory Animal Monitoring System (CLAMS). Weight, energy expenditure and food consumption were monitored.
(A) Percent original weight over an eight-day diet regimen on CID or IRD. Data shown represents two pooled independent experiments (n = 11 mice per group).
(B) Hourly energy expenditure and corresponding area under the curve analysis for mice fed CID or IRD for eight days in CLAMS. White and black/gray boxes/shaded regions represent light/dark cycles, respectively. Data shown represents one independent experiment (n = five to six mice per group).
(C) Principal component analysis of energy expenditure in (B) for CID and IRD fed mice in light/dark cycles. Ellipses are indicative of 95% confidence intervals. Data shown represents analysis of one independent experiment (n = five to six mice per group).
(D and E) Linear regression analysis of daily energy expenditure as a function of daily lean body mass (LBM) over an eight day period and corresponding (E) ANCOVA-predicted EEJ at group average LBM of 15.589 g. Dotted vertical line in (D) represents group average lean mass (CID linear regression, R2 = 0.1467, F = 7.735, dF = 1, 45, p = 0.0079, Y = 568.2∗X + 30,500; IRD linear regression, R2 = 0.1396, F = 5.841, dF = 1, 36, p = 0.0208, Y = 519.6∗X + 27,524; pooled slope = 534.5) Data shown represents analysis of one independent experiment (n = five to six mice per group).
(F) Average daily food consumption of mice fed CID or IRD. Data shown represents two pooled independent experiments (n = 11 mice per group). (G-K) C57BL/6 male mice were fed CID ad libitum (ad libitum), IRD ad libitum (ad libitum), or CID pair fed matched to average historical iron food consumption values.
(G) Percent original body weight for dietary regimens shown. Data shown represents two pooled independent experiments (n = 9 mice per group). Blue asterisks denote comparisons between CID ad libitum and CID pair fed. Gray asterisks denote comparisons between CID pair fed and IRD ad libitum. Peach asterisks denote comparisons between CID ad libitum and IRD ad libitum.
(H) Hourly energy expenditure and corresponding area under the curve analysis for mice on three dietary regimens. White and black/gray boxes/shaded regions represent light/dark cycles, respectively. Data shown represents one independent experiment (n = 4 mice per group).
(I) Principal component analyses of energy expenditure in (H) for light/dark cycles of three dietary regimens. Ellipses are indicative of 95% confidence intervals. Data shown represents analyses of one independent experiment (n = 4 mice per group).
(J) Regression-based analysis of absolute daily EEJ against daily lean mass over an eight day period and corresponding.
(K) ANCOVA-predicted EEJ at group average lean mass of 15.359 g. Dotted vertical line in regression plot represents group average lean mass (CID ad libitum linear regression, R2 = 0.2913, F = 13.98, dF = 1,34, p = 0.0007, Y = 646.6∗X + 38,295; CID pair fed linear regression, R2 = 0.2585, F = 11.85, dF = 1,34, p = 0.0015, Y = 906.9∗X + 29,379; IRD ad libitum linear regression, R2 = 0.2204, F = 9.61, dF = 1,34, p = 0.0039, Y = 1982∗X + 11,641; pooled slope = 1159). Data shown represents analysis of one independent experiment (n = 4 mice per group).
(L and M) Circulating levels of (L) Lipase and (M) total cholesterol in serum on day 9 of diet regimen. Data shown represents one independent experiment (n = 5 mice per group).
(N) Oral lipid tolerance test of mice fed CID or IRD for eight days. Data shown represents analysis of two pooled independent experiments (n = 10 mice per group)All CLAMS data plotted in zeitgeber time. Data represent mean ± SEM; CID ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Related to Figure S1.
Notably, mice fed IRD eat significantly less food during the first 48 h post diet initiation, with no significant difference in food consumption between the dietary groups by day 3 post diet initiation (Figure 1F). To determine if this initial food aversion response in IRD-fed mice was responsible for the whole-body wasting, we performed a pairwise feeding analysis. Pair fed mice on CID had a steep decrease in body mass within the first 48 h of the diet regimen and increased body mass thereafter; whereas mice fed IRD ad libitum exhibited continual weight loss over the 8 days diet regimen (Figures 1G and S1G). Thus, the initial reduced feeding alone does not account for the continual whole body wasting caused by the acute course of excess dietary iron. Mice fed IRD also exhibited comparable body temperature and activity levels as pair fed mice on CID (Figures S1H, S1I, and S1J). We next analyzed energy expenditure of mice of the three dietary regimens in the CLAMS and found that mice fed IRD had a significant decrease in energy expenditure relative to both groups of mice fed CID ad libitum and pair fed (Figures 1H, 1I, and S1K). Consistently, PCA revealed distinct clusters of energy expenditure between the three dietary regimens during the light cycles (Figure 1I). We performed regression-based analyses of daily energy expenditure as a function of daily TBM and daily LBM for our pairwise feeding conditions to determine if dietary regimens caused a mass-independent effect on energy expenditure (Figures 1J, 1K, and S1L). Consistent with our previous analyses of CID and IRD, we found that the relationship between TBM and energy expenditure significantly depended on diet (F = 41.21 df = 2, 102, p < 0.0001) and energy expenditure increased significantly with TBM in all dietary regimens (Figure S1L). Daily energy expenditure increased significantly as a function of daily LBM and the influence of LBM on energy expenditure was equivalent for mice on the three dietary regimens (Figure 1J). We observed that IRD decreased energy expenditure as a function of LBM relative to mice pair fed CID or given CID ad libitum (Figure 1K), demonstrating that IRD does not cause negative energy balance through reduced energy intake or through increasing energy expenditure.
Energy intake is a function of both food intake and the subsequent nutrient absorption by the gastrointestinal tract. Pancreatic-derived enzymes are essential for digestion and absorption of dietary macronutrients and excess dietary iron can contribute to dysfunction of exocrine pancreas (Kimita and Petrov, 2020). Consistent with this, we observed a significant reduction in circulating levels of lipase in mice fed IRD, suggesting that IRD disrupts pancreatic production of lipase and subsequent ability to digest triglyceride and fats into free fatty acids and glycerol for uptake in the intestines (Figure 1L). As lipase inhibition is also known to reduce cholesterol absorption (Erdmann et al., 2004; Ke et al., 2020), we saw a corresponding decrease in circulating total cholesterol levels in mice fed IRD (Figure 1M). To address if IRD reduced the ability of mice to absorb dietary lipids, we performed an oral lipid tolerance test (OLTT) in mice fed CID or IRD for eight days and found that mice fed IRD exhibited a significant reduction in their ability to absorb orally administered lipids(Figure 1N). Finally, mice fed IRD excreted more calories in their stool and had decreased net calorie absorption from diets relative to mice fed CID (Figure S1M). Taken together, our data show that IRD causes negative energy balance by causing intestinal nutrient malabsorption.
Dietary iron increases lipid utilization, lipid mobilization, and wasting of fat energy stores
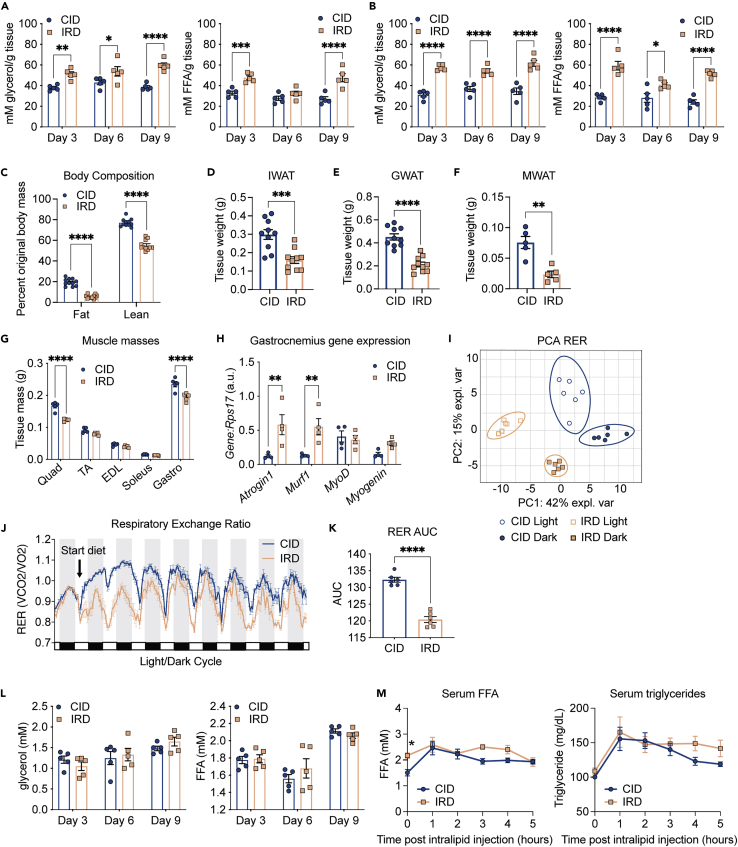
Negative energy balance will cause mobilization of energy substrates from endogenous stores. Mobilization of the energy stores in white adipose tissue (WAT) involves the liberation of triacylglycerol from lipid droplets stored in adipose tissue that provides free fatty acids (FFA) and glycerol to other tissues and organs. Using an ex vivo lipolysis assay, we found that inguinal WAT (IWAT) and gonadal WAT (GWAT) from IRD-fed mice released significantly higher levels of free nonesterified fatty acids (FFA) and free glycerol compared to mice fed CID over the course of the dietary regimen (Figures 2A and 2B). Mobilization of fat stores can result in loss of body fat mass. To determine the extent of fat energy store depletion, we utilized MRI (MRI) to analyze fat and lean body composition of mice fed CID or IRD for eight days. We found that the whole body wasting and increased lipid mobilization in mice fed IRD was associated with wasting of fat energy stores (Figure 2C). We also measured masses of representative WAT pads and found a significant reduction in the mass of IWAT, GWAT, and mesenteric WAT (MWAT) in mice fed IRD (Figures 2D–2F). MRI and direct measurement of hindlimb muscles that represent varied myofiber composition and the heart revealed that mice fed IRD also exhibit wasting of skeletal muscle but not cardiac muscle (Figures 2C, 2G, and S2A, and S2B). Further, we observed significant upregulation of two muscle-specific atrogenes, Atrogin-1 and Murf-1, in gastrocnemius muscles of mice fed IRD— a hallmark of skeletal muscle wasting (Figure 2H) (Baehr et al., 2011; Bodine et al., 2001; Gomes et al., 2001). We also found upregulation of Myogenin in gastrocnemius muscles from IRD fed mice, which is associated with Type I myofiber development (Figure 2H) (Hughes et al., 1999). Consistent with our findings, body composition analysis of mice from our pairwise feeding experiments confirmed that fat and lean body composition were significantly reduced in the IRD ad libitum group relative to pair fed mice on CID (Figure S2C). Similarly, we found that mice fed IRD ad libitum had the greatest decrease in IWAT mass and hindlimb muscle masses after eight days on the diet regimen (Figures S2D and S2E). In support of IRD-induced skeletal muscle wasting, we also observed the greatest upregulation of atrogene expression in gastrocnemius muscles from mice fed IRD ad libitum (Figure S2F). Taken together, our data demonstrate that dietary iron causes lipid mobilization that results in adipose tissue wasting, as well as muscle wasting, that is independent of the initial food aversion response.
Figure 2.
Excess dietary iron increases lipid utilization, lipid mobilization and wasting of fat energy stores
Six-week old C57BL/6 males were provided control (CID) or 2% carbonyl iron diet (IRD) for 3, 6, or 9 days
(A and B) Ex vivo lipolysis assay measuring glycerol and free fatty acids (FFA) released from (A) IWAT (B) and GWAT. Data shown represents one independent experiment (n = 5 mice per group).
(C) Body composition analyses using EchoMRI of mice fed CID or IRD for 6 days. Fat and lean mass were normalized to original body mass. Data shown represents two pooled independent experiments (n = 11 mice per group).
(D–F) Tissue masses of (D) IWAT (E) GWAT and (F) MWAT from mice fed CID or IRD for 6 days. Data shown for D,E represents two pooled independent experiments (n = 10 mice per group). Data shown for F represents one independent experiment (n = 5 mice per group).
(G) Muscle masses from hindlimb of mice fed CID or IRD for 8 days (Quadricep; Quad, tibialis anterior; TA, extensor digitorum longus; EDL, soleus, and gastrocnemius; Gastro). Data shown represents one independent experiment (n = 5 mice per group).
(H) Gene expression in gastrocnemius of mice fed CID or IRD for 8 days. Gene expression was normalized to housekeeping expression of Rps17. Data shown represents one independent experiment (n = 4 mice per group).
(I–K) Mice were housed in the CLAMS and given CID or IRD. The respiratory exchange ratio over the course of the experiment was determined. (I) Principal component analysis for the respiratory exchange ratio (RER) of CID and IRD fed mice in light/dark cycles. Ellipses are indicative of 95% confidence intervals. Data shown represents analyses of one independent experiment (n = 6 mice per group). (J) Average hourly respiratory exchange ratio of mice fed CID or IRD (RER) calculated as ratio of ml CO2 OUT (respired):ml O2 IN (inhaled). Data shown represents one independent experiment (n = 6 mice per group). (K) Area under the curve analysis for RER for periods of matched food consumption.
(L) Circulating levels of glycerol and FFA in serum of fasted CID or IRD mice on days 3, 6, and 9 of diet time course. Data shown represents one independent experiment (n = 5 mice per group).
(M) Intraperitoneal lipid tolerance test (i.p. LTT) performed on fasted mice fed CID or IRD for nine days. Data shown represents one independent experiment (n = 5 mice per group). Data represent mean ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Related to Figure S2.
We hypothesized that the increased lipid mobilization was indicative of IRD causing a shift toward lipid utilization. To determine how IRD influences energy substrate preference, we measured respiratory exchange ratio (RER) by housing mice fed CID or IRD in the CLAMS. An RER close to one is indicative of carbohydrate utilization, whereas an RER closer to 0.7 is indicative of lipid substrate utilization (Speakman, 2013). Using PCA, we found that mice from the different dietary conditions exhibited distinct clustering during their day and night cycles, indicating that there were distinct RE-Rs for each dietary condition and their respective light/dark cycles (Figure 2I). We found that IRD-fed mice exhibited a dramatic shift toward lipid utilization (Figure 2J). Increased lipid utilization is characteristic during periods of fasting when mice will draw from white adipose tissue (WAT) stores (Rosen and Spiegelman, 2006; Schmidt-Nielsen, 1997). Therefore, we performed an RER area under the curve analysis for days 3–8, when food consumption was comparable between CID and IRD-fed mice to analyze energy substrate utilization independent of differences in food consumption (Figure 2K). We found that the IRD induced an increase in lipid utilization independent of the fasted state. We performed RER analyses for the pair fed experimental cohort and found that IRD induced a significant shift toward lipid utilization relative to pair fed mice fed CID—further demonstrating that IRD increases reliance on lipids as an energy substrate (Figures S2G and S2I). Interestingly, the increased reliance on lipids as an energy source did not result in a decrease in circulating lipids or glycerol in fasted mice fed CID or IRD over the course of the experiment (Figure 2L). Consistently, mice fed IRD had a comparable rise and clearance of FFA and triglycerides in a lipid tolerance test performed on day 9 post diet initiation, suggesting that although IRD induces a shift toward lipid utilization, this does not increase rates of lipid turnover (Figure 2M). Taken together, our data demonstrate that dietary iron excess causes negative net energy balance, inducing a global shift toward lipid utilization that is fueled by mobilization of endogenous fat energy stores.
An acute course of excess dietary iron does not induce insulin resistance
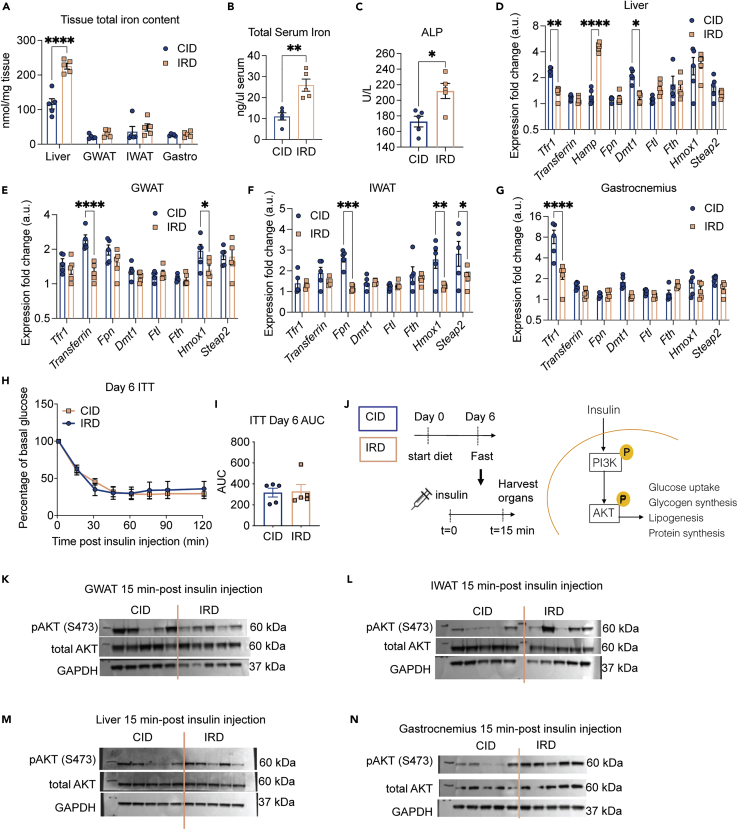
Iron metabolism can influence glucose homeostasis by altering insulin sensitivity. We previously demonstrated in C3H/HeJ mice that an acute course of dietary iron caused transient insulin resistance (IR) during infection with an enteric pathogen and that this was associated with iron overload in WAT (Sanchez et al., 2018). In C57BL/6 mice it was previously shown that long-term administration of dietary iron causes IR associated with an increase in hepatic and adipose tissue iron levels as well as an increase in serum iron (Dongiovanni et al., 2013). We found that in C57BL/6 mice, an acute course of IRD resulted in significantly higher levels of iron in the liver and serum, but not other metabolic tissues including the visceral fat (GWAT), subcutaneous fat (IWAT), and muscle (gastrocnemius) (Figures 3A and 3B). The livers of IRD fed mice were significantly smaller than that of CID ad libitum fed mice. However CID pair fed mice had comparable liver size as IRD mice indicating that the reduced food consumption amount rather than hepatic iron overload caused a reduction in liver size (Figures S3A and S3B). IRD fed mice exhibited significantly elevated ALP levels but no difference in AST levels, and only a modest and insignificant increase in ALT levels (Figure 3C and Figures S3C and S3D), suggesting that IRD may cause liver damage. Consistent with a systemic iron overloading response, IRD induced hepcidin (Hamp) expression in liver and significantly reduced expression of genes involved with iron uptake (Tfr1, Dmt1) (Figures 3D and S3E). Both GWAT and IWAT showed reduced expression of heme oxygenase-1 (Hmox1) which is considered to be a marker of iron overload and inflammation in adipose tissue (Moreno-Navarrete et al., 2017). IRD caused a reduction in transferrin expression in GWAT and reduced ferroportin (Fpn) expression in IWAT—suggesting that both visceral and subcutaneous adipose tissues depots respond to dietary iron overload (Figures 3E, 3F, S3F, and S3G). IRD induced a modest reduction in gastrocnemius expression of iron uptake genes (Dmt1, Hmox1, Steap2) and a significant reduction in transferrin receptor 1 (Tfr1) (Figures 3G and S3H).
Figure 3.
An acute course of dietary iron does not cause insulin resistance
Six-week old C57BL/6 males were provided control (CID) or 2% carbonyl iron diet (IRD).
(A and B) Total iron (ferric and ferrous) content in (A) tissues and (B) serum from mice fed CID or IRD for 6 days. Data shown represents one independent experiment (n = 5 mice per group).
(C) Circulating levels of alkaline phosphatase (ALP) in mice fed CID or IRD for nine days. Data shown represents one independent experiment (n = 5 mice per group).
(D–G) Gene expression in (D) liver, (E) GWAT, (F) IWAT and (G) gastrocnemius muscles of mice 6 days post diet initiation. Genes were normalized to expression of housekeeping gene Rps17 and subsequently normalized to the lowest normalized Gene:Rps17 value within each tissue for each respective gene. Data shown represents one independent experiment (n = 5 mice per group).
(H and I) Insulin tolerance test (ITT) normalized to basal glucose levels and (I) corresponding area under the curve (AUC) analysis performed on day 6 post diet initiation. Data shown represents one independent experiment (n = 5 mice per group).
(J) Diagram for experimental setup to analyze insulin signaling. Following a 6 h fast on day 6 of the diet time course, mice were i.p. injected with insulin and liver, fat pads, and muscle was harvested 15 min post insulin injection to analyze tissue-specific insulin signaling.
(K–N) Western blot analyses for AKT protein activation in protein extracts from (K) GWAT, (L) IWAT, (M) liver, and (N) gastrocnemius muscle from hindlimb. Data shown represents one independent experiment (n = 5 mice per group). Data represent mean ± SEM ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Related to Figures S3–S5.
We next asked whether an acute course of IRD could affect insulin sensitivity in C57BL/6 mice. We performed an insulin tolerance test (ITT) on days 3, 6, and 9 post-diet initiation on mice fed CID and IRD. We found no difference in insulin sensitivity between mice fed the different diets at any of the examined time points (Figures 3H, 3I, S4A–S4J). It has been previously demonstrated that iron overload can lead to tissue-specific insulin resistance; notably, Dongiovanni and colleagues observed insulin resistance in visceral WAT but not subcutaneous WAT in mice fed excess dietary iron (Dongiovanni et al., 2013; Sanchez et al., 2018). Therefore, we examined activation of insulin signaling in the GWAT and IWAT, as well as the liver and gastrocnemius muscle in CID and IRD fed mice. Mice fed CID and IRD for six days were injected with insulin and tissues were harvested 15 min post-insulin injection and processed to measure activation of AKT, a central kinase in the intracellular insulin signaling cascade (Figure 3J). We found no differences in phosphorylated AKT in the GWAT from mice in either dietary condition (Figures 3K, S5A, and S5B). Interestingly, we found a trend toward increased phosphorylation of AKT in the IWAT, liver, and gastrocnemius muscles from mice fed IRD (Figures 3L–3N and S5C–S5H). Thus, our model of acute dietary iron enrichment in C57BL/6 mice causes changes in organismal lipid physiology that is not associated with insulin resistance.
Iron-induced lipid mobilization and adipose tissue wasting is dependent on fat-specific ATGL activity
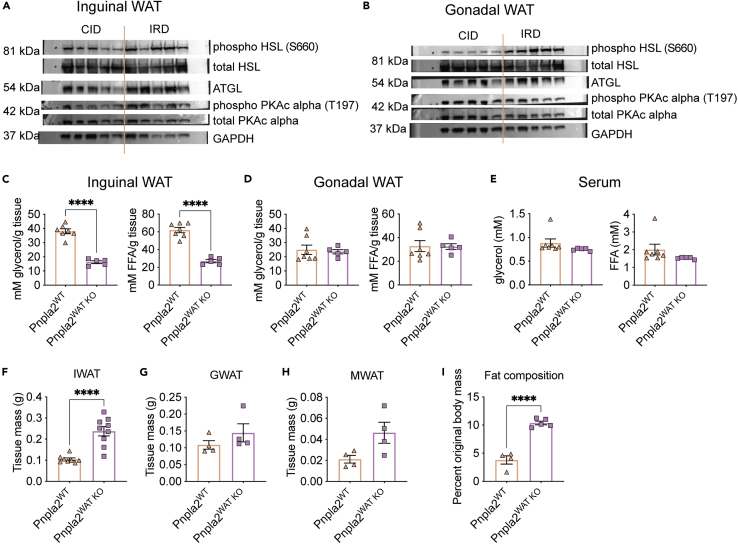
The bulk of lipid mobilization from adipose tissue is mediated through lipolysis. In canonical adipose tissue lipolysis, triglycerides stored in lipid droplets are hydrolyzed by adipose triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL) to produce free glycerol and fatty acids to be supplied into circulation and fuel peripheral tissue metabolism (Haemmerle et al., 2006; Zimmermann et al., 2004). ATGL is the rate limiting lipase present on lipid droplets that is responsible for hydrolyzing triacylglycerol to diacylglycerol. We found that mice fed an IRD exhibited elevated levels of ATGL protein in both subcutaneous (IWAT) and visceral adipose tissue (GWAT) (Figures 4A, 4B, S6, and S7). ATGL activity is directly regulated by its coactivator CGI-58, which is regulated by protein kinase A (PKA). Activated PKA will phosphorylate Perilipin-1 that is conjugated with CGI-58 on lipid droplet surfaces, thereby liberating CGI-58 to activate ATGL (Gruber et al., 2010; Lass et al., 2006). It was previously demonstrated that adipose tissue cultures exposed to iron rich media and human sera (transferrin rich) upregulate lipolysis in a PKA-independent manner, suggesting that iron-induced lipolysis may be regulated in a noncanonical manner, though a mechanism was not determined (Rumberger et al., 2004). Contrary to previous in vitro observations, we found a significant increase in the levels of phosphorylated PKAc in IWAT and GWAT of IRD fed mice, indicating that iron diet leads to increased activation of PKA in WAT in vivo (Figures 4A, 4B, S6, and S7). Finally, IRD led to an increase in the activation of the downstream lipase, HSL, in IWAT and GWAT (Figures 4A, 4B, S6, and S7). Taken together, our data demonstrate that an acute course of excess dietary iron leads to increased activation of the regulators of canonical adipose lipolysis in both subcutaneous and visceral WAT.
Figure 4.
Iron induced lipid mobilization and adipose tissue wasting is dependent on fat specific ATGL activity
(A and B) Six-week old C57BL/6 males were given control (CID) or 2% carbonyl iron diet (IRD) for 3 days. Following a 6-h fast, mice were sacrificed and fat pads were harvested for analysis. (A) Western blot analysis for activated proteins in the canonical lipolysis pathway in (A) IWAT and (B) GWAT protein extracts. (HSL/phospho-HSL Ser563, hormone sensitive lipase; ATGL, Adipose triglyceride lipase; PKA-c, cAMP dependent protein kinase catalytic subunit alpha; GAPDH, Glyceraldehyde3-phosphate dehydrogenase). n = 5 mice per condition.
(C and D) Pnpla2;Fabp4 cre+ (Pnpla2WAT KO) and Pnpla2;Fabp4 cre- (Pnpla2WT) male littermates between six and eight-weeks old were given 2% carbonyl iron diet (IRD) for 3 days and ex vivo lipolysis assays were performed following a 6 h fast. Glycerol and free fatty acids (FFA) released from (C) IWAT (D) and GWAT. n = five to seven mice per condition.
(E) Circulating levels of glycerol and FFA in serum of Pnpla2WAT KO and Pnpla2WT littermates fed IRD for 6 days n = five to seven mice per condition.
(F–H) Raw mass of fat pads from Pnpla2WAT KO and Pnpla2WT littermates fed IRD for five days (F) IWAT (G) GWAT (H) MWAT. For (F) n = eight to nine mice per condition and represent two experiments combined. For (G-H) n = 4 mice per condition.
(I) Body fat composition of Pnpla2WAT KO and Pnpla2WT littermates fed IRD for 5 days using EchoMRI. Fat mass was normalized to original total body mass. n = four to five mice per condition. Data represent mean ± SEM; ∗∗∗∗p < 0.0001. Panels 4A-E, G-I represent one independent experiment and panel 5F represents two pooled experiments. Related to Figures S6 and S7.
To test if ATGL-dependent lipolysis in adipose tissue is necessary for mediating IRD-induced lipid mobilization and adipose tissue wasting, we generated a transgenic mouse model with the gene that encodes ATGL, Pnpla2, specifically deleted from adipose tissue (floxed Pnpla2 x Fab4 cre). Using an ex vivo lipolysis assay, we found that the IWAT from IRD-fed mice lacking adipose-specific ATGL (floxed Pnpla2;Fab4 cre+, designated Pnpla2WAT KO) secreted significantly less FFA and glycerol than wild type mice fed IRD (floxed Pnpla2;Fab4 cre-, designated Pnpla2WT) (Figure 4C). Notably, we observed comparable levels of secreted FFA and glycerol from GWAT and no difference in circulating levels of FFA or glycerol between Pnpla2WAT KO and Pnpla2WT mice fed IRD (Figures 4D and 4E). The apparent difference in rates of lipid mobilization between subcutaneous and visceral adipose tissue may reflect a preferential pattern of organismal lipid storage across fat depots in response to dietary iron excess (Luong et al., 2019; Schoettl et al., 2018; Tchkonia et al., 2013). Consistent with this, Pnpla2WAT KO mice were protected from IRD-induced IWAT wasting, but were only moderately protected from wasting of visceral fat pads, GWAT and MWAT (Figures 4F–4H). Though we observed differences in the pattern of subcutaneous and visceral fat pad wasting, body composition analyses showed that Pnpla2WAT KO mice had significantly more total fat mass relative to Pnpla2WT littermates fed IRD (Figure 4I). Together, these data suggest that ATGL activity is necessary for IRD-induced lipolysis, lipid mobilization, and fat wasting primarily in subcutaneous adipose tissue.
Dietary iron induced adipose-specific ATGL activity protects from wasting of lean energy stores
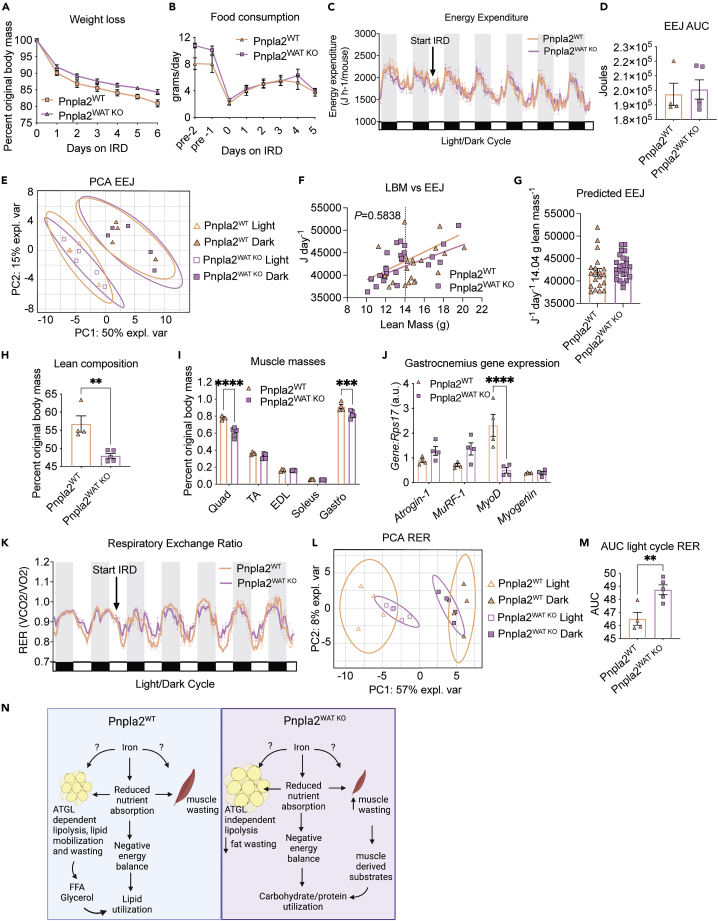
Having established a role for adipose tissue specific ATGL in IRD-mediated lipid mobilization and fat wasting, we next asked if ATGL activity in fat was required for the changes in energy balance we observed in IRD-fed mice (Figure 1). Despite being protected from wasting of subcutaneous fat and having a higher body fat composition (Figures 4F and 4I), Pnpla2WAT KO displayed comparable total body wasting to that exhibited by their wild type littermates when fed dietary iron (Figures 5A and S8A). This suggests that mice lacking ATGL activity in adipose tissue are not protected from the negative energy balance caused by IRD. Consistent with this, we found that food consumption was comparable between Pnpla2WAT KO and their wildtype littermates when fed IRD (Figure 5B). Our analyses with the CLAMS showed that energy expenditure of Pnpla2WT and Pnpla2WAT KO mice was comparable under normal chow conditions (Figures 5C and S8B). Following the introduction of IRD, Pnpla2WT and Pnpla2WAT KO mice showed equivalent changes in energy expenditure (Figures 5C, 5D, and S8B). When we examined the light/dark cycle EEJ for Pnpla2WT and Pnpla2WAT KO mice by PCA, we saw no influence of genotype on EEJ for light or dark cycles when animals were fed IRD (Figure 5E). Furthermore, we found that Pnpla2WT and Pnpla2WAT KO mice fed IRD displayed comparable daily EEJ relative to daily TBM and daily LBM over the course of the experiment (Figures 5F, 5G, S8C, and S8D). Analysis of body temperature and activity over the course of the diet regimen revealed no difference in body temperature, total activity levels, or differences in light/dark cycle activity levels between genotypes (Figures S8E–S8H). Taken together, our results demonstrate that ATGL activity in adipose tissue is not necessary for the effects of dietary iron on net energy balance, and does not change energy expenditure, activity levels, basal metabolism or food consumption when animals are fed an acute course of IRD.
Figure 5.
Dietary iron induced adipose-specific ATGL activity protects from wasting of lean energy stores
Littermate Pnpla2;Fabp4 cre- (Pnpla2WT) and Pnpla2;Fabp4 cre+ (Pnpla2WAT KO) males between six and eight-weeks old were housed in comprehensive laboratory animal monitoring system (CLAMS) metabolic cages for 6 days. Mice were provided a 2% carbonyl iron diet (IRD) after a brief acclimation period. Daily measurements were taken for body mass and food intake.
(A) Percent original weight of Pnpla2WT and Pnpla2WAT KO mice fed IRD. n = eight to nine mice per condition. Data shown represents two pooled independent experiments.
(B) Daily average food intake of Pnpla2WT and Pnpla2WAT KO mice before and during IRD feeding period. Data shown represents three pooled independent experiments (n = 5–10 mice per group and per timepoint. For some timepoints, food was shredded by some mice and was not weighable for those animals at those timepoints).
(C and D) Average hourly energy expenditure (EEJ) calculated from VO2 and VCO2 (mL/h) using the modified Weir equation and corresponding (D) area under the curve analysis for EEJ. Data shown represents one independent experiment (n = four to five mice per group).
(E) Principal component analysis of average EEJ from light/dark cycles from Pnpla2WT and Pnpla2WAT KO mice fed IRD. Ellipses are indicative of 95% confidence intervals. Data shown represents analysis of one independent experiment (n = four to five mice per group).
(F and G) Daily energy expenditure as a function of daily lean body mass (LBM) over a five day period and corresponding (G) ANCOVA-predicted EEJ at group average LBM of 14.04 g. Dotted vertical line in regression plot represents group average lean mass (Pnpla2WT linear regression R2 = 0.1645, F = 3.545, dF = 1, 18, p = 0.076, Y = 795.3∗X+ 30,861; Pnpla2WAT KO linear regression R2 = 0.4920, F = 22.28, dF = 1, 23, p < 0.0001 Y = 1041∗X+ 28,522; pooled slope = 949.6) Data shown represents analysis of one independent experiment (n = four to five mice per group).
(H) Lean body composition of Pnpla2WT and Pnpla2WAT KO mice fed IRD using Echo MRI. Lean mass was normalized to original total body mass. Data shown represents one independent experiment (n = four to five mice per group).
(I) Muscle masses from hindlimb of Pnpla2WT and Pnpla2WAT KO mice fed IRD for five days (Quadricep; Quad, tibialis anterior; TA, extensor digitorum longus; EDL, soleus, and gastrocnemius; Gastro). Data shown represents one independent experiment (n = four to five mice per group).
(J) Gene expression in gastrocnemius of Pnpla2WT and Pnpla2WAT KO mice fed IRD for six days. Gene expression is normalized to Rps17. Data shown represents one independent experiment (n = 4 mice per group).
(K and ) Respiratory exchange ratio (RER) and (L) Principal component analysis of average RER from light/dark cycles from Pnpla2WT and Pnpla2WAT KO mice fed IRD. Ellipses are indicative of 95% confidence intervals. Data shown represents analysis of one independent experiment (n = four to five mice per group).
(M) Area under the curve (AUC) analysis of RER for light cycle periods of CLAMS monitoring following introduction of IRD. Data shown represents one independent experiment (n = four to five mice per group).
(N) Model of dietary iron regulation of energy balance and ATGL-dependent lipid mobilization. All CLAMS data plotted in zeitgeber time. White/black boxes on the X axis represent light/dark cycles of 24-h day. AUC of CLAMS analyses taken from total average values per mouse. Data represent mean ± SEM; ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Related to Figure S8.
We hypothesized that Pnpla2WAT KO mice undergo a compensatory increase in lean energy store wasting that can account for the whole body weight loss driven by IRD. Consistent with our hypothesis, we found that Pnpla2WAT KO mice exhibited increased wasting of lean tissue stores compared to their Pnpla2WT littermates fed IRD (Figure 5H). These differences in body composition were specific to mice fed IRD because mice lacking ATGL in adipose tissue and their wild type littermates exhibited comparable body composition when fed control chow (Figure S8I). Direct measurement of hindlimb muscle masses similarly showed that Pnpla2WAT KO mice fed IRD had decreased mass of individual muscle groups compared to their wild type littermates fed IRD (Figure 5I). As muscle mass is regulated by protein synthesis and degradation cascades in muscle, we analyzed the expression of both atrogenes, Atrogin-1 and Murf-1, and myogenesis factors, MyoD and myogenin in gastrocnemius of IRD-fed mice. We observed increased expression of both Atrogin-1 and Murf-1, and significant downregulation of MyoD expression in Pnpla2WAT KO muscles, suggesting that both atrophic factors and lack of myogenesis signaling may contribute to enhanced muscle wasting in Pnpla2WAT KO fed IRD (Figure 5J).
Excess dietary iron in wild type mice causes increased lipid utilization (Figure 2). We hypothesized that the reduced lipid mobilization and increased lean wasting in IRD-fed Pnpla2WAT KO mice would result in a decrease in global lipid utilization. Consistent with this, we observed modest separation in RER values between Pnpla2WAT KO and Pnpla2WT mice following introduction of IRD (Figure 5K). Principal component analysis of the light/dark cycle RER values from Pnpla2WAT KO and Pnpla2WT mice showed that the pattern of clustering is based on the light vs. dark cycle and independent of genotype (Figure 5L). Therefore, we analyzed AUC of RER by light/dark cycles and observed a significant increase in Pnpla2WAT KO RER values during the light cycle following introduction of IRD (Figure 5M). This suggests that during the diurnal “fasting” state, Pnpla2WAT KO mice fed IRD utilized less lipid substrates relative to Pnpla2WT littermates (Figure 5M). Thus, ATGL activity in adipose tissue protects from lean energy store wasting by promoting lipolysis and lipid mobilization in response to IRD-induced negative energy balance (Figure 5N).
Discussion
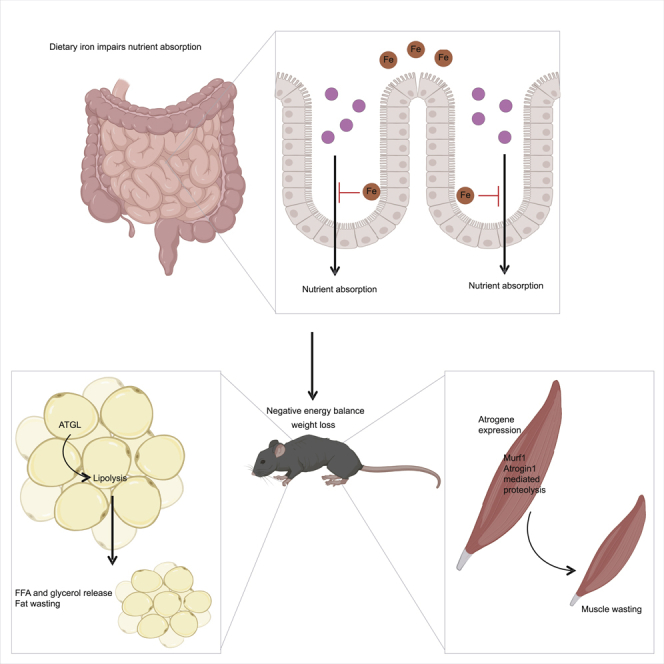
Organismal macronutrient metabolism is intimately linked to micronutrient metabolism. Although a role for iron in regulating lipid metabolism is well appreciated, the mechanistic underpinnings of these regulatory processes are unknown. Here, using an acute course of dietary iron excess, we demonstrate that iron causes profound changes in whole organismal metabolism including nutrient malabsorption and lipid mobilization, resulting in a negative energy balance. We demonstrate that lipid mobilization is dependent on ATGL activity in adipose tissue and that this lipid mobilization protects from cachexia. Although our data support a model that the systemic effects of iron and nutrient absorption lead to ATGL activation, lipolysis, and lipid mobilization, it is formally possible that in addition to these systemic effects iron also directly acts on adipocytes to induce ATGL activity (Figure 5N) ((Rumberger et al., 2004).
We found that an acute course of excess dietary iron caused reduced energy expenditure in mice, and we propose that this is because of a hypometabolic state triggered by the reduced nutrient absorption caused by IRD. Nutrient absorption is largely dependent on proper digestion of macronutrients in the duodenum via digestive enzymes secreted by the exocrine pancreas (Kimita and Petrov, 2020). Iron overload disorders that lead to deposition of iron in parenchymal tissues, like liver and pancreas, are associated with metabolic syndrome and reduced levels of circulating pancreatic-derived digestive enzymes (Gullo et al., 1993; Horne et al., 1997; Kimita and Petrov, 2020). It has been proposed that pancreatic insufficiency caused by iron loading may contribute to malabsorption issues observed in individuals with iron overload disorders (Horne et al., 1997).
Our finding that IRD decreases serum levels of lipase is consistent with experimental and clinical models of iron-overload induced pancreatic dysfunction and atrophy of exocrine pancreatic acinar cells. Further, we observed reduced cholesterol levels in mice fed IRD, which is consistent with pharmacologic inhibition of lipase activity (Erdmann et al., 2004; Ke et al., 2020). In further support of IRD-induced pancreatic insufficiency, lipid absorption was severely blunted in mice fed IRD and we observed decreased net calorie absorption in mice fed IRD. Though we predict that IRD-induced nutrient malabsorption extends to insufficient digestion of proteins and carbohydrates as well, we did not measure levels of other pancreatic-derived digestive enzymes in the current study. It is important to note that while our study implicates iron-induced pancreatic insufficiency in reduced nutrient uptake, iron metabolism can just as likely affect nutrient absorption and turnover through a systemic mechanism independent of digestive function. For example, our lab previously showed that excess dietary iron can indirectly affect luminal macronutrient content through inducing insulin resistance (Sanchez et al., 2018). Furthermore, iron can also influence the microbial landscape of the gut and contribute to gastrointestinal inflammation, compromise barrier integrity, and regulate iron metabolism at the local level to control infection (Bessman et al., 2020; Yilmaz and Li, 2018). Excess iron is known to drive expansion of facultative anaerobes like Enterobacteriaceae, thus displacing and driving competition against Lactobacillus and Firmicutes (Yilmaz and Li, 2018). Moreover, it was recently demonstrated that iron supplementation can similarly influence microbiome composition independent of the route of administration (La Carpia et al., 2019). Altogether, these findings highlight the diverse role of micronutrient metabolism on nutritional status beyond nutrient absorption and digestive function.
It has been previously reported that when fed a chronic 16-week course of dietary iron, C57BL/6 mice develop IR specifically in visceral adipose depots as well as impaired organismal insulin tolerance (Dongiovanni et al., 2013). In our study, we used an acute course of dietary iron and found that C57BL/6 mice did not develop IR in adipose tissue, skeletal muscle or liver, and also showed no changes in organismal insulin tolerance. The discrepancies between our study and that of Dongiovanni et al. may be because of the length of the feeding regimens and suggests that in C57BL/6 mice that the IR caused by excess dietary iron may be largely influenced by length of the dietary iron regimen. In a mouse model of infectious colitis using C3H/HeJ, an acute course of dietary iron excess resulted in IR in visceral adipose tissue and changes in organismal glucose tolerance (Sanchez et al., 2018), suggesting that the ability of a short course of dietary iron to cause IR may be mouse strain specific. It is well established that inbred mouse strains have differences in iron physiology including basal differences in tissue iron content. C75BL/6 mice in particular, have a dampened iron-overloading response following introduction to an iron rich diet (Clothier et al., 2000; Dupic, 2002; Leboeuf et al., 1995; Morse et al., 1999). When considering the physiological differences in iron handling between C3H and C57BL/6 mice that could contribute to differential manifestations in IR development, it is important to note that C57BL/6 mice lack functional Nramp1 while C3H have a viable Nramp1 allele. Though Nramp1 does not appear to be directly involved in tissue iron loading, it has been shown to be important for heme recycling and influence hepcidin expression. Failure to release heme-derived iron from macrophages dampens hepcidin signaling—leading to increased basal iron uptake. Mice lacking Nramp1 accumulate iron in liver and spleen following erythrophagocytotic stimuli in vivo (Soe-Lin et al., 2009). The differences in the studies of Sanchez et al. and our work also suggest that the effects of excess dietary iron on IR may be dependent on the disease state of the host, and that under conditions in which homeostasis is disrupted, as observed during infection, the short course of excess iron may be sufficient to induce IR. Future studies addressing the role of Nramp1 in mediating the IR response to dietary iron in different mouse strains and an understanding of the role of acute dietary iron excess in disease states are needed.
Subcutaneous fat (SAT) is considered to have greater flux in terms of storage and mobilization capacity compared to visceral fat (VAT). Specifically, excess lipids are preferentially stored in SAT, and mobilized from SAT to accommodate negative energy balance (Luong et al., 2019; Schoettl et al., 2018; Tchkonia et al., 2013). We found that an acute course of dietary iron induces wasting and mobilization of SAT and VAT indiscriminately and to a similar degree, suggesting that dietary iron excess leads to lipid mobilization events in an unconventional pattern that operates in response to negative energy balance. Curiously, we found that mice lacking adipose-specific ATGL are protected from iron-induced fat wasting in SAT, but not VAT. We demonstrated that IWAT and GWAT have comparable levels of ATGL protein expression and activation of the PKA signaling cascade upon introduction of IRD. Therefore, the difference in fat wasting between SAT and VAT cannot be attributed to differences in ATGL expression or activation through the PKA cascade. Though PKA activation is considered the canonical signaling cascade for ATGL activation, other pathways can alter ATGL activity including PKC, MAP kinase, PI3K, NF-κB, and lipid peroxidation resulting from Fenton and Haber Weiss reactions (Rejholcova et al., 1988; Rumberger et al., 2004; Winterbourn, 1995; Yang and Stockwell, 2016). Indeed, in an in vitro study utilizing visceral adipocytes from rats, iron induced lipolysis independent of the PKA lipolytic cascade, and was proposed to occur through increased levels of lipid peroxides (Rumberger et al., 2004). In the current study, our data supports a model where iron induces lipolysis in SAT via the canonical PKA-ATGL cascade; however, we propose that VAT is more sensitive to iron-derived lipolytic stimuli such as lipid peroxides or ROS.
We found that an acute course of dietary iron excess caused cachexia with IRD-fed animals exhibiting wasting of muscles with varied oxidative capacity and myofiber composition, suggesting that iron induces muscle wasting in an indiscriminate manner. Cachexia is dependent on the transcriptional upregulation of two muscle specific E3 ubiquitin ligases, Murf-1 and Atrongin-1 (Baehr et al., 2011; Bodine et al., 2001; Gomes et al., 2001). In agreement with this, we found that iron-induced cachexia was associated with the transcriptional upregulation of Murf-1 and Atrogin-1 in wasting hindlimb muscles. Iron-induced muscle wasting was more severe in IRD-fed animals lacking ATGL function in adipose tissue. We propose that this increase in the cachectic response in ATGLWATKO mice is necessary to supply muscle-derived substrates to meet the iron-induced heightened energetic demands when lipid mobilization is impaired. In support of this, we observed that ATGLWATKO mice fed IRD displayed a significant shift toward carbohydrate utilization during light cycles in metabolic cages. Our finding is consistent with previous reports highlighting a diurnal pattern of WAT lipolysis driven by circadian clock transcription factors CLOCK and BMAL (both in vivo and ex vivo). Light cycles represent the diurnal murine “fasting cycle” where they preferentially utilize lipids for energy, which is dependent on circadian regulation of HSL and ATGL in WAT (Duncan et al., 2007; Shostak et al., 2013). We propose that ATGLWATKO mice fed an IRD undergo increased gluconeogenesis during a prolonged period of inactivity—a process that relies on supply of muscle-derived amino acid substrates (Wu et al., 2012), to meet the increased energetic demands induced by the diet. Interestingly, we found upregulation of Myogenin in gastrocnemius muscles from IRD-fed mice, which is a muscle composed of both Type I and Type II myofibers. As Myogenin expression is associated with Type I myofiber development, this may represent adaptive remodeling of muscles to accommodate increased reliance on lipids/beta-oxidation (Hughes et al., 1999; Wu et al., 2012), which may be necessary to meet the energetic demands caused by the acute course of dietary iron excess. In summary, this work defines the mechanistic basis for dietary iron-induced lipid mobilization in response to negative energy balance and highlights the multifaceted role of iron regulation of organismal metabolism.
Limitations of the study
Our work was performed in mice and there are considerable differences between human and murine iron metabolism; therefore, it is difficult to estimate how the iron content in our study would translate to human diets. For instance, because of the small body mass and increased relative food consumption of mice, it’|'s estimated that mice turnover half of their body’s iron stores daily and because meat is not a staple of mouse diets, exhibit poor heme absorption relative to humans(Coffey and Ganz, 2017). Moreover, common inbred strains exhibit substantial variability in iron homeostasis and responsivity to fluctuations in iron status (Clothier et al., 2000; Dupic, 2002; Fleming et al., 2001)
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit mAb AKT | Cell Signaling | Cat#:9272S |
| Rabbit mAB Phospho-AKT (Ser473)(193H12) | Cell Signaling | Cat#:4058S |
| Rabbit mAb GAPDH XP (D16H11) | Cell Signaling | Cat#:2118S |
| Anti-Rabbit IgG HRP-linked Ab | Cell Signaling | Cat#: 7074S |
| Rabbit mAB ATGL | Cell Signaling | Cat#:2439S |
| Rabbit mAB phospho-HSL(Ser660) | Cell Signaling | Cat#:4126S |
| Rabbit mAB HSL | Cell Signaling | Cat#: 18381T |
| Rabbit mAB PKA-C | Cell Signaling | Cat#: 5842T |
| Rabbit mAB phospho-PKA-C (Thr197) | Cell Signaling | Cat#: 5661T |
| Chemicals, peptides and recombinant proteins | ||
| Tissue extraction reagent II | Invitrogen | Cat#:FN0081 |
| Trans-Blot® Turbo™ Midi 0.2 μm Nitrocellulose Transfer Packs | BioRad | Cat# 1704159 |
| SYBR Green | BioRad | Cat# 1725125 |
| Humulin R regular insulin u-100 HI-210 | Lily | Cat# 07-843-8053 |
| Halt Protease and Phosphatase inhibitor cocktail (100x) | Fischer Scientific | Cat# 78440 |
| Nupage Tris-Acetate SDS Running Buffer | ThermoFischer Scientific | Cat# LA0041 |
| Nupage 7% Tris-Acetate Gel 1.0mm X 12 well | ThermoFischer Scientific | Cat#EA03552BOX |
| Nupage LDS Sample Buffer (4x) | Invitrogen | Cat#: NP0007 |
| Protease Inhibitor Cocktail (100x) | Sigma | Cat# P8340 |
| Ponceau | Sigma | Cat#:G7021 |
| D-(+)-Glucose | Sigma | Cat#:G7021 |
| Sodium Chloride | Fisher Chemical | Cat#: S271-3 |
| Dulbecco’s Phosphate Buffered Saline | Gibco | Cat#: 14190-144 |
| Bovine Serum Albumin, heat shock treated | Fisher Bioreagents | Cat#:BP1600100 |
| Tris, Hydrochloride, ULTROL Grade | Millipore Sigma | Cat#:1185-53-1 |
| Tween20 | Sigma-Aldrich | Cat#:P1379-500ml |
| Potassium Phosphate Monobasic | Fischer Scientific | Cat#:P285-500 |
| Magnesium Sulfate | Fischer Scientific | Cat#: M63-500 |
| Calcium Chloride | Sigma | Cat#:C1016-500 |
| Hepes | Sigma | Cat#:3375 |
| Sodium Bicarbonate | Tocris Bioscience | Cat#:3152 |
| Albumin, Bovine Serum, Low Fatty Acid, Low Endotoxin, Low IgG (NZ) (BSA) | US Biological Life Sciences | Cat#: A1311 |
| Sodium Dodecyl Sulfate (SDS), Lauryl | ThermoFischer Scientific | Cat#: 28365 |
| Glycine | Millipore Sigma | Cat#: 410225-250G |
| Glycerol | Fischer Scientific | Cat#: BP229-1 |
| UltraPure distilled water | Invitrogen | Cat#: 10977-015 |
| Sodium Deoxycholate | Sigma | Cat#: D6750-100 |
| NP-40 | Sigma | Cat#: 492018-50ML |
| Intralipid | Sigma | Cat#:I141-100ml |
| Olive oil | Sigma | Cat#: O1514-500ML |
| Indicating Drierite | Drierite | Cat#:23005 |
| Critical commercial assays | ||
| Superscript IV Reverse Transcriptase | Invitrogen | Cat# 18090010 |
| RNase OUT | Invitrogen | Cat#: 10777019 |
| Trizol Reagent | Invitrogen | Cat#: 15596026 |
| RNeasy Mini Kit | Qiagen | Cat#: 74104 |
| RNase-Free DNase Set | Qiagen | Cat#: 79254 |
| Wako HR series NEFA-HR(2) Color reagent A | WAKO Fuji Film | Cat#: 999-34691 |
| Wako HR Series NEFA-HR(2) Solvent A | WAKO Fuji Film | Cat#: 995-34791 |
| Wako HR Series NEFA-HR(2) Color Reagent B | WAKO Fuji Film | Cat#: 991-34891 |
| Wako HR Series NEFA-HR(2) Solvent B | WAKO Fuji Film | Cat#: 993-35191 |
| Multi-Calibrator Lipid | WAKO Fuji Film | Cat#: 464-01601 |
| Triglyceride M Enzyme Color A (L-Type) | WAKO Fuji Film | Cat#: 992-02892 |
| Triglyceride M Enzyme Color B (L-Type) | WAKO Fuji Film | Cat#: 998-02992 |
| Free Glycerol Reagent | Sigma | Cat# F6428-40ml |
| Iron Assay Kit (Colormetric) | Abcam | Cat#: ab83366 |
| OneBlock Wes protein stain | Prometheus Protein Biology Products | Cat# 20-311 |
| Pierce BCA Protein Assay Kit | Fischer Scientific | Cat# 23225 |
| ProSignal Dura | Prometheus Protein Biology Products | Cat# 20-301 |
| 2% carboyl iron enriched chow | Envigo | Cat# TD.08714 |
| Control iron chow | Envigo | Cat# TD.99398 |
| Experimental models: organisms/strains | ||
| Mouse: C57BL/6 | The Jackson Laboratory | 000664 |
| Mouse: B6.Cg-Tg(Fabp4-cre)1Rev/J | The Jackson Laboratory | 005069 |
| Mouse: B6N.129S-Pnpla2tm1Eek/J | The Jackson Laboratory | 02478 |
| Software and algorithms | ||
| SoftMax Pro 5.4.5 Microplate Data Acquisition and Analysis Software | Molecular devices | SMP5ACAD |
| QuantStudio5 Design and Analysis software v1.5.0 | Applied Biosystems | A28140 |
| Image Lab Software 5.2.1 | BioRad | 12012931 |
| Other | ||
| Ohaus® - CS200 - Portable Scale (discontinued) | Ohaus | CS200 |
| VERSAmax Microplate reader | Molecular devices | VERSAMAX |
| Nova Max glucose test strips | Novacares | Cat# 43437 |
| Nova Max plus glucose meter | Novacares | Cat# 43435 |
| EchoMRI™-100H Body Composition Analysis | EchoMRI | ECHOMRI-100H |
| BeadMill24 benchtop bead-based tissue homogenizer | Fischer Scientific | 15-340-163 |
| QuantStudio5 Real-Time 384-well | Applied Biosystems | A28140 |
| Gel doc XR +Gel documentation System | BioRad | 1708195 |
| Digi-sense Type J/K/T Thermocouple meter | Digi-sense | Cat#:20250-91 |
| NanoDrop™ One/OneC Microvolume UV-Vis Spectrophotometer | ThermoFisher Scientific | ND-ONE-W |
| CLAMS | Columbus Instruments | CLAMS-12M-A(XZ)-CTENC54 |
| BD Microtainers SST | BD | Cat#:202204030 |
| Mini Gel Tank | Invitrogen | Cat#: A25977 |
| Microvette® Capillary Blood Collection Tubes | Sarstedt | Cat#: 16.440.100 |
| Parr 6100 Calorimeter | Parr Instrument Company | SKU#:PAR#6100Series |
Resource availability
Lead contact
Further information and requests for resources and reagents can be directed to the lead contact, Janelle Ayres (jayres@salk.edu)
Materials availability
This study did not generate new or unique resources or reagents. All reagents are commercially available.
Experimental model and subject details
Mice
Six week-old male C57BL/6 mice were purchased from Jackson Laboratories and housed in our facility to acclimate for 2 days prior to experimentation. To study ATGL-dependent lipolysis, we performed crosses with B6N.129S-Pnpla2tm1Eek/J mice to B6.Cg-Tg(Fabp4-cre)1Rev/J both from Jackson labs to generate Pnpla2;Fabp4 cre+ and Pnpla2;Fabp4 cre- mice. For experiments, 6-8 week old male littermates were used. All animal experiments were done in accordance with The Salk Institute Animal Care and Use Committee and performed in our AALAC-certified vivarium.
Method details
Mouse diets and pairwise feeding
Mice were fed a 2% carbonyl iron supplemented diet or a control diet from Envigo. Daily food intake was determined by measuring mass of food pellets daily from single-housed mice. For pair feeding experiments, iron and control diets were supplied ad libitum while the third pair fed group of mice were given control diets that were weight-matched to the historical daily food intake of a mouse given iron diet ad libitum.
Body composition measurements
Total body fat and lean mass were measured using an EchoMRI machine. Total fat or total lean mass(g) was normalized to total body mass(g) to determine the percent body composition of fat and lean tissues. Fat pad masses were measured by dissecting and weighing subcutaneous fat pads (inguinal WAT), visceral fat pads (gonadal WAT), and mesenteric WAT post mortem. Muscles from hindlimb were dissected and weighed postmortem. The following muscles were harvested to represent muscles of varied myofiber composition: Quadricep (primarily fast twitch), Tibialis anterior (fast/slow twitch), extensor digitorum longus (fast/slow twitch), Soleus (slow twitch), gastrocnemius (fast/slow twitch).
Metabolic phenotyping by indirect calorimetry
Mice were singly housed in metabolic cages from the Columbus Instruments Comprehensive Lab Animal Monitoring System (CLAMS) where O2 consumption, CO2 production, and activity data were collected. Food consumption was measured by weighing food pellets daily in order to determine calorie intake. Mice were removed from metabolic cages daily to measure body mass with a scale and body composition using EchoMRI. Energy Expenditure was calculated using the modified Weir equation (modified for systems lacking urine collection):
| EEJ(Joules/hour) = 15.818 J/ml ∗ VO2 + 5.176 J/ml ∗VCO2 |
where VO2 and VCO2 are in units of ml/hour. Respiratory Exchange Ratio (RER) was determined by dividing VCO2/VO2.
Insulin tolerance tests
Animals were fasted for 6 hours in fresh cages with ad libitum access to water. Blood glucose measurements were taken using a Nova Max plus by making small cuts to the tips of tails using a sterile razor blade and gently squeezing the tail from base to tip. Insulin was administered according to body mass (1U/kg) in a single intraperitoneal injection at time = 0 minutes. Blood glucose measurements were taken every 15 minutes for the first hour post insulin injection and then again at 90 and 120 minutes post injection. Blood glucose measurements were normalized to their time = 0 fasted blood glucose levels to generate a curve representing the percent drop in glucose levels from basal.
Lipid tolerance test
Animals were fasted for 9 hours in fresh cages with ad libitum access to water on day 9 of dietary regimens. Mice were injected intraperitoneally with 10μL/g of Intralipid. Blood was collected at 0, 1, 2, 3, 4, and 5 hours post injection in Microvettes by tail bleed. The microvettes were centrifuged at 6000 RPM for 20 minutes and serum was harvested for quantifying non esterified free fatty acids (Wako) and total triglycerides (Fujifilm L-Type Triglyceride M kit) and following the manufacturers protocol.
Oral lipid tolerance test
Animals were fasted for 14-16 hours overnight in fresh cages with ad libitum access to water on day 8 of dietary regimens. Mice were orally administered 250 ul of olive oil by gavage. Blood was collected at 0, 1, 2, 3, 4, and 5 hours following administration of oil in Microvettes by tail bleed. The microvettes were centrifuged at 6000 RPM for 20 minutes and serum was harvested for quantifying non esterified free fatty acids (Wako) total triglycerides using the (Fujifilm L-Type Triglyceride M kit) and following the manufacturers protocol.
Bomb calorimetry
Mice were co-housed (5 mice per cage) in Pure-o’Cel bedding and provided ad libitum access to water and control iron diet (CID) or 2% carbonyl iron diet (IRD). Body mass and food mass was measured daily to determine daily food intake (calorie intake). All bedding was collected daily and stored in sealed bags at −20°C until further processing. Fecal pellets were collected from bedding and desiccated in an airtight chamber with drierite for ≥20 hours. Fecal pellets were combined from two cages per day per respective diets. Combined desiccated fecal pellets were weighed and ground using mortar and pestle. ddH2O was mixed into ground feces 50 ul at a time to reach a mealy-pasty consistency. Parr 6100 Calorimeter instruments and accessories were used to determine caloric content of fecal matter thereafter. Fecal mixture was used to form a compacted cylindrical pellet and placed in a brass dish inside bomb containing 1 ml ddH2O and twisted ignition string touching fecal pellet. Bomb was filled with O2, sealed and carefully inserted into 2000 g of room temperature deionized H2O. Ignition leads were connected to bomb and calorimetry chamber was closed for testing. Calculations were performed according to the Parr instrument manual to determine calories/gram of fecal matter. Net caloric absorption was determined by subtracting total fecal caloric content from pooled daily caloric intake from respective diets.
Insulin signaling experiments and western blot
To examine insulin signaling in tissues, mice were fasted for 6 hours in fresh cages with ad libitum access to water and injected with insulin as described above (insulin tolerance test). Injections were performed in 15-minute staggered intervals. Exactly 15 minutes following insulin injection, mice were sacrificed and liver, muscle and fat pads were harvested, immediately flash frozen in liquid nitrogen, and stored at −80°C. Tissues were subsequently processed for western blot analyses as described below.
Western blot
Snap frozen tissues were ground into fine powder using ceramic mortar and pestles equilibrated in liquid nitrogen. Powder was homogenized in BeadMill24 with a ceramic bead in Tissue extraction reagent or adipose tissue protein extraction buffer with phosphatase and protease inhibitors added (Adipose tissue protein lysis buffer: 50mM Tris pH 7.5, 150mM NaCl, 1% NP-40, 0.5% Sodium Deoxycholate, 5% Glycerol). Lysates were purified by three consecutive centrifugations at 4°C to remove debris and fat and quantified using Pierce BCA protein assay kit. Samples were loaded into a 7% Tris-acetate gel following a 10-minute sonication, and a 10-minute boil at 70°C. Gels were ran for 150V for 60 minutes in Invitrogen Mini Gel Tank and transferred to nitrocellulose blot using the BioRad Turbo Transblot system for 10 minutes at 25V (1.3 A). Blots were stained with OneBlock Wes protein stain and cut into strips according to size of target protein. Strips were then washed in TBST (20mM Tris, 150mM NaCl, 0.1% Tween20 w/v) and blocked with 5% BSA in TBST for 1 hour at room temperature. Blots were incubated with primary antibodies overnight at 4°C on an orbital shaker (p-AKT, AKT, ATGL, p-HSL, HSL, p-PKAc, PKAc, GAPDH; see Catalog details in materials table). Blots were washed 3 × 10 minutes with TBST and incubated with anti-rabbit for 1 hour at room temperature with Anti-Rabbit IgG HRP-linked antibodies with gentle shaking and subsequently washed 3 × 10 minutes with TBST. Blots were developed and imaged using ProSignal Dura chemiluminescence in the BioRad Gel Doc XR system. Densitometry analyses were performed suing the ImageLab Software and phosphorylated proteins were normalized to total protein level and subsequently to housekeeping protein, GAPDH. Phosphorylated target proteins were probed/measured first, and the same membrane was used to measure total levels of that respective protein using the following stripping protocol: Blot was covered with stripping buffer for 5 mins and discarded. Blot was covered with fresh stripping buffer again for 5 minutes and discarded. Blot was washed two times with PBS for 10 minutes by gentle shaking. Blot was washed two times with TBST for 5 minutes by gentle shaking. After final wash, blot was blocked with 5% BSA for 1 hour at room temperature and used for subsequent antibody incubations as described above. (Mild stripping buffer (500ml): 7.5 glycine, 5 ml Tween20, 0.5g sodium dodecyl sulfate, pH 2.2)
Gene expression analyses
Tissues were flash frozen in liquid nitrogen immediately following harvest from sacrificed animals. Tissues were ground into a powder using ceramic mortar and pestles equilibrated in liquid nitrogen. Powder was added to Trizol reagent and homogenized with a ceramic bead in a bead beater. Chloroform was added to homogenate and centrifuged to separate the organic and aqueous layers. Aqueous layer was carefully transferred to fresh nuclease-free tubes where isopropanol was added and mixture was left to precipitate at −20°C for at least 1 hour. Isopropanol/aqueous layer solution was added to a Qiagen RNeasy column and RNeasy protocol was followed from this step, including the removal of genomic DNA using Qiagen’s on-column DNase kit. RNA was eluted in nuclease-free H2O and quantified using a Nanodrop Spectrophotometer. cDNA was synthesized using SuperScript IV using ∼200 ng RNA and oligo dT for mammalian cDNA. Real time quantitative PCR was performed using SYBR green Mix on QuantStudio 5 from Applied Biosystems. Relative standard curves method was used to analyze gene expression relative to a pooled sample dilution series. Rps17 was used as an endogenous housekeeping control for relative normalization. Annealing temp of 60°C was used for all RT-qPCR reactions. The primers used for gene expression analyses are listed in Table 1.
Table 1.
Primers used for gene expression analysis
| Gene target | Forward 5’->3′ | Reverse 5’->3′ |
|---|---|---|
| Atrogin-1 | CAGCTTCGTGAGCGACCTC | GGCAGTCGAGAAGTCCAGTC |
| MuRF-1 | AGTGTCCATGTCTGGAGG TCGTTT | ACTGGAGCACTCCTGCTTGTAGAT |
| MyoD | CAGTGGCGACTCAGATGC | CGCCTCACTGTAGTAGGCG |
| Myogenin | GCAATGCACTGGAGTTCG | GTCCACGATGGACGTAAGG |
| Rps17 | CGCCATTATCCCCAGCAAG | TGTCGGGATCCACCTCAATG |
| Fpn | CCCTGCTCTGGCTGTAAAAG | TAGGAGACCCATCCATCTCG |
| Dmt1 | CAATGGAATAGGCTGGAGGA | CCCAATGCAATCAAACACTG |
| Tfr1 | TCGCTTATATTGGGCAGACC | ATAAGGGGCTGGCAGAAACT |
| Transferrin | GCGCATTCAAGTGTCTGAAA | GAGCCACAACAGCATGAGAA |
| Ftl | GGGCCTCCTACACCTACCTC | CTCCTGGGTTTTACCCCATT |
| Fth | CTGAATGCAATGGAGTGTGC | TCTTGCGTAAGTTGGTCACG |
| Hmox1 | ACGCATATACCCGCTACCTG | CCAGAGTGTTCATTCGAGCA |
| Steap2 | ATGCCAGAAACCAGCAGAGT | TGCAAAGAAGAAGCTCAGCA |
| Hepcidin | TTGCGATACCAATGCAGAAG | GGATGTGGCTCTAGGCTATG |
| Genotyping primers | ||
| cre recombinase | CGCGCTCTGGCAGTAAAAACTATC | CCCACCGTCAGTACGTGAGATATC |
Ex vivo lipolysis assay
Mice were fed control or 2% carbonyl iron diets for a period of 3, 6, and 9 days and fasted for 6 hours in fresh cages with ad libitum access to water. Inguinal and gonadal white adipose tissue pads were harvested, cut into 2 ∼30-50 mg pieces and placed into separate wells in a 24 well plate with 300 μl of ice-cold PBS. Following the completion of tissue harvest, all fat pads were gently dabbed on a paper towel, cut with scissors in a central latitudinal location to introduce freshly cut exposed tissue, and carefully transferred to 24 well plates containing 300 μl of room-temperature Krebs-Ringer bicarbonate Hepes buffer ph 7.4 (120mM NaCl2, 4mM KH2PO4, 1mM MgSO4, 0.75 mM CaCl2, 30 mM Hepes pH 7.4, 10mM NaHCO3; solution was sterilized and stored at 4°C until day of ex vivo lipolysis assay when fatty acid free bovine serum albumin (to 2% wbv) and D-glucose (5mM) was added. Plates were incubated for 4 hours in a 37°C tissue culture incubator and supernatants were transferred to fresh tubes and frozen at −20°C until they were used to quantify free fatty acids and glycerol content as described below.
Free fatty acid, glycerol and triglyceride assays
Serum from fasted mice and supernatant from ex vivo lipolysis assay were added to a 96 well plate according to the Wako Fuji Film protocol and reagents were added and incubated according to the respective FFA or glycerol Wako protocols. Standards were generated using a dilution series of glycerol or non-esterified fatty acids (WAKO). A 1:2 dilution series from 1mM to 0.03125mM was used to generate the FFA standard curve. The standard curve for glycerol used the following series in mM: 8, 7, 6, 5, 4, 3, 2, 1, 0.5, 0.25. Triglyceride standards were generated from multi-calibrator lipids in the following series (mg/dL): 48, 96, 144, 192, 240, 288, 384, 480. To determine FFA and glycerol content, 5 μl of samples and standards were used. To determine triglyceride content, 4 μl of samples were used. Following methods outlined in manufacturer’s protocols, samples were quantified from standard curve analyses using a 96-well VERSAmax microplate reader and SoftMax Pro software. FFA and glycerol levels from ex vivo lipolysis assays were normalized to recorded tissue masses.
Iron assay for determining tissue iron content
Total iron content was determined using colorimetric Iron Assay from Abcam according to manufacturer’s protocol. For serum, blood was collected via cardiac puncture in microtainers and centrifuged at 6000 rpm for 20 minutes and serum was stored at −80°C. Tissues were flash frozen in liquid nitrogen immediately following harvest from sacrificed animals. Tissues were ground into a powder using ceramic mortar and pestles equilibrated in liquid nitrogen. 10–30 mg of tissue powder and 10 ul serum was suspended in Iron assay buffer and gently homogenized by pipetting. Tissue samples were centrifuged at 13000 rpm for 10 minutes to remove insoluble materials and supernatant was used for iron content quantification according to manufacturer’s protocol in 96-well plates. A standard curve was generated using the following dilution series (nmol/well): 0.5, 1.0, 1.5, 2.0, 2.5, 4, 6, 8, 10, 20, 40. Samples were quantified using a 96-well VERSAmax microplate reader and SoftMax Pro software. Iron content was normalized to respective tissue masses.
Principal component analyses
To determine the light/dark cycle and diet relationships between samples, EEJ, RER and total activity spreadsheets were transformed to represent hourly blocks of the light/dark cycle during periods of matched food consumption; this was the primary derived data used for PCA (Jolliffe, 2005). Principal component analysis was computed using the mixOmics R package (v6.14.1) with 10 components considered in the calculations (Rohart et al., 2017) . Eigenvalues of the variance-covariance matrix were calculated, and the data centered and scaled (standardized) to account for heterogeneity in variances across the variables. Multivariate analysis plots were generated using the R function plotIndiv (Plot of Individuals). The first two principal components were projected onto the sample space with the ellipses representing 95% confidence intervals.
Linear regression and ANCOVA
For linear regression analyses of energy expenditure based on total or lean mass, the daily energy expenditure for each mouse was determined by summing J/hr per 24 hour periods preceding total and lean mass measurements. Daily energy expenditure (J/day) was plotted as a function of total body mass or lean mass for each mouse over the course of the experimental window for respective dietary groups. GraphPad Prism 9.0e was used to perform regression analyses. Simple linear regression was performed for each dietary group and linear regression lines were compared to determine if the slopes and y intercepts were significantly different between dietary groups. ANCOVA analyses were performed when slopes between dietary group linear regression lines were not significantly different and the pooled slope was used to calculate an adjusted daily energy expenditure at the group mean mass. Group mean mass was determined by calculating the mean total or lean body mass for the groups presented in linear regression plots. ANCOVA adjusted daily energy expenditure was determined by using the following equation: (actual J/day)-[(average group LBM or TBM (g))-(actual TBM or LBM (g))]∗pooled slope (J day−1 per g change in mass). The NIDDK Mouse Metabolic Phenotyping Centers (MMPC, www.mmpc.org) and Energy Expenditure Analysis page (http://www.mmpc.org/shared/regression.aspx) was used as a reference to guide the EE ANCOVA analysis done for this work. NIDDK MMPC is supported by grants DK076169 and DK115255.
Clinical pathology analysis
Six week old C57BL/6 male mice were provided ad libitum access to water and control iron diet (CID) or 2% iron rich diet (IRD) for 8 days. On day 9 of dietary regimens, animals were fasted for 6 hours in fresh cages with ad libitum access to water prior to sacrifice. Blood was collected via cardiac puncture in BD microtainers and centrifuged at 6000 rpm for 20 minutes to collect serum. Serum was stored at −80°C. Serums were sent to IDEXX Laboratories for quantification of ALP, AST, ALT, lipase and cholesterol levels.
Quantification and statistical analysis
All variables were analyzed using GraphPad Prism 9.0e. Categorical variables are expressed as numbers and percentages, and quantitative variables as means ± standard error of the mean (SEM). Where data is shown as dot plots, each dot represents a single mouse or biological replicate. One-way analysis of variance (ANOVA) with a post Tukey’s test or Two-way ANOVA were used as appropriate. Student’s two tailed t-test was performed to evaluate the differences between two independent groups or paired samples as appropriate. A P value <0.05 was used to reject the null hypothesis. All experiments used an n of 3 or more and represent 1-3 independent experiments. Further information regarding specific P values, n used in each experiment, and how data are presented can be found in the figure legends.
Acknowledgments
This work was supported by NIH awards DP1 AI144249 and R01AI114929 (J.S.A.), and the NOMIS Foundation (J.S.A.). Figure 5N and the graphical abstract were generated using images from BioRender. The authors would like to thank Kaito Kikuchi for providing guidance and assistance for analyses of metabolic data and Daniela Michel-Romo for providing technical support in a pair fed animal study.
Author contributions
ARR – experimental design, data analysis and wrote the manuscript
AM – data analysis
JSA – conceptualization, experimental design, data analysis and wrote the paper
Declaration of interests
JSA holds an adjunct appointment at UC San Diego and is a member of the Kenneth Rainin Foundation scientific advisory board.
Published: March 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103941.
Supplemental information
Data and code availability
-
•
Original western blot images are included in the supplemental information
-
•
Any additional information required to reanalyze the data reported in this paper is available form the lead contact upon request
References
- Argilés J.M., Fontes-Oliveira C.C., Toledo M., López-Soriano F.J., Busquets S. Cachexia: a problem of energetic inefficiency. J. Cachexia Sarcopenia Muscle. 2014;5:279–286. doi: 10.1007/s13539-014-0154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehr L.M., Furlow J.D., Bodine S.C. Muscle sparing in muscle RING finger 1 null mice: response to synthetic glucocorticoids. J. Physiol. 2011;589:4759–4776. doi: 10.1113/jphysiol.2011.212845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessman N.J., Mathieu J.R.R., Renassia C., Zhou L., Fung T.C., Fernandez K.C., Austin C., Moeller J.B., Zumerle S., Louis S., et al. Dendritic cell–derived hepcidin sequesters iron from the microbiota to promote mucosal healing. Science. 2020;368:186–189. doi: 10.1126/science.aau6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine S.C., Latres E., Baumhueter S., Lai V.K., Nunez L., Clarke B.A., Poueymirou W.T., Panaro F.J., Na E., Dharmarajan K., et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Brownlie T., Utermohlen V., Hinton P.S., Haas J.D. Tissue iron deficiency without anemia impairs adaptation in endurance capacity after aerobic training in previously untrained women. Am. J. Clin. Nutr. 2004;79:437–443. doi: 10.1093/ajcn/79.3.437. [DOI] [PubMed] [Google Scholar]
- Carrero J.J., Stenvinkel P., Cuppari L., Ikizler T.A., Kalantar-Zadeh K., Kaysen G., Mitch W.E., Price S.R., Wanner C., Wang A.Y.M., et al. Etiology of the protein-energy wasting syndrome in chronic kidney disease: a consensus statement from the International Society of Renal Nutrition and Metabolism (ISRNM) J. Ren. Nutr. 2013;23:77–90. doi: 10.1053/j.jrn.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Clothier B., Robinson S., Akhtar R.A., Francis J.E., Peters T.J., Raja K., Smith A.G. Genetic variation of basal iron status, ferritin and iron regulatory protein in mice: potential for modulation of oxidative stress. Biochem. Pharmacol. 2000;59:115–122. doi: 10.1016/s0006-2952(99)00306-8. [DOI] [PubMed] [Google Scholar]
- Coffey R., Ganz T. Iron homeostasis: an anthropocentric perspective. J. Biol. Chem. 2017;292:12727–12734. doi: 10.1074/jbc.R117.781823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degerman E., Landström T.R., Wijkander J., Holst L.S., Ahmad F., Belfrage P., Manganiello V. Phosphorylation and activation of hormone-sensitive adipocyte phosphodiesterase type 3B. Methods. 1998;14:43–53. doi: 10.1006/meth.1997.0564. [DOI] [PubMed] [Google Scholar]
- Devasthali S.D., Gor-deuk V.R., Brittenham G.M., Bravo J.R., Hughes M.A., Keating L.J. Bioavailability of carbonyl iron: a randomized, double-blind study. Eur. J. Haematol. 1991;46:272–278. doi: 10.1111/j.1600-0609.1991.tb01538.x. [DOI] [PubMed] [Google Scholar]
- Dobner J., Kaser S. Body mass index and the risk of infection - from underweight to obesity. Clin. Microbiol. Infect. 2018;24:24–28. doi: 10.1016/j.cmi.2017.02.013. [DOI] [PubMed] [Google Scholar]
- Dongiovanni P., Ruscica M., Rametta R., Recalcati S., Steffani L., Gatti S., Girelli D., Cairo G., Magni P., Fargion S., et al. Dietary iron overload induces visceral adipose tissue insulin resistance. Am. J. Pathol. 2013;182:2254–2263. doi: 10.1016/j.ajpath.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Duncan R.E., Ahmadian M., Jaworski K., Sarkadi-Nagy E., Sul H.S. Regulation of lipolysis in adipocytes. Annu. Rev. Nutr. 2007;27:79–101. doi: 10.1146/annurev.nutr.27.061406.093734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupic F. Duodenal mRNA expression of iron related genes in response to iron loading and iron deficiency in four strains of mice. Gut. 2002;51:648–653. doi: 10.1136/gut.51.5.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann J., Lippl F., Klose G., Schusdziarra V. Cholesterol lowering effect of dietary weight loss and orlistat treatment – efficacy and limitations. Aliment. Pharmacol. Ther. 2004;19:1173–1179. doi: 10.1111/j.1365-2036.2004.01966.x. [DOI] [PubMed] [Google Scholar]
- Fleming R.E., Holden C.C., Tomatsu S., Waheed A., Brunt E.M., Britton R.S., Bacon B.R., Roopenian D.C., Sly W.S. Mouse strain differences determine severity of iron accumulation in Hfe knockout model of hereditary hemochromatosis. Proc. Natl. Acad. Sci. 2001;98:2707–2711. doi: 10.1073/pnas.051630898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Li Z., Gabrielsen J.S., Simcox J.A., Lee S., Jones D., Cooksey B., Stoddard G., Cefalu W.T., McClain D.A. Adipocyte iron regulates leptin and food intake. J. Clin. Invest. 2015;125:3681–3691. doi: 10.1172/JCI81860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes M.D., Lecker S.H., Jagoe R.T., Navon A., Goldberg A.L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. U S A. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A., Cornaciu I., Lass A., Schweiger M., Poeschl M., Eder C., Kumari M., Schoiswohl G., Wolinski H., Kohlwein S.D., et al. The N-terminal region of comparative gene identification-58 (CGI-58) is important for lipid droplet binding and activation of adipose triglyceride lipase. J. Biol. Chem. 2010;285:12289–12298. doi: 10.1074/jbc.M109.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullo L., Corcioni E., Brancati C., Bria M., Pezzilli R., Sprovieri G. Morphologic and functional evaluation of the exocrine pancreas in p-Thalassemia major. Pancreas. 1993;5:176–180. doi: 10.1097/00006676-199303000-00007. [DOI] [PubMed] [Google Scholar]
- Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312:734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- Horne W.I., Tandler B., Dubick M.A., Niemelä O., Brittenham G.M., Tsukamoto H. Iron overload in the rat pancreas following portacaval shunting and dietary iron supplementation. Exp. Mol. Pathol. 1997;64:90–102. doi: 10.1006/exmp.1997.2212. [DOI] [PubMed] [Google Scholar]
- Hughes S.M., Chi M.M.-Y., Lowry O.H., Gundersen K. Myogenin induces a shift of enzyme activity from glycolytic to oxidative metabolism in muscles of transgenic mice. J. Cell Biol. 1999;145:633–642. doi: 10.1083/jcb.145.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Enomoto H., Tajima S., Izawa-Ishizawa Y., Kihira Y., Ishizawa K., Tomita S., Tsuchiya K., Tamaki T. Dietary iron restriction inhibits progression of diabetic nephropathy in db/db mice. Am. J. Physiol. Ren. Physiol. 2013;304:F1028–F1036. doi: 10.1152/ajprenal.00473.2012. [DOI] [PubMed] [Google Scholar]
- Jolliffe I. In: Encyclopedia of Statistics in Behavioral Science. Everitt B.S., Howell D.C., editors. John Wiley & Sons, Ltd); 2005. Principal component analysis; pp. 1580–1584. [Google Scholar]
- Ke J., An Y., Cao B., Lang J., Wu N., Zhao D. Orlistat-induced gut microbiota modification in obese mice. Evid. Based Complement. Alternat. Med. 2020;2020:1–9. doi: 10.1155/2020/9818349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimita W., Petrov M.S. Iron metabolism and the exocrine pancreas. Clin. Chim. Acta. 2020;511:167–176. doi: 10.1016/j.cca.2020.10.013. [DOI] [PubMed] [Google Scholar]
- La Carpia F., Wojczyk B.S., Annavajhala M.K., Rebbaa A., Culp-Hill R., D’Alessandro A., Freedberg D.E., Uhlemann A.-C., Hod E.A. Transfusional iron overload and intravenous iron infusions modify the mouse gut microbiota similarly to dietary iron. NPJ Biofilms Microbiomes. 2019;5:1–11. doi: 10.1038/s41522-019-0097-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J.G., Gorkiewicz G., Zechner R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Lawless J.W., Latham M.C., Stephenson L.S., Kinoti S.N., Pertet A.M. Iron supplementation improves appetite and growth in anemic Kenyan primary school children. J. Nutr. 1994;124:645–654. doi: 10.1093/jn/124.5.645. [DOI] [PubMed] [Google Scholar]
- Leboeuf R.C., Tolson D., Heinecke J.W. Dissociation between tissue iron concentrations and transferrin saturation among inbred mouse strains. J. Lab. Clin. Med. 1995;126:128–136. [PubMed] [Google Scholar]
- Luong Q., Huang J., Lee K.Y. Deciphering white adipose tissue heterogeneity. Biology. 2019;8:E23. doi: 10.3390/biology8020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Navarrete J.M., Ortega F., Rodríguez A., Latorre J., Becerril S., Sabater-Masdeu M., Ricart W., Frühbeck G., Fernández-Real J.M. HMOX1 as a marker of iron excess-induced adipose tissue dysfunction, affecting glucose uptake and respiratory capacity in human adipocytes. Diabetologia. 2017;60:915–926. doi: 10.1007/s00125-017-4228-0. [DOI] [PubMed] [Google Scholar]
- Morse A.C., Beard J.L., Jones B.C. A genetic developmental model of iron deficiency: biological aspects. Proc. Soc. Exp. Biol. Med. Soc. Exp. Biol. Med. 1999;220:147–152. doi: 10.1046/j.1525-1373.1999.d01-22.x. [DOI] [PubMed] [Google Scholar]
- Niepel D., Klag T., Malek N.P., Wehkamp J. Practical guidance for the management of iron deficiency in patients with inflammatory bowel disease. Ther. Adv. Gastroenterol. 2018;11 doi: 10.1177/1756284818769074. 1756284818769074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill S., O’Driscoll L. Metabolic syndrome: a closer look at the growing epidemic and its associated pathologies. Obes. Rev. 2015;16:1–12. doi: 10.1111/obr.12229. [DOI] [PubMed] [Google Scholar]
- Rausch V., Sala V., Penna F., Porporato P.E., Ghigo A. Understanding the common mechanisms of heart and skeletal muscle wasting in cancer cachexia. Oncogenesis. 2021;10:1–13. doi: 10.1038/s41389-020-00288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejholcova M., Wilhelm J., Svoboda P. Lipid peroxidation inhibits norepinephrine-stimulated lipolysis in rat adipocytes. Reduction of beta-adreno-ceptor number. Biochem. Biophys. Res. Commun. 1988;150:802–810. doi: 10.1016/0006-291x(88)90462-7. [DOI] [PubMed] [Google Scholar]
- Rohart F., Gautier B., Singh A., Lê Cao K.-A. mixOmics: an R package for ’omics feature selection and multiple data integration. PLoS Comput. Biol. 2017;13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen E.D., Spiegelman B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumberger J.M., Peters T., Burrington C., Green A. Transferrin and iron contribute to the lipolytic effect of serum in isolated adipocytes. Diabetes. 2004;53:2535–2541. doi: 10.2337/diabetes.53.10.2535. [DOI] [PubMed] [Google Scholar]
- Ryan B.J., Pelt D.W.V., Guth L.M., Ludzki A.C., Gioscia-Ryan R.A., Ahn C., Foug K.L., Horowitz J.F. Plasma ferritin concentration is positively associated with in vivo fatty acid mobilization and insulin resistance in obese women. Exp. Physiol. 2018;103:1443–1447. doi: 10.1113/EP087283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez K.K., Chen G.Y., Schieber A.M.P., Redford S.E., Shokhirev M.N., Leblanc M., Lee Y.M., Ayres J.S. Cooperative metabolic adaptations in the host can favor asymptomatic infection and select for attenuated virulence in an enteric pathogen. Cell. 2018;175:146–158.e15. doi: 10.1016/j.cell.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Nielsen K. Cambridge University Press; 1997. Animal Physiology: Adaptation and Environment. [Google Scholar]
- Schoettl T., Fischer I.P., Ussar S. Heterogeneity of adipose tissue in development and metabolic function. J. Exp. Biol. 2018;221:jeb162958. doi: 10.1242/jeb.162958. [DOI] [PubMed] [Google Scholar]
- Shostak A., Meyer-Kovac J., Oster H. Circadian regulation of lipid mobilization in white adipose tissues. Diabetes. 2013;62:2195–2203. doi: 10.2337/db12-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soe-Lin S., Apte S.S., Andriopoulos B., Andrews M.C., Schranzhofer M., Kahawita T., Garcia-Santos D., Ponka P. Nramp1 promotes efficient macrophage recycling of iron following erythrophagocytosis in vivo. Proc. Natl. Acad. Sci. 2009;106:5960–5965. doi: 10.1073/pnas.0900808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman J.R. Measuring energy metabolism in the mouse – theoretical, practical, and analytical considerations. Front. Physiol. 2013;4:34. doi: 10.3389/fphys.2013.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman B.M., Flier J.S. Obesity and the regulation of energy balance. Cell. 2001;104:531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- Stoltzfus R.J., Chway H.M., Montresor A., Tielsch J.M., Jape J.K., Albonico M., Savioli L. Low dose daily iron supplementation improves iron status and appetite but not anemia, whereas quarterly anthelminthic treatment improves growth, appetite and anemia in Zanzibari preschool children. J. Nutr. 2004;134:348–356. doi: 10.1093/jn/134.2.348. [DOI] [PubMed] [Google Scholar]
- Tchkonia T., Thomou T., Zhu Y., Karagiannides I., Pothoulakis C., Jensen M.D., Kirkland J.L. Mechanisms and metabolic implications of regional differences among fat depots. Cell Metab. 2013;17:644–656. doi: 10.1016/j.cmet.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tune J.D., Goodwill A.G., Sassoon D.J., Mather K.J. Cardiovascular consequences of metabolic syndrome. Transl. Res. 2017;183:57–70. doi: 10.1016/j.trsl.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijkander J., Landström T.R., Manganiello V., Belfrage P., Degerman E. Insulin-induced phosphorylation and activation of phosphodiesterase 3B in rat adipocytes: possible role for protein kinase B but not mitogen-activated protein kinase or p70 S6 kinase∗. Endocrinology. 1998;139:219–227. doi: 10.1210/endo.139.1.5693. [DOI] [PubMed] [Google Scholar]
- Winterbourn C.C. Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol. Lett. 1995;82–83:969–974. doi: 10.1016/0378-4274(95)03532-x. [DOI] [PubMed] [Google Scholar]
- Wu J.W., Wang S.P., Casavant S., Moreau A., Yang G.S., Mitchell G.A. Fasting energy homeostasis in mice with adipose deficiency of desnutrin/adipose triglyceride lipase. Endocrinology. 2012;153:2198–2207. doi: 10.1210/en.2011-1518. [DOI] [PubMed] [Google Scholar]
- Yang W.S., Stockwell B.R. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz B., Li H. Gut microbiota and iron: the crucial actors in health and disease. Pharmaceuticals. 2018;11:98. doi: 10.3390/ph11040098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook J.-S., Zhou M., Jaekwon L., Chung S. Iron deficiency anemia (IDA) promotes visceral obesity due to defective adipose tissue browning (OR09-07-19) Curr. Dev. Nutr. 2019;3:1829. [Google Scholar]
- Zimmermann R., Strauss J.G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Original western blot images are included in the supplemental information
-
•
Any additional information required to reanalyze the data reported in this paper is available form the lead contact upon request