Abstract
Regulatory T cells (Tregs) are a subset of CD4+ T lymphocytes known to dampen the host immune response against cancer cells. Within the tumor microenvironment, Tregs are potent facilitators of immune tolerance, and a higher proportion of Tregs compared to cytotoxic T cells predicts a worse outcome in most solid tumors. We studied the association between Treg density, and cancer biology and clinical outcome in colorectal cancer (CRC). We used xCell to estimate intratumoral Tregs in total of 898 CRC patients in the Cancer Genome Atlas (TCGA) and GCE39582 cohorts. High-Treg CRCs enriched immune response-related gene sets; inflammatory response, IFN-γ and IFN-α response, IL2/IL6 signaling, and allograft rejection, and had significantly high infiltration of CD8, CD4, M1 and M2 macrophage, and dendritic cells in both cohorts. While high-Treg CRCs enriched multiple pro-cancer signaling pathways compared to low-Treg CRCs, such as Epithelial Mesenchymal Transition, K-ras, Hypoxia, TGF-β, TNF-α, and angiogenesis, Treg infiltration was surprisingly associated with earlier CRC stage in TCGA. Notably, in two separate cohorts a higher proportion of Tregs predicted an improved response to chemotherapy. In the GSE28702 cohort, metastatic CRCs with more Tregs showed a significantly better response to mFOLFOX6 versus low-Treg CRC metastases (88.9% response vs. 16.7%, P<0.001). In the GSE72970 cohort, high-Treg CRCs were found to have a 68.8% response to FOLFOX/FOLFIRI without bevacizumab, compared to 44% response in the low-Treg CRCs. Additionally, high-Treg CRCs were associated with increased expression of immune checkpoint molecules PD-L1/PD-L2, CTLA4, TIGIT and BTLA, implying susceptibility to immunotherapy. We also found that CRCs with higher proportions of Tregs were associated with lower amounts of three microorganisms in the tumor: Lachnoclostridium, flavivirus, and Ornithobacterium. In conclusion, we show that amount of Treg in the tumor is a predictor of host immune response and chemotherapy response in CRC.
Keywords: Treg, colorectal cancer, gene expression, metastasis, FOLFOX, survival, treatment response, tumor microenvironment, xCell
Introduction
Colorectal cancer (CRC) ranks number three among the world’s most common cancers [1]. Despite the fact that CRC-specific mortality has declined by implementation of population screening and development of new treatments [2], approximately 22% of patients are found to have metastatic disease at the time of diagnosis, and 5-year overall survival for these patients is dismal at 14% [3]. Therefore, there is ongoing interest in new predictive biomarkers that identify which metastatic CRC will respond to treatments.
The progression of any given CRC is influenced by several factors, to include tumor biology, the tumor microenvironment (TME) and host immune response, and likely the gut microbiome. The dynamic role of the TME in influencing cancer progression and metastasis is now widely described, and the TME includes not only tumor cells, but also stromal and infiltrating inflammatory/immune cells [4]. Regulatory T cells (Tregs), identified as transcription factor FOXP3 positive CD4+ T lymphocytes, suppress the activation, proliferation, and function of numerous immune cells, which is essential in the maintenance of self-tolerance and immune homeostasis, however, unfavorable in cancer progression [5,6]. Tregs have been shown to develop an immunosuppressive TME by suppression of antitumor immune response in many types of solid cancer. The number of Tregs has been shown to associate with worse survival in many cancer types including cervical, melanoma, and breast cancer [7]. On the other hand, increased number of tumor-infiltrating lymphocytes that include Tregs in TME were shown to associate with favorable survival in CRC patients [7-15]. This may be due in part to heterogeneity within Tregs, which has been previously described [16].
CRC with microsatellite instability (MSI) have been classically identified pathologically by high infiltration of inflammatory lymphocytes and is known to associate with relatively better survival than CRC without MSI [17-19]. Interestingly, while MSI-High CRC are densely infiltrated with cytotoxic T cells as would be expected due to their immunogenicity, studies have also shown that they have a high density of Tregs, which may explain why the primary tumors of MSI-High CRCs grow to a large size [20-22]. Similarly, tumors with a higher mutational burden tend to be MSI-High, and are associated with longer progression-free survival after treatment with chemotherapy and bevacizumab compared to tumors with lower mutational burden [23]. Therefore, in this paper we investigate the association between Treg density and tumor characteristics such as MSI and tumor mutational burden.
The gut microbiome has been implicated in the initiation and development of CRC [24]. Studies have shown that gut bacteria may elicit an inflammatory reaction which promotes tumor progression, and the bacteria Fusobacterium nucleatum secretes a FadA adhesin, which stimulates CRC tumor growth via E-cadherin/β-catenin signaling [25,26]. The balance between Tregs and pro-inflammatory T helper 17 (Th17) cells has been shown to be linked to gut microbiota [27], and thus we examine the relationship between Treg density and the gut microbiome in this study.
Finally, Tregs have been identified as an important therapeutic target in the treatment of several cancers [28]. Controlling immunosuppression by Tregs is thought to be essential for more effective cancer treatment. We previously reported that breast cancer with a high fraction of Tregs was significantly associated with better response to neoadjuvant chemotherapy [29]. Therefore, we evaluated the relationship between Treg density and response to chemotherapy using data from two cohorts.
In this study, we used xCell algorithm to assess the relative percentages of infiltrating immune cells in comparison to stromal cells within the TME using transcriptome data of bulk tumor. The aim of the study is to investigate the clinical relevance of the fraction of Tregs within the colorectal cancer TME using xCell score with mRNA expression data across several CRC cohorts.
Materials and methods
Colorectal cancer patient cohorts
We used The Cancer Genome Atlas (TCGA, n=456), and the GSE39582 (n=443) [30] cohorts, which have a large number of CRC samples with clinical and gene expression data, for the main analysis in study. Clinical, gene expression, and tumor microbiome in the TCGA cohort were obtained from the cBio Cancer Genomic portal as previously described [31-34]. Clinical and gene expression data in the GSE39582 cohort was obtained from the Gene Expression Omnibus (GEO) repository. Other GEO data from the GSE28702 (n=83) [35] and GSE72970 (n=143) [36-38] cohorts were obtained to investigate the association of Tregs in tumor with drug response.
Gene set enrichment analysis
To investigate biological function, Gene Set Enrichment Analyses (GSEA) (Java version 4.0) [36] was used with MSigDB Hallmark gene sets [37], as we previously reported [39-43]. A false discovery rate (FDR) of 0.25 was used to statistical significance, as recommended by the GSEA software.
Composition of immune cells and Treg infiltration fraction
The xCell score, a bioinformatics algorithm with transcriptome data in tumor, was used as infiltration fraction of Tregs in CRC as previously described [29,44]. The algorithm was also used to predict immune and stromal cells composition in the TCGA and GSE39582 cohorts. The top tertile level of Tregs were determined as high Tregs groups within cohorts.
Others
All analyses were performed using the R software (version 4.0.1, R Project for Statistical Computing). Mann-Whitney U, Kruskal-Wallis, and Fisher’s exact test were used to group comparison. The Kaplan-Meier method with log-rank test was used to survival analyses. Boxplots were used to depict median and inter-quartile level values.
Results
High-Treg CRC demonstrated enrichment of several pro-cancer-related gene sets
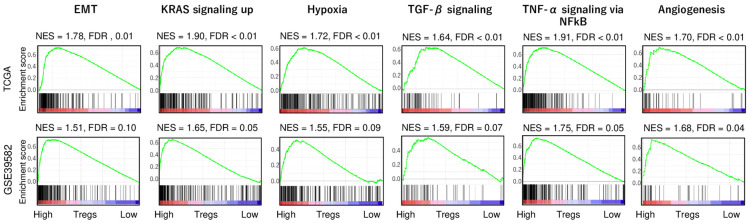
To investigate which hallmark of cancer is associated with Treg infiltration, gene set enrichment analysis (GSEA) with Hallmark collection was conducted as we previously reported [34,45-48]. Interestingly, high-Treg CRC enriched several cancer aggravating gene sets, including epithelial mesenchymal transition (EMT), KRAS signaling up, hypoxia, TGF-β signaling, TNF-α signaling via NFkB, and coagulation, consistently in both TCGA and GSE38582 cohorts (Figure 1). These results suggest that a high Treg fraction was significantly associated with high activity of cancer aggravating pathways in CRC.
Figure 1.
Gene Set Enrichment Assay (GSEA) with significantly enriched gene sets to the high-Treg CRC patients in the TCGA and GSE39582 cohorts. Cancer aggravating gene sets (EMT, KRAS signaling up, hypoxia, TGF-β signaling, TNF-α signaling via NFkB, and coagulation) with false discovery rate (FDR) and normalized enrichment score (NES). As recommended by the GSEA software, we defined statistical significance by an FDR of less than 0.25.
Treg fraction was not associated with tumor mutation load in CRC
Next, we investigated the relationship between Tregs and mutation load, which is associated with improved survival after treatment with immune checkpoint inhibitors across multiple cancer types [49]. There were no significant differences in mutation load-related score, fraction altered, non-silent mutation, single nucleotide variants (SNV) and indel neoantigens, between the low- and high-Treg group, except for silent mutation (Figure 2, P=0.030) in the TCGA cohort. These results suggest that the fraction of Tregs was not related to mutation load in CRC.
Figure 2.
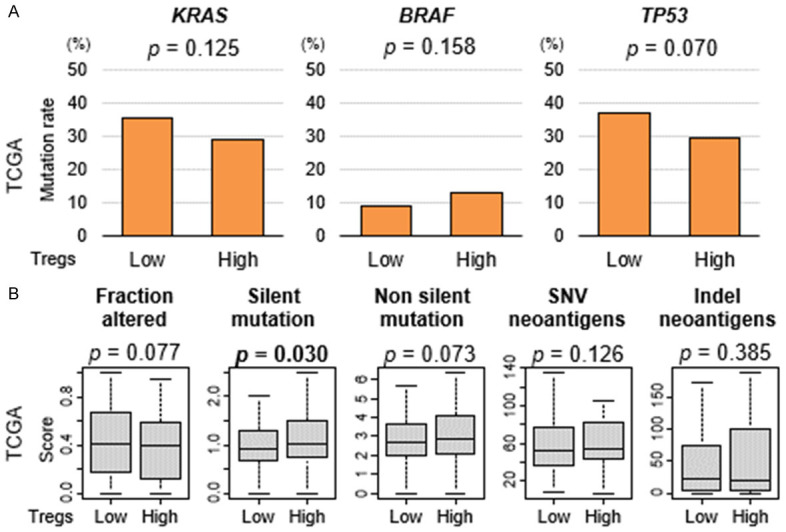
A. Association of the fraction of Tregs with mutation load in CRC. B. Boxplots comparing high and low fraction of Tregs groups by mutation load-related score; fraction altered, silent and non-silent mutation, single nucleotide variants (SNV) and indel neoantigens, in the TCGA cohort. Mann-Whitney U test was used to perform the analysis.
Treg fraction was not associated with clinical aggressiveness or microsatellite instability in CRC
To investigate the clinical relevance of Tregs in CRC, we studied the association of Tregs with clinical parameters, including American Joint Committee on Cancer (AJCC) stage in two cohorts, and microsatellite instability (MSI) status in the TCGA cohort. Advanced AJCC stage was significantly associated with low level of Tregs in the TCGA cohort (Figure 3A, P=0.005), which was not validated by GSE39582 cohort. MSI status did not show association with Tregs in the TCGA (Figure 3B, P=0.210). These results showed no consistent results about the association between Tregs and clinical parameters in two different cohorts.
Figure 3.
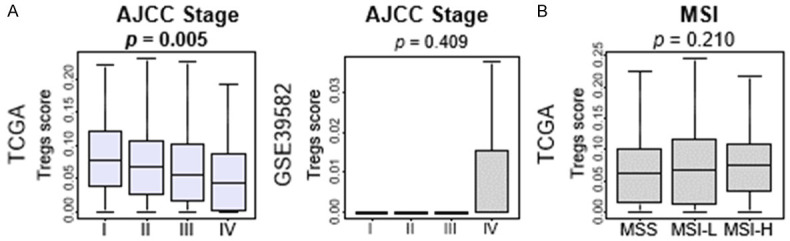
Association between levels of Tregs and clinical factors in CRC. A. Boxplots of Treg fractions by AJCC stage in the TCGA and GSE39582 cohort. B. Boxplots of Treg fractions by MSI in the TCGA cohort.
High-Treg CRC enriched several immune response-related gene sets
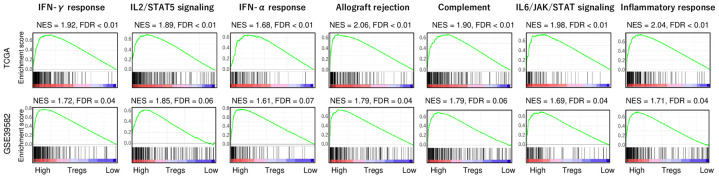
Given the discrepancy that high-Treg CRC enriched cancer aggravating gene sets, but was also associated with less advanced cancer stage, we speculated that there may be another mechanism that prevent these tumors from worsening. To this end, we conducted GSEA with Hallmark immune response-related gene sets between low and high Treg CRC in the two cohorts. We found that high-Treg CRC significantly enriched immune response-related gene sets, including interferon (IFN)-γ response, IFN-α response, IL2/STAT5 signaling, allograft rejection, complement, IL6/JAK/STAT signaling, and inflammatory response (Figure 4; all false discovery rate (FDR) <0.01, and normalized enrichment score (NES) >1.60). These results were all validated in a second cohort. These results suggest that high-Treg CRCs are significantly associated with high level of immune response.
Figure 4.
Gene Set Enrichment Assay (GSEA) of immune response-related gene sets by high vs. low Treg CRC in the TCGA and GSE39582 cohorts. Interferon (IFN)-γ response, IL2/STAT5 signaling, IFN-α response, allograft rejection, complement, IL6/JAK/STAT3 signaling, and inflammatory response gene sets with false discovery rate (FDR) and normalized enrichment score (NES). As recommended by the GSEA software, FDR of 0.25 defined statistical significance.
High-Treg CRC was significantly associated with high fractions of anti-cancer immune cells
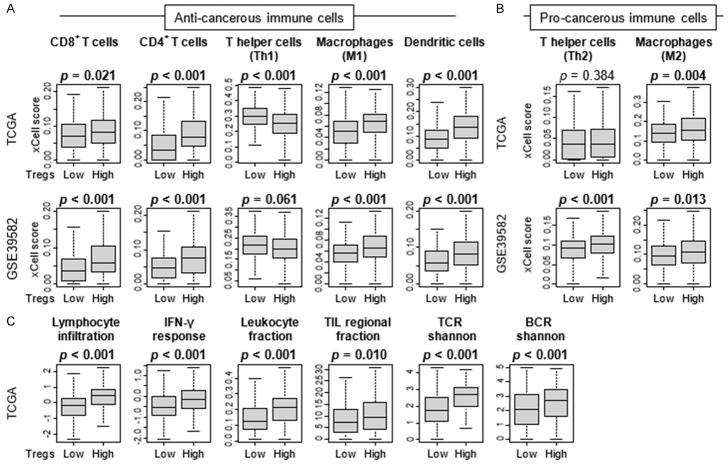
Since high-Treg CRC was associated with high level of immune response, it was of interest to investigate what types of immune cells infiltrate in the tumor immune microenvironment (TME) of the high-Treg CRC. To this end, we compared the subpopulation of immune cells between high and low Tregs groups using xCell algorithm. CRC with high Tregs was significantly infiltrated with multiple anti-cancer immune cells, including CD8+ and CD4+ T cells, M1 macrophages, and dendritic cells consistently in both TCGA and GSE39582 cohorts (Figure 5A). M2 macrophages infiltrated also in high-Treg CRC in both cohorts (Figure 5B). T helper type2 (Th2) cells were significantly higher in high-Treg groups in the GSE39582, but this was not validated by TCGA cohort (Figure 5B). Furthermore, CRCs with high Tregs were significantly associated with high immune fraction-related scores, including lymphocyte infiltration, leukocyte fraction, tumor infiltrating lymphocytes (TIL) regional fraction, and T cell receptor (TCR) and B cell receptor (BCR) Shannon, calculated by Thorsson et al. (Figure 5C). These results suggest that high-Treg CRCs were significantly associated with high fraction of immune cells, especially anti-cancer immune cells.
Figure 5.
Association of the fraction of Tregs with infiltrating of immune cells in CRC immune microenvironment. Boxplots comparing high and low fraction of Tregs groups by infiltrating immune cells, (A) anti-cancer immune cells; CD8+ T cells, CD4+ T cells, T helper type 1 (Th1) cells, M1 macrophages, and dendritic cells, and (B) pro-cancer immune cells; T helper type 2 (Th2) cells and M2 macrophages in the TCGA and GSE39582 cohorts. (C) Boxplots comparing high and low fraction of Tregs groups by immune fraction-related score; lymphocyte infiltration, leukocyte fraction, tumor infiltrating lymphocytes (TIL) regional fraction, and T cell receptor (TCR) and B cell receptor (BCR) Shannon, in the TCGA cohort. The top tertile cut-off and Mann-Whitney U test was used to perform the analysis.
High-Treg CRC was associated with better response to chemotherapy and higher expression of immune checkpoint molecules
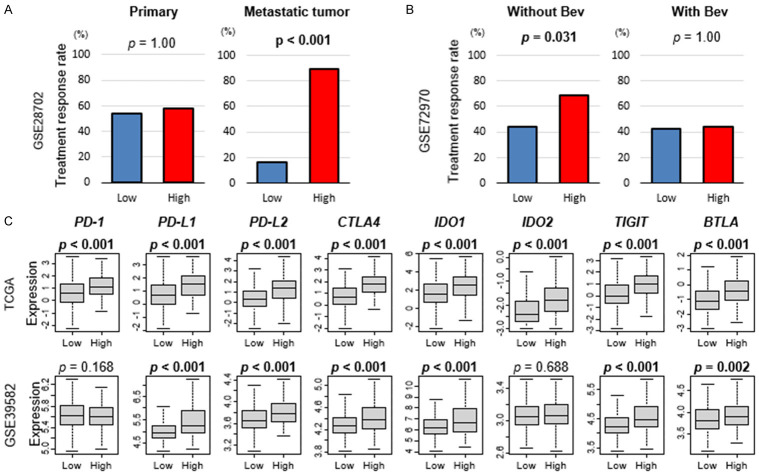
To study the clinical relevance of Treg infiltration in CRC, we investigated the association of the Tregs with CRC drug response using GSE28702 and GSE72970 cohorts, which include response to chemotherapy data. We found that, although there was no association with Treg infiltration and chemotherapy response in the primary tumor, high Treg infiltration was significantly associated with high response rate to mFOLFOX6 chemotherapy regimen in metastatic CRC (Figure 6A; P>0.99 and P<0.001, respectively). Further, high-Treg primary CRC was significantly associated with high response rate to chemotherapy without bevacizumab, but not with bevacizumab (Figure 6B).
Figure 6.
Association of Tregs with drug response in CRC. A. Bar plots of response rate to mFOLFOXC6 by low and high Tregs groups in primary and metastatic CRC in the GSE28702 cohort. B. Bar plots of response rate to chemotherapy with/without bevacizumab (Bev) by low and high Tregs groups in the GSE72970 cohort. C. Boxplots comparing low and high Tregs group with expression levels of immune checkpoint molecules (programmed death-1; PD-1, programmed death ligand 1; PD-L1, programmed death ligand 2; PD-L2, cytotoxic T-lymphocyte-associated protein 4; CTLA4, indoleamine dioxygenase 1; IDO1, T cell immunoreceptor with Ig and ITIM domains; TIGIT, B- and T-lymphocyte attenuator; BTLA) in CRC in the TCGA and GSE39582 cohorts. Mann-Whitney U test was used to perform the analysis.
We next investigated the association of Tregs with immune checkpoint molecules (ICM) gene expressions in CRC. High-Treg CRC was significantly associated with high expression of ICM genes; programmed death-1 (PD-1), programmed death ligand 1 and 2 (PD-L1/PD-L2), cytotoxic T-lymphocyte-associated protein 4 (CTLA4), indoleamine dioxygenase 1 (IDO1), T cell immunoreceptor with Ig and ITIM domains (TIGIT), and B- and T-lymphocyte attenuator (BTLA), in the TCGA cohort (Figure 6C; all P<0.001). These results were validated by GSE39582, except for PD-1.
High-Treg CRC showed a trend toward association with low amounts of three tumor microorganisms
Since tumor microbiomes have been reported to affect CRC growth [16], we investigated the association between tumor microbiome and infiltration of Tregs. Although adjusted P-value did not show significance, high-Treg CRC tended to be highly associated with low amount of three tumor microorganisms, Lachnoclostridium (Figure 7; log FC =0.448, P<0.001), flavivirus (log FC =0.479, P<0.001), and Ornithobacterium (log FC=0.290, P<0.001), in the TCGA cohort (Figure 7; all adjusted P=0.055).
Figure 7.
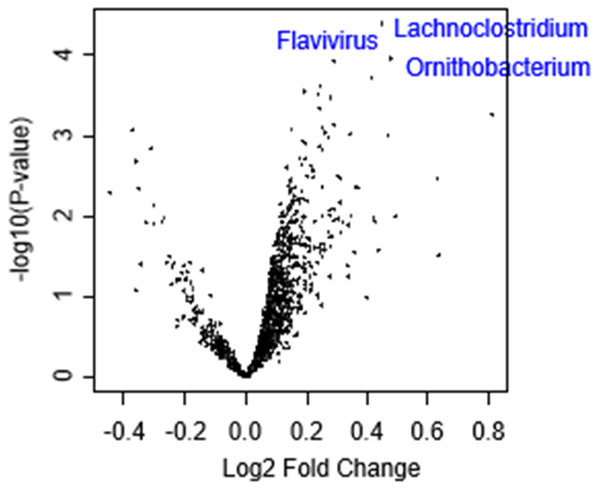
Association of Tregs with microbiome in CRC. Volcano plot showing the differentially expressed mRNAs of gut microbiomes between low and high Tregs CRC groups. X-axes: log2 fold change (FC), Y-axes: -log10 P-value from limma analysis. Mann-Whitney U test was used to perform the analysis.
Discussion
In this study, we used a validated computer algorithm, xCell [50], to stratify several large cohorts of CRCs into two groups based on high versus low proportions of Tregs. We studied the correlation between intratumoral Treg density and cancer biology including host immune response, ICM expression, and the tumor microbiome by comparing gene expression between the two groups. We further examined two CRC cohorts which contained data on response to neoadjuvant chemotherapy using mFOLFOX6 and bevacizumab, in order to explore the association between Treg infiltration and response to treatment.
With regards to cancer biology, we found that high-Treg CRC enriched several gene sets associated with cancer progression: EMT, KRAS signaling, hypoxia, TGF-β signaling, TNF-α signaling via NFkB, and coagulation. This was not an unexpected finding, as in many cancers, a high density of Tregs is correlated with poor clinical outcome [7]. However, this did not translate to clinical aggressiveness in high-Treg CRCs. We found that intratumoral Treg density was in fact inversely correlated with AJCC stage in the TCGA cohort (P=0.005), and there was no statistically significant association between Treg density and AJCC stage in the GSE39582 cohort. Another aspect of CRC biology that we investigated in this study was the association between MSI and Treg infiltration. Approximately 10-15% of CRCs demonstrate high MSI due to deficiencies in DNA mismatch repair [22], as opposed to the majority of CRCs which develop due to chromosomal instability. MSI-High CRCs have been clinically characterized by a more favorable stage-adjusted survival. Several studies have suggested that the MSI stimulates a more robust host immune response, a theory which is supported by the finding of increased infiltration of MSI-High CRC by cytotoxic T cells, while microsatellite-stable (MSS) tumors tend to demonstrate increased levels of Tregs [51]. However, we found no correlation between MSI and Treg density in the TCGA cohort.
The lack of association between Treg density and cancer biology may be explained in part by the findings related to the host immune response. We were surprised to find that high-Treg CRCs demonstrated increased expression of genes for the inflammatory response compared to low-Treg CRCs, including IFN-γ response, IFN-α response, IL2/STAT5 signaling, allograft rejection, complement, and IL6/JAK/STAT signaling. Additionally, CRCs with a high proportion of Tregs were found to also contain infiltrates high in anti-cancer immune cells, such as CD8+ and CD4+ T cells, M1 macrophages, and dendritic cells. High-Treg CRCs also demonstrated high immune fraction scores, including lymphocyte infiltration, leukocyte fraction, TIL regional fraction, and TCR and BCR Shannon. This immune fraction score was described by Thorsson et al. in 2018 and describes various patterns of tumor immune infiltrates [52]. The increased expression of inflammatory response genes and the marked host immune response in high-Treg CRCs runs contrary to the known function of Tregs as target cell suppressors via direct contact with effector T cells and via cytokine signaling [5]. The relationship between chronic inflammation, the immune response, and the development and progression of CRC is complex. It is well known that chronic intestinal inflammation such as occurs in inflammatory bowel disease is a risk factor for the development of CRC and NSAIDs have been shown to decrease the risk [53]. A possible explanation for the positive association between Treg density and clinical outcome in CRC, as opposed to cancers that develop in relatively sterile environments, is the role of gut flora in chronic inflammation and therefore tumor development. It has been proposed that Tregs in the gut suppress the inflammatory response to bacteria, which in turn decreases carcinogenesis [21].
Importantly, we found that a high proportion of Tregs predicted response to mFOLFOX6 in metastatic CRC in the GSE28702 cohort. 88.9% of CRC metastases with a high Treg density had a response to chemotherapy, while only 16.7% of low-Treg metastases responded (P<0.001). However, Treg density was not correlated with response to chemotherapy in the primary tumors. In the GSE72970 cohort which included patients treated with FOLFOX, FOLFIRI, or FOLFIRI + bevacizumab, there was a significantly higher response to treatment with FOLFOX or FOLFIRI in high-Treg CRCs. CRCs with high density of Tregs were found to have a 68.8% response to chemotherapy, while low-Treg CRCs had a 44% response (P=0.031). There was no difference in response to treatment with FOLFIRI + bevacizumab regardless of Treg density, however this cohort may have been underpowered to detect a difference as there were only 22 patients in this treatment arm. The other aspects that we evaluated which is related to Tregs as a predictor of treatment response were the tumor mutational burden, and expression of immune checkpoint molecules. A higher tumor mutational load is associated with improved survival after treatment with immune checkpoint inhibitors (ICIs) across multiple cancer types including CRC [49,54]. Therefore, we compared mutational load in high- and low-Treg CRCs but did not find a statistically significant difference between the two groups. However, we did find that high-Treg CRCs were associated with significantly higher expression of the immune checkpoint molecules PD-L1, PD-L2, CTLA4, IDO1, TIGIT, and BTLA in both the TCGA and GSE39582 cohorts. ICMs have now been widely described as therapeutic targets; for example, PD-L1 is an FDA-approved biomarker for guiding immune checkpoint therapy in several types of cancers [55]. Our findings suggest that a higher proportion of Tregs within a tumor may also predict response to immunotherapy.
There are some limitations to this study, which are primarily related to the inability to minimize the heterogeneity of the samples. For example, in CRC, several studies have shown that primary tumor location affects tumor aggressiveness, metastatic potential, and overall survival [8,56,57]. Due in part to the different embryologic origin of the right and left colon, proximal and distal colon cancers have been shown to have differences in histology, rate of MSI, tumor stage at diagnosis, and risk of death [56]. Berntsson et al. found that the proportions between Tregs and cytotoxic T cells were only associated with metastatic rate in right-sided tumors, suggesting that the dynamics of the TME may vary based on tumor sidedness in CRCs. In our cohorts, we were not able to stratify the samples by primary tumor location due to lack of this information. Additionally, Tregs themselves demonstrate significant heterogeneity; while Tregs are defined by the expression of FOXP3, their quantitative expression of FOXP3 can vary. Saito et al. found that the prognosis of CRCs correlated specifically with the level of expression of FOXP3, rather than the proportion of Tregs overall [16]. More recently, Szeponik et al. stratified intratumoral Tregs in colon tumors by CD39 expression and found that patients with high expression of CD39 by infiltrating Tregs had worse outcomes [58]. As we utilized tumor gene expression rather than protein analyses, we were unable to stratify Tregs based on protein expression. Finally, we had no control over the spatial location of the biopsy in relation to the entire tumor. Other studies have previously noted that ratios of various T-lymphocytes vary significantly between the epithelial, intratumoral and peritumoral tissues [9,53], and this may have affected our results. A final limitation of this study is the absence of our own data to confirm these findings, which would have afforded more control over the biopsy sampling, although at the expense of a smaller sample size.
The current study is one of the first to investigate the clinical relevance of Tregs in CRC patients using transcriptome associated with response to therapy data. The benefits of using computational algorithm such as xCell include the large sample size available to us due to utilizing a transcriptome rather than individually collecting samples for immunohistochemistry or flow cytometry. Additionally, interpretation of these methods can be subjective [15], while xCell analysis produces a quantifiable value for Treg density, allowing us to accurately stratify our cohorts into two groups. Finally, an important benefit of utilizing transcriptome is the ability to simultaneously examine multiple aspects of the TME, including not only cancer gene expression, but also the host immune response and microbial infiltrate. The characterization of the intratumoral microbiome is a relatively new area of research which may yield therapeutic benefits. Given the myriad of interactions that influence CRC progression, this global view of the TME helps to explain some of the reported discordance between Treg density and clinical outcomes in CRC.
In conclusion, we found that high-Treg CRC enriched not only cancer aggravating gene sets, such as EMT, KRAS, TGF-β and angiogenesis, but also immune response-related gene sets, which is consistent with high infiltration of anti-cancerous immune cells. High Treg density was a predictor of response to chemotherapy in metastatic CRC.
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Benning TM, Dellaert BG, Dirksen CD, Severens JL. Preferences for potential innovations in non-invasive colorectal cancer screening: a labeled discrete choice experiment for a Dutch screening campaign. Acta Oncol. 2014;53:898–908. doi: 10.3109/0284186X.2013.877159. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Li S, Liu Y, Zhang C, Li H, Lai B. Metastatic patterns and survival outcomes in patients with stage IV colon cancer: a population-based analysis. Cancer Med. 2020;9:361–373. doi: 10.1002/cam4.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010;10:490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- 6.Savage PA, Malchow S, Leventhal DS. Basic principles of tumor-associated regulatory T cell biology. Trends Immunol. 2013;34:33–40. doi: 10.1016/j.it.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shang B, Liu Y, Jiang SJ, Liu Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: a systematic review and meta-analysis. Sci Rep. 2015;5:15179. doi: 10.1038/srep15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berntsson J, Svensson MC, Leandersson K, Nodin B, Micke P, Larsson AH, Eberhard J, Jirström K. The clinical impact of tumour-infiltrating lymphocytes in colorectal cancer differs by anatomical subsite: a cohort study. Int J Cancer. 2017;141:1654–1666. doi: 10.1002/ijc.30869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Chen Z. The effect of immune microenvironment on the progression and prognosis of colorectal cancer. Med Oncol. 2014;31:82. doi: 10.1007/s12032-014-0082-9. [DOI] [PubMed] [Google Scholar]
- 10.Chew A, Salama P, Robbshaw A, Klopcic B, Zeps N, Platell C, Lawrance IC. SPARC, FOXP3, CD8 and CD45 correlation with disease recurrence and long-term disease-free survival in colorectal cancer. PLoS One. 2011;6:e22047. doi: 10.1371/journal.pone.0022047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Correale P, Rotundo MS, Botta C, Del Vecchio MT, Tassone P, Tagliaferri P. Tumor infiltration by chemokine receptor 7 (CCR7)(+) T-lymphocytes is a favorable prognostic factor in metastatic colorectal cancer. Oncoimmunology. 2012;1:531–532. doi: 10.4161/onci.19404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanke T, Melling N, Simon R, Sauter G, Bokemeyer C, Lebok P, Terracciano LM, Izbicki JR, Marx AH. High intratumoral FOXP3(+) T regulatory cell (Tregs) density is an independent good prognosticator in nodal negative colorectal cancer. Int J Clin Exp Pathol. 2015;8:8227–8235. [PMC free article] [PubMed] [Google Scholar]
- 13.Kim M, Grimmig T, Grimm M, Lazariotou M, Meier E, Rosenwald A, Tsaur I, Blaheta R, Heemann U, Germer CT, Waaga-Gasser AM, Gasser M. Expression of Foxp3 in colorectal cancer but not in Treg cells correlates with disease progression in patients with colorectal cancer. PLoS One. 2013;8:e53630. doi: 10.1371/journal.pone.0053630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waniczek D, Lorenc Z, Snietura M, Wesecki M, Kopec A, Muc-Wierzgoń M. Tumor-associated macrophages and regulatory T cells infiltration and the clinical outcome in colorectal cancer. Arch Immunol Ther Exp (Warsz) 2017;65:445–454. doi: 10.1007/s00005-017-0463-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuo C, Xu Y, Ying M, Li Q, Huang L, Li D, Cai S, Li B. FOXP3+ Tregs: heterogeneous phenotypes and conflicting impacts on survival outcomes in patients with colorectal cancer. Immunol Res. 2015;61:338–347. doi: 10.1007/s12026-014-8616-y. [DOI] [PubMed] [Google Scholar]
- 16.Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E, Nagase H, Nishimura J, Yamamoto H, Takiguchi S, Tanoue T, Suda W, Morita H, Hattori M, Honda K, Mori M, Doki Y, Sakaguchi S. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679–684. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 17.Buckowitz A, Knaebel HP, Benner A, Blaker H, Gebert J, Kienle P, von Knebel Doeberitz M, Kloor M. Microsatellite instability in colorectal cancer is associated with local lymphocyte infiltration and low frequency of distant metastases. Br J Cancer. 2005;92:1746–1753. doi: 10.1038/sj.bjc.6602534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frey DM, Droeser RA, Viehl CT, Zlobec I, Lugli A, Zingg U, Oertli D, Kettelhack C, Terracciano L, Tornillo L. High frequency of tumor-infiltrating FOXP3(+) regulatory T cells predicts improved survival in mismatch repair-proficient colorectal cancer patients. Int J Cancer. 2010;126:2635–2643. doi: 10.1002/ijc.24989. [DOI] [PubMed] [Google Scholar]
- 19.Prall F, Duhrkop T, Weirich V, Ostwald C, Lenz P, Nizze H, Barten M. Prognostic role of CD8+ tumor-infiltrating lymphocytes in stage III colorectal cancer with and without microsatellite instability. Hum Pathol. 2004;35:808–816. doi: 10.1016/j.humpath.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 20.Bauer K, Nelius N, Reuschenbach M, Koch M, Weitz J, Steinert G, Kopitz J, Beckhove P, Tariverdian M, von Knebel Doeberitz M, Kloor M. T cell responses against microsatellite instability-induced frameshift peptides and influence of regulatory T cells in colorectal cancer. Cancer Immunol Immunother. 2013;62:27–37. doi: 10.1007/s00262-012-1303-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim KJ, Lee KS, Cho HJ, Kim YH, Yang HK, Kim WH, Kang GH. Prognostic implications of tumor-infiltrating FoxP3+ regulatory T cells and CD8+ cytotoxic T cells in microsatellite-unstable gastric cancers. Hum Pathol. 2014;45:285–293. doi: 10.1016/j.humpath.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Michel S, Benner A, Tariverdian M, Wentzensen N, Hoefler P, Pommerencke T, Grabe N, von Knebel Doeberitz M, Kloor M. High density of FOXP3-positive T cells infiltrating colorectal cancers with microsatellite instability. Br J Cancer. 2008;99:1867–1873. doi: 10.1038/sj.bjc.6604756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee DW, Han SW, Bae JM, Jang H, Han H, Kim H, Bang D, Jeong SY, Park KJ, Kang GH, Kim TY. Tumor mutation burden and prognosis in patients with colorectal cancer treated with adjuvant fluoropyrimidine and oxaliplatin. Clin Cancer Res. 2019;25:6141–6147. doi: 10.1158/1078-0432.CCR-19-1105. [DOI] [PubMed] [Google Scholar]
- 24.Kordahi MC, Stanaway IB, Avril M, Chac D, Blanc MP, Ross B, Diener C, Jain S, McCleary P, Parker A, Friedman V, Huang J, Burke W, Gibbons SM, Willis AD, Darveau RP, Grady WM, Ko CW, DePaolo RW. Genomic and functional characterization of a mucosal symbiont involved in early-stage colorectal cancer. Cell Host Microbe. 2021;29:1589–1598. e1586. doi: 10.1016/j.chom.2021.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burns MB, Lynch J, Starr TK, Knights D, Blekhman R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med. 2015;7:55. doi: 10.1186/s13073-015-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen S, He L, Zhong Z, Zhao R, Weng S, Mi H, Liu F. Stigmasterol restores the balance of Treg/Th17 cells by activating the butyrate-PPARγ axis in colitis. Front Immunol. 2021;12:741934. doi: 10.3389/fimmu.2021.741934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plitas G, Rudensky AY. Regulatory T cells in cancer. Annu Rev Cancer Biol. 2020;4:459–477. [Google Scholar]
- 29.Oshi M, Asaoka M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Zsiros E, Ishikawa T, Endo I, Takabe K. Abundance of regulatory T cell (Treg) as a predictive biomarker for neoadjuvant chemotherapy in triple-negative breast cancer. Cancers (Basel) 2020;12:3038. doi: 10.3390/cancers12103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D, Ayadi M, Kirzin S, Chazal M, Flejou JF, Benchimol D, Berger A, Lagarde A, Pencreach E, Piard F, Elias D, Parc Y, Olschwang S, Milano G, Laurent-Puig P, Boige V. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayanan S, Kawaguchi T, Peng X, Qi Q, Liu S, Yan L, Takabe K. Tumor Infiltrating lymphocytes and macrophages improve survival in microsatellite unstable colorectal cancer. Sci Rep. 2019;9:13455. doi: 10.1038/s41598-019-49878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narayanan S, Kawaguchi T, Yan L, Peng X, Qi Q, Takabe K. Cytolytic activity score to assess anticancer immunity in colorectal cancer. Ann Surg Oncol. 2018;25:2323–2331. doi: 10.1245/s10434-018-6506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi H, Asaoka M, Yan L, Rashid OM, Oshi M, Ishikawa T, Nagahashi M, Takabe K. Biologically aggressive phenotype and anti-cancer immunity counterbalance in breast cancer with high mutation rate. Sci Rep. 2020;10:1852. doi: 10.1038/s41598-020-58995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuji S, Midorikawa Y, Takahashi T, Yagi K, Takayama T, Yoshida K, Sugiyama Y, Aburatani H. Potential responders to FOLFOX therapy for colorectal cancer by Random Forests analysis. Br J Cancer. 2012;106:126–132. doi: 10.1038/bjc.2011.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherradi S, Ayrolles-Torro A, Vezzo-Vie N, Gueguinou N, Denis V, Combes E, Boissiere F, Busson M, Canterel-Thouennon L, Mollevi C, Pugniere M, Bibeau F, Ychou M, Martineau P, Gongora C, Del Rio M. Antibody targeting of claudin-1 as a potential colorectal cancer therapy. J Exp Clin Cancer Res. 2017;36:89. doi: 10.1186/s13046-017-0558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cherradi S, Martineau P, Gongora C, Del Rio M. Claudin gene expression profiles and clinical value in colorectal tumors classified according to their molecular subtype. Cancer Manag Res. 2019;11:1337–1348. doi: 10.2147/CMAR.S188192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Del Rio M, Mollevi C, Bibeau F, Vie N, Selves J, Emile JF, Roger P, Gongora C, Robert J, Tubiana-Mathieu N, Ychou M, Martineau P. Molecular subtypes of metastatic colorectal cancer are associated with patient response to irinotecan-based therapies. Eur J Cancer. 2017;76:68–75. doi: 10.1016/j.ejca.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 39.Oshi M, Katsuta E, Yan L, Ebos JML, Rashid OM, Matsuyama R, Endo I, Takabe K. A novel 4-gene score to predict survival, distant metastasis and response to neoadjuvant therapy in breast cancer. Cancers (Basel) 2020;12:1148. doi: 10.3390/cancers12051148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oshi M, Tokumaru Y, Asaoka M, Yan L, Satyananda V, Matsuyama R, Matsuhashi N, Futamura M, Ishikawa T, Yoshida K, Endo I, Takabe K. M1 macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci Rep. 2020;10:16554. doi: 10.1038/s41598-020-73624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oshi M, Angarita FA, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. High expression of NRF2 is associated with increased tumor-infiltrating lymphocytes and cancer immunity in er-positive/her2-negative breast cancer. Cancers (Basel) 2020;12:3856. doi: 10.3390/cancers12123856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oshi M, Patel A, Le L, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. G2M checkpoint pathway alone is associated with drug response and survival among cell proliferation-related pathways in pancreatic cancer. Am J Cancer Res. 2021;11:3070–3084. [PMC free article] [PubMed] [Google Scholar]
- 43.Chouliaras K, Oshi M, Asaoka M, Tokumaru Y, Khoury T, Endo I, Ishikawa T, Takabe K. Increased intratumor heterogeneity, angiogenesis and epithelial to mesenchymal transition pathways in metaplastic breast cancer. Am J Cancer Res. 2021;11:4408–4420. [PMC free article] [PubMed] [Google Scholar]
- 44.Gandhi S, Oshi M, Murthy V, Repasky EA, Takabe K. Enhanced thermogenesis in triple-negative breast cancer is associated with pro-tumor immune microenvironment. Cancers (Basel) 2021;13:2559. doi: 10.3390/cancers13112559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okano M, Oshi M, Butash AL, Katsuta E, Tachibana K, Saito K, Okayama H, Peng X, Yan L, Kono K, Ohtake T, Takabe K. Triple-negative breast cancer with high levels of annexin A1 expression is associated with mast cell infiltration, inflammation, and angiogenesis. Int J Mol Sci. 2019;20:4197. doi: 10.3390/ijms20174197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okano M, Oshi M, Butash AL, Asaoka M, Katsuta E, Peng X, Qi Q, Yan L, Takabe K. Estrogen receptor positive breast cancer with high expression of androgen receptor has less cytolytic activity and worse response to neoadjuvant chemotherapy but better survival. Int J Mol Sci. 2019;20:2655. doi: 10.3390/ijms20112655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sporn JC, Katsuta E, Yan L, Takabe K. Expression of microRNA-9 is associated with overall survival in breast cancer patients. J Surg Res. 2019;233:426–435. doi: 10.1016/j.jss.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 48.Tokumaru Y, Katsuta E, Oshi M, Sporn JC, Yan L, Le L, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. High expression of miR-34a associated with less aggressive cancer biology but not with survival in breast cancer. Int J Mol Sci. 2020;21:3045. doi: 10.3390/ijms21093045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, Kaley TJ, Kendall SM, Motzer RJ, Hakimi AA, Voss MH, Russo P, Rosenberg J, Iyer G, Bochner BH, Bajorin DF, Al-Ahmadie HA, Chaft JE, Rudin CM, Riely GJ, Baxi S, Ho AL, Wong RJ, Pfister DG, Wolchok JD, Barker CA, Gutin PH, Brennan CW, Tabar V, Mellinghoff IK, DeAngelis LM, Ariyan CE, Lee N, Tap WD, Gounder MM, D’Angelo SP, Saltz L, Stadler ZK, Scher HI, Baselga J, Razavi P, Klebanoff CA, Yaeger R, Segal NH, Ku GY, DeMatteo RP, Ladanyi M, Rizvi NA, Berger MF, Riaz N, Solit DB, Chan TA, Morris LGT. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drescher KM, Sharma P, Lynch HT. Current hypotheses on how microsatellite instability leads to enhanced survival of Lynch syndrome patients. Clin Dev Immunol. 2010;2010:170432. doi: 10.1155/2010/170432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS Cancer Genome Atlas Research Network. Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The immune landscape of cancer. Immunity. 2018;48:812–830. e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pages F, Galon J, Dieu-Nosjean MC, Tartour E, Sautes-Fridman C, Fridman WH. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29:1093–1102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 54.Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, Peters S. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nimmagadda S. Quantifying PD-L1 expression to monitor immune checkpoint therapy: opportunities and challenges. Cancers (Basel) 2020;12:3173. doi: 10.3390/cancers12113173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner MC, Becerra D, Sun Z, Watson J, Leung K, Migaly J, Mantyh CR, Blazer DG. The side of the primary tumor affects overall survival in colon adenocarcinoma: an analysis of the national cancer database. Tech Coloproctol. 2019;23:537–544. doi: 10.1007/s10151-019-01997-w. [DOI] [PubMed] [Google Scholar]
- 57.Loupakis F, Yang D, Yau L, Feng S, Cremolini C, Zhang W, Maus MK, Antoniotti C, Langer C, Scherer SJ, Muller T, Hurwitz HI, Saltz L, Falcone A, Lenz HJ. Primary tumor location as a prognostic factor in metastatic colorectal cancer. J Natl Cancer Inst. 2015;107:dju427. doi: 10.1093/jnci/dju427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szeponik L, Ahlmanner F, Sundstrom P, Rodin W, Gustavsson B, Bexe Lindskog E, Wettergren Y, Quiding-Järbrink M. Intratumoral regulatory T cells from colon cancer patients comprise several activated effector populations. BMC Immunol. 2021;22:58. doi: 10.1186/s12865-021-00449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]