Abstract
Lymphangiogenesis, the generation of new lymphatic vessels from existing ones, results from the dynamic interactions of lymphatic endothelial cells and the tumor microenvironment (TME). It is well known that lymphangiogenesis occurs during the initial stage of metastasis in various types of malignant tumors. However, it is currently not used as a biomarker partially because gold standard method to quantify it is labor and cost intensive. We hypothesized that the quantity of intratumoral lymphatic endothelial cells (iLECs) in the TME is an indicator of lymphangiogenesis and a predictor of metastatic potential and overall survival in breast cancer. We analyzed a total of 4145 breast cancer patients from the Cancer Genome Atlas (TCGA) and GSE96058 by quantifying their iLECs using the xCell algorithm, and correlated these scores with patient survival, tumor grade, and cancer stage. We also assessed various pro- and anti-cancer gene sets for each tumor to characterize tumor behavior and aggressiveness. As we expected, high-iLEC breast cancer demonstrated enriched lymphoangiogenesis and angiogenesis gene sets and was associated with increased expressions of related genes. Also enriched were inflammatory response and immune response-related gene sets; IL2/STAT5 pathway, IL6/JAK/STAT3 pathway, TNFα pathway, allograft rejection, and complement as well as cancer stemness related gene sets like Notch signaling, Hedgehog signaling, epithelial mesenchymal transition, and Wnt beta-catenin signaling. Tumors with high-iLEC showed higher proportions of stromal cells and fewer anti-cancer immune cells. On the other hand, iLEC score did not correlate with patient survival or lymph node metastasis. Surprisingly, breast cancers with fewer iLECs demonstrated enriched E2F Targets, G2M Checkpoint, MYC Targets v1, and MTORC1 signaling which are cancer cell proliferation-related gene sets and exhibited an abundance of pro-cancer immune cells. The amount of iLEC correlated inversely with Ki67 expression and histological grade, which is in agreement that low-iLEC breast cancer was associated with enhanced cancer cell proliferation. In conclusion, while iLECs can be used as a surrogate for lymphangiogenesis in breast cancer, low-iLEC tumors also exhibit features which correspond to aggressive tumor biology, which may explain why the amount of iLECs was not associated with patient survival in our cohorts.
Keywords: Angiogenesis, GSEA, GSVA, lymphangiogenesis, lymph endothelial cell, tumor microenvironment, xCell
Introduction
Lymphangiogenesis, the generation of new lymphatic vessels from existing lymphatics, is a key step in breast cancer metastasis [1-3]. The complex cytokine-mediated interactions between malignant cells and the surrounding non-cancerous cells (such as stromal and immune cells) ultimately determine the rate of tumor growth and metastasis, or lack thereof. Therefore, it is essential to assess cancer biology in the context of the entire tumor microenvironment (TME), rather than focusing on single interactions [4]. Lymphatic vessels were historically believed to play only a passive role in cancer metastasis, but experimental and clinicopathological research published in the previous decade has shown that lymphangiogenic growth factors, often derived from tumor cells, facilitate metastasis by promoting the formation of new lymphatics and enlarging existing lymphatic vessels, thus enabling the entry of tumor cells into the lymphatic vasculature [5-7]. The clinical relevance of this process has long been recognized, as cancer metastasis to regional and sentinel lymph nodes is a poor prognostic factor in human cancer [8-10].
Lymphatic endothelial cells (LECs) are the primary component of lymphatic vessels and play an active role in tumor metastasis by proliferating and migrating in response to stimulation by cancer cells. There are multiple documented examples of the bidirectional interaction between LECs and cancer cells. For example, our group has previously reported that cancer cells express sphingosine kinase 1 that generate sphingosine-1-phosphate (S1P), which is released out to the extracellular space [11,12] and bind to S1P receptor 1 (S1P1) and 3 (S1P3) on LECs and blood endothelial cells that induce lymphangiogenesis and angiogenesis [13-20].
It has long been known that lymphangiogenesis is induced to resolve excessive inflammation, and since chronic inflammation is associated with cancer progression, lymphangiogenesis may underlie this mechanism [4]. Inflammatory cytokines such as IL-1β and TNF-α have also been reported to promote tumor lymphangiogenesis and lymph node metastasis in a dependent manner on inflammatory cells such as infiltrating M2 macrophages [21,22]. In addition to recruiting leukocytes, IL-1β secreted by tumor-associated macrophages (TAMs) also directly stimulates proliferation and migration of LECs [23]. IL-6 increases lymphangiogenesis in oral squamous cell carcinoma by inducing the production of VEGF-C [24].
Given the known association between lymphangiogenesis and cancer metastasis, logic would dictate that an increased number of infiltrating LECs should predict metastatic potential and an unfavorable clinical outcome. Indeed, in several studies, tumor lymphangiogenesis has been shown to increase the risk of future metastasis and affect survival in breast cancer patients [2,25,26]. However, the literature remains controversial, and several other studies have failed to find an association between lymphatic vessel abundance and malignant clinicopathological factors [27-29]. This may be due in part to the high variability and lack of standardization in the methodologies used to quantify LECs. For example, the microscopic evaluation of lymphatic vessels by counting them in tissue sections is by its nature qualitative and subjective, in contrast with an assay which can be objectively quantified [9]. Additionally, there are several lymphatic vessel markers used in immunostaining, each of which may bind to LECs at different stages of their growth and development, and sometimes even to vascular endothelium and myoepithelial cells [30]. Finally, lymphatic vessel density estimations are subject to variability based on biopsy technique, as different samples will over- or underestimate the density depending on whether the sample is taken from the center or periphery of the tumor. Here, we utilize a bulk tumor transcriptome and a previously validated bioinformatic analysis technique to digitally dissect the tumor microenvironment and simultaneously assess several TME interactions that potentially contribute to the clinical outcome. In particular, we hypothesized that the quantity of intratumoral lymphatic endothelial cells (iLECs) in the TME is an indicator of lymphangiogenesis, and thus may predict patient prognosis and metastatic potential.
Materials and methods
Breast cancer patient cohorts
Two publicly available datasets were collected from online sources and analyzed in this study. RNA-seq data and clinical information of 1076 primary breast cancer patients from the Cancer Genome Atlas (TCGA) were evaluated as an exploring cohort [31], then 3036 primary breast cancer patients from GSE96058 were analyzed as a validation cohort [32,33]. A total of 4145 patients were included in the analysis. TCGA data was collected from cBiopotal, and GSE96058 was collected from the Gene Expression Omnibus (GEO) database, as described in our previous paper [34-37]. All RNA seq data utilized in this study were normalized and annotated to gene symbols from transcriptomics by the time of initial publication and followed by log2 conversion [31,33]. All patient information included in both cohorts was de-identified; therefore, Institutional Review Board approval was not required for this study.
Intratumoral lymph endothelial cell score
Intra-tumoral lymphocyte content as well as other intratumor immune and stromal cell fractions was calculated using the xCell web tool (https://xcell.ucsf.edu/), as we have previously reported [38-41]. xCell is an algorithm for enumerating cell subsets from the transcriptome reported by Aran et al. in 2017 [42]. It integrates the deconvolution approaches used in CIBERSORT [43], which is the most common method to dissect the tumor microenvironment using gene expression profiles, with the gene signature-based comparison method from Gene set enrichment analysis (GSEA) [44]. This algorithm estimates cell type fractions by comparing 489 gene signatures corresponding to 64 cell types, including adaptive and innate immune cells, hematopoietic progenitor cells, epithelial cells, and extracellular matrix cells, with the input bulk tumor gene expression data set.
Gene set enrichment analysis
GSEA java application v4.1.0 was used for function analysis, which is a freely available tool provided by Broad Institute [44]. GSEA is the most common method for examining differences in pathways due to differences in gene expression between two groups. The method ranks the expression of gene members of a predefined gene set to determine whether these genes are randomly distributed within a cohort or biased towards one of the two groups. The enrichment score reflects the degree to which the gene set is biased towards one group. This means that the group expresses the biology represented by the ‘enriched’ gene set. We divided the cohort into high and low median iLEC groups and used the Hallmark and PID gene sets from the MSigDB collection [45,46]. To compare results across gene sets, we used a normalized enrichment score (NES) adjusted for the correlation between the gene set and the expression dataset. Larger NES values indicate whether the pathway represented by each gene set is over- or under-enriched in each phenotype. Because we evaluated more than 30 gene sets, we interpreted False Discovery Rate (FDR) q-values of 0.25 or less as statistically significant, based on Broad Institute recommendations, as previously described [47-50].
Cancer immunity and mutation score estimation
Cytolytic activity (CYT) was measured using the geometric mean of granzyme A and Perforin 1 expression levels [51,52]. Interferon (IFN)-Response, tumor infiltrating lymphocyte (TIL) Regulation Fraction, Lymphocyte Infiltration Signature Score, Leukocyte Fraction, TCR richness, and BCR richness were all determined using the methodology outlined by Thorsson et al. [53].
Others
Analyses were conducted using R (version 4.0.1), Bioconductor (version 3.13), or Microsoft Excel (version 16 for Windows). The following packages were used in this study: survival 3.2-11, survAUC 1.0-5, SummarizedExperiment 1.22.0, GenomicRanges 1.44.0, GenomeInfoDb 1.28.2, S4Vectors 0.30.0, MatrixGenerics 1.4.3, Biobase 2.52.0, greyzoneSurv 1.0, RcmdrPlugin.EZR 1.54, RcmdrMisc 2.7-1. ggplot2 3.3.5, backports 1.2.1, tidyverse 1.3.1, and GEOquery 2.60.0. In all analyses in which tumors were categorized into two groups, high- or low-iLEC, the median was used as the cut-off point. All P-values were calculated by two-sided statistical tests and the cut-off for statistical significance was set at 0.05. Overall survival (OS) was defined as the length of time from completion of treatment to death. Disease-specific survival (DSS) was also defined as the length of time from completion of treatment to death but excluding patients who died from causes other than breast cancer. Disease-free survival (DFS) was defined as the length of time from completion of treatment to recurrence. For survival analysis, the Kaplan-Meier method with a log rank test was used. Statistical comparisons between groups were made using Mann-Whitney U test for two groups or Kruskal-Wallis test for multiple groups. Box-and-whiskers plots were used to display the median values and the interquartile range.
Results
Infiltration of lymphatic endothelial cells is associated with lymphangiogenesis and angiogenesis
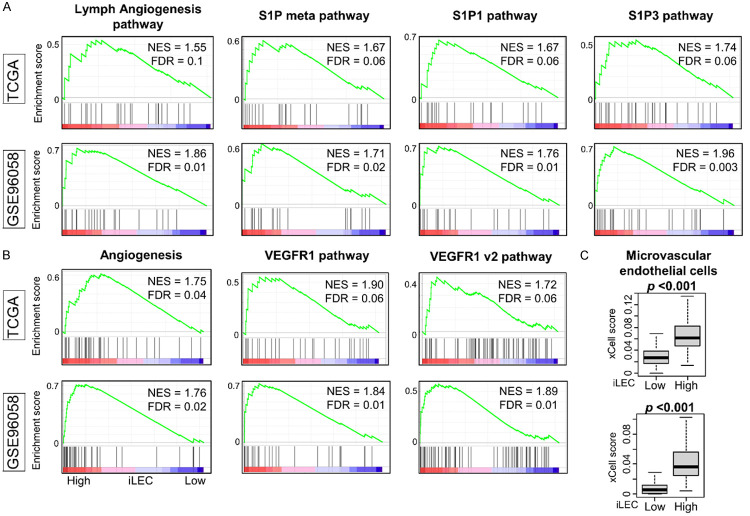
First, we investigated whether xCell-defined intratumoral lymphatic endothelial cells (iLEC) are an accurate measure of lymphangiogenesis. iLEC-rich breast cancer demonstrated enriched lymphangiogenesis-related gene sets: S1P, S1P1, and S1P3 pathways (Figure 1A, all FDR<0.25). These tumors also exhibited enriched angiogenesis-related gene sets: VEGF1 v1 and v2 pathway (Figure 1B, all FDR<0.25), in gene set enrichment analysis (GSEA) of Hallmark collection and The Pathway Interaction Database (PID) gene sets. These gene sets were all expressed to a significantly greater degree in high-iLEC breast cancers, indicating that the high-iLEC group exhibited lymphangiogenesis and angiogenesis biology. Furthermore, breast cancers with high iLEC scores were associated with high numbers of microvascular endothelial cells (Figure 1C, P<0.001). These results demonstrate that iLEC quantity directly correlates with lymphangiogenesis and angiogenesis in the TME.
Figure 1.
Gene set enrichment analysis (GSEA) of intratumor lymph endothermal cell (iLEC) high breast cancer and the relation with microvascular endothelial cells in the TCGA and GSE96058 cohorts. (A) Lymphangiogenesis related gene sets and (B) Angiogenesis rerated gene sets from Hallmark and PID gene sets that significantly enriched to high-iLEC breast cancer in both cohorts are shown. All patients were divided into two groups relative to the median iLEC. (C) Boxplots of microvascular endothelial cells in the TCGA cohort. Mann-Whitney U test was used to determine statistical significance.
The quantity of intratumoral lymphatic endothelial cells (iLEC) does not affect prognosis and does not necessarily predict lymph node metastasis
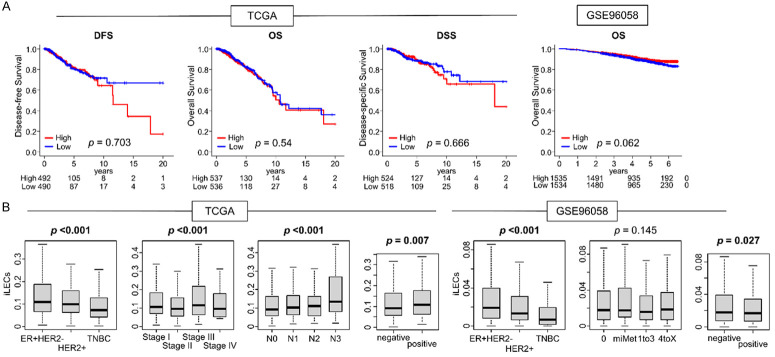
Based on previous reports that lymphangiogenesis and angiogenesis are associated with a negative impact on survival, we next investigated the impact of iLECs on survival in both cohorts. Contrary to our hypothesis, there was no significant difference between high and low-iLEC breast cancer patients with regards to disease-free survival (DFS), disease-specific survival (DSS), and overall survival (OS) within TCGA cohort, nor the OS within the GSE96058 cohort (Figure 2A). Additionally, we investigated the theory that a higher quantity of iLECs within a tumor may predict lymph node metastasis and a higher cancer stage. A higher iLEC score correlated with a higher N category in TCGA (Figure 2B, left P<0.01); however, this was not validated in GSE96058 (Figure 2B, right P<0.01). iLEC scores were significantly higher in the estrogen receptor (ER)-positive/epidermal growth factor receptor 2 (HER2)-negative subtype, while HER2-positive tumors had intermediate scores, and the iLEC score was consistently lowest in triple negative (TNBC) within both cohorts (Figure 2B, both P<0.01). These results were somewhat unexpected given that TNBCs are generally associated with more aggressive behavior and a poorer prognosis, and we had hypothesized that tumors with a higher iLEC score would demonstrate more aggressive features.
Figure 2.
Survival and clinical relevance between iLEC and breast cancer patients in both TCGA and GSE96058 cohort. A. Kaplan-Meier survival curves of the DFS, DSS and OS of iLEC High and Low patients of whole breast cancer of the TCGA cohort, and OS of the GSE96058 cohort. High groups are indicated by red lines, low groups by blue lines. Median was defined as a cut-off, and two groups were compared using a log-rank test to calculate P-values. B. Boxplots of amount of iLEC compared by breast cancer subtype; ER positive/HER2-negative, HER2 positive, and TNBC, pathological stage, and lymph node status in the TCGA and GSE96058 breast cancer cohort. Kruskal-Wallis and Mann-Whitney U tests were used to calculate P-values. DSS; disease-specific survival, ER; estrogen receptor, HER2; human epidermal growth factor receptor 2, OS; overall survival, PR; progesterone receptor, TNBC; triple negative breast cancer.
High-iLEC breast cancer demonstrated enriched cancer stemness related gene sets and a higher proportion of stromal elements compared to low-iLEC tumors
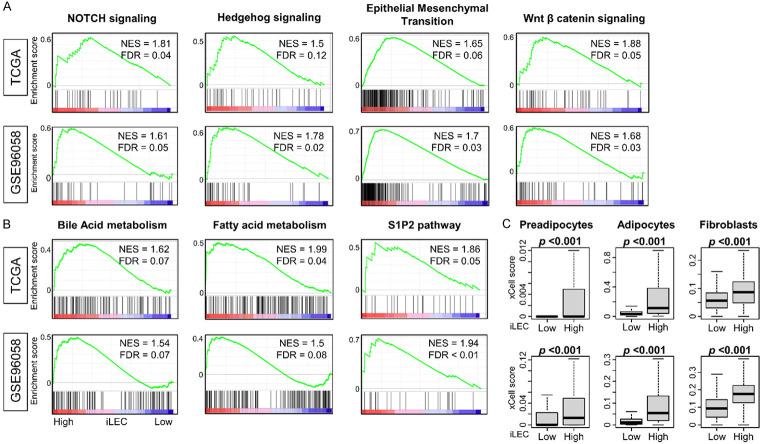
Given the established association between lymphangiogenesis and metastatic potential, we investigated which biological features, represented in gene sets, are “enriched”, i.e., biased towards high iLEC breast cancers in order to further investigate the lack of correlation between a high-iLEC score and overall survival. As we had hypothesized, we found that high-iLEC breast cancer indeed demonstrated enriched cancer stemness related gene sets such as Notch signaling, Hedgehog signaling, epithelial mesenchymal transition, and Wnt beta-catenin signaling in Hallmark gene set of GSEA (Figure 3A, all FDR<0.25). Furthermore, bile acid and fatty acid metabolism and S1P2 pathway were also enriched (Figure 3B, all FDR<0.25). Finally, the intra-tumoral proportion of stromal cells (preadipocytes, adipocytes, and fibroblasts), was consistently higher in high-iLEC breast cancer across both cohorts. Given that increased stromal components within a tumor are known to impede host immune responses and contribute to tumor growth, these findings are consistent with aggressive features in high-iLEC tumors.
Figure 3.
GSEA of intratumor lymph endothermal cell (iLEC) high breast cancer and the relation with stromal cells in the TCGA and GSE96058 cohorts. (A) Cancer stemness related gene set or (B) metabolism and S1P2 pathway from Hallmark and PID gene sets that significantly enriched to high iLEC breast cancer in both cohorts are shown. All patients were divided into two groups relative to the median iLEC. (C) Boxplots of stromal cells in the TCGA and GSE96058 cohort. Mann-Whitney U test was used to determine statistical significance.
High iLEC breast cancer enriched several immune response-related gene sets and was associated with a higher overall cytolytic activity
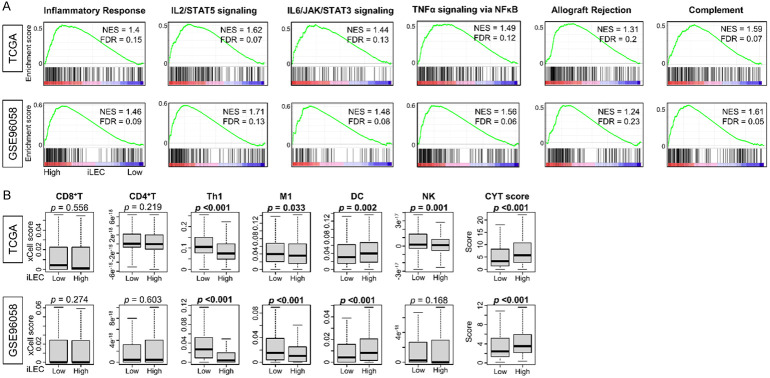
We wondered why high iLEC scores were not associated with worse clinical outcomes, considering that they accurately predicted both increased lymphangiogenesis and metastatic potential, and speculated that there may be other mechanisms at play. For example, we previously reported that breast cancers with high mutation rates do demonstrate aggressive features, but also stimulate a more pronounced host response, which offsets the aggressive phenotype and ultimately results in similar survival rates compared to low mutation breast cancers [54]. Indeed, in our analysis we found that high-iLEC breast cancer enriched various immune response-related gene sets: inflammatory response, IL2/STAT5 pathway, IL6/JAK/STAT3 pathway, TNFα pathway, allograft rejection, and complement compared with low-iLEC breast cancer consistently in both TCGA and GSE96058 cohorts (Figure 4A, all FDR<0.25). Cytolytic activity (CYT) score, which reflects overall anti-cancer immunity, was significantly higher in high-iLEC compared with low-iLEC breast cancer across both cohorts (Figure 4B, P<0.001). However, infiltration of anti-cancer immune cells such as CD8+ T-cell, CD4+ T-cell, M1 Macrophage (M1), and natural killer T-cell (NK) was not significantly elevated in high-iLEC compared with low-iLEC breast cancer (Figure 4B). Dendritic cells (DC) were more abundant in high-iLEC breast cancers, while type 1 T helper cells (Th1) were conversely more abundant in low iLEC breast cancer consistently in both cohorts (Figure 4B, P<0.001). Therefore, while a high-iLEC score did not indicate an increased infiltration of the tumor by specific immune cells, the associated increased CYT score demonstrates an overall stronger host immune response against high-iLEC tumors.
Figure 4.
Relationship between high iLEC breast cancer and cancer immunity. A. Immne-related gene sets from Hallmark gene set that significantly enriched to high iLEC breast cancer in both cohorts are shown. All patients were divided into two groups relative to the median iLEC. B. Boxplots of anti-cancer immune cells and Cytolytic activity (CYT) score in the TCGA and GSE96058 cohort was shown. Mann-Whitney U test was used to determine statistical significance.
Low iLEC breast cancer was strongly associated with cancer cell proliferation
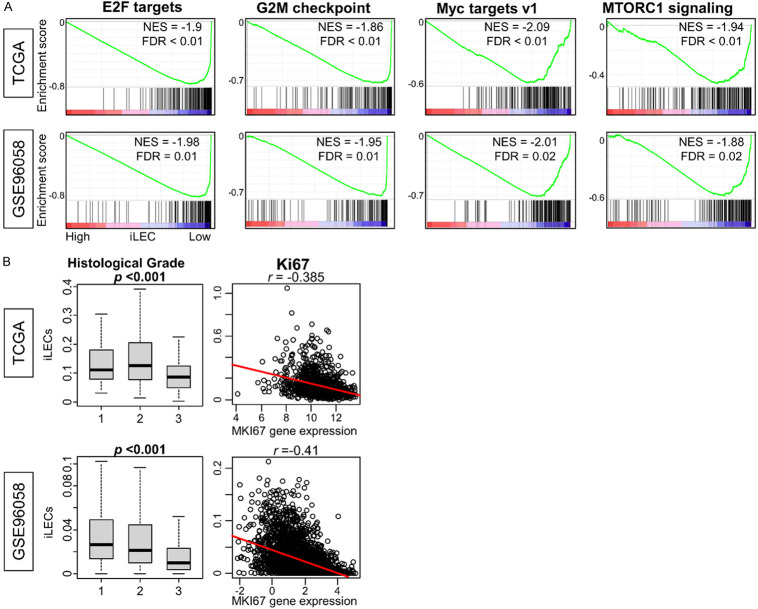
In light of the lack of correlation between iLEC score and clinical outcome, we next investigated the biology of this group by focusing on a gene set that is biased towards low iLEC breast cancer in GSEA. Surprisingly, we found that low-iLEC tumors were enriched in cell proliferation-related gene sets, including E2F Targets, G2M Checkpoint, MYC Targets v1, and MTORC1 signaling compared with high-iLEC breast cancer (Figure 5A, all FDR<0.25). These were the only gene sets enriched to low-iLEC breast cancer, unlike high-iLEC tumors which enriched a diverse group of gene sets. Furthermore, low-iLEC breast cancer was associated with higher histological grade, and the quantity of iLECs was consistently inversely correlated with the expression of MKI67 across both cohorts (Figure 5B, P<0.001 and r<-0.38). In other words, low-iLEC breast cancer was associated with aggressive and highly proliferative biology.
Figure 5.
Gene sets enriched in low iLEC breast cancer and their association with tumor grade. A. Cancer proreferation gene sets from Hallmark gene set that significantly enriched to high iLEC breast cancer in both cohorts are shown. All patients were divided into two groups relative to the median iLEC. B. Box plot of histological grade vs. total amount of iLECs and correlation with MKI67. Kruskal-Wallis test was used to determine statistical significance. Correlation coefficients were derived from the Spearman rank test.
Breast cancers with low iLEC content were associated with high mutation frequency, intratumor heterogeneity, with tumor antigenicity and also with high infiltration of pro-cancer immune cells
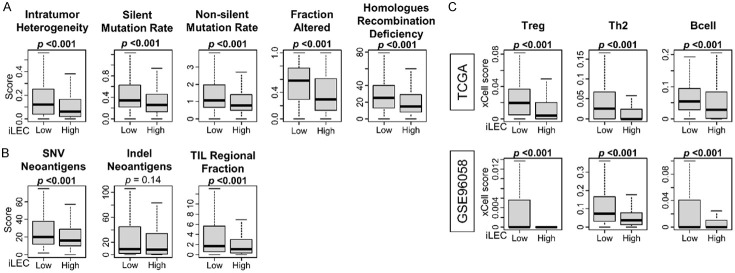
Since highly proliferative tumors are characterized by a high mutation rate, which in turn generates strong host immunogenicity [54-56], we hypothesized that the same trend would be observed in breast cancers with low iLEC content. As expected, low-iLEC breast cancer was significantly associated with high intratumor heterogeneity, silent and non-silent mutation rate, Fraction Altered, and Homologues Recombination Deficiency (HRD) based on calculations by Torsson et al. in the TCGA cohort (Figure 6A, all P<0.001). At the same time, low-iLEC breast cancer was associated with high single nucleotide variant (SNV) neoantigen, indel neoantigen, and tumor-infiltrating lymphocytes (TIL) regional infiltration (Figure 6B, all P<0.01). We further found that low-iLEC cancer was more frequently infiltrated by pro-cancer immune cells, such as regulatory T-cells (Treg), type 2 T helper cells (Th2), and B cells (Figure 6C, all P<0.001).
Figure 6.
Association between amount of iLEC in breast cancer and intratumor heterogeneity and mutation rate, and also the immunogenicity and immune cell invasion. Box plots show (A) intratumor heterogeneity, silent and nonsilent mutation rate, fraction altered, and homologous recombination deficiency, and (B) single nucleotide variant (SNV) and Indel neoantigens and tumor-infiltrating lymphocytes (TIL) regional fraction based on scoring by Thorrson et al. using the TCGA cohort. (C) Boxplot shows difference between high and low iLEC breast cancer in the amount of pro-cancer related immune cell infiltration in both TCGA and GSE96058 cohort. Mann-Whitney U test was used to determine statistical significance.
Discussion
In this study, we used high-powered breast cancer patient cohorts that are associated with RNA-sequence and clinical data and demonstrated that the quantity of intertumoral lymphatic endothelial cells (iLECs) is an indicator of lymphangiogenesis and characterized the differences in cancer biology and host immune response between high- and low-iLEC tumors. First, we demonstrated that the quantity of iLECs positively correlated with lymphangiogenesis-related gene sets and angiogenesis-related gene sets across two independent large breast cancer cohorts, confirming that this score accurately indicates lymphangiogenesis. Microvascular endothelial cells that take part in angiogenesis [57] were also predominant in tumors with high iLEC. Surprisingly, there was no difference in survival between high- and low-iLEC breast cancer, nor was there a statistically significant association between iLEC score and lymph node metastasis across both cohorts. Since lymphangiogenesis is a known prerequisite for cancer progression, we then sought to investigate other factors which affect tumor growth and metastasis; namely, cancer biology and the host immune response. Breast cancers with high iLEC scores enriched cancer stemness-related gene sets and metabolic pathways such as bile acid metabolism, fatty acid metabolism, and S1P2 pathway that we have previously shown to play a key role in fat metabolism [58-61]. Stromal cells, such as preadipocytes, adipocytes, and fibroblasts, were also more prominent in the high-iLEC group, a feature generally associated with tumor growth. We also found that immune-related gene sets were enriched and overall cytolytic activity was elevated in the high-iLEC group of both cohorts. These results suggest that while high iLEC breast cancer is actually associated with genes that predispose to metastasis and tumor growth, these features are offset by a more robust host immune response. Unexpectedly, several cell proliferation-related gene sets were significantly enriched in low-iLEC breast cancers, which demonstrated high Ki67 expression, advanced histological grade, and aggressive cancer subtypes. Low-iLEC breast cancer was also found to be strongly associated with intratumor heterogeneity, silent and non-silent mutation rates, fraction altered, and HRD, which all indicate aggressive cancer biology. This is offset by an IFN-γ response, an increased proportion of TIL, and pro-cancer immune cells such as Th2 cells, Treg cells, and B cells.
Cancer stemness-related gene sets such as Notch signaling, Hedgehog signaling, epithelial-mesenchymal transition (EMT), and Wnt beta-catenin signaling, were enriched in high-iLEC breast cancer. LECs not only promote lymphangiogenesis and act as a conduit of cancer cells but also confer chemotaxis on tumors, involving several chemokines and their receptors. Expression of CCL21 in LECs can promote cancer cell invasion into lymphatic vessels through a CCR7-dependent mechanism [62,63]. Furthermore, LECs have been found to stimulate cancer stem cells along the CXCL-12-CXCL4 axis and promote remodeling that promotes cancer cell survival in perivascular and metastatic regions, contributing to the formation of the so-called “pre-metastatic niche” [64]. LECs are reported to be involved in EMT by inducing migration of CCR7-expressing cancer cells into the pre-metastatic niche and by promoting invasion of tumor cells into lymphatic vessels in several ways [63,65,66].
Breast cancers with high iLECs were found to have a high proportion of stromal cells such as adipocytes and fibroblasts, which is associated with unfavorable chronic inflammation [67]. In fact, the immune-related gene set inducing chronic inflammation, such as the IL-6 and TNFα pathways, was enriched in breast cancer with high iLEC, which is consistent with previous reports. While it is related todetrimental inflammation, LECs upregulate CCL21 expression during inflammation and promote dendritic cells (DCs) entry into lymphatic vessels by interaction with CCR7 expressed on the surface of DC [68]. In the current study, breast cancer with high iLEC had higher DC infiltration consistently in both cohorts. Infiltrated DCs in lymphatic vessels promote T cell priming and induce antigen-specific immune responses. On the other hand, increased lymphangiogenesis has been suggested to promote the suppressive function of LECs, directly inhibiting activated CD8+ T cells and inducing immune tolerance to tumor lymphatics [69]. It appears that LECs balance cancer immunity toward either immunogenicity or tolerance by removing tumor antigens from lymphatic vessels and lymph nodes, or by suppressing anti-tumor immunity by pre-administering tumor antigens [4,70].
We were surprised to find that the quantity of iLECs did not correlate with frequency of metastasis or with survival. However, previous reports have also pointed out that increased lymphangiogenesis and expression of lymphangiogenic growth factors are not necessarily associated with higher rates of lymph node metastasis and poorer prognosis. Williams et al. reported that proliferative images of lymphatic vessels are not observed even in areas of active angiogenesis in early breast cancer and that the diffusing lymphatic network was not related to lymph node metastasis [71]. Sipos et al. also reported that high lymphatic vessel density did not always result in increased lymph node metastasis in a nude mouse model of pancreatic ductal adenocarcinoma [72]. These studies indicate that the role of lymphatic vessels in metastasis may differ depending on their location within the tumor; peritumoral lymphatic vessels may contribute more than those in the center of tumor [73]. To this end, the quantity of peritumoral lymphatics, often quantified as microvessel density, is essential for cancer invasion. However, patient tissue samples tend to be obtained from central parts of the tumor. For instance, only the patient tumor samples with more than 60% of cancer cell nuclei were allowed to be included in TCGA [33], and patient tumor specimens were taken from the most substantive part of the tumor determined by the pathologist at the time of collection in GSE96058 [31]. Therefore, it is reasonable to assume that the amount of lymphatic endothelial cells quantified in this study reflects the cells in the center of the tumor, thus we labeled them intratumoral LECs. Hypothetically, when cancer cells proliferate excessively, the cell density and stromal pressure within the mass increase, accordingly, inhibiting the infiltration of additional stromal cells, including LECs. Our results showed that low-iLEC breast cancers enriched multiple cancer proliferation-related gene sets and were associated with a higher histological grade and Ki67, which all indicate that tumors in this group are indeed highly proliferative. Overall, both high- and low-iLEC groups demonstrate certain features of aggressive cancer biology, counterbalanced by the host immune response, ultimately resulting in no difference in patient survival. A benefit of using a transcriptome based in silico analysis is the opportunity to delve into the complex interactions in the TME. As can be seen in our analysis, several factors besides lymphangiogenesis determine the degree to which a tumor enlarges, invades, or metastasizes, which may help to explain the controversies that exist within the literature.
There are several inevitable limitations in this study that need to be noted. First, this is a retrospective study using publicly available large patient cohorts that have both transcriptomes and clinical data. TCGA and GSE96058 lack detailed treatment information for each patient and have selection bias, in that they do not necessarily reflect the latest treatments and their effectiveness due to the time frame in which they were collected. We estimated the amount of iLECs by analyzing the tumors’ transcriptomic profile with a computational algorithm, thus relying on the accuracy of the gene signature enrichment method which may have discrepancies compared to direct measurement. It is critical to note that what we measured in this study are intratumoral LECs, in contrast to previous similar studies of lymphangiogenesis which assessed peri-tumoral LECs as lymphatic micro-vessel density, which we were unable to conduct due to lack of access to pathology slides of our cohorts. Finally, our study does not include any in vitro nor in vivo experiments to prove the mechanisms due to our capability. Our results help to generate hypotheses and associations, but the underlying mechanisms will need to be proven by future experiments.
In conclusion, breast cancer high in iLECs demonstrates increased lymphangiogenesis but it was not translated to an increased lymph node metastasis nor decreased overall survival, and aggressive cancer biology was observed in both high and low iLEC breast cancer. These results may help to explain why a direct correlation has not thus far been clearly made between lymphangiogenesis, tumor behavior, and clinical outcome.
Acknowledgements
This research was supported by National Institutes of Health, USA grant number R37CA248018, R01CA250412, R01CA251545, R01EB029596, as well as US Department of Defense BCRP grant number W81XWH-19-1-0674 and W81XWH-19-1-0111 to K.T.
Disclosure of conflict of interest
None.
References
- 1.Mikada M, Sukhbaatar A, Miura Y, Horie S, Sakamoto M, Mori S, Kodama T. Evaluation of the enhanced permeability and retention effect in the early stages of lymph node metastasis. Cancer Sci. 2017;108:846–852. doi: 10.1111/cas.13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stacker SA, Williams SP, Karnezis T, Shayan R, Fox SB, Achen MG. Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat Rev Cancer. 2014;14:159–172. doi: 10.1038/nrc3677. [DOI] [PubMed] [Google Scholar]
- 3.Mukhopadhyay P, Ramanathan R, Takabe K. S1P promotes breast cancer progression by angiogenesis and lymphangiogenesis. Breast Cancer Manag. 2015;4:241–244. doi: 10.2217/bmt.15.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He M, He Q, Cai X, Chen Z, Lao S, Deng H, Liu X, Zheng Y, Liu X, Liu J, Xie Z, Yao M, Liang W, He J. Role of lymphatic endothelial cells in the tumor microenvironment-a narrative review of recent advances. Transl Lung Cancer Res. 2021;10:2252–2277. doi: 10.21037/tlcr-21-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jalkanen S, Salmi M. Lymphatic endothelial cells of the lymph node. Nat Rev Immunol. 2020;20:566–578. doi: 10.1038/s41577-020-0281-x. [DOI] [PubMed] [Google Scholar]
- 6.Lee BS, Jang JY, Seo C, Kim CH. Crosstalk between head and neck cancer cells and lymphatic endothelial cells promotes tumor metastasis via CXCL5-CXCR2 signaling. FASEB J. 2021;35:e21181. doi: 10.1096/fj.202001455R. [DOI] [PubMed] [Google Scholar]
- 7.Yeo KP, Angeli V. Bidirectional crosstalk between lymphatic endothelial cell and T cell and its implications in tumor immunity. Front Immunol. 2017;8:83. doi: 10.3389/fimmu.2017.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuttle TM. Technical advances in sentinel lymph node biopsy for breast cancer. Am Surg. 2004;70:407–413. [PubMed] [Google Scholar]
- 9.Nagahashi M, Ramachandran S, Rashid OM, Takabe K. Lymphangiogenesis: a new player in cancer progression. World J Gastroenterol. 2010;16:4003–4012. doi: 10.3748/wjg.v16.i32.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashid OM, Takabe K. Sentinel lymph node biopsy for breast cancer: our technique and future directions in lymph node staging. J Nucl Med Radiat Ther. 2012;2012:005. doi: 10.4172/2155-9619.S2-005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada A, Nagahashi M, Aoyagi T, Huang WC, Lima S, Hait NC, Maiti A, Kida K, Terracina KP, Miyazaki H, Ishikawa T, Endo I, Waters MR, Qi Q, Yan L, Milstien S, Spiegel S, Takabe K. ABCC1-exported sphingosine-1-phosphate, produced by sphingosine kinase 1, shortens survival of mice and patients with breast cancer. Mol Cancer Res. 2018;16:1059–1070. doi: 10.1158/1541-7786.MCR-17-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagahashi M, Takabe K, Terracina KP, Soma D, Hirose Y, Kobayashi T, Matsuda Y, Wakai T. Sphingosine-1-phosphate transporters as targets for cancer therapy. Biomed Res Int. 2014;2014:651727. doi: 10.1155/2014/651727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuchida J, Nagahashi M, Takabe K, Wakai T. Clinical impact of sphingosine-1-phosphate in breast cancer. Mediators Inflamm. 2017;2017:2076239. doi: 10.1155/2017/2076239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katsuta E, Yan L, Nagahashi M, Raza A, Sturgill JL, Lyon DE, Rashid OM, Hait NC, Takabe K. Doxorubicin effect is enhanced by sphingosine-1-phosphate signaling antagonist in breast cancer. J Surg Res. 2017;219:202–213. doi: 10.1016/j.jss.2017.05.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takabe K, Spiegel S. Export of sphingosine-1-phosphate and cancer progression. J Lipid Res. 2014;55:1839–1846. doi: 10.1194/jlr.R046656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang WC, Nagahashi M, Terracina KP, Takabe K. Emerging role of sphingosine-1-phosphate in Inflammation, cancer, and lymphangiogenesis. Biomolecules. 2013;3:408–434. doi: 10.3390/biom3030408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takabe K, Yamada A, Rashid OM, Adams BJ, Huang WC, Aoyagi T, Nagahashi M. Twofer anti-vascular therapy targeting sphingosine-1-phosphate for breast cancer. Gland Surg. 2012;1:80–83. doi: 10.3978/j.issn.2227-684X.2012.07.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoyagi T, Nagahashi M, Yamada A, Takabe K. The role of sphingosine-1-phosphate in breast cancer tumor-induced lymphangiogenesis. Lymphat Res Biol. 2012;10:97–106. doi: 10.1089/lrb.2012.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagahashi M, Abe M, Sakimura K, Takabe K, Wakai T. The role of sphingosine-1-phosphate in inflammation and cancer progression. Cancer Sci. 2018;109:3671–3678. doi: 10.1111/cas.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagahashi M, Ramachandran S, Kim EY, Allegood JC, Rashid OM, Yamada A, Zhao R, Milstien S, Zhou H, Spiegel S, Takabe K. Sphingosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Cancer Res. 2012;72:726–735. doi: 10.1158/0008-5472.CAN-11-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji H, Cao R, Yang Y, Zhang Y, Iwamoto H, Lim S, Nakamura M, Andersson P, Wang J, Sun Y, Dissing S, He X, Yang X, Cao Y. TNFR1 mediates TNF-alpha-induced tumour lymphangiogenesis and metastasis by modulating VEGF-C-VEGFR3 signalling. Nat Commun. 2014;5:4944. doi: 10.1038/ncomms5944. [DOI] [PubMed] [Google Scholar]
- 22.Watari K, Shibata T, Kawahara A, Sata K, Nabeshima H, Shinoda A, Abe H, Azuma K, Murakami Y, Izumi H, Takahashi T, Kage M, Kuwano M, Ono M. Tumor-derived interleukin-1 promotes lymphangiogenesis and lymph node metastasis through M2-type macrophages. PLoS One. 2014;9:e99568. doi: 10.1371/journal.pone.0099568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weichand B, Popp R, Dziumbla S, Mora J, Strack E, Elwakeel E, Frank AC, Scholich K, Pierre S, Syed SN, Olesch C, Ringleb J, Oren B, Doring C, Savai R, Jung M, von Knethen A, Levkau B, Fleming I, Weigert A, Brune B. S1PR1 on tumor-associated macrophages promotes lymphangiogenesis and metastasis via NLRP3/IL-1beta. J Exp Med. 2017;214:2695–2713. doi: 10.1084/jem.20160392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van der Auwera I, Van Laere SJ, Van den Eynden GG, Benoy I, van Dam P, Colpaert CG, Fox SB, Turley H, Harris AL, Van Marck EA, Vermeulen PB, Dirix LY. Increased angiogenesis and lymphangiogenesis in inflammatory versus noninflammatory breast cancer by real-time reverse transcriptase-PCR gene expression quantification. Clin Cancer Res. 2004;10:7965–7971. doi: 10.1158/1078-0432.CCR-04-0063. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S, Zhang D, Gong M, Wen L, Liao C, Zou L. High lymphatic vessel density and presence of lymphovascular invasion both predict poor prognosis in breast cancer. BMC Cancer. 2017;17:335. doi: 10.1186/s12885-017-3338-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura Y, Yasuoka H, Tsujimoto M, Imabun S, Nakahara M, Nakao K, Nakamura M, Mori I, Kakudo K. Lymph vessel density correlates with nodal status, VEGF-C expression, and prognosis in breast cancer. Breast Cancer Res Treat. 2005;91:125–132. doi: 10.1007/s10549-004-5783-x. [DOI] [PubMed] [Google Scholar]
- 27.Gao J, Knutsen A, Arbman G, Carstensen J, Franlund B, Sun XF. Clinical and biological significance of angiogenesis and lymphangiogenesis in colorectal cancer. Dig Liver Dis. 2009;41:116–122. doi: 10.1016/j.dld.2008.07.315. [DOI] [PubMed] [Google Scholar]
- 28.Kazama S, Kitayama J, Watanabe T, Nagawa H. Expression pattern of vascular endothelial growth factor-C in human colorectal normal mucosa and neoplastic mucosa. Hepatogastroenterology. 2004;51:391–395. [PubMed] [Google Scholar]
- 29.Miyazaki T, Okada N, Ishibashi K, Ogata K, Ohsawa T, Ishiguro T, Nakada H, Yokoyama M, Matsuki M, Kato H, Kuwano H, Ishida H. Clinical significance of plasma level of vascular endothelial growth factor-C in patients with colorectal cancer. Jpn J Clin Oncol. 2008;38:839–843. doi: 10.1093/jjco/hyn106. [DOI] [PubMed] [Google Scholar]
- 30.Kong LL, Yang NZ, Shi LH, Zhao GH, Zhou W, Ding Q, Wang MH, Zhang YS. The optimum marker for the detection of lymphatic vessels. Mol Clin Oncol. 2017;7:515–520. doi: 10.3892/mco.2017.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brueffer C, Vallon-Christersson J, Grabau D, Ehinger A, Hakkinen J, Hegardt C, Malina J, Chen Y, Bendahl PO, Manjer J, Malmberg M, Larsson C, Loman N, Ryden L, Borg A, Saal LH. Clinical value of RNA sequencing-based classifiers for prediction of the five conventional breast cancer biomarkers: a report from the population-based multicenter Sweden Cancerome Analysis Network-Breast Initiative. JCO Precis Oncol. 2018;2:PO.17.00135. doi: 10.1200/PO.17.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saal LH, Vallon-Christersson J, Hakkinen J, Hegardt C, Grabau D, Winter C, Brueffer C, Tang MH, Reutersward C, Schulz R, Karlsson A, Ehinger A, Malina J, Manjer J, Malmberg M, Larsson C, Ryden L, Loman N, Borg A. The Sweden Cancerome Analysis Network-Breast (SCAN-B) Initiative: a large-scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med. 2015;7:20. doi: 10.1186/s13073-015-0131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okano M, Oshi M, Butash AL, Katsuta E, Tachibana K, Saito K, Okayama H, Peng X, Yan L, Kono K, Ohtake T, Takabe K. Triple-negative breast cancer with high levels of annexin A1 expression is associated with mast cell infiltration, inflammation, and angiogenesis. Int J Mol Sci. 2019;20:4197. doi: 10.3390/ijms20174197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oshi M, Katsuta E, Yan L, Ebos JML, Rashid OM, Matsuyama R, Endo I, Takabe K. A novel 4-gene score to predict survival, distant metastasis and response to neoadjuvant therapy in breast cancer. Cancers (Basel) 2020;12:1148. doi: 10.3390/cancers12051148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chouliaras K, Tokumaru Y, Asaoka M, Oshi M, Attwood KM, Yoshida K, Ishikawa T, Takabe K. Prevalence and clinical relevance of tumor-associated tissue eosinophilia (TATE) in breast cancer. Surgery. 2021;169:1234–1239. doi: 10.1016/j.surg.2020.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oshi M, Angarita FA, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. High expression of NRF2 is associated with increased tumor-infiltrating lymphocytes and cancer immunity in ER-positive/HER2-negative breast cancer. Cancers (Basel) 2020;12:3856. doi: 10.3390/cancers12123856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le L, Tokumaru Y, Oshi M, Asaoka M, Yan L, Endo I, Ishikawa T, Futamura M, Yoshida K, Takabe K. Th2 cell infiltrations predict neoadjuvant chemotherapy response of estrogen receptor-positive breast cancer. Gland Surg. 2021;10:154–165. doi: 10.21037/gs-20-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oshi M, Asaoka M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Zsiros E, Ishikawa T, Endo I, Takabe K. Abundance of regulatory T cell (Treg) as a predictive biomarker for neoadjuvant chemotherapy in triple-negative breast cancer. Cancers (Basel) 2020;12:3038. doi: 10.3390/cancers12103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oshi M, Asaoka M, Tokumaru Y, Yan L, Matsuyama R, Ishikawa T, Endo I, Takabe K. CD8 T cell score as a prognostic biomarker for triple negative breast cancer. Int J Mol Sci. 2020;21:6968. doi: 10.3390/ijms21186968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gandhi S, Oshi M, Murthy V, Repasky EA, Takabe K. Enhanced thermogenesis in triple-negative breast cancer is associated with pro-tumor immune microenvironment. Cancers (Basel) 2021;13:2559. doi: 10.3390/cancers13112559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the pathway interaction database. Nucleic Acids Res. 2009;37:D674–679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokumaru Y, Asaoka M, Oshi M, Katsuta E, Yan L, Narayanan S, Sugito N, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. High expression of microRNA-143 is associated with favorable tumor immune microenvironment and better survival in estrogen receptor positive breast cancer. Int J Mol Sci. 2020;21:3213. doi: 10.3390/ijms21093213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tokumaru Y, Katsuta E, Oshi M, Sporn JC, Yan L, Le L, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. High expression of miR-34a associated with less aggressive cancer biology but not with survival in breast cancer. Int J Mol Sci. 2020;21:3045. doi: 10.3390/ijms21093045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tokumaru Y, Oshi M, Patel A, Tian W, Yan L, Matsuhashi N, Futamura M, Yoshida K, Takabe K. Organoids are limited in modeling the colon adenoma-carcinoma sequence. Cells. 2021;10:488. doi: 10.3390/cells10030488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oshi M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Endo I, Takabe K. Degree of early estrogen response predict survival after endocrine therapy in primary and metastatic ER-positive breast cancer. Cancers (Basel) 2020;12:3557. doi: 10.3390/cancers12123557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Narayanan S, Kawaguchi T, Yan L, Peng X, Qi Q, Takabe K. Cytolytic activity score to assess anticancer immunity in colorectal cancer. Ann Surg Oncol. 2018;25:2323–2331. doi: 10.1245/s10434-018-6506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takeshita T, Yan L, Asaoka M, Rashid O, Takabe K. Late recurrence of breast cancer is associated with pro-cancerous immune microenvironment in the primary tumor. Sci Rep. 2019;9:16942. doi: 10.1038/s41598-019-53482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS Cancer Genome Atlas Research Network. Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The immune landscape of cancer. Immunity. 2019;51:411–412. doi: 10.1016/j.immuni.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi H, Asaoka M, Yan L, Rashid OM, Oshi M, Ishikawa T, Nagahashi M, Takabe K. Biologically aggressive phenotype and anti-cancer immunity counterbalance in breast cancer with high mutation rate. Sci Rep. 2020;10:1852. doi: 10.1038/s41598-020-58995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tokumaru Y, Oshi M, Katsuta E, Yan L, Satyananda V, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. KRAS signaling enriched triple negative breast cancer is associated with favorable tumor immune microenvironment and better survival. Am J Cancer Res. 2020;10:897–907. [PMC free article] [PubMed] [Google Scholar]
- 56.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Katz MHG, Takabe K. High G2M pathway score pancreatic cancer is associated with worse survival, particularly after margin-positive (R1 or R2) resection. Cancers (Basel) 2020;12:2871. doi: 10.3390/cancers12102871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oshi M, Satyananda V, Angarita FA, Kim TH, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, Takabe K. Angiogenesis is associated with an attenuated tumor microenvironment, aggressive biology, and worse survival in gastric cancer patients. Am J Cancer Res. 2021;11:1659–1671. [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Aoki H, Yang J, Peng K, Liu R, Li X, Qiang X, Sun L, Gurley EC, Lai G, Zhang L, Liang G, Nagahashi M, Takabe K, Pandak WM, Hylemon PB, Zhou H. The role of sphingosine 1-phosphate receptor 2 in bile-acid-induced cholangiocyte proliferation and cholestasis-induced liver injury in mice. Hepatology. 2017;65:2005–2018. doi: 10.1002/hep.29076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagahashi M, Yuza K, Hirose Y, Nakajima M, Ramanathan R, Hait NC, Hylemon PB, Zhou H, Takabe K, Wakai T. The roles of bile acids and sphingosine-1-phosphate signaling in the hepatobiliary diseases. J Lipid Res. 2016;57:1636–1643. doi: 10.1194/jlr.R069286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagahashi M, Takabe K, Liu R, Peng K, Wang X, Wang Y, Hait NC, Wang X, Allegood JC, Yamada A, Aoyagi T, Liang J, Pandak WM, Spiegel S, Hylemon PB, Zhou H. Conjugated bile acid-activated S1P receptor 2 is a key regulator of sphingosine kinase 2 and hepatic gene expression. Hepatology. 2015;61:1216–1226. doi: 10.1002/hep.27592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Studer E, Zhou X, Zhao R, Wang Y, Takabe K, Nagahashi M, Pandak WM, Dent P, Spiegel S, Shi R, Xu W, Liu X, Bohdan P, Zhang L, Zhou H, Hylemon PB. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology. 2012;55:267–276. doi: 10.1002/hep.24681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verastegui E, Zlotnik A. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 63.Shields JD, Emmett MS, Dunn DB, Joory KD, Sage LM, Rigby H, Mortimer PS, Orlando A, Levick JR, Bates DO. Chemokine-mediated migration of melanoma cells towards lymphatics--a mechanism contributing to metastasis. Oncogene. 2007;26:2997–3005. doi: 10.1038/sj.onc.1210114. [DOI] [PubMed] [Google Scholar]
- 64.Kim M, Koh YJ, Kim KE, Koh BI, Nam DH, Alitalo K, Kim I, Koh GY. CXCR4 signaling regulates metastasis of chemoresistant melanoma cells by a lymphatic metastatic niche. Cancer Res. 2010;70:10411–10421. doi: 10.1158/0008-5472.CAN-10-2591. [DOI] [PubMed] [Google Scholar]
- 65.Issa A, Le TX, Shoushtari AN, Shields JD, Swartz MA. Vascular endothelial growth factor-C and C-C chemokine receptor 7 in tumor cell-lymphatic cross-talk promote invasive phenotype. Cancer Res. 2009;69:349–357. doi: 10.1158/0008-5472.CAN-08-1875. [DOI] [PubMed] [Google Scholar]
- 66.Boyle ST, Ingman WV, Poltavets V, Faulkner JW, Whitfield RJ, McColl SR, Kochetkova M. The chemokine receptor CCR7 promotes mammary tumorigenesis through amplification of stem-like cells. Oncogene. 2016;35:105–115. doi: 10.1038/onc.2015.66. [DOI] [PubMed] [Google Scholar]
- 67.Tokumaru Y, Oshi M, Katsuta E, Yan L, Huang JL, Nagahashi M, Matsuhashi N, Futamura M, Yoshida K, Takabe K. Intratumoral adipocyte-high breast cancer enrich for metastatic and inflammation-related pathways but associated with less cancer cell proliferation. Int J Mol Sci. 2020;21:5744. doi: 10.3390/ijms21165744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Forster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8:362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 69.Roberts EW, Broz ML, Binnewies M, Headley MB, Nelson AE, Wolf DM, Kaisho T, Bogunovic D, Bhardwaj N, Krummel MF. Critical role for CD103(+)/CD141(+) dendritic cells bearing CCR7 for tumor antigen trafficking and priming of T cell immunity in Melanoma. Cancer Cell. 2016;30:324–336. doi: 10.1016/j.ccell.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stachura J, Wachowska M, Kilarski WW, Guc E, Golab J, Muchowicz A. The dual role of tumor lymphatic vessels in dissemination of metastases and immune response development. Oncoimmunology. 2016;5:e1182278. doi: 10.1080/2162402X.2016.1182278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams CS, Leek RD, Robson AM, Banerji S, Prevo R, Harris AL, Jackson DG. Absence of lymphangiogenesis and intratumoural lymph vessels in human metastatic breast cancer. J Pathol. 2003;200:195–206. doi: 10.1002/path.1343. [DOI] [PubMed] [Google Scholar]
- 72.Sipos B, Kojima M, Tiemann K, Klapper W, Kruse ML, Kalthoff H, Schniewind B, Tepel J, Weich H, Kerjaschki D, Kloppel G. Lymphatic spread of ductal pancreatic adenocarcinoma is independent of lymphangiogenesis. J Pathol. 2005;207:301–312. doi: 10.1002/path.1840. [DOI] [PubMed] [Google Scholar]
- 73.Hu X, Luo J. Heterogeneity of tumor lymphangiogenesis: progress and prospects. Cancer Sci. 2018;109:3005–3012. doi: 10.1111/cas.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]