Abstract
Blood-brain barrier (BBB) damage can be a result of central nervous system (CNS) diseases and may be a cause of CNS deterioration. However, there are still many unknowns regarding effective and targeted therapies for maintaining BBB integrity during ischemia/reperfusion (I/R) injury. In this study, we demonstrate that the circular RNA of FoxO3 (circ-FoxO3) promotes autophagy via mTORC1 inhibition to attenuate BBB collapse under I/R. Upregulation of circ-FoxO3 and autophagic flux were detected in brain microvessel endothelial cells in patients with hemorrhagic transformation and in mice models with middle cerebral artery occlusion/reperfusion. In vivo and in vitro studies indicated that circ-FoxO3 alleviated BBB damage principally by autophagy activation. Mechanistically, we found that circ-FoxO3 inhibited mTORC1 activity mainly by sequestering mTOR and E2F1, thus promoting autophagy to clear cytotoxic aggregates for improving BBB integrity. These results demonstrate that circ-FoxO3 plays a novel role in protecting against BBB damage, and that circ-FoxO3 may be a promising therapeutic target for neurological disorders associated with BBB damage.
Keywords: ischemic stroke, hemorrhagic transfromation, blood-brain barrier, circular RNA FoxO3, autophagy, mTORC1 complex, E2F1, proteins aggregates
Graphical abstract
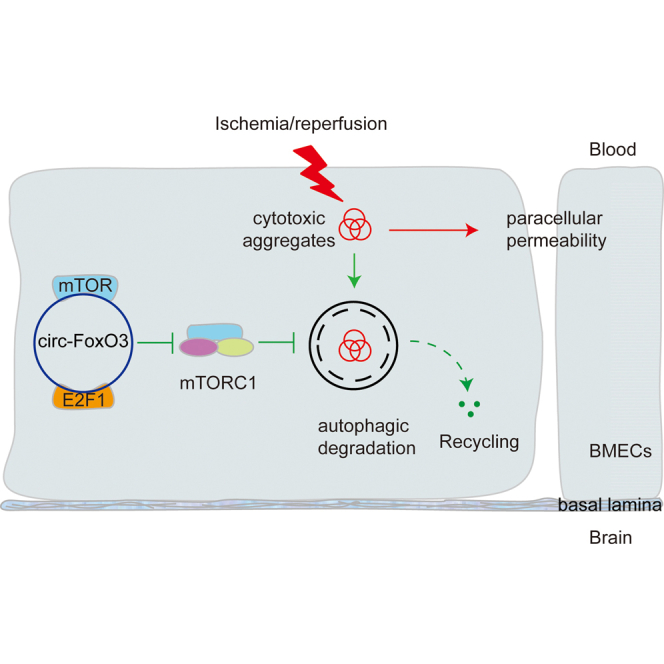
Upregulated circ-FoxO3 and autophagy activation were detected in BBB after ischemic/reperfusion. circ-FoxO3 alleviated BBB damage by sequestering mTOR and E2F1 to activate autophagy. This study reveals that circ-FoxO3 serves a promising therapeutic target for disorders with BBB damage.
Introduction
The blood-brain barrier (BBB) is a highly organized multicellular structure composed of capillaries formed by self-fusion of brain microvascular endothelial cells (BMECs) through intact tight junctions, as well as peripheral pericytes and astrocytes surrounding the capillaries. The BBB is generally considered to be the gatekeeper that helps to maintain the fragile homeostasis of the central nervous system (CNS) through segregation from the systemic circulation.1 BBB disruption can occur as the result of CNS diseases and may be the cause of secondary brain injuries, including hemorrhage and brain edema.2 In the pathological process of ischemic stroke, failure of blood flow caused by sudden blood vessel occlusion results in shrinkage of the BMECs, which causes translocation of tight junction proteins (TJs) from the membrane into the cytosol. This increases BBB permeability, which makes it difficult to avoid secondary brain damage after reperfusion therapy. Due to the critical role of BBB in cerebral protection, a thorough understanding of the endogenous mechanisms for maintaining BBB integrity may be a potential therapeutic strategy for treating neurological disorders including stroke.
Circular RNAs (circRNAs) endogenously express non-coding RNAs, and serve as biomarkers for multiple disorders.3 Although the function of circRNAs is still not fully understood, they are known to exert a role in sequestering microRNAs (miRNAs) or RNA-binding proteins (RBPs) to undergo transcriptional and post-transcriptional regulation. This is mainly due to their covalently closed loops, which help them resist degradation by RNase, and effectively maintain their biological functions.4 Recently, multiple circRNAs have been reported to be abundantly expressed in the brain.5 These molecules take part in neurovascular functions, such as neurodevelopment6 and degenerative diseases.7 Notably, circRNAs protect BBB integrity by targeting miRNAs against ischemic stroke8 and bacterial meningitis.9 Our previous study revealed that the number of circRNAs was differentially expressed in the blood of mice after middle cerebral artery occlusion (MCAO) and in patients with acute ischemic stroke.10 This indicated that some of the blood circRNAs might function as biomarkers for ischemic stroke. Although many circRNAs are associated with CNS disorders, their involvement in neurovascular integrity and pathological processes require further investigation.
Intriguingly, recent literature has shown that circRNAs interact with autophagy-related miRNAs11,12 or proteins13 for regulation of different pathological processes. Autophagy is originally considered a self-protective mechanism of cells to overcome the injury from external stimuli. This is because autophagy eliminates the damaged organelles and proteins, and converts them into nutrients for promoting cell survival in various pathological processes.14 Our previous studies showed that autophagy protected BBB integrity against hypoxic injury by blocking the depolarization of Claudin5 (Cldn5)15 and was regulated by mammalian target of rapamycin complex 1 (mTORC1) under low-serum conditions.16 However, the mechanism of autophagy regulation in the pathological process of BBB damage needs to be further explored. circRNAs have been reported as key regulators of autophagy-related regulatory networks in cancer17 and metabolic disease.18 Typically, circRNAs are able to activate astrocytes via the regulation of autophagy,11 and to promote cell survival via the inhibition of mTORC1 in oral squamous cell carcinoma.19 Thus, we hypothesized that circRNAs are involved in the regulation of autophagy in the pathological process of the BBB in the setting of vascular disorders.
circ-FoxO3 is highly abundant compared with other circRNAs in the mammalian brain.20 It is reported to be down- or upregulated in cancer, and to act with a wide range of regulatory functions by forming a complex with proteins or miRNA.21,22 Specifically, circ-FoxO3 facilitated cardiac senescence via interaction with ID-1, E2F1, FAK, and HIF1α,23 and bond to p21 and CDK2 to retard cell-cycle progression.24 In addition, it could act as a sponge of miR-138-5p to promote glioblastoma progression.25 Although circ-FoxO3 showed functions in the pathological process of multiple diseases, there were still many unknowns about how circFoxO3 was involved in repair after damage caused by neurological disorders, especially in maintaining BBB integrity.
In this study, we showed that circ-FoxO3 was upregulated in BMECs and exerted a role in the alleviation of BBB damage after ischemia/reperfusion (I/R). Further analysis revealed that circ-FoxO3 activated autophagy by interacting with mTOR and E2F1, thus triggering autophagic degradation of cytotoxic aggregates. These results provide a novel insight into circ-FoxO3 as a potential therapeutic target in disorders associated with BBB damage.
Results
circ-FoxO3 is upregulated and autophagy is activated in BBB after I/R
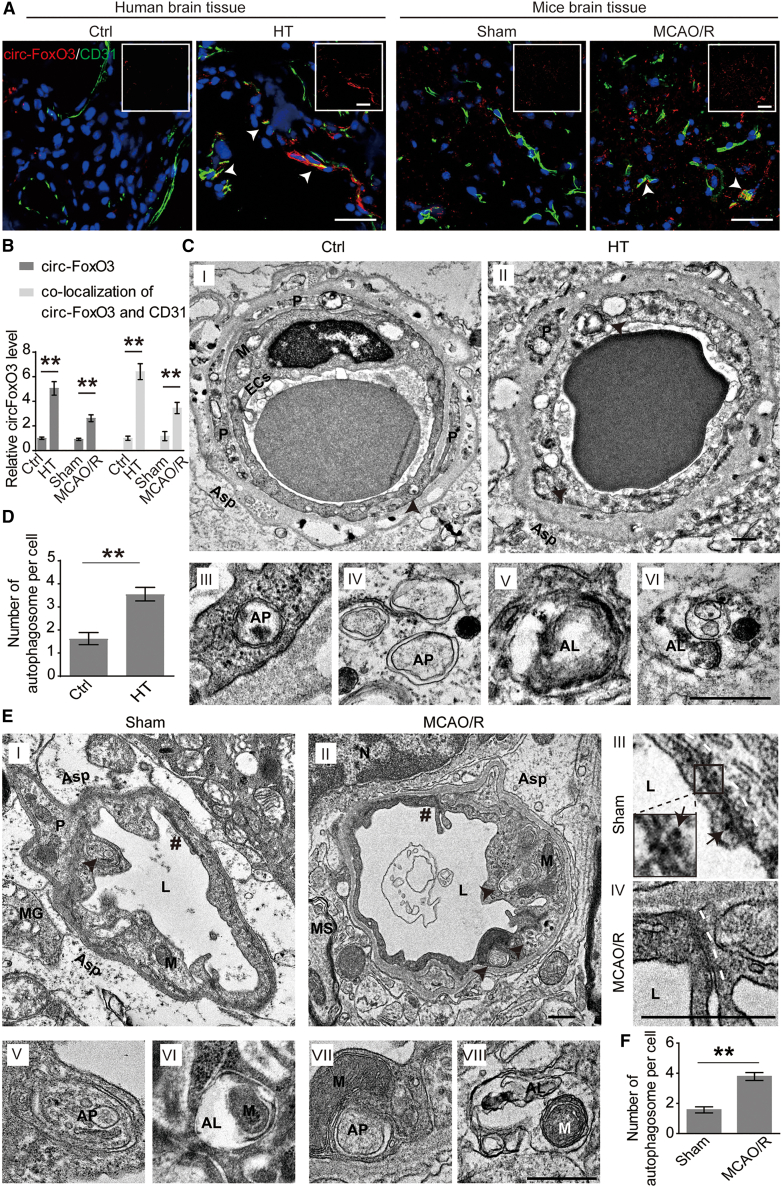
circ-FoxO3 is formed by back-splicing of exon 2 of the FoxO3 gene. It is highly conserved in human and mice species. We comparatively analyzed the homology of circ-FoxO3 in humans (hsa_circ_0006404) and mice (mmu_circ_0002207). Their alignment was found to be 90.87% identical (Figure S1A). More importantly, they both possessed a similar junction loop (Figure S1B, upper panel). This was validated by Sanger sequencing in brain tissues of patients with hemorrhagic transformation (HT) and MCAO and reperfusion (MCAO/R) mice (Figure S1B, lower panel). The qRT-PCR assay was conducted with specially designed divergent and convergent primers, and the results showed that circ-FoxO3, rather than FoxO3 mRNA, resisted RNase R digestion (Figure S2A). In addition, we confirmed the enrichment of circ-FoxO3 in brain tissues at the site of intracerebral hemorrhage from patients with HT and the peri-infarct area of MCAO/R mice (Figure S2B). We also found that circ-FoxO3 exerted differential expression in multiple tissues of normal male mice (Figure S2C). Furthermore, upregulated circ-FoxO3 co-localized with CD31 (an endothelial marker) in Figures 1A and 1B, and with GFAP (an astrocyte marker) but not with PDGFR-β (a pericyte marker) in Figures S2D and S2E. These results indicated that circ-FoxO3 is upregulated in brain tissues after I/R, especially in BMECs and astrocytes.
Figure 1.
circ-FoxO3 is upregulated and autophagy is activated in the BBB after cerebral I/R injury
(A and B) Images of circ-FoxO3 and CD31 and their quantification (integrated optical density) in brain tissues at the site of intracerebral hemorrhage from patients with HT and the peri-infarct area of MCAO/R mice. Ctrl group: brain tissues from non-stroke patients (six females and six males). HT group: patients with hemorrhagic transformation, six females and eight males. Sham group: male mice without ligation of the arteries and insertion of nylon monofilaments, n = 6. MCAO/R group: male mice under MCAO and reperfusion for 2 h, respectively, n = 6. White arrowheads indicate the co-localization of circ-FoxO3 and CD31. Scale bars, 50 μm. (C) The ultra-structures of the BBB in human. The cross-section of the BBB from patients with glioma and patients with HT were imaged in I and II, respectively. The autophagosome-like vacuoles were enlarged in III to VI. Scale bar, 500 nm. (D) Quantitative number of autophagosome-like vacuoles in the BMECs from patients with HT. n (Ctrl group): three females and three males. n (HT group): three females and four males. (E) Ultra-structure of BBB in mice. The cross-section of BBB from sham and MCAO/R mice was presented in I and II, respectively. Cell-cell junctions marked # in I and II was enlarged in III and IV, respectively. White dotted line: cell-cell junctions. Arrowheads indicate desmosomes. Scale bar, 500 nm. The autophagosome-like vacuoles enlarged into V to VIII. Scale bar, 500 nm. (F) Quantitative number of autophagosome-like vacuoles in the BMECs from male MCAO/R mice. n = 6. ECs, endothelial cells. Black arrowheads, autophagosome-like vacuoles; L, lumen; #, cell-cell junction; P, pericyte; Asp, astrocyte processes; MG, microglia; MS, medullary sheath; M, mitochondria; N, nucleus; AP, autophagosome; AL, autolysosome; MCAO/R, middle cerebral artery occlusion for 2 h followed by reperfusion for 2 h. p values indicate a two-tailed unpaired Student's t test. Data are provided as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
To analyze the pathological features of BBB after I/R, we performed transmission electron microscopy (TEM) to investigate the ultra-structure of BBB in brain tissues from patients with HT and MCAO/R mice. We observed that the BMECs showed significant shrinkage (Figure 1C) and failure of cell-cell junction (Figure 1E) indicating that the BBB may have been damaged. Interestingly, a marked increase in the number of autophagic vacuoles was found in BMECs (Figures 1C–1F), which implied that autophagy was activated in the BBB of patients with HT and of male mice after MCAO/R. Sex difference has been demonstrated to be associated with prognosis in stroke.26,27 In this study, we further performed the same experiments in female mice and obtained similar results to male mice (Figure S3). Of note, we observed the engulfment of mitochondria by autophagosome-like vacuoles in BMECs of female mice with MCAO/R (Figure S3CV). Since ischemia-damaged mitochondria were responsible for secondary damage caused by redox dysfunction,28 our observations suggest that autophagy might contribute to intracellular homeostasis of BBB.
Overall, these findings show that upregulated circ-FoxO3 and activated autophagy are found in the pathological process of BBB in patients with HT and in MCAO/R mice.
Both circ-FoxO3 and autophagy attenuate BBB impairment after I/R
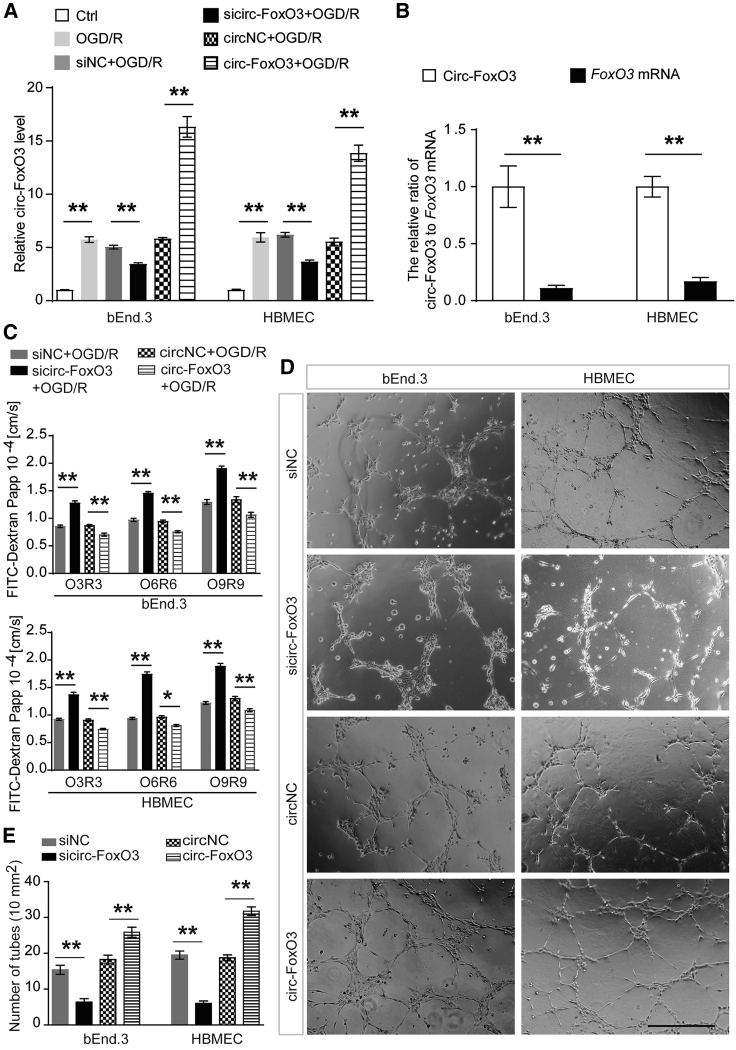
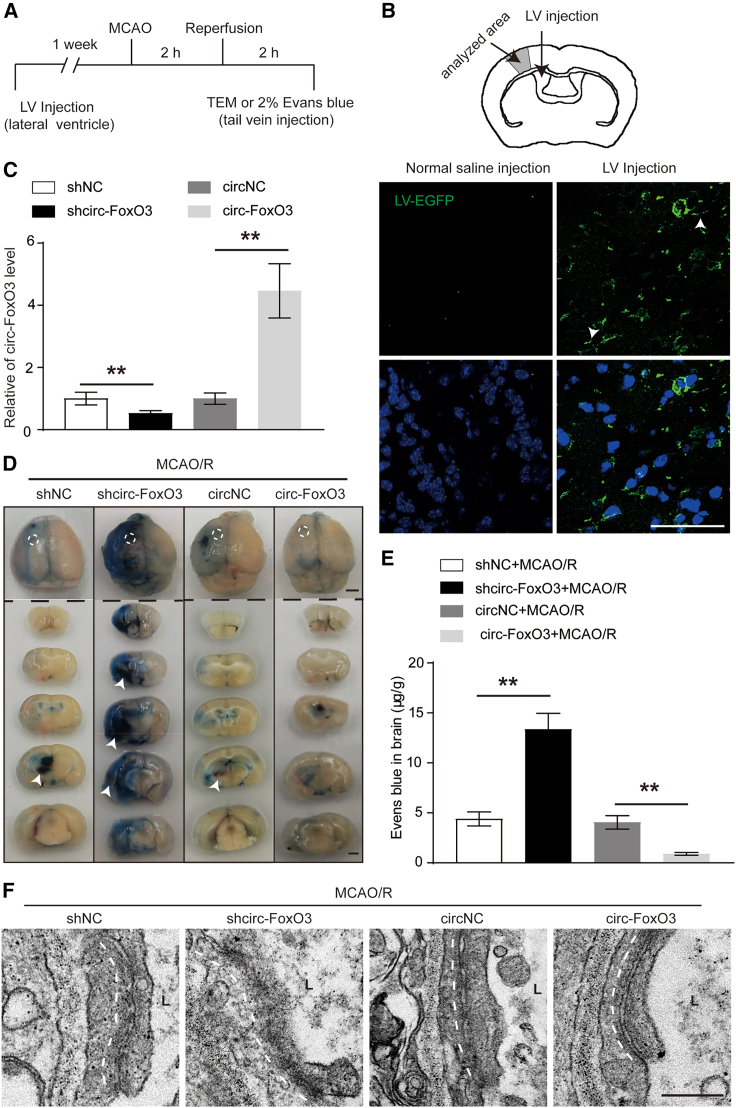
To explore the role of circ-FoxO3 on BBB integrity after I/R, we analyzed BBB permeability in vitro and in vivo. Firstly, we confirmed the upregulated circ-FoxO3 in oxygen-glucose deprivation/reoxygenation (OGD/R)-mediated BMECs (Figures S4A–S4C). This was in line with the results of the analysis of brain tissues from patients with HT and in MCAO/R mice (Figures 1A, 1B, S3A, and S3B). Subsequently, circ-FoxO3 knockdown using siRNA (siRNA1, siRNA2, siRNA3) or shRNA (shRNA1, shRNA2, shRNA3) was found to enhance the permeability of the endothelial cells (Figures S4D and S4E). This indicated that upregulated circ-FoxO3 induced by OGD/R might be involved in the maintenance of BBB integrity. To further support this idea, we first used sicirc-FoxO3 (siRNA1) or LV-circ-FoxO3 (Figures S5A–S5D) transfection to either down- or upregulate circ-FoxO3 in BMECs. As expected, BMECs with circ-FoxO3 knockdown or overexpression were generated (Figure 2A). Of note, the transduction of LV-circ-FoxO3 contributed to a much higher level of circ-FoxO3 than the level of FoxO3 mRNA (Figure 2B). These results suggested that overexpression of circ-FoxO3 via transduction of LV-circ-FoxO3 mainly altered circ-FoxO3 expression, but not too much for FoxO3 mRNA. Next, we analyzed the role of circ-FoxO3 on endothelial cell permeability. The results confirmed that downregulated circ-FoxO3 enhanced the OGD/R-induced permeability, but it could be alleviated by circ-FoxO3 overexpression (Figure 2C). In addition, circ-FoxO3 knockdown suppressed tube formation, while circ-FoxO3 overexpression promoted tube formation in BMECs (Figures 2D and 2E). These data indicate that circ-FoxO3 may play a protective role in maintaining BBB integrity after OGD/R injury. To confirm this result in vivo, we measured the BBB integrity in mice that were subjected to MCAO/R 1 week after intracerebroventricular injection of LV-shcirc-FoxO3 (Figures S5E and S5F) or LV-circ-FoxO3. The flowchart of the experimental procedures is shown in Figure 3A. In the cortex surrounding lateral ventricles, the lentiviral particles encoding EGFP (LV-shcirc-FoxO3 or LV-circ-FoxO3) were observed in brain tissues 1 week after injection (Figure 3B). Notably, part of the EGFP signal was detected in the vascular walls (white arrowheads in Figure 3B), which suggested that circ-FoxO3 might be expressed in BMECs. Moreover, qRT-PCR results indicated that the circ-FoxO3 level in the cortex surrounding lateral ventricles was regulated by LV-shcirc-FoxO3 or LV-circ-FoxO3 (Figure 3C). Next, the role of circ-FoxO3 in BBB permeability was analyzed by Evans blue extravasation in male mice after MCAO/R. Consistent with the results in vitro, downregulated circ-FoxO3 promoted the infiltration of Evans blue across BBB, while circ-FoxO3 overexpression inhibited it (Figures 3D and 3E). In addition, the deformation of the cell-cell junction induced by MCAO/R was exacerbated by shcirc-FoxO3, but was alleviated by overexpressed circ-FoxO3 (Figure 3F). These results demonstrate that circ-FoxO3 is required for the maintenance of BBB integrity after ischemic stroke.
Figure 2.
circ-FoxO3 inhibits OGD/R-induced endothelial barrier permeability
The siRNA targeting of circ-FoxO3 or LV-circ-FoxO3 was transfected into the BMECs (bEnd.3 and HBMEC). (A) The relative level of circ-FoxO3 or FoxO3 mRNA was measured by qRT-PCR in BMECs. n = 3. (B) The level of circ-FoxO3 and FoxO3 mRNA was compared in BMECs with circ-FoxO3 overexpression. n = 4. (C) The infiltration of FITC-dextran (10 kDa) across the monolayer of BMECs was measured to determine the role of circ-FoxO3 on the properties of the endothelial cell barrier. n = 6. (D) Representative photomicrographs of tube formation in BMECs with circ-FoxO3 knockdown or overexpression. n = 6. Scale bar, 200 μm. (E) Quantification of the number of tubes in (D). The p values indicate one-way ANOVA with Dunnett's multiple comparisons test. OGD/R, oxygen-glucose deprivation/reoxygenation; O3/R3, oxygen-glucose deprivation for 3 h and reoxygenation for 3 h (O6/R6 and O9/R9 indicate the same for 6 and 9 h, respectively). Data are provided as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
Figure 3.
circ-FoxO3 attenuates BBB damage in MCAO/R mice
(A) The flowchart of the experimental procedures in mice. (B) The schematic diagram of LV injection (top panel) and the localization of LV in brain tissues of mice (lower panel). White arrowhead indicates that the LV-EGFP-shcirc-FoxO3 or LV-EGFP-circ-FoxO3 localized in vessels. Scale bar, 50 μm. (C) The expression of circ-FoxO3 in the peri-infarct area of mice, n = 4. (D) Representative images of Evans blue extravasation into entire brains of mice with circ-FoxO3 knockdown or overexpression. The white dotted lines in the top panels highlight the injection site of LV. n = 5. Scale bar, 2 mm. (E) Vascular permeability was detected by measurement of Evans blue extravasation in brain tissue using spectrophotometry at 610 nm. n = 5. (F) The cell-cell junctions were imaged by TEM analysis in brain tissue after circ-FoxO3 knockdown or overexpression. n = 5. Scale bar, 250 nm. L, lumen; MCAO/R, middle cerebral artery occlusion for 2 h and followed by reperfusion for 2 h. p values indicate one-way ANOVA with Dunnett's multiple comparisons test. All data are presented as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
To investigate the role of autophagy on endothelial cell integrity, we investigated endothelial cell permeability and the localization of Cldn5 and ZO-1 in BMECs (bEnd.3 and HBMEC) after co-treatment with OGD/R and an autophagy inhibitor (chloroquine [CQ]) or enhancer (Rapacylin, Rapa). The results showed that autophagy attenuated the enhancement of permeability (Figures S6A and S6B), reduced the aggregation of Cldn5 and ZO-1 (yellow arrowheads) in the cytoplasm, and inhibited the loss of Cldn5 and ZO-1 (white arrowheads) on the membrane (Figures S6C and S6D). This might be explained by the fact that autophagy-mediated aggregation clearance contributed to the alleviation of redox dysfunction and reduction of peroxidation products, which protected membrane protein polarity against peroxidation damage.15
circ-FoxO3 attenuates BBB damage via autophagy activation
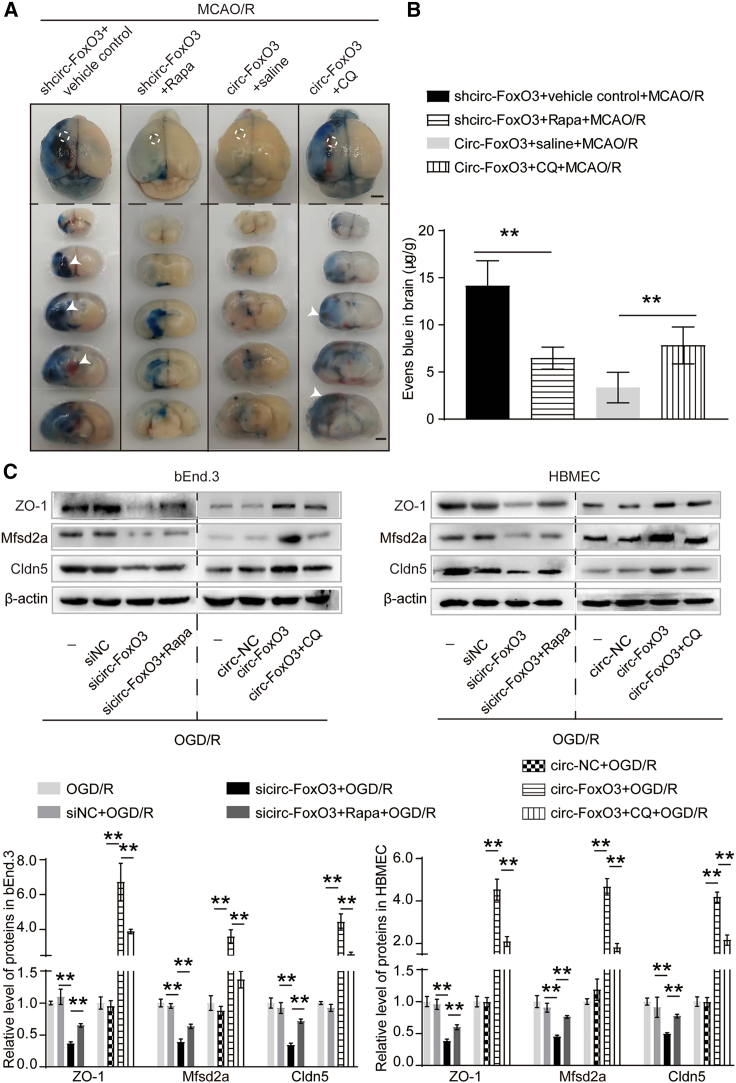
Previous studies have revealed that circRNAs regulate autophagy in cancers13 and ischemic stroke.11 In this study, we aimed to determine whether circ-FoxO3 reduced BBB impairment by inducing autophagy. We measured BBB permeability and its related proteins levels (ZO-1, Msfd2a, and Cldn5) in BBB models with circ-FoxO3 knockdown or upregulation. In these BMECs and mice, Rapa (autophagy activator) or CQ (autophagy inhibitor) was administered before OGD/R or MCAO/R. The Evans blue extravasation results showed that BBB permeability in the mice with circ-FoxO3 knockdown was partly reduced after Rapa pretreatment, but it was strengthened by CQ in mice with circ-FoxO3 overexpression (Figures 4A and 4B). Strikingly, cerebral hemorrhage (four of five mice) was observed when autophagy was inhibited by CQ. This is shown by the white arrowheads in Figure 4A. In addition, the in vitro results confirmed that the permeability of the endothelial barrier formed by bEnd.3 or HBMEC was enhanced when circ-FoxO3 was downregulated. This was partly reversed after use of Rapa to activate autophagy. Similarly, the low permeability formerly maintained by circ-FoxO3 was increased after autophagy was inhibited by CQ (Figure S7). These data indicate that the role of circ-FoxO3 in maintaining the low permeability of BBB is mainly dependent on autophagy activation. Furthermore, autophagy has been confirmed to play an important role in regulating the level of BBB-related proteins (ZO-1, Msfd2a, and Cldn5) by circ-FoxO3. Specifically, enhanced autophagy by Rapa inhibited the reduction of BBB-related proteins induced by circ-FoxO3 knockdown. Suppression of autophagy blocked the effect of upregulated circ-FoxO3 on promoting BBB-related protein levels (Figure 4C). Overall, these results reveal that the reduction of BBB damage by circ-FoxO3 is in part due to autophagy activation during I/R.
Figure 4.
The role of circ-FoxO3 in alleviation of BBB damage depends on autophagy
(A and B) Evans blue extravasation was detected in male mice with circ-FoxO3 knockdown or upregulation that were pretreated with Rapa or CQ for 1 week before MCAO/R. CQ, chloroquine, 5 mg/kg/daily. Rapa, rapamycin, 0.75 mg/kg/daily. Vehicle control, saline containing 5% polyethylene glycol 400 and 5% Tween 80. White arrowheads indicate cerebral hemorrhage. Four of five mice displayed cerebral hemorrhage. Scale bar, 2 mm. n = 5. (C) The level of BBB-related proteins such as ZO-1, Mfsd2a, and Cldn5 in BMECs (bEnd.3 and HBMEC) with circ-FoxO3 knockdown or overexpression was analyzed by immunoblotting when cells were pretreated with CQ (or Rapa) before OGD/R. Rapa, 50 nmol/L; CQ, 30 μmol/L. n = 4. p values indicate one-way ANOVA with Dunnett's multiple comparisons test. MCAO/R, middle cerebral artery occlusion for 2 h followed by reperfusion for 2 h; OGD/R, oxygen-glucose deprivation for 3 h followed by reoxygenation for 3 h. All data are presented as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
circ-FoxO3 promotes autophagy activation
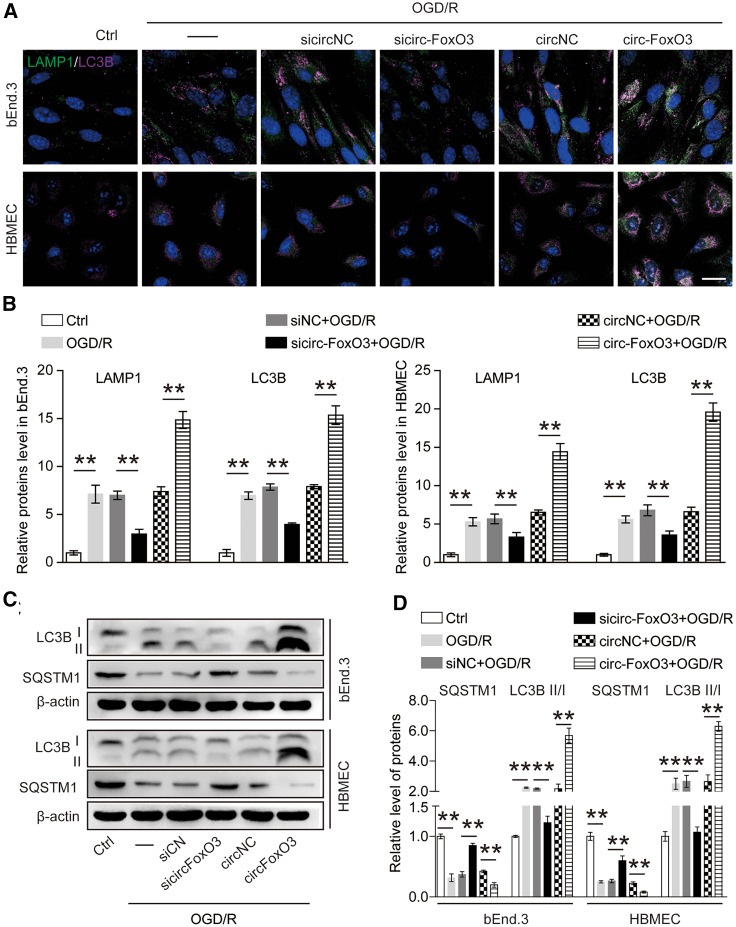
Next, we asked whether circ-FoxO3 could modulate autophagy. To test this, the level of autophagy-related proteins was quantified by immunofluorescence (IF) and immunoblotting in BMECs with knockdown or upregulation of circ-FoxO3. IF results indicated that the levels of LAMP1 and LC3B were reduced in the circ-FoxO3 knockdown group, but were increased in the circ-FoxO3 overexpression group (Figures 5A and 5B). Immunoblotting analysis revealed that knockdown of circ-FoxO3 could inhibit the ratio of LC3B-II/LC3B-I and promote the level of SQSTM1 in BMECs after OGD/R. When circ-FoxO3 was overexpressed, the ratio of LC3B-II/LC3B-I was increased while the level of SQSTM1 declined (Figures 5C and 5D). In addition, we performed TEM to visualize changes in the number of autophagic vacuoles in BMECs with circ-FoxO3 knockdown or upregulation after OGD/R. We found that the number of autophagic vacuoles was significantly reduced after the knockdown of circ-FoxO3, while this number was increased when circ-FoxO3 was upregulated (Figure S8). In addition, circ-FoxO3 was found to inhibit the fluorescent clusters of SQSTM1 in BMECs that were successively transfected with LV-EGFP-circ-FoxO3 and Ad-mCherry-SQSTM1 (Figure S9A). To further confirm the blocking role of down-expressed circ-FoxO3 on autophagy, siRNA and shRNA were used to reduce circ-FoxO3 expression in bEnd.3, and their effects on the rate of LC3B-II/LC3B-I and SQSTM1 were compared. Immunoblotting results demonstrated that both siRNA- and shRNA-mediated knockdown of circ-FoxO3 suppressed the LC3B-II/LC3B-I ratio and increased the SQSMT1 level, indicating that circ-FoxO3 knockdown inhibited autophagy (Figure S9B). The above results support the idea that circ-FoxO3 is an autophagy-related circRNA.
Figure 5.
circ-FoxO3 promotes autophagy in OGD/R-treated BMECs
Autophagy was detected in the monolayer of endothelial cells (bEnd3 and HBMEC) with circ-FoxO3 knockdown or overexpression after OGD/R. (A) Representative images show LAMP1 and LC3B staining. (B) The quantitative integrated optical density of LAMP1 and LC3B in (A). n = 6. Scale bar, 10 μm. (C) Immunoblotting analysis of LC3B II/I and SQSTM1 levels. OGD/R, oxygen-glucose deprivation for 3 h followed by reoxygenation for 3 h. (D) Quantitative results of the bands in (C). n = 4. p values indicate one-way ANOVA with Dunnett's multiple comparisons test. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
circ-FoxO3 interacts with mTOR to inhibit mTORC1 activity
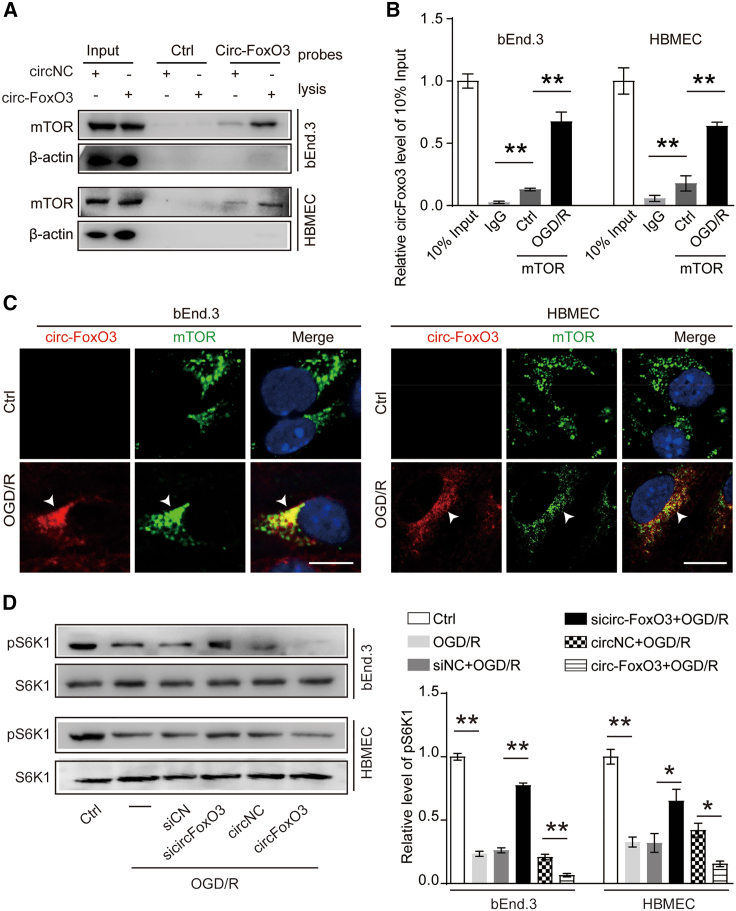
To determine the target of circ-FoxO3 that promotes autophagy, biotinylated RNA-protein pull-down and liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed in this study. As shown in Figure S10, after identification by LC-MS/MS, the proteins pulled down by circ-FoxO3 were mapped to autophagy-related pathways using the R package KEGGREST.29 The results found that mTOR was a common protein in the autophagy-related pathways. To further analyze the interaction between circ-FoxO3 and mTOR, immunoblotting was used to analyze mTOR levels after the biotinylated circ-FoxO3 pull-down assay. The results validated that mTOR was pulled down in the lysates of BMECs with circ-FoxO3 upregulation (Figure 6A). In addition, circ-FoxO3 was detected in the reciprocal pull-down assay (Figure 6B). These findings indicated the interaction between circ-FoxO3 and mTOR, which was further confirmed by co-localization analysis in BMECs after OGD/R (Figure 6C). It is believed that mTOR associates with Raptor and mLST8 to form a complex named mTORC1,30 which is involved in the negative regulation of autophagy.31 The activity of mTORC1 can be determined by the phosphorylation level of S6K, its downstream effector of mTORC1.32 Importantly, our previous studies have shown that autophagy induced by low serum can ameliorate BBB damage via mTORC1 inhibition.16 Mounting evidence has demonstrated that circRNAs competitively interact with and block RBPs.33 We then asked whether circ-FoxO3 could inhibit mTORC1 activity by competitively sequestering mTOR. To test this idea, we measured the level of phosphorylated S6K1 in BMECs with knockdown or upregulation of circ-FoxO3 after OGD/R. We found that circ-FoxO3 knockdown increased S6K1 phosphorylation levels, while circ-FoxO3 overexpression showed the opposite effect (Figure 6D). Together, the activated autophagy induced by circ-FoxO3 may be dependent on mTORC1 inhibition caused by the interaction between circ-FoxO3 and mTOR.
Figure 6.
circ-FoxO3 interacts with mTOR and inhibits mTORC1 activity
(A) The biotin-labeled circ-FoxO3 probe was incubated in lysates of BMECs with circ-FoxO3 overexpression, and mTOR was identified using an immunoblotting assay. (B) circ-FoxO3 was immunoprecipitated with anti-mTOR-coated magnetic beads, and then detected by qRT-PCR. n = 4. (C) Representative images showed co-localization of circ-FoxO3 and mTOR in BMECs (bEnd.3 and HBMEC). Scale bar, 10 μm. n = 6. White arrowheads, co-localization of circ-FoxO3 and mTOR. (D) The activity of the mTORC1 complex was determined by immunoblotting assay to measure the phosphorylation of S6k1. n = 4. p values indicate one-way ANOVA with Dunnett's multiple comparisons test. OGD/R, oxygen-glucose deprivation for 3 h followed by reoxygenation for 3 h. Data are provided as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
circ-FoxO3 interacts with E2F1 to trigger autophagy via mTORC1 inhibition
The activation of mTORC1 can be driven by the transcriptional regulator E2F1. This process requires E2F1 to enter the nucleus and bind to DNA for activation of PFKFB3 transcription, which then facilitates the translocation of mTORC1 to lysosomes with the assistance of PFKFB3.34,35 In addition, E2F1 was found to interact with circ-FoxO3 in senescent myocardial cells.23 In this study, we first explored whether circ-FoxO3 formed a complex with E2F1 in OGD/R-treated BMECs. LC-MS/MS results and RNA-binding protein immunoprecipitation (RIP) demonstrated the interaction between circ-FoxO3 and E2F1 (Figures 7A and S10B). These results were further confirmed by the co-localization between circ-FoxO3 and E2F1 in the cytosol of BMECs (Figure 7B). Of note, a reduced level of E2F1 was captured and analyzed in the nucleus after OGD/R (Figures 7B and 7C), which indicated that E2F1 activity was inhibited in the nucleus. These results were consistent with the previous report showing that the interaction of circ-FoxO3 and E2F1 in the cytosol blocks the entry of E2F1 to the nucleus and thus prevents its transcriptional regulation.23 To mimic the role of sequestrated E2F1 on autophagy activity, we knocked down E2F1 by siRNA and assessed its response on mTORC1 activity, SQSTM1 level, and the ratio of LC3B-II/LC3B-I in OGD/R-treated BMECs. Decreased levels of PFKFB3 and P-S6K1 were observed, indicating that the translocation of mTORC1 to lysosomes was blocked and mTORC1 was inactivated. In addition, a reduced level of SQSTM1 was accompanied by an increased ratio of LC3B-II/LC3B-I (Figures 7D and 7E). These results suggest that reduction of E2F1 inhibits mTORC1 activity and promotes autophagy, and that circ-FoxO3-mediated autophagy activation depends on its sequestration of E2F1 for mTORC1 inhibition. Our working model for the novel role of circ-FoxO3 is depicted in Figure 7F.
Figure 7.
circ-FoxO3 sequesters E2F1 to inhibit mTOCR1 activity and promotes autophagy
(A) circ-FoxO3 was immunoprecipitated with anti-E2F1-coated magnetic beads, and then detected using qRT-PCR. n = 3. p values indicate one-way ANOVA with Dunnett's multiple comparisons test. (B) The localization of circ-FoxO3 and E2F1 in BMECs (bEnd.3 and HBMEC) was determined by FISH and IF. n = 5. Scale bar, 10 μm. The dotted line is the boundary of the nucleus. White arrowheads show the co-localization of circ-FoxO3 and E2F1 in cytosol. Yellow arrowheads indicate the E2F1 in the nucleus. (C) Quantitative integrated optical density of E2F1 in nucleus in (B). p values indicate a two-tailed unpaired Student's t test. (D) The levels of E2F1, PFKFB3, p-S6K1, S6K1, SQSTM1, and LC3B were measured by western blotting. n = 3. (E) Quantitative results of the bands in (D). n = 3. p values indicate one-way ANOVA with Dunnett's multiple comparisons test. (F) The schematic diagram of circ-FoxO3 in the attenuation of BBB damage during I/R. OGD/R, oxygen-glucose deprivation for 3 h followed by reoxygenation for 3 h. I/R, ischemia/reperfusion. Data are provided as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
Discussion
This study provides a novel insight into the role of circ-FoxO3 in the attenuation of BBB damage via inhibiting mTORC1 to promote autophagy. Upregulated circ-FoxO3 and autophagy activation were detected in brain tissues from patients with HT and MCAO/R mice. We found that circ-FoxO3 plays a role in protection against OGD/R-stimulated endothelial barrier damage and MCAO/R-induced BBB collapse in mice, in an autophagy-dependent manner. Mechanistically, we found that circ-FoxO3 exerts a role in the sequestration of mTOR and E2F1, which may be required for inhibition of mTORC1 activity to promote autophagy.
The BBB is an adjustable, physiological interface between systemic circulation and brain parenchyma. It regulates and protects the internal milieu of the CNS against circulating drugs, toxins, and xenobiotics. BBB collapse is a common complication associated with neurological disorders, and accelerates the pathological process of cognitive and motor impairment.36 While BBB protection is recognized as a promising therapeutic protection against secondary brain injury.37 Our previous studies have documented that autophagy selectively scavenges the hypoxia-induced accumulation of caveolin-1 and Cldn5 to inhibit the caveolae-mediated endocytosis of Cldn5 on membranes and reduce toxicity of protein aggregates in cytoplasm, which is important for maintaining BBB integrity.15 Recent results have provided new insights into the role of non-coding RNA in the diagnosis and therapy of multiple disorders through regulating autophagy.19,38 Our previous findings suggest that differentially expressed blood-derived circRNAs are associated with acute ischemic stroke.10 However, whether or how the repair response of circRNAs occurs in neurovascular disorders was still unclear.
circ-FoxO3 is evolutionarily conserved between humans and mice and possesses the same splice site in both species. Emerging evidence has indicated that circ-FoxO3 acts as a sponge for miRNAs and RBPs to participate in cancers24,25 and senescence.23 In this study, we verified that circ-FoxO3 was upregulated in BMECs of HT patients, and in mice after MCAO/R. Ultra-structural analysis revealed BBB damage and simultaneously provided a surprising finding that autophagy was activated in BMECs. Although sex differences in stroke presentation have been identified in recent research,26,27 upregulated circ-FoxO3 and autophagy activation were found in both genders. It can be deduced that this event is a common phenomenon in ischemic stroke and is independent of gender differences. These results indicate that circ-FoxO3 may be linked to autophagy in the pathological process of ischemic stroke-induced BBB collapse.
A substantial number of studies have revealed that upregulated circRNAs are usually accompanied by the deterioration of vascular endothelial barriers39 and cardiovascular pathology40 in vascular disorders. Our findings suggest that upregulated circ-FoxO3 contributes to the maintenance of endothelial barrier integrity. Furthermore, we found that circ-FoxO3 is essential for the self-connection of BMECs in mice, as demonstrated by the observation that the cell-cell junction collapses after circ-FoxO3 knockdown, while this collapse is ameliorated by circ-FoxO3 overexpression. These results indicate that upregulated circ-FoxO3 conferred a protective effect on BBB integrity. This was consistent with previous reports that some highly expressed circRNAs induced by external stimuli exhibit biological functions for self-protection.41,42 We believe that circ-FoxO3 could act as a therapeutic target for BBB-related disorders. Because of the dynamically balanced role of circRNAs in pathology development,43 we determined the time course of circ-FoxO3-mediated protection of the endothelial barrier, and found that it lasts from 3 to 9 h. We also found that autophagy lessens BBB impairment, partly by clearing the proteins aggregates and inhibiting the loss of Cldn5 and ZO-1 on the membrane. This could be explained by the reduction of aggregates or damaged mitochondria mediated by autophagy, which alleviates the oxidative damage to the membrane protein, promoting cell survival.15 This result is similar to the previous finding that cell death induced by Tau aggregation in Alzheimer's disease is associated with autophagy dysfunction and could be mitigated by enhancing autophagy.44
Previous studies have shown that non-coding RNA takes part in neurological processes via the modulation of autophagy,45,46 and indicate that autophagy might be the target of non-coding RNA in some physiological or pathological processes. Our in vivo and in vitro studies reveal that autophagy is required for circ-FoxO3 to maintain low permeability and protein homeostasis in the BBB models with I/R. Further results demonstrate that circ-FoxO3 promotes autophagy in microvascular endothelial cells after OGD/R.
Mechanistically, LC-MS/MS and pull-down assays demonstrate that circ-FoxO3 interacts with autophagy-related mTOR in BMECs after I/R, which provides a new insight into the formation of this complex between circ-FoxO3 and mTOR. Mounting evidence has suggested that circRNAs exert their biological properties via sequestration of miRNA or RBPs.4 Our results reveal that circ-FoxO3 interacts with mTOR and inhibits mTORC1 activity in OGD/R-treated BMECs, which promote autophagy activation. In addition, we find that circ-FoxO3 plays a role in the sequestration of E2F1, which has been also reported to function in regulating cardiac senescence.23 Transcription factor E2F1 is a regulatory factor that triggers mTORC1 activation.35 Although the mechanism of mTORC1 activation is not completely understood, the translocation of mTORC1 to lysosomes is recognized to be one of the key events. This process is dependent on the association of PFKFB3 which could be induced by E2F1. Remarkably, this is a complex gene activation process independent of the AMPK pathway.34 In addition, E2F1 regulates the localization of v-ATPase on lysosomes, which helps mTORC1 target lysosomes.47 Therefore, we speculate that the inhibition of mTORC1 might also be achieved when E2F1 is sequestrated by circ-FoxO3. In this study, pull-down assays and co-localization analysis clearly demonstrated the interaction between circ-FoxO3 and E2F1 in the cytosol, and a reduced E2F1 level in the nucleus, implying that E2F1 activity was inhibited. Furthermore, we showed that, after the E2F1 expression level was reduced, mTORC1 activity was suppressed, as demonstrated by the decreased levels of PFKFB3 and p-S6K1, resulting in autophagy activation. Although circ-FoxO3 is confirmed to interact with mTOR and E2F1 in this study, our findings do not rule out the possibility that other molecules that interact with circ-FoxO3 may be involved in autophagy activation.
In summary, circ-FoxO3 is upregulated and attenuates BBB damage by enhancing autophagy under I/R. Mechanistically, circ-FoxO3 interacts with mTOR and E2F1 to suppress mTORC1 activity, which promotes autophagy activity. This study provides a novel insight into circ-FoxO3 in attenuating BBB damage and reveals its novel function in regulating autophagy via mTORC1 inhibition during cerebral I/R. The autophagy-related circ-FoxO3 might serve as a potential therapeutic target for neurological disorders associated with BBB damage.
Materials and methods
Human brain tissues samples
Full ethical approval for this study was obtained from the Medical Ethics Committee of the First Affiliated Hospital of Jinan University (KY-2020-059). Samples of brain tissue were obtained from patients with HT (n = 17 with 7 females and 10 males, aged from 45 to 55 years). The control group of brain tissue was prepared from non-stroke patients with primary glioblastomas, which was far from the lesion site and had to be removed due to the requirement of the surgical approach (n = 15 with 7 females and 8 males, aged from 50 to 58 years).
A focal ischemic stroke model
All animal protocols were approved by the Institutional Animal Care and Use Committee of Jinan University (IACUC-20201028-03). Mice (C57BL/6J, male and female, 22–25 g) underwent surgery according to our previously described MCAO/R surgery.15 The mice underwent MCAO and reperfusion for 2 h, respectively. Mice in the sham-operated group were subjected to identical operations with the experimental group except for ligation of the arteries and insertion of the nylon monofilaments. For analysis of sex differences in the presentation of stroke, this study investigated the expression of circ-FoxO3 and the ultra-structure of BBB in mice (female, n = 5; male, n = 5) with MCAO/R. To explore the role of autophagy in BBB integrity, tail intravenous injection with autophagy enhancer (rapamycin [Rapa] 0.75 mg/kg, dissolved in saline containing 5% polyethylene glycol 400, and 5% Tween 80) or autophagy inhibitor (CQ, 5 mg/kg, dissolved in saline) was conducted daily in assigned mice for 1 week prior to MCAO/R.
Plasmid construction and RNA interference
circ-FoxO3 overexpression vectors were designed by Sangon Biotech (Shanghai, China). The full-length of circ-FoxO3 (hsa_circ_0006404 and mmu_circ_0002207) was considered as “intron” and inserted in EGFP. Subsequently, this sequence was enclosed into the plasmid (RSV-HIV LTR-RRE-hEF1a-MCS-Puro) provided by Sangon Biotech. Shcirc-FoxO3 sequence (shRNA1, 2, 3) was synthesized and ligated into plasmid (LTR-RRE-hU6-MCS-EGFP-Puro) provided by Sangon Biotech. The mock vectors served as control. These lentiviral vectors were transfected into HEK293T cells, and the viral supernatant was collected for the next study. The sequences of interference with circ-FoxO3 (siRNA1, 2, 3) designed for both bEnd.3 and HBMEC are listed in Table S1.
Microinjection of lentivirus
The microinjection was slightly modified in accordance with a previous description.48 In brief, shcirc-FoxO3-EGFP lentivirus (shcirc-FoxO3), normal control of shRNA lentivirus (shNC), circ-FoxO3-EGFP lentivirus (circ-FoxO3), and normal control circRNA lentivirus (circNC), were microinjected into the left lateral ventricle under the following microinjection coordinates: anteroposterior, –1.2 mm; lateral, 1.2 mm; and ventral, 2.1 mm. One week after microinjection, the mice underwent MCAO/R and divided into four groups: shNC + MCAO/R, shcirc-FoxO3 + MCAO/R, circNC + MCAO/R, and circ-FoxO3 + MCAO/R. The shRNA sequences (shRNA1, 2, 3) used in this study are listed in Table S1.
Cell culture and treatment
BMECs (bEnd.3 or HBMEC obtained from Bioleaf Biotech) were used to establish the in vitro model of BBB in this study. To generate an oxygen-glucose deprivation/reoxygenation (OGD/R) model, the confluent monolayer of bEnd.3 or HBMEC was exposed, in glucose-free medium, to low oxygen-containing gas (94% N2, 5% CO2, and 1% O2) for 3 h, and then the medium was replaced with fresh medium containing 10% fetal bovine serum (FBS) in a CO2 incubator (5% CO2 and 95% air) for an additional 3 h. For autophagy analysis, the monolayer of bEnd.3 or HBMEC was pretreated with 50 nmol/L Rapa or 30 μmol/L CQ in a medium containing 10% FBS for 2 h, respectively, and then incubated in the medium with OGD/R and combined with similar concentrations of Rapa or CQ, respectively. The circ-FoxO3 knockdown cells were generated by transfection with siRNA (or LV-shcirc-FoxO3), and the upregulated circ-FoxO3 was obtained by transfection with LV-circ-FoxO3. Autophagic flux was monitored in BMECs with circ-FoxO3 overexpression after transfection with Ad-mCherry-SQSTM1 (Beyotime Biotechnology, Shanghai, China).
Endothelial function assay
Paracellular permeability was measured to reflect the barrier property of BBB in vivo and in vitro. Evans blue extravasation was conducted in C57BL/6J mice to evaluate cerebrovascular integrity. In brief, Evans blue (2% in saline, 4 mL/kg) was injected into the lateral tail vein. After circulation for 10 min, the mice were anesthetized with isopentane and successively perfused through the left ventricle with ice-cold saline and 4% paraformaldehyde (PFA). Subsequently, the brains were harvested and dissected to image the extravasation of Evans blue. To qualify the extravasated Evans blue, the brain tissues with extravasated Evans blue were homogenized in 1,000 μL of PBS and then centrifuged for 30 min (15,000 × g) to gather the supernatant. Five hundred microliters of 50% trichloroacetic acid was added into the supernatant and placed at 4°C for 12 h. The mixture was centrifuged (15,000 × g) for 30 min at 4°C, and the supernatant was collected to be measured using a spectrophotometer (Thermo Fisher Scientific, 3020) at 610 nm. The paracellular permeability of the endothelial monolayer of bEnd.3 or HBMEC was assessed according to our previous study.15
A tube formation assay was applied to analyze endothelial cell function in this study. In brief, BMECs with circ-FoxO3 knockdown or upregulation were seeded in 48-well plates (1 × 105 cells per well) coated with 150 μL Matrigel (BD Biosciences, 356234). After 10 h incubation, the tubes were obtained using the EVOS FL Auto Imaging System (Invitrogen).
Fluorescence in situ hybridization
The circ-FoxO3 conjugated with CY5 used for in situ hybridization was designed and synthesized by Sangon Biotech according to a previous study.23 Tissues or the BMECs (bEnd.3 or HBMEC) were washed twice with cold 1× DEPC PBS, and then were fixed with 4% PFA for 20 min. Permeability was performed for the tissues or BMECs with 0.25% Triton in PBS for 15 min. The samples were incubated in hybridization solution (Thermo Fisher Scientific, AM8670) containing 50 nmol/L CY5-labeled circ-FoxO3 probes at 55°C for 3 h after prehybridization in hybridization solution for 1 h at 37°C. Subsequently, the samples were washed with 2× SSC (Sigma-Aldrich, S6639) at 42°C and incubated in blocking buffer (0.5% BSA in PBST) before incubation with anti-CD31 antibody (Abcam, ab24590), PDGFR-β (CST, 3169), GFAP (CST, 3670), anti-mTOR (CST, 2983), anti-E2F1 (Abcam, ab112580), anti-SQSTM1/p62 (Abcam, ab56416), Alex Fluro 488-conjugated goat anti-mouse or rabbit IgG (Jackson Laboratory, 115-095-003; CST, 4412), and DAPI. Finally, the samples were captured using a Leica TCS SPII 5 confocal microscope (Leica, Solms, Germany). The sequences of probes are listed in Figure S1B.
IF staining of and immunoblotting
The expression and localization of TJs and autophagy-related proteins were analyzed by IF staining. The protocol was performed with primary antibodies, such as Cldn5 (Invitrogen, 35-2500), ZO-1 (Invitrogen, 40-2200), LC3B (Sigma-Aldrich, L7543), LAMP1 (CST, 9091), and second antibody, including Alex Fluro 488-conjugated goat anti-mouse IgG (Jackson, 115-095-003) or Alex Fluro 647-conjugated goat anti-rabbit IgG (Jackson Laboratory, 111-605-003). Images were obtained using a Leica TCS SPII 5 confocal microscope (Leica). For immunoblotting, equal amounts of protein were separated by dodecyl sulfate-polyacrylamide gel electrophoresis and subsequently transferred to polyvinylidene fluoride membranes (Millipore, ISEQ00010). The membranes were incubated in TBST containing the primary antibody of Cldn5 (Invitrogen, 35-2500), ZO-1 (Invitrogen, 40-2200), Msfd2a (Abcam, ab117618), LC3B (Sigma-Aldrich, L7543), SQSTM1/p62 (Abcam, ab56416), E2F1 (Abcam, ab112580), p-p70S6K/p-S6K1 (Thr389, CST, 9234), p70S6K/S6K1 (CST, 2708), PFKFB3 (Proteintech, 13763-1-AP), and subsequently with horseradish peroxidase-conjugated secondary antibody. After washing, the immunoreactive bands were observed and captured using the Gel-Imaging System (Tanon 4600, Shanghai, China). The siE2F1 (Santa Cruz, sc-35247; Santa Cruz, sc-29297) was used to interfere with E2F1 in BMECs.
TEM
Autophagic vacuoles in brain tissues or BMECs were imaged by TEM as described previously.15 In brief, the samples were fixed with 2.5% glutaraldehyde and 1% osmium tetroxide in 0.2 mol/L sodium phosphate buffer for 12 h. After being dehydrated in graded ethanol (50%, 75%, 95%), the samples were cut into 70-nm-thick sections and stained with uranyl acetate and lead citrate. The autophagic vacuoles were imaged using a JEM-1400 electron microscope (JEM, Tokyo, Japan).
RNA treatment with RNase R and PCR
For samples requiring linear RNA depletion, total RNA (2 μg) was treated with or without 3 U/mg RNase R (Epicentre, RNR07250) at 37°C for 20 min. The RT-PCR was performed using FastStart Essential DNA Green Master (Roche Diagnostics, Risch-Rotkreuz, Switzerland) in an iCycler iQ5 Real-Time PCR Detection System (Bio-Rad). Relative expression of circ-FoxO3 or FoxO3 was normalized to GAPDH and performed using the comparative CT method (2−ΔΔCT). Primer sequences were designed to target (Table S2).
RIP
BMECs (bEnd.3 or HBMEC) with 3 × 107 cells were lysed in 500 μL co-IP buffer (Beyotime Biotechnology), and incubated with 5 μg of mTOR (CST, 2983) or E2F1 (Abcam, ab112580) at room temperature for 1.5 h. Subsequently, each sample was added to 100 μL magnetic beads and then incubated overnight on a rotating wheel at 4°C. After washing and centrifugation, the pellets were re-suspended in 0.5 mL TRI Reagent. The pull-down RNA was treated with or without RNase R. After being reverse-transcribed into cDNA, the level of circ-FoxO3 was analyzed by quantitative RT-PCR.
RNA pull-down assay
RNA pull-down assays were conducted as described previousy.24 In brief, BMECs (bEnd.3 or HBMEC) with 3 × 107 cells were lysed in 500 μL co-IP buffer (Beyotime Biotechnology). The lysates were incubated with biotinylated DNA oligo probes (3 μg) for 2 h before being mixed with 50 μL streptavidin C1 magnetic beads (Invitrogen, 65602) for another 3 h. After washing with co-IP buffer, the bound proteins in the beads were analyzed by LC-MS/MS and immunoblotting. The sequences of probes are listed in Figure S1B.
Statistical analysis
The results are presented as means ± standard error. All experiments were performed at least three times. The mean of the groups was compared using Student's t test (for two groups) or one-way ANOVA followed by Tukey's test (for three or more groups) with Prism 5 software (GraphPad software). Ninety-five percent confidence intervals of data analyses were acquired for in vivo and in vitro experiments. ∗p < 0.05 and ∗∗p < 0.01, respectively.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (grant nos. 82171344, 82171316, 81671167, and 81971121). We thank Niu He for the project management assistance.
Author contributions
Z.Y., A.X., and K.L. conceptualized the experiments. Z.Y., C.H., X.W., W.L., X.H., Y.L., J.Z., and Z.W. conducted the experiments and analyzed the data. Z.Y. wrote the manuscript. C.K.T. edited and proofread the manuscript. D.L. and C.K.T. provided relevant suggestions. Y.L. collected patient samples.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.11.004.
Contributor Information
Keshen Li, Email: likeshen1971@126.com.
Anding Xu, Email: tlil@jnu.edu.cn.
Supplemental information
References
- 1.Kaplan L., Chow B.W., Gu C. Neuronal regulation of the blood–brain barrier and neurovascular coupling. Nat. Rev. Neurosci. 2020;21:416–432. doi: 10.1038/s41583-020-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lublinsky S., Major S., Kola V., Horst V., Santos E., Platz J., Sakowitz O., Scheel M., Dohmen C., Graf R., et al. Early blood-brain barrier dysfunction predicts neurological outcome following aneurysmal subarachnoid hemorrhage. EBioMedicine. 2019;43:460–472. doi: 10.1016/j.ebiom.2019.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gokool A., Loy C.T., Halliday G.M., Voineagu I. Circular RNAs: the brain transcriptome comes full circle. Trends Neurosci. 2020;43:752–766. doi: 10.1016/j.tins.2020.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehta S.L., Dempsey R.J., Vemuganti R. Role of circular RNAs in brain development and CNS diseases. Prog. Neurobiol. 2020;186:101746. doi: 10.1016/j.pneurobio.2020.101746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venø M.T., Hansen T.B., Venø S.T., Clausen B.H., Grebing M., Finsen B., Holm I.E., Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dube U., Del-Aguila J.L., Li Z., Budde J.P., Jiang S., Hsu S., Ibanez L., Fernandez M.V., Farias F., Norton J., et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat. Neurosci. 2019;22:1903–1912. doi: 10.1038/s41593-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai Y., Yuan Z., Han B., Yang L., Chen X., Huang R., Wu F., Chao J., Liu P., Hu G., et al. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J. Neurosci. 2017;38:32–50. doi: 10.1523/JNEUROSCI.1348-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang R., Chen J., Xu B., Yang B., Fu J., Xiao S., Tan C., Chen H., Wang X. circ_2858 helps blood-brain barrier disruption by increasing VEGFA via sponging miR-93-5p during Escherichia coli meningitis. Mol. Ther-Nucl. Acids. 2020;4:708–721. doi: 10.1016/j.omtn.2020.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu D., Ho E.S., Mai H., Zang J., Liu Y., Li Y., Yang B., Ding Y., Tsang C.K., Xu A. Identification of blood circular RNAs as potential biomarkers for acute ischemic stroke. Front. Neurosci. 2020;14:81. doi: 10.3389/fnins.2020.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han B., Zhang Y., Zhang Y., Bai Y., Chen X., Huang R., Wu F., Leng S., Chao J., Zhang J.H., et al. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. 2018;14:1164–1184. doi: 10.1080/15548627.2018.1458173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gan X., Zhu H., Jiang X., Obiegbusi S.C., Yong M., Long X., Hu J. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol. Cancer. 2020;19:45. doi: 10.1186/s12943-020-01163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du W.W., Yang W., Li X., Awan F.M., Yang Z., Ling F., Juanjuan L., Li F., Peng C., Krylov S.N. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829–5842. doi: 10.1038/s41388-018-0369-y. [DOI] [PubMed] [Google Scholar]
- 14.Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z., Lin P., Chen B., Zhang X., Xiao W., Wu S., Huang C., Feng D., Zhang W., Zhang J. Autophagy alleviates hypoxia-induced blood-brain barrier injury via regulation of CLDN5 (claudin 5) Autophagy. 2021;17:3048–3067. doi: 10.1080/15548627.2020.1851897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Z., Huang C., Wu Y., Chen B., Zhang W., Zhang J. Autophagy protects the blood-brain barrier through regulating the dynamic of claudin-5 in short-term starvation. Front. Physiol. 2019;10:2. doi: 10.3389/fphys.2019.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang G., Ling Y., Mehrpour M., Saw P.E., Liu Z., Tan W., Tian Z., Zhong W., Lin W., Luo Q., et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol. Cancer. 2020;19:65. doi: 10.1186/s12943-020-01152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin X., Gao J., Zheng R., Yu M., Ren Y., Yan T., Huang Y., Li Y. Antagonizing circRNA_002581–miR-122–CPEB1 axis alleviates NASH through restoring PTEN–AMPK–mTOR pathway regulated autophagy. Cell Death Dis. 2020;11:123. doi: 10.1038/s41419-020-2293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao L., Dou Z.C., Ren W.H., Li S.M., Liang X., Zhi K.Q. CircCDR1as upregulates autophagy under hypoxia to promote tumor cell survival via AKT/ERK/mTOR signaling pathways in oral squamous cell carcinomas. Cell Death Dis. 2019;10:745. doi: 10.1038/s41419-019-1971-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R., et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 21.Xing Y., Zha W.J., Li X.M., Li H., Gao F., Ye T., Du W.Q., Liu Y.C. Circular RNA circ-Foxo3 inhibits esophageal squamous cell cancer progression via the miR-23a/PTEN axis. J. Cell. Biochem. 2020;121:2595–2605. doi: 10.1002/jcb.29481. [DOI] [PubMed] [Google Scholar]
- 22.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du W.W., Yang W., Chen Y., Wu Z.K., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 24.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S., Liao K., Miao Z., Wang Q., Miao Y., Guo Z., Qiu Y., Chen B., Ren L., Wei Z., et al. CircFOXO3 promotes glioblastoma progression by acting as a competing endogenous RNA for NFAT5. Neuro-oncology. 2019;21:1284–1296. doi: 10.1093/neuonc/noz128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sohrabji F., Park M.J., Mahnke A.H. Sex differences in stroke therapies. J. Neurosci. Res. 2017;95:681–691. doi: 10.1002/jnr.23855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carcel C., Wang X., Sandset E.C., Delcourt C., Arima H., Lindley R., Hackett M.L., Lavados P., Robinson T.G., Muñoz Venturelli P., et al. Sex differences in treatment and outcome after stroke: pooled analysis including 19,000 participants. Neurology. 2019;93:e2170–e2180. doi: 10.1212/WNL.0000000000008615. [DOI] [PubMed] [Google Scholar]
- 28.Lesnefsky E.J., Chen Q., Tandler B., Hoppel C.L. Mitochondrial dysfunction and myocardial ischemia-reperfusion: implications for novel therapies. Annu. Rev. Pharmacol. Toxicol. 2017;57:535–565. doi: 10.1146/annurev-pharmtox-010715-103335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenenbaum D., Maintainer B. 2021. KEGGREST: Client-Side REST Access to the Kyoto Encyclopedia of Genes and Genomes (KEGG). R Package Version 1.32.0. [Google Scholar]
- 30.Aylett C.H., Sauer E., Imseng S., Boehringer D., Hall M.N., Ban N., Maier T. Architecture of human mTOR complex 1. Science. 2016;351:48–52. doi: 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- 31.Odle R.I., Walker S.A., Oxley D., Kidger A.M., Balmanno K., Gilley R., Okkenhaug H., Florey O., Ktistakis N.T., Cook S.J. An mTORC1-to-CDK1 Switch maintains autophagy suppression during mitosis. Mol. Cell. 2020;77:228–240.e7. doi: 10.1016/j.molcel.2019.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Csibi A., Lee G., Yoon S.O., Tong H., Ilter D., Elia I., Fendt S.M., Roberts T.M., Blenis J. The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B-dependent control of c-Myc translation. Curr. Biol. 2014;24:2274–2280. doi: 10.1016/j.cub.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Almacellas E., Pelletier J., Manzano A., Gentilella A., Ambrosio S., Mauvezin C., Tauler A. Phosphofructokinases axis controls glucose-dependent mTORC1 activation driven by E2F1. Iscience. 2019;20:434–448. doi: 10.1016/j.isci.2019.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Real S., Meo-Evoli N., Espada L., Tauler A. E2F1 regulates cellular growth by mTORC1 signaling. PLoS One. 2011;6:e16163. doi: 10.1371/journal.pone.0016163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nation D.A., Sweeney M.D., Montagne A., Sagare A.P., D’Orazio L.M., Pachicano M., Sepehrband F., Nelson A.R., Buennagel D.P., Harrington M.G., et al. Blood–brain barrier breakdown is an early biomarker of human cognitive dysfunction. Nat. Med. 2019;25:270–276. doi: 10.1038/s41591-018-0297-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan J., Qu M., Li Y., Wang L., Zhang L., Wang Y., Tang Y., Tian H.L., Zhang Z., Yang G.Y. MicroRNA-126-3p/-5p overexpression attenuates blood-brain barrier disruption in a mouse model of middle cerebral artery occlusion. Stroke. 2020;51:619–627. doi: 10.1161/STROKEAHA.119.027531. [DOI] [PubMed] [Google Scholar]
- 38.Horos R., Büscher M., Kleinendorst R., Alleaume A.M., Tarafder A.K., Schwarzl T., et al. The small non-coding vault RNA1-1 acts as a riboregulator of autophagy. Cell. 2019;176:1054–1067.e12. doi: 10.1016/j.cell.2019.01.030. [DOI] [PubMed] [Google Scholar]
- 39.Xu Y., Leng K., Yao Y., Kang P., Liao G., Han Y., Shi G., Ji D., Huang P., Zheng W., et al. A novel circular RNA, circ-CCAC1, contributes to CCA progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology. 2021;73:1419–1435. doi: 10.1002/hep.31493. [DOI] [PubMed] [Google Scholar]
- 40.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A., et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boeckel J.-N., Jaé N., Heumüller A.W., Chen W., Boon R.A., Stellos K., Zeiher A.M., John D., Uchida S., Dimmeler S. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ. Res. 2015;117:884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- 42.Yang Z.G., Awan F.M., Du W.W., Zeng Y., Lyu J., Wu D., Gupta S., Yang W., Yang B.B. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol. Ther. 2017;25:2062–2074. doi: 10.1016/j.ymthe.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall I.F., Climent M., Quintavalle M., Farina F.M., Schorn T., Zani S., Carullo P., Kunderfranco P., Civilini E., Condorelli G., Elia L. Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ. Res. 2018;124:498–510. doi: 10.1161/CIRCRESAHA.118.314240. [DOI] [PubMed] [Google Scholar]
- 44.Silva J.M., Rodrigues S., Sampaio-Marques B., Gomes P., Neves-Carvalho A., Dioli C., Soares-Cunha C., Mazuik B.F., Takashima A., Ludovico P., et al. Dysregulation of autophagy and stress granule-related proteins in stress-driven Tau pathology. Cell Death Differ. 2019;26:1411–1427. doi: 10.1038/s41418-018-0217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X., Mao R., Su W., Yang X., Geng Q., Guo C., Wang Z., Wang J., Kresty L.A., Beer D.G., et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy. 2020;16:659–671. doi: 10.1080/15548627.2019.1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barangi S., Hayes A.W., Reiter R., Karimi G. The therapeutic role of long non-coding RNAs in human diseases: a focus on the recent insights into autophagy. Pharmacol. Res. 2019;142:22–29. doi: 10.1016/j.phrs.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 47.Meo-Evoli N., Almacellas E., Massucci F.A., Gentilella A., Ambrosio S., Kozma S.C., Thomas G., Tauler A. V-ATPase: a master effector of E2F1-mediated lysosomal trafficking, mTORC1 activation and autophagy. Oncotarget. 2015;6:28057. doi: 10.18632/oncotarget.4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rashnonejad A., Chermahini G.A., Gündüz C., Onay H., Aykut A., Durmaz B., Baka M., Su Q., Gao G., Zknay F. Fetal gene therapy using single injection of recombinant AAV9 rescued SMA phenotype in mouse. Mol. Ther. 2019;27:2123–2133. doi: 10.1016/j.ymthe.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.